
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol. , 18 June 2021
Sec. Signaling
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.653522
This article is part of the Research Topic With the Eyes on Non-Coding RNAs View all 13 articles
The retina is among the most metabolically active tissues with high-energy demands. The peculiar distribution of mitochondria in cells of retinal layers is necessary to assure the appropriate energy supply for the transmission of the light signal. Photoreceptor cells (PRs), retinal pigment epithelium (RPE), and retinal ganglion cells (RGCs) present a great concentration of mitochondria, which makes them particularly sensitive to mitochondrial dysfunction. To date, visual loss has been extensively correlated to defective mitochondrial functions. Many mitochondrial diseases (MDs) show indeed neuro-ophthalmic manifestations, including retinal and optic nerve phenotypes. Moreover, abnormal mitochondrial functions are frequently found in the most common retinal pathologies, i.e., glaucoma, age-related macular degeneration (AMD), and diabetic retinopathy (DR), that share clinical similarities with the hereditary primary MDs. MicroRNAs (miRNAs) are established as key regulators of several developmental, physiological, and pathological processes. Dysregulated miRNA expression profiles in retinal degeneration models and in patients underline the potentiality of miRNA modulation as a possible gene/mutation-independent strategy in retinal diseases and highlight their promising role as disease predictive or prognostic biomarkers. In this review, we will summarize the current knowledge about the participation of miRNAs in both rare and common mitochondria-mediated eye diseases. Definitely, given the involvement of miRNAs in retina pathologies and therapy as well as their use as molecular biomarkers, they represent a determining target for clinical applications.
Mitochondria are key players in different cellular processes, and their dysfunction contributes to the pathogenesis of neurodegenerative disorders (NDs), including many retinal diseases. To date, a connection between vision and defective mitochondrial functions has been extensively described (Yu-Wai-Man et al., 2011; Gueven et al., 2017). Mitochondrial diseases (MDs) are a heterogeneous group of rare disorders caused by mutations in nuclear or mitochondrial genes that affect proteins essential for mitochondrial structure and function. Although they are highly genetically and clinically heterogeneous, several MDs, such as Leber hereditary optic neuropathy (LHON), autosomal dominant optic atrophy (ADOA), and neuropathy, ataxia, and retinitis pigmentosa (NARP), show some form of vision impairment and can be classified as primary mitochondrial eye diseases (PMEDs). Moreover, mitochondrial dysfunctions represent a common denominator and a common cause of neuronal death involved in the pathogenesis of many NDs due to mutations in genes encoding non-mitochondrial proteins or characterized by more complex pathogenetic events (Niyazov et al., 2016).
The great concentration of mitochondria in metabolically active tissues with high-energy demands, such as the retina, makes them particularly sensitive to mitochondrial dysfunction. The retina comprises different cell types organized in layers that form neuronal circuits working in parallel and in combination to produce a complex visual output (Figure 1) (Carrella et al., 2020). The outer nuclear layer (ONL) is composed of photoreceptor cells (PRs), subdivided into rods and cones. They synapse with interneurons of the inner nuclear layer (INL), namely, bipolar cells, amacrine cells, and horizontal cells, which in turn contact RGCs in the RGC layer. Retinal layers show a peculiar distribution of mitochondria to guarantee the energy supply for the conversion and propagation of the light signal (Figure 1). PRs, which capture photons and generate electrophysiological signals, display many mitochondria in the inner segment. In RPE, mitochondria are located at the basal region, that is, in contact with PRs. Instead, in the inner retina, mitochondria are predominantly concentrated in the unmyelinated proximal axons of RGCs, which transmit visual information to the brain. It is thus not surprising that the most common retinal disorders, i.e., glaucoma, age-related macular degeneration (AMD), and diabetic retinopathy (DR), show mitochondrial dysfunction and share some clinical similarities with PMEDs (Carelli et al., 2004; Yu-Wai-Man et al., 2011; Gueven et al., 2017; Ferrington et al., 2020). Interestingly, many studies also reported vision impairment and retinal abnormalities in the majority of Alzheimer’s and Parkinson’s disease patients and animal models, highlighting the involvement of mitochondrial anomalies in the development of visual defects (Colligris et al., 2018; Indrieri et al., 2020b; Marrocco et al., 2020; Mirzaei et al., 2020).
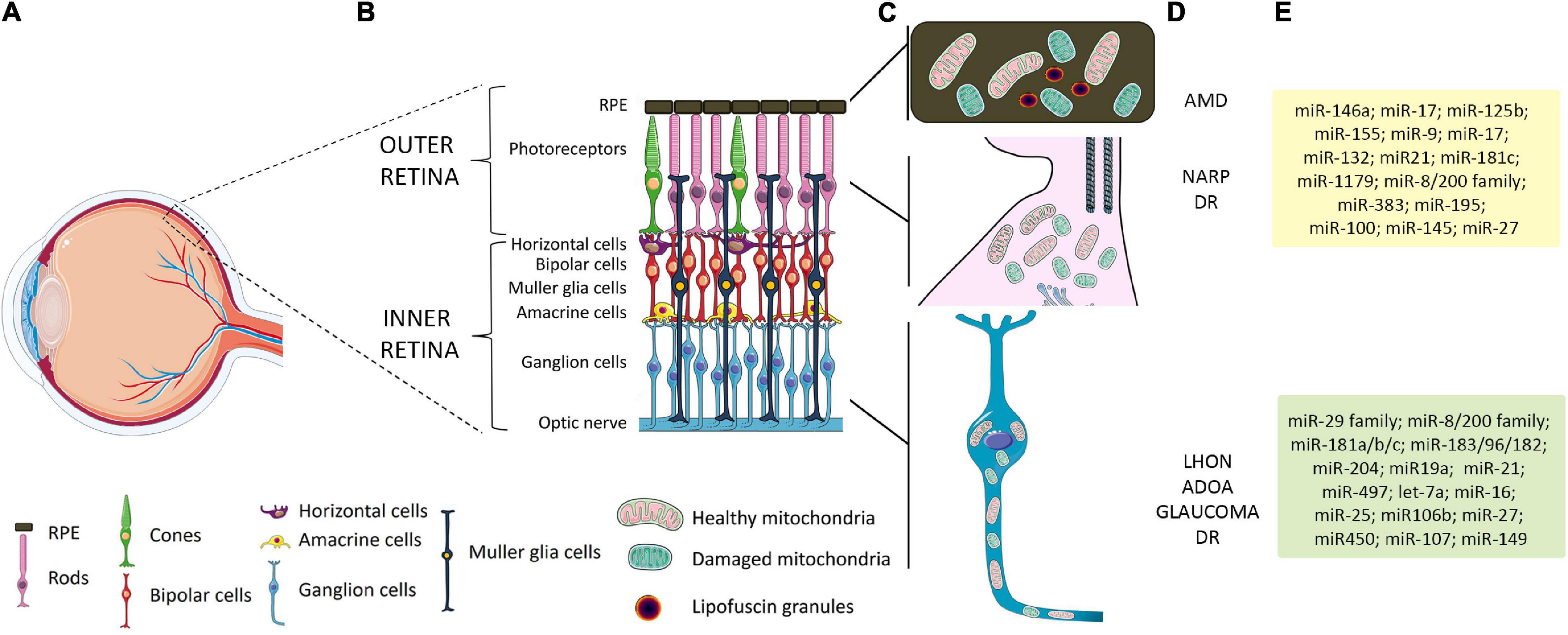
Figure 1. Schematic representation of retinal layers, mitochondrial distribution, and related retinal pathologies and miRNAs. (A) Schematic representation of the human eye. The box identifies the position of the retinal layers in the posterior part of the eye. (B) Magnification of the retina and retinal layers in healthy conditions. (C) A peculiar distribution of mitochondria is graphically represented: in RPE, which overlays the PRs, mitochondria are located at the basal region; in PRs, a high number of mitochondria are present in the inner segment; in the inner retina, mitochondria are predominantly concentrated in the unmyelinated proximal axons of RGCs. (D) Primary or secondary defect in mitochondrial functionality in RPE and PRs has been linked to AMD, NARP, and other forms of visual loss, such as DR. Primary or secondary impairment in mitochondrial function leading to RGC degeneration is considered a major cause of retinal diseases such as LHON, ADOA, glaucoma, and DR. (E) List of miRNAs expressed in specific cell retinal layers whose function or alteration in expression has been linked to mitochondrial dysfunction and retinal diseases. Yellow box shows relevant miRNAs related to AMD and DR; green box, LHON and glaucoma-related miRNAs. RPE, retinal pigment epithelium; PRs, photoreceptors; RGCs, retinal ganglion cells; AMD, age-related macular degeneration; NARP, neuropathy, ataxia, and retinitis pigmentosa; DR, diabetic retinopathy; LHON, Leber hereditary optic neuropathy; ADOA, autosomal dominant optic atrophy.
MicroRNAs (miRNAs) are a class of non-coding RNAs able to post-transcriptionally regulate gene expression through a powerful mechanism of sequence-specific recognition. Each miRNA is predicted to recognize about 200 mRNA targets, guaranteeing a pleiotropic fine-tuning of correlated transcripts that confers robustness to pathway regulation (Bartel, 2018).
Soon, their relevant role in different retina pathologies (Karali and Banfi, 2019; Zuzic et al., 2019) and the possibility to exploit their modulation as a possible gene/mutation-independent strategy for these disorders became evident (Carrella et al., 2020). The extensive genetic heterogeneity of many inherited retinal disorders, including PMEDs, indeed represents a significant limitation to the development and application of gene-replacement therapy in most of patients. Moreover, gene replacement cannot be applied in disorders caused by gain-of-function mutations and too complex multifactorial diseases such as AMD, glaucoma, and DR (Carrella et al., 2020). In this respect, miRNAs represent interesting therapeutic targets able to regulate common dysregulated pathways underlying retinal damage.
Moreover, dysregulated miRNA expression profiles in retinal degeneration models and in patients indicate that they may represent reliable biomarkers for the diagnosis of these disorders or to predict the onset and the progression of the disease, and the evaluation of the response to treatments. Circulating miRNAs and exosomal miRNAs can be indeed easily detected, thus representing promising disease predictive/diagnostic/prognostic biomarkers (Saxena et al., 2015; Palfi et al., 2016; Anasagasti et al., 2018).
MicroRNAs can localize to different subcellular compartments (i.e., mitochondria, endoplasmic reticulum, and exosomes) (Leung, 2015); and an increasing interest is growing about miRNAs, called MitomiRs, that regulate mitochondrial function. MitomiRs can be divided into two subgroups: those binding to nuclear-transcribed mRNA encoding mitochondrial proteins and those imported into mitochondria targeting mitochondrial-encoded mRNAs (Purohit and Saini, 2021). Moreover, some MitomiRs (i.e., miR-1974, miR-1977, and miR-1978) may be transcribed by the mitochondrial DNA (mtDNA) (Bandiera et al., 2011); however, more data are necessary to validate these findings.
