
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 20 December 2021
Sec. Signaling
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.794754
This article is part of the Research Topic DNA Repair Machinery in Innate and Adaptive Immunity View all 10 articles
As the most prevalent internal modification in mRNA, N6-methyladenosine (m6A) plays broad biological functions via fine-tuning gene expression at the post-transcription level. Such modifications are deposited by methyltransferases (i.e., m6A Writers), removed by demethylases (i.e., m6A Erasers), and recognized by m6A binding proteins (i.e., m6A Readers). The m6A decorations regulate the stability, splicing, translocation, and translation efficiency of mRNAs, and exert crucial effects on proliferation, differentiation, and immunologic functions of immunocytes, such as T lymphocyte, B lymphocyte, dendritic cell (DC), and macrophage. Recent studies have revealed the association of dysregulated m6A modification machinery with various types of diseases, including AIDS, cancer, autoimmune disease, and atherosclerosis. Given the crucial roles of m6A modification in activating immunocytes and promoting DNA repair in cells under physiological or pathological states, targeting dysregulated m6A machinery holds therapeutic potential in clinical application. Here, we summarize the biological functions of m6A machinery in immunocytes and the potential clinical applications via targeting m6A machinery.
While RNA modification was first identified in 1970s, it becomes a research focus in recent years. It broadly exists in different species, including fungi (Bodi et al., 2015), plants (Yue et al., 2019), and animals (Yoon et al., 2017; Xia et al., 2018). During the past decades, researchers have found that RNA methylation is a widespread modification in coding sequence and non-coding sequence (Huang et al., 2020), most of which are located at the amine group outside ring, special nitrogen and carbon positions of purine and pyrimidine, and the oxygen atom of the 2′-OH moiety (Liu and Jia, 2014). If classified by the modified position, RNA methylation mainly consists of N6-methyladenosine (m6A), 5-methylcytosine (m5C), N7-methylguanosine (m7G), etc., among which m7G cap at the 5′end of RNA sequence has been rigorously studied for decades (Devarkar et al., 2016; Pandolfini et al., 2019). However, m6A modification, representing the most abundant modification, needs further study.
As reported, m6A modifications are localized in the conversed DRACH motifs (D = G/A/U, R = G/A, H = A/U/C). The distribution of m6A is usually in the coding and 3′ untranslated regions, especially enriched in the upstream of stop codon in mRNA (Roundtree et al., 2017). Recent researches find that m6A modification is also an important biological mark of endogenous circular RNA (circRNA) (Chen et al., 2019). Moreover, m6A modification in lncRNA can regulate the efficiency of glycolysis (Liu J et al., 2019) or promote oncogenesis (Chen et al., 2020). Since m6A modification is dynamic and reversible, the biological function and molecular mechanism of m6A modification have become a research hotspot in many medical fields.
The most momentous breakthrough in this field is the discovery of the m6A machinery involved in m6A modification, including “Writers,” “Erasers” and “Readers,” performing the function of methyltransferase, demethylase, and recognizing the m6A structure, respectively. They dynamically regulate the homeostasis of m6A and its functions in cells.
With the function of forming m6A structure, “Writers” protein is a 1 MDa complex composed of multiple subunits, containing Methyltransferase like-3 (METTL3) (Liu et al., 2014), Methyltransferase like-14 (METTL14) (Weng et al., 2018), Wilm’s Tumor 1-associating protein (WTAP) (Ping et al., 2014; Sorci et al., 2018), etc. METTL3 is responsible for catalyzing the transfer of methyl group with the support of S-adenosyl-methionine (SAM) in many types of RNA including mRNA and miRNA, while METTL14 is a catalytic cofactor capable of recognizing and binding the target mRNA. WTAP is in charge of recruiting the targeting RNA and locating the METTL3/METTL14/WTAP complex into the nuclear speckles, which is relevant to the prognosis and cisplatin resistance (Ma et al., 2021) of cancer and the infiltration of T lymphocyte within tumors (Li H et al., 2020). New subunits, termed RBM15/RBM15B (Knuckles et al., 2018), KIAA1429 (Lan et al., 2019), ZFP217 (Song et al., 2019), and ZC3H3 (Silla et al., 2020), have been identified, and their functions involve in the recruitment, m6A modification of mRNA or lncRNA, and regulation of the m6A catalytic efficiency. Different types of “Writers” may interact with each other, as a result of which may influence the progression of some diseases such as colorectal cancer (Chen H et al., 2021).
The m6A structure can be erased by the “Erasers” protein. Fat mass and obesity-associated protein (FTO) (Jia et al., 2011) was supposed to be the first demethylase discovered, whose existence confirmed the reversibility of m6A modification. FTO and the second identified “Erasers” called AlkB Homolog 5 (ALKBH5) (Zheng et al., 2013) jointly counter the m6A modification of “Writers,” thus maintaining the homeostasis of m6A level in cells, whereas the distribution of the two proteins are tissue-specific. The amino acid sequence HXDXnH and RXXXXXR (X = any amino acid) with demethylase activity are contained in their mutual AlkB domain. Both of them remove the m6A methylation from mRNA with the Fe (II)/α-ketoglutarate-dependent dioxygenase (Fedeles et al., 2015). The demonstration of “Writers” and “Erasers” initiates a new branch, namely, m6A research, in the field of epigenetics. Recent studies on ALKBH5 gradually elucidate its multiple functions in disease progressing and therapeutic efficacy, including CD4+ T cell pathogenicity in autoimmunity (Zhou et al., 2021), anti PD-1 response in tumor treatment (Li N et al., 2020), glucocorticoid resistance in T-cell acute lymphoblastic leukemia cell treatment (Gong et al., 2021), etc.
The level of m6A in cells is dynamically modulated by “Writers” and “Erasers,” while “Readers” can recognize the m6A structure and regulate the subsequent cell processes such as translation and stability of mRNA. The YTH domain-containing family is the first confirmed component of “Readers,” characterized by the YTH domain at C terminus. YTHDF1∼3 and YTHDC1∼2 (Kasowitz et al., 2018; Zhou et al., 2020) are identified as m6A binding proteins, among which researches concerning YTHDF are more detailed. Generally speaking, the aforementioned m6A “Readers” proteins have the same function of binding the m6A-modified mRNA with the consensus YTH domain at C terminus, while YTHDF1 promote translation by binding the m6A at translation initiation site (Zhuang et al., 2019); YTHDF2, characterized by the P/Q/N-rich domain at N terminus, recruits the CCR4-NOT deadenylase complex and brings the target mRNA to cytoplasmic P bodies (Du et al., 2016), resulting in the destabilization of mRNA (Paris et al., 2019); YTHDF3 is also related to mRNA decay, but it is regarded to have a synergistic effect on YTHDF1 and YTHDF2 (Ni et al., 2019). In contrast to YTHDF, additional Readers such as insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) (Hanniford et al., 2020) can uniquely stabilize the target mRNA, while the eukaryotic initiation factor 3 (eIF3) (Meyer et al., 2015; Wolf et al., 2020) can promote cap-independent translation of mRNA with 5′-UTR m6A modified. Moreover, other m6A Readers like ELAVL1 (Zhang et al., 2017) are being studied recently.
Immunocytes play a crucial role in a variety of bioprocesses, such as recognizing and presenting the pathogen and immune response, whose depletion or dysfunction is the important pathological basis of tumorigenesis, viral infection, and autoimmune diseases, etc. Previous researches focused on the function of m6A in cancer cells, including endometrial cancer (Liu et al., 2018), breast cancer (Cai et al., 2018), bladder cancer (Cheng et al., 2019), hepatocellular cancer (Zhao X et al., 2018), nasopharyngeal cancer (Zhang et al., 2018), glioblastoma stem cells (Cui et al., 2017), acute myeloid leukemia (Cui et al., 2017), etc. Nevertheless, recent m6A researches on T lymphocyte, B lymphocyte, DC, and macrophage broaden our cognition towards the human immune system, which are the latest achievements of m6A modification.
T lymphocyte is the executant of human adaptive immune system, which is related to antitumor immunity and autoimmune diseases. Furthermore, T lymphocyte is likely to have interaction with neural stem cells, thus inhibiting its proliferation and resulting in age-related brain disease (Dulken et al., 2019); the abnormal level of m5C, another form of RNA methylation, in CD4+ T lymphocytes may have a potential link with the pathogenesis of systemic lupus erythematosus (SLE) (Guo G et al., 2020). According to the differences in function and phenotype, T lymphocyte can be divided into subtypes including naïve T cell (Tn), cytotoxic T cell, regulatory T cell (Treg), helper T cell (Th), etc., which is mainly driven by the stimulation of inflammation factor such as interleukin (IL) and tumor necrosis factor (TNF). Recently, bile acid has been proved to be an accessional regulator of Th17 (Hang et al., 2019) and Treg differentiation. The newly discovered subtype termed exhausted T cell (Tex) along with its key transcription factor called thymocyte selection-associated high mobility group box (TOX) (Scott et al., 2019) is a breakthrough in antitumor study, although the connection between Tex and m6A remains to be explored.
