
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 03 June 2021
Sec. Signaling
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.661718
This article is part of the Research Topic A Hippo's View: From Molecular Basis to Translational Medicine View all 11 articles
The Hippo pathway is highly conserved from Drosophila to mammals. As a key regulator of cell proliferation, the Hippo pathway controls tissue homeostasis and has a major impact on tumorigenesis. The originally defined core components of the Hippo pathway in mammals include STK3/4, LATS1/2, YAP1/TAZ, TEAD, VGLL4, and NF2. However, for most of these genes, mutations and copy number variations are relatively uncommon in human cancer. Several other recently identified upstream and downstream regulators of Hippo signaling, including FAT1, SHANK2, Gq/11, and SWI/SNF complex, are more commonly dysregulated in human cancer at the genomic level. This review will discuss major genomic events in human cancer that enable cancer cells to escape the tumor-suppressive effects of Hippo signaling.
The Hippo signaling pathway is highly conserved through evolution. The core components of the pathway were originally identified in Drosophila. Their orthologous genes in mammals were found later (Ma et al., 2019; Snigdha et al., 2019). A large number of studies have shown that the Hippo pathway controls organ size mainly by responding to cell contact and various mechanical signals. The Hippo pathway also responds to cell polarity and G protein-coupled receptor (GPCR) signals. Given that loss of contact inhibition is one of the major hallmarks of human cancer, dysregulation of the Hippo pathway, which enables cancer cells to overcome contact inhibition, should be common in human cancers.
Gene dysregulation in human cancer can occur at various levels, including gene mutation/copy number abnormality, DNA methylation, over/under-expression, and post-translational modifications. Comparing to other types of dysregulations, mutation, and copy number abnormality data are more tractable and concrete. Therefore, this review will focus on mutation and copy number abnormality of the Hippo pathway components in human cancers.
The originally defined core components of the Hippo pathway include neurofibromin 2 (NF2), serine/threonine kinase 3/4 (STK3/4, originally called MST1/2), large tumor suppressor kinase 1/2 (LATS1/2), Yes1-associated transcriptional regulator (YAP1), tafazzin (TAZ), and TEA domain transcription factor (TEADS). When the Hippo pathway is activated by upstream signals, STK3/4 and Salvador Family WW domain containing protein 1 (SAV1) form a heterodimer through their C-terminal SARAH domain. Subsequently, STK3/4 phosphorylates LATS1/2, which then phosphorylates and inhibits the downstream substrate YAP1. Phosphorylated YAP1 is sequestered by 14-3-3 protein in the cytoplasm and/or degraded by the ubiquitination process.
When the Hippo pathway is inactivated, YAP1 is dephosphorylated and translocates to the nucleus, where YAP1/TAZ binds to TEAD, inducing target gene expression and promoting cell proliferation (Figure 1A). Vestigial-like family member4 (VGLL4) competitively inhibits the interaction of YAP1 and TEAD, providing another level of regulation on Hippo signaling output.
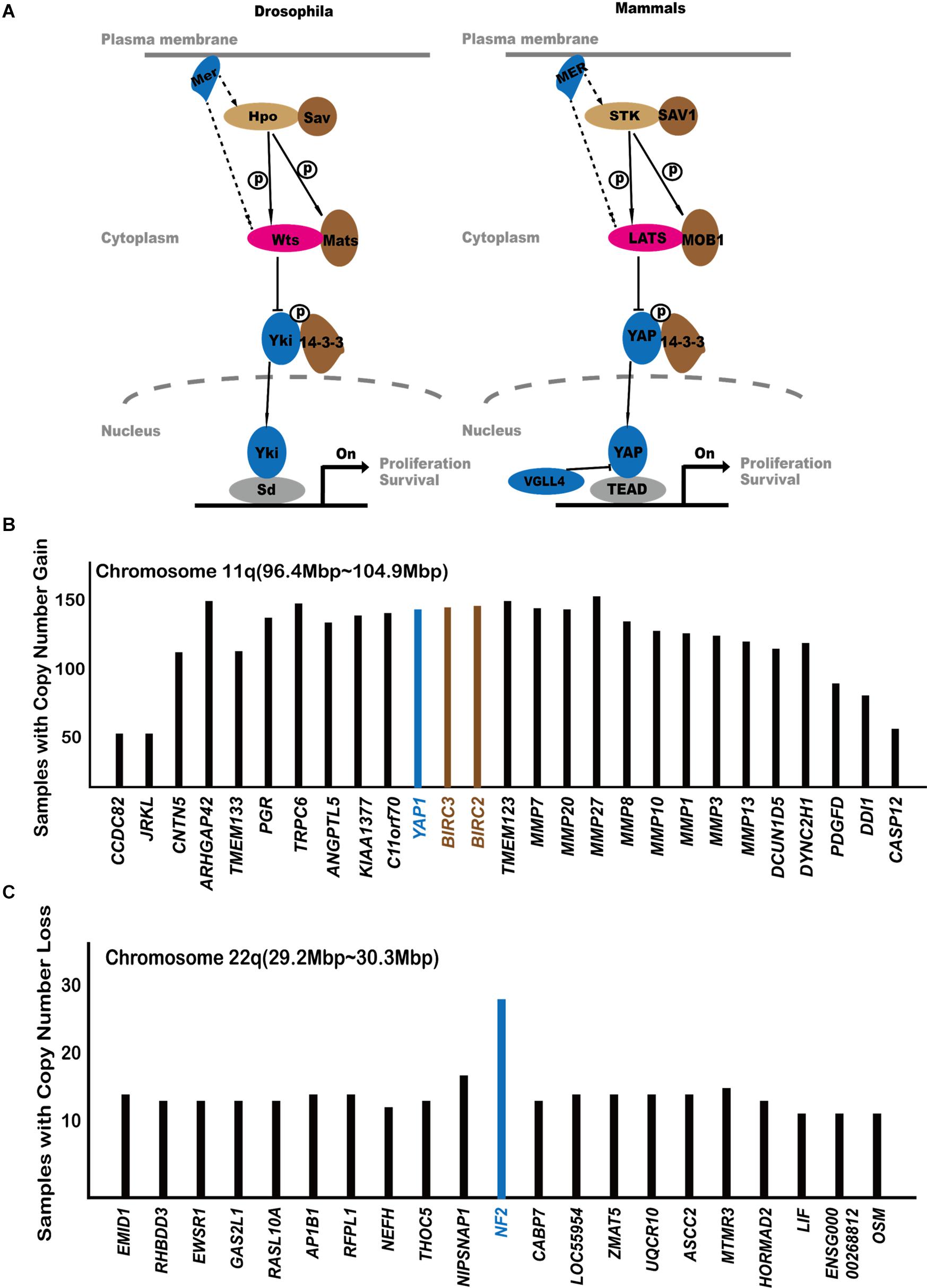
Figure 1. The central players of the Hippo pathway. (A) The originally established core components of the Hippo pathway. (B) Analysis of COSMIC gene amplification data with regard to the 11q22 amplicon. Genes are arranged according to their locations on chromosome 11q22. The Y axis shows how many cancer samples in COSMIC database exhibit gene amplification of each gene. If a gene is located at the amplification peak position, it is more likely to be a cancer-driving event. (C) Analysis of COSMIC gene amplification data with regard to the 22q deleted region.
