
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 26 July 2021
Sec. Epigenomics and Epigenetics
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.694673
This article is part of the Research TopicRNA Modifications and EpitranscriptomicsView all 30 articles
Colorectal cancer and gastric cancer are the most prevalent gastrointestinal malignancies worldwide, and early detection of these cancers is crucial to reduce their incidence and mortality. RNA methylation plays an important regulatory role in a variety of physiological activities, and it has drawn great attention in recent years. Methylated adenosine (A) modifications such as N6-methyladenosine (m6A), N1-methyladenosine (m1A), 2′-O-methyladenosine (Am), N6,2′-O-dimethyladenosine (m6Am), and N6,N6-dimethyladenosine (m62A) are typical epigenetic markers of RNA, and they are closely correlated to various diseases including cancer. Serum is a valuable source of biofluid for biomarker discovery, and determination of these adenosine modifications in human serum is desirable since they are emerging biomarkers for detection of diseases. In this work, a targeted quantitative analysis method using hydrophilic interaction liquid chromatography–tandem mass spectrometry (HILIC-MS/MS) was developed and utilized to analyze these methylated adenosine modifications in serum samples. The concentration differences between the healthy volunteers and cancer patients were evaluated by Mann–Whitney test, and receiver operator characteristic (ROC) curve analysis was performed to access the potential of these nucleosides as biomarkers. We demonstrated the presence of the m6Am in human serum for the first time, and we successfully quantified the concentrations of A, m6A, m1A, and m6Am in serum samples from 99 healthy controls, 51 colorectal cancer patients, and 27 gastric cancer patients. We found that the levels of m6A and m6Am in serum were both increased in colorectal cancer or gastric cancer patients, compared to that in healthy controls. These results indicate that m6A and m6Am in serum may act as potential biomarkers for early detection and prognosis of colorectal cancer and gastric cancer. In addition, the present work will stimulate investigations on the effects of adenosine methylation on the initiation and progression of colorectal cancer and gastric cancer.
Colorectal cancer and gastric cancer are two common malignancies that are the second and third causes of cancer-related deaths all over the world, respectively (Brenner et al., 2014; Siegel et al., 2017; Bray et al., 2018). The incidence and mortality of these two types of cancer have been rising due to the difficulties in early detection. Detection of these cancers in the early stage has the advantage of improving survival rate and reducing costs of patients. Currently, the screening and diagnosis of colorectal cancer and gastric cancer mainly depend on results of colonoscopy and gastroscopy, respectively, which are both invasive to patients, leading to low compliance. From this point of view, it is essential to discover novel non-invasive biomarkers for early detection of colorectal cancer and gastric cancer to elongate the survival time and decrease the pains of patients.
Posttranscriptional modifications of RNA play crucial regulatory roles in a number of physiological activities (Feinberg, 2018; Bates, 2020). Until now, more than 170 chemical modifications of RNA have been identified, and these modifications are involved in the regulation of RNA functions (Jonkhout et al., 2017; Ontiveros et al., 2019). Among these chemical modifications, RNA methylation modifications have attracted great attention since accumulating evidences have been obtained to confirm RNA methylation as a novel layer of epigenetic alteration. Although methylation modifications in RNA had been identified in the 1970s (Dubin and Taylor, 1975; Perry et al., 1975), understanding of the functions of RNA methylation was limited until the identification of regulatory proteins such as fat mass and obesity-associated protein (FTO) (Jia et al., 2011). Through their unique regulatory proteins including “writers,” “erasers,” and “readers,” RNA methylation modifications display distinct features associated with many key cellular functions such as splicing, stability, and translation of RNA (Boo and Kim, 2020; Shi et al., 2020). And it has been revealed that RNA methylation participates in the initiation and progression of a number of diseases including cancer (Fang et al., 2021).
As a typical product of RNA methylation, N6-methyladenosine (m6A) is the most predominant modification in mRNA (Chen et al., 2019). The level of m6A is dynamic and reversible, and this is ascribed to the regulation of m6A methyltransferases and demethylases (Dominissini et al., 2012; Meyer and Jaffrey, 2017). The aberrant level of m6A modification is closely related to tumorigenesis and development (Lan et al., 2019; Li et al., 2019; Zhang et al., 2019; Liu et al., 2020; Wang et al., 2020a, b; Jiang et al., 2021). In recent years, other methylated adenosine modifications such as N1-methyladenosine (m1A), 2′-O-methyladenosine (Am), N6,2′-O-dimethyladenosine (m6Am), and N6,N6-dimethyladenosine (m62A) have also been revealed to play critical roles in the pathogenesis of various cancers (Safra et al., 2017; Sendinc et al., 2019; Chen et al., 2020a; Dong and Cui, 2020; Alriquet et al., 2021). Therefore, these methylated adenosine modifications have great potential to be indicators for early detection of diseases.
Serum contains a large number of biomolecules, and it is a preferred body fluid in the realm of biomarker discovery (Djukovic et al., 2010; Chen et al., 2011; Huang et al., 2018; Ottosson et al., 2019; Stepien et al., 2021). In the past decades, multiple analytical approaches have been utilized for the analysis of modified nucleosides (Beale et al., 2018; Farrokhi et al., 2018; Pero-Gascon et al., 2018; Guo et al., 2019). Compared with other analytical techniques, liquid chromatography tandem mass spectrometry (LC–MS/MS) is more favored to be used for biomarker discovery due to its great advantages in selectivity, sensitivity, accuracy, and high throughput. The analysis time could be largely shortened when ultra-performance liquid chromatography (UPLC) was used, and thus, UPLC–MS/MS becomes the preferred method for the determination of large-scale clinical samples (Guo et al., 2016, 2018a,b, 2020; Chen et al., 2020b; Zheng et al., 2020; Su et al., 2021). In addition, hydrophilic interaction liquid chromatography (HILIC) has the advantages of higher sensitivity because the organic solvent-rich mobile phase is more volatile and can enhance desolvation and ionization efficiency in the ion source. In the present study, a fast, sensitive, simple, and reliable HILIC–MS/MS method for targeted quantitative detection of methylated adenosine modifications including A, m6A, m1A, Am, m6Am, and m62A in human serum was established. By using the developed method, we revealed the presence of m6Am in human serum for the first time and quantified A, m6A, m1A, and m6Am in serum from 51 colorectal cancer patients, 27 gastric cancer patients, and 99 healthy volunteers. In addition, we carried out statistical analysis to compare the differences of these nucleosides between cancer patients and healthy controls and evaluated the potential of these adenosine modifications as biomarkers for early detection of colorectal cancer and gastric cancer.
