
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 08 April 2021
Sec. Stem Cell Research
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.655145
 Yuting Fu1,2
Yuting Fu1,2 Fangyuan Liu1,2
Fangyuan Liu1,2 Shuo Cao1,2
Shuo Cao1,2 Jia Zhang1,2
Jia Zhang1,2 Huizhi Wang1,2
Huizhi Wang1,2 Baojiang Wu1,2
Baojiang Wu1,2 Yongli Song1,2
Yongli Song1,2 Shuguang Duo3
Shuguang Duo3 Xihe Li1,2,4
Xihe Li1,2,4 Siqin Bao1,2*
Siqin Bao1,2*3-hydroxybutyrate dehydrogenase-2 (Bdh2), a short-chain dehydrogenase, catalyzes a rate-limiting step in the biogenesis of the mammalian siderophore, playing a key role in iron homeostasis, energy metabolism and apoptosis. However, the function of Bdh2 in embryonic stem cells (ESCs) remains unknown. To gain insights into the role of Bdh2 on pluripotency and cell fate decisions of mouse ESCs, we generated Bdh2 homozygous knockout lines for both mouse advanced embryonic stem cell (ASC) and ESC using CRISPR/Cas9 genome editing technology. Bdh2 deficiency in both ASCs and ESCs had no effect on expression of core pluripotent transcription factors and alkaline phosphatase activity, suggesting dispensability of Bdh2 for self-renewal and pluripotency of ESCs. Interestingly, cells with Bdh2 deficiency exhibited potency of endoderm differentiation in vitro; with upregulated endoderm associated genes revealed by RNA-seq and RT-qPCR. We further demonstrate that Bdh2 loss inhibited expression of multiple methyltransferases (DNMTs) at both RNA and protein level, suggesting that Bdh2 may be essentially required to maintain DNA methylation in ASCs and ESCs. Overall, this study provides valuable data and resources for understanding how Bdh2 regulate earliest cell fate decision and DNA methylation in ASCs/ESCs.
Mouse embryonic stem cells (ESCs), derived from the inner cell mass (ICM) of blastocysts, can be maintained by self-renewal or differentiate into the three germ layers in embryonic tissues (Evans and Kaufman, 1981). Inhibitors of Mek1/2 (PD0325901, PD) and Gsk3β (CHIR99021, CH), enhanced the derivation of ESCs and promoted ground-state pluripotency (Ying et al., 2008), which was defined as “naïve pluripotency” when LIF was added (2i/L-ESCs) (Nichols and Smith, 2009). The balance of ESCs self-renewal and differentiation is tightly regulated and accurately orchestrated through the modulation of chromatin structures associated with transcriptional regulation (Young, 2011; Tee and Reinberg, 2014; Novo et al., 2016). Three germ layers can be acquired in vitro by inducing spontaneous and irreversible differentiation through aggregation of stem cells known as embryoid bodies (Posfai et al., 2014; Chen et al., 2016). However, it remains elusive how ESCs exit pluripotency and undergo lineage differentiation.
Guo et al. (2006) identified a novel cytosolic 3-hydroxybutyrate dehydrogenase-2 (Bdh2) as a short-chain dehydrogenase/reductase family member, originally named as DHRS6. It has been reported Bdh2 catalyzes a rate-limiting step in the biogenesis of the mammalian siderophore 2, 5-dihydroxybenzoic acid (2, 5-DHBA), which identified as an iron-binding molecule in mammalian cells (Devireddy et al., 2010). In cultured mammalian cells, as well as in developing zebrafish embryos, depletion of the mammalian siderophore by inhibiting expression of Bdh2 results in abnormal accumulation of intracellular iron and heme deficiency (Devireddy et al., 2010; Davuluri et al., 2016). Bdh2 null mice developed microcytic anemia and tissue iron overload, especially in the spleen, and exogenous supplementation with 2,5-DHBA alleviates splenic iron overload in Bdh2 null mice (Liu et al., 2014a). In addition, Bdh2 can also play a physiological role in ketone body metabolism, tricarboxylic acid cycle and iron-limiting innate immunity (Guo et al., 2006; Zughaier et al., 2014a, b). Furthermore, Yang et al. (2013) reported that Bdh2 acts as an anti-apoptotic factor, through a caspase-3 independent pathway. As a result, Bdh2 may be an independent poor prognosis marker of acute myeloid leukemia by affecting apoptosis (Yang et al., 2013). Overall, Bdh2 may be a multifunctional gene in mammalian cells. However, the function of Bdh2 in mouse ESCs and the potential mechanisms involved have not been reported.
We recently showed that the advanced embryonic stem cells (ASCs) represent an intermediate state between naïve (ESCs) and primed pluripotency such as epiblast stem cells (EpiSCs) (Bao et al., 2018). We observed a differential level of Bdh2 expression between ASCs and ESCs. In order to investigate the precise role of Bdh2 in the differentiation of ESCs, bi-allelic Bdh2-knockout ASCs and ESCs were generated by CRISPR/Cas9. Our results reveal that Bdh2 plays distinct roles in the regulation of primitive endoderm differentiation in vitro, despite the pluripotency is maintained in Bdh2-knockout ASCs/ESCs. Bdh2-knockout ASCs/ESCs display unique molecular features, which implies the function of Bdh2 in the maintenance of self-renewal, pluripotency and differentiation.
Advanced embryonic stem cells and Bdh2-knockout ASCs were cultured in ABCL culture medium comprised Activin A (20 ng/mL, R & D systems), BMP4 (50 ng/mL, R & D systems), CHIR99021 (3 μM, Miltenyi Biotec) and leukemia inhibitory factor (1000 U ml–1, Millipore) added into basic N2B27 medium including 50% Neurobasal (Gibco), 50% DMEM/F12 (Gibco), 2 mM GlutaMax (Gibco), 1 × non-essential amino acids (NEAA, Gibco), 1 × Penicillin/Streptomycin (Gibco), 0.1 mM β-mercaptoethanol (Gibco), 0.005% (25 mg) BSA (Gibco) supplemented with 0.5 × N2 (Gibco) and 0.5 × B27 (Gibco). ESCs and Bdh2-knockout ESCs were cultured in 2i/L medium consisting of basic N2B27 medium supplemented with PD0325901 (1 μM, Miltenyi Biotec), CHIR99021 (3 μM, Miltenyi Biotec) and leukemia inhibitory factor (1000 U mL–1, Millipore). Green fluorescence indicated that GFP expression of the reporter was under the control of Oct4 promoter and distal enhancer. The colonies could stably passage by Accutase (Life technology) regularly at every 2–3 days. All using plates were coated by fibronectin (1 mg/mL in PBS, Millipore) at least 0.5 h before use.
The guide RNAs (gRNAs) were designed using CRISPOR1 online tool and were synthesized (Sangon Biotech). The sgRNA was cloned PX459 plasmid that was linearized with BbsI restriction enzyme (Thermo Fisher Scientific). Then, plasmids containing sgRNA were transfected into ASCs and ESCs with lipofectamine 2000 (Invitrogen). Transfected cells were treated with 0.6 μg/μL of puromycin at 48 h post-transfection. Then, the selected cells were subjected to fibronectin-coated 96-well plate to obtain a single cell colony. Approximately after 6 days of colony formation, each single colony was picked and expanded. To confirm the specificity of targeting, genomic DNA was extracted from individual clones by TIANamp Genomic DNA Kit (TIANGEN Biotech) and was sequenced.
Before staining, cells were placed in 4-well plates and washed with 1 × PBS, then fixed in 4% paraformaldehyde at room temperature for 30 min and washed with 1 × PBS again followed by adding AP staining solution. AP staining solution was prepared as following: gently mixed 50 μL sodium nitrite solution with 50 μL FRV-alkaline solution and placed the mixture at 37°C for 3 min, next added 2.25 mL H2O and 50 μL naphthol-As-BI alkaline solution into the mixture, finally mixed gently and incubated staining solution with fixed cells in the dark for overnight.
The tested cells were incubated with 0.2 μg/mL colchicine in culture medium for 2 h and dissociated by using Accutase, then centrifuged at 1500 r/min for 5 min to collect the tested cells. The cells were gently resuspended in 8 ml 0.075 mol/L KCL (Sigma) and incubated in 37°C water bath kettle for 40 min for hypotonic treatment. Stationary liquid (methanol: glacial acetic acid = 3: 1) of 1 mL was subsequently added to the resuspended cells and mixed gently followed by centrifuging at 1000 r/min for 10 min. After discarding supernatant, the cells were mixed gently in 8 mL stationary liquid and incubated in 37°C water bath kettle for 30 min for cell fixation, which repeated twice. Then resuspended the cells with 0.5 mL stationary liquid and dripped the resuspended cells on ice cold glass slides, followed by drying the glass slides for 1 h in 70°C drying oven. The glass slides were stained in Giemsa (Sigma) for 10 min and washed by distilled water, then air-dried following analyzed by LUCIA Cytogenetics.
Total RNA was extracted by Rneasy Mini Kit (Qiagen) and cDNA was isolated by GoScript Reverse Transcription System (Promega). RT-qPCR reactions were set up using the SYBR FAST Universal qPCR kit (KAPA). Relative expression values were normalized to Gapdh expression and data was performed using Ct computing method 2–ΔΔCt. Each experiment was performed in technical triplicate. A list of primers used in Supplementary Table 1. Significance between different groups was determined using T-test, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. The sequences of primers used are listed in Supplementary Table 3.
Cells were plated onto 6-well plates at 3 × 105 per well and incubated. Subconfluent cells (about 6 × 106) were collected with Accutase, washed three times with cold PBS, and lysed in buffer that contained 20 mM Tris (pH 8.0), 137 mM NaCl, 100 g/L glycerol, 50 g/L Triton X-100, and 4 g/L EDTA; 1 μL PMSF (0.1 M) and 10 μL phosphatase inhibitor (10 g/L) were added per 1 mL lysis buffer immediately before use. The cell lysates were put on ice for 15 min and centrifuged at 12,000 rpm at 4°C for 10 min, and the supernatant was transferred to new tubes. The concentrations of the lysates were measured by Coomassie Plus (Bradford) Assay (Thermo Scientific). Equal amounts (40 μg) of protein were electrophoresed on 12% (w/v) sodium dodecylsulfate polyacrylamide gels. Proteins were transferred to Hybond-polyvinylidene difluoride membranes (Amersham) and incubated with the primary antibodies overnight at 4°C and peroxidase-conjugated secondary antibodies at room temperature for 1 h. Enhanced chemiluminescence (ECL) (Amersham) was used to detect the signals.
