- 1The Francis Crick Institute, London, United Kingdom
- 2Imperial College London, London, United Kingdom
Inside the nucleus, chromatin is functionally organized and maintained as a complex three-dimensional network of structures with different accessibility such as compartments, lamina associated domains, and membraneless bodies. Chromatin is epigenetically and transcriptionally regulated by an intricate and dynamic interplay of molecular processes to ensure genome stability. Phase separation, a process that involves the spontaneous organization of a solution into separate phases, has been proposed as a mechanism for the timely coordination of several cellular processes, including replication, transcription and DNA repair. Telomeres, the repetitive structures at the end of chromosomes, are epigenetically maintained in a repressed heterochromatic state that prevents their recognition as double-strand breaks (DSB), avoiding DNA damage repair and ensuring cell proliferation. In pluripotent embryonic stem cells, telomeres adopt a non-canonical, relaxed epigenetic state, which is characterized by a low density of histone methylation and expression of telomere non-coding transcripts (TERRA). Intriguingly, this telomere non-canonical conformation is usually associated with chromosome instability and aneuploidy in somatic cells, raising the question of how genome stability is maintained in a pluripotent background. In this review, we will explore how emerging technological and conceptual developments in 3D genome architecture can provide novel mechanistic perspectives for the pluripotent epigenetic paradox at telomeres. In particular, as RNA drives the formation of LLPS, we will consider how pluripotency-associated high levels of TERRA could drive and coordinate phase separation of several nuclear processes to ensure genome stability. These conceptual advances will provide a better understanding of telomere regulation and genome stability within the highly dynamic pluripotent background.
Introduction
Eukaryotic genomes are dynamic, non-randomly organized structures within the nucleus. A complex and highly hierarchical three-dimensional network of structures organizes chromatin into active/inactive compartments, membraneless bodies, lamina associated domains, protein- or RNA-mediated loops, enhancer–promoter contacts, and chromatin regions with differential accessibility. This complex chromatin architecture is established by epigenetic and transcriptional mechanisms and is spatially and temporally tightly regulated, to ensure the maintenance and viability of cellular functions. Chromatin architecture also segregates the large repetitive and gene-poor domains of the genome, like centromeres and telomeres, into constitutive heterochromatin domains characterized by condensed chromatin fibers, high levels of DNA and histone methylation, and transcriptional repression of the underlying DNA sequences (García-Cao et al., 2004; Benetti et al., 2007b; Bickmore and van Steensel, 2013). Constitutive heterochromatin is critical for chromosome segregation and integrity, and changes to the heterochromatic state are commonly associated with aging and cancer (Villeponteau, 1997; Janssen et al., 2018; Valencia and Kadoch, 2019).
Telomeres are nucleoprotein structures formed at the end of chromosomes by the assembly of the shelterin complex (formed by TRF1, TRF2, POT1, TPP1, TIN2 and Rap1) at the TTAGGG telomeric repeats (de Lange, 2005; Martínez and Blasco, 2011). The heterochromatic state is critical for telomere integrity, as deletion of HMTases (SUV39H1/2, SUV4-20H1/2) or DNA methyltransferases (DNMT3A/B, and DNMT1) results in defective telomere function, increased telomere length, and chromosome instability (García-Cao et al., 2004; Gonzalo et al., 2005, 2006). Together with the shelterin complex, the heterochromatic state ensures that telomeres are not recognized as double-strand breaks (DSB), avoiding DNA damage repair and maintaining genome integrity. Paradoxically, in mouse pluripotent embryonic stem cells (mESCs), telomeres adopt a non-canonical epigenetic state that is usually associated with chromosome instability and aneuploidy in somatic cells (Peters et al., 2001; García-Cao et al., 2004), and is characterized by less compaction, low density of histone-methylation and increased TERRA - the telomeric transcripts (Marion et al., 2009; Wong et al., 2009, 2010). Here, we review how the pluripotent nuclear environment of mESCs adopts unique molecular features that contribute or even require a non-canonical telomeric chromatin to safeguard genomic stability (de Lange, 2005; Martínez and Blasco, 2011).
The Unique Pluripotent Nuclear Environment
mESCs derived from the inner-cell mass (ICM) of early blastocysts retain self-renewal and pluripotent capacity, being able to differentiate into any type of cell. However, the self-renewal and high proliferative capacities expose mESCs to high levels of DNA replication stress (Ahuja et al., 2016). Critically, mutations acquired during early stages of embryonic development must be promptly repaired to prevent chromosomal defects, infertility, or embryonic lethality (Choi et al., 2020). mESCs exploit distinct molecular and biological signatures, like higher proliferative rates, unique cell-cycle composition and checkpoints and better competence for genomic stability maintenance (Boheler, 2009; Boroviak et al., 2015; Ahuja et al., 2016; Vitale et al., 2017).
The Pluripotent Chromatin Architecture
The chromatin of mESCs has an unusual configuration with open 10 nm chromatin fibers widely dispersed throughout the nucleoplasm, including at constitutive heterochromatin domains (Meshorer et al., 2006; Ahmed et al., 2010; Fussner et al., 2010). Chromatin dispersion is conserved through the cell-cycle, as native mitotic chromosomes purified from mESCs are less condensed than those isolated from other cell-types (Djeghloul et al., 2020), and mESCs contain about 30% less histones than differentiated cells (Karnavas et al., 2014). Dispersed chromatin is also present at the early mouse blastocyst (E3.5) but not in the subsequent stages (E5.5) of development (Ahmed et al., 2010). Thus, this chromatin configuration is considered an architectural hallmark of pluripotency, thought to contribute to pluripotency plasticity by ensuring a transcriptionally permissive and accessible genome (Gaspar-Maia et al., 2011; Cavalli and Misteli, 2013; Hassan-Zadeh et al., 2017).
Constitutive heterochromatin rapidly compacts upon mESCs differentiation and in embryo development (Efroni et al., 2008; Wen et al., 2009; Ahmed et al., 2010; Fussner et al., 2010; Figure 1). Forced compaction of heterochromatin domains by disruption of epigenetic regulators (such as Chd1, esBAF complex, Padi4 or H3K9me3 methyltransferases) affects both self-renewal and differentiation potential of mESCs (Meshorer et al., 2006; Gaspar-Maia et al., 2009; Lessard and Crabtree, 2010; Christophorou et al., 2014). Equally, disrupting the pluripotency network by depleting Nanog, a key pluripotency transcription factor, impacts the chromatin structure and organization of euchromatin and heterochromatin in mESCs (Novo et al., 2016, 2018). Consistently, forcing heterochromatin decompaction with inhibitors of DNA methyltransferase or histone deacetylases improves the efficiency of somatic cell reprogramming to a pluripotent state (Huangfu et al., 2008; Mikkelsen et al., 2008; Soufi et al., 2012; Sridharan et al., 2013). These findings suggest that changes to the heterochromatin state may be adverse to the pluripotent state.
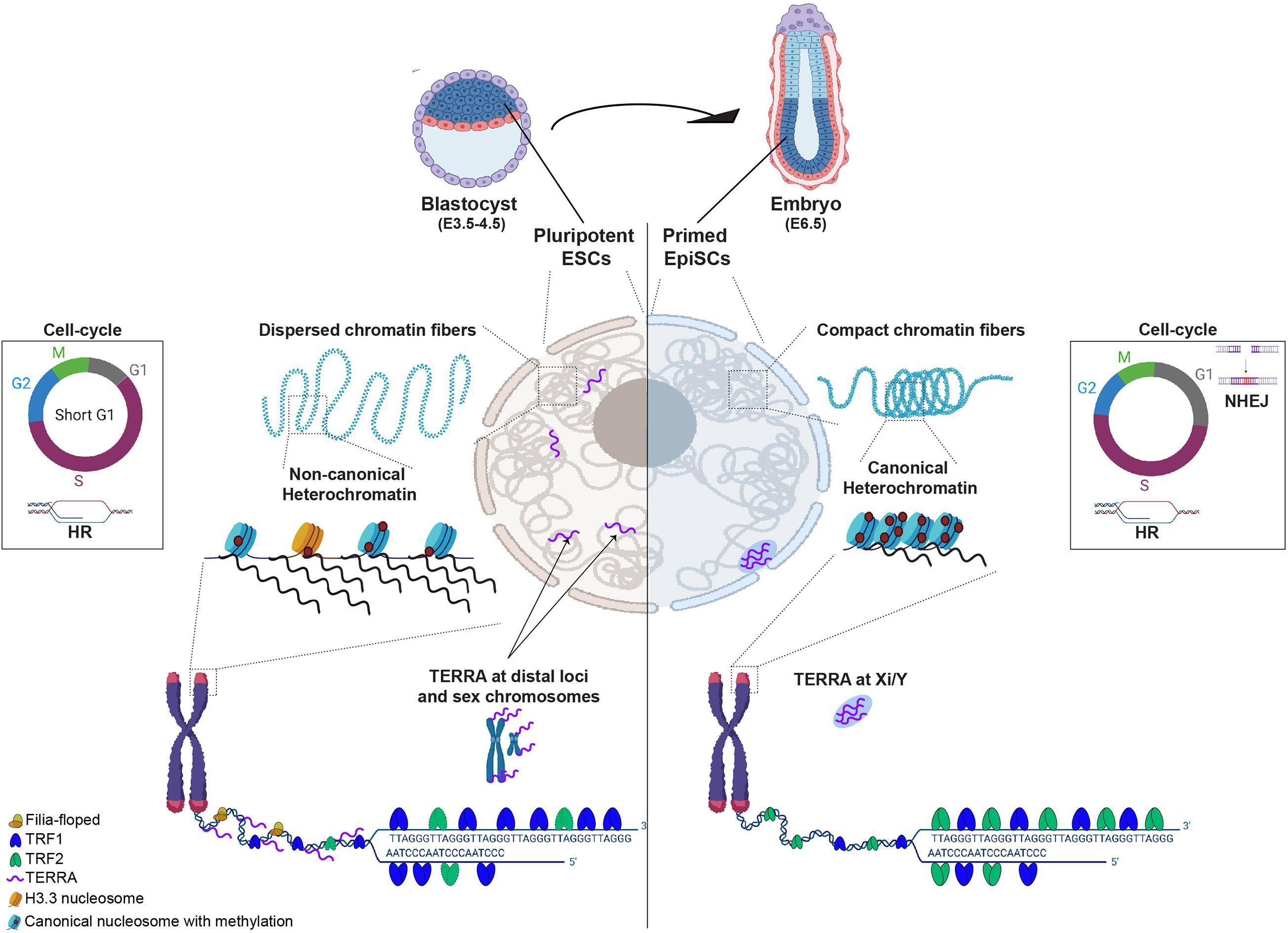
Figure 1. Diagram schematizing differences between pluripotent (cells from blastocyst E3.5-4.5 or ESCs in culture) and cells that exited pluripotency and are primed to differentiate (epiblast at E6.5 embryos). In pluripotent ESCs (left nuclear panel), chromatin fibers are dispersed and heterochromatin is maintained in a non-canonical epigenetic state, characterized by low level of compaction and histone methylation, incorporation of histone H3.3 and high levels of transcription. TRF1, a core shelterin component is highly expressed and TRF2, although present, is not required for telomere protection in ESCs. Filia-floped complexes are recruited to telomeres to mitigate stalled replication forks and telomere transcripts (TERRA) locate at sex chromosomes but also at other distal genomic loci. ESCs have a short G1 phase of the cell-cycle, favoring the use of homologous recombination (HR) to repair DNA damage. In cells that exited pluripotency (in Epiblast of E6.5), chromatin fibers become more dense and heterochromatin adopts a canonical state, characterized by high levels of histone methylation, compaction and transcription silencing. At telomeres, TRF2 becomes crucial for telomere protection and TERRA levels are reduced and redistributed to the inactive sex chromosome. The G1 phase of the cell-cycle becomes longer and cells start using either HR or non-homologous end joining (NHEJ) for DNA repair.
