- 1Department of Hematology, The Seventh Affiliated Hospital, Sun Yat-sen University, Shenzhen, China
- 2Shenzhen Lansi Institute of Artificial Intelligence in Medicine, Shenzhen, China
- 3The First Affiliated Hospital of Shenzhen University, Health Science Center, Shenzhen Second People’s Hospital, Shenzhen, China
- 4RNA Biomedical Institute, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China
- 5Key Laboratory of Stem Cells and Tissue Engineering, Zhongshan School of Medicine, Sun Yat-sen University, Ministry of Education, Guangzhou, China
Bone marrow mesenchymal stem cells (MSCs) are widely used clinically due to their versatile roles in multipotency, immunomodulation, and hematopoietic stem cell (HSC) niche function. However, cellular heterogeneity limits MSCs in the consistency and efficacy of their clinical applications. Metabolism regulates stem cell function and fate decision; however, how metabolites regulate the functional heterogeneity of MSCs remains elusive. Here, using single-cell RNA sequencing, we discovered that fatty acid pathways are involved in the regulation of lineage commitment and functional heterogeneity of MSCs. Functional assays showed that a fatty acid metabolite, butyrate, suppressed the self-renewal, adipogenesis, and osteogenesis differentiation potential of MSCs with increased apoptosis. Conversely, butyrate supplement significantly promoted HSC niche factor expression in MSCs, which suggests that butyrate supplement may provide a therapeutic approach to enhance their HSC niche function. Overall, our work demonstrates that metabolites are essential to regulate the functional heterogeneity of MSCs.
Introduction
Mesenchymal stem cells (MSCs) are multipotent fibroblast colony-forming cells, which can differentiate into adipocytes, chondrocytes, and osteocytes (Friedenstein et al., 1974; Horwitz et al., 2005; Uccelli et al., 2008; Bianco et al., 2013; Zhou et al., 2014). Bone marrow MSCs generate osteoblasts and provide the major source of bone formation during development and regeneration after bone damage (Kassem et al., 2008; Ye et al., 2012; Valenti et al., 2016; Pajarinen et al., 2019). Furthermore, bone marrow MSCs secrete multiple growth factors to support hematopoietic stem cells (HSCs) for their maintenance and regeneration (Mendez-Ferrer et al., 2010; Zhou et al., 2017). Recent works show that bone marrow MSCs also contribute to immunomodulation against infection and autoimmune diseases as well as tissue repair, such as skin and blood vessels (Sun et al., 2009; Zhang et al., 2015; Shook et al., 2020).
The versatile functions of MSCs enable increasing clinical applications of MSCs to treat diseases such as graft versus host disease (GvHD), Crohn’s disease, heart failure, bone marrow failure syndrome, and osteogenesis imperfecta and bone fractures (Galipeau and Sensebe, 2018; Andrzejewska et al., 2019; Martin et al., 2019; Yin et al., 2019). MSC infusion is also applied for facilitating implantation after HSC transplantation (Mendez-Ferrer et al., 2010; Zhou et al., 2017). However, the diverse therapeutic targets challenge the clinical trials of MSCs in which the cell functional heterogenicity, culture methods, and expansion levels could potentially influence the therapeutic consistency and limit clinic efficacy of MSCs (Pattappa et al., 2013; Moll et al., 2014; Yuan et al., 2019).
The cellular metabolism profile controls stem cell fates in self-renewal, lineage commitment, and terminal differentiation (Ryall et al., 2015a; Teslaa and Teitell, 2015; Garcia-Prat et al., 2017). Metabolites, such as acetyl-coenzyme A (Wang et al., 2009), α-ketoglutarate (Hwang et al., 2016), S-adenosylmethionine, and S-adenosylhomocysteine (Shyh-Chang et al., 2013), regulate the proliferation and differentiation of embryonic stem cells (ESCs). The successful reprogramming of induced pluripotent stem cells (iPSCs) requires a metabolic shift from oxidative phosphorylation to anaerobic glycolysis (Yoshida et al., 2009; Folmes et al., 2011). Muscle stem cells reside in an aerobic niche near capillaries (Christov et al., 2007), and transition into committed progenitors is accompanied by a switch from fatty acid (FA) oxidation to glycolysis (Ryall et al., 2015b). Furthermore, metabolic status regulates MSCs in cell fate determination and multifunction maintenance. Bone marrow MSCs reside in a hypoxic niche and rely on anaerobic glycolysis to maintain their self-renewal and multipotency and evade senescence (Ito and Suda, 2014). Glutaminase, the key enzyme in glutamine metabolism, and 5-methoxytryptophan, a tryptophan metabolite, promote osteogenic and suppress adipogenic MSC differentiation (Chang et al., 2017; Yu et al., 2019). On the contrary, arachidonic FA induces MSC adipogenesis but inhibits osteogenesis in human MSC cultures (Casado-Diaz et al., 2013). Unsaturated FAs, such as linoleic and oleic acids, inhibit MSC proliferation and induce the expression of angiogenesis mediators, such as IL-6, IL-8, VEGF, and nitric oxide (Smith et al., 2012).
Recent works show that butyrate, a natural short-chain fatty acid (SCFA) produced by mammalian intestinal microbiota, can inhibit histone deacetylase (HDAC) activity and impairs intestinal epithelial stem/progenitors in wound repair in vivo (Kaiko et al., 2016) but promotes iPSC reprogramming efficiency (Liang et al., 2010). However, the role of butyrate in regulating MSCs remains elusive. Here, using single-cell RNA sequencing (scRNA-seq), we identified FA pathways that are involved in lineage commitment of MSCs, and further functional assays prove that metabolite butyrate alters MSC cell fate in self-renewal, apoptosis, and HSC niche factor expression.
Materials and Methods
Mice
C57BL/6 mice were obtained from the Jackson Laboratory and were maintained in the C57BL/6 background. All animal experiments were performed according to protocols approved by the institutional animal care and use committee.
Bone Marrow Digestion
Bone marrow digestion was performed as described with small changes (Zhou et al., 2014). In brief, intact bone marrow from mice at the age of 6–8 weeks were flushed from mouse femora and tibiae and subjected to two rounds of enzymatic digestion at 37°C for 20 min each. The digestion buffer contained 0.2 mg/ml liberase (Roche) and 200 μg/ml DNAse I (Roche) in 1 × HBSS with calcium and magnesium. The digested marrow cells then underwent red blood cell lysis using 0.16 M ammonium chloride solution.
Flow Cytometry and Cell Sorting
For cell sorting and analysis, monoclonal antibodies to CD45 (30-F11, Biolegend), Ter-119 (TER-119, eBioscience), PDGFRα (APA5, eBioscience), CD31 (MEC13.3, Biolegend), CD51 (RMV-7, Biolegend), and 7AAD (Biolegend) were used where indicated. Cell sorting was performed using a cell sorter (MoFlo Astrios). Cell analysis was performed on a flow cytometer (Attune NxT, Thermo Fisher).
scRNA-seq
Sorted CD45–Ter-119–CD31–PDGFα+CD51+ single cells were processed through the Chromium Single Cell Platform using the Chromium Single Cell 3′ Library and Gel Bead Kit v3 (10X Genomics, PN-1000075) and the Chromium Single Cell B Chip Kit (10X Genomics, PN-1000074) as the manufacturer’s protocol. In brief, 15,000 cells were loaded onto the chromium instrument to generate single-cell barcoded droplets. Cells were lysed and barcoded reverse transcription of RNA was occurred. Libraries were prepared by following the amplification, fragmentation, adaptor, and index attachment and then sequenced on an Illumina NovaSeq platform. The scRNA-seq data generated in this study are deposited in GEO (GSE1670351).
scRNA-seq Data Processing
The scRNA-seq reads were aligned to the mm10 reference genomes, and unique molecular identifier (UMI) counts were obtained by CellRanger 3.0.2. Normalization, dimensionality reduction, and clustering were performed with the Seurat 3.0 R package (Butler et al., 2018) on RStudio, and 3005 of 11,888 cells with Ptprc (CD45) expression were excluded to remove potential contamination. Cells were filtered to have > 200 and < 7000 detected genes and less than 5% of total UMIs mapping to the mitochondrial genome. Data set normalization was performed by dividing the UMI counts per gene by the total UMI counts in the corresponding cell and log-transforming, followed by the results scaling and centering. Cells underwent dimensionality reduction with the t-stochastic neighboring embedding (tSNE) method and partition-based graph abstraction (PAGA) using scanpy (Wolf et al., 2018). Integration of published MSC and immune cell scRNA-seq with our data was performed by Seurat with the function SCTransform() and Harmony algorithm. Signature genes of each cluster were obtained using the Seurat function FindMarkers with the Wilcox test. All correlations were calculated based on values with the function cor() and the parameter “method = ‘spearman’.” The pseudotime trajectory was analyzed by monocle2 based on the Seurat clustering (Qiu et al., 2017). A signature gene heat map was generated by pheatmap R packages. GOChord plots and GOClust plots were generated by GOplot R packages. Gene set enrichment analysis (GSEA) was performed using the gsea R package (Subramanian et al., 2005). Gene lists were preranked by the fold change values of the differential expression analysis using Seurat. GSEA plots were generated by enrichplot R packages. Gene sets were obtained from the gene ontology database as indicated. Signature genes feature plots and violin plots were generated with Seurat R packages.
CFU-F Assay and MSC in vitro Differentiation Assay
For the CFU-F assay, freshly isolated marrow cells were plated at a density of 5 × 105 cells/well in six-well plates with DMEM (Corning) plus 20% fetal bovine serum (Gibco), 10 μM ROCK inhibitor (Selleck), and 1% penicillin/streptomycin (Hyclone). Cell cultures were maintained at 37°C, 5% O2, and 5% CO2 chambers. CFU-F colonies were counted after 7 days of culture by staining with crystal violet (Sangon). For the in vitro differentiation assay, we enzyme-digested CFU-Fs and subcloned them into six-well plates at a density of 1 × 105 cells/well. Cells then underwent adipogenic (7 days) or osteogenic (14 days) differentiation with StemPro Differentiation Kits (Gibco). Adipogenic differentiation was quantified by Oil Red O staining (Sigma). Osteogenic differentiation was quantified by Alizarin Red S (Sigma). In indicated groups, butyrate (500 nM, 5 μM, and 500 μM as indicated), SAHA (Vorinostat, 1 μM), Z-VAD-FMK (30 μM), or Necrostatin-1 (45 μM) were supplemented into the culture medium.
