- 1Department of Clinical Pharmacy, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Clinical Research Center, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 3Department of General Surgery, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
Recently, N6-methyl-adenosine (m6A) ribonucleic acid (RNA) modification, a critical and common internal RNA modification in higher eukaryotes, has generated considerable research interests. Extensive studies have revealed that non-coding RNA m6A modifications (e.g. microRNAs, long non-coding RNAs, and circular RNAs) are associated with tumorigenesis, metastasis, and other tumour characteristics; in addition, they are crucial molecular regulators of cancer progression. In this review, we discuss the relationship between non-coding RNA m6A modification and cancer progression from the perspective of various cancers. In particular, we focus on important mechanisms in tumour progression such as proliferation, apoptosis, invasion and metastasis, tumour angiogenesis. In addition, we introduce clinical applications to illustrate more vividly that non-coding RNA m6A modification has broad research prospects. With this review, we aim to summarize the latest insights and ideas into non-coding RNA m6A modification in cancer progression and targeted therapy, facilitating further research.
1 Introduction
In eukaryotic cells, some non-coding ribonucleic acids (ncRNAs), such as microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), usually do not encode proteins but perform their respective biological functions at the RNA level. Generally, ncRNAs have long been thought to be post-transcriptional regulators of gene expression, but RNA modifications have rendered it possible for ncRNAs to generate new regulatory effects on gene expression. N6-methyl-adenosine (m6A), methylated at the N6 position of adenosine, is a reversible epigenetic RNA modification that modulates splicing, degradation, and other biological processes of RNAs (Fazi and Fatica 2019; Zhou et al., 2020). During the process from DNA to RNA, adenylate undergoes methylation modification at the sixth position under the action of methyltransferase (“writer”) METTL3/14, WTAP, KIAA1429, RBM15, and ZC3H13. Next, the bases are demethylated by demethyltransferase (“eraser”) FTO and ALKBH. Finally, these methylated RNA base sites require specific enzymes (“readers”) to accomplish the purpose of embellishing ncRNAs (Sun et al., 2019). The molecular mechanisms of m6A modification involved in the regulation of ncRNA gene expression have been reported previously (Erson-Bensan and Begik 2017; Fazi and Fatica 2019; He et al., 2020; Zhang et al., 2020d), and more interestingly, the abundance of m6A modification and the expression of m6A regulators are also regulated by ncRNAs.
The genes encoding miRNA are transcribed as long transcripts, called primary miRNA (pri-miRNA) in the nucleus. Pri-miRNA undergoes nuclear cleavage mediated by Drosha-DGCR8 complex, to form precursor miRNA (pre-miRNA), which is then transported to cytoplasm via Exportin-5 and sheared by RNase Ⅲ endonuclease Dicer to short RNA fragments, namely miRNA (Vishnoi and Rani 2017). Studies have shown that pri-miRNAs methylated by m6A are more likely to be identified by molecules that are responsible for miRNA maturation, thus accelerating the initiation, splicing, transportation and other processes of miRNA biogenesis (Alarcón et al., 2015a; Alarcón et al., 2015b; Chen et al., 2020c) (see Figure 1A). For example, the m6A “writer” METTL3, marked in pri-miRNA, has been reported to interact with DGCR8 and to positively promote the splicing and maturation of miR-25 (Alarcón et al., 2015b), miR-143-3p (Wang et al., 2019a), pri-miR-221/miR-222 (Han et al., 2019a), pri-miR-1246 (Peng et al., 2019), and miR-25-3p (Zhang et al., 2019a). Further, the m6A “reader” HNRNPC can directly bind pri-miR-21 to increase miR-21 expression (Park et al., 2012), whereas HNRNPA2B1 interacts with lncRNA LINC01234 to indirectly modulate miR-106b-5p maturation (Chen et al., 2020d). Inconsistent with the above findings, a study has shown that HNRNPA2B1 overexpression downregulates miR-29a-3p, miR-29b-3p, miR-222, and inversely upregulates miR-1266-5p, miR-1268a, and miR-671-3p (Klinge et al., 2019). Notably, the methyltransferase NSun2 blocks the splicing of pri-miR-125b2 and interferes with miR-125b cleavage, which is in contrast to the action of methyltransferase METTL3/14 (Yang et al., 2015; Yuan et al., 2014). These data suggest the extraordinary complexity and diversity of m6A modification in miRNA biosynthesis.
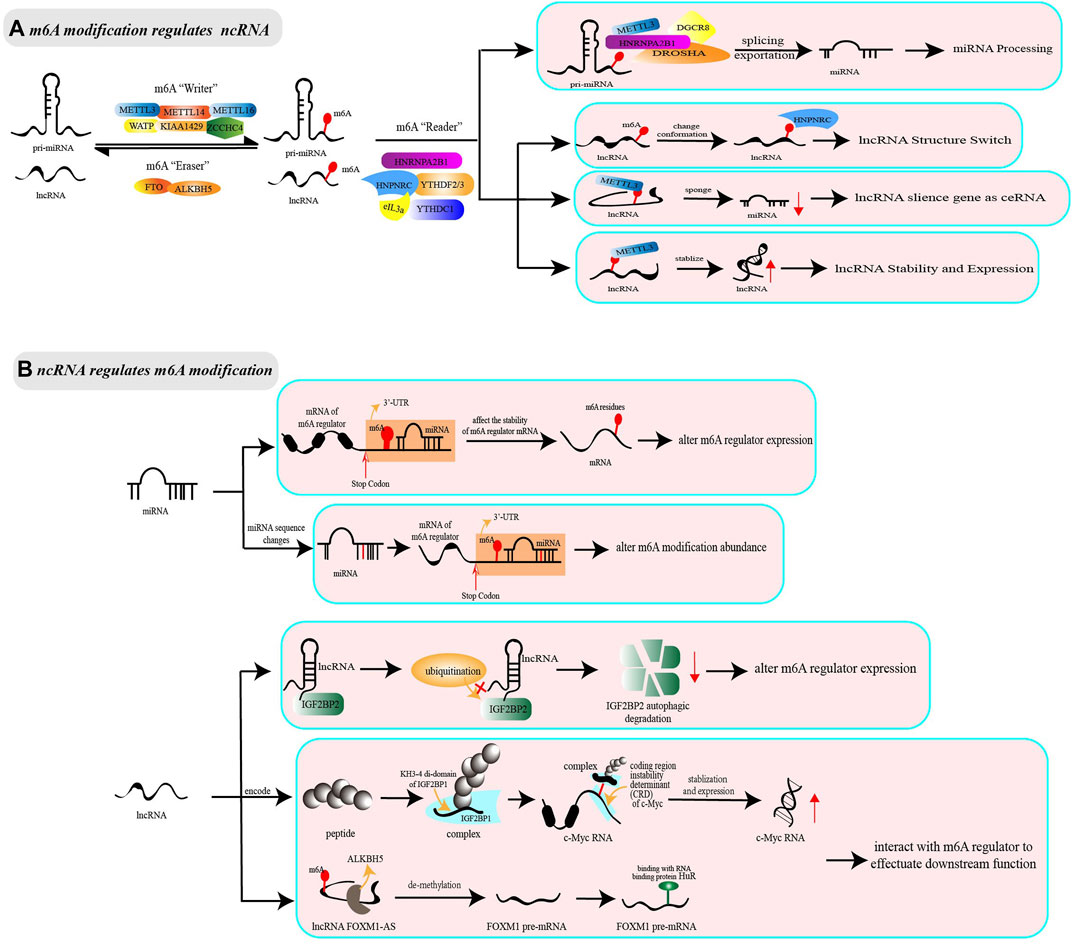
FIGURE 1. The interaction between non-coding RNA and m6A modification. The adenosine(A) bases reside in non-coding RNA could be methylated by methyltransferase complex (“Writer”) comprised of METTL3/METTL14/WATP and other regulator cofactors. The non-coding RNA with m6A modification resides could by recognized by m6A binding proteins (“Reader,” e.g. NRNPA2B1, HNRNPC, YTHDF) to effectuate downstream functions, or be reversibly erased by demethyltransferase (“Eraser,” e.g. FTO, ALKBH5). (A) m6A modification regulates ncRNA. During the processing from pri-miRNA to miRNA, the presence of m6A modification can regulate the splicing and exportation, facilitating the maturation of miRNA. Besides, m6A modification can play extraordinary complex and diversity roles in lncRNA. The lncRNA marked m6A writer proteins could change conformation to bind to proteins, leading to that m6A modification is regarded as lncRNA structure switch and facilitate the RNA-protein interaction. Secondly, the lncRNA which is methylated by m6A writer proteins have the effect on sponging miRNA as ceRNA, greatly augmenting the RNA-RNA interaction. For lncRNA itself, m6A modification could regulate transcript stabilization and expression level to activate or participate in subsequent functions. (B) ncRNA regulates m6A modification. Non-coding RNA could regulate m6A modification from the perspective of expression level and function. MiRNA integrate into the mRNA 3′-UTR region of m6A regulator cofactors, which alters the expression level of m6A regulators and indirectly influences on the abundance of m6A modification. Differently, lncRNA can alter the expression level of m6A regulators by affecting the stability and degradation of mRNA. Notably, lncRNA or RNA-binding regulatory peptide that is encoded by lncRNA (e.g. LncRNA LINC00266-1) has the capability to bind with m6A regulator to strengthen m6A recognition, therefore affecting the stabilization and expression of the downstream target mRNA. Anti-sense lncRNA could recruit m6A eraser proteins to decrease m6A abundance on the sense mRNA to active downstream effects (e.g. lncRNA FOXM1-AS and m6A eraser ALKBH5).
LncRNA is a kind of ncRNA with a length of more than 200 nucleotides, but was initially considered as “transcriptional noise” because its biological functions were unknown (Yang et al., 2020b). With further research, increasing associated machineries of lncRNA have been found. The current studies indicate that m6A modification can act as a structural “switch” to change the conformation of lncRNA, participate in the ceRNA model for silencing miRNA or affect the stability and expression of lncRNA (see Figure 1A). Specifically, m6A-modified lncRNA is demonstrated to have hairpins that are more suitable for conformation in the RNA-HNRNPC complex, suggesting that m6A modification acts as a “switch” to trigger the combination of lncRNA and HNRNPC (Liu et al., 2015; Zhou et al., 2016). In addition, m6A modification can facilitate lncRNA to act as endogenous RNAs (ceRNAs) and then sponge downstream miRNA by improving the stability of lncRNA transcript or reducing RNA degradation (Jin et al., 2019; Wang et al., 2020a). Notably, m6A modification contributes to the function of lncRNAs, as well as affects their expression level. It is reported that YTHDF1 knockdown downregulates LINC00278-sORF1 translation without changing the m6A modification level. METTL3, METTL14, and WTAP knockdown reduces the m6A modification level and LINC00278-sORF1 translation, while ALKBH5 increases them (Wu et al., 2020).
The interaction between m6A modification and ncRNAs is reciprocal, which means that ncRNAs can also regulate m6A modification in biological processes. Some miRNAs can target the mRNA of m6A regulators and integrate into the 3′-UTR region of mRNA to alter their stability and expression, thus indirectly affecting m6A modification abundance. Further, lncRNA can affect the stability and degradation of m6A-related enzymes or combine with them to form complexes, thus facilitating a regulatory effect on the downstream target mRNA of m6A regulators (see Figure 1B). For example, miR-33a, miR-600, miRNA let-7g, miR-744-5p, and miRNA-145 are shown to decrease the expression of METTL3, HNRNPC or YTHDF2 at both the mRNA and protein levels (Cai et al., 2018; Du et al., 2017; Kleemann et al., 2018; Wei et al., 2019; Yang et al., 2017). Furthermore, the IGF2BP2 mRNA is regulated by lncRNA LINRIS, which is not only responsible for maintaining IGF2BP2 mRNA stability but also prevents its degradation (Wang et al., 2019b). Except for changing the expression level of m6A regulators, ncRNA can change m6A abundance via a sequence-dependent manner. For example, the decrease of endonuclease Dicer leads to aberrant miRNA expression, and modulates the binding of m6A-related enzymes to mRNAs that contain miRNA binding sites, thus downregulating the abundance of m6A modification without affecting the expression of m6A regulators, METTL3, FTO, and ALKBH5 (Chen et al., 2015).
So far, several studies have enriched our understanding of the interactions between ncRNA and m6A modification, and indicated that aberrant expression of m6A regulators and m6A modification on ncRNAs can alter normal biological processes. Notably, this abnormal biological change caused by m6A modification makes ncRNAs involved in tumorigenesis and cancer progression, associating with cell proliferation and apoptosis, invasion and metastasis, cell stemness, drug resistance and other mechanisms that enhance the malignancy of cells and the difficulty of cancer therapies. Therefore, the role of ncRNA m6A modification in various cancers is worthy of further study to better understand the related mechanisms, contributing to provide insights for early cancer diagnosis, outcome prediction and cancer treatment strategies. In this review, we discuss the relationship between ncRNA m6A modification and cancer progression from the perspective of various cancers. In particular, we focus on important mechanisms in tumour progression and introduce potential clinical applications to illustrate more vividly that ncRNA m6A modification has broad research prospects in cancer. We aim to summarize the latest insights into ncRNA m6A modification in cancer progression and targeted therapy, facilitating further research.