Chromatographic-grade methanol was obtained from J.T. Baker (Radnor, PA, United States). Acetonitrile of HPLC grade was bought from Merck KGaA (Darmstadt, Germany). A, m6A, m1A, Am, m6Am, m62A, 13C5-adenosine ([13C5]A), D3-N6-methyladenosine ([D3]m6A), D3-N1-methyladenosine ([D3]m1A), and D3-N6-2′-O-dimethyladenosine ([D3]m6Am were gained from Toronto Research Chemical (Toronto, ON, Canada). Acetic acid (CH3COOH) was purchased from Fluka (Muskegon, MI, United States). Ammonium acetate and malic acid were bought from Sigma Aldrich (St Louis, MO, United States). Water was purified by using a Milli-Q water purification apparatus (Millipore, Milford, MA, United States).
Targeted quantitative analyses of adenosine and methylated adenosine were carried out on an Acquity UPLC system (Waters, Milford, MA, United States) coupled with a 4000 QTRAP mass spectrometer (AB SCIEX, Foster City, CA, United States). A Waters BEH HILIC column (2.1 × 100 mm, 1.7 μm) was implemented for chromatographic separation at room temperature. Mass spectrometer was operated in electrospray ionization (ESI) positive ion mode, and data were acquired by using multiple-reaction monitoring (MRM) mode. Data acquisition and processing were controlled by Analyst 1.6.3 software.
This research was approved by the Ethics Committee of the Second Affiliated Hospital, Zhejiang University School of Medicine (SAHZU). A total of 99 healthy controls (mean age of 50.3 ± 7.8 years, range 33–73 years) without serious infections or cancers, 51 patients with colorectal cancer (mean age of 63.2 ± 10.6 years, range 28–79 years), and 27 patients with gastric cancer (mean age of 65.9 ± 8.5 years, range 48–80 years) were recruited. The information of volunteers is shown in Table 1. All the subjects with cancers were confirmed by pathologist and did not undergo any type of therapy. Before participation, informed consent was provided by all volunteers. Then, the serum samples were collected in the early morning and reserved at −80°C until analysis.
Serum samples of 100 μl were placed into a 1.5-ml centrifuge tube and spiked with 10 μl of isotope-labeled internal standards (IS) after being thawed in ice. In order to perfectly remove the protein, 330 μl of prechilled methanol/acetonitrile (2:1, v/v) was added, and the mixture was vortexed for 1 min. After being placed at −20°C for 2 h, the obtained mixture was centrifuged at 13,000 rpm at 4°C for 15 min. Subsequently, 352 μl of supernatant was evaporated to dryness under vacuum. Then, the dried samples were redissolved in 80 μl of acetonitrile/water (9:1, v/v). After vortexing for 10 s, ultrasonication for 15 s, and centrifuging at 13,000 rpm for 15 min at 4°C, in that order, 70 μl of the supernatant fraction was moved into the vial for HILIC–MS/MS detection.
The mobile phases were (A) H2O containing 10 mM ammonium acetate and 0.2% acetic acid and (B) acetonitrile containing 2 mM ammonium acetate, 0.2% acetic acid, and 0.05 mM malic acid. The desired sample separation was achieved by the optimized LC gradient program as follows: 0 min, 95% B; 3 min, 94% B; 3.5 min, 60% B; 5.5 min, 60% B; 6 min, 94% B; and 12.5 min, 94% B. The injection volume was 5 μl, and each sample was measured twice. The samples were maintained at 4°C. To minimize interference of the mass spectrometer, a switching valve was used, and the eluents from the column during 1.5–3.8 and 4.8–5.6 min were introduced into the ion source. The ion spray voltage was optimized as 5.5 kV. The ion source temperature (TEM) was set at 550°C. The pressure of ion source gas 1 (GS1) and ion source gas 2 (GS2) were both set at 50 psi. The curtain gas (CUR) was set at 40 psi. The dwell time was 45 ms for each ion transition.
For targeted quantitative analysis, the transitions of m/z 268.1→136.0 (A), m/z 282.1→150.0 (m6A and m1A), m/z 282.1→136.0 (Am), m/z 296.1→150.0 (m6Am), and m/z 296.1→164.0 (m62A) were monitored. For isotope-labeled internal standards, the transitions of m/z 273.1→136.0 ([13C5]A), m/z 285.1→153.0 ([D3]m6A and [D3]m1A), and m/z 299.1→153.0 ([D3]m6Am) were monitored. The optimized MRM parameters for these nucleosides including declustering potential (DP), entrance potential (EP), collision energy (CE), and collision cell exit potential (CXP) are listed in Supplementary Table 1.
In order to establish the calibration curves, different concentrations of the working solutions of nucleoside standards mixed with internal standards (IS) were made. The final concentrations of IS were as follows: [13C5]A 25 nM, [D3]m6A 2.5 nM, [D3]m1A 125 nM, and [D3]m6Am 1.25 nM. By measuring the peak area ratio of the nucleosides to the corresponding IS (y), against the concentration of the analyte (x), the linearities were constructed as y = ax + b. In addition, the limit of detection (LOD) and limit of quantification (LOQ) were obtained by analyzing the concentrations of the standard solutions with signal-to-noise ratios equal to 3 and 10, respectively.
For the purpose of evaluating extraction recovery, serum samples were spiked with standards at three different levels of A (1, 5, and 50 nM), m6A (1, 5, and 50 nM), m1A (10, 100, and 500 nM), and m6Am (0.1, 1, and 5 nM). After addition of the IS solution, the serum samples were processed and measured as mentioned above. The recovery (R) was estimated by using the following formula: recovery = (concentration in spiked sample − concentration in original sample)/spiked concentration × 100%.
In order to evaluate intra-day and inter-day precision, the quality control (QC) samples at three different levels of A (1, 10, and 75 nM), m6A (1, 10, and 75 nM), m1A (10, 250, and 750 nM), and m6Am (0.1, 2.5, and 20 nM) were measured on the same day and three consecutive days, respectively. The accuracy was acquired by comparing the obtained concentration to the theoretical value.
