- 1Guangxi Academy of Medical Sciences and the People’s Hospital of Guangxi Zhuang Autonomous Region, Nanning, China
- 2State Key Laboratory for Conservation and Utilization of Subtropical Agro-8 Bioresources, Guangxi University, Nanning, China
- 3 School of Animal Science and Technology, Guangxi University, Nanning, China
Mammary epithelial cells are the only cells of mammary glands with lactation capacity. They are closely related to mammary development and milk yield. Our earlier studies showed that the transformation of goat fibroblasts into induced mammary epithelial cells (iMECs) was closely correlated with SMAD3 overexpression. Therefore, we further explored the role of SMAD3 on iMECs reprogramming in this study. The SMAD3 gene was overexpressed in goat ear fibroblasts using the tetracycline-induced expression method. The outcomes demonstrated that goat ear fibroblasts can be converted into iMECs by overexpressing the SMAD3 gene. In contrast, it was discovered that SMAD3 downregulation by RNA interference significantly decrease the reprogramming efficiency of iMECs. These results show that SMAD3 plays a key regulatory role in the reprogramming of iMECs. Surprisingly, we also found a parabolic relationship between SMAD3 expression level and iMECs reprogramming efficiency, and that the reprogramming efficiency was maximum when the addition of doxycycline concentration was 5 μg/ml. In light of this, our findings may offer new perspectives on the regulatory mechanism governing mammary epithelial cell fate in goats as well as a fresh approach to studying mammary development and differentiation in vitro.
Introduction
The development of multicellular organisms is a delicate and complex process, which is dynamically regulated to maintain long-term coordination and stability in the body. Different cell groups have their unique steady state and undergo cell differentiation along established developmental trajectories. Cells hardly ever alter their course and develop into other cell types in the natural condition. An earlier investigation shown that overexpression the transcription factor MyoD can reprogram mouse embryonic fibroblasts into myoblasts (Davis et al., 1987). The conventional notion of cell differentiation was put to the test by this study, which showed that cells have plasticity and that their identity may be modified. The gene expression profile of somatic cells can be modified by external factors induction, and subsequently, somatic cells have stimulated the potential to achieve other cell fates bypassing the pluripotent cell stage. This phenomenon is known as lineage reprogramming or transdifferentiation, which can be regulated by signaling pathways, epigenetic modifications, and cellular metabolic processes (Bannister and Kouzarides, 2011; Yoo et al., 2011; Li et al., 2013; Iwafuchi-Doi and Zaret, 2014; Soufi et al., 2015).
In recent years, transdifferentiation has been achieved by adding particular transcription factors or miRNAs into a variety of cells, including cardiomyocytes (Ieda et al., 2010; Wada et al., 2013), hepatocytes (Huang et al., 2014; Zhu et al., 2014), neural stem cells (Sheng et al., 2012) and neurons (Kim et al., 2011; Wang et al., 2013), astrocytes (Caiazzo et al., 2015), endothelial cells (Han et al., 2014), pancreatic β cells (Li et al., 2014), hematopoietic stem cells (Batta et al., 2014), and macrophages (Bussmann et al., 2009). As a non-viral, non-integrating form, small molecule compounds have the unique advantages of rapid cell entry through high permeability, relatively clear targets, and relatively controlled effects, showing great potential in transdifferentiation studies. To date, various studies have reported the use of small molecule compounds alone for transdifferentiation to generate neural stem cells (Han et al., 2016; Zheng et al., 2016), neurons (Cheng et al., 2015; Li et al., 2015; Wenxiang et al., 2015), endothelial cells (Wang et al., 2016), cardiomyocytes (Fu et al., 2015) and hepatocytes (Wang et al., 2016).
Currently, we have identified the combination of five small molecule compounds (1 μM TTNPB (B), 10 μM Forskolin (F), 10 μM RepSox (R), 10 μM Tranylcypromine (T) and 500 μg/ml VPA (V), BFRTV), which can induce reprogramming in goat ear fibroblasts (GEFs) to give rise to chemically induced MECs (CiMECs) (Zhang et al., 2021). The molecular mechanisms behind the growth, development, and lactation of the mammary gland can be studied using goat mammary epithelial cells. High-quality iMECs were generated by small molecule compounds induction and serve as a cellular platform to study mammary gland-related biological mechanisms in vitro. We next determined that the essential small molecule for this reprogramming was RepSox (R), an inhibitor of TGF receptor 1 (TGFβR1). Our previous findings also indicated that overexpression of SMAD3, one of the TGFβR1 downstream genes, may be crucial for the generation of induced mammary epithelial cells (iMECs). As a key transcription factor mediating TGFβ signaling, SMAD3 plays a key role in a variety of biological processes and performs multiple functions, including growth arrest, differentiation, apoptosis, and epithelial-mesenchymal transition (EMT) (Millet and Zhang, 2007; Padua and Massagué, 2009; Ikushima and Miyazono, 2010; Massagué, 2012). SMAD3 is involved in the development and differentiation of mammary epithelial cells (Yang et al., 2002; Yoo et al., 2009), but the precise mechanism of SMAD3-mediated reprogramming have not been thoroughly explored. Therefore, in the present study, we further elucidated the function of SMAD3 in the reprogramming of iMECs and investigated the relationship between SMAD3 expression level and reprogramming efficiency, complementing the reprogramming mechanism of iMECs in goats while providing insights into mammary epithelial cell fate determination.
Materials and methods
Plasmids and cells
The pLVX-TetOne-Puro, psPAX2, and pMD2.G plasmids were purchased from Hunan Fenghui Biotechnology Co. The pUC57 2As, pLVX-IRES-ZsGreen1, and pSicoR-Ef1a-mCherry plasmids were pre-constructed and preserved by the School of Animal Science and Technology, Guangxi University. HEK-293T cells were preserved by the School of Animal Science and Technology, Guangxi University.
Cell culture and induction medium
Goat ear fibroblasts (GEFs) were cultured in high glucose DMEM (GIBCO) supplemented with 10% fetal bovine serum (FBS, GIBCO) (Fibroblasts culture medium, FM).
