- Cell and Molecular Biology of Cancer Program, Sanford Burnham Prebys, La Jolla, CA, United States
While organelles are individual compartments with specialized functions, it is becoming clear that organellar communication is essential for maintaining cellular homeostasis. This cooperation is carried out by various interactions taking place on the membranes of organelles. The membranes themselves contain a multitude of proteins and lipids that mediate these connections and one such class of molecules facilitating these relations are the phospholipids. There are several phospholipids, but the focus of this perspective is on a minor group called the phosphoinositides and specifically, phosphatidylinositol 4,5-bisphosphate (PI-4,5-P2). This phosphoinositide, on intracellular membranes, is largely generated by the non-canonical Type II PIPKs, namely, Phosphotidylinositol-5-phosphate-4-kinases (PI5P4Ks). These evolutionarily conserved enzymes are emerging as key stress response players in cells. Further, PI5P4Ks have been shown to modulate pathways by regulating organelle crosstalk, revealing roles in preserving metabolic homeostasis. Here we will attempt to summarize the functions of the PI5P4Ks and their product PI-4,5-P2 in facilitating inter-organelle communication and how they impact cellular health as well as their relevance to human diseases.
Introduction
Lipids are essential components of cellular membranes. The type and the amount of lipids vary within the membranes at the organism, cell, as well as the organellar level. The lipid composition, quantitative and qualitative, of the membranes defines not only their physical characteristics but also, their biological properties. The majority of the membrane lipids are grouped into glycerophospholipids (GPLs), sphingolipids, and sterols (van Meer et al., 2008). GPLs are characterized by a glycerol backbone which forms the link between the head group that consists of a phosphate and an alcohol and the tail which consists of varying lengths of fatty acid chains (Harayama and Riezman, 2018). One such group of GPLs are the phosphoinositols (PIs).
PIs derive their name from the inositol head group, with the most predominant fatty acyl chains seen in the PIs in tissues being the stearoyl and arachidonoyl-acyl chains (Harayama and Riezman, 2018). PIs give rise to seven different species of phosphoinositides by the phosphorylation and dephosphorylation at the 3,4 and 5 position of the inositol moieties by the precise actions of specific kinases and phosphatases (Balla, 2013; Burke, 2018; Palamiuc et al., 2020). While found in minute amounts as compared to the other lipids on membranes, PIs play essential roles in regulating various cellular functions such as cytoskeletal remodeling, vesicular budding, membrane dynamics to regulating ion channels and signaling pathways. This is carried out by recruitment and interaction with, as well as activation of proteins, in a spatial and temporal manner (Falkenburger et al., 2010). Every species of phosphoinositide has its own cohort of interacting partners. This, along with the fact that membranes differ in their lipid composition, allows for distinct functional entities. In this review, we will mainly focus on one particular PI species, phosphatidylinositol-4,5-bisphosphate (PI-4,5-P2).
PI-4,5-P2, along with phosphotidylinositol-4-phosphate (PI-4-P), accounts for the bulk of all PIs (Di Paolo and De Camilli, 2006) and are generated by two distinct families of PI kinases. Initially these enzymes were identified as the Type I and Type II PI4P-kinases (Phosphatidylinositol-4-phosphate kinases) based on their biochemical properties and immunochemical cross-reactivities (Bazenet et al., 1990; Ling et al., 1989). It was later discovered that the Type II kinases actually produced PI-4,5-P2 by phosphorylating the 4-position of phosphatidylinositol 5-phosphate (PI-5-P), a previously unknown PI at time and interestingly the last PI to be discovered in higher organisms (Rameh et al., 1997), whereas the Type I kinases phosphorylated the 5-position of phosphatidylinositol 4-phosphate (PI-4-P) to produce the same PI-4,5-P2 species. Thus, giving rise to the current nomenclature of the PI families as the Type I kinases or PI4P5Ks (Phosphatidylinositol 4-phosphate 5-kinases) and the Type II or PI5P4Ks (Phosphatidylinositol 5-phosphate 4-kinases). Moreover, in mammalian cells, both kinase families have three distinct isoforms each, namely, α, β, and γ (Bulley et al., 2015; Clarke and Irvine, 2012; Clarke and Irvine, 2013). Apart from having very diverse immunological and catalytic functions, these kinases also generate PI-4,5-P2 on different cellular membranes (Table 1) (Balla, 2013).
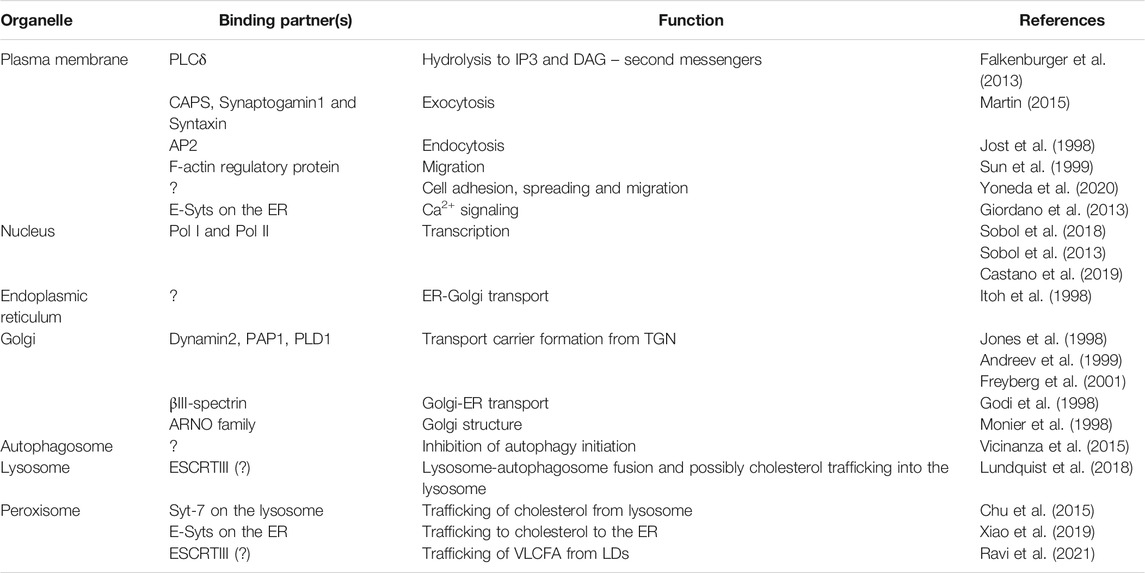
TABLE 1. PI-4,5-P2 at the organelles: Binding partners and functions. PI-4,5-P2 interacting partners and organellar regulation. The table highlights some of the major interacting partners of PI-4,5-P2 on the plasma membrane, nucleus and other organelles and the functions they regulate. While the PM and nucleus functions are attributed to the type I kinases (PI4P5Ks), Golgi and ER have been shown to have roles regulated by PI-4,5-P2 generates by both family of kinases. Whereas autophagosome, lysosome and peroxisome interactions are carried out predominantly by PI-4,5-P2 generated by PI5P4Ks (Type II kinases).
