- 1Hunan Key Laboratory of Medical Epigenomics, Department of Dermatology, The Second Xiangya Hospital of Central South University, Changsha, China
- 2Department of Nephrology, First Medical Center of Chinese PLA General Hospital, Nephrology Institute of the Chinese People’s Liberation Army, State Key Laboratory of Kidney Diseases, National Clinical Research Center for Kidney Diseases, Beijing Key Laboratory of Kidney Disease Research, Beijing, China
Systemic lupus erythematosus (SLE) is an autoimmune disease with complicated clinical manifestations. Although our understanding of the pathogenesis of SLE has greatly improved, the understanding of the pathogenic mechanisms of SLE is still limited by disease heterogeneity, and targeted therapy is still unavailable. Substantial evidence shows that RNA methylation plays a vital role in the mechanisms of the immune response, prompting speculation that it might also be related to the occurrence and development of SLE. RNA methylation has been a hot topic in the field of epigenetics in recent years. In addition to revealing the modification process, relevant studies have tried to explore the relationship between RNA methylation and the occurrence and development of various diseases. At present, some studies have provided evidence of a relationship between RNA methylation and SLE pathogenesis, but in-depth research and analysis are lacking. This review will start by describing the specific mechanism of RNA methylation and its relationship with the immune response to propose an association between RNA methylation and SLE pathogenesis based on existing studies and then discuss the future direction of this field.
Introduction
Systemic lupus erythematosus (SLE) is a common chronic systemic autoimmune disease that mainly affects young or middle-aged women, with a male-to-female incidence ratio of approximately 1:9 (Stojan and Petri, 2018); men are affected by more severe renal injury (Ramírez Sepúlveda et al., 2019). The incidence rates for SLE range from 0.9 to 7.4 per 100,000 persons/year and differ by sex, region and population (Gergianaki et al., 2017; Stojan and Petri, 2018; Tanaka et al., 2020). For the Asia Pacific region, a meta-analysis showed that the incidence rates of SLE (per 100,000 per year) ranged from 0.9 to 8.4 (Tanaka et al., 2020).
Both environmental factors and genetic susceptibility play vital roles in SLE development. Collectively, evidence has shown that the sustained production of autoantibodies, complement activation, immune complex deposition, neutrophil extracellular traps, lymphocyte signaling and inflammation are all the key mechanisms underlying the loss of tolerance and tissue damage (Tsokos et al., 2016). Patients with SLE can present with diverse organ involvement, such as skin, musculoskeletal, kidney, heart, and lung involvement (Smith and Gordon, 2010; Tsokos et al., 2016). Moreover, patients with SLE have a high overall risk of malignancy (Choi et al., 2017). At present, the early diagnosis of SLE is still based on clinical symptoms and signs, a laboratory examination and biopsy. A recent study compared three classification criteria for evaluating SLE: the European League Against Rheumatism (EULAR)/American College of Rheumatology (ACR)-2019 (Aringer et al., 2019), Systemic Lupus International Collaborating Clinics (SLICC)-2012 (Petri et al., 2012) and ACR-1997 criteria (Hochberg, 1997). An early SLE cohort study found that using these three evaluation criteria might miss or delay the classification and diagnosis of a significant subset of patients with moderate/severe SLE. The general therapeutic approach mainly relies on glucocorticoids and immunosuppressive drugs (ISDs) (Kuhn et al., 2015) with unavoidable side effects, and thus targeted therapy remains an urgent need. Although some progress has been achieved in the early diagnosis and treatment of SLE, the prognosis of patients with SLE is still not optimistic, and the medical cost is high. Therefore, further in-depth exploration of the pathogenesis of SLE is very important for its early diagnosis and the search for new therapeutic targets.
In recent years, an increasing number of scholars have focused their attention on the field of epigenetics, such as DNA methylation, which has led to a better understanding of the pathogenesis of SLE (Xiao and Zuo, 2016). However, with advances in RNA methylation research, scholars have found that RNA methylation is very important in the immune response and is closely related to SLE. RNA, an important intermediate product of gene expression, is also regulated by chemical modification after transcription and during translation through a process collectively referred to as epitranscriptomics (Saraceno et al., 2016). Similar to chemical modifications of DNA and histones, chemical modifications of RNA also regulate gene expression through mechanisms such as changing the structural properties of RNA or changing the affinity of mRNA for ribosomes. The discovery, continuous exploration, and in-depth study of chemical modifications in RNA have led epigenetic research to a new level and provided a new direction for further exploring the pathogenesis of diseases and developing new therapeutic strategies.
Therefore, this article will start by describing RNA methylation, summarize the application of RNA methylation in immunology, and speculate on the relationship between RNA methylation and the pathogenesis of SLE to provide more ideas for future research.
The Specific Mechanism of RNA Methylation
Chemical modifications of cellular RNAs are natural and abundant. The dynamic nature and increasing number of RNA modifications provide new possibilities for rapidly altering gene expression to adapt to specific environments. Due to the development of genomic approaches, various modifications on RNA have been identified and investigated. As of 2017, more than 150 distinct chemical modifications on RNA have been detected (Boccaletto et al., 2018).
N6-methyladenosine (m6A) is the most widespread and investigated modification of mammalian mRNA and has a broad range of critical functions in development (Kasowitz et al., 2018), cancer (Ma S. et al., 2019), and viral infection (Williams et al., 2019). Pseudouridine (Ψ) was the first structurally modified nucleoside identified in the 1950s, and is known as the fifth nucleoside (Cohn and Volkin, 1951). Ψ generates an extra hydrogen bond donor at the newly formed N1 position, which increases the structural stability of the RNA and translation efficiency and accuracy (McKenney et al., 2017). The presence of N1mΨ (N1-methyl-pseudouridine (N1mΨ) in mRNA favors ribosome recycling on the same mRNA or de novo ribosome recruitment (Svitkin et al., 2017). 5-Methylcytidine (m5C) has long been studied in DNA. In RNA, m5C levels are 3–10-fold rarer than m6A levels (Legrand et al., 2017). Although the biological function of m5C in eukaryotic mRNA is just beginning to become clear, it is postulated to have a powerful function in regulating cellular processes (Trixl and Lusser, 2019).
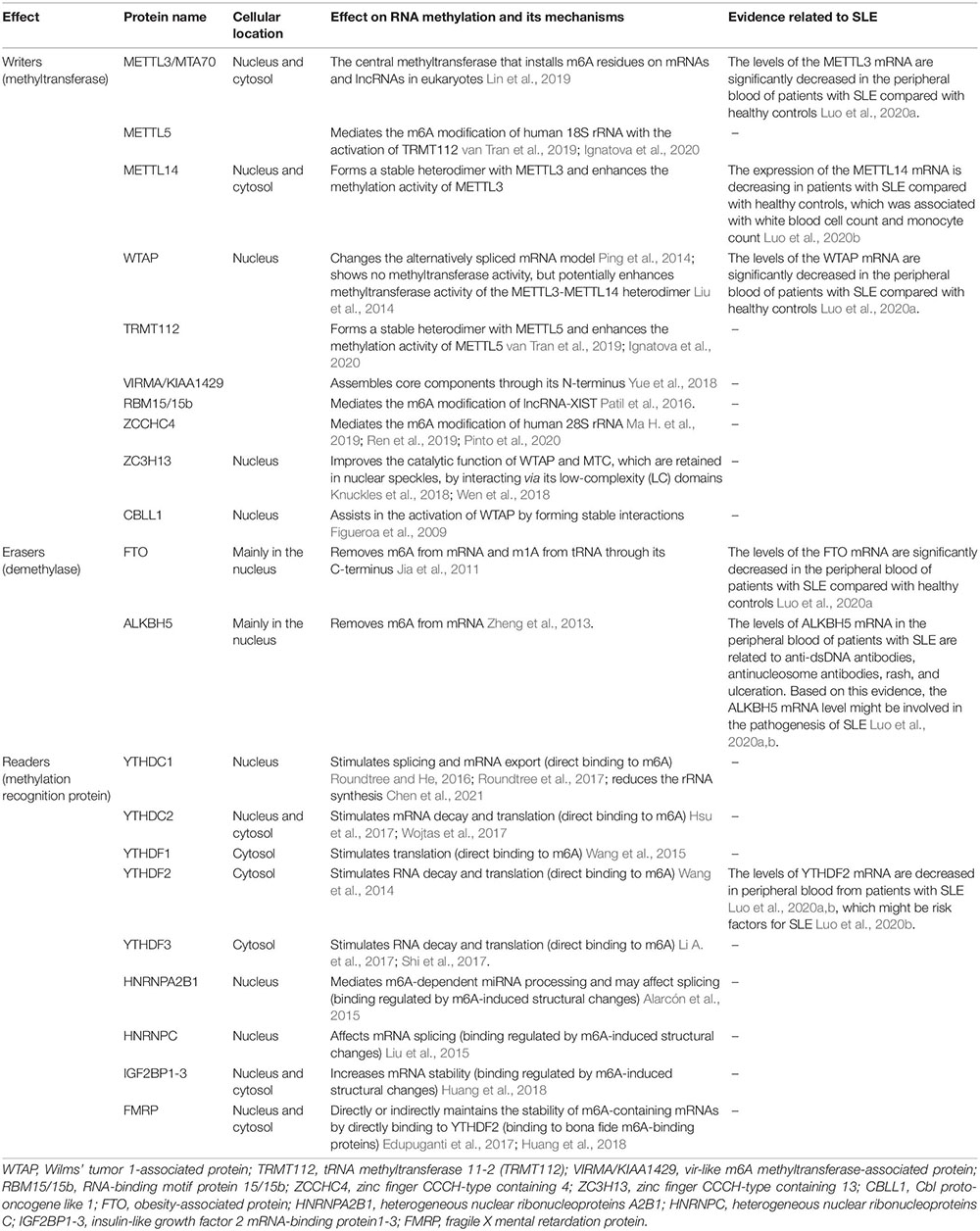
Three types of molecules are involved in RNA methylation: writers, erasers and readers. Writers, namely, methyltransferases, transfer methyl groups to RNA in the form of protein complexes, individual proteins of which might have specific functions or integrate different signals. Erasers, namely, demethylases, erase the RNA methylation modification to convert m6A into RNA. Readers recognize the RNA methylation modification information to guide and participate in the translation and degradation of the modified downstream RNA sequence. In general, RNA methylation is first performed by writer complexes at different RNA sites and then can be demethylated by erasers, which makes RNA methylation a reversible process. The modified base sites are recognized by specific readers to mediate specific biological functions. Readers located in the nucleus might affect mRNA splicing or other nuclear processes, while those located in the cytoplasm might affect the stability, translation, or location of mRNAs. These three types of molecules are indispensable to achieving the regulatory functions of RNA methylation. We list the most well-studied writers, erasers and readers for m6A in Table 1 and m5C in Table 2.
Studies have identified important roles for m6A and m5C in the development and regulation of many organs and systems, especially in the immune system. Therefore, we should focus on the specific mechanism of RNA methylation at m6A and m5C and the potential mechanism in the pathogenesis of SLE.
m6A
The methylation of the sixth position of the RNA adenine ring and occurs in the sequence context Pu[G > A]m6AC[U > A > C](Pu = purine) (Schibler et al., 1977). It was first discovered in 1974 (Desrosiers et al., 1974) and plays a conservative role in the evolution of meiosis and cell differentiation in yeast, plants and mammals (Yue et al., 2015). Although m6A may exist in the primary transcript, in mammals and yeast, m6A is mainly located in genes, namely, the mRNA protein coding region (CDS) near the termination codon and 3′ untranslated region (3′ UTR) (Dominissini et al., 2012; Ke et al., 2015). Coding RNAs and non-coding RNAs, including tRNAs, rRNAs, small nuclear RNAs, microRNA (miRNA) precursors and long non-coding RNAs (lncRNAs), are modified with m6A in a variety of tissues.
m6A Writers
The m6A methyltransferase complex transfers a methyl group from the donor substrate S-adenosyl methionine (SAM) to adenine nucleotides in the recipient RNA subunit (Bokar et al., 1997). The complex consists of METTL3 (also known as MTA70) (Lin et al., 2019), METTL5 (van Tran et al., 2019; Ignatova et al., 2020), METTL14 (Yue et al., 2015; Liu et al., 2016), Wilms’ tumor 1-associated protein (WTAP) (Ping et al., 2014), tRNA methyltransferase 11-2 (TRMT112) (van Tran et al., 2019; Ignatova et al., 2020), vir-like m6A methyltransferase-associated protein (VIRMA, originally known as KIAA1429) (Yue et al., 2018), RNA-binding motif protein 15/15b (RBM15/15b) (Patil et al., 2016), zinc finger CCCH-type containing 4 (ZCCHC4) (Ma H. et al., 2019; Ren et al., 2019; Pinto et al., 2020), zinc finger CCCH-type containing 13 (ZC3H13) (Knuckles et al., 2018; Wen et al., 2018), and Cbl proto-oncogene like 1 (CBLL1) (Figueroa et al., 2009), among which the most common molecular components are METTL3 and METTL14.
