- 1Cheeloo College of Medicine, Shandong University, Jinan, China
- 2Department of Radiation Oncology, Shandong Cancer Hospital and Institute, Shandong First Medical University and Shandong Academy of Medical Sciences, Jinan, China
- 3Department of Radiation Oncology, Huaian No. 2 Hospital, Huaian, China
- 4Shandong Provincial Key Laboratory of Radiation Oncology, Cancer Research Center, Shandong Cancer Hospital and Institute, Shandong First Medical University and Shandong Academy of Medical Sciences, Jinan, China
- 5Department of Oncology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
Transarterial chemoembolization (TACE) has significantly prolonged overall survival (OS) of unresectable hepatocellular carcinoma (HCC) patients. Unfortunately, there are still a portion of patients without therapeutic responses to TACE. Although genome-wide association studies identified multiple HCC susceptibility SNPs, it is still largely unclear how genome-wide identified functional SNPs impacting gene expression contribute to the prognosis of TACE-treated HCC patients. In this study, we developed an integrative functional genomics methodology to identify gene expression-related SNPs significantly contributing to prognosis of TACE-treated HCC patients across the whole genome. Employing integration of data from expression quantitative trait locus (eQTLs) analyses of The Cancer Genome Atlas (TCGA) liver hepatocellular carcinoma (LIHC) as well as the 1000 Genomes project, we successfully annotated 60 gene expression-related SNPs which are associated with OS of the TCGA patients. After genotyping these 60 SNPs in our TACE cohort, we identified four SNPs (rs12574873, rs12513391, rs34597395, and rs35624901) which are significantly associated with OS of HCC patients treated with TACE. For instance, multivariate Cox proportional hazards model indicated that the rs35624901 Deletion.Deletion (Del.Del) genotype carriers had markedly prolonged OS and a 55% decreased death risk compared with individuals with the GG genotype after TACE therapy (p = 8.3 × 10–5). In support of this, the rs35624901 Del.Del genotype is correlated to higher expression of RAG1, a key T-/B-cell deficiency regulator. Our findings reported the first evidence supporting the prognostic value of four eQTL SNPs in TACE-treated HCC patients. Importantly, our data implicated that antitumor immunity might contribute to TACE efficiency for unresectable HCC patients.
Introduction
Hepatocellular carcinoma (HCC) accounts for ninety percent of primary liver tumors and is one of the deadliest malignancies worldwide, especially in Asia (Llovet et al., 2016; Ferlay et al., 2019). Hepatitis due to infection from hepatitis B virus and/or hepatitis C virus, intakes of aflatoxin B1, heavy alcohol drinking, and smoking has been reported to be well-established risk factors of HCC (Llovet et al., 2016; Craig et al., 2019). Most HCC patients are commonly diagnosed at advanced stages and have poor overall survival (OS; Llovet et al., 2016; Ferlay et al., 2019). For these unresectable HCC patients, transarterial chemoembolization (TACE) has been widely used in clinic. TACE promotes ischemic necrosis of tumors by blocking the arterial supply of the tumor and simultaneously delivering a cytotoxic chemotherapeutic agent. The key randomized controlled clinical trials demonstrated that TACE improved OS of unresectable HCC patients (Llovet et al., 2002; Lo et al., 2002). For instance, in Asian patients with unresectable HCC, transarterial Lipiodol chemoembolization is an effective treatment form and could significantly prolong OS (Lo et al., 2002), while not all unresectable HCC patients could benefit from TACE treatment and some even showed rapid disease progression after TACE (Llovet et al., 2002; Lo et al., 2002). Therefore, identification of novel biomarkers for patient-tailored TACE is urgently needed.
Single nucleotide polymorphisms (SNPs) are the most common type of genetic variants in human. Genome-wide association studies (GWAS) identified multiple SNPs associated with HCC susceptibility and prognosis (Zhang et al., 2010; Kumar et al., 2011; Miki et al., 2011; Li et al., 2012). Interestingly, most GWAS-identified HCC-risk or HCC-prognostic SNPs might function via regulating gene expression due to their location in non-coding regions of human genome (Hindorff et al., 2009). As a result, interpreting mechanisms of SNP-regulated gene expression is crucial for understanding the biological nature of these HCC-related SNPs (Gong et al., 2018). As a powerful tool to disclose detailed impacts of functional SNPs in cancers, expression quantitative trait locus (eQTL) analysis links SNP genotypes to gene expression in various tissues (Grundberg et al., 2012; Westra et al., 2013; Zhu et al., 2016). Although candidate gene studies declared the value of SNPs as prognostic biomarkers for HCC treated with TACE (Zhao et al., 2012; Tang et al., 2014; Yu et al., 2014; Yuan et al., 2014; Wu et al., 2015; Liu et al., 2016; Qiu and Liu, 2016), it is still unclear how genome-wide identified functional SNPs impacting gene expression contribute to prognosis of HCC patients undergoing TACE. Therefore, we systematically screened gene expression-related genetic variants via survival-associated eQTLs in The Cancer Genome Atlas (TCGA) liver hepatocellular carcinoma (LIHC) patients. There are 825 candidate SNPs with minor allele frequency (MAF) ≥5% in Han Chinese populations according to the 1,000 Genomes project. We then genotyped 60 candidate genetic variants in the Jiangsu HCC TACE cohort and identified four SNPs which significantly contributed to the prognosis of HCC patients after TACE in Chinese Han population.
Materials and Methods
SNP Selection
The gene expression-related genetic variants associated with OS of LIHC patients in TCGA were identified using the PancanQTL database1. Both cis-eQTLs and trans-eQTLs in HCC tissues were included in the current study. SNPs with MAF less than 0.05 in Han Chinese populations were excluded using information from the 1,000 Genomes project database.
Study Subjects
This study consisted of 273 Stage III or IV HCC Han Chinese patients treated with TACE. The detailed characteristics of all patients have been reported previously (Liu et al., 2021). All HCC patients were recruited at Huaian No. 2 Hospital (Huaian, Jiangsu Province, China) between January 1999 and January 2013. All patients had histologically confirmed HCC with clinical data available. All subjects had no history of other cancers. Within a week after diagnosis, all patients underwent TACE as the first-line therapy. For most HCC cases, both oxaliplatin and doxorubicin were used for TACE. OS was calculated from the date of diagnosis to the date of death for any cause. At recruitment, written informed consent was obtained from each patient. This study was approved by the Institutional Review Board of Huaian No.2 Hospital. The methods were carried out in accordance with the approved guidelines.
