
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 01 April 2021
Sec. Cell Growth and Division
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.653801
This article is part of the Research Topic Cytoskeletal Dynamics and Mechanics in Cell Growth, Division, Differentiation and Aging View all 12 articles
 Tomomi Kiyomitsu*
Tomomi Kiyomitsu* Susan Boerner
Susan BoernerThe nuclear mitotic apparatus (NuMA) protein is well conserved in vertebrates, and dynamically changes its subcellular localization from the interphase nucleus to the mitotic/meiotic spindle poles and the mitotic cell cortex. At these locations, NuMA acts as a key structural hub in nuclear formation, spindle assembly, and mitotic spindle positioning, respectively. To achieve its variable functions, NuMA interacts with multiple factors, including DNA, microtubules, the plasma membrane, importins, and cytoplasmic dynein. The binding of NuMA to dynein via its N-terminal domain drives spindle pole focusing and spindle positioning, while multiple interactions through its C-terminal region define its subcellular localizations and functions. In addition, NuMA can self-assemble into high-ordered structures which likely contribute to spindle positioning and nuclear formation. In this review, we summarize recent advances in NuMA’s domains, functions and regulations, with a focus on human NuMA, to understand how and why vertebrate NuMA participates in these functions in comparison with invertebrate NuMA-related proteins.
The nuclear mitotic apparatus (NuMA) protein was initially identified as a non-histone chromatin protein in human cell lines (Lydersen et al., 1980) and named after its localization pattern to both the interphase nucleus and mitotic spindle poles (Lydersen and Pettijohn, 1980; Figure 1A). Since NuMA’s dynamic translocation from the nucleus to the spindle poles was different from previously characterized nuclear components, NuMA was regarded as a novel class of nuclear protein, involved in both mitosis and nuclear reformation (Compton and Cleveland, 1994). Over the last 40 years, NuMA has been extensively studied in mammalian cultured cells, Xenopus egg extracts, and other vertebrate models (Cleveland, 1995; Sun and Schatten, 2006; Radulescu and Cleveland, 2010). One of the key findings early on was that NuMA interacts with cytoplasmic dynein to tether microtubules to spindle poles (Merdes et al., 1996). Later studies confirmed and expanded upon this result, positioning NuMA as a mitotic dynein adaptor, as described below.
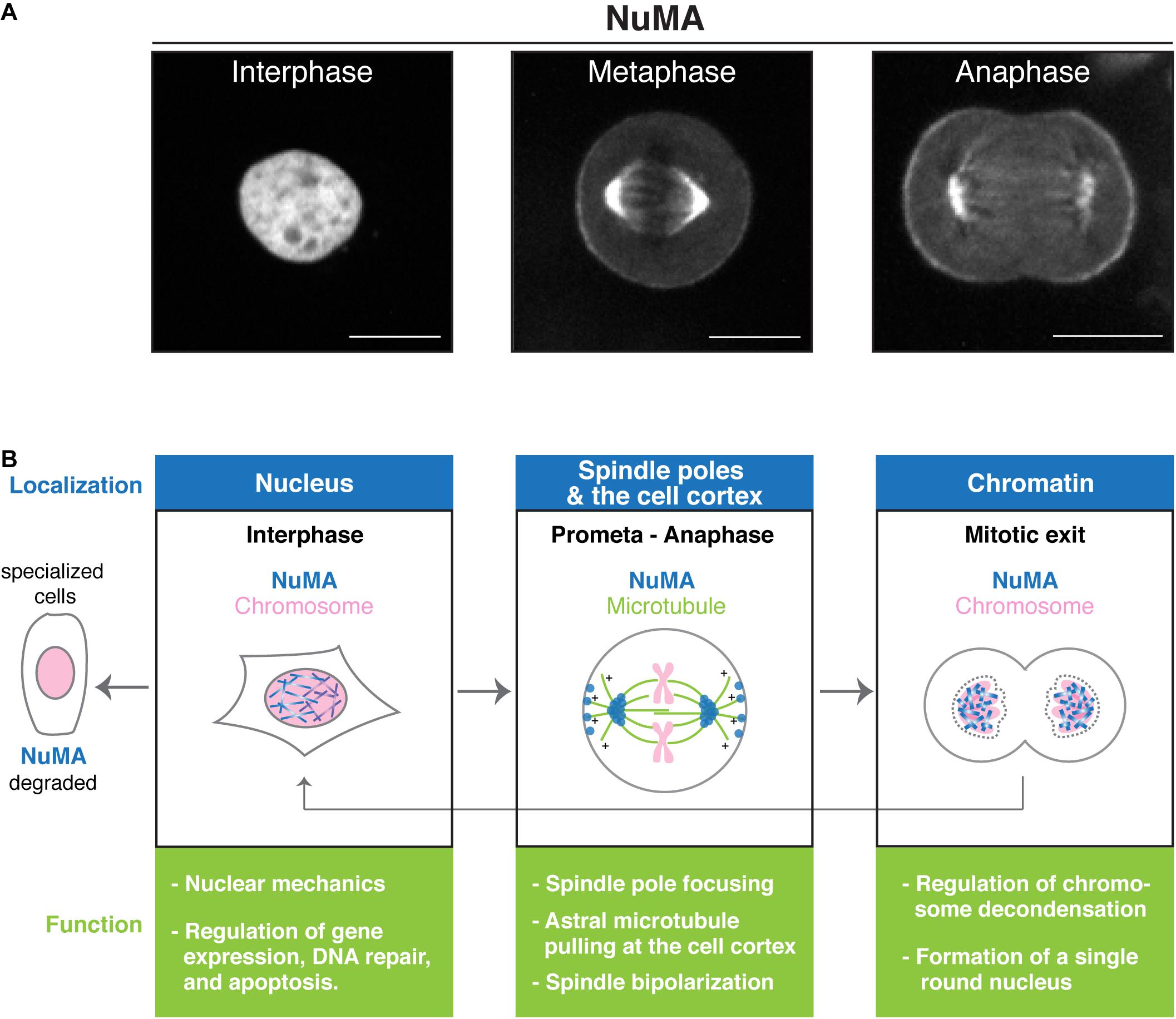
Figure 1. Nuclear mitotic apparatus (NuMA’s) functions during the cell cycle. (A) Images of endogenous NuMA fused with mClover in human HCT116 cells (Okumura et al., 2018). NuMA accumulates in the nucleus in interphase, but mainly localizes at the spindle poles in metaphase. NuMA is also targeted to the cell cortex near the spindle poles during metaphase, and the level increases during anaphase (Collins et al., 2012; Kiyomitsu and Cheeseman, 2013; Kotak et al., 2013; Seldin et al., 2013; Zheng et al., 2014). Scale bars = 10 μm. (B) Schematic showing how NuMA (blue) dynamically changes its subcellular localization during the cell cycle. Chromosomes and microtubules are shown in pink and green, respectively. Key functions of NuMA at these locations are summarized.
Another important finding was that NuMA interacts with leucine/glycine/asparagine-repeat-containing protein (LGN) to form the evolutionarily conserved NuMA/LGN/Gαi complex at the mitotic cell cortex (Du and Macara, 2004). This study led to the discovery that NuMA plays a conserved role at the mitotic cell cortex for spindle positioning like C. elegans LIN-5 (Lorson et al., 2000; Srinivasan et al., 2003) and Drosophila Mud [Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006; The first publication is Lorson et al. (2000)]. Building on this, and other pioneering work on asymmetric cell division in C. elegans and Drosophila (Galli and van den Heuvel, 2008; Gonczy, 2008; Siller and Doe, 2009), further studies have established the conceptual framework that cortical cues converge on a conserved ternary complex, NuMA/LGN/Gαi in vertebrates, Lin-5/GPR-1/2/Gα in C. elegans, and Mud/Pins/Gα in Drosophila, that recruits and activates dynein to position the spindle in asymmetric division (Lechler and Fuchs, 2005; Poulson and Lechler, 2010; Morin and Bellaiche, 2011; Williams et al., 2011). In symmetrically-dividing vertebrate cells, the NuMA/LGN/Gαi complex is also involved in recruiting dynein and controlling spindle position and orientation (Figure 1A; Woodard et al., 2010; Peyre et al., 2011; Collins et al., 2012; Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012; Kiyomitsu, 2019).
Interestingly, the cortical function seems to be most conserved in NuMA-related proteins. For example, LIN-5 and Mud localize at both spindle poles and the cell cortex, but only serve an essential function at the cell cortex for spindle positioning and are non-essential for bipolar spindle assembly (Lorson et al., 2000; Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006). In addition, yeast Num1 was recently proposed as a functional homolog of NuMA based on its functional similarities at the cell cortex (Greenberg et al., 2018). Intriguingly, plants lack a homolog of NuMA, as well as cytoplasmic dynein (Yamada and Goshima, 2017), suggesting that plants have developed alternative mechanisms to control nuclear formation, spindle assembly and positioning.
In the last 10 years, many functional domains of human NuMA were identified, providing useful information to understand NuMA’s functions and regulations at the molecular level. In this review, we focus on vertebrate NuMA because several features, such as nuclear localization, appear to be specific to this group. We begin with an overview of the localization and the structural domains of the human NuMA protein, and then discuss how vertebrate NuMA participates in spindle assembly, spindle positioning and nuclear formation.
