- 1Curriculum in Genetics and Molecular Biology, The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 2Department of Biology, The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 3Integrative Program for Biological and Genome Sciences, The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
Proper number and placement of meiotic crossovers is vital to chromosome segregation, with failures in normal crossover distribution often resulting in aneuploidy and infertility. Meiotic crossovers are formed via homologous repair of programmed double-strand breaks (DSBs). Although DSBs occur throughout the genome, crossover placement is intricately patterned, as observed first in early genetic studies by Muller and Sturtevant. Three types of patterning events have been identified. Interference, first described by Sturtevant in 1915, is a phenomenon in which crossovers on the same chromosome do not occur near one another. Assurance, initially identified by Owen in 1949, describes the phenomenon in which a minimum of one crossover is formed per chromosome pair. Suppression, first observed by Beadle in 1932, dictates that crossovers do not occur in regions surrounding the centromere and telomeres. The mechanisms behind crossover patterning remain largely unknown, and key players appear to act at all scales, from the DNA level to inter-chromosome interactions. There is also considerable overlap between the known players that drive each patterning phenomenon. In this review we discuss the history of studies of crossover patterning, developments in methods used in the field, and our current understanding of the interplay between patterning phenomena.
Meiotic Recombination
Crossovers are generally avoided during repair of DNA double-strand breaks (DSBs) in mitotically proliferating cells, presumably because they can lead to loss of heterozygosity or to chromosome rearrangement (when occurring between non-allelic repetitive sequences) (reviewed in Andersen and Sekelsky, 2010). To avoid crossovers, non-crossover outcomes are promoted through the actions of helicases that disassemble recombination intermediates (reviewed in Huselid and Bunting, 2020). In contrast, in meiotic cells DSBs are induced enzymatically and repaired to ensure that some become crossovers, which lead to chiasmata that promote accurate segregation of homologous chromosomes (Hawley, 1988). This change in outcome is achieved through the addition of numerous meiosis-specific elaborations to DSB repair.
The first step in homology-directed repair of DSBs is resection of the DSB ends (Figure 1) (reviewed in Heyer et al., 2010). In miotic cells, the recombinase Rad51 promotes strand exchange with a homologous duplex that is used as a template for synthesis; in meiosis, the meiosis-specific Rad51 paralog Dmc1 is used instead of or in conjunction with Rad51 (reviewed in Shinohara and Shinohara, 2004). As in mitotic DSB repair, dissociation of the nascent strand from the template allows completion of repair by synthesis-dependent strand annealing (SDSA), a pathway thought to generate most or all non-crossovers (Allers and Lichten, 2001; Marsolier-Kergoat et al., 2018). In the major meiotic crossover pathway, the template strand that is displaced by synthesis anneals to the other resected end of the broken chromatid, and further synthesis and ligation leads to a double-Holliday junction (dHJ) structure, which is resolved into crossover products (Figure 1).
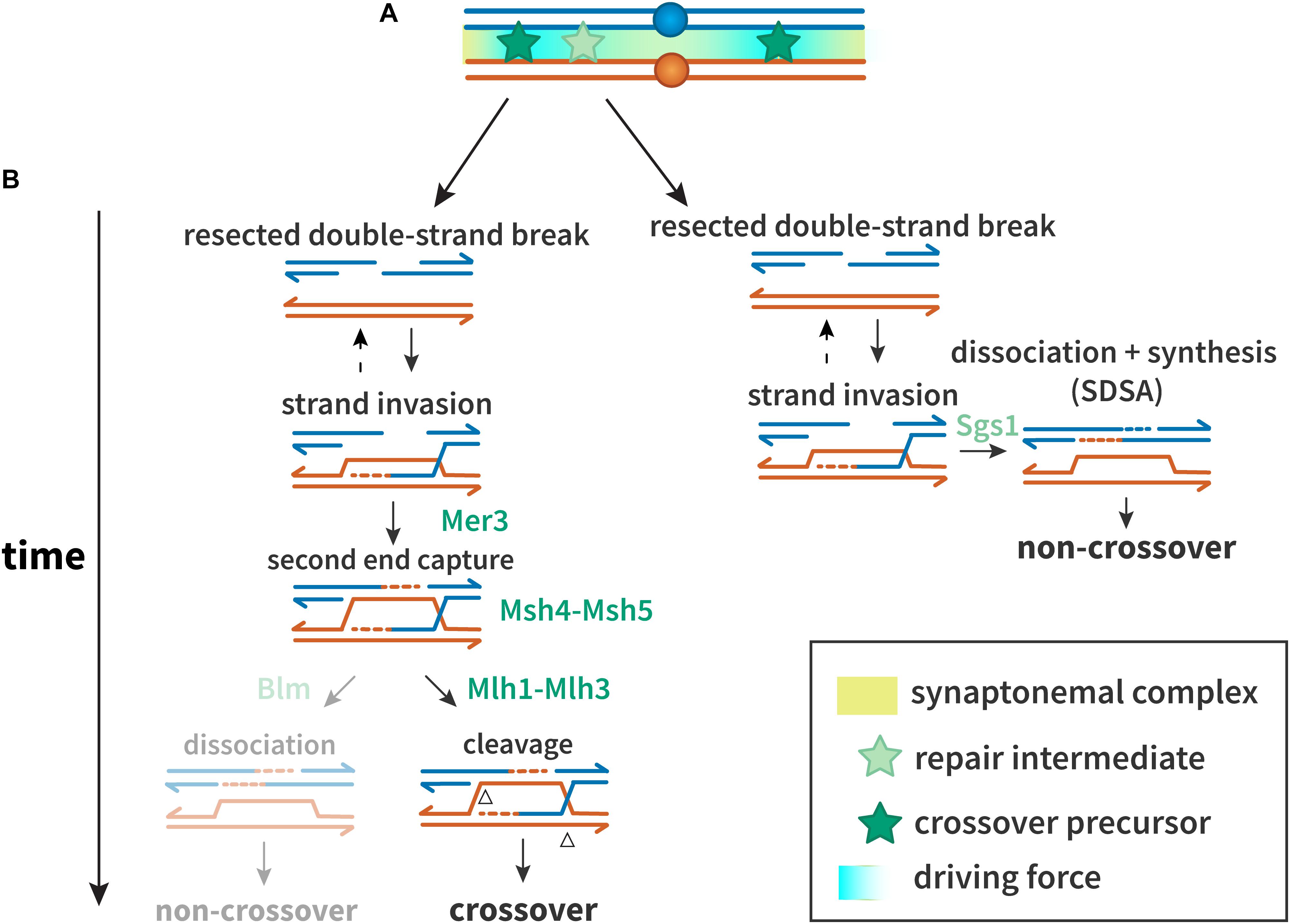
Figure 1. Proteins and mechanisms of crossover interference. (A) A driving force (teal), potentially protein aggregation or mechanical stress, results in designation of crossover precursors (stars, dark green) at spaced intervals. Repair intermediates that do not experience sufficient driving force (stars, light green) are not so designated, and become non-crossovers. (B) Interference at the DNA level. Crossover precursors are processed through the class I crossover pathway, involving the ZMM proteins. Mer3 promotes second end capture, and Msh4–Msh5 clamp to and stabilize branched molecules to prevent their dissociation and to recruit Msh1–3. Intermediates are resolved by Mlh1–Mlh3 to generate crossovers. Non-crossovers in yeast are thought to be exclusively processed through the synthesis-dependent strand annealing (SDSA) pathway, promoted by Sgs1. In other organisms, the Sgs1 ortholog Blm may also promote formation of non-crossovers after second end capture.
A defining feature of the major meiotic crossover pathway is dependence on a group of proteins referred to as ZMM (Zip, Msh, and Mer). In Saccharomyces cerevisiae, the ZMMs consist of at least seven players: Zip1, Zip2, Zip3, Zip4/Spo22, Spo16, Mer3, Msh4, and Msh5. Zip1 is a structural component of the synaptonemal complex (SC), a proteinaceous structure that assembles between homologous chromosomes during prophase I (Sym et al., 1993). Msh4 and Msh5, homologs of the Escherichia coli mismatch repair protein MutS, form a heterodimer termed MutSγ, which is thought to promote crossing over by stabilizing pre-crossover intermediates to block SDSA and/or promote formation of dHJs (Snowden et al., 2004; Jessop et al., 2006; Oh et al., 2007).
Recent studies have identified an important role for proteolysis of ZMM proteins in promoting crossovers in meiosis. Ahuja et al. (2017) found that the proteasome is necessary for chromosomes to pair and for crossover-designated DSBs to effectively be repaired as crossovers. The proteasome is recruited to chromosomes by Zip1 and Zip3. SUMO modification mediated by the meiotic E3 ligases RNF212 and HEI10 is thought to act like a checkpoint in mouse, pausing recombination by inducing degradation of various recombination factors (Rao et al., 2017). Likewise, Msh4 has been shown to be targeted by a degron for ubiquitin-independent proteolysis (He et al., 2020). This degron is under control of the kinase Cdc7, and phosphorylation of the degron permits Msh4–Msh5 to promote crossovers.
The final steps in crossover formation are also modified in meiosis. In mitotic cells, it is believed that the primary pathway for processing dHJs involves unwinding Blm helicase and decatenation by Topoisomerase 3α, a process that generates only non-crossovers (Plank et al., 2006). An alternative is resolution of the Holliday junctions by structure-selective endonucleases, which can produce crossover or non-crossover products (Wyatt and West, 2014). In meiosis, however, most or all dHJs are processed to generate crossovers (Allers and Lichten, 2001), through the action of a complex containing MutLγ, a heterodimer of Mlh1 and Mlh3, and other proteins (Zakharyevich et al., 2012; Cannavo et al., 2020; Kulkarni et al., 2020).
The meiotic DSB repair process outlined above ensures that some DSBs will be repaired as crossovers. However, the determination of which become crossovers and which are repaired as non-crossovers is highly regulated to ensure that all chromosome pairs receive at least one crossover, that crossovers are in positions that facilitate segregation, and that crossovers are excluded from regions where they might impede segregation (Koehler et al., 1996b). The phenomena that achieve this are collectively referred to as “crossover patterning.” Below, we discuss various meiotic crossover patterning phenomena – interference, assurance, and suppression (Figure 2), from initial recognition to current understanding.
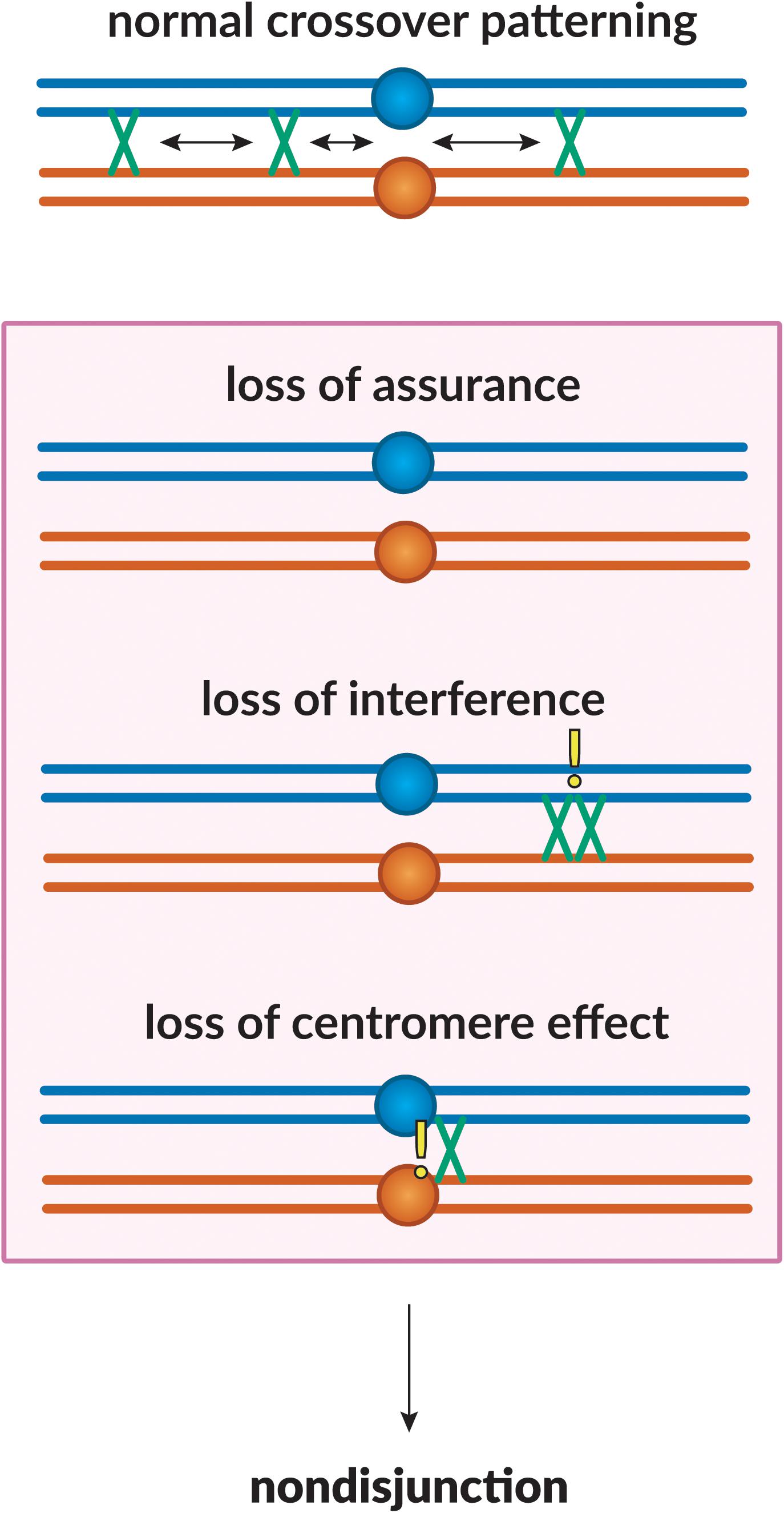
Figure 2. Crossover patterning phenomena. The proper placement of crossovers along the chromosome is governed by three patterning phenomena. Homologous chromosomes are shown in blue and orange, with crossovers between them shown in green. Loss of assurance results in a lack of crossing-over between a pair of homologs; loss of interference results in two crossovers being placed in close proximity to one another; loss of the centromere effect results in centromere-proximal crossovers. These phenomena can lead to a failure in proper chromosome segregation, leading to non-disjunction.
Interference
History
Crossover interference was originally observed by Sturtevant when constructing the first linkage map in Drosophila melanogaster in 1913 (Sturtevant, 1913). His observation that one crossover makes the occurrence of another less likely was based on the results of counting double crossovers (DCOs) between six sex-linked factors, where he observed that DCOs between two adjacent intervals were much lower in frequency than should be expected by chance. This phenomenon, termed interference by Muller, was later shown by Sturtevant to be an intra-chromosomal process (Sturtevant, 1915).
Muller later showed that interference is limited to crossovers on the same chromosome, with a crossover on one chromosome having no effect on crossing over on another (Muller, 1916a). Muller further noted, just as Sturtevant had, that the reach of interference seemed to depend on the distance of the intervals being considered, with longer intervals showing less interference than shorter ones. Weinstein (Weinstein, 1918) confirmed this by showing that interference decreases with distances up to 46 map units (a measure of recombination in D. melanogaster equivalent to centiMorgans) from the initial crossover.
The possibility of chromatid interference was also considered (Figure 3): When there are two crossovers on the same chromosome arm, do the chromatids involved in one affect which are used in the other? This question was first investigated by Emerson and Beadle (1933). They found that in Drosophila the first crossover did not seem to influence which chromatids were involved in the second, suggesting no chromatid interference. The same result was also shown in both budding and fission yeast (Mortimer and Fogel, 1974; Munz, 1994), though not in Neurospora crassa (Perkins, 1962). Nonetheless, most studies have assumed no chromatid interference (Mather, 1935; Chakraborty et al., 2017).
