
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 19 September 2022
Sec. Membrane Traffic and Organelle Dynamics
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.1008078
The second step of eukaryotic lipid-linked oligosaccharide (LLO) biosynthesis is catalyzed by the conserved ALG13/ALG14 heterodimeric UDP-N-acetylglucosamine transferase (GnTase). In humans, mutations in ALG13 or ALG14 lead to severe neurological disorders with a multisystem phenotype, known as ALG13/14-CDG (congenital disorders of glycosylation). How these mutations relate to disease is unknown because to date, a reliable GnTase assay for studying the ALG13/14 complex is lacking. Here we describe the development of a liquid chromatography/mass spectrometry-based quantitative GnTase assay using chemically synthesized GlcNAc-pyrophosphate-dolichol as the acceptor and purified human ALG13/14 dimeric enzyme. This assay enabled us to demonstrate that in contrast to the literature, only the shorter human ALG13 isoform 2, but not the longer isoform 1 forms a functional complex with ALG14 that participates in LLO synthesis. The longer ALG13 isoform 1 does not form a complex with ALG14 and therefore lacks GnTase activity. Importantly, we further established a quantitative assay for GnTase activities of ALG13- and ALG14-CDG variant alleles, demonstrating that GnTase deficiency is the cause of ALG13/14-CDG phenotypes.
Asparagine (N)-linked glycosylation is a posttranslational modification that directly affects the structure and function of glycoproteins, thus influencing a multitude of essential biological processes (Takahashi et al., 2016; Loke et al., 2016; Dwyer and Esko, 2016; Esmail and Manolson, 2021). Protein N-glycosylation begins with the assembly of a lipid-linked oligosaccharide (LLO) precursor whose synthesis starts on the cytosolic side of the endoplasmic reticulum (ER) membrane and finishes in its lumen (Supplementary Figure S1) (Aebi, 2013). The sequential addition of sugars to dolichol pyrophosphate (PDol) at the ER membrane by the ALG (asparagine-linked glycosylation) glycosyltransferases (GTases) produce a LLO containing fourteen sugars, including two GlcNAcs (Gn), nine mannoses (M) and three glucoses (G) (Li et al., 2019). Once assembled, the G3M9Gn2 oligosaccharide is transferred from the lipid to nascent proteins by oligosaccharyltransferase (Cherepanova et al., 2016; Shrimal and Gilmore, 2019). In humans, defects in LLO synthesis or transfer to proteins cause inherited metabolic diseases known as congenital disorders of glycosylation (CDG) that manifest as a broad range of multisystem disorders or even embryonic lethality (Jaeken, 2020; Pascoal et al., 2020). Based on recent statistical data, the number of identified ALG-CDGs is rapidly increasing, underscoring the significance of LLO synthesis to human disease (Sosicka et al., 2021).
Fourteen ALG genes are required for N-glycosylation in the ER (Schwarz and Aebi, 2011; Aebi, 2013). Among them, the ALG13 and ALG14 are unique in that the enzymes they encode interact to form a heterodimeric UDP-N-acetylglucosamine transferase (GnTase). Unlike other ALG GTases which may function alone, ALG13/14 GnTase only possesses activity when they interact with each other (Gao et al., 2004; Noffz et al., 2009). This dimeric ALG13/14 catalyzes the second step of LLO synthesis, transferring a β1,4-linked N-acetylglucosamine (GlcNAc) from UDP-GlcNAc to GlcNAc-pyrophosphate-dolichol (Gn-PDol) (Supplementary Figure S2) (Bickel et al., 2005; Chantret et al., 2005; Gao et al., 2005). ALG13/14 GnTase is widely conserved in eukaryotes. Most of our knowledge about the function of ALG13/14 GnTase comes from biochemical and genetic analyses in S. cerevisiae. Yeast Alg13 (yAlg13) is a small, soluble cytosolic protein that contains the GnTase catalytic subunit, while Alg14 (yAlg14) is a membrane protein that recruits soluble yAlg13 to the cytosolic face of the ER. Both yAlg13 and yAlg14 are essential for viability (Bickel et al., 2005; Chantret et al., 2005; Gao et al., 2005) and yAlg13 is inactive unless bound to yAlg14 (Bickel et al., 2005; Chantret et al., 2005). Because of its complex structure, in which the active heterodimer consists of both a hydrophilic and hydrophobic subunit, the development of an in vitro quantitative assay for studying the enzymatic properties of ALG13/14 GnTase has been hindered for a long time. Recently, liquid chromatography/mass spectrometry (LC–MS)-based techniques have been used for developing in vitro quantitative assays to study the enzymatic activity of ALG GTases (Li et al., 2017a; Li et al., 2017b; Wang et al., 2022; Xiang et al., 2022), but as yet, these have not been tested for their suitability for ALG13/14.
Phylogenetic analysis of ALG14-ALG13 homologs revealed their ancient origin in eukaryotes (Meyer et al., 2022). While ALG14 is structurally similar across eukaryotes, ALG13 has evolved differently: fungal and plant cells only possess a short form of the ALG13 protein, while vertebrates have multiple isoforms. In humans, four ALG13 isoforms generated by alternative splicing have been described (https://www.uniprot.org-ALG13, identifier: Q9NP73). Among them, isoform 1 (ALG13-iso1) and 2 (ALG13-iso2) are found in all mammals. ALG13-iso1 is the longest isoform of ALG13, consisting of 1,137 amino acids, while ALG13-iso2 is shorter, consisting of 165 amino acids. This short ALG13-iso2 shows more than 30% sequence similarity with animal, fungi and plant (Supplementary Figure S3). Co-expression of human ALG13-iso2 and ALG14 complemented the growth defect of yeast alg13 and alg14 mutant cells, indicating ALG13-iso2 is catalytically active when complexed with ALG14 (Gao et al., 2005). Studies have shown that it is possible to completely delete the ALG13-iso1 in a mouse model, but not the isoform 2 (Gao et al., 2019), suggesting the ALG13-iso2 is essential for glycosylation. On the other hand, ALG13-iso1 knockout mice have an increased susceptibility to epileptic seizures (Gao et al., 2019; Huo et al., 2020). Thus the biological function of ALG13-iso1 as it relates to glycosylation remains undefined.
Pathogenic mutations in human ALG13 or ALG14 cause severe neurological disorders with a multisystem phenotype. ALG13 variants underlie infantile-onset developmental and epileptic encephalopathy (DEE) (Timal et al., 2012; Ng et al., 2020; Alsharhan et al., 2021; Datta et al., 2021), while ALG14 gene mutants present as early lethal neurodegeneration with myasthenic and myopathic features, also known as congenital myasthenic syndrome (CMS) (Judith et al., 2013; Schorling et al., 2017; Datta et al., 2021; Katata et al., 2022). Given the essential role of ALG13/14 GnTase in ER N-glycosylation, it has been assumed that these phenotypes fall under the umbrella of CDG. Surprisingly, however, unlike other ALG-CDGs, most ALG13- and ALG14-CDG variants show a normal glycosylation pattern on transferrin, the commonly used biomarker for CDG (Schorling et al., 2017; Alsharhan et al., 2021). Thus direct evidence linking ALG13/14 GnTase activity to the DEE or CMS phenotypes remains elusive.
In this work, we co-expressed human ALG13 and ALG14 in E. coli and purified recombinant ALG13/14 complex from a membrane fraction. Using chemically synthesized Gn-PDol as acceptor substrate, we confirmed the GnTase activity of recombinant ALG13/14. We used this purified enzyme to study the kinetic properties of ALG13/14 GnTase with an in vitro quantitative assay utilizing LC–MS. Furthermore, this in vitro assay system was used to compare the GnTase activities of the different human ALG13 isoforms. Our results confirmed that ALG13-iso2, but not ALG13-iso1, forms a dimeric complex with ALG14 on the cytosolic face of the ER membrane and contributes to GnTase activity for LLO synthesis. We also applied this assay to demonstrate the GnTase activities of ALG13- and ALG14-CDG variants. These variants display severe enzymatic defects in vitro, thus providing evidence for a genetic link between GnTase activity and ALG13-related DEE or ALG14-related CMS.
The plasmids used in this study and their important features are described in Table 1. Standard molecular biological techniques were used for plasmid construction, and mutations that altered various amino acids (see Table 2) were introduced using overlapping PCR with mutagenic primers (BGI, Shenzhen, China) and verified by DNA sequence analysis.
Human embryonic kidney cells, HEK293, were cultured in DMEM supplemented with 10% FBS (FCS, Biological Industries, Beit Haemek, Israel), penicillin and streptomycin (1 μg/ml) in a humidified 5% CO2 atmosphere at 37°C.