PI-4,5-P2 Functions on Cellular Membranes and Correlations for Organellar Cooperation
With PI-4-P being 10 times more abundant than PI-5-P on the plasma membrane (PM), PI-4,5-P2 generated here is predominantly by the action of Type I PIPKs or PI4P5Ks and constitute only 1–3% of the total lipid content on the membrane (King et al., 1987; King et al., 1989; McLaughlin et al., 2002). As a substrate, PI-4,5-P2 can be hydrolyzed to IP3 (inositol 1,4,5-trisphosphate) and DAG (diacylglycerol), by the activity of phospholipase C (PLC), which in turn serve as second messengers in various intracellular signaling cascades (Falkenburger et al., 2013). PI-4,5-P2 is also a precursor for PI-4-P and PI-3,4,5-P3 both of which play a role themselves in signaling and membrane dynamics (Auger et al., 1989; Varnai et al., 2006).
Additionally, PI-4,5-P2 plays a major role in membrane remodeling and trafficking. As will be discussed in the later sections, this function is essential not only at the level of the PM but is also a feature in key inter-organellar events mediated by these lipids to regulate cellular and metabolic homeostasis. At the PM, fusion and fission cycles can lead to exocytosis and endocytosis, both of which are invariably dependent on PI-4,5-P2. These are mediated by the ability of PI-4,5-P2 to bind various proteins via structured basic regions such as pleckstrin homology (PH) and C2-domains (Balla, 2013). PI-4,5-P2 is also a requisite for Ca2+ dependent PM-Endoplasmic Reticulum (ER) interaction by binding C2-domain containing E-Syts (Extended-synaptogamins) that are embedded in the ER membrane (Giordano et al., 2013). Interestingly, for a lipid that is quantitatively low on the PM, PI-4,5-P2 has shown to be a central mediator of a multitude of cellular functions and not surprisingly, loss of or mutation in kinases and phosphatases that regulate PI-4,5-P2 levels can lead to various diseased states (Pendaries et al., 2003; McCrea and De Camilli, 2009). Even though the bulk of the PI-4,5-P2 in the cell is generated by the Type I kinases, here we are focusing on the small but significant pool of PI-4,5-P2 that is created by the action of the Type II kinases, which to date is under appreciated yet is emerging to be fundamental for many essential cellular and metabolic events. The role of Type I kinases and their product is discussed in detail in another review (Katan and Cockcroft, 2020). Figure 1 and Table 1 also highlight some of the interactions mediated by PI-4,5-P2 generated via the activity of the Type I kinases.
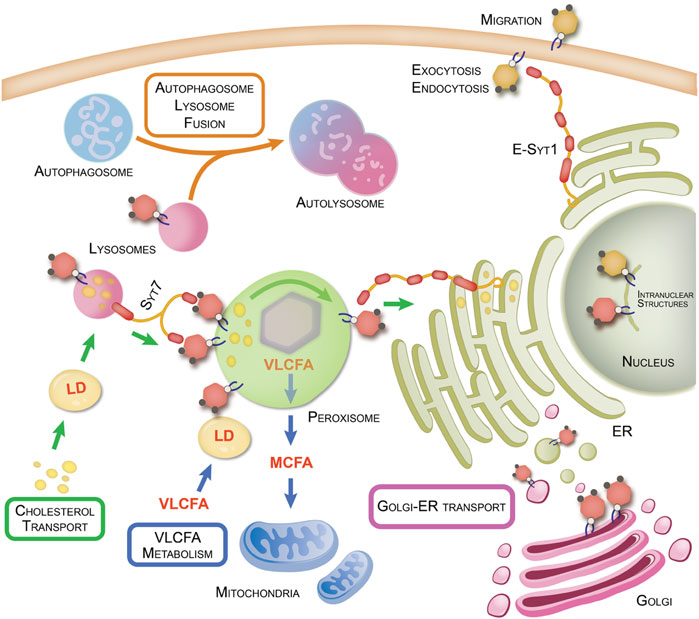
FIGURE 1. PI-4,5-P2 regulation of organellar interactions and cellular metabolism. Organelle-organelle interactions within the cell are key to regulating various cellular processes and transport of nutrients and materials to maintain metabolic homeostasis. Disruption of these interactions can lead to various diseased states such as cancer and neurodegenerative disorders. Here we summarize the various functions regulated by PI4P5K-generated PI-4,5-P2 (yellow hexagons) and PI5P4K-generated PI-4,5-P2 (red hexagons). This figure also highlights organellar interactions regulated by PI5P4Ks and their products as well the proteins involved and the metabolic pathways impacted by such interactions. Refer also to Table 1 for details of PI-4,5-P2 interactors and functions on the plasma membrane.
With the Type I or PI4P5Ks being considered the main pathway for PI-4,5-P2 synthesis and PI-4-P being more abundant compared to the minor phosphoinositide, PI-5-P, the Type II or PI5P4Ks were relegated to the role of merely regulating the level of this lipid in the cells. However, recent studies have eloquently shown that PI-4,5-P2 generated by PI5P4Ks are not just by-products but have the capacity to modulate cellular metabolism by regulating organellar functions (Hu et al., 2018; Xiao et al., 2019; Ravi et al., 2021). Of the three isoforms described, PI5P4Kα is the most active and PI5P4Kγ the least active (Clarke and Irvine, 2013; Giudici et al., 2016). As a substrate, PI-5-P is also the most elusive of the phosphoinositides, with its separation and identification in the cell made difficult due to technical limitations. Since its discovery almost 25 years ago (Rameh et al., 1997), new roles and localization in the cellular compartments is being constantly uncovered. Studies have shown that the levels of PI-5-P change in response to various stimuli such as insulin, oxidative stress, bacterial infection etc. and in turn regulate numerous cellular functions such as cell signaling, vesicular transport and even play a role in the nucleus (Hasegawa et al., 2017; Ghosh et al., 2019).
