- Institute for Cell Biology (Cancer Research), Medical Faculty, University of Duisburg-Essen, Essen, Germany
Stem cells display a unique cell type within the body that has the capacity to self-renew and differentiate into specialized cell types. Compared to pluripotent stem cells, adult stem cells (ASC) such as mesenchymal stem cells (MSCs) and hematopoietic stem cells (HSCs) exhibit restricted differentiation capabilities that are limited to cell types typically found in the tissue of origin, which implicates that there must be a certain code or priming determined by the tissue of origin. HOX genes, a subset of homeobox genes encoding transcription factors that are generally repressed in undifferentiated pluripotent stem cells, emerged here as master regulators of cell identity and cell fate during embryogenesis, and in maintaining this positional identity throughout life as well as specifying various regional properties of respective tissues. Concurrently, intricate molecular circuits regulated by diverse stem cell-typical signaling pathways, balance stem cell maintenance, proliferation and differentiation. However, it still needs to be unraveled how stem cell-related signaling pathways establish and regulate ASC-specific HOX expression pattern with different temporal-spatial topography, known as the HOX code. This comprehensive review therefore summarizes the current knowledge of specific ASC-related HOX expression patterns and how these were integrated into stem cell-related signaling pathways. Understanding the mechanism of HOX gene regulation in stem cells may provide new ways to manipulate stem cell fate and function leading to improved and new approaches in the field of regenerative medicine.
Introduction
Stem cells are undifferentiated cells that represent unique cell types within the body. Based on their self-renewal capacity (multiply themselves) and their differentiation potential (develop into specialized cells), stem cells are clearly essential for tissue growth and maintenance. The many different types of stem cells are formed at different times in life, and differ in the places in the body where they persist (Heins et al., 2004; Snippert and Clevers, 2011; Alvarez et al., 2012). Embryonic stem cells (ESCs) exist only at the earliest stages of development (4–7 days after fertilization) and can be extracted from the inner cell mass of the blastocyst (Evans and Kaufman, 1981; Thomson et al., 1998). ESCs are known as pluripotent stem cells, which are able to self-renew and to give rise to all cell types of the three embryonic germ layers (ectoderm, mesoderm, endoderm). Under appropriate laboratory conditions ESCs can be grown in the undifferentiated state and potentially proliferate indefinitely. As postnatal derivates of ESCs, various types of tissue-specific so-called adult stem cells (ASCs) appear during fetal development and remain as “primitive cells” in a specialized environment called niche in our bodies throughout life. ASCs are found in almost every tissue, e.g., in umbilical cord, placenta, bone marrow, larger blood vessels, lung, skin, and fat tissue (Hsu and Fuchs, 2012; Klein, 2016; Redondo et al., 2017; Cable et al., 2020; Klein, 2021). These non-reproductive “somatic” stem cells can be divided into hematopoietic stem cells (HSCs; blood stem cells), mesenchymal stem cells (MSCs) and epithelial stem cells (EpSCs) as well as neural stem cells (NSCs) (Dulak et al., 2015; Cable et al., 2020). ASCs have a high proliferative potential and -depending on their tissue of origin- the capacity to differentiate into various cell types. Compared to ESCs, the differentiation potential of ASCs is restricted, meaning multipotency (Kaebisch et al., 2015; Zakrzewski et al., 2019).
As tissue-resident stem cells, ASCs generally were important orchestrators of normal tissue homeostasis with the potential to suppress inflammation by direct or indirect cellular communications resulting in immune cell education and thus disease-specific microenvironment regulation (Bernardo and Fibbe, 2013; Planat-Benard et al., 2021). In healthy conditions, ASCs are initially considered quiescent until “activation” by organ demands, e.g. following a pathological trigger, which induces proliferation and/or migration out of their niche, and finally fosters tissue regeneration in a paracrine fashion or by differentiation to replace damaged cells (Rumman et al., 2015; Klein, 2016; Clevers and Watt, 2018; Klein, 2021; Urban and Cheung, 2021). Thus, ASCs are essential for the constant renewal of all tissues maintaining organ structure and function. Due to the enormous regenerative potential furthermore, the transplantation of isolated and in vitro expanded ASCs and especially MSCs has established itself as a possible strategy for the therapy of a large number of diseases, including graft-versus-host disease, lung injury, and bone diseases and defects (Cheung et al., 2020; Fu et al., 2021; Klein, 2021). The safety and feasibility of such stem cell based-based therapeutic approaches has already been confirmed in numerous clinical studies and is still the subject of current studies. However, while the exact mechanism of MSC action remains elusive, numerous preclinical studies are already showing ways to increase the therapeutic efficacy, particularly of MSCs, through specific modulations to produce exogenous stem cells with superior repair capabilities (Hu and Li, 2018; Gardin et al., 2020; Ocansey et al., 2020). Besides an inherent tropism towards inflammatory sites, MSCs also exhibit a natural tumor-trophic migration ability, suggesting that exogenous MSCs could be exploited as pathotropic delivery vehicles when loaded with bioactive anticancer drugs (Ding et al., 2021; Sentek and Klein, 2021; Takayama et al., 2021). Within these scenarios, a continuous presence of the cells does not seem to be necessary as the therapeutic potential of exogenously applied ASCs is mainly based on a “hit/kiss and run” mechanism. Recent studies also suggest that release of paracrine factors from MSCs is accomplished via so-called extracellular vesicles (EVs) such as exosomes and microvesicles (Witwer et al., 2019; Massa et al., 2020). As carriers of the whole information panel characterizing the use of ASCs for regenerative purposes, these ASC- and particularly MSC-derived EVs thus hold advantages as non-self-replicating “entities” for therapeutic applications. However, the quality of ASC-derived EVs from different sources and across batches was shown to be difficult and moreover inconsistent, which severely restricts respective quality control and management (Kou et al., 2022). In a recently published and very elegant work, the development of ‘NANOBIOME’, namely NANOmetric BIO-banked MSC-derived Exosome, is specifically proposed here for biobanking of EVs secreted by MSCs for their easy and available storage and distribution, since the use of ASC-derived EVs in particular circumvents specific biobanking problems, i.e. technical problems and regulatory concerns that have so far limited the use, for example, of MSC banking for rapid regenerative applications (Codispoti et al., 2018). Thus, regarding prospects of basic research and clinical applications, protocol standardization, including precise evaluation of samples in terms of harvesting rate, characterization as well as pre-clinical parameter assessment are worthy of attention and exploration (Codispoti et al., 2018). Within these challenges, the limited secretion of EVs from ASCs maybe a bottleneck for efficient exosome production and application, which in turn would require efficient pre-treatment strategies of ASCs for boosting the biogenesis and secretion of ASC-derived EVs (Wang et al., 2020).
Several growth factors as well as small molecules generally orchestrate the cells signaling important for stem cell maintenance and cell-type specific functions, and signaling alterations can affect the cellular features of stem cells, particularly induce differentiation resulting in the conversion of stem cells into appropriate progenitor cells, which in turn give rise to associated cell types (Figure 1). An improved understanding of these signaling pathways and key signaling molecules for the promotion of homogenous stem cell populations and/or targeted differentiation of stem cells is not only crucial to improve stem cell function as internal repair systems of the body; the potentially manipulation of core gene networks could even improve culture conditions of respective stem cells that might allow refined generation of desired stem cell cultures and tissues with refined functional features for regenerative approaches. Signals that influence stem cell specialization processes, including the determination of cellular identity, are intimately linked to gene expression networks (Wells and Choi, 2019; Zakrzewski et al., 2019). Here, the HOX genes and HOX-regulated signaling pathways have evolved to become an integral regulator of stem cell identity and cell fate. This review therefore summarizes the latest insights how HOX gene expressions are regulated in stem cells and how HOX genes orchestrate stem cell maintenance, differentiation, and thus function.
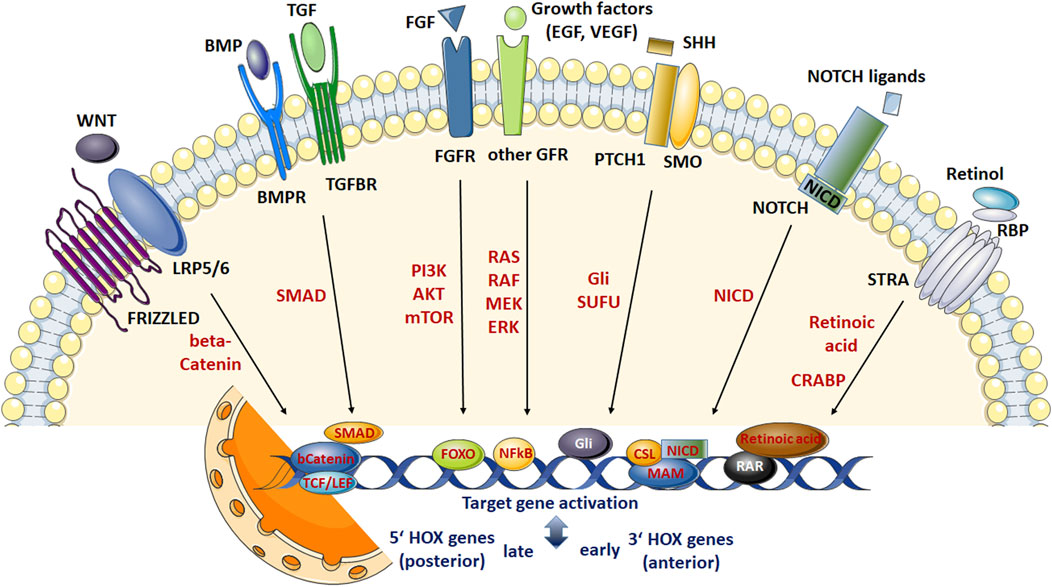
FIGURE 1. Scheme summarizing the most relevant signaling networks regulating stem cell fate. Development of adult stem cells (ASCs) as well as stem cell maintenance, proliferation, differentiation and survival require complex interactions between diverse molecular signaling pathways and downstream transduction molecules. These pathways include WNT signaling, signaling by multi-functional growth factors that belong to the transforming growth factor (TGF) beta superfamily, fibroblast growth factor (FGF) signaling, sonic hedgehog (SHH), NOTCH, and retinoic acid (RA) signaling pathways. WNT ligand binding to the receptor complex consisting of FRIZZLED and low-density lipoprotein receptor-related proteins 5/6 (LPR 5/6) results in intracellular β-catenin stabilization enabling nuclear translocation. Nuclear β-catenin then elicits gene expression changes (including HOX genes) through the T-cell factor (TCF)/lymphoid enhancer factor (LEF) family of transcription factors. Signaling pathways initiated by TGFβ ligands are transduced through cell surface receptor complexes resulting in (type I; BMPR, TGFR) receptor phosphorylation and serine-threonine phosphorylations of (effector) SMAD transcription factorsmeaning activation. Following nuclear translocation (in complexes formed with SMAD4) target gene transcriptions (including HOX genes) are activated. SMADs further can interactwith β-catenin and LEF/TCF transcriptional regulators enablingWNTsignaling in a TGFβ-dependentmanner. FGF ligands, and FGF receptors (FGFR) -like other growth factor receptors including epidermal and vascular endothelial growth factor receptors- lead to autophosphorylation of the (intracellular) protein tyrosine kinase domains and activation of various effectors such as RAS/RAF/mitogen-activated protein kinase (MAPK) and phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/AKT/mTOR and RAS/RAF/MAPKs finally mediating NF-kB (nuclear factor “kappa-light-chain-enhancer” of activated B-cells) or FOXO (Forkhead Box O)-dependent gene expressions (including HOX genes). Other signal transducers (Phospholipase C Gamma, PLC-γ) and activators of transcription (STAT) pathways can also be activated, which intersect and synergize with other signaling pathways, e.g.,WNT, RA and TGFβ signaling (not shown). SHHsignals through a receptor complex that includes the G-protein-coupled receptor smoothened (SMO) and the (twelve-pass)membrane protein patched 1 (PTCH1). In response to SHH ligand binding, SUFU (suppressor of fused) binding, and thus cytoplasmic sequestrations of GLI (glioma-associated oncogene family members) transcription factors become inhibited, leading to GLI stabilization and nuclear translocation resulting in SHH signal transduction, namely transcriptional activation of SHH target genes (includingHOX genes). NOTCH receptor activation results in NOTCH cleavage (through a cascade of proteolytic cleavages by ADAM metalloprotease and γ-secretase) releasing the intracellular domain of the receptor (NICD). NICD translocates to the nucleus, displaces corepressor complex, and recruits coactivators finally forming a ternary complex with the DNA binding protein CSL and the transcriptional coactivator Mastermind (MAM) to activate transcription of Notch target genes (including HOX genes). NICD can also activate the NF-κB transcription factor and thus cooperatewith growth factor signaling. All-trans RA and other active retinoids generated fromvitamin A (retinol)mediate their action by binding toRAreceptors (RAR), nuclear receptors acting as transcription factors, which are bound to DNA as a heterodimer with the retinoid X receptor (RXR) in regions called retinoic acid response elements (RAREs). The multi-transmembrane protein STRA6 was shown to mediate mediates vitamin A uptake from plasma retinol binding protein 4 (RBP4). DNA interaction of RA following nuclear transport by CRABP (cellular retinoic acid binding protein) induces transcription of genes encoding transcription factors and signaling proteins that further modify gene expression particularly early HOX genes, e.g., HOXA1 with sequential activation of the clustered HOX genes in an anterior-posterior order that resembles their positions in the chromosomal cluster. RA can also activate FOXOtranscription factors and thus cooperate with growth factor, particularly FGF signaling. Generally, it is assumed that anteriorHOXgenes locatedmore at the 3′ end of a chromosome are preferentially activated by RA pathways, while activation of posterior 5′ HOX genes are preferred by BMP and WNT signaling.
