
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 14 September 2021
Sec. Molecular and Cellular Pathology
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.720688
This article is part of the Research Topic Identification of New Molecular Mechanisms of Bone Disease View all 14 articles
Saul–Wilson syndrome (SWS) is a rare, skeletal dysplasia with progeroid appearance and primordial dwarfism. It is caused by a heterozygous, dominant variant (p.G516R) in COG4, a subunit of the conserved oligomeric Golgi (COG) complex involved in intracellular vesicular transport. Our previous work has shown the intracellular disturbances caused by this mutation; however, the pathological mechanism of SWS needs further investigation. We sought to understand the molecular mechanism of specific aspects of the SWS phenotype by analyzing SWS-derived fibroblasts and zebrafish embryos expressing this dominant variant. SWS fibroblasts accumulate glypicans, a group of heparan sulfate proteoglycans (HSPGs) critical for growth and bone development through multiple signaling pathways. Consistently, we find that glypicans are increased in zebrafish embryos expressing the COG4p.G516R variant. These animals show phenotypes consistent with convergent extension (CE) defects during gastrulation, shortened body length, and malformed jaw cartilage chondrocyte intercalation at larval stages. Since non-canonical Wnt signaling was shown in zebrafish to be related to the regulation of these processes by glypican 4, we assessed wnt levels and found a selective increase of wnt4 transcripts in the presence of COG4p.G516R. Moreover, overexpression of wnt4 mRNA phenocopies these developmental defects. LGK974, an inhibitor of Wnt signaling, corrects the shortened body length at low concentrations but amplifies it at slightly higher concentrations. WNT4 and the non-canonical Wnt signaling component phospho-JNK are also elevated in cultured SWS-derived fibroblasts. Similar results from SWS cell lines and zebrafish point to altered non-canonical Wnt signaling as one possible mechanism underlying SWS pathology.
Saul–Wilson syndrome (SWS) is a rare skeletal dysplasia characterized by profound short stature and distinctive craniofacial features such as prominent forehead, prominent eyes, and micrognathia (Saul and Wilson, 1990; Ferreira et al., 2020). Recently, we defined a specific heterozygous COG4 substitution (p.G516R) as the molecular basis of this rare form of primordial dwarfism (Ferreira et al., 2018). COG4 is one of the eight subunits of the conserved oligomeric Golgi (COG) complex regulating protein trafficking and Golgi homeostasis (Ungar et al., 2002). Biallelic pathogenic variants in COG4 and other COG subunits cause multiple human congenital disorders of glycosylation (CDGs) (D’Souza et al., 2020). COG4-CDG individuals have a very severe, usually lethal, phenotype with dysmorphia, neurological and intellectual disabilities, and altered N-glycosylation with an almost total loss of COG4 (Reynders et al., 2009; Ng et al., 2011). However, SWS subjects show very different features, since their N-glycans and their intellectual and neurological features appear normal (Ferreira et al., 2018). At the cellular level, the COG4p.G516R variant disrupted protein trafficking by accelerating brefeldin-A (BFA)-induced retrograde transport and delaying anterograde transport, causing the collapse of the Golgi stacks. This interrupted bidirectional trafficking between the ER and the Golgi and altered decorin, a proteoglycan (Ferreira et al., 2018), indicate that modified proteoglycans may be involved in the pathogenesis of SWS.
Proteoglycans play critical roles in multiple cell processes at the cellular, tissue, and organismal levels, and their deficiencies cause bone and connective tissue disorders (Iozzo and Schaefer, 2015; Paganini et al., 2019). Several proteoglycan deficiencies have been studied in zebrafish, a powerful vertebrate model for studying CDGs and skeletal disorders, with some showing a shortened body axis (Zoeller et al., 2009; Cline et al., 2012; Lee et al., 2020; Tonelli et al., 2020; Dietrich et al., 2021). Besides a body axis defect, morpholino-mediated knockdown of zebrafish decorin (dcn) displayed relatively severe defects in body curvature associated with a curved or not fully extended tail (Zoeller et al., 2009). Defects in glypicans, a group of heparan sulfate proteoglycans (HSPGs), can cause abnormal skull and skeletal dysplasia in both humans (Simpson–Golabi–Behmel syndrome: glypican 3, GPC3; Keipert syndrome: glypican 4, GPC4) and zebrafish (Knypek: Gpc4) (LeClair et al., 2009; Tenorio et al., 2014). Interestingly, studies of knypek (kny/gpc4) mutant zebrafish demonstrated that Gpc4 deficiency causes chondrocyte stacking and intercalation defects in Meckel’s cartilage, which is not seen in dcn morphants or chondroitin sulfate proteoglycan 4 (cspg4)-deficient zebrafish (Topczewski et al., 2001; Zoeller et al., 2009; Hu et al., 2012; Lee et al., 2020). kny adult zebrafish also show craniofacial defects including a smaller head, domed skull, and shorter jawbones, reminiscent of some of the clinical features of SWS individuals (Topczewski et al., 2001; LeClair et al., 2009). Studies also found that optimized expression of gpc4 can suppress the defects caused by Wnt11f2 (formerly known as Wnt11/Silberblick; Postlethwait et al., 2019) deficiency, indicating the role of Gpc4 in the Wnt signaling pathway, probably as a Wnt coreceptor (LeClair et al., 2009). It is worth noting that both absence and high expression of gpc4 lead to the loss of ability to suppress Wnt11f2 deficiency (Topczewski et al., 2001). Presumably, abnormalities arise when either these receptors or ligands are outside the optimal ratio. Taken together, the similarity between kny mutant zebrafish craniofacial defects and some of the SWS individuals’ clinical features encouraged us to use zebrafish as a vertebrate model to explore the underlying pathological mechanism of SWS.
The zebrafish Cog4 is 72% identical to human COG4, and the amino acid corresponding to the SWS mutation site is conserved across multiple vertebrate species.1 Zebrafish that lack the Cog4 protein show phenotypes consistent with the clinical features of COG4-CDG individuals, including defective synthesis of N- and O-linked glycans and decreased glycosphingolipid complexity (Clement et al., 2019). SWS individuals show very different features compared to COG4-CDG individuals, and SWS cells show accelerated BFA-induced retrograde trafficking in contrast to COG4-CDG cells. Considering these facts, a zebrafish model for the SWS-specific variant is highly desired to investigate phenotypic features and the possible pathogenesis of this heterozygous mutation in COG4.
In this study, we utilize SWS-derived fibroblasts and a zebrafish system to test a specific heterozygous COG4 substitution (p.G516R), which is causal for SWS. We assessed a broader category of proteoglycans and found a consistent increase of glypican level in SWS-derived fibroblasts and zebrafish expressing human COG4p.G516R variant. Further studies on non-canonical Wnts revealed that the presence of COG4p.G516R specifically elevated the wnt4 transcript, but not wnt5a, wnt5b, or wnt11f2. Overexpression of wnt4 phenocopies the COG4p.G516R zebrafish, and the Wnt inhibitor LGK974 suppresses the defects caused by the expression of COG4p.G516R. These findings suggest that disrupted Wnt signaling is one possible mechanism underlying the pathogenesis of SWS.
As one of the major components of extracellular matrix (ECM) and cell membrane proteins, proteoglycans comprise a large, heterogeneous group including HSPGs and chondroitin sulfate proteoglycans (CSPGs). Decorin is a predominant proteoglycan in human skin, covalently linked with one glycosaminoglycan chain (GAG), which requires normal function of the Golgi for its posttranslational modification. Our results showing abnormal decorin modification encouraged us to study other proteoglycans in SWS cells. As a first step in determining which proteoglycan may change, we analyzed the core proteins of HSPGs using an antibody (3G10) against HS-stubs (a heparan sulfate neo-epitope) that appear only after heparinase III digestion. As shown in Figure 1A, glypicans, syndecan 1, and syndecan 2 were the most prevalent cell surface HSPGs in dermal fibroblasts. Compared to syndecans, glypicans showed consistent increases in all three SWS cell lines with an average twofold elevation (Figure 1B). We performed qPCR to assess whether transcript abundance is also altered. There are six glypicans in humans, and among those, glypicans 1, 4, and 6 are present in multiple tissues, while glypicans 3 and 5 are restricted to the ovary and brain. Glypican 2 is specifically expressed in the nervous system during embryonic development (Thul and Lindskog, 2018; Guo et al., 2020). As seen in Figure 1C, glypicans 1, 4, 5, and 6 were detectable in dermal fibroblasts; however, we did not observe a significant change in any of their transcript abundance (Figure 1D). We hypothesize that this increase of glypicans results from SWS COG4-dependent abnormal trafficking or turnover, instead of transcriptional regulation. A similar strategy was applied to study CSPGs. Chondroitinase ABC was used to digest CSPGs followed by immunoblotting against the ΔDi-6S. There was a general decrease of CSPG core proteins in three out of four SWS-derived cell lines (Supplementary Figure 1). Considering that glypicans show the most prominent difference between controls and SWS-derived cells and the phenotypes of gpc4 mutant zebrafish, we focused on glypicans in our study.
