
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 14 April 2021
Sec. Stem Cell Research
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.649605
This article is part of the Research Topic Understanding the Role of Cancer Stem Cells in Tumor Formation and Metastasis View all 4 articles
Chromatin-enriched RNAs (cheRNAs) constitute a special class of long noncoding RNAs (lncRNAs) that are enriched around chromatin and function to activate neighboring or distal gene transcription. Recent studies have shown that cheRNAs affect chromatin structure and gene expression by recruiting chromatin modifiers or acting as bridges between distal enhancers and promoters. The abnormal transcription of cheRNAs plays an important role in the occurrence of many diseases, particularly tumors. The critical effect of cancer stem cells (CSCs) on the formation and development of tumors is well known, but the function of cheRNAs in tumorigenesis, especially in CSC proliferation and stemness maintenance, is not yet fully understood. This review focuses on the mechanisms of cheRNAs in epigenetic regulation and chromatin conformation and discusses the way cheRNAs function in CSCs to deepen the understanding of tumorigenesis and provide novel insight to advance tumor-targeting therapy.
As one of the main causes of death worldwide, cancer impedes the increase in people’s life expectancy. It is estimated that approximately 9.6 million people died of cancer in 2018 (Bray et al., 2018). The reasons of tumor formation are complex and involve many factors, which also increases the difficulty of tumor therapy. Studies have shown that somatic copy-number changes (Beroukhim et al., 2010), epigenetic changes (Kundaje et al., 2015), and mutations in noncoding regions (Melton et al., 2015) are deeply associated with tumorigenesis. Compared to protein-coding genes, those noncoding genes account for more than 98% of the entire human genome, in which mutations may lead to tumorigenesis. Genome-wide association analysis (GWAS) indicates that noncoding genome mutations are relevant to a variety of diseases, including tumors (Maurano et al., 2012).
According to transcriptome analysis, human cells generate a large number of transcripts, of which over 68% were classified as long noncoding RNAs (lncRNAs; Iyer et al., 2015). LncRNAs constitute a class of functional RNA molecules with a length of more than 200 nucleotides and lacking protein coding ability. They are usually located in the nucleus and enriched in chromatin and specific subnuclear compartments (Sun et al., 2018). As a unique subgroup of lncRNAs, chromatin-enriched RNAs (cheRNAs) that are generally transcribed by RNA polymerase II and enriched around chromatin, and can transcriptionally activate neighboring genes (Werner and Ruthenburg, 2015). CheRNA can be regarded as a new kind of enhancer RNA, but their strand length, histone modification markers, and strand-specificity are different (Gayen and Kalantry, 2017). CheRNAs are lineage specific, which can stimulate gene transcription in cis by promoting contacts to a downstream enhancer (Werner et al., 2017). In addition, cheRNAs function at multiple chromatin levels, including chromatin modification (Khalil et al., 2009), chromosome inactivation (Brown et al., 1992), nuclear structure changes (Bergmann and Spector, 2014; Quinodoz and Guttman, 2014; Engreitz et al., 2016), and transcription (Ponting et al., 2009; Vance and Ponting, 2014). Recent studies have shown that transcription disorders associated with cheRNAs are deeply relevant to the progression of a number of tumors (Gupta et al., 2010; Martens-Uzunova et al., 2014; Huarte, 2015; Schmitt and Chang, 2016). Considering the importance of cheRNA regulation at different chromatin levels, it is necessary to study its effects on tumorigenesis.
Tumors comprise a complicated group of heterogeneous cells, other than a collection of a single type of cloned tumor cell. Due to the influence of heredity, epigenetics, and the complex tumor microenvironment, tumor cells show a high degree of heterogeneity (Magee et al., 2012; Nguyen et al., 2012; Meacham and Morrison, 2013). Tumor heterogeneity is the major factor characterizing tumor metastasis and recurrence, and it is also a key barrier to tumor therapy (Meacham and Morrison, 2013; Saygin et al., 2019). Hence, it is assumed that certain types of cells in tumors are responsible for maintaining tumor growth and metastasis. Generally, the cells presenting the greatest tumor propagating potential could be regarded as the cancer stem cells (CSCs; Greaves and Maley, 2012; Beck and Blanpain, 2013). CSCs hold the properties of stem cells, such as self-renewal capacity, tumor differentiation potential, and stem gene expression (Kreso and Dick, 2014; Batlle and Clevers, 2017; Suva and Tirosh, 2020). The CSC model provides an explanation suggesting that a small number of CSCs can maintain the heterogeneity and occurrence of tumors and differentiate into cells with distinct hierarchical structures (Reya et al., 2001; Shackleton et al., 2009; Kreso and Dick, 2014). In 1994, John Dick et al. provided a model for CSC research in acute myeloid leukemia (AML; Lapidot et al., 1994; Bonnet and Dick, 1997). To date, CSCs have been identified in a variety of tumor tissues, including breast cancer (Al-Hajj et al., 2003), colon cancer (Ricci-Vitiani et al., 2007), brain tumors (Singh et al., 2004), pancreatic cancer (Hermann et al., 2007), melanoma (Schatton et al., 2008), gastric cancer (Takaishi et al., 2008), and liver cancer (Sell and Leffert, 2008). Previous studies have shown that CSCs play significant roles in tumor growth and migration (Peitzsch et al., 2017). Shimokawa et al. (2017) found that Lgr5+ colon CSCs had the potential for self-renewal and differentiation, driving the growth of colon cancer, and tumorigenesis can be regressed once the CSCs are lost. Pancreatic cancer is a malignant tumor with a high fatality rate, partly due to its persistent metastasis, in which pancreatic CSCs play crucial roles. Furthermore, the CD133+ and CXCR4+ pancreatic CSCs control the metastasis of pancreatic cancer, and their ablation can lead to the loss of the pancreatic cancer metastatic phenotype (Hermann et al., 2007). This review describes chromatin-related cheRNAs as the anchor point, trying to clarify the cheRNA-induced imbalance in CSCs and tumors at the chromatin level, thereby providing a new perspective for understanding tumorigenesis and tumor-targeting therapy.
Nucleosome remodeling is a key factor in gene expression, and chromatin remodeling agents are involved in this process (Segal and Widom, 2009). ATP-dependent chromatin remodelers can be divided into four main families: switch/sucrose non-fermentable (SWI/SNF), ISWI, CHD, and INO80 (Portela and Esteller, 2010; You and Jones, 2012). A growing number of studies have clarified that chromatin remodeling agents are deeply connected with the occurrence of tumors (Clapier and Cairns, 2009; Ho and Crabtree, 2010; Hargreaves and Crabtree, 2011; Wilson and Roberts, 2011). CheRNA-mediated nucleosome localization has been reported in the Arabidopsis, yeast, and animals, indicating that this biological process has universal significance (Marina et al., 2013; Zhu et al., 2013; Zhao et al., 2016). Similarly, cheRNA-mediated nucleosome localization also plays a critical role in tumorigenesis. The cheRNA SChLAP1 is highly expressed in prostate cancer, which promotes the invasion and metastasis of prostate cancer cells by antagonizing the SWI/SNF complex and then affecting the localization of nucleosomes (Prensner et al., 2013). In view of the key roles of CSCs in tumorigenesis and metastasis, it is significant to understand their own regulatory mechanisms. Therefore, we emphasize the essential part of cheRNA in CSC proliferation and stemness maintenance. The cheRNA HAND2-AS1 is highly expressed in liver stem cells. By recruiting the INO80 chromatin remodeling complex to the promoter region of BMPR1A, HAND2-AS1 maintains the self-renewal of liver stem cells and promotes the occurrence of liver cancer (Wang et al., 2019b). Besides, the cheRNA lncTCF7 recruits the chromatin remodeling complex SWI/SNF to the promoter region of TCF7 to activate the Wnt signaling pathway, thereby promoting the self-renewal of liver CSCs (Wang et al., 2015). As a member of the CHD family, the Mi-2/NuRD complex plays a critical role in genome stability and tumor biology (Clapier and Cairns, 2009; Lai and Wade, 2011; Xia et al., 2017). The Mi-2/NuRD complex is recruited by cheRNA LncHDAC2 to repress PTCH1 transcription, which contributes to enhancing the Hedgehog signaling pathway and advancing the self-renewal of liver CSCs (Wu et al., 2019b). Figure 1A depicts a schematic diagram of cheRNAs regulating nucleosome positioning. Currently, cheRNAs are known to mainly regulate the conformation of different chromatin regions by recruiting or rejecting chromatin remodeling complexes, thus further changing gene expression. However, it remains unclear whether other RNA or protein factors are involved in this process, and therefore, the potential roles of these factors need to be further explored.
