
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 21 July 2021
Sec. Developmental Epigenetics
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.642931
This article is part of the Research Topic Impact on Embryonic Development of Chromatin Remodeling Alterations View all 9 articles
 Feng Zhang1,2*†
Feng Zhang1,2*† Xiong Zhao3†
Xiong Zhao3† Runmin Jiang4†
Runmin Jiang4† Yuying Wang1
Yuying Wang1 Xinli Wang3
Xinli Wang3 Yu Gu1
Yu Gu1 Longyong Xu5
Longyong Xu5 Jing Ye1
Jing Ye1 Charlie Degui Chen5
Charlie Degui Chen5 Shuangping Guo1
Shuangping Guo1 Dawei Zhang3*
Dawei Zhang3* Daqing Zhao6*
Daqing Zhao6*Body axial patterning develops via a rostral-to-caudal sequence and relies on the temporal colinear activation of Hox genes. However, the underlying mechanism of Hox gene temporal colinear activation remains largely elusive. Here, with small-molecule inhibitors and conditional gene knockout mice, we identified Jmjd3, a subunit of TrxG, as an essential regulator of temporal colinear activation of Hox genes with its H3K27me3 demethylase activity. We demonstrated that Jmjd3 not only initiates but also maintains the temporal collinear expression of Hox genes. However, we detected no antagonistic roles between Jmjd3 and Ezh2, a core subunit of PcG repressive complex 2, during the processes of axial skeletal patterning. Our findings provide new insights into the regulation of Hox gene temporal collinear activation for body axial patterning in mice.
The development of the mammalian body axis is a continuous process via a rostral-to-caudal sequence (Deschamps and Duboule, 2017; Mongera et al., 2019). The morphological characteristics of each part of the body axis are mainly determined by a “Hox code” (Kessel and Gruss, 1991; Alexander et al., 2009; Wellik, 2009). Hox genes were first discovered in Drosophila (Lewis, 1978). Mammals have 39 Hox genes, which are divided into four clusters. Each cluster locates on different chromosomes and has 9–11 Hox genes. According to the sequence homologies and their position within the cluster, the Hox genes are divided into 1–13 paralog groups (PGs) (Casaca et al., 2014). The function of Hox genes in the same PG is similar and can be mutually replaced (Wellik, 2009). The expression of Hox genes in mammals is characterized by spatial collinearity, which means that the sequence of the Hox gene cluster from 3′ to 5′ on chromosomes corresponds to the expression domain of Hox genes via a rostral-to-caudal sequence on the body axis (Deschamps and van Nes, 2005; Montavon and Soshnikova, 2014; Deschamps and Duboule, 2017). Thus, the inactivation of 3′ Hox genes leads to the altered identities in the rostral side of the body axis, while the loss of 5′ Hox genes causes the morphological change of caudal segments (Deschamps and van Nes, 2005; Montavon and Soshnikova, 2014; Deschamps and Duboule, 2017). Interestingly, establishment of spatial collinear Hox gene expression relies on the temporal collinear activation of Hox genes in mice (Deschamps and van Nes, 2005; Montavon and Soshnikova, 2014; Deschamps and Duboule, 2017). In other words, Hox genes become activated in a time order corresponding to their location within the clusters (Montavon and Soshnikova, 2014; Deschamps and Duboule, 2017; Krumlauf, 2018). At E7.2, the 3′ Hox genes are firstly expressed in the posterior primitive streak, and over time, the expression of Hox genes gradually transited to the more 5′ Hox13 PGs at E9.5 (Deschamps and van Nes, 2005; Soshnikova and Duboule, 2009; Montavon and Soshnikova, 2014; Deschamps and Duboule, 2017). Synchronization with the initiation order of Hox gene activation and Hox gene expression domains start to spread rostrally from the primitive streak, then the expression of 3′ Hox genes spreads to the more rostral side, while the 5′ Hox genes were expressed in the more caudal segments (Deschamps and van Nes, 2005; Montavon and Soshnikova, 2014; Deschamps and Duboule, 2017). Temporal collinear abnormalities of Hox genes will lead to corresponding morphological alterations of the Hox gene that spatially expresses the domain via a rostral-to-caudal sequence on the body axis. For example, deletion of the Hoxc8 enhancer causes a transient delay in the initial transcription of the Hoxc8 gene and results in homeotic transformations, which phenocopies the axial defects of the Hoxc8-null mutant in mice (Juan and Ruddle, 2003). On the contrary, a precocious expression of Hox13 genes causes premature arrest of posterior axial growth (Young et al., 2009). Therefore, the accurately spatiotemporal expression of Hox genes is essential for correct body axis patterning.