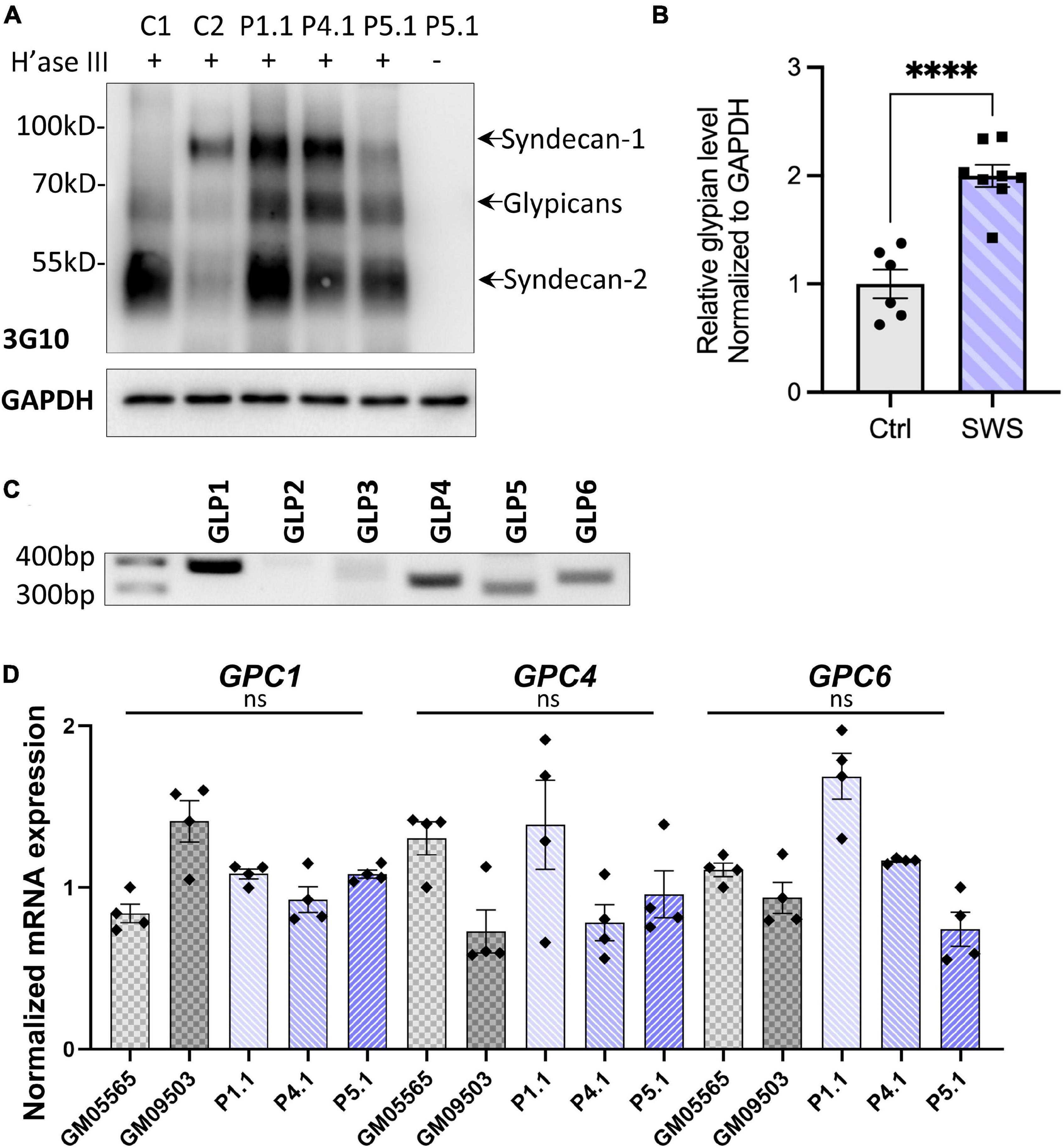
Figure 1. SWS-derived fibroblasts show altered HSPGs and glypicans after heparinase III (H’ase III) digestion. (A) Western blotting of ΔHS-stub using 3G10 antibody following heparinase III digestion of three SWS-derived fibroblasts and two control fibroblasts. C1 and C2 are control fibroblasts. C1, GM08429; C2, GM08680. P1.1, P4.1, and P5.1 are SWS-derived fibroblasts. (B) Quantitation assay of glypican band density in (A) and two other replicates. The data are presented as mean ± SEM. An unpaired two-tailed t-test was used. ****p < 0.0001. (C) Agarose gel of six human glypicans after reverse-transcript PCR. (D) qPCR of three dominant glypicans in SWS-derived fibroblasts. The relative glypican level was normalized to GAPDH. The graphs represent the 2–ΔΔCt values. Experiments were performed in triplicates with similar results.
To study the impact of the SWS variant on skeletal development, we used zebrafish as a vertebrate model. Since SWS is a dominant disorder, we overexpressed the human SWS allele in developing zebrafish embryos. Human COG4WT and COG4p.G516R mRNAs or DNA constructs, as shown in Figure 2A, were injected into one-cell-stage embryos. The presence of human COG4 in zebrafish was confirmed by an antibody specifically recognizing human COG4 protein (Figures 2B,C). Lacking a zebrafish Cog4 antibody makes it impossible to determine the relative expression level of COG4p.G516R; therefore, we include COG4WT as an injected control to ensure comparable expression of COG4p.G516R in zebrafish. Both COG4WT and COG4p.G516R were expressed at a very similar level (Figures 2B,C), and no adverse effects were seen in embryos expressing COG4WT compared to uninjected siblings.
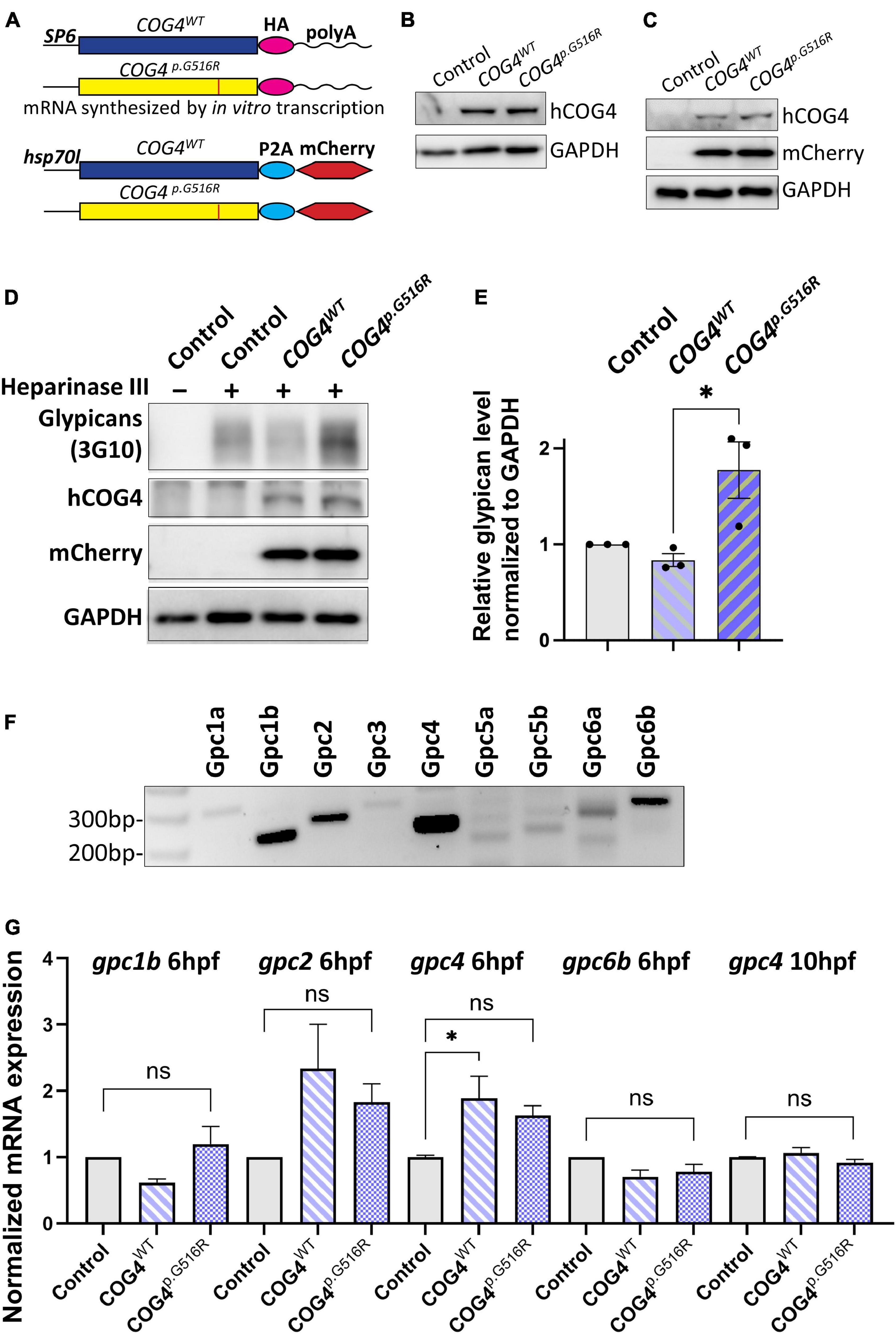
Figure 2. Expression of human COG4p.G516R in zebrafish increases the protein level of glypicans. (A–C) Expression of human COG4WT and COG4p.G516R in zebrafish after mRNA or DNA injection. (A) The scheme of COG4 constructs for in vitro transcription (top) and DNA injection (bottom). (B) Western blot at 24 hpf to detect the presence of COG4 after mRNA injection. (C) Western blot at 48 hpf to detect COG4 after DNA injection; heat shock was performed at 24 hpf for 2 h at 38°C. (D–G) Glypican analysis in zebrafish. (D) Western blotting of ΔHS-stub using 3G10 antibody following heparinase III digestion of control and embryos injected with COG4WT or COG4p.G516R mRNA at 3 dpf. (E) Quantitation assay of glypican band density in (D) and two more replicates. The data are presented as mean ± SEM. An unpaired two-tailed t-test was used. ∗p < 0.05. (F) mRNA expression of glypican genes by RT-PCR using cDNA from control embryos at 6 hpf. (G) qPCR analyses of highly expressed glypicans in control and zebrafish embryos injected with human COG4WT or COG4p.G516R mRNA. The relative glypican level was normalized to β-actin. The graphs represent the 2–ΔΔCt values. One-way ANOVA with Tukey’s multiple comparison tests was applied. ns, not significant; ∗p < 0.05. Experiments were performed in triplicates with similar results.
We first checked glypican proteins using the same strategy as in SWS-derived cells. Interestingly, we found increased glypicans in embryos expressing COG4p.G516R but not COG4WT at 3 dpf (days post-fertilization) (Figures 2D,E), consistent with the observation in SWS-derived fibroblasts. Zebrafish have 10
glypicans expressed at different developmental stages (Gupta and Brand, 2013). At 6 hpf (hours post-fertilization), we detected five glypicans, namely, Gpc1b, Gpc2, Gpc4, Gpc6a, and Gpc6b, by RT-PCR (Figure 2F), followed by qPCR to compare their transcript levels. We found that the gpc2 and gpc4 transcript levels are elevated in both COG4WT and COG4p.G516R embryos compared to the uninjected control, but there is no significant difference between COG4WT and COG4p.G516R (Figure 2G). At 10 hpf, the transcript level of endogenous gpc4 fell to a level comparable to that of the control, with no distinction between COG4WT and COG4p.G516R (Figure 2G). The gpc2 mRNA level at 10 hpf was barely detectable.