Genotyping
A total of 60 candidate SNPs were genotyped in Jiangsu TACE cohort and 58 candidate SNPs were successfully genotyped (Table 1). Genotypes of these fifty-eight polymorphisms were determined using the iPLEX Sequenom MassARRAY platform (Sequenom) as previously reported (Li Y. et al., 2020; Li Z. et al., 2020). A 15% random sample was reciprocally tested and the reproducibility was 100%.
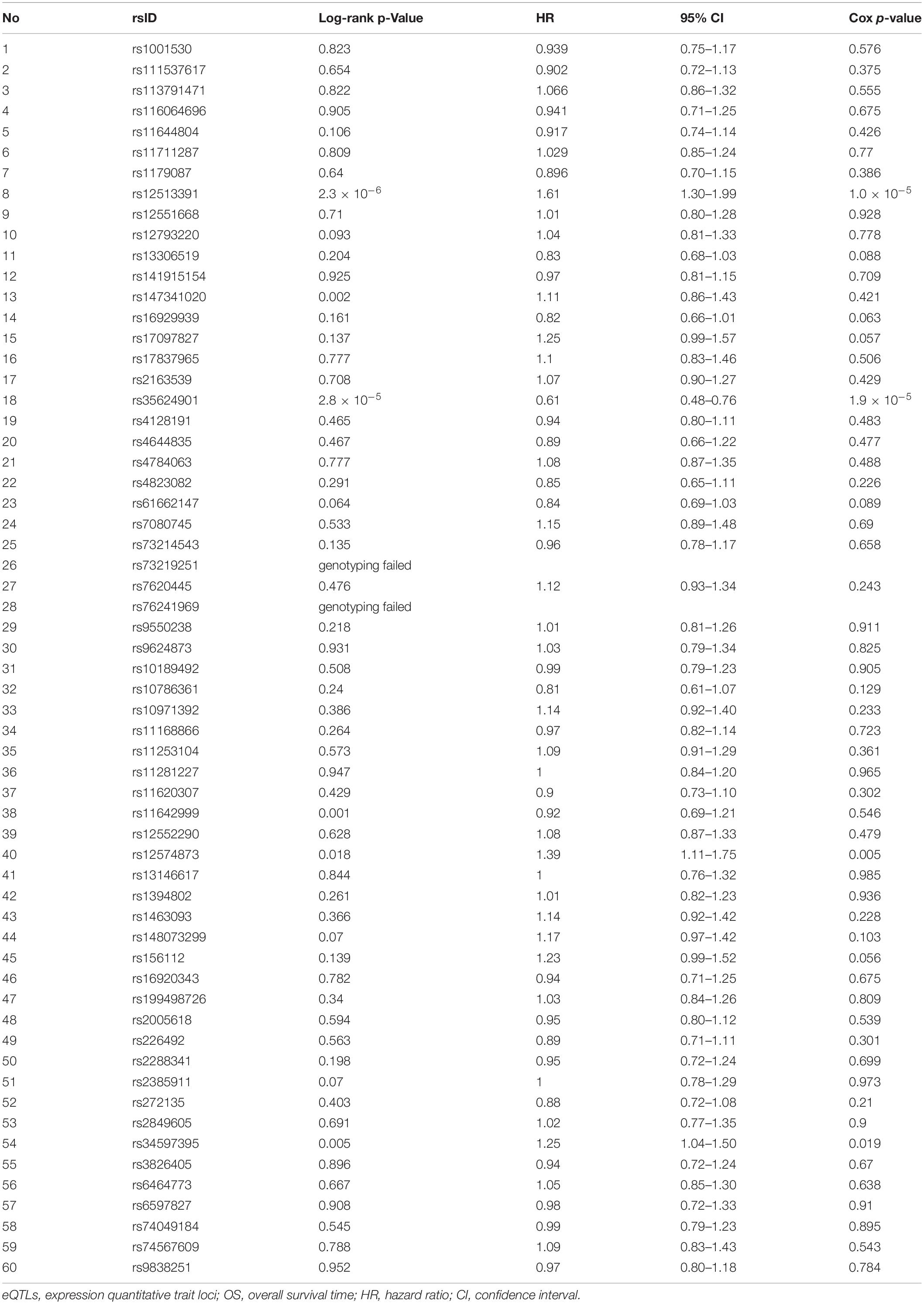
Table 1. Log-rank and Cox-regression analyses of 60 PancanQTL-identified candidate genetic variants for OS in the Jiangsu cohort.
Statistics
Survival curves were compared between different genotypes using the log-rank test. The impacts of genotypes on OS were examined using the Kaplan–Meier method. Multivariate prognostic factors for OS were analyzed using Cox regression analyses. Hazard ratios (HRs) and 95% confidence intervals (CIs) for the association between the candidate SNPs and death risk of HCC patients were adjusted for age of onset, sex, smoking status, drinking status, hepatitis history, stage, and HCC family history, where they were appropriate. A p-value of less than 0.05 was used as the criterion of statistical significance. All statistical tests were two-sided. All analyses were performed with GraphPad or SPSS software package.
Results
Genome-Wide Identification of eQTLs SNPs Associated With Survival of the TCGA LIHC Patients
As summarized in Figure 1, we conducted a genome-wide screening of gene expression-related genetic polymorphisms (eQTLs SNPs) which are significantly associated with OS of the TCGA LIHC patients. First, a total of 1,286 LIHC survival-associated eQTLs SNPs were identified using the PancanQTL database (all log-rank p < 4.9 × 10–4; all eQTLs p < 1.7 × 10–4). Next, according to the 1,000 Genomes project, 463 genetic variants were excluded due to their MAFs less than 5% in Han Chinese populations. A total of 763 genetic polymorphisms were then excluded based on linkage disequilibrium (LD) pruning and genotyping feasibility via Sequenom MassARRAY. For LD pruning, we only included one genetic variant with the lowest log-rank p-value when the pairwise r2 ≥ 0.8 within one gene. Eight SNPs could not be genotyped via Sequenom MassARRAY due to the presence of highly homologous DNA sequences in human genome. In particular, one of the important steps during Sequenom MassARRAY is to amplify a DNA fragment containing the candidate SNP using PCR. However, specific primers cannot be designed for PCR due to highly homologous DNA sequences around the SNP that exist in other gene loci of the human genome. This screening finally resulted in a total of 60 eQTLs SNPs associated with OS of the TCGA LIHC patients (Table 1).
