- 1Department of Plastic Surgery, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Department of Thyroid and Breast Surgery, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
As one of the most frequently occurring malignancies in women, breast cancer (BC) is still an enormous threat to women all over the world. The high mortality rates in BC patients are associated with BC recurrence, metastatic progression to distant organs, and therapeutic resistance. Circular RNAs (circRNAs), belonging to the non-coding RNAs (ncRNAs), are connected end to end to form covalently closed single-chain circular molecules. CircRNAs are widely found in different species and a variety of human cells, with the features of diversity, evolutionary conservation, stability, and specificity. CircRNAs are emerging important participators in multiple diseases, including cardiovascular disease, inflammation, and cancer. Recent studies have shown that circRNAs are involved in BC progress by regulating gene expression at the transcriptional or post-transcriptional level via binding to miRNAs then inhibiting their function, suggesting that circRNAs may be potential targets for early diagnosis, treatment, and prognosis of BC. Herein, in this article, we have reviewed and summarized the current studies about the biogenesis, features, and functions of circRNAs. More importantly, we emphatically elucidate the pivotal functions and mechanisms of circRNAs in BC growth, metastasis, diagnosis, and drug resistance. Deciphering the complex networks, especially the circRNA-miRNA target gene axis, will endow huge potentials in developing therapeutic strategies for combating BC.
Introduction
Breast cancer (BC) is the most frequently occurring malignancy and the leading cause of cancer-related death among women worldwide. In the last decade, despite therapies, such as surgery, radiation, chemotherapy, endocrine therapy, targeted therapy, and immunotherapy, have achieved certain curative effect, the incidence rates for BC have been rising over the world (Trimboli et al., 2020). The high mortality rates in BC patients are associated with recurrence and metastatic progression to distant organs, including the bone, lung, brain, and liver. In addition, as the resistance of disseminated tumor cells to existing therapeutic agents, the resistance of therapies is also emerging as an urgent issue (Jiang et al., 2020). Therefore, to improve the BC diagnosis and therapy with efficiency, it is imperative to explore the molecular mechanisms of BC pathogenesis.
Nowadays, genetic mutations and epigenetic modifications are playing increasingly significant roles in the BC process. Epigenetic modifications mainly include DNA methylation, histone modifications, and non-coding RNA (ncRNA) (Wu et al., 2020). It is well-established that the occurrence and development of BC are intricately regulated by a variety of ncRNAs, including long non-coding RNA (lncRNA), microRNA (miRNA), and circular RNA (circRNA) (Xu et al., 2020). Notably, as a member of short endogenous ncRNA, circRNA is predominantly derived from gene exons with a closed-loop structure (Xie et al., 2020). This circular structure differs from linear RNA in that it lacks the exposed 5′-terminal cap and 3′-polyadenylate tail structure, reflecting the ability of the circRNA to tolerate exonuclease digestion. CircRNAs are widely found in different species and a variety of human cells, with the features of diversity, evolutionary conservation, stability, and specificity. CircRNAs have been proven to possess a wide range of biological effects, especially functioning to sponge miRNAs as competing endogenous RNA (ceRNA) to offset the impact of miRNAs (Han et al., 2020). In recent years, the chelating protein or transcription regulation of circRNA function has also been gradually reported and attracted attention. Generally speaking, circRNAs play an important role in gene regulation, and their changes in abundance are involved in shaping many disease initiations and processes.
With the progress of the research, it is gradually recognized that circRNA dysregulation is closely related to the occurrence, initiation, and progression of multiple cancer types. It is found that circRNA expression is dysregulated in BC tissue and is closely related to the occurrence and development of BC. For example, Wang J. et al. (2019) identified a total of 480 dysregulated circRNAs in exosomes from metastatic patients compared with localized patients, and these dysregulated circRNAs were enriched in eight pathways. Innovative and novel bioinformatics technologies have revealed the ceRNA crosstalk of circRNAs. Shi et al. (2018) collected four paired BC samples for the human circRNA array to construct a genome-wide circRNA profile of BC. The findings showed that 715 circRNAs were upregulated and 440 circRNAs were downregulated, while hsa_circRNA_005230 was upregulated 12.2-fold and hsa_circRNA_406225 was downregulated 12.4-fold. Zhao et al. (2019) selected a total of seven differentially expressed circRNAs, 27 differentially expressed miRNAs, and 102 differentially expressed mRNAs for the construction of the ceRNA network of BC. Additionally, the hsa_circ_0000519 was involved in the circRNA-miRNA-hub gene network in the pathogenesis of BC. These findings have driven circRNAs as a potential target for improving BC treatment. At the same moment, circRNAs are also regarded to be diagnosis and prognosis biomarkers.
Based on the above, the presentation of circRNA in BC is complex and diverse, and is the key point for BC diagnosis and therapy. Herein, this article has reviewed the recent existing studies on the biogenesis, feature, and function classification of circRNAs. More importantly, it focuses on the expression features, impacts, and mechanisms of the reported circRNAs in BC growth and metastasis. It also emphasizes the diagnostic value of circRNAs in BC. The in-depth understanding of the circRNA mechanisms in BC development will provide better insight into circRNA-associated tumorigenesis and screen novel strategies for combating BC.
Biogenesis and Feature
Since Sanger et al. (1976) firstly discovered that viroids were composed of covalently closed circRNAs in 1976, a variety of circRNAs have been found in different species such as drosophila, mouse, nematode, and human. With the advancement of high-throughput sequencing technology, emerging evidence indicates that circRNAs are widespread in mammals and their functions have been further explored (Qu et al., 2015). It is gradually recognized that circRNAs are closely related to the occurrence and development of diabetes, hypertension, myocardial infarction, tumor, ischemic attack, osteoporosis, and other diseases (Fang et al., 2019). Based on their compositions, circRNAs can be classified into three types: exonic circRNAs (ecircRNAs), circular intronic RNAs (ciRNAs), and exon-intronic circRNAs (EIciRNAs). Based on their positions and their adjacent mRNAs, circRNAs can be classified as ecircRNAs, intronic circRNAs (ciRNAs), antisense circRNAs, sense overlapping circRNAs, and intergenic circRNAs (Li et al., 2019).
CircRNA formation is a variable cyclization mode to generate different types of circRNAs according to different splice sites, mainly including three mechanisms that permit the back-splicing reaction: exon skipping, direct back-splicing, and dimerization of RNA-binding proteins (RBPs) (Tran et al., 2020) (Figure 1A). Exon skipping is also known as lariat-driven circularization. The pre-mRNA is partially folded in the transcription process, making the 3′-SD of the downstream exon connect with the 5′-SA of the upstream exon, resulting in the exon skipping and forming an RNA lariat containing both exons and introns. The ecircRNAs are further formed with the removal of introns (Geng et al., 2018) (Figure 1B). Direct back-splicing is the main form of ecircRNA and EIciRNA production, in which the flanking intron complementary sequences of pre-mRNAs form a lasso by direct base pairing. The ecircRNAs or EIciRNAs are formed through removing or retaining introns (Figure 1C). RBP-dependent circRNA formation: RBPs bind to the sequence motifs of upstream and downstream in introns on both sides of the exon and act as the bridge to draw the distance between the flanking introns to form circRNA (Figure 1D). The formation of ciRNAs: the pre-mRNA can be spliced into an RNA lasso containing a 2′,5′-phosphodiester, resulted in the formation of ciRNAs after cutting off the 3′-tail (Figure 1E). The intergenic circRNA formation pattern: the circRNA consists of two intronic circRNA fragments flanked by GT-AG splicing signals, which act as the splice donor and splice acceptor of the circular junction to eventually form an intergenic circRNA (Nedoluzhko et al., 2020) (Figure 1F).
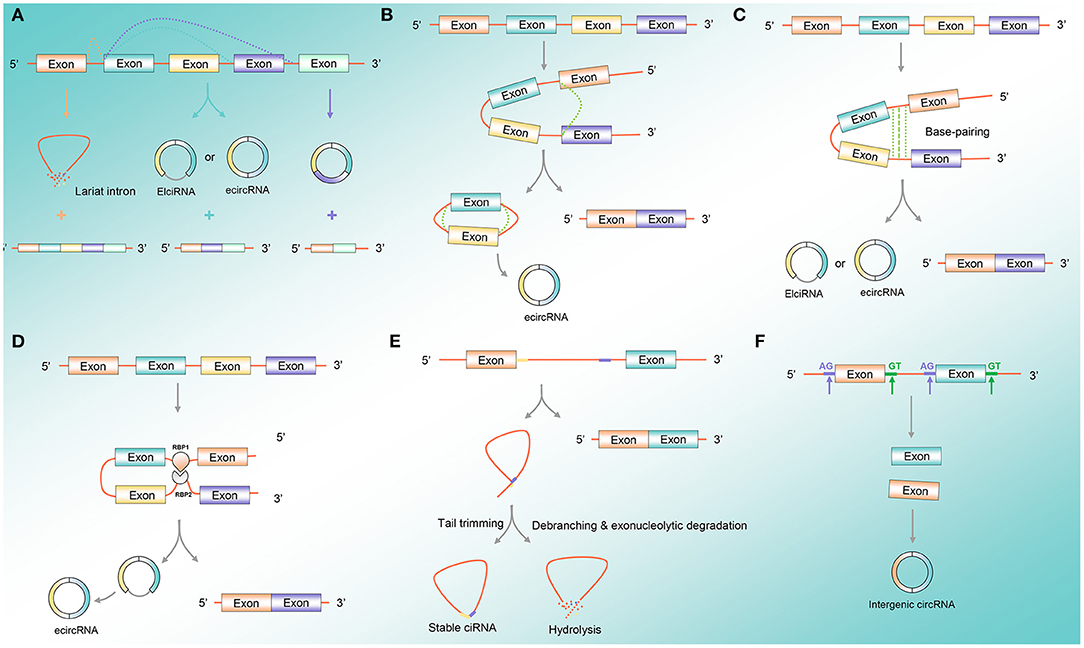
Figure 1. CircRNA formation pattern. (A) Variable cyclization mode: circRNA formation is a variable cyclization mode to generate different types of circRNAs according to different splice sites. (B) Exon skipping: the pre-mRNA is partially folded during the transcription process, so that the 3′-SD of the downstream exon is connected to the 5′-SA of the upstream exon, causing the exon to skip and form an RNA lariat containing both exons and introns. (C) Direct back-splicing: direct back-splicing is the main form of ecircRNA and EIciRNA production, where the flanking intron complementary sequences of pre-mRNAs form a lasso by direct base pairing. (D) RBP-dependent circRNA formation: RBPs bind to the sequence motifs of upstream and downstream in introns on both sides of the exon and act as the bridge to draw the distance between the flanking introns to form circRNA. (E) CiRNA formation pattern: the pre-mRNA can be spliced into an RNA lasso containing a 2′,5′-phosphodiester, resulted in the formation of ciRNAs after cutting off the 3′-tail. (F) Intergenic circRNA formation pattern: the circRNA contains two intronic circRNA fragments flanked by GT-AG splicing signals, acting as the splice donor and splice acceptor of the circular junction to finally form an intergenic circRNA. Abbreviations: circRNA, circular RNA; ecircRNA, exonic circRNA; EIciRNA, exon-intronic circRNA; RBP, RNA-binding protein; ciRNA, circular intronic RNA.
CircRNAs are endogenous molecules that display high enrichment, diversity, evolutionary conservation, relative stability, and specificity. CircRNAs also exhibit tissue-specific and developmental phase-specific expression. (i) Diversity: At present, there are many kinds and quantities of circRNAs. CircRNAs are widely expressed in protozoa, fungi, flies, plants, mammals, and humans, and more than 160,000 species of circRNAs have been identified in eukaryotes (Chen et al., 2016). CircRNAs are also present in various body fluids and are widely distributed in tissues and organs. In some special cases, circRNAs are transcribed in a single cell in greater quantities than related linear RNAs, such as mRNAs. (ii) Evolutionary conservation: The expression profiles of circRNAs are highly evolutionary conserved across species not only in mammals but also in flies, plants, and zebrafish, which are relatively far away in evolution (Barrett and Salzman, 2016). The conservation may imply the important role of circRNAs in biological functions. The study of Jeck et al. (2013) discovered that circRNAs were conserved and non-random products of RNA splicing, related to ALU repeats, and involved in regulating gene expression. Venø et al. (2015) also found that approximately 20% of porcine splice sites were involved in circRNA productions between mice and humans with conservative functions. (iii) Stability: The 5′- and 3′-ends are connected end to end to form the covalently closed circular molecule (Hansen et al., 2013), structurally lacking the easily degraded poly-A tail of 3′-end in structure (Greene et al., 2017). Moreover, circRNAs obtain high stability via RNA folding (Zhang et al., 2013). Interesting, due to the lack of the 2–5 chain of the RNA lasso characteristic structure, ciRNAs are resistant to RNA debranching enzymes and can exist stably. Thus, circRNAs can avoid RNA degradation pathways. The stability of circRNAs endows their potential to serve as enriched and identifiable ncRNA markers, hence bringing innovative directions in circRNA-based research (Zhou et al., 2019). (iv) Specificity: In disparate cells and tissues, circRNAs possess specific expression profiles, which change spatiotemporally with the development of the cell cycle (Zhang et al., 2015). Importantly, circRNAs differ significantly in normal and pathological tissues.