E. coli Rosetta cells (DE3, Merck, NJ, United States) harboring both pET26b-pelB-FLAG-ALG13-iso2 and pCDFDuet-6His-ALG14 plasmids were cultured in 200 ml of Terrific-Broth medium (TB, 1.2% tryptone, 2.4% yeast extract and 0.5% glycerol) at 37°C to an OD600 of 1.0 and then cooled to 16°C. After adding 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG, Sangon Biotech, Shanghai, China), the cells were incubated for additional 18 h–24 h at 16°C to induce protein expression. The harvested cells were resuspended in 15 ml lysis buffer (150 mM NaCl, 50 mM Tris/HCl, pH 8.0) and homogenized by sonication. The cell lysate was centrifuged at 4,000×g to remove debris, and the supernatant was collected (this supernatant was used as the “crude extract”). To obtain the membrane fraction containing ALG13/14 complex, the crude extract was further centrifuged at 12,000×g to collect precipitated membranes. Any remaining cytosolic ALG13 was removed by washing the membranes with 15 ml lysis buffer three times. This washed membrane fraction was then solubilized by resuspension in 5 ml lysis buffer containing 1% Triton X-100 and incubated on ice for 15 min to obtain the detergent extract. This detergent extract was used for purification of the recombinant ALG13/14 complex following the procedure described below. This detergent extract was also used for the co-immunoprecipitation experiment presented in Figure 2A.
The detergent extract prepared from the E. coli cells co-expressing FLAG-ALG13-iso2 and His-ALG14 was used to purify the recombinant ALG13/14 complex. First, His-tagged ALG14 proteins, including the recombinant FLAG-ALG13-iso2/His-ALG14 complex in detergent extract, were trapped by a HisTrap HP affinity column (GE Healthcare Life Sciences). Their eluted solution was then incubated with anti-FLAG Affinity Matrix (Sigma–Aldrich, MO, United States) to enrich the ALG13/14 complex and remove the free His-ALG14. After elution with FLAG peptide, the purified ALG13/14 complex was analyzed by native PAGE and SDS–PAGE followed by silver staining to confirm the purity. Native PAGE was performed using a Blue/Clear Native PAGE Electrophoresis Kit (Real-time, Beijing, China) according to the manufacturer’s instructions.
HEK293 cells were seeded in 6-well plate and transfection was performed when the cell density was 80%–90%. Transfection was performed using Lipofectamine 2,000 (Thermo Fisher Scientific, MA, United States) according to the manufacturer’s instructions. 4 μg plasmid (when co-transfect two plasmids, 2 μg per plasmid) and 5 μL lipofectamine 2000 were diluted using 250 μl OPTI-MEM respectively, after incubated at room temperature for 5 min, then mixed and incubated for another 20 min. The mixed reagent was added in HEK293 cells which grown in 2 ml DMEM medium with 10% FBS. Changed the mixture to DMEM medium with 10% FBS after 8 h and harvested the cell after 72 h.
To prepare the protein extract, 1 × 106 cells were solubilized in HEPES buffer with 1% NP-40 (25 mM HEPES, pH 7.4, 150 mM NaCl, 1%, 1 mM PMSF and 1 × Protein inhibitor cocktail) and incubated on ice for 30 min. Cell lysates were centrifuged at 15,000×g for 10 min at 4°C. The resulting supernatants were collected as the protein extracts. To prepare the membrane fraction which contain endogenous ALG13/14, HEK293 cells (1 × 106 cells) were resuspended in 1 ml of hypotonic buffer (10 mM Tris/HCl, pH 7.4, 1.5 mM MgCl2, 10 mM KCl, protease inhibitor), followed by incubation on ice for 30 min. Cells were then homogenized by passing them 36 times through a homogenizer. Nuclei and cell debris were pelleted by centrifugation at 2,500×g for 5 min at 4°C. The supernatant was collected and centrifuged again at 12,000×g for 30 min at 4°C. The obtained membrane fraction (equivalent to 30 μg membrane protein) was used as the enzyme fraction of ALG13/14 GnTase in studies that characterized the suitability of synthetic Gn-PDol (C95) as substrate, as presented in Figure 1C.
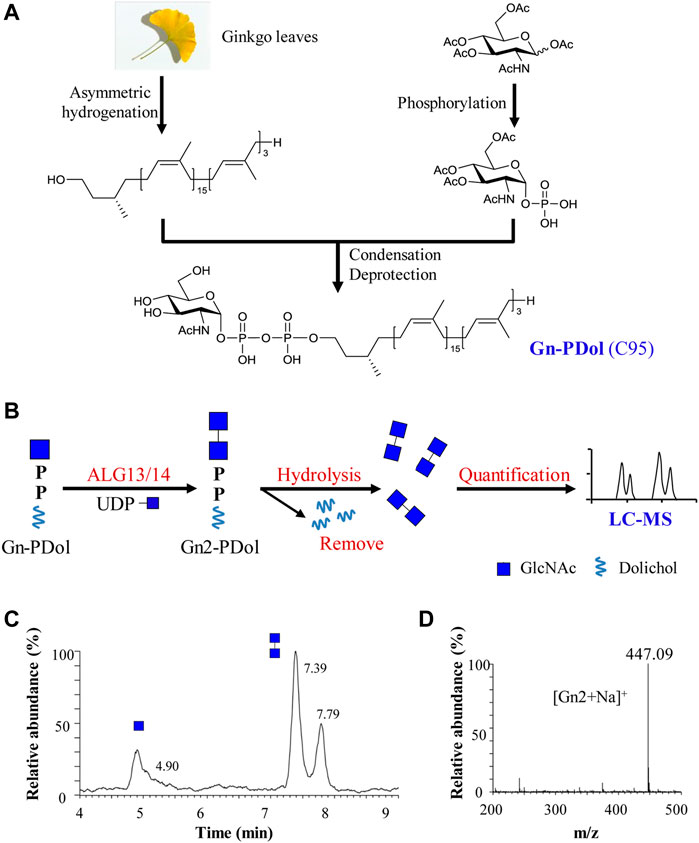
FIGURE 1. Preparation and feasibility test of lipid-linked acceptor GlcNAc-PP-Dol (Gn-PDol, C95). (A) The semisynthetic strategy of lipid-linked acceptor Gn-PDol (C95); (B) Work flow of an in vitro quantitative GnTase assay for ALG13/14 complex; (C) The UPLC chromatogram of glycans released from ALG13/14 reaction mixture. Reaction was performed using HEK293 cells membrane fraction with the standard reaction mixture and incubated for 1 h. Peaks eluted earlier represent Gn and eluted later represent Gn2; (D) ESI-MS spectra of peaks eluted at 7.39 and 7.79 min in UPLC correspond to Gn2 ([Gn2+Na]+).
E. coli co-expressing FLAG-ALG13-iso2 and 6His-ALG14 were cultured, and crude extracts were prepared as described above. Ten micrograms of this crude extract was separated by 12% SDS–PAGE analysis and transferred to a PVDF membrane (Bio-Rad, CA, United States). Immunoblotting was performed with an anti-His mouse antibody (TransGen Biotech, Beijing, China) and anti-FLAG mouse monoclonal antibody (TransGen Biotech, Beijing, China) as the primary antibody and the anti-mouse IgG-horseradish peroxidase (HRP) conjugate (TransGen Biotech, Beijing, China) as the secondary antibody followed by chemiluminescence (ECL) detection (Bio-Rad, CA, United States). For co-immunoprecipitation, 6His-ALG14 or FLAG-ALG13-iso2 was immunoprecipitated with Ni-NTA His binding resin (TransGen Biotech, Beijing, China) or anti-FLAG Affinity Matrix (Sigma–Aldrich, St. Louis, MO, United States) as described elsewhere (Gao et al., 2005). After separation by 12% SDS–PAGE, the precipitated proteins were transferred to an Immobilon-PVDF membrane. The membrane was incubated with an anti-FLAG mouse monoclonal antibody or anti-His mouse monoclonal antibody, washed, and incubated with anti-mouse IgG–HRP conjugate. Immunoreactive bands were visualized by ECL.
Similarly, HEK293 cells co-expressing HA-ALG13-iso1/FLAG-ALG14 or HA-ALG13-iso2/FLAG-ALG14 were cultured, and their protein extracts were prepared as described above. Ten μg of each protein extract was analyzed using western blot. Immunoblotting was performed with anti-FLAG mouse monoclonal antibody (Sigma-Aldrich, MO, United States) and anti-HA rabbit polyclonal antibody (Cell Signaling Technology, MA, United States) as the primary antibody and the anti-mouse IgG-HRP conjugate or the anti-rabbit IgG-HRP conjugate (TransGen Biotech, Beijing, China) as the secondary antibody. Immunoreactive bands were visualized by ECL. For co-immunoprecipitation assays, protein extracts were immunoprecipitated with anti-HA rabbit polyclonal antibody followed by incubation with 40 μl Protein A + G Agarose (Beyotime Shanghai, China) for 3 h to pull down the HA-tagged ALG13-iso1 or ALG13-iso2 protein. After washing 3 times with HEPES buffer, the immunoprecipitate was solubilized, separated by SDS-PAGE and analyzed by western blotting with anti-FLAG mouse monoclonal antibody to detect the co-immunoprecipitated FLAG-ALG14.
For the purification of ALG13-iso1 and ALG13-iso2, HEK293 cells (3 × 106 cells) expressing HA-ALG13-iso1 or HA-ALG13-iso2 were harvested and the protein extract was prepared as described above. The protein extract was incubated with anti-HA rabbit polyclonal antibody and 40 μl Protein A + G Agarose for 4 h to enrich the HA-tagged ALG13-iso1 or ALG13-iso2 protein. After washing the agarose by PBS for 3 times, the agarose-bound immunoprecipitated proteins were divided into two parts. One part was immunoblotted with anti-ALG13 rabbit polyclonal antibody (Proteintech, Wuhan, China) to detect HA-ALG13 while the other part was employed to test the GnTase activity.