The initiation of body axis patterning is proposed by signal molecules. For example, embryos of Wnt3 inactivation do not express any Hox genes (Liu et al., 1999). Pre-gastrulation embryos exposed to the Wnt agonist precociously express Hoxa1 and Hoxb1 (Neijts et al., 2016). Homozygous Wnt-3a mutant mice exhibit homeotic transformations in the vertebrae along their entire body axis (Ikeya and Takada, 2001). These results suggest that Wnt3 is required for the Hox gene activation and body axis patterning. After initiation of expression, the transcriptional pattern of Hox genes is maintained by the antagonistic complex Polycomb (PcG) and Trithorax (TrxG) (Geisler and Paro, 2015; Piunti and Shilatifard, 2016). PcG generally maintain Hox gene repression, while TrxG counteract PcG and maintain the active expression state of Hox genes (Mallo and Alonso, 2013; Montavon and Soshnikova, 2014). PcG genes were originally identified in Drosophila. Mutations in PcG genes cause ectopic Hox gene expression, resulting in posterior homeotic transformations (Simon et al., 1992; Montavon and Soshnikova, 2014). TrxG genes were discovered to suppress the homeotic phenotypes displayed by PcG mutants (Kennison and Tamkun, 1988). Biochemical studies have revealed that PcG and TrxG form multiprotein complexes containing both histone methyltransferase and demethylases activities, respectively. For example, Ezh2 (Kmt6b) is a core subunit of PcG repressive complex 2 (PRC2) and inhibits Hox gene transcription by establishment of repressed histone modification H3K27 trimethylation (H3K27me3) with its histone methyltransferase activity (Cao et al., 2002; Czermin et al., 2002; Kuzmichev et al., 2002; Muller et al., 2002). H3K27me3 can be removed by histone demethylases Utx (Kdm6a) and Jmjd3 (Kdm6b) (Agger et al., 2007; De Santa et al., 2007; Hong et al., 2007; Lan et al., 2007), which belong to the TrxG group for the biochemical association with Mll2 or RbBP5 (De Santa et al., 2007; Lee et al., 2007). Interestingly, a ChIP-seq study found that the temporal collinear activation of Hox genes was accompanied with a sequential elimination of repressive marker H3K27me3 on the Hox genes (Soshnikova and Duboule, 2009; Noordermeer et al., 2014). Using high-resolution chromatin conformation capture methodology, Noordermeer et al. detected newly activated Hox genes progressively clustering into a transcriptionally active compartment from a transcriptionally inactive Hox gene cluster after transcription starts; thus, the Hox gene clusters switch to a bimodal 3D organization. This transition in spatial configurations coincides with the dynamical erase of H3K27me3 on the Hox gene clusters (Noordermeer et al., 2011, 2014). However, to date a rare epigenetic regulator for temporal collinear activation of Hox genes has been identified. Moreover, whether PcG and TrxG complexes antagonistically adjust the temporary collinear activation of Hox genes is not strictly examined in mammals.
Due to obvious morphological differences of each part, axial skeleton is an ideal model to study the relation between Hox gene expression and morphogenesis of the body axis (Wellik, 2009). Here, with the axial bone as a model, combined with conditional genetic deletion and biochemical techniques, we identified Jmjd3 as an essential epigenetic regulator of Hox gene temporal colinear activation for mouse body axial patterning. Furthermore, we detected no dynamical interplay between Jmjd3 and Ezh2 during the process of axial skeletal patterning in mice.
The temporal collinear activation of mouse Hox genes occurs at the embryonic stage of E7.2–E9.5 days (Deschamps and van Nes, 2005; Soshnikova and Duboule, 2009; Deschamps and Duboule, 2017). To identify the epigenetic regulators of Hox gene temporal collinear activation, we profiled the expression of 17 histone methyltransferases and 28 histone demethylases in E7.5, E8.5, and E9.5 embryonic tissues. Among these tissues, E7.5 tissue was from the whole mouse embryos, while E8.5 and E9.5 tissues were from the trunk of embryos, as previously described (Soshnikova and Duboule, 2009). Compared with E7.5 embryos, real-time reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) revealed that, except for a few silenced genes, most histone methyltransferases were significantly activated in E9.5 embryos instead of either E7.5 or E8.5 embryonic tissues (Supplementary Figure 1A). Because most Hox genes had been activated at E9.5 days (Deschamps and van Nes, 2005; Soshnikova and Duboule, 2009; Deschamps and Duboule, 2017), these results indicated that most histone methyltransferases might not be essential for the temporal collinear activation of Hox genes. Interestingly, although half of the 28 histone demethylases genes were silenced, Kdm6b (Jmjd3) was significantly successively activated during the E7.5–E9.5 stages (Supplementary Figure 1B). Jmjd3 is a potent H3K27me3 demethylase (Agger et al., 2007; De Santa et al., 2007; Lan et al., 2007). A previous report showed that the temporal collinear activation of Hox genes was accompanied with the progressive elimination of H3K27me3 on Hox genes (Soshnikova and Duboule, 2009). Therefore, these results suggest that Jmjd3 is a potential candidate for the regulation of Hox gene temporal collinear activation.
