
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 13 May 2021
Sec. Stem Cell Research
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.671841
This article is part of the Research TopicFibroblast Growth Factors and Stem Cells in Regenerative Pharmacology and Anti-Aging InterventionView all 21 articles
 Xuran Chu1,2,3
Xuran Chu1,2,3 Sara Taghizadeh2,3
Sara Taghizadeh2,3 Ana Ivonne Vazquez-Armendariz3,4
Ana Ivonne Vazquez-Armendariz3,4 Susanne Herold3,4
Susanne Herold3,4 Lei Chong5
Lei Chong5 Chengshui Chen2
Chengshui Chen2 Jin-San Zhang1,2*
Jin-San Zhang1,2* Elie El Agha3,4*
Elie El Agha3,4* Saverio Bellusci1,2,3*
Saverio Bellusci1,2,3*Fgf10 is a key gene during development, homeostasis and repair after injury. We previously reported a knock-in Fgf10Cre–ERT2 line (with the Cre-ERT2 cassette inserted in frame with the start codon of exon 1), called thereafter Fgf10Ki–v1, to target FGF10Pos cells. While this line allowed fairly efficient and specific labeling of FGF10Pos cells during the embryonic stage, it failed to target these cells after birth, particularly in the postnatal lung, which has been the focus of our research. We report here the generation and validation of a new knock-in Fgf10Cre–ERT2 line (called thereafter Fgf10Ki–v2) with the insertion of the expression cassette in frame with the stop codon of exon 3. Fgf10Ki−v2/+ heterozygous mice exhibited comparable Fgf10 expression levels to wild type animals. However, a mismatch between Fgf10 and Cre expression levels was observed in Fgf10Ki–v2/+ lungs. In addition, lung and limb agenesis were observed in homozygous embryos suggesting a loss of Fgf10 functional allele in Fgf10Ki–v2 mice. Bioinformatic analysis shows that the 3′UTR, where the Cre-ERT2 cassette is inserted, contains numerous putative transcription factor binding sites. By crossing this line with tdTomato reporter line, we demonstrated that tdTomato expression faithfully recapitulated Fgf10 expression during development. Importantly, Fgf10Ki–v2 mouse is capable of significantly targeting FGF10Pos cells in the adult lung. Therefore, despite the aforementioned limitations, this new Fgf10Ki–v2 line opens the way for future mechanistic experiments involving the postnatal lung.
The fibroblast growth factor (FGF) family consisting of 22 members is divided into three groups: the paracrine FGF group signaling through FGFR and heparin-sulfate proteoglycans, the endocrine FGF group signaling through FGFR with Klotho family of proteins as co-receptors, and the intracellular FGF group involved in FGFR independent signaling (Ornitz and Itoh, 2001). The FGF7 subgroup which contains FGF3, 7, 10, 22 belongs to the paracrine FGF group. These growth factors interact mostly with the FGFR2b receptor. FGF10 in particular has been shown to play important roles during development, homeostasis and repair after injury (Yuan et al., 2018). In the lung, it plays a crucial role in regulating branching morphogenesis (Jones et al., 2020). Genetic deletion of either Fgf10 or its predominant receptor Fgfr2b leads to agenesis of both the limb and the lung, specific portions of the gut, the pancreas as well as the mammary, lacrimal and salivary glands (Min et al., 1998; Sekine et al., 1999; Ohuchi et al., 2000; Mailleux et al., 2002; Jaskoll et al., 2005; Parsa et al., 2008). During homeostasis, Fgfr2b signaling has been shown to be critical for the regeneration of the incisors in mice as well as for the maintenance of the terminal end buds in the mammary gland (Parsa et al., 2008, 2010). Lineage tracing of FGF10Pos during development indicated that these cells serve as progenitors for lipofibroblast as well as vascular and airway smooth muscle cells (El Agha et al., 2014).
In the context of the repair process, Fgf10 deletion in peribronchial mesenchymal cells leads to impaired repair following injury to the bronchial epithelium using naphthalene (Volckaert et al., 2011; Moiseenko et al., 2020). On the other hand, overexpression of Fgf10 reduces the severity of lung fibrosis in bleomycin-induced mice (Gupte et al., 2009). Given these diverse biological activities, it is important to generate and validate mouse knock-in lines allowing to monitor the localization, fate and status of FGF10Pos cells during development, homeostasis and repair after injury.
We have previously generated a Fgf10Cre–ERT2 knock-in mouse line, called thereafter Fgf10KI–v1 mice, to monitor the fate of FGF10Pos cells after tamoxifen (Tam) administration (El Agha et al., 2012). In these mice, the Tam-inducible Cre recombinase (Cre-ERT2-IRES-YFP) was inserted in frame with the start codon of the endogenous Fgf10 gene. Fgf10KI–v1 corresponds to a loss-of-function allele for Fgf10 as evidenced by our observation that Fgf10KI–v1/KI–v1 homozygous embryos die at birth from multi-organ agenesis, including the lung. In the Fgf10KI–v1/+ lungs, the expression of Cre gradually decreases to almost undetectable levels postnatally, rendering the monitoring of FGF10Pos cells postnatally impossible. This is likely due to the deletion of intronic sequences containing key transcription factor binding sites at the insertion site of the Cre-expression cassette.
In order to circumvent this problem, we therefore generated a new Cre-ERT2 knock-in line (named Fgf10KI–v2) by targeting the 3′UTR of the endogenous Fgf10 gene. We here provide experimental evidence for the validation of these mice. Besides a PCR-based strategy to genotype the Fgf10KI–v2 allele, we have also established a qPCR-based approach to monitor the expression levels of Fgf10 and Cre at different developmental stages in the lung. Fgf10KI–v2/KI–v2 homozygous embryos have been generated to check for developmental defects. Fgf10KI–v2/+ lines were crossed with the tdTomatoflox mice to validate, at two distinct embryonic stages, the expression patterns of tdTomato in previously known domains of Fgf10 expression. Importantly, we validated the use of these mice in the adult stages to target FGF10Pos cells in the lung. Flow cytometry analysis and immunofluorescence staining were carried out to further characterize the contribution of these cells to the lipofibroblast lineage. Bioinformatic analysis of the insertion site of the Cre-ERT2 cassette in the 3′UTR was also carried out. Altogether, our results indicate that the new Fgf10Cre–ERT2 line can be successfully used to target FGF10Pos cells both in embryonic and adult stages.
Two pairs of primers were used to determine the genotype of Fgf10Ki–v2 knock-in mice. Primer P1 (5′-AACACC TCTGCTCACTTCCTC-3′); and primer P2 (5′-AGGGTCCACC TTCCGCTTTT-3′) were used to detect the knock-in allele (252 bp band) whereas primer P3 (5′-GCAGGCAAA TGTATGTGGCA-3′) and primer P4 (5′-TGCTTGCGTGTCT TACTGCT-3′) were used to detect the wild-type allele (580 bp band). The PCR program consists of a denaturation step at 94°C for 3 min, followed by 34 cycles of denaturation (94°C for 1 min), annealing (60°C for 30 s) and extension steps (72°C for 300 s). The program ends with a completion step at 4°C for infinity hold. Each PCR tube contains 4.3 μL of H2O, Taq DNA Polymerase in 5.5 μL of Qiagen Master Mix (QIAGEN, Hilden, Germany), 10 pmol of each primer, and 50 ng of genomic DNA in a final volume of 11 μL.
All mice were kept under specific pathogen free (SPF) conditions with unlimited food and water. tdTomatoflox/flox reporter mice were purchased from Jackson lab (B6; 129S6-Gt(ROSA)26Sortm9(CAG–tdTomato)Hze/J, ref 007905). Embryonic day 0.5 (E0.5) was assigned to the day when a vaginal plug was detected. Animal experiments were approved by the Regierungspraesidium Giessen (approval number RP GI20/10-Nr. G47/2019). Tamoxifen stock solution was prepared by dissolving tamoxifen powder (Sigma, T5648-5G) in corn oil at a concentration of 20 mg/mL at room temperature and stored in −20°C. Adult mice received 3 successive intraperoneal (IP) injections of tamoxifen (0.25 mg/g body weight) before analysis. Pregnant mice received a single IP injection of tamoxifen (0.1 mg/g body weight) and pups also received a single subcutaneous injection of tamoxifen (0.2 mg/pup) before analysis. Dissected mice were examined using Leica M205 FA fluorescent stereoscope (Leica, Wetzlar, Germany) and images were acquired using Leica DFC360 FX camera. Figures were assembled in Adobe Photoshop and Illustrator.
Freshly isolated embryos and lungs were lysed, and RNA was extracted using RNeasy kit (74106, Qiagen, Hilden, Germany). One microgram of RNA was used for cDNA synthesis using Quantitect Reverse Transcription kit (205311, Qiagen). Primers and probes for Fgf10, Cre, and β2-Microglobulin (B2M) were designed using NCBI Primer-BLAST1. More details about the used primers and probes can be found in Supplementary Table 1. Quantitative real-time PCR (qPCR) was performed using LightCycler 480 real-time PCR machine (Roche Applied Science). Samples were run in doublets using B2M as a reference gene and the delta Ct method was used to calculate the relative quantification. GraphPad Prism 7.0 software was used to generate and analyze data. Statistical analyses were performed using Student’s t-test (for comparing two groups) or One-way ANOVA (for comparing three or more groups). Data were considered significant if P < 0.05.
