
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 24 December 2021
Sec. Molecular and Cellular Pathology
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.797072
This article is part of the Research Topic Various-Scale Identification of Cognitive Impairments and Emotional Disorders View all 9 articles
Epidemiological and clinical studies have suggested comorbidity between schizophrenia and several neurodegenerative disorders. However, little is known whether there exists shared genetic architecture. To explore their relationship from a genetic and transcriptomic perspective, we applied polygenic and linkage disequilibrium-informed methods to examine the genetic correlation between schizophrenia and amyotrophic lateral sclerosis (ALS), Parkinson’s disease, Alzheimer’s disease and frontotemporal dementia. We further combined genome-wide association summary statistics with large-scale transcriptomic datasets, to identify putative shared genes and explore related pathological tissues. We identified positive and significant correlation between schizophrenia and ALS at genetic (correlation 0.22; 95% CI: 0.16–0.28; p = 4.00E-04) and transcriptomic (correlation 0.08; 95% CI: 0.04–0.11; p = 0.034) levels. We further demonstrated that schizophrenia- and ALS-inferred gene expression overlap significantly in four tissues including skin, small intestine, brain cortex and lung, and highlighted three genes, namely GLB1L3, ZNHIT3 and TMEM194A as potential mediators of the correlation between schizophrenia and ALS. Our findings revealed overlapped gene expression profiles in specific tissues between schizophrenia and ALS, and identified novel potential shared genes. These results provided a better understanding for the pleiotropy of schizophrenia, and paved way for future studies to further elucidate the molecular drivers of schizophrenia.
Schizophrenia is a chronic and severe mental disorder characterized by continuous or relapsing episodes of psychosis with major symptoms like hallucinations and delusions (Owen et al., 2016). Usually, schizophrenia is considered as a neurodevelopmental disorder attributed to alterations in prenatal-to-early adolescent development. However, emerging evidence from neuropathological and longitudinal studies of schizophrenia also supports a neurodegenerative hypothesis. For example, schizophrenia was suggested as associated with accumulated protein insolubility and ubiquitination in a way similar to that presented in cases of neurodegenerative diseases (Nucifora et al., 2019). Meanwhile, abnormal white matter (WM) integrity as indicated by reduced fractional anisotropy at the onset and during chronic states of schizophrenia was observed in patients compared with controls, and the difference might increase over time (Kochunov and Hong, 2014). Mapping the trajectory of WM changes along the lifespan could provide novel insight into the biological nature of the factors underlying WM deficits (Cetin-Karayumak et al., 2020). The accelerated decline with aging also supported the neurodegenerative model. Even though, such a hypothesis was still debated due to various factors such as the absence of characteristics of neurodegeneration in schizophrenia, like gliosis (Gupta and Kulhara, 2010). Therefore, the link between schizophrenia and neurodegeneration might be complex and worth further exploration.
Clinically, growing evidence suggested that schizophrenia was linked to several neurodegenerative diseases. Amyotrophic lateral sclerosis (ALS), classically described as a motor system disorder, has been reported to co-exist with a variety of mental symptoms like psychosis and dementia, while disturbances in motor neuron function at both central and peripheral levels existed in schizophrenia (Crayton et al., 1977; Crayton and Meltzer, 1979; Zucchi et al., 2019). Meanwhile, a recent genetic study identified genetic pleiotropy between ALS and schizophrenia using the conditional false discovery rate method (McLaughlin et al., 2017). Both schizophrenia and Parkinson’s disease (PD) were closely related to dopaminergic function, with dopaminergic system imbalance or dysfunction as a common cause (Salamone et al., 2016). In clinical practice, several cases with both schizophrenia and PD have been reported, suggesting potential commodities between the two disorders (Habermeyer et al., 2008; Gadit, 2011). Psychotic symptoms like delusions and hallucinations which were characteristics of schizophrenia, were also frequent in Alzheimer’s disease (AD), affecting about 40–60% of individuals with AD. And AD with psychosis was found inversely associated with polygenic risk for schizophrenia (DeMichele-Sweet et al., 2018; Creese et al., 2019). Frontotemporal dementia (FTD) is a disorder of frontal lobe function, while recent research has demonstrated changes in the frontal and temporal lobes in schizophrenia as well. Meanwhile, schizophrenia and FTD patients presented with some similar clinical symptoms, and molecular evidence has been proposed for potential shared pathophysiologic mechanisms (Cooper and Ovsiew, 2013). Taken together, schizophrenia was closely correlated with these neurodegenerative disorders and might share common pathogenesis with them.
Recently, the integration of large-scale functional genomic data with genome-wide association study (GWAS) results has been proposed to characterize the functional effects of associated variants (Li et al., 2019). Transcriptome-wide association study (TWAS) is a powerful approach to research the genetic architecture of complex traits like schizophrenia, by building a model to impute gene expression levels from genotypes using samples with matched genotypes and gene expression data in given tissues (Hu et al., 2019). In this context, we explored the genetic correlation between schizophrenia and four neurodegenerative diseases including ALS, PD, AD, and FTD, and identified a positive and significant correlation between schizophrenia and ALS. We further conducted transcriptome-wide expression analysis and identified specific tissues and genes with overlapping expression profiles between schizophrenia and ALS.
We investigated the genetic correlation between schizophrenia (Lam et al., 2019) and four neurodegenerative disorders, namely ALS (Nicolas et al., 2018), PD (Nalls et al., 2019), AD (Lambert et al., 2013) and FTD (Ferrari et al., 2014) based on GWAS summary statistics. Details of the summary data from all GWAS used in the current study were listed in Supplementary Table S1. The study design like the collection of samples, quality control procedures and imputation methods have been described in each publication. The relevant institutional review boards or ethics committees approved the research protocol of each GWAS, and all participants gave written informed consent.