Both Jmjd3 and Utx are potent H3K27me3 demethylases. Previously, Utx was demonstrated to be required for the body axis patterning of zebrafish and Drosophila (Lan et al., 2007; Copur and Muller, 2013, 2018). To test the roles of Utx and Jmjd3 during body axis patterning in mice, we conditionally delete Utx or Jmjd3 in uncommitted mesenchymal cells of mice, respectively. Skeleton assays revealed, in addition to the fusions of C1 and C2 and sternal asymmetries, that no obvious homeotic transformations in the vertebrae along the entire body axis in Utxfl/Y;Prx1-Cre (0/25) or Utxfl/fl;Prx1-Cre (0/22) mice (Supplementary Figure 2 and Table 1) was detected. This indicated Utx is not required for mouse axial skeletal patterning. However, the phenotypes of Jmjd3fl/fl;Prx1-Cre mice were very similar to those of Jmjd3fl/fl;EIIa-Cre mice as previously described for E18.5 (Zhang et al., 2015). Both embryos have dwarfism with thoracic kyphosis (Supplementary Figure 2; Zhang et al., 2015). The complete fusions of C1 and C2 (14/14) were detected in Jmjd3fl/fl;Prx1-Cre mice. Homeotic transformations in the vertebrae along the whole-body axis in Jmjd3fl/fl;Prx1-Cre mice were visibly detected, including anterior transformation of C2 to C1 (an extra ventral tubercle shared by fused C1 and C2, 14/14), T1 to C7 (T1 without ribs or with incomplete ribs, 7/14), T3 to T2 (longest spinous process on T3 instead of T2,3/14), L1 to T13 (L1 with a pairs of complete or incomplete ribs, 14/14), and S1 to T6 (S1 without bilateral or unilateral iliac bones, 10/14) (Supplementary Figure 2 and Table 1). In addition, some anterior transformations, such as L1 to T13 (7/10), were also detected in Jmjd3fl/+;Prx1-Cre embryos, indicating the dose effect of Jmjd3 on axial skeletal patterning (Supplementary Figure 2 and Table 1). Consistently, RNA-seq assays indicated that Jmjd3 loss in uncommitted mesenchymal cells mildly decreased the expression of Hox3–5 PGs but markedly reduced the mRNA level of Hox6–10 PGs compared to WT in E8.5 littermates (Supplementary Figures 3, 4 and Supplementary Table 2). Therefore, these results indicated that Jmjd3 rather than Utx is required for mouse body axis patterning.
Jmjd3 deletion in uncommitted mesenchymal cells leads to the homeotic transformation in the vertebrae along the whole-body axis (Supplementary Figure 2 and Table 1). To test whether Jmjd3 temporally regulates the axial skeletal patterning with its H3K27me3 demethylase activity, we tested the role of GSK-J4, an inhibitor of Jmjd3 (Ntziachristos et al., 2014), on body axis patterning in vivo. One dose of GSK-J4 (50 mg/kg) treatment at E8.5 induced only anterior transformation of T8 to T7 (26/26) at the middle part of the trunk but not at the cervical, lumbar, and sacral segments of E18.5 mice (Figures 1A,B and Table 2). GSK-J4 treatment at E9.5 produced homeotic transformations of L1 to T13 (15/15) and S1 to T6 (14/15) in the caudal half of the vertebral column of E18.5 mice (Figures 1A,B and Table 2). GSK-J4 treatment at E10.5 produced no obvious homeotic transformations in E18.5 mouse embryos (Figures 1A,B and Table 2). These were consistent with previous reports that Hox genes of the middle part or 5′ end of the Hox gene cluster were activated at E8.5 or E9.5, respectively (Deschamps et al., 1999; Kmita and Duboule, 2003; Deschamps and van Nes, 2005; Soshnikova and Duboule, 2009). GSK-J4 treatment at E8.5 or E9.5 induced anterior transformation at the corresponding part of axial skeletal patterning, strongly supporting that Jmjd3 temporally regulates the axial patterning of mice with its H3K27me3 demethylase activity.