In the Hippo signaling pathway, STK3/4 and LATS1/2 play antiproliferative roles and should act as tumor suppressors. However, according to the Catalogs of Somatic Mutations in Cancers (COSMIC) database1, genetic dysregulation of these genes is rare for most types of human cancers. This may, in part, be explained by gene redundancy (Meng et al., 2015; Plouffe et al., 2016). For example, mutation or deletion of LATS1 alone will not enable escape from the Hippo pathway’s anti-proliferative effect since LATS2 is still intact in cells. Therefore, single genetic events impacting STK3/4 and LATS1/2 will be insufficient to drive cancer formation. To fully destroy LATS1/2 activity, it will require four inactivating events to destroy both copies of LATS1 and LATS2. Therefore, from a cancer development point of view, achieving YAP1 activation through inactivating Hippo kinases may not be an easy route.
In addition to gene redundancy, other possibilities may also have contributed to the low mutation rates of Hippo kinases in cancer. For example, (Moroishi et al., 2016) showed that although inactivation of LATS1 and LATS2 led to enhanced anchorage-independent cell growth in vitro, it also caused increased tumor immunogenicity and tumor regression in vivo. This highlights the dual functions of LATS1/2 in cancer and reflects the crucial role of Hippo kinases in regulating tissue homeostasis.
As the key component of the Hippo pathway, hyperactivation of YAP1 is widespread in cancers (Harvey et al., 2013; Mo et al., 2014) as evidenced from immunostaining of YAP1 in human cancer samples. These studies found that YAP1 is commonly enriched in the nucleus in tumors while residing in the cytoplasm in normal tissues. The percentage of cells with nuclear YAP1 staining in hepatocellular carcinomas, ovarian cancers, and non-small-cell lung cancers is 60, 15, and 65%, respectively (Harvey et al., 2013).
Yes1 Associated Transcriptional Regulator is not frequently mutated in human cancers. Although the YAP1 S127A activating mutant is commonly used in cellular studies and tumor models, there is no corresponding active point mutation of YAP1 enriched in human cancer2. As discussed below, the increased activity of YAP1 in human cancers may be attributed to YAP1 gene amplification and gene fusion as well as the dysregulation of other components of the Hippo pathway.
(Zender et al., 2006) first noticed that the chromosome 9qA1 region, containing Yap1 gene, is amplified in mouse liver cancer cells. They also showed that overexpression of YAP1 can promote tumor formation. The corresponding amplified chromosome region in humans is 11q22 (Zender et al., 2006). This raised the possibility that the amplification of YAP1 may contribute to cancer development. Figure 1B shows the gene amplification status in the 11q22 region, including YAP1. YAP1 localized within the amplification peak region, suggesting for an oncogenic role. However, it is worth noticing that two adjacent genes, baculoviral IAP repeat containing 2/3 (BIRC2/3), are also amplified in human cancers. BIRC2/3 are anti-apoptotic proteins and promote cancer cell survival. It remains unclear whether the 11q22 amplicon utilizes all three genes to promote human cancer.
An interesting question is why YAP1 amplification is not a more frequent event in human cancers. In the COSMIC database, the number of human cancer samples amplifying YAP1 is 148. In comparison, such numbers for other major oncogenes are MYC = 968, EGFR = 514, ERBB2 = 358, SKP2 = 317, and MDM2 = 324. Cellular studies demonstrated that YAP1 rapidly induces the expression of LATS2, thus forming a negative feedback loop that self-limits its activity (Moroishi et al., 2015). The YAP1 S127A mutant, which escapes negative regulation by LATS kinases, is more effective at inducing tumor and has been used in many cancer models (Zhang S. et al., 2017; Min et al., 2019; Smith et al., 2021). It is possible that, due to such a negative feedback mechanism, simple overexpression of wild-type YAP1 may not be sufficient to trigger long-lasting proliferative events, which may partially explain the relative lower frequencies of YAP1 amplification in human cancers. On the other hand, it is possible that, similar to the case of LATS1/2 inactivation (Moroishi et al., 2016), YAP1 activity might also trigger additional events that negatively affect tumorigenesis.
