- 1Department of Gastrointestinal Surgery, Yuncheng Central Hospital, Yuncheng, China
- 2Department of Hepatopancreatobiliary Surgery, Yuncheng Central Hospital, Yuncheng, China
- 3Department of Pediatrics, Yuncheng Central Hospital, Yuncheng, China
- 4Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Center for Cancer Bioinformatics, Peking University Cancer Hospital and Institute, Beijing, China
- 5Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Gastrointestinal Cancer Center, Peking University Cancer Hospital and Institute, Beijing, China
MUC4, a transmembrane mucin, plays important roles in epithelial renewal and differentiation. Recent studies suggest that MUC4 has been implicated in pancreatic cancer pathogenesis and is expressed in various normal and cancer tissues. The underlying features of MUC4 across various cancer types may allow us to ensure appropriate treatment and patient monitoring. However, the contributions of MUC4 to pan-cancer have not been well characterized. In this study, we investigated the expression pattern and prognostic value of MUC4 across multiple databases. We further explored genomic and epigenetic alterations of MUC4, its association with proliferation and metastasis, and the correlation with immune infiltration in different cancers. Our results characterized the distinct expression profile and prognostic values of MUC4 in pan-cancer. Through examining its association with genomic alteration, tumor proliferation, and metastasis, as well as tumor infiltration, we revealed multiple function effects of MUC4. MUC4 may influence prognosis, proliferation, metastasis, and immune response in opposite directions. In conclusion, our findings suggested the necessity to more carefully evaluate MUC4 as a biomarker and therapeutic target and develop the new antibodies for cancer detection and intervention.
Introduction
In general, mucins are a family of glycosylated proteins, which are expressed by epithelial cells and provide protection and lubrication to epithelial surfaces (Kufe, 2009). However, aberrant expression of mucins occurs in various cancers and has been implicated in cancer progression and prognosis (Bhatia et al., 2019).
MUC4, a transmembrane mucin, is localized on chromosome band 3q29. Human MUC4 was first identified in 1991 from a tracheal library (Carraway et al., 2009). MUC4 is synthesized as two subunits: MUC4α and MUC4β. MUC4α contains a tandem-repeat domain altering glycosylation and epitope multiplicity, a nidogen-like domain, and an adhesion-associated domain. MUC4β consists of a von Willebrand factor-type D domain and three epidermal growth factor (EGF)-like domains. On the basis of the specific structure, MUC4 was suggested to modulate HER2/ERBB2 signaling and play a critical role in cancer.
Normally, MUC4 is expressed in the salivary glands, trachea and bronchioles, reproductive tract, colon, and mammary epithelium. In the past few years, many studies have reported that MUC4 is aberrantly produced in a variety of cancers, including lung, breast, pancreatic, prostate, ovarian, and bladder, and functionally links to tumor initiation, metastasis, and interaction of tumor cells with the components of the tumor microenvironment. The available evidence indicates that MUC4 is overexpressed in pancreatic cancer and contributes to the aggressiveness and metastasis of pancreatic cancer (Gautam et al., 2020; Sagar et al., 2021). MUC4 is also overexpressed in ovarian cancer and promotes the pathobiology and aggressiveness of ovarian cancer cells (Ponnusamy et al., 2011; Bae et al., 2017). MUC4 also plays a pivotal role in intestinal cell proliferation during tumorigenesis (Das et al., 2016). Rowson-Hodel et al. have provided considerable evidence that aberrantly expressed MUC4 can lead to the metastatic efficiency of breast cancer (Rowson-Hodel et al., 2018). In contrast, MUC4 expression was associated with improved survival and decreased recurrence in squamous cell carcinoma of the upper aerodigestive tract (Weed et al., 2004). These results suggested that the role of MUC4 appears to be complicated depending on the particular cancer and cell context. Thus, the expression and function of MUC4 in human tumors remain unclear and need to be analyzed in detail.
In our study, we conducted a comprehensive and profound bioinformatics analysis of MUC4 expression and correlation with prognosis and immune infiltration in cancer patients through Oncomine (Rhodes et al., 2004), TIMER2.0 (Li et al., 2020), PrognoScan (Mizuno et al., 2009), Kaplan–Meier plotter (Nagy et al., 2021), GEPIA2 (Tang et al., 2019), UALCAN (Chandrashekar et al., 2017), TISIDB (Ru et al., 2019), cBioPortal (Gao et al., 2013), and CVCDAP (Guan et al., 2020). Our findings may elucidate its significant function in cancer pathogenesis and prospective uses in cancer diagnosis and prognosis and as a target for cancer immunotherapy.
Materials and Methods
Differential Expression Analysis
We compared mRNA expression levels of MUC4 between normal and tumor tissues of each cancer type using Oncomine and GEPIA2. In Oncomine, a t-test was used to calculate the p-value, and the threshold was set as a p-value of 0.0001 and a fold change of 2. In GEPIA2, we used analysis of variance (ANOVA) for differential expression analysis and considered genes as differentially expressed genes with a fold change >2 and an FDR < 0.01. We also used GEPIA2 to assess the differential expression between different stages across independent cancer types by t-test and defined p-value <0.05 as significant.
Survival Analysis
We performed overall survival analysis based on MUC4 expression using PrognoScan and Kaplan–Meier Plotter. PrognoScan employed a univariate Cox regression model to find the optimal cut point in continuous gene expression measurement without prior biological knowledge or assumption and calculate the minimum p-value and hazard ratios with 95% confidence intervals for grouping patients. The Kaplan–Meier plotter split all patients into high- or low-expression groups according to the median value of MUC4 and used the log-rank test for hypothesis testing. We also used cBioPortal to evaluate the prognosis of MUC4 alteration. p-value <0.05 was considered significant.
Characterize the Alteration and Methylation Profile in Pan-Cancer
We explored the genomic alteration frequency including non-synonymous mutation and copy number alteration of MUC4 in the TCGA PanCancer Atlas dataset via cBioPortal. We obtained the promoter DNA methylation level of normal and tumor samples from UALCAN. Significance of difference was estimated by t-test. A p-value under 0.05 was considered statistically significant.
Construct a PPI Network and Estimate the Associations Between MUC4 and Tumor Proliferation and Metastasis
We carried out PPI analysis with STRING, and the max number of interactions to show was 10. The proliferation marker ki67 was used to reflect tumor proliferation across tumor samples. We used CVCDAP to evaluate the correlation between MUC4 and MKI67 by Pearson’s correlation and considered |R| > 0.3 and p-value <0.05 to indicate significance. We also used CVCDAP to divide tumor samples into a high and low group by the median expression value of MUC4 for each cancer type and calculate the epithelial–mesenchymal transition (EMT) enrichment score by GSEA. The gene set of EMT was collected from MSigDB. FDR < 0.25 was set as the threshold for screening.
