- 1Laboratory of Tissue Engineering, College of Life Sciences, Northwest University, Xi’an, China
- 2Provincial Key Laboratory of Biotechnology of Shaanxi, Northwest University, Xi’an, China
- 3Key Laboratory of Resource Biology and Biotechnology in Western China, Ministry of Education, School of Medicine, Northwest University, Xi’an, China
Phosphatidylinositol 3-kinase (PI3K) signaling plays a central role in various biological processes, and its abnormality leads to a broad spectrum of human diseases, such as cancer, fibrosis, and immunological disorders. However, the mechanisms by which PI3K signaling regulates the behavior of stem cells during regeneration are poorly understood. Planarian flatworms possess abundant adult stem cells (called neoblasts) allowing them to develop remarkable regenerative capabilities, thus the animals represent an ideal model for studying stem cells and regenerative medicine in vivo. In this study, the spatiotemporal expression pattern of Djpi3k, a PI3K ortholog in the planarian Dugesia japonica, was investigated and suggests its potential role in wound response and tissue regeneration. A loss-of-function study was conducted using small molecules and RNA interference technique, providing evidence that PI3K signaling is required for blastema regrowth and cilia maintenance during planarian regeneration and homeostasis. Interestingly, the mitotic and apoptotic responses to amputation are substantially abated in PI3K inhibitor-treated regenerating animals, while knockdown of Djpi3k alleviates the mitotic response and postpones the peak of apoptotic cell death, which may contribute to the varying degrees of regenerative defects induced by the pharmacological and genetic approaches. These observations reveal novel roles for PI3K signaling in the regulation of the cellular responses to amputation during planarian regeneration and provide insights for investigating the disease-related genes in the regeneration-competent organism in vivo.
Introduction
Phosphatidylinositol 3-kinase is a ubiquitous protein kinase that participates in multiple cellular processes, including cell proliferation, apoptosis, differentiation, and migration (Madsen, 2020). The catalytic subunit of the kinase converts phosphatidylinositol 4,5-biphosphate into phosphatidylinositol 3,4,5-triphosphate, which recruits phosphoinositide-dependent protein kinase-1 to phosphorylates and activates its downstream effector Akt serine/threonine kinase (Chen et al., 2013). It is now well established from a variety of studies that PI3K plays a crucial role in cancer, diabetes, and neurodegenerative diseases (Thorpe et al., 2015; Huang et al., 2018; Xu et al., 2020). Intriguingly, it has been found that chemical inhibitors against PI3K catalytic subunit, such as LY294002 and A66, either alone or in combination with other therapeutics, are effectively used in preclinical studies for the treatment of related diseases (Courtney et al., 2010; Jamieson et al., 2011).
Animal regeneration is an essential biological phenomenon, and its abnormalities are closely related to the occurrence of many diseases (Scimone et al., 2020). Therefore, it is of great significance to the investigation of regeneration processes and regulation mechanisms. In recent years, there has been a shred of increasing evidence suggesting the important role of the PI3K pathway in animal regeneration. In hydra, PI3K participates in the regulation of head formation via activating the early head-specific genes (Manuel et al., 2006). Biliary epithelial cells (BECs) are facultative liver stem cells that give rise to hepatocytes following injury, the positive role of the PI3K pathway in BEC-driven liver regeneration has been demonstrated in the zebrafish hepatocyte ablation model (Jung et al., 2020). For amphibian limb regeneration, PI3K facilitates the formation of limb blastema by controlling the reentry of mesodermal cells into the cell cycle in the Xenopus laevis froglet (Suzuki et al., 2007); several components of the PI3K pathway have been identified to be closely associated with Cynops orientalis limb regeneration by using an iTRAQ-based proteomic approach (Tang et al., 2017). In mice, PI3K activation promotes sensory axon regeneration (Saijilafu et al., 2013) and hepatic regenerative response (Jackson et al., 2008). Despite extensive research on PI3K, the mechanisms by which PI3K signaling regulates the regenerative process have yet to be thoroughly elucidated.
The planarian Dugesia japonica, a species of triclad flatworm that inhabits the freshwater bodies of East Asia, has long been an excellent model organism for the study of regeneration (Cebrià et al., 2002; Yazawa et al., 2009; Yujia et al., 2019). The remarkable regenerative ability of planarians derives from an abundance of adult stem cells, also known as neoblasts, that are distributed throughout the mesenchyme and account for 20–30% of all planarian cells (Wagner et al., 2011). During normal tissue turnover and regeneration after injury, neoblasts serve as the only mitotically active cells in the adult to give rise to all types of planarian cells, including neurons, muscle, and epidermal cells (Reddien and Sánchez Alvarado, 2004; Hayashi et al., 2006). Therefore, planarian Dugesia japonica represents an emerging model for unraveling the mechanisms of PI3K signaling involved in regeneration events.
In this study, we identified and characterized a planarian PI3K-like gene, Djpi3k. The expression pattern of Djpi3k was profiled in intact and regenerating planarians. Functional analysis using small molecular inhibitors LY294002/A66 or RNA interference (RNAi) technique showed that PI3K signaling is required for planarian regeneration and homeostasis. We also found that inhibition of PI3K signaling attenuates the mitotic and apoptotic responses to amputation. Our work reveals the role and mechanism of PI3K signaling in the regulation of planarian regeneration.
Materials and Methods
Planarian Culture
A clonal strain of the planarian Dugesia japonica was used in all experiments (Wang et al., 2021). The colony was maintained in autoclaved stream water in the dark at 20°C. Planarians 6–8 mm in length were starved for at least one week. Two regenerating fragments were obtained via transverse amputation at the anterior of the pharynx.
Bioinformatic Analysis
DNAMAN software (7.0) was used to analyze the obtained cDNA and deduced the amino acid sequence of Djpi3k. The conserved domains of Djpi3k protein were predicted using the NCBI Conserved Domain Database (Lu et al., 2020) and the SMART database (Letunic et al., 2021). The ProtParam online tool1 was used to calculate the molecular weight and isoelectric point (pI) of Djpi3k protein. Through the neighbor-joining (NJ) method, a phylogenetic tree was constructed via MEGA software (7.0), and further annotation was conducted by the Evolview tool using a Pfam database (He et al., 2016).
Reverse Transcription and Quantitative Real-Time PCR Analysis
At 0 h, 4 h, 8 h, 12 h, 1 days, 3 days, 5 days, 7 days post-amputation, head- and tail-regenerating tissues were harvested and grinded using a homogenizer, respectively. For each replicate, 5–10 planarians were homogenized together, and total RNA was isolated using RNAiso Plus reagent (Takara, China) according to the standard protocol. Next, cDNA was synthesized from 1 μg of total RNA using Transcriptor First Strand cDNA Synthesis Kit (Roche, Switzerland). The qRT-PCR analysis was performed using SYBR Premix Ex Taq (TaKaRa, China) and a CFX96TM Real-Time PCR Detection System (Bio-Rad, United States) with the following parameters: 95°C for 10 s; followed by 40 cycles at 95°C for 10 s, 60°C for 10 s, and 72°C for 10 s. The β-actin housekeeping gene was selected as an internal reference. The relative gene expression of each gene was calculated using the 2–ΔΔCT method. PCR primers used in this study were shown in Supplementary Table 1.
Treatment With Chemical Inhibitors
The PI3K-specific inhibitor LY294002 (Abmole Bioscience, United States) or A66 (Abmole Bioscience, United States) was dissolved in DMSO and used at a series of gradient concentrations. Planarians were allowed to regenerate in water supplemented with LY294002 or A66 immediately after amputation until the indicated period of regeneration. For homeostasis experiments, intact animals were maintained in water supplemented with 30 μM LY294002 for 20 days.
RNAi Experiment
RNAi was conducted by oral delivery of bacterially expressed dsRNA as described previously (Newmark et al., 2003; Zeng et al., 2013). Briefly, two fragments against the Class I PI3K accessory and catalytic domains of the Djpi3k gene were introduced into the L4440 vector (BioVector NTCC, China) between two convergent T7 promoters. The RNAi vectors were transformed into an RNaseIII-deficient E. coli strain HT115 (BioVector NTCC, China) to generate dsRNA by IPTG induction (0.4 mM, Amresco, United States). Then animals were fed four rounds with a mixture of the dsRNA-expressing bacteria and liver paste. Animals fed with HT115 bacteria carrying a fragment encoding EGFP served as a negative control. Three days after the last feeding, the RNAi efficiency was examined by whole-mount in situ hybridization (WISH) and qRT-PCR.
Whole-Mount in situ Hybridization
Whole-mount in situ hybridization was performed using a slightly modified protocol previously described (Pearson et al., 2009). Digoxigenin-labeled antisense RNA probes were synthesized using the in vitro RNA labeling kit (Roche, Germany). Prior to 5% N-Acetyl Cysteine treatment and 4% paraformaldehyde fixation, planarians were treated with 2% HCl to kill and remove mucus for 5 min. After bleaching and rehydration, specimens were treated with proteinase K (10 mg/mL, Merck, Germany) and fixed in 4% paraformaldehyde at room temperature. Hybridizations with the labeled probe in the concentration of 1 ng/μL were performed at 56°C for 16 h after pre-hybridization. Next, specimens were washed with preheated (to 56°C) gradient solutions. Blocking and incubation of the anti-Digoxigenin-AP antibody (1:5,000, Roche, Germany) were done overnight at 4°C. Following 5-Bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium (Roche, Germany) development to detect the Dig-labeled riboprobe.