We first applied linkage disequilibrium (LD) score regression in specifically expressed genes (LDSC-SEG) to identify disease-relevant tissues and cell types by analyzing gene expression data together with GWAS summary statistics (Finucane et al., 2018). LDSC-SEG identifies tissues in which genes with increased expression are enriched in single-nucleotide polymorphisms (SNPs) that tag a large amount of heritability. Data for LDSC-SEG was prepared as described earlier (Finucane et al., 2018). Gene expression data from the following sources were analyzed: 1) RNA sequencing (RNA-seq) read counts from Genotype-Tissue Expression (GTEx) v6p (Gamazon et al., 2018). Genes for which fewer than four samples had at least one read count per million, as well as samples for which fewer than 100 genes had at least one read count per million were removed. 2) GeneChip expression array data from mouse forebrain sorted cells downloaded in the Gene Expression Omnibus (GEO) (GSE9566) (Cahoy et al., 2008). 3) Publicly available gene expression data from the ImmGen project (Heng and Painter, 2008) downloaded in GEO (GSE15907, GSE37448). A p value below 9.43E-04 (0.05/53), 0.017 (0.05/3), and 1.71E-04 (0.05/292) was considered statistically significant after the Bonferroni correction for analysis using the GTEx, GSE9566 and ImmGen data respectively.
We estimated the genetic correlation between schizophrenia and each neurodegenerative disorder using LDSC (Bulik-Sullivan et al., 2015) and GNOVA (Lu et al., 2017) with default parameters. The LDSC method quantifies the contribution of each SNP by examining the relationship between GWAS summary statistics and linkage disequilibrium (Abecasis et al., 2012). Compared with LDSC, GNOVA corrects for sample overlap and provides greater statistical power as well as higher estimation accuracy. We ran LDSC and GNOVA on SNPs in both diseases together with reference data derived from the 1,000 Genomes Project European population. Genetic correlation and p values were corrected for sample overlap for GNOVA. A p value below 0.0025 (0.05/20) was considered statistically significant after the Bonferroni correction.
We investigated whether the genetic overlap between schizophrenia and each neurodegenerative disease was mediated by shared regulation of gene expression. We generated tissue-specific, disease-inferred gene expression profiles using TWAS software with default parameters (Gusev et al., 2016). TWAS integrates gene expression measurements with GWAS summary statistics to identify genes whose cis-regulated expression is associated with complex traits (Gusev et al., 2016). Briefly, TWAS leverages large-scale RNA-seq data to impute tissue-specific gene expression levels from GWAS summary statistics, which can further be used to identify novel associated genes. RNA-seq data from 48 tissues in the GTEx v7 reference panel (Battle et al., 2017) were used to generate all disease-inferred gene expression profiles. Summary statistics of all the SNPs from each GWAS were utilized. Furthermore, we estimated the overlap between the disease-inferred gene expression with RHOGE (Mancuso et al., 2017), using TWAS results with nominal association (p < 0.05). RHOGE estimates the genetic correlation between two traits which can be attributed to cis-expression quantitative trait loci (eQTL) as represented by different trait-inferred gene expression profiles. The major histocompatibility complex (MHC) region was excluded due to its complex LD structure. A p value below 0.0010 (0.05/48) was considered statistically significant after the Bonferroni correction.
We used UTMOST to infer the genes for schizophrenia and ALS underlying the overlapping disease-inferred gene expression across tissues (Hu et al., 2019). UTMOST combines multiple single-tissue associations into a powerful metric to quantify the overall gene-trait association and is suggested to achieve improved accuracy than single-tissue strategy. First, we generated the single-tissue expression model using GWAS summary statistics and matched expression data from all tissues in GTEx with default parameters. Then we performed a cross-tissue test for each gene to summarize single-tissue association statistics to quantify the overall gene-trait association. We identified transcriptome-wide significant gene-disease associations using a false discovery rate (FDR) threshold of 0.001.
To determine whether the identified risk genes were differentially expressed in tissues from patients with ALS or schizophrenia compared with healthy controls, we analyzed the gene expression of the target genes in whole blood from 233 ALS patients and 508 controls (GEO accession number GSE8397, GPL6947), and neurons from human induced pluripotent stem cell (hiPSC) in 12 schizophrenia patients and 12 controls (GEO accession number GSE25673). Data were prepared and analyzed using GEO2R (Barrett et al., 2013), and a p value below 0.05 was considered significant after adjusting for multiple testing.
To better understand how the genetic variants affected disease risk, we used LDSC-SEG to identify tissues or cell types likely to be relevant to the etiology of each disease. When applying to 53 tissues from the GTEx project, we detected significant enrichment in 13 brain-related tissues for schizophrenia, and nominal enrichment in brain-related tissues for PD (Figure 1). In contrast, there is less enrichment of SNP heritability near genes expressed in brain-related tissues for ALS, AD and FTD. Moreover, we applied LDSC-SEG to expression data from sorted primary mouse CNS cells (Cahoy et al., 2008). We found significant enrichment in neuron (p = 2.11E-05) for schizophrenia, as well as nominal enrichment in oligodendrocyte (p = 0.037) and neuron (p = 0.049) for PD (Supplementary Figure S1). Finally, considering that the immune dysfunction was closely related to the pathophysiology of schizophrenia (Müller and Schwarz, 2010), we applied LDSC-SEG on an atlas of 292 mouse immunological cell types (Heng and Painter, 2008) to assess the cell types where signal enrichment was specifically expressed. Though no significant association was identified, a marked enrichment in myeloid cell types was observed for ALS, PD, and AD, while schizophrenia was relatively enriched in T cells (Figure 1). Altogether, these results suggested that schizophrenia was associated with gene expression in brain-related tissues, neurons and T cells, while the other neurodegenerative disorders were less enriched in these tissues and cell types.
