- 1Shaanxi Key Laboratory of Ischemic Cardiovascular Disease, and Brain Disorders, Institute of Basic and Translational Medicine, Xi’an Medical University, Xi’an, China
- 2College of Medical Technology, Xi’an Medical University, Xi’an, China
Bladder cancer (BC) is the most common malignant tumor in the urinary system, and its early diagnosis is conducive to improving clinical prognosis and prolonging overall survival time. However, few biomarkers with high sensitivity and specificity are used as diagnostic markers for BC. Multiple long non-coding RNAs (lncRNAs) are abnormally expressed in BC, and play key roles in tumorigenesis, progression and prognosis of BC. In this review, we summarize the expression, function, molecular mechanisms and the clinical significance of lncRNAs on bladder cancer. There are more than 100 dysregulated lncRNAs in BC, which are involved in the regulation of proliferation, cell cycle, apoptosis, migration, invasion, metabolism and drug resistance of BC. Meanwhile, the molecular mechanisms of lncRNAs in BC was explored, including lncRNAs interacting with DNA, RNA and proteins. Additionally, the abnormal expression of thirty-six lncRNAs is closely associated with multiple clinical characteristics of BC, including tumor size, metastasis, invasion, and drug sensitivity or resistance of BC. Furthermore, we summarize some potential diagnostic and prognostic biomarkers of lncRNA for BC. This review provides promising novel biomarkers in early diagnosis, prognosis and monitoring of BC based on lncRNAs.
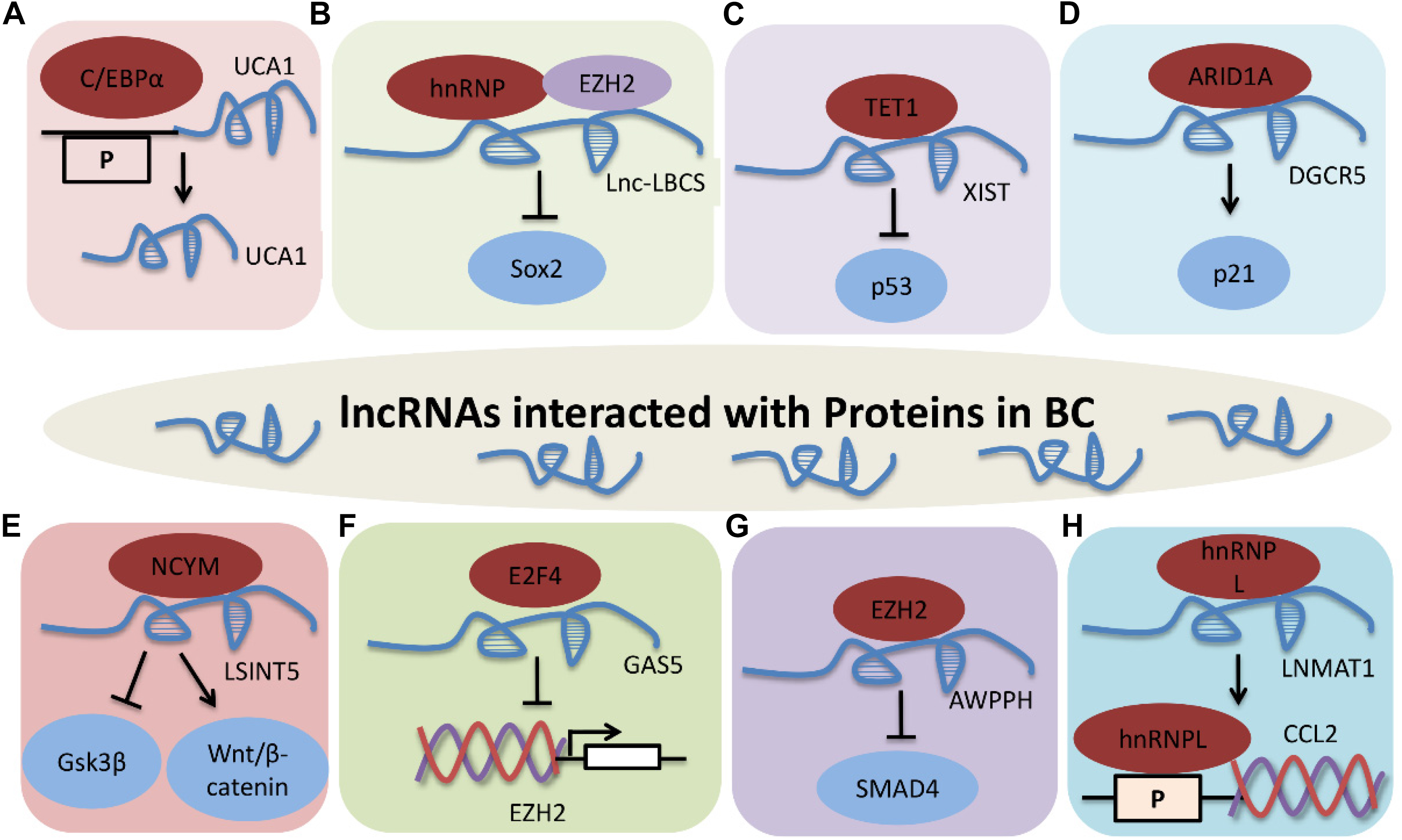
Graphical Abstract. LncRNAs interacted with proteins in BC. (A) UCA1 interacts with C/EBPα. (B) Lnc-LBCS directly binds to hnRNPK and EZH2, and inhibits SOX2 transcription. (C) XIST-TET1-p53 pathway in BC. (D) DGCR5 promotes P21 transcription via interacting with ARID1A. (E) LSINCT5 promotes tumor progression by interacting with NCYM, and inhibiting GSK3β activity and promoting Wnt/β-catenin signaling activation. (F) GAS5 inhibits EZH2 transcription by interacting with E2F4. (G) AWPPH promotes cell proliferation, autophagy, and migration through binding to SMAD4 via EZH2. P indicates promoter. (H) LNMAT1 regulates CCL2 expression through interaction with hnRNPL. Protein-RNA interactions are important aspects of many cellular functions, and lncRNAs are involved in modulating BC through different molecular mechanisms, including binding to one or more protein partners.
Highlights
- The abnormal expression of 100 lncRNAs is involved in regulating of proliferation, cell cycle, apoptosis, migration, invasion, metabolism and drug resistance of bladder cancer.
- The abnormal expression of thirty-six lncRNAs is closely related to multiple clinical parameters, including tumor size, metastasis, invasion, tumor lymph node metastasis stage, diagnosis and prognosis of bladder cancer.
- LncRNAs regulate bladder cancer by interacting with DNA, RNA and proteins.
- This review will provide novel strategies and biomarkers for diagnosis, prognosis and monitoring of bladder cancer base on lncRNAs.
Introduction
Bladder cancer (BC) is one of the most common malignant tumors in the urinary system, and its incidence is increasing. It is estimated that 81,400 new cases and 17,980 deaths of BC in the United States in 2020 (Siegel et al., 2020). Clinically, 75–80% of BC patients exhibit moderately differentiated non-muscle-invasive bladder cancer (NMIBC) at the initial diagnosis, and the remaining cases are muscle-invasive bladder cancer (MIBC). The standard treatment of NMIBC includes transurethral resection of bladder tumor (TUR-Bt), but high recurrence rate of BC after TUR-Bt and a subsequent tumor progression. The 5-year survival rate of patients with NMIBC is approximately 90%, while the 5-year survival rate of MIBC patients is only 20–40%, due to rapid disease progression and some early symptoms.
Long non-coding RNA (lncRNA) is defined as non-protein-coding RNA molecule longer than 200 nucleotides (Iyer et al., 2015). LncRNA plays an important regulatory role in epigenetic modification, transcription and post-transcriptional processing. Recent findings highlight that lncRNA contributes to the regulation of multiple signaling pathways in BC. LncRNA urethral cancer associated 1 (UCA1) was cloned from two human bladder transitional cell carcinoma cell lines. UCA1 is highly expressed in BC, and promotes progression, glycolysis, glutamine metabolism, migration and invasion of BC through the UCA1-mTOR/STAT3/miR-143/HK2 and UCA1/miR-16/GLS2 signaling network (Li et al., 2014, 2015). Increasing evidence suggests that lncRNA exert their roles during the biological processes of tumorigenesis, tumor proliferation, differentiation, apoptosis, invasion, migration and stemness (Zhang et al., 2013; Nicolas, 2017; Zhang and Tang, 2018).
In this review, we focused on the expression, function and related genes or signaling pathways of lncRNAs, as well as the molecular mechanisms of lncRNAs and their clinical significance for diagnosis and prognosis in BC, which can provide new strategies and novel biomarkers for the diagnosis and treatment of BC based on lncRNAs.
Importance of Novel Biomarkers for BC
At present, diagnostic tests for BC include cystoscopy, urine cytology, FDA-approved markers, and non-FDA-approved markers. Four urinary FDA-approved commercial urinary biomarker tests are as follows: (1) bladder tumor antigen stat (BTA stat) and BTA TRAK; (2) Nuclear Matrix Protein 22 (NMP22) BC Test and NMP22 BladderChek; (3) uCyt+ assay; and (4) UroVysion assay. Several new biomarkers at the genomic, transcriptomic, and epigenetic levels have been developed, including: (1) ADXBLADDER, a novel ELISA test for Mini-chromosome maintenance 5; (2) Cxbladder Monitor; (3) XPERT BC, measure mRNA levels of ABL1, CRH, IGF2, UPK 1B, and ANXA 10; (4) UroMark assay; and (5) AssureMDX, test of the DNA methylation of OTX1, ONECUT2, and TWIST1 and somatic statuses of FGFR3, TERT, and HRAS in urine (Dangle et al., 2009; Tabayoyong and Kamat, 2018). Due to their poor sensitivity and specificity, cystoscopy and urine cytology are still used as the gold standard for the diagnosis of BC. Cystoscopy is the primary method for the diagnosis and monitoring of BC, and is not only invasive, entailing complications such as urinary tract infection or hematuria, but is also expensive (Casadevall et al., 2017). The urine cytology test is used to detect exfoliated malignant cells under the microscope, and is a non-invasive examination. It has high sensitivity and specificity for detection of high-grade BC; however, its sensitivity is only 4–31% for low-grade malignant tumor. In addition, false positive rate is as high as 12% due to inflammation and urothelial heterogeneity (Burchardt et al., 2000).
Early diagnosis of BC is essential for improvement of the prognosis and overall survival of patients. Given the current limited diagnostic methods, it is important to develop novel specific biomarkers as non-invasive tools that will facilitate accurate diagnosis and highly personalized treatment.