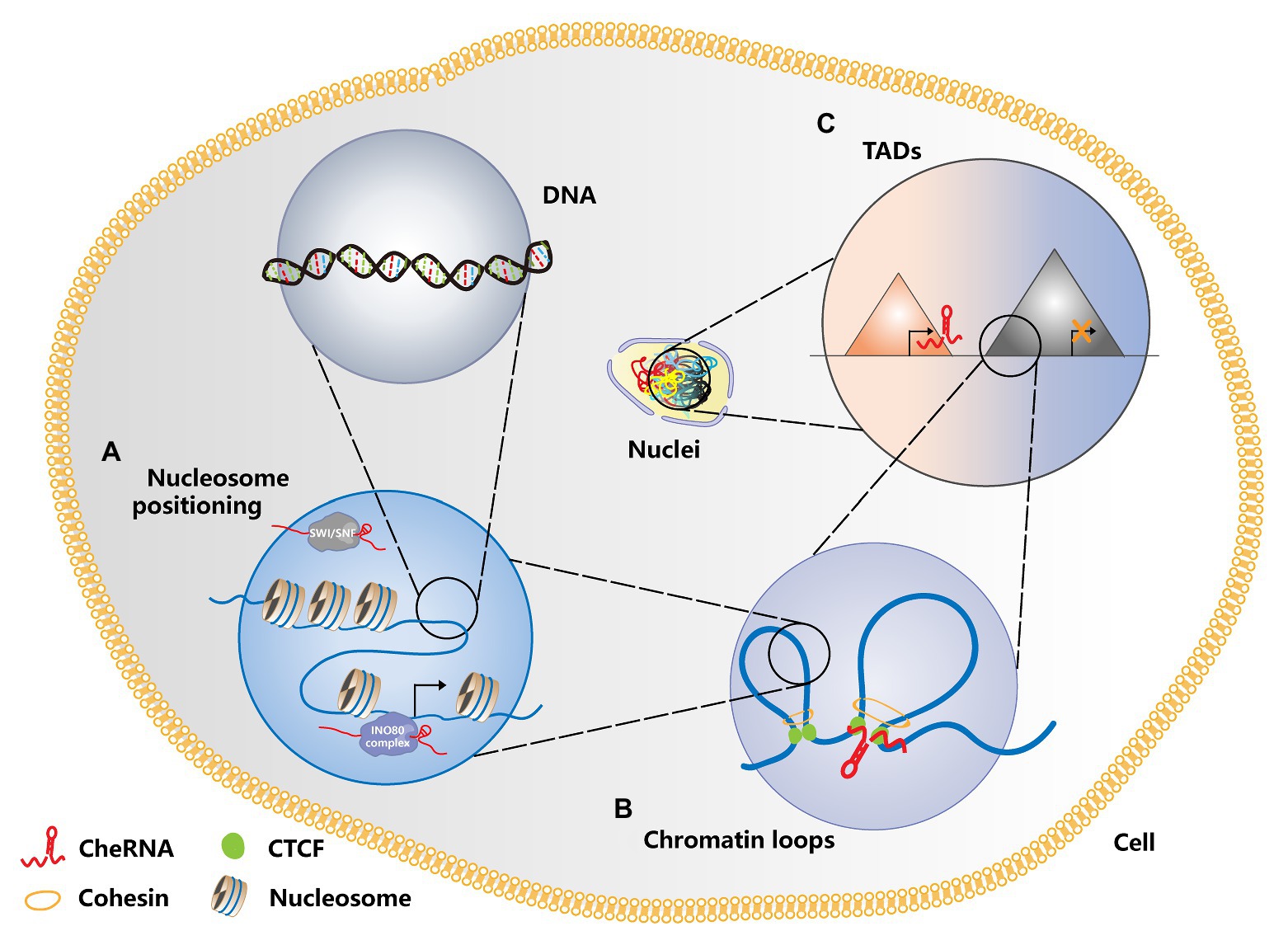
Figure 1. Schematic diagram of cheRNAs regulating chromosome conformation. (A) CheRNAs antagonize the SWI/SNF complex and prevent its binding to nucleosomes, thereby changing the nucleosome positioning and regulating gene transcription. In contrast, cheRNA recruits the INO80 complex, disaggregates the nucleosomes in the gene promoter region, and activates gene transcription. (B) CheRNAs can participate in the formation of chromatin loops in the promoter and enhancer regions, activate gene expression, and promote tumorigenesis. (C) Whether cheRNAs are transcribed or not can change the topologically associating domain (TAD) conformation of relevant regions, thereby regulating gene expression patterns and affecting tumor progression. TADS are represented by different triangles. The red one and the gray one, respectively shows the TADs where cheRNAs are transcribed or not transcribed.
Numerous researches have shown that chromatin conformation is closely correlated with gene expression regulation, especially the long-distance ones (Sexton and Cavalli, 2015; Uhler and Shivashankar, 2017). The most common mode of long-distance chromatin interaction is that of promoters and distal enhancers (Li et al., 2012; Sanyal et al., 2012). These chromatin-loop-mediated promoter and enhancer interactions also hold essential roles in the formation of induced pluripotent stem cells (Zhang et al., 2013). In addition to the most common proteins, such as CTCF, the SMC complex, and Cohesin, recent studies have found that cheRNAs can also modulate the chromatin loop formation of an active enhancer region and its target promoter (Orom and Shiekhattar, 2013; Yang et al., 2013; Trimarchi et al., 2014). The highly expressed cheRNAs PRNCR1 (also known as PCAT8) and PCGEM1 in prostate cancer can bind to the androgen receptor and mediate the cyclization of the promoters of androgen receptor binding enhancers and their target genes to promote the development of tumors (Yang et al., 2013). In T-cell acute lymphoblastic leukemia (T-ALL), the Notch-regulated cheRNA LUNAR1 binds to the enhancer of IGF1R, activates the promoter of IGF1R in cis, maintains a high level of mRNA expression, and promotes the growth of T-ALL (Trimarchi et al., 2014). The function of cheRNA in the formation of chromatin loops is explained in Figure 1B. These studies indicated that cheRNAs promote the circularization of enhancer and promoter interactions in different regions by recruiting chromatin regulators, thereby affecting the expression of downstream genes. However, it is still unclear that how cheRNAs participate in the formation of chromatin loops, and further in-depth studies are needed to determine the mechanism.
