
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 13 October 2021
Sec. Molecular and Cellular Oncology
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.748925
Solute carrier family 7 member 11 (SLC7A11), glutathione peroxidase 4 (GPX4), and apoptosis inducing factor mitochondria associated 2 (AIFM2) are the key regulators in ferroptosis. However, the expression patterns and prognostic roles of these genes in pan-cancer are still largely unclear. The expression patterns and prognostic roles of SLC7A11, GPX4, and AIFM2 and the relationships between the expression levels of these genes and immune infiltration levels in pan-cancer were analyzed by using TIMER, gene expression profiling interactive analysis (GEPIA), Oncomine, and Kaplan–Meier databases. Our results showed that both SLC7A11 and GPX4 were overexpressed in colorectal cancer, and SLC7A11 was overexpressed in lung cancer. High levels of SLC7A11 and AIFM2 were significantly linked with the shortened disease-free survival and overall survival (OS) in adrenocortical carcinoma (ACC), respectively. And high expression of SLC7A11, GPX4, and AIFM2 were significantly correlated with the shortened OS of acute myeloid leukemia patients. In esophageal carcinoma (ESCA), GPX4 expression was significantly associated with the infiltration of macrophage and myeloid dendritic cell, and AIFM2 expression was significantly associated with the infiltration of CD4+ T cell. Importantly, GPX4 expression was positively correlated with the expression levels of monocyte markers (CD14 and CD115) and M2 macrophage markers (VSIG4 and MS4A4A) both in ESCA and in head and neck squamous cell carcinoma (HNSC). In summary, SLC7A11, GPX4, and AIFM2 are dysregulated in many types of cancers, and are candidate prognostic biomarkers for many types of cancers, and can be used to evaluate the infiltration of immune cells in tumor tissues.
Ferroptosis is a new form of regulated cell death induced by iron-dependent lipid peroxidation and with the characteristic of oxidative perturbations of the intracellular microenvironment (Chen et al., 2021). A recent study suggested that ferroptosis might be developed as a natural and promising therapy against cancer (Wang et al., 2021a). However, up to now, the expression pattern and prognostic value of key regulators of ferroptosis especially the correlation between the expression levels of these genes and immune infiltration in pan-cancer are largely unclear.
Solute carrier family 7 member 11 (SLC7A11), the catalytic subunit of a heterodimeric cystine/glutamate antiporter (System Xc–), transports extracellular cystine into cells for glutathione biosynthesis and is identified as a key regulator of ferroptosis. Knockout or knockdown of SLC7A11 markedly suppressed the growth, survival, and tumor formation of cancer cells by inducing ferroptosis (Daher et al., 2019; Lim et al., 2019; Wang et al., 2020). Overexpression of SLC7A11 was detected in KRAS-mutant lung adenocarcinoma (LUAD) and positively correlated with cancer progression. Genetic depletion or pharmacological inhibition with sulfasalazine or HG106 markedly induced the cell death of KRAS-mutant cancer cells in vitro and tumor growth suppression in vivo (Hu et al., 2020). Immunotherapy-activated CD8+ T cells secreted interferon gamma (IFNγ), and then downregulated SLC7A11 in tumor cells and finally promoted cancer cell lipid peroxidation and ferroptosis (Wang et al., 2019). Combination of radiotherapy and immunotherapy could enhance the lipid oxidation and ferroptosis of cancer cells by synergistically targeting and suppressing SLC7A11 (Lang et al., 2019).
Glutathione peroxidase 4 (GPX4), an antioxidant enzyme and a key regulator of ferroptosis, could catalyze and reduce lipid peroxides (Yang et al., 2014). Knockdown of GPX4 could induce the ferroptosis of oral cancer cells (Fukuda et al., 2021). Inhibiting GPX4 by resibufogenin (RB) induced ferroptotic cancer cell death and suppressed tumor growth in vivo (Shen et al., 2021). GPX4 was reported to be involved in the metformin, miR-324-3p, and loss of PTPN18 or SRSF9 induced ferroptosis (Deng et al., 2021; Hou et al., 2021; Wang et al., 2021b,c). Apoptosis inducing factor mitochondria associated 2 (AIFM2, also known as PRG3 or FSP1), a traditional apoptotic regulator, was identified as a new ferroptosis regulator and found to block the RSL3-, sorafenib-, and erastin-induced ferroptosis of cancer cells, and inhibition of AIFM2-dependent pathway enhanced the antitumor activity of sorafenib in mouse model (Bersuker et al., 2019; Doll et al., 2019; Dai et al., 2020).
In the present study, we comprehensively analyzed the mRNA levels of SLC7A11, GPX4, and AIFM2, and the correlations between the mRNA levels of these three genes and prognosis in pan-cancer using databases including Oncomine, gene expression profiling interactive analysis (GEPIA), TIMER, and Kaplan–Meier Plotter. We further investigated the relationships between the expression levels of SLC7A11, GPX4, and AIFM2 and immune infiltration levels in pan-cancer by using the TIMER database.
Kaplan--Meier Plotter1 was used to evaluate the prognostic roles of SLC7A11, GPX4, and AIFM2 in 21 different cancers using 7,462 cancer samples in overall survival (OS) analysis and 4,420 cancer samples in relapse-free survival (RFS) analysis (Nagy et al., 2021). The best cutoff was automatically selected by the database program.
TIMER database2 was used to analyze the expression patterns of SLC7A11, GPX4, and AIFM2, and the correlation between the expression levels of SLC7A11, GPX4, or AIFM2 and immune infiltration, and the correlation between the expression levels of SLC7A11, GPX4, or AIFM2 and markers of different subsets of immune cells [including CD8+ T cells, CD4+ T cells, B cells, tumor-associated macrophages (TAMs), monocytes, M1 macrophages, M2 macrophages, neutrophils, dendritic cells (DCs), natural killer (NK) cells, and several types of T cells including T helper 1 (Th1), Th2, follicular helper T (Tfh), Th17, regulatory T (Tregs), and exhausted T cells] in pan-cancer including more than 10,000 samples from The Cancer Genome Atlas (TCGA) (Li et al., 2016, 2017; Li T. et al., 2020).
The GEPIA database3 was used to evaluate the prognostic roles of SLC7A11, GPX4, and AIFM2 in pan-cancer including 9,736 tumors and 8,587 normal samples from the TCGA and the GTEx projects (Tang et al., 2017).
The mRNA expression levels of SLC7A11, GPX4, and AIFM2 in several cancers such as breast cancer, colorectal cancer (CRC), and so on were analyzed by using Oncomine database4 (Rhodes et al., 2007).
The expression patterns and prognostic roles of SLC7A11, GPX4, and AIFM2 were analyzed by using TIMER, GEPIA, Oncomine, and Kaplan–Meier databases. The correlation between the expression levels of SLC7A11, GPX4, or AIFM2 and immune infiltration, and the correlation between the expression levels of SLC7A11, GPX4, or AIFM2 and markers of different subsets of immune cells were analyzed by using TIMER database. p-Value < 0.05 was regarded as statistically significant.
We evaluated the mRNA expression levels of SLC7A11, GPX4, and AIFM2 in TCGA RNA-seq data by analyzing TIMER database. The results in Figure 1 showed that all of the three genes were significantly higher in esophageal carcinoma (ESCA, head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), liver hepatocellular carcinoma (LIHC), LUAD, stomach adenocarcinoma (STAD), and uterine corpus endometrial carcinoma (UCEC). Very importantly, both SLC7A11 and GPX4 were overexpressed in CRC including colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) and upregulated in pan-kidney cohort including kidney renal clear cell carcinoma (KIRC), KICH, and kidney renal papillary cell carcinoma (KIRP) comparing with adjacent normal tissues. We also found that SLC7A11 was overexpressed both in LUAD and lung squamous cell carcinoma (LUSC).