Correlation Between MUC4 and Immune Infiltration
We employed TIMER2.0 for correlation between MUC4 and tumor immune infiltration. Via TIMER2.0, we analyzed the correlation between MUC4 and six immune cells, including CD8+ and CD4+ T cells, B cells, macrophages, neutrophils, and dendritic cells, in KIRC and PAAD. We also explored the relationships between MUC4 and immune gene markers in KIRC and PAAD. The association was generated with tumor purification adjusted. We used TISIDB to calculate the correlation between expression of MUC4 and abundance of immunomodulators and chemokines. The correlation was statistically assessed by Spearman’s correlation. p-value <0.05 was considered significant.
Results
Differential Expression of MUC4
To understand the differences in MUC4 expression between human cancer and normal tissues, MUC4 expression was explored via Oncomine. Our results revealed that MUC4 expression is upregulated or downregulated in different types of cancer (Figure 1A). Compared to normal tissues, the expression of MUC4 was significantly higher in bladder cancer, cervical cancer, lung cancer, and pancreatic cancer. In contrast, the expression of MUC4 was lower in colorectal cancer, head and neck cancer, prostate cancer. Of interest, MUC4 was upregulated in two datasets while it was downregulated in four datasets in kidney cancer. Similarly, MUC4 was upregulated in one dataset while it was downregulated in one dataset in sarcoma.
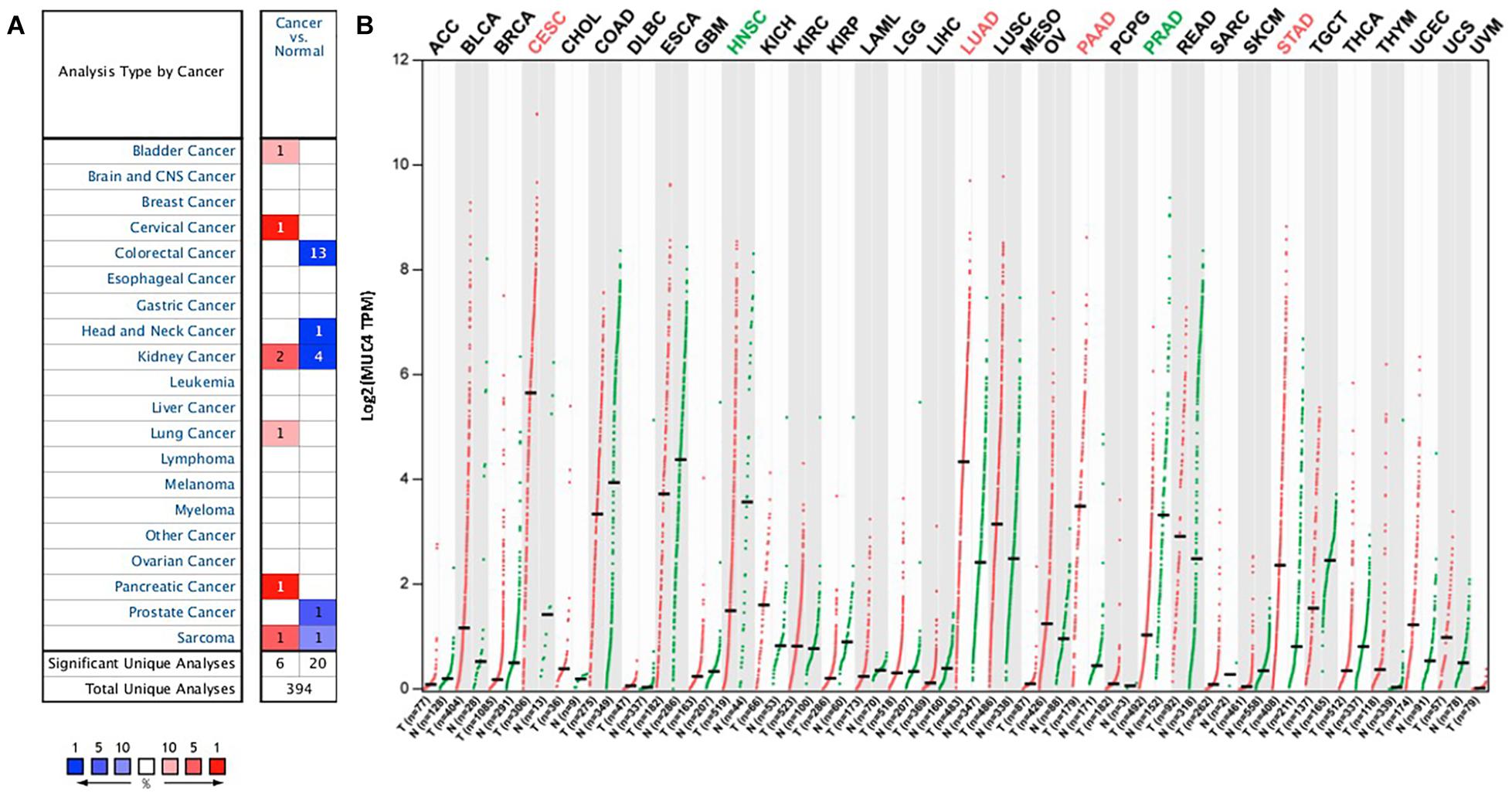
Figure 1. MUC4 mRNA expression levels in pan-cancer. (A) Upregulated or downregulated expression of MUC4 in tumor tissues, compared with normal tissues of each cancer type in Oncomine. Red signifies the gene’s overexpression in the analyses represented by that cell in the table; blue represents the gene’s underexpression in those analyses. Intensity of color signifies the best rank of that gene in those analyses. The number in each cell represents the number of analyses that meet your thresholds within those analyses and cancer types. (B) MUC4 expression profile across all tumor samples and paired normal tissues in GEPIA2. Each dot represents a distinct tumor or normal sample.
We further confirmed the differential gene expression between tumor samples and normal tissues using TCGA datasets in GEPIA2 (Figure 1B). Compared to normal tissues, the expression of MUC4 was significantly higher in CESC, LUAD, PAAD, and STAD, while the expression of MUC4 was lower in HNSC and PRAD.
Prognostic Analysis of MUC4
In cancer research, the relevance of MUC4 to clinical outcome may suggest the potential pathogenesis of disease and stimulate further researches. The impact of MUC4 on overall survival was evaluated through PrognoScan (Figure 2A). The results showed that high expression of MUC4 was associated with a poor prognosis in ovarian cancer (HR = 1.14, p = 0.0335), brain cancer (HR = 1.42, p = 0.0312), and lung cancer (HR = 1.25, p = 0.0335). However, in breast cancer (HR = 0.83, p = 0.0014) and colorectal cancer (HR = 0.02, p = 0.0456), increased expression of MUC4 was significantly correlated with good survival.

Figure 2. Overall survival curves comparing the high and low expression of MUC4 and pathological stage plot across different cancer types. (A,B) Kaplan–Meier plot in PrognoScan panel (A) and Kaplan–Meier Plotter panel (B). The X-axis represents time, and the Y-axis represents survival rate. Ninety-five percent confidence intervals for each group are also indicated by dotted lines from PrognoScan. (C) Expression violin plot based on patient pathological stage in GEPIA2. HR, hazard ratio.