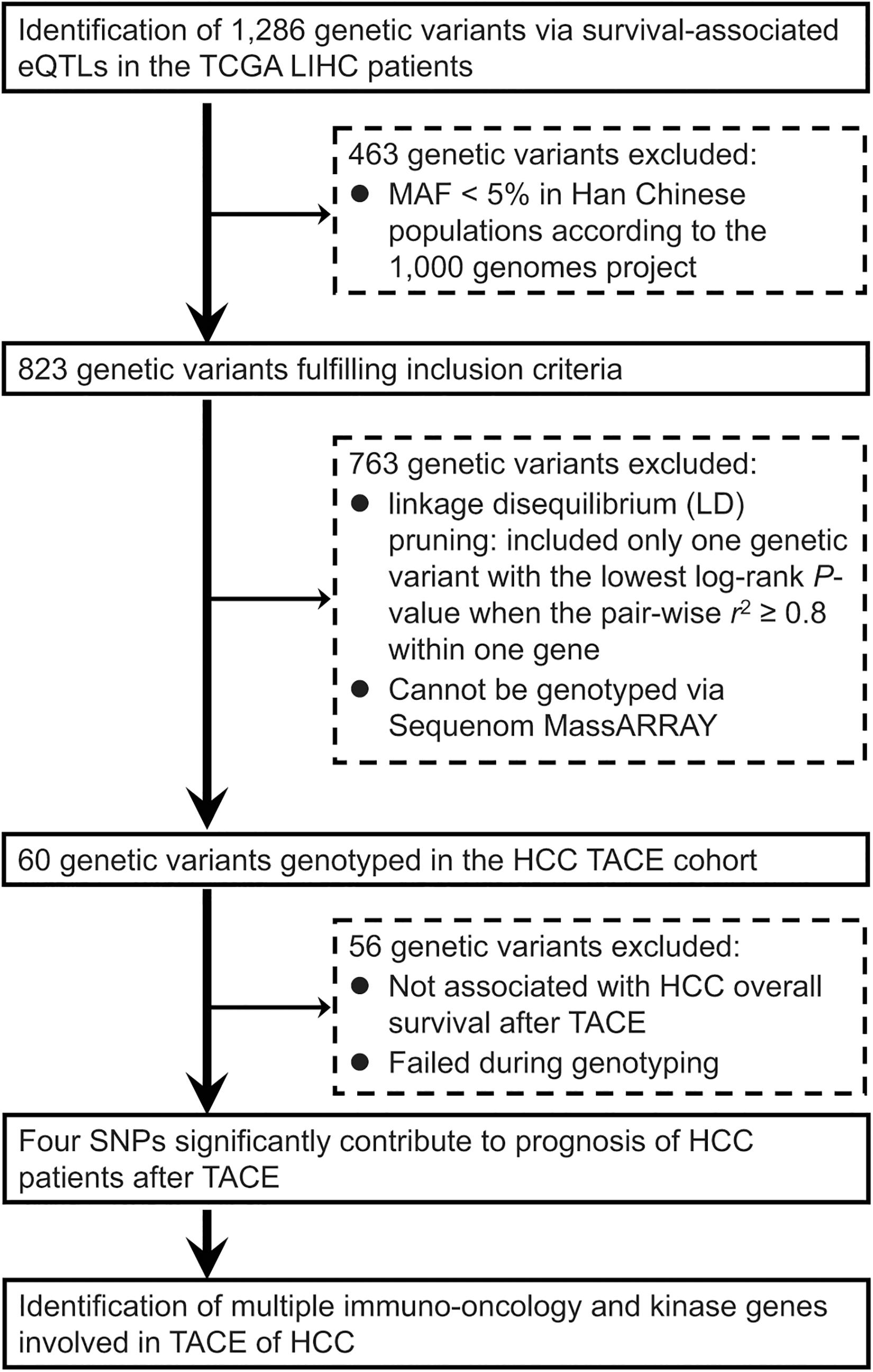
Figure 1. Flowchart of an integrative functional genomics methodology to identify survival-related expression quantitative trait locus (eQTL) genetic variants involved in the transarterial chemoembolization (TACE) treatment of hepatocellular carcinoma (HCC) across the whole genome.
Characteristics of HCC Patient Undergoing TACE
The median age of HCC patients treated with TACE was 56 years (range, 25–81 years). There were 232 males (85.0%), 75 smokers (27.5%), 77 drinkers, 169 individuals with hepatitis history (61.9%), and 18 patients with HCC family history (6.6%). Among these patients, 72 (26.4%) had stage III diseases and 201 (73.6%) had stage IV diseases. At the final analysis, all patients died and the median OS time was 20 months (range, 1–105 months).
Effects of eQTLs Genetic Polymorphisms on OS of TACE-Treated HCC Patients
Among 60 candidate SNPs, only four genetic variants (rs12574873, rs12513391, rs34597395, and rs35624901) are significantly associated with OS of HCC patients treated with TACE (rs12574873: log-rank p = 0.018, unadjusted Cox p = 0.005; rs12513391: log-rank p = 2.3 × 10–6, unadjusted Cox p = 1.0 × 10–5; rs34597395: log-rank p = 0.005, unadjusted Cox p = 0.019; rs35624901: log-rank p = 2.8 × 10–5, unadjusted Cox p = 1.9 × 10–5) (Table 2). As shown in Figure 2A, TACE-treated HCC patients with the rs12574873 CA or AA genotype had a lower survival than the cases with the CC genotype (log-rank p = 0.018). Similarly, the HCC cases with the rs12513391 CT or TT genotype had a shorter survival time than the patients carrying the CC genotype (log-rank p < 0.001) after TACE treatment (Figure 2B). Although there were no obvious differences for OS time between rs34597395 CC and CA genotypes, Kaplan–Meier survival estimates indicated that the rs34597395 AA genotype carriers had a lower survival than the HCC cases carrying the CC or CA genotype after TACE (log-rank p = 0.005) (Figure 2C). Moreover, TACE-treated HCC patients with the rs35624901 Deletion.Deletion (Del.Del) genotype had markedly prolonged survival time than the individuals carrying the TT or T.Del genotype (log-rank p < 0.001) (Figure 2D).