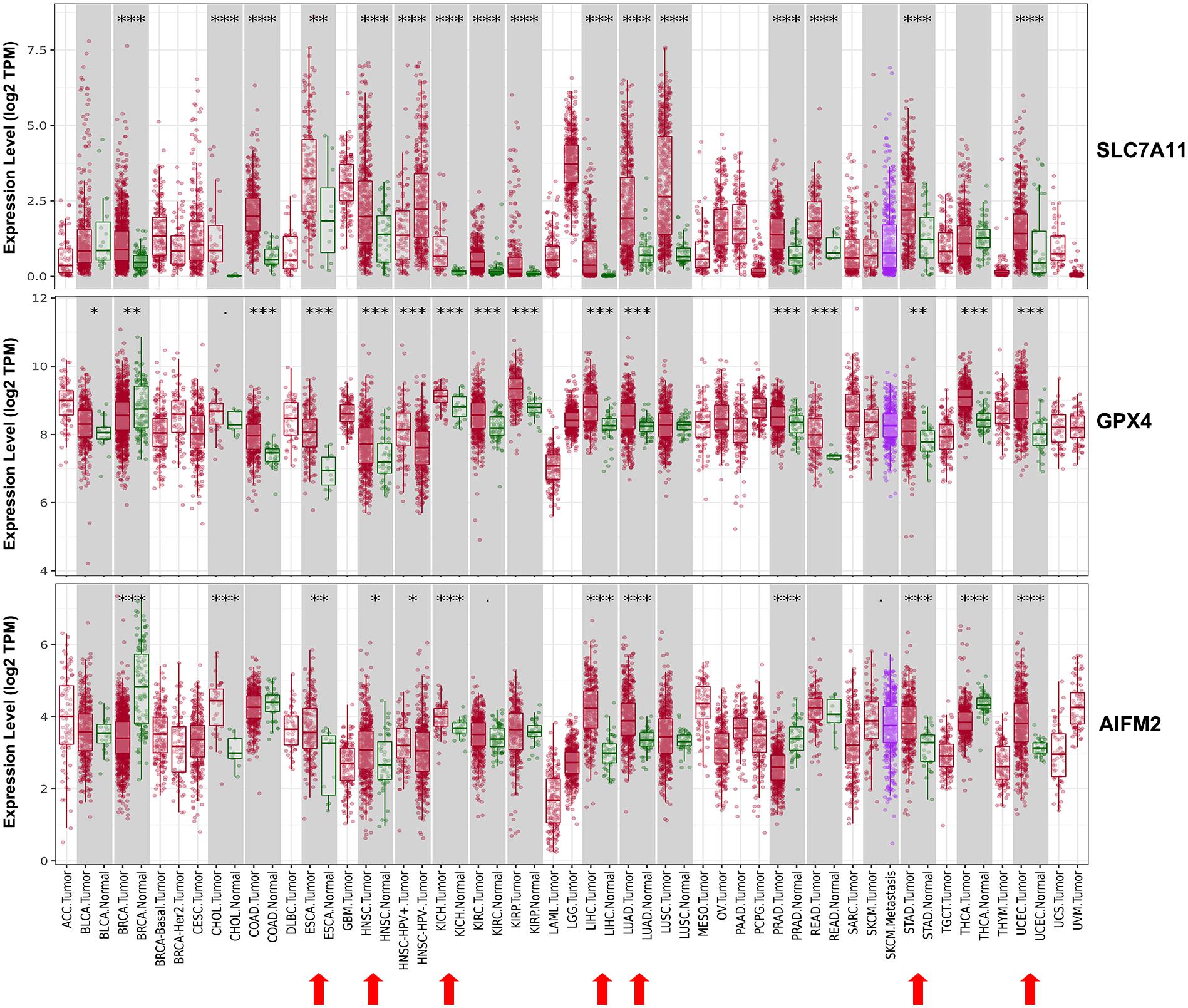
Figure 1. Expression patterns of SLC7A11, GPX4, and AIFM2 in pan-cancer. The expression patterns of SLC7A11, GPX4, and AIFM2 were analyzed by using TIMER database. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
We also analyzed the mRNA levels of SLC7A11, GPX4, and AIFM2 in pan-cancer using Oncomime database. The results showed that SLC7A11 was higher in CRC, esophageal cancer, head and neck cancer, kidney cancer, liver cancer, lung cancer, pancreatic cancer, and uterus cancer, and lower in lymphoma and ovarian cancer (Figure 2 and Supplementary Table 1). The expression of GPX4 was higher in CRC, gastric cancer, lymphoma and melanoma, and lower in sarcoma. AIFM2 was overexpressed in lymphoma and under-expressed in breast cancer and CRC. Interestingly, both SLC7A11 and GPX4 were overexpressed in CRC (in 15 and 6 datasets, respectively, Figure 2 and Supplementary Table 1), and these findings suggested that anti-ferroptosis might be the characteristic of CRC cells.
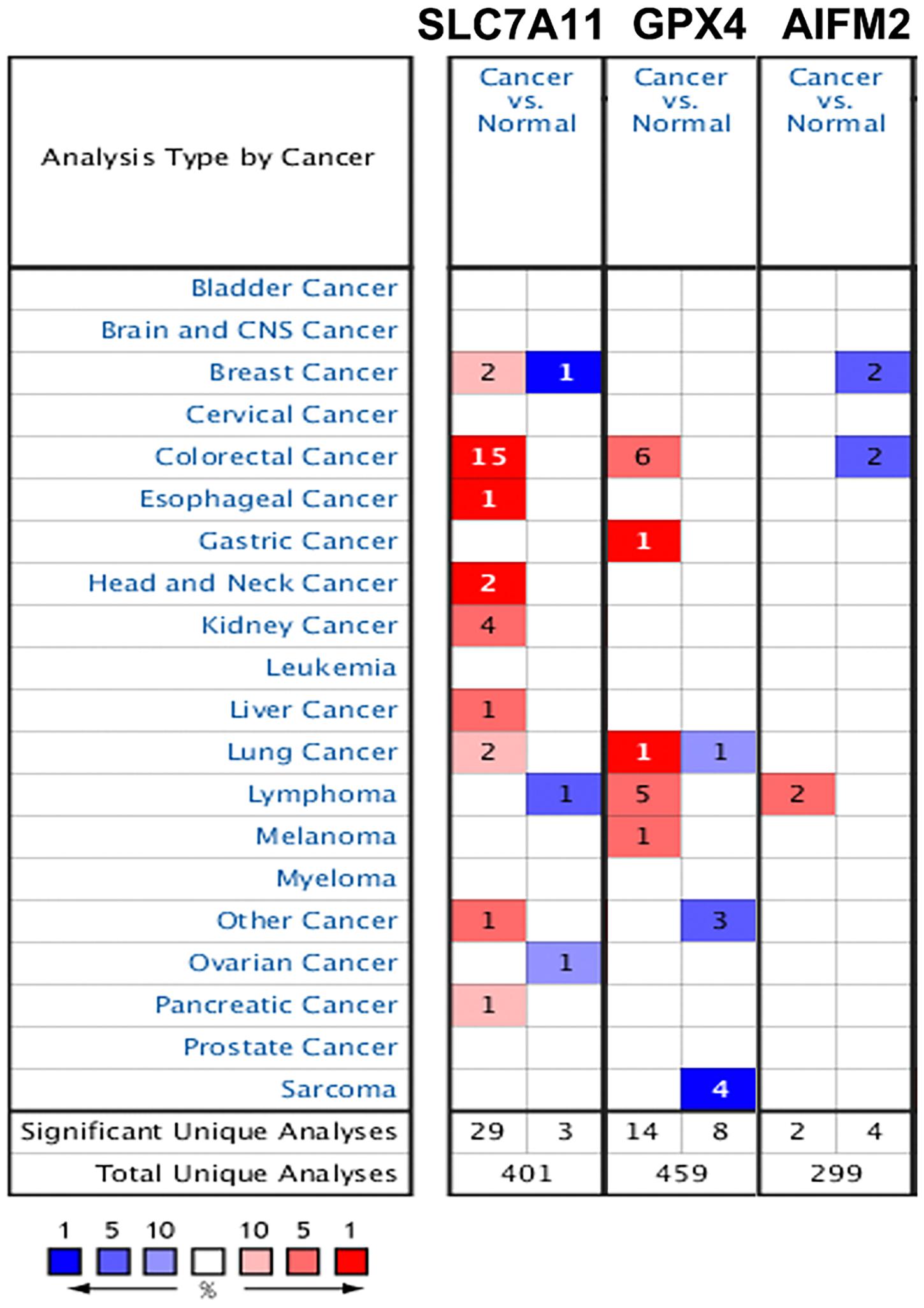
Figure 2. Expression patterns of SLC7A11, GPX4, and AIFM2 in pan-cancer. The expression patterns of SLC7A11, GPX4, and AIFM2 were analyzed by using Oncomine database. %, gene rank percentile (%).