One of the first biochemical studies of cellular compartments showed enrichment of PI-5-P at both the Golgi and the PM (Sarkes and Rameh, 2010). The product of its phosphorylation, PI-4,5-P2 has been detected on the stacked cisternae of the Golgi and its role has been studied in connection with maintaining the structural and functional integrity of the Golgi. PI-4,5-P2-mediated interactions with various proteins at the Golgi is essential for its structural organization, formation of carriers from the trans-Golgi network (TGN) as well as Golgi-ER transport (De Matteis et al., 2002; De Matteis and D'Angelo, 2007). Moreover, PI5P4Ks are functionally involved in various signaling events at the Golgi (Mackey et al., 2014). In fact, PI5P4Kγ has been localized to the ER suggesting PI-4,5-P2 synthesis at this organelle by these kinases (Itoh et al., 1998) and with PI-4,5-P2 playing a role in ER-Golgi transport, it remains to be seen whether and what percent of the lipid is generated by PI5P4Ks (De Matteis et al., 2002). However, this is contradictory to the studies demonstrating that the gamma isoform has very little inherent kinase function in vitro (Clarke et al., 2008). The mechanism by which PI5P4Kγ functions, and the role it plays in regulating cellular processes, remains to be explored further.
Interestingly, PI-5-P has also been found in the nucleus where it not only serves as a substrate for PI5P4Ks at that location but has been implicated in various nuclear outputs (Poli et al., 2019). PI5P4Kβ is the only isoform that has a unique nuclear localization signal that allows it to be targeted to the nucleus. However, the PI5P4Ks are dimeric enzymes and therefore the beta isoform has been shown to dimerize with the alpha, directing it into the nucleus as well (Bultsma et al., 2010). While PI-4,5-P2 has been shown to have nuclear functions such as the involvement with Pol I and Pol II mediated transcription as well as associating with splicing compartments, these functions have mostly been attributed to the canonical Type I PI4P5Ks. Thus far, the main function of the Type II PI5P4Ks at the nucleus seems to be quelling the starvation-induced increase in levels of PI-5-P. Details of the role of PI-4,5-P2 at the organelles mentioned thus far are discussed in Tan X et al. (2015) Also, see Table 1 for various interacting proteins of PI-4,5-P2 and the roles played in inter-organellar communication.
Vicinanza et al., show that PI-5-P is essential for autophagosome formation and overexpression studies showed that PI5P4Kγ localizes to the autophagosomes more frequently than both the PI5P4Kα and β isoforms. Knockdown of the kinases leads to the increase in the autophagosome formation, indicating that PI-4,5-P2 generation attenuates autophagosome biogenesis (Vicinanza et al., 2015). On the contrary another study showed that ATG16L1 interacts with PI-4,5-P2 on the PM to activate this process (Ravikumar et al., 2010). PI5P4Ks have also been shown to localize to the lysosome (Lundquist et al., 2018). Further, the loss of PI5P4Kα and PI5P4Kβ is sufficient to prevent the fusion of autophagosomes with lysosomes, which inhibits the process of autophagy and leads to accumulation of autophagosomes. This function of the PI5P4Ks requires a concomitant loss of p53 or a similar cellular stress (Lundquist et al., 2018). Together, these data indicate an important role for PI-4,5-P2 in the process of autolysosome formation. Moreover, a recent study showed that the endosomal sorting complex required for transport (ESCRT) is essential for lysosomal membrane repair (Gupta et al., 2021). These complexes are involved in membrane remodeling, which follows fusion events such as those between organelles. Interestingly, ESCRT complex proteins interact specifically with PI-4,5-P2 (McCullough et al., 2015), which could explain the importance of the lipid on the lysosomal membrane and its role in autophagy. For an in-depth discussion of PI-4,5-P2, and other phosphoinositide in autophagy, refer to Palamiuc et al. (2020).
Studies have also shown the localization of PI5P4Kα and the role of the PI5P4Ks in general at the peroxisomes. Peroxisomes, though highly essential for regulating various metabolic functions, are not well studied in mammalian cells. In the recent years, peroxisomes, their actions, as well as their role in regulating lipid metabolism have come to the forefront, bringing with them the PI5P4Ks and their product, PI-4,5-P2 into the spotlight.
PI5P4Ks as Key Regulators of Peroxisomal Functions by Sustaining PI-4,5-P2 Homeostasis
Peroxisomes are single-membraned organelles with multifaceted functions, ranging from ether lipid biosynthesis to fatty acid (FA) oxidation and reactive oxygen species (ROS) metabolism (He et al., 2021). They respond to metabolic and environmental cues, putting them at the center of various signaling nodes in the cell. Also, because of their roles, peroxisomes interact with and regulate other organellar functions such as the lysosomes, lipid droplets (LDs) and the mitochondria (He et al., 2021). Multiple studies have placed PI-4,5-P2 at the peroxisomes (Jeynov et al., 2006; Chu et al., 2015; Ravi et al., 2021). Furthermore, recent work has showed that this pool of PI-4,5-P2 is generated by PI5P4Kα (Hu et al., 2018). We, in a recent publication, were the first to physically localize PI5P4Kα to the peroxisome in the mouse prostate tissue (Ravi et al., 2021). Further, using imaging techniques, we showed the localization of PI-4,5-P2 to the peroxisomes, confirming the lipid blot data from Chu et al. We also categorically demonstrated that the knockout of the two most active isoforms of the Type II kinases, namely, PI5P4Kα and PI5P4Kβ were sufficient to deplete the peroxisomal pool of PI-4,5-P2 and this can be rescued by adding back the wild-type (WT) but not by the kinase-dead PI5P4Kα (Ravi et al., 2021).
