- 1Center for Gene Regulation in Health and Disease, Department of Biological, Geological, and Environmental Sciences, College of Sciences and Health Professions, Cleveland State University, Cleveland, OH, United States
- 2Case Comprehensive Cancer Center, Case Western Reserve University, Cleveland, OH, United States
- 3Department of Inflammation and Immunity, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, United States
- 4Center for RNA Science and Therapeutics, Case Western Reserve University, Cleveland, OH, United States
Telomeres, the nucleoprotein complexes at chromosome ends, are well-known for their essential roles in genome integrity and chromosome stability. Yet, telomeres and subtelomeres are frequently less stable than chromosome internal regions. Many subtelomeric genes are important for responding to environmental cues, and subtelomeric instability can facilitate organismal adaptation to extracellular changes, which is a common theme in a number of microbial pathogens. In this review, I will focus on the delicate and important balance between stability and plasticity at telomeres and subtelomeres of a kinetoplastid parasite, Trypanosoma brucei, which causes human African trypanosomiasis and undergoes antigenic variation to evade the host immune response. I will summarize the current understanding about T. brucei telomere protein complex, the telomeric transcript, and telomeric R-loops, focusing on their roles in maintaining telomere and subtelomere stability and integrity. The similarities and differences in functions and underlying mechanisms of T. brucei telomere factors will be compared with those in human and yeast cells.
Introduction
As eukaryotic cells have evolved to have linear chromosomes, so has telomere evolved to play a key role in maintaining genome integrity and chromosome stability. Telomeres are the nucleoprotein complexes at linear chromosome ends. The telomere sequence, structure, and telomere-associated proteins play essential roles in proper telomere length maintenance, chromosome end protection, and regulation of subtelomeric gene expression (Ottaviani et al., 2008; Sobinoff and Pickett, 2017; de Lange, 2018; Laberthonnière et al., 2019). Nevertheless, recent studies in unicellular protozoan parasites and fungi suggest that subtle telomere and subtelomere instability can be beneficial for individual organism to adapt to challenging growth environment in the short term and can contribute to species evolution in the long term. In this review, I will first briefly summarize key telomere functions, then describe the relationship between telomere stability and antigenic variation in a protozoan parasite Trypanosoma brucei, focusing on similar and different challenges faced by T. brucei telomere proteins and those in mammalian and yeast cells. Last I will discuss potential benefit of limited telomere stability, especially in a number of microbial pathogens.
Telomeres Are Essential for Genome Stability
Telomeres in many eukaryotes consist of simple repetitive TG-rich sequences (Podlevsky et al., 2008). All vertebrates, a small number of insects, a few species of plants and amoeba, and several kinetoplastids including T. brucei and Leishmania have telomeres with a sequence of perfect TTAGGG repeats (Podlevsky et al., 2008). Telomeres are mostly double stranded, but a terminal single-stranded overhang structure has been observed in many organisms, including vertebrates (150–400 nt long) and yeasts (12–40 nt long) (Wellinger et al., 1993; Makarov et al., 1997). There is also a single-stranded TG-rich 3′ overhang structure at the end of T. brucei telomere, although it appears to be very short (∼12 nt long) (Sandhu and Li, 2011, Sandhu and Li, 2017). This terminal G-overhang can invade the duplex telomere region and form the T-loop structure, which has been observed in human, mouse, chicken, T. brucei, ciliates, common garden pea, Caenorhabditis elegans, and Kluyveromyces lactis (Griffith et al., 1999; Murti and Prescott, 1999; Munoz-Jordan et al., 2001; Cesare et al., 2003, 2008; Nikitina and Woodcock, 2004; Raices et al., 2008). The T-loop structure buries the telomere G-overhang, which suppresses ATM activation at mammalian telomeres (Van Ly et al., 2018). In addition, telomere binding proteins can also help protect the telomere by inhibiting Non-Homologous End Joining (NHEJ) (Smogorzewska et al., 2002; Celli and de Lange, 2005; Deng et al., 2009a; Pardo and Marcand, 2005), Homologous Recombination (HR) (Wang et al., 2004; Guo et al., 2007; Sfeir et al., 2010; Chen et al., 2011), and Microhomology-Mediated End Joining (MMEJ) (Rai et al., 2010; Sfeir and de Lange, 2012) at the telomere.
Naked telomere DNA is not only vulnerable to nucleolytic degradation but also resembles a DNA double-strand break (DSB) product. Therefore, without the protection from telomere associated proteins, DNA damage response machinery is recruited to the telomere and repair processes are attempted. In fact, it is well known that removal of key telomere proteins results in chromosomes end-to-end fusions in mammalian and yeast cells (van Steensel et al., 1998; Ferreira and Cooper, 2001; Celli and de Lange, 2005; Pardo and Marcand, 2005). The resulting dicentric chromosomes can initiate the so-called breakage-fusion-bridge (BFB) cycle (de Lange, 2018): when dicentric chromosomes are pulled to opposite poles of the dividing cell, anaphase bridges form, which is frequently followed by another round of chromosome breaks and subsequent end-fusions. BFB is a severe genome instability factor and can induce loss of heterozygosity, non-reciprocal translocations, and gene amplification (Maciejowski and de Lange, 2017). In human cells, anaphase bridges can be resolved by TREX1, a cytoplasmic 3′ exonuclease, to form single-stranded DNA, which can eventually result in chromothripsis (a process where a chromosome region is broken in a single step into many fragments followed by haphazard repair) (Maciejowski et al., 2015). In addition, APOBEC- (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) mediated kataegis (clustered C > T and C > G changes at TpC dinucleotides) can occur with chromothripsis (Maciejowski et al., 2015). Importantly, chromothripsis and kataegis have been observed in many tumor types (Cleal and Baird, 2020), and chromoanagenesis (including chromothripsis, chromoplexy, and chromoanasynthesis) has been recognized as an important mechanism of genome instability that can contribute to tumorigenesis (Cleal and Baird, 2020; Pellestor et al., 2021). Therefore, proper telomere protection is critical for genome integrity and chromosome stability, and a key function of the telomere associated factors is to prevent the natural chromosome ends from being recognized as DNA damage sites (de Lange, 2018).
Chromosome end protection relies on a number of telomere proteins to be loaded onto the telomere DNA directly through DNA binding activities or indirectly through protein-protein interactions. Therefore, telomere DNA serves as a docking site for the telomere binding proteins and proper telomere maintenance is a pre-requisite for a stable telomere. In most eukaryotes, the 3′ end of the G-rich telomere strand can be extended by telomerase, a specialized reverse transcriptase, through de novo DNA synthesis, which relies on its intrinsic RNA to provide the template sequence (Greider and Blackburn, 1985, 1987; Shay and Wright, 2019). With the help from the CST telomere complex (CTC1/STN1/TEN1 in mammals and Cdc13/Stn1/Ten1 in budding yeast), the C-rich telomere strand can be subsequently filled-in by primase-polymerase alpha (normally involved in lagging strand synthesis) (Feng et al., 2017; Stewart et al., 2018). The telomerase activity counteracts the “end replication problem” due to the inability of conventional DNA polymerases to fully replicate the ends of linear DNA molecules (Greider and Blackburn, 1987, 1989). In telomerase-negative cells, DNA recombination [including the break-induced replication (BIR)], and rolling circle DNA replication can serve as mechanisms to amplify telomere and subtelomere sequences, achieving the goal of telomere maintenance (Zhang and Zou, 2020).
In many organisms including T. brucei, both budding and fission yeasts, and human cells, telomeres form a heterochromatic structure that exerts a repressive effect on transcription of genes located at subtelomeric regions (Gottschling et al., 1990; Nimmo et al., 1994; Horn and Cross, 1997; Baur et al., 2001; Ottaviani et al., 2008; Yang et al., 2009; Tennen et al., 2011; Pandya et al., 2013; Robin et al., 2014; Laberthonnière et al., 2019). This repressive effect is position-dependent, where in general stronger effects are observed for genes located closer to the telomere, hence the name telomere position effect or telomeric silencing (Renauld et al., 1993). On the other hand, at least at some chromosome ends, the telomere sequence is transcribed into a long, non-coding RNA called TElomere Repeat-containing RNA (TERRA) in a number of organisms including T. brucei (Rudenko and Van der Ploeg, 1989; Damasceno et al., 2017; Nanavaty et al., 2017; Saha et al., 2019, 2021), several other kinetoplastids and Plasmodium falciparum (Rudenko and Van der Ploeg, 1989; Damasceno et al., 2017; Morea et al., 2021), human (Azzalin et al., 2007), mouse (Schoeftner and Blasco, 2008), fission (Bah et al., 2012) and budding yeasts (Luke et al., 2008), and birds (Solovei et al., 1994). TERRA exhibits a propensity to form an R-loop structure with the telomere DNA (Toubiana and Selig, 2018). Both TERRA and telomeric R-loop have been shown to regulate telomerase-dependent and recombination-mediated telomere maintenance and also play a role in chromosome end protection (Toubiana and Selig, 2018).
Maintaining Telomere/Subteloemere Stability and Antigenic Variation in Trypanosoma brucei
Trypanosoma brucei is a protozoan parasite that belongs to the Euglenozoa phylum and the Kinetoplastea class and diverged from the mammals in the evolutionary tree more than 500 million years ago. T. brucei has linear chromosomes (van der Ploeg et al., 1984). The T. brucei telomere complex is also essential for maintaining genome stability in this unicellular organism (Li et al., 2005; Jehi et al., 2014a, b; Nanavaty et al., 2017; Afrin et al., 2020a), although the detailed underlying mechanisms are not exactly the same as that in mammalian and yeast cells (see below). Interestingly, T. brucei harbors important virulence genes encoding its major surface antigen at subtelomeres (de Lange and Borst, 1982; Hertz-Fowler et al., 2008; Müller et al., 2018), and the telomere and subtelomere stability has been shown to influence the parasite’s pathogenesis mechanism (Boothroyd et al., 2009; Hovel-Miner et al., 2012; Jehi et al., 2014a, b; Nanavaty et al., 2017; Afrin et al., 2020a). A better understanding of how these parasites maintain their genome stability and how they evade the host immune response will help their eventual elimination.
Antigenic Variation in T. brucei
Trypanosoma brucei causes human African trypanosomiasis (HAT). Its close relatives, Trypanosoma cruzi and Leishmania, also cause debilitating Chagas disease and leishmaniasis, respectively, in humans. These kinetoplastids are important human parasites that collectively affect more than 10 million people world-wide (WHO, 2015). However, few drugs are available to treat these diseases effectively and safely with easy administering. In addition, drug resistance cases have been observed (WHO, 2015).
While proliferating in the extra-cellular space of its mammalian host, bloodstream form (BF) T. brucei expresses variant surface glycoprotein, VSG, as its major surface antigen. ∼10 million VSG proteins are packed densely on the surface of each T. brucei cell, masking a number of invariant surface molecules from the host immune surveillance (Morrison et al., 2009). Although T. brucei has a large VSG gene pool (>2,500 VSG genes and pseudogenes, Figures 1A–D) (Cross et al., 2014), VSGs are monoallelically expressed exclusively from subtelomeric polycistronic transcription units (PTUs) called VSG expression sites (ESs) (Hertz-Fowler et al., 2008; Müller et al., 2018). Each ES typically contains a single functional VSG as the last gene, which is flanked by upstream 70 bp repeats and downstream telomere repeats (Hertz-Fowler et al., 2008). To evade the host immune response, T. brucei regularly expresses immunologically distinct VSGs on the cell surface (Barry and McCulloch, 2001), although VSG switching can happen without any immune selection (Doyle et al., 1980; Myler et al., 1985). VSG switching is sometimes a transcriptional switch (in situ) but frequently mediated by DNA recombination (Myler et al., 1984b), where a previously silent VSG gene is recombined into the active ES to replace the originally active VSG (Figure 1E). In gene conversion (GC) events, the originally active VSG is lost and the donor VSG is duplicated (Robinson et al., 1999), while in reciprocal crossover (CO) events, the originally active and silent VSGs simply exchange places without any loss of genetic information (Rudenko et al., 1996; Figure 1E). Since VSG 3′ UTRs contain a common 14 nt sequence (Cross et al., 2014; Ridewood et al., 2017), the VSG 3′UTR (sometimes together with the downstream telomere sequence) and the 70 bp repeat located upstream of nearly all VSG genes can provide homologous sequences for efficient DNA recombination (Sima et al., 2019), and the DNA recombination-mediated VSG switching has been observed to occur more frequently than in situ switching in many studies (Cross et al., 1998; Robinson et al., 1999; Boothroyd et al., 2009; Kim and Cross, 2010, 2011; Hovel-Miner et al., 2012; Glover et al., 2013; Jehi et al., 2014a, b; Nanavaty et al., 2017). Many proteins involved in DNA replication, recombination, and DNA damage repair are important for VSG switching (McCulloch et al., 2015). HR can be efficiently initiated with DSBs (Haber, 2018). Indeed, introducing a DSB at the active VSG vicinity can induce a 250-fold higher VSG switching rate (Boothroyd et al., 2009; Glover et al., 2013). This is likely why the extremely short telomere downstream of the active VSG in telomerase negative cells (Hovel-Miner et al., 2012) and depletion of several telomere proteins that diminish telomere integrity/stability lead to increased VSG switching frequencies (Jehi et al., 2014a, b, 2016; Nanavaty et al., 2017; Afrin et al., 2020a).
