
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Cell Dev. Biol. , 30 April 2021
Sec. Stem Cell Research
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.670654
This article is part of the Research Topic Hematopoiesis: Learning from in vitro and in vivo Models View all 10 articles
A correction has been applied to this article in:
Corrigendum: Mutation of Gemin5 causes defective hematopoietic stem/progenitor cells proliferation in zebrafish embryonic hematopoiesis
 Xiaofen Liu1
Xiaofen Liu1 Wenjuan Zhang2
Wenjuan Zhang2 Changbin Jing2
Changbin Jing2 Lei Gao2
Lei Gao2 Cong Fu2
Cong Fu2 Chunguang Ren2
Chunguang Ren2 Yimei Hao2
Yimei Hao2 Mengye Cao2
Mengye Cao2 Ke Ma2,3*
Ke Ma2,3* Weijun Pan1,2*
Weijun Pan1,2* Dantong Li1,2*
Dantong Li1,2*Fate determination and expansion of Hematopoietic Stem and Progenitor Cells (HSPCs) is tightly regulated on both transcriptional and post-transcriptional level. Although transcriptional regulation of HSPCs have achieved a lot of advances, its post-transcriptional regulation remains largely underexplored. The small size and high fecundity of zebrafish makes it extraordinarily suitable to explore novel genes playing key roles in definitive hematopoiesis by large-scale forward genetics screening. Here, we reported a novel zebrafish mutant line gemin5cas008 with a point mutation in gemin5 gene obtained by ENU mutagenesis and genetic screening, causing an earlier stop codon next to the fifth WD repeat. Gemin5 is an RNA-binding protein with multifunction in post-transcriptional regulation, such as regulating the biogenesis of snRNPs, alternative splicing, stress response, and translation control. The mutants displayed specific deficiency in definitive hematopoiesis without obvious defects during primitive hematopoiesis. Further analysis showed the impaired definitive hematopoiesis was due to defective proliferation of HSPCs. Overall, our results indicate that Gemin5 performs an essential role in regulating HSPCs proliferation.
In vertebrates, embryonic development of blood system involves two waves of hematopoiesis: the primitive hematopoiesis and the definitive hematopoiesis. In definitive wave, the fate of Hematopoietic Stem and Progenitor Cells (HSPCs) is determined in the aorta-gonad-mesonephros (AGM) region, and then generating all blood lineages throughout the life (Galloway and Zon, 2003). Ontogeny from HSPCs fate determination to expansion and then terminal differentiation of types of blood cells requires an orchestrated regulation of transcriptional networks and signaling pathways, however, its underlying mechanism remains largely unknown.
Recently, zebrafish as a wonderful vertebrate model system, has become popular in hematopoiesis research. With high fecundity and small size, zebrafish extremely suits large-scale forward genetics screening (Gao et al., 2015). Taking the benefit of the external fertilization, we can observe and manipulate on zebrafish embryos as early as the one-cell stage. In addition, definitive hematopoiesis is highly conserved between zebrafish and mammals (Song et al., 2004; Gore et al., 2018). In zebrafish, HSPCs emerge from the ventral wall of the dorsal aorta, equals to the AGM in mammals, through endothelium to hematopoietic transition (Kissa and Herbomel, 2010). Then they migrate and colonize in the caudal hematopoietic tissue (CHT), which is similar to mammalian fetal liver, for HSPCs rapid expansion and the differentiation of erythroid and myeloid cells (Ciau-Uitz et al., 2014). By 4 days post-fertilization (dpf), the HSPCs migrate to the kidney marrow, analogous to the bone marrow of mammals (Murayama et al., 2006). Although the main hematopoietic sites are different in zebrafish and mammals, both of them share the same major blood cell types arising from common hematopoietic lineages (Traver et al., 2003), which means findings of blood development in zebrafish could be translated to mammalian systems.
Normal function of HSPCs is sustained through an orchestrated regulation at transcriptional and post-transcriptional level. Major advances have been achieved in the understanding of transcriptional regulation of HSPCs; however, it remains underexplored for the post-transcriptional regulation. RNA-binding proteins (RBPs) take control of post-transcriptional regulation in hematopoiesis through modulating RNA properties including RNA splicing, localization, degradation, or translation (de Rooij et al., 2019). It has been reported RBPs have essential roles in regulating cell fate determination and cell proliferation in stem cell biology (Grech and von Lindern, 2012; de Rooij et al., 2019). Gemin5, an RBP with multidomain, consists of the N-terminal tryptophan-aspartic acid (WD) repeat domains, a dimerization domain in the middle, and a non-canonical RNA-binding domain in the C-terminal region (Martinez-Salas et al., 2020). Its WD repeat motifs recognize the small nuclear RNAs (snRNAs), allowing assembly of the survival of motor neurons (SMN) complex into small nuclear ribonucleoproteins (snRNPs; Pineiro et al., 2015; Martinez-Salas et al., 2020). Structure analyses show the dimerization module of Gemin5 acts a fundamental part in its architecture and essential for its activity (Martinez-Salas et al., 2020). The non-canonical RNA-binding domain allows Gemin5 to interact with its mRNA targets and play a key role in translation. Research on Gemin5 is emerging but far from enough to understand the consequence of its multifunction.