Cells for immunofluorescence assays were washed with PBS and fixed in 4% paraformaldehyde for 30 min at room temperature, and following permeabilized with 0.1% Triton X-100 (Sigma) and 1% BSA in PBS for 30 min. Then incubated cells with primary antibody at 4°C overnight. The cells were subsequently washed three times in 1% BSA, 0.1% Triton X-100 in PBS for 5 min per wash, and incubated with secondary antibody for 1 h at room temperature in the dark, then washed once for 5 min in 1% BSA, 0.1% Triton X-100 in PBS and twice for 5 min in PBS. The cells were then mounted in Vectashield with DAPI (Vector Laboratories) The samples were observed by laser microscope (Nikon, Tokyo, Japan). The anti-bodies used are listed in Supplementary Table 5.
Cells were dissociated into single cells using Accutase and collected using centrifugation. After being incubated with Hochest33342 for 1 hr at 37°C in the dark, the cell suspension was filtered using a cell strainer (FACSAria II, BD Biosciences) to remove large clumps of cells. The cells were then performed flow cytometry (FACSAria II, BD Company). Data analysis was performed using FlowJo software.
To generate chimeric embryo, donor cells (12∼15 cells/blastocyst) were microinjected into the ICR mice blastocoel cavity using a piezo-assisted micromanipulator attached to an inverted microscope and recover the injected blastocysts in KSOM medium (Millipore), then transplant the chimeric blastocysts into the uterus of pseudopregnant ICR female mice at 2.5 dpc (days post coitus). The chimeric embryos were collected at E13.5.
Total RNA of ASCs and Bdh2-knockout ASCs was isolated by Rneasy Mini Kit (Qiagen). cDNA was synthesized from purified RNA templates. The cDNA libraries were sequenced using Illumina HiSeq10×. The sequencing-obtained raw image data were transformed into sequence data, called raw data or raw reads, which were deposited in the GEO2 database under the GEO submission number of GSE158256. Raw data was firstly processed to remove reads with more than 20% low-quality bases and to remove adaptors. Then the clean data were mapped to the reference genome (GRCm38/mm10) with Hisat2(v2.0.5) with default settings. FeatureCounts (v1.5.0-p3) was used for reads counting, and then FPKM (Fragments Per Kilobase of transcript sequence per Millions) of each gene was calculated to estimate gene expression levels. DEGs in different samples were determined using the DEseq2 R package (v1.16.1) with fold change ≥ 1 and adj p-value ≤ 0.05. Heatmaps of selected genes were plotted by using the pheatmap R package (v1.0.12). GO and KEGG enrichment analysis was implemented by the clusterProfiler R package. Gene Set Variation Analysis was performed by using the GSVA R package (v1.32.0) and limma package (v3.40.6) with fold change ≥ 0.6 and adj p-value ≤ 0.05. GSEA analysis was performed with the GSEA software (v4.0.3) developed by the Broad Institute.
Cells on fibronectin-coated plate were incubated with Accutase for 3 min at 37°C till the colonies were completely disaggregated. The colonies were resuspended in EB medium: DMEM/F12 + 20% Knockout serum replacement, KSR (Gibco) + 1% GlutaMax (Gibco) + 1% NEAA (Gibco) + 1% β-mercaptoethanol (Gibco). The colonies were cultured in suspension for up to 7 days at a concentration of 1 × 105 cells/mL.
For primitive endoderm (PrE) differentiation, ASCs and ESCs were differentiated to naïve PrE over the course of 4–8 days by plating 3.7–5.3 × 104 cells cm–2 onto gelatinized plates in ESC medium for 24 h. And then cells were grown in RPMI 1640 plus Glutamax (Gibco, 61870036) supplemented with 1 μM Retinoic Acid (Sigma), 3 μM CHIR99021, 20 ng/mL Activin A and 10 ng/mL leukemia inhibitory factor. The medium was changed daily. ESCs differentiated to PrE cells within 4–8 days depending on the cell line and conditions.
Descriptive statistics were generated for all quantitative data, expressed as mean ± SD. The mean ± SD values were calculated from three samples per group and three technical replicas for per samples. Differences in means between control and Bdh2-knockout groups were determined by Student’s T-test. All statistical analysis were performed using GraphPad Prism, v.5.0 (GraphPad Software, CA, United States).
All animal experiments were performed in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at Inner Mongolia University, China.
Given that Bdh2 is known to regulate iron homeostasis, energy metabolism and apoptosis (Yang et al., 2013; Liang et al., 2019; Liu et al., 2014b, 2020), we hypothesized that Bdh2 may play a role in the regulation of self-renewal, pluripotency or differentiation in ESCs. To investigate these questions, we first analyzed its expression across different state mouse ESC lines in previously published RNAseq datasets of ESC, EpiSC and ASC (Bao et al., 2018). ASCs represents an intermediate state between naïve ESC and primed pluripotency such as EpiSC, which is highly potent and has higher levels of DNA methylation (Bao et al., 2018; Wu et al., 2020). We observed Bdh2 was expressed at lower levels in ground/naïve state ESCs; in contrast, higher level expression was seen in advanced ESCs with the highest levels in ASCs (Supplementary Figure 1A). To confirm this finding, both RT-qPCR and Western-blot analysis were performed and showed that Bdh2 expression was significantly higher in ASCs compared with ESCs (Supplementary Figures 1B,C).
CRISPR/Cas9 system was applied to target the CDS1 of the Bdh2 gene using two sgRNAs near the ATG codon (Figure 1A). The sequence of two sgRNAs was listed in Table S1. A total of 98 ASCs and 101 ESCs single colonies were picked for characterization of editing events. PCR was performed, and electrophoresis and Sanger sequencing of PCR products revealed eight ASCs and two ESCs that carried nucleotide deletions. The sequence of primers used in the PCR was listed in Supplementary Table 2. Out of the eight ASC lines, seven were Bdh2–/– and one was Bdh2+/–; all the two ESC lines were Bdh2–/– (Supplementary Figures 1D–F). One of the ASCs possessed a bi-allelic 56-nucleotide (nt) deletion, as well as one of the ESCs possessed a bi-allelic 46-nt deletion (Supplementary Figures 1G,H). The establishment rate of Bdh2-knockout ASCs and ESCs were 8.2 and 2%, respectively (Supplementary Table 3).
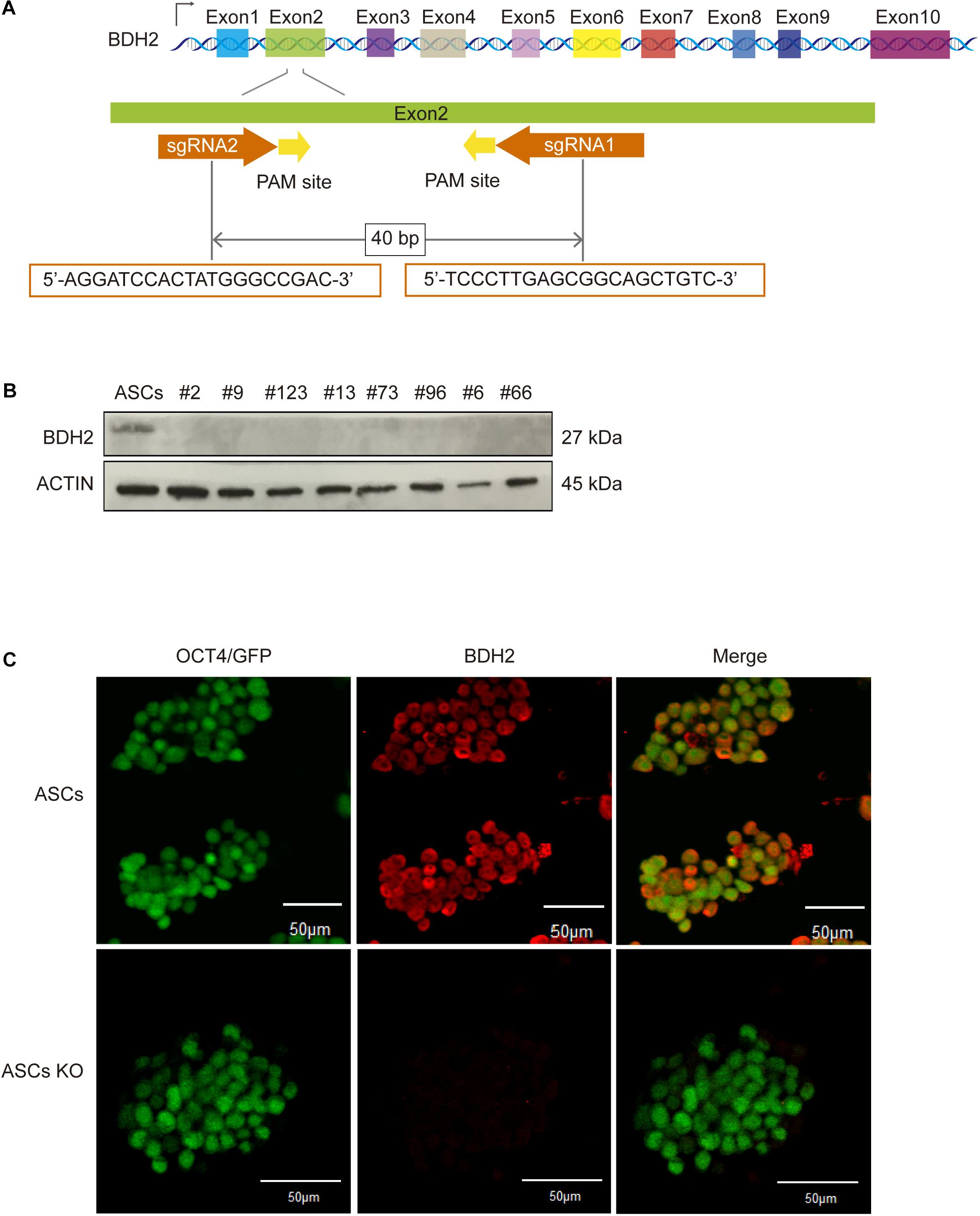
Figure 1. Generation and identification of bi-allelic Bdh2-knockout mouse advanced pluripotent stem cells (ASCs). (A) Schematic of the exon of Bdh2 gene and location of gRNAs. (B) Western blotting was used to detect the expression of BDH2 in the above eight knockout cells. (C) Immunostaining for BDH2 in ASCs and bi-allelic 56-nt clone. nt, nucleotide; PCR, polymerase chain reaction. Scale bars, 50 μm.