Recent studies also identified YAP1 gene fusion events in several kinds of rare cancers, such as supratentorial (ST) ependymoma (Pajtler et al., 2015, 2019), epithelioid hemangioendothelioma, cervical squamous cell carcinoma (Hu et al., 2018), endocervical adenocarcinoma (Antonescu et al., 2013), and other cancers (Picco et al., 2019; Sekine et al., 2019; Sievers et al., 2020). The fusion protein products of YAP1 in these cancers include YAP1-MAMLD1, YAP1-FAM118B, YAP1-TFE3, and YAP1-SS18. Szulzewsky et al. (2020) demonstrated that these fusion proteins are resistant to negative Hippo pathway regulation and stay in the nucleus. In addition, the fusion proteins of YAP1 are also more stable and escape from degradation.
As the final executor of Hippo signaling, the transcription factor TEAD has very low transcriptional activity without the binding of YAP1 (Ma et al., 2019). TEAD genes are not known to be localized in gene amplification peaks in human cancers. Mutations of TEAD genes are also rare in human cancer, and there are few literatures reporting the functional mutations of TEAD genes (Huh et al., 2019).
Neurofibromin 2 (NF2) is a well-established tumor suppressor gene. It encodes Merlin, ortholog of the Drosophila Merlin protein, which encodes a FERM domain-containing protein. Studies of mouse and tumor patients showed that the inactivation of NF2 is an important cause of cancer. Hamaratoglu et al. (2006) firstly found that NF2 inhibits tumor development by regulating the Hippo pathway. Many reports demonstrated that YAP1 is dephosphorylated and activated with the loss of NF2, whereas the proliferation effect of NF2 loss can be eliminated by YAP1 knockout (Zhang et al., 2010). Hong et al. (2020) found that the lipid binding ability of Merlin is critical for its function in activating the Hippo pathway, which further clarifies the function and mechanism of NF2 (Yin et al., 2013). Several other mouse model studies also demonstrated that deletion of NF2 promotes tumor development (Giovannini et al., 2000; Kalamarides et al., 2002).
Hereditary loss of function mutations of NF2 causes type 2 neurofibromatosis, a disorder characterized by neoplastic growth in the nervous system (Yin et al., 2013). Somatic loss-of-function NF2 mutations are also found in many other kinds of cancers such as mesotheliomas and bladder, thyroid, and skin cancer. The COSMIC database indicates that NF2 mutations are highly enriched for nonsense mutations3. NF2 is also located at a deletion peak in cancer samples (Figure 1C). This indicates that gene mutation and gene deletion are both common means of NF2 loss of function in human cancers.
Vestigial Like Family Member 4 can inhibit organ overgrowth and cancer formation caused by YAP1 dysregulation in both human and Drosophila (Guo et al., 2013; Zhang et al., 2014). Jiao et al. (2014) found that VGLL4 competes with YAP1 to bind TEADs. Such an event will inhibit gene transcription by YAP1–TEADs and suppress cell proliferation (Zhang Y. et al., 2017).
Vestigial Like Family Member 4 has been described as a tumor suppressor in many cancers (Jiao et al., 2014; Zhang et al., 2014; Zhang Y. et al., 2017; Gallagher et al., 2020). For example, (Zhang et al., 2014) found that the VGLL4 expression level in mouse and human lung tumor specimen is significantly lower than in normal tissue. Overexpression of VGLL4 inhibits the progression of lung cancer in mice (Zhang et al., 2014). The findings of Jiao et al. (2014); Zhang Y. et al. (2017) in gastric cancer and breast cancer also support the idea that VGLL4 inhibits tumor progression.
Although VGLL4 is rarely mutated in human cancer, it is located at the short arm (3p) of chromosome 3, which is lost in many types of cancer (Williamson, 2002; Cancer Genome Atlas Research Network, 2013; Jonasch et al., 2020; Nidorf et al., 2020; Shaikh et al., 2020). The Cancer Genome Atlas (TCGA) Research Network found that around 90% of clear cell renal carcinoma patients exhibit loss of one or both copies of chromosome 3p. Chromosome 3p loss is also commonly observed in lung and stomach cancers. The rate of both 3p arm loss is lower in human cancer, typically around 10–20% in renal clear cell carcinoma. It is worth noticing that, through 3p loss, cancer cells also delete other important tumor suppressors such as VHL, SETD2,BAP1, and PBRM1. These events may also promote cancer development independent of VGLL4.
In addition to the afore-mentioned core components of the Hippo pathway, several recent studies identified new regulators of Hippo signaling. Some of these new regulators are also prominently dysregulated in human cancers, providing additional routes for cancer cells to escape from Hippo signaling.
FAT atypical cadherin 1 (FAT1) is a transmembrane protein, homologous to the tumor-suppressor genes fat and kujelei (also known as fat2) in Drosophila. By analyzing the TCGA database, (Martin et al., 2018) found that FAT1 shows a high-frequency mutation in many types of cancer. The study revealed that the cytoplasmic domain of FAT1 can activate the Hippo pathway by recruiting Hippo components such as NF2, STK3/4, and LATS1/2 to the cell membrane, forming a “kinase signalome” (Martin et al., 2018). In addition, other researchers also found that FAT1 regulates the Hippo pathway by YAP1 and TAZ (Ahmed et al., 2015; Li et al., 2018). One recent study showed that deletion of FAT1 in mouse epithelial cells induces epithelial–mesenchymal transition of epithelial cells and promote tumorigenesis in mice (Pastushenko et al., 2021).
According to the TCGA database, FAT1 is frequently mutated in many types of cancer (Katoh, 2012; Ahmed et al., 2015; Zhang et al., 2016). FAT1 exhibits 25.2 and 25.3% mutation rates in head and neck cancer and uterine corpus endometrial carcinoma, respectively, according to the TCGA database. Analysis of the COSMIC database indicated that FAT1 is located at a gene deletion peak in tumor samples, further suggesting a tumor suppressor role for FAT1 (Figure 2A).