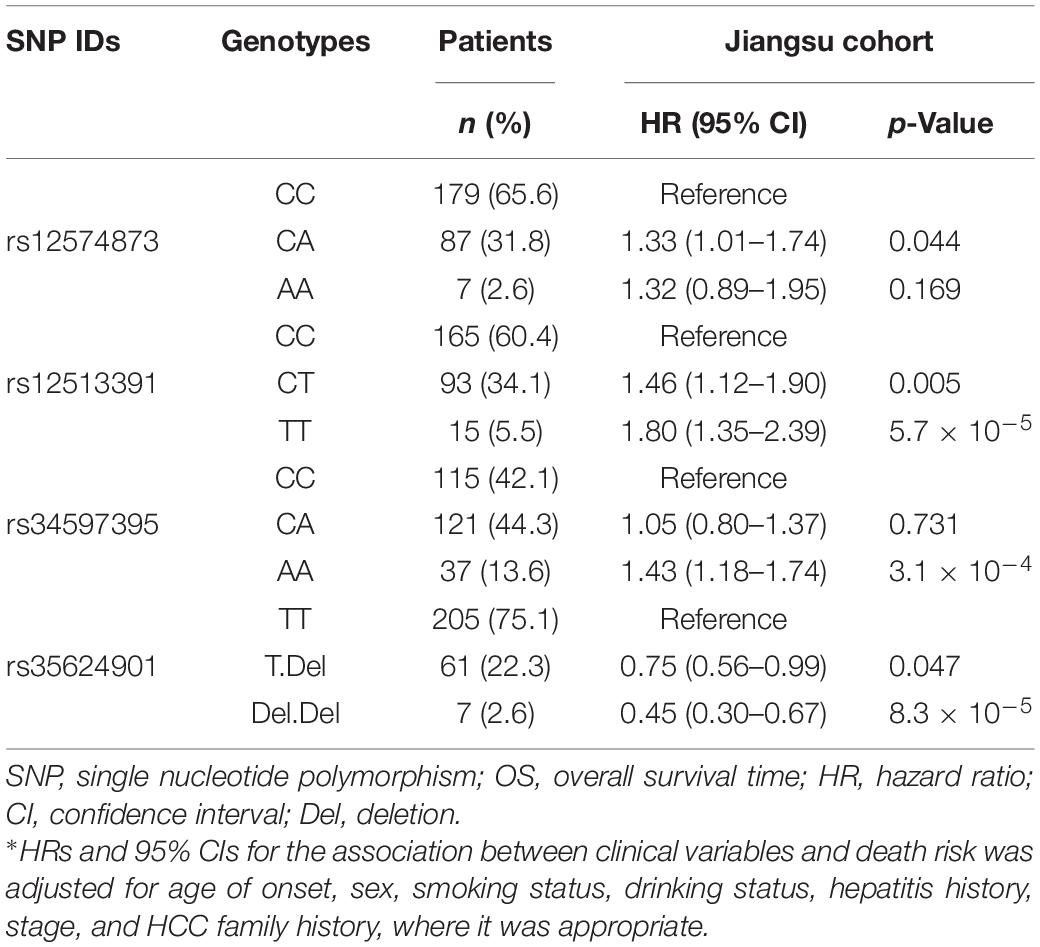
Table 2. Multivariate Cox-regression analyses of PancanQTL-identified genetic variants for OS in the Jiangsu cohort.
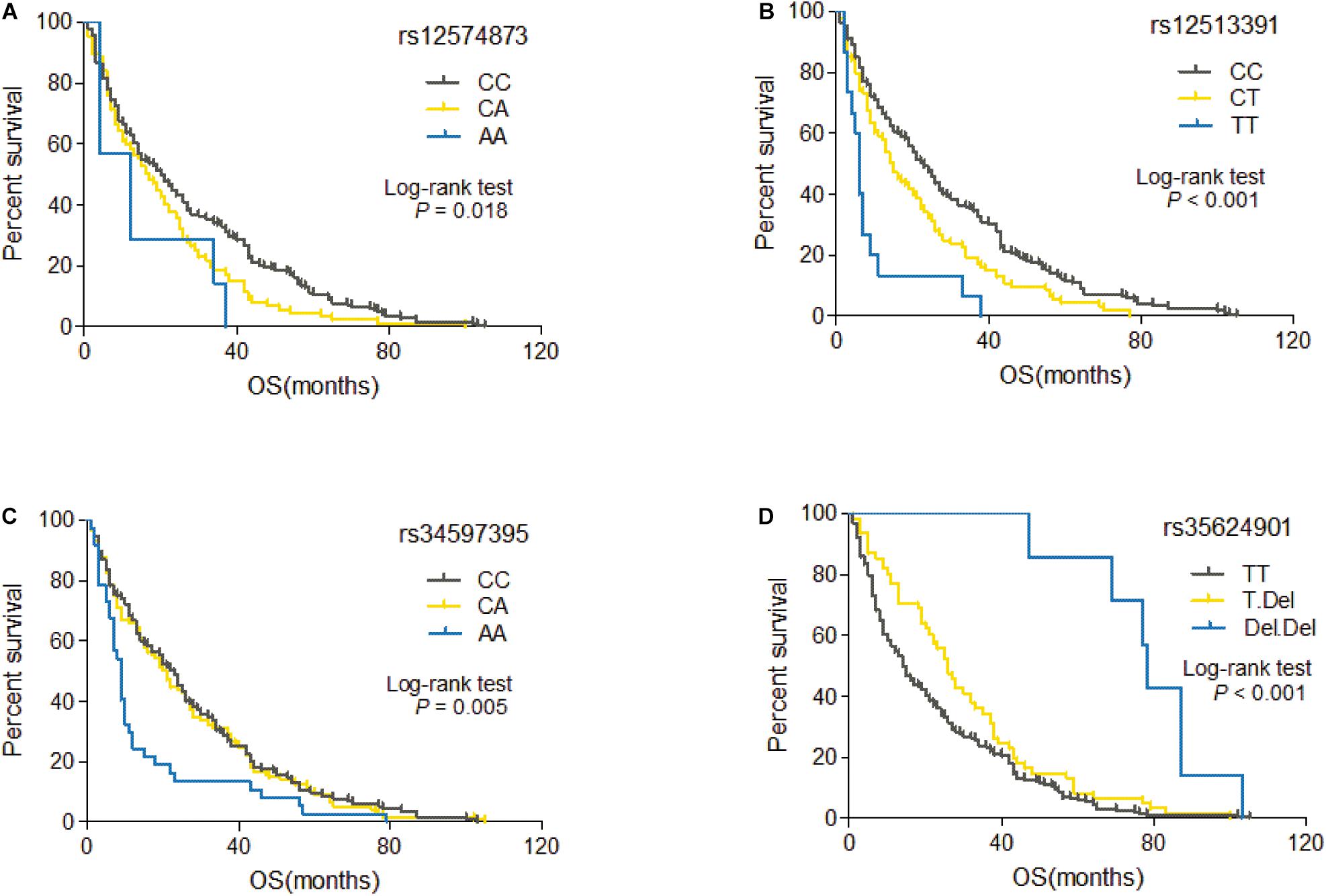
Figure 2. Kaplan–Meier curve of OS for TACE-treated HCC patients with different genotypes of rs12574873 (A), rs12513391 (B), rs34597395 (C), or rs35624901 (D).
