- 1Department of Biotherapy, State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu, China
- 2Department of Thoracic Oncology, West China Hospital, Sichuan University, Chengdu, China
- 3West China School of Medicine, Sichuan University, Chengdu, China
- 4Department of Pharmacy, State Key Laboratory of Biotherapy and Cancer Center, National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University, Chengdu, China
In recent years, immunotherapy has showed fantastic promise in pioneering and accelerating the field of cancer therapy and embraces unprecedented breakthroughs in clinical practice. The clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 9 (CRISPR-Cas9) system, as a versatile gene-editing technology, lays a robust foundation to efficiently innovate cancer research and cancer therapy. Here, we summarize recent approaches based on CRISPR/Cas9 system for construction of chimeric antigen receptor T (CAR-T) cells and T cell receptor T (TCR-T) cells. Besides, we review the applications of CRISPR/Cas9 in inhibiting immune checkpoint signaling pathways and highlight the feasibility of CRISPR/Cas9 based engineering strategies to screen novel cancer immunotherapy targets. Conclusively, we discuss the perspectives, potential challenges and possible solutions in this vivid growing field.
Introduction
In recent years, cancer immunotherapy, including immune checkpoint blockades and adoptive T cell therapy (ACT), has experienced incredible success in various types of cancer. Immunotherapies mainly function by relieving tumor-induced immunosuppression and re-boosting anti-cancer immunity (O’Donnell et al., 2019). Immune checkpoint blockades, such as anti-PD1/PD-L1 antibody and anti-CTLA-4 antibody, inhibit immunosuppressive signals and promote T cells reinvigoration. Adoptive T cell therapy, including tumor infiltrating lymphocytes (TILs) therapy, transgenic T cell receptor (TCR)-T cell therapy and chimeric antigen receptors (CAR)-T cell therapy, functions by increasing the number of tumor-reactive T cells and directly against tumor cells. Although significant and durable clinical responses have been resulted from cancer immunotherapy in certain cancer types, unfortunately, most patients fail to benefit from immunotherapy due to intrinsic and adaptive tumor resistance. Therefore, additional and novel immunotherapies are in urgent need.
CRISPR/Cas9, as a versatile gene-editing technology, has been extensively applied in cancer research. Since its first application as a genome-editing tool in mammalian cells in 2013, the use of CRISPR/Cas9 system has been rapidly expanded owing to its high flexibility and efficiency (Cong et al., 2013). CRISPR/Cas9 has been widely used in establishing cancer models (Platt et al., 2014; Tuveson and Clevers, 2019), verifying essential genes as druggable targets (Evers et al., 2016; Tzelepis et al., 2016), investigating the mechanism of drug resistance (Pettitt et al., 2018; Wei L. et al., 2019), comprehensively understanding the function of gene non-coding regions (Zhu et al., 2016; Esposito et al., 2019), and so on. An in-depth discussion of CRISPR/Ca9 in cancer research is has been recently reviewed in detail elsewhere (Zhan et al., 2019), which will not be covered in this review. The combination of CRISPR/Cas9 and cancer immunotherapy, the two revolutionary technologies in cancer research and treatment, may further broaden the application of immunotherapy to more cancer patients. In this review, we summarized recent developments of CRISPR/Cas9 technology in cancer immunotherapy, involving the construction of CAR-T cells, designing of TCR-T cells, inhibiting immune checkpoint signaling pathways, and screening for new druggable targets in immunotherapy.
Application of CRISPR/Cas9 System in CAR-T Cell Immunotherapy
Recently, genetically modified T cells that express chimeric antigen receptors (CAR T-cells) have shown unprecedented efficacy in hematological malignancies. A CAR structure is comprised of three parts: an extracellular antigen recognizing domain usually a single-chain variable fragment (scFv) derived from an antibody, a transmembrane domain, and an intracellular signaling transduction domain containing CD3ζ chain with or without costimulatory molecules (Figure 1). One of the most successful clinical trials is applying anti-CD19 CAR-T cells in B-cell malignancy (Maude et al., 2014). Because of the striking clinical efficacy of CAR-T cells, Kymriah and Yescarta, two types of anti-CD19 CAR-T cells, have been approved by the U.S. Food and Drug Administration (FDA) to treat pediatric/young adult B lymphoblastic leukemia (B-ALL) and adult diffuse large B cell lymphoma (DLBCL). In July 2020, Tecartus was approved for the treatment of adult patients with mantle cell lymphoma (MCL). In February 2021, Breyanzi, the fourth CAR-T therapy for adults with relapsed and refractory large B cell lymphoma, was approved by US FDA. Till now, a large number of registered clinical trials are springing up. Novel constructions and various applications of CAR-T cells are developed at a rapid pace (Jackson et al., 2016; Zhang C. et al., 2017; Labanieh et al., 2018).
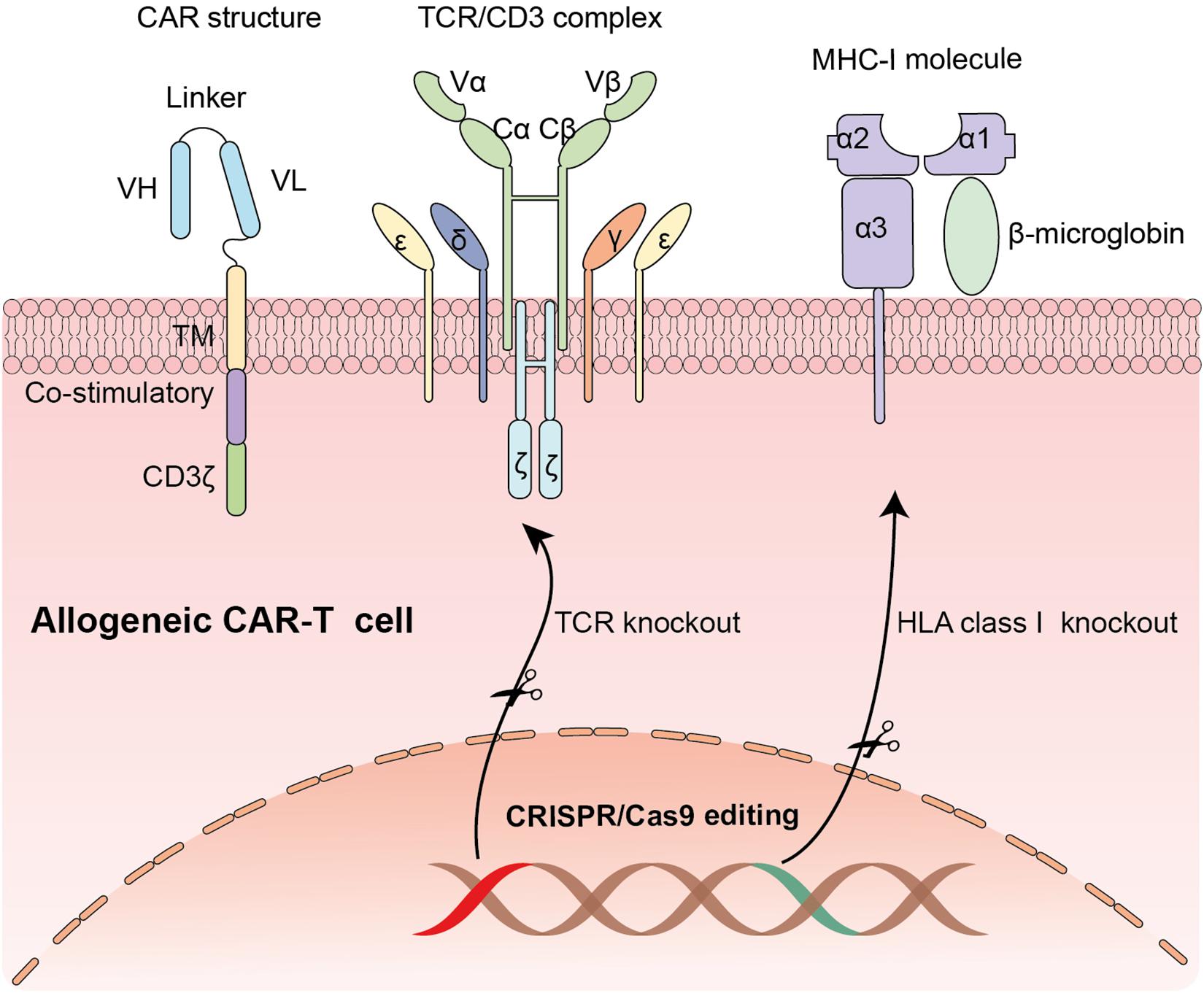
Figure 1. The application of CRISPR/Cas9 system in editing CAR-T cells: Knocking out endogenous TCR locus and MHC-1 molecule to generate universal CAR-T cells.
Despite such impressive clinical results have been achieved, plenty of patients are unable to benefit from T-cell therapy due to several reasons. First, the personalized approach of manufacturing T-cells is time-consuming and costly, which impedes many patients, especially with rapidly progressive diseases, to make the most of this immunotherapy. Second, during the production process, it is hard to generate enough high-quality T-cells from lymphopenic patients in poor condition. Even if patients get enough immune cells, these cells may fail to complete the whole manufacturing process. Moreover, the risk of manufacturing T cells always exists (Torikai and Cooper, 2016). A patient with B cell leukemia was reported relapsing 9 months after receiving anti-CD19 CAR T-cell infusion due to unintentionally transduction of CAR gene into a single leukemic B cell (Ruella et al., 2018). Finally, heterogeneity among autologous CAR-T products contributes to unpredictable and variable clinical activity.
