
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 11 October 2021
Sec. Epigenomics and Epigenetics
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.753931
This article is part of the Research TopicEpigenomic and Epitranscriptomic Basis of Development and Human DiseaseView all 14 articles
Musculoskeletal degenerative diseases (MSDDs) are pathological conditions that affect muscle, bone, cartilage, joint and connective tissue, leading to physical and functional impairments in patients, mainly consist of osteoarthritis (OA), intervertebral disc degeneration (IDD), rheumatoid arthritis (RA) and ankylosing spondylitis (AS). Long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) are novel regulators of gene expression that play an important role in biological regulation, involving in chondrocyte proliferation and apoptosis, extracellular matrix degradation and peripheral blood mononuclear cell inflammation. Research on MSDD pathogenesis, especially on RA and AS, is still in its infancy and major knowledge gaps remain to be filled. The effects of lncRNA/circRNA-miRNA-mRNA axis on MSDD progression help us to fully understand their contribution to the dynamic cellular processes, provide the potential OA, IDD, RA and AS therapeutic strategies. Further studies are needed to explore the mutual regulatory mechanisms between lncRNA/circRNA regulation and effective therapeutic interventions in the pathology of MSDD.
Musculoskeletal degenerative diseases (MSDDs) are pathological conditions that affect muscle, bone, cartilage, joint and connective tissue, leading to physical and functional impairment in patients (Chen Y. et al., 2017; Huo et al., 2018). With the acceleration of the global aging process, the prevalence of MSDD is increasing. This is a huge challenge for patients and healthcare workers, and adds to the global healthcare burden (Li and Chen, 2019). The main MSDD consists of osteoarthritis (OA), intervertebral disc degeneration (IDD), rheumatoid arthritis (RA), and ankylosing spondylitis (AS) (Vinatier et al., 2016; Huo et al., 2018; Loef et al., 2018). OA is a chronic age-related MSDD, featuring for subchondral bone thickening, articular cartilage degradation, and osteophyte formation (Loeser et al., 2012; Hunter and Bierma-Zeinstra, 2019). IDD is also age-related and is caused by progressive degeneration of the disk (Yang S. et al., 2020), causing loss of disk height, reduced hydration and decreased potential to absorb load (Samartzis et al., 2011; Cooper et al., 2016). RA is an autoimmune disease characterized by aggressive arthritis that can lead to joint deformities and loss of function (Smolen et al., 2016). AS, a rare but clear cause of chronic back pain, is an inflammatory disease involving the spine, sacroiliac joints and other joints (Taurog et al., 2016). OA and IDD became mainly responsible for MSDD. Their common character is the broken dynamic equilibrium between catabolism and anabolism in the extracellular matrix (ECM). On the one hand, chondrocytes is only resident cells in the articular system, the ECM degeneration in OA is leaded by chondrocytes’ catabolic and abnormal differentiation (Zhou Z.B. et al., 2019). Cartilage cellularity is reduced in OA because of chondrocyte death. On the other hand, ECM breakdown and abnormal matrix synthesis in IDD is responsible by nucleus pulposus (NP) cells, which are predominant cells in the NP tissue (Fontana et al., 2015). Excessive apoptosis of NP cells could accelerate IDD progression (Zhao et al., 2006). Meanwhile, endplate cartilage degeneration is another risk factor of IDD (Iwakura et al., 2013) due to its irreplaceable nutrition supplement of intervertebral disk (Yuan et al., 2015). Although multiple factors are involved in the pathogenesis of MSDD (Li and Chen, 2019), the development of molecular mechanism of MSDD is still poor. Thus, it is urgent to discover new biomarkers to optimize MSDD early diagnosis and treatment.
With the development of sequencing technology, recent advances have shown that about 98% of the human genome is composed of non-coding RNAs (ncRNAs). In the past, ncRNAs were thought to act as ‘evolutionary junk.’ However, an increasing amount of evidence reported that ncRNAs play an important role in biological regulation (Beermann et al., 2016; Vieira et al., 2018). The main types of ncRNAs include long non-coding RNA (lncRNA), circular RNA (circRNA) and microRNA (miRNA) (Beermann et al., 2016). Recently, extensive evidence suggested that ncRNAs play a vital role in the development of MSDD (Chen W.K. et al., 2017; Yu and Sun, 2018; Wang J. et al., 2019). Moreover, circRNA and lncRNA can interact with miRNA to further regulate downstream target mRNA in the MSDD and play regulatory roles in numerous biological functions, such as proliferation, apoptosis and inflammation. In this review, we focused on the role of lncRNA/circRNA-miRNA-mRNA axis in the development of MSDD and further explored related molecular mechanism of MSDD.
MicroRNAs are encoded by endogenous genes, are approximately 20 nucleotides in length and are non-coding single-stranded RNA molecules (Beermann et al., 2016). Since they were first described in Caenorhabditis elegans, the number of miRNAs that have been found in mammals increased (Lee et al., 1993). miRNA is evolutionarily conserved and regulates gene expression at the post-transcriptional level by interfering with mRNA translation and degradation (Zhang et al., 2020b). With the iteration of gene chip and sequencing technology, numerous miRNAs have been found to play important roles in MSDD and can be used as biomarkers for clinical diagnosis and treatment (Satoshi Yamashita, 2012; Seeliger et al., 2016; Moran-Moguel et al., 2018). lncRNAs refers to non-protein-coding transcripts with the lengths of more than 200 nucleotides (Beermann et al., 2016). According to the position of the protein-encoding genes in the genome, lncRNAs are divided into five types, namely, intronic, intergenic, bidirectional, sense and antisense (Wang J. et al., 2019). More and more evidence argues that lncRNAs can act as an enhancer or suppressor to regulate the immune response at the epigenetic level, function as scaffold molecules through interactions with RNA-binding proteins in chromatin remodeling complexes (Mathy and Chen, 2017), and then, are involved in many cellular and biological processes in MSDD, such as proliferation, apoptosis, differentiation, inflammation and ECM degradation (Jiang et al., 2017; Li Z. et al., 2018; Abbasifard et al., 2020). Thus, it is important to develop lncRNA as a biomarker and therapeutic target for MSDD.
In recent years, extensive evidence has shown that lncRNAs can interact with miRNAs through several post-transcriptional mechanisms, and the four mechanisms of interaction are as follows. (1) lncRNAs act as miRNA sponges. The lncRNA that can prevent miRNA from acting on mRNA is called competing endogenous RNAs (ceRNAs). These lncRNAs have similar miRNA targets, and they can act as sponges of miRNA, thereby reducing the expression of miRNA and enhancing the translation of target mRNA. lncRNA AK048451 was first considered as an endogenous sponge of miR-489 that can combine with and inhibit the expression of miR-489 (Huang, 2018). (2) Several lncRNAs could directly compete with miRNAs to bind with mRNAs, thereby removing the regulatory roles of miRNAs on mRNAs. For example, lncRNA BACE1AS competes with miR-485-5p to combine with BACE1 mRNA. Thus, the degradation of BACE1 induced by miR-485-5p was inhibited (Faghihi et al., 2010). (3) miRNAs aim at lncRNAs to decrease the stability of lncRNAs and affect the abundance of lncRNAs. It has been verified that lncRNA-p21 was modulated by miRNA let-7b. Upregulation of let-7b promoted the degradation of RNA, leading to the instability of lncRNA-p21 (Deng et al., 2016). (4) Several lncRNAs could generate miRNAs. For instance, lncRNA H19 can generate miR-675 (Deng et al., 2016). To achieve a better understanding of the molecular mechanisms in MSDD progression, in-depth studies about the effects of lncRNAs and their potential downstream miRNA regulators have been performed in recent years.
As endogenous RNAs, circRNAs are characterize by covalent loop structures without 5′–3′ polarity nor a polya- denylated tail (Zhou et al., 2018). Different from linear RNA, circRNAs are inherently conserved due to their closed covalent structure and resistance to exonuclides; they are considered to be stable in exosomes (Haque and Harries, 2017). circRNAs are classified into four types according to their origin, namely, exonic circRNAs, exon-intron circRNAs, intronic circRNAs and intergenic circRNAs (Deng et al., 2016). A growing number of studies indicate that circRNAs exist miRNA complementary binding sites to interact with miRNAs, thereby playing regulatory roles in diseases and effecting in many biological processes, such as inflammation, apoptosis and ECM degradation, by participating in the modulation of transcriptional and post-transcriptional levels (Rong et al., 2017; Verduci et al., 2019). The mechanisms included circRNAs acting as miRNAs sponges and miRNAs regulating circRNAs (Kulcheski et al., 2016). For instance, the circAnks1a could regulate VEGFB (vascular endothelial growth factor-B) expression to suppress the excitability of spinal cord by sponging miR-324-3p in neuropathic pain (Zhang S.B. et al., 2019). Pan et al. (2019) elucidated that the miR-1224 could mediate circRNA-Filip1l expression through regulating Ubr5 in the spinal cord of chronic inflammatory pain mice. Although circRNAs are generally considered as ncRNAs because of non-linear structure, several circRNAs, such as CircFBXW7 (Ye et al., 2019) and Circ-EGFR (Liu et al., 2021), are proved to have translation functions due to its translatable open reading frame containing a start codon. The cap-independent translation pathway is thought to be the main mechanism of circRNA translation to encode protein (He et al., 2021). Combined with the above explanation, currently known that circRNAs can interact with proteins or act as miRNA sponges and regulate the expression of upstream gene to participate in the process of diseases development. In recent years, circRNAs have become a research hotspot in MSDD and showed great potential as biomarkers and therapeutic targets (Li H.Z. et al., 2018; Lei B. et al., 2019; Wu et al., 2019).
In the past decade, quite number of studies have shown that the interaction between lnRNAs and miRNAs is involved in the multiple biological processes of OA, such as inflammation, proliferation, apoptosis, autophagy, cell viability and ECM degradation (Table 1). The major interaction mechanism between lncRNA and miRNA in OA was that lncRNAs as ceRNAs acts as miRNAs sponges. Wang Q. et al. (2017) reported that the expressions of lncRNA OPN and NEAT1 significantly increased, whereas that of miR-181c decreased. According to luciferase assays, miR-181c could combine with NEAT1 and 3′UTR of OPN in synoviocytes, leading to NEAT1 competing with OPN for binding with miR-181c and further enhancing the level of OPN. Chen Y. et al. (2020) showed that lncRNA HOTAIR (HOX transcript antisense intergenic RNA) and mRNA PTEN (phosphatase and tensin homolog) was significantly increased in the OA mice, whereas miR-20b decreased. HOTAIR was involved in the process of apoptosis and ECM degradation by sponging miR-20b and regulating the downstream target PTEN. Lu and Zhou (2020) revealed that lncRNA00662 was downregulated in the cartilage of OA rats. The expression of miR-15b-5p was negative with lncRNA00662, whereas the expression of GPR120 was positively correlated with lncRNA00662. lncRNA00662 regulated GPR120 in apoptosis by serving as a sponge for miR-15b-5p. Sun P. et al. (2020) also studied the effect of XIST on OA patients and showed that XIST upregulated SGTB and inhibited the depression on SGTB induced by miR-142-5p through sponging miR-142-5p. Another study reported that the level of lnc00623 and HRAS was downregulated, whereas miR-101 was increased in OA tissues compared with normal tissues (Lü et al., 2020). Based on luciferase reporter, miR-101 could combine with lnc00623 and HRAS. lnc00623 sponges miR-101 through competing with HRAS, thereby preventing the miR-101-induced depression on HRAS. Some other lncRNAs act as miRNAs sponges in OA and more detailed information is presented in Table 1.
