
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 25 January 2022
Sec. Signaling
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.759354
This article is part of the Research Topic Signaling Molecules Involved in Sperm Development, Maturation and Fertilization View all 7 articles
 Pedro O. Corda1
Pedro O. Corda1 Joana Vieira Silva1,2,3
Joana Vieira Silva1,2,3 Sara C. Pereira3
Sara C. Pereira3 Alberto Barros2,4,5
Alberto Barros2,4,5 Marco G. Alves3
Marco G. Alves3 Margarida Fardilha1*
Margarida Fardilha1*Currently, two conventional freezing techniques are used in sperm cryopreservation: slow freezing (SF) and rapid freezing (RF). Despite the protocolar improvements, cryopreservation still induces significant alterations in spermatozoon that are poorly understood. Here, available proteomic data from human cryopreserved sperm was analyzed through bioinformatic tools to unveil key differentially expressed proteins (DEPs) that can be used as modulation targets or quality markers. From the included proteomic studies, 160 and 555 DEPs were collected for SF and RF groups, respectively. For each group, an integrative network was constructed using gene ontology and protein-protein interaction data to identify key DEPs. Among them, arylsulfatase A (ARSA) was highlighted in both freezing networks, and low ARSA levels have been associated with poor-sperm quality. Thus, ARSA was selected for further experimental investigation and its levels were assessed in cryopreserved samples by western blot. ARSA levels were significantly decreased in RF and SF samples (∼31.97 and ∼39.28%, respectively). The bioinformatic analysis also revealed that the DEPs were strongly associated with proteasomal and translation pathways. The purposed bioinformatic approach allowed the identification of potential key DEPs in freeze-thawed human spermatozoa. ARSA has the potential to be used as a marker to assess sperm quality after cryopreservation.
Sperm cryopreservation is a key procedure in reproductive medicine worldwide. This technique has been used extensively to preserve male fertility in situations that can lead to testicular dysfunction and ejaculatory failures such as some pathological scenarios (malignant cancers and severe spinal cord injuries), corrective surgeries, or chemo-/radio-therapies (Di Santo et al., 2012; Tournaye et al., 2014; Tomlinson et al., 2015). Also, sperm cryopreservation has proved to be useful in several male infertile conditions, such as azoospermia and oligozoospermia (Di Santo et al., 2012). Nowadays, two conventional freezing techniques are used in sperm cryopreservation: slow freezing (SF) and rapid freezing (RF) (World Health Organization, 2010). In SF, the sperm sample is progressively cooled in a period of 2–4 h until it reaches -100°C (World Health Organization, 2010; Di Santo et al., 2012). In contrast, sperm samples frozen through a RF protocol are exposed to nitrogen vapors (-80°C) during 10–15 min before being placed in liquid nitrogen (-196°C) (World Health Organization, 2010; Di Santo et al., 2012). Although the efforts made in the last decades, the current cryopreservation protocols are still far from ideal inducing significant deleterious structural and molecular alterations in spermatozoa (Hezavehei et al., 2018; Rienzi et al., 2018; Kumar et al., 2019).
The primary cause of cryoinjury is water phase changes in low temperatures that lead to ice crystal formation (intra- and/or extracellular). In RF, water movement is impaired which leads to intracellular ice crystal formation (Said et al., 2010; Di Santo et al., 2012). On the other hand, when the cooling rate is too slow, there is water efflux to the extracellular environment which contributes to the formation of extracellular ice crystals and intracellular osmotic pressure increase (Said et al., 2010; Di Santo et al., 2012). In both situations, there is cell membranes disruption, organelle damage and, consequently, cell viability loss (Said et al., 2010; Di Santo et al., 2012; Jang et al., 2017; Hezavehei et al., 2018). Moreover, the cooling temperatures induce changes in the phase transition of lipid layers and membrane proteins’ function (Quinn, 1985; Talaei et al., 2010). The cryoinjuries are not limited to the freezing steps and may occur during the thawing process due to ice melting or recrystallization (Verheyen et al., 1993). Sperm cryopreservation also induces changes in mitochondrial membrane fluidity (O’Connell et al., 2002). The mitochondrial membrane potential is compromised, and therefore the ATP synthesis is reduced. This event may explain the motility reduction of thawed spermatozoa since mitochondrial ATP is one of the main sources of energy in sperm (O’Connell et al., 2002; Amaral et al., 2013). Additionally, mitochondrial membrane changes induce ROS release and promote spermatozoon oxidative damages such as membranes impairment and destabilization of axonemal structures (Saleh and Agarwal, 2002; Talaei et al., 2010). The activity of the antioxidant enzymes is decreased during cryopreservation which contributes to an increase in ROS damage (Lasso et al., 1994; Banihani and Alawneh, 2019). High ROS concentrations and the failure of antioxidant activity lead to the activation of sperm apoptotic pathways and, consequently, the release of apoptosis-inducing factors (Martin et al., 2007; Said et al., 2010; Karabulut et al., 2018). Together, those changes significantly reduce sperm motility, viability, acrosome integrity, and fertilization ability of freeze-thawed spermatozoa (Kumar et al., 2019). Despite the known consequences of sperm cryopreservation, the molecular mechanisms responsible remain largely uncharacterized.
The emergence of proteomic techniques brought a new perspective on the study of sperm physiology, allowing the evaluation of the protein profile in certain physiological/pathological contexts and the molecular understanding of complex processes such as motility (Xu et al., 2012b; Cao et al., 2018; De Rose et al., 2018). To date, few proteomic studies have been carried out to understand the proteomic changes underlying sperm cryopreservation. Most of them were performed in animal models such as bovine and pig (Parrilla et al., 2019; Ryu et al., 2019). In 2014, Wang and colleagues published the first proteomic study using cryopreserved human spermatozoa (Wang et al., 2014). They compared the proteomic changes between freeze-thawed spermatozoa and fresh controls from normozoospermic donors. Twenty-seven differentially expressed proteins (DEPs) were identified in the cryopreserved samples, being most of them associated with sperm motility, acrosome integrity, capacitation, viability, and mitochondrial activity (Wang et al., 2014).
The present work aims to identify key DEPs in human spermatozoa after SF and RF that can be used as modulation targets or quality biomarkers. For that purpose, proteomic data was collected from the available studies that compared freeze-thawed human spermatozoa with fresh controls, and, through a bioinformatic workflow, a set of proteins that seem to be relevant among the DEPs was isolated. From those proteins, arylsulfatase A (ARSA)—a sperm membrane protein—was investigated as a potential marker for quality assessment in cryopreserved sperm samples.
An exhaustive literature search was conducted in the PubMed database to identify proteomic studies that compare changes in cryopreserved vs fresh controls human spermatozoa. Only studies published in English (or at least with English abstract) until May 18th, 2020, were considered. From those, only the ones that meet the following inclusion criteria were selected: 1) use of ejaculated human samples from normozoospermic healthy donors; 2) use of fresh samples as control; 3) cryopreservation performed through slow or rapid freezing methods; 4) thawing process performed at temperatures equal or higher than 23°C; 5) protein extraction from whole spermatozoa in the sample; 6) quantitative proteomics; and 6) a false discovery rate (FDR) < 5% for protein identification.
From each proteomic study selected, only DEPs (p-value <0.05) were collected. Considering the two conventional freezing methods, proteins were divided into two groups: SF and RF. Based on the fold-change ratio (comparison of protein levels between control and freeze-thawed samples in each study), proteins were also tagged as increased or decreased. To avoid redundancy, all DEPs were mapped in the UniProt database (downloaded on May 26th, 2020) and annotated using the UniProtKB/Swiss-Prot accession number. Then, the UniProt lists were cross-compared with human spermatozoon proteome (Santiago et al., 2019) through a Venn diagram analysis using the JVenn tool (Bardou et al., 2014). Only proteins with a reviewed status and previously reported in human spermatozoon were used for further analysis. An additional Venn diagram analysis was performed to identify common proteins among rapid and slow freezing groups.