In cultured human cells, NuMA accumulates in the nucleus in interphase and at the spindle poles and cell cortex during mitosis (Figure 1A). NuMA’s spindle-pole localization is most likely conserved in all cell types, including meiotic cells (Taimen et al., 2004; Alvarez Sedo et al., 2011; Kolano et al., 2012), but its nuclear and cortical localization may vary in different developmental contexts: nuclear NuMA is degraded in some specialized cells, such as smooth and skeletal muscle fibers (Merdes and Cleveland, 1998; Figure 1B left). Furthermore, cortical NuMA targeting is differentially regulated between symmetric and asymmetric division in mouse epidermal cells (Poulson and Lechler, 2010).
Human NuMA is a large (∼238 kDa) protein that consists of N-terminal and C-terminal globular domains and a central long coiled-coil domain (Compton et al., 1992; Yang et al., 1992; Figure 2A). Full length NuMA expressed in E. coli forms a homo-dimer using the coiled-coil region (Harborth et al., 1995; Forth et al., 2014). Purified NuMA shows a long rod-shaped structure that has globular ends and a central long (∼210 nm) α-helical domain that appears more or less flexible (Harborth et al., 1995, 1999; Figure 2A). The globular domains interact with many factors, whereas the central region has structural and likely intramolecular regulatory roles as described below.
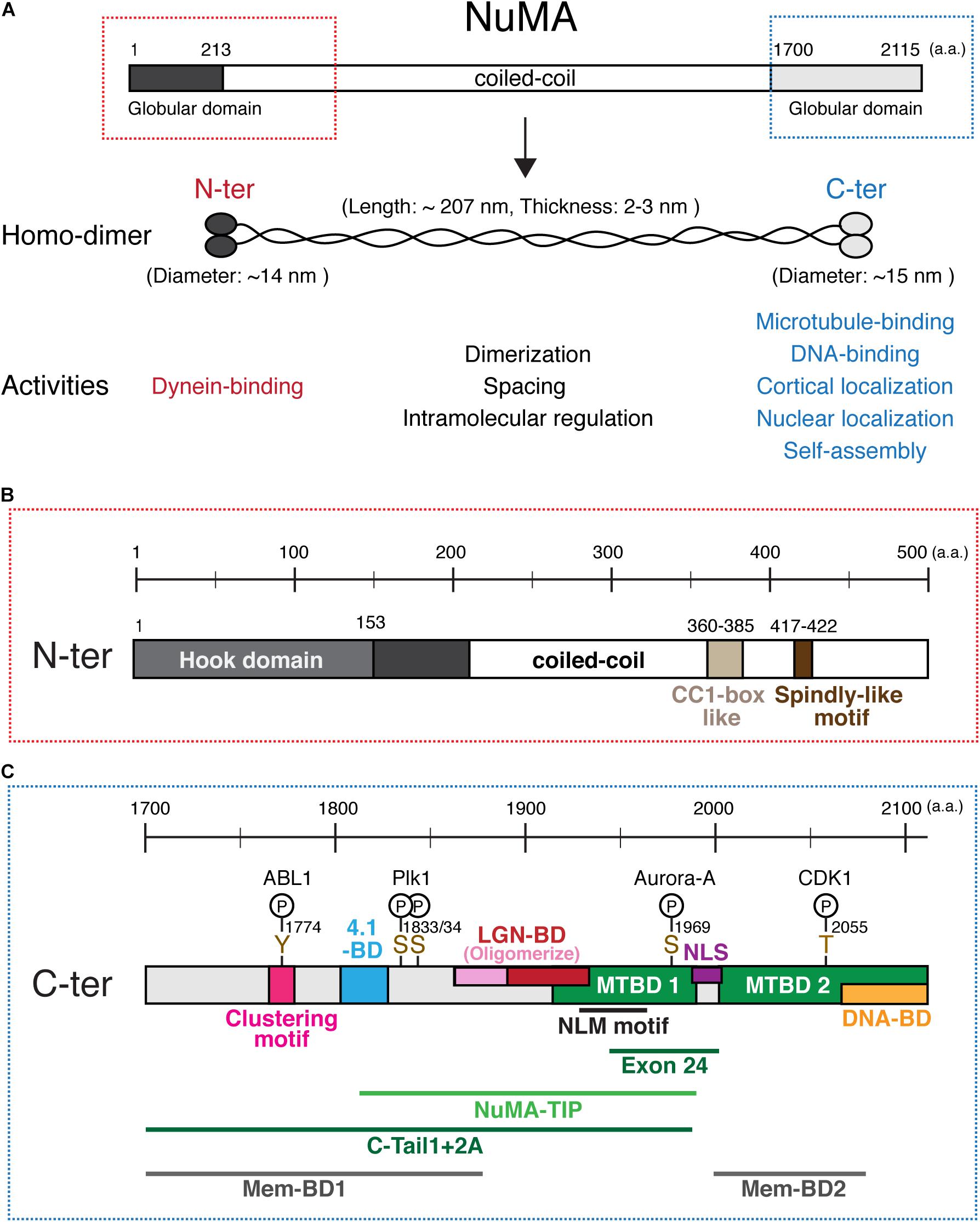
Figure 2. NuMA’s domain structure. (A) Diagrams of full length NuMA. Human NuMA isoform 1 (NP_006176) consists of 2,115 amino acids (a.a.) (Yang et al., 1992), whereas NuMA isoform 2 (NP_001273490) is lacking 14 a.a. at the 1,539–1,552 region (Compton et al., 1992). In this review, we refer to isoform 1. NuMA forms a homodimer through its central coiled-coil region. The central rod-shaped structure has an average length of ∼207 nm and a thickness of ∼2–3 nm, while N-terminal and C-terminal globular structures show a diameter of ∼14 and ∼15 nm, respectively (Harborth et al., 1995). (B) Domains in the N-terminal region. (C) Domains, motifs, phosphorylation sites, and functional regions in the C-terminal region of NuMA. See the text and Table 1 for details. NuMA1944–2003, corresponding to either exon 22 for mice or exon 24 for human, was deleted in Silk et al. (2009); Kolano et al. (2012), and Tsuchiya et al. (2021), instead of depleting the complete MTBD1.
The N-terminal region is required to interact with cytoplasmic dynein and dynactin complexes during mitosis (Kotak et al., 2012). A recent structural study revealed that NuMA1–153 contains a Hook domain that directly interacts with dynein light intermediate chain (LIC) 1 and 2 (Renna et al., 2020; Figure 2B; all domains are listed in Table 1). The authors also identified a CC1-box like motif, NuMA360–385, adjacent to the Hook domain which forms part of the binding interface between NuMA and LIC (Renna et al., 2020; Figure 2B). In addition, NuMA417–422 contains a Spindly-like motif which is well conserved in vertebrates (Okumura et al., 2018; Tsuchiya et al., 2021), and may interact with the dynactin point-end complex (Gama et al., 2017; Lee et al., 2020). As NuMA1–505, but not NuMA1–413 and NuMA214–705, is sufficient for dynein recruitment to the cell cortex (Okumura et al., 2018), multiple interaction sites in the N-terminal region appear to be important to stably interact with the dynein-dynactin complex during mitosis. These studies indicate that NuMA acts as a dynein activating adaptor (Lee et al., 2020), but in contrast to other established dynein adaptors such as BICD2 (McKenney et al., 2014; Schlager et al., 2014), its ability to form a ternary complex with purified dynein and dynactin and activate dynein motility in vitro has not been shown; post translational modifications may be required to form dynein–dynactin–NuMA complexes during mitosis.
On the other hand, the interphase role of the N-terminal region of NuMA is not clear. Although both N-terminal and C-terminal globular domains contain several S/TPXX motifs which are supposed to contribute to DNA binding (Suzuki, 1989), the N-terminal NuMA fragments do not show clear DNA or chromatin binding compared to NuMA’s C-terminal (Serra-Marques et al., 2020). However, the N-terminal domain may contribute to the lattice formation of NuMA oligomers in the nucleus (Gueth-Hallonet et al., 1998; Harborth et al., 1999), as described below.
Electron micrographs of recombinant human NuMA indicate that the central region forms a long flexible rod-shaped structure (Harborth et al., 1995; Figure 2A). One of most important functions of the central region is homo-dimerization. In vitro studies suggest that either the N-terminal (199–432 or 1–400 a.a) or C-terminal part (670–1700 a.a.) of the central coiled-coil is sufficient to form a homo-dimer (Harborth et al., 1995; Forth et al., 2014). Dimerization is most likely critical for most NuMA functions, including dynein binding via its N-terminal Hook domain (Renna et al., 2020) and microtubule cross-linking activities via the C-terminal domain (Forth et al., 2014).
The predicted long coiled-coil region of vertebrate NuMA proteins, which exceeds 1,000 a.a. in length, is likely to perform other important functions. Transient overexpression of wild type NuMA in HeLa cells induced a regular nuclear lattice structure which has quasi-hexagonal organization (Gueth-Hallonet et al., 1998). Interestingly, an addition or deletion in the coiled-coil domain changed the spacing of the hexagons, suggesting that the central coiled-coil defines the length of the nuclear lattice structure (Gueth-Hallonet et al., 1998; Harborth et al., 1999).
Recently, Serra-Marques et al. (2020) found that the long coiled-coil is also required for the formation of a single, round nucleus, and that this role is independent from NuMA’s role in spindle formation. In addition, the authors revealed that a small portion of the coiled-coil213–705 is sufficient to prevent NuMA’s C-terminal region from binding chromosomes during late metaphase and anaphase (Serra-Marques et al., 2020).