2 Functions of ncRNA m6A Modification in Different Types of Cancer
2.1 Lung Cancer and ncRNA m6A Modification
Lung cancer, also known as primary bronchial lung cancer, refers to malignant tumour originating in bronchial mucous epithelium or alveolar epithelium, and is mainly classified into adenocarcinoma, squamous cell carcinoma, large cell carcinoma and small-cell lung cancer. Due to the huge difference between small cell carcinoma and other types in biological behaviour, treatment prognosis and other aspects, lung cancer other than small cell lung cancer is collectively referred to as non-small-cell lung cancer (NSCLC) (Yousef and Tsiani 2017; Inamura 2018). During the progression of NSCLC, LINC01234 is reported to be an oncogenic lncRNA that interacts with m6A “reader” HNRNPA2B1. Overexpression of LINC01234 combined with HNRNPA2B1 recruits DGCR8 and facilitates the processing of several miRNA precursors, including pri-miR-106b. Interestingly, activated c-Myc by miR-106b-5p can bind to the LINC01234 promoter to create a positive feedback loop (Chen et al., 2020d). Contrast to interaction, METTL3 accelerates the splicing of the precursor and generates mature miR-143-3p, regulating the expression of oncogene miR-143-3p (Wang et al., 2019a). Similarly, the miR-107/LAST2 axis is regulated by the m6A “eraser” ALKBH5 in an HuR-dependent manner to decrease YAP activity and inhibit tumour growth (Jin et al., 2020); lncRNA MALAT1 is stabilized by the METTL3/YTHDF1 complex and its RNA level is increased with high levels of m6A modification (Jin et al., 2019). METTL3 has been shown to promote the expression of some crucial oncoproteins and facilitate tumour proliferation, apoptosis, and invasion in human lung cancer (Du et al., 2017). However, miR-600 and miR-33a inhibit the expression of METTL3, reversing its positive effects on NSCLC progression and playing the role of tumour suppressor genes (Du et al., 2017; Wei et al., 2019). In summary, the effect of m6A modification on cancers may be completely opposite under the regulation of ncRNAs, suggesting the relationship between ncRNA and m6A modification is complex and variable in cancer progression.
2.2 Liver Cancer and ncRNA m6A Modification
Primary carcinoma of the liver has multifactorial pathogenesis and is a malignant tumour occurring in hepatocytes or intrahepatic bile duct epithelial cells, including hepatocellular carcinoma (HCC), hepatobiliary carcinoma and hepatic sarcoma (Gao et al., 2020). HCC occurs in liver cells and is the main histological subtype of the hepatic malignancy (accounting for more than 90% cases of primary carcinoma of the liver). Overwhelming evidence has proved the regulatory roles of ncRNAs related to liver carcinogenesis, and their relationship can be summed up as “frenemy” which is means a kind of love-hate relationship (Wong et al., 2018). The miR-186, which plays the role of anti-hepatoblastoma, is weakly expressed in HCC tissues, and its overexpression inhibits cell aggressive characteristics; however, the increase of its direct target METTL3 reverses the inhibitory effect (Cui et al., 2020). In addition, another study has shown that METTL3 can improve the expression of c-Myc by increasing m6A modification, whereas miR-338-5p inhibit the expression of METTL3 to interfere with the lung cancer progression (Wu et al., 2021). The m6A “writer” METTL14 positively modulate pri-miRNA-126 maturation by interacting with DGCR8 to suppress the metastatic potential of HCC cells. Similar to the interaction of miR-186 and METTL3, the overexpression of pri-miRNA-126 inversely inhibits the repressing effect of METTL14 (Ma et al., 2017).
Thus, hostility alters their normal biological functions, while the friendship represents a positive correlation between ncRNAs and m6A regulators. With the help of m6A modification mediated by METTL3/METTL14, lncRNA LINC00958 (Zuo et al., 2020)and circ-SORE (Xu et al., 2020a)are upregulated because of enhancement of transcript stability. The m6A “reader” IGF2BP1 plays an oncogenic role in HCC progression, but the decrease of lncRNA LIN28B-AS can deplete the expression of IGF2BP1-dependent mRNAs, such as IGF2 and Myc, thereby inhibiting HCC cell proliferation, migration, and invasion (Zhang et al., 2020c). In addition to influencing the expression of molecules, m6A modification and ncRNA also affect their binding to downstream molecules. KIAA1429 is a member of m6A methyltransferases, and GATA3 mRNA is a direct downstream target of KIAA1429-methylation, facilitating the degradation of GATA3. Strikingly, KIAA1429 preferentially induces GATA3 pre-mRNA in a targeted manner under the guidance of the antisense gene lncRNA GATA3-AS to maintain their roles in cancer progression (Lan et al., 2019). In addition, m6A modification targets the 3ʹ-UTR of YAP, which induces the repression effect of miR-582-3p, although whether the m6A regulator is primarily responsible for this essential part of the interaction is unclear until now (Zhang et al., 2018). The complexity of m6A modification and ncRNAs suggest that it is necessary to further study the mechanisms and effects of their crosstalk in liver cancer, thus using the stable and significant indicators to assist early diagnosis and treatment.
2.3 Gastric Cancer and ncRNA m6A Modification
Gastric cancer (GC) is a malignant tumour of digestive system occurring from gastric mucosal epithelium and glandular epithelium (Karimi et al., 2014), accounting for the second largest malignant tumour in China. In GC tissues, it is found that MEETL3 promotes the maturation of pri-miR-17-92 (Sun et al., 2020b), and m6A-modified motif-assisted miR-660 to reduce oncogene E2F3 activity (He and Shu 2019). Besides, METTL3 can upregulate the expression of target mRNA SEC62 by facilitating the stabilizing effect of IGF2BP1 on SEC62, but this positive regulation could be inhibited by miR-4429 (He et al., 2019). In contrast, lncRNA ARHGAP5-AS1 is responsible for recruiting METTLE3 to stabilise ARHGAP5 mRNA in the cytoplasm (Zhu et al., 2019). Unlike enhancing downstream target mRNA stability, METTL3 has been reported to combine with the m6A “reader” YTHDF2 to promote the degradation of PTEN mRNA and increase tumour malignancy, and subtly, the oncogenic lncRNA LINC00470 serves as an accelerator in this process (Yan et al., 2020). Many studies have reported that aberrant expressions of lncRNA (Sun et al., 2016), miRNA (Shin and Chu 2014), and circRNA (Shan et al., 2019) play important roles in GC progression and could be regarded as diagnostic/prognostic markers and chemotherapeutic tools. Compared with the complex change of m6A modification in liver cancer, most ncRNAs and protein genes related to m6A are upregulated in GC tissues, suggesting that the m6A-related risk score might be informative for risk assessment and prognostic stratification (Guan et al., 2020).
2.4 Bladder Cancer and ncRNA m6A Modification
Bladder cancer is a morbid malignancy of the urinary tract, and 95% of bladder tumors originate from epithelial tissue. Notably, the symptoms of bladder cancer mimic those of a urinary tract infection, increasing the diagnosis difficulty and delaying timely therapy (Li et al., 2019c). Accumulating evidence implies that the abnormal m6A modification of ncRNA is related to tumorigenicity and poor prognosis in bladder cancer. Oncogenic METTL3 facilitates DGCR8 to recognise pri-miRNA and accelerate the maturation of pri-miR221/222, which is targeted to PTEN (Han et al., 2019a). Differently from the direct regulation, the m6A “eraser” FTO regulates the expression of MALAT/miR-384/MAL2 axis by catalysing MALAT1 demethylation in an m6A-dependent manner, indicating the potential of FTO as a diagnostic and prognostic biomarker (Tao et al., 2021). Low expression of METTL14 and decreased global m6A abundance in bladder cancer tissue are also associated with the clinical severity and outcomes of bladder cancer. METTL14 knockout promotes the proliferation, self-renewal, metastasis, and initiation of tumour cells, while overexpression plays an adverse role (Gu et al., 2019). These m6A-related molecules may play important roles in the clinical diagnosis and treatment of bladder cancer in the future.
2.5 Breast Cancer and ncRNA m6A Modification
Breast cancer is an epithelial malignant tumour originating from the terminal ductal lobule unit of the breast. The breast epithelial cells have unlimited replicative potential and other malignant characteristics because of various carcinogenic factors (Nagini 2017). The hostile hypoxic microenvironment is a major reason for the rapid expansion of cancer cells. It has been reported that the lncRNA KB-1980E6.3 that is associated with hypoxia can recruit m6A “reader” IGF2BP1 to stabilize c-Myc mRNA and enhance the malignant characteristics (Zhu et al., 2021). In endocrine-resistant LCC9 breast cancer cells, HNRNPA2B1 expression is higher than that in parental and tamoxifen-sensitive cells, and this increase alters the transcriptome and expression of miRNAs, because HNRNPA2B1 is a “reader” of the m6A mark in pri-miRNAs and is responsible for promoting DROSHA processing of pre-miRNAs (Klinge et al., 2019). Similarly, m6A methyltransferase METTL14 has been demonstrated to be significantly increased in breast cancer tissues with the function of reshaping miRNA profile of cancer cell lines. It has been verified that hsa-miR-146a-5p is a differentially expressed miRNA, modulated by METTL14-induced m6A modification, and thus affects the migration and invasion of breast cancer cells (Yi et al., 2020). In addition, METTL14 is regulated by the oncogenic lncRNA LNC942 to facilitate cell growth and cancer progression. Mechanistically, LNC942 recruits the METTL14 protein and elevated post-transcriptional METTL14-mediated m6A modification levels, thereby stabilising the expression of downstream targets (Sun et al., 2020a). In human epidermal growth factor receptor 2 (HER2)-positive breast cancer, it is reported that m6A “eraser” FTO inhibits miR-181b-3p and upregulates the expression of oncogenic ARL5B (Xu et al., 2020b). Overall, more extensive and in-depth researches on the aberrant m6A modification of ncRNA for the carcinogenesis and development of breast cancer are required.
2.6 Colorectal Cancer and ncRNA m6A Modification
Carcinoma of large intestine is a malignant gastrointestinal tract tumour that originates from large intestine mucous membrane epithelium and gland with a high metastasis and reoccurrence rate, including colon cancer and carcinoma of the rectum (Muller et al., 2016). Most researchers focus on the relationship between ncRNA m6A modification and metastasis of colorectal cancer (CRC). A novel lncRNA RP11-138 J23.1 (RP11) is demonstrated to positively regulate epithelial-mesenchymal transition (EMT) in CRC cells. Mechanistically, the expression of RP11 is upregulated in an m6A modification-dependent manner; then, the complex composed by RP11 and HNRNPA2B1 prevents Zeb1 degradation, triggering the dissemination of cancer cells (Wu et al., 2019). LncRNA GAS5 promotes YAP ubiquitin-mediated degradation, but YTHDF3 alleviates this effect via lncRNA GAS5 decay, confirming that YTHDF3 is not only a novel target of YAP but also a significant player in YAP signalling by facilitating lncRNA GAS5 degradation (Ni et al., 2019). Overexpression of METTL14 is also reported to correlate with the YAP pathway and can suppress CRC cell growth and metastasis via the miR-375/YAP1 pathway (Chen et al., 2020b). Besides, the expression of oncogene lncRNA XIST negatively correlates with YTHDF2 and METTL14 in CRC tissues because of the m6A modification (Yang et al., 2020c). In contrast, YTHDC1 recognises circNSUN2 and export it to the cytoplasm, eventually participating in cancer progression by forming the cir-cNSun2/IGF2BP2/HMGA2 complex (Chen et al., 2019a). In addition, the oncogene lncRNA LINC00460 combines with IGF2BP2 and upregulates the expression of HMGA1 mRNA and increase its stability, participating in cancer development (Hou et al., 2021). The m6A “reader” YTH protein family and IGF2BP2 have been widely reported in colorectal cancer, and therefore it is reasonable to speculate that the m6A “reader” may play an important role in the progression of colorectal cancer, which deserves deeply study.