Whole-Mount Immunostaining
The planarians were sacrificed in ice-cold 2% HCl for 2–5 min and fixed in 4% paraformaldehyde, then rinsed in 4°C methanol for 1 h and bleached in methanol containing 6% H2O2 overnight. After rehydration, specimens were blocked in 0.6% BSA and 0.45% fish gelatin diluted with PBSTx for 1 h at room temperature and incubated sequentially in primary antibody concentrations were used as follows: anti-phospho-Histone H3 (anti-H3P, 1:250, R&D Systems, United States); anti-Acetylated Tubulin (anti-Ac-Tubulin, 1:500, Sigma, United States); anti-SYNORF1 (1:50, Developmental Studies Hybridoma Bank, United States). Secondary antibody concentrations were: goat-anti-mouse IgG (1:200, Millipore, United States), and goat anti-rabbit IgG (Alexa Fluor488) (1:1,000, Abcam, United Kingdom). The signals were photographed and fluorescent images were captured and analyzed with LEICA SP8 confocal microscope and ImageJ software.
TUNEL Assay
A whole-mount TUNEL assay using the planarians was performed as described previously (Pellettieri et al., 2010). After fixation, the planarians were permeabilized in 1% SDS for 20 min and bleached overnight in bleached in methanol containing 6% H2O2 overnight. Next, specimens were incubated with the terminal transferase enzyme diluted in label solution (Roche, Germany) for 4 h at 37°C, and then rinsed in PBSTB (PBST with 0.25% BSA), and these animals were incubated in anti–digoxigenin-rhodamine diluted in blocking solution for 4 h at room temperature. DAPI was used to stain nuclei. The signals were photographed and fluorescent images were captured and analyzed with LEICA SP8 confocal microscope and ImageJ software.
Measurements of Planaria and Image Processing
Planarian live and WISH images were acquired with Canon EOS 600D and OLYMPUS SZX10 microscopes, respectively. Z-stacks of fluorescently labeled samples were acquired with a LEICA SP8 confocal microscope and processed with the ImageJ package Fiji. For quantification of H3P-positive or TUNEL-positive cells, stacks with equal numbers of z-sections were taken and cells were counted in the indicated areas using ImageJ package Fiji. To quantify cilia density, the fluorescence intensity of the Ac-tubulin signal was measured and normalized to DAPI fluorescence. To assess blastema formation, the length of the regenerated blastema was measured using ImageJ package Fiji and normalized to the whole-body length of the trunk. The ratio of blastema/whole-body length of small molecules treated animals or Djpi3k(RNAi) animals were compared with control animals at each time point.
Statistical Analysis
Values were expressed as mean ± SEM. The statistical differences were calculated using the Student’s t-test in SPSS 21.0 software. Statistical significance is indicated as follows: ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ns, not significant.
Results
Characterization of a PI3K-Like Gene in Planarian
A planarian PI3K-like gene, Djpi3k, was cloned from Dugesia japonica and contains an ORF of 3,075 bp. The predicted Djpi3k protein is composed of 1,024 amino acid residues and shares conserved functional domains with the catalytic subunit of Class I PI3K, including p85-binding domain, Ras-binding domain, C2 domain, PI3K accessory, and catalytic domains (Supplementary Figure 1). The molecular weight of the predicted Djpi3k protein was estimated to be 256.21 kDa, and the pI was estimated at 4.98.
The catalytic subunits of Class I PI3K includes four highly homologous p110 isoforms (p110α,p110β, p110γ, and p110δ) encoded by genes PIK3CA, PIK3CB, PIK3CG, and PIK3CD, respectively (Zhao and Vogt, 2008). To investigate which subgroup the Djpi3k is classified, 30 genes encoding the p110 catalytic isoforms from 13 representative species were selected for phylogenetic analysis. As shown in Figure 1, all selected genes were mainly categorized into four clusters corresponding to PIK3CA (blue branch), PIK3CB (yellow branch), PIK3CG (green branch), and PIK3CD (red branch), among which Djpi3k (A0A222C681_DUGJA) was classified into the PIK3CA subgroup as well as another planarian ortholog (D2DJU4_SCHMD) from Schmidtea mediterranea. Domain annotations for each sequence were determined using the Pfam database (Mistry et al., 2021), and the predicted domains were consistent with the results from other databases (Supplementary Figure 1).
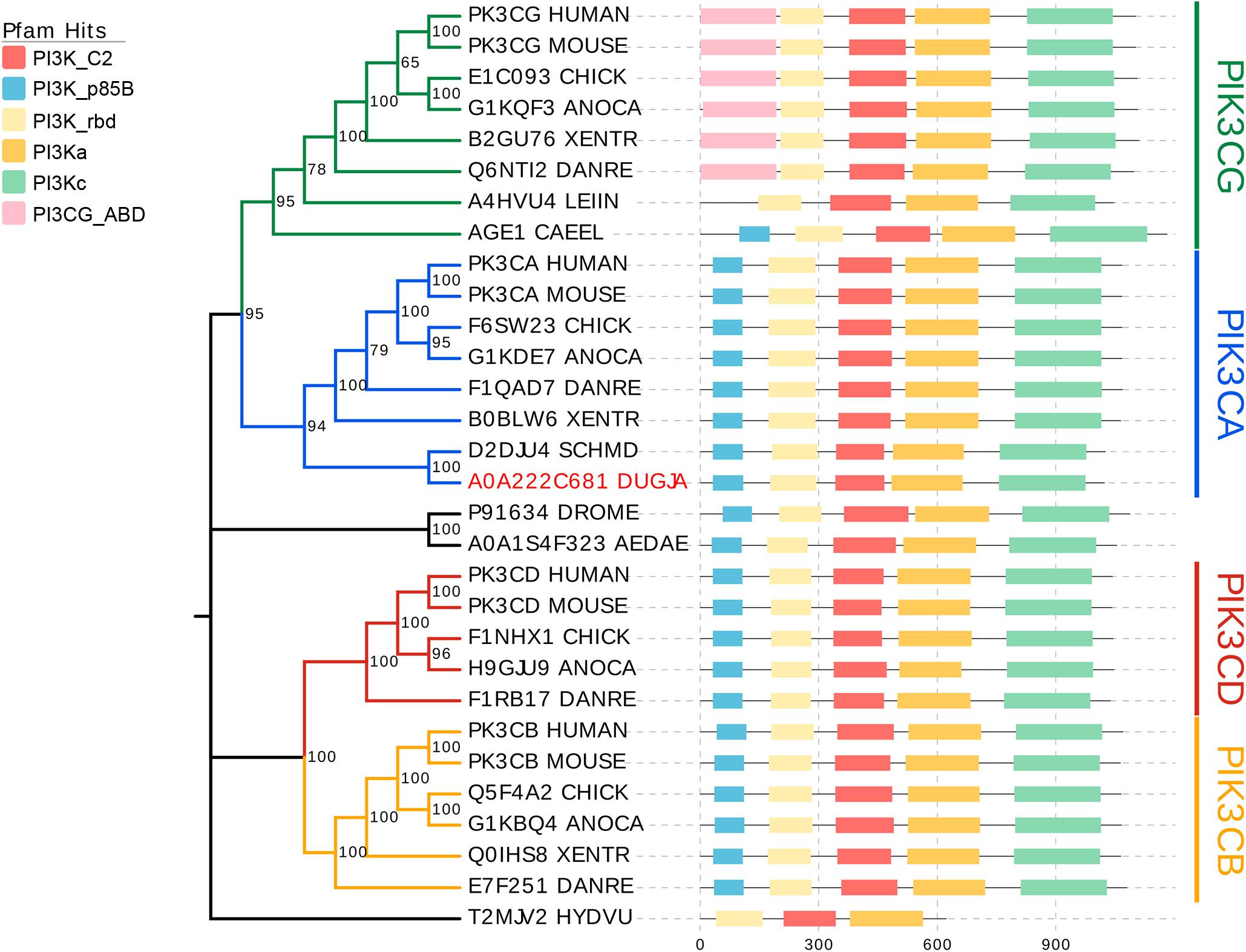
Figure 1. Phylogenetic analysis of catalytic PI3Ks subunits. All selected genes encoding the catalytic subunits of Class I PI3K are mainly categorized into four clusters corresponding to PIK3CA (blue), PIK3CB (yellow), PIK3CD (red), and PIK3CG (green). Domain annotations for each sequence were determined using the Pfam database (Mistry et al., 2021). C2, C2 domain; p85B, p85-binding domain; rbd, Ras-binding domain; PI3Ka, PI3K accessory domain; PI3Kc, PI3K catalytic domain; PIK3CG_ABD, PIK3 catalytic subunit gamma adaptor-binding domain.
Expression Pattern of Djpi3k During Planarian Regeneration
To characterize the expression pattern of Djpi3k during planarian regeneration, animals were cut anterior of the pharynx and allowed to regenerate for 7 days (Figure 2A), samples at different regenerating time were fixed and subjected to WISH. The specification of the Djpi3k probe was confirmed in intact animals using a Djpi3k sense probe as a negative control and a neoblast marker gene DjpiwiA as a positive control (Supplementary Figures 2A,B). The WISH results showed that Djpi3k was expressed immediately after amputation at the wound surface, suggesting that the gene was in response to injury/tissue absence. High levels of Djpi3k expression were detected in the newly-regenerated blastema and pharynx-forming regions during regeneration. The Djpi3k level decreased gradually as the regeneration became complete (Figure 2B). The dynamic change of Djpi3k expression during planarian regeneration was also analyzed using qRT-PCR analysis during anterior and posterior regeneration (Figure 2C). The qRT-PCR results indicated that Djpi3k levels were upregulated following amputation, peaked at 1 day post amputation (dpa), and then fell to basal level until the regeneration was complete, which was consistent with the results of WISH analysis.