BFTV induction medium contained Neurobasal Medium (GIBCO), KnockOut DMEM-F12 (GIBCO), KSR (GIBCO), 100 × N2 (GIBCO), 50 × B27 (GIBCO) supplements, 1% GlutaMAX (GIBCO), we refer to it as N2B27-based medium; and supplemented with four small molecule cocktail, 1 μM TTNPB (B), 10 μM Forskolin (F), 10 μM Tranylcypromine (T) and 500 μg/ml VPA (V); R induction medium was supplemented with 10 μM RepSox(R) in N2B27-basal medium.
Plasmid construction
The goat SMAD family member 3 (SMAD3) mRNA sequence (GenBank Accession No. XM_013966776.2), pUC57 2As, and pLVX-IRES-ZsGreen1 plasmids were used to design the primers listed in Table 1. SMAD3, P2A, and ZsGreen1 fragments carrying homologous arms were amplified by RT-PCR and spliced into SMAD3-P2A-ZsGreen1 by overlap extension PCR (SOE PCR). SMAD3-P2A-ZsGreen1 inserted into the EcoRI/BamHI sites of pLVX-TetOne-Puro (pLVX; empty vector). Then, the doxycycline (Dox)-induced overexpression of SMAD3 lentiviral plasmid (pLVX-TetOne-SMAD3-P2A-ZsGreen1-Puro) was constructed. Construction of the lentiviral plasmid was confirmed by bacterial liquid PCR, double enzyme digestion, and sequencing. (See Supplementary Material for details of this part of the reaction system and procedures).
Three interference targets were designed for the mRNA sequence of goat SMAD3, as listed in Table 1. The oligo DNA was annealed to synthesize shRNA1/2/3-SMAD3 and the nonsense shRNA (NC; negative control, a disrupted and meaningless sequence was inserted into the vector to exclude inserted sequence from potentially interfering with the experiment results). The goat SMAD3 gene lentiviral interference vector (pSicoR-Ef1a-mCherry-shRNA1/2/3-SMAD3) and its NC control vector (pSicoR-Ef1a-mCherry-shRNA1/2/3-SMAD3) were constructed by cleaving the pSicoR-Ef1a-mCherry vector (pSicoR; empty vector) with HpaI and Xhol enzymes, ligating the shRNA to linearized vector using T4 DNA ligase. Bacterial liquid PCR and sequencing provided proof that the lentiviral interference plasmids had been successfully created. The oligo DNA fragments were synthesized by Sangon Biotech, China.
Isolation and culture of goat ear fibroblasts
Goat’s ears were disinfected and dehaired, and small pieces of ear margin tissue were dissected. The tissues were washed once with 75% (v/v) alcohol and then three times with PBS (containing 2% penicillin-streptomycin). The tissues were then cut into 1-mm3 sections and plated in a 100-mm dish. The dish was placed in a carbon dioxide (CO2) incubator (37°C, 5% CO2). When the tissue mass reached translucence, FM was added. Once the confluence of GEF reached 80%–90%, it was passaged.
Screening of suitable concentration of puromycin
The GEFs were cultured in 24-well plates at a density of 1 × 104 cells/well in FM and washed with PBS after 48 h, replace with the new culture medium. Different concentrations of puromycin (1, 2, 3, 4, 5 μg/ml) were added to the cell culture medium, three replicate wells were treated with each concentration and changed daily to a fresh cell culture medium containing the corresponding concentration of puromycin.
Lentivirus infection
Lentiviral recombinant plasmid/vector (2.5 μg) was cotransfected together with the pMD2.G (1 μg) and psPAX2 (1.5 μg) plasmids into HEK-293T cells using Lipofectamine 3,000 (Invitrogen) in 6-cm dishes and Dox was added to the culture medium. The viral supernatant was collected after transfection for 48–72 h at 37°C and 5% CO2. The supernatant was centrifuged (2,000 rpm for 10 min at 4°C), filtered through 0.45-μm filters, and then placed onto GEFs supplemented with 8 ng/μL polybrene (Solarbio) and Dox. Two days later, microscopic images, evaluated the efficiency of viral infection, and the infected cells were collected, counted, and plated onto dishes (2.5 × 105 cells for a 6-cm dish) with FM addition of puromycin (3 μg/ml).
Generation of SMAD3 overexpression-induced mammary epithelial cells (SMAD3-iMECs)
Dox-induced overexpression of SMAD3 cells (GEFs-SMAD3) seeded at a density of 5 × 104 cells per well in a 4-well plate or at 5 × 105 cells per 60-mm dish and cultured with fibroblasts culture medium in an incubator at 37°C in a humidified atmosphere of 5% CO2. After 8 h, the FM was replaced by BFTV, and Dox was added. The culture was continued for 8 days, and the induction medium was refreshed every 2 days. GEFs were induced by R induction medium to generate R-CiMECs acted as positive control cells.
Real-time fluorescence quantitative PCR
Total RNA was isolated using TRIzol Reagent (Servicebio, G3013) and cDNA synthesis was performed using a HiScript® III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme, R223-01). RT-qPCR was performed using ChamQ Universal SYBR® qPCR Master Mix (Vazyme, Q711-02/03). The data were analyzed using the 2−ΔΔCT method. The primer sequences for RT-qPCR in this study are listed in Table 2. The expression of genes was normalized to that of the internal control gene GAPDH, and the expression fold change for the samples was calculated relative to the expression of the fibroblasts (GEFs).
Saturated oil red O staining
The cells were fixed with 4% paraformaldehyde for 10 min, and then the cells were stained for 15 min with oil red O staining solution (Solarbio, China, G1262). The cells were then rinsed with 60% isopropanol for 15–30 s and washed three times with sterile water. Mayer’s hematoxylin was added to stain the nuclei for 20 s, and the cells were washed with sterile water three times.
Immunofluorescence staining
After fixation with 4% paraformaldehyde (Sigma) at room temperature for 30 min, the cells were permeabilized with 1% Triton X-100 and blocked with 10% donkey serum at 37°C for one and a half hours. Primary antibodies were incubated with the cells at the appropriate dilutions at 4°C overnight in blocking buffer. The next day, the cells were probed with respective secondary antibodies for 1 h in the dark at room temperature. Nuclei were stained with Hoechst 33342. Antibody details are listed in the Supplemental Material.