The mechanism by which lncRNA and miRNA act on IDD that has been most studied is as follows: lncRNA acts as the sponge of miRNA to modulate target genes (Figure 1). Xi et al. (2017) demonstrated that lncRNA HCG18 was upregulated in the IDD and plays the sponge roles of miR-146a-5p in NP cells. HCG18 is involved in the progression of cell proliferation and apoptosis in NP cells via the miR-146a-5p/TARF6/NF-κB axis. Compared with normal NP tissues, lncRNA SNHG1 (small nucleolar RNA host gene 1) expression was boosted and miR-326, a target gene of SNHG1, was reduced in IDD samples (Tan et al., 2018). Moreover, miR-326 could directly bind with Cyclin D1 (CCND1), and the level of CCND1 in the NP cells markedly increased. Thus, Tan et al. (2018) observed that SNHG1 modulates NP cells proliferation via sponging miR-326 and further regulating CCND1. Another study reported that lncRNA H19 was upregulated in the IDD tissues and could activate Wnt/β-catenin signaling pathway (Wang et al., 2018d). Moreover, miR-326 could directly bind with Cyclin D1 (CCND1), and the level of CCND1 in the NP cells markedly increased. Thus, Tan et al. (2018) observed that SNHG1 modulates NP cells proliferation via sponging miR-326 and further regulating CCND1. Another study reported that lncRNA H19 was upregulated in the IDD tissues and could activate Wnt/β-catenin signaling pathway (Shao et al., 2019). Another research suggested that LINC00641 level increased in NP tissues, whereas miR-153-3p level decreased. ATG5 (autophagy-related gene 5) was a downstream gene of miR-153-3p and upregulated in NP cells (Wang J. et al., 2019). Moreover, LINC00641 could sponge miR-153-3p, and thereby regulate the level of ATG5, cell death and the progression of IDD. Yang Y. et al. (2019) elucidated that lncRNA lincRNA-SLC20A1 (SLC20A1) was overexpressed in IDD patients, and SLC20A1 could induce ECM degradation via sponging miR-31-5p and further modulating the downstream target gene MMP3. Another study established that the level of lncRNA PART1 and mRNA matrix metallopeptidase 2 (MMP2) in NP tissues were significantly higher than those in the control groups, whereas the levels of miR-93 were lower (Gao et al., 2020). Through dual-luciferase reporter assay, they proved that PART1 acts as miR-93 sponges in NP tissues and cells to suppress the expression of miR-93 and to further regulate MMP2. Zheng et al. (2020) showed that MALAT1 was reduced in NP cells, and upregulation of MALAT1 could relieve cell proliferation and apoptosis in vitro and inhibit the degree of INN in vivo. Moreover, they found that MALAT1 plays pivotal roles in IDD through sponging miR-503, and thereby modulate downstream MAPK signaling pathways.
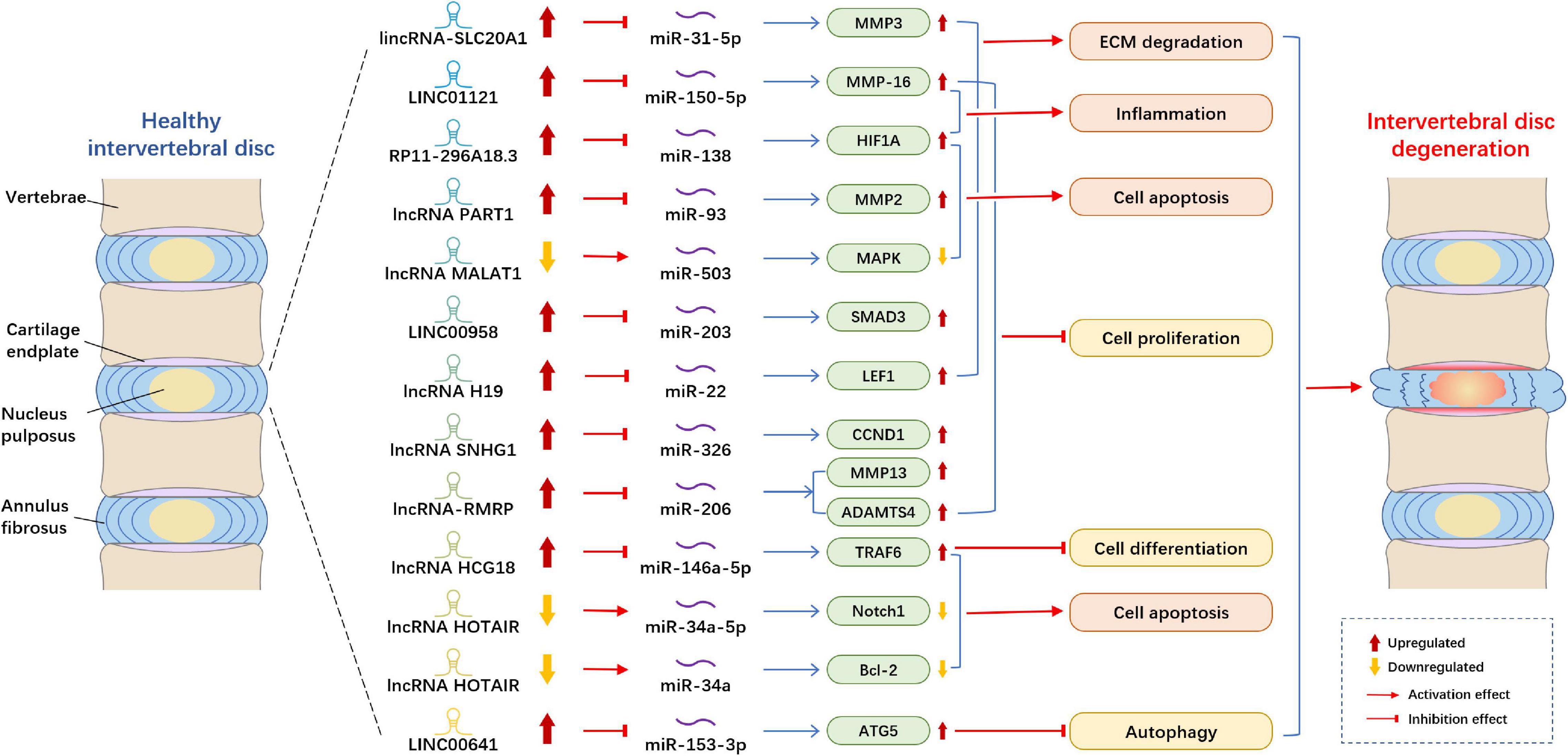
Figure 1. Example of altered lncRNA expression patterns and their biological effects in intervertebral disk degeneration. lncRNA, long non-coding RNA; ECM, extracellular matrix; MMP, matrix metallopeptidase; MAPK, mitogen-activated protein kinase; SMAD3, SMAD family member 3; LEF1, lymphoid enhancing factor-1; CCND1, cyclin D1; ADAMTS4, A disintegrin and metalloproteinase with thrombospondin motifs 4; TRAF6, tumor necrosis factor receptor-associated factor 6; Notch1, Notch Receptor 1; Bcl-2, B cell lymphoma 2; ATG5, autophagy-related gene 5.
Several studies indicated that lncRNAs plays roles in IDD by modulating miRNA and their target genes. Wang et al. (2018b) showed that the level of lncRNA-RMRP in degenerated NP tissues was higher than that in normal NP tissues, whereas the expression of miR-206 was lower. They indicated that lncRNA-RMRP could promote cell proliferation via modulating miR-206, thereby regulating downstream target gene MMP13 and ADAMTS4. lncRNA HOTAIR was downregulated in NP tissues and cells, whereas miR-34a expression was negatively correlated with HOTAIR and the expression of Bcl-2 was positively connected with HOTAIR (Yu et al., 2018). HOTAIR could inhibit NP cell apoptosis through regulating miR-34a/Bcl-2 axis. A study found that LINC00958 and mRNA SMAD3 were upregulated in NP tissues, whereas miR-203 was downregulated. Ectopic expression of miR-203 could suppress cell growth and ECM degradation (Zhao et al., 2019). Therefore, LINC00958 participates in the cell process by regulating miR-203 and SMAD3. Another study reported that the expression levels of LINC01121 and MMP-16 significantly increased in NP cells, whereas the level of miR-150-5p decreased (Chen X. et al., 2020). They demonstrated that LINC01121 could enhance the cell process of IDD, such as cell growth, ECM degradation and inflammation by regulating miR-150-5p and MMP-16.
In RA disease, the most studied mechanism of lncRNA and miRNA is that lncRNA acts as the miRNA sponge to modulate downstream genes (Table 2). lncRNA PVT1 (plasmacytoma variant translocation 1) and SCUBE2 (signal peptide-CUB-EGF-like containing protein 2) were upregulated, whereas miR-543 was downregulated in synovial tissues of RA rats and patients (Wang et al., 2020). Wang et al. (2020) found that the overexpression of PVT1 or the suppression of miR-543 elevated the level of SCUBE2. Moreover, the knockdown of PVT1 could suppress proliferation and induce apoptosis of RA through hindering the expression of SCUBE2 by sponging miR-543 (Wang et al., 2020). lncRNA LINC-PINT (long intergenic non-protein encoding long-chain RNA p53-induced transcript) was reduced in RA tissues and cells (Wang and Zhao, 2020). Through bioinformatics techniques and RNA Binding Protein Immunoprecipitation (RIP) assay, they found that miR-155-5p could interact with LINC-PINT, and SOCS1 was the target mRNA of miR-155-5p. LINC-PINT could inhibit cell proliferation and invasion via sponging miR-155-5p and regulating the level of SOCS1. Yan et al. (2019) revealed that the level of lncRNA HIX003209 in the peripheral blood mononuclear cells (PBMCs) and macrophages of RA samples and the expression of TLR4 was positively correlated with HIX003209. lncRNA HIX003209 directly targeted miR-6089 and was involved in the regulation of inflammation through acting as miR-6089 sponge via the TLR4/NF-κB signaling pathway.
That lncRNA acts as the sponge of miRNA to modulate target genes is the most studied mechanism of lncRNA and miRNA acting on AS (Table 2). Li Y. et al. (2020) reported the role of MEG3 (maternally expressed gene 3) in the inflammation of AS. They observed that the expression level of MEG3 in the serum of AS patients was significantly downregulated compared with that in normal people, and MEG3 could inhibit inflammatory responses. However, the expression of miR-146a was upregulated in the AS patients and miR-146a could directly bind with MEG3 (Li Y. et al., 2020). Li Y. et al. (2020) assumed that MEG3 may played a vital role in the repression of inflammation factors in AS through sponging miR-146a, thereby exploring a novel potential treatment target for AS patients. Zhang et al. (2020f) found that lncRNA H19 was highly expressed in the AS patients and elevated the expression level of IL-17A and IL-23 inflammation factors. H19 could directly modulate miR-22-5p and miR-675-5p, and VDR (vitamin D receptor) was the target mRNAs of these two miRNAs. Among them, the level of miR-22-5p was negatively correlated with H19, while miR-675-5p and VDR was positively with H19 in AS patients. H19 plays regulatory roles in inflammatory reaction in AS through binding with VDR by sponging miR-22-5p and interacting with miR-675-5p (Zhang et al., 2020f).