METTL3 is considered the central methyltransferase because of its ability to bind SAM, and it is highly conserved in eukaryotes (Lin et al., 2019). The discovery of METTL3 initiated research on the relationship between m6A and cellular physiology. Differences in the METTL3 expression levels reflect changes in the total m6A level. METTL3 can be modified by SUMO1 at lysine residues K177, K211, K212, and K215, significantly repressing the methyltransferase activity of METTL3 (Du et al., 2018). SUMOylation is a reversible posttranslational modification process that attaches small ubiquitin-like modifiers to protein substrates (Hay, 2005). Moreover, sentrin/SUMO-SPECIFIC PROTEASE 7 is significantly upregulated in patients with SLE (Cui Y. et al., 2017), and levels of the METTL3 mRNA are significantly decreased in the peripheral blood of patients with SLE compared with healthy controls (Luo et al., 2020a). Thus, we speculate that SUMOylation might play roles in reducing the expression of METTL3 in patients with SLE.
METTL14 is highly homologous to METTL3. It forms a stable heterodimer with METTL3, enhancing the methylation activity of METTL3. Together, the heterodimer of METTL3-METTL14 forms the catalytic core of the m6A methyltransferase complex (Yue et al., 2015; Liu et al., 2016). Sanna Bystrom found that the expression profile of the METTL14 protein was altered in patients with multiple sclerosis (Byström et al., 2014). Qing Luo et al. observed decreased expression of the METTL14 mRNA in patients with SLE compared with healthy controls (p < 0.001), which was associated with white blood cell count and monocyte count (Luo et al., 2020b). Evidence also shows that the production of type 1 interferon (IFN I), the most important cytokine involved in SLE pathogenesis, its production triggered by dsDNA or human cytomegalovirus is controlled by cellular METTL14 and ALKBH5 demethylases (Rubio et al., 2018). METTL14 depletion increases both the production and stability of the nascent IFN β1 mRNA in response to dsDNA (Rubio et al., 2018). This phenomenon represents a potential mechanism by which METTL14 participates in the development of autoimmune diseases, especially SLE.
WTAP is also a core component of the m6A methyltransferase complex that interacts with METTL3 and METTL14 (Ping et al., 2014). The intracellular m6A abundance was markedly decreased when WTAP was knocked out compared with METTL3 or METTL14, which might result from changes in the alternative splicing of the mRNAs to which WTAP binds (Ping et al., 2014). Liu et al. found that the m6A level in polyadenylated RNA was decreased by ∼30, ∼40, and ∼50% in HeLa cells with knockdown of cellular METTL3, METTL14, and WTAP, respectively (Liu et al., 2014). Moreover, WTAP itself showed no methyltransferase activity but dramatically enhanced the methyltransferase activity when interacting with the METTL3-METTL14 heterodimer (Liu et al., 2014). WTAP is upregulated in many tumors, where it functions as an oncogene by interacting with different proteins involved in RNA processing (Sorci et al., 2018).
m6A Erasers
The m6A demethylase is responsible for the selective removal of SAM from the adenine nucleotides of RNA to regulate gene expression and cell fate. Fat mass and obesity-associated protein (FTO) was the first RNA demethylase discovered, and its C-terminal structure demethylates mRNA, mainly in the nucleus (Jia et al., 2011; Bartosovic et al., 2017). Several studies have shown that FTO might not have a physiological function toward m6A because the FTO knockout transcriptome does not contain an increased number of m6A sites (Hess et al., 2013; Mauer et al., 2017; Garcia-Campos et al., 2019). Instead, FTO appears to function in specific tissues or under specific conditions. For example, FTO regulates the expression of oncogenes, namely, an ∼20% increase, by reducing the m6A modification in mRNA to enhance leukemic oncogene-mediated cell transformation and leukemogenesis (Li Z. et al., 2017). As shown in the study by Qing Luo, FTO expression positively correlates with SLE in patients. The mRNA levels of FTO in the peripheral blood of patients with SLE are significantly decreased compared with those of healthy controls (Luo et al., 2020a).
ALKBH5, the second RNA eraser identified, was shown to affect mouse spermatogenesis (Zheng et al., 2013). ALKBH5 is primarily colocalized with nuclear speckles and affects mRNA export and RNA metabolism in a demethylation-dependent manner (Zheng et al., 2013). Rubio et al. (2018) found that IFN I production triggered by dsDNA or human cytomegalovirus is affected by ALKBH5. ALKBH5 depletion reduces nascent IFN β1 mRNA production without detectably influencing IFN β1 mRNA decay (Rubio et al., 2018). Moreover, the levels of ALKBH5 mRNA in the peripheral blood of patients with SLE are related to anti-dsDNA antibodies, antinucleosome antibodies, rash and ulceration (Luo et al., 2020a). Based on this evidence, the ALKBH5 mRNA level might be involved in the pathogenesis of SLE (Luo et al., 2020a).
m6A Readers
The downstream function of the presence of m6A on mRNA molecules is closely related to the recognition and binding of m6A-methylated recognition proteins, termed m6A readers. Various m6A readers have been identified, but their mechanism is similar: m6A readers recognize and bind the RNA decorated by m6A. The regulatory function of m6A is achieved by enhancing or weakening the recruitment of different RNA-binding proteins (RBPs) to target mRNAs or directly inducing secondary structural changes in target mRNAs to influence the interaction between RNAs and RBPs (Adhikari et al., 2016; Wu B. et al., 2017).
The most important m6A recognition protein in eukaryotes is the YTH domain-containing family proteins, which comprise the conserved C-terminal RNA recognition and binding domain YTH and the N-terminal variable region. This protein family is also considered the most primitive m6A reader, falling into three classes: YTHDC1, YTHDC2, and the YTHDF family (Zhang et al., 2010). Among them, YTHDF1, YTHDF2, and YTHDF3, which exist in humans, have been studied more extensively and have different cellular localizations but similar functions (Liao et al., 2018; Patil et al., 2018). Other recognized m6A recognition proteins include heterogeneous nuclear ribonucleoprotein A2B1 (HNRNPA2B1) (Alarcón et al., 2015), heterogeneous nuclear ribonucleoproteins C (HNRNPC) (Liu et al., 2015), fragile X mental retardation protein (FMRP) (Edupuganti et al., 2017) and insulin-like growth factor 2 mRNA-binding protein1-3 (IGF2BP1-3) (Huang et al., 2018).
Effects of m6A Methylation
The m6A modification on RNA regulates the transcription of genes to achieve functional regulation at the cellular or tissue level, which is mainly achieved by the functions of various m6A methyltransferases, m6A demethylases and m6A recognition proteins. m6A is mainly modified on mRNA, and thus the main effects of the m6A modification include pre-mRNA shearing, the stability of mRNA, nuclear transfer of mRNA, and translation of mRNA, thus achieving the regulation of mRNA function (Meyer et al., 2015; Gan et al., 2019; Zhang et al., 2019a). For example, protein translation typically begins with the recruitment of the 43S ribosomal complex to the 5′ cap of mRNAs by a cap-binding complex. Evidence has shown that mRNAs containing m6A in their 5′ UTR are translated in a cap-independent manner (Meyer et al., 2015).
In addition, m6A also modifies non-coding RNAs, such as rRNA, miRNAs, and lncRNAs. m6A methylation of rRNA is dispensable in cell growth and ribosome biogenesis, but plays important roles in increasing translation efficiency and cell proliferation and differentiation (van Tran et al., 2019). Increasing translation efficiency is achieved by the regulation of the kinetics of translation rather than rRNA processing (van Tran et al., 2019; Pinto et al., 2020). Some researchers an increasing polysome/monosome ratio via m6A methylation of rRNA (Ma H. et al., 2019), but this phenomenon has not been widely recognized in other studies (van Tran et al., 2019; Pinto et al., 2020). The positive effects on cell proliferation and differentiation have been shown by knocking down the m6A writers of rRNA (Ma H. et al., 2019; Ignatova et al., 2020; Xing et al., 2020). High m6A methylation levels in rRNA have also been found in cancer (Ma H. et al., 2019). Meanwhile, METTL3 may play a positive role in prerRNA processing by influencing relevant protein expression or modifying snoRNA (Sergeeva et al., 2020). The regulation of m6A methylation is also be embodied in premiRNA processing and lncRNA processing. Meanwhile, by sequence pairing with mRNAs containing miRNA target sites, miRNAs regulate the binding of METTL3 to target RNAs, leading to an increase in the m6A modification (Fazi and Fatica, 2019).
m5C
m5C is the best-understood epigenetic modification of DNA (Jones, 2012) and is also present in RNAs that are more diverse and complex (Gilbert et al., 2016) The m5C methyltransferase catalyzes the methylation of the 5th position of the cytosine pyrimidine ring at a specific site (Schosserer et al., 2016). The function of m5C has been unclear for many years. However, with the continuous development of high-throughput sequencing technology, the specific locations and related functions of m5C have gradually become clear. Researchers have found that m5C is distributed across coding RNA sequences and all types of non-coding RNAs, such as tRNA and rRNA (Squires et al., 2012; Gilbert et al., 2016). Moreover, the m5C modification of RNA is an important regulator of many aspects of gene expression, including RNA processing and degradation, ribosomal assembly, translation, and RNA stability (Tuorto et al., 2012; Blanco et al., 2016; Nakano et al., 2016).
m5C Writers
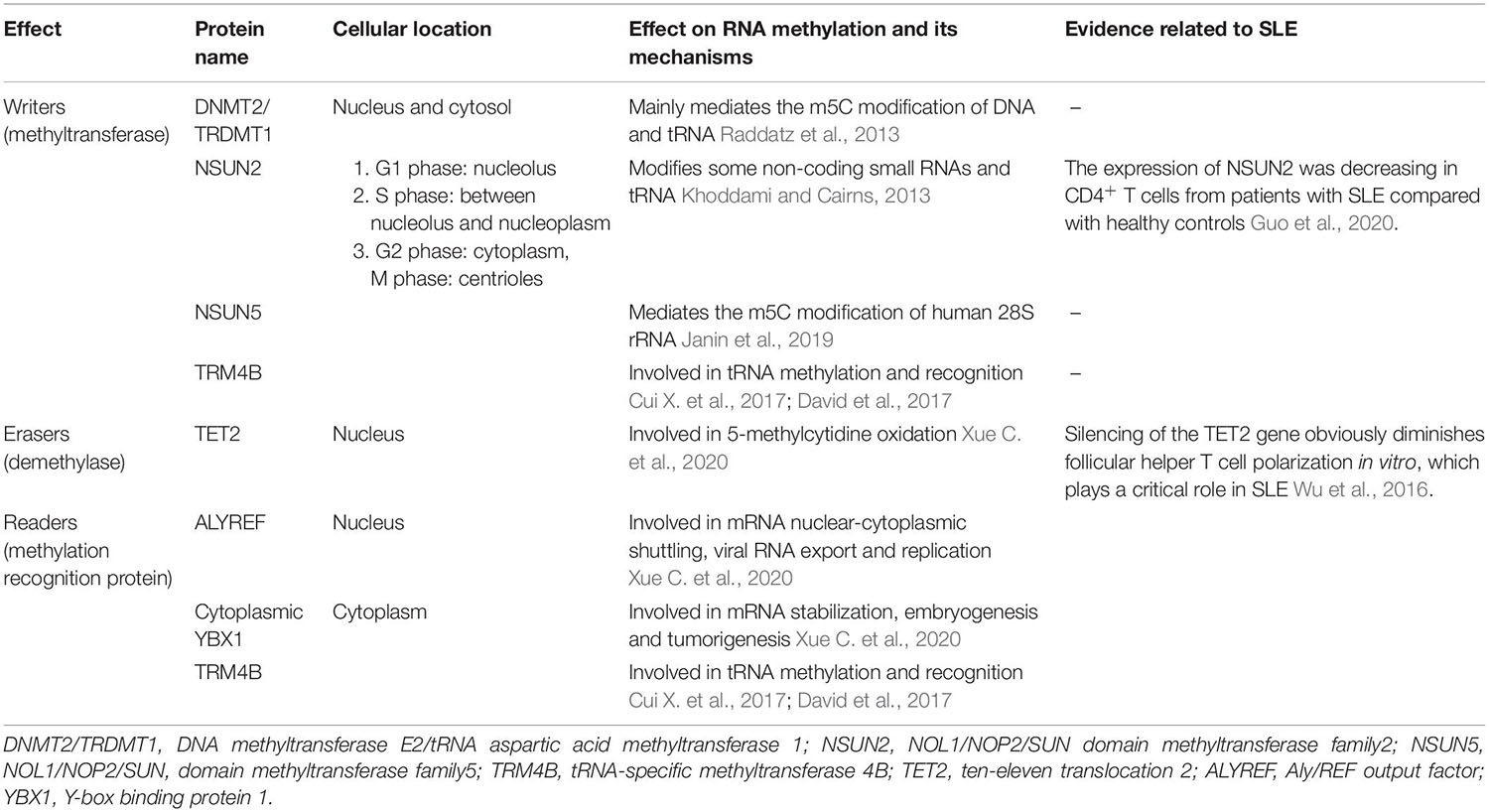
Among higher eukaryotes, the most frequently studied m5C methyltransferases are DNA methyltransferase E2/tRNA aspartic acid methyltransferase 1 (DNMT2/TRDMT1) (Tang et al., 2003) and the NOL1/NOP2/SUN domain methyltransferase family (NSUN) (Bohnsack et al., 2019). Mechanistically, both DNMT2 and NSUN form covalent intermediates through the interaction of a cysteine and cytosine of the target RNA to promote nucleophilic attack at C5 of the pyrimidine ring by SAM to form the m5C modification. However, the difference is that a single cysteine in DNMT2 molecules forms a covalent intermediate, with cytosine Myc-induced SUN domain-containing protein (Misu/NSUN2) molecules formed by two cysteine and cytosine in the covalent intermediate (King and Redman, 2002).