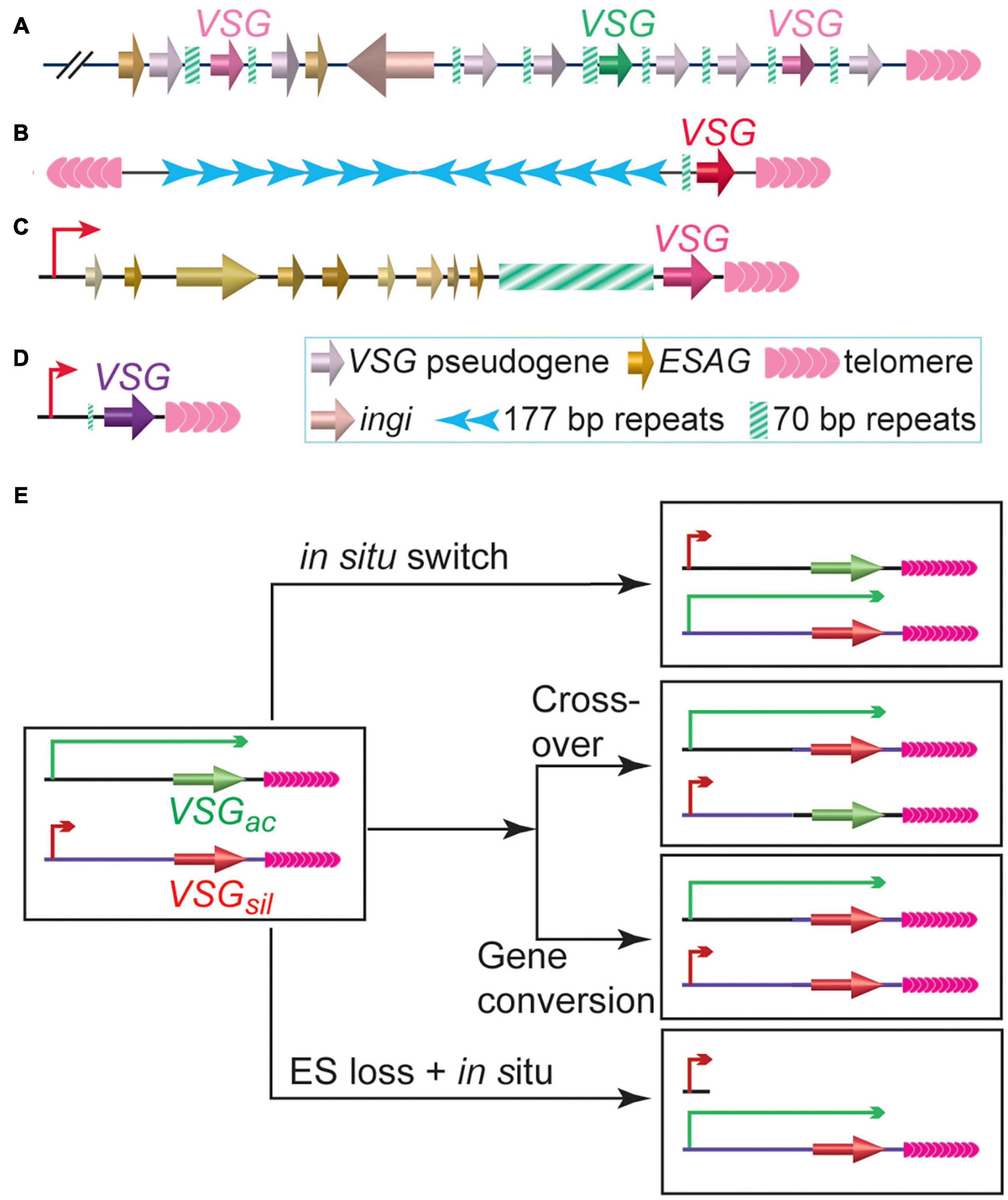
Figure 1. Trypanosoma brucei has a large VSG gene pool. (A) A representative subtelomeric VSG gene array. (B) A typical minichromosome with a subtelomeric VSG gene. (C) A representative bloodstream form VSG expression site, which is a polycistronic transcription unit and can be expressed while T. brucei proliferates in its mammalian host. (D) A representative metacyclic VSG expression site, which is a monocistronic transcription unit and can be expressed while T. brucei resides in the salivary gland of its insect vector. (E) Major VSG switching pathways. VSGac stands for the originally active VSG. VSGsil stands for an originally silent VSG.
T. brucei Telomeres Face Different Telomere Instability Threats Than Those in Mammalian and Yeast Cells
Major pathways for DSB repair include HR, NHEJ, and MMEJ (Ceccaldi et al., 2016). HR has a high fidelity to repair DSBs, seldom introducing mutations (Haber, 2018). However, HR requires a homologous sequence as the repair template, which is usually only feasible in the late S/G2 phase during vegetative growth (Haber, 2018). NHEJ does not require a homologous sequence and can repair DSBs efficiently during G1 (Chang et al., 2017). MMEJ requires very short sequence homology (one or more bps) but frequently introduces insertion-deletions or even translocations and other chromosome rearrangements and is considered an error-prone DNA repair pathway (Sfeir and Symington, 2015; Seol et al., 2018). MMEJ events have been detected in T. brucei (Burton et al., 2007; Glover et al., 2008, 2011), although whether MMEJ can mediate VSG switching or cause chromosome end fusions in T. brucei is currently unknown.
In mammalian cells, Shelterin [the core telomere protein complex including TRF1 (Chong et al., 1995), TRF2 (Bilaud et al., 1997; Broccoli et al., 1997b), Repressor Activator Protein 1 (RAP1) (Li et al., 2000), TIN2 (Kim et al., 1999), POT1 (Baumann and Cech, 2001; Loayza and de Lange, 2003), and TPP1 (Houghtaling et al., 2004; Liu et al., 2004; Ye et al., 2004)] not only inhibits ATM and ATR activation (Karlseder et al., 1999; Celli and de Lange, 2005; Wu et al., 2006; Denchi and de Lange, 2007; Guo et al., 2007; Hockemeyer et al., 2007; Gong and de Lange, 2010; Takai et al., 2011; Frescas and de Lange, 2014; Van Ly et al., 2018), suppresses NHEJ (Smogorzewska et al., 2002; Doksani et al., 2013; Arnoult et al., 2017; de Lange, 2018) and HR (Wang et al., 2004; Wu et al., 2006; Palm et al., 2009; Sfeir et al., 2010) at the telomere, but also prevents nucleolytic degradation (Sfeir and de Lange, 2012; Lottersberger et al., 2013; Kibe et al., 2016) and suppresses MMEJ (Rai et al., 2010; Sfeir and de Lange, 2012; Mateos-Gomez et al., 2015). In addition, TERRA is transcribed by RNA Polymerase II at many telomeres from subtelomeric CpG islands-containing promoters in human cells (Nergadze et al., 2009; Deng et al., 2012; Porro et al., 2014a; Diman et al., 2016; Feretzaki and Lingner, 2017). Telomeric R-loops are detectable in human primary cells (Shiromoto et al., 2021) and HeLa cells (Feretzaki et al., 2020), while ALT cancer cells and ICF syndrome cells, in which TERRA is transcribed at a higher than normal level (Yehezkel et al., 2008; Arora et al., 2014), appear to have more telomeric R-loops (Arora et al., 2014; Min et al., 2017; Nguyen et al., 2017; Sagie et al., 2017). R-loops have been shown to be a genome instability factor (García-Muse and Aguilera, 2019; Brambati et al., 2020; Hegazy et al., 2020). Elevated TERRA and telomeric R-loop levels at very short telomeres have been shown to induce telomere HR in human cells (Graf et al., 2017), and telomeric R-loops in ALT cancer cells are important for HR-mediated telomere maintenance (Arora et al., 2014). Surprisingly, TRF2 facilitates telomeric R-loop formation and TRF1 antagonizes this function (Lee et al., 2018), suggesting that the telomeric R-loop-induced telomere DNA damage is not a major threat to human telomere integrity. Telomere proteins in yeasts also suppresses NHEJ and HR: Saccharomyces cerevisiae RAP1 suppresses NHEJ (Pardo and Marcand, 2005), and Schizosaccharomyces pombe RAP1 appears to suppress HR induced by a high level of telomeric R-loop in cells lacking the telomerase recruitment factor Ccq1 (Hu et al., 2019).
Interestingly, T. brucei does not have the NHEJ machinery (Burton et al., 2007), as the T. brucei genome lacks the DNA ligase IV that is essential for the NHEJ pathway, and no NHEJ events have been observed in this parasite. Therefore, T. brucei telomeres are not threatened by the NHEJ-mediated chromosome end-to-end fusions. However, HR events have been frequently observed at subtelomeric regions, where HR is clearly one of the major pathways of VSG switching (McCulloch et al., 2015). Therefore, telomere HR can be an important instability factor in T. brucei. In addition, telomeric R-loop-induced telomere DNA damage can be a great threat to telomere/subtelomere integrity in T. brucei and induce subtelomeric HR events (Jehi et al., 2014a; Nanavaty et al., 2017; Briggs et al., 2018; Saha et al., 2021). TERRA has been detected in T. brucei (Rudenko and Van der Ploeg, 1989; Damasceno et al., 2017; Nanavaty et al., 2017; Saha et al., 2019, 2021). T. brucei TERRA transcription has several unique features (Saha et al., 2021). First, TERRA appears to be transcribed only from the active VSG-adjacent telomere, as the polycistronic transcript including the active VSG (but not silent VSG or a VSG-free subtelomere) and TERRA sequences can be detected by RT-PCR (Damasceno et al., 2017; Nanavaty et al., 2017; Saha et al., 2021). Most T. brucei cells have 1–3 nuclear TERRA foci, and in cells that have 2–3 TERRA foci (∼39% of G1 and 57–63% S and G2/M cells), frequently only the brightest TERRA focus is co-localized with the telomere (Saha et al., 2021). Second, TERRA is transcribed by RNA Polymerase I in T. brucei, as it is not sensitive to α-Amanitin (Rudenko and Van der Ploeg, 1989), and treating cells with an RNA Polymerase I inhibitor, BMH-21, for only 15 min can abolish >92% of TERRA (Saha et al., 2021). The RNA Polymerase I-mediated TERRA transcription is apparently at a very high level and can be better appreciated when TbTRF is depleted, where a single TERRA focus is frequently observed in the nucleus, and the size of the TERRA focus can be nearly as big as the nucleolus (Saha et al., 2021). The single TERRA transcription site presumably also helps to increase local TERRA concentration at the active telomere, which promotes telomeric R-loop formation. Indeed, telomeric R-loops are readily detectable in WT T. brucei cells (Nanavaty et al., 2017; Saha et al., 2021). Intriguingly, the active ES-adjacent telomere (but not silent telomeres) frequently experiences large truncations (Bernards et al., 1983), suggesting that TERRA transcription and/or telomeric R-loops formed at the active telomere promote telomere instability. Hence, T. brucei telomere faces a great threat from telomeric R-loop-induced telomere/subtelomere DNA damage. Our recent studies further indicate that suppressing the telomeric R-loop level is an important end protection function of T. brucei telomere proteins (see below) (Nanavaty et al., 2017; Saha et al., 2021). Furthermore, introducing a DSB at the active VSG vicinity induces many more DNA recombination-mediated VSG switching events (Boothroyd et al., 2009; Glover et al., 2013), suggesting that telomeric R-loop-induced telomere and subtelomeric DNA damage can be repaired by HR. A direct link between elevated amount of telomeric R-loop, increased amount of DNA damage at the telomere/subtelomere, and many more VSG GC-mediated VSG switchings has been established in TbRAP1-depleted cells, where overexpression of RNaseH1 that specifically degrades RNA in the RNA:DNA hybrid suppresses all three phenotypes (Nanavaty et al., 2017). Therefore, T. brucei telomere proteins have an important role to suppress the telomeric R-loop level and telomere HR.
It is important to note that a certain degree of telomere and subtelomere plasticity is beneficial to T. brucei, as all VSG genes are located at subtelomere regions (Müller et al., 2018), and HR is an important means of VSG diversification and a major pathway of VSG switching (Myler et al., 1984a; McCulloch et al., 2015). Indeed, our studies have shown that telomere and subtelomere instability contributes to increased VSG switching frequencies (Benmerzouga et al., 2013; Jehi et al., 2014a, b; Nanavaty et al., 2017; Afrin et al., 2020a; Saha et al., 2021). In the case of TbTRF and TbRAP1, their roles in suppression of the telomeric R-loop level help maintain telomere and subtelomere integrity and suppress VSG switching frequency (see below). Therefore, these telomere proteins have a delicate job to balance the genome stability and plasticity at T. brucei telomeres and subtelomeres.
Shelterin Homologs in T. brucei
In mammalian cells, Shelterin associates with the telomere tightly and plays indispensable roles in telomere end protection and telomere length regulation (de Lange, 2005, 2018). Several Shelterin homologs have been identified in T. brucei (Li et al., 2005; Yang et al., 2009; Jehi et al., 2014b). Here I will focus on different mechanisms underlying TbTRF and TbRAP1’s functions when compared to their mammalian and yeast homologs.
T. brucei TRF vs. Mammalian TRF1/2
TbTRF was identified as the duplex TTAGGG repeat binding factor in T. brucei (Figure 2; Li et al., 2005). Its duplex telomere binding activity resides in the C-terminal Myb domain but it does not bind single stranded DNA (Li et al., 2005), which is similar to its mammalian homologs TRF1 and TRF2 (Zhong et al., 1992; Bilaud et al., 1997; Broccoli et al., 1997b). TbTRF associates with the telomere chromatin, and is almost always co-localized with the telomere as shown in telomere FISH combined with TbTRF IF experiments (Li et al., 2005). Mammalian TRF1 and TRF2 both have a TRF Homology (TRFH) domain in the N-terminal half of the protein (Broccoli et al., 1997b), which is responsible for TRF homodimerization (Fairall et al., 2001; Chen et al., 2008). In addition, human TRF1 has an acidic N-terminus (Broccoli et al., 1997a), while TRF2 has a basic N-terminal GAR domain (Broccoli et al., 1997b; Mitchell et al., 2009). TbTRF also has a TRFH domain that mediates homodimerization, although the TbTRFH domain only presents limited sequence and structure homology with its mammalian counterparts (Li et al., 2005).