2.7 Other Cancers and ncRNA m6A Modification
Abnormal m6A modification of ncRNAs is also associated with the progression of other cancers. In clear cell renal cell carcinoma, Gu et al. find a novel DNA methylation-deregulated and RNA m6A reader-cooperating lncRNA (DMDRMR), which interacts with IGF2BP3 to stabilize target genes and plays a carcinogenic role (Gu et al., 2021). In thyroid cancer, lncRNA MALAT1 competitively binds to miR-204, upregulates IGF2BP2 and enhances MYC expression in m6A-dependent manner, conferring a stimulatory effect on cancer progression (Ye et al., 2021). In osteosarcoma, ALKBH5 inhibits the degradation of lncRNA PVT1 and leads to the overexpression of lncRNA PVT1. Overexpressed PVT1 can suppress the binding of YTHDF2 with PVT1 to promote cancer cell proliferation (Chen et al., 2020a). This combination of the m6A “reader” and lncRNA, which affects cancer cell proliferation, also occurs in pancreatic cancer. IGF2BP2 and lncRNA DANCR have been reported to interact and promote the cancer stemness-like properties of pancreatic cancer. The adenosine at 664 of DANCR is modified by N6-methyladenosine, and IGF2BP2 serves as a reader for this structural change to stabilise DANCR RNA and promote cell proliferation (Hu et al., 2020). In pancreatic duct epithelial cells, oncogenic miR-25-3p is correlated with poor prognosis in patients with pancreatic cancer. One study has found that NF-κB-associated protein induced by m6A modification facilitates excessive pri-miR-25 maturation under the environment of cigarette smoke condensate; then it activates the oncogenic AKT-p70S6K signalling and provokes malignancy of pancreatic cancer (Zhang et al., 2019a). In head and neck squamous cell carcinoma, oncogenic LNCAROD affects the degree of tumour malignancy. METTL3-and METTL14-induced dysregulated m6A modification could account for the aberrant expression of LNCAROD and lead to the malignant behaviour of tumour cells (Ban et al., 2020). Moreover, Zheng et al. propose that m6A is highly enriched in oncogenic lncRNA FAM225A and enhances RNA stability, contributing to cell proliferation, migration, invasion, and metastasis (Zheng et al., 2019). Similarly, m6A modification improves the transcript stability of lncRNA RHPN1-AS1 and upregulates its expression in epithelial ovarian cancer tissues, facilitating RHPN1-AS1 to act as an oncogene (Wang et al., 2020a). In glioblastoma, it has been found that 13 central m6A methylation regulators are associated with the clinical and molecular phenotype, suggesting that m6A regulators are important participants in malignant progression (Du et al., 2020b). A more specific example is HNRNPC, which directly binds to pri-miR-21 and promotes its expression, playing a role in the metastatic potential of the glioblastoma cell lines. The experimental results show that the expression levels of HNRNPC are higher in highly aggressive cancer cells and are elevated along with the brain tumour grade (Park et al., 2012). In the cancers listed above, the abundance of m6A modification and the enzymes involved in it can affect ncRNA maturation and activation and change their biological functions, which in turn can impact the progression of various cancers (see Figure 2).
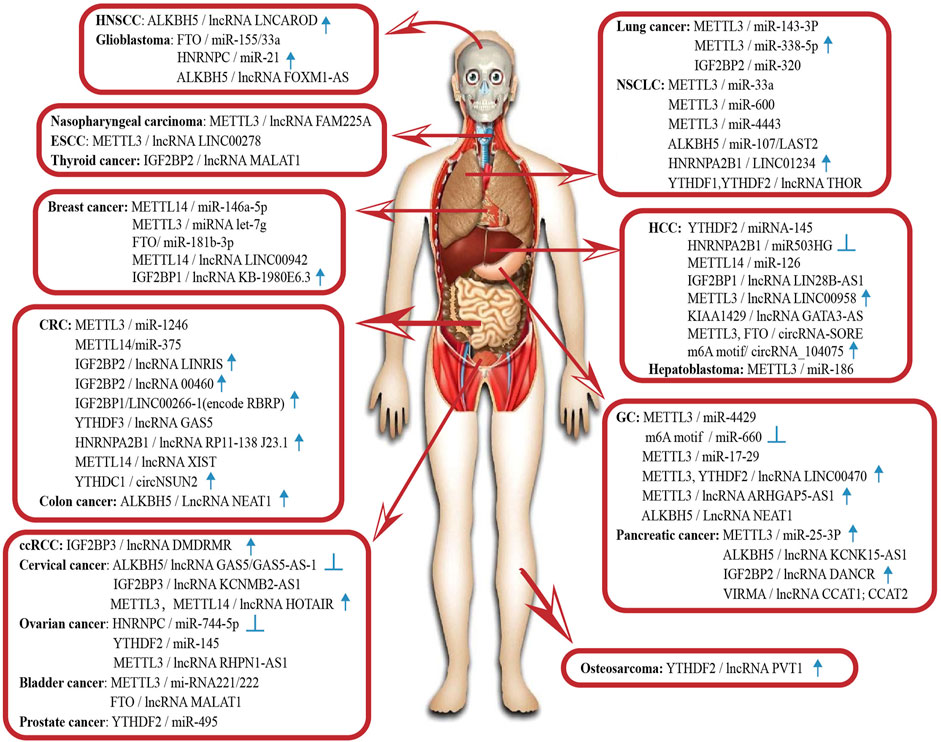
FIGURE 2. Abnormal m6A modification of non-coding RNA in human cancers. HNSCC, head and neck squamous cell carcinoma; ESCC, esophageal squamous cell carcinoma; HCC, hepatocellular carcinoma; CRC, colorectal cancer; NSCLC, non-small-cell lung carcinoma; GC, gastric cancer; ccRCC, clear cell renal cell carcinoma. The icons represent the ncRNAs that can be regarded as potential diagnostic and prognostic biomarkers. ↑ High expression is positively associated with poor prognosis. High expression is negatively associated with poor prognosis.
3 Mechanisms of ncRNA m6A Modification in Cancer Progression
The ncRNA and m6A regulators show significant changes in different cancers owing to their interactions, which cause abnormal biological functions and thereby affect cancer progression (see Figure 3). Among urological cancers (Wang et al., 2020c), METTL3 is overexpressed in bladder cancer (Xie et al., 2020)and prostate cancer (Yuan et al., 2020), but shows low expression in kidney cancer (Tao et al., 2020). METTL14 is downregulated in kidney and bladder cancers, playing a tumour suppressive role. Surprisingly, even though METTL3 and METTL14 show different expression levels and functions in urological cancers, they both participate in cancer progression through cell growth- and cell death-related pathways (Tao et al., 2020). Therefore, understanding ncRNA m6A modification from the perspective of cancer progression mechanisms will provide more promising ideas and methods to interfere with cancer progression (see Table 1).
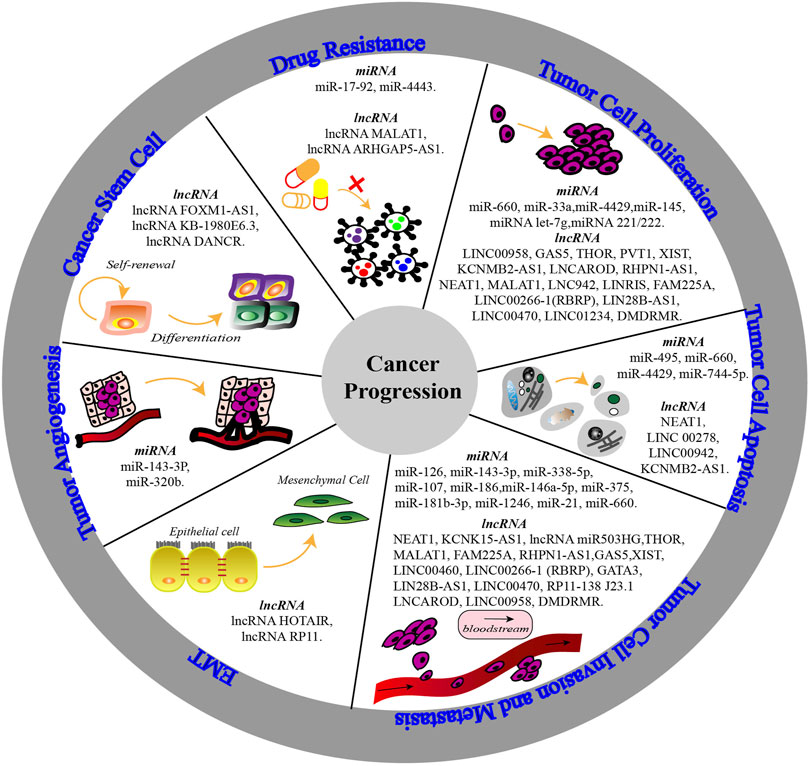
FIGURE 3. The mechanisms of non-coding RNA (miRNA & lncRNA) m6A modification involved in cancer progression. The biological functions of non-coding RNAs are altered by abnormal m6A modification and the interaction with m6A regulators. Thereby, non-coding RNAs participate in tumor cell proliferation, apoptosis, invasion and metastasis, epithelial-mesenchymal transition (EMT), tumor angiogenesis, cancer stemness and drug resistance to affect cell characteristics and cancer progression.

TABLE 1. The biological function and mechanism of ncRNAs and m6A modification involved in cancer progression.
3.1 Tumour Cell Proliferation
Many studies have reported that m6A modification of ncRNAs influences tumorigenesis and cell proliferation; for example, ectopic expression of miR-660 directly binds to oncogene E2F3 to affect cell proliferation owing to the m6A motif in the region of E2F3 3ʹ-UTR (He and Shu 2019). The stabilisation of oncogene SEC62 mRNA could be enhanced by the m6A modification of METTL3 and IGF2BP1; however, it was demonstrated that miR-4429 targets and suppresses METTL3 to repress SEC62 and realise an anti-proliferative effect (He et al., 2019). In addition, the overexpression of miR-145 inhibits cancer cell proliferation, while this suppression is impaired by the overexpression of YTHDF2, resulting in a crucial crosstalk between miR-145 and YTHDF2 by forming a double-negative feedback loop (Li et al., 2020a). Carcinogenic lncRNA THOR has been reported to be related to m6A modification for the first time. YTHDF1 and YTHDF2 can regulate the stability of THOR by reading the m6A motifs, thereby strengthening cell proliferation and helping THOR to realise its oncogenic function (Liu et al., 2020a). In terms of the m6A “reader,” METTL3-induced m6A methylation reduces lncRNA RHPN1-AS1 degradation to help it serve as a ceRNA and activate the PI3K/AKT pathway (Wang et al., 2020a). Similarly, LNC942, lncRNA XIST and lncRNA FAM225A can affect cell proliferation and cancer progression under the m6A “writer”-mediated m6A modification (Sun et al., 2020a; Yang et al., 2020c; Zheng et al., 2019).
With the participation of m6A modification, both lncRNA THOR and lncRNA RHPN1-AS1, regarded as oncogenes, can promote tumorigenesis and cell proliferation, while it is worth noting that LINC00266-1 itself cannot promote tumorigenesis but RBRP encoded by it is a potential oncogene in colorectal cancer. LncRNA LINC00266-1 encodes a 71-amino acid peptide named “RNA-binding regulatory peptide” (RBRP), which binds to the RNA-binding proteins, including the m6A “reader” IGF2BP1. The existence and expression of natural endogenous RBRP has been verified in colorectal, breast, and ovarian cancers and nasopharyngeal carcinoma. RBRP, considered as a regulatory subunit of a m6A “reader,” strengthens m6A recognition by IGF2BP1 on targeted RNAs through the essential G19 residue and promotes tumorigenesis (Zhu et al., 2020a).
3.2 Tumour Cell Apoptosis
To investigate the function of miRNA m6A modification in cell apoptosis, regions upstream and downstream of miRNA 675 m6A modification sites in the H19 locus are mutated with a system of adenine base editors, showing that mutation facilitates cell apoptosis by downregulating the expression of H19 (Hao et al., 2020). Gu et al. (2018) also believed that m6A-modified miRNAs regulated the pathway closely related to cell apoptosis, as verified in arsenite-transformed cells. In prostate cancer, YTHDF2 is regarded as a target of miR-495 to recognise MOB3B m6A modification and inhibit its expression. At the same time, the miR-495 promoter interacts with overexpressed oncogenic KDM5A and restrains the expression of miR-495. It has been reported that the progression of prostate cancer can be motivated by the activation of the KDM5A/miRNA-495/YTHDF2/m6A-MOB3B axis by influencing the apoptosis of cancer cells (Du et al., 2020a). YTHDF2 is also considered as a direct target gene of miR-145 in epithelial ovarian cancer, and its overexpression enhances cell apoptosis (Li et al., 2020a). Besides, cigarette smoke decreases the m6A modification of LINC00278 and the translation of YY1BM encoded by LINC00278, and these dysregulations inhibit the apoptosis of cancer cells (Wu et al., 2020). In addition, overexpressed lncRNA KCNMB2-AS1 plays the role of ceRNA and upregulates the oncogene IGF2BP3 by sponging miR-130b-5p and miR-4294. IGF2BP3 binds to KCNMB2-AS1 m6A sites, forming a positive regulatory loop composed of KCNMB2-AS1 and IGF2BP3 to inhibit cell apoptosis and induce proliferation (Zhang et al., 2020e).