Studies focusing on m6A in T lymphocyte mark the initiation of m6A study in adaptive immune field. Li H-B et al. (2017) found that 5 weeks after transplant of wild type Tn, Rag−/− mice develop colitis due to the differentiation of Tn into effector T cell, whereas Rag−/− mice with Mettl3−/− transplanted exhibit no sign of similar symptoms and no T cell infiltration or inflammation inside spleen and colon can be observed. FACS shows the dysfunction of Mettl3−/− Tn differentiation. Molecular biology studies indicate that Soc1, Soc3, and Cish mRNA are stabilized owing to the lack of m6A modification; afterwards, elevated SOCS protein inhibits the phosphorylation of STAT5, then the IL-7 mediated JAK-STAT5 pathway will be blocked, and thus the differentiation of Tn is suffocated. However, because Mettl3−/− strengthen ERK and APK pathway simultaneously, no obvious increase in T cell apoptosis can be observed. Follow-up study (Tong et al., 2018) found that Tn homeostasis of Mettl3f/f; CD4-Cre mice can be destroyed and have colitis 3 months after being born, because Treg’s suppression of effector T cell is faulted. During this process, genetic depletion of Mettl3 reduces the m6A modification of Socs mRNA, then stabilizes mRNA, and upregulates its expression. High level SOCS protein inhibits IL-2-STAT5 pathway, resulting in the dysfunction of Treg. Additionally, it is demonstrated that Treg can strengthen type-II DC’s ability of presenting antigen (Binnewies et al., 2019), enhance antitumor response, and improve prognosis of checkpoint blockade such as PD-1 block immunotherapy (shown in Figure 1).
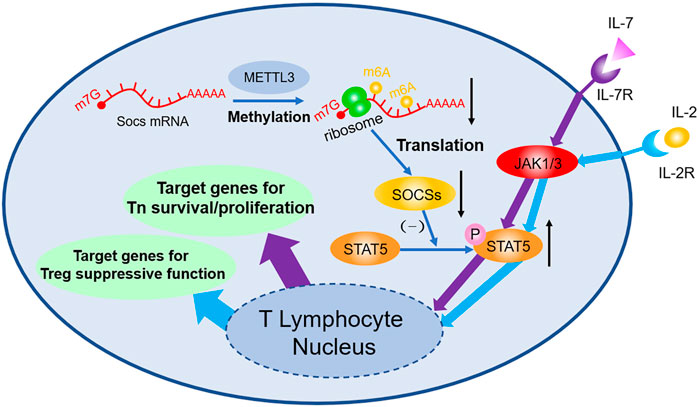
FIGURE 1. m6A decoration contributes to the regulation of Tn differentiation and Treg function both through JAK-STAT5 pathway. m6A modifications in Soc1, Soc3, and Cish mRNA accelerate their degradation, thus decreasing the expression of SOCS. As a consequence, through the JAK/STAT5 pathway, IL-7 induced differentiation of Tn and IL-2 induced suppressive function of Treg are both influenced afterwards.
Another study (Lu et al., 2020) unveils the function of Mettl14 in T lymphocytes. In this research, CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice have found to develop spontaneous colitis due to the increased level in Th1 cytokines, such as IFN-γ and TNF-α. Follow-up studies show that RORγt expression in Mettl14 deficient Tregs is downregulated compared to the wild-type Tregs and the induction efficacy of Mettl14 deficient Tn to iTreg is obviously impaired. As a consequence, both the reduction of iTregs, whose function has reported to be controlling the experimental colitis, and the dysfunctional Mettl14 deficient Tregs lead to the development of spontaneous colitis. With the function of METTL3 in T follicular helper cell differentiation being clarified recently (Yao et al., 2021), plenty of evidences have persuade us that m6A may play an irreplaceable role in all subtypes of T cells.
Furthermore, in acute myeloid leukemia (AML), FTO inhibition can lead to downregulation of leukocyte immunoglobulin-like receptor subfamily B member 4 (LILRB4), render AML cells vulnerable to activated T cells, and simultaneously overcome hypomethylating agent (HMA)-induced immune evasion. FTO’s function in anti-tumor immunity, especially its function in T cells, can be a promising therapeutic strategy in the field of m6A research (Su et al., 2020).
In addition, when CD4+ T cell is infected by the Human Immunodeficiency Virus (HIV), both HIV RNA and intrinsic RNA will be obviously increased (Lichinchi et al., 2016) independent of virus replication. Only gp120 will upregulate the level of m6A without influencing the expression of Writers and Erasers (Tirumuru and Wu, 2019). The m6A modification position of HIV RNA mainly distributes at 3′-UTR (Kennedy et al., 2016), which is directly involved in the regulation of viral mRNA nuclear export and protein synthesis. The m6A modification machinery, such as YTHDF (Tirumuru et al., 2016), METTL3/METTL14, and ALKBH5 (Lichinchi et al., 2016), act as a crucial regulator of this process. The m6A modification of HIV RNA will affect the expression of viral proteins including p55 (the product of gene gag) and Rev, consequently impacting the viral infectivity and replication, while suppressing the expression of IFN-I in monocytic cells and macrophages at the same time (Chen S et al., 2021). Therefore, antibodies neutralizing gp120 or CD4+ probably have the potential to counter HIV. Besides, Fu et al. (2019) for the first time applied HIV transgenic rats for m6A research and elucidated the role of m6A modification in mRNA in chronic HIV diseases, especially neurologic disorder (shown in Figure 2).
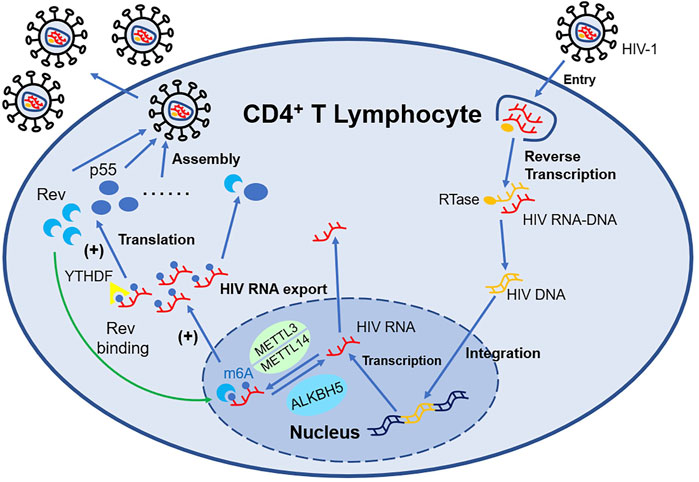
FIGURE 2. m6A-associated proteins in HIV-infected T lymphocytes regulate the expression of HIV mRNA. METTL3/METTL14 can install m6A decoration on HIV mRNA, while ALKBH5 can remove such modification. YTHDF1-3 recognize the m6A structure and influence the expression of HIV mRNA such as Rev and p55. With the support of Rev, HIV mRNA modified by m6A can export nucleus more easily, which promote the replication of HIV.
In conclusion, the effect of YTHDF2 on virus remains to be ascertained or can be bidirectional (Toro-Ascuy et al., 2016; Lu W et al., 2018); YTHDF3 weakens the viral infectivity and inhibits the viral replication. Uniquely, YTHDF3 can be incorporated into the virion and still keep its antiviral activation, but the HIV protease can cleave the virion containing YTHDF3. This mechanism prevents HIV from being killed thoroughly and provides a new thought for HIV treatment (Jurczyszak et al., 2020).
All the researches above reveal that m6A along with associated protein can alter the stability of mRNA and regulate nuclear export and translation of mRNA, thus influencing the bioprocess of T cell with different phenotype and promoting the progression of certain diseases.
B lymphocyte is involved in the humoral immunity by producing antibodies. Applying bioengineering technology to design special improbable immunogen can induce the synthesis of antibodies with high affinity, which have the potential to treat virus infection, such as HIV (Saunders et al., 2019). A recent clinical study suggests that m6A modification is closely related to the oncogenesis and progress of mantle cell lymphoma (Zhang W et al., 2019). Mantle cell lymphocyte is a kind of non-hodgkin B cell lymphoma characterized by aggressive phenotype and rapid rate of progression. After analysis of 123 samples of clinical patients, the hazard ratios of YTHDF3, METTL3, FTO, METTL14, ALKBH5, YTHDF2, and WTAP are below 1, while those of YTHDF1, KIAA1429, and ELAVL1 are above 1, among which the maximum is ELAVL while the minimum is YTHDF3, implying that ELAVL and YTHDF3 might be the most important regulators of mantle cell lymphoma. Moreover, “m6A index” is proposed to evaluate the prognosis of patients. Without much available biology research data, this statistical study directs a path for the following m6A research concerning B lymphocyte.
Recent studies have shown that the deletion of Mettl14 can decrease the m6A level in developing B cells and inhibit some important processes, such as the IL-7-induced Pro-B cell proliferation and the transition to the Large Pre-B Stage, which depends on the function of YTHDF2 (Zheng et al., 2020) (shown in Figure 3). However, the Large-Pre-B-to-Small-Pre-B Transition depends on METTL14, but is independent of YTHDF1 or YTHDF2 (shown in Figure 3).
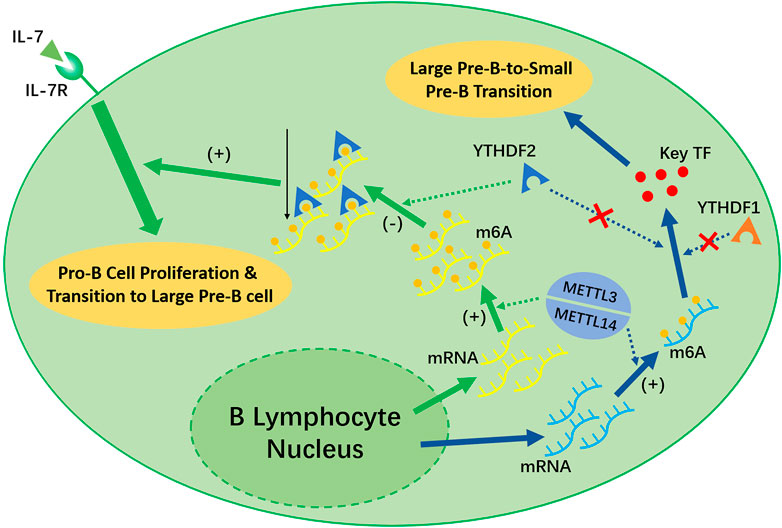
FIGURE 3. METTL3/METTL14 complex and YTHDF1 is necessary for the early development of B lymphocytes. METTL14 plays an unreplaceable role in the IL-7 induced Pro-B lymphocyte proliferation, Pro-B-to-Large-Pre-B transition, and Large-Pre-B-to-Small-Pre-B transition. YTHDF2 will recognize the m6A modification afterwards and decrease the transcripts as a result, which promote early B lymphocyte development. Notably, YTHDF2 only facilitate in the first two process. The Large-Pre-B-to Small-Pre-B Transition is independent of YTHDF1 or YTHDF2.