At the mitotic cell cortex, the coiled-coil region of NuMA706–1699 is required to generate proper spindle pulling forces when NuMA constructs are targeted to the membrane during mitosis (Okumura et al., 2018). However, the coiled-coil region is dispensable for bipolar spindle formation (Hueschen et al., 2017). At the cell cortex, the long coiled-coil may be used to separate dynein from the actin-rich cell cortex and/or to increase the efficiency of astral microtubule capture by cortical NuMA-dynein complexes during mitosis (Kiyomitsu, 2019).
The C-terminal globular region of NuMA1700–2115 contains several important domains that determine its localization and function (Figure 2C). First, this region contains two microtubule binding domains (MTBDs); here we refer to them as MTBD11914–1985 (Du et al., 2002) and MTBD22002–2115 (Gallini et al., 2016; Chang et al., 2017; Figure 2C). Although MTBD1 has weaker microtubule-binding affinity than MTBD2 in vitro (Chang et al., 2017), MTBD1 contains the conserved NuMA–LIN-5–Mud (NLM) motif1922–1957 (Siller et al., 2006) and acts to establish and maintain spindle-pole focusing (Figure 3A) in mouse fibroblasts (Silk et al., 2009), mouse meiosis I spindle (Kolano et al., 2012), and human HCT116 cells (Tsuchiya et al., 2021).
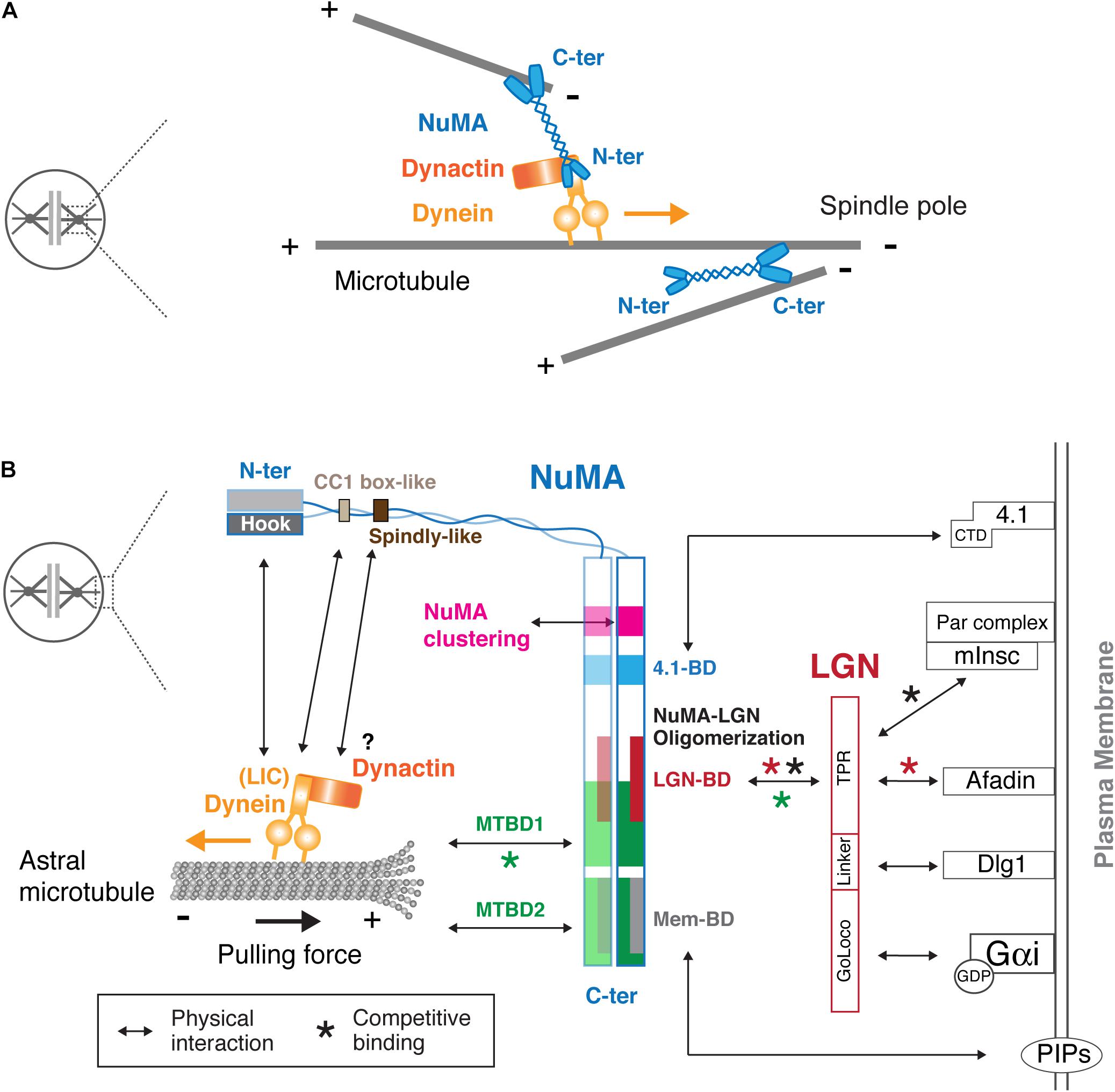
Figure 3. Physical interaction map of NuMA at the spindle pole and the mitotic cell cortex. (A) Models of the spindle pole focusing function of NuMA. Using two microtubule-binding C-terminal globular domains, the NuMA homodimer bundles and crosslinks microtubules around the spindle pole region (Radulescu and Cleveland, 2010; Forth et al., 2014). In addition, NuMA recognizes the minus-ends of spindle microtubules and recruits the dynein-dynactin complex, which transports NuMA-bound spindle microtubules toward poles, resulting in microtubule focusing at the poles (Hueschen et al., 2017). See text for details. (B) NuMA interacts with the cortical proteins and plasma membrane through its C-terminal region, whereas it binds to dynein and dynactin via its N-terminal region. Arrows indicate physical interactions. As indicated by red asterisks, NuMA competes with Afadin for LGN binding. In addition, NuMA competes with mInsc for LGN binding during asymmetric cell division (black asterisks). The MTBD1 overlaps LGN–BD, and thus LGN binding to NuMA inhibits the microtubule binding activity of MTBD1 (green asterisks). LGN consists of TPR, Linker and GoLoco motifs, and each motif interacts with Afadin (Carminati et al., 2016), Dlg1 (Saadaoui et al., 2014), and the GDP-bound form of Gαi (Jia et al., 2012), respectively. NuMA/LGN/Gαi constitutes a conserved core pathway for cortical dynein recruitment. The C-terminal domain (CTD) of band 4.1 proteins interacts with the NuMA C-terminal domain, and is sufficient to rescue cortical NuMA enrichment in anaphase in LGN and 4.1 co-depleted cells (Kiyomitsu and Cheeseman, 2013). See text for details.
In some cells, such as mouse keratinocytes, MTBD1 is not required for spindle pole focusing, but is required for spindle orientation (Seldin et al., 2016). Full length NuMA accumulates at the minus-end of microtubules, and the NuMA1701–1981 fragment, called C-Tail1 + 2A, is sufficient for minus-end recognition (Hueschen et al., 2017). In addition, NuMA’s dimerized C-terminal fragment that crosslinks two parallel microtubules tends to move in the minus-end direction under forces (Forth et al., 2014). However, intriguingly, the NuMA1811–1985 fragment, called NuMA-TIP, appears to preferentially accumulate at the curling microtubule ends, and can remain attached to the depolymerizing microtubule plus-end (Seldin et al., 2016). The authors propose that this unique property of MTBD1 may be important to facilitate the interaction of astral microtubule plus-ends with NuMA at the cell cortex during spindle orientation (Seldin et al., 2016). However, in human cells, MTBD2, but not MTBD1, is required for spindle pulling activity when NuMA constructs are targeted to the cell cortex (Okumura et al., 2018). Interestingly, MTBD2 can bind both the microtubule lattice and tubulin dimers (Pirovano et al., 2019), indicating that MTBD2 may act not only for astral microtubule binding, but also for regulating plus-end dynamics of astral microtubules during the cortical pulling-force generation. How and when the MTBDs come into play during spindle pole focusing and orientation likely depends on the cellular context (Borgal and Wakefield, 2018).
NuMA’s C-terminal domain also defines its cortical localization during mitosis by binding to LGN, band 4.1 proteins and the plasma membrane. Whereas LGN is targeted to the cell cortex by binding to GDP-bound Gαi through its C-terminal GoLoco motif, LGN binds and links NuMA to the cell cortex using its N-terminal TPR motif (Figure 3B). LGN and Gαi are indispensable for cortical localization of NuMA in metaphase (Du and Macara, 2004; Woodard et al., 2010; Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012). Zhu et al. (2011b) identified the NuMA1900–1926 peptide as the minimal region required to bind to the inner groove of LGNTPR. This LGN-binding domain partially overlaps with MTBD1 (Figure 2C), and LGN binding thus inhibits the microtubule binding activity of NuMA in vitro (Du et al., 2002; Figure 3, green asterisk). Recently, Pirovano et al. (2019) revealed that a longer NuMA1861–1928 region forms a hetero-hexamer with LGN7–367, in which an extended NuMA1861–1880 region hooks onto an adjacent LGN to form oligomers. Consistently, expression of a longer NuMA1788–1925 fragment is targeted to the LGN-localizing metaphase cell cortex, whereas a shorter one, NuMA1892–1925, is not (Seldin et al., 2013).