Modulation of miRNAs has been recently applied as therapy to different disorders reaching preclinical and clinical stages (Bajan and Hutvagner, 2020). However, investigations on the role of miRNAs, and specifically MitomiRs, in mitochondrial-mediated disorders are few. In this review, we will summarize the current knowledge about the involvement of miRNAs in mitochondria-mediated eye diseases, including both rare PMEDs and the most common retinal disorders, i.e., glaucoma, AMD, and DR. In particular, their role in retina pathologies and therapy, as well as their role as biomarkers in these disorders, will be analyzed, highlighting their huge potential in clinical medicine.
Leber hereditary optic neuropathy is one of the most frequent PMEDs with a prevalence of between 1/15,000 and 1/50,000 people worldwide. LHON is an organ-specific disease, characterized by death of RGCs leading to degeneration of the optic nerve (ON) and bilateral or unilateral loss of vision, which typically occurs between the ages of 20 and 40 (Meyerson et al., 2015). It shows maternal inheritance, and it results more commonly in men, with variable disease penetrance. Approximately 95% of LHON cases are associated with three mtDNA point mutations (m.11778G > A, m.3460G > A, and m.14484T > C) that primarily affect mitochondrial respiratory chain (MRC) complex I genes (ND1, ND4, and ND6) (Yu-Wai-Man et al., 2002; Newman, 2005).
The molecular mechanism underlying death of RGCs is still not clear, even if it has been correlated to a reduction of ATP, an increase of reactive oxygen species (ROS) production due to defective MRC, and a significantly impaired mitophagy (Sharma et al., 2019).
With a prevalence of 1/10,000–1/35,000, ADOA is the most common form of PMEDs due to nuclear DNA mutations. Bilaterally symmetric progressive deterioration of the central visual acuity, ON pallor, dyschromatopsia, and blindness are the main symptoms, usually beginning in childhood (Fraser et al., 2010; Yu-Wai-Man et al., 2014). As in LHON, the disease primarily affects the RGCs and their axons, even if the ADOA progression with age is highly variable (Lenaers et al., 2012). In about 50–60% of the cases, patients harbor mutations in the OPA1 (Alexander et al., 2000; Ferre et al., 2005). In addition, other mutated genes include OPA2, OPA3, OPA4, OPA5, OPA8, WFS1, and SSBP1 (Finsterer et al., 2018; Piro-Megy et al., 2020).
OPA1 is a crucial component of the mitochondrial fusion machinery and also controls crista biogenesis and remodeling, impacting apoptosis and mitochondrial respiration (Cogliati et al., 2016). In accordance, Opa1 deficiency induces a significant fragmentation of the mitochondrial network and impairs ON structure and visual function in a mouse model of ADOA (Davies et al., 2007).
Recently, the role of autophagy in the regulation of mitochondrial distribution in axons of RGC and in visual loss in an ADOA mouse model (Zaninello et al., 2020) has also been demonstrated, indicating an important patho-mechanism contribution of mitophagy.
Neuropathy, ataxia, and retinitis pigmentosa is an inherited neurologic/metabolic syndrome whose clinical hallmarks are (i) sensory neuropathy including progressive motor weakness and lethargy, (ii) ataxia, which affects the balance and coordination, and (iii) ophthalmologic findings including retinitis pigmentosa, optic atrophy, and eye movement disorders. Usually, the retina defects worsen over time, leading to severe vision loss and blindness. NARP typically begins in childhood or early adulthood. The clinical expression of the NARP syndrome is very variable, and the predominant ocular manifestation is characterized by an initial RPE degeneration and a rod/cone dysfunction in different families (Gelfand et al., 2011).
neuropathy, ataxia, and retinitis pigmentosa results from mtDNA heteroplasmic mutations in ATP6 gene (predominantly m.8993T > G/C), coding for the mitochondrial ATP synthase subunit 6 (Holt et al., 1990; Duno et al., 2013; Miyawaki et al., 2015). ATP synthase impairment affects oxidative phosphorylation, causing energy deprivation and overproduction of ROS (Nijtmans et al., 2001; Baracca et al., 2007).
With about 70 million patients worldwide, glaucoma is the leading cause of blindness and a major economic burden (Quigley and Broman, 2006). The term glaucoma describes a group of complex multifactorial diseases characterized by ON damage and loss of RGCs resulting in progressive loss of vision. Age, genetics, and elevated intraocular pressure (IOP) are prominent risk factors; however, about one-third of cases have ON degeneration despite IOP in the normal range (Almasieh and Levin, 2017).
Interestingly, similarities between glaucoma and PMEDs have been described, and defects in mitochondria have been connected to glaucomatous neurodegeneration (Kong et al., 2009; Sundaresan et al., 2015; Shim et al., 2016; Williams et al., 2017; Singh et al., 2018; Tribble et al., 2019). Moreover, mutations in OPTN encoding for the mitophagy adaptor protein optineurin (Rezaie et al., 2002; Wong and Holzbaur, 2014), TBK1 encoding the serine threonine protein kinase TANK-binding kinase 1 involved in autophagy (Sears et al., 2019), and OPA1 (Aung et al., 2002; Yu-Wai-Man et al., 2010; Guo et al., 2012) have been associated with glaucoma, thus highlighting a crucial role for mitochondrial dynamics and mitophagy pathways in glaucoma pathogenesis (Ito and Di Polo, 2017). In addition, numerous studies reported mtDNA mutations, decrease in the mitochondrial respiratory activity, and oxidative stress in glaucoma patients and in animal models of this disease (Lee et al., 2011; Tseng et al., 2015; Hondur et al., 2017; Williams et al., 2017; Tribble et al., 2019). Notably, mitochondrial dysfunction can be detected before RGC death occurs in glaucoma animal models (Kong et al., 2009; Williams et al., 2017; Singh et al., 2018), suggesting a primary effect for mitochondrial abnormalities in glaucoma onset.
Mitochondrial dysfunction has been implicated in the pathophysiology of several age-related diseases including those that involve PRs and RPE cells (Lukiw et al., 2012; Lefevere et al., 2017; Ferrington et al., 2020). Aging and oxidative stress have been recognized as primary risk factors for AMD (Liang and Godley, 2003; Jarrett and Boulton, 2012; Lefevere et al., 2017), a complex degenerative and progressive disease.
There are two forms of AMD: the “wet” form that is associated with abnormal growth of blood vessels into the retina and the “dry” form with primary pathogenic event involving RPE degeneration causing PR cell death (Liang and Godley, 2003). RPE cells engulf photoreceptor outer segments (POSs) that are shed daily during renewal of PRs. RPE accumulation of lipofuscin, a product of POS turnover, has been hypothesized to be the primary source of ROS responsible for oxidative damage of the RPE resulting in impaired metabolism and apoptosis (Liang and Godley, 2003; Vives-Bauza et al., 2008; Jarrett and Boulton, 2012). Several studies have provided evidence that impaired autophagy (Mitter et al., 2014; Hyttinen et al., 2017) and mitochondrial dysfunction (Barron et al., 2001; Feher et al., 2006), in both RPE and PRs, exacerbate oxidative stress and contribute to the pathogenesis of AMD.
Diabetic retinopathy represents one of the most common slow-progressing microvascular complications of diabetes. In diabetic patients, damaged blood vessel of the retina leads to retinal detachment and reduction in the visual field and blindness (Frank, 2004). The retinal neurodegeneration is associated with retinal electrophysiological dysfunction and thinning of RGC and PR layers (Carbonell et al., 2019). Accelerated apoptosis of both neuronal and vascular cells (Mizutani et al., 1996; Barber et al., 2011) indicates apoptotic cell death as a contributing process to DR.
Although the detailed mechanisms of action in the development of DR are still unknown, involvement of mitochondrial dysfunctions with ROS formation and a decrease of the mitochondrial fusion protein mitofusin 2 (Mfn2) have been found in experimental models of this retinopathy (Eshaq et al., 2014; Duraisamy et al., 2019).
As reported before, an increasing number of miRNAs have been shown to be involved in the regulation of mitochondrial metabolism, although there is no evidence, to date, that mitochondrial disorders affect their expression or are directly caused by their dysregulation. Recently, miR-181a and miR-181b (miR-181a/b) were shown to directly target genes involved in mitochondrial biogenesis and function, and ROS detoxification (Indrieri et al., 2019). Inactivation of miR-181a/b leads to increased levels of mitochondrial biogenesis and mitophagy leading to a significant amelioration of the disease phenotype in LHON mouse models. These data suggest that miR−181a/b may represent gene−independent therapeutic targets for mitochondrial-related eye diseases (Indrieri et al., 2019). In accordance with the pervasive and pleiotropic roles of the miR-181 family (Indrieri et al., 2020a), miR-181c might be associated with vascular proliferation in high glucose diabetic-like environment (Qing et al., 2014; Zitman-Gal et al., 2014).
Large-scale studies have been performed to identify glaucoma-relevant miRNAs (Li et al., 2009; Liu et al., 2018; Hindle et al., 2019). Among the 159 miRNAs identified, many were differentially expressed in the aqueous humor (AH) and/or tear of glaucoma patients and controls. MiRNA-29 family controls extracellular matrix (ECM) homeostasis in trabecular meshwork (TM) cells, by negatively regulating collagens, fibrillins, and elastin (Luna et al., 2009; Villarreal et al., 2011). Moreover, a specific crosstalk between TGFβ, whose alteration are often observed in glaucoma, and miR-29 levels highlighted miR-29-family implication in glaucoma (Luna et al., 2011). The expression profile of miR-8/miR-200 family is upregulated in transgenic mice carrying a mutation in OPTN (Chi et al., 2010; Gao et al., 2016). Moreover, miR-200c can decrease trabecular contraction and IOP by regulating genes associated with TM cell contraction regulation (Luna et al., 2012). The miR-183/96/182 cluster is highly expressed in retina and implicated in several aspects of retinal cell development and maintenance (Amini-Farsani and Asgharzade, 2020). In particular, miR-182 was found to be the most abundant miRNA also in the axons of developing RGC where it regulates axon guidance (Bellon et al., 2017). Interestingly, a case–control study conducted on patients with primary open-angle glaucoma (POAG) concludes that the carriers of polymorphism in miR-182 and CDKN2B genes have an increased risk of developing POAG (Moschos et al., 2020). MiR-204 caused reduced expression of FOXC1, implicated in glaucoma development, and its target genes (Paylakhi et al., 2013). Moreover, it has been shown that in ON injury, miR-204 can downregulate GAP43, which plays an important role in axonal growth and in experimental chronic glaucomatous injury (Huang et al., 2013; Wang et al., 2018). Moreover, overexpression of miR-19a augments axon regeneration via miR-19a–PTEN axis, underscoring the therapeutic potential of local administration of miRNAs via intravitreal injection (Mak et al., 2020). Another interesting miRNA is miR-21, whose inhibition in a model of ON crush promotes axonal regeneration and RGC survival and function (Li H. J. et al., 2018; Li et al., 2019). In the retina of rats with advanced nerve damage induced by elevated IOP, eight miRNAs were significantly downregulated as compared with those in controls (miR-181c, miR-497, miR-204, let-7a, miR-29b, miR-16, miR-106b, and miR-25) and miR-27a was significantly upregulated. Observed miRNA level alterations caused enrichment of targets associated with ECM/cell proliferation, immune system, and regulation of apoptosis (Jayaram et al., 2015). Several miRNAs have been also found to be released in extracellular space in glaucomatous AH. Released miRNAs include miR-21 (apoptosis), miR-450 (cell aging and maintenance of contractile tone), miR-107 (nestin expression and apoptosis), and miR-149 (endothelia and ECM homeostasis) (Tanaka et al., 2014; Izzotti et al., 2015).