Circular RNAs acting as miRNA sponges is the one of the most studied mechanisms (Figure 2). Compared with normal cartilage, circRNA-CER (circRNA_100876) was overexpressed and increased with IL-1 (interleukin-1) and TNF-α (tumor necrosis factor-alpha) in OA chondrocytes. circRNA-CER regulated matrix-degrading matrix metalloproteinase (MMP)-13 expression to participated in the process of chondrocyte ECM degradation by sponging miR-136 (Liu et al., 2016b). According to the research of Zhou et al. (2018), overexpressed circRNA_Atp9b sponge miR-138-5p and then mediate ECM catabolism and inflammation to regulates OA progression in chondrocytes by targeting MMP13. circ_0136474 was also verified by the research of Li et al. (2019f) to sponge miR-127-5p to regulate MMP13 in human OA chondrocytes, then, it suppressed cell proliferation and enhanced cell apoptosis during OA progression. The results were in line with those obtained in a study performed by Zhou Z.B. et al. (2019), who found that circRNA.33186/miR-127-5p/MMP13 axis contributes to OA pathogenesis. Furthermore, circSERPINE2 overexpression could slow down the pace of human chondrocytes apoptosis and promote ECM anabolism by sponging miR-1271-5p and thereby targeting ERG (E26 transformation-specific-related gene) to alleviate OA (Shen et al., 2019). In OA blood samples, the downregulation of ciRS-7 and the upregulation of miR-7 were observed (Zhou X. et al., 2019). ciRS-7 was verified to act as a miR-7 sponge to mediate OA progression. Increased cirM3 expression in OA cartilage tissue and cells could serve as a sponge of miR-296-5p to slow down the proliferation and differentiation of OA chondrocytes, thus involving in regulating the occurrence and development of OA chondrocytes (Ni et al., 2020). The overexpression of circRNA-CDR1as regulated OA progression via reducing Col II level but increased IL-6 and MMP13 contents to modulate inflammation and ECM metabolism by sponging miR-641 (Zhang et al., 2020d).
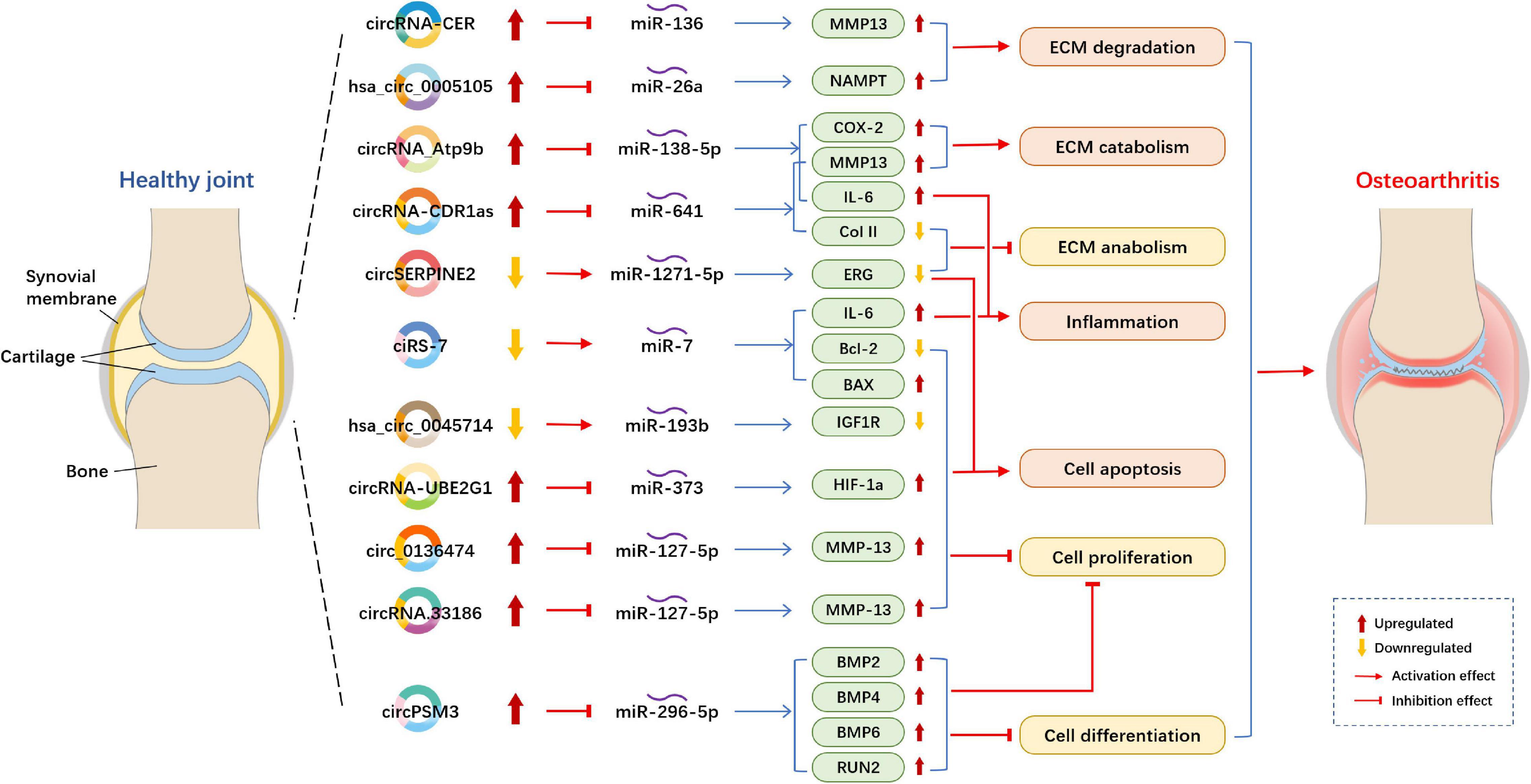
Figure 2. Example of altered circRNA expression patterns and their biological effects in osteoarthritis. circRNA, circular RNA; ECM, extracellular matrix; MMP, matrix metallopeptidase; NAMPT, Nicotinamide phosphoribosyltransferase; COX-2, cyclooxygenase-2; IL-6, interleukin-6; Col II, type II collagen; ERG, E26 transformation-specific-related gene; BAX, BCL2 associated X, apoptosis regulator; Bcl-2, B cell lymphoma 2; IGF1R, insulin-like growth factor 1 receptor; HIF, hypoxia inducible factor; BMP, bone morphogenetic protein.
Several circRNA studies showed that circRNAs act as ceRNAs to competitively bind to miRNAs in OA. Hsa_circ_0045714 expression was downregulated (Liu et al., 2016b; Li B.F. et al., 2017). Furthermore, Li B.F. et al. (2017) determined that hsa_circ_0045714 promoted the expression of miR-193b target gene IGF1R (insulin-like growth factor 1 receptor) to regulate chondrocytes proliferation, apoptosis and ECM synthesis. Otherwise, hsa_circ_0005105 expression is significantly enhanced in OA chondrocytes and can promote ECM degradation by mediating the expression of miR-26a target NAMPT (Nicotinamide phosphoribosyltransferase) (Wu et al., 2017). In the lipopolysaccharide (LPS)-induced OA cell model, the expression levels of circRNA-UBE2G1 was significantly increased and bound to miR-373 as ceRNAs to aggravate the OA progression by targeting hypoxia-inducible factor (HIF)-1a (Chen G. et al., 2020).
Over the past years, some circRNAs have merged as molecular drivers to serve as miRNA sponges or ceRNAs in circRNA/miRNA/mRNA networks in the pathogenesis of IDD (Figure 3). Compared with normal NP tissues, circVMA21 (hsa_circ_0091702) was downregulated in NP tissues and NP cells in IDD and alleviated NP cell apoptosis by targeting miR-200c and XIAP (X linked inhibitor-of-apoptosis protein) (Cheng et al., 2018). Similarly, circ-GRB10 was downregulated during IDD progression, and competitively bound to miR-328-5p to regulate NP cell apoptosis by targeting erb-b2 receptor tyrosine kinase 2 (ERBB2) in the ErbB signaling pathway (Guo et al., 2018). circRNA_104670 was selected via microarray analysis because of its large multiplier expression in IDD tissues (Song et al., 2018). A study reported that circRNA_104670 acted as a ceRNA that binds to miR-17-3p, downregulated circRNA_104670-suppressed MMP-2 expression through circRNA_104670/miR-17-3p/MMP-2 axis, reduced cell apoptosis and increased ECM formation. According to another microarray assay made by Wang et al. (2018a), they selected circ-4099 among 72 upregulated circRNAs in degenerated NP tissues for further analysis. They demonstrated that circ-4099 competitively sponged miR-616-5p, which reversed the suppression of Sox9 by miR-616-5p. Wang et al. (2018c) verified that circSEMA4B was downregulated in IDD specimens, and circSEMA4B served as a miR-431 sponge to compete with SFRP1 or GSK-3β, which are two inhibitory regulators of Wnt signaling, for miR-431 binding, thereby alleviating IL-1β-induced degenerative process in NP cells. circRNA-CIDN was downregulated during IDD progression and bound to miR-34a-5p as a miRNA sponge. Upregulation of miR-34a-5p repressed SIRT1 (silent mating type information regulation 2 homolog 1) to enhance the compression-induced damage of NP cells (Xiang et al., 2020). circERCC2 was also downregulated in IDD NP tissues and NP cells. Furthermore, circERCC2 was associated with the alleviation of IDD through miR-182-5p/SIRT1 axis by activating mitophagy and inhibiting apoptosis (Xie et al., 2019). The expression of circ-FAM169A in IDD samples was significantly upregulated with enhanced ECM catabolism and suppressed ECM anabolism in NP cells. The overexpressed circ-FAM169A competitively bound to miR-583, thus upregulating BTRC (an inducer of the NF-κB signaling pathway) (Guo et al., 2020). circ-FAM169A promoted IDD development via miR-583/BTRC signaling. In addition, circ_001653 could be another novel therapeutic target for IDD that functions by regulating miR-486-3p expression to upregulate CEMIP (cell migration-inducing hyaluronan binding protein) (Cui and Zhang, 2020). circ_001653 downregulation could potentially promote cell proliferation and ECM synthesis through the miR486-3p/CEMIP axis.