Next, we analyzed the prognostic values of SLC7A11, GPX4, and AIFM2 in pan-cancer using GEPIA database. We found that high expression of SLC7A11, GPX4, and AIFM2 were significantly linked with the shortened disease-free survival in adrenocortical carcinoma (ACC), respectively, and high expression of SLC7A11 and AIFM2 were significantly associated with shortened OS in ACC respectively (Figures 3A–I). And high expression of SLC7A11, GPX4, and AIFM2 were significantly correlated with the shortened OS of acute myeloid leukemia (LAML) patients (Figures 3A–C,J–L). Interestingly, we further found that high expression of SLC7A11 were significantly associated with shortened OS and disease-free survival of KIRP, LIHC, and MESO (mesothelioma), and high expression of GPX4 was significantly correlated with shortened OS and disease-free survival of STAD (Figures 4A–H).
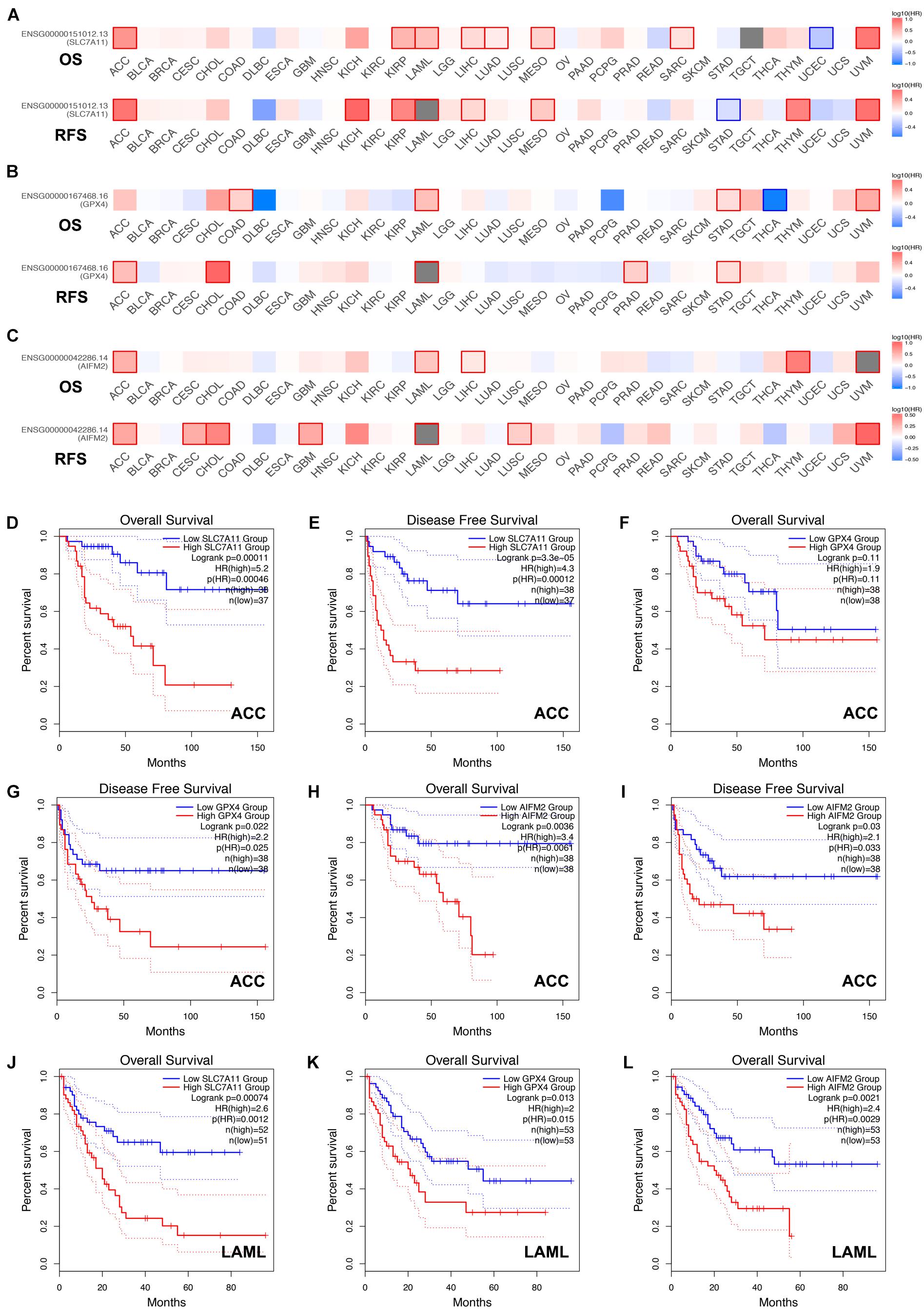
Figure 3. Prognostic roles of SLC7A11, GPX4, and AIFM2 in pan-cancer. (A–C) Prognostic pattern of SLC7A11, GPX4, and AIFM2 in pan-cancer were analyzed by using GEPIA database. (D–I) The correlations between SLC7A11, GPX4, or AIFM2 and OS or DFS of ACC patients were evaluated by using GEPIA database. (J–L) The correlations between SLC7A11, GPX4, or AIFM2 and OS of LAML patients were evaluated by using GEPIA database.