DNMT2 is a widely conserved member of the eukaryotic cytokine-5-DNA methyltransferase protein family, which is widely distributed in the nucleus and cytosol (Tang et al., 2003). Although DNMT2 mainly mediates m5C methylation in DNA, many studies have recently shown that DNMT2 could mediate the m5C modification of tRNA (Raddatz et al., 2013; Genenncher et al., 2018). According to recent studies, DNMT2 mainly mediates the methylation of tRNA at C38 in eukaryotic cells (Schaefer et al., 2010), and DNMT2-mediated RNA methylation has been detected in some eukaryotic organisms, such as zebrafish (Rai et al., 2007) and Drosophila (Schaefer et al., 2010). DNMT2-mediated RNA methylation exerts an important effect on organ differentiation and environmental tolerance (Schaefer et al., 2010). Evidence has shown that DNMT2 is required for an efficient innate immune response in Drosophila, which is possibly mediated by RNA methylation (Durdevic et al., 2013).
NSUN2 is a member of the protein family containing the NOL1/NOP2/SUN domains, which are mainly located in the nucleus. NSUN2 modifies some non-coding small RNAs and mRNAs in addition to tRNA (Khoddami and Cairns, 2013; Li Q. et al., 2017; Yang et al., 2017). A recent study showed substantially reduced NSUN2 expression levels in CD4+ T cells from patients with SLE compared with healthy controls, which might be due to RNA methylation (Guo et al., 2020).
tRNA-specific methyltransferase 4B (TRM4B) is also an important m5C methyltransferase, and it has been proven to be relevant to tRNA methylation and root growth in Arabidopsis (Cui X. et al., 2017; David et al., 2017), but this result still needs to be confirmed in mammalian cells.
m5C Erasers
The modification of m5C is reversible. Recent studies identified the m5C demethylase ten-eleven translocation 2 (TET2) (Wu and Zhang, 2011; Xue M. et al., 2020). Silencing of the TET2 gene obviously diminishes follicular helper T cell polarization in vitro, which plays a critical role in SLE. Because TET2 functions in both DNA demethylation and RNA demethylation, the specific mechanism of m5C in SLE deserves further study (Wu et al., 2016).
m5C Readers
To date, three m5C-binding proteins have been identified as m5C readers: Aly/REF output factor (ALYREF, an mRNA transport adaptor) (Yang et al., 2017), cytoplasmic Y-box binding protein 1 (YBX1), and TRM4B (Cui X. et al., 2017; Chen X. et al., 2019). The ALYREF-dependent pathway potentially represents one of the main mechanisms for the selective export of m5C-modified mRNAs in mammals (Yang et al., 2017).
Effects of m5C Methylation
Similar to m6A methylation, the effect of m5C is mainly achieved by influencing the process of protein translation. Studies have shown that m5C induces ribonuclease activity to promote tRNA degradation and affect protein translation; moreover, the modification of m5C on rRNA also affects protein translation (Burgess et al., 2015), and some experiments have suggested that m5C affects the stability of mRNA (Hussain et al., 2013). Although a variety of molecules are modified with m5C, in eukaryotes, the m5C modification is mainly detected on tRNA (Squires et al., 2012). With the exception of tRNALeu CAA, the m5C modification of tRNA mainly occurred outside anticodon rings (Chan et al., 2012). Therefore, the main effect of the CAA m5C modification is to regulate the translation efficiency by affecting CAA oscillation (Chan et al., 2012), while the m5C modification outside the anti-codon ring mainly affects the structure and stability of tRNA for the purpose of regulation (Vare et al., 2017; Janin et al., 2019). By regulating protein translation, m5C also plays a role in many normal physiological processes and abnormal diseases.
Potential Links Between RNA Methylation and Sle
RNA Methylation in T Cells
T cells are regarded as a central component of the pathogenesis of SLE (Tsokos et al., 2016). T cell homeostasis is the key process in maintaining the T cell pool size, and its imbalanced state is essential in the pathogenesis of SLE (Oster et al., 2019; Katsuyama et al., 2020). Evidence has shown that the m6A modification plays an important role in maintaining T cell homeostasis. Li HB and colleagues found that the m6A modification controls the differentiation of naïve T cells (Li H.-B. et al., 2017). Conditioned knockout of the Mettl3 gene in mouse CD4+ T cells reduces the m6A methylation level in naïve T cells, which leads to an increase in Th2 cells and a decrease in Th1 and Th17 cells, but has no effect on cell apoptosis or TCR-mediated proliferation (Li H.-B. et al., 2017). Researchers have inferred that naïve T cells lacking METTL3 or METTL14 does not undergo homeostatic amplification and remain naïve, mainly because SOCS family genes (Socs1, Socs3, and Cis) with less m6A modification exhibit slower mRNA decay and increased protein expression levels (Li H.-B. et al., 2017). Therefore, the increased SOCS family activity inhibits the IL-7/STAT5 signaling pathway and activates the TCR/ERK/AKT pathway, which inhibits T cell proliferation and differentiation (Palmer and Restifo, 2009; Li H.-B. et al., 2017). Afterward, RNA methylation was also shown to regulate T cell homeostasis through a repressive loss of function of regulatory T cells (Tregs) (Tong et al., 2018). The authors generated Mettl3f/f; Foxp3Cre mice to determine the role of m6A methylation in T cell homeostasis in vivo. Inflammatory Th1 and Th7 responses were significantly increased in spleen and peripheral lymph nodes compared with wild-type mice at 60 days after birth (Tong et al., 2018). In Mettl3–/– Tregs, the reduction in m6A levels increases the mRNA stability of SOCS genes, including Cish, Socs1, Socs2, Socs3, and Asb2, and increased levels of SOCS proteins block signal transduction from the IL2-STAT5 pathway, which is essential for Treg function and stability (Shi et al., 2018; Tong et al., 2018). Thus, a loss of Treg function eventually leads to excessive inflammation, inhibiting the function of Tregs and playing an important role in regulating T cell homeostasis (Li and Rudensky, 2016; Tong et al., 2018). In addition to METTL3, METTL14 deficiency in T cells also induces unbalanced T cell homeostasis. A METTL14 deficiency in T cells induces spontaneous colitis in mice, manifesting as increased inflammatory cell infiltration and cytokine production from Th1 and Th17 cells (Lu T. X. et al., 2020). The Mettl14 deficiency also caused impaired induction of the differentiation of naïve T cells into induced Tregs (Lu T. X. et al., 2020). Therefore, the m6A modification exerts a positive regulatory effect on T cell differentiation and development and is one of the important regulatory mechanisms of adaptive immunity, as shown in Figure 1.
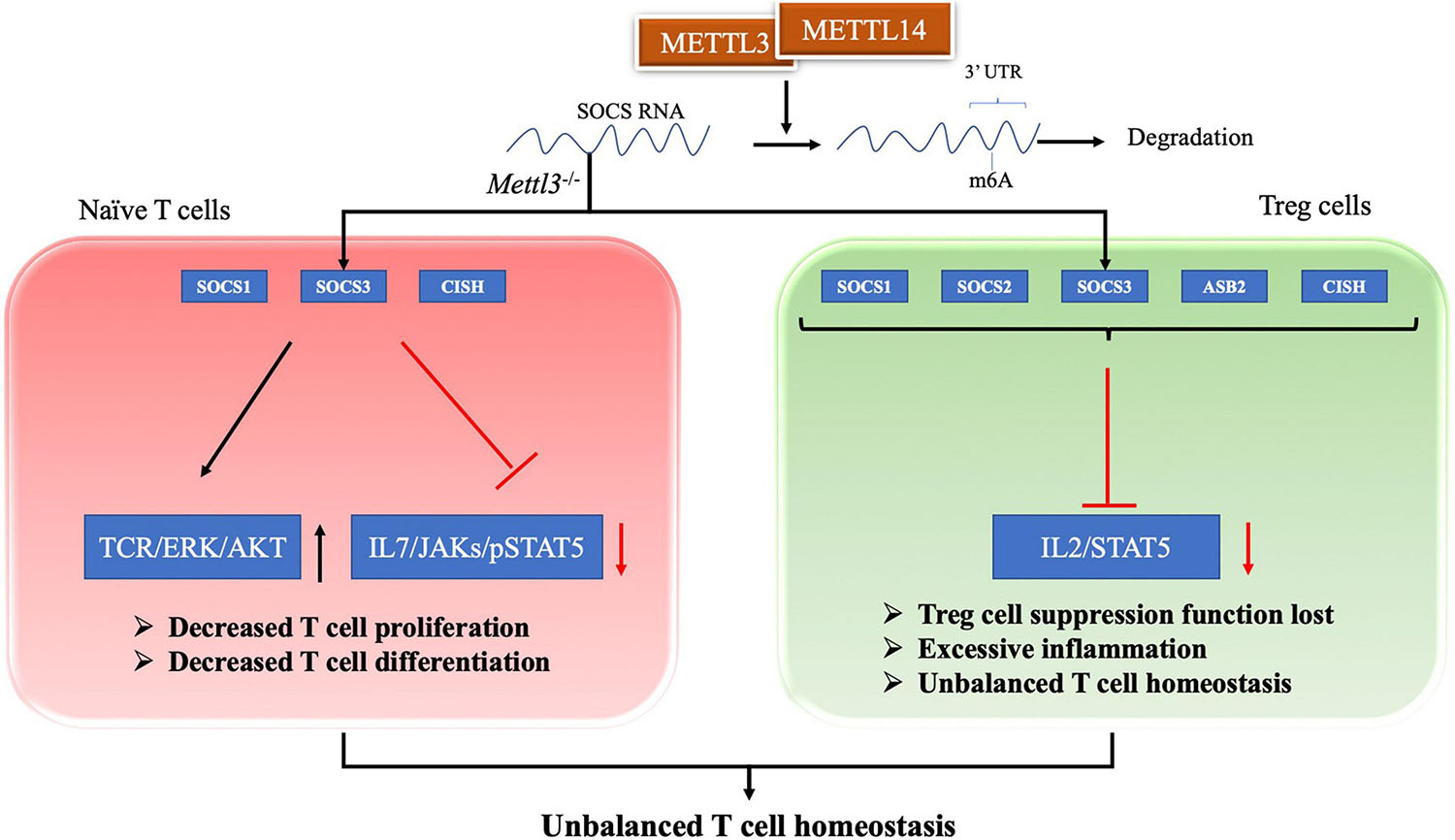
Figure 1. RNA methylation in T cell homeostasis. For naive T cell, SOCS family genes with m6A modification exhibit slower mRNA decay and increased protein expression levels. The increased SOCS family activity inhibits the IL-7/STAT5 signaling pathway and activates the TCR/ERK/AKT pathway, which lead to decreased T cell proliferation and differentiation. For Treg cell, reduction in m6A enhances the mRNA stability of SOCS genes and the proteins levels. The increased SOCS family activity blocks signal transduction of the IL2-STAT5 pathway, which lead to Treg cell suppression function lost. These two evidences shows that m6A modification plays an important role in maintaining T cell homeostasis.