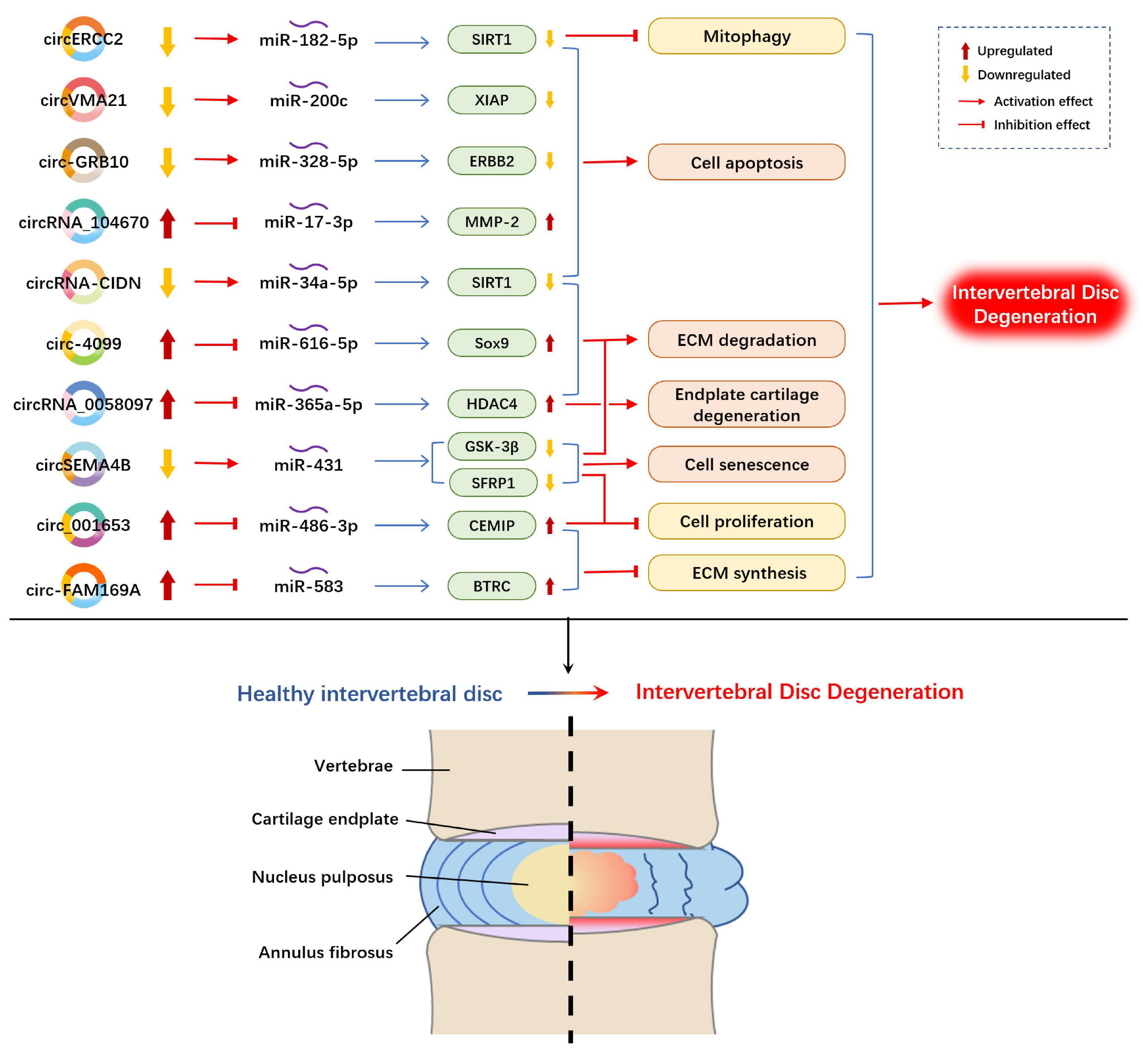
Figure 3. Example of altered circRNA expression patterns and their biological effects in intervertebral disk degeneration. circRNA, circular RNA; ECM, extracellular matrix; SIRT1, silent mating type information regulation 2 homolog 1; XIAP, X linked inhibitor of-apoptosis protein; ERBB2, erb-b2 receptor tyrosine kinase 2; MMP, matrix metallopeptidase; Sox9, SRY-Box transcription factor 9; HDAC4, histone deacetylase 4; GSK-3β, glycogen synthase kinase-3β; SFRP1, secreted frizzled-related protein 1; CEMIP, cell migration-inducing hyaluronan binding protein; BTRC, beta-transducin repeat-containing protein.
Human NP tissues and human endplate tissues were collected to detect differentially expressed circRNAs during IDD progression. Xiao et al. (2020) induced circRNA expression profile changes in endplate chondrocytes, and results reported that 17 circRNAs were upregulated and 12 circRNAs were downregulated (with fold changes higher than 1.5). circRNA_0058097 was selected for further analysis. circRNA_0058097 increased the expression of HDAC4 (histone deacetylase 4) by sponging miR-365a-5p, which intensified the morphological changes of endplate chondrocytes, and aggravated endplate cartilage and ECM degradation.
Rheumatoid arthritis and AS are both characterized by chronic inflammatory disease (van der Heijde et al., 2019; Zhou Y. et al., 2019). However, only a limited number of studies have been conducted on circRNAs in RA and AS (Table 3). Li B. et al. (2018) identified circRNAs in RA synovial tissues and suggested that hsa_circ_0001859 regulated ATF2 expression by competitively sponging miR-204/211. Knockdown of hsa_circ_0001859 suppressed ATF2 expression and decreased inflammatory activity. Hsa_circ_0001859/miR-204/211/ATF2 axis may be used as an approach for treating RA. Another circRNA/miRNA/mRNA network study in RA was conducted by Yang J. et al. (2020), They reported that circRNA_09505 is upregulated in PBMCs from RA patients and mice. The knockdown of circRNA_09505 inhibits macrophage proliferation and alleviates arthritis and inflammation. miR-6089 functions as a ceRNA that is being competitively sponged by circRNA_09505 to regulated macrophage inflammatory response. Furthermore, circRNA_09505 was detected to promote AKT1 expression, which is a direct target of miR-6089, to mediate IκBα/NF-κB signaling pathway. To sum up, circRNA_09505 can sponge miR-6089 and regulate inflammation via miR-6089/AKT1/NF-κB axis in arthritis mice model. Combined with RNA-seq data and RT-qPCR validation of PBMCs from RA patients, the results of Ouyang et al. (2017) showed several upregulated circRNAs (circRNA_101873, circRNA_003524, circRNA_104871, and circRNA_103047), and Wen et al. (2020) proved three upregulated hsa-circRNAs (hsa_circ_0001200, hsa_circ_0001566, and hsa_circ_0003972) and one downregulated hsa_circRNAs (hsa_circ_0008360), but without downstream gene detection to establish circRNA/miRNA/mRNA networks.
At present, studies on circRNA and miRNA interaction mechanism on AS are lacking. The roles of circRNAs in AS remain unclear. Only one profiling and bioinformatics analysis showed differentially expressed circRNAs in AS patients (sampled form spinal ligament tissues), reported the presence of 57 upregulated circRNAs and 66 downregulated circRNAs in AS spinal ligament tissues (Kou et al., 2020).
Taken together, the study about the interactions among circRNA, miRNA and mRNA in RA and AS may have a great clinical prospect.
Recent advances in gene expression of lncRNAs and circRNAs, coupled with the ability to interact with the miRNA, mRNA or signaling pathway, have started to expose the different molecular consequence associated with RNA transcriptions and the roles they play in the development of MSDDs (including OA, IDD, RA, and AS) that involve chondrocyte proliferation and apoptosis, ECM degradation and PBMCs inflammation. The effects of ncRNA/circRNA-miRNA-mRNA axis on MSDD progression elucidated their contribution to the dynamic cellular processes and provided the potential OA, IDD, RA and AS therapeutic strategies. The altered expression of lncRNAs or circRNAs refers to diverse biological processes of MSDD, thereby indicating that lncRNAs/circRNAs may be developed as biomarkers and therapeutic targets. Despite the large numbers of ncRNAs, including lncRNAs and circRNAs, determined to be differentially expressed during these pathogenic processes, only a small portion of them has been elucidated. Research on MSDD pathogenesis, especially on RA and AS, is still in its infancy and major knowledge gaps remain to be filled. Therefore, the interactions among lncRNA/circRNA, miRNA and mRNA in MSDD to present the potential pathogenesis is required. Further studies are needed to explore the mutual regulatory mechanisms between lncRNA/circRNA regulation and effective therapeutic interventions in the pathology of MSDD.
X-QW and P-JC: conceptualization and methodology. J-BG, XS, Y-MC, and ZY: investigation. Y-LZ and GS: writing – original draft preparation and writing – review and editing. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (81871844); Shuguang Program supported by Shanghai Education Development Foundation and Shanghai Municipal Education Commission (18SG48); the Shanghai Municipal Commission of Health and Family Planning (201840346); the Shanghai Key Lab of Human Performance (Shanghai University of Sport) (11DZ2261100); and Shanghai Frontiers Science Research Base of Exercise and Metabolic Health.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbasifard, M., Kamiab, Z., Bagheri-Hosseinabadi, Z., and Sadeghi, I. (2020). The role and function of long non-coding RNAs in osteoarthritis. Exp. Mol. Pathol. 114:104407. doi: 10.1016/j.yexmp.2020.104407
Ai, D., and Yu, F. (2019). LncRNA DNM3OS promotes proliferation and inhibits apoptosis through modulating IGF1 expression by sponging MiR-126 in CHON-001 cells. Diagn. Pathol. 14:106.
Beermann, J., Piccoli, M. T., Viereck, J., and Thum, T. (2016). Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 96, 1297–1325. doi: 10.1152/physrev.00041.2015
Cao, L., Wang, Y., Wang, Q., and Huang, J. (2018). LncRNA FOXD2-AS1 regulates chondrocyte proliferation in osteoarthritis by acting as a sponge of miR-206 to modulate CCND1 expression. Biomed. Pharmacother. 106, 1220–1226. doi: 10.1016/j.biopha.2018.07.048
Chen, G., Liu, T., Yu, B., Wang, B., and Peng, Q. (2020). CircRNA-UBE2G1 regulates LPS-induced osteoarthritis through miR-373/HIF-1a axis. Cell Cycle 19, 1696–1705. doi: 10.1080/15384101.2020.1772545
Chen, H., Qi, J., Bi, Q., and Zhang, S. (2017). Expression profile of long noncoding RNA (HOTAIR) and its predicted target miR-17-3p in LPS-induced inflammatory injury in human articular chondrocyte C28/I2 cells. Int. J. Clin. Exp. Pathol. 10, 9146–9157.
Chen, K., Zhu, H., Zheng, M. Q., and Dong, Q. R. (2019). LncRNA MEG3 inhibits the degradation of the extracellular matrix of chondrocytes in osteoarthritis via targeting miR-93/TGFBR2 axis. Cartilage 1947603519855759. doi: 10.1177/1947603519855759
CrossRef Full Text | [Published online ahead of print] | PubMed Abstract | Google Scholar
Chen, W. K., Yu, X. H., Yang, W., Wang, C., He, W. S., Yan, Y. G., et al. (2017). lncRNAs: novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 50:e12313. doi: 10.1111/cpr.12313
Chen, X., Li, Z., Xu, D., and Li, S. (2020). LINC01121 induced intervertebral disc degeneration via modulating miR-150-5p/MMP16 axis. J. Gene Med. 22:e3231.
