- 1Department of Hepatobiliary Surgery, Affiliated Hospital of Guangdong Medical University, Zhanjiang, China
- 2Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences & Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, China
- 3Department of Hepatobiliary Surgery, The Second Affiliated Hospital of Guangdong Medical University, Zhanjiang, China
- 4Department of General Surgery, Dongguan Liaobu Hospital, Dongguan, China
- 5Center for Life Sciences, School of Life Sciences, Yunnan University, Kunming, China
The calcium binding protein S100 family in humans contains 21 known members, with each possessing a molecular weight between 10 and 14 kDa. These proteins are characterized by a unique helix-loop-helix EF hand motif, and often form dimers and multimers. The S100 family mainly exists in vertebrates and exerts its biological functions both inside cells as a calcium sensor/binding protein, as well as outside cells. S100A11, a member of the S100 family, may mediate signal transduction in response to internal or external stimuli and it plays various roles in different diseases such as cancers, metabolic disease, neurological diseases, and vascular calcification. In addition, it can function as chemotactic agent in inflammatory disease. In this review, we first detail the discovery of S100 proteins and their structural features, and then specifically focus on the tissue and organ expression of S100A11. We also summarize its biological activities and roles in different disease and signaling pathways, providing an overview of S100A11 research thus far.
Introduction
In 1965, in order to explore the unique proteins in the nervous system, American scientist Blake W. Moore, from Washington University School of Medicine, compared protein maps from brain and liver tissues of cattle. He found, through starch gel electrophoresis experiments, that one protein band migrated faster than the others, and that this protein band was only present in the brain tissue protein map (Moore, 1965; Moore and McGregor, 1965). After purification, it was found that this type of protein could be dissolved in a saturated ammonium sulfate solution, therefore this protein was named S100 protein. Subsequently, Blake W. Moore further confirmed that S100 protein was found in the brain tissues of 17 species including pigs, rats, mice, rabbits, hamsters, guinea pigs, dogs, monkeys, humans, turkeys, hawks, alligators, snakes, turtles, pompano fish, and red snapper. Data from rats demonstrated that the content of this protein in the brain is 1,000–10,000 times higher than in other tissues, hence the S100 protein was identified as a neural protein (Moore, 1965). Much later, it was shown that the S100 proteins identified by Moore were actually S100A1 and S100B, and that they were not only expressed in nervous tissue, but also were widely expressed in multiple tissues and cell types of vertebrates (Donato, 1999; Donato et al., 2009).
S100A11 is a member of the S100 protein family, and its cDNA cloning and protein purification was first done by Todoroki et al. (1991) in October, 1991. They reported that S100A11 has a molecular weight of 13 kDa, and possesses two EF-hand domains, which can bind calcium ions and undergo conformational changes. At the same time, they found the same protein in the aorta of cattle and the lungs of rabbits, and eventually named it calgizzarin. In December of the same year, Ohta et al. (1991) also isolated a protein with a molecular weight of about 11 kDa from the heart of pigs, and then cloned it. The cDNA was expressed in vitro, and subsequent amino acid sequence analysis showed that the protein had two EF-hand domains, with 40.9 and 37.5% homology with the known S100A1 and S100B proteins, respectively. The tissue expression pattern of this protein is apparently different from other known S100 proteins, which are highly expressed in lung and kidney tissues, but relatively low in liver and brain. Thus, they considered the protein to be a new S100 protein and named it S100C. Later, research found that calgizzarin and S100C are actually the same protein, and it is now collectively referred to as S100A11.
The Family Members of S100 Proteins and Their Structures
At present, a total of 21 S100 proteins have been identified in humans (Bresnick et al., 2015), with their molecular weight varying between 10 and 14 kDa. There are also 21 corresponding genes that constitute the S100 gene family. 14 of the 21 genes are located in the q21 region of chromosome 1 (Eckert et al., 2004). S100 proteins exhibit high similarity with conserved EF-hand domains and share extensive structural homology (Figure 1A). Most S100 proteins exist as homo or heterodimers, and can bind calcium ions through their two EF-hand domains (Donato, 2003), in which one is a non-canonical EF-hand in the N-terminus and the other is a canonical EF-hand in the C-terminus, respectively (Figure 1A). Once bound to calcium ions, the conformation of the S100 proteins changes, which consequently makes them available to bind to targeting proteins and perform their corresponding biological functions. For example, they can play multiple roles in buffering calcium ion concentration (S100G) or sensing and transmitting calcium ion signals (other S100 proteins), they can also regulate enzyme activity, participate in energy metabolism, or regulate cell proliferation and differentiation (Donato et al., 2013). In addition, some S100 proteins can also be secreted outside the cell in an autocrine or paracrine manner, thereby acting as a signaling molecule to activate their corresponding receptors to participate in innate and acquired immune responses (Donato et al., 2013).
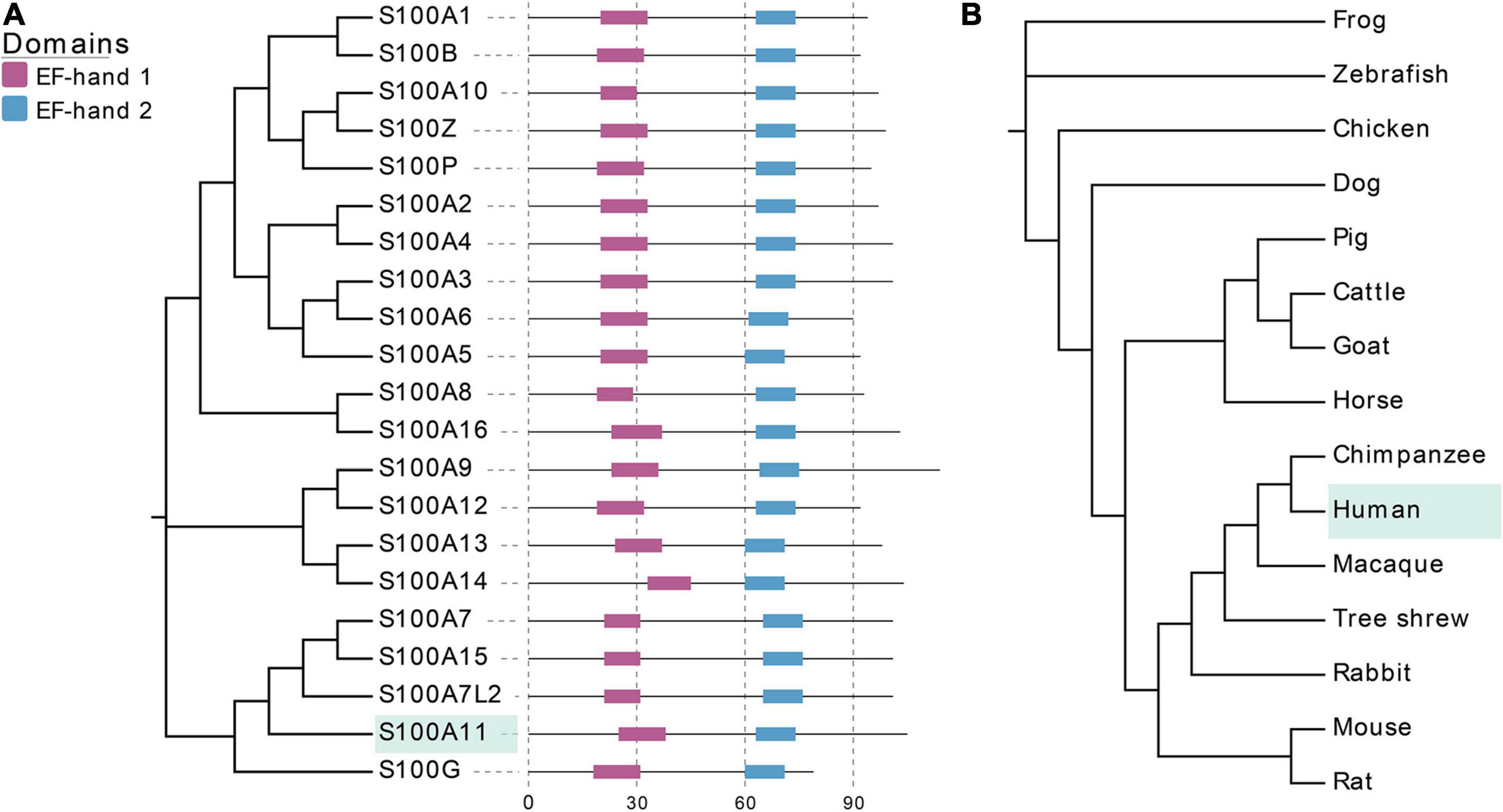
Figure 1. EF-hand domains of S100 family proteins and evolutionary relationships of S100A11 in different species. (A) The amino acid sequences of S100 proteins are from human, which can be found at UniProt (www.uniprot.org/). EF-hand 1 represents the non-canonical EF-hand, EF-hand 2 represents the canonical EF-hand. (B) The cladogram of CDS sequences of S100A11 gene in typical species of different families. The CDS sequences data are from NCBI (www.ncbi.nlm.nih.gov/).
Thus far, there is no evidence for S100 proteins being present in invertebrates, though the S100 proteins are highly conserved among the vertebrates. Kraemer et al. (2008) studied the structural and functional diversification of S100 proteins in the teleost fish, which has the earliest S100 genes in terms of evolution. They found that several mammalian S100 genes have counterparts in teleost. Using S100A11 as an example, an evolutionary tree was ranked from Osteichthyes to Amphibian, Aves, and Mammalia (Figure 1B). A CDS sequence comparison of human S100 genes shows that S100A11 has a higher sequence similarity with S100G and S100A7L2 than the other family members (Figure 1A). Like several other members of the family, S100A11 can also form a homodimer (Figure 2A), which was verified by Réty et al. (2000) and Hung et al. (2012) in pig and in human, respectively. The structure of S100A11 homodimer with calcium ions (Réty et al., 2000) is shown in Figure 2B, which exhibits the locations of calcium ions in the S100A11 structure.