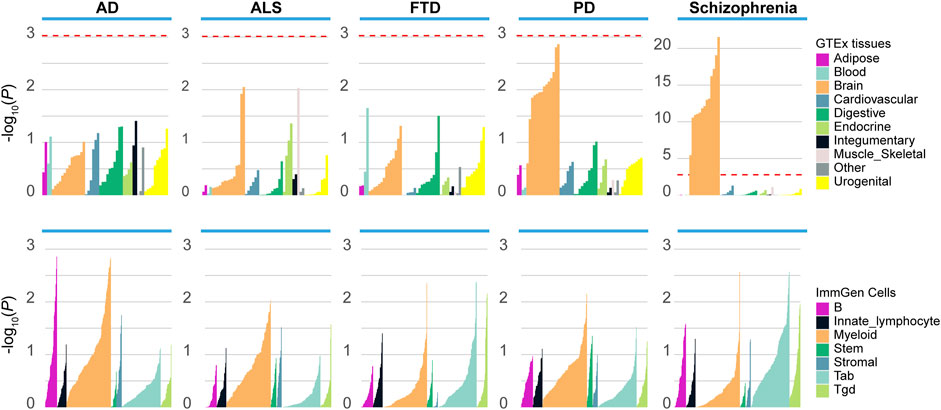
FIGURE 1. Enrichment of tissues and cell types in schizophrenia and neurodegenerative diseases. Top panel: Regions of the genome with specific expression in central nervous system (CNS) tissues are highly enriched for schizophrenia and PD, among 53 tissues obtained from the Genotype-Tissue expression project (GTEx). Bottom panel: LDSC-SEG analysis using 292 immune cells from the ImmGen Consortium. ALS, amyotrophic lateral sclerosis; PD, Parkinson’s disease; AD, Alzheimer’s disease; FTD, frontotemporal dementia. The red dashed line denotes the significance level after the Bonferroni correction.
We detected a significant and positive genetic correlation between ALS and schizophrenia using both LDSC (genetic correlation: 0.22; p = 4.00E-04) and GNOVA (genetic correlation 0.13, p = 6.83E-05) methods after the Bonferroni correction (Table 1). No significant genetic correlation was observed between the other diseases.
To investigate whether the genetic overlap between schizophrenia and ALS was mediated by shared regulation of gene expression, we generated tissue-specific gene expression profiles using TWAS, and further estimated the overlap between the different disease-inferred gene expression profiles using RHOGE. We identified a positive overlap between ALS- and schizophrenia-inferred gene expression profiles in a joint analysis of the GTEx v7 reference panel (gene expression correlation: 0.08; 95% CI: 0.04–0.11; p = 0.034). Analyzing the genetic correlation in each tissue separately, we observed positive and significant overlap in not-sun-exposed skin (gene expression correlation: 0.57, 95% CI: 0.48–0.67; p = 2.97E-08), small intestine tissues (gene expression correlation: 0.75; 95% CI: 0.61–0.89; p = 7.07E-05), lung (gene expression correlation: 0.38; 95% CI: 0.28–0.49; p = 6.29E-04) and brain cortex (gene expression correlation: 0.46; 95% CI: 0.33–0.58; p = 6.63E-04) (Figure 2; Table 2). The overall expression overlap was even more significant between schizophrenia and ALS when considering only these four tissues together (gene expression correlation: 0.39; 95% CI: 0.32–0.46; p = 2.42E-07). For the other three neurodegenerative disorders, only skeletal muscle was found with overlapped expression profiles between FTD and schizophrenia (Supplementary Figures S2–S4; Supplementary Table S2).
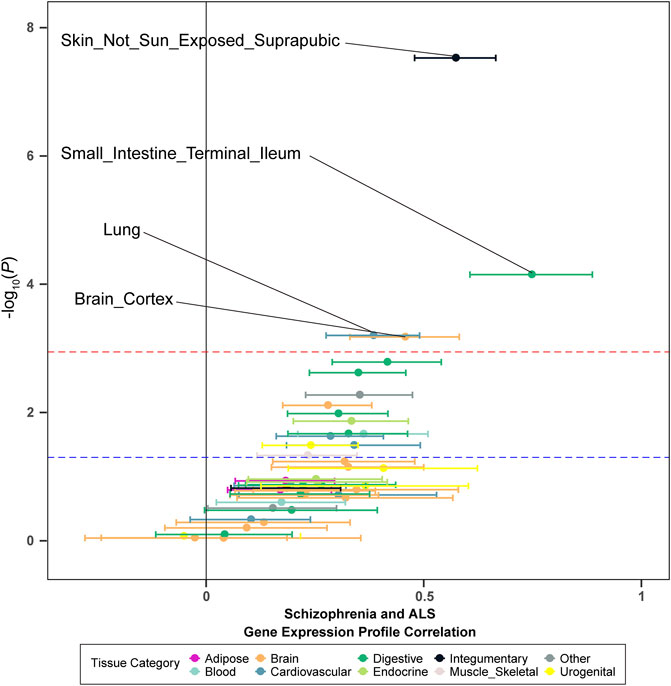
FIGURE 2. Tissue-specific disease-inferred gene expression profile correlation between schizophrenia and ALS. The red dashed line demarks the multiple-test corrected threshold (p = 0.05/48), while the blue dotted line demarks the nominal threshold (p = 0.05).

TABLE 2. Disease-inferred gene expression profile overlap between ALS and schizophrenia in GTEx v7 reference panel tissues.
To highlight the genes whose expression was commonly regulated by schizophrenia and ALS risk variants, we generated cross-tissue disease-associated genes using UTMOST. In the results of ALS, known ALS genes like SCFD1 (p = 2.35E-07), G2E3 (p = 5.26E-07), B4GALNT1 (p = 1.41E-06) and so on showed significant association. In the results of schizophrenia, known schizophrenia genes like WBP1L (p = 2.24E-14), TMTC1 (p = 3.75E-14), ZNF804A (p = 8.50E-14), TAOK2 (4.82E-12) and so on showed significant association. Combining the cross-tissue summary metric results, we identified 3 genes passing the FDR threshold for both schizophrenia and ALS, namely ZNHIT3, GLB1L3 and TMEM194A (Figure 3; Supplementary Table S3). We further analyzed the expression profiles of the three genes using available expression data from GEO. As a result, we found that ZNHIT3 (p = 1.20E-13) and TMEM194A (p = 5.12E-07) were differentially expressed between ALS patients and healthy controls in whole blood, and ZNHIT3 (p = 0.02) was differentially expressed in hiPSC-derived neurons between schizophrenia patients and controls.