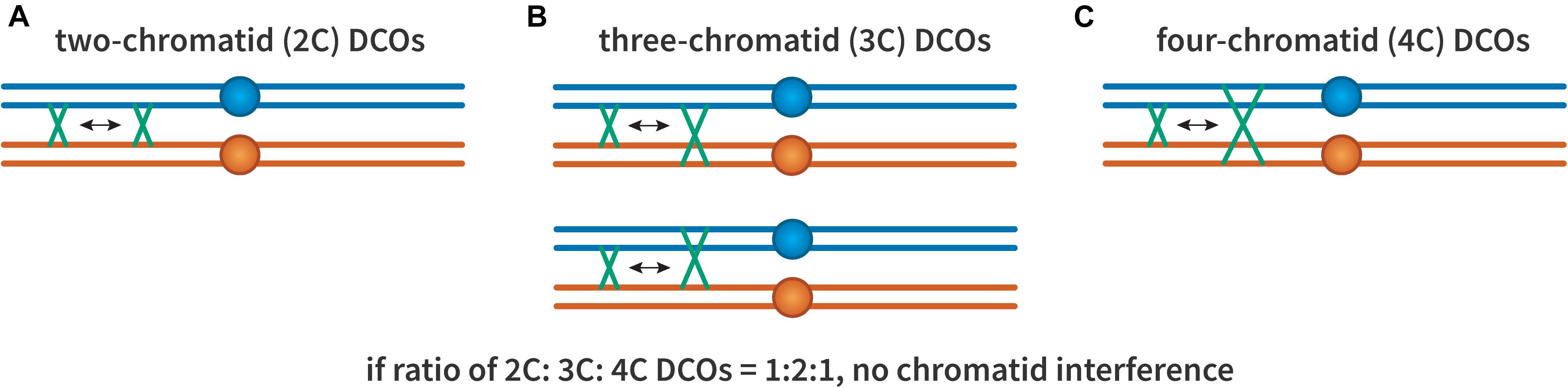
Figure 3. Chromatid interference. Each panel illustrates two crossovers on one bivalent, with each line representing one double-stranded DNA chromatid. The first (leftmost) crossover is the same in all cases, occurring between the two inner chromatids. (A) A 2-chromatid DCO. (B) Two possible 3-chromatid DCO configurations. (C) A 4-chromatid DCO. If there is no chromatid interference, the ratio of 2:3:4 should be 1:2:1.
Interfering and Non-interfering Crossovers
Not all crossovers participate in interference. Based on studies of recombination mutants in Caenorhabditis elegans, Zalevsky et al. (1999) proposed the existence of two meiotic recombination pathways. In most organisms, the primary meiotic crossover pathway (described above and in Figure 1) produces crossovers that are subject to interference, while other pathways produce crossovers that are indifferent to interference (de los Santos et al., 2003; Argueso et al., 2004; Börner et al., 2004; Higgins et al., 2004; de Boer et al., 2006; Holloway et al., 2008). This results in some crossovers participating in interference and others neither contributing to nor responding to interference. These have been called classes I and II crossovers, respectively.
Class II crossovers were initially identified as being generated independently of MutSγ (Zalevsky et al., 1999; de los Santos et al., 2003), but the class II designation has been used in different ways. For example, when a blockage occurs in the primary pathway due to a missing component, backup mechanisms will complete repair (De Muyt et al., 2012; Zakharyevich et al., 2012; Kohl and Sekelsky, 2013). These backup pathways, like mitotic DSB repair, are not directed toward a crossover outcome. Nonetheless, crossovers do occur in some instances, but this requires structure-selective nucleases such as Mus81–Mms4 rather than MutLγ (de los Santos et al., 2003; Argueso et al., 2004; Berchowitz et al., 2007; Holloway et al., 2008). Some consider these to be class II crossovers because they come from a different pathway, but if the blockage happens after crossover designation has occurred and interference has been enforced, these crossovers may exhibit interference.
The proteins involved and percentage of crossovers attributed to classes I and II differ among organisms. In budding yeast, only about 60% of meiotic crossovers come from the class I pathway (de los Santos et al., 2003; Medhi et al., 2016). Unique features of this pathway are similar in Arabidopsis and mouse, with ZMM proteins and MutLγ playing central roles (Falque et al., 2007; Higgins et al., 2008; Lu et al., 2008; Svetlanov et al., 2008); however, class I crossovers comprise 75–90% of all meiotic crossovers in Arabidopsis (Berchowitz et al., 2007; Higgins et al., 2008) and more than 90% of crossovers in mouse meiosis (Baker et al., 1996; Holloway et al., 2008).
In Drosophila, 90–100% of crossovers come from a class I pathway (Baker and Carpenter, 1972), but Dmc1, Msh4, and Msh5 are absent from fly genomes (Sekelsky, 2017). Kohl et al. (2012) hypothesized that crossover-designated recombination intermediates are instead stabilized by the MCM-like proteins Mei-217, Mei-218 and Rec. Furthermore, the class I resolvase function of Mlh1–Mlh3 is replaced by the XPF ortholog Mei-9 and its partners Ercc1 and the Slx4 ortholog Mus312 (Sekelsky et al., 1995; Yıldız et al., 2002; Radford et al., 2005). All crossovers in C. elegans are processed through the ZMM-based class I pathway (Zalevsky et al., 1999), though it also uses resolvases other than MutLγ (Saito et al., 2009; Agostinho et al., 2013; O’Neil et al., 2013). Schizosaccharomyces pombe appears to lack most of the specialized features of the class I pathway, including ZMM proteins, and most crossovers are dependent on Mus81 and exhibit little or no interference (Smith et al., 2003; Fowler et al., 2018).
Measures of Interference
Interference can be measured based on genetic distance (recombination frequencies between genetic loci) or physical distance (chromosome axis length or distance in base-pairs between crossovers from whole-genome sequencing). The main methods in use are described below.
Coefficient of Coincidence
Although development of the genetic map by Sturtevant (1913, 1915) and Bridges (1915) was integral to understanding interference, a better measure to directly measure interference was soon developed. In 1916, Muller defined coincidence as the occurrence of a crossover in each of two adjacent intervals (i.e., a double-crossover, DCO) (Muller, 1916b). Muller measured strength of interference through the ratio between the number of observed DCOs and the number of DCOs expected if the two intervals are independent (no interference). This measure is now referred to as the coefficient of coincidence (CoC). Interference (I) is represented as 1 - CoC. If CoC is one, then I is zero and there is no interference between the two intervals in question; if CoC is between zero and one there is interference between the two intervals (negative interference is also possible but not considered here). This method is often used to calculate strength of interference based on counts of crossovers in specific intervals, as in genetic studies.
Interference in Tetrads
The use of fungi as genetic model organisms allows recovery of all products of meiosis in tetrads or octads, making it possible to calculate interference with just two markers. If a strain that is heterozygous for two linked markers, say A B on one homolog and a b on the other, goes through meiosis to produce a tetrad with four spores, three outcomes are possible. Parental ditype (two A B spores and two a b spores) results when there has not been a crossover between the two loci or a 2-strand DCO (Figure 3). Non-parental ditype (two A b spores and two a B spores) results from a 4-strand DCO, and tetratype (A B, A b, a B, a b) results from a single crossover or a 3-strand DCO. Papazian (1952) developed a simple equation to calculate interference from the observed numbers of each class. Although widely used, these formulas are applicable only to datasets within certain parameters, and determining statistical significance between datasets if difficult. Stahl (2008) published a method termed “The Better Way,” that overcomes these limitations.
Cytological Measures of Interference
Chiasma interference has been measured cytologically in humans and other organisms. Hulten (1974) measured the length of each chromosome and counted chiasmata in chromosome spreads of a human testicular sample. Hulten (1974) mapped the distribution of chiasmata on chromosome arms and determined interference based on the mean crossover count compared to the variance of the data. Jones (1984) noted that this measurement is flawed in that it does not consider the position of crossovers along the chromosome. He expanded on this approach by arbitrarily dividing the chromosome arms into even intervals to calculate CoC using chiasmata from cytological data. Hulten later developed the Chiasma Interference Map (CHIM), in which chiasmata are marked along the chromosome and chiasmata that occurred on the same chromosome are joined by a loop. The strength of interference can be visualized via the size and quantity of the loops relative to the total number of chiasmata, with longer loops indicating stronger interference (Hulten, 2011).
Fluorescence microscopy allows measurement of interference based on other cytological markers. MLH1 marks sites of crossovers in mammalian meiosis, and distances between foci along a bivalent can be used to estimate interference (Froenicke et al., 2002). Zip3 has been used as a similar marker in budding yeast (Zhang et al., 2014b). These measurements of interference are unique in counting only class I crossovers, whereas other methods include both class I and class II crossovers in calculations of interference. It is also notable that these methods mark crossover sites during pachytene, while chiasmata are apparent later, during diakinesis and metaphase I.
Measuring Interference Using Whole-Genome Sequencing
In whole-genome genotyping or sequencing, the markers (e.g., single nucleotide polymorphisms) are so dense that each interval is too small to have enough crossovers for CoC analysis. One method to circumvent this problem is to divide the chromosome into arbitrary intervals of equal length and use these to calculate CoC or to fit a gamma distribution (Broman et al., 2002; Anderson et al., 2015). Another is to compare the average distance between crossovers on chromosomes with multiple crossovers (Miller et al., 2016).
Modeling Interference Data
A more recent way to compare interference between datasets is to make use of computational models that simulate interference, such as the beam-film model (discussed below) (White et al., 2017). The beam-film program can fit experimental data to a model and contains a parameter (L) that reflects the strength of interference.
It should be noted that these methods differ in profound ways. Chromosome length is measured in recombination frequency in the genetic methods, in meiotic chromosome axis length in cytological methods, and in DNA sequence length in sequence-based methods. Also, counting of MLH1 or Zip1 foci includes only class I crossovers, but other methods include both class I and class II crossovers. The beam-film model can accommodate date from genetic, cytological, or sequence studies, and has a parameter to include class II crossovers.
Factors Influencing Interference
Temperature
The influence of temperature on crossover frequencies was first studied by Plough, Stern, and Graubard in the early 20th century (Plough, 1917a,b; Stern, 1926; Graubard, 1934). Schweitzer (1935) analyzed Graubard’s data and concluded that temperature does not affect interference in Drosophila, and that the average distance between two crossovers does not change at different temperatures. However, a more recent study in A. thaliana showed that increased temperatures lead to an increase in overall crossover frequencies through the formation of additional class I crossovers, suggesting that physical measures of interference are decreasing (De Storme and Geelen, 2020).
Chromosome Size
Small chromosomes exhibit higher rates of crossing over per physical unit than larger chromosomes (Kaback et al., 1989; Mortimer et al., 1989; Riles et al., 1993), and the smallest chromosome in S. cerevisiae shows increased recombination when split into two smaller segments (Link and Olson, 1991). Kaback et al. (1999) observed that interference increased with the size of the chromosome. Intervals of the same cytological length showed greater interference when relocated to larger chromosomes, leading the authors to conclude that the size dependency of meiotic recombination rates is mediated at least in part by crossover interference. Kaback et al. (1999) hypothesized that the recombination machinery initiates crossover formation on larger chromosomes earlier than on smaller ones because they are larger targets. This leads to an interference signal “spreading” along the chromosome on either side of the crossover, rendering many areas of the larger chromosomes unable to form another crossover and leading to smaller chromosomes having less competition for the recombination machinery. The authors proposed that when interference begins to act on larger chromosomes there is more of some rate-limiting component available per unit of recombination-proficient DNA. This results in the remaining rate-limiting component promoting crossing over at greater rates on smaller chromosomes. Thus, the size dependency of recombination rates is explained by larger chromosomes initiating crossing over earlier and having longer interference tracts. Many rate-limiting factors in the recombination machinery have been identified (Reynolds et al., 2013; Qiao et al., 2014; Ziolkowski et al., 2017; He et al., 2020).
In contradiction to this hypothesis, Stahl et al. (2004) showed that shorter chromosomes had more class II crossovers than longer chromosomes, suggesting that the chromosomal size dependence of recombination rates isn’t due to changes in the strength of interference, but is instead due to a difference in the proportion of class I versus class II crossovers.
More recently, Murakami et al. (2020) have shown that in short chromosomes of budding yeast there is a greater recruitment of DSB proteins, indicating a higher density of DSBs. Their data also showed that shorter chromosomes tend to stay in a DSB-competent state for longer; however, no inferences can be made about how this may affect the process of interference in these chromosomes, as it is possible, based on the study by Stahl et al. (2004) that the extra DSBs are being repaired as class II crossovers.
Synaptonemal Complex
The synaptonemal complex (SC) was first suggested to be important for crossover interference when it was reported that Aspergillus nidulans and S. pombe, two organisms that do not have an SC, also do not exhibit interference (Moens, 1969; Snow, 1979; Egel-Mitani et al., 1982; Bähler et al., 1993; Munz, 1994). Concomitant defects in interference and synapsis were also seen in as1 and asb mutants in tomato (Moens, 1969; Havekes et al., 1994). Further, organisms heterozygous for chromosomal translocations do not display interference in the region of the rearrangement, where SC continuity is presumably disrupted (Arana et al., 1987; Parker, 1987).
Studies in budding yeast suggest that the relationship between SC and interference is more complex. Zip1 is a component of the SC, and zip1 mutants fail to build SC (Sym et al., 1993). Based on tetrad analysis in zip1 mutants, Sym and Roeder (Sym and Roeder, 1994) concluded that interference requires the SC, and suggested that Zip1 could be the polymer that diffuses outward from crossover sites as in the model polymerization model proposed by King and Mortimer (see below). This requirement of Zip1 and its orthologs for crossover interference has also been shown by numerous other studies (Hayashi et al., 2010; Libuda et al., 2013; Duroc et al., 2014; Voelkel-Meiman et al., 2015; Rog et al., 2017; Capilla-Perez et al., 2021). However, it is important to note that since Zip1 mutants are defective in class I crossover formation, the apparent reduction in interference may be due to the remaining crossovers being class II. Chua and Roeder (1997) further supported the idea that the SC plays a role in interference when they showed that budding yeast mutants lacking the telomere-associated meiotic protein TAM1, also known as Ndj1 and required for telomeric clustering during prophase I as well as proper homolog pairing and disjunction (Conrad et al., 1997; Trelles-Sticken et al., 2000), had defects in both chromosome synapsis and crossover interference. They attributed this to TAM1 playing a role in homolog pairing, leading to defects in alignment and synapsis.
In contrast, Fung et al. (2004) observed that synapsis initiation complexes (SIC), identified as Zip2 foci, exhibit interference, suggesting that crossing over happens at these initiation sites, as had been suggested earlier by Egel (1978). Since SICs assemble prior to SC formation and are still present in a zip1 mutant where SC is absent, Fung et al. (2004) concluded that interference in budding yeast can act independently of synapsis. This independence has also been shown in Sordaria (Zickler et al., 1992). A 2006 study showed substantial interference between MSH4 foci in mice with defective SCs, although the data did not allow determination of whether MLH1 foci interfere in the absence of the SC (de Boer et al., 2006). It is also important to note that zip1 mutants often undergo delays in prophase (Sym et al., 1993), which may be true in the tam1 mutant as well, suggesting that the SC’s apparent role in interference could be due to cell cycle defects. Thus, the role of the synaptonemal complex in interference remains enigmatic.