Chen, Y., Huang, J., Tang, C., Chen, X., Yin, Z., Heng, B. C., et al. (2017). Small molecule therapeutics for inflammation-associated chronic musculoskeletal degenerative diseases: past, present and future. Exp. Cell Res. 359, 1–9. doi: 10.1016/j.yexcr.2017.07.027
Chen, Y., Zhang, L., Li, E., Zhang, G., Hou, Y., Yuan, W., et al. (2020). Long-chain non-coding RNA HOTAIR promotes the progression of osteoarthritis via sponging miR-20b/PTEN axis. Life Sci. 253:117685. doi: 10.1016/j.lfs.2020.117685
Cheng, X., Zhang, L., Zhang, K., Zhang, G., Hu, Y., Sun, X., et al. (2018). Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann. Rheum. Dis. 77, 770–779. doi: 10.1136/annrheumdis-2017-212056
Chu, P., Wang, Q., Wang, Z., and Gao, C. (2019). Long non-coding RNA highly up-regulated in liver cancer protects tumor necrosis factor-alpha-induced inflammatory injury by down-regulation of microRNA-101 in ATDC5 cells. Int. Immunopharmacol. 72, 148–158. doi: 10.1016/j.intimp.2019.04.004
Cooper, N. A., Scavo, K. M., Strickland, K. J., Tipayamongkol, N., Nicholson, J. D., Bewyer, D. C., et al. (2016). Prevalence of gluteus medius weakness in people with chronic low back pain compared to healthy controls. Eur. Spine J. 25, 1258–1265. doi: 10.1007/s00586-015-4027-6
Cui, S., and Zhang, L. (2020). circ_001653 silencing promotes the proliferation and ECM synthesis of NPCs in IDD by downregulating miR-486-3p-mediated CEMIP. Mol. Ther. Nucleic Acids 20, 385–399. doi: 10.1016/j.omtn.2020.01.026
Deng, K. Y., Wang, H., Guo, X. Q., and Xia, J. Z. (2016). The cross talk between long, non-coding RNAs and microRNAs in gastric cancer. Acta Biochim. Biophys. Sin. 48, 111–116. doi: 10.1093/abbs/gmv120
Faghihi, M. A., Zhang, M., Huang, J., Modarresi, F., Van Der Brug, M. P., Nalls, M. A., et al. (2010). Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 11:R56.
Fan, X., Yuan, J., Xie, J., Pan, Z., Yao, X., Sun, X., et al. (2018). Long non-protein coding RNA DANCR functions as a competing endogenous RNA to regulate osteoarthritis progression via miR-577/SphK2 axis. Biochem. Biophys. Res. Commun. 500, 658–664. doi: 10.1016/j.bbrc.2018.04.130
Fontana, G., See, E., and Pandit, A. (2015). Current trends in biologics delivery to restore intervertebral disc anabolism. Adv. Drug Deliv. Rev. 84, 146–158. doi: 10.1016/j.addr.2014.08.008
Gao, D., Hao, L., and Zhao, Z. (2020). Long non-coding RNA PART1 promotes intervertebral disc degeneration through regulating the miR-93/MMP2 pathway in nucleus pulposus cells. Int. J. Mol. Med. 46, 289–299.
Gao, Y., Zhao, H., and Li, Y. (2019). LncRNA MCM3AP-AS1 regulates miR-142-3p/HMGB1 to promote LPS-induced chondrocyte apoptosis. BMC Musculoskelet. Disord. 20:605. doi: 10.1186/s12891-019-2967-4
Guo, W., Mu, K., Zhang, B., Sun, C., Zhao, L., Dong, Z. Y., et al. (2020). The circular RNA FAM169A functions as a competitive endogenous RNA and regulates intervertebral disc degeneration by targeting miR-583 and BTRC. Cell Death Dis. 11:315.
Guo, W., Zhang, B., Mu, K., Feng, S. Q., Dong, Z. Y., Ning, G. Z., et al. (2018). Circular RNA GRB10 as a competitive endogenous RNA regulating nucleus pulposus cells death in degenerative intervertebral disk. Cell Death Dis. 9:319.
Haque, S., and Harries, L. W. (2017). Circular RNAs (circRNAs) in health and disease. Genes 8:353. doi: 10.3390/genes8120353
He, B., and Jiang, D. (2020). HOTAIR-induced apoptosis is mediated by sponging miR-130a-3p to repress chondrocyte autophagy in knee osteoarthritis. Cell Biol. Int. 44, 524–535. doi: 10.1002/cbin.11253
He, L., Man, C., Xiang, S., Yao, L., Wang, X., and Fan, Y. (2021). Circular RNAs’ cap-independent translation protein and its roles in carcinomas. Mol. Cancer 20:119.
Hu, J., Wang, Z., Shan, Y., Pan, Y., Ma, J., and Jia, L. (2018). Long non-coding RNA HOTAIR promotes osteoarthritis progression via miR-17-5p/FUT2/β-catenin axis. Cell Death Dis. 9:711.
Hu, Y., Li, S., and Zou, Y. (2019). Knockdown of LncRNA H19 relieves LPS-induced damage by modulating miR-130a in osteoarthritis. Yonsei. Med. J. 60, 381–388. doi: 10.3349/ymj.2019.60.4.381
Huang, B., Yu, H., Li, Y., Zhang, W., and Liu, X. (2019). Upregulation of long noncoding TNFSF10 contributes to osteoarthritis progression through the miR-376-3p/FGFR1 axis. J. Cell Biochem. 120, 19610–19620. doi: 10.1002/jcb.29267
Huang, Y. (2018). The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J. Cell Mol. Med. 22, 5768–5775. doi: 10.1111/jcmm.13866
Huo, J. Z., Ji, X. H., Su, Z. Y., Shang, P., and Gao, F. (2018). Association of ADAMTS4 and ADAMTS5 polymorphisms with musculoskeletal degenerative diseases: a systematic review and meta-analysis. Biosci. Rep. 38:BSR20181619.
Iwakura, T., Inui, A., and Reddi, A. H. (2013). Stimulation of superficial zone protein accumulation by hedgehog and Wnt signaling in surface zone bovine articular chondrocytes. Arthritis Rheum. 65, 408–417.
Ji, Q., Qiao, X., Liu, Y., Wang, D., and Yan, J. (2020). Silencing of long-chain non-coding RNA GAS5 in osteoarthritic chondrocytes is mediated by targeting the miR-34a/Bcl-2 axis. Mol. Med. Rep. 21, 1310–1319.
Ji, Y., Fang, Q. Y., Wang, S. N., Zhang, Z. W., Hou, Z. J., Li, J. N., et al. (2020). Lnc-RNA BLACAT1 regulates differentiation of bone marrow stromal stem cells by targeting miR-142-5p in osteoarthritis. Eur. Rev. Med. Pharmacol. Sci. 24, 2893–2901.
Jiang, S. D., Lu, J., Deng, Z. H., Li, Y. S., and Lei, G. H. (2017). Long noncoding RNAs in osteoarthritis. Joint Bone Spine 84, 553–556.
Kou, J., Liu, G., Liu, X., Li, T., Wei, Y., Sun, Y., et al. (2020). Profiling and bioinformatics analysis of differentially expressed circRNAs in spinal ligament tissues of patients with ankylosing spondylitis. BioMed Res. Int. 2020:7165893.
Kulcheski, F. R., Christoff, A. P., and Margis, R. (2016). Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J. Biotechnol. 238, 42–51. doi: 10.1016/j.jbiotec.2016.09.011
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. doi: 10.1016/0092-8674(93)90529-y
Lei, B., Xuan, X. Y., and Fan, W. P. (2019). Progress in research on role of CircRNA in autoimmune diseases. Chin. J. Biol. 32, 347–350.
Lei, J., Fu, Y., Zhuang, Y., Zhang, K., and Lu, D. (2019). LncRNA SNHG1 alleviates IL-1β-induced osteoarthritis by inhibiting miR-16-5p-mediated p38 MAPK and NF-κB signaling pathways. Biosci. Rep. 39:BSR20191523.
Li, B., and Chen, D. (2019). Degenerative musculoskeletal diseases: pathology and treatments. J. Orthop. Translat. 17, 1–2. doi: 10.1016/j.jot.2019.05.001
Li, B., Li, N., Zhang, L., Li, K., Xie, Y., Xue, M., et al. (2018). Hsa_circ_0001859 Regulates ATF2 expression by functioning as an MiR-204/211 sponge in human rheumatoid arthritis. J. Immunol. Res. 2018:9412387.
Li, B. F., Zhang, Y., Xiao, J., Wang, F., Li, M., Guo, X. Z., et al. (2017). Hsa_circ_0045714 regulates chondrocyte proliferation, apoptosis and extracellular matrix synthesis by promoting the expression of miR-193b target gene IGF1R. Hum. Cell 30, 311–318. doi: 10.1007/s13577-017-0177-7
Li, C., Pan, S., Song, Y., Li, Y., and Qu, J. (2019a). Silence of lncRNA MIAT protects ATDC5 cells against lipopolysaccharides challenge via up-regulating miR-132. Artif. Cells Nanomed. Biotechnol. 47, 2521–2527. doi: 10.1080/21691401.2019.1626410
Li, D., Sun, Y., Wan, Y., Wu, X., and Yang, W. (2020). LncRNA NEAT1 promotes proliferation of chondrocytes via down-regulation of miR-16-5p in osteoarthritis. J. Gene Med. 22:e3203.
Li, H., Xie, S., Li, H., Zhang, R., and Zhang, H. (2019b). LncRNA MALAT1 mediates proliferation of LPS treated-articular chondrocytes by targeting the miR-146a-PI3K/Akt/mTOR axis. Life Sci. 254:116801. doi: 10.1016/j.lfs.2019.116801
Li, H. Z., Lin, Z., Xu, X. H., Lin, N., and Lu, H. D. (2018). The potential roles of circRNAs in osteoarthritis: a coming journey to find a treasure. Biosci. Rep. 38:BSR20180542.
Li, L., Lv, G., Wang, B., and Kuang, L. (2020). XIST/miR-376c-5p/OPN axis modulates the influence of proinflammatory M1 macrophages on osteoarthritis chondrocyte apoptosis. J. Cell Physiol. 235, 281–293. doi: 10.1002/jcp.28968
Li, X., Huang, T. L., Zhang, G. D., Jiang, J. T., and Guo, P. Y. (2019c). LncRNA ANRIL impacts the progress of osteoarthritis via regulating proliferation and apoptosis of osteoarthritis synoviocytes. Eur. Rev. Med. Pharmacol. Sci. 23, 9729–9737.