To gain insight into the molecular role of DEPs, a Gene Ontology (GO) enrichment analysis for biological process, molecular function, and the cellular compartment was performed for each group. This analysis was made through the ClueGO plugin (version 2.5.5) in Cytoscape (version 3.7.2) (Shannon et al., 2003; Trindade et al., 2018). Moreover, a KEGG pathway enrichment analysis was performed using the same plugin. The increased and decreased proteins of each group were uploaded in separated clusters to identify associated terms. The default settings were used in the SF group analysis. For the RF group, a more restrictive GO range, between 7 and 12, was used, and the GO term perfusion option was selected. Default settings were used in KEGG pathways analyses for both groups. Only GO and KEGG terms with a p-value <0.05 were considered.
To identify potential key DEPs in SF and RF groups, two independent integrative networks were built using protein-protein interaction (PPI) and enrichment analyses data. The String database (v11.0) was used to collect PPIs among the DEPs in slow and rapid freezing groups (Szklarczyk et al., 2019). Only experimental data with a high confidence score (0.700) were considered. For each freezing group, the 10 most significant biological processes, cellular compartments, and KEGG terms were included with the respectively associated proteins. Furthermore, common terms of both freezing groups were also used in the network construction. Network visualization was performed using Cytoscape. To identify the protein hubs of each network, the NetworkAnalyzer tool was used to compute the network topological properties (Shannon et al., 2003; Assenov et al., 2008). The top 10 proteins with the highest node degree were considered network hubs. For each protein hub, a search was conducted in DisGeNET (v7.0) and PubMed databases to identify an association between the protein and poor sperm quality or male (in)fertility.
Human sperm samples were collected at the Centre for Reproductive Genetics Professor Alberto Barros after approval by the Joint Ethics Committee CHUP/ICBAS (2021/CE/P002 [P342/CETI/ICBAS]). All procedures were conducted following the ethical guidelines for human samples research. Donors signed an informed consent allowing the use of the samples for scientific purposes. The Centre for Reproductive Genetics A. Barros infertility clinic’s procedure is under the provisions of the National Medically Assisted Procreation Act (Law of 2017) and overseen by the National Council for Medically Assisted Procreation (CNPMA-2018).
Semen samples were obtained from healthy male donors. Only sperm samples from normozoospermic donors (n = 15) were included in this study (mean age of 39.3 ± 5.20 years old). The semen samples were collected by masturbation into sterile containers, after an abstinence period of 2–7 days. Following complete liquefaction, basic semen analyses were performed according to World Health Organization (WHO)’s guidelines (World Health Organization, 2010). Then, each semen sample was divided equally into three aliquots: control, RF, and SF. For control samples, viability and motility were analyzed immediately.
Sperm cryopreservation was performed following the WHO guidelines (World Health Organization, 2010). The RF and SF semen samples were cryopreserved with Sperm Freezing Medium (Origio, Hovedstaden, Denmark) according to the manufacturer’s instructions. Briefly, Sperm Freezing Medium was added, slowly and dropwise, to each rapid and slow freezing semen sample (1:1, v/v), and then carefully mixed. The solution was maintained at room temperature (RT) for 10 min and subsequently transferred to cryovials. RF cryovials were placed at a -80°C refrigerator freezer for 30 min. SF cryovials were placed at a -20°C refrigerator freezer for 30 min and then transferred to a -80°C freezer for 30 min. In the end, SF and RF cryovials were immersed in liquid nitrogen (-196°C). After 8 days, the frozen samples were thawed by being placed in a pre-warmed water bath (23°C) for 15 min. The post-thawed parameters of motility and vitality were analyzed.
Spermatozoa viability was evaluated through the eosin-nigrosine staining, as previously describe (Carrageta et al., 2020). Briefly, spermatozoa suspensions were mixed with an equal volume of 0.5% eosin-nigrosine stain and smeared onto a glass microscope slide; 200 spermatozoa were counted per replicate with the use of a light microscope. White spermatozoa were considered viable whereas pink-stained spermatozoa were considered non-viable.
Sperm total motility and progressive motility were assessed using a Makler counting chamber (Sefi Medical Instruments, Haifa, Israel), according to the manufacturer’s recommendations. A 10 µL of sperm suspension was placed in the center of the chamber and sperm motility was observed in 20 squares using an optical microscope. A total of 100 spermatozoa were counted per sample.
Control and freeze-thawed semen samples were washed twice in sterile phosphate-buffered saline (PBS) by centrifugation (500x g, 5 min at 37°C) to remove the seminal plasma and cryoprotectant solution. Next, spermatozoa pellets were lysate in 1% sodium dodecyl sulfate (SDS; Acros Organics, Belgium) solution for 15 min on ice, followed by sonication (3 bursts, 5 s each with ice-cooling between bursts) (Naz, 1999). The lysates were centrifugated 16,000x g for 15 min at 4°C and the supernatant, corresponding to sperm soluble protein, was collected and stored at -20°C for subsequent use. For each sample, protein concentration was measured using bicinchoninic acid (BCA) assay (Pierce Biotechnology, Waltham, Massachusetts, United States) following the manufacturer’s instruction. The final absorbance was measured at 562 nm in a microplate reader (TECAN, Genius, Männedorf, Switzerland).
To investigate the impact of cryopreservation in ARSA levels, sperm extracts corresponding to 7.5 µg of protein were resolved 10% SDS-polyacrylamide gel electrophoresis, and proteins were electro-transferred onto nitrocellulose membranes (Amersham, 0.45 µm). For loading control, membranes were stained with Ponceau S solution (Sigma Aldrich; 0.1% [w/v] in 5% acetic acid) for 15 min at RT with slow agitation. Membranes were washed with distilled water until protein bands were well defined and analyzed using densitometer GS-800. To remove the Ponceau S staining, the membrane was washed with 1x tris buffered saline containing 0.1% Tween 20 (1x TBST). Then, non-specific protein-binding sites on membranes were blocked for an hour with 5% bovine serum albumin (BSA)/TBST followed by incubation of the primary antibodies for 1 h at RT. Primary antibodies were used as follows: mouse anti-ARSA (Santa Cruz Biotechnology, Germany, catalog number: sc-365176) at 1:500; rabbit anti-β-tubulin (Proteintech Europe, UK, catalog number: 10094-1-AP) at 1:1000. After primary incubation, membranes were washed twice in 1x TBST for 10 min at RT and then incubated with appropriate secondary antibody: IRDye®680RD anti-rabbit (catalog number: 926–68,071) and IRDye®800CW anti-mouse (catalog number: 926–32,210). Both were diluted at the final concentration of 1:10,000 and purchased from LI-COR Biosciences (US). After the secondary incubation, membranes were washed three times for 10 min with 1x TBST and immunodetected using Odyssey Infrared Imaging System (LI-COR Biosciences, US). Ponceau and blot results were analyzed in the Image Studio Lite Version 5.2.5 Software (LI-COR Biosciences, Lincoln, Nebraska, United States). Bands’ intensity was normalized to Ponceau staining.
The statistical analysis was performed using GraphPad Prism (v8.0.1). Datasets were tested for normal distribution using the Shapiro-Wilk’s test. Since normality was not observed, the Mann-Whitney U test was used to compare vitality, motility, and protein levels in each freezing condition to the control group. Values of p < 0.05 were considered statistically significant. All experimental data are presented as mean ± standard deviation (SD). For ARSA levels, statistical power was assessed using the GPower (v3.1) tool, through the posthoc power analysis option for Wilcoxon-Mann-Whitney tests (Mayr et al., 2007).
From the search conducted in the PubMed database, only five proteomic studies which used samples from normozoospermic healthy donors were identified (Wang et al., 2014; Bogle et al., 2017; Gholami et al., 2018; Fu et al., 2019; Li et al., 2019). From those, the Wang et al. (Wang et al., 2014) study was excluded since the thawed samples were washed through a Percoll gradient. The Gholami et al. (Gholami et al., 2018) study was also excluded because only the sperm tails were analyzed. Therefore, for the present analysis, only the three remaining studies (Bogle et al., 2017; Fu et al., 2019; Li et al., 2019), which fit the defined inclusion criteria, were considered eligible. In the study of Bogle et al. (Bogle et al., 2017), two methods of thawing were performed: 1) samples placed in a 23°C bath (15–20 s) and then placed on ice (∼0°C); or 2) samples placed and maintained in a 23°C bath. Thus, only the set of proteins identified in samples thawed by the second method were considered for subsequent analysis.