3.3 Tumour Cell Invasion and Metastasis
Some researchers have proposed a correlation between m6A and invasion, metastasis, such as the overexpression of FTO, which promotes the migration and invasion of GC cell lines (Xu et al., 2017). ALKBH5 is involved in mediating methylation reversal, and it is reported that ALKBH5-demethylated lncRNA NEAT1 is overexpressed in GC and colon cancer cells, promoting invasion and metastasis (Guo et al., 2020; Zhang et al., 2019b). Yang et al. also determined that the downregulation of ALKBH5 in colon cancer is associated with tumour inhibition. Overexpression of ALKBH5 restrains the invasion and metastasis of colon cancer cells (Yang et al., 2020a). In addition, ALKBH5 demethylates lncRNA KCNK15-AS1 and downregulates its expression in pancreatic cancer tissues, inhibiting cell motility and invasion (He et al., 2018). These interesting roles of ALKBH5 in different cancers warrant further study. The m6A modification level is significantly reduced in HCC tissue, especially in metastatic tissues. It was confirmed that the main factor is the downregulation of METTL14, which mediates miRNA maturation and alters the metastatic capacity of HCC (Ma et al., 2017). LncRNA miR503HG is also reduced in HCC tissues. Further investigation suggested that HNRNPA2B1 degradation is promoted by miR503HG, suppressing the NF-κB pathway and facilitating the invasion and metastasis of HCC cells (Wang et al., 2018). In other tumours, METTL3-methylated regulates the MALAT1-miR-1914-3p-YAP axis and increases YAP activity to enhance drug resistance and metastasis (Jin et al., 2019). METTL3-mediated methylation can also enhance the stability of lncRNA FAM225A, which regulates the expression of ITGB3 by binding to miR-590-3p and miR-1275 as ceRNA, and thus activates the FAK/PI3K/AKT signalling pathway and promotes the invasion and migration of cancer cells (Zheng et al., 2019). Moreover, the carcinogenic lncRNA RHPN1-AS1 and lncRNA THOR have also been found to be associated with cell viability and mobility by ectopic m6A modification, thereby affecting cancer cell invasion and metastasis (Liu et al., 2020a; Wang et al., 2020a).
3.4 Epithelial-Mesenchymal Transition
During epithelial-mesenchymal transition (EMT) progression, the loss of E-cadherin causes cells to acquire mobility and the capacity (Castosa et al., 2018; Ribatti et al., 2020). It has previously been reported that the decrease of METTL3 inhibited the lung cancer cell morphological conversion induced by TGF-β, and augmented the expression changes of EMT-related marker genes, indicating the importance of m6A modification in EMT (Wanna-Udom et al., 2020). As for ncRNA m6A modification, it has been proposed that m6A-methylated lncRNA HOTAIR could upregulate EMT genes and increase the aggressiveness of tumour cells (Peng et al., 2016). In addition, lncRNA RP11 was reported to positively regulate the migration, invasion, and EMT of colorectal cancer cells. It is believed that m6A methylation increases nuclear accumulation and participates in the upregulation of RP11, triggering the spread of colorectal cancer cells (Wu et al., 2019). By analysing the cancer cells and clinical samples, Kandimalla et al. established the RNAMethyPro-a gene expression signature, which is correlated with EMT-related prognosis genes (Kandimalla et al., 2019). The specific mechanisms between ncRNA m6A modification and EMT deserve further study.
3.5 Tumour Angiogenesis
Angiogenesis is regarded as a significant step in tumour development, and some of the genes associated with EMT and tumour angiogenesis are m6A target genes involved in tumorigenesis; they are very sensitive to m6A modification. Previously, RNA functional analysis confirm that METTL14 and ALKBH5 can regulate the expression of each other, inhibit the demethylation activity of YTHDF3, and induce aberrant m6A modification to play roles in tumour angiogenesis and metastasis (Panneerdoss et al., 2018). In lung cancer, the anti-cancer gene miR-320b downregulates the expression of IGF2BP2 and thymidine kinase 1 (TK1), thus suppressing angiogenesis and lung cancer growth (Ma et al., 2021). During the process of lung cancer brain metastasis, upregulated miR-143-3p promoted by METTL3-methylation facilitates miRNA splicing and biogenesis, interacts with VASH1 and increases invasion capability and angiogenesis (Wang et al., 2019a). In brain metastasis of breast cancer, transcripts associated with brain metastasis are enriched by YTHDF3 and promote cancer cells to interact with cells in the tumour microenvironment, benefiting angiogenesis and metastasis (Chang et al., 2020).
3.6 Cancer Stem Cells
Cancer stem cells maintain the vitality of the cellular population through self-renewal and infinite proliferation, which are essential for cancer initiation and metastasis. Experimental analysis has shown that ALKBH5 facilitates the proliferation and tumorigenicity of cancer stem cells by interacting with lncRNA FOXM1-AS (Zhang et al., 2017). Moreover, the identified m6A “reader” YTHDF2 can reduce the half-life of diverse m6A transcripts, including the tumour necrosis factor receptor (Tnfrsf2), leading to negative effects on the overall integrity of leukemic stem cell function. Under YTHDF2 depletion, Tnfrsf2 is upregulated to facilitate cell apoptosis in leukemic stem cells (Paris et al., 2019). In addition to leukemic stem cells, YTHDF2 is associated with the liver cancer stem cell phenotype (Zhang et al., 2020b). In breast cancer, a recent study showed that lncRNA KB-1980E6.3 can augment cancer cell self-renewal and maintain the stemness of cancer cells under a hypoxic microenvironment, indicating that targeting the lncRNA KB-1980E6.3/IGF2BP1/c-Myc axis may be a promising therapy for refractory hypoxic tumours (Zhu et al., 2021).
3.7 Drug Resistance
Drug resistance that develops during conventional drug therapy is one of the significant reasons for chemotherapy failure in cancer. As the main component of methylase involved in m6A modification, METTL3 has been widely reported to be associated with drug resistance. In NSCLC, the METTL3/YTHDF3 complex enhances the stability of lncRNA MALAT1, regulating the MALAT1-miR-1914-3p-YAP axis to increase YAP expression and induce drug resistance (Jin et al., 2019); METTL3, as a direct target gene of miR-4443 in tumour exosomes, is involved in regulating FSP1 expression and interferes with the ferroptosis induced by cisplatin, resulting in drug resistance (Song et al., 2021). Furthermore, in GC cells, METTL3 is recruited by lncRNA ARHGAP5-AS1 to stabilize ARHGAP5 mRNA and promotes chemoresistance (Zhu et al., 2019), and consistent with this study, Sun et al. find that elevated METTL3 and levels of m6A modification facilitate the maturation of pri-miR-17-92 to the miR-17-92 cluster, which activates the AKT/mTOR pathway. High levels of METTL3 and the miR-17-92 cluster then enhance the sensitivity of cancer cells to the mTOR inhibitor, everolimus (Sun et al., 2020b).
In tamoxifen-resistant LCC9 breast cancer cells, studies have determined that overexpressed m6A “reader” HNRNPA2B1 can upregulate and downregulate different miRNAs simultaneously and affect the downstream signalling pathways of these miRNAs. Transient overexpression of HNRNPA2B1 reduces the sensitivity of cancer cells to 4-hydroxytamoxifen and fulvestrant, suggesting the potential role of HNRNPA2B1 in endocrine-resistance (Klinge et al., 2019). Oncogene circRNA-SORE is upregulated in sorafenib-resistant HCC cells, acting as ceRNA to sponge miR-103a-2-5p and miR-660-3p, competitively activate the Wnt/β-catenin pathway, and induce sorafenib resistance (Xu et al., 2020a). However, further study shows that interfering the expression of circRNA-SORE has effects on improving sorafenib efficacy, which represents a promising pharmaceutical intervention targeted circRNA-SORE in sorafenib-treated HCC patients. Besides, recent studies have shown that depletion or inhibition of FTO suppresses the expression of immune checkpoint genes and attenuates the self-renewal ability of cancer stem cells, thus overcoming their immune escape (Su et al., 2020). Another study indicated that the m6A modification induced by ‘eraser’ FTO augments melanoma growth, whereas FTO inhibition increases the sensitivity of cancer cells to anti-PD-1 immunotherapy, suggesting that the immune checkpoint molecular inhibitors represented by PD-1/PD-L1 can play an important role with the help of FTO inhibitors (Yang et al., 2019). Although these two studies were based on the demethylation effect of FTO on mRNAs, it is reasonable to believe that FTO inhibitors can also inhibit ncRNA-related targets or signal pathways with the deepening of studies on the crosstalk of FTO and ncRNAs. These findings provide in-depth insights into chemoresistance and support the therapeutic potential of ncRNA m6A modification, especially in combination with existing drugs to target refractory malignant tumours.
4 Clinical Application of ncRNA m6A Modification in Cancers
4.1 m6A Regulators and Abnormally Modified ncRNAs as Potential Diagnostic and Prognostic Biomarkers
The interaction between ncRNAs and m6A modification suggests that they may provide new ideas for the clinical treatment of cancer and even become prognostic biomarkers or therapeutic targets for cancer therapy. Zhang et al. constructed an m6A-score model to quantify m6A modification patterns in cancers. By analysing tumour microenvironment phenotypes and 5-year survival rates in patients with low and high m6A-score subgroups, it was found that patients with low m6A-scores show significant therapeutic advantages, suggesting a crosstalk between m6A modification and tumour microenvironment diversity and complexity (Zhang et al., 2020a). In HCC tissues, m6A methyltransferases WATP and KIAA1429 are related to prognosis (Chen et al., 2019b; Liu et al., 2020b), and METTL3, YTHDF2, ZC3H13 and ALKBH5 are considered independent prognostic factors for overall survival; of these, only METTL3 is an independent prognostic factor for recurrence-free survival (Li et al., 2019b; Liu et al., 2021). In CRC, depletion of METTL14 is associated with poor prognosis (Chen et al., 2020b; Yang et al., 2020c), which is different from the methylated enzymes METTTL3 and KIAA1429 in liver cancer. Notably, m6A “reader” ALKBH5 is associated with TNM stage, tumour size, lymph node metastasis (Tang et al., 2020), and can be regarded as a prognostic indicator in colon cancer (Guo et al., 2020). ALKBH5 is also believed to be a novel biomarker and independent prognostic factor in pancreatic cancer and NSCLC (Tang et al., 2020; Zhu et al., 2020b). Further, studies have revealed that FTO expression is closely correlated with low differentiation, peritumoral lymphovascular invasion, lymph node metastasis and is positively correlated with TNM stage in HCC, GC, HER2-positive breast cancer and bladder cancer, which makes FTO a possible biomarker for diagnostic and prognostic purposes (Cui et al., 2020; Tao et al., 2021; Xu et al., 2017; Xu et al., 2020b). Notably, high expression of HNRNPC in glioblastoma enhances cell invasiveness by regulating the miR-21/PDCD4 axis, and the level of HNRNPC is reported to increase with the increasing grade of brain tumour (Park et al., 2012). Bioinformatics analysis and validation experiments further confirmed the value of high expression HNRNPC in the prognosis of GBM patients at the expression level (Wang et al., 2020b). Interestingly, Ying et al. have identified m6A “reader” HNRNPC genes in the Chinese population and discussed the relationship between single nucleotide polymorphisms and susceptibility to pancreatic cancer, providing a completely novel idea on studying the role of m6A modification in tumorigenesis (Ying et al., 2021). Besides, several studies have analyzed the m6A regulators expression differences and their correlations in normal tissue and tumour tissue based on the clinical data of TCGA and GTEx databases, and it is found that the differential expression of some m6A regulators are closely associated with the risk factors of prognosis in hematologic system tumours and endocrine-system-related tumours (Li et al., 2020b; Zhang et al., 2021). Based on TCGA, GTEx and GEO, a novel risk signature has been constructed to evaluate the relationship between m6A regulators and survival rate of patients in uterine corpus endometrial carcinoma and uterine carcinosarcoma, further inspiring us to combine the bioinformatics with the study of m6A modification (Zou et al., 2021). Taken together, focusing on the expression of m6A regulators and their prognostic value provides us with a potential experimental avenue. It will have good clinical significance to directly or indirectly measure the expression level of m6A, the degree of tumours malignancy and the prognosis of patients by using tumour samples of patients.
ncRNAs modified by abnormal m6A modification could also be regarded as biomarkers, but these ncRNAs show a high degree of specificity and difference in various cancers. A m6A-lncRNA co-expression network has been constructed in primary glioblastoma to identify four m6A-related prognostic lncRNAs: MIR9-3HG, LINC00900, MIR155HG, and LINC00515 (Wang et al., 2021). Using a similar approach, LINC00152, LINC00265, and nine other lncRNAs are documented as biomarkers for predicting the overall survival of lower-grade glioma patients (Tu et al., 2020). In HCC, high expressed circRNA_104075 acts as a ceRNA to sponge miR-582-3p and upregulate YAP expression. The significant changes in downstream pathways caused by circRNA_104075 render it possible to be a new diagnostic marker (Zhang et al., 2018). LncRNA00266-1-encoded peptides RBRP bind to m6A “reader” IGF2BP1 to facilitate tumorigenesis, and the patients with high level of RBRP display an unfavourable prognosis (Zhu et al., 2020a); high expression of LINC00958 also can independently predict poor overall survival in patients with HCC. The good news is to take advantage of the role of LINC00958 as a biomarker, Zuo et al. constructed a novel drug delivery system, a PLGA-based nanoplatform filled with si-LINC00958, with the advantages of controlled release, tumour targeting, and better safety and effectiveness (Zuo et al., 2020). Differently, as a prognostic indicator, lncRNA miR503HG is significantly decreased in HCC tissues, and is positively associated with overall survival and time to recurrence (Wang et al., 2018).