qPCR
For qPCR, CFU cells were dissociated in Trizol (Magen), and RNA was extracted following the manufacture’s instruction. RNA was reverse transcribed into cDNA using the TransScript All-in-One First-Strand cDNA Synthesis kit (Transgene). Quantitative PCR was performed using a Bio-Rad CFX 96 touch. The primers for Runx2 were 5′ GACTGTGGTTACCGTCATGGC 3′ (forward) and 5′ ACTTGGTTTTTCATAACAGCGGA 3′ (reverse). The primers for Ocn were 5′ CAGACACCATGAGGAC CATC 3′ (forward) and 5′ GGACTGAGGCTCTGTGAGT 3′ (reverse). The primers for Col1a1 were 5′ GCTCCTCTTAGGG GCCACT 3′ (forward) and 5′ ATTGGGGACCCTTAGGCCAT 3′ (reverse). The primers for Fabp4 were 5′ AAGGTGAAGA GCATCATAACCCT 3′ (forward) and 5′ TCACGCCTTTC ATAACACATTCC 3′ (reverse). The primers for Adiponectin were 5′ TGTTCCTCTTAATCCTGCCCA 3′ (forward) and 5′ CCAACCTGCACAAGTTCCCTT 3′ (reverse). The primers for Cebpa were 5′ GCGGGAACGCAACAACATC 3′ (forward) and 5′ GTCACTGGTCAACTCCAGCAC 3′ (reverse). The primers for Cmyc were 5′ ATGCCCCTCAACGTGAACTTC 3′ (forward) and 5′ GTCGCAGATGAAATAGGGCTG 3′ (reverse). The primers for Ccnb1 were 5′ AAGGTGCCTGT GTGTGAACC 3′ (forward) and 5′ GTCAGCCCCATCATCT GCG 3′ (reverse). The primers for Ccnd1 were 5′ GCGTACC CTGACACCAATCTC 3′ (forward) and 5′ CTCCTCTTCGCAC TTCTGCTC 3′ (reverse). The primers for Bcl2 were 5′ GTCG CTACCGTCGTGACTTC 3′ (forward) and 5′ CAGACATGCA CCTACCCAGC 3′ (reverse). The primers for Bax were 5′ TG AAGACAGGGGCCTTTTTG 3′ (forward) and AATTCGCCG GAGACAC TCG 3′ (reverse). The primers for Bak1 were 5′ CAACCCCGAGATGGACAACTT 3′ (forward) and 5′ CGTAG CGCCGGTTAATATCAT 3′ (reverse). The primers for Bid were 5′ GCCGAGCACATCACAGACC 3′ (forward) and 5′ TGGCAATGTTGTGGATGATTTCT 3′ (reverse). The primers for Kitl were 5′ AGGAACGGAACAGAAAGG 3′ (forward) and 5′ GTCGGATAGACTTCACTTGG 3′ (reverse). The primers for Angpt1 were 5′ CACATAGGGTGCAGCAACCA 3′ (forward) and 5′ CGTCGTGTTCTGAAGAATGA 3′ (reverse). The primers for Cxcl12 were 5′ AGGTTCTTATTTCACGG CTTGT 3′ (forward) and 5′ TGGGTGCTGAGACCTTTGAT 3′ (reverse). The primers for Jag2 were 5′ CAATGACACCACTC CAGATGAG 3′ (forward) and 5′ GGCCAAAGAAGTCGT TGCG 3′ (reverse).
Cell Counting Kit-8 (CCK-8) and Lactate Dehydrogenase (LDH) Activity Assay
Freshly isolated marrow cells were plated at a density of 2 × 104 cells/well in a 96-well plate and cultured with or without butyrate, ZVAD, or Nec-1 as indicated for 24 h in 37°C, 5% O2, and 5% CO2 chambers. For the CCK-8 assay, 10 μl CCK-8 reagent (Solarbio) was added in wells and continued incubated for 4 h in chambers. The optical density was measured at 450 nm using a microplate reader. LDH activities were performed following the manufacturer’s instructions (Njjcbio). Cell death frequency was calculated as LDH release into cell culture medium dividing LDH in the total cell lysate.
Statistical Analyses
Data are presented as mean ± s.e.m. For multiple comparisons analysis, data were analyzed by repeated-measures one-way ANOVA followed by Dunnett’s test. The difference was considered statistically significant if p < 0.05, and  P < 0.05,
P < 0.05, 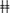 P < 0.01,
P < 0.01,  P < 0.001. For other experiments, except for scRNA-seq analysis, data were analyzed by two-tailed Student’s t test. The difference was considered statistically significant if p < 0.05, and ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
P < 0.001. For other experiments, except for scRNA-seq analysis, data were analyzed by two-tailed Student’s t test. The difference was considered statistically significant if p < 0.05, and ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Results
Single-Cell Atlas Identifies the Heterogeneity of MSCs
To explore the heterogeneity of bone marrow MSCs, we applied droplet-based scRNA-seq with non-hematopoietic (CD45–Ter-119–), non-endothelial (CD31–) bone marrow cells that express MSC markers (PDGFα+CD51+) (Morikawa et al., 2009; Yue et al., 2016; Boulais et al., 2018; Lee et al., 2018) (Figures 1A,B). We successfully detected a total of 8883 cells with an average of ∼2000 genes per cell in bone marrow MSCs (Supplementary Figure 1A). We then employed t-SNE and identified 11 clusters in MSCs (Figure 1C). Results of PAGA (Figure 1D) and enriched gene ontology (GO) (Figure 1E) annotated the 11 clusters into six populations, including pre-MSCs, adipogenic MSCs, chondrogenic MSCs, osteogenic cells, angiogenic MSCs, and immunomodulating MSCs (Figure 1F). Pre-MSCs, constituted of clusters 6 and 8, enriched pathways in protein transport, nuclear transport, and ribosome biogenesis, which are critical for MSC lineage commitment (Herencia et al., 2012; Chen et al., 2015). Adipogenic MSCs, chondrogenic MSCs, and osteogenic cells, constituted of clusters 1 and 5, cluster 3, and clusters 4 and 9, enriched pathways in fat cell differentiation, chondrocyte development, and bone development, respectively. Angiogenic MSCs were constituted of cluster 2, enriching pathways in endothelial and epithelial cell migration. Immunomodulating MSCs were constituted by clusters 7, 10, and 11, which enriched pathways associated with leukocyte proliferation and myeloid cell homeostasis (Figure 1E). Furthermore, these immunomodulating MSCs overlapped with other MSC subpopulations and published bone marrow MSCs (Baryawno et al., 2019; Leimkuhler et al., 2020) but not with immune cells (Choi et al., 2019) (Figure 1G). In line with this, immunomodulating MSCs did not express any immune cell markers, such as T cells, B cells, NK cells, and macrophages (Supplementary Figure 1B). This ruled out the potential contamination of immune cells in this immunomodulating MSC population. Furthermore, adipogenic MSCs, chondrogenic MSCs, and osteogenic cells were referred to as lineage-committed MSCs. Consistently, our unsupervised trajectory analysis by Monocle 2 showed that the pre-MSCs clustered in the root of the trajectory, and adipogenic, chondrogenic, osteogenic, and angiogenic MSCs clustered in different branches (Figure 1H).
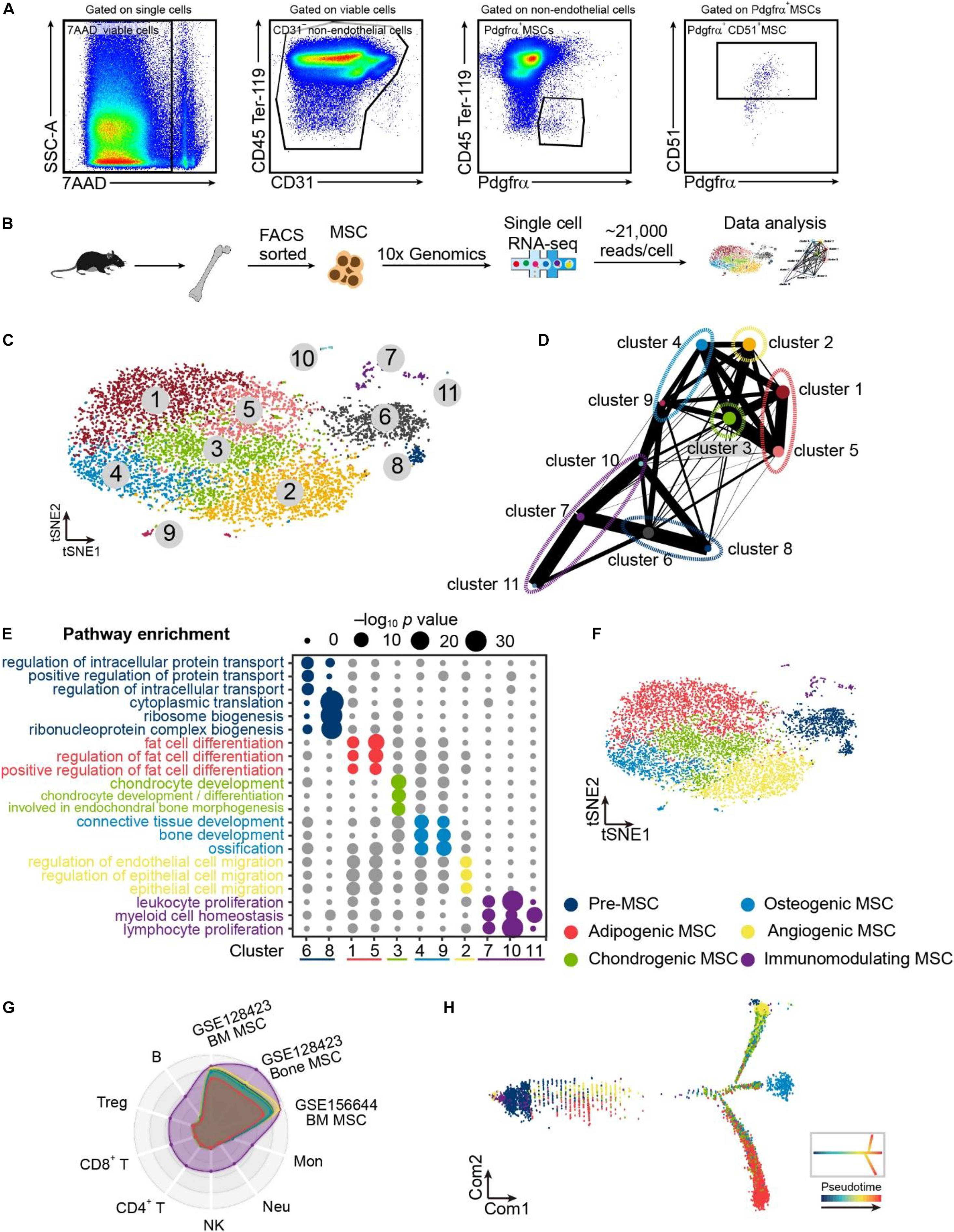
Figure 1. Single-cell atlas identifies heterogeneous MSC populations. (A) Flow cytometry gating for isolating bone marrow MSCs. (B) Schematic depicting the strategy of cell isolation, MSC sorting, single-cell RNA sequencing, and data analysis. (C) Clustering 8883 cells from MSCs by tSNE. (D) Results of PAGA. Each node represents a cluster, and edges show the connectivity between clusters. The size of nodes indicates the number of cells in each cluster, and the thickness of the edges denotes the connectivity from low (thin) to high (thick). (E) Eleven clusters annotating by gene sets of GO terms based on their signatures and dividing into pre-, adipogenic, chondrogenic, osteogenic, angiogenic, and immunomodulating MSCs. (F) Clustering 8883 cells from bone marrow MSCs annotating six populations inferred from PAGA results and enriched pathways of marker genes. (G) Radar chart showing the spearman rho between the MSC scRNA-seq result in this study compared with the published MSC and immune cell scRNA-seq results. Each ring represents one population in (F). (H) Developmental pseudotime of MSCs. The inset depicts the schema of pseudotime of MSCs.
Overall, using scRNA-seq, we identified pre-MSCs and lineage-committed MSC clusters and revealed their potential regulatory mechanisms in MSC lineage commitment.