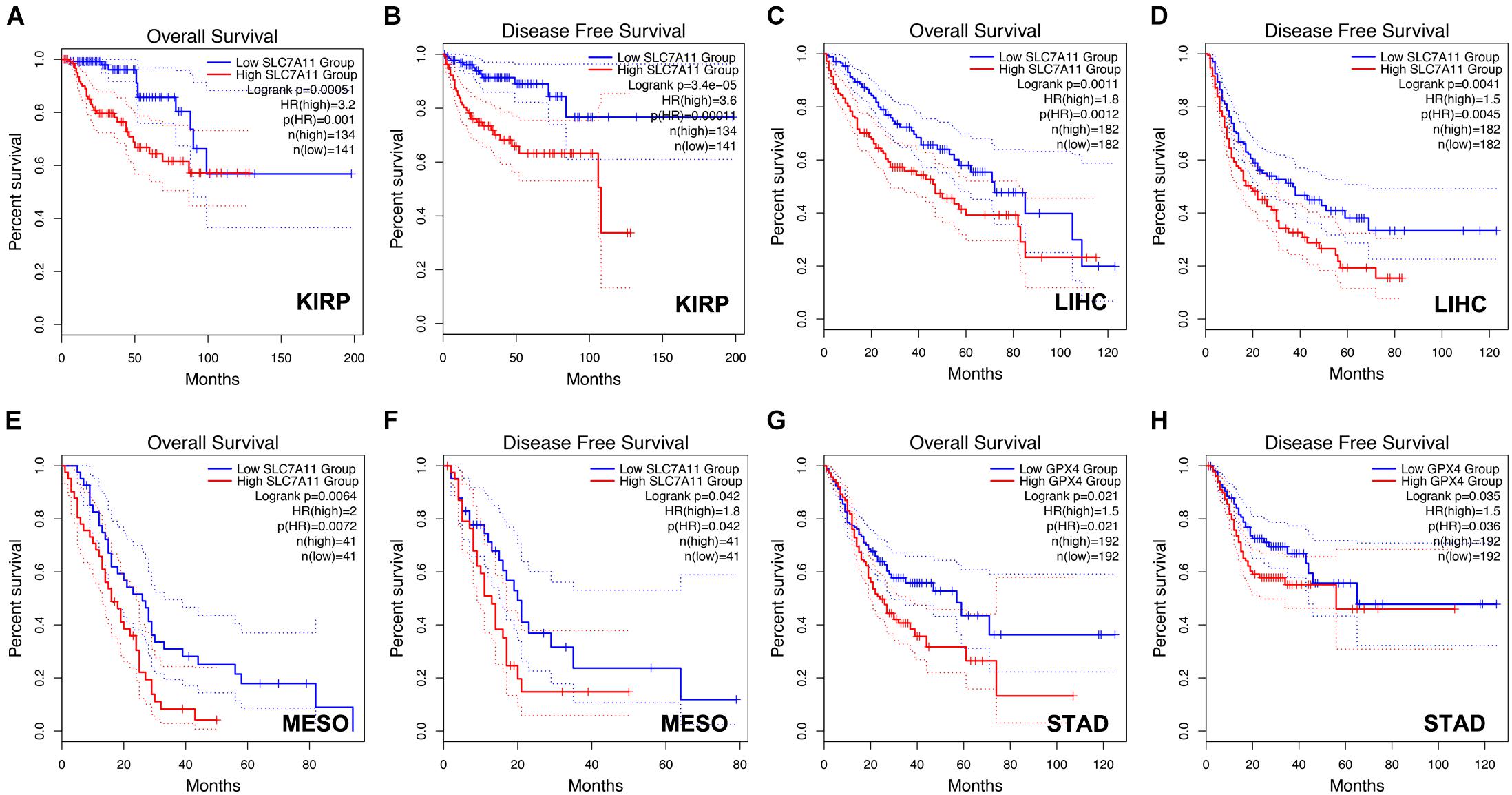
Figure 4. Prognostic role of SLC7A11 in KIRP, LIHC, and MESO and prognostic role of GPX4 in STAD. (A–F) The correlations between SLC7A11 and OS or DFS of KIRP, LIHC, and MESO patients were evaluated by using GEPIA database. (G,H) The correlations between GPX4 and OS or DFS of STAD patients were evaluated by using GEPIA database.
We also analyzed the prognostic values of SLC7A11, GPX4, and AIFM2 in pan-cancer using Kaplan–Meier database. High expression of SLC7A11 was significantly correlated with the poor OS and RFS in bladder carcinoma, breast cancer, kidney renal papillary cell carcinoma, liver hepatocellular carcinoma, and pancreatic ductal adenocarcinoma (Table 1). Importantly, the higher mRNA levels of SLC7A11, GPX4, and AIFM2 were all significantly linked with the good OS in UCEC, and both higher expression of SLC7A11 and AIFM2 were significantly correlated with the poor OS and RFS in liver hepatocellular carcinoma (Table 1). Different from SLC7A11 in bladder carcinoma, the high expression of AIFM2 was significantly associated with the good OS and RFS (Table 1).
We further analyzed the correlation between the expression levels of SLC7A11, GPX4, or AIFM2 and immune infiltration in ESCA, HNSC, COAD, READ, STAD, and LUAD in which at least two of the three ferroptosis-associated genes were overexpressed. We observed that high expression levels of GPX4 and AIFM2 were significantly associated with tumor purity in ESCA (R = 0.25, p = 6.93e-04; R = 0.279, p = 1.43e-04) and in HNSC, respectively (R = 0.223, p = 5.37e-07; R = 0.205, p = 4.41e-06; Figure 5). In ESCA, the level of SLC7A11 was significantly associated with the infiltration level of CD4+ T cell (R = −0.211, p = 4.40e-03), and the level of GPX4 was significantly correlated with the infiltration level of macrophage (R = 0.256, p = 5.21e-04) and DC (R = 0.254, p = 5.93e-04), and the level of AIFM2 was markedly linked with the infiltration level of CD4+ T cell (R = 0.232, p = 1.71e-03). In HNSC, the level of GPX4 was significantly correlated with the infiltration level of B cell (R = −0.38, p = 2.41e-18), CD8+ T cell (R = −0.402, p = 1.42e-20), CD4+ T cell (R = 0.356, p = 3.69e-16), and DC (R = 0.2, p = 7.43e-06), and the level of AIFM2 was significantly associated with the infiltration level of B cell (R = −0.425, p = 5.91e-23), CD8+ T cell (R = −0.436, p = 2.71e-24), and CD4+ T cell (R = 0.41, p = 2.51e-21; Figure 5).
