
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 08 September 2021
Sec. Developmental Epigenetics
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.699621
This article is part of the Research TopicLegacies of Epigenetic PerturbationsView all 5 articles
Mitophagy is a specialized autophagic pathway responsible for the selective removal of damaged or dysfunctional mitochondria by targeting them to the autophagosome in order to maintain mitochondria quality. The role of mitophagy in tumorigenesis has been conflicting, with the process both supporting tumor cell survival and promoting cell death. Cancer cells may utilize the mitophagy pathway to augment their metabolic requirements and resistance to cell death, thereby leading to increased cell proliferation and invasiveness. This review highlights major regulatory pathways of mitophagy involved in cancer. In particular, we summarize recent progress regarding how nuclear-encoded long non-coding RNAs (lncRNAs) function as novel epigenetic players in the mitochondria of cancer cells, affecting the malignant behavior of tumors by regulating mitophagy. Finally, we discuss the potential application of regulating mitophagy as a new target for cancer therapy.
Mitochondria play a central role in cellular bioenergetics, regulating essential biochemical reactions that generate adenosine triphosphate (ATP) and reactive oxygen species (ROS; Friedman and Nunnari, 2014; Spinelli and Haigis, 2018; Missiroli et al., 2020). Abnormalities in respiratory chain structure or mutations of mitochondrial genome DNA can interfere with normal mitochondrial functions, leading to an imbalance in calcium ion homeostasis, ROS production, and apoptosis. Mitochondria quality control (MQC) is a critical mechanism to maintain mitochondrial health and mitochondrial homeostasis (Pickles et al., 2018), including mitochondria biogenesis, fission and fusion, and mitophagy (Wai and Langer, 2016). Mitophagy, a specialized form of autophagy that degrades dysfunctional mitochondria, serves as the primary mechanism to regulate mitochondrial functions, including energy metabolism, metabolic reprogramming, and mitochondria self-repair and renewal (Pickles et al., 2018; Roca-Portoles and Tait, 2021). Mitophagy is activated by hypoxia, metabolic stress, and mitochondrial depolarization through regulatory pathways involving PINK1/Parkin, BNIP3/NIX, and FUNDC1 (Poole and Macleod, 2021).
The concept of mitochondrial degradation was first proposed by Margaret and Warren Lewis as early as 1915 (Lewis and Lewis, 1915). Later studies using electron microscopy demonstrated the presence of mitochondria within lysosomes (Clark, 1957). However, this morphological observation was not followed up with functional experiments until the landmark studies of Okamoto et al. (2009), who identified ATG32 as a critical receptor of mitophagy in yeast, a finding which won the Nobel Prize in 2016. Since then, studies on mitophagy have dramatically increased (Figure 1; Montava-Garriga and Ganley, 2020).
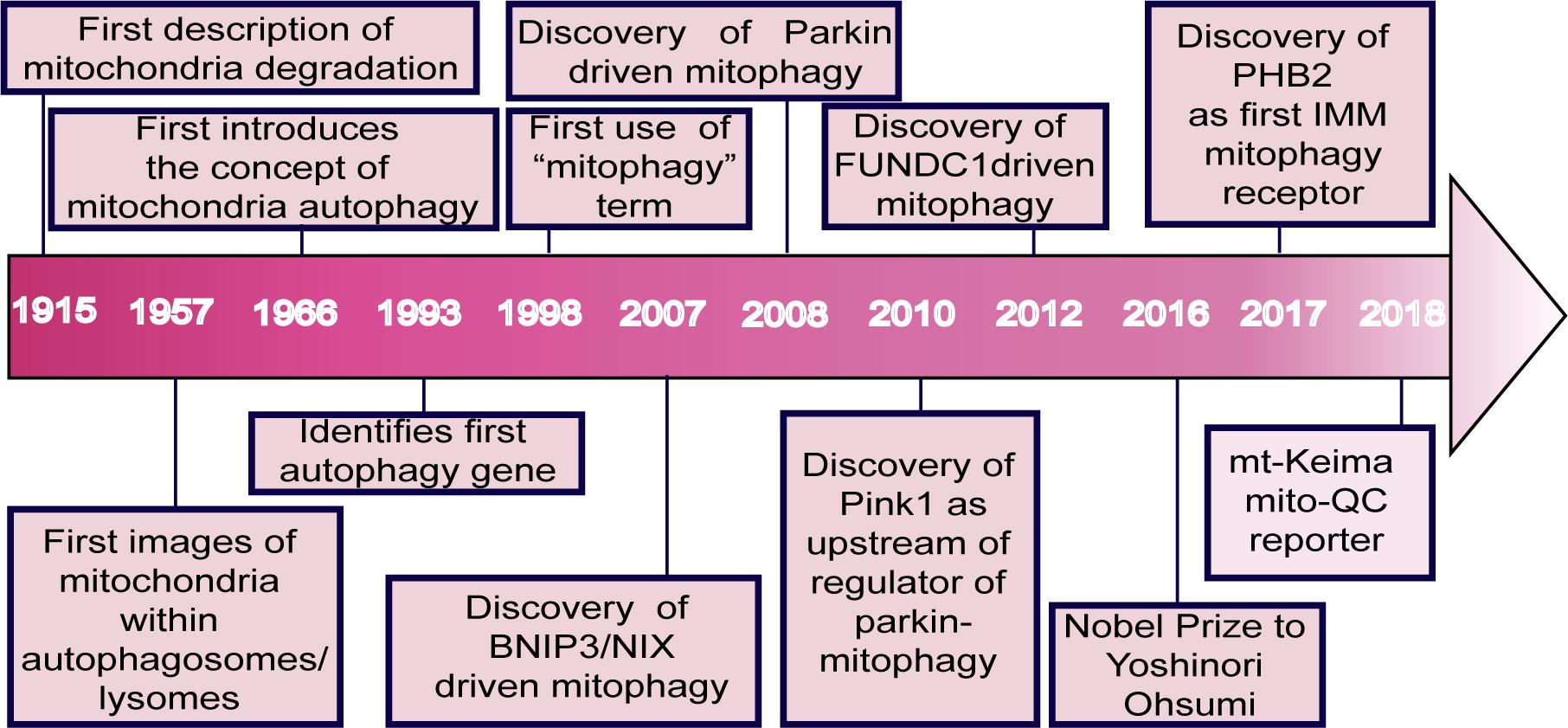
Figure 1. Major events in the field of mitophagy. The idea of mitochondrial degradation was first suggested by Lewis and Margaret in 1915. With the use of electron microscopy, the presence of mitochondria within vesicles and lysosomes in rat tissues was observed in 1957. de Duve postulated the concept of cellular autophagy in 1966. Ohsumi’s lab identified the first autophagy gene in 1993 (Tsukada and Ohsumi, 1993). In 1998, the term mitophagy was first used by Klionsky and Scott (Scott and Klionsky, 1998). BNIP3L was shown to play a vital role in mitophagy during mammalian erythrocyte differentiation in 2007 (Schweers et al., 2007). Parkin and PINK1 were characterized in 2008 (Narendra et al., 2008) and 2010 (Matsuda et al., 2010), respectively. Chen et al. discovered the mitophagy receptor FUNDC1 in 2012. Ohsumi won the Nobel Prize for his groundbreaking work in the field of autophagy in 2016. Wei et al. (2017) found the IMM protein, prohibitin 2 (PHB2), as a crucial mitophagy receptor in 2017. In 2018, Juliette et al. used a fluorescent pH-biosensor system to generate the mito-QC reporter for assessing basal mitophagy in vitro and in vivo (Lee J. J. et al., 2018). A fluorescent probe system called mt-Keima was also developed, which uses a pH-sensitive protein with a pH-dependent shift in fluorescence excitation to assess mitophagy in cells and tissues.
The role of mitophagy in tumorigenesis has recently been studied in detail. Since mitophagy leads to the degradation of dysfunctional mitochondria and decreased ROS production, mitophagy was initially thought to be related to tumor suppression. However, it has been shown that mitophagy actually plays a complicated role in tumor growth. Mitophagy is vital for metabolic remodeling within tumor cells and for modulating interactions among tumor cells, but its role is complex and depends on the type and stage of the tumor.
Recent studies have shown that lncRNAs, a class of long non-coding RNA (lncRNA) molecules with a length of >200 bp, have emerged as critical regulators in cancer. Many lncRNAs are transcribed from the human genome, and a number of them are involved in the regulation of mitophagy and mitophagy-associated drug resistance. LncRNAs can regulate various cellular functions, including mitochondrial metabolic reprogramming, to meet the needs of tumor metabolism. Therefore, it is important to understand the relationship between lncRNAs and mitophagy regulation in cancer. Exploiting the newly emerging knowledge of the lncRNA-mitophagy-cancer axis may provide novel targets for cancer therapy. In this review, we will summarize recent progress on the role of lncRNAs in malignancy, demonstrating the importance of mitochondria-associated lncRNAs in cancer metabolism, apoptosis, and mitophagy (Dong et al., 2017; Zhao et al., 2018; Anand and Pandi, 2021).
The main physiological function of mitophagy is to ensure the recycling of old and damaged organelles through continuous mitochondrial housekeeping (Palikaras et al., 2018). Little is known about steady-state mitophagy levels (also known as basal mitophagy) as mitophagy is typically examined under stress-induced conditions. Cells undergo basal mitophagy as part of routine mitochondrial maintenance (McWilliams et al., 2016). In tissues that consume large amounts of ATP, such as the brain, skeletal muscle, heart, liver and kidney, mitochondrial biogenesis is actively maintained to meet the cells’ metabolic requirements. When cells are switched from an energy-consuming state to a stable state, they tend to activate mitophagy in order to reduce mitochondrial mass as an adaptation to changes in cell metabolism. Proper homeostasis of mitochondrial mass is therefore critical to maintaining biological resilience (Onishi and Okamoto, 2021). Mitophagy is also induced during cell fate determination, such as erythrocyte differentiation (Schweers et al., 2007; Sandoval et al., 2008), cardiomyocyte maturation (Gong et al., 2015; Gottlieb and Bernstein, 2015; Kuznetsov et al., 2020; Morales et al., 2020), the developmental transitions of muscle tissue (Sin et al., 2016), and stem cell pluripotency (Vazquez-Martin et al., 2016; Xiang et al., 2017). In addition, mitophagy regulates the elimination of sperm mitochondria, thereby avoiding the inheritance of paternal mitochondrial DNA (mtDNA; Rojansky et al., 2016; Song et al., 2016). Since mitophagy plays a crucial role in maintaining mitochondrial homeostasis (Bin-Umer et al., 2014), it is not surprising that defective, inadequate, or excessive mitophagy can result in pathological conditions (Palikaras et al., 2017), such as neurodegenerative disease (Um and Yun, 2017; Tran and Reddy, 2020; Wang et al., 2021), cardiovascular disease (Billia et al., 2011; Zhang W. et al., 2016; Zhang et al., 2017; Chang et al., 2020), metabolic disorders (He et al., 2021; Pang et al., 2021; Zia et al., 2021), inflammation (Sliter et al., 2018; Lee et al., 2020), liver disease (Ke, 2020; Kouroumalis et al., 2021), aging (Richard et al., 2013; Cornelissen et al., 2018), and cancer.