Few dysregulated miRNAs in multiple studies have been identified in the blood and vitreous humor of AMD patients. The serum profiles of patients with both wet and dry AMD have shown differences and partial overlap in several miRNAs (Szemraj et al., 2015; Berber et al., 2017), reflecting the difficulty of reducing biomarkers for AMD to one common group (Natoli and Fernando, 2018). A group of dysregulated miRNAs were reported in mouse models of distinct AMD features and demonstrated some similarities with the human AMD findings, including miR-146a, miR-9, miR-17, miR-125b, and miR-155 (Lukiw et al., 2012; Berber et al., 2017; Natoli and Fernando, 2018; Pogue and Lukiw, 2018; Martinez and Peplow, 2021). Those miRNAs can be considered as potential biomarkers and as possible therapeutic targets for AMD. MiR-146a has been found in the plasma (Ertekin et al., 2014; Menard et al., 2016) and retinas (Bhattacharjee et al., 2016) of AMD patients and was modulated in human monocytes stimulated with lipopolysaccharide (Taganov et al., 2006). MiR-146a and miR-9 are upregulated by NF-κB and present indirect correlation with complement factor H (CFH) levels, a key repressor of the innate immune response and a key player in AMD pathogenesis, indicating their modulation as a therapeutic strategy (Lukiw et al., 2012). MiR-17, a regulator of angiogenesis (Doebele et al., 2010) and anti-apoptotic genes as well (Song et al., 2015), is upregulated in an oxidative-induced retina model, an oxidative stress model in RPE cells, and neovascularization AMD plasma. MiR-155 has a role in angiogenesis, complement activation, and inflammation, making it a candidate for therapeutic interventions for AMD. The expression of miR-155 is also induced by AMD-related inflammatory cytokines (O’Connell et al., 2007). In an animal model of AMD, miR-155 has been shown to be upregulated in correlation with increased cell death and inflammation (Saxena et al., 2015), and its downregulation reduced retinal neovascularization (Zhuang et al., 2015). In addition, miR-155 depletion correlates with decreased levels of the mitochondrial translocator protein (TSPO), a selective marker of microglia in their highly reactive state (Yan et al., 2015). Interestingly, miR-146a and miR-155 recognize an overlapping 3′ UTR in CFH, to which both miRNAs may interact (Lukiw et al., 2012).
Several miRNAs, related to DR, are involved in vasculature regulation (miR-126, miR-200b, and miR-31), chronic inflammation pathway (miR-146, miR-155, miR-132, and miR-21), and oxidative stress (miR-21, miR-181c, miR-1179, and miR-8/miR-200 family); other miRNAs present altered expression in DR, but their role is not yet defined (Wu et al., 2012; Andreeva and Cooper, 2014; Mastropasqua et al., 2014; Pusparajah et al., 2016; Shafabakhsh et al., 2019). MiR-383 presents an increased expression in hyperglycemic conditions and targets the mitochondrial peroxiredoxin 3 involved in ROS detoxification and apoptosis (Li et al., 2013). Indeed, miR-383 inhibition diminished ROS and cell death in RPE treated with high glucose (Jiang et al., 2017), representing one of the major keys for the treatment of DR. The expression of miR-451a was found downregulated in diabetic conditions. MiR-451a mimic overexpression showed a protective effect on mitochondrial function in diabetic conditions, probably via the downregulation of activating transcription factor 2 (ATF2) and its downstream target genes CyclinA1, CyclinD1, and MMP2, providing new perspectives for developing effective therapies for proliferative DR (Shao et al., 2019). In both experimental and human diabetes, miR-34a showed increased expression. It promotes mitochondrial dysfunction and retinal microvascular endothelial cell senescence by suppressing the SIRT1–PGC-1α axis as well as the mitochondrial antioxidants TrxR2 and SOD2 (Thounaojam et al., 2019). MiR-195 acts as a regulator for Mfn2, which is reduced in the retina of diabetic patients and is involved in maintaining mitochondrial morphology, fusion, and ROS metabolism (Sugioka et al., 2004; Zheng and Xiao, 2010). Oxidative stress-induced overexpression of miR-195 can result in the downregulation of Mfn2 leading to tube formation and to increased blood–retinal barrier permeability, which are two common pathogenic events of DR (Zhang et al., 2017). Therefore, miR-195 could be considered as a potential therapeutic target for DR (Zhong and Kowluru, 2011). Another miRNA increased upon oxidative stress is miR-100, able to downregulate AKT pathway, extracellular-signal regulated kinase pathway, and TrkB pathway (Kong et al., 2014). MiR-145 overexpression reduced ROS production and increased the activity of SOD (Hui and Yin, 2018). Finally, miR-27 reduces ROS generation and downregulates the P13K/AKT/mTOR signaling pathway by inhibition of Nox2 (Li J. et al., 2018) implicated in ROS induction and neovascularization (Chan et al., 2013, 2015).
Overall, the positive effect of miR-19a, miR-204, and miR-21 modulation on glaucoma murine models, as well as downregulation of miR-155 in AMD mice, highlights the possibility of their rapid translation into clinical application as therapeutic molecules for these eye diseases (bold miRNAs in Table 1). However, other preclinical validation steps are required for most of the previously mentioned miRNAs, thus underlining the need and importance of this emerging field of research.
Systematic expression profiling of miRNAs in retinal cells could be of benefit to identify possible involvement of their function in specific retinal cell types, in physiological and pathological conditions. Although novel strategies are under development to study miRNA expression in single-cell transcriptomic conditions (Liu and Shomron, 2021), there are no data reported for such analysis in the retina. However, systematic analysis of miRNA expression and variability in the mouse (Soundara Pandi et al., 2013) and human neural retina and RPE/choroid tissues (Karali et al., 2016) have been reported. Interestingly, among the top 30 expressed miRNAs in retina are reported several miRNAs that present a role in mitochondrial-mediated eye diseases (i.e., miR-181a/b, miR-182, miR-183, miR-204, let-7a, miR-9, miR-96, miR-125b, miR-100, and miR-181c; see Table 1). Notably, many of the miRNAs here described and associated with mitochondria-mediated eye diseases can be classified as MitomiR (Purohit and Saini, 2021) (Table 1) since they regulate important transcripts impacting different mitochondrial pathways (Table 2), thus suggesting an additional possible role of these miRNAs in the pathogenesis and therapy of these disorders.
MicroRNAs are promising therapeutic tools due to their capability to simultaneously modulate multiple pathways involved in disease pathogenesis and progression. Moreover, they also represent a class of interesting molecules useful as disease predictive/prognostic biomarkers. Indeed, several miRNAs (let-7a, miR-450, miR-107, miR-204, miR-21, and miR-149 for glaucoma; miR-17 and miR-125b for AMD; miR-126, miR-146a, miR-155, miR-132, miR-21, and miR-34a/c for DR) differentially expressed in body fluids (i.e., serum, plasma, and vitreous liquid or tears) of eye diseases associated with mitochondria dysfunctions (EDAMDs) human patients may be already considered as clinically relevant biomarkers (bold miRNAs in Table 1).
Recently, an increasing interest is growing about MitomiRs, which regulate mitochondrial function. As described before, many MitomiRs have been linked to mitochondria-mediated eye diseases, including both rare PMEDs and common retinal diseases (Tables 1, 2). Due to the genetic heterogeneity that characterizes PMEDs and to the big complexity that underlies the most common retinal disorders (e.g., glaucoma, AMD, and DR), no effective treatments are still available. For the above-mentioned reasons, miRNA-based gene/mutation-independent therapeutic strategies may represent a great promise. By targeting common dysregulated pathways that play a key effector role in retinal damage (e.g., mitochondrial dysfunction, oxidative stress, inflammation, and neovascularization), miRNA modulation can protect retinal cells regardless of the primary etiology of the addressed disorder. Considering that the retina is an easily accessible tissue, we believe that the potential application of miRNA therapeutics in retinal disorders could rapidly move to the clinic.
SC and AI conceived the study. SC, FM, and AI wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the BrightFocus Foundation (grant no. M2020184 to SC) and the Telethon Foundation (grant no. AIMTX16 to AI).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Italian Telethon Foundation for continuous support.
Alexander, C., Votruba, M., Pesch, U. E., Thiselton, D. L., Mayer, S., Moore, A., et al. (2000). OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26, 211–215. doi: 10.1038/79944
Almasieh, M., and Levin, L. A. (2017). Neuroprotection in glaucoma: animal models and clinical trials. Annu. Rev. Vis. Sci. 3, 91–120. doi: 10.1146/annurev-vision-102016-061422
Amini-Farsani, Z., and Asgharzade, S. (2020). The impact of miR-183/182/96 gene regulation on the maturation, survival, and function of photoreceptor cells in the retina. J. Comp. Neurol. 528, 1616–1625. doi: 10.1002/cne.24833
Anasagasti, A., Ezquerra-Inchausti, M., Barandika, O., Munoz-Culla, M., Caffarel, M. M., Otaegui, D., et al. (2018). Expression profiling analysis reveals key microrna-mrna interactions in early retinal degeneration in retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 59, 2381–2392. doi: 10.1167/iovs.18-24091
Andreeva, K., and Cooper, N. G. (2014). MicroRNAs in the neural retina. Int. J. Genom. 2014:165897. doi: 10.1155/2014/165897
Aung, T., Ocaka, L., Ebenezer, N. D., Morris, A. G., Krawczak, M., Thiselton, D. L., et al. (2002). A major marker for normal tension glaucoma: association with polymorphisms in the OPA1 gene. Hum. Genet. 110, 52–56. doi: 10.1007/s00439-001-0645-7
Bai, X. Y., Ma, Y., Ding, R., Fu, B., Shi, S., and Chen, X. M. (2011). miR-335 and miR-34a promote renal senescence by suppressing mitochondrial antioxidative enzymes. J. Am. Soc. Nephrol. 22, 1252–1261. doi: 10.1681/ASN.2010040367
Bajan, S., and Hutvagner, G. (2020). RNA-based therapeutics: from antisense oligonucleotides to miRNAs. Cells 9:137. doi: 10.3390/cells9010137
Bandiera, S., Ruberg, S., Girard, M., Cagnard, N., Hanein, S., Chretien, D., et al. (2011). Nuclear outsourcing of RNA interference components to human mitochondria. PLoS One 6:e20746. doi: 10.1371/journal.pone.0020746
Baracca, A., Sgarbi, G., Mattiazzi, M., Casalena, G., Pagnotta, E., Valentino, M. L., et al. (2007). Biochemical phenotypes associated with the mitochondrial ATP6 gene mutations at nt8993. Biochim. Biophys. Acta 1767, 913–919. doi: 10.1016/j.bbabio.2007.05.005
Barbato, A., Iuliano, A., Volpe, M., D’Alterio, R., Brillante, S., Massa, F., et al. (2021). Integrated genomics identifies miR-181/TFAM pathway as a critical driver of drug resistance in melanoma. Int. J. Mol. Sci. 22:1801. doi: 10.3390/ijms22041801
Barber, A. J., Gardner, T. W., and Abcouwer, S. F. (2011). The significance of vascular and neural apoptosis to the pathology of diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 52, 1156–1163. doi: 10.1167/iovs.10-6293
Barron, M. J., Johnson, M. A., Andrews, R. M., Clarke, M. P., Griffiths, P. G., Bristow, E., et al. (2001). Mitochondrial abnormalities in ageing macular photoreceptors. Invest. Ophthalmol. Vis. Sci. 42, 3016–3022.
