- Department of Biochemistry and Molecular Biology, School of Basic Medicine, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
During oocyte maturation and the oocyte-to-embryo transition, key developmental regulators such as RNA-binding proteins coordinate translation of particular messenger RNA (mRNAs) and related developmental processes by binding to their cognate maternal mRNAs. In the nematode Caenorhabditis elegans, these processes are regulated by a set of CCCH zinc finger proteins. Oocyte maturation defective-1 (OMA-1) and OMA-2 are two functionally redundant CCCH zinc finger proteins that turnover rapidly during the first embryonic cell division. These turnovers are required for proper transition from oogenesis to embryogenesis. A gain-of-function mutant of OMA-1, oma-1(zu405), stabilizes and delays degradation of OMA-1, resulting in delayed turnover and mis-segregation of other cell fate determinants, which eventually causes embryonic lethality. We performed a large-scale forward genetic screen to identify suppressors of the oma-1(zu405) mutant. We show here that multiple alleles affecting functions of various anaphase promoting complex/cyclosome (APC/C) subunits, including MAT-1, MAT-2, MAT-3, EMB-30, and FZY-1, suppress the gain-of-function mutant of OMA-1. Transcriptome analysis suggested that overall transcription in early embryos occurred after introducing mutations in APC/C genes into the oma-1(zu405) mutant. Mutations in APC/C genes prevent OMA-1 enrichment in P granules and correct delayed degradation of downstream cell fate determinants including pharynx and intestine in excess-1 (PIE-1), posterior segregation-1 (POS-1), muscle excess-3 (MEX-3), and maternal effect germ-cell defective-1 (MEG-1). We demonstrated that only the activator FZY-1, but not FZR-1, is incorporated in the APC/C complex to regulate the oocyte-to-embryo transition. Our findings suggested a genetic relationship linking the APC/C complex and OMA-1, and support a model in which the APC/C complex promotes P granule accumulation and modifies RNA binding of OMA-1 to regulate the oocyte-to-embryo transition process.
Introduction
Early embryogenesis is a fundamental cellular process during development in metazoans. The specification of early cell fates and developmental patterns are usually determined at this stage. The process of early embryogenesis is governed by maternal mRNA and proteins loaded into eggs during oogenesis (Moore, 2005; Schier, 2007; Farley and Ryder, 2008). Maternal messenger RNA (mRNAs) are translationally repressed when loaded into eggs and their translations are regulated by a set of maternal loaded RNA-binding proteins (Schier, 2007; Rosario et al., 2017). During the oocyte-to-embryo transition, RNA-binding proteins coordinate translation of particular mRNAs and their corresponding developmental processes, such as anterior–posterior axis formation and cell fate specification (Newport and Kirschner, 1982; Edgar and Schubiger, 1986; Powell-Coffman et al., 1996; Oh and Houston, 2017). Shortly after, transcription from the zygotic genome begins and control of embryonic development is shifted from maternal to zygotic gene products. Meanwhile, primordial germ cells (PGCs) are specified during early embryogenesis. PGCs are kept transcriptionally quiescent until they need to proliferate, which also requires maternal supplied factors such as RNA-binding proteins (Forbes and Lehmann, 1998; Dansereau and Lasko, 2008).
The oocyte-to-embryo transition and early embryogenesis is rapid and dynamic in Caenorhabditis elegans, which provides a powerful model system to study these processes (Robertson and Lin, 2015). After fertilization, the C. elegans zygote undergoes several rounds of asymmetric divisions to produce particular early blastomeres that produce differentiated descendants and germ line cells. Specifically, during the first four rounds of asymmetric divisions, each division generates a somatic blastomere and a germline blastomere, named P1, P2, P3, and P4 (Figure 1A; Matova and Cooley, 2001; Rose and Gonczy, 2014). Later, P4 divides once to produce the PGCs, Z2, and Z3 (Wang and Seydoux, 2013). In order to ensure accurate completion of this process, maternal loaded mRNAs and RNA-binding proteins are needed to follow some special spatial and temporal localization patterns. More importantly, these RNA-binding proteins must have the ability to select their own targets from a large variety of maternal mRNAs.
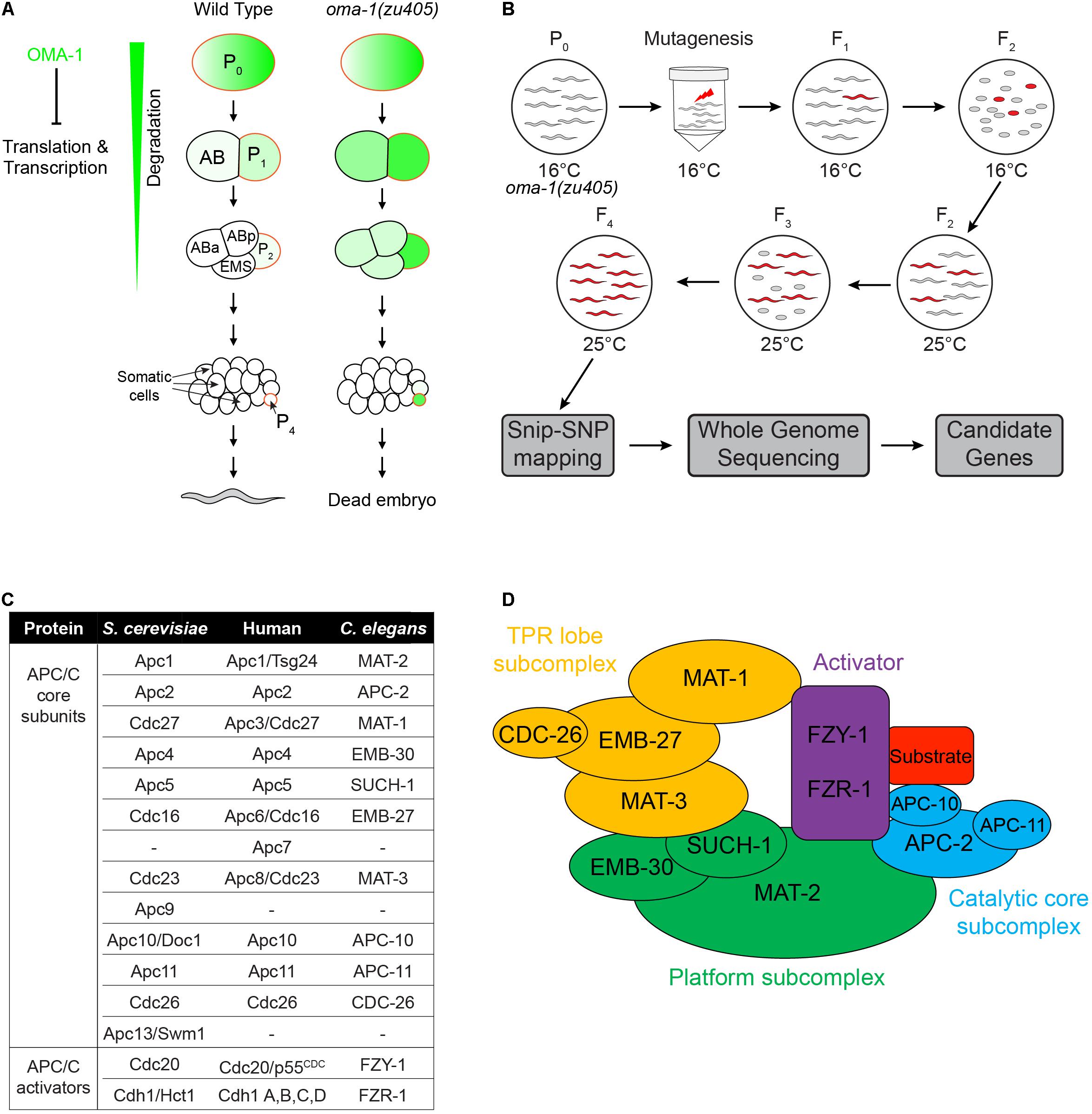
Figure 1. Schematic of the suppressor genetic screen to identify factors involved in OMA-1-regulated early embryogenesis in C. elegans. (A) Early embryonic development from a 1-cell to an ∼20-cell stage embryo in the wild type and oma-1(zu405) mutant, highlighting the expression of OMA-1 in P0 and P1 cells and delayed degradation in P2 and P4 cells. (B) Suppressor screen schematic. Synchronized oma-1(zu405) L4 animals were exposed to the mutagen, and phenotypes were scored in F3 and F4 at 25°C. Snip-SNP mapping and whole genome sequencing were performed to determine candidate genes. (C) Homologs of APC/C genes in Saccharomyces cerevisiae, humans, and C. elegans. It should be noted that not all homologs of APC/C genes in S. cerevisiae and humans are identified in C. elegans. (D) Schematic structure of the APC/C complex. The APC/C is generally divided into three subcomplexes and one activator.
Previous studies have identified a set of RNA-binding proteins that bind to specific mRNAs in different blastomere cells and at different developmental stages (Lee and Schedl, 2006). Among these RNA-binding proteins, oocyte maturation proteins OMA-1 and OMA-2, two closely related and functionally redundantly CCCH zinc finger proteins, play unique roles in oocyte maturation and the oocyte-to-embryo transition (Detwiler et al., 2001; Kaymak and Ryder, 2013). OMA-1/2 protein expression starts in developing oocytes and peaks in maturated oocytes and newly fertilized eggs. After the first embryonic division, OMA-1/2 proteins are rapidly degraded and are barely detectable in 4-cell stage embryos. Rapid degradation of OMA-1/2 proteins is required to avoid embryonic lethality (Lin, 2003). Degradation of oocyte maturation defective-1 (OMA-1) is regulated by DYRK Kinase MBK-2. During the oocyte-to-embryo transition, MBK-2 directly phosphorylates OMA-1 and promotes rapid turnover of OMA-1 (Shirayama et al., 2006). A gain of function mutation, P240L, in OMA-1 reduces MBK-2’s ability to phosphorylate OMA-1, resulting in increased protein stability and embryonic lethality (Shirayama et al., 2006). OMA-1/2 have been proposed to regulate the oocyte-to-embryo transition by targeting specific maternal mRNAs and repressing their translation (Lin, 2003). Previous studies have identified several targets of OMA-1/2, such as zif-1 and nos-2 (Jadhav et al., 2008; Guven-Ozkan et al., 2010). nos-2 is a nanos homolog that is required for development of PGCs (Jadhav et al., 2008), while zif-1 encodes a subunit of E3 ubiquitin ligase, which is required to restrict downstream cell fate determinants to proper localization through proteolysis (DeRenzo et al., 2003). One direct target of ZIF-1 is pharynx and intestine in excess-1 (PIE-1), another CCCH zinc finger protein, which is an essential germ cell fate determinant and it segregates with the germ cell lineage (DeRenzo et al., 2003). PIE-1 is accumulated in the nuclei of germline blastomeres to inhibit mRNA transcription. Hyper stabilized OMA-1 by the point mutation P240L causes delayed expression of ZIF-1 and abnormal distribution of PIE-1 from only being in germline blastomeres to being in somatic cells (Lin, 2003). Besides PIE-1, other cell fate determinants, including MEX-1, muscle excess-3 (MEX-3), MEX-5, and posterior segregation-1 (POS-1), which are also RNA-binding proteins and essential for embryogenesis, are incorrectly segregated and localized if OMA-1 is not properly degraded during the oocyte-to-embryo transition (Lin, 2003). OMA-1 and OMA-2 not only bind maternal mRNAs to repress translation, but also bind TATA box-binding protein (TBP)-associated factor TAF-4, a component that is essential for assembly of a functional TFIID and RNA polymerase II pre-initiation complex and to sequester TAF-4 in the cytoplasm in P0 and P1 germline blastomeres (Guven-Ozkan et al., 2008). MBK-2 directed phosphorylation promotes degradation of OMA-1 and allows TAF-4 to enter the nuclei of somatic cells to initiate zygotic transcription (Guven-Ozkan et al., 2008).
Although the key events of OMA-1/2 mediated oocyte maturation and oocyte-to-embryo transition are understood, the molecular mechanisms regulating OMA-1/2-mRNA binding, especially in the situation when OMA-1 is not properly degraded, are poorly understood. In this study, we designed a forward genetic screen to identify suppressors of the gain-of-function mutation P240L in OMA-1 (Lin, 2003). Using this unbiased genetic screen, we discovered that the anaphase promoting complex/cyclosome (APC/C) complex, especially when binding to activator Cdc20, participates in OMA-1 mediated oocyte maturation and the oocyte-to-embryo transition.