Function Classification of circRNAs
CircRNAs are ubiquitous in eukaryotic cells, and their functions are diverse including adsorption of miRNAs, interaction with RBPs, regulation of transcription, involvement in translation, and formation of pseudogenes. In cancer, circRNA function as tumor promoters or suppressors by acting as miRNA sponges (Figure 2).
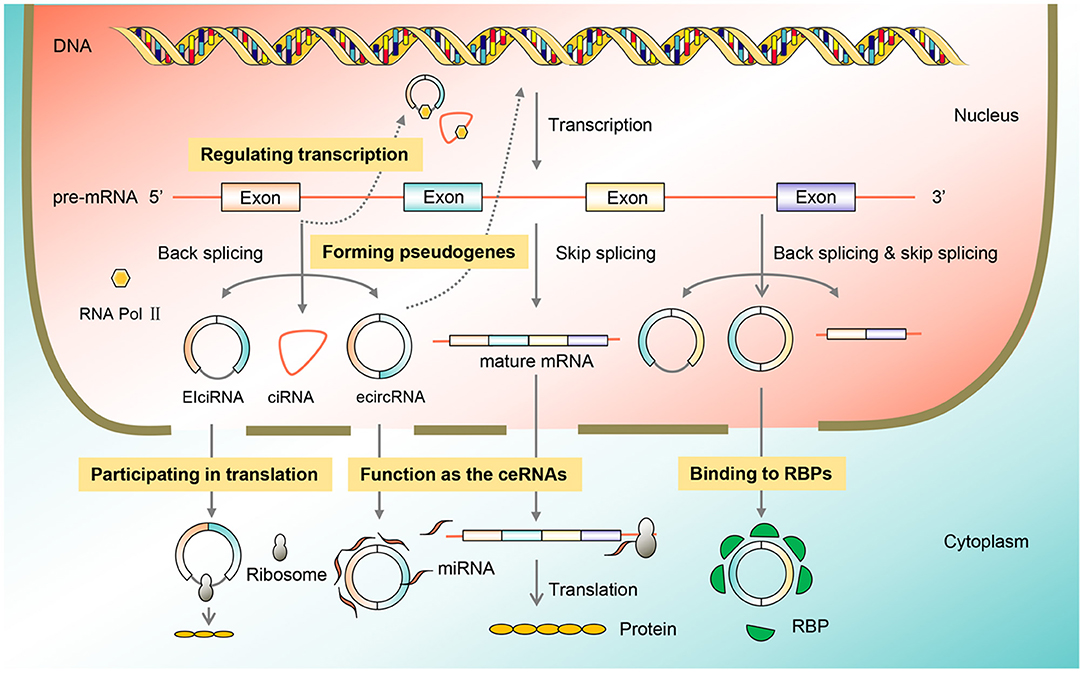
Figure 2. Function classification of circRNAs. CircRNAs are omnipresent in eukaryotic cells, and their functions include acting as the ceRNAs, binding to the RBP, regulating transcription, participating in translation, and forming pseudogenes, thus participating in the process of BC. BC, breast cancer; circRNA, circular RNA; RBP, RNA-binding protein.
Acting as the ceRNAs
The majority of endogenous circRNAs located in the cytoplasm have been identified to have a huge miRNA-binding capacity. miRNAs are transcripts that contained a complementary sequence with target mRNAs, resulting in a substantial alteration of mRNA activity. The most studied function of circRNAs is miRNA sponging, also known as ceRNAs, which can enhance downstream gene expression and regulate cell function and related disease development (Chao et al., 1998). Numerous studies have reported that CDR1as, also called ciRS-7, contained more than 70 conserved miR-7 binding sites (Memczak et al., 2013). The ciRS-7 competitively binds to miR-7 via the ceRNA mechanism, affecting miR-7 activity, thus playing a significant role in gastric cancer, colorectal cancer, and other cancer types (Pan et al., 2018). As the most vital circRNA mechanism, ceRNAs are involved in the occurrence and growth of tumors through circRNA axis-associated signaling pathways (Wang et al., 2017).
Binding to RBPs
RBPs are a class of proteins that bind to RNA and help regulate important processes, such as cell function, cell communication, localization, particularly in the post-transcriptional regulation of RNA. Due to the crucial role in circRNA splicing, processing, folding, stabilization, and localization, RBPs are essential for the generation of circRNAs (Legnini et al., 2017). CircRNAs can bind to different RBPs to exert potential roles such as inhibiting the function of proteins, facilitating the formation of protein complexes, and permitting the interaction between different proteins (Tran et al., 2020). For instance, circ-Foxo3 can form a complex with p21 protein and cyclin-dependent kinases 2 (CDK2), inhibit CDK2 function, block the transformation from the G1 phase to S phase, thereby holding back the proliferation of cancer cells (Cao et al., 2020).
Regulating Transcription
Some specific circRNAs, including EIciRNAs, intronic circRNAs, and circRNAs composed of exons and introns, are regulators in transcriptional and post-transcriptional levels. EIciRNAs are a type of circRNAs that circulate between exons and introns, and retain between exons, also known as exon-intron circRNA. EIciRNA can modulate the expression of genes by interacting with U1 small nuclear RNA (snRNA). Mechanistically, EIciRNA-U1 snRNA compounds may interact with RNA polymerase II future complex to promote gene expression (Sun et al., 2019).
Participating in Translation
CircRNAs have long been defined as untranslatable ncRNAs because of their lack of cap structure and poly tail, which are required for linear mRNA translation (Li et al., 2017). Chen and Sarnow (1995) first proposed the hypothesis that circRNA transcripts could be translated in vitro. With the in-depth studies, most circRNAs are endogenous and are located primarily in the cytoplasm without translation function, but some circRNAs in the cytoplasm containing start codons, exons, and internal ribosome entry sites (IRES) can bind to initiation factors or ribosomes for translation. As proven by the study of Zhang M. et al. (2018), circ-SHPRH contained the open reading frame (ORF), which could be driven by IRES to translate functional protein SHPRH-146aa, leading to the inhibited proliferation of U251 and U373 glioblastoma cells.
Forming Pseudogenes
Pseudogenes abound in the human genome and are traditionally regarded as useless “junk genes.” However, recent studies have verified that pseudogenes play a key regulatory role at the level of DNA, RNA, and proteins, thus participating in different physiological and pathophysiological states (Lou et al., 2020). Some studies have indicated that many pseudogenes are de-regulated in cancer, as well as that circRNAs can form pseudogenes through reverse transcription (Ruan et al., 2020). Based on an RNA-sequencing resource from 293 samples including 13 types of cancer and normal tissues, Kalyana-Sundaram et al. (2012) confirmed that at least 33 pseudogenes might all come from circRFWD2. Braicu et al. (2019) verified that a kind of pseudogene RPL13AP17 could act as a tumor suppressor to inhibit the development of lung cancer. The correlation between circRNAs and pseudogenes is not yet fully understood. The role of pseudogene-derived circRNAs in a variety of cancers and the underlying ceRNA mechanism may provide some key clues to identify potential therapeutic targets.
The Roles and Mechanisms of circRNAs in BC
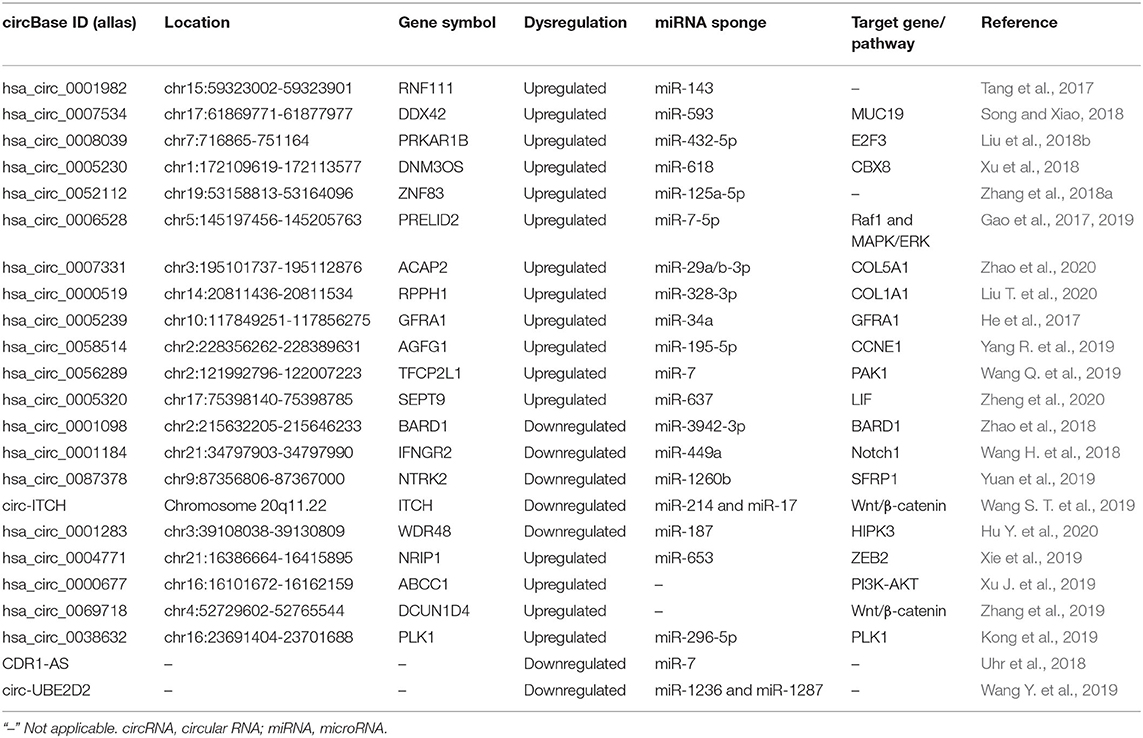
As a novel class of epigenetic regulators, circRNAs are attracting greater attention and are known to function either as oncogenic or anticancer genes in BC tumor growth and metastasis. In addition, circRNAs have many miRNA binding sites, thereby sponging miRNAs to suppress miRNAs binding to target genes, and subsequently affecting tumor growth (Table 1).
CircRNAs in BC Growth
The Promoting Effect of circRNAs in BC Growth
As circRNA CDR1-AS was able to specifically sponge human miR-7, Uhr et al. (2018) further emphasized that CDR1-AS was differentially expressed among the BC cell line subtypes with higher CDR1-AS levels in the luminal and normal-like subtypes. Tang et al. (2017) screened the circRNA expression profiles in BC tissue using circRNA microarray analysis and found that hsa_circ_0001982 was overexpressed in BC tissue and cell lines. Through dual-luciferase reporter assay, miR-143 was verified to be the target of hsa_circ_0001982. The inhibition of hsa_circ_0001982 could restrain BC cell proliferation and invasion and promote apoptosis by targeting miR-143. Song and Xiao (2018) observed the markedly upregulated expression of hsa_circ_0007534 in BC tissues and cell lines. Hsa_circ_0007534 knockdown could suppress BC cell proliferation, colony formation, and invasion and induce apoptosis in BC cells via acting as a miR-593 sponge to increase MUC19 expression. Xie et al. (2019) performed high-throughput circRNA sequencing to detect the differentially expressed circRNAs and confirmed that hsa_circ_0004771 and Zinc finger E-box-binding homeobox 2 (ZEB2) expression levels were upregulated and positively correlated in BC tumor tissues. Mechanistically, hsa_circ_0004771 functioned as a sponge of miR-653 and then downregulated its expression. miR-653 could inhibit ZEB2 expression via binding to its 3′-UTR. Intriguingly, the knockdown of hsa_circ_0004771 and ZEB2 served as equally authentic of miR-653 mimics to induce growth inhibition and apoptosis in BC cells. Liu et al. (2018b) distinguished a new circRNA hsa_circ_0008039 with a high expression level in BC tissues. Hsa_circ_0008039 knockdown distinctly inhibited the proliferation and migration in BC. Mechanistic analyses indicated that hsa_circ_0008039 acted as a sponge of miR-432-5p to improve E2F3 expression.