In consideration of matrix effect evaluation, a slope comparison method was applied. By adding standard solutions and IS to the serum and pure solvent, respectively, different calibration curves were subsequently established, and their slopes were compared. The slope ratio of the calibration curve established in serum to that in pure solvent was described as the matrix effect.
Statistical analyses were performed by utilizing the SPSS statistics 24.0 software (IBM, Armonk, NY, United States). The concentration differences between healthy volunteers and cancer patients were assessed by the Mann–Whitney test, and a p-value less than 0.05 was significant. Receiver operating characteristic (ROC) analysis was used to assess the ability of these nucleosides to discriminate cancer patients from healthy controls.
In order to achieve excellent chromatographic separation and obtain good chromatographic peak shape with appropriate retention, chromatographic conditions including the type of column and the composition of mobile phase were optimized. The chemical structures of adenosine and its methylated modifications are illustrated in Figure 1. Since m1A is positively charged, the retention of m1A on a C18 column is very weak. It is eluted from the C18 column quickly, and a symmetric peak shape is hard to obtain. As a complementary tool, HILIC is widely used for the separation of compounds with high polarity, and we found that m1A can be retained well on the HILIC column. Besides, a high proportion of organic solvent was used for HILIC separation, which could enhance the ionization efficiency of targets, leading to the improvement of detection sensitivity. Therefore, the BEH HILIC column (2.1 × 100 mm, 1.7 μm) was selected for subsequent analysis. In our previous study, we found that malic acid could enhance the detection of methylated nucleosides in HILIC–MS/MS (Guo et al., 2018a). Hence, malic acid was added into the mobile phase for the analysis of methylated adenosine modifications. As illustrated in Figure 2, the analysis can be finished within 6 min, and target nucleosides were perfectly separated, which suggested that the analytical method is quick, has high throughput, and is fit for large clinical samples.
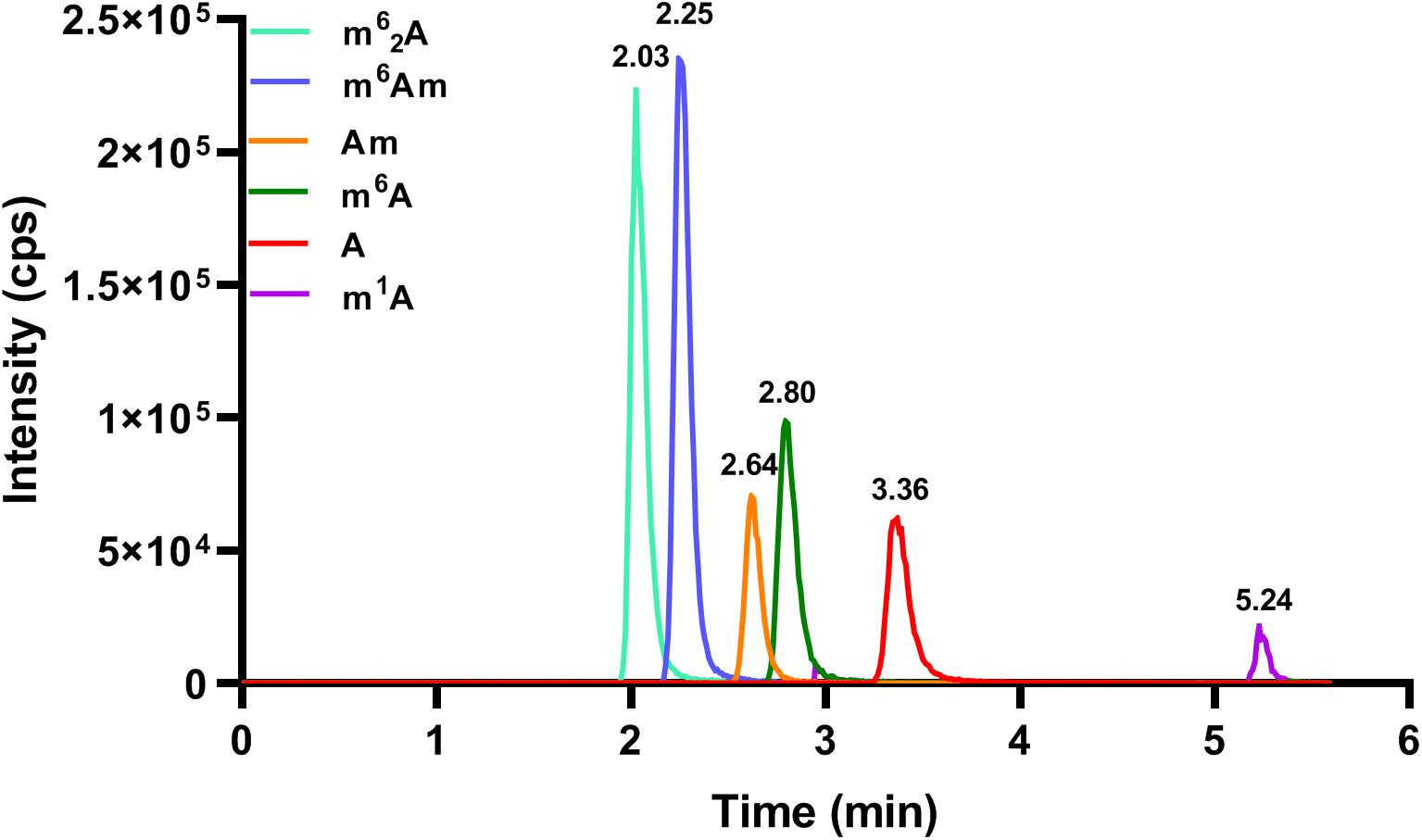
Figure 2. The MRM chromatograms of A, m6A, m1A, m6Am, Am, and m62A standards. The concentration of each nucleoside standard was 25 nM, and the injection volume was 5.0 μl.