The standard reaction mixture for ALG13/14 GnTase assay contained in a volume of 50 μl: 28 mM Tris/HCl, pH 7.5, 0.3% NP-40, 23% glycerol, 40 μM Gn-PDol, 2 mM UDP-GlcNAc (Sigma-Aldrich, MO, United States) and protein. After incubation at 37°C, the reaction was stopped by heating to 100°C. The specific amount of protein included and reaction times are described in detail in each section. The donor concentration using 2 mM, which was 50 folds than the substrate Gn-PDol concentration. ALG13/14 can only transfer one GlcNAc from UDP-GlcNAc to Gn-PDol, so the donor concentration was excess which could facilitate the reaction.
After terminating the reaction, glycans were released by adding 50 μl of HCl (40 mM) to the reaction and shifting the temperature to 100°C for 1 h. The hydrolyzed glycans were purified as described (Li et al., 2017a; Li et al., 2017b; Xu et al., 2018; Li et al., 2019; Xiang et al., 2022). The desalted sample was injected into a Dionex Ultimate 3,000 UPLC (Thermo Scientific, MA, United States) and separated using a Waters Acquity UPLC BEH Amide Column (1.7 μm 2.1 × 150 mm) at a flow rate of 0.2 ml/min. The UPLC conditions were same as previous report while the ESI-MS range was adjusted to 100–1,500 m/z (positive mode). The oligosaccharide transfer rate was calculated using the peak intensity in LC-ESI-MS through Xcalibur (Version 2.0, Thermo Scientific, MA, United States).
Enzymatic assays were performed using purified protein with the final concentration as 0.3 ng/μl (Figures 3B–E) or 0.2 ng/μl (Figure 3F) under the standard conditions for 15 min as described above. The optimized reaction conditions were as follows: temperature from 10°C to 60°C, pH from 4.0 mM to 10.0 mM, 10 mM ions (Co2+, Mn2+ and Mg2+, respectively) or 10 mM EDTA for depletion conditions. The apparent Km and kcat values for Gn-PDol (20 μM–100 μM) were calculated by nonlinear regression curve fitting (GraphPad Prism 8.0, GraphPad Software Inc.). The donor specificity of ALG13/14 was confirmed using different donor (UDP-Glc, UDP-GlcNAc and GDP-Man) with same concentration (2 mM).
The stereochemistry of the linkage between two GlcNAc residues in Gn2-PDol (C95), the product of ALG13/14 GnTase, was confirmed by using yeast Alg1 (yAlg1) MTase. Purified recombinant His-yAlg1ΔTM and GDP-Man were added to the recombinant ALG13/14 reaction system, followed by 1 h of incubation at 30°C. The products were analyzed by LC–MS as described above.
The yeast strain XGY186 (as in W303a; ∆alg13::His+5; ∆alg14::Kan+ and GAL1/10pr-3HA-yALG14/yALG13-FLAG::trp1), contains a deletion of ALG13 and ALG14 in the chromosome and a GAL1/10 promoter-driven yALG13 and yALG14. This strain was used to test for the ability of human ALG13 different isoforms and ALG14 to complement growth in the presence of glucose. The URA3-marked YEp352GAPII-3HA-ALG13-iso1 or YEp352GAPII-3HA-ALG13-iso2 plasmid was co-transformed with the LEU2-marked YEp351GAPII-ALG14-FLAG into XGY186 and selected on SG (-Ura-Leu) plates. Transformants were cultured in medium containing glucose, followed by growth on solid YPA medium (1% yeast extract, 2% peptone, 50 mg/L adenine) supplemented with 2% glucose (YPAD) or 2% galactose (YPAG) at 30°C for 2 days.
The enzymatic properties, kinetic analysis and CDG variants relative activity experiments were independently repeated three times. Sample size, mean, and kinetic standard deviation (SD) value are provided in the figure, figure legend and table footnote. Kinetic parameters were determined by nonlinear regression fitted to Michaelis-Menten equation in GraphPad Prism 8.0.
The human ALG13/14 (ALG13/14) complex catalyzes the addition of a β1,4-linked GlcNAc to Gn-PDol to produce Gn2-PDol in the DLO synthetic pathway. We aimed to establish an in vitro assay system for quantitatively detecting UDP-GlcNAc transferase (GnTase) activity. We previously described an LC–MS-based quantitative assay for LLO mannosyltransferases (Li et al., 2019) and this assay was modified for quantitative measurements of ALG13/14 GnTase activity with a suitable Gn-PDol acceptor substrate. Unlike other ALG GTases, which can use phytanyl-linked oligosaccharides as their acceptor substrates for enzymatic studies (Li et al., 2017a; Li et al., 2017b; Xu et al., 2018; Li et al., 2019; Xiang et al., 2022), ALG13/14 GnTase does not recognize GlcNAc-pyrophosphate-phytanyl as the acceptor (data not shown). Therefore, we tried to prepare its natural Gn-PDol acceptor using semisynthetic methods. Since the lipid tails of the DLO in mammalian cells usually contain 17–21 isoprene units (C85–C105), we synthesized a Gn-PDol (C95) substrate that contains 19 isoprene units of lipid tails (Figure 1A). A polyprenol (C95) obtained from ginkgo leaves was applied to the asymmetric hydrogenation to generate dolichol, which was coupled to phosphorylated GlcNAc to obtain the target substrate Gn-PDol (C95, Figure 1A). The product structure was characterized by mass and nuclear magnetic resonance (NMR) analyses. The detailed synthetic procedures and data will be published elsewhere.
To test if this Gn-PDol (C95) is a suitable substrate for ALG13/14 GnTase, the HEK293 cells membrane fraction (equivalent to 30 μg membrane protein, which was confirmed contain endogenous ALG13 and ALG14 using anti-ALG13 antibody and anti-ALG14 antibody, data not shown) was prepared and applied to a standard reaction mixture with a final volume of 50 μl, in which Gn-PDol (C95) and UDP-GlcNAc were used as the acceptor and donor substrates (see Material and methods for details). Hydrochloric acid was added to hydrolyze the saccharide moieties, followed by purification and detection using LC–MS for quantitative analysis (Figure 1B). Two group peaks appeared on UPLC (Figure 1C). The corresponding ESI-MS analysis showed that the first peak eluted at 4.90 min was [Gn + H]+, and the mass peak at m/z 222.12 (Supplementary Figure S4) was hydrolyzed from the substrate Gn-PDol. The second group peak eluted at 7.39 min and 7.79 min was [Gn2+Na]+, with the mass peak m/z: 447.09, which was hydrolyzed from the product Gn2-PDol (Figures 1C,D). The two peaks of Gn2 have been proven to be glycan anomeric isomers, which were designated alpha (α) and beta (β) (Li et al., 2017a). The conversion rate was quantified by calculating the peaks intensity of Gn2 (two peaks)/peak intensity of (Gn + Gn2), which was 79.8% in Figure 1C. The boiled membrane fraction was applied to the standard reaction mixture as negative control, only one peak representing Gn was detected (data not shown). These results demonstrated the production of Gn2-PDol, indicating that Gn-PDol (C95) is a suitable substrate for ALG13/14 GnTase.
Although neither ALG13 nor ALG14 alone can rescue yeast alg13 or alg14 mutants, when co-expressed, human ALG13 (ALG13-iso2) and ALG14 can rescue the growth defect caused by either the loss of yeast ALG13 or ALG14 (Gao et al., 2005). These results demonstrated the formation of an active human ALG13/14 GnTase complex in yeast cells. Thus, to prepare active recombinant ALG13/14 complex, we chose to co-express human ALG13-iso2 and ALG14 in E. coli. Using N-FLAG- or His-tags, ALG13-iso2 and ALG14 were cloned into the pET26b and pCDFDuet expression vectors, respectively (Table 1). The resulting plasmids pET26b-pelB-FLAG-ALG13-iso2 and pCDFDuet-6His-ALG14 were transformed into E. coli Rosetta (DE3) cells. After induction with IPTG, the expression of both FLAG-ALG13-iso2 and His-ALG14 proteins was confirmed using anti-FLAG or anti-His antibodies (Figure 2A, left panel). Co-immunoprecipitation assays confirmed the formation of the ALG13/14 complex (Figure 2A, right panel). Immunoprecipitation of His-ALG14 with Ni-NTA His binding resin brought down the FLAG-ALG13-iso2 protein, which was detected by anti-FLAG antibody, and vice versa. These results suggested that ALG13/14 form a heterodimer in E. coli.