PI-4,5-P2, as mentioned earlier, can bind to a wide range of proteins with PH- or C2- domains to mediate diverse cellular functions. Similarly, as shown by Chu et al., peroxisomal PI-4,5-P2 interacts with the C2 domain containing Synaptogamin VII (Syt7) on the lysosomal membrane to regulate cholesterol trafficking from the lysosome to the peroxisome. Their work revealed a previously unappreciated role for peroxisomes in cholesterol transport. While NPC1 (Niemann-Pick disease, type C1) on lysosomes and ABCD1 (ATP Binding Cassette Subfamily D Member 1) on peroxisomes are indispensable for the formation of lysosome-peroxisome membrane contact (LPMC), Syt7 allows for the stabilization of these sites by binding to PI-4,5-P2 on the peroxisome. Knockdown on Syt7 leads to accumulation of cholesterol in the lysosomes of the cells, uncovering a crucial role for the LPMC in cholesterol transport (Chu et al., 2015). In their following paper, they further demonstrated that knockdown of PI5P4Kα but not PI5P4Kβ or PI5P4Kγ leads to accumulation cholesterol in the lysosome, similar to the loss of Syt7 (Hu et al., 2018). Notably, we showed that the β isoform of the kinases is also able to rescue, albeit to a lesser extent than the α isoform, PI-4,5-P2 on peroxisomal membranes (Ravi et al., 2021). This correlates with the fact that PI5P4Kα is the most catalytically active of the isoforms and also may explain why Hu et al., did not observe a marked increase in cholesterol accumulation in the lysosome upon PI5P4Kβ or PI5P4Kγ knockdown. They also go on to show that while the loss of PI5P4Kα is sufficient to regulate peroxisomal PI-4,5-P2 it does not seem to affect lysosomal PI-4,5-P2 (Hu et al., 2018). This supports the role of the PI5P4Ks in autophagosome-lysosome fusion, which requires the loss of both the α and the β isoforms (Lundquist et al., 2018). Since both lysosomes and peroxisomes are sites of localization for the PI5P4Ks, it was also elegantly demonstrated in vitro that, peroxisomes and not lysosomes isolated from PI5P4Kα knockdown affect LPMC formation, conclusively showing the role of PI-4,5-P2 on the peroxisomes in this process (Chu et al., 2015). Further, similar to the PM-ER interaction, the PI-4,5-P2 on the peroxisomes also binds E-Syts on the ER, to traffic cholesterol. This lysosome-peroxisome-ER transport explains a previously not understood mechanism about the how exogenous cholesterol is trafficked from lysosomes to the ER (Xiao et al., 2019).
Other than cholesterol metabolism, peroxisomes are also sites of breakdown of very long chain fatty acids (VLCFAs) to medium chain fatty acids (MCFAs), known as peroxisomal β-oxidation (Poirier et al., 2006). Our study has shown a role for the PI5P4Ks in peroxisomal β-oxidation as seen by the change in peroxisomal gene signature upon loss of α and β isoforms (Ravi et al., 2021). For this process to occur, the peroxisomes need to take up VLCFAs, which are stored in the form of LDs. Concisely, peroxisome-LD interactions, which as with the LPMC formation, require ABCD1 on peroxisomal membranes and M1 Spastin on LDs, following which the FAs are trafficked across the membranes of the organelles (Chang et al., 2019). While loss of the PI5P4Ks does not affect the peroxisome’s interaction with LDs, they are no longer able to take up the FAs. This phenotype can be rescued by adding back the WT but not the kinase-dead PI5P4Kα, indicating that PI-4,5-P2 is essential for the trafficking event (Ravi et al., 2021). Chang et al., showed that upon tethering LDs to the peroxisomes, M1 Spastin is also responsible for recruiting ESCRT-III proteins to the surface, which is essential for FA uptake (Chang et al., 2019). Interestingly, as previously mentioned, ESCRT-III complex proteins preferentially associate with PI-4,5-P2. This could provide a possible explanation for the involvement of PI-4,5-P2 in regulating FA trafficking. The chain shortened FAs from peroxisomal β-oxidation are then utilized by mitochondria to break them down into carbon dioxide and water, in a process that generates ATP (Fransen et al., 2017). Studies have shown that peroxisomal storage and biogenesis disorders lead to mitochondrial dysfunction (Lismont et al., 2015; Tanaka et al., 2019). Similarly, knockdown of the PI5P4Ks leads to major structural and functional defects in the mitochondria downstream of peroxisome dysregulation (Ravi et al., 2021). These two organelles not only play a key role in lipid metabolism, but in redox mechanisms as well, tying them together in maintaining cellular homeostasis and PI-4,5-P2 generated by the PI5P4Ks is surfacing to be crucial in maintaining this balance. See Figure 1 and Table 1.
Health and Disease: PI5P4Ks Emerge as Exciting Targets
It is not surprising, considering the wide-ranging role PI-4,5-P2 plays, that PI5P4Ks will have such a drastic effect on cellular signaling and metabolism. Depending on the disease state, the activity of PI5P4Ks can be leveraged to bring about a change in the system. While targeting a kinase might seem like a daunting task, PI5P4Ks are a more feasible target. This is predominantly because their substrate, PI-5-P, mainly accumulates under conditions of stress. Our previous work demonstrated that PI5P4Ks are essential for tumor formation upon loss of p53, suggesting the PI5P4Ks are attractive targets for p53 mutant cancers (Emerling et al., 2013) and become relevant in regulating autophagosome-lysosome fusion under these conditions (Lundquist et al., 2018). Further, we have shown elevated expression of the PI5P4Ks in breast tumors compared to normal breast tissue (Emerling et al., 2013) and other studies have shown the impact of the kinases in breast cancer (Luoh et al., 2004; Keune et al., 2013). Similarly, PI5P4Ks have been demonstrated to be upregulated in several cancer subtypes, including glioblastomas, AML, and sarcomas (Fiume et al., 2015; Jude et al., 2015; Zhang et al., 2018; Lima et al., 2019; Shin et al., 2019; Ravi et al., 2021). Moreover, we recently illustrated the requirement of the PI5P4Ks for not only the establishment of sarcoma tumors but also in maintaining them, possibly through their role in regulating peroxisome-mitochondrial interplay (Ravi et al., 2021). While in diseases such as cancer, it might be necessary to inhibit the PI5P4Ks, other diseases/conditions might benefit from enhancing their activity. Accordingly, inhibiting autophagy is an appealing strategy in cancer therapeutics, whereas the reverse is true from an ageing standpoint. Studies have shown that PI-4,5-P2 levels are often lowered in neurodegenerative disorders, such as Alzheimer’s disease (Arancio, 2008). Lysosomal and peroxisomal storage disorders, encompass a large group of metabolic diseases, which are associated with the inability of the cells to properly process their cargo, such as cholesterol. These could serve as prime targets where we could possibly exploit the enzymatic function of the PI5P4Ks to our advantage. All things considered, in the recent past, PI5P4Ks and their product PI-4,5-P2 have risen from insignificance to being without a doubt one of the key metabolic sensors and regulators within the cell as well as pivotal players for inter-organelle communication necessary for cell survival. With drugs being developed against these kinases (Davis et al., 2013; Clarke et al., 2015; Al-Ramahi et al., 2017; Kitagawa et al., 2017; Manz et al., 2020; Sivakumaren et al., 2020; Chen et al., 2021), targeting them in the near future in various diseases is looking brighter.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author Contributions
AR and BE wrote the manuscript. LP and AR designed the artwork for the figure and table.