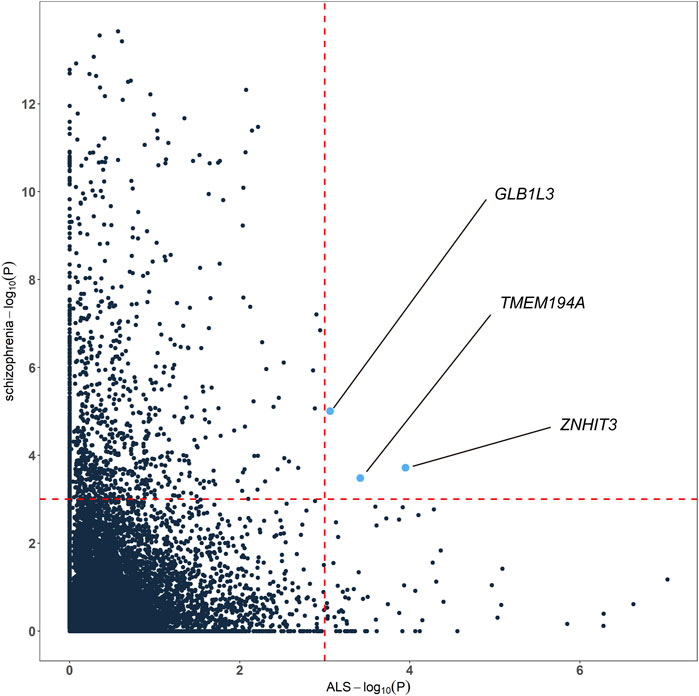
FIGURE 3. Genes associated with both schizophrenia and ALS cross tissues. The red dashed lines denotes the false discovery rate (FDR) threshold of 0.001.
Leveraging summary statistics from large-scale GWAS, public transcriptomic datasets and the polygenic modeling for complex traits, we investigated the overlapping genetic architecture between schizophrenia and four neurodegenerative disorders. We identified a significant genetic correlation between schizophrenia and ALS, and further located three genes with overlapped gene expression profiles across tissues. Our results provided evidence for shared genetic architecture between schizophrenia and ALS.
Both schizophrenia and neurodegenerative disorders are complex diseases affected by multiple genetic and environmental factors. Previous multi-variant genetic analyses investigating schizophrenia and ALS focused specifically on GWAS summary statistics and thus have missed a substantial proportion of the transcriptome architecture (McLaughlin et al., 2017). By combining GWAS summary statistics and large-scale transcriptome data, we observed a positive correlation between schizophrenia and ALS at both genetic and transcriptomic levels. We further identified overlapped expression profiles in specific tissues including not-sun-exposed skin, small intestine, brain cortex and lung, which were hard to detect using only genetic data. Meanwhile, we also noticed that schizophrenia and PD were both specifically expressed in brain-related tissues using LDSC-SEG, but no genome-wide genetic correlation was found, suggesting different genetic background between PD and schizophrenia. To support this hypothesis, we estimated local SNP heritability using ρ-HESS(Shi et al., 2017), which could estimate and visualize the local SNP heritability from GWAS summary statistics. We could see that the SNP heritability for PD and schizophrenia was enriched in different regions across the chromosomes, suggesting potential diverse genetic architecture (Supplementary Figure S5). In addition, PD symptoms could be alleviated with dopamine receptor agonists, whereas schizophrenia was commonly treated with dopamine receptor antagonists. And dopaminergic treatment used for PD was a particular risk factor for psychoses in PD (Ecker et al., 2009), suggesting differing underlying molecular mechanism.
Although both schizophrenia and ALS mainly affected the central nervous system, growing evidence has indicated that other tissues and organs might be involved in their pathogenesis as well. The brain cortex is primarily responsible for thinking, perceiving and motor activity, and has been shown to play important roles in neurodegenerative and neuropsychiatric disorders. It was noticed that both schizophrenia and ALS patients had a thinner cerebral cortex compared to healthy individuals on average (Walhout et al., 2015; van Erp et al., 2018). Meanwhile, A number ALS risk genes were differentially expressed in motor cortex of ALS patients compared with controls (Aronica et al., 2015). Clinically, both schizophrenia and ALS patients could present cognitive impairment correlated with pathologic and radiographic changes in the cerebral cortex (Woolley and Jonathan, 2008). In the developing embryo, both the brain and skin arise from the same ectodermal germ layer. Researchers have reported that protein aggregates found in the brains of patients with neurodegenerative disorders like ALS, PD and AD appeared in the dermal layers as well (Widmer et al., 2006). Meanwhile, ALS patients have been reported with dermatological conditions such as bullous pemphigoid and decreased collagen, and because of such changes, the skin has been proposed as a source of diagnostic markers for ALS (Watanabe et al., 1987; Kolde et al., 1996; Paré and Gros-Louis, 2017). In addition, increased expression of TDP-43 was found in skin of ALS patients compared with controls (Suzuki et al., 2010). Patients with schizophrenia showed increased rates of skin disorders as well, like psoriasis and atopic dermatitis (McPhie et al., 2020). Previous study found that some messenger RNA and microRNA were differentially expressed between schizophrenia patients and controls, and scalp hair follicles were also suggested as a beneficial genetic biomarker resource for schizophrenia (Maekawa et al., 2015). However, it remains unclear about the exact nature of the association between gene expression changes in skin and the two disorders. Further studies are warranted to elucidate the underlying pathways.
Emerging epidemiological evidence has suggested that intestinal homeostasis and the gut microbiome play essential roles in neurological diseases, and potential function of gut microbiome and metabolites were identified in the ALS model and ALS patients as well (Wu et al., 2015; Blacher et al., 2019; Zeng et al., 2020). Similarly, it was found that patients with schizophrenia had a decreased microbiome α-diversity index and marked disturbances of gut microbial composition versus healthy controls (Zheng et al., 2019). Consistent with these observations, our findings support the involvement of intestinal tissues in the pathogenesis of ALS and schizophrenia, though the mechanism of such relation is still unclear. Recent evidence suggested that the gut microbiota could modulate brain function via the “microbiota-gut-brain” axis (Hsiao et al., 2013; Dinan and Cryan, 2015), and it was also proposed that gastrointestinal function could impact the brain via immune system pathways (Severance et al., 2015). Future explorations are warranted to provide deeper understandings. Besides, we identified lung tissue with overlapped expression profiles. Although patients with schizophrenia or ALS both presented lung dysfunction and respiratory problems (Javad Mousavi et al., 2014), whether they were related to gene expression in lung tissue and how they were correlated has not been explained yet. Previous genetic enrichment analysis between schizophrenia and several cancers identified polygenic enrichment between schizophrenia and lung cancer, and functional analysis identified downstream pleiotropic effects on gene-expression in lung and brain tissues as well (Zuber et al., 2018). Further explorations are necessary to elucidate how lung tissues were involved in schizophrenia.