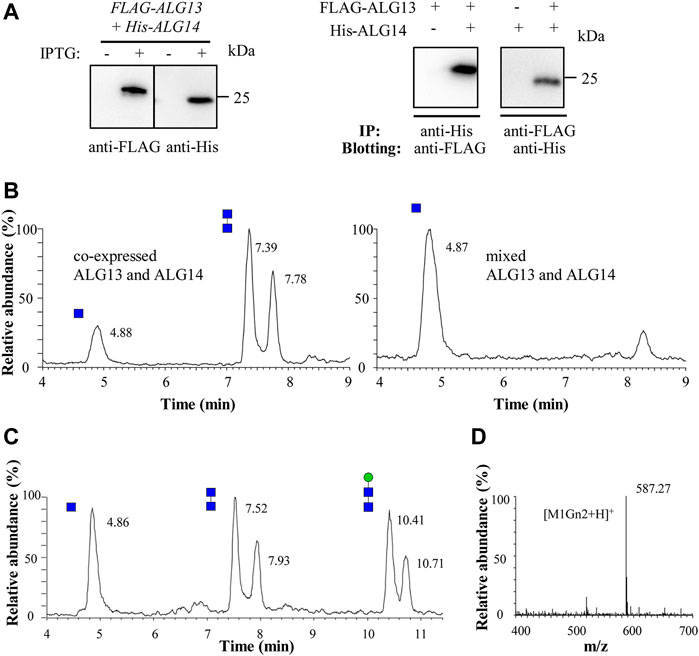
FIGURE 2. Recombinant ALG13/14 complex catalyzes the formation of Gn2-PDol. (A) Western blot and co-immunoprecipitation analysis of recombinant co-expressed ALG13 and ALG14 in E. coli. The crude extract prepared from E. coli cells was analyzed by western blot with anti-FLAG antibody or with anti-His antibody (left panel). The co-immunoprecipitation performed using Ni-NTA His binding resin or anti-FLAG Affinity Matrix, and immunoblotted with anti-FLAG antibody or anti-His antibody (right panel); (B) The UPLC chromatogram of glycans released from different reaction mixture. Reaction was performed using crude extract from co-expressed strain (5 μg total protein) or two different single expressed strain (5 μg total protein from each strain); (C) The UPLC chromatogram of glycans released from the recombinant ALG13/14 reaction mixture which add recombinant yAlg1 for additional 1 h. Peaks eluted at 10.41 min and 10.71 min represent Man-Gn2 (M1Gn2); (D) ESI-MS spectra of peaks eluted at 10.41 min and 10.71 min in UPLC correspond to M1Gn2 ([M1Gn2+H]+).
To determine if these recombinant proteins are active, GnTase activity of crude extracts was measured. Total protein (5 μg) prepared from E. coli that co-expressed ALG13-iso2 and ALG14 was incubated with the standard reaction mixture containing Gn-PDol (C95) and UDP-GlcNAc for 1 h, followed by detection of Gn2 product using LC–MS. The Gn2 product can be detected as two peaks at retention times of 7.39 min and 7.78 min (Figure 2B, left panel) with a mass of [Gn2+Na]+ (data not shown). When extracts prepared from two different E. coli strains (5 μg total protein from each strain), which individually expressed ALG13-iso2 and ALG14 were mixed and assayed for activity under the same conditions, no product peaks were detected (Figure 2B, right panel). These results indicate that the GnTase activity of the ALG13/14 complex cannot be obtained by mixing the individual recombinant ALG13 and ALG14 proteins. Instead, they imply that complex formation occurs in vivo and once formed, is stable.
To further confirm the stereochemistry of the GlcNAc-GlcNAc linkage in the newly formed product, we asked if it could be recognized as a substrate by yAlg1, the mannosyltransferase that catalyzes the reaction following ALG13/14 GnTase. yAlg1, adds β1,4-Man to GlcNAc(β1,4)GlcNAc-PDol (Gn2-PDol) with strict substrate specificity (Li et al., 2017a). To test this, recombinant yAlg1 was added to ALG13/14 GnTase reaction buffer with 2 mM GDP-mannose (GDP-Man) and incubated for another 1 h. After incubation, peaks corresponding to Man-Gn2 were observed at 10.41 min and 10.71 min, with a mass peak m/z of 587.27 (Figures 2C,D). The Man-Gn2 also was proved as glycan anomeric isomers. The ability of yAlg1 to produce this product implies the presence a β1,4 linkage in the GlcNAc-GlcNAc (Gn2) product of ALG13/14 GnTase. Taken together, our results demonstrated that co-expression of human ALG13-iso2 and ALG14 genes in E. coli can form an enzymatically active GnTase complex.
To further study its enzymatic properties, recombinant human ALG13/14 complex from E. coli harboring both pET26b-pelB-FLAG-ALG13-iso2 and pCDFduet-6His-ALG14 plasmids was purified as described in “Material and methods”. The purified protein was calculated as 2.6 mg/ml (0.1 ml), the concentration exhibiting a 500-fold compared with E. coli membrane extract. Native-PAGE analysis, in which the complex remains associated, revealed a single band with a molecular weight of approximately 45 kDa–66 kDa, while denaturing SDS–PAGE analysis revealed two bands, the individual ALG13-iso2 (25 kDa) and ALG14 (22 kDa) proteins (Figure 3A). Together, these results suggested that the purified recombinant ALG13/14 complex exists as a heterodimer.
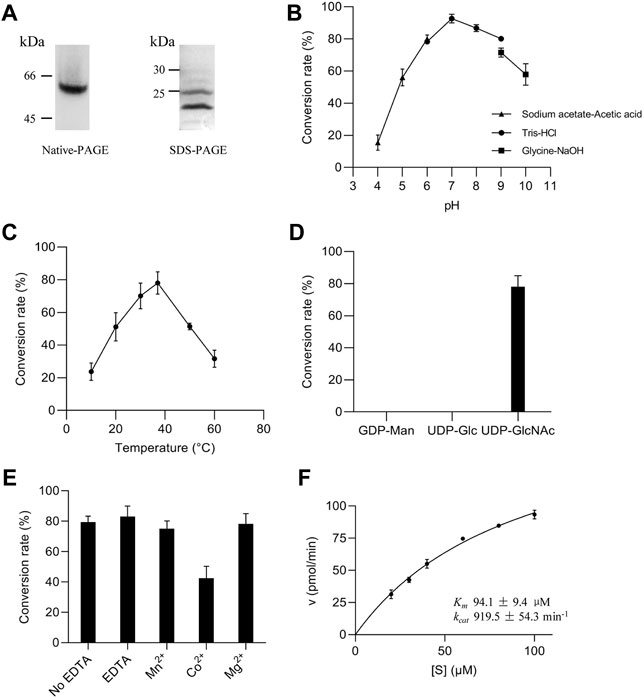
FIGURE 3. Enzymatic properties of ALG13/14 GnTase. (A) Native PAGE and SDS-PAGE analysis of purified heteromeric ALG13/14 complex; (B) Optimal pH was examined using various buffers, including sodium acetate-acetic acid buffer (pH 4.0, 5.0 and 6.0); Tris/HCl buffer (pH 6.0, 7.0, 8.0 and 9.0); glycine/NaOH buffer (pH 9.0 and 10.0); (C) Optimal temperature was evaluated at indicated temperatures (10°C, 20°C, 30°C, 37°C, 50°C and 60°C); (D) The specificity of the nucleotide sugar donor was studied by performing the reaction with 2 mM GDP-Man, UDP-Glc, or UDP-GlcNAc; (E) Divalent cation dependency was examined using 10 mM ions (Co2+, Mn2+ and Mg2+, respectively) or 10 mM EDTA for depletion conditions; (F) The Km (94.1 μM ± 9.4 μM) and kcat (919.5 min−1 ± 54.3 min−1) values for the substrate Gn-PDol whose concentration ranged from 20 μM to 100 μM, were calculated by nonlinear regression with a constant concentration of 2 mM UDP-GlcNAc. Each data point represents the mean ± SD value calculated from three independent experiments.
Purified ALG13/14 was used to optimize reaction conditions (see Material and methods for details). As shown in Figures 3B–E, using 0.3 ng/μl recombinant proteins, our experiments demonstrated that the optimum pH and reaction temperature were 7.0°C and 37°C, respectively (Figures 3B,C). Moreover, of the three nucleotide sugars tested as donor substrates (UDP-GlcNAc, GDP-Man, and UDP-Glc), only UDP-GlcNAc was specifically recognized by the ALG13/14 complex (Figure 3D). Since additional divalent metal cations were required for most glycosyltransferase activities, the effects of various divalent metal ions on GnTase activity of ALG13/14 were tested. There were no improved activities confirmed in the presence of Mn2+, Mg2+ and EDTA, whereas Co2+ even inhibited the activity to some extent (Figure 3E). These results suggested that ALG13/14 GnTase does not require divalent metal ions for its enzyme activity. Furthermore, to investigate the kinetics of the recombinant ALG13/14, the rate of conversion to Gn2-PDol by purified recombinant ALG13/14 (0.2 ng/μl) was measured for the Gn-PDol (C95) acceptor from 20 mM to 100 mM with a fixed concentration of 2 mM UDP-GlcNAc as the donor substrate. The Km value was calculated as 94.1 μM, and the kcat value was 919.5 pmol/min by nonlinear regression fitted to Michaelis–Menten (Figure 3F).