Generation of Universal Allogeneic CAR-T Cells by CRISPR/Cas9
To overcome the barriers of limiting wide application of CAR-T cell therapy, multiple strategies have been developed. One of the most feasible and durable approaches is to generate allogeneic universal CAR-T cells from healthy donors (Depil et al., 2020). Compared with autologous CAR-T cells, “off-the shelf” allogeneic CAR-T cells have many potential advantages, including immediate availability of cryopreserved CAR-T cells for patients in urgent need, enough quantity for first infusion or re-dosing and possible standardization of CAR-T cells producing (Depil et al., 2020). Whereas considering the presence of endogenous HLA and TCR on donor’s T lymphocytes, the biggest challenge of universal products is the potential risk of alloreactivity (host versus graft response) and graft-versus-host disease (GVHD) (Figure 1). With the advances of Gene editing technology, eradication of endogenous TCR can be achieved. Hiroki Torikai and colleagues generated CD19 specific CAR-T cells with disruption of endogenous TCR via zinc finger nucleases (ZFNs) to reduce graft-versus-host responses. The genetically modified CAR-T cells showed an expected specificity of CD19 antigen with no responding to TCR stimulation (Torikai et al., 2012). Similarly, transcription activator-like effector nucleases (TALENs), another widely used gene-editing tool, have also been used in producing universal CAR-T cells by knocking out αβ chains of TCR (Poirot et al., 2015). However, generation of fully allogeneic CAR-T cells requires simultaneous knockout of TCR and HLA molecules and transduction of CAR. An efficient and precise gene-editing technique with the unique capability to achieve multiplexed genome engineering is needed.
Compared with ZFNs and TALENs, CRISPR/Cas9 has more applications in producing allogeneic CAR-T cell due to its excellent flexibility and high effectiveness. CRISPR/Cae9 system can simultaneously and efficiently knock out multiple gene loci. TCR– HLA class I–,Fas–TCR– HLA class I–,PD1– TCR– HLA class I– allogeneic universal T cells can be easily yielded via a one-shot CRISPR protocol by incorporation of multiple guide RNAs in a CAR lentiviral vector (Ren et al., 2017b; Choi et al., 2019). Ren et al. used CRISPR/Cas9 to generate CAR-T cells simultaneously deficient in endogenous TCR, HLA-I and PD-1, which shows potent antitumor activity in vitro and in animal models (Ren et al., 2017a). Besides, a uniform CAR expression was generated by inserting a CD19 specific CAR into the T-cell receptor α constant (TRAC) locus via CRISPR/Cas9 genome editing (Eyquem et al., 2017). The edited cells vastly outperformed the conventional CAR-T cells with enhanced anti-tumor activity in vitro and in mouse models with acute lymphoblastic leukemia (Eyquem et al., 2017). While the safety and efficacy of CRISPR/Cas9-edited universal CAR T cells in vivo needs to be further tested in clinical studies. Currently, eight relevant clinical trials are going on (Table 1).
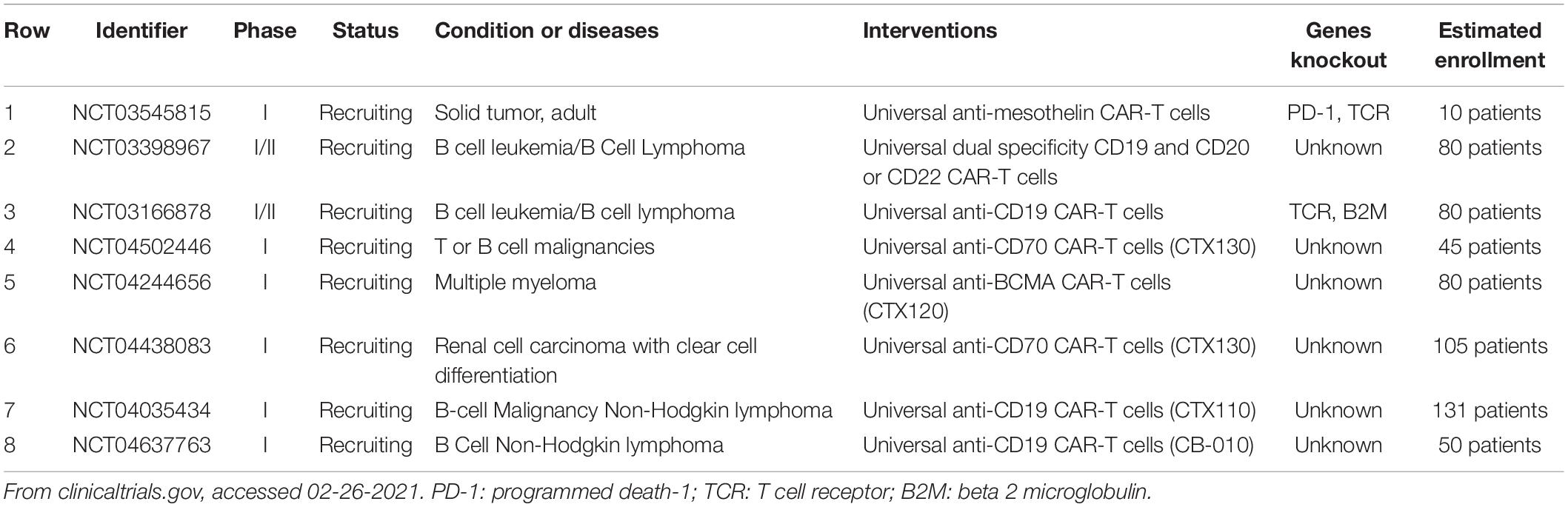
Table 1. Registered clinical trials using CRISPR/Cas9 modified universal CAR-T cells for treatment of malignancies.
Enhancing CAR-T Cell Function via CRISPR/Cas9
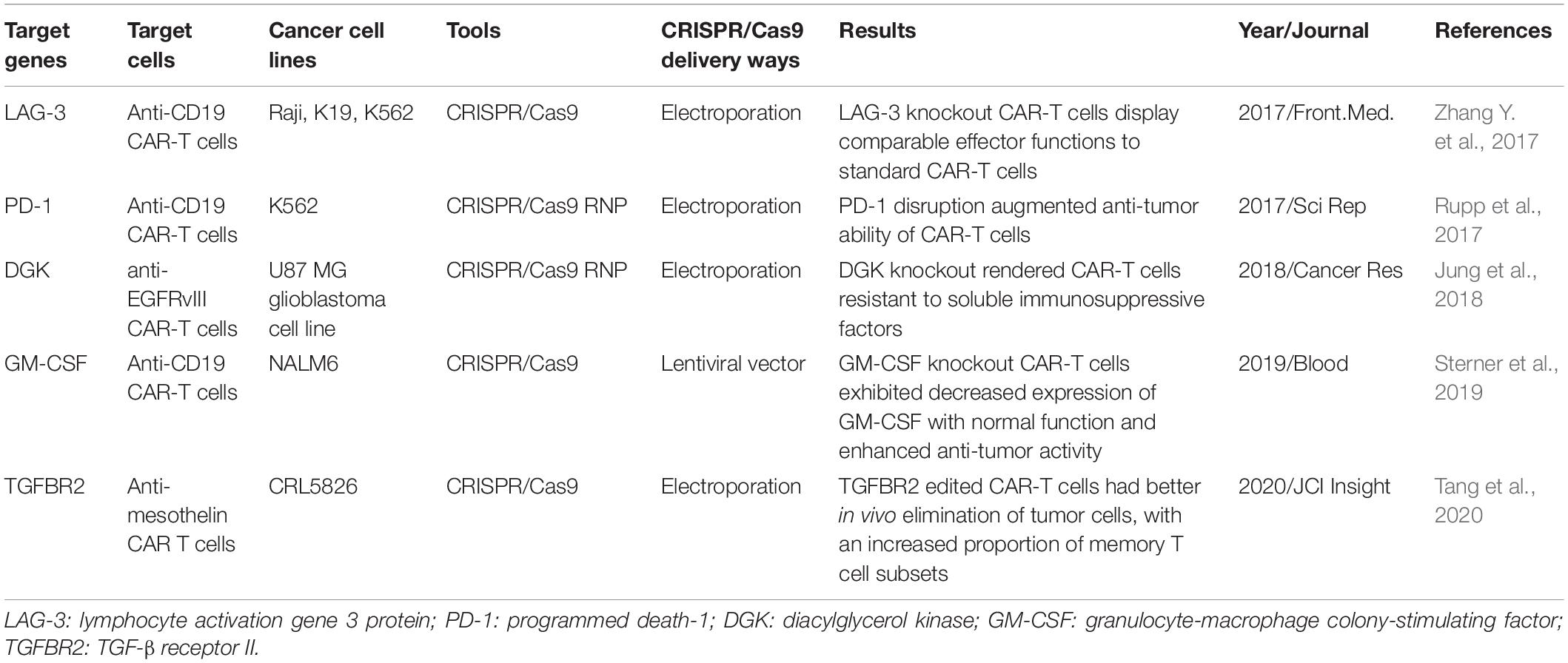
Despite remarkable success have been achieved in the treatment of hematological malignancies, CAR-T adoptive cell therapy have been floundered in many patients, to some extent due to immunosuppressive tumor microenvironment and T cell exhaustion (Cherkassky et al., 2016). Because of the established role of co-inhibitory molecules, such as PD-1, CTLA-4, LAG-3 and TIM-3 in T cells dysfunction, CRISPR/Cas9 system has also been applied to disrupt these inhibitory genes to enhance CAR-T cell function (Table 2 and Figure 2). CRISPR/Cas9-mediated PD-1 depletion was proven to augment the ability of CAR-T cells in killing tumor cells in vitro and clearing PD-L1+ tumor xenografts in vivo (Rupp et al., 2017; Choi et al., 2019). In addition to co-inhibitory genes, Diacylglycerol Kinase (DGK) ablation in CAR-T cells resulting in improvement of anti-tumor immunity (Jung et al., 2018). Knocking out granulocyte-macrophage colony-stimulating factor (GM-CSF) gene was demonstrated to enhance CAR-T cells function as well as reduce the risk of cytokine release syndrome (CRS) and inflammation (Sterner et al., 2019). Studies have also confirmed that knocking down the endogenous TGF-β receptor II (TGFBR2) in CAR T cells with CRISPR/Cas9 technology could decrease the exhaustion of CAR-T cells and increase solid tumor-killing efficacy both in vitro and in vivo (Tang et al., 2020). Moreover, eradicating CD7 and TRAC in CAR T cells by CRISPR/Cas9 increased the efficacy to treat T cell acute lymphoblastic leukemia (T-ALL) (Cooper et al., 2018).