Xu et al. (2018) confirmed that circ_0005230 was upregulated in BC tissue specimens and cell lines, which was associated with adverse phenotypes in BC patients. Data in vitro and in vivo verified the promoting effect of circ_0005230 on cell growth, cell migratory, and invasive capacities. Regarding the mechanism, circ_0005230 could play a carcinogenic role in BC by sponging miR-618 to increase CBX8 expression. Hsa_circ_0052112, which was significantly higher in MDA-MB-231 cells than that in MCF-7 cells, might promote the migration and invasion of BC cells via directly sponging to miR-125a-5p and possibly upregulating ZNF83 expression (Zhang et al., 2018b). Xu J. et al. (2019) found that hsa_circ_001569 expression was significantly upregulated in both BC tissues and cell lines while hsa_circ_001569 might contribute to the progression of BC by modulating the PI3K-AKT pathway. Karedath et al. (2019) identified and characterized a circRNA derived from the ANKRD12 gene, termed as circANKRD12, which was abundantly expressed in breast and ovarian cancer cells. Manipulating the levels of circANKRD12 induced a strong phenotypic change by significantly regulating cell cycle, invasion, and migration and altering the metabolism in cancer cells (Karedath et al., 2019). Circ_0006528 could facilitate DNA synthesis and cell proliferation, invasion, and migration in BC cells. For the mechanism investigation, circ_0006528 could sponge miR-7-5p to promote Raf1 expression, which activated the MAPK/ERK signaling pathway (Gao et al., 2019). Zhao et al. (2020) applied bioinformatics analysis to finding that circACAP2 expression was elevated in BC tissues and could lead to the malignant phenotype of BC cells. Mechanistically, circACAP2 induced BC proliferation and motility by sponging miR-29a/b-3p and regulating COL5A1. Wang Y. et al. (2019) found that circ-UBE2D2 was a high expressional biomarker closely associated with aggressive clinical features and dismal prognosis. Circ-UBE2D2 could significantly suppress the proliferation, migration, and invasion of BC cells by binding to miR-1236 and miR-1287, thus modulating the expression of their respective target genes. Besides, the delivery of cholesterol-conjugated si-circ-UBE2D2 oligonucleotides significantly repressed tumor growth in vivo. Liu T. et al. (2020) analyzed that higher expression of hsa_circRNA_002178 led to the worse prognosis in BC by competitively harboring miR-328-3p and then increasing COL1A1 expression. In vivo experiments further demonstrated the suppression of tumor growth and inflammation by inhibiting hsa_circRNA_002178 or upregulating miR-328-3p.
Some specific circRNAs are tumor promoters in the malignant transformation of triple-negative breast cancer (TNBC). He et al. (2017) conducted circRNA microarrays and observed that circGFRA1 was abnormally upregulated in TNBC. Upregulated circGFRA1 was associated with poorer survival via sponging miR-34a to modulate GFRA1 expression. CircRNA_069718 was a tumor oncogenic circRNA in TNBC progression. CircRNA_069718 inhibition reduced the expression levels of Wnt/β-catenin pathway-related genes, including β-catenin, c-myc, and cyclin D1 (Zhang et al., 2019). In four paired TNBC tissues and para-cancerous tissues using RNA sequencing, Yang R. et al. (2019) found a total of 354 circRNAs were differentially expressed with 47 upregulated and 307 downregulated expressions. Among them, circAGFG1 was the most upregulated circRNA spliced from AGFG1 and was correlated with clinical stage, pathological grade, and poor prognosis of TNBC patients. CircAGFG1 could act as a ceRNA for sponging miR-195-5p to alleviate the inhibition of miR-195-5p on CCNE1, thus accelerating the cell division. CircPLK1 was significantly upregulated in TNBC and associated with worse survivals. The knockdown of circPLK1 efficiently inhibited the cell growth and invasion in vitro as well as tumor occurrence and metastasis in vivo. Therefore, the ceRNA mechanism of the circPLK1-miR-296-5p-PLK1 axis regulated the tumor progression in TNBC (Kong et al., 2019). Wang Q. et al. (2019) conducted a human circRNA microarray in three pairs of TNBC tissues and adjacent non-cancerous tissues to screen circRNA expression patterns in TNBC. The results confirmed that circ-TFCP2L1 was markedly upregulated in TNBC tissues and cells and led to the shorter disease-free survival for TNBC patients. Circ-TFCP2L1 induced the proliferation and migration of TNBC by sponging miR-7 so as to decrease PAK1 expression. Mechanistic studies revealed that circSEPT9 could sponge miR-637 and activate the leukemia inhibitory factor (LIF)/STAT3 signaling pathway involved in TNBC progression to modulate the expression of LIF (Zheng et al., 2020). In addition, E2F1 and EIF4A3 might improve the biogenesis of circSEPT9.
The Inhibiting Effect of circRNAs in BC Growth
Breast cancer stem cells (BCSCs) are defined as a small subset of cancer cells with the properties of increasing self-renewal, multilineage differentiation, and tumor maintenance, all of which are considered contributions to the BC progression, metastasis, and recurrence. It was of note that Yan et al. (2017) proposed the work to prove the evidence of the circRNA profiles and circRNA/miRNA interplays in BCSCs. They found 27 aberrantly expressed circRNAs, of which circVRK1 could suppress BCSC proliferation and self-renewal capacity. In BC cell MCF-7 with the treatment of tetrachlorodibenzo-p-dioxin (TCDD), circRNA_BARD1 was upregulated to suppress cell proliferation, block cell cycle, and induce cell apoptosis (Zhao et al., 2018). Moreover, circRNA_BARD1 was overexpressed by TCDD and then suppressed BC tumorigenesis via miR-3942-3p/BARD1 axis both in vivo and in vitro. Wang H. et al. (2018) conducted circRNA microarray analysis to investigate abnormally expressed circRNAs in BC tissue and confirmed that circRNA-000911 was downregulated in BC cells. CircRNA-000911 could restrain cell proliferation, migration, and invasion and facilitate the apoptosis of BC cells via capturing miR-449a to improve Notch1 expression and inhibiting nuclear factor-κB (NF-κB) signaling. By systematically profiling circRNA expression patterns, Yuan et al. (2019) observed 3,653 differentially expressed circRNAs in estrogen receptor (ER)-positive BC compared with adjacent non-tumor tissues. Among them, has_circ_0072304, has_circ_0087378, and has_circ_0087377 were verified to be downregulated in ER-positive BC. Besides, the has_circ_0087378-miR-1260b-SFRP1 axis was considered to be a key regulatory pathway in BC tumorigenesis (Yuan et al., 2019). Wang S. T. et al. (2019) reported that circ-ITCH was significantly downregulated in TNBC and correlated with poor prognosis. Furthermore, circ-ITCH could bind miR-214 and miR-17 to increase its expression of ITCH linear isoform, thereby inactivating Wnt/β-catenin signaling and then blocking the expression of downstream genes to inhibit the proliferation of BC cells (Wang S. T. et al., 2019). Sang et al. (2019) conducted RNA sequencing on a large number of clinical samples and screened their circRNA expression profiles and demonstrated that hsa_circ_0025202 was significantly decreased in BC tissues and could restrain the proliferation and metastasis of BC cells. Hu Y. et al. (2020) observed that the circRNA-0001283 expression was downregulated in BC tissue samples. CircRNA-0001283 ectopic expression remarkably suppressed BC cell viability and invasion and promoted apoptosis in BC cells via decreasing the expression of miR-187 and then positively regulating HIPK3 expression, which might provide targets for developing novel therapeutic strategies for BC.
CircRNAs in BC Metastasis
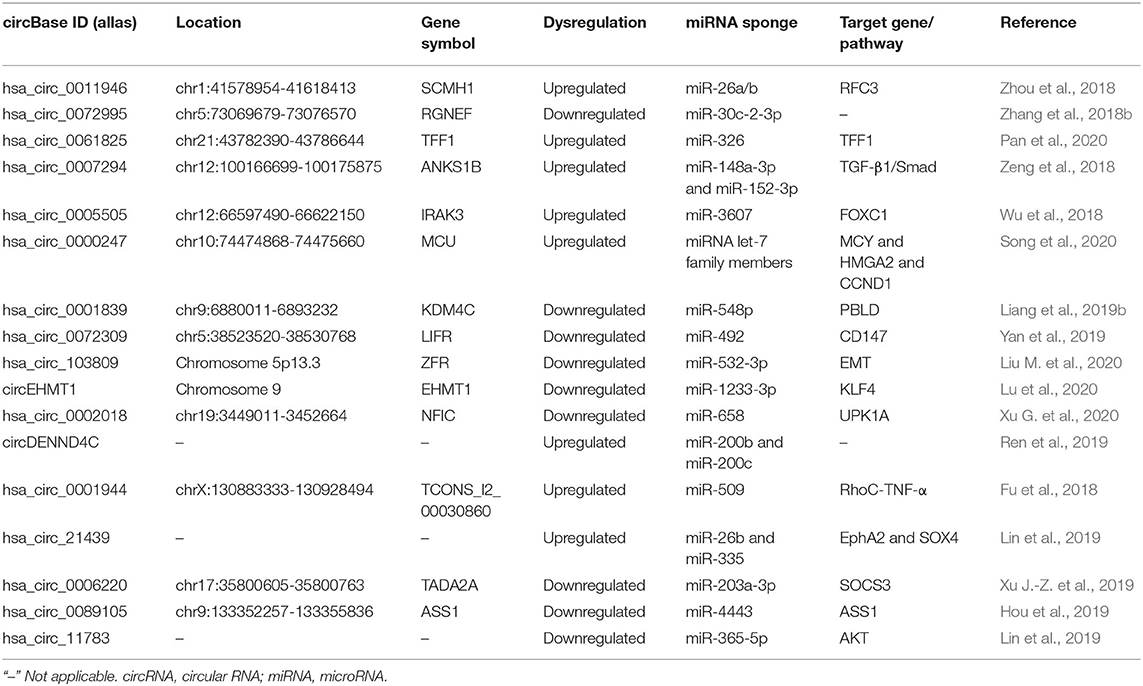
Metastasis is the result of multiple steps, including local infiltration of tumor cells through the surrounding extracellular matrix (ECM) and stromal cell layer, interpolation, and survival in the circulation, extravasation, and seeding to distant places and the beginning of metastasis and colonization. CircRNAs play vital roles in metastasis and cancer progression (Table 2). For instance, Li Z. et al. (2020) indicated that a total of 4,370 circRNAs in general expression profiling, with 2,375 upregulated circRNAs and 1,995 downregulated circRNAs in these BC/adjacent tissues. Hsa_circ_0069094, hsa_circ_0079876, hsa_circ_0017650, and hsa_circ_0017526 were promising candidate targets, as they were associated with tumor-node metastasis (TNM) staging, lymph node infiltration, and Ki67.
The Promoting Effect of circRNAs in BC Metastasis
Elucidation of the underlying mechanisms of circRNA-promoting metastasis is desperately needed to offer novel therapeutic strategies for patients with metastatic BC. Cancer metastasis is a complex and multistep process. Zhou et al. (2018) utilized high-throughput sequencing to uncover the 152 differentially expressed circRNAs in BC tissues and cell lines and found that hsa_circ_0011946 was significantly upregulated. Whereas, the inhibition of hsa_circ_0011946 significantly suppressed the migration and invasion of MCF-7 cells. Possibly, hsa_circ_0011946 sponged miR-26a/b to directly target replication factor C subunit 3 (RFC3) and knockdown of hsa_circ_0011946 could inhibit RFC3 mRNA and protein expression thus promoting cell migration and invasion, as confirmed by bioinformatics analysis and validation. Hsa_circ_0072995 was generated from the ARHGEF28 gene and also might be a novel biomarker for BC (Zhang et al., 2018b). Hsa_circ_0072995 could promote BC cell migration and invasion by capturing miR-30c-2-3p. Besides, the overexpression of hsa_circ_0072995 upregulated ARHGEF28 expression, which activated the activity of FAK and RhoC, enhancing migration and invasion capability of BC cells. Zhang et al. (2019) investigated the potential regulatory mechanism of circRNA_069718 in BC and found that circRNA_069718 expression was increased in BC tissues and cell lines and its high expression was significantly correlated with advanced TNM, lymph node metastasis, and poor overall survival ability in BC patients. Interestingly, circDENND4C was a HIF1A-associated circRNA and was highly expressed in hypoxia conditions to promote BC cell proliferation. Silencing circDENND4C suppressed glycolysis, migration, and invasion in BC cells under hypoxia possibly by promoting the expression miR-200b and miR-200c (Ren et al., 2019). This work highlighted circDENND4C as a novel prognostic and therapeutic target for BC. Pan et al. (2020) observed that circ-TFF1 and TFF1 were both overexpressed and positively correlated with each other in BC. Circ-TFF1 could promote BC cell proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT) in vitro and facilitate tumor growth in vivo via sponging miR-326 to increase TFF1 expression.
Some circRNAs possess a promoting impact on BC lung metastasis. Zeng et al. (2018) reported that circANKS1B feedback loop promoted BC invasion and metastasis both in vitro and mouse lung metastasis model by inducing EMT, with no effect on BC growth. Mechanistically, circANKS1B abundantly sponged miR-148a-3p and miR-152-3p to increase the expression of USF1, which could transcriptionally upregulate TGF-β1 expression, thereby activating TGF-β1/Smad signaling for enhancing EMT (Zeng et al., 2018). Wu et al. (2018) verified an oncogenic circRNA derived from the IRAK3 gene locus, named circIRAK3, which was closely related to the poor prognosis of BC patients with recurrence to distant organs. Besides, circIRAK3 was significantly upregulated in metastatic BC cells and could promote cell migration, invasion, and lung metastasis by the sponge of miR-3607 to regulate FOXC1 expression, thus triggering a positive-feedback loop. CircHMCU exerted oncogenic functions including promoting the migration and invasion, via efficiently harboring the let-7 family to increase the expression of MYC, HMGA2, and CCND1. Further in vivo studies showed the overexpression of circHMCU resulted in the rapid proliferation and lung metastasis of BC (Song et al., 2020).