Freshly dissected lung were washed with Hanks’ balanced salt solution (HBSS, 14175-095, Thermo Fisher) and kept on ice. Sharp blades were used to cut the lung into small pieces and digested with 0.5% collagenase Type IV in HBSS (17104019, Life Technologies, Invitrogen) for 45 min at 37°C. Lung homogenates were then passed through 18, 21, and 24G needles followed by 70 and 40 μm cell strainers (542070 and 542040, Greiner Bio-one International). Lung homogenates were centrifuged at 200g at 4°C for 10 min and cell pellets were then re-suspended in HBSS. 20 (μL of sample is taken as an unstained control. Antibodies against CD45 (103114, APC-conjugated; 1:50), CD31 (102409, APC-conjugated; 1:50), EPCAM (APC-Cy7-conjugated; 1:50) and SCA1 (108120, Pacific blue-conjugated; 1:50) (all from Biolegend) as well as LipidTOX stain (FITC-conjugated, 1:200) (H34350, Life Technologies, Invitrogen) were applied for 30 min on ice in the dark. Samples were then washed with 1 mL of HBSS at 4°C for 5 min. FACSAria III cell sorter (BD Biosciences) was used to carry out the FACS measurements and sorting. Endogenous tdTomato signal was detected through PE channel. Gates were set up according to the unstained controls.
National Center of Biotechnology Information (https://www.ncbi.nlm.nih.gov/gene/) and rVista (rVista; http://rvista.dcode.org) were used to find the murine Fgf10 sequence and the identification of putative transcription factor binding site (TFBS) was done by using PROMO software2. The list of putative TFBS located in the area including exon 3 and 3′UTR was further compared with previously identified transcription factors expressed in the lung mesenchyme (Herriges et al., 2012).
129Sv ES cells were electroporated with a targeting vector containing the first 3 kb of exon 3 of the Fgf10 open reading frame (Figures 1A,B). Immediately downstream of the stop codon of exon 3 is the F2A sequence encoding for the self-cleaving peptide, followed by the coding sequence of eYFP, the self-cleaving peptide sequence T2A, the tamoxifen-inducible form of Cre recombinase (Cre-ERT2) (Feil et al., 1997), and the Neomycin-resistance gene (Neo), respectively. Resistant ES cell clones were selected, screened by PCR and then verified by Southern blotting. Selected ES clones were injected into C57BL/6J blastocysts to generate chimeric pups (Figure 1C). Chimeras were then crossed with C57BL/6J mice ubiquitously expressing Flp recombinase to generate heterozygous Fgf10Ki–v2 knock-in mice where the Neo cassette was totally excised (Figure 1D). Genotyping strategy with primers P1/P2 (with P1 located just before the STOP codon and P2 being part of the F2A sequence) to detect the Fgf10Ki–v2 mutant allele and P3/P4 (located before and after the STOP codon in exon 3, respectively) to detect the Fgf10+ wild type allele (Figure 1E).
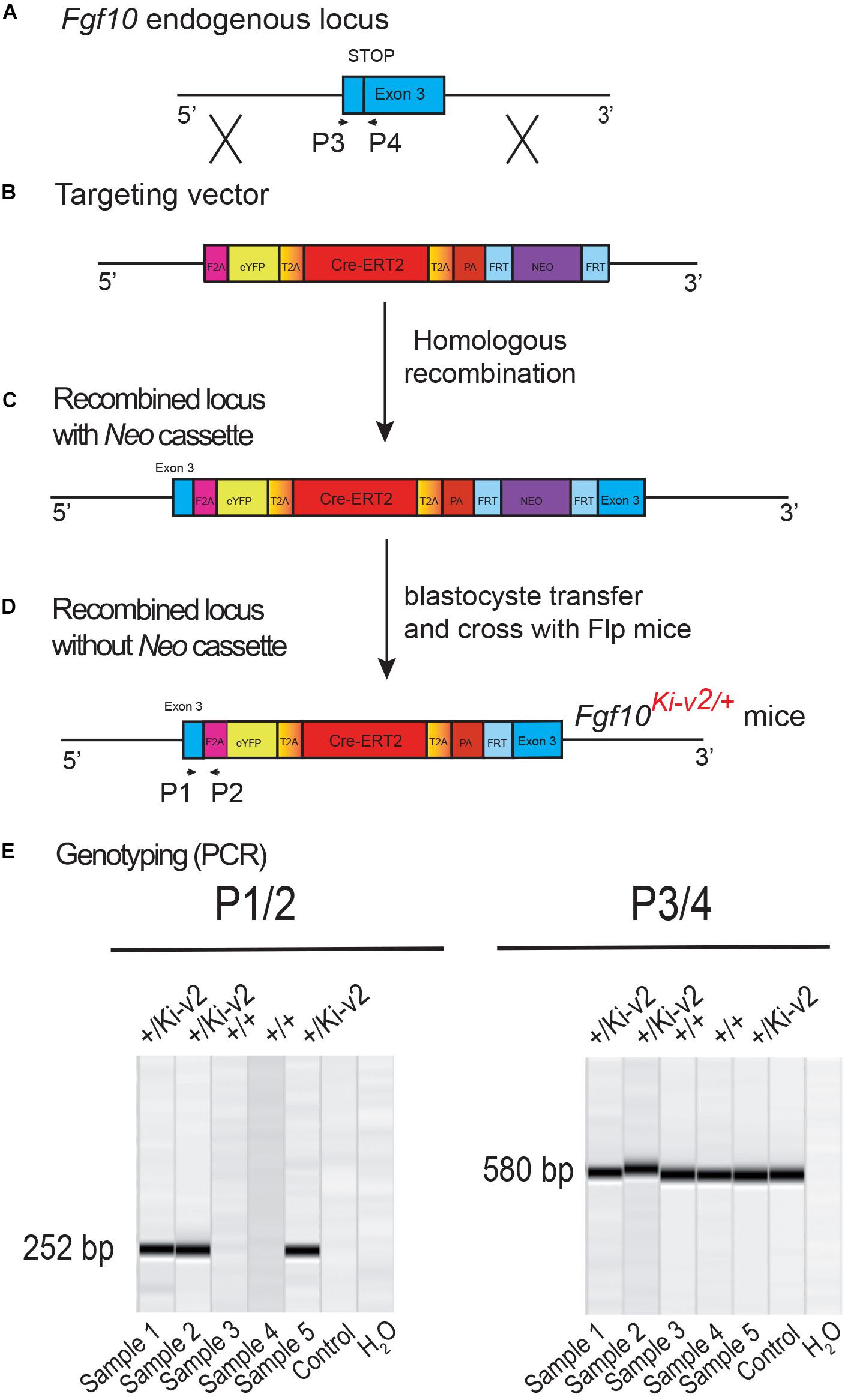
Figure 1. Generation and genotyping of the novel Fgf10Ki-v2 line. (A,B) Homologous recombination was carried out to insert the F2A-eYFP-T2A-Cre-ERT2- T2A-PA-NEO construct in frame with the stop codon of exon 3 of the mouse Fgf10 gene. Neomycin resistance coding gene was used for the positive selection. (C,D) Recombined ES cell clones were treated with flipase to remove the Neo cassette and blastocyste transfer of the selected ES cells was carried out to generate chimera animals. (E) PCR strategy to genotype mutant and wild type animals. Primers 1 and 2 were used for the detection of the mutant Fgf10Ki-v2 allele (252 bp) and Primers 3 and 4 were used for the detection of the wild type Fgf10+ allele (580 bp).
Our initial design of the novel Fgf10Ki–v2 knock-in line targeting the 3′UTR was conceived to allow normal expression of Fgf10. We carried out the initial validation for Fgf10 expression in Fgf10Ki–v2/+ vs. Fgf10+/+ (WT) in the lung of embryonic and postnatal mice isolated at different time-points (Figure 2A). Our results indicated that Fgf10 expression level in Fgf10Ki–v2/+ lungs is comparable to the one observed in the Fgf10+/+ lungs at all these time-points (Figure 2B). Next, we compared Fgf10 vs. Cre expression in Fgf10Ki–v2/+ lungs at different time-points (Figure 2C). Our results indicate a lower level of Cre compared to Fgf10 at all these time-points (Figure 2D). This difference between Cre and Fgf10 expression in Fgf10Ki–v2/+ lungs suggests that the insertion of the Cre-ERT2 cassette in the 3′UTR disrupted the expression of the endogenous Fgf10 gene produced from the recombined allele. Together with Fgf10 expression in Fgf10Ki–v2/+ vs. Fgf10+/+ (WT), this result suggests that Fgf10 expression from the non-recombined allele in Fgf10Ki–v2/+ lungs is increased to compensate the loss of Fgf10 expression from the recombined allele.