By merging data from the selected studies (Bogle et al., 2017; Fu et al., 2019; Li et al., 2019), 173 and 621 DEPs in sperm cryopreserved through slow and rapid freezing methods, respectively, were identified (Supplementary Table S1). In the SF group, 160 proteins had a reviewed status and were previously identified in human spermatozoon, of which 95 were decreased and 65 were increased (Figure 1A). In the RF group, there were 555 human sperm-reviewed proteins (Figure 1B). From those, 366 were decreased, 179 proteins were increased, and 10 proteins were described as increased and decreased (blue highlight in Supplementary Table S1). These 10 proteins were not considered in further analysis. Then, a cross-comparison was performed to identify common proteins in the SF and RF groups (Figure 1C, yellow highlight in Supplementary Table S1). One increased and 36 decreased proteins were identified in both cryopreservation groups. One protein (Epididymal sperm-binding protein 1, ELSPBP1) was described as increased in the RF group and as decreased in the SF group.
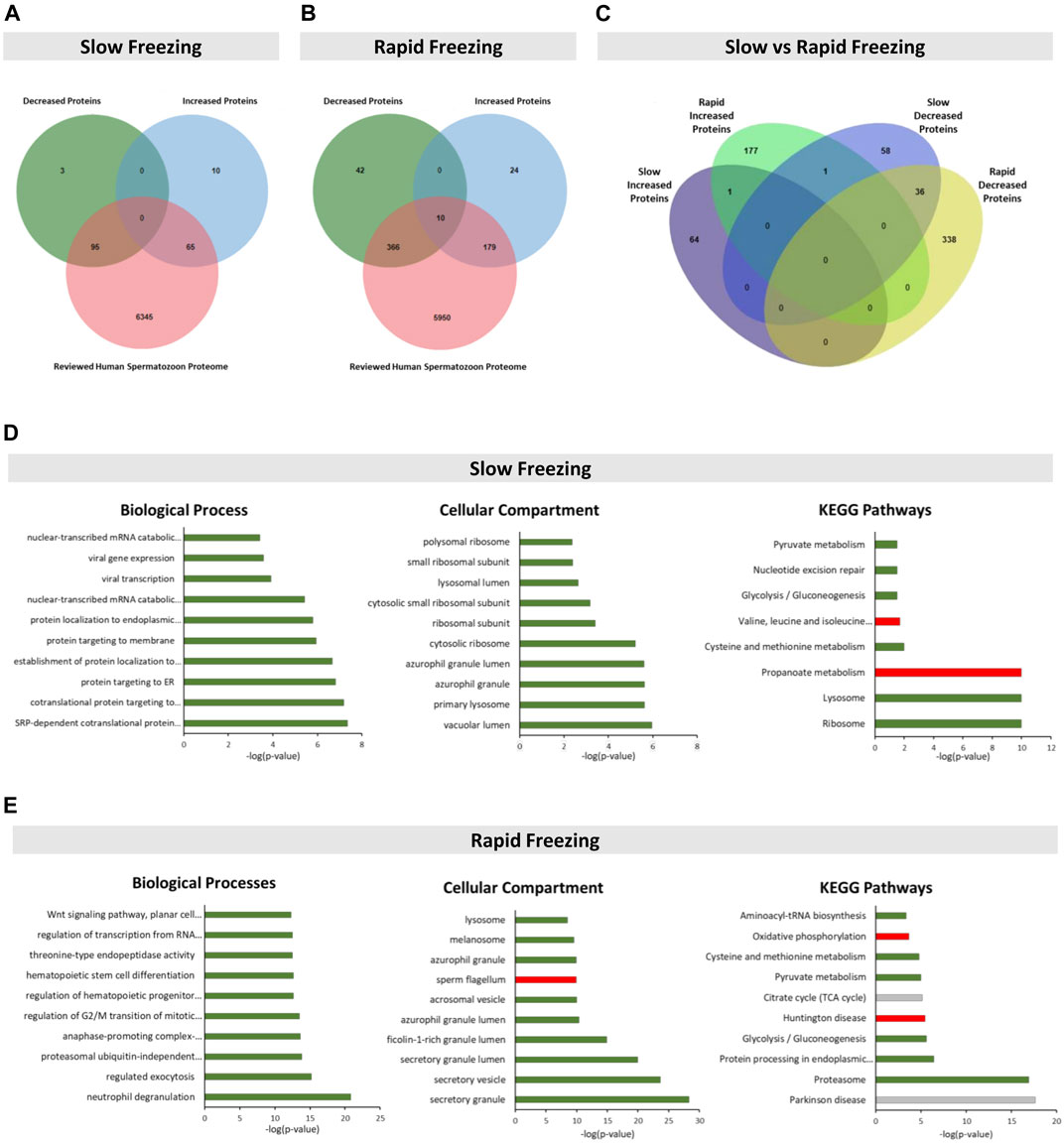
FIGURE 1. Identification and enrichment analysis of the DEPs in slow and rapid freezing groups. (A,B) Venn’s diagram illustrates the cross-comparison performed between DEPs of each freezing group and the reviewed human spermatozoon proteome. For the subsequent analyses, only DEPs reviewed and previously described in human spermatozoon were considered. In Venn’s diagram of the rapid freezing group (B), the 10 DEPs that were described as increased and decreased were not considered. (C) Venn’s diagram shows the common DEPs between slow and rapid freezing groups (D,E) Top 10 significant terms for biological processes, cellular components, and KEGG pathways related with DEPs of slow and rapid freezing groups, respectively. Enrichment analyses were performed using the ClueGO plugin through Cytoscape. In each graph, the bar color represents a specific association as follows: green for decreased proteins; red for increased proteins; and grey for non-specific associations. The significant terms were identified by Bonferroni’s step-down corrected p-value (p < 0.01).
DEPs in the SF group were significantly enriched with GO terms related to translation, protein targeting, viral infection, antigen processing, and metabolic processes (Figure 1D, Supplementary Table S2). On the other hand, in the RF group, DEPs were overrepresented with GO terms associated with exocytosis, proteasome, cell differentiation, cell cycle, metabolic processes, signaling pathways, protein processing and trafficking, translation, and mRNA processing (Figure 1E, Supplementary Table S2). For the RF group, biological processes and cellular compartment terms related to spermatozoon were also identified: “binding of sperm to zona pellucida”, “sperm-egg recognition”, “sperm capacitation”, “acrosomal vesicle” (also identified in the slow freezing group), “sperm flagellum”, “sperm fibrous sheath”, “acrosomal membrane”, “sperm principal piece” and “sperm connecting piece” (Figure 1E, Supplementary Table S2). The KEGG pathways enrichment analyses revealed that proteins from the SF group were associated with the ribosome, lysosome, and metabolic pathways, whereas proteins from RF were related to the proteasome, protein processing in the endoplasmic reticulum, signaling and metabolic pathways (Figures 1D,E, Supplementary Table S2).
The GO and KEGG terms of both groups were cross-compared to find common terms among them (highlight in blue in Supplementary Table S2). Eight common GO terms (“nuclear-transcribed mRNA catabolic process, nonsense-mediated decay”, “translational initiation”, “protein targeting to ER”, “SPR-dependent cotranslational protein targeting to membrane”, “azurophil granule lumen”, “acrosomal vesicle”, “azurophil granule” and “small ribosomal subunit”) and three common KEGG terms (“Glycolysis/Gluconeogenesis”, “Pyruvate metabolism” and “Cysteine and methionine metabolism”) were found. Those common terms were specifically related to the decreased proteins in both freezing groups.
An integrative network for each freezing group was constructed using PPI and enrichment analysis data (Figure 2). In the String database, 54 and 607 PPIs were collected for the slow and rapid freezing groups, respectively. Noteworthy, for both groups, only PPIs with a high confidence score were selected to ensure highly probable interactions among the proteins in the cell. Based on node degree, a measure that indicates the number of connections of each node, top 10 hubs in each network was selected (represented with bigger circles in Figure 2 and Table 1). Most of the hubs in the SF network were cytoplasmic ribosomal proteins (Table 1). On the other hand, 11 hubs were identified in the RF (PSMC2, PSMC3, PSMD12, PSMD7 had the same node degree) and all hubs were proteasomal proteins (Table 1). Among SF hubs, the RPS28, RPS19, RPLP1, and ARSA proteins were also identified as decreased proteins in the RF group. None of the hubs has been described in the DISGENET database. ARSA was previously reported as decreased in poor quality sperm samples and male infertility-related conditions (Table 1). An additional search was performed in available proteomic studies carried out on human sperm (control condition vs male infertility/poor-sperm quality) to observe if any of the hubs had previously been identified as differentially expressed.