The Definition, Features, and Classification of lncRNAs
Of the 3 billion base pairs in the human genome, only 1.5% are protein-coding sequences, and the rest are non-protein-coding sequences. There are many types of ncRNA, including short-stranded ncRNA and lncRNA. A greater proportion of lncRNA encoded by the human genome has tissue or cell type specificity, and is even related to specific stages of biological development. The definition of lncRNA was first proposed by Okazaki Y in 2002, mainly due to their discovery of a large number of lncRNA transcripts in large-scale sequencing studies of mouse cDNA libraries (Okazaki et al., 2002). LncRNA is defined as RNA longer than 200 nucleotides that not translated into proteins.
Anatomy of the global features of lncRNA suggests that lncRNA has some typical features, as follows: (1) lncRNA is usually larger than 200 nucleotides, (2) most of them have the eukaryotic structure of mRNA, the structure of promoter and polyA tail after RNA splicing, (3) contains few and long exons, (4) low expression, (5) less evolutionally conserved.
LncRNA has a variety of classifications according to its different characteristics. Currently, lncRNAs are mainly classified relying on four characteristics, including the location within the genome, the effect on the DNA sequence, the mechanisms of function, and targeting mechanisms of lncRNAs. According to the position of lncRNA in the genome and protein-coding genes, it is usually classified into five categories: sense lncRNA, antisense lncRNA, bidirectional lncRNA, intragenic lncRNA, and intergenic lncRNA. Based on the effect of lncRNA on DNA sequence, it can be divided into cis-lncRNAs and trans-lncRNAs. According to function mechanisms of lncRNA, it is fall into transcriptional regulation, post-transcriptional regulation, and translational regulation. Depending on the targeting mechanism of lncRNAs, it can be divided into signal, decoy, guide and scaffold (Ma et al., 2013).
Expression and Role of lncRNAs in BC
To date, 120,353 human annotated lncRNAs have been collected by the LNCipedia database, and more than 70,000 published human and mouse lncRNAs have been annotated by NON-CODE (Zhang et al., 2013). In general, lncRNAs show more tissue-specific patterns and higher expression variability than protein-coding genes in cell lines and tissues. A large fraction of lncRNAs preferentially localize in the nucleus, some are located in the cytoplasm, and a few are found both in the cytoplasm and the nucleus (Ponting et al., 2009). Nuclear lncRNAs are involved in diverse functions, such as histone modifications, chromatin remodeling, modulation of gene expression in cis or trans. Cytoplasmic lncRNAs can perform cytoplasmic functions, including post-transcriptional modification of mRNAs, interaction with proteins and miRNAs sponging.
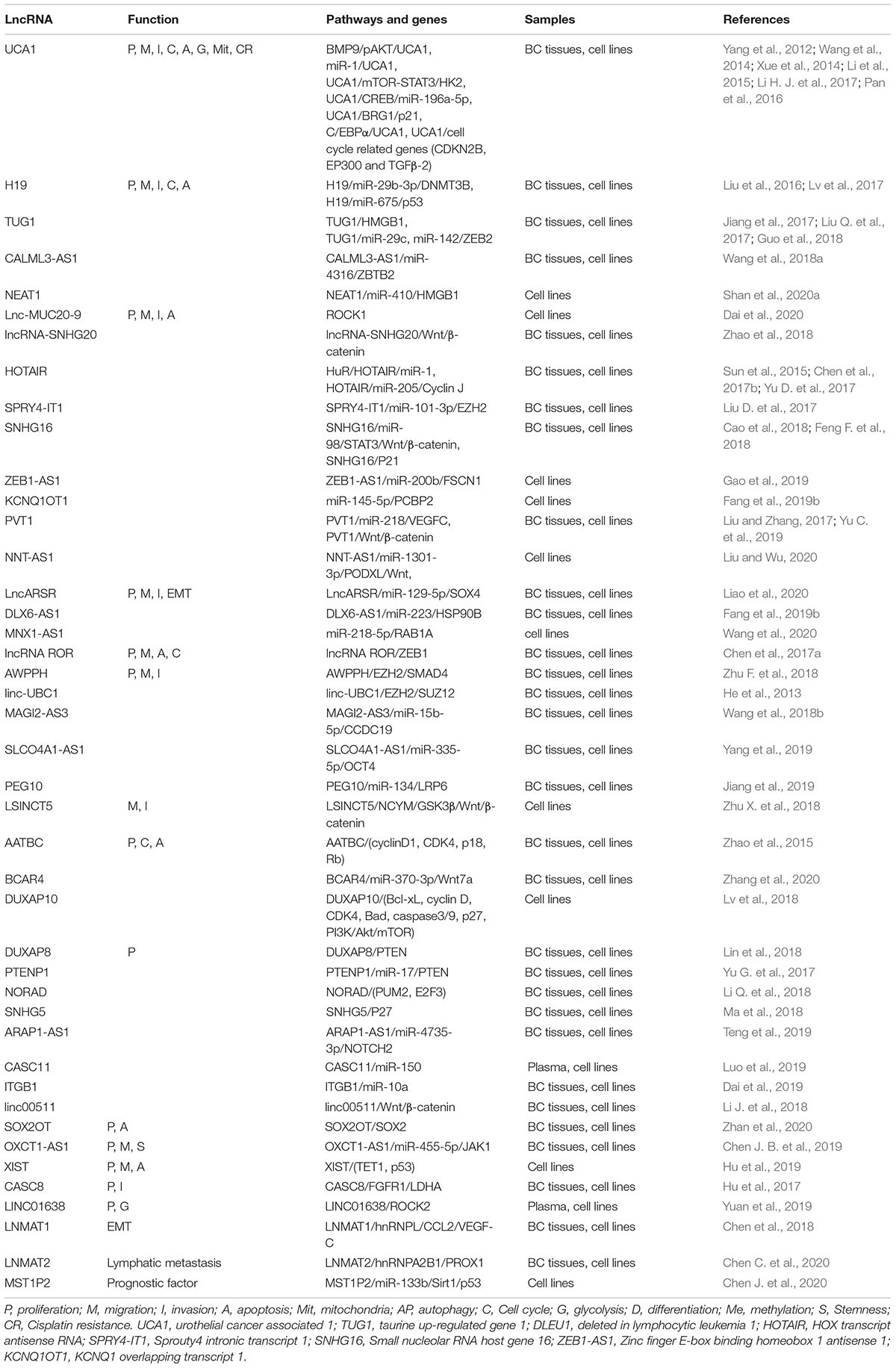
Upregulated lncRNAs in BC
There are more than 100 dysregulated lncRNAs engaged in regulation of various biological functions of BC, such as cell proliferation, apoptosis, and metastasis. As shown in Table 1, forty-three upregulated lncRNAs involved in proliferation, migration, invasion, and cell cycle of BC, such as UCA1, long non-coding RNA 19 (H19), taurine up-regulated gene 1 (TUG1), and Calmodulin Like 3 Antisense RNA 1 (CALML3-AS1). Cytoplasmic lncRNAs can serve as oncogenes. UCA1, which is overexpressed in bladder cancer, specifically induces Glutaminase 2 (GLS2) by sponging miR-16, and can also activate AKT by recruiting E1A Binding Protein P300 (EP300) and lead to bladder cancer cell growth. UCA1 has a further oncogenic function by enhancing mTOR/STAT3/HK2 signaling pathway, which promotes Warburg effect of bladder cancer (Li et al., 2014). Additionally, UCA1 is modulated by upstream molecules, such as bone morphogenetic protein 9 (BMP9), which then promotes tumor progression of bladder cancer.
LncRNAs can also be oncogene by regulating signaling molecules in the nucleus. Linc-UBC1 is upregulated and preferentially localizes in nucleus, which aggravates BC progression through altering polycomb repressive complex 2 (PRC2) complex localization by interacting with enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) and SUZ12 polycomb repressive complex 2 subunit (SUZ12) (He et al., 2016). Similar to lncRNA upregulated in bladder cancer 1 (linc-UBC1), associated with poor prognosis of hepatocellular carcinoma (AWPPH) and non-coding RNA activated by DNA damage (NORAD) play regulatory roles in nucleus of bladder cancer cells.
A few lncRNAs can act as both cytoplasmic lncRNA and nuclear lncRNA. H19 promoter region can bind to CCCTC-binding factor (CTCF) and insulin like growth factor 2 (IGF2), and thereby regulating bladder cancer development in nucleus of BC cells (Takai et al., 2001). It was also reported that H19 promotes epithelial-mesenchymal transition and metastasis of bladder cancer via H19/miR-29b/DNMT3B signaling pathway in cytoplasm (Lv et al., 2017).