The APC/C complex is an E3 ubiquitin ligase that targets proteins for degradation during cell cycles, particularly during mitotic exit and the onset of anaphase (Harper et al., 2002; Peters, 2002, 2006; Satyanarayana and Kaldis, 2009; Pines, 2011). The APC/C complex is a large complex containing 11–13 subunits in different organisms, including a catalytic core of a Cullin subunit Apc2 and a Ring domain protein Apc11. The substrate recognition is specified by the activators Cdc20 and Cdh1 (Deshaies, 1999; Passmore, 2004; Pintard et al., 2004). The main substrate of the APC/CCdc20 complex is securin, whose degradation leads to activation of separase and promotion of anaphase onset (Fang et al., 1999; Lara-Gonzalez et al., 2017), whereas the main substrate of APC/CCdh1 is cyclin B, whose degradation leads to downregulation of CDK activity and is required for mitotic exit (Fang et al., 1998, 1999; Blanco et al., 2000; Listovsky et al., 2000; Lara-Gonzalez et al., 2017). All APC/C subunits are highly conserved from yeast to humans, and the molecular architecture of the APC/C complex has been elucidated recently (Dube et al., 2005; Passmore et al., 2005; Herzog et al., 2009; Uzunova et al., 2012; Primorac and Musacchio, 2013; Chang et al., 2014). Subunits of the APC/C complex can be grouped into three subcomplexes, including the platform, the catalytic core and the tetratricopeptide repeat (TPR) lobe (Figures 1C,D). Functions of APC/C during the oocyte-to-embryo transition have been studied previously, focusing on the release of meiosis arrest at the beginning of the oocyte-to-embryo transition (Marangos et al., 2007; Verlhac et al., 2010; Whitfield et al., 2013). Meiosis arrest happens during oocyte maturation in most animals, mainly because of the presence of cyclin B. The APC/C complex is activated upon fertilization and targets cyclin B for degradation to allow meiosis to proceed (Blanco et al., 2001; Pesin and Orr-Weaver, 2007; Swan and Schupbach, 2007; Radford et al., 2008). However, whether the APC/C complex plays other roles during oocyte maturation and the oocyte-to-embryo transition is still unclear.
In this study, we found that, although OMA-1 is not the direct target of the APC/C complex, the APC/C complex may participate in a pathway that regulates the association between OMA-1 and its cognate mRNA targets. We showed that mutations in multiple APC/C genes suppress embryonic lethality of the oma-1(zu405) strain, which has the gain-of-function mutation P240L in OMA-1 to impedes oocyte maturation. Transcriptome analysis of early embryos indicated gene expressions in oma-1(zu405) early embryos are similar to the wild type strain after introducing mutations in APC/C genes. We also showed that, although the APC/C complex does not regulate the degradation of OMA-1, it enhances P granule localization of OMA-1, where OMA-1 binds to its mRNA targets. This implies that the APC/C complex may modify the binding ability of OMA-1 to its cognate targets. Finally, proper segregation of some key cell fate determinants, including PIE-1, POS-1, MEX-3, and maternal effect germ-cell defective-1 (MEG-1), are corrected in the oma-1(zu405) mutant after introducing mutations in APC/C genes.
Results
A Suppressor Genetic Screen to Identify Factors Involved in OMA-1-Regulated Early Embryogenesis
In oma-1(zu405) embryos, the C blastomere is transformed to the ethyl methanesulfonate (EMS) blastomere fate at a restrictive temperature, resulting in temperature sensitive embryonic lethality (Figure 1A; Lin, 2003). We performed the suppressor screen to identify mutations rescuing embryonic lethality of the oma-1(zu405) strain. During the screen, F2 progenies of EMS/N-ethyl-N-nitrosourea (ENU) treated oma-1(zu405) worms were cultured at 25°C to determine whether F3 eggs were hatched or not. All viable F3 worms were collected for further study (Figure 1B). A total of 0.6 × 105 F2s (i.e., 1.2 × 105 genomes) were screened and 23 mutant strains were isolated. We used snip-SNPs as molecular markers to roughly map mutations in different chromosomes, then we performed whole genome sequencing (WGS) for oma-1(zu405) and all mutants, and analyzed the mutated genes with reference to the corresponding mapped chromosomes (Figure 1B). By comparing mutated genes in different strains, we identified three strains that had independent mutations in mat-1, two strains that had independent mutations in mat-3, and four strains that had independent mutations in fzy-1. Interestingly, MAT-1, MAT3, and FZY-1 all belong to subunits of the APC/C complex (Kitagawa et al., 2002; Shakes et al., 2003; Garbe et al., 2004). APC/C is a complex that has E3 ubiquitin ligase activity and plays a central role in the metaphase to anaphase transition and chromosome segregation steps during mitosis. To date, 12 subunits have been identified in yeast, 11 subunits have been identified in humans, and 10 subunits have been identified in C. elegans (Figure 1C; Furuta et al., 2000; Golden et al., 2000; Davis et al., 2002; Peters, 2002; Rappleye et al., 2002; Shakes et al., 2003; Passmore, 2004; Yeong, 2004; Pines, 2006). These subunits are organized into three functionally related subcomplexes: the platform (MAT-2/Apc1, EMB-30/Apc4, and SUCH-1/Apc5), the TPR lobe (MAT-1/Apc3, EMB-27/Apc6, MAT-3/Apc8, and CDC-26/Cdc26), and the catalytic core (APC-2/Apc2, APC-10/Apc10, and APC-11/Apc-11). There are also two activators, FZY-1/Cdc20 and FZR-1/Cdh1, that determine the target specificity of APC/C (Figure 1D).
Variation of Mutations in Coding Genes of Subunits of the Anaphase Promoting Complex/Cyclosome Complex as Functional Genetic Alternations in Mutants From Ethyl Methanesulfonate Screens
We next asked whether other genes coding APC/C subunits have mutations in these strains. Eventually we identified one strain that had a mutation in mat-2 and one strain that had a mutation in emb-30 (Figure 2A). All mutations we identified in APC/C genes are recessive, since only homozygous, but not heterozygous, mutants suppressed lethality of oma-1(zu405).
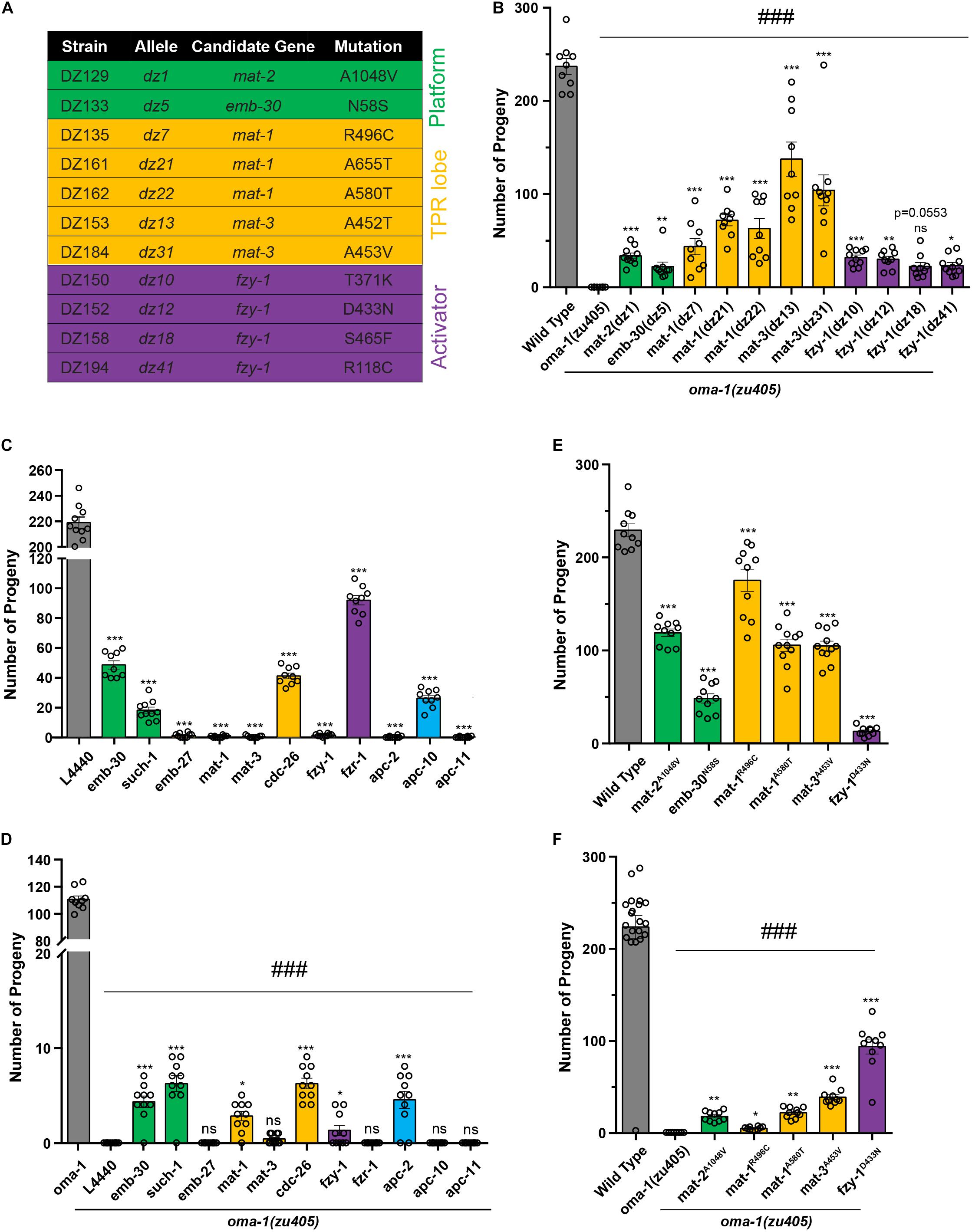
Figure 2. Mutations in APC/C genes suppressed lethality of the oma-1(zu405) mutant. (A) Summary of potential mutations in APC/C genes identified from the suppressor screen. Genes encoding for platform subunits, TRP lob subunits, and activators are highlighted in green, yellow, and purple, respectively. (B) Brood size of mutants identified from the suppressor screen of the oma-1(zu405) mutant. (C) Brood size of strains treated with RNAi of corresponding APC/C genes in the wild type N2. L4440 expressed an empty RNAi vector, which served as the control. (D) Brood size of strains treated with RNAi of corresponding APC/C genes in the oma-1(zu405) mutant. L4440 expressed an empty RNAi vector, which served as the control. (E) Brood size of strains contacting point mutations in APC/C genes generated by the CRISPR/Cas9 gene editing system. It should be noted that these point mutations were identified in mutants from the suppressor screen. (F) Brood size of oma-1(zu405) combined with corresponding point mutations in APC/C genes. The results are presented as the average ± S.D. with individual values plotted. *p < 0.05; **p < 0.01; ***, ###p < 0.001; ns: not significant. The p-values were calculated by 1-way ANOVA followed by Dunnett’s multiple comparisons. N = 10–15.
To confirm whether these candidate genes are functional genes that suppress lethality of oma-1(zu405), we performed two sets of experiments and investigated animal viability by scoring brood size: (1) RNAi treatment of candidate genes in oma-1(zu405), and (2) regeneration of exactly the same mutations of candidate genes in oma-1(zu405) identified in EMS screens using the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) gene editing system. We first scored brood size for all strains that contained the APC/C related alleles from our forward genetic suppressor screen (Figure 2B). oma-1(zu405) has a very tight phenotype of absolutely no progenies that can escape the lethality phenotype at 25°C (Lin, 2003). Although not all alleles restored the brood size of oma-1(zu405) to the wild type, they all suppressed lethality of oma-1(zu405) and restored the brood size from ∼20 to ∼140 offspring per animal. When wild type worms were treated with mat-1, mat-2, mat-3, emb-30, and fzy-1 RNAi, we found that knockdown of mat-1, mat-3, and fzy-1 caused lethality of wild type worms, while knockdown of mat-2 and emb-30 only had a mild effect on the viability of wild type animals (Figure 2C). Previous studies indicate that known alleles of these genes are either lethal or temperature sensitive lethal (Furuta et al., 2000; Golden et al., 2000; Kitagawa et al., 2002), suggesting RNAi of mat-2 and emb-30 did not completely deplete functional proteins. To assess RNAi efficiency, we measured mRNA levels of corresponding genes by quantitative real-time PCR (qRT-PCR), and found that, in RNAi treated strains, mRNA levels were knocked down to 15–40% of wild type RNA levels (Supplementary Figure 1A). In addition, we treated a strain expressing OMA-1::GFP (see below) with oma-1 RNAi, and found that the OMA-1 protein level was reduced to ∼20% of the wild type protein level (Supplementary Figure 1B). We next scored brood size after treating the oma-1(zu405) strain with mat-1, mat-2, mat-3, emb-30, and fzy-1 RNAi. We found all RNAi treatments restored fertility in oma-1(zu405) to very low levels. RNAi treatment of emb-30 on oma-1(zu405) produced the largest brood size, which was only about 5 (Figure 2D).
Next, we reconstituted exact mutations in the APC/C genes identified in EMS screens by the CRISPR/Cas9 gene editing system. The mutations we regenerated included mat-1R496C, mat-1A580T, mat-2A1048V, mat-3A453V, emb-30N58S, and fzy-1D433N (Supplementary Figure 1C). We first generated these mutations in the wild type N2 background, and we found strains introduced by these mutations had relatively large brood sizes, from the smallest of 12 (fzy-1D433N) to the largest of more than 175 (mat-1R496C) (Figure 2E). Considering other known mutants of these genes are lethal, these data suggest that mutants we identified contain hypomorph alleles of APC/C genes. We crossed these CRISPR strains with oma-1(zu405) and scored brood sizes in double mutants. All regenerated mutations by CRISPR suppressed lethality of oma-1(zu405) to similar levels of EMS mutants of corresponding genes (Figure 2F). These results demonstrated that mutations in key APC/C subunits are the basis for the phenotypes observed in the mutants we isolated from the genetic screen.
It should be noted that the CRISPR generated fzy-1D433N mutant only had a very small brood size; however, the fzy-1D433N;oma-1(zu405) double mutant had a brood size of more than 90. This means that fzy-1D433N and oma-1(zu405) mutually suppress each other’s lethality phenotype, which further implied that OMA-1-regulated the embryogenesis pathway and this pathway and the APC/C complex have strong genetic interactions.