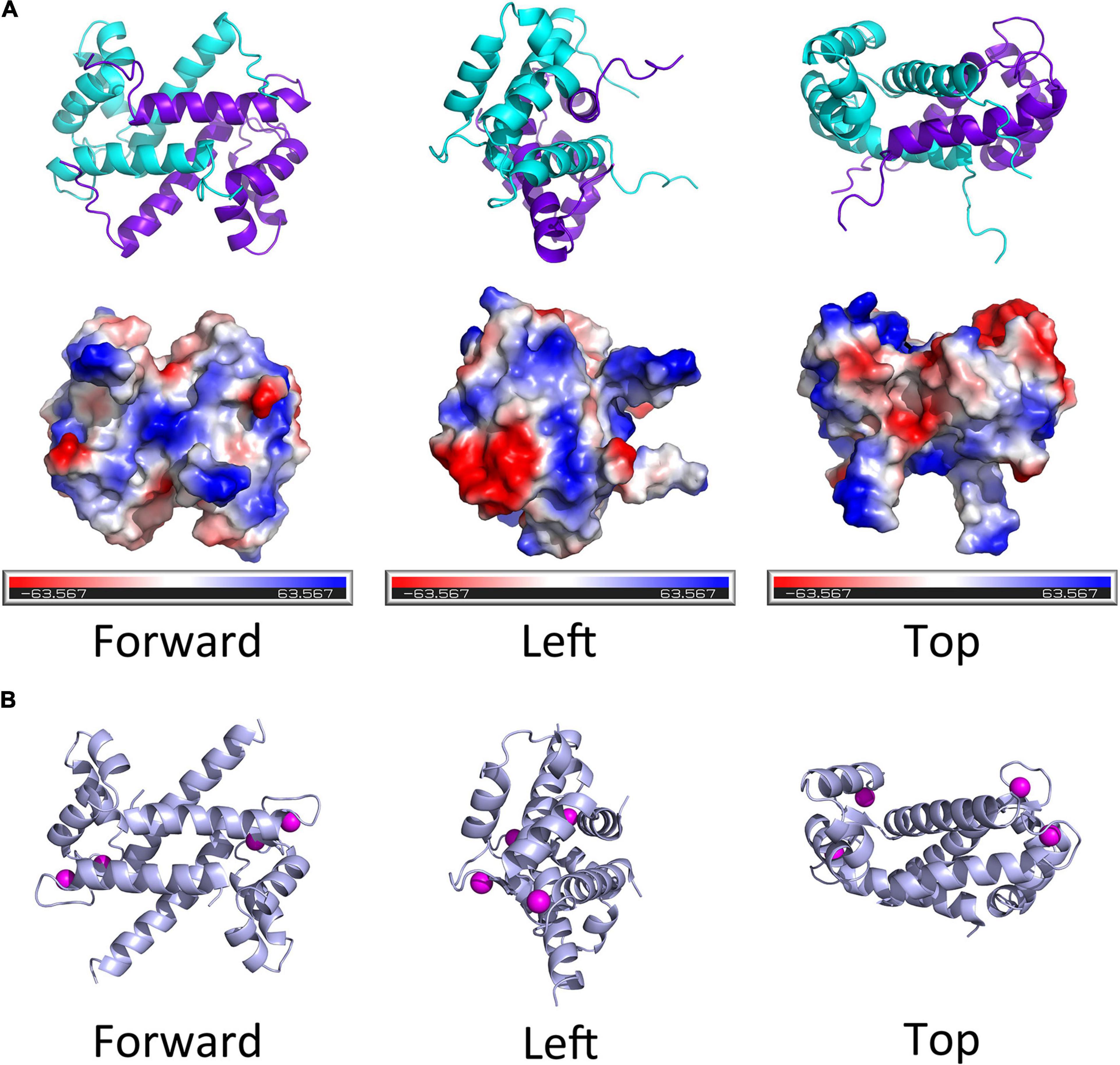
Figure 2. The diagram of spatial structure of S100A11 protein. (A) Top panel, the 3D structure of human S100A11 homodimer. Bottom panel, the surface charge profile of 3D structure of S100A11 homodimer shown in the same orientation as top panel, the surfaces with negative charge, positive charge and hydrophobic are colored in red, blue and white, respectively. The PDB file that used to make the 3D structure of human S100A11 was from RCSB PDB (www.rcsb.org/structure/2LUC), the 3D structures were made by PyMOL software. (B) The pig (Sus scrofa) S100A11 homodimer with the calcium ions. The calcium ions are shown as pink balls. The PDB file that used to make the 3D structure of porcine S100A11 was from RCSB PDB (www.rcsb.org/structure/1qls), the 3D structures were made by PyMOL software.
Tissue and Cell-Type Specific Expression and Location of S100A11
The expressions of S100A11, like other S100 proteins, is tissue and cell-type specific. In human, S100A11 is ubiquitously expressed in various tissues (He et al., 2009). In 1999, using Northern blot, Inada et al. (1999) showed that the expression level of S100A11 is the highest in the placenta, followed by the heart, lungs, pancreas, and kidneys, and less in skeletal muscle, liver, and brain. However, following the development of sequencing technology, a more comprehensive tissue expression profile of S100A11 has been established. As shown in Figure 3A, human S100A11 is highly expressed in skin, spleen, lung, kidney, sWAT (subcutaneous white adipose tissue), and stomach; it is moderately expressed in the small intestine, heart, and pancreas; while it is less expressed in the liver, brain, and muscle. Likewise, in the mouse, S100A11 is highly expressed in the sWAT, lung, and kidney; it is moderately expressed in the stomach, spleen, heart, and small intestine; and it is less expressed in the brain and liver (Figure 3B).
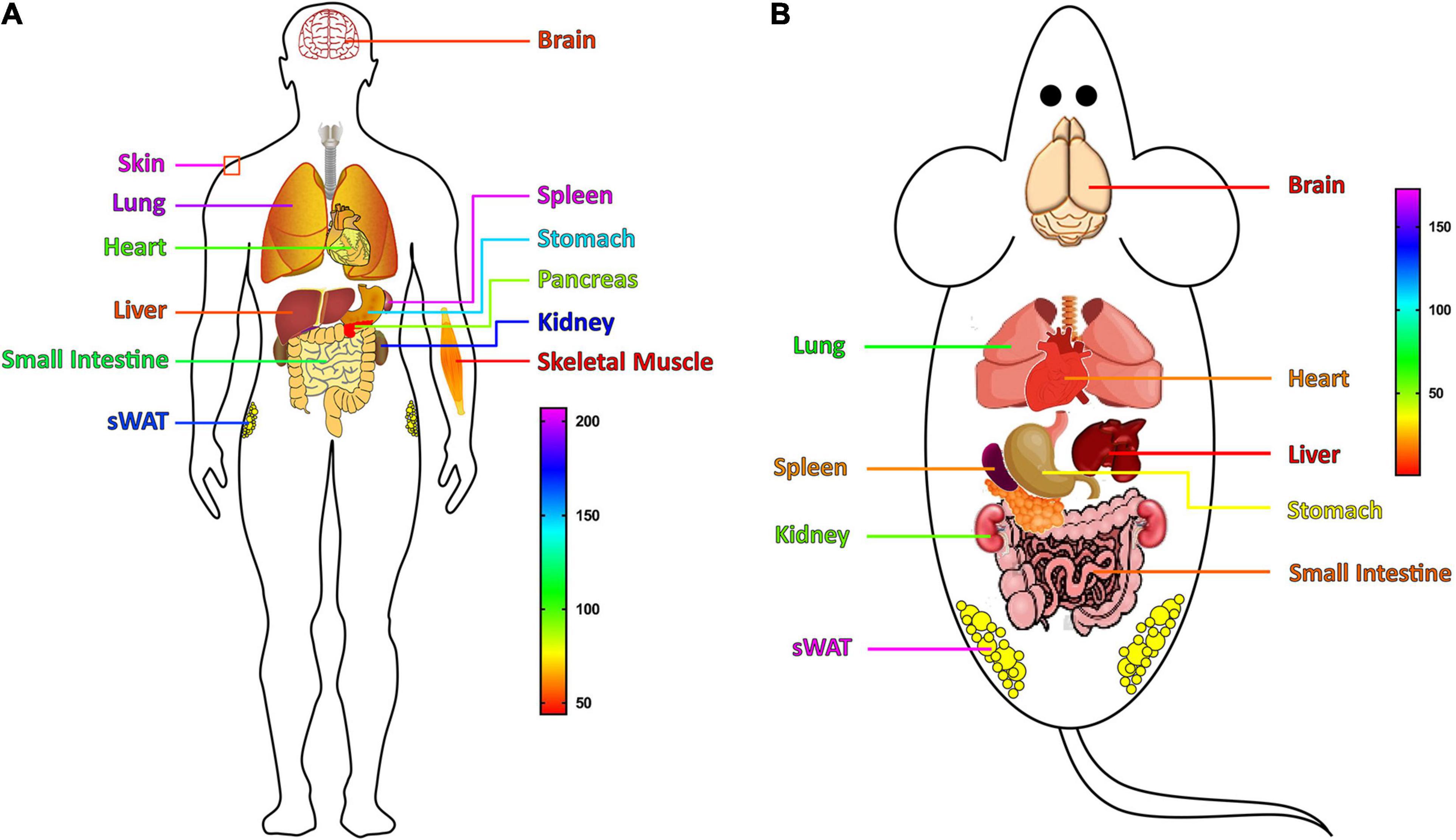
Figure 3. The tissues expression profile of the S100A11 gene. (A) The expression profile of the S100A11 gene in human tissues. S100A11 is highly expressed in human skin, spleen, and lung, and its expression is lower in liver, brain, and skeletal muscle. The expression data are from GeneCards (www.genecards.org/). (B) The expression profile of the S100A11 gene in mouse tissues. S100A11 is highly expressed in mouse sWAT, lung, and kidney, and it has a lower distribution in the small intestine, brain, and liver. The expression data are from NCBI (www.ncbi.nlm.nih.gov/). sWAT, subcutaneous white adipose tissue.
The cellular distribution of S100A11 can vary in different tissues and it can also vary under distinct physiological conditions. For example, in the cells of normal kidney tissue, S100A11 is mainly distributed in the nucleus. In the cells of invasive cervical squamous cell carcinoma of the uterine cervix, S100A11 is distributed in both the nucleus and the cytoplasm. In cells of serous adenocarcinoma of the ovary, S100A11 is mainly distributed in the nucleus, with only a small amount in the cytoplasm. However, in the cells of invasive breast carcinoma of ductal no special type, S100A11 shows a strong distribution in the cytoplasm, with only a weak amount in the nucleus (Cross et al., 2005).
The dynamic distribution of S100A11 in cells also has been investigated. In human keratinocytes, S100A11 is uniformly distributed in the cytoplasm under normal conditions, but when stimulated with a high concentration of calcium ions (0.3 mmol/L), it interacts with another protein, Annexin I, to accumulate on the inner membrane side of the cell membrane with the help of tubulin. This process does not rely on the classical Golgi/ER export pathway. Through subsequent exosome generation at the plasma membrane, S100A11 can be secreted from the cell. Now extracellularly localized, S100A11 can act as a signal molecule to bind the RAGE (receptor for advanced glycation end products) receptor (Broome and Eckert, 2004). Overall however, based on the sum of this data, we can summarize that S100A11 can be distributed in the nucleus, cytoplasmic matrix, and extracellular region, which is cell type and tissue state dependent.
The Biological Activities of S100A11
Current studies have found that S100A11 has the following main functions: (1) Regulation of enzyme activity. In smooth muscle, S100A11 can interact with actin filaments and inhibit ATPase activity in the presence of calcium ions (Zhao et al., 2000). (2) Regulation of cell growth. Using human keratinocytes as example, when the cells are stimulated by calcium ion or TGFβ, S100A11 will be phosphorylated by PKCα and then enter the nucleus to promote the expression of the p21 gene, which further inhibits keratinocyte proliferation by inhibiting cdk2 (Sakaguchi et al., 2003, 2004), this process is illustrated in Figure 4. Conversely, down-regulation of S100A11 can lead to a decrease in the protein level of p21. It has been shown that the down-regulation of S100A11 caused a decrease in phosphorylation of AKT, which then activated GSK3. Activated GSK3 can phosphorylate threonine at position 57 in p21, which causes the degradation of p21 protein in a ubiquitination-independent manner. Therefore, the down-regulation of S100A11 promotes the turnover of p21 through the PI3K/AKT signaling pathway, thereby promoting the cell proliferation process (Foertsch et al., 2013). In addition, other pathways that are involved in the regulation of cell proliferation and growth by S100A11 are summarized in Figure 5. (3) Inducing apoptosis. Studies in human tumor cell lines, such as melanoma, pancreatic cancer, breast cancer, and lung cancer, have found that a 19 amino acid peptide from the N-terminus of the S100A11 protein can promote the translocation of apoptosis-inducing factor (AIF) from the cytoplasm to the nucleus. AIF in the nucleus triggers chromatin condensation and DNA fragmentation, thereby inducing apoptosis of tumor cells (Makino et al., 2004). (4) Participating in the inflammatory response (Figure 6). During the occurrence of osteoarthritis, CXCL8 (C-X-C Motif Chemokine Ligand 8) and TNF-α (Tumor necrosis factor α) can induce the expression of S100A11 and promote its release to the extracellular space to form a dimer. This dimer then binds to the receptor RAGE, promoting the development of osteoarthritis through the p38 signaling pathway (Cecil et al., 2005; He et al., 2009).