We examined zebrafish body length at different stages to assess the developmental phenotypes caused by COG4p.G516R expression. At the end of gastrulation, embryos expressing the COG4p.G516R variant exhibit shorter axis extension (Figure 3A), suggesting an abnormal convergent extension (CE) movement (Topczewski et al., 2001). Embryos expressing COG4WT developed normally (Figure 3A). We tracked these injected embryos at later stages and found that expression of COG4p.G516R causes a shortened anterior–posterior (AP) body axis (Figure 3B) by an average of 18% at 3 dpf and 10% at 6 dpf (Figure 3C).

Figure 3. Expression of human COG4p.G516R impairs zebrafish early development and chondrocyte intercalation. (A,B) Expression of COG4p.G516R in zebrafish causes gastrulation defects and shortened body axis. (A) Lateral view of representative embryos. Anterior to the top. Compared to the control construct hsp70l:mCherry and COG4WT, embryos expressing COG4p.G516R show an axis extension defect at 10 hpf. White arrow points to the head region, and the black arrow points to the tailbud. Expression of COG4p.G516R mRNA causes similar results. (C) Graphs show the measured body length of each group at 3 and 6 dpf. The data are presented as mean ± SD. One-way ANOVA with Tukey’s multiple comparison tests was applied. ****p < 0.0001; ns, not significant. (D) Expression of COG4p.G516G causes craniofacial abnormalities. Ventral view of representative Meckel’s cartilage of zebrafish larvae at 4 dpf after WGA staining and imaged by a confocal microscope. Dotted circular lines highlight chondrocyte cell shape and their relative configuration with each other. (E) Graphical representation of the length-to-width ratio of chondrocytes in the region of interest, yellow box in (D). Individual cell length-to-width ratio was measured in three representative Meckel’s cartilage images of each group. The data are presented as mean ± SEM. One-way ANOVA with Tukey’s multiple comparison tests was applied. ****p < 0.0001; ns, not significant. Experiments were performed in triplicates with similar results.
To investigate whether the presence of COG4p.G516R in zebrafish impacts chondrocyte development, zebrafish at 4 dpf were stained with wheat germ agglutinin (WGA), a lectin binding to glycoproteins in cartilage ECM to visualize chondrocyte morphology. In embryos expressing COG4p.G516R, Meckel’s cartilage was deformed, shown as defects in chondrocyte stacking and elongation (Figure 3D). Compared to COG4WT embryos, more rounded chondrocytes were present in Meckel’s cartilage (Figure 3E). These defects can be observed as late as 7 dpf, as shown by Alcian blue staining (Supplementary Figure 2). Chondrocyte stacking and intercalation problems further confirmed that abnormal glypican levels could be one of the pathogenetic mechanisms involved in SWS. Other phenotypes seen in COG4p.G516R-injected embryos also include abnormal, stunted fin, and cyclopia (Supplementary Figures 3A,B).
Glypican 4 plays an essential role in gastrulation movements and contributes to craniofacial morphogenesis probably through planar cell polarity (PCP)/non-canonical Wnt signaling (Topczewski et al., 2001; Sisson et al., 2015). Therefore, we assessed the expression of non-canonical wnts by qPCR and their spatial–temporal transcription pattern by whole-mount in situ hybridization. We found that COG4p.G516R mRNA-injected embryos contain more wnt4 compared to controls (Figure 4A). In contrast, no significant changes were detected in a few other non-canonical wnts, such as wnt5b and wnt11f2. Also, the upregulation of wnt4 shows a dose-dependent response to the amount of COG4p.G516R mRNA injected (Supplementary Figure 4). This elevated transcript level of wnt4 was further confirmed by whole-mount in situ hybridization (Figure 4B). At 6 hpf, wnt4 was not detectable in either control or COG4WT mRNA-injected embryos, but it was in COG4p.G516R-mRNA injected embryos. At 10 hpf, wnt4 is confined to the hindbrain in control and COG4WT embryos, but COG4p.G516R embryos had both increased and spatially expanded expression of wnt4. The wnt11f2 transcript level did not change, but the distribution pattern was significantly altered. In uninjected control and COG4WT embryos, wnt11f2 was restricted to the dorsal midline. However, its expression pattern is dispersed in COG4p.G516R embryos. No measurable difference was found for β-catenin protein, a marker of canonical Wnt signaling between COG4p.G516R and controls (Supplementary Figure 6).
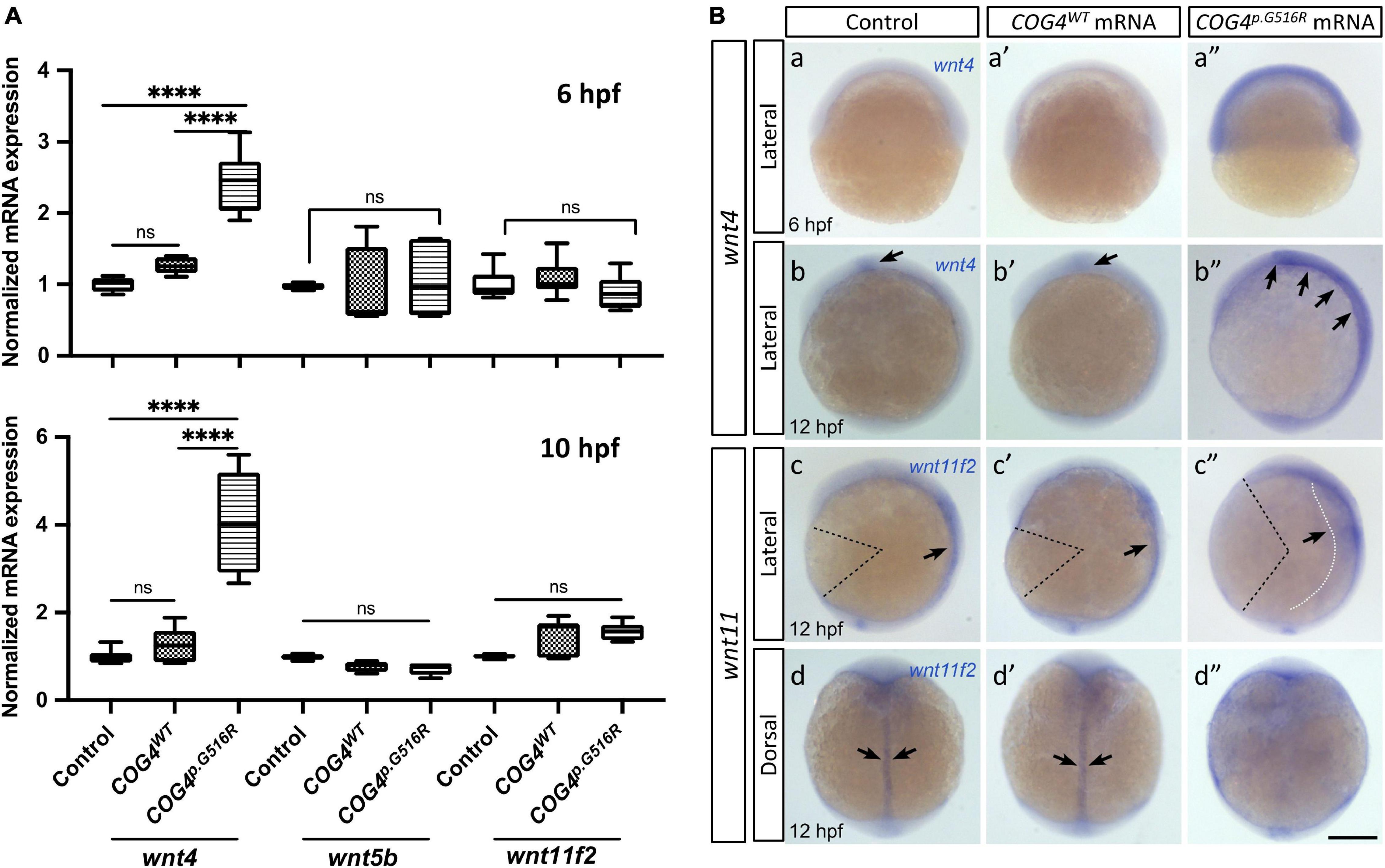
Figure 4. Expression of human COG4p.G516R elevates the wnt4 transcript in zebrafish embryos. (A) Quantitative PCR analyses of selective non-canonical Wnt pathway ligands in zebrafish embryos injected with human COG4WT or COG4p.G516R mRNA. At 6 hpf, wnt4 expression is significantly upregulated in embryos injected with COG4p.G516R mRNA, but not in those injected with COG4WT mRNA. Similar upregulation of wnt4 transcripts is observed at 10 hpf as well. Bar graphs represent average gene expression relative to the housekeeping gene β-actin. The data are presented as mean ± SD. One-way ANOVA with Tukey’s multiple comparison tests was used. ****p < 0.0001; ns, not significant. (B) Representative images of whole-mount in situ hybridization from control and treated embryos. Whole-mount in situ hybridization analyses demonstrate that wnt4 expression is elevated in embryos injected with COG4p.G516R mRNA (a″, at 6 hpf; b″ at 12 hpf), but there are no obvious changes in embryos injected with human COG4WT mRNA (a′ and b′). (b, b’) Arrows point to the restricted wnt4 expression domain in the hindbrain of zebrafish embryos at 12 hpf; (b″) arrows point to expanded expression of wnt4 in the hindbrain region. At 12 hpf, wnt11f2 expression intensity is not significantly changed in embryos injected with COG4p.G516R mRNA (c″, d″), compared to either control siblings (c, d) or human COG4WT mRNA-injected embryos (c′, d′). The dotted black color lines landmark the degree of angle between the head and tail, which is much more increased in embryos injected with COG4p.G516R mRNA, suggesting defects in extension movement during gastrulation. Arrows in c, c′ and d, d″ indicate that wnt11f2 is restricted to the dorsal midline; however, its expression pattern is dispersed in COG4p.G516R mRNA-injected embryos, suggesting that the convergence movement is impaired (c″, white dotted line and arrow; d″, there is no clear midline expression). Scale bars: 200 μm. Experiments were performed in triplicates with similar results.