As the canonical human isoform, the longer ALG13-iso1 contains not only an N-terminal glycosyltransferase 28 domain, but also several other domains including an ovarian tumor deubiquitinase domain (Ng et al., 2020). Compared to isoform 1, the short ALG13-iso2 lacks a large portion of the C-terminal region and uses an alternate 3′ terminal exon. The resulting iso2 protein shares N-terminal 127 amino acids with isoform 1, which contains the catalytic domain, and possesses a distinct C-terminal region consisting of 38 amino acids (Figure 4A). Among the ALG13 isoforms, only isoforms 1 and 2 contain the catalytic domain of ALG13/14 GnTase. It has been reported that ALG13-iso2 forms an active GnTase complex with ALG14 (Gao et al., 2005; Gao et al., 2008) (Figures 2, 3), but to date, no functional studies have ruled out that ALG13-iso1 also has catalytic activity.
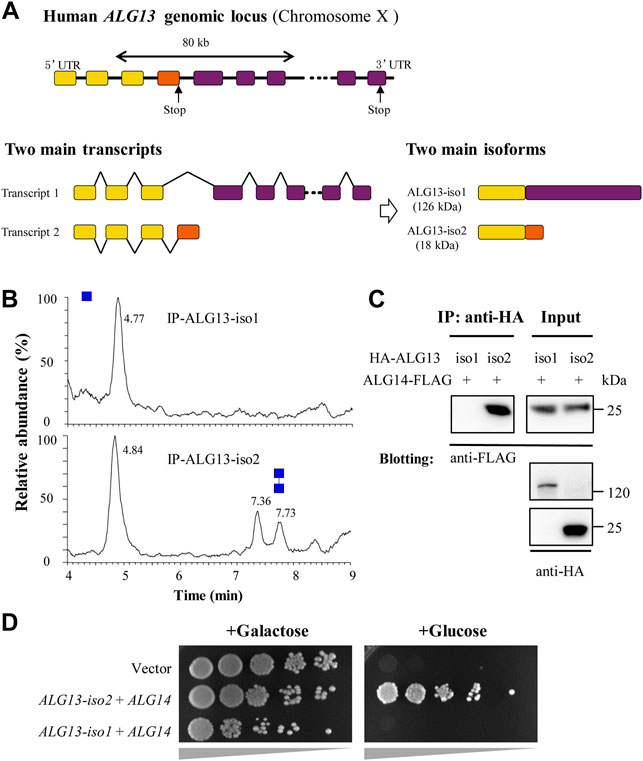
FIGURE 4. Human ALG13 isoform 1 fails to form GnTase complex with ALG14. (A) The gene map of ALG13-iso1 and ALG13-iso2. The two isoforms transcript from ALG13 gene and share a same N-terminal while the C-terminal substantially different; (B) The UPLC chromatogram of glycans released from the reaction of ALG13 isoforms. HA-tagged ALG13 isoforms were expressed in HEK293 cell and enriched by using anti-HA agarose; (C) Co-immunoprecipitation analysis of ALG13 isoforms and ALG14. HA-tagged ALG13 isoforms and FLAG-tagged ALG14 were co-transfected in HEK293 cells, the co-immunoprecipitation performed using anti-HA agarose, and immunoblotted with anti-FLAG antibody; (D) Yeast complementary assay of ALG13 isoforms. An ALG13/14 deficient yeast strain XGY186 was grown under Gal promoter driving yAlg13 and yAlg14. ALG13 isoform and ALG14 were transfected and culture in YPA plate with galactose or glucose and incubated at 30°C for 2 days. iso: isoform.
To compare their GnTase activity, HA-tagged ALG13-iso1 protein was over-expressed alone in HEK293 and analyzed for enzymatic activity in vitro (Table 2). Cultured HEK293 cells (3 × 106 cells) expressing HA-ALG13-iso1 were harvested and resuspended in 300 μl of HEPES buffer containing 1% Triton X-100 (see Material and methods for detailed composition). After incubation for 30 min on ice to solubilize membrane proteins, HA-ALG13-iso1 was immunoprecipitated from the cell lysate with anti-HA antibody and Protein A + G Agarose beads. The collected agarose beads were resuspended in 40 μL of reaction buffer. One half of this mixture (20 µl) was incubated with our standard reaction mixture to detect GnTase activity while the other half was subjected to SDS–PAGE analysis and immunoblotted with anti-ALG13 rabbit antibodies to confirm the expression and enrichment of HA-ALG13-iso1 proteins (data not shown). In parallel, HA-ALG13-iso2 was also expressed in HEK 293 cells and immunoprecipitated with anti-HA agarose (Table 2) (Gao et al., 2005). This pulled down ALG13-iso2 was selected as the positive control for GnTase activity because it can form an active GnTase complex with ALG14 in the ER membrane (Gao et al., 2005). Unexpectedly, by this assay HA-ALG13-iso1 did not show any GnTase activity, while HA-ALG13-iso2 could catalyze the formation of Gn2-PDol under the same reaction conditions (Figure 4B).
Because heterodimer formation is essential for ALG13/14 GnTase activity (Gao et al., 2008), we sought to determine the efficiency of complex formation between human ALG13-iso1 and ALG14 in HEK293 cells using co-immunoprecipitation assays (see Material and methods). HA-tagged ALG13-iso1 was co-expressed with FLAG-tagged ALG14 in HEK293 cells (Table 2). After extraction of the membrane proteins with 1% Triton X-100, HA-ALG13-iso1 was pulled down by anti-HA agarose. FLAG-ALG14 that co-precipitated with the HA-ALG13 was measured by immunoblotted with anti-FLAG antibodies. This experiment demonstrated that FLAG-tagged ALG14 completely failed to coprecipitate with HA-ALG13-iso1, although it efficiently interacted with HA-tagged ALG13-iso2 (Figure 4C). These experiments demonstrated that, unlike the shorter isoform 2, ALG13-iso1 does not display GnTase activity nor form a heterometric complex with ALG14.
To confirm the above results, a yeast complementation assay was performed to test whether ALG13-iso1 has any detectable in vivo GnTase activity. HA-ALG13-iso1 and ALG14-FLAG were co-expressed in yeast XGY186 strain (Table 2), in which both ALG13 and ALG14 chromosomal loci are deleted. The survival of this strain depends on the galactose-driven promoter ALG13/14 (GAL1pr-ALG13/14), which can be shut off by shifting to growth in medium containing glucose. To determine if ALG13-iso1 and ALG14 have in vivo activity, we measured their ability to rescue growth of this yeast strain in the presence of galactose versus glucose. As shown in Figure 4D, HA-ALG13-iso1 and ALG14-FLAG co-expression did not rescue the growth defect caused by the inhibition of yeast Alg13/14; in contrast, HA-ALG13-iso2 and ALG14-FLAG co-expression rescued the growth defect. Consistent with our in vitro results (Figure 4B), this complementation assay further confirmed that ALG13-iso1 does not possess GnTase activity in vivo or in vitro.
To date, a total of sixteen ALG13-CDG and nine ALG14-CDG missense mutations have been identified from patients (de Ligt et al., 2012; Timal et al., 2012; Esposito et al., 2013; Judith et al., 2013; Smith-Packard et al., 2015; Dimassi et al., 2016; Kobayashi et al., 2016; Bastaki et al., 2017; Hamici et al., 2017; Schorling et al., 2017; Kvarnung et al., 2018; Ng et al., 2020; Alsharhan et al., 2021; Datta et al., 2021; Palombo et al., 2021; Gang et al., 2022; Katata et al., 2022). Among these ALG13-CDG mutations, eight are located in the N-terminal region of ALG13 that is shared by isoform 1 and 2 (Figure 5A), while the remaining seven mutations are located in regions unique to ALG13-iso1, and only one mutation is located in short C-terminal of isoform 2 (data not shown). Based on the results from our study on ALG13 isoforms (Figures 1, 4), we predicted that the eight ALG13-CDG mutations located in the common N-terminal region may affect GnTase activity. To test this idea, we developed a semiquantitative assay to measure the effect of ALG13-CDG and ALG14-CDG mutations on GnTase activity.