To optimize the MRM parameters, a peristaltic pump was used, and the standard solution was directly infused into the mass spectrometer. For A, [M + H]+ ion at m/z 268.1 was observed in full-scan ESI-MS. Abundant [M + H]+ ions at m/z 282.1 were both observed for m6A and Am; for m6Am and m62A, [M + H]+ ions at m/z 296.1 were both observed; and for m1A, M+ ion at m/z 282.1 was observed. Then, we carried out collision-induced dissociation (CID) experiments. According to the results of CID, for m6A, m1A, and m6Am, the most abundant fragment ions were all at m/z 150.0. For A and Am, the most abundant fragment ions were both at m/z 136.0, and for m62A, the most abundant fragment ion was at m/z 164.0. From this point of view, the ion transitions m/z 268.1→136.0 (A), m/z 282.1→150.0 (m6A and m1A), m/z 282.1→136.0 (Am), m/z 296.1→150.0 (m6Am), and m/z 296.1→164.0 (m62A) were utilized for quantitative determination. Similarly, the ion transitions m/z 273.1→136.0 ([13C5]A), m/z 285.1→153.0 ([D3]m6A and [D3]m1A), and m/z 299.1→153.0 ([D3]m6Am) were monitored. Additionally, mass spectrometry parameters containing DP, EP, CE, CXP, ion source temperature (TEM), ion spray voltage, ion source gas 1 (GS1), ion source gas 2 (GS2), and curtain gas (CUR) were optimized to enhance the sensitivity and are summarized in Supplementary Table 1. Under these optimized conditions, the limit of detection (LOD) value can reach 0.0025 nM (A), 0.01 nM (m6A), 0.25 nM (m1A), and 0.01 nM (m6Am), which are lower than the LOD values reported before (Djukovic et al., 2010; Chen et al., 2011; Guo et al., 2019), indicating that the analytical method has admirable sensitivity.
For method validation, we investigated the linearity, recovery, LOD, LOQ, intra-day and inter-day precision, and matrix effects. As shown in Table 2, the calibration curve of each analyte showed excellent linearity (R2 > 0.999) within a wide analytical range. The data of LOD and the LOQ of the analytes are also illustrated in Table 2, which indicated that the sensitivity of the developed method was superb. As for the matrix effect, calibration curves were also constructed in serum extracts and in pure solvent. The slope ratio values for A, m6A, m1A, and m6Am were 92.5, 96.9, 98.5, and 93.3%, respectively, which implied that the matrix had no interferences in this study.
As shown in Supplementary Table 3, recoveries ranged from 98.9 to 109.4%, indicating that satisfactory recovery was obtained. As shown in Supplementary Table 4, the intra-day precision value was within 6.0%, and the inter-day precision value was within 8.8%. The accuracy of the intra-day and inter-day analysis was in range of 89.0 to 105.5%. These data demonstrated that satisfactory reproducibility and accuracy were determined.
Moreover, to check the system stability after numbers of injections, QC samples were analyzed every 20 serum samples, and the accuracy and retention time were monitored. The results showed that the equipment system had an outstanding stability during measurement. In a word, all these results mentioned above revealed that the developed HILIC-MS/MS method is quick, sensitive, accurate, reliable, and reproducible. And it can meet the requirement for the targeted quantitative analysis of these nucleosides in serum samples.
By applying the developed HILIC-MS/MS method, we investigated the contents of these nucleosides in serum samples collected from 51 colorectal cancer patients, 27 gastric cancer patients, and 99 healthy controls. As demonstrated in Figure 3, the presence of A, m6A, m1A, and m6Am could be unquestionably confirmed in all serum samples since the retention time of these four nucleosides was identical to that of their corresponding internal standard, whereas Am and m62A were not detected in all the serum samples due to their trace amounts in serum. The levels of A, m6A, m1A, and m6Am in serum samples were calculated according to the calibration curves, and the detailed concentrations were presented in Supplementary Table 2. In summary, the concentrations of A, m6A, m1A, and m6Am in human serum ranged from 1.95 to 34.19, 2.24 to 9.74, 115.16 to 215.77, and 0.16 to 2.56 nM, respectively.
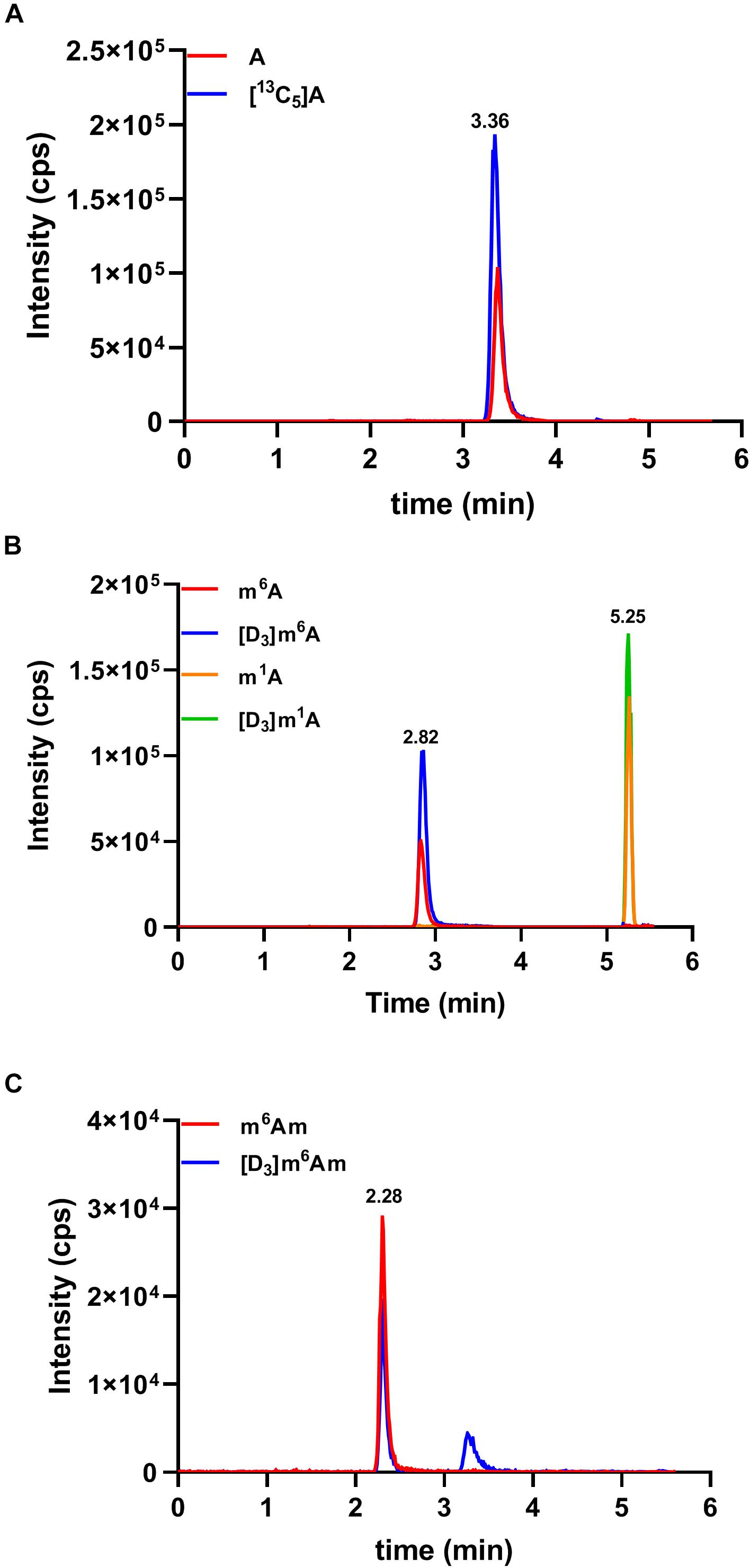
Figure 3. Representative MRM chromatograms of (A) A, (B) m6A and m1A, (C) m6Am, and spiked isotope-labeled internal standards in a serum sample.