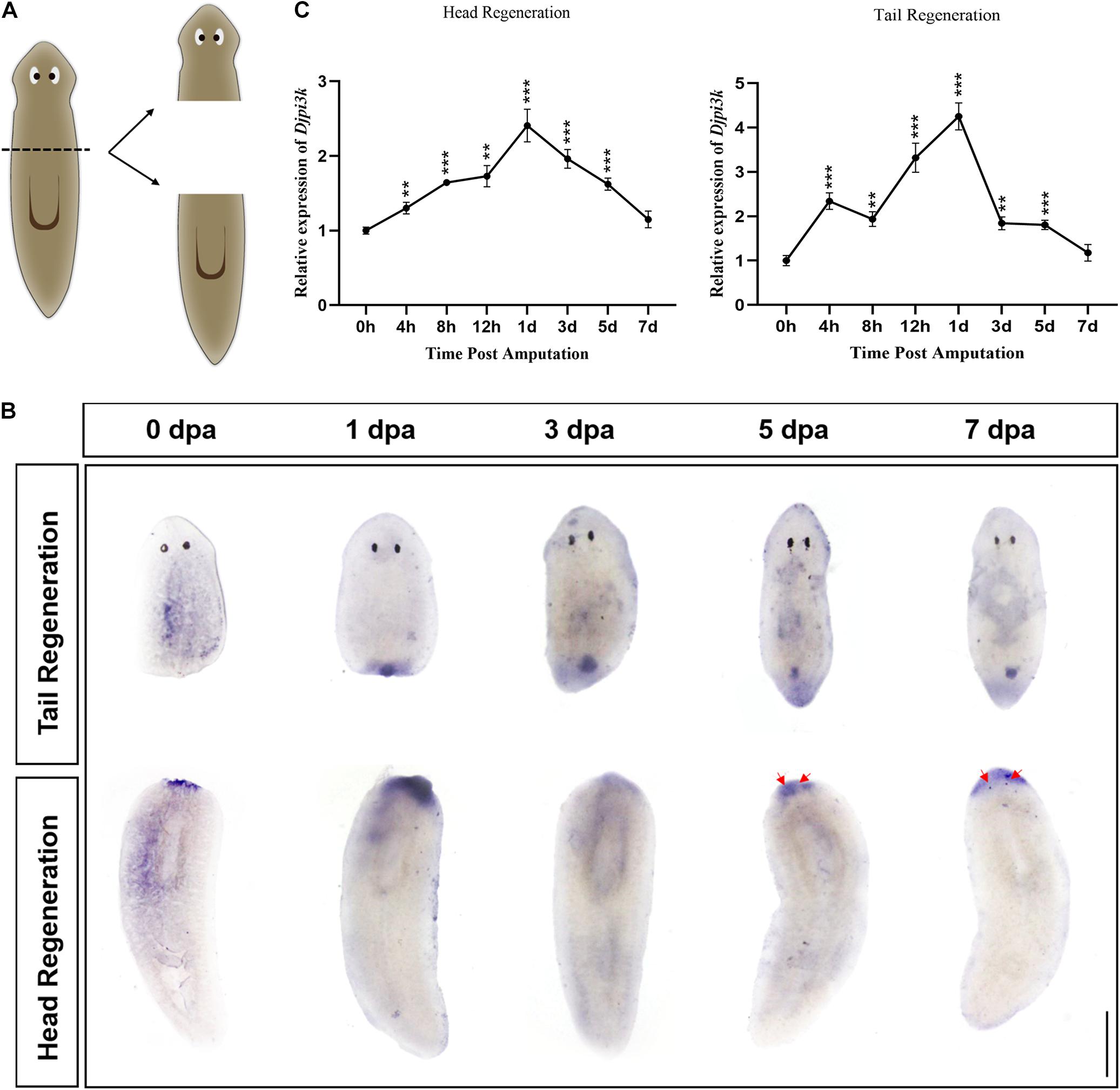
Figure 2. Expression pattern of Djpi3k during planarian regeneration. (A) Schematic representation of the general sampling set-up. (B) Spatiotemporal expression pattern of Djpi3k during planarian regeneration assessed by whole-mount in situ hybridization. Newly-regenerated photoreceptors are indicated with red arrows. Scale bar = 1 mm. (C) Fold change in mRNA expression levels of Djpi3k during planarian head and tail regeneration. All values are relative to the expression level of Djpi3k mRNA at 0 h. **P < 0.01 and ***P < 0.001, by the Student’s t-test.
Inhibition of PI3K Signaling Impairs Planarian Regeneration and Homeostasis
To investigate the role of PI3K signaling in planarian regeneration, we inactivated the PI3K signaling by using LY294002, a selective PI3K inhibitor (Walker et al., 2000). Animals were amputated and exposed to a gradient concentration of LY294002 for 7 days. Treatment with 45 μM LY294002 caused damage to the epidermis followed soon by lysis, while animals survived by treatment with a moderate concentration at 30 μM (Supplementary Figure 3). However, both head and tail regeneration were largely impaired after 30 μM LY294002 treatment (Figure 3A). The LY294002 treated fragments only regrew marginal blastemas at 7 dpa, which were significantly smaller than those of the control animals (Figure 3B). In addition, no eyes were formed in the regenerating tail fragments at 7 dpa when the control animals had regenerated their eyes properly (Figure 3A). A similar inhibitory effect was observed in the animals treated with A66 (80 μM, Supplementary Figures 4A,B), a highly specific and selective p110α inhibitor (Jamieson et al., 2011). The planarian brain is mainly composed of four structurally and functionally different regions including mechanosensory neurons (yellow), lateral branch neurons (blue, nine asterisks), two main lobes (red), and photosensory neurons (green) (Agata and Umesono, 2008). We tested whether planarian brain regeneration was disturbed after treatment of LY294002 using an anti-SYNORF1 antibody that recognizes brain structures. The immunostaining results revealed that PI3K inhibition caused apparent defects in brain regeneration at 7 dpa, mainly manifested as incomplete main lobes, disappeared lateral branch neurons, and the brain commissure of the two lobes not connected in the top (Figure 3C). These data indicate that PI3K signaling is required for appropriate regeneration in planarians.
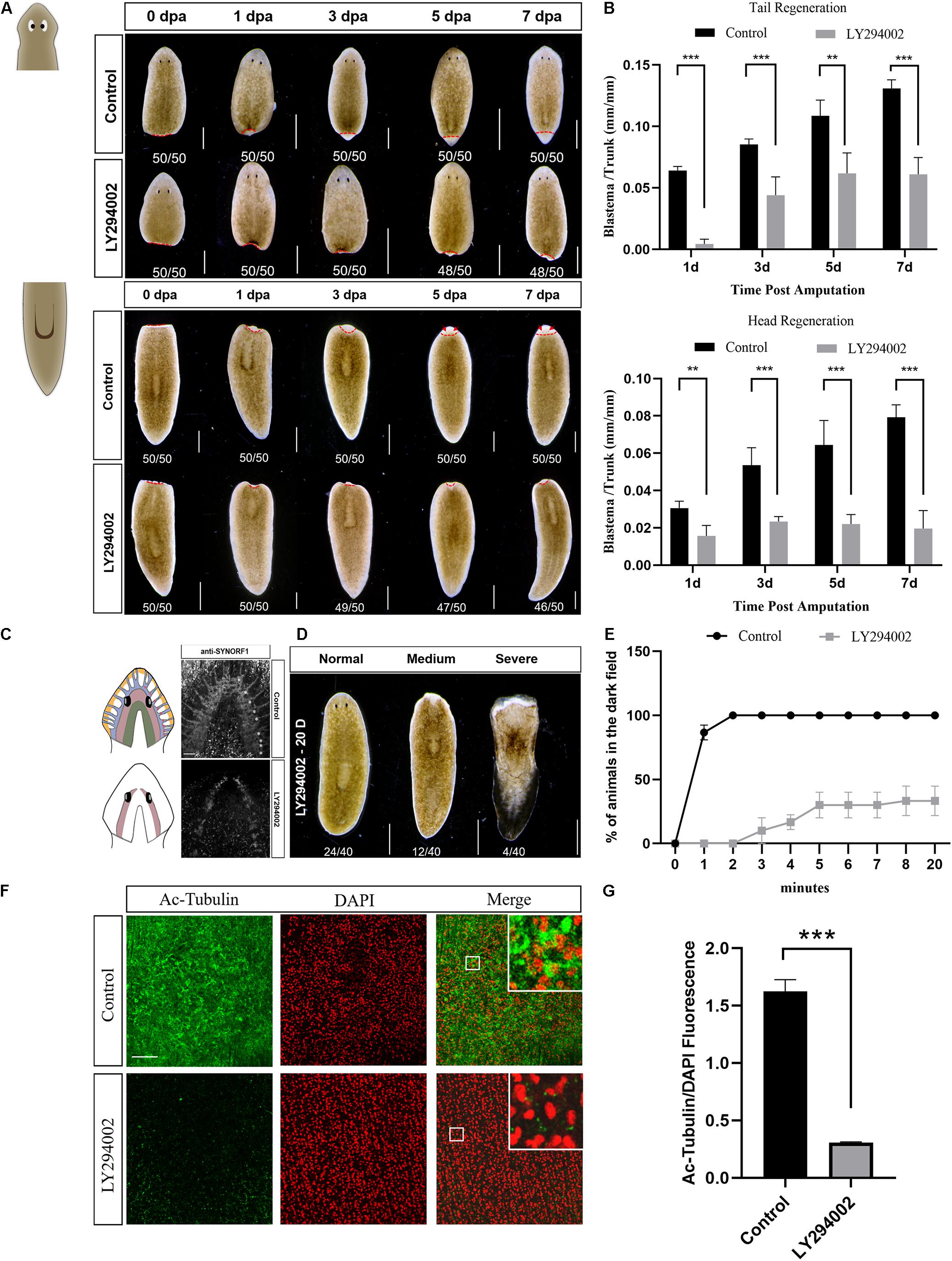
Figure 3. Effect of the PI3K inhibitor LY294002 on planarian regeneration and homeostasis. (A) Dorsal images of control and LY294002-treated head and tail fragments at 0, 1, 3, 5, and 7 days post amputation. Newly-regenerated photoreceptors are indicated with red arrows. Scale bar = 1 mm. (B) Quantification analysis of blastema growth. The length of the regenerated blastema was measured and normalized to the whole-body length of the trunk. **P < 0.01 and ***P < 0.001, by the Student’s t-test. (C) Cartoon and immunostaining of the regenerated CNS at 7 dpa using an anti-SYNORF1 antibody in control (top) and LY294002-treated animals (bottom). Lateral branch neurons are indicated with asterisks. Scale bar = 100 μm. (D) Representative phenotypes induced by a 20-day treatment of LY294002. Scale bar = 1 mm. (E) Planarian locomotion is hampered by LY294002 treatment. Gliding ability is evaluated by calculating the proportion of planarians that reach the dark zone on the side opposite to light. (F) Immunostaining of the ventral cilia using an anti-Ac-Tubulin antibody (green). DAPI is used to label the nucleus (red). Magnified images are shown in the right corner. Scale bar = 100 μm. (G) Quantification analysis of cilia density using the green/red fluorescence intensity ratio. ***P < 0.001, by the Student’s t-test.