The same phenomenon occurs in GC and cervical cancer. High expression of lncRNA ARHGAP5-AS1 (Zhu et al., 2019)and lncRNA LINC00470 (Yan et al., 2020) is related to the poor prognosis of patients with GC, whereas low expression of miR-660 is strongly linked with a large tumour size, severe lymph node metastasis, advanced TNM stage, and poor outcome (He and Shu 2019). In cervical cancer, lncRNA HOTAIR and lncRNA GAS5 show opposite effects on forecasting the clinical states of patients, high expression of HOTAIR indicates a high degree of malignancy, whereas lncRNA GAS5 is downregulated in cancer tissues, indicating that low GAS5 expression suggests poor prognosis (Peng et al., 2016). The above examples show that under m6A modification, multiple ncRNAs with significant changes may appear in the same cancer, which makes them have great potential for future prognostic diagnosis. However, accuracy cannot be determined solely using existing studies, and is worth further experimental exploration.
Surprisingly, lncRNA LINRIS (Wang et al., 2019b), lncRNA RP11 (Wu et al., 2019), lncRNA LINC00460 (Hou et al., 2021), lncRNA NEAT1 (Guo et al., 2020), and circRNA NSUN2 (Chen et al., 2019a) are all upregulated in patients with CRC and poor overall survival, indicating their promising role as critical prognostic markers and therapeutic targets. In prostate cancer, lncRNA CCAT1 and lncRNA CCAT2 can be regarded as a group variable to independently predict the prognosis of patients (Barros-Silva et al., 2020). Further, evidence confirms that potential prognostic markers also exist in other cancers, such as LINC01234 in NSCLC (Chen et al., 2020d), miR-744-5p in ovarian cancer (Kleemann et al., 2018), lncRNA LNCAROD in head and neck squamous cell carcinoma (Ban et al., 2020), lncRNA PVT1 in osteosarcoma (Chen et al., 2020a), miR-25-3p (Zhang et al., 2019a)and lncRNA DANCR (Hu et al., 2020)in pancreatic cancer (see Figure 2). Notably, several recent studies have used data models to analyse differentially expressed ncRNAs in some cancers to identify potential prognostic or diagnostic markers, such as in ovarian cancer (Li et al., 2021), adrenocortical carcinoma (Jin et al., 2021), kidney renal clear cell carcinoma (Yu et al., 2021), and colorectal cancer (Zuo et al., 2021). This new approach can explore the relationship between ncRNAs and different cancers on a large scale, as well as provide ideas and directions for further molecular mechanism research. To effectuate the role of ncRNAs and m6A regulators as biomarkers, the remarkable relationship between the expression level of m6A and prognosis needs to be explored more specifically. Demonstrating or testing the changes in m6A levels based on existing data in various cancers to evaluate clinical significance maybe more convincing.
4.2 m6A Regulators as Drug Targets to Participate in Cancer Therapy
Increasing attention has been focused on m6A regulators in hope of finding new treatments. For instance, a study shows that the new sodium channel blocker MV1035 can disturb migration and invasion characteristics by targeting ALKBH5 in glioblastoma (Malacrida et al., 2020). The potential of ALKBH as a drug target has been demonstrated by screening ALKBH blockers from newly synthesised anthraquinone derivatives (Huang et al., 2015). As an important demethylase, FTO has been identified to play an oncogenic role in NSCLC, ovarian cancer, and acute myeloid leukemia, associating with cancer cell proliferation, cancer stem cell maintenance and other malignant characteristics (Huang et al., 2020a; Huang et al., 2019; Li et al., 2019a). In recent years, researchers have used FTO as a target to develop FTO inhibitors, hoping to make new breakthroughs in cancer treatment. It is well known that meclofenamic acid, R-2-hydroxyglutarate, and MO-I-500 inhibit the activity of FTO and display anti-cancer activity (Huang et al., 2015; Singh et al., 2016; Su et al., 2018). Huang et al. find that artificially developed FTO inhibitors FB23 and FB23-2 can directly bind to FTO, selectively inhibit the m6A demethylase activity of FTO, and significantly inhibit cell proliferation in human acute myeloid leukaemia (Huang et al., 2019). Su et al. screen out two small-molecule FTO inhibitors, CS1 and CS2, and showed strong anti-cancer effects in multiple types of cancers. According to a previous study, the self-renewal of leukaemia stem cells can be attenuated and the immune response can be reprogrammed by CS1/2 (Su et al., 2020). A recent trial of FTO inhibitors shows that the ethylester form of meclofenamic acid (MA2) can negatively regulate Myc-miR-155/23a cluster-MXI1 feedback circuit and augment the efficacy of the chemotherapy drug temozolomide (TMZ) on inhibiting proliferation of malignant glioma cells (Xiao et al., 2020). In addition, the protease inhibitor nafamostat mesilate (NM) has been shown to inhibit the growth and metastasis of colorectal cancer and has anti-cancer effects in pancreatic cancer and lung cancer (Han et al., 2019b). Han et al. find that the activity of FTO can be inhibited by NM, indicating that NM might play a role in interfering with cancer progression as an FTO inhibitor. In addition, some biotech or pharmaceutic companies get down to developing high-efficiency small-molecule inhibitors, targeting the m6A regulators, in particular molecules like METTL3 and FTO that vary significantly in many cancers (Huang et al., 2020b). Undoubtedly, it provides a new choice for the effective management of a variety of cancers whether used alone or in combination with anti-tumour drugs, especially under the circumstance of chemotherapy drug resistance.
5 Conclusion and Prospects
Multiple regulation of various “writers” and “erasers” allows m6A modification in a dynamic and reversible manner, and significantly interacts with ncRNAs. Numerous studies have focused on the role of abnormal ncRNA m6A modifications in cancer progression. Aberrant m6A-modified ncRNAs and alteration of m6A regulators are closely related to cellular proliferation, apoptosis, invasion and metastasis, as well as other key steps of tumorigenesis and cancer progression. Undoubtedly, the cancers listed in this review form only a small part of the disease, and mainly include mainstream cancers and ncRNAs from recent years. Therefore, from the perspective of m6A modification, further research is needed to understand whether ncRNAs can participate in tumorigenesis and progression in other cancers, whether the same ncRNA plays the same role in different cancers (e.g. lncRNA MALAT1 and RBRP encoded by lncRNA LINC00266-1), and how different ncRNAs come into effect when they are involved in the same cancer; more research are also needed to explore the underlying mechanisms, especially in the field of stemness, reprogramming of tumour immune microenvironment and angiogenesis, and to illustrate the effects of ncRNA and m6A modification on cancer initiation, promotion and progression. Accordingly, the overall abundance of m6A modification may not be of vital significance; instead, some specific transcripts or genes play significant roles in biological function and cancer progression (most of them play the role in promoting cancer). Therefore, increased attention should be given to the significant changes in specific molecules, improving their clinical value as diagnostic/prognostic biomarkers. Further, studies now mainly focus on the cellular level, whereas fewer studies have been conducted in vivo. In clinical applications, several studies have elucidated the effect of ncRNAs in cancer prognosis based on m6A-Seq, m6A score, and designing data models, which has revealed molecules that could serve as biomarkers or therapeutic targets, and have greatly enriched the research in this field. However, applying these molecules in developing pharmaceutical preparations and in clinical diagnosis is still challenging. Overall, given the importance of ncRNA m6A modification and m6A regulators in various cancers, we consider that they are promising diagnostic and prognostic biomarkers contributing to the prediction of recurrence and survival, even serving as potential drug targets for therapeutic interventions in cancer treatment. Nevertheless, more specific mechanisms, more in-depth theories, and closer to practical applications of clinical diagnosis and treatment require further exploration.
Author Contributions
D-HC and J-GZ drafted the manuscript. C-XW participated in interpretation. QL participated in the design of the manuscript and helped modifying the manuscript.
Funding
This work was supported by the subject of the National Key R&D Program of China (No. 2017YFC0909900) and the Clinical Research Plan of SHDC (No. SHDC2020CR4076).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CRC, colorectal cancer; ccRCC, clear cell renal cell carcinoma; ceRNA, endogenous ribonucleic acid; circRNA, circular ribonucleic acid; EMT, epithelial-mesenchymal transition; ESCC, esophageal squamous cell carcinoma; GC, gastric cancer; HCC, hepatocellular carcinoma; HNSCC, head and neck squamous cell carcinoma; lncRNA, long non-coding ribonucleic acid; miRNA, micro ribonucleic acid; m6A, N6-methyl-adenosine; ncRNA, non-coding ribonucleic acid; NM, nafamostat mesilate; NSCLC, non-small cell lung cancer; RBRP, RNA-binding regulatory peptide; 3ʹ-UTR, 3ʹ-untranslated region.
References
Alarcón, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015a). HNRNPA2B1 Is a Mediator of m6A-dependent Nuclear RNA Processing Events. Cell 162, 1299–1308. doi:10.1016/j.cell.2015.08.011
Alarcón, C. R., Lee, H., Goodarzi, H., Halberg, N., and Tavazoie, S. F. (2015b). N6-methyladenosine marks Primary microRNAs for Processing. Nature 519, 482–485. doi:10.1038/nature14281
Ban, Y., Tan, P., Cai, J., Li, J., Hu, M., Zhou, Y., et al. (2020). LNCAROD Is Stabilized by m6A Methylation and Promotes Cancer Progression via Forming a Ternary Complex with HSPA1A and YBX1 in Head and Neck Squamous Cell Carcinoma. Mol. Oncol. 14, 1282–1296. doi:10.1002/1878-0261.12676
Barros-Silva, D., Lobo, J., Guimarães-Teixeira, C., Carneiro, I., Oliveira, J., Martens-Uzunova, E. S., et al. (2020). VIRMA-dependent N6-Methyladenosine Modifications Regulate the Expression of Long Non-coding RNAs CCAT1 and CCAT2 in Prostate Cancer. Cancers 12, 771. doi:10.3390/cancers12040771
Cai, X., Wang, X., Cao, C., Gao, Y., Zhang, S., Yang, Z., et al. (2018). HBXIP-elevated Methyltransferase METTL3 Promotes the Progression of Breast Cancer via Inhibiting Tumor Suppressor Let-7g. Cancer Lett. 415, 11–19. doi:10.1016/j.canlet.2017.11.018
Castosa, R., Martinez-Iglesias, O., Roca-Lema, D., Casas-Pais, A., Díaz-Díaz, A., Iglesias, P., et al. (2018). Hakai Overexpression Effectively Induces Tumour Progression and Metastasis In Vivo. Sci. Rep. 8, 3466. doi:10.1038/s41598-018-21808-w
Chang, G., Shi, L., Ye, Y., Shi, H., Zeng, L., Tiwary, S., et al. (2020). YTHDF3 Induces the Translation of m6A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell 38, 857–871. e7. doi:10.1016/j.ccell.2020.10.004
Chen, R.-X., Chen, X., Xia, L.-P., Zhang, J.-X., Pan, Z.-Z., Ma, X.-D., et al. (2019a). N6-methyladenosine Modification of circNSUN2 Facilitates Cytoplasmic export and Stabilizes HMGA2 to Promote Colorectal Liver Metastasis. Nat. Commun. 10, 4695. doi:10.1038/s41467-019-12651-2
Chen, S., Zhou, L., and Wang, Y. (2020a). ALKBH5-mediated m6A Demethylation of lncRNA PVT1 Plays an Oncogenic Role in Osteosarcoma. Cancer Cell Int 20, 34. doi:10.1186/s12935-020-1105-6
Chen, T., Hao, Y.-J., Zhang, Y., Li, M.-M., Wang, M., Han, W., et al. (2015). m6A RNA Methylation Is Regulated by MicroRNAs and Promotes Reprogramming to Pluripotency. Cell Stem Cell 16, 289–301. doi:10.1016/j.stem.2015.01.016
Chen, X., Xu, M., Xu, X., Zeng, K., Liu, X., Sun, L., et al. (2020b). METTL14 Suppresses CRC Progression via Regulating N6-methyladenosine-dependent Primary miR-375 Processing. Mol. Ther. 28, 599–612. doi:10.1016/j.ymthe.2019.11.016
Chen, Y., Lin, Y., Shu, Y., He, J., and Gao, W. (2020c). Interaction between N6-Methyladenosine (m6A) Modification and Noncoding RNAs in Cancer. Mol. Cancer 19, 94. doi:10.1186/s12943-020-01207-4
Chen, Y., Peng, C., Chen, J., Chen, D., Yang, B., He, B., et al. (2019b). WTAP Facilitates Progression of Hepatocellular Carcinoma via m6A-HuR-dependent Epigenetic Silencing of ETS1. Mol. Cancer 18, 127. doi:10.1186/s12943-019-1053-8
Chen, Z., Chen, X., Lei, T., Gu, Y., Gu, J., Huang, J., et al. (2020d). Integrative Analysis of NSCLC Identifies LINC01234 as an Oncogenic lncRNA that Interacts with HNRNPA2B1 and Regulates miR-106b Biogenesis. Mol. Ther. 28, 1479–1493. doi:10.1016/j.ymthe.2020.03.010
Cui, X., Wang, Z., Li, J., Zhu, J., Ren, Z., Zhang, D., et al. (2020). Cross Talk between RNA N6‐methyladenosine Methyltransferase‐like 3 and miR‐186 Regulates Hepatoblastoma Progression through Wnt/β‐catenin Signalling Pathway. Cell Prolif 53, e12768. doi:10.1111/cpr.12768
Du, C., Lv, C., Feng, Y., and Yu, S. (2020a). Activation of the KDM5A/miRNA-495/YTHDF2/m6A-MOB3B axis Facilitates Prostate Cancer Progression. J. Exp. Clin. Cancer Res. 39, 223. doi:10.1186/s13046-020-01735-3
Du, J., Hou, K., Mi, S., Ji, H., Ma, S., Ba, Y., et al. (2020b). Malignant Evaluation and Clinical Prognostic Values of m6A RNA Methylation Regulators in Glioblastoma. Front. Oncol. 10, 208. doi:10.3389/fonc.2020.00208
Du, M., Zhang, Y., Mao, Y., Mou, J., Zhao, J., Xue, Q., et al. (2017). MiR-33a Suppresses Proliferation of NSCLC Cells via Targeting METTL3 mRNA. Biochem. Biophysical Res. Commun. 482, 582–589. doi:10.1016/j.bbrc.2016.11.077
Erson-Bensan, A. E., and Begik, O. (2017). m6A Modification and Implications for microRNAs. Mirna 6, 97–101. doi:10.2174/2211536606666170511102219
Fazi, F., and Fatica, A. (2019). Interplay Between N6-Methyladenosine (m6A) and Non-coding RNAs in Cell Development and Cancer. Front. Cell Dev. Biol. 7, 116. doi:10.3389/fcell.2019.00116
Gao, Y. X., Yang, T. W., Yin, J. M., Yang, P. X., Kou, B. X., Chai, M. Y., et al. (2020). Progress and Prospects of Biomarkers In Primary Liver Cancer (Review). Int. J. Oncol. 57, 54–66. doi:10.3892/ijo.2020.5035
Gu, C., Wang, Z., Zhou, N., Li, G., Kou, Y., Luo, Y., et al. (2019). Mettl14 Inhibits Bladder TIC Self-Renewal and Bladder Tumorigenesis through N6-Methyladenosine of Notch1. Mol. Cancer 18, 168. doi:10.1186/s12943-019-1084-1
Gu, S., Sun, D., Dai, H., and Zhang, Z. (2018). N6-methyladenosine Mediates the Cellular Proliferation and Apoptosis via microRNAs in Arsenite-Transformed Cells. Toxicol. Lett. 292, 1–11. doi:10.1016/j.toxlet.2018.04.018
Gu, Y., Niu, S., Wang, Y., Duan, L., Pan, Y., Tong, Z., et al. (2021). DMDRMR-mediated Regulation of m6A-Modified CDK4 by m6A Reader IGF2BP3 Drives ccRCC Progression. Cancer Res. 81, 923–934. doi:10.1158/0008-5472.CAN-20-1619
Guan, K., Liu, X., Li, J., Ding, Y., Li, J., Cui, G., et al. (2020). Expression Status And Prognostic Value of M6A-Associated Genes in Gastric Cancer. J. Cancer 11, 3027–3040. doi:10.7150/jca.40866
Guo, T., Liu, D. F., Peng, S. H., and Xu, A. M. (2020). ALKBH5 Promotes colon Cancer Progression by Decreasing Methylation of the lncRNA NEAT1. Am. J. Transl Res. 12, 4542–4549.