Breast cancer brain metastases (BCBM) often cause neurological impairments by affecting cognitive and sensory functions, which occupy a proportion of diagnosed metastatic BC and confer an extremely poor prognosis. Identifying the mechanism of key circRNAs of BCBM is a prerequisite for unlocking BCBM to predict, diagnose, and future innovative treatments. Fu et al. (2018) performed RNA-sequencing to identify the circRNA expression profiles of BCBM cell line 231-BR in comparison with non-specific metastatic MDA-MB-231. The result verified that 215 upregulated and 191 downregulated circRNAs, among which 90% differentially expressed circRNAs were generated from the exonic regions of host genes and demonstrated that the most enriched pathways of the differentially expressed circRNAs were related to the RNA degradation and axon guidance. It hypothesized that has_circ_0001944 might be involved in BCBM through sponging up miR-509 and interfering with its downstream targets, thereby forming the has_circ_0001944/miR-509/RhoC-TNF-α axis (Fu et al., 2018). Lin et al. (2019) reported that hsa_circ_21439 was verified to be the most significantly upregulated in breast cancer liver metastases (BCLM) tissues and cells. The has_circ_21439 would regulate the process of BCLM through has_circ_21439/miR-26b/EphA2 axis and/or has_circ_21439/miR-335/SOX4 axis.
The Inhibiting Effect of CircRNAs in BC Metastasis
By screening the circRNA profiles in a large cohort of BC patients, Xu J.-Z. et al. (2019) distinguished 235 differentially expressed circRNAs including the two differentially expressed circTADA2As, circTADA2A-E6, and circTADA2A-E5/E6 in BC. Especially, circTADA2A-E6 possessed the tumor-suppressor capability in vitro through sponging a miR-203a-3p to restore the expression of SOCS3, resulting in a less aggressive oncogenic phenotype. Hou et al. (2019) confirmed that circASS1 was significantly downregulated in highly aggressive BC cell line MDA-MB-231 cells compared with MCF-7. CircASS1 suppressed invasion and migration of BC cells by harboring miR-4443 and upregulating the ASS1 expression, implying the tumor-suppressive role of circASS1 in BC metastasis. Liang et al. (2019a) identified a fresh circBMPR2, which was significantly reduced in human BC tissues with metastasis and was negatively associated with the motility of BC. Functionally, the circBMPR2 knockdown effectively enhanced cell proliferation, migration, and invasion (Liang et al., 2019a). In the following studies, they further characterized a novel KDM4C gene-derived circRNA circKDM4C which was decreased in BC tissues of a cohort of 219 BC patients (Liang et al., 2019b). The lower circKDM4C expression was associated with poor prognosis and metastasis in BC, showing the tumor suppressor function of circKDM4C in BC metastasis. Yan et al. (2019) analyzed that the downregulated hsa_circ_0072309 significantly suppressed the proliferation, migration, and invasion of BC cells in vitro. In vivo assays showed that hsa_circ_0072309 ectopic expression inhibited BC growth via the hsa_circ_0072309-miR-492 axis. Liu M. et al. (2020) discovered that circRNA_103809 was downregulated in BC tissues, and circRNA_103809 expression level was closely related to the distant metastasis size, TNM stage, HER-2 status, and overall survival time. In vitro experiments indicated that circRNA_103809 overexpression could obviously suppress the EMT pathway to inhibit BC cell proliferation and metastasis via harboring miR-532-3p.
In the study of lung metastasis, Lu et al. (2020) reported that circEHMT1 was decreased in human BC tissues, and the overexpression of circEHMT1 inhibited lung metastasis of BC in vitro and in vivo, emphasizing the importance of circEHMT1 in inhibiting BC metastatic potential by reducing MMP2 levels through modulating circEHMT1/miR-1233-3p/KLF4 axis. Xu G. et al. (2020) demonstrated that circNFIC was the most downregulated circRNA in lung metastatic tissues and that low levels of circNFIC were associated with poor outcome of BC by Kaplan-Meier survival analysis. Moreover, both in vivo and in vitro, circNFIC functioned as a ceRNA to upregulate UPK1A expression by decoying miR-658, possibly resulting in suppressing BC cell proliferation and migration to the lung (Xu G. et al., 2020). In BCLM and cells, Lin et al. (2019) used RNA sequence to identify the altered expression profile of circRNAs, showing that the downregulated hsa_circ_11783 exerted the functions in BCLM by hsa_circ_11783/miR-365-5p/AKT axis by further bioinformatics exploration.
Diagnostic Value of circRNAs in BC
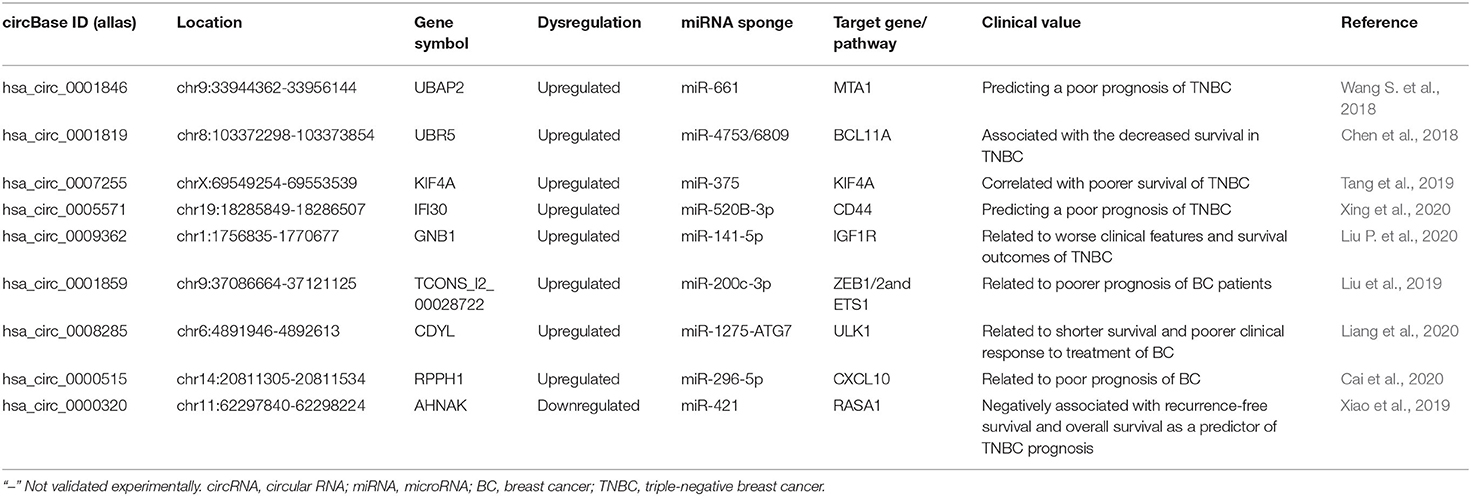
Given the unique characteristics of expression, stability, and enriched miRNA binding sites, circRNAs have been endowed with great potential as tumor diagnostic biomarkers and therapeutic targets in BC (Greene et al., 2017) (Table 3). These identified circRNA modules provide novel insights into the potentially BC-associated circRNAs and will contribute to the clinical applications of circRNA for BC diagnosis, treatment, and prognosis in the future. Therefore, identifying changes of circRNA expression in body fluids may open a novel avenue for the non-invasive BC diagnosis.
CircRNA as Potential Prognosis Biomarkers in BC
Identification and elucidation of the effective potential diagnostic and prognostic value of circRNA biomarkers are essential for clinical decision-making. Numerous studies have shown that circRNAs present differential expressions in BC and are linked to the pathogenesis and prognosis-related biomarkers. The integration of circRNAs, miRNAs, DNA methylation, and gene expression data to identify sponge circRNAs is crucial for revealing the DNA methylation-mediated regulation of sponge circRNAs in cancer progression. Gu et al. (2019) revealed a complex DNA methylation-mediated regulation of circRNA sponges, and 10 sponge circRNA host genes were potential prognostic biomarkers of BC. Ma et al. (2020) conducted a weighted gene co-expression network analysis with the differentially expressed miRNAs and mRNAs in BC from The Cancer Genome Atlas (TCGA) database to identify the key modules associated with the carcinogenesis of BC. By database co-screening, has_circ_0083373, hsa_circ_0083374, and hsa_circ_0083375 were involved in the pathogenesis and development of BC, mechanistically through regulating DLC1 expression via has-mir-511. Yang et al. (2020) used RNA-sequencing, qPCR, and bioinformatic analysis to explore potential mechanisms involving differentially expressed circRNAs in the serum exosomes and tissues of BC patients. Compared with paracancerous normal tissues, hsa-circRNA-0005795 and hsa-circRNA-0088088 were significantly different both in serum exosomes and tissues and might function as ceRNAs with 11 and 8 circRNA-miRNA interactions, respectively (Yang et al., 2020). The overwhelming majority of circRNAs, which had been reported, showed upregulated expression levels in BC. Therefore, the novel circRNA biomarkers are warranted to be discovered for the early detection and prognosis for BC (Tomar et al., 2020).
The Upregulated CircRNAs as Prognosis Biomarkers
TNBC is the most aggressive BC subtype, exhibiting a strong proliferative capacity, increased recurrence, and decreased survival rate. Revealing the role of dysregulated circRNAs will be critical for understanding TNBC pathogenesis and improving the prognosis. Wang S. et al. (2018) observed that circ-UBAP2 with high expression was closely correlated with larger tumor size, lymph node metastasis, advanced TNM stage, and unfavorable prognosis. In vitro and in vivo, circ-UBAP2 could enhance the proliferation and migration of TNBC cells and suppress apoptosis by sponging miRNA-661 to enhance the oncogene MTA1 expression. Likewise, circANKS1B was notably upregulated in TNBC compared with non-TNBC tissues and cell lines, which was closely associated with lymph node metastasis and advanced clinical stage, serving as an independent risk factor for overall survival of BC patients (Zeng et al., 2018). Chen et al. (2018) found that circEPSTI1 was significantly upregulated and a high level of circEPSTI1 was associated with the decreased survival in TNBC patients. Their experiments showed that circEPSTI1 bound to miR-4753 and miR-6809 as the miRNA sponge to enhance BCL11A expression and then inhibited TNBC cell proliferation and promoted apoptosis, thus presuming that circEPSTI1 might be an independent prognostic marker for survival in TNBC patients. Tang et al. (2019) explored the circRNA profiles by circRNA microarray and observed that circKIF4A was overexpressed in BC cells and TNBC tissues. CircKIF4A promoted the proliferation and migration of TNBC cells via sponging miR-375 to modulate KIF4A expression. Thus, circKIF4A might act as a prognostic and therapeutic target for TNBC and positively correlated with poorer survival of TNBC. Through quantitative real-time polymerase chain reaction (qRT-PCR) and in situ hybridization, Xing et al. (2020) observed that circIFI30 was upregulated in TNBC tissues and could remarkably enhance TNBC cell malignant biological properties by acting as the miR-520b-3p sponge to improve CD44 expression. The increased circIFI30 was positively associated with the clinical TNM stage, pathological grade, and poor prognosis of TNBC patients, thus circIFI30 could be a new diagnostic/prognostic marker and therapeutic target for TNBC patients (Xing et al., 2020). Liu P. et al. (2020) characterized circGNB1 via analyzing the circRNA microarray profiling and found that circGNB1 was overexpressed in TNBC cell lines. Knockdown of circGNB1 obviously inhibited cell proliferation, migration, and tumor growth via the circGNB1-miR-141-5p-IGF1R axis. Consequently, circGNB1 might be potential for BC prognosis as that high circGNB1 expression was related to worse clinical features and survival outcomes (Liu P. et al., 2020). Zheng et al. (2020) studied the circRNA expression patterns in four pairs of TNBC tissues and paracancerous normal tissues using RNA-sequencing. The results indicated that circSEPT9 was upregulated in TNBC tissues, which was positively related to the advanced clinical stage and poor prognosis (Zheng et al., 2020).
Liu et al. (2019) systematically investigated the circRNAs and found hsa_circ_001783 was a top-ranked circRNA in both BC cells and tissues. The higher level of hsa_circ_001783 was closely related to heavier tumor burden and poorer prognosis of BC patients. Moreover, hsa_circ_001783 promoted the progression of BC cells by sponging miR-200c-3p to improve the expression of ZEB1, ZEB2, and ETS1. Therefore, hsa_circ_001783 might act as a new prognostic and therapeutic target for BC (Liu et al., 2019). Yang L. et al. (2019) explored a novel increased circ_0103552, with clinical severity and dismal prognosis. Furthermore, circ_0103552 could promote cell growth, clone-forming ability, migration, and invasion and suppress apoptotic cells. Further mechanistic studies revealed that circ_0103552 could directly sponge miR-1236 to perform oncogenic activities in BC cells (Yang L. et al., 2019). Liang et al. (2020) screened autophagy correlated circRNAs in BC tissues with high- and low-autophagic levels by circRNA deep sequencing. The results showed that circCDYL improved the autophagic level in BC cells by the miR-1275-ATG7/ULK1 axis and promoted the malignant progression of BC cells in vitro and in vivo. Clinically, increased circCDYL in the BC tissues and serum was related to higher tumor burden, shorter survival, and poorer clinical response to treatment (Liang et al., 2020). Cai et al. (2020) gathered in 340 BC tissues and identified hsa_circ_0000515 which was overexpressed and related to the poor prognosis of BC. Knockdown of hsa_circ_0000515 disrupted cell cycle progression, cell proliferation, and invasion, weakened inflammatory response, and decreased the pro-angiogenetic potential of BC cells (Cai et al., 2020). Regarding the mechanism, hsa_circ_0000515 could sponge miR-296-5p to hinder it from inhibiting CXCL10 expression.