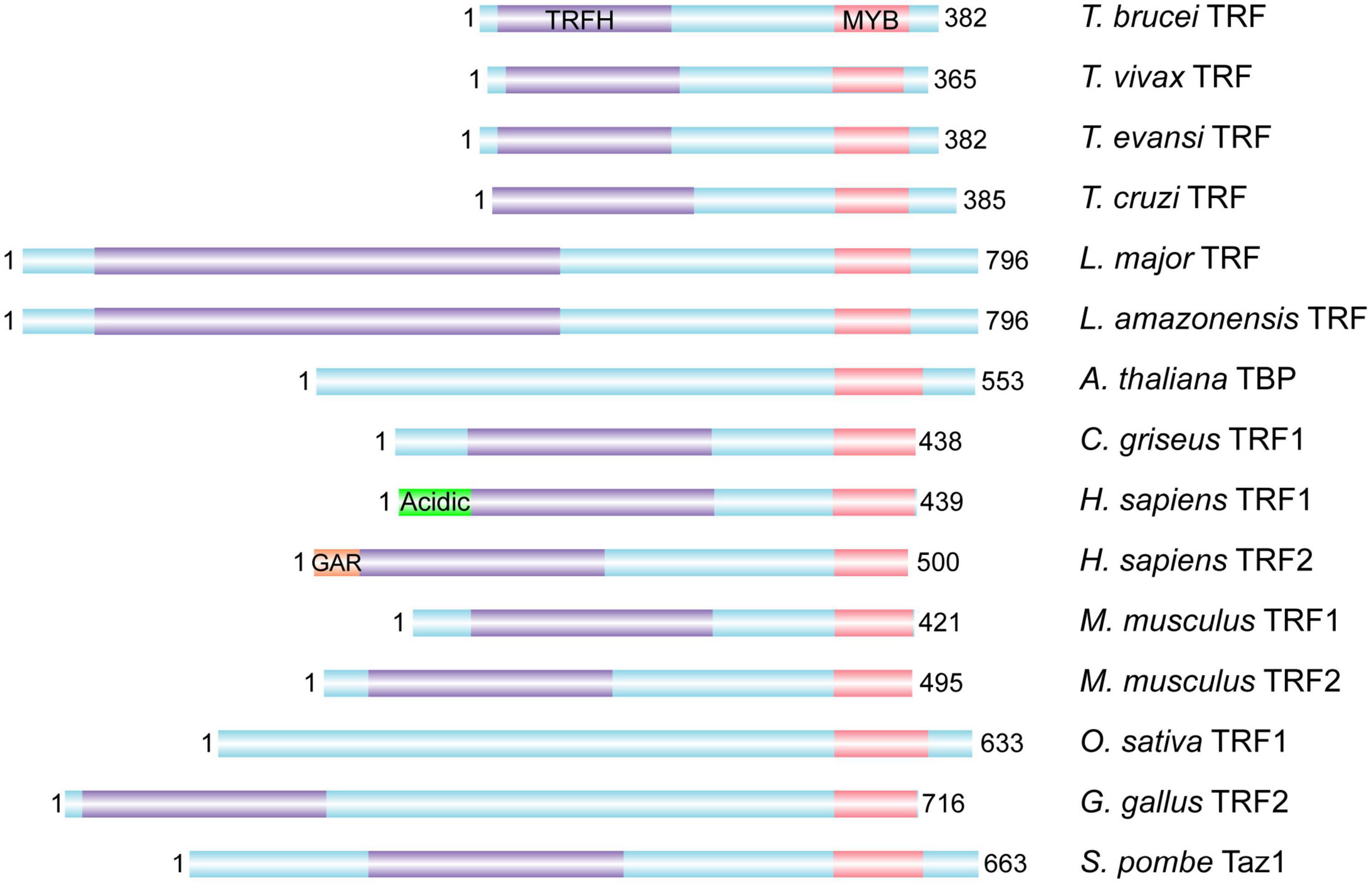
Figure 2. TRF homologs from various organisms. The DNA binding Myb domain, the homodimerization TRFH domain, the N-terminal acidic domain of human TRF1 and basic GAR domain of human TRF2 are marked whenever identified. In addition, Leishmania amazonensis TRF has been identified and shown to associate with the telomere (da Silva et al., 2010). T. brucei, Trypanosoma brucei; T. vivax, Trypanosoma vivax; T. evansi, Trypanosoma evansi; T. cruzi, Trypanosoma cruzi; L. major, Leishmania major; L. amazonensis, Leishmania amazonensis; A. thaliana, Arabidopsis thaliana; C. griseus, Cricetulus griseus; H. sapiens, Homo sapiens; M. musculus, Mus musculus; O. sativa, Oryza sativa; G. gallus, Gallus gallus; S. pombe, Schizosaccharomyces pombe.
It is impossible to tell whether TbTRF is more homologous to TRF1 or TRF2 based solely on sequence analysis, as TbTRF lacks an N-terminal domain (Figure 2; Li et al., 2005). Depletion of TbTRF leads to a loss of the telomere 3′ overhang structure (Li et al., 2005), while removal of TRF2 from the telomere results in the same phenotype (van Steensel et al., 1998), indicating that TbTRF and TRF2 have the same function in maintaining the telomere terminal structure. On the other hand, TRF2 facilitates telomeric R-loop formation while TRF1 suppresses this effect (Lee et al., 2018), and TbTRF also suppresses the telomeric R-loop level (Saha et al., 2021), indicating that TbTRF and TRF1 have similar effects on telomeric R-loop. Recently, it has been shown that human TRF2 at the telomere is sufficient and necessary for the T-loop formation (Doksani et al., 2013; Timashev and de Lange, 2020), which in turn suppresses ATM activation (Van Ly et al., 2018). The T-loop structure has been observed in T. brucei (Munoz-Jordan et al., 2001). However, it is unknown whether TbTRF is required to establish/maintain the T-loop structure. In addition to suppression of ATM activation, TRF2 also prevents the NHEJ-mediated chromosome end-to-end fusions (Karlseder et al., 1999; Smogorzewska et al., 2002; Celli and de Lange, 2005), and its N-terminal basic domain suppresses HR-mediated telomere recombinations (Wang et al., 2004).
In T. brucei, although NHEJ is absent, telomeres and subtelomeres are fragile (Glover et al., 2013), and HR is the major mechanism of VSG switching (Navarro and Cross, 1996; Kim and Cross, 2010; Benmerzouga et al., 2013; Jehi et al., 2014a, b; Nanavaty et al., 2017; da Silva et al., 2018). Therefore, HR is a major DNA damage response pathway in T. brucei (Glover et al., 2008) and posts realistic threat to telomere and subtelomere stability. Indeed, in TbTRF-depleted cells, the γH2A level is increased (Saha et al., 2021), where γH2A is the C-terminal phosphorylated H2A that is deposited to the chromatin at DNA damage sites in T. brucei (Glover and Horn, 2012). Furthermore, a transient depletion of TbTRF leads to an increased number of VSG switching events, most of which involving the loss of the originally active ES (Jehi et al., 2014a). As expected, TbTRF’s role in maintaining telomere and subtelomere stability requires its telomere DNA binding activity (Jehi et al., 2014a). Unexpectedly, depletion of TbTRF results in an increased amount of TERRA (Saha et al., 2021). TbTRF does not affect TERRA’s half-life (Saha et al., 2021). Rather, a higher level of the polycistronic transcript containing the TERRA sequence and the active VSG sequence is detected upon TbTRF depletion, suggesting that TbTRF normally suppresses TERRA transcription. It is possible that TbTRF’s binding to the telomere DNA directly hinders RNA Polymerase I-mediated TERRA transcription. On the other hand, although TbTRF depletion does not derepress silent polycistronic BF VSG ESs (Figure 1; Yang et al., 2009), it does derepress subtelomeric monocistronic metacyclic VSG ESs (Figure 1; Saha et al., 2021), indicating that TbTRF is likely important for telomeric silencing, but its effect spreads only to a short distance from the telomere. TbTRF’s telomeric silencing function presumably contributes to TERRA suppression. In addition, the amount of telomeric R-loops is increased upon depletion of TbTRF (Saha et al., 2021). Overexpression of RNaseH1 reduces the telomeric R-loop level in TbTRF-depleted cells and the amount of DNA damage, confirming that more telomeric R-loops cause more telomere DNA damage upon TbTRF depletion (Saha et al., 2021). Therefore, these observations indicate that TbTRF helps maintain telomere integrity through suppressing the levels of TERRA and telomeric R-loop (Saha et al., 2021), which is similar to TRF1 but different from TRF2 (Lee et al., 2018).
Depletion of human TRF2 also results in an increased level of TERRA (Porro et al., 2014b). In addition, TRF2 binds TERRA predominantly through its N-terminal GAR domain (Deng et al., 2009b; Mei et al., 2021; Figure 2). TbTRF also has a TERRA binding activity, which surprisingly resides in its Myb domain (Saha et al., 2021). Most interestingly, a TbTRF Myb domain point mutant that loses its telomere DNA binding activity (Jehi et al., 2014a) binds TERRA more strongly (Saha et al., 2021). In addition, TbTRF exhibits a slightly stronger affinity to the duplex telomere DNA than TERRA in in vitro competition binding assays (Saha et al., 2021). Therefore, the telomere DNA binding and TERRA-binding activities of TbTRF may have overlapping nucleic acid interaction interfaces, which is clearly different from how human TRF 2 binds TERRA and telomere DNA (Bilaud et al., 1997; Broccoli et al., 1997b; Deng et al., 2009b; Mei et al., 2021).
The fact that TbTRF has both TERRA and a ds(TTAGGG)n binding activities provides an additional possible mechanism how TbTRF regulates the level of the telomeric R-loop. Significantly more TbTRF-depleted cells (in G1, S, or G2/M phases) than WT cells have only one TERRA focus (Saha et al., 2021), suggesting that TbTRF helps recruit TERRA away from its transcription site. In this case, the TERRA and ds(TTAGGG)n binding activities of TbTRF can help transport TERRA to TTAGGG repeats other than its transcription site, as TbTRF also has a homodimerization function (Li et al., 2005). Although TbTRF can theoretically bind all telomeres, it is expected that the actively transcribed telomere region is mostly free of TbTRF due to the high-level RNA Polymerase I-mediated transcription, similar to the situation in the active ES, which is deprived of nucleosomes (Figueiredo and Cross, 2010; Stanne and Rudenko, 2010). Translocation of TERRA away from its transcription site will effectively limit TERRA accumulation at a single telomere, significantly reducing the chance of telomeric R-loop formation. Therefore, TbTRF may suppress the telomeric R-loop level through suppressing of TERRA transcription and promoting TERRA translocation. On the other hand, human TRF1 and TRF2 have an opposite effect and restrict TERRA’s translocation away from its transcription site (Feretzaki et al., 2020). The key telomeric functions of human TRF1/2 and TbTRF are compared in Table 1.
RAP1 Homologs in Vertebrates, Yeasts, and T. brucei
Saccharomyces cerevisiae RAP1 is the first protein identified to directly bind telomere DNA among yeast and vertebrate telomere proteins (Shore and Nasmyth, 1987; Longtine et al., 1989; Conrad et al., 1990). RAP1 is also one of the most conserved telomere proteins (Figure 3), with its homologs identified in kinetoplastids (Yang et al., 2009), yeasts (Shore and Nasmyth, 1987; Kanoh and Ishikawa, 2001; Wahlin and Cohn, 2002; Yu et al., 2010; Steinberg-Neifach and Lue, 2015), and vertebrates (Li et al., 2000; Tan et al., 2003).
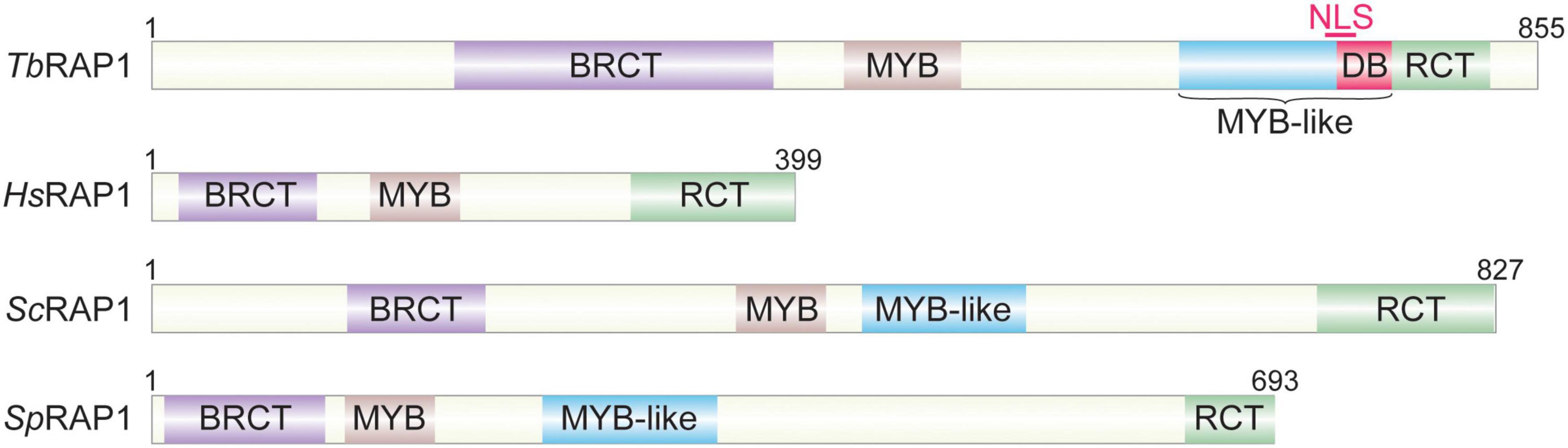
Figure 3. Repressor activator protein 1 (RAP1) homologous. Tb, Trypanosoma brucei; Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe.
The functions of yeast RAP1 homologs in telomeric silencing and telomere length regulation have been extensively studied (Lustig et al., 1990, 1996; Hardy et al., 1992; Kyrion et al., 1992, 1993; Liu et al., 1994; Cockell et al., 1995; Liu and Lustig, 1996; Marcand et al., 1997; Wotton and Shore, 1997; Kanoh and Ishikawa, 2001). In addition, ScRAP1 prevents NHEJ-mediated telomere end-to-end fusions (Pardo and Marcand, 2005). On the other hand, the functions of mammalian RAP1 homologs have been somewhat hard to define. Initial studies in telomerase positive cancer cell lines indicated that human RAP1 is involved in telomere length regulation (Li et al., 2000; Li and de Lange, 2003). Subsequently, in vitro biochemical study and engineered tethering of human RAP1 to the telomere both indicated that RAP1 is capable of suppressing NHEJ at the telomere (Bae and Baumann, 2007; Sarthy et al., 2009). However, TALEN-mediated deletion of the human RAP1 exon 2 in a number of cell lines showed that hRAP1 is not required for telomere length regulation or suppression of telomere end-to-end fusions (Kabir et al., 2014). Recently, it has been shown that human RAP1 is required to inhibit NHEJ-mediated telomere fusions at critically short telomeres (Lototska et al., 2020). On the other hand, conditional knockout of mouse RAP1 or expression of a mouse TRF2 mutant that does not interact with RAP1 (so that RAP1 is not recruited to the telomere) showed that mouse RAP1 coordinates with TRF2 N-terminal basic domain to suppress telomere HR (Sfeir et al., 2010; Rai et al., 2016).
Trypanosoma brucei RAP1 was identified as a TbTRF-interacting factor (Yang et al., 2009). TbRAP1 was the first telomere protein that has been shown to be essential for VSG monoallelic expression, as depletion of TbRAP1 by RNAi or conditional knockout of TbRAP1 result in derepression of essentially all subtelomeric VSG genes in T. brucei (Yang et al., 2009; Pandya et al., 2013; Afrin et al., 2020b). The silent ES-linked VSGs are derepressed up to several thousand folds, which represents the most dramatic VSG derepression phenotype among T. brucei mutants that affect VSG silencing (Figueiredo et al., 2008; Yang et al., 2009; Alsford and Horn, 2012; Tiengwe et al., 2012; Benmerzouga et al., 2013; Kim et al., 2013; Cestari and Stuart, 2015; Glover et al., 2016; Reynolds et al., 2016; Schulz et al., 2016; Briggs et al., 2018, 2019; Faria et al., 2019; Kim, 2019). In addition, depletion of TbRAP1 leads to an increased amount of DNA damage at the telomere and subtelomere and more VSG switching events (Nanavaty et al., 2017; Afrin et al., 2020a, b). Interestingly, depletion of TbRAP1 also leads to elevated levels of TERRA and telomeric R-loops (Nanavaty et al., 2017). Overexpression of an ectopic allele of RNaseH1 in the TbRAP1-depleted cells suppresses the increased amount of telomeric R-loop and DNA damage and the increased VSG switching frequency phenotypes, while the TERRA level in these cells is still higher than that in WT cells (Nanavaty et al., 2017). Therefore, the function of TbRAP1 in maintaining telomere/subtelomere integrity relies on its repressive effects on TERRA and telomeric R-loop. The TERRA molecules in TbRAP1-depleted cells are much longer than that in WT cells (Nanavaty et al., 2017), suggesting that TbRAP1 can block the transcription elongation of RNA Polymerase I along the telomere by associating with the telomere chromatin. These observations indicate that TbRAP1 suppresses telomeric and subtelomeric HR by suppression of the telomeric R-loop level. A recent study in fission yeast suggests that SpRAP1 has a similar suppressive effect on telomeric R-loop and telomeric HR (Hu et al., 2019). Therefore, the function of RAP1 homologs in suppressing telomere HR appears to be conserved, although there is no direct evidence showing that mammalian RAP1 suppresses the telomeric R-loop level.