The pathogenesis of diffuse large B-cell lymphoma (DLBCL), which is the most common subtype of lymphoma derived from B lymphocytes, has been illustrated to have a link with upregulated METTL3 and the m6A level of the mRNA of pigment epithelium-derived factor (Cheng et al., 2020), while PIWI-interacting RNAs have been identified to function in this process recently (Han et al., 2021). The association between B lymphocytes and other m6A methylation-related proteins remains to be explored.
Besides, Kaposi’s sarcoma (KS) is evidently associated with infection of HIV and Kaposi’s sarcoma-associated herpesvirus (KSHV). KSHV shows strong lymphotropic and invades B cells in the circulation (Myoung and Ganem, 2011). During this process, m6A Reader protein YTHDF2 plays a positive role in KSHV replication (Hesser et al., 2018). However, inconsistent with its feature in vivo, KSHV exhibits weaken infectivity and proliferation in B cells lines in vitro, so the role of m6A and associated protein in the oncogenesis of KS remains to be explored.
As the bridge between the innate and adaptive immune, DCs function as antigen-presenting cells and can also produce VEGF-α for the recruitment of neutrophil to control cutaneous bacterial infections (Janela et al., 2019). Therefore, DCs play a core role in the eradication of pathogen and inducement of immune tolerance. It has been evidenced that the dysfunction of DC activation is involved in the progression of multiple inflammation, cancer, and autoimmune diseases. Tyrosine kinase AXL can induce the expression of PD-1. IFN-γ and IL-4 can respectively promote and inhibit the production of IL-12; thus, blockade of IL-4 receptor can strengthen antitumor response by expanding the infiltration of T lymphocyte at tumor position (Maier et al., 2020). Though the association between the suppression of DCs and extracellular m6A-modified RNA has been confirmed for decades (Karikó et al., 2005), studies concerning m6A in DCs are still at its infant stage.
Wang H et al. (2019) found that total level of m6A in DCs is increasing parallel with its maturation. The distribution of m6A is mainly located in NLR, TNF, and NF-κB pathways, which are responsible for the induction of co-stimulatory factors and pro-inflammation factors which promote maturation of DCs. METTL3 was involved in this physiological process. Distinct from most of the previous laboratory findings, the fundamental mechanism is the upregulation of translation efficiency, but not the stability of mRNA (Wang H et al., 2019) (shown in Figure 4).
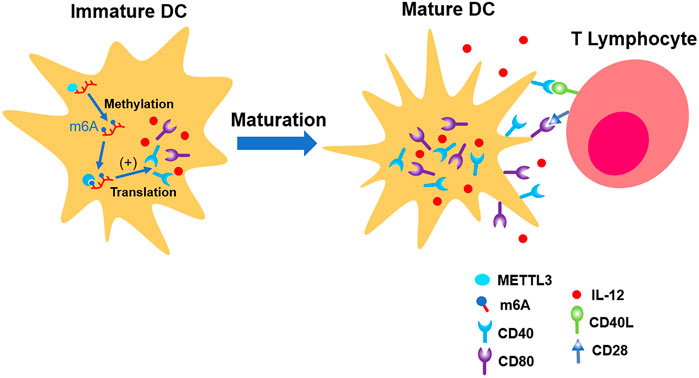
FIGURE 4. METTL3 up-regulates the expression of CD40, CD80, and IL-12, which promote the maturation of immature DC. In the end, its ability of presenting antigen and interaction with T lymphocytes are strengthened.
Han et al. (2019) discovered the connection between upregulated translation and YTHDF1. The depletion of YTHDF1 in DCs will limit the expression of lysosomal protease, which decelerates the degradation of antigen, thus improving DCs’ ability of presenting antigen and activating CD8+ T cell. This suggests a new mechanism of immune escape as well as an important reason for weak antitumor immune response in certain clinical cases (Han et al., 2019) (shown in Figure 5).
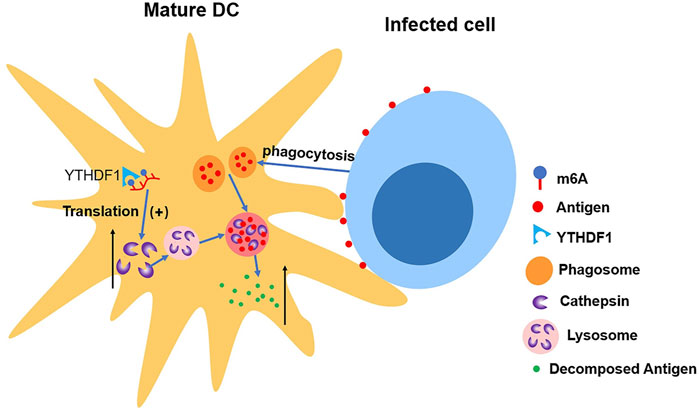
FIGURE 5. YTHDF1 increases the level of cathepsin in mature DCs, which accelerates the cleavage of antigens in phagosome, making DCs’ ability of presenting antigen impaired.
Macrophage is a kind of innate immune cell dwelling in various tissues with multiple subtypes, which can be classified into classically activated macrophages (M1) and alternatively activated macrophages (M2). Early researches have implied the possibility of m6A weakening immune response (Durbin et al., 2016). However, recent researches indicate that m6A modification plays an important role in antivirus, negative feedback control of macrophage activation (Du et al., 2020), and the polarization of macrophages. Roles of m6A Writers, Erasers, and Readers in macrophages are summarized in Table 1.
In detail, “Writers” METTL3 catalyzes the m6A methylation at coding sequence (CDS) and 3′-UTR of STAT1 mRNA, facilitating the polarization of M1, but having opposite impact on M2 (Liu Y et al., 2019). Another study has also illustrated METTL3’s function in promoting M1 differentiation, which afterwards benefits bone marrow mesenchymal stem cells (Lei et al., 2021). Moreover, METTL3 can methylate hnRNPA2B1 and Cgas, Ifi16, and Sting mRNA simultaneously, and then the affinity of hnRNPA2B1 to three mRNA above is improved, which ultimately increases the production of IFN-β and amplifies the immune response to DNA virus (Wang L et al., 2019). In addition, METTL3 deficiency have proved to impede the activation of macrophages through TLR4 signaling pathway by stabilizing Irakm transcripts (Tong et al., 2021). As for METTL14, there is also indirect evidence indicating its function in suppressing CD8+ T cell dysfunction and tumor growth (Dong et al., 2021).
After knockout of “Erasers” FTO, the polarization of both M1 and M2 can be inhibited, during which the expression of STAT1 in M1 was decreased, while the degradation of STAT6 and PPAR-γ mRNA is increased (Gu et al., 2020). Furthermore, ALKBH5 demethylate DDX46-binded Mavs, Traf3, and Traf6 mRNAs, leading to the retention of these mRNAs inside nuclear, indirectly inhibiting translation and reducing the production of IFN. In the end, antiviral activation of macrophages is weakened (Zheng et al., 2017).
Knockdown of “Readers” YTHDF2 can stimulate the expression of STAT1 (Huangfu et al., 2020) and PPAR-γ mRNA (Gu et al., 2020), while forced expression of YTHDF2 could destabilize MAP2K4 and MAP4K4 mRNA and activate the NF-κB and MAPK pathway (Yu et al., 2019), which will facilitate LPS-induced osteoclastogenesis and inflammatory response (Fang et al., 2021). YTHDF3 was regarded as a negative regulator of antivirus. With the assistance of PABP1 and elF4G2, YTHDF3 can bind the translation initiation site of FOXO3 mRNA and promote translation. As a result, the expression of IFN-stimulated genes is inhibited, which weakens the immunity response to Vesicular stomatitis virus (VSV) (Zhang Y et al., 2019). Moreover, another reader IGF2BP2 has also proved to be associated with the phenotypic activation of macrophage (Wang X et al., 2021).
Researches focusing on m6A modification in macrophages reveal its association with some diseases. The occurrence of atherosclerosis (AS) has proved to be linked with m6A modification. During this progress, ox-LDL induces the expression of DDX5 in macrophages and limits the function of METTL3 which transfers the methyl group to macrophages scavenger receptor A (MSR1) mRNA. Ultimately, MSR1 mRNA is stabilized, and more MSR1 is synthesized. Uptake of more lipids further facilitates the formation of foam cells, resulting in the progression of AS (Zhao W et al., 2018). In another study, Zhang X et al. (2021) has also identified the function of METTL3 in promoting ox-LDL-induced inflammation and mitochondrial dysfunction by methylating peroxisome proliferator-activated receptor-γ coactivator 1-alpha (PGC-1α) mRNA with the assistance of YTHDF2. Intriguingly, m5C, another form of RNA methylation, can deteriorate AS induced by hyper-homocysteinemia (Wang et al., 2017), while acute coronary syndrome, whose main pathological basis is AS, is also related to the m6A modification of circ-0029589 in macrophage (Guo M et al., 2020).