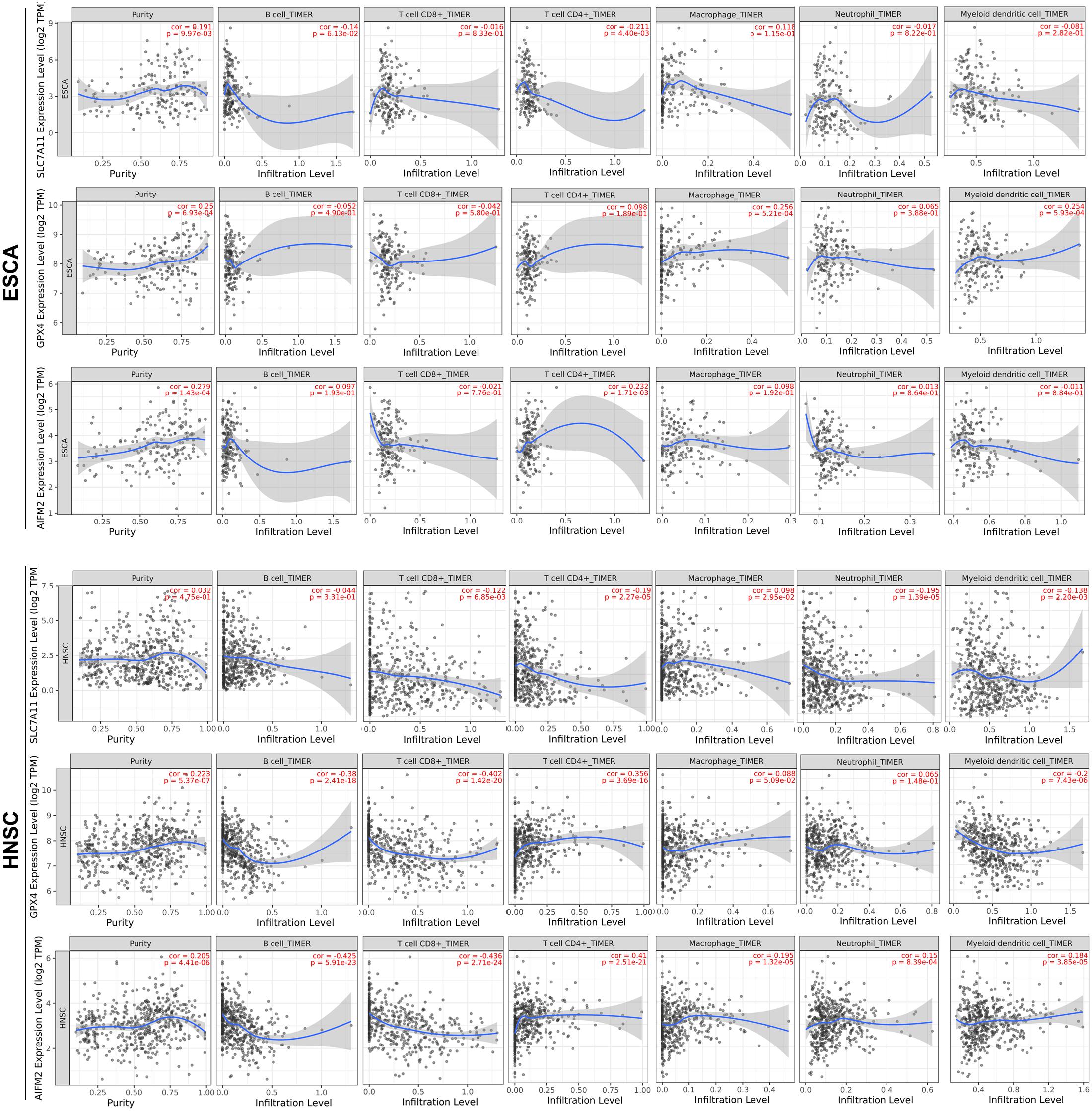
Figure 5. Correlation analysis of SLC7A11, GPX4, or AIFM2 and infiltration levels of immune cells in ESCA and HNSC. The correlation analysis of SLC7A11, GPX4, or AIFM2 and infiltration levels of immune cells in ESCA and HNSC was performed by using TIMER database.
In COAD, the expression of SLC7A11 was significantly correlated with the infiltration level of CD8+ T cell (R = 0.229, p = 1.32e-04). In READ, the expression of SLC7A11 were significantly associated with the infiltration level of neutrophil (R = 0.295, p = 4.73e-03). In STAD, the expression of SLC7A11 was significantly associated with the infiltration level of CD4+ T cell and macrophage, respectively (R = −0.259, p = 3.13e-07; R = −0.212, p = 3.09e-05). In LUAD, the expression of SLC7A11 was significantly associated with the infiltration level of DC (R = −0.206, p = 4.01e-06), and the expression of AIFM2 was significantly associated with the infiltration level of macrophage, neutrophil, and DC, respectively (R = −0.248, p = 2.34e-08; R = −0.201, p = 6.89e-06; R = −0.218, p = 1.01e-06; Supplementary Figures 1, 2).
By analyzing the gene expression data of immune markers in the TIMER database, we investigated the association between the levels of SLC7A11, GPX4, and AIFM2 and the status of tumor-infiltrating immune cells in ESCA, HNSC, COAD, READ, STAD, and LUAD. The CD8+ T cells, CD4+ T cells, B cells, TAMs, monocytes, M1 macrophages, M2 macrophages, neutrophils, DCs, NK cells, and several types of T cells including Th1, Th2, Tfh, Th17, Tregs, and exhausted T cells were analyzed in our study. The correlation analysis in the present study was adjusted for purity because of the influence of tumor purity on the immune infiltration.
In ESCA, GPX4 expression was significantly correlated with the expression of markers of immune cells (at least two significant correlated markers in one cell type) such as monocyte markers, CD14 (R = 0.299, p = 4.59e-05) and CD115 (R = 0.252, p = 6.57e-04); TAM markers, CCL2 (R = 0.341, p = 2.84e-06) and IL10 (R = 0.288, p = 8.65e-05); M1 macrophage markers, iNOS (R = −0.29, p = 7.63e-05), IL6 (R = 0.293, p = 6.57e-05), and CD64 (R = 0.29, p = 7.74e-05); M2 macrophage markers, VSIG4 (R = 0.307, p = 2.73e-05) and MS4A4A (R = 0.307, p = 2.73e-05); and DC markers, BDCA-1 (R = 0.244, p = 9.68e-04) and BDCA-4 (R = 0.26, p = 4.14e-04; Table 2). In HNSC, GPX4 expression was significantly correlated with the expression of markers of immune cells (at least two significant correlated markers in one cell type) such as B cell markers, CD19 (R = 0.207, p = 3.51e-06) and CD27 (R = 0.248, p = 2.52e-08); monocyte markers, CD14 (R = 0.205, p = 4.74e-06) and CD115 (R = 0.236, p = 1.14e-07); M1 macrophage markers, iNOS (R = 0.293, p = 3.65e-11) and CD64 (R = 0.282, p = 1.79e-10); M2 macrophage markers, VSIG4 (R = 0.217, p = 1.12e-06) and MS4A4A (R = 0.248, p = 2.44e-08); and T cell exhaustion markers, CTLA4 (R = 0.223, p = 5.83e-07) and TIM-3 (R = 0.259, p = 5.35e-09; Table 2). Interestingly, both in ESCA and HNSC, the expression of GPX4 was significantly positively associated with the monocyte markers, CD14 and CD115, and the M2 macrophage markers, VSIG4 and MS4A4A.
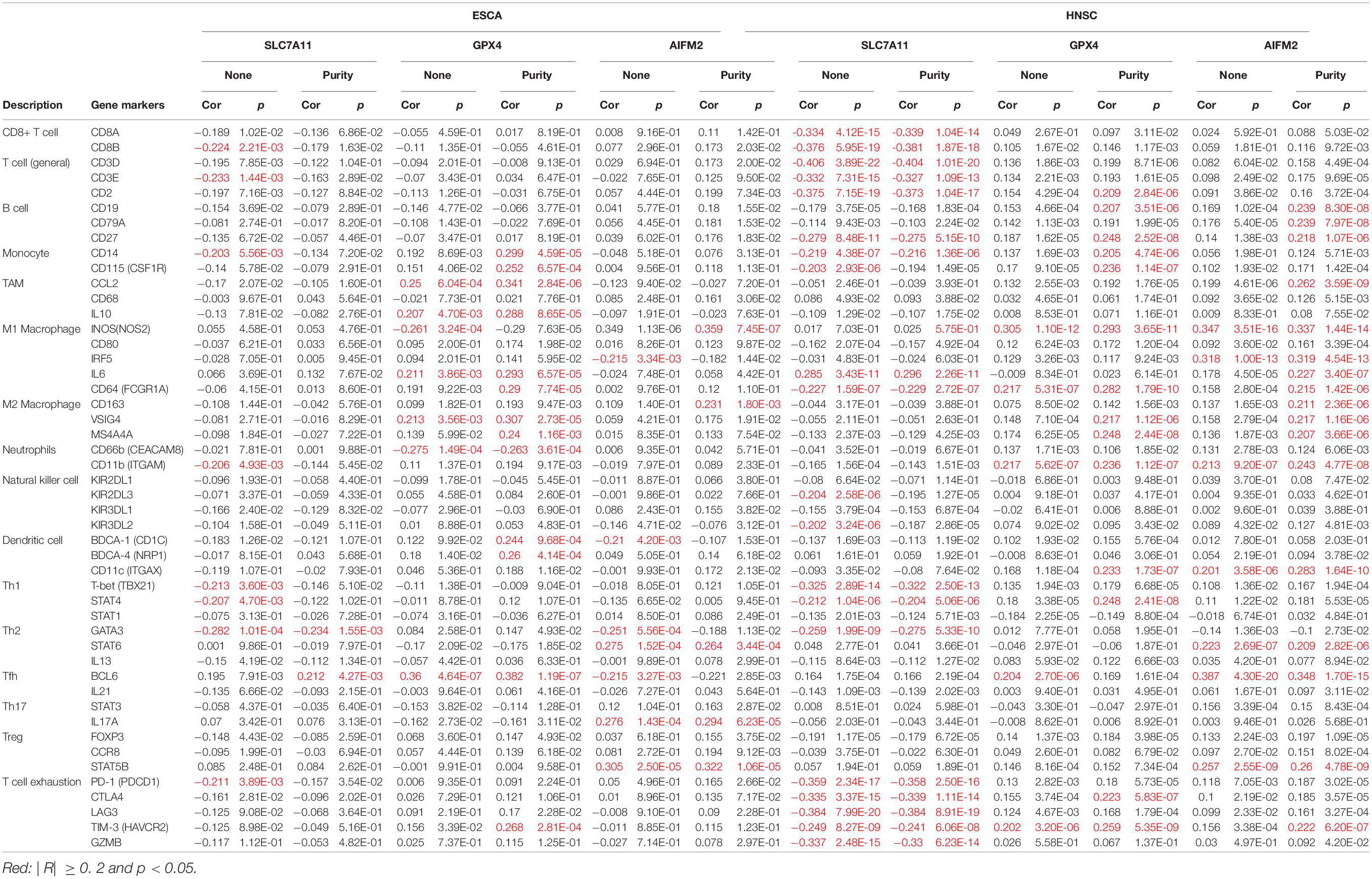
Table 2. Correlation between the expression levels of SLC7A11, GPX4, or AIFM2 and markers of different subsets of immune cells in ESCA and HNSC.