All telomere functions of RAP1 homologs depend on their telomere association. However, RAP1 homologs achieve this goal through different mechanisms. ScRAP1 has both a Myb domain and a Myb-Like motif in the central region of the protein (Figure 3), which were confirmed to contain the duplex DNA binding activity (Konig et al., 1996), recognizing a consensus sequence 5′ ACACCCAYACAYY 3′ (where Y represents a pyrimidine) (Graham and Chambers, 1994). However, this sequence-specific duplex DNA binding activity was only identified in budding yeast RAP1 homologs (Konig et al., 1996; Rhodin et al., 2006; Yu et al., 2010). Human RAP1 is only recruited to the telomere through its interaction with TRF2, while itself does not directly bind the telomere DNA (Li et al., 2000; Loayza and de Lange, 2003). Similarly, SpRAP1 is also recruited to the telomere through its interaction with TAZ1 (Kanoh and Ishikawa, 2001), the duplex telomere DNA binding factor in S. pombe and the functional homolog of mammalian TRF1/2 (Cooper et al., 1997; Li et al., 2000). It was originally hypothesized that TbRAP1 was also recruited to the telomere via its interaction with TbTRF (Yang et al., 2009). However, TbRAP1 still associates with the telomere chromatin in TbTRF-depleted cells (Afrin et al., 2020a), and TbRAP1’s telomere association does not require its Myb domain (Yang et al., 2009; Afrin et al., 2020a), even though Myb motifs frequently have DNA binding activities (Ogata et al., 1994). Rather, we recently identified both dsDNA and ssDNA binding activities in TbRAP1 (Afrin et al., 2020a). Both activities are electrostatic-based and require a positively charged 737RKRRR741 patch that overlaps with the nuclear localization signal (NLS) in the Myb-Like domain of TbRAP1 (Figure 3; Yang et al., 2009; Afrin et al., 2020a). Proteomic studies detected phosphorylated S742 and S744 residues in T. brucei cells (Nett et al., 2009; Urbaniak et al., 2013). Interestingly, the phospho-mimic S742DS744D mutation of TbRAP1 disrupts most of its dsDNA binding activity but retains most of its ssDNA binding activity (Afrin et al., 2020a). In addition, TbRAP1-S742DS744D is no longer associated with the telomere chromatin while TbRAP1-S742AS744A is still located at the telomere, suggesting that phosphorylation of the two 737RKRRR741-adjacent S residues can remove TbRAP1 from the telomere (Afrin et al., 2020a). Furthermore, VSG silencing and telomere integrity are disrupted in all TbRAP1 mutants that do not associate with the telomere chromatin, further suggesting that phosphorylation of the two S residues can regulate VSG silencing and telomere integrity in T. brucei cells (Afrin et al., 2020a). The TbRAP1’s DNA binding activities are quite different from that of ScRAP1. ScRAP1’s duplex DNA binding activity is sequence specific and depends on its Myb and Myb-Like domains (Konig et al., 1996). So far no protein modification has been shown to regulate ScRAP1’s DNA binding activity. On the other hand, TbRAP1 has both dsDNA and ssDNA binding activities, which are sequence non-specific, and phospho-mimicking mutation of S742 and S744 can disrupt its dsDNA binding activity (Afrin et al., 2020a). Therefore, although the main functions of RAP1 homologs at the telomere are conserved from kinetoplastids to mammals, the underlying mechanisms are clearly different among different RAP1 homologs. The key telomeric functions of human, yeast, and T. brucei RAP1 homologs are compared in Table 2.
The fact that TbTRF and TbRAP1 are essential for maintaining telomere integrity and stability indicates that these telomere proteins have conserved essential functions as their yeast and mammalian homologs. However, recent findings indicate that the underlying mechanisms of how TbTRF and TbRAP1 achieve their goals are quite different from those in human and yeast cells. First, T. brucei does not have the NHEJ machinery. Therefore, neither TbTRF nor TbRAP1 needs to suppress any NHEJ-mediated telomere fusions. Second, telomeric R-loops have been shown to be an important factor contributing to telomere damage if not controlled at a WT level in T. brucei (Nanavaty et al., 2017; Briggs et al., 2018; Saha et al., 2021). While both TbTRF and TbRAP1 suppress the level of telomeric R-loop (Nanavaty et al., 2017; Saha et al., 2021), whether mammalian RAP1 homologs have the same function is unknown, and human TRF2 actually stimulates the formation of telomeric R-loop, which is antagonized by human TRF1 (Lee et al., 2018). The unique features of TbRAP1 (such as its DNA binding activities) and TbTRF (such as its Myb-mediated TERRA binding activity) telomere functions can be targeted as a means to eliminate the parasites from its mammalian host. In addition, TbRAP1 and TbTRF only have limited sequence homology with their mammalian counterparts in their functional domains (Li et al., 2005; Yang et al., 2009), making it more feasible to specifically target the parasite telomere proteins without affecting their mammalian homologs.
Subtelomere Plasticity Benefits Antigenic Variation
As discussed earlier, telomere integrity and stability is important for genome integrity. Subtelomere stability is also important for organism health and fitness. In humans, unstable subtelomeres are frequently associated with various diseases. For example, reduced copy number of polymorphic macrosatellite repeat D4Z4 at chromosome 4q subtelomere has long been associated with facioscapulohumeral muscular dystrophy (FSHD) (van der Maarel et al., 2007; Daxinger et al., 2015). Submicroscopic deletion of subtelomeric 6p25 has been recognized as a clinically identifiable syndrome (DeScipio, 2007), and deletion of the EHMT1 gene at the chromosome 9q subtelomere leads to the Kleefstra syndrome (Stewart and Kleefstra, 2007; Bonati et al., 2019). A recent study further indicates that 3–16% of syndromic intellectual disability cases are caused by cryptic subtelomeric abnormalities (Soares et al., 2019). Subtelomere integrity and stability is also important in T. brucei, as VSG is essential (Sheader et al., 2005), and all VSG genes are located at subtelomeres (de Lange and Borst, 1982; Hertz-Fowler et al., 2008; Müller et al., 2018). DSBs in the active VSG gene vicinity are generally poorly tolerated: introducing an artificial DSB (an I-SceI cut) within or near the active VSG gene leads to death in more than 80% of the cells (Glover et al., 2013). Inefficient repair of the I-SceI cut due to continued I-SceI expression may contribute to the catastrophic consequence. Nevertheless, the location of the damage site appears to be a critical factor, as inducing the same I-SceI cut in a silent ES is much better tolerated (Glover et al., 2013). However, maintaining subtelomere stability can be challenging. Subtelomeres often consist of duplicated sequence blocks near the ends of multiple chromosomes and are highly dynamic with very heterogeneous sequences, sizes, and copy numbers (Pryde et al., 1997; Mefford and Trask, 2002; Li, 2012). Increased rates of sister chromatid exchange have been observed at human chromosome ends by cytological studies (Rudd et al., 2007), and human subtelomeres are hot spots of interchromosomal recombination and segmental duplications (Linardopoulou et al., 2005). High polymorphism in the subtelomere is frequently observed among different chromosome ends and individuals in humans (Ambrosini et al., 2007; Young et al., 2017), yeast (Pryde et al., 1997; Quispe et al., 2017), fly (Anderson et al., 2008), plant (Kuo et al., 2006), and fungal pathogens (Farman, 2007; Schmid-Siegert et al., 2017). Similarly, it has been shown that T. brucei subtelomere is a fragile site (Glover et al., 2013). T. brucei homologous megabase chromosome pairs often differ greatly in size (Melville et al., 2000) due to different sizes of subtelomeric ESs and VSG gene arrays, telomere, and repetitive chromosomal regions (Melville et al., 1999). In fact, two-thirds of the size polymorphisms are due to variations in subtelomeric regions, while chromosomal core regions, containing all essential genes, are relatively stable (Callejas et al., 2006). Therefore, maintaining subtelomere stability is important yet challenging.
On the other hand, subtelomere plasticity can be beneficial to the organism to a certain extent. For example, some human olfactory receptor (OR) genes encoding olfactory receptors are located at subtelomeres, and changes in subtelomere regions may contribute to the diversity of the OR gene family (Trask et al., 1998). Subtelomeres also have important functions for microbial pathogens, where genes with roles in niche adaptation are frequently enriched (Underwood et al., 1996; Zhang et al., 1997; De Las Penas et al., 2003; Farman, 2007). Subtelomeric plasticity and relative frequent subtelomere recombination in these microbial pathogens is expected to help increase the diversity of their major surface antigen and enhance the effectiveness of evading host immune responses. As described above, in T. brucei, removal of telomere proteins (such as TbTRF, TbTIF2–a TbTRF-interacting factor and the human TIN2 homolog, and TbRAP1) leads to telomere instability and increased VSG switching rates (Jehi et al., 2014a, b; Nanavaty et al., 2017; Afrin et al., 2020a). Subtelomere instability has also been frequently observed in P. falciparum that causes malaria in humans. At the erythrocyte stage, P. falciparum infects the host red blood cells (RBCs) and expresses PfEMP1 on the host RBC surface, which is important for adhering the infected RBCs to the endothelial lining of host blood vessels so that the infected RBCs will not be eliminated by the host immune system (Hviid and Jensen, 2015; Wahlgren et al., 2017). Additional parasite proteins including RIFIN and STEVOR are also expressed on host RBC membrane, facilitating interaction of parasite-infected RBCs and other host cells (Wahlgren et al., 2017). The var, rif, and stevor gene families that encode PfEMP1, RIFIN, and STEVOR, respectively, are mostly located at subtelomeric regions (Rubio et al., 1996; Cheng et al., 1998). P. falciparum regularly switches to express different var, rif, and stevor genes to evade the host immune attack (Wahlgren et al., 2017). In addition, var gene expression is strictly monoallelic (Voss et al., 2006). Importantly, recent studies showed that P. falciparum subtelomeres frequently have HR events that contributes to divergence of var gene families (Calhoun et al., 2017), and this subtelomere plasticity is enhanced when a DSB is introduced at the vicinity (Calhoun et al., 2017; Zhang et al., 2019). In the pathogenic yeast Pneumocystis jirovecii that causes pneumonia in immunodeficient patients, genes encoding its major surface antigen, MSG, are also located at subtelomeric loci (Keely et al., 2005) and are expressed in a monoallelic fashion (Kutty et al., 2001). There is only one subtelomeric MSG ES (Kutty et al., 2001), and antigenic variation is achieved through recombining a silent MSG gene into the active ES and creating novel mosaic MSG genes though recombination (Stringer, 2007; Kutty et al., 2008; Schmid-Siegert et al., 2017). Even in non-pathogenic microbial organisms, subtelomere plasticity can be beneficial for the organism to better adapt to their living environment. In K. lactis, genes encoding β-galactosidase are located at subtelomeres, and variation in these genes allows yeast to better cope with different nutrition (Mason and McEachern, 2018). Interestingly, it has been observed that mild telomere dysfunction that does not induce global genome instability leads to an increased variation of the subtelomere β-galactosidase-coding genes, while severe telomere dysfunction causes complete deletion of these genes (Mason and McEachern, 2018). Therefore, telomere and subtelomere plasticity to a certain extent may not be deleterious but even beneficial to improve the organism’s adaptation to various environmental growth conditions.
Strategies of Tolerating Mild Telomere/Subtelomere Damage in T. brucei
If telomere/subtelomere plasticity and mild telomere damage is beneficial to T. brucei, the parasite may have evolved ways to encourage it. Indeed, T. brucei does not appear to have a stringent DNA damage surveillance mechanism. The initial hint comes from the study where an I-SceI site was targeted to the junction of telomere and sub-telomere downstream a silent ES in telomerase null cells (Glover et al., 2007). After induction of the I-SceI endonuclease, the marked telomere and the upstream silent VSG gene were lost, yet the cells did not go into cell cycle arrest (Glover et al., 2007). Subsequently, it was shown that as short as 40 bp of telomere DNA downstream of a silent ES can be stably maintained in a telomerase-independent manner without iliciting cell cycle arrest (Dreesen and Cross, 2006). In addition, DSBs near silent VSGs are much better tolerated than those in the active VSG vicinity (Glover and Horn, 2014). Similarly, a single induced I-SCE I cut in T. brucei genome failed to activate cell cycle checkpoint (Glover et al., 2019). Therefore, as long as the active VSG gene (and essential genes at chromosome core regions) is not damaged and VSG synthesis is normal, individual DSB in T. brucei genome, particularly those at telomere/subtelomere vicinity is well-tolerated.
On the other hand, T. brucei does have cell cycle arrest mechanisms in response to telomere defects. For example, depletion of TbTRF leads to an acute G2/M cell cycle arrest (Li et al., 2005), while depletion of TbRAP1 results in cell growth arrest with a decrease in the S phase population and an increase in the G2/M population (Yang et al., 2009). Interestingly, depletion of TbTRF caused an increased amount of telomere DNA damage (Saha et al., 2021) and depletion of TbRAP1 results in an increased amount of both telomere and subtelomere DNA damage (Nanavaty et al., 2017), and it is expected that the damage occurs at multiple chromosome ends rather than at a single telomere. Hence, a much higher level of DNA damage occurs in TbTRF- and TbRAP1-depleted cells than a single I-SceI cut. Therefore, T. brucei appears to be able to tolerate a very small amount of DNA damage, but still guard against DNA damage of a global scale. Presumably, this will allow more genome plasticity, particularly at the subtelomere regions to facilitate a more effective antigenic variation.