Western blot
Cells were lysed in denaturing lysis buffer containing protease inhibitors (RIPA, Solarbio, Beijing, China) for 30 min on ice and centrifuged (12,000 rpm) for 10 min at 4°C. Protein concentrations in the lysates were determined by a BCA protein assay kit (Solarbio, Beijing, China). Approximately 30 μg of protein was separated by 12% SDS-PAGE and transferred to a nitrocellulose filter membrane that was blocked with 5% nonfat dried milk in Tris-buffered saline containing 0.05% Tween 20 (pH 7.6) for 1 h at 25°C. Subsequently, the membranes were incubated overnight at 4°C with primary antibodies. Then, the blots were incubated with secondary antibodies for 1 h at room temperature. After washing three times with TBST, immunostaining was visualized using Western blotting detection reagents (Bio-Rad, United States). Antibody details are listed in the Supplemental Material.
Quantification and statistical analysis
Independent pavement-like cell colonies were formed on day 4 of induction, and this compact pavement-like cell colonies were defined as primary colonies. Reprogramming efficiency (cell colony formation rate) was determined by the percentage of the number of independent primary colonies to the number of seeded fibroblasts. Efficiency (%) = No. of primary colonies/No. of seeded cells × 100%.
Statistical analyses for differential gene expression (Figures 3C, 4C, 5A) and comparison of reprogramming efficiency (Figures 3B,E, 4B,E) were performed in GraphPad. Significance was calculated with Student’s t test or one-way ANOVA. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
Results
Overexpression and interference of SMAD3 gene in goat fibroblasts
To investigate the function of the goat SMAD3 gene in the reprogramming process of induced mammary epithelial cells, we regulated the expression level of SMAD3 gene in goat fibroblasts. A Dox-induced eukaryotic expression vector for goat SMAD3 overexpression, pLVX-TetOne-SMAD3-Puro, was successfully created through a series of PCR experiments (Figures 1A–E). In this vector, the expression of the P2A-linked green fluorescent protein ZsGreen1 allowed us to estimate the expression of SMAD3 in an approximate manner. We also obtained the eukaryotic expression vector pSicoR-Ef1a-mCherry-shRNA-SMAD3, which targets the knockdown of the SMAD3 gene in goats (Figures 1F–H).
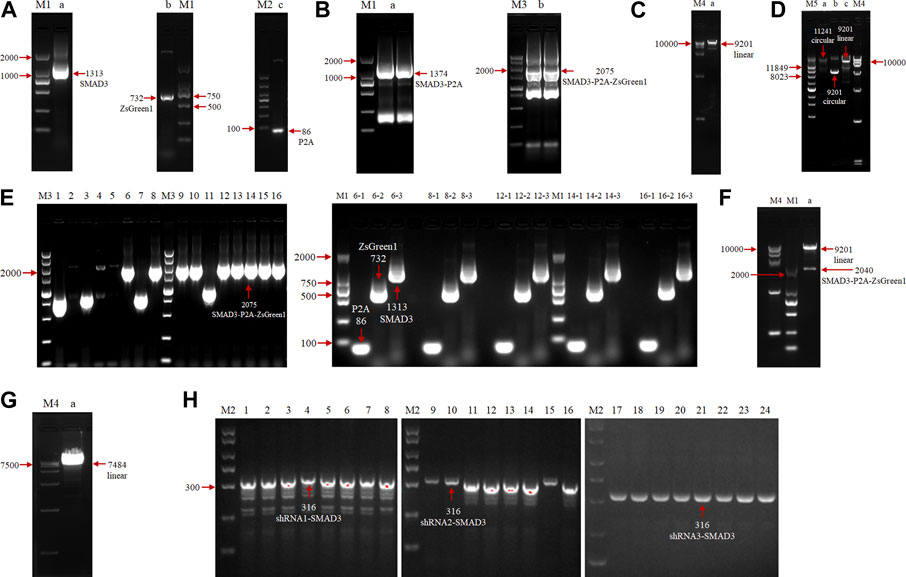
FIGURE 1. Agarose gel electrophoresis analysis of overexpression and construction of eukaryotic expression vectors for the interfering goat SMAD3 gene. (A) PCR amplification of SMAD3, P2A, and ZsGreen1 sequences, respectively. M: marker (M1: DL2000, M2: DL1000); lane a: SMAD3 gene, 1313 bp; lane b: ZsGreen1 sequence, 732 bp; lane c: P2A sequence, 86 bp. (B) SOE PCR ligation of SMAD3, P2A, and ZsGreen1 fragments. M: marker (M3: DL5000); lane a: SMAD3-P2A fragment, 1374 bp; lane b: SMAD3-P2A-ZsGreen1 fragment, 2075 bp (C) Enzymatic cleavage of the pLVX-TetOne-Puro vector. M: marker (M4: DL15000); lane a: linearized pLVX-TetOne-Puro vector, 9201 bp. (D) Homologous recombination reaction. M: marker (M5: Supercoiled DNA Ladder Marker); lane a: recombination product, 11241 bp; lane b: cyclic pLVX-TetOne-Puro vector, 9201 bp; lane c: linearized pLVX-TetOne-Puro vector, 9201 bp. (E) PCR identification of recombinant product transformed bacterial solution. Lanes 1–16: first bacterial fluid PCR bands of 16 monoclonal colonies, positive colony product (SMAD3-P2A-ZsGreen1) size 2075 bp; lanes 6/8/12/14/16–1/2/3: 6, 8, 12, 14, 16 positive colonies second bacterial fluid PCR bands, positive colonies all can produce 86 bp (P2A), 732 bp (ZsGreen1) and 1313 bp (SMAD3) bands, respectively. (F) Plasmid double digestion identification. Lane a: positive plasmids were digested by EcoRI and BamHI to become linearized pLVX-TetOne-Puro vector as well as SMAD3-P2A-ZsGreen1 bands (without homologous arms), size 9201 and 2040 bp, respectively. (G) Enzymatic cleavage of the pSicoR-Ef1a-mCherry vector. Lane a: linearized pSicoR-Ef1a-mCherry vector, 7484 bp. (H) PCR identification of ligated products transformed bacterial solution. Lanes 1–24: bacterial fluid PCR bands of 8 monoclonal colonies each of 3 interfering target sequences, positive colonies products all 316 bp in size (shRNA1/2/3-SMAD3).