Bellon, A., Iyer, A., Bridi, S., Lee, F. C. Y., Ovando-Vazquez, C., Corradi, E., et al. (2017). miR-182 regulates Slit2-mediated axon guidance by modulating the local translation of a specific mRNA. Cell Rep. 18, 1171–1186. doi: 10.1016/j.celrep.2016.12.093
Berber, P., Grassmann, F., Kiel, C., and Weber, B. H. (2017). An eye on age-related macular degeneration: the role of MicroRNAs in disease pathology. Mol. Diagn. Ther. 21, 31–43. doi: 10.1007/s40291-016-0234-z
Bhattacharjee, S., Zhao, Y., Dua, P., Rogaev, E. I., and Lukiw, W. J. (2016). microRNA-34a-mediated down-regulation of the microglial-enriched triggering receptor and phagocytosis-sensor TREM2 in age-related macular degeneration. PLoS One 11:e0150211. doi: 10.1371/journal.pone.0150211
Caravia, X. M., Fanjul, V., Oliver, E., Roiz-Valle, D., Moran-Alvarez, A., Desdin-Mico, G., et al. (2018). The microRNA-29/PGC1alpha regulatory axis is critical for metabolic control of cardiac function. PLoS Biol. 16:e2006247. doi: 10.1371/journal.pbio.2006247
Carbonell, M., Alonso, N., Castelblanco, E., Real, J., Ramirez-Morros, A., Simo, R., et al. (2019). Assessment of inner retinal layers and choroidal thickness in Type 1 diabetes mellitus: a cross-sectional study. J. Clin. Med. 8:1412. doi: 10.3390/jcm8091412
Carelli, V., Ross-Cisneros, F. N., and Sadun, A. A. (2004). Mitochondrial dysfunction as a cause of optic neuropathies. Prog. Retin. Eye Res. 23, 53–89. doi: 10.1016/j.preteyeres.2003.10.003
Carrella, S., Indrieri, A., Franco, B., and Banfi, S. (2020). Mutation-independent therapies for retinal diseases: focus on gene-based approaches. Front. Neurosci. 14:588234. doi: 10.3389/fnins.2020.588234
Catuogno, S., Cerchia, L., Romano, G., Pognonec, P., Condorelli, G., and de Franciscis, V. (2013). miR-34c may protect lung cancer cells from paclitaxel-induced apoptosis. Oncogene 32, 341–351. doi: 10.1038/onc.2012.51
Chan, E. C., Liu, G. S., and Dusting, G. J. (2015). Redox mechanisms in pathological angiogenesis in the retina: roles for NADPH oxidase. Curr. Pharm. Des. 21, 5988–5998. doi: 10.2174/1381612821666151029111127
Chan, E. C., van Wijngaarden, P., Liu, G. S., Jiang, F., Peshavariya, H., and Dusting, G. J. (2013). Involvement of Nox2 NADPH oxidase in retinal neovascularization. Invest. Ophthalmol. Vis. Sci. 54, 7061–7067. doi: 10.1167/iovs.13-12883
Cheng, M., Liu, L., Lao, Y., Liao, W., Liao, M., Luo, X., et al. (2016). MicroRNA-181a suppresses parkin-mediated mitophagy and sensitizes neuroblastoma cells to mitochondrial uncoupler-induced apoptosis. Oncotarget 7, 42274–42287. doi: 10.18632/oncotarget.9786
Chi, Z. L., Akahori, M., Obazawa, M., Minami, M., Noda, T., Nakaya, N., et al. (2010). Overexpression of optineurin E50K disrupts Rab8 interaction and leads to a progressive retinal degeneration in mice. Hum. Mol. Genet. 19, 2606–2615. doi: 10.1093/hmg/ddq146
Cimmino, A., Calin, G. A., Fabbri, M., Iorio, M. V., Ferracin, M., Shimizu, M., et al. (2005). miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U.S.A. 102, 13944–13949. doi: 10.1073/pnas.0506654102
Cogliati, S., Enriquez, J. A., and Scorrano, L. (2016). Mitochondrial cristae: where beauty meets functionality. Trends Biochem. Sci. 41, 261–273. doi: 10.1016/j.tibs.2016.01.001
Colligris, P., Perez de Lara, M. J., Colligris, B., and Pintor, J. (2018). Ocular manifestations of Alzheimer’s and other neurodegenerative diseases: the prospect of the eye as a tool for the early diagnosis of Alzheimer’s disease. J. Ophthalmol. 2018:8538573. doi: 10.1155/2018/8538573
Das, S., Ferlito, M., Kent, O. A., Fox-Talbot, K., Wang, R., Liu, D., et al. (2012). Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 110, 1596–1603. doi: 10.1161/CIRCRESAHA.112.267732
Davies, V. J., Hollins, A. J., Piechota, M. J., Yip, W., Davies, J. R., White, K. E., et al. (2007). Opa1 deficiency in a mouse model of autosomal dominant optic atrophy impairs mitochondrial morphology, optic nerve structure and visual function. Hum. Mol. Genet. 16, 1307–1318. doi: 10.1093/hmg/ddm079
Doebele, C., Bonauer, A., Fischer, A., Scholz, A., Reiss, Y., Urbich, C., et al. (2010). Members of the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood 115, 4944–4950. doi: 10.1182/blood-2010-01-264812
Dong, J., Zhao, Y. P., Zhou, L., Zhang, T. P., and Chen, G. (2011). Bcl-2 upregulation induced by miR-21 via a direct interaction is associated with apoptosis and chemoresistance in MIA PaCa-2 pancreatic cancer cells. Arch. Med. Res. 42, 8–14. doi: 10.1016/j.arcmed.2011.01.006
Duno, M., Wibrand, F., Baggesen, K., Rosenberg, T., Kjaer, N., and Frederiksen, A. L. (2013). A novel mitochondrial mutation m.8989G>C associated with neuropathy, ataxia, retinitis pigmentosa - the NARP syndrome. Gene 515, 372–375. doi: 10.1016/j.gene.2012.12.066
Duraisamy, A. J., Mohammad, G., and Kowluru, R. A. (2019). Mitochondrial fusion and maintenance of mitochondrial homeostasis in diabetic retinopathy. Biochim. Biophys. Acta Mol. Basis Dis. 1865, 1617–1626. doi: 10.1016/j.bbadis.2019.03.013
Duroux-Richard, I., Roubert, C., Ammari, M., Presumey, J., Grun, J. R., Haupl, T., et al. (2016). miR-125b controls monocyte adaptation to inflammation through mitochondrial metabolism and dynamics. Blood 128, 3125–3136. doi: 10.1182/blood-2016-02-697003
Eades, G., Yang, M., Yao, Y., Zhang, Y., and Zhou, Q. (2011). miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J. Biol. Chem. 286, 40725–40733. doi: 10.1074/jbc.M111.275495
Ertekin, S., Yildirim, O., Dinc, E., Ayaz, L., Fidanci, S. B., and Tamer, L. (2014). Evaluation of circulating miRNAs in wet age-related macular degeneration. Mol. Vis. 20, 1057–1066.
Eshaq, R. S., Wright, W. S., and Harris, N. R. (2014). Oxygen delivery, consumption, and conversion to reactive oxygen species in experimental models of diabetic retinopathy. Redox Biol. 2, 661–666. doi: 10.1016/j.redox.2014.04.006
Fan, F., Zhuang, J., Zhou, P., Liu, X., and Luo, Y. (2017). MicroRNA-34a promotes mitochondrial dysfunction-induced apoptosis in human lens epithelial cells by targeting Notch2. Oncotarget 8, 110209–110220. doi: 10.18632/oncotarget.22597
Feher, J., Kovacs, I., Artico, M., Cavallotti, C., Papale, A., and Balacco Gabrieli, C. (2006). Mitochondrial alterations of retinal pigment epithelium in age-related macular degeneration. Neurobiol. Aging 27, 983–993. doi: 10.1016/j.neurobiolaging.2005.05.012
Feng, X., Jiang, J., Shi, S., Xie, H., Zhou, L., and Zheng, S. (2016). Knockdown of miR-25 increases the sensitivity of liver cancer stem cells to TRAIL-induced apoptosis via PTEN/PI3K/Akt/Bad signaling pathway. Int. J. Oncol. 49, 2600–2610. doi: 10.3892/ijo.2016.3751
Ferre, M., Amati-Bonneau, P., Tourmen, Y., Malthiery, Y., and Reynier, P. (2005). eOPA1: an online database for OPA1 mutations. Hum. Mutat. 25, 423–428. doi: 10.1002/humu.20161
Ferrington, D. A., Fisher, C. R., and Kowluru, R. A. (2020). Mitochondrial defects drive degenerative retinal diseases. Trends Mol. Med. 26, 105–118. doi: 10.1016/j.molmed.2019.10.008
Finsterer, J., Mancuso, M., Pareyson, D., Burgunder, J. M., and Klopstock, T. (2018). Mitochondrial disorders of the retinal ganglion cells and the optic nerve. Mitochondrion 42, 1–10. doi: 10.1016/j.mito.2017.10.003
Fraser, J. A., Biousse, V., and Newman, N. J. (2010). The neuro-ophthalmology of mitochondrial disease. Surv. Ophthalmol. 55, 299–334. doi: 10.1016/j.survophthal.2009.10.002
Gao, L., Jiang, B. O., Lei, D., Zhou, X., and Yuan, H. (2016). Expression profiling of microRNAs in optineurin (E50K) mutant transgenic mice. Biomed. Rep. 4, 193–196. doi: 10.3892/br.2015.565
Garzon, R., Heaphy, C. E., Havelange, V., Fabbri, M., Volinia, S., Tsao, T., et al. (2009). MicroRNA 29b functions in acute myeloid leukemia. Blood 114, 5331–5341. doi: 10.1182/blood-2009-03-211938
Gelfand, J. M., Duncan, J. L., Racine, C. A., Gillum, L. A., Chin, C. T., Zhang, Y., et al. (2011). Heterogeneous patterns of tissue injury in NARP syndrome. J. Neurol. 258, 440–448. doi: 10.1007/s00415-010-5775-1
Gueven, N., Nadikudi, M., Daniel, A., and Chhetri, J. (2017). Targeting mitochondrial function to treat optic neuropathy. Mitochondrion 36, 7–14. doi: 10.1016/j.mito.2016.07.013
Guo, Y., Chen, X., Zhang, H., Li, N., Yang, X., Cheng, W., et al. (2012). Association of OPA1 polymorphisms with NTG and HTG: a meta-analysis. PLoS One 7:e42387. doi: 10.1371/journal.pone.0042387
Heggermont, W. A., Papageorgiou, A. P., Quaegebeur, A., Deckx, S., Carai, P., Verhesen, W., et al. (2017). Inhibition of MicroRNA-146a and overexpression of its target dihydrolipoyl succinyltransferase protect against pressure overload-induced cardiac hypertrophy and dysfunction. Circulation 136, 747–761. doi: 10.1161/CIRCULATIONAHA.116.024171
Hindle, A. G., Thoonen, R., Jasien, J. V., Grange, R. M. H., Amin, K., Wise, J., et al. (2019). Identification of candidate miRNA biomarkers for glaucoma. Invest. Ophthalmol. Vis. Sci. 60, 134–146. doi: 10.1167/iovs.18-24878
Holt, I. J., Harding, A. E., Petty, R. K., and Morgan-Hughes, J. A. (1990). A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am. J. Hum. Genet. 46, 428–433.