Mitophagy is induced during stress to mediate metabolic adjustments to an external challenge. Mitophagy is highly regulated by various signaling pathways that can be roughly classified as ubiquitin (Ub)-dependent and Ub-independent mitophagy. The PINK1/Parkin pathway relies on a Ub-dependent mitophagy mechanism (Greene et al., 2012; Yamano and Youle, 2013; Harper et al., 2018). After PINK1-mediated phosphorylation, the E3 ligase PARKIN is translocated to mitochondria, where it induces the ubiquitination of the outer membrane of mitochondria. The mitophagy cargo adaptors p62, OPTN, and NDP52 bind to the phospho-Ub chains and interact with processed LC3 to target mitochondria for phagosome degradation (Figure 2A; Trempe et al., 2013; Kazlauskaite et al., 2014; Koyano et al., 2014). The inner mitochondrial membrane (IMM) protein PHB2 is a crucial mitophagy receptor that is required for Parkin-induced mitophagy (Wei et al., 2017; De Falco et al., 2020). BNIP3, BNIP3-like (BNIP3L) or NIX proteins and FUNDC1, on the other hand, function via Ub-independent mitophagy (Schweers et al., 2007; Liu et al., 2012; Wu et al., 2014). After hypoxia or other stress stimulation, BNIP3 and NIX can form stable homodimers in the outer mitochondrial membrane and recruit LC3 to induce mitophagy (Figure 2B; Rogov et al., 2017; Marinković et al., 2021; Springer et al., 2021). Both pathways cooperate to ensure efficient mitophagy (Lee et al., 2011; Zhang T. et al., 2016).
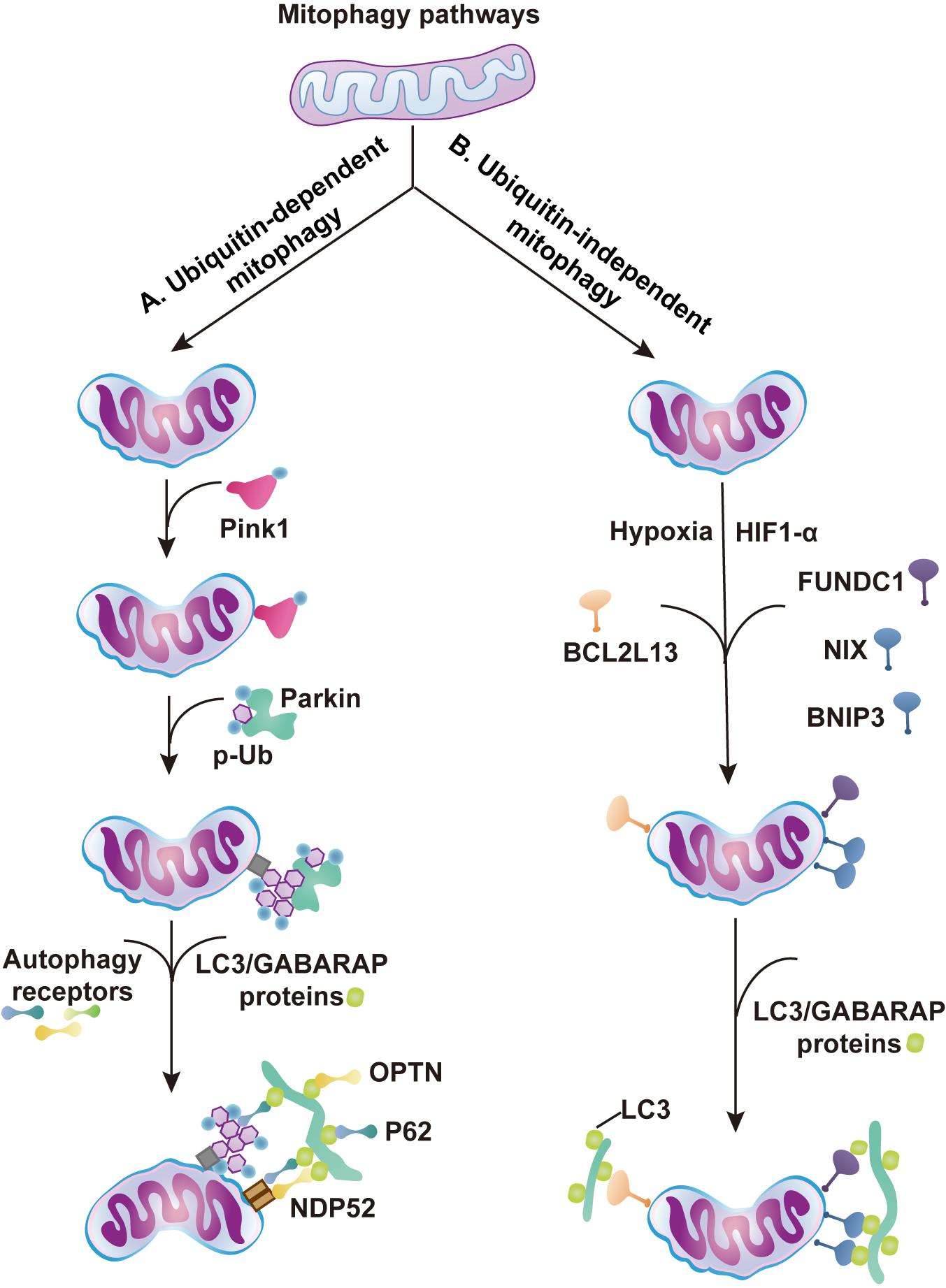
Figure 2. Major regulatory pathways of mitophagy. The schematic diagram summarizes the ubiquitin (Ub)-dependent and independent mitophagy pathways. (A) Ub-dependent mitophagy. (1) Aberrations in mitochondrial membrane potential leads to stabilization and activation of PINK1 on OMM. (2) The PINK1 kinase phosphorylates basal Ub and the cytosolic Ub ligase Parkin. Parkin is recruited to the mitochondria and activated by phospho-Ub and PINK1 phosphorylation. (3) Generation of Ub chains recruit mitophagy receptors, such as OPTN, NDP52, or p62. (4) Autophagosome machinery is recruited by mitophagy receptors. Mitophagy receptors then interact with LC3/GABARAP proteins coating the forming phagophore. (B) Ub-independent mitophagy. (1) Hypoxia stimulates HIF1α expression, and FUNDC1, NIX, and BNIP3 accumulate on the OMM and interact with LC3 to mediate mitophagy. (2) BCL2L13 interacts with LC3 proteins to mediate mitophagy (Murakawa et al., 2015).
The PINK1/Parkin pathway activates mitophagy in a Ub-dependent manner in cancer. As compared with non-transgenic controls, Parkin-knockout (KO) mice exhibited a significantly lower tumor burden of melanoma, including tumor size and lung metastases (Lee Y. S. et al., 2018). PINK1 was upregulated in esophageal squamous cell carcinoma (ESCC) tissues from patients who received chemotherapy (Yamashita et al., 2017). ESCC patients with high CD44 expression were more likely to have distant metastases and poor prognosis. Interestingly, mitophagy is increased in CD44 high-expressing cells (Whelan et al., 2017). Depletion of Parkin or inhibition of autophagy with chloroquine reduced CD44 high-expressing cells both in vitro and in vivo. Collectively, these studies suggest that PINK1/Parkin-dependent mitophagy may play an essential role in promoting the development of tumors.
BNIP3 plays a pro-tumorigenic role by driving aggressive features in melanoma cells, including migration, clonal growth, cell survival, and vasculogenic mimicry (Maes et al., 2014). Knockdown of baseline BNIP3 in melanoma cells caused accumulation of actin stress fibers and membrane ruffles and induced aberrations in cytoskeletal structures. Thus, BNIP3 maintains the plasticity of the actin cytoskeleton, accounting for cell migration and cancer progression. By analyzing immunohistochemical scores, Jiang et al. (2018) found that elevated expression of BNIP3 was found in 31.9% (15/47) of patients with uveal melanoma. High expression of BNIP3 was associated with deeper scleral invasion and lower cancer survival. These studies suggest that BNIP3/NIX-dependent mitophagy may play a pro-tumorigenic role in the development of cancer.
FUNDC1 also plays a critical role in hypoxia-induced mitophagy. Hui et al. (2019) found that FUNDC1 was upregulated in tumor tissues of laryngeal cancer patients in parallel with lipid peroxidation. Treatment of laryngeal cancer cells with low doses of hydrogen peroxide upregulated FUNDC1 through the ERK1/2 signal pathway and promoted proliferation of laryngeal cancer cells. Similarly, FUNDC1 was also upregulated in cervical cancer tissues, and the FUNDC1 expression score was an independent factor to determine overall and disease-free survival. Knockdown of FUNDC1 reduced the proliferation of cervical cancer cells and enhanced cell sensitivity to cisplatin and radiotherapy (Hou et al., 2017).
Recent studies have shown that mitophagy plays an important role in the metabolic transition of tumors and the maintenance of phenotype of cancer stem cells (CSCs). Continuous reprogramming of cellular metabolism is a critical milestone event in cancer (Pavlova and Thompson, 2016; Chen et al., 2020; Ohshima and Morii, 2021). Metabolic alterations lead to enhanced uptake and utilization of glucose and amino acid nutrients from a nutrient-deficient environment and increased use of glycolysis/TCA cycle intermediates for macromolecular biosynthesis. Cancer cell metabolism is governed by the Warburg effect: aerobic glycolysis with the increased rate of glycolysis and lactate production despite exposure to ambient oxygen. It is now clear that mitophagy is an important cellular mechanism that facilitates the metabolic switch to a glycolytic phenotype (Naik et al., 2019). Both canonical and non-canonical mitophagy contribute to alterations of bioenergetics and the metabolome, enhancing cellular developmental capability. Metabolic transformation of tumor cells, such as increased aerobic glycolysis or reduced oxidative phosphorylation, is controlled by altered expression and activity of several key enzymes involved in metabolism (Pavlova and Thompson, 2016). Mitophagy may play a critical role in this reprogramming by eliminating dysfunctional and damaged mitochondria. The turnover of mitochondria activates mitochondrial biogenesis to generate new mitochondrial mass that more effectively responds to nutrient stress (Drake et al., 2017). Hypoxia-inducible factor 1 (HIF-1) is a key factor that coordinates mitophagy and mitochondria turnover by regulating gene expression. In response to chronic hypoxia, HIF-1 initiates mitophagy by regulating the expression of mitophagy receptors BNIP3 and BNIP3L. The reduction of mitochondrial mass decreases total oxygen consumption of tumor cells and enhances cell survival under hypoxic stress (Vara-Perez et al., 2019; Figure 3).