We next examined the effect of PI3K inhibition on planarian homeostasis, a self-regulating process allowing for tissue maintenance during adult cell turnover. After a 20-day treatment with LY294002, slight ulceration of the epidermis was observed in quite a few animals (12/40), and tissue lysis could be found in several animals (4/40) (Figure 3D). Although more than half of the animals appeared no morphological abnormality after LY294002 treatment, their locomotion was considerably hampered. When exposed to light, the control animals moved quickly away from the light and reached the dark field within 2 min. On the contrary, LY294002 treated animals twisted forward, flipped over, or stayed still during their traveling toward the dark field. No animals reached the dark field until 3 min post light exposure, and two-third of the animals never arrived at the dark field even at 20 min after light exposure (Figure 3E and Supplementary Videos 1, 2). Plane gliding is mediated by synchronous cilia movement on the ventral surface of planarians (Rink et al., 2009). To verify the possibility that LY294002-induced impaired gliding ability was caused by cilia defect, we used whole-mount staining with the anti-Ac-tubulin antibody to visualize the integrity of ciliary epithelial cells in the ventral epithelium. The LY294002 treated animals displayed poor staining in cilia that were severely shortened, whereas the control animals showed dense coverage of cilia (Figures 3F,G). Our results suggest that impaired motility in the Djpi3k inhibition phenotype is most likely due to insufficient density of ventral epithelial cells. Thus, these results indicated that PI3K activation is required for planarian homeostasis, especially cilia maintenance.
RNAi has been widely used to study regeneration-associated genes in planarians (Sánchez Alvarado and Newmark, 1999; Newmark et al., 2003; Rouhana et al., 2013). A bacterial feeding protocol (Newmark et al., 2003; Zeng et al., 2013) was employed in this study to investigate the role of Djpi3k in planarian regeneration and homeostasis. RNAi efficiency was confirmed by WISH and qRT-PCR analysis (Figures 4A,B). Djpi3k(RNAi) animals also exhibited limited regenerative capacity compared with the control animals (Figures 4C,D). Different from the LY294002 treatment, RNAi-mediated PI3K inhibition led to relatively mild phenotypes and a lower proportion of animals with regeneration defects (27/50 vs 46/50 in head regeneration, 29/50 vs 48/50 in tail regeneration). A similar situation occurs in tissue homeostasis, decreased cilia density was observed in Djpi3k(RNAi) animals, nevertheless, the integrity of the ciliated structure was maintained (Figures 4E,F). Taken together, these data suggest that PI3K signaling is required for planarian regeneration and tissue maintenance.
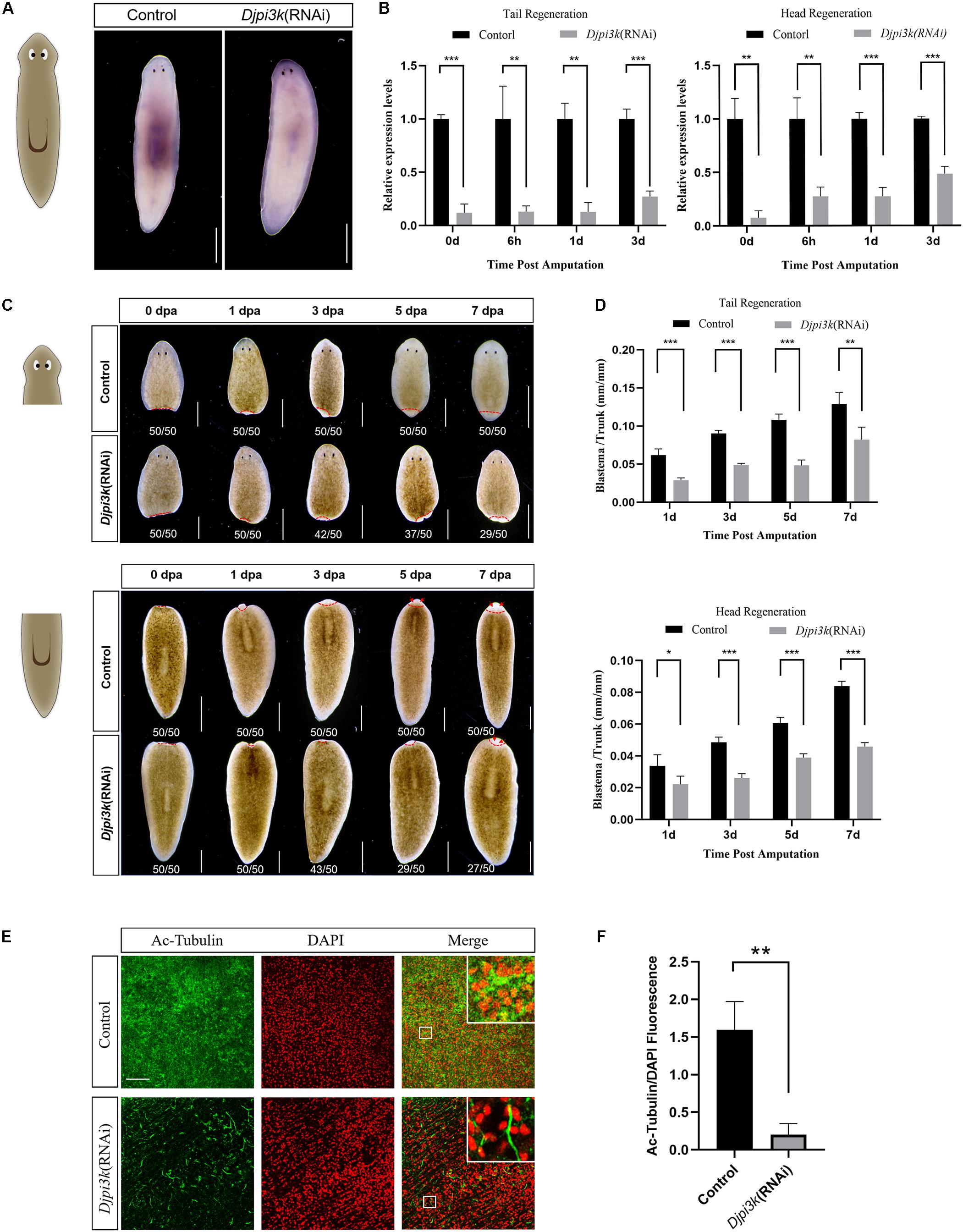
Figure 4. Effect of Djpi3k RNAi knockdown on planarian regeneration and homeostasis. (A) RNAi efficiency was verified in intact animals by WISH using an antisense Djpi3k probe. Scale bar = 1 mm. (B) RNAi efficiency was examined during planarian regeneration by qRT-PCR assay. **P < 0.01 and ***P < 0.001, by the Student’s t-test. (C) Dorsal images of regenerating trunk fragments from the control and Djpi3k (RNAi) animals at 0, 1, 3, 5, and 7 days post amputation. Newly-regenerated photoreceptors are indicated with red arrows. Scale bar = 1 mm. (D) Quantification analysis of blastema growth. The length of the regenerated blastema was measured and normalized to the whole-body length of the trunk. *P < 0.05, **P < 0.01, and ***P < 0.001, by the Student’s t-test. (E) Immunostaining of the ventral cilia using an anti-Ac-Tubulin antibody (green). DAPI is used to label the nucleus (red). Magnified images are shown in the right corner. Scale bar = 100 μm. (F) Quantification analysis of cilia density using the green/red fluorescence intensity ratio. **P < 0.01, by the Student’s t-test.