Previously, the eukaryotic genome was generally regarded as a one-dimensional entity composed of linear DNA sequences. However, with the development of chromatin conformation capture technology, people have gradually realized that chromatin constitutes a complex, ordered three-dimensional spatial structure. The double helix of DNA and histone octamers is packaged into nucleosomes, which are further folded to form chromatin fibers (Bonev and Cavalli, 2016). Chromatin fibers form rings, and multiple chromatin rings are assembled to form chromosome domains or topologically associating domains (TADs), which are megabases (Dixon et al., 2012; Nora et al., 2012). The interaction between multiple TADs leads to the formation of chromosomal compartments, and chromatin compartments further form chromatin territories (Bonev and Cavalli, 2016). A large number of studies have shown that TADs have major roles in gene expression (Andrey et al., 2013), disease occurrence (Kloetgen et al., 2020), and mammalian embryonic development (Ke et al., 2017). TAD disorders are deeply relevant to tumorigenesis. The depletion of CTCF and Cohesin weakens the formation of TADs, indicating that they can participate in the formation of the TAD boundary (Phillips-Cremins et al., 2013; Wutz et al., 2017). Studies have revealed that cheRNAs can participate in changes in the TAD boundary during immunoglobulin class transition (Rothschild et al., 2020). In the same manner, there are relevant studies in tumors. The cheRNA HOTTIP was upregulated in AML, and its knockout inhibited the expression of genes related to leukogenesis. Mechanistically, HOTTIP can alter HOXA-driven TAD and transcription program, leading to the occurrence of AML (Luo et al., 2019). In addition, HOTTIP modulates the stemness maintenance of pancreatic CSCs by enhancing the Wnt/β-catenin pathway (Fu et al., 2017). Patients with high HOTTIP expression had shorter survival than those with low expression. Furthermore, the high expression of HOTTIP is correlated with lymph node metastasis, indicating that HOTTIP may be a biomarker for lymph node metastasis in patients with pancreatic cancer and provide a basis for pancreatic cancer targeting therapy. Figure 1C shows whether transcription of cheRNA can alter TAD conformation. Changes in TAD boundaries are very important for gene expression regulation. Previous studies have indicated that CTCF occupies most of TAD boundaries and usually function with cohesion complex. However, little is known about the involvement of cheRNAs in the regulation of the TAD boundary, which should be paid more attention to so that we can discover new models of gene expression regulation. Table 1 summarizes the regulatory role of RNA in chromatin conformation in tumors and CSCs.
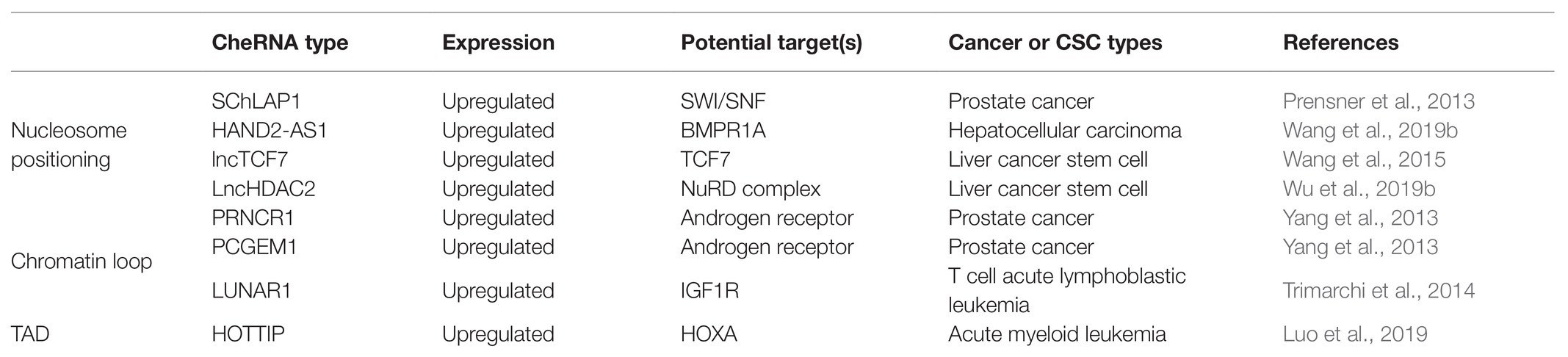
Table 1. Summary of chromatin-enriched RNAs (cheRNAs) modulate chromosome conformation in cancers and cancer stem cells (CSCs).
Epigenetic modification is the covalent modification of DNA, histones, or nucleosomes that does not affect the coding sequence of the genome, but can change the expression of genes (Egger et al., 2004; Allis and Jenuwein, 2016). The dysregulation of the epigenetic regulatory network can lead to inappropriate transcription or silencing of certain genes, thereby inducing tumorigenesis (Jones and Baylin, 2002; Egger et al., 2004). CheRNA-mediated epigenetic modification is involved in the development of a variety of cancers.
The hypomethylation status of the tumor genome was one of the first epigenetic factors to be studied (Feinberg and Vogelstein, 1983). DNA methylation is usually regarded as a mark of epigenetic silencing, and its dysregulation is deeply related to the occurrence of many tumors (Egger et al., 2004; Jones and Baylin, 2007; Chin et al., 2008; Muzny et al., 2012). DNA methylation prevents other DNA-binding proteins from binding to the genome at a specific site, leading to transcriptional silencing. In recent years, studies have illustrated that cheRNAs can also participate in DNA methylation modification, which in turn affects gene expression. The cheRNA LUCAT1 promotes the proliferation and metastasis of esophageal squamous cell carcinoma (ESCC) by increasing the stability of DNA methyltransferase 1 (DNMT1) and increases the methylation level of tumor suppressor factors, leading to the proliferation and metastasis of ESCC (Yoon et al., 2018). In colon cancer, the expression of cheRNA DACOR1 is inhibited, and induction of DACOR1 leads to the remethylation of CpG islands at FOS and JUN promoter sites, thereby affecting the development of colon cancer (Somasundaram et al., 2018). In addition, the cheRNA can also participate in DNA demethylation. CheRNA TARID recruits the DNA demethylation regulator GADD45A to the promoter region of TCF21 and mediates its demethylation to activate the expression of the tumor suppressor TCF21 (Arab et al., 2014). Furthermore, in CSCs, cheRNA can also participate in the formation of DNA methylation. CheRNA DLX6-AS1 downregulates the expression of the tumor suppressor CADM1 by increasing the methylation level of its promoter region, thus activating the STAT3 signaling pathway to promote the self-renewal of liver CSCs (Wu et al., 2019a). Figure 2A illustrates how cheRNAs regulate the DNA methylation level of the promoter region to affect gene transcription. Besides, other studies have shown that the occupation of cheRNA in the promoter region can suppress gene expression and regulate the progression of CSC. By competitively binding IL-6 promoter with NF-κB, lnc-DILC inhibits the autocrine IL-6/STAT3 signaling, therefore suppressing liver CSC expansion (Wang et al., 2016b). Clinical analysis shows that generally hepatocellular carcinoma patients have lower levels of lnc-DILC, while some patients with higher levels of lnc-DILC have lower recurrence rate and longer survival time. This research indicates that lnc-DILC may be a potential prognostic marker of hepatocellular carcinoma, and it can be utilized as a potential therapeutic target for liver CSCs.