Beyond the four tissues identified with overlapped expression profiles, we also located three genes underlying the overlapped disease-inferred gene expression across tissues, including GLB1L3, ZNHIT3 and TMEM194A. GLB1L3 was widely expressed in the central nervous system. It was involved in several important metabolic processes such as carbohydrate metabolic process and beta-galactosidase activity, while carbohydrate metabolism has been implicated in both schizophrenia and ALS (Joardar et al., 2017; Bryll et al., 2020). In addition, it was indicated that copy number variants in the GLB1L3 were potentially associated with schizophrenia (Levinson et al., 2011). ZNHIT3 encodes a nuclear zinc finger protein which is implicated in transcriptional regulation and metal ion binding activity. Metal ions within the cellular environment were suggested to play a potential role in formation and deposition of toxic protein aggregates in ALS (Sirangelo and Iannuzzi, 2017), and metal ions might also influence the efficacy of treatment of schizophrenia with antipsychotics (Sussulini et al., 2018). In addition, recent summary statistic-based Mendelian randomization (SMR) analysis which combines disease-SNP association and expression-SNP association results identified ZNHIT3 as a risk gene for ALS (Benyamin et al., 2017; Du et al., 2018). Meanwhile, cellular knockdown of ZNHIT3 in mouse cerebellar granule cells sensitized the neurons to death, suggesting its potential role in neuron survival (Anttonen et al., 2017). TMEM194A is a novel inner nuclear membranes protein expressed across the tissues. We did not find direct evidence for the role of TMEM194A in ALS and schizophrenia. Further replication was necessary to explore its potential involvement in the pathogenesis of ALS and schizophrenia.
Our study has several strengthes and limitations worth noting. By combining GWAS summary statistics and large-scale transcriptomic datasets using a TWAS approach, we identified specific tissues with overlapped gene expression profiles between schizophrenia and ALS, as well as three risk genes underlying the correlation. Such method could characterize the functional effects of associated variants for diseases, and get results which cannot be obtained using only genetic data. Moreover, the current results have clinical implications. Since we combined GWAS summary statistics from different but related diseases, the findings may increase our understanding of the pathogenetic mechanisms influenced by pleiotropic genes and facilitate novel treatment strategies in clinical trials. However, the GWASs used in the current study were mostly performed on participants of European ancestry, thus the findings of shared genetic architecture might be biased and not applicable to other populations. Meanwhile, there was a potential sample overlap in the original GWAS. Such overlap might bring some bias to the statistical analysis.
In conclusion, by integrating GWAS summary data and large-scale transcriptomic data, we identified genetic correlation between schizophrenia and ALS. Moreover, we identified four tissues with overlapped expression profiles which might be involved in the pathogenesis of schizophrenia and ALS, and located three novel risk genes underlying the overlapping disease-inferred gene expression across tissues. These findings provided novel insights into the shared genetic background between schizophrenia and neurodegenerative disorders and might help better understand the etiology of schizophrenia.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
CL and HS conceived the study. CL performed the statistical analyses and prepared the drafted manuscript. TY and RO assisted in the statistical analysis. CL, TY, RO, and HS contributed to writing and editing of the manuscript. All authors reviewed and approved the final manuscript.
This research was supported by the funding of the National Natural Science Foundation of China (Grant No. 81901294), the National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University (Grant No. Z20192006), the Post-Doctor Research Project, West China Hospital, Sichuan University (Grant No. 18HXBH037), the Sichuan Science and Technology Program (Grant No. 2021YJ0415) and the Science Foundation of Chengdu Science and Technology Bureau (2019-YF05-00307-SN). The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.797072/full#supplementary-material
AD, Alzheimer’s disease; ALS, amyotrophic lateral sclerosis; eQTL, expression quantitative trait loci; FDR, false discovery rate; FTD, frontotemporal dementia; GWAS, genome-wide association study; LD, linkage disequilibrium; MHC, major histocompatibility complex; PD, Parkinson’s disease; SNP, single nucleotide polymorphism; TWAS, transcriptome-wide association study.