Multivariate Cox proportional hazards model showed that the rs12574873, rs12513391, and rs34597395 SNPs were significantly associated with increased death risk of TACE-treated HCC patients, and rs35624901 markedly contributed to decreased death risk (Table 2). Compared with the CC genotype, the rs12574873 CA genotype was significantly associated with elevated death risk (HR = 1.33, 95% CI = 1.01–1.74, p = 0.044). The rs12513391 CT or TT genotype was significantly associated with increased death risk after TACE treatment compared with the CC genotype (the CT genotype: HR = 1.46, 95% CI = 1.12–1.90, p = 0.005; the TT genotype: HR = 1.80, 95% CI = 1.35–2.39, p = 5.7 × 10–5). Similarly, TACE-treated HCC patients with the rs34597395 AA genotype had a 43% increased death risk compared with patients carrying the CC genotype (95% CI = 1.18–1.74, p = 3.1 × 10–4). On the contrary, the rs35624901 T.Del or Del.Del genotype carriers had a 25% or 55% decreased death risk compared with individuals with the GG genotype after TACE therapy (95% CI = 0.56–0.99, p = 0.047; 95% CI = 0.30–0.67, p = 8.3 × 10–5).
Impacts of Four eQTLs SNPs on Gene Expression and OS of TCGA LIHC Patients
For eQTLs analyses, the rs12574873 AA genotype is evidently associated with increased ENDOD1 gene expression in TCGA LIHC patients compared to the CC or CA genotype (p = 3.9 × 10–5) (Figure 3A and Table 3). Interestingly, in these patients, the rs12513391 TT genotype is associated with both decreased MAN2A1 gene expression and elevated TMEM232 gene expression compared to the TT or CT genotype (for MAN2A1: p = 2.8 × 10–5; for TMEM232: p = 6.6 × 10–5) (Figure 3B and Table 3). Moreover, compared to the CC or CA genotype, the rs34597395 AA genotype is significantly associated with decreased PAK4 gene expression in LIHC patients (p = 1.0 × 10–8) (Figure 3C and Table 3). On the contrary, there is obviously increased RAG1 gene expression in the rs35624901 Del.Del genotype carriers compared with that in individuals with the TT or T.Del genotype (p = 1.6 × 10–4) (Figure 3D and Table 3).
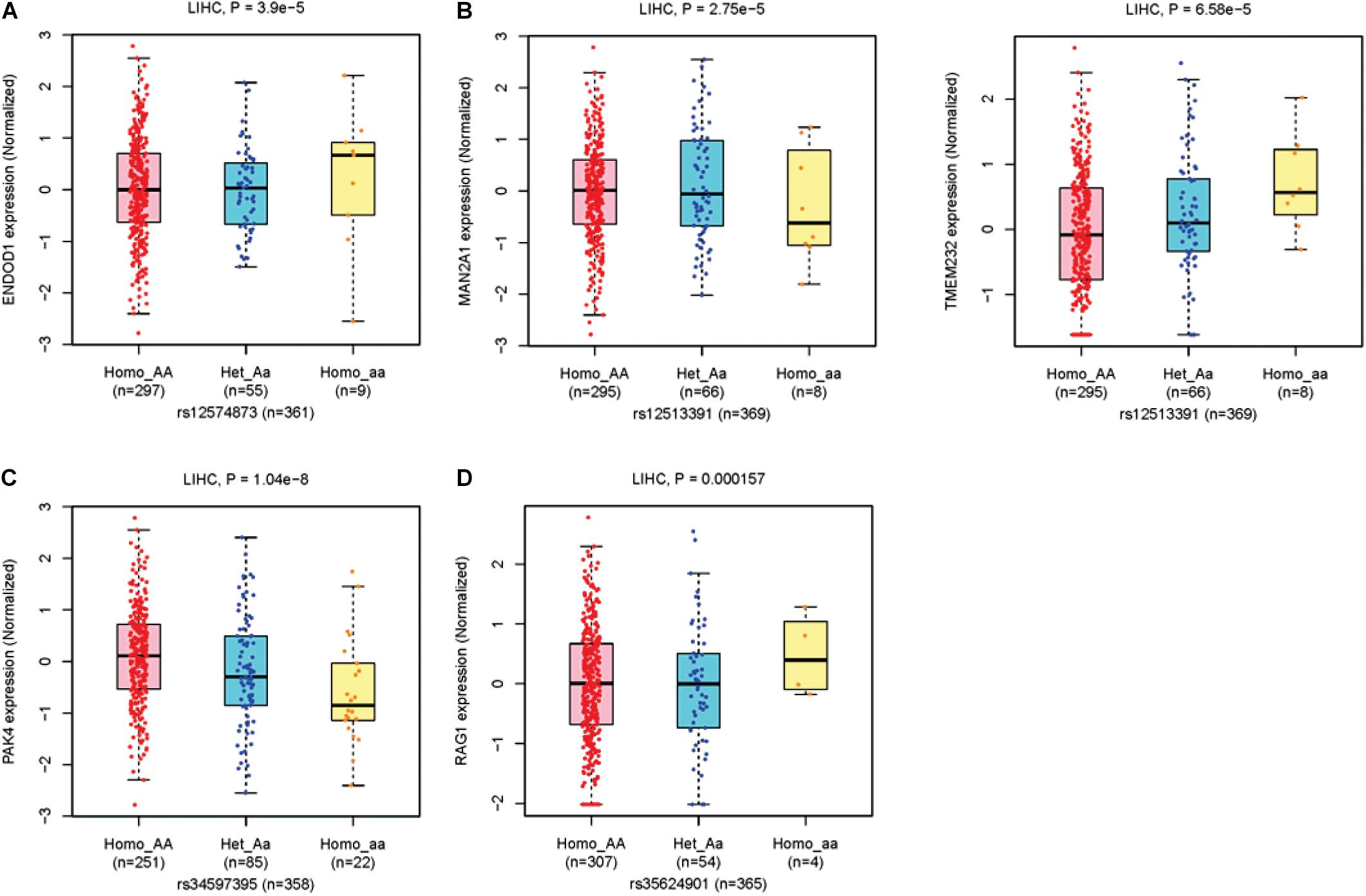
Figure 3. The boxplots of TCGA LIHC eQTLs. (A) The ENDOD1 expression levels grouped by different genotypes of rs12574873. (B) The MAN2A1 (left panel) and TMEM232 (right panel) expression levels grouped by different genotypes of rs12513391. (C) The PAK4 expression levels grouped by different genotypes of rs34597395. (D) The RAG1 expression levels grouped by different genotypes of rs35624901.