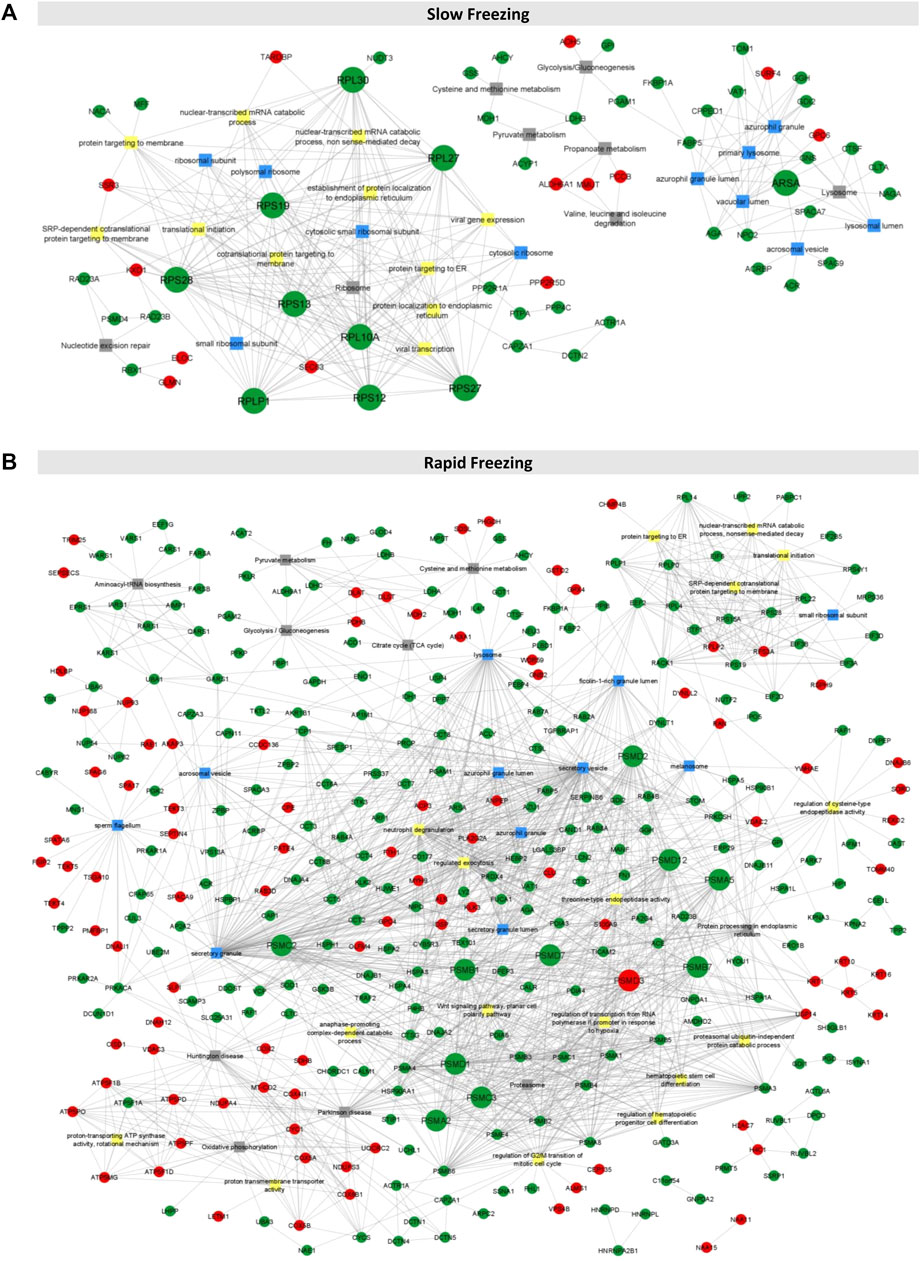
FIGURE 2. Integrative networks for rapid and slow freezing groups. (A, B) The integrative networks constructed for slow and rapid freezing groups using information from enrichment analyses (top 10 biological processes, cellular compartment, and KEGG terms as well as common terms to both groups and sperm-related terms) and PPIs data. Only connected proteins were represented (N = 60 and N = 344 for slow and rapid freezing, respectively). Each node corresponds to a DEP (represented by the gene name), being the red nodes increased proteins and the green nodes decreased proteins. The squares represent GO or KEGG terms (yellow squares for biological processes, blue squares for the cellular compartment, and grey squares for KEGG pathways). The nodes with bigger sizes represent the proteins with higher degree.
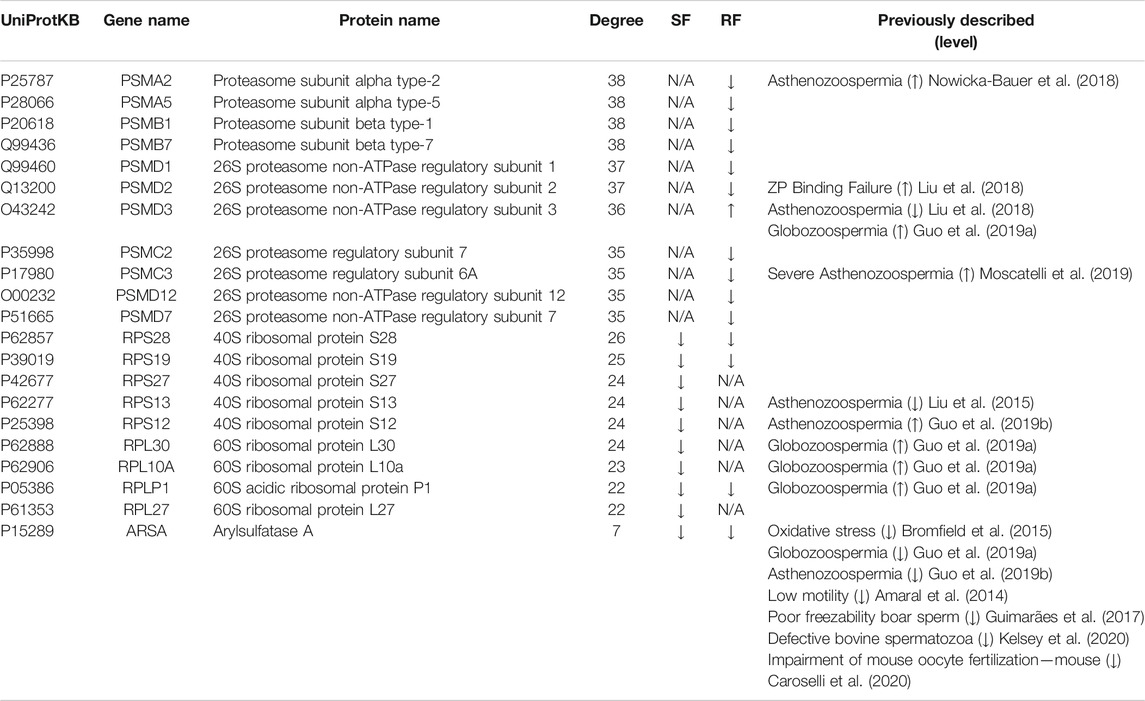
TABLE 1. Protein hubs identified in each integrative protein-protein interaction network: Slow Freezing (SF) and Rapid Freezing (RF). Node degree (computed in the respective network) and expression level (up or down) for each protein are indicated. Previous studies concerning expression levels of these proteins in conditions associated with poor-sperm quality are also indicated (studies were performed in human spermatozoa, except those where the specie is indicated). ZP, Zona Pellucida; N/A, not applicable.