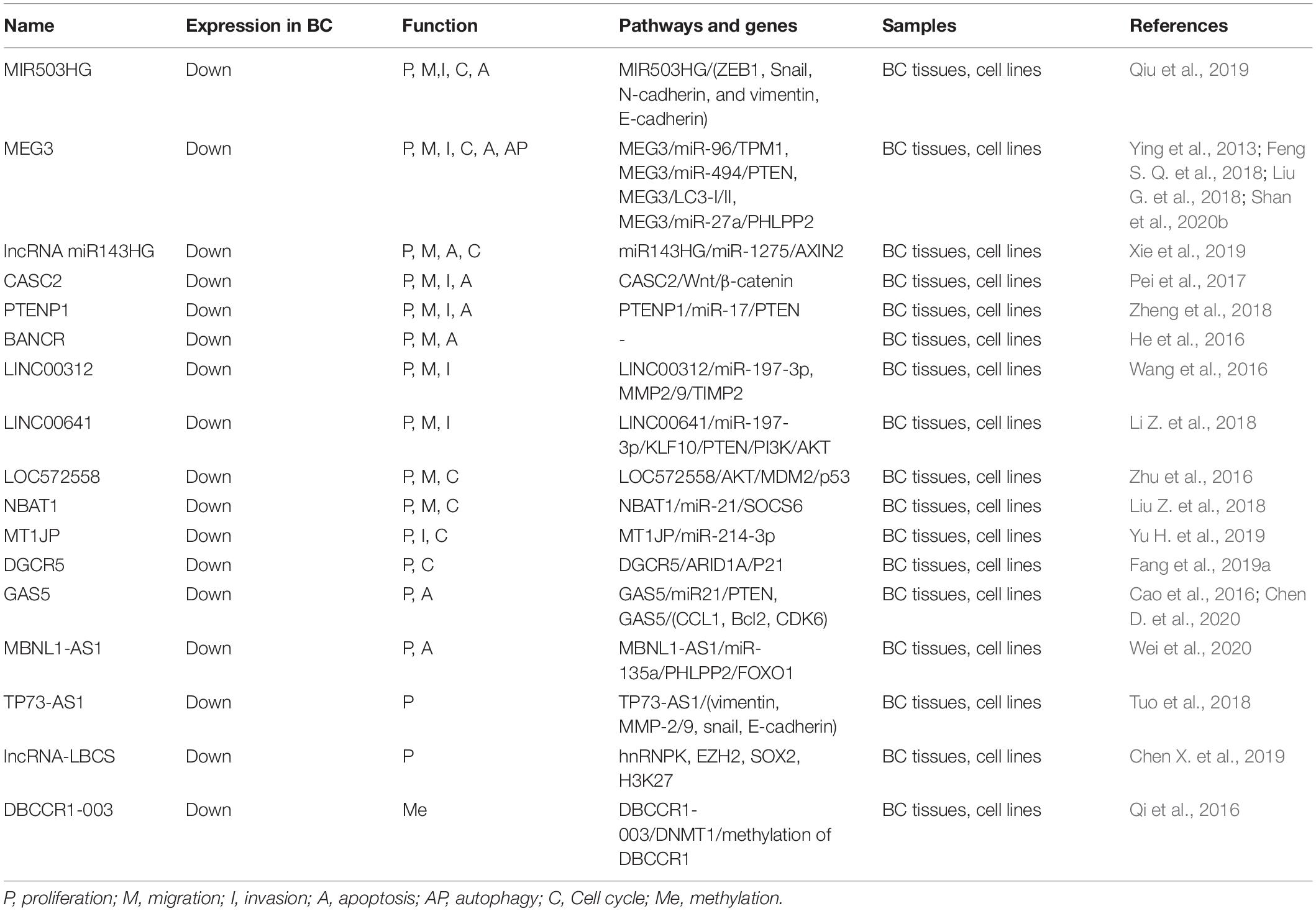
Downregulated lncRNAs in BC
As shown in Table 2, seventeen downregulated lncRNAs involved in regulation of BC. Among them, MIR503 host gene (MIR503HG), LncRNA maternally expressed gene 3 (MEG3), lncRNA miR143 host gene (MIR5143HG), lncRNA cancer susceptibility candidate 2 (CASC2) (Pei et al., 2017), phosphatase and tensin homolog pseudogene 1 (PTENP1) (Zheng et al., 2018) and BRAF-activated non-coding RNA (BANCR) (He et al., 2016) are involved in regulating cell proliferation, migration and apoptosis. In addition, MIR503HG is engaged in the regulation of cell invasion and cycle, MEG3 is involved in regulating cell invasion, cycle and autophagy, lncRNA miR143HG regulates cell cycle, and CASC2 and PTENP1 medicate cell invasion. LINC00312 and LINC00641 are participated in regulating cell proliferation, migration and invasion (Li Z. et al., 2018). LOC572558 and NBAT1 are involved in modulating proliferation, migration and cell cycle (Zhu et al., 2016; Liu and Zhang, 2017; Liu Z. et al., 2018). The nuclear lncRNA deleted in bladder cancer chromosome region 1-003 (DBCCR1-003) can suppress DBCCR1 transcription by methylation of promoter region of DBCCR1 in T24 cells. The cytoplasmic lncRNA MEG3 inhibits bladder cancer progression by regulating multiple signaling axis, such as miR-96/TPM1, miR-27a/PHLPP2 and miR-494/PTEN.
Functional Mechanisms of lncRNAs on BC
Gene expression is modulated by lncRNAs at various levels. The mechanistic characterization of lncRNAs on bladder cancer is not well documented. It is partly due to the fact that lncRNAs harbor more cell type specificity and tissue type specificity, and generally low expression level. In spite of lncRNAs are less conversed among different species, there are similarities in functional mechanisms in bladder cancer. In this section, we highlight uncover molecular mechanisms from the following three aspects: lncRNAs by interacting with DNA, RNA and proteins.
LncRNAs Affect BC by Interacting With DNA
LncRNAs may anchor to promoters or regulators of target DNA to regulate gene expression in BC (Figure 1). LncRNA DBCCR1-003 as a tumor suppressor, derived from the DBCCR1 locus. DBCCR1-003 modulates the expression of DBCCR1 via DNA methylation in bladder cancer tissues and T24 cell lines. DBCCR1-003 can bind to DNA methyltransferase 1 (DNMT1), but the expression of DNMT1 remains unchanged when DBCCR1-003 is overexpressed in T24 cells. The results reveal that lncRNA DBCCR1-003 can modulate the expression of DBCCR1 by binding to DNMT1 and inhibiting DNMT1-mediated methylation of DBCCR1 in BC (Qi et al., 2016).
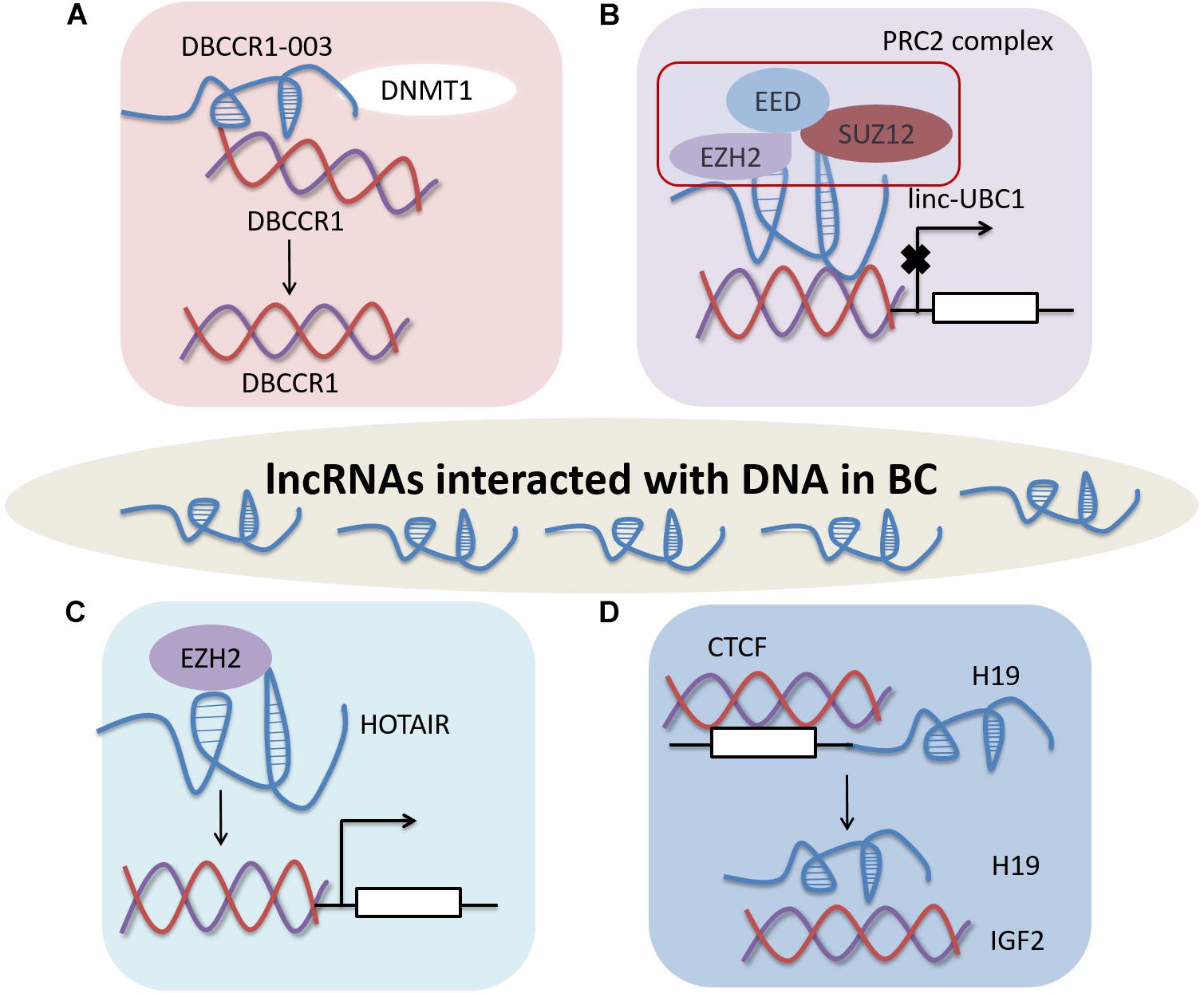
Figure 1. LncRNAs affect BC by interacting with DNA. (A) DBCCR1-003 regulates the expression of DBCCR1 by binding to DNMT1 and inhibiting DNMT1-mediated methylation of DBCCR1. (B) Linc-UBC1 modulating the PRC2 complex by combining with EZH2 and SUZ12. (C) HOTAIR modulates gene expression by binding to EZH2. (D) CTCF regulates expression of H19 by methylating the H19 promoter in human BC.
Linc-UBC1 can physically bind to EZH2 and SUZ12, key component of PRC2 complex. Some PRC2 target genes, such as Cyclin D2 (CCND2), plasminogen activator inhibitor type 2 (SERPINB2), Bone morphogenetic protein 2 (BMP2), Kruppel-like factor 4 (KLF4) and Homeobox A5 (HOXA5), are downregulated after knockdown of linc-UBC1. Moreover, H3K27 trimethylation in the promoter regions of CCND2, SERPINB2, the expression levels of BMP2 and HOXA5 decreased with the low expression of linc-UBC1. These results indicate that linc-UBC1 may partly modulate PRC2 complex localization by regulating histone H3K27 trimethylation with EZH2 and SUZ12 (He et al., 2013).
LncRNA HOX transcript antisense intergenic RNA (HOTAIR) modulates gene expression by binding to enzymes, such as EZH2, to inhibit histone modifications. HOTAIR mostly depends on EZH2, which is significantly increased in BC. HOTAIR originates from the HOX locus, which regulates HOX gene expression, differentiation and tissue homeostasis (Heubach et al., 2015). Additionally, the expression of HOTAIR is regulated by EZH2 after reduction of EZH2 in 5637 cells (Mónica et al., 2015).
Human H19 promoter contains large amount of methylcytosine in the CTCF binding site of and is abnormal hypomethylated in human bladder cancer. The sixth CTCF-binding site may serve as a significant regulatory domain, thereby switching H19 or IGF2 expression in human bladder cancer (Takai et al., 2001). Up to now, only a small number of lncRNAs has been demonstrated to participate in the regulation of BC through interacting with DNA.
LncRNAs Affect BC Through Interacting With RNA
Accumulating data indicate that both coding and non-coding RNAs can regulate one another by competing miRNA binding, known as competing endogenous RNAs (ceRNAs) (Salmena et al., 2011). ceRNAs can decoy miRNAs and protect their target mRNAs from degradation (Kartha and Subramanian, 2014). According to the ceRNA hypothesis, lncRNAs may exert their biological effects in BC by sponging miRNAs, which would in turn modulate the effect of miRNAs on their targets (Figure 2).