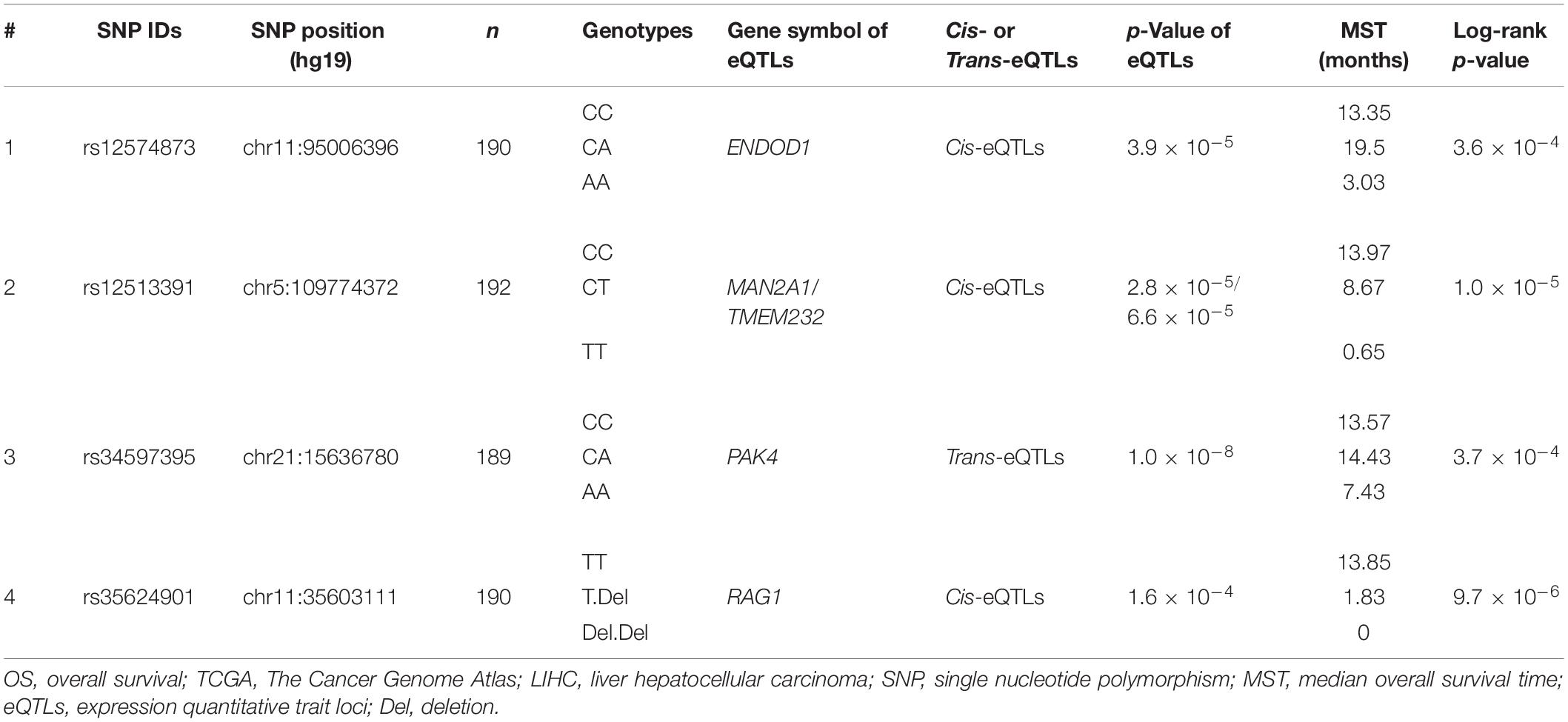
Table 3. PancanQTL-identified genetic variants significantly associated with OS of TCGA LIHC patients.
Evident associations between these four genetic variants and OS of TCGA LIHC patients are also observed (Table 3). The median overall survival time (MST) of LIHC patients carrying the rs12574873 CC, CA, or AA genotype is 13.35, 19.5, or 3.03 months (log-rank p = 3.6 × 10–4). Similarly, MST of individuals with the rs12513391CC, CT, or TT genotype is 13.97, 8.67, or 0.65 months (log-rank p = 1.0 × 10–5). There is also markedly short OS time for HCC cases with the rs34597395 AA genotype compared to patients with the CC or CA genotype (MST AA vs. CC: 7.43 vs. 13.57 months; MST AA vs. CA: 7.43 vs. 14.43 months). Additionally, the TCGA LIHC patients carrying the T.Del genotype have a lower survival than the cases with the TT genotype (log-rank p = 9.7 × 10–6).