Pluripotency and DNA Damage Repair
The pluripotent chromatin architecture is conducive to DNA Damage Repair (DDR), but pluripotent cells adopted strategies to minimize accumulation of DNA mutations and preserve genome stability (Tichy and Stambrook, 2008; Wyles et al., 2014). DDR mechanisms, such as mismatch repair (MMR), base excision repair (BER), nucleotide excision repair (NER), non-homologous end joining (NHEJ), and homologous recombination (HR) repair different types of DNA damage by arresting the cell-cycle at the G1-, S-, or G2/M-phase checkpoints.
In mESCs, cyclins A and CDK1/2 are highly expressed and there is an increased transcription of S-phase genes by hyperphosphorylation of retinoblastoma (Rb), forcing a rapid entry in S-phase (Tsai et al., 2008; Kalaszczynska et al., 2009). This results in an unusually short G1 phase (Tichy et al., 2010), which mESCs compensate for by bypassing the G1/S cell-cycle checkpoint (van der Laan et al., 2013; Kareta et al., 2015; Soufi and Dalton, 2016). Instead, the intra-S and G2 checkpoints are critical for mESCs and consequently the HR pathway is favored for efficiently and accurately repairing DNA double-strand breaks (DSBs) (Tichy et al., 2010; Momčilović et al., 2011; Figure 1). HR proteins, including RAD51, RAD52 and RAD54, are constitutively expressed through the entire cell-cycle (Choi et al., 2017), and the HR process could suffice in efficiently repairing aberrant DNA in mESCs (Yoon et al., 2014; Choi et al., 2017). As mESCs differentiate, expression of HR factors steadily decreases (Choi et al., 2018). Finally, since the strength of the DDR response depends on chromatin compaction levels (Murga et al., 2007), the elevated chromatin accessibility in mESCs can also contribute to a stronger DDR response and genome stability (Murga et al., 2007; Ahuja et al., 2016). In the case of excessive damage, increased mitochondria priming and hyper-sensitivity to apoptosis can remove cells from the mESCs proliferating pool (Roos and Kaina, 2006; Stambrook and Tichy, 2010; Dumitru et al., 2012; Liu et al., 2013).
Heterochromatin is permeable to DNA repair mechanisms (Kallimasioti-Pazi et al., 2018), and so a preference for HR repair in mESCs poses a challenge for these domains due to their repetitive nature and essential function in genome integrity. For example, DNA repair factors like 53BP1 can bind deprotected telomeres, increase their mobility and foster contact with other telomeres, leading to telomere fusions (Dimitrova et al., 2008). However, 53BP1 foci only appear upon irradiation in mESCs, and telomere hyper-recombination is prevented by the telomere-associated protein Rif1 (Dan et al., 2014). Thus, mESCs can exploit alternative mechanisms to compensate for high proliferative rates and to ensure heterochromatin integrity.
Pluripotency and DNA Replication
DNA replication is essential to the self-renewal and pluripotency capacities of mESCs, whilst conferring an opportunity to alter chromatin with incorporation of new histones or by spatially reorganizing pre-existent histone modifications (McNairn and Gilbert, 2003). Conversely, replication also exposes chromatin to mutations and copy number abnormalities, which could compromise embryonic survival (De and Michor, 2011; Hodgkinson et al., 2012; Schuster-Böckler and Lehner, 2012). Due to the relatively short G1 phase, mESCs are unable to complete DDR before moving to the S-phase (Hyka-Nouspikel et al., 2012; Choi et al., 2017), leading to accumulation of ssDNA gaps and formation of DSBs at stalled replication forks and to accumulation of γH2AX (Chuykin et al., 2008; Banáth et al., 2009; Choi et al., 2020; Blakemore et al., n.d.). Despite rapid proliferation rates and elevated replication stress (Banáth et al., 2009; Ahuja et al., 2016), mESCs have surprisingly low mutation rates. In culture, mESCs display a 1,000-fold lower mutation rate than their isogenically-matched mouse embryonic fibroblasts (Tichy and Stambrook, 2008; Wyles et al., 2014). Therefore, pluripotent mESCs may be more efficient than differentiated cells in resolving replication stress. Indeed, dormant origins can be fired in mESCs to ensure completion of DNA replication under replication stress (Ge et al., 2015). Also, HR factors are constitutively expressed through the cell-cycle, which can facilitate their rapid recruitment to stalled forks in mESCs (Burhans and Weinberger, 2007; Petermann et al., 2010). Indeed, RAD51 depletion in mESCs causes G2/M-phase arrest and replication fork collapse (Petermann et al., 2010). Finally, mESCs use unique protein complexes, like Filia-Floped (a mESC specific regulator of genomic stability and a factor essential for the maternal-zygotic transition, respectively), which scaffold and amplify DDR signaling response at stalled replication forks (Zhao et al., 2015; Figure 1).
In sum, pluripotent cells acquired mechanisms to balance for high proliferative rates without compromising genome integrity, including at heterochromatin domains.
Pluripotency and Telomeres
Telomere Length and Pluripotency
Long telomeres are essential for self-renewal and high proliferative capacities in embryogenesis. Two waves of telomere elongation occur during early embryonic development. Through early cleavage stages, parental telomere length is reset and telomeres are elongated by a recombination-based mechanism known as the Alternative Lengthening of Telomeres (ALT) pathway (Schaetzlein et al., 2004; Liu et al., 2007; Varela et al., 2011; Dang-Nguyen et al., 2013). Telomerase activity becomes detectable at the morula-blastocyst transition, when it is thought to stabilize telomere length, and its reverse transcriptase component (TERT) becomes repressed during cellular differentiation, as embryonic development progresses (Holt et al., 1996; Schaetzlein et al., 2004). In humans, telomerase activity is regulated by the alternative splicing of TERT that ensures telomerase repression in somatic cells (Penev et al., 2021).
Short telomeres affect mESCs pluripotency and pose a barrier to an efficient reprogramming process (Zhang et al., 2015). Telomerase deficient mESCs with critically short telomeres are unable to differentiate, as they retain DNA hypomethylation and altered H3K27me3 enrichment at pluripotency promoters, like Nanog and Oct4 (Pucci et al., 2013; Criqui et al., 2020). Telomere length is also influenced by subtelomeric DNA methylation: hypomethylation facilitates recombination-mediated telomere lengthening, while hypermethylation correlates with shorter telomeres (Gonzalo et al., 2006; Yang et al., 2016). These studies strongly support a function for telomere length in regulating the differentiation capacity of mESCs, underscoring the importance of telomere length maintenance in embryonic development.
TBX3, a pluripotency factor required for self-renewal of mESCs and iPSCs (Han et al., 2010; Lu et al., 2011), activates Zscan4 expression, a 2-cell embryo marker. ZSCAN4 enables heterochromatin decondensation and subtelomeric DNA demethylation in mESCs, promoting telomere elongation by HR (Falco et al., 2007; Zalzman et al., 2010; Macfarlan et al., 2012; Dan et al., 2013; Nakai-Futatsugi and Niwa, 2016) and DNA repair (Akiyama et al., 2015; Eckersley-Maslin et al., 2016; Dan et al., 2017). mESCs expressing Zscan4 (1–5% of mESCs in serum-culture conditions (Macfarlan et al., 2012)) are characterized by global DNA hypomethylation, histone hyperacetylation, and transcription of heterochromatin domains (pericentromeres, telomeres, and retrotransposons) (Akiyama et al., 2015; Eckersley-Maslin et al., 2016). Interestingly, exogenously induced replication stress in mESCs activates the DNA damage sensor ATR and the transcriptionally signature of 2-cell state, including upregulation of Zscan4 (Zalzman et al., 2010; Zhang et al., 2016; De Iaco et al., 2017; Hendrickson et al., 2017; Whiddon et al., 2017; Atashpaz et al., 2020). Therefore, an interdependence of distinct pluripotent networks, telomere biology and DNA repair ensure genomic integrity in mESCs.