Interference Models
Since the discovery of crossover interference, several models have been proposed to describe this phenomenon. We briefly describe the most influential models below, in chronological order; additional discussions can be found in Berchowitz and Copenhaver (2010) and Otto and Payseur (2019).
Polymerization Model
The polymerization model proposed by King and Mortimer (King and Mortimer, 1990) hypothesized that early recombination structures are randomly distributed across the chromosome, and that once crossing over is initiated at the positions where these structures attach, a polymerization reaction is triggered which would then inhibit crossovers in neighboring regions by preventing the binding of other early structures to the SC. This polymerization reaction extends bidirectionally from each crossover site, explaining why interference decreases with distance from the original crossover. King and Mortimer also put forth a computational simulation of the model that they were able to fit well to Drosophila and budding yeast crossover data. While no polymer has been identified that fits this model, it agrees well with an earlier interference model proposed by Egel (1978) suggesting that crossover interference is a result of synapsis. Maguire (1977) put forth the idea that establishment of crossover sites may occur before synapsis is initiated, and that while the SC may be important for the process of crossing over, its “deployment” along the chromosome may have another function. Based on her arguments, Egel argued that if crossover sites are formed before synapsis and are also points of nucleation for the SC, it is this formation and zippering of the SC that prevents crossovers in neighboring sites. According to this model, crossovers cannot form in chromosomal regions that have already synapsed, and only regions that are yet to synapse retain the ability to form a crossover (Maguire, 1977).
This model would seem to be incompatible with interference in Drosophila and C. elegans, where SC appears to be complete before recombination is initiated (Dernburg et al., 1998; McKim et al., 1998). However, the SC is not a static structure. This was first recognized in the phenomenon of synaptic adjustment, wherein rearranged chromosomes that have different lengths initially appear to synapse based on homology, but then adjust so that the two chromosomes in each bivalent are equal lengths (reviewed in Moses et al., 1984). Studies in C. elegans found that crossovers locally alter SC, possibly to a form that is not permissive to additional crossovers (Libuda et al., 2013; Machovina et al., 2016). As discussed by Otto and Payseur (2019), these observations fit well with the proposal that interference propagates through the SC.
Counting Model
Like earlier models of interference based on a renewal process, Foss et al. (1993) proposed a model in which recombination intermediates that are randomly distributed along the chromosome can either become crossovers or non-crossovers, but intermediates are “counted” by a recombinase in such a way that two crossovers need to be separated by a certain number of non-crossovers. Although unable to explain interference data in S. cerevisiae (Foss and Stahl, 1995), a mathematic implementation of the counting model fit satisfactorily when tested against crossover data from other species and incorporated class II crossovers (Copenhaver et al., 2002; Housworth and Stahl, 2003; Stahl et al., 2004).
Beam-Film Model
The beam-film model of Kleckner et al. (2004) proposes that chromosomes behave like elastic beams covered on one face by a thin film that has flaws along its edges. When the chromosome is subjected to forces that cause the beam to expand, it does so to a greater degree than the film. This leads to the stretching of the film, which puts mechanical stress on it, resulting in the flaws cracking. When a crack is formed at one of the flaws, tensile stress in neighboring regions is relieved, with the release dissipating for some distance. Cracks may still occur at flaws on the film that are outside the reach of the stress relief perpetuating from the initial crack. This is applied to meiotic recombination by considering crossover precursors, such as DSBs or other intermediates, as the flaws on the film, with those that crack under mechanical stress being designated to become crossovers. Interference is explained by a crossover being able to prevent others in its vicinity by relieving the tensile stress on nearby flaws, consequently preventing them from cracking as well. A mathematical model based on beam-film can fit crossover distribution data from several species (Zhang et al., 2014a).
Compartmentalized Signaling Model
Studies in C. elegans suggested that the SC has liquid-like properties and Rog et al. (2017) proposed that this might help to explain crossover patterning. Zhang et al. (2018) showed that a set of four RING finger proteins, ZHP1-4, are part of a signaling network that functions within the SC to select early recombination intermediates for crossover designation. In C. elegans, there is always one crossover per pair of homologous chromosomes, so this signaling pathway designates only one crossover per compartment, with each SC being one compartment. Zhang et al. (2018) suggest that this may occur if both positive and negative activities within the SC to set up a wave of crossover designation potential.
Spatial Cluster Model
Fowler et al. (2018) proposed a model for crossover interference through DSB interference, in which DSB hotspots cluster within a chromosomal region of approximately 200 kb, with a limit of one DSB per cluster. This model fits the distance dependency of interference, as longer distances are more likely to span multiple clusters that act independently. However, this model seems to suggest that non-crossovers are also subject to interference, which does not appear to be the case (e.g., Miller et al., 2016). Additionally, since DSBs within each cluster are formed randomly, this model would suggest that crossovers forming at the boundaries of two neighboring clusters would not interfere. Another criticism of this model has been that it is based on studies in S. pombe, an organism in which interference had been reported to be absent (Mortimer and Fogel, 1974; Munz, 1994; Fowler et al., 2018). Fowler et al. (2018) provide evidence for weak crossover interference and suggest that interference in S. pombe occurs through clusters encompassing only one homolog instead of both, implying that clusters are distributed independently along each homolog, leading to efficient DSB interference without crossover interference.
Mutations That Disrupt Interference
Genetic studies have identified many mutations that have an apparent effect on interference, but interpretation of these is often complicated. First, changes in interference would be expected to be inversely correlated with crossover number (e.g., weaker interference would be expected to increase crossover number), but changes in crossover number can occur without changes in interference. One important way this can happen is through loss of class I crossovers or increase in class II crossovers. Indeed, many mutations that appear to alter interference affect the absolute or relative numbers of class I and class II.
A good example is the sgs1 mutant of S. cerevisiae. Oh et al. (2007) reported that crossover interference is reduced in sgs1 mutants, which might be interpreted as evidence that Sgs1 plays a role in establishing interference; however, Zip2 foci, which mark sites designated to become class I crossovers, are normal in sgs1 mutants, suggesting that normal interference is established (Fung et al., 2004). The solution to this apparent paradox came from subsequent studies that revealed that all crossovers in sgs1 mutants require structure-selective endonucleases other than MutLγ (De Muyt et al., 2012; Zakharyevich et al., 2012). Thus, designation of crossover sites may occur normally, with interference, but maturation of these into crossovers is defective. Unlike MutLγ, other structure-selective endonucleases are not biased toward crossover resolution. This should result in a twofold decrease in class I crossovers. At the same time, loss of the anti-crossover activity of Sgs1 in the class II pathway results in an increase in those crossovers. Therefore, the reduction in interference reported by Oh et al. (2007) does not reveal a role for Sgs1 in the process of interference, but rather a change in ability to complete formation of class I crossovers combined with an increase in class II crossovers.
One protein that does appear to have a direct role in interference is Topoisomerase II in S. cerevisiae. Zhang et al. (2014b) analyzed distances between Zip3 foci in top2 mutants and found that they were decreased relative to wild type, while total Zip3 foci were increased in the mutants. They hypothesized that topoisomerase II is required to relieve torsional stress at the site of crossovers. SUMOylation of Top2 by Ubc9 and subsequent ubiquitination by Slx5/8 is required for this function, and absence of Slx5 or Slx8 yields the same phenotype as in top2 mutants. The sirtuin Sir2 physically interacts with Slx5/8 and activates its SUMO-targeted ubiquitin ligase (STUbL) activity to perform ubiquitination, and is also required for interference, as is Red1, another substrate of Ubc9.
Crossover Assurance
History
In most organisms, achiasmate chromosomes are rare (Hillers and Villeneuve, 2003). The first observation that all chromosomes exhibit at least one chiasma was made by Darlington and Dark (1932). They studied recombination in a grasshopper species with large variations in chromosome size. If crossovers were distributed randomly among the genome, smaller chromosomes would be less likely to experience at least one crossover, while larger chromosomes would be more likely to experience multiple crossovers. However, Darlington and Dark found that all chromosomes had at least one chiasma, indicating that there is a process that ensures at least one inter-homolog crossover on all chromosomes. Darlington hypothesized that chiasmata must be important for proper segregation of chromosomes (Darlington, 1937). Mather (1937) also graphed the number of chiasmata as a function of chromosome length and showed a minimum of one chiasma regardless of how short a chromosome is. He noted that the first chiasma is formed “irrespective of the length of the chromosome” and that additional chiasmata, if any, are subject to interference.
Another observation of assurance was made by Owen (1949). Callan and Montalenti (1947) studied the mosquito Culex pipiens and found what they interpreted as evidence for interference between intervals on different chromosome arms. This was unexpected, as interference between intervals separated by the centromere had not been found in any organisms prior. Owen reanalyzed their data and determined that interference need not cross the centromere in Culex if it is assumed that formation of a first crossover per bivalent is more favored than formation of subsequent crossovers. He defined this as a “primary chiasma” or “obligatory chiasma,” and reasoned that it is not on the same footing with other chiasmata, as it is required for synapsis and proper chromosome segregation. The reason behind the apparent cross-centromere interference in Culex was the short chromosome arms in this organism, resulting in bivalents that frequently had just one crossover, making it appear as if that crossover suppresses crossovers on the other arm. Owen’s interpretation held for an additional mosquito species studied by Callan and Montalenti, Theobaldia longiareolata, in which apparent cross-centromere interference was not observed due to its longer chromosome arms with a greater average number of chiasmata per bivalent. Jones (1984), noting that the distribution of chiasmata between chromosomes of similar sizes is more dispersed than would be expected if events were placed randomly, suggested that the lack of achiasmate chromosomes or univalents indicates that there must be one obligate chiasma per bivalent.
The Obligate Crossover
The function of the crossover assurance mechanism is to ensure the generation of at least one crossover per bivalent. In recombination-dependent meiotic programs, a crossover is required between each pair of homologs to ensure a chiasma that will promote proper chromosome segregation at the end of meiosis I (Darlington, 1937; Hawley, 1988). A dramatic example of how crossovers promote segregation comes from studies of mammalian sex chromosomes. Koller and Darlington (1934) found that an obligate crossover was always formed between the X and Y chromosomes in rat. This conclusion was disputed for several decades due to the observation that in most mammals the X and Y associate end-to-end and do not exhibit visible chiasmata. However, Burgoyne (1982) later discovered via electron microscopy that a very distal chiasma forms between the X and Y to promote pairing. Burgoyne proposed an “X–Y crossover model” that suggests that there is a region of genetic homology between the X and Y chromosome (the “pseudoautosomal region”) in mammals that must experience an obligatory crossover during meiosis for proper chromosome segregation. All genes distal to the obligate crossover will be transmitted to both male and female offspring, behaving in pedigrees like autosomal genes.
The obligate crossover between the X and Y chromosomes must occur proximal to the sex determining region of the Y chromosome, Sry, which activates the male transcriptional program. Ashley (1994) used fluorescence in situ hybridization (FISH) to study how the X and Y chromosomes segregate into sperm, and determined that aneuploid products were infrequent, indicating that spermatocytes that experience XY non-disjunction fail to progress and differentiate into sperm. Hinch et al. (2014) later confirmed that PRDM9 binding sites, which coincide with recombination hotspots, are found in the human pseudoautosomal region between the X and Y chromosomes, PAR1. Intriguingly, recombination appears to drive sequence evolution more strongly in the pseudoautosomal region than on the autosomes. Recently, Papanikos et al. (2019) found that mouse ANKRD31 is essential to promoting double-strand break assurance, especially in the pseudoautosomal region. ANKRD31 deficiency leads to loss of an obligate crossover between the X and Y chromosome as well as consequent chromosome missegregation.
Exceptions to the requirement for an obligate crossover are seen in Drosophila, in which the X chromosomes do not experience a crossover in 10–15% of meioses and chromosome 4 never has crossovers. Nonetheless, these chromosomes segregate correctly in >99% of meioses due to an achiasmate segregation system (Hawley and Theurkauf, 1993), though the X still segregates with higher fidelity when it has at least one crossover (Koehler et al., 1996a).
Mechanisms of Assurance
Crossover assurance is enforced on multiple levels (Figure 4): assurance that sufficient DSBs are formed to generate an obligate crossover per chromosome, assurance of inter-homolog recombination bias, and assurance that a crossover is implemented by enforcing pro-crossover recombination pathways. These mechanisms are covered below.
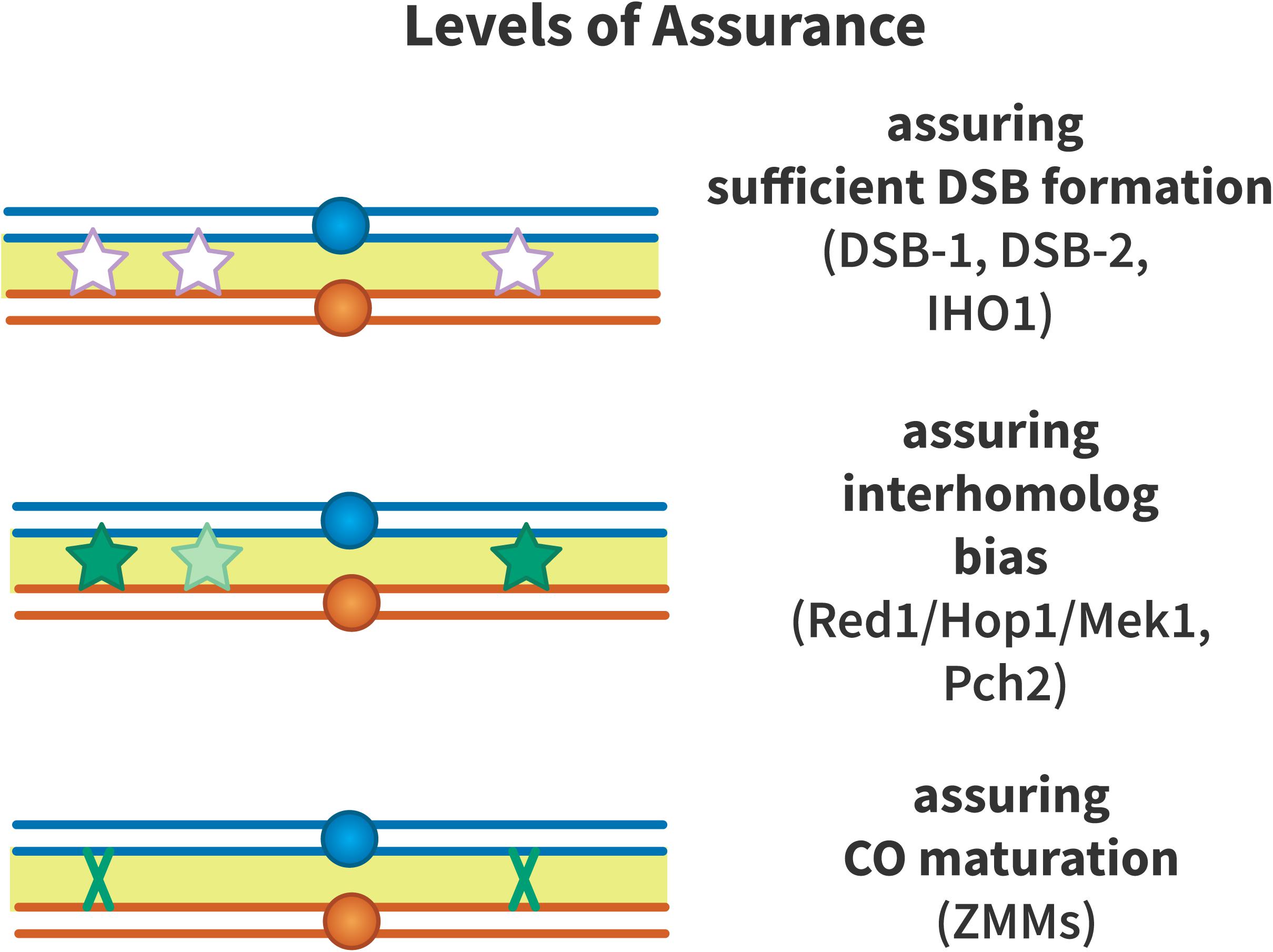
Figure 4. Levels of crossover assurance. Assurance is enforced at three levels. DSB-1 and DSB-2 form a checkpoint in C. elegans that ensures that sufficient double-strand breaks (DSBs, white stars) are formed to generate an obligate crossover. In mouse, IHO1 plays a similar role. From this pool of DSBs, inter-homolog recombination is promoted by Red1, Hop1, Mek1, and Pch2 to ensure that recombination intermediates (light green stars) engage with the homologous chromosome to form crossover precursors (dark green stars). The driving force to generate crossover precursors may be aggregation of ZMM proteins at recombination nodules or mechanical stress that must be relieved by crossover formation. Efficient crossover maturation (dark green X) is ensured by the ZMM proteins that define the class I crossover pathway.