We next evaluated whether there were differences in the concentrations of these nucleosides between healthy controls and cancer patients. In serum samples, the concentrations of A, m6A, m1A, and m6Am from healthy volunteers were in the range of 3.20–17.86, 2.24–7.73, 115.16–211.44, and 0.16–2.28 nM, respectively, and the average concentrations were 7.98 ± 3.01, 4.51 ± 1.08, 154.58 ± 21.10, and 0.63 ± 0.37 nM, respectively (n = 99). The concentrations of A, m6A, m1A, and m6Am in serum from patients with colorectal cancer were in the range of 2.41–27.94, 2.64–9.24, 117.45–215.77, and 0.17–2.56 nM, respectively, and the average concentrations were 8.47 ± 6.30, 5.57 ± 1.67, 158.62 ± 24.79, and 1.13 ± 0.53 nM, respectively (n = 51). For patients with gastric cancers, the concentrations of A, m6A, m1A, and m6Am in serum were in the range of 1.95–34.19, 4.94–9.74, 116.84–209.92, and 0.24–2.15 nM, respectively, and the average concentrations were 11.53 ± 8.55, 6.93 ± 1.38, 156.83 ± 27.07, and 0.91 ± 0.57 nM, respectively (n = 27). As illustrated in Figure 4, it is apparent that the levels of m6A and m6Am in serum were intensely increased in patients with colorectal cancer or gastric cancers, compared to those in healthy controls (colorectal cancer: p < 0.001 for m6A and p < 0.0001 for m6Am; gastric cancer: p < 0.0001 for m6A and p < 0.05 for m6Am). However, there was no difference in the levels of A and m1A between healthy controls and colorectal cancer or gastric cancer patients (p > 0.05).
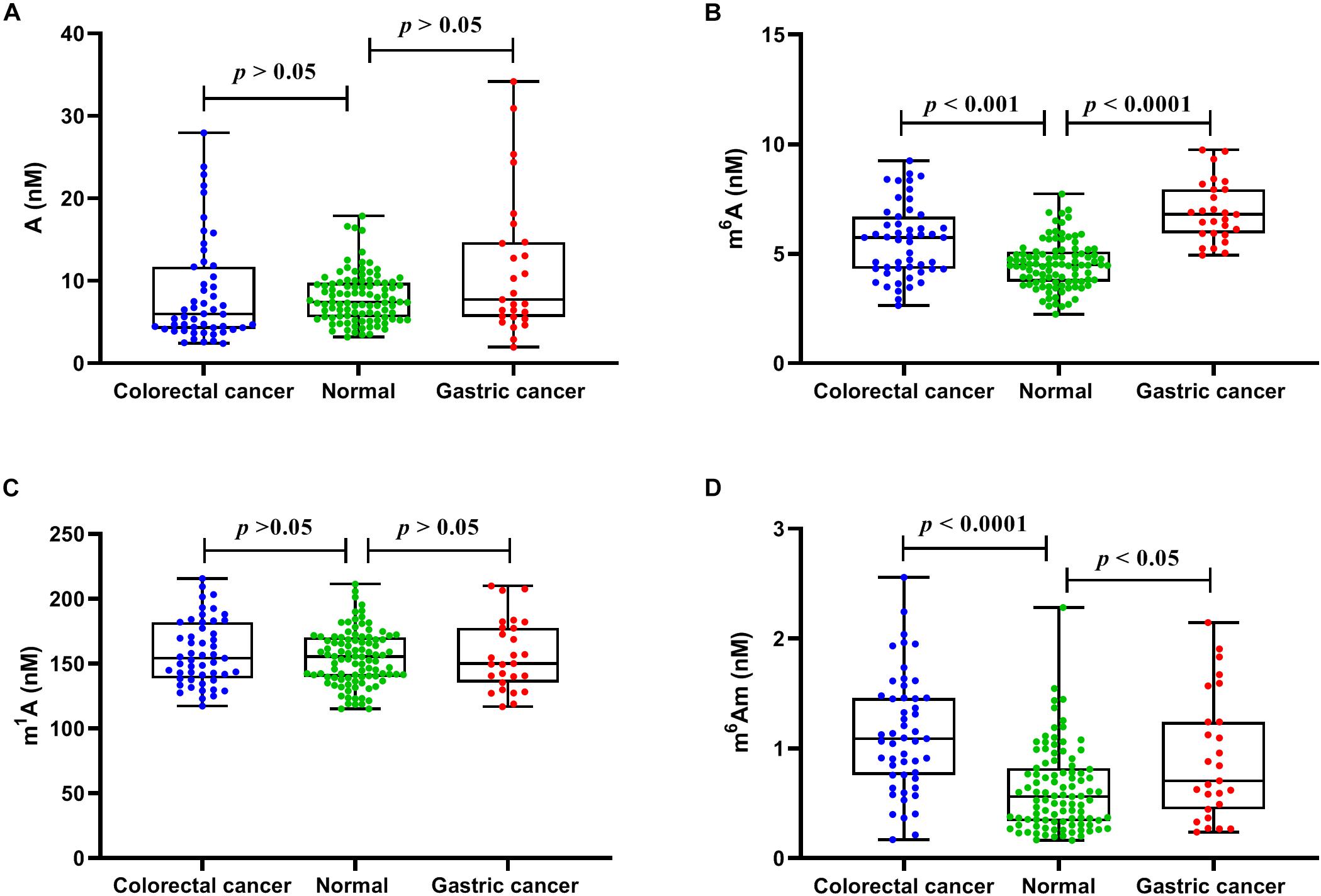
Figure 4. The measured concentrations of (A) A, (B) m6A, (C) m1A, and (D) m6Am in serum samples and statistical analysis.