Since ENU-mutagenesis combined with forward genetic screening is a powerful approach to unbiasedly uncover novel genetic and molecular pathways, we conducted it in zebrafish model organism to dissect the sophisticated mechanisms of definitive hematopoiesis (Gao et al., 2015; Jia et al., 2015). We first generated mutants by mutagen ethylnitrosourea (ENU). Then we did whole mount in situ hybridization (WISH) on the embryos of F2 families with myb probe, a crucial marker and transcription factor of HSPCs. We obtained a new mutant cas008 showing severe hematopoietic defects. Then we accomplished positional cloning and located the mutation in gene gemin5. Furthermore, we found abnormal definitive hematopoiesis was due to defective proliferation of HSPCs. To sum up, we reported a zebrafish mutant family carrying Gemin5 point mutation with severe definitive hematopoiesis defects due to HSPCs proliferation failure.
The zebrafish facility and studies were approved by the Animal Research Advisory Committee of Shanghai Institute of Nutrition and Health, CAS. Zebrafish were kept in accordance with the guidelines of the Institutional Animal Care and Use Committee (Li et al., 2018).
The transgenic zebrafish line Tg(myb: eGFP; North et al., 2007) was described previously. For the forward genetics screen, Wild-type (WT) TU zebrafish line was treated with ENU, Sigma (Gao et al., 2015; Jia et al., 2015). For positional cloning, we first outcrossed heterozygous mutantcas008 in Tu background with wild-type in polymorphic WIK background and then mapped the mutantcas008 allele in Tu background as described previously (Bahary et al., 2004).
Based on the myb expression in zebrafish embryos at 5 dpf, the mutantcas008 line was identified among numerous ENU-mutagenized F2 families. We carried on the ENU screen and positional cloning as mentioned before (Bahary et al., 2004; Gao et al., 2015; Jia et al., 2015). By bulk segregation analysis with sequence length polymorphism (SSLP) markers, the mutation was first mapped to chromosome 21. To narrow down the genetic interval, fine mapping was performed and identified the mutation flanked by two SSLP markers, zK189O20 and zC153M12. Then, we cloned the cDNAs of candidate genes in the range and sequenced from siblings and mutants separately. Finally, we sequenced genomic DNA of individual mutant embryos to confirm the putative mutation.
We cloned the zebrafish Gemin5 (accession number: ZDB-GENE-031112-9) and inserted it into the Tol2 backbone between the promoter UAS and p2A (primer F: atgcacgaaagacatctgcc; primer R: gtgtatgagtctcctcacag) (Schematic diagram in Figure 2C). For transient transgene, Tol2 transposase mRNA (40 pg) and the Gemin5 construct or Tol2 vector (40 pg) were microinjected into one-cell-stage embryos (Suster et al., 2009).
The myb, ae1-globin, lyz, mpx, rag1, gata1, pu.1, and kdrl probe were transcribed by polymerase T3 or T7 (Ambion) with Digoxigenin RNA Labeling Mix (Roche) in vitro (Li et al., 2018). WISH was carried out as described before using NBT/BCIP (Sigma) as substrates. To detect HSPCs proliferation, we performed fluorescence whole mount in situ hybridization (FISH) and pH3 immunofluorescence double-staining as described previously (Gao et al., 2015; Jia et al., 2015). We first stained embryos with cy3 (TSA system, Perkin Elmer), and used anti-pH3 (ser10) antibody (1:500; Santa Cruz) for pH3 immunofluorescence staining, and then imaged by Olympus FV1000 Fluoview scanning confocal microscope. With in situ Cell Death Detection Kit and TMR Red Kit (Roche), TUNEL assay was conducted according to manufacturer’s instruction. Embryos in Tg(myb:eGFP) background were fixed with 4% PFA. After pretreatment, the embryos were stained at 37°C for 2 h using the TUNEL Kit (100 μl, enzyme: labeling solution = 1:9). Next, the immunostaining of eGFP was performed with incubation of anti-GFP (Rabbit, 1:500, Invitrogen) and secondary antibody goat-Alexa Fluore488-conjugated anti-rabbit (1:500, Invitrogen). Then, the embryos were imaged by confocal microscope Olympus FV1000 Fluoview scanning.
Images of zebrafish immunofluorescence staining (Figures 3A,B and Supplementary Figure 3A) were taken by Olympus FV1000 Fluoview scanning confocal microscope. 1% low-melt agarose was used to mount the embryos. These confocal images were captured by UPLSAPO 20X objective. Scale bars, 50 μm.
Data were analyzed with the two-tailed Student’s t-test using the software Graphpad Prism 8. Error values in the plots were calculated by standard error of the mean (SEM). In this study, all data were repeated for at least twice.
To explore novel genes which played essential roles in definitive hematopoiesis, we used zebrafish model organism to perform a large-scale forward genetics screen (Gao et al., 2015; Jia et al., 2015). First of all, we generated mutants by crossing zebrafish previously treated with ENU mutagen. Then we tested myb expression by WISH on the embryos of F2 families. As a key transcription factor and marker of HSPCs, myb was expressed in all hematopoietic tissues of sibling embryos at 5 dpf, including CHT, thymus and kidney tested by WISH (Figure 1A). While mutantcas008 displayed dramatically decreased myb expression in CHT, thymus and kidney suggesting defective definitive hematopoiesis (Figure 1A′). The cell marker of downstream hematopoietic lineages, including ae1-globin (erythrocyte marker), lyz (macrophage marker), mpx (granulocyte marker), and rag1 (lymphocyte marker) were also examined. Comparing with siblings, mutantcas008 expressed decreased ae1-globin and barely expressed lyz, mpx, and rag1 (Figures 1B–E,B′–E′). All these results showed definitive HSPCs in the CHT are severely disrupted and downstream differentiation of HSPCs were blocked in mutantcas008 embryos.