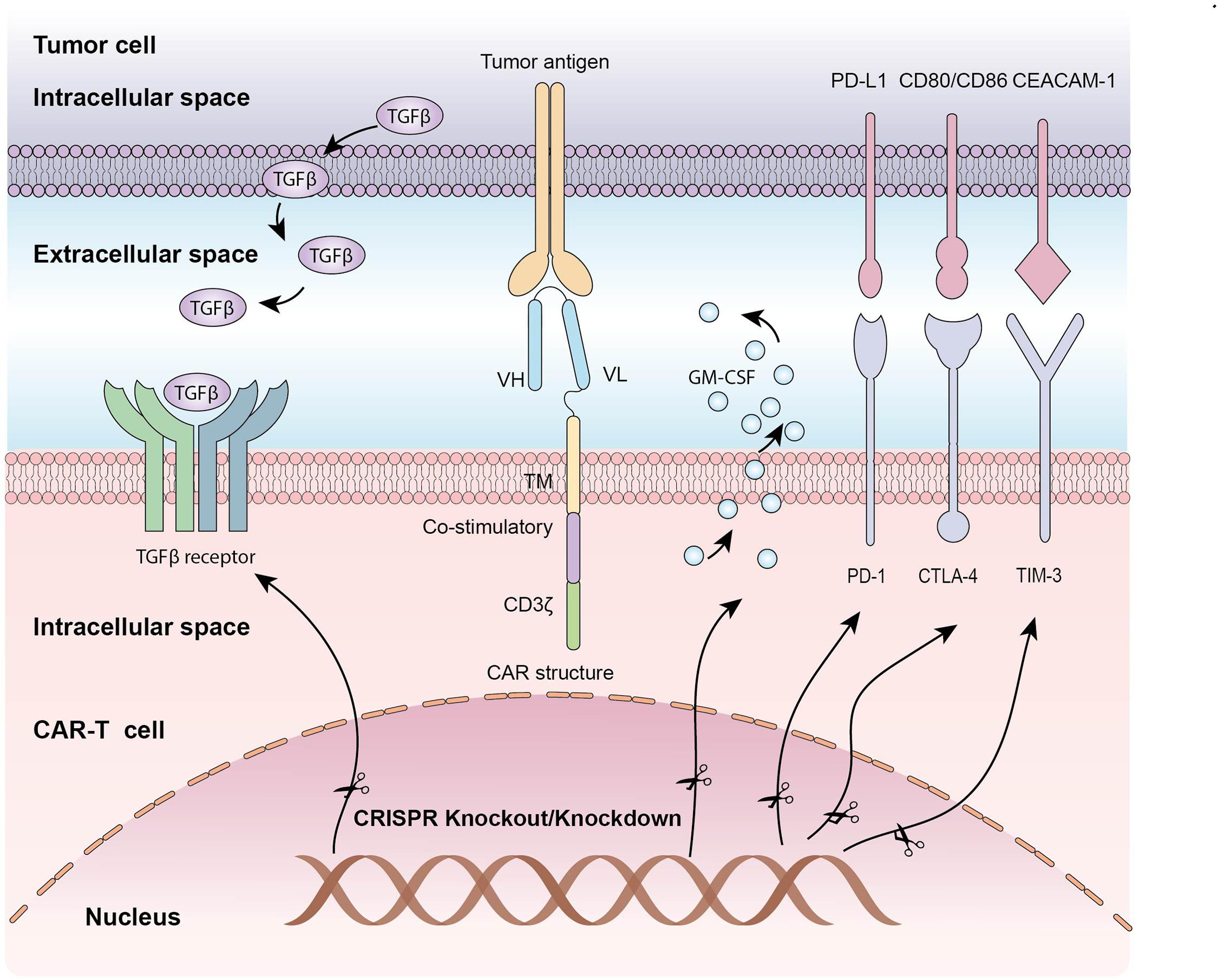
Figure 2. The application of CRISPR/Cas9 system in editing CAR-T cells: Knocking out inhibitory molecules to enhance function of CAR-T cells.
Application of CRISPR/Cas9 System in T Cell Receptor (Tcr)-Based Adoptive T Cell Therapy
It is known that CAR-T cells play limited roles in solid tumors. The main causes include lack of tumor specific antigens, heterogeneity of tumor antigens and tumor microenvironment suppression. Compared with CAR-T immunotherapy, engineered TCR-T cell therapy holds greater promise for targeting a wider range of antigens and thereby enlarge the scope to treat cancers (Morris and Stauss, 2016). The T cell receptor is an antigen recognition structure expressed on the membrane of T cells. The TCR is a heterodimer consisting of TCRα chain and TCR β chain. Both chains contain a variable antigen binding region, extracellular constant region and transmembrane region. The constant regions of TCRα/β chain and CD3 chains (γ, δ, ζ, ε) form the TCR/CD3 complex (Figure 3). The TCR/CD3 complex recognizes tumor antigens in a major histocompatibility complex (MHC)-dependent manner. Natural TCR specificities of tumor infiltrating lymphocytes (TILs) have been successfully exploited as adoptive cell therapy, and remarkable clinical responses have been achieved in several solid tumors, such as melanoma (Rosenberg et al., 1988), cholangiocarcinoma (Tran et al., 2014), breast cancer (Zacharakis et al., 2018) and papillomavirus-associated cervical cancer (Doran et al., 2019). While adoptive transfer of T cells with engineered tumor-specific TCRs has also shown promising therapeutic potential in several cancers, including melanoma (Robbins et al., 2011; Chodon et al., 2014), sarcoma (Robbins et al., 2011), and multiple myeloma (Mastaglio et al., 2017).
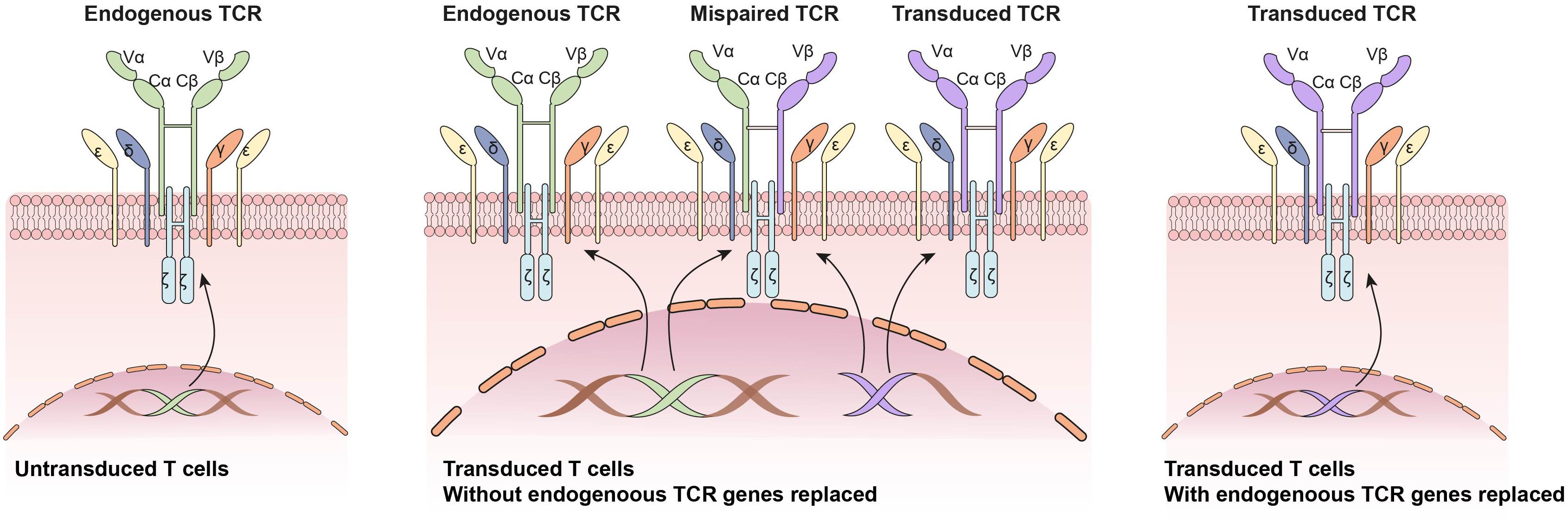
Figure 3. Application of CRISPR/Cas9 system in engineered T cell receptor (TCR)-based adoptive T cell therapy: Knocking out endogenous TCRs to avoid mixed TCR dimer formation and improve transduced TCR expression.
One of the major issues related to the generation of TCR-T is the pre-existing endogenous TCRs on the recipient T cells. The endogenous TCRs competes with transgenic TCRs for CD3 association and surface expression. Besides, there is also potential for mixed TCR dimer formation by mispairing between endogenous and transgenic TCRs. To circumvent these problems, CRISPR/Cas9 system was applied to replace endogenous TCRs α and β genes with artificial tumor-specific TCR sequence (Mastaglio et al., 2017; Legut et al., 2018; Morton et al., 2020; Figure 3). Knocking out endogenous TCRs leads to improved expression and function of transgenic TCRs compared with conventional TRC-T cells (Legut et al., 2018). Besides, anti-tumor responses of CRISPR/Cas9 modified T cells was enhanced in animal models (Roth et al., 2018). To reduce the risk of mispairing, another strategy is to transduce a stabilized Vα/Vβ single-chain TCRs (Sc-TCRs) (Aggen et al., 2012). The combination of Sc-TCRs transduction and CRISPR disruption was reported to almost completely eliminate TCR mispairing (Xue et al., 2020). The first clinical trial testing the safety and feasibility of CRISPR/Cas9 editing TCR-T cells in patients with refractory cancer was reported in 2020 (Stadtmauer et al., 2020). Patients’ autologous T cells were isolated and engineered by lentiviral transduction to express an TCR-specific for NY-ESO-1 and LAGE-1. Then the endogenous TCRs and PD-1 genes were disrupted by CRISPR/Cas9 system. The CRISPR modified T cells were expanded in vitro and re-fused to three patients. Two patients experienced a stable disease, and the other patient experienced disease progression (Stadtmauer et al., 2020).
In addition to knock out endogenous TCR genes, CRISPR/Cas9 system has also been used to identify a TCR that recognized and killed cancer cells in MHC-independent manner (via monomorphic MHC class I related protein, MR1) (Crowther et al., 2020).