It is interesting to note that T. brucei does not have the NHEJ machinery (Burton et al., 2007). This helps avoid deleterious chromosome end fusion products that frequently result from telomere defects in human and yeast cells. Several telomere proteins have been shown to suppress subtelomere HR events (Jehi et al., 2014a, b, 2016; Nanavaty et al., 2017). Therefore, mild telomere protein defects, if tolerated, can enhance antigenic variation by allowing more subtelomere HR events. In fact, we have recently identified TbRAP1-S742AS744A and TbRAP1-S265AS586AS742AS744AT752A mutants that only exhibit a very mild growth defect but increase VSG switching rates (Afrin et al., 2020a). The lack of NHEJ machinery likely helps to achieve this middle ground where mild telomere protein defects and subtly increased subtelomere plasticity can enhance antigenic variation without affecting global genome stability.
Conclusion Remarks
Although the telomere complex is essential for genomic integrity and chromosome stability in all eukaryotic cells studied, recent discoveries indicate that the detailed mechanisms of the “chromosome end protection” can have different features in different organisms. First, the telomere proteins can face different threats that cause genome instability. Hence, studying telomere biology in pathogenic kinetoplastids yields invaluable information on how telomere proteins suppress telomeric and subtelomeric DNA recombination events, which represent a minor pathway compared to NHEJ in human and yeast cells. Second, recent findings illustrate that telomere protein homologs in different organisms can achieve the same goals using distinct mechanisms, which sheds light on telomere protein evolution and provides potential targets for future development of anti-parasite agents. Third, a better understanding of the balance between stability and plasticity at the telomere and subtelomere in pathogenic eukaryotic micro-organisms will help us to better appreciate how eukaryotic cells adapt to different living conditions and evolve to better survive the environment.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This work was supported by an NIH R01 grant AI066095 to BL. The publication cost is partly supported by GRHD at CSU.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Afrin, M., Gaurav, A. K., Yang, X., Pan, X., Zhao, Y., and Li, B. (2020a). TbRAP1 has an unusual duplex DNA binding activity required for its telomere localization and VSG silencing. Sci. Adv. 6:eabc4065. doi: 10.1126/sciadv.abc4065
Afrin, M., Kishmiri, H., Sandhu, R., Rabbani, M. A. G., and Li, B. (2020b). Trypanosoma brucei RAP1 has essential functional domains that are required for different protein interactions. mSphere 5:e00027-20. doi: 10.1128/mSphere.00027-20
Alsford, S., and Horn, D. (2012). Cell-cycle-regulated control of VSG expression site silencing by histones and histone chaperones ASF1A and CAF-1b in Trypanosoma brucei. Nucleic Acids Res. 40, 10150–10160. doi: 10.1093/nar/gks813
Ambrosini, A., Paul, S., Hu, S., and Riethman, H. (2007). Human subtelomeric duplicon structure and organization. Genome Biol. 8:R151. doi: 10.1186/gb-2007-8-7-r151
Anderson, J. A., Song, Y. S., and Langley, C. H. (2008). Molecular population genetics of Drosophila subtelomeric DNA. Genetics 178, 477–487. doi: 10.1534/genetics.107.083196
Arnoult, N., Correia, A., Ma, J., Merlo, A., Garcia-Gomez, S., Maric, M., et al. (2017). Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature 549, 548–552. doi: 10.1038/nature24023
Arora, R., Lee, Y., Wischnewski, H., Brun, C. M., Schwarz, T., and Azzalin, C. M. (2014). RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 5:5220. doi: 10.1038/ncomms6220
Azzalin, C. M., Reichenbach, P., Khoriauli, L., Giulotto, E., and Lingner, J. (2007). Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801. doi: 10.1126/science.1147182
Bae, N. S., and Baumann, P. (2007). A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol. Cell. 26, 323–334. doi: 10.1016/j.molcel.2007.03.023
Bah, A., Wischnewski, H., Shchepachev, V., and Azzalin, C. M. (2012). The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res. 40, 2995–3005. doi: 10.1093/nar/gkr1153
Barry, J. D., and McCulloch, R. (2001). Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 49, 1–70. doi: 10.1016/s0065-308x(01)49037-3
Baumann, P., and Cech, T. R. (2001). Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292, 1171–1175. doi: 10.1126/science.1060036
Baur, J. A., Zou, Y., Shay, J. W., and Wright, W. E. (2001). Telomere position effect in human cells. Science 292, 2075–2077. doi: 10.1126/science.1062329
Benmerzouga, I., Concepcion-Acevedo, J., Kim, H. S., Vandoros, A. V., Cross, G. A. M., Klingbeil, M. M., et al. (2013). Trypanosoma brucei Orc1 is essential for nuclear DNA replication and affects both VSG silencing and VSG switching. Mol. Microbiol. 87, 196–210. doi: 10.1111/mmi.12093
Bernards, A., Michels, P. A. M., Lincke, C. R., and Borst, P. (1983). Growth of chromosome ends in multiplying trypanosomes. Nature 303, 592–597. doi: 10.1038/303592a0
Bilaud, T., Brun, C., Ancelin, K., Koering, C. E., Laroche, T., and Gilson, E. (1997). Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17, 236–239. doi: 10.1038/ng1097-236
Bonati, M. T., Castronovo, C., Sironi, A., Zimbalatti, D., Bestetti, I., Crippa, M., et al. (2019). 9q34.3 microduplications lead to neurodevelopmental disorders through EHMT1 overexpression. Neurogenetics 20, 145–154. doi: 10.1007/s10048-019-00581-6
Boothroyd, C. E., Dreesen, O., Leonova, T., Ly, K. I., Figueiredo, L. M., Cross, G. A. M., et al. (2009). A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature 459, 278–281. doi: 10.1038/nature07982
Brambati, A., Zardoni, L., Nardini, E., Pellicioli, A., and Liberi, G. (2020). The dark side of RNA:DNA hybrids. Mutat. Res. 784:108300. doi: 10.1016/j.mrrev.2020.108300
Briggs, E., Crouch, K., Lemgruber, L., Hamilton, G., Lapsley, C., and McCulloch, R. (2019). Trypanosoma brucei ribonuclease H2A is an essential R-loop processing enzyme whose loss causes DNA damage during transcription initiation and antigenic variation. Nucleic Acids Res. 47, 9180–9197. doi: 10.1093/nar/gkz644
Briggs, E., Crouch, K., Lemgruber, L., Lapsley, C., and McCulloch, R. (2018). Ribonuclease H1-targeted R-loops in surface antigen gene expression sites can direct trypanosome immune evasion. PLoS Genet 14:e1007729. doi: 10.1371/journal.pgen.1007729
Broccoli, D., Chong, L., Oelmann, S., Fernald, A. A., Marziliano, N., van Steensel, B., et al. (1997a). Comparison of the human and mouse genes encoding the telomeric protein. TRF1: chromosomal localization, expression and conserved protein domains. Hum. Mol. Genet. 6, 69–76. doi: 10.1093/hmg/6.1.69
Broccoli, D., Smogorzewska, A., Chong, L., and de Lange, T. (1997b). Human telomeres contain two distinct Myb-related proteins. TRF1 and TRF2. Nat. Genet. 17, 231–235. doi: 10.1038/ng1097-231
Burton, P., McBride, D. J., Wilkes, J. M., Barry, J. D., and McCulloch, R. (2007). Ku heterodimer-independent end joining in Trypanosoma brucei cell extracts relies upon sequence microhomology. Eukaryot. Cell 6, 1773–1781. doi: 10.1128/EC.00212-07
Calhoun, S. F., Reed, J., Alexander, N., Mason, C. E., Deitsch, K. W., and Kirkman, L. A. (2017). Chromosome end repair and genome stability in Plasmodium falciparum. mBio 8:e00547-17. doi: 10.1128/mBio.00547-17
Callejas, S., Leech, V., Reitter, C., and Melville, S. (2006). Hemizygous subtelomeres of an African trypanosome chromosome may account for over 75% of chromosome length. Genome Res. 16, 1109–1118. doi: 10.1101/gr.5147406
Ceccaldi, R., Rondinelli, B., and D’Andrea, A. D. (2016). Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26, 52–64. doi: 10.1016/j.tcb.2015.07.009
Celli, G. B., and de Lange, T. (2005). DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7, 712–718. doi: 10.1038/ncb1275
Cesare, A. J., Groff-Vindman, C., Compton, S. A., McEachern, M. J., and Griffith, J. D. (2008). Telomere loops and homologous recombination-dependent telomeric circles in a Kluyveromyces lactis telomere mutant strain. Mol. Cell. Biol. 28, 20–29. doi: 10.1128/MCB.01122-07
Cesare, A. J., Quinney, N., Willcox, S., Subramanian, D., and Griffith, J. D. (2003). Telomere looping in P. sativum (common garden pea). Plant J. 36, 271–279. doi: 10.1046/j.1365-313x.2003.01882.x
Cestari, I., and Stuart, K. (2015). Inositol phosphate pathway controls transcription of telomeric expression sites in trypanosomes. Proc. Natl. Acad. Sci. U S A. 112, E2803–E2812. doi: 10.1073/pnas.1501206112
Chang, H. H. Y., Pannunzio, N. R., Adachi, N., and Lieber, M. R. (2017). Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18, 495–506. doi: 10.1038/nrm.2017.48
Chen, Y., Rai, R., Zhou, Z. R., Kanoh, J., Ribeyre, C., Yang, Y., et al. (2011). A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat Struct Mol Biol. 18, 213–221. doi: 10.1038/nsmb.1974
Chen, Y., Yang, Y., van Overbeek, M., Donigian, J. R., Baciu, P., de Lange, T., et al. (2008). A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science 319, 1092–1096. doi: 10.1126/science.1151804
Cheng, Q., Cloonan, N., Fischer, K., Thompson, J., Waine, G., Lanzer, M., et al. (1998). stevor and rif are Plasmodium falciparum multicopy gene families which potentially encode variant antigens. Mol. Biochem. Parasitol. 97, 161–176. doi: 10.1016/s0166-6851(98)00144-3
Chong, L., van Steensel, B., Broccoli, D., Erdjument-Bromage, H., Hanish, J., Tempst, P., et al. (1995). A human telomeric protein. Science 270, 1663–1667.
Cleal, K., and Baird, D. M. (2020). Catastrophic endgames: emerging mechanisms of telomere-driven genomic instability. Trends Genet. 36, 347–359. doi: 10.1016/j.tig.2020.02.001
Cockell, M., Palladino, F., Laroche, T., Kyrion, G., Liu, C., Lustig, A. J., et al. (1995). The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J. Cell Biol. 129, 909–924. doi: 10.1083/jcb.129.4.909
Conrad, M. N., Wright, J. H., Wolf, A. J., and Zakian, V. A. (1990). RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell 63, 739–750. doi: 10.1016/0092-8674(90)90140-a
Cooper, J. P., Nimmo, E. R., Allshire, R. C., and Cech, T. R. (1997). Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385, 744–747. doi: 10.1038/385744a0
Cross, G. A. M., Kim, H. S., and Wickstead, B. (2014). Capturing the variant surface glycoprotein repertoire (the VSGnome) of Trypanosoma brucei lister 427. Mol. Biochem. Parasitol. 195, 59–73. doi: 10.1016/j.molbiopara.2014.06.004
Cross, M., Taylor, M. C., and Borst, P. (1998). Frequent loss of the active site during variant surface glycoprotein expression site switching in vitro in Trypanosoma brucei. Mol. Cell. Biol. 18, 198–205. doi: 10.1128/mcb.18.1.198
da Silva, M. S., Hovel-Miner, G. A., Briggs, E. M., Elias, M. C., and McCulloch, R. (2018). Evaluation of mechanisms that may generate DNA lesions triggering antigenic variation in African trypanosomes. PLoS Pathog 14:e1007321. doi: 10.1371/journal.ppat.1007321
da Silva, M. S., Perez, A. M., da Silveira, R. C., de Moraes, C. E., Siqueira-Neto, J. L., Freitas, L. H., et al. (2010). The Leishmania amazonensis TRF (TTAGGG repeat-binding factor) homologue binds and co-localizes with telomeres. BMC Microbiol. 10:136. doi: 10.1186/1471-2180-10-136
Damasceno, J. D., Silva, G., Tschudi, C., and Tosi, L. R. (2017). Evidence for regulated expression of Telomeric Repeat-containing RNAs (TERRA) in parasitic trypanosomatids. Mem. Inst. Oswaldo Cruz 112, 572–576. doi: 10.1590/0074-02760170054
Daxinger, L., Tapscott, S. J., and van der Maarel, S. M. (2015). Genetic and epigenetic contributors to FSHD. Curr. Opin. Genet. Dev. 33, 56–61. doi: 10.1016/j.gde.2015.08.007
de Lange, T. (2005). Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110. doi: 10.1101/gad.1346005
de Lange, T. (2018). Shelterin-Mediated telomere protection. Annu. Rev. Genet. 52, 223–247. doi: 10.1146/annurev-genet-032918-021921
de Lange, T., and Borst, P. (1982). Genomic environment of the expression-linked extra copies of genes for surface antigens of Trypanosoma brucei resembles the end of a chromosome. Nature 299, 451–453. doi: 10.1038/299451a0
De Las Penas, A., Pan, S. J., Castano, I., Alder, J., Cregg, R., and Cormack, B. P. (2003). Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 17, 2245–2258. doi: 10.1101/gad.1121003
Denchi, E. L., and de Lange, T. (2007). Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448, 1068–1071. doi: 10.1038/nature06065
Deng, Y., Guo, X., Ferguson, D. O., and Chang, S. (2009a). Multiple roles for MRE11 at uncapped telomeres. Nature 460, 914–918. doi: 10.1038/nature08196
Deng, Z., Norseen, J., Wiedmer, A., Riethman, H., and Lieberman, P. M. (2009b). TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 35, 403–413. doi: 10.1016/j.molcel.2009.06.025
Deng, Z., Wang, Z., Stong, N., Plasschaert, R., Moczan, A., Chen, H. S., et al. (2012). A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 31, 4165–4178. doi: 10.1038/emboj.2012.266
DeScipio, C. (2007). The 6p subtelomere deletion syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 145C, 377–382. doi: 10.1002/ajmg.c.30156