The deletions generated in the ASC and ESC lines are frameshift mutations which result in a premature stop codon. Several studies indicate that mRNAs with premature stop codon are eliminated in the cells by nonsense-mediated mRNA decay pathway (Danckwardt et al., 2002; Frischmeyer et al., 2002). To verify whether Bdh2 was not expressed in these edited ASC and ESC lines, RT-qPCR, western blot and Immunostaining were employed. The results showed that Bdh2 mRNAs were knocked down to 10–40% in the eight Bdh2-knockout ASCs (Supplementary Figure 1I). Importantly, BDH2 protein was barely detectable in all the Bdh2- knockout ASCs (Figures 1B,C). Similarly, Bdh2 mRNAs were also knocked down to 30-50% in two Bdh2- knockout ESCs (Supplementary Figure 1J), and the protein levels of BDH2 were undetectable in the two Bdh2-knockout ESCs (Supplementary Figures 1K,L).
Taken together, multiple Bdh2-knockout ASC and ESC lines were generated; ASCs with the bi-allelic 56 nt-deletion and ESCs with bi-allelic 46 nt-deletion were selected for further studies, in which the Bdh2 protein translation was successfully abolished.
Bdh2-knockout ASCs and ESCs exhibited normal morphology similar to wide type (WT) ASCs and ESCs (Figures 2A–C and Supplementary Figures 2A,B). Both WT and Bdh2-knockout ASCs/ESCs were alkaline phosphatase (AP)-positive colonies, indicating that deletion of Bdh2 had no effect on self-renewal potential of the ESCs (Figure 2D and Supplementary Figure 2C). To understand the characteristics of Bdh2-knockout ASCs/ESCs, we next determined the expression levels of several pluripotency markers. Both the mRNA and protein of these genes, OCT4, SOX2, and NANOG, were expressed at similar levels in WT and Bdh2-knockout ASCs/ESCs (Figures 2E,F and Supplementary Figures 2D,E). Interestingly, Bdh2-knockout ASCs propagated more quickly than ASCs, as shown in cell growth curves (Figure 2G). However, Bdh2 deficiency did not affect the proliferation of ESCs (Supplementary Figure 2F).
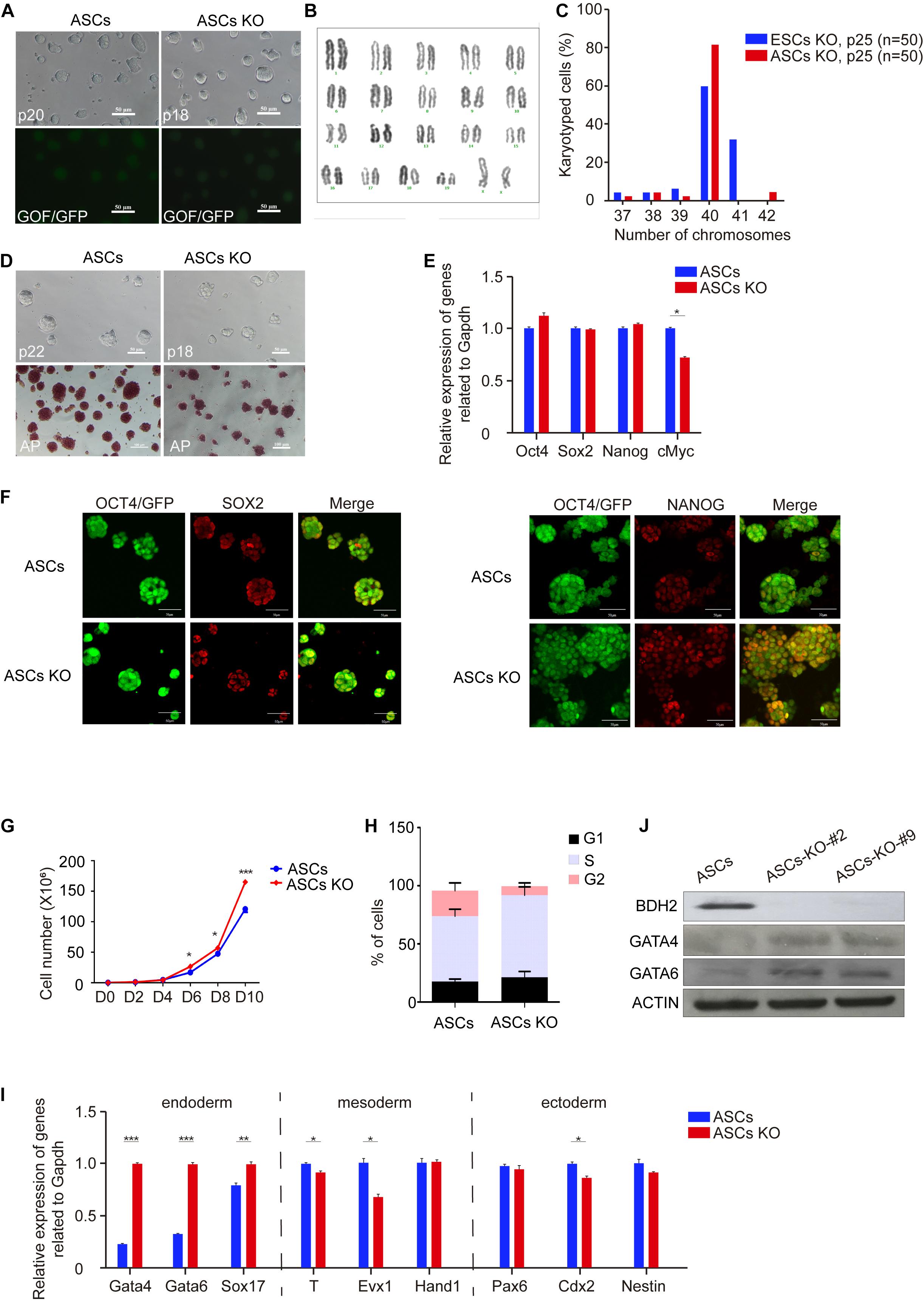
Figure 2. Characteristics of Bdh2-knockout ASCs. (A) Morphology of Bdh2-knockout ASCs. Here, we use ASCs with GOF/GFP reporter. Scale bars, 50 μm. (B) Karyotyping of Bdh2-knockout ASCs (P25). (C) Distribution of chromosome number in Bdh2-knockout ASCs (P25) and Bdh2-knockout ESCs (P25). (D) Alkaline phosphatase (AP) staining on Bdh2-knockout ASCs and ASCs. Scale bars, 50 μm. (E) Real-time PCR analysis of pluripotency-associated genes expression in the Bdh2-knockout ASCs, ASCs were used as control. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001. (F) Immunostaining of SOX2 and NANOG in Bdh2-knockout ASCs and ASCs. Scale bars, 50 μm. (G) Cell proliferation curves in Bdh2-knockout ASCs and ASCs. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001. (H) Cell cycle analysis of Bdh2-knockout ASCs and ASCs. The percentage of cells in G1, S, and G2 phase were determined by flow cytometry. Values are means of three independent experiments. Error bars indicate mean ± SD. (I) Real-time PCR analysis of endoderm, mesoderm, and ectoderm-associated genes expression in the Bdh2-knockout ASCs, ASCs were used as control. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001. (J) Western blotting was used to detect the expression of BDH2, GATA4, and GATA6 in Bdh2-knockout ASCs and ASCs.
To confirm these findings, we performed cell cycle analysis. ESCs have a unique cell cycle structure, including long S phase and remarkably short G1 and G2 phase (White and Dalton, 2005). The percentages of cells in G1, S, and G2 phase were determined by flow cytometry. As expected, Bdh2 deficiency promoted cell growth of ASCs as judged by an increase in the percentage of Bdh2-knockout ASCs in S phase compared to ASCs (Figure 2H). However, Bdh2-knockout has little impact on cell cycle of ESCs (Supplementary Figure 2G). Collectively, these findings were consistent with the cell proliferation results (Figure 2G and Supplementary Figure 2F).
To address whether Bdh2 deficiency modulates the expression of gene markers for three germ layers, i.e., endoderm, mesoderm and ectoderm, RT-qPCR was performed. Here, the RT-qPCR analysis data showed that the expression of endoderm marker Gata4, Gata6, and Sox17 was significantly upregulated in Bdh2-knockout ASCs. In contrast, expression of mesoderm genes Brachyury (T), Evx1 and trophectoderm gene Cdx2 was significantly decreased (Figure 2I). Similarly, the expression of Gata4 and Gata6 was found to be significantly upregulated in Bdh2-knockout ESCs, while Brachyury (T) and Cdx2 transcripts significantly was decreased in Bdh2-knockout ESCs (Supplementary Figure 2H). These findings were confirmed by Western Blot and Immunoflourance staining against GATA4, GATA6, SOX17 (Figure 2J and Supplementary Figures 2I,J).
Taken together, our results indicate that Bdh2 deficiency in ASCs and ESCs did not affect the pluripotency and identity of mouse ESCs. Interestingly, Bdh2 deficiency promoted proliferation of ASCs but not ESCs by affecting the cell cycle phase distribution. More importantly, deletion of Bdh2 increased the expression of endoderm marker genes while decreasing the expression of some mesoderm and ectoderm genes. These results suggest that Bdh2 plays distinct roles in regulating stem cell fate.
To gain further molecular insights into Bdh2-knockout ASCs, we determined global gene expression profiles using RNA sequencing (RNA-Seq). To examine differences between Bdh2-knockout ASCs and WT ASCs, we analyzed differentially expressed genes using DEseq2 R package (adj p-value < 0.05, fold change |log2Ratio| ≥ 0). There were 5673 differentially expressed genes in Bdh2-knockout ASCs compared with WT ASCs; 3175 genes were significantly upregulated, including major endoderm lineage markers (Gata4, Gata6, Sox17, Sox7, Pdgfra), and 2498 genes were significantly downregulated which are mainly related to metabolic processes (Figure 3A). It is interesting to note that the primitive endoderm markers (Gata4, Gata6, Sox7) and endoderm transcription factor genes (Sox17) were highly expressed in Bdh2-knockout ASCs compared to WT ASCs (Figure 3B), in line with the RT-qPCR and Western Blot results. Together, these data suggest that Bdh2 may influence the early differentiation of ESCs.