Telomere-Associated Proteins
Unexpectedly, telomere binding protein 2, TRF2, a key mediator of telomere protection and core component of the shelterin complex, is dispensable for telomere protection in pluripotent mESCs and in early embryonic development (Markiewicz-Potoczny et al., 2021; Ruis et al., 2021). TRF2 protects and stabilizes telomere structure by binding abnormal DNA conformations that arise at stalled replication forks (like branched DNA, positive DNA supercoils, or G-quadruplexes), suppressing ATM activation and recruiting RTEL1 helicase and other enzymes to remove the blockades (Denchi and de Lange, 2007; Sarek et al., 2016; Mendez-Bermudez et al., 2018). In most cells, TRF2 loss leads to telomere deprotection and fusion via NHEJ (Denchi and de Lange, 2007) but telomeres remain surprisingly protected in mESCs that lack TRF2, despite fully functional ATM and NHEJ pathways (Markiewicz-Potoczny et al., 2021; Ruis et al., 2021). This extraordinary feature of pluripotent telomeres is lost upon differentiation, when TRF2 assumes its fully protective role.
Another core component of the shelterin complex, TRF1, is a direct transcriptional target of the key pluripotent factor Oct3/4 and is upregulated in pluripotent cells (Boué et al., 2010; Schneider et al., 2013; Figure 1). TRF1 deletion causes embryonic lethality at the blastocyst stage, around E5 (Karlseder et al., 2003; Schneider et al., 2013). Interestingly, TRF2 embryonic lethality occurs much later than TRF1, at E13.5 (Karlseder et al., 2003; Celli and de Lange, 2005), reinforcing the preferential requirement for TRF1 in the pluripotent stages of embryonic development. In most cells, TRF1 promotes DNA replication by blocking HR at telomeres (Karlseder et al., 2003; Sfeir et al., 2009; Porreca et al., 2020). However, TRF1 depletion in induced pluripotent stem cells (iPSCs) leads to genome-wide expression and epigenetic changes through TERRA-mediated Polycomb recruitment to pluripotent and cell-fate genes (Marión et al., 2019).
Thus, pluripotent cells coordinate the pluripotency network, telomere proteins and DNA repair to ensure genome integrity.
Telomere Chromatin State in Pluripotent Cells
The non-canonical heterochromatin (low density of H3K9me3 and H4K20me3 and increased expression) of pluripotent telomeres may enable access for recombination or telomerase to modulate telomere length (Benetti et al.,2007a,b). As mESCs exit pluripotency, heterochromatin shifts to a canonical state (Azuara et al., 2006; Meshorer and Misteli, 2006). NANOG, a core pluripotent transcription factor, regulates the non-canonical heterochromatin at pericentromeres in mESCs (Novo et al., 2016). Deletion of Nanog in mESCs induces a canonical state at pericentromeres, impacts pericentromeric transcription and nuclear architecture and results in genetic instability (Novo et al., 2020). Similarly, interfering with epigenetic factors regulating telomere chromatin in mESCs results in telomere dysfunction and instability (Peters et al., 2001; García-Cao et al., 2004; Benetti et al., 2007b; Dang-Nguyen et al., 2013). Together, these findings suggest that a non-canonical heterochromatin is a requirement for genetic stability in pluripotency.
The ATRX/DAXX chaperone complex deposits histone H3.3, typically associated with active/open chromatin, at telomeres and pericentromeres in mESCs and embryonic germ cells, but not in differentiated cells (Goldberg et al., 2010; Ratnakumar et al., 2012; Clynes et al., 2015; Figure 1). ATRX and H3.3 levels at telomeres decrease as mESCs differentiate (Wong et al., 2009; Lewis et al., 2010), whilst the repressive marks H4K20me3 and H3K9me3 increase (Marion et al., 2009). ATRX knockdown causes telomere dysfunction and up-regulation of TERRA (Goldberg et al., 2010; Wong et al., 2010), and facilitates ALT features at telomeres, in murine cells (Lovejoy et al., 2012). Similarly, H3.3 depletion induces DNA damage and telomeric sister chromatid exchange (Udugama et al., 2015). Importantly, ATRX/DAXX mutations are associated with the ALT mechanism, characterized by telomeres with a relaxed heterochromatin state and high TERRA expression (Lovejoy et al., 2012). However, increased TERRA expression upon ATRX depletion is only observed in murine cells, as ATRX depletion in human cells is insufficient to increase telomeric expression (Episkopou et al., 2014). Despite shared features between ALT + and mESCs telomeres (low H3K9me3 density and compaction and increased TERRA) (Arnoult et al., 2012; Episkopou et al., 2014; Eid et al., 2015), the role of ATRX at telomeres likely depends on cellular context and might also be species-specific.
In human cells, the loss of Tousled-like kinases 1 and 2 (TLK1/2, histone deposition regulators), lead to chromatin decompaction and increased genome accessibility, particularly at heterochromatin domains. Importantly, chromatin decompaction induces heterochromatin expression and ALT features at telomeres (Segura-Bayona et al., 2020), suggesting that telomeres are highly susceptible to chromatin changes. As epigenetic features can impact telomere biology in a cellular-dependent context (Novo et al., 2013), the implications of distinct chromatin states at telomeres need to be further elucidated in different cellular backgrounds.
Telomere Transcripts in Pluripotent Cells
Telomeric RNA is transcribed by RNA polymerase II moving toward the telomere, from promoters located at subtelomeres, and is composed of G-rich repeats with heterogeneous size (200 bp to several kilobases) (Deng et al., 2012a). In humans, TERRA is transcribed from subtelomeric promoters at most chromosomes ends and stays associated with telomeres (Azzalin and Lingner, 2008; Schoeftner and Blasco, 2008; Zhang et al., 2009; Le et al., 2013; Feretzaki et al., 2019). In mice, TERRA predominantly originates from the pseudoautosomal PAR locus, but TERRA from chromosomes 18q, 2 and X have also been found (López de Silanes et al., 2014; Viceconte et al., 2021). TERRA transcription is sensitive to subtelomeric DNA methylation (Feretzaki et al., 2019). In mESCs, TERRA is enriched at both sex chromosomes and relocates to the heterochromatic sex chromosomes (Y or Xi) during differentiation (Schoeftner et al., 2009; Zhang et al., 2009; Deng et al., 2012a). Despite different origins, and consequently composition, murine and human TERRA share many interacting proteins (including shelterin complex, Bloom helicase, chromatin remodeling factor and DNA replication proteins) (Scheibe et al., 2013; Viceconte et al., 2021), suggesting similar functions. Live-imaging studies showed that TERRA molecules are confined to the telomeric region, forming clusters that may scaffold the nucleation of telomere-associated proteins, as shown for hnRNAP1 or for telomerase (Deng et al., 2012b; Cusanelli et al., 2013; Yamada et al., 2016; Avogaro et al., 2018).
Interestingly, TRF2-TERRA interactions were proposed to mediate telomere heterochromatin in human cells (Deng et al., 2009). As TRF2 appears to be dispensable for mESCs (Markiewicz-Potoczny et al., 2021; Ruis et al., 2021), it is probable that TERRA has a distinct function in murine pluripotent cells. Indeed, most TERRA locates and regulates expression of distal intergenic and intronic regions in the mESCs genome (Chu et al., 2017; Figure 1). However, TERRA depletion in mESCs induces telomere dysfunction, indicating that TERRA is nevertheless important for mouse telomeric integrity (Chu et al., 2017). Importantly, there is conflicting evidence from RNA-FISH vs sequencing-based technologies for TERRA location in mESCs. Thus, elucidating this technical divergence is essential to understand TERRA function and better elucidate how telomere higher-ordered structure impacts genome stability and pluripotency.
A Golden Thread: Phase-Separation
Phase-separation is based on the spontaneous organization of a solution into two-phases with different densities (Berry et al., 2015; Banani et al., 2017; Boeynaems et al., 2018). The biophysical properties of molecules and their modulation by the surrounding environment enables membraneless compartmentalization and subsequent concentration of biochemical reactions within the cell (Alberti et al., 2019; Gibson et al., 2019). The multivalency of interactions between DNA/RNA molecules and intrinsically disordered regions of proteins can promote liquid–liquid phase separation (LLPS) (Kato et al., 2012; Lin et al., 2015; Hnisz et al., 2017; Langdon et al., 2018). Phase-separation contributes to distinct cellular functions, like stress sensing (Munder et al., 2016; Riback et al., 2017) or increased biochemical kinetics by confining molecules into a compartment (Case et al., 2019). Furthermore, LLPS has been implicated in several nuclear processes, including nucleoli formation, transcription elongation, super-enhancer activity and binding of transcription factors to DNA (Feric et al., 2016; Hnisz et al., 2017; Larson et al., 2017; Strom et al., 2017; Boehning et al., 2018; Boija et al., 2018; Cho et al., 2018; Lu et al., 2018; Sabari et al., 2018; Case et al., 2019; Trivedi et al., 2019; Huo et al., 2020).
Phase-separation has also been proposed to regulate heterochromatin (Larson et al., 2017; Strom et al., 2017; Trivedi et al., 2019; Huo et al., 2020; Novo et al., 2020). The heterochromatin protein 1 (HP1α) is thought to induce LLPS formation at heterochromatin and reinforce the heterochromatic environment by binding to H3K9me3, increasing nucleosome compaction and repressing transcription by exclusion of transcription factors and RNA polymerase (Feng and Michaels, 2015; Sanulli et al.,2019a,b). Similarly, heterochromatin regions interspersed along the chromosome arms can loop and interact in the three-dimensional space (Dernburg et al., 1996; Lee and Karpen, 2017), further supporting the coalescence of multiple condensates into a larger one. CBX2, a subunit of the canonical PRC1 complex responsible for DNA compaction, can also phase-separate both in vitro and in vivo (Plys et al., 2019; Tatavosian et al., 2019).