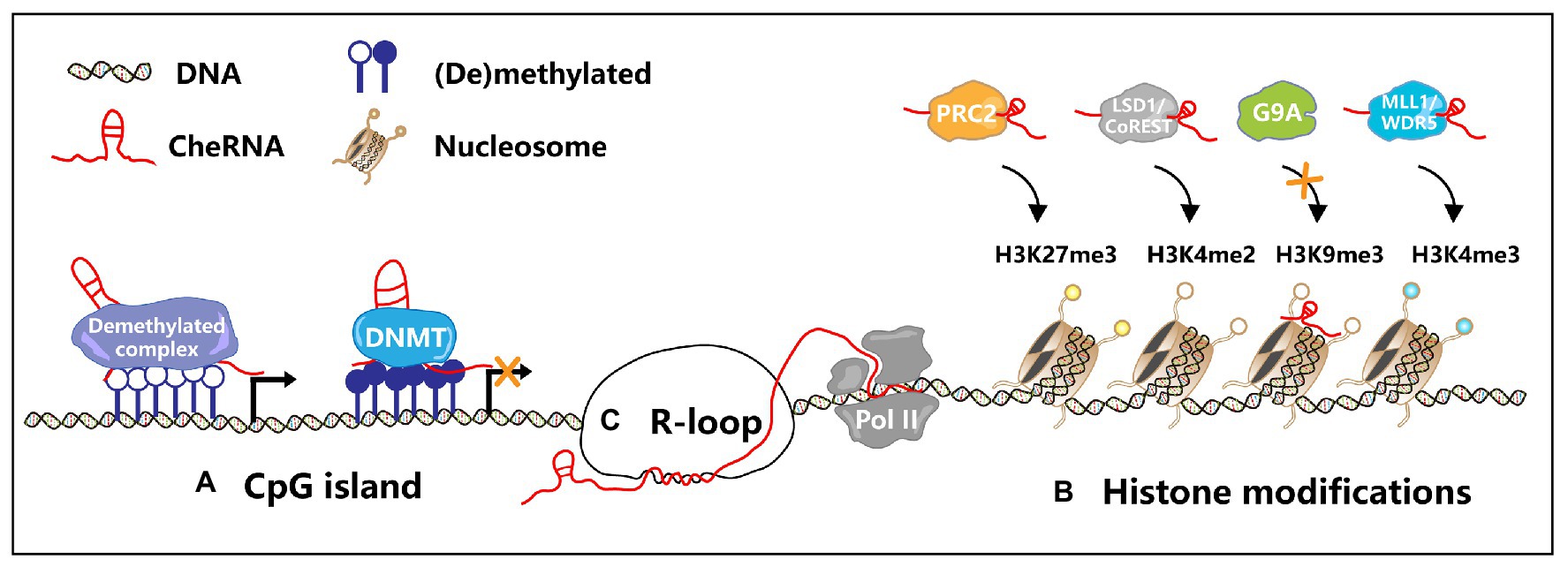
Figure 2. Transcription regulation by cheRNAs. (A) In the CpG island region, cheRNAs recruit DNA methyltransferase (DNMT) or demethylated complex to regulate gene transcription. (B) CheRNAs recruit PRC2 and MLL1/WDR5 complexes, mediate specific regions H3K27me3 and H3K4me3, and then inhibit or activate gene expression. Similarly, cheRNAs recruit the LSD1/CoREST complex and promote H3K4me2 demethylation. On the contrary, cheRNAs repel G9A, inhibit H3K9 trimethylation, and activate gene expression. (C) The antisense cheRNAs form R-loops in the gene promoter region, maintain the open state of chromatin, and promote the gene transcription.
Abnormal histone modification patterns are involved in many aspects of tumorigenesis. The cheRNA HOTAIR acts as a scaffold for the histone modification complexes PRC2 and LSD1/CoREST/REST, thereby coupling histone H3K27 methylation and H3K4 demethylation (Tsai et al., 2010). The abnormal transcription of cheRNA ROR blocks the binding of histone methyltransferase G9A to the oncogene TESC promoter region, and thus promotes TESC expression by inhibiting histone H3K9 methylation, leading to tumorigenesis (Fan et al., 2015). The cheRNA ZEB1-AS1 was shown to promote the occurrence of prostate cancer via recruiting histone methyltransferase MLL1 to the promoter region of ZEB1, inducing H3K4me3 modification, and thus activating ZEB1 transcription (Su et al., 2017). Moreover, the cheRNA GAS8-AS1 forms a complex with MLL1 and WDR5 to facilitate the formation of H3K4me3 in the GAS8 promoter region, thus enhancing the activity of RNA polymerase II, promoting the expression of GAS8, and accelerating the occurrence of liver cancer (Pan et al., 2018). In vitro and in vivo experiments have shown that inhibition of cheRNA 00511 represses cell proliferation and migration, whereas cheRNA 00511 upregulation enhances tumorigenesis via serving as a scaffold for the EZH2/PRC2 complex to target p57 H3K27 methylation in NSCLC cell lines (Sun et al., 2016a). Gastric cancer-related lncRNA 1 (GClnc1) is upregulated in gastric cancer and is associated with gastric cancer occurrence, metastasis, and poor prognosis. GClnc1 recruits WDR5 (a histone methyltransferase) to the promoter region of the superoxide dismutase 2 mitochondrial (SOD2, a mitochondrial enzymes) and KAT2A (a histone acetyltransferase), which in turn promotes H3K4 trimethylation and H3K9 acetylation levels, thereby promoting the expression of SOD2 (Sun et al., 2016b). High expression of cheRNA GClnc1 leads to lager tumor volume, higher vascular infiltration, and lower survival rate of patients, indicating that it can be regarded as a biomarker for gastric cancer. Uveal melanoma (UM) is one of the most common malignant tumors in the eyes of adults. Studies have shown that cheRNA CANT1 can bind to the promoter regions of JPX and FTX and further increase the expression of the H3K4me3 to impede UM tumorigenesis (Xing et al., 2017). Conversely, cheRNA PAUPAR modulates HES1 transcription via repressing H3K4me3, which significantly reduces UM growth and metastasis (Ding et al., 2016).
In colon CSCs, the cheRNA Lnc34a simultaneously recruits HDAC1 and Dnmt3a, modulates histone deacetylation and DNA methylation in the miR-34a promoter region, suppresses Lnc34a expression, and promotes colon CSC proliferation and self-renewal (Wang et al., 2016a). Furthermore, inhibition of cheRNA HotairM modulates HOXA1 epigenetic silencing via histone H3K27 trimethylation, which promotes the self-renewal of colorectal CSCs through the HOXA1-Nanog signal loop (Li et al., 2020). About cheRNAs modulate histone modification to alter gene transcription is depicted in Figure 2B. It has been reported that cheRNAs induce epigenetic modification in numerous tumors and CSCs, indicating that this biological process is of great significance for tumorigenesis.