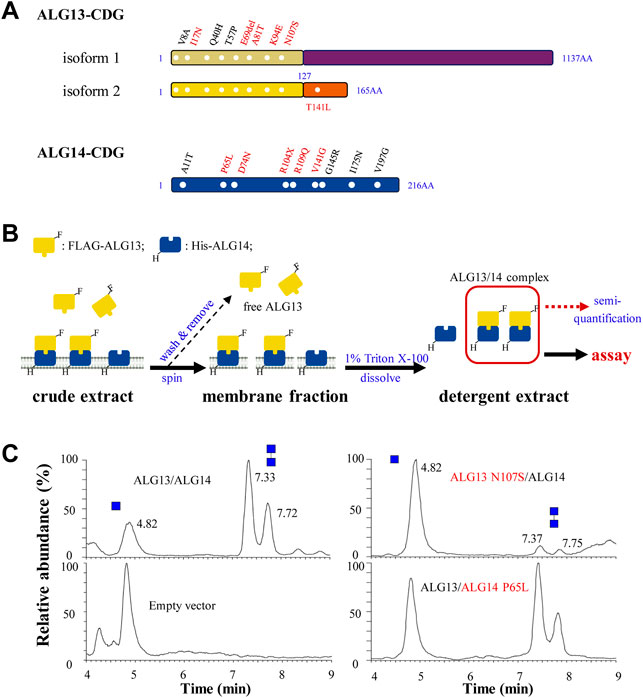
FIGURE 5. The semi-quantitative assay for ALG13- and ALG14-CDG mutants. (A) ALG13- and ALG14-CDG missense mutations distribution diagram. Six mutations in ALG13-iso2 and five mutations in ALG14 (labeled in red) were constructed; (B) Schematic diagram of ALG13/14 quantification. E. coli cells was broke by sonication and centrifuged to observe the membrane fraction, after washing 3 times by lysis buffer, the membrane fraction was solubilized and the detergent extract was used for GnTase activity assay, and the active ALG13/14 complex amount was semi-quantified using FLAG-ALG13 amount by anti-FLAG antibody; (C) The UPLC chromatogram of glycans released from the reaction of wild type ALG13/14, ALG13 N107S/ALG14 and ALG13/ALG14 P65L. Reaction included ALG13/ALG14 (0.1 ng/μl FLAG-ALG13 semi-quantified by western-blot) with the standard reaction mixture for 15 min.
We first chose ALG13 N107S and ALG14 P65L as candidates for this analysis as these two mutations have been identified in multiple CDG patients (see Figures 5A,B). These mutations were introduced into plasmids and co-expressed as FLAG (ALG13) or 6His (ALG14) tagged alleles in E. coli to produce strains co-expressing wild type (WT) ALG13 + WT ALG14, WT ALG13 + mutant ALG14 P65L or mutant ALG13 N107S and WT ALG14 (see Materials and Methods). Membrane fractions containing the ALG13 N107S/ALG14 or ALG13/ALG14 P65L complex were collected and solubilized in 5 ml lysis buffer with 1% Triton X-100 (Figure 5B, see Material and methods for details). These detergent extracts were analyzed with anti-FLAG or anti-His antibodies to detect the expression of the ALG13/ALG14 complex (Supplementary Figure S5A). To quickly quantify the concentration of the ALG13/ALG14 complex in the extract, a standard curve method was used, in which purified recombinant yeast Alg13 was used as the standard (Supplementary Figure S5B).
To compare the GnTase activity of the WT and CDG mutants, detergent extracts containing 0.1 ng/μl of ALG13 protein were incubated with the standard reaction mixture at 37°C for 15 min, followed by LC-MS analysis. The use of E. coli membrane which is absent from endogenous ALG13/14 would retain the enzyme activity and avoid the laborious protein purification steps. As shown in Figure 5C, WT ALG13/ALG14 complex GnTase catalyzed the production of Gn2-PDol (C95) with a conversion rate of 74%, while the conversion rate catalyzed by ALG13 N107S/ALG14 or ALG13/ALG14 P65L complex was 6% or 59%, respectively. These results demonstrated that ALG13 N107S and ALG14 P65L CDG mutations diminished the GnTase activity of ALG13/ALG14.
Similar to ALG13 N107S and ALG14 P65L, GnTase activities in several other ALG13- and ALG14-CDG mutants (Figure 5A, marked with red) were tested. Plasmids containing FLAG-tagged ALG13-iso2 I17N, E69del, A81T, K94E or T141L were transformed into E. coli cells and co-expressed with WT 6His-tagged ALG14. Similarly, plasmids containing 6His-tagged ALG14 R104X, D74N, R109Q or V141G were co-expressed with WT FLAG-tagged ALG13. After confirming protein expression of each ALG13-iso2 and ALG14 variant (Supplementary Figure S5C,E), detergent extracts prepared from each of these E. coli strains were assayed for GnTase activity. For a more rigorous comparison with WT ALG13/ALG14 GnTase, the relative activity of each mutation was introduced to show the remaining activity of the ALG13/ALG14 complex mutants. For example, as shown in Table 3, the relative activities were defined using the conversion rate of the mutants/the conversion rate of the WT, and ALG13 N107S/ALG14 and ALG13/ALG14 P65L were calculated as 8% and 80%, respectively. Notably, all of the ALG13/ALG14-CDG mutants that were tested displayed reduced GnTase activity despite the fact that most patients carrying these mutants had a normal N-linked glycosylation phenotype as judged by CDT analysis (Table 2) (Judith et al., 2013; Gadomski et al., 2017; Alsharhan et al., 2021). In conclusion, our results confirmed the GnTase deficiency of these ALG13/ALG14-CDG variants.
In the second step of the LLO synthetic pathway, the ALG13/14 complex catalyzes the transfer of GlcNAc from UDP-GlcNAc to Gn-PDol, resulting in the formation of Gn2-PDol. This reaction is essential for viability (Gao et al., 2005). Given its conserved role in LLO synthesis, it is reasonable to speculate that ALG13/14 GnTase affects proper N-glycosylation in all eukaryotes. However, contrary to other ALG-CDGs, most reported ALG13- and ALG14-CDG individuals showed essentially normal N-glycosylation (Judith et al., 2013; Schorling et al., 2017; Ng et al., 2020; Alsharhan et al., 2021). This unexpected phenomenon raised doubt about whether or how these ALG13/14-CDG mutations affect the formation of Gn2-PDol in ER LLO synthesis. To verify the causal relationship between ALG13/14 GnTase activity and DEE or CMS phenotypes of their related CDGs, we tried to develop an in vitro quantitative assay that directly detects GnTase activity using a recombinant ALG13/14 complex. Our assay system not only enabled us to study the kinetic properties of ALG13/14 GnTase but was also sensitive enough to discriminate the GnTase activities of various ALG13-or 14-CDG mutants.
In previous studies, we successfully used phytanyl-instead of dolichol-linked oligosaccharides as acceptor substrates for enzymatic studies of various ALG GTases (Li et al., 2017a; Li et al., 2017b; Ram Rez et al., 2017; Xu et al., 2018; Li et al., 2019; Xiang et al., 2022). However, when GlcNAc-PP-phytanyl was tested as the acceptor, unlike its yeast Alg13/14, counterpart human ALG13/14 failed to transfer GlcNAc to GlcNAc-PP-phytanyl (data not shown), suggesting a species-dependent substrate specificity of ALG13/14 GnTase. Therefore, in this study, we chose a natural Gn-PDol (C95) that contains 19 isoprene units of lipid tails as the acceptor substrate for the human ALG13/14 GnTase assay. In addition to determining the acceptor substrate, several other useful hints from previous studies were beneficial for development of the in vitro GnTase assay. For instance, we knew the formation of the ALG13/14 complex is critical for its GnTase activity (Gao et al., 2008). Moreover, there was no glycosylation modification reported or predicted in databases (https://www.uniprot.org, Q9NP73 and Q96F25; https://www.phosphosite.org) for ALG13 and ALG14. In this study, we tried to purify the active ALG13/14 complex from the E. coli membrane fraction by co-expression of ALG13 and ALG14. In contrast, GnTase activity was not detected by simply mixing the individual recombinant ALG13 and ALG14 proteins (Figure 1B). Using the purified ALG13/14 complex, we were able to determine the optimum conditions for the ALG13/14 GnTase reaction and study its kinetic properties (Figure 3). It is noteworthy that unlike other ALG GTases, ALG13/14 GnTase does not require the coordination of metal ions for its activity (Figure 3), demonstrating a metal-independent GTase in the LLO pathway.
We used this in vitro GnTase assay to study the splicing variants of human ALG13. Our results demonstrated that the shortest ALG13-iso2 isoform forms the active human ALG13/14 GnTase required for GlcNAc(β1,4)GlcNAc-PP-Dol (Gn2-PDol) production while the long isoform 1, which failed to interact with ALG14 and therefore lacked GnTase activity (Figures 4B,C). This interpretation was supported not only by a direct detection of GnTase activity of HA-tagged ALG13-iso1 immunoprecipitated from the HEK293 cell lysate but also by a complementation test of co-expressed human ALG13 and ALG14 in an ALG13- and ALG14-deficient yeast strain. In these experiments, ALG13-iso1 showed no GnTase activity either alone or in combination with ALG14 (Figures 4B,D), suggesting it cannot form a complex with ALG14, probably due to the lack of the C-terminus α-helical structure. As reported (Gao et al., 2008), the C-terminus α-helix of ALG13-iso2 is required for the interaction with ALG14 to obtain the enzymatic activity, which was supported by the decrease (26%) of a pathogenic mutation in the C-terminus of ALG13 (T141L, Table 2). Bioinformatic and phylogenetic analysis indicated that homologs of ALG13-iso2 are widely conserved among kingdoms of Fungi, Plantae, Animalia, and even in Protist, while ALG13-iso1 homologs are only found in Aves and Mammalia classes of the animal kingdom (data not shown). These findings further support our conclusion that isoform 2 is the only ALG13 isoform required for N-glycosylation. This raises the question of what is the function of ALG13-iso1. Besides containing the N-terminal GnTase catalytic domain, human ALG13-iso1 also contains an ovarian tumor deubiquitinase domain with predicted active sites in its C-terminal region. It has been suggested that ALG13 ovarian tumor deubiquitinase domain may catalyze ubiquitination of certain protein targets, such as GTases in the LLO pathway (Ng et al., 2020). It will be of interest to determine whether or not ALG13-iso1 regulates the activity of one of these GTases, or even ALG13-iso2, via its deubiquitinase activity. Nevertheless, in this study, we confirmed that ALG13-iso1 is not directly involved in ALG13/14 GnTase activity.