Interestingly, the fusion of DNA repair foci into larger clusters was observed in both euchromatin and heterochromatin, which facilitates a rapid but transient recruitment and concentration of repair factors restricted to the damaged region (Aten et al., 2004; Kruhlak et al., 2006; Chiolo et al., 2011; Krawczyk et al., 2012; Aymard et al., 2017; Caridi et al., 2017). Similarly, recruitment of polyADP-ribosylation (PARylation) at DDR foci promotes LLPS (Altmeyer et al., 2015; Duan et al., 2019). Also, the DNA repair protein 53BP1 forms LLPS promoted by non-coding RNA (Binz et al., 2006; Kilic et al., 2019; Pessina et al., 2019). Finally, RAD52 liquid-like condensates formed at different DSB sites can fuse, and mutants unable to form these condensates show limited fusion in vitro and increased genome instability in vivo (Oshidari et al., 2020). Thus, the ability to phase-separate DNA repair sites could ensure genome stability by restricting access of DDR factors to the damage site (Altmeyer et al., 2015; Patel et al., 2015; Banani et al., 2017). Importantly, DSB repair within constitutive heterochromatin actively decompacts and relocates the damaged locus to the nuclear periphery for HR repair, whilst preventing spurious recombination (Chiolo et al., 2011; Jakob et al., 2011; Janssen et al., 2016; Tsouroula et al., 2016). Whether phase-separation contributes to this mobility, whilst isolating heterochromatin from the surrounding nuclear environment remains to be elucidated.
One hallmark of ALT + cells is telomere clustering at promyelocytic leukemia (PML) bodies, known as ALT-associated PML Bodies (APBs) (Yeager et al., 1999; Heaphy et al., 2011). APBs contain telomeres and many proteins involved in DNA replication, repair, and recombination processes. Interestingly, PML bodies form membraneless organelles by LLPS, mediated by multivalent interactions between SUMO (Small Ubiquitin-like MOdifier) and SIM (SUMO-Interacting Motif) motifs in PML and other proteins (Banani et al., 2017). An elegant study mimicked APBs by engineering polySUMO/polySIM condensates targeted to telomeres in telomerase-positive cells. In the presence of BLM and RAD52, polySUMO/polySIM induce telomere clustering and rapidly recapitulate the ALT phenotype (C-circles, heterogeneous telomere length, and complex telomere structures) (Min et al., 2019). Indeed, telomere clustering seems to depend on the liquid properties of APB condensates, rather than their chemical composition (Zhang et al., 2020).
Pluripotent telomeres may also be able to cluster at PML bodies (Gauchier et al., 2019) but whether phase-separation is involved is still unknown. Also, ATRX recruits HP1a to telomeres in mESCs (Wong et al., 2010; He et al., 2015), where it may mediate HP1a LLPS formation. Interestingly, pericentromeric domains form LLPS condensates in early embryonic development (Strom et al., 2017) and in mESCs (Huo et al., 2020; Novo et al., 2020) but collapse into ‘ordered collapsed globules’ in differentiated cells (Erdel et al., 2020). Thus, the material state of heterochromatin associated with pluripotency seems to facilitate LLPS formation, and may be regulated in different cellular contexts. Importantly, these differences stress the crucial need to characterize phase-separation in many different systems to better understand the mechanisms governing phase-separation.
Discussion
Telomere maintenance relies on the coordinated crosstalk involving the telomeric structure, TERRA and nuclear processes (such as replication, transcription, repair, etc.). Telomere end protection is critical for genome stability and cell proliferation and the mechanisms involved are fairly conserved across cellular backgrounds and species. Thus, it is striking that a core shelterin component, TRF2, is redundant for telomere protection in pluripotent mESCs. Additionally, as pluripotent cells acquired special features to compensate for the unique nuclear environment, it is paramount to further explore the mechanisms governing telomere maintenance in pluripotent cells.
One of the hallmarks of pluripotent telomeres is high TERRA levels, which may have pluripotent-specific roles, like shown by the regulation of pluripotent gene expression in iPSCs. Furthermore, TERRA molecules can originate from different genomic locations and have different sizes. Thus, regulation of TERRA properties (sequence composition; length; levels) may affect its function by modulating (i) its ability to recruit heterochromatinization factors; (ii) competition with yet unknown cell-specific proteins at telomeres and/or iii) the molecular substrate available for weak multivalent interactions that can affect the material state of telomeres. Strong evidence supports a role for RNA molecules to act as a regulatory elements of LLPS condensate formation, size and constitution (reviewed in Palikyras and Papantonis (2019). Thus, the orchestrated interplay between the pluripotency network, telomeres and DNA repair in mESCs could rely on LLPS to balance the accessible chromatin whilst maintaining genome integrity.
Phase-separation presents an attractive model for harmonizing genome compartmentalization and the diverse biochemical reactions occurring in the nucleus by enabling a spatial and timely unification of nuclear processes through functional concentration of chromatin, RNA/proteins and relevant cellular factors in membraneless compartments. Importantly, as it depends on weak multivalency interactions, phase-separated condensates can dynamically engage in coalescence/fission events to isolate or expose specific chromatin domains, enabling different processes such as replication, transcription or heterochromatin to concomitantly occur within the nuclear environment. Many important questions are left to address and still much to be elucidated, particularly how phase-separation is regulated and how mechanistically promotes cellular functions. Critical open questions are (i) what are the signaling triggers that promote phase-separation; (ii) how the nuclear environment modulates distinct condensates at the same loci and at the same phase of the cell-cycle (for example, transcription vs. heterochromatin aggregates)? New tools that can regulate phase separation in live cells are starting to emerge and will undoubtedly probe cellular functions and the functional possibilities enabled by phase-separation.
Author Contributions
CN wrote the manuscript and prepared the figure.
Funding
This work was funded by Tommys National Miscarriage Centre and The Francis Crick Institute, which receives its core funding from Cancer Research UK (FC0010048), the UK Medical Research Council (FC0010048), and the Wellcome Trust (FC0010048).
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
CN is grateful to Phillip Bennett, Véronique Azuara, and Simon Boulton for their support. Figure was created with BioRender.com.
References
Ahmed, K., Dehghani, H., Rugg-Gunn, P., Fussner, E., Rossant, J., and Bazett-Jones, D. P. (2010). Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One 5:e10531. doi: 10.1371/journal.pone.0010531
Ahuja, A. K., Jodkowska, K., Teloni, F., Bizard, A. H., Zellweger, R., Herrador, R., et al. (2016). A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat. Commun. 7:10660.
Akiyama, T., Xin, L., Oda, M., Sharov, A. A., Amano, M., and Piao, Y., et al. (2015). Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells. DNA Res. 22, 307–318. doi: 10.1093/dnares/dsv013
Alberti, S., Gladfelter, A., and Mittag, T. (2019). Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434. doi: 10.1016/j.cell.2018.12.035
Altmeyer, M., Neelsen, K. J., Teloni, F., Pozdnyakova, I., Pellegrino, S., Grøfte, M., et al. (2015). Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6:8088.
Arnoult, N., Van Beneden, A., and Decottignies, A. (2012). Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α. Nat. Struct. Mol. Biol. 19, 948–956. doi: 10.1038/nsmb.2364
Atashpaz, S., Samadi Shams, S., Gonzalez, J. M., Sebestyén, E., Arghavanifard, N., Gnocchi, A., et al. (2020). ATR expands embryonic stem cell fate potential in response to replication stress. eLife 9:e54756. doi: 10.7554/eLife.54756
Aten, J. A., Stap, J., Krawczyk, P. M., van Oven, C. H., Hoebe, R. A., Essers, J., et al. (2004). Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 303, 92–95. doi: 10.1126/science.1088845
Avogaro, L., Querido, E., Dalachi, M., Jantsch, M. F., Chartrand, P., and Cusanelli, E. (2018). Live-cell imaging reveals the dynamics and function of single-telomere TERRA molecules in cancer cells. RNA Biol. 15, 787–796.
Aymard, F., Aguirrebengoa, M., Guillou, E., Javierre, B. M., Bugler, B., Arnould, C., et al. (2017). Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes. Nat. Struct. Mol. Biol. 24, 353–361. doi: 10.1038/nsmb.3387
Azuara, V., Perry, P., Sauer, S., Spivakov, M., Jørgensen, H. F., John, R. M., et al. (2006). Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8, 532–538. doi: 10.1038/ncb1403
Azzalin, C. M., and Lingner, J. (2008). Telomeres: the silence is broken. Cell Cycle 7, 1161–1165. doi: 10.4161/cc.7.9.5836
Banani, S. F., Lee, H. O., Hyman, A. A., and Rosen, M. K. (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298. doi: 10.1038/nrm.2017.7
Banáth, J. P., Bañuelos, C. A., Klokov, D., MacPhail, S. M., Lansdorp, P. M., and Olive, P. L. (2009). Explanation for excessive DNA single-strand breaks and endogenous repair foci in pluripotent mouse embryonic stem cells. Exp. Cell Res. 315, 1505–1520. doi: 10.1016/j.yexcr.2008.12.007
Benetti, R., García-Cao, M., and Blasco, M. A. (2007a). Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat. Genet. 39, 243–250. doi: 10.1038/ng1952
Benetti, R., Gonzalo, S., Jaco, I., Schotta, G., Klatt, P., Jenuwein, T., et al. (2007b). Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J. Cell Biol. 178, 925–936. doi: 10.1083/jcb.200703081
Berry, J., Weber, S. C., Vaidya, N., Haataja, M., and Brangwynne, C. P. (2015). RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl. Acad. Sci. U.S.A. 112, E5237–E5245.
Bickmore, W. A., and van Steensel, B. (2013). Genome architecture: domain organization of interphase chromosomes. Cell 152, 1270–1284. doi: 10.1016/j.cell.2013.02.001
Binz, S. K., Dickson, A. M., Haring, S. J., and Wold, M. S. (2006). Functional assays for replication protein A (RPA). Methods Enzymol. 409, 11–38. doi: 10.1016/s0076-6879(05)09002-6
Blakemore, D., Vilaplana, N., Almaghrabi, R., Gonzalez, E., Moya, M., Ward, C., et al. (n.d.). MYBL2 Regulates ATM to Control Replication Initiation and Prevent Replication Stress in Pluripotent Stem Cells. doi: 10.1101/2020.06.04.131276
Boehning, M., Dugast-Darzacq, C., Rankovic, M., Hansen, A. S., Yu, T., Marie-Nelly, H., et al. (2018). RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 25, 833–840. doi: 10.1038/s41594-018-0112-y
Boeynaems, S., Alberti, S., Fawzi, N. L., Mittag, T., Polymenidou, M., Rousseau, F., et al. (2018). Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28, 420–435. doi: 10.1016/j.tcb.2018.02.004
Boheler, K. R. (2009). Stem cell pluripotency: a cellular trait that depends on transcription factors, chromatin state and a checkpoint deficient cell cycle. J. Cell. Physiol. 221, 10–17. doi: 10.1002/jcp.21866
Boija, A., Klein, I. A., Sabari, B. R., Dall’Agnese, A., Coffey, E. L., Zamudio, A. V., et al. (2018). Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842.e16–1855.e16.