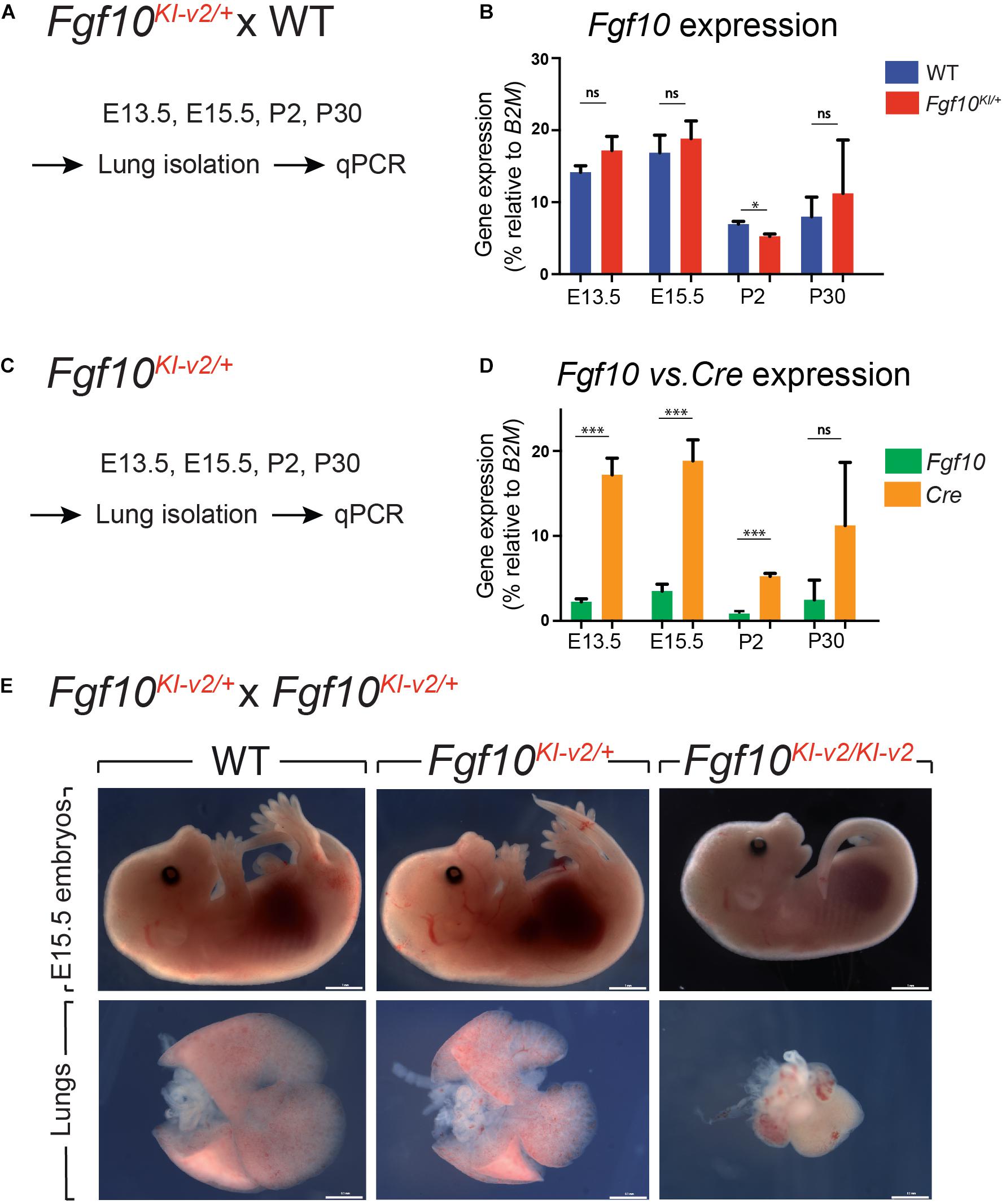
Figure 2. Fgf10Ki-v2 is a loss-of-function allele. (A) Fgf10Ki-v2/+ and Fgf10+/+ mice were crossed and lungs were isolated at E13.5, E15.5, P2, and P30 for qPCR analysis. (B) qPCR results for Fgf10 expression in Fgf10+/+ and Fgf10Ki-v2/+ at these time points. (C,D) Lungs were isolated from Fgf10Ki-v2/+ at different time points and the expression levels of Fgf10 and Cre were analyzed by qPCR. (E) Fgf10Ki-v2/+ animals were crossed to generate Fgf10+/+, Fgf10Ki-v2/+, and Fgf10Ki-v2/Ki-v2 embryos at E13.5. Note the absence of limbs and lungs in the Fgf10Ki-v2/Ki-v2 embryos. Scale bar in (E): whole embryo: 1 mm; dissected lungs: 0.1 mm. *P ≤ 0.05, ***P ≤ 0.001.
To determine whether the insertion of Cre-ERT2 in the endogenous Fgf10 locus led to loss of function of Fgf10, Fgf10Ki–v2 heterozygous animals were self-crossed and embryos were harvested at E15.5. Fgf10Ki–v2/Ki–v2 homozygous embryos suffered from lung and limb agenesis, which is consistent with complete loss of function of Fgf10 (Figure 2E). Analysis of Fgf10 expression by qPCR at that stage indicated a drastic reduction in Fgf10 expression in Fgf10Ki–v2/Ki–v2 embryo (n = 1) compared to Fgf10Ki–v2/+ or WT lungs (Supplementary Figure 1). We therefore conclude that the Fgf10Ki–v2 allele corresponds to a Fgf10 loss-of-function allele.
In order to test the recombinase activity of Cre-ERT2, Fgf10Ki–v2/+ heterozygous mice were crossed with tdTomatoflox/flox reporter mice. Pregnant mice received a single intraperitoneal (IP) injection of tamoxifen at E11.5 (Figure 3A) or E15.5 (Figure 4A). Embryos were harvested at E18.5. No fluorescent signal was observed in Fgf10+/+; tdTomatoflox/+ embryos (Figures 3B, 4B; n = 4) indicating absence of recombination in control embryos and lack of leakiness of the tdTomatoflox allele. By contrast, tamoxifen treatment at E11.5 led to a strong fluorescent signal in the limbs, stomach, cecum, colon and lungs of Fgf10Ki–v2/+; tdTomatoflox/+ embryos (n = 3). In the limb, the labeled cells were more abundant in the digit tip area, known to express high level of Fgf10 (Danopoulos et al., 2013). Along the gastro-intestinal tract, labeled cells were located in the anterior part of the stomach as well as in duodenum (data not shown) which are both reported to express high level of Fgf10 (Lv et al., 2019). A similar observation was made in the cecum and the distal colon (Lv et al., 2019). Throughout the lung, we found a robust tdTomato expression with a higher expression in the interlobular septa. This is similar to what was observed with the previously validated Fgf10LacZ reporter line and Fgf10KI–v1/+ line (Mailleux et al., 2005; El Agha et al., 2012, 2014). Interestingly, in the trachea, no labeled cells were observed in this experimental condition (Figure 3C). Additionally, tamoxifen treatment at E15.5 revealed strong fluorescent signal in the pinna of the developing ear as well as in the trachea and in between the cartilage rings (Figure 4C). These two additional expression domains are consistent with sites of Fgf10 expression (Sala et al., 2011; Zhang et al., 2020). We therefore conclude that Cre expression reflects Fgf10 expression and that this line can be used to target FGF10Pos cells.
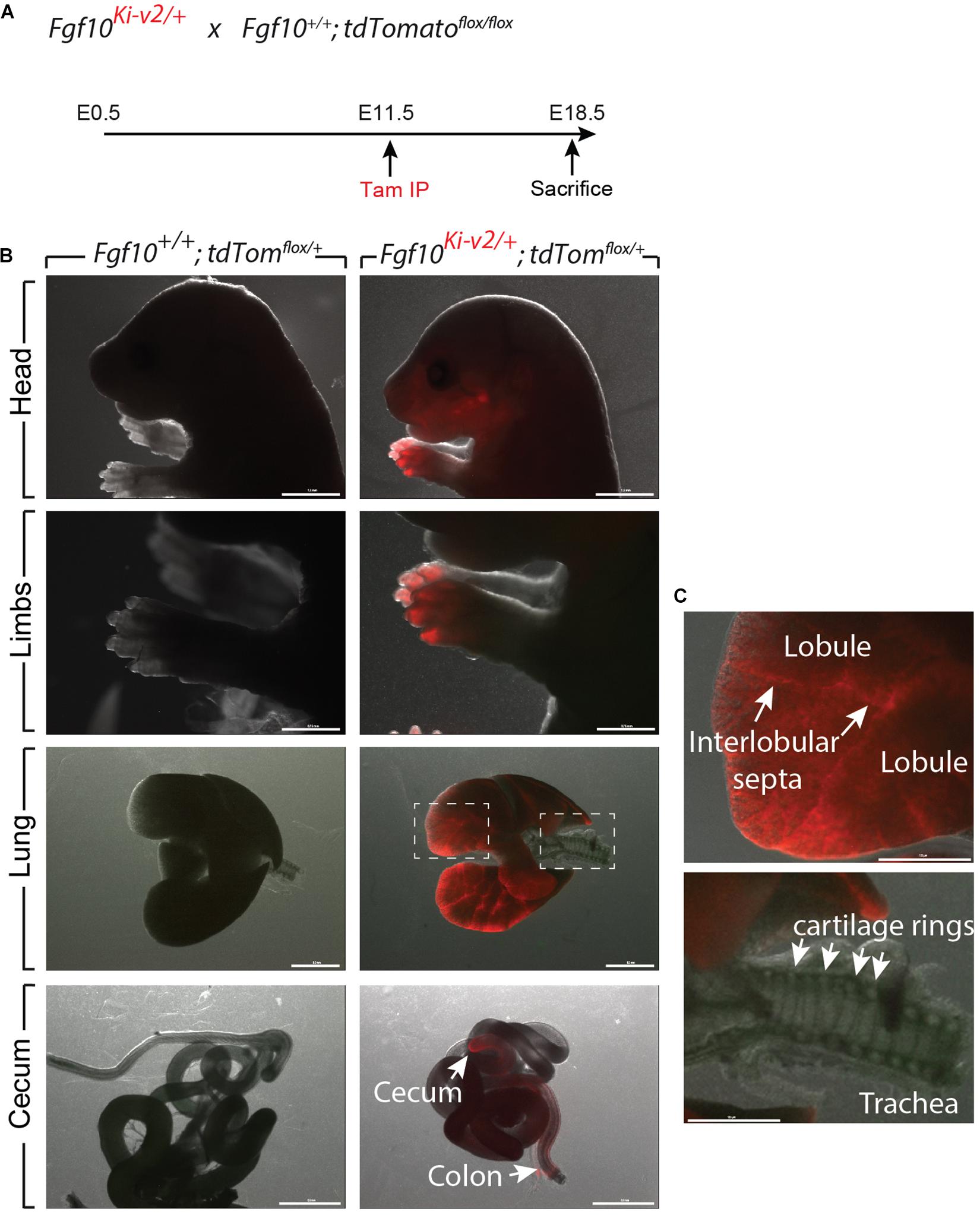
Figure 3. Validation of the labeling of FGF10Pos cells at E11.5. (A) Fgf10Ki-v2/+ were crossed with Fgf10+/+; tdTomflox/flox mice. Pregnant females received a single IP injection of tamoxifen when the embryos were at E11.5 and sacrificed at E18.5. (B) Head, limbs, lung and cecum of Fgf10+/+; tdTomflox/+ and Fgf10Ki-v2/+; tdTomflox/+ embryos are shown. Note the absence of fluorescence in the Fgf10+/+; tdTomflox/+ indicating that the non-recombined LoxP-Stop-LoxP-tdTomato allele is not leaky. (C) Higher magnification of lung and trachea showing enriched tdTomato expression in the interlobular septa and the lack of tdTomato expression between the cartilage rings, respectively. Scale bar in (B): head: 1.5 mm, Limb: 0.75 mm, Lung: 0.5 mm, cecum: 0.5 mm. Scale bar in (C): 125 μm.