Inhibition of Immune Checkpoint Signaling Pathway
In the past decade, immunotherapy has revolutionized cancer treatment by activating immune system to eliminate malignant tumor cells (Mellman et al., 2011; Page et al., 2014; Zarour, 2016). Although tumor cells can be immunogenic, and the presence of CD8+ tumor infiltrating lymphocytes was regarded as a positive prognostic factor in multiple solid tumors (Azimi et al., 2012; Bremnes et al., 2016; Pollari et al., 2018), the immune system often fails to eradicate tumor cells in vivo. The paradoxical coexistence of tumor-reactive T cells and tumor cells may arise from T cell exhaustion due to persistent antigen exposure and immunosuppressive factors in the tumor microenvironment (Schietinger and Greenberg, 2014; Pauken and Wherry, 2015; Wherry and Kurachi, 2015; Zarour, 2016). Gene profiling and phenotypical studies in human and mice have shown that exhausted tumor-infiltrating T lymphocytes typically express high level of inhibitory receptors including cytotoxic T lymphocyte antigen-4 (CTLA-4, CD152), programmed death-1 (PD-1, CD279), lymphocyte activation gene 3 protein (LAG3), T cell immunoglobulin domain and mucin domain-containing protein 3 (TIM-3, HAVCR2), 2B4 (CD244), CD160, TIGIT, and many other inhibitory molecules (Fourcade et al., 2012; Qin et al., 2019). In recent years, these immune checkpoint molecules have arisen a wide attention in cancer research due to their critical roles in anti-tumor immunity.
Once antigens are recognized by TCR/CD3 complex, CD28, as a co-stimulatory molecule, can strongly amplify TCR signals to activate T cells upon binding to CD80 (B7-1) or CD86 (B7-2) (Krummel and Allison, 1995; Rowshanravan et al., 2018). Whereas, CTLA-4, as a homologous receptor of CD28, is expressed exclusively on both CD4 + and CD8 + T cells and mediates an opposing function in T cell activation by delivering inhibitory signals upon binding to the same ligands as CD28 (Krummel and Allison, 1995). Targeting CTLA-4 by monoclonal antibody or genetic tool increases the availability of ligands to CD28, allowing to enhance T cell activation. Accordingly, Ipilimumab, a recombinant human IgG1 monoclonal antibody to blockade CTLA-4, was developed to boost patents’ immunity to eliminate tumor cells (Leach et al., 1996; Hodi et al., 2010). Ipilimumab, used as a single agent or in combination with gp100 peptide vaccine, as compared with gp100 vaccine alone, improved overall survival of patients with metastatic melanoma (Hodi et al., 2010). It was approved by US FDA for the treatment of metastatic melanoma in March 2011. Anti-CTLA-4 inhibitors have also been extensively investigated in many other cancers, such as non-small cell lung cancer, breast cancer, prostate cancer, and so on (Boutros et al., 2016; Page et al., 2016; Beer et al., 2017). In addition to using antibody to blockade CTLA-4, CRISPR/Cas9 has been applied to knockout CTLA-4. Shi et al. (2017) and Zhang et al. (2019) knocked out CTLA-4 in cytotoxic T lymphocytes (CTLs) using CRISPR/Cas9 system, and found that CTLA-4 disruption, as compared with control groups, could increase TNF-α and IFN-γ secretion and enhance anti-tumor activity of CTLs.
PD-1/PD-L1 immune checkpoint axis is another T cell function regulating pathway, upon TCR binding to MHC-peptides presented by antigen presenting cells (Keir et al., 2008). It has been found that PD-1 is expressed on a variety of activated immune cells, including T cells, monocytes, dendritic cells and so on (Alsaab et al., 2017). While its ligands PD-L1 and PD-L2 are expressed on tumor cells and antigen presenting cells. The interaction between PD-1 and PD-L1 suppresses T cell activation and function, and results in T cell exhaustion (Alsaab et al., 2017; Goodman et al., 2017). Inhibiting PD-1/PD-L1signal pathway using monoclonal antibody can reinvigorate T cell function. And anti-PD-1 antibody, nivolumab and pembrolizumab, have shown impressive anti-tumor responses in diverse malignancies, such as melanoma, non-small cell lung cancer, head and neck squamous cell carcinoma, and metastatic urothelial carcinoma (Brahmer et al., 2015; Robert et al., 2015; Ferris et al., 2016; Sharma et al., 2017). Therefore, the disruption of inhibitory genes may be a potential alternative to blockade immunotherapy. CRISPR/Cas9 has recently been applied to disrupt PD-1 expression on human peripheral blood T cells, CAR-T cells, and antigen specific cytotoxic T lymphocytes (CTLs) (Su et al., 2016, 2017; Ren et al., 2017a; Rupp et al., 2017; Shao et al., 2017; Guo et al., 2018; Zhang et al., 2018; Nakazawa et al., 2020). Su et al. (2016, 2017) reported the knockout of PD-1 by electroporation of plasmids encoding CRISPR/Cas9 system was technically feasible and did not affect the viability of T cell in vitro. Besides, IFN-γ secretion and anti-tumor cytotoxicity of gene modified T cells was enhanced. A recently completed clinical trial have reported that PD-1 disrupted T cells by CRISPR/Cas9 were safe and feasible, but lack of efficacy in patients with non-small cell lung cancer (Lu et al., 2020). Another trail evaluating the safety of PD-1 knockout T cell based on CRISPR/Cas9 system in patients with advanced esophageal cancer (NCT03081715) has also been registered and completed. While three other registered trials applying the same concept of PD-1 disrupted T cells in the treatment of muscle-invasive bladder cancer (NCT02863913), castration resistant prostate cancer (NCT02867345) and renal cell carcinoma (NCT02867332) have been withdrawn.
In addition to PD-1 knockout, disruption of PD-L1 in tumor cells has also been demonstrated to improve the efficacy of cancer immunotherapy. Tu et al. designed a novel type of weak acidity-responsive nanoparticles featured with CRISPR/Cas9-Cdk5 plasmid (Cas9-Cdk5) and paclitaxel (PTX). PTX encapsulated in the nanoparticles could induce immunogenic cells death and reduce suppressive immune cells. While the CRISPR/Cas9 plasmids could specifically target cyclin-dependent kinase 5 gene to mediate PD-L1 attenuation on tumor cells, so as to enhance anti-tumor immune response (Tu et al., 2020). Besides, another study conducted by Zhao et al. (2020) constructed a photoswitched CRISPR/Cas9 system to target PD-L1 gene. Under light irradiation, this system could efficiently knock out PD-L1 gene not only in bulk cancer cells but cancer stem-like cells as well.
Novel Targets Screening for Cancer Immunotherapy
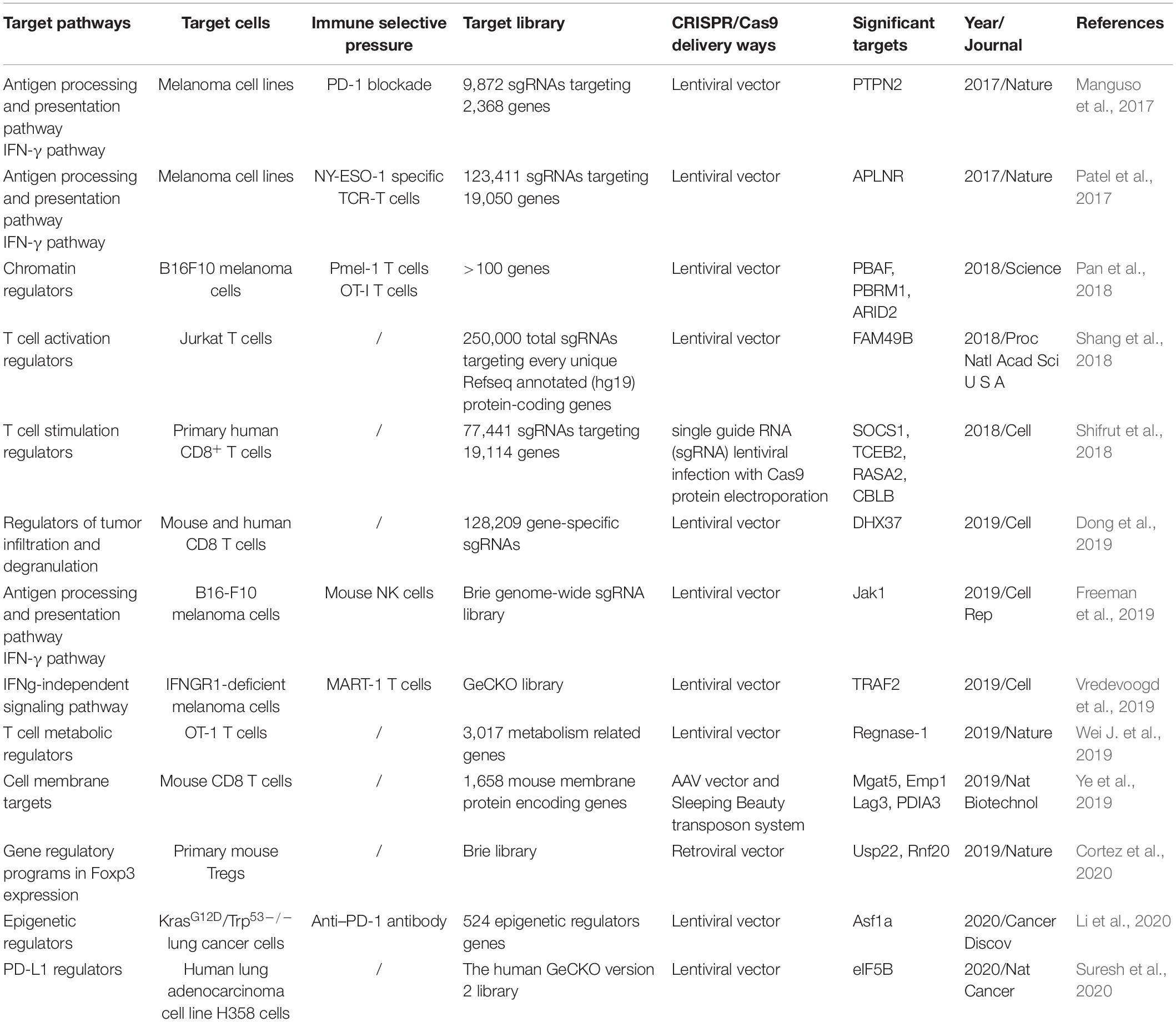
Tumor mutations can give rise to neoantigens and elicit anti-tumor immunity. However, genetic alterations can also induce immunotherapy resistance (Patel et al., 2017). For instance, tumor cells deficient in interferon-receptor signaling (JAK1/2) and antigen-presenting pathway (beta-2-microglobulin, B2M) have been reported to be associated with resistance to PD-1 blockade immunotherapy (Zaretsky et al., 2016). CRISPR/Cas9 system as a formidable large-scale gene screening toolkit, is developed to identify critical genes and new targets in cancer treatment (Liu et al., 2020). Various strategies of CRISPR screen for novel immunotherapy (IO) targets discovery have been used, including targeting antigen processing and presentation, IFN-γ pathway, TNF signaling, epigenetic regulators, and PD-L1 regulators in tumor cells, and metabolic regulators in T cells (Table 3).