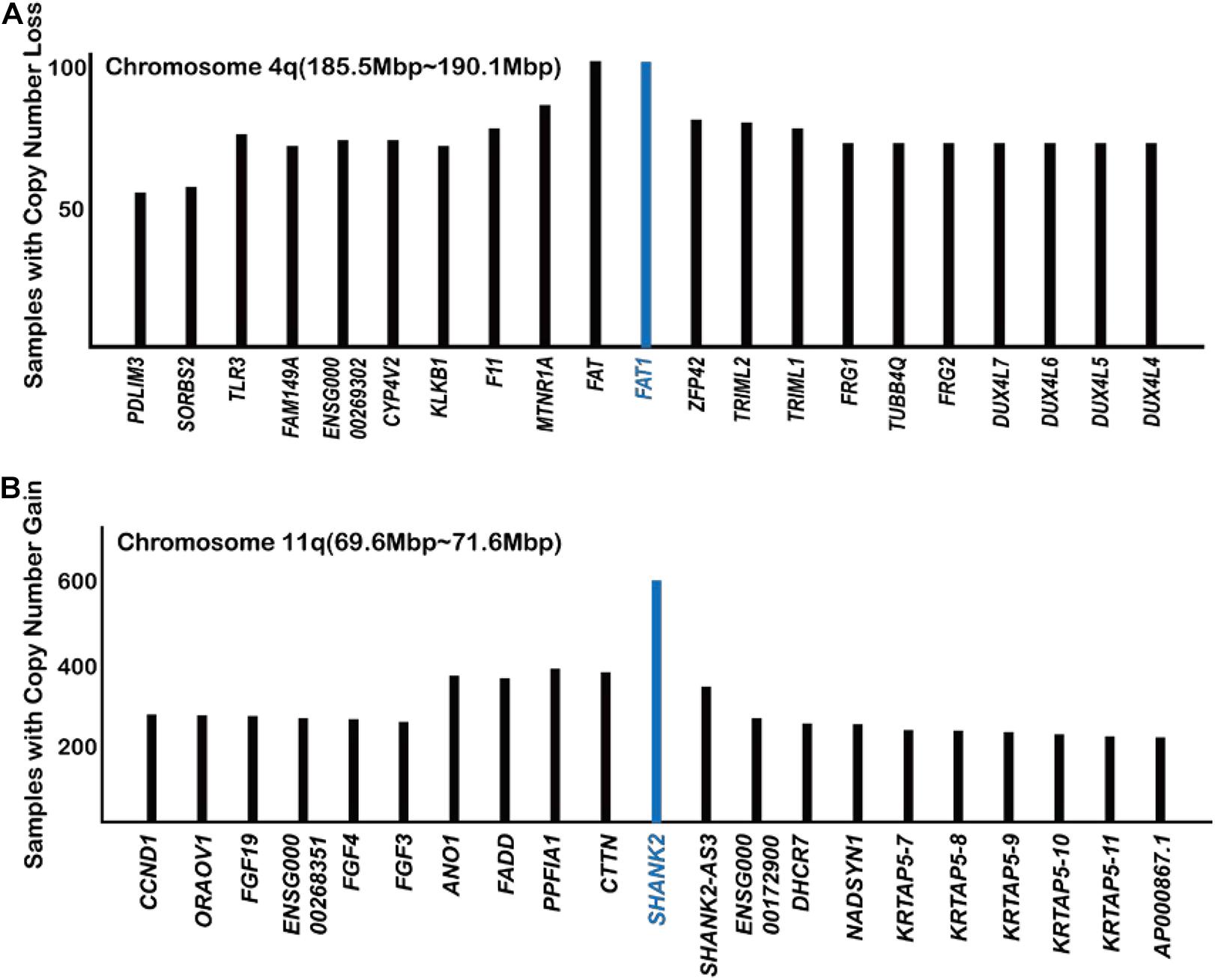
Figure 2. Analysis of COSMIC gene copy number data with regard to FAT1 (A) and SHANK2 (B) (https://cancer.sanger.ac.uk/cosmic/).
The SH3 and multiple ankyrin repeat domains (SHANK) protein family has been studied in the field of neuroscience mostly. As a scaffold protein, SHANK promotes synapse formation and balances synaptic transmission. In a recent study (Xu et al., 2020), found that the overexpression of SHANK2 ortholog in Drosophila suppresses Hippo signaling, causing an overgrowth of wings and eyes. In human cells, overexpression of SHANK2 also caused sustained YAP1 activity even at high cell density and promoted tumor formation in mice. SHANK2 inhibits the LATS1/2-mediated phosphorylation of YAP1 by competing for binding to rho guanine nucleotide exchange factor 7 (ARHGEF7), which is an activator protein of LATS1/2 (Heidary Arash et al., 2014). Analysis of the COSMIC database also showed that SHANK2 is the most frequently amplified gene in the 11q13 amplicon (Figure 2B; Xu et al., 2020). It is worth noticing that the number of cancer samples with SHANK2 amplifications way exceeds those with YAP1 amplification, FAT1 deletion, and NF2 deletion (Figures 1B, 2), indicating a previously unnoticed role of SHANK2 as a major oncogene.
The G protein subunit alpha Q (GNAQ) and G protein subunit alpha 11 (GNA11) genes encode Gq and G11 proteins, respectively, which play an essential role in GPCR signaling pathway. Yu et al. (2014) found that the mutant Gq/G11 can activate the YAP1 protein. In addition, inhibition of YAP1 can block the proliferation of Gq/G11 mutant cells These results imply that the pro-proliferation effect of mutant Gq/G11 depends on the function of YAP1.
Previous studies (Van Raamsdonk et al., 2009, 2010) showed that overexpression of mutant Gq/G11 causes normal melanocyte transformation, whereas knockdown of Gq/G11 blocks tumor formation in xenograft experiments. Data from the TCGA database indicated that the percentage of GNAQ and GNA11 gene mutation is 50 and 43.8%, respectively, in uveal melanoma patients, suggesting a major involvement of the Hippo pathway for this type of cancer.