Depletion Functions of Other Anaphase Promoting Complex/Cyclosome Subunits Repressed oma-1(zu405) Lethality
Since 10 subunits and two activators of APC/C have been identified in C. elegans, we asked whether other components of APC/C suppress lethality of oma-1(zu405). We treated oma-1(zu405) with RNAi for apc-2, such-1, emb-27, apc-10, apc-11, cdc-26, and fzr-1, and then scored brood sizes. We found knockdown of apc-2, such-1, and cdc-26 restored the brood size of oma-1(zu405) from 0 to around 6, which was very similar to RNAi treatment for APC/C genes identified by genetic screens, while knockdown of other genes, including emb-27, apc-10, apc-11, and fzr-1, did not suppress oma-1(zu405) (Figures 2C,D). Considering the observation that RNAi treatment of APC/C genes we identified by genetic screen only mildly suppress oma-1(zu405), but hypomorph alleles of these genes efficiently restored viability of oma-1(zu405) (Figures 2C,D), we suspected that apc-2, such-1, and cdc-26 could also have the same genetic interactions with oma-1(zu405). RNAi of emb-27 and apc-11 caused complete lethality of wild type animals, indicating RNAi of these two genes were efficient. Although mRNA levels of fzy-1 and apc-10 were reduced to similar mRNAs levels of other APC/C genes after RNAi treatments (Supplementary Figure 1A), we cannot completely rule out that RNAi of fzr-1 and apc-10 depleted all function proteins because fzr-1 RNAi and apc-10 RNAi animals are not lethal. Among all genes that were investigated, fzy-1 and fzr-1 encoded the only two activators of the APC/C complex in C. elegans, which were homologs of yeast Cdc20 and Cdh1, respectively (Fay et al., 2002; Kitagawa et al., 2002). The main substrate of APC/CCdc20 is securin, which inhibits separase and prevents chromosome segregation, and the main substrate of APC/CCdh1 is cyclin B, which when removed can promote metaphase to anaphase during mitosis. It is interesting that only mutants of fzy-1, but not fzr-1, suppressed lethality of oma-1(zu405), indicating the APC/CCdc20 complex plays a particular role in the oma-1-regulated oocyte-to-zygote transition and oocyte maturation processes. Since among all mutants of APC/C genes, fzy-1D433N restored the brood size of oma-1(zu405) the best, we next used fzy-1D433N as the representative mutant of APC/C genes in the following studies.
Depletion of Anaphase Promoting Complex/Cyclosome Subunits Partially Correct Embryonic Transcriptomes in the oma-1(zu405) Mutant
To better characterize how mutations in APC/C complex suppress oma-1(zu405) lethality, we investigated transcriptome changes in fzy-1D433N;oma-1(zu405) double mutants versus the oma-1(zu405) single mutant. For this purpose, we isolated early embryos of corresponding strains, extracted total RNA, and performed mRNA sequencing (mRNA-seq) analysis. The isolated embryos were all from synchronized early young adults, and almost all were ∼30-cell or earlier stage embryos. We compared transcriptomes of oma-1(zu405) and fzy-1D433N;oma-1(zu405) to the wild type strain N2. Differentially expressed genes for each comparison were counted and identified by DESeq2. We identified 1,222 mRNAs that were upregulated and 4,656 mRNAs that were downregulated in the oma-1(zu405) mutant compared to the wild type N2 strain (| log2(fold change)| > 2, p < 0.05, Benjamini-Hochberg adjusted p-value). Based on the same criteria, mis-regulated expressions of mRNAs were largely corrected to normal levels, with only 266 mRNAs that were upregulated and eight mRNAs that were downregulated in the fzy-1D433N;oma-1(zu405) double mutant (Figure 3A). Venn diagrams showed that 192 mRNAs were commonly upregulated in both oma-1(zu405) and the fzy-1D433N;oma-1(zu405) mutants, while only six mRNAs were commonly downregulated (Figure 3B). These mRNA-seq data indicated mutations in APC/C genes can in general restore the overall disordered transcriptome in the oma-1(zu405) mutant to the wild type.
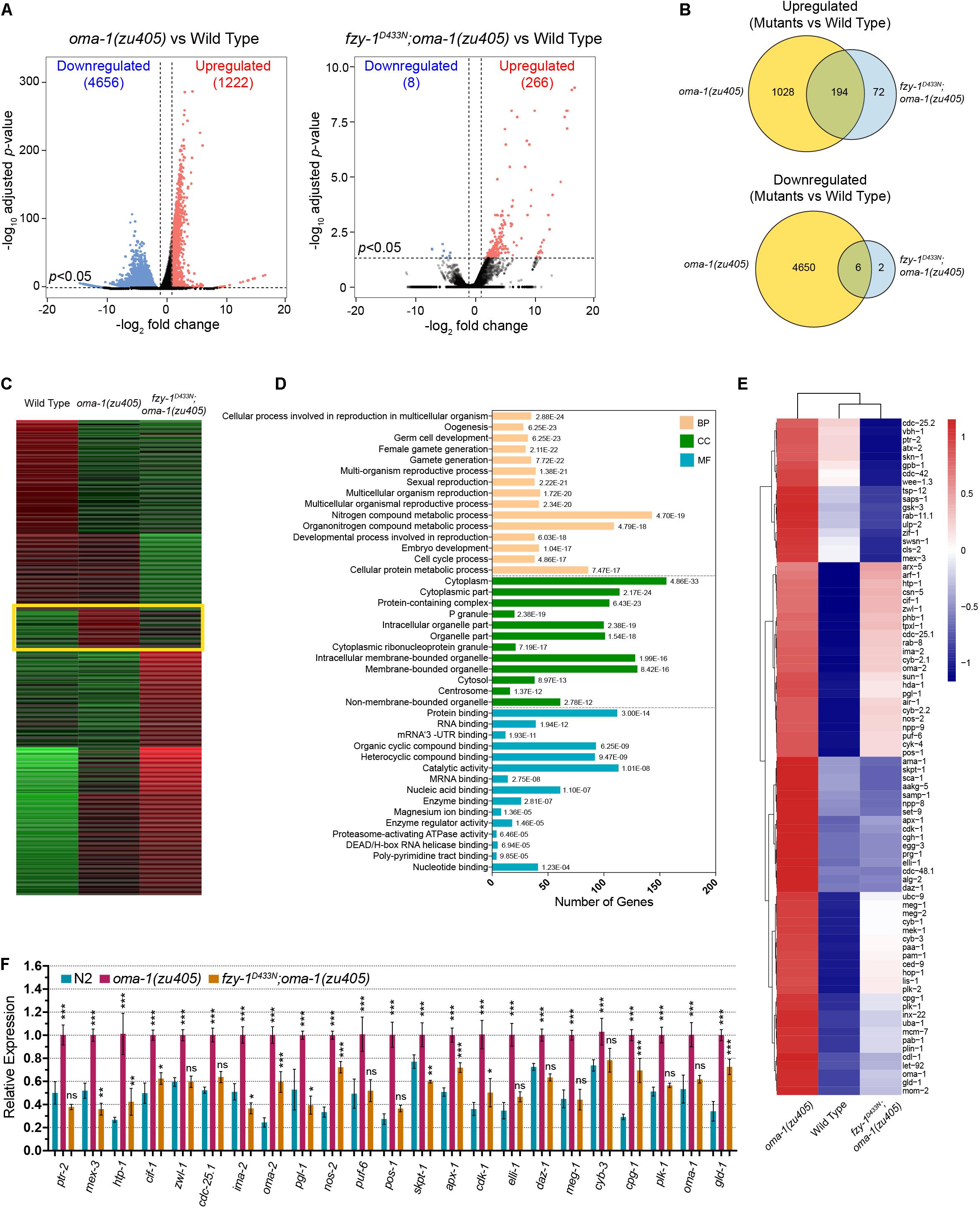
Figure 3. Comparative transcriptome analysis of oma-1(zu405), fzy-1D433N;oma-1(zu405), and the wild type. (A) Volcano plots of all differentially expressed genes detected in the mRNA-seq data in oma-1(zu405) (left) and fzy-1D433N;oma-1(zu405) (right), compared to the wild type. Criteria of deferential expression was | log2(fold change)| > 2, and Benjamini-Hochberg adjusted p-value < 0.05. (B) Venn diagrams depict the number of genes that are upregulated (or downregulated) only in oma-1(zu405) or fzy-1D433N;oma-1(zu405) or in both. (C) K-means cluster analysis of differentially expressed genes. The group of genes highlighted in the yellow rectangle show similar expression levels in fzy-1D433N;oma-1(zu405) and wild type strains. Expression levels were calculated as RPKM-normalized values. Each row represents a differentially expressed gene. (D) GO enrichment analysis of genes within the yellow rectangle in panel (C). Category distribution of the 318 differentially expressed genes is shown for three GO domains: biological process (BP), molecular function (MF), and cellular component (CC). (E) The heatmap shows differential expression of 80 genes, which were assigned to the terms of germ cell development, embryo development, protein binding, and RNA binding in panel (D). (F) Quantitative real-time PCR measurements of expression levels of selected 23 genes from (E) in the indicated strains. The results are presented as the average ± S.D. *p < 0.05; **p < 0.01; ***p < 0.001; ns: not significant. The p-values were calculated by 2-way ANOVA followed by Dunnett’s multiple comparisons.
To better understand our mRNA-seq data, we used k-means clustering to assign gene expression levels into groups, and aimed to identify which group of genes were most affected when introducing fzy-1D433N into the oma-1(zu405) mutant. One group containing 318 genes was particularly notable, since in this group, expression levels of genes were similar in the wild type and fzy-1D433N;oma-1(zu405) mutant, implying expression of this group of genes is best representative of the overall gene expression levels (Figure 3C). We next performed Gene Ontology (GO) enrichment analysis for these 318 genes and determined the most significant terms in biological process (BP), cellular compartment (CC), and molecular function (MF). The terms were ordered from most enriched (up) to least enriched (bottom) in each category (Figure 3D). Interestingly, the terms assigned to biological processes with the most enrichment were involved in germ cells and embryo development; the terms assigned to CCs included protein containing complexes, P granules, and cytoplasmic ribonucleoprotein (RNP) granules; and the terms assigned into CCs were involved in protein and nucleic acid binding, especially mRNA and mRNA 3′UTR binding (Figure 3D). The data from the GO enrichment analysis further implicated mutations in APC/C genes promote normal embryonic development of the oma-1(zu405) strain, which may be caused by regulating cellular distributions of OMA-1 and its downstream cell fate determinants, including RNA and transcription factor-binding proteins.
We chose 80 genes that were assigned to the terms of germ cell development, embryo development, protein binding, and RNA binding and drew a heatmap of expression levels of these genes. These genes are among the genes with the most relevant functions during OMA-1-regulated embryogenesis and have the most dramatic differential expression levels, including zif-1, pos-1, meg-1, meg-2, and skn-1. Again, the heatmap showed that introducing fzy-1D433N could change expression levels of these genes in the oma-1(zu405) mutant to wild type (Figure 3E). Finally, we chose 23 genes from these 80 genes and performed qRT-PCR to evaluate expression levels. qRT-PCR data confirmed that changes in gene expression levels in oma-1(zu405), fzy-1D433N;oma-1(zu405), and wild type strains were consistent with mRNA-seq data (Figure 3F).
Depletion of Anaphase Promoting Complex/Cyclosome Subunits Corrected Defects of the OMA-1 Distribution in the oma-1(zu450) Mutant
To investigate whether APC/C mutants suppress lethality of oma-1(zu405) by regulating cellular distribution of OMA-1, we created in situ knock-in strains expressing OMA-1::GFP and OMA-1P240L::GFP using the CRISPR/Cas9 gene editing system. Brood size of OMA-1::GFP strain was similar as the untagged strain either in wild type or oma-2 mutant background, indicating tagging green fluorescent protein (GFP) into C-terminal OMA-1 did not affect OMA-1 function (Supplementary Figure 1D). As in previous studies, OMA-1::GFP accumulated in oocytes and very early embryos and started to be degraded after the first embryonic cell division and disappeared after 4-cell embryos (Figures 4A,B). OMA-1P240L::GFP fluorescence was still detected easily in 4-cell embryos and signals in P2 cells were stronger than those in somatic cells. OMA-1P240L::GFP fluorescence was eventually only detectable in P4 cells (Figures 4A,B).