PI3K Regulates Mitotic and Apoptotic Responses During Planarian Regeneration
PI3K signaling participates in multiple biological processes associated with cell growth and proliferation (Madsen, 2020). Neoblasts represent the only proliferative cell population in planarians and are therefore the source of new cells for normal tissue turnover, growth, and regeneration (Wagner et al., 2011). We checked the effect of PI3K inhibition on the mitotic activity of neoblasts during planarian regeneration using the anti-H3P antibody, which labels cells in the G2/M phase (Peiris et al., 2012, 2016). In intact planarians, the H3P-positive cells were detected distributed throughout the body except in the head region anterior to the eyes and the pharynx region (Supplementary Figure 2C), which was consistent with the distribution patterns of planarian neoblasts reported in previous studies (Newmark and Sánchez Alvarado, 2000; Salvetti et al., 2000) and further verified the specificity of the H3P staining results in our study. For regeneration studies, the number of the H3P-positive cells in the control animals reached the first peak of mitosis at 6 hpa and the second peak at 3 dpa, which was consistent with previous studies (Wenemoser and Reddien, 2010). However, the number of the H3P-positive cells was dramatically decreased after LY294002 treatment, making the mitotic response vanished at the early stage of planarian regeneration (Figures 5A,B). Knockdown of Djpi3k also reduced the neoblast number, the mitotic response of Djpi3k(RNAi) animals was alleviated but still existed (Supplementary Figures 5A,B). Moreover, the expression pattern of Djmcm2, a neoblast marker (Salvetti et al., 2000), was evaluated before and after PI3K inactivation. The qRT-PCR results showed that the expression levels of Djmcm2 could reflect the mitotic trends as manifested by H3P staining, and were substantially suppressed after LY294002/A66 treatment or Djpi3k knockdown (Figure 5C and Supplementary Figures 4C, 5C). Our results suggest that PI3K signaling is required to maintain the appropriate number of proliferating neoblast during tissue renewal in adult planarians.
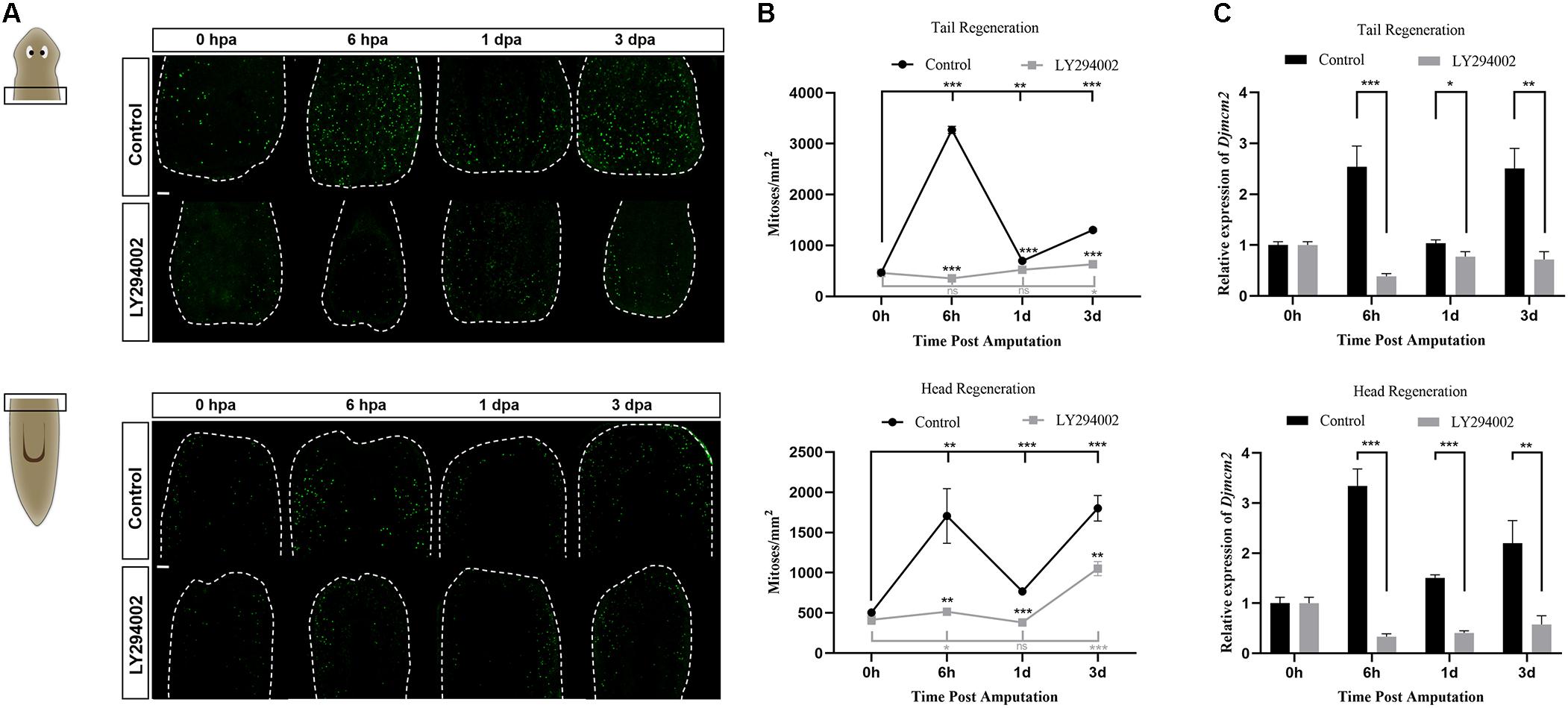
Figure 5. Effect of the PI3K inhibitor LY294002 on neoblast proliferation during planarian regeneration. (A) Representative images showing mitotic activity (green dots, H3P positive cells) at 0 h, 6 h, 1 days, and 3 days post amputation. Scale bar = 100 μm. (B) Quantification of the average number of H3P positive cells/mm2 in the regenerating trunk fragments at indicated time point. *P < 0.05, **P < 0.01, ***P < 0.001, and ns, not significant, by the Student’s t-test. (C) Relative expression levels of Djmcm2 gene in regenerating head and tail fragments. The expression level of Djmcm2 at 0 hpa was set to one. *P < 0.05, **P < 0.01, and ***P < 0.001, by the Student’s t-test.
Apoptosis, a prominent form of cell death in planarians, facilitates cell turnover during homeostasis and regeneration (Pellettieri et al., 2010). To investigate the possible role of PI3K signaling in apoptotic cell death during planarian regeneration, the spatiotemporal distribution of apoptosis was evaluated by TUNEL staining. The results revealed that the number of apoptotic cells in both the head and tail fragments was substantially decreased throughout the regeneration period after LY294002 treatment (Figure 6A). The system-wide cell death expected at 3 dpa was also suppressed by LY294002 treatment (Figure 6A and left panel of Figure 6B). It was noted that the localized cell death response at 6 hpa in control animals was also largely impaired by LY294002 treatment (Figure 6B, right panel). Similarly, cell death in Djpi3k(RNAi) animals failed to localize at the amputation site as is expected at 6 hpa (Supplementary Figures 6A,B). Nevertheless, Djpi3k(RNAi) animals showed a delayed apoptotic peak at 1 dpa during head and tail regeneration. Moreover, the expression pattern of Djbax, a pro-apoptotic marker gene, was evaluated by qRT-PCR before and after PI3K inhibition, and the results were consistent with the TUNEL staining results (Figure 6C and Supplementary Figures 4D, 6C). These findings indicate that PI3K signaling is essential for controlling wound-induced apoptotic cell death during tissue regeneration. Taken together, we propose Djpi3k functions as a regulator of proliferation and apoptosis during large-scale tissue regeneration in planarians.
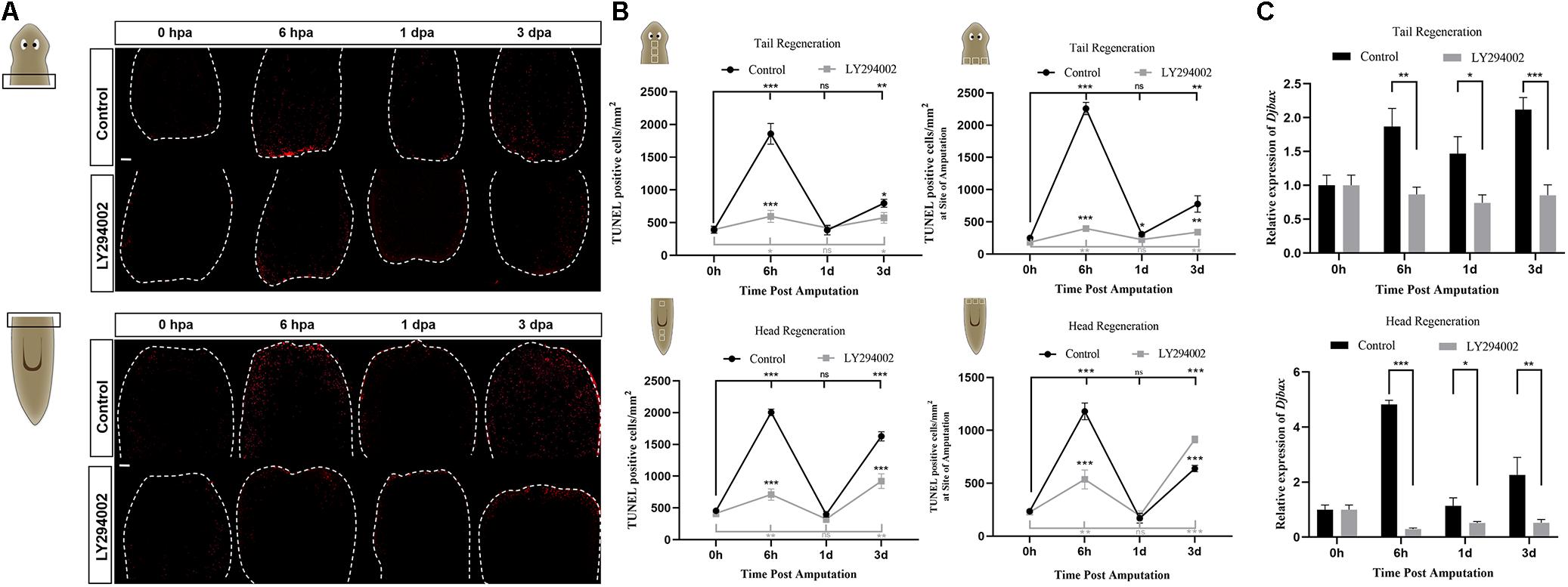
Figure 6. Effect of the PI3K inhibitor LY294002 on apoptotic cell death during planarian regeneration. (A) Immunostaining of TUNEL-positive nuclei of trunk fragments at 0 h, 6 h, 1 days, and 3 days post amputation. Scale bar = 100 μm. (B) Quantification of the average number of TUNEL-positive cells/mm2 in the whole trunk fragments (left panel) or at the amputation sites (right). *P < 0.05, **P < 0.01, ***P < 0.001, and ns, not significant, by the Student’s t-test. (C) Relative expression levels of Djbax gene in regenerating head and tail fragments. The expression level of Djbax at 0 hpa was set to one. *P < 0.05, **P < 0.01, ***P < 0.001, and ns, not significant, by the Student’s t-test.