Boroviak, T., Loos, R., Lombard, P., Okahara, J., Behr, R., Sasaki, E., et al. (2015). Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev. Cell 35, 366–382. doi: 10.1016/j.devcel.2015.10.011
Boué, S., Paramonov, I., Barrero, M. J., and Izpisúa Belmonte, J. C. (2010). Analysis of human and mouse reprogramming of somatic cells to induced pluripotent stem cells. What is in the plate? PLoS One 5:e12664. doi: 10.1371/journal.pone.0012664
Burhans, W. C., and Weinberger, M. (2007). DNA replication stress, genome instability and aging. Nucleic Acids Res. 35, 7545–7556. doi: 10.1093/nar/gkm1059
Caridi, P. C., Delabaere, L., Zapotoczny, G., and Chiolo, I. (2017). And yet, it moves: nuclear and chromatin dynamics of a heterochromatic double-strand break. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372:20160291. doi: 10.1098/rstb.2016.0291
Case, L. B., Ditlev, J. A., and Rosen, M. K. (2019). Regulation of transmembrane signaling by phase separation. Annu. Rev. Biophys. 48, 465–494. doi: 10.1146/annurev-biophys-052118-115534
Cavalli, G., and Misteli, T. (2013). Functional implications of genome topology. Nat. Struct. Mol. Biol. 20, 290–299. doi: 10.1038/nsmb.2474
Celli, G. B., and de Lange, T. (2005). DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7, 712–718. doi: 10.1038/ncb1275
Chiolo, I., Minoda, A., Colmenares, S. U., Polyzos, A., Costes, S. V., and Karpen, G. H. (2011). Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144, 732–744. doi: 10.1016/j.cell.2011.02.012
Cho, W.-K., Spille, J.-H., Hecht, M., Lee, C., Li, C., Grube, V., et al. (2018). Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415. doi: 10.1126/science.aar4199
Choi, E.-H., Yoon, S., and Kim, K. P. (2018). Combined ectopic expression of homologous recombination factors promotes embryonic stem cell differentiation. Mol. Therapy 26, 1154–1165. doi: 10.1016/j.ymthe.2018.02.003
Choi, E.-H., Yoon, S., Koh, Y. E., Seo, Y.-J., and Kim, K. P. (2020). Maintenance of genome integrity and active homologous recombination in embryonic stem cells. Exp. Mol. Med. 52, 1220–1229. doi: 10.1038/s12276-020-0481-2
Choi, E.-H., Yoon, S., Park, K.-S., and Kim, K. P. (2017). The homologous recombination machinery orchestrates post-replication DNA repair during self-renewal of mouse embryonic stem cells. Sci. Rep. 7:11610.
Christophorou, M. A., Castelo-Branco, G., Halley-Stott, R. P., Oliveira, C. S., Loos, R., Radzisheuskaya, A., et al. (2014). Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature 507, 104–108. doi: 10.1038/nature12942
Chu, H.-P., Cifuentes-Rojas, C., Kesner, B., Aeby, E., Lee, H.-G., Wei, C., et al. (2017). TERRA RNA antagonizes ATRX and protects telomeres. Cell 170, 86.e16–101.e16.
Chuykin, I. A., Lianguzova, M. S., Pospelova, T. V., and Pospelov, V. A. (2008). Activation of DNA damage response signaling in mouse embryonic stem cells. Cell Cycle 7, 2922–2928. doi: 10.4161/cc.7.18.6699
Clynes, D., Jelinska, C., Xella, B., Ayyub, H., Scott, C., Mitson, M., et al. (2015). Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 6:7538.
Criqui, M., Qamra, A., Chu, T. W., Sharma, M., Tsao, J., Henry, D. A., et al. (2020). Telomere dysfunction cooperates with epigenetic alterations to impair murine embryonic stem cell fate commitment. eLife 9:e47333. doi: 10.7554/eLife.47333
Cusanelli, E., Romero, C. A. P., and Chartrand, P. (2013). Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol. Cell. 51, 780–791. doi: 10.1016/j.molcel.2013.08.029
Dan, J., Li, M., Yang, J., Li, J., Okuka, M., Ye, X., et al. (2013). Roles for Tbx3 in regulation of two-cell state and telomere elongation in mouse ES cells. Sci. Rep. 3:3492.
Dan, J., Liu, Y., Liu, N., Chiourea, M., Okuka, M., Wu, T., et al. (2014). Rif1 maintains telomere length homeostasis of ESCs by mediating heterochromatin silencing. Dev. Cell 29, 7–19. doi: 10.1016/j.devcel.2014.03.004
Dan, J., Rousseau, P., Hardikar, S., Veland, N., Wong, J., Autexier, C., et al. (2017). Zscan4 inhibits maintenance DNA methylation to facilitate telomere elongation in mouse embryonic stem cells. Cell Rep. 20, 1936–1949. doi: 10.1016/j.celrep.2017.07.070
Dang-Nguyen, T. Q., Haraguchi, S., Furusawa, T., Somfai, T., Kaneda, M., Watanabe, S., et al. (2013). Downregulation of histone methyltransferase genes SUV39H1 and SUV39H2 increases telomere length in embryonic stem-like cells and embryonic fibroblasts in pigs. J. Reprod. Dev. 59, 27–32. doi: 10.1262/jrd.2012-118
De Iaco, A., Planet, E., Coluccio, A., Verp, S., Duc, J., and Trono, D. (2017). DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 49, 941–945. doi: 10.1038/ng.3858
de Lange, T. (2005). Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110. doi: 10.1101/gad.1346005
De, S., and Michor, F. (2011). DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat. Biotechnol. 29, 1103–1108. doi: 10.1038/nbt.2030
Denchi, E. L., and de Lange, T. (2007). Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448, 1068–1071. doi: 10.1038/nature06065
Deng, Z., Norseen, J., Wiedmer, A., Riethman, H., and Lieberman, P. M. (2009). TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 35, 403–413. doi: 10.1016/j.molcel.2009.06.025
Deng, Z., Wang, Z., Stong, N., Plasschaert, R., Moczan, A., Chen, H.-S., et al. (2012a). A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 31, 4165–4178. doi: 10.1038/emboj.2012.266
Deng, Z., Wang, Z., Xiang, C., Molczan, A., Baubet, V., Conejo-Garcia, J., et al. (2012b). Formation of telomeric repeat-containing RNA (TERRA) foci in highly proliferating mouse cerebellar neuronal progenitors and medulloblastoma. J. Cell Sci. 125(Pt 18), 4383–4394.
Dernburg, A. F., Broman, K. W., Fung, J. C., Marshall, W. F., Philips, J., Agard, D. A., et al. (1996). Perturbation of nuclear architecture by long-distance chromosome interactions. Cell 85, 745–759. doi: 10.1016/s0092-8674(00)81240-4
Dimitrova, N., Chen, Y.-C. M., Spector, D. L., and de Lange, T. (2008). 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456, 524–528. doi: 10.1038/nature07433
Djeghloul, D., Patel, B., Kramer, H., Dimond, A., Whilding, C., Brown, K., et al. (2020). Identifying proteins bound to native mitotic ESC chromosomes reveals chromatin repressors are important for compaction. Nat. Commun. 11:4118.
Duan, Y., Du, A., Gu, J., Duan, G., Wang, C., Gui, X., et al. (2019). PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res. 29, 233–247. doi: 10.1038/s41422-019-0141-z
Dumitru, R., Gama, V., Fagan, B. M., Bower, J. J., Swahari, V., Pevny, L. H., et al. (2012). Human embryonic stem cells have constitutively active Bax at the Golgi and are primed to undergo rapid apoptosis. Mol. Cell. 46, 573–583. doi: 10.1016/j.molcel.2012.04.002
Eckersley-Maslin, M. A., Svensson, V., Krueger, C., Stubbs, T. M., Giehr, P., Krueger, F., et al. (2016). MERVL/Zscan4 Network Activation Results in Transient Genome-wide DNA Demethylation of mESCs. Cell Rep. 17, 179–192. doi: 10.1016/j.celrep.2016.08.087
Efroni, S., Duttagupta, R., Cheng, J., Dehghani, H., Hoeppner, D. J., Dash, C., et al. (2008). Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2, 437–447.
Eid, R., Demattei, M.-V., Episkopou, H., Augé-Gouillou, C., Decottignies, A., Grandin, N., et al. (2015). Genetic inactivation of ATRX leads to a decrease in the amount of telomeric cohesin and level of telomere transcription in human glioma cells. Mol. Cell. Biol. 35, 2818–2830. doi: 10.1128/mcb.01317-14
Episkopou, H., Draskovic, I., Van Beneden, A., Tilman, G., Mattiussi, M., Gobin, M., et al. (2014). Alternative Lengthening of Telomeres is characterized by reduced compaction of telomeric chromatin. Nucleic Acids Res. 42, 4391–4405. doi: 10.1093/nar/gku114
Erdel, F., Rademacher, A., Vlijm, R., Tünnermann, J., Frank, L., Weinmann, R., et al. (2020). Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol. Cell. 78, 236.e16–249.e16.