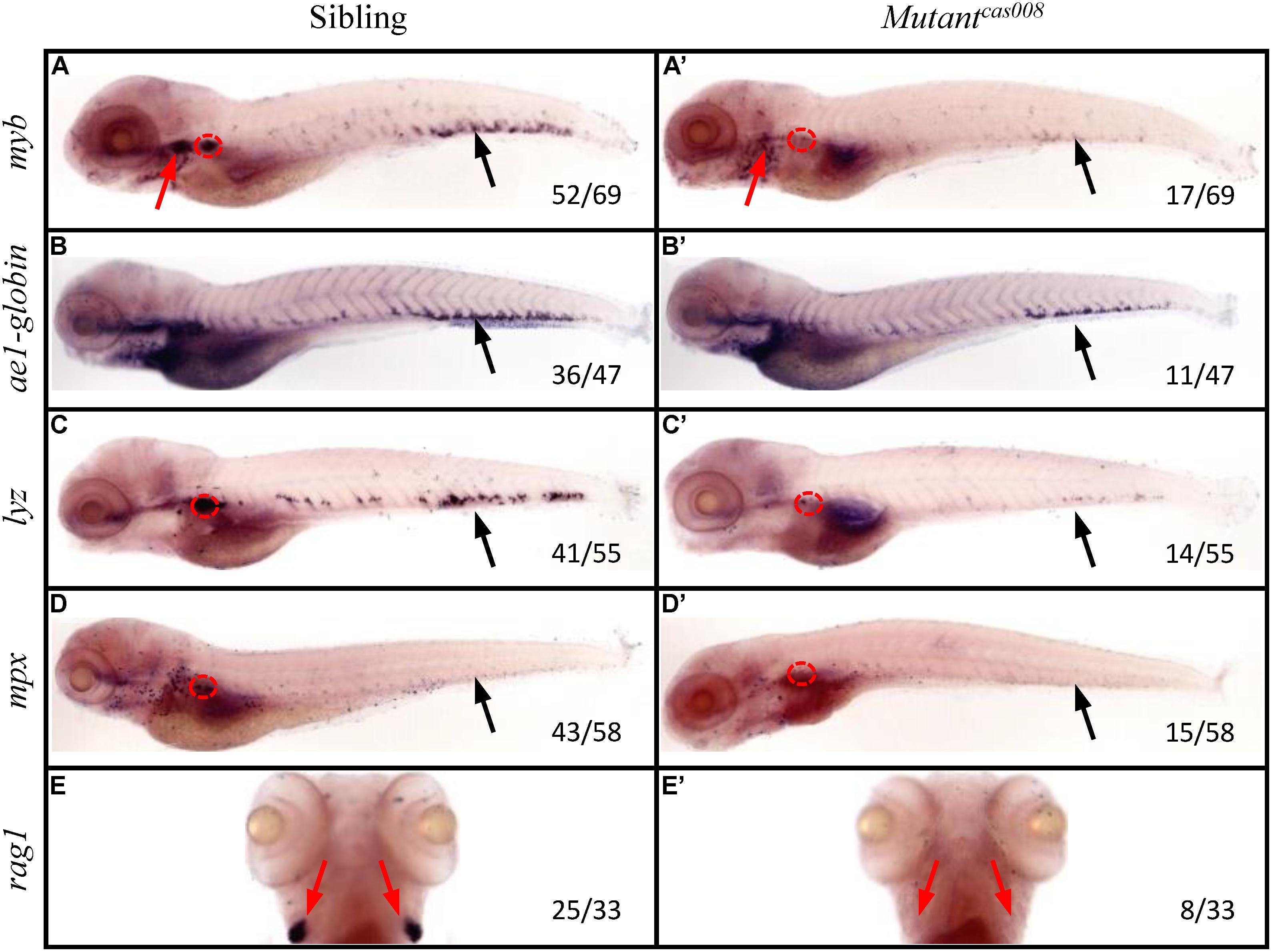
Figure 1. The mutantcas008 line showed defective definitive hematopoiesis. WISH analysis of myb (A,A′), ae1-globin (B,B′), lyz (C,C′), mpx (D,D′), and rag1 (E,E′) expression in sibling (A–E) and mutantcas008 (A′–E′) embryos at 5 dpf. The penetrance of the indicated phenotype is shown in the bottom right of each panel. Black arrows indicate the position of CHT, while red arrows and circles show the position of thymus and kidney, respectively.
Furthermore, we tested the primitive hematopoiesis in mutantcas008 by WISH. The results showed the expression of primitive hematopoietic cell markers, including gata1 (erythrocyte progenitor marker) and pu.1 (myeloid progenitor marker; Supplementary Figures 1A,B,A′,B′) were identical between siblings and mutantcas008 at 22 hpf. As mentioned, in the developing dorsal aorta HSPCs emerge from endothelial cells during 33–54 hpf (Kissa and Herbomel, 2010), thus vascular morphogenesis is essential for HSPCs initiation and maintenance (Burns et al., 2009). The expression of the pan-endothelial cell marker kdrl at 26 hpf before HSPCs emergence was examined and it was intact in mutantcas008 as well (Supplementary Figures 1C,C′). We also examined the expression of runx1 at 36 hpf, required for HSPCs emergence and showed no defects in mutantcas008 comparing with sibling embryos (Supplementary Figures 1D,D′). Taken together, we obtained mutantcas008 displayed specific deficiency in definitive hematopoiesis without obvious defects during primitive hematopoiesis, vasculogenesis and HSPCs emergence.