Evidence related to the role of m5C in T cells is limited. The m5C modification stabilizes mRNA expression, leading to an imbalance in Th17/Treg differentiation, which may reveal the potential mechanism of m5C in SLE pathology (Guo et al., 2020). The specific mechanism is worth further exploration.
RNA Methylation in Dendritic Cells
Dendritic cells (DCs) play important roles in the innate and adaptive immune responses, as well as in the progression of SLE (McHugh, 2019; Xiao et al., 2020). A study revealed that DCs exposed to m6A- or m5C-modified nucleosides expressed significantly fewer cytokines and activation markers than those treated with unmodified RNA (Karikó et al., 2005). Mammalian total RNA, but not bacterial RNA, is abundant in modified nucleosides (Karikó et al., 2005). The innate immune system selectively responds to bacteria or necrotic tissue by detecting RNA lacking modification (Karikó et al., 2005). However, METTL3-mediated m6A methylation in DCs is required for the activation and function of DCs. METTL3 induces the phenotypic and functional maturation of DCs, and the expression of CD40, CD80, and IL-12 and the ability to stimulate T cells both in vitro and in vivo are promoted in Mettl3KO DCs (Wang H. et al., 2019). As shown in the study by Han D., knockdown of YTHDF1, an m6A-binding protein, in classic DCs enhanced the antitumor immune response in vivo by enhancing the cross-presentation of tumor antigens and the cross-priming of CD8+ T cells (Han et al., 2019). Thus, RNA methylation also regulates the function of DCs in the immune response.
RNA Methylation in Inflammation
RNA methylation is also an essential mechanism in the process of inflammation, and it is a key component of the pathogenesis of SLE (Frangou et al., 2019). The dynamic regulation of these inflammatory factors contributes to the susceptibility to SLE but is more strongly implicated in the loss of tolerance and end organ effects (Hu et al., 2015). Patients with SLE usually present increased serum levels of inflammatory factors, such as IL-6, TNF-α, and IL-1β, which are related to various immune processes in SLE pathogenesis (Wu Y. et al., 2017; Paradowska-Gorycka et al., 2019; Uzrail et al., 2019). For example, the elevated levels of IL-6 and TNF-α expression in PBMCs from patients with SLE are closely associated with the Th1/Th2/Th17 inflammatory response, which is positively correlated with SLE disease activity (De la Cruz-Mosso et al., 2018). Increased levels of IL-6, IL-10, and TNF-α are consistent with B cell proliferation and activation in patients with SLE (Fleischer et al., 2015). Thus, RNA methylation participates in SLE pathogenesis by regulating inflammatory factor expression, but further evidence is needed. To date, in vitro and in vivo studies have shown that RNA methylation regulates the occurrence of inflammation through several signaling pathways.
First, the level of RNA methylation regulates the expression of inflammatory factors through the MyD88 pathway. METTL3 knockdown decreases the lipopolysaccharide-induced expression of inflammatory cytokines, including IL-6, IL-8, GRO, Gro-α and RANTES (Feng et al., 2018; Zhang et al., 2019b), by facilitating the expression of MyD88S, a splice variant of myeloid differential protein-88 (MyD88) (Feng et al., 2018). Moreover, RNA methylation regulates inflammation by influencing the NF-κB and MAPK signaling pathways through effects on the phosphorylation of related molecules. METTL3 knockdown decreases the phosphorylation of IKKα/β, p65 and IκBα in the NF-κB signaling pathway and p38, ERK, and JNK in the MAPK signaling pathway in LPS-induced inflammation (Feng et al., 2018; Zhang et al., 2019b). Analogously, YTHDF2 knockdown increases the phosphorylation of p65, p38 and ERK1/2 in the NF-κB and MAPK signaling pathways (Yu et al., 2019). In addition, RNA methylation of the forkhead box O (FOXO) mRNA is also involved in the inflammation process. METTL14 and METTL3 separately increase m6A methylation of the FOXO1 and FOXO3 mRNAs and increase their stability via the interaction of YTHDF1 (Jian et al., 2020; Lin et al., 2020). YTHDF3 promotes FOXO3 translation by binding to the translation initiation region of the FOXO3 mRNA (Zhang et al., 2019c). As an important transcription factor, FOXO1 promotes the expression of VCAM-1 and ICAM-1 by directly binding to their promoter regions, which leads to endothelial inflammation and atherosclerosis development (Jian et al., 2020). The FOXO3 levels in B cells from patients with SLE are inversely correlated with disease activity and reduced in patients with elevated anti-dsDNA Ab levels (Ottens et al., 2018).
The regulatory effect on these inflammasome pathways in different autoimmune diseases has been reported. In vitro and in vivo experiments using samples from patients with rheumatoid arthritis (RA) revealed significantly elevated METTL3 levels in patients with RA that played an important role in LPS-induced inflammation in macrophages via the NF-κB signaling pathway (Wang J. et al., 2019). In the development of osteoarthritis (OA), METTL3 expression increases and subsequently regulates inflammation via the NF-κB signaling pathway and the degradation of extracellular matrix (ECM) (Liu et al., 2019). These findings not only provide new insights into the pathogenesis of SLE but also facilitate the identification of therapeutic targets and provide new directions for future research. For example, oleuropein (OL) regulates the activation of the JAK/STAT, MAPK, NF-κB and NLRP3 inflammasome pathways and exert its therapeutic effect on patients with SLE (Castejon et al., 2019).
Role of RNA Methylation in IFN I Production
IFN I is a vital component of the antiviral innate immune response and is also the most important cytokine involved in SLE pathogenesis. We outline the studies that revealed the role of RNA methylation in IFN I production to determine the potential correlation between RNA methylation and SLE.
First, the m6A modification plays an important role in the regulation of IFN I production during virus recognition (Figure 2). The dead-box (DDX) helicase family plays an important role in identifying viral nucleic acids and regulating downstream pathways (Parvatiyar et al., 2012). DDX3 and DDX46 recruit and interact with ALKBH5 via the DEAD helicase domain (Shah et al., 2017; Zheng et al., 2017). The complex of DDX46 and ALKBH5 demethylates m6A-modified antiviral transcripts, which increases antiviral transcript (Mavs, Traf3, and Traf6) retention in the nucleus and decreases the expression of these proteins and IFN I (Zheng et al., 2017). In addition to DDX46, the function of HNRNPA2B1, a DNA virus sensor, in activating the TANK-binding kinase 1–interferon regulatory factor 3 (TBK1–IRF3) pathway and subsequent IFN-α/β production is also regulated by the m6A modification (Wang L. et al., 2019). In the cytoplasm, HNRNPA2B1 activates the TBK1-IRF3 pathway by binding to CGAS, IFI16, and STING to promote IFN I production (Wang L. et al., 2019). In this process, an RNA writer (METTL3) promotes, while an RNA eraser (FTO) inhibits, the function of HNRNPA2B1 (Wang L. et al., 2019). The m6A modification influences the function of retinoic acid-induced gene I (RIG-I). RIG-I plays a key role in recognizing viral infection, and its activated conformer engages the adaptor mitochondrial antiviral signaling protein (MAVS) to mediate the activation of transcription factors and interferon-stimulated gene (ISG) (Durbin et al., 2016). RNAs containing modified nucleotides interrupt signaling at early steps of the RIG-I-like innate immune activation pathway, and nucleotide modifications with similar chemical structures are organized into classes that suppress or evade innate immune signaling (Durbin et al., 2016). This result is consistent with the mechanism we discussed above in DCs, in which the innate immune system may selectively respond to invading pathogenic nucleic acids by detecting RNA lacking modification (Karikó et al., 2005). For example, human metapneumovirus (HMPV) RNA is modified by m6A, which promotes HMPV replication and gene expression. However, if the m6A modification of HMPV RNA is demethylated, the production of IFN I is increased by the high expression of RIG-I (Lu M. et al., 2020). Thus, if viruses or other invading pathogens acquire m6A in their RNA to mimic cellular RNA, they might avoid detection by the innate immune system (Lu M. et al., 2020).
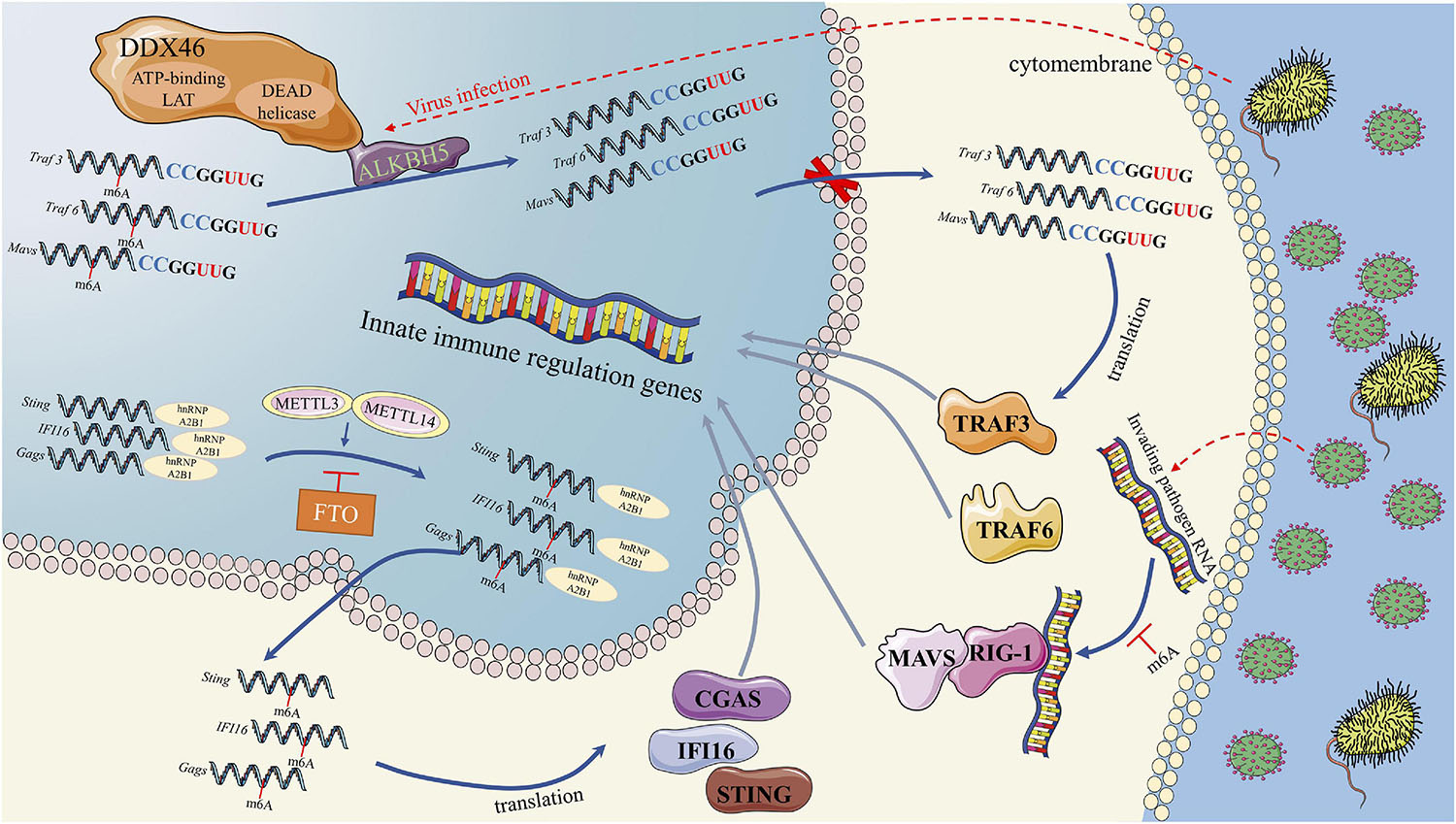
Figure 2. RNA methylation in Interferon production in process of virus recognition. RNA methylation in Interferon production were mainly through three pathways. First, DDX46 and ALKBH5 complex could demethylate m6A-modified antiviral transcripts, which lead to antiviral transcript retention in the nucleus and decreases the IFN I protein level. Second, HNRNPA2B1 could activates the TBK1-IRF3 pathway by binding to CGAS, IFI16, and STING to promote IFN I production. METTL3 and FTO could regulate the function of HNRNPA2B1 by m6A methylation. Third, m6A modified transcripts could interrupts the RIG-I like innate immune activation, which could mediate the activation of transcription factors and interferon-stimulated genes.