Signaling networks regulating stem cell fate
The formation of tissues and organs from naïve stem and progenitor cells is controlled by combinatorial signaling of certain pathways in distinct temporal windows to progressively direct embryonic cells through a series of fate decisions into specific tissue lineages. Thus, stem cell functions in adults from quiescence through “activation” resulting in self-renewal, mobilization or differentiation require tightly control by these signaling networks and complex cross-talks between the different signaling pathways (Blank et al., 2008; Denner et al., 2010; Tanabe, 2015). Fibroblast growth factor (FGF) and leukemia inhibitory factor mediated signaling turned out to be of critical importance in regulating key transcriptional regulators of pluripotency, including sex determining region Y box 2 (SOX2), octamer-binding transcription factor (OCT) 3/4, krueppel-like factor 4 (KLF4), NANOG, and c-MYC (Coutu and Galipeau, 2011; Theunissen and Jaenisch, 2014; Tanabe, 2015; Mossahebi-Mohammadi et al., 2020). Signaling pathways that further regulate cell fate decisions particularly stem cell maintenance, self-renewal and differentiation, include: (i) Wingless related integration site (WNT) molecules supposed to act as niches factors maintaining a self-renewing state (Nusse, 2008; Van Camp et al., 2014), (ii) bone morphogenetic proteins (BMPs) that belong to the transformation growth factor beta (TGFβ) superfamily critically controlling differentiation (including epithelial and mesenchymal transitions) and cell death (Wagner, 2007; Sakaki-Yumoto et al., 2013), (iii) sonic hedgehog (SHH) signaling that is an important player for ASC maintenance (Petrova and Joyner, 2014), (iv) NOTCH signaling known to be responsible for maintaining balance between the different cell fates (Chiba, 2006; Wang et al., 2009), as well as (v) small molecules such as retinoic acid (RA) that orchestrate FGF signaling to drive differentiation, and thus decisively impact on balancing of stem cell quiescence and activation (Stavridis et al., 2010; Gudas and Wagner, 2011) (Figure 1). These signaling pathways in turn modulate the expression of homeotic selector (HOX) transcription factor genes, known as master regulators of cellular fate, in a spatio-temporal manner, finally balancing stem cell maintenance, proliferation, differentiation, and survival. However, it remains be unraveled how these signaling pathways and decisive molecular pathway candidates here mediate full colinear HOX activation and enable deterministic patterning of diverse cell-type specific HOX profiles, which could potentially be used to recapitulate HOX expression profiles e.g., for the generation of target cells or for an on-site manipulation of HOX signaling alterations in diseased states.
Homeotic selector transcription factors as master regulators of (tissue-) specific stem cells identities.
Establishing homeotic selector gene expression
HOX genes comprise a family of highly conserved, regulatory genes that encode transcription factors, which control the activity of other, functionally contiguous genes in the context of morphogenesis. Beside a number of other gene families that include position-relevant information for body construction (e.g., PAX and T-BOX genes), the main task of HOX genes is structuring the embryo along the body longitudinal axis (Figure 1). HOX genes are named after the homeobox, a evolutionarily conserved 180 nucleotide pairs long sequence encoding for the homeodomain (Figure 2A), a DNA binding motif allowing respective HOX proteins to negatively or positively regulate target gene expressions (Krumlauf, 1994; Ruddle et al., 1994). In vertebrates, HOX genes were found to be arranged in four clusters (A–D) with the known 39 human HOX genes being located on chromosome 7q, 17q, 12q and 2q (Ruddle et al., 1994). The order of the HOX genes on a chromosome corresponds to the time expressing the body sections controlled by them and follows HOX gene expression along the anterior-posterior axis (Figure 2B). This means that HOX genes controlling the development of terminal body segments are also at the end of this gene group on the chromosome (Harding et al., 1985; Kmita and Duboule, 2003; Seo et al., 2004). During embryonic development HOX genes become activated following the temporal sequence in 3′-5′direction (temporal collinearity) and correlating with the gene position (spatial collinearity) (Gaunt, 2015; Durston, 2019a).
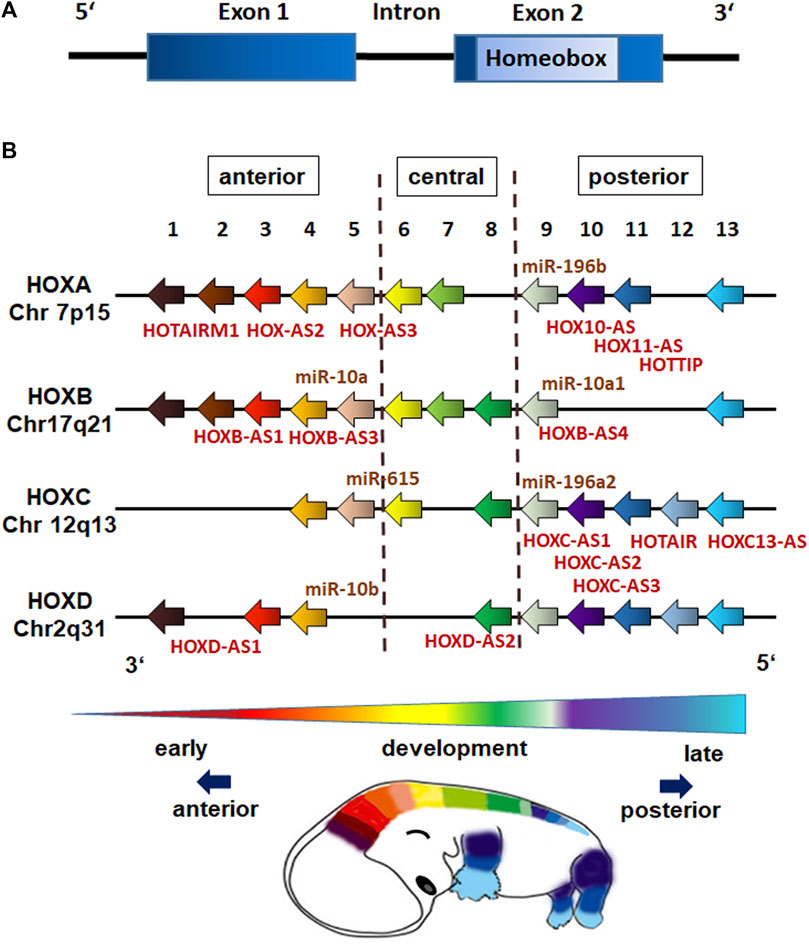
FIGURE 2. HOX gene structure and genome organization (schematic representation). (A) HOX genes are comprised of one intron separating two exons with the second exon having a 120-nucleotide sequence encoding for the 60 amino acid DNA-binding domain known as the homeobox (homeodomain). (B) The 39 human HOX genes are clustered into the four HOX families HOXA, HOXB, HOXC, and HOXD with each family consisting of nine to eleven paralogous genes (assigned by numbers based on sequence similarity and cluster position), which are responsible for the anterior-posterior specification of body segments. The position of non-coding RNAs that are interspersed within the coding HOX genes are marked (miR,microRNAs; AS, antisense RNAs). HOX gene expressions exhibit spatial and temporal collinearity: nested domains of HOX genes are generated with anteriorly HOX expressions operating earlier in development and posteriorly HOX expressions occurring later.
HOX gene expression is generally coordinated by transcriptional regulation of HOX genes from earlier segmentation genes and cross-regulatory interactions among HOX genes (Morata and Kerridge, 1982; Irish et al., 1989). 3′ anterior HOX genes are expressed first and can activate expression of 5′posterior HOX genes, a regulation called posterior induction (PI) (Durston, 2019b). Posterior HOX genes in turn are able to suppress anterior ones at functional level, a phenomenon called posterior prevalence (PP) (Akam, 1987). The newer concept of posterior dominance (PD) nowadays describes the regulation of HOX expressions at mRNA level (Durston, 2019b). HOX proteins bind to small AT rich base regions, often with the same TAAT core sequence (Noyes et al., 2008). HOX-DNA-binding specificities are further achieved by cooperatively binding cofactors such as TALE homeodomain cofactor proteins PBX and MEIS, and by HOX collaborators, namely proteins that bind in parallel to HOX-targeted cis-regulatory modules (CRMs) (Mann et al., 2009; Slattery et al., 2011; Sanchez-Higueras et al., 2019). An integration of growth factors and signaling molecules such as RA, FGFs, BMPs and WNTs, which are expressed in gradients along the embryonic axis further determine the restricted domains of HOX expression (Diez del Corral and Storey, 2004; Darras et al., 2018; Nolte et al., 2019; Afzal and Krumlauf, 2022). Small non-coding RNAs (ncRNAs) such as microRNAs and the long ncRNAs (lncRNA) can further impact on HOX gene expressions (Figure 2B). LncRNAs for example were shown to determine an epigenetic profile of HOX loci by association with Polycomb Group (PcG) and Trithorax Group (ThrxG) proteins (Rinn et al., 2007). The lncRNA HOTAIR, which is located within the HOXC locus, takes part in silencing of HOXD genes by interacting with polycomb repressive complex 2 (PRC2) and thus complex formation finally leading to histone H3 lysine 27 trimethylation (H3K27me3) of the HOXD locus to repress genes (Rinn et al., 2007). In contrast, the lncRNA HOTTIP that is located at the 5′end of the HOXA locus was shown to coordinate the activation of various 5′ HOXA genes (Wang et al., 2011). Mechanistically, HOTTIP directly binds to the adapter protein WDR5, which is a component of the mixed lineage leukemia complex methylating lysine 4 of histone H3 across the HOXA locus leading to histone H3 lysine 4 trimethylation (H3K4), and thus gene activation (Wang et al., 2011). Generally, histone H3 methylations leading to an active H3K4 and a repressive H3K27 mark (with H3K27me3 being dominant over the H3K4me3) decisively regulate HOX expression patterns. During gastrulation, trimethylation of H3K27 is induced by PRC2, which recruits PRC1 leading to inhibition of chromatin remodeling activity and chromatin condensation to maintain silencing of HOX genes (Rea et al., 2000; Soshnikova and Duboule, 2008). Demethylation of H3K27 in turn can induce the expression of the hitherto repressed (lineage-specific) HOX genes (Bernstein et al., 2006; Soshnikova and Duboule, 2008). These chromosomal domains marked either by an active H3K4 or silent H3K27 turned out to be crucial to establish an epigenetic memory (including the HOX genes) of the cellular identity in stem cells and moreover following differentiation (Ringrose and Paro, 2007; Kim et al., 2010; Ezziane, 2012). The simultaneous expression of a certain combination of HOX genes (termed the “HOX-code”) further on was presented to be tissue-specific (Kessel and Gruss, 1991), as HOX genes impose positional identity to developing tissues (Mallo et al., 2010). In adults, this segmental or particularly HOX memory persists in downstream tissue-resident stem cells (Kamkar et al., 2016; Smith et al., 2019). Accordingly, stem cells exhibit heterogeneous but characteristic HOX expression profiles that are highly specific for their anatomical origin, and are maintained during differentiation (Ackema and Charite, 2008; Liedtke et al., 2010; Klein et al., 2013; Kamkar et al., 2016; Smith et al., 2019). Together with the fact that nested domains of HOX expression arise in part through the ability of HOX clusters to integrate and respond to certain signaling gradients, it is important to understand these interactions, and how decisive regulatory mechanisms through which signaling pathways coordinately control the precise HOX expression patterns that are required for specifying diverse stem cell features including morphology (Afzal and Krumlauf, 2022).
The homeotic selector code
Homeotic selector gene expression in pluripotent stem cells
Pluripotent stem cells are characterized by the capabilities to self-renew and to differentiate into the three primary germ cell layers during early embryogenesis, and therefore to generate all cells of the adult body. Within humans, OCT4, SOX2 and the homeobox transcription factor NANOG are considered to be the core nuclear transcription factors that regulate pluripotency, particularly in ESCs (Chambers et al., 2003; Boyer et al., 2005; Young, 2011). Other ESC characteristic markers, which are used to define, characterize and to isolate ESCs are the cell surface antigens stage-specific embryonic antigen-1 (SSEA1/CD15), SSEA3, SSEA4, as well as TRA-1-60 (Podocalyxin/TRA-1-81), and signal pathway-related intracellular markers like the signal transducer SMAD2/3, the TGFβ/Activin/Nodal signal pathway, transcription activators like the WNT/β-catenin signaling pathway, as well as the enzymatic marker alkaline phosphatase (Zhao et al., 2012). As an “in vitro alternative”, nonembryonic, so-called induced pluripotent stem cells (iPSCs) reprogrammed from somatic cells (Takahashi and Yamanaka, 2006; Takahashi et al., 2007) have been established, which have the same pluripotent characteristics than ESCs but lack ethical concerns raised about the use of embryos for ESC isolation (Choi et al., 2015; De Los Angeles et al., 2015; Volarevic et al., 2018).
As HOX genes are not expressed before gastrulation, HOX genes were not found to be transcribed in ESCs (Deschamps and Duboule, 2017). Pluripotent stem cells in general were shown to harbor an active epigenetic repression of HOX genes in this undifferentiated, naïve state (Ezziane, 2012; Luo et al., 2019; Smith et al., 2019). Herein, HOX gene promoters often exhibit both H3K4me3 and H3K27me3 marks, and thereby HOX genes remain in a dormant state waiting for activation (Soshnikova and Duboule, 2008). Upon differentiation, histone demethylases such as UTX (lysine-specific demethylase 6A, known as ubiquitously transcribed tetratricopeptide repeat, X chromosome) and JMJD3 (Jumonji domain-containing protein-3) are recruited to remove repressive marks on H3K27 from HOX lineage-specific genes, while transcriptional repression of HOX genes being specific for other lineages are maintained by PcGs (Agger et al., 2007; Riising et al., 2014). Similarly, so-called unrestricted somatic stem cells (USSCs) from human cord blood that exhibit a broad ecto-, meso-, and endodermal differentiation potential were characterized by the absence of HOX expressions (Liedtke et al., 2010; Santourlidis et al., 2011). Although USSCs lack presence of the major stem cell factors OCT4, SOX2, and NANOG, an epigenetic state in between that of an ESC and of a terminally differentiated cell was reported enabling USSCs to exhibit differentiation and reprogramming cues typical for pluripotent stem cells (Santourlidis et al., 2011). In the course of the differentiation process tendencies for HOX profiles become then more prominent and refined. However, it needs still to be unraveled which HOX code exactly commits pluripotent stem cells to a certain lineage. The recent discoveries of HOX profiles specific to ASCs, thus orchestrating multipotent stem cells through differentiation and tissue-specific maturation processes are now summarized.
Homeotic selector gene expression in adult stem cells
The positional identity that emerged from HOX gene expression patterns during embryonic development was found to be maintained in many adult tissues, particularly in ASCs, suggesting that the topographic specificity of these HOX codes as an intrinsic property is maintained during differentiation, and thus provides a mechanism for imposing cell identity and fate restriction (Yamamoto et al., 2003; Smith et al., 2019). Indeed, ASCs were found to exhibit characteristic HOX expression signatures that are heterogeneous but highly specific for their anatomical origin (Figure 3). The persistent expression of specific HOX genes in many adult tissues indicates that, in addition to the cellular identity, the function of these cells and respective organ tissues containing them is also determined by HOX genes meaning that HOX genes were co-opted for location-specific functions (Wellik, 2009; Krumlauf and Ahn, 2013; Quinonez and Innis, 2014). Due to the large number of findings in the ASCs, the following focus concerning stem cell-type specific HOX gene expression pattern and how this is related to stem cell function is rather on mesodermal stem cells: MSCs, HSCs and endothelial progenitor cells (EPCs).
