- 1The Mina and Everard Goodman Faculty of Life Sciences, Bar-Ilan University, Ramat Gan, Israel
- 2Department of Molecular Biology, Cell Biology and Biochemistry, Brown University, Providence, RI, United States
The development of multicellular organisms and the uniqueness of each cell are achieved by distinct transcriptional programs. Multiple processes that regulate gene expression converge at the core promoter region, an 80 bp region that directs accurate transcription initiation by RNA polymerase II (Pol II). In recent years, it has become apparent that the core promoter region is not a passive DNA component, but rather an active regulatory module of transcriptional programs. Distinct core promoter compositions were demonstrated to result in different transcriptional outputs. In this mini-review, we focus on the role of the core promoter, particularly its downstream region, as the regulatory hub for developmental genes. The downstream core promoter element (DPE) was implicated in the control of evolutionarily conserved developmental gene regulatory networks (GRNs) governing body plan in both the anterior-posterior and dorsal-ventral axes. Notably, the composition of the basal transcription machinery is not universal, but rather promoter-dependent, highlighting the importance of specialized transcription complexes and their core promoter target sequences as key hubs that drive embryonic development, differentiation and morphogenesis across metazoan species. The extent of transcriptional activation by a specific enhancer is dependent on its compatibility with the relevant core promoter. The core promoter content also regulates transcription burst size. Overall, while for many years it was thought that the specificity of gene expression is primarily determined by enhancers, it is now clear that the core promoter region comprises an important regulatory module in the intricate networks of developmental gene expression.
Introduction
The uniqueness of each cell type and its various developmental roles in multicellular organisms are largely achieved by distinct transcriptional programs. Transcription initiation is composed of a series of highly conserved, coordinated and complex steps (reviewed in Thomas and Chiang, 2006; Fuda et al., 2009; Danino et al., 2015; Koster et al., 2015). The core promoter region encompasses the ± 40 bp region relative to the transcription start site (TSS; defined as the + 1 or first ribonucleotide of a transcript). This critical regulatory region, which is required for accurate initiation of transcription by Pol II, is often referred to as the “gateway to transcription” (Heintzman and Ren, 2007; Juven-Gershon and Kadonaga, 2010). Since transcriptional regulation by Pol II determines proper organismal growth and development, alterations in transcriptional regulation may result in a variety of phenotypes, including cell fate changes and lethality (Levine and Tjian, 2003; Ohler and Wassarman, 2010; Bolt and Duboule, 2020). Genome-wide analysis of mammalian promoters led to the discovery that transcription initiation may occur in two distinct modes, namely focused and dispersed, with single or multiple start sites, respectively (Carninci et al., 2006). Recent studies suggest that transcription initiation is more intricate, likely involving combinations between the “focused” and “dispersed” modes (Kadonaga, 2012; Lenhard et al., 2012; Frith and Fantom Consortium, 2014). Drosophila was instrumental in deciphering the identity and function of core promoter composition. The transcription machinery, as well as its regulatory principles, are largely conserved in evolution. Thus, although most of the data discussed here is largely based on findings in Drosophila, many of the underlying principles are applicable to a wider array of species.
Importantly, the regulation of transcription initiation by Pol II occurs at both the DNA (promoter) and the protein (basal transcription machinery) levels. Recent reviews focused on the structure of polymerases, the components of the general/basal transcription machinery and the preinitiation complex (Cramer, 2019a,b; Roeder, 2019; Schier and Taatjes, 2020). In this mini-review, we will highlight the function of the core promoter as the regulatory hub that recruits specific transcription factors to control key developmental gene expression programs.
Core Promoter Composition
Although the core promoter was previously considered a universal component that works similarly for all protein-coding genes, it is now established that core promoters are divergent in their architecture and function, and each core promoter is rather unique (Sandelin et al., 2007; Muller and Tora, 2014; Roy and Singer, 2015). The core promoter may contain one or more short DNA sequences, termed core promoter elements or motifs, which contribute to its function. The major sequence elements identified in Drosophila and human focused promoters are illustrated in Figure 1 and reviewed in detail in Danino et al. (2015); Vo Ngoc et al. (2017b), and Haberle and Stark (2018).
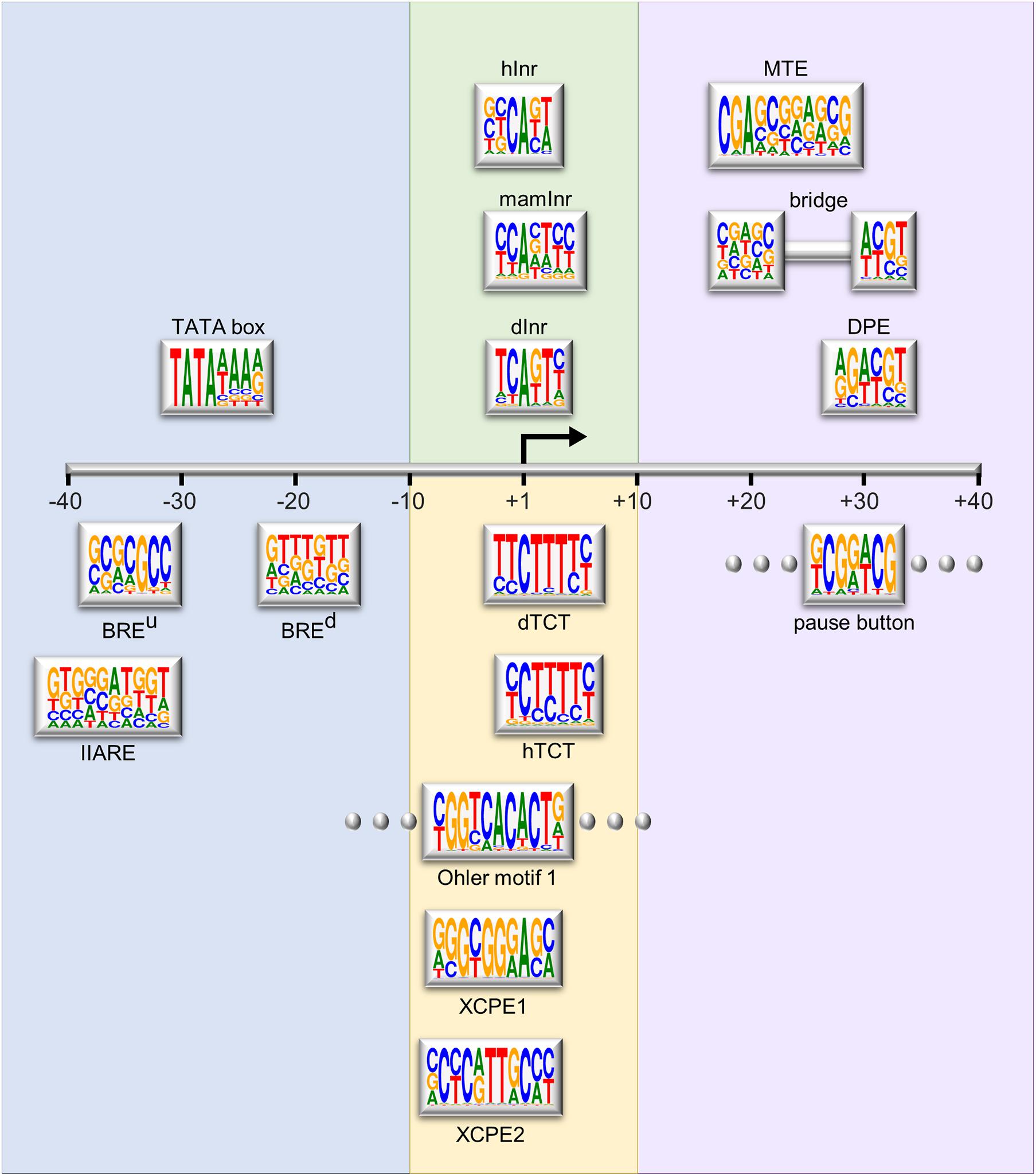
Figure 1. Schematic representation of the major core promoter elements. The region of the core promoter area (–40 to +40 relative to the TSS) is illustrated to scale. The Logo representation for each element is based on the position weight matrix (PWM) derived from functional sequences (Sloutskin et al., 2015), or as provided in the primary reference. Logos were generated using WebLogo (Crooks et al., 2004). Rectangles with flanking dots represent elements that are not strictly spaced. Background colors represent the grouping of the elements based on their position relative to the A+1 or functional similarity. The depicted elements are: upstream and downstream TFIIB recognition elements (BREu and BREd, respectively, Lagrange et al., 1998; Deng and Roberts, 2005); TFIIA recognition element (IIARE, Wang et al., 2017); TATA box (Goldberg, 1979); Drosophila initiator (dInr, Lo and Smale, 1996); mammalian initiator (mamInr, Smale and Baltimore, 1989); focused human initiator (hInr, Vo Ngoc et al., 2017a); Drosophila and human polypyrimidine transcription initiation (dTCT and hTCT, respectively, Parry et al., 2010); Ohler motif 1 (Ohler et al., 2002); X core promoter element 1 and 2 (XCPE1 and XCPE2, respectively, Tokusumi et al., 2007; Anish et al., 2009); downstream core promoter element (DPE, Burke and Kadonaga, 1996, 1997; Kutach and Kadonaga, 2000); motif ten element (MTE, Ohler et al., 2002; Lim et al., 2004); bridge (Theisen et al., 2010); pause button (PB, Hendrix et al., 2008).