However, given the complicated cortical protein network, additional mechanisms might contribute to cortical NuMA–LGN targeting and stability. In fact, it has been shown that cortical NuMA–LGN is affected by disrupting cortical actin networks (Kaji et al., 2008; Carminati et al., 2016) or their regulators (Machicoane et al., 2014; Kschonsak and Hoffmann, 2018). Recently, Carminati et al. (2016) demonstrated that F-actin binding protein Afadin, which also binds LGN competitively with NuMA, is required to facilitate NuMA–LGN complex formation at the metaphase cell cortex (Figure 3B). On the other hand, Dlg1, that directly interacts with the phosphorylated LGN linker region (Zhu et al., 2011a) is also required for cortical LGN and NuMA localization (Saadaoui et al., 2014).
In asymmetrically dividing epithelial or mammary stem cells, the situation is more complex. Par3-binding mInsc (mammalian homolog of Inscuteable) and NuMA compete for binding to LGNTPR, with mInsc showing a more than fivefold higher affinity (Culurgioni et al., 2011; Yuzawa et al., 2011; Zhu et al., 2011b). Previously, mInsc-bound LGN-Gαi was supposed to be transferred to NuMA, but Cukurgioni et al. recently reported that the Inscuteable-LGN tetramer is so stable that LGN cannot be dissociated from Inscuteable by NuMA (Culurgioni et al., 2018). The authors proposed that the Inscuteable-LGN tetramer generates a localized pool of Gαi-GTP molecules, which upon GTP-hydrolysis recruits a distinct population of LGN that subsequently recruits NuMA and dynein to orient the spindle (Culurgioni et al., 2018).
Although the LGN pathways are critical for cortical NuMA localization during metaphase in both symmetric and asymmetric divisions (Morin and Bellaiche, 2011), recent studies revealed that NuMA can be targeted to the anaphase cell cortex independently of LGN in symmetrically dividing mammalian cells (Collins et al., 2012; Kiyomitsu and Cheeseman, 2013; Kotak et al., 2013; Seldin et al., 2013; Zheng et al., 2014). Band 4.1 proteins link the plasma membrane to the actin cytoskeleton (Baines et al., 2014), and interact with NuMA via their C-terminal domain (CTD) and NuMA’s 4.1-binding domain (1802–1824) (Mattagajasingh et al., 2009; Figure 3B). This NuMA-4.1 interaction appears to be important for the cortical stability of NuMA in metaphase keratinocytes (Seldin et al., 2013), but also has an important role for anaphase NuMA localization: double depletion of LGN and 4.1 proteins eliminates cortical NuMA in anaphase human cells (Kiyomitsu and Cheeseman, 2013). Importantly, this phenotype was rescued by the expression of the membrane-targeted C-terminal domain (CTD) of band 4.1 (Kiyomitsu and Cheeseman, 2013), which does not bind to actin, suggesting that 4.1 proteins contribute to anaphase NuMA localization independently of LGN and cortical actin structures. How 4.1 proteins regulate NuMA remains unclear; one possibility is that they increase cortical NuMA retention by linking it with the cell cortex and/or transferring NuMA to the plasma membrane (as described below). In addition to these cortical proteins, other proteins may be involved in cortical NuMA recruitment in developmental contexts. For example, NuMA’s C-terminal region interacts with Disheveled, which controls spindle orientation during Zebrafish gastrulation (Segalen et al., 2010).
Importantly, NuMA can directly interact with the plasma membrane. The C-terminal region contains two membrane-binding domains, NuMA1699–1876 (Kotak et al., 2014) and NuMA1996–2074 (Zheng et al., 2014), which overlap with 4.1-BD and MTBD2, respectively, (Figure 2C) and are referred to as Mem-BD1 and 2 in this review (Figure 2C). Both membrane-binding domains preferentially bind to phosphorylated forms of phosphatidylinositol (PIPs) and are required for efficient cortical accumulation of NuMA (Kotak et al., 2014; Zheng et al., 2014). Mem-BD2, which partially overlaps with the DNA-binding domain (Figure 2C), is required for proper chromosome separation during anaphase (Zheng et al., 2014).
In contrast to invertebrate NuMA-related proteins, vertebrate NuMA proteins analyzed so far localize in the nucleus (Lydersen and Pettijohn, 1980; Compton et al., 1992; Yang et al., 1992; Merdes et al., 1996). Previously, human NuMA was reported to interact with defined DNA sequences called matrix attached regions (MARs) in vitro (Luderus et al., 1994). Recently, two studies demonstrated that the C-terminal region of human NuMA interacts with DNA in vitro and chromatin in cells (Rajeevan et al., 2020; Serra-Marques et al., 2020). Rajeevan et al. (2020) showed that the C-terminus NuMA2058–2115 fragment is sufficient to bind DNA in vitro, and the basic amino acids within the region are critical for its interaction with chromatin in cells (Figure 2C). When endogenous NuMA was replaced with mutated versions lacking its DNA-binding ability, cells showed improper chromosome decondensation during mitotic exit and an abnormal nuclear shape (Rajeevan et al., 2020), suggesting that NuMA–DNA interactions are critical for proper regulation of chromosome decondensation during nuclear reformation.
NuMA has a nuclear localization signal/sequence (NLS) between the MTBDs in its C-terminal region (Tang et al., 1994; Figure 2C). When Chang et al. (2017) solved the crystal structure of the importin-α-NuMA-C-terminus complex, they found that NuMA–NLS exhibits a novel, non-classic interaction mode with importin-α, and that importin-β sterically inhibits NuMA’s MTBD2 in vitro. The NLS sequence is well conserved from H. sapiens to X. laevis (Chang et al., 2017), but not in C. elegans LIN-5, or drosphila Mud (Lorson et al., 2000; Siller et al., 2006). In fish, the highly conserved KR, H and KK residues of the NLS, which interact with the minor-, linker and the major-NLS-binding site on importin-α, respectively (Chang et al., 2017), are not identical and several amino acids are inserted between the H and KK residues (Tsuchiya et al., 2021); yet the Zebrafish NuMA C-terminal region can be targeted to the nucleus (Segalen et al., 2010). It would be interesting to understand whether NuMA localizes to the nucleus in other fish species, and why the NLS was acquired in vertebrates.
The C-terminal globular domain has another key feature that facilitates NuMA’s self-assembly into oligomers (Harborth et al., 1999). In vitro, 10–12 NuMA homo-dimers assemble through its C-terminal region to form multi-arm oligomers. Each oligomer has a central clustered core with projected arms, and may be connected to create 3D nuclear architecture during interphase (Harborth et al., 1999). NuMA’s punctate signals at the cell cortex are most likely a result of its oligomerization/clustering. This clustering activity is attributed to a well-conserved 10 a.a sequence of NuMA1768–1777 (Okumura et al., 2018). Mutant analyses indicated that NuMA’s clustering is required for spindle pulling and spindle orientation at metaphase (Okumura et al., 2018), and for spindle bipolarization during prometaphase in acentrosomal human cells (Chinen et al., 2020), but is dispensable for spindle pole focusing (Okumura et al., 2018). It will be exciting to see what kind of structures are actually generated in cells by NuMA’s clustering activity both in mitosis and interphase. Especially, it is important to understand how this clustering activity synergetically functions with NuMA–LGN oligomer formation to organize high-ordered functional structures that capture and pull on astral microtubules at the mitotic cell cortex (Pirovano et al., 2019; Figure 3).
Since NuMA’s C-terminal domain binds to microtubules, the plasma membrane, and chromatin (Figure 2C), these interactions must be regulated throughout the cell cycle. In fact, the well conserved threonine at 2055 (T2055) is phosphorylated by CDK during metaphase (Compton and Luo, 1995; Kotak et al., 2013; Seldin et al., 2013), which inhibits NuMA’s cortical association and thus promotes microtubule-binding. CDK-based phosphorylation of NuMA is also critical for releasing NuMA from the chromosomes at mitotic entry (Rajeevan et al., 2020). During anaphase, NuMA is dephosphorylated by the PPP2CA–B55gamma–PPP2R1B complex (Keshri et al., 2020), which dissociates NuMA from the spindle poles (Gehmlich et al., 2004) and promotes its cortical association.
The microtubule binding activity of NuMA is vital for its mitotic and meiotic functions (Silk et al., 2009; Kolano et al., 2012; Tsuchiya et al., 2021). Thus, importins that sterically inhibit C-terminal microtubule-binding (Chang et al., 2017) must be released during mitosis. Previously, it was demonstrated that the NuMA-importin interaction is disrupted by Ran-GTP binding to importin-β in Xenopus egg extracts (Nachury et al., 2001; Wiese et al., 2001), and that Ran-GTP is essential for acentrosomal spindle assembly in female meiosis (Dumont et al., 2007; Holubcova et al., 2015; Drutovic et al., 2020). However, recent evidence suggests that Ran-GTP is not essential for mitotic spindle assembly and mitotic progression in chicken DT40 (Furuta et al., 2016) and human HCT116 cells (Tsuchiya et al., 2021). In addition, Ran-GTP is not required to activate NuMA and TPX2 in HCT116 cells (Tsuchiya et al., 2021), suggesting that additional parallel pathways exist to activate these proteins at a distance from chromosomes (Wei et al., 2015; Eibes et al., 2018; Brownlee and Heald, 2019). As cellular concentrations of Ran-GTP are variable across cell types and organisms (Kalab et al., 2006; Hasegawa et al., 2013), and the size of spindles and cells change dramatically during early embryonic divisions (Courtois et al., 2012; Levy and Heald, 2012), it would be important to determine how NuMA is spatiotemporally activated by Ran and other factors during mitosis and meiosis.