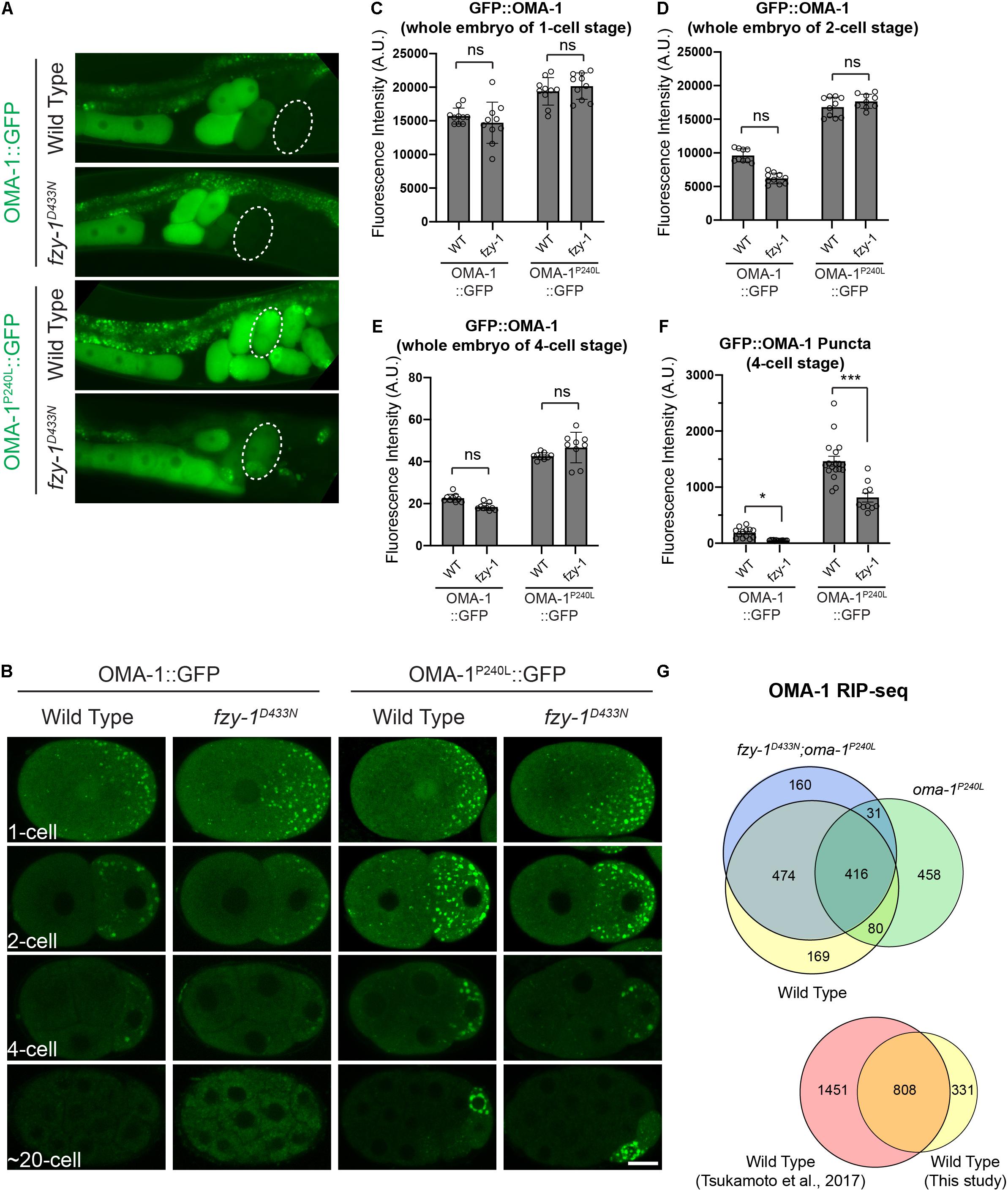
Figure 4. fzy-1D433N mutant affected the OMA-1 distribution in early embryos. (A) Fluorescent micrographs show the localization of OMA-1::GFP and OMA-1P240L::GFP in intact gonads in the wild type and fzy-1D433N mutant. 4-cell stage embryos are indicted by dotted white circles. (B) Fluorescent micrographs show the localization of OMA-1::GFP and OMA-1P240L::GFP in 1-cell, 2-cell, 4-cell, and ∼20-cell stage embryos in the wild type and fzy-1D433N mutant. Scale bar: 10 microns. (C) Quantification of overall OMA-1::GFP fluorescence in 1-cell stage embryos. (D) Quantification of overall OMA-1::GFP fluorescence in 2-cell stage embryos. (E) Quantification of overall OMA-1::GFP fluorescence in 4-cell stage embryos. (F) Quantification of OMA-1::GFP fluorescence enriched in granules in 4-cell stage embryos. The results are presented as the average ± S.D. with individual values plotted. *p < 0.05; ***p < 0.001. The p-values were calculated by 2-way ANOVA followed by Dunnett’s multiple comparisons. N = 8–12. (G) Above: Venn diagram showing the overlap between OMA-1- and OMA-1P240L-associated RNAs in wild type and fzy-1D433N mutant with a twofold enrichment cutoff. Bottom: Venn diagram showing the overlap between OMA-1-associated RNAs identified in this study and a previous study (Tsukamoto et al., 2017).
Next, we introduced fzy-1D433N into OMA-1::GFP and OMA-1P240L::GFP transgenic strains to investigate whether OMA-1P240L::GFP is properly degraded in early embryos of the fzy-1D433N mutant. To rule out the possibility that introducing fzy-1D433N caused changes of overall OMA-1 protein levels, we compared OMA-1::GFP levels in wild type and fzy-1D433N embryos at the same developmental stage. By measuring fluorescent signals within whole embryos, we found that fzy-1D433N did not affect overall OMA-1 expression at any developmental stages we investigated (Figures 4C–E). We found fzy-1D433N mutants affected the distribution of wild type OMA-1::GFP in early embryos (Figures 4A,B). Especially in 2- and 4-cell embryos, OMA-1::GFP was less localized into the puncta-like structure P granules (Figure 4B and see below). Quantification in the 4-cell embryos showed reduced P granule localization was statistically significant (Figure 4F). In addition, OMA-1P240L::GFP was also altered when APC/C mutants were introduced. Although OMA-1P240L:GFP fluorescence was still easily detected in 2-, 4-, or 20-cell embryos, its accumulation in P granules was reduced compared to OMA-1P240L::GFP in the APC/C wild type cells (Figure 4B and see below).
Previous studies showed that OMA-1 accumulates at P granules in early embryos (Shimada et al., 2006). Considering untranslated maternal mRNAs accumulate at P granules during early embryogenesis (Noble et al., 2008), and OMA-1 binds to maternal mRNA to repress translation, it is reasonable to determine whether the APC/C complex regulates OMA-1 accumulation at P granules. We used mRuby::PGL-1 as the P granule marker, since PGL-1 is a typical component of P granules (Kawasaki et al., 1998). We first confirmed P granule accumulation of OMA-1 by showing that OMA-1::GFP and mRuby::PGL-1 were co-localized at P lineage cells in the wild type embryos (Figures 5A,C). By introducing the fzy-1D433N mutant, as we observed above, GFP:OMA-1 accumulated less to mRuby::PGL-1 labeled P granules, and Pearson’s correlation coefficients (PCCs) also indicated the reduction of co-localization between OMA-1::GFP and mRuby::PGL-1 in fzy-1D433N mutant (Figures 5A,C). Notably, highly enriched OMA-1P240L:GFP in P cells was also co-localized with mRuby::PGL-1, suggesting OMA-1P240L::GFP was indeed accumulated at P granules (Figure 5B). Surprisingly, we did not observe an obvious reduction of co-localization between OMA-1P240L:GFP and mRuby::PGL-1 in fzy-1D433N mutant (Figure 5C). It is possible that PGL-1 was also delocalized to P granules in OMA-1P240L::GFP genetic background. Another possibility is that mRuby::PGL-1 is overexpressed in this strain (Yang et al., 2014), which does not fully behave like the endogenous PGL-1 under certain circumstances. All the data above indicated APC/C mutants suppress oma-1(zu405) not by promoting degradation of OMA-1P240L proteins, but possibly by restricting localization of OMA-1P240L at P granules in early embryos. Less accumulation of OMA-1P240L at P granules may reflect that OMA-1P240L bound to different groups of mRNAs in APC/C mutants.
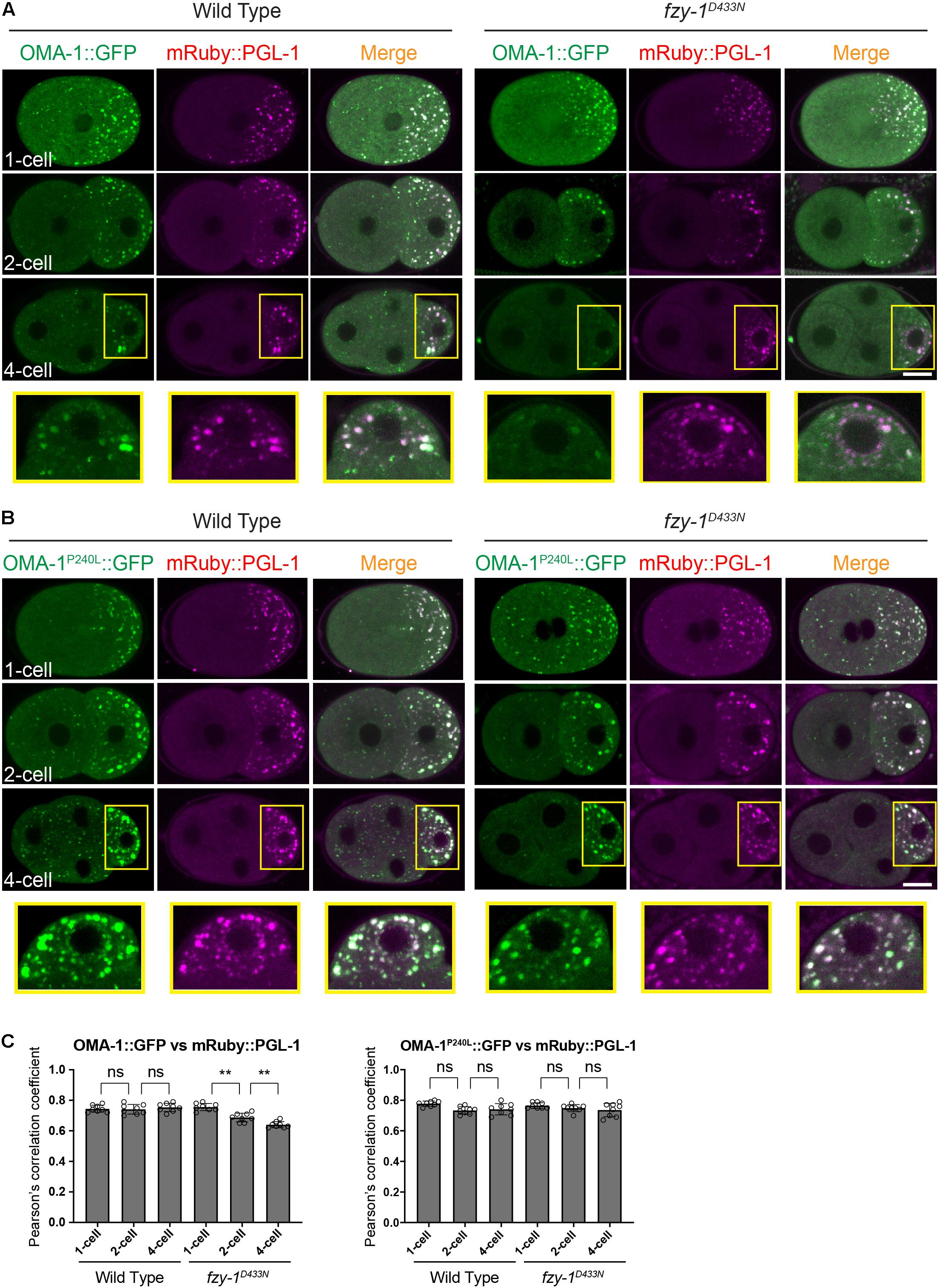
Figure 5. OMA-1::GFP and OMA-1P240L::GFP are localized to P granules in early embryos. (A) Fluorescent micrographs show the co-localization of OMA-1::GFP and mRuby::PGL-1 in P granules in 1-cell, 2-cell, and 4-cell stage embryos in the wild type and fzy-1D433N mutant. Regions within yellow rectangles are enlarged, turned 90 degrees counterclockwise, and displayed at the bottom. (B) Fluorescent micrographs show the co-localization of OMA-1P240L:GFP and mRuby::PGL-1 in P granules in 1-cell, 2-cell, and 4-cell stage embryos in the wild type and fzy-1D433N mutant. Regions within the yellow rectangles are enlarged, turned 90 degrees counterclockwise, and displayed at the bottom. Scale bar: 10 microns. (C) Pearson’s correlation coefficient to evaluate the extent of co-localization between OMA-1::GFP and mRuby::PGL-1. The results are presented as the average ± S.D. **p < 0.01; ns: not significant. The p-values were calculated by 2-way ANOVA followed by Dunnett’s multiple comparisons. N = 8.
Depletion of Anaphase Promoting Complex/Cyclosome Subunits Affected Association Between OMA-1 and mRNAs
To investigate whether APC/C complex regulate OMA-1-mRNA association, we immunopurified OMA-1::GFP by GFP nanobody and sequenced OMA-1-associated mRNAs in duplicate from young adults of oma-1:gfp, oma-1P240L:gfp, and fzy-1D433N;oma-1P240L:gfp strains. We also sequenced total RNAs in corresponding strains as input controls. OMA-1-associated mRNAs were identified using DESeq2 with a cutoff of | log2(fold change)| > 1 and Benjamini-Hochberg adjusted p-value < 0.05. Eventually, 1139 mRNAs were identified as OMA-1-associated mRNAs. To validate our analysis, we referred to published OMA-1 RIP-seq data (Tsukamoto et al., 2017), and found that 808/1139 (71%) mRNAs identified in our study were identified as OMA-1-assoicated mRNAs previously (Figure 4G), indicating our OMA-1 RIP-seq analysis was reliable. Based on the same criteria, 985 and 1081 mRNAs were identified as OMA-1P240L-associated mRNAs in the wild type and fzy-1D433N mutant, respectively. OMA-1 and OMA-1P240L shared 496 mRNAs (44% of OMA-1- and 50% of OMA-1P240L-associated mRNAs), whereas OMA-1 and OMA-1P240L in fzy-1D433N background shared 890 mRNAs (78% of OMA-1- and 82% of OMA-1P240L-assoicated mRNAs). Our data indicated that depletion of APC/C subunits affected OMA-1-RNA association, and turned mRNAs associated with OMA-1P240L to be more similar to mRNAs associated with wild type OMA-1. It should be noted that numbers of mRNAs bound to OMA-1 and OMA-1P240L were almost the same, implying that although mutation P240L altered OMA-1 binding affinity to untranslated mRNAs, the overall number of mRNAs bound to OMA-1 was largely unchanged. Developmental progresses during embryogenesis are largely determined by OMA-1 and OMA-1P240L specifically associated RNAs.