Hondur, G., Goktas, E., Yang, X., Al-Aswad, L., Auran, J. D., Blumberg, D. M., et al. (2017). Oxidative stress-related molecular biomarker candidates for glaucoma. Invest. Ophthalmol. Vis. Sci. 58, 4078–4088. doi: 10.1167/iovs.17-22242
Houzelle, A., Dahlmans, D., Nascimento, E. B. M., Schaart, G., Jorgensen, J. A., Moonen-Kornips, E., et al. (2020). MicroRNA-204-5p modulates mitochondrial biogenesis in C2C12 myotubes and associates with oxidative capacity in humans. J. Cell Physiol. 235, 9851–9863. doi: 10.1002/jcp.29797
Hu, G., Zhao, X., Wang, J., Lv, L., Wang, C., Feng, L., et al. (2018). miR-125b regulates the drug-resistance of breast cancer cells to doxorubicin by targeting HAX-1. Oncol. Lett. 15, 1621–1629. doi: 10.3892/ol.2017.7476
Hu, H., Hone, E. A., Provencher, E. A. P., Sprowls, S. A., Farooqi, I., Corbin, D. R., et al. (2020). MiR-34a interacts with cytochrome c and shapes stroke outcomes. Sci. Rep. 10:3233. doi: 10.1038/s41598-020-59997-y
Huang, C., Cen, L. P., Liu, L., Leaver, S. G., Harvey, A. R., Cui, Q., et al. (2013). Adeno-associated virus-mediated expression of growth-associated protein-43 aggravates retinal ganglion cell death in experimental chronic glaucomatous injury. Mol. Vis. 19, 1422–1432.
Hui, Y., and Yin, Y. (2018). MicroRNA-145 attenuates high glucose-induced oxidative stress and inflammation in retinal endothelial cells through regulating TLR4/NF-kappaB signaling. Life Sci. 207, 212–218. doi: 10.1016/j.lfs.2018.06.005
Hwang, T. I. S., Lin, J.-F., Lin, Y.-C., Chen, H.-E., Chou, K.-Y., and Tsai, T.-F. (2016). miR-204 acts as a tumor suppressor in human bladder cancer cell T24 by targeting antiapoptotic BCL2. Urol. Sci. 27, 101–108. doi: 10.1016/j.urols.2016.02.004
Hyttinen, J. M. T., Blasiak, J., Niittykoski, M., Kinnunen, K., Kauppinen, A., Salminen, A., et al. (2017). DNA damage response and autophagy in the degeneration of retinal pigment epithelial cells-Implications for age-related macular degeneration (AMD). Ageing Res. Rev. 36, 64–77. doi: 10.1016/j.arr.2017.03.006
Indrieri, A., Carrella, S., Carotenuto, P., Banfi, S., and Franco, B. (2020a). The pervasive role of the miR-181 family in development, neurodegeneration, and cancer. Int. J. Mol. Sci. 21:2092. doi: 10.3390/ijms21062092
Indrieri, A., Carrella, S., Romano, A., Spaziano, A., Marrocco, E., Fernandez-Vizarra, E., et al. (2019). miR-181a/b downregulation exerts a protective action on mitochondrial disease models. EMBO Mol. Med. 11:e8734. doi: 10.15252/emmm.201708734
Indrieri, A., Pizzarelli, R., Franco, B., and De Leonibus, E. (2020b). Dopamine, alpha-synuclein, and mitochondrial dysfunctions in parkinsonian eyes. Front. Neurosci. 14:567129. doi: 10.3389/fnins.2020.567129
Ito, Y. A., and Di Polo, A. (2017). Mitochondrial dynamics, transport, and quality control: a bottleneck for retinal ganglion cell viability in optic neuropathies. Mitochondrion 36, 186–192. doi: 10.1016/j.mito.2017.08.014
Iwai, N., Yasui, K., Tomie, A., Gen, Y., Terasaki, K., Kitaichi, T., et al. (2018). Oncogenic miR-96-5p inhibits apoptosis by targeting the caspase-9 gene in hepatocellular carcinoma. Int. J. Oncol. 53, 237–245. doi: 10.3892/ijo.2018.4369
Izzotti, A., Ceccaroli, C., Longobardi, M. G., Micale, R. T., Pulliero, A., La Maestra, S., et al. (2015). Molecular damage in glaucoma: from anterior to posterior eye segment. The MicroRNA role. Microrna 4, 3–17. doi: 10.2174/2211536604666150707124640
Jarrett, S. G., and Boulton, M. E. (2012). Consequences of oxidative stress in age-related macular degeneration. Mol. Aspects Med. 33, 399–417. doi: 10.1016/j.mam.2012.03.009
Jayaram, H., Cepurna, W. O., Johnson, E. C., and Morrison, J. C. (2015). MicroRNA expression in the glaucomatous retina. Invest. Ophthalmol. Vis. Sci. 56, 7971–7982. doi: 10.1167/iovs.15-18088
Jiang, Y., Sang, Y., and Qiu, Q. (2017). microRNA-383 mediates high glucose-induced oxidative stress and apoptosis in retinal pigment epithelial cells by repressing peroxiredoxin 3. Am. J. Transl. Res. 9, 2374–2383.
Jing, X., Yang, J., Jiang, L., Chen, J., and Wang, H. (2018). MicroRNA-29b regulates the mitochondria-dependent apoptotic pathway by targeting bax in doxorubicin cardiotoxicity. Cell Physiol. Biochem. 48, 692–704. doi: 10.1159/000491896
Kang, T., Lu, W., Xu, W., Anderson, L., Bacanamwo, M., Thompson, W., et al. (2013). MicroRNA-27 (miR-27) targets prohibitin and impairs adipocyte differentiation and mitochondrial function in human adipose-derived stem cells. J. Biol. Chem. 288, 34394–34402. doi: 10.1074/jbc.M113.514372
Kao, Y. Y., Chou, C. H., Yeh, L. Y., Chen, Y. F., Chang, K. W., Liu, C. J., et al. (2019). MicroRNA miR-31 targets SIRT3 to disrupt mitochondrial activity and increase oxidative stress in oral carcinoma. Cancer Lett. 456, 40–48. doi: 10.1016/j.canlet.2019.04.028
Karali, M., and Banfi, S. (2019). Non-coding RNAs in retinal development and function. Hum. Genet. 138, 957–971. doi: 10.1007/s00439-018-1931-y
Karali, M., Persico, M., Mutarelli, M., Carissimo, A., Pizzo, M., Singh Marwah, V., et al. (2016). High-resolution analysis of the human retina miRNome reveals isomiR variations and novel microRNAs. Nucleic Acids Res. 44, 1525–1540. doi: 10.1093/nar/gkw039
Kim, J., Fiesel, F. C., Belmonte, K. C., Hudec, R., Wang, W. X., Kim, C., et al. (2016). miR-27a and miR-27b regulate autophagic clearance of damaged mitochondria by targeting PTEN-induced putative kinase 1 (PINK1). Mol. Neurodegener. 11:55. doi: 10.1186/s13024-016-0121-4
Kong, G. Y., Van Bergen, N. J., Trounce, I. A., and Crowston, J. G. (2009). Mitochondrial dysfunction and glaucoma. J. Glaucoma 18, 93–100. doi: 10.1097/IJG.0b013e318181284f
Kong, N., Lu, X., and Li, B. (2014). Downregulation of microRNA-100 protects apoptosis and promotes neuronal growth in retinal ganglion cells. BMC Mol. Biol. 15:25. doi: 10.1186/s12867-014-0025-1
Lee, H., Tak, H., Park, S. J., Jo, Y. K., Cho, D. H., and Lee, E. K. (2017). microRNA-200a-3p enhances mitochondrial elongation by targeting mitochondrial fission factor. BMB Rep. 50, 214–219. doi: 10.5483/bmbrep.2017.50.4.006
Lee, M. R., Mantel, C., Lee, S. A., Moon, S. H., and Broxmeyer, H. E. (2016). MiR-31/SDHA axis regulates reprogramming efficiency through mitochondrial metabolism. Stem Cell Rep. 7, 1–10. doi: 10.1016/j.stemcr.2016.05.012
Lee, S., Van Bergen, N. J., Kong, G. Y., Chrysostomou, V., Waugh, H. S., O’Neill, E. C., et al. (2011). Mitochondrial dysfunction in glaucoma and emerging bioenergetic therapies. Exp. Eye Res. 93, 204–212. doi: 10.1016/j.exer.2010.07.015
Lefevere, E., Toft-Kehler, A. K., Vohra, R., Kolko, M., Moons, L., and Van Hove, I. (2017). Mitochondrial dysfunction underlying outer retinal diseases. Mitochondrion 36, 66–76. doi: 10.1016/j.mito.2017.03.006
Lenaers, G., Hamel, C., Delettre, C., Amati-Bonneau, P., Procaccio, V., Bonneau, D., et al. (2012). Dominant optic atrophy. Orphanet. J. Rare Dis. 7:46. doi: 10.1186/1750-1172-7-46
Leung, A. K. L. (2015). The whereabouts of microRNA actions: cytoplasm and beyond. Trends Cell Biol. 25, 601–610. doi: 10.1016/j.tcb.2015.07.005
Li, G., Luna, C., Qiu, J., Epstein, D. L., and Gonzalez, P. (2009). Alterations in microRNA expression in stress-induced cellular senescence. Mech. Ageing Dev. 130, 731–741. doi: 10.1016/j.mad.2009.09.002
Li, H., Liu, J., Wang, Y., Fu, Z., Huttemann, M., Monks, T. J., et al. (2017). MiR-27b augments bone marrow progenitor cell survival via suppressing the mitochondrial apoptotic pathway in Type 2 diabetes. Am. J. Physiol. Endocrinol. Metab. 313, E391–E401. doi: 10.1152/ajpendo.00073.2017
Li, H. J., Pan, Y. B., Sun, Z. L., Sun, Y. Y., Yang, X. T., and Feng, D. F. (2018). Inhibition of miR-21 ameliorates excessive astrocyte activation and promotes axon regeneration following optic nerve crush. Neuropharmacology 137, 33–49. doi: 10.1016/j.neuropharm.2018.04.028
Li, H. J., Sun, Z. L., Pan, Y. B., Sun, Y. Y., Xu, M. H., and Feng, D. F. (2019). Inhibition of miRNA-21 promotes retinal ganglion cell survival and visual function by modulating Muller cell gliosis after optic nerve crush. Exp. Cell Res. 375, 10–19. doi: 10.1016/j.yexcr.2019.01.009
Li, J., Hui, L., Kang, Q., and Li, R. (2018). Down-regulation of microRNA-27b promotes retinal pigment epithelial cell proliferation and migration by targeting Nox2. Pathol. Res. Pract. 214, 925–933. doi: 10.1016/j.prp.2018.05.025
Li, K. K., Pang, J. C., Lau, K. M., Zhou, L., Mao, Y., Wang, Y., et al. (2013). MiR-383 is downregulated in medulloblastoma and targets peroxiredoxin 3 (PRDX3). Brain Pathol. 23, 413–425. doi: 10.1111/bpa.12014
Li, P., Shen, M., Gao, F., Wu, J., Zhang, J., Teng, F., et al. (2017). An antagomir to microRNA-106b-5p ameliorates cerebral ischemia and reperfusion injury in rats via inhibiting apoptosis and oxidative stress. Mol. Neurobiol. 54, 2901–2921. doi: 10.1007/s12035-016-9842-1
Li, R., Yan, G., Li, Q., Sun, H., Hu, Y., Sun, J., et al. (2012). MicroRNA-145 protects cardiomyocytes against hydrogen peroxide (H(2)O(2))-induced apoptosis through targeting the mitochondria apoptotic pathway. PLoS One 7:e44907. doi: 10.1371/journal.pone.0044907
Liang, F. Q., and Godley, B. F. (2003). Oxidative stress-induced mitochondrial DNA damage in human retinal pigment epithelial cells: a possible mechanism for RPE aging and age-related macular degeneration. Exp. Eye Res. 76, 397–403. doi: 10.1016/s0014-4835(03)00023-x
Liu, W., and Shomron, N. (2021). Analysis of microRNA regulation in single cells. Methods Mol. Biol. 2243, 339–354. doi: 10.1007/978-1-0716-1103-6_18
Liu, Y., Chen, Y., Wang, Y., Zhang, X., Gao, K., Chen, S., et al. (2018). microRNA profiling in glaucoma eyes with varying degrees of optic neuropathy by using next-generation sequencing. Invest. Ophthalmol. Vis. Sci. 59, 2955–2966. doi: 10.1167/iovs.17-23599
Lu, Z., Li, S., Zhao, S., and Fa, X. (2016). Upregulated miR-17 regulates hypoxia-mediated human pulmonary artery smooth muscle cell proliferation and apoptosis by targeting mitofusin 2. Med. Sci. Monit. 22, 3301–3308. doi: 10.12659/msm.900487
Lukiw, W. J., Surjyadipta, B., Dua, P., and Alexandrov, P. N. (2012). Common micro RNAs (miRNAs) target complement factor H (CFH) regulation in Alzheimer’s disease (AD) and in age-related macular degeneration (AMD). Int. J. Biochem. Mol. Biol. 3, 105–116.