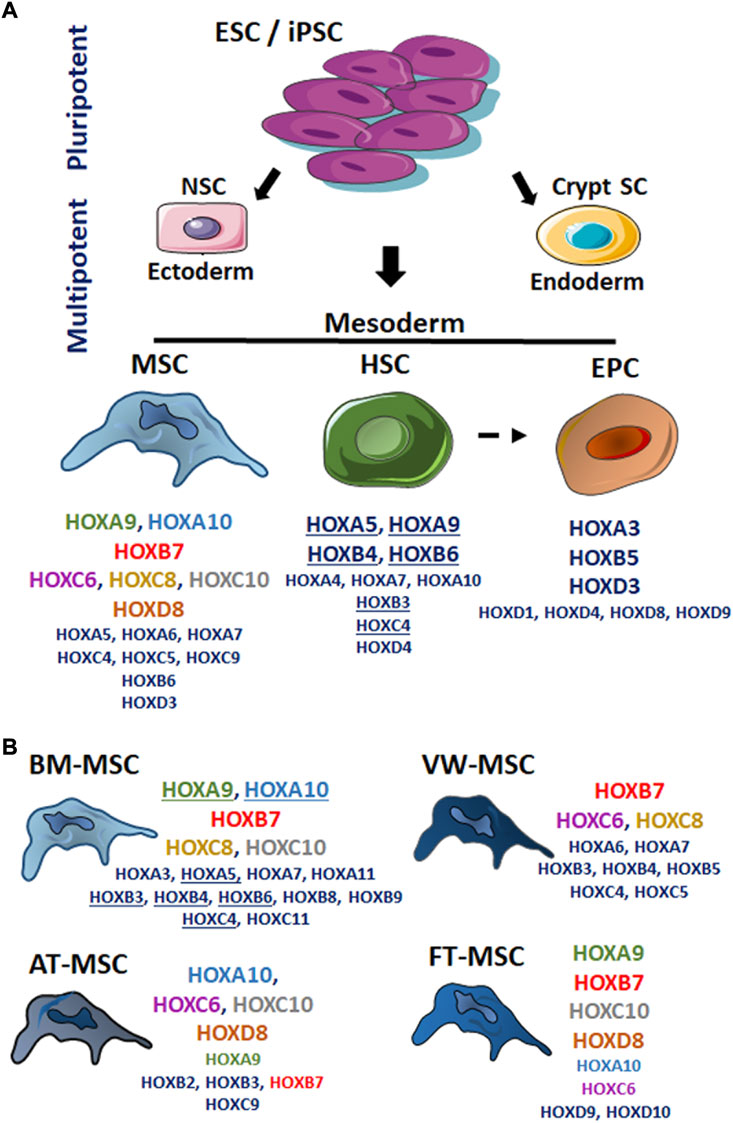
FIGURE 3. The HOX code of mesodermal stem cells. Stem cells derived from different tissues present patterns of HOX gene expression (“the HOX code”) that mirrors their developmental origin. According to the fact that HOX genes are not expressed before gastrulation, HOX genes were not found to be transcribed in non-differentiated, pluripotent stem cells (SC) due to active epigenetic repression of HOX genes. In cells, particularly stem cells lying at equivalent anteroposterior positions but in distinct embryonic germ layers, HOX proteins have distinct regulatory activities. (A) The reported HOX expression pattern for the mesodermal-derived adult stem cell types were listened: mesenchymal stem cell (MSC), hematopoietic stem cell (HSC) and endothelial progenitor cell (EPC). (For details: see main text.) (B) HOX expression pattern for bone marrow (BM)-, vascular wall (VW-), adipose tissue (AT)-, and fetal tissue (FT; summarizing umbilical cord, placenta and amniotic fluid MSCs)-MSCs were separately shown. Capital letters name more frequently identified HOX genes potentially representing the cell type-specific HOX code, whereas smaller letters designate individual additionally identified HOX genes above it. Same colors highlight same HOX genes that are common in different MSC types; underlined HOX genes emphasize similar HOX genes between BM-MSCs and HSCs. ESC, embryonic stem cell; iPSC, induced pluripotent stem cell; NSC neural stem cell.
Homeotic selector gene expression in mesenchymal stem cells
Human MSCs can be isolated and expanded from almost every organ, preferred however from bone marrow, peripheral blood and various neonatal birth-associated tissues, blood vessels, and adipose tissue. Although these MSCs were phenotypically highly similar, MSC cultures are generally quite heterogenous. According to the minimal MSC criteria (as defined by the International Society of Cell Therapy) in vitro expanded MSC must adhere on plastic, express the MSC markers CD73, CD90 and CD105 while lacking expression of hematopoietic and endothelial lineage markers, and exhibit the capability of in vitro differentiation into adipocyte, osteoblast, and chondrocyte lineages (Dominici et al., 2006; Viswanathan et al., 2019).
Compared gene expression profiling of MSCs derived from bone marrow (BM), adipose tissue and cord blood identified 25 well-characterized genes including the two HOX genes HOXA5 and HOXB6 in all MSC preparations, irrespective of origin and culture conditions (Wagner et al., 2006). However, none of these genes alone was specific for MSCs and the definition of a unique marker, particularly a HOX gene or a HOX gene constellation, for MSCs remained elusive (Wagner et al., 2006; Ho et al., 2008). Investigations using murine MSCs (identified as colony forming unit-fibroblasts) from different organs firstly revealed that MSCs of different origins -although phenotypically highly similar- can be distinguished by their specific topographic HOX code depending on the tissue of origin (Ackema and Charite, 2008). Consistent with the assumption that the HOX code is an intrinsic property of ASCs, it was hypothesized that the MSC-type specific HOX code is part of a “blueprint” required for the tissue-specific (regenerative) action of MSCs (Ackema and Charite, 2008). Accordingly, it was suggested that MSC-specific HOX expression patterns as “biological fingerprint” could be used to distinguish human MSC populations of functionally distinct tissues (Liedtke et al., 2010). HOXA9, HOXB7, HOXC10 and HOXD8 (and to a lesser extend HOXA10 and HOXC6) were defined as potential molecular markers, which are highly expressed in fetal MSC derived from cord blood (CB-MSCs; compared to USSC) (Jansen et al., 2010; Liedtke et al., 2010). The HOXA cluster here turned out to be highly methylated, whereas the HOXB-D clusters are less methylated indicating that HOX genes from these clusters (HOXB6,7, HOXC4-10, HOXD3,4) are involved in CB-MSC maintenance and function (Liedtke et al., 2010). BM-MSCs showed a highly similar HOX code compared to CB-MSCs identifying HOXD9 and HOXD10 (being absent in BM-MSC) as decisive HOX genes enabling distinction (Liedtke et al., 2010). HOXC10 (being present in Decidua-derived MSCs) was further defined as a potential marker to distinguish amnion- and decidua-derived MSCs (Hwang et al., 2009). And HOXC10 (together with T-box 15 transcription factor) seemed to be a fundamental developmental transcription factor in adipose tissue-derived MSCs (AT-MSCs) (AD-MSCs) (Onate et al., 2013; Miklosz et al., 2022). Furthermore, increased expressions of HOXA10, HOXC6, HOXD8, and to a lesser extend of HOXA9, HOXB2, HOXB3, HOXC9, HOXC10 were reported in these cells (Kouidhi et al., 2015). BM-MSCs, as one of the commonly used sources for MSCs, usually show increased expressions of HOXA9, HOXA10, HOXB4, HOXB7, HOXC8, HOXC10, and HOXD8 (Coenen et al., 2015). By comparing different human BM-MSCs, namely MSCs isolated from bone marrow isolated from iliac crest, sternum, and vertebrae, it was shown that BM-MSCs at all express HOX genes in high numbers (35/39), with the different BM-MSC populations expressing specific sets of HOX genes in increased levels (Picchi et al., 2013). HOXC10 and HOXC11 were found to be upregulated in iliac crest-MSCs, HOXB8 and HOXB9 in vertebrae-MSCs, and HOXA3, HOXA5, HOXB6, HOXB7, HOXB9, HOXC4 as well as HOXC8 in sternum-MSCs (Picchi et al., 2013). Besides, so-called vascular wall-derived MSCs (VW-MSCs), which are located within the vascular stem cell niche, in the so-called “vasculogenic zone” of the blood vessel wall, were characterized by increased expression levels of HOXB7, HOXC6 and HOXC8; but also HOXA6, HOXA7, HOXB3, HOXB4 and HOXB5 as well as HOXC4 and HOXC5 expression can be detected (Klein et al., 2013). Even lung resident MSCs (LR-MSCs) were shown to express the VW-MSC specific HOX code of HOXB7, HOXC6 and HOXC8 (together with HOXB5), as these cells were found to be predominately located with the vascular adventitial stem cell niche (Klein, 2021; Steens et al., 2021). Accordingly, LR-MSCs turned out to be phenotypically and functionally indistinguishable from VW-MSC, a fact that further highlights the HOX code as master regulator of cellular identity.
Taken together, a distinct expression of HOXA9 and HOXA10, HOXB7, HOXC6, HOXC8, and HOXC10, as well as HOXD8 can frequently be observed in MSCs derived from different human tissues, and thus in certain combinations- potentially represent the MSC-type specific HOX code (Figure 3). It can also be stated that mainly central HOX genes, or HOX genes that are in the immediate vicinity of the central cluster region are expressed within tissue-specific MSCs. But besides their central role in defining segment identity, ensuring that the right structures are formed in the right body places, the activity HOX genes as well as their expression levels control segment-specific structures and cell types. Herein, other less frequently reported HOX genes, e.g. HOXA5, HOXA6 and HOXA7, HOXB6, HOXC4, HOXC5, and HOXC9, as well as HOXD3 might be important contributors to these actions.
The most common and reliable way to identify MSCs (in addition to surface marker analysis), is to verify their trilineage differentiation potential into adipocytes, osteoblasts, and chondrocytes in vivo and in vitro. HOX genes were of functional importance in regulating these differentiation processes [reviewed in (Seifert et al., 2015)]. As revealed from early mouse studies for example, the HOX candidate HOXC8 was shown to critically regulate the progression of cells along the chondrogenic differentiation pathway (Yueh et al., 1998). Particularly in MSCs, HOXC8 expressions were shown to be upregulated during chondrogenic differentiation and a fostered HOXC8 expression caused an enhanced expression of chondrogenic markers, promoted the chondrogenic differentiation and the formation of cartilage clumps (Yang et al., 2020a), while osteogenic differentiation was suppressed (Yang et al., 2020b). A forced expression of HOXC8 was even associated with adipogenesis inhibition (Mori et al., 2012). As another example, HOXA11 and HOXD11 functioned in regulating (the early steps) of chondrocyte differentiation (Gross et al., 2012). Loss of function studies further revealed that HOX11 impairments in MSCs of the bone marrow (and periosteum) at adult stages caused defects in differentiation, leading to an overall deficit in the cartilage production and thus defects in endochondral ossification (Rux et al., 2016). And in certain MSC types, namely in regional skeletal MSCs, which arose from the earliest stages of skeletal development, HOX11 expression turned out to be decisive for their self-renewal and for functioning as progenitors for osteoblasts, chondrocytes and adipocytes throughout lifetime (Pineault et al., 2019). Particularly HOXA cluster genes seem to play a central role here, because HOXA cluster negative cord blood MSCs failed to differentiate properly into the chondro-osteogenic lineages with the HOX candidates HOXA2 and HOXA10 being of special meaning (Liedtke et al., 2017). Herein, HOXA10 in BM-MSCs was already shown enabling RUNX2 activation, a central regulator of osteogenesis (Hassan et al., 2007). Mechanistically HOXA10 mediated chromatin hyperacetylation and H3K4 methylation of the bone-related RUNX2 P1 promoter by interacting through a HOX core motif, that caused activation of early osteogenic genes through the chromatin remodeling (Hassan et al., 2007). HOXC10 was further identified as a direct target of the lncRNA lncHOXC-AS3, which stabilized HOXC10 expression in BM-MSCs thereby regulating osteogenesis (Li et al., 2019).
Thus, it is clear that HOX genes exert MSC-specific functions particularly concerning the trilineage potential along mesodermal lineages. However, the observed differences of HOX genes in regulating similar processes maybe based in the difference of HOX codes orchestrating different but phenotypical highly similar MSC types, which in turn dependent of the tissue of origin. Interestingly, HOXB7 turned out to be an “universal MSC” HOX gene as it was reported to be expressed in all the different types of MSCs (Liedtke et al., 2010; Klein et al., 2013; Foppiani et al., 2019). An induced expression of HOXB7 was identified as a master player driving proliferation and differentiation of human BM- and AT-MSCs (Foppiani et al., 2019; Casari et al., 2021), and accounts for the high proliferation and sprouting potential of VW-MSCs (Klein et al., 2013; Klein, 2016). However, rather than accounting for stem cell type-specific functions, HOX genes specify various regional properties of a tissue along the rostral-caudal axis by regulating a wide range of cellular activities such as proliferation and differentiation, cellular adhesion, migration and invasion, as well as cell death (Parrish et al., 2009). The persisting HOX expression profiles of the different tissue-specific MSCs strongly suggest that those specific HOX profiles are of particular importance for the functioning of tissues throughout adult life with resident HOX-expressing MSCs as leading regulators of embedding stroma renewal and regeneration (Kulebyakina and Makarevich, 2020). Following a rough classification along the cranial-caudal direction, HOX1-4 paralogues were shown to act predominately in cranial tissues, followed by HOX5-6 in subsequent upper sternal tissues, and HOX7-8 in lower sternal and abdominal tissues, whereas the paralogues HOX9-13 represent HOX genes building up HOX codes in caudal body parts and extremities (Kulebyakina and Makarevich, 2020). An example for the association of HOX gene expression within the organization of tissue-specific structures and features comes from VW-MSCs (Klein et al., 2013). Under normal tissue homeostasis (“quiesence”), when residing in the adventitial vasculogenic stem cell niche, these cells express the HOX pattern HOXB7, HOXC6 and HOXC8 at high level that were shown to suppress the expression of transgelin (TAGLN), a TGFβ-inducible gene-inducible gene found to be essentiell for early smooth muscle cell differentiation. Upon commitment, VW-MSCs progress through tissue-specific differentiation events, where a silencing of the VW-specific HOX genes altered the TAGLN promotor methylation sites finally causing increased TAGLN expression, which induced then VW-MSC differentiation into tissue-typical smooth muscle cells (Klein et al., 2013).