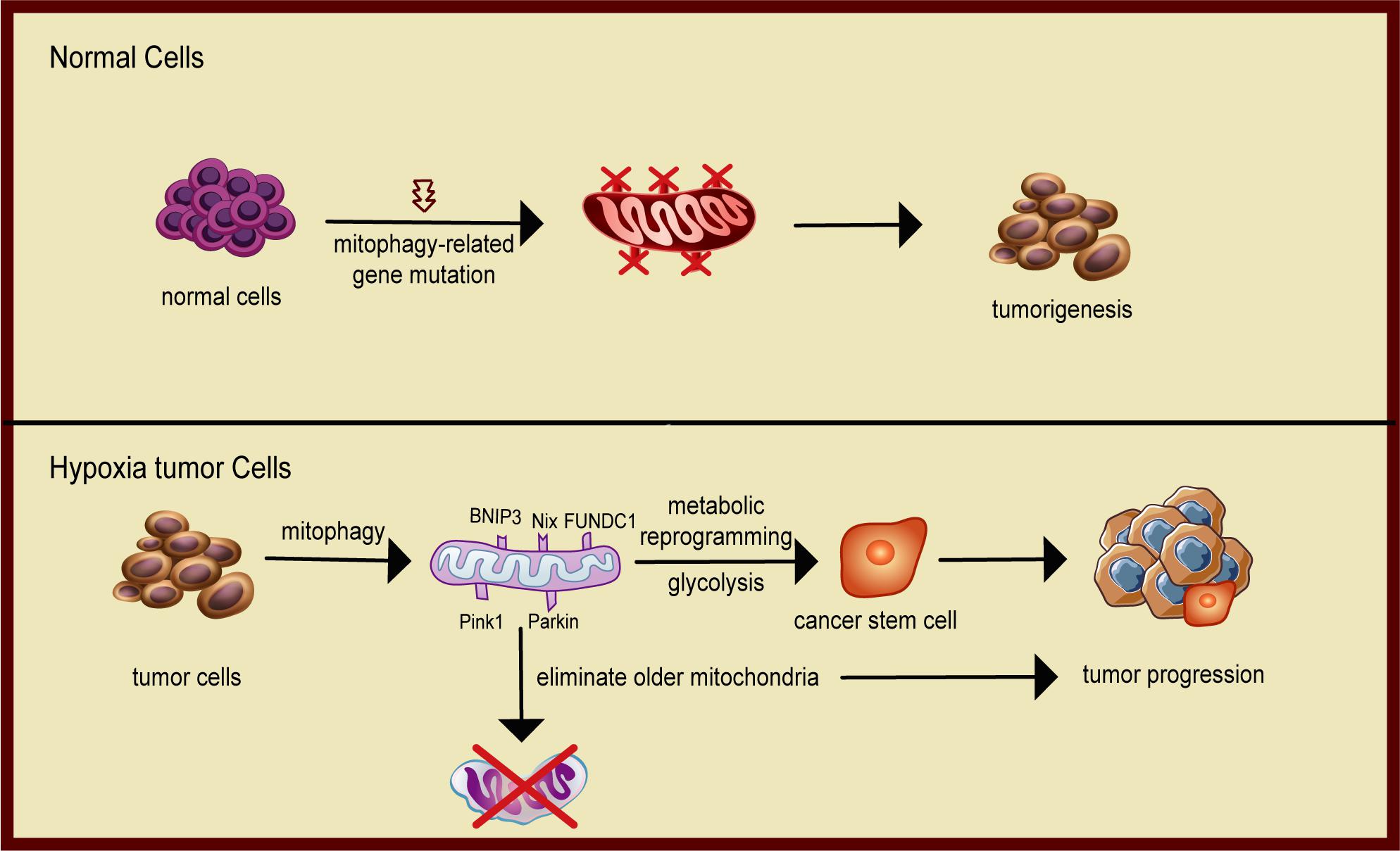
Figure 3. Schematic diagram of mitophagy promoting tumor progression. In normal cells, mitophagy eliminates damaged mitochondria and protects the normal metabolism of the cells. When mitophagy is inhibited, cells are prone to carcinogenesis. In solid tumors, cancer cells face metabolic stress from limited oxygen and nutrient availability. Mitophagy removes dysfunctional mitochondria that cannot metabolically handle the nutrient stress and maintain the cancer stem cell phenotype, hence facilitating tumor progression.
Mitophagy is also required for self-renewal and maintenance of stemness (Figure 3). CSCs exhibit enhanced potential for metastatic spread and contribute to the resistance to anticancer therapies. CSCs have unique metabolic signatures and heavily depend on the coordination of mitochondrial oxidative phosphorylation and mitophagy to maintain CSC phenotypes, including glycolytic reprogramming, epithelial-to-mesenchymal transition, survival after chemotherapeutic challenge, and increased formation of ROS (Sessions and Kashatus, 2021). Liu et al. (2017) reported that mitophagy was a positive regulator of hepatic CSCs. Active p53 binds to the promoter of NANOG and prevents OCT4 and SOX2 transcription factors from activating that stemness gene, leading to a reduction in hepatoma stem cells. When mitophagy is activated, p53 is phosphorylated at serine-392 by PINK1, a mitophagy-associated kinase, and is degraded by a mitophagy-dependent mechanism. Thus, mitophagy maintains hepatic CSCs by targeting the activities of tumor suppressor p53. Katajisto et al. (2015) found that mitophagy may play a critical role in regulating stemness by isolating and eliminating old mitochondria during asymmetric division of stem cells. Collectively, these studies demonstrate that mitophagy plays a positive role in the metabolic transition of tumors and the maintenance of phenotype of CSCs.
Mitophagy is a multi-pathway-regulated and multi-step process to degrade dysfunctional and damaged mitochondria. As a result, the role of mitophagy in tumor progression is very complex and depends on the type and stage of the tumor, as well as the specific mitophagy component molecules examined. Chourasia et al. (2015) reported that BNIP3 is most commonly deleted in patients with triple-negative breast cancer (TNBC). Similarly, BNIP3 functioned as a tumor suppressor in a MMTV-PyMT mouse mammary tumor model. Lack of Bnip3 caused an increment in tumor growth, invasiveness, and lung metastases. The role of BNIP3-dependent mitophagy, however, is controversial, partly depending on alternative pre-mRNA splicing of BNIP3. Normal cells express the full-length BNIP3, which promotes cell death. However, human adenocarcinoma cells preferentially express the truncated BNIP3 variant Bnip3Δex that lacks exon 3. The truncated Bnip3Δex3 isoform promotes tumor survival (Gang et al., 2015). It is also noteworthy that the move toward glycolytic metabolism upregulates the expression of the truncated Bnip3Δex3 isoform. Thus, BNIP3-mediated mitophagy may have a complex role in tumor growth and metastasis, depending on tumor type, alternative gene splicing regulation, and metabolic activity in cancer cells (Ferro et al., 2020).
Some studies suggested that PINK1 and PARK2 may function as tumor suppressors. Li C. et al. (2018) showed that expression of PARK2 was associated with improved survival in patients with pancreatic cancer. Using a mutant Kras-driven spontaneous pancreatic tumorigenesis model, they found that genetic deletion of Parkin or Pink1 promoted the development of pancreatic tumorigenesis. PARK2 is a component in the p53 tumor suppressor pathway. Lack of PARK2 induced the Warburg effect with increased glycolysis and ROS production, potentially enhancing tumorigenesis (Zhang et al., 2011). FUNDC1 is a mitophagy receptor that mediates mitophagy in response to hypoxia. Li W. et al. (2019) showed that FUNDC1 was upregulated in human hepatocellular carcinoma (HCC) tissues. Using a hepatocyte-specific Fundc1-knockout mouse model, they demonstrated that Fundc1 depletion enhanced the development and progression of HCC through the mitophagy-inflammasome pathway.
Overall, most mitophagy receptors or regulators are involved in cancer, but whether they function as tumor promoters or suppressors depends on tumor type and tumor microenvironment. Consistent with this dual role of mitophagy in tumors, mitophagy pathway proteins may be either overexpressed or downregulated in cancer (Table 1).
Mitochondria and the nucleus contain distinct genomes. Mitochondria participate in crucial cellular processes involved in energy harvesting, intermediate metabolism, and apoptosis. Consequently, mitochondria must communicate and coordinate precisely with the nuclear genome to ensure proper cellular function and energy homeostasis (Barcena et al., 2018; English et al., 2020). This mitochondrial-nuclear crosstalk is coordinated by anterograde (from the nucleus to mitochondria) and retrograde (from mitochondria to nucleus) signals, ensuring that cells maintain homeostasis under basal conditions while enabling adaptation to various stressors (Noh et al., 2016; Quiros et al., 2016; Singh et al., 2017). In response to mitochondrial stressors, retrograde signals induce specific nuclear expression of proteins that migrate into the mitochondria to resolve these perturbations.
Recent studies suggest that lncRNAs may function as novel retrograde and anterograde signal molecules in this mitochondria-nuclear crosstalk (Dong et al., 2017; Zhao et al., 2018). LncRNAs are encoded not only from the nuclear genome but also from the mitochondrial genome (mtDNA). LncRNAs encoded by the mitochondrial genome can be exported into the cytoplasm and nucleus, where they can have pleiotropic interactions. Similarly, lncRNAs synthesized from the nuclear genome can be shuttled into the mitochondria, where they can modulate gene transcription or mitochondrial metabolism (Rackham et al., 2011; Noh et al., 2016).
The mitochondrial genome codes for 13 mRNAs, 22 tRNAs, and 2 rRNAs, producing proteins that participate in oxidative phosphorylation. In addition, several mtDNA-encoded lncRNAs have recently been characterized, including lncND5, lncND6, and lncCyt B (Rackham et al., 2011), and their expression is cell- and tissue-specific, suggesting an important role in the regulation of mitochondrial gene expression. Together, at least 18 mitochondria-associated ncRNAs have been identified, and they play critical roles in manipulating mitochondrial functions, altering metabolic reprogramming, mitochondrial genome transcription, stress signal transmission, and mitochondria-associated apoptosis (Zhao et al., 2018).
Using RNA-FISH with MitoTracker staining, Zhao et al. (2019) examined the mitochondrial localization of lncRNAs, and found that some nuclear genome-transcribed lncRNAs, like the oncogenic lncRNA MALAT1, were aberrantly translocated to the mitochondria of HCC cells; in normal liver cells, this lncRNA was found predominantly in the nucleus. MALAT1 acts as an anterograde signal to regulate mitochondrial function in HepG2 cells. In contrast, the mitochondria-encoded lncRNA lncCytB acts as a retrograde signal in this mitochondria-nuclear crosstalk. This is in a sharp contrast to normal hepatic HL7702 cells, where lncCytB was primarily distributed in the mitochondria. Thus, aberrant shuttling of lncRNAs, whether nuclear-encoded or mitochondria-encoded, may be involved in the regulation of metabolic reprogramming in cancer cells.
The role of lncRNAs as crucial regulators of the Warburg effect in cancer has recently been investigated (Shankaraiah et al., 2018; Cruz-Gil et al., 2020; Liu et al., 2021). LncRNA EPB41L4A-AS1, a downstream target of p53, was downregulated in several human cancers, including breast cancer, and its downregulation was associated with poor survival. Depletion of EPB41L4A-AS1 expression induced the Warburg effect and increased aerobic glycolysis and glutamine metabolism through the HIF-1α pathway (Liao et al., 2019). FILNC1 (FoxO-induced lncRNA-1), an energy stress-induced lncRNA, was downregulated in renal cancer. Under glucose starvation, FILNC1-depleted cells exhibited enhanced glucose uptake and lactate production, probably through the c-Myc network (Xiao et al., 2017). LncRNA Ftx, derived from the X-inactivation center on the X chromosome, was overexpressed in HCC tissues, and its expression was associated with malignant clinicopathological characteristics in HCC patients. Ftx was shown to promote the Warburg effect and enhance tumor progression through the PPARγ pathway (Li X. et al., 2018). p21, a well-known p53-inducible lncRNA, is also a key regulator of the Warburg effect and is essential for hypoxia-induced glycolysis (Yang et al., 2014). While these latter two lncRNAs are not located in the mitochondria, we believe that a wide variety of mitochondrial-associated lncRNAs play important roles in controlling energy production as well as mtDNA, providing potent therapeutic targets and diagnostic markers for cancer.