In zebrafish, wnt4 overexpression has been studied at earlier embryonic stages and was found to inhibit cell movements without altering cell fates (Ungar et al., 1995; Topczewski et al., 2001). Thus, we evaluated the phenotypes of wnt4 overexpression at a later embryonic stage. At 4 dpf, wnt4 mRNA-injected embryos showed shortened body length in response to the dosage of wnt4 injected (Figure 5A). Interestingly, we also found abnormal chondrocyte stacking in Meckel’s cartilage (Figure 5B) and cyclopia (Figure 5C). These data show that overexpression of wnt4 phenocopies shortened body length and malformed chondrocyte intercalation in COG4p.G516R-injected zebrafish.
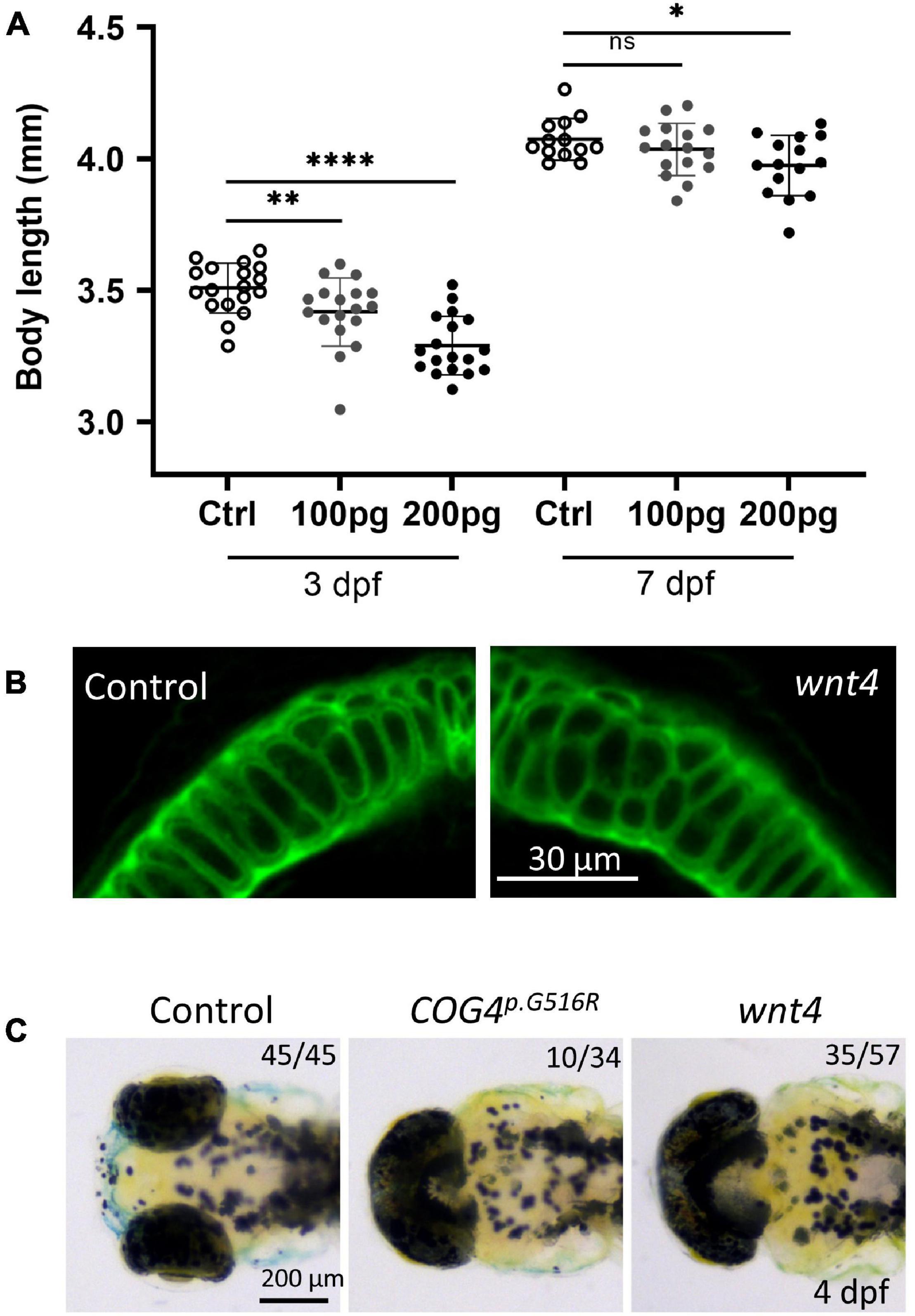
Figure 5. Overexpression of wnt4 phenocopies zebrafish embryos injected with the COG4p.G516G mRNA. (A) Graphs show the measured body length of each group after wnt4 mRNA injection at 3 and 7 dpf. The data are presented as mean ± SD. An unpaired two-tailed t-test was used. ****p < 0.0001; ∗∗p < 0.01; ∗p < 0.05; ns, not significant. (B) Overexpression of wnt4 causes abnormal chondrocyte stacking and intercalation at Meckel’s cartilage. Ventral view of representative Meckel’s cartilage of zebrafish larvae after wnt4 injection at 4 dpf following WGA staining and imaged by a confocal microscope. (C) Overexpression of wnt4 causes cyclopia as expression of the COG4p.G516G variant. Dorsal view of representative images of cyclopia compared to control. Two hundred picograms of wnt4 or COG4p.G516G mRNA was used per embryo. Experiments were performed in triplicates with similar results.
We hypothesize that the increased abundance of wnt4 contributes to the pathogenetic mechanism for SWS. Therefore, reducing Wnt4 activity may suppress the developmental defects caused by the presence of COG4p.G516R. LGK974 is a pharmacological inhibitor of WNT porcupine O-acyltransferase (PORCN), which affects palmitoylation and secretion of Wnts. After optimizing the treatment procedure (Figure 6A), we found that 0.05–0.2 μM of LGK974 significantly shortened the body length of control siblings (Supplementary Figure 5) without causing optic cup morphogenesis and shorter tail induced by high concentrations of LGK974 (Eckert et al., 2019). A 24-h incubation with low concentrations (0.05 and 0.1 μM) of LGK974 restored the shortened body length caused by COG4p.G516R expression at 4 dpf (Figure 6B). Higher concentrations (0.15 and 0.2 μM) of LGK974 further shortened the body length in COG4p.G516R injected zebrafish, showing that there is a narrow optimal range for Wnt activity. These data suggest that the imbalance of Wnt signaling may contribute to SWS pathogenesis. We also examined the chondrocyte morphology in Meckel’s cartilage and found that 0.05 and 0.1 μM of LGK974 are sufficient to suppress the deformed cartilage caused by COG4p.G516R expression. Up to 0.1 μM LGK974 does not cause significant defects in control chondrocytes (Figures 6C,D).
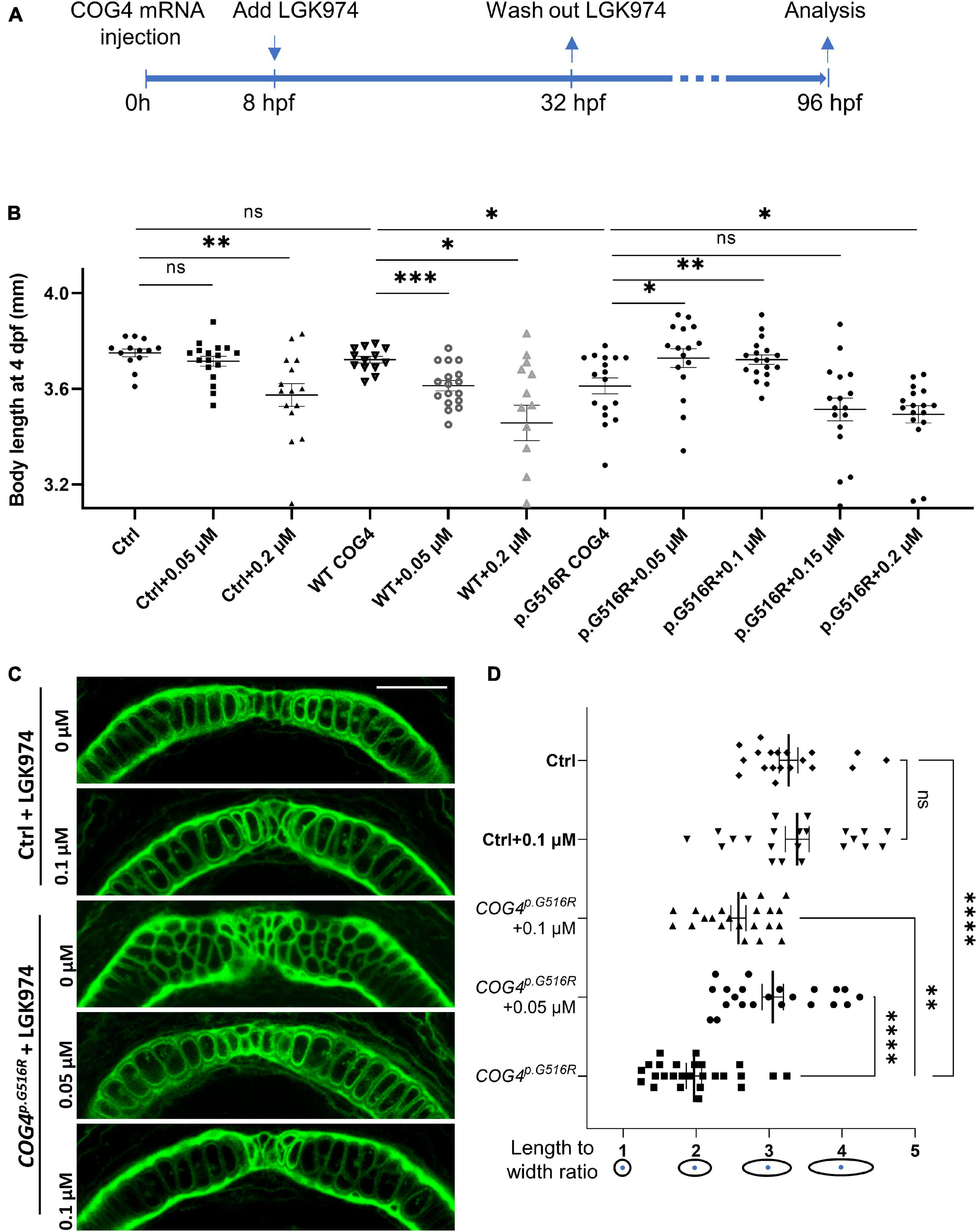
Figure 6. LGK974 treatment suppresses shortened body length and chondrocyte defect caused by of COG4p.G516G expression in zebrafish. (A) Scheme of the LGK974 treatment procedure. (B) Graphs show the measured body length of each group at 4 dpf. The data are presented as mean ± SEM. An unpaired two-tailed t-test was used. ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; ns, not significant. (C) Ventral view of representative Meckel’s cartilage of control and COG4p.G516G-injected embryos with or without LGK974 treatment at 4 dpf following WGA staining and imaged by a confocal microscope. (D) Graphs show the length-to-width ratio of chondrocytes in Meckel’s cartilage in (C) and two more representative Meckel’s cartilage images of each group. The data are presented as mean ± SEM. One-way ANOVA with Tukey’s multiple comparison tests was applied. ****p < 0.0001; ∗∗p < 0.01; ns, not significant. Experiments were performed in two biological replicates with similar results.