Importantly, the core promoter serves as the scaffold for the assembly of the pre-initiation complex (PIC), which is comprised of Pol II, the basal transcription factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) and promoter DNA. Multiple core promoter elements were identified as the regions bound by distinct PIC components. The upstream and downstream TFIIB recognition elements (BREu and BREd, respectively) are bound by TFIIB (Lagrange et al., 1998; Deng and Roberts, 2005). The TFIIA recognition element (IIARE) was recently described (Wang et al., 2017; Wang J. et al., 2020).
The DPE, motif ten element (MTE) and bridge downstream core promoter elements (see below) serve as docking sites for TFIID, which initiates PIC formation by binding to the core promoter. TFIID is composed of TATA box-binding protein (TBP) and 13–14 TBP-associated factors (TAFs) (recently reviewed in Antonova et al., 2019; Bhuiyan and Timmers, 2019; Patel et al., 2020). The first identified and perhaps most well-known core promoter element is the TATA box (Goldberg, 1979), bound by TBP and originally discovered in Drosophila histone genes. Both the TATA box and TBP are conserved from archaea to humans (Reeve, 2003). Core promoters were previously classified as having or lacking a TATA-box, yet only a minority of metazoan promoters contain a TATA-box (Gershenzon and Ioshikhes, 2005; Kim et al., 2005; Dikstein, 2011). Thus, TATA-less promoters still require TFIID binding presumably through other core promoter elements.
The initiator (Inr) element, which encompasses the TSS (Corden et al., 1980; Smale and Baltimore, 1989), is the most prevalent core promoter motif within Drosophila focused promoters (FitzGerald et al., 2006; Gershenzon et al., 2006). It is bound by the TAF1 and TAF2 subunits of TFIID (Kaufmann and Smale, 1994; Verrijzer et al., 1995; Chalkley and Verrijzer, 1999; Wu et al., 2001; Louder et al., 2016). Functional initiator motifs were first defined in Drosophila and mammalian species, and more recently in focused human promoters (Vo Ngoc et al., 2017a). The TCT is a polypyrimidine transcription initiation motif that is conserved from Drosophila to humans, enriched in ribosomal protein genes and proteins involved in translational regulation (Hariharan and Perry, 1990; Perry, 2005; Parry et al., 2010). This example highlights the importance of specific core promoter elements for distinct functional transcriptional systems. The X core promoter element 1 and 2 (XCPE1 and XCPE2, respectively), identified around the TSSs of the hepatitis B virus X gene, regulate transcription of a minority of human Pol II promoters (Tokusumi et al., 2007; Anish et al., 2009). While most of the above mentioned core promoter elements are associated with focused promoters, several core promoter motifs are associated with dispersed initiation (Rach et al., 2009; Hoskins et al., 2011). Specifically, Ohler motifs 1, 6, and 7 were computationally identified as over-represented motifs in Drosophila core promoters (Ohler et al., 2002). Motif 1 binding protein (M1BP) specifically binds Ohler motif 1, located in the vicinity of the TSS, was identified and biochemically characterized (Li and Gilmour, 2013), providing the experimental validation of the original predictions. Another element associated with dispersed initiation is the DNA-replication-related element (DRE), which was discovered along with its protein binding factor, DREF (Hirose et al., 1993). Notably, a universal core promoter element combination does not exist and novel core promoter motifs are still being discovered.
The DPE motif is the most well characterized downstream motif, precisely located at + 28 to + 33 relative to the A+1 of the Inr (Burke and Kadonaga, 1996, 1997; Kutach and Kadonaga, 2000). The MTE was identified as an overrepresented core promoter sequence located immediately upstream of the DPE (positions + 18 to + 29; Ohler et al., 2002), and was experimentally characterized as a functional regulatory element (Lim et al., 2004). The MTE and DPE partially overlap, and together encompass three functional sub-regions located at nucleotides + 18 to + 22, + 27 to + 29 and + 30 to + 33 downstream of the A+1 (Theisen et al., 2010). The bridge motif was defined as a combination of only the first and third functional sub-regions (bridge I and II, positions + 18 to + 22 and + 30 to + 33, respectively). It is a rare yet functional, core promoter element. Interestingly, recent reviews present the MTE as a bipartite element, replacing the originally identified bridge motif (Vo Ngoc et al., 2017b, 2019). The MTE, DPE and bridge elements are exclusively dependent on the presence of a functional initiator with a strict spacing requirement, and are typically enriched in TATA-less promoters (Danino et al., 2015; Vo Ngoc et al., 2019). The MTE and DPE are likely bound by TAF1, TAF2, and TAF7 subunits of TFIID (Verrijzer et al., 1995; Louder et al., 2016; Patel et al., 2020).
The partially overlapping elements (MTE, DPE and bridge motifs) were discovered in Drosophila and characterized independently, and the exact interplay between them was originally not resolved. Bioinformatic examination of the core promoter composition revealed that functional DPE motifs are typically accompanied by the bridge motif (Shir-Shapira et al., 2019). The transcriptional output of Fushi tarazu (Ftz) target genes is mostly dependent on the DPE motif, with contribution of the bridge I motif. We therefore envision bridge I as an “auxiliary” element, which supports the function of DPE-dependent transcription but not sufficient for fully restoring it upon DPE loss. Potentially, this could promote robustness, where a strong DPE accompanied by a bridge motif ensures proper docking of TFIID for accurate transcriptional activity. Notably, sequence biases in positions + 17, + 19, and + 24 within DPE promoters were reported (Kutach and Kadonaga, 2000; Arnold et al., 2017), suggesting that additional downstream nucleotide positions could contribute to transcriptional output. A recent comprehensive analysis using machine learning resulted in the identification of a 19 bp DPE-like stretch, termed downstream promoter region (DPR), which contributes to the transcriptional output of many human promoters (Vo Ngoc et al., 2020). Taken together, we propose to relate to the downstream core promoter region as a single transcriptional unit comprised of distinct promoter elements. The DPE motif might serve as the heart of this regulatory hub that helps recruit TFIID downstream of the TSS.
The DPE Motif is Associated With Developmental Programs in Drosophila
Core promoter composition is not a passive component of the DNA, but rather an active regulatory module of transcriptional output (Juven-Gershon et al., 2006; Zabidi et al., 2015). A genome-wide comparison of promoter activity throughout embryogenesis in 5 Drosophila species spanning 25–50 million years of evolution, indicated that distinct core promoter elements are associated with different developmental times. Maternally expressed oocyte genes are enriched for DRE and Ohler-1/5/6/7 motifs, whereas zygotic genes expressed through embryogenesis are enriched for Inr, MTE and DPE motifs; larval-related transcripts are enriched for TATA box elements (Batut and Gingeras, 2017). These observations highlight the role of specific core promoter motifs to comprise an additional regulatory dimension of the expression pattern required throughout different developmental stages.