In addition, we examined the potential effects of MUC4 on prognosis across different cancer types via the Kaplan–Meier plotter (Figure 2B). Our study revealed that the poor prognosis of ovarian cancer (HR = 1.42, 95% CI: 1.02–1.72, p = 0.0312), pancreatic ductal adenocarcinoma (HR = 1.55, 95% CI: 1.02–2.36, p = 0.039), thymoma (HR = 4.58, 95% CI: 0.93–22.53, p = 0.041), and bladder cancer (HR = 1.36, 95% CI: 1.01–1.83, p = 0.04) was related to the high expression of MUC4. Furthermore, the increased expression of MUC4 was related with prolonged overall survival in kidney renal clear cell carcinoma (HR = 0.67, 95% CI: 0.5–0.91, p = 0.0099). These findings revealed that the expression of MUC4 has important significance in the prognosis in pan-cancer and can be used as a prognostic factor. To further understand the relevance of MUC4 expression in cancer, we used the TCGA database to study the relationship between MUC4 expression and pathological stage via GEPIA2 (Figure 2C). The MUC4 expression profile observed in KIRC and TGCT may suggest a link between the level of MUC4 and the tumor stage.
Alteration Frequency and Methylation Level of MUC4
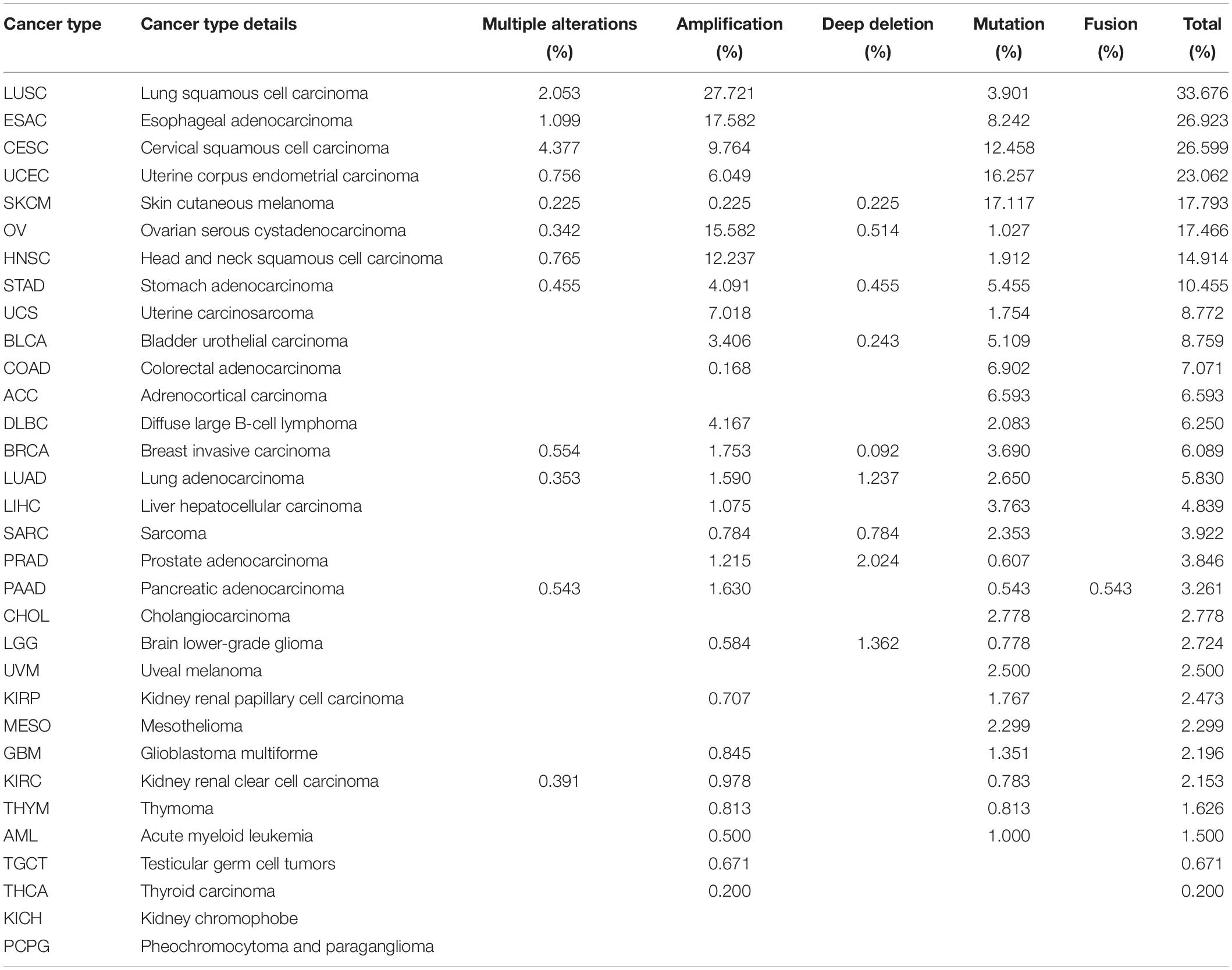
To identify the mechanism by which MUC4 impacts survival, we used cBioPortal to explore alteration frequencies, including mutation, fusion, amplification, deep deletion, and multiple alterations, of MUC4 in different cancer types (Figure 3A and Table 1). Results showed that the top five cancer types with more total mutations were LUSC (33.676%), ESCA (26.923%), CESC (26.599%), UCEC (23.062%), and SKCM (17.793%). For specific alteration types, amplifications of MUC4 were enriched in LUSC (27.721%), ESAC (17.582%), OV (15.582%), HNSC (12.237%), and CESC (9.764%), while deep deletions were enriched in PRAD (2.024%), LGG (1.362%), LUAD (1.237%), SARC (0.784%), and OV 0.514%). SKCM (17.117%), UCEC (16.257%), CESC (12.458%), ESCA (8.242%), and COAD (6.902%) were the top five cancer types with more mutation frequencies. We also found one MUC4-PCYT1A fusion in PAAD (0.543%). Based on these results, we further studied the correlation of MUC4 alteration with prognosis in top five cancer types and found the prognostic value of MUC4 alteration. The results are summarized in Figure 3B. Altered MUC4 is significantly associated with a good prognosis in LUSC (disease-specific survival, p = 6.8973-03) and UCEC (disease-free survival, p = 0.0107).

Figure 3. Genomic alteration and methylation of MUC4 across different cancer types. (A) Alteration frequency of MUC4 across different cancer types in cBioPortal. (B) The prognostic value of MUC4 alteration in LUSC and UCEC. (C) MUC4 promoter methylation level in tumor and normal samples of the TCGA database in UALCAN. The beta value indicates level of DNA methylation ranging from 0 (unmethylated) to 1 (fully methylated). Different beta value cutoffs have been considered to indicate hyper-methylation [beta value: 0.7–0.5] or hypo-methylation [beta-value: 0.3–0.25].