The dysfunction of m6A in macrophages is also a pathogenesis factor of autoimmune diseases. Wang J et al. (2019) found that in patients with rheumatoid arthritis, METTL3 in macrophages is obviously improved and positively associated with CRP and ESR. Moreover, lipopolysaccharide (LPS) can stimulate the expression of METTL3 in macrophages and then slack the immune response to inflammation through NF-κB pathway (Wang J et al., 2019). Other autoimmune diseases such as osteoarthritis (Liu Q et al., 2019) and SLE (Li et al., 2018) show possibility of having connection with m6A dysfunction in macrophages.
Given the identification of aberrant m6A modification in various diseases, targeting m6A machinery in specific cells can be regarded as a new treatment for viral infection, cancer, and autoimmune diseases. However, changes in m6A levels in different diseases are lack of consistency, so treatment targeting m6A should be supposed to modulating m6A level to normal level, instead of simply accelerating or decelerating m6A modification (Wang et al., 2018).
Plenty of researches have confirmed the correlation between m6A and DNA repair in different situations. Zhang et al. have reported the METTL3-m6A-YTHDC1 axis which promotes the double-strand breaks by modulating DNA-RNA hybrid accumulation (Zhang et al., 2020). Other researches indicate that accumulation of the three-stranded R-loops, formed by RNA: DNA hybrid and single stranded DNA, is regulated by YTHDF2 (Abakir et al., 2020), METTL3, and tonicity-responsive enhancer binding protein (TonEBP) (Kang et al., 2021). More surprisingly, the arginine substrates of METTL14 itself at intrinsically disordered C terminus can also be methylated. It proved to be an initial signal of interaction between METTL14 and RNA polymerase II, which will afterwards implement the m6A modification of target mRNA. Subsequent studies have confirmed that METTL14 arginine methylation is associated with the enhanced translation of DNA repair genes (Wang Z et al., 2021). Recent studies even clarified the correlation between DNA damage repair and m6A-modified retrotransposable element (RTE) RNAs, in which intronic Long Interspersed Element-1 (LINE-1) interacts with the hosting gene transcription, resulting in the downregulation of its expression (Xiong et al., 2021). KIAA1429 was also recently discovered to have close links with the modulation of response to cisplatin in germ cell tumor by interfering with DNA damage response (Miranda-Gonçalves et al., 2021).
As for Erasers, the homologs of AlkB (ALKBHs), which originally function as repair proteins in E. coli, are also important regulators of DNA repair and m6A modification at the same time in mammalian cells (Müller et al., 2017; Müller et al., 2018). Recently, the ERK/JNK/ALKBH5-PTMs/m6A axis has been reported to participate in the regulation of ROS-induced DNA damage response, in which progress IGF2BP also plays a part in extending mRNA half time (Yu et al., 2021). METTL3 and FTO can jointly regulate the m6A modification in RNA at DNA damage sites induced by ultraviolet. The function of DNA polymerase κ (Pol κ), which is the key DNA repair enzyme, require the catalytic activity of METTL3, implying the m6A modification directs the recruitment of Pol κ to the DNA damage sites (Xiang et al., 2017). Furthermore, METTL14 has proved to play a tumor-suppressive role in ultraviolet-induced skin tumorigenesis (Yang et al., 2021). All the m6A-associated proteins which regulate DNA repair are summarized in Table 2.
In conclusion, since m6A, along with its associated proteins, plays an important role in the pathogenesis and facilitating DNA repair, attaching m6A-targeted therapy to traditional chemo- or radiotherapy may improve the prognosis of some diseases such as carcinoma through two mechanisms.
Abnormally elevated m6A level is the feature of most malignant tumor, so developing new drugs inhibiting m6A modification is the most fundamental idea to treat these diseases. Actually, this kind of drugs has been developed for decades. 3-Deaza-Adenosine (DAA) with its analogue can block S-adenosylhomocysteine (SAH) hydrolase, which results in the accumulation of SAH and feedback suppression of SAM (Chiang, 1998). So DAA can indirectly inhibit the m6A modification of mRNA. However, since DAA can suppress the m6A modification in many physiological or pathological processes, there will be many unexpected side effects, such as the prevention of T lymphocyte activation, hypotensive effect, and activation of gene expression. At present, DAA is mainly used for the treatment of AIDS (Kennedy et al., 2016).
Moreover, some diseases like acute myeloid leukemia (AML) are characterized by the aberrant decrease of m6A (Li Z et al., 2017). Rhein (Chen et al., 2012), curcumin (Lu N et al., 2018), meclofenamic acid (Huang et al., 2015), and Saikosaponin-D (Sun et al., 2021) can inhibit the function of FTO by binding the active site of FTO or m6A position of mRNA. Two emerging small molecules targeting FTO demethylase called FB32 and FB32-2, which can dramatically inhibit the progression of AML cells in vitro and in vivo, have been developed recently (Huang et al., 2019). Coupled with the latest advancement called STM2457, which is a highly potent and selective first-in-class catalytic inhibitor of METTL3 (Yankova et al., 2021), we can foresee that the era of treating AML or other diseases featured by m6A level unbalance using m6A-targeted method is coming.
Although solid researches data is unavailable, the regulatory function of m6A modification to immunocytes makes it possible for m6A to be a new target for immunotherapy. It has been demonstrated that m6A is a regulator of Tn differentiation, T lymphocyte homeostasis (Li H-B et al., 2017), and suppressive function of Tregs (Tong et al., 2018). Recent studies have found that loss of METTL3 in myeloid cells reprograms the macrophages and increases Treg infiltration into tumors by influencing the YTHDF1-mediated translation of SPRED2 (Yin et al., 2021). Therefore, inhibiting mRNA m6A modification of Tregs at tumor site or myeloid cells can motivate the antitumor activation of CD8+ T cell, which can be a promising immunotherapy. Meanwhile, inhibiting mRNA m6A modification in Tn can reduce the formation of effector T cell, which is helpful for the treatment for autoimmune diseases. Furthermore, regulating the expression of METTL3 (Wang H et al., 2019) and YTHDF1 to a suitable level can improve DCs’ ability of presenting tumor neoantigen. More importantly, depletion of YTHDF1 and blockade of checkpoint have a synergistic effect on strengthening antitumor immunity (Han et al., 2019), and ALKBH5 (Li N et al., 2020) and METTL3/14 (Wang et al., 2020) have also proved to regulate anti PD-1 response. So as for patients resistant to the PD-1 immunotherapy, targeting m6A can be a new alternative treatment. Since m6A decoration of viral double-stranded RNA can also downregulate the innate sensing pathway of antiviral response (Qiu et al., 2021), immunotherapy targeting m6A is bound to be a promising therapy for various diseases including viral infections, autoimmune disorders, cancers, etc.
As the most abundant post-transcriptional mRNA modification in mammals, m6A is involved in the occurrence of several diseases. Recently, an enormous amount of m6A related studies in immunocytes highlight the fact that targeting m6A can be a promising new treatment strategy for viral infection, cancer, and autoimmune diseases. However, in different cell lines, diseases, even different types of the same diseases, the changes of m6A level, as well as functions of three m6A-associated enzymes lack consistency. Moreover, m6A modification is widely involved in a variety of cellular processes and medication targeting m6A is not selective. All these reasons indicate that clinical treatment via targeting m6A modification may be not safe enough. Thus, there is unmet need to develop more sophisticated techniques for m6A detection (Dai et al., 2018; Castellanos-Rubio et al., 2019). Moreover, detailed studies on disease mechanisms are required to realize the clinical application of m6A-targeting treatment. Pharmaceutical researches on drugs with high selectivity or combination of existing drugs and targeted drug delivery system can also promote the accurate treatment of diseases through the m6A-targeting method. Some recent experimental results have indicated the promising prospects of this field (Zhu et al., 2021). Last but not least, m6A modification is supposed to be highly relevant to gut microbiota (Jabs et al., 2020), heat shock proteins (Feng et al., 2020), sepsis (Sun et al., 2020; Xing et al., 2021), and pulmonary hypertension (Pan et al., 2020) and even peripheral nerve injury (Zhang L et al., 2021). These studies can provide valuable experimental basis for development of new treatments.