Once activated, the NuMA homo-dimer binds to microtubules independently of dynein (Heald et al., 1997; Hueschen et al., 2017) and cross-links two microtubules using its two C-terminal globular domains (Figure 3A; Forth et al., 2014). This microtubule crosslinking function may be facilitated at the spindle poles by binding to other microtubule associated proteins such as Rae1 (Wong et al., 2006), Eg5 (Iwakiri et al., 2013), and dynein/dynactin complexes (Merdes et al., 1996, 2000). On the other hand, when new microtubules are created in the spindle, NuMA is targeted to their minus-ends and subsequently forms a complex with dynein and dynactin; when this complex binds adjacent microtubules and moves toward their minus end, it pulls the NuMA-bound microtubules along, resulting in a focused spindle pole (Hueschen et al., 2017; Figure 3A).
At the poles, NuMA is phosphorylated by Aurora-A kinase at S1969, which leads to its dynamic mobility from the spindle poles to the cell cortex during metaphase (Gallini et al., 2016; Kotak et al., 2016; Figure 2C). NuMA is also phosphorylated by Polo-like kinase 1 (Plk1) at SS1833/34 (Kettenbach et al., 2011), which promotes NuMA’s turnover rate at both spindle poles and the cell cortex, and when inhibited results in NuMA’s accumulation at both locations (Sana et al., 2018). Plk1 localizes at the spindle poles, but also accumulates at kinetochores of misaligned chromosomes which locally diminish cortical LGN when they are located near the cell cortex (Tame et al., 2016). Since NuMA and LGN are inter-dependent (Du and Macara, 2004), the kinetochore-localized Plk1 may also target NuMA, which in turn reduces LGN. However, artificial membrane tethering of Plk1 dissociated dynein, but not LGN, from the cell cortex (Kiyomitsu and Cheeseman, 2012). In addition, when the immunoprecipitated GFP–LGN complexes were incubated with Plk1, Plk1 dissociated dynein and dynactin, but not NuMA, from LGN (Kiyomitsu and Cheeseman, 2012). Therefore, kinetochore- or centrosome-localized Plk1 may down-regulate the NuMA–LGN interaction synergetically in cooperation with other factors derived from chromosomes or centrosomes. Another kinase, ABL1, phosphorylates the well-conserved Y1774 residue of NuMA to control spindle orientation (Matsumura et al., 2012). Y1774 is located in the clustering motif (Okumura et al., 2018), but it remains unknown whether ABL1 regulates NuMA’s clustering activity.
In addition to these kinases, Ran-GTP gradients negatively regulate cortical NuMA–LGN localization near chromosomes in a distance dependent manner (Kiyomitsu and Cheeseman, 2012). Although molecular mechanisms by which Ran-GTP eliminates the NuMA–LGN complex from the cell cortex remain unclear, the cortical patterning created by chromosome-derived Ran-GTP is sufficient to explain why the mitotic spindle orients along its interphase cell axis (Dimitracopoulos et al., 2020). NuMA’s continuous exclusion from the equatorial region of the cell cortex during anaphase is dependent on the signals downstream of the centralspindlin complexes (Kotak et al., 2014).
Several lines of evidence indicate that NuMA acts as a non-essential nucleoskeletal element in interphase (Zeng et al., 1994; Merdes and Cleveland, 1998; Harborth et al., 1999), which is nicely reviewed by Radulescu and Cleveland (2010). However, it is difficult to understand this role separately from the several roles it plays during mitosis since mitotic errors cause abnormal nuclei. Recently, Serra-Marques et al. (2020) nicely demonstrated that NuMA’s contribution to building a single, round nucleus is independent from its mitotic functions by inducing mitotic exit in NuMA KO Rpe1 cells without spindles (Figure 1B). Furthermore, they showed that NuMA keeps the decondensing chromosome mass compact during mitotic exit (Serra-Marques et al., 2020). Although the precise mechanisms are still unclear, evidence suggests that NuMA, like barrier-to-auto integration factor (BAF) at the chromosome ensemble surface (Samwer et al., 2017), may offer structural support throughout the nucleus by cross-linking chromosomes and preventing the nuclear envelop from penetrating into the chromosome mass, which results in a decrease of multinucleation and abnormally shaped nuclei during mitotic exit (Serra-Marques et al., 2020). Alternatively, NuMA may coordinate chromosome compaction and nuclear envelop assembly using its binding abilities to both chromosomes and membrane (Serra-Marques et al., 2020), as well as its binding to importins which can recruit membrane vesicles and nucleoporins (Lu et al., 2012).
In addition to its roles during mitotic exit, NuMA promotes the nucleus’ mechanical robustness (Serra-Marques et al., 2020) and could contribute to several interphase events, including chromatin organization (Abad et al., 2007), gene expression (Harborth et al., 2000; Ohata et al., 2013), DNA repair (Vidi et al., 2014; Moreno et al., 2019), and apoptosis (Gueth-Hallonet et al., 1997; Kivinen et al., 2005; Lin et al., 2007). Since chromatin architecture is highly dynamic during different phases of the cell cycle (Shoaib et al., 2020), acute protein depletion technologies, such as auxin-inducible degron (AID) (Natsume et al., 2016; Yesbolatova et al., 2020; Tsuchiya et al., 2021), would be useful to precisely understand the interphase functions of NuMA in future studies.
Vertebrate NuMA, and its related proteins in invertebrates, have been extensively studied using many techniques and model organisms. Over the past 10 years, much light has been shed on this multi-functional protein, revealing key domains, modifications and binding partners. However, we still do not know how NuMA contributes to spindle pole focusing, spindle positioning or nuclear formation at the molecular and structural level. In the next 10 years, it would be especially important to visualize the functional structures of NuMA and its complexes using biochemical reconstitution, high-resolution imaging and in situ structural analyses. In addition, most reported interactions and functions are not sufficiently validated in a physiological condition. Since many functions discussed in this review appear to be specific in vertebrates, vertebrate developmental models would be useful to obtain a comprehensive understanding of the diverse functions of NuMA. Furthermore, optogenetic manipulation of NuMA, or its related proteins, would serve as a powerful tool to control spindle position and orientation (Fielmich et al., 2018; Okumura et al., 2018), and could reveal physiological roles of division orientation and daughter cell size during development (Jankele et al., 2021).
Many key questions about NuMA remain to be answered. (1) How does NuMA recognize microtubule minus-ends and depolymerizing microtubule plus-ends? (2) How are dynein–dynactin–NuMA complexes formed during mitosis and meiosis and regulated at the spindle poles? (3) How are different cortical NuMA complexes assembled at the mitotic cell cortex and spatiotemporally regulated by intrinsic and extrinsic signals? (4) When does NuMA start to localize at the mitotic cell cortex to control spindle positioning during early vertebrate development? (5) What kinds of high-ordered structures are created by NuMA at the cell cortex and interphase nucleus to generate cortical spindle pulling forces and a mechanically robust nucleus, respectively? (6) How is NuMA dissociated from importins in a Ran-independent manner? (7) Why did NuMA acquire an NLS in vertebrates? (8) How has the NuMA gene evolved to achieve different functions in different organisms? Addressing these questions must provide new, exciting insights not only to advance our knowledge about nucleus formation, spindle assembly and spindle positioning, but also to understand how complex human cell architecture evolved.
TK wrote the first draft, acquired the fund. TK and SB revised manuscript, figures, and Table. SB got the images of Figure 1A. Both authors contributed to the article and approved the submitted version.
This work was supported by a grant from the Okinawa Institute of Science and Technology Graduate University, Japan.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Euikyung Yu for assistance in preparing the Table. We also thank the reviewers for their critical and constructive comments.
Abad, P. C., Lewis, J. I, Mian, S., Knowles, D. W., Sturgis, J., Badve, S., et al. (2007). NuMA influences higher order chromatin organization in human mammary epithelium. Mol. Biol. Cell 18, 348–361. doi: 10.1091/mbc.e06-06-0551
Alvarez Sedo, C., Schatten, H., Combelles, C. M., and Rawe, V. Y. (2011). The nuclear mitotic apparatus (NuMA) protein: localization and dynamics in human oocytes, fertilization and early embryos. Mol. Hum. Reprod. 17, 392–398. doi: 10.1093/molehr/gar009
Baines, A. J., Lu, H. C., and Bennett, P. M. (2014). The Protein 4.1 family: hub proteins in animals for organizing membrane proteins. Biochim. Biophys. Acta 1838, 605–619. doi: 10.1016/j.bbamem.2013.05.030
Borgal, L., and Wakefield, J. G. (2018). Context-dependent spindle pole focusing. Essays Biochem. 62, 803–813. doi: 10.1042/ebc20180034
Bowman, S. K., Neumuller, R. A., Novatchkova, M., Du, Q., and Knoblich, J. A. (2006). The Drosophila NuMA homolog mud regulates spindle orientation in asymmetric cell division. Dev. Cell 10, 731–742. doi: 10.1016/j.devcel.2006.05.005
Brownlee, C., and Heald, R. (2019). Importin alpha partitioning to the plasma membrane regulates intracellular scaling. Cell 176, 805–815.e808.