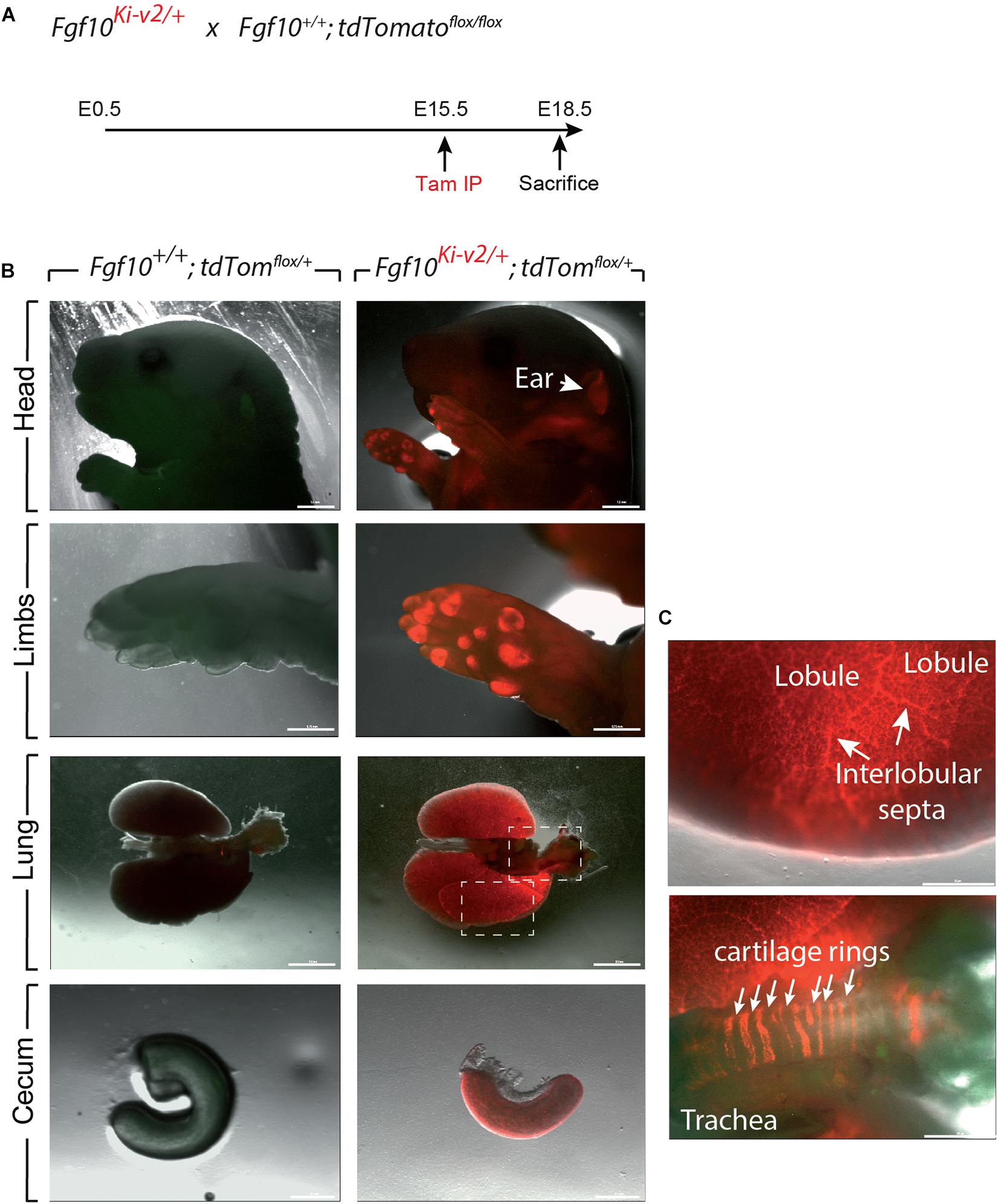
Figure 4. Validation of the labeling of FGF10Pos cells at E15.5. (A) Fgf10Ki-v2/+ were crossed with Fgf10+/+; tdTomflox/flox mice. Pregnant females received a single IP injection of tamoxifen when the embryos were at E15.5 and sacrificed at E18.5. (B) Head, limbs, lung and cecum of Fgf10+/+; tdTomflox/+ and Fgf10Ki-v2/+; tdTomflox/+ embryos are shown. Note the expression in the external ear (arrow). (C) Higher magnification of lung and trachea showing enriched tdTomato expression in the interlobular septa and the presence of tdTomato expression between the cartilage rings, respectively. Scale bar in (B): head: 1.5 mm, Limb: 0.75 mm, Lung: 0.5 mm, Cecum: 0.1 mm. Scale bar in (C): Lung 25 μm, Trachea 125 μm.
Using the previously generated Fgf10Ki–v1/+ line, we demonstrated that FGF10Pos cells labeled postnatally strongly contribute to the lipofibroblast (LIF) lineage but not the smooth muscle cell (SMC) lineage. In particular, they do not contribute in a major way to the ACTA2Pos secondary crest myofibroblasts (SCMF) which are abundant during the first 2–3 weeks during alveologenesis which takes place from postnatal day 5 (P5) to -P28 (El Agha et al., 2014). To confirm this observation with the new Fgf10Ki–v2/+ line, we labeled FGF10Pos cells at P4 and examined the status of the labeled cells at P21, 1 week before the end of the alveologenesis phase (Figure 5A). Analysis of the whole lung by fluorescence stereomicroscopy indicated a much higher number of labeled cells in the Fgf10Ki–v2; tdTomatoflox/+ lung compared with the Fgf10Ki–v1; tdTomatoflox/+ lung (Figure 5B). Quantification of tdTomPos cells indicated that a higher percentile of tdTomPos/DAPI is observed on sections of Fgf10Ki–v2; tdTomatoflox/+ vs. Fgf10Ki–v1; tdTomatoflox/+ (4.7% ± 0.6% vs. 1.5% ± 0.2%, n = 2) thereby confirming the fluorescence stereomicroscopy results (Figure 5C). LipidTOX staining of these lungs was used to visualize LIFs (Figure 5D). Quantification of this staining indicated that 62.6% ± 5.0% (n = 2) of the total tdTomPos are LTPos and that 27.0% ± 4.4% (n = 2) of the LTPos derive from tdTomPos cells. These data are in line with our results obtained with the previous Fgf10Ki–v1/+ line. Immunofluorescence (IF) for ACTA2 on these lungs was also carried out (Figures 5E,F). First, we quantified the number of ACTA2Pos/tdTomPos present in the respiratory airway (Figure 5E) and alveolar space (Supplementary Figure 2). ACTA2Pos cells in the respiratory airway during alveologenesis mark secondary crest myofibroblasts (SCMF). We found 4.7% ± 0.9% (n = 2) tdTomPosACTA2Pos/tdTomPos indicating in our experimental conditions, a minimal commitment of the FGF10Pos cells to the SCMF lineage. Second, we identified peribronchial tdTomPos cells both in longitudinal and cross sections of the bronchi (Figure 5F). Airway smooth muscle cells express ACTA2, display the typical bundle-like circular shape and are located in close proximity to the bronchial epithelium. tdTomPos cells are located close to ACTA2Pos-ASMCs but are nevertheless negative for ACTA2. A similar observation was made for the perivascular tdTomPos cells (data not shown).