In hepatoma cells, the nuclear genome-encoded lncRNA MALAT1 is abnormally transported into the mitochondria. Using an RNA reverse transcription-associated trap sequencing (RAT-seq) technology, Zhao et al. (2021) profiled the target binding sites of MALAT1 in the mitochondrial genome. They showed that mitochondrial MALAT1 lncRNA molecules were able to bind to multiple targets in mtDNA, including COX2, which encodes a subunit of the cytochrome c oxidase complex responsible for electron transfer in the mitochondrial respiratory chain. Depletion of the mitochondria-enriched MALAT1, either using shRNA or LwaCas13a-BN-MLS mitochondrial RNA targeting, altered mtDNA CpG methylation and induced multiple abnormalities in mitochondrial function and energy metabolism. Their findings suggest that lncRNA MALAT1 may function as a novel nucleus-mitochondria epigenetic messenger. By shuttling between the nucleus and mitochondria, it epigenetically regulates mitochondrial metabolism in hepatoma cells.
Hypoxia is common inducer of mitophagy, and CSCs can adapt to the hypoxic tumor microenvironment through mitophagy and maintain their stemness. In addition, CSCs can use mitophagy to maintain their own self-renewal ability. The expression of HIFs after hypoxia can directly or indirectly stimulate the maintenance of stemness markers, such as OCT4, SOX2, etc (Zaarour et al., 2021). LncRNA XIST can increase the sensitivity of breast CSCs to chemotherapy drugs by regulating KMT2C (Huang et al., 2020). In liver cancer, lncRNA HDAC2 promotes the proliferation and self-renewal of liver CSCs by activating the Hedgehog signaling pathway (Wu J. et al., 2019).
Exosomal lncRNAs are key factors that mediate extracellular communication in the tumor microenvironment. DOCK9-as2 is an exosomal lncRNA, and its down-regulation reduces the proliferation, migration, invasion, epithelial-mesenchymal (EMT), and stemness of papillary thyroid carcinoma cells (Dai et al., 2020). In both in vivo and in vitro studies, Gao et al. (2021) demonstrated that exosomal lncRNA UCA1 promoted self-renewal and differentiation of CD133 + cervical cancer cells through the micro-rRNA-122-5P/SOX2 axis. Silencing of UCA1 reduced cell proliferation and invasion. Similarly, depletion of lncRNA HotairM1 promotes self-renewal of CSCs through the HOXA1-Nanog loop (Li F. et al., 2020). These observations suggest that the crosstalk between hypoxia-driven HIFs, lncRNAs, and mitophagy plays a critical role in maintaining the stemness of CSCs.
LncRNAs are involved in the regulation of autophagy as well as autophagy-associated drug resistance. LncRNA HULC is highly upregulated in liver cancer and other malignancies. HULC overexpression is associated with poor overall survival and metastasis in cancers (Ding et al., 2019; Ghafouri-Fard et al., 2020). Knockdown of HULC suppresses autophagy and reduces cisplatin resistance by targeting downstream protein FoxM1 in drug-resistant gastric cancer cells (Xin et al., 2019). Another study reported that HULC triggered autophagy by stabilizing Sirt1, attenuating the sensitivity of HCC cells to chemotherapy (Xiong et al., 2017). Sun et al. (2017) found that lncRNA HOTAIR regulated cisplatin-resistance and autophagy by targeting Beclin-1, MDR, and P-GP in endometrial cancer cells. Finally, Liu C. et al. (2020) showed that lncRNA RMST was overexpressed in clinical glioma samples. RMST inhibited autophagy in glioma cells by inducing SUMO1 modification at K333 of FUS. SUMOylation of FUS promotes the degradation of ATG4D, a regulator of autophagy. More advances in the regulation of autophagy by lncRNAs in tumors are summarized in Table 2.
The crosstalk between autophagy and lncRNA may play a vital role in tumor progression. Using TCGA and CGGA databases, Xu et al. (Luan et al., 2019) analyzed 988 diffuse glioma patients and found that these patients were divided into two clusters with different prognostic outcomes based on the autophagy-related lncRNAs (ARLs) score. The ARLs signatures constructed by the authors showed good accuracy in predicting the prognosis of glioma patients. The ARL score was significantly elevated in the malignant subtype of glioma, and a high ARL score suggested a poor prognosis. A high ARL score indicated high infiltration of macrophages and neutrophils, thus serving as a promising prognostic biomarker for glioma patients. Using similar data analysis approaches, Li et al. (2021) found 11 ARLs in association with breast cancer prognosis. These 11 ARLs may become potential targets for autophagy-related targeted therapy.
The mechanism underlying the role of lncRNAs in the regulation of mitophagy remains unclear. Zhao et al. (2021) showed that the nuclear genome-encoded lncRNA MALAT1 acts as a new epigenetic messenger by shuttling to the mitochondria, where it regulates mitophagy (Figure 4). Knockdown of MALAT1 exhibited a significant decrease in mitophagy events. Mitophagy proteins, particularly PINK1, P62, NDP52, BNIP3, and LC3, were significantly downregulated in MALAT1-depleted HCC cells. This study proved for the first time that the lncRNA encoded by the nuclear genome might act as a new epigenetic player and regulate mitochondrial metabolism through mitophagy. In a second study, Xiang et al. (2021) observed the overexpression of PINT87, a peptide encoded by p53-induced transcript LINC-PINT, in senescent HCC cells. Overexpression of PINT87 inhibited mitophagy by directly binding to FOXM1 and blocking the transcription of PHB2, a crucial IMM receptor for Parkin-induced mitophagy.
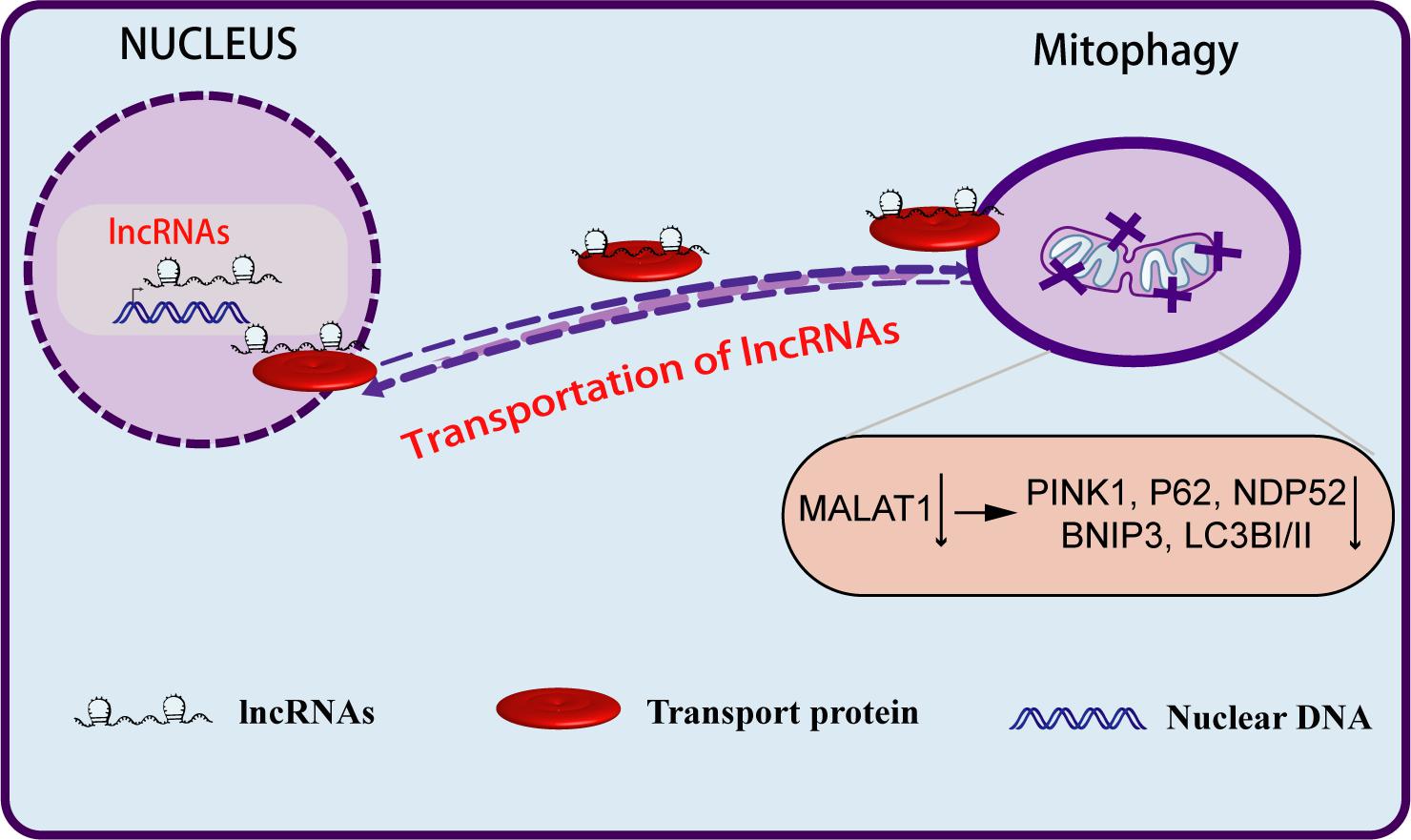
Figure 4. Long non-coding RNAs regulate the biological behavior of tumor cells through mitophagy. LncRNA MALAT1, after being transcribed from the nuclear genome, can be aberrantly transported to the mitochondria, where it epigenetically regulates mitochondrial functions and mitophagy. Depletion of MALAT1 downregulates mitophagy markers, particularly PINK1, P62, BNIP3, NDP52, and LC3.
Many anti-cancer drugs directly or indirectly cause damage to the mitochondria. Thus, the removal of dysfunctional mitochondria by mitophagy may alter their overall effectiveness (Bishop and Bradshaw, 2018). The induction of mitophagy during anti-tumor therapy may alter its cytotoxicity, leading to treatment resistance. ARIH1, an E3 Ub ligase belonging to the RING-between-RING (RBR) family, is overexpressed in many cancer cells. As a key regulator of mitophagy, ARIH1 activates the PINK1-dependent mitophagy that promotes the resistance to cisplatin chemotherapy (Villa et al., 2017). There is a tight regulatory crosstalk between apoptosis, mitophagy, endoplasmic reticulum stress, and mitochondrial dynamics/biogenesis in determining the fate of cells in chemotherapy (Abdrakhmanov et al., 2019). Stimulation of mitophagy with the protonophore carbonyl cyanide-m chlorophenylhydrazone (CCCP) inhibited the apoptosis induced by cisplatin, particularly at the high dose in HCT116 cells. On the other hand, inhibition of mitophagy enhanced the apoptotic response.
TMZ-POH, a novel conjugation analog of alkylating anticancer agent temozolomide, impaired mitophagy flux in non-small cell lung cancer (NSCLC) cells by inducing lysosomal dysfunction and hampering autophagosome-lysosome fusion. In radiotherapy, inhibition of mitophagy by TMZ-POH sensitizes cancer cells to irradiation-induced apoptosis (Chang et al., 2018). PINK1/Parkin-mediated mitophagy plays a critical role in hypoxia-induced radioresistance. Inhibition of this mitophagy pathway by a hypoxia-targeting p53 fusion protein, consisting of p53, TAT, and HIF-1α minimum oxygen-dependent degradation domain, sensitizes cancer cells in response to radiotherapy both in vitro and in vivo (Zheng et al., 2015). Collectively, these data support the pro-survival role of mitophagy in chemo/radiotherapy.