Abecasis, G. R., Auton, A., Brooks, L. D., DePristo, M. A., Durbin, R. M., Handsaker, R. E., et al. (2012). An Integrated Map of Genetic Variation from 1,092 Human Genomes. Nature 491 (7422), 56–65. doi:10.1038/nature11632
Anttonen, A.-K., Laari, A., Kousi, M., Yang, Y. J., Jääskeläinen, T., Somer, M., et al. (2017). ZNHIT3 Is Defective in PEHO Syndrome, a Severe Encephalopathy with Cerebellar Granule Neuron Loss. Brain 140 (5), 1267–1279. doi:10.1093/brain/awx040
Aronica, E., Baas, F., Iyer, A., ten Asbroek, A. L. M. A., Morello, G., and Cavallaro, S. (2015). Molecular Classification of Amyotrophic Lateral Sclerosis by Unsupervised Clustering of Gene Expression in Motor Cortex. Neurobiol. Dis. 74, 359–376. doi:10.1016/j.nbd.2014.12.002
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., et al. (2013). NCBI GEO: Archive for Functional Genomics Data Sets-Update. Nucleic Acids Res. 41 (Database issue), D991–D995. doi:10.1093/nar/gks1193
Battle, A., Brown, C. D., Engelhardt, B. E., Montgomery, S. B., Battle, A., Brown, C. D., et al. (2017). Genetic Effects on Gene Expression across Human Tissues. Nature 550 (7675), 204–213. doi:10.1038/nature24277
Benyamin, B., He, J., Zhao, Q., Gratten, J., Garton, F., Leo, P. J., et al. (2017). Cross-ethnic Meta-Analysis Identifies Association of the GPX3-TNIP1 Locus with Amyotrophic Lateral Sclerosis. Nat. Commun. 8 (1), 611. doi:10.1038/s41467-017-00471-1
Blacher, E., Bashiardes, S., Shapiro, H., Rothschild, D., Mor, U., Dori-Bachash, M., et al. (2019). Potential Roles of Gut Microbiome and Metabolites in Modulating ALS in Mice. Nature 572 (7770), 474–480. doi:10.1038/s41586-019-1443-5
Bryll, A., Skrzypek, J., Krzyściak, W., Szelągowska, M., Śmierciak, N., Kozicz, T., et al. (2020). Oxidative-Antioxidant Imbalance and Impaired Glucose Metabolism in Schizophrenia. Biomolecules 10 (3), 384. doi:10.3390/biom10030384
Bulik-Sullivan, B., Finucane, H. K., Finucane, H. K., Anttila, V., Gusev, A., Day, F. R., et al. (2015). An Atlas of Genetic Correlations across Human Diseases and Traits. Nat. Genet. 47 (11), 1236–1241. doi:10.1038/ng.3406
Cahoy, J. D., Emery, B., Kaushal, A., Foo, L. C., Zamanian, J. L., Christopherson, K. S., et al. (2008). A Transcriptome Database for Astrocytes, Neurons, and Oligodendrocytes: a New Resource for Understanding Brain Development and Function. J. Neurosci. 28 (1), 264–278. doi:10.1523/jneurosci.4178-07.2008
Cetin-Karayumak, S., Di Biase, M. A., Chunga, N., Reid, B., Somes, N., Lyall, A. E., et al. (2020). White Matter Abnormalities across the Lifespan of Schizophrenia: a Harmonized Multi-Site Diffusion MRI Study. Mol. Psychiatry 25 (12), 3208–3219. doi:10.1038/s41380-019-0509-y
Cooper, J. J., and Ovsiew, F. (2013). The Relationship between Schizophrenia and Frontotemporal Dementia. J. Geriatr. Psychiatry Neurol. 26 (3), 131–137. doi:10.1177/0891988713490992
Crayton, J. W., and Meltzer, H. Y. (1979). Degeneration and Regeneration of Motor Neurons in Psychotic Patients. Biol. Psychiatry 14 (5), 803–819.
Crayton, J. W., Meltzer, H. Y., and Goode, D. J. (1977). Motoneuron Excitability in Psychiatric Patients. Biol. Psychiatry 12 (4), 545–561.
Creese, B., Vassos, E., Vassos, E., Bergh, S., Athanasiu, L., Johar, I., et al. (2019). Examining the Association between Genetic Liability for Schizophrenia and Psychotic Symptoms in Alzheimer's Disease. Transl Psychiatry 9 (1), 273. doi:10.1038/s41398-019-0592-5
DeMichele-Sweet, M. A. A., Weamer, E. A., Klei, L., Vrana, D. T., Hollingshead, D. J., Seltman, H. J., et al. (2018). Genetic Risk for Schizophrenia and Psychosis in Alzheimer Disease. Mol. Psychiatry 23 (4), 963–972. doi:10.1038/mp.2017.81
Dinan, T. G., and Cryan, J. F. (2015). The Impact of Gut Microbiota on Brain and Behaviour. Curr. Opin. Clin. Nutr. Metab. Care 18 (6), 552–558. doi:10.1097/mco.0000000000000221
Du, Y., Wen, Y., Guo, X., Hao, J., Wang, W., He, A., et al. (2018). A Genome-wide Expression Association Analysis Identifies Genes and Pathways Associated with Amyotrophic Lateral Sclerosis. Cell Mol Neurobiol 38 (3), 635–639. doi:10.1007/s10571-017-0512-2
Ecker, D., Unrath, A., Kassubek, J., and Sabolek, M. (2009). Dopamine Agonists and Their Risk to Induce Psychotic Episodes in Parkinson's Disease: a Case-Control Study. BMC Neurol. 9, 23. doi:10.1186/1471-2377-9-23
Ferrari, R., Hernandez, D. G., Nalls, M. A., Rohrer, J. D., Ramasamy, A., Kwok, J. B., et al. (2014). Frontotemporal Dementia and its Subtypes: a Genome-wide Association Study. Lancet Neurol. 13 (7), 686–699. doi:10.1016/s1474-4422(14)70065-1
Finucane, H. K., Reshef, Y. A., Reshef, Y. A., Anttila, V., Slowikowski, K., Gusev, A., et al. (2018). Heritability Enrichment of Specifically Expressed Genes Identifies Disease-Relevant Tissues and Cell Types. Nat. Genet. 50 (4), 621–629. doi:10.1038/s41588-018-0081-4
Gadit, A. (2011). Schizophrenia and Parkinson's Disease: Challenges in Management. Case Rep. 2011, bcr1120115108. doi:10.1136/bcr.11.2011.5108
Gamazon, E. R., Segrè, A. V., Segrè, A. V., van de Bunt, M., Wen, X., Xi, H. S., et al. (2018). Using an Atlas of Gene Regulation across 44 Human Tissues to Inform Complex Disease- and Trait-Associated Variation. Nat. Genet. 50 (7), 956–967. doi:10.1038/s41588-018-0154-4
Gusev, A., Ko, A., Shi, H., Bhatia, G., Chung, W., Penninx, B. W. J. H., et al. (2016). Integrative Approaches for Large-Scale Transcriptome-wide Association Studies. Nat. Genet. 48 (3), 245–252. doi:10.1038/ng.3506
Habermeyer, B., Kneifel, S., Lotz-Bläuer, I., and Müller-Spahn, F. (2008). Psychosis in a Case of Schizophrenia and Parkinson's Disease. Jnp 20 (3), 373–375. doi:10.1176/jnp.2008.20.3.373
Heng, T. S. P., Painter, M. W., Painter, M. W., Elpek, K., Lukacs-Kornek, V., Mauermann, N., et al. (2008). The Immunological Genome Project: Networks of Gene Expression in Immune Cells. Nat. Immunol. 9 (10), 1091–1094. doi:10.1038/ni1008-1091
Hsiao, E. Y., McBride, S. W., Hsien, S., Sharon, G., Hyde, E. R., McCue, T., et al. (2013). Microbiota Modulate Behavioral and Physiological Abnormalities Associated with Neurodevelopmental Disorders. Cell 155 (7), 1451–1463. doi:10.1016/j.cell.2013.11.024
Hu, Y., Li, M., Li, M., Lu, Q., Weng, H., Wang, J., et al. (2019). A Statistical Framework for Cross-Tissue Transcriptome-wide Association Analysis. Nat. Genet. 51 (3), 568–576. doi:10.1038/s41588-019-0345-7
Javad Mousavi, S. A., Zamani, B., Shahabi Shahmiri, S., Rohani, M., Shahidi, G. A., Mostafapour, E., et al. (2014). Pulmonary Function Tests in Patients with Amyotrophic Lateral Sclerosis and the Association between These Tests and Survival. Iran J. Neurol. 13 (3), 131–137.