Single-Cell Atlas Identifies Pre-MSCs and Lineage-Committed MSCs
Our scRNA-seq data shows that pre-MSCs significantly enriched stemness genes (such as Hp1bp3 and Baz1b) and FA metabolic genes (such as Eci2 and Pam) (Satani et al., 2003; Dutta et al., 2014; van Weeghel et al., 2014; Zanella et al., 2019). Conversely, lineage-committed MSCs dominantly enriched differentiation genes (such as Col27a1 and Jund) and proliferation genes (such as Fgfr1 and Mafb) (Naito et al., 2005; Naba et al., 2017; Ardizzone et al., 2020; Guo et al., 2020) (Figure 2A). This is consistent with previous reports that multipotent MSCs are quiescent (Mendez-Ferrer et al., 2010; Zhou et al., 2014). We further found that pre-MSCs highly enriched ribosome biogenesis and cellular respiration-associated genes, such as Rps24, Rpl35a, and Ndufb3 (Choesmel et al., 2008; Narla et al., 2011; Alston et al., 2016) (Figures 2B,C). These indicate that energy metabolism and protein synthesis control are essential for stem cell maintenance (Teslaa and Teitell, 2015; Wanet et al., 2015; Blanco et al., 2016; Sanchez et al., 2016). However, lineage-committed MSCs highly enriched proliferation, lineage commitment, and apoptosis genes, such as Kmt2e, Sox9, and Acvr1 (Chakkalakal et al., 2012; Zhang et al., 2017; Liu C.F. et al., 2018) (Figures 2D,E).
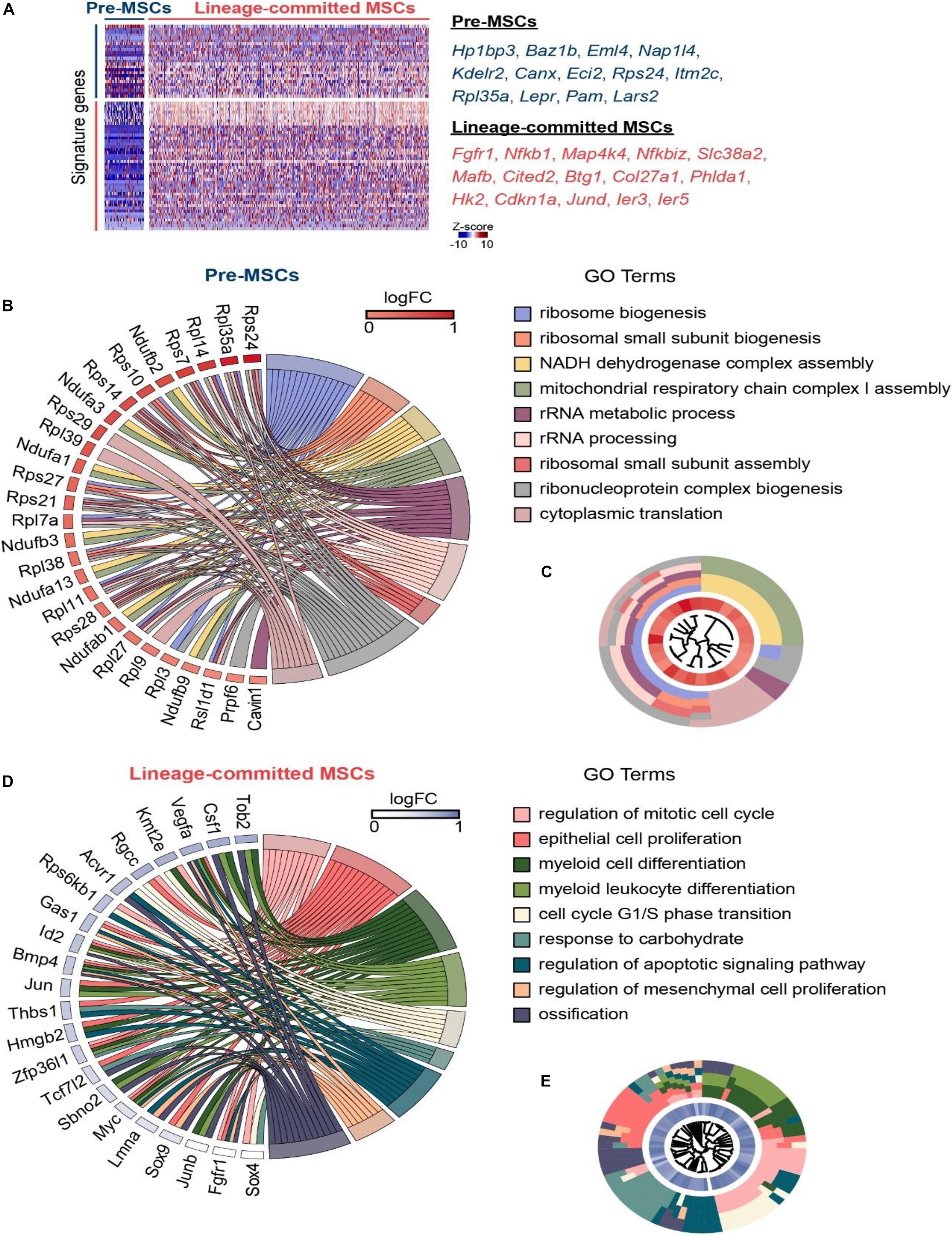
Figure 2. Single-cell atlas identifies pre-MSCs and lineage-committed MSCs. (A) Heat map of signature genes in pre- or lineage-committed MSCs; exemplar genes are listed on right. Columns denote cells; rows denote genes. (B) GOChord plot showing the association between statistically significant genes in pre-MSCs and their associated GO terms; the genes are associated via ribbons to their assigned terms. White-to-red coding next to the selected genes indicates the gene expression fold change. The outer ring shows the assigned functional terms. (C) GOCluster plot showing the clustering of the pre-MSC signature gene expression profiles. The inner ring shows the color-coded expression fold changes, and the outer ring shows the assigned functional terms as indicated in (B). (D) GOChord plot showing the association between statistically significant genes in lineage-committed MSCs and their associated GO terms; the genes are associated via ribbons to their assigned terms. White-to-blue coding next to the selected genes indicates the gene expression fold change. The outer ring shows the assigned functional terms. (E) GOCluster plot showing the clustering of the lineage-committed MSC signature gene expression profiles. The inner ring shows the color-coded expression fold changes, and the outer ring shows the assigned functional terms as indicated in (D).
Overall, these findings indicate the distinguished regulation mechanisms for multipotent pre-MSCs and lineage-committed MSCs.
The FA Metabolic Process Regulates MSC Lineage Commitment
To explore the underlying mechanism in regulating different MSC clusters, we performed GSEA on our scRNA-seq data between pre-MSCs and lineage-committed MSCs using gene sets in the GO database (Ashburner et al., 2000; The Gene Ontology Consortium, 2019). As expected, pre-MSCs had much less osteoblast cell differentiation (GO: 0001649) and fat cell differentiation (GO: 0045444) genes compared with lineage-committed MSCs (NES = –1.60 and –1.56, respectively, Figures 3A,B). Furthermore, we discovered that pre-MSCs are relatively quiescent as they had much less activated cell cycle (GO: 0045787) genes (NES = –1.45, Figure 3C). We next investigated how metabolites regulate pre-MSCs and lineage-committed MSCs in their stem cell fate decision. The GSEAs showed that pre-MSCs had less activated genes under the carbohydrate metabolic process (GO: 0005975) compared with lineage-committed MSCs (NES = –1.28) but were not significantly different under glycogen (GO: 0005977) or amino acid metabolic process (GO: 0006520) (Figures 3D,E). However, pre-MSCs significantly enriched genes under the FA metabolic process (GO: 0006631) (NES = 1.61, Figure 3F). We also confirmed that FA metabolic genes are much enriched in pre-MSCs compared with lineage-committed MSCs, such as Pam, Cyp1b1, and Lpl (Satani et al., 2003; Rozovski et al., 2015; Bushkofsky et al., 2016) (Figure 3G). These indicate that the FA metabolic process might contribute to the regulation of pre-MSCs and lineage-committed MSCs through their distinguished metabolic profile.
Figure 3. The FA metabolic process regulates MSC lineage commitment. (A,B) GSEA evaluating enrichment of osteoblast (A) or fat cell differentiation genes (B) in pre- and lineage-committed MSCs. Normalized enrichment score and adjusted p-value were calculated by permutation tests. (C) GSEA evaluating enrichment of cell cycle genes in pre- and lineage-committed MSCs. Normalized enrichment score and adjusted p-value were calculated by permutation tests. (D,E) GSEA evaluating enrichment of carbohydrate (D), glycogen, and amino acid metabolic process genes (E) in pre- and lineage-committed MSCs. Normalized enrichment score and adjusted p value were calculated by permutation tests. (F) GSEA evaluating enrichment of FA metabolic process genes in pre- and lineage-committed MSCs. Normalized enrichment score and adjusted p-value were calculated by permutation tests. (G) Feature plots and violin plots showing the selected fatty acid metabolic process genes expression in 8883 cells.
Overall, our scRNA-seq data indicate a novel potential role of MSCs responding to FA treatment, which could be applied to in vitro culture.
Supplement of Butyrate Suppresses Self-Renewal and Differentiation Potential of Bone Marrow MSCs
To investigate whether supplement butyrate influences the maintenance of MSCs during in vitro culture, we performed a bone marrow MSC CFU-F assay and found that supplement of butyrate at biological serum concentration (500 nM) (Tyagi et al., 2018) or higher concentration (5 μM and 500 μM) effectively reduced the CFU-F clone numbers compared with vehicle control (33.5%, 31.7%, and 60.8% decreased, respectively; Figures 4A,B). These show that butyrate suppressed MSC proliferation and self-renewal potential.