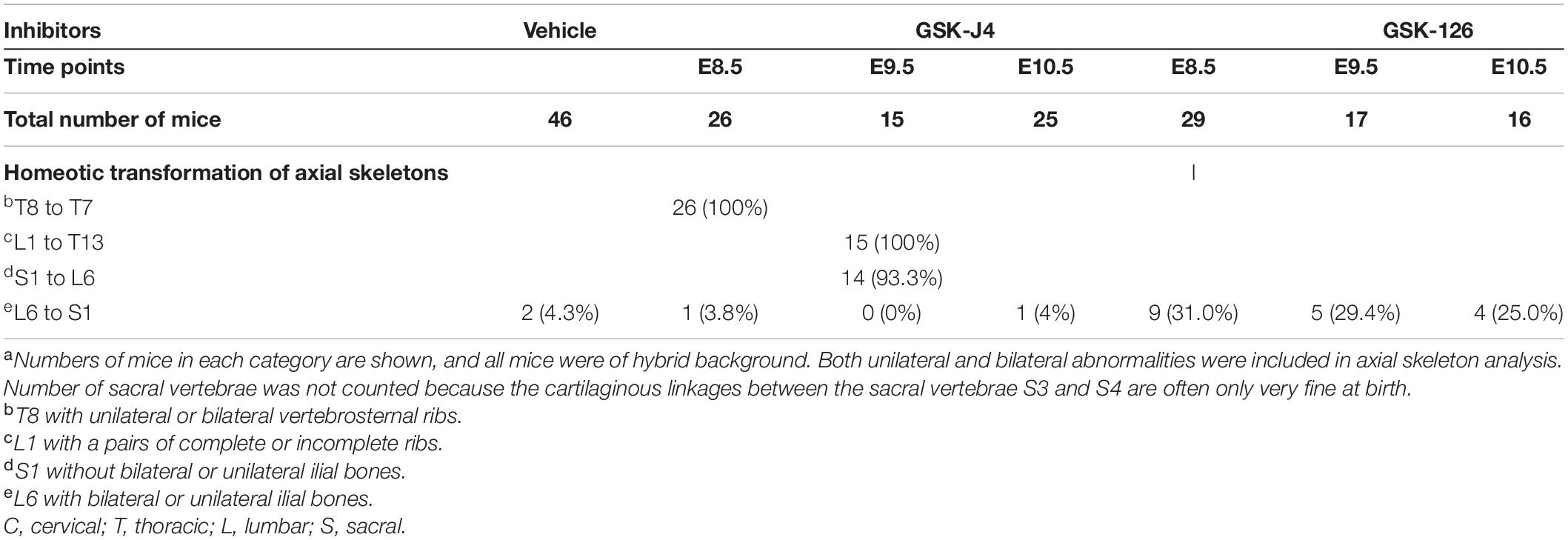
Table 2. Skeletal analysis of E18.5 mouse embryos after treatment by GSK-J4 (50 mg/kg) or GSK-126 (100 mg/kg) at the indicated time pointsa.
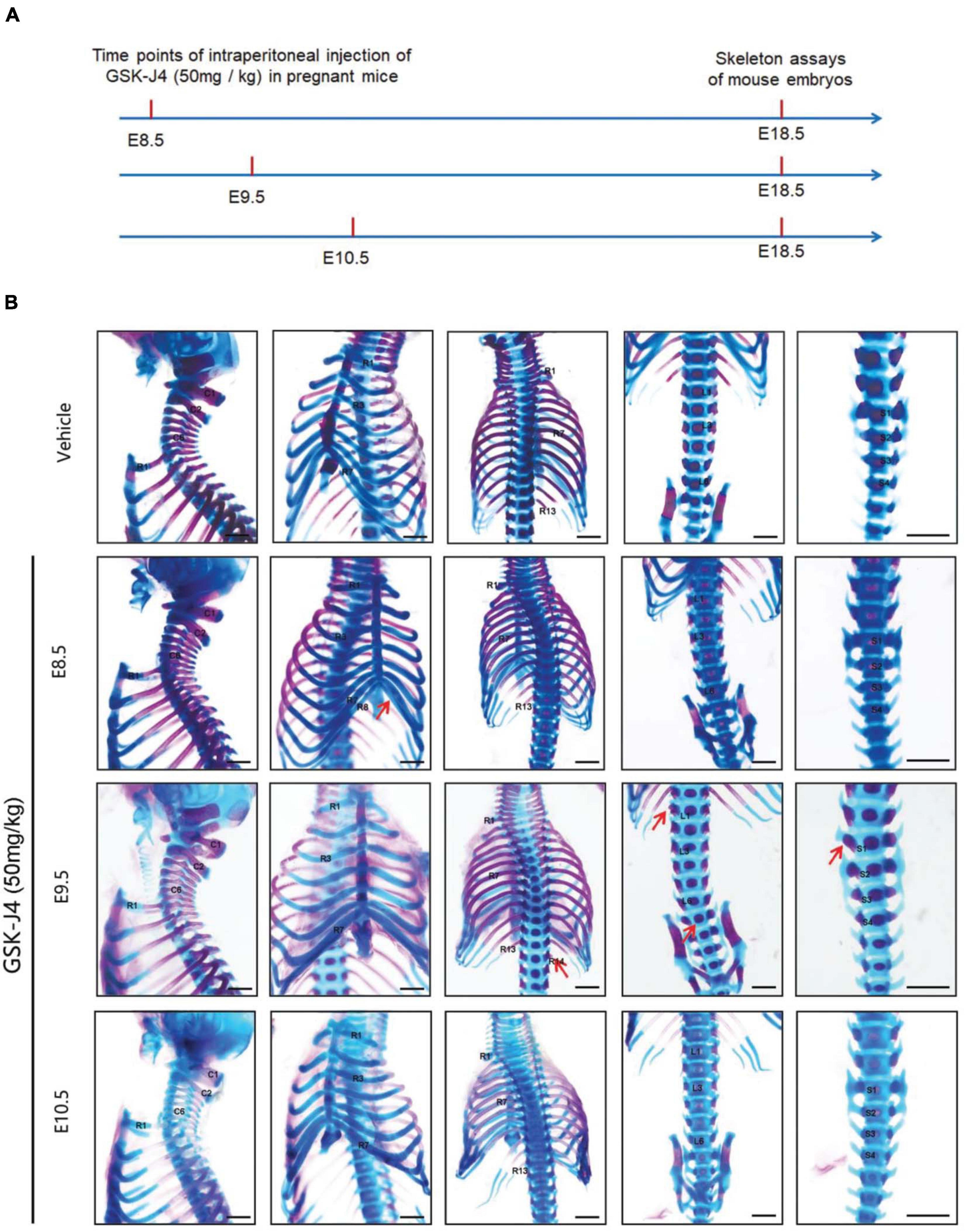
Figure 1. Homeotic transformations by GSK-J4 treatment at the time points of E8.5, E9.5, or E10.5. (A) Experimental design for studying the roles of GSK-J4 on body axis patterning. (B) Embryos had been exposed to GSK-J4 at the time points of E8.5, E9.5, or E10.5 as indicated. Skeletal phenotypes were prepared by removing forelimbs and hindlimbs. The red arrow indicates homeotic transformations of axial skeleton induced by GSK-J4. Frequencies of homeotic transformations are given in Table 1. The cervical (C), rib (R), lumbar (L), and sacral (S) vertebrae are numbered. For schematic explanation, see Supplementary Figure 9. Scale bar, 1 mm.
In murine embryos, Hox genes at the 3′ end of Hox clusters are first activated at E7.2 (Deschamps and Wijgerde, 1993; Forlani et al., 2003) and the last Hox genes at the 5′ end of Hox clusters complete the transcription at around E9.5 (Deschamps et al., 1999; Kmita and Duboule, 2003; Deschamps and van Nes, 2005; Soshnikova and Duboule, 2009). Subsequently, the transcriptional states of Hox genes at each segment are memorized along the rostral-to-caudal axis and ultimately determine body axial patterning (Kmita and Duboule, 2003; Deschamps and van Nes, 2005). A previous investigation indicated that the temporal collinear activation of Hox genes was accompanied with a sequential elimination of H3K27me3 on the Hox genes during the process of body axis patterning (Soshnikova and Duboule, 2009; Noordermeer et al., 2014). To test whether Jmjd3 regulates the Hox gene temporal collinear activation with its H3K27me3 demethylase activity, we firstly examined the mRNA level of Hox genes after GSK-J4 treatment at different time points. Consistent with the anterior transformation of T8 to T7 in the middle part instead of the cervical, lumbar, and sacral segments of mouse embryos induced by GSK-J4 at E8.5 (Figures 1A,B and Table 2), GSK-J4 treatment at E8.0 significantly reduced the mRNA level of Hox8–9 PGs, which are responsible for the establishment of the T7 and T8 patterns (Favier and Dolle, 1997; Wellik, 2009), but did not affect the mRNA levels of other Hox PGs, such as Hox1–6 PGs in E8.5 mouse embryos (Figure 2A and Supplementary Figure 5A). Correspondingly, GSK-J4 treatment at E9.0 significantly reduced the mRNA level of Hox11–13 PGs (Supplementary Figures 5A, 6A) in E9.5 mouse embryos, which was consistent with the GSK-J4 treatment at E9.5 which produced anterior transformations of the vertebrae only at the caudal part of the vertebral column, including L1 to T13 and S1 to T6 (Figures 1A,B and Table 2). To investigate the reason for the mRNA level reduction on Hox8–9 PGs in E8.5 embryos or on Hox11–13 PGs in E9.5 embryos by GSK-J4 treatment, we examined the H3K27me3 and Jmjd3 levels on the Hox genes by ChIP-qPCR assays. GSK-J4 treatment at E8.0 or E9.0 significantly increased the level of H3K27me3 on the gene body of Hox8–9 PGs in E8.5 embryos (Figure 2B and Supplementary Figures 5A, 13) or Hox10–13 PGs in E9.5 embryos, respectively (Supplementary Figures 5A, 6B, 