Discussion
PI3K signaling is one of the most important and conserved signaling pathways in eukaryotic cells and is closely related to animal regeneration (Chen et al., 2013). In the present study, the planarian Dugesia japonica was employed to study the potential role of PI3K signaling in the regulation of the regeneration process. Our results demonstrated that PI3K plays a crucial role in maintaining proper mitotic and apoptotic responses to amputation, which might be essential for planarian regeneration.
According to sequence homology and substrate specificity, PI3Ks are mainly divided into three categories: Class I, II, and III (Thorpe et al., 2015). The catalytic subunits of Class I PI3K includes four highly homologous p110 isoforms (p110α,p110β, p110γ, and p110δ) encoded by genes PIK3CA, PIK3CB, PIK3CG, and PIK3CD, respectively (Zhao and Vogt, 2008). The results of conserved domain prediction and phylogenetic analysis suggested Djpi3k is highly homologous to the PIK3CA gene, which encodes the best-characterized Class I PI3K catalytic isoform p110α. The ubiquitously expressed p110α is crucial for organismal growth and development, with pleiotropic functions including self-renewal of stem cells, tissue maintenance, and metabolic regulation (Madsen, 2020). Our phylogenetic results demonstrate that the majority of the PI3K catalytic isoforms in each subgroup share similarities in sequence homology and conserved domain pattern. Nevertheless, the PI3K-like genes derived from Caenorhabditis elegans (AGE1_CAEEL) and Leishmania infantum (A4HVU4_LEIIN), lacking the agonist binding domain (ABD) though, were also clustered into the PIK3CG subgroup probably due to the sequence homology. Surprisingly, the hydra and arthropod PI3K-like genes were not clustered into any of the four sub-categories, probably because of the low similarity in domain pattern and sequence homology. Interestingly, the amphibian Xenopus tropicalis does not contain a p110 delta isoform, which is quite different from other chordates possessing four p110 isoforms. These findings suggest that the combined utilization of phylogenetic analysis and associated annotations provide a powerful tool to organize genes of interest into sub-categories.
The precise modulation of PI3K signaling is critical for the growth and development of the organism, while uncontrolled activation of PI3K leads to pathological phenomena including tumorigenesis (Lien et al., 2017). In this study, the expression pattern of Djpi3k was characterized and suggests it appears to be an early wound response gene. Wound-induced genes, including a Schmidtea mediterranea pi3k like gene, have been well described in planarians (Wenemoser et al., 2012) and clustered based on their onset and offset times into three groups: early, late, and sustained (Wurtzel et al., 2015). The early cluster genes, such as egr-l 1, ston, and heat shock protein genes, are immediately induced post amputation and fast to decay. Conversely, the late and sustained cluster genes, such as runt-1 and inhibin-1, also exhibit rapid induction but last for days, which resembles the expression pattern of Djpi3k characterized in this study. It is now well accepted that a generic response triggered by different types of injuries leads to mere wound healing or regeneration initiation depending on if tissue loss occurs (Owlarn et al., 2017). During planarian regeneration, Djpi3k is highly expressed in newly-regenerated blastema and pharynx-forming regions, implying that it can somehow sense the tissue context. Further research should be undertaken to investigate what signals the Djpi3k expression is triggered by and how PI3K signaling translates the generic wound into regeneration.
To investigate the role of PI3K signaling in planarian regeneration and homeostasis, a loss-of-function study was first conducted using small molecules. LY294002 treatment leads to regeneration defects in planarians manifested as regrowing marginal blastemas and incomplete brain structures. Tissue homeostasis is also disturbed by long-term treatment of LY294002, one of the hallmarks is the impaired cilia-dependent gliding ability. In accordance with the present results, previous studies have demonstrated that other components of the PI3K pathway play crucial roles in planarian regeneration and tissue maintenance. Akt, as a main downstream target of PI3K, regulates neoblast biology and cell death response during tissue repair in planarian Schmidtea mediterranea. Knockdown of Smed-Akt also impairs the maintenance of adult tissues including the nervous and excretory systems and ciliated structures (Peiris et al., 2016). Similarly, systemic inhibition of Target of Rapamycin (TOR) signaling leads to restricted proliferative capacity of neoblasts and tissue degeneration. Intriguingly, though blastema formation is hampered by Smed-TOR(RNAi), regeneration of CNS and visual neurons takes place within pre-existing tissue (Peiris et al., 2012). On the contrary, a phosphoinositide 3-kinase-related kinase (PIKK) family member SMG-1 acts antagonistically with mTOR signaling and works as a brake on the initial response to injury. Loss of SMG-1 leads to hyperactive responses to injury and subsequent growth that continues out of control (González-Estévez et al., 2012). Tumor suppressor gene PTEN is a predominant negative regulator of PI3K signaling (Song et al., 2012). The hallmarks of RNAi phenotype after silencing of Smed-PTEN-1 and Smed-PTEN-2 are hyperproliferation of neoblasts, tissue disorganization, and a significant accumulation of postmitotic cells with impaired differentiation capacity (Oviedo et al., 2008), the latter of which also accords with the earlier observations in Pten-ablated mouse embryonic stem cells (Di Cristofano et al., 1998). These findings suggest that PI3K signaling is required for planarian regeneration and tissue homeostasis.
During planarian regeneration, neoblasts undergo extensive cell division to regenerate lost or damaged tissue (Wenemoser and Reddien, 2010), and apoptotic cell death coordinates with cell division to drive adult tissue renewal and repair (Pellettieri et al., 2010). Thus, we further checked the effect of PI3K inhibition on cell proliferation and apoptotic cell death during planarian regeneration. The mitotic and apoptotic responses to amputation are substantially abated throughout the regeneration period after LY294002 or A66 treatment, which may contribute to the inhibitor-induced regeneration defects. To verify the specificity of these small molecules, RNAi experiments were also conducted. One interesting finding is that Djpi3k(RNAi) animals exhibit similar but relatively mild phenotypes in morphology compared with the inhibitor-treated animals. For neoblast proliferation, LY294002 completely vanishes planarian mitotic response to amputation at the early regenerative stage (Figure 5B), while a weak but significant mitotic response still exists in the regenerating Djpi3k(RNAi) animals (Supplementary Figure 5B). For cell death, it is unexpectedly to note that LY294002 acts to “blunt” planarian apoptotic response while knockdown of Djpi3k acts more likely to “postpone” the onset of the first wave of apoptotic cell death. These findings suggest that the different phenotypes between PI3K inhibitor-treated animals and Djpi3k(RNAi) animals may be attributed to the different extent of inhibition against PI3K activity.
Taken together, we propose that PI3K signaling is required for planarian regeneration and homeostasis (Figure 7). A loss-of-function assay using small molecules or RNAi technique suggests PI3K plays crucial roles in tissue regeneration, cilia maintenance, neoblast proliferation, and apoptotic cell death. Our findings complement those of earlier studies relative to the planarian PI3K signaling pathway and provide insights for investigating the disease-related genes in the regeneration-competent organism in vivo.