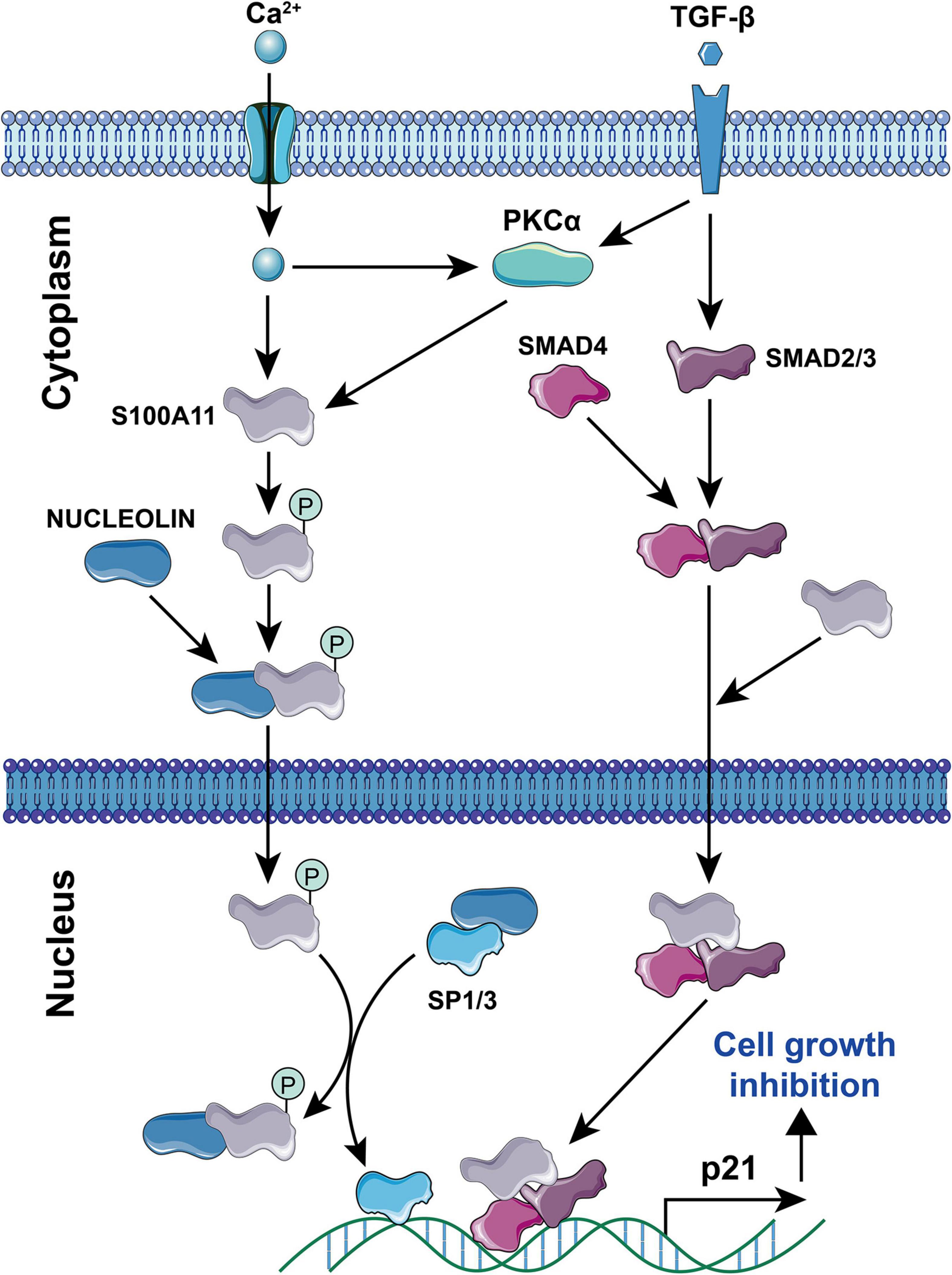
Figure 4. Signaling pathways for Ca2+ and TGF-β-induced S100A11-mediated cell growth inhibition. Elemental Ca2+, or PKCα induced by Ca2+/TGF-β can promote the phosphorylation of S100A11. Phosphorylated S100A11 then translocates to the nucleus through binding to NUCLEOLIN. In the nucleus, S100A11 competes with Sp1/3 for binding to NUCLEOLIN. This then releases free Sp1/3 to induce p21 expression. Moreover, S100A11 can bind to SMAD2/3 and SMAD4, that has been stimulated by TGF-β, to form a complex. Subsequently, this complex migrates to the nucleus to induce p21 expression. In either case, increasing levels of p21 results in cell growth inhibition. The circled “P” represents phosphorylation.
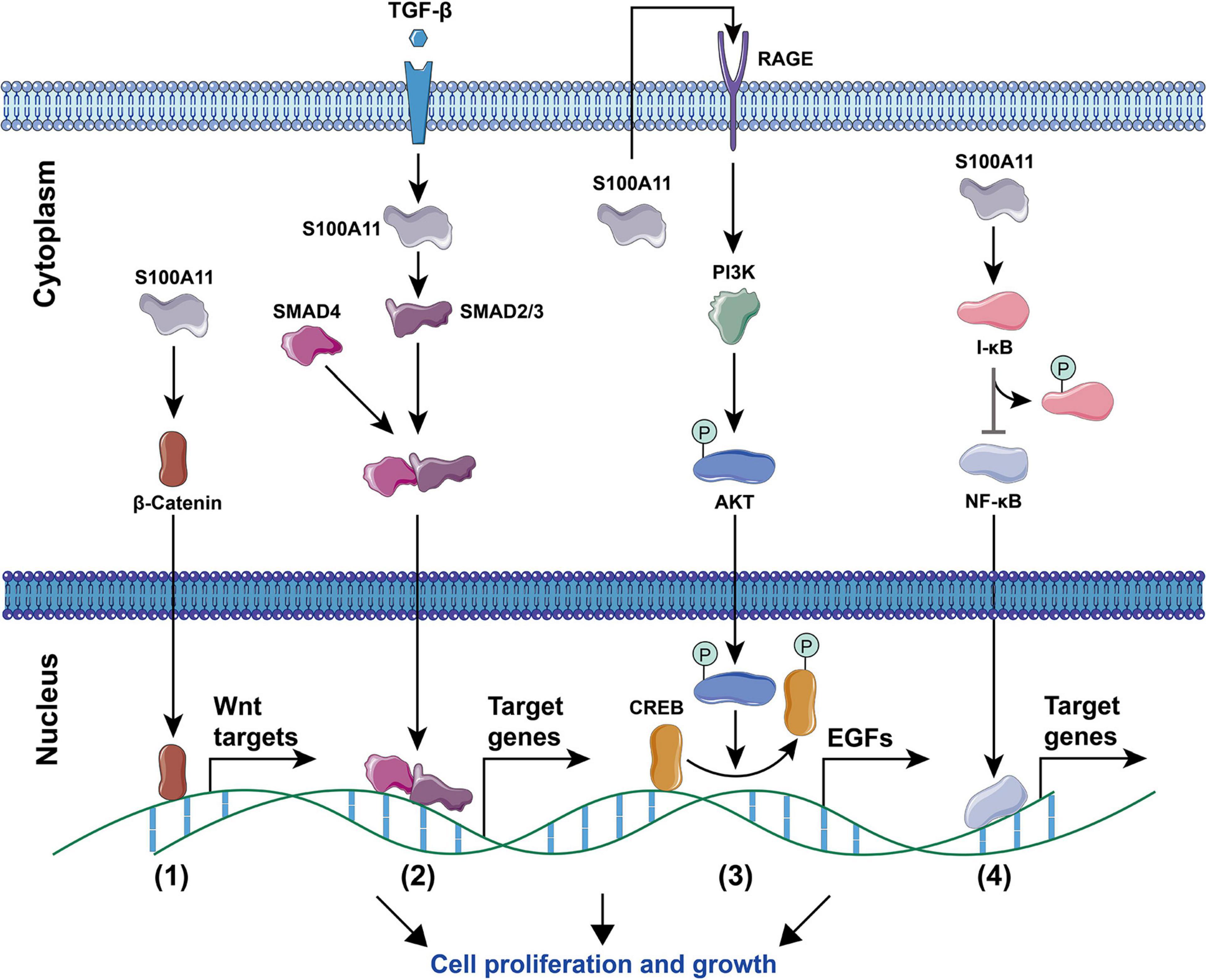
Figure 5. Signaling pathways for S100A11-mediated cell proliferation and growth. (1) The Wnt signaling pathway. S100A11 accelerates the entry of β-Catenin into the nucleus, thereby regulating the downstream Wnt target genes. (2) The TGF-β signaling pathway. Under the stimulation of TGF-β, S100A11 promotes the SMAD2/3 to form a complex with SMAD4, the complex then enters into the nucleus and controls the expression of target genes. (3) The PI3K/AKT signaling pathway. S100A11 is first secreted outside the cell and then binds to the RAGE receptor to induce the expression of EGF genes through the Akt signaling pathway. CREB, cAMP response element-binding protein; EGFs, epidermal growth factors. (4) The NF-κB signaling pathway. S100A11 promotes the phosphorylation of I-κB, which then leads to the activation and nuclear migration of NF-κB to regulate the expression of target genes. All of the above four processes ultimately promote cell proliferation and growth. The circled “P” represents phosphorylation.
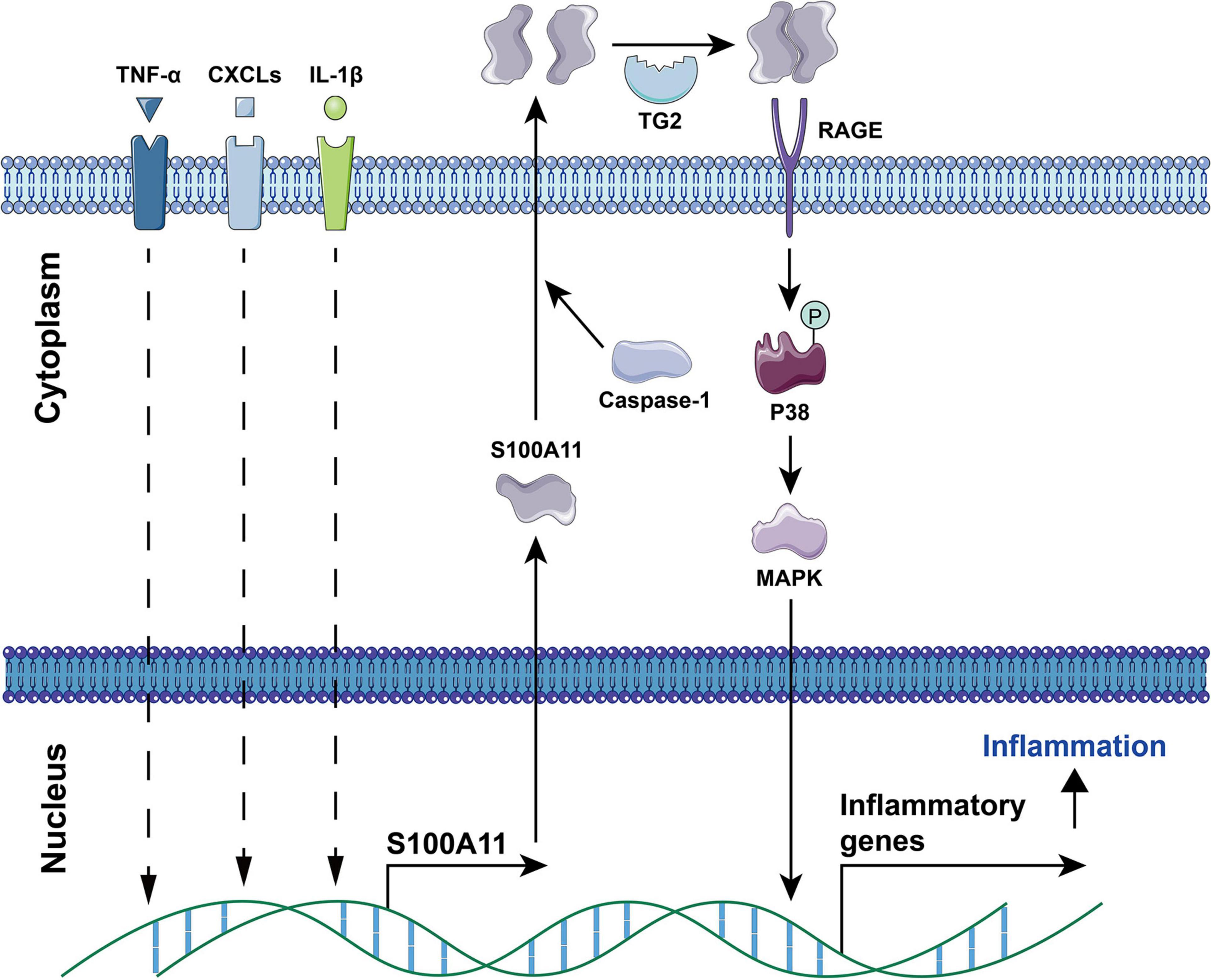
Figure 6. The involvement of S100A11 in TNF-α, CXCLs, and IL-1β induced inflammation. TNF-α, CXCL8, and IL-1β induce the expression of S100A11. Subsequently, S100A11 is secreted through a Caspase-1 dependent manner. Extracellular S100A11 is dimerized under the catalysis of TG2, and then binds to the RAGE receptor to induce the expression of inflammatory genes through the p38 MAPK signaling pathway, which causes inflammation progression. CXCLs, CXC chemokine ligands; TG2, transglutaminase 2. The circled “P” represents phosphorylation.