Diman, A., Boros, J., Poulain, F., Rodriguez, J., Purnelle, M., Episkopou, H., et al. (2016). Nuclear respiratory factor 1 and endurance exercise promote human telomere transcription. Sci. Adv. 2:e1600031. doi: 10.1126/sciadv.1600031
Doksani, Y., Wu, J. Y., de Lange, T., and Zhuang, X. (2013). Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 155, 345–356. doi: 10.1016/j.cell.2013.09.048
Doyle, J. J., Hirumi, H., Hirumi, K., Lupton, E. N., and Cross, G. A. M. (1980). Antigenic variation in clones of animal-infective Trypanosoma brucei derived and maintained in vitro. Parasitology 80, 359–369. doi: 10.1017/s0031182000000810
Dreesen, O., and Cross, G. A. M. (2006). Telomerase-independent stabilization of short telomeres in Trypanosoma brucei. Mol. Cell. Biol. 26, 4911–4919. doi: 10.1128/MCB.00212-06
Fairall, L., Chapman, L., Moss, H., de Lange, T., and Rhodes, D. (2001). Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell 8, 351–361. doi: 10.1016/s1097-2765(01)00321-5
Faria, J., Glover, L., Hutchinson, S., Boehm, C., Field, M. C., and Horn, D. (2019). Monoallelic expression and epigenetic inheritance sustained by a Trypanosoma brucei variant surface glycoprotein exclusion complex. Nat. Commun. 10:3023. doi: 10.1038/s41467-019-10823-8
Farman, M. L. (2007). Telomeres in the rice blast fungus Magnaporthe oryzae: the world of the end as we know it. FEMS Microbiol. Lett. 273, 125–132. doi: 10.1111/j.1574-6968.2007.00812.x
Feng, X., Hsu, S. J., Kasbek, C., Chaiken, M., and Price, C. M. (2017). CTC1-mediated C-strand fill-in is an essential step in telomere length maintenance. Nucleic Acids Res. 45, 4281–4293. doi: 10.1093/nar/gkx125
Feretzaki, M., and Lingner, J. (2017). A practical qPCR approach to detect TERRA, the elusive telomeric repeat-containing RNA. Methods 114, 39–45. doi: 10.1016/j.ymeth.2016.08.004
Feretzaki, M., Pospisilova, M., Valador Fernandes, R., Lunardi, T., Krejci, L., and Lingner, J. (2020). RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature 587, 303–308. doi: 10.1038/s41586-020-2815-6
Ferreira, M. G., and Cooper, J. P. (2001). The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell 7, 55–63. doi: 10.1016/s1097-2765(01)00154-x
Figueiredo, L. M., and Cross, G. A. M. (2010). Nucleosomes are depleted at the VSG expression site transcribed by RNA polymerase I in African trypanosomes. Eukaryot. Cell 9, 148–154. doi: 10.1128/EC.00282-09
Figueiredo, L. M., Janzen, C. J., and Cross, G. A. M. (2008). A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 6:e161. doi: 10.1371/journal.pbio.0060161
Frescas, D., and de Lange, T. (2014). Binding of TPP1 protein to TIN2 protein is required for POT1a,b protein-mediated telomere protection. J. Biol. Chem. 289, 24180–24187. doi: 10.1074/jbc.M114.592592
García-Muse, T., and Aguilera, A. (2019). R loops: from physiological to pathological roles. Cell 179, 604–618. doi: 10.1016/j.cell.2019.08.055
Glover, L., Alsford, S., Beattie, C., and Horn, D. (2007). Deletion of a trypanosome telomere leads to loss of silencing and progressive loss of terminal DNA in the absence of cell cycle arrest. Nuc Acids Res. 35, 872–880. doi: 10.1093/nar/gkl1100
Glover, L., Alsford, S., and Horn, D. (2013). DNA break site at fragile subtelomeres determines probability and mechanism of antigenic variation in African trypanosomes. PLoS Pathog 9:e1003260. doi: 10.1371/journal.ppat.1003260
Glover, L., and Horn, D. (2012). Trypanosomal histone gammaH2A and the DNA damage response. Mol. Biochem. Parasitol. 183, 78–83. doi: 10.1016/j.molbiopara.2012.01.008
Glover, L., and Horn, D. (2014). Locus-specific control of DNA resection and suppression of subtelomeric VSG recombination by HAT3 in the African trypanosome. Nucleic Acids Res. 42, 12600–12613. doi: 10.1093/nar/gku900
Glover, L., Hutchinson, S., Alsford, S., and Horn, D. (2016). VEX1 controls the allelic exclusion required for antigenic variation in trypanosomes. Proc. Natl. Acad. Sci. U S A. 113, 7225–7230. doi: 10.1073/pnas.1600344113
Glover, L., Jun, J., and Horn, D. (2011). Microhomology-mediated deletion and gene conversion in African trypanosomes. Nucleic Acids Res. 39, 1372–1380. doi: 10.1093/nar/gkq981
Glover, L., McCulloch, R., and Horn, D. (2008). Sequence homology and microhomology dominate chromosomal double-strand break repair in African trypanosomes. Nucleic Acids Res. 36, 2608–2618. doi: 10.1093/nar/gkn104
Glover, L., Marques, C. A., Suska, O., and Horn, D. (2019). Persistent DNA Damage Foci and DNA Replication with a Broken Chromosome in the African Trypanosome. mBio 10:e01252-19. doi: 10.1128/mBio.01252-19
Gong, Y., and de Lange, T. (2010). A Shld1-controlled POT1a provides support for repression of ATR signaling at telomeres through RPA exclusion. Mol. Cell. 40, 377–387. doi: 10.1016/j.molcel.2010.10.016
Gottschling, D. E., Aparicio, O. M., Billington, B. L., and Zakian, V. A. (1990). Position effect at S. cerevisiae telomeres: reversible repression of pol II transcription. Cell 63, 751–762. doi: 10.1016/0092-8674(90)90141-z
Graf, M., Bonetti, D., Lockhart, A., Serhal, K., Kellner, V., Maicher, A., et al. (2017). Telomere length determines TERRA and R-Loop regulation through the cell cycle. Cell 170, 72–85.e14. doi: 10.1016/j.cell.2017.06.006
Graham, I. R., and Chambers, A. (1994). Use of a selection technique to identify the diversity of binding sites for the yeast RAP1 transcription factor. Nucleic Acids Res. 22, 124–130. doi: 10.1093/nar/22.2.124
Greider, C. W., and Blackburn, E. H. (1985). Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43, 405–413. doi: 10.1016/0092-8674(85)90170-9
Greider, C. W., and Blackburn, E. H. (1987). The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 51, 887–898. doi: 10.1016/0092-8674(87)90576-9
Greider, C. W., and Blackburn, E. H. (1989). A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337, 331–337. doi: 10.1038/337331a0
Griffith, J. D., Comeau, L., Rosenfield, S., Stansel, R. M., Bianchi, A., Moss, H., et al. (1999). Mammalian telomeres end in a large duplex loop. Cell 97, 503–514. doi: 10.1016/s0092-8674(00)80760-6
Guo, X., Deng, Y., Lin, Y., Cosme-Blanco, W., Chan, S., He, H., et al. (2007). Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 26, 4709–4719. doi: 10.1038/sj.emboj.7601893
Haber, J. E. (2018). DNA repair: the search for homology. Bioessays 40:e1700229. doi: 10.1002/bies.201700229
Hardy, C. F., Sussel, L., and Shore, D. (1992). A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 6, 801–814. doi: 10.1101/gad.6.5.801
Hegazy, Y. A., Fernando, C. M., and Tran, E. J. (2020). The balancing act of R-loop biology: the good, the bad, and the ugly. J. Biol. Chem. 295, 905–913. doi: 10.1074/jbc.REV119.011353
Hertz-Fowler, C., Figueiredo, L. M., Quail, M. A., Becker, M., Jackson, A., Bason, N., et al. (2008). Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One 3:e3527. doi: 10.1371/journal.pone.0003527
Hockemeyer, D., Palm, W., Else, T., Daniels, J. P., Takai, K. K., Ye, J. Z., et al. (2007). Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat. Struct. Mol. Biol. 14, 754–761. doi: 10.1038/nsmb1270
Horn, D., and Cross, G. A. M. (1997). Position-dependent and promoter-specific regulation of gene expression in Trypanosoma brucei. EMBO J. 16, 7422–7431. doi: 10.1093/emboj/16.24.7422
Houghtaling, B. R., Cuttonaro, L., Chang, W., and Smith, S. (2004). A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 14, 1621–1631. doi: 10.1016/j.cub.2004.08.052
Hovel-Miner, G. A., Boothroyd, C. E., Mugnier, M., Dreesen, O., Cross, G. A. M., and Papavasiliou, F. N. (2012). Telomere length affects the frequency and mechanism of antigenic variation in Trypanosoma brucei. PLoS Pathog 8:e1002900. doi: 10.1371/journal.ppat.1002900
Hu, Y., Bennett, H. W., Liu, N., Moravec, M., Williams, J. F., Azzalin, C. M., et al. (2019). RNA-DNA hybrids support recombination-based telomere maintenance in fission yeast. Genetics 213, 431–447. doi: 10.1534/genetics.119.302606
Hviid, L., and Jensen, A. T. (2015). PfEMP1 - a parasite protein family of key importance in Plasmodium falciparum malaria immunity and pathogenesis. Adv. Parasitol. 88, 51–84. doi: 10.1016/bs.apar.2015.02.004
Iglesias, N., Redon, S., Pfeiffer, V., Dees, M., Lingner, J., and Luke, B. (2011). Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep. 12, 587–593. doi: 10.1038/embor.2011.73
Irie, H., Yamamoto, I., Tarumoto, Y., Tashiro, S., Runge, K. W., and Ishikawa, F. (2019). Telomere-binding proteins Taz1 and Rap1 regulate DSB repair and suppress gross chromosomal rearrangements in fission yeast. PLoS Genet. 15:e1008335. doi: 10.1371/journal.pgen.1008335
Jehi, S. E., Li, X., Sandhu, R., Ye, F., Benmerzouga, I., Zhang, M., et al. (2014a). Suppression of subtelomeric VSG switching by Trypanosoma brucei TRF requires its TTAGGG repeat-binding activity. Nucleic Acids Res. 42, 12899–12911. doi: 10.1093/nar/gku942
Jehi, S. E., Wu, F., and Li, B. (2014b). Trypanosoma brucei TIF2 suppresses VSG switching by maintaining subtelomere integrity. Cell Res. 24, 870–885. doi: 10.1038/cr.2014.60
Jehi, S. E., Nanavaty, V., and Li, B. (2016). Trypanosoma brucei TIF2 and TRF suppress VSG switching using overlapping and independent mechanisms. PLoS One 11:e0156746. doi: 10.1371/journal.pone.0156746
Kabir, S., Hockemeyer, D., and de Lange, T. (2014). TALEN gene knockouts reveal no requirement for the conserved human shelterin protein Rap1 in telomere protection and length regulation. Cell Rep. 9, 1273–1280. doi: 10.1016/j.celrep.2014.10.014
Kanoh, J., and Ishikawa, F. (2001). spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 11, 1624–1630. doi: 10.1016/s0960-9822(01)00503-6
Karlseder, J., Broccoli, D., Dai, Y., Hardy, S., and de Lange, T. (1999). p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 283, 1321–1325. doi: 10.1126/science.283.5406.1321
Keely, S. P., Renauld, H., Wakefield, A. E., Cushion, M. T., Smulian, A. G., Fosker, N., et al. (2005). Gene arrays at Pneumocystis carinii telomeres. Genetics 170, 1589–1600. doi: 10.1534/genetics.105.040733
Kibe, T., Zimmermann, M., and de Lange, T. (2016). TPP1 blocks an ATR-Mediated resection mechanism at telomeres. Mol. Cell. 61, 236–246. doi: 10.1016/j.molcel.2015.12.016
Kim, H. S. (2019). Genome-wide function of MCM-BP in Trypanosoma brucei DNA replication and transcription. Nucleic Acids Res. 47, 634–647. doi: 10.1093/nar/gky1088
Kim, H. S., and Cross, G. A. M. (2010). TOPO3alpha influences antigenic variation by monitoring expression-site-associated VSG switching in Trypanosoma brucei. PLoS Pathog 6:e1000992. doi: 10.1371/journal.ppat.1000992
Kim, H. S., and Cross, G. A. M. (2011). Identification of Trypanosoma brucei RMI1/BLAP75 homologue and its roles in antigenic variation. PLoS One 6:e25313. doi: 10.1371/journal.pone.0025313
Kim, H. S., Park, S. H., Gunzl, A., and Cross, G. A. (2013). MCM-BP is required for repression of life-cycle specific genes transcribed by RNA polymerase I in the mammalian infectious form of Trypanosoma brucei. PLoS One 8:e57001. doi: 10.1371/journal.pone.0057001
Kim, S. H., Kaminker, P., and Campisi, J. (1999). TIN2, a new regulator of telomere length in human cells. Nat. Genet. 23, 405–412. doi: 10.1038/70508
Konig, P., Giraldo, R., Chapman, L., and Rhodes, D. (1996). The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell 85, 125–136. doi: 10.1016/s0092-8674(00)81088-0
Kuo, H. F., Olsen, K. M., and Richards, E. J. (2006). Natural variation in a subtelomeric region of Arabidopsis: implications for the genomic dynamics of a chromosome end. Genetics 173, 401–417. doi: 10.1534/genetics.105.055202
Kutty, G., Ma, L., and Kovacs, J. A. (2001). Characterization of the expression site of the major surface glycoprotein of human-derived Pneumocystis carinii. Mol. Microbiol. 42, 183–193. doi: 10.1046/j.1365-2958.2001.02620.x
Kutty, G., Maldarelli, F., Achaz, G., and Kovacs, J. A. (2008). Variation in the major surface glycoprotein genes in Pneumocystis jirovecii. J. Infect. Dis. 198, 741–749. doi: 10.1086/590433
Kyrion, G., Boakye, K. A., and Lustig, A. J. (1992). C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol. Cell Biol. 12, 5159–5173. doi: 10.1128/mcb.12.11.5159-5173.1992
Kyrion, G., Liu, K., Liu, C., and Lustig, A. J. (1993). RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 7, 1146–1159. doi: 10.1101/gad.7.7a.1146
Laberthonnière, C., Magdinier, F., and Robin, J. D. (2019). Bring it to an end: does telomeres size matter. Cells 8:30. doi: 10.3390/cells8010030
Lee, Y. W., Arora, R., Wischnewski, H., and Azzalin, C. M. (2018). TRF1 participates in chromosome end protection by averting TRF2-dependent telomeric R loops. Nat. Struct. Mol. Biol. 25, 147–153. doi: 10.1038/s41594-017-0021-5
Li, B. (2012). Telomere components as potential therapeutic targets for treating microbial pathogen infections. Front. Oncol. 2:156. doi: 10.3389/fonc.2012.00156
Li, B., and de Lange, T. (2003). Rap1 affects the length and heterogeneity of human telomeres. Mol. Biol. Cell 14, 5060–5068. doi: 10.1091/mbc.e03-06-0403
Li, B., Espinal, A., and Cross, G. A. M. (2005). Trypanosome telomeres are protected by a homologue of mammalian TRF2. Mol. Cell. Biol. 25, 5011–5021. doi: 10.1128/mcb.25.12.5011-5021.2005
Li, B., Oestreich, S., and de Lange, T. (2000). Identification of human Rap1: implications for telomere evolution. Cell 101, 471–483.