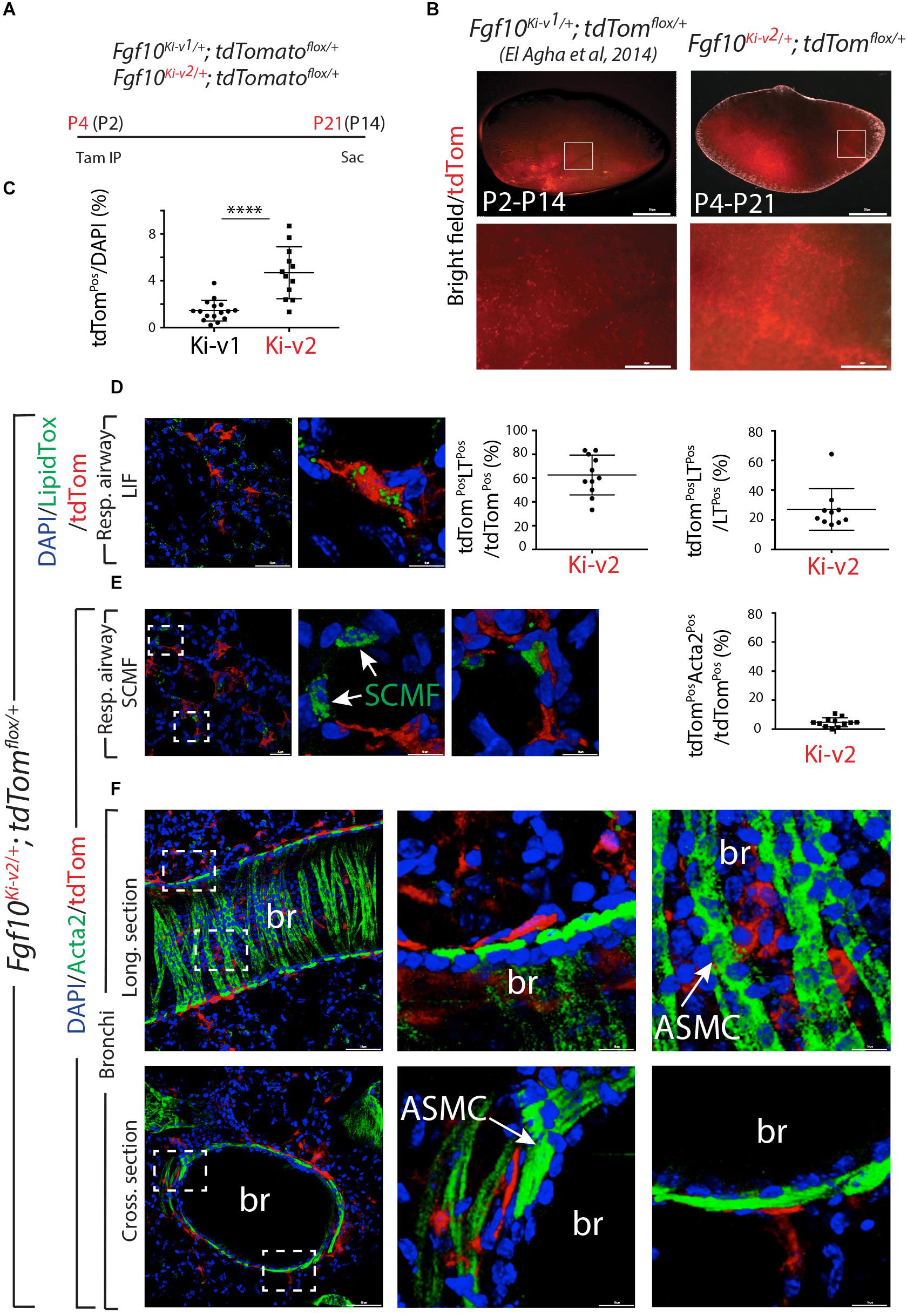
Figure 5. FGF10Pos cells labeled after birth do not contribute significantly to secondary crest myofibroblasts during alveologenesis. (A) FGF10Pos cells in Fgf10Ki-v2/+; tdTomatoflox/+ pups were labeled in vivo at P4 and analyzed at P21. We also used previously generated Fgf10Ki-v1/+; tdTomatoflox/+ samples labeled between P2 and P14. (B) Whole-mount fluorescence images of Fgf10Ki-v2/+; tdTomflox/+ and Fgf10Ki-v1/+; tdTomflox/+ lungs showing more abundant labeled cells in Fgf10Ki-v2/+ vs. Fgf10Ki-v1/+ lungs. (C) ACTA2 IF on Fgf10Ki-v2/+; tdTomflox/+ lungs shows little contribution of FGF10Pos cells to SCMF (ACTA2PostdTomPos/ACTA2Pos). (D) Quantification of tdTomPos cells. Br: bronchi. Scale bar in (B): low magnification: 0.5 mm, High magnification: 50 μm. Scale bar in (D): low magnification: 50 μm, High magnification: 10 μm. Scale bar in (E): Llow magnification: 25 μm, High magnification: 10 μm. Scale bar in (F): Low magnification: 50 μm, High magnification: 10 μm. ****P ≤ 0.0001.
Two months old Fgf10K−v2/+; tdTomatoflox/flox mice were treated with Tam IP or oil at day 1 (D61), 3 (D63), and 5(D65) and the lungs were collected at day 7 (D67) (Figure 6A). No fluorescent signal was observed in oil-treated Fgf10K−v2/+; tdTomatoflox/flox mice, indicating that the line is not leaky. By contrast, a solid signal was found in Tam-treated Fgf10Ki–v2/+; tdTomatoflox/flox lungs. A weak signal was detected in Tam-treated Fgf10Ki–v1/+; tdTomatoflox/flox lungs as described in a previously study (El Agha et al., 2014; Figure 6B). Flow cytometry analysis was also conducted to quantify the total number of tdTomPos cells in both conditions as well as their identity (El Agha et al., 2014; Figure 6C). Only 0.3% tdTomPos cells over total number of cells were detected in Fgf10Ki–v1/+; tdTomatoflox/flox. This number is in line with the previously reported 0.1% (El Agha et al., 2014) and confirms that the Fgf10Ki–v1/+ line is not efficient to target FGF10Pos cells in the adult lung. By contrast, we observed 5.8% of tdTomPos cells over total cells in Fgf10Ki–v2/+ lungs. Further analysis showed that these cells were mostly CD31NegCD45NegEPCAMNeg cells (85%) identifying them as resident mesenchymal cells (rMC). 14.3% of the tdTomPos cells were also SCA1High, a functional marker of the rMC subpopulation capable of sustaining the self-renewal of AT2 stem cells in the alveolosphere organoid model (Taghizadeh et al., 2021). Of note 53% of the SCA1High were LipidTOXPos identifying them as lipofibroblasts (LIFs). Altogether these results suggest that tdTomPos cells are heterogeneous and comprise a significant percentile of LIFs as previously reported (El Agha et al., 2014).
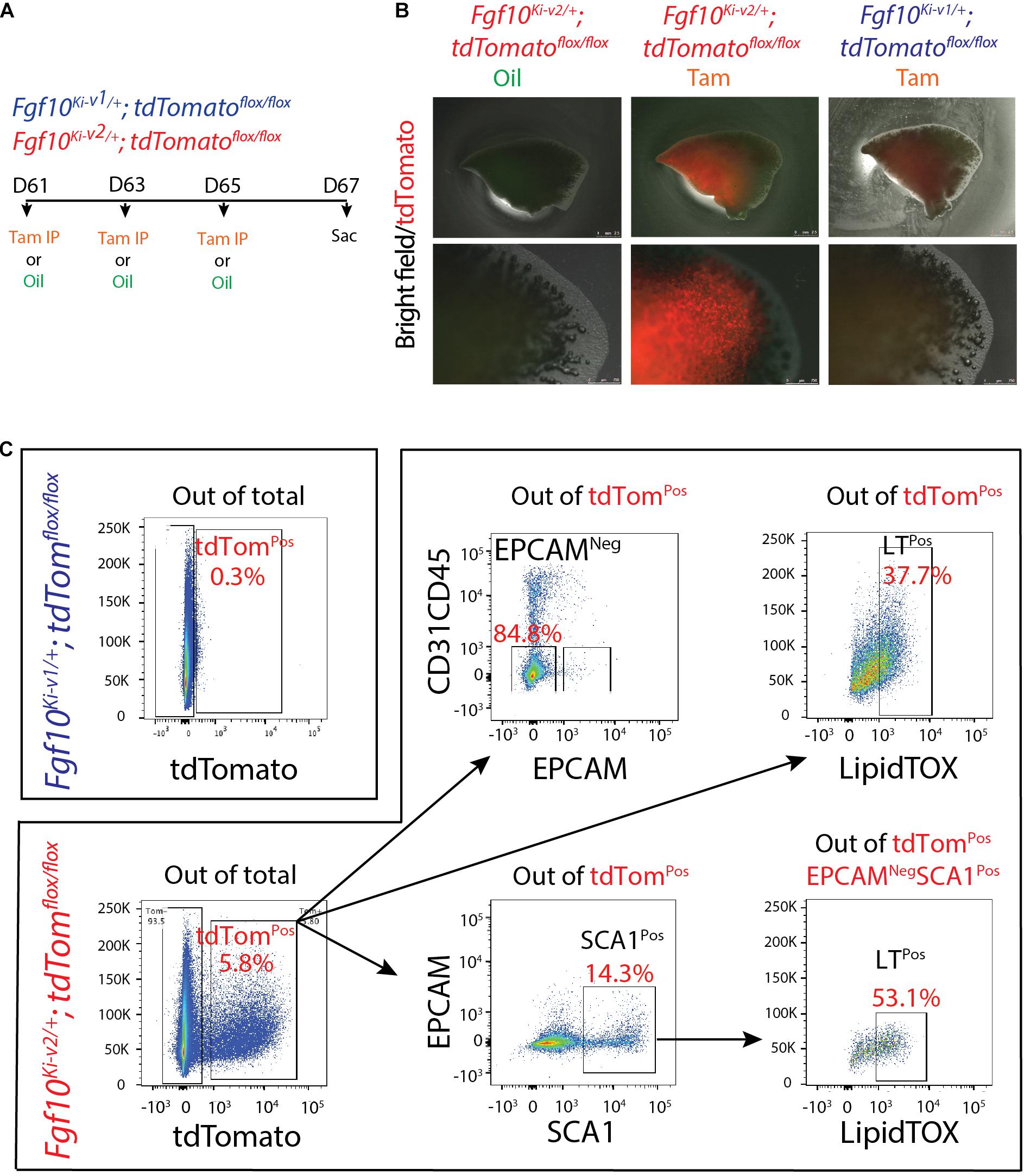
Figure 6. Fgf10Ki-v2/+; tdTomflox/+ adult lungs display enhanced number of tdTomPos cells compared with to the Fgf10Ki-v1/+; tdTomflox/+ adult lungs. (A) 2-months-old; Fgf10Ki-v1/+; tdTomflox/+ and Fgf10Ki-v2/+; tdTomflox/+ mice received 3 Tam IPs or oil at P61, P63 and P65 and were sacrificed at P67. (B) Whole-mount fluorescence images of oil-treated Fgf10Ki-v2/+; tdTomflox/+, Tam-treated Fgf10Ki-v2/+; tdTomflox/+ and Tam-treated Fgf10Ki-v1/+; tdTomflox/+ lungs at P67. Higher magnification of the lungs are shown in the lower panel. (C) Flow cytometry analysis of Fgf10Ki-v2/+; tdTomflox/flox lung homogenate. Scale bar in (B): low magnification: 2.5 mm, High magnification: 0.75 mm.