To target antigen processing and presentation and IFN-γ pathway, Manguso et al. created a library consisting of 9,872 sgRNAs targeting 2368 genes, then delivered the library into B16 melanoma cells via lentiviral infection. After transduction, tumor cells were purified and implanted into mice that were then treated with immunotherapy, so as to apply immune selective pressure on the transduced tumor cells. In parallel, the library-transduced tumor cells were transplanted into Tcra-/- mice in order to generate animal models without immune selective pressure. After about 12–14 days, tumor tissues were collected. The library representation in tumors from wild type mice or Tcra-/- mice were compared. In this study, they found that deletion of protein tyrosine phosphatase PTPN2 sensitized tumor cells to immunotherapy by enhancing IFN-γ signaling pathway (Manguso et al., 2017). Patel et al. (2017) transduced NY-ESO-1+ Mel624 melanoma cells with a genome-scale CRISPR library of 123,411 sgRNAs, and constructed NY-ESO-1 antigen specific TCR-T cells. Then the transduced melanoma cells were co-incubated with NY-ESO-1 antigen specific TCR-T cells. They found antigen presentation and IFN-γ pathway related genes, such as HLA-A, B2M, TAP1, TAP2, and TAPBP were among the most critical genes in the screen. Besides, the functional loss of APLNR reduced the efficacy of immunotherapy by interacting with JAK1 and modulating IFN-γ responses in tumors (Patel et al., 2017).
By targeting TNF signaling, metabolic protein 2-aminoethanethiol dioxygenase (Ado) was found to modulate sensitivity of tumor cells to TNF released by cytotoxic T cells (Kearney et al., 2018). Vredevoogd et al. reported that selectively inactivation of TNF pathway, such as the ablation of TRAF2,sensitized tumor cells to T cell attack (Vredevoogd et al., 2019). To identify critical epigenetic regulators in anti-tumor immunity, a sgRNA library focused on epigenetic regulators were constructed and CRISPR screening was performed in a KrasG12D/Trp53–/– lung adenocarcinoma model. Loss of the histone chaperone Asf1a was found to sensitize tumors to anti-PD-1treatment (Li et al., 2020). In order to identify important regulators of PD-L1 expression. Suresh et al. (2020) performed a genome wide loss-of-function CRISPR screen in human lung cancer cells. They demonstrated that heme biosynthesis pathway was a key regulator of PD-L1 expression, and impairment of heme production would activate integrated stress response (ISR), resulting in enhanced PD-L1 expression. While the translation initiation factor eIF5 was required in ISR-dependent PD-L1 expression. Targeting eIF5 might be potentially therapeutic. In addition to targeting genes in tumor cells, Jun Wei and colleagues constructed two sub-libraries of sgRNAs targeting 3,017 metabolism related genes in T cells to investigate the roles of metabolism-associated factors in T cell immunity. They demonstrated that CD8 T cells could be reprogramed to long-lived effector cells by knocking out Regnase-1. Regnase-1 null T cells showed remarkable improvement of therapeutic efficacy against mouse melanoma and leukemia (Wei L. et al., 2019). Newly potential immunotherapeutic targets which have been identified by CRISPR/Cas9 screening technology are summarized in Table 3.
Perspectives and Challenges
Perspectives: CRISPR/Cas9 and TILs Based Adoptive Cell Therapy
The presence of tumor-infiltrating lymphocytes (TILs) in the tumor microenvironment is regarded as an adaptive anti-tumor immune response. About 67% TILs grown from metastatic melanoma were identified to be tumor specific (Goff et al., 2010). Due to the natural TCR specificity to tumor antigens, TILs have been exploited as an adoptive cell therapy (ACT) to treat cancers. Tumors were resected and fragmented into small pieces at about 1-2 mm3, or enzymatically digested as single cell suspension. TILs were isolated and expanded in vitro at the presence of interleukin-2 (IL-2) to large numbers, and then adoptively transfer to patients (Dudley et al., 2003). The clinical responses with TIL therapy were firstly demonstrated in melanoma with objective response rate (ORR) up to 72%, in which 40% patients experienced durable clinical responses and 10-20% patients reached complete remission (CR) (Rosenberg et al., 1994, 2011). The encouraging clinical responses of TILs based ACT in melanoma have stimulated researchers to conduct studies in other types of solid cancers. Three out of nine patients with metastatic cervical cancer experienced objective tumor responses (with two complete remission and one partial remission) after receiving a single infusion of autologous TILs (Stevanovic et al., 2015). Besides, ACT with TILs targeting tumor neoantigens encoded by mutated genes has achieved substantial objective clinical responses in patients with metastatic cholangiocarcinoma (Tran et al., 2014), breast cancer (Zacharakis et al., 2018), and colorectal cancer (Tran et al., 2016).
Despite promising clinical outcome have been achieved, the majority of patients with epithelial cancers did not respond to TIL therapy. The impairment and exhaustion of TILs may account for the poor responses, as TILs may be in “progenitor exhausted” state before being expanded ex vivo. T cells get into terminal differentiated state after rapid expansion. Studies have found that longer duration of responses to immune checkpoint blockades could be noticed in patients with melanoma who had a higher percentage of progenitor exhausted TILs (Miller et al., 2019). Progenitor exhausted TILs can respond to checkpoint blockades, while exhausted TILs cannot. TIL phenotype analysis has also revealed that less differentiated CD39–CD69– stem-like TIL phenotype was associated with complete tumor responses and longer TIL persistence in patients who received TIL therapy (Krishna et al., 2020). Thus reversing dysfunctional T cells state and retaining stem-like TIL phenotype by using CRISPR/Cas9 gene-editing tool may further improve the efficacy of TIL therapy. Metabolism-associated factor Regnase-I was found to be a negative regulator of T cell anti-tumor responses. Knocking out Regnase-I reprogrammed T cells to long-lived effector cells with better infiltration and persistence in tumor microenvironment, and improved ACT therapeutic efficacy (Wei L. et al., 2019). Another study identified that Gata-3, a zinc-finger transcription factor, drove CD8+ TILs dysfunction. Disrupting Gata-3 in naïve CD8+ T cells improved T cells anti-tumor function (Singer et al., 2016). However, studies on engineering TILs to enhance T cells function and proliferative activity by using CRISPR/Cas9 system were rarely reported. Only zinc finger nuclease was once reported to target the gene encoding human PD-1 in melanoma tumor-infiltrating lymphocytes. The PD-1 knockout TILs could be expanded into clinical scale. In addition, the edited TILs showed enhanced ex vivo effector function and a significantly increased cytokine releasing compared to unedited TILs (Beane et al., 2015).
Challenges: CRISPR/Cas9 Technology in Clinical Translation
Although CRISPR/Cas9 system have shown immense potential for improving efficacy of immunotherapy, several concerns related to safety and efficacy impede its translation to clinical applications.
Firstly, clinical efficacy of ACT based cancer immunotherapy, including CAR-T cells, TCR-T cells and TILs therapy, is dependent on adequate T cells for refusion. However, most of CRISPR-engineered T cells for clinical trials were transduced by electroporation which might result in cell damage and impeding T cell proliferation ex vivo (Lu et al., 2020; Song et al., 2021). Therefore, more safe and efficient delivery ways such as viral vectors are in urgent exploration. In addition, direct delivery in situ or in vivo may be an alternative option that is worthy to develop (Song et al., 2021). However, the immunogenicity of cas9 proteins may be another challenge that constrain the clinical translation of CRISPR/Cas9 system. Anti-SaCas9 (Cas9 from S. aureus) and anti-SpCas9 (Cas9 from S. pyogenes) antibodies were detected in 78% and 58% of donors, respectively. Moreover, 78% and 67% of donors possess T cells against SaCas9 and SpCas9 protein, respectively (Charlesworth et al., 2019). This showed that there were pre-existing adaptive immune responses to Cas9 proteins in human which may cause adverse effects when treating patients with CRISPR/Cas9 system. Whereas no potential rejections were noted in clinical trial (Stadtmauer et al., 2020). Some researchers point out that the pre-existing immune responses to Cas9 proteins do not appear to be an obstacle to clinical application of CRISPR/Cas9 system (Stadtmauer et al., 2020).
Secondly, although various approaches have been reported to improve gRNA design (Bin Moon et al., 2018; Matson et al., 2019; Han et al., 2020) and increase the specificity of Cas enzyme (Kleinstiver et al., 2016; Slaymaker et al., 2016), the risk of off-target effects, resulting from non-specific cutting and further leading to unwanted mutations, still remains a major obstacle to translation of CRISPR/Cas9 system to clinical therapeutic use (Cook and Ventura, 2019). Whereas, in clinical trials, no significant or only a small number of off-target sites, chromosomal rearrangements or long-range deletions were actually detected. Moreover, the frequency of off-target sites declined over time (Roth et al., 2018; Xu et al., 2019; Lu et al., 2020). But experience with more patients and longer follow-up are needed to further validate the safety and feasibility. The commonly used off-target effects detecting methods including the T7 Endonuclease I (T7E1) mutation mismatch assay, deep sequencing, and whole genome sequencing are not perfect (Ren and Zhao, 2017; Li et al., 2019). More sensitive, accurate and practical approaches are needed to identify off-target mutations. However, the past decades have witnessed the introduction and development of high-throughput sequencing and multi-omics analysis. The combination of multi-omics analysis may help detect and understand causal mutations, functions of these genes and affected cellular and signaling pathways, so as to predict the potential and significance of off-target effects.