SWI/SNF is a multi-subunit ATP-dependent chromosome remodeling complex. The SWI/SNF complex plays key roles in regulating gene expression and tissue homeostasis. Mutations of the subunits of this complex are detected in a variety of human malignancies (Kadoch et al., 2013; Shain and Pollack, 2013; Lou et al., 2020).
Increasing evidence indicated that the SWI/SNF complex inhibits tumor development (Weissman and Knudsen, 2009; Wilson and Roberts, 2011; Shain and Pollack, 2013; Ribeiro-Silva et al., 2019). A large number of studies showed that the deletion of components of SWI/SNF promotes tumor development in mouse models (Klochendler-Yervin et al., 2000; Roberts et al., 2000; Glaros et al., 2007, 2008). In a recent study, (Chang et al., 2018) found that the ARID1A subunit of the SWI/SNF complex binds to and inactivates YAP1. Therefore, once the SWI/SNF complex is inactivated by various mutations in cancer cells, YAP1 will be released and promote carcinogenesis. In addition, the SWI/SNF complex may also inhibit cancer development by maintaining genome stability (Ribeiro-Silva et al., 2019). The statistical results of Ribeiro-Silva et al. (2019) indicate that mutations in genes encoding for SWI/SNF subunits are found in approximately 20% of all human cancers of various types. This may constitute one of the most frequent routes of Hippo dysregulation in human cancer.
The Hippo pathway, by responding to cell density and maintaining cell–cell contact, is a crucial barrier for tumor development. When the Hippo pathway is dysregulated, cells will acquire the potential for uncontrolled proliferation, promoting cancer formation. Although mutations and/or copy number abnormalities directly impacting the core Hippo kinases are relatively rare in human cancers, cancer cells manage to escape from Hippo regulation by means of other upstream and downstream Hippo regulators, including FAT1, SHANK2, SWI/SNF, Gq/11, VGLL4, etc. In this review, we focused on Hippo dysregulation in human cancers at the genomic level. It is also worth noticing that other oncogenic events, for example, IDH1 mutation, can also affect the Hippo pathway components through gene hypermethylation (Gu et al., 2020).
Several recent studies also identified other important regulators of the Hippo pathway, including the RAP family of small GTPase, MST4, and others (Meng et al., 2018; An et al., 2020). These genes do not appear to be frequently dysregulated at the genomic level in human cancers, possibly due to gene redundancy or other reasons. For example, all three RAP2 genes (Rap2A/B/C) need to be simultaneously knocked out to cause YAP1 nuclear localization (Meng et al., 2018). Therefore, these genes are not included in this review. Figure 3 summarizes the major cancer players of the Hippo pathway and their frequency of genetic dysregulation in various forms of human cancers. Such findings will help bring a clearer view of Hippo pathway dysregulation in cancers as well as point to potential precision medicine approaches targeting YAP1 activity for cancer therapy.
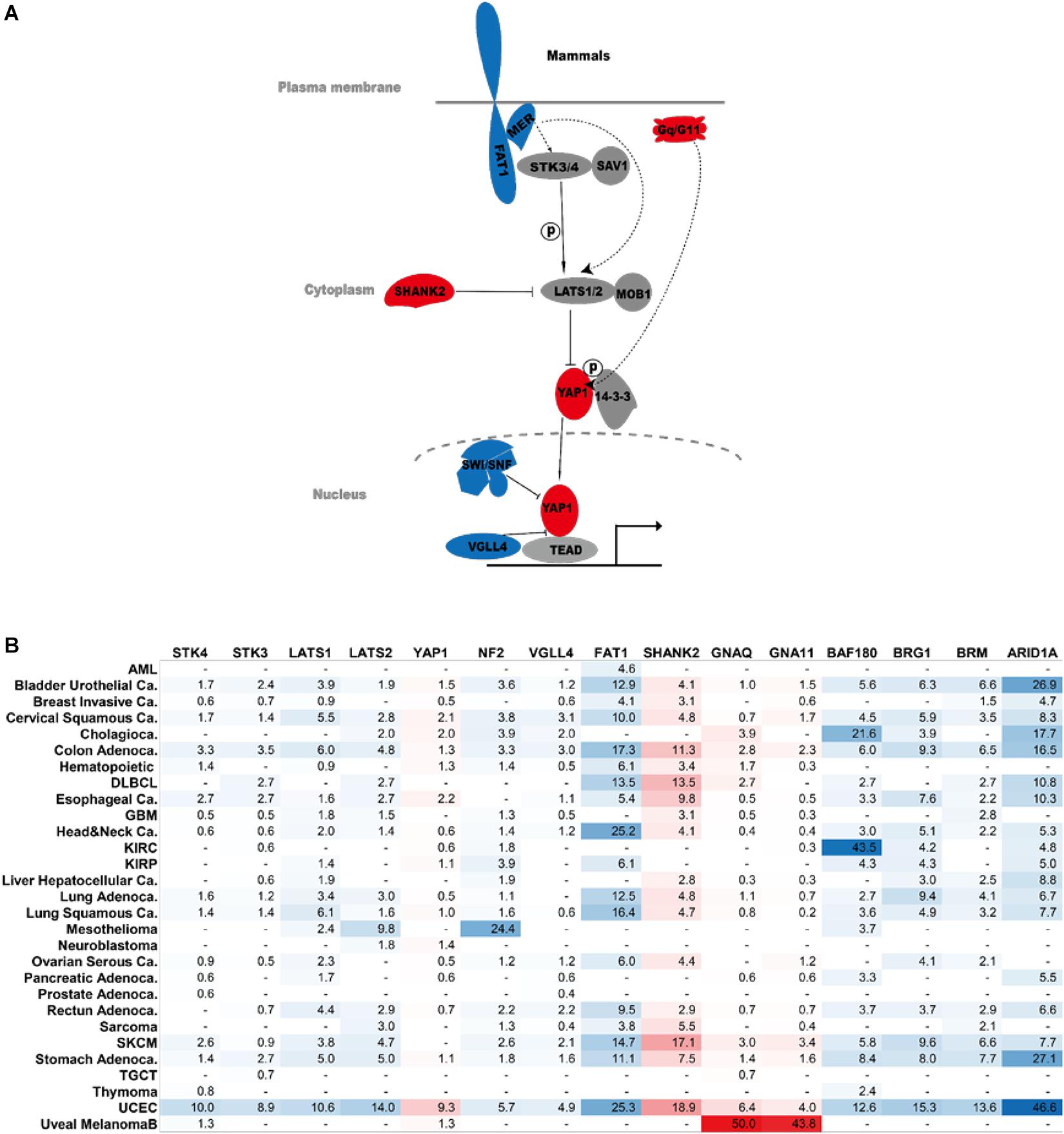
Figure 3. Hippo components in human cancer. (A) Major cancer players of the Hippo signaling pathway. Genes in red are oncogenes. Genes in blue are tumor suppressors. (B) Percentage of genetic abnormalities of the indicated Hippo pathway components in human cancer. The analysis was performed on The Cancer Genome Atlas dataset. AML, acute myeloid leukemia; DLBCL, diffuse large B cell lymphoma; GBM, glioblastoma multiforme; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; SKCM, skin cutaneous melanoma; TGCT, testicular germ cell tumors; UCEC, uterine corpus endometrial carcinoma.