We selected the lentivirus-mediated method to infect GEFs after lentiviral packaging of the target plasmid in order to increase transfection efficiency and obtain cell lines with long-term stable overexpression and interference with the goat SMAD3 gene. The results showed that the infection efficiency of pLVX-TetOne-SMAD3 lentivirus was close to 60% (Figure 2A), while the infection efficiency of pSicoR-Ef1a-mCherry-shRNA1/2/3-SMAD3 lentivirus virus could reach 80%–90% (Figure 2B). In addition, the outcomes of puromycin screening suggested that 3 μg/ml could be used as the ideal dose to utilize for cell selection (Figure 2C; Table 3).
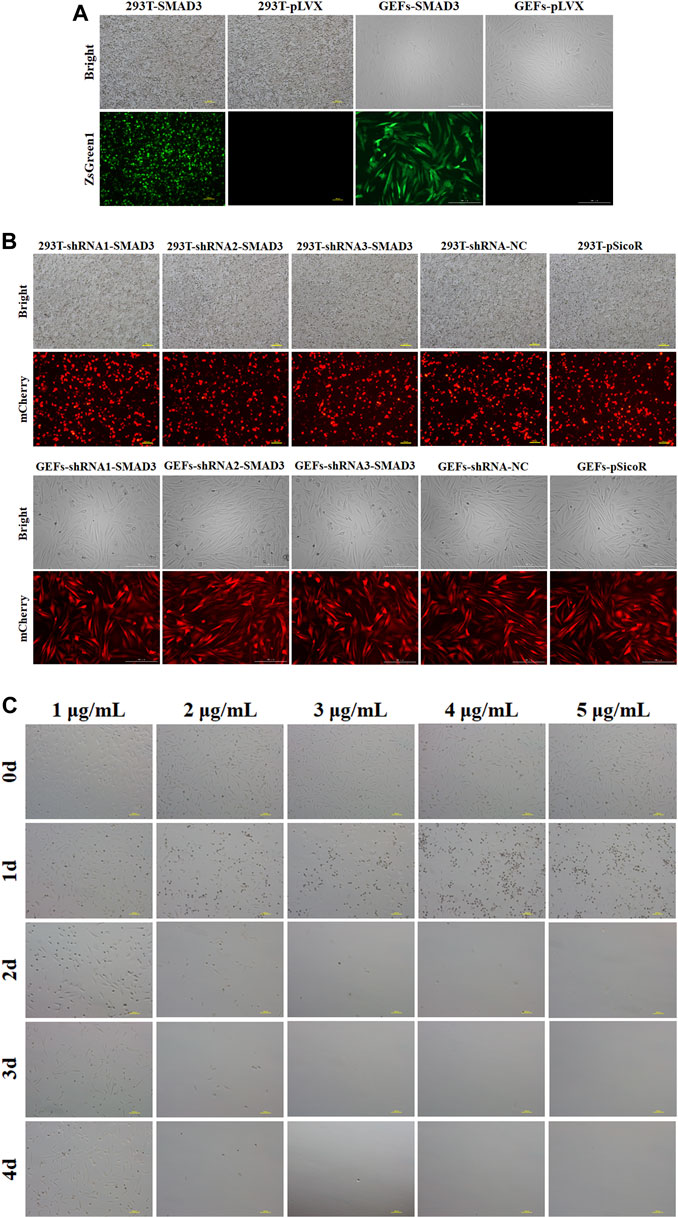
FIGURE 2. Stable overexpression and interference of goat SMAD3 gene cell lines (GEFs-SMAD3 and GEFs-shRNA-SMAD3) were obtained. (A) Fluorescence rate of pLVX-TetOne-SMAD3-Puro packaged lentivirus and virus supernatant infected goat fibroblasts for 48 h. Yellow scale bar, 100 μm, white scale bar, 200 μm. (B) Fluorescence rate of pSicoR-Ef1a-mCherry-shRNA-SMAD3 packaged lentivirus and virus supernatant infected goat fibroblasts for 48 h. Yellow scale bar, 100 μm, white scale bar, 200 μm. (C) Screening of suitable concentrations of puromycin. Scale bar, 100 μm.
The reprogramming efficiency of induced mammary epithelial cell tends to increase and then decrease with increasing SMAD3 expression levels
Subsequently, we determined the effects of different concentrations of doxycycline (Dox) on inducing GEFs-SMAD3 to reprogram into SMAD3-iMECs. Under the addition of various Dox concentrations (2.5, 5, 7.5, and 10 g/ml), GEFs-SMAD3 allows incremental epithelization from the initial typical fibroblasts during the reprogramming process. Independent and compact epithelial cell-like colonies were formed on day 4 of the reprogramming process (Figure 3A), but the state and number of colonies varied depending on the Dox concentration groups. At a Dox concentration of 5 g/ml, the reprogramming efficiency was 3.31%, which was the highest among the various groups (Figure 3B). However, GEFs-pLVX cells as the empty control group failed to form any independent and compact epithelial cell colonies under the same culture conditions. The results also showed that the expression levels of SMAD3 increased when the concentration of Dox increased, peaked at a concentration of 7.5 μg/ml but decreased at a concentration of 10 μg/ml (Figure 3C). The aforementioned findings thus indicated that there was a parabolic rather than linear relationship between the reprogramming efficiency of iMECs and the expression levels of SMAD3. Therefore, we decided 5 g/ml was the optimal concentration of Dox for subsequent induction assays. As induction time increased, GEFs-SMAD3 underwent epithelialization and formed small epithelial cell-like aggregates; followed by the independent paver-like clones formed, and the clonal area gradually increased and eventually stabilized (Figure 3D). Compared to the GEFs-pLVX, the reprogramming efficiency of GEFs-SMAD3 was greatly enhanced, but slightly lower than the positive control R-CiMECs (3% vs. 4.68%) (Figure 3E).