Han, J., Wang, J.-z., Yang, X., Yu, H., Zhou, R., Lu, H.-C., et al. (2019a). METTL3 Promote Tumor Proliferation of Bladder Cancer by Accelerating Pri-miR221/222 Maturation in m6A-dependent Manner. Mol. Cancer 18, 110. doi:10.1186/s12943-019-1036-9
Han, X., Wang, N., Li, J., Wang, Y., Wang, R., and Chang, J. (2019b). Identification of Nafamostat Mesilate as an Inhibitor of the Fat Mass and Obesity-Associated Protein (FTO) Demethylase Activity. Chemico-Biological Interactions 297, 80–84. doi:10.1016/j.cbi.2018.10.023
Hao, J., Li, C., Lin, C., Hao, Y., Yu, X., Xia, Y., et al. (2020). Targeted point Mutations of the m6A Modification in miR675 Using RNA-Guided Base Editing Induce Cell Apoptosis. Biosci. Rep. 40. doi:10.1042/bsr20192933
He, H., Wu, W., Sun, Z., and Chai, L. (2019). MiR-4429 Prevented Gastric Cancer Progression through Targeting METTL3 to Inhibit m6A-Caused Stabilization of SEC62. Biochem. Biophysical Res. Commun. 517, 581–587. doi:10.1016/j.bbrc.2019.07.058
He, R.-Z., Jiang, J., and Luo, D.-X. (2020). The Functions of N6-Methyladenosine Modification in lncRNAs. Genes Dis. 7, 598–605. doi:10.1016/j.gendis.2020.03.005
He, X., and Shu, Y. (2019). RNA N6-Methyladenosine Modification Participates in miR-660/E2F3 axis-mediated Inhibition of Cell Proliferation in Gastric Cancer. Pathol. - Res. Pract. 215, 152393. doi:10.1016/j.prp.2019.03.021
He, Y., Hu, H., Wang, Y., Yuan, H., Lu, Z., Wu, P., et al. (2018). ALKBH5 Inhibits Pancreatic Cancer Motility by Decreasing Long Non-coding RNA KCNK15-AS1 Methylation. Cell Physiol Biochem 48, 838–846. doi:10.1159/000491915
Hou, P., Meng, S., Li, M., Lin, T., Chu, S., Li, Z., et al. (2021). LINC00460/DHX9/IGF2BP2 Complex Promotes Colorectal Cancer Proliferation and Metastasis by Mediating HMGA1 mRNA Stability Depending on m6A Modification. J. Exp. Clin. Cancer Res. 40, 52. doi:10.1186/s13046-021-01857-2
Hu, X., Peng, W.-X., Zhou, H., Jiang, J., Zhou, X., Huang, D., et al. (2020). IGF2BP2 Regulates DANCR by Serving as an N6-Methyladenosine Reader. Cell Death Differ 27, 1782–1794. doi:10.1038/s41418-019-0461-z
Huang, H., Wang, Y., Kandpal, M., Zhao, G., Cardenas, H., Ji, Y., et al. (2020a). FTO-dependent N6-Methyladenosine Modifications Inhibit Ovarian Cancer Stem Cell Self-Renewal by Blocking cAMP Signaling. Cancer Res. 80, 3200–3214. doi:10.1158/0008-5472.Can-19-4044
Huang, H., Weng, H., and Chen, J. (2020b). m6A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell 37, 270–288. doi:10.1016/j.ccell.2020.02.004
Huang, Y., Su, R., Sheng, Y., Dong, L., Dong, Z., Xu, H., et al. (2019). Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 35, 677–691. e10. doi:10.1016/j.ccell.2019.03.006
Huang, Y., Yan, J., Li, Q., Li, J., Gong, S., Zhou, H., et al. (2015). Meclofenamic Acid Selectively Inhibits FTO Demethylation of m6A over ALKBH5. Nucleic Acids Res. 43, 373–384. doi:10.1093/nar/gku1276
Inamura, K. (2018). Update on Immunohistochemistry for the Diagnosis of Lung Cancer. Cancers 10, 72. doi:10.3390/cancers10030072
Jin, D., Guo, J., Wu, Y., Du, J., Yang, L., Wang, X., et al. (2019). m6A mRNA Methylation Initiated by METTL3 Directly Promotes YAP Translation and Increases YAP Activity by Regulating the MALAT1-miR-1914-3p-YAP axis to Induce NSCLC Drug Resistance and Metastasis. J. Hematol. Oncol. 12, 135. doi:10.1186/s13045-019-0830-6
Jin, D., Guo, J., Wu, Y., Yang, L., Wang, X., Du, J., et al. (2020). m6A Demethylase ALKBH5 Inhibits Tumor Growth and Metastasis by Reducing YTHDFs-Mediated YAP Expression and Inhibiting miR-107/LATS2-Mediated YAP Activity in NSCLC. Mol. Cancer 19, 40. doi:10.1186/s12943-020-01161-1
Jin, Y., Wang, Z., He, D., Zhu, Y., Hu, X., Gong, L., et al. (2021). Analysis of m6A-Related Signatures in the Tumor Immune Microenvironment and Identification of Clinical Prognostic Regulators in Adrenocortical Carcinoma. Front. Immunol. 12, 637933. doi:10.3389/fimmu.2021.637933
Kandimalla, R., Gao, F., Li, Y., Huang, H., Ke, J., Deng, X., et al. (2019). RNAMethyPro: a Biologically Conserved Signature of N6-Methyladenosine Regulators for Predicting Survival at Pan-Cancer Level. Npj Precis. Onc. 3, 13. doi:10.1038/s41698-019-0085-2
Karimi, P., Islami, F., Anandasabapathy, S., Freedman, N. D., and Kamangar, F. (2014). Gastric Cancer: Descriptive Epidemiology, Risk Factors, Screening, and Prevention. Cancer Epidemiol. Biomarkers Prev. 23, 700–713. doi:10.1158/1055-9965.EPI-13-1057
Kleemann, M., Schneider, H., Unger, K., Sander, P., Schneider, E. M., Fischer-Posovszky, P., et al. (2018). MiR-744-5p Inducing Cell Death by Directly Targeting HNRNPC and NFIX in Ovarian Cancer Cells. Sci. Rep. 8, 9020. doi:10.1038/s41598-018-27438-6
Klinge, C. M., Piell, K. M., Tooley, C. S., and Rouchka, E. C. (2019). HNRNPA2/B1 Is Upregulated in Endocrine-Resistant LCC9 Breast Cancer Cells and Alters the miRNA Transcriptome when Overexpressed in MCF-7 Cells. Sci. Rep. 9, 9430. doi:10.1038/s41598-019-45636-8
Lan, T., Li, H., Zhang, D., Xu, L., Liu, H., Hao, X., et al. (2019). KIAA1429 Contributes to Liver Cancer Progression through N6-methyladenosine-dependent post-transcriptional Modification of GATA3. Mol. Cancer 18, 186. doi:10.1186/s12943-019-1106-z
Li, J., Zhu, L., Shi, Y., Liu, J., Lin, L., and Chen, X. (2019b). m6A Demethylase FTO Promotes Hepatocellular Carcinoma Tumorigenesis via Mediating PKM2 Demethylation. Am. J. Transl Res. 11, 6084–6092.