Additionally, as circHMCU was upregulated in high metastatic cell lines compared with parental cell lines and in BC tissues compared with normal tissues, circHMCU was related to poor BC prognosis and could be used as a novel biomarker in the diagnosis and prognosis of BC (Song et al., 2020). Gao et al. (2019) analyzed a cohort of 97 patients and observed that circ_0006528 expression was upregulated in BC tissues and was significantly related to advanced TNM stage and poor prognosis. The multivariate assay showed that hsa_circ_001569 expression was an independent prognostic factor for 5-year overall survival (Xu J. et al., 2019). This study indicated that hsa_circ_001569 upregulation was associated with BC lymph-node metastasis, clinical stage, and poor prognosis. Smid et al. (2019) demonstrated an independent circRNA pool of CNOT2, CREBBP, and RERE in primary BC. The circCNOT2 could be detectable in cell-free RNA from plasma and exhibited clinical potential for aromatase inhibitor therapy because circCNOT2 levels were predictive of progression-free survival time to aromatase inhibitor therapy in advanced BC patients.
The Downregulated CircRNAs as Prognosis Biomarkers
Circ-ITCH is a tumor repressor and was markedly downregulated in TNBC tissues and cell lines and correlated with poor prognosis (Wang S. T. et al., 2019). Circ-ITCH improved its expression via sponging miR-214 and miR-17, which in turn inactivated the Wnt/β-catenin signal and then inhibited the expression of downstream genes. Circ-ITCH was a promising prognostic biomarker in TNBC, and its restoration could well be a successful strategy in TNBC (Wang S. T. et al., 2019). Xiao et al. (2019) confirmed that circAHNAK1 was markedly downregulated in TNBC cell lines, which was negatively associated with recurrence-free survival and overall survival as a predictor of TNBC prognosis. The mechanistic analysis showed that circAHNAK1 inhibited proliferation and metastasis of TNBC via acting as the miR-421 sponge to promote RASA1 expression. CircEHMT1 was downregulated in human BC tissues compared with adjacent normal breast tissues (Lu et al., 2020). The AUC value of circEHMT1 was very close to 1, suggesting that circEHMT1 was related to a better prognosis of human BC.
CircRNA-Based Computational Methods for BC Detection
Increasing evidence indicates that circRNAs are broadly in the occurrence and development of BC. However, due to the complex mechanisms, it is expensive, difficult, and time consuming to discover the new circRNA-disease associations by biological experiments. Therefore, it is an increasingly urgent need to utilize computational methods to predict new circRNA disease associations. Based on existing bioinformatics approaches, Nair et al. (2016) developed a comprehensive workflow called Circ-Seq, which was applied to RNA-sequencing data from BC cell lines. Notably, normal-adjacent tissues in ER-positive subtype had higher numbers of circRNAs than tumor samples in TCGA, and the number of circRNAs in normal-adjacent samples of the ER+ subtype was inversely correlated to the risk-of-relapse proliferation (ROR-P) score for proliferating genes, suggesting that the circRNA frequency might be a marker for cell proliferation in BC. Tarrero et al. (2018) developed a novel computational tool, named CircHunter, to exploit RNA-sequencing data to define a compilation of exonic circRNAs, more accurately. With this mean, a subset of MCF-7 circRNAs was specific to tumor vs. normal tissue, while some circRNAs could distinguish Luminal from other tumor subtypes, thus suggesting that circRNAs could be exploited as novel biomarkers and drug targets for BC. Lei et al. (2018) proposed a new computational path-weighted method Path-Weighed Method for Predicting CircRNA-Disease Associations (PWCDA), showing that circPVT1 could be worked as a miRNA spouse to regulate miRNA by moderating let-7 activity in BC. Their computational method in terms of different validation measures was verified to be reliable and useful to predict potential circRNA-disease associations.
According to a similar method, Wang S. et al. (2019) developed a systematic approach to identify circRNA modules in the BC context through integrating circRNAs, mRNAs, miRNAs, and pathway data based on a non-negative matrix factorization (NMF) algorithm. Employing the systemic pipeline in 33 BC RNA-sequencing data with tumor and normal samples, 13 circRNA modules were identified in BC, containing 80 circRNAs, 2,703 genes, 63 miRNAs, and 1,318 pathways. After screening by functional enrichment analysis, 9 circRNA modules potentially associated with BC were obtained. Within them, one circRNA hsa_circ_0006528 had been recognized as a known disease circRNA. Lei and Fang (2019) proposed a machine learning-based computational model named Gradient Boosting Decision Tree with multiple biological data to predict circRNA-disease associations (GBDTCDA). With this method, the results of those predicting circRNA-disease associations in BC such as circMYO9B and circRNA_100984 were validated to be consistent with the database circ2Disease and circRNADisease. Deng et al. (2019) also established a computational method named KATZCPDA, which was based on the KATZ method and the integrations among circRNAs, proteins, and diseases. By using the KATZCPDA calculation model, BC was ranked at the top of the list of associated diseases. Among the illnesses associated with hsa_circ_0011946, hsa_circ_0001982, hsa_circ_0001785, hsa_circ_0001785, and hsa_circ_0002113, BC was ranked 6th, 6th, 6th, 6th, and 8th, respectively. As detailed in the database, hsa_circ_0001982 had been verified to be involved in the BC cell proliferation and invasion by targeting miR-143.
In 2020, Ge et al. (2020) also invented a computational method named as LLCDC to efficiently predict circRNAs related to BC, showing that 19 of the top 20 potential circRNAs were validated by updating the circRNADisease database and recent biological experiments. Li M. et al. (2020) presented a new method called Speedup Inductive Matrix Completion for CircRNA-Disease Associations prediction (SIMCCDA) to predict circRNA-disease associations. They found that the top 29 predicted candidates were associated with BC in related studies. For instance, hsa_circ_0001875 was upregulated and hsa_circ_0006054 was significantly downregulated in BC tissues (Li M. et al., 2020). Coincidentally, Wang et al. (2020) also proposed a computational method called GCNCDA. In the case study experiments on diseases including BC, glioma, and colorectal cancer, about 16, 15, and 17 of the top 20 candidate circRNAs, respectively, with the highest prediction scores were verified by relevant literature and databases, suggesting that GCNCDA was effective in predicting potential circRNA-disease associations.
These identified circRNA modules provide novel insights into the potentially BC-associated circRNAs. Combining these systematic methods will benefit the clinical applications of circRNA biomarkers for BC diagnosis, treatment, and prognosis in the future.
CircRNAs in BC Drug Resistance
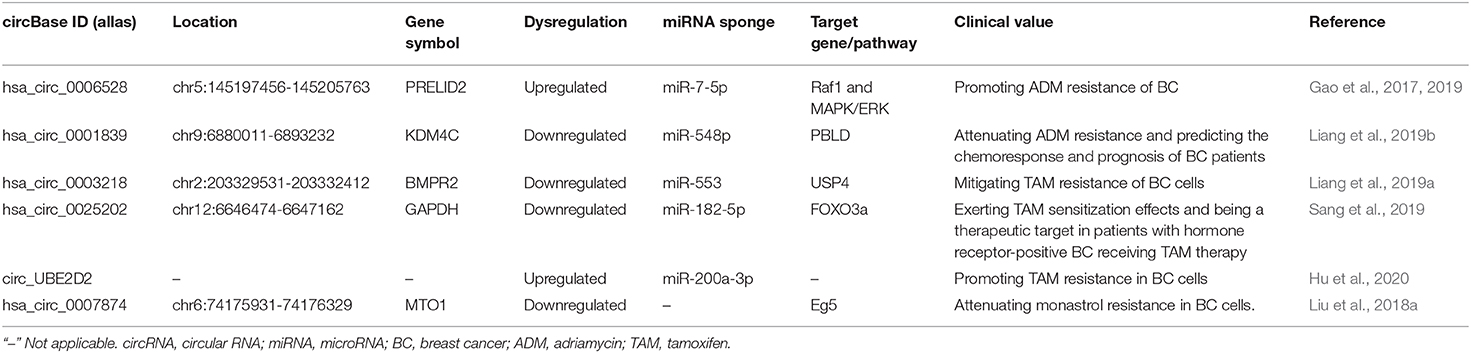
Currently, chemotherapy plays an indispensable role in the therapy of BC for which it can control and minimize lesions before the operation and prevent recurrence and metastasis after the operation. However, in the process of chemotherapy, patients are prone to drug resistance which hinders the treatment progress. Therefore, it is urgent to slow down or eliminate the chemotherapy resistance of BC and improve the cure rate and quality of life of BC patients. Considering the crucial roles of circRNAs in the chemotherapy resistance of BC therapy, more and more studies are focusing on addressing this obstacle (Table 4). In fact, the resistance-related circRNAs are promising therapeutic and diagnostic biomarkers to increase the chemosensitivity and predict chemotherapeutic agent effectiveness in the individual with resistance thereby improving the clinical management of BC. Adriamycin (ADM), tamoxifen (TAM), and monastrol are the most reported resistant drugs associated with circRNAs in recent studies in the chemotherapy of BC patients.
ADM is a cell cycle non-specific antibiotic with a broad antitumor spectrum, which can inhibit the synthesis of DNA and RNA to inhibit tumor growth. ADM is one of the most effective chemotherapy drugs for BC. Nevertheless, the efficacy of ADM is reduced by drug resistance. By circRNA microarray expression profiles and PCR, Gao et al. (2017) found that hsa_circ_0006528 was obviously higher in ADM-resistant human BC tissues and cell lines (MCF-7/ADM and MDA-MB-231/ADM) than that in the ADM-sensitive groups. Additionally, they also discovered a regulatory role of the hsa_circ_0006528-miR-7-5p-Raf1 axis in ADM-resistant BC, strongly inferring that circRNAs participated in BC chemoresistance and further verification and functional analysis were needed (Gao et al., 2019). The circKDM4C was able to regulate BC cell proliferation, metastasis, and ADM resistance both in vitro and in vivo, mechanistically through sponging miR-548p to increase the PBLD expression (Liang et al., 2019b). CircKDM4C might serve as a biomarker to predict the chemoresponse and prognosis of BC patients, and the approach of targeting the circKDM4C axis might be beneficial for BC treatment (Liang et al., 2019b). Although TAM is a commonly used endocrine therapy for the treatment of advanced BC, TAM resistance remains an inevitable clinical issue for many patients. CircBMPR2 knockdown promoted TAM resistance of BC cells by inhibiting TAM-induced apoptosis, whereas circBMPR2 overexpression led to decreased TAM resistance. Mechanistically, circBMPR2 could inhibit the progression and TAM resistance of BC by sponging miR-553 to upregulate ubiquitin-specific protease 4 (USP4) expression (Liang et al., 2019a). Therefore, circBMPR2 may serve as a potential therapeutic and prognostic predictor for BC patients. Sang et al. (2019) characterized a novel hsa_circ_0025202, which could exert tumor inhibition and TAM sensitization effects via the miR-182-5p/FOXO3a axis in vitro and in vivo. This result confirmed that hsa_circ_0025202 could be a therapeutic target in patients with hormone receptor-positive BC receiving TAM therapy. Additionally, circ_UBE2D2 was upregulated in TAM-resistant BC tissues and cell lines while circ_UBE2D2 deletion mitigated TAM resistance in BC cells (Hu et al., 2020). Circ_UBE2D2 was also significantly loaded in resistant cell-derived exosomes and could be transferred to parental cells. Thus, exosome-mediated transfer of circ_UBE2D2 reinforced the resistance of BC to TAM by binding to miR-200a-3p. Monastrol is the prototype of the anti-actin drug with high efficiency and cell permeability, but its drug resistance limits the application in BC therapy. By using a human circRNA microarray, Liu et al. (2018a) discovered the downregulation of circRNA-MTO1 in monastrol-resistant BC cells. CircRNA-MTO1 could repress Eg5-mediated cellular viability and promote chemosensitivity by sequestering TRAF4 from binding to Eg5 protein. The regulatory pattern of circRNA-MTO1 revealed a regulatory mechanism on circRNA-MTO1 controlling cell viability and monastrol resistance in BC cells. Targeting these newly identified circRNAs may help develop potential novel therapies for BC patients.
Conclusion
Altogether, the above studies have demonstrated the aberrantly altered expression and biogenesis of circRNAs in different subtypes and stage-specific manners of BC. The fundamental functions of various circRNAs can act as either tumor promoters or suppressors in BC, involving in proliferation, metastasis, and chemotherapy resistance via regulating the gene expression (Tran et al., 2020). Most of the circRNA functions in BC are reported to be achieved by miRNA sponge to cause a substantial change in the downstream miRNA activity. These ceRNA functions manifest as circRNA-miRNA-mRNA axis mechanisms (Li et al., 2020). Elucidating these circRNA roles and mechanisms will eventually facilitate the clinical application of circRNAs in the diagnosis, therapy, and prognosis evaluation.