We wondered whether MUC4 was differentially methylated between tumor and normal samples, and we used UALCAN to compare their methylation level in dependent cancer types. We found that methylation levels were different between normal and tumor tissues in 16 cancer types (Figure 3C). In LUSC (p < 1e-12), ESCA (p = 1.864e-02), BRCA (p = 3.769e-12), LUAD (p = 1.248e-11), KIRP (p = 6.893e-09), PRAD (p = 1.624e-12), SARC (p < 1e-12), KIRC (p < 1e-12), LIHC (p < 1e-12), UCEC (p = 1.658e-12), TGCT (p < 1e-12), HNSC (p = 1.624e-12), BLCA (p = 1.934e-12), and PCPG (p < 1e-12), MUC4 was lowly methylated in tumor samples, while in READ (p = 2.688e-02) and COAD (p = 2.105e-03), MUC4 was highly methylated in tumor samples.
Functional Effects of MUC4 Associated With Proliferation and Metastasis
To suspect the network of predicted associations for MUC4 and proteins with 10 best-scoring hits, we performed the PPI analysis and found that there were interactions between MUC4 and MUC16 (score = 0.986), MUC1 (score = 0.986), MUC6 (score = 0.984), MUC20 (score = 0.979), MUC13 (score = 0.979), MUC7 (score = 0.978), MUC15 (score = 0.971), MUC21 (score = 0.964), GRLANT6 (score = 0.957), and B3GNT5 (score = 0.947) (Figure 4A). All these proteins are critical in O-glycan processing, maintaining the gastrointestinal epithelium, and regulation of cell adhesion.
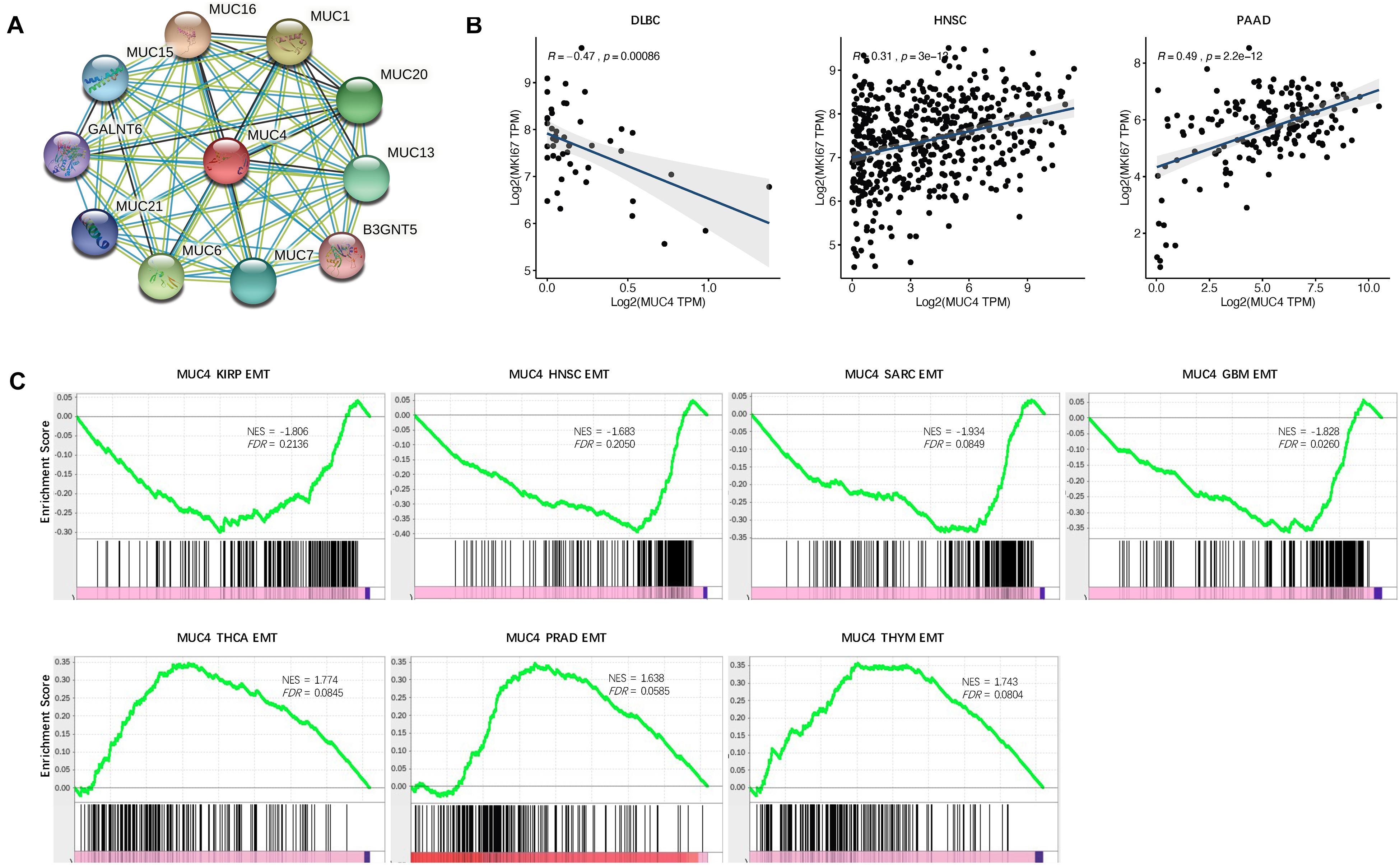
Figure 4. Functional effects of MUC4. (A) PPI network. Network nodes represent proteins and edges represent protein–protein associations, including both functional and physical protein associations. Line thickness indicates the strength of data support. (B) Correlation of MUC4 with proliferation in CVCDAP. (C) Enrichment of MUC4 in EMT in CVCDAP. Values of NES denote positive and negative enrichment. EMT, epithelial–mesenchymal transition.
Cell proliferation is one of the significant hallmarks of cancer. To characterize the functional roles of MUC4 in cell proliferation, we calculated the Rs between MUC4 and the well-known proliferation marker ki67 across cancer types. We identified a total of three significant associations (Figure 4B). MUC4 negatively correlated with cell proliferation in DLBC (R = −0.47, p = 0.00086) and positively correlated with cell proliferation in HMSC (R = 0.31, p = 3e-13) and PAAD (R = 0.49, p = 2.2e-12).