WL and NS contributed to the design of the topic. MZ and JZ searched and organized the relevant papers. MZ and JZ wrote the first draft of the manuscript. MZ drew the figures and tables in the manuscript. WL and NS contributed to the polish of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by Medical Research Project of Health Commission of Heibei Province. Development of a novel HER2-targeting CAR-T and its efficacy and safety in solid tumors (No. 20200037). Natural Science Foundation of Hebei Province. GM-CSF rich exosomes from local X-ray irradiated breast tumors mediate lung metastasis by enhancing MDSC recruitment (No. H2020206424).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abakir, A., Giles, T. C., Cristini, A., Foster, J. M., Dai, N., Starczak, M., et al. (2020). N6-methyladenosine Regulates the Stability of RNA:DNA Hybrids in Human Cells. Nat. Genet. 52 (1), 48–55. doi:10.1038/s41588-019-0549-x
Binnewies, M., Mujal, A. M., Pollack, J. L., Combes, A. J., Hardison, E. A., Barry, K. C., et al. (2019). Unleashing Type-2 Dendritic Cells to Drive Protective Antitumor CD4+ T Cell Immunity. Cell 177 (3), 556–571. doi:10.1016/j.cell.2019.02.005
Bodi, Z., Bottley, A., Archer, N., May, S. T., and Fray, R. G. (2015). Yeast m6A Methylated mRNAs Are Enriched on Translating Ribosomes during Meiosis, and under Rapamycin Treatment. PLoS One 10, e0132090. doi:10.1371/journal.pone.0132090
Cai, X., Wang, X., Cao, C., Gao, Y., Zhang, S., Yang, Z., et al. (2018). HBXIP-elevated Methyltransferase METTL3 Promotes the Progression of Breast Cancer via Inhibiting Tumor Suppressor Let-7g. Cancer Lett. 415, 11–19. doi:10.1016/j.canlet.2017.11.018
Castellanos-Rubio, A., Santin, I., Olazagoitia-Garmendia, A., Romero-Garmendia, I., Jauregi-Miguel, A., Legarda, M., et al. (2019). A Novel RT-QPCR-Based Assay for the Relative Quantification of Residue Specific m6A RNA Methylation. Sci. Rep. 9 (1), 4220. doi:10.1038/s41598-019-40018-6
Chen, B., Ye, F., Yu, L., Jia, G., Huang, X., Zhang, X., et al. (2012). Development of Cell-Active N6-Methyladenosine RNA Demethylase FTO Inhibitor. J. Am. Chem. Soc. 134 (43), 17963–17971. doi:10.1021/ja3064149
Chen, H., Yao, J., Bao, R., Dong, Y., Zhang, T., Du, Y., et al. (2021). Cross-talk of Four Types of RNA Modification Writers Defines Tumor Microenvironment and Pharmacogenomic Landscape in Colorectal Cancer. Mol. Cancer 20 (1), 29. doi:10.1186/s12943-021-01322-w
Chen, S., Kumar, S., Espada, C. E., Tirumuru, N., Cahill, M. P., Hu, L., et al. (2021). N6-methyladenosine Modification of HIV-1 RNA Suppresses Type-I Interferon Induction in Differentiated Monocytic Cells and Primary Macrophages. Plos Pathog. 17, e1009421. doi:10.1371/journal.ppat.1009421
Chen, S., Zhou, L., and Wang, Y. (2020). ALKBH5-mediated m6A Demethylation of lncRNA PVT1 Plays an Oncogenic Role in Osteosarcoma. Cancer Cel Int 20, 34. doi:10.1186/s12935-020-1105-6
Chen, Y. G., Chen, R., Ahmad, S., Verma, R., Kasturi, S. P., Amaya, L., et al. (2019). N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol. Cel 76 (1), 96–109. doi:10.1016/j.molcel.2019.07.016
Cheng, M., Sheng, L., Gao, Q., Xiong, Q., Zhang, H., Wu, M., et al. (2019). The m6A Methyltransferase METTL3 Promotes Bladder Cancer Progression via AFF4/NF-Κb/MYC Signaling Network. Oncogene 38 (19), 3667–3680. doi:10.1038/s41388-019-0683-z
Cheng, Y., Fu, Y., Wang, Y., and Wang, J. (2020). The m6A Methyltransferase METTL3 Is Functionally Implicated in DLBCL Development by Regulating m6A Modification in PEDF. Front. Genet. 11, 955. doi:10.3389/fgene.2020.00955
Chiang, P. K. (1998). Biological Effects of Inhibitors of S-Adenosylhomocysteine Hydrolase. Pharmacol. Ther. 77 (2), 115–134. doi:10.1016/s0163-7258(97)00089-2
Cui, Q., Shi, H., Ye, P., Li, L., Qu, Q., Sun, G., et al. (2017). m 6 A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cel Rep. 18 (11), 2622–2634. doi:10.1016/j.celrep.2017.02.059
Dai, T., Pu, Q., Guo, Y., Zuo, C., Bai, S., Yang, Y., et al. (2018). Analogous Modified DNA Probe and Immune Competition Method-Based Electrochemical Biosensor for RNA Modification. Biosens. Bioelectron. 114, 72–77. doi:10.1016/j.bios.2018.05.018
Devarkar, S. C., Wang, C., Miller, M. T., Ramanathan, A., Jiang, F., Khan, A. G., et al. (2016). Structural Basis for m7G Recognition and 2′-O-Methyl Discrimination in Capped RNAs by the Innate Immune Receptor RIG-I. Proc. Natl. Acad. Sci. USA 113 (3), 596–601. doi:10.1073/pnas.1515152113
Dong, L., Chen, C., Zhang, Y., Guo, P., Wang, Z., Li, J., et al. (2021). The Loss of RNA N6-Adenosine Methyltransferase Mettl14 in Tumor-Associated Macrophages Promotes CD8+ T Cell Dysfunction and Tumor Growth. Cancer Cell 39 (7), 945–957. doi:10.1016/j.ccell.2021.04.016
Du, H., Zhao, Y., He, J., Zhang, Y., Xi, H., Liu, M., et al. (2016). YTHDF2 Destabilizes m6A-Containing RNA through Direct Recruitment of the CCR4-Not Deadenylase Complex. Nat. Commun. 7, 12626. doi:10.1038/ncomms12626
Du, J., Liao, W., Liu, W., Deb, D. K., He, L., Hsu, P. J., et al. (2020). N6-Adenosine Methylation of Socs1 mRNA Is Required to Sustain the Negative Feedback Control of Macrophage Activation. Developmental Cel 55 (6), 737–753. doi:10.1016/j.devcel.2020.10.023
Dulken, B. W., Buckley, M. T., Navarro Negredo, P., Saligrama, N., Cayrol, R., Leeman, D. S., et al. (2019). Single-cell Analysis Reveals T Cell Infiltration in Old Neurogenic Niches. Nature 571 (7764), 205–210. doi:10.1038/s41586-019-1362-5
Durbin, A. F., Wang, C., Marcotrigiano, J., and Gehrke, L. (2016). RNAs Containing Modified Nucleotides Fail to Trigger RIG-I Conformational Changes for Innate Immune Signaling. mBio 7, e00833–16. doi:10.1128/mBio.00833-16
Fang, C., He, M., Li, D., and Xu, Q. (2021). YTHDF2 Mediates LPS-Induced Osteoclastogenesis and Inflammatory Response via the NF-Κb and MAPK Signaling Pathways. Cell Signal. 85, 110060. doi:10.1016/j.cellsig.2021.110060
Fedeles, B. I., Singh, V., Delaney, J. C., Li, D., and Essigmann, J. M. (2015). The AlkB Family of Fe(II)/α-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and beyond. J. Biol. Chem. 290 (34), 20734–20742. doi:10.1074/jbc.r115.656462
Feng, Y., Hu, Y., Hou, Z., Sun, Q., Jia, Y., and Zhao, R. (2020). Chronic Corticosterone Exposure Induces Liver Inflammation and Fibrosis in Association with m6A-Linked post-transcriptional Suppression of Heat Shock Proteins in Chicken. Cell Stress and Chaperones 25 (1), 47–56. doi:10.1007/s12192-019-01034-7
Fu, Y., Zorman, B., Sumazin, P., Sanna, P. P., and Repunte-Canonigo, V. (2019). Epitranscriptomics: Correlation of N6-Methyladenosine RNA Methylation and Pathway Dysregulation in the hippocampus of HIV Transgenic Rats. PLoS One 14, e0203566. doi:10.1371/journal.pone.0203566
Gong, H., Liu, L., Cui, L., Ma, H., and Shen, L. (2021). ALKBH5‐mediated m6A‐demethylation of USP1 Regulated T‐cell Acute Lymphoblastic Leukemia Cell Glucocorticoid Resistance by Aurora B. Mol. Carcinogenesis 60 (9), 644–657. doi:10.1002/mc.23330
Gu, X., Zhang, Y., Li, D., Cai, H., Cai, L., and Xu, Q. (2020). N6-methyladenosine Demethylase FTO Promotes M1 and M2 Macrophage Activation. Cell Signal. 69, 109553. doi:10.1016/j.cellsig.2020.109553
Guo, G., Wang, H., Shi, X., Ye, L., Yan, K., Chen, Z., et al. (2020). Disease Activity-Associated Alteration of mRNA M5 C Methylation in CD4+ T Cells of Systemic Lupus Erythematosus. Front. Cel Dev. Biol. 8, 430. doi:10.3389/fcell.2020.00430
Guo, M., Yan, R., Ji, Q., Yao, H., Sun, M., Duan, L., et al. (2020). IFN Regulatory Factor-1 Induced Macrophage Pyroptosis by Modulating m6A Modification of Circ_0029589 in Patients with Acute Coronary Syndrome. Int. Immunopharmacology 86, 106800. doi:10.1016/j.intimp.2020.106800