Furthermore, ROC analysis was performed to evaluate the ability of these nucleosides to distinguish cancer patients from healthy controls. As demonstrated in Figure 5, for healthy volunteers and colorectal cancer patients, the area under the curve (AUC) for m6A and m6Am was 0.679 and 0.791, respectively. And for healthy controls and gastric cancer patients, the AUC was 0.931 and 0.647 for m6A and m6Am, respectively. These results indicated a correlation between the levels of m6A and m6Am in serum and the incidence of colorectal cancer and gastric cancer. Interestingly, we found that the AUC of m6Am was higher than that of m6A in colorectal cancer, whereas the AUC of m6A was higher than that of m6Am in gastric cancer, implying that m6Am and m6A were more effective indicators of colorectal cancer and gastric cancer, respectively. Colorectal cancer and gastric cancer are of a high incidence worldwide and has awfully high mortality, and early detection is extremely necessary. The results of this study revealed that the increase of m6A and m6Am in serum might have great potential to be novel non-invasive biomarkers for early detection of colorectal cancer and gastric cancer.
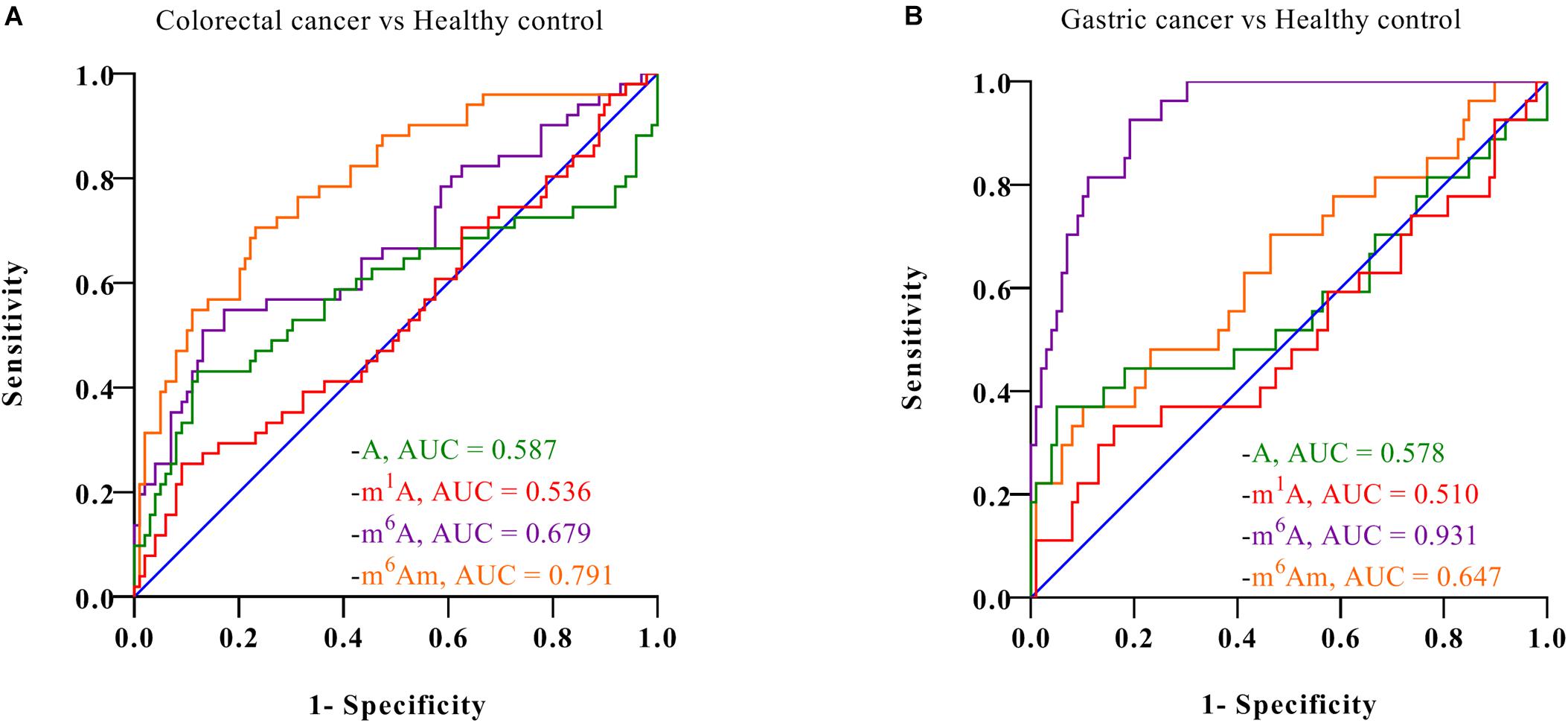
Figure 5. ROC analysis for (A) colorectal cancer vs. healthy controls and (B) gastric cancer vs. healthy controls.
In the present study, we developed a robust, sensitive, and trustworthy HILIC–MS/MS method for targeted quantitative analysis of A, m6A, m1A, Am, m6Am, and m62A in human serum. By applying the established method, we successfully revealed, for the first time, the presence of m6Am in human serum and quantified the concentrations of A, m6A, m1A, and m6Am in 177 serum samples from three groups, namely, healthy volunteers, colorectal cancer patients, and gastric cancer patients. We found that the amount of m6A and m6Am were significantly higher in colorectal cancer or gastric cancer patients than those in healthy volunteers. Our data indicated that m6A and m6Am may serve as potential non-invasive biomarkers for early detection of colorectal cancer and gastric cancer. Furthermore, these results suggested that these methylated adenosine modifications might play important regulatory roles in the pathogenesis and progression of cancer.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the Institutional Review Board of Medical Research, The Second Affiliated Hospital, Zhejiang University School of Medicine. The patients/participants provided their written informed consent to participate in this study.
CG and YY designed the study. YiH, CG, and ZF performed the experiments. ZF and JM collected the serum samples. YiH and CG analyzed and interpreted the data and wrote the manuscript. YaH and SZ edited the manuscript. All authors commented and approved the final manuscript.