Linardopoulou, E. V., Williams, E. M., Fan, Y., Friedman, C., Young, J. M., and Trask, B. J. (2005). Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature 437, 94–100. doi: 10.1038/nature04029
Liu, C., and Lustig, A. J. (1996). Genetic analysis of Rap1p/Sir3p interactions in telomeric and HML silencing in Saccharomyces cerevisiae. Genetics 143, 81–93. doi: 10.1093/genetics/143.1.81
Liu, C., Mao, X., and Lustig, A. J. (1994). Mutational analysis defines a C-terminal tail domain of RAP1 essential for telomeric silencing in Saccharomyces cerevisiae. Genetics 138, 1025–1040. doi: 10.1093/genetics/138.4.1025
Liu, D., Safari, A., O’Connor, M. S., Chan, D. W., Laegeler, A., Qin, J., et al. (2004). PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 6, 673–680. doi: 10.1038/ncb1142
Loayza, D., and de Lange, T. (2003). POT1 as a terminal transducer of TRF1 telomere length control. Nature 424, 1013–1018. doi: 10.1038/nature01688
Longtine, M. S., Wilson, N. M., Petracek, M. E., and Berman, J. (1989). A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr. Genet. 16, 225–239. doi: 10.1007/bf00422108
Lototska, L., Yue, J. X., Li, J., Giraud-Panis, M. J., Songyang, Z., Royle, N. J., et al. (2020). Human RAP1 specifically protects telomeres of senescent cells from DNA damage. EMBO Rep. 21:e49076. doi: 10.15252/embr.201949076
Lottersberger, F., Bothmer, A., Robbiani, D. F., Nussenzweig, M. C., and de Lange, T. (2013). Role of 53BP1 oligomerization in regulating double-strand break repair. Proc. Natl. Acad. Sci. U S A. 110, 2146–2151. doi: 10.1073/pnas.1222617110
Luke, B., Panza, A., Redon, S., Iglesias, N., Li, Z., and Lingner, J. (2008). The Rat1p 5’ to 3’ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell 32, 465–477. doi: 10.1016/j.molcel.2008.10.019
Lustig, A. J., Kurtz, S., and Shore, D. (1990). Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250, 549–553. doi: 10.1126/science.2237406
Lustig, A. J., Liu, C., Zhang, C., and Hanish, J. P. (1996). Tethered sir3p nucleates silencing at telomeres and internal loci in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 2483–2495. doi: 10.1128/mcb.16.5.2483
Maciejowski, J., and de Lange, T. (2017). Telomeres in cancer: tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 18, 175–186. doi: 10.1038/nrm.2016.171
Maciejowski, J., Li, Y., Bosco, N., Campbell, P. J., and de Lange, T. (2015). Chromothripsis and kataegis induced by telomere crisis. Cell 163, 1641–1654. doi: 10.1016/j.cell.2015.11.054
Makarov, V. L., Hirose, Y., and Langmore, J. P. (1997). Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88, 657–666. doi: 10.1016/s0092-8674(00)81908-x
Marcand, S., Gilson, E., and Shore, D. (1997). A protein-counting mechanism for telomere length regulation in yeast. Science 275, 986–990. doi: 10.1126/science.275.5302.986
Marión, R. M., Montero, J. J., López, de Silanes, I., Graña-Castro, O., Martínez, P., et al. (2019). TERRA regulate the transcriptional landscape of pluripotent cells through TRF1-dependent recruitment of PRC2. eLife 8:e44656. doi: 10.7554/eLife.44656
Mason, J. M. O., and McEachern, M. J. (2018). Mild telomere dysfunction as a force for altering the adaptive potential of subtelomeric genes. Genetics 208, 537–548. doi: 10.1534/genetics.117.300607
Mateos-Gomez, P. A., Gong, F., Nair, N., Miller, K. M., Lazzerini-Denchi, E., and Sfeir, A. (2015). Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257. doi: 10.1038/nature14157
McCulloch, R., Morrison, L. J., and Hall, J. P. J. (2015). DNA recombination strategies during antigenic variation in the African trypanosome. Microbiol. Spectr. 3:MDNA3-0016-2014.. doi: 10.1128/microbiolspec.MDNA3-0016-2014
Mefford, H. C., and Trask, B. J. (2002). The complex structure and dynamic evolution of human subtelomeres. Nat. Rev. Genet. 3, 91–102. doi: 10.1038/nrg727
Mei, Y., Deng, Z., Vladimirova, O., Gulve, N., Johnson, F. B., Drosopoulos, W. C., et al. (2021). TERRA G-quadruplex RNA interaction with TRF2 GAR domain is required for telomere integrity. Sci. Rep. 11, 1–14.
Melville, S. E., Gerrard, C. S., and Blackwell, J. M. (1999). Multiple causes of size variation in the diploid megabase chromosomes of African trypanosomes. Chromosome Res. 7, 191–203.
Melville, S. E., Leech, V., Navarro, M., and Cross, G. A. M. (2000). The molecular karyotype of the megabase chromosomes of Trypanosoma brucei stock 427. Mol. Biochem. Parasitol. 111, 261–273. doi: 10.1016/s0166-6851(00)00316-9
Min, J., Wright, W. E., and Shay, J. W. (2017). Alternative lengthening of telomeres mediated by mitotic DNA synthesis engages break-induced replication processes. Mol. Cell. Biol. 37:e00226-17. doi: 10.1128/MCB.00226-17
Mitchell, T. R., Glenfield, K., Jeyanthan, K., and Zhu, X. D. (2009). Arginine methylation regulates telomere length and stability. Mol. Cell. Biol. 29, 4918–4934. doi: 10.1128/MCB.00009-09
Morea, E. G. O., Vasconcelos, E. J. R., Alves, C. S., Giorgio, S., Myler, P. J., Langoni, H., et al. (2021). Exploring TERRA during Leishmania major developmental cycle and continuous in vitro passages. Int. J. Biol. Macromol. 174, 573–586. doi: 10.1016/j.ijbiomac.2021.01.192
Morrison, L. J., Marcello, L., and McCulloch, R. (2009). Antigenic variation in the African trypanosome: molecular mechanisms and phenotypic complexity. Cell Microbiol. 11, 1724–1734. doi: 10.1111/j.1462-5822.2009.01383.x
Müller, L. S. M., Cosentino, R. O., Förstner, K. U., Guizetti, J., Wedel, C., Kaplan, N., et al. (2018). Genome organization and DNA accessibility control antigenic variation in trypanosomes. Nature 563, 121–125. doi: 10.1038/s41586-018-0619-8
Munoz-Jordan, J. L., Cross, G. A. M., de Lange, T., and Griffith, J. D. (2001). t-loops at trypanosome telomeres. EMBO J. 20, 579–588. doi: 10.1093/emboj/20.3.579
Murti, K. G., and Prescott, D. M. (1999). Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proc. Natl. Acad. Sci. U S A. 96, 14436–14439. doi: 10.1073/pnas.96.25.14436
Myler, P., Nelson, R. G., Agabian, N., and Stuart, K. (1984a). Two mechanisms of expression of a variant antigen gene of Trypanosoma brucei. Nature 309, 282–284. doi: 10.1038/309282a0
Myler, P. J., Allison, J., Agabian, N., and Stuart, K. (1984b). Antigenic variation in African trypanosomes by gene replacement or activation of alternative telomeres. Cell 39, 203–211. doi: 10.1016/0092-8674(84)90206-x
Myler, P. J., Allen, A. L., Agabian, N., and Stuart, K. (1985). Antigenic variation in clones of Trypanosoma brucei grown in immune-deficient mice. Infect. Immun. 47, 684–690. doi: 10.1128/iai.47.3.684-690.1985
Nanavaty, V., Sandhu, R., Jehi, S. E., Pandya, U. M., and Li, B. (2017). Trypanosoma brucei RAP1 maintains telomere and subtelomere integrity by suppressing TERRA and telomeric RNA:DNA hybrids. Nucleic Acids Res. 45, 5785–5796. doi: 10.1093/nar/gkx184
Navarro, M., and Cross, G. A. M. (1996). DNA rearrangements associated with multiple consecutive directed antigenic switches in Trypanosoma brucei. Mol. Cell. Biol. 16, 3615–3625. doi: 10.1128/mcb.16.7.3615
Nergadze, S. G., Farnung, B. O., Wischnewski, H., Khoriauli, L., Vitelli, V., Chawla, R., et al. (2009). CpG-island promoters drive transcription of human telomeres. RNA 15, 2186–2194. doi: 10.1261/rna.1748309
Nett, I. R., Martin, D. M., Miranda-Saavedra, D., Lamont, D., Barber, J. D., Mehlert, A., et al. (2009). The phosphoproteome of bloodstream form Trypanosoma brucei, causative agent of African sleeping sickness. Mol. Cell. Proteomics 8, 1527–1538. doi: 10.1074/mcp.M800556-MCP200
Nguyen, D. T. T., Voon, H. P. J., Xella, B., Scott, C., Clynes, D., Babbs, C., et al. (2017). The chromatin remodelling factor ATRX suppresses R-loops in transcribed telomeric repeats. EMBO Rep. 18, 914–928. doi: 10.15252/embr.201643078
Nikitina, T., and Woodcock, C. L. (2004). Closed chromatin loops at the ends of chromosomes. J. Cell Biol. 166, 161–165. doi: 10.1083/jcb.200403118
Nimmo, E. R., Cranston, G., and Allshire, R. C. (1994). Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J. 13, 3801–3811. doi: 10.1002/j.1460-2075.1994.tb06691.x
Ogata, K., Morikawa, S., Nakamura, H., Sekikawa, A., Inoue, T., Kanai, H., et al. (1994). Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79, 639–648. doi: 10.1016/0092-8674(94)90549-5
Ottaviani, A., Gilson, E., and Magdinier, F. (2008). Telomeric position effect: from the yeast paradigm to human pathologies? Biochimie 90, 93–107. doi: 10.1016/j.biochi.2007.07.022
Palm, W., Hockemeyer, D., Kibe, T., and de Lange, T. (2009). Functional dissection of human and mouse POT1 proteins. Mol. Cell. Biol. 29, 471–482. doi: 10.1128/MCB.01352-08
Pandya, U. M., Sandhu, R., and Li, B. (2013). Silencing subtelomeric VSGs by Trypanosoma brucei RAP1 at the insect stage involves chromatin structure changes. Nucleic Acids Res. 41, 7673–7682. doi: 10.1093/nar/gkt562
Pardo, B., and Marcand, S. (2005). Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 24, 3117–3127. doi: 10.1038/sj.emboj.7600778
Pellestor, F., Gaillard, J. B., Schneider, A., Puechberty, J., and Gatinois, V. (2021). Chromoanagenesis, the mechanisms of a genomic chaos. Semin. Cell Dev. Biol. doi: 10.1016/j.semcdb.2021.01.004 [Epub ahead of print].
Podlevsky, J. D., Bley, C. J., Omana, R. V., Qi, X., and Chen, J. J. (2008). The telomerase database. Nucleic Acids Res. 36, D339–D343. doi: 10.1093/nar/gkm700
Porro, A., Feuerhahn, S., Delafontaine, J., Riethman, H., Rougemont, J., and Lingner, J. (2014a). Functional characterization of the TERRA transcriptome at damaged telomeres. Nat. Commun. 5:5379. doi: 10.1038/ncomms6379
Porro, A., Feuerhahn, S., and Lingner, J. (2014b). TERRA-reinforced association of LSD1 with MRE11 promotes processing of uncapped telomeres. Cell Rep. 6, 765–776. doi: 10.1016/j.celrep.2014.01.022
Pryde, F. E., Gorham, H. C., and Louis, E. J. (1997). Chromosome ends: all the same under their caps. Curr. Opin. Genet. Dev. 7, 822–828. doi: 10.1016/s0959-437x(97)80046-9
Quispe, X., Tapia, S. M., Villarroel, C., Oporto, C., Abarca, V., García, V., et al. (2017). Genetic basis of mycotoxin susceptibility differences between budding yeast isolates. Sci. Rep. 7:9173. doi: 10.1038/s41598-017-09471-z
Rai, R., Chen, Y., Lei, M., and Chang, S. (2016). TRF2-RAP1 is required to protect telomeres from engaging in homologous recombination-mediated deletions and fusions. Nat. Commun. 7:10881. doi: 10.1038/ncomms10881
Rai, R., Zheng, H., He, H., Luo, Y., Multani, A., Carpenter, P. B., et al. (2010). The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J. 29, 2598–2610. doi: 10.1038/emboj.2010.142
Raices, M., Verdun, R. E., Compton, S. A., Haggblom, C. I., Griffith, J. D., Dillin, A., et al. (2008). C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell 132, 745–757. doi: 10.1016/j.cell.2007.12.039
Renauld, H., Aparicio, O. M., Zierath, P. D., Billington, B. L., Chhablani, S. K., and Gottschling, D. E. (1993). Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7, 1133–1145. doi: 10.1101/gad.7.7a.1133
Reynolds, D., Hofmeister, B. T., Cliffe, L., Alabady, M., Siegel, T. N., Schmitz, R. J., et al. (2016). Histone H3 variant regulates RNA polymerase ii transcription termination and dual strand transcription of siRNA loci in Trypanosoma brucei. PLoS Genet 12:e1005758. doi: 10.1371/journal.pgen.1005758
Rhodin, J., Astromskas, E., and Cohn, M. (2006). Characterization of the DNA binding features of Saccharomyces castellii Cdc13p. J. Mol. Biol. 355, 335–346. doi: 10.1016/j.jmb.2005.10.078
Ridewood, S., Ooi, C. P., Hall, B., Trenaman, A., Wand, N. V., Sioutas, G., et al. (2017). The role of genomic location and flanking 3’UTR in the generation of functional levels of variant surface glycoprotein in Trypanosoma brucei. Mol. Microbiol. 106, 614–634. doi: 10.1111/mmi.13838
Robin, J. D., Ludlow, A. T., Batten, K., Magdinier, F., Stadler, G., Wagner, K. R., et al. (2014). Telomere position effect: regulation of gene expression with progressive telomere shortening over long distances. Genes Dev. 28, 2464–2476. doi: 10.1101/gad.251041.114
Robinson, N. P., Burman, N., Melville, S. E., and Barry, J. D. (1999). Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Mol. Cell Biol. 19, 5839–5846. doi: 10.1128/mcb.19.9.5839
Rubio, J. P., Thompson, J. K., and Cowman, A. F. (1996). The var genes of Plasmodium falciparum are located in the subtelomeric region of most chromosomes. EMBO J. 15, 4069–4077. doi: 10.1002/j.1460-2075.1996.tb00780.x
Rudd, M. K., Friedman, C., Parghi, S. S., Linardopoulou, E. V., Hsu, L., and Trask, B. J. (2007). Elevated rates of sister chromatid exchange at chromosome ends. PLoS Genet 3:e32. doi: 10.1371/journal.pgen.0030032
Rudenko, G., McCulloch, R., Dirksmulder, A., and Borst, P. (1996). Telomere exchange can be an important mechanism of variant surface glycoprotein gene switching in Trypanosoma brucei. Mol. Biochem. Parasitol. 80, 65–75. doi: 10.1016/0166-6851(96)02669-2
Rudenko, G., and Van der Ploeg, L. H. (1989). Transcription of telomere repeats in protozoa. EMBO J. 8, 2633–2638. doi: 10.1002/j.1460-2075.1989.tb08403.x
Sagie, S., Toubiana, S., Hartono, S. R., Katzir, H., Tzur-Gilat, A., Havazelet, S., et al. (2017). Telomeres in ICF syndrome cells are vulnerable to DNA damage due to elevated DNA:RNA hybrids. Nat. Commun. 8:14015. doi: 10.1038/ncomms14015
Saha, A., Gaurav, A. K., Pandya, U. M., Afrin, M., Sandhu, R., Nanavaty, V., et al. (2021). TbTRF suppresses the TERRA level and regulates the cell cycle-dependent TERRA foci number with a TERRA binding activity in its C-terminal Myb domain. Nucleic Acids Res. doi: 10.1093/nar/gkab401 Online ahead of print.