The E. coli expression system we used does not contain any endogenous interfering GTase activities and therefore has proven very useful for production and analysis of recombinant ALG proteins involved in LLO synthesis (Li et al., 2017a; Li et al., 2017b; Ram Rez et al., 2017; Li et al., 2019). Expression level of ALG13/14 in E. coli was approximately 100 folds than that in HEK293 cells. Therefore, we chose E. coli expression system to obtain larger amount of ALG13/14 for studying the enzymatic properties and synthesizing N-glycans in vitro. This property enabled a convenient semiquantitative GnTase assay, in which the membrane fraction of E. coli co-expressing various combinations of human ALG13 and ALG14 alleles were incubated with purified Gn-PDol (C95) acceptor (Figure 5). By omitting the purification step, this assay provided a quick way to test the GnTase activity of ALG13/14-CDG variants. Pathogenic mutants in ALG13 cause an X-linked congenital disorder of glycosylation with variable clinical phenotypes. Due to the lack of a suitable assay system, the effects of ALG13-CDG mutations on N-glycosylation were not yet understood. Using the ALG13 c. 320A>G (p.N107S) mutation as an example, more than 40 patients, including three males with this recurring mutation, were reported to have clear intellectual disabilities. However, all of the tested ALG13 N107S patients presented normal or only subtle deficiencies in N-glycan modifications using isoelectric focusing of transferrin or ESI-QTOF/MS assays (Smith-Packard et al., 2015; Dimassi et al., 2016; Bastaki et al., 2017; Hamici et al., 2017; Ng et al., 2020; Alsharhan et al., 2021). Our semiquantitative assay demonstrated that the ALG13 N107S/ALG14 complex possesses 8% relative activity compared with WT ALG13/14 GnTase (Figure 5; Table 2). The discrepancy between previously published results and those described here demonstrating severe deficiencies of ALG13 N107S emphasizes the importance of having a convenient and sensitive assay for discriminating the N-glycosylation abnormalities in patients with ALG13/14 mutations. Our results confirmed GnTase deficiency as the cause of ALG13/14-CDG. The in vitro assay we describe will open new avenues for better understanding N-glycosylation and its relationship to the pathogenicity and clinical severity of CDG.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
C-DW, SX, SC, and Z-HC performed experiments and analyzed data. X-DG, ND and NW proposed and supervised the project and wrote the manuscript. All authors confirmed and edited the manuscript.
This work was supported by grants-in-aid from the National Natural Science Foundation of China (21807048; 31971216; 22077053), Jiangsu Planned Projects for Postdoctoral Research Funds (2020Z167), Shandong Provincial Major Scientific and Technological Innovation Project (2019JZZY011006), Top-notch Academic Programs Project of Jiangsu Higher Education Institutions, Program of Introducing Talents of Discipline to Universities (No. 111-2–06), Special fund for Zaozhuang Excellence agglomeration project to X-DG and Qing Lan Project of Jiangsu Province to NW.
We thank Drs. Hideki Nakanishi, Morihisa Fujita, Zijie Li and Ganglong Yang for discussion. We are grateful to Haili Liu for the arrangement of the data collection facilities.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.1008078/full#supplementary-material
ALG, Asparagine-linked glycosylation; CDG, congenital disorders of glycosylation; CDT, carbohydrate-deficient transferrin; CMS, congenital myasthenic syndromes; DEE, developmental and epileptic encephalopathy; ER, endoplasmic reticulum; G, glucoses; GTases, glycosyltransferases; Gn, GlcNAc; GnTase, UDP-N-acetylglucosamine transferase; GlcNAc, N-acetylglucosamine; Gn-PDol, GlcNAc-PP-Dol; GDP-Man, GDP-Mannose; LC–MS, liquid chromatography/mass spectrometry; LLO, lipid-linked oligosaccharide; M, mannoses; PDol, dolichol pyrophosphate; SDS–PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; UDP-Glc, UDP-glucose; WT, wild type.
Aebi, M. (2013). N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833, 2430–2437. doi:10.1016/j.bbamcr.2013.04.001
Alsharhan, H., He, M., Edmondson, A. C., Daniel, E. J. P., Chen, J., Donald, T., et al. (2021). ALG13 X-linked intellectual disability: New variants, glycosylation analysis, and expanded phenotypes. J. Inherit. Metab. Dis. 44, 1001–1012. doi:10.1002/jimd.12378
Bastaki, F., Bizzar, S., Hamici, S., Nair, P., Mohamed, M., Saif, F., et al. (2017). Single-center experience of N-linked congenital disorders of glycosylation with a summary of molecularly characterized cases in arabs. Ann. Hum. Genet. 82, 35–47. doi:10.1111/ahg.12220
Bickel, T., Lehle, L., Schwarz, M., Aebi, M., and Jakob, C. A. (2005). Biosynthesis of lipid-linked oligosaccharides in Saccharomyces cerevisiae: Alg13p and Alg14p form a complex required for the formation of GlcNAc(2)-PP-dolichol. J. Biol. Chem. 280, 34500–34506. doi:10.1074/jbc.M506358200
Chantret, I., Dancourt, J., Barbat, A., and Moore, S. E. (2005). Two proteins homologous to the N- and C-terminal domains of the bacterial glycosyltransferase Murg are required for the second step of dolichyl-linked oligosaccharide synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 280, 9236–9242. doi:10.1074/jbc.M413941200
Cherepanova, N., Shrimal, S., and Gilmore, R. (2016). N-linked glycosylation and homeostasis of the endoplasmic reticulum. Curr. Opin. Cell Biol. 41, 57–65. doi:10.1016/j.ceb.2016.03.021
Datta, A. N., Bahi-Buisson, N., Bienvenu, T., Buerki, S. E., Gardiner, F., Cross, J. H., et al. (2021). The phenotypic spectrum of X-linked, infantile onset ALG13-related developmental and epileptic encephalopathy. Epilepsia 62, 325–334. doi:10.1111/epi.16761
de Ligt, J., Willemsen, M. H., van Bon, B. W., Kleefstra, T., Yntema, H. G., Kroes, T., et al. (2012). Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 367 (20), 1921–1929. doi:10.1056/NEJMoa1206524
Dimassi, S., Labalme, A., Ville, D., Calender, A., Mignot, C., Boutry-Kryza, N., et al. (2016). Whole-exome sequencing improves the diagnosis yield in sporadic infantile spasm syndrome. Clin. Genet. 89, 198–204. doi:10.1111/cge.12636
Dwyer, C. A., and Esko, J. D. (2016). Glycan susceptibility factors in autism spectrum disorders. Mol. Asp. Med. 51, 104–114. doi:10.1016/j.mam.2016.07.001
Esmail, S., and Manolson, M. F. (2021). Advances in understanding N-glycosylation structure, function, and regulation in health and disease. Eur. J. Cell Biol. 100, 151186. doi:10.1016/j.ejcb.2021.151186
Esposito, T., Lea, R. A., Maher, B. H., Moses, D., Cox, H. C., Magliocca, S., et al. (2013). Unique X-linked familial FSGS with co-segregating heart block disorder is associated with a mutation in the NXF5 gene. Hum. Mol. Genet. 22, 3654–3666. doi:10.1093/hmg/ddt215
Gadomski, T. E., Bolton, M., Alfadhel, M., Dvorak, C., Ogunsakin, O. A., Nelson, S. L., et al. (2017). ALG13-CDG in a male with seizures, normal cognitive development, and normal transferrin isoelectric focusing. Am. J. Med. Genet. A 173, 2772–2775. doi:10.1002/ajmg.a.38377
Gang, Q., Bettencourt, C., Brady, S., Holton, J. L., Healy, E. G., Mcconville, J., et al. (2022). Genetic defects are common in myopathies with tubular aggregates. Ann. Clin. Transl. Neurol. 9, 4–15. doi:10.1002/acn3.51477
Gao, P., Wang, F., Huo, J., Wan, D., Zhang, J., Niu, J., et al. (2019). ALG13 deficiency associated with increased seizure susceptibility and severity. Neuroscience 409, 204–221. doi:10.1016/j.neuroscience.2019.03.009
Gao, X. D., Moriyama, S., Miura, N., Dean, N., and Nishimura, S. (2008). Interaction between the C termini of Alg13 and Alg14 mediates formation of the active UDP-N-acetylglucosamine transferase complex. J. Biol. Chem. 283, 32534–32541. doi:10.1074/jbc.M804060200