Author Contributions
XO and QM wrote the first draft of the manuscript. WY completed the references collection and drew the figures. XM and ZH reviewed and revised the original manuscript. All authors commented on previous versions of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aggen, D. H., Chervin, A. S., Schmitt, T. M., Engels, B., Stone, J. D., Richman, S. A., et al. (2012). Single-chain VαVβ T-cell receptors function without mispairing with endogenous TCR chains. Gene Ther. 19, 365–374. doi: 10.1038/gt.2011.104
Alsaab, H. O., Sau, S., Alzhrani, R., Tatiparti, K., Bhise, K., Kashaw, S. K., et al. (2017). PD-1 and PD-L1 checkpoint signaling inhibition for cancer immunotherapy: mechanism, combinations, and clinical outcome. Front. Pharmacol. 8:561. doi: 10.3389/fphar.2017.00561
Azimi, F., Scolyer, R. A., Rumcheva, P., Moncrieff, M., Murali, R., McCarthy, S. W., et al. (2012). Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J. Clin. Oncol. 30, 2678–2683. doi: 10.1200/jco.2011.37.8539
Beane, J. D., Lee, G., Zheng, Z., Mendel, M., Abate-Daga, D., Bharathan, M., et al. (2015). Clinical scale zinc finger nuclease-mediated gene editing of PD-1 in tumor infiltrating lymphocytes for the treatment of metastatic melanoma. Mol. Ther. 23, 1380–1390. doi: 10.1038/mt.2015.71
Beer, T. M., Kwon, E. D., Drake, C. G., Fizazi, K., Logothetis, C., Gravis, G., et al. (2017). Randomized, double-blind, phase III trial of ipilimumab versus placebo in asymptomatic or minimally symptomatic patients with metastatic chemotherapy-naive castration-resistant Prostate cancer. J. Clin. Oncol. 35, 40–47. doi: 10.1200/jco.2016.69.1584
Bin Moon, S., Lee, J. M., Kang, J. G., Lee, N. E., Ha, D. I., Kim, D. Y., et al. (2018). Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3’-overhang. Nat. Commun. 9:3651.
Boutros, C., Tarhini, A., Routier, E., Lambotte, O., Ladurie, F. L., Carbonnel, F., et al. (2016). Safety profiles of anti-CTLA-4 and anti-PD-1 antibodies alone and in combination. Nat. Rev. Clin. Oncol. 13, 473–486. doi: 10.1038/nrclinonc.2016.58
Brahmer, J., Reckamp, K. L., Baas, P., Crino, L., Eberhardt, W. E., Poddubskaya, E., et al. (2015). Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med. 373, 123–135.
Bremnes, R. M., Busund, L. T., Kilvaer, T. L., Andersen, S., Richardsen, E., Paulsen, E. E., et al. (2016). The role of tumor-infiltrating lymphocytes in development, progression, and prognosis of non-small cell Lung cancer. J. Thorac. Oncol. 11, 789–800. doi: 10.1016/j.jtho.2016.01.015
Charlesworth, C. T., Deshpande, P. S., Dever, D. P., Camarena, J., Lemgart, V. T., Cromer, M. K., et al. (2019). Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 25, 249–254. doi: 10.1038/s41591-018-0326-x
Cherkassky, L., Morello, A., Villena-Vargas, J., Feng, Y., Dimitrov, D. S., Jones, D. R., et al. (2016). Human CAR T cells with cell-intrinsic PD-1 checkpoint blockade resist tumor-mediated inhibition. J. Clin. Invest. 126, 3130–3144. doi: 10.1172/jci83092
Chodon, T., Comin-Anduix, B., Chmielowski, B., Koya, R. C., Wu, Z., Auerbach, M., et al. (2014). Adoptive transfer of MART-1 T-cell receptor transgenic lymphocytes and dendritic cell vaccination in patients with metastatic melanoma. Clin. Cancer Res. 20, 2457–2465.
Choi, B. D., Yu, X., Castano, A. P., Darr, H., Henderson, D. B., Bouffard, A. A., et al. (2019). CRISPR-Cas9 disruption of PD-1 enhances activity of universal EGFRvIII CAR T cells in a preclinical model of human glioblastoma. J. Immunother. Cancer 7:304.
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science (New York, N.Y.) 339, 819–823.
Cook, P. J., and Ventura, A. (2019). Cancer diagnosis and immunotherapy in the age of CRISPGenes. Genes Chromosomes Cancer 58, 233–243. doi: 10.1002/gcc.22702
Cooper, M. L., Choi, J., Staser, K., Ritchey, J. K., Devenport, J. M., Eckardt, K., et al. (2018). An “off-the-shelf” fratricide-resistant CAR-T for the treatment of T cell hematologic malignancies. Leukemia 32, 1970–1983. doi: 10.1038/s41375-018-0065-5
Cortez, J. T., Montauti, E., Shifrut, E., Gatchalian, J., Zhang, Y., Shaked, O., et al. (2020). CRISPR screen in regulatory T cells reveals modulators of Foxp3. Nature 582, 416–420. doi: 10.1038/s41586-020-2246-4
Crowther, M. D., Dolton, G., Legut, M., Caillaud, M. E., Lloyd, A., Attaf, M., et al. (2020). Genome-wide CRISPR-Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat. Immunol. 21, 178–185. doi: 10.1038/s41590-019-0578-8
Depil, S., Duchateau, P., Grupp, S. A., Mufti, G., and Poirot, L. (2020). ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 19, 185–199. doi: 10.1038/s41573-019-0051-2
Dong, M. B., Wang, G., Chow, R. D., Ye, L., Zhu, L., Dai, X., et al. (2019). Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T Cells. Cell 178, 1189–1204.e23.
Doran, S. L., Stevanovic, S., Adhikary, S., Gartner, J. J., Jia, L., Kwong, M. L. M., et al. (2019). T-Cell receptor gene therapy for human papillomavirus-associated epithelial cancers: a first-in-human, Phase I/II Study. J. Clin. Oncol. 37, 2759–2768. doi: 10.1200/jco.18.02424
Dudley, M. E., Wunderlich, J. R., Shelton, T. E., Even, J., and Rosenberg, S. A. (2003). Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J. Immunotherapy 26, 332–342. doi: 10.1097/00002371-200307000-00005
Esposito, R., Bosch, N., Lanzós, A., Polidori, T., Pulido-Quetglas, C., and Johnson, R. (2019). Hacking the cancer genome: profiling therapeutically actionable long non-coding rnas using CRISPR-Cas9 screening. Cancer Cell 35, 545–557. doi: 10.1016/j.ccell.2019.01.019
Evers, B., Jastrzebski, K., Heijmans, J. P., Grernrum, W., Beijersbergen, R. L., and Bernards, R. (2016). CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat. Biotechnol. 34, 631–633. doi: 10.1038/nbt.3536
Eyquem, J., Mansilla-Soto, J., Giavridis, T., van der Stegen, S. J., Hamieh, M., Cunanan, K. M., et al. (2017). Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117. doi: 10.1038/nature21405
Ferris, R. L., Blumenschein, G. Jr., Fayette, J., Guigay, J., Colevas, A. D., Licitra, L., et al. (2016). Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 375, 1856–1867.
Fourcade, J., Sun, Z., Pagliano, O., Guillaume, P., Luescher, I. F., Sander, C., et al. (2012). CD8(+) T cells specific for tumor antigens can be rendered dysfunctional by the tumor microenvironment through upregulation of the inhibitory receptors BTLA and PD-1. Cancer Res. 72, 887–896. doi: 10.1158/0008-5472.can-11-2637
Freeman, A. J., Vervoort, S. J., Ramsbottom, K. M., Kelly, M. J., Michie, J., Pijpers, L., et al. (2019). Natural killer cells suppress T Cell-Associated tumor immune evasion. Cell Rep. 28, 2784–2794.e5.
Goff, S. L., Smith, F. O., Klapper, J. A., Sherry, R., Wunderlich, J. R., Steinberg, S. M., et al. (2010). Tumor infiltrating lymphocyte therapy for metastatic melanoma: analysis of tumors resected for TIL. J. Immunother. 33, 840–847. doi: 10.1097/cji.0b013e3181f05b91
Goodman, A., Patel, S. P., and Kurzrock, R. (2017). PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat. Rev. Clin. Oncol. 14, 203–220. doi: 10.1038/nrclinonc.2016.168
Guo, X., Jiang, H., Shi, B., Zhou, M., Zhang, H., Shi, Z., et al. (2018). Disruption of PD-1 enhanced the anti-tumor activity of chimeric antigen receptor T cells against hepatocellular carcinoma. Front. Pharmacol. 9:1118. doi: 10.3389/fphar.2018.01118
Han, H. A., Pang, J. K. S., and Soh, B. S. (2020). Mitigating off-target effects in CRISPR/Cas9-mediated in vivo gene editing. J. Mol. Med. (Berl) 98, 615–632. doi: 10.1007/s00109-020-01893-z
Hodi, F. S., O’Day, S. J., McDermott, D. F., Weber, R. W., Sosman, J. A., Haanen, J. B., et al. (2010). Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723.