Han, D., Liu, J., Chen, C., Dong, L., Liu, Y., Chang, R., et al. (2019). Anti-tumour Immunity Controlled through mRNA m6A Methylation and YTHDF1 in Dendritic Cells. Nature 566 (7743), 270–274. doi:10.1038/s41586-019-0916-x
Han, H., Fan, G., Song, S., Jiang, Y., Qian, C. a., Zhang, W., et al. (2021). piRNA-30473 Contributes to Tumorigenesis and Poor Prognosis by Regulating m6A RNA Methylation in DLBCL. Blood 137 (12), 1603–1614. doi:10.1182/blood.2019003764
Hang, S., Paik, D., Yao, L., Kim, E., Trinath, J., Lu, J., et al. (2019). Bile Acid Metabolites Control TH17 and Treg Cell Differentiation. Nature 576 (7785), 143–148. doi:10.1038/s41586-019-1785-z
Hanniford, D., Ulloa-Morales, A., Karz, A., Berzoti-Coelho, M. G., Moubarak, R. S., Sánchez-Sendra, B., et al. (2020). Epigenetic Silencing of CDR1as Drives IGF2BP3-Mediated Melanoma Invasion and Metastasis. Cancer Cell 37 (1), 55–70. doi:10.1016/j.ccell.2019.12.007
Hesser, C. R., Karijolich, J., Dominissini, D., He, C., and Glaunsinger, B. A. (2018). N6-methyladenosine Modification and the YTHDF2 Reader Protein Play Cell Type Specific Roles in Lytic Viral Gene Expression during Kaposi's Sarcoma-Associated Herpesvirus Infection. Plos Pathog. 14, e1006995. doi:10.1371/journal.ppat.1006995
Huang, H., Weng, H., and Chen, J. (2020). m6A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell 37 (3), 270–288. doi:10.1016/j.ccell.2020.02.004
Huang, Y., Su, R., Sheng, Y., Dong, L., Dong, Z., Xu, H., et al. (2019). Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 35 (4), 677–691. doi:10.1016/j.ccell.2019.03.006
Huang, Y., Yan, J., Li, Q., Li, J., Gong, S., Zhou, H., et al. (2015). Meclofenamic Acid Selectively Inhibits FTO Demethylation of m6A over ALKBH5. Nucleic Acids Res. 43 (1), 373–384. doi:10.1093/nar/gku1276
Huangfu, N., Zheng, W., Xu, Z., Wang, S., Wang, Y., Cheng, J., et al. (2020). RBM4 Regulates M1 Macrophages Polarization through Targeting STAT1-Mediated Glycolysis. Int. Immunopharmacology 83, 106432. doi:10.1016/j.intimp.2020.106432
Jabs, S., Biton, A., Bécavin, C., Nahori, M.-A., Ghozlane, A., Pagliuso, A., et al. (2020). Impact of the Gut Microbiota on the m6A Epitranscriptome of Mouse Cecum and Liver. Nat. Commun. 11 (1), 1344. doi:10.1038/s41467-020-15126-x
Janela, B., Patel, A. A., Lau, M. C., Goh, C. C., Msallam, R., Kong, W. T., et al. (2019). A Subset of Type I Conventional Dendritic Cells Controls Cutaneous Bacterial Infections through VEGFα-Mediated Recruitment of Neutrophils. Immunity 50 (4), 1069–1083. doi:10.1016/j.immuni.2019.03.001
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 7 (12), 885–887. doi:10.1038/nchembio.687
Jurczyszak, D., Zhang, W., Terry, S. N., Kehrer, T., Bermúdez González, M. C., McGregor, E., et al. (2020). HIV Protease Cleaves the Antiviral m6A Reader Protein YTHDF3 in the Viral Particle. Plos Pathog. 16, e1008305. doi:10.1371/journal.ppat.1008305
Kang, H. J., Cheon, N. Y., Park, H., Jeong, G. W., Ye, B. J., Yoo, E. J., et al. (2021). TonEBP Recognizes R-Loops and Initiates m6A RNA Methylation for R-Loop Resolution. Nucleic Acids Res. 49 (1), 269–284. doi:10.1093/nar/gkaa1162
Karikó, K., Buckstein, M., Ni, H., and Weissman, D. (2005). Suppression of RNA Recognition by Toll-like Receptors: the Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity 23 (2), 165–175. doi:10.1016/j.immuni.2005.06.008
Kasowitz, S. D., Ma, J., Anderson, S. J., Leu, N. A., Xu, Y., Gregory, B. D., et al. (2018). Nuclear m6A Reader YTHDC1 Regulates Alternative Polyadenylation and Splicing during Mouse Oocyte Development. Plos Genet. 14, e1007412. doi:10.1371/journal.pgen.1007412
Kennedy, E. M., Bogerd, H. P., Kornepati, A. V. R., Kang, D., Ghoshal, D., Marshall, J. B., et al. (2016). Posttranscriptional M 6 A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host & Microbe 19 (5), 675–685. doi:10.1016/j.chom.2016.04.002
Knuckles, P., Lence, T., Haussmann, I. U., Jacob, D., Kreim, N., Carl, S. H., et al. (2018). Zc3h13/Flacc Is Required for Adenosine Methylation by Bridging the mRNA-Binding Factor Rbm15/Spenito to the m6A Machinery Component Wtap/Fl(2)d. Genes Dev. 32 (5-6), 415–429. doi:10.1101/gad.309146.117
Lan, T., Li, H., Zhang, D., Xu, L., Liu, H., Hao, X., et al. (2019). KIAA1429 Contributes to Liver Cancer Progression through N6-methyladenosine-dependent post-transcriptional Modification of GATA3. Mol. Cancer 18 (1), 186. doi:10.1186/s12943-019-1106-z
Lei, H., He, M., He, X., Li, G., Wang, Y., Gao, Y., et al. (2021). METTL3 Induces Bone Marrow Mesenchymal Stem Cells Osteogenic Differentiation and Migration through Facilitating M1 Macrophage Differentiation. Am. J. Transl Res. 13 (5), 4376–4388.
Li, H.-B., Tong, J., Zhu, S., Batista, P. J., Duffy, E. E., Zhao, J., et al. (2017). m6A mRNA Methylation Controls T Cell Homeostasis by Targeting the IL-7/STAT5/SOCS Pathways. Nature 548 (7667), 338–342. doi:10.1038/nature23450
Li, H., Su, Q., Li, B., Lan, L., Wang, C., Li, W., et al. (2020). High Expression of WTAP Leads to Poor Prognosis of Gastric Cancer by Influencing Tumour‐associated T Lymphocyte Infiltration. J. Cel Mol Med 24 (8), 4452–4465. doi:10.1111/jcmm.15104
Li, L.-J., Fan, Y.-G., Leng, R.-X., Pan, H.-F., and Ye, D.-Q. (2018). Potential Link between M 6 A Modification and Systemic Lupus Erythematosus. Mol. Immunol. 93, 55–63. doi:10.1016/j.molimm.2017.11.009
Li, N., Kang, Y., Wang, L., Huff, S., Tang, R., Hui, H., et al. (2020). ALKBH5 Regulates Anti-PD-1 Therapy Response by Modulating Lactate and Suppressive Immune Cell Accumulation in Tumor Microenvironment. Proc. Natl. Acad. Sci. USA 117 (33), 20159–20170. doi:10.1073/pnas.1918986117
Li, Z., Weng, H., Su, R., Weng, X., Zuo, Z., Li, C., et al. (2017). FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N 6 -Methyladenosine RNA Demethylase. Cancer Cell 31 (1), 127–141. doi:10.1016/j.ccell.2016.11.017
Lichinchi, G., Gao, S., Saletore, Y., Gonzalez, G. M., Bansal, V., Wang, Y., et al. (2016). Dynamics of the Human and Viral m6A RNA Methylomes during HIV-1 Infection of T Cells. Nat. Microbiol. 1, 16011. doi:10.1038/nmicrobiol.2016.11
Liu, J., Eckert, M. A., Harada, B. T., Liu, S.-M., Lu, Z., Yu, K., et al. (2018). m6A mRNA Methylation Regulates AKT Activity to Promote the Proliferation and Tumorigenicity of Endometrial Cancer. Nat. Cel Biol 20 (9), 1074–1083. doi:10.1038/s41556-018-0174-4
Liu, J., and Jia, G. (2014). Methylation Modifications in Eukaryotic Messenger RNA. J. Genet. Genomics 41 (1), 21–33. doi:10.1016/j.jgg.2013.10.002
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat. Chem. Biol. 10 (2), 93–95. doi:10.1038/nchembio.1432
Liu, J., Zhang, X., Chen, K., Cheng, Y., Liu, S., Xia, M., et al. (2019). CCR7 Chemokine Receptor-Inducible Lnc-Dpf3 Restrains Dendritic Cell Migration by Inhibiting HIF-1α-Mediated Glycolysis. Immunity 50 (3), 600–615. doi:10.1016/j.immuni.2019.01.021
Liu, Q., Li, M., Jiang, L., Jiang, R., and Fu, B. (2019). METTL3 Promotes Experimental Osteoarthritis Development by Regulating Inflammatory Response and Apoptosis in Chondrocyte. Biochem. Biophysical Res. Commun. 516 (1), 22–27. doi:10.1016/j.bbrc.2019.05.168
Liu, Y., Liu, Z., Tang, H., Shen, Y., Gong, Z., Xie, N., et al. (2019). The N6-Methyladenosine (m6A)-Forming Enzyme METTL3 Facilitates M1 Macrophage Polarization through the Methylation of STAT1 mRNA. Am. J. Physiol. Cel Physiol 317, C762. doi:10.1152/ajpcell.00212.2019
Lu, N., Li, X., Yu, J., Li, Y., Wang, C., Zhang, L., et al. (2018). Curcumin Attenuates Lipopolysaccharide-Induced Hepatic Lipid Metabolism Disorder by Modification of M6 A RNA Methylation in Piglets. Lipids 53 (1), 53–63. doi:10.1002/lipd.12023
Lu, T. X., Zheng, Z., Zhang, L., Sun, H.-L., Bissonnette, M., Huang, H., et al. (2020). A New Model of Spontaneous Colitis in Mice Induced by Deletion of an RNA m6A Methyltransferase Component METTL14 in T Cells. Cell Mol. Gastroenterol. Hepatol. 10 (4), 747–761. doi:10.1016/j.jcmgh.2020.07.001