The R-loop consists of the hybrid chain formed by RNA and DNA and a replaced single-stranded DNA, which is generally produced during DNA replication or transcription (Santos-Pereira and Aguilera, 2015). Previously, people usually believed that the R-loop was a byproduct of transcription, while recent studies have shown that the R-loop plays a vital role in gene expression and genome stability. Generally, R-loops can be categorized as a physiological R-loop or a pathological R-loop. Physiological R loops occur in normal biological processes, such as B cell immunoglobulin class switching (Yu et al., 2003), mitochondrial DNA replication, or Escherichia coli ColE1 plasmid replication (Aguilera and Garcia-Muse, 2012). The excessive accumulation of R-loops and pathological R-loops affects DNA replication, transcription, and genome stability (Aguilera and Garcia-Muse, 2012; Santos-Pereira and Aguilera, 2015; Sollier and Cimprich, 2015; Garcia-Muse and Aguilera, 2019). The formation of the R-loop is closely associated with the occurrence of a variety of human diseases and may be responsible for the imbalance of chromatin homeostasis in tumors (Bhatia et al., 2014; Santos-Pereira and Aguilera, 2015).
The recent finding is that the production of R-loops is deeply relevant to cheRNAs, especially antisense cheRNAs. CheRNA-mediated R-loop formation has been reported in Arabidopsis thaliana (Sun et al., 2013), yeast (Cloutier et al., 2016), and mammalian cells (Boque-Sastre et al., 2015). Similarly, cheRNA-mediated R-loop formation also contributes to tumorigenesis (Figure 2C). In colon cancer, the antisense cheRNA VIM-AS1 forms an R-loop near the VIM transcription start site, which upregulates the expression of VIM by maintaining the open state of chromatin (Boque-Sastre et al., 2015). Besides, the antisense cheRNA TARID forms an R-loop in the promoter region of the tumor suppressor TCF21. GADD45A binds to the R-loop and recruits the DNA demethylase TET1 to modulate the expression of TCF21 (Arab et al., 2019). Although cheRNAs are involved in the formation of R-loops in tumor cells, how cheRNA-mediated R-loops function in CSCs remains unclear. Further exploration of the function and mechanism of R-loops induced by cheRNAs in CSCs is needed, which might inspire novel insights into tumor therapy. The role of cheRNAs in transcription regulation is generalized in Table 2.
The mechanism of tumorigenesis is complicated, and the imbalance of chromatin homeostasis plays a crucial role in it (Yu and Ren, 2017). Our previous researches showed that the aberrant open chromatin status at chr12p13.3 induces a novel cheRNA GAU1 and cis activates the expression of oncogene GALNT8 by recruiting the transcription elongation factor TCEA1, which accelerates tumor progression (Chai et al., 2018). On the contrary, cheRNAs can also affect chromatin homeostasis in many ways, such as nucleosome localization, TAD conformation, and R-loop formation. With the application of transcriptomics and computational biology, more and more cheRNAs are found in tumors and CSCs. Numerous studies have shown that cheRNAs function in tumor growth, metastasis, drug resistance, angiogenesis, and CSC stemness maintenance (Gutschner and Diederichs, 2012; Xiong et al., 2016). In summary, more researches have drawn attention to the role of cheRNAs participating chromatin regulation in tumors and CSCs.
With high tissue and cell specificity, cheRNAs make it possible to become a biomarker or therapeutic target. At present, cheRNA PCA3 has been used in the clinical diagnosis of prostate cancer for its high expression, providing a reliable basis for the treatment of prostate cancer (Lee et al., 2011). Moreover, specifically targeting cheRNA MALAT1 by antisense oligonucleotides (ASOs) can significantly reduce tumor volume, induce cell differentiation, and inhibit metastasis in a mouse mammary carcinoma model (Arun et al., 2016; Amodio et al., 2018). It is expected that more newly discovered cheRNAs will be useful in clinical diagnosis and tumor therapy with the development of advanced technologies.
However, challenges are obvious before cheRNAs can be applied in tumor targeted therapy on a large scale. Firstly, cheRNAs modulate chromatin homeostasis in many ways, but the mechanism of its binding to chromatin remains unclear. In addition to directly binding with protein and DNA, is there any other mechanism of cheRNA locating around chromatin? Secondly, RNA molecules have shortcomings, such as low transfection efficiency, off-target, and short half-life, which may greatly impact the drug delivery efficiency. Therefore, finding a more suitable drug delivery method is a priority. It has been proposed to use nanoparticles to improve the efficiency of targeted drug delivery (Wang et al., 2019a). In conclusion, clarifying cheRNA with its expression, structure, and mechanism is significant for the understanding of tumorigenesis, and it can also provide new insights for tumor diagnosis and targeting therapy.
JZ and TD designed and drafted the manuscript. HZ discussed, revised, and approved the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Plan (2017YFE0196300), the National Natural Science Foundation of China (Grant 31870748), the ShuGuang Project of Shanghai Municipal Education Commission and Shanghai Education Development Foundation (17SG19), the Outstanding Yong Medical Scholar of Shanghai Municipal Commission of Health and Family Planning (2017YQ067), and the Outstanding Yong Scholar Grant of Tongji University (PA2019000239), and the Startup Funding of Frontier Science Research Center for Stem Cells & Shanghai East Hospital of Tongji University (DFRC2019003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aguilera, A., and Garcia-Muse, T. (2012). R loops: from transcription Byproducts to threats to genome stability. Mol. Cell 46, 115–124. doi: 10.1016/j.molcel.2012.04.009
Al-Hajj, M., Wicha, M. S., Benito-Hernandez, A., Morrison, S. J., and Clarke, M. F. (2003). Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 100, 3983–3988. doi: 10.1073/pnas.0530291100
Allis, C. D., and Jenuwein, T. (2016). The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500. doi: 10.1038/nrg.2016.59
Amodio, N., Raimondi, L., Juli, G., Stamato, M. A., Caracciolo, D., Tagliaferri, P., et al. (2018). MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 11:63. doi: 10.1186/s13045-018-0606-4
Andrey, G., Montavon, T., Mascrez, B., Gonzalez, F., Noordermeer, D., Leleu, M., et al. (2013). A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340:1234167. doi: 10.1126/science.1234167
Arab, K., Karaulanov, E., Musheev, M., Trnka, P., Schafer, A., Grummt, I., et al. (2019). GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat. Genet. 51, 217–223. doi: 10.1038/s41588-018-0306-6