For example, cancers that dysregulate the Hippo pathway may be more sensitive to suppression of YAP1 activity. On the one hand, the degradation of YAP1 may be a choice for cancer treatment. Proteolysis-targeting chimera (PROTAC) technology could provide us with a useful method for YAP1 degradation (Li and Song, 2020). On the other hand, VGLL4’s ability to disrupt YAP1–TEADs interaction indicates that peptide mimics of VGLL4 could potentially be useful for suppressing cancers driven by Hippo pathway dysregulation. Newly identified oncogenes in the Hippo pathway such as Gq/G11 and SHANK2 may also represent potential PROTAC targets for cancer treatment.
All authors wrote the manuscript. ZH prepared Figures.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the National Science and Technology Major Project of the Ministry of Science and Technology of China (Grant No. 2017YFA0504503) and the Strategic Priority Research Program of Chinese Academy of Sciences (Grant No. XDB19000000).
Ahmed, A. F., de Bock, C. E., Lincz, L. F., Pundavela, J., Zouikr, I., and Sontag, E., et al. (2015). FAT1 cadherin acts upstream of Hippo signalling through TAZ to regulate neuronal differentiation. Cell Mol. Life. Sci. 72, 4653–4669 doi: 10.1007/s00018-015-1955-6
An, L., Nie, P., Chen, M., Tang, Y., Zhang, H., Guan, J., et al. (2020). MST4 kinase suppresses gastric tumorigenesis by limiting YAP activation via a non-canonical pathway. J. Exp. Med. 217:e20191817
Antonescu, C. R., Le Loarer, F., Mosquera, J. M., Sboner, A., Zhang, L., Chen, C. L., et al. (2013). Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer 52, 775–784 doi: 10.1002/gcc.22073
Cancer Genome Atlas Research Network (2013). Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 doi: 10.1038/nature12222
Chang, L., Azzolin, L., Di Biagio, D., Zanconato, F., Battilana, G., and Lucon Xiccato, R., et al. (2018). The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature 563, 265–269 doi: 10.1038/s41586-018-0658-1
Gallagher, K. M., Roderick, J. E., Tan, S. H., Tan, T. K., Murphy, L., Yu, J., et al. (2020). ESRRB regulates glucocorticoid gene expression in mice and patients with acute lymphoblastic leukemia. Blood Adv. 4, 3154–3168.
Giovannini, M., Robanus-Maandag, E., van der Valk, M., Niwa-Kawakita, M., Abramowski, V, Goutebroze, L., et al. (2000). Conditional biallelic Nf2 mutation in the mouse promotes manifestations of human neurofibromatosis type 2. Genes Dev. 14, 1617–1630
Glaros, S., Cirrincione, G. M., Muchardt, C., Kleer, C. G., Michael, C. W., and Reisman, D. (2007). The reversible epigenetic silencing of BRM: implications for clinical targeted therapy. Oncogene 26, 7058–7066. doi: 10.1038/sj.onc.1210514
Glaros, S., Cirrincione, G. M., Palanca, A., Metzger, D., and Reisman, D. (2008). Targeted knockout of BRG1 potentiates lung cancer development. Cancer Res. 68, 3689–3696 doi: 10.1158/0008-5472.can-07-6652
Gu, Y., Wang, Y., Wang, Y., Luo, J., Wang, X., Ma, M., et al. (2020). Hypermethylation of LATS2 promoter and its prognostic value in IDH-mutated low-grade gliomas. Front. Cell Dev. Biol. 8:586581
Guo, T., Lu, Y., Li, P., Yin, M. X., Lv, D., and Zhang, W. (2013). A novel partner of Scalloped regulates Hippo signaling via antagonizing Scalloped-Yorkie activity. Cell Res. 23, 1201–1214 doi: 10.1038/cr.2013.120
Hamaratoglu, F., Willecke, M., Kango-Singh, M., Nolo, R., Hyun, E., Tao, C., et al. (2006). The tumour-suppressor genes NF2/Merlin and expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8, 27–36 doi: 10.1038/ncb1339
Harvey, K. F., Zhang, X., and Thomas, D. M. (2013). The hippo pathway and human cancer. Nat. Rev. Cancer 13, 246–257
Heidary Arash, E., Song, K. M., Song, S., Shiban, A., and Attisano, L. (2014). Arhgef7 promotes activation of the Hippo pathway core kinase Lats. EMBO J. 33, 2997–3011 doi: 10.15252/embj.201490230