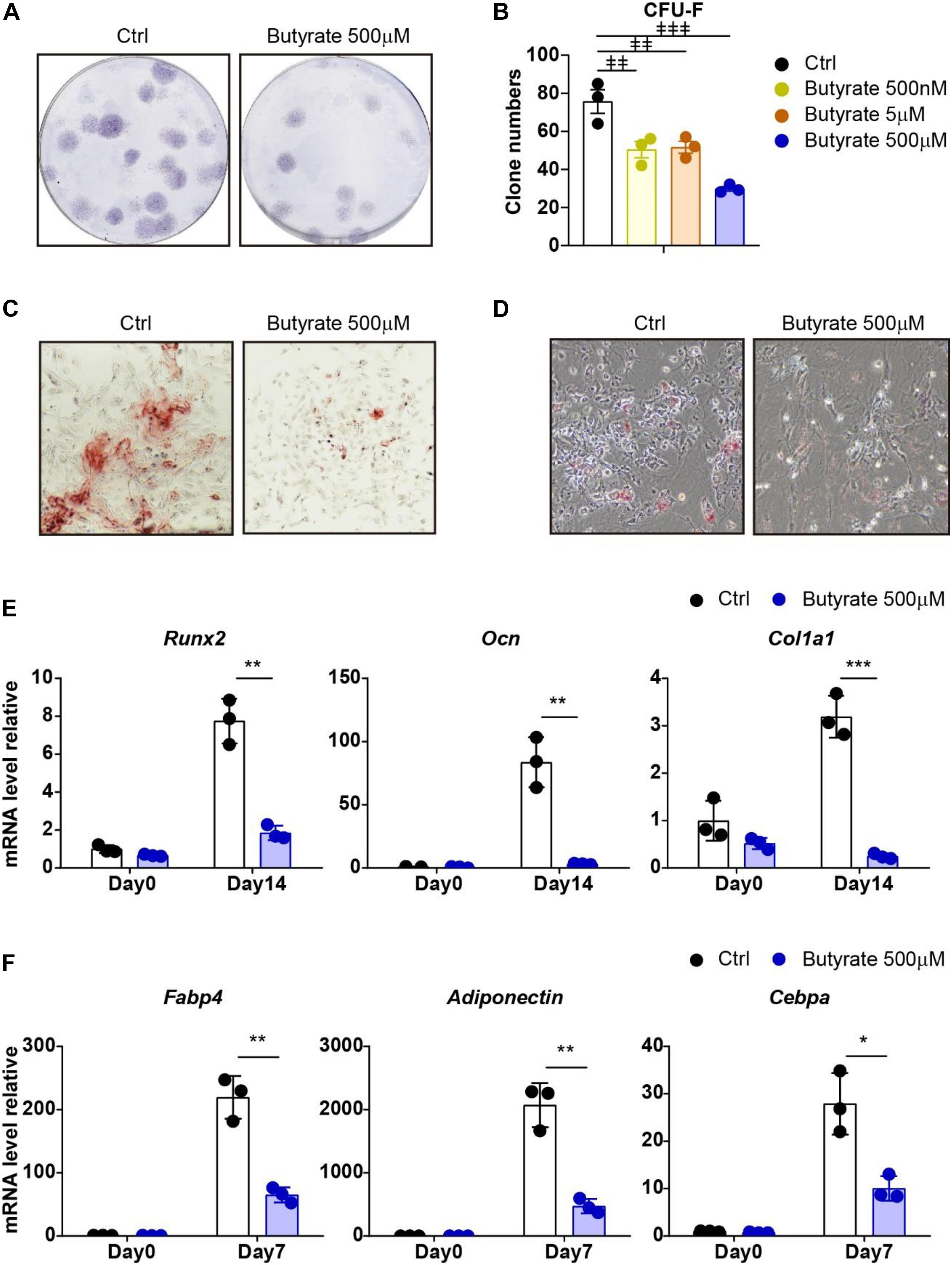
Figure 4. Supplement of butyrate suppresses self-renewal and differentiation potential of MSCs. (A) Representative wells of CFU-F colonies stained with crystal violet (5 × 105 bone marrow cells were initially plated). (B) CFU-F colony numbers after butyrate treatment as indicated doses or with vehicle control treatment as indicated (5 × 105 bone marrow cells were initially plated). (C) Osteogenic differentiation of MSCs with or without butyrate treatment. Alizarin red S staining of MSCs cultured in osteogenic differentiation medium at day 14. (D) Adipogenic differentiation of MSCs with or without butyrate treatment. Oil Red O staining of MSCs cultured in adipogenic differentiation medium at day 7. (E) Quantitative RT-PCR of transcript levels of osteogenic genes (normalized to day 0 control group). (F) Quantitative RT-PCR of transcript levels of adipogenic genes (normalized to day 0 control group). Repeated-measures one-way ANOVA followed by Dunnett’s test for multiple comparisons in (B). 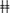 p < 0.01,
p < 0.01,  p < 0.001. Two-tailed Student’s t tests were used to assess statistical significance in (E,F). *p < 0.05, **p < 0.01, ***p < 0.001.
p < 0.001. Two-tailed Student’s t tests were used to assess statistical significance in (E,F). *p < 0.05, **p < 0.01, ***p < 0.001.
We next asked whether butyrate impairs the self-renewal potential of MSCs due to enhanced differentiation. To this aim, we performed an ex vivo differentiation assay to examine the osteogenesis and adipogenesis capacities of MSCs. Interestingly, butyrate-treated MSCs have reduced osteogenic and adipogenic differentiation ability (Figures 4C,D). Consistently, the block of osteogenic differentiation ability was confirmed by multiple osteogenic-specific marker genes, such as Runx2, Ocn, and Col1a1, which were markedly increased after osteogenic differentiation (7.75-fold, 83.7-fold, and 3.19-fold increased, respectively, compared with undifferentiated MSCs) in control MSCs but were inhibited in butyrate-treated MSCs (76.1%, 96.2%, and 92.3% decreased, respectively, compared with differentiated control MSCs, Figures 4C,E). Furthermore, the adipogenic-specific marker genes, such as Fabp4, Adiponectin, and Cebpa, were increased in control MSCs (219-fold, 2071-fold, and 27.9-fold increased, respectively, compared with undifferentiated MSCs) but were dramatically compromised in butyrate-treated MSCs (70.3%, 77.2%, and 64.0% decreased, respectively, compared with differentiated control MSCs; Figures 4D,F).
Overall, our data show that supplement of butyrate during in vitro culture impaired MSCs in their self-renewal potential and differentiation abilities.
Supplement of Butyrate Triggers Apoptosis but Promotes HSC Niche Factor Expression in Bone Marrow MSCs
To investigate the potential mechanisms of butyrate treatment on MSCs, we performed 7AAD cell death staining to examine whether butyrate treatment influences their survival. We found that butyrate induced cell death in MSCs at a high concentration (500 μM, 2.67-fold increased) but not at low concentration (5 μM) or biological serum concentration (500 nM) (Figures 5A,B). We next asked whether butyrate induced MSC death through apoptosis or necroptosis and found that apoptosis inhibitor Z-VAD-FMK (ZVAD) but not necroptosis inhibitor Necrostatin-1 (Nec-1) completely blocked butyrate-induced cell death in MSCs (87.4%) (Figures 5A,B). Furthermore, we found that butyrate-induced cell growth inhibition (38.5% decreased) was also markedly rescued by ZVAD (53.6%) (Figure 5C). The consistent results were also observed in an LDH assay in which butyrate treatment increased the LDH activity (1.79-fold increased) and could be rescued by ZVAD (75.3%) (Figure 5D). These demonstrate that butyrate supplement attenuates the survival pathway and further triggers apoptosis in MSCs.
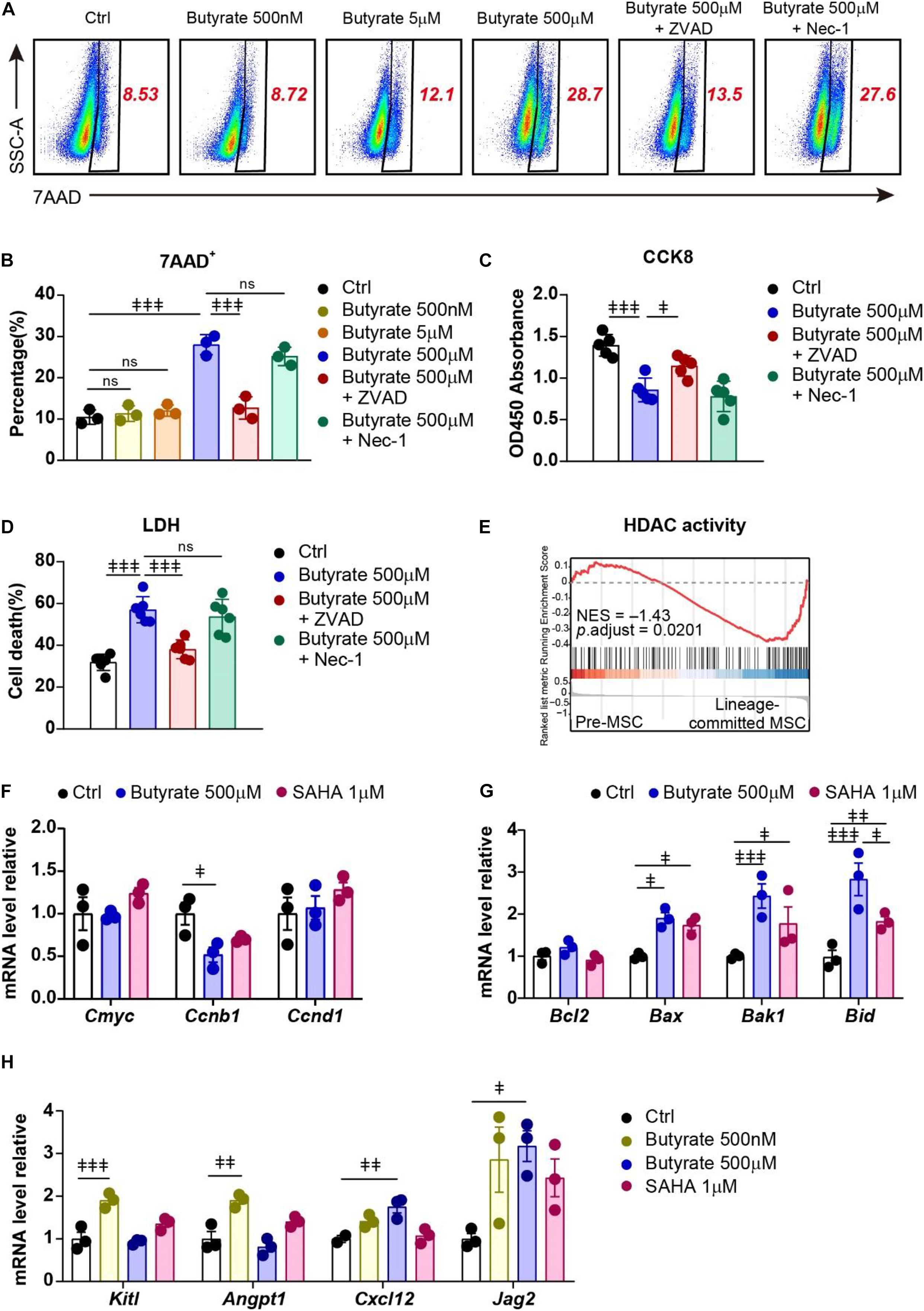
Figure 5. Supplement of butyrate triggers apoptosis but promotes HSC niche factor expression in MSCs. (A) Representative flow cytometry plots of 7AAD staining of MSCs under different treatments as indicated. The numbers in the plots denote the percentage of gated cells. (B) Percentage of 7AAD+ MSCs under different treatments as indicated. (C) CCK-8 assay showing MSC viability under different treatments as indicated. (D) LDH activity showing MSC death frequency under different treatments as indicated. (E) GSEA evaluating enrichment of genes associated with HDAC activity in pre- and lineage-committed MSCs. Normalized enrichment score and adjusted p value were calculated by permutation tests. (F) Quantitative RT-PCR of transcript levels of proliferation gene in MSCs with indicated treatments (normalized to control group). (G) Quantitative RT-PCR of transcript levels of apoptosis genes in MSCs with indicated treatments (normalized to control group). (H) Quantitative RT-PCR of transcript levels of HSC niche factor genes in MSCs with indicated treatments (normalized to control group). Repeated-measures one-way ANOVA followed by Dunnett’s test for multiple comparisons in (B–D,F–H). ns, non-significant;  p < 0.05,
p < 0.05, 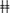 p < 0.01,
p < 0.01,  p < 0.001.
p < 0.001.