S100A11 was also shown to have a role in the organization of early endosomes. Seemann et al. (1997) reported that the deletion of 14 amino acid residues in the C-terminal of the S100A11 protein abolished the interaction between S100A11 and Annexin A1 (ANXA1), which then restricted the endosomal localization of this complex and restrained the structural organization of the endosome membranes. Since ANXA1 has been linked to the internal vesiculation process and structural organization in multivesicular endosomes by localizing on the membranes, binding with S100A11 could modulate the physiological properties of ANXA1 through phosphorylation by protein kinase C (Fava and Cohen, 1984; Futter et al., 1993; Seemann et al., 1996). Thus, physiologically, this complex could possibly hold the membranes in place for endosome fusion to occur or could organize the compartment of endosome. In addition, the interaction of S100A11 with ANXA1 promotes the transport of cholesterol from the endoplasmic reticulum to the endosome by mediating membrane contact between these two organelles. The transport of cholesterol is a process that is necessary for the formation of intraluminal vesicles within the endosome, which then can regulate the degradation of the epidermal growth factor receptor (EGFR). Therefore, S100A11 also plays an important role in the regulation of EGFR signaling (Ringerike et al., 2002; Eden et al., 2016; Tan et al., 2016). Moreover, S100A11 was shown to form a complex with ANXA1 to enhance lysosomal targeting of the EGFR, which then led to interruption of EGF signal and degradation of EGFR. In this case, the complex functions as a physical scaffold to support the sorting of EGFR into multivesicular bodies (Poeter et al., 2013).
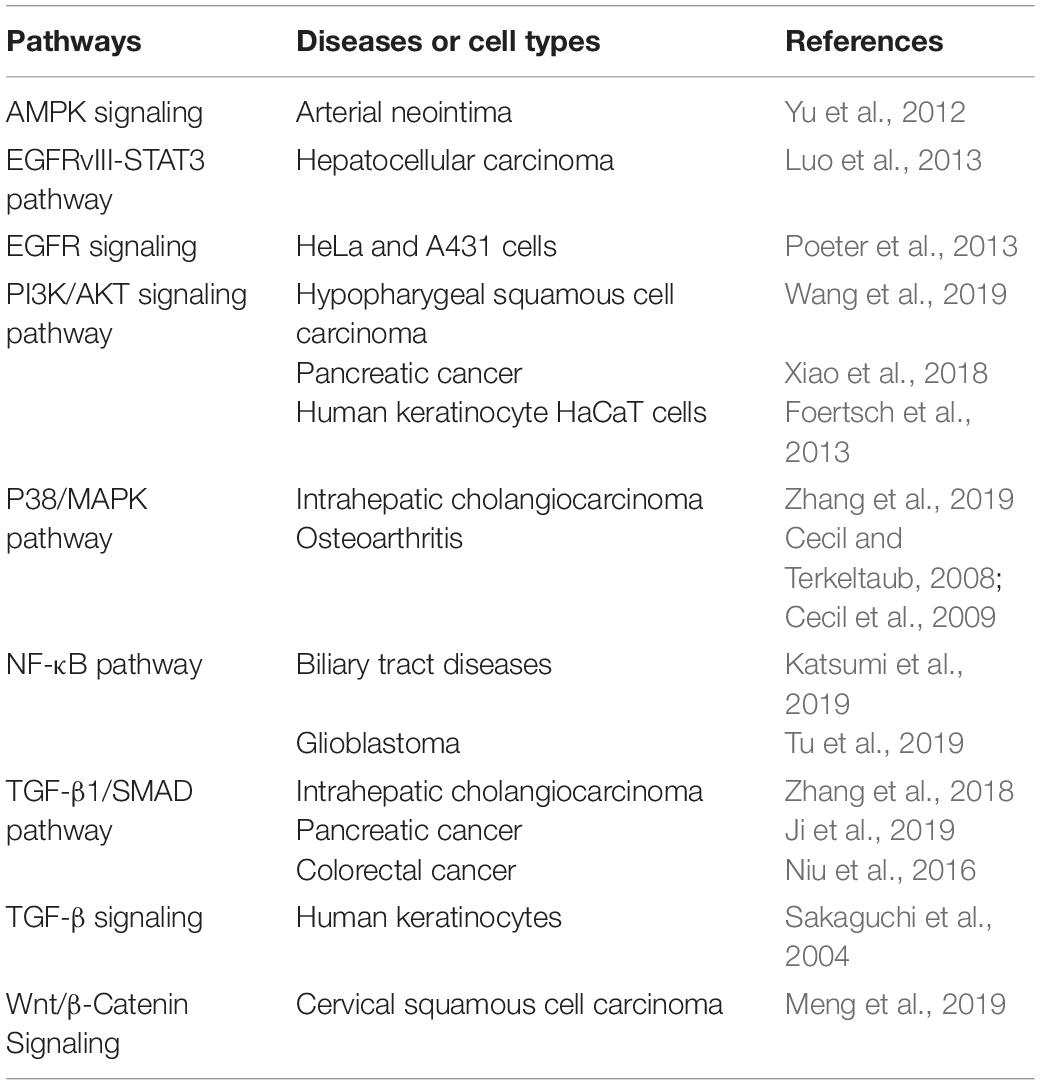
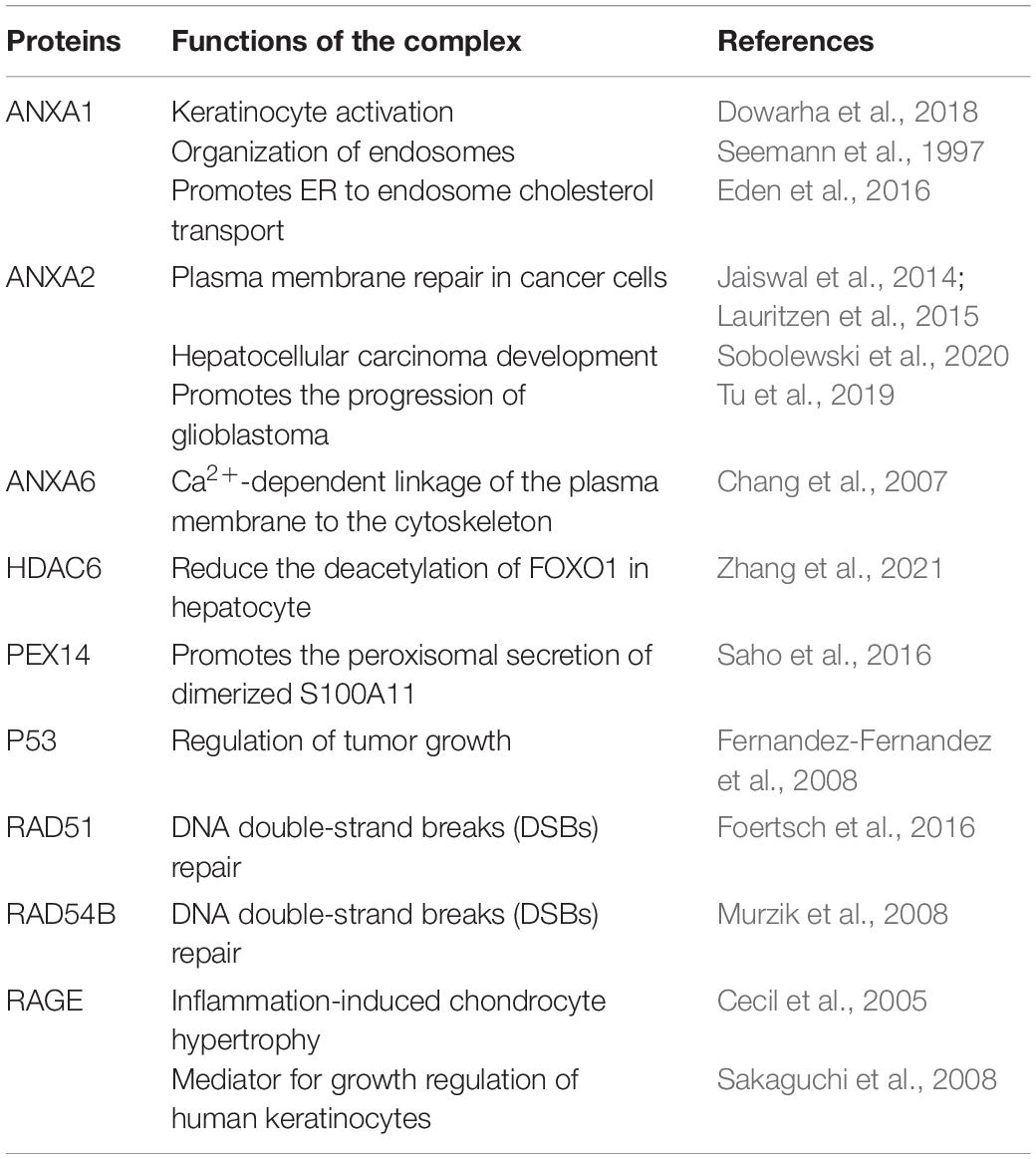
The Roles of S100A11 in Diseases
Dysregulation of S100A11 has been shown to be involved in various human diseases via different signaling pathways. A summation of these results is detailed in Table 1. In addition, several studies have shown that S100A11 can form a complex with specific proteins (Table 2) in some diseases. Saho et al. (2016) showed that in mesothelioma cells, S100A11, lacking the classical secretory signal, can be dimerized and secreted through the peroxisome and thus play a key role in mesothelioma progression in a tumor microenvironment. Moreover, extracellular vesicles (EVs), such as exosomes and microvesicles (Raposo and Stoorvogel, 2013), have been shown to display marked potential in the organ-to-organ communications (Thomou et al., 2017). A recent study reported that in both human and mouse, cholangiocyte cell lines could release EVs containing S100A11 to induce bone marrow-derived macrophages to express pro-inflammatory cytokines (Katsumi et al., 2019).