We carried out positional cloning to elucidate the mechanism underlying the hematopoietic failure in mutantcas008. According to the previously reported procedure (Gao et al., 2015; Jia et al., 2015), by bulk segregation analysis (BSA) the mutation was first mapped to chromosome 21. Then a high-resolution mapping approach was performed and it identified the mutation was flanked by two linked SSLP markers, zK189O20 and zC153M12. There were four candidate genes in the flanked region (Supplementary Figure 2A). Among them, we identified a C to T nonsense mutation causing an immediate stop codon in gemin5 in mutantcas008 by sequencing cDNA of the four genes (NC_007132.7: g.36404684 C > T, Figure 2A). Next, the result was verified by genomic sequencing. The earlier stop codon caused by this mutation was next to the end of the fifth WD repeats domain of Gemin5 protein (XP_001339880.4:p. Gln303_His1440del, Figure 2B). Hence, we changed the name of mutantcas008 into NC_007132.7: g.36404684 C > T, gemin5cas008 for short. To further confirm our results, we performed a rescue experiment by employing a Tol2 transposase-mediated transgenic approach to express wild-type (WT) gemin5 gene in gemin5cas008 embryos. In the construction of transgenic plasmid, the ubiquitin promoter drove ubiquitous expression of wildtype gemin5, followed by P2A peptide-mCherry fusion protein to indicate transgenic efficiency (P2A peptide allows self-cleavage to avoid influencing Gemin5 function). This plasmid element was flanked by Tol2 arms (Figure 2C). After co-injection of the plasmid and Tol2 transposase mRNA into one-cell stage gemin5cas008 embryos, ubiquitous expression of wild-type gemin5 could rescue gemin5cas008 phenotype to a large extend at 6 dpf (Figure 2C). Taken together, the nonsense mutation in gemin5 caused the defective hematopoietic phenotypes in gemin5cas008.
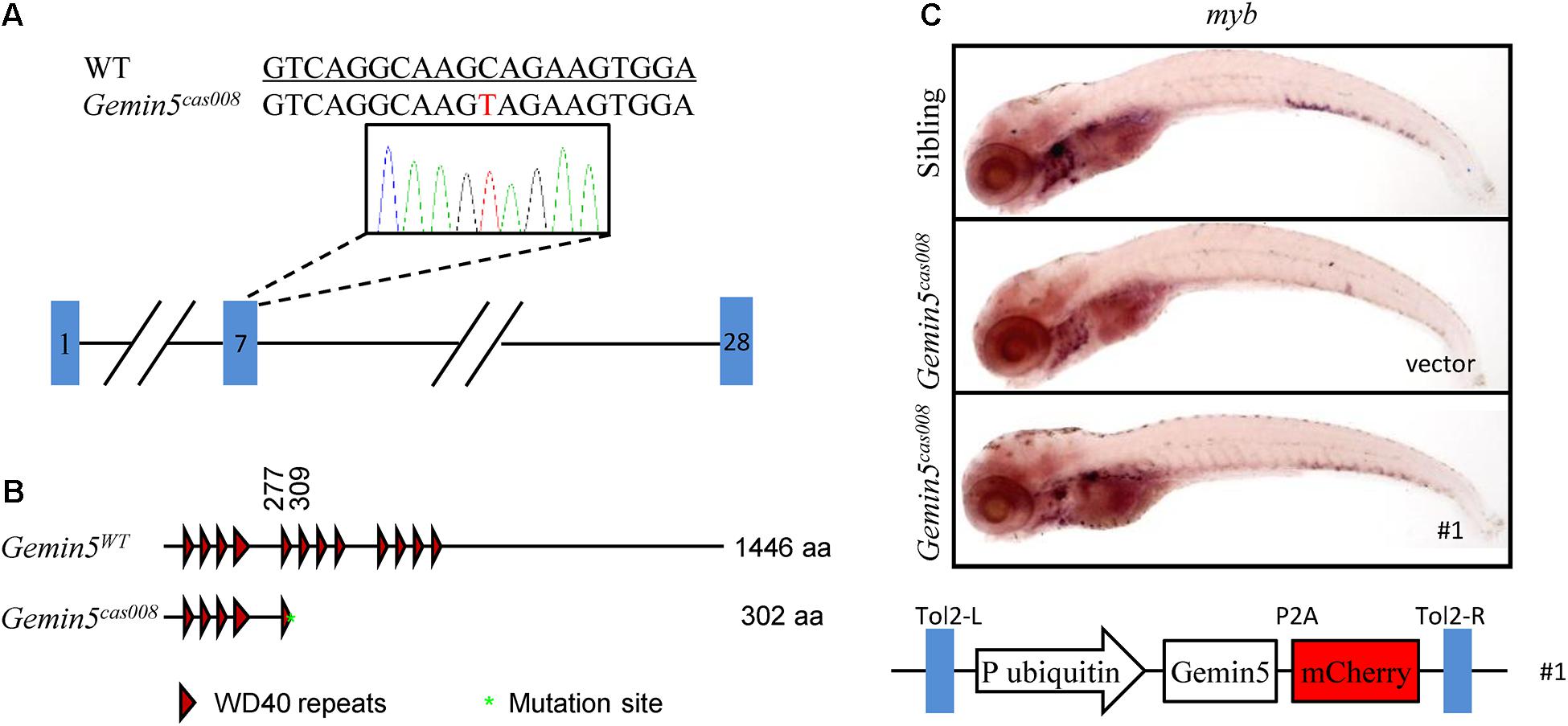
Figure 2. The mutation was mapped to a point mutation in gene gemin5. (A) The alignment of WT (underlined) and NC_007132.7: g.36404684 C > T (gemin5008) sequences are listed. The mutation by ENU is indicated in red (a C to T nonsense mutation causes an immediate stop codon in gemin5 in mutantcas008). (B) According to the stop codon in the genome, SMART software was used to predict the structure of Gemin5WT and XP_001339880.4:p.Gln303_His1440del (Gemin5008) presumed protein. The molecular sizes of the presumed protein are indicated on the right. (C) WISH analysis with myb probe in the CHT region (at 6 dpf) of sibling and gemin5008 with transient transgenesis of vector (only plasmid backbone) or ubi: gemin5 (#1, The construction of the plasmid is shown below).