Funding
This work is supported by grants from the NCI and ACS (R01 CA237536 and RSG-20-064-01-TBE) to BE.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Al-Ramahi, I., Giridharan, S. S. P., Chen, Y. C., Patnaik, S., Safren, N., Hasegawa, J., et al. (2017). Inhibition of PIP4Kγ Ameliorates the Pathological Effects of Mutant Huntingtin Protein. Elife 6, e29123. doi:10.7554/eLife.29123
Andreev, J., Simon, J.-P., Sabatini, D. D., Kam, J., Plowman, G., Randazzo, P. A., et al. (1999). Identification of a New Pyk2 Target Protein with Arf-GAP Activity. Mol. Cel Biol 19, 2338–2350. doi:10.1128/mcb.19.3.2338
Auger, K. R., Serunian, L. A., Soltoff, S. P., Libby, P., and Cantley, L. C. (1989). PDGF-dependent Tyrosine Phosphorylation Stimulates Production of Novel Polyphosphoinositides in Intact Cells. Cell 57, 167–175. doi:10.1016/0092-8674(89)90182-7
Balla, T. (2013). Phosphoinositides: Tiny Lipids with Giant Impact on Cell Regulation. Physiol. Rev. 93, 1019–1137. doi:10.1152/physrev.00028.2012
Bazenet, C. E., Ruano, A. R., Brockman, J. L., and Anderson, R. A. (1990). The Human Erythrocyte Contains Two Forms of Phosphatidylinositol-4-Phosphate 5-kinase Which Are Differentially Active toward Membranes. J. Biol. Chem. 265, 18012–18022. doi:10.1016/s0021-9258(18)38264-4
Bulley, S. J., Clarke, J. H., Droubi, A., Giudici, M.-L., and Irvine, R. F. (2015). Exploring Phosphatidylinositol 5-phosphate 4-kinase Function. Adv. Biol. Regul. 57, 193–202. doi:10.1016/j.jbior.2014.09.007
Bultsma, Y., Keune, W.-J., and Divecha, N. (2010). PIP4Kβ Interacts with and Modulates Nuclear Localization of the High-Activity PtdIns5P-4-Kinase Isoform PIP4Kα. Biochem. J. 430, 223–235. doi:10.1042/bj20100341
Burke, J. E. (2018). Structural Basis for Regulation of Phosphoinositide Kinases and Their Involvement in Human Disease. Mol. Cel 71, 653–673. doi:10.1016/j.molcel.2018.08.005
Castano, E., Yildirim, S., Faberova, V., Krausova, A., Ulicna, L., Paprckova, D., et al. (2019). Nuclear Phosphoinositides-Versatile Regulators of Genome Functions. Cells 8, 649. doi:10.3390/cells8070649
Chang, C.-L., Weigel, A. V., Ioannou, M. S., Pasolli, H. A., Xu, C. S., Peale, D. R., et al. (2019). Spastin Tethers Lipid Droplets to Peroxisomes and Directs Fatty Acid Trafficking through ESCRT-III. J. Cel Biol 218, 2583–2599. doi:10.1083/jcb.201902061
Chen, S., Chandra Tjin, C., Gao, X., Xue, Y., Jiao, H., Zhang, R., et al. (2021). Pharmacological Inhibition of PI5P4Kalpha/beta Disrupts Cell Energy Metabolism and Selectively Kills P53-Null Tumor Cells. Proc. Natl. Acad. Sci. U S A. 118, e2002486118. doi:10.1073/pnas.2002486118
Chu, B.-B., Liao, Y.-C., Qi, W., Xie, C., Du, X., Wang, J., et al. (2015). Cholesterol Transport through Lysosome-Peroxisome Membrane Contacts. Cell 161, 291–306. doi:10.1016/j.cell.2015.02.019
Clarke, J. H., Emson, P. C., and Irvine, R. F. (2008). Localization of Phosphatidylinositol Phosphate Kinase IIγ in Kidney to a Membrane Trafficking Compartment within Specialized Cells of the Nephron. Am. J. Physiology-Renal Physiol. 295, F1422–F1430. doi:10.1152/ajprenal.90310.2008
Clarke, J. H., Giudici, M.-L., Burke, J. E., Williams, R. L., Maloney, D. J., Marugan, J., et al. (2015). The Function of Phosphatidylinositol 5-phosphate 4-kinase γ (PI5P4Kγ) Explored Using a Specific Inhibitor that Targets the PI5P-Binding Site. Biochem. J. 466, 359–367. doi:10.1042/bj20141333
Clarke, J. H., and Irvine, R. F. (2013). Evolutionarily Conserved Structural Changes in Phosphatidylinositol 5-phosphate 4-kinase (PI5P4K) Isoforms Are Responsible for Differences in Enzyme Activity and Localization. Biochem. J. 454, 49–57. doi:10.1042/bj20130488
Clarke, J. H., and Irvine, R. F. (2012). “Phosphatidylinositol 5-phosphate 4-kinase,” in Encyclopedia of Signaling Molecules. Editor S. Choi (New York, NY: Springer New York), 1369–1377.