The DPE motif was implicated in the control of developmental gene regulatory networks (GRNs) (Lenhard et al., 2012; Zehavi et al., 2014b), specifically during anterior-posterior and dorsal-ventral axes formation, including in mesoderm development in Drosophila melanogaster (Figure 2A). The homeotic (Hox) genes specify segment identity along the anterior-posterior axis of the developing embryo in all multicellular animals. All of the Drosophila Hox gene promoters lack TATA box elements, and the majority of them contain functional DPE motifs (Juven-Gershon et al., 2008). Importantly, the DPE is necessary in vivo for transcriptional regulation of the Antp P2 promoter within the developing Drosophila melanogaster embryo (Zehavi et al., 2014b). The Drosophila dorsal-ventral developmental GRN includes multiple genes that are activated by different nuclear concentrations of the Dorsal transcription factor along the dorsal-ventral axis. This GRN is dependent on the presence of the DPE motif; over two-thirds of Dorsal target genes contain DPE sequence motifs, which is significantly higher than the proportion of DPE-containing promoters in the genome. Multiple Dorsal target genes are evolutionarily conserved and functionally dependent on the DPE (Zehavi et al., 2014a). Furthermore, the observed expression levels of hybrid enhancer-promoter constructs recapitulates the levels detected for the relevant core promoter, and not the enhancer (Zehavi et al., 2014b). fushi tarazu (ftz) is a pair-rule gene orchestrating the segmentation phase of Drosophila embryonic development, expressed along the anterior-posterior axis (reviewed in Gehring and Hiromi, 1986). Ftz target genes are enriched for functional Inr + bridge + DPE combinations, conserved within Drosophila species (Shir-Shapira et al., 2019). Using CRISPR/Cas9, the in vivo contribution of the DPE motif to the regulation of the endogenous tinman gene in the developing Drosophila embryo was recently tested. Following mutation of tinman’s endogenous DPE motif using a co-CRISPR strategy, the endogenous tinman RNA levels were reduced twofold at 2–4 h of embryonic development (Levi et al., 2020). While the in vivo compatibility of distinct enhancers to their cognate promoters was previously demonstrated to determine expression levels during Drosophila development (Li and Noll, 1994; Merli et al., 1996), this is the first evidence for the contribution of a specific core promoter motif to transcriptional regulation of an endogenous gene. Furthermore, bioinformatics analysis revealed that Drosophila DPE-containing promoters are enriched for GO terms associated with embryonic development of the heart, circulatory system, skeletal muscle, peripheral nervous system, digestive system, renal system and reproductive system (Sloutskin et al., 2015). Thus, we propose that the DPE acronym, originally defined based on its downstream position, may be indicative of its function in Drosophila as a Developmental core Promoter Element.
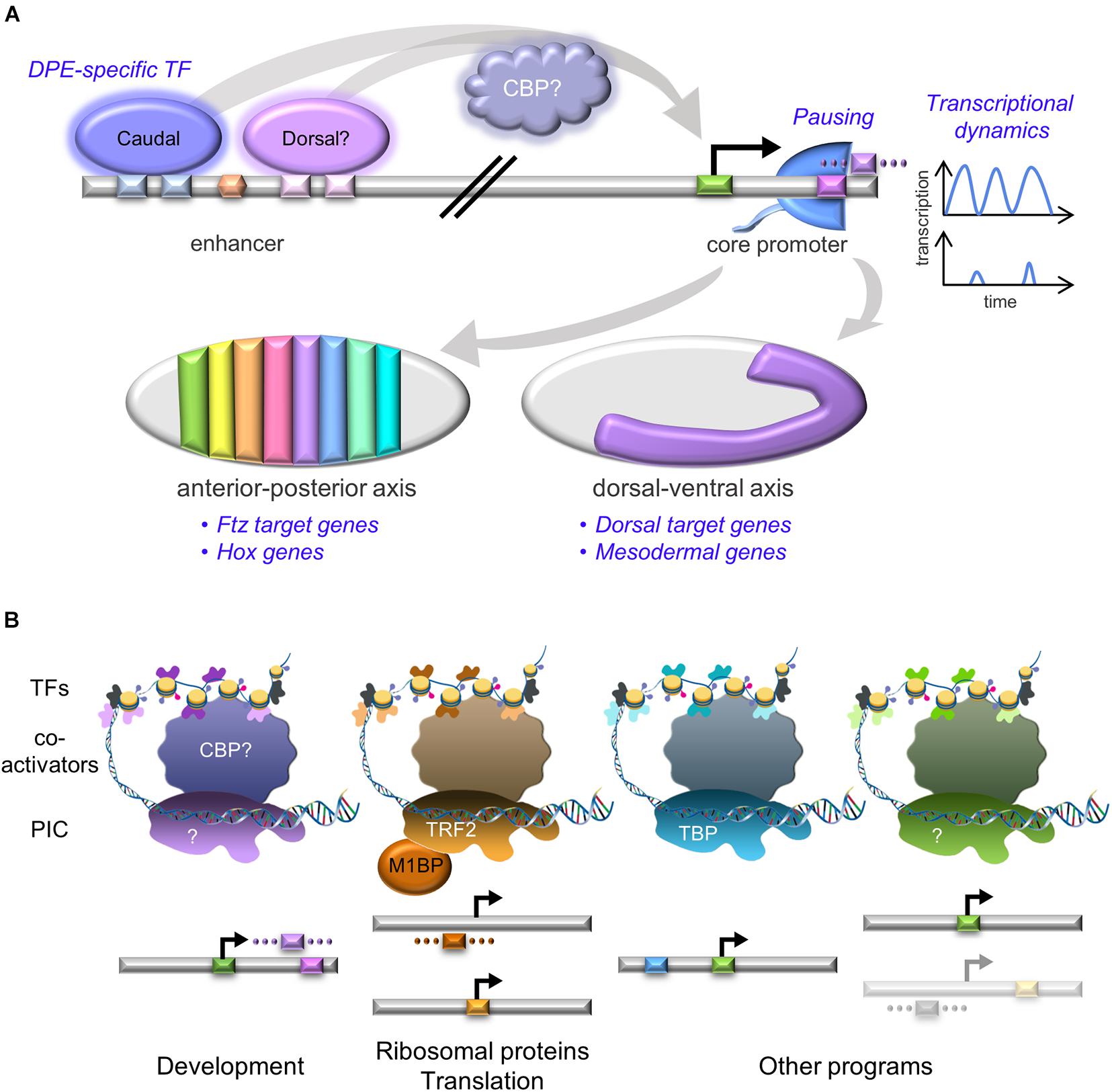
Figure 2. Unique core promoter assemblies are associated with diverse transcriptional programs. (A) DPE-containing core promoters are enriched in developmental programs. DPE-specific transcription factors (e.g., Caudal and perhaps Dorsal) are bound to enhancers, preferentially activating DPE-containing promoters. This regulation might be mediated by the CBP co-activator. Pol II promoter-proximal pausing (Pol II cartoon, blue) and transcriptional dynamics (blue graphs) comprise additional regulatory levels of developmental promoters. The graphs represent fluorescently-measured transcription intensities used to model bursting. All the above regulatory layers are then integrated to accomplish the developmental-related transcriptional program, resulting in gene expression that governs both anterior-posterior and dorsal-ventral axes of the developing Drosophila embryo, depicted as gray ovals (traditionally oriented with anterior to the left and dorsal side facing up). Left- stages 13–14 embryo, colored stripes represent distinct segment identity, based on specific Hox gene expression. Right- stage 8 embryo, purple region corresponds to mesoderm. CBP- CREB-binding protein, Ftz- Fushi Tarazu. (B) Distinct core promoter elements are associated with specific transcriptional programs. The core promoter elements are color-coded as the background colors in Figure 1. The faded core promoter represents promoters containing putative, yet undiscovered motifs. Transcription is initiated by the binding of the pre-initiation complex (PIC) to core promoter DNA. The identity of the PIC components likely differs between the various transcriptional programs, resembling the compatibility of each promoter type with the relevant basal transcription factors. Co-activators mediate PIC recruitment following binding of transcription factors (TFs) to enhancer regions, which contain binding sites for sequence-specific TFs. Some of these TFs may preferentially activate promoters containing specific core promoter elements (colored according to the core promoter composition). The diagram is not to scale. TBP- TATA box-binding protein; TRF2- TBP-related factor 2; M1BP- motif 1 binding protein; CBP- CREB-binding protein. DNA framework was modified from an image of the National Human Genome Research Institute Image Gallery.
Pol II Promoter-Proximal Pausing
The core promoter not only plays a role in Pol II recruitment to initiate transcription, it also regulates Pol II promoter-proximal pausing following transcription initiation. Pausing of transcriptionally engaged Pol II when the transcript is ∼25 nucleotides long was discovered in the Drosophila heat shock protein 70 (hsp70) promoter (Gilmour and Lis, 1986; Rougvie and Lis, 1988). The development and application of genome-wide techniques directly measuring nascent RNA revealed that pausing of Pol II when the transcript is 20–60 nucleotides long is a common feature of Drosophila and mammalian genes under tight temporal control (reviewed in Core and Adelman, 2019; Lis, 2019; Wissink et al., 2019). In general, pausing extent and timing are regulated via nucleosome positioning (Gilchrist et al., 2010; Luse et al., 2020). Pol II pausing is now considered a major step regulating promoters that are required to elicit precise and rapid transcription, such as developmental and signal-responsive processes (reviewed in Gaertner and Zeitlinger, 2014; Mayer et al., 2017; Core and Adelman, 2019).