Arab, K., Park, Y. J., Lindroth, A. M., Schafer, A., Oakes, C., Weichenhan, D., et al. (2014). Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol. Cell 55, 604–614. doi: 10.1016/j.molcel.2014.06.031
Arun, G., Diermeier, S., Akerman, M., Chang, K. C., Wilkinson, J. E., Hearn, S., et al. (2016). Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 30, 34–51. doi: 10.1101/gad.270959.115
Batlle, E., and Clevers, H. (2017). Cancer stem cells revisited. Nat. Med. 23, 1124–1134. doi: 10.1038/nm.4409
Beck, B., and Blanpain, C. (2013). Unravelling cancer stem cell potential. Nat. Rev. Cancer 13, 727–738. doi: 10.1038/nrc3597
Bergmann, J. H., and Spector, D. L. (2014). Long non-coding RNAs: modulators of nuclear structure and function. Curr. Opin. Cell Biol. 26, 10–18. doi: 10.1016/j.ceb.2013.08.005
Beroukhim, R., Mermel, C. H., Porter, D., Wei, G., Raychaudhuri, S., Donovan, J., et al. (2010). The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905. doi: 10.1038/nature08822
Bhatia, V., Barroso, S. I., Garcia-Rubio, M. L., Tumini, E., Herrera-Moyano, E., and Aguilera, A. (2014). BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511, 362–365. doi: 10.1038/nature13374
Bonev, B., and Cavalli, G. (2016). Organization and function of the 3D genome. Nat. Rev. Genet. 17, 661–678. doi: 10.1038/nrg.2016.112
Bonnet, D., and Dick, J. E. (1997). Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3, 730–737. doi: 10.1038/nm0797-730
Boque-Sastre, R., Soler, M., Oliveira-Mateos, C., Portela, A., Moutinho, C., Sayols, S., et al. (2015). Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl. Acad. Sci. U. S. A. 112, 5785–5790. doi: 10.1073/pnas.1421197112
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Brown, C. J., Hendrich, B. D., Rupert, J. L., Lafreniere, R. G., Xing, Y., Lawrence, J., et al. (1992). The human Xist gene - analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71, 527–542. doi: 10.1016/0092-8674(92)90520-M
Chai, P., Jia, R., Jia, R., Pan, H., Wang, S., Ni, H., et al. (2018). Dynamic chromosomal tuning of a novel GAU1 incing driver at chr12p13.32 accelerates tumorigenesis. Nucleic Acids Res. 46, 6041–6056. doi: 10.1093/nar/gky366
Chin, L., Meyerson, M., Aldape, K., Bigner, D., Mikkelsen, T., VandenBerg, S., et al. (2008). Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068. doi: 10.1038/nature07385
Clapier, C. R., and Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304. doi: 10.1146/annurev.biochem.77.062706.153223
Cloutier, S. C., Wang, S. W., Ma, W. K., Al Husini, N., Dhoondia, Z., Ansari, A., et al. (2016). Regulated formation of lncRNA-DNA hybrids enables faster transcriptional induction and environmental adaptation. Mol. Cell 61, 393–404. doi: 10.1016/j.molcel.2015.12.024
Ding, X., Wang, X., Lin, M., Xing, Y., Ge, S. F., Jia, R. B., et al. (2016). PAUPAR lncRNA suppresses tumourigenesis by H3K4 demethylation in uveal melanoma. FEBS Lett. 590, 1729–1738. doi: 10.1002/1873-3468.12220
Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., et al. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. doi: 10.1038/nature11082
Egger, G., Liang, G. N., Aparicio, A., and Jones, P. A. (2004). Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463. doi: 10.1038/nature02625
Engreitz, J. M., Ollikainen, N., and Guttman, M. (2016). Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756–770. doi: 10.1038/nrm.2016.126
Fan, J., Xing, Y., Wen, X., Jia, R., Ni, H., He, J., et al. (2015). Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis. Genome Biol. 16:139. doi: 10.1186/s13059-015-0705-2
Feinberg, A. P., and Vogelstein, B. (1983). Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301, 89–92. doi: 10.1038/301089a0
Fu, Z. Q., Chen, C. H., Zhou, Q. B., Wang, Y. X., Zhao, Y., Zhao, X. H., et al. (2017). LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 410, 68–81. doi: 10.1016/j.canlet.2017.09.019
Garcia-Muse, T., and Aguilera, A. (2019). R loops: from physiological to pathological roles. Cell 179, 604–618. doi: 10.1016/j.cell.2019.08.055
Gayen, S., and Kalantry, S. (2017). Chromatin-enriched lncRNAs: a novel class of enhancer RNAs. Nat. Struct. Mol. Biol. 24, 556–557. doi: 10.1038/nsmb.3430
Greaves, M., and Maley, C. C. (2012). Clonal evolution in cancer. Nature 481, 306–313. doi: 10.1038/nature10762
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–U1148. doi: 10.1038/nature08975
Gutschner, T., and Diederichs, S. (2012). The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 9, 703–719. doi: 10.4161/rna.20481
Hargreaves, D. C., and Crabtree, G. R. (2011). ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 21, 396–420. doi: 10.1038/cr.2011.32
Hermann, P. C., Huber, S. L., Herrler, T., Aicher, A., Ellwart, J. W., Guba, M., et al. (2007). Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 1, 313–323. doi: 10.1016/j.stem.2007.06.002
Ho, L., and Crabtree, G. R. (2010). Chromatin remodelling during development. Nature 463, 474–484. doi: 10.1038/nature08911
Huarte, M. (2015). The emerging role of IncRNAs in cancer. Nat. Med. 21, 1253–1261. doi: 10.1038/nm.3981
Iyer, M. K., Niknafs, Y. S., Malik, R., Singhal, U., Sahu, A., Hosono, Y., et al. (2015). The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208. doi: 10.1038/ng.3192
Jones, P. A., and Baylin, S. B. (2002). The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3, 415–428. doi: 10.1038/nrg816
Jones, P. A., and Baylin, S. B. (2007). The epigenomics of cancer. Cell 128, 683–692. doi: 10.1016/j.cell.2007.01.029
Ke, Y. W., Xu, Y. A., Chen, X. P., Feng, S. K., Liu, Z. B., Sun, Y. Y., et al. (2017). 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell 170, 367–381.e20. doi: 10.1016/j.cell.2017.06.029
Khalil, A. M., Guttman, M., Huarte, M., Garber, M., Raj, A., Morales, D. R., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U. S. A. 106, 11667–11672. doi: 10.1073/pnas.0904715106
Kloetgen, A., Thandapani, P., Ntziachristos, P., Ghebrechristos, Y., Nomikou, S., Lazaris, C., et al. (2020). Three-dimensional chromatin landscapes in T cell acute lymphoblastic leukemia. Nat. Genet. 52, 388–400. doi: 10.1038/s41588-020-0602-9
Kreso, A., and Dick, J. E. (2014). Evolution of the cancer stem cell model. Cell Stem Cell 14, 275–291. doi: 10.1016/j.stem.2014.02.006
Kundaje, A., Meuleman, W., Ernst, J., Bilenky, M., Yen, A., Heravi-Moussavi, A., et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. doi: 10.1038/nature14248
Lai, A. Y., and Wade, P. A. (2011). Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer 11, 588–596. doi: 10.1038/nrc3091
Lapidot, T., Sirard, C., Vormoor, J., Murdoch, B., Hoang, T., Cacerescortes, J., et al. (1994). A cell initiating human acute myeloid-Leukemia after transplantation into SCID mice. Nature 367, 645–648. doi: 10.1038/367645a0