In HNSC, the expression of SLC7A11 was negatively correlated with CD8+ T cell markers, CD8A (R = −0.339, p = 1.04e-14) and CD8B (R = −0.381, p = 1.87e-18); T cell (general) markers, CD3D (R = −0.404, p = 1.01e-20), CD3E (R = −0.327, p = 1.09e-13), and CD2 (R = −0.373, p = 1.04e-17); B cell marker, CD27 (R = −0.275, p = 5.15e-10); monocyte marker, CD14 (R = −0.216, p = 1.36e-06); M1 macrophage marker, CD64 (R = −0.229, p = 2.72e-07); Th1 markers, T-bet (R = −0.322, p = 2.50e-13) and STAT4 (R = −0.204, p = 5.06e-06); Th2 marker, GATA3 (R = −0.275, p = 5.33e-10); and T cell exhaustion markers, PD-1 (R = −0.358, p = 2.50e-16), CTLA4 (R = −0.339, p = 1.11e-14), LAG3 (R = −0.384, p = 8.91e-19), TIM-3 (R = −0.241, p = 6.06e-08), and GZMB (R = −0.33, p = 6.23e-14; Table 2). In HNSC, the expression of AIFM2 was significantly correlated with the expression of markers of immune cells (at least two significant correlated markers in one cell type) such as B cell, CD19 (R = 0.239, p = 8.30e-08), CD79A (R = 0.239, p = 7.97e-08), and CD27 (R = 0.218, p = 1.07e-06); M1 macrophage markers, iNOS (R = 0.337, p = 1.44e-14), IRF5 (R = 0.319, p = 4.54e-13), IL6 (R = 0.227, p = 3.40e-07), and CD64 (R = 0.215, p = 1.42e-06); and M2 macrophage markers, CD163 (R = 0.211, p = 2.36e-06), VSIG4 (R = 0.217, p = 1.16e-06), and MS4A4A (R = 0.207, p = 3.66e-06; Table 2). Very interestingly, the expression of AIFM2 was significantly associated with the expression of STAT6 and STAT5B both in ESCA and HNSC.
In READ, SLC7A11 expression was significantly correlated with the expression of markers of immune cells (at least two significant correlated markers in one cell type) such as monocyte markers, CD14 (R = −0.286, p = 6.48e-04) and CD115 (R = −0.2, p = 1.84e-02); and DC markers, BDCA-1 (R = −0.232, p = 6.04e-03) and BDCA-4 (R = 0.256, p = 2.31e-03; Supplementary Table 2). GPX4 expression was associated with the expression of marker immune cells (at least two significant correlated markers in one cell type) such as monocyte markers, CD14 (R = 0.339, p = 4.58e-05) and CD115 (R = 0.269, p = 1.36e-03); M1 macrophage markers, iNOS (R = −0.242, p = 4.08e-03) and CD64 (R = 0.334, p = 6.00e-05); and DC markers, BDCA-1 (R = 0.218, p = 9.79e-03) and BDCA-4 (R = −0.205, p = 1.53e-02; Supplementary Table 2).
In LUAD, SLC7A11 expression was significantly correlated with the expression of markers of immune cells (at least two significant correlated markers in one cell type) such as M1 macrophage markers, CD80 (R = −0.217, p = 1.20e-06) and IRF5 (R = −0.246, p = 3.25e-08); and DC markers, BDCA-1 (R = −0.346, p = 2.62e-15) and BDCA-4 (R = −0.227, p = 3.48e-07). GPX4 expression was significantly correlated with the expression of markers of immune cells (at least two significant correlated markers in one cell type) such as M1 macrophage markers, CD80 (R = −0.229, p = 2.85e-07) and CD64 (R = −0.204, p = 5.23e-06); and M2 macrophage markers, VSIG4 (R = −0.216, p = 1.36e-06) and MS4A4A (R = −0.234, p = 1.44e-07; Supplementary Table 2).
In COAD and STAD, there were no correlations between SLC7A11/GPX4/AIFM2 with the expression of markers of immune cells (at least two significant correlated markers in one cell type).
Very interestingly, the expression of SLC7A11 was significantly positively associated with the level of STAT1 in COAD, READ, and STAD (R = 0.256, p = 1.77e-07; R = 0.337, p = 5.07e-05; R = 0.21, p = 3.78e-05). The expression of GPX4 was significantly negatively associated with the level of STAT3 in COAD, READ, and LUAD (R = −0.217, p = 1.02e-05; R = −0.288, p = 5.98e-04; R = −0.229, p = 2.77e-07).
Recent studies reported that therapy-resistant tumor cells were vulnerable to ferroptosis; therefore, ferroptosis induction will be a potential therapeutic method for tumors especially drug-resistant tumors (Li B. et al., 2020; Jiang et al., 2021). Our study found that both SLC7A11 and GPX4 were overexpressed in CRC, and SLC7A11 was highly expressed in lung cancer by analyzing TIMER and Oncomine databases. These findings were consistent with the published literatures. SLC7A11 and GPX4 were highly expressed in CRC cell lines and CRC tissues compared with paired normal tissues, and downregulation of SLC7A11 by talaroconvolutin or downregulation of GPX4 by apatinib or inactivation of GPX4 by resibufogenin could induce ferroptosis of CRC cells (Yagublu et al., 2011; Ma et al., 2015; Xia et al., 2020; Shen et al., 2021; Tian et al., 2021). SLC7A11 was overexpressed in non-small cell lung cancer (NSCLC) tissues and its overexpression was significantly correlated with poor prognosis of NSCLC patients (Ji et al., 2018). Inhibition of SLC7A11 by sulforaphane or YTHDC2 (a m6A reader) induced the ferroptosis of lung cancer cells and suppressed the progression of lung cancer (Hu et al., 2020; Iida et al., 2021; Ma et al., 2021). Interestingly, our results also showed that AIFM2 was overexpressed in ESCA and HNSC; however, up to now, the roles and mechanisms of AIFM2 in the progression of ESCA and HNSC were still unknown.