Gao, X. D., Nishikawa, A., and Dean, N. (2004). Physical interactions between the Alg1, Alg2, and Alg11 mannosyltransferases of the endoplasmic reticulum. Glycobiology 14, 559–570. doi:10.1093/glycob/cwh072
Gao, X. D., Tachikawa, H., Sato, T., Jigami, Y., and Dean, N. (2005). Alg14 recruits Alg13 to the cytoplasmic face of the endoplasmic reticulum to form a novel bipartite UDP-N-acetylglucosamine transferase required for the second step of N-linked glycosylation. J. Biol. Chem. 280, 36254–36262. doi:10.1074/jbc.M507569200
Hamici, S., Bastaki, F., and Khalifa, M. (2017). Exome sequence identified a c.320A > G ALG13 variant in a female with infantile epileptic encephalopathy with normal glycosylation and random X inactivation: Review of the literature. Eur. J. Med. Genet. 60 (10), 541–547. doi:10.1016/j.ejmg.2017.07.014
Huo, J., Ren, S., Gao, P., Wan, D., Rong, S., Li, X., et al. (2020). ALG13 participates in epileptogenesis via regulation of GABA(A) receptors in mouse models. Cell Death Discov. 6, 87. doi:10.1038/s41420-020-00319-6
Jaeken, J. (2020). Congenital disorders of glycosylation: A multi-genetic disease family with multiple subcellular locations. J. Mother Child. 24, 14–20. doi:10.34763/jmotherandchild.20202402si.2005.000004
Judith, C., Katsiaryna, B., Debbie, H., Salih, M. A., Sarah, F., Nicola, C., et al. (2013). Congenital myasthenic syndromes due to mutations in ALG2 and ALG14. Brain 136, 944–956. doi:10.1093/brain/awt010
Katata, Y., Uneoka, S., Saijyo, N., Aihara, Y., Miyazoe, T., Koyamaishi, S., et al. (2022). The longest reported sibling survivors of a severe form of congenital myasthenic syndrome with the ALG14 pathogenic variant. Am. J. Med. Genet. A 188, 1293–1298. doi:10.1002/ajmg.a.62629
Kobayashi, Y., Tohyama, J., Kato, M., Akasaka, N., Magara, S., Kawashima, H., et al. (2016). High prevalence of genetic alterations in early-onset epileptic encephalopathies associated with infantile movement disorders. Brain Dev. 38, 285–292. doi:10.1016/j.braindev.2015.09.011
Kvarnung, M., Taylan, F., Nilsson, D., Anderlid, B. M., Malmgren, H., Lagerstedt-Robinson, K., et al. (2018). Genomic screening in rare disorders: New mutations and phenotypes, highlighting ALG14 as a novel cause of severe intellectual disability. Clin. Genet. 94, 528–537. doi:10.1111/cge.13448
Li, S. T., Lu, T. T., Xu, X. X., Ding, Y., Gao, X. D., Kitajima, T., et al. (2019). Reconstitution of the lipid-linked oligosaccharide pathway for assembly of high-mannose N-glycans. Nat. Commun. 10, 1813. doi:10.1038/s41467-019-09752-3
Li, S. T., Wang, N., Xu, S., Yin, J., Gao, X. D., Dean, N., et al. (2017a). Quantitative study of yeast Alg1 beta-1, 4 mannosyltransferase activity, a key enzyme involved in protein N-glycosylation. Biochim. Biophys. Acta. Gen. Subj. 1861, 2934–2941. doi:10.1016/j.bbagen.2016.09.023
Li, S. T., Wang, N., Xu, X. X., Fujita, M., Gao, X. D., Kitajima, T., et al. (2017b). Alternative routes for synthesis of N-linked glycans by Alg2 mannosyltransferase. FASEB J. 32, 2492–2506. doi:10.1096/fj.201701267R
Loke, I., Kolarich, D., Packer, N. H., and Thaysen-Andersen, M. (2016). Emerging roles of protein mannosylation in inflammation and infection. Mol. Asp. Med. 51, 31–55. doi:10.1016/j.mam.2016.04.004
Meyer, B. H., Adam, P. S., Wagstaff, B. A., Kolyfetis, G. E., Probst, A. J., Albers, S. V., et al. (2022). Agl24 is an ancient archaeal homolog of the eukaryotic N-glycan chitobiose synthesis enzymes. Elife 11, e67448. doi:10.7554/eLife.67448
Ng, B. G., Eklund, E. A., Shiryaev, S. A., Dong, Y. Y., Abbott, M. A., Asteggiano, C., et al. (2020). Predominant and novel de novo variants in 29 individuals with ALG13 deficiency: Clinical description, biomarker status, biochemical analysis, and treatment suggestions. J. Inherit. Metab. Dis. 43, 1333–1348. doi:10.1002/jimd.12290
Noffz, C., Keppler-Ross, S., and Dean, N. (2009). Hetero-oligomeric interactions between early glycosyltransferases of the dolichol cycle. Glycobiology 19, 472–478. doi:10.1093/glycob/cwp001
Palombo, F., Piccolo, B., Saccani, E., Fiorini, C., Capristo, M., Caporali, L., et al. (2021). A novel ALG14 missense variant in an alive child with myopathy, epilepsy, and progressive cerebral atrophy. Am. J. Med. Genet. A 185, 1918–1921. doi:10.1002/ajmg.a.62153
Pascoal, C., Francisco, R., Ferro, T., Dos Reis Ferreira, V., Jaeken, J., and Videira, P. A. (2020). CDG and immune response: From bedside to bench and back. J. Inherit. Metab. Dis. 43, 90–124. doi:10.1002/jimd.12126
Ram Rez, A., Boilevin, J., Lin, C. W., Gan, B. H., Locher, K. P., Aebi, M., et al. (2017). Chemo-enzymatic synthesis of lipid-linked GlcNAc2Man5 oligosaccharides using recombinant Alg1, Alg2 and Alg11 proteins. Glycobiology 27, 726–733. doi:10.1093/glycob/cwx045
Schorling, D. C., Rost, S., Lefeber, D. J., Brady, L., Kirschner, J., Korinthenberg, R., et al. (2017). Early and lethal neurodegeneration with myasthenic and myopathic features: A new ALG14-CDG. Neurology 89, 657–664. doi:10.1212/WNL.0000000000004234
Schwarz, F., and Aebi, M. (2011). Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 21, 576–582. doi:10.1016/j.sbi.2011.08.005
Shrimal, S., and Gilmore, R. (2019). Oligosaccharyltransferase structures provide novel insight into the mechanism of asparagine-linked glycosylation in prokaryotic and eukaryotic cells. Glycobiology 29, 288–297. doi:10.1093/glycob/cwy093
Smith-Packard, B., Myers, S. M., and Williams, M. S. (2015). Girls with seizures due to the c.320A>G variant in ALG13 do not show abnormal glycosylation pattern on standard testing. JIMD Rep. 22, 95–98. doi:10.1007/8904_2015_416
Sosicka, P., Ng, B. G., and Freeze, H. H. (2021). Chemical therapies for congenital disorders of glycosylation. ACS Chem. Biol. acschembio.1c00601. doi:10.1021/acschembio.1c00601
Takahashi, M., Kizuka, Y., Ohtsubo, K., Gu, J., and Taniguchi, N. (2016). Disease-associated glycans on cell surface proteins. Mol. Asp. Med. 51, 56–70. doi:10.1016/j.mam.2016.04.008
Timal, S., Hoischen, A., Lehle, L., Adamowicz, M., Huijben, K., Sykut-Cegielska, J., et al. (2012). Gene identification in the congenital disorders of glycosylation type I by whole-exome sequencing. Hum. Mol. Genet. 21, 4151–4161. doi:10.1093/hmg/dds123
Wang, N., Li, S. T., Xiang, M. H., and Gao, X. D. (2022). Alg mannosyltransferases: From functional and structural analyses to the lipid-linked oligosaccharide pathway reconstitution. Biochim. Biophys. Acta. Gen. Subj. 1866, 130112. doi:10.1016/j.bbagen.2022.130112
Xiang, M. H., Xu, X. X., Wang, C. D., Chen, S., Xu, S., Xu, X. Y., et al. (2022). Topological and enzymatic analysis of human Alg2 mannosyltransferase reveals its role in lipid-linked oligosaccharide biosynthetic pathway. Commun. Biol. 5, 117. doi:10.1038/s42003-022-03066-9
Keywords: N-glycosylation, lipid-linked oligosaccharide (LLO), ALG glycosyltransferases, ALG13/14 UDP-N-acetylglucosamine transferase, ALG13 isoforms, congenital disorders of glycosylation (CDG)
Citation: Wang C-D, Xu S, Chen S, Chen Z-H, Dean N, Wang N and Gao X-D (2022) An in vitro assay for enzymatic studies on human ALG13/14 heterodimeric UDP-N-acetylglucosamine transferase. Front. Cell Dev. Biol. 10:1008078. doi: 10.3389/fcell.2022.1008078
Received: 31 July 2022; Accepted: 30 August 2022;
Published: 19 September 2022.
Edited by:
Ashutosh Pandey, Baylor College of Medicine, United StatesReviewed by:
Yoshinao Wada, Osaka Women’s and Children’s Hospital, JapanCopyright © 2022 Wang, Xu, Chen, Chen, Dean, Wang and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Dong Gao, eGRnYW9AaXBlLmFjLmNu; Ning Wang, d2FuZ25pbmdAamlhbmduYW4uZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.