Carminati, M., Gallini, S., Pirovano, L., Alfieri, A., Bisi, S., and Mapelli, M. (2016). Concomitant binding of Afadin to LGN and F-actin directs planar spindle orientation. Nat. Struct. Mol. Biol. 23, 155–163. doi: 10.1038/nsmb.3152
Chang, C. C., Huang, T. L., Shimamoto, Y., Tsai, S. Y., and Hsia, K. C. (2017). Regulation of mitotic spindle assembly factor NuMA by Importin-beta. J. Cell Biol. 216, 3453–3462. doi: 10.1083/jcb.201705168
Chinen, T., Yamamoto, S., Takeda, Y., Watanabe, K., Kuroki, K., Hashimoto, K., et al. (2020). NuMA assemblies organize microtubule asters to establish spindle bipolarity in acentrosomal human cells. EMBO J. 39:e102378.
Cleveland, D. W. (1995). NuMA: a protein involved in nuclear structure, spindle assembly, and nuclear re-formation. Trends Cell Biol. 5, 60–64. doi: 10.1016/s0962-8924(00)88947-3
Collins, E. S., Balchand, S. K., Faraci, J. L., Wadsworth, P., and Lee, W. L. (2012). Cell cycle-regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol. Biol. Cell 23, 3380–3390. doi: 10.1091/mbc.e12-02-0109
Compton, D. A., and Cleveland, D. W. (1994). NuMA, a nuclear protein involved in mitosis and nuclear reformation. Curr. Opin. Cell Biol. 6, 343–346. doi: 10.1016/0955-0674(94)90024-8
Compton, D. A., and Luo, C. (1995). Mutation of the predicted p34cdc2 phosphorylation sites in NuMA impair the assembly of the mitotic spindle and block mitosis. J. Cell Sci. 108(Pt 2), 621–633.
Compton, D. A., Szilak, I., and Cleveland, D. W. (1992). Primary structure of NuMA, an intranuclear protein that defines a novel pathway for segregation of proteins at mitosis. J. Cell Biol. 116, 1395–1408. doi: 10.1083/jcb.116.6.1395
Courtois, A., Schuh, M., Ellenberg, J., and Hiiragi, T. (2012). The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J. Cell Biol. 198, 357–370. doi: 10.1083/jcb.201202135
Culurgioni, S., Alfieri, A., Pendolino, V., Laddomada, F., and Mapelli, M. (2011). Inscuteable and NuMA proteins bind competitively to Leu-Gly-Asn repeat-enriched protein (LGN) during asymmetric cell divisions. Proc. Natl. Acad. Sci. U.S.A. 108, 20998–21003. doi: 10.1073/pnas.1113077108
Culurgioni, S., Mari, S., Bonetti, P., Gallini, S., Bonetto, G., Brennich, M., et al. (2018). Insc:LGN tetramers promote asymmetric divisions of mammary stem cells. Nat. Commun. 9:1025.
Dimitracopoulos, A., Srivastava, P., Chaigne, A., Win, Z., Shlomovitz, R., Lancaster, O. M., et al. (2020). Mechanochemical crosstalk produces cell-intrinsic patterning of the cortex to orient the mitotic spindle. Curr. Biol. 30, 3687–3696.e3684.
Drutovic, D., Duan, X., Li, R., Kalab, P., and Solc, P. (2020). RanGTP and importin beta regulate meiosis I spindle assembly and function in mouse oocytes. EMBO J. 39:e101689.
Du, Q., and Macara, I. G. (2004). Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 119, 503–516. doi: 10.1016/j.cell.2004.10.028
Du, Q., Taylor, L., Compton, D. A., and Macara, I. G. (2002). LGN blocks the ability of NuMA to bind and stabilize microtubules. A mechanism for mitotic spindle assembly regulation. Curr. Biol. 12, 1928–1933. doi: 10.1016/s0960-9822(02)01298-8
Dumont, J., Petri, S., Pellegrin, F., Terret, M. E., Bohnsack, M. T., Rassinier, P., et al. (2007). A centriole- and RanGTP-independent spindle assembly pathway in meiosis I of vertebrate oocytes. J. Cell Biol. 176, 295–305. doi: 10.1083/jcb.200605199
Eibes, S., Gallisa-Sune, N., Rosas-Salvans, M., Martinez-Delgado, P., Vernos, I., and Roig, J. (2018). Nek9 phosphorylation defines a new role for TPX2 in Eg5-dependent centrosome separation before nuclear envelope breakdown. Curr. Biol. 28, 121–129.e124.
Fielmich, L. E., Schmidt, R., Dickinson, D. J., Goldstein, B., Akhmanova, A., and van den Heuvel, S. (2018). Optogenetic dissection of mitotic spindle positioning in vivo. Elife 7:e38198.
Forth, S., Hsia, K. C., Shimamoto, Y., and Kapoor, T. M. (2014). Asymmetric friction of nonmotor MAPs can lead to their directional motion in active microtubule networks. Cell 157, 420–432. doi: 10.1016/j.cell.2014.02.018
Furuta, M., Hori, T., and Fukagawa, T. (2016). Chromatin binding of RCC1 during mitosis is important for its nuclear localization in interphase. Mol. Biol. Cell 27, 371–381. doi: 10.1091/mbc.e15-07-0497
Galli, M., and van den Heuvel, S. (2008). Determination of the cleavage plane in early C. elegans embryos. Annu. Rev. Genet. 42, 389–411. doi: 10.1146/annurev.genet.40.110405.090523
Gallini, S., Carminati, M., De Mattia, F., Pirovano, L., Martini, E., and Oldani, A. I, et al. (2016). NuMA phosphorylation by aurora-A orchestrates spindle orientation. Curr. Biol. 26, 458–469. doi: 10.1016/j.cub.2015.12.051
Gama, J. B., Pereira, C., Simoes, P. A., Celestino, R., Reis, R. M., Barbosa, D. J., et al. (2017). Molecular mechanism of dynein recruitment to kinetochores by the Rod-Zw10-Zwilch complex and Spindly. J. Cell Biol. 216, 943–960. doi: 10.1083/jcb.201610108
Gehmlich, K., Haren, L., and Merdes, A. (2004). Cyclin B degradation leads to NuMA release from dynein/dynactin and from spindle poles. EMBO Rep. 5, 97–103. doi: 10.1038/sj.embor.7400046
Gonczy, P. (2008). Mechanisms of asymmetric cell division: flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9, 355–366. doi: 10.1038/nrm2388
Greenberg, S. R., Tan, W., and Lee, W. L. (2018). Num1 versus NuMA: insights from two functionally homologous proteins. Biophys. Rev. 10, 1631–1636. doi: 10.1007/s12551-018-0472-x
Gueth-Hallonet, C., Wang, J., Harborth, J., Weber, K., and Osborn, M. (1998). Induction of a regular nuclear lattice by overexpression of NuMA. Exp. Cell Res. 243, 434–452. doi: 10.1006/excr.1998.4178
Gueth-Hallonet, C., Weber, K., and Osborn, M. (1997). Cleavage of the nuclear matrix protein NuMA during apoptosis. Exp. Cell Res. 233, 21–24. doi: 10.1006/excr.1997.3557
Harborth, J., Wang, J., Gueth-Hallonet, C., Weber, K., and Osborn, M. (1999). Self assembly of NuMA: multiarm oligomers as structural units of a nuclear lattice. EMBO J. 18, 1689–1700. doi: 10.1093/emboj/18.6.1689
Harborth, J., Weber, K., and Osborn, M. (1995). Epitope mapping and direct visualization of the parallel, in-register arrangement of the double-stranded coiled-coil in the NuMA protein. EMBO J. 14, 2447–2460. doi: 10.1002/j.1460-2075.1995.tb07242.x
Harborth, J., Weber, K., and Osborn, M. (2000). GAS41, a highly conserved protein in eukaryotic nuclei, binds to NuMA. J. Biol. Chem. 275, 31979–31985. doi: 10.1074/jbc.m000994200
Hasegawa, K., Ryu, S. J., and Kalab, P. (2013). Chromosomal gain promotes formation of a steep RanGTP gradient that drives mitosis in aneuploid cells. J. Cell Biol. 200, 151–161. doi: 10.1083/jcb.201206142
Heald, R., Tournebize, R., Habermann, A., Karsenti, E., and Hyman, A. (1997). Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138, 615–628. doi: 10.1083/jcb.138.3.615
Holubcova, Z., Blayney, M., Elder, K., and Schuh, M. (2015). Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science 348, 1143–1147. doi: 10.1126/science.aaa9529
Hueschen, C. L., Kenny, S. J., Xu, K., and Dumont, S. (2017). NuMA recruits dynein activity to microtubule minus-ends at mitosis. Elife 6:e29328.
Iwakiri, Y., Kamakura, S., Hayase, J., and Sumimoto, H. (2013). Interaction of NuMA protein with the kinesin Eg5: its possible role in bipolar spindle assembly and chromosome alignment. Biochem. J. 451, 195–204. doi: 10.1042/bj20121447
Izumi, Y., Ohta, N., Hisata, K., Raabe, T., and Matsuzaki, F. (2006). Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat. Cell Biol. 8, 586–593. doi: 10.1038/ncb1409
Jankele, R., Jelier, R., and Gonczy, P. (2021). Physically asymmetric division of the C. elegans zygote ensures invariably successful embryogenesis. Elife 10:e61714.