Thus, modulation of cell-type specific HOX expression levels as well as respective patterns might offer potential druggable targets for innovative on-site tissue regeneration approaches or for the improved generation of stem cells in vitro, particularly of MSCs with superior repair capabilities. Within that scenario, enforcing trans-differentiation of pluripotent stem cells or even other somatic cell types towards MSCs by ectopic expression of HOX genes as MSC-specific transcription factors turned out to be a straight forward approach to generate MSCs in vitro in huge numbers desirable especially for regenerative purposes (Steens and Klein, 2018; Abdal Dayem et al., 2019). The in vitro generation of vascular wall-typical MSCs from (murine) iPSCs, based on a VW-wall MSC-specific HOX code was already reported (Klein et al., 2013; Steens et al., 2017; Steens et al., 2020a). A lentiviral vector expressing a small set of identified (human) VW-MSCs-specific HOX genes, namely HOXB7, HOXC6 and HOXC8 was here used to directly program murine iPSCs into MSCs, which then displayed classical MSC characteristics, both in vitro and in vivo. As HOX selector genes are highly conserved throughout evolution, it is assumed that forced expression of this HOX code also leads to MSC differentiation from human iPSCs, although the final proof for human iPSCs remains to be shown (Steens et al., 2017; Steens et al., 2020a). It could be shown accordingly, that the same triple combination of HOX genes as VW-MSC-specific gene code was sufficient to directly convert human skin fibroblasts towards MSCs, and thus directing cell fate conversion bypassing pluripotency (Steens et al., 2020b). The introduced HOX-code in primary human skin fibroblasts could further be linked to an increased colony formation and trilineage differentiation potential as (mesenchymal) stem cell characteristics (Steens et al., 2020b). However, it remains to be clarified whether the triple HOX combination approach of HOXB7, HOXC6 and HOXC8 actually retains the VW-MSC-specific master regulatory function to generate VW-MSCs, or whether the induced expression of individual (HOXB7, HOXC6 or HOX candidates already could serve as MSC-specific key transcription factor. Corresponding investigations could also shed light on which MSC characteristic(s) depend(s) on which introduced HOX gene.
Homeotic selector expressions in hematopoietic stem cells and endothelial progenitor cells
HSCs can be classically found in the bone marrow and the peripheral blood being prerequisite for sustained hematopoietic reconstitution and the formation of blood cells. Whereas HSCs were known to be derived from mesodermal precursor cells called hemangioblasts during embryogenesis, HSC in adults (as identified phenotypically by tyrosine-protein kinase c-KIT/CD117 and stem cell antigen-1 (SCA1) expressions, while being lineage negative and by exhibiting functional hemangioblast activity) serve as a rich source for circulating EPCs in addition to the generation of blood cells (Cogle and Scott, 2004). During adult life, EPCs have been defined by their cell surface expression of the hematopoietic marker proteins CD133 and CD34 and the endothelial marker vascular endothelial growth factor receptor-2 (VEGFR2), and their capacity to incorporate into sites of neovascularization prior in situ differentiation into endothelial cells; thus, being decisive for new vessel formation (Asahara and Kawamoto, 2004; Ishige-Wada et al., 2016; Yoder, 2018). HSCs as well as their derived hematopoietic progenitors were known to be characterized by HOX genes in a pattern characteristic of the lineage and stage of differentiation of the cell (He et al., 2011), although total HOX expression levels of HSCs were suggested to be rather low (Bijl et al., 2006; Ackema and Charite, 2008).
The vigorous ability of HSC to produce huge numbers of lineage differentiated cells includes erythrocytes, (mega-) karyocytes, innate and acquired immune cells (Haas et al., 2018). And hematopoiesis is critically impacted by HOX gene expressions: anterior HOX genes (comprising the numbers 1–6 of the 3′ region) are maximally expressed in the more primitive HSCs, whereas posterior HOX genes (comprising the numbers 7–13 of the 5′ region) become prominent in the committed progenitors (Sauvageau et al., 1994; He et al., 2011). Quantification of HOX gene expression levels in the different hematopoietic cell populations isolated from human peripheral blood revealed that all these cells generally express HOXA and HOXC cluster genes at significantly higher levels compared to expression levels HOXB and HOXD cluster genes (10–100-fold lower) (Morgan and Whiting, 2008). High HOXA5 expression levels for example were suggested to account for hematopoietic lineage determination being able to promote differentiation along myelopoietic lineages (Fuller et al., 1999; Argiropoulos and Humphries, 2007). HOXB3, HOXB4, and HOXA9 were shown to play a crucial role for the presence of HSCs, as a combined deficiency in these HOX genes fostered severe effects on hematopoietic organs (Magnusson et al., 2007). Furthermore, the HOX candidates HOXA9, HOXB4, and HOXB6 were highly expressed in HSCs (Seifert et al., 2015; Bhatlekar et al., 2018). Particularly high HOXB4 expression levels seemed to be important for most potent HSCs, namely long-term reconstitution HSCs (Wang et al., 2005; Fan et al., 2012; Forrester and Jackson, 2012). A forced HOXB4 expression was shown to generate HSCs in vitro, and to account for the proliferative response of long-term repopulating HSCs as well as for HSC maintenance (Brun et al., 2004; Pilat et al., 2005). And, the other way around, a reduction of HOXB4 led to a reduction of HSC proliferation (and differentiation) and, to a reduction of neighboring HOX genes in an 3′to 5′order namely of HOXB2, HOXB3, HOXB5 (Brun et al., 2004). Similarly, HOXA4 and the HOX paralog group 4 at all were shown to be decisive for a functional HSC phenotype (Iacovino et al., 2009). Besides, HOXA5, HOXA9, and HOXA10 were identified as three of seven transcription factors (together with ERG, LCOR, RUNX1 and SPI1) to convert “pre-differentiated” tissue, namely haemogenic endothelium cells into HSCs (Sugimura et al., 2017; Fidanza and Forrester, 2021). Studies from the in vitro generation of HSCs using human pluripotent stem cells further revealed that HOXA cluster genes were significantly downregulated in (CD34-positive) HSCs that were incapable of long-term engraftment and repopulation, strongly indicating that HOXA genes critically regulate definitive hematopoiesis (Ng et al., 2016). Particularly, increased HOXA5 and HOXA7 expression levels were associated with the repopulation activity (Dou et al., 2016). In accordance with the 3′ to 5′ HOX direction, the HOX candidate HOXA9 then together with the ETS-family transcription factor ERG were further shown to re-specify lineage-restricted (CD34− and CD45-positive) HSC precursors derived from human iPSCs into primitive (CD34-positive and CD38-negative) HSCs (Doulatov et al., 2013). Thus, stimulation of HOXA gene expression potentially improves HSC maintenance generating self-renewing HSCs from pluripotent stem cells (Collins and Thompson, 2018).
Decreases in HOX gene expressions were then observed upon HSC differentiation in a manner that seems to follow their anterior-posterior position with anterior HOX genes being downregulated earlier than posterior HOX genes (Guo et al., 2003; He et al., 2011). Generally, HOXB and single HOXC cluster genes were associated to hematopoietic cells with erythroid features, and HOXA cluster genes with myeloid features (Lawrence et al., 1996; He et al., 2011). Within that scenario, HOXA5 and HOXA9 were shown to be involved in the proliferation and differentiation of HSCs to common myeloid progenitors, with HOXA9 also regulating the differentiation of HSCs into common lymphoid progenitors (Bhatlekar et al., 2018). During the differentiation (of pre-B cells) into B cells, HOXB3 was found to be a relevant factor, while HOXC3 and HOXC4 crucially impacted on erythroid lineage differentiation (Bhatlekar et al., 2018). HOXA5 and HOXC8 further seemed to regulate erythroid differentiation of megakaryocyte-erythrocyte progenitors, with HOXA7 being expressed during megakaryocyte differentiation (Bhatlekar et al., 2018).
Conclusively, HOX gene expression does not only specify HSC identity, HOX genes significantly impact in HSC function as well as in stages of hematopoietic differentiation; and dysregulation of these HOX genes were associated with a number of leukocyte malignancies (as discussed elsewhere: (McGonigle et al., 2008; Alharbi et al., 2013)). However, a precise activation of indicated HOX genes following stimulation of different pathways orchestrating HSC characteristics were not reported up to now (Demirci et al., 2020). Thus, the sequential activation pattern of HOX genes following signaling induced HSC differentiation remains to be unraveled.
Although a number of transcriptomic profiles and EPCs characterization are available, the role of HOX genes in EPCs known to be involved in neovascularization processes remains nearly completely elusive (Medina et al., 2010; Wu et al., 2018; Abdelgawad et al., 2021). This may be due, at least in part, to the fact that many different cell subtypes are consistently grouped together under the term “EPC”, which in turn would argue in favor of setting minimum criteria for defining EPCs, e.g., detailed immunophenotyping and/or potency assays, similar to defining minimal criteria for characterizing MSCs (Medina et al., 2017). However, several HOX transcription factors were already associated with target gene expression known to promote the differentiation of mature endothelial cells, which might indicate that also EPCs exhibit a specific HOX code. HOXB5 might be an EPC HOX code candidate, as HOXB5 was involved in the in vitro differentiation of embryonic precursor cells towards the endothelial lineage (Wu et al., 2003). Endothelial cell differentiation of ESCs further showed that HOXA3 and HOXD3 are immediately expressed when differentiation is induced, whereas HOXA5 and HOXD10 are expressed in more mature and adult endothelial cells (Bahrami et al., 2011). Accordingly, high expressions of HOXD3 together with HOXD1, HOXD4, HOXD8 and HOXD9 were reported for less mature blood-derived outgrowth endothelial cells (Toshner et al., 2014). HOXD3 was further linked to endothelial activation (from a resting to an angiogenic state) (Boudreau et al., 1997), and HOXB3 seemed to be required for subsequent capillary morphogenesis (Myers et al., 2000). And HOXA13 was found to be essential for placental vascular patterning and labyrinth endothelial specification (Shaut et al., 2008). HOX genes involvement in endothelial differentiation was further revealed when the HOX expression profiles of BM-MSCs were investigated following endothelial differentiation (Chung et al., 2009). The expression patterns of the four HOX genes HOXA7, HOXB3, HOXA3, and HOXB13, significantly changed during endothelial cell differentiation with expression levels of HOXA7 and HOXB3 becoming increased, whereas those of HOXA3 and HOXB13 became decreased. According to the central role of HOXA9 for the cellular identity of HSCs, HOXA9 was shown to mediate maturation of endothelial cells and being a master switch to regulate the expression of typical endothelial-committed genes such as endothelial nitric oxide synthase, VEGFR2, and VE-cadherin (Rossig et al., 2005). HOXA9-deficiency was further reported to result in reduced EPCs numbers and thereby impaired the postnatal neovascularization capacity (Rossig et al., 2005). Likewise, reduced HOXA9 expression levels were estimated in CD34-positive cells of hypertensive patients, an effect that correlated with reduced EPC numbers (Pirro et al., 2007). EPCs isolated from umbilical cord blood showed that HOXD9 seemed to be required for EPC maintenance (Iordache et al., 2015). Thus, several HOX genes, especially HOX genes from HOXA and HOXD cluster, take part in EPC maintenance, endothelial cell differentiation and function, however an EPC-specific HOX code could not be defined up to now. It seems that within endothelial differentiation processes a complex interaction of various HOX genes are important. EPC or endothelial cell-specific HOX codes, and how respective HOX genes then contribute to EPC-endothelial cell functions as well as to the proper functioning of the adult vasculature need to be unraveled.
Signaling networks regulating stem cell properties by modulating homeotic selector gene regulation
The diverse molecular signaling pathways and dependent transduction molecules known to orchestrate various stem cell characteristics regulate and control HOX gene expression. In addition to the involvement of HOX genes in the positional identity of stem cells along the stem cell hierarchy, HOX genes decisively function in all stem cell characteristics: self-renewal and maintenance, proliferation and survival, migration and invasion and lineage specification (differentiation). However, less is known how the main signaling pathways operating in stem cells regulate HOX expressions, and how single HOX transcription factors in turn impact on stem cell-related signaling pathways, and thus account for certain stem cell functions. Now we summarize the recent knowledge how the important stem cell-related signaling pathways such as WNT, BMP, FGF, SHH, NOTCH and RA, integrate HOX genes for maintaining cellular identity and regulation of differentiation.
Generally, during human embryogenesis, more 3′ located (anterior) HOX genes (HOX1-8) become activated and regulate paraxial mesoderm development with the formation of embryonic primordial segments called somites by modulating cell ingression into the primitive streak (Iimura and Pourquie, 2006). Subsequently activated more 5′ located (posterior) HOX genes (HOX10-13) limit mesoderm ingression by repressing WNT signaling, and mediating body axis termination. (Young et al., 2009; Denans et al., 2015). Concerning the signaling it is known that the paraxial regions of the mesoderm become specified following the action of BMPs, members of the FGF family and WNT molecules. Cross-regulation of these pathways, e.g., by the WNT effector molecule NOGGIN that inhibits BMP signaling, is not only important for the paraxial mesoderm specification but also for maintaining this identity (Aulehla and Pourquie, 2010; Budjan et al., 2022). As a model of human somitogenesis, paraxial mesoderm organoids were developed by differentiating human pluripotent stem cells towards paraxial mesoderm using a combination of WNT signaling activation together with BMP inhibition and following FGF2 treatment (Budjan et al., 2022). Thereby, the sequential activation pattern of the HOX genes was elegantly recapitulated. HOXA1 expression was early detected following WNT activation and BMP inhibition (with 24 h), followed by other cervical (HOX1-HOX5 paralogues) and thoracic (HOX6-HOX9 paralogues) HOX genes within the next 48–72 h, to HOXD9, a lumbosacral HOX gene in the somite-stage organoids of day 4–5, which was 24–48 h after the successive FGF2 treatment (Budjan et al., 2022). Concerning the mechanism it was revealed that WNT molecules activate HOX gene expression in a temporally collinear way as WNT-dependent enhancers, namely interaction regions with HOXA1 called “HOXA developmental early side (ADES)” that are located within the posterior region. WNT signals (particularly WNT3) caused thereby removal of H3K27 repressive marks on the 3′ region of the HOXA cluster that in turn promotes transcription of HOXA genes (Neijts et al., 2016). Subsequently to initial WNT-induced activation of 3′ HOX genes, coactivated caudal type homeobox genes (Cdx genes) were shown to act as crucial effectors for expression induction of central and 5′ HOX genes in a collinear manner (Neijts et al., 2017).