Gemin5, as an RBP with multidomain consists of the N-terminal WD repeat domains, a dimerization domain in the middle, and a non-canonical RNA-binding domain in the C-terminal region (Martinez-Salas et al., 2020). Its WD repeat motifs recognize the snRNAs, allowing assembly of the SMN complex into snRNPs (Pineiro et al., 2015; Martinez-Salas et al., 2020). Structure analyses show the dimerization module of Gemin5 is fundamental for its architecture and plays an essential role in its activity (Martinez-Salas et al., 2020). The non-canonical RNA-binding domain allows Gemin5 to interact with its mRNA targets and play a key role in translation. Research on Gemin5 is emerging but far from enough to understand the consequence of its multifunction, including its canonical role in the biogenesis of snRNPs, and its control on alternative splicing and translation, etc. (Pineiro et al., 2015).
To elucidate the reason of HSPCs abrogation, we first examined HSPCs proliferation during hematopoietic development by combination of fluorescence in situ hybridization (FISH) and phospho-histone 3 (pH3) immunostaining. We calculated the percentage of proliferative HSPCs (pH3 + myb+) in total HSPCs (myb+). At both 4 and 5 dpf, the percentage of proliferative HSPCs (pH3+ myb+) in the CHT in gemin5cas008 was significantly decreased (Figures 3A–D, 4 dpf P = 0.0012, and 5 dpf P < 0.0001). Next, we examined the apoptosis of HSPCs by terminal-transferase dUTP Nick End Labeling (TUNEL) assays in transgenic background Tg(myb:eGFP) and calculated the percentage of apoptotic HSPCs (TUNEL+ GFP+) in total HSPCs (GFP+). Apoptotic signals of HSPCs in the CHT region had no significant difference between sibling and gemin5cas008 at 4 dpf (Supplementary Figures 3A,B, P = 0.1430), which indicated there was no excessive HSPCs apoptosis in mutant gemin5cas008. In brief, impaired definitive hematopoiesis in gemin5cas008 was due to defective proliferation of HSPCs.

Figure 3. Decreased HSPCs proliferation in mutant gemin5cas008. (A,B) Representative confocal images of double staining of pH3 protein (green) and myb RNA (red) in the CHT at 4 dpf (A) and 5 dpf (B). Arrows point at proliferative HSPCs. Scale bars, 50 μm. (C,D) Percentage of proliferative HSPCs (pH3+myb+)/ total HSPCs (myb+) in sibling and gemin5cas008 embryos at 4 dpf (C) and 5 dpf (D). (C) t = 4.202, df = 12, and P = 0.0012. (D) t = 10.96, df = 10, and P < 0.0001.
In brief, we obtained a zebrafish mutant family with Gemin5 point mutation. Mutant gemin5cas008 showed no obvious defects during primitive hematopoiesis or vasculogenesis. However, the reductive HSPCs proliferation caused severe definitive hematopoiesis failure in the gemin5cas008.
The basis of stem cells’ self-maintenance and differentiation could be described as genetically programmed and integrated circuits (Yuan and Muljo, 2013). In the hematopoietic system, major advances have been achieved in elucidating the regulatory networks which orchestrate gene expression programs and direct cell-fate determinations. Among them, much work has focused on elucidating signaling pathways and transcriptional programs. However, the post-transcriptional regulation of HSPCs is far from well-explored. This “RNA centric” level of regulation precisely and quickly tunes protein expression by regulating splicing, localization, degradation, or translation of mRNA (de Rooij et al., 2019). As one of the key players, RBPs perform their function by interaction with existent transcriptional networks in the cell. They interact with their target mRNAs which are based on RNA sequence, strongly influencing their downstream processing. It has been reported different changes of RBPs will cause the disruption of hematopoietic homeostasis (Yuan and Muljo, 2013). For instance, the RBP Rbm15 regulates splicing of Mpl-TR, the truncated isoform of the c-Mpl receptor, which affects thrombopoietin signaling and subsequently influences HSCs quiescence and proliferation (Xiao et al., 2015). The oncogenic fusion gene BCR-ABL markedly increased the expression of the RBPs La/SSB, multiple hnRNPs and TLS/FUS and interfered mRNA processing, nuclear-cytoplasmic export and translation, resulting in the unbalance of HSCs proliferation and differentiation (Perrotti and Calabretta, 2002).