Li, J., Han, Y., Zhang, H., Qian, Z., Jia, W., Gao, Y., et al. (2019a). The m6A Demethylase FTO Promotes the Growth of Lung Cancer Cells by Regulating the m6A Level of USP7 mRNA. Biochem. Biophysical Res. Commun. 512, 479–485. doi:10.1016/j.bbrc.2019.03.093
Li, J., Wu, L., Pei, M., and Zhang, Y. (2020a). YTHDF2, a Protein Repressed by miR-145, Regulates Proliferation, Apoptosis, and Migration in Ovarian Cancer Cells. J. Ovarian Res. 13, 111. doi:10.1186/s13048-020-00717-5
Li, K., Luo, H., Luo, H., and Zhu, X. (2020b). Clinical and Prognostic Pan-Cancer Analysis of m6A RNA Methylation Regulators in Four Types of Endocrine System Tumors. Aging 12, 23931–23944. doi:10.18632/aging.104064
Li, Q., Ren, C.-C., Chen, Y.-N., Yang, L., Zhang, F., Wang, B.-J., et al. (2021). A Risk Score Model Incorporating Three m6A RNA Methylation Regulators and a Related Network of miRNAs-m6A Regulators-m6A Target Genes to Predict the Prognosis of Patients with Ovarian Cancer. Front. Cell Dev. Biol. 9, 703969. doi:10.3389/fcell.2021.703969
Li, Q., Wang, H., Peng, H., Huang, Q., Huyan, T., Huang, Q., et al. (2019c). MicroRNAs: Key Players in Bladder Cancer. Mol. Diagn. Ther. 23, 579–601. doi:10.1007/s40291-019-00410-4
Liu, G.-M., Zeng, H.-D., Zhang, C.-Y., and Xu, J.-W. (2021). Identification of METTL3 as an Adverse Prognostic Biomarker in Hepatocellular Carcinoma. Dig. Dis. Sci. 66, 1110–1126. doi:10.1007/s10620-020-06260-z
Liu, H., Xu, Y., Yao, B., Sui, T., Lai, L., and Li, Z. (2020a). A Novel N6-Methyladenosine (m6A)-dependent Fate Decision for the lncRNA THOR. Cell Death Dis 11, 613. doi:10.1038/s41419-020-02833-y
Liu, J., Sun, G., Pan, S., Qin, M., Ouyang, R., Li, Z., et al. (2020b). The Cancer Genome Atlas (TCGA) Based m6A Methylation-Related Genes Predict Prognosis in Hepatocellular Carcinoma. Bioengineered 11, 759–768. doi:10.1080/21655979.2020.1787764
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N6-methyladenosine-dependent RNA Structural Switches Regulate RNA-Protein Interactions. Nature 518, 560–564. doi:10.1038/nature14234
Ma, J. Z., Yang, F., Zhou, C. C., Liu, F., Yuan, J. H., Wang, F., et al. (2017). METTL14 Suppresses the Metastatic Potential of Hepatocellular Carcinoma by Modulating N 6 ‐methyladenosine‐dependent Primary MicroRNA Processing. Hepatology 65, 529–543. doi:10.1002/hep.28885
Ma, Y.-S., Shi, B.-W., Guo, J.-H., Liu, J.-B., Yang, X.-L., Xin, R., et al. (2021). microRNA-320b Suppresses HNF4G and IGF2BP2 Expression to Inhibit Angiogenesis and Tumor Growth of Lung Cancer. Carcinogenesis 42, 762–771. doi:10.1093/carcin/bgab023
Malacrida, A., Rivara, M., Di Domizio, A., Cislaghi, G., Miloso, M., Zuliani, V., et al. (2020). 3D Proteome-wide Scale Screening and Activity Evaluation of a New ALKBH5 Inhibitor in U87 Glioblastoma Cell Line. Bioorg. Med. Chem. 28, 115300. doi:10.1016/j.bmc.2019.115300
Müller, M. F., Ibrahim, A. E. K., and Arends, M. J. (2016). Molecular Pathological Classification of Colorectal Cancer. Virchows Arch. 469, 125–134. doi:10.1007/s00428-016-1956-3
Nagini, S. (2017). Breast Cancer: Current Molecular Therapeutic Targets and New Players. Acamc 17, 152–163. doi:10.2174/1871520616666160502122724
Ni, W., Yao, S., Zhou, Y., Liu, Y., Huang, P., Zhou, A., et al. (2019). Long Noncoding RNA GAS5 Inhibits Progression of Colorectal Cancer by Interacting with and Triggering YAP Phosphorylation and Degradation and Is Negatively Regulated by the m6A Reader YTHDF3. Mol. Cancer 18, 143. doi:10.1186/s12943-019-1079-y
Panneerdoss, S., Eedunuri, V. K., Yadav, P., Timilsina, S., Rajamanickam, S., Viswanadhapalli, S., et al. (2018). Cross-talk Among Writers, Readers, and Erasers of m6A Regulates Cancer Growth and Progression. Sci. Adv. 4, eaar8263. doi:10.1126/sciadv.aar8263
Paris, J., Morgan, M., Campos, J., Spencer, G. J., Shmakova, A., Ivanova, I., et al. (2019). Targeting the RNA m6A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell 25, 137–148. e6. doi:10.1016/j.stem.2019.03.021
Park, Y. M., Hwang, S. J., Masuda, K., Choi, K.-M., Jeong, M.-R., Nam, D.-H., et al. (2012). Heterogeneous Nuclear Ribonucleoprotein C1/C2 Controls the Metastatic Potential of Glioblastoma by Regulating PDCD4. Mol. Cell Biol 32, 4237–4244. doi:10.1128/mcb.00443-12
Peng, L., Yuan, X., Jiang, B., Tang, Z., and Li, G.-C. (2016). LncRNAs: Key Players and Novel Insights into Cervical Cancer. Tumor Biol. 37, 2779–2788. doi:10.1007/s13277-015-4663-9
Peng, W., Li, J., Chen, R., Gu, Q., Yang, P., Qian, W., et al. (2019). Upregulated METTL3 Promotes Metastasis of Colorectal Cancer via miR-1246/SPRED2/MAPK Signaling Pathway. J. Exp. Clin. Cancer Res. 38, 393. doi:10.1186/s13046-019-1408-4
Ribatti, D., Tamma, R., and Annese, T. (2020). Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Translational Oncol. 13, 100773. doi:10.1016/j.tranon.2020.100773
Shan, C., Zhang, Y., Hao, X., Gao, J., Chen, X., and Wang, K. (2019). Biogenesis, Functions and Clinical Significance of circRNAs in Gastric Cancer. Mol. Cancer 18, 136. doi:10.1186/s12943-019-1069-0
Shin, V. Y., and Chu, K. M. (2014). MiRNA as Potential Biomarkers and Therapeutic Targets for Gastric Cancer. Wjg 20, 10432–10439. doi:10.3748/wjg.v20.i30.10432
Singh, B., Kinne, H. E., Milligan, R. D., Washburn, L. J., Olsen, M., and Lucci, A. (2016). Important Role of FTO in the Survival of Rare Panresistant Triple-Negative Inflammatory Breast Cancer Cells Facing a Severe Metabolic Challenge. PLoS One 11, e0159072. doi:10.1371/journal.pone.0159072
Song, Z., Jia, G., Ma, P., and Cang, S. (2021). Exosomal miR-4443 Promotes Cisplatin Resistance in Non-small Cell Lung Carcinoma by Regulating FSP1 m6A Modification-Mediated Ferroptosis. Life Sci. 276, 119399. doi:10.1016/j.lfs.2021.119399
Su, R., Dong, L., Li, C., Nachtergaele, S., Wunderlich, M., Qing, Y., et al. (2018). R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m6A/MYC/CEBPA Signaling. Cell 172, 90–105. doi:10.1016/j.cell.2017.11.031
Su, R., Dong, L., Li, Y., Gao, M., Han, L., Wunderlich, M., et al. (2020). Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell 38, 79–96. e11. doi:10.1016/j.ccell.2020.04.017
Sun, T., Wu, R., and Ming, L. (2019). The Role of m6A RNA Methylation in Cancer. Biomed. Pharmacother. 112, 108613. doi:10.1016/j.biopha.2019.108613
Sun, T., Wu, Z., Wang, X., Wang, Y., Hu, X., Qin, W., et al. (2020a). LNC942 Promoting METTL14-Mediated m6A Methylation in Breast Cancer Cell Proliferation and Progression. Oncogene 39, 5358–5372. doi:10.1038/s41388-020-1338-9
Sun, W., Yang, Y., Xu, C., Xie, Y., and Guo, J. (2016). Roles of Long Noncoding RNAs in Gastric Cancer and Their Clinical Applications. J. Cancer Res. Clin. Oncol. 142, 2231–2237. doi:10.1007/s00432-016-2183-7
Sun, Y., Li, S., Yu, W., Zhao, Z., Gao, J., Chen, C., et al. (2020b). N6-methyladenosine-dependent Pri-miR-17-92 Maturation Suppresses PTEN/TMEM127 and Promotes Sensitivity to Everolimus in Gastric Cancer. Cell Death Dis 11, 836. doi:10.1038/s41419-020-03049-w
Tang, B., Yang, Y., Kang, M., Wang, Y., Wang, Y., Bi, Y., et al. (2020). m6A Demethylase ALKBH5 Inhibits Pancreatic Cancer Tumorigenesis by Decreasing WIF-1 RNA Methylation and Mediating Wnt Signaling. Mol. Cancer 19, 3. doi:10.1186/s12943-019-1128-6
Tao, L., Mu, X., Chen, H., Jin, D., Zhang, R., Zhao, Y., et al. (2021). FTO Modifies the m6A Level of MALAT and Promotes Bladder Cancer Progression. Clin. Translational Med. 11, e310. doi:10.1002/ctm2.310
Tao, Z., Zhao, Y., and Chen, X. (2020). Role of Methyltransferase-like Enzyme 3 and Methyltransferase-like Enzyme 14 in Urological Cancers. PeerJ 8, e9589. doi:10.7717/peerj.9589
Tu, Z., Wu, L., Wang, P., Hu, Q., Tao, C., Li, K., et al. (2020). N6-Methylandenosine-Related lncRNAs Are Potential Biomarkers for Predicting the Overall Survival of Lower-Grade Glioma Patients. Front. Cell Dev. Biol. 8, 642. doi:10.3389/fcell.2020.00642
Vishnoi, A., and Rani, S. (2017). MiRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol. Biol. 1509, 1–10. doi:10.1007/978-1-4939-6524-3_1
Wang, H., Deng, Q., Lv, Z., Ling, Y., Hou, X., Chen, Z., et al. (2019a). N6-methyladenosine Induced miR-143-3p Promotes the Brain Metastasis of Lung Cancer via Regulation of VASH1. Mol. Cancer 18, 181. doi:10.1186/s12943-019-1108-x
Wang, H., Liang, L., Dong, Q., Huan, L., He, J., Li, B., et al. (2018). Long Noncoding RNA miR503HG, a Prognostic Indicator, Inhibits Tumor Metastasis by Regulating the HNRNPA2B1/NF-κB Pathway in Hepatocellular Carcinoma. Theranostics 8, 2814–2829. doi:10.7150/thno.23012
Wang, J., Ding, W., Xu, Y., Tao, E., Mo, M., Xu, W., et al. (2020a). Long Non-coding RNA RHPN1-AS1 Promotes Tumorigenesis and Metastasis of Ovarian Cancer by Acting as a ceRNA against miR-596 and Upregulating LETM1. Aging 12, 4558–4572. doi:10.18632/aging.102911
Wang, L.-c., Chen, S.-h., Shen, X.-l., Li, D.-c., Liu, H.-y., Ji, Y.-l., et al. (2020b). M6A RNA Methylation Regulator HNRNPC Contributes to Tumorigenesis and Predicts Prognosis in Glioblastoma Multiforme. Front. Oncol. 10, 536875. doi:10.3389/fonc.2020.536875
Wang, W., Li, J., Lin, F., Guo, J., and Zhao, J. (2021). Identification of N6-Methyladenosine-Related lncRNAs for Patients with Primary Glioblastoma. Neurosurg. Rev. 44, 463–470. doi:10.1007/s10143-020-01238-x
Wang, X., Xie, H., Ying, Y., Chen, D., and Li, J. (2020c). Roles of N 6 ‐methyladenosine (M 6 A) RNA Modifications in Urological Cancers. J. Cell Mol Med 24, 10302–10310. doi:10.1111/jcmm.15750
Wang, Y., Lu, J.-H., Wu, Q.-N., Jin, Y., Wang, D.-S., Chen, Y.-X., et al. (2019b). LncRNA LINRIS Stabilizes IGF2BP2 and Promotes the Aerobic Glycolysis in Colorectal Cancer. Mol. Cancer 18, 174. doi:10.1186/s12943-019-1105-0
Wanna-Udom, S., Terashima, M., Lyu, H., Ishimura, A., Takino, T., Sakari, M., et al. (2020). The m6A Methyltransferase METTL3 Contributes to Transforming Growth Factor-Beta-Induced Epithelial-Mesenchymal Transition of Lung Cancer Cells through the Regulation of JUNB. Biochem. Biophysical Res. Commun. 524, 150–155. doi:10.1016/j.bbrc.2020.01.042
Wei, W., Huo, B., and Shi, X. (2019). miR-600 Inhibits Lung Cancer via Downregulating the Expression of METTL3. Cmar Vol. 11, 1177–1187. doi:10.2147/cmar.S181058
Wong, C.-M., Tsang, F. H.-C., and Ng, I. O.-L. (2018). Non-coding RNAs in Hepatocellular Carcinoma: Molecular Functions and Pathological Implications. Nat. Rev. Gastroenterol. Hepatol. 15, 137–151. doi:10.1038/nrgastro.2017.169
Wu, H., Li, F., and Zhu, R. (2021). miR-338-5p Inhibits Cell Growth and Migration via Inhibition of the METTL3/m6A/c-Myc Pathway in Lung Cancer. Acta Biochim. Biophys. Sin (Shanghai) 53, 304–316. doi:10.1093/abbs/gmaa170