Li, X., Yu, M., Chen, L., Sun, T., Wang, H., Zhao, L., et al. (2019d). LncRNA PMS2L2 protects ATDC5 chondrocytes against lipopolysaccharide-induced inflammatory injury by sponging miR-203. Life Sci. 217, 283–292. doi: 10.1016/j.lfs.2018.12.020
Li, Y., Li, S., Luo, Y., Liu, Y., and Yu, N. (2017). LncRNA PVT1 regulates chondrocyte apoptosis in osteoarthritis by acting as a sponge for miR-488-3p. DNA Cell Biol. 36, 571–580. doi: 10.1089/dna.2017.3678
Li, Y., Li, Z., Li, C., Zeng, Y., and Liu, Y. (2019e). Long noncoding RNA TM1P3 is involved in osteoarthritis by mediating chondrocyte extracellular matrix degradation. J. Cell Biochem. 120, 12702–12712. doi: 10.1002/jcb.28539
Li, Y., Zhang, S., Zhang, C., and Wang, M. (2020). LncRNA MEG3 inhibits the inflammatory response of ankylosing spondylitis by targeting miR-146a. Mol. Cell Biochem. 466, 17–24. doi: 10.1007/s11010-019-03681-x
Li, Y. F., Li, S. H., Liu, Y., and Luo, Y. T. (2017). Long noncoding RNA CIR promotes chondrocyte extracellular matrix degradation in osteoarthritis by acting as a sponge for Mir-27b. Cell Physiol. Biochem. 43, 602–610. doi: 10.1159/000480532
Li, Z., Li, X., Jiang, C., Qian, W., Tse, G., Chan, M. T. V., et al. (2018). Long non-coding RNAs in rheumatoid arthritis. Cell Prolif. 51:e12404.
Li, Z., Yuan, B., Pei, Z., Zhang, K., Ding, Z., Zhu, S., et al. (2019f). Circ_0136474 and MMP-13 suppressed cell proliferation by competitive binding to miR-127-5p in osteoarthritis. J. Cell Mol. Med. 23, 6554–6564. doi: 10.1111/jcmm.14400
Liang, J., Xu, L., Zhou, F., Liu, A. M., Ge, H. X., Chen, Y. Y., et al. (2018). MALAT1/miR-127-5p regulates osteopontin (OPN)-mediated proliferation of human chondrocytes through PI3K/Akt pathway. J. Cell Biochem. 119, 431–439. doi: 10.1002/jcb.26200
Liu, C., Ren, S., Zhao, S., and Wang, Y. (2019). LncRNA MALAT1/MiR-145 adjusts IL-1β-induced chondrocytes viability and cartilage matrix degradation by regulating ADAMTS5 in human osteoarthritis. Yonsei Med. J. 60, 1081–1092. doi: 10.3349/ymj.2019.60.11.1081
Liu, F., Liu, X., Yang, Y., Sun, Z., Deng, S., Jiang, Z., et al. (2020). NEAT1/miR-193a-3p/SOX5 axis regulates cartilage matrix degradation in human osteoarthritis. Cell Biol Int 44, 947–957. doi: 10.1002/cbin.11291
Liu, G., Wang, Y., Zhang, M., and Zhang, Q. (2019). Long non-coding RNA THRIL promotes LPS-induced inflammatory injury by down-regulating microRNA-125b in ATDC5 cells. Int. Immunopharmacol. 66, 354–361. doi: 10.1016/j.intimp.2018.11.038
Liu, Q., Hu, X., Zhang, X., Dai, L., Duan, X., Zhou, C., et al. (2016a). The TMSB4 pseudogene LncRNA functions as a competing endogenous RNA to promote cartilage degradation in human osteoarthritis. Mol. Ther. 24, 1726–1733. doi: 10.1038/mt.2016.151
Liu, Q., Zhang, X., Hu, X., Dai, L., Fu, X., Zhang, J., et al. (2016b). Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘Sponge’ in human cartilage degradation. Sci. Rep. 6:22572.
Liu, Y., Li, Z., Zhang, M., Zhou, H., Wu, X., Zhong, J., et al. (2021). Rolling-translated EGFR variants sustain EGFR signaling and promote glioblastoma tumorigenicity. Neuro. Oncol. 23, 743–756. doi: 10.1093/neuonc/noaa279
Liu, Y., Lin, L., Zou, R., Wen, C., Wang, Z., and Lin, F. (2018). MSC-derived exosomes promote proliferation and inhibit apoptosis of chondrocytes via lncRNA-KLF3-AS1/miR-206/GIT1 axis in osteoarthritis. Cell Cycle 17, 2411–2422. doi: 10.1080/15384101.2018.1526603
Loef, M., Van Beest, S., Kroon, F. P. B., Bloem, J. L., Dekkers, O. M., Reijnierse, M., et al. (2018). Comparison of histological and morphometrical changes underlying subchondral bone abnormalities in inflammatory and degenerative musculoskeletal disorders: a systematic review. Osteoarthritis Cartilage 26, 992–1002. doi: 10.1016/j.joca.2018.05.007
Loeser, R. F., Goldring, S. R., Scanzello, C. R., and Goldring, M. B. (2012). Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum. 64, 1697–1707. doi: 10.1002/art.34453
Lu, C., Li, Z., Hu, S., Cai, Y., and Peng, K. (2019). LncRNA PART-1 targets TGFBR2/Smad3 to regulate cell viability and apoptosis of chondrocytes via acting as miR-590-3p sponge in osteoarthritis. J. Cell Mol. Med. 23, 8196–8205. doi: 10.1111/jcmm.14690
Lü, G., Li, L., Wang, B., and Kuang, L. (2020). LINC00623/miR-101/HRAS axis modulates IL-1β-mediated ECM degradation, apoptosis and senescence of osteoarthritis chondrocytes. Aging (Albany NY) 12, 3218–3237. doi: 10.18632/aging.102801
Lu, M., and Zhou, E. (2020). Long noncoding RNA LINC00662-miR-15b-5p mediated GPR120 dysregulation contributes to osteoarthritis. Pathol. Int. 70, 155–165. doi: 10.1111/pin.12875
Lu, Z., Luo, M., and Huang, Y. (2019). lncRNA-CIR regulates cell apoptosis of chondrocytes in osteoarthritis. J. Cell. Biochem. 120, 7229–7237. doi: 10.1002/jcb.27997
Luo, X., Wang, J., Wei, X., Wang, S., and Wang, A. (2020). Knockdown of lncRNA MFI2-AS1 inhibits lipopolysaccharide-induced osteoarthritis progression by miR-130a-3p/TCF4. Life Sci. 240:117019. doi: 10.1016/j.lfs.2019.117019
Mao, G., Kang, Y., Lin, R., Hu, S., Zhang, Z., Li, H., et al. (2019). Long non-coding RNA HOTTIP promotes CCL3 expression and induces cartilage degradation by sponging miR-455-3p. Front. Cell Dev. Biol. 7:161. doi: 10.3389/fcell.2019.00161
Mathy, N. W., and Chen, X. M. (2017). Long non-coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J. Biol. Chem. 292, 12375–12382. doi: 10.1074/jbc.r116.760884
Mohammadi, A., Kelly, O. B., Filice, M., Kabakchiev, B., Smith, M. I., and Silverberg, M. S. (2018). Differential expression of microRNAs in peripheral blood mononuclear cells identifies autophagy and TGF-Beta-related signatures aberrantly expressed in inflammatory bowel disease. J. Crohns Colitis 12, 568–581. doi: 10.1093/ecco-jcc/jjy010
Moran-Moguel, M. C., Petarra-Del Rio, S., Mayorquin-Galvan, E. E., and Zavala-Cerna, M. G. (2018). Rheumatoid arthritis and miRNAs: a critical review through a functional view. J. Immunol. Res. 2018:2474529.
Ni, J. L., Dang, X. Q., and Shi, Z. B. (2020). CircPSM3 inhibits the proliferation and differentiation of OA chondrocytes by targeting miRNA-296-5p. Eur. Rev. Med. Pharmacol. Sci. 24, 3467–3475.
Ouyang, Q., Wu, J., Jiang, Z., Zhao, J., Wang, R., Lou, A., et al. (2017). Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell Physiol. Biochem. 42, 651–659. doi: 10.1159/000477883
Pan, L., Liu, D., Zhao, L., Wang, L., Xin, M., and Li, X. (2018). Long noncoding RNA MALAT1 alleviates lipopolysaccharide-induced inflammatory injury by upregulating microRNA-19b in murine chondrogenic ATDC5 cells. J. Cell Biochem. 119, 10165–10175.
Pan, Z., Li, G. F., Sun, M. L., Xie, L., Liu, D., Zhang, Q., et al. (2019). MicroRNA-1224 splicing circularRNA-Filip1l in an Ago2-dependent manner regulates chronic inflammatory pain via targeting Ubr5. J. Neurosci. 39, 2125–2143. doi: 10.1523/jneurosci.1631-18.2018
Park, S., Lee, M., Chun, C. H., and Jin, E. J. (2019). The lncRNA, Nespas, Is associated with osteoarthritis progression and serves as a potential new prognostic biomarker. Cartilage 10, 148–156. doi: 10.1177/1947603517725566
Qi, K., Lin, R., Xue, C., Liu, T., Wang, Y., Zhang, Y., et al. (2019). Long non-coding RNA (LncRNA) CAIF is downregulated in osteoarthritis and inhibits LPS-induced interleukin 6 (IL-6) upregulation by downregulation of MiR-1246. Med. Sci. Monit. 25, 8019–8024. doi: 10.12659/msm.917135
Rong, D., Sun, H., Li, Z., Liu, S., Dong, C., Fu, K., et al. (2017). An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget 8, 73271–73281. doi: 10.18632/oncotarget.19154
Samartzis, D., Karppinen, J., Mok, F., Fong, D. Y., Luk, K. D., and Cheung, K. M. (2011). A population-based study of juvenile disc degeneration and its association with overweight and obesity, low back pain, and diminished functional status. J. Bone Joint Surg. Am. 93, 662–670. doi: 10.2106/jbjs.i.01568
Satoshi Yamashita, H. A. (2012). miRNA functions in arthritis. Curr. Rheumatol. Rev. 8, 98–102. doi: 10.2174/157339712802083786
Seeliger, C., Balmayor, E. R., and Van Griensven, M. (2016). miRNAs related to skeletal diseases. Stem Cells Dev. 25, 1261–1281. doi: 10.1089/scd.2016.0133
Shao, T., Hu, Y., Tang, W., Shen, H., Yu, Z., and Gu, J. (2019). The long noncoding RNA HOTAIR serves as a microRNA-34a-5p sponge to reduce nucleus pulposus cell apoptosis via a NOTCH1-mediated mechanism. Gene 715:144029. doi: 10.1016/j.gene.2019.144029
Shen, H., Wang, Y., Shi, W., Sun, G., Hong, L., and Zhang, Y. (2018). LncRNA SNHG5/miR-26a/SOX2 signal axis enhances proliferation of chondrocyte in osteoarthritis. Acta Biochim. Biophys. Sin. (Shanghai) 50, 191–198. doi: 10.1093/abbs/gmx141
Shen, S., Wu, Y., Chen, J., Xie, Z., Huang, K., Wang, G., et al. (2019). CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Ann. Rheum. Dis. 78, 826–836. doi: 10.1136/annrheumdis-2018-214786
Song, J., Ahn, C., Chun, C. H., and Jin, E. J. (2014). A long non-coding RNA, GAS5, plays a critical role in the regulation of miR-21 during osteoarthritis. J. Orthop. Res. 32, 1628–1635. doi: 10.1002/jor.22718
Song, J., Wang, H. L., Song, K. H., Ding, Z. W., Wang, H. L., Ma, X. S., et al. (2018). CircularRNA_104670 plays a critical role in intervertebral disc degeneration by functioning as a ceRNA. Exp. Mol. Med. 50:94.