Hong, A. W., Meng, Z., Plouffe, S. W., Lin, Z., Zhang, M., and Guan, K. L. (2020). Critical roles of phosphoinositides and NF2 in Hippo pathway regulation. Genes Dev. 34, 511–525 doi: 10.1101/gad.333435.119
Hu, X., Wang, Q., Tang, M., Barthel, F., Amin, S., and Yoshihara, K., et al. (2018). TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res. 46, D1144–D1149
Huh, H. D., Kim, D. H., Jeong, H. S., and Park, H. W. (2019). Regulation of TEAD transcription factors in cancer biology. Cells 8:600 doi: 10.3390/cells8060600
Jiao, S., Wang, H., Shi, Z., Dong, A., Zhang, W., Song, X., et al. (2014). A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell 25, 166–180 doi: 10.1016/j.ccr.2014.01.010
Jonasch, E., Walker, C. L., and Rathmell, W. K. (2020). Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat. Rev. Nephrol. 17, 245–261 doi: 10.1038/s41581-020-00359-2
Kadoch, C., Hargreaves, D. C., Hodges, C., Elias, L., Ho, L., and Ranish, J. (2013). Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592–601 doi: 10.1038/ng.2628
Kalamarides, M., Niwa-Kawakita, M., Leblois, H., Abramowski, V., Perricaudet, M., and Janin, A. (2002). Nf2 gene inactivation in arachnoidal cells is rate-limiting for meningioma development in the mouse. Genes Dev. 16, 1060–1065 doi: 10.1101/gad.226302
Katoh, M. (2012). Function and cancer genomics of FAT family genes (review). Int. J. Oncol. 41, 1913–1918 doi: 10.3892/ijo.2012.1669
Klochendler-Yervin, A., Fiette, L., Barra, J., Muchardt, C., Babinet, C., and Yaniv, M. (2000). The murine SNF5/NIN1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 15, 500–506 doi: 10.1093/embo-reports/kvd129
Li, X., and Song, Y. (2020). Proteolysis-targeting chimera (PROTAC) for targeted protein degradation and cancer therapy. J. Hematol. Oncol. 13:50
Li, Z., Razavi, P., Li, Q., Toy, W., Liu, B., Ping, C., et al. (2018). Loss of the FAT1 tumor suppressor promotes resistance to CDK4/6 inhibitors via the hippo pathway. Cancer Cell 34, 893–905 e898
Lou, Y., Liu, L., and Qiu, Y. (2020). Clinical outcomes and plasma concentrations of Baloxavir Marboxil and favipiravir in COVID-19 patients: an exploratory randomized, controlled trial. medRxiv [Preprint] doi: 10.1101/2020.04.29.20085761.
Ma, S., Meng, Z., Chen, R., and Guan, K. L. (2019). The hippo pathway: biology and pathophysiology. Annu. Rev. Biochem. 88, 577–604 doi: 10.1146/annurev-biochem-013118-111829
Martin, D., Degese, M. S., Vitale-Cross, L., Iglesias-Bartolome, R., Valera, J. L. C., Wang, Z., et al. (2018). Assembly and activation of the Hippo signalome by FAT1 tumor suppressor. Nat. Commun. 9:2372
Meng, Z., Moroishi, T., Mottier-Pavie, V., Plouffe, S. W., Hansen, C. G., Hong, A. W., et al. (2015). MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat. Commun. 6:8357
Meng, Z., Qiu, Y., Lin, K. C., Kumar, A., Placone, J. K., Fang, C., et al. (2018). RAP2 mediates mechanoresponses of the Hippo pathway. Nature 560, 655–660. doi: 10.1038/s41586-018-0444-0
Min, Q., Molina, L., Li, J., Adebayo Michael, A. O., Russell, J. O., Preziosi, M. E., et al. (2019). β-Catenin and yes-associated protein 1 cooperate in hepatoblastoma pathogenesis. Am. J. Pathol. 189, 1091–1104 doi: 10.1016/j.ajpath.2019.02.002
Mo, J. S., Park, H. W., and Guan, K. L. (2014). The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 15, 642–656 doi: 10.15252/embr.201438638
Moroishi, T., Hayashi, T., Pan, W. W., Fujita, Y., Holt, M. V., Qin, J., et al. (2016). The hippo pathway kinases LATS1/2 suppress cancer immunity. Cell 167, 1525.e17–1539.e17
Moroishi, T., Park, H. W., Qin, B., Chen, Q., Meng, Z., Plouffe, S. W., et al. (2015). A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev. 29, 1271–1284 doi: 10.1101/gad.262816.115
Nidorf, S. M., Fiolet, A. T. L., Mosterd, A., Eikelboom, J. W., Schut, A., Opstal, T. S. J., et al. (2020). Colchicine in patients with chronic coronary disease. N. Engl. J. Med. 383, 1838–1847.