Joardar, A., Manzo, E., and Zarnescu, D. C. (2017). Metabolic Dysregulation in Amyotrophic Lateral Sclerosis: Challenges and Opportunities. Curr. Genet. Med. Rep. 5 (2), 108–114. doi:10.1007/s40142-017-0123-8
Kochunov, P., and Hong, L. E. (2014). Neurodevelopmental and Neurodegenerative Models of Schizophrenia: white Matter at the center Stage. Schizophrenia Bull. 40 (4), 721–728. doi:10.1093/schbul/sbu070
Kolde, G., Bachus, R., and Ludolph, A. C. (1996). Skin Involvement in Amyotrophic Lateral Sclerosis. The Lancet 347 (9010), 1226–1227. doi:10.1016/s0140-6736(96)90737-0
Kulhara, P., and Gupta, S. (2010). What Is Schizophrenia: A Neurodevelopmental or Neurodegenerative Disorder or a Combination of Both? A Critical Analysis. Indian J. Psychiatry 52 (1), 21–27. doi:10.4103/0019-5545.58891
Lam, M., Chen, C. Y., Chen, C.-Y., Li, Z., Martin, A. R., Bryois, J., et al. (2019). Comparative Genetic Architectures of Schizophrenia in East Asian and European Populations. Nat. Genet. 51 (12), 1670–1678. doi:10.1038/s41588-019-0512-x
Lambert, J. C., Ibrahim-Verbaas, C. A., Harold, D., Naj, A. C., Sims, R., Bellenguez, C., et al. (2013). Meta-analysis of 74,046 Individuals Identifies 11 New Susceptibility Loci for Alzheimer's Disease. Nat. Genet. 45 (12), 1452–1458. doi:10.1038/ng.2802
Levinson, D. F., Duan, J., Oh, S., Wang, K., Sanders, A. R., Shi, J., et al. (2011). Copy Number Variants in Schizophrenia: Confirmation of Five Previous Findings and New Evidence for 3q29 Microdeletions and VIPR2 Duplications. Ajp 168 (3), 302–316. doi:10.1176/appi.ajp.2010.10060876
Li, Y. I., Wong, G., Humphrey, J., and Raj, T. (2019). Prioritizing Parkinson's Disease Genes Using Population-Scale Transcriptomic Data. Nat. Commun. 10 (1), 994. doi:10.1038/s41467-019-08912-9
Lu, Q., Li, B., Ou, D., Erlendsdottir, M., Powles, R. L., Jiang, T., et al. (2017). A Powerful Approach to Estimating Annotation-Stratified Genetic Covariance via GWAS Summary Statistics. Am. J. Hum. Genet. 101 (6), 939–964. doi:10.1016/j.ajhg.2017.11.001
Maekawa, M., Yamada, K., Toyoshima, M., Ohnishi, T., Iwayama, Y., Shimamoto, C., et al. (2015). Utility of Scalp Hair Follicles as a Novel Source of Biomarker Genes for Psychiatric Illnesses. Biol. Psychiatry 78 (2), 116–125. doi:10.1016/j.biopsych.2014.07.025
Mancuso, N., Shi, H., Goddard, P., Kichaev, G., Gusev, A., and Pasaniuc, B. (2017). Integrating Gene Expression with Summary Association Statistics to Identify Genes Associated with 30 Complex Traits. Am. J. Hum. Genet. 100 (3), 473–487. doi:10.1016/j.ajhg.2017.01.031
McLaughlin, R. L., Schijven, D., Schijven, D., van Rheenen, W., van Eijk, K. R., O’Brien, M., et al. (2017). Genetic Correlation between Amyotrophic Lateral Sclerosis and Schizophrenia. Nat. Commun. 8, 14774. doi:10.1038/ncomms14774
McPhie, M. L., Bridgman, A. C., and Kirchhof, M. G. (2020). A Review of Skin Disease in Schizophrenia. Dermatology 237 (2), 248–261. doi:10.1159/000508868
Muller, N., and J. Schwarz, M. (2010). The Role of Immune System in Schizophrenia. Cir 6 (3), 213–220. doi:10.2174/157339510791823673
Nalls, M. A., Blauwendraat, C., Vallerga, C. L., Heilbron, K., Bandres-Ciga, S., Chang, D., et al. (2019). Identification of Novel Risk Loci, Causal Insights, and Heritable Risk for Parkinson's Disease: a Meta-Analysis of Genome-wide Association Studies. Lancet Neurol. 18 (12), 1091–1102. doi:10.1016/s1474-4422(19)30320-5
Nicolas, A., Kenna, K. P., Renton, A. E., Ticozzi, N., Faghri, F., Chia, R., et al. (2018). Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron 97 (6), 1268–e6. doi:10.1016/j.neuron.2018.02.027
Nucifora, L. G., MacDonald, M. L., Lee, B. J., Peters, M. E., Norris, A. L., Orsburn, B. C., et al. (2019). Increased Protein Insolubility in Brains from a Subset of Patients with Schizophrenia. Ajp 176 (9), 730–743. doi:10.1176/appi.ajp.2019.18070864
Owen, M. J., Sawa, A., and Mortensen, P. B. (2016). Schizophrenia. The Lancet 388 (10039), 86–97. doi:10.1016/s0140-6736(15)01121-6
Paré, B., and Gros-Louis, F. (2017). Potential Skin Involvement in ALS: Revisiting Charcot's Observation - a Review of Skin Abnormalities in ALS. Rev. Neurosci. 28 (5), 551–572. doi:10.1515/revneuro-2017-0004