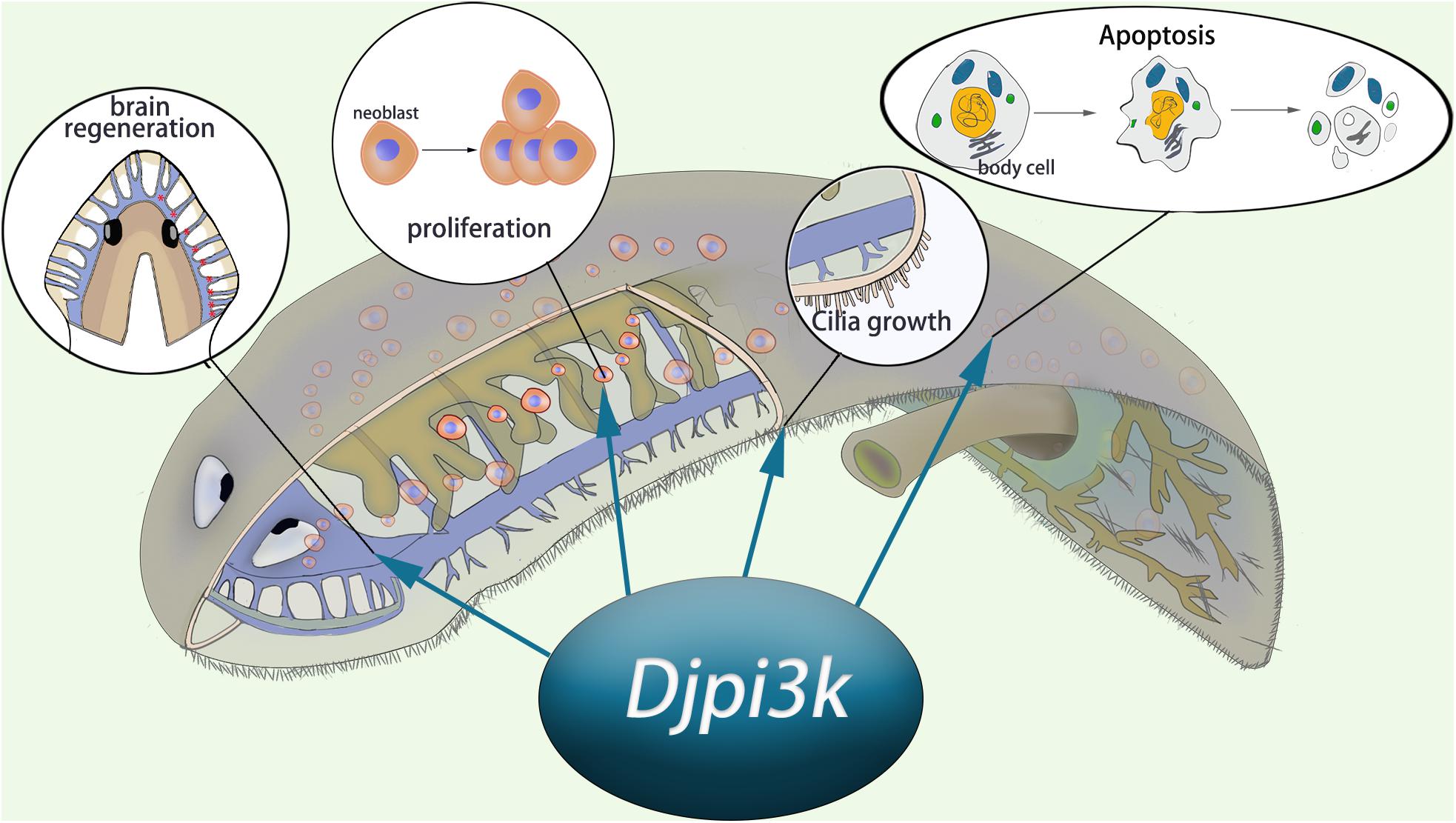
Figure 7. Schematic representation of the essential role of Djpi3k in planarian regeneration and tissue maintenance.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
YY and FC designed and supervised the study. HZ, HL, LL, and GY performed the experiments. QX, WW, and XW analyzed the data. YY and HZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31900330 and 31771055) and the Natural Science Basic Research Plan in Shaanxi Province of China (2017JQ3021).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Shoutao Zhang for helpful suggestions and Qingnan Tian for technical assistance. We would also like to thank Ziyi Shen for her assistance in confocal microscopy and photography.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.649656/full#supplementary-material
Supplementary Figure 1 | Conserved domain prediction of Djpi3k protein. The conserved domains of Djpi3k protein were predicted using the SMART database (A) and the NCBI Conserved Domain Database (B). The predicted Class I PI3K catalytic domain and accessory domain were labeled with red stars.
Supplementary Figure 2 | Specificity verification of the WISH and H3P immunostaining analyses in intact animals. Whole-mount in situ hybridization with sense and anti-sense Djpi3k (A) or DjpiwiA (B) probes, respectively. Scale bar = 1 mm. (C) Distribution of proliferative neoblasts in an intact planarian detected by immunostaining using an anti-H3P antibody. Photoreceptors are indicated with white stars and the pharynx region is outlined with white dashed lines. Scale bar = 200 μm.
Supplementary Figure 3 | Representative regenerative phenotypes induced by a 7-day treatment of a concentration gradient of LY2940027. Scale bar = 1 mm.
Supplementary Figure 4 | Effect of the PI3K inhibitor A66 on planarian regeneration. (A) Dorsal images of control and A66-treated head and tail fragments at 0, 1, 3, 5, and 7 days post amputation. Newly-regenerated photoreceptors are indicated with red arrows. Scale bar = 1 mm. (B) Quantification analysis of blastema growth. The length of the regenerated blastema was measured and normalized to the whole-body length of the trunk. ∗P < 0.05 and ∗∗∗P < 0.001, by the Student’s t-test. (C,D) Effect of the PI3K inhibitor A66 on Djmcm2 (C) and Djbax (D) expression during planarian regeneration. The expression level of Djmcm2 or Djbax at 0 hpa was set to one. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ns, not significant, by the Student’s t-test.
Supplementary Figure 5 | Effect of Djpi3k RNAi knockdown on neoblast proliferation during planarian regeneration. (A) Representative images showing mitotic activity (green dots, H3P positive cells) at 0 h, 6 h, 1 day, and 3 days post amputation. Scale bar = 100 μm. (B) Quantification of the average number of H3P positive cells/mm2 in the regenerating trunk fragments at indicated time point. ∗∗P < 0.01, ∗∗∗P < 0.001, and ns, not significant, by the Student’s t-test. (C) Relative expression levels of Djmcm2 gene in regenerating head and tail fragments. The expression level of Djmcm2 at 0 hpa was set to one. ∗∗P < 0.01 and ∗∗∗P < 0.001, by the Student’s t-test.
Supplementary Figure 6 | Effect of Djpi3k RNAi knockdown on apoptotic cell death during planarian regeneration. (A) Immunostaining of TUNEL-positive nuclei of trunk fragments at 0 h, 6 h, 1 days, and 3 days post amputation. Scale bar = 100 μm. (B) Quantification of the average number of TUNEL-positive cells/mm2 in the whole trunk fragments (left panel) or at the amputation sites (right). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ns, not significant, by the Student’s t-test. (C) Relative expression levels of Djbax gene in regenerating head and tail fragments. The expression level of Djbax at 0 hpa was set to one. ∗∗P < 0.01, ∗∗∗P < 0.001, and ns, not significant, by the Student’s t-test.
Supplementary Video 1 | Representative video showing the gliding ability of control animals. All control animals moved quickly away from the light and reached the dark field within 2 min when exposed to light.
Supplementary Video 2 | Representative video showing the gliding ability of LY294002-treated animals. Two-third of the LY294002-treated animals never arrived at the dark field even at 20 min post light exposure, exhibiting photophobic but impaired locomotion behaviors including twisting, flipping, and staying still.
Abbreviations
Dj, Dugesia japonica; PI3K, phosphatidylinositol 3-kinase; dsRNA, double-stranded RNA; RNAi, RNA interference; ORF, open reading frame; pI, isoelectric point; WISH, whole-mount in situ hybridization; qRT-PCR, quantitative Real-time PCR; hpa/dpa, hours/days post amputation; H3P, phosphorylated histone H3; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling.