FIGURE 3. Effect of SMAD3 expression on reprogramming efficiency of goat induced mammary epithelial cells. (A) The ability of cells to form independent clones within 4 days under different concentrations of Dox induction. Scale bar, 100 μm. (B) Reprogramming efficiency of goat induced mammary epithelial cells under different concentrations of Dox treatment (Efficiency (%) = No. of primary colonies/No. of seeded cells × 100%). (C) Changes in SMAD3 gene expression levels in goat fibroblasts induced by different concentrations of Dox. (Due to the specificity of the vector, there was a small leakage of expression at Dox = 0 μg/ml) n = 3 biological replicates. Data are represented as the mean ± SEM. *p < 0.05, ***p < 0.001 (Student’s t test). (D) Morphological changes during overexpression of SMAD3-induced reprogramming of goat fibroblasts into mammary epithelial cells (Dox = 5 μg/ml). Scale bar, 100 μm. (E) Overexpression of SMAD3 significantly improves the reprogramming efficiency of goat induced mammary epithelial cells (Dox = 5 μg/ml). Scale bar, 100 μm. n = 3 biological replicates. Data are represented as the mean ± SEM. ***p < 0.001 (one-way ANOVA).
The SMAD3 gene plays a key role in the acquisition of induced mammary epithelial cells in goats
We found that addition of SMAD3 inhibitor Halofuginone (HF) significantly reduced the number of iMEC colonies formed (Figures 4A,B). Then, we further applied RNA interference (RNAi) to down-regulate expression of SMAD3, and the results revealed that all three of the interfering target sequences could significantly decrease the expression level of SMAD3 gene (Figure 4C). In order to determine whether reduced SMAD3 gene expression affects RepSox-induced reprogramming of goat fibroblasts into mammary epithelial cells, we selected GEFs-shRNA2-SMAD3, which has the lowest SMAD3 expression level. The results demonstrated that SMAD3 downregulation severely affected RepSox-induced colonies formed (Figure 4D), and decrease the reprogramming efficiency to 0.85% from 3.56% (Figure 4E). The conclusion drawn from these findings is that SMAD3 plays an important role in the conversion of fibroblasts into iMECs. Moreover, these findings provide further evidence that the RepSox-induced reprogramming of iMECs may be mediated through the regulation of SMAD3.
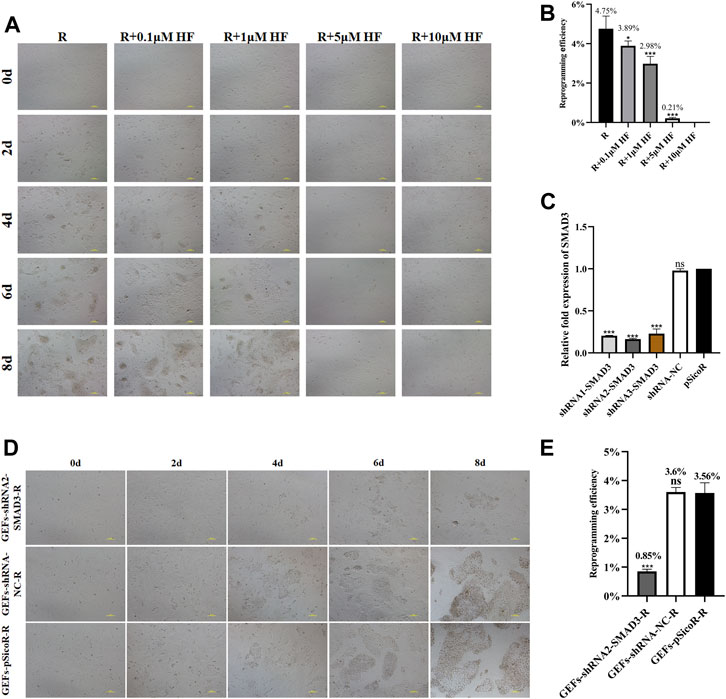
FIGURE 4. SMAD3 downregulation impedes the reprogramming process of induced mammary epithelial cells in goats. (A) Effect of different concentrations of HF on the reprogramming process of induced mammary epithelial cells in goats. Scale bar, 100 μm. (B) Inhibitory effect of different concentrations of HF on the reprogramming efficiency of induced mammary epithelial cells in goats. n = 3 biological replicates. Data are represented as the mean ± SEM. *p < 0.05, ***p < 0.001 (one-way ANOVA). (C) Expression levels of SMAD3 gene in three interfering target sequences in the corresponding cell lines (GEFs-shRNA1/2/3-SMAD3). n = 3 biological replicates. Data are represented as the mean ± SEM. ***p < 0.001 (one-way ANOVA). (D) Effect of interfering with SMAD3 expression on the reprogramming process of induced mammary epithelial cells in goats. Scale bar, 100 μm. (E) Interfering with SMAD3 expression significantly reduces the reprogramming efficiency of induced mammary epithelial cells in goats. Scale bar, 100 μm; n = 3 biological replicates. Data are represented as the mean ± SEM. ***p < 0.001 (one-way ANOVA).
SMAD3-iMECs have similar biological properties to R-CiMECs
Finally, the biological characteristics of SMAD3-iMECs were identified. SMAD3-iMECs and R-CiMECs (positive control cells) strongly expressed the mammary epithelial cell marker genes KRT18 and LTF, as well as the mammary development-related genes MSX2 and GATA3, in comparison to the GEFs (negative control group cells). However, the fibroblast-related marker genes FBN1, COL3A1, and VIMENTIN were significantly downregulated (Figure 5A). An important feature of MECs is the ability to secrete milk fat. R-CiMECs and SMAD3-iMECs secreted lipid droplets of different sizes around the cytoplasm, which were stained red by saturated oil red O staining (Figure 5B), confirming their ability to secrete lipids. Furthermore, our immunofluorescence staining results showed that R-CiMECs and SMAD3-iMECs significantly expressed the epithelial cell-specific antigens CDH1, EpCAM, and KRT18, but not the fibroblasts-specific antigen VIMENTIN (Figure 5C). Western blot analysis revealed that SMAD3-iMECs expressed the lactating state mammary epithelial cell-specific proteins LTF and αs2-CSN, and the protein expression trend was similar to R-CiMECs (Figure 5D). These results indicated that SMAD3-iMECs with lactation function has similar biological properties to R-CiMECs.