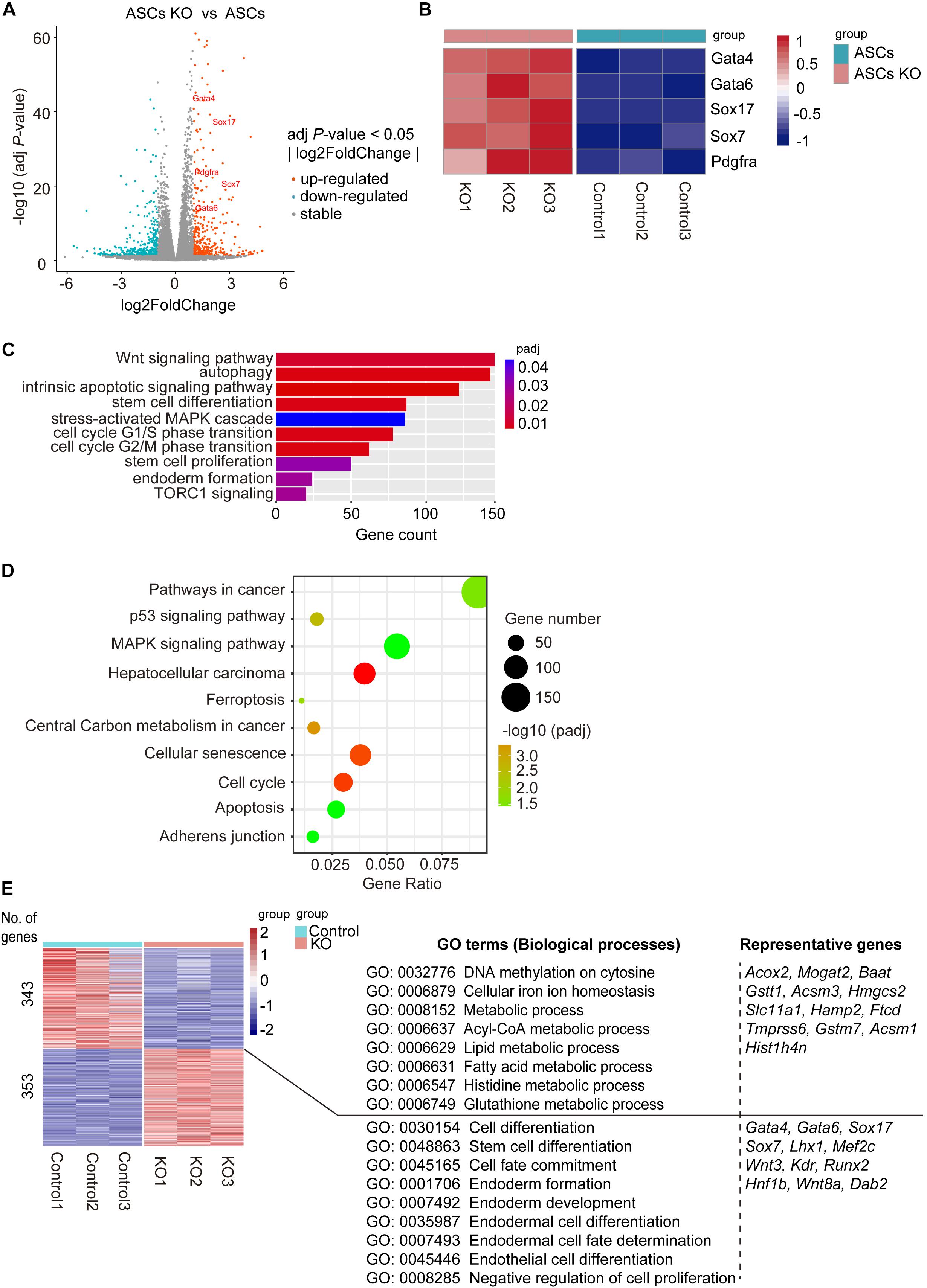
Figure 3. Analysis of molecular features of Bdh2-knockout ASCs. (A) Volcano plot of differentially expressed genes for Bdh2-knockout ASCs versus ASCs. (B) Heatmap showing scaled expression of endoderm-associated genes. (C) Gene Ontology (GO) analysis of significantly enriched “biological processes” (BP) of the differentially expressed genes (DEGs) for Bdh2-knockout ASCs versus ASCs described in (A). (D) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of DEGs for Bdh2-knockout ASCs versus ASCs described in (A). (E) Heatmap showing scaled expression values of a total of 696 DEGs [mean log2 (normalized read counts) > 2, log2 (fold change) ≥ 1, adjusted p value < 0.05] in Bdh2-knockout ASCs versus ASCs. Significantly enriched GO terms and representative genes in each cluster are listed on the right.
Genes ontology (GO) analysis suggested that GO terms such as “stem cell differentiation,” “endoderm formation,” “stem cell proliferation,” “cell cycle,” “autophagy,” “intrinsic apoptotic signaling pathway” and a number of metabolic process terms in biological process were enriched in the differentially expressed genes between Bdh2-knockout ASCs and WT ASCs (Figure 3C and Supplementary Figure 3A). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis showed the top 10 enriched pathways (Figure 3D), including “ferroptosis,” “cell cycle,” “apoptosis,” “adherens junction,” “pathways in cancer,” etc. When cutoff of adj p-value < 0.05, fold change |log2Ratio| ≥ 1 was applied, there were 696 differentially expressed genes. Upregulated genes were mainly enriched in such “GO-biological processes” as “negative regulation of cell proliferation,” “cell fate commitment,” and “endoderm formation,” with representative genes as Gata4, Gata6, Sox17, Sox7, Wnt3, Runx2. The downregulated genes were primarily enriched in DNA methylation (“DNA methylation on cytosine”) and metabolism (“cellular iron ion homeostasis,” “Lipid metabolic process,” “histidine metabolic process,” “glutathione metabolic process,” and “Acyl-CoA metabolic process”), with representative genes as Acox2, Mogat2, Gsttl1, Ftcd, Histlh4n (Figure 3E and Supplementary Figure 3B).
In line with the GO analysis, GSEA analysis revealed enrichment for cell fate specification and several metabolic processes (Supplementary Figure 3C). It is worth noting that adhesion-related genes, enriched in such GO terms as “adherens junction,” “focal adhesion,” and “gap junction,” were all down-regulated in Bdh2-knockout ASCs (Supplementary Figure 3D). Taken together, Bdh2 may regulate adhesion of ASCs.
The RT-qPCR was performed on key genes and showed high concordance with the RNA-Seq data (Figure 2I and Supplementary Figure 2H). Overall, these results showed that the knockout of Bdh2 altered the ASCs with particular molecular features. In short, the endoderm lineage markers were significantly upregulated, while the genes associated with various metabolic processes were significantly downregulated in Bdh2-knockout ASCs.
It was previously reported that ASCs have higher DNA methylation levels compared with ESCs (Bao et al., 2018; Wu et al., 2020). To investigate if loss of Bdh2 altered the DNA methylation levels, we examined the mRNA and protein levels of methyltransferases in Bdh2-knockout ASCs/ESCs and WT ASCs/ESCs. RT-qPCR analysis demonstrated that Dnmt3a, Dnmt3b, and Dnmt3l were significantly decreased in Bdh2-knockout ASCs/ESCs compared to WT ASCs/ESCs (Figure 4A and Supplementary Figure 4A). Furthermore, the protein expression of DNMT3A and DNMT3B by Western Blot were undetectable or much lower in the Bdh2 knockout lines, consistent with the results of the transcript levels (Figure 4B and Supplementary Figure 4B), however DNMT3L protein levels were comparable. In line with DNMT3A and DNMT3B, the expression of DNA methyltransferases, DNMT1, was also significantly markedly decreased in Bdh2-knockout ASCs/ESCs (Figure 4B and Supplementary Figure 4B). Immunofluorescence staining of DNMT3A also confirmed its downregulation in Bdh2-knockout ASCs/ESCs as well (Figure 4C and Supplementary Figure 4C).