Cancers
S100A11 is involved in many types of cancers and it plays a distinct role depending on the specific tumor type. A summation of these results is detailed in Table 3. In most cancers, S100A11 is highly expressed and correlated to tumor promotion and progression. Studies have found that in renal cell carcinoma, which is prevalent in North America, the expression level of S100A11 is positively correlated to the degree of cancer progression and tumor size, and negatively correlated to the disease-free survival rate of patients (Gabril et al., 2016). In addition, in prostate and breast cancer, the expression of S100A11 is significantly associated with high pathologic cancer stage, suggesting that S100A11 is involved in cancer development and progression (Rehman et al., 2004; Cross et al., 2005). Sobolewski et al. (2020) found that overexpression of S100A11 leads to high-grade hepatocellular carcinoma and poor prognosis by promoting cancer cell proliferation and migration. They therefore suggested that S100A11 be considered as an oncogenic factor (Sobolewski et al., 2020). In these cases, the up-regulation of S100A11 significantly promotes the proliferation, migration, and tissue invasion of tumor cells by activating different signal pathways (Meng et al., 2019; Mitsui et al., 2019). A S100A11/ANXA2 complex has also been shown to be able to reseal the plasma membrane if it has become damaged due to the stress of metastatic tissue invasion (Jaiswal et al., 2014; Sobolewski et al., 2020).
In tumors with a high expression of S100A11, such as in intrahepatic cholangiocarcinoma, the silencing of S100A11 can inhibit TGF-β1-induced cell migration, invasion, and epithelial-mesenchymal transition (EMT). Upon silencing of S100A11, the phosphorylation levels of SMAD2/3, induced by TGF-β, also are decreased. Therefore, conversely, one of the reasons for the invasivity of this type of tumor may be that S100A11 induces phosphorylation of SMAD2/3 through TGF-β, which then accelerates the process of tumor cell migration, tissue infiltration, and EMT (Zhang et al., 2018).
On the other hand, in bladder cancer, S100A11 is thought to function as a tumor suppressor. Memon et al. (2005) found that the mRNA and protein levels of S100A11 were all down-regulated in patients with bladder cancer. In addition, a negative correlation between S100A11 expression and bladder cancer progression was observed, and loss of S100A11 was associated with poor survival in bladder cancer patients (Memon et al., 2005). At the same time, research in fibroblasts found that when normal cells shift to an immortal state, the expression level of S100A11 is also reduced (Sakaguchi et al., 2000). The underlying mechanism may be related to the function of S100A11 being able to be phosphorylated, with the phosphorylated S100A11 entering the nucleus and inducing the p16 and p21 proteins to inhibit cell DNA synthesis (Sakaguchi et al., 2000, 2001). Taken altogether, these studies suggest that the level of S100A11 expression, the signaling pathways that S100A11 is involved in, and the posttranscriptional modification status of S100A11 protein contribute to the different roles of S100A11 in different tumors.
Furthermore, in a recent study on human cancers, S100A11 was found to be highly enriched in the proteomic profile of extracellular vesicles and particles (EVPs) in human pancreatic adenocarcinoma. It was identified as a tumor-derived EVP protein involved in eliciting immune responses, suggesting that S100A11 is also involved in tumor immunity (Hoshino et al., 2020).
Metabolic Disease
Type 2 diabetes (T2D) is an increasing global health problem, and the gene regulation in human pancreatic islets under T2D is complicated. Using global genomic and transcriptomic analysis of human pancreatic islets from 89 donors, Fadista et al. (2014) found that S100A11 positively associated with HbA1c. HbA1c is one of the genes that can influence glucose metabolism, and it plays an important role in the pathogenesis of T2D (Fadista et al., 2014). In addition to T2D, non-alcoholic fatty liver disease (NAFLD) is another metabolic disease mainly induced by high energy consumption, for example a high fat or high fructose diet. Zhou et al. (2017) found that in a mouse NAFLD model generated by a western diet (40% fat and 0.2% cholesterol) treatment, the expression of S100A11 was significantly upregulated in the liver. Another study using a mouse model of non-alcoholic steatohepatitis treated with a high fat/high cholesterol diet (20% fat and 1% cholesterol) showed that S100A11 increased in the liver in a time-dependent manner (Oh et al., 2013).
According to the latest study in our lab (Zhang et al., 2021), it was confirmed that S100A11 was significantly upregulated in the liver of a tree shrew NAFLD model (Zhang et al., 2015, 2016). Following this result, we overexpressed S100A11 in Hepa 1-6 and human Hep 3B cell lines and found that overexpression resulted in enhanced lipid(s) accumulation. More importantly, overexpression of S100A11 in mice liver also accelerated the hepatic lipids deposition. Mechanistically, our results indicate that exogenous dietary lipids promote liver S100A11 expression, which then competitively interacts with HDAC6 to block the binding between HDAC6 and FOXO1. Loss of HDAC6 then promotes the acetylation of FOXO1, consequently activating autophagy and lipogenesis pathways, thereby accelerating liver lipid accumulation (Figure 7). Coincidentally, Teng et al. (2021) also reported that S100A11 could contribute to hepatic steatosis through RAGE-mediated AKT-mTOR signaling. In this study the experimental phenotype is consistent with ours, but the hypothesized molecular mechanism is different (Teng et al., 2021). In any case, these studies do show that S100A11 can affect autophagy and lipid metabolism, and that it plays an important and complex role in fatty liver disease.
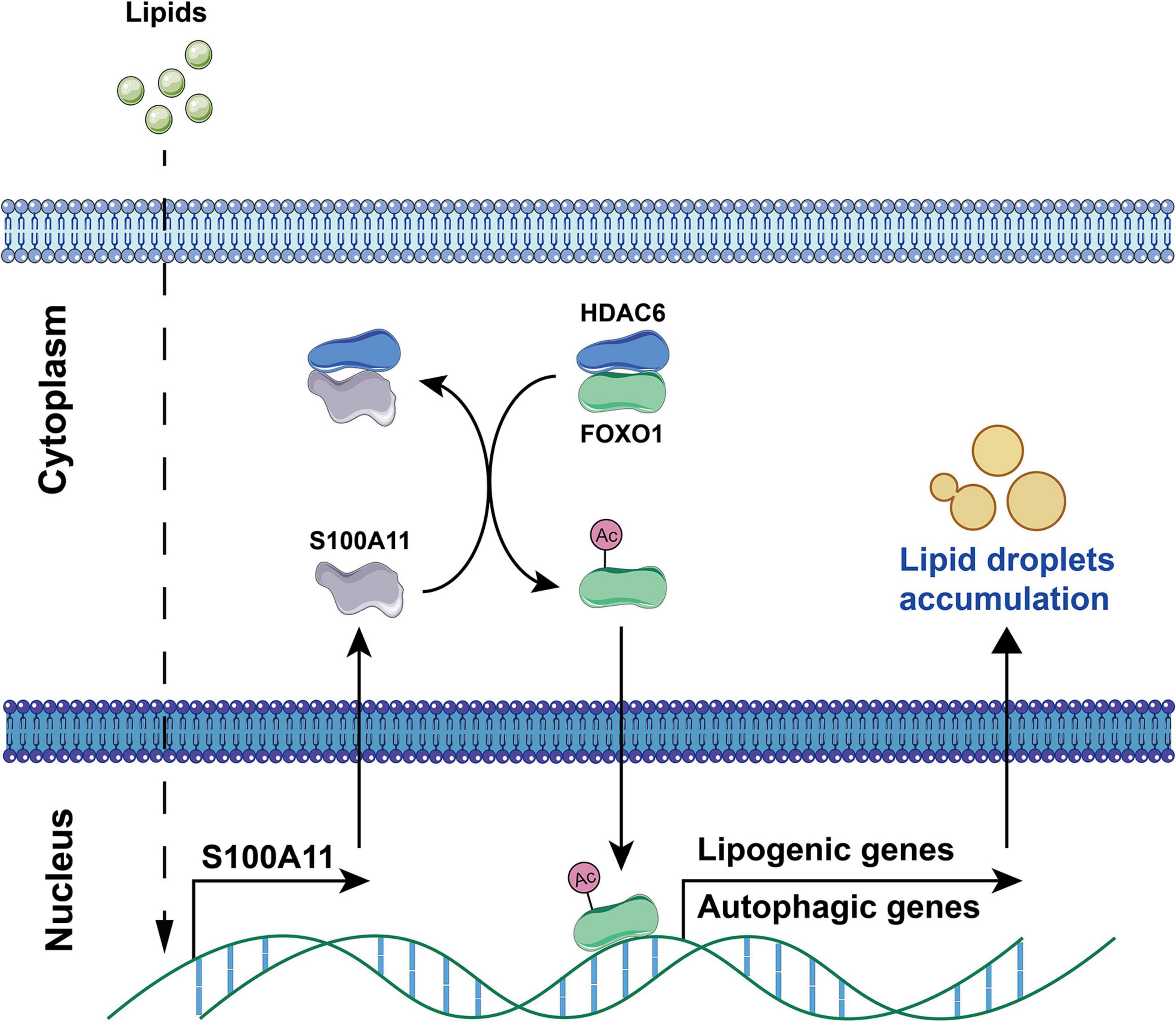
Figure 7. S100A11-HDAC6-FOXO1 axis in the regulation of autophagy and lipogenesis in liver induced by high fat diet. Dietary lipids induce the overexpression of S100A11. S100A11 then competes with FOXO1 for binding to HDAC6, which releases and promotes the acetylation of FOXO1. The acetylated FOXO1 activates autophagy and lipogenesis pathways leading to lipid droplet accumulation. HDAC6, histone deacetylase 6; FOXO1, forkhead box protein O1. The circled “Ac” represents acetylation.
Although there are relatively few studies on S100A11 in metabolic diseases, these published works indicate that S100A11 may play an important role in the development and progression of other metabolic diseases, not only T2D and NAFLD.
Neurological Diseases
S100A11 has also been shown to be an important factor in neurological diseases. Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease characterized by a degeneration of motor neurons in the spinal cord and motor cortex. Studies have shown that S100A11 was significantly enriched in ALS where it functions as a calcium sensor and modulator to promote vulnerability to neurodegeneration (Alexianu et al., 1994; Iridoy et al., 2018). In another severe neurological disease, autoimmune encephalitis, S100A11 was also found to be up-regulated and hypomethylated. Along with S100A6, in this state specific S100 family members may facilitate B lymphocyte infiltration into the central nervous system through the blood-brain barrier (Tsai et al., 2019). However, it has been suggested that S100A11 does not always play a role in pathogenicity. Ischemia-reperfusion can directly lead to cell death. This can subsequently result in many neurological conditions. ANXA1 nuclear translocation has long been recognized as a pathogenic factor through inducing neuronal apoptosis (Li et al., 2016). In a mouse model of ischemic stroke, S100A11 exerts neuroprotective effects, and upregulation of S100A11 protected against neuronal apoptosis by interaction with ANXA1 through the nuclear translocation signal (NTS) of ANXA1, thereby blocking ANXA1 nuclear translocation (Xia et al., 2018). According to these studies, like its role in tumors, S100A11 can also have a dual role in neurological diseases.
Vascular Calcification
Vascular calcification is a highly prevalent vascular pathophenotype that has been associated with aging, atherothrombotic cardiovascular disease, diabetes mellitus, and chronic kidney disease (Leopold, 2015). In a mouse model of chronic kidney disease, S100A11 and its receptor, RAGE (receptor for advanced glycation end products), were both up-regulated. S100A11/RAGE signaling activates ERK1/2 MAPK, which further activates mTORC1 and the ER stress-Unfolded Protein Response to induce vascular calcification (Panda et al., 2018). Coincidentally, in the ectonucleotide pyrophosphatase/phosphodiesterase 1 knockout (Enpp1–/–) mice model of arterial calcification, it was shown that S100A11 was also highly expressed in the aorta. Mechanistically, dependent upon RAGE, S100A11 induced cartilage-specific collagen IX/XI expression to promote ectopic chondrogenic differentiation and calcification in the aorta (Cecil and Terkeltaub, 2011). In addition to accelerated calcification, S100A11 also plays a role in neointima formation in response to arterial injury. S100A11 positive cells were significantly enriched in injured arteries. Furthermore, S100A11/RAGE signaling mediates vascular remodeling by regulation of the AMPK pathway via liver kinase B1 and STAT3 (Yu et al., 2012). Thus, what we know about the function of S100A11 in vascular calcification is largely dependent on its receptor, RAGE.