Lu, W., Tirumuru, N., St. Gelais, C., Koneru, P. C., Liu, C., Kvaratskhelia, M., et al. (2018). N6-Methyladenosine-binding Proteins Suppress HIV-1 Infectivity and Viral Production. J. Biol. Chem. 293 (34), 12992–13005. doi:10.1074/jbc.ra118.004215
Ma, H., Shen, L., Yang, H., Gong, H., Du, X., and Li, J. (2021). m6A Methyltransferase Wilms' Tumor 1‐associated Protein Facilitates Cell Proliferation and Cisplatin Resistance in NK/T Cell Lymphoma by Regulating Dual‐specificity Phosphatases 6 Expression via m6A RNA Methylation. IUBMB Life 73 (1), 108–117. doi:10.1002/iub.2410
Maier, B., Leader, A. M., Chen, S. T., Tung, N., Chang, C., LeBerichel, J., et al. (2020). A Conserved Dendritic-Cell Regulatory Program Limits Antitumour Immunity. Nature 580 (7802), 257–262. doi:10.1038/s41586-020-2134-y
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5′ UTR m6A Promotes Cap-independent Translation. Cell 163 (4), 999–1010. doi:10.1016/j.cell.2015.10.012
Miranda-Gonçalves, V., Lobo, J., Guimarães-Teixeira, C., Barros-Silva, D., Guimarães, R., Cantante, M., et al. (2021). The Component of the m6A Writer Complex VIRMA Is Implicated in Aggressive Tumor Phenotype, DNA Damage Response and Cisplatin Resistance in Germ Cell Tumors. J. Exp. Clin. Cancer Res. 40 (1), 268. doi:10.1186/s13046-021-02072-9
Müller, T. A., Struble, S. L., Meek, K., and Hausinger, R. P. (2018). Characterization of Human AlkB Homolog 1 Produced in Mammalian Cells and Demonstration of Mitochondrial Dysfunction in ALKBH1-Deficient Cells. Biochem. Biophysical Res. Commun. 495 (1), 98–103. doi:10.1016/j.bbrc.2017.10.158
Müller, T. A., Tobar, M. A., Perian, M. N., and Hausinger, R. P. (2017). Biochemical Characterization of AP Lyase and m6A Demethylase Activities of Human AlkB Homologue 1 (ALKBH1). Biochemistry 56 (13), 1899–1910. doi:10.1021/acs.biochem.7b00060
Myoung, J., and Ganem, D. (2011). Infection of Lymphoblastoid Cell Lines by Kaposi's Sarcoma. J. Virol. 85 (19), 9767–9777. doi:10.1128/jvi.05136-11
Ni, W., Yao, S., Zhou, Y., Liu, Y., Huang, P., Zhou, A., et al. (2019). Long Noncoding RNA GAS5 Inhibits Progression of Colorectal Cancer by Interacting with and Triggering YAP Phosphorylation and Degradation and Is Negatively Regulated by the m6A Reader YTHDF3. Mol. Cancer 18 (1), 143. doi:10.1186/s12943-019-1079-y
Pan, Y. Y., Yang, J. X., Xu, Y. F., and Mao, W. (2020). Yin Yang‐1 Suppresses CD40 ligand‐CD40 Signaling‐mediated Anti‐inflammatory Cytokine Interleukin‐10 Expression in Pulmonary Adventitial Fibroblasts by Promoting Histone H3 Tri‐methylation at Lysine 27 Modification on Interleukin‐10 Promoter. Cell Biol Int 44 (7), 1544–1555. doi:10.1002/cbin.11351
Pandolfini, L., Barbieri, I., Bannister, A. J., Hendrick, A., Andrews, B., Webster, N., et al. (2019). METTL1 Promotes Let-7 MicroRNA Processing via m7G Methylation. Mol. Cel 74 (6), 1278–1290. doi:10.1016/j.molcel.2019.03.040
Paris, J., Morgan, M., Campos, J., Spencer, G. J., Shmakova, A., Ivanova, I., et al. (2019). Targeting the RNA m6A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell 25 (1), 137–148. doi:10.1016/j.stem.2019.03.021
Ping, X.-L., Sun, B.-F., Wang, L., Xiao, W., Yang, X., Wang, W.-J., et al. (2014). Mammalian WTAP Is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res 24 (2), 177–189. doi:10.1038/cr.2014.3
Qiu, W., Zhang, Q., Zhang, R., Lu, Y., Wang, X., Tian, H., et al. (2021). N6-methyladenosine RNA Modification Suppresses Antiviral Innate Sensing Pathways via Reshaping Double-Stranded RNA. Nat. Commun. 12 (1), 1582. doi:10.1038/s41467-021-21904-y
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA Modifications in Gene Expression Regulation. Cell 169 (7), 1187–1200. doi:10.1016/j.cell.2017.05.045
Saunders, K. O., Wiehe, K., Tian, M., Acharya, P., Bradley, T., Alam, S. M., et al. (2019). Targeted Selection of HIV-specific Antibody Mutations by Engineering B Cell Maturation. Science 366, e7199. doi:10.1126/science.aay7199
Scott, A. C., Dündar, F., Zumbo, P., Chandran, S. S., Klebanoff, C. A., Shakiba, M., et al. (2019). TOX Is a Critical Regulator of Tumour-specific T Cell Differentiation. Nature 571 (7764), 270–274. doi:10.1038/s41586-019-1324-y
Silla, T., Schmid, M., Dou, Y., Garland, W., Milek, M., Imami, K., et al. (2020). The Human ZC3H3 and RBM26/27 Proteins Are Critical for PAXT-Mediated Nuclear RNA Decay. Nucleic Acids Res. 48 (5), 2518–2530. doi:10.1093/nar/gkz1238
Song, T., Yang, Y., Wei, H., Xie, X., Lu, J., Zeng, Q., et al. (2019). Zfp217 Mediates m6A mRNA Methylation to Orchestrate Transcriptional and post-transcriptional Regulation to Promote Adipogenic Differentiation. Nucleic Acids Res. 47 (12), 6130–6144. doi:10.1093/nar/gkz312
Sorci, M., Ianniello, Z., Cruciani, S., Larivera, S., Ginistrelli, L. C., Capuano, E., et al. (2018). METTL3 Regulates WTAP Protein Homeostasis. Cell Death Dis 9 (8), 796. doi:10.1038/s41419-018-0843-z
Su, R., Dong, L., Li, Y., Gao, M., Han, L., Wunderlich, M., et al. (2020). Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell 38 (1), 79–96. doi:10.1016/j.ccell.2020.04.017
Sun, K., Du, Y., Hou, Y., Zhao, M., Li, J., Du, Y., et al. (2021). Saikosaponin D Exhibits Anti-leukemic Activity by Targeting FTO/m6A Signaling. Theranostics 11 (12), 5831–5846. doi:10.7150/thno.55574
Sun, X., Dai, Y., Tan, G., Liu, Y., and Li, N. (2020). Integration Analysis of m6A-SNPs and eQTLs Associated with Sepsis Reveals Platelet Degranulation and Staphylococcus aureus Infection Are Mediated by m6A mRNA Methylation. Front. Genet. 11, 7. doi:10.3389/fgene.2020.00007
Tirumuru, N., Zhao, B. S., Lu, W., Lu, Z., He, C., and Wu, L. (2016). N(6)-methyladenosine of HIV-1 RNA Regulates Viral Infection and HIV-1 Gag Protein Expression. eLife 5, e15528. doi:10.7554/eLife.15528
Tirumuru, N., and Wu, L. (2019). HIV-1 Envelope Proteins Up-Regulate N6-Methyladenosine Levels of Cellular RNA Independently of Viral Replication. J. Biol. Chem. 294 (9), 3249–3260. doi:10.1074/jbc.ra118.005608
Tong, J., Wang, X., Liu, Y., Ren, X., Wang, A., Chen, Z., et al. (2021). Pooled CRISPR Screening Identifies m6A as a Positive Regulator of Macrophage Activation. Sci. Adv. 7, eabd4742. doi:10.1126/sciadv.abd4742
Tong, J., Cao, G., Zhang, T., Sefik, E., Amezcua Vesely, M. C., Broughton, J. P., et al. (2018). m6A mRNA Methylation Sustains Treg Suppressive Functions. Cel Res 28 (2), 253–256. doi:10.1038/cr.2018.7
Toro-Ascuy, D., Rojas-Araya, B., Valiente-Echeverría, F., and Soto-Rifo, R. (2016). Interactions between the HIV-1 Unspliced mRNA and Host mRNA Decay Machineries. Viruses 8 (11), 320. doi:10.3390/v8110320
Wang, H., Hu, X., Huang, M., Liu, J., Gu, Y., Ma, L., et al. (2019). Mettl3-mediated mRNA m6A Methylation Promotes Dendritic Cell Activation. Nat. Commun. 10 (1), 1898. doi:10.1038/s41467-019-09903-6
Wang, J., Yan, S., Lu, H., Wang, S., and Xu, D. (2019). METTL3 Attenuates LPS-Induced Inflammatory Response in Macrophages via NF-Κb Signaling Pathway. Mediators Inflamm. 2019, 3120391. doi:10.1155/2019/3120391
Wang, L., Hui, H., Agrawal, K., Kang, Y., Li, N., Tang, R., et al. (2020). m6 A RNA Methyltransferases METTL3/14 Regulate Immune Responses to Anti-PD-1 Therapy. EMBO J. 39, e104514. doi:10.15252/embj.2020104514
Wang, L., Wen, M., and Cao, X. (2019). Nuclear hnRNPA2B1 Initiates and Amplifies the Innate Immune Response to DNA Viruses. Science 365, eaav0758. doi:10.1126/science.aav0758
Wang, N., Tang, H., Wang, X., Wang, W., and Feng, J. (2017). Homocysteine Upregulates interleukin-17A Expression via NSun2-Mediated RNA Methylation in T Lymphocytes. Biochem. Biophysical Res. Commun. 493 (1), 94–99. doi:10.1016/j.bbrc.2017.09.069
Wang, S., Chai, P., Jia, R., and Jia, R. (2018). Novel Insights on m6A RNA Methylation in Tumorigenesis: a Double-Edged Sword. Mol. Cancer 17 (1), 101. doi:10.1186/s12943-018-0847-4