Pajtler, K. W., Wei, Y., Okonechnikov, K., Silva, P. B. G., Vouri, M., Zhang, L., et al. (2019). YAP1 subgroup supratentorial ependymoma requires TEAD and nuclear factor I-mediated transcriptional programmes for tumorigenesis. Nat. Commun. 10:3914
Pajtler, K. W., Witt, H., Sill, M., Jones, D. T., Hovestadt, V., and Kratochwil, F., et al. (2015). Molecular classification of ependymal tumors across All CNS compartments, histopathological grades, and age groups. Cancer Cell 27, 728–743 doi: 10.1016/j.ccell.2015.04.002
Pastushenko, I., Mauri, F., Song, Y., de Cock, F., Meeusen, B., Swedlund, B., et al. (2021). Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature 589, 448–455 doi: 10.1038/s41586-020-03046-1
Picco, G., Chen, E. D., Alonso, L. G., Behan, F. M., Goncalves, E., Bignell, G., et al. (2019). Functional linkage of gene fusions to cancer cell fitness assessed by pharmacological and CRISPR-Cas9 screening. Nat. Commun. 10:2198
Plouffe, S. W., Meng, Z., Lin, K. C., Lin, B., Hong, A. W., Chun, J. V., et al. (2016). Characterization of Hippo pathway components by gene inactivation. Mol. Cell 64, 993–1008 doi: 10.1016/j.molcel.2016.10.034
Ribeiro-Silva, C., Vermeulen, W., and Lans, H. (2019). SWI/SNF: complex complexes in genome stability and cancer. DNA Repair 77, 87–95 doi: 10.1016/j.dnarep.2019.03.007
Roberts, C. W., Galusha, S. A., McMenamin, E., Fletcher, C. M., and Orkin, S. H. (2000). Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc. Natl. Acad. Sci. U. S. A. 97, 13796–13800 doi: 10.1073/pnas.250492697
Sekine, S., Kiyono, T., Ryo, E., Ogawa, R., Wakai, S., and Ichikawa, H. (2019). Recurrent YAP1-MAML2 and YAP1-NUTM1 fusions in poroma and porocarcinoma. J. Clin. Invest. 129, 3827–3832 doi: 10.1172/jci126185
Shaikh, M. H., Barrett, J. W., Khan, M. I., Kim, H. A. J., Zeng, P. Y. F., and Mymryk, J. S. (2020). Chromosome 3p loss in the progression and prognosis of head and neck cancer. Oral Oncol. 109:104944 doi: 10.1016/j.oraloncology.2020.104944
Shain, A. H., and Pollack, J. R. (2013). The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One 8:e55119. doi: 10.1371/journal.pone.0055119
Sievers, P., Chiang, J., Schrimpf, D., Stichel, D., Paramasivam, N., Sill, M., et al. (2020). YAP1-fusions in pediatric NF2-wildtype meningioma. Acta Neuropathol. 139, 215–218 doi: 10.1007/s00401-019-02095-9
Smith, J. L., Rodríguez, T. C., Mou, H., Kwan, S. Y., Pratt, H., Zhang, X. O., et al. (2021). YAP1 withdrawal in hepatoblastoma drives therapeutic differentiation of tumor cells to functional hepatocyte-like cells. Hepatology 73, 1011–1027 doi: 10.1002/hep.31389
Snigdha, K., Gangwani, K. S., Lapalikar, G. V., Singh, A., and Kango-Singh, M. (2019). Hippo signaling in cancer: lessons from drosophila models. Front. Cell Dev. Biol. 7:85
Szulzewsky, F., Arora, S., Hoellerbauer, P., King, C., Nathan, E., Chan, M., et al. (2020). Comparison of tumor-associated YAP1 fusions identifies a recurrent set of functions critical for oncogenesis. Genes Dev. 34, 1051–1064 doi: 10.1101/gad.338681.120
Van Raamsdonk, C. D., Bezrookove, V., Green, G., Bauer, J., Gaugler, L., O’Brien, J. M., et al. (2009). Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 457, 599–602 doi: 10.1038/nature07586
Van Raamsdonk, C. D., Griewank, K. G., Crosby, M. B., Garrido, M. C., Vemula, S., Wiesner, T., et al. (2010). Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 363, 2191–2199
Weissman, B., and Knudsen, K. E. (2009). Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Cancer Res. 69, 8223–8230. doi: 10.1158/0008-5472.can-09-2166
Williamson, S. R. (2002). Clear cell papillary renal cell carcinoma: an update after 15 years. Pathology 53, 109–119 doi: 10.1016/j.pathol.2020.10.002
Wilson, B. G., and Roberts, C. W. (2011). SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11, 481–492 doi: 10.1038/nrc3068
Xu, L., Li, P., Hao, X., Lu, Y., Liu, M., and Song, W. et al. (2020). SHANK2 is a frequently amplified oncogene with evolutionarily conserved roles in regulating Hippo signaling. Protein Cell 12, 174–193 doi: 10.1007/s13238-020-00742-6
Yin, F., Yu, J., Zheng, Y., Chen, Q., Zhang, N., and Pan, D. (2013). Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell 154, 1342–1355 doi: 10.1016/j.cell.2013.08.025
Yu, F. X., Luo, J., Mo, J. S., Liu, G., Kim, Y. C., Meng, Z., et al. (2014). Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell 25, 822–830 doi: 10.1016/j.ccr.2014.04.017
Zender, L., Spector, M. S., Xue, W., Flemming, P., Cordon-Cardo, C., Silke, J., et al. (2006). Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 125, 1253–1267 doi: 10.1016/j.cell.2006.05.030
Zhang, N., Bai, H., David, K. K., Dong, J., Zheng, Y., Cai, J., et al. (2010). The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell 19, 27–38 doi: 10.1016/j.devcel.2010.06.015
Zhang, S., Chen, Q., Liu, Q., Li, Y., Sun, X., Hong, L., et al. (2017). Hippo signaling suppresses cell ploidy and tumorigenesis through Skp2. Cancer Cell 31, 669–684.e667
Zhang, W., Gao, Y., Li, P., Shi, Z., Guo, T., and Li, F., et al. (2014). VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res. 24, 331–343 doi: 10.1038/cr.2014.10
Zhang, X., Liu, J., Liang, X., Chen, J., Hong, J., Li, L., et al. (2016). History and progression of fat cadherins in health and disease. Onco. Targets Ther. 9, 7337–7343 doi: 10.2147/ott.s111176
Keywords: hippo deficiency, cancer formation, copy number abberation, gene mutation, cancer genome
Citation: He Z, Li R and Jiang H (2021) Mutations and Copy Number Abnormalities of Hippo Pathway Components in Human Cancers. Front. Cell Dev. Biol. 9:661718. doi: 10.3389/fcell.2021.661718
Received: 31 January 2021; Accepted: 12 April 2021;
Published: 03 June 2021.
Edited by:
Zhaocai Zhou, Fudan University, ChinaReviewed by:
Fa-Xing Yu, Fudan University, ChinaCopyright © 2021 He, Li and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai Jiang, aGFpQHNpYmNiLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.