Butyrate is shown to regulate gene expression in blood cells through inhibition of HDAC (Song et al., 2011; Akimova et al., 2012; Arpaia et al., 2013). Interestingly, our scRNA-seq data revealed that lineage-committed MSCs had enriched HDAC activity (GO:0000118, GO:0004407, GO:0042826) compared to premature MSCs (NES = –1.43; Figure 5E). This indicates that butyrate might regulate MSC lineage commitment through influencing their gene expression network. Indeed, we found that butyrate treatment reduced Ccnb1 expression in MSCs (48.1% decreased; Figure 5F), which might explain the butyrate-induced growth inhibition (Pines and Hunter, 1989). Butyrate treatment also induced upregulation of multiple apoptotic genes, including Bax, Bak1, and Bid (Youle and Strasser, 2008) (1.91-fold, 2.43-fold, and 2.83-fold increased, respectively; Figure 5G), which is consistent with increased apoptosis in butyrate-treated MSCs (Figures 5A,B). Furthermore, the downregulation of apoptosis genes upon butyrate treatment was in line with another HDAC inhibitor, suberoylanilide hydroxamic acid (SAHA) (Butler et al., 2002; Yin et al., 2018) (1.74-fold, 2.43-fold, and 1.83-fold increased, respectively; Figure 5G), suggesting that butyrate-induced MSC self-renewal potentially through HDAC inhibition.
As bone marrow MSCs play a pivotal role in maintaining HSCs through producing multiple growth factors (Crane et al., 2017; Pinho and Frenette, 2019), we next investigated how butyrate treatment influences MSCs in their HSC niche function. We found that butyrate significantly increased HSC niche factor expression, including Kitl and Angpt1 at biological serum concentration (500 nM) (1.90-fold and 1.90-fold, respectively) and Cxcl12, Jag2 at a higher concentration (500 μM) (1.75-fold and 3.17-fold, respectively; Figure 5H) (Greenbaum et al., 2013; Guo et al., 2017; Aprile et al., 2020). Interestingly, the HSC niche-inducing effect of butyrate was more robust compared to SAHA (Kitl 1.35-fold, Angpt1 1.40-fold, Cxcl12 1.07-fold, and Jag2 2.43-fold increased, respectively; Figure 5H). This indicates that metabolite butyrate is a robust HSC niche factor expression booster for MSCs during in vitro culture.
Overall, our data show that butyrate supplement dichotomously regulates MSCs in their self-renewal and HSC niche function potentially through altering the MSC metabolic status and inhibiting HDAC activity.
Discussion
MSCs are present in all organs and tissues, and MSCs are a highly heterogenous subset (Uccelli et al., 2008). The heterogeneity of MSCs is closely related to their clinical utilities and also determines the barriers in transferring MSC capacities into the clinic (Costa et al., 2020). The functional heterogeneity of MSCs has been indicated in previous studies. For example, IL-17+ MSCs have enhanced antibacterial effects but with reduced immunosuppressive function compared with bulk MSCs due to altered NFκB-TGF-β signaling (Yang et al., 2013). Studies using RNA fluorescence in situ hybridization (FISH) (Cote et al., 2016) and fluorescent probes (Li et al., 2016) indicate that canonical markers are tenuously linked to the differentiated phenotypes, and it is difficult to use single markers to predict functional potential. Recent advanced single-cell studies have further explored the heterogeneity of bone marrow MSCs with distinct differentiation potential (Baryawno et al., 2019; Leimkuhler et al., 2020). Furthermore, researchers also identified an IL-10 regulated metabolically active mature adipocyte subtype from subcutaneous adipose tissue (Rajbhandari et al., 2019) and Runx2+/Gli1+ cells in the adult mouse incisor, which maintains Gli1+ MSCs (Chen et al., 2020).
In our work, we performed 10x scRNA-seq on bone marrow non-hematopoietic (CD45–, Ter-119–), non-endothelial (CD31–), and PDGFRα+ CD51+ MSCs. In line with previous studies (Pacini and Petrini, 2014; Baccin et al., 2020; Leimkuhler et al., 2020), we identified lineage-committed MSCs, including adipogenic, osteogenic, chondrogenic, angiogenic, and immunomodulating MSCs. Furthermore, we identified a pre-MSC population, which enriched prelineage commitment genes involved in protein transport, nuclear transport, and ribosome biogenesis pathways, such as Rps24, Rpl35a, and Ndufb3 (Choesmel et al., 2008; Narla et al., 2011; Alston et al., 2016) and clustering at the root of the unsupervised pseudotime trajectory.
The metabolic profile determines the functional heterogeneity of MSCs (Costa et al., 2020). Genetic inhibition of mitochondrial complex III in human MSCs and murine adipocyte precursor cells impacts adipocyte differentiation (Tormos et al., 2011; Joffin et al., 2021). Glutamine metabolism regulates proliferation and osteoblast–adipocyte lineage determination (Yu et al., 2019). Among the diverse tissues from which MSCs could be isolated, the most common source tissue is the bone marrow (Yin et al., 2019). To meet the clinical requirement for MSCs in cell therapy, MSCs have to undergo a rapid cultural expansion and long-term cryopreservation, which are largely different compared with their biological microenvironment (Yuan et al., 2019). The metabolic profile determines MSC cell fate and heterogeneity (Pattappa et al., 2013; Moll et al., 2014; Costa et al., 2020). Interestingly, we discovered that pre-MSCs, despite the reduced expression of proliferation and differentiation genes, enriched genes in FA metabolic process compared with lineage-committed MSCs. This finding indicates that supplement of metabolites, such as FA in MSC during in vitro culture, may impact MSC functional heterogeneity.
Recent studies indicated that butyrate, one of the metabolites produced in the healthy intestinal lumen (Jacobi and Odle, 2012), enhances the effect of parathyroid hormone (PTH) to support bone formation (Fan et al., 2017; Li et al., 2020; Pacifici, 2020). Furthermore, butyrate promotes Treg cell regeneration and differentiation, which stimulates bone formation by activating Wnt signaling in osteoblasts (Arpaia et al., 2013; Furusawa et al., 2013; Tyagi et al., 2018). In our study, we discovered that supplement of butyrate suppressed the self-renewal capacity and differentiation potential toward osteoblasts and adipocytes in MSCs.
HDACs, a series of critical transcriptional cofactors modulating gene expression by deacetylating histones and transcription factors, participate in stemness maintenance, lineage commitment determination, cell differentiation, and proliferation as well as other activities in normal hematopoiesis (Akimova et al., 2012; Cortiguera et al., 2019). Homozygous deletion of HDAC3 in Prrx1-expressing cells reduced chondrocyte and osteoblast differentiation in vitro (Feigenson et al., 2017). HDAC inhibitors exhibit antitumor activity for multiple myeloma (Song et al., 2011; Santo et al., 2012) and B cell lymphoma (Cortiguera et al., 2019) by suppressing cells survival and differentiation and inducing apoptosis. Butyrate is one of the extensively studied HDAC inhibitors (Steliou et al., 2012). Butyrate blocks the activity of class I and II HDACs and increases histone acetylation globally in multiple types of cells, including CD8+ T cells, B cells, hepatocytes, and some tumor cell lines, such as MCF-7 (human breast cancer cells) and HCT116 (human colon carcinoma cells) (Candido et al., 1978; Davie, 2003; Fellows et al., 2018; Luu et al., 2018; Ji et al., 2019; Sanchez et al., 2020). In our work, we discovered that butyrate supplement reduced MSC proliferation and differentiation abilities at biological serum butyrate concentration without cell death. Butyrate treatment reduced the expression of proliferation gene Ccnb1 but upregulated apoptotic genes, including Bax, Bak1, and Bid, which might explain the butyrate-induced growth inhibition. Furthermore, we found that lineage-committed MSCs enriched more genes associated with HDAC activity than pre-MSCs did. HDACs support cell growth for multiple tumor cells, including diffuse large B cell lymphoma, lung adenocarcinoma, and breast cancer (Gupta et al., 2012; Lapierre et al., 2016; Wang et al., 2016); therefore, HDAC inhibitors are clinically used for cancer treatment (Khan and La Thangue, 2012; Li and Seto, 2016). Our findings show that, unlike quiescent pre-MSCs, lineage-committed MSCs are more sensitive to HDAC inhibition, potentially due to their high proliferation potential.
Our data show that a high dose (500 μM) of butyrate treatment increased apoptosis in MSCs and also increased their HSC niche function. MSCs engulfed apoptotic bodies to enhance their differentiation ability and ameliorate the ovariectomy-induced osteopenia through activation of the Wnt/β-catenin pathway (Liu D. et al., 2018). Moreover, MSC apoptosis is related to enhanced osteoblast differentiation (Schaub et al., 2019), and apoptotic MSCs have enhanced immunosuppression activity when infused in patients with GvHD (Galleu et al., 2017; Burnham et al., 2020). These findings inspired us that the apoptosis of MSC might benefit their tissue regeneration or immunomodulation functions, which is consistent with our observation that a high dose of butyrate supplement increased apoptosis and enhanced their HSC niche function. Inhibition of HDAC2 and 3 promotes the proliferation of hematopoietic stem and progenitor cells (HSPCs) (Dhoke et al., 2016; Wang et al., 2020), but whether HDAC inhibition alters the HSC niche remains unclear. In our work, we demonstrate that in vitro supplement of butyrate-promotes HSC niche factor expression in MSCs, including Kitl, Angpt1, Cxcl12, and Jag2. As bone marrow MSCs produce SCF (Kitl), CXCL12 (Cxcl12) or Angiopoietin-1 (Angpt1) and endothelial cells secrete Jagged-2 (Jag2), which are critical for HSC maintenance (Greenbaum et al., 2013; Zhou et al., 2015; Guo et al., 2017; Comazzetto et al., 2019), our observation indicates that butyrate supplement may enhance HSC niche function potentially through inhibiting HDAC in MSCs. Overall, our findings indicate the possibility that the application of butyrate in MSC culture can amplify their HSC niche function and shed light on MSC treatments for patients with ineffective hematopoiesis and patients who underwent HSC transplantation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GEO (GSE167035, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE167035).
Ethics Statement
The animal study was reviewed and approved by the Sun Yat-sen University.
Author Contributions
JX performed the scRNA-seq data analysis and the functional assays, generated figures, and wrote the manuscript. QL, SX, and QX contributed to the scRNA-seq library construction and functional assays. YZ, YL, and LY contributed to the functional data analysis. JW performed and analyzed the scRNA-seq. LJ, LM, DL, and MZ supervised the project. All authors contributed to the article and approved the submitted version.