Depletion of Anaphase Promoting Complex/Cyclosome Subunits Correct Disrupted Distribution of Downstream Cell Fate Determinants of OMA-1, Including PIE-1, POS-1, MEX-3, and MEG-1
During embryogenesis, one of the mRNA targets OMA-1/2 binds to is zif-1. Binding of OMA-1/2 represses translation of zif-1 (Guven-Ozkan et al., 2010). ZIF-1 encodes a subunit of E3 ligase whose direct substrate is PIE-1, an important cell fate determinant that segregates with the germ lineage for germ cell development during embryogenesis (Tenenhaus et al., 1998, 2001). Degradation of PIE-1 in somatic cells is delayed in oma-1(zu405) mutant embryos, possibly by delayed expression of ZIF-1. Degradation of several other maternal cell fate determinants, including MEX-1, MEX-3, MEX-5, POS-1, and MEG-1, was also delayed in oma-1(zu405) mutant embryos (Lin, 2003). To test whether APC/C mutants suppress delayed degradation of these cell fate determinants in the oma-1(zu405) strain, we investigated the distribution of CRISPR-mediated knock-in transgenic PIE-1::GFP, POS-1:GPF, and GFP::MEG-1, and a MosSCI-mediated single copy inserted transgenic GFP::MEX-3 in the oma-1(zu405) mutant and double mutants containing mutations of selected APC/C genes.
PIE-1 is a CCCH zinc finger protein that binds to the 3′UTR of maternal mRNAs to serve as an essential regulator of germ cell fate. Previous studies showed that PIE-1 is accumulated at P cells; i.e., germline precursor cells (Tenenhaus et al., 1998; Oldenbroek et al., 2013). We also observed that PIE-1:GFP asymmetrically localized at the posterior pole in 1-cell embryos so that PIE-1::GFP fluorescence in the anterior AB cell was weaker than that in the posterior P1 cell in 2-cell embryos. PIE-1::GFP further accumulated in P1, P2, and P4 cells as granule-like structures and in the nucleus, and at the same time, PIE-1:GFP in the somatic cells (i.e., non-P lineage cells) was rapidly degraded to levels below detection threshold (Figure 6A). On the other hand, in the oma-1(zu405) mutant, degradation of PIE-1::GFP in somatic cells was severely delayed, resulting in obvious nuclear located PIE-1::GFP signals in somatic cells at ∼20-cell embryos, although fluorescence in somatic cells was weaker than in P cells (Figure 6A). fzy-1D433N, mat-1A580T, or mat-2A1048V single mutants did not affect the dynamic distribution pattern of PIE-1::GFP in wild type animals. However, in the oma-1(zu405) mutant, fzy-1D433N, mat-1A580T, or mat-2A1048V mutants all promoted degradation of PIE-1::GFP to various levels. Specifically, somatic cell accumulation of PIE-1::GFP completely disappeared in fzy-1D433N;oma-1(zu405) and mat-1A580T;oma-1(zu405) double mutants, but only weak suppression of delayed degradation of PIE-1::GFP was shown in the mat-2A1048V;oma-1(zu405) double mutant since trace fluorescence signals were still detectable in some of the soma cells (Figure 6A). It should be noted that PIE-1::GFP only accumulated at the nucleus but not at cytoplasmic granules in somatic cells. The delayed degradation of nuclear PIE-1::GFP in the oma-1(zu405) mutant was also suppressed by APC/C mutants.
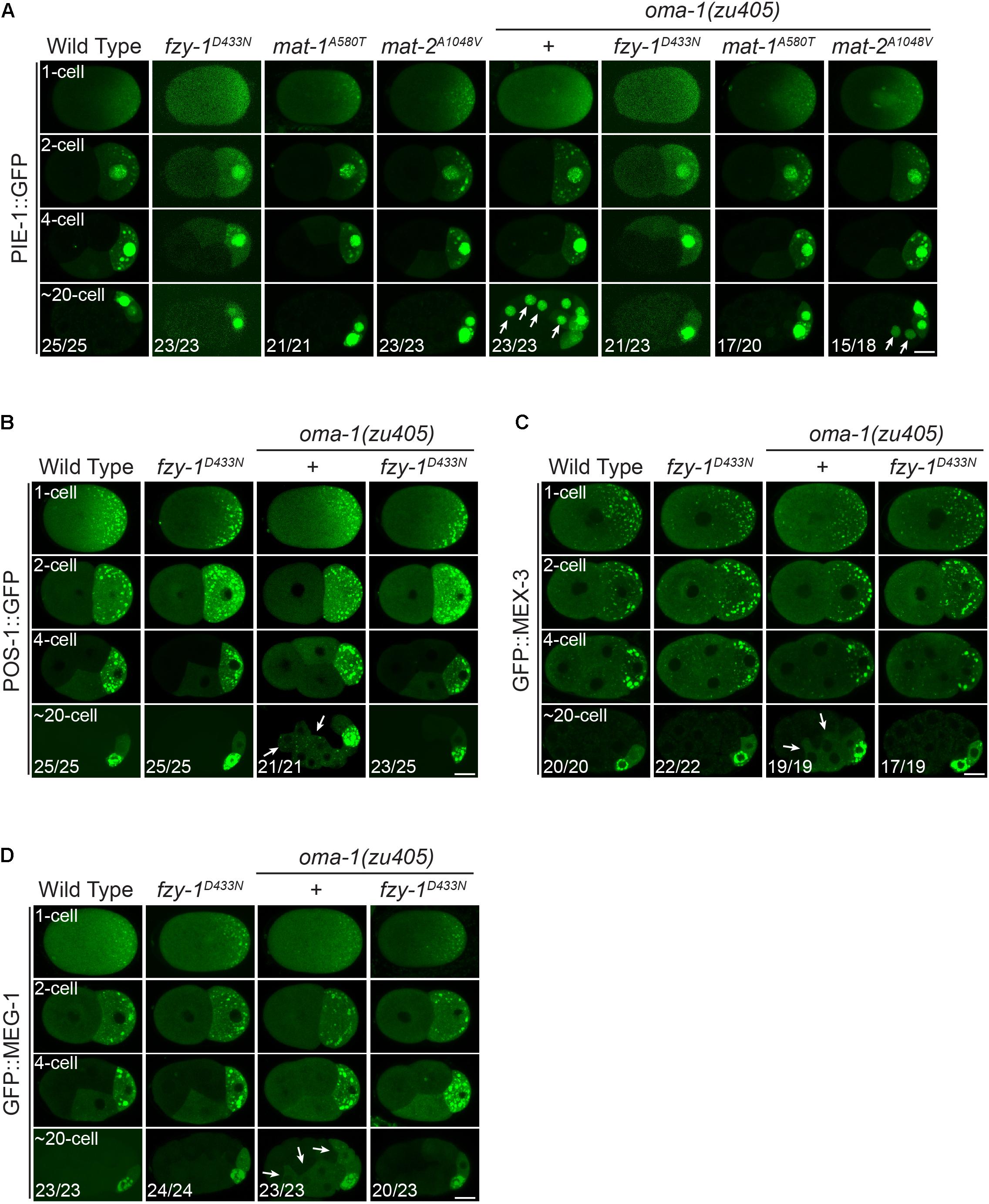
Figure 6. Mutants of APC/C genes correct the disrupted distribution of downstream cell fate determinants of OMA-1 in the oma-1(zu405) mutant. (A) Fluorescent micrographs show the localization of PIE-1::GFP in 1-cell, 2-cell, 4-cell, and ∼20-cell stage embryos in the wild type, fzy-1D433N, mat-1A580T, and mat-2A1048V mutants, with or without oma-1(zu405) in the background. Arrows indicate abnormal localization of PIE-1::GFP in somatic cells. (B) Fluorescent micrographs show the localization of POS-1::GFP in 1-cell, 2-cell, 4-cell, and ∼20-cell stage embryos in the wild type and fzy-1D433N mutants, with or without oma-1(zu405) in the background. Arrows indicate abnormal localization of POS-1::GFP in somatic cells. (C) Fluorescent micrographs show the localization of GFP::MEX-3 in 1-cell, 2-cell, 4-cell, and ∼20-cell stage embryos in the wild type and fzy-1D433N mutants, with or without oma-1(zu405) in the background. Arrows indicate abnormal localization of GFP::MEX-3 in somatic cells. (D) Fluorescent micrographs show the localization of GFP::MEG-1 in 1-cell, 2-cell, 4-cell, and ∼20-cell stage embryos in the wild type and fzy-1D433N mutants, with or without oma-1(zu405) in the background. Arrows indicate abnormal localization of GFP::MEG-1 in somatic cells. All phenotypes in 1-, 2-, and 4-cell embryos were 100% penetrant. Scale bar: 10 microns.
Similar to PIE-1, POS-1 is another CCCH zinc finger protein and cell fate determinant for germ cells, whose proper localization is disrupted in the oma-1(zu405) mutant (Lin, 2003; Farley et al., 2008). As with PIE-1::GFP, POS-1::GFP asymmetrically accumulates in granules at the posterior pole of 1-cell embryos, and eventually accumulates at P4 in ∼20-cell embryos after several rounds of asymmetric cell division (Figure 6B). In the oma-1(zu405) mutant, POS-1::GFP was still detectable in somatic cells rather than only in P4 in ∼20-cell embryos. Introduction of fzy-1D433N in oma-1(zu405) eliminated the remaining POS-1::GFP in somatic cells, recovering the defective distribution of POS-1::GFP to the wild type (Figure 6B).
We also investigated two other cell fate determinants; i.e., GFP::MEX-3 and GFP::MEG-1, which are essential for germ cell development. In the wild type, GFP::MEX-3 and GFP::MEG-1 only accumulated at P4 in ∼20-cell embryos after several rounds of asymmetric cell division. In the oma-1(zu405) mutant, GFP::MEX-3 and GFP::MEG-1 remained in the somatic cells, and most were cytoplasmic and a small portion accumulated at granules (Figures 6C,D). fzy-1D433N could fully remove the abnormal somatic remaining GFP::MEX-3 and GFP::MEG-1 in the oma-1(zu405) mutant. In the fzy-1D433N;oma-1(zu405) double mutant, localization of GFP::MEX-3 and GFP::MEG-1 behaved as in the wild type animals (Figures 6C,D).
All these results together indicated that disrupting APC/C functions could rescue delayed degradation of downstream cell fate determinants of OMA-1 caused by abnormal prolonged accumulation of OMA-1 protein during embryogenesis in the oma-1(zu405) mutant. This further suggested APC/C may regulate the binding ability of OMA-1 to RNAs or to proteins so that even OMA-1 is not degraded properly in the oma-1(zu405) mutant; it loses the ability to repress transcription or translation in early embryos. However, we cannot completely rule out the other possibility that APC/C indirectly regulates pathways that could bypass functions of OMA-1 to promote proper embryogenesis.
Discussion
We report here that from a forward genetic suppressor screen for embryonic lethality of the oma-1(zu405) strain, mutations in five APC/C genes, represented by 11 alleles, were isolated (Figure 2A). We expanded our analysis to all APC/C subunits and found that subunits in all subcomplexes of APC/C participate in OMA-1 related oocyte maturation and the oocyte-to-embryo transition, including EMB-30/Apc4 and SUCH-1/Apc5 in the platform, MAT-1/Apc3, and CDC-26/Cdc26 in the TPR lobe, APC-2/Apc2 in the catalytic core, and activator FZY-1/Cdc20. Because depletion of APC/C functions usually cause lethality, we cannot rule out the role of EMB-27/Apc6, MAT-3/Apc8, and APC-11/Apc11 in these developmental processes (Figures 2C,D). One exception is that although depleting APC-10/Apc10 and FZR-1/Cdh1 did not cause lethality, oma-1(zu405) was not suppressed under these treatments. This indicated only the activator FZY-1/Cdc20, but not FZR-1/Cdh1, is incorporated in the APC/C complex to regulate early embryogenesis. Subunits from all subcomplexes participating suppression of oma-1(zu405) lethality indicates that the intact APC/C complex, but not any particular subunit or subcomplex, is needed to regulate oocyte maturation and the oocyte-to-embryo transition.
Many known alleles of APC/C genes are from screens for phenotypes of either abnormal embryogenesis (emb genes) or metaphase-to-anaphase transition defect (mat genes). Therefore, these alleles often cause lethality of animals. It should be noted the alleles identified in this study are viable and healthy, indicating they are hypomorph alleles of APC/C genes. Notably, nearly all mutations identified by our screens are located in protein-protein interaction domains of various subunits in APC/C complex. For example, the mutation of the platform subunit EMB-30, N58S, is in the WD40 domain; mutations of the TRP lob subunit MAT-1, R496C, A580T, and R655T, are all in the TRP repeats; mutations of another TRP lob subunit MAT-3, R452T, and A453V, are both in the TRP repeats; mutations of the co-activator subunit FZY-1, T371K, D433N, and S465F, are all in the WD40 domain. This provides a possible explanation that why those alleles are hypomorph. Mutations in theses alleles just affected amino acid residues within protein-protein interaction domains, which may change the overall stability of the APC/C complex and cause perturbation, but not complete depletion, of APC/C functions. APC-2 and APC-11 constitute the core catalytic module, and we did not identify any viable alleles of apc-2 and apc-11, indicating that either any mutations in APC-2 or APC-11 would greatly affect functions of the APC/C complex, or our screen is not saturated. The hypomorph alleles are valuable reagents to study functions and molecular mechanisms of the APC/C complex, especially during the developmental stages of early embryogenesis.