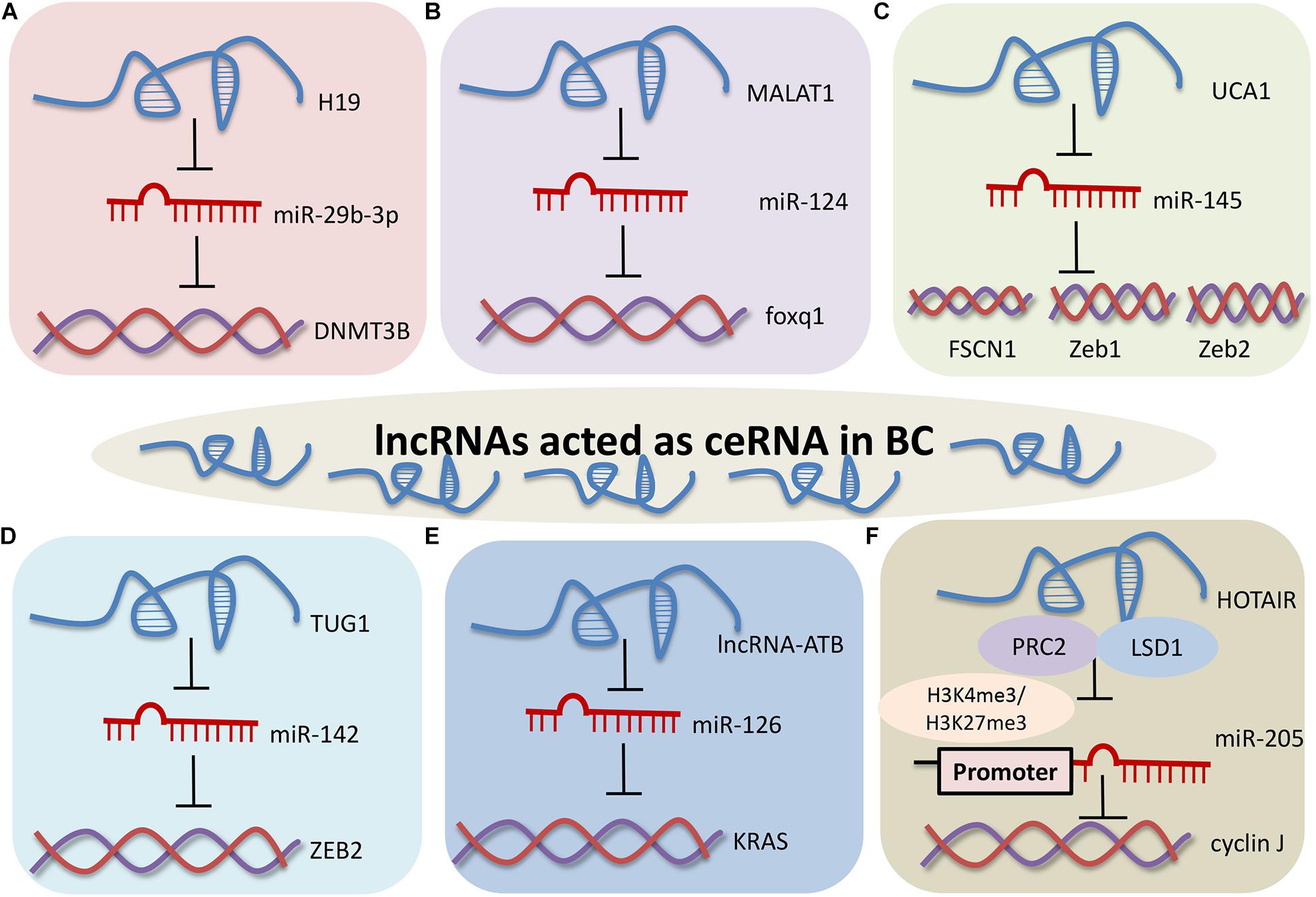
Figure 2. LncRNAs exert their biological effects by sponging miRNAs in BC. (A) H19 regulated EMT and metastasis of BC through the H19/miR-29b-3p/DNMT3B axis. (B) MALAT1 promoted development of BC through the MALAT1/miR-124/foxq1 signaling pathway. (C) UCA1 enhanced BC migration and invasion through the UCA1-miR-145/FSCN1/Zeb1/Zeb2 network. (D) TUG1 enhanced proliferation of BC through the TUG1/miR-142/ZEB2 axis. (E) LncRNA-ATB positively regulated BC through the lncRNA-ATB-miR-126/KRAS. (F) The HOTAIR/miR-205/cyclin J signaling pathway in BC.
H19 regulates the expression level of DNMT3B protein and epithelial-mesenchymal transition in bladder cancer. H19 can directly combine with the seed region of miR-29b-3p to sequester H19 by dual luciferase reporter assay, FISH and RIP assay. The expression of DNMT3B is repressed by miR-29b-3p in T24 cells. Collectively, H19 regulates the epithelial-mesenchymal transition and metastasis of BC through the H19/miR-29b-3p/DNMT3B axis (Lv et al., 2017). There is significant negative correlation between metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and miR-124 in bladder transitional cell carcinoma tissues and normal mucosa samples. Foxq1, putative target gene of miR-124, is positively correlated with the expression of MALAT1 in bladder transitional cell carcinoma. Upregulation of MALAT1 promotes proliferation, migration, and invasion of bladder transitional cell carcinoma through the MALAT1/miR-124/foxq1 signaling pathway (Jiao et al., 2018). The exons 2 and 3 of UCA1 harbor miR-145 binding site and can suppress the expression of miR-145. ZEB1/2 and FSCN1 are engaged in migration and invasion of bladder cancer, and are the direct target genes of miR-145. These data suggest that UCA1 enhances BC migration and invasion through the miR-145-ZEB1/2-FSCN1 network (Xue et al., 2016). TUG1 and ZEB2 act as oncogene in bladder cancer. TUG1 contains the binding sites of miR-142, and TUG1 inhibits miR-142 expression in T24 and BIU-87 cells. These results imply that TUG1 enhances the proliferation and inhibits apoptosis of BC cells via the TUG1/miR-142/ZEB2 axis (Liu Q. et al., 2017). LncRNA-activated by transforming growth factor-β (lncRNA-ATB) can promote cell proliferation, migration and invasion by capturing miR-126 in T24 cells. KRAS is a direct target gene of miR-126 and is inversed regulated by miR-126. These results indicate that lncRNA-ATB promotes cell proliferation, invasion and migration by decoying miR-126 in bladder cancer (Zhai and Xu, 2018).
Unlike sponge miRNAs, HOTAIR can repress miR-205 transcription by recruiting PRC2 and lysine-specific demethylase 1 (LSD1), and then recruiting H3K4me3 and H3K27me3 in miR-205 promoter region in HCV29 cells. Cyclin J is a putative downstream target gene of miR-205 in bladder cancer. These findings suggest that HOTAIR participates in the regulation of proliferation, migration and invasion of bladder tumor cells through the HOTAIR/miR-205/cyclin J signaling pathway (Sun et al., 2015).
LncRNAs Affect BC by Interacting With Proteins
Protein-RNA interactions are important aspects of many cellular functions, and lncRNAs are involved in modulating BC through different molecular mechanisms, including binding to one or more protein partners (Figure 3). UCA1 is upregulated by C/EBPα through the combination of core promoter region of UCA1 with C/EBPα in BLZ-211, 5637 and T24 cells. C/EBPα enhances UCA1 transcription was detected by qRT-PCR and luciferase activity. These results indicate that UCA1 increases cell viability and inhibits cell apoptosis through the UCA1 core promoter interacting with C/EBPα (Xue et al., 2014).
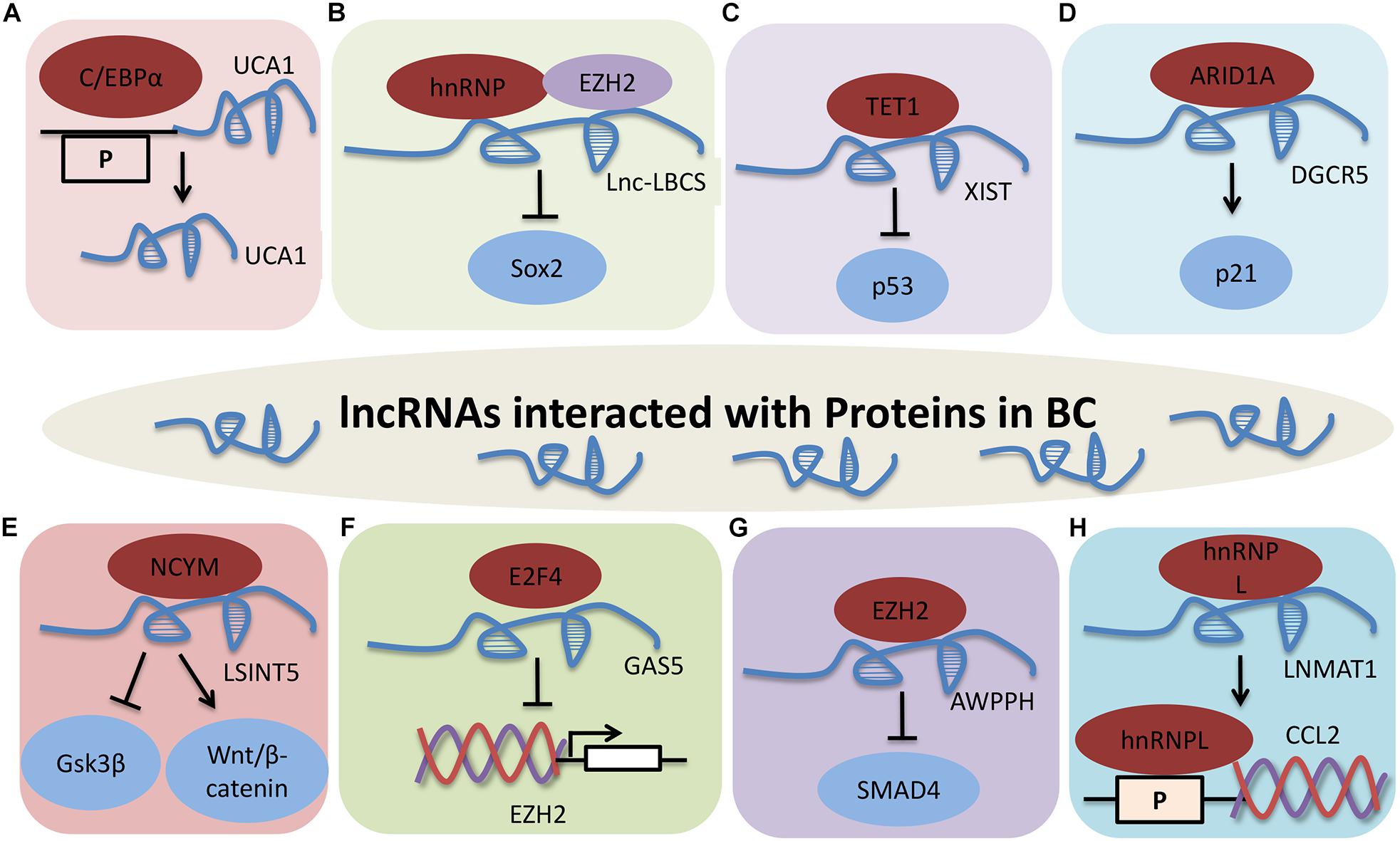
Figure 3. LncRNAs that interact with proteins in BC. (A) UCA1 interacts with C/EBPα. (B) Lnc-LBCS directly binds to hnRNPK and EZH2, and inhibits SOX2 transcription. (C) XIST-TET1-p53 pathway in BC. (D) DGCR5 promotes P21 transcription by interacting with ARID1A. (E) LSINCT5 promotes tumor progression by interacting with NCYM, and inhibiting GSK3β activity and promoting Wnt/β-catenin signaling activation. (F) GAS5 inhibits EZH2 transcription by interacting with E2F4. (G) AWPPH promotes cell proliferation, autophagy, and migration through binding to SMAD4 via EZH2. P indicates the promoter. (H) LNMAT1 regulates CCL2 expression through interacting with hnRNPL.