In addition, the m6A modification of ISGs is logical. The m6A machinery, such as METTL3, YTHDF2, and YTHDF3, has been reported to regulate IFN β levels via the m6A modification of ISGs (Winkler et al., 2019a, b; Zhang et al., 2019c). YTHDF2 assists ISG20 in degrading HBV transcripts by selectively recognizing and binding HBV transcripts with the m6A modification (Imam et al., 2020), as the m6A modification participates in IFN α-induced viral RNA degradation. A study conducted by Rubio R. M. et al. revealed that the writer METTL14 and eraser ALKBH5 negatively regulate IFN β production through the m6A modification of the coding sequence and the 3′ untranslated region of the IFN β mRNA (Rubio et al., 2018).
Methylation of host non-coding RNAs also exerts an effect. The m6A modification of lncRNAs plays an important role in antiviral innate immunity based on IFN I signaling (Wang et al., 2020). Studies have revealed that m6A on circular RNA (circRNA) inhibits innate immunity (Chen Y. G. et al., 2019). CircRNAs lacking the m6A modification directly activate the RNA pattern recognition receptor RIG-I in the presence of lysine-63-linked polyubiquitin chains to cause filamentation of the adaptor protein MAVS and activation of the downstream transcription factor IRF3, potentially inducing IFN I production (Chen Y. G. et al., 2019).
Role of RNA Methylation in the Antiviral Immune Response
RNA methylation also plays an important role in the antiviral immune response, which is regulated by two systems: the innate immune response and adaptive immune response. In addition to IFN I production, the m6A modification has also been shown to regulate the innate immune response through other mechanisms. Toll-like receptors (TLRs) recognize viral RNA, which is an important information transmission process in the innate immune response (Karikó et al., 2005). Methylated RNA destroys the stability of double-stranded RNA bodies (Yang et al., 2020), which restrains the activation of TLRs (Kierzek and Kierzek, 2003).
RNA methylation is also important in specific antiviral immune responses. For example, in HIV-infected T cells, both host and viral RNA methylation levels are elevated (Lichinchi et al., 2016). Knockdown of METTL3 or METTL14 alone results in reduced viral replication, while ALKBH5 silencing results in significantly increased viral replication, indicating that m6A methylation exerts a positive regulatory effect on HIV replication (Lichinchi et al., 2016). YTHDF3 overexpressed in CD4+ T cells recognizes the m6A modification on HIV and limits its infection (Jurczyszak et al., 2020). YTHDF1-3 also restrains HIV-1 reverse transcriptase (Tirumuru et al., 2016). In HIV-1-infected cells, 56 genes modified by m6A methylation were identified, 19 of which are related to HIV replication and mainly encode functional proteins. The m6A modification of these 56 genes is mainly involved in the T cell response to viral infection by changing their RNA metabolism (Lichinchi et al., 2016). At the same time, the m5C modification also plays a role in antiviral immunity. Experiments have proven that m5C regulates the splicing of HIV-1 mRNA and posttranscriptional function, which affects the expression of viral genes (Courtney et al., 2019). In a direct homologous RNAi screen of host factors regulating HIV-1 replication, the m5C methyltransferase NOP2/NSUN1 was found to limit HIV-1 replication in the nucleus. NOP2 is associated with the HIV-1 5′ LTR and interacts with the HIV-1 TAR RNA through its competition with HIV-1 Tat protein and contributes to the methylation of TAR m5C, which also proves that m5C methylation promotes HIV-1 transcription and virus entry into the incubation period (Kong et al., 2020). RNA chemical modification plays an important role not only in the specific immune response to HIV but also in the responses to other viruses, such as Kaposi’s sarcoma-associated herpes virus (KSHV), and m6A has become one of the new targets for the treatment of KSHV (Ye, 2017; Tan and Gao, 2018).
RNA Methylation in Autoimmune Responses
SLE is a typical systemic autoimmune disease characterized by various autoantibodies, a loss of tolerance and tissue damage. While less evidence is available for the regulation of m6A in the autoimmune response, the role of m6A in autoimmunity is non-negligible. In patients with SLE, the levels of ALKBH5 mRNA are associated with anti-dsDNA and antinucleosome antibodies, which are typical autoantibodies detected in patients with SLE (Luo et al., 2020a). IL-17 participates in various autoimmune diseases and in autoantibody production in patients with SLE (Ma et al., 2020). Wang et al. (2017) found that NSun2 methylates the IL-17A mRNA at cytosine C466 in vitro and in vivo, which promotes the translation of IL-17. Evidence has shown that m6A RNA methylation participates in coeliac disease, a complex autoimmune disorder (Olazagoitia-Garmendia et al., 2021). Higher m6A methylation in the 5′ UTR of the XPO1 RNA results in increased XPO1 protein concentrations that lead to nuclear factor kappa B activation and inflammation (Wang et al., 2017). Although we did not obtain direct evidence for the relationship between RNA methylation and autoantibodies in patients with SLE, these findings suggested that the topic deserves in-depth study.
RNA Methylation and Organ Damage in Patients With Sle
Evidence shows that the loss of tolerance and tissue damage are distinct processes. Tissue effects might be major contributors to organ damage in patients with SLE independent of the effects of blood cells (Tsokos et al., 2016). These effects might also be mediated by RNA methylation.
Nephritis
Renal tubulointerstitial fibrosis is one of the typical features of chronic kidney disease (CKD). In individuals with lupus nephritis, interstitial fibrosis is associated with CKD progression (Morales et al., 2021). A mouse fibrotic kidney disease model induced by UUO exhibited significant increases in the total m6A level, and ALKBH5, the eraser, suppressed fibrosis in this model (Ning et al., 2020). In an in vitro study, overexpression of METTL3 and METTL14 increased m6A levels and subsequently increased p53 mRNA and protein levels in cisplatin-treated HK2 cells (Zhou et al., 2019).
Skin
Damage to stem cells in follicles might be one process leading to permanent loss of follicles in patients with SLE or cutaneous lupus erythematosus (CLE) (Al-Refu and Goodfield, 2009), which might also be regulated by RNA methylation. RNA methyltransferase (NSUN2) is required to balance stem cell self-renewal and differentiation in skin (Blanco et al., 2011). Compared with wild-type mice, an increase in the number of quiescent bulge stem cells was observed in Misu–/– mice, along with delayed exit from the bulge, increased self-renewal, and aberrant hair cycling (Blanco et al., 2011).
Bone
Mesenchymal stem cells originally isolated from the bone marrow stoma are multipotent and possess strong immunomodulatory activities that interact with multiple immune cells, including DCs, neutrophils, NK cells, T cells, and B cells. Bone marrow-derived mesenchymal stem cells (BM-MSCs) from patients with SLE exhibit defective immune regulation, which might contribute to the imbalance between Treg and Th17 cells in patients with SLE (Geng et al., 2020). On the other hand, the capacity of osteogenic differentiation of BM-MSCs from patients with SLE is reduced compared with cells from healthy controls, which contributes to osteoporosis in patients with SLE (Tang et al., 2013). This outcome might be due to increased IFN β production and activation of the NF-κB pathway in BM-MSCs from patients with SLE (Tang et al., 2013; Gao et al., 2020). According to recent studies, METTL3 silencing reduces mA methylation levels and inhibits the osteogenic differentiation of bone marrow-derived mesenchymal stem cells (Wu et al., 2018; Yan et al., 2020). Thus, the METTL3-mediated m6A modification might induce the dysfunction of BM-MSCs in patients with SLE.
Conclusion
We discussed the role of RNA methylation in the pathogenesis of SLE, including innate immunity and adaptive immunity. Although the evidence has generally indicated a potential relationship between RNA methylation and SLE, few studies have interpreted a direct relationship between them. Therefore, in the next step, we must investigate the direct relationship by quantitatively measuring m6A levels and their association with these existing mechanisms to establish a stronger causal link. Moreover, we investigated the function of RNA methylation in a cohort with SLE to obtain more biomarkers for the diagnosis, treatment, and prognosis of SLE and related complications. Further in-depth research on RNA methylation may clarify the pathogenesis of SLE and provide additional insights into diagnostic and therapeutic strategies.
Author Contributions
XLV and XLI analyzed the literature and studies and wrote the manuscript. MZ, HW, and WZ assisted with constructing figures and polishing the language. QL and XC revised the manuscript. All authors listed have made substantial, direct and intellectual contributions to the work, and approved the article for publication.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 81830019, 82030097, 81830097, and 81700629), the Non-profit Central Research Institute Fund of the Chinese Academy of Medical Sciences (No. 2020-RC320-003), the Key Project for International and Regional Cooperation in Science and Technology Innovation of Hunan Province (No. 2019WK2081), the Beijing Municipal Natural Science Foundation (No. 7202188), and the Big Data Program from Chinese PLA General Hospital (No. 2019MBD-053).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adhikari, S., Xiao, W., Zhao, Y.-L., and Yang, Y.-G. (2016). m(6)A: Signaling for mRNA splicing. RNA Biol. 13, 756–759. doi: 10.1080/15476286.2016.1201628
Alarcón, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015). HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell 162, 1299–1308. doi: 10.1016/j.cell.2015.08.011
Al-Refu, K., and Goodfield, M. (2009). Hair follicle stem cells in the pathogenesis of the scarring process in cutaneous lupus erythematosus. Autoimmun. Rev. 8, 474–477. doi: 10.1016/j.autrev.2008.12.015
Aringer, M., Costenbader, K., Daikh, D., Brinks, R., Mosca, M., Ramsey-Goldman, R., et al. (2019). 2019 European League Against Rheumatism/American College of Rheumatology classification criteria for systemic lupus erythematosus. Ann. Rheum. Dis. 78, 1151–1159. doi: 10.1136/annrheumdis-2018-214819
Bartosovic, M., Molares, H. C., Gregorova, P., Hrossova, D., Kudla, G., and Vanacova, S. (2017). N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 45, 11356–11370. doi: 10.1093/nar/gkx778
Blanco, S., Bandiera, R., Popis, M., Hussain, S., Lombard, P., Aleksic, J., et al. (2016). Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340. doi: 10.1038/nature18282
Blanco, S., Kurowski, A., Nichols, J., Watt, F. M., Benitah, S. A., and Frye, M. (2011). The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 7:e1002403. doi: 10.1371/journal.pgen.1002403
Boccaletto, P., Machnicka, M. A., Purta, E., Piatkowski, P., Baginski, B., Wirecki, T. K., et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307. doi: 10.1093/nar/gkx1030
Bohnsack, K. E., Hobartner, C., and Bohnsack, M. T. (2019). Eukaryotic 5-methylcytosine (m(5)C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes 10:genes10020102. doi: 10.3390/genes10020102
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G., and Rottman, F. M. (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247.
Burgess, A. L., David, R., and Searle, I. R. (2015). Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae. BMC Plant Biol. 15:199. doi: 10.1186/s12870-015-0580-8
Byström, S., Ayoglu, B., Häggmark, A., Mitsios, N., Hong, M.-G., Drobin, K., et al. (2014). Affinity proteomic profiling of plasma, cerebrospinal fluid, and brain tissue within multiple sclerosis. J. Proteome Res. 13, 4607–4619. doi: 10.1021/pr500609e
Castejon, M. L., Sánchez-Hidalgo, M., Aparicio-Soto, M., Montoya, T., Martín-LaCave, I., Fernández-Bolaños, J. G., et al. (2019). Dietary oleuropein and its new acyl-derivate attenuate murine lupus nephritis through HO-1/Nrf2 activation and suppressing JAK/STAT, NF-κB, MAPK and NLRP3 inflammasome signaling pathways. J. Nutr. Biochem. 74:108229. doi: 10.1016/j.jnutbio.2019.108229
Chan, C. T. Y., Pang, Y. L. J., Deng, W., Babu, I. R., Dyavaiah, M., Begley, T. J., et al. (2012). Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 3:937. doi: 10.1038/ncomms1938
Chen, C., Liu, W., Guo, J., Liu, Y., Liu, X., Liu, J., et al. (2021). Nuclear m(6)A reader YTHDC1 regulates the scaffold function of LINE1 RNA in mouse ESCs and early embryos. Protein Cell 2021, 837–838. doi: 10.1007/s13238-021-00837-8
Chen, X., Li, A., Sun, B.-F., Yang, Y., Han, Y.-N., Yuan, X., et al. (2019). 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 21, 978–990. doi: 10.1038/s41556-019-0361-y
Chen, Y. G., Chen, R., Ahmad, S., Verma, R., Kasturi, S. P., Amaya, L., et al. (2019). N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol. Cell 76, 96.e–109.e. doi: 10.1016/j.molcel.2019.07.016
Choi, M. Y., Flood, K., Bernatsky, S., Ramsey-Goldman, R., and Clarke, A. E. (2017). A review on SLE and malignancy. Best Pract. Res. Clin. Rheumatol. 31, 373–396. doi: 10.1016/j.berh.2017.09.013
Cohn, W. E., and Volkin, E. (1951). Nucleoside-5′-Phosphates from Ribonucleic Acid. Nature 167, 483–484.