To assess whether a similar mechanism was at play in human cells, we assayed the transcript abundance of WNT4 and other non-canonical WNTs in SWS individuals’ fibroblasts. Interestingly, both WNT4 transcript and protein levels were increased in SWS individual fibroblasts (Figures 7A–C). Neither WNT5a, WNT5b, nor WNT11 showed consistent significant differences in three SWS-derived cell lines (Figure 7A). We further detected downstream gene expression in the non-canonical pathway and found increased JNK phosphorylation (pJNK) in SWS cells compared to controls (Figures 7B,C), indicating an elevated non-canonical Wnt signaling. We further examined Wnt signaling markers in zebrafish and found that p-Rac1/cdc42, a marker for non-canonical Wnt pathway, was elevated in embryos expressing COG4p.G516R, but not those of COG4WT. A similar increase of p-Rac1/cdc42 was also seen in wnt4 overexpression embryos (Supplementary Figure 6). β-Catenin, a marker for the canonical Wnt pathway, did not change (Figure 7B).
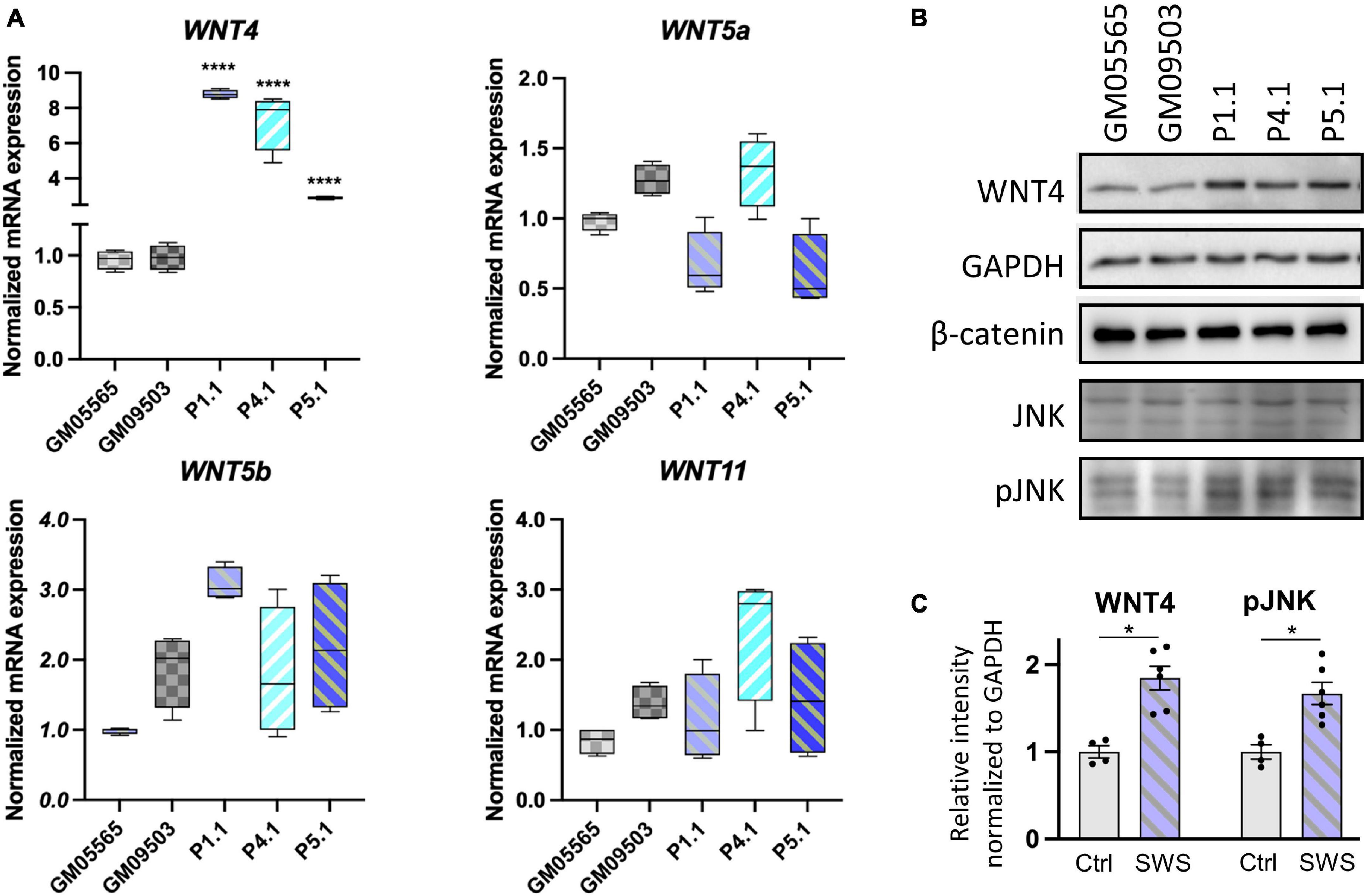
Figure 7. Non-canonical WNTs and related component level in SWS-derived cells. (A) qPCR analyses of selective non-canonical Wnt pathway components in SWS-derived fibroblasts. GAPDH was used as an internal control. The graphs represent the 2–ΔΔCt values. Unpaired two-tailed t-test was applied for comparison of each SWS fibroblast with two controls. ****p < 0.0001. Experiments were performed in triplicates with similar results. (B) Western blotting of a few Wnt pathway components. GM05565 and GM09503 are control fibroblasts. P1.1, P4.1, and P5.1 are SWS-derived fibroblasts. (C) Quantitation assay of WNT4 and pJNK band density in (B) and the other replicates. The data are presented as mean ± SEM. An unpaired two-tailed t-test was used. ∗p < 0.05.
In this paper, we utilized SWS-derived fibroblasts and zebrafish, a vertebrate model to study SWS. In SWS-derived cells, the specific p.G516R amino acid substitution in COG4 selectively affected proteoglycans instead of global glycosylation. Increased glypicans were seen in both SWS-derived cells and COG4p.G516R-injected zebrafish embryos. Glypicans are glycosylphosphatidylinositol-anchored proteins (GPI-APs) that consist of a conserved core glycan, phosphatidylinositol, glycan side chains, and a protein moiety (Kinoshita, 2020). A recent study found that subunits of the COG complex affect proteoglycan turnover and GAG chain polymerization (Adusumalli et al., 2021). Studies also found that glycosylation changes of a cell surface protein could affect the protein’s half-life (Ohtsubo et al., 2005). Therefore, the increased glypicans are probably due to impaired protein trafficking or turnover caused by COG4p.G516R rather than changes in glypican transcript abundance. In previous gpc4 zebrafish studies, Topczewski demonstrated that Gpc4 regulates cellular movements during gastrulation by potentiating Wnt11f2 signaling and chondrocyte behavior independent of core Wnt pathway molecules (Topczewski et al., 2001; Sisson et al., 2015). Interestingly, low expression of gpc4 suppressed Wnt11f2 defects, but high overexpression of gpc4 inhibited the rescue, indicating a requirement for a fine balance between Gpc4 and Wnt11f2 to ensure normal development. This is also true for COG4 function, since more than twofold accumulation of COG4 leads to similar protein trafficking defects as seen in COG4-deficient cells (Reynders et al., 2009). Therefore, COG4WT is an essential control in this zebrafish study to confirm that the expression of COG4 is not excessive to impair its normal function.