Inflammatory Diseases
In osteoarthritis, inflammation promoted chondrocyte differentiation always contributes to the disease progression. Studies have shown that S100A11 can be released by the chondrocyte after stimulation by IL-1β. Once secreted, S100A11 is then covalently crosslinked into a dimer through the catalysis of transglutaminase 2 (Cecil and Terkeltaub, 2008). The dimerized S100A11 acquires the capacity to promote chondrocyte activation through the p38/MAPK pathway, which then accelerates the development of osteoarthritis (Cecil et al., 2005; Cecil and Terkeltaub, 2008; Figure 6). By contrast, rheumatoid arthritis is a chronic systemic autoimmune disease. Andres Cerezo et al. (2017) found that rheumatoid arthritis patients have increased levels of S100A11 in their synovial tissue and synovial fluid but not in serum. The increased amount of S100A11 can stimulate the production of the pro-inflammatory cytokine IL-6 by peripheral blood mononuclear cells and synovial fibroblasts, suggesting an association between S100A11, inflammation, and disease activity in rheumatoid arthritis patients (Andres Cerezo et al., 2017). Further, a recent study (Navrátilová et al., 2021) uncovered that, under rheumatoid arthritis conditions, S100A11 could be released by neutrophils. The extracellular S100A11 then enhanced the secretion of IL-6 and TNF by neutrophils and subsequently aggravated the inflammatory response.
S100A11 also has functions in other inflammatory diseases. CCL2 is a crucial chemokine required for host resistance to parasites. It has been recently reported that after infection by Toxoplasma gondii, human monocytes detect S100A11 protein, which has been released from infected cells in a caspase-1-dependent manner. S100A11 binding to RAGE induces the production of the chemokine CCL2 to resist further Toxoplasma gondii infection. Therefore S100A11 plays a role of innate immune sensor in this infection (Safronova et al., 2019). In addition, studies have shown that circulating S100A11 can be used as a biomarker of myositis (Andrés Cerezo et al., 2019) and infective endocarditis (Thuny et al., 2012). While all these studies give strong evidence for the importance of S100A11 in inflammatory diseases, further research is necessary to determine if this would be a useful clinical target to ameliorate inflammation.
Future Perspectives and Challenges
The S100 protein family constitutes the largest subgroup of the EF-hand family of Ca2+-binding proteins, and S100A11 is one of these proteins. To date, a large number of studies have shown that S100A11 has biological functions such as regulating cell growth, enzyme activity, and the inflammatory response. As well, it is involved in the regulatory process of cancers, metabolic diseases, neurological diseases, vascular calcification, and inflammatory diseases. Taken together, all the evidences cited in this review indicate that S100A11 is an important molecule involved in many diverse aspects of cellular functions.
The studies show that S100A11 is involved in many different biological processes, for which we speculate that this may be related to the characteristics of S100A11. First, as a protein that can bind calcium ions, S100A11 may perform distinct functions due to binding with or without calcium ions. Secondly, S100A11 is a protein with small molecular weight that can be secreted extracellularly and can also be transported in the blood by means of exosomes, extracellular vesicles, or as a covalently bound dimer. Thus, it may be involved in the cross-talk among and between cells as well as organs. This may also account for its functional diversity. Ultimately, as a protein that can enter the nucleus, S100A11 can directly or indirectly affect the expression of different genes and thus exert various functions.
As far as the current research is concerned, our understanding of S100A11 is very limited, and there are still many challenges to be overcome in the future. First of all, it is not clear how S100A11 is regulated at the level of transcription, translation, or the status of post-translational modification(s) under physiological and/or pathological conditions. Secondly, although several studies have shown that S100A11 is involved in certain signaling pathways and can form complexes with proteins in some diseases, nevertheless, the specific molecular mechanism of S100A11 function in these diseases is still not clear. Thirdly, most of the studies on S100A11 are mainly focused on cancers, such as breast cancer or colorectal cancer, implying that S100A11 is expected to be developed as a biomarker or therapeutic target for tumors. However, S100A11 plays a dual opposing role for tumor promotion and tumor inhibition in the progress of different cancers, while, again, the underlying molecular mechanisms are still unknown. Clearly, further research is needed to study the dichotomy of S100A11 in different cancers. Furthermore, S100A11 has long been regarded as a secretory protein and biomarker for some cancers. Whether S100A11 is always secreted in the form of extracellular vesicle cargo in different organs and diseases, and if during this process whether the S100A11 protein is modified after secretion is unknown. Fourthly, S100A11 has a clear functionality in osteoarthritis. However, many questions concerning its functionality have not yet been answered, such as the pathway through which inflammatory factors (TNF-α, CXCLs, and IL-1β) induce the expression of S100A11 and release it from cartilage, the region where S100A11 interacts with RAGE, and ultimately how the signal is transduced to activate the P38 phosphorylation pathway. Finally, there is very little research on S100A11 in other areas, especially in metabolic diseases, which are widely prevalent worldwide. Therefore, the role of S100A11 in metabolic diseases deserves much greater consideration in the future.
The application of modern biology and bioinformatics tools to study the mechanistic and functional interaction between S100A11 and its related proteins, as well as deciphering its intrinsic cell signal transduction pathways in-depth in multiple cell types, will contribute to the clinical diagnosis, prevention, and treatment of S100A11 related diseases in the future.
Author Contributions
LZ conceived and wrote the original draft. TZ collected data of the CDS sequences and amino acid sequences of S100 proteins as well as the data of expression profile of human and mouse S100A11 genes. HM provided guidance for the draft. BL provided advice and supervision, and revised the manuscript. All the authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81700520, U1702288, U1702287, 31671230, 91857113, and 31860323), the Ministry of Science and Technology of the People’s Republic of China (2018YFA0800700), the Yunnan Applied Basic Research Projects (2017FA007, 2018FB117, and 2019FY003021), and the Key Projects of Dongguan Science and Technology Bureau (2018507150391621).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alexianu, M. E., Ho, B. K., Mohamed, A. H., La Bella, V., Smith, R. G., and Appel, S. H. (1994). The role of calcium-binding proteins in selective motoneuron vulnerability in amyotrophic lateral sclerosis. Ann. Neurol. 36, 846–858. doi: 10.1002/ana.410360608
Anania, M. C., Miranda, C., Vizioli, M. G., Mazzoni, M., Cleris, L., Pagliardini, S., et al. (2013). S100A11 overexpression contributes to the malignant phenotype of papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 98, E1591–E1600. doi: 10.1210/jc.2013-1652
Andrés Cerezo, L., Hulejová, H., Šumová, B., Kropáčková, T., Kryštůfková, O., Klein, M., et al. (2019). Pro-inflammatory S100A11 is elevated in inflammatory myopathies and reflects disease activity and extramuscular manifestations in myositis. Cytokine 116, 13–20. doi: 10.1016/j.cyto.2018.12.023
Andres Cerezo, L., Sumova, B., Prajzlerova, K., Veigl, D., Damgaard, D., Nielsen, C. H., et al. (2017). Calgizzarin (S100A11): a novel inflammatory mediator associated with disease activity of rheumatoid arthritis. Arthritis Res. Ther. 19:79. doi: 10.1186/s13075-017-1288-y
Bresnick, A. R., Weber, D. J., and Zimmer, D. B. (2015). S100 proteins in cancer. Nat. Rev. Cancer. 15, 96–109. doi: 10.1038/nrc3893
Broome, A. M., and Eckert, R. L. (2004). Microtubule-dependent redistribution of a cytoplasmic cornified envelope precursor. J. Invest. Dermatol. 122, 29–38. doi: 10.1046/j.0022-202X.2003.22105.x
Cecil, D. L., and Terkeltaub, R. (2008). Transamidation by transglutaminase 2 transforms S100A11 calgranulin into a procatabolic cytokine for chondrocytes. J. Immunol. 180, 8378–8385. doi: 10.4049/jimmunol.180.12.8378
Cecil, D. L., and Terkeltaub, R. A. (2011). Arterial calcification is driven by RAGE in Enpp1-/- mice. J. Vasc. Res. 48, 227–235. doi: 10.1159/000318805
Cecil, D. L., Appleton, C. T., Polewski, M. D., Mort, J. S., Schmidt, A. M., Bendele, A., et al. (2009). The pattern recognition receptor CD36 is a chondrocyte hypertrophy marker associated with suppression of catabolic responses and promotion of repair responses to inflammatory stimuli. J. Immunol. 182, 5024–5031. doi: 10.4049/jimmunol.0803603
Cecil, D. L., Johnson, K., Rediske, J., Lotz, M., Schmidt, A. M., and Terkeltaub, R. (2005). Inflammation-induced chondrocyte hypertrophy is driven by receptor for advanced glycation end products. J. Immunol. 175, 8296–8302. doi: 10.4049/jimmunol.175.12.8296
Chang, N., Sutherland, C., Hesse, E., Winkfein, R., Wiehler, W. B., Pho, M., et al. (2007). Identification of a novel interaction between the Ca(2+)-binding protein S100A11 and the Ca(2+)- and phospholipid-binding protein annexin A6. Am. J. Physiol. Cell Physiol. 292, C1417–C1430. doi: 10.1152/ajpcell.00439.2006
Cross, S. S., Hamdy, F. C., Deloulme, J. C., and Rehman, I. (2005). Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: S100A6, S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology 46, 256–269. doi: 10.1111/j.1365-2559.2005.02097.x
Donato, R. (1999). Functional roles of S100 proteins, calcium-binding proteins of the EF-hand type. Biochim. Biophys. Acta. 1450, 191–231. doi: 10.1016/S0167-4889(99)00058-0
Donato, R. (2003). Intracellular and extracellular roles of S100 proteins. Microsc. Res. Tech. 60, 540–551. doi: 10.1002/jemt.10296
Donato, R., Cannon, B. R., Sorci, G., Riuzzi, F., Hsu, K., Weber, D. J., et al. (2013). Functions of S100 proteins. Curr. Mol. Med. 13, 24–57.