Davis, M. I., Sasaki, A. T., Shen, M., Emerling, B. M., Thorne, N., Michael, S., et al. (2013). A Homogeneous, High-Throughput Assay for Phosphatidylinositol 5-phosphate 4-kinase with a Novel, Rapid Substrate Preparation. PLoS One 8, e54127. doi:10.1371/journal.pone.0054127
De Matteis, M. A., and D'Angelo, G. (2007). The Role of the Phosphoinositides at the Golgi Complex. Biochem. Soc. Symp. 74, 107–116. doi:10.1042/bss2007c10
De Matteis, M. A., Godi, A., and Corda, D. (2002). Phosphoinositides and the Golgi Complex. Curr. Opin. Cel Biol. 14, 434–447. doi:10.1016/s0955-0674(02)00357-5
Di Paolo, G., and De Camilli, P. (2006). Phosphoinositides in Cell Regulation and Membrane Dynamics. Nature 443, 651–657. doi:10.1038/nature05185
Emerling, B. M., Hurov, J. B., Poulogiannis, G., Tsukazawa, K. S., Choo-Wing, R., Wulf, G. M., et al. (2013). Depletion of a Putatively Druggable Class of Phosphatidylinositol Kinases Inhibits Growth of P53-Null Tumors. Cell 155, 844–857. doi:10.1016/j.cell.2013.09.057
Falkenburger, B. H., Dickson, E. J., and Hille, B. (2013). Quantitative Properties and Receptor reserve of the DAG and PKC branch of Gq-Coupled Receptor Signaling. J. Gen. Physiol. 141, 537–555. doi:10.1085/jgp.201210887
Falkenburger, B. H., Jensen, J. B., Dickson, E. J., Suh, B.-C., and Hille, B. (2010). SYMPOSIUM REVIEW: Phosphoinositides: Lipid Regulators of Membrane Proteins. J. Physiol. 588, 3179–3185. doi:10.1113/jphysiol.2010.192153
Fiume, R., Stijf-Bultsma, Y., Shah, Z. H., Keune, W. J., Jones, D. R., Jude, J. G., et al. (2015). PIP4K and the Role of Nuclear Phosphoinositides in Tumour Suppression. Biochim. Biophys. Acta (Bba) - Mol. Cel Biol. Lipids 1851, 898–910. doi:10.1016/j.bbalip.2015.02.014
Fransen, M., Lismont, C., and Walton, P. (2017). The Peroxisome-Mitochondria Connection: How and Why. Int. J. Mol. Sci. 18, 1126. doi:10.3390/ijms18061126
Freyberg, Z., Sweeney, D., Siddhanta, A., Bourgoin, S., Frohman, M., and Shields, D. (2001). Intracellular Localization of Phospholipase D1 in Mammalian Cells. MBoC 12, 943–955. doi:10.1091/mbc.12.4.943
Ghosh, A., Sharma, S., Shinde, D., Ramya, V., and Raghu, P. (2019). A Novel Mass Assay to Measure Phosphatidylinositol-5-Phosphate from Cells and Tissues. Biosci. Rep. 39, BSR20192502. doi:10.1042/BSR20192502
Giordano, F., Saheki, Y., Idevall-Hagren, O., Colombo, S. F., Pirruccello, M., Milosevic, I., et al. (2013). PI(4,5)P2-Dependent and Ca2+-Regulated ER-PM Interactions Mediated by the Extended Synaptotagmins. Cell 153, 1494–1509. doi:10.1016/j.cell.2013.05.026
Giudici, M.-L., Clarke, J. H., and Irvine, R. F. (2016). Phosphatidylinositol 5-phosphate 4-kinase γ (PI5P4Kγ), a Lipid Signalling enigma. Adv. Biol. Regul. 61, 47–50. doi:10.1016/j.jbior.2015.11.007
Godi, A., Santone, I., Pertile, P., Devarajan, P., Stabach, P. R., Morrow, J. S., et al. (1998). ADP Ribosylation Factor Regulates Spectrin Binding to the Golgi Complex. Proc. Natl. Acad. Sci. 95, 8607–8612. doi:10.1073/pnas.95.15.8607
Gupta, S., Yano, J., Mercier, V., Htwe, H. H., Shin, H. R., Rademaker, G., et al. (2021). Lysosomal Retargeting of Myoferlin Mitigates Membrane Stress to Enable Pancreatic Cancer Growth. Nat. Cel Biol 23, 232–242. doi:10.1038/s41556-021-00644-7
Harayama, T., and Riezman, H. (2018). Understanding the Diversity of Membrane Lipid Composition. Nat. Rev. Mol. Cel Biol 19, 281–296. doi:10.1038/nrm.2017.138
Hasegawa, J., Strunk, B. S., and Weisman, L. S. (2017). PI5P and PI(3,5)P2: Minor, but Essential Phosphoinositides. Cell Struct. Funct. 42, 49–60. doi:10.1247/csf.17003
He, A., Dean, J. M., and Lodhi, I. J. (2021). Peroxisomes as Cellular Adaptors to Metabolic and Environmental Stress. Trends Cel Biol. 31, 656–670. doi:10.1016/j.tcb.2021.02.005
Hu, A., Zhao, X.-T., Tu, H., Xiao, T., Fu, T., Wang, Y., et al. (2018). PIP4K2A Regulates Intracellular Cholesterol Transport through Modulating PI(4,5)P2 Homeostasis. J. Lipid Res. 59, 507–514. doi:10.1194/jlr.m082149
Itoh, T., Ijuin, T., and Takenawa, T. (1998). A Novel Phosphatidylinositol-5-Phosphate 4-Kinase (Phosphatidylinositol-Phosphate Kinase IIγ) Is Phosphorylated in the Endoplasmic Reticulum in Response to Mitogenic Signals. J. Biol. Chem. 273, 20292–20299. doi:10.1074/jbc.273.32.20292
Jeynov, B., Lay, D., Schmidt, F., Tahirovic, S., and Just, W. W. (2006). Phosphoinositide Synthesis and Degradation in Isolated Rat Liver Peroxisomes. FEBS Lett. 580, 5917–5924. doi:10.1016/j.febslet.2006.09.058
Jones, S. M., Howell, K. E., Henley, J. R., Cao, H., and McNiven, M. A. (1998). Role of Dynamin in the Formation of Transport Vesicles from the Trans-golgi Network. Science 279, 573–577. doi:10.1126/science.279.5350.573
Jost, M., Simpson, F., Kavran, J. M., Lemmon, M. A., and Schmid, S. L. (1998). Phosphatidylinositol-4,5-bisphosphate Is Required for Endocytic Coated Vesicle Formation. Curr. Biol. 8, 1399–1404. doi:10.1016/s0960-9822(98)00022-0