LncRNA low expressed in Bladder Cancer Stem cells (Lnc-LBCS) is markedly downregulated in bladder cancer tissues, and inhibits the self-renewal, tumor initiation, and tumor sphere formation of BC. Lnc-LBCS can act as a scaffold to activate the formation of hnRNPK-EZH2 complex by directly binding to heterogeneous nuclear ribonucleoprotein K (hnRNPK) and EZH2. Lnc-LBCS medicates SOX2 expression by forming hnRNPK-EZH2-SOX2 complex triplexes. Collectively, lnc-LBCS directly binds to hnRNPK and EZH2 and acts as a scaffold to induce complex formation to inhibit SOX2 transcription by regulating H3K27me3 (Chen X. et al., 2019).
LncRNA X-inactive specific transcript (XIST) facilitates cell growth and supresses cell apoptosis by reducing p53 expression in T24 and 5637 cells. XIST can bind to ten-eleven translocation 1 (TET1), and TET1 can enhance p53 expression by combing with the promoter region of p53. These data show that XIST regulates cell proliferation, migration and apoptosis via the XIST/TET1/p53 pathway in BC (Hu et al., 2019).
LncRNA DiGeorge syndrome critical region gene 5 (DGCR5) is significantly downregulated in bladder cancer. Overexpression of DGCR5 represses cell proliferation, cell cycle progression, migration, invasion, and EMT while promotes cell apoptosis of bladder cancer. DGCR5 is require for AT-rich interaction domain 1A (ARID1A)-induced P21 expression in T24 and SW780 cells. This study demonstrate that DGCR5 promotes P21 transcription by interacting with ARID1A in BC (Fang et al., 2019a).
Long stress-induced non-coding transcript 5 (LSINCT5) directly interacts with NCYM, and inhibits GSK3β activity, leading to Wnt/β-catenin signaling activation and promoting EMT in bladder cancer cell lines (Zhu X. et al., 2018). High GAS5 expression is correlated with poor prognosis in BC, and GAS5 effectively inhibits EZH2 transcription by binding and recruiting E2F4 to EZH2 promoter region, to promote BC cell apoptosis (Wang et al., 2018c). LncRNA AWPPH is highly expressed in BC and induces cell proliferation, autophagy, and migration, and restrains cell apoptosis by suppressing SMAD4 and EZH2 (Zhu F. et al., 2018).
Long non-coding RNA lymph node metastasis associated transcript 1 (LNMAT1) promotes bladder cancer cells to lymph node and lymphangiogenesis in vivo. Chemokine (C-C motif) ligand 2 (CCL2) is necessary for LNMAT1-mediated lymph node metastasis, and is regulated by the LNMAT1-CCL2 promoter through directly interacting with hnRNPL in UM-UC-3 cells. Taken together, LNMAT1 regulates CCL2 expression through physically interaction with hnRNPL in BC (Chen et al., 2018).
LncRNAs Could Be Promising Diagnostic or Prognostic Biomarker in BC
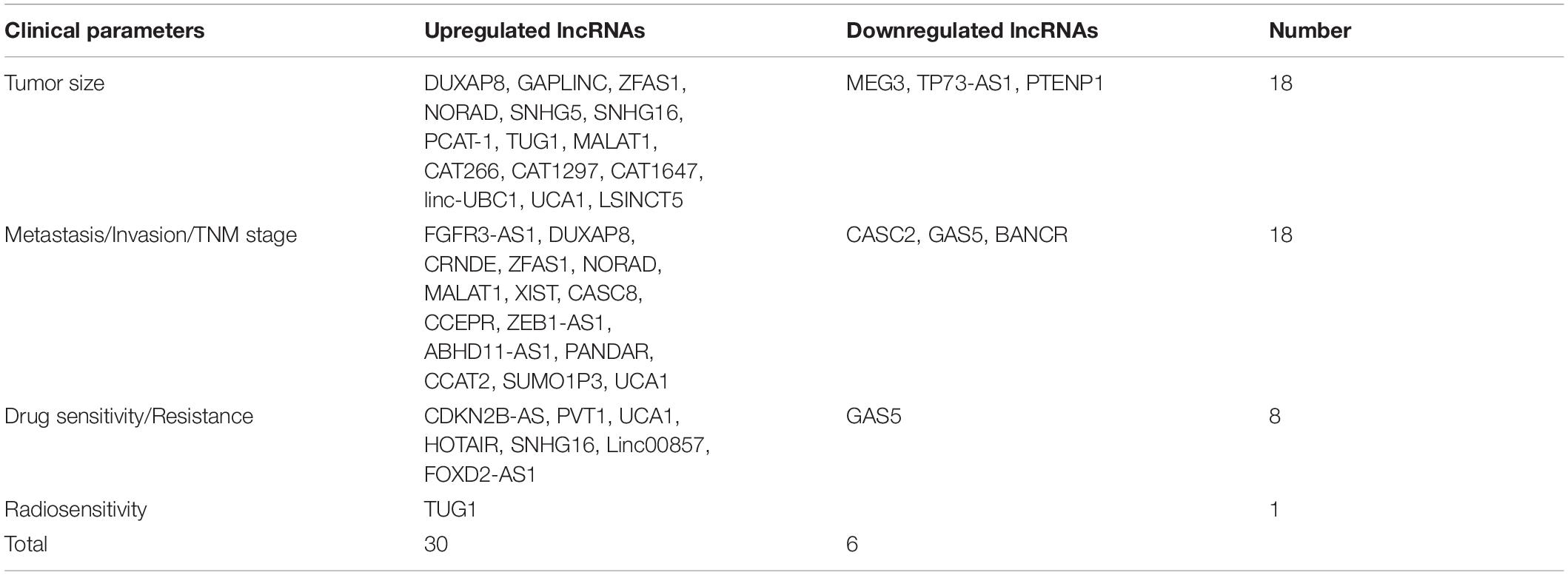
In recent years, accumulating evidence has demonstrated that abnormal expression of lncRNAs has crucial clinical significance for the diagnosis of BC, and is closely related to multiple clinical characteristics of BC, including tumor size, metastasis, invasion, tumor node metastasis (TNM) stage, diagnosis and prognosis (Table 3).
As shown in Table 3, eighteen lncRNAs are related to tumor size, including double homeobox A pseudogene 8 (DUXAP8), gastric adenocarcinoma predictive long intergenic non-coding RNA (GAPLINC), zinc finger antisense 1 (ZFAS1), P73 antisense RNA 1?T (TP73-AS1), non-coding RNA-activated by DNA damage (NORAD), small nucleolar RNA host gene 5 (SNHG5), small nucleolar RNA host gene 16 (SNHG16), prostate cancer associated transcript-1 (PCAT-1), TUG1, MALAT1, CAT266, CAT1297, CAT1647, linc-UBC1, UCA1, MEG-3, LSINCT5, and PTENP1. 18 lncRNAs are implicated in metastasis, invasion and TNM stage, including FGFR3 antisense transcript 1 (FGFR3-AS1), DUXAP8, lncRNA colorectal neoplasia differentially expressed (lncRNA CRNDE), ZFAS1, NORAD, MALAT1, XIST, cancer susceptibility candidate 8 (CASC8), cervical carcinoma expressed PCNA regulatory lncRNA (CCEPR), zinc finger E-box binding homeobox 1 antisense 1 (ZEB1-AS1), CASC2, ABHD11 antisense RNA 1 (ABHD11-AS1), growth arrest specific transcript 5 (GAS5), BANCR, p21-associated ncRNA DNA damage-activated (PANDA), CCAT2, small ubiquitin-like modifier 1 pseudogene 3 (SUMO1P3) and UCA1. There are 8 lncRNAs associated with drug sensitivity/resistance, including CDKN2B antisense RNA 1 (CDKN2B-AS), lncRNA plasmacytoma variant translocation 1 (lncRNA PVT1), GAS5, UCA1, HOTAIR, SNHG16, Linc00857, and forkhead box D2 antisense 1 (FOXD2-AS1). Only lncRNA TUG1 contributes to radiosensitivity in bladder cancer.
In view of low expression, cell-specificity and tissue-specificity, lncRNAs represent favorable features to act as diagnostic or prognostic indicators for BC. Considering the low expression of lncRNA, the upregulated lncRNAs are more conducive to diagnosis for bladder cancer. Therefore, we selected potential clinical biomarkers among 30 up-regulated lncRNAs.
There are fifteen upregulated lncRNAs that contribute to tumor size of bladder cancer, including DUXAP8, GAPLINC, ZFAS1, NORAD, SNHG5, SNHG16, PCAT-1, TUG1, MALAT1, CAT266, CAT1297, CAT1647, linc-UBC1, UCA1 and LSINCT5. As shown in Supplementary Table 1, only three upregulated lncRNAs of MALAT1, SNHG16 and UCA1 have complete diagnostic data. The sensitivity of MALAT1, SNHG16 and UCA1 is 0.567, 0.642, 0.83; the specificity of MALAT1, SNHG16 and UCA1 is 0.675, 0.650, 0.86; the ROC curve AUC of MALAT1, SNHG16 and UCA1 used for bladder cancer is 0.635, 0.679, 0.86 (Duan et al., 2016; Ding et al., 2021). Taken together, UCA1 has relatively high sensitivity, specificity and AUC, and it can be regarded as the most potential diagnostic biomarker for bladder cancer.
There are fifteen upregulated lncRNAs significantly associated with over survival of BC patients, including FGFR3-AS1, DUXAP8, CRNDE, ZFAS1, NORAD, MALAT1, XIST, CASC8, CCEPR, ZEB1-AS1, ABHD11-AS1, PANDAR, CCAT2, SUMO1P3, and UCA1. As reported by Su et al. (2018) only MALAT1 is an independent prognostic biomarker for bladder cancer, suggesting that MALAT1 can be considered as a possible prognostic indicator of BC.