Pol II pausing is a major regulator of Drosophila morphogenesis, coordinating the synchronization of gene expression that is crucial for proper mesoderm development (Lagha et al., 2013). Analysis of core promoters of paused genes revealed that they are GC rich, and one-fourth of them contain a pause button (PB) motif (Hendrix et al., 2008). The PB motif consensus sequence “KCGRWCG” is enriched in the downstream core promoter, from + 1 to + 60 relative to the TSS. Moreover, the PB closely resembles the DPE, and can be located in close proximity to it. The exact interplay between the DPE and the PB, which is not strictly positioned relative to the Inr, awaits further investigation. GAGA factor binding sites and Ohler motif 1, as well as the transcription factors that specifically interact with them, also play a role in the regulation of pausing (Li and Gilmour, 2013). The precise mapping of Pol II on nascent RNA using PRO-seq demonstrated that the strength and position of sequence elements directly regulate pausing levels (Kwak et al., 2013). ChIP-nexus analysis demonstrated that downstream promoter sequences contribute to pausing stability, and a G at the + 2 position has an important role in stabilizing Pol II pausing (Shao et al., 2019). Remarkably, TFIID itself is required for establishing Pol II pausing, and depletion of its TAF1 and TAF2 subunits, but not of TBP, resulted in increased Pol II pause release and transcription re-initiation (Fant et al., 2020). Notably, some developmental promoters are paused, yet are not expressed in specific tissues (Gaertner et al., 2012), suggesting the involvement of other, tissue-specific, transcription factors in their regulation. These findings highlight the potential contribution of the downstream core promoter region to Pol II pausing, a critical regulatory aspect of transcriptional and developmental gene regulation.
Diversity in TAFs and TBP Family Members is Associated With Different Core Promoter Compositions and Developmental Programs
Similar to core promoter sequence diversity, PIC composition is not universal, but rather promoter-dependent (illustrated in Figure 2B; Sikorski and Buratowski, 2009). This diversity is manifested by distinct TFIID complexes that contain paralogous TBP and tissue-specific TAF variants, a feature shared across metazoan organisms (Davidson, 2003; Freiman, 2009; Goodrich and Tjian, 2010; Ohler and Wassarman, 2010; Jones, 2014; Levine et al., 2014; Gupta et al., 2017). Depletion of TBP, TBP-related factor 2 (TRF2) and other TAFs using RNAi in Drosophila demonstrated the importance of TAF9, TAF5, and TRF2 to establish neural stem cell identity, emphasizing the role of specialized basal transcription machinery in developmental gene regulation (Neves and Eisenman, 2019). Tissue-specific complexes can also facilitate the discovery of additional core promoter elements. Indeed, the analysis of cell type-specific transcriptional programs during Drosophila spermatogenesis revealed the enrichment of novel downstream core promoter sequences bound by the spermatocyte-specific tMAC complex (Lu et al., 2020).
Such precise expression and function of tissue-specific TAF subunits is seen in diverse aspects of mammalian differentiation. Taf4b, Taf7l, and Taf9b are coordinately expressed in a highly germ cell-specific manner during mouse embryonic germ cell development (Gura et al., 2020). Interestingly, TAF7l and TAF9b are also required for regulating mouse adipocyte and neuronal differentiation, respectively (Zhou et al., 2014; Herrera et al., 2014). Selective expression and use of subsets of TAFs establishes a transcriptional program required for human embryonic stem cell (ESCs) self-renewal (Maston et al., 2012). Knockdown of multiple TFIID subunits affects the pluripotent circuitry in mouse ESCs and inhibits reprogramming of fibroblasts. Conversely, ectopic expression of mouse TAF4 greatly enhanced the reprogramming efficiency of mouse embryonic fibroblasts to induced pluripotent stem cells (Pijnappel et al., 2013). These data highlight the adaptability of selective assemblies of TAFs and their core promoter target sequences as key hubs for driving pluripotency and differentiation across highly divergent organisms.
The basal transcription factor TBP-related factor 2 (TRF2) is essential for embryonic development of C. elegans, D. melanogaster, D. rerio, and X. laevis (Dantonel et al., 2000; Kaltenbach et al., 2000; Veenstra et al., 2000; Muller et al., 2001; Kopytova et al., 2006). While TRF2 does not bind TATA-box-containing promoters, TRF2-containing complexes specifically interact with DPE-containing promoters and regulate TCT-containing ribosomal protein genes, among other gene classes (Bashirullah et al., 2007; Isogai et al., 2007; Oyama et al., 2013; Kedmi et al., 2014; Wang et al., 2014). Interestingly, M1BP was shown to interact with TRF2 and to regulate the expression of ribosomal protein genes (Baumann and Gilmour, 2017).
TRF2 emerged from TBP via gene duplication in the last common ancestor before the evolutionary split between bilaterians and non-bilaterian species (Duttke et al., 2014). Since the TRF2 protein lacks 3 of the 4 phenylalanine residues required for TATA box-binding, it may have evolved to support transcriptional programs dependent on TATA-less promoters, which are involved in the generation of the three germ layers (endoderm, mesoderm and ectoderm). Notably, DPE-containing genes are highly enriched for GO terms associated with embryonic development and particularly in mesoderm formation (Zehavi et al., 2014a; Sloutskin et al., 2015), suggesting that TRF2-based transcriptional programs might support mesoderm formation via the DPE. Moreover, TATA box and DPE-containing promoters can be regulated in an antagonist manner through TBP and NC2 with Mot1 binding, respectively (Hsu et al., 2008; van Werven et al., 2008). Interestingly, multiple Xenopus genes involved in mesoderm and organizer specification are TBP family-insensitive (Gazdag et al., 2016). The existence of multiple TBP family members with distinct characteristics, and of TBP family-insensitive developmental programs, are additional manifestations of the diversity in the transcriptional programs that support embryonic development through the downstream core promoter.
Core Promoter-Enhancer Specificity, Similarity and Transcriptional Dynamics
While for many years it was thought that the specificity of gene expression is provided by enhancers, it is now clear that the core promoter region plays a key role in regulating gene- and cell type-specific transcription (Zehavi et al., 2014a; Zabidi and Stark, 2016). Distinct enhancers that preferentially work with either DPE- or TATA box-dependent Drosophila promoters have been identified (Butler and Kadonaga, 2001). Using a Drosophila cell culture-based reporter assay, developmental gene promoters were activated to higher levels than housekeeping gene promoters, with the Inr and DPE motifs contributing toward greater enhancer responsiveness than the TATA box (Arnold et al., 2017). Different TFs can have specific preferences for binding and activating promoters, based on their core promoter composition (Haberle et al., 2019). It was recently shown that the tMAC spermatocyte-specific complex binds novel promoter-proximal sequence motifs and opens the local chromatin to promote transcription initiation from downstream alternative promoters, indicating that proximal promoter composition may influence progression of differentiation programs as robustly as enhancers (Lu et al., 2020).
Drosophila Caudal, as well as mouse Caudal-related homeobox (Cdx) proteins (Cdx1, Cdx2, and Cdx4), which are key regulators of embryonic development and differentiation, activate transcription with a preference for a DPE relative to a TATA box (Juven-Gershon et al., 2008; Shir-Shapira et al., 2015). It is the unique combination of the Drosophila CREB-binding protein (dCBP) co-activator and Caudal that enables core promoter-preferential activation. Notably, dCBP co-occupies genomic loci in Drosophila embryos that are bound by Dorsal (Holmqvist et al., 2012; Holmqvist and Mannervik, 2013), in line with the involvement of CBP in Dorsal-target gene expression during embryogenesis (Akimaru et al., 1997) and with the enrichment of DPE motifs in mesodermal genes (Zehavi et al., 2014a). Tethering nuclease deficient Cas9 (dCas9) fused to either the Synergistic Activation Mediator (SAM) or the CBP HAT domain to the same promoters resulted in different activation levels for different promoters, supporting the concept of activator-promoter compatibility (Sajwan and Mannervik, 2019).
There is a large body of evidence supporting the similarity between promoters and enhancers, as many enhancers are transcribed and basal transcription machinery components are recruited to enhancers (Koch et al., 2011; Andersson et al., 2014; Core et al., 2014; Scruggs et al., 2015). Transcription of non-coding enhancer RNAs (eRNAs) is a common phenomenon discovered in human cell lines, correlated with the corresponding promoter’s strength (reviewed in Andersson et al., 2015). Both the existence of eRNA and the correlation to transcriptional output was confirmed in Drosophila embryos as well (Mikhaylichenko et al., 2018). The similarity between promoters and enhancers is reviewed in detail in Andersson and Sandelin (2020). Furthermore, core promoter sequences are necessary for proper enhancer function in human K562 cells, and can be used to predict and modulate enhancer activity (Tippens et al., 2020). A systematic dissection of distinct core promoter elements that function within transcribed enhancers will be instrumental.