Funding
We would like to thank the National Key Research and Development Program of China (2017YFA0103403, 2018YFA0107200, and 2017YFA0105500), the Key Research and Development Program of Guangdong Province (2019B020 234002), Shenzhen Foundation of Science and Technology (JCYJ20170818103626421 and JCYJ20190806164009212), NSFC (81822001, 31871467, 81900101, 81800164, 81870127, and 82000838), Guangdong Innovative and Entrepreneurial Research Team Program (2016ZT06S029 and 2019ZT08Y485), Sanming Project of Medicine in Shenzhen (No. SZSM201911004), Natural Science Foundation of Guangdong Province (2018A030313497 and 2018A030313033), and 111 Project (B13037) for their generous support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.653308/full#supplementary-material
Footnotes
References
Akimova, T., Beier, U. H., Liu, Y., Wang, L., and Hancock, W. W. (2012). Histone/protein deacetylases and T-cell immune responses. Blood 119, 2443–2451. doi: 10.1182/blood-2011-10-292003
Alston, C. L., Howard, C., Olahova, M., Hardy, S. A., He, L., Murray, P. G., et al. (2016). A recurrent mitochondrial p.Trp22Arg NDUFB3 variant causes a distinctive facial appearance, short stature and a mild biochemical and clinical phenotype. J Med Genet 53, 634–641. doi: 10.1136/jmedgenet-2015-103576
Andrzejewska, A., Lukomska, B., and Janowski, M. (2019). Concise Review: Mesenchymal Stem Cells: From Roots to Boost. Stem Cells 37, 855–864. doi: 10.1002/stem.3016
Aprile, A., Gulino, A., Storto, M., Villa, I., Beretta, S., Merelli, I., et al. (2020). Hematopoietic stem cell function in beta-thalassemia is impaired and is rescued by targeting the bone marrow niche. Blood 136, 610–622. doi: 10.1182/blood.2019002721
Ardizzone, A., Scuderi, S. A., Giuffrida, D., Colarossi, C., Puglisi, C., Campolo, M., et al. (2020). Role of Fibroblast Growth Factors Receptors (FGFRs) in Brain Tumors, Focus on Astrocytoma and Glioblastoma. Cancers (Basel) 12, 825. doi: 10.3390/cancers12123825
Arpaia, N., Campbell, C., Fan, X., Dikiy, S., van der Veeken, J., deRoos, P., et al. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. doi: 10.1038/nature12726
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29. doi: 10.1038/75556
Baccin, C., Al-Sabah, J., Velten, L., Helbling, P. M., Grunschlager, F., Hernandez-Malmierca, P., et al. (2020). Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat Cell Biol 22, 38–48. doi: 10.1038/s41556-019-0439-6
Baryawno, N., Przybylski, D., Kowalczyk, M. S., Kfoury, Y., Severe, N., Gustafsson, K., et al. (2019). A Cellular Taxonomy of the Bone Marrow Stroma in Homeostasis and Leukemia. Cell 177, doi: 10.1016/j.cell.2019.04.040 ∗∗1915-1932 e1916
Bianco, P., Cao, X., Frenette, P. S., Mao, J. J., Robey, P. G., Simmons, P. J., et al. (2013). The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nature Medicine 19, 35–42. doi: 10.1038/nm.3028
Blanco, S., Bandiera, R., Popis, M., Hussain, S., Lombard, P., Aleksic, J., et al. (2016). Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340. doi: 10.1038/nature18282
Boulais, P. E., Mizoguchi, T., Zimmerman, S., Nakahara, F., Vivie, J., Mar, J. C., et al. (2018). The Majority of CD45(-) Ter119(-) CD31(-) Bone Marrow Cell Fraction Is of Hematopoietic Origin and Contains Erythroid and Lymphoid Progenitors. Immunity 49, doi: 10.1016/j.immuni.2018.08.019 ∗∗627-639 e626
Burnham, A. J., Daley-Bauer, L. P., and Horwitz, E. M. (2020). Mesenchymal stromal cells in hematopoietic cell transplantation. Blood Adv 4, 5877–5887. doi: 10.1182/bloodadvances.2020002646
Bushkofsky, J. R., Maguire, M., Larsen, M. C., Fong, Y. H., and Jefcoate, C. R. (2016). Cyp1b1 affects external control of mouse hepatocytes, fatty acid homeostasis and signaling involving HNF4alpha and PPARalpha. Arch Biochem Biophys 597, 30–47. doi: 10.1016/j.abb.2016.03.030
Butler, A., Hoffman, P., Smibert, P., Papalexi, E., and Satija, R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. doi: 10.1038/nbt.4096
Butler, L. M., Zhou, X., Xu, W. S., Scher, H. I., Rifkind, R. A., Marks, P. A., et al. (2002). The histone deacetylase inhibitor SAHA arrests cancer cell growth, up-regulates thioredoxin-binding protein-2, and down-regulates thioredoxin. Proc Natl Acad Sci U S A 99, 11700–11705. doi: 10.1073/pnas.182372299
Candido, E. P., Reeves, R., and Davie, J. R. (1978). Sodium butyrate inhibits histone deacetylation in cultured cells. Cell 14, 105–113. doi: 10.1016/0092-8674(78)90305-7
Casado-Diaz, A., Santiago-Mora, R., Dorado, G., and Quesada-Gomez, J. M. (2013). The omega-6 arachidonic fatty acid, but not the omega-3 fatty acids, inhibits osteoblastogenesis and induces adipogenesis of human mesenchymal stem cells: potential implication in osteoporosis. Osteoporos Int 24, 1647–1661. doi: 10.1007/s00198-012-2138-z
Chakkalakal, S. A., Zhang, D., Culbert, A. L., Convente, M. R., Caron, R. J., Wright, A. C., et al. (2012). An Acvr1 R206H knock-in mouse has fibrodysplasia ossificans progressiva. J Bone Miner Res 27, 1746–1756. doi: 10.1002/jbmr.1637
Chang, T. C., Hsu, M. F., Shih, C. Y., and Wu, K. K. (2017). 5-methoxytryptophan protects MSCs from stress induced premature senescence by upregulating FoxO3a and mTOR. Sci Rep 7, 11133. doi: 10.1038/s41598-017-11077-4
Chen, F., Lin, X., Xu, P., Zhang, Z., Chen, Y., Wang, C., et al. (2015). Nuclear Export of Smads by RanBP3L Regulates Bone Morphogenetic Protein Signaling and Mesenchymal Stem Cell Differentiation. Mol Cell Biol 35, 1700–1711. doi: 10.1128/MCB.00121-15
Chen, S., Jing, J., Yuan, Y., Feng, J., Han, X., Wen, Q., et al. (2020). Runx2+ Niche Cells Maintain Incisor Mesenchymal Tissue Homeostasis through IGF Signaling. Cell Rep 32, 108007. doi: 10.1016/j.celrep.2020.108007
Choesmel, V., Fribourg, S., Aguissa-Toure, A. H., Pinaud, N., Legrand, P., Gazda, H. T., et al. (2008). Mutation of ribosomal protein RPS24 in Diamond-Blackfan anemia results in a ribosome biogenesis disorder. Hum Mol Genet 17, 1253–1263. doi: 10.1093/hmg/ddn015
Choi, J., Baldwin, T. M., Wong, M., Bolden, J. E., Fairfax, K. A., Lucas, E. C., et al. (2019). Haemopedia RNA-seq: a database of gene expression during haematopoiesis in mice and humans. Nucleic Acids Res 47, D780–D785. doi: 10.1093/nar/gky1020
Christov, C., Chretien, F., Abou-Khalil, R., Bassez, G., Vallet, G., Authier, F. J., et al. (2007). Muscle satellite cells and endothelial cells: close neighbors and privileged partners. Mol Biol Cell 18, 1397–1409. doi: 10.1091/mbc.e06-08-0693
Comazzetto, S., Murphy, M. M., Berto, S., Jeffery, E., Zhao, Z., and Morrison, S. J. (2019). Restricted Hematopoietic Progenitors and Erythropoiesis Require SCF from Leptin Receptor+ Niche Cells in the Bone Marrow. Cell Stem Cell 24, doi: 10.1016/j.stem.2018.11.022 ∗∗477-486 e476
Cortiguera, M. G., Garcia-Gaipo, L., Wagner, S. D., Leon, J., Batlle-Lopez, A., and Delgado, M. D. (2019). Suppression of BCL6 function by HDAC inhibitor mediated acetylation and chromatin modification enhances BET inhibitor effects in B-cell lymphoma cells. Sci Rep 9, 16495. doi: 10.1038/s41598-019-52714-4
Costa, L. A., Eiro, N., Fraile, M., Gonzalez, L. O., Saa, J., Garcia-Portabella, P., et al. (2020). Functional heterogeneity of mesenchymal stem cells from natural niches to culture conditions: implications for further clinical uses. Cell Mol Life Sci. 78, 447–467. doi: 10.1007/s00018-020-03600-0
Cote, A. J., McLeod, C. M., Farrell, M. J., McClanahan, P. D., Dunagin, M. C., Raj, A., et al. (2016). Single-cell differences in matrix gene expression do not predict matrix deposition. Nat Commun 7, 10865. doi: 10.1038/ncomms10865
Crane, G. M., Jeffery, E., and Morrison, S. J. (2017). Adult haematopoietic stem cell niches. Nat Rev Immunol 17, 573–590. doi: 10.1038/nri.2017.53
Davie, J. R. (2003). Inhibition of histone deacetylase activity by butyrate. J Nutr 133(7 Suppl), 2485S–2493S. doi: 10.1093/jn/133.7.2485S
Dhoke, N. R., Kalabathula, E., Kaushik, K., Geesala, R., Sravani, B., and Das, A. (2016). Histone deacetylases differentially regulate the proliferative phenotype of mouse bone marrow stromal and hematopoietic stem/progenitor cells. Stem Cell Res 17, 170–180. doi: 10.1016/j.scr.2016.07.001
Dutta, B., Ren, Y., Hao, P., Sim, K. H., Cheow, E., Adav, S., et al. (2014). Profiling of the Chromatin-associated Proteome Identifies HP1BP3 as a Novel Regulator of Cell Cycle Progression. Mol Cell Proteomics 13, 2183–2197. doi: 10.1074/mcp.M113.034975
Fan, Y., Hanai, J. I., Le, P. T., Bi, R., Maridas, D., DeMambro, V., et al. (2017). Parathyroid Hormone Directs Bone Marrow Mesenchymal Cell Fate. Cell Metab 25, 661–672. doi: 10.1016/j.cmet.2017.01.001
Feigenson, M., Shull, L. C., Taylor, E. L., Camilleri, E. T., Riester, S. M., van Wijnen, A. J., et al. (2017). Histone Deacetylase 3 Deletion in Mesenchymal Progenitor Cells Hinders Long Bone Development. J Bone Miner Res 32, 2453–2465. doi: 10.1002/jbmr.3236
Fellows, R., Denizot, J., Stellato, C., Cuomo, A., Jain, P., Stoyanova, E., et al. (2018). Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat Commun 9, 105. doi: 10.1038/s41467-017-02651-5
Folmes, C. D. L., Nelson, T. J., Martinez-Fernandez, A., Arrell, D. K., Lindor, J. Z., Dzeja, P. P., et al. (2011). Somatic Oxidative Bioenergetics Transitions into Pluripotency-Dependent Glycolysis to Facilitate Nuclear Reprogramming. Cell Metabolism 14, 264–271. doi: 10.1016/j.cmet.2011.06.011