As described above, it should be considered that lncRNAs can serve as feasible diagnostic and prognostic biomarkers. However, new lncRNA biomarkers used for clinical diagnosis must previously be carried out extensive studies in cells and clinical specimen. A single lncRNA may not be a reliable indicator for accurate and timely detection of bladder cancer. In this regard, additional systematic studies on lncRNAs will facilitate the diagnostic and therapeutic performance for bladder cancer.
As the current literature on lncRNA and bladder cancer pays more attention to the correlation between lncRNA and bladder cancer, there is still no certain lncRNA for clinical diagnosis, prognosis and subsequent therapy of bladder cancer. It help to combination of traditional indicators with novel lncRNA biomarkers for diagnosis and prognosis of bladder cancer.
Discussion
Over the past decade, accumulating findings have uncovered that lncRNAs are crucial for initiation and progression of bladder cancer. Up to now, standard bladder cancer biomarkers are still very rare, owing to lack of the high sensitivity and specificity, and high costs of use. Due to the high recurrence and poor prognosis of bladder cancer, even after effective transurethral resection and systemic treatment, it is require to develop novel biomarkers for the early diagnosis and prognosis of bladder cancer. In this review, we summarize the expression, function, molecular mechanisms of lncRNAs, and the clinical significance of lncRNAs in the diagnosis and prognosis of bladder cancer. The molecular mechanisms of lncRNAs in bladder cancer have been explored, including lncRNAs interacting with DNA, RNA and proteins.
Circulating lncRNAs can be enriched in urine supernatant and plasma of bladder cancer, which may have a preferable potential for new bladder cancer tests. It has been suggested that aberrant expression of thirty-six lncRNAs is closely related to multiple clinical characteristics of bladder cancer. In the light of low expression, less evolutionally conserved, the upregulated lncRNAs represent favorable features to act as diagnostic or prognostic indicators for BC. Therefore, we screened potential clinical biomarkers among thirty up-regulated lncRNAs. Dissection of fifteen up-regulated lncRNAs associated with tumor size of bladder cancer, UCA1 has relatively high sensitivity, specificity and AUC, and it can be considered as the most potential diagnostic biomarker for bladder cancer.
Importantly, novel lncRNA biomarkers for clinical diagnosis must previously be carried out large studies in cells and clinical specimen. In this regard, extensive systematic studies on lncRNAs will facilitate the diagnostic and therapeutic performance for bladder cancer. It is worth noting that there is currently no any lncRNA can be applied to the specific diagnosis, prognosis and treatment of bladder cancer. In addition to mRNAs, microRNAs, circular RNAs and exosomes also play important roles in the development of BC. Our present knowledge indicates that combination of mRNAs, microRNAs and lncRNAs would hopefully better for improvement of early diagnosis and prognosis in bladder cancer.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
H-JL, XG, and Z-KL were responsible for the framework and the writing of manuscript, and analysis of references. WQ, C-XH, and LX were mainly dedicated to searching and sorting of references. XZ and DZ were focused on the reference analysis. H-LC was mainly responsible for the revision and polishing of the manuscript. All authors have made great contributions.
Funding
This work was supported by the National Natural Science Foundation of China (U1932130, 31700699, and 81502249), the Chunhui Program of the Ministry of Education (Chunhui Program13), the Shaanxi Provincial Department of Education Key Scientific Research Project (18JS103), the Program of Shaanxi Provincial Science and Technology Department (2017ZDXM-SF-029 and 2020JQ-880), the Program of Shaanxi Administration of Traditional Chinese Medicine (2019-ZZ-ZY009), the Key Program of Weiyang District Bureau of Science, Technology and Industry Information Technology (202031 and 201928), the Program of Shaanxi Provincial Research Center for the Project of Prevention and Treatment of Respiratory Diseases (2017GCKF01), and the Program of Xi’an Medical University (2015RCYJ01, 2018GJFY09, and 202011840023).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.672679/full#supplementary-material
References
Burchardt, M., Burchardt, T., Shabsigh, A., De La Taille, A., Benson, M. C., and Sawczuk, I. (2000). Current concepts in biomarker technology for bladder cancers. Clin. Chem. 46, 595–605. doi: 10.1093/clinchem/46.5.595
Cao, Q., Wang, N., Qi, J., Gu, Z., and Shen, H. (2016). Long non-coding RNA-GAS5 acts as a tumor suppressor in bladder transitional cell carcinoma via regulation of chemokine (C-C motif) ligand 1 expression. Mol. Med. Rep. 13, 27–34. doi: 10.3892/mmr.2015.4503
Cao, X., Xu, J., and Yue, D. (2018). LncRNA-SNHG16 predicts poor prognosis and promotes tumor proliferation through epigenetically silencing p21 in bladder cancer. Cancer Gene Ther. 25, 10–17. doi: 10.1038/s41417-017-0006-x
Casadevall, D., Kilian, A. Y., and Bellmunt, J. (2017). The prognostic role of epigenetic dysregulation in bladder cancer: a systematic review. Cancer Treat. Rev. 61, 82–93. doi: 10.1016/j.ctrv.2017.10.004
Chen, C., He, W., Huang, J., Wang, B., Li, H., Cai, Q., et al. (2018). LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat. Commun. 9:3826.
Chen, C., Luo, Y., He, W., Zhao, Y., Kong, Y., Liu, H., et al. (2020). Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J. Clin. Invest. 130, 404–421. doi: 10.1172/jci130892
Chen, D., Guo, Y., Chen, Y., Guo, Q., Chen, J., Li, Y., et al. (2020). LncRNA growth arrest-specific transcript 5 targets miR-21 gene and regulates bladder cancer cell proliferation and apoptosis through PTEN. Cancer Med. 9, 2846–2858. doi: 10.1002/cam4.2664
Chen, J. B., Zhu, Y. W., Guo, X., Yu, C., Liu, P. H., Li, C., et al. (2019). Microarray expression profiles analysis revealed lncRNA OXCT1-AS1 promoted bladder cancer cell aggressiveness via miR-455-5p/JAK1 signaling. J. Cell. Physiol. 234, 13592–13601. doi: 10.1002/jcp.28037
Chen, J., Li, Y., Li, Z., and Cao, L. (2020). LncRNA MST1P2/miR-133b axis affects the chemoresistance of bladder cancer to cisplatin-based therapy via Sirt1/p53 signaling. J. Biochem. Mol. Toxicol. 34:e22452.
Chen, X., Xie, R., Gu, P., Huang, M., Han, J., Dong, W., et al. (2019). Long Noncoding RNA LBCS inhibits self-renewal and chemoresistance of bladder cancer stem cells through epigenetic silencing of SOX2. Clin. Cancer Res. 25, 1389–1403. doi: 10.1158/1078-0432.ccr-18-1656
Chen, Y., Peng, Y., Xu, Z., Ge, B., Xiang, X., Zhang, T., et al. (2017a). LncROR promotes bladder cancer cell proliferation, migration, and epithelial-mesenchymal transition. Cell Physiol. Biochem. 41, 2399–2410. doi: 10.1159/000475910
Chen, Y., Xie, H., Zou, Y., Lai, X., Ma, L., Liu, Y., et al. (2017b). Tetracycline-controllable artificial microRNA-HOTAIR + EZH2 suppressed the progression of bladder cancer cells. Mol. Biosyst. 13, 1597–1607. doi: 10.1039/c7mb00202e
Dai, L., Chai, C. M., Shen, T. Y., Tian, Y., Shang, Z. Q., and Niu, Y. J. (2019). LncRNA ITGB1 promotes the development of bladder cancer through regulating microRNA-10a expression. Eur. Rev. Med. Pharmacol. Sci. 23, 6858–6867.
Dai, R., Zhou, Y., Chen, Z., Zou, Z., Pan, Z., Liu, P., et al. (2020). Lnc-MUC20-9 binds to ROCK1 and functions as a tumor suppressor in bladder cancer. J. Cell. Biochem. 121, 4214–4225. doi: 10.1002/jcb.29626
Dangle, P. P., Zaharieva, B., Jia, H., and Pohar, K. S. (2009). Ras-MAPK pathway as a therapeutic target in cancer–emphasis on bladder cancer. Recent Pat. Anticancer Drug Discov. 4, 125–136. doi: 10.2174/157489209788452812
Ding, Z., Ying, W., He, Y., Chen, X., Jiao, Y., Wang, J., et al. (2021). lncRNA-UCA1 in the diagnosis of bladder cancer: a meta-analysis. Medicine 100:e24805. doi: 10.1097/md.0000000000024805
Duan, W., Du, L., Jiang, X., Wang, R., Yan, S., Xie, Y., et al. (2016). Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget 7, 78850–78858. doi: 10.18632/oncotarget.12880
Fang, C., He, W., Xu, T., Dai, J., Xu, L., and Sun, F. (2019a). Upregulation of lncRNA DGCR5 correlates with better prognosis and inhibits bladder cancer progression via transcriptionally facilitating P21 expression. J. Cell. Physiol. 234, 6254–6262. doi: 10.1002/jcp.27356
Fang, C., Xu, L., He, W., Dai, J., and Sun, F. (2019b). Long noncoding RNA DLX6-AS1 promotes cell growth and invasiveness in bladder cancer via modulating the miR-223-HSP90B1 axis. Cell Cycle 18, 3288–3299. doi: 10.1080/15384101.2019.1673633
Feng, F., Chen, A., Huang, J., Xia, Q., Chen, Y., and Jin, X. (2018). Long noncoding RNA SNHG16 contributes to the development of bladder cancer via regulating miR-98/STAT3/Wnt/β-catenin pathway axis. J. Cell. Biochem. 119, 9408–9418. doi: 10.1002/jcb.27257
Feng, S. Q., Zhang, X. Y., Fan, H. T., Sun, Q. J., and Zhang, M. (2018). Up-regulation of LncRNA MEG3 inhibits cell migration and invasion and enhances cisplatin chemosensitivity in bladder cancer cells. Neoplasma 65, 925–932. doi: 10.4149/neo_2018_180125n55
Gao, R., Zhang, N., Yang, J., Zhu, Y., Zhang, Z., Wang, J., et al. (2019). Long non-coding RNA ZEB1-AS1 regulates miR-200b/FSCN1 signaling and enhances migration and invasion induced by TGF-β1 in bladder cancer cells. J. Exp. Clin. Cancer Res. 38:111.
Guo, P., Zhang, G., Meng, J., He, Q., Li, Z., and Guan, Y. (2018). Upregulation of long noncoding RNA TUG1 promotes bladder cancer cell proliferation, migration, and invasion by inhibiting miR-29c. Oncol. Res. 26, 1083–1091. doi: 10.3727/096504018x15152085755247
He, A., Liu, Y., Chen, Z., Li, J., Chen, M., Liu, L., et al. (2016). Over-expression of long noncoding RNA BANCR inhibits malignant phenotypes of human bladder cancer. J. Exp. Clin. Cancer Res. 35:125.