RA (a vitamin A metabolite)-dependent signaling is another central signaling pathway being essential for early embryonic development and the promotion of stem cell lineage specifications in pluripotent stem cells (Soprano et al., 2007; Gudas and Wagner, 2011). Studying the kinetics of transcriptional and epigenomic patterning responses of RA in ESCs revealed that RA plays even an essential role in HOX gene regulation by binding to specific retinoic acid receptors (NR1Bs) at RA responsive DNA elements (RAREs) located within the HOX clusters finally erasing H3K27me3 from repressed HOX genes during ESC differentiation (Kashyap et al., 2011). RA stimulation generally induces activation and recruitment of demethylases to HOX genes, which in turn become demethylated thereby promoting HOX expression and thus differentiation (Shahhoseini et al., 2013). Genes at the 3′ end of the complexes (e.g., the paralogs HOXA1, HOXB1, and HOXD1) display herein a higher responsiveness to RA while genes at the 5’ end (the paralogs HOXA13, HOXB13, HOXC13, and HOXD13) are more responsive to FGF signaling (Nolte et al., 2019). FGF signaling was shown to interact antagonistically with RA signaling during posterior development, finally resulting in morphogen gradients within the anterior-posterior-axis with high RA concentrations at the anterior and high FGF concentrations at the posterior end, which lead to an increased expression of the HOX genes HOXB9 and HOXA7 (Kumar et al., 2021).
Thus, stem cell fate decisions into specific tissue lineages require distinct temporal windows for the known stem cell-related signaling pathways and respective cross-regulations (Rankin et al., 2018). On the one hand, FGF-related signaling was shown to orchestrate the pluripotent stage (Mossahebi-Mohammadi et al., 2020). FGF family members, namely FGF2 and FGF4 were shown to function as intrinsic regulators of pluripotent stem cell self-renewal and survival by promoting MAPK and AKT signaling pathways. Particularly MAPK signaling seems to be required, whereas PI3K/AKT signals increase as pluripotency gets restricted (Lee et al., 2019; Mossahebi-Mohammadi et al., 2020). Moreover, FGF-related signaling being essential for pluripotency maintenance, especially FGF4 expression, depends on the presence of the pluripotency transcription factors OCT4 (and SOX2). On the other hand, FGF-related signaling, particularly through FGF2, has even the potential to induce lineage differentiation of pluripotent cells (Steens and Klein, 2018; Ma et al., 2019). An FGF2 activation was found to activate the downstream kinase Src, a non-receptor tyrosine kinase that in turn activates MEK1/2 (Mitogen-activated protein kinase 1) resulting in differentiation and/or increased proliferation particularly of mesodermal ASCs (Ma et al., 2019). Very recently it was shown that respective processes here were linked to HOX gene regulation (Tiana et al., 2022). The pluripotency factor OCT4 turned out to be important for maintaining HOX genes silent in the stage of pluripotency. Upon lineage commitment however, OCT4 switches from a repressor to an activator being required for the proper activation of HOX genes. Especially the genes of the HOXB cluster are coordinately regulated by OCT4 binding sites located at the 3′ end of the cluster (Tiana et al., 2022).
Concerning the time of induced HOX expressions, so how extrinsic factors influence the tempo of HOX expressions, it was recently shown that the progressive activation of HOX genes in pluripotent stem cells is controlled by a dynamic increase in FGF signaling (Mouilleau et al., 2021). Herein, an increase in FGF pathway activity was successfully associated with the sequential activation of HOX genes; and cells differentiated under accelerated HOX induction (Mouilleau et al., 2021). Combination of “cell type-specific” growth factors then are able to foster lineage specifications and thus further modulation of HOX expressions. Pluripotent stem cell differentiation towards mesodermal stem cells for example can be initiated by the induction of mesodermal differentiation, achieved by TGFβ/Activin/Nodal signaling inhibition (Mahmood et al., 2010). Subsequent treatment with cell type-specific growth factors particularly FGF2 and/or platelet-derived growth factor A/B, two known potent inducers of MSC differentiation under defined cell-culture conditions foster then MSCs differentiation (Steens and Klein, 2018; Abdal Dayem et al., 2019). For HSCs, VEGF together with hematopoietic cytokine cocktails as type-specific growth factors were generally used to drive hematopoiesis following mesoderm specification of pluripotent stem cells by the use of WNT agonists and BMP4 (Sturgeon et al., 2014; Alsayegh et al., 2019). However, reports are lacking, which follow the timely expression of mesodermal stem cell-related HOX genes induced by extrinsic signals. In contrast, it has become well established that deregulated signaling pathways and thus abnormal signal transduction as selective characteristics common to many cancers, are associated with deregulation of HOX genes. Increasing numbers of reports here revealed important molecular signaling interactions, mainly due to up-regulations of certain HOX candidates that finally result in cancer (stem cell) overpopulation, limited differentiation, and cellular disorganization of respective cancer tissues. For example, in HOXB7-overexpressing tumors, which were shown to be enriched in gene signatures classically characterizing ASCs as well as pluripotent stem cells, the pluripotency factor LIN28B was identified that sustained the expansion of a subpopulation of cells with stem cell characteristics (Monterisi et al., 2018). As another example, HOXC8 was shown to function as transcriptional activator of TGFβ signaling, finally mediating increased proliferation, anchorage-independent growth and migration of lung cancer cells (Liu et al., 2018). A more systematic evaluation of all human HOX gene target pathways being involved in biological processes associated with the cancer hallmarks revealed then, that five HOX genes (HOXC4, HOXB2, HOXB3, HOXC6, and HOXA13) and their respective targets contribute to sustained proliferative signaling (Brotto et al., 2020). As cancers are commonly characterized by an abnormal signal transduction associated with increased proliferation (and loss of apoptosis), the identification of the signal(s) that cause upregulation of certain HOX genes is still missing. It is generally assumed that a tumor-specific loss of control over the spatiotemporal expression pattern and levels compared to the expression pattern of related normal tissues occurs. Induced “tumor” HOX genes can also occur due to so-called gene dominance, when respective HOX gene expressions in normal tissues are missing. In addition, epigenetic changes in regulatory regions of HOX genes can lead to a loss of normal control (Abate-Shen, 2002; Brotto et al., 2020). The deregulated expression of HOX gene candidates however turned out to be cancer-type dependent and even show variations in respective clinical responses. Thus, unraveling the driving forces of HOX gene dysregulations may foster the development of new and additional therapeutic targets to improve cancer therapy. Within these scenarios, data sharing through the so-called blockchain technology is becoming increasingly necessary, which would improve not only the insights of researchers and clinicians; it could also serve as a suitable basis for an efficient and effective evidence-based decision-making process in therapeutic approaches, as reasoned from other diseases (Dubovitskaya et al., 2020; Fusco et al., 2020). For the current state of knowledge on the roles of HOX genes in human cancer the following review articles were recommended (Bhatlekar et al., 2018; Smith et al., 2019; Paco et al., 2020; Feng et al., 2021).
Conclusion
The positional identity provided by HOX gene expression that arose during development and being maintained in ASCs can be linked to a limited number of stem cell typical signaling pathways in the different ASCs. On the one hand, HOX gene expression pattern (“HOX codes”) become more and more established in ASCs, particularly for MSCs. The HOX genes HOXA9 and HOXA10, HOXB7, HOXC6, HOXC8, and HOXC10, as well as HOXD8 in certain combinations potentially built up the MSC-type specific HOX codes as these HOX candidates are frequently observed in human MSCs derived from tissues different sources. Among them, HOXB7 was identified as a master player driving MSC proliferations, whereas the HOXC cluster candidates were shown to be involved in the (trilineage) differentiation capabilities. HOXA9 and HOXA10 were further identified as relevant candidates contributing to the ASC phenotype of HSCs, and being regulators of hematopoietic differentiation, whereas the additional HSC HOX code candidate HOXB4 decisively impacts on HSC maintenance and expansion. At the same time there are known extrinsic factors that can be used to generate ASCs at least in vitro; but how these signaling factors definitely account for a given HOX identity, and what is the respective timing of these factors remains to be unraveled. Genetically engineered HOX gene expressions hold great promise to manipulate and improve stem cell functions, particularly to promote differentiation into desired lineages, which in turn would allow then manufacturing exogenous stem cells or tissues with superior repair capabilities in vitro, particularly for regenerative purposes. Modified, mostly induced gene expression of individual HOX genes and particularly their overexpression can be functionally linked to tumor development and progression. However, it is not yet understood how a HOX candidate, which is widely expressed in many tissues, regulates different target genes in a cell-type-specific manner, and which interactions with additional co-activators, co-repressors and sequence-specific transcription factors predominate that are involved in, and thus could presumably be used to, model cell type-specific transcription outcomes. The identification of crucial HOX upstream regulators for individual HOX genes or HOX gene “partners” as well as their respective targets in a given cell specification process remain urgently needed, which in turn would allow generating desired ASCs without the use of genetic material that generally limits their clinical uses. An improved understanding of HOX gene functions and their respective regulations in ASCs would even foster the development of innovative strategies for a potential on-site manipulation of these cells, or particularly their dysregulated HOX gene expression as found in many cancers, directly within their endogenous niche.
Author contributions
JS and DK wrote and edited the manuscript.
Funding
This work was supported by IFORES of the Medical Faculty of the University Duisburg-Essen (Essen, Germany), and by the Brigitte and Konstanze Wegener-Stiftung (Düsseldorf, Germany).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abate-Shen, C. (2002). Deregulated homeobox gene expression in cancer: Cause or consequence? Nat. Rev. Cancer 2, 777–785. doi:10.1038/nrc907
Abdal Dayem, A., Lee, S. B., Kim, K., Lim, K. M., Jeon, T. I., Seok, J., et al. (2019). Production of mesenchymal stem cells through stem cell reprogramming. Ijms 20, 1922. doi:10.3390/ijms20081922
Abdelgawad, M. E., Desterke, C., Uzan, G., and Naserian, S. (2021). Single-cell transcriptomic profiling and characterization of endothelial progenitor cells: New approach for finding novel markers. Stem Cell Res. Ther. 12, 145. doi:10.1186/s13287-021-02185-0
Ackema, K. B., and Charité, J. (2008). Mesenchymal stem cells from different organs are characterized by distinct TopographicHoxCodes. Stem Cells Dev. 17, 979–992. doi:10.1089/scd.2007.0220
Afzal, Z., and Krumlauf, R. (2022). Transcriptional regulation and implications for controlling hox gene expression. Jdb 10, 4. doi:10.3390/jdb10010004
Agger, K., Cloos, P. A., Christensen, J., Pasini, D., Rose, S., Rappsilber, J., et al. (2007). UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731–734. doi:10.1038/nature06145
Akam, M. (1987). The molecular basis for metameric pattern in the Drosophila embryo. Development 101, 1–22. doi:10.1242/dev.101.1.1
Alharbi, R. A., Pettengell, R., Pandha, H. S., and Morgan, R. (2013). The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia 27, 1000–1008. doi:10.1038/leu.2012.356
Alsayegh, K., Cortés-Medina, L. V., Ramos-Mandujano, G., Badraiq, H., and Li, M. (2019). Hematopoietic differentiation of human pluripotent stem cells: HOX and GATA transcription factors as master regulators. Cg 20, 438–452. doi:10.2174/1389202920666191017163837
Alvarez, C. V., Garcia-Lavandeira, M., Garcia-Rendueles, M. E., Diaz-Rodriguez, E., Garcia-Rendueles, A. R., Perez-Romero, S., et al. (2012). Defining stem cell types: Understanding the therapeutic potential of ESCs, ASCs, and iPS cells. J. Mol. Endocrinol. 49, R89–R111. doi:10.1530/JME-12-0072
Argiropoulos, B., and Humphries, R. K. (2007). Hox genes in hematopoiesis and leukemogenesis. Oncogene 26, 6766–6776. doi:10.1038/sj.onc.1210760