The decrease in Fgf10 expression in Fg10Ki–v2 mice (Figure 2D) suggested that important transcription factor binding sites (TFBS) were impacted by the genetic manipulation in the 3′UTR of the Fgf10 gene. We determined the identity of TFBS located at proximity of the 3′UTR of the Fgf10 gene using an online TFBS prediction tool. We compared these TFBS with previously published TF expressed in the lung mesenchyme (Herriges et al., 2012). We found several key TFBS matching the previously reported TF expression in the lung such as Hoxa5, Pou3f1, Pou2f1, Foxm1, and Meis1 (Figure 7). Interestingly, all these transcription factors appear to play a functional role in the lung. Mutant Hoxa5 mice display decreased surfactant production and disrupted tracheal cartilage, leading to respiratory distress and low survival rate at birth (Aubin et al., 1997; Kinkead et al., 2004; Mandeville et al., 2006). Pou3F1, also known as Oct6 is primarily expressed in neural cells. Pou3F1 deletion caused lethality at birth due to respiratory distress (Bermingham et al., 1996, 2002; Ghazvini et al., 2002). The deletion of the other related transcription factor, Pou2f1, is associated with smaller body size of embryos and full lethality at birth (Wang et al., 2004). Foxm1 expression plays a crucial role in both the epithelium and the mesenchyme. Conditional inactivation of Foxm1 in the lung mesenchyme leads to increased smooth muscles around the proximal airways and reduced pulmonary microvasculature (Kim et al., 2005). In the lung epithelium, Foxm1 conditional inactivation causes reduction in sacculation and delayed differentiation of alveolar epithelial type I cells (Kalin et al., 2008). Knockout of Meis1 caused lethality during the embryonic stage around E14.5 due to microvascular and hematopoietic defects in the lung (Hisa et al., 2004).
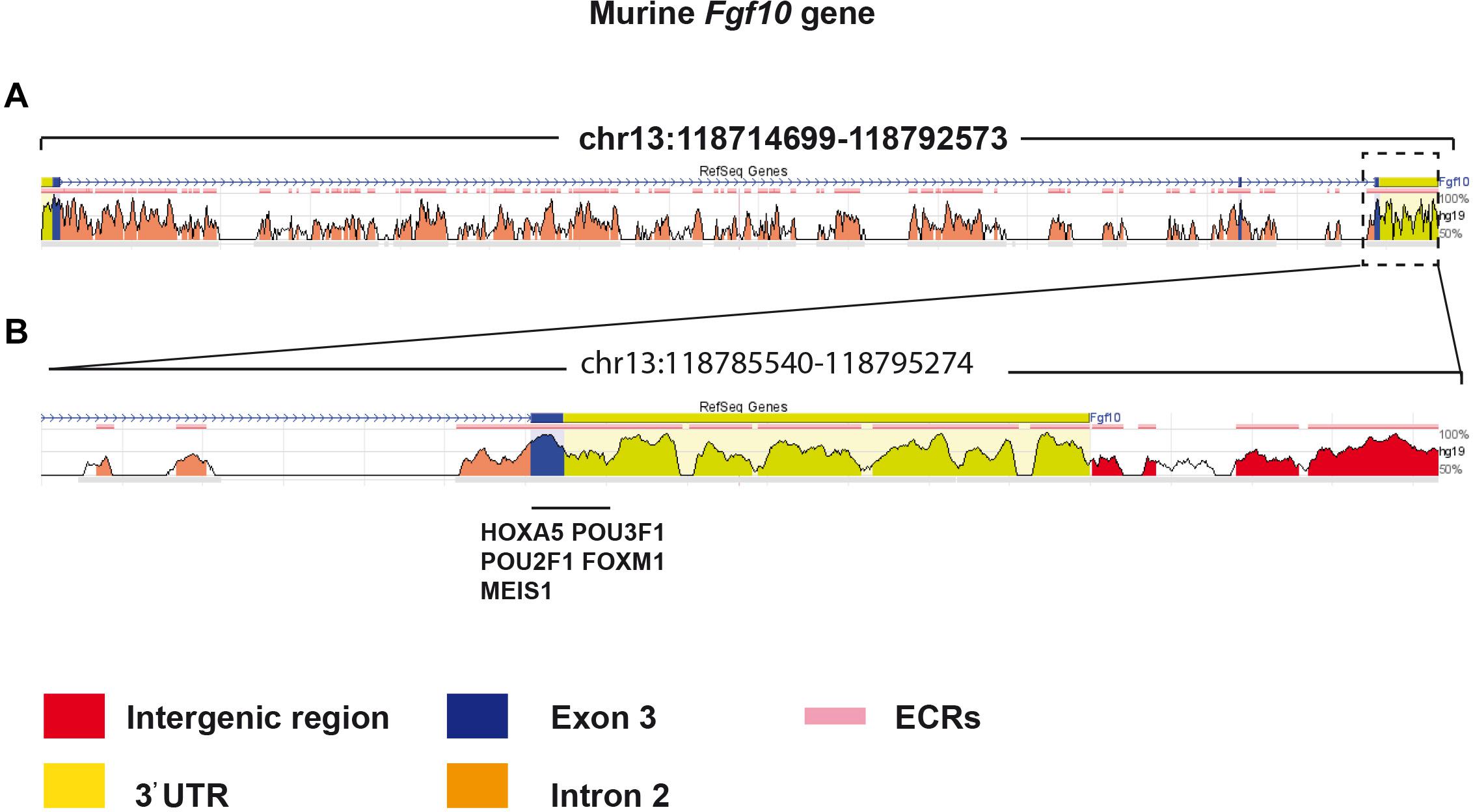
Figure 7. Bioinformatic analysis identified potential transcriptional factors binding sites in the 3′UTR of Exon 3. (A) The murine Fgf10 gene is made of 3 exons. (B) High magnification of Exon 3 and the associated 3′UTR. Several putative important transcriptional factor binding sites were found in this region.
FGF10 is an essential morphogen underlying the developmental process of multiple organs including the lung. FGF10 signaling is also crucial during homeostasis and in the process of injury/repair in the adult lung. FGF10 dysregulation in human has been implicated in some major respiratory diseases, such as bronchopulmonary dysplasia (BPD), Idiopathic pulmonary fibrosis (IPF) and chronic obstructive pulmonary disease (COPD) (Yuan et al., 2018). For example, increased FGF10 expression level in IPF patients has been found (El Agha et al., 2017). However, FGF10 expression is inversely correlated to the disease progression with higher levels in stable IPF vs. lower level in end-stage IPF. Higher FGF10 expression in the early, stable stage of IPF is most likely correlated with the repair process. Insufficient FGF10 level in prematurely newborn infants is associated with arrested lung development at the saccular stage (Prince, 2018). Fgf10 deficiency in a newborn mouse model of hyperoxia-induced BPD led to drastic increase in lethality associated with abnormal alveolar epithelial type 2 (AT2) cell differentiation as well as surfactant production (Chao et al., 2017).