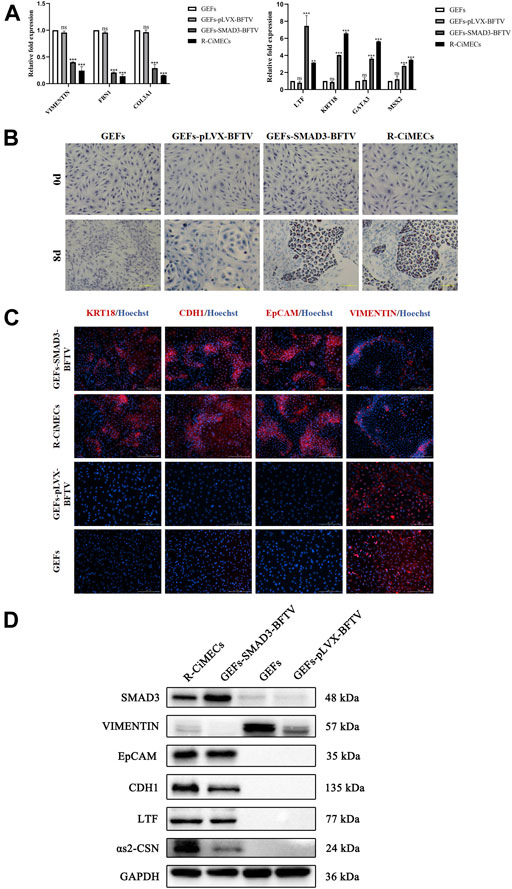
FIGURE 5. Biological characterization of SMAD3-iMECs. (A) RT-qPCR was performed to detect changes in the expression of fibroblast-related marker genes (VIMENTIN, COL3A1, and FBN1), MECs-related marker genes (LTF and KRT18), and MECs development-related genes (MSX2 and GATA3) after induced mammary epithelial cell reprogramming in goats. n = 3 biological replicates. Data are represented as the mean ± SEM. **p < 0.01, ***p < 0.001 (one-way ANOVA). (B) Saturated oil red O staining was performed to identify SMAD3-iMECs with similar lipid droplet secretion functions as R-CiMECs. Scale bar, 100 μm. (C) Immunofluorescence staining was performed to detect the expression of fibroblasts marker antigen (VIMENTIN) and epithelial cell marker antigen (EpCAM, CDH1, and KRT18) in SMAD3-iMECs and R-CiMECs. Scale bar, 200 μm. (D) Western blot detection of SMAD3-iMECs and R-CiMECs with similar specific marker protein expression patterns (Fibroblasts marker protein: VIMENTIN; epithelial cell marker protein: CDH1 and EpCAM; mammary epithelial cell marker protein: LTF and αs2-CSN).
Discussion
Our previous report (Zhang et al., 2021) demonstrated that goat fibroblasts could be chemically induced to reprogram into mammary epithelial cells (CiMECs) by exposure to small molecule compounds (BFRTV). We further identified that RepSox (R), an inhibitor of TGFβR1, was the essential small molecule for this reprogramming, and revealed that R may act on CiMECs generation through the regulatory effects of SMAD3. In this study, we further proved that SMAD3 overexpression induced the conversion of goat fibroblasts into iMECs, and confirmed that SMAD3 plays a significant role in the iMECs reprogramming process. The regulatory mechanism of mammary epithelial cell fate may be better understood as a result of our discoveries.
The TGF-β/SMAD pathway is essential for the normal development of mammary epithelial tissue. Mammary epithelial cells exhibit significant levels of Smad3 expression. In mice, Smad3 deletion causes mammary development abnormalities secondary to ovarian insufficiency (Yang et al., 2002). Constitutive active Smad2/Smad3 is a broad-spectrum enhancer in the somatic reprogramming process regulated by transcription factors, which increases the efficiency of somatic reprogramming. Smad3 interacts with coactivators and reprogramming factors, combined with the OCT4 target motif, and significantly improves the efficiency of iPSCs generation by the Yamanaka factor (Ruetz et al., 2017). Smad2/Smad3 also significantly improved the efficiency of CEBPα-mediated transformation of B cells into macrophages (Ruetz et al., 2017). Additionally, Smad3 can directly reprogram spermatogonial stem cells into hepatocyte-like cells with mature hepatocyte morphology, phenotype, and function through the ERK1/2 and Smad2/3 signaling pathways (Zhang et al., 2013). Therefore, SMAD3 plays an important role in cellular reprogramming. Surprisingly, in this study, we not only demonstrated that SMAD3 overexpression can initiate and complete the reprogramming of iMECs, but also that the reprogramming efficiency of iMECs shows a parabolic relationship with SMAD3 expression levels. Moreover, the parabolic relationship might indicate that SMAD3 expression level is the key to achieving mammary epithelial cell fate in other species.
The primary cell type in the mammary gland is the mammary epithelial cells, which are crucial for lactation synthesis (Richert et al., 2000). Numerous signaling factors regulate the development and differentiation of mammary epithelial cells, among which Gata3 is a key transcription factor for luminal epithelial cell differentiation in the mammary gland (Kouros-Mehr et al., 2006; Siegel and Muller, 2010); targeted knockout of Gata3 in the mammary gland results in severely impaired mammary gland development and inability to determine and maintain the fate of luminal epithelial cells (Asselin-Labat et al., 2007; Watson et al., 2011). In addition, Msx2 is a transcription factor necessary for mammary gland developmental differentiation, knockout of Msx2 at E16.5 leads to a stagnation of mammary bud development, which is comparable to the mammary phenotype of conditional knockout Gata3 mice (Phippard et al., 1996; Satokata et al., 2000). Our work discovered that GATA3 and MSX2 gene expression was upregulated in SMAD3-iMECs compared to fibroblasts, implying that SMAD3 overexpression may activate the expression of mammary gland development-related genes such as GATA3 and MSX2, which are involved in cell fate transformation. Last but not least, our findings showed that SMAD3-iMECs shared similar characterization with R-CiMECs, including MEC-specific markers and milk-secreting functions. Thus, these SMAD3-iMECs contained secretory luminal epithelial cells can secrete milk. It may suggest that SMAD3-iMECs have the capacity to generate mammary organoids for investigating mammary gland development and obtaining “culture dish milk” in vitro.