Metastasis is the major cause of death among cancer patients. Recent studies have heralded that EMT plays a critical role in metastasis (Heerboth et al., 2015; Mittal, 2018). To investigate the functional roles of MUC4 in metastasis, we assessed their enrichment associated with EMT through GSEA. We identified seven significant MUC4 enrichments (FDR < 0.25) (Figure 4C). MUC4 was negatively enriched in four cancer types, including KIRP (NES = −1.806, FDR = 0.2136), HNSC (NES = −1.683, FDR = 0.205), SARC (NES = −1.934, FDR = 0.0849), and GBM (NES = −1.828, FDR = 0.026). In contrast, MUC4 was positively enriched in three cancer types, including THCA (NES = 1.774, FDR = 0.0845), PRAD (NES = 1.638, FDR = 0.0585), and THYM (NES = 1.743, FDR = 0.0804).
Correlation Between MUC4 and Immune Infiltration Level
Given the distinctive roles of MUC4 in immunomodulation during cancer progression and metastasis (Yang et al., 2020; Peng et al., 2021), we used TIMER2.0 to investigate the impact of the expression of MUC4 on tumor immune infiltration levels. The detailed results are shown in Figure 5. According to the TIMER2.0 results, we identified MUC4 expression has weak positive relevance with tumor purity and the immune-infiltrating levels of CD4+ T cells (R = 0.147, p = 1.55e-03) and B cells (R = 0.119, p = 1.07e-02) but a negative correlation with dendritic cells (R = −0.127, p = 6.19e-03) in KIRC. In PAAD, MUC4 expression has significant positive correlations with the immune-infiltrating levels of B cells (R = 0.27, p = 3.6e-04) but no significant correlation with the infiltrating levels of CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells.
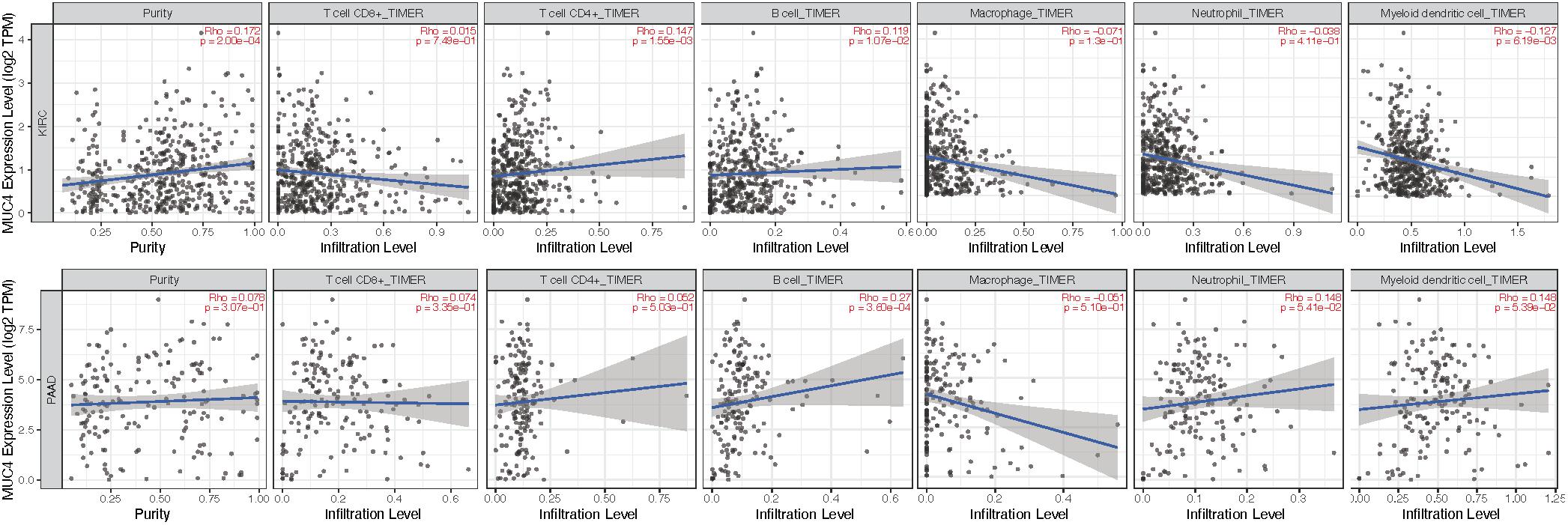
Figure 5. Coefficients for correlation between MUC4 and immune cell infiltration of KIRC and PAAD in TIMER2.0.
Moreover, we assessed the relevance between MUC4 and immune marker genes to clarify the mechanism of MUC4 in immune regulation in cancers (Figure 6). After adjusting the correlations by purity, we found that MUC4 expression has a positive correlation with NOS2 (M1) (R = 0.201, p = 1.39e-05) and MRC1 (M2) (R = 0.127, p = 6.18e-03) but negative relevance to CD19 (B cell) (R = −0.117, p = 1.17e-02), CD86 (tumor-associated macrophages, TAM) (R = −0.132, p = 4.41e-03), ROS1 (M1) (R = −0.121, p = 9.2e-03), and CD14 (monocyte) (R = −0.188, p = 5.04e-05) in KIRC. MUC4 expression in PAAD has a weak to moderate positive correlation with the expression of gene marker sets of TAM (HLA-G) (R = 0.174, p = 2.27e-02) and M1 (ROS1) (R = 0.219, p = 3.92e-03) but a negative correlation with M2 (ARG1) (R = −0.159, p = 3.78e-02). We also found that the relevance of MUC4 expression to other gene marker sets was not significant, such as B cell (CD19, MS4A1, CD38), TAM (CD80 and CD86), M1 (NOS2), M2 (MRC1), and monocyte (CD14 and FCGR3B). We further studied the correlations between MUC4 and three kinds of immunomodulators and chemokines in UALCAN. The detailed results are described in Figure 7.