13). Therefore, the mRNA level reduction on Hox8–9 PGs in E8.5 embryos or on Hox11–13 PGs in E9.5 embryos by GSK-J4 treatment was associated with the increased level of H3K27me3 on corresponding Hox genes. That is, Jmjd3 regulated Hox gene temporal collinear activation with its H3K27me3 demethylase activity. In addition, at E8.5 embryos, the signal of Jmjd3 was detected on Hox1–9 PGs but not Hox10–13 PGs by ChIP-qPCR assays (Figure 2C and Supplementary Figure 13). At E9.5, the signal of Jmjd3 was detected on the whole Hox1–13 PGs (Supplementary Figures 6C, 13). These results indicated that Jmjd3 bound to Hox genes with a temporal collinear manner and was consistent with Hox gene temporal collinear activation.
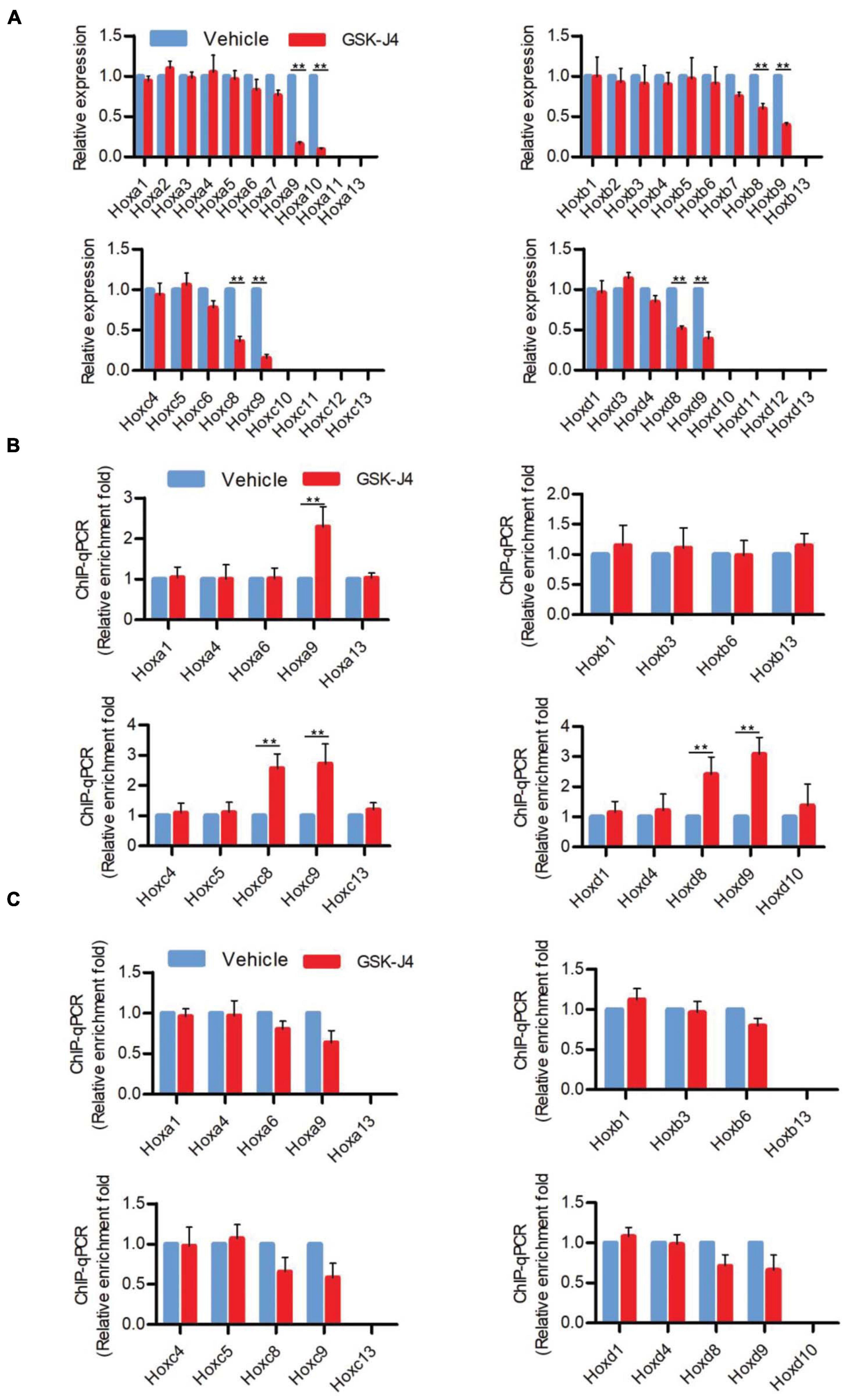
Figure 2. GSK-J4 inhibits transcription of Hox genes by blocking the H3K27me3 demethylase activity of Jmjd3 in E8.5 mouse embryos. (A) The mRNA levels of Hox genes were determined by RT-qPCR in E8.5 embryos, which were treated by vehicle or GSK-J4 (50 mg/kg) 12 h before. (B,C) The ChIP-qPCR assays with antibodies specific for H3K27me3 (B) or Jmjd3 (C) on the Hox genes in E8.5 embryos, which were exposed to vehicle or GSK-J4 (50 mg/kg) 12 h before. Bars indicate triplicate PCR reactions ± SD. Representative results of three independent experiments are shown. *p < 0.05, **p < 0.01.
Axial skeletal formation starts from the differentiation of condensed mesenchymal cells to cartilaginous templates (Karsenty, 2008; Long and Ornitz, 2013). To test whether chondrogenic cells are essential for Jmjd3-mediating axial skeletal patterning in mice, Jmjd3fl/fl;Col2a1-CreERT2 mice were