Asahara, T., and Kawamoto, A. (2004). Endothelial progenitor cells for postnatal vasculogenesis. Am. J. Physiology-Cell Physiology 287, C572–C579. doi:10.1152/ajpcell.00330.2003
Aulehla, A., and Pourquie, O. (2010). Signaling gradients during paraxial mesoderm development. Cold Spring Harb. Perspect. Biol. 2, a000869. doi:10.1101/cshperspect.a000869
Bahrami, S. B., Veiseh, M., Dunn, A. A., and Boudreau, N. J. (2011). Temporal changes in Hox gene expression accompany endothelial cell differentiation of embryonic stem cells. Cell Adhesion Migr. 5, 133–141. doi:10.4161/cam.5.2.14373
Bernardo, M. E., and Fibbe, W. E. (2013). Mesenchymal stromal cells: Sensors and switchers of inflammation. Cell Stem Cell 13, 392–402. doi:10.1016/j.stem.2013.09.006
Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. doi:10.1016/j.cell.2006.02.041
Bhatlekar, S., Fields, J. Z., and Boman, B. M. (2018). Role of HOX genes in stem cell differentiation and cancer. Stem Cells Int. 2018, 1–15. doi:10.1155/2018/3569493
Bijl, J., Thompson, A., Ramirez-Solis, R., Krosl, J., Grier, D. G., Lawrence, H. J., et al. (2006). Analysis of HSC activity and compensatory Hox gene expression profile in Hoxb cluster mutant fetal liver cells. Blood 108, 116–122. doi:10.1182/blood-2005-06-2245
Blank, U., Karlsson, G., and Karlsson, S. (2008). Signaling pathways governing stem-cell fate. Blood 111, 492–503. doi:10.1182/blood-2007-07-075168
Boudreau, N., Andrews, C., Srebrow, A., Ravanpay, A., and Cheresh, D. A. (1997). Induction of the angiogenic phenotype by Hox D3. J. Cell Biol. 139, 257–264. doi:10.1083/jcb.139.1.257
Boyer, L. A., Lee, T. I., Cole, M. F., Johnstone, S. E., Levine, S. S., Zucker, J. P., et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956. doi:10.1016/j.cell.2005.08.020
Brotto, D. B., Siena, A. D. D., de Barros, B., Carvalho, S., Muys, B. R., Goedert, L., et al. (2020). Contributions of HOX genes to cancer hallmarks: Enrichment pathway analysis and review. Tumour Biol. 42, 101042832091805. doi:10.1177/1010428320918050
Brun, A. C., Björnsson, J. M., Magnusson, M., Larsson, N., Leveén, P., Ehinger, M., et al. (2004). Hoxb4-deficient mice undergo normal hematopoietic development but exhibit a mild proliferation defect in hematopoietic stem cells. Blood 103, 4126–4133. doi:10.1182/blood-2003-10-3557
Budjan, C., Liu, S., Ranga, A., Gayen, S., Pourquié, O., and Hormoz, S. (2022). Paraxial mesoderm organoids model development of human somites. Elife 11, e68925. doi:10.7554/eLife.68925
Cable, J., Fuchs, E., Weissman, I., Jasper, H., Glass, D., Rando, T. A., et al. (2020). Adult stem cells and regenerative medicine-a symposium report. Ann. N.Y. Acad. Sci. 1462, 27–36. doi:10.1111/nyas.14243
Casari, G., Resca, E., Giorgini, A., Candini, O., Petrachi, T., Piccinno, M. S., et al. (2021). Microfragmented adipose tissue is associated with improved ex vivo performance linked to HOXB7 and b-FGF expression. Stem Cell Res. Ther. 12, 481. doi:10.1186/s13287-021-02540-1
Chambers, I., Colby, D., Robertson, M., Nichols, J., Lee, S., Tweedie, S., et al. (2003). Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655. doi:10.1016/s0092-8674(03)00392-1
Cheung, T. S., Bertolino, G. M., Giacomini, C., Bornhäuser, M., Dazzi, F., and Galleu, A. (2020). Mesenchymal stromal cells for graft versus host disease: Mechanism-based biomarkers. Front. Immunol. 11, 1338. doi:10.3389/fimmu.2020.01338
Chiba, S. (2006). Concise review: Notch signaling in stem cell systems. Stem Cells 24, 2437–2447. doi:10.1634/stemcells.2005-0661
Choi, J., Lee, S., Mallard, W., Clement, K., Tagliazucchi, G. M., Lim, H., et al. (2015). A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat. Biotechnol. 33, 1173–1181. doi:10.1038/nbt.3388
Chung, N., Jee, B. K., Chae, S. W., Jeon, Y. W., Lee, K. H., and Rha, H. K. (2009). HOX gene analysis of endothelial cell differentiation in human bone marrow-derived mesenchymal stem cells. Mol. Biol. Rep. 36, 227–235. doi:10.1007/s11033-007-9171-6
Clevers, H., and Watt, F. M. (2018). Defining adult stem cells by function, not by phenotype. Annu. Rev. Biochem. 87, 1015–1027. doi:10.1146/annurev-biochem-062917-012341
Codispoti, B., Marrelli, M., and Paduano, F. (2018). NANOmetric BIO-banked MSC-derived exosome (NANOBIOME) as a novel approach to regenerative medicine. J. Clin. Med. 7. doi:10.3390/jcm7100357
Coenen, C., Liedtke, S., and Kogler, G. (2015). RNA amplification protocol leads to biased polymerase chain reaction results especially for low-copy transcripts of human bone marrow-derived stromal cells. PLoS One 10, e0141070. doi:10.1371/journal.pone.0141070
Cogle, C. R., and Scott, E. W. (2004). The hemangioblast: Cradle to clinic. Exp. Hematol. 32, 885–890. doi:10.1016/j.exphem.2004.07.014
Collins, E. M., and Thompson, A. (2018). HOX genes in normal, engineered and malignant hematopoiesis. Int. J. Dev. Biol. 62, 847–856. doi:10.1387/ijdb.180206at
Coutu, D. L., and Galipeau, J. (2011). Roles of FGF signaling in stem cell self-renewal, senescence and aging. Aging 3, 920–933. doi:10.18632/aging.100369
Darras, S., Fritzenwanker, J. H., Uhlinger, K. R., Farrelly, E., Pani, A. M., Hurley, I. A., et al. (2018). Anteroposterior axis patterning by early canonical Wnt signaling during hemichordate development. PLoS Biol. 16, e2003698. doi:10.1371/journal.pbio.2003698
De Los Angeles, A., Ferrari, F., Xi, R., Fujiwara, Y., Benvenisty, N., Deng, H., et al. (2015). Hallmarks of pluripotency. Nature 525, 469–478. doi:10.1038/nature15515
del Corral, R., and Storey, K. G. (2004). Opposing FGF and retinoid pathways: A signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. Bioessays 26, 857–869. doi:10.1002/bies.20080
Demirci, S., Leonard, A., and Tisdale, J. F. (2020). Hematopoietic stem cells from pluripotent stem cells: Clinical potential, challenges, and future perspectives. Stem Cells Transl. Med. 9, 1549–1557. doi:10.1002/sctm.20-0247
Denans, N., Iimura, T., and Pourquié, O. (2015). Hox genes control vertebrate body elongation by collinear Wnt repression. Elife 4. doi:10.7554/eLife.04379
Denner, L., Howe, M., and Urban, R. J. (2010). “Signaling pathways regulating growth and differentiation of adult stem cells,” in Handbook of cell signaling. Editors R. A. Bradshaw, and E. A. Dennis. Second Edition (San Diego: Academic Press), 2743–2751. doi:10.1016/b978-0-12-374145-5.00320-x
Deschamps, J., and Duboule, D. (2017). Embryonic timing, axial stem cells, chromatin dynamics, and the Hox clock. Genes Dev. 31, 1406–1416. doi:10.1101/gad.303123.117
Ding, Y., Wang, C., Sun, Z., Wu, Y., You, W., Mao, Z., et al. (2021). Mesenchymal stem cells engineered by nonviral vectors: A powerful tool in cancer gene therapy. Pharmaceutics 13, 913. doi:10.3390/pharmaceutics13060913
Dominici, M., Le Blanc, K., Mueller, I., Slaper-Cortenbach, I., Marini, F., Krause, D., et al. (2006). Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8, 315–317. doi:10.1080/14653240600855905
Dou, D. R., Calvanese, V., Sierra, M. I., Nguyen, A. T., Minasian, A., Saarikoski, P., et al. (2016). Medial HOXA genes demarcate haematopoietic stem cell fate during human development. Nat. Cell Biol. 18, 595–606. doi:10.1038/ncb3354
Doulatov, S., Vo, L. T., Chou, S. S., Kim, P. G., Arora, N., Li, H., et al. (2013). Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell 13, 459–470. doi:10.1016/j.stem.2013.09.002
Dubovitskaya, A., Novotny, P., Xu, Z., and Wang, F. (2020). Applications of blockchain technology for data-sharing in oncology: Results from a systematic literature review. Oncology 98, 403–411. doi:10.1159/000504325
Dulak, J., Szade, K., Szade, A., Nowak, W., and Józkowicz, A. (2015). Adult stem cells: Hopes and hypes of regenerative medicine. Acta Biochim. Pol. 62, 329–337. doi:10.18388/abp.2015_1023
Durston, A. J. (2019a). Vertebrate hox temporal collinearity: Does it exist and what is it's function? Cell Cycle 18, 523–530. doi:10.1080/15384101.2019.1577652
Durston, A. J. (2019b). What are the roles of retinoids, other morphogens, and Hox genes in setting up the vertebrate body axis? Genesis 57, e23296. doi:10.1002/dvg.23296
Evans, M. J., and Kaufman, M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156. doi:10.1038/292154a0
Fan, R., Bonde, S., Gao, P., Sotomayor, B., Chen, C., Mouw, T., et al. (2012). Dynamic HoxB4-regulatory network during embryonic stem cell differentiation to hematopoietic cells. Blood 119, e139–e147. doi:10.1182/blood-2011-12-396754
Feng, Y., Zhang, T., Wang, Y., Xie, M., Ji, X., Luo, X., et al. (2021). Homeobox genes in cancers: From carcinogenesis to recent therapeutic intervention. Front. Oncol. 11, 770428. doi:10.3389/fonc.2021.770428
Fidanza, A., and Forrester, L. M. (2021). Progress in the production of haematopoietic stem and progenitor cells from human pluripotent stem cells. J. Immunol. Regen. Med. 13, 100050. doi:10.1016/j.regen.2021.100050
Foppiani, E. M., Candini, O., Mastrolia, I., Murgia, A., Grisendi, G., Samarelli, A. V., et al. (2019). Impact of HOXB7 overexpression on human adipose-derived mesenchymal progenitors. Stem Cell Res. Ther. 10, 101. doi:10.1186/s13287-019-1200-6
Forrester, L. M., and Jackson, M. (2012). Mechanism of action of HOXB4 on the hematopoietic differentiation of embryonic stem cells. Stem Cells 30, 379–385. doi:10.1002/stem.1036
Fu, J., Wang, Y., Jiang, Y., Du, J., Xu, J., and Liu, Y. (2021). Systemic therapy of MSCs in bone regeneration: A systematic review and meta-analysis. Stem Cell Res. Ther. 12, 377. doi:10.1186/s13287-021-02456-w
Fuller, J. F., McAdara, J., Yaron, Y., Sakaguchi, M., Fraser, J. K., and Gasson, J. C. (1999). Characterization of HOX gene expression during myelopoiesis: Role of HOX A5 in lineage commitment and maturation. Blood 93, 3391–3400. doi:10.1182/blood.v93.10.3391.410k26_3391_3400
Fusco, A., Dicuonzo, G., Dell’Atti, M., and Tatullo, M. (2020). Dell'attiBlockchain in healthcare: Insights on COVID-19. Ijerph 17, 7167. doi:10.3390/ijerph17197167
Gardin, C., Bosco, G., Ferroni, L., Quartesan, S., Rizzato, A., Tatullo, M., et al. (2020). Hyperbaric oxygen therapy improves the osteogenic and vasculogenic properties of mesenchymal stem cells in the presence of inflammation in vitro. Ijms 21, 1452. doi:10.3390/ijms21041452
Gaunt, S. J. (2015). The significance of Hox gene collinearity. Int. J. Dev. Biol. 59, 159–170. doi:10.1387/ijdb.150223sg
Gross, S., Krause, Y., Wuelling, M., and Vortkamp, A. (2012). Hoxa11 and Hoxd11 regulate chondrocyte differentiation upstream of Runx2 and Shox2 in mice. PLoS One 7, e43553. doi:10.1371/journal.pone.0043553
Gudas, L. J., and Wagner, J. A. (2011). Retinoids regulate stem cell differentiation. J. Cell. Physiol. 226, 322–330. doi:10.1002/jcp.22417
Guo, Y., Lübbert, M., and Engelhardt, M. (2003). CD34 − hematopoietic stem cells: Current concepts and controversies. Stem Cells 21, 15–20. doi:10.1634/stemcells.21-1-15
Haas, S., Trumpp, A., and Milsom, M. D. (2018). Causes and consequences of hematopoietic stem cell heterogeneity. Cell Stem Cell 22, 627–638. doi:10.1016/j.stem.2018.04.003
Harding, K., Wedeen, C., McGinnis, W., and Levine, M. (1985). Spatially regulated expression of homeotic genes in Drosophila. Science 229, 1236–1242. doi:10.1126/science.3898362
Hassan, M. Q., Tare, R., Lee, S. H., Mandeville, M., Weiner, B., Montecino, M., et al. (2007). HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol. Cell. Biol. 27, 3337–3352. doi:10.1128/MCB.01544-06
He, H., Hua, X., and Yan, J. (2011). Epigenetic regulations in hematopoietic Hox code. Oncogene 30, 379–388. doi:10.1038/onc.2010.484
Heins, N., Englund, M. C., Sjöblom, C., Dahl, U., Tonning, A., Bergh, C., et al. (2004). Derivation, characterization, and differentiation of human embryonic stem cells. Stem Cells 22, 367–376. doi:10.1634/stemcells.22-3-367
Ho, A. D., Wagner, W., and Franke, W. (2008). Heterogeneity of mesenchymal stromal cell preparations. Cytotherapy 10, 320–330. doi:10.1080/14653240802217011
Hsu, Y. C., and Fuchs, E. (2012). A family business: Stem cell progeny join the niche to regulate homeostasis. Nat. Rev. Mol. Cell Biol. 13, 103–114. doi:10.1038/nrm3272
Hu, C., and Li, L. (2018). Preconditioning influences mesenchymal stem cell propertiesin vitroandin vivo. J. Cell. Mol. Med. 22, 1428–1442. doi:10.1111/jcmm.13492
Hwang, J. H., Seok, O. S., Song, H. R., Jo, J. Y., and Lee, J. K. (2009). HOXC10 as a potential marker for discriminating between amnion- and decidua-derived mesenchymal stem cells. Cloning Stem Cells 11, 269–279. doi:10.1089/clo.2008.0068
Iacovino, M., Hernandez, C., Xu, Z., Bajwa, G., Prather, M., and Kyba, M. (2009). A conserved role for Hox paralog group 4 in regulation of hematopoietic progenitors. Stem Cells Dev. 18, 783–792. doi:10.1089/scd.2008.0227
Iimura, T., and Pourquié, O. (2006). Collinear activation of Hoxb genes during gastrulation is linked to mesoderm cell ingression. Nature 442, 568–571. doi:10.1038/nature04838
Iordache, F., Constantinescu, A., Andrei, E., and Maniu, H. (2015). Histone acetylation regulates the expression of HoxD9 transcription factor in endothelial progenitor cells. Rom. J. Morphol. Embryol. 56, 107–113.