Luna, C., Li, G., Huang, J., Qiu, J., Wu, J., Yuan, F., et al. (2012). Regulation of trabecular meshwork cell contraction and intraocular pressure by miR-200c. PLoS One 7:e51688. doi: 10.1371/journal.pone.0051688
Luna, C., Li, G., Qiu, J., Epstein, D. L., and Gonzalez, P. (2009). Role of miR-29b on the regulation of the extracellular matrix in human trabecular meshwork cells under chronic oxidative stress. Mol. Vis. 15, 2488–2497.
Luna, C., Li, G., Qiu, J., Epstein, D. L., and Gonzalez, P. (2011). Cross-talk between miR-29 and transforming growth factor-betas in trabecular meshwork cells. Invest. Ophthalmol. Vis. Sci. 52, 3567–3572. doi: 10.1167/iovs.10-6448
Mak, H. K., Yung, J. S. Y., Weinreb, R. N., Ng, S. H., Cao, X., Ho, T. Y. C., et al. (2020). MicroRNA-19a-PTEN axis is involved in the developmental decline of axon regenerative capacity in retinal ganglion cells. Mol. Ther. Nucleic Acids 21, 251–263. doi: 10.1016/j.omtn.2020.05.031
Marchi, S., Lupini, L., Patergnani, S., Rimessi, A., Missiroli, S., Bonora, M., et al. (2013). Downregulation of the mitochondrial calcium uniporter by cancer-related miR-25. Curr. Biol. 23, 58–63. doi: 10.1016/j.cub.2012.11.026
Marrocco, E., Indrieri, A., Esposito, F., Tarallo, V., Carboncino, A., Alvino, F. G., et al. (2020). alpha-synuclein overexpression in the retina leads to vision impairment and degeneration of dopaminergic amacrine cells. Sci. Rep. 10:9619. doi: 10.1038/s41598-020-66497-6
Martinez, B., and Peplow, P. V. (2021). MicroRNAs as diagnostic and prognostic biomarkers of age-related macular degeneration: advances and limitations. Neural Regen. Res. 16, 440–447. doi: 10.4103/1673-5374.293131
Mastropasqua, R., Toto, L., Cipollone, F., Santovito, D., Carpineto, P., and Mastropasqua, L. (2014). Role of microRNAs in the modulation of diabetic retinopathy. Prog. Retin. Eye Res. 43, 92–107. doi: 10.1016/j.preteyeres.2014.07.003
Menard, C., Rezende, F. A., Miloudi, K., Wilson, A., Tetreault, N., Hardy, P., et al. (2016). MicroRNA signatures in vitreous humour and plasma of patients with exudative AMD. Oncotarget 7, 19171–19184. doi: 10.18632/oncotarget.8280
Meyerson, C., Van Stavern, G., and McClelland, C. (2015). Leber hereditary optic neuropathy: current perspectives. Clin. Ophthalmol. 9, 1165–1176. doi: 10.2147/OPTH.S62021
Mirzaei, N., Shi, H., Oviatt, M., Doustar, J., Rentsendorj, A., Fuchs, D. T., et al. (2020). Alzheimer’s retinopathy: seeing disease in the eyes. Front. Neurosci. 14:921. doi: 10.3389/fnins.2020.00921
Mitter, S. K., Song, C., Qi, X., Mao, H., Rao, H., Akin, D., et al. (2014). Dysregulated autophagy in the RPE is associated with increased susceptibility to oxidative stress and AMD. Autophagy 10, 1989–2005. doi: 10.4161/auto.36184
Miyawaki, T., Koto, S., Ishihara, H., Goto, Y., Nishino, I., Kanda, F., et al. (2015). [A case of neurologic muscle weakness, ataxia, and retinitis pigmentosa (NARP) syndrome with a novel mitochondrial mutation m.8729 G>A]. Rinsho Shinkeigaku 55, 91–95. doi: 10.5692/clinicalneurol.55.91
Mizutani, M., Kern, T. S., and Lorenzi, M. (1996). Accelerated death of retinal microvascular cells in human and experimental diabetic retinopathy. Clin. Invest. 97, 2883–2890. doi: 10.1172/JCI118746
Mohamed, J. S., Hajira, A., Pardo, P. S., and Boriek, A. M. (2014). MicroRNA-149 inhibits PARP-2 and promotes mitochondrial biogenesis via SIRT-1/PGC-1alpha network in skeletal muscle. Diabetes 63, 1546–1559. doi: 10.2337/db13-1364
Moschos, M. M., Dettoraki, M., Karekla, A., Lamprinakis, I., Damaskos, C., Gouliopoulos, N., et al. (2020). Polymorphism analysis of miR182 and CDKN2B genes in Greek patients with primary open angle glaucoma. PLoS One 15:e0233692. doi: 10.1371/journal.pone.0233692
Mott, J. L., Kobayashi, S., Bronk, S. F., and Gores, G. J. (2007). mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 26, 6133–6140. doi: 10.1038/sj.onc.1210436
Muluhngwi, P., Alizadeh-Rad, N., Vittitow, S. L., Kalbfleisch, T. S., and Klinge, C. M. (2017). The miR-29 transcriptome in endocrine-sensitive and resistant breast cancer cells. Sci. Rep. 7:5205. doi: 10.1038/s41598-017-05727-w
Natoli, R., and Fernando, N. (2018). MicroRNA as therapeutics for age-related macular degeneration. Adv. Exp. Med. Biol. 1074, 37–43. doi: 10.1007/978-3-319-75402-4_5
Newman, N. J. (2005). Hereditary optic neuropathies: from the mitochondria to the optic nerve. Am. J. Ophthalmol. 140, 517–523. doi: 10.1016/j.ajo.2005.03.017
Nijtmans, L. G., Henderson, N. S., Attardi, G., and Holt, I. J. (2001). Impaired ATP synthase assembly associated with a mutation in the human ATP synthase subunit 6 gene. J. Biol. Chem. 276, 6755–6762. doi: 10.1074/jbc.M008114200
Nishi, H., Ono, K., Iwanaga, Y., Horie, T., Nagao, K., Takemura, G., et al. (2010). MicroRNA-15b modulates cellular ATP levels and degenerates mitochondria via Arl2 in neonatal rat cardiac myocytes. J. Biol. Chem. 285, 4920–4930. doi: 10.1074/jbc.M109.082610
Niyazov, D. M., Kahler, S. G., and Frye, R. E. (2016). Primary mitochondrial disease and secondary mitochondrial dysfunction: importance of distinction for diagnosis and treatment. Mol. Syndromol. 7, 122–137. doi: 10.1159/000446586
O’Connell, R. M., Taganov, K. D., Boldin, M. P., Cheng, G., and Baltimore, D. (2007). MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 104, 1604–1609. doi: 10.1073/pnas.0610731104
Ouyang, Y. B., Lu, Y., Yue, S., and Giffard, R. G. (2012). miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion 12, 213–219. doi: 10.1016/j.mito.2011.09.001
Palfi, A., Hokamp, K., Hauck, S. M., Vencken, S., Millington-Ward, S., Chadderton, N., et al. (2016). microRNA regulatory circuits in a mouse model of inherited retinal degeneration. Sci. Rep. 6:31431. doi: 10.1038/srep31431
Paylakhi, S. H., Moazzeni, H., Yazdani, S., Rassouli, P., Arefian, E., Jaberi, E., et al. (2013). FOXC1 in human trabecular meshwork cells is involved in regulatory pathway that includes miR-204, MEIS2, and ITGbeta1. Exp. Eye Res. 111, 112–121. doi: 10.1016/j.exer.2013.03.009
Piro-Megy, C., Sarzi, E., Tarres-Sole, A., Pequignot, M., Hensen, F., Quiles, M., et al. (2020). Dominant mutations in mtDNA maintenance gene SSBP1 cause optic atrophy and foveopathy. J. Clin. Invest. 130, 143–156. doi: 10.1172/JCI128513
Pogue, A. I., and Lukiw, W. J. (2018). Up-regulated Pro-inflammatory MicroRNAs (miRNAs) in Alzheimer’s disease (AD) and age-related macular degeneration (AMD). Cell. Mol. Neurobiol. 38, 1021–1031. doi: 10.1007/s10571-017-0572-3
Purohit, P. K., and Saini, N. (2021). Mitochondrial microRNA (MitomiRs) in cancer and complex mitochondrial diseases: current status and future perspectives. Cell. Mol. Life Sci 78, 1405–1421. doi: 10.1007/s00018-020-03670-0
Pusparajah, P., Lee, L. H., and Abdul Kadir, K. (2016). Molecular markers of diabetic retinopathy: potential screening tool of the future? Front. Physiol. 7:200. doi: 10.3389/fphys.2016.00200
Qing, S., Yuan, S., Yun, C., Hui, H., Mao, P., Wen, F., et al. (2014). Serum miRNA biomarkers serve as a fingerprint for proliferative diabetic retinopathy. Cell. Physiol. Biochem. 34, 1733–1740. doi: 10.1159/000366374
Quigley, H. A., and Broman, A. T. (2006). The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol. 90, 262–267. doi: 10.1136/bjo.2005.081224
Quinones-Lombrana, A., and Blanco, J. G. (2015). Chromosome 21-derived hsa-miR-155-5p regulates mitochondrial biogenesis by targeting Mitochondrial Transcription Factor A (TFAM). Biochim. Biophys. Acta 1852, 1420–1427. doi: 10.1016/j.bbadis.2015.04.004
Ragusa, M., Caltabiano, R., Russo, A., Puzzo, L., Avitabile, T., Longo, A., et al. (2013). MicroRNAs in vitreus humor from patients with ocular diseases. Mol. Vis. 19, 430–440.