It is worth noting that now this field has still embraced some challenges. Firstly, circRNAs are usually excavated based on RNA sequencing, bioinformatic analyses, and PCR experiments. However, there is a lack of a uniform standard for circRNA naming (Guo et al., 2020). In addition, in some studies, only a small sample size was collected to identify and quantify circRNAs, accompanied by the relatively low abundance of circRNAs, making it possible to acquire the false negatives or inexact expression profiles. Secondly, most of the current researches are still focusing on the exploration of targets and preliminary mechanism verification, but still lacking further mechanism exploration, such as the impact on tumor biogenesis, tumor immunology, and metabolic immune characteristics (Wilusz, 2019). Therefore, more supplementary studies are needed to investigate the molecular mechanisms of their biological functions in BC. Thirdly, generally speaking, the endogenous circRNAs can regulate gene expression via binding to miRNAs to inhibit their function in parental cells. It is very important that the circRNA carried by exosomes is shown to transfer to recipient cells, thus playing a gene silencing effect to fine-tune target expression and affecting the environment surrounding the tumor (Shao and Lu, 2020). As exosomes are capable of protecting their cargoes from degradation with high stability in serum and blood, the pluripotent circRNAs encapsulated in exosomes are of more stable peculiarities as biomarkers in BC detection, compared with circRNAs in the cytoplasm. Therefore, exosomal circRNAs are excellent candidates for BC diagnosis and therapy.
Lastly, the circRNAs are of great value in the translation of clinical applications. Because circRNAs are novel ncRNAs, it also emphasizes the clinical significance in diagnosis and treatment. However, most of the existing studies have ascertained the inference and potential of its clinical significance, rather than directly using the target to guide clinical practice (Yin and Liu, 2020). CircRNAs have been recognized as valuable sources of biomarkers and as an auxiliary diagnosis method for the diagnosis and prognosis of BC. Therefore, identifying circRNA disorders in body fluids may be helpful for non-invasive BC diagnosis, but it should be identified and characterized through experiments in the future (Szilágyi et al., 2020). In another aspect, inhibiting overexpressed, pro-tumorigenic circRNAs or sensitizing downregulated, tumor-suppressive circRNAs will be superb strategies for BC therapy. In general, large-scale actual clinical trials in diagnosis and treatment are needed to promote clinical applications of circRNA.
In conclusion, circRNAs are crucial orchestrators in BC growth, metastasis, and drug resistance and are promising biomarkers in BC diagnosis and prognosis (Figure 3). The in-depth elucidation of the relationship between circRNAs and BC will benefit for combating BC.
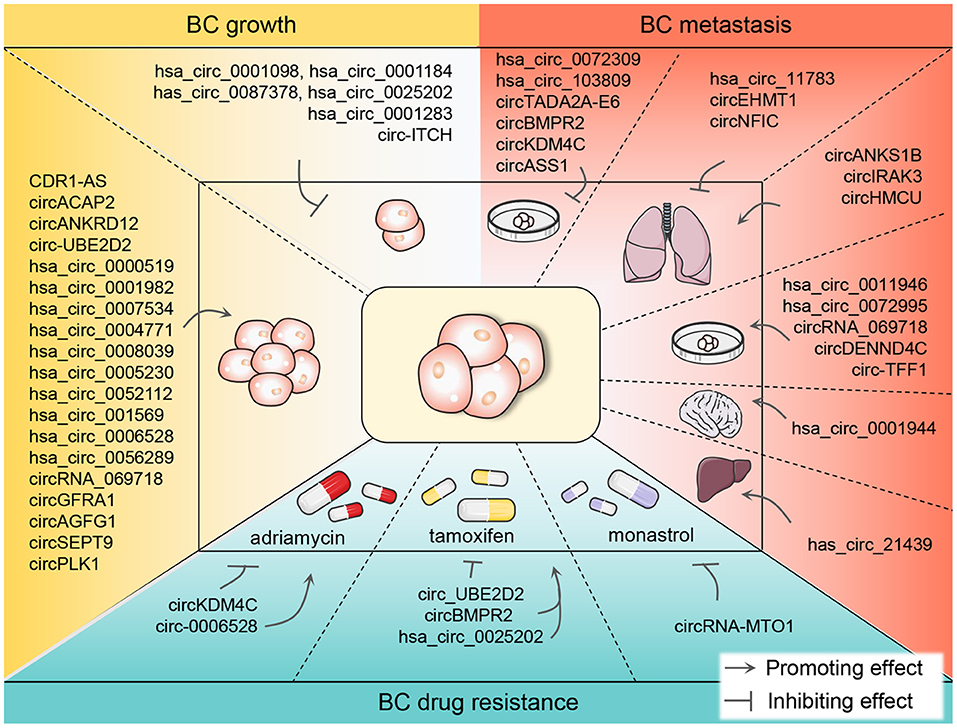
Figure 3. The promoting or inhibiting impacts of differentially expressed circRNAs on the growth, metastasis, and drug resistance of BC. CircRNAs are known to function either as oncogenic or anticancer genes in BC tumor growth and metastasis, promoting BC cell-level metastasis and even the metastasis to the lung, liver, and brain. Currently, chemotherapy plays an indispensable role in the therapy of BC. However, in the process of chemotherapy, patients are prone to drug resistance which hinders the treatment progress. ADM, TAM, and monastrol are the most reported resistant drugs associated with circRNAs in recent studies in the chemotherapy of BC patients. BC, breast cancer; circRNA, circular RNA; ADM, adriamycin; TAM, tamoxifen.
Author Contributions
XH, TX, and WH performed the literature search and wrote the manuscript. XL, YWu, and QZ conceived the project and revised the manuscript. YT, DW, YWa, CZ, YY, MX, WL, and MW edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by China GuangHua Science and Technology Foundation (grant number 2019JZXM001) and Wuhan Science and Technology Bureau (grant number 2020020601012241).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
BC, breast cancer; ncRNA, non-coding RNA; lncRNA, long non-coding RNA; circRNA, circular RNA; miRNA, microRNA; ceRNA, competing endogenous RNA; ecircRNA, exonic circRNA; ciRNA, circular intronic RNA; EIciRNA, exon-intronic circRNA; RBP, RNA-binding protein; CDK2, cyclin-dependent kinases 2; snRNA, small nuclear RNA; IRES, internal ribosome entry sites; ORF, open reading frame; ZEB2, zinc finger E-box-binding homeobox 2; TNBC, triple-negative breast cancer; LIF, leukemia inhibitory factor; BCSC, breast cancer stem cell; TCDD, tetrachlorodibenzo-p-dioxin; NF-κB, nuclear factor-κB; ER, estrogen receptor; ECM, extracellular matrix; TNM, tumor-node metastasis; EMT, epithelial-mesenchymal transition; BCBM, breast cancer brain metastases; BCLM, breast cancer liver metastases; TCGA, The Cancer Genome Atlas; qRT-PCR, quantitative real-time polymerase chain reaction; ROR-P, risk-of-relapse proliferation; PWCDA, Path-Weighed Method for Predicting CircRNA-Disease Associations; NMF, non-negative matrix factorization; GBDTCDA, Gradient Boosting Decision Tree with multiple biological data to predict circRNA-disease associations; SIMCCDA, Speedup Inductive Matrix Completion for CircRNA-Disease Associations prediction; USP4, ubiquitin-specific protease 4; ADM, adriamycin; TAM, tamoxifen.
References
Barrett, S. P., and Salzman, J. (2016). Circular RNAs: analysis, expression and potential functions. Development 143, 1838–1847. doi: 10.1242/dev.128074
Braicu, C., Zimta, A.-A., Harangus, A., Iurca, I., Irimie, A., Coza, O., et al. (2019). The function of non-coding RNAs in lung cancer tumorigenesis. Cancers (Basel). 11:605. doi: 10.3390/cancers11050605
Cai, F., Fu, W., Tang, L., Tang, J., Sun, J., Fu, G., et al. (2020). Hsa_circ_0000515 is a novel circular RNA implicated in the development of breast cancer through its regulation of the microRNA-296-5p/CXCL10 axis. FEBS J. 283, 861–883. doi: 10.1111/febs.15373
Cao, L., Wang, M., Dong, Y., Xu, B., Chen, J., Ding, Y., et al. (2020). Circular RNA circRNF20 promotes breast cancer tumorigenesis and Warburg effect through miR-487a/HIF-1α/HK2. Cell Death Dis. 11:145. doi: 10.1038/s41419-020-2336-0
Chao, C. W., Chan, D. C., Kuo, A., and Leder, P. (1998). The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol. Med. 4, 614–628. doi: 10.1007/BF03401761
Chen, B., Wei, W., Huang, X., Xie, X., Kong, Y., Dai, D., et al. (2018). circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics 8, 4003–4015. doi: 10.7150/thno.24106
Chen, C., and Sarnow, P. (1995). Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268, 415–417. doi: 10.1126/science.7536344
Chen, Y., Li, C., Tan, C., and Liu, X. (2016). Circular RNAs: a new frontier in the study of human diseases. J. Med. Genet. 53, 359–365. doi: 10.1136/jmedgenet-2016-103758
Deng, L., Zhang, W., Shi, Y., and Tang, Y. (2019). Fusion of multiple heterogeneous networks for predicting circRNA-disease associations. Sci. Rep. 9:9605. doi: 10.1038/s41598-019-45954-x
Fang, S., Pan, J., Zhou, C., Tian, H., He, J., Shen, W., et al. (2019). Circular RNAs serve as novel biomarkers and therapeutic targets in cancers. Curr. Gene Ther. 19, 125–133. doi: 10.2174/1566523218666181109142756
Fu, B., Zhang, A., Li, M., Pan, L., Tang, W., An, M., et al. (2018). Circular RNA profile of breast cancer brain metastasis: identification of potential biomarkers and therapeutic targets. Epigenomics 10, 1619–1630. doi: 10.2217/epi-2018-0090
Gao, D., Qi, X., Zhang, X., Fang, K., Guo, Z., and Li, L. (2019). hsa_circRNA_0006528 as a competing endogenous RNA promotes human breast cancer progression by sponging miR-7-5p and activating the MAPK/ERK signaling pathway. Mol. Carcinog. 58, 554–564. doi: 10.1002/mc.22950
Gao, D., Zhang, X., Liu, B., Meng, D., Fang, K., Guo, Z., et al. (2017). Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics 9, 1175–1188. doi: 10.2217/epi-2017-0055
Ge, E., Yang, Y., Gang, M., Fan, C., and Zhao, Q. (2020). Predicting human disease-associated circRNAs based on locality-constrained linear coding. Genomics 112, 1335–1342. doi: 10.1016/j.ygeno.2019.08.001
Geng, Y., Jiang, J., and Wu, C. (2018). Function and clinical significance of circRNAs in solid tumors. J. Hematol. Oncol. 11:98. doi: 10.1186/s13045-018-0643-z
Greene, J., Baird, A.-M., Brady, L., Lim, M., Gray, S. G., McDermott, R., et al. (2017). Circular RNAs: biogenesis, function and role in human diseases. Front. Mol. Biosci. 8:38. doi: 10.3389/fmolb.2017.00038
Gu, Y., Ci, C., Zhang, X., Su, M., Lv, W., Chen, C., et al. (2019). Prediction of circRNAs based on the DNA methylation-mediated feature sponge function in breast cancer. Front. Bioeng. Biotechnol. 7:365. doi: 10.3389/fbioe.2019.00365
Guo, Z., Cao, Q., Zhao, Z., and Song, C. (2020). Biogenesis, features, functions, and disease relationships of a specific circular RNA: CDR1as. Aging Dis. 11, 1009–1020. doi: 10.14336/AD.2019.0920
Han, T.-S., Hur, K., Cho, H.-S., and Ban, H. S. (2020). Epigenetic associations between lncRNA/circRNA and miRNA in hepatocellular carcinoma. Cancers (Basel). 12:2622. doi: 10.3390/cancers12092622
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi: 10.1038/nature11993
He, R., Liu, P., Xie, X., Zhou, Y., Liao, Q., Xiong, W., et al. (2017). circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J. Exp. Clin. Cancer Res. 36:145. doi: 10.1186/s13046-017-0614-1
Hou, J., Xu, Z., Zhong, S., Zhang, H., Jiang, L., Chen, X., et al. (2019). Circular RNA circASS1 is downregulated in breast cancer cells MDA-MB-231 and suppressed invasion and migration. Epigenomics 11, 199–213. doi: 10.2217/epi-2017-0167
Hu, K., Liu, X., Li, Y., Li, Q., Xu, Y., Zeng, W., et al. (2020). Exosomes mediated transfer of Circ_UBE2D2 enhances the resistance of breast cancer to tamoxifen by binding to MiR-200a-3p. Med. Sci. Monit. 26:e922253. doi: 10.12659/MSM.922253
Hu, Y., Guo, F., Zhu, H., Tan, X., Zhu, X., Liu, X., et al. (2020). Circular RNA-0001283 suppresses breast cancer proliferation and invasion via MiR-187/HIPK3 axis. Med. Sci. Monit. 26:e921502. doi: 10.12659/MSM.921502
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. doi: 10.1261/rna.035667.112