Jackson, H. J., Rafiq, S., and Brentjens, R. J. (2016). Driving CAR T-cells forward. Nat. Rev. Clin. Oncol. 13, 370–383. doi: 10.1038/nrclinonc.2016.36
Jung, I. Y., Kim, Y. Y., Yu, H. S., Lee, M., Kim, S., and Lee, J. (2018). CRISPR/Cas9-Mediated knockout of DGK improves antitumor activities of human T Cells. Cancer Res. 78, 4692–4703. doi: 10.1158/0008-5472.can-18-0030
Kearney, C. J., Vervoort, S. J., Hogg, S. J., Ramsbottom, K. M., Freeman, A. J., Lalaoui, N., et al. (2018). Tumor immune evasion arises through loss of TNF sensitivity. Sci. Immunol. 3:eaar3451. doi: 10.1126/sciimmunol.aar3451
Keir, M. E., Butte, M. J., Freeman, G. J., and Sharpe, A. H. (2008). PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 26, 677–704. doi: 10.1146/annurev.immunol.26.021607.090331
Kleinstiver, B. P., Pattanayak, V., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Zheng, Z., et al. (2016). High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495. doi: 10.1038/nature16526
Krishna, S., Lowery, F. J., Copeland, A. R., Bahadiroglu, E., Mukherjee, R., Jia, L., et al. (2020). Stem-like CD8 T cells mediate response of adoptive cell immunotherapy against human cancer. Science (New York, N.Y.) 370, 1328–1334. doi: 10.1126/science.abb9847
Krummel, M. F., and Allison, J. P. (1995). CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med. 182, 459–465. doi: 10.1084/jem.182.2.459
Labanieh, L., Majzner, R. G., and Mackall, C. L. (2018). Programming CAR-T cells to kill cancer. Nat. Biomed. Eng. 2, 377–391. doi: 10.1038/s41551-018-0235-9
Leach, D. R., Krummel, M. F., and Allison, J. P. (1996). Enhancement of antitumor immunity by CTLA-4 blockade. Science (New York, N.Y.) 271, 1734–1736. doi: 10.1126/science.271.5256.1734
Legut, M., Dolton, G., Mian, A. A., Ottmann, O. G., and Sewell, A. K. (2018). CRISPR-mediated TCR replacement generates superior anticancer transgenic T cells. Blood 131, 311–322. doi: 10.1182/blood-2017-05-787598
Li, F., Huang, Q., Luster, T. A., Hu, H., Zhang, H., Ng, W. L., et al. (2020). in vivo epigenetic CRISPR screen identifies Asf1a as an immunotherapeutic target in kras-mutant Lung adenocarcinoma. Cancer Discovery 10, 270–287. doi: 10.1158/2159-8290.cd-19-0780
Li, J., Hong, S., Chen, W., Zuo, E., and Yang, H. (2019). Advances in detecting and reducing off-target effects generated by CRISPR-mediated genome editing. J. Genet. Genom. 46, 513–521. doi: 10.1016/j.jgg.2019.11.002
Liu, D., Zhao, X., Tang, A., Xu, X., Liu, S., Zha, L., et al. (2020). CRISPR screen in mechanism and target discovery for cancer immunotherapy. Biochim. et Biophys. Acta. Rev. on Cancer 1874:188378. doi: 10.1016/j.bbcan.2020.188378
Lu, Y., Xue, J., Deng, T., Zhou, X., Yu, K., Deng, L., et al. (2020). Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 26, 732–740.
Manguso, R. T., Pope, H. W., Zimmer, M. D., Brown, F. D., Yates, K. B., Miller, B. C., et al. (2017). In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418. doi: 10.1038/nature23270
Mastaglio, S., Genovese, P., Magnani, Z., Ruggiero, E., Landoni, E., Camisa, B., et al. (2017). NY-ESO-1 TCR single edited stem and central memory T cells to treat multiple myeloma without graft-versus-host disease. Blood 130, 606–618. doi: 10.1182/blood-2016-08-732636
Matson, A. W., Hosny, N., Swanson, Z. A., Hering, B. J., and Burlak, C. (2019). Optimizing sgRNA length to improve target specificity and efficiency for the GGTA1 gene using the CRISPR/Cas9 gene editing system. PLoS One 14:e0226107. doi: 10.1371/journal.pone.0226107
Maude, S. L., Frey, N., Shaw, P. A., Aplenc, R., Barrett, D. M., Bunin, N. J., et al. (2014). Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 371, 1507–1517.
Mellman, I., Coukos, G., and Dranoff, G. (2011). Cancer immunotherapy comes of age. Nature 480, 480–489. doi: 10.1038/nature10673
Miller, B. C., Sen, D. R., Al Abosy, R., Bi, K., Virkud, Y. V., LaFleur, M. W., et al. (2019). Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336. doi: 10.1038/s41590-019-0312-6
Morris, E. C., and Stauss, H. J. (2016). Optimizing T-cell receptor gene therapy for hematologic malignancies. Blood 127, 3305–3311. doi: 10.1182/blood-2015-11-629071
Morton, L. T., Reijmers, R. M., Wouters, A. K., Kweekel, C., Remst, D. F. G., Pothast, C. R., et al. (2020). Simultaneous deletion of endogenous TCRαβ for TCR gene therapy creates an improved and safe cellular therapeutic. Mol. Ther. 28, 64–74. doi: 10.1016/j.ymthe.2019.10.001
Nakazawa, T., Natsume, A., Nishimura, F., Morimoto, T., Matsuda, R., Nakamura, M., et al. (2020). Effect of CRISPR/Cas9-Mediated PD-1-Disrupted primary human third-generation CAR-T cells targeting EGFRvIII on in vitro human glioblastoma cell growth. Cells 9:998. doi: 10.3390/cells9040998
O’Donnell, J. S., Teng, M. W. L., and Smyth, M. J. (2019). Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol. 16, 151–167. doi: 10.1038/s41571-018-0142-8
Page, D. B., Postow, M. A., Callahan, M. K., Allison, J. P., and Wolchok, J. D. (2014). Immune modulation in cancer with antibodies. Annu. Rev. Med. 65, 185–202. doi: 10.1146/annurev-med-092012-112807
Page, D. B., Yuan, J., Redmond, D., Wen, Y. H., Durack, J. C., Emerson, R., et al. (2016). Deep sequencing of T-cell receptor DNA as a biomarker of clonally expanded TILs in breast cancer after immunotherapy. Cancer Immunol. Res. 4, 835–844. doi: 10.1158/2326-6066.cir-16-0013
Pan, D., Kobayashi, A., Jiang, P., Ferrari, de Andrade, L., Tay, R. E., et al. (2018). A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 359, 770–775. doi: 10.1126/science.aao1710
Patel, S. J., Sanjana, N. E., Kishton, R. J., Eidizadeh, A., Vodnala, S. K., Cam, M., et al. (2017). Identification of essential genes for cancer immunotherapy. Nature 548, 537–542.
Pauken, K. E., and Wherry, E. J. (2015). Overcoming T cell exhaustion in infection and cancer. Trends Immunol. 36, 265–276. doi: 10.1016/j.it.2015.02.008
Pettitt, S. J., Krastev, D. B., Dréan, A., Song, F., Aleksandrov, R., and Harrell, M. I. (2018). Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat. Commun. 9:1849.
Platt, R. J., Chen, S., Zhou, Y., Yim, M. J., Swiech, L., Kempton, H. R., et al. (2014). CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455. doi: 10.1016/j.cell.2014.09.014
Poirot, L., Philip, B., Schiffer-Mannioui, C., Le Clerre, D., Chion-Sotinel, I., Derniame, S., et al. (2015). Multiplex genome-Edited T-cell manufacturing platform for “Off-the-Shelf”. adoptive T-cell immunotherapies. Cancer Res. 75, 3853–3864. doi: 10.1158/0008-5472.can-14-3321
Pollari, M., Brück, O., Pellinen, T., Vähämurto, P., Karjalainen-Lindsberg, M. L., Mannisto, S., et al. (2018). PD-L1(+) tumor-associated macrophages and PD-1(+) tumor-infiltrating lymphocytes predict survival in primary testicular lymphoma. Haematologica 103, 1908–1914. doi: 10.3324/haematol.2018.197194
Qin, S., Xu, L., Yi, M., Yu, S., Wu, K., and Luo, S. (2019). Novel immune checkpoint targets: moving beyond PD-1 and CTLA-4. Mol. Cancer 18:155.
Ren, J., Liu, X., Fang, C., Jiang, S., June, C. H., and Zhao, Y. (2017a). Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin. Cancer Res. 23, 2255–2266. doi: 10.1158/1078-0432.ccr-16-1300
Ren, J., Zhang, X., Liu, X., Fang, C., Jiang, S., June, C. H., et al. (2017b). A versatile system for rapid multiplex genome-edited CAR T cell generation. Oncotarget 8, 17002–17011. doi: 10.18632/oncotarget.15218
Ren, J., and Zhao, Y. (2017). Advancing chimeric antigen receptor T cell therapy with CRISPR/Cas9. Protein Cell 8, 634–643. doi: 10.1007/s13238-017-0410-x
Robbins, P. F., Morgan, R. A., Feldman, S. A., Yang, J. C., Sherry, R. M., Dudley, M. E., et al. (2011). Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J. Clin. Oncol. 29, 917–924. doi: 10.1200/jco.2010.32.2537
Robert, C., Long, G. V., Brady, B., Dutriaux, C., Maio, M., Mortier, L., et al. (2015). Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 372, 320–330.
Rosenberg, S. A., Packard, B. S., Aebersold, P. M., Solomon, D., Topalian, S. L., Toy, S. T., et al. (1988). Use of tumor-infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. a preliminary report. New Eng. J. Med. 319, 1676–1680.
Rosenberg, S. A., Yang, J. C., Sherry, R. M., Kammula, U. S., Hughes, M. S., Phan, G. Q., et al. (2011). Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res. 17, 4550–4557. doi: 10.1158/1078-0432.ccr-11-0116
Rosenberg, S. A., Yannelli, J. R., Yang, J. C., Topalian, S. L., Schwartzentruber, D. J., Weber, J. S., et al. (1994). Treatment of patients with metastatic melanoma with autologous tumor-infiltrating lymphocytes and interleukin 2. J. Natl. Cancer Inst. 86, 1159–1166.
Roth, T. L., Puig-Saus, C., Yu, R., Shifrut, E., Carnevale, J., Li, P. J., et al. (2018). Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 559, 405–409.
Rowshanravan, B., Halliday, N., and Sansom, D. M. (2018). CTLA-4: a moving target in immunotherapy. Blood 131, 58–67. doi: 10.1182/blood-2017-06-741033
Ruella, M., Xu, J., Barrett, D. M., Fraietta, J. A., Reich, T. J., Ambrose, D. E., et al. (2018). Induction of resistance to chimeric antigen receptor T cell therapy by transduction of a single leukemic B cell. Nat. Med. 24, 1499–1503.
Rupp, L. J., Schumann, K., Roybal, K. T., Gate, R. E., Ye, C. J., Lim, W. A., et al. (2017). CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci. Rep. 7:737.
Schietinger, A., and Greenberg, P. D. (2014). Tolerance and exhaustion: defining mechanisms of T cell dysfunction. Trends Immunol. 35, 51–60. doi: 10.1016/j.it.2013.10.001
Shang, W., Jiang, Y., Boettcher, M., Ding, K., Mollenauer, M., Liu, Z., et al. (2018). Genome-wide CRISPR screen identifies FAM49B as a key regulator of actin dynamics and T cell activation. Proc. Natl. Acad. Sci. U S A. 115, E4051–E4060.