Rao, G., Dwivedi, S. K. D., Zhang, Y., Dey, A., Shameer, K., Karthik, R., et al. (2020). MicroRNA-195 controls MICU1 expression and tumor growth in ovarian cancer. EMBO Rep. 21:e48483. doi: 10.15252/embr.201948483
Rezaie, T., Child, A., Hitchings, R., Brice, G., Miller, L., Coca-Prados, M., et al. (2002). Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science 295, 1077–1079. doi: 10.1126/science.1066901
Rivetti di Val Cervo, P., Lena, A. M., Nicoloso, M., Rossi, S., Mancini, M., Zhou, H., et al. (2012). p63-microRNA feedback in keratinocyte senescence. Proc. Natl. Acad. Sci. U.S.A. 109, 1133–1138. doi: 10.1073/pnas.1112257109
Saxena, K., Rutar, M. V., Provis, J. M., and Natoli, R. C. (2015). Identification of miRNAs in a model of retinal degenerations. Invest. Ophthalmol. Vis. Sci. 56, 1820–1829. doi: 10.1167/iovs.14-15449
Sears, N. C., Boese, E. A., Miller, M. A., and Fingert, J. H. (2019). Mendelian genes in primary open angle glaucoma. Exp. Eye Res. 186:107702. doi: 10.1016/j.exer.2019.107702
Shafabakhsh, R., Aghadavod, E., Mobini, M., Heidari-Soureshjani, R., and Asemi, Z. (2019). Association between microRNAs expression and signaling pathways of inflammatory markers in diabetic retinopathy. J. Cell. Physiol. 234, 7781–7787. doi: 10.1002/jcp.27685
Shao, Y., Dong, L. J., Takahashi, Y., Chen, J., Liu, X., Chen, Q., et al. (2019). miRNA-451a regulates RPE function through promoting mitochondrial function in proliferative diabetic retinopathy. Am. J. Physiol. Endocrinol. Metab. 316, E443–E452. doi: 10.1152/ajpendo.00360.2018
Sharma, L. K., Tiwari, M., Rai, N. K., and Bai, Y. (2019). Mitophagy activation repairs Leber’s hereditary optic neuropathy-associated mitochondrial dysfunction and improves cell survival. Hum. Mol. Genet. 28, 422–433. doi: 10.1093/hmg/ddy354
Shen, L., Chen, L., Zhang, S., Du, J., Bai, L., Zhang, Y., et al. (2016). MicroRNA-27b regulates mitochondria biogenesis in myocytes. PLoS One 11:e0148532. doi: 10.1371/journal.pone.0148532
Shim, M. S., Takihara, Y., Kim, K. Y., Iwata, T., Yue, B. Y., Inatani, M., et al. (2016). Mitochondrial pathogenic mechanism and degradation in optineurin E50K mutation-mediated retinal ganglion cell degeneration. Sci. Rep. 6:33830. doi: 10.1038/srep33830
Singh, L. N., Crowston, J. G., Lopez Sanchez, M. I. G., Van Bergen, N. J., Kearns, L. S., Hewitt, A. W., et al. (2018). Mitochondrial DNA variation and disease susceptibility in primary open-angle glaucoma. Invest. Ophthalmol. Vis. Sci. 59, 4598–4602. doi: 10.1167/iovs.18-25085
Singh, R., and Saini, N. (2012). Downregulation of BCL2 by miRNAs augments drug-induced apoptosis–a combined computational and experimental approach. J. Cell Sci. 125(Pt 6), 1568–1578. doi: 10.1242/jcs.095976
Song, S., Seo, H. H., Lee, S. Y., Lee, C. Y., Lee, J., Yoo, K. J., et al. (2015). MicroRNA-17-mediated down-regulation of apoptotic protease activating factor 1 attenuates apoptosome formation and subsequent apoptosis of cardiomyocytes. Biochem. Biophys. Res. Commun. 465, 299–304. doi: 10.1016/j.bbrc.2015.08.028
Soundara Pandi, S. P., Chen, M., Guduric-Fuchs, J., Xu, H., and Simpson, D. A. (2013). Extremely complex populations of small RNAs in the mouse retina and RPE/choroid. Invest. Ophthalmol. Vis. Sci. 54, 8140–8151. doi: 10.1167/iovs.13-12631
Su, Q., Xu, Y., Cai, R., Dai, R., Yang, X., Liu, Y., et al. (2021). miR-146a inhibits mitochondrial dysfunction and myocardial infarction by targeting cyclophilin D. Mol. Ther. Nucleic Acids 23, 1258–1271. doi: 10.1016/j.omtn.2021.01.034
Sugioka, R., Shimizu, S., and Tsujimoto, Y. (2004). Fzo1, a protein involved in mitochondrial fusion, inhibits apoptosis. J. Biol. Chem. 279, 52726–52734. doi: 10.1074/jbc.M408910200
Sundaresan, P., Simpson, D. A., Sambare, C., Duffy, S., Lechner, J., Dastane, A., et al. (2015). Whole-mitochondrial genome sequencing in primary open-angle glaucoma using massively parallel sequencing identifies novel and known pathogenic variants. Genet. Med. 17, 279–284. doi: 10.1038/gim.2014.121
Szemraj, M., Bielecka-Kowalska, A., Oszajca, K., Krajewska, M., Gos, R., Jurowski, P., et al. (2015). Serum MicroRNAs as potential biomarkers of AMD. Med. Sci. Monit. 21, 2734–2742. doi: 10.12659/MSM.893697
Taganov, K. D., Boldin, M. P., Chang, K. J., and Baltimore, D. (2006). NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 103, 12481–12486. doi: 10.1073/pnas.0605298103
Tanaka, H., Sasayama, T., Tanaka, K., Nakamizo, S., Nishihara, M., Mizukawa, K., et al. (2013). MicroRNA-183 upregulates HIF-1alpha by targeting isocitrate dehydrogenase 2 (IDH2) in glioma cells. J. Neurooncol. 111, 273–283. doi: 10.1007/s11060-012-1027-9
Tanaka, Y., Tsuda, S., Kunikata, H., Sato, J., Kokubun, T., Yasuda, M., et al. (2014). Profiles of extracellular miRNAs in the aqueous humor of glaucoma patients assessed with a microarray system. Sci. Rep. 4:5089. doi: 10.1038/srep05089
Thounaojam, M. C., Jadeja, R. N., Warren, M., Powell, F. L., Raju, R., Gutsaeva, D., et al. (2019). MicroRNA-34a (miR-34a) mediates retinal endothelial cell premature senescence through mitochondrial dysfunction and loss of antioxidant activities. Antioxidants (Basel) 8:328. doi: 10.3390/antiox8090328
Tribble, J. R., Vasalauskaite, A., Redmond, T., Young, R. D., Hassan, S., Fautsch, M. P., et al. (2019). Midget retinal ganglion cell dendritic and mitochondrial degeneration is an early feature of human glaucoma. Brain Commun. 1:fcz035. doi: 10.1093/braincomms/fcz035
Tseng, H. C., Riday, T. T., McKee, C., Braine, C. E., Bomze, H., Barak, I., et al. (2015). Visual impairment in an optineurin mouse model of primary open-angle glaucoma. Neurobiol. Aging 36, 2201–2212. doi: 10.1016/j.neurobiolaging.2015.02.012
Tsujimoto, T., Mori, T., Houri, K., Onodera, Y., Takehara, T., Shigi, K., et al. (2020). miR-155 inhibits mitophagy through suppression of BAG5, a partner protein of PINK1. Biochem. Biophys. Res. Commun. 523, 707–712. doi: 10.1016/j.bbrc.2020.01.022
Villarreal, G. Jr., Oh, D. J., Kang, M. H., and Rhee, D. J. (2011). Coordinated regulation of extracellular matrix synthesis by the microRNA-29 family in the trabecular meshwork. Invest. Ophthalmol. Vis. Sci. 52, 3391–3397. doi: 10.1167/iovs.10-6165
Vives-Bauza, C., Anand, M., Shiraz, A. K., Magrane, J., Gao, J., Vollmer-Snarr, H. R., et al. (2008). The age lipid A2E and mitochondrial dysfunction synergistically impair phagocytosis by retinal pigment epithelial cells. J. Biol. Chem. 283, 24770–24780. doi: 10.1074/jbc.M800706200
Wang, C., You, Q., Cao, X., Guo, H., Gao, X., and Peng, X. (2017). Micro RNA-19a suppresses interleukin-10 in peripheral B cells of patients with diabetic retinopathy. Am. J. Transl. Res. 9, 1410–1417.