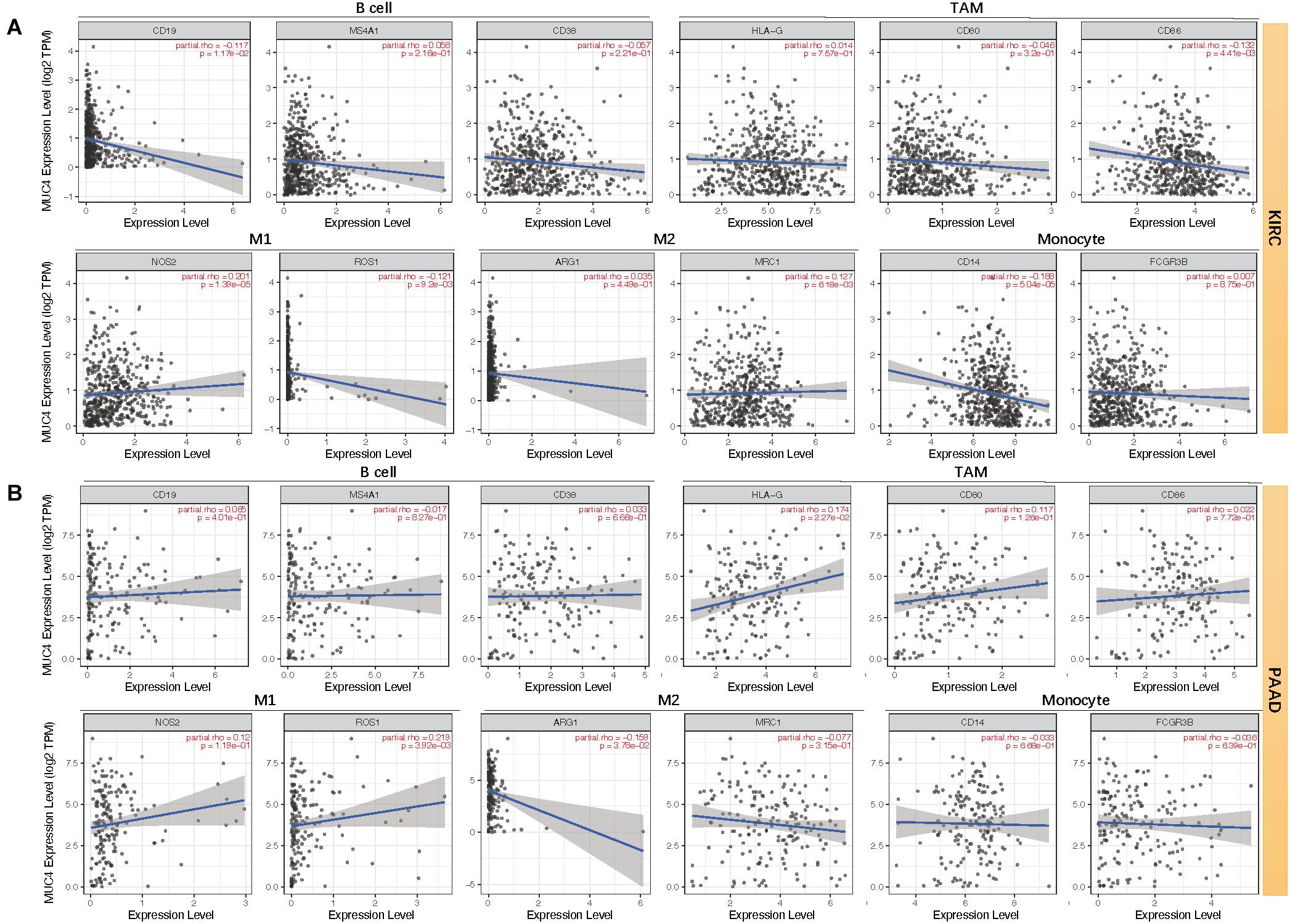
Figure 6. Correlation between mRNA expression of MUC4 and immune markers of KIRC (A) and PAAD (B) in TIMER2.0. TAM, tumor-associated macrophages; M1, type-1 macrophage; M2, type-2 macrophage.
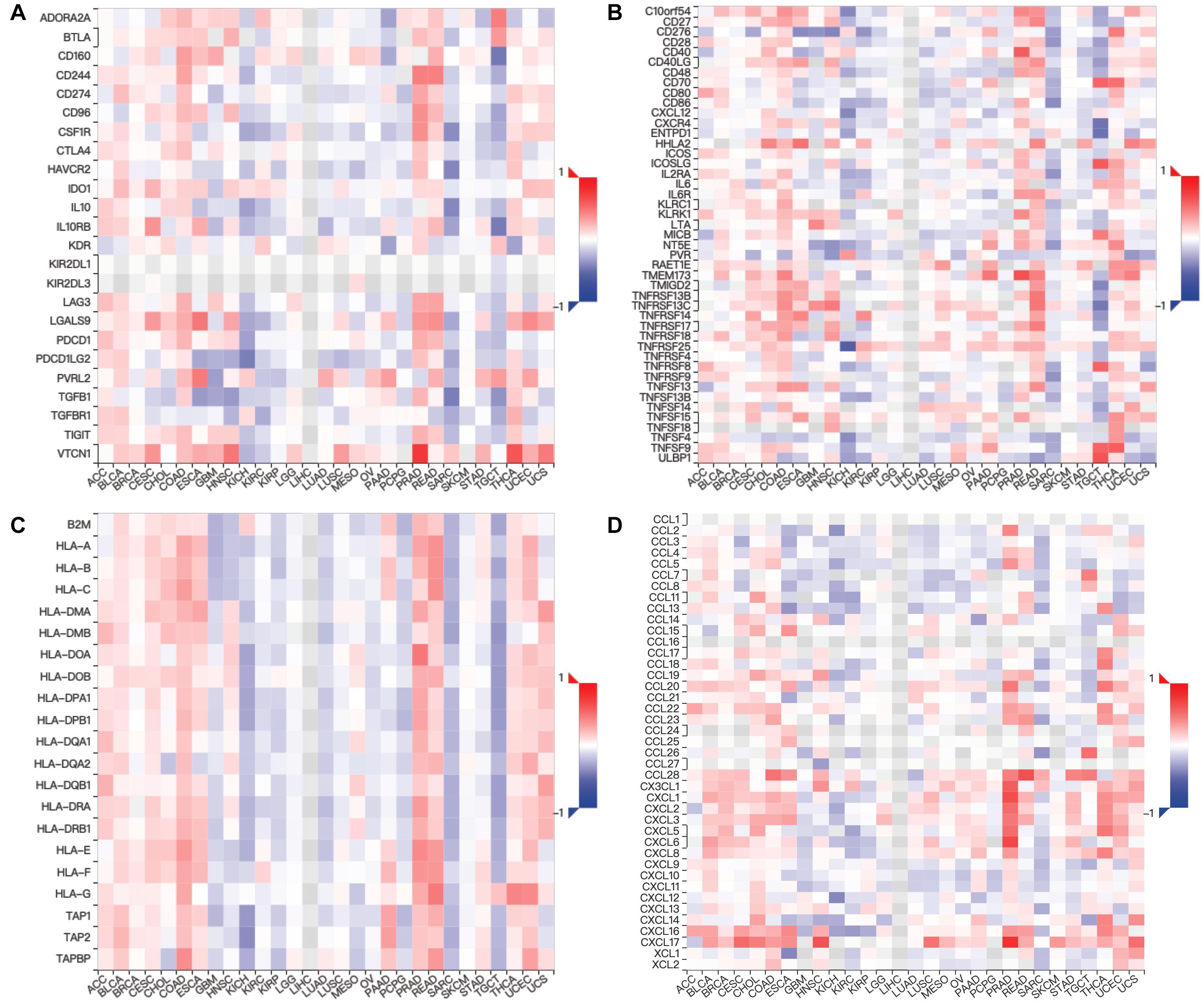
Figure 7. Relations between expression of MUC4 and three kinds of immunomodulators and chemokines in TISIDB. Spearman correlations between expression of MUC4 and immunoinhibitors (A), immunostimulators (B), MHCs (C), and chemokines (D) across human cancers.