Discussion
Hepatocellular carcinoma is a common kind of malignancy worldwide, especially in Asia. It remains difficult to cure HCC since patients were frequently diagnosed at advanced tumor stages (Llovet et al., 2016; Ferlay et al., 2019). Currently, there is no single treatment applicable to all HCC cases. TACE has been proven to be an effective palliative treatment for unresectable HCC (Llovet et al., 2002; Lo et al., 2002). In Asian patients, TACE resulted in a marked tumor response, and the 1-year survival was significantly better in the TACE group than in the control group (57 vs. 32%) (Lo et al., 2002). However, there were still 43% unresectable HCC patients who had progressive disease during TACE treatment in the first year (Lo et al., 2002). This suggests that further studies are required to identify biomarkers for patient-tailored therapy targeting individual genetic background correlated to the prognosis of HCC patients who received TACE.
Through a genome-wide eQTL strategy, we successfully identified four gene expression-related genetic variants which are significantly associated with OS of unresectable HCC patients treated with TACE. For instance, the rs35624901 Del.Del genotype is correlated to higher RAG1 expression levels as well as prolonged OS of TACE-treated HCC patients. RAG1 is critical to initiate the V(D)J recombination which ensures appropriate assembly of antigen receptor genes in the correct cell lineage and in the proper developmental order (Schatz and Ji, 2011). Loss-of-function of RAG1 due to nonsense mutations or gene knock-out abrogates T- and B-lymphocyte receptor formation prevents the maturation of T- and B-lymphocytes and leads to severe combined immunodeficiency (SCID; Schatz and Ji, 2011). RAG1 plays a critical role in HCC development. Upon treatment of the chemical carcinogen diethylnitrosamine (DEN), progression of hepatic tumors was strikingly enhanced in T-/B cell-deficient Rag1(-/-) mice (Schneider et al., 2012). Rag1(-/-) mice showed markedly enhanced growth in the number and mass of hepatic tumors (Schneider et al., 2012). Consistently, we observed that TACE-treated HCC patients with low RAG1 expression due to carrying the TT or T.Del genotype showed shorter OS compared to the Del.Del genotype carriers. This is biologically plausible. It has been reported that TACE did not significantly modify numbers of tumor-infiltrating lymphocytes (TILs) in HCC tissues as compared with the untreated condition in patients who received surgery (Craciun et al., 2020). Therefore, TACE-treated HCC patients with low RAG1 expression may show low antitumor immune responses, rapid tumor progression, and thus, short OS time.
We enrolled 232 males (85.0%) in the current HCC TACE cohort. Consistently, we observed more male HCC patients in the two most famous randomized controlled trials on the role of TACE in HCC treatment (Llovet et al., 2002; Lo et al., 2002). In the study, for the first time, it was indicated that TACE improved the survival of stringently selected patients with unresectable HCC; there were 30 males (81%) in the Embolization group, 32 males (80%) in the Chemoembolization group, and 23 males (66%) in the Control group (Llovet et al., 2002). Similarly, in the Asian randomized controlled trial, there were 36 males (90%) in the Chemoembolization group and 34 males (87%) in the Control group (Lo et al., 2002).
In summary, our results reported the first evidence supporting the prognostic value of four eQTL genetic polymorphisms in HCC patients who received TACE therapy. Importantly, our integrative functional genomics indicated that T-/B-cell development-related RAG1 may impact TACE efficiency via regulating antitumor immunity in HCC, and RAG1 may be a target to improve therapeutic strategy for unresectable HCC patients.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
The studies involving human participants were reviewed and approved by this study was approved by the Institutional Review Board of Huaian No. 2 Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
NZ designed the study. YX performed the experiments. NZ, YX, and LZha performed data analyses. MY, LZho, BW, TW, and JZ were responsible for the collection of specimens. NZ and LZha supervised the project. NZ and YX contributed to drafting the manuscript. All authors have read and approved the final submitted manuscript.
Funding
This study was supported by the Program of Science and Technology for the youth innovation team in universities of Shandong Province (2020KJL001) and the National Natural Science Foundation of China (82173070, 31871306, 82103291, and 82172825).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Craciun, L., de Wind, R., Demetter, P., Lucidi, V., Bohlok, A., Michiels, S., et al. (2020). Retrospective analysis of the immunogenic effects of intra-arterial locoregional therapies in hepatocellular carcinoma: a rationale for combining selective internal radiation therapy (SIRT) and immunotherapy. BMC Cancer 20:135. doi: 10.1186/s12885-020-6613-1
Craig, P., Young, S., and Golzarian, J. (2019). Current trends in the treatment of hepatocellular carcinoma with transarterial embolization: variability in technical aspects. Cardiovasc. Intervent. Radiol. 42, 1322–1328. doi: 10.1007/s00270-019-02232-7
Ferlay, J., Colombet, M., Soerjomataram, I., Mathers, C., Parkin, D. M., Pineros, M., et al. (2019). Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 144, 1941–1953. doi: 10.1002/ijc.31937
Gong, J., Mei, S., Liu, C., Xiang, Y., Ye, Y., Zhang, Z., et al. (2018). PancanQTL: systematic identification of cis-eQTLs and trans-eQTLs in 33 cancer types. Nucleic Acids Res. 46, D971–D976.
Grundberg, E., Small, K. S., Hedman, A. K., Nica, A. C., Buil, A., Keildson, S., et al. (2012). Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44, 1084–1089.
Hindorff, L. A., Sethupathy, P., Junkins, H. A., Ramos, E. M., Mehta, J. P., Collins, F. S., et al. (2009). Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U. S. A. 106, 9362–9367. doi: 10.1073/pnas.0903103106
Kumar, V., Kato, N., Urabe, Y., Takahashi, A., Muroyama, R., Hosono, N., et al. (2011). Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat. Genet. 43, 455–458. doi: 10.1038/ng.809
Li, S., Qian, J., Yang, Y., Zhao, W., Dai, J., Bei, J. X., et al. (2012). GWAS identifies novel susceptibility loci on 6p21.32 and 21q21.3 for hepatocellular carcinoma in chronic hepatitis B virus carriers. PLoS Genet. 8:e1002791. doi: 10.1371/journal.pgen.1002791
Li, Y., Zhang, N., Zhang, L., Song, Y., Liu, J., Yu, J., et al. (2020). Oncogene HSPH1 modulated by the rs2280059 genetic variant diminishes EGFR-TKIs efficiency in advanced lung adenocarcinoma. Carcinogenesis 41, 1195–1202. doi: 10.1093/carcin/bgaa069
Li, Z., Song, Y., Xu, Y., Shen, Y., Zhang, N., Yang, M., et al. (2020). Identification of Leukocyte telomere length-related genetic variants contributing to predisposition of Esophageal Squamous Cell Carcinoma. J. Cancer 11, 5025–5031.