Lee, G. L., Dobi, A., and Srivastava, S. (2011). Prostate cancer: diagnostic performance of the PCA3 urine test. Nat. Rev. Urol. 8, 123–124. doi: 10.1038/nrurol.2011.10
Li, G. L., Ruan, X. A., Auerbach, R. K., Sandhu, K. S., Zheng, M. Z., Wang, P., et al. (2012). Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98. doi: 10.1016/j.cell.2011.12.014
Li, F., Xu, Y., Xu, X., Ge, S., Zhang, F., Zhang, H., et al. (2020). lncRNA HotairM1 depletion promotes self-renewal of cancer stem cells through HOXA1-Nanog regulation loop. Mol. Ther. Nucleic Acids 22, 456–470. doi: 10.1016/j.omtn.2020.09.008
Luo, H. C., Zhu, G. Q., Xu, J. F., Lai, Q., Yan, B. W., Guo, Y., et al. (2019). HOTTIP lncRNA promotes hematopoietic stem cell self-renewal leading to AML-like disease in mice. Cancer Cell 36, 645–659.e8. doi: 10.1016/j.ccell.2019.10.011
Magee, J. A., Piskounova, E., and Morrison, S. J. (2012). Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell 21, 283–296. doi: 10.1016/j.ccr.2012.03.003
Marina, D. B., Shankar, S., Natarajan, P., Finn, K. J., and Madhani, H. D. (2013). A conserved ncRNA-binding protein recruits silencing factors to heterochromatin through an RNAi-independent mechanism. Genes Dev. 27, 1851–1856. doi: 10.1101/gad.226019.113
Martens-Uzunova, E. S., Bottcher, R., Croce, C. M., Jenster, G., Visakorpi, T., and Calin, G. A. (2014). Long noncoding RNA in prostate, bladder, and kidney cancer. Eur. Urol. 65, 1140–1151. doi: 10.1016/j.eururo.2013.12.003
Maurano, M. T., Humbert, R., Rynes, E., Thurman, R. E., Haugen, E., Wang, H., et al. (2012). Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195. doi: 10.1126/science.1222794
Meacham, C. E., and Morrison, S. J. (2013). Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337. doi: 10.1038/nature12624
Melton, C., Reuter, J. A., Spacek, D. V., and Snyder, M. (2015). Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat. Genet. 47, 710–716. doi: 10.1038/ng.3332
Muzny, D. M., Bainbridge, M. N., Chang, K., Dinh, H. H., Drummond, J. A., Fowler, G., et al. (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337. doi: 10.1038/nature11252
Nguyen, L. V., Vanner, R., Dirks, P., and Eaves, C. J. (2012). Cancer stem cells: an evolving concept. Nat. Rev. Cancer 12, 133–143. doi: 10.1038/nrc3184
Nora, E. P., Lajoie, B. R., Schulz, E. G., Giorgetti, L., Okamoto, I., Servant, N., et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385. doi: 10.1038/nature11049
Orom, U. A., and Shiekhattar, R. (2013). Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 154, 1190–1193. doi: 10.1016/j.cell.2013.08.028
Pan, W. T., Zhang, N. S., Liu, W. J., Liu, J. B., Zhou, L. Q., Liu, Y., et al. (2018). The long noncoding RNA GAS8-AS1 suppresses hepatocarcinogenesis by epigenetically activating the tumor suppressor GAS8. J. Biol. Chem. 293, 17154–17165. doi: 10.1074/jbc.RA118.003055
Peitzsch, C., Tyutyunnykova, A., Pantel, K., and Dubrovska, A. (2017). Cancer stem cells: the root of tumor recurrence and metastases. Semin. Cancer Biol. 44, 10–24. doi: 10.1016/j.semcancer.2017.02.011
Phillips-Cremins, J. E., Sauria, M. E. G., Sanyal, A., Gerasimova, T. I., Lajoie, B. R., Bell, J. S. K., et al. (2013). Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153, 1281–1295. doi: 10.1016/j.cell.2013.04.053
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. doi: 10.1016/j.cell.2009.02.006
Portela, A., and Esteller, M. (2010). Epigenetic modifications and human disease. Nat. Biotechnol. 28, 1057–1068. doi: 10.1038/nbt.1685
Prensner, J. R., Iyer, M. K., Sahu, A., Asangani, I. A., Cao, Q., Patel, L., et al. (2013). The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 45, 1392–1398. doi: 10.1038/ng.2771
Quinodoz, S., and Guttman, M. (2014). Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 24, 651–663. doi: 10.1016/j.tcb.2014.08.009
Reya, T., Morrison, S. J., Clarke, M. F., and Weissman, I. L. (2001). Stem cells, cancer, and cancer stem cells. Nature 414, 105–111. doi: 10.1038/35102167
Ricci-Vitiani, L., Lombardi, D. G., Pilozzi, E., Biffoni, M., Todaro, M., Peschle, C., et al. (2007). Identification and expansion of human colon-cancer-initiating cells. Nature 445, 111–115. doi: 10.1038/nature05384
Rothschild, G., Zhang, W., Lim, J., Giri, P. K., Laffleur, B., Chen, Y., et al. (2020). Noncoding RNA transcription alters chromosomal topology to promote isotype-specific class switch recombination. Sci. Immunol. 5:eaay5864. doi: 10.1126/sciimmunol.aay5864
Santos-Pereira, J. M., and Aguilera, A. (2015). R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 16, 583–597. doi: 10.1038/nrg3961
Sanyal, A., Lajoie, B. R., Jain, G., and Dekker, J. (2012). The long-range interaction landscape of gene promoters. Nature 489, 109–U127. doi: 10.1038/nature11279
Saygin, C., Matei, D., Majeti, R., Reizes, O., and Lathia, J. D. (2019). Targeting cancer stemness in the clinic: from hype to hope. Cell Stem Cell 24, 25–40. doi: 10.1016/j.stem.2018.11.017
Schatton, T., Murphy, G. F., Frank, N. Y., Yamaura, K., Waaga-Gasser, A. M., Gasser, M., et al. (2008). Identification of cells initiating human melanomas. Nature 451, 345–U311. doi: 10.1038/nature06489
Schmitt, A. M., and Chang, H. Y. (2016). Long noncoding RNAs in cancer pathways. Cancer Cell 29, 452–463. doi: 10.1016/j.ccell.2016.03.010
Segal, E., and Widom, J. (2009). What controls nucleosome positions? Trends Genet. 25, 335–343. doi: 10.1016/j.tig.2009.06.002
Sell, S., and Leffert, H. L. (2008). Liver cancer stem cells. J. Clin. Oncol. 26, 2800–2805. doi: 10.1200/JCO.2007.15.5945
Sexton, T., and Cavalli, G. (2015). The role of chromosome domains in shaping the functional genome. Cell 160, 1049–1059. doi: 10.1016/j.cell.2015.02.040
Shackleton, M., Quintana, E., Fearon, E. R., and Morrison, S. J. (2009). Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell 138, 822–829. doi: 10.1016/j.cell.2009.08.017
Shimokawa, M., Ohta, Y., Nishikori, S., Matano, M., Takano, A., Fujii, M., et al. (2017). Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature 545, 187–192. doi: 10.1038/nature22081
Singh, S. K., Hawkins, C., Clarke, I. D., Squire, J. A., Bayani, J., Hide, T., et al. (2004). Identification of human brain tumour initiating cells. Nature 432, 396–401. doi: 10.1038/nature03128
Sollier, J., and Cimprich, K. A. (2015). Breaking bad: R-loops and genome integrity. Trends Cell Biol. 25, 514–522. doi: 10.1016/j.tcb.2015.05.003
Somasundaram, S., Forrest, M. E., Moinova, H., Cohen, A., Varadan, V., LaFramboise, T., et al. (2018). The DNMT1-associated lincRNA DACOR1 reprograms genome-wide DNA methylation in colon cancer. Clin. Epigenetics 10:127. doi: 10.1186/s13148-018-0555-3
Su, W., Xu, M., Chen, X., Chen, N., Gong, J., Nie, L., et al. (2017). Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol. Cancer 16:142. doi: 10.1186/s12943-017-0711-y