Steck, E., Boeuf, S., Gabler, J., Werth, N., Schnatzer, P., Diederichs, S., et al. (2012). Regulation of H19 and its encoded microRNA-675 in osteoarthritis and under anabolic and catabolic in vitro conditions. J. Mol. Med. (Berl) 90, 1185–1195. doi: 10.1007/s00109-012-0895-y
Sun, P., Wu, Y., Li, X., and Jia, Y. (2020). miR-142-5p protects against osteoarthritis through competing with lncRNA XIST. J. Gene Med. 22:e3158.
Sun, T., Yu, J., Han, L., Tian, S., Xu, B., Gong, X., et al. (2018). Knockdown of long non-coding RNA RP11-445H22.4 alleviates LPS-induced injuries by regulation of MiR-301a in osteoarthritis. Cell Physiol. Biochem. 45, 832–843. doi: 10.1159/000487175
Sun, Y., Kang, S., Pei, S., Sang, C., and Huang, Y. (2020). MiR93-5p inhibits chondrocyte apoptosis in osteoarthritis by targeting lncRNA CASC2. BMC Musculoskelet. Disord. 21:26. doi: 10.1186/s12891-019-3025-y
Tan, F., Wang, D., and Yuan, Z. (2020). The fibroblast-like synoviocyte derived exosomal long non-coding RNA H19 alleviates osteoarthritis progression through the miR-106b-5p/TIMP2 axis. Inflammation 43, 1498–1509. doi: 10.1007/s10753-020-01227-8
Tan, H., Zhao, L., Song, R., Liu, Y., and Wang, L. (2018). The long noncoding RNA SNHG1 promotes nucleus pulposus cell proliferation through regulating miR-326 and CCND1. Am. J. Physiol. Cell Physiol. 315, C21–C27.
Tang, L., Ding, J., Zhou, G., and Liu, Z. (2018). LncRNA-p21 promotes chondrocyte apoptosis in osteoarthritis by acting as a sponge for miR-451. Mol. Med. Rep. 18, 5295–5301.
Tang, L. P., Ding, J. B., Liu, Z. H., and Zhou, G. J. (2018). LncRNA TUG1 promotes osteoarthritis-induced degradation of chondrocyte extracellular matrix via miR-195/MMP-13 axis. Eur. Rev. Med. Pharmacol. Sci. 22, 8574–8581.
Taurog, J. D., Chhabra, A., and Colbert, R. A. (2016). Ankylosing spondylitis and axial spondyloarthritis. N. Engl. J. Med. 374, 2563–2574.
Tian, F., Wang, J., Zhang, Z., and Yang, J. (2020). LncRNA SNHG7/miR-34a-5p/SYVN1 axis plays a vital role in proliferation, apoptosis and autophagy in osteoarthritis. Biol. Res. 53:9.
van der Heijde, D., Braun, J., Deodhar, A., Baraliakos, X., Landewé, R., Richards, H. B., et al. (2019). Modified stoke ankylosing spondylitis spinal score as an outcome measure to assess the impact of treatment on structural progression in ankylosing spondylitis. Rheumatology (Oxford, England) 58, 388–400.
Verduci, L., Strano, S., Yarden, Y., and Blandino, G. (2019). The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol. Oncol. 13, 669–680. doi: 10.1002/1878-0261.12468
Vieira, A. S., Dogini, D. B., and Lopes-Cendes, I. (2018). Role of non-coding RNAs in non-aging-related neurological disorders. Braz. J. Med. Biol. Res. 51, e7566.
Vinatier, C., Merceron, C., and Guicheux, J. (2016). Osteoarthritis: from pathogenic mechanisms and recent clinical developments to novel prospective therapeutic options. Drug Discov. Today 21, 1932–1937. doi: 10.1016/j.drudis.2016.08.011
Wang, A., Hu, N., Zhang, Y., Chen, Y., Su, C., Lv, Y., et al. (2019). MEG3 promotes proliferation and inhibits apoptosis in osteoarthritis chondrocytes by miR-361-5p/FOXO1 axis. BMC Med. Genomics 12:201. doi: 10.1186/s12920-019-0649-6
Wang, G., Bu, X., Zhang, Y., Zhao, X., Kong, Y., Ma, L., et al. (2017). LncRNA-UCA1 enhances MMP-13 expression by inhibiting miR-204-5p in human chondrocytes. Oncotarget 8, 91281–91290. doi: 10.18632/oncotarget.20108
Wang, H., He, P., Pan, H., Long, J., Wang, J., Li, Z., et al. (2018a). Circular RNA circ-4099 is induced by TNF-α and regulates ECM synthesis by blocking miR-616-5p inhibition of Sox9 in intervertebral disc degeneration. Exp. Mol. Med. 50:27.
Wang, J., Kong, X., Hu, H., and Shi, S. (2020). Knockdown of long non-coding RNA PVT1 induces apoptosis of fibroblast-like synoviocytes through modulating miR-543-dependent SCUBE2 in rheumatoid arthritis. J. Orthop. Surg. Res. 15:142.
Wang, J., Yan, S., Yang, J., Lu, H., Xu, D., and Wang, Z. (2019). Non-coding RNAs in rheumatoid arthritis: from bench to bedside. Front. Immunol. 10:3129.
Wang, J., and Zhao, Q. (2020). LncRNA LINC-PINT increases SOCS1 expression by sponging miR-155-5p to inhibit the activation of ERK signaling pathway in rheumatoid arthritis synovial fibroblasts induced by TNF-α. Int. Immunopharmacol. 84:106497. doi: 10.3389/fimmu.2019.03129
Wang, Q., Wang, W., Zhang, F., Deng, Y., and Long, Z. (2017). NEAT1/miR-181c regulates osteopontin (OPN)-mediated synoviocyte proliferation in osteoarthritis. J. Cell Biochem. 118, 3775–3784. doi: 10.1002/jcb.26025
Wang, T., Liu, Y., Wang, Y., Huang, X., Zhao, W., and Zhao, Z. (2019). Long non-coding RNA XIST promotes extracellular matrix degradation by functioning as a competing endogenous RNA of miR-1277-5p in osteoarthritis. Int. J. Mol. Med. 44, 630–642.
Wang, X., Peng, L., Gong, X., Zhang, X., Sun, R., and Du, J. (2018b). LncRNA-RMRP promotes nucleus pulposus cell proliferation through regulating miR-206 expression. J. Cell Mol. Med. 22, 5468–5476. doi: 10.1111/jcmm.13817
Wang, X., Wang, B., Zou, M., Li, J., Lü, G., Zhang, Q., et al. (2018c). CircSEMA4B targets miR-431 modulating IL-1β-induced degradative changes in nucleus pulposus cells in intervertebral disc degeneration via Wnt pathway. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 3754–3768. doi: 10.1016/j.bbadis.2018.08.033
Wang, X., Zou, M., Li, J., Wang, B., Zhang, Q., Liu, F., et al. (2018d). LncRNA H19 targets miR-22 to modulate H(2) O(2) -induced deregulation in nucleus pulposus cell senescence, proliferation, and ECM synthesis through Wnt signaling. J. Cell Biochem. 119, 4990–5002. doi: 10.1002/jcb.26738
Wang, Y., Cao, L., Wang, Q., Huang, J., and Xu, S. (2019). LncRNA FOXD2-AS1 induces chondrocyte proliferation through sponging miR-27a-3p in osteoarthritis. Artif. Cells Nanomed. Biotechnol. 47, 1241–1247. doi: 10.1080/21691401.2019.1596940
Wang, Z., Chi, X., Liu, L., Wang, Y., Mei, X., Yang, Y., et al. (2018e). Long noncoding RNA maternally expressed gene 3 knockdown alleviates lipopolysaccharide-induced inflammatory injury by up-regulation of miR-203 in ATDC5 cells. Biomed. Pharmacother. 100, 240–249. doi: 10.1016/j.biopha.2018.02.018
Wang, Z., Hao, J., and Chen, D. (2019). Long noncoding RNA nuclear enriched abundant transcript 1 (NEAT1) regulates proliferation, apoptosis, and inflammation of chondrocytes via the miR-181a/glycerol-3-phosphate dehydrogenase 1-Like (GPD1L) axis. Med. Sci. Monit. 25, 8084–8094. doi: 10.12659/msm.918416
Wen, J., Liu, J., Zhang, P., Jiang, H., Xin, L., Wan, L., et al. (2020). RNA-seq reveals the circular RNA and miRNA expression profile of peripheral blood mononuclear cells in patients with rheumatoid arthritis. Biosci. Rep. 40:BSR20193160.
Wu, Y., Zhang, Y., Zhang, Y., and Wang, J. J. (2017). CircRNA hsa_circ_0005105 upregulates NAMPT expression and promotes chondrocyte extracellular matrix degradation by sponging miR-26a. Cell Biol. Int. 41, 1283–1289. doi: 10.1002/cbin.10761
Wu, Y. G., Lu, X. X., Shen, B., and Zeng, Y. (2019). The Therapeutic potential and role of miRNA, lncRNA, and circRNA in osteoarthritis. Curr. Gene Ther. 19, 255–263. doi: 10.2174/1566523219666190716092203
Xi, Y., Jiang, T., Wang, W., Yu, J., Wang, Y., Wu, X., et al. (2017). Long non-coding HCG18 promotes intervertebral disc degeneration by sponging miR-146a-5p and regulating TRAF6 expression. Sci. Rep. 7:13234.
Xiang, Q., Kang, L., Wang, J., Liao, Z., Song, Y., Zhao, K., et al. (2020). CircRNA-CIDN mitigated compression loading-induced damage in human nucleus pulposus cells via miR-34a-5p/SIRT1 axis. EBioMedicine 53:102679. doi: 10.1016/j.ebiom.2020.102679
Xiao, L., Ding, B., Xu, S., Gao, J., Yang, B., Wang, J., et al. (2020). circRNA_0058097 promotes tension-induced degeneration of endplate chondrocytes by regulating HDAC4 expression through sponge adsorption of miR-365a-5p. J. Cell. Biochem. 121, 418–429. doi: 10.1002/jcb.29202
Xiao, Y., Yan, X., Yang, Y., and Ma, X. L. (2019). Downregulation of long noncoding RNA HOTAIRM1 variant 1 contributes to osteoarthritis via regulating miR-125b/BMPR2 axis and activating JNK/MAPK/ERK pathway. Biomed. Pharmacother. 109, 1569–1577. doi: 10.1016/j.biopha.2018.10.181
Xie, L., Huang, W., Fang, Z., Ding, F., Zou, F., Ma, X., et al. (2019). CircERCC2 ameliorated intervertebral disc degeneration by regulating mitophagy and apoptosis through miR-182-5p/SIRT1 axis. Cell Death Dis. 10:751.
Xu, J., and Xu, Y. (2017). The lncRNA MEG3 downregulation leads to osteoarthritis progression via miR-16/SMAD7 axis. Cell Biosci. 7:69.