Discussion
Past research has shown the critical roles of mucins in tumorigenesis (van Putten and Strijbis, 2017; Maeda et al., 2018; Ganguly et al., 2020; Liu et al., 2020), which indicated their prospective usefulness in cancer diagnosis, prognosis, and therapy (Nabavinia et al., 2017; Aithal et al., 2018; Guo et al., 2018; Lucchetta et al., 2019; Pothuraju et al., 2020). Previous studies have raised hope that MUC4 can be a good candidate marker for pancreatic malignancy (Trabbic et al., 2019; Gautam et al., 2020). MUC4 could also be used in combination with MUC16 for detection of advanced ovarian cancer (Chauhan et al., 2006). However, it remains unclear whether MUC4 can be characterized as a friend or foe across the cancer types (Jonckheere and Van Seuningen, 2018). To address this issue, we performed an integrative analysis about MUC4 to understand its effect on survival and immunomodulation, which is necessary to develop the MUC4-based cancer therapy. Through a comprehensive analysis in large-scale datasets, we identified consistent expression levels of MUC4 in pan-cancer using independent datasets via Oncomine and GEPIA2, which showed that MUC4 expression compared with normal samples was upregulated in cervical cancer, lung cancer, and pancreatic cancer, while it was downregulated in head and neck cancer and prostate cancer. We analyzed its prognostic significance statistically via PrognoScan and Kaplan–Meier plotter. Increased MUC4 expression was significantly correlated with prolonged survival time in breast cancer, colorectal cancer, and kidney renal cell carcinoma, while it was associated with poor survival in ovarian cancer, brain cancer, lung cancer, pancreatic cancer, thymoma, bladder carcinoma. Overall, these findings strongly suggest that MUC4 can serve as a prognostic biomarker in pan-cancer.
Furthermore, we explored the mechanism by which MUC4 influences prognosis. We depicted global alterations and epigenetic regulation of MUC4 across multiple cancer types, which showed that genomic alteration was an unfavorable factor for overall survival in LUSC and UCEC. As we expected, alteration in mRNA level and genomic level may influence tumor-malignant traits through different mechanisms. A previous study showed a high correlation between hypomethylation status and mRNA expression MUC4, and patients with MUC4 hypomethylation correlated with poor prognosis in pancreatic cancer (Yokoyama et al., 2016). Yamada et al. reported that the mRNA expression of MUC4 negatively correlated with its DNA methylation status at promoter regions in human lung cancer cell lines (Yamada et al., 2009), which was consistent with our results.
The effects on protein network, tumor growth, and metastasis were also concerned. Highly expressed MUC4 was correlated with MKI67 expression and negatively enriched with EMT in HNSC, which was consistent with its expression profile. In HNSC, upregulated MUC4 expression was enriched with EMT, which was identical with its poor prognosis. In addition, we focused on the functional roles on the tumor-associated microenvironment, especially in KIRC and PAAD. Our results demonstrated that the correlation with immune gene markers was not always the same as the overall trend (the relationships with immune cells). For example, in PAAD, B cells significantly correlated with MUC4 expression; however, gene markers of B cell had no relation to MUC4 expression. These discrepant implications on immune response and survival need further experiments for validation.
When considered together, our data demonstrated that MUC4 expression and promoter methylation status are potential prognosis biomarkers for lung cancer. MUC4 can be explored in pancreatic cancer as an early diagnostic tool. Thus, these findings in our study may provide new avenues for understanding the biological characterization of MUC4 and make sense in the design of therapeutic strategies.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author Contributions
X-PG and XG led the bioinformatic and biostatistical data analysis. X-PG and J-JD collected the literature, wrote the manuscript, and made the figures. XG and TX edited and made significant revisions to the manuscript. XG contributed to the study design and project supervision. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Research Seed Funds from Yuncheng Central Hospital and fellowship of China Postdoctoral Science Foundation (2021M692873).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aithal, A., Rauth, S., Kshirsagar, P., Shah, A., Lakshmanan, I., Junker, W. M., et al. (2018). MUC16 as a novel target for cancer therapy. Expert Opin. Ther. Targets 22, 675–686.
Bae, J. S., Lee, J., Park, Y., Park, K., Kim, J. R., Cho, D. H., et al. (2017). Attenuation of MUC4 potentiates the anticancer activity of auranofin via regulation of the Her2/Akt/FOXO3 pathway in ovarian cancer cells. Oncol. Rep. 38, 2417–2425. doi: 10.3892/or.2017.5853
Bhatia, R., Gautam, S. K., Cannon, A., Thompson, C., Hall, B. R., Aithal, A., et al. (2019). Cancer-associated mucins: role in immune modulation and metastasis. Cancer Metastasis Rev. 38, 223–236. doi: 10.1007/s10555-018-09775-0
Carraway, K. L., Theodoropoulos, G., Kozloski, G. A., and Carothers Carraway, C. A. (2009). Muc4/MUC4 functions and regulation in cancer. Future Oncol. 5, 1631–1640. doi: 10.2217/fon.09.125
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B., et al. (2017). UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19, 649–658. doi: 10.1016/j.neo.2017.05.002
Chauhan, S. C., Singh, A. P., Ruiz, F., Johansson, S. L., Jain, M., Smith, L. M., et al. (2006). Aberrant expression of MUC4 in ovarian carcinoma: diagnostic significance alone and in combination with MUC1 and MUC16 (CA125). Mod. Pathol. 19, 1386–1394. doi: 10.1038/modpathol.3800646
Das, S., Rachagani, S., Sheinin, Y., Smith, L. M., Gurumurthy, C. B., Roy, H. K., et al. (2016). Mice deficient in Muc4 are resistant to experimental colitis and colitis-associated colorectal cancer. Oncogene 35, 2645–2654. doi: 10.1038/onc.2015.327
Ganguly, K., Rauth, S., Marimuthu, S., Kumar, S., and Batra, S. K. (2020). Unraveling mucin domains in cancer and metastasis: when protectors become predators. Cancer Metastasis Rev. 39, 647–659. doi: 10.1007/s10555-020-09896-5
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:l1.
Gautam, S. K., Kumar, S., Dam, V., Ghersi, D., Jain, M., and Batra, S. K. (2020). MUCIN-4 (MUC4) is a novel tumor antigen in pancreatic cancer immunotherapy. Semin. Immunol. 47:101391. doi: 10.1016/j.smim.2020.101391
Guan, X., Cai, M., Du, Y., Yang, E., Ji, J., and Wu, J. (2020). CVCDAP: an integrated platform for molecular and clinical analysis of cancer virtual cohorts. Nucleic Acids Res. 48, W463–W471.
Guo, M., You, C., and Dou, J. (2018). Role of transmembrane glycoprotein mucin 1 (MUC1) in various types of colorectal cancer and therapies: current research status and updates. Biomed. Pharmacother. 107, 1318–1325. doi: 10.1016/j.biopha.2018.08.109
Heerboth, S., Housman, G., Leary, M., Longacre, M., Byler, S., Lapinska, K., et al. (2015). EMT and tumor metastasis. Clin. Transl. Med. 4:6.