Double-Strand Break Assurance
Meiotic DSBs are formed in excess relative to crossovers, likely to ensure that at least one crossover will be formed per homolog pair. A crossover assurance checkpoint involving the paralogs DSB-1 and DSB-2 was identified in C. elegans (Stamper et al., 2013). DSB-1 localizes to chromosomes during the time of DSB formation. When crossover formation is impaired, DSB-1 persists on chromosomes, suggesting that the time in which DSB formation is permitted is extended in dsb-1 mutants. Failure to form an obligate crossover is sufficient for this phenotype to appear. DSB-2 similarly localizes to chromatin as DSBs form and disappears as RAD-51 foci appear, which mark early recombination intermediates. Association of DSB-2 with chromosomes is dependent on phosphorylation of the SUN domain protein SUN-1 and loading of RAD-51 at DSBs. Like DSB-1, DSB-2 persists on chromosomes when DSBs fail to form or recombination intermediates are not repaired (Rosu et al., 2013).
Double-strand breaks numbers are modulated by at least four different pathways in mouse (Dereli et al., 2021). DSBs regulate the number of SPO11-auxiliary protein complexes by activating the DNA damage response kinase ATM. DSBs additionally reduce IHO1 in their direct vicinity. IHO1 (homolog of yeast Mer2), a HORMAD1-interacting protein and SPO11 auxiliary protein, forms a chromatin-binding complex with MEI4 and REC114 that is required for DSB formation in mice (Dereli et al., 2021). In mice, DSB-dependent homologous recombination facilitates pairing and synapsis, which results in loss of IHO1, HORMAD1 and SPO11 complexes (Dereli et al., 2021). Lastly, DSBs along with the DNA damage response kinases ATM, ATR and PRKDC globally deplete IHO1 from chromosome axes. These pathways likely act to ensure that sufficient DSBs are generated to make an obligate crossover without creating a vast surplus of DSBs that threaten genomic integrity.
Double-strand break formation in yeast is regulated to ensure recombination on shorter chromosomes. The Spo11 accessory proteins Rec114 and Mer2 associate earlier and dissociate later from short chromosomes compared to longer chromosomes (Murakami et al., 2020). The mechanism that promotes this bias toward short chromosomes is unclear, but Rec114 and Mer2 accumulation are influenced by replication timing, while their dissociation is triggered by homolog pairing. ZMM-mutant budding yeast also generate higher numbers of DSBs than wild-type, and this control of DSB number is genetically separable from the pathway that connects DSB formation to meiotic progression (Thacker et al., 2014). These results suggest that homolog pairing, mediated by ZMM proteins, reduces DSB formation to prevent unproductive DSBs between paired homologs that are already engaging in inter-homolog recombination.
Inter-Homolog Strand Exchange Bias
An obligate crossover must occur between homologous chromosomes to ensure proper chromosome segregation. Mechanisms that promote inter-homolog recombination over recombination between sister chromatids have been described. In yeast, Hop1 and Red1 function structurally in axial/lateral elements and function jointly with Mek1 to enforce a barrier to inter-sister recombination. The chromosome axis protein Hop1 is phosphorylated in response to DSBs, and then triggers dimerization of the meiosis-specific kinase Mek1, which phosphorylates proteins that limit inter-sister recombination (Niu et al., 2005, 2007). One target of Mek1 is Hed1, a meiosis-specific protein that binds Rad51 and suppresses its activity (Tsubouchi and Roeder, 2006; Busygina et al., 2008). Mek1 also phosphorylates the Rad51 binding partner Rad54, reducing Rad51’s activity (Niu et al., 2009). In organisms that use the meiosis-specific strand exchange protein Dmc1, Dmc1 is required for interhomolog bias (Brown et al., 2015), and thus Rad51 activity is inhibited to allow Dmc1 to function as the major meiotic strand exchange protein (Callender et al., 2016).
Pch2 is a highly conserved ATPase that has been implicated in many processes in meiosis. In yeast, Pch2 has been found to control association between Zip3, which localizes to crossover precursors, and the chromosome axis proteins Hop1/Red1. Although Pch2 is not required to generate crossovers at normal levels, pch2 mutants appear to have reduced interference and lack crossover assurance (Chakraborty et al., 2017). Joshi et al. (2009) suggested that Pch2 remodels the chromosome axis into an array of crossover control modules that interact over a long range to ensure that there is an obligate chiasma and that crossovers are maximally spaced. C. elegans pch-2 mutants lose access to recombination with the homologous chromosome early, suggesting that PCH-2 is required to maintain interhomolog recombination bias (Deshong et al., 2014).
Robust Crossover Designation and Maturation
Shinohara et al. (2008) studied null mutants of the ZMM gene SPO16 and found that, while crossovers are reduced in these mutants, the residual crossovers that are observed display interference. Furthermore, Spo16 interacts with Zip4 via co-immunoprecipitation, and assembly of Spo16 foci depends on Zip1 and Zip3, but not on Msh4. Shinohara et al. proposed that the ZMMs may consist of two sub-assemblies: one that enforces interference by stabilizing crossover precursors at the DNA level, and one that promotes crossovers at these sites to generate assurance.
In mouse, the E3 ligases RNF212 and HEI10 generate crossovers by SUMOylating recombination proteins (Reynolds et al., 2013; Qiao et al., 2014). These factors are thought to act as a checkpoint that pauses recombination by altering the turnover time of meiotic recombination proteins such as DMC1 and MutSγ. This may stall intermediates at an early step in recombination to ensure that the appropriate pro-crossover factors are present to establish crossover designation (Prasada Rao et al., 2017).
In C. elegans, DSBs are preferentially repaired as COs in the absence of inhibitory effects from other recombination precursors (Rosu et al., 2011). Rosu et al. (2011) used excision of the Mos1 transposon to make a targeted DSB in spo-11 mutants, in which no meiotic breaks are formed. They found that nearly all interhomolog repair events in these mutants were COs.
Cytoskeletal forces have also been shown to promote synapsis, recombination, and crossover assurance through signaling via SUN- and KASH-domain proteins. In Arabidopsis, the kinesin AtPSS1 is required for crossover assurance (Duroc et al., 2014). In Atpss1 mutants, some chromosomes lack an obligate crossover, while the number of total meiotic crossovers is comparable to wild type. This kinesin directly interacts with the KASH-domain proteins WIP1 and WIP2. Ndj1/Tam1, a protein that tethers telomeres to the nuclear envelope, stabilizes interactions between homologous chromosomes and thus may promote inter-homolog recombination (Conrad et al., 1997; Chua and Roeder, 1997).
In addition to crossover designation, it is important to ensure effective crossover maturation to generate the obligate crossover. Wang et al. (2017) modeled human oogenesis using the beam-film framework and found that a lower crossover maturation efficiency could explain the higher number of achiasmate bivalents in oocytes.
Crossover Suppression
History
The final patterning event we will discuss is crossover suppression associated with specific chromosomal features. Most notably, suppression occurs both in the pericentromeric region and telomeric regions. Centromeric suppression is thought to be crucial for proper chromosome segregation in humans, budding yeast, fission yeast, and D. melanogaster (Koehler et al., 1996a; Lamb et al., 1996; Rockmill et al., 2006; Smith and Nambiar, 2020). Two models have been proposed to explain how proximal crossovers promote non-disjunction (Orr-Weaver, 1996). First, proximal crossovers may disrupt sister-chromatid cohesion at the centromere. Since proximal cohesion needs to be maintained until separation of sisters at anaphase II, disruption may lead to premature separation of sister chromatids (PSSC). The second model is the converse: Because proximal cohesion is not released at anaphase I, a proximal crossover causes the homologs to become entangled, resulting in both segregating to the same daughter. One or both suggestions may account for the existence of mechanisms that reduce or eliminate centromere-proximal crossovers. It may be that disruption of cohesion and PSSC is more important in organisms with point centromeres and relatively small regions of pericentromeric cohesion, but entanglement is more of a risk in organisms in which centromeres are embedded in large blocks of heterochromatin are more expansive pericentromeric cohesion.
The first observation of centromere-proximal crossover suppression, now known as the centromere effect, was made by Beadle (1932), when he observed a decrease in crossing over in medial regions of a Drosophila chromosome when they were moved closer to the centromere in a translocation stock. Beadle studied a translocation that attached the right half of 3R to the centromere (or “spindle fiber” as he called it) of chromosome 4. In homozygous translocation flies, the percent crossing over in the intervals now closest to the 4 centromere dropped dramatically, while crossing over within more distant intervals was not affected. Beadle concluded that the decrease in crossovers in these two regions was a result of them having been moved closer to the chromosome 4 centromere.
In a 1936 review, Mather presented more evidence for the idea that the centromere exerts control over the distribution of crossing over (Mather, 1936). He used recombination data from other researchers to show that while genetic loci are well spaced out in the middle of the chromosome (as in the case of the telocentric X) or chromosome arm (as in the case of metacentric 2 and 3), a clear crowding of loci can be seen around the centromere. This is indicative of low recombination frequency near the centromere, which led Mather to speculate that crossing over in a particular region is a function of its distance from the centromere.
Factors Influencing the Centromere Effect
Temperature
Harold Plough was the first to study the effects of environmental changes on crossing over (Plough, 1917a). His data showed that while starvation and food type did not affect crossover frequencies in Drosophila, temperature did. When studying the regions between chromosome 2 markers close to and flanking the centromere, he observed an increase in crossover percentages at both higher (31 C) and lower (13 C) temperatures, which he attributed to a physical change that was occurring in chromatin structure. This increase in centromere-proximal crossover frequencies at high temperatures was later shown to hold for chromosomes 1 and 3 as well (Plough, 1921; Stern, 1926). Mather (1939) showed that heterochromatin was responsible for crossing over being highly sensitive to temperature, and speculated that variations caused by other environmental factors could also be a result of their effect on heterochromatin. A more recent study also found that increasing temperature leads to an increase in proximal crossovers in barley, a species in which crossovers are primarily distal under typical temperature conditions (Phillips et al., 2015).
Maternal Age
Bridges (1915) showed that maternal age can influence crossing over in Drosophila when he observed that older females showed a decrease in pericentromeric crossover percentages on chromosome 2. Later, he showed that this decrease was most drastic in the center of metacentric chromosome 3, decreasing as loci further from the centromere were considered (Bridges, 1927). Thus, Bridges showed in 1915, nearly two decades before Beadle’s data on suppression of crossing over near the centromere, that crossover percentages between centromere-proximal markers were as low as 5%. Although it would have been impossible for Bridges to have noticed this centromere-proximal reduction in crossing over as he had no knowledge of the physical distances between these markers, this data is possibly the first evidence of the centromere effect.
Heterochromatin
Mather (1939) studied how both the centromere and heterochromatin influenced crossing over in the Drosophila X chromosome using various inversion stocks in which markers that were ordinarily centromere proximal were moved to more distal positions and vice versa. His results led him to confirm his earlier belief that crossover frequencies in euchromatic regions are dependent on their distance from the centromere. Mather also observed some amount of crossing over in heterochromatic regions that had been moved farther away from the centromere and speculated that these levels may also be dependent on distance from the centromere. Baker (1958) did not agree with these findings, and thought that the crossovers Mather reported were most likely in adjacent euchromatin since the markers he’d used did not delineate heterochromatin very precisely. Baker supported this claim by showing virtually no crossing over in a Drosophila virilis stock in which a large chunk of chromosome 5 heterochromatin had been moved to the tip of chromosome 3 via reciprocal translocation. Since then, it has been widely believed that crossing over in heterochromatic regions is uncommon, if not absent (Carpenter and Baker, 1982; Szauter, 1984). It was also thought that pericentromeric heterochromatin was causing crossover suppression near the centromere, possibly by restricting access to recombination proteins (Westphal and Reuter, 2002), as structural differences in the synaptonemal complex are seen in the two types of chromatin (Carpenter, 1975; Stack, 1984).
Westphal and Reuter (2002), investigated the role of chromatin in centromeric crossover suppression by studying the effects of dominant suppressor-of-variegation mutations on crossover frequencies. These mutations are thought to cause changes in heterochromatin structure, consequently leading to suppression of heterochromatin-mediated gene silencing. The authors hypothesized that this may result in a change in crossover patterning in heterochromatin as well as flanking euchromatin. In support of this hypothesis, of the 46 mutations they tested, 16 increased crossing over in pericentromeric heterochromatin. A 2012 study in A. thaliana also showed the same dependence of crossover frequency on chromatin states, when a DNA hypomethylated mutant that showed pericentromeric transcriptional activity also showed an increase in centromere proximal crossovers (Yelina et al., 2012). However, this study, along with others that looked at hypomethylated Arabidopsis mutants, observed no change in crossover suppression in pericentromeric regions (Colomé-Tatché et al., 2012; Melamed-Bessudo and Levy, 2012; Mirouze et al., 2012). This was a surprising result as pericentric heterochromatin is hypermethylated in wild-type plants and would have been expected to show increased rates of recombination under conditions of DNA methylation loss. The authors of these studies propose that lowering DNA methylation levels may be making chromatin that is already open further accessible to crossover formation.
Despite considerable evidence that heterochromatin plays an important role in suppressing centromere-proximal crossovers, there is also support for the hypothesis that distance from the centromere is just as, if not more, critical. Mather (1939) showed in Drosophila that a euchromatic interval moved farther from the centromere but closer to a larger length of heterochromatin showed less crossover suppression than an interval moved closer to the centromere but near a smaller length of heterochromatin. He concluded from this that the centromere fiber effect is more a consequence of centric action than proximity to heterochromatin. Lindsley and Sandler (1977) showed that despite having the largest amount of pericentromeric heterochromatin, the Drosophila X chromosome had significantly higher crossing over in proximal euchromatin than either autosome. Their data also suggested that the centromere effects of each chromosome in Drosophila remained roughly the same despite them having different amounts of heterochromatin. This has been corroborated by Yamamoto and Miklos (1978), who also showed that crossing over in the proximal euchromatin of Drosophila X chromosomes with varying extents of heterochromatin deletions depended more on the distance from the centromere than on amount of heterochromatin. They suggested that heterochromatin acts only as a passive spacer between the centromere and euchromatin, implying that chromosome structure and mechanics, as well as DNA content, would be crucial in defining the centromere’s control over crossing over in an organism. However, they decidedly acknowledge that when combined with other data on recombination being genetically controlled (Catcheside, 1977; Lindsley and Sandler, 1977), crossover suppression near the centromere appears to be mediated by a complex system with multiple facets of control.