Sun, Q. W., Csorba, T., Skourti-Stathaki, K., Proudfoot, N. J., and Dean, C. (2013). R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340, 619–621. doi: 10.1126/science.1234848
Sun, Q. Y., Hao, Q. Y., and Prasanth, K. V. (2018). Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. 34, 142–157. doi: 10.1016/j.tig.2017.11.005
Sun, T. T., He, J., Liang, Q., Ren, L. L., Yan, T. T., Yu, T. C., et al. (2016b). LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 6, 784–801. doi: 10.1158/2159-8290.Cd-15-0921
Sun, C. C., Li, S. J., Li, G., Hua, R. X., Zhou, X. H., and Li, D. J. (2016a). Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol. Ther. Nucleic Acids 5:14. doi: 10.1038/mtna.2016.94
Suva, M. L., and Tirosh, I. (2020). The glioma stem cell model in the era of single-cell genomics. Cancer Cell 37, 630–636. doi: 10.1016/j.ccell.2020.04.001
Takaishi, S., Okumura, T., and Wang, T. C. (2008). Gastric cancer stem cells. J. Clin. Oncol. 26, 2876–2882. doi: 10.1200/JCO.2007.15.2603
Trimarchi, T., Bilal, E., Ntziachristos, P., Fabbri, G., Dalla-Favera, R., Tsirigos, A., et al. (2014). Genome-wide mapping and characterization of notch-regulated long noncoding RNAs in acute leukemia. Cell 158, 593–606. doi: 10.1016/j.cell.2014.05.049
Tsai, M. C., Manor, O., Wan, Y., Mosammaparast, N., Wang, J. K., Lan, F., et al. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693. doi: 10.1126/science.1192002
Uhler, C., and Shivashankar, G. V. (2017). Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat. Rev. Mol. Cell Biol. 18, 717–727. doi: 10.1038/nrm.2017.101
Vance, K. W., and Ponting, C. P. (2014). Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 30, 348–355. doi: 10.1016/j.tig.2014.06.001
Wang, L., Bu, P., Ai, Y., Srinivasan, T., Chen, H. J., Xiang, K., et al. (2016a). A long non-coding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric division. elife 5:e14620. doi: 10.7554/eLife.14620
Wang, W. T., Han, C., Sun, Y. M., Chen, T. Q., and Chen, Y. Q. (2019a). Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. Oncol. 12:55. doi: 10.1186/s13045-019-0748-z
Wang, Y. Y., He, L., Du, Y., Zhu, P. P., Huang, G. L., Luo, J. J., et al. (2015). The long noncoding RNA IncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 16, 413–425. doi: 10.1016/j.stem.2015.03.003
Wang, X., Sun, W., Shen, W., Xia, M., Chen, C., Xiang, D., et al. (2016b). Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J. Hepatol. 64, 1283–1294. doi: 10.1016/j.jhep.2016.01.019
Wang, Y., Zhu, P., Luo, J., Wang, J., Liu, Z., Wu, W., et al. (2019b). LncRNA HAND2-AS1 promotes liver cancer stem cell self-renewal via BMP signaling. EMBO J. 38:e101110. doi: 10.15252/embj.2018101110
Werner, M. S., and Ruthenburg, A. J. (2015). Nuclear fractionation reveals thousands of chromatin-tethered noncoding RNAs adjacent to active genes. Cell Rep. 12, 1089–1098. doi: 10.1016/j.celrep.2015.07.033
Werner, M. S., Sullivan, M. A., Shah, R. N., Nadadur, R. D., Grzybowski, A. T., Galat, V., et al. (2017). Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription. Nat. Struct. Mol. Biol. 24, 596–603. doi: 10.1038/nsmb.3424
Wilson, B. G., and Roberts, C. W. M. (2011). SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11, 481–492. doi: 10.1038/nrc3068
Wu, D. M., Zheng, Z. H., Zhang, Y. B., Fan, S. H., Zhang, Z. F., Wang, Y. J., et al. (2019a). Down-regulated lncRNA DLX6-AS1 inhibits tumorigenesis through STAT3 signaling pathway by suppressing CADM1 promoter methylation in liver cancer stem cells. J. Exp. Clin. Cancer Res. 38:237. doi: 10.1186/s13046-019-1239-3
Wu, J., Zhu, P., Lu, T., Du, Y., Wang, Y., He, L., et al. (2019b). The long non-coding RNA LncHDAC2 drives the self-renewal of liver cancer stem cells via activation of hedgehog signaling. J. Hepatol. 70, 918–929. doi: 10.1016/j.jhep.2018.12.015
Wutz, G., Varnai, C., Nagasaka, K., Cisneros, D. A., Stocsits, R. R., Tang, W., et al. (2017). Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599. doi: 10.15252/embj.201798004
Xia, L., Huang, W., Bellani, M., Seidman, M. M., Wu, K., Fan, D., et al. (2017). CHD4 has oncogenic functions in initiating and maintaining epigenetic suppression of multiple tumor suppressor genes. Cancer Cell 31, 653–668.e7. doi: 10.1016/j.ccell.2017.04.005
Xing, Y., Wen, X. Y., Ding, X., Fan, J. Y., Chai, P. W., Jia, R. B., et al. (2017). CANT1 lncRNA triggers efficient therapeutic efficacy by correcting aberrant incing cascade in malignant uveal melanoma. Mol. Ther. 25, 1209–1221. doi: 10.1016/j.ymthe.2017.02.016
Xiong, X. D., Ren, X., Cai, M. Y., Yang, J. W., Liu, X., and Yang, J. M. (2016). Long non-coding RNAs: an emerging powerhouse in the battle between life and death of tumor cells. Drug Resist. Updat. 26, 28–42. doi: 10.1016/j.drup.2016.04.001
Yang, L. Q., Lin, C. R., Jin, C. Y., Yang, J. C., Tanasa, B., Li, W. B., et al. (2013). lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 500, 598–602. doi: 10.1038/nature12451
Yoon, J. H., You, B. H., Park, C. H., Kim, Y. J., Nam, J. W., and Lee, S. K. (2018). The long noncoding RNA LUCAT1 promotes tumorigenesis by controlling ubiquitination and stability of DNA methyltransferase 1 in esophageal squamous cell carcinoma. Cancer Lett. 417, 47–57. doi: 10.1016/j.canlet.2017.12.016
You, J. S., and Jones, P. A. (2012). Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell 22, 9–20. doi: 10.1016/j.ccr.2012.06.008
Yu, K. F., Chedin, F., Hsieh, C. L., Wilson, T. E., and Lieber, M. R. (2003). R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4, 442–451. doi: 10.1038/ni919
Yu, M., and Ren, B. (2017). The three-dimensional organization of mammalian genomes. Annu. Rev. Cell Dev. Biol. 33, 265–289. doi: 10.1146/annurev-cellbio-100616-060531
Zhang, H., Jiao, W. W., Sun, L., Fan, J. Y., Chen, M. F., Wang, H., et al. (2013). Intrachromosomal looping is required for activation of endogenous pluripotency genes during reprogramming. Cell Stem Cell 13, 30–35. doi: 10.1016/j.stem.2013.05.012
Zhao, Z. L., Dammert, M. A., Grummt, I., and Bierhoff, H. (2016). lncRNA-induced nucleosome repositioning reinforces transcriptional repression of rRNA genes upon hypotonic stress. Cell Rep. 14, 1876–1882. doi: 10.1016/j.celrep.2016.01.073
Keywords: chromatin-enriched RNA, tumor, cancer stem cell, chromatin loop, R-loop
Citation: Zhang J, Ding T and Zhang H (2021) Insight Into Chromatin-Enriched RNA: A Key Chromatin Regulator in Tumors. Front. Cell Dev. Biol. 9:649605. doi: 10.3389/fcell.2021.649605
Received: 05 January 2021; Accepted: 18 March 2021;
Published: 14 April 2021.
Edited by:
Jin Qian, Stanford University, United StatesReviewed by:
Yiting Li, St. Francis Medical Center, United StatesCopyright © 2021 Zhang, Ding and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: He Zhang, emhhbmdoZUB0b25namkuZWR1LmNu
†These authors share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.