Shao, J., Xu, Q., Su, S., Meng, F., Zou, Z., Chen, F., et al. (2017). Engineered cells for costimulatory enhancement combined with IL-21 enhance the generation of PD-1-disrupted CTLs for adoptive immunotherapy. Cell Immunol. 320, 38–45. doi: 10.1016/j.cellimm.2017.09.003
Sharma, P., Retz, M., Siefker-Radtke, A., Baron, A., Necchi, A., Bedke, J., et al. (2017). Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol. 18, 312–322. doi: 10.1016/s1470-2045(17)30065-7
Shi, L., Meng, T., Zhao, Z., Han, J., Zhang, W., Gao, F., et al. (2017). CRISPR knock out CTLA-4 enhances the anti-tumor activity of cytotoxic T lymphocytes. Gene 636, 36–41. doi: 10.1016/j.gene.2017.09.010
Shifrut, E., Carnevale, J., Tobin, V., Roth, T. L., Woo, J. M., Bui, C. T., et al. (2018). Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell 175, 1958–1971.e15.
Singer, M., Wang, C., Cong, L., Marjanovic, N. D., Kowalczyk, M. S., Zhang, H., et al. (2016). A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T Cells. Cell 166, 1500–1511.e9.
Slaymaker, I. M., Gao, L., Zetsche, B., Scott, D. A., Yan, W. X., and Zhang, F. (2016). Rationally engineered Cas9 nucleases with improved specificity. Science (New York, N.Y.) 351, 84–88. doi: 10.1126/science.aad5227
Song, X., Liu, C., Wang, N., Huang, H., He, S., Gong, C., et al. (2021). Delivery of CRISPR/Cas systems for cancer gene therapy and immunotherapy. Adv. Drug Deliv. Rev. 168, 158–180. doi: 10.1016/j.addr.2020.04.010
Stadtmauer, E. A., Fraietta, J. A., Davis, M. M., Cohen, A. D., Weber, K. L., Lancaster, E., et al. (2020). CRISPR-engineered T cells in patients with refractory cancer. Science (New York, N.Y.) 367:eaba7365.
Sterner, R. M., Sakemura, R., Cox, M. J., Yang, N., Khadka, R. H., Forsman, C. L., et al. (2019). GM-CSF inhibition reduces cytokine release syndrome and neuroinflammation but enhances CAR-T cell function in xenografts. Blood 133, 697–709. doi: 10.1182/blood-2018-10-881722
Stevanovic, S., Draper, L. M., Langhan, M. M., Campbell, T. E., Kwong, M. L., Wunderlich, J. R., et al. (2015). Complete regression of metastatic cervical cancer after treatment with human papillomavirus-targeted tumor-infiltrating T cells. J. Clin. Oncol. 33, 1543–1550. doi: 10.1200/jco.2014.58.9093
Su, S., Hu, B., Shao, J., Shen, B., Du, J., Du, Y., et al. (2016). CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci. Rep. 6:20070.
Su, S., Zou, Z., Chen, F., Ding, N., Du, J., Shao, J., et al. (2017). CRISPR-Cas9-mediated disruption of PD-1 on human T cells for adoptive cellular therapies of EBV positive gastric cancer. Oncoimmunology 6:e1249558. doi: 10.1080/2162402x.2016.1249558
Suresh, S., Chen, B., Zhu, J., Golden, R. J., Lu, C., Evers, B. M., et al. (2020). eIF5B drives integrated stress response-dependent translation of PD-L1 in lung cancer. Nat. Cancer 1, 533–545. doi: 10.1038/s43018-020-0056-0
Tang, N., Cheng, C., Zhang, X., Qiao, M., Li, N., Mu, W., et al. (2020). TGF-(inhibition via CRISPR promotes the long-term efficacy of CAR T cells against solid tumors. JCI Insight 5:e133977.
Torikai, H., and Cooper, L. J. (2016). Translational implications for Off-the-shelf immune cells expressing chimeric antigen receptors. Mol. Ther. 24, 1178–1186. doi: 10.1038/mt.2016.106
Torikai, H., Reik, A., Liu, P. Q., Zhou, Y., Zhang, L., Maiti, S., et al. (2012). A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood 119, 5697–5705. doi: 10.1182/blood-2012-01-405365
Tran, E., Robbins, P. F., Lu, Y. C., Prickett, T. D., Gartner, J. J., Jia, L., et al. (2016). T-Cell transfer therapy targeting mutant KRAS in Cancer. N. Engl. J. Med. 375, 2255–2262.
Tran, E., Turcotte, S., Gros, A., Robbins, P. F., Lu, Y. C., Dudley, M. E., et al. (2014). Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science (New York, N.Y.) 344, 641–645. doi: 10.1126/science.1251102
Tu, K., Deng, H., Kong, L., Wang, Y., Yang, T., Hu, Q., et al. (2020). Reshaping tumor immune microenvironment through acidity-responsive nanoparticles featured with CRISPR/Cas9-Mediated programmed death-ligand 1 attenuation and chemotherapeutics-induced immunogenic cell death. ACS Appl. Mater. Interfaces 12, 16018–16030. doi: 10.1021/acsami.9b23084
Tuveson, D., and Clevers, H. (2019). Cancer modeling meets human organoid technology. Science (New York, N.Y.) 364, 952–955. doi: 10.1126/science.aaw6985
Tzelepis, K., Koike-Yusa, H., De Braekeleer, E., Li, Y., Metzakopian, E., Dovey, O. M., et al. (2016). A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 17, 1193–1205. doi: 10.1016/j.celrep.2016.09.079
Vredevoogd, D. W., Kuilman, T., Ligtenberg, M. A., Boshuizen, J., Stecker, K. E., de Bruijn, B., et al. (2019). Augmenting immunotherapy impact by lowering tumor TNF cytotoxicity threshold. Cell 178, 585–599.e15.
Wei, J., Long, L., Zheng, W., Dhungana, Y., Lim, S. A., Guy, C., et al. (2019). Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature 576, 471–476. doi: 10.1038/s41586-019-1821-z
Wei, L., Lee, D., Law, C. T., Zhang, M. S., Shen, J., Chin, D. W., et al. (2019). Genome-wide CRISPR/Cas9 library screening identified PHGDH as a critical driver for Sorafenib resistance in HCC. Nat. Commun. 10:4681.
Wherry, E. J., and Kurachi, M. (2015). Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15, 486–499. doi: 10.1038/nri3862
Xu, L., Wang, J., Liu, Y., Xie, L., Su, B., Mou, D., et al. (2019). CRISPR-Edited stem cells in a patient with HIV and acute lymphocytic leukemia. N. Engl. J. Med. 381, 1240–1247. doi: 10.1056/nejmoa1817426
Xue, S. A., Chen, Y., Voss, R. H., Kisan, V., Wang, B., Chen, K. K., et al. (2020). Enhancing the expression and function of an EBV-TCR on engineered T cells by combining Sc-TCR design with CRISPR editing to prevent mispairing. Cell Mol. Immunol. 17, 1275–1277. doi: 10.1038/s41423-020-0396-9
Ye, L., Park, J. J., Dong, M. B., Yang, Q., Chow, R. D., Peng, L., et al. (2019). In vivo CRISPR screening in CD8 T cells with AAV-Sleeping Beauty hybrid vectors identifies membrane targets for improving immunotherapy for glioblastoma. Nat. Biotechnol. 37, 1302–1313. doi: 10.1038/s41587-019-0246-4
Zacharakis, N., Chinnasamy, H., Black, M., Xu, H., Lu, Y. C., Zheng, Z., et al. (2018). Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat. Med. 24, 724–730. doi: 10.1038/s41591-018-0040-8
Zaretsky, J. M., Garcia-Diaz, A., Shin, D. S., Escuin-Ordinas, H., Hugo, W., Hu-Lieskovan, S., et al. (2016). Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 375, 819–829.
Zarour, H. M. (2016). Reversing T-cell dysfunction and exhaustion in Cancer. Clin. Cancer Res. 22, 1856–1864. doi: 10.1158/1078-0432.ccr-15-1849
Zhan, T., Rindtorff, N., Betge, J., Ebert, M. P., and Boutros, M. (2019). CRISPR/Cas9 for cancer research and therapy. Semin. Cancer Biol. 55, 106–119.
Zhang, C., Peng, Y., Hublitz, P., Zhang, H., and Dong, T. (2018). Genetic abrogation of immune checkpoints in antigen-specific cytotoxic T-lymphocyte as a potential alternative to blockade immunotherapy. Sci. Rep. 8: 5549.
Zhang, W., Shi, L., Zhao, Z., Du, P., Ye, X., Li, D., et al. (2019). Disruption of CTLA-4 expression on peripheral blood CD8 + T cell enhances anti-tumor efficacy in bladder cancer. Cancer Chemother. Pharmacol. 83, 911–920. doi: 10.1007/s00280-019-03800-x
Zhang, Y., Zhang, X., Cheng, C., Mu, W., Liu, X., Li, N., et al. (2017). CRISPR-Cas9 mediated LAG-3 disruption in CAR-T cells. Front. Med. 11:554–562. doi: 10.1007/s11684-017-0543-6
Zhao, L., Luo, Y., Huang, Q., Cao, Z., and Yang, X. (2020). Photo-Enhanced CRISPR/Cas9 system enables robust PD-L1 gene disruption in cancer cells and cancer stem-like cells for efficient cancer immunotherapy. Small 2: e2004879.
Keywords: CRISPR/Cas9, immunotherapy, TCR-T, tumor-infiltrating lymphocytes, CAR-T
Citation: Ou X, Ma Q, Yin W, Ma X and He Z (2021) CRISPR/Cas9 Gene-Editing in Cancer Immunotherapy: Promoting the Present Revolution in Cancer Therapy and Exploring More. Front. Cell Dev. Biol. 9:674467. doi: 10.3389/fcell.2021.674467
Received: 01 March 2021; Accepted: 16 April 2021;
Published: 20 May 2021.
Edited by:
Dali Li, East China Normal University, ChinaReviewed by:
Sabarish Ramachandran, Texas Tech University Health Sciences Center, United StatesYu Wang, Shenzhen University, China
Copyright © 2021 Ou, Ma, Yin, Ma and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuelei Ma, ZHJtYXh1ZWxlaUBnbWFpbC5jb20=; Zhiyao He, aGV5YW9kZUAxNjMuY29t; emhpeWFvaGVAc2N1LmVkdS5jbg==
†These authors have contributed equally to this work
 Xuejin Ou
Xuejin Ou Qizhi Ma
Qizhi Ma Wei Yin
Wei Yin Xuelei Ma
Xuelei Ma Zhiyao He
Zhiyao He