More recently, Hartmann et al. (2019) observed that when a large block of heterochromatin was inserted into chromosome 2R of Drosophila, there was no significant crossover suppression in adjacent intervals, suggesting that the centromere effect does not arise from an innate property of heterochromatin. They also went on to show that pericentromeric crossover suppression seems to be through two mechanisms: complete suppression in the densely staining, highly repetitive alpha heterochromatin, and the centromere effect that exhibits a distance-dependent suppression that extends far into euchromatic sequences. This is consistent with Yamamoto and Miklos’ conclusion that suppression of crossovers in the vicinity of the centromere has multiple causes.
Synaptonemal Complex
Carpenter (1975) observed using electron microscopy that the structure and morphology of the SC is different in the pericentromeric heterochromatin as compared to more distal regions on the chromosome that are euchromatic. This was further confirmed by her observation that the SC of the largely heterochromatic chromosome 4 in Drosophila usually had only heterochromatic morphology. Combined with her observation that all recombination nodules in her experiment were found in distal chromosomal regions that had euchromatic SC morphology, this raises the possibility that the SC could be mediating the centromere effect.
Stack conducted an electron microscopic study of meiotic chromosomes from mice and two angiospermous plants and showed that the SC is shorter and more under-represented in the heterochromatic regions of meiotic chromosomes, suggesting a reason for the lack of crossing over in heterochromatin (Stack, 1984). In addition to confirming Carpenter’s observations of SC structure being different in euchromatin versus heterochromatin, he also showed that heterochromatic SC is more densely enclosed in compact chromatin, which he implied could sterically inhibit essential recombination enzymes from accessing it.
Mechanistic Insights Into the Centromere Effect
Although much is known about various factors influencing centromere-proximal crossover suppression, the exact mechanism behind the centromere effect is still unknown. One hypothesis is that DSB formation is suppressed near the centromere. This is supported by a 2006 study in Drosophila that showed that the DSB cytological marker γ-His2Av failed to colocalize with HP1, a marker for heterochromatin (Mehrotra and McKim, 2006). As colocalization was observed in irradiated flies, DSBs were believed to be excluded from heterochromatin in wild-type meiotic cells. Further support for this proposal comes from a study in S. pombe showing that the pericentric cohesin complex actively represses pericentric DSB formation (Nambiar and Smith, 2016) (Figure 4). The mechanism of action proposed involves the heterochromatin protein Swi6 recruiting Rec8-Psc3 – mitotic cohesin complex subunits – to pericentric regions. Both Swi6 and Psc3 function to keep out Rec11, a meiotic cohesin complex protein, and prevent it from binding Rec8. This ensures that Rec11 is unable to recruit linear element protein Rec10, leading to the protein responsible for creating DSBs, Spo11, remaining inactive. This leads to the suppression of DSB formation and therefore meiotic recombination. Furthermore, the authors suggest that a molecular mechanism involving different cohesin complexes acting at pericentromeric heterochromatic regions to suppress recombination is a mechanism conserved across those species in which heterochromatin is found at the pericentromere.
Spo11 itself has been observed to play a role in crossover patterning, when a 2018 study in Arabidopsis determined that a Spo11 hypomorph that showed a proportional reduction in the number of DSBs along the chromosome was able to disproportionately decrease the number of crossovers in pericentromeric regions (Xue et al., 2018) (Figure 5). However, studies in budding yeast that mapped DSBs using whole-genome methods and ssDNA accumulation have shown that although DSBs are suppressed near the centromere, the length of chromosome along which suppression is observed, originally thought to be 20 kb (Gerton et al., 2000), and later 8–10 kb (Buhler et al., 2007), is really only 1–3 kb (Pan et al., 2011). Considerable DSB activity was seen in pericentromeric regions, including hotspots within 10 kb of the centromere (Blitzblau et al., 2007). Further support for the presence of DSBs in pericentromeric regions comes from Symington and Petes (1988), who showed, using strains that were heterozygous for restriction sites within the centromeric sequence, that there are widespread gene conversion (GC) events occurring at centromeric regions in yeast. Symington and Petes (1988) concluded that in budding yeast, the rates of centromeric gene conversion events during meiosis are similar to non-centromeric regions, and since GC events, while leading only to non-crossovers, still arise from DSB repair, these results show that DSBs are indeed formed in centromeric regions. More recently, a 2015 study threw some light on the molecular mechanism behind pericentromeric crossover suppression in budding yeast by showing that the kinetochore Ctf19 complex is important in two ways: It prevents DSB formation within a 6 kb region of the centromere and also suppresses inter-homolog recombination within the pericentromere (Vincenten et al., 2015). The authors suggest that prevention of DSB formation is through regulating recruitment of chromosomal axis proteins/local chromatin organization, and that crossover suppression is through recruitment of cohesins at pericentromeric regions, which they speculate promotes inter-sister repair over inter-homolog recombination. These cohesins were proposed by Vincenten et al. (2015) to recruit Zip1, an SC protein that has been implicated in promoting pericentromeric inter-sister repair at the expense of inter-homolog repair (Chen et al., 2008).
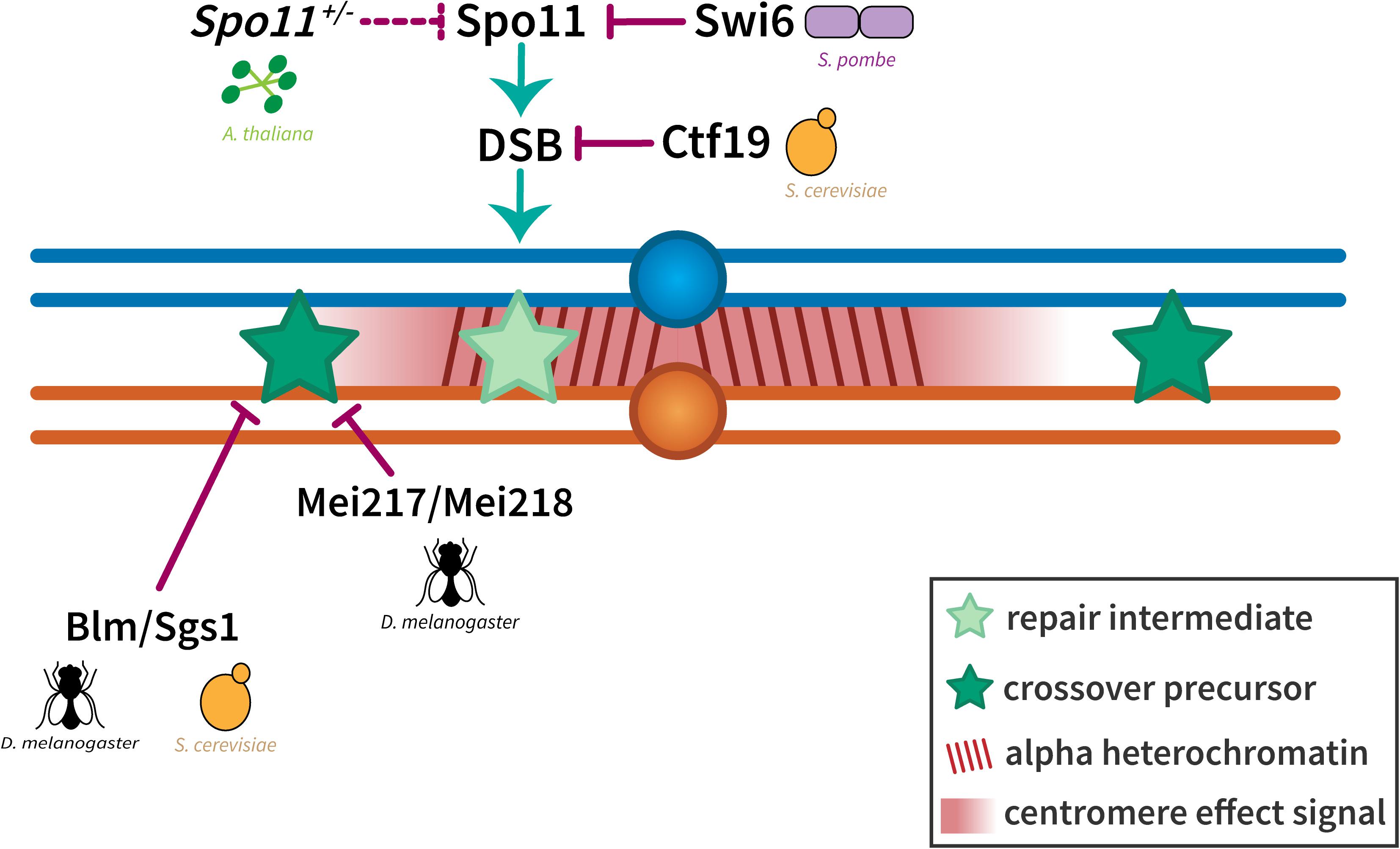
Figure 5. Suppression of crossovers near the centromere. The exclusion of crossovers near the centromere seems to be enforced at two levels. Crossover suppression in the highly repetitive alpha heterochromatin has been shown to be a result of the prevention of DSBs either through local chromatin organization (in S. cerevisiae) or the inactivation of Spo11 (in S. pombe and A. thaliana). Crossover suppression in the less repetitive beta heterochromatin and the proximal euchromatin has been shown to be regulated through Bloom syndrome helicase/Sgs1 (in D. melanogaster and S. cerevisiae) and dependent on distance from the centromere (represented by a centromere effect signal shown in red). Homologous chromosomes are shown in blue and orange with dark red lines in the pericentromeric region representing alpha heterochromatin. Light and dark green stars represent repair intermediates and crossover precursors, respectively.
Similar results were later also seen in Drosophila when Comeron et al. (2012) detected gene conversion in regions of low or no crossing over, and showed that non-crossover recombination events are more uniformly distributed along the genome than crossovers were. In a subsequent study, Miller et al. (2016) found that non-crossover gene conversion events are not sensitive to the centromere effect. Both studies were able to evaluate non-crossover repair products by detecting changes in single nucleotide polymorphisms in unique-sequence centromere-proximal regions.
Sgs1, the budding yeast ortholog of Blm helicase, was shown by Rockmill et al. (2006) to be an important factor in preventing precocious separation of sister chromatids. Their study showed that the most common cause of spore inviability in budding yeast is aneuploidy arising from PSSC, which is often associated with, and promoted by, crossing over in centromere-proximal regions, further supporting the suggestion that proximal crossovers lead to non-disjunction.
In the experiments of Brand et al. (2018) described above, in which Drosophila mauritiana Mei-217 and Mei-218 were put into D. melanogaster, the researchers noted that crossovers were especially elevated in telomere- and centromere-proximal regions. Furthermore, they saw an increase in crossovers in the interval that spans the centromere in chromosome 2, leading them to conclude that the Mei-217 and Mei-218 proteins may be involved in centromeric and telomeric crossover suppression.
It is also possible that the mechanisms behind the centromere effect are primarily structural, and that the number of centromeres is able to influence the strength of the effect. In support of this argument, Redfield showed in studies of triploid crossing over that there was a 1.5- and 3-fold increase in crossover frequencies at the center of chromosomes 2 and 3 – where the centromere is located - when compared to diploid females (Redfield, 1930, 1932). This increase in crossover frequencies decreased with distance from the centromere, which suggests that an increased total number of centromeres could be acting as a molecular sponge and “soaking up” a CE signal, ultimately leading to a weaker centromere effect.
While some genes involved in facilitating the centromere effect have come to light in the past few years, there is much that is still unknown. It will be important to look closely at whether the mechanisms behind the centromere effect are genetic or structural, how this effect relates to other patterning events, and how crossover suppression in centromeric regions is different from suppression in telomeric regions.
Interplay Between Patterning Phenomena
Recent work has suggested that crossover assurance, interference, and suppression may be interconnected or part of an overarching crossover control mechanism. Interactions between crossover patterning phenomena are highlighted below.
Interference and the Centromere
The earliest suggestion that the centromere can impede interference comes from Muller’s 1916 paper where he speculated that coincidence values on the second chromosome would be different if crossovers on the same chromosomal arm are compared to crossovers on either side of the centromere even when separated by the same distance in both cases (Muller, 1916a). He suggested that this could be due to chromosomes bending in the middle, or due to differences in structure at centromeric regions. In support of Muller’s finding, Kikkawa (1932) showed an increase in coincidence values in the central regions of the chromosome, which led him to conclude that crossover interference in the sections spanning the centromere is weaker than elsewhere on the chromosome. Graubard (1934) also showed that crossovers on one chromosomal arm do not interfere with crossovers on the other. These findings imply that the centromere can block an interference ‘signal.’ However, this block does not seem to be complete, as Graubard also observed that coincidence values between two small, adjacent intervals on either side of the centromere do not equal one (as they should for zero interference) but display less interference than intervals of the same size that are located on the same chromosomal arm. He concluded that coincidence values in a particular interval are not affected by the presence of either a terminal or central centromere but instead depend on the length of the interval being considered.
These conclusions from studies in insects were extended to Neurospora by Bole-Gowda et al. (1962), who demonstrated through tetrad analysis that there is little to no interference between crossovers on either side of the centromere. However, more than 30 years later, Colombo and Jones (1997) showed using previously published statistical analyses of chiasma data from grasshoppers that interference seemed to be able to act across the centromere when coincidence values were calculated, or distances from the nearest chiasma and the centromere were correlated. This led them to conclude that interference is blind to the presence of a centromere. Later, Kaback et al. (1999) also showed that in S. cerevisiae recombination on one chromosome arm is affected by changes in the size of the other. They concluded that this is indicative of the interference ‘signal’ having the ability to pass through the centromere, since their results suggest that crossover interference in budding yeast increases with the size of the chromosome. They explained their stance by pointing out that the model in which the centromere blocks interference ignores the fact that centromeric markers have greater physical separation and is based only on genetic mapping data. They proposed that in centromere-proximal regions, particularly in organisms that have large amounts of pericentromeric heterochromatin, interference appears to be blocked by the centromere because it has a much greater distance to cover. However, this does not seem to be the case across species, as Fowler et al., who in 2018 proposed the DSB hotspot clustering model of interference in S. pombe, speculated that the reason an interference ‘signal’ is unable to travel across the centromere in certain organisms is due to DSB hotspot clusters not being formed in pericentromeric regions (Fowler et al., 2018). Further, since Drosophila and other metazoa differ greatly from yeast in their chromatin structure, sequence, and even centromeric SC morphology, the role that the centromere plays in relaying or blocking an interference signal may well be different across these species.
In a 2012 study in A. thaliana, Yelina et al. (2012) showed increased centromere-proximal crossovers in a methyltransferase1 (met1) mutant with hypomethylated DNA. However, they observed no difference when the total number of crossovers in met1 mutants was compared to wild type, which they took as an indication that crossover interference and crossover homeostasis were acting to remodel the changed crossover landscape in the hypomethylated mutant to compensate for the increased centromere-proximal crossovers. Similarly, an Arabidopsis study by Xue et al. (2018) showed that pericentromeric crossover frequencies fell in SPO-11 hypomorphs, where DSB numbers are drastically reduced. The authors suggest that this could allow medial crossover frequencies to rise, which in turn could inhibit pericentromeric crossovers through interference.
Hatkevich et al. (2017) showed that D. melanogaster mutants deficient for Bloom syndrome helicase lost crossover interference and crossover suppression in centromeric regions. They went on to speculate that interference and the centromere effect may be interdependent, with the centromere effect reinforcing interference by ensuring that most crossovers happen medially along the chromosome. However, they also state that this could be unlikely as the D. melanogaster telomere effect, or the suppression of crossovers in the telomeric regions, is not as strong as it should be for the two crossover suppression events to be coordinately reinforcing interference. The experiments of Brady et al. (2018) discussed above also provide a link between crossover suppression in pericentromeric regions and interference. However, Brady et al. (2018) found that flies mutant for mei-41, which encodes the Drosophila ortholog of ATR kinase, lose class I crossovers but appear to retain suppression of centromere-proximal crossovers. The authors proposed that crossover suppression is established earlier than other patterning processes.