Falco, G., Lee, S.-L., Stanghellini, I., Bassey, U. C., Hamatani, T., and Ko, M. S. H. (2007). Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev. Biol. 307, 539–550. doi: 10.1016/j.ydbio.2007.05.003
Feng, W., and Michaels, S. D. (2015). Accessing the inaccessible: the organization, transcription, replication, and repair of heterochromatin in plants. Annu. Rev. Genet. 49, 439–459. doi: 10.1146/annurev-genet-112414-055048
Feretzaki, M., Renck Nunes, P., and Lingner, J. (2019). Expression and differential regulation of human TERRA at several chromosome ends. RNA 25, 1470–1480. doi: 10.1261/rna.072322.119
Feric, M., Vaidya, N., Harmon, T. S., Mitrea, D. M., Zhu, L., Richardson, T. M., et al. (2016). Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697. doi: 10.1016/j.cell.2016.04.047
Fussner, E., Ahmed, K., Dehghani, H., Strauss, M., and Bazett-Jones, D. P. (2010). Changes in chromatin fiber density as a marker for pluripotency. Cold. Spring Harb. Symp. Quant. Biol. 75, 245–249. doi: 10.1101/sqb.2010.75.012
García-Cao, M., O’Sullivan, R., Peters, A. H. F. M., Jenuwein, T., and Blasco, M. A. (2004). Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 36, 94–99. doi: 10.1038/ng1278
Gaspar-Maia, A., Alajem, A., Meshorer, E., and Ramalho-Santos, M. (2011). Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12, 36–47. doi: 10.1038/nrm3036
Gaspar-Maia, A., Alajem, A., Polesso, F., Sridharan, R., Mason, M. J., Heidersbach, A., et al. (2009). Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460, 863–868. doi: 10.1038/nature08212
Gauchier, M., Kan, S., Barral, A., Sauzet, S., Agirre, E., Bonnell, E., et al. (2019). SETDB1-dependent heterochromatin stimulates alternative lengthening of telomeres. Sci. Adv. 5:eaav3673. doi: 10.1126/sciadv.aav3673
Ge, X. Q., Han, J., Cheng, E.-C., Yamaguchi, S., Shima, N., Thomas, J.-L., et al. (2015). Embryonic stem cells license a high level of dormant origins to protect the genome against replication stress. Stem Cell Rep. 5, 185–194. doi: 10.1016/j.stemcr.2015.06.002
Gibson, B. A., Doolittle, L. K., Schneider, M. W. G., Jensen, L. E., Gamarra, N., Henry, L., et al. (2019). Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470.e21–484.e21.
Goldberg, A. D., Banaszynski, L. A., Noh, K.-M., Lewis, P. W., Elsaesser, S. J., Stadler, S., et al. (2010). Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691.
Gonzalo, S., García-Cao, M., Fraga, M. F., Schotta, G., Peters, A. H. F. M., Cotter, S. E., et al. (2005). Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat. Cell Biol. 7, 420–428. doi: 10.1038/ncb1235
Gonzalo, S., Jaco, I., Fraga, M. F., Chen, T., Li, E., Esteller, M., et al. (2006). DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 8, 416–424. doi: 10.1038/ncb1386
Han, J., Yuan, P., Yang, H., Zhang, J., Soh, B. S., Li, P., et al. (2010). Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature 463, 1096–1100. doi: 10.1038/nature08735
Hassan-Zadeh, V., Rugg-Gunn, P., and Bazett-Jones, D. P. (2017). DNA methylation is dispensable for changes in global chromatin architecture but required for chromocentre formation in early stem cell differentiation. Chromosoma 126, 605–614. doi: 10.1007/s00412-017-0625-x
He, Q., Kim, H., Huang, R., Lu, W., Tang, M., Shi, F., et al. (2015). The Daxx/Atrx complex protects tandem repetitive elements during DNA hypomethylation by promoting H3K9 trimethylation. Cell Stem Cell 17, 273–286. doi: 10.1016/j.stem.2015.07.022
Heaphy, C. M., de Wilde, R. F., Jiao, Y., Klein, A. P., Edil, B. H., Shi, C., et al. (2011). Altered telomeres in tumors with ATRX and DAXX mutations. Science 333:425.
Hendrickson, P. G., Doráis, J. A., Grow, E. J., Whiddon, J. L., Lim, J.-W., Wike, C. L., et al. (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 49, 925–934. doi: 10.1038/ng.3844
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K., and Sharp, P. A. (2017). A phase separation model for transcriptional control. Cell 169, 13–23. doi: 10.1016/j.cell.2017.02.007
Hodgkinson, A., Chen, Y., and Eyre-Walker, A. (2012). The large-scale distribution of somatic mutations in cancer genomes. Hum. Mutat. 33, 136–143. doi: 10.1002/humu.21616
Holt, S. E., Shay, J. W., and Wright, W. E. (1996). Refining the telomere-telomerase hypothesis of aging and cancer. Nat. Biotechnol. 14, 836–839. doi: 10.1038/nbt0796-836
Huangfu, D., Maehr, R., Guo, W., Eijkelenboom, A., Snitow, M., Chen, A. E., et al. (2008). Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 26, 795–797. doi: 10.1038/nbt1418
Huo, X., Ji, L., Zhang, Y., Lv, P., Cao, X., Wang, Q., et al. (2020). The nuclear matrix protein SAFB cooperates with major satellite RNAs to stabilize heterochromatin architecture partially through phase separation. Mol. Cell. 77, 368.e7–383.e7.
Hyka-Nouspikel, N., Desmarais, J., Gokhale, P. J., Jones, M., Meuth, M., Andrews, P. W., et al. (2012). Deficient DNA damage response and cell cycle checkpoints lead to accumulation of point mutations in human embryonic stem cells. Stem Cells 30, 1901–1910. doi: 10.1002/stem.1177
Jakob, B., Splinter, J., Conrad, S., Voss, K.-O., Zink, D., Durante, M., et al. (2011). DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 39, 6489–6499. doi: 10.1093/nar/gkr230
Janssen, A., Breuer, G. A., Brinkman, E. K., van der Meulen, A. I., Borden, S. V., van Steensel, B., et al. (2016). A single double-strand break system reveals repair dynamics and mechanisms in heterochromatin and euchromatin. Genes Dev. 30, 1645–1657. doi: 10.1101/gad.283028.116
Janssen, A., Colmenares, S. U., and Karpen, G. H. (2018). Heterochromatin: guardian of the genome. Annu. Rev. Cell Dev. Biol. 34, 265–288. doi: 10.1146/annurev-cellbio-100617-062653
Kalaszczynska, I., Geng, Y., Iino, T., Mizuno, S.-I., Choi, Y., Kondratiuk, I., et al. (2009). Cyclin A is redundant in fibroblasts but essential in hematopoietic and embryonic stem cells. Cell 138, 352–365. doi: 10.1016/j.cell.2009.04.062
Kallimasioti-Pazi, E. M., Thelakkad Chathoth, K., Taylor, G. C., Meynert, A., Ballinger, T., Kelder, M. J. E., et al. (2018). Heterochromatin delays CRISPR-Cas9 mutagenesis but does not influence the outcome of mutagenic DNA repair. PLoS Biol. 16:e2005595. doi: 10.1371/journal.pbio.2005595
Kareta, M. S., Gorges, L. L., Hafeez, S., Benayoun, B. A., Marro, S., Zmoos, A.-F., et al. (2015). Inhibition of pluripotency networks by the Rb tumor suppressor restricts reprogramming and tumorigenesis. Cell Stem Cell 16, 39–50. doi: 10.1016/j.stem.2014.10.019
Karlseder, J., Kachatrian, L., Takai, H., Mercer, K., Hingorani, S., Jacks, T., et al. (2003). Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol. Cell. Biol. 23, 6533–6541. doi: 10.1128/mcb.23.18.6533-6541.2003
Karnavas, T., Pintonello, L., Agresti, A., and Bianchi, M. E. (2014). Histone content increases in differentiating embryonic stem cells. Front. Physiol. 5:330. doi: 10.3389/fphys.2014.00330
Kato, M., Han, T. W., Xie, S., Shi, K., Du, X., Wu, L. C., et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767. doi: 10.1016/j.cell.2012.04.017
Kilic, S., Lezaja, A., Gatti, M., Bianco, E., Michelena, J., Imhof, R., et al. (2019). Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 38:e101379.
Krawczyk, P. M., Borovski, T., Stap, J., Cijsouw, T., ten Cate, R., Medema, J. P., et al. (2012). Chromatin mobility is increased at sites of DNA double-strand breaks. J. Cell Sci. 125(Pt 9), 2127–2133.
Kruhlak, M. J., Celeste, A., Dellaire, G., Fernandez-Capetillo, O., Müller, W. G., McNally, J. G., et al. (2006). Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 172, 823–834. doi: 10.1083/jcb.200510015
Langdon, E. M., Qiu, Y., Ghanbari Niaki, A., McLaughlin, G. A., Weidmann, C. A., Gerbich, T. M., et al. (2018). mRNA structure determines specificity of a polyQ-driven phase separation. Science 360, 922–927. doi: 10.1126/science.aar7432
Larson, A. G., Elnatan, D., Keenen, M. M., Trnka, M. J., Johnston, J. B., Burlingame, A. L., et al. (2017). Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240. doi: 10.1038/nature22822
Le, P. N., Maranon, D. G., Altina, N. H., Battaglia, C. L. R., and Bailey, S. M. (2013). TERRA, hnRNP A1, and DNA-PKcs interactions at human telomeres. Front. Oncol. 3:91. doi: 10.3389/fonc.2013.00091
Lee, Y. C. G., and Karpen, G. H. (2017). Pervasive epigenetic effects of euchromatic transposable elements impact their evolution. eLife 6:e25762. doi: 10.7554/eLife.25762
Lessard, J. A., and Crabtree, G. R. (2010). Chromatin regulatory mechanisms in pluripotency. Annu. Rev. Cell Dev. Biol. 26, 503–532. doi: 10.1146/annurev-cellbio-051809-102012
Lewis, P. W., Elsaesser, S. J., Noh, K.-M., Stadler, S. C., and Allis, C. D. (2010). Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. U.S.A. 107, 14075–14080. doi: 10.1073/pnas.1008850107
Lin, Y., Protter, D. S. W., Rosen, M. K., and Parker, R. (2015). Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell. 60, 208–219. doi: 10.1016/j.molcel.2015.08.018
Liu, J. C., Guan, X., Ryan, J. A., Rivera, A. G., Mock, C., Agrawal, V., et al. (2013). High mitochondrial priming sensitizes hESCs to DNA-damage-induced apoptosis. Cell Stem Cell 13, 483–491. doi: 10.1016/j.stem.2013.07.018
Liu, L., Bailey, S. M., Okuka, M., Muñoz, P., Li, C., Zhou, L., et al. (2007). Telomere lengthening early in development. Nat. Cell Biol. 9, 1436–1441.