Salamone, J. D., Yohn, S. E., López-Cruz, L., San Miguel, N., and Correa, M. (2016). Activational and Effort-Related Aspects of Motivation: Neural Mechanisms and Implications for Psychopathology. Brain 139 (Pt 5), 1325–1347. doi:10.1093/brain/aww050
Severance, E. G., Prandovszky, E., Castiglione, J., and Yolken, R. H. (2015). Gastroenterology Issues in Schizophrenia: Why the Gut Matters. Curr. Psychiatry Rep. 17 (5), 27. doi:10.1007/s11920-015-0574-0
Shi, H., Mancuso, N., Spendlove, S., and Pasaniuc, B. (2017). Local Genetic Correlation Gives Insights into the Shared Genetic Architecture of Complex Traits. Am. J. Hum. Genet. 101 (5), 737–751. doi:10.1016/j.ajhg.2017.09.022
Sirangelo, I., and Iannuzzi, C. (2017). The Role of Metal Binding in the Amyotrophic Lateral Sclerosis-Related Aggregation of Copper-Zinc Superoxide Dismutase. Molecules 22 (9), 1429. doi:10.3390/molecules22091429
Sussulini, A., Erbolato, H. M., Pessôa, G. d. S., Arruda, M. A. Z., Steiner, J., and Martins-de-Souza, D. (2018). Elemental Fingerprinting of Schizophrenia Patient Blood Plasma before and after Treatment with Antipsychotics. Eur. Arch. Psychiatry Clin. Neurosci. 268 (6), 565–570. doi:10.1007/s00406-017-0836-4
Suzuki, M., Mikami, H., Watanabe, T., Yamano, T., Yamazaki, T., Nomura, M., et al. (2010). Increased Expression of TDP-43 in the Skin of Amyotrophic Lateral Sclerosis. Acta Neurol. Scand. 122 (5), 367–372. doi:10.1111/j.1600-0404.2010.01321.x
van Erp, T. G. M., Walton, E., Hibar, D. P., Schmaal, L., Jiang, W., Glahn, D. C., et al. (2018). Cortical Brain Abnormalities in 4474 Individuals with Schizophrenia and 5098 Control Subjects via the Enhancing Neuro Imaging Genetics through Meta Analysis (ENIGMA) Consortium. Biol. Psychiatry 84 (9), 644–654. doi:10.1016/j.biopsych.2018.04.023
Walhout, R., Westeneng, H.-J., Verstraete, E., Hendrikse, J., Veldink, J. H., van den Heuvel, M. P., et al. (2015). Cortical Thickness in ALS: towards a Marker for Upper Motor Neuron Involvement. J. Neurol. Neurosurg. Psychiatry 86 (3), 288–294. doi:10.1136/jnnp-2013-306839
Watanabe, S., Yamada, K., Ono, S., and Ishibashi, Y. (1987). Skin Changes in Patients with Amyotrophic Lateral Sclerosis: Light and Electron Microscopic Observations. J. Am. Acad. Dermatol. 17 (6), 1006–1012. doi:10.1016/s0190-9622(87)70290-4
Widmer, R., Ziaja, I., and Grune, T. (2006). Protein Oxidation and Degradation during Aging: Role in Skin Aging and Neurodegeneration. Free Radic. Res. 40 (12), 1259–1268. doi:10.1080/10715760600911154
Woolley, S. C., and Jonathan, S. (2008). Cognitive and Behavioral Impairment in Amyotrophic Lateral Sclerosis. Phys. Med. Rehabil. Clin. North America 19 (3), 607–617. doi:10.1016/j.pmr.2008.04.002
Wu, S., Yi, J., Zhang, Y.-g., Zhou, J., and Sun, J. (2015). Leaky Intestine and Impaired Microbiome in an Amyotrophic Lateral Sclerosis Mouse Model. Physiol. Rep. 3 (4), e12356. doi:10.14814/phy2.12356
Zeng, Q., Shen, J., Chen, K., Zhou, J., Liao, Q., Lu, K., et al. (2020). The Alteration of Gut Microbiome and Metabolism in Amyotrophic Lateral Sclerosis Patients. Sci. Rep. 10 (1), 12998. doi:10.1038/s41598-020-69845-8
Zheng, P., Zeng, B., Liu, M., Chen, J., Pan, J., Han, Y., et al. (2019). The Gut Microbiome from Patients with Schizophrenia Modulates the Glutamate-Glutamine-GABA Cycle and Schizophrenia-Relevant Behaviors in Mice. Sci. Adv. 5 (2), eaau8317. doi:10.1126/sciadv.aau8317
Zuber, V., Jönsson, E. G., Frei, O., Witoelar, A., Thompson, W. K., Schork, A. J., et al. (2018). Identification of Shared Genetic Variants between Schizophrenia and Lung Cancer. Sci. Rep. 8 (1), 674. doi:10.1038/s41598-017-16481-4
Keywords: TWAS, genetic correlation, schizophrenia, neurodegeneration, expression profiles
Citation: Li C, Yang T, Ou R and Shang H (2021) Overlapping Genetic Architecture Between Schizophrenia and Neurodegenerative Disorders. Front. Cell Dev. Biol. 9:797072. doi: 10.3389/fcell.2021.797072
Received: 18 October 2021; Accepted: 06 December 2021;
Published: 24 December 2021.
Edited by:
Renjie Chai, Southeast University, ChinaReviewed by:
Johnathan Cooper-Knock, The University of Sheffield, United KingdomCopyright © 2021 Li, Yang, Ou and Shang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huifang Shang, aGZzaGFuZzIwMDJAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.