Xue, H., Yu, P., Wang, W. Z., Niu, Y. Y., and Li, X. (2020). The reduced lncRNA NKILA inhibited proliferation and promoted apoptosis of chondrocytes via miR-145/SP1/NF-κB signaling in human osteoarthritis. Eur. Rev. Med. Pharmacol. Sci. 24, 535–548.
Yan, S., Wang, P., Wang, J., Yang, J., Lu, H., Jin, C., et al. (2019). Long non-coding RNA HIX003209 promotes inflammation by sponging miR-6089 via TLR4/NF-κB signaling pathway in rheumatoid arthritis. Front. Immunol. 10:2218. doi: 10.3389/fimmu.2019.02218
Yang, J., Cheng, M., Gu, B., Wang, J., Yan, S., and Xu, D. (2020). CircRNA_09505 aggravates inflammation and joint damage in collagen-induced arthritis mice via miR-6089/AKT1/NF-κB axis. Cell Death Dis. 11:833.
Yang, Q., Li, X., Zhou, Y., Fu, W., Wang, J., and Wei, Q. (2019). A LINC00341-mediated regulatory pathway supports chondrocyte survival and may prevent osteoarthritis progression. J. Cell Biochem. 120, 10812–10820. doi: 10.1002/jcb.28372
Yang, S., Zhang, F., Ma, J., and Ding, W. (2020). Intervertebral disc ageing and degeneration: The antiapoptotic effect of oestrogen. Ageing Res. Rev. 57:100978. doi: 10.1016/j.arr.2019.100978
Yang, Y., Zhong, Z., Zhao, Y., Ren, K., and Li, N. (2019). LincRNA-SLC20A1 (SLC20A1) promotes extracellular matrix degradation in nucleus pulposus cells in human intervertebral disc degeneration by targeting the miR-31-5p/MMP3 axis. Int. J. Clin. Exp. Pathol. 12, 3632–3643.
Ye, F., Gao, G., Zou, Y., Zheng, S., Zhang, L., Ou, X., et al. (2019). circFBXW7 inhibits malignant progression by sponging miR-197-3p and encoding a 185-aa protein in triple-negative breast cancer. Mol. Ther. Nucleic Acids 18, 88–98. doi: 10.1016/j.omtn.2019.07.023
Ying, H., Wang, Y., Gao, Z., and Zhang, Q. (2019). Long non-coding RNA activated by transforming growth factor beta alleviates lipopolysaccharide-induced inflammatory injury via regulating microRNA-223 in ATDC5 cells. Int. Immunopharmacol. 69, 313–320. doi: 10.1016/j.intimp.2019.01.056
Yu, C., Shi, D., Li, Z., Wan, G., and Shi, X. (2019). Long noncoding RNA CHRF exacerbates IL-6-induced inflammatory damages by downregulating microRNA-146a in ATDC5 cells. J. Cell Physiol. 234, 21851–21859. doi: 10.1002/jcp.28749
Yu, C. X., and Sun, S. (2018). An emerging role for circular RNAs in osteoarthritis. Yonsei Med. J. 59, 349–355. doi: 10.3349/ymj.2018.59.3.349
Yu, Y., Zhang, X., Li, Z., Kong, L., and Huang, Y. (2018). LncRNA HOTAIR suppresses TNF-α induced apoptosis of nucleus pulposus cells by regulating miR-34a/Bcl-2 axis. Biomed. Pharmacother. 107, 729–737. doi: 10.1016/j.biopha.2018.08.033
Yuan, W., Che, W., Jiang, Y.-Q., Yuan, F.-L., Wang, H.-R., Zheng, G.-L., et al. (2015). Establishment of intervertebral disc degeneration model induced by ischemic sub-endplate in rat tail. Spine J. 15, 1050–1059. doi: 10.1016/j.spinee.2015.01.026
Zhang, G., Wu, Y., Xu, D., and Yan, X. (2016). Long noncoding RNA UFC1 promotes proliferation of chondrocyte in osteoarthritis by acting as a sponge for miR-34a. DNA Cell Biol. 35, 691–695. doi: 10.1089/dna.2016.3397
Zhang, H., Li, J., Shao, W., and Shen, N. (2020a). LncRNA CTBP1-AS2 is upregulated in osteoarthritis and increases the methylation of miR-130a gene to inhibit chondrocyte proliferation. Clin. Rheumatol. 39, 3473–3478. doi: 10.1007/s10067-020-05113-4
Zhang, L., Wu, H., Zhao, M., Chang, C., and Lu, Q. (2020b). Clinical significance of miRNAs in autoimmunity. J. Autoimmun. 109:102438. doi: 10.1016/j.jaut.2020.102438
Zhang, L., Zhang, P., Sun, X., Zhou, L., and Zhao, J. (2018). Long non-coding RNA DANCR regulates proliferation and apoptosis of chondrocytes in osteoarthritis via miR-216a-5p-JAK2-STAT3 axis. Biosci. Rep. 38:BSR20181228.
Zhang, P., Sun, J., Liang, C., Gu, B., Xu, Y., Lu, H., et al. (2020c). lncRNA IGHCγ1 Acts as a ceRNA to regulate macrophage inflammation via the miR-6891-3p/TLR4 Axis in osteoarthritis. Mediators Inflamm. 2020:9743037.
Zhang, S. B., Lin, S. Y., Liu, M., Liu, C. C., Ding, H. H., Sun, Y., et al. (2019). CircAnks1a in the spinal cord regulates hypersensitivity in a rodent model of neuropathic pain. Nat. Commun. 10:4119.
Zhang, W., Zhang, C., Hu, C., Luo, C., Zhong, B., and Yu, X. (2020d). Circular RNA-CDR1as acts as the sponge of microRNA-641 to promote osteoarthritis progression. J. Inflamm. (Lond) 17, 8.
Zhang, X., Huang, C. R., Pan, S., Pang, Y., Chen, Y. S., Zha, G. C., et al. (2020e). Long non-coding RNA SNHG15 is a competing endogenous RNA of miR-141-3p that prevents osteoarthritis progression by upregulating BCL2L13 expression. Int. Immunopharmacol. 83:106425. doi: 10.1016/j.intimp.2020.106425
Zhang, X., Ji, S., Cai, G., Pan, Z., Han, R., Yuan, Y., et al. (2020f). H19 Increases IL-17A/IL-23 releases via regulating VDR by interacting with miR675-5p/miR22-5p in ankylosing spondylitis. Mol. Ther. Nucleic Acids 19, 393–404. doi: 10.1016/j.omtn.2019.11.025
Zhang, X., Liu, X., Ni, X., Feng, P., and Wang, Y. (2019). Long non-coding RNA H19 modulates proliferation and apoptosis in osteoarthritis via regulating miR-106a-5p. J. Biosci. 44:128.
Zhang, Y., Ma, L., Wang, C., Wang, L., Guo, Y., and Wang, G. (2020g). Long noncoding RNA LINC00461 induced osteoarthritis progression by inhibiting miR-30a-5p. Aging (Albany NY) 12, 4111–4123. doi: 10.18632/aging.102839
Zhang, Y., Wang, F., Chen, G., He, R., and Yang, L. (2019). LncRNA MALAT1 promotes osteoarthritis by modulating miR-150-5p/AKT3 axis. Cell Biosci. 9:54.
Zhao, C. Q., Jiang, L. S., and Dai, L. Y. (2006). Programmed cell death in intervertebral disc degeneration. Apoptosis 11, 2079–2088. doi: 10.1007/s10495-006-0290-7
Zhao, K., Zhang, Y., Yuan, H., Zhao, M., and Zhao, D. (2019). Long noncoding RNA LINC00958 accelerates the proliferation and matrix degradation of the nucleus pulposus by regulating miR-203/SMAD3. Aging (Albany NY) 11, 10814–10825. doi: 10.18632/aging.102436
Zhao, Y., Zhao, J., Guo, X., She, J., and Liu, Y. (2018). Long non-coding RNA PVT1, a molecular sponge for miR-149, contributes aberrant metabolic dysfunction and inflammation in IL-1β-simulated osteoarthritic chondrocytes. Biosci. Rep. 38:BSR20180576.
Zheng, H., Wang, T., Li, X., He, W., Gong, Z., Lou, Z., et al. (2020). LncRNA MALAT1 exhibits positive effects on nucleus pulposus cell biology in vivo and in vitro by sponging miR-503. BMC Mol. Cell Biol. 21:23.
Zhi, L., Zhao, J., Zhao, H., Qing, Z., Liu, H., and Ma, J. (2020). Downregulation of LncRNA OIP5-AS1 induced by IL-1b aggravates osteoarthritis via regulating miR-29b-3p/PGRN. Cartilage 1947603519900801. doi: 10.1177/1947603519900801
CrossRef Full Text [Published online ahead of print] | PubMed Abstract | Google Scholar
Zhou, X., Jiang, L., Fan, G., Yang, H., Wu, L., Huang, Y., et al. (2019). Role of the ciRS-7/miR-7 axis in the regulation of proliferation, apoptosis and inflammation of chondrocytes induced by IL-1β. Int. Immunopharmacol. 71, 233–240. doi: 10.1016/j.intimp.2019.03.037
Zhou, Y., Li, S., Chen, P., Yang, B., Yang, J., Liu, R., et al. (2019). MicroRNA-27b-3p inhibits apoptosis of chondrocyte in rheumatoid arthritis by targeting HIPK2. Artif. Cells Nanomed. Biotechnol. 47, 1766–1771. doi: 10.1080/21691401.2019.1607362
Zhou, Z. B., Du, D., Huang, G. X., Chen, A., and Zhu, L. (2018). Circular RNA Atp9b, a competing endogenous RNA, regulates the progression of osteoarthritis by targeting miR-138-5p. Gene 646, 203–209. doi: 10.1016/j.gene.2017.12.064
Zhou, Z. B., Huang, G. X., Fu, Q., Han, B., Lu, J. J., Chen, A. M., et al. (2019). circRNA.33186 contributes to the pathogenesis of osteoarthritis by sponging miR-127-5p. Mol. Ther. 27, 531–541. doi: 10.1016/j.ymthe.2019.01.006
Keywords: degenerative musculoskeletal disorders, aging, age-related disease, non-coding RNAs, miRNA, circRNA, lncRNA
Citation: Zheng Y-L, Song G, Guo J-B, Su X, Chen Y-M, Yang Z, Chen P-J and Wang X-Q (2021) Interactions Among lncRNA/circRNA, miRNA, and mRNA in Musculoskeletal Degenerative Diseases. Front. Cell Dev. Biol. 9:753931. doi: 10.3389/fcell.2021.753931
Received: 05 August 2021; Accepted: 22 September 2021;
Published: 11 October 2021.
Edited by:
Zhao-Qian Teng, Institute of Zoology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Lei Zhao, University of Wisconsin-Madison, United StatesCopyright © 2021 Zheng, Song, Guo, Su, Chen, Yang, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pei-Jie Chen, Y2hlbnBlaWppZUBzdXMuZWR1LmNu; Xue-Qiang Wang, d2FuZ3h1ZXFpYW5nQHN1cy5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.