López de Silanes, I., Graña, O., De Bonis, M. L., Dominguez, O., Pisano, D. G., and Blasco, M. A. (2014). Identification of TERRA locus unveils a telomere protection role through association to nearly all chromosomes. Nat. Commun. 5:4723.
Lovejoy, C. A., Li, W., Reisenweber, S., Thongthip, S., Bruno, J., de Lange, T., et al. (2012). Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 8:e1002772. doi: 10.1371/journal.pgen.1002772
Lu, H., Yu, D., Hansen, A. S., Ganguly, S., Liu, R., Heckert, A., et al. (2018). Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323. doi: 10.1038/s41586-018-0174-3
Lu, R., Yang, A., and Jin, Y. (2011). Dual functions of T-box 3 (Tbx3) in the control of self-renewal and extraembryonic endoderm differentiation in mouse embryonic stem cells. J. Biol. Chem. 286, 8425–8436. doi: 10.1074/jbc.m110.202150
Macfarlan, T. S., Gifford, W. D., Driscoll, S., Lettieri, K., Rowe, H. M., Bonanomi, D., et al. (2012). Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63. doi: 10.1038/nature11244
Marión, R. M., Montero, J. J., López de Silanes, I., Graña-Castro, O., Martínez, P., Schoeftner, S., et al. (2019). TERRA regulate the transcriptional landscape of pluripotent cells through TRF1-dependent recruitment of PRC2. eLife 8:e44656. doi: 10.7554/eLife.44656
Marion, R. M., Strati, K., Li, H., Tejera, A., Schoeftner, S., Ortega, S., et al. (2009). Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell 4, 141–154. doi: 10.1016/j.stem.2008.12.010
Markiewicz-Potoczny, M., Lobanova, A., Loeb, A. M., Kirak, O., Olbrich, T., Ruiz, S., et al. (2021). TRF2-mediated telomere protection is dispensable in pluripotent stem cells. Nature 589, 110–115. doi: 10.1038/s41586-020-2959-4
Martínez, P., and Blasco, M. A. (2011). Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat. Rev. Cancer 11, 161–176. doi: 10.1038/nrc3025
McNairn, A. J., and Gilbert, D. M. (2003). Epigenomic replication: linking epigenetics to DNA replication. BioEssays 25, 647–656. doi: 10.1002/bies.10305
Mendez-Bermudez, A., Lototska, L., Bauwens, S., Giraud-Panis, M.-J., Croce, O., Jamet, K., et al. (2018). Genome-wide control of heterochromatin replication by the telomere capping protein TRF2. Mol. Cell 70, 449.e5–461.5e.
Meshorer, E., and Misteli, T. (2006). Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol Cell Biol. 7, 540–546.
Meshorer, E., Yellajoshula, D., George, E., Scambler, P. J., Brown, D. T., and Misteli, T. (2006). Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell 10, 105–116. doi: 10.1016/j.devcel.2005.10.017
Mikkelsen, T. S., Hanna, J., Zhang, X., Ku, M., Wernig, M., Schorderet, P., et al. (2008). Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55. doi: 10.1038/nature07056
Min, J., Wright, W. E., and Shay, J. W. (2019). Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev. 33, 814–827. doi: 10.1101/gad.324905.119
Momčilović, O., Navara, C., and Schatten, G. (2011). Cell cycle adaptations and maintenance of genomic integrity in embryonic stem cells and induced pluripotent stem cells. Results Probl. Cell Differ. 53, 415–458. doi: 10.1007/978-3-642-19065-0_18
Munder, M. C., Midtvedt, D., Franzmann, T., Nüske, E., Otto, O., Herbig, M., et al. (2016). A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife 5:e09347. doi: 10.7554/eLife.09347
Murga, M., Jaco, I., Fan, Y., Soria, R., Martinez-Pastor, B., Cuadrado, M., et al. (2007). Global chromatin compaction limits the strength of the DNA damage response. J. Cell Biol. 178, 1101–1108. doi: 10.1083/jcb.200704140
Nakai-Futatsugi, Y., and Niwa, H. (2016). Zscan4 is activated after telomere shortening in mouse embryonic stem cells. Stem Cell Rep. 6, 483–495. doi: 10.1016/j.stemcr.2016.02.010
Novo, C. L., Javierre, B.-M., Cairns, J., Segonds-Pichon, A., Wingett, S. W., Freire-Pritchett, P., et al. (2018). Long-range enhancer interactions are prevalent in mouse embryonic stem cells and are reorganized upon pluripotent state transition. Cell Rep. 22, 2615–2627. doi: 10.1016/j.celrep.2018.02.040
Novo, C. L., Tang, C., Ahmed, K., Djuric, U., Fussner, E., Mullin, N. P., et al. (2016). The pluripotency factor Nanog regulates pericentromeric heterochromatin organization in mouse embryonic stem cells. Genes Dev. 30, 1101–1115. doi: 10.1101/gad.275685.115
Novo, C. L., Wong, E., Hockings, C., Poudel, C., Sheekey, E., Walker, S., et al. (2020). Satellite repeat transcripts modulate heterochromatin condensates and safeguard chromosome stability in mouse embryonic stem cells. bioRxiv [Preprint] doi: 10.1101/2020.06.08.139642
Novo, C., Arnoult, N., Bordes, W.-Y., Castro-Vega, L., Gibaud, A., Dutrillaux, B., et al. (2013). The heterochromatic chromosome caps in great apes impact telomere metabolism. Nucleic Acids Res. 41, 4792–4801. doi: 10.1093/nar/gkt169
Oshidari, R., Huang, R., Medghalchi, M., Tse, E. Y. W., Ashgriz, N., Lee, H. O., et al. (2020). DNA repair by Rad52 liquid droplets. Nat. Commun. 11:695.
Palikyras, S., and Papantonis, A. (2019). Modes of phase separation affecting chromatin regulation. Open Biol. 9:190167. doi: 10.1098/rsob.190167
Patel, A., Lee, H. O., Jawerth, L., Maharana, S., Jahnel, M., Hein, M. Y., et al. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077. doi: 10.1016/j.cell.2015.07.047
Penev, A., Bazley, A., Shen, M., Boeke, J. D., Savage, S. A., and Sfeir, A. (2021). Alternative splicing is a developmental switch for hTERT expression. Mol. Cell 81, 2349.e6–2360.e6. doi: 10.1016/j.molcel.2021.03.033
Pessina, F., Giavazzi, F., Yin, Y., Gioia, U., Vitelli, V., Galbiati, A., et al. (2019). Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat. Cell Biol. 21, 1286–1299. doi: 10.1038/s41556-019-0392-4
Petermann, E., Orta, M. L., Issaeva, N., Schultz, N., and Helleday, T. (2010). Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 37, 492–502. doi: 10.1016/j.molcel.2010.01.021
Peters, A. H., O’Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schöfer, C., et al. (2001). Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337. doi: 10.1016/s0092-8674(01)00542-6
Plys, A. J., Davis, C. P., Kim, J., Rizki, G., Keenen, M. M., Marr, S. K., et al. (2019). Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 33, 799–813. doi: 10.1101/gad.326488.119
Porreca, R. M., Herrera-Moyano, E., Skourti, E., Law, P. P., Gonzalez Franco, R., Montoya, A., et al. (2020). TRF1 averts chromatin remodelling, recombination and replication dependent-break induced replication at mouse telomeres. eLife 9:e49817. doi: 10.7554/eLife.49817
Pucci, F., Gardano, L., and Harrington, L. (2013). Short telomeres in ESCs lead to unstable differentiation. Cell Stem Cell 12, 479–486. doi: 10.1016/j.stem.2013.01.018
Ratnakumar, K., Duarte, L. F., LeRoy, G., Hasson, D., Smeets, D., Vardabasso, C., et al. (2012). ATRX-mediated chromatin association of histone variant macroH2A1 regulates α-globin expression. Genes Dev. 26, 433–438. doi: 10.1101/gad.179416.111
Riback, J. A., Katanski, C. D., Kear-Scott, J. L., Pilipenko, E. V., Rojek, A. E., Sosnick, T. R., et al. (2017). Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168, 1028.e19–1040.e19.
Roos, W. P., and Kaina, B. (2006). DNA damage-induced cell death by apoptosis. Trends Mol. Med. 12, 440–450. doi: 10.1016/j.molmed.2006.07.007
Ruis, P., Van Ly, D., Borel, V., Kafer, G. R., McCarthy, A., Howell, S., et al. (2021). TRF2-independent chromosome end protection during pluripotency. Nature 589, 103–109. doi: 10.1038/s41586-020-2960-y
Sabari, B. R., Dall’Agnese, A., Boija, A., Klein, I. A., Coffey, E. L., Shrinivas, K., et al. (2018). Coactivator condensation at super-enhancers links phase separation and gene control. Science 361:eaar3958. doi: 10.1126/science.aar3958
Sanulli, S., Gross, J. D., and Narlikar, G. J. (2019a). Biophysical properties of HP1-mediated heterochromatin. Cold. Spring Harb. Symp. Quant. Biol. 84, 217–225. doi: 10.1101/sqb.2019.84.040360
Sanulli, S., Trnka, M. J., Dharmarajan, V., Tibble, R. W., Pascal, B. D., Burlingame, A. L., et al. (2019b). HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575, 390–394. doi: 10.1038/s41586-019-1669-2
Sarek, G., Vannier, J.-B., Panier, S., Petrini, J. H. J., and Boulton, S. J. (2016). TRF2 recruits RTEL1 to telomeres in S phase to promote T-loop unwinding. Mol. Cell. 61, 788–789. doi: 10.1016/j.molcel.2016.02.016
Schaetzlein, S., Lucas-Hahn, A., Lemme, E., Kues, W. A., Dorsch, M., Manns, M. P., et al. (2004). Telomere length is reset during early mammalian embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 8034–8038. doi: 10.1073/pnas.0402400101
Scheibe, M., Arnoult, N., Kappei, D., Buchholz, F., Decottignies, A., Butter, F., et al. (2013). Quantitative interaction screen of telomeric repeat-containing RNA reveals novel TERRA regulators. Genome Res. 23, 2149–2157. doi: 10.1101/gr.151878.112
Schneider, R. P., Garrobo, I., Foronda, M., Palacios, J. A., Marión, R. M., Flores, I., et al. (2013). TRF1 is a stem cell marker and is essential for the generation of induced pluripotent stem cells. Nat. Commun. 4:1946.