This study was supported by the National Key R&D Program of China (2016YFC1302803), Key R&D Program of Zhejiang Province (2021C03125), Natural Science Foundation of Zhejiang Province (LY19B050007), and National Natural Science Foundation of China (21402172).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.694673/full#supplementary-material
Alriquet, M., Calloni, G., Martinez-Limon, A., Delli Ponti, R., Hanspach, G., Hengesbach, M., et al. (2021). The protective role of m1A during stress-induced granulation. J. Mol. Cell. Biol. 12, 870–880. doi: 10.1093/jmcb/mjaa023
Bates, S. E. (2020). Epigenetic therapies for cancer. N. Engl. J. Med. 383, 650–663. doi: 10.1056/NEJMra1805035
Beale, D. J., Pinu, F. R., Kouremenos, K. A., Poojary, M. M., Narayana, V. K., Boughton, B. A., et al. (2018). Review of recent developments in GC–MS approaches to metabolomics-based research. Metabolomics 14:152. doi: 10.1007/s11306-018-1449-2
Boo, S. H., and Kim, Y. K. (2020). The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 52, 400–408. doi: 10.1038/s12276-020-0407-z
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer. J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Brenner, H., Kloor, M., and Pox, C. P. (2014). Colorectal cancer. Lancet. 383, 1490–1502. doi: 10.1016/s0140-6736(13)61649-9
Chen, F., Xue, J., Zhou, L., Wu, S., and Chen, Z. (2011). Identification of serum biomarkers of hepatocarcinoma through liquid chromatography/mass spectrometry-based metabonomic method. Anal. Bioanal. Chem. 401, 1899–1904. doi: 10.1007/s00216-011-5245-3
Chen, H., Gu, L., Orellana, E. A., Wang, Y., Guo, J., Liu, Q., et al. (2020a). METTL4 is an snRNA m6Am methyltransferase that regulates RNA splicing. Cell. Res. 30, 544–547. doi: 10.1038/s41422-019-0270-4
Chen, Q., Hu, Y., Fang, Z., Ye, M., Li, J., Zhang, S., et al. (2020b). Elevated levels of oxidative nucleic acid modification markers in urine from gastric cancer patients: quantitative analysis by ultra performance liquid chromatography-tandem mass spectrometry. Front. Chem. 8:606495. doi: 10.3389/fchem.2020.606495
Chen, Y. G., Chen, R., Ahmad, S., Verma, R., Kasturi, S. P., Amaya, L., et al. (2019). N6-methyladenosine modification controls circular RNA immunity. Mol. Cell. 76, 96–109. doi: 10.1016/j.molcel.2019.07.016
Djukovic, D., Baniasadi, H. R., Kc, R., Hammoud, Z., and Raftery, D. (2010). Targeted serum metabolite profiling of nucleosides in esophageal adenocarcinoma. Rapid. Commun. Mass. Spectrom. 24, 3057–3062. doi: 10.1002/rcm.4739
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 485, 201–206. doi: 10.1038/nature11112
Dong, Z., and Cui, H. (2020). The emerging roles of RNA modifications in glioblastoma. Cancers. (Basel) 12:736. doi: 10.3390/cancers12030736
Dubin, D. T., and Taylor, R. H. (1975). The methylation state of poly a-containing messenger RNA from cultured hamster cells. Nucleic. Acids. Res. 2, 1653–1668. doi: 10.1093/nar/2.10.1653
Fang, Z., Hu, Y., Hu, J., Huang, Y., Zheng, S., and Guo, C. (2021). The crucial roles of N6-methyladenosine m6A modification in the carcinogenesis and progression of colorectal cancer. Cell Biosci 11:72. doi: 10.1186/s13578-021-00583-8
Farrokhi, V., Chen, X., and Neubert, H. (2018). Protein turnover measurements in human serum by serial immunoaffinity LC-MS/MS. Clin. Chem. 64, 279–288. doi: 10.1373/clinchem.2017.272922
Feinberg, A. P. (2018). The key role of epigenetics in human disease prevention and mitigation. N. Engl. J. Med. 378, 1323–1334. doi: 10.1056/NEJMra1402513
Guo, C., Chen, Q., Chen, J., Yu, J., Hu, Y., Zhang, S., et al. (2020). 8-Hydroxyguanosine as a possible RNA oxidative modification marker in urine from colorectal cancer patients: evaluation by ultra performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. B. Analyt. Technol. Biomed. Life. Sci. 1136:121931. doi: 10.1016/j.jchromb.2019.121931
Guo, C., Li, X., Wang, R., Yu, J., Ye, M., Mao, L., et al. (2016). Association between oxidative DNA damage and risk of colorectal cancer: sensitive determination of urinary 8-hydroxy-2′-deoxyguanosine by UPLC-MS/MS analysis. Sci. Rep. 6:32581. doi: 10.1038/srep32581
Guo, C., Xie, C., Chen, Q., Cao, X., Guo, M., Zheng, S., et al. (2018a). A novel malic acid-enhanced method for the analysis of 5-methyl-2′-deoxycytidine, 5-hydroxymethyl-2′-deoxycytidine, 5-methylcytidine and 5-hydroxymethylcytidine in human urine using hydrophilic interaction liquid chromatography-tandem mass spectrometry. Analytica. Chimica. Acta. 1034, 110–118. doi: 10.1016/j.aca.2018.06.081
Guo, C., Xie, C., Ding, P., Qin, G., Mo, W., Cao, X., et al. (2018b). Quantification of glycocholic acid in human serum by stable isotope dilution ultra performance liquid chromatography electrospray ionization tandem mass spectrometry. J. Chromatogr. B. Analyt. Technol. Biomed. Life. Sci. 1072, 315–319. doi: 10.1016/j.jchromb.2017.11.037
Guo, M., Liu, D., Sha, Q., Geng, H., Liang, J., and Tang, D. (2019). Succinic acid enhanced quantitative determination of blood modified nucleosides in the development of diabetic nephropathy based on hydrophilic interaction liquid chromatography mass spectrometry. J. Pharm. Biomed. Anal. 164, 309–316. doi: 10.1016/j.jpba.2018.10.042