Donato, R., Sorci, G., Riuzzi, F., Arcuri, C., Bianchi, R., Brozzi, F., et al. (2009). S100B’s double life: Intracellular regulator and extracellular signal. Biochim. Biophys. Acta. 1793, 1008–1022. doi: 10.1016/j.bbamcr.2008.11.009
Dowarha, D., Chou, R. H., and Yu, C. (2018). S100B as an antagonist to interfere with the interface area flanked by S100A11 and RAGE V domain. ACS Omega 3, 9689–9698. doi: 10.1021/acsomega.8b00922
Eckert, R. L., Broome, A.-M., Ruse, M., Robinson, N., Ryan, D., and Lee, K. (2004). S100 proteins in the epidermis. J. Invest. Dermatol. 123, 23–33. doi: 10.1111/j.0022-202X.2004.22719.x
Eden, E. R., Sanchez-Heras, E., Tsapara, A., Sobota, A., Levine, T. P., and Futter, C. E. (2016). Annexin A1 tethers membrane contact sites that mediate ER to endosome cholesterol transport. Dev. Cell 37, 473–483. doi: 10.1016/j.devcel.2016.05.005
Fadista, J., Vikman, P., Laakso, E. O., Mollet, I. G., Esguerra, J. L., Taneera, J., et al. (2014). Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc. Natl. Acad. Sci. U.S.A. 111, 13924–13929. doi: 10.1073/pnas.1402665111
Fava, R. A., and Cohen, S. (1984). Isolation of a calcium-dependent 35-kilodalton substrate for the epidermal growth factor receptor/kinase from A-431 cells. J. Biol. Chem. 259, 2636–2645. doi: 10.1016/S0021-9258(17)43401-6
Fernandez-Fernandez, M. R., Rutherford, T. J., and Fersht, A. R. (2008). Members of the S100 family bind p53 in two distinct ways. Protein Sci. 17, 1663–1670. doi: 10.1110/ps.035527.108
Foertsch, F., Szambowska, A., Weise, A., Zielinski, A., Schlott, B., Kraft, F., et al. (2016). S100A11 plays a role in homologous recombination and genome maintenance by influencing the persistence of RAD51 in DNA repair foci. Cell Cycle 15, 2766–2779. doi: 10.1080/15384101.2016.1220457
Foertsch, F., Teichmann, N., Kob, R., Hentschel, J., Laubscher, U., and Melle, C. (2013). S100A11 is involved in the regulation of the stability of cell cycle regulator p21(CIP1/WAF1) in human keratinocyte HaCaT cells. FEBS J. 280, 3840–3853. doi: 10.1111/febs.12378
Futter, C. E., Felder, S., Schlessinger, J., Ullrich, A., and Hopkins, C. R. (1993). Annexin I is phosphorylated in the multivesicular body during the processing of the epidermal growth factor receptor. J. Cell Biol. 120, 77–83. doi: 10.1083/jcb.120.1.77
Gabril, M., Girgis, H., Scorilas, A., Rotondo, F., Wala, S., Bjarnason, G. A., et al. (2016). S100A11 is a potential prognostic marker for clear cell renal cell carcinoma. Clin. Exp. Metast. 33, 63–71. doi: 10.1007/s10585-015-9758-6
Guo, A.-J., Wang, F.-J., Ji, Q., Geng, H.-W., Yan, X., Wang, L.-Q., et al. (2021). Proteome analyses reveal S100A11, S100P and RBM25 are tumor biomarkers in colorectal cancer. Proteom. Clin. Appl. 15:e2000056. doi: 10.1002/prca.202000056
He, H., Li, J., Weng, S., Li, M., and Yu, Y. (2009). S100A11: diverse function and pathology corresponding to different target proteins. Cell Biochem. Biophys. 55, 117–126. doi: 10.1007/s12013-009-9061-8
Hoshino, A., Kim, H. S., Bojmar, L., Gyan, K. E., Cioffi, M., Hernandez, J., et al. (2020). Extracellular vesicle and particle biomarkers define multiple human cancers. Cell 182, 1044–1061.e1018. doi: 10.1016/j.cell.2020.07.009
Hung, K.-W., Chang, Y.-M., and Yu, C. (2012). NMR structure note: the structure of human calcium-bound S100A11. J. Biomol. NMR 54, 211–215. doi: 10.1007/s10858-012-9661-2
Inada, H., Naka, M., Tanaka, T., Davey, G. E., and Heizmann, C. W. (1999). Human S100A11 exhibits differential steady-state RNA levels in various tissues and a distinct subcellular localization. Biochem. Biophys. Res. Commun. 263, 135–138. doi: 10.1006/bbrc.1999.1319
Iridoy, M. O., Zubiri, I., Zelaya, M. V., Martinez, L., Ausin, K., Lachen-Montes, M., et al. (2018). Neuroanatomical quantitative proteomics reveals common pathogenic biological routes between amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Int. J. Mol. Sci. 20:4. doi: 10.3390/ijms20010004
Jaiswal, J. K., Lauritzen, S. P., Scheffer, L., Sakaguchi, M., Bunkenborg, J., Simon, S. M., et al. (2014). S100A11 is required for efficient plasma membrane repair and survival of invasive cancer cells. Nat. Commun. 5, 3795–3795. doi: 10.1038/ncomms4795
Ji, J., Zhao, L., Wang, X., Zhou, C., Ding, F., Su, L., et al. (2004). Differential expression of S100 gene family in human esophageal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 130, 480–486. doi: 10.1007/s00432-004-0555-x
Ji, Y. F., Li, T., Jiang, F., Ni, W. K., Guan, C. Q., Liu, Z. X., et al. (2019). Correlation between S100A11 and the TGF-beta1/SMAD4 pathway and its effects on the proliferation and apoptosis of pancreatic cancer cell line PANC-1. Mol. Cell. Biochem. 450, 53–64. doi: 10.1007/s11010-018-3372-2
Jiang, Y., Sun, A., Zhao, Y., Ying, W., Sun, H., Yang, X., et al. (2019). Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature 567, 257–261. doi: 10.1038/s41586-019-0987-8
Kanamori, T., Takakura, K., Mandai, M., Kariya, M., Fukuhara, K., Sakaguchi, M., et al. (2004). Increased expression of calcium-binding protein S100 in human uterine smooth muscle tumours. Mol. Hum. Reprod. 10, 735–742. doi: 10.1093/molehr/gah100
Katsumi, T., Guicciardi, M. E., Azad, A., Bronk, S. F., Krishnan, A., and Gores, G. J. (2019). Activated cholangiocytes release macrophage-polarizing extracellular vesicles bearing the DAMP S100A11. Am. J. Physiol. Cell Physiol. 317, C788–C799. doi: 10.1152/ajpcell.00250.2019
Kraemer, A. M., Saraiva, L. R., and Korsching, S. I. (2008). Structural and functional diversification in the teleost S100 family of calcium-binding proteins. BMC Evol. Biol. 8:48. doi: 10.1186/1471-2148-8-48
Lauritzen, S. P., Boye, T. L., and Nylandsted, J. (2015). Annexins are instrumental for efficient plasma membrane repair in cancer cells. Semin. Cell Dev. Biol. 45, 32–38. doi: 10.1016/j.semcdb.2015.10.028
Leopold, J. A. (2015). Vascular calcification: mechanisms of vascular smooth muscle cell calcification. Trends Cardiovasc. Med. 25, 267–274. doi: 10.1016/j.tcm.2014.10.021
Li, X., Zhao, Y., Xia, Q., Zheng, L., Liu, L., Zhao, B., et al. (2016). Nuclear translocation of annexin 1 following oxygen-glucose deprivation-reperfusion induces apoptosis by regulating Bid expression via p53 binding. Cell Death Dis. 7:e2356. doi: 10.1038/cddis.2016.259
Liu, Y., Han, X., and Gao, B. (2015). Knockdown of S100A11 expression suppresses ovarian cancer cell growth and invasion. Exp. Ther. Med. 9, 1460–1464. doi: 10.3892/etm.2015.2257
Luo, X., Xie, H., Long, X., Zhou, M., Xu, Z., Shi, B., et al. (2013). EGFRvIII mediates hepatocellular carcinoma cell invasion by promoting S100 calcium binding protein A11 expression. PLoS One 8:e83332. doi: 10.1371/journal.pone.0083332
Makino, E., Sakaguchi, M., Iwatsuki, K., and Huh, N. H. (2004). Introduction of an N-terminal peptide of S100C/A11 into human cells induces apoptotic cell death. J. Mol. Med. (Berl.) 82, 612–620. doi: 10.1007/s00109-004-0560-1
Memon, A. A., Sorensen, B. S., Meldgaard, P., Fokdal, L., Thykjaer, T., and Nexo, E. (2005). Down-regulation of S100C is associated with bladder cancer progression and poor survival. Clin. Cancer Res. 11, 606–611.
Meng, M., Sang, L., and Wang, X. (2019). S100 calcium binding protein A11 (S100A11) promotes the proliferation, migration and invasion of cervical cancer cells, and activates Wnt/beta-catenin signaling. Onco. Targets. Ther. 12, 8675–8685. doi: 10.2147/ott.s225248
Mitsui, Y., Tomonobu, N., Watanabe, M., Kinoshita, R., Sumardika, I. W., Youyi, C., et al. (2019). Upregulation of mobility in pancreatic cancer cells by secreted S100A11 through activation of surrounding fibroblasts. Oncol. Res. 27, 945–956. doi: 10.3727/096504019x15555408784978
Moore, B. W. (1965). A soluble protein characteristic of the nervous system. Biochem. Biophys. Res. Commun. 19, 739–744. doi: 10.1016/0006-291x(65)90320-7
Moore, B. W., and McGregor, D. (1965). Chromatographic and electrophoretic fraction of soluble proteins of brain and liver. J. Biol. Chem. 240, 1647–1653. doi: 10.1016/S0021-9258(18)97483-1
Murzik, U., Hemmerich, P., Weidtkamp-Peters, S., Ulbricht, T., Bussen, W., Hentschel, J., et al. (2008). Rad54B targeting to DNA double-strand break repair sites requires complex formation with S100A11. Mol. Biol. Cell 19, 2926–2935. doi: 10.1091/mbc.E07-11-1167
Navrátilová, A., Bečvář, V., Baloun, J., Damgaard, D., Nielsen, C. H., Veigl, D., et al. (2021). S100A11 (calgizzarin) is released via NETosis in rheumatoid arthritis (RA) and stimulates IL-6 and TNF secretion by neutrophils. Sci. Rep. 11:6063. doi: 10.1038/s41598-021-85561-3
Niu, Y., Shao, Z., Wang, H., Yang, J., Zhang, F., Luo, Y., et al. (2016). LASP1-S100A11 axis promotes colorectal cancer aggressiveness by modulating TGFbeta/Smad signaling. Sci. Rep. 6:26112. doi: 10.1038/srep26112
Oh, H. Y., Shin, S. K., Heo, H. S., Ahn, J. S., Kwon, E. Y., Park, J. H., et al. (2013). Time-dependent network analysis reveals molecular targets underlying the development of diet-induced obesity and non-alcoholic steatohepatitis. Genes Nutr. 8, 301–316. doi: 10.1007/s12263-012-0322-6
Ohta, H., Sasaki, T., Naka, M., Hiraoka, O., Miyamoto, C., Furuichi, Y., et al. (1991). Molecular cloning and expression of the cDNA coding for a new member of the S100 protein family from porcine cardiac muscle. FEBS Lett. 295, 93–96. doi: 10.1016/0014-5793(91)81393-m
Ohuchida, K., Mizumoto, K., Ohhashi, S., Yamaguchi, H., Konomi, H., Nagai, E., et al. (2006). S100A11, a putative tumor suppressor gene, is overexpressed in pancreatic carcinogenesis. Clin. Cancer Res. 12, 5417–5422. doi: 10.1158/1078-0432.ccr-06-0222
Oue, N., Hamai, Y., Mitani, Y., Matsumura, S., Oshimo, Y., Aung, P. P., et al. (2004). Gene expression profile of gastric carcinoma: identification of genes and tags potentially involved in invasion, metastasis, and carcinogenesis by serial analysis of gene expression. Cancer Res. 64, 2397–2405. doi: 10.1158/0008-5472.can-03-3514
Panda, D. K., Bai, X., Sabbagh, Y., Zhang, Y., Zaun, H. C., Karellis, A., et al. (2018). Defective interplay between mTORC1 activity and endoplasmic reticulum stress-unfolded protein response in uremic vascular calcification. Am. J. Physiol. Renal Physiol. 314, F1046–F1061. doi: 10.1152/ajprenal.00350.2017