High-precision methodologies enabled the examination of transcriptional dynamics, which demonstrated the existence of transcriptional bursts (Suter et al., 2011; Bothma et al., 2014). Burst size and initiation are controlled by the release of the paused Pol II molecules, while the recruitment rate remains the same across different promoters (Bartman et al., 2019; Cao et al., 2020). Both core promoter and TFIID compositions were shown to affect the magnitude of the initial transcriptional burst following the induction of cultured Drosophila cells with Cu (Pennington et al., 2013). The combination of advanced live-imaging techniques with quantitative analysis of transcriptional strength in Drosophila embryos revealed that enhancers control burst frequency (Fukaya et al., 2016). Interestingly, it was recently suggested that enhancer-promoter distance contributes to the regulation of gene activity by changing the size and the timing of transcriptional bursting in the developing Drosophila embryo (Yokoshi et al., 2020). Transcriptional dynamics is dictated not only by enhancers, but also by core promoter composition. Different core promoter sequences display divergent transcriptional dynamics profiles, observed in Drosophila embryos using MS2-based reporters (Fukaya et al., 2016). The TATA box and Inr elements affect burst size in mouse embryonic stem cells and in human fibroblasts (Larsson et al., 2019), whereas burst frequency is subject to regulation by enhancers and activators (Larsson et al., 2019; Wang Y. et al., 2020). The effects of endogenous core promoter motifs, including downstream elements, on transcriptional bursting remain to be determined.
Concluding Remarks
We envision the core promoter composition as an additional regulatory dimension of the complex developmental GRNs. The DPE acronym, originally defined based on its position, may be indicative of its function as a Developmental core Promoter Element. Notably, the downstream core promoter region is transcribed, thus one could speculate that it may exert a regulatory effect at the RNA level. The downstream core promoter region is an important regulatory tier of gene expression, likely conserved and diversified among metazoans. Thus, identification and characterization of downstream motifs in additional mammalian species would likely uncover the regulatory roles of mammalian downstream core promoters and their protein binding factors. Differential core promoter composition may provide a nuanced regulatory switch that recruits the relevant basal transcription factors at the right time during embryonic development to more precisely control the timing of developmental transcriptional programs. Being able to directly visualize and measure transcriptional dynamics will uncover novel insights regarding the functions, mechanisms and definitions of promoters and enhancers, which regulate the complex process known as “transcription.”
Author Contributions
TJ-G and RF conceptualized the mini-review. AS and HS-S wrote the first draft of the manuscript. AS prepared the figures. AS and TJ-G finalized the review with input from all authors.
Funding
The analysis of the basal transcription machinery and core promoter regulation of pluripotency and differentiation in the Juven-Gershon and Freiman labs was supported by the United States-Israel Binational Science Foundation Grant No. 2019285. Research in the Juven-Gershon lab was supported by the grant from the Israeli Science Foundation (1234/17) to TJ-G. Research in the Freiman lab was supported by the grant from the United States National Institutes of Health/National Institute of Child Health and Human Development (HD09184) to RF.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Jim Kadonaga, Diana Ideses, Orit Adato, and Yehuda M. Danino for critically reading this manuscript. We apologize to authors whose work could not be cited due to space limitations.
References
Akimaru, H., Hou, D. X., and Ishii, S. (1997). Drosophila CBP is required for dorsal-dependent twist gene expression. Nat. Genet. 17, 211–214. doi: 10.1038/ng1097-211
Andersson, R., Gebhard, C., Miguel-Escalada, I., Hoof, I., Bornholdt, J., Boyd, M., et al. (2014). An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461. doi: 10.1038/nature12787
Andersson, R., and Sandelin, A. (2020). Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 21, 71–87. doi: 10.1038/s41576-019-0173-8
Andersson, R., Sandelin, A., and Danko, C. G. (2015). A unified architecture of transcriptional regulatory elements. Trends Genet. 31, 426–433. doi: 10.1016/j.tig.2015.05.007
Anish, R., Hossain, M. B., Jacobson, R. H., and Takada, S. (2009). Characterization of transcription from TATA-less promoters: identification of a new core promoter element XCPE2 and analysis of factor requirements. PLoS One 4:e5103. doi: 10.1371/journal.pone.0005103
Antonova, S. V., Boeren, J., Timmers, H. T. M., and Snel, B. (2019). Epigenetics and transcription regulation during eukaryotic diversification: the saga of TFIID. Genes Dev. 33, 888–902. doi: 10.1101/gad.300475.117
Arnold, C. D., Zabidi, M. A., Pagani, M., Rath, M., Schernhuber, K., Kazmar, T., et al. (2017). Genome-wide assessment of sequence-intrinsic enhancer responsiveness at single-base-pair resolution. Nat. Biotechnol. 35, 136–144. doi: 10.1038/nbt.3739
Bartman, C. R., Hamagami, N., Keller, C. A., Giardine, B., Hardison, R. C., Blobel, G. A., et al. (2019). Transcriptional burst initiation and polymerase pause release are key control points of transcriptional regulation. Mol. Cell 73, 519–532.e4. doi: 10.1016/j.molcel.2018.11.004
Bashirullah, A., Lam, G., Yin, V. P., and Thummel, C. S. (2007). dTrf2 is required for transcriptional and developmental responses to ecdysone during Drosophila metamorphosis. Dev. Dyn. 236, 3173–3179. doi: 10.1002/dvdy.21350
Batut, P. J., and Gingeras, T. R. (2017). Conserved noncoding transcription and core promoter regulatory code in early Drosophila development. Elife 6:e29005. doi: 10.7554/eLife.29005
Baumann, D. G., and Gilmour, D. S. (2017). A sequence-specific core promoter-binding transcription factor recruits TRF2 to coordinately transcribe ribosomal protein genes. Nucleic Acids Res. 45, 10481–10491. doi: 10.1093/nar/gkx676
Bhuiyan, T., and Timmers, H. T. M. (2019). Promoter recognition: putting TFIID on the spot. Trends Cell Biol. 29, 752–763. doi: 10.1016/j.tcb.2019.06.004
Bolt, C. C., and Duboule, D. (2020). The regulatory landscapes of developmental genes. Development 147:dev171736. doi: 10.1242/dev.171736
Bothma, J. P., Garcia, H. G., Esposito, E., Schlissel, G., Gregor, T., and Levine, M. (2014). Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc. Natl. Acad. Sci. U.S.A. 111, 10598–10603. doi: 10.1073/pnas.1410022111
Burke, T. W., and Kadonaga, J. T. (1996). Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 10, 711–724. doi: 10.1101/gad.10.6.711
Burke, T. W., and Kadonaga, J. T. (1997). The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAF(II)60 of Drosophila. Genes Dev. 11, 3020–3031. doi: 10.1101/gad.11.22.3020
Butler, J. E., and Kadonaga, J. T. (2001). Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15, 2515–2519. doi: 10.1101/gad.924301
Cao, Z., Filatova, T., Oyarzun, D. A., and Grima, R. (2020). A stochastic model of gene expression with polymerase recruitment and pause release. Biophys. J. 119, 1002–1014. doi: 10.1016/j.bpj.2020.07.020
Carninci, P., Sandelin, A., Lenhard, B., Katayama, S., Shimokawa, K., Ponjavic, J., et al. (2006). Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38, 626–635. doi: 10.1038/ng1789
Chalkley, G. E., and Verrijzer, C. P. (1999). DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J. 18, 4835–4845. doi: 10.1093/emboj/18.17.4835
Corden, J., Wasylyk, B., Buchwalder, A., Sassone-Corsi, P., Kedinger, C., and Chambon, P. (1980). Promoter sequences of eukaryotic protein-coding genes. Science 209, 1406–1414. doi: 10.1126/science.6251548
Core, L., and Adelman, K. (2019). Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 33, 960–982. doi: 10.1101/gad.325142.119
Core, L. J., Martins, A. L., Danko, C. G., Waters, C. T., Siepel, A., and Lis, J. T. (2014). Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 46, 1311–1320. doi: 10.1038/ng.3142
Cramer, P. (2019a). Eukaryotic transcription turns 50. Cell 179, 808–812. doi: 10.1016/j.cell.2019.09.018
Cramer, P. (2019b). Organization and regulation of gene transcription. Nature 573, 45–54. doi: 10.1038/s41586-019-1517-4
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Danino, Y. M., Even, D., Ideses, D., and Juven-Gershon, T. (2015). The core promoter: at the heart of gene expression. Biochim. Biophys. Acta 1849, 1116–1131. doi: 10.1016/j.bbagrm.2015.04.003
Dantonel, J. C., Quintin, S., Lakatos, L., Labouesse, M., and Tora, L. (2000). TBP-like factor is required for embryonic RNA polymerase II transcription in C. elegans. Mol. Cell 6, 715–722.