Among the hubs described in the prior section, ARSA was chosen for experimental investigation since it was decreased in both freezing groups and low ARSA levels have been consistently associated with poor sperm quality and male infertility-related conditions (Table 1). To assess the ARSA levels in freeze-thawed spermatozoa, sperm protein extracts were analyzed by western blot. The ARSA levels were significantly reduced after RF (∼31.97%) and SF (∼39.28%) samples (Figures 3A,B). The post hoc power analysis revealed that differences between control and RF and SF groups yielded a statistical power of 75,32% and 74,63%, respectively. No significant alterations were observed in β-tubulin levels (Figures 3A,C). Cell viability and sperm motility were also evaluated. As expected, cell viability was significantly decreased in freeze-thawed spermatozoa (Figure 3D). Total and progressive motilities were significantly decreased in RF and SF (Figures 3E,F, respectively) and the percentage of immotile sperm was increased (Figure 3G).
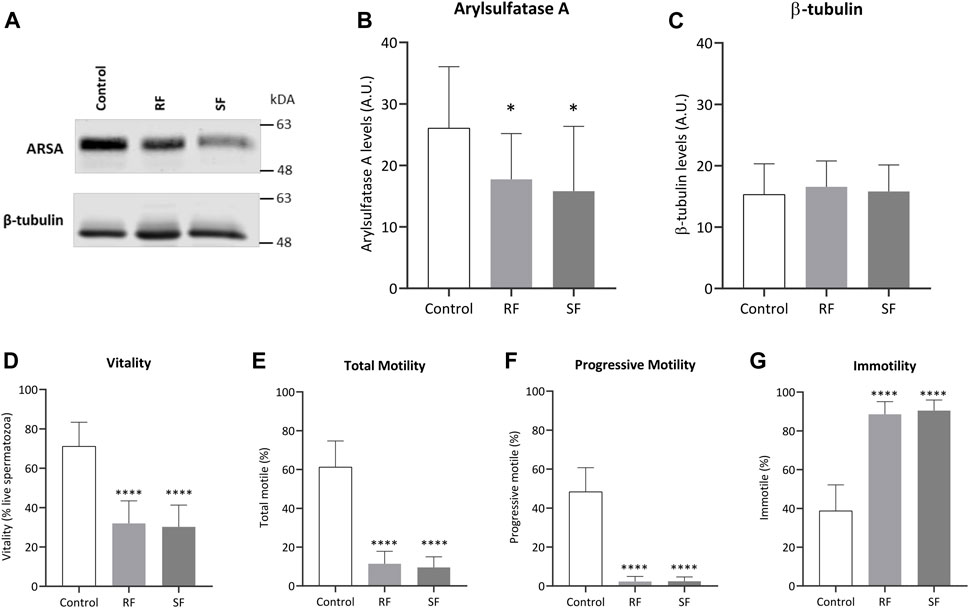
FIGURE 3. Impact of cryopreservation on ARSA levels and sperm parameters. (A) Representative image of ARSA (62 kDa) and β-tubulin (50 kDa) levels in ejaculated human spermatozoa, from normozoospermic men (n = 15), before and after rapid and slow freezing. Whole-cell lysates were separated by SDS-PAGE (B,C) Quantification of ARSA and β-tubulin levels, respectively (n = 15). For each condition, the band intensity was normalized to Ponceau staining. (D) Sperm viability was assessed through the eosin-nigrosine staining. The impact of the freezing method was also observed in sperm (E) total motility, (F) progressive motility, and (G) immotility. In all graphs, bars represent the mean values, and the error bars correspond to the standard deviation. Statistically significant findings compared with control are indicated with a *p < 0.05 or ****p < 0.0001. RF, rapid freezing; SF, slow freezing.
Cryopreservation induces significant cellular changes in spermatozoon that are extensively described though poorly understood from a molecular point of view (Hezavehei et al., 2018; Rienzi et al., 2018; Kumar et al., 2019). In recent years, proteomic analysis has become an important tool to identify DEPs allowing the molecular understanding of certain physiological and pathological contexts (Li et al., 2017; Agarwal et al., 2020). Still, one of the major limitations of examining single proteomic studies is to draw robust conclusions concerning potential markers and altered pathways in a specific condition. To date, the present study is the first to merge all the available proteomic data from human cryopreserved sperm to unveil key DEPs that can be potential cryopreservation markers or modulation targets.
From the included proteomic studies (Bogle et al., 2017; Fu et al., 2019; Li et al., 2019), decreased and increased DEPs were collected for the two conventional freezing techniques used in sperm cryopreservation: slow freezing (SF) and rapid freezing (RF). Decreased protein levels are easily explained by the cryopreservation process, which leads to protein degradation, plasmatic membrane damage, and osmotic stress induced by the cryoprotectant. Antagonistically, the underlying mechanisms for increased proteins levels are less clear. First, the protein increased levels are not induced by the cryoprotectant used, since included studies only used egg-yolk-free mediums (Bogle et al., 2017; Fu et al., 2019; Li et al., 2019). Wang and colleagues (Wang et al., 2014) conjectured that post-translational modifications (PTMs), such as protein phosphorylation, could be the reason behind the protein increment. In fact, cryopreservation appears to induce a set of differential protein-tyrosine phosphorylation patterns (Naresh and Atreja, 2015). Nonetheless, despite PTMs being identified as perturbations in protein abundance (Larsen et al., 2006), shotgun proteomics (the same approach used in the included studies) has various limitations in PTMs analysis and enrichment protocols are necessary to a proper PTMs study (Agarwal et al., 2020; Virág et al., 2020). Thus, further investigation should be performed to understand why some proteins are increased after cryopreservation.
Integrative networks using GO and KEGG pathways enrichment analyses, and PPI data allowed the identification of relevant proteins in each freezing group network. In the SF network, nine of the 10 hubs were cytoplasmic ribosomal proteins (RPS28, RPS19, RPS27, RPS13, RPS12, RPL30, RPL10A, RPLP1, and RPL27). Despite mammalian spermatozoa has been considered as transcriptionally and translationally inherent, some studies reported the existence of de novo protein synthesis under capacitation conditions (Gur and Breitbart, 2006, 2008; Li et al., 2010; Rajamanickam et al., 2017; Zhu et al., 2019). The protein translation occurrence appears to be important for the replacement of some degraded proteins during capacitation events and its impairment was associated with a reduction in motility, actin polymerization, acrosomal reaction, and in vitro fertilization (Gur and Breitbart, 2006). In the light of current knowledge, no available studies are referring neither the occurrence of translation activity in freeze-thawed spermatozoa nor the association between low levels of ribosomal proteins with poor-sperm quality. Nevertheless, the GO and KEGG pathways analyses revealed that decreased proteins from the SF group were strongly associated with translation-related terms, suggesting that translation pathways were negatively affected by cryopreservation. Further approaches should be performed to assess the translation activity in freeze-thawed sperm and the impact of translation impairment after cryopreservation.
By contrast, in the RF network, all hubs were proteasomal proteins (PSMA2, PSMA5, PSMB1, PSMB7, PSMD1, PSMD2, PSMD3, PSMC2, PSMC3, PSMD12, and PSMD7), which are components of the 26S proteasome. The 26S proteasome is a multi-catalytic protease with a high affinity for ubiquitinated proteins and has a crucial role in protein degradation and turnover in all living organisms (Livneh et al., 2016). In mammalian spermatozoa, 26S proteasome has been implicated in sperm capacitation and fertilization events. Recent evidence showed that 26S proteasome activity is increased during the early onset of capacitation, through activating phosphorylation by protein kinase A (Kerns et al., 2016). Sperm active proteasome participates in protein degradation and modification “helping” the plasma membrane and acrosome remodulation, acrosomal exocytosis, and sperm-zona pellucida (ZP) interaction (Kerns et al., 2016). If proteasomal proteins are decreased, the 26S proteasome pathway could be affected leading to capacitation failure and consequently fertilization inability of freeze-thawed spermatozoa. In fact, decreased proteasomal activity was already associated with poor-sperm quality (Rawe et al., 2008; Rosales et al., 2011). Additionally, the enrichment analyses revealed a significant association between decreased DEPs from the RF group and proteasome pathway, corroborating this hypothesis. Thus, this study presents the first evidence that the proteasomal pathway could be a good target for future studies in sperm cryopreservation.