He, W., Cai, Q., Sun, F., Zhong, G., Wang, P., Liu, H., et al. (2013). linc-UBC1 physically associates with polycomb repressive complex 2 (PRC2) and acts as a negative prognostic factor for lymph node metastasis and survival in bladder cancer. Biochim. Biophys. Acta 1832, 1528–1537. doi: 10.1016/j.bbadis.2013.05.010
Heubach, J., Monsior, J., Deenen, R., Niegisch, G., Szarvas, T., Niedworok, C., et al. (2015). The long noncoding RNA HOTAIR has tissue and cell type-dependent effects on HOX gene expression and phenotype of urothelial cancer cells. Mol. Cancer 14:108.
Hu, B., Shi, G., Li, Q., Li, W., and Zhou, H. (2019). Long noncoding RNA XIST participates in bladder cancer by downregulating p53 via binding to TET1. J. Cell. Biochem. 120, 6330–6338. doi: 10.1002/jcb.27920
Hu, R., Zhong, P., Xiong, L., and Duan, L. (2017). Long noncoding RNA cancer susceptibility candidate 8 suppresses the proliferation of bladder cancer cells via regulating glycolysis. DNA Cell Biol. 36, 767–774. doi: 10.1089/dna.2017.3785
Iyer, M. K., Niknafs, Y. S., Malik, R., Singhal, U., Sahu, A., Hosono, Y., et al. (2015). The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208.
Jiang, F., Qi, W., Wang, Y., Wang, W., and Fan, L. (2019). lncRNA PEG10 promotes cell survival, invasion and migration by sponging miR-134 in human bladder cancer. Biomed. Pharmacother. 114:108814. doi: 10.1016/j.biopha.2019.108814
Jiang, H., Hu, X., Zhang, H., and Li, W. (2017). Down-regulation of LncRNA TUG1 enhances radiosensitivity in bladder cancer via suppressing HMGB1 expression. Radiat. Oncol. 12:65.
Jiao, D., Li, Z., Zhu, M., Wang, Y., Wu, G., and Han, X. (2018). LncRNA MALAT1 promotes tumor growth and metastasis by targeting miR-124/foxq1 in bladder transitional cell carcinoma (BTCC). Am. J. Cancer Res. 8, 748–760.
Kartha, R. V., and Subramanian, S. (2014). Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front. Genet. 5:8.
Li, C., Cui, Y., Liu, L. F., Ren, W. B., Li, Q. Q., Zhou, X., et al. (2017). High expression of long noncoding RNA MALAT1 indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Clin. Genitourinary Cancer 15, 570–576. doi: 10.1016/j.clgc.2017.05.001
Li, H. J., Li, X., Pang, H., Pan, J. J., Xie, X. J., and Chen, W. (2015). Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Jpn. J. Clin. Oncol. 45, 1055–1063. doi: 10.1093/jjco/hyv132
Li, H. J., Sun, X. M., Li, Z. K., Yin, Q. W., Pang, H., Pan, J. J., et al. (2017). LncRNA UCA1 promotes mitochondrial function of bladder cancer via the MiR-195/ARL2 signaling pathway. Cell Physiol. Biochem. 43, 2548–2561. doi: 10.1159/000484507
Li, J., Li, Y., Meng, F., Fu, L., and Kong, C. (2018). Knockdown of long non-coding RNA linc00511 suppresses proliferation and promotes apoptosis of bladder cancer cells via suppressing Wnt/β-catenin signaling pathway. Biosci. Rep. 38:BSR20171701.
Li, Q., Li, C., Chen, J., Liu, P., Cui, Y., Zhou, X., et al. (2018). High expression of long noncoding RNA NORAD indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Urologic Oncol. 36, 310.e315–310.e322.
Li, Z., Hong, S., and Liu, Z. (2018). LncRNA LINC00641 predicts prognosis and inhibits bladder cancer progression through miR-197-3p/KLF10/PTEN/PI3K/AKT cascade. Biochem. Biophys. Res. Commun. 503, 1825–1829. doi: 10.1016/j.bbrc.2018.07.120
Li, Z., Li, X., Wu, S., Xue, M., and Chen, W. (2014). Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci. 105, 951–955. doi: 10.1111/cas.12461
Liao, C., Long, Z., Zhang, X., Cheng, J., Qi, F., Wu, S., et al. (2020). LncARSR sponges miR-129-5p to promote proliferation and metastasis of bladder cancer cells through increasing SOX4 expression. Int. J. Biol. Sci. 16, 1–11. doi: 10.7150/ijbs.39461
Lin, M. G., Hong, Y. K., Zhang, Y., Lin, B. B., and He, X. J. (2018). Mechanism of lncRNA DUXAP8 in promoting proliferation of bladder cancer cells by regulating PTEN. Eur. Rev. Med. Pharmacol. Sci. 22, 3370–3377.
Liu, C., Chen, Z., Fang, J., Xu, A., Zhang, W., and Wang, Z. (2016). H19-derived miR-675 contributes to bladder cancer cell proliferation by regulating p53 activation. Tumour Biol. 37, 263–270. doi: 10.1007/s13277-015-3779-2
Liu, D., Li, Y., Luo, G., Xiao, X., Tao, D., Wu, X., et al. (2017). LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. 388, 281–291. doi: 10.1016/j.canlet.2016.12.005
Liu, G., Zhao, X., Zhou, J., Cheng, X., Ye, Z., and Ji, Z. (2018). Long non-coding RNA MEG3 suppresses the development of bladder urothelial carcinoma by regulating miR-96 and TPM1. Cancer Biol. Ther. 19, 1039–1056. doi: 10.1080/15384047.2018.1480279
Liu, Q., Liu, H., Cheng, H., Li, Y., Li, X., and Zhu, C. (2017). Downregulation of long noncoding RNA TUG1 inhibits proliferation and induces apoptosis through the TUG1/miR-142/ZEB2 axis in bladder cancer cells. OncoTargets Ther. 10, 2461–2471. doi: 10.2147/ott.s124595
Liu, Y., and Wu, G. (2020). NNT-AS1 enhances bladder cancer cell growth by targeting miR-1301-3p/PODXL axis and activating Wnt pathway. Neurourol. Urodyn. 39, 547–557. doi: 10.1002/nau.24238
Liu, Z., and Zhang, H. (2017). LncRNA plasmacytoma variant translocation 1 is an oncogene in bladder urothelial carcinoma. Oncotarget 8, 64273–64282. doi: 10.18632/oncotarget.19604
Liu, Z., Xie, D., and Zhang, H. (2018). Long noncoding RNA neuroblastoma-associated transcript 1 gene inhibits malignant cellular phenotypes of bladder cancer through miR-21/SOCS6 axis. Cell Death Dis. 9:1042.
Luo, H., Xu, C., Le, W., Ge, B., and Wang, T. (2019). lncRNA CASC11 promotes cancer cell proliferation in bladder cancer through miRNA-150. J. Cell. Biochem. 120, 13487–13493. doi: 10.1002/jcb.28622
Lv, M., Zhong, Z., Huang, M., Tian, Q., Jiang, R., and Chen, J. (2017). lncRNA H19 regulates epithelial-mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1887–1899. doi: 10.1016/j.bbamcr.2017.08.001
Lv, X. Y., Ma, L., Chen, J. F., Yu, R., Li, Y., Yan, Z. J., et al. (2018). Knockdown of DUXAP10 inhibits proliferation and promotes apoptosis in bladder cancer cells via PI3K/Akt/mTOR signaling pathway. Int. J. Oncol. 52, 288–294.
Ma, L., Bajic, V. B., and Zhang, Z. (2013). On the classification of long non-coding RNAs. RNA Biol. 10, 924–933.
Ma, Z., Xue, S., Zeng, B., and Qiu, D. (2018). SNHG5lncRNA is associated with poor prognosis of bladder cancer and promotes bladder cancer cell proliferation through targeting. Oncol. Lett. 15, 1924–1930.
Mónica, M. F., Andrew, F., Marta, D., Cristina, S., Carolina, R., Maria, F., et al. (2015). Analysis of the Polycomb-related lncRNAs HOTAIR and ANRIL in bladder cancer. Clin Epigenet. 7:109.
Nicolas, F. E. (2017). Role of ncRNAs in development, diagnosis and treatment of human cancer. Recent Pat. Anticancer Drug Discov. 12, 128–135. doi: 10.2174/1574892812666170105113415
Okazaki, Y., Furuno, M. K., and Kasukawa, T. (2002). Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420, 563–573. doi: 10.1038/nature01266
Pan, J., Li, X., Wu, W., Xue, M., Hou, H., Zhai, W., et al. (2016). Long non-coding RNA UCA1 promotes cisplatin/gemcitabine resistance through CREB modulating miR-196a-5p in bladder cancer cells. Cancer Lett. 382, 64–76. doi: 10.1016/j.canlet.2016.08.015
Pei, Z., Du, X., Song, Y., Fan, L., Li, F., Gao, Y., et al. (2017). Down-regulation of lncRNA CASC2 promotes cell proliferation and metastasis of bladder cancer by activation of the Wnt/β-catenin signaling pathway. Oncotarget 8, 18145–18153. doi: 10.18632/oncotarget.15210
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. doi: 10.1016/j.cell.2009.02.006
Qi, D., Li, J., Que, B., Su, J., Li, M., Zhang, C., et al. (2016). Long non-coding RNA DBCCR1-003 regulate the expression of DBCCR1 via DNMT1 in bladder cancer. Cancer Cell Int. 16:81.
Qiu, F., Zhang, M. R., Zhou, Z., Pu, J. X., and Zhao, X. J. (2019). lncRNA MIR503HG functioned as a tumor suppressor and inhibited cell proliferation, metastasis and epithelial-mesenchymal transition in bladder cancer. J. Cell. Biochem. 120, 10821–10829. doi: 10.1002/jcb.28373
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell 146, 353–358. doi: 10.1016/j.cell.2011.07.014
Shan, G., Tang, T., Xia, Y., and Qian, H. J. (2020a). Long non-coding RNA NEAT1 promotes bladder progression through regulating miR-410 mediated HMGB1. Biomed. Pharmacother. 121:109248. doi: 10.1016/j.biopha.2019.109248
Shan, G., Tang, T., Xia, Y., and Qian, H. J. (2020b). MEG3 interacted with miR-494 to repress bladder cancer progression through targeting PTEN. J. Cell. Physiol. 235, 1120–1128. doi: 10.1002/jcp.29025
Siegel, R. L., Miller, K. D., and Jemal, A. (2020). Cancer statistics, 2020. Cancer J. Clin. 70, 7–30.