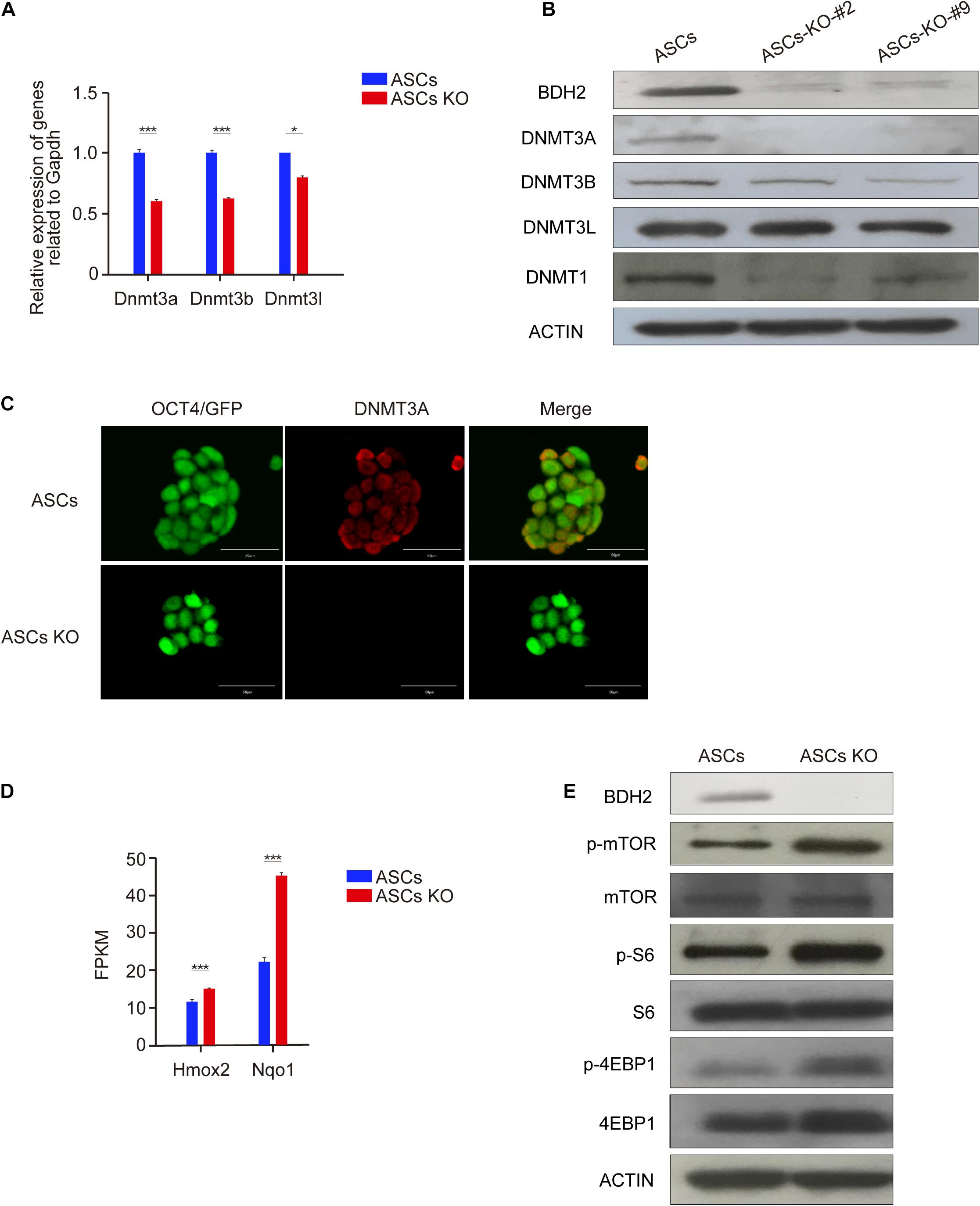
Figure 4. Bdh2 knockout regulates DNA methylation in ASCs. (A) Real-time PCR analysis of DNA methyltransferase genes expression (Dnmt3a, Dnmt3b, and Dnmt3l) in the Bdh2-knockout ASCs, ASCs were used as control. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, ***p < 0.001. (B) Western blotting analysis for BDH2, DNMT3A, DNMT3B, DNMT3L, and DNMT1 in two Bdh2-knockout ASCs. (C) Immunostaining of DNMT3A in Bdh2-knockout ASCs and ASCs. Scale bars, 50 μm. (D) Expression levels of oxidative stress-related key genes were tested by RNA-Seq analysis. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, ***p < 0.001. (E) The effect of Bdh2 knockout on mTOR signaling pathway in ASCs was detected by Western blotting.
Zhang et al. (2017) previously found that DNA demethylation was mediated by activation of mTORC1 signaling via inhibiting DNMT1 and DNMT3A expression. Interestingly, mTORC1 signaling can be activated by oxidative stress (Liu et al., 2014c). Given that that Bdh2 deficiency was reported to increase oxidative stress (Liu et al., 2014c), we speculated that the decreased expression of DNA methyltransferases was related to increased oxidative stress and mTORC1 signaling activity. In support of this notion, RNA-Seq analysis revealed that oxidative stress-related genes, heme oxygenase 2 (Hmox2) and NAD(P)H dehydrogenase quinone 1 (Nqo1), were significantly upregulated in Bdh2-knockout ASCs (Figure 4D). As expected, Western Blot analysis showed that mTORC1 signaling was increased in Bdh2-knockout ASCs compared with WT ASCs, in particular the phosphorylation of mTOR protein and its downstream effector proteins, p-S6 and p-4EBP1 (Figure 4E).
Taken together, these results indicate that loss of BDH2 suppress expression of multiple methyltransferases in ASCs, and increased phosphorylation of mTOR and several of the downstream effectors.
To address if Bdh2 knockout impacts differentiation of ASCs/ESCs in vitro, we assessed embryoid body (EB) formation using WT and Bdh2-knockout ASC/ESC lines. Suspension culture allows for the formation of cell clusters during spontaneous differentiation (or embryoid bodies; EBs), and can mimic cellular interactions reminiscent of development processes in vivo (Boheler et al., 2002).
RT-qPCR analysis of trilineage markers showed the endoderm markers Gata4, Gata6 and Sox17 increased in the early stages of Bdh2-knockout ASC derived EBs. In contrast the mesodermal markers, Brachyury (T) and Evx1 were downregulated in Bdh2 deficient cells compared with WT cells. Similarly, the ectodermal markers, Pax6, Cdx2, and Nestin, were also downregulated in Bdh2-knockout EBs (Figure 5A). We also assessed EBs derived from ESCs, and observed a similar trend (Supplementary Figure 5A). Collectively, our data showed that absence of Bdh2 in ASCs/ESCs potentiate exit from the pluripotency state and differentiate into derivatives of the three germ layers with bias toward endoderm with compromised formation or the maintenance of mesoderm and ectoderm.
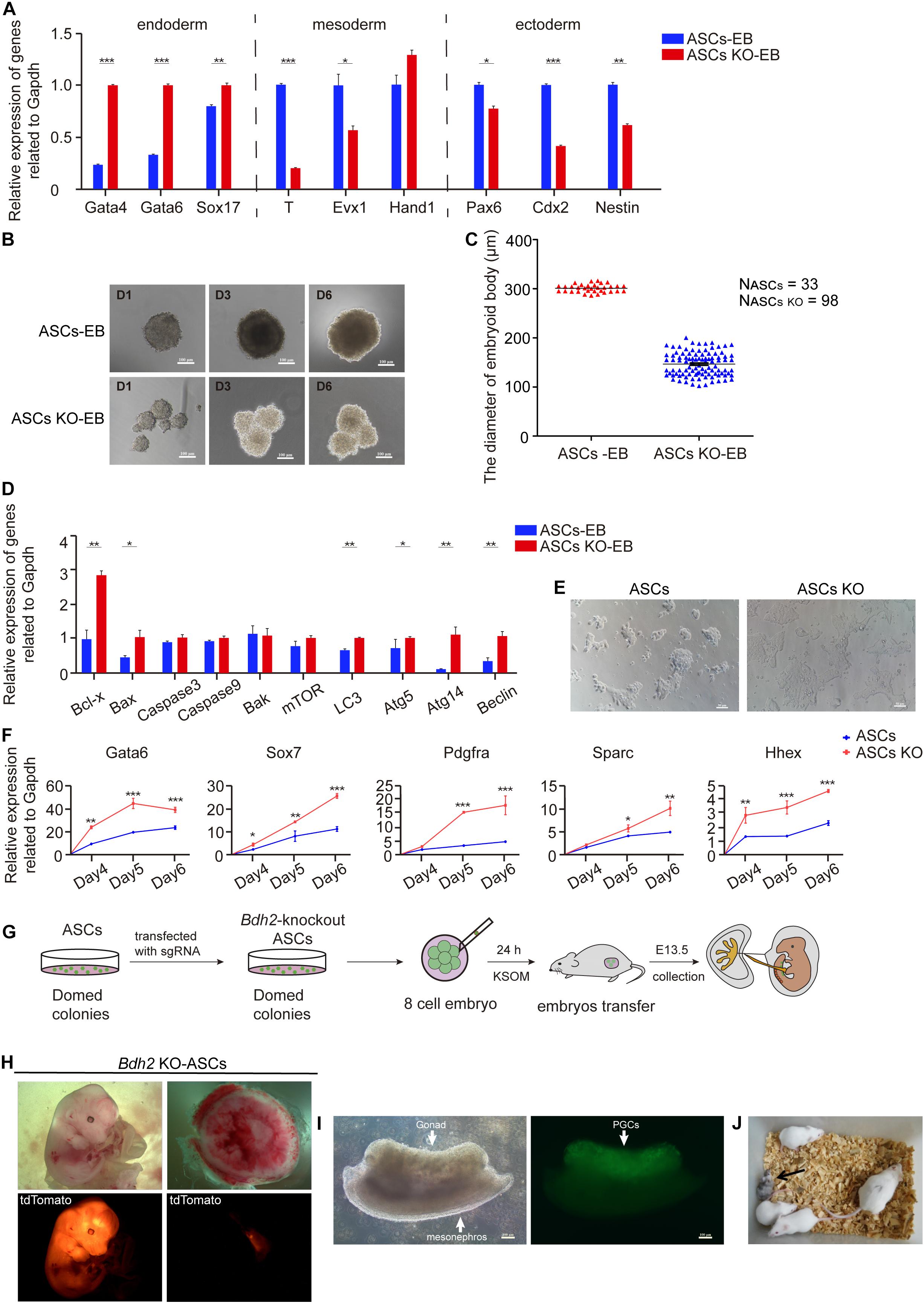
Figure 5. Differentiated and developmental potency of Bdh2-knockout ASCs. (A) Real-time PCR analysis of endoderm, mesoderm and ectoderm-associated genes expression in the embryoid bodies of Bdh2-knockout ASCs, embryoid bodies of ASCs were used as control. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001. (B) Morphology of embryoid bodies induction by Bdh2-knockout ASCs and ASCs in Day1, Day3, and Day6. Scale bars, 100 μm. (C) Statistics of the number and diameter of embryoid bodies formed by Bdh2-knockout ASCs and ASCs, respectively. N, represents the number of EB spheres. (D) Real-time PCR analysis of apoptotic and autophagy-associated genes expression in the embryoid bodies of Bdh2-knockout ASCs, embryoid bodies of ASCs were used as control. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001. (E) Representative bright-field images from Bdh2-knockout ASCs and ASCs differentiated for 5 days toward the primitive endoderm lineage. Scale bars, 50 μm. (F) Real-time PCR analysis of key markers after 4/5/6 days of directed differentiation toward primitive endoderm. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001. (G) Schematic illustrating chimera production from Bdh2-knockout ASCs. (H) Chimeras (E13.5) generated with Bdh2-knockout ASCs (multiple cells). ASCs carried tdTomato+/GOF+ reporter. (I) Germline transmission of Bdh2-knockout ASCs in E13.5 chimeras. PGCs were shown by GOF/GFP+ cells (arrow). Scale bars, 100 μm. (J) A chimeric pup generated by injecting 15∼20 Bdh2-knockout ASCs in ICR host blastocysts.