RNA interference (RNAi) treatment of mat-1, fzy-1, or apc-2 caused complete lethality in wild type animals, but not in the oma-1(zu405) strain, which suggests that, not only depleting the functions of these APC/C genes suppresses lethality of oma-1(zu405), but oma-1(zu405) also suppresses lethality of mutations in these APC/C genes (Figures 2C,D). Another example is that the mutant with the hypomorph allele fzy-1D433N has a brood size of around 10, but fzy-1D433N;oma-1(zu405) has a brood size of more than 90 (Figures 2E,F). Since oma-1(zu405) is a gain-of-function allele, these mutual suppression effects may arise from accordant influences on oocyte maturation and the oocyte-to-embryo transition by the APC/C complex and OMA-1. Whether the APC/C complex regulates early embryogenesis through the OMA-1 related pathway needs more investigation, but our results provided some evidence to suggest that the APC/C complex may impair the RNA binding ability of OMA-1 (see below).
Our data showed that OMA-1 and OMA-1P240L were less localized to P granules after introducing mutations in APC/C genes (Figures 4, 5). Previous studies suggested during early embryogenesis, untranslated maternal mRNAs are enriched in P granules and are released from P granules at specific developmental stages to exhibit corresponding functions (Spike et al., 2014; Lee et al., 2020; Parker et al., 2020; Aoki et al., 2021). Since we and others also found OMA-1 is enriched in P granules in 1-, 2-, and 4-cell stage embryos, considering OMA-1 binds to 3′UTR to repress translation of maternal mRNAs, it is reasonable to suggest that OMA-1 and its cognate mRNAs are associated with each other in P granules, and delocalization of OMA-1 from P granules indicates disassociation of OMA-1 from its cognate mRNAs (Figure 7). Our RIP-seq data also support this model. OMA-1 and OMA-1P240L associate with different groups of mRNAs, and depletion of APC/C subunits turned mRNAs associated with OMA-1P240L to be more similar to mRNAs associated with OMA-1. Due to technical difficulties, whole worms, rather than embryos, were used in this study to perform RIP-seq. Further validation of this model may need direct investigation of OMA-1-RNA bindings in defined developmental stage (i.e., 1-, 2-, and 4-cell embryos) during embryogenesis.
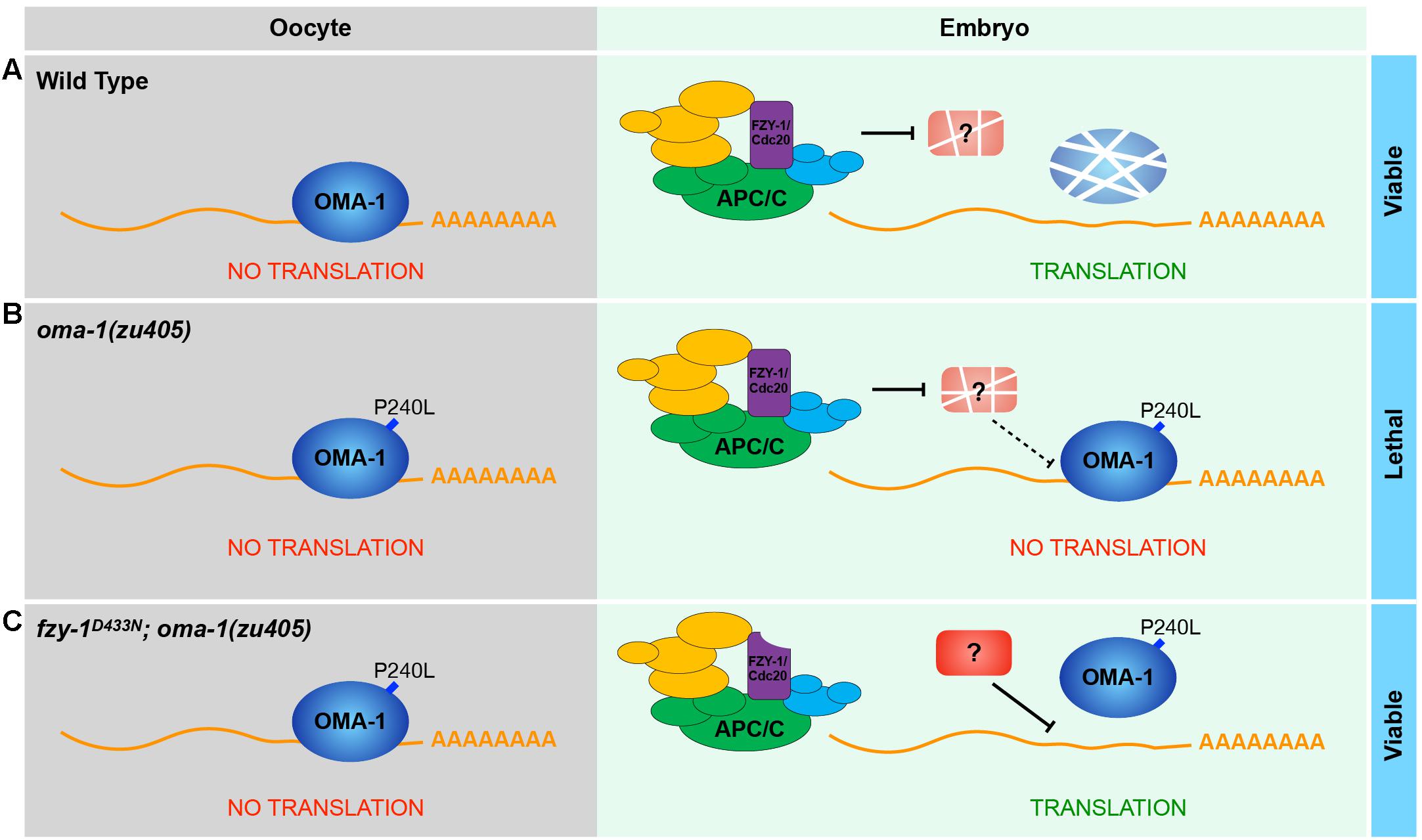
Figure 7. A proposed model to show how APC/C complex and OMA-1 are coordinated to regulate the oocyte-to-embryo transition. (A) In the wild type, OMA-1 binds to 3′UTR of its cognate mRNA to repress translation and undergoes rapid turnover in the oocyte-to-embryo transition. Maternal mRNAs can be translated normally in early embryos. (B) In the oma-1(zu405) mutant, the gain-of-function mutation P240L stabilizes OMA-1, causing OMA-1 continuously binds to its targets and repress their translation in early embryos, which eventually leads to embryonic lethality. (C) When the function of APC/C is perturbed (fzy-1D433N as an example), its substrates are not degraded. In this situation, P240L mutation still stabilizes OMA-1, however, the association between OMA-1P240L and mRNA is weakened, which is probably regulated a potential APC/C substrate.
Because the APC/C complex is a E3 ubiquitin ligase that participates in proteasome mediated proteolysis, one key plan is to identify direct targets of the APC/C complex during developmental processes of oocyte maturation and the oocyte-to-embryo transition. Only the APC/CCdc20 complex is involved in these processes, and the well-known target of the APC/CCdc20 complex is securin. Degradation of securin leads to anaphase onset. If securin is the direct substrate of the APC/CCdc20 complex, RNAi treatment of the fzy-1D433N;oma-1(zu405) strain with ify-1, the gene encoding securin homolog in C. elegans (Kitagawa et al., 2002), could switch phenotypes from the fzy-1D433N;oma-1(zu405) double mutant to the oma-1(zu405) single mutant. However, because RNAi of ify-1 itself leads to embryos arresting at the one-cell stage [(Kitagawa et al., 2002) and data not shown], it is impossible to test whether securin is the direct substrate of the APC/CCdc20 complex during early embryogenesis. It is possible that OMA-1 has a binding partner that prevents mRNA binding of OMA-1 and is the direct substrate of the APC/CCdc20 complex. We performed OMA-1 immunoprecipitation followed by mass spectrometry. Again, depletion of nearly all potential OMA-1 binding partners caused lethality of animals, which excluded the possibility to direct test whether OMA-1 binding partners can serve as targets of the APC/CCdc20 complex and their roles on OMA-1-RNA binding. Systematic analysis of binding partners and substrates of the APC/CCdc20 complex may be helpful to identify which proteins are direct targets of the APC/CCdc20 complex during OMA-1 regulated oocyte maturation and the oocyte-to-embryo transition.
Materials and Methods
Caenorhabditis elegans Strains
Animals were grown on standard nematode growth media (NGM) plates seeded with the Escherichia coli OP50 strain at 16, 20, or 25°C where indicated. Some of the C. elegans strains used in this study were purchased from the Caenorhabditis Genetics Center (CGC). Other strains were obtained through genetic screens or generated by the CRISPR/Cas9 gene editing system in this study. The strains that were used are listed in Supplementary Table 1.
RNA Interference Treatment
RNA interference experiments were performed as described previously (Timmons and Fire, 1998). Monoclonal bacteria of HT115 (DE3) were inoculated and incubated overnight in LB medium supplemented with 100 μg/mL ampicillin at 37°C. Fresh cultures were seeded on NGM plates supplemented with 100 μg/mL ampicillin and 1 mM IPTG, and incubated at room temperature for 24 h before use. RNAi feeding bacterial strains were obtained from the C. elegans RNAi Collection (Ahringer; Source BioScience). L1–L3 stage animals were fed RNAi feed bacteria expressing corresponding double-stranded RNA at the normal culture temperature for 24 h, then shifted to 25°C for phenotype scoring. HT115 (DE3) expressing empty RNAi vector L4440 was used as the control.
Forward Genetic Suppressor Screen
Forward genetic screens were performed as described (Brenner, 1974). EMS and ENU were combined to serve as mutagens in forward genetic screens (final concentration: 47 mM of EMS and 0.98 mM of ENU). Generally, oma-1(zu405) animals were treated with mutagens and mutants that could survive at 25°C were isolated. Synchronized L4 animals of the oma-1(zu405) strain were collected and incubated with EMS/ENU mutagens for 4 h at 16°C on a head-to-toe rotator. Animals were recovered and washed with M9 buffer (22 mM KH2PO4, 42 mM Na2HPO4, and 86 mM NaCl) five times and cultured at 16°C to adults. Animals were then bleached and synchronized F1s were collected. F1 progeny were cultured at 16°C with a density of around 300 animals per plate. After laying eggs, F1s were washed away with M9 buffer, and only F2 eggs were retained on plates. F2 was transferred to 25°C after reaching the L1 or L2 stage for further culture. Plates with significant amounts of F3 and F4 progenies were retained and mutants from those plates were isolated.
Mapping and Cloning Candidate Genes Identified From the Suppressor Screen
We first sequenced the oma-1 locus to exclude any mutation in oma-1 coding sequences in each mutant strain. Mutants were crossed with the Hawaiian strain CB4856 and the Snip-SNP strategy was used to determine the genetic location of candidate genes, as described previously (Minevich et al., 2012). Next, genomic DNA was extracted from the mutants and oma-1(zu405) strain, and WGS was performed. WGS data from mutants and oma-1(zu405) were compared using homemade scripts. Combining the methods described above, candidate genes were determined. Reconstruction of candidate mutations were generated by the CRISPR/Cas9 gene editing system to eventually confirm the candidate genes.
Brood Size Analysis
Single hermaphrodite L4s (P0s) were placed onto individual freshly seeded NGM plates and allowed to grow for 24 h at 25°C. P0 adults were transferred to new NGM plates every 24 h until they no longer laid eggs. All the F1 progeny on each plate were counted. The brood size of each P0 animal was the total sum of F1s for all plates where the P0 animal laid eggs. 10–15 P0 animals were used to calculated the brood size for each strain. Data were analyzed by 1-way ANOVA followed by Dunnett’s multiple comparisons.
CRISPR/Cas9 Gene Editing
Single guide RNAs (sgRNAs) were designed with the online tool CHOPCHOP.1 sgRNA sequences used in this study are listed in Supplementary Table 2. Two strategies were used to obtain sgRNA. (1) The DNA sequences encoding sgRNAs were cloned into pDD162 by overlapping polymerase chain reaction (PCR) using appropriate primers and pDD162 as the PCR template (Dickinson et al., 2013). Overlapping PCR products were inserted into pDD162 linearized with SpeI/BsrBI digestion with a ClonExpress Ultra One Step Cloning Kit (Vazyme #C115-01). (2) sgRNAs were synthesized and purified in vitro with a HiScribe Quick T7 High Yield RNA Synthesis Kit (New England Biolabs #E2050).
To generate the donor construct for gfp knock-in into the oma-1 locus, 500 bp upstream and downstream DNA of the oma-1 3′UTR and GFP coding sequences were amplified by PCR using N2 genomic DNA or plasmids containing GFP coding sequences as templates. Overlapping PCR fragments were linearized with HindIII/KpnI digestion with a ClonExpress Ultra One Step Cloning Kit (Vazyme #C115-01) and inserted into pUC19. Single-stranded oligonucleotides (ssODN) with 30 bp upstream and 30 bp downstream homology sequences and desired mutations were used as donors to introduce point mutations.