Davidson, I. (2003). The genetics of TBP and TBP-related factors. Trends Biochem. Sci. 28, 391–398. doi: 10.1016/S0968-0004(03)00117-8
Deng, W., and Roberts, S. G. (2005). A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 19, 2418–2423. doi: 10.1101/gad.342405
Dikstein, R. (2011). The unexpected traits associated with core promoter elements. Transcription 2, 201–206. doi: 10.4161/trns.2.5.17271
Duttke, S. H., Doolittle, R. F., Wang, Y. L., and Kadonaga, J. T. (2014). TRF2 and the evolution of the bilateria. Genes Dev. 28, 2071–2076. doi: 10.1101/gad.250563.114
Fant, C. B., Levandowski, C. B., Gupta, K., Maas, Z. L., Moir, J., Rubin, J. D., et al. (2020). TFIID enables RNA polymerase II promoter-proximal pausing. Mol. Cell 78, 785.e–793.e. doi: 10.1016/j.molcel.2020.03.008
FitzGerald, P. C., Sturgill, D., Shyakhtenko, A., Oliver, B., and Vinson, C. (2006). Comparative genomics of Drosophila and human core promoters. Genome Biol. 7:R53. doi: 10.1186/gb-2006-7-7-r53
Freiman, R. N. (2009). Specific variants of general transcription factors regulate germ cell development in diverse organisms. Biochim. Biophys. Acta 1789, 161–166.
Frith, M. C. and Fantom Consortium (2014). Explaining the correlations among properties of mammalian promoters. Nucleic Acids Res. 42, 4823–4832. doi: 10.1093/nar/gku115
Fuda, N. J., Ardehali, M. B., and Lis, J. T. (2009). Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461, 186–192. doi: 10.1038/nature08449
Fukaya, T., Lim, B., and Levine, M. (2016). Enhancer control of transcriptional bursting. Cell 166, 358–368. doi: 10.1016/j.cell.2016.05.025
Gaertner, B., Johnston, J., Chen, K., Wallaschek, N., Paulson, A., Garruss, A. S., et al. (2012). Poised RNA polymerase II changes over developmental time and prepares genes for future expression. Cell Rep. 2, 1670–1683. doi: 10.1016/j.celrep.2012.11.024
Gaertner, B., and Zeitlinger, J. (2014). RNA polymerase II pausing during development. Development 141, 1179–1183. doi: 10.1242/dev.088492
Gazdag, E., Jacobi, U. G., van Kruijsbergen, I., Weeks, D. L., and Veenstra, G. J. (2016). Activation of a T-box-Otx2-Gsc gene network independent of TBP and TBP-related factors. Development 143, 1340–1350. doi: 10.1242/dev.127936
Gehring, W. J., and Hiromi, Y. (1986). Homeotic genes and the homeobox. Annu. Rev. Genet. 20, 147–173. doi: 10.1146/annurev.ge.20.120186.001051
Gershenzon, N. I., and Ioshikhes, I. P. (2005). Synergy of human Pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics 21, 1295–1300. doi: 10.1093/bioinformatics/bti172
Gershenzon, N. I., Trifonov, E. N., and Ioshikhes, I. P. (2006). The features of Drosophila core promoters revealed by statistical analysis. BMC Genomics 7:161. doi: 10.1186/1471-2164-7-161
Gilchrist, D. A., Dos Santos, G., Fargo, D. C., Xie, B., Gao, Y., Li, L., et al. (2010). Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143, 540–551. doi: 10.1016/j.cell.2010.10.004
Gilmour, D. S., and Lis, J. T. (1986). RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol. Cell. Biol. 6, 3984–3989. doi: 10.1128/mcb.6.11.3984-3989.1986
Goldberg, M. L. (1979). Sequence Analysis of Drosophila Histone Genes. Ph.D. Thesis. Stanford, CA: Stanford University.
Goodrich, J. A., and Tjian, R. (2010). Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat. Rev. Genet. 11, 549–558. doi: 10.1038/nrg2847
Gupta, K., Watson, A. A., Baptista, T., Scheer, E., Chambers, A. L., Koehler, C., et al. (2017). Architecture of TAF11/TAF13/TBP complex suggests novel regulation properties of general transcription factor TFIID. Elife 6:e30395. doi: 10.7554/eLife.30395
Gura, M. A., Mikedis, M. M., Seymour, K. A., de Rooij, D. G., Page, D. C., and Freiman, R. N. (2020). Dynamic and regulated TAF gene expression during mouse embryonic germ cell development. PLoS Genet. 16:e1008515. doi: 10.1371/journal.pgen.1008515
Haberle, V., Arnold, C. D., Pagani, M., Rath, M., Schernhuber, K., and Stark, A. (2019). Transcriptional cofactors display specificity for distinct types of core promoters. Nature 570, 122–126. doi: 10.1038/s41586-019-1210-7
Haberle, V., and Stark, A. (2018). Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 19, 621–637. doi: 10.1038/s41580-018-0028-8
Hariharan, N., and Perry, R. P. (1990). Functional dissection of a mouse ribosomal protein promoter: significance of the polypyrimidine initiator and an element in the TATA-box region. Proc. Natl. Acad. Sci. U.S.A. 87, 1526–1530. doi: 10.1073/pnas.87.4.1526
Heintzman, N. D., and Ren, B. (2007). The gateway to transcription: identifying, characterizing and understanding promoters in the eukaryotic genome. Cell. Mol. Life Sci. 64, 386–400. doi: 10.1007/s00018-006-6295-0
Hendrix, D. A., Hong, J. W., Zeitlinger, J., Rokhsar, D. S., and Levine, M. S. (2008). Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc. Natl. Acad. Sci. U.S.A. 105, 7762–7767. doi: 10.1073/pnas.0802406105
Herrera, F. J., Yamaguchi, T., Roelink, H., and Tjian, R. (2014). Core promoter factor TAF9B regulates neuronal gene expression. Elife 3:e02559. doi: 10.7554/eLife.02559
Hirose, F., Yamaguchi, M., Handa, H., Inomata, Y., and Matsukage, A. (1993). Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase alpha and proliferating cell nuclear antigen. J. Biol. Chem. 268, 2092–2099.
Holmqvist, P. H., Boija, A., Philip, P., Crona, F., Stenberg, P., and Mannervik, M. (2012). Preferential genome targeting of the CBP co-activator by Rel and Smad proteins in early Drosophila melanogaster embryos. PLoS Genet. 8:e1002769. doi: 10.1371/journal.pgen.1002769
Holmqvist, P. H., and Mannervik, M. (2013). Genomic occupancy of the transcriptional co-activators p300 and CBP. Transcription 4, 18–23. doi: 10.4161/trns.22601
Hoskins, R. A., Landolin, J. M., Brown, J. B., Sandler, J. E., Takahashi, H., Lassmann, T., et al. (2011). Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 21, 182–192. doi: 10.1101/gr.112466.110
Hsu, J. Y., Juven-Gershon, T., Marr, M. T. II, Wright, K. J., Tjian, R., and Kadonaga, J. T. (2008). TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 22, 2353–2358. doi: 10.1101/gad.1681808
Isogai, Y., Keles, S., Prestel, M., Hochheimer, A., and Tjian, R. (2007). Transcription of histone gene cluster by differential core-promoter factors. Genes Dev. 21, 2936–2949. doi: 10.1101/gad.1608807
Juven-Gershon, T., Cheng, S., and Kadonaga, J. T. (2006). Rational design of a super core promoter that enhances gene expression. Nat. Methods 3, 917–922.
Juven-Gershon, T., and Kadonaga, J. T. (2010). Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339, 225–229.
Juven-Gershon, T., Hsu, J.-Y., and Kadonaga, J. T. (2008). Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes Dev. 22, 2823–2830. doi: 10.1101/gad.1698108
Kadonaga, J. T. (2012). Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip. Rev. Dev. Biol. 1, 40–51. doi: 10.1002/wdev.21
Kaltenbach, L., Horner, M. A., Rothman, J. H., and Mango, S. E. (2000). The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol. Cell 6, 705–713.
Kaufmann, J., and Smale, S. T. (1994). Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev. 8, 821–829. doi: 10.1101/gad.8.7.821
Kedmi, A., Zehavi, Y., Glick, Y., Orenstein, Y., Ideses, D., Wachtel, C., et al. (2014). Drosophila TRF2 is a preferential core promoter regulator. Genes Dev. 28, 2163–2174. doi: 10.1101/gad.245670.114
Kim, T. H., Barrera, L. O., Zheng, M., Qu, C., Singer, M. A., Richmond, T. A., et al. (2005). A high-resolution map of active promoters in the human genome. Nature 436, 876–880. doi: 10.1038/nature03877
Koch, F., Fenouil, R., Gut, M., Cauchy, P., Albert, T. K., Zacarias-Cabeza, J., et al. (2011). Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat. Struct. Mol. Biol. 18, 956–963. doi: 10.1038/nsmb.2085
Kopytova, D. V., Krasnov, A. N., Kopantceva, M. R., Nabirochkina, E. N., Nikolenko, J. V., Maksimenko, O., et al. (2006). Two isoforms of Drosophila TRF2 are involved in embryonic development, premeiotic chromatin condensation, and proper differentiation of germ cells of both sexes. Mol. Cell. Biol. 26, 7492–7505. doi: 10.1128/MCB.00349-06
Koster, M. J., Snel, B., and Timmers, H. T. (2015). Genesis of chromatin and transcription dynamics in the origin of species. Cell 161, 724–736. doi: 10.1016/j.cell.2015.04.033
Kutach, A. K., and Kadonaga, J. T. (2000). The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 20, 4754–4764. doi: 10.1128/mcb.20.13.4754-4764.2000
Kwak, H., Fuda, N. J., Core, L. J., and Lis, J. T. (2013). Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339, 950–953. doi: 10.1126/science.1229386
Lagha, M., Bothma, J. P., Esposito, E., Ng, S., Stefanik, L., Tsui, C., et al. (2013). Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell 153, 976–987. doi: 10.1016/j.cell.2013.04.045
Lagrange, T., Kapanidis, A. N., Tang, H., Reinberg, D., and Ebright, R. H. (1998). New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 12, 34–44.