Multiple studies have shown the role of glypicans in regulating Wnt signaling. In zebrafish, Gpc4 has been reported as a positive modulator of non-canonical Wnt signaling during zebrafish gastrulation (Topczewski et al., 2001). Studies in both Drosophila and Xenopus found that glypicans can modulate the distribution of Wnt through binding the palmitoleate on Wnts and thus contribute to the release of signaling-competent Wnt (Franch-Marro et al., 2005; Yan et al., 2009; McGough et al., 2020). Among two groups of glypicans, Dally and Dally-like (Dlp) subfamilies, only Dlp has this palmitoleate-binding activity, which mainly includes Gpc4 and Gpc6 (McGough et al., 2020). Studies in chick embryos demonstrated that Gpc4 in the neural crest enhances Wnt1/Wnt3a signaling and wnt11 expression on the dorsomedial lip (Serralbo and Marcelle, 2014). Our data also suggest the role of glypicans in regulating Wnt signaling. We hypothesize that the accumulation of glypican(s) could induce the transcriptional upregulation of wnt4. However, lacking a specific Gpc4 antibody in zebrafish makes it difficult to confirm whether Gpc4 is the major player in causing these phenotypes seen in COG4p.G516R-injected embryos. Simply overexpressing gpc4 in zebrafish seems to show no significant defects (LeClair et al., 2009). We speculate that the abnormalities seen in COG4p.G516R-injected embryos are due to disrupted glypican trafficking or turnover rather than changes in glypican expression. Further studies on how glypican and Wnt4 crosstalk is yet to be demonstrated. More studies are ongoing in our lab to further address these questions. Whereas glypicans are the most impaired proteoglycans caused by the heterozygous variant in COG4 (p.G516R), we could not rule out that other proteoglycans may also contribute to the pathogenesis of SWS. Although knockdown of dcn and cspg4 in zebrafish does not show chondrocyte intercalation problems, they may cause a shorter body axis or other types of cartilage malformations (Zoeller et al., 2009; Lee et al., 2020).
Besides increased glypicans, it is interesting that we also found an elevated wnt4 transcript in SWS-derived cells and COG4p.G516R-injected zebrafish embryos. Studies have shown that the cell surface receptor could directly regulate WNT4 expression in chondrocytes (Dennis et al., 2020). Therefore, we hypothesize that the increased WNT4 is probably a compensatory transcriptional response to increased glypicans. Wnts are a group of secreted glycoproteins involved in cell–cell signaling (Niehrs, 2012; Willert and Nusse, 2012). To date, 22 Wnts have been identified in vertebrates (the Wnt homepage: http://web.stanford.edu/group/nusselab/cgi-bin/wnt/). Among those, Wnt4 has shown important roles in embryonic development, skeletal/bone regeneration, and sex determination with additional developmental defects in a cell-specific and tissue-specific manner (Matsui et al., 2005; Bernard and Harley, 2007; Chang et al., 2007; Rao et al., 2019; Yang et al., 2020). Most importantly, Wnt4 plays a pivotal role in regulating chondrocyte differentiation. In chick limb studies, Wnt4 expression is found in joint cells and cartilage, and its misexpression accelerates maturation of chondrocytes, causing shortened long bones (Kawakami et al., 1999; Hartmann and Tabin, 2000; Church et al., 2002). An in vitro study also found that Wnt4 blocks the initiation of chondrogenesis and accelerates terminal chondrocyte differentiation (Church et al., 2002). Misexpression of Zwnt4 (zebrafish wnt4) and Xwnt4 (Xenopus wnt4) in zebrafish causes shortened trunk and tail (Ungar et al., 1995) and abnormal chondrocyte intercalation in Meckel’s cartilage (our data), confirming Wnt4 is involved in regulation of chondrocyte behavior, like gpc4 and wnt5b (Sisson et al., 2015). In mice, Wnt4 defects led to short-limb dwarfism (Dennis et al., 2020). Most relevant, conditional expression of Wnt4 in mice also causes dwarfism with small skeletons, dome-shaped skulls, and small jaws (Lee and Behringer, 2007). Both Wnt4 defects and overexpression cause dwarfism, indicating Wnt4 functions within a narrow range for normal development, and bidirectional disturbance causes developmental abnormalities.
LGK974 is a PORCN inhibitor that blocks PORCN-dependent palmitoylation and secretion of WNT family ligands (Liu et al., 2013). Currently, it is being evaluated as an anticancer agent against a broad range of diseases associated with deviant Wnt signaling (Liu et al., 2013; Zhang and Lum, 2016). LGK974 has been used to inhibit canonical and non-canonical signaling in multiple studies as a WNT modifier (Han et al., 2017; Katoh, 2017; Lutze et al., 2019). In our study, titrating LGK974 allowed us to regulate the amount of Wnt during development. At low concentrations, it suppressed and nearly normalized the shortened body length caused by the presence of COG4p.G516R, probably by inhibiting Wnt4 (or other Wnts) palmitoylation and secretion. This rescue experiment not only supports altered Wnt signaling as one of the possible pathogenesis mechanisms contributing to SWS but also suggests a potential therapeutic strategy for SWS individuals.
In our SWS zebrafish model, the specific heterozygous dominant variant in COG4 (p.G516R) shows elevated glypicans and wnt4, along with developmental defects. Compared to COG4-KO zebrafish (Ferreira et al., 2018; Clement et al., 2019), it is impressive that the single mutation in COG4 is sufficient for shortened body length and abnormal chondrocyte stacking. However, there are still some differences between COG4-KO and COG4p.G516R zebrafish; for example, the single variant in COG4 does not decrease the overall GAG amount shown in COG4-KO zebrafish by Alcian blue staining. COG4p.G516R larvae also show cyclopia and a distinct pectoral fin phenotype compared to COG4-KO zebrafish.
In summary, using SWS-derived fibroblasts and a zebrafish model, we demonstrated that the specific dominant variant COG4p.G516R causes the accumulation of GPI-anchored glypicans, which most likely involves COG4-dependent altered trafficking or turnover of these proteins. In zebrafish, the presence of COG4p.G516R also elevates wnt4 transcript and causes chondrocyte morphogenesis defects, which might explain the short stature and distinctive craniofacial features in SWS individuals. WNT4 and non-canonical Wnt signaling component pJNK are also elevated in cultured SWS-derived fibroblasts. We further demonstrate that the Wnt inhibitor LGK974 could suppress the defects caused by COG4p.G516R in zebrafish. These results from SWS-derived cell lines and zebrafish point to altered non-canonical Wnt signaling as one possible mechanism underlying SWS pathology. How increased COG4p.G516R leads to elevated WNT4 in SWS cells and zebrafish is still unknown. Transgenic and CRISPR-knock-in SWS zebrafish lines are under development to address these issues.
Dermal primary fibroblasts derived from healthy controls (GM08429, GM08680, GM03349, GM05565, and GM09503) were obtained from Coriell Institute for Medical Research (Camden, NJ). Each SWS-derived fibroblast line was obtained by the referring clinician and grown via a clinical laboratory service and then sent to us with consent through an approved IRB.
Fibroblasts were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) containing 1 g/L glucose supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% antibiotic–antimycotic (Life Technologies, Carlsbad, CA, United States).
All zebrafish experiments were performed in accordance with the protocols approved by SBP IACUC. Zebrafish were maintained under standard laboratory conditions at 28.5°C. Embryos were staged according to Kimmel et al. (1995).
For glypicans analysis, fibroblasts were treated with 5 mU/ml heparinase III in a serum-free medium for 1 h at 37°C. Zebrafish larvae were homogenized and treated with 25 mU heparinase III in H buffer (20 mM Tris-HCl, pH 7.0, 0.1 mg/ml BSA, and 4 mM CaCl2) for 1.5 h at 37°C. Samples were harvested using SDS lysis buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, and 10% glycerol) supplemented with protease and phosphatase inhibitors (Sigma-Aldrich) as previously described (Ferreira et al., 2018). For analysis of PCP pathway components, 2 × 104 fibroblast cells were seeded in six-well plates and harvested after 2 days using SDS lysis buffer. Equal amounts of denatured proteins were separated via SDS–polyacrylamide gel electrophoresis followed by transfer and antibody inoculation as described previously (Tambe et al., 2019). Antibodies used were Δ-heparan sulfate (AMSBIO, F69-3G10), chondroitin 6 sulfate (Millipore, MAB2035), GAPDH (Invitrogen, MA5-15738), WNT4 (R&D, MAB4751), mCherry (Rockland, 600-401-P16), COG4 (provided by Dr. Daniel Ungar, University of York, United Kingdom), β-catenin (Santa Cruz, sc-7963), JNK (sc-7345), pJNK (sc-293136), and p-Rac1/cdc42 (Cell Signaling, #2461).
Total RNA was extracted from cells or zebrafish embryos using TRIzolTM (Thermo Fisher, 15596081) reagent according to the manufacturer protocol. cDNA was synthesized using a QuantiTect Reverse Transcription Kit (QIAGEN, 205311). qPCR and data analysis were performed as described previously (Tambe et al., 2019). Briefly, primer pairs targeting genes of interest were designed using NCBI Primer-BLAST and available upon request. qPCRs were performed with the PowerUp SYBR®Green PCR Master Mix (Thermo Fisher, A25742). The standardized cycle conditions were applied in Applied Biosystems 7900HT Fast Real-Time PCR System. SDS2.3 software was used to analyze expression data of reference genes. The mRNA levels were normalized to the levels of housekeeping genes, GAPDH for fibroblasts and β-actin for zebrafish, and 2–ΔΔCt values were calculated and compared.
Whole-mount immunofluorescence was performed as previously described (Alexander et al., 1998), using lectin WGA (Vector Laboratories, FL1021) against the cell membrane. Fluorescence images were acquired using an LSM 510 confocal microscope (Zeiss, Germany) with a × 40 water objective. Digital images were processed with Adobe Creative Suites.
Full-length human COG4 was cloned into the pCS2 + vector from plasmid hCOG4-siR-3myc in AAZ6 (a gift from Professor Vladimir V. Lupashin) using an In-Fusion® HD Cloning Kit (TaKaRa Bio, 638909) with primers 5′-ATGGGAACCAAGATGGCGGA-3′, 5′-TTACAGGCGCAG CCTCTTGATAT-3′, 5′-ATCAAGAGGCTGCGCCTGTAATCA AGGCCTCTCGAGCCTCT-3′, and 5′-TCCGCCATCTTGGTT CCCATATTCGAATCGATGGGATCCT-3′. SWS point mutation of G to A was generated using Q5® Site-Directed Mutagenesis Kit (NEB, E0554S) with primers 5′-CATCCAGCGCa GGGTGACAAG-3′ and 5′-TCCTGGAAGGTGGTGGCA-3′. WT COG4 and SWS COG4 mRNAs (COG4WT and COG4p.G516R) were synthesized using the Invitrogen mMESSAGE mMACHINE SP6 Transcription Kit (Thermo Fisher, AM1340) following restriction enzyme NotI digestion and purified using a MEGAclearTM Transcription Clean-Up Kit (Thermo Fisher, AM1908). One hundred picograms of mRNA was injected into each zebrafish embryo unless stated otherwise. As a complementing strategy, Hsp70l:COG4-P2A-mCherry, cryaa:dsRED, and SWS COG4 were generated by cloning the coding sequence for WT COG4 and SWS COG4 downstream of the Hsp70l promoter in the parent vector hsp70l:-LateSV40pA, cryaa:dsRED-RabBGpA, and I-Sce vector (a gift from Dr. Joseph Lancman). I-SceI meganuclease (NEB, R0694) was co-injected with 20 pg of the constructs per embryo.