FGF10 also performs a key function for the repair of the bronchial epithelium after injury (Volckaert et al., 2011). Our knowledge about the sources of FGF10 in this context has been evolving. FGF10 was first described to be expressed by airway smooth muscle cells (ASMCs) (Volckaert et al., 2011), whereas more recent work identified a peribronchiolar mesenchymal population capable of producing FGF10 during the repair process, which is not derived from the ASMCs (Moiseenko et al., 2020).
In COPD, the conducting airway epithelium undergoes massive remodeling causing an irreversible airway obstruction (Decramer et al., 2012). Interestingly, we have reported that the FGF10-HIPPO epithelial mesenchymal crosstalk also maintains and recruits lung basal stem cells in the conducting airways (Volckaert et al., 2017). While transient Fgf10 expression by ASMCs is critical for proper airway epithelial regeneration in response to injury, sustained FGF10 secretion by the ASMC niche, in response to chronic ILK/HIPPO inactivation, results in pathological changes in airway architecture resembling the abnormalities seen in COPD. The inhibition of FGF10/FGFR2b signaling may therefore be an interesting approach to treat chronic obstructive airway lung diseases. Conversely, the opposite situation might occur in the respiratory airways in that destruction of the alveolar compartments resulting in emphysema may be due to insufficient FGF signaling. Interestingly, recombinant FGF7 has been reported to induce de novo-alveologenesis in the elastase model of emphysema in mice (Yildirim et al., 2010).
The previous Fgf10Ki–V1 model was mainly used to trace the FGF10Pos cells during embryonic development. A near complete loss of the labeling capacity of FGF10Pos cells during postnatal stages limited its utilization in the analysis of their cell fate in adult lung homeostasis and during the process of injury/repair. In order to overcome the limitations of the Fgf10Ki–V1 line, we generated and validated this new knock-in Fgf10Ki–V2 line. Upon crossing with a tdTomato reporter line, we demonstrated that the tdTomato expression domain faithfully reproduced the previously reported the Fgf10 expression pattern (El Agha et al., 2012), and a more robust labeling of FGF10Pos cells was achieved in the postnatal stages in spite of a mismatch between Cre and Fgf10 expression, which could be explained by the disruption of critical TFBS located in the 3′UTR of the Fgf10 gene. Therefore, this line will be a valuable tool to further define mesenchymal cell populations in the adult lung contributing to the repair process after injury. Combined crosses with existing or novel Dre-ERT2 recombinase driver lines may allow to capture subpopulations of FGF10Pos cells/lineages based on the expression of two markers (Jones et al., 2019). The main FGF10Pos subpopulation is represented by the lipid-containing alveolar interstitial fibroblasts (lipofibroblasts or LIFs). More and more studies have acknowledged LIFs as an essential piece of the AT2 stem cell niche in the rodent lungs. Despite the fact that LIFs were initially believed to only assist AT2 cells in surfactant production during neonatal life, recent studies have shown that these cells are important for self-renewal and differentiation of AT2 stem cells during adulthood (Barkauskas et al., 2013). In spite of the increasing interests in lipofibroblast biology, little is known about their cellular origin or the molecular pathways that control their formation during embryonic development. We have shown that in the developing mouse lung, FGF10Pos cells labeled at E11.5 or E15.5 are progenitors for LIFs (El Agha et al., 2014). In addition, FGF10 is also essential for the differentiation of these progenitors into the LIF lineage (Al Alam et al., 2015). We have also reported the existence of FGF10Pos-LIF as well as FGF10Neg-LIFs (Al Alam et al., 2015). The difference between these two populations is still unclear and will require further studies. In the context of bleomycin-induced lung fibrosis, in vivo lineage tracing indicates that LIFs transdifferentiate into activated myofibroblast during fibrosis formation and that a significant proportion of the labeled activated myofibroblasts transdifferentiate back to LIFs during fibrosis resolution (El Agha et al., 2017).
In conclusion, we have successfully generated a new Fgf10Cre–ERT2 line with enhanced labeling efficiency of FGF10Pos cells postnatally. This line, which displays normal expression of Fgf10 in Fgf10Cre–ERT2/+, avoids many developmental defects linked to deficient Fgf10 expression. Therefore, it paves the way for performing cell-autonomous based studies to investigate the role of these FGF10Pos cells as well as associated signaling pathways during lung development and disease.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Animal experiments were reviewed and approved by the Regierungspraesidium Giessen (approval number RP GI/47- 2019).
XC, ST, AIV-A, and LC performed the experiments. SH, CC, and J-SZ contributed to methodology. EEA and SB conceived the study. XC and SB wrote the manuscript. J-SZ, EEA, and SB edited the manuscript. All authors contributed to the article and approved the submitted version.
EEA acknowledges the support of the Institute for Lung Health (ILH), the German Research Foundation (DFG; EL 931/4-1, KFO309 P7, and SFB CRC1213-project A04), the Cardio-Pulmonary Institute (CPI, EXC 2026, Project ID: 390649896) and the German Center for Lung Research (DZL). SB was supported by the CPI and by grants from the DFG (BE4443/1-1, BE4443/4-1, BE4443/6-1, KFO309 P7, and SFB1213-projects A02 and A04). CC was supported by the Interventional Pulmonary Key Laboratory of Zhejiang Province, the Interventional Pulmonology Key Laboratory of Wenzhou City, the Interventional Pulmonology Innovation Subject of Zhejiang Province, the National Nature Science Foundation of China (81570075 and 81770074), Zhejiang Provincial Natural Science Foundation (LZ15H010001), Zhejiang Provincial Science Technology Department Foundation (2015103253), and the National Key Research and Development Program of China (2016YFC1304000).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.671841/full#supplementary-material
Supplementary Figure 1 | Comparative Fgf10 expression in Fgf10+/+ and Fgf10Ki–v2/+ lungs vs. Fgf10Ki–v2/Ki–v2 embryos at E15.5. qPCR was used to determine Fgf10 expression.
Supplementary Figure 2 | Co-expression of ACTA2 and tdTomato in FGF10Pos cells. Fgf10Ki–v1; tdTomatoflox/+ pups received one injection of Tam subcutaneously at P2 and were analyzed at P14.
Al Alam, D., El Agha, E., Sakurai, R., Kheirollahi, V., Moiseenko, A., Danopoulos, S., et al. (2015). Evidence for the involvement of fibroblast growth factor 10 in lipofibroblast formation during embryonic lung development. Development 142, 4139–4150. doi: 10.1242/dev.109173
Aubin, J., Lemieux, M., Tremblay, M., Bérard, J., and Jeannotte, L. (1997). Early postnatal lethality in Hoxa-5 mutant mice is attributable to respiratory tract defects. Dev. Biol. 192, 432–445. doi: 10.1006/dbio.1997.8746
Barkauskas, C. E., Cronce, M. J., Rackley, C. R., Bowie, E. J., Keene, D. R., Stripp, B. R., et al. (2013). Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025–3036.
Bermingham, J. R. Jr., Scherer, S. S., O’Connell, S., Arroyo, E., Kalla, K. A., Powell, F. L., et al. (1996). Tst-1/Oct-6/SCIP regulates a unique step in peripheral myelination and is required for normal respiration. Genes Dev. 10, 1751–1762. doi: 10.1101/gad.10.14.1751
Bermingham, J. R. Jr., Shumas, S., Whisenhunt, T., Sirkowski, E. E., O’Connell, S., Scherer, S. S., et al. (2002). Identification of genes that are downregulated in the absence of the POU domain transcription factor pou3f1 (Oct-6. Tst-1, SCIP) in sciatic nerve. J. Neurosci. 22, 10217–10231. doi: 10.1523/jneurosci.22-23-10217.2002
Chao, C. M., Yahya, F., Moiseenko, A., Tiozzo, C., Shrestha, A., Ahmadvand, N., et al. (2017). Fgf10 deficiency is causative for lethality in a mouse model of bronchopulmonary dysplasia. J. Pathol. 241, 91–103.
Danopoulos, S., Parsa, S., Al Alam, D., Tabatabai, R., Baptista, S., Tiozzo, C., et al. (2013). Transient Inhibition of FGFR2b-ligands signaling leads to irreversible loss of cellular beta-catenin organization and signaling in AER during mouse limb development. PLoS One 8:e76248. doi: 10.1371/journal.pone.0076248
Decramer, M., Janssens, W., and Miravitlles, M. (2012). Chronic obstructive pulmonary disease. Lancet 379, 1341–1351.
El Agha, E., Al Alam, D., Carraro, G., MacKenzie, B., Goth, K., De Langhe, S. P., et al. (2012). Characterization of a novel fibroblast growth factor 10 (Fgf10) knock-in mouse line to target mesenchymal progenitors during embryonic development. PLoS One 7:e38452. doi: 10.1371/journal.pone.0038452
El Agha, E., Herold, S., Al Alam, D., Quantius, J., MacKenzie, B., Carraro, G., et al. (2014). Fgf10-positive cells represent a progenitor cell population during lung development and postnatally. Development 141, 296–306. doi: 10.1242/dev.099747
El Agha, E., Moiseenko, A., Kheirollahi, V., De Langhe, S., Crnkovic, S., Kwapiszewska, G., et al. (2017). Two-way conversion between lipogenic and myogenic fibroblastic phenotypes marks the progression and resolution of lung fibrosis. Cell Stem Cell 20, 261.e3–273.e3.