Courtney, D. G., Tsai, K., Bogerd, H. P., Kennedy, E. M., Law, B. A., Emery, A., et al. (2019). Epitranscriptomic Addition of m(5)C to HIV-1 Transcripts Regulates Viral Gene Expression. Cell Host Microbe 26, 217.e–227.e. doi: 10.1016/j.chom.2019.07.005
Cui, X., Liang, Z., Shen, L., Zhang, Q., Bao, S., Geng, Y., et al. (2017). 5-Methylcytosine RNA Methylation in Arabidopsis Thaliana. Mol. Plant 10, 1387–1399. doi: 10.1016/j.molp.2017.09.013
Cui, Y., Yu, H., Zheng, X., Peng, R., Wang, Q., Zhou, Y., et al. (2017). SENP7 Potentiates cGAS Activation by Relieving SUMO-Mediated Inhibition of Cytosolic DNA Sensing. PLoS Pathog. 13:e1006156. doi: 10.1371/journal.ppat.1006156
David, R., Burgess, A., Parker, B., Li, J., Pulsford, K., Sibbritt, T., et al. (2017). Transcriptome-Wide Mapping of RNA 5-Methylcytosine in Arabidopsis mRNAs and Noncoding RNAs. Plant Cell 29, 445–460. doi: 10.1105/tpc.16.00751
De la Cruz-Mosso, U., García-Iglesias, T., Bucala, R., Estrada-García, I., González-López, L., Cerpa-Cruz, S., et al. (2018). MIF promotes a differential Th1/Th2/Th17 inflammatory response in human primary cell cultures: Predominance of Th17 cytokine profile in PBMC from healthy subjects and increase of IL-6 and TNF-α in PBMC from active SLE patients. Cell Immunol. 324, 42–49. doi: 10.1016/j.cellimm.2017.12.010
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U S A. 71, 3971–3975. doi: 10.1073/pnas.71.10.3971
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Du, Y., Hou, G., Zhang, H., Dou, J., He, J., Guo, Y., et al. (2018). SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res. 46, 5195–5208. doi: 10.1093/nar/gky156
Durbin, A. F., Wang, C., Marcotrigiano, J., and Gehrke, L. (2016). RNAs Containing Modified Nucleotides Fail To Trigger RIG-I Conformational Changes for Innate Immune Signaling. MBio 7:16. doi: 10.1128/mBio.00833-16
Durdevic, Z., Hanna, K., Gold, B., Pollex, T., Cherry, S., Lyko, F., et al. (2013). Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep. 14, 269–275. doi: 10.1038/embor.2013.3
Edupuganti, R. R., Geiger, S., Lindeboom, R. G. H., Shi, H., Hsu, P. J., Lu, Z., et al. (2017). N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878. doi: 10.1038/nsmb.3462
Fazi, F., and Fatica, A. (2019). Interplay Between N (6)-Methyladenosine (m(6)A) and Non-coding RNAs in Cell Development and Cancer. Front. Cell Dev. Biol. 7:116. doi: 10.3389/fcell.2019.00116
Feng, Z., Li, Q., Meng, R., Yi, B., and Xu, Q. (2018). METTL3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells. J. Cell. Mol. Med. 22, 2558–2568. doi: 10.1111/jcmm.13491
Figueroa, A., Kotani, H., Toda, Y., Mazan-Mamczarz, K., Mueller, E.-C., Otto, A., et al. (2009). Novel roles of hakai in cell proliferation and oncogenesis. Mol. Biol. Cell 20, 3533–3542. doi: 10.1091/mbc.e08-08-0845
Fleischer, V., Sieber, J., Fleischer, S. J., Shock, A., Heine, G., Daridon, C., et al. (2015). Epratuzumab inhibits the production of the proinflammatory cytokines IL-6 and TNF-α, but not the regulatory cytokine IL-10, by B cells from healthy donors and SLE patients. Arthritis Res. Ther. 17:185. doi: 10.1186/s13075-015-0686-2
Frangou, E., Vassilopoulos, D., Boletis, J., and Boumpas, D. T. (2019). An emerging role of neutrophils and NETosis in chronic inflammation and fibrosis in systemic lupus erythematosus (SLE) and ANCA-associated vasculitides (AAV): Implications for the pathogenesis and treatment. Autoimmun. Rev. 18, 751–760. doi: 10.1016/j.autrev.2019.06.011
Gan, H., Hong, L., Yang, F., Liu, D., Jin, L., and Zheng, Q. (2019). Progress in epigenetic modification of mRNA and the function of m6A modification. Sheng Wu Gong Cheng Xue Bao 35, 775–783. doi: 10.13345/j.cjb.180416
Gao, L., Liesveld, J., Anolik, J., Mcdavid, A., and Looney, R. J. (2020). IFNβ signaling inhibits osteogenesis in human SLE bone marrow. Lupus 29, 1040–1049. doi: 10.1177/0961203320930088
Garcia-Campos, M. A., Edelheit, S., Toth, U., Safra, M., Shachar, R., Viukov, S., et al. (2019). Deciphering the “m(6)A Code” via Antibody-Independent Quantitative Profiling. Cell 178, 731.e–747.e. doi: 10.1016/j.cell.2019.06.013
Genenncher, B., Durdevic, Z., Hanna, K., Zinkl, D., Mobin, M. B., Senturk, N., et al. (2018). Mutations in Cytosine-5 tRNA Methyltransferases Impact Mobile Element Expression and Genome Stability at Specific DNA Repeats. Cell Rep. 22, 1861–1874. doi: 10.1016/j.celrep.2018.01.061
Geng, L., Tang, X., Wang, S., Sun, Y., Wang, D., Tsao, B. P., et al. (2020). Reduced Let-7f in Bone Marrow-Derived Mesenchymal Stem Cells Triggers Treg/Th17 Imbalance in Patients With Systemic Lupus Erythematosus. Front. Immunol. 11:233. doi: 10.3389/fimmu.2020.00233
Gergianaki, I., Fanouriakis, A., Repa, A., Tzanakakis, M., Adamichou, C., Pompieri, A., et al. (2017). Epidemiology and burden of systemic lupus erythematosus in a Southern European population: data from the community-based lupus registry of Crete, Greece. Ann. Rheum. Dis. 76, 1992–2000. doi: 10.1136/annrheumdis-2017-211206
Gilbert, W. V., Bell, T. A., and Schaening, C. (2016). Messenger RNA modifications: Form, distribution, and function. Science 352, 1408–1412. doi: 10.1126/science.aad8711
Guo, G., Wang, H., Shi, X., Ye, L., Yan, K., Chen, Z., et al. (2020). Disease Activity-Associated Alteration of mRNA m5C Methylation in CD4+ T Cells of Systemic Lupus Erythematosus. Front. Cell Dev. Biol. 8:430. doi: 10.3389/fcell.2020.00430
Han, D., Liu, J., Chen, C., Dong, L., Liu, Y., Chang, R., et al. (2019). Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature 566, 270–274. doi: 10.1038/s41586-019-0916-x
Hay, R. T. (2005). SUMO: a history of modification. Mol. Cell 18, 1–12. doi: 10.1016/j.molcel.2005.03.012
Hess, M. E., Hess, S., Meyer, K. D., Verhagen, L. A. W., Koch, L., Brönneke, H. S., et al. (2013). The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16, 1042–1048. doi: 10.1038/nn.3449
Hochberg, M. C. (1997). Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 40:1725. doi: 10.1002/art.1780400928
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127. doi: 10.1038/cr.2017.99
Hu, S., Ke, S., Wang, W., Ran, H., Chen, M., Zhang, F., et al. (2015). A single fas gene mutation changes lupus onset, severity, location, and molecular abnormalities in mice. Curr. Mol. Med. 15, 380–385. doi: 10.2174/1566524015666150505162638
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295. doi: 10.1038/s41556-018-0045-z
Hussain, S., Tuorto, F., Menon, S., Blanco, S., Cox, C., Flores, J. V., et al. (2013). The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol. Cell Biol. 33, 1561–1570. doi: 10.1128/MCB.01523-12
Ignatova, V. V., Stolz, P., Kaiser, S., Gustafsson, T. H., Lastres, P. R., Sanz-Moreno, A., et al. (2020). The rRNA m(6)A methyltransferase METTL5 is involved in pluripotency and developmental programs. Genes Dev. 34, 715–729. doi: 10.1101/gad.333369.119
Imam, H., Kim, G.-W., Mir, S. A., Khan, M., and Siddiqui, A. (2020). Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified Hepatitis B Virus transcripts. PLoS Pathog. 16:e1008338. doi: 10.1371/journal.ppat.1008338
Janin, M., Ortiz-Barahona, V., de Moura, M. C., Martínez-Cardús, A., Llinàs-Arias, P., Soler, M., et al. (2019). Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. 138, 1053–1074. doi: 10.1007/s00401-019-02062-4
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi: 10.1038/nchembio.687
Jian, D., Wang, Y., Jian, L., Tang, H., Rao, L., Chen, K., et al. (2020). METTL14 aggravates endothelial inflammation and atherosclerosis by increasing FOXO1 N6-methyladeosine modifications. Theranostics 10, 8939–8956. doi: 10.7150/thno.45178
Jones, P. A. (2012). Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492. doi: 10.1038/nrg3230
Jurczyszak, D., Zhang, W., Terry, S. N., Kehrer, T., Bermúdez González, M. C., McGregor, E., et al. (2020). HIV protease cleaves the antiviral m6A reader protein YTHDF3 in the viral particle. PLoS Pathog. 16:e1008305. doi: 10.1371/journal.ppat.1008305
Karikó, K., Buckstein, M., Ni, H., and Weissman, D. (2005). Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175. doi: 10.1016/j.immuni.2005.06.008
Kasowitz, S. D., Ma, J., Anderson, S. J., Leu, N. A., Xu, Y., Gregory, B. D., et al. (2018). Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 14:e1007412. doi: 10.1371/journal.pgen.1007412
McKenney, K. M., Rubio, M. A. T., and Alfonzo, J. D (2017). The Evolution of Substrate Specificity by tRNA Modification Enzymes. Enzymes 41, 51–88.
Katsuyama, T., Martin-Delgado, I. J., Krishfield, S. M., Kyttaris, V. C., and Moulton, V. R. (2020). Splicing factor SRSF1 controls T cell homeostasis and its decreased levels are linked to lymphopenia in systemic lupus erythematosus. Rheumatology 59, 2146–2155. doi: 10.1093/rheumatology/keaa094
Ke, S., Alemu, E. A., Mertens, C., Gantman, E. C., Fak, J. J., Mele, A., et al. (2015). A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29, 2037–2053. doi: 10.1101/gad.269415.115
Khoddami, V., and Cairns, B. R. (2013). Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 31, 458–464. doi: 10.1038/nbt.2566
Kierzek, E., and Kierzek, R. (2003). The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 31, 4472–4480. doi: 10.1093/nar/gkg633
King, M. Y., and Redman, K. L. (2002). RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry 41, 11218–11225. doi: 10.1021/bi026055q
Knuckles, P., Lence, T., Haussmann, I. U., Jacob, D., Kreim, N., Carl, S. H., et al. (2018). Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 32, 415–429. doi: 10.1101/gad.309146.117
Kong, W., Biswas, A., Zhou, D., Fiches, G., Fujinaga, K., Santoso, N., et al. (2020). Nucleolar protein NOP2/NSUN1 suppresses HIV-1 transcription and promotes viral latency by competing with Tat for TAR binding and methylation. PLoS Pathog. 16:e1008430. doi: 10.1371/journal.ppat.1008430
Kuhn, A., Bonsmann, G., Anders, H.-J., Herzer, P., Tenbrock, K., and Schneider, M. (2015). The Diagnosis and Treatment of Systemic Lupus Erythematosus. Dtsch. Arztebl. Int. 112, 423–432. doi: 10.3238/arztebl.2015.0423
Legrand, C., Tuorto, F., Hartmann, M., Liebers, R., Jacob, D., Helm, M., et al. (2017). Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 27, 1589–1596. doi: 10.1101/gr.210666.116
Li, A., Chen, Y.-S., Ping, X.-L., Yang, X., Xiao, W., Yang, Y., et al. (2017). Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 27, 444–447. doi: 10.1038/cr.2017.10
Li, H.-B., Tong, J., Zhu, S., Batista, P. J., Duffy, E. E., Zhao, J., et al. (2017). m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548, 338–342. doi: 10.1038/nature23450
Li, M. O., and Rudensky, A. Y. (2016). T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat. Rev. Immunol. 16, 220–233. doi: 10.1038/nri.2016.26
Li, Q., Li, X., Tang, H., Jiang, B., Dou, Y., Gorospe, M., et al. (2017). NSUN2-Mediated m5C Methylation and METTL3/METTL14-Mediated m6A Methylation Cooperatively Enhance p21 Translation. J. Cell Biochem. 118, 2587–2598. doi: 10.1002/jcb.25957
Li, Z., Weng, H., Su, R., Weng, X., Zuo, Z., Li, C., et al. (2017). FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 31, 127–141. doi: 10.1016/j.ccell.2016.11.017
Liao, S., Sun, H., and Xu, C. (2018). YTH Domain: A Family of N(6)-methyladenosine (m(6)A) Readers. Genomics Proteom. Bioinform. 16, 99–107. doi: 10.1016/j.gpb.2018.04.002
Lichinchi, G., Gao, S., Saletore, Y., Gonzalez, G. M., Bansal, V., Wang, Y., et al. (2016). Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 1:16011. doi: 10.1038/nmicrobiol.2016.11
Lin, S., Liu, J., Jiang, W., Wang, P., Sun, C., Wang, X., et al. (2019). METTL3 Promotes the Proliferation and Mobility of Gastric Cancer Cells. Open Med. 14, 25–31. doi: 10.1515/med-2019-0005
Lin, Z., Niu, Y., Wan, A., Chen, D., Liang, H., Chen, X., et al. (2020). RNA m(6) A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 39:e103181. doi: 10.15252/embj.2019103181
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Liu, K., Ding, Y., Ye, W., Liu, Y., Yang, J., Liu, J., et al. (2016). Structural and Functional Characterization of the Proteins Responsible for N(6)-Methyladenosine Modification and Recognition. Curr. Protein Pept. Sci. 17, 306–318.