These results suggest that it is possible for the centromere effect and interference to be independent events operating through entirely different mechanisms that happen to require some of the same factors to proceed correctly. However, it remains possible that the two events are established through the same mechanism, involving a crossover suppression signal traveling outward from the centromere/existing crossovers, mediated by some common proteins but separated temporally during meiosis.
Interference and Assurance
As pointed out by Wang et al. (2015), the need for crossover assurance is obvious, since at least one chiasma is necessary to promote accurate segregation of homologs, but whether there is a selective advantage of having interference is less clear. Several possible functions for interference have been suggested. It has been suggested that crossovers near one another may lead to a lack of sufficient cohesion to stabilize the bivalent (Fowler et al., 2018), but this should only be the case for two-chromatid DCOs. A study in C. elegans found that interference plays a key role in ensuring proper chromosome segregation by limiting the number of crossovers per homolog (Hollis et al., 2020); however, this may be a problem unique to organisms without defined centromeres. Interference may serve to limit the number of crossovers, but in Arabidopsis, increasing crossovers by three–sixfold has no apparent negative effect on fertility or chromosome segregation (Crismani et al., 2012; Girard et al., 2015). Wang et al. (2015) proposed that spacing out crossovers strikes a balance between the advantages of generating new combinations of alleles and the disadvantages of breaking up co-adapted combinations.
Several observations suggest that the strength of interference can be subject to selection. Brand et al. (2018) found that replacing D. melanogaster Mei-217 and Mei-218 with the D. mauritiana orthologs captured many of the crossover differences between these species, including reduced interference. They also reported strong evidence for positive selection within mei-218 in both species, suggesting this might be associated with changes in interference and therefore in crossover numbers. Gorlov and Gorlova (2001) simulated a cost-benefit analysis of recombination and suggested that natural selection balances, through changes in interference, the positive and negative effects of recombination. The selective pressures that might drive higher or lower crossover rates are unknown.
There are also special cases where selection may operate on the strength of interference. In C. elegans, chromosomes are holocentric in somatic cells, but in meiosis the crossover produces an asymmetry that results in one end being selected for assembly of the kinetochore (Albertson and Thomson, 1993; Martinez-Perez et al., 2008). Two crossovers on the same bivalent may disrupt this process, leading to selection for absolute interference (one crossover per bivalent). Another possible case is suggested by studies of autopolyploids that arise from intraspecies whole-genome duplication. In early stages of allopolyploidy, homologous chromosomes form quadrivalents, and multiple chiasmata can lead to missegregation (Yant et al., 2013). Bomblies et al. (2016) proposed that increases to the strength of interference might promote more accurate meiotic segregation by reducing the number of chiasmata.
Another function for interference might be to promote assurance. The phenomenon of crossover homeostasis might support a dependency relationship between assurance and interference. Martini et al. (2006) studied budding yeast mutants with hypomorphic alleles of spo11 and found that reduced DSB levels did not lead to the same reductions in crossover levels, suggesting an assurance mechanism that is buffered to some degree against different numbers of precursors. Homeostasis might involve a mechanism to produce a set number of crossovers per meiosis, within some range. This could explain the interchromosomal effect in Drosophila, where the presence of structural rearrangements on one chromosome both prevent it from crossing over with its homolog and lead to increased crossovers on other chromosomes (Lucchesi and Suzuki, 1968; Crown et al., 2018). If a set number of crossovers must be achieved, interference would limit the number of crossovers on larger chromosomes, forcing smaller chromosomes to cross over.
Finally, it is possible that interference and homeostasis are merely byproducts of the process through which a precursor is designated to become a crossover. In the beam-film model, mechanical stress provides a driving force to generate designation of an obligate crossover, and the resulting distance-dependent release of stress leads to both spacing of crossovers (interference) and homeostasis (Wang et al., 2015). The compartmentalized signaling model proposed by Zhang et al. (2018) is similar in this regard.
Conclusion
Although they have been traditionally defined as distinct phenomena, there is a great deal of overlap between the proteins and potentially the mechanisms underlying crossover interference, assurance, and the centromere effect. It is possible that a common mechanism is responsible for all three phenomena, whether that is aggregation of pro-crossover factors, mechanical stress, or another driving force that limits crossovers to specific regions of the genome. It is also apparent that the currently known and proposed mechanisms for crossover patterning operate at a wide range of scales, from the DNA level to the chromosome and inter-chromosome levels. Future work may focus on illuminating the connection between these mechanisms to better understand the overarching regulation of crossover patterning.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
NP and CT were supported by the National Institute of General Medical Sciences under award 1T32GM135128. JS was supported in part by the National Institute of General Medical Sciences under award 1R35GM118127.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Agostinho, A., Meier, B., Sonneville, R., Jagut, M., Woglar, A., Blow, J., et al. (2013). Combinatorial regulation of meiotic Holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet. 9:e1003591. doi: 10.1371/journal.pgen.1003591
Ahuja, J. S., Sandhu, R., Mainpal, R., Lawson, C., Henley, H., Hunt, P. A., et al. (2017). Control of meiotic pairing and recombination by chromosomally tethered 26S proteasome. Science 355, 408–411. doi: 10.1126/science.aaf4778
Albertson, D. G., and Thomson, J. N. (1993). Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1, 15–26. doi: 10.1007/bf00710603
Allers, T., and Lichten, M. (2001). Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57. doi: 10.1016/s0092-8674(01)00416-0
Andersen, S. L., and Sekelsky, J. (2010). Meiotic versus mitotic recombination: two different routes for double-strand break repair: the different functions of meiotic versus mitotic DSB repair are reflected in different pathway usage and different outcomes. Bioessays 32, 1058–1066. doi: 10.1002/bies.201000087
Anderson, C. M., Oke, A., Yam, P., Zhuge, T., and Fung, J. C. (2015). Reduced crossover interference and increased ZMM-independent recombination in the absence of Tel1/ATM. PLoS Genet. 11:e1005478. doi: 10.1371/journal.pgen.1005478
Arana, P., Santos, J. L., and Henriques-Gil, N. (1987). Interference relationships in grasshopper reciprocal translocation heterozygotes. Heredity 59, 85–93. doi: 10.1038/hdy.1987.99
Argueso, J. L., Wanat, J., Gemici, Z., and Alani, E. (2004). Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics 168, 1805–1816. doi: 10.1534/genetics.104.032912
Bähler, J., Wyler, T., Loidl, J., and Kohli, J. (1993). Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J. Cell Biol. 121, 241–256. doi: 10.1083/jcb.121.2.241
Baker, B. S., and Carpenter, A. T. C. (1972). Genetic analysis of sex chromosomal meiotic mutants in Drosophila melanogaster. Genetics 71, 255–286. doi: 10.1093/genetics/71.2.255
Baker, S. M., Plug, A. W., Prolla, T. A., Bronner, C. E., Harris, A. C., Yao, X., et al. (1996). Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13, 336–342. doi: 10.1038/ng0796-336
Beadle, G. W. (1932). A possible influence of the spindle fibre on crossing-over in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 18, 160–165. doi: 10.1073/pnas.18.2.160
Berchowitz, L. E., and Copenhaver, G. P. (2010). Genetic interference: don’t stand so close to me. Curr. Genomics 11, 91–102. doi: 10.2174/138920210790886835
Berchowitz, L. E., Francis, K. E., Bey, A. L., and Copenhaver, G. P. (2007). The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 3:e132. doi: 10.1371/journal.pgen.0030132
Blitzblau, H. G., Bell, G. W., Rodriguez, J., Bell, S. P., and Hochwagen, A. (2007). Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr. Biol. 17, 2003–2012. doi: 10.1016/j.cub.2007.10.066
Bole-Gowda, B. N., Perkins, D. D., and Strickland, W. N. (1962). Crossing-over and interference in the centromere region of linkage group I of Neurospora. Genetics 47, 1243–1252. doi: 10.1093/genetics/47.9.1243
Bomblies, K., Jones, G., Franklin, C., Zickler, D., and Kleckner, N. (2016). The challenge of evolving stable polyploidy: Could an increase in “crossover interference distance” play a central role? Chromosoma 125, 287–300. doi: 10.1007/s00412-015-0571-4
Börner, G. V., Kleckner, N., and Hunter, N. (2004). Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45. doi: 10.1016/s0092-8674(04)00292-2
Brady, M. M., McMahan, S., and Sekelsky, J. (2018). Loss of Drosophila Mei-41/ATR alters meiotic crossover patterning. Genetics 208, 579–588. doi: 10.1534/genetics.117.300634
Brand, C. L., Cattani, M. V., Kingan, S. B., Landeen, E. L., and Presgraves, D. C. (2018). Molecular evolution at a meiosis gene mediates species differences in the rate and patterning of recombination. Curr. Biol. 28, 1289–1295.e4.
Bridges, C. B. (1915). A linkage variation in Drosophila. J. Exp. Zool. 19, 1–21. doi: 10.1002/jez.1400190102
Bridges, C. B. (1927). The relation of the age of the female to crossing over in the third chromosome of Drosophila melanogaster. J. Gen. Physiol. 8, 689–700. doi: 10.1085/jgp.8.6.689
Broman, K. W., Rowe, L. B., Churchill, G. A., and Paigen, K. (2002). Crossover interference in the mouse. Genetics 160, 1123–1131. doi: 10.1093/genetics/160.3.1123
Brown, M. S., Grubb, J., Zhang, A., Rust, M. J., and Bishop, D. K. (2015). Small Rad51 and Dmc1 complexes often co-occupy both ends of a meiotic DNA double strand break. PLoS Genet. 11:e1005653. doi: 10.1371/journal.pgen.1005653
Buhler, C., Borde, V., and Lichten, M. (2007). Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 5:e324. doi: 10.1371/journal.pbio.0050324
Burgoyne, P. S. (1982). Genetic homology and crossing over in the X and Y chromosomes of mammals. Hum. Genet. 61, 85–90. doi: 10.1007/bf00274192
Busygina, V., Sehorn, M. G., Shi, I. Y., Tsubouchi, H., Roeder, G. S., and Sung, P. (2008). Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 22, 786–795. doi: 10.1101/gad.1638708
Callan, H. G., and Montalenti, G. (1947). Chiasma interference in mosquitoes. J. Genet. 48, 119–134. doi: 10.1007/bf02989374
Callender, T. L., Laureau, R., Wan, L., Chen, X., Sandhu, R., Laljee, S., et al. (2016). Mek1 down regulates Rad51 activity during yeast meiosis by phosphorylation of Hed1. PLoS Genet. 12:e1006226. doi: 10.1371/journal.pgen.1006226
Cannavo, E., Sanchez, A., Anand, R., Ranjha, L., Hugener, J., Adam, C., et al. (2020). Regulation of the MLH1-MLH3 endonuclease in meiosis. Nature 586, 618–622. doi: 10.1038/s41586-020-2592-2
Capilla-Perez, L., Durand, S., Hurel, A., Lian, Q., Chambon, A., Taochy, C., et al. (2021). The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 118:e2023613118. doi: 10.1073/pnas.2023613118
Carpenter, A. T. C. (1975). Electron microscopy of meiosis in Drosophila melanogaster females. I. Structure, arrangement, and temporal change of the synaptonemal complex in wild-type. Chromosoma 51, 157–182. doi: 10.1007/bf00319833
Carpenter, A. T. C., and Baker, B. S. (1982). On the control of the distribution of meiotic exchange in Drosophila melanogaster. Genetics 101, 81–89. doi: 10.1093/genetics/101.1.81
Chakraborty, P., Pankajam, A. V., Lin, G., Dutta, A., Krishnaprasad, G. N., Tekkedil, M. M., et al. (2017). Modulating crossover frequency and interference for obligate crossovers in Saccharomyces cerevisiae meiosis. G3 7, 1511–1524. doi: 10.1534/g3.117.040071
Chen, S. Y., Tsubouchi, T., Rockmill, B., Sandler, J. S., Richards, D. R., Vader, G., et al. (2008). Global analysis of the meiotic crossover landscape. Dev. Cell 15, 401–415. doi: 10.1016/j.devcel.2008.07.006
Chua, P. R., and Roeder, G. S. (1997). Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev. 11, 1786–1800. doi: 10.1101/gad.11.14.1786
Colombo, P. C., and Jones, G. H. (1997). Chiasma interference is blind to centromeres. Heredity 79(Pt 2), 214–227. doi: 10.1038/hdy.1997.145
Colomé-Tatché, M., Cortijo, S., Wardenaar, R., Morgado, L., Lahouze, B., Sarazin, A., et al. (2012). Features of the Arabidopsis recombination landscape resulting from the combined loss of sequence variation and DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 109, 16240–16245. doi: 10.1073/pnas.1212955109
Comeron, J. M., Ratnappan, R., and Bailin, S. (2012). The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8:e1002905. doi: 10.1371/journal.pgen.1002905
Conrad, M. N., Dominguez, A. M., and Dresser, M. E. (1997). Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science 276, 1252–1255. doi: 10.1126/science.276.5316.1252
Copenhaver, G. P., Housworth, E. A., and Stahl, F. W. (2002). Crossover interference in Arabidopsis. Genetics 160, 1631–1639. doi: 10.1093/genetics/160.4.1631
Crismani, W., Girard, C., Froger, N., Pradillo, M., Santos, J. L., Chelysheva, L., et al. (2012). FANCM limits meiotic crossovers. Science 336, 1588–1590. doi: 10.1126/science.1220381
Crown, K. N., Miller, D. E., Sekelsky, J., and Hawley, R. S. (2018). Local inversion heterozygosity alters recombination throughout the genome. Curr. Biol. 28, 2984–2990.e3.
Darlington, C. D. (1937). Recent Advances in Cytology, 2nd Edn. Philadelphia, PA: P. Blakiston’s Son & Co. Inc.
Darlington, C. D., and Dark, S. O. S. (1932). The origin and behaviour of chiasmata, II. Stenobothrus parallelus. Cytologia 3, 169–185. doi: 10.1508/cytologia.3.169
de Boer, E., Stam, P., Dietrich, A. J., Pastink, A., and Heyting, C. (2006). Two levels of interference in mouse meiotic recombination. Proc. Natl. Acad. Sci. U.S.A. 103, 9607–9612. doi: 10.1073/pnas.0600418103
de los Santos, T., Hunter, N., Lee, C., Larkin, B., Loidl, J., and Hollingsworth, N. M. (2003). The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164, 81–94. doi: 10.1093/genetics/164.1.81
De Muyt, A., Jessop, L., Kolar, E., Sourirajan, A., Chen, J., Dayani, Y., et al. (2012). BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46, 43–53. doi: 10.1016/j.molcel.2012.02.020
De Storme, N., and Geelen, D. (2020). High temperatures alter cross-over distribution and induce male meiotic restitution in Arabidopsis thaliana. Commun Biol. 3:187.