In addition to the gene expression differences, Bdh2-knockout EBs presented with a smaller size but a larger number as early as day 2 when compared to the one spherical EBs derived from WT ASCs/ESCs (Figure 5B and Supplementary Figure 5B,C). To further investigate this difference, pendant-drop method was used to form embryoid body. While the WT ASCs/ESCs only formed one EB sphere per drop, there were three times more in the number of EBs derived from Bdh2-knockout ASCs (Figure 5C and Supplementary Figure 5D). The diameter of EBs derived from Bdh2-knockout ASCs was around 150 μm at day 5, while the diameter of EBs derived from WT ASCs was around 300 μm (Figure 5C). Similarly, the number of EBs derived from Bdh2-knockout ESCs was about five times as much as the number of EBs derived from WT ESCs. The diameter of EBs derived from Bdh2-knockout ESCs was around 100 μm at day 5, while the diameter of EBs derived from WT ESCs was around 300 μm (Supplementary Figure 5D). More importantly, Bdh2-knockout EB spheres seem to be more incompact, which may due to the down-regulation of adhesion-related genes in Bdh2-knockout cells (Supplementary Figure 3D).
Interestingly, when Bdh2-knockout ASCs/ESCs were differentiated in vitro, we observed a striking increase of cell apoptosis at day 3 of differentiation morphologically. We therefore measured the mRNA levels of apoptosis and autophagy-associated genes by RT-qPCR. The results showed that Bax, Bcl-x, LC3, Atg5, Atg14, and Beclin in Bdh2-knockout EBs were significantly increased compared to the WT ASCs/ESCs EBs (Figure 5D and Supplementary Figure 5E), and suggest that cells from Bdh2-knockout ASCs/ESCs EBs were markedly more sensitive to apoptosis. These results are in agreement with the findings of Yang et al. (2013) that Bdh2 acts as an anti-apoptotic factor.
We then decided to probe the potential of Bdh2-knockout ASCs/ESCs for a different cell fate controlled by inductive cues. Smith demonstrated mESCs do not generate endoderm cells directly, as only late epiblast cells have this capacity. As a result, germ-layer specification can be only attained after transition through different pluripotent states (Smith, 2017). mESCs, however, can directly give rise to primitive endoderm. Thus, we decided to test the capacity of Bdh2-knockout ASCs/ESCs for this differentiation by utilizing a protocol based on defined culture conditions (Anderson et al., 2017; Ortmann et al., 2020). The differentiation efficiency was quantified by measuring the expression of PrE markers, Gata6, Sox7, Pdgfrα, Sparc, and Hhex. Interestingly, Bdh2-knockout ASCs/ESCs were more able to produce cells with distinctive morphology expressing Gata6, Sox7, Pdgfrα, Sparc, and Hhex (Figures 5E,F and Supplementary Figures 5F,G) compared to WT ASCs/ESCs. These analyses confirm that Bdh2 deficiency influence the capacity of mESCs to directly generate differentiated progenies.
To further evaluate the role of Bdh2 in mESC differentiation, we set to investigate the identity and developmental potential of Bdh2-knockout ASCs in vivo. Bdh2-knockout ASCs were transfected with H2B tdTomato plasmid to perform chimera test by injecting them into 8-cell stage mouse embryos, and the chimeric embryos were transplanted into 2.5 dpc pseudopregnancy female mice (Figure 5G). We analyzed their contribution to chimeric embryos at E13.5 and found that tdTomato-positive Bdh2-knockout ASCs contributed robustly to the embryo, and slightly contributed to the yolk sac, as well as placental labyrinth (Figure 5H). Furthermore, Bdh2-knockout ASCs were able to contribute to germline lineages (Figure 5I) and full-term chimeras (Figure 5J). These data suggest that the Bdh2-knockout ASCs have the pluripotential to contribute to embryo, and generate full-term pups.
Taken together, our data demonstrate that loss of Bdh2 did not affect the pluripotency of ASCs in vivo and Bdh2 deficient ASCs have the capacity to chimeric mice and contribute to germline lineages. However, analysis of germ layer formation in vitro suggests that the Bdh2 deficiency promoted a biased differentiation toward endoderm.
In this study, we generated Bdh2-knockout ASCs and ESCs using CRISPR/Cas9 system and analyzed the molecular and cellular consequences of Bdh2 deficiency in ASCs and ESCs. Our results demonstrate Bdh2-knockout ASCs/ESCs were capable to maintain pluripotency and contribute to chimeric embryos. Interestingly, Bdh2 deficiency promoted ASCs/ESCs to a biased differentiation toward primitive endoderm lineage. We demonstrate that loss of Bdh2 suppressed expression of multiple methyltransferase such as DNMT1, DNMT3A and DNMT3B, together with increased phosphorylation of mTOR and multiple effectors (Figure 6).
In here, we used two types of ESCs with distinct states: one was ASCs generated by our lab (Bao et al., 2018; Wu et al., 2020), which is an advanced state with a unique molecular feature and developmental potential; another was ground state-2i/L-ESCs. In here, we found Bdh2-knochout ASCs and ESCs showed typical morphology of WT ASCs/ESCs, and expressed pluripotent genes Oct4, Sox2, and Nanog. These data indicated that Bdh2 is not essential to maintain pluripotency. Notably, Bdh2-knockout ASCs showed increased proliferation with larger fraction of cells in S-phase compared to WT ASCs. We noticed that the percentage of cells in G1 phase appears to be increased in Bdh2-knockout ASCs compared to ASCs, as well as with the long S phase (Figure 2H). Soufi and Dalton reviewed that there exists a strong connection between the cell cycle and mechanisms required for executing cell fate decisions, and that G1 phase represents a period of differentiation competency (Soufi and Dalton, 2016). As pluripotent stem cells commit to one of the three embryonic germ layers, their progeny acquire an extended G1 phase, resulting in increased cell division times. The endoderm and mesoderm commitment occur in early G1 phase (Soufi and Dalton, 2016), thus, the increased G1 phase in Bdh2-KO cells may explain at least partly, their biased differentiation toward endoderm lineage (Figure 2G). Interestingly, both Bdh2 deficient ESCs and ASCs showed increased expression of endoderm genes such as Gata4, Gata6, and Sox17 in both mRNA and protein level, and decreased expression of several mesoderm and ectoderm related genes such as Brachyury (T), Evx1 and Cdx2. These data suggest that Bdh2 is involved in the proper control of germ layer specific transcripts for cell fate decision. Wnt and Nodal signaling play a critical role in endoderm formation (Scheibner et al., 2019), our study show that Wnt signaling was the top pathway enriched in the Bdh2-deficient ASCs; it would be interesting to investigate how Bdh2 regulates Wnt signaling, which in turn controls cell fate decision during germ layer differentiation.
Differential methylation levels are critically associated with establishment and maintenance of the pluripotent state, as well as in the differentiation process in ESCs (Petell et al., 2016; McLaughlin et al., 2019; van Mierlo et al., 2019), methylation regulation has been an area of intense investigation in ESC biology. Our data demonstrate that Bdh2 deficient ASCs showed markedly reduced expression of multiple methytransferases such as DNMT1, DNMT3A, and DNMT3B suggesting loss of Bdh2 may cause hypomethylation in ESCs. Recently, Zhao et al. (2018) found that knockdown of Bdh2 expression caused increased DNA hydroxymethylation and decreased DNA methylation in CD4+ T cells, proposing the association between Bdh2 and DNA demethylation in the pathogenesis of systemic lupus erythematosus, and they revealed that the Bdh2 knockdown induced DNA hypomethylation was associated with increasing intracellular iron in CD4+ T cells. However, from multiple studies, it was suggested that DNA hypomethylation induced by Bdh2 deficiency can be caused by increased oxidative stress, and consequent activation of mTORC1 signaling, thereby mediating DNA hypomethylation via inhibiting the expression of DNMT1 and DNMT3A (Liu et al., 2014c; Zhang et al., 2017). Our results are consistent with this mechanism; knockout of Bdh2 induced the expression of oxidative related genes, and the activation of mTORC1 signaling. DNA transmethylases (DNMTs), DNMT3A, DNMT3B, and DNMT1, were also nicely downregulated in Bdh2-knochout ASCs/ESCs, suggesting Bdh2 may be essentially required to control DNA methylation in ASCs/ESCs. However, the molecular mechanisms on how Bdh2 regulates DNA methylation need to be dissected in further detail.
It is noteworthy that morphological differences between EB spheres derived from Bdh2-knockout and WT ASCs/ESCs were remarkable; Bdh2-knockout EB spheres seemed to be more incompact compared to the WT EBs. We propose two explanations for this observed difference. Firstly, Bdh2 plays distinct roles in the regulation of primitive endoderm-like stem cell. It was reported that cellular adhesion of primitive endoderm (PrE) showed difference with epiblast (Stanton et al., 2019). Interestingly, we found downregulation of adhesion-related genes in Bdh2-knockout cells (Supplementary Figure 3D), which may explain the formation of smaller EBs in Bdh2-knockout ASCs/ESCs. Secondly, Bdh2-knockout cells were more apoptotic, at least some of the cells. Previous studies have demonstrated that oxidative stress is responsible for triggering apoptosis and autophagy in tumors (Sato et al., 2004; Kaminskyy et al., 2012; Fernandez-Gil et al., 2019; Zhao et al., 2019), and influences numerous biological processes, including development and differentiation (Meier et al., 2000; Fuchs and Steller, 2011). In our study, a striking increase of cell apoptosis was observed at day 3 of in vitro differentiation (Figure 5B and Supplementary Figures 5B,C) with much more obvious apoptosis in Bdh2-knockout EBs than in WT EBs, which was in line with the report that Bdh2 acts as an anti-apoptotic factor (Yang et al., 2013). Indeed, apoptosis and autophagy-associated genes such as Bcl-x, Bax, LC3, Atg5, Atg14, and Beclin were significantly increased in Bdh2-knockout EBs compared to the WT ASCs/ESCs EBs. Furthermore, activated mTORC1 signaling in Bdh2-knockout cells (Figure 4E) may also be responsible for control of cellular status, as mTOR kinase is a key regulator of homeostasis, ribosome synthesis, various metabolic pathways and autophagy which are known to finely regulates the balance between self-renewal and differentiation in stem cells (Wullschleger et al., 2006; Han et al., 2008; Endo et al., 2009).