Among the hubs identified, arylsulfatase A (ARSA) had decreased levels in both freezing groups and thus it was selected for subsequent investigation after cryopreservation. ARSA is a lysosomal enzyme present in sperm membrane that interacts, before gametes fusion, with sulfated sugar residues of ZP-glycoproteins and, consequently, contributes to sperm binding and penetration of the ZP (Xu et al., 2012a). Together with hyaluronidase PH-20 (SPAM1) and Heat shock-related 70 kDa protein 2 (HSPA2), they form a protein-membrane complex being only expressed on sperm surface after capacitation (Redgrove et al., 2012, 2013; Gómez-Torres et al., 2021). ARSA was identified in proteomic studies of cryopreserved human sperm (Fu et al., 2019; Li et al., 2019). Still, this is the first study to highlight ARSA potential as a marker to assess sperm quality after cryopreservation and to perform further experimental validation. ARSA levels were analyzed in freeze-thawed human sperm samples, being significantly decreased after both SF and RF cryopreservation. This observation suggests that cryopreservation methods induce ARSA degradation. As expected, both methods also induced a drastic decrease in sperm viability and motility. Low ARSA levels were previously reported after cryopreservation in boar sperm with poor freezability and defective bovine sperm (Guimarães et al., 2017; Kelsey et al., 2020). Besides cryopreservation, ARSA reduced levels were observed in sperm exposed to oxidative stress inducers (Bromfield et al., 2015), sperm with low motility (Amaral et al., 2014), and globozoospermia (Guo et al., 2019a) and asthenozoospermia (Guo et al., 2019b) conditions. Together, these findings suggest ARSA as a potential good biomarker to access sperm quality after cryopreservation and to distinguish between good- and poor-sperm. Curiously, the oxidative stress effects in ARSA expression levels appeared to be minimized in sperm cells treated with penicillamine (Bromfield et al., 2015). Caroselli and colleagues showed that ARSA could be reversibly inhibited in mouse sperm and its inhibition significantly impaired the in vitro oocyte fertilization (Caroselli et al., 2020). Therefore, considering the ARSA key role in sperm-oocyte interaction (Xu H. et al., 2012), future studies should be performed to prevent ARSA degradation and increase the success of cryopreservation protocols.
The present work presents three main limitations. Considering the importance of cryopreservation protocols in reproductive medicine, there is a very limited number of proteomic studies on human sperm available. Second, there is limited knowledge in the databases for a cell type as specialized as the sperm cell. Most signaling pathways have been thoroughly investigated in somatic cells. However, they are poorly characterized in male germ cells. This negatively affects the bioinformatic analyses performed in this study. The last limitation concerns the small number of samples included in experimental validation. Despite the high statistical power yielded, the validation can be strengthened by increasing the number of men recruited.
To summarize, the integration of the available proteomic data, using a bioinformatic workflow, proved to be very useful to identify a set of key proteins that are differentially expressed in freeze-thawed human spermatozoa and to highlight ARSA for further experimental validation. For the first time, ARSA levels were assessed in freeze-thawed human sperm, being significantly decreased in both freezing groups. Thus, ARSA has the potential to be a good marker to evaluate sperm quality after cryopreservation. On the other hand, the enrichment analysis also allowed to recognize, for the first time, two altered pathways (proteasomal and translation pathways) on cryopreserved human sperm that could be investigated in future research.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by the National Council for Medically Assisted Procreation. The patients/participants provided their written informed consent to participate in this study.
PC—conception, study design, execution, analysis, critical discussion, figure preparation, and manuscript drafting; JS—conception, study design, execution, analysis, critical discussion, and manuscript revision; SP—study execution; AB–study execution; MA—study design, critical discussion, and manuscript revision; MF—conception, study design, critical discussion, and manuscript revision.
This work was supported by “MEDISIS IDEAS” an initiative integrated in the “MEDISIS - Promotion of the involvement of key actors in the Centro Region for the transfer of knowledge and the creation of new partnerships in the areas of Systems Medicine”, co-financed by PORTUGAL 2020, through CENTRO 2020. This study was also supported by Institute for Biomedicine—iBiMED (UIDB/04501/2020 and POCI-0145-FEDER-007628) and by individual grants from the Foundation for Science and Technology (FCT) of the Portuguese Ministry of Science and Higher Education to PC (2020.10111.BD), JVS (SFRH/BPD/123155/2016) and SP (2021.05487.BD). Unit for Multidisciplinary Research in Biomedicine (UMIB) is funded by FCT (grant numbers UIDB/00215/2020, and UIDP/00215/2020), and ITR - Laboratory for Integrative and Translational Research in Population Health (LA/P/0064/2020).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Professor Doctor Vera Afreixo (CIDMA, Department of Mathematics, University of Aveiro) for her scientific advice.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.759354/full#supplementary-material
ARSA, Arylsulfatase A; DEPs, Differentially Expressed Proteins; GO, Gene Ontology; PPI, Protein-Protein Interaction; RF, Rapid Freezing; SF, Slow Freezing; ZP, Zona-Pellucida.
Agarwal, A., Panner Selvam, M. K., and Baskaran, S. (2020). Proteomic Analyses of Human Sperm Cells: Understanding the Role of Proteins and Molecular Pathways Affecting Male Reproductive Health. Ijms 21, 1621. doi:10.3390/ijms21051621
Amaral, A., Lourenço, B., Marques, M., and Ramalho-Santos, J. (2013). Mitochondria Functionality and Sperm Quality. Reproduction 146, R163–R174. doi:10.1530/REP-13-0178
Amaral, A., Paiva, C., Attardo Parrinello, C., Estanyol, J. M., Ballescà, J. L., Ramalho-Santos, J., et al. (2014). Identification of Proteins Involved in Human Sperm Motility Using High-Throughput Differential Proteomics. J. Proteome Res. 13, 5670–5684. doi:10.1021/pr500652y
Assenov, Y., Ramírez, F., Schelhorn, S.-E., Lengauer, T., and Albrecht, M. (2008). Computing Topological Parameters of Biological Networks. Bioinformatics 24, 282–284. doi:10.1093/bioinformatics/btm554
Banihani, S. A., and Alawneh, R. F. (2019). Human Semen Samples with High Antioxidant Reservoir May Exhibit Lower post-cryopreservation Recovery of Sperm Motility. Biomolecules 9, 111. doi:10.3390/biom9030111
Bardou, P., Mariette, J., Escudié, F., Djemiel, C., and Klopp, C. (2014). Jvenn: an Interactive Venn Diagram Viewer. BMC Bioinformatics 15, 293–297. doi:10.1186/1471-2105-15-293
Bogle, O. A., Kumar, K., Attardo-Parrinello, C., Lewis, S. E. M., Estanyol, J. M., Ballescà, J. L., et al. (2017). Identification of Protein Changes in Human Spermatozoa throughout the Cryopreservation Process. Andrology 5, 10–22. doi:10.1111/andr.12279
Bromfield, E. G., Aitken, R. J., Anderson, A. L., McLaughlin, E. A., and Nixon, B. (2015). The Impact of Oxidative Stress on Chaperone-Mediated Human Sperm-Egg Interaction. Hum. Reprod. 30, 2597–2613. doi:10.1093/humrep/dev214
Cao, X., Cui, Y., Zhang, X., Lou, J., Zhou, J., Bei, H., et al. (2018). Proteomic Profile of Human Spermatozoa in Healthy and Asthenozoospermic Individuals. Reprod. Biol. Endocrinol. 16, 4–11. doi:10.1186/s12958-018-0334-1