Cancer stem cells are known to be drug-resistant. To study the role of mitophagy in CSC-mediated drug resistance, Yan et al. (2017) isolated CSCs carrying the CD133+/CD44+ marker from human colorectal cancer cells that were resistant to doxorubicin. Treatment with doxorubicin significantly upregulated the BNIP3L mitophagy pathway. Inhibition of this mitophagy pathway significantly enhanced the sensitivity to doxorubicin in CSCs. Thus, mitophagy may contribute to drug resistance in CSCs.
To examine the mechanisms underlying chemoresistance and its correlation with stemness, Naik et al. (2018) established cisplatin−resistant oral squamous cell carcinoma cells with CSC-like features. They found that mitophagy flux was significantly higher in cisplatin-resistant oral cancer cells than that in their parental counterparts, suggesting that mitophagy is responsible for chemoresistance in oral cancer. Inhibition of autophagy effectively downregulated the stemness and inhibited chemoresistance.
Long non-coding RNAs may promote tumor cell survival and reduce sensitivity of cancer cells to chemo- and radiotherapy by regulating mitophagy. Thus, targeting lncRNAs may be an appropriate approach to render cancer cells sensitive to chemotherapy. Cai et al. (2019) found that lncRNA GBCDRlnc1 (gallbladder cancer drug resistance-associated lncRNA1), a key regulator of chemotherapy resistance, is upregulated in gallbladder cancer. Depletion of this lncRNA inhibited autophagy and enhanced the sensitivity of gallbladder cancer cells to doxorubicin. Their findings established that targeting the chemoresistant driver GBCDRlnc1 might be an attractive therapeutic approach for the treatment of advanced gallbladder cancer.
Long non-coding RNA H19 plays an important role in cell proliferation, metastasis, and chemotherapy resistance. Wang J. et al. (2019) showed that H19 was significantly upregulated in breast cancer cell lines and tumor tissues that were resistant to tamoxifen treatment. Silencing H19 significantly inhibited autophagy and sensitized tumor cells to tamoxifen in vitro and in vivo. In contrast, overexpression of H19 triggered autophagy in tamoxifen-sensitive cells and recapitulated the characteristics of tamoxifen resistance. Another study with lncRNA KCNQ1OT1 in colon cancer patients found that KCNQ1OT1 enhanced the chemotherapy resistance of colon cancer by activating autophagy through the miR-34a-Atg4B axis (Li Y. et al., 2019). Thus, targeting lncRNAs to regulate autophagy in tumors may be a promising therapeutic approach.
Disruption of the mitochondria-nuclear crosstalk network is a key event in many human diseases, including cancer (Mello et al., 2019). As critical regulatory components of this mitochondria-nuclear crosstalk, lncRNAs are also often dysregulated, including aberrant expression and shuttling between the nucleus and mitochondria. For example, the nuclear-encoded MALAT1 can be shuttled to mitochondria, where it regulates the mitochondrial functions and mitophagy (Zhao et al., 2021). Thus, targeting a component of the dysregulated lncRNA-mitochondria-nuclear network may provide an ideal method to combat some malignancies. Novel targets include the nuclear expression of the lncRNA, nuclear-mitochondria shuttling, lncRNA binding to mitochondria, and downstream signals of the mitophagy pathway.
Targeting mitophagy may present a new approach to develop anti-cancer therapies. However, currently we know very little about the precise mechanisms underlying the regulation of mitophagy in human tumors. Thus, it is critical to identify the role of mitophagy as well as the key regulatory components of each mitophagy pathway in tumor progression. More importantly, we must accurately define specific mitophagy regulators involved in radio- and chemoresistance in order to develop a precision medicine approach to target a specific component or a key regulator in mitophagy, rather than using inhibitors to target general mitophagy. A more comprehensive understanding of the key regulators of mitophagy in tumor progression may guide the development of novel therapeutics to treat some cancers (Bishop and Bradshaw, 2018). At the same time, it is also critical to understand the extent to which normal cells can tolerate these mitophagy inhibitors, so that drugs targeting mitophagy will not damage normal cells. More importantly, we should identify key regulatory factors that aberrantly regulate mitophagy in cancer cells to improve the precision of anti-tumor therapy.
The roles of mitochondria-associated lncRNAs in mitophagy are just beginning to be explored. As epigenetic regulatory factors, lncRNAs play important roles in intracellular environmental homeostasis, including mitophagy. Notably, some nucleus/mitochondria-shuttling lncRNAs are involved in the regulation of cancer metabolic reprogramming and the stemness maintenance of CSCs. After being transported to mitochondria, the nuclear-encoded lncRNAs not only epigenetically regulate mitochondrial metabolism, but also play a vital role in apoptosis and mitophagy. Therefore, precisely targeting mitophagy-related lncRNAs in cancer may become a very promising and attractive strategy for future tumor therapy with fewer toxic and side effects.
Since lncRNAs regulate mitophagy through multiple signaling pathways and mechanisms, more studies are needed before we target mitophagy-associated lncRNAs for precision medicine in cancer. For example, how are these nuclear lncRNAs transported into mitochondria? Can we interrupt this mitochondria-nuclear transportation to target mitophagy? What are the targets of these lncRNAs in mitochondria? How do these lncRNAs regulate mitophagy? A greater understanding of lncRNAs in this mitochondria-nuclear crosstalk network may help us develop novel lncRNA-based therapeutic approaches for malignancies that currently have few curative options.
YL wrote the manuscript. J-FH, JC, and WL supervised and funded the project. J-FH and AH edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key R&D Program of China (2018YFA0106902), the National Natural Science Foundation of China (82050003, 81900701, 31430021, 81874052, 81672275, 31871297, 81670143, 81900701, and 32000431), the Key Project of Chinese Ministry of Education grant (311015), the National Basic Research Program of China (973 Program; 2015CB943303), the Nation Key Research and Development Program of China grant (2016YFC13038000), Research on Chronic Non-communicable Diseases Prevention and Control of National Ministry of Science and Technology (2016YFC1303804), the National Health Development Planning Commission Major Disease Prevention and Control of Science and Technology Plan of Action, Cancer Prevention and Control (ZX-07-C2016004), the Natural Science Foundation of Jilin Province (20200801046GH, 20150101176JC, 20180101117JC, and 20130413010GH), the Provincial Science Fund of Jilin Province Development and Reform Commission (2014N147 and 2017C022), the 10th Youth Fund of First Hospital of Jilin University (JDYY 102019034), California Institute of Regenerative Medicine (CIRM) grant (RT2-01942), and the Biomedical Research Service of the Department of Veterans Affairs (BX002905).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdrakhmanov, A., Kulikov, A. V., Luchkina, E. A., Zhivotovsky, B., and Gogvadze, V. (2019). Involvement of mitophagy in cisplatin-induced cell death regulation. Biol. Chem. 400, 161–170. doi: 10.1515/hsz-2018-0210
Agnihotri, S., Golbourn, B., Huang, X., Remke, M., Younger, S., Cairns, R. A., et al. (2016). PINK1 is a negative regulator of growth and the Warburg effect in Glioblastoma. Cancer Res. 76, 4708–4719. doi: 10.1158/0008-5472.can-15-3079
Anand, A., and Pandi, G. (2021). Noncoding RNA: an insight into chloroplast and mitochondrial gene expressions. Life 11:49. doi: 10.3390/life11010049
Bacon, A. L., Fox, S., Turley, H., and Harris, A. L. (2007). Selective silencing of the hypoxia-inducible factor 1 target gene BNIP3 by histone deacetylation and methylation in colorectal cancer. Oncogene 26, 132–141. doi: 10.1038/sj.onc.1209761
Barcena, C., Mayoral, P., and Quiros, P. M. (2018). Mitohormesis, an Antiaging paradigm. Int. Rev. Cell Mol. Biol. 340, 35–77. doi: 10.1016/bs.ircmb.2018.05.002
Billia, F., Hauck, L., Konecny, F., Rao, V., Shen, J., and Mak, T. W. (2011). PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc. Natl. Acad. Sci. U.S.A. 108, 9572–9577. doi: 10.1073/pnas.1106291108
Bin-Umer, M. A., McLaughlin, J. E., Butterly, M. S., McCormick, S., and Tumer, N. E. (2014). Elimination of damaged mitochondria through mitophagy reduces mitochondrial oxidative stress and increases tolerance to trichothecenes. Proc. Natl. Acad. Sci. U.S.A. 111, 11798–11803. doi: 10.1073/pnas.1403145111
Bishop, E., and Bradshaw, T. D. (2018). Autophagy modulation: a prudent approach in cancer treatment? Cancer Chemother. Pharmacol. 82, 913–922. doi: 10.1007/s00280-018-3669-6
Cai, Q., Wang, S., Jin, L., Weng, M., Zhou, D., Wang, J., et al. (2019). Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol. Cancer 18:82.
Chang, M., Song, X., Geng, X., Wang, X., Wang, W., Chen, T. C., et al. (2018). Temozolomide-Perillyl alcohol conjugate impairs Mitophagy flux by inducing lysosomal dysfunction in non-small cell lung Cancer cells and sensitizes them to irradiation. J. Exp. Clin. Cancer Res. 37:250.
Chang, X., Zhang, W., Zhao, Z., Ma, C., Zhang, T., Meng, Q., et al. (2020). Regulation of mitochondrial quality control by natural drugs in the treatment of cardiovascular diseases: potential and advantages. Front. Cell Dev. Biol. 8:616139. doi: 10.3389/fcell.2020.616139
Chen, J. F., Wu, P., Xia, R., Yang, J., Huo, X. Y., Gu, D. Y., et al. (2018). STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol. Cancer 17:6.
Chen, X., Chen, S., and Yu, D. (2020). Metabolic reprogramming of chemoresistant cancer cells and the potential significance of metabolic regulation in the reversal of cancer chemoresistance. Metabolites 10:289. doi: 10.3390/metabo10070289
Chen, X., Gong, J., Zeng, H., Chen, N., Huang, R., Huang, Y., et al. (2010). MicroRNA145 targets BNIP3 and suppresses prostate cancer progression. Cancer Res. 70, 2728–2738. doi: 10.1158/0008-5472.can-09-3718
Chourasia, A. H., Tracy, K., Frankenberger, C., Boland, M. L., Sharifi, M. N., Drake, L. E., et al. (2015). Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 16, 1145–1163. doi: 10.15252/embr.201540759
Clark, S. L. Jr. (1957). Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. J. Biophys. Biochem. Cytol. 3, 349–362. doi: 10.1083/jcb.3.3.349
Cornelissen, T., Vilain, S., Vints, K., Gounko, N., Verstreken, P., and Vandenberghe, W. (2018). Deficiency of parkin and PINK1 impairs age-dependent mitophagy in Drosophila. eLife 7:e35878.
Cruz-Gil, S., Fernandez, L. P., Sanchez-Martinez, R., Gomez de Cedron, M., Ramirez, et al. (2020). Non-coding and regulatory RNAs as epigenetic remodelers of fatty acid homeostasis in cancer. Cancers 12:2890. doi: 10.3390/cancers12102890
Dai, W., Jin, X., Han, L., Huang, H., Ji, Z., Xu, X., et al. (2020). Exosomal lncRNA DOCK9-AS2 derived from cancer stem cell-like cells activated Wnt/β-catenin pathway to aggravate stemness, proliferation, migration, and invasion in papillary thyroid carcinoma. Cell Death Dis. 11:743.