Full-length zebrafish wnt4 was cloned from cDNA into the pCS2 + vector using an In-Fusion® HD Cloning Kit with the following primers: 5′-CCCATCGATTCGAATATGTCATCG GAGTATTTGATAAGGT-3′, 5′-CTCGAGAGGCCTTGATCA CCGACACGTGTGCAT-3′, 5′-TCAAGGCCTCTCGAGCCT CT-3′, and 5′-ATTCGAATCGATGGGATCCTGCA-3′. wnt4 mRNA synthesis and purification were performed as described above. One hundred picograms of mRNA was injected into each zebrafish embryo unless mentioned otherwise. For antisense RNA probe synthesis, the full length of wnt4 including 3′-UTR was cloned into the pGEM-T easy vector (Promega, A1360) by primers 5′-ATGTCATCGGAGTATTTGATAAGG-3′ and 5′-AGTCTTTGACACAGCATATATTTC-3′ from cDNA. After verifying the insertion direction by sequencing, the antisense RNA probe was then synthesized with SP6 RNA polymerase following ApaI digestion.
Standard whole-mount in situ hybridization was performed as described previously (Thisse and Thisse, 2008). INT/BCIP (175 μg/ml; Roche) was used as alkaline phosphatase substrates. The following molecular markers were used: wnt4 and wnt11f2 (a gift from Dr. Diane Sepich, previously used in Makita et al., 1998; Marlow et al., 2002).
LGK974 (Cayman Chemical, No. 14072) was dissolved in DMSO to make a 10 μM stock. Different concentrations of LGK974 were added to the control or injected zebrafish groups at 8 hpf for 24 h. DMSO of 0.01% was used as a vehicle.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank: NM_015386.3, NM_001040387.1 (https://www.ncbi.nlm.nih.gov/nuccore/NM_015386.3).
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) protocols of Sanford Burnham Prebys Medical Discovery Institute.
Z-JX, X-XZ, PDSD, and HHF conceptualized the project, designed the experiments, and oversaw all studies. Z-JX, X-XZ, MT, and BGN performed the experiments and analyzed the data with HHF. Z-JX and X-XZ prepared the figures. HHF and PDSD contributed to research funding. Z-JX and X-XZ wrote the manuscript with coauthors. All authors reviewed and contributed to editing the manuscript.
This work was supported by the Rocket Fund (HHF), R01DK99551 (HHF), and R01DK124583 (PDSD). Z-JX was a Rocket Fund Fellow.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Heather Flanagan-Steet for providing reagents and guidance and for critical reading of the manuscript. We thank Yu Yamaguchi for critical discussion. We thank Diane Sepich for providing the wnt11f2 DNA construct for in situ probe synthesis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.720688/full#supplementary-material
Supplementary Figure 1 | Western blots of CSPG following chondroitinase ABC digestion to generate CS-stubs. C1 and C2 are control fibroblasts. C1, GM0849; C2, GM0860. P1.1, P4.1, and P5.1 are SWS-derived fibroblasts.
Supplementary Figure 2 | Expression of COG4p.G516R in zebrafish shows abnormal chondrocyte intercalation of Meckel’s cartilage at the late developmental stage. Ventral view of representative Meckel’s cartilage of zebrafish larvae at 7 dpf after Alcian blue staining, imaged by a light dissecting microscope. Dotted circular lines highlight cell shape and their relative configuration with each other.
Supplementary Figure 3 | Expression of COG4p.G516R in zebrafish causes cyclopia, stunted fin, and abnormal ceratohyal cartilage in zebrafish. (A) Expression of COG4p.G516G causes cyclopia in zebrafish. Dorsal view of representative images of cyclopia compared to control and COG4WT. Two hundred picograms of COG4WT or COG4p.G516G mRNA was injected into each embryo. (B) Ventral view of representative zebrafish larvae at 6 dpf after Alcian blue staining, imaged by a light dissecting microscope. Twenty picograms of DNA was injected into each embryo, and heat shock was performed at 24 and 46 hpf for 1 h each at 38°C.
Supplementary Figure 4 | wnt4 upregulation shows a dose-dependent response to COG4p.G516R mRNA. Bar graphs represent quantitative PCR analyses of wnt4 at 6 hpf after injection of different amounts of COG4p.G516R mRNA. wnt4 expression was normalized to β-actin. ****p < 0.0001; ∗∗p < 0.01; ns, not significant.
Supplementary Figure 5 | Dose-dependent response analysis of the Wnt inhibitor LGK974 in zebrafish. (A) Body length at 3 dpf after LGK974 treatment. (B) Representative images after LGK974 treatment at 3 dpf. The data are presented as mean. ****p < 0.0001.
Supplementary Figure 6 | Expression of COG4p.G516R and wnt4 elevated non-canonical Wnt signaling in zebrafish. Samples for Western blots were collected at 6 hpf. The p-Rac1/cdc42 level increased in embryos injected with COG4p.G516R mRNA, but not with COG4WT mRNA. Similar elevation of p-Rac1/cdc42 was also seen in wnt4 overexpression embryos. β-Catenin did not change.
Adusumalli, R., Asheim, H. C., Lupashin, V., Blackburn, J. B., and Prydz, K. (2021). Proteoglycan synthesis in conserved oligomeric Golgi subunit deficient HEK293T cells is affected differently, depending on the lacking subunit. Traffic 22, 230–239. doi: 10.1111/tra.12804
Alexander, J., Stainier, D. Y., and Yelon, D. (1998). Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev. Genet. 22, 288–299. doi: 10.1002/(sici)1520-6408(1998)22:3<288::aid-dvg10>3.0.co;2-2
Bernard, P., and Harley, V. R. (2007). Wnt4 action in gonadal development and sex determination. Int. J. Biochem. Cell Biol. 39, 31–43. doi: 10.1016/j.biocel.2006.06.007
Chang, J., Sonoyama, W., Wang, Z., Jin, Q., Zhang, C., Krebsbach, P. H., et al. (2007). Noncanonical Wnt-4 signaling enhances bone regeneration of mesenchymal stem cells in craniofacial defects through activation of p38 MAPK. J. Biol. Chem. 282, 30938–30948. doi: 10.1074/jbc.m702391200
Church, V., Nohno, T., Linker, C., Marcelle, C., and Francis-West, P. (2002). Wnt regulation of chondrocyte differentiation. J. Cell Sci. 115, 4809–4818. doi: 10.1242/jcs.00152
Clement, A., Blanco-Sanchez, B., Peirce, J. L., and Westerfield, M. (2019). Cog4 is required for protrusion and extension of the epithelium in the developing semicircular canals. Mech. Dev. 155, 1–7. doi: 10.1016/j.mod.2018.09.003
Cline, A., Gao, N., Flanagan-Steet, H., Sharma, V., Rosa, S., Sonon, R., et al. (2012). A zebrafish model of PMM2-CDG reveals altered neurogenesis and a substrate-accumulation mechanism for N-linked glycosylation deficiency. Mol. Biol. Cell 23, 4175–4187. doi: 10.1091/mbc.e12-05-0411
Dennis, E. P., Edwards, S. M., Jackson, R. M., Hartley, C. L., Tsompani, D., Capulli, M., et al. (2020). CRELD2 is a novel LRP1 chaperone that regulates Noncanonical WNT signaling in skeletal development. J. Bone Miner. Res. 35, 1452–1469. doi: 10.1002/jbmr.4010
Dietrich, K., Fiedler, I. A., Kurzyukova, A., Lopez-Delgado, A. C., McGowan, L. M., Geurtzen, K., et al. (2021). Skeletal biology and disease modeling in Zebrafish. J. Bone Miner. Res. 36, 436–458.
D’Souza, Z., Taher, F. S., and Lupashin, V. V. (2020). Golgi inCOGnito: from vesicle tethering to human disease. Biochim. Biophys. Acta Gen. Subj. 1864:129694. doi: 10.1016/j.bbagen.2020.129694
Eckert, P., Knickmeyer, M. D., Schutz, L., Wittbrodt, J., and Heermann, S. (2019). Morphogenesis and axis specification occur in parallel during optic cup and optic fissure formation, differentially modulated by BMP and Wnt. Open Biol. 9:180179. doi: 10.1098/rsob.180179
Ferreira, C. R., Xia, Z. J., Clement, A., Parry, D. A., Davids, M., Taylan, F., et al. (2018). A recurrent de novo heterozygous COG4 substitution leads to saul-wilson syndrome, disrupted vesicular trafficking, and Altered Proteoglycan Glycosylation. Am. J. Hum. Genet. 103, 553–567. doi: 10.1016/j.ajhg.2018.09.003
Ferreira, C. R., Zein, W. M., Huryn, L. A., Merker, A., Berger, S. I., Wilson, W. G., et al. (2020). Defining the clinical phenotype of Saul-Wilson syndrome. Genet. Med. 22, 857–866.
Franch-Marro, X., Marchand, O., Piddini, E., Ricardo, S., Alexandre, C., and Vincent, J. P. (2005). Glypicans shunt the Wingless signal between local signalling and further transport. Development 132, 659–666. doi: 10.1242/dev.01639
Guo, M., Zhang, H., Zheng, J., and Liu, Y. (2020). Glypican-3: a new target for diagnosis and treatment of hepatocellular Carcinoma. J. Cancer 11, 2008–2021. doi: 10.7150/jca.39972
Gupta, M., and Brand, M. (2013). Identification and expression analysis of zebrafish glypicans during embryonic development. PLoS One 8:e80824. doi: 10.1371/journal.pone.0080824
Han, T., Schatoff, E. M., Murphy, C., Zafra, M. P., Wilkinson, J. E., Elemento, O., et al. (2017). R-Spondin chromosome rearrangements drive Wnt-dependent tumour initiation and maintenance in the intestine. Nat. Commun. 8:15945.