Larsson, A. J. M., Johnsson, P., Hagemann-Jensen, M., Hartmanis, L., Faridani, O. R., Reinius, B., et al. (2019). Genomic encoding of transcriptional burst kinetics. Nature 565, 251–254. doi: 10.1038/s41586-018-0836-1
Lenhard, B., Sandelin, A., and Carninci, P. (2012). Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 13, 233–245. doi: 10.1038/nrg3163
Levi, T., Sloutskin, A., Kalifa, R., Juven-Gershon, T., and Gerlitz, O. (2020). Efficient in vivo introduction of point mutations using ssODN and a Co-CRISPR approach. Biol. Proced. Online 22:14. doi: 10.1186/s12575-020-00123-7
Levine, M., Cattoglio, C., and Tjian, R. (2014). Looping back to leap forward: transcription enters a new era. Cell 157, 13–25. doi: 10.1016/j.cell.2014.02.009
Levine, M., and Tjian, R. (2003). Transcription regulation and animal diversity. Nature 424, 147–151. doi: 10.1038/nature01763
Li, J., and Gilmour, D. S. (2013). Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor. EMBO J. 32, 1829–1841. doi: 10.1038/emboj.2013.111
Li, X., and Noll, M. (1994). Compatibility between enhancers and promoters determines the transcriptional specificity of gooseberry and gooseberry neuro in the Drosophila embryo. EMBO J. 13, 400–406.
Lim, C. Y., Santoso, B., Boulay, T., Dong, E., Ohler, U., and Kadonaga, J. T. (2004). The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 18, 1606–1617. doi: 10.1101/gad.1193404
Lis, J. T. (2019). A 50 year history of technologies that drove discovery in eukaryotic transcription regulation. Nat. Struct. Mol. Biol. 26, 777–782. doi: 10.1038/s41594-019-0288-9
Lo, K., and Smale, S. T. (1996). Generality of a functional initiator consensus sequence. Gene 182, 13–22. doi: 10.1016/s0378-1119(96)00438-6
Louder, R. K., He, Y., Lopez-Blanco, J. R., Fang, J., Chacon, P., and Nogales, E. (2016). Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531, 604–609. doi: 10.1038/nature17394
Lu, D., Sin, H. S., Lu, C., and Fuller, M. T. (2020). Developmental regulation of cell type-specific transcription by novel promoter-proximal sequence elements. Genes Dev. 34, 663–677. doi: 10.1101/gad.335331.119
Luse, D. S., Parida, M., Spector, B. M., Nilson, K. A., and Price, D. H. (2020). A unified view of the sequence and functional organization of the human RNA polymerase II promoter. Nucleic Acids Res. 48, 7767–7785. doi: 10.1093/nar/gkaa531
Maston, G. A., Zhu, L. J., Chamberlain, L., Lin, L., Fang, M., and Green, M. R. (2012). Non-canonical TAF complexes regulate active promoters in human embryonic stem cells. Elife 1:e00068. doi: 10.7554/eLife.00068
Mayer, A., Landry, H. M., and Churchman, L. S. (2017). Pause & go: from the discovery of RNA polymerase pausing to its functional implications. Curr. Opin. Cell Biol. 46, 72–80. doi: 10.1016/j.ceb.2017.03.002
Merli, C., Bergstrom, D. E., Cygan, J. A., and Blackman, R. K. (1996). Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev. 10, 1260–1270. doi: 10.1101/gad.10.10.1260
Mikhaylichenko, O., Bondarenko, V., Harnett, D., Schor, I. E., Males, M., Viales, R. R., et al. (2018). The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 32, 42–57. doi: 10.1101/gad.308619.117
Muller, F., Lakatos, L., Dantonel, J., Strahle, U., and Tora, L. (2001). TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr. Biol. 11, 282–287.
Muller, F., and Tora, L. (2014). Chromatin and DNA sequences in defining promoters for transcription initiation. Biochim. Biophys. Acta 1839, 118–128. doi: 10.1016/j.bbagrm.2013.11.003
Neves, A., and Eisenman, R. N. (2019). Distinct gene-selective roles for a network of core promoter factors in Drosophila neural stem cell identity. Biol. Open 8:bio042168. doi: 10.1242/bio.042168
Ohler, U., Liao, G. C., Niemann, H., and Rubin, G. M. (2002). Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3:RESEARCH0087.
Ohler, U., and Wassarman, D. A. (2010). Promoting developmental transcription. Development 137, 15–26.
Oyama, T., Sasagawa, S., Takeda, S., Hess, R. A., Lieberman, P. M., Cheng, E. H., et al. (2013). Cleavage of TFIIA by Taspase1 activates TRF2-specified mammalian male germ cell programs. Dev. Cell 27, 188–200. doi: 10.1016/j.devcel.2013.09.025
Parry, T. J., Theisen, J. W. M., Hsu, J.-Y., Wang, Y.-L., Corcoran, D. L., Eustice, M., et al. (2010). The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes Dev. 24, 2013–2018. doi: 10.1101/gad.1951110
Patel, A. B., Greber, B. J., and Nogales, E. (2020). Recent insights into the structure of TFIID, its assembly, and its binding to core promoter. Curr. Opin. Struct. Biol. 61, 17–24. doi: 10.1016/j.sbi.2019.10.001
Pennington, K. L., Marr, S. K., Chirn, G. W., and Marr, M. T. II (2013). Holo-TFIID controls the magnitude of a transcription burst and fine-tuning of transcription. Proc. Natl. Acad. Sci. U.S.A. 110, 7678–7683. doi: 10.1073/pnas.1221712110
Perry, R. P. (2005). The architecture of mammalian ribosomal protein promoters. BMC Evol. Biol. 5:15. doi: 10.1186/1471-2148-5-15
Pijnappel, W. W., Esch, D., Baltissen, M. P., Wu, G., Mischerikow, N., Bergsma, A. J., et al. (2013). A central role for TFIID in the pluripotent transcription circuitry. Nature 495, 516–519. doi: 10.1038/nature11970
Rach, E. A., Yuan, H. Y., Majoros, W. H., Tomancak, P., and Ohler, U. (2009). Motif composition, conservation and condition-specificity of single and alternative transcription start sites in the Drosophila genome. Genome Biol. 10:R73. doi: 10.1186/gb-2009-10-7-r73
Reeve, J. N. (2003). Archaeal chromatin and transcription. Mol. Microbiol. 48, 587–598. doi: 10.1046/j.1365-2958.2003.03439.x
Roeder, R. G. (2019). 50+ years of eukaryotic transcription: an expanding universe of factors and mechanisms. Nat. Struct. Mol. Biol. 26, 783–791. doi: 10.1038/s41594-019-0287-x
Rougvie, A. E., and Lis, J. T. (1988). The RNA polymerase II molecule at the 5’ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54, 795–804. doi: 10.1016/s0092-8674(88)91087-2
Roy, A. L., and Singer, D. S. (2015). Core promoters in transcription: old problem, new insights. Trends Biochem. Sci. 40, 165–171. doi: 10.1016/j.tibs.2015.01.007
Sajwan, S., and Mannervik, M. (2019). Gene activation by dCas9-CBP and the SAM system differ in target preference. Sci. Rep. 9:18104. doi: 10.1038/s41598-019-54179-x
Sandelin, A., Carninci, P., Lenhard, B., Ponjavic, J., Hayashizaki, Y., and Hume, D. A. (2007). Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 8, 424–436. doi: 10.1038/nrg2026
Schier, A. C., and Taatjes, D. J. (2020). Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 34, 465–488. doi: 10.1101/gad.335679.119
Scruggs, B. S., Gilchrist, D. A., Nechaev, S., Muse, G. W., Burkholder, A., Fargo, D. C., et al. (2015). Bidirectional transcription arises from two distinct hubs of transcription factor binding and active chromatin. Mol. Cell 58, 1101–1112. doi: 10.1016/j.molcel.2015.04.006
Shao, W., Alcantara, S. G., and Zeitlinger, J. (2019). Reporter-ChIP-nexus reveals strong contribution of the Drosophila initiator sequence to RNA polymerase pausing. Elife 8:e41461. doi: 10.7554/eLife.41461
Shir-Shapira, H., Sharabany, J., Filderman, M., Ideses, D., Ovadia-Shochat, A., Mannervik, M., et al. (2015). Structure-function analysis of the Drosophila melanogaster caudal transcription factor provides insights into core promoter-preferential activation. J. Biol. Chem. 290, 17293–17305. doi: 10.1074/jbc.M114.632109
Shir-Shapira, H., Sloutskin, A., Adato, O., Ovadia-Shochat, A., Ideses, D., Zehavi, Y., et al. (2019). Identification of evolutionarily conserved downstream core promoter elements required for the transcriptional regulation of Fushi tarazu target genes. PLoS One 14:e0215695. doi: 10.1371/journal.pone.0215695
Sikorski, T. W., and Buratowski, S. (2009). The basal initiation machinery: beyond the general transcription factors. Curr. Opin. Cell Biol. 21, 344–351. doi: 10.1016/j.ceb.2009.03.006
Sloutskin, A., Danino, Y. M., Orenstein, Y., Zehavi, Y., Doniger, T., Shamir, R., et al. (2015). ElemeNT: a computational tool for detecting core promoter elements. Transcription 6, 41–50. doi: 10.1080/21541264.2015.1067286
Smale, S. T., and Baltimore, D. (1989). The “initiator” as a transcription control element. Cell 57, 103–113.