Su, G., He, Q., and Wang, J. (2018). Clinical values of long non-coding RNAs in bladder cancer: a systematic review. Front. Physiol. 9:652.
Sun, X., Du, P., Yuan, W., Du, Z., Yu, M., Yu, X., et al. (2015). Long non-coding RNA HOTAIR regulates cyclin J via inhibition of microRNA-205 expression in bladder cancer. Cell Death Dis. 6:e1907. doi: 10.1038/cddis.2015.269
Tabayoyong, W., and Kamat, A. M. (2018). Current use and promise of urinary markers for urothelial cancer. Curr. Urol. Rep 19:96.
Takai, D., Gonzales, F. A., Tsai, Y. C., Thayer, M. J., and Jones, P. A. (2001). Large scale mapping of methylcytosines in CTCF-binding sites in the human H19 promoter and aberrant hypomethylation in human bladder cancer. Hum. Mol. Genet. 10, 2619–2626. doi: 10.1093/hmg/10.23.2619
Teng, J., Ai, X., Jia, Z., Wang, K., Guan, Y., and Guo, Y. (2019). Long non-coding RNA ARAP1-AS1 promotes the progression of bladder cancer by regulating miR-4735-3p/NOTCH2 axis. Cancer Biol. Ther. 20, 552–561. doi: 10.1080/15384047.2018.1538613
Tuo, Z., Zhang, J., and Xue, W. (2018). LncRNA TP73-AS1 predicts the prognosis of bladder cancer patients and functions as a suppressor for bladder cancer by EMT pathway. Biochem. Biophys. Res. Commun. 499, 875–881. doi: 10.1016/j.bbrc.2018.04.010
Wang, F., Zu, Y., Huang, W., Chen, H., Xie, H., and Yang, Y. (2018a). LncRNA CALML3-AS1 promotes tumorigenesis of bladder cancer via regulating ZBTB2 by suppression of microRNA-4316. Biochem. Biophys. Res. Commun. 504, 171–176. doi: 10.1016/j.bbrc.2018.08.150
Wang, F., Zu, Y., Zhu, S., Yang, Y., Huang, W., Xie, H., et al. (2018b). Long noncoding RNA MAGI2-AS3 regulates CCDC19 expression by sponging miR-15b-5p and suppresses bladder cancer progression. Biochem. Biophys. Res. Commun. 507, 231–235. doi: 10.1016/j.bbrc.2018.11.013
Wang, J., Xing, H., Nikzad, A. A., Liu, B., Zhang, Y., Li, S., et al. (2020). Long noncoding RNA MNX1 antisense RNA 1 exerts oncogenic functions in bladder cancer by regulating miR-218-5p/RAB1A Axis. J. Pharmacol. Exp. Ther. 372, 237–247. doi: 10.1124/jpet.119.262949
Wang, M., Guo, C., Wang, L., Luo, G., Huang, C., Li, Y., et al. (2018c). Long noncoding RNA GAS5 promotes bladder cancer cells apoptosis through inhibiting EZH2 transcription. Cell Death Dis. 9:238.
Wang, X., Gong, Y., Jin, B., Wu, C., Yang, J., Wang, L., et al. (2014). Long non-coding RNA urothelial carcinoma associated 1 induces cell replication by inhibiting BRG1 in 5637 cells. Oncol. Rep. 32, 1281–1290. doi: 10.3892/or.2014.3309
Wang, Y. Y., Wu, Z. Y., Wang, G. C., Liu, K., Niu, X. B., Gu, S., et al. (2016). LINC00312 inhibits the migration and invasion of bladder cancer cells by targeting miR-197-3p. Tumour Biol. 37, 14553–14563. doi: 10.1007/s13277-016-5303-8
Wei, X., Yang, X., Wang, B., Yang, Y., Fang, Z., Yi, C., et al. (2020). LncRNA MBNL1-AS1 represses cell proliferation and enhances cell apoptosis via targeting miR-135a-5p/PHLPP2/FOXO1 axis in bladder cancer. Cancer Med. 9, 724–736. doi: 10.1002/cam4.2684
Xie, H., Huang, H., Huang, W., Xie, Z., Yang, Y., and Wang, F. (2019). LncRNA miR143HG suppresses bladder cancer development through inactivating Wnt/β-catenin pathway by modulating miR-1275/AXIN2 axis. J. Cell. Physiol. 234, 11156–11164. doi: 10.1002/jcp.27764
Xue, M., Li, X., Wu, W., Zhang, S., Wu, S., Li, Z., et al. (2014). Upregulation of long non-coding RNA urothelial carcinoma associated 1 by CCAAT/enhancer binding protein α contributes to bladder cancer cell growth and reduced apoptosis. Oncol. Rep. 31, 1993–2000. doi: 10.3892/or.2014.3092
Xue, M., Pang, H., Li, X., Li, H., Pan, J., and Chen, W. (2016). Long non-coding RNA urothelial cancer-associated 1 promotes bladder cancer cell migration and invasion by way of the hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci. 107, 18–27. doi: 10.1111/cas.12844
Yang, C., Li, X., Wang, Y., Zhao, L., and Chen, W. (2012). Long non-coding RNA UCA1 regulated cell cycle distribution via CREB through PI3-K dependent pathway in bladder carcinoma cells. Gene 496, 8–16. doi: 10.1016/j.gene.2012.01.012
Yang, Y., Wang, F., Huang, H., Zhang, Y., Xie, H., and Men, T. (2019). lncRNA SLCO4A1-AS1 promotes growth and invasion of bladder cancer through sponging miR-335-5p to upregulate OCT4. OncoTargets Ther. 12, 1351–1358. doi: 10.2147/ott.s191740
Ying, L., Huang, Y., Chen, H., Wang, Y., Xia, L., Chen, Y., et al. (2013). Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol. Biosyst. 9, 407–411. doi: 10.1039/c2mb25386k
Yu, C., Longfei, L., Long, W., Feng, Z., Chen, J., Chao, L., et al. (2019). LncRNA PVT1 regulates VEGFC through inhibiting miR-128 in bladder cancer cells. J. Cell. Physiol. 234, 1346–1353. doi: 10.1002/jcp.26929
Yu, D., Zhang, C., and Gui, J. (2017). RNA-binding protein HuR promotes bladder cancer progression by competitively binding to the long noncoding HOTAIR with miR-1. OncoTargets Ther. 10, 2609–2619. doi: 10.2147/ott.s132728
Yu, G., Ou, Z. Y., Tao, Q. Y., Wan, G. Y., Lu, Z. H., and Lang, B. (2017). Role of lncRNA PTENP1 in tumorigenesis and progression of bladder cancer and the molecular mechanism. Nan fang yi ke da xue xue bao. 37, 1494–1500.
Yu, H., Wang, S., Zhu, H., and Rao, D. (2019). LncRNA MT1JP functions as a tumor suppressor via regulating miR-214-3p expression in bladder cancer. J. Cell. Physiol. doi: 10.1002/jcp.28274 Online ahead of print.
Yuan, S., Luan, X., Han, G., Guo, K., Wang, S., and Zhang, X. (2019). LINC01638 lncRNA mediates the postoperative distant recurrence of bladder cancer by upregulating ROCK2. Oncol. Lett. 18, 5392–5398.
Zhai, X., and Xu, W. (2018). Long noncoding RNA ATB promotes proliferation, migration, and invasion in bladder cancer by suppressing MicroRNA-126. Oncol. Res. 26, 1063–1072. doi: 10.3727/096504018x15152072098476
Zhan, Y., Chen, Z., He, S., Gong, Y., He, A., Li, Y., et al. (2020). Long non-coding RNA SOX2OT promotes the stemness phenotype of bladder cancer cells by modulating SOX2. Mol. Cancer 19:25.
Zhang, Q., Su, M., Lu, G., and Wang, J. (2013). The complexity of bladder cancer: long noncoding RNAs are on the stage. Mol. Cancer 12:101. doi: 10.1186/1476-4598-12-101
Zhang, R., Wang, J., Jia, E., Zhang, J., Liu, N., and Chi, C. (2020). lncRNA BCAR4 sponges miR-370-3p to promote bladder cancer progression via Wnt signaling. Int. J. Mol. Med. 45, 578–588.
Zhang, Y., and Tang, L. (2018). The application of lncRNAs in cancer treatment and diagnosis. Recent Pat. Anticancer Drug Discov. 13, 292–301. doi: 10.2174/1574892813666180226121819
Zhao, F., Lin, T., He, W., Han, J., Zhu, D., Hu, K., et al. (2015). Knockdown of a novel lincRNA AATBC suppresses proliferation and induces apoptosis in bladder cancer. Oncotarget 6, 1064–1078. doi: 10.18632/oncotarget.2833
Zhao, Q., Gao, S., Du, Q., and Liu, Y. (2018). Long non-coding RNA SNHG20 promotes bladder cancer via activating the Wnt/β-catenin signalling pathway. Int. J. Mol. Med. 42, 2839–2848.
Zheng, R., Du, M., Wang, X., Xu, W., Liang, J., Wang, W., et al. (2018). Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol. Cancer 17:143.
Zhu, F., Zhang, X., Yu, Q., Han, G., Diao, F., Wu, C., et al. (2018). LncRNA AWPPH inhibits SMAD4 via EZH2 to regulate bladder cancer progression. J. Cell. Biochem. 119, 4496–4505. doi: 10.1002/jcb.26556
Zhu, X., Li, Y., Zhao, S., and Zhao, S. (2018). LSINCT5 activates Wnt/β-catenin signaling by interacting with NCYM to promote bladder cancer progression. Biochem. Biophys. Res. Commun. 502, 299–306. doi: 10.1016/j.bbrc.2018.05.076
Keywords: bladder cancer (BC), long non-coding RNAs (lncRNAs), biomarker, early diagnosis, tumor development
Citation: Li H-J, Gong X, Li Z-K, Qin W, He C-X, Xing L, Zhou X, Zhao D and Cao H-L (2021) Role of Long Non-coding RNAs on Bladder Cancer. Front. Cell Dev. Biol. 9:672679. doi: 10.3389/fcell.2021.672679
Received: 26 February 2021; Accepted: 17 June 2021;
Published: 04 August 2021.
Edited by:
Massimiliano Berretta, University of Messina, ItalyReviewed by:
Qingbin Cui, University of Toledo, United StatesFlaviana Marzano, Institute of Biomembranes and Bioenergetics, Italy
Copyright © 2021 Li, Gong, Li, Qin, He, Xing, Zhou, Zhao and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui-Ling Cao, Y2FvaHVpbGluZ19qenNAeGl5aS5lZHUuY24=
†These authors have contributed equally to this work
 Hui-Jin Li1†
Hui-Jin Li1† Wei Qin
Wei Qin Lu Xing
Lu Xing Xin Zhou
Xin Zhou Dong Zhao
Dong Zhao Hui-Ling Cao
Hui-Ling Cao