In this study, we obtained a zebrafish mutant family with Gemin5 point mutation through a large-scale forward genetics screen. Gemin5 is an RBP described as a peripheral component of the SMN complex, first identified in 2002 (Gubitz et al., 2002). In eukaryotic cells, the SMN complex acts as an assemblyosome, mediating the biogenesis and function of spliceosomal snRNPs. It is intensive in nuclear Gems (Gemini of Cajal bodies), though it is dispersed throughout the cytoplasm. The SMN complex is composed of nine members, including SMN, Gemins2–8 and the unr-interacting protein (unrip) in human (Martinez-Salas et al., 2020). Gemin5 comprises WD repeat domains at the N-terminal, recognizing the snRNAs, delivering them to the SMN complex and allowing assembly of the SMN complex and snRNAs into snRNPs in the cytoplasm. Gemin5 is predominantly distributed in the cell cytoplasm (Gubitz et al., 2002). However, a large fraction of Gemin5 is found outside of the SMN complex (Battle et al., 2007), which suggests there may be additional functions of Gemin5. In accordance with this, it has been detected Gemin5 and the SMN complex colocalize in nuclear gems, rather than in nuclear Cajal bodies. Gemin5 may participate in tumor cell motility through the alternative splicing process. For instance, analyze the global mRNA splicing profile of the MDA-MB-435 tumor cell line at metastatic state or suppression state, showing a distinct splicing profile between different cellular states in dependence on Gemin5 (Lee et al., 2008, 2009). In addition, it has been reported that Gemin5 may interact with signal recognition particle or ribosome, and regulate the translation or trans-splicing of several mRNAs. More recently, structural analyses have unveiled there is a robust dimerization module in Gemin5, helping understand the functional role of the middle region (Moreno-Morcillo et al., 2020). The dimerization module of Gemin5 is fundamental in its architecture and essential for its activity. At the C-terminal region, it has a non-canonical RNA-binding site (RBS) with two parts named as RBS1 and RBS2, not only determining the interaction with IRES (viral internal ribosome entry site) of the FMDV (foot-and-mouth disease virus), but also harboring the capacity to downregulate translation (Fernandez-Chamorro et al., 2014). In sum, Gemin5 plays an essential role in the snRNPs biogenesis. Moreover, with the analysis of its structure, its function involved in alternative splicing, stress response and translation control has been uncovered. Although genetic evidence shown defective Gemin5 might link to neuron degeneration, its role in other tissues/organs remains unknown.
In mutant gemin5cas008, we identified a C to T nonsense mutation causing an earlier stop codon next to the end of the fifth WD repeats domain of Gemin5 protein (Figures 2A,B). Mutant gemin5cas008 had intact primitive hematopoiesis, while showed specific deficiency in definitive hematopoiesis. FISH and pH3 immunostaining results showed the percentage of proliferative HSPCs dramatically decreased in gemin5cas008 (Figures 3A–D). However, the percentage of apoptotic HSPCs had no significant difference (Supplementary Figures 3A,B). In accordance, inhibiting p53 dependent apoptosis pathway by p53 MO or bcl2 mRNA injection at one-cell stage could not rescue the phenotype of gemin5cas008 (similar experiments could rescue another mutant due to increased apoptosis in Gao et al., 2015). So, we concluded that impaired definitive hematopoiesis in gemin5cas008 was due to defective proliferation of HSPCs. It has been reported that in human cells, Gemin5 interact with snRNAs through the WD repeat domain. Particularly, RNA-mediated radical probing and mass spectrometry results showed the fifth WD repeat contributes to the interaction (Lau et al., 2009). In addition, the establishment of the interaction depends on the structural integrity of this domain, for even a point mutation at the W286 residue (human) would disrupt the interaction shown by mutational experiment. This residue in zebrafish Gemin5 orthologs (W270) is conserved and might be preserved in mutant gemin5cas008. Considering the intact primitive hematopoiesis and specific deficient definitive hematopoiesis, we speculate the truncated Gemin5 protein can still recognize snRNAs through its residual domain for there is no large-scale transcriptional splicing disorder in the cell. However, the efficiency of the interaction may be reduced, and its target may even be biased toward certain snRNAs, eventually leading to the appearance of specific transcription and splicing disorders, thereby targeting the hematopoietic process.
In addition, Gemin5 has cap-binding capacity in the WD repeat domains and may influence translation initiation in a cap-dependent manner (Pineiro et al., 2015). W286 and F338 (human), as the putative cap-interacting residues, are identified by combination of the protein homology/analogy recognition engine (PHYRE) algorithm and site-directed mutagenesis. These sites are conserved in zebrafish (W270 and F322), and W270 might be preserved while F322 was not in the mutant. As mentioned, truncated Gemin5 protein lacked C-terminal which interacts with IRES and regulates transition. It’s reasonable to believe the regulation of Gemin5 in translation initiation was partially preserved in the mutants. The defective Gemin5 might still maintain various basic functions to let the mutants survive early development without obvious defects, however, the biological processes during definitive hematopoiesis were beyond the ability of truncated Gemin5 and drove abnormal development. Based on reported researches about the regulation of Gemin5 on the ASK1-JNK1 signaling pathway (Kim et al., 2007) and its cooperation with nuclear receptor to transmit ecdysone signaling (Gates et al., 2004), we speculate some relatively sensitive signal pathways of HSPCs or the transcriptional regulatory network required for proliferation depending on Gemin5 were specifically disrupted in mutant gemin5cas008.