Caroselli, S., Zwergel, C., Pirolli, A., Sabatino, M., Xu, Z., Kirsch, G., et al. (2020). Discovery of the First Human Arylsulfatase A Reversible Inhibitor Impairing Mouse Oocyte Fertilization. ACS Chem. Biol. 15, 1349–1357. doi:10.1021/acschembio.9b00999
Carrageta, D. F., Guerra-Carvalho, B., Sousa, M., Barros, A., Oliveira, P. F., Monteiro, M. P., et al. (2020). Mitochondrial Activation and Reactive Oxygen-Species Overproduction during Sperm Capacitation Are Independent of Glucose Stimuli. Antioxidants 9, 750. doi:10.3390/antiox9080750
De Rose, M. B., Piccolomini, M. M., Soares Belo, A. S., Borges, E., and Filho, F. F. (2018). Proteomics in Human Reproduction. Ppl 25, 420–423. doi:10.2174/0929866525666180412164602
Di Santo, M., Tarozzi, N., Nadalini, M., and Borini, A. (2012). Human Sperm Cryopreservation: Update on Techniques, Effect on DNA Integrity, and Implications for ART. Adv. Urol. 2012, 1–12. doi:10.1155/2012/854837
Fu, L., An, Q., Zhang, K., Liu, Y., Tong, Y., Xu, J., et al. (2019). Quantitative Proteomic Characterization of Human Sperm Cryopreservation: Using Data-independent Acquisition Mass Spectrometry. BMC Urol. 19, 1–9. doi:10.1186/s12894-019-0565-2
Gholami, D., Ghaffari, S. M., Shahverdi, A., Sharafi, M., Riazi, G., Fathi, R., et al. (2018). Proteomic Analysis and Microtubule Dynamicity of Human Sperm in Electromagnetic Cryopreservation. J. Cel. Biochem. 119, 9483–9497. doi:10.1002/jcb.27265
Gómez-Torres, M. J., Huerta-Retamal, N., Robles-Gómez, L., Sáez-Espinosa, P., Aizpurua, J., Avilés, M., et al. (2021). Arylsulfatase A Remodeling during Human Sperm In Vitro Capacitation Using Field Emission Scanning Electron Microscopy (FE-SEM). Cells 10, 222. doi:10.3390/cells10020222
Guimarães, D. B., Barros, T. B., van Tilburg, M. F., Martins, J. A. M., Moura, A. A., Moreno, F. B., et al. (2017). Sperm Membrane Proteins Associated with the Boar Semen Cryopreservation. Anim. Reprod. Sci. 183, 27–38. doi:10.1016/j.anireprosci.2017.06.005
Guo, Y., Jiang, J., Zhang, H., Wen, Y., Zhang, H., Cui, Y., et al. (2019a). Proteomic Analysis of Dpy19l2‐Deficient Human Globozoospermia Reveals Multiple Molecular Defects. Prot. Clin. Appl. 13, 1900007–1900009. doi:10.1002/prca.201900007
Guo, Y., Jiang, W., Yu, W., Niu, X., Liu, F., Zhou, T., et al. (2019b). Proteomics Analysis of Asthenozoospermia and Identification of Glucose-6-Phosphate Isomerase as an Important Enzyme for Sperm Motility. J. Proteomics 208, 103478. doi:10.1016/j.jprot.2019.103478
Gur, Y., and Breitbart, H. (2006). Mammalian Sperm Translate Nuclear-Encoded Proteins by Mitochondrial-type Ribosomes. Genes Dev. 20, 411–416. doi:10.1101/gad.367606
Gur, Y., and Breitbart, H. (2008). Protein Synthesis in Sperm: Dialog between Mitochondria and Cytoplasm. Mol. Cell Endocrinol. 282, 45–55. doi:10.1016/j.mce.2007.11.015
Hezavehei, M., Sharafi, M., Kouchesfahani, H. M., Henkel, R., Agarwal, A., Esmaeili, V., et al. (2018). Sperm Cryopreservation: A Review on Current Molecular Cryobiology and Advanced Approaches. Reprod. BioMedicine Online 37, 327–339. doi:10.1016/j.rbmo.2018.05.012
Jang, T. H., Park, S. C., Yang, J. H., Kim, J. Y., Seok, J. H., Park, U. S., et al. (2017). Cryopreservation and its Clinical Applications. Integr. Med. Res. 6, 12–18. doi:10.1016/J.IMR.2016.12.001
Karabulut, S., Demiroğlu-Zergeroğlu, A., Yılmaz, E., Kutlu, P., and Keskin, İ. (2018). Effects of Human Sperm Cryopreservation on Apoptotic Markers in Normozoospermic and Non-normozoospermic Patients. Zygote 26, 308–313. doi:10.1017/S0967199418000254
Kelsey, K. M., Zigo, M., Thompson, W. E., Kerns, K., Manandhar, G., Sutovsky, M., et al. (2020). Reciprocal Surface Expression of Arylsulfatase A and Ubiquitin in normal and Defective Mammalian Spermatozoa. Cell Tissue Res 379, 561–576. doi:10.1007/s00441-019-03144-1
Kerns, K., Morales, P., and Sutovsky, P. (2016). Regulation of Sperm Capacitation by the 26S Proteasome: An Emerging New Paradigm in Spermatology1. Biol. Reprod. 94, 1–9. doi:10.1095/biolreprod.115.136622
Kumar, A., Prasad, J. K., Srivastava, N., and Ghosh, S. K. (2019). Strategies to Minimize Various Stress-Related Freeze-Thaw Damages during Conventional Cryopreservation of Mammalian Spermatozoa. Biopreservation and Biobanking 17, 603–612. doi:10.1089/bio.2019.0037
Larsen, M. R., Trelle, M. B., Thingholm, T. E., and Jensen, O. N. (2006). Analysis of Posttranslational Modifications of Proteins by Tandem Mass Spectrometry. Biotechniques 40, 790–798. doi:10.2144/000112201
Lasso, L., Alvarez, G., and June, M. (1994). Mechanism of Superoxide Cells during Cryopreservation. J. Androl. 15, 255–265. doi:10.1002/j.1939-4640.1994.tb00444.x
Li, C.-H., Cheng, Y.-W., Liao, P.-L., Yang, Y.-T., and Kang, J.-J. (2010). Chloramphenicol Causes Mitochondrial Stress, Decreases ATP Biosynthesis, Induces Matrix Metalloproteinase-13 Expression, and Solid-Tumor Cell Invasion. Toxicol. Sci. 116, 140–150. doi:10.1093/toxsci/kfq085
Li, S., Ao, L., Yan, Y., Jiang, J., Chen, B., Duan, Y., et al. (2019). Differential Motility Parameters and Identification of Proteomic Profiles of Human Sperm Cryopreserved with Cryostraw and Cryovial. Clin. Proteom 16, 1–14. doi:10.1186/s12014-019-9244-2
Li, X., Wang, W., and Chen, J. (2017). Recent Progress in Mass Spectrometry Proteomics for Biomedical Research. Sci. China Life Sci. 60, 1093–1113. doi:10.1007/s11427-017-9175-2
Liu, X., Liu, G., Liu, J., Zhu, P., Wang, J., Wang, Y., et al. (2018). ITRAQ-based Analysis of Sperm Proteome from Normozoospermic Men Achieving the rescue-ICSI Pregnancy after the IVF Failure. Clin. Proteom 15, 1–15. doi:10.1186/s12014-018-9203-3
Liu, Y., Guo, Y., Song, N., Fan, Y., Li, K., Teng, X., et al. (2015). Proteomic Pattern Changes Associated with Obesity-Induced Asthenozoospermia. Andrology 3, 247–259. doi:10.1111/andr.289
Livneh, I., Cohen-Kaplan, V., Cohen-Rosenzweig, C., Avni, N., and Ciechanover, A. (2016). The Life Cycle of the 26S Proteasome: From Birth, through Regulation and Function, and onto its Death. Cell Res 26, 869–885. doi:10.1038/cr.2016.86
Martin, G., Cagnon, N., Sabido, O., Sion, B., Grizard, G., Durand, P., et al. (2007). Kinetics of Occurrence of Some Features of Apoptosis during the Cryopreservation Process of Bovine Spermatozoa. Hum. Reprod. 22, 380–388. doi:10.1093/humrep/del399
Mayr, S., Erdfelder, E., Buchner, A., and Faul, F. (2007). A Short Tutorial of GPower. Tqmp 3, 51–59. doi:10.20982/tqmp.03.2.p051
Moscatelli, N., Lunetti, P., Braccia, C., Armirotti, A., Pisanello, F., De Vittorio, M., et al. (2019). Comparative Proteomic Analysis of Proteins Involved in Bioenergetics Pathways Associated with Human Sperm Motility. Ijms 20, 3000. doi:10.3390/ijms20123000
Naresh, S., and Atreja, S. K. (2015). The Protein Tyrosine Phosphorylation during In Vitro Capacitation and Cryopreservation of Mammalian Spermatozoa. Cryobiology 70, 211–216. doi:10.1016/j.cryobiol.2015.03.008
Naz, R. K. (1999). Involvement of Protein Serine and Threonine Phosphorylation in Human Sperm Capacitation1. Biol. Reprod. 60, 1402–1409. doi:10.1095/biolreprod60.6.1402
Nowicka-Bauer, K., Lepczynski, A., Ozgo, M., Kamieniczna, M., Fraczek, M., Stanski, L., et al. (2018). Sperm Mitochondrial Dysfunction and Oxidative Stress as Possible Reasons for Isolated Asthenozoospermia. J. Physiol. Pharmacol. 69, 403–417. doi:10.26402/jpp.2018.3.05
O'Connell, M., McClure, N., and Lewis, S. E. M. (2002). The Effects of Cryopreservation on Sperm Morphology, Motility and Mitochondrial Function. Hum. Reprod. 17, 704–709. doi:10.1093/humrep/17.3.704
Organization, W. H. (2010). WHO Laboratory Manual for the Examination and Processing of Human Semen. 5th ed. Geneva, Switzerland: World Health Organization.