Feil, R., Wagner, J., Metzger, D., and Chambon, P. (1997). Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem. Biophys. Res. Commun. 237, 752–757. doi: 10.1006/bbrc.1997.7124
Ghazvini, M., Mandemakers, W., Jaegle, M., Piirsoo, M., Driegen, S., Koutsourakis, M., et al. (2002). A cell type-specific allele of the POU gene Oct-6 reveals Schwann cell autonomous function in nerve development and regeneration. Embo J. 21, 4612–4620. doi: 10.1093/emboj/cdf475
Gupte, V. V., Ramasamy, S. K., Reddy, R., Lee, J., Weinreb, P. H., Violette, S. M., et al. (2009). Overexpression of fibroblast growth factor-10 during both inflammatory and fibrotic phases attenuates bleomycin-induced pulmonary fibrosis in mice. Am. J. Respir. Crit. Care Med. 180, 424–436. doi: 10.1164/rccm.200811-1794oc
Herriges, J. C., Yi, L., Hines, E. A., Harvey, J. F., Xu, G., Gray, P. A., et al. (2012). Genome-scale study of transcription factor expression in the branching mouse lung. Dev. Dyn. 241, 1432–1453. doi: 10.1002/dvdy.23823
Hisa, T., Spence, S. E., Rachel, R. A., Fujita, M., Nakamura, T., Ward, J. M., et al. (2004). Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. Embo J. 23, 450–459. doi: 10.1038/sj.emboj.7600038
Jaskoll, T., Abichaker, G., Witcher, D., Sala, F. G., Bellusci, S., Hajihosseini, M. K., et al. (2005). FGF10/FGFR2b signaling plays essential roles during in vivo embryonic submandibular salivary gland morphogenesis. BMC Dev. Biol. 5:11. doi: 10.1186/1471-213X-5-11
Jones, M., Zhang, J.-S., and Bellusci, S. (2019). Bronchioalveolar stem cells vindicated! Biotarget 3:4. doi: 10.21037/biotarget.2019.04.01
Jones, M. R., Chong, L., and Bellusci, S. (2020). Fgf10/Fgfr2b signaling orchestrates the symphony of molecular, cellular, and physical processes required for harmonious airway branching morphogenesis. Front. Cell Dev. Biol. 8:620667. doi: 10.3389/fcell.2020.620667
Kalin, T. V., Wang, I. C., Meliton, L., Zhang, Y., Wert, S. E., Ren, X., et al. (2008). Forkhead Box m1 transcription factor is required for perinatal lung function. Proc. Natl. Acad. Sci. U.S.A. 105, 19330–19335. doi: 10.1073/pnas.0806748105
Kim, I. M., Ramakrishna, S., Gusarova, G. A., Yoder, H. M., Costa, R. H., and Kalinichenko, V. V. (2005). The forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J. Biol. Chem. 280, 22278–22286. doi: 10.1074/jbc.m500936200
Kinkead, R., LeBlanc, M., Gulemetova, R., Lalancette-Hébert, M., Lemieux, M., Mandeville, I., et al. (2004). Respiratory adaptations to lung morphological defects in adult mice lacking Hoxa5 gene function. Pediatr. Res. 56, 553–562. doi: 10.1203/01.pdr.0000139427.26083.3d
Lv, Y. Q., Wu, J., Li, X. K., Zhang, J. S., and Bellusci, S. (2019). Role of FGF10/FGFR2b signaling in mouse digestive tract development, repair and regeneration following injury. Front. Cell Dev. Biol. 7:326.
Mailleux, A. A., Kelly, R., Veltmaat, J. M., De Langhe, S. P., Zaffran, S., Thiery, J. P., et al. (2005). Fgf10 expression identifies parabronchial smooth muscle cell progenitors and is required for their entry into the smooth muscle cell lineage. Development 132, 2157–2166. doi: 10.1242/dev.01795
Mailleux, A. A., Spencer-Dene, B., Dillon, C., Ndiaye, D., Savona-Baron, C., Itoh, N., et al. (2002). Role of FGF10/FGFR2b signaling during mammary gland development in the mouse embryo. Development 129, 53–60.
Mandeville, I., Aubin, J., LeBlanc, M., Lalancette-Hébert, M., Janelle, M. F., Tremblay, G. M., et al. (2006). Impact of the loss of Hoxa5 function on lung alveogenesis. Am. J. Pathol. 169, 1312–1327. doi: 10.2353/ajpath.2006.051333
Min, H., Danilenko, D. M., Scully, S. A., Bolon, B., Ring, B. D., Tarpley, J. E., et al. (1998). Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 12, 3156–3161. doi: 10.1101/gad.12.20.3156
Moiseenko, A., Vazquez-Armendariz, A. I., Kheirollahi, V., Chu, X., Tata, A., Rivetti, S., et al. (2020). Identification of a repair-supportive mesenchymal cell population during airway epithelial regeneration. Cell Rep. 33:108549. doi: 10.1016/j.celrep.2020.108549
Ohuchi, H., Hori, Y., Yamasaki, M., Harada, H., Sekine, K., Kato, S., et al. (2000). FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem. Biophys. Res. Commun. 277, 643–649. doi: 10.1006/bbrc.2000.3721
Parsa, S., Kuremoto, K.-I., Seidel, K., Tabatabai, R., MacKenzie, B., Yamaza, T., et al. (2010). Signaling by FGFR2b controls the regenerative capacity of adult mouse incisors. Development 137, 3743–3752. doi: 10.1242/dev.051672
Parsa, S., Ramasamy, S. K., De Langhe, S., Gupte, V. V., Haigh, J. J., Medina, D., et al. (2008). Terminal end bud maintenance in mammary gland is dependent upon FGFR2b signaling. Dev. Biol. 317, 121–131. doi: 10.1016/j.ydbio.2008.02.014
Prince, L. S. (2018). FGF10 and human lung disease across the life spectrum. Front. Genet. 9:517. doi: 10.3389/fgene.2018.00517
Sala, F. G., Del Moral, P.-M., Tiozzo, C., Alam, D. A., Warburton, D., Grikscheit, T., et al. (2011). FGF10 controls the patterning of the tracheal cartilage rings via Shh. Development 138, 273–282. doi: 10.1242/dev.051680
Sekine, K., Ohuchi, H., Fujiwara, M., Yamasaki, M., Yoshizawa, T., Sato, T., et al. (1999). Fgf10 is essential for limb and lung formation. Nat. Genet. 21, 138–141. doi: 10.1038/5096
Taghizadeh, S., Heiner, M., Wilhelm, J., Herold, S., Chen, C., Zhang, J., et al. (2021). Characterization in mice of the stromal niche maintaining AT2 stem cell self-renewal in homeostasis and disease. bioRxiv [Preprint]. doi: 10.1101/2021.01.28.428090
Volckaert, T., Dill, E., Campbell, A., Tiozzo, C., Majka, S., Bellusci, S., et al. (2011). Parabronchial smooth muscle constitutes an airway epithelial stem cell niche in the mouse lung after injury. J. Clin. Invest. 121, 4409–4419. doi: 10.1172/jci58097
Volckaert, T., Yuan, T., Chao, C. M., Bell, H., Sitaula, A., Szimmtenings, L., et al. (2017). Fgf10-hippo epithelial-mesenchymal crosstalk maintains and recruits lung basal stem cells. Dev. Cell 43, 48.e5–59.e5.
Wang, V. E., Schmidt, T., Chen, J., Sharp, P. A., and Tantin, D. (2004). Embryonic lethality, decreased erythropoiesis, and defective octamer-dependent promoter activation in Oct-1-deficient mice. Mol. Cell Biol. 24, 1022–1032. doi: 10.1128/mcb.24.3.1022-1032.2004
Yildirim, A. O., Muyal, V., John, G., Müller, B., Seifart, C., Kasper, M., et al. (2010). Palifermin induces alveolar maintenance programs in emphysematous mice. Am. J. Respir. Crit. Care Med. 181, 705–717. doi: 10.1164/rccm.200804-573oc
Yuan, T., Volckaert, T., Chanda, D., Thannickal, V. J., and De Langhe, S. P. (2018). Fgf10 signaling in lung development, homeostasis, disease, and repair after injury. Front. Genet. 9:418. doi: 10.3389/fgene.2018.00418
Keywords: Fgf10, knock-in Cre line, lipofibroblast, adult lung, lineage tracing
Citation: Chu X, Taghizadeh S, Vazquez-Armendariz AI, Herold S, Chong L, Chen C, Zhang J-S, El Agha E and Bellusci S (2021) Validation of a Novel Fgf10Cre–ERT2 Knock-in Mouse Line Targeting FGF10Pos Cells Postnatally. Front. Cell Dev. Biol. 9:671841. doi: 10.3389/fcell.2021.671841
Received: 24 February 2021; Accepted: 25 March 2021;
Published: 13 May 2021.
Edited by:
Zhouguang Wang, Albert Einstein College of Medicine, United StatesReviewed by:
Helen P. Makarenkova, The Scripps Research Institute, United StatesCopyright © 2021 Chu, Taghizadeh, Vazquez-Armendariz, Herold, Chong, Chen, Zhang, El Agha and Bellusci. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-San Zhang, emhhbmcuamluc2FuQFdNVS5lZHUuY24=; Elie El Agha, RWxpZS5FbC1BZ2hhQGlubmVyZS5tZWQudW5pLWdpZXNzZW4uZGU=; Saverio Bellusci, c2F2ZXJpby5iZWxsdXNjaUBpbm5lcmUubWVkLnVuaS1naWVzc2VuLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.