Questions remain open regarding how Gemin5 regulates hematopoiesis. First of all, the location and the protein level of the mutant Gemin5 protein need to be determined. Secondly, HSPC-specific re-expression of WT Gemin5 in gemin5cas008 need to be carried out to elucidate its requirement in a stem cell or niche cell manner. Lack of zebrafish Gemin5 antibody and lack of efficient HSPC-specific promoter for transgenesis need be overcome in future study. Thirdly, the pathogenic mechanism downstream of the mutated Gemin5 needs to be elucidated. It needs to be described which signal pathways of HSPCs or which transcriptional regulatory network required for proliferation depending on Gemin5 are essential. It will also be interesting to figure out the function of diverse regions of Gemin5 in the regulation of HSPCs. At last, it is worth identifying whether there are human hematopoietic diseases caused by Gemin5 mutations and what extent of Gemin5 involved in human diseases in blood system. It is very likely that the extend of other components in the SMN complex, as well as other RBPs, need be studied in hematopoiesis.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by the Animal Research Advisory Committee of Shanghai Institute of Nutrition and Health, CAS.
XL, KM, WP, and DL designed the experiments. XL and KM carried on the experiments and analyzed data. CJ, LG, CF, and CR assisted with ENU screening and positional cloning. WZ, YH, and MC assisted with imaging with Olympus FV1000 Fluoview scanning confocal microscope. All the authors provided ideas and discussions throughout the project. WP and DL wrote the manuscript. KM, WP, and DL supervised the project.
The National Key R&D Program of China (2018YFA0800203), The National Science Fund for Distinguished Young Scholars (Grant No. 31925014), Chinese Academy of Sciences (CAS) Research Program of Frontier Sciences (ZDBS-LY-SM010); China National Postdoctoral Program for Innovative Talents (Grant No. BX20200347), and Shanghai Post-doctoral Excellence Program (Grant No. 2020505).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Yi Chen, Yi Jin, Min Deng, Mei Dong, Peili Ni, and Yan Hao for zebrafish husbandry and technical support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.670654/full#supplementary-material
Bahary, N., Davidson, A., Ransom, D., Shepard, J., Stern, H., Trede, N., et al. (2004). The zon laboratory guide to positional cloning in zebrafish. Methods Cell. Biol. 77, 305–329. doi: 10.1016/s0091-679x(04)77017-x
Battle, D. J., Kasim, M., Wang, J., and Dreyfuss, G. (2007). SMN-independent subunits of the SMN complex. Identification of a small nuclear ribonucleoprotein assembly intermediate. J. Biol. Chem. 282, 27953–27959. doi: 10.1074/jbc.m702317200
Burns, C. E., Galloway, J. L., Smith, A. C., Keefe, M. D., Cashman, T. J., Paik, E. J., et al. (2009). A genetic screen in zebrafish defines a hierarchical network of pathways required for hematopoietic stem cell emergence. Blood 113, 5776–5782. doi: 10.1182/blood-2008-12-193607
Ciau-Uitz, A., Monteiro, R., Kirmizitas, A., and Patient, R. (2014). Developmental hematopoiesis: ontogeny, genetic programming and conservation. Exp. Hematol. 42, 669–683. doi: 10.1016/j.exphem.2014.06.001
de Rooij, L., Chan, D. C. H., Keyvani Chahi, A., and Hope, K. J. (2019). Post-transcriptional regulation in hematopoiesis: RNA binding proteins take control (1). Biochem. Cell. Biol. 97, 10–20.
Fernandez-Chamorro, J., Pineiro, D., Gordon, J. M., Ramajo, J., Francisco-Velilla, R., Macias, M. J., et al. (2014). Identification of novel non-canonical RNA-binding sites in Gemin5 involved in internal initiation of translation. Nucleic Acids Res. 42, 5742–5754.
Galloway, J. L., and Zon, L. I. (2003). Ontogeny of hematopoiesis: examining the emergence of hematopoietic cells in the vertebrate embryo. Curr. Top Dev. Biol. 53, 139–158. doi: 10.1016/s0070-2153(03)53004-6
Gao, L., Li, D., Ma, K., Zhang, W., Xu, T., Fu, C., et al. (2015). TopBP1 governs hematopoietic stem/progenitor cells survival in zebrafish definitive hematopoiesis. PLoS Genet. 11:e1005346. doi: 10.1371/journal.pgen.1005346
Gates, J., Lam, G., Ortiz, J. A., Losson, R., and Thummel, C. S. (2004). Rigor mortis encodes a novel nuclear receptor interacting protein required for ecdysone signaling during drosophila larval development. Development 131, 25–36. doi: 10.1242/dev.00920
Gore, A. V., Pillay, L. M., Venero Galanternik, M., and Weinstein, B. M. (2018). The zebrafish: a fintastic model for hematopoietic development and disease. Wiley Interdiscip. Rev. Dev. Biol. 7:e312. doi: 10.1002/wdev.312
Grech, G., and von Lindern, M. (2012). The role of translation initiation regulation in haematopoiesis. Comp. Funct. Genom. 2012:576540.