By analyzing the GEPIA database, we found that high expression of SLC7A11, GPX4, and AIFM2 were correlated with the shortened disease-free survival in ACC respectively, and high expression of SLC7A11 and AIFM2 were significantly associated with shortened overall survival in ACC respectively. Previous studies have reported that human ACC cells NCI-H295R cell line was sensitive to ferroptosis induction, and the mRNA level of SLC7A11 was higher in ACCs than in normal adrenal glands (nAGs) and its low expression was significantly associated with good OS of ACC patients (Belavgeni et al., 2019; Weigand et al., 2020). However, the functional roles and regulatory mechanisms of SLC7A11, GPX4, and AIFM2 in the ferroptosis of ACC cells were still unclear. Our study also revealed that high expression of SLC7A11, GPX4, and AIFM2 were significantly correlated with the lower overall survival of LAML. A previous study reported that combination of APR-246 and inactivation of SLC7A11/GPX4 could synergistically promote the ferroptosis of AML cells both in in vivo and ex vivo experiments (Birsen et al., 2021). However, how SLC7A11 and GPX4 were regulated in LAML and whether AIFM2 was involved in the regulation of ferroptosis of AML cells were still largely unknown. Therefore, future studies should focus on the roles of ferroptosis in the progression of ACC and LAML, especially the regulatory mechanisms of SLC7A11 or GPX4 or AIFM2 in ferroptosis of ACC and LAML cells.
Although a previous study reported that GPX4 could prevent Treg cells from lipid peroxidation and ferroptosis (Xu et al., 2021), whether SLC7A11, GPX4, and AIFM2 participated in the regulation of ferroptosis of other immune cells and tumor progression is still largely unknown. Our study demonstrated that in ESCA, GPX4 expression was significantly associated with the infiltration of macrophage and myeloid DC, and AIFM2 expression was significantly associated with the infiltration of CD4+ T cell. These findings suggested that GPX4 and AIFM2 might be involved in the progression of esophageal cancer probably by regulating the ferroptosis of different immune cells. Our results also showed that the expression levels of GPX4 and AIFM2 were correlated with the infiltration of B cell, CD8+ T cell, and CD4+ T cell in HNSC, respectively. This suggested that GPX4 and AIFM2 played important roles in regulating tumor immunity by affecting the ferroptosis of B cell, CD8+ T cell, and CD4+ T cell in HNSC. Our study also reported that the expression of AIFM2 was significantly correlated with the expression of markers of B cell including CD19, CD79A, and CD27.
Interestingly, our study also indicated that GPX4 expression was positively correlated with the expression levels of monocyte markers including CD14 and CD115 and M2 macrophage markers including VSIG4 and MS4A4A both in ESCA and in HNSC. Hsieh et al. reported that zero-valent-iron nanoparticle (ZVI-NP) could promote the shift from pro-tumor M2 macrophages to anti-tumor M1 macrophages and finally inhibit the tumor progression (Hsieh et al., 2021). Our results suggested that activation of M2 macrophages in ESCA and HNSC might be correlated with high expression of GPX4.
Our results suggest that SLC7A11, GPX4, and AIFM2 are dysregulated in many types of cancers, and are candidate prognostic biomarkers for many types of cancers, and can be used to evaluate the infiltration of immune cells in tumor tissues. Future studies should focus on the regulation of ferroptosis of tumor cells and immune cells in different types of cancers.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Z-ZS and HT analyzed the data. Z-ZS, HT, Z-WF, S-JS, and JB wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was funded by the National Natural Science Foundation of China (Nos. 81802794 and 81760526), Yunnan Provincial Research Foundation for Basic Research, China (Nos. 2018FB135 and 202001AT070028), and the Key Laboratory for Oxidative Stress Damage and Defense in University of Yunnan Province (2018).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.748925/full#supplementary-material
Belavgeni, A., Bornstein, S. R., von Massenhausen, A., Tonnus, W., Stumpf, J., Meyer, C., et al. (2019). Exquisite sensitivity of adrenocortical carcinomas to induction of ferroptosis. Proc. Natl. Acad. Sci. U.S.A. 116, 22269–22274. doi: 10.1073/pnas.1912700116
Bersuker, K., Hendricks, J. M., Li, Z., Magtanong, L., Ford, B., Tang, P. H., et al. (2019). The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575, 688–692. doi: 10.1038/s41586-019-1705-2
Birsen, R., Larrue, C., Decroocq, J., Johnson, N., Guiraud, N., Gotanegre, M., et al. (2021). APR-246 induces early cell death by ferroptosis in acute myeloid leukemia. Haematologica Online ahead of print doi: 10.3324/haematol.2020.259531
Chen, X., Kang, R., Kroemer, G., and Tang, D. (2021). Broadening horizons: the role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. 18, 280–296. doi: 10.1038/s41571-020-00462-0
Daher, B., Parks, S. K., Durivault, J., Cormerais, Y., Baidarjad, H., Tambutte, E., et al. (2019). Genetic ablation of the cystine transporter xCT in PDAC cells inhibits mTORC1, growth, survival, and tumor formation via nutrient and oxidative stresses. Cancer Res. 79, 3877–3890. doi: 10.1158/0008-5472.CAN-18-3855
Dai, E., Zhang, W., Cong, D., Kang, R., Wang, J., and Tang, D. (2020). AIFM2 blocks ferroptosis independent of ubiquinol metabolism. Biochem. Biophys. Res. Commun. 523, 966–971. doi: 10.1016/j.bbrc.2020.01.066
Deng, S. H., Wu, D. M., Li, L., Liu, T., Zhang, T., Li, J., et al. (2021). miR-324-3p reverses cisplatin resistance by inducing GPX4-mediated ferroptosis in lung adenocarcinoma cell line A549. Biochem. Biophys. Res. Commun. 549, 54–60. doi: 10.1016/j.bbrc.2021.02.077
Doll, S., Freitas, F. P., Shah, R., Aldrovandi, M., da Silva, M. C., Ingold, I., et al. (2019). FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698. doi: 10.1038/s41586-019-1707-0
Fukuda, M., Ogasawara, Y., Hayashi, H., Okuyama, A., Shiono, J., Inoue, K., et al. (2021). Down-regulation of Glutathione Peroxidase 4 in Oral Cancer Inhibits Tumor Growth Through SREBP1 Signaling. Anticancer Res. 41, 1785–1792. doi: 10.21873/anticanres.14944
Hou, Y., Cai, S., Yu, S., and Lin, H. (2021). Metformin induces ferroptosis by targeting miR-324-3p/GPX4 axis in breast cancer. Acta Biochim. Biophys. Sin. 53, 333–341. doi: 10.1093/abbs/gmaa180
Hsieh, C. H., Hsieh, H. C., Shih, F. S., Wang, P. W., Yang, L. X., Shieh, D. B., et al. (2021). An innovative NRF2 nano-modulator induces lung cancer ferroptosis and elicits an immunostimulatory tumor microenvironment. Theranostics 11, 7072–7091. doi: 10.7150/thno.57803