Footnotes
References
Agata, K., and Umesono, Y. (2008). Brain regeneration from pluripotent stem cells in planarian. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 2071–2078. doi: 10.1098/rstb.2008.2260
Cebrià, F., Kobayashi, C., Umesono, Y., Nakazawa, M., Mineta, K., Ikeo, K., et al. (2002). FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature 419, 620–624. doi: 10.1038/nature01042
Chen, J., Crawford, R., Chen, C., and Xiao, Y. (2013). The key regulatory roles of the PI3K/Akt signaling pathway in the functionalities of mesenchymal stem cells and applications in tissue regeneration. Tissue Eng. Part B Rev. 19, 516–528. doi: 10.1089/ten.TEB.2012.0672
Courtney, K. D., Corcoran, R. B., and Engelman, J. A. (2010). The PI3K pathway as drug target in human cancer. J. Clin. Oncol. 28, 1075–1083. doi: 10.1200/jco.2009.25.3641
Di Cristofano, A., Pesce, B., Cordon-Cardo, C., and Pandolfi, P. P. (1998). Pten is essential for embryonic development and tumour suppression. Nat. Genet. 19, 348–355. doi: 10.1038/1235
González-Estévez, C., Felix, D. A., Smith, M. D., Paps, J., Morley, S. J., James, V., et al. (2012). SMG-1 and mTORC1 act antagonistically to regulate response to injury and growth in planarians. PLoS Genet. 8:e1002619. doi: 10.1371/journal.pgen.1002619
Hayashi, T., Asami, M., Higuchi, S., Shibata, N., and Agata, K. (2006). Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev. Growth Differ. 48, 371–380. doi: 10.1111/j.1440-169X.2006.00876.x
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W. H., and Hu, S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44, W236–W241. doi: 10.1093/nar/gkw370
Huang, X., Liu, G., Guo, J., and Su, Z. (2018). The PI3K/AKT pathway in obesity and type 2 diabetes. Int. J. Biol. Sci. 14, 1483–1496. doi: 10.7150/ijbs.27173
Jackson, L. N., Larson, S. D., Silva, S. R., Rychahou, P. G., Chen, L. A., Qiu, S., et al. (2008). PI3K/Akt activation is critical for early hepatic regeneration after partial hepatectomy. Am. J. Physiol. Gastrointest. Liver Physiol. 294, G1401–G1410. doi: 10.1152/ajpgi.00062.2008
Jamieson, S., Flanagan, J. U., Kolekar, S., Buchanan, C., Kendall, J. D., Lee, W. J., et al. (2011). A drug targeting only p110α can block phosphoinositide 3-kinase signalling and tumour growth in certain cell types. Biochem. J. 438, 53–62. doi: 10.1042/bj20110502
Jung, K., Kim, M., So, J., Lee, S. H., Ko, S., and Shin, D. (2020). Farnesoid X receptor activation impairs liver progenitor cell-mediated liver regeneration via the PTEN-PI3K-AKT-mTOR axis in zebrafish. Hepatology 2020:31679. doi: 10.1002/hep.31679
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Lien, E. C., Dibble, C. C., and Toker, A. (2017). PI3K signaling in cancer: beyond AKT. Curr. Opin. Cell Biol. 45, 62–71. doi: 10.1016/j.ceb.2017.02.007
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Madsen, R. R. (2020). PI3K in stemness regulation: from development to cancer. Biochem. Soc. Trans. 48, 301–315. doi: 10.1042/bst20190778
Manuel, G. C., Reynoso, R., Gee, L., Salgado, L. M., and Bode, H. R. (2006). PI3K and ERK 1-2 regulate early stages during head regeneration in hydra. Dev. Growth Differ. 48, 129–138. doi: 10.1111/j.1440-169X.2006.00847.x
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Newmark, P. A., Reddien, P. W., Cebrià, F., and Sánchez Alvarado, A. (2003). Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc. Natl. Acad. Sci. USA 100, 11861–11865. doi: 10.1073/pnas.1834205100
Newmark, P. A., and Sánchez Alvarado, A. (2000). Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev. Biol. 220, 142–153. doi: 10.1006/dbio.2000.9645
Oviedo, N. J., Pearson, B. J., Levin, M., and Sánchez Alvarado, A. (2008). Planarian PTEN homologs regulate stem cells and regeneration through TOR signaling. Dis. Model. Mech. 1, 131–143. doi: 10.1242/dmm.000117
Owlarn, S., Klenner, F., Schmidt, D., Rabert, F., Tomasso, A., Reuter, H., et al. (2017). Generic wound signals initiate regeneration in missing-tissue contexts. Nat. Commun. 8:2282. doi: 10.1038/s41467-017-02338-x
Pearson, B. J., Eisenhoffer, G. T., Gurley, K. A., Rink, J. C., Miller, D. E., and Sánchez Alvarado, A. (2009). Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev. Dyn. 238, 443–450. doi: 10.1002/dvdy.21849
Peiris, T. H., Ramirez, D., Barghouth, P. G., and Oviedo, N. J. (2016). The Akt signaling pathway is required for tissue maintenance and regeneration in planarians. BMC Dev. Biol. 16:7. doi: 10.1186/s12861-016-0107-z
Peiris, T. H., Weckerle, F., Ozamoto, E., Ramirez, D., Davidian, D., García-Ojeda, M. E., et al. (2012). TOR signaling regulates planarian stem cells and controls localized and organismal growth. J. Cell Sci. 125, 1657–1665. doi: 10.1242/jcs.104711
Pellettieri, J., Fitzgerald, P., Watanabe, S., Mancuso, J., Green, D. R., and Sánchez Alvarado, A. (2010). Cell death and tissue remodeling in planarian regeneration. Dev. Biol. 338, 76–85. doi: 10.1016/j.ydbio.2009.09.015
Reddien, P. W., and Sánchez Alvarado, A. (2004). Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 20, 725–757. doi: 10.1146/annurev.cellbio.20.010403.095114
Rink, J. C., Gurley, K. A., Elliott, S. A., and Sánchez Alvarado, A. (2009). Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science 326, 1406–1410. doi: 10.1126/science.1178712
Rouhana, L., Weiss, J. A., Forsthoefel, D. J., Lee, H., King, R. S., Inoue, T., et al. (2013). RNA interference by feeding in vitro-synthesized double-stranded RNA to planarians: methodology and dynamics. Dev. Dyn. 242, 718–730. doi: 10.1002/dvdy.23950
Saijilafu, Hur, E. M., Liu, C. M., Jiao, Z., Xu, W. L., and Zhou, F. Q. (2013). PI3K-GSK3 signalling regulates mammalian axon regeneration by inducing the expression of Smad1. Nat. Commun. 4:2690. doi: 10.1038/ncomms3690
Salvetti, A., Rossi, L., Deri, P., and Batistoni, R. (2000). An MCM2-related gene is expressed in proliferating cells of intact and regenerating planarians. Dev. Dyn. 218, 603–614. doi: 10.1002/1097-0177(2000)9999:9999<::Aid-dvdy1016>3.0.Co;2-c
Sánchez Alvarado, A., and Newmark, P. A. (1999). Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc. Natl. Acad. Sci. USA 96, 5049–5054. doi: 10.1073/pnas.96.9.5049
Scimone, M. L., Atabay, K. D., Fincher, C. T., Bonneau, A. R., Li, D. J., and Reddien, P. W. (2020). Muscle and neuronal guidepost-like cells facilitate planarian visual system regeneration. Science 368:eaba3203. doi: 10.1126/science.aba3203
Song, M. S., Salmena, L., and Pandolfi, P. P. (2012). The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 13, 283–296. doi: 10.1038/nrm3330
Suzuki, M., Satoh, A., Ide, H., and Tamura, K. (2007). Transgenic Xenopus with prx1 limb enhancer reveals crucial contribution of MEK/ERK and PI3K/AKT pathways in blastema formation during limb regeneration. Dev. Biol. 304, 675–686. doi: 10.1016/j.ydbio.2007.01.019
Tang, J., Yu, Y., Zheng, H., Yin, L., Sun, M., Wang, W., et al. (2017). ITRAQ-based quantitative proteomic analysis of Cynops orientalis limb regeneration. BMC Genomics 18:750. doi: 10.1186/s12864-017-4125-4
Thorpe, L. M., Yuzugullu, H., and Zhao, J. J. (2015). PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 15, 7–24. doi: 10.1038/nrc3860
Wagner, D. E., Wang, I. E., and Reddien, P. W. (2011). Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811–816. doi: 10.1126/science.1203983
Walker, E. H., Pacold, M. E., Perisic, O., Stephens, L., Hawkins, P. T., Wymann, M. P., et al. (2000). Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin. LY294002, quercetin, myricetin, and staurosporine. Mol. Cell 6, 909–919. doi: 10.1016/s1097-2765(05)00089-4
Wang, W., Yu, Y., Liu, H., Zheng, H., Jia, L., Zhang, J., et al. (2021). Protein core fucosylation regulates planarian head regeneration via neoblast proliferation. Front. Cell Dev. Biol. 9:625823. doi: 10.3389/fcell.2021.625823
Wenemoser, D., Lapan, S. W., Wilkinson, A. W., Bell, G. W., and Reddien, P. W. (2012). A molecular wound response program associated with regeneration initiation in planarians. Genes. Dev. 26, 988–1002. doi: 10.1101/gad.187377.112
Wenemoser, D., and Reddien, P. W. (2010). Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev. Biol. 344, 979–991. doi: 10.1016/j.ydbio.2010.06.017
Wurtzel, O., Cote, L. E., Poirier, A., Satija, R., Regev, A., and Reddien, P. W. (2015). A generic and cell-type-specific wound response precedes regeneration in planarians. Dev. Cell 35, 632–645. doi: 10.1016/j.devcel.2015.11.004
Xu, F., Na, L., Li, Y., and Chen, L. (2020). Roles of the PI3K/AKT/mTOR signalling pathways in neurodegenerative diseases and tumours. Cell Biosci. 10:54. doi: 10.1186/s13578-020-00416-0
Yazawa, S., Umesono, Y., Hayashi, T., Tarui, H., and Agata, K. (2009). Planarian Hedgehog/Patched establishes anterior-posterior polarity by regulating Wnt signaling. Proc. Natl. Acad. Sci. USA 106, 22329–22334. doi: 10.1073/pnas.0907464106
Yujia, S., Tingting, G., Jiaxin, L., Saisai, Z., Zhitai, H., Qingnan, T., et al. (2019). Cdc42 regulate the apoptotic cell death required for planarian epidermal regeneration and homeostasis. Int. J. Biochem. Cell Biol. 112, 107–113. doi: 10.1016/j.biocel.2019.05.008
Zeng, A., Li, Y. Q., Wang, C., Han, X. S., Li, G., Wang, J. Y., et al. (2013). Heterochromatin protein 1 promotes self-renewal and triggers regenerative proliferation in adult stem cells. J. Cell Biol. 201, 409–425. doi: 10.1083/jcb.201207172
Keywords: PI3K, planarian, regeneration, stem cell, cellular response
Citation: Zheng H, Liu H, Xu Q, Wang W, Li L, Ye G, Wen X, Chen F and Yu Y (2021) PI3K Plays an Essential Role in Planarian Regeneration and Tissue Maintenance. Front. Cell Dev. Biol. 9:649656. doi: 10.3389/fcell.2021.649656
Received: 05 January 2021; Accepted: 19 July 2021;
Published: 06 August 2021.
Edited by:
Zuoren Yu, Tongji University, ChinaReviewed by:
Qing Jing, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (CAS), ChinaYunfei Qin, Third Affiliated Hospital of Sun Yat-sen University, China
Copyright © 2021 Zheng, Liu, Xu, Wang, Li, Ye, Wen, Chen and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fulin Chen, Y2hlbmZsQG53dS5lZHUuY24=; Yuan Yu, eXV5dWFuQG53dS5lZHUuY24=
†These authors have contributed equally to this work
 Hanxue Zheng
Hanxue Zheng Hongbo Liu
Hongbo Liu Qian Xu
Qian Xu Wenjun Wang
Wenjun Wang Linfeng Li
Linfeng Li Gang Ye
Gang Ye Xiaomin Wen
Xiaomin Wen Fulin Chen
Fulin Chen Yuan Yu
Yuan Yu