Jude, J. G., Spencer, G. J., Huang, X., Somerville, T. D. D., Jones, D. R., Divecha, N., et al. (2015). A Targeted Knockdown Screen of Genes Coding for Phosphoinositide Modulators Identifies PIP4K2A as Required for Acute Myeloid Leukemia Cell Proliferation and Survival. Oncogene 34, 1253–1262. doi:10.1038/onc.2014.77
Katan, M., and Cockcroft, S. (2020). Phosphatidylinositol(4,5)bisphosphate: Diverse Functions at the Plasma Membrane. Essays Biochem. 64, 513–531. doi:10.1042/ebc20200041
Keune, W.-J., Sims, A. H., Jones, D. R., Bultsma, Y., Lynch, J. T., Jirström, K., et al. (2013). Low PIP4K2B Expression in Human Breast Tumors Correlates with Reduced Patient Survival: A Role for PIP4K2B in the Regulation of E-Cadherin Expression. Cancer Res. 73, 6913–6925. doi:10.1158/0008-5472.can-13-0424
King, C. E., Hawkins, P. T., Stephens, L. R., and Michell, R. H. (1989). Determination of the Steady-State Turnover Rates of the Metabolically Active Pools of Phosphatidylinositol 4-phosphate and Phosphatidylinositol 4,5-bisphosphate in Human Erythrocytes. Biochem. J. 259, 893–896. doi:10.1042/bj2590893
King, C. E., Stephens, L. R., Hawkins, P. T., Guy, G. R., and Michell, R. H. (1987). Multiple Metabolic Pools of Phosphoinositides and Phosphatidate in Human Erythrocytes Incubated in a Medium that Permits Rapid Transmembrane Exchange of Phosphate. Biochem. J. 244, 209–217. doi:10.1042/bj2440209
Kitagawa, M., Liao, P.-J., Lee, K. H., Wong, J., Shang, S. C., Minami, N., et al. (2017). Dual Blockade of the Lipid Kinase PIP4Ks and Mitotic Pathways Leads to Cancer-Selective Lethality. Nat. Commun. 8, 2200. doi:10.1038/s41467-017-02287-5
Lima, K., Coelho-Silva, J. L., Kinker, G. S., Pereira-Martins, D. A., Traina, F., Fernandes, P. A. C. M., et al. (2019). PIP4K2A and PIP4K2C Transcript Levels Are Associated with Cytogenetic Risk and Survival Outcomes in Acute Myeloid Leukemia. Cancer Genet. 233-234, 56–66. doi:10.1016/j.cancergen.2019.04.002
Ling, L. E., Schulz, J. T., and Cantley, L. C. (1989). Characterization and Purification of Membrane-Associated Phosphatidylinositol-4-Phosphate Kinase from Human Red Blood Cells. J. Biol. Chem. 264, 5080–5088. doi:10.1016/s0021-9258(18)83702-4
Lismont, C., Nordgren, M., Van Veldhoven, P. P., and Fransen, M. (2015). Redox Interplay between Mitochondria and Peroxisomes. Front. Cel Dev. Biol. 3, 35. doi:10.3389/fcell.2015.00035
Lundquist, M. R., Goncalves, M. D., Loughran, R. M., Possik, E., Vijayaraghavan, T., Yang, A., et al. (2018). Phosphatidylinositol-5-Phosphate 4-Kinases Regulate Cellular Lipid Metabolism by Facilitating Autophagy. Mol. Cel 70, 531–544. doi:10.1016/j.molcel.2018.03.037
Luoh, S.-W., Venkatesan, N., and Tripathi, R. (2004). Overexpression of the Amplified Pip4k2β Gene from 17q11-12 in Breast Cancer Cells Confers Proliferation Advantage. Oncogene 23, 1354–1363. doi:10.1038/sj.onc.1207251
Mackey, A. M., Sarkes, D. A., Bettencourt, I., Asara, J. M., and Rameh, L. E. (2014). PIP4kγ Is a Substrate for mTORC1 that Maintains Basal mTORC1 Signaling during Starvation. Sci. Signal. 7, ra104. doi:10.1126/scisignal.2005191
Manz, T. D., Sivakumaren, S. C., Yasgar, A., Hall, M. D., Davis, M. I., Seo, H.-S., et al. (2020). Structure-Activity Relationship Study of Covalent Pan-Phosphatidylinositol 5-Phosphate 4-Kinase Inhibitors. ACS Med. Chem. Lett. 11, 346–352. doi:10.1021/acsmedchemlett.9b00402
Martin, T. F. J. (2015). PI(4,5)P2-binding Effector Proteins for Vesicle Exocytosis. Biochim. Biophys. Acta 1851, 785–793. doi:10.1016/j.bbalip.2014.09.017
McCrea, H. J., and De Camilli, P. (2009). Mutations in Phosphoinositide Metabolizing Enzymes and Human Disease. Physiology 24, 8–16. doi:10.1152/physiol.00035.2008
McCullough, J., Clippinger, A. K., Talledge, N., Skowyra, M. L., Saunders, M. G., Naismith, T. V., et al. (2015). Structure and Membrane Remodeling Activity of ESCRT-III Helical Polymers. Science 350, 1548–1551. doi:10.1126/science.aad8305
McLaughlin, S., Wang, J., Gambhir, A., and Murray, D. (2002). PIP2 and Proteins: Interactions, Organization, and Information Flow. Annu. Rev. Biophys. Biomol. Struct. 31, 151–175. doi:10.1146/annurev.biophys.31.082901.134259
Monier, S., Chardin, P., Robineau, S., and Goud, B. (1998). Overexpression of the ARF1 Exchange Factor ARNO Inhibits the Early Secretory Pathway and Causes the Disassembly of the Golgi Complex. J. Cel Sci 111 ( Pt 22) (Pt 22), 3427–3436. doi:10.1242/jcs.111.22.3427
Palamiuc, L., Ravi, A., and Emerling, B. M. (2020). Phosphoinositides in Autophagy: Current Roles and Future Insights. FEBS J. 287, 222–238. doi:10.1111/febs.15127
Pendaries, C., Tronchère, H., Plantavid, M., and Payrastre, B. (2003). Phosphoinositide Signaling Disorders in Human Diseases. FEBS Lett. 546, 25–31. doi:10.1016/s0014-5793(03)00437-x