Conclusion
In this study, we demonstrated that the regulatory role of SMAD3 is directly involved in mammary epithelial cell fate determination in goats, which offers novel insights into the regulatory mechanism of the TGFβR1-SMAD3 pathway. These findings may provide an alternative strategy for other species to regulate mammary epithelial cell fate in vitro. However, further experiments are still required to conduct in order to elucidate how SMAD3 expression level regulates mammary epithelial cell fate, and find downstream genes of SMAD3 on regulating reprogramming of iMECs.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
YW and DZ designed and supervised the study. YW, SY, and QL performed the experiments and analyzed the data. YW wrote the paper. BH and DZ revised the manuscript and provided critical comments. All authors read and approved the final manuscript.
Funding
This research is supported by grants from the Chinese National Natural Science Foundation (Grant nos. 31960160 and 32160171) and Guangxi Natural Science Foundation (Grant nos. 2017GXNSFDA198035, AB18221072, and 2018GXNSFAA138148).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.1002874/full#supplementary-material
References
Asselin-Labat, M. L., Sutherland, K. D., Barker, H., Thomas, R., Shackleton, M., Forrest, N. C., et al. (2007). Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 9, 201–209. doi:10.1038/ncb1530
Bannister, A. J., and Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381–395. doi:10.1038/CR.2011.22
Batta, K., Florkowska, M., Kouskoff, V., and Lacaud, G. (2014). Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell Rep. 9, 1871–1884. doi:10.1016/j.celrep.2014.11.002
Bussmann, L. H., Schubert, A., Vu Manh, T. P., De Andres, L., Desbordes, S. C., Parra, M., et al. (2009). A robust and highly efficient immune cell reprogramming system. Cell Stem Cell 5, 554–566. doi:10.1016/j.stem.2009.10.004
Caiazzo, M., Giannelli, S., Valente, P., Lignani, G., Carissimo, A., Sessa, A., et al. (2015). Direct conversion of fibroblasts into functional astrocytes by defined transcription factors. Stem Cell Rep. 4, 25–36. doi:10.1016/j.stemcr.2014.12.002
Cheng, L., Gao, L., Guan, W., Mao, J., Hu, W., Qiu, B., et al. (2015). Direct conversion of astrocytes into neuronal cells by drug cocktail. Cell Res. 25 (11), 1269–1272. doi:10.1038/cr.2015.120
Davis, R. L., Weintraub, H., and Lassar, A. B. (1987). Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000. doi:10.1016/0092-8674(87)90585-X
Fu, Y., Huang, C., Xu, X., Gu, H., Ye, Y., Jiang, C., et al. (2015). Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res. 25, 1013–1024. doi:10.1038/CR.2015.99
Han, J. K., Chang, S. H., Cho, H. J., Choi, S. B., Ahn, H. S., Lee, J., et al. (2014). Direct conversion of adult skin fibroblasts to endothelial cells by defined factors. Circulation 130, 1168–1178. doi:10.1161/CIRCULATIONAHA.113.007727
Han, Y. C., Lim, Y., Duffieldl, M. D., Li, H., Liu, J., Abdul Manaph, N. P., et al. (2016). Direct reprogramming of mouse fibroblasts to neural stem cells by small molecules. Stem Cells Int. 2016, 4304916. doi:10.1155/2016/4304916
Huang, P., Zhang, L., Gao, Y., He, Z., Yao, D., Wu, Z., et al. (2014). Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell 14, 370–384. doi:10.1016/J.STEM.2014.01.003
Ieda, M., Fu, J.-D., Delgado-Olguin, P., Vedantham, V., Hayashi, Y., Bruneau, B. G., et al. (2010). Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386. doi:10.1016/j.cell.2010.07.002
Ikushima, H., and Miyazono, K. (2010). TGFbeta signalling: A complex web in cancer progression. Nat. Rev. Cancer 10, 415–424. doi:10.1038/NRC2853
Iwafuchi-Doi, M., and Zaret, K. S. (2014). Pioneer transcription factors in cell reprogramming. Genes Dev. 28, 2679–2692. doi:10.1101/GAD.253443.114
Kim, J., Efe, J. A., Zhu, S., Talantova, M., Yuan, X., Wang, S., et al. (2011). Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. U. S. A. 108 (19), 7838–7843. doi:10.1073/pnas.1103113108
Kouros-Mehr, H., Slorach, E. M., Sternlicht, M. D., and Werb, Z. (2006). GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 127, 1041–1055. doi:10.1016/j.cell.2006.09.048
Li, K., Zhu, S., Russ, H. A., Xu, S., Xu, T., Zhang, Y., et al. (2014). Small molecules facilitate the reprogramming of mouse fibroblasts into pancreatic lineages. Cell Stem Cell 14, 228–236. doi:10.1016/j.stem.2014.01.006
Li, W., Li, K., Wei, W., and Ding, S. (2013). Chemical approaches to stem cell biology and therapeutics. Cell Stem Cell 13, 270–283. doi:10.1016/J.STEM.2013.08.002
Li, X., Zuo, X., Jing, J., Ma, Y., Wang, J., Liu, D., et al. (2015). Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell 17, 195–203. doi:10.1016/J.STEM.2015.06.003
Massagué, J. (2012). TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630. doi:10.1038/NRM3434
Millet, C., and Zhang, Y. E. (2007). Roles of Smad3 in TGF-β signaling during carcinogenesis. Crit. Rev. Eukaryot. Gene Expr. 17, 281–293. doi:10.1615/CRITREVEUKARGENEEXPR.V17.I4.30
Padua, D., and Massagué, J. (2009). Roles of TGFbeta in metastasis. Cell Res. 19, 89–102. doi:10.1038/CR.2008.316