CRISPR experiments were conducted with co-CRISPR or Cas9 RNP strategies (Dokshin et al., 2018). For the co-CRISPR strategy, DNA mixtures were introduced into the germline of C. elegans young adults by microinjection. Final concentrations of plasmids in the injection mixtures were as follows: 50 ng/mL of pCCM935 unc-22 sgRNA, 50 ng/mL of pDD162 Cas9 + sgRNA, 30 ng/mL of pRF4 rol-6 (dm) and 50 ng/mL of single-stranded DNA (ssDNA) donor oligo (or 50 ng/mL of plasmid-based donor). F1 twitchers or rollers were isolated, followed by genotyping of the desired mutations. When applicable, transgenic strains were outcrossed to remove unc-22 mutations. For the RNP strategy, final concentrations of injection components were as follows: 250 ng/mL of Alt-R Cas9 protein (IDT), 100 ng/μL of Alt-R tracrRNA (IDT), and 56 ng/μL of customized Alt-R crRNA (IDT) or 200 ng/μL of customized Alt-R sgRNA (IDT). All components were mixed and incubated at 37°C for 10 min, then 110 ng/μL of ssDNA donor oligo or 200 ng/μL of dsDNA donor and 40 ng/μL of pRF4 rol-6 (dm) were added. Injection mixtures were introduced into the germline of C. elegans young adults by microinjection. The F1 rollers were isolated and genotyped to identify the desired mutations.
Fluorescence Microscopy
GFP- and mRuby-tagged fluorescent proteins were visualized in living nematodes or dissected embryos by mounting young adult animals on 2% agarose pads with M9 buffer (22 mM KH2PO4, 42 mM Na2HPO4, and 86 mM NaCl) with 10–50 mM levamisole, or mounting one-cell embryos on 2% agarose pads by dissecting gravid hermaphrodites into egg salt buffer (5 mM HEPES pH = 7.4, 118 mM NaCl, 40 mM KCl, 3.4 mM MgCl2, and CaCl2 3.4 mM). Fluorescent images were captured using a Zeiss LSM800 confocal microscope with a Plan-Apochromat 63X/1.4 Oil DIC M27 objective.
The quantification of fluorescent puncta (i.e., OMA-1::GFP and OMA-1P240L::GFP) was performed using ImageJ. Maximum intensity projections of z-series were obtained. Image thresholds were set manually and fluorescent puncta were selected. Integrated intensity of all puncta in a single embryo was measured and summed together. Images of 8–12 embryos were collected and quantified. Data were analyzed by 2-way ANOVA followed by Dunnett’s multiple comparisons.
The quantification of overall fluorescence of OMA-1::GFP and OMA-1P240L::GFP in embryos or germlines was performed using ImageJ. Maximum intensity projections of z-series were obtained. Region of interest (ROIs) was selected and integrated intensity of all pixels within an ROI was measured. Images of 8–12 embryos or germlines were collected and quantified. For quantification of OMA-1::GFP and OMA-1P240L::GFP in embryos, data were analyzed by 2-way ANOVA followed by Dunnett’s multiple comparisons. For quantification of OMA-1::GFP in germlines, data were analyzed by student’s t-test.
For quantitative co-localization analysis, all image manipulations were performed with ImageJ using the Coloc 2 plugin. PCCs were calculated. Data were analyzed by 1-way ANOVA followed by Dunnett’s multiple comparisons. N = 8 for each strains.
Transcriptome Analysis by mRNA-seq
Synchronized L1 animals were cultured at 25°C until they grew into early young adults, which were then collected and washed with M9 buffer (22 mM KH2PO4, 42 mM Na2HPO4, and 86 mM NaCl) several times. Early embryos were obtained by bleaching and washing with M9 buffer five times and ice water for the last time. Total RNA was extracted using the standard method with TRIzol reagent (Invitrogen), and an mRNA library was generated for high throughput sequencing using the VAHTS Universal V8 RNA-seq Library Prep Kit for Illumina (Vazyme #NR605-01). Sequencing reads were generated with an Illumina NovaSeq 6000 system. All trimmed reads were aligned to the reference genome using HISAT2 (Kim et al., 2019). The read counts mapped to each transcript were calculated using HTSeq (Anders et al., 2015), and then normalized to reads per kilobases per million reads (RPKM). The results were used for the analysis of differentially expressed genes. All differentially expressed genes were identified using DESeq2 (Love et al., 2014) with a cutoff of | log2(fold change)| > 2 and Benjamini-Hochberg adjusted p-value < 0.05. k-means clustering analysis of differentially expressed genes was performed using iDEP 0.932 (Ge et al., 2018). GO enrichment analysis of differentially expressed genes was performed with the GO Enrichment Analysis tool at https://www.omicshare.com/tools/.
RNA Immunoprecipitation Sequencing
A total of 100,000 synchronized young adult animals were frozen in liquid nitrogen and stored at −80°C. Pellets were resuspended in equal volumes of immunoprecipitation buffer [20 mM Tris–HCl pH 7.5, 150 mM NaCl, 2.5 mM MgCl2, 0.5% NP-40, 80 U ml–1 RNase Inhibitor (Thermo), 1 mM dithiothreitol, and protease inhibitor cocktail without EDTA (Promega)], and grinded in a glass grinder for 8–10 times. The whole grinding process should not exceed 10 min. Lysates were clarified by spinning down at 15000 rpm, 4°C, for 15 min. Supernatants were incubated with the GFP-Trap magnetic agarose beads (ChromoTek) at 4°C for 1 h. Beads were washed with IP wash buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 2.5 mM MgCl2, 0.5% NP-40, and 1 mM dithiothreitol) six times, and then resuspended in TBS buffer for RNA extraction. Total RNA was extracted using the standard method with TRIzol reagent (Invitrogen). The mRNA library was generated, sequenced and analyzed as transcriptome analysis by mRNA-seq in this study described above to get RPKM values. RPKM values from IP samples and input samples were used for defining OMA-1-associated mRNAs by DESeq2 (Love et al., 2014) with a cutoff of | log2(fold change)| > 1 and Benjamini-Hochberg adjusted p-value < 0.05.
Quantitative Real-Time PCR
RNA was extracted from the early embryos as described above. cDNA was then synthesized using HiScript II Q Select RT SuperMix for qPCR with gDNA wiper (Vazyme #R233-01) according to the manufacturer’s instructions. Real-time PCR was performed on a CFX Connect Thermal Cycler (Bio-Rad) with ChamQ SYBR qPCR Master Mix (Vazyme # Q311-02). Amplification was performed with a two-step reaction at 95°C for 3 min and 40 cycles at 95°C for 15 s and at 60°C for 30 s. A 20 μL PCR mixture included 10 μL ChamQ SYBR qPCR Master Mix, 2 μL primer mix (10 μM each), 3 μL diluted template cDNA, and 5 μL deionized distilled water. The relative fold changes in related genes were normalized to expression levels of actin, and analyzed by 2-way ANOVA followed by Dunnett’s multiple comparisons. Each experiment was repeated four times. The PCR primers used in this study are listed in Supplementary Table 2.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181115.
Author Contributions
YH carried out the majority of experiments and data analysis. XH conducted the bioinformatics analysis. DL conducted some of the crosses and genotyping. YH, ZD, KS, and CH performed the genetic screens. YH and ZD cloned mutated genes. DZ designed the study and wrote the manuscript. YZ and DZ supervised the work. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grants 31771500 and 31922019) and the Academic Frontier Youth Team of Huazhong University of Science and Technology (grant 2018QYTD11) to DZ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Heng-Chi Lee and Shouhong Guang for providing strains and reagents. All confocal microscopy was carried out at the core facility of the department of forensic medicine at Huazhong University of Science and Technology. Some strains used in this study were provided by the Caenorhabditis Genetics Center (CGC). We thank members of the Zhang Laboratory for helpful discussions. We also thank Margaret Lutze for language editing on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.749654/full#supplementary-material
Supplementary Figure 1 | (A) Quantitative real-time PCR measurements of expression levels of indicated APC/C genes in RNAi treated strains. The results are presented as the average ± S.D. ∗∗∗p < 0.001; ns: not significant. The p-values were calculated by 2-way ANOVA followed by Dunnett’s multiple comparisons. (B) Fluorescent micrographs show OMA-1::GFP in germlines in control and oma-1 RNAi treated strains to demonstrate oma-1 RNAi efficiency. Quantification of overall OMA-1::GFP fluorescence in germlines is presented as the average ± S.D. with individual values plotted. ∗∗∗p < 0.001. The p-values were calculated by student’s t-test. N = 8–12. (C) Validation of mutations generated by CRISPR/Cas9 gene editing system by Sanger Sequencing. Wild type sequences are shown at the top and mutated sequences are shown at the bottom for each mutation. Red box, missense mutations causing desired amino acid residue change; blue box, silent mutations for genotyping. (D) Brood size of oma-1(ne3800) and oma-1:gfp strains in wild type and oma-2 RNAi background. The results are presented as the average ± S.D. with individual values plotted. ∗∗∗p < 0.001; ns: not significant. The p-values were calculated by 1-way ANOVA followed by Dunnett’s multiple comparisons. N = 10–15.
Footnotes
References
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Aoki, S. T., Lynch, T. R., Crittenden, S. L., Bingman, C. A., Wickens, M., and Kimble, J. (2021). C. elegans germ granules require both assembly and localized regulators for mRNA repression. Nat. Commun. 12:996. doi: 10.1038/s41467-021-21278-1
Blanco, M. A., Pelloquin, L., and Moreno, S. (2001). Fission yeast mfr1 activates APC and coordinates meiotic nuclear division with sporulation. J. Cell Sci. 114(Pt 11), 2135–2143.
Blanco, M. A., Sanchez-Diaz, A., de Prada, J. M., and Moreno, S. (2000). APC(ste9/srw1) promotes degradation of mitotic cyclins in G(1) and is inhibited by cdc2 phosphorylation. EMBO J. 19, 3945–3955. doi: 10.1093/emboj/19.15.3945
Chang, L. F., Zhang, Z., Yang, J., McLaughlin, S. H., and Barford, D. (2014). Molecular architecture and mechanism of the anaphase-promoting complex. Nature 513, 388–393. doi: 10.1038/nature13543
Dansereau, D. A., and Lasko, P. (2008). The development of germline stem cells in Drosophila. Methods Mol. Biol. 450, 3–26. doi: 10.1007/978-1-60327-214-8_1
Davis, E. S., Wille, L., Chestnut, B. A., Sadler, P. L., Shakes, D. C., and Golden, A. (2002). Multiple subunits of the Caenorhabditis elegans anaphase-promoting complex are required for chromosome segregation during meiosis I. Genetics 160, 805–813.
DeRenzo, C., Reese, K. J., and Seydoux, G. (2003). Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature 424, 685–689. doi: 10.1038/nature01887
Deshaies, R. J. (1999). SCF and cullin/ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. doi: 10.1146/annurev.cellbio.15.1.435
Detwiler, M. R., Reuben, M., Li, X., Rogers, E., and Lin, R. (2001). Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell 1, 187–199. doi: 10.1016/s1534-5807(01)00026-0
Dickinson, D. J., Ward, J. D., Reiner, D. J., and Goldstein, B. (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028–1034. doi: 10.1038/nmeth.2641
Dokshin, G. A., Ghanta, K. S., Piscopo, K. M., and Mello, C. C. (2018). Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics 210, 781–787. doi: 10.1534/genetics.118.301532
Dube, P., Herzog, F., Gieffers, C., Sander, B., Riedel, D., Muller, S. A., et al. (2005). Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Mol. Cell 20, 867–879. doi: 10.1016/j.molcel.2005.11.008
Edgar, B. A., and Schubiger, G. (1986). Parameters controlling transcriptional activation during early Drosophila development. Cell 44, 871–877. doi: 10.1016/0092-8674(86)90009-7
Fang, G., Yu, H., and Kirschner, M. W. (1998). Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell 2, 163–171. doi: 10.1016/s1097-2765(00)80126-4
Fang, G., Yu, H., and Kirschner, M. W. (1999). Control of mitotic transitions by the anaphase-promoting complex. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354, 1583–1590. doi: 10.1098/rstb.1999.0502
Farley, B. M., Pagano, J. M., and Ryder, S. P. (2008). RNA target specificity of the embryonic cell fate determinant POS-1. RNA 14, 2685–2697. doi: 10.1261/rna.1256708
Farley, B. M., and Ryder, S. P. (2008). Regulation of maternal mRNAs in early development. Crit. Rev. Biochem. Mol. Biol. 43, 135–162. doi: 10.1080/10409230801921338
Fay, D. S., Keenan, S., and Han, M. (2002). fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 16, 503–517. doi: 10.1101/gad.952302
Forbes, A., and Lehmann, R. (1998). Nanos and pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125, 679–690.