Jia, M., Li, J., Zhu, J., Wen, W., Zhang, M., and Wang, W. (2012). Crystal structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from Galphai. J. Biol. Chem. 287, 36766–36776. doi: 10.1074/jbc.m112.391607
Kaji, N., Muramoto, A., and Mizuno, K. (2008). LIM kinase-mediated cofilin phosphorylation during mitosis is required for precise spindle positioning. J. Biol. Chem. 283, 4983–4992. doi: 10.1074/jbc.m708644200
Kalab, P., Pralle, A., Isacoff, E. Y., Heald, R., and Weis, K. (2006). Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 440, 697–701. doi: 10.1038/nature04589
Keshri, R., Rajeevan, A., and Kotak, S. (2020). PP2A–B55gamma counteracts Cdk1 and regulates proper spindle orientation through the cortical dynein adaptor NuMA. J. Cell Sci. 133:jcs243857. doi: 10.1242/jcs.243857
Kettenbach, A. N., Schweppe, D. K., Faherty, B. K., Pechenick, D., Pletnev, A. A., and Gerber, S. A. (2011). Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4:rs5. doi: 10.1126/scisignal.2001497
Kivinen, K., Kallajoki, M., and Taimen, P. (2005). Caspase-3 is required in the apoptotic disintegration of the nuclear matrix. Exp. Cell Res. 311, 62–73. doi: 10.1016/j.yexcr.2005.08.006
Kiyomitsu, T. (2019). The cortical force-generating machinery: how cortical spindle-pulling forces are generated. Curr. Opin. Cell Biol. 60, 1–8. doi: 10.1016/j.ceb.2019.03.001
Kiyomitsu, T., and Cheeseman, I. M. (2012). Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 14, 311–317. doi: 10.1038/ncb2440
Kiyomitsu, T., and Cheeseman, I. M. (2013). Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell 154, 391–402. doi: 10.1016/j.cell.2013.06.010
Kolano, A., Brunet, S., Silk, A. D., Cleveland, D. W., and Verlhac, M. H. (2012). Error-prone mammalian female meiosis from silencing the spindle assembly checkpoint without normal interkinetochore tension. Proc. Natl. Acad. Sci. U.S.A. 109, E1858–E1867.
Kotak, S., Afshar, K., Busso, C., and Gonczy, P. (2016). Aurora A kinase regulates proper spindle positioning in C. elegans and in human cells. J. Cell Sci. 129, 3015–3025. doi: 10.1242/jcs.184416
Kotak, S., Busso, C., and Gonczy, P. (2012). Cortical dynein is critical for proper spindle positioning in human cells. J. Cell Biol. 199, 97–110. doi: 10.1083/jcb.201203166
Kotak, S., Busso, C., and Gonczy, P. (2013). NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function. EMBO J. 32, 2517–2529. doi: 10.1038/emboj.2013.172
Kotak, S., Busso, C., and Gonczy, P. (2014). NuMA interacts with phosphoinositides and links the mitotic spindle with the plasma membrane. EMBO J. 33, 1815–1830. doi: 10.15252/embj.201488147
Kschonsak, Y. T., and Hoffmann, I. (2018). Activated ezrin controls MISP levels to ensure correct NuMA polarization and spindle orientation. J. Cell Sci. 131:jcs214544. doi: 10.1242/jcs.214544
Lechler, T., and Fuchs, E. (2005). Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437, 275–280. doi: 10.1038/nature03922
Lee, I. G., Cason, S. E., Alqassim, S. S., Holzbaur, E. L. F., and Dominguez, R. (2020). A tunable LIC1-adaptor interaction modulates dynein activity in a cargo-specific manner. Nat. Commun. 11:5695.
Levy, D. L., and Heald, R. (2012). Mechanisms of intracellular scaling. Annu. Rev. Cell Dev. Biol. 28, 113–135. doi: 10.1146/annurev-cellbio-092910-154158
Lin, H. H., Hsu, H. L., and Yeh, N. H. (2007). Apoptotic cleavage of NuMA at the C-terminal end is related to nuclear disruption and death amplification. J. Biomed. Sci. 14, 681–694. doi: 10.1007/s11373-007-9165-3
Lorson, M. A., Horvitz, H. R., and van den Heuvel, S. (2000). LIN-5 is a novel component of the spindle apparatus required for chromosome segregation and cleavage plane specification in Caenorhabditis elegans. J. Cell Biol. 148, 73–86. doi: 10.1083/jcb.148.1.73
Lu, Q., Lu, Z., Liu, Q., Guo, L., Ren, H., Fu, J., et al. (2012). Chromatin-bound NLS proteins recruit membrane vesicles and nucleoporins for nuclear envelope assembly via importin-alpha/beta. Cell Res. 22, 1562–1575. doi: 10.1038/cr.2012.113
Luderus, M. E., den Blaauwen, J. L., de Smit, O. J., Compton, D. A., and van Driel, R. (1994). Binding of matrix attachment regions to lamin polymers involves single-stranded regions and the minor groove. Mol. Cell. Biol. 14, 6297–6305. doi: 10.1128/mcb.14.9.6297
Lydersen, B. K., Kao, F. T., and Pettijohn, D. (1980). Expression of genes coding for non-histone chromosomal proteins in human-Chinese hamster cell hybrids. An electrophoretic analysis. J. Biol. Chem. 255, 3002–3007. doi: 10.1016/s0021-9258(19)85842-8
Lydersen, B. K., and Pettijohn, D. E. (1980). Human-specific nuclear protein that associates with the polar region of the mitotic apparatus: distribution in a human/hamster hybrid cell. Cell 22, 489–499. doi: 10.1016/0092-8674(80)90359-1
Machicoane, M., de Frutos, C. A., Fink, J., Rocancourt, M., Lombardi, Y., Garel, S., et al. (2014). SLK-dependent activation of ERMs controls LGN-NuMA localization and spindle orientation. J. Cell Biol. 205, 791–799. doi: 10.1083/jcb.201401049
Matsumura, S., Hamasaki, M., Yamamoto, T., Ebisuya, M., Sato, M., Nishida, E., et al. (2012). ABL1 regulates spindle orientation in adherent cells and mammalian skin. Nat. Commun. 3:626.
Mattagajasingh, S. N., Huang, S. C., and Benz, E. J. Jr. (2009). Inhibition of protein 4.1 R and NuMA interaction by mutagenization of their binding-sites abrogates nuclear localization of 4.1 R. Clin. Transl. Sci. 2, 102–111. doi: 10.1111/j.1752-8062.2008.00087.x
McKenney, R. J., Huynh, W., Tanenbaum, M. E., Bhabha, G., and Vale, R. D. (2014). Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345, 337–341. doi: 10.1126/science.1254198
Merdes, A., and Cleveland, D. W. (1998). The role of NuMA in the interphase nucleus. J. Cell Sci. 111(Pt 1), 71–79.
Merdes, A., Heald, R., Samejima, K., Earnshaw, W. C., and Cleveland, D. W. (2000). Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149, 851–862. doi: 10.1083/jcb.149.4.851
Merdes, A., Ramyar, K., Vechio, J. D., and Cleveland, D. W. (1996). A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87, 447–458. doi: 10.1016/s0092-8674(00)81365-3
Moreno, N. S., Liu, J., Haas, K. M., Parker, L. L., Chakraborty, C., Kron, S. J., et al. (2019). The nuclear structural protein NuMA is a negative regulator of 53BP1 in DNA double-strand break repair. Nucleic Acids Res. 47, 10475. doi: 10.1093/nar/gkz802
Morin, X., and Bellaiche, Y. (2011). Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev. Cell 21, 102–119. doi: 10.1016/j.devcel.2011.06.012
Nachury, M. V., Maresca, T. J., Salmon, W. C., Waterman-Storer, C. M., Heald, R., and Weis, K. (2001). Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell 104, 95–106. doi: 10.1016/s0092-8674(01)00194-5
Natsume, T., Kiyomitsu, T., Saga, Y., and Kanemaki, M. T. (2016). Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 15, 210–218. doi: 10.1016/j.celrep.2016.03.001
Ohata, H., Miyazaki, M., Otomo, R., Matsushima-Hibiya, Y., Otsubo, C., Nagase, T., et al. (2013). NuMA is required for the selective induction of p53 target genes. Mol. Cell. Biol. 33, 2447–2457. doi: 10.1128/mcb.01221-12
Okumura, M., Natsume, T., Kanemaki, M. T., and Kiyomitsu, T. (2018). Dynein-Dynactin-NuMA clusters generate cortical spindle-pulling forces as a multi-arm ensemble. Elife 7:e36559.
Peyre, E., Jaouen, F., Saadaoui, M., Haren, L., Merdes, A., Durbec, P., et al. (2011). A lateral belt of cortical LGN and NuMA guides mitotic spindle movements and planar division in neuroepithelial cells. J. Cell Biol. 193, 141–154. doi: 10.1083/jcb.201101039
Pirovano, L., Culurgioni, S., Carminati, M., Alfieri, A., Monzani, S., Cecatiello, V., et al. (2019). Hexameric NuMA:LGN structures promote multivalent interactions required for planar epithelial divisions. Nat. Commun. 10:2208.