Jiang, W., Rixiati, Y., Huang, H., Shi, Y., Huang, C., and Jiao, B. (2020). Asperolide A prevents bone metastatic breast cancer via the PI3K/AKT/mTOR/c-Fos/NFATc1 signaling pathway. Cancer Med. 9, 8173–8185. doi: 10.1002/cam4.3432
Kalyana-Sundaram, S., Kumar-Sinha, C., Shankar, S., Robinson, D. R., Wu, Y.-M., Cao, X., et al. (2012). Expressed pseudogenes in the transcriptional landscape of human cancers. Cell 149, 1622–1634. doi: 10.1016/j.cell.2012.04.041
Karedath, T., Ahmed, I., Al Ameri, W., Al-Dasim, F. M., Andrews, S. S., Samuel, S., et al. (2019). Silencing of ANKRD12 circRNA induces molecular and functional changes associated with invasive phenotypes. BMC Cancer 19:565. doi: 10.1186/s12885-019-5723-0
Kong, Y., Yang, L., Wei, W., Lyu, N., Zou, Y., Gao, G., et al. (2019). circPLK1 sponges miR-296-5p to facilitate triple-negative breast cancer progression. Epigenomics 11, 1163–1176. doi: 10.2217/epi-2019-0093
Legnini, I., Di Timoteo, G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., et al. (2017). circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66, 22.e9–37.e9. doi: 10.1016/j.molcel.2017.02.017
Lei, X., and Fang, Z. (2019). GBDTCDA: predicting circRNA-disease associations based on gradient boosting decision tree with multiple biological data fusion. Int. J. Biol. Sci. 15, 2911–2924. doi: 10.7150/ijbs.33806
Lei, X., Fang, Z., Chen, L., and Wu, F.-X. (2018). PWCDA: path weighted method for predicting circRNA-disease associations. Int. J. Mol. Sci. 19:3410. doi: 10.3390/ijms19113410
Li, L.-J., Leng, R.-X., Fan, Y.-G., Pan, H.-F., and Ye, D.-Q. (2017). Translation of noncoding RNAs: focus on lncRNAs, pri-miRNAs, and circRNAs. Exp. Cell Res. 361, 1–8. doi: 10.1016/j.yexcr.2017.10.010
Li, M., Liu, M., Bin, Y., and Xia, J. (2020). Prediction of circRNA-disease associations based on inductive matrix completion. BMC Med. Genomics 13:42. doi: 10.1186/s12920-020-0679-0
Li, W., Yang, X., Shi, C., and Zhou, Z. (2020). Hsa_circ_002178 promotes the growth and migration of breast cancer cells and maintains cancer stem-like cell properties through regulating miR-1258/KDM7A axis. Cell Transplant. 29:096368972096017. doi: 10.1177/0963689720960174
Li, Z., Chen, Z., Hu, G., Zhang, Y., Feng, Y., Jiang, Y., et al. (2020). Profiling and integrated analysis of differentially expressed circRNAs as novel biomarkers for breast cancer. J. Cell. Physiol. 235, 7945–7959. doi: 10.1002/jcp.29449
Li, Z., Ruan, Y., Zhang, H., Shen, Y., Li, T., and Xiao, B. (2019). Tumor-suppressive circular RNAs: mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 110, 3630–3638. doi: 10.1111/cas.14211
Liang, G., Ling, Y., Mehrpour, M., Saw, P. E., Liu, Z., Tan, W., et al. (2020). Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol. Cancer 19:65. doi: 10.1186/s12943-020-01152-2
Liang, Y., Song, X., Li, Y., Ma, T., Su, P., Guo, R., et al. (2019a). Targeting the circBMPR2/miR-553/USP4 axis as a potent therapeutic approach for breast cancer. Mol. Ther. Nucleic Acids 17, 347–361. doi: 10.1016/j.omtn.2019.05.005
Liang, Y., Song, X., Li, Y., Su, P., Han, D., Ma, T., et al. (2019b). circKDM4C suppresses tumor progression and attenuates doxorubicin resistance by regulating miR-548p/PBLD axis in breast cancer. Oncogene 38, 6850–6866. doi: 10.1038/s41388-019-0926-z
Lin, X., Hong, S., Chen, J., Chen, W., and Wu, Z. (2019). The potential targets for metastases: a study on altered circular RNA profile in breast cancer liver metastases. Epigenomics 11, 1237–1250. doi: 10.2217/epi-2019-0099
Liu, M., Luo, C., Dong, J., Guo, J., Luo, Q., Ye, C., et al. (2020). circRNA_103809 suppresses the proliferation and metastasis of breast cancer cells by sponging microRNA-532-3p (miR-532-3p). Front. Genet. 11:485. doi: 10.3389/fgene.2020.00485
Liu, P., Zou, Y., Li, X., Yang, A., Ye, F., Zhang, J., et al. (2020). circGNB1 facilitates triple-negative breast cancer progression by regulating miR-141-5p-IGF1R Axis. Front. Genet. 11:193. doi: 10.3389/fgene.2020.00193
Liu, T., Ye, P., Ye, Y., Lu, S., and Han, B. (2020). Circular RNA hsa_circRNA_002178 silencing retards breast cancer progression via microRNA-328-3p-mediated inhibition of COL1A1. J. Cell. Mol. Med. 24, 2189–2201. doi: 10.1111/jcmm.14875
Liu, Y., Dong, Y., Zhao, L., Su, L., and Luo, J. (2018a). Circular RNA-MTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis. Int. J. Oncol. 53, 1752–1762. doi: 10.3892/ijo.2018.4485
Liu, Y., Lu, C., Zhou, Y., Zhang, Z., and Sun, L. (2018b). Circular RNA hsa_circ_0008039 promotes breast cancer cell proliferation and migration by regulating miR-432-5p/E2F3 axis. Biochem. Biophys. Res. Commun. 502, 358–363. doi: 10.1016/j.bbrc.2018.05.166
Liu, Z., Zhou, Y., Liang, G., Ling, Y., Tan, W., Tan, L., et al. (2019). Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 10:55. doi: 10.1038/s41419-018-1287-1
Lou, W., Ding, B., and Fu, P. (2020). Pseudogene-derived lncRNAs and their miRNA sponging mechanism in human cancer. Front. Cell Dev. Biol. 8:85. doi: 10.3389/fcell.2020.00085
Lu, M., Wu, Y., Zeng, B., Sun, J., Li, Y., Luo, J., et al. (2020). circEHMT1 inhibits metastatic potential of breast cancer cells by modulating miR-1233-3p/KLF4/MMP2 axis. Biochem. Biophys. Res. Commun. 526, 306–313. doi: 10.1016/j.bbrc.2020.03.084
Ma, X., Liu, C., Gao, C., Li, J., Zhuang, J., Liu, L., et al. (2020). circRNA-associated ceRNA network construction reveals the circRNAs involved in the progression and prognosis of breast cancer. J. Cell. Physiol. 235, 3973–3983. doi: 10.1002/jcp.29291
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi: 10.1038/nature11928
Nair, A. A., Niu, N., Tang, X., Thompson, K. J., Wang, L., Kocher, J.-P., et al. (2016). Circular RNAs and their associations with breast cancer subtypes. Oncotarget 7, 80967–80979. doi: 10.18632/oncotarget.13134
Nedoluzhko, A., Sharko, F., Rbbani, M. G., Teslyuk, A., Konstantinidis, I., and Fernandes, J. M. O. (2020). CircParser: a novel streamlined pipeline for circular RNA structure and host gene prediction in non-model organisms. PeerJ 8:e8757. doi: 10.7717/peerj.8757
Pan, G., Mao, A., Liu, J., Lu, J., Ding, J., and Liu, W. (2020). Circular RNA hsa_circ_0061825 (circ-TFF1) contributes to breast cancer progression through targeting miR-326/TFF1 signalling. Cell Prolif. 53, 1–14. doi: 10.1111/cpr.12720
Pan, H., Li, T., Jiang, Y., Pan, C., Ding, Y., Huang, Z., et al. (2018). Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J. Cell. Biochem. 119, 440–446. doi: 10.1002/jcb.26201
Qu, S., Yang, X., Li, X., Wang, J., Gao, Y., Shang, R., et al. (2015). Circular RNA: a new star of noncoding RNAs. Cancer Lett. 365, 141–148. doi: 10.1016/j.canlet.2015.06.003
Ren, S., Liu, J., Feng, Y., Li, Z., He, L., Li, L., et al. (2019). Knockdown of circDENND4C inhibits glycolysis, migration and invasion by up-regulating miR-200b/c in breast cancer under hypoxia. J. Exp. Clin. Cancer Res. 38:388. doi: 10.1186/s13046-019-1398-2
Ruan, Y., Li, Z., Shen, Y., Li, T., Zhang, H., and Guo, J. (2020). Functions of circular RNAs and their potential applications in gastric cancer. Expert Rev. Gastroenterol. Hepatol. 14, 85–92. doi: 10.1080/17474124.2020.1715211
Sang, Y., Chen, B., Song, X., Li, Y., Liang, Y., Han, D., et al. (2019). circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol. Ther. 27, 1638–1652. doi: 10.1016/j.ymthe.2019.05.011
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A. 73, 3852–3856. doi: 10.1073/pnas.73.11.3852
Shao, Y., and Lu, B. (2020). The crosstalk between circular RNAs and the tumor microenvironment in cancer metastasis. Cancer Cell Int. 20:448. doi: 10.1186/s12935-020-01532-0
Shi, P., Sun, J., He, B., Song, H., Li, Z., Kong, W., et al. (2018). Profiles of differentially expressed circRNAs in esophageal and breast cancer. Cancer Manag. Res. 10, 2207–2221. doi: 10.2147/CMAR.S167863
Smid, M., Wilting, S. M., Uhr, K., Rodríguez-González, F. G., de Weerd, V., Prager-Van der Smissen, W. J. C., et al. (2019). The circular RNome of primary breast cancer. Genome Res. 29, 356–366. doi: 10.1101/gr.238121.118
Song, L., and Xiao, Y. (2018). Downregulation of hsa_circ_0007534 suppresses breast cancer cell proliferation and invasion by targeting miR-593/MUC19 signal pathway. Biochem. Biophys. Res. Commun. 503, 2603–2610. doi: 10.1016/j.bbrc.2018.08.007
Song, X., Liang, Y., Sang, Y., Li, Y., Zhang, H., Chen, B., et al. (2020). circHMCU promotes proliferation and metastasis of breast cancer by sponging the let-7 family. Mol. Ther. Nucleic Acids 20, 518–533. doi: 10.1016/j.omtn.2020.03.014
Sun, Z., Chen, C., Su, Y., Wang, W., Yang, S., Zhou, Q., et al. (2019). Regulatory mechanisms and clinical perspectives of circRNA in digestive system neoplasms. J. Cancer 10, 2885–2891. doi: 10.7150/jca.31167
Szilágyi, M., Pös, O., Márton, É., Buglyó, G., Soltész, B., Keseru, J., et al. (2020). Circulating cell-free nucleic acids: main characteristics and clinical application. Int. J. Mol. Sci. 21:6827. doi: 10.3390/ijms21186827
Tang, H., Huang, X., Wang, J., Yang, L., Kong, Y., Gao, G., et al. (2019). circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol. Cancer 18:23. doi: 10.1186/s12943-019-0946-x
Tang, Y.-Y., Zhao, P., Zou, T.-N., Duan, J.-J., Zhi, R., Yang, S.-Y., et al. (2017). Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 36, 901–908. doi: 10.1089/dna.2017.3862
Tarrero, L. C., Ferrero, G., Miano, V., De Intinis, C., Ricci, L., Arigoni, M., et al. (2018). Luminal breast cancer-specific circular RNAs uncovered by a novel tool for data analysis. Oncotarget 9, 14580–14596. doi: 10.18632/oncotarget.24522
Tomar, D., Yadav, A. S., Kumar, D., Bhadauriya, G., and Kundu, G. C. (2020). Non-coding RNAs as potential therapeutic targets in breast cancer. Biochim. Biophys. Acta Gene Regul. Mech. 1863:194378. doi: 10.1016/j.bbagrm.2019.04.005
Tran, A. M., Chalbatani, G. M., Berland, L., Cruz De los Santos, M., Raj, P., Jalali, S. A., et al. (2020). A new world of biomarkers and therapeutics for female reproductive system and breast cancers: circular RNAs. Front. Cell Dev. Biol. 8:50. doi: 10.3389/fcell.2020.00050
Trimboli, R. M., Giorgi Rossi, P., Battisti, N. M. L., Cozzi, A., Magni, V., Zanardo, M., et al. (2020). Do we still need breast cancer screening in the era of targeted therapies and precision medicine? Insights Imaging 11:105. doi: 10.1186/s13244-020-00905-3
Uhr, K., Sieuwerts, A. M., de Weerd, V., Smid, M., Hammerl, D., Foekens, J. A., et al. (2018). Association of microRNA-7 and its binding partner CDR1-AS with the prognosis and prediction of 1st-line tamoxifen therapy in breast cancer. Sci. Rep. 8:9657. doi: 10.1038/s41598-018-27987-w