Indeed, our data found Bdh2 deficient ASCs/ESCs could contribute to germ line transmission and full-term pup in vivo; the biased endoderm differentiation in vitro didn’t affect chimeric development. A possible explanation could be that while Bdh2-deficient ASCs/ESCs showed increased endoderm markers, they also express mesoderm and ectoderm genes similar to ASCs/ESCs (Figure 2I and Supplementary Figure 2H).
We generated Bdh2 deficient ASC and ESC lines and show that loss of Bdh2 had no significant impact on the maintenance of self-renewal and the pluripotency of ESCs. Interestingly, our data demonstrate that Bdh2 deficiency affected the formation of EBs and differentiation (primitive endoderm formation) by regulating the expression of related genes. We further demonstrate that Bdh2 loss inhibited expression of multiple methyltransferases, suggesting Bdh2 may be essentially required to maintain DNA methylation which is critically associated with the pluripotent state, as well as in the differentiation process in ESCs. We propose that the bi-allelic Bdh2-knockout ASC and ESC lines would be a valuable laboratory resource to study the role of Bdh2 in the development, as well as a worthwhile tool for earliest cell fate decision and DNA methylation studies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/info/linking.html, GSE158256.
All animal experiments were performed in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at Inner Mongolia University, China.
SB and XL conceived and designed the experiments. YF, SC, JZ, and HW performed the experiments. YF and FL analyzed the data. YF wrote the manuscript. SB, BW, and YS revised the manuscript. All the authors contributed to the article and approved the submitted version.
This study was supported by grants from Major Projects of Inner Mongolia (2020ZD0007).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Juan Li for proof of the manuscript, and also thank Zhigang Wang and Changshan Wang for technical support, and Inner Mongolia Saikexing Institutes of Breeding and Reproductive Biotechnologies in Domestic Animal for instrument support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.655145/full#supplementary-material
Supplementary Figure 1 | Identification of bi-allelic Bdh2-knockout mouse embryonic stem cells (ESCs). (A) Expression of Bdh2 in different cell lines by RNA-Seq analysis. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001. (B) Real-time PCR analysis of Bdh2 expression in ESCs and ASCs. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001. (C) Western blotting was used to detect the expression of BDH2 in ESCs and ASCs. (D) The target region was sequenced to verify deletion in the desired region (a: wild type control; b: the sequence of bi-allelic knockout; c: the sequence of target region before single cell colony formation that showed overlapped pick in the gRNA site). (E) Genomic DNA PCR flanking the gRNAs sites from single cell colonies (1 and 12:DNA ladder; 2: Negative control; 3: Deletion not occurred; 4–9 and 11: Bi-allelic knockout; 10: Deletion occurred in one allele). (F) Genomic DNA PCR flanking the gRNAs sites from single cell colonies (1 and 6: DNA ladder; 2: Negative control; 3: Deletion not occurred; 4 and 5: Bi-allelic knockout). (G) Alignment between ASCs wild type sequence and bi-allelic knockout sequences that showed 56-nt deleted region. (H) Alignment between ESCs wild type sequence and bi-allelic knockout sequences that showed 46-nt deleted region. (I) Real-time PCR analysis of Bdh2 expression in the 7 bi-allelic knockout clones and a one-allelic knockout clone. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001. (J) Real-time PCR analysis of Bdh2 expression in the 2 bi-allelic knockout clones. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001. (K) Western blotting was used to detect the expression of BDH2 in the 2 bi-allelic knockout cells. (L) Immunostaining for BDH2 in ESCs and bi-allelic 46-nt clone.
Supplementary Figure 2 | Characteristics of Bdh2-knockout ESCs. (A) Morphology of Bdh2-knockout ESCs. Here, we use ESCs with GOF/GFP reporter. Scale bars, 50 μm. (B) Karyotyping of Bdh2-knockout ESCs (P25). (C) Alkaline phosphatase (AP) staining on Bdh2-knockout ESCs (P18) and ESCs (P22). Scale bars, 50 μm. (D) Real-time PCR analysis of pluripotency-associated genes expression in the Bdh2-knockout ESCs, ESCs were used as control. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001. (E) Immunostaining of SOX2 and NANOG in Bdh2-knockout ESCs and ESCs. Scale bars, 50 μm. (F) Cell proliferation curves in Bdh2-knockout ESCs and ESCs. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001. (G) Cell cycle analysis of Bdh2-knockout ESCs and ESCs. The percentage of cells in G1, S, and G2 phase were determined by flow cytometry. Values are means of three independent experiments. Error bars indicate mean ± SD. (H) Real-time PCR analysis of endoderm, mesoderm and ectoderm-associated genes expression in the Bdh2-knockout ESCs, ESCs were used as control. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001. (I) Western blotting was used to detect the expression of BDH2, GATA4, and GATA6 in Bdh2-knockout ESCs and ESCs. (J) Immunostaining of GATA6 and SOX17 in Bdh2-knockout ASCs/ESCs and WT ASCs/ESCs. Scale bars, 50 μm.
Supplementary Figure 3 | Molecular features of Bdh2-knockout ASCs. (A) Gene Ontology (GO) analysis of significantly enriched metabolism-related “biological processes” (BP) of the differentially expressed genes (DEGs) for Bdh2-knockout ASCs versus ASCs described in Figure 3A. (B) Heatmap showed up/down-regulated GO-BP terms and KEGG pathways, respectively. (C) Gene Set Enrichment Analysis (GSEA) analysis showing enriched GO-BP terms for up-regulated DEGs and down-regulated DEGs, respectively. (D) Heatmap listed a number of differentially expressed adhesion-related genes in Bdh2-knockout ASCs versus ASCs, enriched in three GO terms, “adhesion junction,” “focal adhesion,” and “gap junction.”
Supplementary Figure 4 | Bdh2 knockout regulates DNA methylation in ESCs. (A) Real-time PCR analysis of DNA methyltransferase genes expression (Dnmt3a, Dnmt3b, and Dnmt3l) in the Bdh2-knockout ESCs, ESCs were used as control. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.01, ***p < 0.001. (B) Western blotting analysis for BDH2, DNMT3A, DNMT3B, DNMT3L, and DNMT1 in two Bdh2-knockout ESCs. (C) Immunostaining of DNMT3A in Bdh2-knockout ESCs and ESCs. Scale bars, 50 μm.
Supplementary Figure 5 | Differentiated potency of Bdh2-knockout ESCs. (A) Real-time PCR analysis of endoderm, mesoderm and ectoderm-associated genes expression in the embryoid bodies of Bdh2-knockout ESCs, embryoid bodies of ESCs were used as control. Data were obtained in triplicate and presented as mean ± SD. p values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001. (B) Morphology of embryoid bodies induction by Bdh2-knockout ASCs and ASCs in Day2, Day3, Day5, and Day7. Scale bars, 100 μm. (C) Morphology of embryoid bodies induction by Bdh2-knockout ESCs and ESCs in Day1∼Day7. Scale bars, 100 μm. (D) Statistics of the number and diameter of embryoid bodies formed by Bdh2-knockout ESCs and ESCs, respectively. N, represents the number of EB spheres. (E) Real-time PCR analysis of apoptotic and autophagy-associated genes expression in the embryoid bodies of Bdh2-knockout ESCs, embryoid bodies of ESCs were used as control. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001. (F) Representative bright-field images from Bdh2-knockout ESCs and ESCs differentiated for 5 days toward the primitive endoderm lineage. Scale bars, 50 μm. (G) Real-time PCR analysis of key markers after 4/5/6 days of directed differentiation toward primitive endoderm. Data were obtained in triplicate and presented as mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.001, ***p < 0.0001.
Anderson, K. G. V., Hamilton, W. B., Roske, F. V., Azad, A., Knudsen, T. E., Canham, M. A., et al. (2017). Insulin fine-tunes self-renewal pathways governing naive pluripotency and extra-embryonic endoderm. Nat. Cell Biol. 19, 1164–1177. doi: 10.1038/ncb3617
Bao, S., Tang, W. W., Wu, B., Kim, S., Li, J., Li, L., et al. (2018). Derivation of hypermethylated pluripotent embryonic stem cells with high potency. Cell Res. 28, 22–34. doi: 10.1038/cr.2017.134
Boheler, K. R., Czyz, J., Tweedie, D., Yang, H. T., Anisimov, S. V., and Wobus, A. M. (2002). Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ. Res. 91, 189–201. doi: 10.1161/01.Res.0000027865.61704.32
Chen, C. Y., Cheng, Y. Y., Yen, C. Y. T., and Hsieh, P. C. H. (2016). Mechanisms of pluripotency maintenance in mouse embryonic stem cells. Cell. Mol. Life Sci. 74, 1805–1817. doi: 10.1007/s00018-016-2438-0
Danckwardt, S., Neu-Yilik, G., Thermann, R., Frede, U., Hentze, M. W., and Kulozik, A. E. (2002). Abnormally spliced beta-globin mRNAs: a single point mutation generates transcripts sensitive and insensitive to nonsense-mediated mRNA decay. Blood 99, 1811–1816.
Davuluri, G., Song, P., Liu, Z., Wald, D., Sakaguchi, T. F., Green, M. R., et al. (2016). Inactivation of 3-hydroxybutyrate dehydrogenase 2 delays zebrafish erythroid maturation by conferring premature mitophagy. Proc. Natl. Acad. Sci. U.S.A. 113, E1460–E1469. doi: 10.1073/pnas.1600077113
Devireddy, L. R., Hart, D. O., Goetz, D. H., and Green, M. R. (2010). A mammalian siderophore synthesized by an enzyme with a bacterial homolog involved in enterobactin production. Cell 141, 1006–1017. doi: 10.1016/j.cell.2010.04.040
Endo, M., Antonyak, M. A., and Cerione, R. A. (2009). Cdc42-mTOR signaling pathway controls Hes5 and Pax6 expression in retinoic acid-dependent neural differentiation. J. Biol. Chem. 284, 5107–5118. doi: 10.1074/jbc.M807745200
Evans, M. J., and Kaufman, M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156.