Dereli, I., Stanzione, M., Olmeda, F., Papanikos, F., Baumann, M., Demir, S., et al. (2021). Four-pronged negative feedback of DSB machinery in meiotic DNA-break control in mice. Nucleic Acids Res. 49, 2609–2628. doi: 10.1093/nar/gkab082
Dernburg, A. F., McDonald, K., Moulder, G., Barstead, R., Dresser, M., and Villeneuve, A. M. (1998). Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94, 387–398. doi: 10.1016/s0092-8674(00)81481-6
Deshong, A. J., Ye, A. L., Lamelza, P., and Bhalla, N. (2014). A quality control mechanism coordinates meiotic prophase events to promote crossover assurance. PLoS Genet. 10:e1004291. doi: 10.1371/journal.pgen.1004291
Duroc, Y., Lemhemdi, A., Larcheveque, C., Hurel, A., Cuacos, M., Cromer, L., et al. (2014). The kinesin AtPSS1 promotes synapsis and is required for proper crossover distribution in meiosis. PLoS Genet. 10:e1004674. doi: 10.1371/journal.pgen.1004674
Egel, R. (1978). Synaptonemal complex and crossing-over: Structural support or interference? Heredity 41, 233–237. doi: 10.1038/hdy.1978.92
Egel-Mitani, M., Olson, L. W., and Egel, R. (1982). Meiosis in Aspergillus nidulans: another example for lacking synaptonemal complexes in the absence of crossover interference. Hereditas 97, 179–187. doi: 10.1111/j.1601-5223.1982.tb00870.x
Emerson, S., and Beadle, G. W. (1933). Crossing-over near the spindle fiber in attached-X chromosomes of Drosophila melangoaster. Z. Indukt. Abstamm. Vererbungsl. 65, 129–140. doi: 10.1007/bf01848844
Falque, M., Mercier, R., Mezard, C., de Vienne, D., and Martin, O. C. (2007). Patterns of recombination and MLH1 foci density along mouse chromosomes: modeling effects of interference and obligate chiasma. Genetics 176, 1453–1467. doi: 10.1534/genetics.106.070235
Foss, E. J., and Stahl, F. W. (1995). A test of a counting model for chiasma interference. Genetics 139, 1201–1209. doi: 10.1093/genetics/139.3.1201
Foss, E., Lande, R., Stahl, F. W., and Steinberg, C. M. (1993). Chiasma interference as a function of genetic distance. Genetics 133, 681–691. doi: 10.1093/genetics/133.3.681
Fowler, K. R., Hyppa, R. W., Cromie, G. A., and Smith, G. R. (2018). Physical basis for long-distance communication along meiotic chromosomes. Proc. Natl. Acad. Sci. U.S.A. 115, E9333–E9342.
Froenicke, L., Anderson, L. K., Wienberg, J., and Ashley, T. (2002). Male mouse recombination maps for each autosome identified by chromosome painting. Am. J. Hum. Genet. 71, 1353–1368. doi: 10.1086/344714
Fung, J. C., Rockmill, B., Odell, M., and Roeder, G. S. (2004). Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116, 795–802. doi: 10.1016/s0092-8674(04)00249-1
Gerton, J. L., DeRisi, J., Shroff, R., Lichten, M., Brown, P. O., and Petes, T. D. (2000). Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 97, 11383–11390. doi: 10.1073/pnas.97.21.11383
Girard, C., Chelysheva, L., Choinard, S., Froger, N., Macaisne, N., Lemhemdi, A., et al. (2015). AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet. 11:e1005369. doi: 10.1371/journal.pgen.1005369
Gorlov, I. P., and Gorlova, O. Y. (2001). Cost-benefit analysis of recombination and its application for understanding of chiasma interference. J. Theor. Biol. 213, 1–8. doi: 10.1006/jtbi.2001.2397
Graubard, M. A. (1934). Temperature effect on interference and crossing over. Genetics 19, 83–94. doi: 10.1093/genetics/19.1.83
Hartmann, M., Umbanhowar, J., and Sekelsky, J. (2019). Centromere-proximal meiotic crossovers in Drosophila melanogaster are suppressed by both highly repetitive heterochromatin and proximity to the centromere. Genetics 213, 113–125. doi: 10.1534/genetics.119.302509
Hatkevich, T., Kohl, K. P., McMahan, S., Hartmann, M. A., Williams, A. M., and Sekelsky, J. (2017). Bloom syndrome helicase promotes meiotic crossover patterning and homolog disjunction. Curr. Biol. 27, 96–102. doi: 10.1016/j.cub.2016.10.055
Havekes, F. W., de Jong, J. H., Heyting, C., and Ramanna, M. S. (1994). Synapsis and chiasma formation in four meiotic mutants of tomato (Lycopersicon esculentum). Chromosome Res. 2, 315–325. doi: 10.1007/bf01552725
Hawley, R. S. (1988). “Exchange and chromosomal segregation in eucaryotes,” in Genetic Recombination, eds R. Kucherlapati and G. R. Smith (Washington, DC: American Society of Microbiology), 497–527.
Hawley, R. S., and Theurkauf, W. E. (1993). Requiem for distributive segregation: achiasmate segregation in Drosophila females. Trends Genet. 9, 310–317. doi: 10.1016/0168-9525(93)90249-h
Hayashi, M., Mlynarczyk-Evans, S., and Villeneuve, A. M. (2010). The synaptonemal complex shapes the crossover landscape through cooperative assembly, crossover promotion and crossover inhibition during Caenorhabditis elegans meiosis. Genetics 186, 45–58. doi: 10.1534/genetics.110.115501
He, W., Prasada Rao, H. B., Tang, S., Bhagwat, N., Kulkarni, D. S., Ma, Y., et al. (2020). Regulated proteolysis of MutSγ controls meiotic crossing over. Mol. Cell 78, 168–183. doi: 10.1016/j.molcel.2020.02.001
Heyer, W. D., Ehmsen, K. T., and Liu, J. (2010). Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113–139. doi: 10.1146/annurev-genet-051710-150955
Higgins, J. D., Armstrong, S. J., Franklin, F. C., and Jones, G. H. (2004). The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 18, 2557–2570. doi: 10.1101/gad.317504
Higgins, J. D., Vignard, J., Mercier, R., Pugh, A. G., Franklin, F. C., and Jones, G. H. (2008). AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J. 55, 28–39. doi: 10.1111/j.1365-313x.2008.03470.x
Hillers, K. J., and Villeneuve, A. M. (2003). Chromosome-wide control of meiotic crossing over in C. elegans. Curr. Biol. 13, 1641–1647. doi: 10.1016/j.cub.2003.08.026
Hinch, A. G., Altemose, N., Noor, N., Donnelly, P., and Myers, S. R. (2014). Recombination in the human pseudoautosomal region PAR1. PLoS Genet. 10:e1004503. doi: 10.1371/journal.pgen.1004503
Hollis, J. A., Glover, M. L., Schlientz, A. J., Cahoon, C. K., Bowerman, B., Wignall, S. M., et al. (2020). Excess crossovers impede faithful meiotic chromosome segregation in C. elegans. PLoS Genet. 16:e1009001. doi: 10.1371/journal.pgen.1009001
Holloway, J. K., Booth, J., Edelmann, W., McGowan, C. H., and Cohen, P. E. (2008). MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 4:e1000186. doi: 10.1371/journal.pgen.1000186
Housworth, E. A., and Stahl, F. W. (2003). Crossover interference in humans. Am. J. Hum. Genet. 73, 188–197. doi: 10.1086/376610
Hulten, M. A. (1974). Chiasma distribution at diakinesis in the normal human male. Hereditas 76, 55–78. doi: 10.1111/j.1601-5223.1974.tb01177.x
Hulten, M. A. (2011). On the origin of crossover interference: a chromosome oscillatory movement (COM) model. Mol. Cytogenet. 4:10. doi: 10.1186/1755-8166-4-10
Huselid, E., and Bunting, S. F. (2020). The regulation of homologous recombination by helicases. Genes 11:498. doi: 10.3390/genes11050498
Jessop, L., Rockmill, B., Roeder, G. S., and Lichten, M. (2006). Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of Sgs1. PLoS Genet. 2:e155. doi: 10.1371/journal.pgen.0020155
Joshi, N., Barot, A., Jamison, C., and Borner, G. V. (2009). Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet. 5:e1000557. doi: 10.1371/journal.pgen.1000557
Kaback, D. B., Barber, D., Mahon, J., Lamb, J., and You, J. (1999). Chromosome size-dependent control of meiotic reciprocal recombination in Saccharomyces cerevisiae: the role of crossover interference. Genetics 152, 1475–1486. doi: 10.1093/genetics/152.4.1475
Kaback, D. B., Steensma, H. Y., and de Jonge, P. (1989). Enhanced meiotic recombination on the smallest chromosome of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 86, 3694–3698. doi: 10.1073/pnas.86.10.3694
Kikkawa, H. (1932). Studies on the mechanism of crossing-over in Drosophila, with special reference to the phenomenon of interference. Cytologia 3, 285–332. doi: 10.1508/cytologia.3.285
King, J. S., and Mortimer, R. K. (1990). A polymerization model of chiasma interference and corresponding computer simulation. Genetics 126, 1127–1138. doi: 10.1093/genetics/126.4.1127
Kleckner, N., Zickler, D., Jones, G. H., Dekker, J., Padmore, R., Henle, J., et al. (2004). A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. U.S.A. 101, 12592–12597.
Koehler, K. E., Boulton, C. L., Collins, H. E., French, R. L., Herman, K. C., Lacefield, S. M., et al. (1996a). Spontaneous X chromosome MI and MII nondisjunction events in Drosophila melanogaster oocytes have different recombinational histories. Nat. Genet. 14, 406–414. doi: 10.1038/ng1296-406
Koehler, K. E., Hawley, R. S., Sherman, S., and Hassold, T. (1996b). Recombination and nondisjunction in humans and flies. Hum. Mol. Genet. 5, 1495–1504. doi: 10.1093/hmg/5.supplement_1.1495
Kohl, K. P., and Sekelsky, J. (2013). Meiotic and mitotic recombination in meiosis. Genetics 194, 327–334. doi: 10.1534/genetics.113.150581
Kohl, K. P., Jones, C. D., and Sekelsky, J. (2012). Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science 338, 1363–1365. doi: 10.1126/science.1228190
Koller, P. C., and Darlington, C. D. (1934). The genetical and mechan-ical properties of the sex-chromosomes. J. Genet. 29, 159–173. doi: 10.1007/bf02982193
Kulkarni, D. S., Owens, S. N., Honda, M., Ito, M., Yang, Y., Corrigan, M. W., et al. (2020). PCNA activates the MutLgamma endonuclease to promote meiotic crossing over. Nature 586, 623–627. doi: 10.1038/s41586-020-2645-6
Lamb, N. E., Freeman, S. B., Savage-Austin, A., Pettay, D., Taft, L., Hersey, J., et al. (1996). Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat. Genet. 14, 400–405. doi: 10.1038/ng1296-400
Libuda, D. E., Uzawa, S., Meyer, B. J., and Villeneuve, A. M. (2013). Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature 502, 703–706. doi: 10.1038/nature12577
Lindsley, D. L., and Sandler, L. (1977). The genetic analysis of meiosis in female Drosophila melanogaster. Philos. Trans. R. Soc. Lond. B Biol. Sci. 277, 295–312. doi: 10.1098/rstb.1977.0019
Link, A. J., and Olson, M. V. (1991). Physical map of the Saccharomyces cerevisiae genome at 110-kilobase resolution. Genetics 127, 681–698. doi: 10.1093/genetics/127.4.681
Lu, X., Liu, X., An, L., Zhang, W., Sun, J., Pei, H., et al. (2008). The Arabidopsis MutS homolog AtMSH5 is required for normal meiosis. Cell Res. 18, 589–599. doi: 10.1038/cr.2008.44
Lucchesi, J. C., and Suzuki, D. T. (1968). The interchromosomal control of recombination. Annu. Rev. Genet. 2, 53–86. doi: 10.1146/annurev.ge.02.120168.000413
Machovina, T. S., Mainpal, R., Daryabeigi, A., McGovern, O., Paouneskou, D., Labella, S., et al. (2016). A surveillance system ensures crossover formation in C. elegans. Curr. Biol. 26, 2873–2884. doi: 10.1016/j.cub.2016.09.007
Maguire, M. P. (1977). Homologous chromosome pairing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 277, 245–258.
Marsolier-Kergoat, M. C., Khan, M. M., Schott, J., Zhu, X., and Llorente, B. (2018). Mechanistic view and genetic control of DNA recombination during meiosis. Mol. Cell 70, 9–20.e6.
Martinez-Perez, E., Schvarzstein, M., Barroso, C., Lightfoot, J., Dernburg, A. F., and Villeneuve, A. M. (2008). Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 22, 2886–2901. doi: 10.1101/gad.1694108
Martini, E., Diaz, R. L., Hunter, N., and Keeney, S. (2006). Crossover homeostasis in yeast meiosis. Cell 126, 285–295. doi: 10.1016/j.cell.2006.05.044
Mather, K. (1935). Reductional and equational separation of the chromosomes in bivalents and multivalents. J. Genet. 30, 53–78. doi: 10.1007/bf02982205
Mather, K. (1936). Competition between bivalents during chiasma formation. Proc. R. Soc. Lond. B 120, 208–227. doi: 10.1098/rspb.1936.0032
Mather, K. (1937). The determination of position in crossing-over. II. The chromosome length-chiasma frequency relation. Cytologia Fujii Jubilee, 514–526. doi: 10.1508/cytologia.fujiijubilaei.514
Mather, K. (1939). Crossing over and heterochromatin in the X chromosome of Drosophila melanogaster. Genetics 24, 413–435. doi: 10.1093/genetics/24.3.413
McKim, K. S., Green-Marroquin, B. L., Sekelsky, J. J., Chin, G., Steinberg, C., Khodosh, R., et al. (1998). Meiotic synapsis in the absence of recombination. Science 279, 876–878. doi: 10.1126/science.279.5352.876
Medhi, D., Goldman, A. S., and Lichten, M. (2016). Local chromosome context is a major determinant of crossover pathway biochemistry during budding yeast meiosis. eLife 5:e19669.
Mehrotra, S., and McKim, K. S. (2006). Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet. 2:e200. doi: 10.1371/journal.pgen.0020200
Melamed-Bessudo, C., and Levy, A. A. (2012). Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, E981–E988.
Miller, D. E., Smith, C. B., Yeganeh Kazemi, N., Cockrell, A. J., Arvanitakis, A. V., Blumenstiel, J. P., et al. (2016). Whole-genome analysis of individual meiotic events in Drosophila melanogaster reveals that noncrossover gene conversions are insensitive to interference and the centromere effect. Genetics 203, 159–171. doi: 10.1534/genetics.115.186486
Mirouze, M., Lieberman-Lazarovich, M., Aversano, R., Bucher, E., Nicolet, J., Reinders, J., et al. (2012). Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 5880–5885. doi: 10.1073/pnas.1120841109
Moens, P. B. (1969). Genetic and cytological effects of three desynaptic genes in the tomato. Can. J. Genet. Cytol. 11, 857–869. doi: 10.1139/g69-101
Mortimer, R. K., Schild, D., Contopoulou, C. R., and Kans, J. A. (1989). Genetic map of Saccharomyces cerevisiae, edition 10. Yeast 5, 321–403.
Mortimer, R. R., and Fogel, S. (1974). “Genetical interference and gene conversion,” in Mechanisms in Recombination, ed. R. Grell (New York, NY: Plenum Press), 263–275. doi: 10.1007/978-1-4684-2133-0_23
Moses, M. J., Dresser, M., and Poorman, P. A. (1984). “Composition and role of the synaptonemal complex,” in Controlling Events in Meiosis, eds C.W. Evans and H. G. Dickinson (Cambridge: Company of Biologists), 245–270.