Friedenstein, A. J., Chailakhyan, R. K., Latsinik, N. V., Panasyuk, A. F., and Keiliss-Borok, I. V. (1974). Stromal cells responsible for transferring the microenvironment of the hemopoietic tissues. Cloning in vitro and retransplantation in vivo. Transplantation 17, 331–340. doi: 10.1097/00007890-197404000-00001
Furusawa, Y., Obata, Y., Fukuda, S., Endo, T. A., Nakato, G., Takahashi, D., et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450. doi: 10.1038/nature12721
Galipeau, J., and Sensebe, L. (2018). Mesenchymal Stromal Cells: Clinical Challenges and Therapeutic Opportunities. Cell Stem Cell 22, 824–833. doi: 10.1016/j.stem.2018.05.004
Galleu, A., Riffo-Vasquez, Y., Trento, C., Lomas, C., Dolcetti, L., Cheung, T. S., et al. (2017). Apoptosis in mesenchymal stromal cells induces in vivo recipient-mediated immunomodulation. Sci Transl Med 9, 416. doi: 10.1126/scitranslmed.aam7828
Garcia-Prat, L., Sousa-Victor, P., and Munoz-Canoves, P. (2017). Proteostatic and Metabolic Control of Stemness. Cell Stem Cell 20, 593–608. doi: 10.1016/j.stem.2017.04.011
Greenbaum, A., Hsu, Y. M., Day, R. B., Schuettpelz, L. G., Christopher, M. J., Borgerding, J. N., et al. (2013). CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 495, 227–230. doi: 10.1038/nature11926
Guo, L., Li, Y., Zhao, C., Peng, J., Song, K., Chen, L., et al. (2020). RECQL4, Negatively Regulated by miR-10a-5p, Facilitates Cell Proliferation and Invasion via MAFB in Ovarian Cancer. Front Oncol 10:524128. doi: 10.3389/fonc.2020.524128
Guo, P., Poulos, M. G., Palikuqi, B., Badwe, C. R., Lis, R., Kunar, B., et al. (2017). Endothelial jagged-2 sustains hematopoietic stem and progenitor reconstitution after myelosuppression. J Clin Invest 127, 4242–4256. doi: 10.1172/JCI92309
Gupta, M., Han, J. J., Stenson, M., Wellik, L., and Witzig, T. E. (2012). Regulation of STAT3 by histone deacetylase-3 in diffuse large B-cell lymphoma: implications for therapy. Leukemia 26, 1356–1364. doi: 10.1038/leu.2011.340
Herencia, C., Martinez-Moreno, J. M., Herrera, C., Corrales, F., Santiago-Mora, R., Espejo, I., et al. (2012). Nuclear translocation of beta-catenin during mesenchymal stem cells differentiation into hepatocytes is associated with a tumoral phenotype. PLoS One 7:e34656. doi: 10.1371/journal.pone.0034656
Horwitz, E. M., Le Blanc, K., Dominici, M., Mueller, I., Slaper-Cortenbach, I., Marini, F. C., et al. (2005). Clarification of the nomenclature for MSC: The International Society for Cellular Therapy position statement. Cytotherapy 7, 393–395. doi: 10.1080/14653240500319234
Hwang, I. Y., Kwak, S., Lee, S., Kim, H., Lee, S. E., Kim, J. H., et al. (2016). Psat1-Dependent Fluctuations in alpha-Ketoglutarate Affect the Timing of ESC Differentiation. Cell Metab 24, 494–501. doi: 10.1016/j.cmet.2016.06.014
Ito, K., and Suda, T. (2014). Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol 15, 243–256. doi: 10.1038/nrm3772
Jacobi, S. K., and Odle, J. (2012). Nutritional factors influencing intestinal health of the neonate. Adv Nutr 3, 687–696. doi: 10.3945/an.112.002683
Ji, X., Zhou, F., Zhang, Y., Deng, R., Xu, W., Bai, M., et al. (2019). Butyrate stimulates hepatic gluconeogenesis in mouse primary hepatocytes. Exp Ther Med 17, 1677–1687. doi: 10.3892/etm.2018.7136
Joffin, N., Paschoal, V. A., Gliniak, C. M., Crewe, C., Elnwasany, A., Szweda, L. I., et al. (2021). Mitochondrial metabolism is a key regulator of the fibro-inflammatory and adipogenic stromal subpopulations in white adipose tissue. Cell Stem Cell doi: 10.1016/j.stem.2021.01.002 ∗∗∗
Kaiko, G. E., Ryu, S. H., Koues, O. I., Collins, P. L., Solnica-Krezel, L., Pearce, E. J., et al. (2016). The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell 165, 1708–1720. doi: 10.1016/j.cell.2016.05.018
Kassem, M., Abdallah, B. M., and Saeed, H. (2008). Osteoblastic cells: differentiation and trans-differentiation. Arch Biochem Biophys 473, 183–187. doi: 10.1016/j.abb.2008.03.028
Khan, O., and La Thangue, N. B. (2012). HDAC inhibitors in cancer biology: emerging mechanisms and clinical applications. Immunol Cell Biol 90, 85–94. doi: 10.1038/icb.2011.100
Lapierre, M., Linares, A., Dalvai, M., Duraffourd, C., Bonnet, S., Boulahtouf, A., et al. (2016). Histone deacetylase 9 regulates breast cancer cell proliferation and the response to histone deacetylase inhibitors. Oncotarget 7, 19693–19708. doi: 10.18632/oncotarget.7564
Lee, D., Taniguchi, N., Sato, K., Choijookhuu, N., Hishikawa, Y., Kataoka, H., et al. (2018). HMGB2 is a novel adipogenic factor that regulates ectopic fat infiltration in skeletal muscles. Sci Rep 8, 9601. doi: 10.1038/s41598-018-28023-7
Leimkuhler, N. B., Gleitz, H. F. E., Ronghui, L., Snoeren, I. A. M., Fuchs, S. N. R., Nagai, J. S., et al. (2020). Heterogeneous bone-marrow stromal progenitors drive myelofibrosis via a druggable alarmin axis. Cell Stem Cell 28, 1–16. doi: 10.1016/j.stem.2020.11.004
Li, B., Menzel, U., Loebel, C., Schmal, H., Alini, M., and Stoddart, M. J. (2016). Monitoring live human mesenchymal stromal cell differentiation and subsequent selection using fluorescent RNA-based probes. Sci Rep 6, 26014. doi: 10.1038/srep26014
Li, J. Y., Yu, M., Pal, S., Tyagi, A. M., Dar, H., Adams, J., et al. (2020). Parathyroid hormone-dependent bone formation requires butyrate production by intestinal microbiota. J Clin Invest 130, 1767–1781. doi: 10.1172/JCI133473
Li, Y., and Seto, E. (2016). HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb Perspect Med 6, e026831. doi: 10.1101/cshperspect.a026831
Liang, G., Taranova, O., Xia, K., and Zhang, Y. (2010). Butyrate promotes induced pluripotent stem cell generation. J Biol Chem 285, 25516–25521. doi: 10.1074/jbc.M110.142059
Liu, C. F., Angelozzi, M., Haseeb, A., and Lefebvre, V. (2018). SOX9 is dispensable for the initiation of epigenetic remodeling and the activation of marker genes at the onset of chondrogenesis. Development 145, 164459. doi: 10.1242/dev.164459
Liu, D., Kou, X., Chen, C., Liu, S., Liu, Y., Yu, W., et al. (2018). Circulating apoptotic bodies maintain mesenchymal stem cell homeostasis and ameliorate osteopenia via transferring multiple cellular factors. Cell Res 28, 918–933. doi: 10.1038/s41422-018-0070-2
Luu, M., Weigand, K., Wedi, F., Breidenbend, C., Leister, H., Pautz, S., et al. (2018). Regulation of the effector function of CD8(+) T cells by gut microbiota-derived metabolite butyrate. Sci Rep 8, 14430. doi: 10.1038/s41598-018-32860-x
Martin, I., Galipeau, J., Kessler, C., Le Blanc, K., and Dazzi, F. (2019). Challenges for mesenchymal stromal cell therapies. Sci Transl Med 11, 480. doi: 10.1126/scitranslmed.aat2189
Mendez-Ferrer, S., Michurina, T. V., Ferraro, F., Mazloom, A. R., Macarthur, B. D., Lira, S. A., et al. (2010). Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466, 829–834. doi: 10.1038/nature09262
Moll, G., Alm, J. J., Davies, L. C., von Bahr, L., Heldring, N., Stenbeck-Funke, L., et al. (2014). Do Cryopreserved Mesenchymal Stromal Cells Display Impaired Immunomodulatory and Therapeutic Properties? Stem Cells 32, 2430–2442. doi: 10.1002/stem.1729
Morikawa, S., Mabuchi, Y., Kubota, Y., Nagai, Y., Niibe, K., Hiratsu, E., et al. (2009). Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J Exp Med 206, 2483–2496. doi: 10.1084/jem.20091046
Naba, A., Clauser, K. R., Mani, D. R., Carr, S. A., and Hynes, R. O. (2017). Quantitative proteomic profiling of the extracellular matrix of pancreatic islets during the angiogenic switch and insulinoma progression. Sci Rep 7, 40495. doi: 10.1038/srep40495
Naito, J., Kaji, H., Sowa, H., Hendy, G. N., Sugimoto, T., and Chihara, K. (2005). Menin suppresses osteoblast differentiation by antagonizing the AP-1 factor. JunD. J Biol Chem 280, 4785–4791. doi: 10.1074/jbc.M408143200
Narla, A., Hurst, S. N., and Ebert, B. L. (2011). Ribosome defects in disorders of erythropoiesis. Int J Hematol 93, 144–149. doi: 10.1007/s12185-011-0776-0
Pacifici, R. (2020). Role of Gut Microbiota in the Skeletal Response to PTH. J Clin Endocrinol Metab 106, 636–645. doi: 10.1210/clinem/dgaa895
Pacini, S., and Petrini, I. (2014). Are MSCs angiogenic cells? New insights on human nestin-positive bone marrow-derived multipotent cells. Front Cell Dev Biol 2:20. doi: 10.3389/fcell.2014.00020
Pajarinen, J., Lin, T., Gibon, E., Kohno, Y., Maruyama, M., Nathan, K., et al. (2019). Mesenchymal stem cell-macrophage crosstalk and bone healing. Biomaterials 196, 80–89. doi: 10.1016/j.biomaterials.2017.12.025
Pattappa, G., Thorpe, S. D., Jegard, N. C., Heywood, H. K., de Bruijn, J. D., and Lee, D. A. (2013). Continuous and uninterrupted oxygen tension influences the colony formation and oxidative metabolism of human mesenchymal stem cells. Tissue Eng Part C Methods 19, 68–79. doi: 10.1089/ten.TEC.2011.0734
Pines, J., and Hunter, T. (1989). Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell 58, 833–846. doi: 10.1016/0092-8674(89)90936-7
Pinho, S., and Frenette, P. S. (2019). Haematopoietic stem cell activity and interactions with the niche. Nat Rev Mol Cell Biol 20, 303–320. doi: 10.1038/s41580-019-0103-9
Qiu, X., Hill, A., Packer, J., Lin, D., Ma, Y.-A., and Trapnell, C. (2017). Single-cell mRNA quantification and differential analysis with Census. Nature Methods 14, 309–315. doi: 10.1038/nmeth.4150
Rajbhandari, P., Arneson, D., Hart, S. K., Ahn, I. S., Diamante, G., Santos, L. C., et al. (2019). Single cell analysis reveals immune cell-adipocyte crosstalk regulating the transcription of thermogenic adipocytes. Elife 8, 49501. doi: 10.7554/eLife.49501