Poeter, M., Radke, S., Koese, M., Hessner, F., Hegemann, A., Musiol, A., et al. (2013). Disruption of the annexin A1/S100A11 complex increases the migration and clonogenic growth by dysregulating epithelial growth factor (EGF) signaling. Biochim. Biophys. Acta 1833, 1700–1711. doi: 10.1016/j.bbamcr.2012.12.006
Raposo, G., and Stoorvogel, W. (2013). Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell. Biol. 200, 373–383. doi: 10.1083/jcb.201211138
Rehman, I., Azzouzi, A. R., Cross, S. S., Deloulme, J. C., Catto, J. W., Wylde, N., et al. (2004). Dysregulated expression of S100A11 (calgizzarin) in prostate cancer and precursor lesions. Hum. Pathol. 35, 1385–1391. doi: 10.1016/j.humpath.2004.07.015
Réty, S., Osterloh, D., Arié, J. P., Tabaries, S., Seeman, J., Russo-Marie, F., et al. (2000). Structural basis of the Ca(2+)-dependent association between S100C (S100A11) and its target, the N-terminal part of annexin I. Structure 8, 175–184. doi: 10.1016/s0969-2126(00)00093-9
Ringerike, T., Blystad, F. D., Levy, F. O., Madshus, I. H., and Stang, E. (2002). Cholesterol is important in control of EGF receptor kinase activity but EGF receptors are not concentrated in caveolae. J. Cell Sci. 115, 1331–1340. doi: 10.1242/jcs.115.6.1331
Safronova, A., Araujo, A., Camanzo, E. T., Moon, T. J., Elliott, M. R., Beiting, D. P., et al. (2019). Alarmin S100A11 initiates a chemokine response to the human pathogen Toxoplasma gondii. Nat. Immunol. 20, 64–72. doi: 10.1038/s41590-018-0250-8
Saho, S., Satoh, H., Kondo, E., Inoue, Y., Yamauchi, A., Murata, H., et al. (2016). Active secretion of dimerized S100A11 induced by the peroxisome in mesothelioma cells. Cancer Microenviron. 9, 93–105. doi: 10.1007/s12307-016-0185-2
Sakaguchi, M., Miyazaki, M., Inoue, Y., Tsuji, T., Kouchi, H., Tanaka, T., et al. (2000). Relationship between contact inhibition and intranuclear S100C of normal human fibroblasts. J. Cell Biol. 149, 1193–1206. doi: 10.1083/jcb.149.6.1193
Sakaguchi, M., Miyazaki, M., Sonegawa, H., Kashiwagi, M., Ohba, M., Kuroki, T., et al. (2004). PKCα mediates TGFβ-induced growth inhibition of human keratinocytes via phosphorylation of S100C/A11. J. Cell. Biol. 164, 979–984. doi: 10.1083/jcb.200312041
Sakaguchi, M., Miyazaki, M., Takaishi, M., Sakaguchi, Y., Makino, E., Kataoka, N., et al. (2003). S100C/A11 is a key mediator of Ca(2+)-induced growth inhibition of human epidermal keratinocytes. J. Cell Biol. 163, 825–835. doi: 10.1083/jcb.200304017
Sakaguchi, M., Sonegawa, H., Murata, H., Kitazoe, M., Futami, J., Kataoka, K., et al. (2008). S100A11, an dual mediator for growth regulation of human keratinocytes. Mol. Biol. Cell 19, 78–85. doi: 10.1091/mbc.e07-07-0682
Sakaguchi, M., Tsuji, T., Inoue, Y., Miyazaki, M., Namba, M., Yamada, H., et al. (2001). Loss of nuclear localization of the S100C protein in immortalized human fibroblasts. Radiat. Res. 155, 208–214. doi: 10.1667/0033-7587(2001)155[0208:lonlot]2.0.co;2
Schaefer, K., Brachwitz, K., Wai, D., Braun, Y., Diallo, R., Korsching, E., et al. (2004). Expression profiling of t(12;22) positive clear cell sarcoma of soft tissue cell lines reveals characteristic up-regulation of potential new marker genes including ERBB3. Cancer Res. 64, 3395–3405. doi: 10.1158/0008-5472.CAN-03-0809
Seemann, J., Weber, K., and Gerke, V. (1997). Annexin I targets S100C to early endosomes. FEBS Lett. 413, 185–190. doi: 10.1016/s0014-5793(97)00911-3
Seemann, J., Weber, K., Osborn, M., Parton, R. G., and Gerke, V. (1996). The association of annexin I with early endosomes is regulated by Ca2+ and requires an intact N-terminal domain. Mol. Biol. Cell 7, 1359–1374. doi: 10.1091/mbc.7.9.1359
Sobolewski, C., Abegg, D., Berthou, F., Dolicka, D., Calo, N., Sempoux, C., et al. (2020). S100A11/ANXA2 belongs to a tumour suppressor/oncogene network deregulated early with steatosis and involved in inflammation and hepatocellular carcinoma development. Gut 2:gutjnl–2019–319019. doi: 10.1136/gutjnl-2019-319019
Stulik, J., Koupilova, K., Osterreicher, J., Knizek, J., Macela, A., Bures, J., et al. (1999). Protein abundance alterations in matched sets of macroscopically normal colon mucosa and colorectal carcinoma. Electrophoresis 20, 3638–3646. doi: 10.1002/(sici)1522-2683(19991201)20:18<3638::aid-elps3638<3.0.co;2-w
Tan, X., Lambert, P. F., Rapraeger, A. C., and Anderson, R. A. (2016). Stress-induced EGFR trafficking: mechanisms, functions, and therapeutic implications. Trends Cell Biol. 26, 352–366. doi: 10.1016/j.tcb.2015.12.006
Tanaka, M., Adzuma, K., Iwami, M., Yoshimoto, K., Monden, Y., and Itakura, M. (1995). Human calgizzarin; one colorectal cancer-related gene selected by a large scale random cDNA sequencing and northern blot analysis. Cancer Lett. 89, 195–200. doi: 10.1016/0304-3835(94)03687-e
Teng, F., Jiang, J., Zhang, J., Yuan, Y., Li, K., Zhou, B., et al. (2021). The S100 calcium-binding protein A11 promotes hepatic steatosis through RAGE-mediated AKT-mTOR signaling. Metabolism 117:154725. doi: 10.1016/j.metabol.2021.154725
Thomou, T., Mori, M. A., Dreyfuss, J. M., Konishi, M., Sakaguchi, M., Wolfrum, C., et al. (2017). Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 542, 450–455. doi: 10.1038/nature21365
Thuny, F., Textoris, J., Amara, A. B., Filali, A. E., Capo, C., Habib, G., et al. (2012). The gene expression analysis of blood reveals S100A11 and AQP9 as potential biomarkers of infective endocarditis. PLoS One 7:e31490–. doi: 10.1371/journal.pone.0031490
Todoroki, H., Kobayashi, R., Watanabe, M., Minami, H., and Hidaka, H. (1991). Purification, characterization, and partial sequence analysis of a newly identified EF-hand type 13-kDa Ca(2+)-binding protein from smooth muscle and non-muscle tissues. J. Biol. Chem. 266, 18668–18673. doi: 10.1016/S0021-9258(18)55115-2
Torres-Cabala, C., Panizo-Santos, A., Krutzsch, H. C., Barazi, H., Namba, M., Sakaguchi, M., et al. (2004). Differential expression of S100C in thyroid lesions. Int. J. Surg. Pathol. 12, 107–115. doi: 10.1177/106689690401200203
Tsai, M. H., Lin, C. H., Tsai, K. W., Lin, M. H., Ho, C. J., Lu, Y. T., et al. (2019). S100A6 promotes B lymphocyte penetration through the blood-brain barrier in autoimmune encephalitis. Front. Genet. 10:1188. doi: 10.3389/fgene.2019.01188
Tu, Y., Xie, P., Du, X., Fan, L., Bao, Z., Sun, G., et al. (2019). S100A11 functions as novel oncogene in glioblastoma via S100A11/ANXA2/NF-κB positive feedback loop. J. Cell. Mol. Med. 23, 6907–6918. doi: 10.1111/jcmm.14574
Wang, C., Lin, C., Tao, Q., Zhao, S., Liu, H., and Li, L. (2019). Evaluation of calcium-binding protein A11 promotes the carcinogenesis of hypopharygeal squamous cell carcinoma via the PI3K/AKT signaling pathway. Am. J. Trans. Res. 11, 3472–3480.
Woo, T., Okudela, K., Mitsui, H., Tajiri, M., Rino, Y., Ohashi, K., et al. (2015). Up-regulation of S100A11 in lung adenocarcinoma-its potential relationship with cancer progression. PLoS One 10:e0142642. doi: 10.1371/journal.pone.0142642
Xia, Q., Li, X., Zhou, H., Zheng, L., and Shi, J. (2018). S100A11 protects against neuronal cell apoptosis induced by cerebral ischemia via inhibiting the nuclear translocation of annexin A1. Cell. Death. Dis. 9:657. doi: 10.1038/s41419-018-0686-7
Xiao, M., Li, T., Ji, Y., Jiang, F., Ni, W., Zhu, J., et al. (2018). S100A11 promotes human pancreatic cancer PANC-1 cell proliferation and is involved in the PI3K/AKT signaling pathway. Oncol. Lett. 15, 175–182. doi: 10.3892/ol.2017.7295
Yu, W., Liu-Bryan, R., Stevens, S., Damanahalli, J. K., and Terkeltaub, R. (2012). RAGE signaling mediates post-injury arterial neointima formation by suppression of liver kinase B1 and AMPK activity. Atherosclerosis 222, 417–425. doi: 10.1016/j.atherosclerosis.2012.04.001
Zhang, L., Wu, X., Liao, S., Li, Y., Zhang, Z., Chang, Q., et al. (2016). Tree shrew (Tupaia belangeri chinensis), a novel non-obese animal model of non-alcoholic fatty liver disease. Biol. Open. 5, 1545–1552. doi: 10.1242/bio.020875
Zhang, L., Zhang, Z., Li, C., Zhu, T., Gao, J., Zhou, H., et al. (2021). S100A11 promotes liver steatosis via FOXO1-mediated autophagy and lipogenesis. Cell. Mol. Gastroenterol. Hepatol. 11, 697–724. doi: 10.1016/j.jcmgh.2020.10.006
Zhang, L., Zhang, Z., Li, Y., Liao, S., Wu, X., Chang, Q., et al. (2015). Cholesterol induces lipoprotein lipase expression in a tree shrew (Tupaia belangeri chinensis) model of non-alcoholic fatty liver disease. Sci. Rep. 5:15970. doi: 10.1038/srep15970
Zhang, M. X., Gan, W., Jing, C. Y., Zheng, S. S., Yi, Y., Zhang, J., et al. (2019). S100A11 promotes cell proliferation via P38/MAPK signaling pathway in intrahepatic cholangiocarcinoma. Mol. Carcinog. 58, 19–30. doi: 10.1002/mc.22903
Zhang, M., Zheng, S., Jing, C., Zhang, J., Shen, H., Xu, X., et al. (2018). S100A11 promotes TGF-beta1-induced epithelial-mesenchymal transition through SMAD2/3 signaling pathway in intrahepatic cholangiocarcinoma. Future Oncol. 14, 837–847. doi: 10.2217/fon-2017-0534
Zhao, X. Q., Naka, M., Muneyuki, M., and Tanaka, T. (2000). Ca(2+)-dependent inhibition of actin-activated myosin ATPase activity by S100C (S100A11), a novel member of the S100 protein family. Biochem. Biophys. Res. Commun. 267, 77–79. doi: 10.1006/bbrc.1999.1918
Keywords: S100 proteins, S100A11, diseases, signaling pathways, protein interaction
Citation: Zhang L, Zhu T, Miao H and Liang B (2021) The Calcium Binding Protein S100A11 and Its Roles in Diseases. Front. Cell Dev. Biol. 9:693262. doi: 10.3389/fcell.2021.693262
Received: 10 April 2021; Accepted: 21 May 2021;
Published: 11 June 2021.
Edited by:
Jesus Chimal-Monroy, National Autonomous University of Mexico, MexicoReviewed by:
Sabrina Reinehr, Ruhr-University Bochum, GermanyEugene Anatolievich Permyakov, Institute for Biological Instrumentation (RAS), Russia
Rosario Donato, University of Perugia, Italy
Copyright © 2021 Zhang, Zhu, Miao and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huilai Miao, NjI3MjI1MzcwQHFxLmNvbQ==; Bin Liang, bGlhbmdiNzNAeW51LmVkdS5jbg==
†These authors share first authorship
 Linqiang Zhang
Linqiang Zhang Tingting Zhu2†
Tingting Zhu2† Bin Liang
Bin Liang