Wang, X., Ji, Y., Feng, P., Liu, R., Li, G., Zheng, J., et al. (2021). The m6A Reader IGF2BP2 Regulates Macrophage Phenotypic Activation and Inflammatory Diseases by Stabilizing TSC1 and PPAR γ. Adv. Sci. 8 (13), 2100209. doi:10.1002/advs.202100209
Wang, Z., Pan, Z., Adhikari, S., Harada, B. T., Shen, L., Yuan, W., et al. (2021). m(6) A Deposition Is Regulated by PRMT1-Mediated Arginine Methylation of METTL14 in its Disordered C-Terminal Region. EMBO J. 40 (5), e106309. doi:10.15252/embj.2020106309
Weng, H., Huang, H., Wu, H., Qin, X., Zhao, B. S., Dong, L., et al. (2018). METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell 22 (2), 191–205. doi:10.1016/j.stem.2017.11.016
Wolf, D. A., Lin, Y., Duan, H., and Cheng, Y. (2020). eIF-Three to Tango: Emerging Functions of Translation Initiation Factor eIF3 in Protein Synthesis and Disease. J. Mol. Cel Biol 12 (6), 403–409. doi:10.1093/jmcb/mjaa018
Xia, H., Zhong, C., Wu, X., Chen, J., Tao, B., Xia, X., et al. (2018). Mettl3 Mutation Disrupts Gamete Maturation and Reduces Fertility in Zebrafish. Genetics 208 (2), 729–743. doi:10.1534/genetics.117.300574
Xiang, Y., Laurent, B., Hsu, C.-H., Nachtergaele, S., Lu, Z., Sheng, W., et al. (2017). RNA m6A Methylation Regulates the Ultraviolet-Induced DNA Damage Response. Nature 543 (7646), 573–576. doi:10.1038/nature21671
Xing, Y., Cheng, D., Shi, C., and Shen, Z. (2021). The Protective Role of YTHDF1-Knock Down Macrophages on the Immune Paralysis of Severe Sepsis Rats with ECMO. Microvasc. Res. 137, 104178. doi:10.1016/j.mvr.2021.104178
Xiong, F., Wang, R., Lee, J.-H., Li, S., Chen, S.-F., Liao, Z., et al. (2021). RNA m6A Modification Orchestrates a LINE-1-Host Interaction that Facilitates Retrotransposition and Contributes to Long Gene Vulnerability. Cel Res 31 (8), 861–885. doi:10.1038/s41422-021-00515-8
Yang, Z., Yang, S., Cui, Y-H., Wei, J., Shah, P., Park, G., et al. (2021). METTL14 Facilitates Global Genome Repair and Suppresses Skin Tumorigenesis. Proc. Natl. Acad. Sci. U S A. 118 (35), e2025948118. doi:10.1073/pnas.2025948118
Yankova, E., Blackaby, W., Albertella, M., Rak, J., De Braekeleer, E., Tsagkogeorga, G., et al. (2021). Small-molecule Inhibition of METTL3 as a Strategy against Myeloid Leukaemia. Nature 593 (7860), 597–601. doi:10.1038/s41586-021-03536-w
Yao, Y., Yang, Y., Guo, W., Xu, L., You, M., Zhang, Y.-C., et al. (2021). METTL3-dependent m6A Modification Programs T Follicular Helper Cell Differentiation. Nat. Commun. 12 (1), 1333. doi:10.1038/s41467-021-21594-6
Yin, H., Zhang, X., Yang, P., Zhang, X., Peng, Y., Li, D., et al. (2021). RNA m6A Methylation Orchestrates Cancer Growth and Metastasis via Macrophage Reprogramming. Nat. Commun. 12 (1), 1394. doi:10.1038/s41467-021-21514-8
Yoon, K.-J., Ringeling, F. R., Vissers, C., Jacob, F., Pokrass, M., Jimenez-Cyrus, D., et al. (2017). Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation. Cell 171 (4), 877–889. doi:10.1016/j.cell.2017.09.003
Yu, F., Wei, J., Cui, X., Yu, C., Ni, W., Bungert, J., et al. (2021). Post-translational Modification of RNA m6A Demethylase ALKBH5 Regulates ROS-Induced DNA Damage Response. Nucleic Acids Res. 49 (10), 5779–5797. doi:10.1093/nar/gkab415
Yu, R., Li, Q., Feng, Z., Cai, L., and Xu, Q. (2019). m6A Reader YTHDF2 Regulates LPS-Induced Inflammatory Response. Ijms 20 (6), 1323. doi:10.3390/ijms20061323
Yue, H., Nie, X., Yan, Z., and Weining, S. (2019). N6‐methyladenosine Regulatory Machinery in Plants: Composition, Function and Evolution. Plant Biotechnol. J. 17 (7), 1194–1208. doi:10.1111/pbi.13149
Zhang, C., Chen, L., Peng, D., Jiang, A., He, Y., Zeng, Y., et al. (2020). METTL3 and N6-Methyladenosine Promote Homologous Recombination-Mediated Repair of DSBs by Modulating DNA-RNA Hybrid Accumulation. Mol. Cel 79 (3), 425–442. doi:10.1016/j.molcel.2020.06.017
Zhang, J., Kong, L., Guo, S., Bu, M., Guo, Q., Xiong, Y., et al. (2017). hnRNPs and ELAVL1 Cooperate with uORFs to Inhibit Protein Translation. Nucleic Acids Res. 45 (5), 2849–2864. doi:10.1093/nar/gkw991
Zhang, L., Hao, D., Ma, P., Ma, B., Qin, J., Tian, G., et al. (2021). Epitranscriptomic Analysis of m6A Methylome after Peripheral Nerve Injury. Front. Genet. 12, 686000. doi:10.3389/fgene.2021.686000
Zhang, P., He, Q., Lei, Y., Li, Y., Wen, X., Hong, M., et al. (2018). m6A-mediated ZNF750 Repression Facilitates Nasopharyngeal Carcinoma Progression. Cel Death Dis 9 (12), 1169. doi:10.1038/s41419-018-1224-3
Zhang, W., He, X., Hu, J., Yang, P., Liu, C., Wang, J., et al. (2019). Dysregulation of N6-Methyladenosine Regulators Predicts Poor Patient Survival in Mantle Cell Lymphoma. Oncol. Lett. 18 (4), 3682–3690. doi:10.3892/ol.2019.10708
Zhang, X., Li, X., Jia, H., An, G., and Ni, J. (2021). The m6A Methyltransferase METTL3 Modifies PGC-1α mRNA Promoting Mitochondrial Dysfunction and oxLDL-Induced Inflammation in Monocytes. J. Biol. Chem. 297, 101058. doi:10.1016/j.jbc.2021.101058
Zhang, Y., Wang, X., Zhang, X., Wang, J., Ma, Y., Zhang, L., et al. (2019). RNA-binding Protein YTHDF3 Suppresses Interferon-dependent Antiviral Responses by Promoting FOXO3 Translation. Proc. Natl. Acad. Sci. USA 116 (3), 976–981. doi:10.1073/pnas.1812536116
Zhao, W., Wang, Z., Sun, Z., He, Y., Jian, D., Hu, X., et al. (2018). RNA Helicase DDX5 Participates in oxLDL-Induced Macrophage Scavenger Receptor 1 Expression by Suppressing mRNA Degradation. Exp. Cel Res. 366 (2), 114–120. doi:10.1016/j.yexcr.2018.03.003
Zhao, X., Chen, Y., Mao, Q., Jiang, X., Jiang, W., Chen, J., et al. (2018). Overexpression of YTHDF1 Is Associated with Poor Prognosis in Patients with Hepatocellular Carcinoma. Cbm 21 (4), 859–868. doi:10.3233/cbm-170791
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C.-M., Li, C. J., et al. (2013). ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cel 49 (1), 18–29. doi:10.1016/j.molcel.2012.10.015
Zheng, Q., Hou, J., Zhou, Y., Li, Z., and Cao, X. (2017). The RNA Helicase DDX46 Inhibits Innate Immunity by Entrapping m6A-Demethylated Antiviral Transcripts in the Nucleus. Nat. Immunol. 18 (10), 1094–1103. doi:10.1038/ni.3830
Zheng, Z., Zhang, L., Cui, X.-L., Yu, X., Hsu, P. J., Lyu, R., et al. (2020). Control of Early B Cell Development by the RNA N6-Methyladenosine Methylation. Cel Rep. 31 (13), 107819. doi:10.1016/j.celrep.2020.107819
Zhou, B., Liu, C., Xu, L., Yuan, Y., Zhao, J., Zhao, W., et al. (2020). N 6 ‐Methyladenosine Reader Protein YT521‐B Homology Domain‐Containing 2 Suppresses Liver Steatosis by Regulation of mRNA Stability of Lipogenic Genes. Hepatology 73 (1), 91–103. doi:10.1002/hep.31220
Zhou, J., Zhang, X., Hu, J., Qu, R., Yu, Z., Xu, H., et al. (2021). m6A Demethylase ALKBH5 Controls CD4+ T Cell Pathogenicity and Promotes Autoimmunity. Sci. Adv. 7, eabg0470. doi:10.1126/sciadv.abg0470
Zhu, X. J., Feng, J. Q., Zheng, M. Z., Yang, Z. R., Zhao, L., Zhang, W., et al. (2021). Metal-Protein Nanoparticles Facilitate Anti-VSV and H1N1 Viruses through the Coordinative Actions on Innate Immune Responses and METTL14. Macromol Biosci. 21, e2000382. doi:10.1002/mabi.202000382
Keywords: m6A (N6-methyladenosine), immunocyte, epigenetics, immunotherapy, DNA repair
Citation: Zhou M, Liu W, Zhang J and Sun N (2021) RNA m6A Modification in Immunocytes and DNA Repair: The Biological Functions and Prospects in Clinical Application. Front. Cell Dev. Biol. 9:794754. doi: 10.3389/fcell.2021.794754
Received: 14 October 2021; Accepted: 01 November 2021;
Published: 20 December 2021.
Edited by:
Teng Ma, Capital Medical University, ChinaReviewed by:
Lei Gao, City of Hope National Medical Center, United StatesCopyright © 2021 Zhou, Liu, Zhang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Sun, c3VubmFudGhlb25lQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.