Hartmann, C., and Tabin, C. J. (2000). Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development 127, 3141–3159. doi: 10.1242/dev.127.14.3141
Hu, G., Codina, M., and Fisher, S. (2012). Multiple enhancers associated with ACAN suggest highly redundant transcriptional regulation in cartilage. Matrix Biol. 31, 328–337.
Iozzo, R. V., and Schaefer, L. (2015). Proteoglycan form and function: a comprehensive nomenclature of proteoglycans. Matrix Biol. 42, 11–55.
Katoh, M. (2017). Canonical and non-canonical WNT signaling in cancer stem cells and their niches: cellular heterogeneity, omics reprogramming, targeted therapy and tumor plasticity (Review). Int. J. Oncol. 51, 1357–1369. doi: 10.3892/ijo.2017.4129
Kawakami, Y., Wada, N., Nishimatsu, S. I., Ishikawa, T., Noji, S., and Nohno, T. (1999). Involvement of Wnt-5a in chondrogenic pattern formation in the chick limb bud. Dev. Growth Differ. 41, 29–40. doi: 10.1046/j.1440-169x.1999.00402.x
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310.
Kinoshita, T. (2020). Biosynthesis and biology of mammalian GPI-anchored proteins. Open Biol. 10:190290. doi: 10.1098/rsob.190290
LeClair, E. E., Mui, S. R., Huang, A., Topczewska, J. M., and Topczewski, J. (2009). Craniofacial skeletal defects of adult zebrafish Glypican 4 (knypek) mutants. Dev. Dyn. 238, 2550–2563. doi: 10.1002/dvdy.22086
Lee, H. H., and Behringer, R. R. (2007). Conditional expression of Wnt4 during chondrogenesis leads to dwarfism in mice. PLoS One 2:e450. doi: 10.1371/journal.pone.0000450
Lee, Y. H., Kawakami, K., HuangFu, W. C., and Liu, I. H. (2020). Chondroitin sulfate proteoglycan 4 regulates zebrafish body axis organization via Wnt/planar cell polarity pathway. PLoS One 15:e0230943. doi: 10.1371/journal.pone.0230943
Liu, J., Pan, S., Hsieh, M. H., Ng, N., Sun, F., Wang, T., et al. (2013). Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc. Natl. Acad. Sci. U.S.A. 110, 20224–20229.
Lutze, G., Haarmann, A., Demanou Toukam, J. A., Buttler, K., Wilting, J., and Becker, J. (2019). Non-canonical WNT-signaling controls differentiation of lymphatics and extension lymphangiogenesis via RAC and JNK signaling. Sci. Rep. 9:4739.
Makita, R., Mizuno, T., Koshida, S., Kuroiwa, A., and Takeda, H. (1998). Zebrafish wnt11: pattern and regulation of the expression by the yolk cell and No tail activity. Mech. Dev. 71, 165–176. doi: 10.1016/s0925-4773(98)00013-6
Marlow, F., Topczewski, J., Sepich, D., and Solnica-Krezel, L. (2002). Zebrafish Rho kinase 2 acts downstream of Wnt11 to mediate cell polarity and effective convergence and extension movements. Curr. Biol. 12, 876–884. doi: 10.1016/s0960-9822(02)00864-3
Matsui, T., Raya, A., Kawakami, Y., Callol-Massot, C., Capdevila, J., Rodriguez-Esteban, C., et al. (2005). Noncanonical Wnt signaling regulates midline convergence of organ primordia during zebrafish development. Genes Dev. 19, 164–175. doi: 10.1101/gad.1253605
McGough, I. J., Vecchia, L., Bishop, B., Malinauskas, T., Beckett, K., Joshi, D., et al. (2020). Glypicans shield the Wnt lipid moiety to enable signalling at a distance. Nature 585, 85–90. doi: 10.1038/s41586-020-2498-z
Ng, B. G., Sharma, V., Sun, L., Loh, E., Hong, W., Tay, S. K., et al. (2011). Identification of the first COG-CDG patient of Indian origin. Mol. Genet. Metab. 102, 364–367. doi: 10.1016/j.ymgme.2010.11.161
Niehrs, C. (2012). The complex world of WNT receptor signalling. Nat. Rev. Mol. Cell Biol. 13, 767–779. doi: 10.1038/nrm3470
Ohtsubo, K., Takamatsu, S., Minowa, M. T., Yoshida, A., Takeuchi, M., and Marth, J. D. (2005). Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell 123, 1307–1321. doi: 10.1016/j.cell.2005.09.041
Paganini, C., Costantini, R., Superti-Furga, A., and Rossi, A. (2019). Bone and connective tissue disorders caused by defects in glycosaminoglycan biosynthesis: a panoramic view. FEBS J. 286, 3008–3032. doi: 10.1111/febs.14984
Postlethwait, J. H., Navajas Acedo, J., and Piotrowski, T. (2019). Evolutionary origin and nomenclature of vertebrate Wnt11-Family Genes. Zebrafish 16, 469–476. doi: 10.1089/zeb.2019.1760
Rao, D. M., Shackleford, M. T., Bordeaux, E. K., Sottnik, J. L., Ferguson, R. L., Yamamoto, T. M., et al. (2019). Wnt family member 4 (WNT4) and WNT3A activate cell-autonomous Wnt signaling independent of porcupine O-acyltransferase or Wnt secretion. J. Biol. Chem. 294, 19950–19966. doi: 10.1074/jbc.ra119.009615
Reynders, E., Foulquier, F., Leao Teles, E., Quelhas, D., Morelle, W., Rabouille, C., et al. (2009). Golgi function and dysfunction in the first COG4-deficient CDG type II patient. Hum. Mol. Genet. 18, 3244–3256. doi: 10.1093/hmg/ddp262
Saul, R. A., and Wilson, W. G. (1990). A “new” skeletal dysplasia in two unrelated boys. Am. J. Med. Genet. 35, 388–393. doi: 10.1002/ajmg.1320350315
Serralbo, O., and Marcelle, C. (2014). Migrating cells mediate long-range WNT signaling. Development 141, 2057–2063. doi: 10.1242/dev.107656
Sisson, B. E., Dale, R. M., Mui, S. R., Topczewska, J. M., and Topczewski, J. (2015). A role of glypican4 and wnt5b in chondrocyte stacking underlying craniofacial cartilage morphogenesis. Mech. Dev. 138(Pt 3), 279–290. doi: 10.1016/j.mod.2015.10.001
Tambe, M. A., Ng, B. G., and Freeze, H. H. (2019). N-Glycanase 1 transcriptionally regulates aquaporins independent of its enzymatic activity. Cell Rep. 29, 4620–4631.e4.
Tenorio, J., Arias, P., Martinez-Glez, V., Santos, F., Garcia-Minaur, S., Nevado, J., et al. (2014). Simpson-Golabi-Behmel syndrome types I and II. Orphanet. J. Rare Dis. 9:138.
Thisse, C., and Thisse, B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69. doi: 10.1038/nprot.2007.514
Thul, P. J., and Lindskog, C. (2018). The human protein atlas: a spatial map of the human proteome. Protein Sci. 27, 233–244.
Tonelli, F., Bek, J. W., Besio, R., De Clercq, A., Leoni, L., Salmon, P., et al. (2020). Zebrafish: a resourceful vertebrate model to investigate skeletal disorders. Front. Endocrinol. 11:489.
Topczewski, J., Sepich, D. S., Myers, D. C., Walker, C., Amores, A., Lele, Z., et al. (2001). The zebrafish glypican knypek controls cell polarity during gastrulation movements of convergent extension. Dev. Cell 1, 251–264. doi: 10.1016/s1534-5807(01)00005-3
Ungar, A. R., Kelly, G. M., and Moon, R. T. (1995). Wnt4 affects morphogenesis when misexpressed in the zebrafish embryo. Mech. Dev. 52, 153–164. doi: 10.1016/0925-4773(95)00386-f
Ungar, D., Oka, T., Brittle, E. E., Vasile, E., Lupashin, V. V., Chatterton, J. E., et al. (2002). Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. J. Cell Biol. 157, 405–415. doi: 10.1083/jcb.200202016
Yan, D., Wu, Y., Feng, Y., Lin, S. C., and Lin, X. (2009). The core protein of glypican Dally-like determines its biphasic activity in wingless morphogen signaling. Dev. Cell 17, 470–481. doi: 10.1016/j.devcel.2009.09.001
Yang, D., Li, Q., Shang, R., Yao, L., Wu, L., Zhang, M., et al. (2020). WNT4 secreted by tumor tissues promotes tumor progression in colorectal cancer by activation of the Wnt/beta-catenin signalling pathway. J. Exp. Clin. Cancer Res. 39, 251.
Zhang, L. S., and Lum, L. (2016). Delivery of the Porcupine Inhibitor WNT974 in Mice. Methods Mol. Biol. 1481, 111–117. doi: 10.1007/978-1-4939-6393-5_12
Keywords: Saul–Wilson syndrome, COG4, glypican, WNT4, zebrafish, early development
Citation: Xia ZJ, Zeng XXI, Tambe M, Ng BG, Dong PDS and Freeze HH (2021) A Dominant Heterozygous Mutation in COG4 Causes Saul–Wilson Syndrome, a Primordial Dwarfism, and Disrupts Zebrafish Development via Wnt Signaling. Front. Cell Dev. Biol. 9:720688. doi: 10.3389/fcell.2021.720688
Received: 04 June 2021; Accepted: 13 August 2021;
Published: 14 September 2021.
Edited by:
Amélie E. Coudert, Université Paris Diderot, FranceReviewed by:
Ross F. Collery, Medical College of Wisconsin, United StatesCopyright © 2021 Xia, Zeng, Tambe, Ng, Dong and Freeze. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin-Xin I. Zeng, c3plbmdAc2JwZGlzY292ZXJ5Lm9yZw==; Hudson H. Freeze, aHVkc29uQHNicGRpc2NvdmVyeS5vcmc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.