Phippard, D. J., Weber-Hall, S. J., Sharpe, P. T., Stuart Naylor, M., Jayatalake, H., Maas, R., et al. (1996). Regulation of Msx-1, Msx-2, Bmp-2 and Bmp-4 during foetal and postnatal mammary gland development. Development 122, 2729–2737. doi:10.1242/DEV.122.9.2729
Richert, M. M., Schwertfeger, K. L., Ryder, J. W., and Anderson, S. M. (2000). An atlas of mouse mammary gland development. J. Mammary Gland. Biol. Neoplasia 5, 227–241. doi:10.1023/a:1026499523505
Ruetz, T., Pfisterer, U., Di Stefano, B., Ashmore, J., Beniazza, M., Tian, T. V., et al. (2017). Constitutively active SMAD2/3 are broad-scope potentiators of transcription-factor-mediated cellular reprogramming. Cell Stem Cell 21, 791–805. doi:10.1016/j.stem.2017.10.013
Satokata, I., Ma, L., Ohshima, H., Bei, M., Ian, W., Nishizawa, K., et al. (2000). Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat. Genet. 24, 391–395. doi:10.1038/74231
Sheng, C., Zheng, Q., Wu, J., Xu, Z., Wang, L., Li, W., et al. (2012). Direct reprogramming of Sertoli cells into multipotent neural stem cells by defined factors. Cell Res. 22, 208–218. doi:10.1038/cr.2011.175
Siegel, P. M., and Muller, W. J. (2010). Transcription factor regulatory networks in mammary epithelial development and tumorigenesis. Oncogene 29, 2753–2759. doi:10.1038/ONC.2010.43
Soufi, A., Garcia, M. F., Jaroszewicz, A., Osman, N., Pellegrini, M., and Zaret, K. S. (2015). Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568. doi:10.1016/J.CELL.2015.03.017
Wada, R., Muraoka, N., Inagawa, K., Yamakawa, H., Miyamoto, K., Sadahiro, T., et al. (2013). Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. U. S. A. 110, 12667–12672. doi:10.1073/PNAS.1304053110/-/DCSUPPLEMENTAL/SM07
Wang, L., Wang, L., Huang, W., Su, H., Xue, Y., Su, Z., et al. (2013). Generation of integration-free neural progenitor cells from cells in human urine. Nat. Methods 10 (1), 84–89. doi:10.1038/nmeth.2283
Wang, Y., Qin, J., Wang, S., Zhang, W., Duan, J., Zhang, J., et al. (2016). Conversion of human gastric epithelial cells to multipotent endodermal progenitors using defined small molecules. Cell Stem Cell 19, 449–461. doi:10.1016/J.STEM.2016.06.006
Watson, C. J., Oliver, C. H., and Khaled, W. T. (2011). Cytokine signalling in mammary gland development. J. Reprod. Immunol. 88, 124–129. doi:10.1016/j.jri.2010.11.006
Wenxiang, H., Binlong, Q., Wuqiang, G., Qinying, W., Min, W., Wei, L., et al. (2015). Direct conversion of normal and alzheimer’s disease human fibroblasts into neuronal cells by small molecules. Cell Stem Cell 17, 204–212. doi:10.1016/J.STEM.2015.07.006
Yang, Y. A., Tang, B., Robinson, G., Hennighausen, L., Brodie, S. G., Deng, C. X., et al. (2002). Smad3 in the mammary epithelium has a nonredundant role in the induction of apoptosis, but not in the regulation of proliferation or differentiation by transforming growth factor-β. Cell Growth Differ. 13 (3), 123. PMC11959813.
Yoo, A. S., Sun, A. X., Li, L., Shcheglovitov, A., Portmann, T., Li, Y., et al. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231. doi:10.1038/NATURE10323
Yoo, Y. A., Kang, M. H., Kim, B. S., Kim, J. S., and Seo, J. H. (2009). Sustained co-cultivation with human placenta-derived MSCs enhances ALK5/Smad3 signaling in human breast epithelial cells, leading to EMT and differentiation. Differentiation. 77, 450–461. doi:10.1016/j.diff.2009.03.003
Zhang, D., Ren, Y., Qin, L., Liu, Q., Wang, G., Sun, L., et al. (2021). Transdifferentiation of goat ear fibroblasts into lactating mammary epithelial cells induced by small molecule compounds. Biochem. Biophys. Res. Commun. 573, 55–61. doi:10.1016/j.bbrc.2021.07.087
Zhang, Z., Gong, Y., Guo, Y., Hai, Y., Yang, H., Yang, S., et al. (2013). Direct transdifferentiation of spermatogonial stem cells to morphological, phenotypic and functional hepatocyte-like cells via the ERK1/2 and Smad2/3 signaling pathways and the inactivation of cyclin A, cyclin B and cyclin e. Cell Commun. Signal. 11, 67. doi:10.1186/1478-811X-11-67
Zheng, J., Choi, K. A., Kang, P. J., Hyeon, S., Kwon, S., Moon, J. H., et al. (2016). A combination of small molecules directly reprograms mouse fibroblasts into neural stem cells. Biochem. Biophys. Res. Commun. 476, 42–48. doi:10.1016/J.BBRC.2016.05.080
Keywords: somatic cell reprogramming, Smad3, induced mammary epithelial cells, cell fate determination, Tet On
Citation: Wu Y, Zhang D, Ye S, Liu Q and Huang B (2022) Parabolic relationship between SMAD3 expression level and the reprogramming efficiency of goat induced mammary epithelial cells. Front. Cell Dev. Biol. 10:1002874. doi: 10.3389/fcell.2022.1002874
Received: 25 July 2022; Accepted: 30 September 2022;
Published: 14 October 2022.
Edited by:
Ruttachuk Rungsiwiwut, Srinakharinwirot University, ThailandReviewed by:
Rajkumar P. Thummer, Indian Institute of Technology Guwahati, IndiaYusheng Liang, School of Medicine, University of Michigan, United States
Feng Su, Shandong Agricultural University, China
Jiangjiang Zhu, Southwest Minzu University, China
Copyright © 2022 Wu, Zhang, Ye, Liu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ben Huang, Ymh1YW5nQGd4YW1zLm9yZy5jbg==, YmVuaHVhbmdAZ3h1LmVkdS5jbg==
†These authors share first authorship
 Yulian Wu
Yulian Wu Dandan Zhang1†
Dandan Zhang1† Quanhui Liu
Quanhui Liu Ben Huang
Ben Huang