D’Amico, A. G., Maugeri, G., Magro, G., Salvatorelli, L., Drago, F., and D’Agata, V. (2015). Expression pattern of parkin isoforms in lung adenocarcinomas. Tumour Biol. 36, 5133–5141. doi: 10.1007/s13277-015-3166-z
De Falco, F., Gentile, I., Cerino, P., Cutarelli, A., Catoi, C., and Roperto, S. (2020). Prohibitin 2 is involved in Parkin-mediated mitophagy in urothelial cells of cattle infected with bovine papillomavirus. Pathogens 9:621. doi: 10.3390/pathogens9080621
Ding, Y., Sun, C., Li, J., Hu, L., Li, M., Liu, J., et al. (2019). The significance of long non-coding RNA HULC in predicting prognosis and metastasis of cancers: a meta-analysis. Pathol. Oncol. Res. 25, 311–318. doi: 10.1007/s12253-017-0351-y
Dong, Y., Yoshitomi, T., Hu, J. F., and Cui, J. (2017). Long noncoding RNAs coordinate functions between mitochondria and the nucleus. Epigenetics Chromatin 10:41.
Drake, L. E., Springer, M. Z., Poole, L. P., Kim, C. J., and Macleod, K. F. (2017). Expanding perspectives on the significance of mitophagy in cancer. Semin. Cancer Biol. 47, 110–124. doi: 10.1016/j.semcancer.2017.04.008
English, J., Son, J. M., Cardamone, M. D., Lee, C., and Perissi, V. (2020). Decoding the rosetta stone of mitonuclear communication. Pharmacol. Res. 161:105161. doi: 10.1016/j.phrs.2020.105161
Ferro, F., Servais, S., Besson, P., Roger, S., Dumas, J. F., and Brisson, L. (2020). Autophagy and mitophagy in cancer metabolic remodelling. Semin. Cell Dev. Biol. 98, 129–138. doi: 10.1016/j.semcdb.2019.05.029
Friedman, J. R., and Nunnari, J. (2014). Mitochondrial form and function. Nature 505, 335–343. doi: 10.1038/nature12985
Gang, H., Dhingra, R., Lin, J., Hai, Y., Aviv, Y., Margulets, V., et al. (2015). PDK2-mediated alternative splicing switches Bnip3 from cell death to cell survival. J. Cell Biol. 210, 1101–1115. doi: 10.1083/jcb.201504047
Gao, Z., Wang, Q., Ji, M., Guo, X., Li, L., and Su, X. (2021). Exosomal lncRNA UCA1 modulates cervical cancer stem cell self-renewal and differentiation through microRNA-122-5p/SOX2 axis. J. Transl. Med. 19:229.
Ghafouri-Fard, S., Esmaeili, M., Taheri, M., and Samsami, M. (2020). Highly upregulated in liver cancer (HULC): an update on its role in carcinogenesis. J. Cell. Physiol. 235, 9071–9079. doi: 10.1002/jcp.29765
Gong, G., Song, M., Csordas, G., Kelly, D. P., Matkovich, S. J., and Dorn, G. W. II (2015). Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science 350:aad2459. doi: 10.1126/science.aad2459
Gottlieb, R. A., and Bernstein, D. (2015). METABOLISM, Mitochondria shape cardiac metabolism. Science 350, 1162–1163. doi: 10.1126/science.aad8222
Greene, A. W., Grenier, K., Aguileta, M. A., Muise, S., Farazifard, R., Haque, M. E., et al. (2012). Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 13, 378–385. doi: 10.1038/embor.2012.14
Guo, J., Ma, Y., Peng, X., Jin, H., and Liu, J. (2019). LncRNA CCAT1 promotes autophagy via regulating ATG7 by sponging miR-181 in hepatocellular carcinoma. J. Cell. Biochem. 120, 17975–17983. doi: 10.1002/jcb.29064
Harper, J. W., Ordureau, A., and Heo, J. M. (2018). Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Biol. 19, 93–108. doi: 10.1038/nrm.2017.129
He, F., Huang, Y., Song, Z., Zhou, H. J., Zhang, H., Perry, R. J., et al. (2021). Mitophagy-mediated adipose inflammation contributes to type 2 diabetes with hepatic insulin resistance. J. Exp. Med. 218:e20201416.
Hou, H., Er, P., Cheng, J., Chen, X., Ding, X., Wang, Y., et al. (2017). High expression of FUNDC1 predicts poor prognostic outcomes and is a promising target to improve chemoradiotherapy effects in patients with cervical cancer. Cancer Med. 6, 1871–1881. doi: 10.1002/cam4.1112
Huang, F., Chen, W., Peng, J., Li, Y., Zhuang, Y., Zhu, Z., et al. (2018). LncRNA PVT1 triggers Cyto-protective autophagy and promotes pancreatic ductal adenocarcinoma development via the miR-20a-5p/ULK1 Axis. Mol. Cancer 17:98.
Huang, R., Zhu, L., and Zhang, Y. (2020). XIST lost induces ovarian cancer stem cells to acquire taxol resistance via a KMT2C-dependent way. Cancer Cell Int. 20:436.
Hui, L., Wu, H., Wang, T. W., Yang, N., Guo, X., and Jang, X. J. (2019). Hydrogen peroxide-induced mitophagy contributes to laryngeal cancer cells survival via the upregulation of FUNDC1. Clin. Transl. Oncol. 21, 596–606. doi: 10.1007/s12094-018-1958-5
Humpton, T. J., Alagesan, B., DeNicola, G. M., Lu, D., Yordanov, G. N., Leonhardt, C. S., et al. (2019). Oncogenic KRAS induces NIX-mediated mitophagy to promote pancreatic cancer. Cancer Discov. 9, 1268–1287. doi: 10.1158/2159-8290.cd-18-1409
Jiang, Z., Yu, F., and Li, M. (2018). Upregulation of BCL2 19 kD Protein-Interacting Protein 3 (BNIP3) is Predictive of Unfavorable Prognosis in Uveal Melanoma. Med. Sci. Monit. 24, 4711–4717. doi: 10.12659/msm.907679
Katajisto, P., Döhla, J., Chaffer, C. L., Pentinmikko, N., Marjanovic, N., Iqbal, S., et al. (2015). Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science 348, 340–343. doi: 10.1126/science.1260384
Kazlauskaite, A., Kondapalli, C., Gourlay, R., Campbell, D. G., Ritorto, M. S., Hofmann, K., et al. (2014). Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, 127–139. doi: 10.1042/bj20140334
Ke, P. Y. (2020). Mitophagy in the pathogenesis of liver diseases. Cells 9:831. doi: 10.3390/cells9040831
Kouroumalis, E., Voumvouraki, A., Augoustaki, A., and Samonakis, D. N. (2021). Autophagy in liver diseases. World J. Hepatol. 13, 6–65.
Koyano, F., Okatsu, K., Kosako, H., Tamura, Y., Go, E., Kimura, M., et al. (2014). Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166. doi: 10.1038/nature13392
Kuznetsov, A. V., Javadov, S., Grimm, M., Margreiter, R., Ausserlechner, M. J., and Hagenbuchner, J. (2020). Crosstalk between mitochondria and cytoskeleton in cardiac cells. Cells 9:222. doi: 10.3390/cells9010222
Lee, J. J., Andreazza, S., and Whitworth, A. J. (2020). The STING pathway does not contribute to behavioural or mitochondrial phenotypes in Drosophila Pink1/parkin or mtDNA mutator models. Sci. Rep. 10:2693.
Lee, J. J., Sanchez-Martinez, A., Martinez Zarate, A., Benincá, C., Mayor, U., Clague, M. J., et al. (2018). Basal mitophagy is widespread in Drosophila but minimally affected by loss of Pink1 or parkin. J. Cell Biol. 217, 1613–1622. doi: 10.1083/jcb.201801044
Lee, Y., Lee, H. Y., Hanna, R. A., and Gustafsson, Å. B. (2011). Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 301, H1924–H1931.
Lee, Y. S., Jung, Y. Y., Park, M. H., Yeo, I. J., Im, H. S., Nam, K. T., et al. (2018). Deficiency of parkin suppresses melanoma tumor development and metastasis through inhibition of MFN2 ubiquitination. Cancer Lett. 433, 156–164. doi: 10.1016/j.canlet.2018.07.007
Lewis, M. R., and Lewis, W. H. (1915). Mitochondria (and other cytoplasmic structures) in tissue cultures. Dev. Dyn. 17, 339–401. doi: 10.1002/aja.1000170304
Li, C., Zhang, Y., Cheng, X., Yuan, H., Zhu, S., Liu, J., et al. (2018). PINK1 and PARK2 suppress pancreatic tumorigenesis through control of mitochondrial iron-mediated immunometabolism. Dev. Cell 46, 441–455.e8.
Li, F., Xu, Y., Xu, X., Ge, S., Zhang, F., Zhang, H., et al. (2020). lncRNA HotairM1 depletion promotes self-renewal of cancer stem cells through HOXA1-nanog regulation loop. Mol. Ther. Nucleic Acids 22, 456–470. doi: 10.1016/j.omtn.2020.09.008
Li, J., Zhai, D. S., Huang, Q., Chen, H. L., Zhang, Z., and Tan, Q. F. (2019). LncRNA DCST1-AS1 accelerates the proliferation, metastasis and autophagy of hepatocellular carcinoma cell by AKT/mTOR signaling pathways. Eur. Rev. Med. Pharmacol. Sci. 23, 6091–6104.
Li, P., He, J., Yang, Z., Ge, S., Zhang, H., Zhong, Q., et al. (2020). ZNNT1 long noncoding RNA induces autophagy to inhibit tumorigenesis of uveal melanoma by regulating key autophagy gene expression. Autophagy 16, 1186–1199. doi: 10.1080/15548627.2019.1659614
Li, W., Li, Y., Siraj, S., Jin, H., Fan, Y., Yang, X., et al. (2019). FUN14 Domain-Containing 1-mediated mitophagy suppresses hepatocarcinogenesis by inhibition of inflammasome activation in mice. Hepatology 69, 604–621. doi: 10.1002/hep.30191
Li, X., Jin, F., and Li, Y. (2021). A novel autophagy-related lncRNA prognostic risk model for breast cancer. J. Cell. Mol. Med. 25, 4–14. doi: 10.1111/jcmm.15980
Li, X., Zhao, Q., Qi, J., Wang, W., Zhang, D., Li, Z., et al. (2018). lncRNA Ftx promotes aerobic glycolysis and tumor progression through the PPARgamma pathway in hepatocellular carcinoma. Int. J. Oncol. 53, 551–566.