Saha, A., Nanavaty, V. P., and Li, B. (2019). Telomere and subtelomere R-loops and antigenic variation in trypanosomes. J. Mol. Biol. 432, 4167–4185. doi: 10.1016/j.jmb.2019.10.025
Sandhu, R., and Li, B. (2017). Telomerase activity is required for the telomere G-overhang structure in Trypanosoma brucei. Sci. Rep. 7:15983. doi: 10.1038/s41598-017-16182-y
Sandhu, R., and Li, B. (2011). Examination of the telomere G-overhang structure in Trypanosoma brucei. J. Vis. Exp. 1959. doi: 10.3791/1959
Sarthy, J., Bae, N. S., Scrafford, J., and Baumann, P. (2009). Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J. 28, 3390–3399. doi: 10.1038/emboj.2009.275
Schmid-Siegert, E., Richard, S., Luraschi, A., Mühlethaler, K., Pagni, M., and Hauser, P. M. (2017). Mechanisms of surface antigenic variation in the human pathogenic fungus Pneumocystis jirovecii. mBio 8:e01470-17. doi: 10.1128/mBio.01470-17
Schoeftner, S., and Blasco, M. A. (2008). Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10, 228–236. doi: 10.1038/ncb1685
Schulz, D., Zaringhalam, M., Papavasiliou, F. N., and Kim, H. S. (2016). Base J and H3.V regulate transcriptional termination in Trypanosoma brucei. PLoS Genet 12:e1005762. doi: 10.1371/journal.pgen.1005762
Seol, J. H., Shim, E. Y., and Lee, S. E. (2018). Microhomology-mediated end joining: good, bad and ugly. Mutat. Res. 809, 81–87. doi: 10.1016/j.mrfmmm.2017.07.002
Sfeir, A., and de Lange, T. (2012). Removal of shelterin reveals the telomere end-protection problem. Science 336, 593–597. doi: 10.1126/science.1218498
Sfeir, A., Kabir, S., van Overbeek, M., Celli, G. B., and de Lange, T. (2010). Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 327, 1657–1661. doi: 10.1126/science.1185100
Sfeir, A., Kosiyatrakul, S. T., Hockemeyer, D., MacRae, S. L., Karlseder, J., Schildkraut, C. L., et al. (2009). Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138, 90–103. doi: 10.1016/j.cell.2009.06.021
Sfeir, A., and Symington, L. S. (2015). Microhomology-Mediated end joining: a back-up survival mechanism or dedicated pathway. Trends Biochem. Sci. 40, 701–714. doi: 10.1016/j.tibs.2015.08.006
Shay, J. W., and Wright, W. E. (2019). Telomeres and telomerase: three decades of progress. Nat. Rev. Genet. 20, 299–309. doi: 10.1038/s41576-019-0099-1
Sheader, K., Vaughan, S., Minchin, J., Hughes, K., Gull, K., and Rudenko, G. (2005). Variant surface glycoprotein RNA interference triggers a precytokinesis cell cycle arrest in African trypanosomes. Proc. Natl. Acad. Sci. U S A. 102, 8716–8721. doi: 10.1073/pnas.0501886102
Shiromoto, Y., Sakurai, M., Minakuchi, M., Ariyoshi, K., and Nishikura, K. (2021). ADAR1 RNA editing enzyme regulates R-loop formation and genome stability at telomeres in cancer cells. Nat. Commun. 12:1654. doi: 10.1038/s41467-021-21921-x
Shore, D., and Nasmyth, K. (1987). Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell 51, 721–732. doi: 10.1016/0092-8674(87)90095-x
Sima, N., McLaughlin, E. J., Hutchinson, S., and Glover, L. (2019). Escaping the immune system by DNA repair and recombination in African trypanosomes. Open Biol. 9:190182. doi: 10.1098/rsob.190182
Smogorzewska, A., Karlseder, J., Holtgreve-Grez, H., Jauch, A., and de Lange, T. (2002). DNA ligase IV-Dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 12:1635. doi: 10.1016/s0960-9822(02)01179-x
Smogorzewska, A., van Steensel, B., Bianchi, A., Oelmann, S., Schaefer, M. R., Schnapp, G., et al. (2000). Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 20, 1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000
Soares, A. R., Soares, G., Mota-Freitas, M., Oliva-Teles, N., and Fortuna, A. M. (2019). Subtelomeric rearrangements: presentation of 21 probands with emphasis on familial cases. Acta Med. Port. 32, 529–535. doi: 10.20344/amp.11466
Sobinoff, A. P., and Pickett, H. A. (2017). alternative lengthening of telomeres: DNA repair pathways converge. Trends Genet. 33, 921–932. doi: 10.1016/j.tig.2017.09.003
Solovei, I., Gaginskaya, E. R., and Macgregor, H. C. (1994). The arrangement and transcription of telomere DNA sequences at the ends of lampbrush chromosomes of birds. Chromosome Res. 2, 460–470. doi: 10.1007/bf01552869
Stanne, T. M., and Rudenko, G. (2010). Active VSG expression sites in Trypanosoma brucei are depleted of nucleosomes. Eukaryot. Cell 9, 136–147. doi: 10.1128/EC.00281-09
Steinberg-Neifach, O., and Lue, N. F. (2015). Telomere DNA recognition in Saccharomycotina yeast: potential lessons for the co-evolution of ssDNA and dsDNA-binding proteins and their target sites. Front. Genet. 6:162. doi: 10.3389/fgene.2015.00162
Stewart, D. R., and Kleefstra, T. (2007). The chromosome 9q subtelomere deletion syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 145C, 383–392. doi: 10.1002/ajmg.c.30148
Stewart, J. A., Wang, Y., Ackerson, S. M., and Schuck, P. L. (2018). Emerging roles of CST in maintaining genome stability and human disease. Front. Biosci. (Landmark Ed) 23:1564–1586. doi: 10.2741/4661
Stringer, J. R. (2007). Antigenic variation in pneumocystis. J. Eukaryot. Microbiol. 54, 8–13. doi: 10.1111/j.1550-7408.2006.00225.x
Takai, K. K., Kibe, T., Donigian, J. R., Frescas, D., and de Lange, T. (2011). Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol. Cell 44, 647–659. doi: 10.1016/j.molcel.2011.08.043
Tan, M., Wei, C., and Price, C. M. (2003). The telomeric protein Rap1 is conserved in vertebrates and is expressed from a bidirectional promoter positioned between the Rap1 and KARS genes. Gene 323, 1–10. doi: 10.1016/j.gene.2003.08.026
Tennen, R. I., Bua, D. J., Wright, W. E., and Chua, K. F. (2011). SIRT6 is required for maintenance of telomere position effect in human cells. Nat. Commun. 2:433. doi: 10.1038/ncomms1443
Tiengwe, C., Marcello, L., Farr, H., Dickens, N., Kelly, S., Swiderski, M., et al. (2012). Genome-wide analysis reveals extensive functional interaction between DNA replication initiation and transcription in the genome of Trypanosoma brucei. Cell Rep. 2, 185–197. doi: 10.1016/j.celrep.2012.06.007
Timashev, L. A., and de Lange, T. (2020). Characterization of t-loop formation by TRF2. Nucleus 11, 164–177. doi: 10.1080/19491034.2020.1783782
Toubiana, S., and Selig, S. (2018). DNA:RNA hybrids at telomeres - when it is better to be out of the (R) loop. FEBS J. 285, 2552–2566. doi: 10.1111/febs.14464
Trask, B. J., Friedman, C., Martin-Gallardo, A., Rowen, L., Akinbami, C., Blankenship, J., et al. (1998). Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum. Mol. Genet. 7, 13–26. doi: 10.1093/hmg/7.1.13
Underwood, A. P., Louis, E. J., Borts, R. H., Stringer, J. R., and Wakefield, A. E. (1996). Pneumocystis carinii telomere repeats are composed of TTAGGG and the subtelomeric sequence contains a gene encoding the major surface glycoprotein. Mol. Microbiol. 19, 273–281. doi: 10.1046/j.1365-2958.1996.374904.x
Urbaniak, M. D., Martin, D. M., and Ferguson, M. A. (2013). Global quantitative SILAC phosphoproteomics reveals differential phosphorylation is widespread between the procyclic and bloodstream form lifecycle stages of Trypanosoma brucei. J. Proteome Res. 12, 2233–2244. doi: 10.1021/pr400086y
van der Maarel, S. M., Frants, R. R., and Padberg, G. W. (2007). Facioscapulohumeral muscular dystrophy. Biochim. Biophys. Acta 1772, 186–194. doi: 10.1016/j.bbadis.2006.05.009
van der Ploeg, L. H., Cornelissen, A. W., Barry, J. D., and Borst, P. (1984). Chromosomes of kinetoplastidia. EMBO J. 3, 3109–3115.
Van Ly, D., Low, R. R. J., Frölich, S., Bartolec, T. K., Kafer, G. R., Pickett, H. A., et al. (2018). Telomere loop dynamics in chromosome end protection. Mol. Cell 71, 510–525.e6. doi: 10.1016/j.molcel.2018.06.025
van Steensel, B., and de Lange, T. (1997). Control of telomere length by the human telomeric protein TRF1. Nature 385, 740–743. doi: 10.1038/385740a0
van Steensel, B., Smogorzewska, A., and de Lange, T. (1998). TRF2 protects human telomeres from end-to-end fusions. Cell 92, 401–413. doi: 10.1016/s0092-8674(00)80932-0
Voss, T. S., Healer, J., Marty, A. J., Duffy, M. F., Thompson, J. K., Beeson, J. G., et al. (2006). A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature 439, 1004–1008. doi: 10.1038/nature04407
Wahlgren, M., Goel, S., and Akhouri, R. R. (2017). Variant surface antigens of Plasmodium falciparum and their roles in severe malaria. Nat. Rev. Microbiol. 15, 479–491. doi: 10.1038/nrmicro.2017.47
Wahlin, J., and Cohn, M. (2002). Analysis of the RAP1 protein binding to homogeneous telomeric repeats in Saccharomyces castellii. Yeast 19, 241–256. doi: 10.1002/yea.816
Wang, R. C., Smogorzewska, A., and de Lange, T. (2004). Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119, 355–368. doi: 10.1016/j.cell.2004.10.011
Wellinger, R. J., Wolf, A. J., and Zakian, V. A. (1993). Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72, 51–60. doi: 10.1016/0092-8674(93)90049-v
Who. (2015). Investing to Overcome the Global Impact of Neglected Tropical Diseases. Third WHO Report on Neglected Tropical Diseases. Geneva: WHO.
Wotton, D., and Shore, D. (1997). Novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11, 748–760. doi: 10.1101/gad.11.6.748
Wu, L., Multani, A. S., He, H., Cosme-Blanco, W., Deng, Y., Deng, J. M., et al. (2006). Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 126, 49–62. doi: 10.1016/j.cell.2006.05.037
Yang, X., Figueiredo, L. M., Espinal, A., Okubo, E., and Li, B. (2009). RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell 137, 99–109. doi: 10.1016/j.cell.2009.01.037
Ye, J. Z., Hockemeyer, D., Krutchinsky, A. N., Loayza, D., Hooper, S. M., Chait, B. T., et al. (2004). POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 18, 1649–1654. doi: 10.1101/gad.1215404
Yehezkel, S., Segev, Y., Viegas-Péquignot, E., Skorecki, K., and Selig, S. (2008). Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum. Mol. Genet. 17, 2776–2789. doi: 10.1093/hmg/ddn177
Young, E., Pastor, S., Rajagopalan, R., McCaffrey, J., Sibert, J., Mak, A. C. Y., et al. (2017). High-throughput single-molecule mapping links subtelomeric variants and long-range haplotypes with specific telomeres. Nucleic Acids Res. 45:e73. doi: 10.1093/nar/gkx017
Yu, E. Y., Yen, W. F., Steinberg-Neifach, O., and Lue, N. F. (2010). Rap1 in Candida albicans: an unusual structural organization and a critical function in suppressing telomere recombination. Mol. Cell. Biol. 30, 1254–1268. doi: 10.1128/MCB.00986-09
Zhang, J. M., and Zou, L. (2020). Alternative lengthening of telomeres: from molecular mechanisms to therapeutic outlooks. Cell Biosci. 10:30. doi: 10.1186/s13578-020-00391-6
Zhang, J. R., Hardham, J. M., Barbour, A. G., and Norris, S. J. (1997). Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89, 275–285. doi: 10.1016/s0092-8674(00)80206-8
Zhang, X., Alexander, N., Leonardi, I., Mason, C., Kirkman, L. A., and Deitsch, K. W. (2019). Rapid antigen diversification through mitotic recombination in the human malaria parasite Plasmodium falciparum. PLoS Biol. 17:e3000271. doi: 10.1371/journal.pbio.3000271
Keywords: telomere, Trypanosoma brucei, genome stability, TRF, RAP1
Citation: Li B (2021) Keeping Balance Between Genetic Stability and Plasticity at the Telomere and Subtelomere of Trypanosoma brucei. Front. Cell Dev. Biol. 9:699639. doi: 10.3389/fcell.2021.699639
Received: 23 April 2021; Accepted: 08 June 2021;
Published: 05 July 2021.
Edited by:
Maria Isabel Nogueira Cano, São Paulo State University, BrazilReviewed by:
Miguel A. Chiurillo, University of Cincinnati, United StatesAmelie Fradet-Turcotte, Laval University, Canada
Catarina de Almeida Marques, University of Glasgow, United Kingdom
Copyright © 2021 Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bibo Li, Yi5saTM3QGNzdW9oaW8uZWR1
 Bibo Li
Bibo Li