Fernandez-Gil, B. I., Guerra-Librero, A., Shen, Y. Q., Florido, J., Martinez-Ruiz, L., Garcia-Lopez, S., et al. (2019). Melatonin enhances cisplatin and radiation cytotoxicity in head and neck squamous cell carcinoma by stimulating mitochondrial ROS generation, apoptosis, and autophagy. Oxid. Med. Cell Longev. 2019:7187128. doi: 10.1155/2019/7187128
Frischmeyer, P. A., van Hoof, A., O’Donnell, K., Guerrerio, A. L., Parker, R., and Dietz, H. C. (2002). An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295, 2258–2261.
Fuchs, Y., and Steller, H. (2011). Programmed cell death in animal development and disease. Cell 147, 742–758. doi: 10.1016/j.cell.2011.10.033
Guo, K., Lukacik, P., Papagrigoriou, E., Meier, M., Lee, W. H., Adamski, J., et al. (2006). Characterization of human DHRS6, an orphan short chain dehydrogenase/reductase enzyme: a novel, cytosolic type 2 R-beta-hydroxybutyrate dehydrogenase. J. Biol. Chem. 281, 10291–10297. doi: 10.1074/jbc.M511346200
Han, J., Wang, B., Xiao, Z., Gao, Y., Zhao, Y., Zhang, J., et al. (2008). Mammalian target of rapamycin (mTOR) is involved in the neuronal differentiation of neural progenitors induced by insulin. Mol. Cell. Neurosci. 39, 118–124. doi: 10.1016/j.mcn.2008.06.003
Kaminskyy, V. O., Piskunova, T., Zborovskaya, I. B., Tchevkina, E. M., and Zhivotovsky, B. (2012). Suppression of basal autophagy reduces lung cancer cell proliferation and enhances caspase-dependent and -independent apoptosis by stimulating ROS formation. Autophagy 8, 1032–1044. doi: 10.4161/auto.20123
Liang, H., Xiong, Z., Li, R., Hu, K., Cao, M., Yang, J., et al. (2019). BDH2 is downregulated in hepatocellular carcinoma and acts as a tumor suppressor regulating cell apoptosis and autophagy. J. Cancer 10, 3735–3745. doi: 10.7150/jca.32022
Liu, J. Z., Hu, Y. L., Feng, Y., Jiang, Y., Guo, Y. B., Liu, Y. F., et al. (2020). BDH2 triggers ROS-induced cell death and autophagy by promoting Nrf2 ubiquitination in gastric cancer. J. Exp. Clin. Cancer Res. 39:123. doi: 10.1186/s13046-020-01620-z
Liu, Z., Ciocea, A., and Devireddy, L. (2014a). Endogenous siderophore 2,5-dihydroxybenzoic acid deficiency promotes anemia and splenic iron overload in mice. Mol. Cell. Biol. 34, 2533–2546. doi: 10.1128/MCB.00231-14
Liu, Z., Reba, S., Chen, W. D., Porwal, S. K., Boom, W. H., Petersen, R. B., et al. (2014b). Regulation of mammalian siderophore 2,5-DHBA in the innate immune response to infection. J. Exp. Med. 211, 1197–1213. doi: 10.1084/jem.20132629
Liu, Z., Velpula, K. K., and Devireddy, L. (2014c). 3-Hydroxybutyrate dehydrogenase-2 and ferritin-H synergistically regulate intracellular iron. FEBS J. 281, 2410–2421. doi: 10.1111/febs.12794
McLaughlin, K., Flyamer, I. M., Thomson, J. P., Mjoseng, H. K., Shukla, R., Williamson, I., et al. (2019). DNA methylation directs polycomb-dependent 3D genome re-organization in naive pluripotency. Cell Rep. 29, 1974.e1976–1985.e1976. doi: 10.1016/j.celrep.2019.10.031
Nichols, J., and Smith, A. (2009). Naive and primed pluripotent states. Cell Stem Cell 4, 487–492. doi: 10.1016/j.stem.2009.05.015
Novo, C. L., Tang, C., Ahmed, K., Djuric, U., Fussner, E., Mullin, N. P., et al. (2016). The pluripotency factor Nanog regulates pericentromeric heterochromatin organization in mouse embryonic stem cells. Genes Dev. 30, 1101–1115. doi: 10.1101/gad.275685.115
Ortmann, D., Brown, S., Czechanski, A., Aydin, S., Muraro, D., Huang, Y., et al. (2020). Naive pluripotent stem cells exhibit phenotypic variability that is driven by genetic variation. Cell Stem Cell 27, 470.e476–481.e476. doi: 10.1016/j.stem.2020.07.019
Petell, C. J., Alabdi, L., He, M., San Miguel, P., Rose, R., and Gowher, H. (2016). An epigenetic switch regulates de novo DNA methylation at a subset of pluripotency gene enhancers during embryonic stem cell differentiation. Nucleic Acids Res. 44, 7605–7617. doi: 10.1093/nar/gkw426
Posfai, E., Tam, O. H., and Rossant, J. (2014). Mechanisms of pluripotency in vivo and in vitro. Curr. Top. Dev. Biol. 107:1e37. doi: 10.1016/B978-0-12-416022-4.00001-9
Sato, T., Machida, T., Takahashi, S., Iyama, S., Sato, Y., Kuribayashi, K., et al. (2004). Fas-mediated apoptosome formation is dependent on reactive oxygen species derived from mitochondrial permeability transition in jurkat cells. J. Immunol. 173, 285–296.
Scheibner, K., Bakhti, M., Bastidas-Ponce, A., and Lickert, H. (2019). Wnt signaling: implications in endoderm development and pancreas organogenesis. Curr. Opin. Cell Biol. 61, 48–55. doi: 10.1016/j.ceb.2019.07.002
Smith, A. (2017). Formative pluripotency: the executive phase in a developmental continuum. Development 144, 365–373. doi: 10.1242/dev.142679
Soufi, A., and Dalton, S. (2016). Cycling through developmental decisions: how cell cycle dynamics control pluripotency, differentiation and reprogramming. Development 143, 4301–4311. doi: 10.1242/dev.142075
Stanton, J.-A. L., Filimonow, K., Saiz, N., Suwińska, A., Wyszomirski, T., Grabarek, J. B., et al. (2019). No evidence of involvement of E-cadherin in cell fate specification or the segregation of Epi and PrE in mouse blastocysts. PLoS One 14:e0212109. doi: 10.1371/journal.pone.0212109
Tee, W. W., and Reinberg, D. (2014). Chromatin features and the epigenetic regulation of pluripotency states in ESCs. Development 141, 2376–2390. doi: 10.1242/dev.096982
van Mierlo, G., Dirks, R. A. M., De Clerck, L., Brinkman, A. B., Huth, M., Kloet, S. L., et al. (2019). Integrative proteomic profiling reveals PRC2-dependent epigenetic crosstalk maintains ground-state pluripotency. Cell Stem Cell 24, 123.e128–137.e128. doi: 10.1016/j.stem.2018.10.017
White, J., and Dalton, S. (2005). Cell cycle control of embryonic stem cells. Stem Cell Rev. 1, 131–138.
Wu, B., Li, L., Li, B., Gao, J., Chen, Y., Wei, M., et al. (2020). Activin A and BMP4 signaling expands potency of mouse embryonic stem cells in serum-free media. Stem Cell Rep. 14, 241–255. doi: 10.1016/j.stemcr.2020.01.004
Wullschleger, S., Loewith, R., and Hall, M. N. (2006). TOR signaling in growth and metabolism. Cell 124, 471–484. doi: 10.1016/j.cell.2006.01.016
Yang, W. C., Tsai, W. C., Lin, P. M., Yang, M. Y., Liu, Y. C., Chang, C. S., et al. (2013). Human BDH2, an anti-apoptosis factor, is a novel poor prognostic factor for de novo cytogenetically normal acute myeloid leukemia. J. Biomed. Sci. 20, 58–73.
Ying, Q. L., Wray, J., Nichols, J., Batlle-Morera, L., Doble, B., Woodgett, J., et al. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519–523. doi: 10.1038/nature06968
Young, R. A. (2011). Control of the embryonic stem cell state. Cell 144, 940–954. doi: 10.1016/j.cell.2011.01.032
Zhang, X., He, X., Li, Q., Kong, X., Ou, Z., Zhang, L., et al. (2017). PI3K/AKT/mTOR signaling mediates valproic acid-induced neuronal differentiation of neural stem cells through epigenetic modifications. Stem Cell Rep. 8, 1256–1269. doi: 10.1016/j.stemcr.2017.04.006
Zhao, M., Li, M. Y., Gao, X. F., Jia, S. J., Gao, K. Q., Zhou, Y., et al. (2018). Downregulation of BDH2 modulates iron homeostasis and promotes DNA demethylation in CD4(+) T cells of systemic lupus erythematosus. Clin. Immunol. 187, 113–121. doi: 10.1016/j.clim.2017.11.002
Zhao, Q., Liu, Y., Zhong, J., Bi, Y., Liu, Y., Ren, Z., et al. (2019). Pristimerin induces apoptosis and autophagy via activation of ROS/ASK1/JNK pathway in human breast cancer in vitro and in vivo. Cell Death Discov. 5:125. doi: 10.1038/s41420-019-0208-0
Zughaier, S. M., Kandler, J. L., and Shafer, W. M. (2014a). Neisseria gonorrhoeae modulates iron-limiting innate immune defenses in macrophages. PLoS One 9:e87688. doi: 10.1371/journal.pone.0087688
Keywords: BDH2, embryonic stem cells, CRISPR/Cas9, endoderm differentiation, DNA methylation
Citation: Fu Y, Liu F, Cao S, Zhang J, Wang H, Wu B, Song Y, Duo S, Li X and Bao S (2021) Bdh2 Deficiency Promotes Endoderm-Biased Early Differentiation of Mouse Embryonic Stem Cells. Front. Cell Dev. Biol. 9:655145. doi: 10.3389/fcell.2021.655145
Received: 18 January 2021; Accepted: 18 March 2021;
Published: 08 April 2021.
Edited by:
Wei Jiang, Wuhan University, ChinaReviewed by:
Chunliang Li, St. Jude Children’s Research Hospital, United StatesCopyright © 2021 Fu, Liu, Cao, Zhang, Wang, Wu, Song, Duo, Li and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siqin Bao, YmFvc3FAaW11LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.