Li, X., Zhou, Y., Yang, L., Ma, Y., Peng, X., Yang, S., et al. (2020). LncRNA NEAT1 promotes autophagy via regulating miR-204/ATG3 and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J. Cell. Physiol. 235, 3402–3413. doi: 10.1002/jcp.29230
Li, Y., Li, C., Li, D., Yang, L., Jin, J., and Zhang, B. (2019). lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in colon cancer by targeting the miR-34a/ATG4B pathway. Oncotargets Ther. 12, 2649–2660. doi: 10.2147/ott.s188054
Liao, M., Liao, W., Xu, N., Li, B., Liu, F., Zhang, S., et al. (2019). LncRNA EPB41L4A-AS1 regulates glycolysis and glutaminolysis by mediating nucleolar translocation of HDAC2. EBioMedicine 41, 200–213. doi: 10.1016/j.ebiom.2019.01.035
Liu, C., Peng, Z., Li, P., Fu, H., Feng, J., Zhang, Y., et al. (2020). lncRNA RMST Suppressed GBM Cell Mitophagy through Enhancing FUS SUMOylation. Mol. Ther. Nucleic Acids 19, 1198–1208. doi: 10.1016/j.omtn.2020.01.008
Liu, F., Ai, F. Y., Zhang, D. C., Tian, L., Yang, Z. Y., and Liu, S. J. (2020). LncRNA NEAT1 knockdown attenuates autophagy to elevate 5-FU sensitivity in colorectal cancer via targeting miR-34a. Cancer Med. 9, 1079–1091. doi: 10.1002/cam4.2746
Liu, K., Lee, J., Kim, J. Y., Wang, L., Tian, Y., Chan, S. T., et al. (2017). Mitophagy controls the activities of tumor suppressor p53 to regulate hepatic cancer stem cells. Mol. Cell 68:281. doi: 10.1016/j.molcel.2017.09.022
Liu, L., Feng, D., Chen, G., Chen, M., Zheng, Q., Song, P., et al. (2012). Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 14, 177–185. doi: 10.1038/ncb2422
Liu, L., Zuo, Z., Lu, S., Wang, L., Liu, A., and Liu, X. (2018). Silencing of PINK1 represses cell growth, migration and induces apoptosis of lung cancer cells. Biomed. Pharmacother. 106, 333–341. doi: 10.1016/j.biopha.2018.06.128
Liu, R., Wang, X., Shen, Y., and He, A. (2021). Long non-coding RNA-based glycolysis-targeted cancer therapy: feasibility, progression and limitations. Mol. Biol. Rep. 48, 2713–2727. doi: 10.1007/s11033-021-06247-7
Luan, F., Chen, W., Chen, M., Yan, J., Chen, H., Yu, H., et al. (2019). An autophagy-related long non-coding RNA signature for glioma. FEBS Open Bio 9, 653–667.
Macher-Goeppinger, S., Keith, M., Hatiboglu, G., Hohenfellner, M., Schirmacher, P., Roth, W., et al. (2017). Expression and functional characterization of the BNIP3 protein in renal cell carcinomas. Transl. Oncol. 10, 869–875. doi: 10.1016/j.tranon.2017.08.008
Maes, H., Van Eygen, S., Krysko, D. V., Vandenabeele, P., Nys, K., Rillaerts, K., et al. (2014). BNIP3 supports melanoma cell migration and vasculogenic mimicry by orchestrating the actin cytoskeleton. Cell Death Dis. 5:e1127. doi: 10.1038/cddis.2014.94
Marinković, M., Šprung, M., and Novak, I. (2021). Dimerization of mitophagy receptor BNIP3L/NIX is essential for recruitment of autophagic machinery. Autophagy 17, 1232–1243. doi: 10.1080/15548627.2020.1755120
Matsuda, N., Sato, S., Shiba, K., Okatsu, K., Saisho, K., Gautier, C. A., et al. (2010). PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221. doi: 10.1083/jcb.200910140
Maugeri, G., D’Amico, A. G., Magro, G., Salvatorelli, L., Barbagallo, G. M., Saccone, S., et al. (2015). Expression profile of parkin isoforms in human gliomas. Int. J. Oncol. 47, 1282–1292. doi: 10.3892/ijo.2015.3105
McWilliams, T. G., Prescott, A. R., Allen, G. F., Tamjar, J., Munson, M. J., Thomson, C., et al. (2016). mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 214, 333–345. doi: 10.1083/jcb.201603039
Mello, T., Simeone, I., and Galli, A. (2019). Mito-nuclear communication in hepatocellular carcinoma metabolic rewiring. Cells 8:417. doi: 10.3390/cells8050417
Missiroli, S., Perrone, M., Genovese, I., Pinton, P., and Giorgi, C. (2020). Cancer metabolism and mitochondria: finding novel mechanisms to fight tumours. EBioMedicine 59:102943. doi: 10.1016/j.ebiom.2020.102943
Montava-Garriga, L., and Ganley, I. G. (2020). Outstanding questions in mitophagy: What we do and do not know. J. Mol. Biol. 432, 206–230. doi: 10.1016/j.jmb.2019.06.032
Morales, P. E., Arias-Durán, C., Ávalos-Guajardo, Y., Aedo, G., Verdejo, H. E., Parra, V., et al. (2020). Emerging role of mitophagy in cardiovascular physiology and pathology. Mol. Aspects Med. 71:100822. doi: 10.1016/j.mam.2019.09.006
Murakawa, T., Yamaguchi, O., Hashimoto, A., Hikoso, S., Takeda, T., Oka, T., et al. (2015). Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat. Commun. 6:7527.
Naik, P. P., Birbrair, A., and Bhutia, S. K. (2019). Mitophagy-driven metabolic switch reprograms stem cell fate. Cell. Mol. Life Sci. 76, 27–43. doi: 10.1007/s00018-018-2922-9
Naik, P. P., Mukhopadhyay, S., Panda, P. K., Sinha, N., Das, C. K., Mishra, R., et al. (2018). Autophagy regulates cisplatin-induced stemness and chemoresistance via the upregulation of CD44, ABCB1 and ADAM17 in oral squamous cell carcinoma. Cell Prolif. 51:e12411.
Narendra, D., Tanaka, A., Suen, D. F., and Youle, R. J. (2008). Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803. doi: 10.1083/jcb.200809125
Noh, J. H., Kim, K. M., Abdelmohsen, K., Yoon, J. H., Panda, A. C., Munk, R., et al. (2016). HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev. 30, 1224–1239.
Ohshima, K., and Morii, E. (2021). Metabolic reprogramming of cancer cells during tumor progression and metastasis. Metabolites 11:28. doi: 10.3390/metabo11010028
Okamoto, K., Kondo-Okamoto, N., and Ohsumi, Y. (2009). Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17, 87–97. doi: 10.1016/j.devcel.2009.06.013
Onishi, M., and Okamoto, K. (2021). Mitochondrial clearance: mechanisms and roles in cellular fitness. FEBS Lett. 595, 1239–1263. doi: 10.1002/1873-3468.14060
Palikaras, K., Daskalaki, I., Markaki, M., and Tavernarakis, N. (2017). Mitophagy and age-related pathologies: development of new therapeutics by targeting mitochondrial turnover. Pharmacol. Ther. 178, 157–174. doi: 10.1016/j.pharmthera.2017.04.005
Palikaras, K., Lionaki, E., and Tavernarakis, N. (2018). Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 20, 1013–1022. doi: 10.1038/s41556-018-0176-2
Pang, B. P. S., Chan, W. S., and Chan, C. B. (2021). Mitochondria Homeostasis and Oxidant/Antioxidant Balance in Skeletal Muscle-Do Myokines Play a Role? Antioxidants 10:179. doi: 10.3390/antiox10020179
Pavlova, N. N., and Thompson, C. B. (2016). The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47. doi: 10.1016/j.cmet.2015.12.006
Pickles, S., Vigié, P., and Youle, R. J. (2018). Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170–R185.
Poole, L. P., and Macleod, K. F. (2021). Mitophagy in tumorigenesis and metastasis. Cell. Mol. Life Sci. 78, 3817–3851. doi: 10.1007/s00018-021-03774-1
Poulogiannis, G., McIntyre, R. E., Dimitriadi, M., Apps, J. R., Wilson, C. H., Ichimura, K., et al. (2010). PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc. Natl. Acad. Sci. U.S.A. 107, 15145–15150. doi: 10.1073/pnas.1009941107
Quiros, P. M., Mottis, A., and Auwerx, J. (2016). Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 17, 213–226. doi: 10.1038/nrm.2016.23
Rackham, O., Shearwood, A. M., Mercer, T. R., Davies, S. M., Mattick, J. S., and Filipovska, A. (2011). Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 17, 2085–2093. doi: 10.1261/rna.029405.111
Richard, V. R., Leonov, A., Beach, A., Burstein, M. T., Koupaki, O., Gomez-Perez, A., et al. (2013). Macromitophagy is a longevity assurance process that in chronologically aging yeast limited in calorie supply sustains functional mitochondria and maintains cellular lipid homeostasis. Aging 5, 234–269. doi: 10.18632/aging.100547
Roca-Portoles, A., and Tait, S. W. G. (2021). Mitochondrial quality control: from molecule to organelle. Cell. Mol. Life Sci. 78, 3853–3866. doi: 10.1007/s00018-021-03775-0
Rogov, V. V., Suzuki, H., Marinković, M., Lang, V., Kato, R., Kawasaki, M., et al. (2017). Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins. Sci. Rep. 7:1131.
Rojansky, R., Cha, M. Y., and Chan, D. C. (2016). Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. eLife 5:e17896.
Sandoval, H., Thiagarajan, P., Dasgupta, S. K., Schumacher, A., Prchal, J. T., Chen, M., et al. (2008). Essential role for Nix in autophagic maturation of erythroid cells. Nature 454, 232–235. doi: 10.1038/nature07006
Schweers, R. L., Zhang, J., Randall, M. S., Loyd, M. R., Li, W., Dorsey, F. C., et al. (2007). NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U.S.A. 104, 19500–19505. doi: 10.1073/pnas.0708818104
Scott, S. V., and Klionsky, D. J. (1998). Delivery of proteins and organelles to the vacuole from the cytoplasm. Curr. Opin. Cell Biol. 10, 523–529. doi: 10.1016/s0955-0674(98)80068-9
Sessions, D. T., and Kashatus, D. F. (2021). Mitochondrial dynamics in cancer stem cells. Cell. Mol. Life Sci. 78, 3803–3816.
Shankaraiah, R. C., Veronese, A., Sabbioni, S., and Negrini, M. (2018). Non-coding RNAs in the reprogramming of glucose metabolism in cancer. Cancer Lett. 419, 167–174. doi: 10.1016/j.canlet.2018.01.048
Sheng, J. Q., Wang, M. R., Fang, D., Liu, L., Huang, W. J., Tian, D. A., et al. (2021). LncRNA NBR2 inhibits tumorigenesis by regulating autophagy in hepatocellular carcinoma. Biomed. Pharmacother. 133:111023. doi: 10.1016/j.biopha.2020.111023
Sin, J., Andres, A. M., Taylor, D. J., Weston, T., Hiraumi, Y., Stotland, A., et al. (2016). Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy 12, 369–380. doi: 10.1080/15548627.2015.1115172
Singh, B., Modica-Napolitano, J. S., and Singh, K. K. (2017). Defining the momiome: promiscuous information transfer by mobile mitochondria and the mitochondrial genome. Semin. Cancer Biol. 47, 1–17. doi: 10.1016/j.semcancer.2017.05.004
Sliter, D. A., Martinez, J., Hao, L., Chen, X., Sun, N., Fischer, T. D., et al. (2018). Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262. doi: 10.1038/s41586-018-0448-9
Song, W. H., Yi, Y. J., Sutovsky, M., Meyers, S., and Sutovsky, P. (2016). Autophagy and ubiquitin-proteasome system contribute to sperm mitophagy after mammalian fertilization. Proc. Natl. Acad. Sci. U.S.A. 113, E5261–E5270.
Spinelli, J. B., and Haigis, M. C. (2018). The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 20, 745–754. doi: 10.1038/s41556-018-0124-1
Springer, M. Z., Poole, L. P., Drake, L. E., Bock-Hughes, A., Boland, M. L., Smith, A. G., et al. (2021). BNIP3-dependent mitophagy promotes cytosolic localization of LC3B and metabolic homeostasis in the liver. Autophagy. doi: 10.1080/15548627.2021.1877469 [Epub ahead of print].