Muller, H. J. (1916b). The mechanism of crossing-over. II. Am. Nat. 50, 284–305. doi: 10.1086/279541
Munz, P. (1994). An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics 137, 701–707. doi: 10.1093/genetics/137.3.701
Murakami, H., Lam, I., Huang, P. C., Song, J., van Overbeek, M., and Keeney, S. (2020). Multilayered mechanisms ensure that short chromosomes recombine in meiosis. Nature 582, 124–128. doi: 10.1038/s41586-020-2248-2
Nambiar, M., and Smith, G. R. (2016). Repression of harmful meiotic recombination in centromeric regions. Semin. Cell Dev. Biol. 54, 188–197. doi: 10.1016/j.semcdb.2016.01.042
Niu, H., Li, X., Job, E., Park, C., Moazed, D., Gygi, S. P., et al. (2007). Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol. Cell. Biol. 27, 5456–5467. doi: 10.1128/mcb.00416-07
Niu, H., Wan, L., Busygina, V., Kwon, Y., Allen, J. A., Li, X., et al. (2009). Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol. Cell 36, 393–404. doi: 10.1016/j.molcel.2009.09.029
Niu, H., Wan, L., Baumgartner, B., Schaefer, D., Loidl, J., and Hollingsworth, N. M. (2005). Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell 16, 5804–5818. doi: 10.1091/mbc.e05-05-0465
O’Neil, N. J., Martin, J. S., Youds, J. L., Ward, J. D., Petalcorin, M. I. R., Rose, A. M., et al. (2013). Joint molecule resolution requires the redundant activities of MUS-81 and XPF-1 during C. elegans meiosis. PLoS Genet. 9:e1003582. doi: 10.1371/journal.pgen.1003582
Oh, S. D., Lao, J. P., Hwang, P. Y., Taylor, A. F., Smith, G. R., and Hunter, N. (2007). BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130, 259–272. doi: 10.1016/j.cell.2007.05.035
Orr-Weaver, T. (1996). Meiotic nondisjunction does the two-step. Nat. Genet. 14, 374–376. doi: 10.1038/ng1296-374
Otto, S. P., and Payseur, B. A. (2019). Crossover interference: shedding light on the evolution of recombination. Annu. Rev. Genet. 53, 19–44. doi: 10.1146/annurev-genet-040119-093957
Owen, A. R. G. (1949). A possible interpretation of the apparent interference across the centromere found by Callan and Montalenti in Culex pipiens. Heredity 3, 357–367. doi: 10.1038/hdy.1949.26
Pan, J., Sasaki, M., Kniewel, R., Murakami, H., Blitzblau, H. G., Tischfield, S. E., et al. (2011). A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144, 719–731. doi: 10.1016/j.cell.2011.02.009
Papanikos, F., Clement, J. A. J., Testa, E., Ravindranathan, R., Grey, C., Dereli, I., et al. (2019). Mouse ANKRD31 regulates spatiotemporal patterning of meiotic recombination initiation and ensures recombination between X and Y sex chromosomes. Mol. Cell 74, 1069–1085.e11.
Papazian, H. P. (1952). The analysis of tetrad data. Genetics 37, 175–188. doi: 10.1093/genetics/37.2.175
Parker, J. S. (1987). Increased chiasma frequency as a result of chromosome rearrangement. Heredity 58, 87–94. doi: 10.1038/hdy.1987.13
Perkins, D. D. (1962). Crossing-over and interference in a multiply marked chromosome arm of Neurospora. Genetics 47, 1253–1274. doi: 10.1093/genetics/47.9.1253
Phillips, D., Jenkins, G., Macaulay, M., Nibau, C., Wnetrzak, J., Fallding, D., et al. (2015). The effect of temperature on the male and female recombination landscape of barley. New Phytol. 208, 421–429. doi: 10.1111/nph.13548
Plank, J. L., Wu, J., and Hsieh, T. S. (2006). Topoisomerase IIIα and Bloom’s helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc. Natl. Acad. Sci. U.S.A. 103, 11118–11123. doi: 10.1073/pnas.0604873103
Plough, H. H. (1917a). The effect of temperature on crossingover in Drosophila. J. Exp. Zool. 24, 147–209. doi: 10.1002/jez.1400240202
Plough, H. H. (1917b). The effect of temperature on linkage in the second chromosome of Drosophila. Proc. Natl. Acad. Sci. U.S.A. 3, 553–555. doi: 10.1073/pnas.3.9.553
Plough, H. H. (1921). Further studies on the effect of temperature on crossing over. J. Exp. Zool. 32, 187–202. doi: 10.1002/jez.1400320202
Prasada Rao, H. B., Qiao, H., Bhatt, S. K., Bailey, L. R., Tran, H. D., Bourne, S. L., et al. (2017). A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355, 403–407.
Qiao, H., Prasada Rao, H. B., Yang, Y., Fong, J. H., Cloutier, J. M., Deacon, D. C., et al. (2014). Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat. Genet. 46, 194–199. doi: 10.1038/ng.2858
Radford, S. J., Goley, E., Baxter, K., McMahan, S., and Sekelsky, J. (2005). Drosophila ERCC1 is required for a subset of MEI-9-dependent meiotic crossovers. Genetics 170, 1737–1745. doi: 10.1534/genetics.104.036178
Rao, H. B., Qiao, H., Bhatt, S. K., Bailey, L. R., Tran, H. D., Bourne, S. L., et al. (2017). A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355, 403–407. doi: 10.1126/science.aaf6407
Redfield, H. (1930). Crossing over in the third chromosomes of triploids of Drosophila melanogaster. Genetics 15, 205–252.
Redfield, H. (1932). A comparison of triploid and diploid crossing over for chromosome II of Drosophila melanogaster. Genetics 17, 137–152.
Reynolds, A., Qiao, H., Yang, Y., Chen, J. K., Jackson, N., Biswas, K., et al. (2013). RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 45, 269–278. doi: 10.1038/ng.2541
Riles, L., Dutchik, J. E., Baktha, A., McCauley, B. K., Thayer, E. C., Leckie, M. P., et al. (1993). Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics 134, 81–150. doi: 10.1093/genetics/134.1.81
Rockmill, B., Voelkel-Meiman, K., and Roeder, G. S. (2006). Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics 174, 1745–1754. doi: 10.1534/genetics.106.058933
Rog, O., Kohler, S., and Dernburg, A. F. (2017). The synaptonemal complex has liquid crystalline properties and spatially regulates meiotic recombination factors. eLife 6:e21455.
Rosu, S., Libuda, D. E., and Villeneuve, A. M. (2011). Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science 334, 1286–1289. doi: 10.1126/science.1212424
Rosu, S., Zawadzki, K. A., Stamper, E. L., Libuda, D. E., Reese, A. L., Dernburg, A. F., et al. (2013). The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS Genet. 9:e1003674. doi: 10.1371/journal.pgen.1003674
Saito, T. T., Youds, J. L., Boulton, S. J., and Colaiácovo, M. P. (2009). Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 5:e1000735. doi: 10.1371/journal.pgen.1000735
Schweitzer, M. D. (1935). An analytical study of crossing over in Drosophila melanogaster. Genetics 20, 497–527. doi: 10.1093/genetics/20.5.497
Sekelsky, J. (2017). DNA repair in Drosophila: mutagens, models, and missing genes. Genetics 205, 471–490. doi: 10.1534/genetics.116.186759
Sekelsky, J. J., McKim, K. S., Chin, G. M., and Hawley, R. S. (1995). The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics 141, 619–627. doi: 10.1093/genetics/141.2.619
Shinohara, A., and Shinohara, M. (2004). Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination. Cytogenet. Genome Res. 107, 201–207. doi: 10.1159/000080598
Shinohara, M., Oh, S. D., Hunter, N., and Shinohara, A. (2008). Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 40, 299–309. doi: 10.1038/ng.83
Smith, G. R., and Nambiar, M. (2020). New solutions to old problems: molecular mechanisms of meiotic crossover control. Trends Genet. 36, 337–346. doi: 10.1016/j.tig.2020.02.002
Smith, G. R., Boddy, M. N., Shanahan, P., and Russell, P. (2003). Fission yeast Mus81.Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics 165, 2289–2293. doi: 10.1093/genetics/165.4.2289
Snow, R. (1979). Maximum likelihood estimation of linkage and interference from tetrad data. Genetics 92, 231–245. doi: 10.1093/genetics/92.1.231
Snowden, T., Acharya, S., Butz, C., Berardini, M., and Fishel, R. (2004). hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15, 437–451. doi: 10.1016/j.molcel.2004.06.040
Stack, S. M. (1984). Heterochromatin, the synaptonemal complex and crossing over. J. Cell Sci. 71, 159–176. doi: 10.1242/jcs.71.1.159
Stahl, F. W. (2008). On the “NPD ratio” as a test for crossover interference. Genetics 179, 701–704. doi: 10.1534/genetics.108.086918
Stahl, F. W., Foss, H. M., Young, L. S., Borts, R. H., Abdullah, M. F., and Copenhaver, G. P. (2004). Does crossover interference count in Saccharomyces cerevisiae? Genetics 168, 35–48. doi: 10.1534/genetics.104.027789
Stamper, E. L., Rodenbusch, S. E., Rosu, S., Ahringer, J., Villeneuve, A. M., and Dernburg, A. F. (2013). Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLoS Genet. 9:e1003679. doi: 10.1371/journal.pgen.1003679
Stern, C. (1926). An effect of temperature and age on crossing-over in the first chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 12, 530–532. doi: 10.1073/pnas.12.8.530
Sturtevant, A. H. (1913). The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J. Exp. Biol. 14, 43–59. doi: 10.1002/jez.1400140104
Sturtevant, A. H. (1915). The behavior of the chromosomes as studied through linkage. Z. Indukt. Abstamm. Vererbungsl. 13, 234–287. doi: 10.1007/bf01792906
Svetlanov, A., Baudat, F., Cohen, P. E., and de Massy, B. (2008). Distinct functions of MLH3 at recombination hot spots in the mouse. Genetics 178, 1937–1945. doi: 10.1534/genetics.107.084798
Sym, M., and Roeder, G. S. (1994). Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell 79, 283–292. doi: 10.1016/0092-8674(94)90197-x
Sym, M., Engebrecht, J. A., and Roeder, G. S. (1993). ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72, 365–378. doi: 10.1016/0092-8674(93)90114-6
Symington, L. S., and Petes, T. D. (1988). Meiotic recombination within the centromere of a yeast chromosome. Cell 52, 237–240. doi: 10.1016/0092-8674(88)90512-0
Szauter, P. (1984). An analysis of regional constraints on exchange in Drosophila melanogaster using recombination-defective meiotic mutants. Genetics 106, 45–71. doi: 10.1093/genetics/106.1.45
Thacker, D., Mohibullah, N., Zhu, X., and Keeney, S. (2014). Homologue engagement controls meiotic DNA break number and distribution. Nature 510, 241–246. doi: 10.1038/nature13120
Trelles-Sticken, E., Dresser, M. E., and Scherthan, H. (2000). Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J. Cell Biol. 151, 95–106. doi: 10.1083/jcb.151.1.95
Tsubouchi, H., and Roeder, G. S. (2006). Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 20, 1766–1775. doi: 10.1101/gad.1422506
Vincenten, N., Kuhl, L. M., Lam, I., Oke, A., Kerr, A. R., Hochwagen, A., et al. (2015). The kinetochore prevents centromere-proximal crossover recombination during meiosis. eLife 4:e10850.
Voelkel-Meiman, K., Johnston, C., Thappeta, Y., Subramanian, V. V., Hochwagen, A., and MacQueen, A. J. (2015). Separable crossover-promoting and crossover-constraining aspects of Zip1 activity during budding yeast meiosis. PLoS Genet. 11:e1005335. doi: 10.1371/journal.pgen.1005335
Wang, S., Hassold, T., Hunt, P., White, M. A., Zickler, D., Kleckner, N., et al. (2017). Inefficient crossover maturation underlies elevated aneuploidy in human female meiosis. Cell 168, 977–989.e17.
Wang, S., Zickler, D., Kleckner, N., and Zhang, L. (2015). Meiotic crossover patterns: obligatory crossover, interference and homeostasis in a single process. Cell Cycle 14, 305–314. doi: 10.4161/15384101.2014.991185
Weinstein, A. (1918). Coincidence of crossing over in Drosophila melanogaster (AMPELOPHILA). Genetics 3, 135–172. doi: 10.1093/genetics/3.2.135
Westphal, T., and Reuter, G. (2002). Recombinogenic effects of suppressors of position-effect variegation in Drosophila. Genetics 160, 609–621. doi: 10.1093/genetics/160.2.609
White, M. A., Wang, S., Zhang, L., and Kleckner, N. (2017). Quantitative modeling and automated analysis of meiotic recombination. Methods Mol. Biol. 1471, 305–323. doi: 10.1007/978-1-4939-6340-9_18
Wyatt, H. D., and West, S. C. (2014). Holliday junction resolvases. Cold Spring Harb. Perspect. Biol. 6:a023192.
Xue, M., Wang, J., Jiang, L., Wang, M., Wolfe, S., Pawlowski, W. P., et al. (2018). The number of meiotic double-strand breaks influences crossover distribution in Arabidopsis. Plant Cell 30, 2628–2638. doi: 10.1105/tpc.18.00531
Yamamoto, M., and Miklos, G. L. (1978). Genetic studies on heterochromatin in Drosophila melanogaster and their implications for the functions of satellite DNA. Chromosoma 66, 71–98. doi: 10.1007/bf00285817
Yant, L., Hollister, J. D., Wright, K. M., Arnold, B. J., Higgins, J. D., Franklin, F. C. H., et al. (2013). Meiotic_ adaptation to genome duplication in Arabidopsis arenosa. Curr. Biol. 23, 2151–2156. doi: 10.1016/j.cub.2013.08.059
Yelina, N. E., Choi, K., Chelysheva, L., Macaulay, M., de Snoo, B., Wijnker, E., et al. (2012). Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet. 8:e1002844. doi: 10.1371/journal.pgen.1002844
Yıldız, Ö., Majumder, S., Kramer, B. C., and Sekelsky, J. (2002). Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol. Cell 10, 1503–1509. doi: 10.1016/s1097-2765(02)00782-7
Zakharyevich, K., Tang, S., Ma, Y., and Hunter, N. (2012). Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149, 334–347. doi: 10.1016/j.cell.2012.03.023
Zalevsky, J., MacQueen, A. J., Duffy, J. B., Kemphues, K. J., and Villeneuve, A. M. (1999). Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153, 1271–1283. doi: 10.1093/genetics/153.3.1271
Zhang, L., Kohler, S., Rillo-Bohn, R., and Dernburg, A. F. (2018). A compartmentalized signaling network mediates crossover control in meiosis. eLife 7:e30789.
Zhang, L., Liang, Z., Hutchinson, J., and Kleckner, N. (2014a). Crossover patterning by the beam-film model: analysis and implications. PLoS Genet. 10:e1004042. doi: 10.1371/journal.pgen.1004042
Zhang, L., Wang, S., Yin, S., Hong, S., Kim, K. P., and Kleckner, N. (2014b). Topoisomerase II mediates meiotic crossover interference. Nature 511, 551–556. doi: 10.1038/nature13442
Zickler, D., Moreau, P. J., Huynh, A. D., and Slezec, A. M. (1992). Correlation between pairing initiation sites, recombination nodules and meiotic recombination in Sordaria macrospora. Genetics 132, 135–148.
Keywords: meiosis, recombination, interference, centromere, crossover assurance
Citation: Pazhayam NM, Turcotte CA and Sekelsky J (2021) Meiotic Crossover Patterning. Front. Cell Dev. Biol. 9:681123. doi: 10.3389/fcell.2021.681123
Received: 15 March 2021; Accepted: 28 June 2021;
Published: 22 July 2021.
Edited by:
Liangran Zhang, Shandong University, ChinaReviewed by:
Neil Hunter, University of California, Davis, United StatesFumiko Esashi, University of Oxford, United Kingdom
Copyright © 2021 Pazhayam, Turcotte and Sekelsky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeff Sekelsky, c2VrZWxza3lAdW5jLmVkdQ==
†These authors have contributed equally to this work
 Nila M. Pazhayam
Nila M. Pazhayam Carolyn A. Turcotte
Carolyn A. Turcotte Jeff Sekelsky
Jeff Sekelsky