Rozovski, U., Grgurevic, S., Bueso-Ramos, C., Harris, D. M., Li, P., Liu, Z., et al. (2015). Aberrant LPL Expression, Driven by STAT3, Mediates Free Fatty Acid Metabolism in CLL Cells. Mol Cancer Res 13, 944–953. doi: 10.1158/1541-7786.MCR-14-0412
Ryall, J. G., Cliff, T., Dalton, S., and Sartorelli, V. (2015a). Metabolic Reprogramming of Stem Cell Epigenetics. Cell Stem Cell 17, 651–662. doi: 10.1016/j.stem.2015.11.012
Ryall, J. G., Dell’Orso, S., Derfoul, A., Juan, A., Zare, H., Feng, X., et al. (2015b). The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell 16, 171–183. doi: 10.1016/j.stem.2014.12.004
Sanchez, C. G., Teixeira, F. K., Czech, B., Preall, J. B., Zamparini, A. L., Seifert, J. R., et al. (2016). Regulation of Ribosome Biogenesis and Protein Synthesis Controls Germline Stem Cell Differentiation. Cell Stem Cell 18, 276–290. doi: 10.1016/j.stem.2015.11.004
Sanchez, H. N., Moroney, J. B., Gan, H., Shen, T., Im, J. L., Li, T., et al. (2020). B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nat Commun 11, 60. doi: 10.1038/s41467-019-13603-6
Santo, L., Hideshima, T., Kung, A. L., Tseng, J. C., Tamang, D., Yang, M., et al. (2012). Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood 119, 2579–2589. doi: 10.1182/blood-2011-10-387365
Satani, M., Takahashi, K., Sakamoto, H., Harada, S., Kaida, Y., and Noguchi, M. (2003). Expression and characterization of human bifunctional peptidylglycine alpha-amidating monooxygenase. Protein Expr Purif 28, 293–302. doi: 10.1016/s1046-5928(02)00684-8
Schaub, T., Gurgen, D., Maus, D., Lange, C., Tarabykin, V., Dragun, D., et al. (2019). mTORC1 and mTORC2 Differentially Regulate Cell Fate Programs to Coordinate Osteoblastic Differentiation in Mesenchymal Stromal Cells. Sci Rep 9, 20071. doi: 10.1038/s41598-019-56237-w
Shook, B. A., Wasko, R. R., Mano, O., Rutenberg-Schoenberg, M., Rudolph, M. C., Zirak, B., et al. (2020). Dermal Adipocyte Lipolysis and Myofibroblast Conversion Are Required for Efficient Skin Repair. Cell Stem Cell 26, doi: 10.1016/j.stem.2020.03.013 ∗∗880-895 e886
Shyh-Chang, N., Locasale, J. W., Lyssiotis, C. A., Zheng, Y., Teo, R. Y., Ratanasirintrawoot, S., et al. (2013). Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339, 222–226. doi: 10.1126/science.1226603
Smith, A. N., Muffley, L. A., Bell, A. N., Numhom, S., and Hocking, A. M. (2012). Unsaturated fatty acids induce mesenchymal stem cells to increase secretion of angiogenic mediators. J Cell Physiol 227, 3225–3233. doi: 10.1002/jcp.24013
Song, W., Tai, Y. T., Tian, Z., Hideshima, T., Chauhan, D., Nanjappa, P., et al. (2011). HDAC inhibition by LBH589 affects the phenotype and function of human myeloid dendritic cells. Leukemia 25, 161–168. doi: 10.1038/leu.2010.244
Steliou, K., Boosalis, M. S., Perrine, S. P., Sangerman, J., and Faller, D. V. (2012). Butyrate histone deacetylase inhibitors. Biores Open Access 1, 192–198. doi: 10.1089/biores.2012.0223
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. doi: 10.1073/pnas.0506580102
Sun, L., Akiyama, K., Zhang, H., Yamaza, T., Hou, Y., Zhao, S., et al. (2009). Mesenchymal stem cell transplantation reverses multiorgan dysfunction in systemic lupus erythematosus mice and humans. Stem Cells 27, 1421–1432. doi: 10.1002/stem.68
Teslaa, T., and Teitell, M. A. (2015). Pluripotent stem cell energy metabolism: an update. EMBO J 34, 138–153. doi: 10.15252/embj.201490446
The Gene Ontology Consortium. (2019). The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47, D330–D338. doi: 10.1093/nar/gky105547(D1):D330-D338
Tormos, K. V., Anso, E., Hamanaka, R. B., Eisenbart, J., Joseph, J., Kalyanaraman, B., et al. (2011). Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab 14, 537–544. doi: 10.1016/j.cmet.2011.08.007
Tyagi, A. M., Yu, M., Darby, T. M., Vaccaro, C., Li, J. Y., Owens, J. A., et al. (2018). The Microbial Metabolite Butyrate Stimulates Bone Formation via T Regulatory Cell-Mediated Regulation of WNT10B Expression. Immunity 49, doi: 10.1016/j.immuni.2018.10.013 ∗∗1116-1131 e1117
Uccelli, A., Moretta, L., and Pistoia, V. (2008). Mesenchymal stem cells in health and disease. Nature Reviews Immunology 8, 726–736. doi: 10.1038/nri2395
Valenti, M. T., Dalle Carbonare, L., and Mottes, M. (2016). Osteogenic Differentiation in Healthy and Pathological Conditions. Int J Mol Sci 18, 41. doi: 10.3390/ijms18010041
van Weeghel, M., Ofman, R., Argmann, C. A., Ruiter, J. P., Claessen, N., Oussoren, S. V., et al. (2014). Identification and characterization of Eci3, a murine kidney-specific Delta3,Delta2-enoyl-CoA isomerase. FASEB J 28, 1365–1374. doi: 10.1096/fj.13-240416
Wanet, A., Arnould, T., Najimi, M., and Renard, P. (2015). Connecting Mitochondria, Metabolism, and Stem Cell Fate. Stem Cells Dev 24, 1957–1971. doi: 10.1089/scd.2015.0117
Wang, J., Alexander, P., Wu, L., Hammer, R., Cleaver, O., and McKnight, S. L. (2009). Dependence of mouse embryonic stem cells on threonine catabolism. Science 325, 435–439. doi: 10.1126/science.1173288
Wang, P., Wang, Z., and Liu, J. (2020). Role of HDACs in normal and malignant hematopoiesis. Mol Cancer 19, 5. doi: 10.1186/s12943-019-1127-7
Wang, Z., Tang, F., Hu, P., Wang, Y., Gong, J., Sun, S., et al. (2016). HDAC6 promotes cell proliferation and confers resistance to gefitinib in lung adenocarcinoma. Oncol Rep 36, 589–597. doi: 10.3892/or.2016.4811
Wolf, F. A., Angerer, P., and Theis, F. J. (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15. doi: 10.1186/s13059-017-1382-0
Yang, R., Liu, Y., Kelk, P., Qu, C., Akiyama, K., Chen, C., et al. (2013). A subset of IL-17(+) mesenchymal stem cells possesses anti-Candida albicans effect. Cell Res 23, 107–121. doi: 10.1038/cr.2012.179
Ye, L., Fan, Z., Yu, B., Chang, J., Al Hezaimi, K., Zhou, X., et al. (2012). Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 11, 50–61. doi: 10.1016/j.stem.2012.04.009
Yin, J. Q., Zhu, J., and Ankrum, J. A. (2019). Manufacturing of primed mesenchymal stromal cells for therapy. Nat Biomed Eng 3, 90–104. doi: 10.1038/s41551-018-0325-8
Yin, L., Liu, Y., Peng, Y., Peng, Y., Yu, X., Gao, Y., et al. (2018). PARP inhibitor veliparib and HDAC inhibitor SAHA synergistically co-target the UHRF1/BRCA1 DNA damage repair complex in prostate cancer cells. J Exp Clin Cancer Res 37, 153. doi: 10.1186/s13046-018-0810-7
Yoshida, Y., Takahashi, K., Okita, K., Ichisaka, T., and Yamanaka, S. (2009). Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell 5, 237–241. doi: 10.1016/j.stem.2009.08.001
Youle, R. J., and Strasser, A. (2008). The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 9, 47–59. doi: 10.1038/nrm2308
Yu, Y., Newman, H., Shen, L., Sharma, D., Hu, G., Mirando, A. J., et al. (2019). Glutamine Metabolism Regulates Proliferation and Lineage Allocation in Skeletal Stem Cells. Cell Metab 29, doi: 10.1016/j.cmet.2019.01.016 ∗∗966-978 e964
Yuan, X. G., Logan, T. M., and Ma, T. (2019). Metabolism in Human Mesenchymal Stromal Cells: A Missing Link Between hMSC Biomanufacturing and Therapy? Frontiers in Immunology 10:977. doi: 10.3389/fimmu.2019.00977
Yue, R., Zhou, B. O., Shimada, I. S., Zhao, Z., and Morrison, S. J. (2016). Leptin Receptor Promotes Adipogenesis and Reduces Osteogenesis by Regulating Mesenchymal Stromal Cells in Adult Bone Marrow. Cell Stem Cell 18, 782–796. doi: 10.1016/j.stem.2016.02.015
Zanella, M., Vitriolo, A., Andirko, A., Martins, P. T., Sturm, S., O’Rourke, T., et al. (2019). Dosage analysis of the 7q11.23 Williams region identifies BAZ1B as a major human gene patterning the modern human face and underlying self-domestication. Sci Adv 5, eaaw7908. doi: 10.1126/sciadv.aaw7908
Zhang, L. J., Guerrero-Juarez, C. F., Hata, T., Bapat, S. P., Ramos, R., Plikus, M. V., et al. (2015). Innate immunity. Dermal adipocytes protect against invasive Staphylococcus aureus skin infection. Science 347, 67–71. doi: 10.1126/science.1260972
Zhang, X., Novera, W., Zhang, Y., and Deng, L. W. (2017). MLL5 (KMT2E): structure, function, and clinical relevance. Cell Mol Life Sci 74, 2333–2344. doi: 10.1007/s00018-017-2470-8
Zhou, B. O., Ding, L., and Morrison, S. J. (2015). Hematopoietic stem and progenitor cells regulate the regeneration of their niche by secreting Angiopoietin-1. Elife 4, e05521. doi: 10.7554/eLife.05521
Zhou, B. O., Yu, H., Yue, R., Zhao, Z., Rios, J. J., Naveiras, O., et al. (2017). Bone marrow adipocytes promote the regeneration of stem cells and haematopoiesis by secreting SCF. Nat Cell Biol 19, 891–903. doi: 10.1038/ncb3570
Keywords: single-cell RNA-seq, butyrate, cell heterogeneity, HSC niche, mesenchymal stem cells
Citation: Xie J, Lou Q, Zeng Y, Liang Y, Xie S, Xu Q, Yuan L, Wang J, Jiang L, Mou L, Lin D and Zhao M (2021) Single-Cell Atlas Reveals Fatty Acid Metabolites Regulate the Functional Heterogeneity of Mesenchymal Stem Cells. Front. Cell Dev. Biol. 9:653308. doi: 10.3389/fcell.2021.653308
Received: 14 January 2021; Accepted: 09 March 2021;
Published: 12 April 2021.
Edited by:
Yan Xu, Third Affiliated Hospital of Sun Yat-sen University, ChinaReviewed by:
Bin-Zhi Qian, The University of Edinburgh, United KingdomChunliang Xu, Albert Einstein College of Medicine, United States
Copyright © 2021 Xie, Lou, Zeng, Liang, Xie, Xu, Yuan, Wang, Jiang, Mou, Lin and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meng Zhao, emhhb20zOEBtYWlsLnN5c3UuZWR1LmNu; Dongjun Lin, bGluZGpAbWFpbC5zeXN1LmVkdS5jbg==; Lisha Mou, bGlzaGFtb3VAZ21haWwuY29t
†These authors have contributed equally to this work
 Jiayi Xie
Jiayi Xie Qi Lou
Qi Lou Yunxin Zeng1†
Yunxin Zeng1† Quanhui Xu
Quanhui Xu Lisha Mou
Lisha Mou Dongjun Lin
Dongjun Lin Meng Zhao
Meng Zhao