Sun, J. L., He, X. S., Yu, Y. H., and Chen, Z. C. (2004). [Expression and structure of BNIP3L in lung cancer]. Chin. J. Cancer 23, 8–14.
Sun, M. Y., Zhu, J. Y., Zhang, C. Y., Zhang, M., Song, Y. N., Rahman, K., et al. (2017). Autophagy regulated by lncRNA HOTAIR contributes to the cisplatin-induced resistance in endometrial cancer cells. Biotechnol. Lett. 39, 1477–1484. doi: 10.1007/s10529-017-2392-4
Tay, S. P., Yeo, C. W., Chai, C., Chua, P. J., Tan, H. M., Ang, A. X., et al. (2010). Parkin enhances the expression of cyclin-dependent kinase 6 and negatively regulates the proliferation of breast cancer cells. J. Biol. Chem. 285, 29231–29238. doi: 10.1074/jbc.m110.108241
Tran, M., and Reddy, P. H. (2020). Defective autophagy and mitophagy in aging and Alzheimer’s Disease. Front. Neurosci. 14:612757. doi: 10.3389/fnins.2020.612757
Trempe, J. F., Sauvé, V., Grenier, K., Seirafi, M., Tang, M. Y., Ménade, M., et al. (2013). Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 340, 1451–1455. doi: 10.1126/science.1237908
Tsukada, M., and Ohsumi, Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174. doi: 10.1016/0014-5793(93)80398-e
Um, J. H., and Yun, J. (2017). Emerging role of mitophagy in human diseases and physiology. BMB Rep. 50, 299–307. doi: 10.5483/bmbrep.2017.50.6.056
Vara-Perez, M., Felipe-Abrio, B., and Agostinis, P. (2019). Mitophagy in cancer: a tale of adaptation. Cells 8:493. doi: 10.3390/cells8050493
Vazquez-Martin, A., Van den Haute, C., Cufí, S., Corominas-Faja, B., Cuyàs, E., Lopez-Bonet, E., et al. (2016). Mitophagy-driven mitochondrial rejuvenation regulates stem cell fate. Aging 8, 1330–1352. doi: 10.18632/aging.100976
Villa, E., Proïcs, E., Rubio-Patiño, C., Obba, S., Zunino, B., Bossowski, J. P., et al. (2017). Parkin-independent mitophagy controls chemotherapeutic response in cancer cells. Cell Rep. 20, 2846–2859. doi: 10.1016/j.celrep.2017.08.087
Wai, T., and Langer, T. (2016). Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab. 27, 105–117. doi: 10.1016/j.tem.2015.12.001
Wang, C. Z., Yan, G. X., Dong, D. S., Xin, H., and Liu, Z. Y. (2019). LncRNA-ATB promotes autophagy by activating Yes-associated protein and inducing autophagy-related protein 5 expression in hepatocellular carcinoma. World J. Gastroenterol. 25, 5310–5322. doi: 10.3748/wjg.v25.i35.5310
Wang, J., Xie, S., Yang, J., Xiong, H., Jia, Y., Zhou, Y., et al. (2019). The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J. Hematol. Oncol. 12:81.
Wang, S. Y., Wang, X., and Zhang, C. Y. (2020). LncRNA SNHG7 enhances chemoresistance in neuroblastoma through cisplatin-induced autophagy by regulating miR-329-3p/MYO10 axis. Eur. Rev. Med. Pharmacol. Sci. 24, 3805–3817.
Wang, X., Cheng, M. L., Gong, Y., Ma, W. J., Li, B., and Jiang, Y. Z. (2020). LncRNA DANCR promotes ATG7 expression to accelerate hepatocellular carcinoma cell proliferation and autophagy by sponging miR-222-3p. Eur. Rev. Med. Pharmacol. Sci. 24, 8778–8787.
Wang, X. L., Feng, S. T., Wang, Z. Z., Chen, N. H., and Zhang, Y. (2021). Role of mitophagy in mitochondrial quality control: mechanisms and potential implications for neurodegenerative diseases. Pharmacol. Res. 165:105433. doi: 10.1016/j.phrs.2021.105433
Wei, Y., Chiang, W. C., Sumpter, R. Jr., Mishra, P., and Levine, B. (2017). Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell 168, 224–238.e10.
Whelan, K. A., Chandramouleeswaran, P. M., Tanaka, K., Natsuizaka, M., Guha, M., Srinivasan, S., et al. (2017). Autophagy supports generation of cells with high CD44 expression via modulation of oxidative stress and Parkin-mediated mitochondrial clearance. Oncogene 36, 4843–4858. doi: 10.1038/onc.2017.102
Wu, J., Zhu, P., Lu, T., Du, Y., Wang, Y., He, L., et al. (2019). The long non-coding RNA LncHDAC2 drives the self-renewal of liver cancer stem cells via activation of Hedgehog signaling. J. Hepatol. 70, 918–929. doi: 10.1016/j.jhep.2018.12.015
Wu, L., Zhang, D., Zhou, L., Pei, Y., Zhuang, Y., Cui, W., et al. (2019). FUN14 domain-containing 1 promotes breast cancer proliferation and migration by activating calcium-NFATC1-BMI1 axis. EBioMedicine 41, 384–394. doi: 10.1016/j.ebiom.2019.02.032
Wu, W., Tian, W., Hu, Z., Chen, G., Huang, L., Li, W., et al. (2014). ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 15, 566–575. doi: 10.1002/embr.201438501
Xiang, G., Yang, L., Long, Q., Chen, K., Tang, H., Wu, Y., et al. (2017). BNIP3L-dependent mitophagy accounts for mitochondrial clearance during 3 factors-induced somatic cell reprogramming. Autophagy 13, 1543–1555. doi: 10.1080/15548627.2017.1338545
Xiang, X., Fu, Y., Zhao, K., Miao, R., Zhang, X., Ma, X., et al. (2021). Cellular senescence in hepatocellular carcinoma induced by a long non-coding RNA-encoded peptide PINT87aa by blocking FOXM1-mediated PHB2. Theranostics 11, 4929–4944. doi: 10.7150/thno.55672
Xiao, Z. D., Han, L., Lee, H., Zhuang, L., Zhang, Y., Baddour, J., et al. (2017). Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat. Commun. 8:783.
Xin, L., Zhou, Q., Yuan, Y. W., Zhou, L. Q., Liu, L., Li, S. H., et al. (2019). METase/lncRNA HULC/FoxM1 reduced cisplatin resistance in gastric cancer by suppressing autophagy. J. Cancer Res. Clin. Oncol. 145, 2507–2517. doi: 10.1007/s00432-019-03015-w
Xiong, H., Ni, Z., He, J., Jiang, S., Li, X., He, J., et al. (2017). LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene 36, 3528–3540. doi: 10.1038/onc.2016.521
Yamano, K., and Youle, R. J. (2013). PINK1 is degraded through the N-end rule pathway. Autophagy 9, 1758–1769. doi: 10.4161/auto.24633
Yamashita, K., Miyata, H., Makino, T., Masuike, Y., Furukawa, H., Tanaka, K., et al. (2017). High expression of the mitophagy-related protein pink1 is associated with a poor response to chemotherapy and a poor prognosis for patients treated with Neoadjuvant Chemotherapy for Esophageal Squamous Cell Carcinoma. Ann. Surg. Oncol. 24, 4025–4032. doi: 10.1245/s10434-017-6096-8
Yan, C., Luo, L., Guo, C. Y., Goto, S., Urata, Y., Shao, J. H., et al. (2017). Doxorubicin-induced mitophagy contributes to drug resistance in cancer stem cells from HCT8 human colorectal cancer cells. Cancer Lett. 388, 34–42. doi: 10.1016/j.canlet.2016.11.018
Yang, F., Zhang, H., Mei, Y., and Wu, M. (2014). Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol. Cell 53, 88–100. doi: 10.1016/j.molcel.2013.11.004
Zaarour, R. F., Azakir, B., Hajam, E. Y., Nawafleh, H., Zeinelabdin, N. A., Engelsen, A. S. T., et al. (2021). Role of hypoxia-mediated autophagy in tumor cell death and survival. Cancers 13:533. doi: 10.3390/cancers13030533
Zhang, C., Lin, M., Wu, R., Wang, X., Yang, B., Levine, A. J., et al. (2011). Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc. Natl. Acad. Sci. U.S.A. 108, 16259–16264. doi: 10.1073/pnas.1113884108
Zhang, T., Xue, L., Li, L., Tang, C., Wan, Z., Wang, R., et al. (2016). BNIP3 protein suppresses PINK1 kinase proteolytic cleavage to promote mitophagy. J. Biol. Chem. 291, 21616–21629. doi: 10.1074/jbc.m116.733410
Zhang, W., Ren, H., Xu, C., Zhu, C., Wu, H., Liu, D., et al. (2016). Hypoxic mitophagy regulates mitochondrial quality and platelet activation and determines severity of I/R heart injury. eLife 5:e21407.
Zhang, W., Siraj, S., Zhang, R., and Chen, Q. (2017). Mitophagy receptor FUNDC1 regulates mitochondrial homeostasis and protects the heart from I/R injury. Autophagy 13, 1080–1081. doi: 10.1080/15548627.2017.1300224
Zhao, Y., Liu, S., Li, X., Zhou, L., Meng, Y., Li, Y., et al. (2019). Aberrant shuttling of long noncoding RNAs during the mitochondria-nuclear crosstalk in hepatocellular carcinoma cells. Am. J. Cancer Res. 9, 999–1008.
Zhao, Y., Sun, L., Wang, R. R., Hu, J. F., and Cui, J. (2018). The effects of mitochondria-associated long noncoding RNAs in cancer mitochondria: new players in an old arena. Crit. Rev. Oncol. 131, 76–82. doi: 10.1016/j.critrevonc.2018.08.005
Zhao, Y., Zhou, L., Li, H., Sun, T., Wen, X., Li, X., et al. (2021). Nuclear-Encoded lncRNA MALAT1 epigenetically controls metabolic reprogramming in hcc cells through the mitophagy pathway. Mol. Ther. Nucleic Acids 23, 264–276. doi: 10.1016/j.omtn.2020.09.040
Zheng, R., Yao, Q., Xie, G., Du, S., Ren, C., Wang, Y., et al. (2015). TAT-ODD-p53 enhances the radiosensitivity of hypoxic breast cancer cells by inhibiting Parkin-mediated mitophagy. Oncotarget 6, 17417–17429. doi: 10.18632/oncotarget.4002
Keywords: mitochondria, mitophagy, cancer metabolism, long non-coding RNA, cancer stem cells, cancer therapy
Citation: Li Y, Li W, Hoffman AR, Cui J and Hu J-F (2021) The Nucleus/Mitochondria-Shuttling LncRNAs Function as New Epigenetic Regulators of Mitophagy in Cancer. Front. Cell Dev. Biol. 9:699621. doi: 10.3389/fcell.2021.699621
Received: 23 April 2021; Accepted: 20 August 2021;
Published: 08 September 2021.
Edited by:
Simão Teixeira da Rocha, University of Lisbon, PortugalReviewed by:
Qi Liao, Ningbo University, ChinaCopyright © 2021 Li, Li, Hoffman, Cui and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ji-Fan Hu, amlmYW5Ac3RhbmZvcmQuZWR1; aHVqaWZhbkBqbHUuZWR1LmNu; Jiuwei Cui, Y3VpandAamx1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.