generated by crossing mice with Jmjd3fl/fl and Col2a1-CreERT2 genotypes. The expressing Cre recombinase in chondrogenic cells of Col2a1-CreERT2 mice can be activated by tamoxifen. Jmjd3fl/fl;Col2a1-CreERT2 pregnant mice were intraperitoneally injected by tamoxifen in a single dose (50 mg/kg) at a time point from E8.5 to E10.5. Interestingly, different from the phenotypes of GSK-J4 treatment at E8.5, Jmjd3 knockout in chondrogenic cells at E8.5 induced more broadness and frequency of axial skeletal abnormalities along the whole-body axis of E18.5 embryos, primarily exhibiting anterior transformation of C2 to C1 (7/18), T1 to C7 (10/18), T3 to T2 (6/18), T8 to T7 (16/18), L1 to T13 (18/18), and S1 to T6 (9/18) (Figures 3A,B and Tables 1, 2). Similarly, different from the phenotypes induced by GSK-J4 treatment at E9.5, Jmjd3 knockout by tamoxifen at E9.5 induced homeotic transformations of T8 to T7 (15/17), L1 to T13 (17/17), and S1 to T6 (13/17) at the thoracic, lumbar, and sacral regions of E18.5 mouse embryos (Figures 3A,B and Tables 1, 2). Lastly, although GSK-J4 treatment at E10.5 induced no visible homeotic transformations along the axial skeleton (Figures 1A,B and Table 2), Jmjd3 conditional deletion by tamoxifen at E10.5 produced, though with less frequency, homeotic transformations of T8 to T7 (3/21) and L1 to T13 (4/21) at the thoracic and lumbar segments of E18.5 embryos (Figures 3A,B and Table 1). These results indicate that Jmjd3 knockout in chondrogenic cells produces more broadness and frequency of homeotic transformations along the whole-body axis than those induced by GSK-J4 at the same time points.