Jonckheere, N., and Van Seuningen, I. (2018). Integrative analysis of the cancer genome atlas and cancer cell lines encyclopedia large-scale genomic databases: MUC4/MUC16/MUC20 signature is associated with poor survival in human carcinomas. J. Transl. Med. 16:259.
Kufe, D. W. (2009). Mucins in cancer: function, prognosis and therapy. Nat. Rev. Cancer 9, 874–885. doi: 10.1038/nrc2761
Li, T., Fu, J., Zeng, Z., Cohen, D., Li, J., Chen, Q., et al. (2020). TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48, W509–W514.
Liu, F., Fu, J., Bergstrom, K., Shan, X., McDaniel, J. M., McGee, S., et al. (2020). Core 1-derived mucin-type O-glycosylation protects against spontaneous gastritis and gastric cancer. J. Exp. Med. 217:e20182325. doi: 10.1084/jem.20182325
Lucchetta, M., da Piedade, I., Mounir, M., Vabistsevits, M., Terkelsen, T., and Papaleo, E. (2019). Distinct signatures of lung cancer types: aberrant mucin O-glycosylation and compromised immune response. BMC Cancer 19:824. doi: 10.1186/s12885-019-5965-x
Maeda, T., Hiraki, M., Jin, C., Rajabi, H., Tagde, A., Alam, M., et al. (2018). MUC1-C induces PD-L1 and immune evasion in triple-negative breast cancer. Cancer Res. 78, 205–215. doi: 10.1158/0008-5472.can-17-1636
Mittal, V. (2018). Epithelial mesenchymal transition in tumor metastasis. Annu. Rev. Pathol. 13, 395–412.
Mizuno, H., Kitada, K., Nakai, K., and Sarai, A. (2009). PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genomics 2:18. doi: 10.1186/1755-8794-2-18
Nabavinia, M. S., Gholoobi, A., Charbgoo, F., Nabavinia, M., Ramezani, M., and Abnous, K. (2017). Anti-MUC1 aptamer: a potential opportunity for cancer treatment. Med. Res. Rev. 37, 1518–1539. doi: 10.1002/med.21462
Nagy, A., Munkacsy, G., and Gyorffy, B. (2021). Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 11:6047.
Peng, L., Li, Y., Gu, H., Xiang, L., Xiong, Y., Wang, R., et al. (2021). Mucin 4 mutation is associated with tumor mutation burden and promotes antitumor immunity in colon cancer patients. Aging 13, 9043–9055. doi: 10.18632/aging.202756
Ponnusamy, M. P., Seshacharyulu, P., Vaz, A., Dey, P., and Batra, S. K. (2011). MUC4 stabilizes HER2 expression and maintains the cancer stem cell population in ovarian cancer cells. J. Ovarian Res. 4:7. doi: 10.1186/1757-2215-4-7
Pothuraju, R., Rachagani, S., Krishn, S. R., Chaudhary, S., Nimmakayala, R. K., Siddiqui, J. A., et al. (2020). Molecular implications of MUC5AC-CD44 axis in colorectal cancer progression and chemoresistance. Mol. Cancer 19:37.
Rhodes, D. R., Yu, J., Shanker, K., Deshpande, N., Varambally, R., Ghosh, D., et al. (2004). ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6. doi: 10.1016/s1476-5586(04)80047-2
Rowson-Hodel, A. R., Wald, J. H., Hatakeyama, J., O’Neal, W. K., Stonebraker, J. R., VanderVorst, K., et al. (2018). Membrane mucin Muc4 promotes blood cell association with tumor cells and mediates efficient metastasis in a mouse model of breast cancer. Oncogene 37, 197–207. doi: 10.1038/onc.2017.327
Ru, B., Wong, C. N., Tong, Y., Zhong, J. Y., Zhong, S. S. W., Wu, W. C., et al. (2019). TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 35, 4200–4202. doi: 10.1093/bioinformatics/btz210
Sagar, S., Leiphrakpam, P. D., Thomas, D., McAndrews, K. L., Caffrey, T. C., Swanson, B. J., et al. (2021). MUC4 enhances gemcitabine resistance and malignant behaviour in pancreatic cancer cells expressing cancer-associated short O-glycans. Cancer Lett. 503, 91–102. doi: 10.1016/j.canlet.2021.01.015
Tang, Z., Kang, B., Li, C., Chen, T., and Zhang, Z. (2019). GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47, W556–W560.
Trabbic, K. R., Whalen, K., Abarca-Heideman, K., Xia, L., Temme, J. S., Edmondson, E. F., et al. (2019). A tumor-selective monoclonal antibody from immunization with a tumor-associated mucin glycopeptide. Sci. Rep. 9:5662.
van Putten, J. P. M., and Strijbis, K. (2017). Transmembrane mucins: signaling receptors at the intersection of inflammation and cancer. J. Innate Immun. 9, 281–299. doi: 10.1159/000453594
Weed, D. T., Gomez-Fernandez, C., Yasin, M., Hamilton-Nelson, K., Rodriguez, M., Zhang, J., et al. (2004). MUC4 and ErbB2 expression in squamous cell carcinoma of the upper aerodigestive tract: correlation with clinical outcomes. Laryngoscope 114(8 Pt 2 Suppl. 101), 1–32. doi: 10.1097/00005537-200408001-00001
Yamada, N., Nishida, Y., Tsutsumida, H., Goto, M., Higashi, M., Nomoto, M., et al. (2009). Promoter CpG methylation in cancer cells contributes to the regulation of MUC4. Br. J. Cancer 100, 344–351. doi: 10.1038/sj.bjc.6604845
Yang, Y., Zhang, J., Chen, Y., Xu, R., Zhao, Q., and Guo, W. (2020). MUC4, MUC16, and TTN genes mutation correlated with prognosis, and predicted tumor mutation burden and immunotherapy efficacy in gastric cancer and pan-cancer. Clin. Transl. Med. 10:e155.
Keywords: mucin, prognosis, tumor immune microenvironment, pan-cancer, bioinformatic analysis
Citation: Gao X-P, Dong J-J, Xie T and Guan X (2021) Integrative Analysis of MUC4 to Prognosis and Immune Infiltration in Pan-Cancer: Friend or Foe? Front. Cell Dev. Biol. 9:695544. doi: 10.3389/fcell.2021.695544
Received: 15 April 2021; Accepted: 07 June 2021;
Published: 16 July 2021.
Edited by:
Simone Patergnani, University of Ferrara, ItalyReviewed by:
Yanqiang Li, Boston Children’s Hospital and Harvard Medical School, United StatesChangzheng He, Capital Medical University, China
Copyright © 2021 Gao, Dong, Xie and Guan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoqing Guan, Z3VhbnhxMTk5MUBvdXRsb29rLmNvbQ==
 Xiao-Peng Gao1
Xiao-Peng Gao1 Xiaoqing Guan
Xiaoqing Guan