Irish, V. F., Martinez-Arias, A., and Akam, M. (1989). Spatial regulation of the Antennapedia and Ultrabithorax homeotic genes during Drosophila early development. EMBO J. 8, 1527–1537. doi:10.1002/j.1460-2075.1989.tb03537.x
Ishige-Wada, M., Kwon, S. M., Eguchi, M., Hozumi, K., Iwaguro, H., Matsumoto, T., et al. (2016). Jagged-1 signaling in the bone marrow microenvironment promotes endothelial progenitor cell expansion and commitment of CD133+ human cord blood cells for postnatal vasculogenesis. PLoS One 11, e0166660. doi:10.1371/journal.pone.0166660
Jansen, B. J., Gilissen, C., Roelofs, H., Schaap-Oziemlak, A., Veltman, J. A., Raymakers, R. A., et al. (2010). Functional differences between mesenchymal stem cell populations are reflected by their transcriptome. Stem Cells Dev. 19, 481–490. doi:10.1089/scd.2009.0288
Kaebisch, C., Schipper, D., Babczyk, P., and Tobiasch, E. (2015). The role of purinergic receptors in stem cell differentiation. Comput. Struct. Biotechnol. J. 13, 75–84. doi:10.1016/j.csbj.2014.11.003
Kamkar, F., Xaymardan, M., and Asli, N. S. (2016). Hox-mediated spatial and temporal coding of stem cells in homeostasis and neoplasia. Stem Cells Dev. 25, 1282–1289. doi:10.1089/scd.2015.0352
Kashyap, V., Gudas, L. J., Brenet, F., Funk, P., Viale, A., and Scandura, J. M. (2011). Epigenomic reorganization of the clustered Hox genes in embryonic stem cells induced by retinoic acid. J. Biol. Chem. 286, 3250–3260. doi:10.1074/jbc.M110.157545
Kessel, M., and Gruss, P. (1991). Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell 67, 89–104. doi:10.1016/0092-8674(91)90574-i
Kim, K., Doi, A., Wen, B., Ng, K., Zhao, R., Cahan, P., et al. (2010). Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290. doi:10.1038/nature09342
Klein, D., Benchellal, M., Kleff, V., Jakob, H. G., and Ergün, S. (2013). Hox genes are involved in vascular wall-resident multipotent stem cell differentiation into smooth muscle cells. Sci. Rep. 3, 2178. doi:10.1038/srep02178
Klein, D. (2021). Lung multipotent stem cells of mesenchymal nature: Cellular basis, clinical relevance, and implications for stem cell therapy. Antioxidants Redox Signal. 35, 204–216. doi:10.1089/ars.2020.8190
Klein, D. (2016). Vascular wall-resident multipotent stem cells of mesenchymal nature within the process of vascular remodeling: Cellular basis, clinical relevance, and implications for stem cell therapy. Stem Cells Int. 2016, 1. doi:10.1155/2016/1905846
Kmita, M., and Duboule, D. (2003). Organizing axes in time and space; 25 years of colinear tinkering. Science 301, 331–333. doi:10.1126/science.1085753
Kou, M., Huang, L., Yang, J., Chiang, Z., Chen, S., Liu, J., et al. (2022). Mesenchymal stem cell-derived extracellular vesicles for immunomodulation and regeneration: A next generation therapeutic tool? Cell Death Dis. 13, 580. doi:10.1038/s41419-022-05034-x
Kouidhi, M., Villageois, P., Mounier, C. M., Ménigot, C., Rival, Y., Piwnica, D., et al. (2015). Characterization of human knee and chin adipose-derived stromal cells. Stem Cells Int. 2015–11. 1. doi:10.1155/2015/592090
Krumlauf, R., and Ahn, Y. (2013). “Hox genes,” in Brenner's encyclopedia of genetics. Editors S. Maloy, and K. Hughes. Second Edition (San Diego: Academic Press), 539–542. doi:10.1016/b978-0-12-374984-0.00742-7
Krumlauf, R. (1994). Hox genes in vertebrate development. Cell 78, 191–201. doi:10.1016/0092-8674(94)90290-9
Kulebyakina, M., and Makarevich, P. (2020). Hox-positive adult mesenchymal stromal cells: Beyond positional identity. Front. Cell Dev. Biol. 8, 624. doi:10.3389/fcell.2020.00624
Kumar, V., Goutam, R. S., Park, S., Lee, U., and Kim, J. (2021). Functional roles of FGF signaling in early development of vertebrate embryos. Cells 10, 2148. doi:10.3390/cells10082148
Lawrence, H. J., Sauvageau, G., Humphries, R. K., and Largman, C. (1996). The role of HOX homeobox genes in normal and leukemic hematopoiesis. Stem Cells 14, 281–291. doi:10.1002/stem.140281
Lee, J., Park, Y. J., and Jung, H. (2019). Protein kinases and their inhibitors in pluripotent stem cell fate regulation. Stem Cells Int. 2019, 1–10. doi:10.1155/2019/1569740
Li, B., Han, H., Song, S., Fan, G., Xu, H., Zhou, W., et al. (2019). HOXC10 regulates osteogenesis of mesenchymal stromal cells through interaction with its natural antisense transcript lncHOXC-AS3. Stem Cells 37, 247–256. doi:10.1002/stem.2925
Liedtke, S., Buchheiser, A., Bosch, J., Bosse, F., Kruse, F., Zhao, X., et al. (2010). The HOX Code as a "biological fingerprint" to distinguish functionally distinct stem cell populations derived from cord blood. Stem Cell Res. 5, 40–50. doi:10.1016/j.scr.2010.03.004
Liedtke, S., Sacchetti, B., Laitinen, A., Donsante, S., Klöckers, R., Laitinen, S., et al. (2017). Low oxygen tension reveals distinctHOXcodes in human cord blood-derived stromal cells associated with specific endochondral ossification capacitiesin vitroandin vivo. J. Tissue Eng. Regen. Med. 11, 2725–2736. doi:10.1002/term.2167
Liu, H., Zhang, M., Xu, S., Zhang, J., Zou, J., Yang, C., et al. (2018). HOXC8 promotes proliferation and migration through transcriptional up-regulation of TGFβ1 in non-small cell lung cancer. Oncogenesis 7, 1. doi:10.1038/s41389-017-0016-4
Luo, Z., Rhie, S. K., and Farnham, P. J. (2019). The enigmatic HOX genes: Can we crack their code? Cancers 11, 323. doi:10.3390/cancers11030323
Ma, Y., Kakudo, N., Morimoto, N., Lai, F., Taketani, S., and Kusumoto, K. (2019). Fibroblast growth factor-2 stimulates proliferation of human adipose-derived stem cells via Src activation. Stem Cell Res. Ther. 10, 350. doi:10.1186/s13287-019-1462-z
Magnusson, M., Brun, A. C., Lawrence, H. J., and Karlsson, S. (2007). Hoxa9/hoxb3/hoxb4 compound null mice display severe hematopoietic defects. Exp. Hematol. 35, e1–1421. doi:10.1016/j.exphem.2007.05.011
Mahmood, A., Harkness, L., Schrøder, H. D., Abdallah, B. M., and Kassem, M. (2010). Enhanced differentiation of human embryonic stem cells to mesenchymal progenitors by inhibition of TGF-β/activin/nodal signaling using SB-431542. J. Bone Min. Res. 25, 1216–1233. doi:10.1002/jbmr.34
Mallo, M., Wellik, D. M., and Deschamps, J. (2010). Hox genes and regional patterning of the vertebrate body plan. Dev. Biol. 344, 7–15. doi:10.1016/j.ydbio.2010.04.024
Mann, R. S., Lelli, K. M., and Joshi, R. (2009). Chapter 3 hox specificity. Curr. Top. Dev. Biol. 88, 63–101. doi:10.1016/S0070-2153(09)88003-4
Massa, M., Croce, S., Campanelli, R., Abbà, C., Lenta, E., Valsecchi, C., et al. (2020). Clinical applications of mesenchymal stem/stromal cell derived extracellular vesicles: Therapeutic potential of an acellular product. Diagnostics 10, 999. doi:10.3390/diagnostics10120999
McGonigle, G. J., Lappin, T. R., and Thompson, A. (2008). Grappling with the HOX network in hematopoiesis and leukemia. Front. Biosci. Volume, 4297–4308. doi:10.2741/3006
Medina, R. J., Barber, C. L., Sabatier, F., Dignat-George, F., Melero-Martin, J. M., Khosrotehrani, K., et al. (2017). Endothelial progenitors: A consensus statement on nomenclature. Stem Cells Transl. Med. 6, 1316–1320. doi:10.1002/sctm.16-0360
Medina, R. J., O'Neill, C. L., Sweeney, M., Guduric-Fuchs, J., Gardiner, T. A., Simpson, D. A., et al. (2010). Molecular analysis of endothelial progenitor cell (EPC) subtypes reveals two distinct cell populations with different identities. BMC Med. Genomics 3, 18. doi:10.1186/1755-8794-3-18
Mikłosz, A., Nikitiuk, B. E., and Chabowski, A. (2022). Using adipose-derived mesenchymal stem cells to fight the metabolic complications of obesity: Where do we stand? Obes. Rev. 23, e13413. doi:10.1111/obr.13413
Monterisi, S., Lo Riso, P., Russo, K., Bertalot, G., Vecchi, M., Testa, G., et al. (2018). HOXB7 overexpression in lung cancer is a hallmark of acquired stem-like phenotype. Oncogene 37, 3575–3588. doi:10.1038/s41388-018-0229-9
Morata, G., and Kerridge, S. (1982). The role of position in determining homoeotic gene function in Drosophila. Nature 300, 191–192. doi:10.1038/300191a0
Morgan, R., and Whiting, K. (2008). Differential expression of HOX genes upon activation of leukocyte sub-populations. Int. J. Hematol. 87, 246–249. doi:10.1007/s12185-008-0057-8
Mori, M., Nakagami, H., Rodriguez-Araujo, G., Nimura, K., and Kaneda, Y. (2012). Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biol. 10, e1001314.
Mossahebi-Mohammadi, M., Quan, M., Zhang, J. S., and Li, X. (2020). FGF signaling pathway: A key regulator of stem cell pluripotency. Front. Cell Dev. Biol. 8, 79. doi:10.3389/fcell.2020.00079
Mouilleau, V., Vaslin, C., Robert, R., Gribaudo, S., Nicolas, N., Jarrige, M., et al. (2021). Dynamic extrinsic pacing of the HOX clock in human axial progenitors controls motor neuron subtype specification. Development 148, 194514. doi:10.1242/dev.194514
Myers, C., Charboneau, A., and Boudreau, N. (2000). Homeobox B3 promotes capillary morphogenesis and angiogenesis. J. Cell Biol. 148, 343–352. doi:10.1083/jcb.148.2.343
Neijts, R., Amin, S., van Rooijen, C., and Deschamps, J. (2017). Cdx is crucial for the timing mechanism driving colinear Hox activation and defines a trunk segment in the Hox cluster topology. Dev. Biol. 422, 146–154. doi:10.1016/j.ydbio.2016.12.024
Neijts, R., Amin, S., van Rooijen, C., Tan, S., Creyghton, M. P., de Laat, W., et al. (2016). Polarized regulatory landscape and Wnt responsiveness underlie Hox activation in embryos. Genes Dev. 30, 1937–1942. doi:10.1101/gad.285767.116
Ng, E. S., Azzola, L., Bruveris, F. F., Calvanese, V., Phipson, B., Vlahos, K., et al. (2016). Differentiation of human embryonic stem cells to HOXA+ hemogenic vasculature that resembles the aorta-gonad-mesonephros. Nat. Biotechnol. 34, 1168–1179. doi:10.1038/nbt.3702
Nolte, C., De Kumar, B., and Krumlauf, R. (2019). Hox genes: Downstream "effectors" of retinoic acid signaling in vertebrate embryogenesis. Genesis 57, e23306. doi:10.1002/dvg.23306
Noyes, M. B., Christensen, R. G., Wakabayashi, A., Stormo, G. D., Brodsky, M. H., and Wolfe, S. A. (2008). Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133, 1277–1289. doi:10.1016/j.cell.2008.05.023
Nusse, R. (2008). Wnt signaling and stem cell control. Cell Res. 18, 523–527. doi:10.1038/cr.2008.47
Ocansey, D. K. W., Pei, B., Yan, Y., Qian, H., Zhang, X., Xu, W., et al. (2020). Improved therapeutics of modified mesenchymal stem cells: An update. J. Transl. Med. 18, 42. doi:10.1186/s12967-020-02234-x
Oñate, B., Vilahur, G., Camino-López, S., Díez-Caballero, A., Ballesta-López, C., Ybarra, J., et al. (2013). Stem cells isolated from adipose tissue of obese patients show changes in their transcriptomic profile that indicate loss in stemcellness and increased commitment to an adipocyte-like phenotype. BMC Genomics 14, 625. doi:10.1186/1471-2164-14-625
Paço, A., Aparecida de Bessa Garcia, S., Leitão Castro, J., Costa-Pinto, A. R., and Freitas, R. (2020). Roles of the HOX proteins in cancer invasion and metastasis. Cancers 13, 10. doi:10.3390/cancers13010010
Parrish, M., Nolte, C., and Krumlauf, R. (2009). “Hox genes expression,” in Encyclopedia of neuroscience. Editor L. R. Squire (Oxford: Academic Press), 1221–1231. doi:10.1016/b978-008045046-9.01067-6
Petrova, R., and Joyner, A. L. (2014). Roles for Hedgehog signaling in adult organ homeostasis and repair. Development 141, 3445–3457. doi:10.1242/dev.083691
Picchi, J., Trombi, L., Spugnesi, L., Barachini, S., Maroni, G., Brodano, G. B., et al. (2013). HOXandTALEsignatures specify human stromal stem cell populations from different sources. J. Cell. Physiol. 228, 879–889. doi:10.1002/jcp.24239
Pilat, S., Carotta, S., Schiedlmeier, B., Kamino, K., Mairhofer, A., Will, E., et al. (2005). HOXB4 enforces equivalent fates of ES-cell-derived and adult hematopoietic cells. Proc. Natl. Acad. Sci. U.S.A. 102, 12101–12106. doi:10.1073/pnas.0505624102
Pineault, K. M., Song, J. Y., Kozloff, K. M., Lucas, D., and Wellik, D. M. (2019). Hox11 expressing regional skeletal stem cells are progenitors for osteoblasts, chondrocytes and adipocytes throughout life. Nat. Commun. 10, 3168. doi:10.1038/s41467-019-11100-4
Pirro, M., Schillaci, G., Menecali, C., Bagaglia, F., Paltriccia, R., Vaudo, G., et al. (2007). Reduced number of circulating endothelial progenitors and HOXA9 expression in CD34+ cells of hypertensive patients. J. Hypertens. 25, 2093–2099. doi:10.1097/HJH.0b013e32828e506d
Planat-Benard, V., Varin, A., and Casteilla, L. (2021). MSCs and inflammatory cells crosstalk in regenerative medicine: Concerted actions for optimized resolution driven by energy metabolism. Front. Immunol. 12, 626755. doi:10.3389/fimmu.2021.626755
Quinonez, S. C., and Innis, J. W. (2014). Human HOX gene disorders. Mol. Genet. Metabolism 111, 4–15. doi:10.1016/j.ymgme.2013.10.012
Rankin, S. A., McCracken, K. W., Luedeke, D. M., Han, L., Wells, J. M., Shannon, J. M., et al. (2018). Timing is everything: Reiterative Wnt, BMP and RA signaling regulate developmental competence during endoderm organogenesis. Dev. Biol. 434, 121–132. doi:10.1016/j.ydbio.2017.11.018
Rea, S., Eisenhaber, F., O'Carroll, D., Strahl, B. D., Sun, Z. W., Schmid, M., et al. (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599. doi:10.1038/35020506