Schoeftner, S., and Blasco, M. A. (2008). Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10, 228–236. doi: 10.1038/ncb1685
Schoeftner, S., Blanco, R., Lopez de Silanes, I., Muñoz, P., Gómez-López, G., Flores, J. M., et al. (2009). Telomere shortening relaxes X chromosome inactivation and forces global transcriptome alterations. Proc. Natl. Acad. Sci. U.S.A. 106, 19393–19398. doi: 10.1073/pnas.0909265106
Schuster-Böckler, B., and Lehner, B. (2012). Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature 488, 504–507. doi: 10.1038/nature11273
Segura-Bayona, S., Villamor-Payà, M., Attolini, C. S.-O., Koenig, L. M., Sanchiz-Calvo, M., Boulton, S. J., et al. (2020). Tousled-like kinases suppress innate immune signaling triggered by alternative lengthening of telomeres. Cell Rep. 32:107983. doi: 10.1016/j.celrep.2020.107983
Sfeir, A., Kosiyatrakul, S. T., Hockemeyer, D., MacRae, S. L., Karlseder, J., Schildkraut, C. L., et al. (2009). Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138, 90–103. doi: 10.1016/j.cell.2009.06.021
Soufi, A., and Dalton, S. (2016). Cycling through developmental decisions: how cell cycle dynamics control pluripotency, differentiation and reprogramming. Development 143, 4301–4311. doi: 10.1242/dev.142075
Soufi, A., Donahue, G., and Zaret, K. S. (2012). Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151, 994–1004. doi: 10.1016/j.cell.2012.09.045
Sridharan, R., Gonzales-Cope, M., Chronis, C., Bonora, G., McKee, R., Huang, C., et al. (2013). Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat. Cell Biol. 15, 872–882. doi: 10.1038/ncb2768
Stambrook, P. J., and Tichy, E. D. (2010). Preservation of genomic integrity in mouse embryonic stem cells. Adv. Exp. Med. Biol. 695, 59–75. doi: 10.1007/978-1-4419-7037-4_5
Strom, A. R., Emelyanov, A. V., Mir, M., Fyodorov, D. V., Darzacq, X., and Karpen, G. H. (2017). Phase separation drives heterochromatin domain formation. Nature 547, 241–245. doi: 10.1038/nature22989
Tatavosian, R., Kent, S., Brown, K., Yao, T., Duc, H. N., Huynh, T. N., et al. (2019). Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 294, 1451–1463. doi: 10.1074/jbc.ra118.006620
Tichy, E. D., and Stambrook, P. J. (2008). DNA repair in murine embryonic stem cells and differentiated cells. Exp. Cell Res. 314, 1929–1936. doi: 10.1016/j.yexcr.2008.02.007
Tichy, E. D., Pillai, R., Deng, L., Liang, L., Tischfield, J., Schwemberger, S. J., et al. (2010). Mouse embryonic stem cells, but not somatic cells, predominantly use homologous recombination to repair double-strand DNA breaks. Stem Cells Dev. 19, 1699–1711. doi: 10.1089/scd.2010.0058
Trivedi, P., Palomba, F., Niedzialkowska, E., Digman, M. A., Gratton, E., and Stukenberg, P. T. (2019). The inner centromere is a biomolecular condensate scaffolded by the chromosomal passenger complex. Nat. Cell Biol. 21, 1127–1137. doi: 10.1038/s41556-019-0376-4
Tsai, S.-Y., Opavsky, R., Sharma, N., Wu, L., Naidu, S., Nolan, E., et al. (2008). Mouse development with a single E2F activator. Nature 454, 1137–1141. doi: 10.1038/nature07066
Tsouroula, K., Furst, A., Rogier, M., Heyer, V., Maglott-Roth, A., Ferrand, A., et al. (2016). Temporal and spatial uncoupling of DNA double strand break repair pathways within mammalian heterochromatin. Mol. Cell. 63, 293–305. doi: 10.1016/j.molcel.2016.06.002
Udugama, M., Chang, M., Chan, F. T., Tang, F. L., Pickett, M. C., R McGhie, H. A., et al. (2015). Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res. 43, 10227–10237.
Valencia, A. M., and Kadoch, C. (2019). Chromatin regulatory mechanisms and therapeutic opportunities in cancer. Nat. Cell Biol. 21, 152–161. doi: 10.1038/s41556-018-0258-1
van der Laan, S., Tsanov, N., Crozet, C., and Maiorano, D. (2013). High Dub3 expression in mouse ESCs couples the G1/S checkpoint to pluripotency. Mol. Cell. 52, 366–379. doi: 10.1016/j.molcel.2013.10.003
Varela, E., Schneider, R. P., Ortega, S., and Blasco, M. A. (2011). Different telomere-length dynamics at the inner cell mass versus established embryonic stem (ES) cells. Proc. Natl. Acad. Sci. U.S.A. 108, 15207–15212. doi: 10.1073/pnas.1105414108
Viceconte, N., Loriot, A., Lona Abreu, P., Scheibe, M., Fradera Sola, A., Butter, F., et al. (2021). PAR-TERRA is the main contributor to telomeric repeat-containing RNA transcripts in normal and cancer mouse cells. RNA 27, 106–121. doi: 10.1261/rna.076281.120
Villeponteau, B. (1997). The heterochromatin loss model of aging. Exp. Gerontol. 32, 383–394. doi: 10.1016/s0531-5565(96)00155-6
Vitale, I., Manic, G., De Maria, R., Kroemer, G., and Galluzzi, L. (2017). DNA damage in stem cells. Mol. Cell. 66, 306–319.
Wen, B., Wu, H., Shinkai, Y., Irizarry, R. A., and Feinberg, A. P. (2009). Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 41, 246–250. doi: 10.1038/ng.297
Whiddon, J. L., Langford, A. T., Wong, C.-J., Zhong, J. W., and Tapscott, S. J. (2017). Conservation and innovation in the DUX4-family gene network. Nat. Genet. 49, 935–940. doi: 10.1038/ng.3846
Wong, L. H., McGhie, J. D., Sim, M., Anderson, M. A., Ahn, S., Hannan, R. D., et al. (2010). ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 20, 351–360. doi: 10.1101/gr.101477.109
Wong, L. H., Ren, H., Williams, E., McGhie, J., Ahn, S., Sim, M., et al. (2009). Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 19, 404–414. doi: 10.1101/gr.084947.108
Wyles, S. P., Brandt, E. B., and Nelson, T. J. (2014). Stem cells: the pursuit of genomic stability. Int. J. Mol. Sci. 15, 20948–20967. doi: 10.3390/ijms151120948
Yamada, T., Yoshimura, H., Shimada, R., Hattori, M., Eguchi, M., Fujiwara, T. K., et al. (2016). Spatiotemporal analysis with a genetically encoded fluorescent RNA probe reveals TERRA function around telomeres. Sci. Rep. 6:38910.
Yang, J., Guo, R., Wang, H., Ye, X., Zhou, Z., Dan, J., et al. (2016). Tet enzymes regulate telomere maintenance and chromosomal stability of mouse ESCs. Cell Rep. 15, 1809–1821. doi: 10.1016/j.celrep.2016.04.058
Yeager, T. R., Neumann, A. A., Englezou, A., Huschtscha, L. I., Noble, J. R., and Reddel, R. R. (1999). Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59, 4175–4179.
Yoon, S.-W., Kim, D.-K., Kim, K. P., and Park, K.-S. (2014). Rad51 regulates cell cycle progression by preserving G2/M transition in mouse embryonic stem cells. Stem Cells Dev. 23, 2700–2711. doi: 10.1089/scd.2014.0129
Zalzman, M., Falco, G., Sharova, L. V., Nishiyama, A., Thomas, M., Lee, S.-L., et al. (2010). Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464, 858–863. doi: 10.1038/nature08882
Zhang, H., Zhao, R., Tones, J., Liu, M., Dilley, R. L., Chenoweth, D. M., et al. (2020). Nuclear body phase separation drives telomere clustering in ALT cancer cells. Mol. Biol. Cell 31, 2048–2056. doi: 10.1091/mbc.e19-10-0589
Zhang, L.-F., Ogawa, Y., Ahn, J. Y., Namekawa, S. H., Silva, S. S., and Lee, J. T. (2009). Telomeric RNAs mark sex chromosomes in stem cells. Genetics 182, 685–698. doi: 10.1534/genetics.109.103093
Zhang, Q., Dan, J., Wang, H., Guo, R., Mao, J., Fu, H., et al. (2016). Tcstv1 and Tcstv3 elongate telomeres of mouse ES cells. Sci. Rep. 6:19852.
Zhang, W., Li, J., Suzuki, K., Qu, J., Wang, P., Zhou, J., et al. (2015). Aging stem cells. A werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 348, 1160–1163.
Keywords: telomeres, pluripotency, phase separation, chromatin, nuclear architecture, epigenetics
Citation: Novo CL (2021) A Tale of Two States: Pluripotency Regulation of Telomeres. Front. Cell Dev. Biol. 9:703466. doi: 10.3389/fcell.2021.703466
Received: 30 April 2021; Accepted: 08 June 2021;
Published: 08 July 2021.
Edited by:
Marcelo Santos Da Silva, São Paulo State University, BrazilReviewed by:
Pascal Chartrand, Université de Montréal, CanadaDean Harvey Betts, Western University, Canada
Copyright © 2021 Novo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Clara Lopes Novo, Y2xhcmEubm92b0Bjcmljay5hYy51aw==
 Clara Lopes Novo
Clara Lopes Novo