Parrilla, I., Perez-Patiño, C., Li, J., Barranco, I., Padilla, L., Rodriguez-Martinez, H., et al. (2019). Boar Semen Proteomics and Sperm Preservation. Theriogenology 137, 23–29. doi:10.1016/j.theriogenology.2019.05.033
Quinn, P. J. (1985). A Lipid-phase Separation Model of Low-Temperature Damage to Biological Membranes. Cryobiology 22, 128–146. doi:10.1016/0011-2240(85)90167-1
Rajamanickam, G. D., Kastelic, J. P., and Thundathil, J. C. (2017). Content of Testis-specific Isoform of Na/K-ATPase (ATP1A4) Is Increased during Bovine Sperm Capacitation through Translation in Mitochondrial Ribosomes. Cel Tissue Res 368, 187–200. doi:10.1007/s00441-016-2514-7
Rawe, V. Y., Diaz, E. S., Abdelmassih, R., Wojcik, C., Morales, P., Sutovsky, P., et al. (2008). The Role of Sperm Proteasomes during Sperm Aster Formation and Early Zygote Development: Implications for Fertilization Failure in Humans. Hum. Reprod. 23, 573–580. doi:10.1093/humrep/dem385
Redgrove, K. A., Anderson, A. L., McLaughlin, E. A., O'Bryan, M. K., Aitken, R. J., and Nixon, B. (2013). Investigation of the Mechanisms by Which the Molecular Chaperone HSPA2 Regulates the Expression of Sperm Surface Receptors Involved in Human Sperm-Oocyte Recognition. Mol. Hum. Reprod. 19, 120–135. doi:10.1093/molehr/gas064
Redgrove, K. A., Nixon, B., Baker, M. A., Hetherington, L., Baker, G., Liu, D.-Y., et al. (2012). The Molecular Chaperone HSPA2 Plays a Key Role in Regulating the Expression of Sperm Surface Receptors that Mediate Sperm-Egg Recognition. PLoS One 7, e50851. doi:10.1371/journal.pone.0050851
Rienzi, L., Iussig, B., Dovere, L., Fabozzi, G., Cimadomo, D., and Ubaldi, F. (2018). Perspectives in Gamete and Embryo Cryopreservation. Semin. Reprod. Med. 36, 253–264. doi:10.1055/s-0038-1677463
Rosales, O., Opazo, C., Diaz, E. S., Villegas, J. V., Sanchez, R., and Morales, P. (2011). Proteasome Activity and Proteasome Subunit Transcripts in Human Spermatozoa Separated by a Discontinuous Percoll Gradient. Andrologia 43, 106–113. doi:10.1111/j.1439-0272.2009.01029.x
Ryu, D.-Y., Song, W.-H., Pang, W.-K., Yoon, S.-J., Rahman, M. S., and Pang, M.-G. (2019). Freezability Biomarkers in Bull Epididymal Spermatozoa. Sci. Rep. 9, 1–9. doi:10.1038/s41598-019-49378-5
Said, T. M., Gaglani, A., and Agarwal, A. (2010). Implication of Apoptosis in Sperm Cryoinjury. Reprod. BioMedicine Online 21, 456–462. doi:10.1016/j.rbmo.2010.05.011
Saleh, R. A., and Agarwal, A. (2002). Oxidative Stress and Male Infertility: From Research Bench to Clinical Practice. J. Androl. 23, 737–752. doi:10.1002/j.1939-4640.2002.tb02324.x
Santiago, J., Silva, J. V., and Fardilha, M. (2019). First Insights on the Presence of the Unfolded Protein Response in Human Spermatozoa. Ijms 20, 5518. doi:10.3390/ijms20215518
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 13, 2498–2504. doi:10.1101/gr.1239303.metabolite
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-wide Experimental Datasets. Nucleic Acids Res. 47, D607–D613. doi:10.1093/nar/gky1131
Talaei, T., Esmaeelpour, T., Aekiyash, F., and Bahmanpour, S. (2010). Effects of Cryopreservation on Plasma Membrane Glycoconjugates of Human Spermatozoa. Iran. J. Reprod. Med. 8, 119–124.
Tomlinson, M., Meadows, J., Kohut, T., Haoula, Z., Naeem, A., Pooley, K., et al. (2015). Review and Follow-Up of Patients Using a Regional Sperm Cryopreservation Service: Ensuring that Resources Are Targeted to Those Patients Most in Need. Andrology 3, 709–716. doi:10.1111/andr.12045
Tournaye, H., Dohle, G. R., and Barratt, C. L. R. (2014). Fertility Preservation in Men with Cancer. The Lancet 384, 1295–1301. doi:10.1016/S0140-6736(14)60495-5
Trindade, F., Ferreira, R., Magalhães, B., Leite-Moreira, A., Falcão-Pires, I., and Vitorino, R. (2018). How to Use and Integrate Bioinformatics Tools to Compare Proteomic Data from Distinct Conditions? A Tutorial Using the Pathological Similarities between Aortic Valve Stenosis and Coronary Artery Disease as a Case-Study. J. Proteomics 171, 37–52. doi:10.1016/j.jprot.2017.03.015
Verheyen, G., Pletincx, I., and Van Steirteghem, A. (1993). Andrology: Effect of Freezing Method, Thawing Temperature and post-thaw Dilution/washing on Motility (CASA) and Morphology Characteristics of High-Quality Human Sperm. Hum. Reprod. 8, 1678–1684. doi:10.1093/oxfordjournals.humrep.a137912
Virág, D., Dalmadi-Kiss, B., Vékey, K., Drahos, L., Klebovich, I., Antal, I., et al. (2020). Current Trends in the Analysis of Post-translational Modifications. Chromatographia 83, 1–10. doi:10.1007/s10337-019-03796-9
Wang, S., Wang, W., Xu, Y., Tang, M., Fang, J., Sun, H., et al. (2014). Proteomic Characteristics of Human Sperm Cryopreservation. Proteomics 14, 298–310. doi:10.1002/pmic.201300225
Xu, H., Liu, F., Srakaew, N., Koppisetty, C., Nyholm, P.-G., Carmona, E., et al. (2012). Sperm Arylsulfatase a Binds to mZP2 and mZP3 Glycoproteins in a Nonenzymatic Manner. Reproduction 144, 209–219. doi:10.1530/REP-11-0338
Xu, W., Hu, H., Wang, Z., Chen, X., Yang, F., Zhu, Z., et al. (2012). Proteomic Characteristics of Spermatozoa in Normozoospermic Patients with Infertility. J. Proteomics 75, 5426–5436. doi:10.1016/j.jprot.2012.06.021
Keywords: sperm cryopreservation, proteomic analysis, bioinformatic tools, ARSA, biomarker candidates
Citation: Corda PO, Silva JV, Pereira SC, Barros A, Alves MG and Fardilha M (2022) Bioinformatic Approach to Unveil Key Differentially Expressed Proteins in Human Sperm After Slow and Rapid Cryopreservation. Front. Cell Dev. Biol. 9:759354. doi: 10.3389/fcell.2021.759354
Received: 16 August 2021; Accepted: 23 December 2021;
Published: 25 January 2022.
Edited by:
Souvik Dey, Manipal Academy of Higher Education (MAHE), IndiaReviewed by:
Eva Tvrda, Slovak University of Agriculture, SlovakiaCopyright © 2022 Corda, Silva, Pereira, Barros, Alves and Fardilha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Margarida Fardilha, bWZhcmRpbGhhQHVhLnB0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.