Wu, S., Zhang, L., Deng, J., Guo, B., Li, F., Wang, Y., et al. (2020). A Novel Micropeptide Encoded by Y-Linked LINC00278 Links Cigarette Smoking and AR Signaling in Male Esophageal Squamous Cell Carcinoma. Cancer Res. 80, 2790–2803. doi:10.1158/0008-5472.Can-19-3440
Wu, Y., Yang, X., Chen, Z., Tian, L., Jiang, G., Chen, F., et al. (2019). m6A-induced lncRNA RP11 Triggers the Dissemination of Colorectal Cancer Cells via Upregulation of Zeb1. Mol. Cancer 18, 87. doi:10.1186/s12943-019-1014-2
Xiao, L., Li, X., Mu, Z., Zhou, J., Zhou, P., Xie, C., et al. (2020). FTO Inhibition Enhances the Antitumor Effect of Temozolomide by Targeting MYC-miR-155/23a Cluster-MXI1 Feedback Circuit in Glioma. Cancer Res. 80, 3945–3958. doi:10.1158/0008-5472.CAN-20-0132
Xie, H., Li, J., Ying, Y., Yan, H., Jin, K., Ma, X., et al. (2020). METTL3/YTHDF2 M6 A axis Promotes Tumorigenesis by Degrading SETD7 and KLF4 mRNAs in Bladder Cancer. J. Cell Mol Med 24, 4092–4104. doi:10.1111/jcmm.15063
Xu, D., Shao, W., Jiang, Y., Wang, X., Liu, Y., and Liu, X. (2017). FTO Expression Is Associated with the Occurrence of Gastric Cancer and Prognosis. Oncol. Rep. 38, 2285–2292. doi:10.3892/or.2017.5904
Xu, J., Wan, Z., Tang, M., Lin, Z., Jiang, S., Ji, L., et al. (2020a). N6-methyladenosine-modified CircRNA-SORE Sustains Sorafenib Resistance in Hepatocellular Carcinoma by Regulating β-catenin Signaling. Mol. Cancer 19, 163. doi:10.1186/s12943-020-01281-8
Xu, Y., Ye, S., Zhang, N., Zheng, S., Liu, H., Zhou, K., et al. (2020b). The FTO/miR‐181b‐3p/ARL5B Signaling Pathway Regulates Cell Migration and Invasion in Breast Cancer. Cancer Commun. 40, 484–500. doi:10.1002/cac2.12075
Yan, J., Huang, X., Zhang, X., Chen, Z., Ye, C., Xiang, W., et al. (2020). LncRNA LINC00470 Promotes the Degradation of PTEN mRNA to Facilitate Malignant Behavior in Gastric Cancer Cells. Biochem. Biophysical Res. Commun. 521, 887–893. doi:10.1016/j.bbrc.2019.11.016
Yang, L., Ma, Y., Han, W., Li, W., Cui, L., Zhao, X., et al. (2015). Proteinase-activated Receptor 2 Promotes Cancer Cell Migration through RNA Methylation-Mediated Repression of miR-125b. J. Biol. Chem. 290, 26627–26637. doi:10.1074/jbc.M115.667717
Yang, P., Wang, Q., Liu, A., Zhu, J., and Feng, J. (2020a). ALKBH5 Holds Prognostic Values and Inhibits the Metastasis of Colon Cancer. Pathol. Oncol. Res. 26, 1615–1623. doi:10.1007/s12253-019-00737-7
Yang, S., Wei, J., Cui, Y.-H., Park, G., Shah, P., Deng, Y., et al. (2019). m6A mRNA Demethylase FTO Regulates Melanoma Tumorigenicity and Response to Anti-PD-1 Blockade. Nat. Commun. 10, 2782. doi:10.1038/s41467-019-10669-0
Yang, X., Liu, M., Li, M., Zhang, S., Hiju, H., Sun, J., et al. (2020b). Epigenetic Modulations of Noncoding RNA: a Novel Dimension of Cancer Biology. Mol. Cancer 19, 64. doi:10.1186/s12943-020-01159-9
Yang, X., Zhang, S., He, C., Xue, P., Zhang, L., He, Z., et al. (2020c). METTL14 Suppresses Proliferation and Metastasis of Colorectal Cancer by Down-Regulating Oncogenic Long Non-coding RNA XIST. Mol. Cancer 19, 46. doi:10.1186/s12943-020-1146-4
Yang, Z., Li, J., Feng, G., Gao, S., Wang, Y., Zhang, S., et al. (2017). MicroRNA-145 Modulates N6-Methyladenosine Levels by Targeting the 3′-Untranslated mRNA Region of the N6-Methyladenosine Binding YTH Domain Family 2 Protein. J. Biol. Chem. 292, 3614–3623. doi:10.1074/jbc.M116.749689
Ye, M., Dong, S., Hou, H., Zhang, T., and Shen, M. (2021). Oncogenic Role of Long Noncoding RNAMALAT1 in Thyroid Cancer Progression through Regulation of the miR-204/IGF2BP2/m6A-MYC Signaling. Mol. Ther. - Nucleic Acids 23, 1–12. doi:10.1016/j.omtn.2020.09.023
Yi, D., Wang, R., Shi, X., Xu, L., Yilihamu, Y. e., and Sang, J. (2020). METTL14 Promotes the Migration and Invasion of Breast Cancer Cells by Modulating N6-methyladenosine and hsa-miR-146a-5p E-xpression. Oncol. Rep. 43, 1375–1386. doi:10.3892/or.2020.7515
Ying, P., Li, Y., Yang, N., Wang, X., Wang, H., He, H., et al. (2021). Identification of Genetic Variants in m6A Modification Genes Associated with Pancreatic Cancer Risk in the Chinese Population. Arch. Toxicol. 95, 1117–1128. doi:10.1007/s00204-021-02978-5
Yousef, M., and Tsiani, E. (2017). Metformin in Lung Cancer: Review of in Vitro and in Vivo Animal Studies, Cancers 9, 45. doi:10.3390/cancers9050045
Yu, J., Mao, W., Sun, S., Hu, Q., Wang, C., Xu, Z., et al. (2021). Identification of an m6A-Related lncRNA Signature for Predicting the Prognosis in Patients with Kidney Renal Clear Cell Carcinoma. Front. Oncol. 11, 663263. doi:10.3389/fonc.2021.663263
Yuan, S., Tang, H., Xing, J., Fan, X., Cai, X., Li, Q., et al. (2014). Methylation by NSun2 Represses the Levels and Function of microRNA 125b. Mol. Cell Biol. 34, 3630–3641. doi:10.1128/MCB.00243-14
Yuan, Y., Du, Y., Wang, L., and Liu, X. (2020). The M6A Methyltransferase METTL3 Promotes the Development and Progression of Prostate Carcinoma via Mediating MYC Methylation. J. Cancer 11, 3588–3595. doi:10.7150/jca.42338
Zhang, B., Wu, Q., Li, B., Wang, D., Wang, L., and Zhou, Y. L. (2020a). m6A Regulator-Mediated Methylation Modification Patterns and Tumor Microenvironment Infiltration Characterization in Gastric Cancer. Mol. Cancer 19, 53. doi:10.1186/s12943-020-01170-0
Zhang, C., Huang, S., Zhuang, H., Ruan, S., Zhou, Z., Huang, K., et al. (2020b). YTHDF2 Promotes the Liver Cancer Stem Cell Phenotype and Cancer Metastasis by Regulating OCT4 Expression via m6A RNA Methylation. Oncogene 39, 4507–4518. doi:10.1038/s41388-020-1303-7
Zhang, J., Bai, R., Li, M., Ye, H., Wu, C., Wang, C., et al. (2019a). Excessive miR-25-3p Maturation via N6-Methyladenosine Stimulated by Cigarette Smoke Promotes Pancreatic Cancer Progression. Nat. Commun. 10, 1858. doi:10.1038/s41467-019-09712-x
Zhang, J., Guo, S., Piao, H.-y., Wang, Y., Wu, Y., Meng, X.-y., et al. (2019b). ALKBH5 Promotes Invasion and Metastasis of Gastric Cancer by Decreasing Methylation of the lncRNA NEAT1. J. Physiol. Biochem. 75, 379–389. doi:10.1007/s13105-019-00690-8
Zhang, J., Hu, K., Yang, Y.-q., Wang, Y., Zheng, Y.-f., Jin, Y., et al. (2020c). LIN28B-AS1-IGF2BP1 Binding Promotes Hepatocellular Carcinoma Cell Progression. Cell Death Dis 11, 741. doi:10.1038/s41419-020-02967-z
Zhang, L., Hou, C., Chen, C., Guo, Y., Yuan, W., Yin, D., et al. (2020d). The Role of N6-Methyladenosine (m6A) Modification in the Regulation of circRNAs. Mol. Cancer 19, 105. doi:10.1186/s12943-020-01224-3
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m 6 A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 31, 591–606. e6. doi:10.1016/j.ccell.2017.02.013
Zhang, X., Xu, Y., Qian, Z., Zheng, W., Wu, Q., Chen, Y., et al. (2018). circRNA_104075 Stimulates YAP-dependent Tumorigenesis through the Regulation of HNF4a and May Serve as a Diagnostic Marker in Hepatocellular Carcinoma. Cell Death Dis 9, 1091. doi:10.1038/s41419-018-1132-6
Zhang, X., Zhong, L., Zou, Z., Liang, G., Tang, Z., Li, K., et al. (2021). Clinical and Prognostic Pan-Cancer Analysis of N6-Methyladenosine Regulators in Two Types of Hematological Malignancies: A Retrospective Study Based on TCGA and GTEx Databases. Front. Oncol. 11, 623170. doi:10.3389/fonc.2021.623170
Zhang, Y., Wang, D., Wu, D., Zhang, D., and Sun, M. (2020e). Long Noncoding RNA KCNMB2-AS1 Stabilized by N6-Methyladenosine Modification Promotes Cervical Cancer Growth through Acting as a Competing Endogenous RNA. Cell Transpl. 29, 096368972096438. doi:10.1177/0963689720964382
Zheng, Z.-Q., Li, Z.-X., Zhou, G.-Q., Lin, L., Zhang, L.-L., Lv, J.-W., et al. (2019). Long Noncoding RNA FAM225A Promotes Nasopharyngeal Carcinoma Tumorigenesis and Metastasis by Acting as ceRNA to Sponge miR-590-3p/miR-1275 and Upregulate ITGB3. Cancer Res. 79, 4612–4626. doi:10.1158/0008-5472.Can-19-0799
Zhou, K. I., Parisien, M., Dai, Q., Liu, N., Diatchenko, L., Sachleben, J. R., et al. (2016). N6-Methyladenosine Modification in a Long Noncoding RNA Hairpin Predisposes its Conformation to Protein Binding. J. Mol. Biol. 428, 822–833. doi:10.1016/j.jmb.2015.08.021
Zhou, Y., Kong, Y., Fan, W., Tao, T., Xiao, Q., Li, N., et al. (2020). Principles of RNA Methylation and Their Implications for Biology and Medicine. Biomed. Pharmacother. 131, 110731. doi:10.1016/j.biopha.2020.110731
Zhu, L., Zhu, Y., Han, S., Chen, M., Song, P., Dai, D., et al. (2019). Impaired Autophagic Degradation of lncRNA ARHGAP5-AS1 Promotes Chemoresistance in Gastric Cancer. Cell Death Dis 10, 383. doi:10.1038/s41419-019-1585-2
Zhu, P., He, F., Hou, Y., Tu, G., Li, Q., Jin, T., et al. (2021). A Novel Hypoxic Long Noncoding RNA KB-1980E6.3 Maintains Breast Cancer Stem Cell Stemness via Interacting with IGF2BP1 to Facilitate C-Myc mRNA Stability. Oncogene 40, 1609–1627. doi:10.1038/s41388-020-01638-9
Zhu, S., Wang, J.-Z., Chen, D., He, Y.-T., Meng, N., Chen, M., et al. (2020a). An Oncopeptide Regulates m6A Recognition by the m6A Reader IGF2BP1 and Tumorigenesis. Nat. Commun. 11, 1685. doi:10.1038/s41467-020-15403-9
Zhu, Z., Qian, Q., Zhao, X., Ma, L., and Chen, P. (2020b). N6-methyladenosine ALKBH5 Promotes Non-small Cell Lung Cancer Progress by Regulating TIMP3 Stability. Gene 731, 144348. doi:10.1016/j.gene.2020.144348
Zou, Z., Zhou, S., Liang, G., Tang, Z., Li, K., Tan, S., et al. (2021). The Pan-Cancer Analysis of the Two Types of Uterine Cancer Uncovered Clinical and Prognostic Associations with m6A RNA Methylation Regulators. Mol. Omics 17, 438–453. doi:10.1039/d0mo00113a
Zuo, L., Su, H., Zhang, Q., Wu, W.-y., Zeng, Y., Li, X.-m., et al. (2021). Comprehensive Analysis of lncRNAs N6-Methyladenosine Modification in Colorectal Cancer. Aging 13, 4182–4198. doi:10.18632/aging.202383
Keywords: m6A RNA modification, non-coding RNA, tumorigenesis mechanism, cancer therapy, epigenetics
Citation: Chen D-H, Zhang J-G, Wu C-X and Li Q (2021) Non-Coding RNA m6A Modification in Cancer: Mechanisms and Therapeutic Targets. Front. Cell Dev. Biol. 9:778582. doi: 10.3389/fcell.2021.778582
Received: 17 September 2021; Accepted: 06 December 2021;
Published: 22 December 2021.
Edited by:
Xiao Zhu, Guangdong Medical University, ChinaReviewed by:
Apiwat Mutirangura, Chulalongkorn University, ThailandAntonino Zito, Massachusetts General Hospital and Harvard Medical School, United States
Copyright © 2021 Chen, Zhang, Wu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuan-Xing Wu, ZG9jX3pmQDE2My5jb20=; Qin Li, bGlxaW4wNjI2QGhvdG1haWwuY29t
†These authors have contributed equally to this work
 Da-Hong Chen
Da-Hong Chen Ji-Gang Zhang
Ji-Gang Zhang Chuan-Xing Wu
Chuan-Xing Wu Qin Li
Qin Li