Huang, J., Weinstein, S. J., Moore, S. C., Derkach, A., Hua, X., Liao, L. M., et al. (2018). Serum metabolomic profiling of all-cause mortality: a prospective analysis in the alpha-tocopherol, beta-carotene cancer prevention (ATBC) study cohort. Am. J. Epidemiol. 187, 1721–1732. doi: 10.1093/aje/kwy017
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi: 10.1038/nchembio.687
Jiang, X., Liu, B., Nie, Z., Duan, L., Xiong, Q., Jin, Z., et al. (2021). The role of m6A modification in the biological functions and diseases. Signal. Transduct. Target. Ther. 6:74. doi: 10.1038/s41392-020-00450-x
Jonkhout, N., Tran, J., Smith, M. A., Schonrock, N., Mattick, J. S., and Novoa, E. M. (2017). The RNA modification landscape in human disease. RNA. 23, 1754–1769. doi: 10.1261/rna.063503.117
Lan, Q., Liu, P. Y., Haase, J., Bell, J. L., Huttelmaier, S., and Liu, T. (2019). The critical role of RNA m6A methylation in cancer. Cancer. Res. 79, 1285–1292. doi: 10.1158/0008-5472.CAN-18-2965
Li, T., Hu, P. S., Zuo, Z., Lin, J. F., Li, X., Wu, Q. N., et al. (2019). METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer. 18:112. doi: 10.1186/s12943-019-1038-7
Liu, T., Yang, S., Sui, J., Xu, S. Y., Cheng, Y. P., Shen, B., et al. (2020). Dysregulated N6-methyladenosine methylation writer METTL3 contributes to the proliferation and migration of gastric cancer. J. Cell. Physiol. 235, 548–562. doi: 10.1002/jcp.28994
Meyer, K. D., and Jaffrey, S. R. (2017). Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell. Dev. Biol. 33, 319–342. doi: 10.1146/annurev-cellbio-100616-060758
Ontiveros, R. J., Stoute, J., and Liu, K. F. (2019). The chemical diversity of RNA modifications. Biochem. J. 476, 1227–1245. doi: 10.1042/BCJ20180445
Ottosson, F., Smith, E., Gallo, W., Fernandez, C., and Melander, O. (2019). Purine metabolites and carnitine biosynthesis intermediates are biomarkers for incident type 2 diabetes. J. Clin. Endocrinol. Metab. 104, 4921–4930. doi: 10.1210/jc.2019-00822
Pero-Gascon, R., Sanz-Nebot, V., Berezovski, M. V., and Benavente, F. (2018). Analysis of circulating microRNAs and their post-transcriptional modifications in cancer serum by on-line solid-phase extraction-capillary electrophoresis-mass spectrometry. Anal. Chem. 90, 6618–6625. doi: 10.1021/acs.analchem.8b00405
Perry, R. P., Kelley, D. E., Friderici, K., and Rottman, F. (1975). The methylated constituents of L cell messenger RNA: Evidence for an unusual cluster at the 5′ terminus. Cell. 4, 387–394. doi: 10.1016/0092-8674(75)90159-2
Safra, M., Sas-Chen, A., Nir, R., Winkler, R., Nachshon, A., Bar-Yaacov, D., et al. (2017). The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551, 251–255. doi: 10.1038/nature24456
Sendinc, E., Valle-Garcia, D., Dhall, A., Chen, H., Henriques, T., Navarrete-Perea, J., et al. (2019). PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Mol. Cell. 75, 620–630. doi: 10.1016/j.molcel.2019.05.030
Shi, H., Chai, P., Jia, R., and Fan, X. (2020). Novel insight into the regulatory roles of diverse RNA modifications: re-defining the bridge between transcription and translation. Mol. Cancer. 19:78. doi: 10.1186/s12943-020-01194-6
Siegel, R. L., Miller, K. D., Fedewa, S. A., Ahnen, D. J., Meester, R. G. S., Barzi, A., et al. (2017). Colorectal cancer statistics, 2017. CA. Cancer. J. Clin. 67, 177–193. doi: 10.3322/caac.21395
Stepien, M., Keski-Rahkonen, P., Kiss, A., Robinot, N., Duarte-Salles, T., Murphy, N., et al. (2021). Metabolic perturbations prior to hepatocellular carcinoma diagnosis: Findings from a prospective observational cohort study. Int. J. Cancer. 148, 609–625. doi: 10.1002/ijc.33236
Su, X., Li, X., Wang, H., and Cai, Z. (2021). Simultaneous determination of methionine cycle metabolites, urea cycle intermediates and polyamines in serum, urine and intestinal tissue by using UHPLC-MS/MS. Talanta 224:121868. doi: 10.1016/j.talanta.2020.121868
Wang, Q., Chen, C., Ding, Q., Zhao, Y., Wang, Z., Chen, J., et al. (2020a). METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 69, 1193–1205. doi: 10.1136/gutjnl-2019-319639
Wang, T., Kong, S., Tao, M., and Ju, S. (2020b). The potential role of RNA N6-methyladenosine in cancer progression. Mol. Cancer. 19:88. doi: 10.1186/s12943-020-01204-7
Zhang, Y., Kang, M., Zhang, B., Meng, F., Song, J., Kaneko, H., et al. (2019). m6A modification-mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5. Mol. Cancer. 18:185. doi: 10.1186/s12943-019-1116-x
Keywords: colorectal cancer, gastric cancer, RNA methylation, methylated adenosine, hydrophilic interaction liquid chromatography-tandem mass spectrometry, biomarker
Citation: Hu Y, Fang Z, Mu J, Huang Y, Zheng S, Yuan Y and Guo C (2021) Quantitative Analysis of Methylated Adenosine Modifications Revealed Increased Levels of N6-Methyladenosine (m6A) and N6,2′-O-Dimethyladenosine (m6Am) in Serum From Colorectal Cancer and Gastric Cancer Patients. Front. Cell Dev. Biol. 9:694673. doi: 10.3389/fcell.2021.694673
Received: 13 April 2021; Accepted: 21 June 2021;
Published: 26 July 2021.
Edited by:
Jia Meng, Xi’an Jiaotong-Liverpool University, ChinaReviewed by:
Tongxing Song, Huazhong Agricultural University, ChinaCopyright © 2021 Hu, Fang, Mu, Huang, Zheng, Yuan and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Yuan, eXVhbnlpbmcxOTk5QHpqdS5lZHUuY24=; Cheng Guo, Y2hlbmdfZ3VvQHpqdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.