Poirier, Y., Antonenkov, V. D., Glumoff, T., and Hiltunen, J. K. (2006). Peroxisomal β-oxidation-A Metabolic Pathway with Multiple Functions. Biochim. Biophys. Acta (Bba) - Mol. Cel Res. 1763, 1413–1426. doi:10.1016/j.bbamcr.2006.08.034
Poli, A., Zaurito, A. E., Abdul-Hamid, S., Fiume, R., Faenza, I., and Divecha, N. (2019). Phosphatidylinositol 5 Phosphate (PI5P): From behind the Scenes to the Front (Nuclear) Stage. Int. J. Mol. Sci. 20, 2080. doi:10.3390/ijms20092080
Rameh, L. E., Tolias, K. F., Duckworth, B. C., and Cantley, L. C. (1997). A New Pathway for Synthesis of Phosphatidylinositol-4,5-Bisphosphate. Nature 390, 192–196. doi:10.1038/36621
Ravi, A., Palamiuc, L., Loughran, R. M., Triscott, J., Arora, G. K., Kumar, A., et al. (2021). PI5P4Ks Drive Metabolic Homeostasis through Peroxisome-Mitochondria Interplay. Develop. Cel 56, 1661–1676. doi:10.1016/j.devcel.2021.04.019
Ravikumar, B., Moreau, K., Jahreiss, L., Puri, C., and Rubinsztein, D. C. (2010). Plasma Membrane Contributes to the Formation of Pre-autophagosomal Structures. Nat. Cel Biol 12, 747–757. doi:10.1038/ncb2078
Sarkes, D., and Rameh, L. E. (2010). A Novel HPLC-Based Approach Makes Possible the Spatial Characterization of Cellular PtdIns5P and Other Phosphoinositides. Biochem. J. 428, 375–384. doi:10.1042/bj20100129
Shin, Y. J., Sa, J. K., Lee, Y., Kim, D., Chang, N., Cho, H. J., et al. (2019). PIP4K2A as a Negative Regulator of PI3K in PTEN-Deficient Glioblastoma. J. Exp. Med. 216, 1120–1134. doi:10.1084/jem.20172170
Sivakumaren, S. C., Shim, H., Zhang, T., Ferguson, F. M., Lundquist, M. R., Browne, C. M., et al. (2020). Targeting the PI5P4K Lipid Kinase Family in Cancer Using Covalent Inhibitors. Cel Chem. Biol. 27, 525–537. doi:10.1016/j.chembiol.2020.02.003
Sobol, M., Krausová, A., Yildirim, S., Kalasová, I., Fáberová, V., Vrkoslav, V., et al. (2018). Nuclear Phosphatidylinositol 4,5-bisphosphate Islets Contribute to Efficient RNA Polymerase II-dependent Transcription. J. Cel Sci 131, jcs211094. doi:10.1242/jcs.211094
Sobol, M., Yildirim, S., Philimonenko, V. V., Marášek, P., Castaño, E., and Hozák, P. (2013). UBF Complexes with Phosphatidylinositol 4,5-bisphosphate in Nucleolar Organizer Regions Regardless of Ongoing RNA Polymerase I Activity. Nucleus 4, 478–486. doi:10.4161/nucl.27154
Sun, H. Q., Yamamoto, M., Mejillano, M., and Yin, H. L. (1999). Gelsolin, a Multifunctional Actin Regulatory Protein. J. Biol. Chem. 274, 33179–33182. doi:10.1074/jbc.274.47.33179
Tan, X., Thapa, N., Choi, S., and Anderson, R. A. (2015). Emerging Roles of PtdIns(4,5)P2 - beyond the Plasma Membrane. J. Cel Sci 128, 4047–4056. doi:10.1242/jcs.175208
Tanaka, H., Okazaki, T., Aoyama, S., Yokota, M., Koike, M., Okada, Y., et al. (2019). Peroxisomes Control Mitochondrial Dynamics and the Mitochondrion-dependent Apoptosis Pathway. J. Cel Sci 132, jcs224766. doi:10.1242/jcs.224766
van Meer, G., Voelker, D. R., and Feigenson, G. W. (2008). Membrane Lipids: where They Are and How They Behave. Nat. Rev. Mol. Cel Biol 9, 112–124. doi:10.1038/nrm2330
Varnai, P., Thyagarajan, B., Rohacs, T., and Balla, T. (2006). Rapidly Inducible Changes in Phosphatidylinositol 4,5-bisphosphate Levels Influence Multiple Regulatory Functions of the Lipid in Intact Living Cells. J. Cel Biol 175, 377–382. doi:10.1083/jcb.200607116
Vicinanza, M., Korolchuk, V. I., Ashkenazi, A., Puri, C., Menzies, F. M., Clarke, J. H., et al. (2015). PI(5)P Regulates Autophagosome Biogenesis. Mol. Cel 57, 219–234. doi:10.1016/j.molcel.2014.12.007
Xiao, J., Luo, J., Hu, A., Xiao, T., Li, M., Kong, Z., et al. (2019). Cholesterol Transport through the Peroxisome-ER Membrane Contacts Tethered by PI(4,5)P2 and Extended Synaptotagmins. Sci. China Life Sci. 62, 1117–1135. doi:10.1007/s11427-019-9569-9
Yoneda, A., Kanemaru, K., Matsubara, A., Takai, E., Shimozawa, M., Satow, R., et al. (2020). Phosphatidylinositol 4,5-bisphosphate Is Localized in the Plasma Membrane Outer Leaflet and Regulates Cell Adhesion and Motility. Biochem. Biophysical Res. Commun. 527, 1050–1056. doi:10.1016/j.bbrc.2020.05.040
Keywords: phosphoinositides, phosphotidylinositol-5-phosphate-4-kinases, PI-4, 5-P2, organelle, metabolism, peroxisomes, lipids
Citation: Ravi A, Palamiuc L and Emerling BM (2022) Crucial Players for Inter-Organelle Communication: PI5P4Ks and Their Lipid Product PI-4,5-P2 Come to the Surface. Front. Cell Dev. Biol. 9:791758. doi: 10.3389/fcell.2021.791758
Received: 08 October 2021; Accepted: 24 November 2021;
Published: 07 January 2022.
Edited by:
Joe Costello, University of Exeter, United KingdomReviewed by:
Masashi Maekawa, Keio University, JapanRaghu Padinjat, National Centre for Biological Sciences, India
Copyright © 2022 Ravi, Palamiuc and Emerling. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brooke M. Emerling, YmVtZXJsaW5nQHNicGRpc2NvdmVyeS5vcmc=
 Archna Ravi
Archna Ravi Brooke M. Emerling
Brooke M. Emerling