Suter, D. M., Molina, N., Gatfield, D., Schneider, K., Schibler, U., and Naef, F. (2011). Mammalian genes are transcribed with widely different bursting kinetics. Science 332, 472–474. doi: 10.1126/science.1198817
Theisen, J. W., Lim, C. Y., and Kadonaga, J. T. (2010). Three key subregions contribute to the function of the downstream RNA polymerase II core promoter. Mol. Cell. Biol. 30, 3471–3479. doi: 10.1128/MCB.00053-10
Thomas, M. C., and Chiang, C. M. (2006). The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178.
Tippens, N. D., Liang, J., Leung, A. K., Wierbowski, S. D., Ozer, A., Booth, J. G., et al. (2020). Transcription imparts architecture, function and logic to enhancer units. Nat. Genet. 52, 1067–1075. doi: 10.1038/s41588-020-0686-2
Tokusumi, Y., Ma, Y., Song, X., Jacobson, R. H., and Takada, S. (2007). The new core promoter element XCPE1 (X core promoter element 1) directs activator-, mediator-, and TATA-binding protein-dependent but TFIID-independent RNA polymerase II transcription from TATA-less promoters. Mol. Cell. Biol. 27, 1844–1858. doi: 10.1128/MCB.01363-06
van Werven, F. J., van Bakel, H., van Teeffelen, H. A., Altelaar, A. F., Koerkamp, M. G., Heck, A. J., et al. (2008). Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev. 22, 2359–2369. doi: 10.1101/gad.1682308
Veenstra, G. J., Weeks, D. L., and Wolffe, A. P. (2000). Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science 290, 2312–2315. doi: 10.1126/science.290.5500.2312
Verrijzer, C. P., Chen, J. L., Yokomori, K., and Tjian, R. (1995). Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell 81, 1115–1125. doi: 10.1016/s0092-8674(05)80016-9
Vo Ngoc, L., Cassidy, C. J., Huang, C. Y., Duttke, S. H., and Kadonaga, J. T. (2017a). The human initiator is a distinct and abundant element that is precisely positioned in focused core promoters. Genes Dev. 31, 6–11. doi: 10.1101/gad.293837.116
Vo Ngoc, L., Huang, C. Y., Cassidy, C. J., Medrano, C., and Kadonaga, J. T. (2020). Identification of the human DPR core promoter element using machine learning. Nature 585, 459–463. doi: 10.1038/s41586-020-2689-7
Vo Ngoc, L., Kassavetis, G. A., and Kadonaga, J. T. (2019). The RNA polymerase II core promoter in Drosophila. Genetics 212, 13–24. doi: 10.1534/genetics.119.302021
Vo Ngoc, L., Wang, Y. L., Kassavetis, G. A., and Kadonaga, J. T. (2017b). The punctilious RNA polymerase II core promoter. Genes Dev. 31, 1289–1301. doi: 10.1101/gad.303149.117
Wang, J., Shi, K., Wu, Z., Zhang, C., Li, Y., Deng, H., et al. (2020). Disruption of the interaction between TFIIAalphabeta and TFIIA recognition element inhibits RNA polymerase II gene transcription in a promoter context-dependent manner. Biochim. Biophys. Acta Gene Regul. Mech. 1863:194611. doi: 10.1016/j.bbagrm.2020.194611
Wang, J., Zhao, S., He, W., Wei, Y., Zhang, Y., Pegg, H., et al. (2017). A transcription factor IIA-binding site differentially regulates RNA polymerase II-mediated transcription in a promoter context-dependent manner. J. Biol. Chem. 292, 11873–11885. doi: 10.1074/jbc.M116.770412
Wang, Y., Qi, J., Shao, J., and Tang, X. Q. (2020). Signaling mechanism of transcriptional bursting: a technical resolution-independent study. Biology 9:339. doi: 10.3390/biology9100339
Wang, Y. L., Duttke, S. H., Chen, K., Johnston, J., Kassavetis, G. A., Zeitlinger, J., et al. (2014). TRF2, but not TBP, mediates the transcription of ribosomal protein genes. Genes Dev. 28, 1550–1555. doi: 10.1101/gad.245662.114
Wissink, E. M., Vihervaara, A., Tippens, N. D., and Lis, J. T. (2019). Nascent RNA analyses: tracking transcription and its regulation. Nat. Rev. Genet. 20, 705–723. doi: 10.1038/s41576-019-0159-6
Wu, C. H., Madabusi, L., Nishioka, H., Emanuel, P., Sypes, M., Arkhipova, I., et al. (2001). Analysis of core promoter sequences located downstream from the TATA element in the hsp70 promoter from Drosophila melanogaster. Mol. Cell. Biol. 21, 1593–1602. doi: 10.1128/MCB.21.5.1593-1602.2001
Yokoshi, M., Segawa, K., and Fukaya, T. (2020). Visualizing the role of boundary elements in enhancer-promoter communication. Mol. Cell 78, 224.e–235.e. doi: 10.1016/j.molcel.2020.02.007
Zabidi, M. A., Arnold, C. D., Schernhuber, K., Pagani, M., Rath, M., Frank, O., et al. (2015). Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518, 556–559. doi: 10.1038/nature13994
Zabidi, M. A., and Stark, A. (2016). Regulatory enhancer-core-promoter communication via transcription factors and cofactors. Trends Genet. 32, 801–814. doi: 10.1016/j.tig.2016.10.003
Zehavi, Y., Kuznetsov, O., Ovadia-Shochat, A., and Juven-Gershon, T. (2014a). Core promoter functions in the regulation of gene expression of Drosophila dorsal target genes. J. Biol. Chem. 289, 11993–12004. doi: 10.1074/jbc.M114.550251
Zehavi, Y., Sloutskin, A., Kuznetsov, O., and Juven-Gershon, T. (2014b). The core promoter composition establishes a new dimension in developmental gene networks. Nucleus 5, 298–303. doi: 10.4161/nucl.29838
Keywords: development, transcriptional regulation, core promoter, core promoter elements/motifs, Hox genes, dorsal-ventral axis, mesoderm formation, basal transcription machinery
Citation: Sloutskin A, Shir-Shapira H, Freiman RN and Juven-Gershon T (2021) The Core Promoter Is a Regulatory Hub for Developmental Gene Expression. Front. Cell Dev. Biol. 9:666508. doi: 10.3389/fcell.2021.666508
Received: 10 February 2021; Accepted: 25 August 2021;
Published: 10 September 2021.
Edited by:
Mark Rebeiz, University of Pittsburgh, United StatesReviewed by:
Ingrid Lohmann, Heidelberg University, GermanyArthur Günzl, UCONN Health, United States
Copyright © 2021 Sloutskin, Shir-Shapira, Freiman and Juven-Gershon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tamar Juven-Gershon, dGFtYXIuZ2Vyc2hvbkBiaXUuYWMuaWw=
 Anna Sloutskin
Anna Sloutskin Hila Shir-Shapira1
Hila Shir-Shapira1 Richard N. Freiman
Richard N. Freiman Tamar Juven-Gershon
Tamar Juven-Gershon