Furuta, T., Tuck, S., Kirchner, J., Koch, B., Auty, R., Kitagawa, R., et al. (2000). EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol. Biol. Cell 11, 1401–1419. doi: 10.1091/mbc.11.4.1401
Garbe, D., Doto, J. B., and Sundaram, M. V. (2004). Caenorhabditis elegans lin-35/Rb, efl-1/E2F and other synthetic multivulva genes negatively regulate the anaphase-promoting complex gene mat-3/APC8. Genetics 167, 663–672. doi: 10.1534/genetics.103.026021
Ge, S. X., Son, E. W., and Yao, R. (2018). iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics 19:534. doi: 10.1186/s12859-018-2486-6
Golden, A., Sadler, P. L., Wallenfang, M. R., Schumacher, J. M., Hamill, D. R., Bates, G., et al. (2000). Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 151, 1469–1482. doi: 10.1083/jcb.151.7.1469
Guven-Ozkan, T., Nishi, Y., Robertson, S. M., and Lin, R. (2008). Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4. Cell 135, 149–160. doi: 10.1016/j.cell.2008.07.040
Guven-Ozkan, T., Robertson, S. M., Nishi, Y., and Lin, R. (2010). zif-1 translational repression defines a second, mutually exclusive OMA function in germline transcriptional repression. Development 137, 3373–3382. doi: 10.1242/dev.055327
Harper, J. W., Burton, J. L., and Solomon, M. J. (2002). The anaphase-promoting complex: it’s not just for mitosis any more. Genes Dev. 16, 2179–2206. doi: 10.1101/gad.1013102
Herzog, F., Primorac, I., Dube, P., Lenart, P., Sander, B., Mechtler, K., et al. (2009). Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science 323, 1477–1481. doi: 10.1126/science.1163300
Jadhav, S., Rana, M., and Subramaniam, K. (2008). Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development 135, 1803–1812. doi: 10.1242/dev.013656
Kawasaki, I., Shim, Y. H., Kirchner, J., Kaminker, J., Wood, W. B., and Strome, S. (1998). PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94, 635–645. doi: 10.1016/s0092-8674(00)81605-0
Kaymak, E., and Ryder, S. P. (2013). RNA recognition by the Caenorhabditis elegans oocyte maturation determinant OMA-1. J. Biol. Chem. 288, 30463–30472. doi: 10.1074/jbc.M113.496547
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kitagawa, R., Law, E., Tang, L., and Rose, A. M. (2002). The Cdc20 homolog, FZY-1, and its interacting protein, IFY-1, are required for proper chromosome segregation in Caenorhabditis elegans. Curr. Biol. 12, 2118–2123. doi: 10.1016/s0960-9822(02)01392-1
Lara-Gonzalez, P., Kim, T., and Desai, A. (2017). Taming the beast: control of APC/C(Cdc20)-dependent destruction. Cold Spring Harb. Symp. Quant. Biol. 82, 111–121. doi: 10.1101/sqb.2017.82.033712
Lee, C. S., Putnam, A., Lu, T., He, S., Ouyang, J. P. T., and Seydoux, G. (2020). Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. Elife 9:e52896. doi: 10.7554/eLife.52896
Lee, M. H., and Schedl, T. (2006). RNA-binding proteins. WormBook 18, 1–13. doi: 10.1895/wormbook.1.79.1
Lin, R. (2003). A gain-of-function mutation in oma-1, a C. elegans gene required for oocyte maturation, results in delayed degradation of maternal proteins and embryonic lethality. Dev. Biol. 258, 226–239. doi: 10.1016/s0012-1606(03)00119-2
Listovsky, T., Zor, A., Laronne, A., and Brandeis, M. (2000). Cdk1 is essential for mammalian cyclosome/APC regulation. Exp. Cell Res. 255, 184–191. doi: 10.1006/excr.1999.4788
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Marangos, P., Verschuren, E. W., Chen, R., Jackson, P. K., and Carroll, J. (2007). Prophase I arrest and progression to metaphase I in mouse oocytes are controlled by Emi1-dependent regulation of APC(Cdh1). J. Cell Biol. 176, 65–75. doi: 10.1083/jcb.200607070
Matova, N., and Cooley, L. (2001). Comparative aspects of animal oogenesis. Dev. Biol. 231, 291–320. doi: 10.1006/dbio.2000.0120
Minevich, G., Park, D. S., Blankenberg, D., Poole, R. J., and Hobert, O. (2012). CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics 192, 1249–1269. doi: 10.1534/genetics.112.144204
Moore, M. J. (2005). From birth to death: the complex lives of eukaryotic mRNAs. Science 309, 1514–1518. doi: 10.1126/science.1111443
Newport, J., and Kirschner, M. (1982). A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell 30, 675–686. doi: 10.1016/0092-8674(82)90272-0
Noble, S. L., Allen, B. L., Goh, L. K., Nordick, K., and Evans, T. C. (2008). Maternal mRNAs are regulated by diverse P body-related mRNP granules during early Caenorhabditis elegans development. J. Cell Biol. 182, 559–572. doi: 10.1083/jcb.200802128
Oh, D., and Houston, D. W. (2017). RNA localization in the vertebrate oocyte: establishment of oocyte polarity and localized mRNA assemblages. Results Probl. Cell Differ. 63, 189–208. doi: 10.1007/978-3-319-60855-6_9
Oldenbroek, M., Robertson, S. M., Guven-Ozkan, T., Spike, C., Greenstein, D., and Lin, R. (2013). Regulation of maternal Wnt mRNA translation in C. elegans embryos. Development 140, 4614–4623. doi: 10.1242/dev.096313
Parker, D. M., Winkenbach, L. P., Boyson, S., Saxton, M. N., Daidone, C., Al-Mazaydeh, Z. A., et al. (2020). mRNA localization is linked to translation regulation in the Caenorhabditis elegans germ lineage. Development 147:dev186817. doi: 10.1242/dev.186817
Passmore, L. A. (2004). The anaphase-promoting complex (APC): the sum of its parts? Biochem. Soc. Trans. 32(Pt 5), 724–727. doi: 10.1042/BST0320724
Passmore, L. A., Booth, C. R., Venien-Bryan, C., Ludtke, S. J., Fioretto, C., Johnson, L. N., et al. (2005). Structural analysis of the anaphase-promoting complex reveals multiple active sites and insights into polyubiquitylation. Mol. Cell 20, 855–866. doi: 10.1016/j.molcel.2005.11.003
Pesin, J. A., and Orr-Weaver, T. L. (2007). Developmental role and regulation of cortex, a meiosis-specific anaphase-promoting complex/cyclosome activator. PLoS Genet. 3:e202. doi: 10.1371/journal.pgen.0030202
Peters, J. M. (2002). The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell 9, 931–943. doi: 10.1016/s1097-2765(02)00540-3
Peters, J. M. (2006). The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7, 644–656. doi: 10.1038/nrm1988
Pines, J. (2006). Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol. 16, 55–63. doi: 10.1016/j.tcb.2005.11.006
Pines, J. (2011). Cubism and the cell cycle: the many faces of the APC/C. Nat. Rev. Mol. Cell Biol. 12, 427–438. doi: 10.1038/nrm3132
Pintard, L., Willems, A., and Peter, M. (2004). Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 23, 1681–1687. doi: 10.1038/sj.emboj.7600186
Powell-Coffman, J. A., Knight, J., and Wood, W. B. (1996). Onset of C. elegans gastrulation is blocked by inhibition of embryonic transcription with an RNA polymerase antisense RNA. Dev. Biol 178, 472–483. doi: 10.1006/dbio.1996.0232
Primorac, I., and Musacchio, A. (2013). Panta rhei: the APC/C at steady state. J. Cell Biol. 201, 177–189. doi: 10.1083/jcb.201301130
Radford, H. E., Meijer, H. A., and de Moor, C. H. (2008). Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim. Biophys. Acta 1779, 217–229. doi: 10.1016/j.bbagrm.2008.02.002
Rappleye, C. A., Tagawa, A., Lyczak, R., Bowerman, B., and Aroian, R. V. (2002). The anaphase-promoting complex and separin are required for embryonic anterior-posterior axis formation. Dev. Cell 2, 195–206. doi: 10.1016/s1534-5807(02)00114-4
Robertson, S., and Lin, R. (2015). The maternal-to-zygotic transition in C. elegans. Curr. Top. Dev. Biol. 113, 1–42. doi: 10.1016/bs.ctdb.2015.06.001
Rosario, R., Childs, A. J., and Anderson, R. A. (2017). RNA-binding proteins in human oogenesis: balancing differentiation and self-renewal in the female fetal germline. Stem Cell Res. 21, 193–201. doi: 10.1016/j.scr.2017.04.008
Rose, L., and Gonczy, P. (2014). Polarity establishment, asymmetric division and segregation of fate determinants in early C. elegans embryos. WormBook 10, 1–43. doi: 10.1895/wormbook.1.30.2
Satyanarayana, A., and Kaldis, P. (2009). Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 28, 2925–2939. doi: 10.1038/onc.2009.170
Schier, A. F. (2007). The maternal-zygotic transition: death and birth of RNAs. Science 316, 406–407. doi: 10.1126/science.1140693
Shakes, D. C., Sadler, P. L., Schumacher, J. M., Abdolrasulnia, M., and Golden, A. (2003). Developmental defects observed in hypomorphic anaphase-promoting complex mutants are linked to cell cycle abnormalities. Development 130, 1605–1620. doi: 10.1242/dev.00385
Shimada, M., Yokosawa, H., and Kawahara, H. (2006). OMA-1 is a P granules-associated protein that is required for germline specification in Caenorhabditis elegans embryos. Genes Cells 11, 383–396. doi: 10.1111/j.1365-2443.2006.00945.x
Shirayama, M., Soto, M. C., Ishidate, T., Kim, S., Nakamura, K., Bei, Y., et al. (2006). The conserved kinases CDK-1, GSK-3, KIN-19, and MBK-2 promote OMA-1 destruction to regulate the oocyte-to-embryo transition in C. elegans. Curr. Biol. 16, 47–55. doi: 10.1016/j.cub.2005.11.070
Spike, C. A., Coetzee, D., Nishi, Y., Guven-Ozkan, T., Oldenbroek, M., Yamamoto, I., et al. (2014). Translational control of the oogenic program by components of OMA ribonucleoprotein particles in Caenorhabditis elegans. Genetics 198, 1513–1533. doi: 10.1534/genetics.114.168823
Swan, A., and Schupbach, T. (2007). The Cdc20 (Fzy)/Cdh1-related protein, Cort, cooperates with Fzy in cyclin destruction and anaphase progression in meiosis I and II in Drosophila. Development 134, 891–899. doi: 10.1242/dev.02784
Tenenhaus, C., Schubert, C., and Seydoux, G. (1998). Genetic requirements for PIE-1 localization and inhibition of gene expression in the embryonic germ lineage of Caenorhabditis elegans. Dev. Biol. 200, 212–224. doi: 10.1006/dbio.1998.8940
Tenenhaus, C., Subramaniam, K., Dunn, M. A., and Seydoux, G. (2001). PIE-1 is a bifunctional protein that regulates maternal and zygotic gene expression in the embryonic germ line of Caenorhabditis elegans. Genes Dev. 15, 1031–1040. doi: 10.1101/gad.876201
Timmons, L., and Fire, A. (1998). Specific interference by ingested dsRNA. Nature 395:854. doi: 10.1038/27579
Tsukamoto, T., Gearhart, M. D., Spike, C. A., Huelgas-Morales, G., Mews, M., Boag, P. R., et al. (2017). LIN-41 and OMA ribonucleoprotein complexes mediate a translational repression-to-activation switch controlling oocyte meiotic maturation and the oocyte-to-embryo transition in Caenorhabditis elegans. Genetics 206, 2007–2039. doi: 10.1534/genetics.117.203174
Uzunova, K., Dye, B. T., Schutz, H., Ladurner, R., Petzold, G., Toyoda, Y., et al. (2012). APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nat. Struct. Mol. Biol. 19, 1116–1123. doi: 10.1038/nsmb.2412
Verlhac, M. H., Terret, M. E., and Pintard, L. (2010). Control of the oocyte-to-embryo transition by the ubiquitin-proteolytic system in mouse and C. elegans. Curr. Opin. Cell Biol. 22, 758–763. doi: 10.1016/j.ceb.2010.09.003
Wang, J. T., and Seydoux, G. (2013). Germ cell specification. Adv. Exp. Med. Biol. 757, 17–39. doi: 10.1007/978-1-4614-4015-4_2
Whitfield, Z. J., Chisholm, J., Hawley, R. S., and Orr-Weaver, T. L. (2013). A meiosis-specific form of the APC/C promotes the oocyte-to-embryo transition by decreasing levels of the Polo kinase inhibitor matrimony. PLoS Biol. 11:e1001648. doi: 10.1371/journal.pbio.1001648
Yang, H., Vallandingham, J., Shiu, P., Li, H., Hunter, C. P., and Mak, H. Y. (2014). The DEAD box helicase RDE-12 promotes amplification of RNAi in cytoplasmic foci in C. elegans. Curr. Biol. 24, 832–838. doi: 10.1016/j.cub.2014.01.008
Keywords: OMA-1, APC/C, ocyte-to-embryo transition, RNA binding protein, C. elegans
Citation: Hu Y, Hu X, Li D, Du Z, Shi K, He C, Zhang Y and Zhang D (2021) The APC/CFZY–1/Cdc20 Complex Coordinates With OMA-1 to Regulate the Oocyte-to-Embryo Transition in Caenorhabditis elegans. Front. Cell Dev. Biol. 9:749654. doi: 10.3389/fcell.2021.749654
Received: 29 July 2021; Accepted: 27 September 2021;
Published: 15 October 2021.
Edited by:
Huijie Bian, Fourth Military Medical University, ChinaReviewed by:
Eyal Schejter, Weizmann Institute of Science, IsraelNatalia Bulgakova, The University of Sheffield, United Kingdom
Copyright © 2021 Hu, Hu, Li, Du, Shi, He, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donglei Zhang, emhhbmdfZG9uZ2xlaUBodXN0LmVkdS5jbg==; Ying Zhang, enpoYW5neWluZ0BodXN0LmVkdS5jbg==
 Yabing Hu
Yabing Hu Xuewen Hu
Xuewen Hu Donglei Zhang
Donglei Zhang