Poulson, N. D., and Lechler, T. (2010). Robust control of mitotic spindle orientation in the developing epidermis. J. Cell Biol. 191, 915–922. doi: 10.1083/jcb.201008001
Radulescu, A. E., and Cleveland, D. W. (2010). NuMA after 30 years: the matrix revisited. Trends Cell Biol. 20, 214–222. doi: 10.1016/j.tcb.2010.01.003
Rajeevan, A., Keshri, R., Kapoor, S., and Kotak, S. (2020). NuMA interaction with chromatin is vital for proper chromosome decondensation at the mitotic exit. Mol. Biol. Cell 31, 2437–2451. doi: 10.1091/mbc.e20-06-0415
Renna, C., Rizzelli, F., Carminati, M., Gaddoni, C., Pirovano, L., Cecatiello, V., et al. (2020). Organizational principles of the NuMA-dynein interaction interface and implications for mitotic spindle functions. Structure 28, 820–829.e826.
Saadaoui, M., Machicoane, M., di Pietro, F., Etoc, F., Echard, A., and Morin, X. (2014). Dlg1 controls planar spindle orientation in the neuroepithelium through direct interaction with LGN. J. Cell Biol. 206, 707–717. doi: 10.1083/jcb.201405060
Samwer, M., Schneider, M. W. G., Hoefler, R., Schmalhorst, P. S., Jude, J. G., Zuber, J., et al. (2017). DNA cross-bridging shapes a single nucleus from a set of mitotic chromosomes. Cell 170, 956–972.e923.
Sana, S., Keshri, R., Rajeevan, A., Kapoor, S., and Kotak, S. (2018). Plk1 regulates spindle orientation by phosphorylating NuMA in human cells. Life Sci. Alliance 1:e201800223. doi: 10.26508/lsa.201800223
Schlager, M. A., Hoang, H. T., Urnavicius, L., Bullock, S. L., and Carter, A. P. (2014). In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 33, 1855–1868. doi: 10.15252/embj.201488792
Segalen, M., Johnston, C. A., Martin, C. A., Dumortier, J. G., Prehoda, K. E., David, N. B., et al. (2010). The Fz-Dsh planar cell polarity pathway induces oriented cell division via Mud/NuMA in Drosophila and zebrafish. Dev. Cell 19, 740–752. doi: 10.1016/j.devcel.2010.10.004
Seldin, L., Muroyama, A., and Lechler, T. (2016). NuMA-microtubule interactions are critical for spindle orientation and the morphogenesis of diverse epidermal structures. Elife 5:e12504.
Seldin, L., Poulson, N. D., Foote, H. P., and Lechler, T. (2013). NuMA localization, stability, and function in spindle orientation involve 4.1 and Cdk1 interactions. Mol. Biol. Cell 24, 3651–3662. doi: 10.1091/mbc.e13-05-0277
Serra-Marques, A., Houtekamer, R., Hintzen, D., Canty, J. T., Yildiz, A., and Dumont, S. (2020). The mitotic protein NuMA plays a spindle-independent role in nuclear formation and mechanics. J. Cell Biol. 219:e202004202.
Shoaib, M., Nair, N., and Sorensen, C. S. (2020). Chromatin landscaping at mitotic exit orchestrates genome function. Front. Genet. 11:103. doi: 10.3389/fgene.2020.00103
Silk, A. D., Holland, A. J., and Cleveland, D. W. (2009). Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J. Cell Biol. 184, 677–690. doi: 10.1083/jcb.200810091
Siller, K. H., Cabernard, C., and Doe, C. Q. (2006). The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat. Cell Biol. 8, 594–600. doi: 10.1038/ncb1412
Siller, K. H., and Doe, C. Q. (2009). Spindle orientation during asymmetric cell division. Nat. Cell Biol. 11, 365–374. doi: 10.1038/ncb0409-365
Srinivasan, D. G., Fisk, R. M., Xu, H., and van den Heuvel, S. (2003). A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C elegans. Genes Dev. 17, 1225–1239. doi: 10.1101/gad.1081203
Sun, Q. Y., and Schatten, H. (2006). Role of NuMA in vertebrate cells: review of an intriguing multifunctional protein. Front. Biosci. 11:1137–1146. doi: 10.2741/1868
Suzuki, M. (1989). SPXX, a frequent sequence motif in gene regulatory proteins. J. Mol. Biol. 207, 61–84. doi: 10.1016/0022-2836(89)90441-5
Taimen, P., Parvinen, M., Osborn, M., and Kallajoki, M. (2004). NuMA in rat testis–evidence for roles in proliferative activity and meiotic cell division. Exp. Cell Res. 298, 512–520. doi: 10.1016/j.yexcr.2004.05.002
Tame, M. A., Raaijmakers, J. A., Afanasyev, P., and Medema, R. H. (2016). Chromosome misalignments induce spindle-positioning defects. EMBO Rep. 17, 317–325. doi: 10.15252/embr.201541143
Tang, T. K., Tang, C. J., Chao, Y. J., and Wu, C. W. (1994). Nuclear mitotic apparatus protein (NuMA): spindle association, nuclear targeting and differential subcellular localization of various NuMA isoforms. J. Cell Sci. 107(Pt 6), 1389–1402.
Tsuchiya, K., Hayashi, H., Nishina, M., Okumura, M., Sato, Y., Kanemaki, M. T., et al. (2021). Ran-GTP is non-essential to activate NuMA for mitotic spindle-pole focusing but dynamically polarizes HURP near chromosomes. Curr. Biol. 31, 115–127.e113.
Vidi, P. A., Liu, J., Salles, D., Jayaraman, S., Dorfman, G., Gray, M., et al. (2014). NuMA promotes homologous recombination repair by regulating the accumulation of the ISWI ATPase SNF2h at DNA breaks. Nucleic Acids Res. 42, 6365–6379. doi: 10.1093/nar/gku296
Wei, J. H., Zhang, Z. C., Wynn, R. M., and Seemann, J. (2015). GM130 regulates golgi-derived spindle assembly by activating TPX2 and capturing microtubules. Cell 162, 287–299. doi: 10.1016/j.cell.2015.06.014
Wiese, C., Wilde, A., Moore, M. S., Adam, S. A., Merdes, A., and Zheng, Y. (2001). Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science 291, 653–656. doi: 10.1126/science.1057661
Williams, S. E., Beronja, S., Pasolli, H. A., and Fuchs, E. (2011). Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature 470, 353–358. doi: 10.1038/nature09793
Wong, R. W., Blobel, G., and Coutavas, E. (2006). Rae1 interaction with NuMA is required for bipolar spindle formation. Proc. Natl. Acad. Sci. U.S.A. 103, 19783–19787. doi: 10.1073/pnas.0609582104
Woodard, G. E., Huang, N. N., Cho, H., Miki, T., Tall, G. G., and Kehrl, J. H. (2010). Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol. Cell. Biol. 30, 3519–3530. doi: 10.1128/mcb.00394-10
Yamada, M., and Goshima, G. (2017). Mitotic spindle assembly in land plants: molecules and mechanisms. Biology (Basel) 6:6. doi: 10.3390/biology6010006
Yang, C. H., Lambie, E. J., and Snyder, M. (1992). NuMA: an unusually long coiled-coil related protein in the mammalian nucleus. J. Cell Biol. 116, 1303–1317. doi: 10.1083/jcb.116.6.1303
Yesbolatova, A., Saito, Y., Kitamoto, N., Makino-Itou, H., Ajima, R., Nakano, R., et al. (2020). The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice. Nat. Commun. 11:5701.
Yuzawa, S., Kamakura, S., Iwakiri, Y., Hayase, J., and Sumimoto, H. (2011). Structural basis for interaction between the conserved cell polarity proteins Inscuteable and Leu-Gly-Asn repeat-enriched protein (LGN). Proc. Natl. Acad. Sci. U.S.A. 108, 19210–19215. doi: 10.1073/pnas.1110951108
Zeng, C., He, D., and Brinkley, B. R. (1994). Localization of NuMA protein isoforms in the nuclear matrix of mammalian cells. Cell Motil. Cytoskeleton 29, 167–176. doi: 10.1002/cm.970290208
Zheng, Z., Wan, Q., Meixiong, G., and Du, Q. (2014). Cell cycle-regulated membrane binding of NuMA contributes to efficient anaphase chromosome separation. Mol. Biol. Cell 25, 606–619. doi: 10.1091/mbc.e13-08-0474
Zhu, J., Shang, Y., Xia, C., Wang, W., Wen, W., and Zhang, M. (2011a). Guanylate kinase domains of the MAGUK family scaffold proteins as specific phospho-protein-binding modules. EMBO J. 30, 4986–4997. doi: 10.1038/emboj.2011.428
Keywords: NuMA, spindle, nuclear formation, dynein, Ran-GTP
Citation: Kiyomitsu T and Boerner S (2021) The Nuclear Mitotic Apparatus (NuMA) Protein: A Key Player for Nuclear Formation, Spindle Assembly, and Spindle Positioning. Front. Cell Dev. Biol. 9:653801. doi: 10.3389/fcell.2021.653801
Received: 15 January 2021; Accepted: 10 March 2021;
Published: 01 April 2021.
Edited by:
Anne Straube, University of Warwick, United KingdomReviewed by:
Andreas Merdes, Centre de Biologie Intégrative (CBI), FranceCopyright © 2021 Kiyomitsu and Boerner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomomi Kiyomitsu, dG9tb21pLmtpeW9taXRzdUBvaXN0Lmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.