Venø, M. T., Hansen, T. B., Venø, S. T., Clausen, B. H., Grebing, M., Finsen, B., et al. (2015). Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 16:245. doi: 10.1186/s13059-015-0801-3
Wang, H., Xiao, Y., Wu, L., and Ma, D. (2018). Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int. J. Oncol. 52, 743–754. doi: 10.3892/ijo.2018.4265
Wang, J., Zhang, Q., Zhou, S., Xu, H., Wang, D., Feng, J., et al. (2019). Circular RNA expression in exosomes derived from breast cancer cells and patients. Epigenomics 11, 411–421. doi: 10.2217/epi-2018-0111
Wang, L., You, Z.-H., Li, Y.-M., Zheng, K., and Huang, Y.-A. (2020). GCNCDA: a new method for predicting circRNA-disease associations based on Graph Convolutional Network Algorithm. PLOS Comput. Biol. 16:e1007568. doi: 10.1371/journal.pcbi.1007568
Wang, Q., Li, Z., Hu, Y., Zheng, W., Tang, W., Zhai, C., et al. (2019). circ-TFCP2L1 promotes the proliferation and migration of triple negative breast cancer through sponging miR-7 by inhibiting PAK1. J. Mammary Gland Biol. Neoplasia 24, 323–331. doi: 10.1007/s10911-019-09440-4
Wang, S., Li, Q., Wang, Y., Li, X., Wang, R., Kang, Y., et al. (2018). Upregulation of circ-UBAP2 predicts poor prognosis and promotes triple-negative breast cancer progression through the miR-661/MTA1 pathway. Biochem. Biophys. Res. Commun. 505, 996–1002. doi: 10.1016/j.bbrc.2018.10.026
Wang, S., Xia, P., Zhang, L., Yu, L., Liu, H., Meng, Q., et al. (2019). Systematical identification of breast cancer-related circular RNA modules for deciphering circRNA functions based on the non-negative matrix factorization algorithm. Int. J. Mol. Sci. 20:919. doi: 10.3390/ijms20040919
Wang, S. T., Liu, L. B., Li, X. M., Wang, Y. F., Xie, P. J., Li, Q., et al. (2019). circ-ITCH regulates triple-negative breast cancer progression through the Wnt/β-catenin pathway. Neoplasma 66, 232–239. doi: 10.4149/neo_2018_180710N460
Wang, Y., Li, J., Du, C., Zhang, L., Zhang, Y., Zhang, J., et al. (2019). Upregulated circular RNA circ-UBE2D2 predicts poor prognosis and promotes breast cancer progression by sponging miR-1236 and miR-1287. Transl. Oncol. 12, 1305–1313. doi: 10.1016/j.tranon.2019.05.016
Wang, Y., Mo, Y., Gong, Z., Yang, X., Yang, M., Zhang, S., et al. (2017). Circular RNAs in human cancer. Mol. Cancer 16:25. doi: 10.1186/s12943-017-0598-7
Wilusz, J. E. (2019). Circle the wagons: circular RNAs control innate immunity. Cell 177, 797–799. doi: 10.1016/j.cell.2019.04.020
Wu, J., Cheng, J., Zhang, F., Luo, X., Zhang, Z., and Chen, S. (2020). Estrogen receptor α is involved in the regulation of ITGA8 methylation in estrogen receptor-positive breast cancer. Ann. Transl. Med. 8, 993–993. doi: 10.21037/atm-20-5220
Wu, J., Jiang, Z., Chen, C., Hu, Q., Fu, Z., Chen, J., et al. (2018). circIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. 430, 179–192. doi: 10.1016/j.canlet.2018.05.033
Xiao, W., Zheng, S., Zou, Y., Yang, A., Xie, X., Tang, H., et al. (2019). circAHNAK1 inhibits proliferation and metastasis of triple-negative breast cancer by modulating miR-421 and RASA1. Aging (Albany. NY). 11, 12043–12056. doi: 10.18632/aging.102539
Xie, R., Tang, J., Zhu, X., and Jiang, H. (2019). Silencing of hsa_circ_0004771 inhibits proliferation and induces apoptosis in breast cancer through activation of miR-653 by targeting ZEB2 signaling pathway. Biosci. Rep. 39:BSR20181919. doi: 10.1042/BSR20181919
Xie, R., Zhang, Y., Zhang, J., Li, J., and Zhou, X. (2020). The role of circular RNAs in immune-related diseases. Front. Immunol. 11:545. doi: 10.3389/fimmu.2020.00545
Xing, L., Yang, R., Wang, X., Zheng, X., Yang, X., Zhang, L., et al. (2020). The circRNA circIFI30 promotes progression of triple-negative breast cancer and correlates with prognosis. Aging (Albany. NY). 12, 10983–11003. doi: 10.18632/aging.103311
Xu, G., Ye, D., Zhao, Q., He, R., Ma, W., Li, Y., et al. (2020). circNFIC suppresses breast cancer progression by sponging miR-658. J. Cancer 11, 4222–4229. doi: 10.7150/jca.38830
Xu, J., Wang, Y., and Xu, D. (2019). Hsa_circ_001569 is an unfavorable prognostic factor and promotes cell proliferation and metastasis by modulating PI3K-AKT pathway in breast cancer. Cancer Biomarkers 25, 193–201. doi: 10.3233/CBM-182293
Xu, J., Wu, K., Jia, Q., and Ding, X. (2020). Roles of miRNA and IncRNA in triple-negative breast cancer. J. Zhejiang Univ. B 21, 673–689. doi: 10.1631/jzus.B1900709
Xu, J.-Z., Shao, C.-C., Wang, X.-J., Zhao, X., Chen, J.-Q., Ouyang, Y.-X., et al. (2019). circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 10:175. doi: 10.1038/s41419-019-1382-y
Xu, Y., Yao, Y., Leng, K., Ji, D., Qu, L., Liu, Y., et al. (2018). Increased expression of circular RNA circ_0005230 indicates dismal prognosis in breast cancer and regulates cell proliferation and invasion via miR-618/CBX8 signal pathway. Cell. Physiol. Biochem. 51, 1710–1722. doi: 10.1159/000495675
Yan, L., Zheng, M., and Wang, H. (2019). Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Manag. Res. 11, 1033–1041. doi: 10.2147/CMAR.S186857
Yan, N., Xu, H., Zhang, J., Xu, L., Zhang, Y., Zhang, L., et al. (2017). Circular RNA profile indicates circular RNA VRK1 is negatively related with breast cancer stem cells. Oncotarget 8, 95704–95718. doi: 10.18632/oncotarget.21183
Yang, L., Song, C., Chen, Y., Jing, G., and Sun, J. (2019). Circular RNA circ_0103552 forecasts dismal prognosis and promotes breast cancer cell proliferation and invasion by sponging miR-1236. J. Cell. Biochem. 120, 15553–15560. doi: 10.1002/jcb.28822
Yang, R., Xing, L., Zheng, X., Sun, Y., Wang, X., and Chen, J. (2019). The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol. Cancer 18:4. doi: 10.1186/s12943-018-0933-7
Yang, S., Wang, D., Zhou, S., Zhang, Q., Wang, J., Zhong, S., et al. (2020). Identification of circRNA–miRNA networks for exploring an underlying prognosis strategy for breast cancer. Epigenomics 12, 101–125. doi: 10.2217/epi-2019-0058
Yin, K., and Liu, X. (2020). circMMP1 promotes the progression of glioma through miR-433/HMGB3 axis in vitro and in vivo. IUBMB Life 72, 2508–2524. doi: 10.1002/iub.2383
Yuan, C., Zhou, L., Zhang, L., Yin, K., Peng, J., Sha, R., et al. (2019). Identification and integrated analysis of key differentially expressed circular RNAs in ER-positive subtype breast cancer. Epigenomics 11, 297–321. doi: 10.2217/epi-2018-0147
Zeng, K., He, B., Yang, B. B., Xu, T., Chen, X., Xu, M., et al. (2018). The pro-metastasis effect of circANKS1B in breast cancer. Mol. Cancer 17:160. doi: 10.1186/s12943-018-0914-x
Zhang, C., Wu, H., Wang, Y., Zhao, Y., Fang, X., Chen, C., et al. (2015). Expression patterns of circular RNAs from primary kinase transcripts in the mammary glands of lactating rats. J. Breast Cancer 18:235. doi: 10.4048/jbc.2015.18.3.235
Zhang, H., Jiang, L., Hou, J., Zhong, S., Zhou, S., Zhu, L., et al. (2018a). Circular RNA hsa_circ_0052112 promotes cell migration and invasion by acting as sponge for miR-125a-5p in breast cancer. Biomed. Pharmacother. 107, 1342–1353. doi: 10.1016/j.biopha.2018.08.030
Zhang, H.-D., Jiang, L.-H., Hou, J.-C., Zhou, S.-Y., Zhong, S.-L., Zhu, L.-P., et al. (2018b). Circular RNA hsa_circ_0072995 promotes breast cancer cell migration and invasion through sponge for miR-30c-2-3p. Epigenomics 10, 1229–1242. doi: 10.2217/epi-2018-0002
Zhang, J., Xu, H. D., Xing, X. J., Liang, Z. T., Xia, Z. H., and Zhao, Y. (2019). circRNA-069718 promotes cell proliferation and invasion in triple-negative breast cancer by activating Wnt/β-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 5315–5322. doi: 10.26355/eurrev_201906_18198
Zhang, M., Huang, N., Yang, X., Luo, J., Yan, S., Xiao, F., et al. (2018). A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37, 1805–1814. doi: 10.1038/s41388-017-0019-9
Zhang, Y., Zhang, X.-O., Chen, T., Xiang, J.-F., Yin, Q.-F., Xing, Y.-H., et al. (2013). Circular intronic long noncoding RNAs. Mol. Cell 51, 792–806. doi: 10.1016/j.molcel.2013.08.017
Zhao, B., Song, X., and Guan, H. (2020). circACAP2 promotes breast cancer proliferation and metastasis by targeting miR-29a/b-3p-COL5A1 axis. Life Sci. 244, 117179. doi: 10.1016/j.lfs.2019.117179
Zhao, C.-H., Qu, L., Zhang, H., and Qu, R. (2019). Identification of breast cancer-related circRNAs by analysis of microarray and RNA-sequencing data. Medicine (Baltimore). 98:e18042. doi: 10.1097/MD.0000000000018042
Zhao, J., Zou, H., Han, C., Ma, J., Zhao, J., and Tang, J. (2018). Circlular RNA BARD1 (Hsa_circ_0001098) overexpression in breast cancer cells with TCDD treatment could promote cell apoptosis via miR-3942/BARD1 axis. Cell Cycle 17, 2731–2744. doi: 10.1080/15384101.2018.1556058
Zheng, X., Huang, M., Xing, L., Yang, R., Wang, X., Jiang, R., et al. (2020). The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol. Cancer 19:73. doi: 10.1186/s12943-020-01183-9
Zhou, J., Zhang, W.-W., Peng, F., Sun, J.-Y., He, Z.-Y., and Wu, S.-G. (2018). Downregulation of hsa_circ_0011946 suppresses the migration and invasion of the breast cancer cell line MCF-7 by targeting RFC3. Cancer Manag. Res. 10, 535–544. doi: 10.2147/CMAR.S155923
Keywords: breast cancer, circular RNA, microRNA sponge, metastasis, diagnosis, drug resistance
Citation: He X, Xu T, Hu W, Tan Y, Wang D, Wang Y, Zhao C, Yi Y, Xiong M, Lv W, Wu M, Li X, Wu Y and Zhang Q (2021) Circular RNAs: Their Role in the Pathogenesis and Orchestration of Breast Cancer. Front. Cell Dev. Biol. 9:647736. doi: 10.3389/fcell.2021.647736
Received: 30 December 2020; Accepted: 11 February 2021;
Published: 11 March 2021.
Edited by:
Giampaolo Morciano, University of Ferrara, ItalyReviewed by:
Lidia Avalle, University of Turin, ItalyAlessandra Cataldo, Istituto Nazionale dei Tumori (IRCCS), Italy
Copyright © 2021 He, Xu, Hu, Tan, Wang, Wang, Zhao, Yi, Xiong, Lv, Wu, Li, Wu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingrui Li, bGl4aW5ncnVpQHRqaC50am11LmVkdS5jbg==; Yiping Wu, dG9uZ2ppcGxhc3RpY0AxNjMuY29t; Qi Zhang, emhhbmdxaTA2MTcyQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Xiao He
Xiao He Tao Xu
Tao Xu Weijie Hu
Weijie Hu Yufang Tan
Yufang Tan Dawei Wang
Dawei Wang Yichen Wang
Yichen Wang Chongru Zhao
Chongru Zhao Yi Yi
Yi Yi Mingchen Xiong
Mingchen Xiong Wenchang Lv
Wenchang Lv Min Wu
Min Wu Xingrui Li
Xingrui Li Yiping Wu
Yiping Wu Qi Zhang
Qi Zhang