Gubitz, A. K., Mourelatos, Z., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. (2002). Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 277, 5631–5636. doi: 10.1074/jbc.m109448200
Jia, X. E., Ma, K., Xu, T., Gao, L., Wu, S., Fu, C., et al. (2015). Mutation of kri1l causes definitive hematopoiesis failure via PERK-dependent excessive autophagy induction. Cell. Res. 25, 946–962. doi: 10.1038/cr.2015.81
Kim, E. K., Noh, K. T., Yoon, J. H., Cho, J. H., Yoon, K. W., Dreyfuss, G., et al. (2007). Positive regulation of ASK1-mediated c-Jun NH(2)-terminal kinase signaling pathway by the WD-repeat protein Gemin5. Cell. Death Differ. 14, 1518–1528. doi: 10.1038/sj.cdd.4402157
Kissa, K., and Herbomel, P. (2010). Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–125. doi: 10.1038/nature08761
Lau, C. K., Bachorik, J. L., and Dreyfuss, G. (2009). Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat. Struct. Mol. Biol. 16, 486–491. doi: 10.1038/nsmb.1584
Lee, J. H., Horak, C. E., Khanna, C., Meng, Z., Yu, L. R., Veenstra, T. D., et al. (2008). Alterations in Gemin5 expression contribute to alternative mRNA splicing patterns and tumor cell motility. Cancer Res. 68, 639–644. doi: 10.1158/0008-5472.can-07-2632
Lee, J. H., Marshall, J. C., Steeg, P. S., and Horak, C. E. (2009). Altered gene and protein expression by Nm23-H1 in metastasis suppression. Mol. Cell. Biochem. 329, 141–148. doi: 10.1007/s11010-009-0124-3
Li, D., Xue, W., Li, M., Dong, M., Wang, J., Wang, X., et al. (2018). VCAM-1(+) macrophages guide the homing of HSPCs to a vascular niche. Nature 564, 119–124. doi: 10.1038/s41586-018-0709-7
Martinez-Salas, E., Embarc-Buh, A., and Francisco-Velilla, R. (2020). Emerging roles of gemin5: from snRNPs assembly to translation control. Int. J. Mol. Sci. 21:3868. doi: 10.3390/ijms21113868
Moreno-Morcillo, M., Francisco-Velilla, R., Embarc-Buh, A., Fernandez-Chamorro, J., Ramon-Maiques, S., and Martinez-Salas, E. (2020). Structural basis for the dimerization of Gemin5 and its role in protein recruitment and translation control. Nucleic Acids Res. 48, 788–801. doi: 10.1093/nar/gkz1126
Murayama, E., Kissa, K., Zapata, A., Mordelet, E., Briolat, V., Lin, H. F., et al. (2006). Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity 25, 963–975. doi: 10.1016/j.immuni.2006.10.015
North, T. E., Goessling, W., Walkley, C. R., Lengerke, C., Kopani, K. R., Lord, A. M., et al. (2007). Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–1011. doi: 10.1038/nature05883
Perrotti, D., and Calabretta, B. (2002). Post-transcriptional mechanisms in BCR/ABL leukemogenesis: role of shuttling RNA-binding proteins. Oncogene 21, 8577–8583. doi: 10.1038/sj.onc.1206085
Pineiro, D., Fernandez-Chamorro, J., Francisco-Velilla, R., and Martinez-Salas, E. (2015). Gemin5: a multitasking RNA-binding protein involved in translation control. Biomolecules 5, 528–544. doi: 10.3390/biom5020528
Song, H. D., Sun, X. J., Deng, M., Zhang, G. W., Zhou, Y., Wu, X. Y., et al. (2004). Hematopoietic gene expression profile in zebrafish kidney marrow. Proc. Natl. Acad. Sci. U S A 101, 16240–16245. doi: 10.1073/pnas.0407241101
Suster, M. L., Kikuta, H., Urasaki, A., Asakawa, K., and Kawakami, K. (2009). Transgenesis in zebrafish with the tol2 transposon system. Methods Mol. Biol. 561, 41–63. doi: 10.1007/978-1-60327-019-9_3
Traver, D., Paw, B. H., Poss, K. D., Penberthy, W. T., Lin, S., and Zon, L. I. (2003). Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol. 4, 1238–1246. doi: 10.1038/ni1007
Xiao, N., Laha, S., Das, S. P., Morlock, K., Jesneck, J. L., and Raffel, G. D. (2015). Ott1 (Rbm15) regulates thrombopoietin response in hematopoietic stem cells through alternative splicing of c-Mpl. Blood 125, 941–948. doi: 10.1182/blood-2014-08-593392
Keywords: Gemin5, Hematopoietic stem/progenitor cells, cell proliferation, zebrafish, definitive hematopoiesis, forward genetic screening, positional cloning
Citation: Liu X, Zhang W, Jing C, Gao L, Fu C, Ren C, Hao Y, Cao M, Ma K, Pan W and Li D (2021) Mutation of Gemin5 Causes Defective Hematopoietic Stem/Progenitor Cells Proliferation in Zebrafish Embryonic Hematopoiesis. Front. Cell Dev. Biol. 9:670654. doi: 10.3389/fcell.2021.670654
Received: 22 February 2021; Accepted: 13 April 2021;
Published: 30 April 2021.
Edited by:
Yiyue Zhang, South China University of Technology, ChinaReviewed by:
Mugen Liu, Huazhong University of Science and Technology, ChinaCopyright © 2021 Liu, Zhang, Jing, Gao, Fu, Ren, Hao, Cao, Ma, Pan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ke Ma, TUtFMjAwMkAxNjMuY29t; Weijun Pan, d2VpanVucGFuQHNpYnMuYWMuY24=; Dantong Li, ZHRsaUBzaWJzLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.