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564. doi: 10.1038/nature14234
Liu, Q., Li, M., Jiang, L., Jiang, R., and Fu, B. (2019). METTL3 promotes experimental osteoarthritis development by regulating inflammatory response and apoptosis in chondrocyte. Biochem. Biophys. Res. Commun. 516, 22–27. doi: 10.1016/j.bbrc.2019.05.168
Lu, M., Zhang, Z., Xue, M., Zhao, B. S., Harder, O., Li, A., et al. (2020). N(6)-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 5, 584–598. doi: 10.1038/s41564-019-0653-9
Lu, T. X., Zheng, Z., Zhang, L., Sun, H.-L., Bissonnette, M., Huang, H., et al. (2020). A New Model of Spontaneous Colitis in Mice Induced by Deletion of an RNA m(6)A Methyltransferase Component METTL14 in T Cells. Cell. Mol. Gastroenterol. Hepatol. 10, 747–761. doi: 10.1016/j.jcmgh.2020.07.001
Luo, Q., Fu, B., Zhang, L., Guo, Y., Huang, Z., and Li, J. (2020a). Decreased Peripheral Blood ALKBH5 Correlates with Markers of Autoimmune Response in Systemic Lupus Erythematosus. Dis. Markers 2020:8193895. doi: 10.1155/2020/8193895
Luo, Q., Rao, J., Zhang, L., Fu, B., Guo, Y., Huang, Z., et al. (2020b). The study of METTL14, ALKBH5, and YTHDF2 in peripheral blood mononuclear cells from systemic lupus erythematosus. Mol. Genet. Genomic Med. 2020:e1298. doi: 10.1002/mgg3.1298
Ma, H., Wang, X., Cai, J., Dai, Q., Natchiar, S. K., Lv, R., et al. (2019). N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 15, 88–94. doi: 10.1038/s41589-018-0184-3
Ma, K., Du, W., Xiao, F., Han, M., Huang, E., Peng, N., et al. (2020). IL-17 sustains the plasma cell response via p38-mediated Bcl-xL RNA stability in lupus pathogenesis. Cell. Mol. Immunol. 2020, 540–544. doi: 10.1038/s41423-020-00540-4
Ma, S., Chen, C., Ji, X., Liu, J., Zhou, Q., Wang, G., et al. (2019). The interplay between m6A RNA methylation and noncoding RNA in cancer. J. Hematol. Oncol. 12:121. doi: 10.1186/s13045-019-0805-7
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m(6)A(m) in the 5′ cap controls mRNA stability. Nature 541, 371–375. doi: 10.1038/nature21022
McHugh, J. (2019). Newly defined pro-inflammatory DC subset expanded in SLE. Nat. Rev. Rheumatol. 15:637. doi: 10.1038/s41584-019-0311-x
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell 163, 999–1010. doi: 10.1016/j.cell.2015.10.012
Morales, E., Trujillo, H., Bada, T., Alonso, M., Gutiérrez, E., Rodríguez, E., et al. (2021). What is the value of repeat kidney biopsies in patients with lupus nephritis? Lupus 30, 25–34. doi: 10.1177/0961203320965703
Nakano, S., Suzuki, T., Kawarada, L., Iwata, H., Asano, K., and Suzuki, T. (2016). NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat. Chem. Biol. 12, 546–551. doi: 10.1038/nchembio.2099
Ning, Y., Chen, J., Shi, Y., Song, N., Yu, X., Fang, Y., et al. (2020). Genistein Ameliorates Renal Fibrosis Through Regulation Snail via m6A RNA Demethylase ALKBH5. Front. Pharmacol. 11:579265. doi: 10.3389/fphar.2020.579265
Olazagoitia-Garmendia, A., Zhang, L., Mera, P., Godbout, J. K., Sebastian-DelaCruz, M., Garcia-Santisteban, I., et al. (2021). Gluten-induced RNA methylation changes regulate intestinal inflammation via allele-specific XPO1 translation in epithelial cells. Gut 2021:322566. doi: 10.1136/gutjnl-2020-322566
Oster, C., Wilde, B., Specker, C., Sun, M., Kribben, A., Witzke, O., et al. (2019). BTLA Expression on Th1, Th2 and Th17 Effector T-Cells of Patients with Systemic Lupus Erythematosus Is Associated with Active Disease. Int. J. Mol. Sci. 20:ijms20184505. doi: 10.3390/ijms20184505
Ottens, K., Hinman, R. M., Barrios, E., Skaug, B., Davis, L. S., Li, Q.-Z., et al. (2018). Foxo3 Promotes Apoptosis of B Cell Receptor-Stimulated Immature B Cells, Thus Limiting the Window for Receptor Editing. J. Immunol. 201, 940–949. doi: 10.4049/jimmunol.1701070
Palmer, D. C., and Restifo, N. P. (2009). Suppressors of cytokine signaling (SOCS) in T cell differentiation, maturation, and function. Trends Immunol. 30, 592–602. doi: 10.1016/j.it.2009.09.009
Paradowska-Gorycka, A., Roszak, M., Stypinska, B., Lutkowska, A., Walczyk, M., Olesinska, M., et al. (2019). IL-6 and TGF-β gene polymorphisms, their serum levels, as well as HLA profile, in patients with systemic lupus erythematosus. Clin. Exp. Rheumatol. 37, 963–975.
Parvatiyar, K., Zhang, Z., Teles, R. M., Ouyang, S., Jiang, Y., Iyer, S. S., et al. (2012). The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 13, 1155–1161. doi: 10.1038/ni.2460
Patil, D. P., Chen, C.-K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. doi: 10.1038/nature19342
Patil, D. P., Pickering, B. F., and Jaffrey, S. R. (2018). Reading m(6)A in the Transcriptome: m(6)A-Binding Proteins. Trends Cell Biol. 28, 113–127. doi: 10.1016/j.tcb.2017.10.001
Petri, M., Orbai, A.-M., Alarcón, G. S., Gordon, C., Merrill, J. T., Fortin, P. R., et al. (2012). Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 64, 2677–2686. doi: 10.1002/art.34473
Ping, X.-L., Sun, B.-F., Wang, L., Xiao, W., Yang, X., Wang, W.-J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189. doi: 10.1038/cr.2014.3
Pinto, R., Vågbø, C. B., Jakobsson, M. E., Kim, Y., Baltissen, M. P., O’Donohue, M.-F., et al. (2020). The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res. 48, 830–846. doi: 10.1093/nar/gkz1147
Raddatz, G., Guzzardo, P. M., Olova, N., Fantappie, M. R., Rampp, M., Schaefer, M., et al. (2013). Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc. Natl. Acad. Sci. U S A. 110, 8627–8631. doi: 10.1073/pnas.1306723110
Rai, K., Chidester, S., Zavala, C. V., Manos, E. J., James, S. R., Karpf, A. R., et al. (2007). Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 21, 261–266. doi: 10.1101/gad.1472907
Ramírez Sepúlveda, J. I., Bolin, K., Mofors, J., Leonard, D., Svenungsson, E., Jönsen, A., et al. (2019). Sex differences in clinical presentation of systemic lupus erythematosus. Biol. Sex Differ. 10:60. doi: 10.1186/s13293-019-0274-2
Ren, W., Lu, J., Huang, M., Gao, L., Li, D., Wang, G. G., et al. (2019). Structure and regulation of ZCCHC4 in m(6)A-methylation of 28S rRNA. Nat. Commun. 10:5042. doi: 10.1038/s41467-019-12923-x
Roundtree, I. A., and He, C. (2016). Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Trends Genet. 32, 320–321. doi: 10.1016/j.tig.2016.03.006
Roundtree, I. A., Luo, G.-Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 6:31311. doi: 10.7554/eLife.31311
Rubio, R. M., Depledge, D. P., Bianco, C., Thompson, L., and Mohr, I. (2018). RNA m(6) A modification enzymes shape innate responses to DNA by regulating interferon β. Genes Dev. 32, 1472–1484. doi: 10.1101/gad.319475.118
Saraceno, G. E., Caceres, L. G., Guelman, L. R., Castilla, R., Udovin, L. D., Ellisman, M. H., et al. (2016). Consequences of excessive plasticity in the hippocampus induced by perinatal asphyxia. Exp. Neurol. 286, 116–123. doi: 10.1016/j.expneurol.2016.08.017
Schaefer, M., Pollex, T., Hanna, K., Tuorto, F., Meusburger, M., Helm, M., et al. (2010). RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 24, 1590–1595. doi: 10.1101/gad.586710
Schibler, U., Kelley, D. E., and Perry, R. P. (1977). Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J. Mol. Biol. 115, 695–714. doi: 10.1016/0022-2836(77)90110-3
Schosserer, M., Minois, N., Angerer, T. B., Amring, M., Dellago, H., Harreither, E., et al. (2016). Corrigendum: Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat. Commun. 7:11530. doi: 10.1038/ncomms11530
Sergeeva, O., Sergeev, P., Melnikov, P., Prikazchikova, T., Dontsova, O., and Zatsepin, T. (2020). Modification of Adenosine196 by Mettl3 Methyltransferase in the 5′-External Transcribed Spacer of 47S Pre-rRNA Affects rRNA Maturation. Cells 9:cells9041061. doi: 10.3390/cells9041061
Shah, A., Rashid, F., Awan, H. M., Hu, S., Wang, X., Chen, L., et al. (2017). The DEAD-Box RNA Helicase DDX3 Interacts with m(6)A RNA Demethylase ALKBH5. Stem Cells Int. 2017:8596135. doi: 10.1155/2017/8596135
Shi, H., Liu, C., Tan, H., Li, Y., Nguyen, T.-L. M., Dhungana, Y., et al. (2018). Hippo Kinases Mst1 and Mst2 Sense and Amplify IL-2R-STAT5 Signaling in Regulatory T Cells to Establish Stable Regulatory Activity. Immunity 49, 899.e–914.e. doi: 10.1016/j.immuni.2018.10.010
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27, 315–328. doi: 10.1038/cr.2017.15
Smith, P. P., and Gordon, C. (2010). Systemic lupus erythematosus: clinical presentations. Autoimmun. Rev. 10, 43–45. doi: 10.1016/j.autrev.2010.08.016
Sorci, M., Ianniello, Z., Cruciani, S., Larivera, S., Ginistrelli, L. C., Capuano, E., et al. (2018). METTL3 regulates WTAP protein homeostasis. Cell Death Dis. 9:796. doi: 10.1038/s41419-018-0843-z
Squires, J. E., Patel, H. R., Nousch, M., Sibbritt, T., Humphreys, D. T., Parker, B. J., et al. (2012). Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40, 5023–5033. doi: 10.1093/nar/gks144
Stojan, G., and Petri, M. (2018). Epidemiology of systemic lupus erythematosus: an update. Curr. Opin. Rheumatol. 30, 144–150. doi: 10.1097/BOR.0000000000000480
Svitkin, Y. V., Cheng, Y. M., Chakraborty, T., Presnyak, V., John, M., and Sonenberg, N. (2017). N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 45, 6023–6036. doi: 10.1093/nar/gkx135
Tan, B., and Gao, S.-J. (2018). RNA epitranscriptomics: Regulation of infection of RNA and DNA viruses by N(6) -methyladenosine (m(6) A). Rev. Med. Virol. 28:e1983. doi: 10.1002/rmv.1983
Tanaka, Y., O’Neill, S., Li, M., Tsai, I.-C., and Yang, Y.-W. (2020). Systemic Lupus Erythematosus: Targeted literature review of the epidemiology, current treatment and disease burden in the Asia Pacific region. Arthritis Care Res. 2020:24431. doi: 10.1002/acr.24431