Hu, K., Li, K., Lv, J., Feng, J., Chen, J., Wu, H., et al. (2020). Suppression of the SLC7A11/glutathione axis causes synthetic lethality in KRAS-mutant lung adenocarcinoma. J. Clin. Invest. 130, 1752–1766. doi: 10.1172/JCI124049
Iida, Y., Okamoto-Katsuyama, M., Maruoka, S., Mizumura, K., Shimizu, T., Shikano, S., et al. (2021). Effective ferroptotic small-cell lung cancer cell death from SLC7A11 inhibition by sulforaphane. Oncol. Lett. 21:71. doi: 10.3892/ol.2020.12332
Ji, X., Qian, J., Rahman, S. M. J., Siska, P. J., Zou, Y., Harris, B. K., et al. (2018). xCT (SLC7A11)-mediated metabolic reprogramming promotes non-small cell lung cancer progression. Oncogene 37, 5007–5019. doi: 10.1038/s41388-018-0307-z
Jiang, X., Stockwell, B. R., and Conrad, M. (2021). Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 22, 266–282. doi: 10.1038/s41580-020-00324-8
Lang, X., Green, M. D., Wang, W., Yu, J., Choi, J. E., Jiang, L., et al. (2019). Radiotherapy and immunotherapy promote tumoral lipid oxidation and ferroptosis via synergistic repression of SLC7A11. Cancer Discov. 9, 1673–1685. doi: 10.1158/2159-8290.CD-19-0338
Li, B., Severson, E., Pignon, J. C., Zhao, H., Li, T., Novak, J., et al. (2016). Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 17:174. doi: 10.1186/s13059-016-1028-7
Li, B., Yang, L., Peng, X., Fan, Q., Wei, S., Yang, S., et al. (2020). Emerging mechanisms and applications of ferroptosis in the treatment of resistant cancers. Biomed. Pharmacother. 130:110710. doi: 10.1016/j.biopha.2020.110710
Li, T., Fan, J., Wang, B., Traugh, N., Chen, Q., Liu, J. S., et al. (2017). TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77, e108–e110. doi: 10.1158/0008-5472.CAN-17-0307
Li, T., Fu, J., Zeng, Z., Cohen, D., Li, J., Chen, Q., et al. (2020). TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48, W509–W514. doi: 10.1093/nar/gkaa407
Lim, J. K. M., Delaidelli, A., Minaker, S. W., Zhang, H. F., Colovic, M., Yang, H., et al. (2019). Cystine/glutamate antiporter xCT (SLC7A11) facilitates oncogenic RAS transformation by preserving intracellular redox balance. Proc. Ntl. Acad. Sci. U.S.A. 116, 9433–9442. doi: 10.1073/pnas.1821323116
Ma, L., Chen, T., Zhang, X., Miao, Y., Tian, X., Yu, K., et al. (2021). The m(6)A reader YTHDC2 inhibits lung adenocarcinoma tumorigenesis by suppressing SLC7A11-dependent antioxidant function. Redox Biol. 38:101801. doi: 10.1016/j.redox.2020.101801
Ma, M. Z., Chen, G., Wang, P., Lu, W. H., Zhu, C. F., Song, M., et al. (2015). Xc- inhibitor sulfasalazine sensitizes colorectal cancer to cisplatin by a GSH-dependent mechanism. Cancer Lett. 368, 88–96. doi: 10.1016/j.canlet.2015.07.031
Nagy, A., Munkacsy, G., and Gyorffy, B. (2021). Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 11:6047. doi: 10.1038/s41598-021-84787-5
Rhodes, D. R., Kalyana-Sundaram, S., Mahavisno, V., Varambally, R., Yu, J., Briggs, B. B., et al. (2007). Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9, 166–180. doi: 10.1593/neo.07112
Shen, L. D., Qi, W. H., Bai, J. J., Zuo, C. Y., Bai, D. L., Gao, W. D., et al. (2021). Resibufogenin inhibited colorectal cancer cell growth and tumorigenesis through triggering ferroptosis and ROS production mediated by GPX4 inactivation. Anat. Rec. 304, 313–322. doi: 10.1002/ar.24378
Tang, Z., Li, C., Kang, B., Gao, G., Li, C., and Zhang, Z. (2017). GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102. doi: 10.1093/nar/gkx247
Tian, X., Li, S., and Ge, G. (2021). Apatinib promotes ferroptosis in colorectal cancer cells by targeting ELOVL6/ACSL4 signaling. Cancer Manag. Res. 13, 1333–1342. doi: 10.2147/CMAR.S274631
Wang, H., Cheng, Y., Mao, C., Liu, S., Xiao, D., Huang, J., et al. (2021a). Emerging mechanisms and targeted therapy of ferroptosis in cancer. Mol. Ther. 29, 2185–2208. doi: 10.1016/j.ymthe.2021.03.022
Wang, H., Peng, S., Cai, J., and Bao, S. (2021b). Silencing of PTPN18 induced ferroptosis in endometrial cancer cells through p-P38-mediated GPX4/xCT down-regulation. Cancer Manag. Res. 13, 1757–1765. doi: 10.2147/CMAR.S278728
Wang, R., Su, Q., Yin, H., Wu, D., Lv, C., and Yan, Z. (2021c). Inhibition of SRSF9 enhances the sensitivity of colorectal cancer to erastin-induced ferroptosis by reducing glutathione peroxidase 4 expression. Int. J. Biochem. Cell Biol. 134:105948. doi: 10.1016/j.biocel.2021.105948
Wang, L., Liu, Y., Du, T., Yang, H., Lei, L., Guo, M., et al. (2020). ATF3 promotes erastin-induced ferroptosis by suppressing system Xc(). Cell Death Differ. 27, 662–675. doi: 10.1038/s41418-019-0380-z
Wang, W., Green, M., Choi, J. E., Gijon, M., Kennedy, P. D., Johnson, J. K., et al. (2019). CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 569, 270–274. doi: 10.1038/s41586-019-1170-y
Weigand, I., Schreiner, J., Rohrig, F., Sun, N., Landwehr, L. S., Urlaub, H., et al. (2020). Active steroid hormone synthesis renders adrenocortical cells highly susceptible to type II ferroptosis induction. Cell Death Dis. 11:192. doi: 10.1038/s41419-020-2385-4
Xia, Y., Liu, S., Li, C., Ai, Z., Shen, W., Ren, W., et al. (2020). Discovery of a novel ferroptosis inducer-talaroconvolutin A-killing colorectal cancer cells in vitro and in vivo. Cell Death Dis. 11:988. doi: 10.1038/s41419-020-03194-2
Xu, C., Sun, S., Johnson, T., Qi, R., Zhang, S., Zhang, J., et al. (2021). The glutathione peroxidase Gpx4 prevents lipid peroxidation and ferroptosis to sustain Treg cell activation and suppression of antitumor immunity. Cell Rep. 35:109235. doi: 10.1016/j.celrep.2021.109235
Yagublu, V., Arthur, J. R., Babayeva, S. N., Nicol, F., Post, S., and Keese, M. (2011). Expression of selenium-containing proteins in human colon carcinoma tissue. Anticancer Res. 31, 2693–2698.
Keywords: ferroptosis, SLC7A11, GPX4, AIFM2, pan-cancer
Citation: Shi Z-Z, Tao H, Fan Z-W, Song S-J and Bai J (2021) Prognostic and Immunological Role of Key Genes of Ferroptosis in Pan-Cancer. Front. Cell Dev. Biol. 9:748925. doi: 10.3389/fcell.2021.748925
Received: 28 July 2021; Accepted: 09 September 2021;
Published: 13 October 2021.
Edited by:
Giuseppe Palma, Istituto Nazionale Tumori Fondazione G. Pascale, Scientific Institute for Research, Hospitalization and Healthcare (IRCCS), ItalyReviewed by:
Macrina Beatriz Silva Cázares, Autonomous University of San Luis Potosí, MexicoCopyright © 2021 Shi, Tao, Fan, Song and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi-Zhou Shi, emhpemhvdXNoaUBrdXN0LmVkdS5jbg==; Jie Bai, amllYmFpNjYyMDAxQDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.