Liu, J., Wang, D., Zhou, J., Wang, L., Zhang, N., Zhou, L., et al. (2021). N6-methyladenosine reader YTHDC2 and eraser FTO may determine hepatocellular carcinoma prognoses after transarterial chemoembolization. Arch. Toxicol. 95, 1621–1629.
Liu, X., Yu, L., Han, C., Lu, S., Zhu, G., Su, H., et al. (2016). Polymorphisms of HLA-DQB1 predict survival of hepatitis B virus-related hepatocellular carcinoma patients receiving hepatic resection. Clin. Res. Hepatol. Gastroenterol. 40, 739–747.
Llovet, J. M., Real, M. I., Montana, X., Planas, R., Coll, S., Aponte, J., et al. (2002). Arterial embolisation or chemoembolisation versus symptomatic treatment in patients with unresectable hepatocellular carcinoma: a randomised controlled trial. Lancet 359, 1734–1739. doi: 10.1016/s0140-6736(02)08649-x
Llovet, J. M., Zucman-Rossi, J., Pikarsky, E., Sangro, B., Schwartz, M., Sherman, M., et al. (2016). Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2:16018.
Lo, C. M., Ngan, H., Tso, W. K., Liu, C. L., Lam, C. M., Poon, R. T., et al. (2002). Randomized controlled trial of transarterial lipiodol chemoembolization for unresectable hepatocellular carcinoma. Hepatology 35, 1164–1171. doi: 10.1053/jhep.2002.33156
Miki, D., Ochi, H., Hayes, C. N., Abe, H., Yoshima, T., Aikata, H., et al. (2011). Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers. Nat. Genet. 43, 797–800.
Qiu, G. P., and Liu, J. (2016). MicroRNA gene polymorphisms in evaluating therapeutic efficacy after transcatheter arterial chemoembolization for primary hepatocellular carcinoma. Genet. Test. Mol. Biomarkers 20, 579–586. doi: 10.1089/gtmb.2016.0073
Schatz, D. G., and Ji, Y. (2011). Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 11, 251–263. doi: 10.1038/nri2941
Schneider, C., Teufel, A., Yevsa, T., Staib, F., Hohmeyer, A., Walenda, G., et al. (2012). Adaptive immunity suppresses formation and progression of diethylnitrosamine-induced liver cancer. Gut 61, 1733–1743. doi: 10.1136/gutjnl-2011-301116
Tang, Q., Zhao, Y., Wang, Y., and Wei, M. (2014). A genetic variant (rs17251221) in the calcium-sensing receptor relates to hepatocellular carcinoma susceptibility and clinical outcome treated by transcatheter hepatic arterial chemoembolization (TACE) therapy. Med. Oncol. 31:267.
Westra, H. J., Peters, M. J., Esko, T., Yaghootkar, H., Schurmann, C., Kettunen, J., et al. (2013). Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243.
Wu, Y. S., Bao, D. K., Dai, J. Y., Chen, C., Zhang, H. X., Yang, Y., et al. (2015). Polymorphisms in genes of the de novo lipogenesis pathway and overall survival of hepatocellular carcinoma patients undergoing transarterial chemoembolization. Asian Pac. J. Cancer Prev. 16, 1051–1056.
Yu, X., Ge, N., Guo, X., Shen, S., Liang, J., Huang, X., et al. (2014). Genetic variants in the EPCAM gene is associated with the prognosis of transarterial chemoembolization treated hepatocellular carcinoma with portal vein tumor thrombus. PLoS One 9:e93416. doi: 10.1371/journal.pone.0093416
Yuan, P., Wang, S., Zhou, F., Wan, S., Yang, Y., Huang, X., et al. (2014). Functional polymorphisms in the NPAS2 gene are associated with overall survival in transcatheter arterial chemoembolization-treated hepatocellular carcinoma patients. Cancer Sci. 105, 825–832. doi: 10.1111/cas.12428
Zhang, H., Zhai, Y., Hu, Z., Wu, C., Qian, J., Jia, W., et al. (2010). Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat. Genet. 42, 755–758. doi: 10.1038/ng.638
Zhao, B., Lu, J., Yin, J., Liu, H., Guo, X., Yang, Y., et al. (2012). A functional polymorphism in PER3 gene is associated with prognosis in hepatocellular carcinoma. Liver Int. 32, 1451–1459. doi: 10.1111/j.1478-3231.2012.02849.x
Keywords: RAG1, hepatocellular carcinoma, TACE, eQTL, TCGA, genetic polymorphism
Citation: Xu Y, Wang T, Zeng J, Wang B, Zhou L, Yang M, Zhang L and Zhang N (2021) Integrative Functional Genomics Implicated the Key T-/B-Cell Deficiency Regulator RAG1 in Transarterial Chemoembolization of Hepatocellular Carcinoma. Front. Cell Dev. Biol. 9:720791. doi: 10.3389/fcell.2021.720791
Received: 05 June 2021; Accepted: 30 August 2021;
Published: 27 September 2021.
Edited by:
Andrea Casadei Gardini, Vita-Salute San Raffaele University, ItalyReviewed by:
Cheng-Maw Ho, National Taiwan University, TaiwanSabahattin Cömertpay, Kahramanmaraş Sütçü Ýmam University, Turkey
Copyright © 2021 Xu, Wang, Zeng, Wang, Zhou, Yang, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nasha Zhang, d293bnNldmFAMTI2LmNvbQ==; Li Zhang, bHV6aWdhbmdAMTYzLmNvbQ==
†Lead contact
 Yeyang Xu
Yeyang Xu Teng Wang1
Teng Wang1 Liqing Zhou
Liqing Zhou Ming Yang
Ming Yang Li Zhang
Li Zhang Nasha Zhang
Nasha Zhang