Tang, L.-Y., Reddy, M. N., Rasheva, V., Lee, T.-L., Lin, M.-J., Hung, M.-S., et al. (2003). The eukaryotic DNMT2 genes encode a new class of cytosine-5 DNA methyltransferases. J. Biol. Chem. 278, 33613–33616. doi: 10.1074/jbc.C300255200
Tang, Y., Xie, H., Chen, J., Geng, L., Chen, H., Li, X., et al. (2013). Activated NF-κB in bone marrow mesenchymal stem cells from systemic lupus erythematosus patients inhibits osteogenic differentiation through downregulating Smad signaling. Stem Cells Dev. 22, 668–678. doi: 10.1089/scd.2012.0226
Tirumuru, N., Zhao, B. S., Lu, W., Lu, Z., He, C., and Wu, L. (2016). N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife 5:15528. doi: 10.7554/eLife.15528
Tong, J., Cao, G., Zhang, T., Sefik, E., Amezcua Vesely, M. C., Broughton, J. P., et al. (2018). m(6)A mRNA methylation sustains Treg suppressive functions. Cell Res. 28, 253–256. doi: 10.1038/cr.2018.7
Trixl, L., and Lusser, A. (2019). The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip. Rev. RNA 10:e1510. doi: 10.1002/wrna.1510
Tsokos, G. C., Lo, M. S., Costa Reis, P., and Sullivan, K. E. (2016). New insights into the immunopathogenesis of systemic lupus erythematosus. Nat. Rev. Rheumatol. 12, 716–730. doi: 10.1038/nrrheum.2016.186
Tuorto, F., Liebers, R., Musch, T., Schaefer, M., Hofmann, S., Kellner, S., et al. (2012). RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 19, 900–905. doi: 10.1038/nsmb.2357
Uzrail, A. H., Assaf, A. M., and Abdalla, S. S. (2019). Correlations of Expression Levels of a Panel of Genes (IRF5, STAT4, TNFSF4, MECP2, and TLR7) and Cytokine Levels (IL-2, IL-6, IL-10, IL-12, IFN-γ, and TNF-α) with Systemic Lupus Erythematosus Outcomes in Jordanian Patients. Biomed. Res. Int. 2019:1703842. doi: 10.1155/2019/1703842
van Tran, N., Ernst, F. G. M., Hawley, B. R., Zorbas, C., Ulryck, N., Hackert, P., et al. (2019). The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47, 7719–7733. doi: 10.1093/nar/gkz619
Vare, V. Y. P., Eruysal, E. R., Narendran, A., Sarachan, K. L., and Agris, P. F. (2017). Chemical and Conformational Diversity of Modified Nucleosides Affects tRNA Structure and Function. Biomolecules 7:biom7010029. doi: 10.3390/biom7010029
Wang, H., Hu, X., Huang, M., Liu, J., Gu, Y., Ma, L., et al. (2019). Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat. Commun. 10:1898. doi: 10.1038/s41467-019-09903-6
Wang, J., Yan, S., Lu, H., Wang, S., and Xu, D. (2019). METTL3 Attenuates LPS-Induced Inflammatory Response in Macrophages via NF-κB Signaling Pathway. Mediat. Inflamm. 2019:3120391. doi: 10.1155/2019/3120391
Wang, L., Wen, M., and Cao, X. (2019). Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science 365:aav0758. doi: 10.1126/science.aav0758
Wang, N., Tang, H., Wang, X., Wang, W., and Feng, J. (2017). Homocysteine upregulates interleukin-17A expression via NSun2-mediated RNA methylation in T lymphocytes. Biochem. Biophys. Res. Commun. 493, 94–99. doi: 10.1016/j.bbrc.2017.09.069
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. doi: 10.1016/j.cell.2015.05.014
Wang, Y., Wang, Y., Luo, W., Song, X., Huang, L., Xiao, J., et al. (2020). Roles of long non-coding RNAs and emerging RNA-binding proteins in innate antiviral responses. Theranostics 10, 9407–9424. doi: 10.7150/thno.48520
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 69, 1028.e–1038.e. doi: 10.1016/j.molcel.2018.02.015
Williams, G. D., Gokhale, N. S., and Horner, S. M. (2019). Regulation of Viral Infection by the RNA Modification N6-Methyladenosine. Annu. Rev. Virol. 6, 235–253. doi: 10.1146/annurev-virology-092818-015559
Winkler, R., Gillis, E., Lasman, L., Safra, M., Geula, S., Soyris, C., et al. (2019a). m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 20, 173–182. doi: 10.1038/s41590-018-0275-z
Winkler, R., Gillis, E., Lasman, L., Safra, M., Geula, S., Soyris, C., et al. (2019b). Publisher Correction: m6A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 20:243. doi: 10.1038/s41590-019-0314-4
Wojtas, M. N., Pandey, R. R., Mendel, M., Homolka, D., Sachidanandam, R., and Pillai, R. S. (2017). Regulation of m(6)A Transcripts by the 3′→5′ RNA Helicase YTHDC2 Is Essential for a Successful Meiotic Program in the Mammalian Germline. Mol. Cell 68, 374.e–387.e. doi: 10.1016/j.molcel.2017.09.021
Wu, B., Li, L., Huang, Y., Ma, J., and Min, J. (2017). Readers, writers and erasers of N(6)-methylated adenosine modification. Curr. Opin. Struct. Biol. 47, 67–76. doi: 10.1016/j.sbi.2017.05.011
Wu, H., and Zhang, Y. (2011). Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 25, 2436–2452. doi: 10.1101/gad.179184.111
Wu, H., Huang, X., Qiu, H., Zhao, M., Liao, W., Yuan, S., et al. (2016). High salt promotes autoimmunity by TET2-induced DNA demethylation and driving the differentiation of Tfh cells. Sci. Rep. 6:28065. doi: 10.1038/srep28065
Wu, Y., Cai, B., Zhang, J., Shen, B., Huang, Z., Tan, C., et al. (2017). IL-1β and IL-6 Are Highly Expressed in RF+IgE+ Systemic Lupus Erythematous Subtype. J. Immunol. Res. 2017:5096741. doi: 10.1155/2017/5096741
Wu, Y., Xie, L., Wang, M., Xiong, Q., Guo, Y., Liang, Y., et al. (2018). Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat. Commun. 9:4772. doi: 10.1038/s41467-018-06898-4
Xiao, G., and Zuo, X. (2016). Epigenetics in systemic lupus erythematosus. Biomed. Rep. 4, 135–139. doi: 10.3892/br.2015.556
Xiao, Z. X., Hu, X., Zhang, X., Chen, Z., Wang, J., Jin, K., et al. (2020). High salt diet accelerates the progression of murine lupus through dendritic cells via the p38 MAPK and STAT1 signaling pathways. Signal Transduct. Target. Ther. 5:34. doi: 10.1038/s41392-020-0139-5
Xing, M., Liu, Q., Mao, C., Zeng, H., Zhang, X., Zhao, S., et al. (2020). The 18S rRNA m(6) A methyltransferase METTL5 promotes mouse embryonic stem cell differentiation. EMBO Rep. 21:e49863. doi: 10.15252/embr.201949863
Xue, C., Zhao, Y., and Li, L. (2020). Advances in RNA cytosine-5 methylation: detection, regulatory mechanisms, biological functions and links to cancer. Biomark. Res. 8:43. doi: 10.1186/s40364-020-00225-0
Xue, M., Shi, Q., Zheng, L., Li, Q., Yang, L., and Zhang, Y. (2020). Gene signatures of m5C regulators may predict prognoses of patients with head and neck squamous cell carcinoma. Am. J. Transl. Res. 12, 6841–6852.
Yan, G., Yuan, Y., He, M., Gong, R., Lei, H., Zhou, H., et al. (2020). m(6)A Methylation of Precursor-miR-320/RUNX2 Controls Osteogenic Potential of Bone Marrow-Derived Mesenchymal Stem Cells. Mol. Ther. Nucleic Acids 19, 421–436. doi: 10.1016/j.omtn.2019.12.001
Yang, S., Wu, F., Peng, S., Wang, F., Chen, Y., Yuan, Y., et al. (2020). A m(6) A Sensing Method by Its Impact on the Stability of RNA Double Helix. Chem. Biodivers. 17:e2000050. doi: 10.1002/cbdv.202000050
Yang, X., Yang, Y., Sun, B.-F., Chen, Y.-S., Xu, J.-W., Lai, W.-Y., et al. (2017). 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 27, 606–625. doi: 10.1038/cr.2017.55
Ye, F. (2017). RNA N(6)-adenosine methylation (m(6)A) steers epitranscriptomic control of herpesvirus replication. Inflamm. Cell Signal. 4:e1604.
Yu, R., Li, Q., Feng, Z., Cai, L., and Xu, Q. (2019). m6A Reader YTHDF2 Regulates LPS-Induced Inflammatory Response. Int. J. Mol. Sci. 20:ijms20061323. doi: 10.3390/ijms20061323
Yue, Y., Liu, J., and He, C. (2015). RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355. doi: 10.1101/gad.262766.115
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4:10. doi: 10.1038/s41421-018-0019-0
Zhang, C., Fu, J., and Zhou, Y. (2019a). A Review in Research Progress Concerning m6A Methylation and Immunoregulation. Front. Immunol. 10:922. doi: 10.3389/fimmu.2019.00922
Zhang, Y., Gu, X., Li, D., Cai, L., and Xu, Q. (2019b). METTL3 Regulates Osteoblast Differentiation and Inflammatory Response via Smad Signaling and MAPK Signaling. Int. J. Mol. Sci. 21:ijms21010199. doi: 10.3390/ijms21010199
Zhang, Y., Wang, X., Zhang, X., Wang, J., Ma, Y., Zhang, L., et al. (2019c). RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc. Natl. Acad. Sci. U S A. 116, 976–981. doi: 10.1073/pnas.1812536116
Zhang, Z., Theler, D., Kaminska, K. H., Hiller, M., de la Grange, P., Pudimat, R., et al. (2010). The YTH domain is a novel RNA binding domain. J. Biol. Chem. 285, 14701–14710. doi: 10.1074/jbc.M110.104711
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C.-M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. doi: 10.1016/j.molcel.2012.10.015
Zheng, Q., Hou, J., Zhou, Y., Li, Z., and Cao, X. (2017). The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 18, 1094–1103. doi: 10.1038/ni.3830
Keywords: RNA methylation, SLE, m6A, m5C, immune
Citation: Lv X, Liu X, Zhao M, Wu H, Zhang W, Lu Q and Chen X (2021) RNA Methylation in Systemic Lupus Erythematosus. Front. Cell Dev. Biol. 9:696559. doi: 10.3389/fcell.2021.696559
Received: 17 April 2021; Accepted: 28 May 2021;
Published: 07 July 2021.
Edited by:
Wesley H. Brooks, University of South Florida, United StatesReviewed by:
Hong Zan, The University of Texas Health Science Center at San Antonio, United StatesMelek Kechida, Hôpital Universitaire Fattouma Bourguiba, Tunisia
Aijing Liu, Second Hospital of Hebei Medical University, China
Copyright © 2021 Lv, Liu, Zhao, Wu, Zhang, Lu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangmei Chen, eG1jaGVuMzAxQDEyNi5jb20=; Qianjin Lu, cWlhbmx1NTg2MEBjc3UuZWR1LmNu
†ORCID: Xinyi Lv, orcid.org/0000-0002-6939-4293; Xiaomin Liu, orcid.org/0000-0003-1306-5447
 Xinyi Lv
Xinyi Lv Xiaomin Liu
Xiaomin Liu Ming Zhao1
Ming Zhao1 Wuiguang Zhang
Wuiguang Zhang Xiangmei Chen
Xiangmei Chen