Wang, H., Li, J., Chi, H., Zhang, F., Zhu, X., Cai, J., et al. (2015). MicroRNA-181c targets Bcl-2 and regulates mitochondrial morphology in myocardial cells. J. Cell. Mol. Med. 19, 2084–2097. doi: 10.1111/jcmm.12563
Wang, L., Huang, H., Fan, Y., Kong, B., Hu, H., Hu, K., et al. (2014). Effects of downregulation of microRNA-181a on H2O2-induced H9c2 cell apoptosis via the mitochondrial apoptotic pathway. Oxid. Med. Cell Longev. 2014:960362. doi: 10.1155/2014/960362
Wang, N., Yang, W., Xiao, T., Miao, Z., Luo, W., You, Z., et al. (2018). Possible role of miR-204 in optic nerve injury through the regulation of GAP-43. Mol. Med. Rep. 17, 3891–3897. doi: 10.3892/mmr.2017.8341
Wang, Y. G., Li, L., Liu, C. H., Hong, S., and Zhang, M. J. (2014). Peroxiredoxin 3 is resistant to oxidation-induced apoptosis of Hep-3b cells. Clin. Transl. Oncol. 16, 561–566. doi: 10.1007/s12094-013-1117-y
Wei, N., Xiao, L., Xue, R., Zhang, D., Zhou, J., Ren, H., et al. (2016). MicroRNA-9 mediates the cell apoptosis by targeting Bcl2l11 in ischemic stroke. Mol. Neurobiol. 53, 6809–6817. doi: 10.1007/s12035-015-9605-4
Weng, H., Huang, H., Dong, B., Zhao, P., Zhou, H., and Qu, L. (2014). Inhibition of miR-17 and miR-20a by oridonin triggers apoptosis and reverses chemoresistance by derepressing BIM-S. Cancer Res. 74, 4409–4419. doi: 10.1158/0008-5472.CAN-13-1748
Williams, P. A., Harder, J. M., Foxworth, N. E., Cochran, K. E., Philip, V. M., Porciatti, V., et al. (2017). Vitamin B3 modulates mitochondrial vulnerability and prevents glaucoma in aged mice. Science 355, 756–760. doi: 10.1126/science.aal0092
Wong, Y. C., and Holzbaur, E. L. (2014). Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. U.S.A. 111, E4439–E4448. doi: 10.1073/pnas.1405752111
Wu, H., Li, Z., Wang, Y., Yang, P., Li, Z., Li, H., et al. (2016). MiR-106b-mediated Mfn2 suppression is critical for PKM2 induced mitochondrial fusion. Am. J. Cancer Res. 6, 2221–2234.
Wu, J. H., Gao, Y., Ren, A. J., Zhao, S. H., Zhong, M., Peng, Y. J., et al. (2012). Altered microRNA expression profiles in retinas with diabetic retinopathy. Ophthalmic Res. 47, 195–201. doi: 10.1159/000331992
Wu, M. Z., Cheng, W. C., Chen, S. F., Nieh, S., O’Connor, C., Liu, C. L., et al. (2017). miR-25/93 mediates hypoxia-induced immunosuppression by repressing cGAS. Nat. Cell Biol. 19, 1286–1296. doi: 10.1038/ncb3615
Wu, R., Tang, S., Wang, M., Xu, X., Yao, C., and Wang, S. (2016). MicroRNA-497 induces apoptosis and suppresses proliferation via the Bcl-2/Bax-Caspase9-caspase3 pathway and cyclin D2 protein in HUVECs. PLoS One 11:e0167052. doi: 10.1371/journal.pone.0167052
Wu, T., Chen, W., Kong, D., Li, X., Lu, H., Liu, S., et al. (2015). miR-25 targets the modulator of apoptosis 1 gene in lung cancer. Carcinogenesis 36, 925–935. doi: 10.1093/carcin/bgv068
Xie, X., Hu, Y., Xu, L., Fu, Y., Tu, J., Zhao, H., et al. (2015). The role of miR-125b-mitochondria-caspase-3 pathway in doxorubicin resistance and therapy in human breast cancer. Tumour Biol. 36, 7185–7194. doi: 10.1007/s13277-015-3438-7
Xu, C., Shi, L., Chen, W., Fang, P., Li, J., Jin, L., et al. (2017). MiR-106b inhibitors sensitize TRAIL-induced apoptosis in hepatocellular carcinoma through increase of death receptor 4. Oncotarget 8, 41921–41931. doi: 10.18632/oncotarget.16707
Xu, Y., Fang, F., Zhang, J., Josson, S., St Clair, W. H., and St Clair, D. K. (2010). miR-17∗ suppresses tumorigenicity of prostate cancer by inhibiting mitochondrial antioxidant enzymes. PLoS One 5:e14356. doi: 10.1371/journal.pone.0014356
Xue, T., Wei, L., Zha, D. J., Qiu, J. H., Chen, F. Q., Qiao, L., et al. (2016). miR-29b overexpression induces cochlear hair cell apoptosis through the regulation of SIRT1/PGC-1alpha signaling: Implications for age-related hearing loss. Int. J. Mol. Med. 38, 1387–1394. doi: 10.3892/ijmm.2016.2735
Yadav, S., Pandey, A., Shukla, A., Talwelkar, S. S., Kumar, A., Pant, A. B., et al. (2011). miR-497 and miR-302b regulate ethanol-induced neuronal cell death through BCL2 protein and cyclin D2. J. Biol. Chem. 286, 37347–37357. doi: 10.1074/jbc.M111.235531
Yan, L., Lee, S., Lazzaro, D. R., Aranda, J., Grant, M. B., and Chaqour, B. (2015). Single and compound knock-outs of microRNA (miRNA)-155 and its angiogenic gene target CCN1 in mice alter vascular and neovascular growth in the retina via resident microglia. J. Biol. Chem. 290, 23264–23281. doi: 10.1074/jbc.M115.646950
Yao, J., Zhou, E., Wang, Y., Xu, F., Zhang, D., and Zhong, D. (2014). microRNA-200a inhibits cell proliferation by targeting mitochondrial transcription factor A in breast cancer. DNA Cell Biol. 33, 291–300. doi: 10.1089/dna.2013.2132
Yu-Wai-Man, P., Griffiths, P. G., and Chinnery, P. F. (2011). Mitochondrial optic neuropathies - disease mechanisms and therapeutic strategies. Prog. Retin. Eye Res. 30, 81–114. doi: 10.1016/j.preteyeres.2010.11.002
Yu-Wai-Man, P., Stewart, J. D., Hudson, G., Andrews, R. M., Griffiths, P. G., Birch, M. K., et al. (2010). OPA1 increases the risk of normal but not high tension glaucoma. J. Med. Genet. 47, 120–125. doi: 10.1136/jmg.2009.067512
Yu-Wai-Man, P., Turnbull, D. M., and Chinnery, P. F. (2002). Leber hereditary optic neuropathy. J. Med. Genet. 39, 162–169. doi: 10.1136/jmg.39.3.162
Yu-Wai-Man, P., Votruba, M., Moore, A. T., and Chinnery, P. F. (2014). Treatment strategies for inherited optic neuropathies: past, present and future. Eye (Lond.) 28, 521–537. doi: 10.1038/eye.2014.37
Zaninello, M., Palikaras, K., Naon, D., Iwata, K., Herkenne, S., Quintana-Cabrera, R., et al. (2020). Inhibition of autophagy curtails visual loss in a model of autosomal dominant optic atrophy. Nat. Commun. 11:4029. doi: 10.1038/s41467-020-17821-1
Zhang, C., Nie, P., Zhou, C., Hu, Y., Duan, S., Gu, M., et al. (2021). Oxidative stress-induced mitophagy is suppressed by the miR-106b-93-25 cluster in a protective manner. Cell Death Dis. 12:209. doi: 10.1038/s41419-021-03484-3
Zhang, H., Zuo, Z., Lu, X., Wang, L., Wang, H., and Zhu, Z. (2012). MiR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol. Rep. 27, 594–598. doi: 10.3892/or.2011.1530
Zhang, L., Fang, Y., Zhao, X., Zheng, Y., Ma, Y., Li, S., et al. (2021). miR-204 silencing reduces mitochondrial autophagy and ROS production in a murine AD model via the TRPML1-activated STAT3 pathway. Mol. Ther. Nucleic Acids 24, 822–831. doi: 10.1016/j.omtn.2021.02.010
Zhang, R., Garrett, Q., Zhou, H., Wu, X., Mao, Y., Cui, X., et al. (2017). Upregulation of miR-195 accelerates oxidative stress-induced retinal endothelial cell injury by targeting mitofusin 2 in diabetic rats. Mol. Cell Endocrinol. 452, 33–43. doi: 10.1016/j.mce.2017.05.009
Zhang, X., Ji, R., Liao, X., Castillero, E., Kennel, P. J., Brunjes, D. L., et al. (2018). MicroRNA-195 regulates metabolism in failing myocardium via alterations in Sirtuin 3 expression and mitochondrial protein acetylation. Circulation 137, 2052–2067. doi: 10.1161/CIRCULATIONAHA.117.030486
Zhao, D., Chen, Y., Chen, S., Zheng, C., Hu, J., and Luo, S. (2017). MiR-19a regulates the cell growth and apoptosis of osteosarcoma stem cells by targeting PTEN. Tumour Biol. 39:1010428317705341. doi: 10.1177/1010428317705341
Zheng, M., and Xiao, R. P. (2010). Role of mitofusin 2 in cardiovascular oxidative injury. J. Mol. Med. (Berl.) 88, 987–991. doi: 10.1007/s00109-010-0675-5
Zhong, Q., and Kowluru, R. A. (2011). Diabetic retinopathy and damage to mitochondrial structure and transport machinery. Invest. Ophthalmol. Vis. Sci. 52, 8739–8746. doi: 10.1167/iovs.11-8045
Zhou, Y., Jiang, H., Gu, J., Tang, Y., Shen, N., and Jin, Y. (2013). MicroRNA-195 targets ADP-ribosylation factor-like protein 2 to induce apoptosis in human embryonic stem cell-derived neural progenitor cells. Cell Death Dis. 4:e695. doi: 10.1038/cddis.2013.195
Zhu, J. N., Fu, Y. H., Hu, Z. Q., Li, W. Y., Tang, C. M., Fei, H. W., et al. (2017). Activation of miR-34a-5p/Sirt1/p66shc pathway contributes to doxorubicin-induced cardiotoxicity. Sci. Rep. 7:11879. doi: 10.1038/s41598-017-12192-y
Zhu, W., Xu, H., Zhu, D., Zhi, H., Wang, T., Wang, J., et al. (2012). miR-200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer Chemother. Pharmacol. 69, 723–731. doi: 10.1007/s00280-011-1752-3
Zhuang, Z., Xiao, Q., Hu, H., Tian, S. Y., Lu, Z. J., Zhang, T. Z., et al. (2015). Down-regulation of microRNA-155 attenuates retinal neovascularization via the PI3K/Akt pathway. Mol. Vis. 21, 1173–1184.
Zitman-Gal, T., Green, J., Pasmanik-Chor, M., Golan, E., Bernheim, J., and Benchetrit, S. (2014). Vitamin D manipulates miR-181c, miR-20b and miR-15a in human umbilical vein endothelial cells exposed to a diabetic-like environment. Cardiovasc. Diabetol. 13:8. doi: 10.1186/1475-2840-13-8
Keywords: microRNA, retina, mitochondria, mitochondrial diseases, glaucoma, AMD, diabetic retinopathy, MitomiR
Citation: Carrella S, Massa F and Indrieri A (2021) The Role of MicroRNAs in Mitochondria-Mediated Eye Diseases. Front. Cell Dev. Biol. 9:653522. doi: 10.3389/fcell.2021.653522
Received: 14 January 2021; Accepted: 20 May 2021;
Published: 18 June 2021.
Edited by:
Ruth Ashery Padan, Tel Aviv University, IsraelReviewed by:
Salil A. Lachke, University of Delaware, United StatesCopyright © 2021 Carrella, Massa and Indrieri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sabrina Carrella, Y2FycmVsbGFAdGlnZW0uaXQ=; Alessia Indrieri, aW5kcmllcmlAdGlnZW0uaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.