Redondo, P. A., Pavlou, M., Loizidou, M., and Cheema, U. (2017). Elements of the niche for adult stem cell expansion. J. Tissue Eng. 8, 204173141772546. doi:10.1177/2041731417725464
Riising, E. M., Comet, I., Leblanc, B., Wu, X., Johansen, J. V., and Helin, K. (2014). Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol. Cell 55, 347–360. doi:10.1016/j.molcel.2014.06.005
Ringrose, L., and Paro, R. (2007). Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134, 223–232. doi:10.1242/dev.02723
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. doi:10.1016/j.cell.2007.05.022
Rössig, L., Urbich, C., Brühl, T., Dernbach, E., Heeschen, C., Chavakis, E., et al. (2005). Histone deacetylase activity is essential for the expression of HoxA9 and for endothelial commitment of progenitor cells. J. Exp. Med. 201, 1825–1835. doi:10.1084/jem.20042097
Ruddle, F. H., Bartels, J. L., Bentley, K. L., Kappen, C., Murtha, M. T., and Pendleton, J. W. (1994). Evolution of hox genes. Annu. Rev. Genet. 28, 423–442. doi:10.1146/annurev.ge.28.120194.002231
Rumman, M., Dhawan, J., and Kassem, M. (2015). Concise review: Quiescence in adult stem cells: Biological significance and relevance to tissue regeneration. Stem Cells 33, 2903–2912. doi:10.1002/stem.2056
Rux, D. R., Song, J. Y., Swinehart, I. T., Pineault, K. M., Schlientz, A. J., Trulik, K. G., et al. (2016). Regionally restricted hox function in adult bone marrow multipotent mesenchymal stem/stromal cells. Dev. Cell 39, 653–666. doi:10.1016/j.devcel.2016.11.008
Sakaki-Yumoto, M., Katsuno, Y., and Derynck, R. (2013). TGF-β family signaling in stem cells. Biochimica Biophysica Acta (BBA) - General Subj. 1830, 2280–2296. doi:10.1016/j.bbagen.2012.08.008
Sánchez-Higueras, C., Rastogi, C., Voutev, R., Bussemaker, H. J., Mann, R. S., and Hombría, J. C. (2019). In vivo Hox binding specificity revealed by systematic changes to a single cis regulatory module. Nat. Commun. 10, 3597. doi:10.1038/s41467-019-11416-1
Santourlidis, S., Wernet, P., Ghanjati, F., Graffmann, N., Springer, J., Kriegs, C., et al. (2011). Unrestricted somatic stem cells (USSC) from human umbilical cord blood display uncommitted epigenetic signatures of the major stem cell pluripotency genes. Stem Cell Res. 6, 60–69. doi:10.1016/j.scr.2010.08.003
Sauvageau, G., Lansdorp, P. M., Eaves, C. J., Hogge, D. E., Dragowska, W. H., Reid, D. S., et al. (1994). Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc. Natl. Acad. Sci. U.S.A. 91, 12223–12227. doi:10.1073/pnas.91.25.12223
Seifert, A., Werheid, D. F., Knapp, S. M., and Tobiasch, E. (2015). Role ofHoxgenes in stem cell differentiation. Wjsc 7, 583–595. doi:10.4252/wjsc.v7.i3.583
Sentek, H., and Klein, D. (2021). Lung-resident mesenchymal stem cell fates within lung cancer. Cancers 13, 4637. doi:10.3390/cancers13184637
Seo, H. C., Edvardsen, R. B., Maeland, A. D., Bjordal, M., Jensen, M. F., Hansen, A., et al. (2004). Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature 431, 67–71. doi:10.1038/nature02709
Shahhoseini, M., Taghizadeh, Z., Hatami, M., and Baharvand, H. (2013). Retinoic acid dependent histone 3 demethylation of the clusteredHOXgenes during neural differentiation of human embryonic stem cells. Biochem. Cell Biol. 91, 116–122. doi:10.1139/bcb-2012-0049
Shaut, C. A., Keene, D. R., Sorensen, L. K., Li, D. Y., and Stadler, H. S. (2008). HOXA13 Is essential for placental vascular patterning and labyrinth endothelial specification. PLoS Genet. 4, e1000073. doi:10.1371/journal.pgen.1000073
Slattery, M., Riley, T., Liu, P., Abe, N., Gomez-Alcala, P., Dror, I., et al. (2011). Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147, 1270–1282. doi:10.1016/j.cell.2011.10.053
Smith, J., Zyoud, A., and Allegrucci, C. (2019). A Case of Identity: HOX Genes in Normal and Cancer Stem Cells, 11. doi:10.3390/cancers11040512Cancers (Basel)
Snippert, H. J., and Clevers, H. (2011). Tracking adult stem cells. EMBO Rep. 12, 113–122. doi:10.1038/embor.2010.216
Soprano, D. R., Teets, B. W., and Soprano, K. J. (2007). Role of retinoic acid in the differentiation of embryonal carcinoma and embryonic stem cells. Vitam. Horm. 75, 69–95. doi:10.1016/S0083-6729(06)75003-8
Soshnikova, N., and Duboule, D. (2008). Epigenetic regulation ofHox gene activation: The waltz of methyls. Bioessays 30, 199–202. doi:10.1002/bies.20724
Stavridis, M. P., Collins, B. J., and Storey, K. G. (2010). Retinoic acid orchestrates fibroblast growth factor signalling to drive embryonic stem cell differentiation. Development 137, 881–890. doi:10.1242/dev.043117
Steens, J., Klar, L., Hansel, C., Slama, A., Hager, T., Jendrossek, V., et al. (2021). The vascular nature of lung-resident mesenchymal stem cells. Stem Cells Transl. Med. 10, 128–143. doi:10.1002/sctm.20-0191
Steens, J., and Klein, D. (2018). Current strategies to generate human mesenchymal stem cells in vitro. Stem Cells Int. 2018, 1–10. doi:10.1155/2018/6726185
Steens, J., Klump, H., and Klein, D. (2020a). In vitro generation of -typical mesenchymal (VW-MSC) from murine induced pluripotent through VW-MSC-Specific gene transfer. Methods Mol. Biol. 2155, 83–97. doi:10.1007/978-1-0716-0655-1_7
Steens, J., Unger, K., Klar, L., Neureiter, A., Wieber, K., Hess, J., et al. (2020b). Direct conversion of human fibroblasts into therapeutically active vascular wall-typical mesenchymal stem cells. Cell. Mol. Life Sci. 77, 3401–3422. doi:10.1007/s00018-019-03358-0
Steens, J., Zuk, M., Benchellal, M., Bornemann, L., Teichweyde, N., Hess, J., et al. (2017). In vitro generation of vascular wall-resident multipotent stem cells of mesenchymal nature from murine induced pluripotent stem cells. Stem Cell Rep. 8, 919–932. doi:10.1016/j.stemcr.2017.03.001
Sturgeon, C. M., Ditadi, A., Awong, G., Kennedy, M., and Keller, G. (2014). Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat. Biotechnol. 32, 554–561. doi:10.1038/nbt.2915
Sugimura, R., Jha, D. K., Han, A., Soria-Valles, C., da Rocha, E. L., Lu, Y. F., et al. (2017). Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545, 432–438. doi:10.1038/nature22370
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. doi:10.1016/j.cell.2007.11.019
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi:10.1016/j.cell.2006.07.024
Takayama, Y., Kusamori, K., Tsukimori, C., Shimizu, Y., Hayashi, M., Kiyama, I., et al. (2021). Anticancer drug-loaded mesenchymal stem cells for targeted cancer therapy. J. Control. Release 329, 1090–1101. doi:10.1016/j.jconrel.2020.10.037
Tanabe, S. (2015). Signaling involved in stem cell reprogramming and differentiation. World J. Stem Cells 7, 992–998. doi:10.4252/wjsc.v7.i7.992
Theunissen, T. W., and Jaenisch, R. (2014). Molecular control of induced pluripotency. Cell Stem Cell 14, 720–734. doi:10.1016/j.stem.2014.05.002
Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S., et al. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147. doi:10.1126/science.282.5391.1145
Tiana, M., Lopez-Jimenez, E., de Aja, J. S., Barral, A., Victorino, J., Badia-Careaga, C., et al. (2022). Pluripotency factors regulate the onset of Hox cluster activation in the early embryo. Sci. Adv. 8, eabo3583. doi:10.1126/sciadv.abo3583
Toshner, M., Dunmore, B. J., McKinney, E. F., Southwood, M., Caruso, P., Upton, P. D., et al. (2014). Transcript analysis reveals a specific HOX signature associated with positional identity of human endothelial cells. PLoS One 9, e91334. doi:10.1371/journal.pone.0091334
Urbán, N., and Cheung, T. H. (2021). Stem cell quiescence: The challenging path to activation. Development 148, dev165084. doi:10.1242/dev.165084
Van Camp, J. K., Beckers, S., Zegers, D., and Van Hul, W. (2014). Wnt signaling and the control of human stem cell fate. Stem Cell Rev Rep 10, 207–229. doi:10.1007/s12015-013-9486-8
Viswanathan, S., Shi, Y., Galipeau, J., Krampera, M., Leblanc, K., Martin, I., et al. (2019). Mesenchymal stem versus stromal cells: International society for cell & gene therapy (ISCT) mesenchymal stromal cell committee position statement on nomenclature. Cytotherapy 21, 1019–1024. doi:10.1016/j.jcyt.2019.08.002
Volarevic, V., Markovic, B. S., Gazdic, M., Volarevic, A., Jovicic, N., Arsenijevic, N., et al. (2018). Ethical and safety issues of stem cell-based therapy. Int. J. Med. Sci. 15, 36–45. doi:10.7150/ijms.21666
Wagner, T. U. (2007). Bone morphogenetic protein signaling in stem cells − one signal, many consequences. FEBS J. 274, 2968–2976. doi:10.1111/j.1742-4658.2007.05839.x
Wagner, W., Feldmann, R. E., Seckinger, A., Maurer, M. H., Wein, F., Blake, J., et al. (2006). The heterogeneity of human mesenchymal stem cell preparations-Evidence from simultaneous analysis of proteomes and transcriptomes. Exp. Hematol. 34, 536–548. doi:10.1016/j.exphem.2006.01.002
Wang, J., Bonacquisti, E. E., Brown, A. D., and Nguyen, J. (2020). Boosting the biogenesis and secretion of mesenchymal stem cell-derived exosomes. Cells 9, 660. . doi:10.3390/cells9030660
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124. doi:10.1038/nature09819
Wang, L., Menendez, P., Shojaei, F., Li, L., Mazurier, F., Dick, J. E., et al. (2005). Generation of hematopoietic repopulating cells from human embryonic stem cells independent of ectopic HOXB4 expression. J. Exp. Med. 201, 1603–1614. doi:10.1084/jem.20041888
Wang, Z., Li, Y., Banerjee, S., and Sarkar, F. H. (2009). Emerging role of Notch in stem cells and cancer. Cancer Lett. 279, 8–12. doi:10.1016/j.canlet.2008.09.030
Wellik, D. M. (2009). Chapter 9 hox genes and vertebrate axial pattern. Curr. Top. Dev. Biol. 88, 257–278. doi:10.1016/S0070-2153(09)88009-5
Wells, C. A., and Choi, J. (2019). Transcriptional profiling of stem cells: Moving from descriptive to predictive paradigms. Stem Cell Rep. 13, 237–246. doi:10.1016/j.stemcr.2019.07.008
Witwer, K. W., Van Balkom, B. W. M., Bruno, S., Choo, A., Dominici, M., Gimona, M., et al. (2019). Defining mesenchymal stromal cell (MSC)-derived small extracellular vesicles for therapeutic applications. J. Extracell. Vesicles 8, 1609206. doi:10.1080/20013078.2019.1609206
Wu, T. W., Liu, C. C., Hung, C. L., Yen, C. H., Wu, Y. J., Wang, L. Y., et al. (2018). Genetic profiling of young and aged endothelial progenitor cells in hypoxia. PLoS One 13, e0196572. doi:10.1371/journal.pone.0196572
Wu, Y., Moser, M., Bautch, V. L., and Patterson, C. (2003). HoxB5 is an upstream transcriptional switch for differentiation of the vascular endothelium from precursor cells. Mol. Cell. Biol. 23, 5680–5691. doi:10.1128/mcb.23.16.5680-5691.2003
Yamamoto, M., Takai, D., Yamamoto, F., and Yamamoto, F. (2003). Comprehensive expression profiling of highly homologous 39 hox genes in 26 different human adult tissues by the modified systematic multiplex RT-pCR method reveals tissue-specific expression pattern that suggests an important role of chromosomal structure in the regulation of hox gene expression in adult tissues. Gene Expr. 11, 199–210. doi:10.3727/000000003108749071
Yang, H., Cao, Y., Zhang, J., Liang, Y., Su, X., Zhang, C., et al. (2020a). DLX5 and HOXC8 enhance the chondrogenic differentiation potential of stem cells from apical papilla via LINC01013. Stem Cell Res. Ther. 11, 271. doi:10.1186/s13287-020-01791-8
Yang, H., Liang, Y., Cao, Y., Cao, Y., and Fan, Z. (2020b). Homeobox C8 inhibited the osteo‐/dentinogenic differentiation and migration ability of stem cells of the apical papilla via activating KDM1A. J. Cell. Physiology 235, 8432–8445. doi:10.1002/jcp.29687
Yoder, M. C. (2018). Endothelial stem and progenitor cells (stem cells): (2017 grover conference series). Pulm. Circ. 8, 1–9. doi:10.1177/2045893217743950
Young, R. A. (2011). Control of the embryonic stem cell state. Cell 144, 940–954. doi:10.1016/j.cell.2011.01.032
Young, T., Rowland, J. E., van de Ven, C., Bialecka, M., Novoa, A., Carapuco, M., et al. (2009). Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev. Cell 17, 516–526. doi:10.1016/j.devcel.2009.08.010
Yueh, Y. G., Gardner, D. P., and Kappen, C. (1998). Evidence for regulation of cartilage differentiation by the homeobox gene Hoxc-8. Proc. Natl. Acad. Sci. U.S.A. 95, 9956–9961. doi:10.1073/pnas.95.17.9956
Zakrzewski, W., Dobrzyński, M., Szymonowicz, M., and Rybak, Z. (2019). Stem cells: Past, present, and future. Stem Cell Res. Ther. 10, 68. doi:10.1186/s13287-019-1165-5
Keywords: adult stem cell, mesenchymal stem cell, MSC, HSC, HOX code, differentiation, signaling pathways
Citation: Steens J and Klein D (2022) HOX genes in stem cells: Maintaining cellular identity and regulation of differentiation. Front. Cell Dev. Biol. 10:1002909. doi: 10.3389/fcell.2022.1002909
Received: 25 July 2022; Accepted: 25 August 2022;
Published: 13 September 2022.
Edited by:
Marco Tatullo, University of Bari Medical School, ItalyReviewed by:
Salvatore Scacco, S.G. Moscati Hospital of Taranto, ItalyOmar Etienne, INSERM Délégation Paris 6, France
Copyright © 2022 Steens and Klein. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diana Klein, RGlhbmEuS2xlaW5AdWstZXNzZW4uZGU=
 Jennifer Steens
Jennifer Steens Diana Klein
Diana Klein