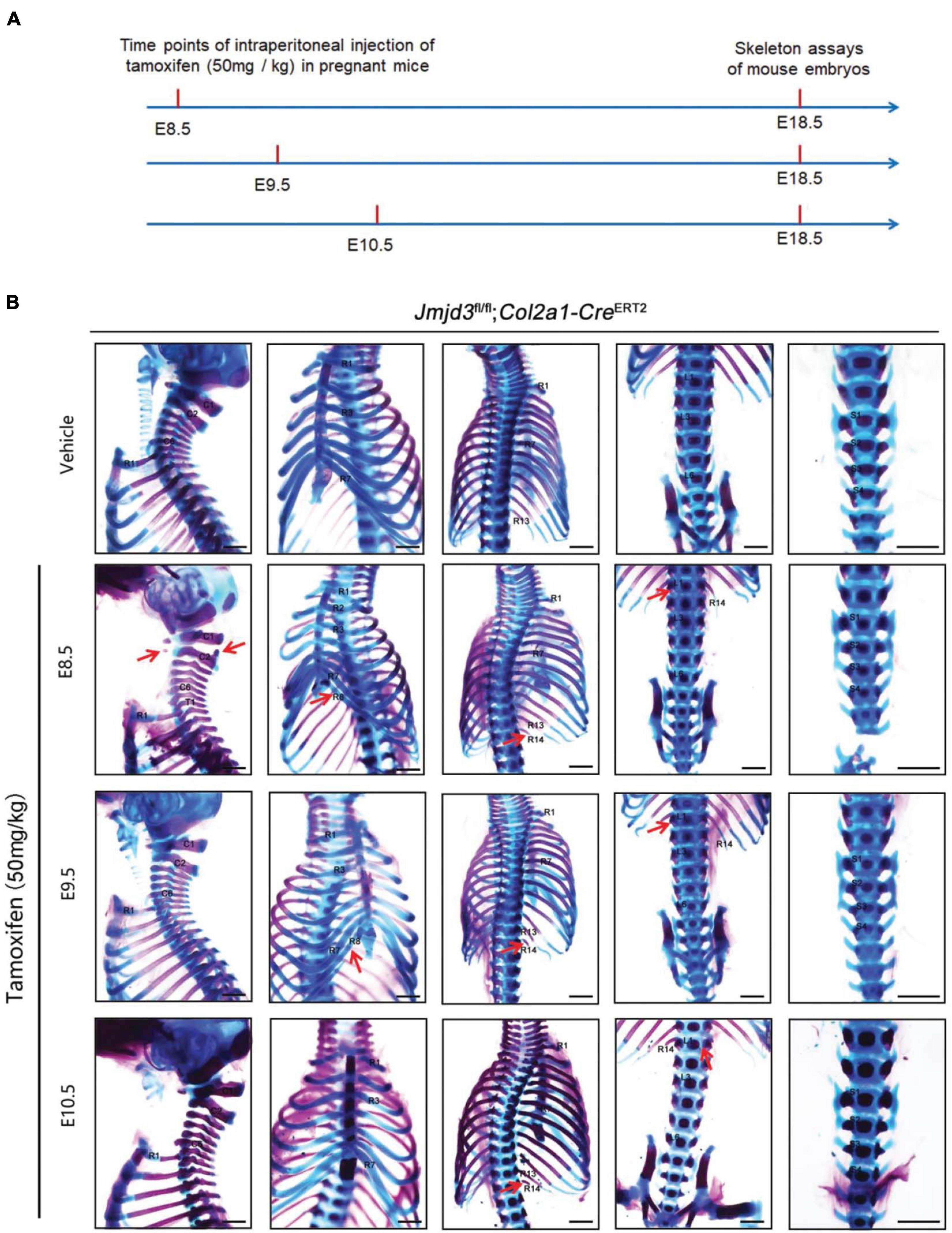
Figure 3. Homeotic transformations by tamoxifen induced Jmjd3 conditional knockout in Col2a1 expressing chondrogenic cells at E8.5, E9.5, and E10.5 days. (A) Experimental design for studying the roles of Jmjd3 in chondrogenic cells on body axis patterning. (B) Skeletal phenotypes were prepared from E18.5 mouse embryos. The red arrow indicates homeotic transformations of axial skeletons. Frequencies of homeotic transformations are given in Table 1. The cervical (C), rib (R), lumbar (L), and sacral (S) vertebrae are numbered. For schematic explanation, see Supplementary Figure 9. Scale bar, 1 mm.
Different from the effects of GSK-J4 treatment, conditional deletion of Jmjd3 at the same time points in chondrogenic cells produced more broadness and frequency of anterior transformation of axial skeleton (Figures 1A,B, 3A,B and Tables 1, 2). Previous results showed that, additionally as an H3K27m3 demethylase, Jmjd3 maintains a gene expression independent of H3K27m3 demethylase activity in differentiated cells (Miller et al., 2010) or during the process of early embryonic development (Shpargel et al., 2014). Therefore, we speculated that Jmjd3 in chondrogenic cells might not only mediate the Hox gene temporal colinear activation but also maintain Hox gene expression after their transcription activation. To test the hypothesis, we examined the effect of Jmjd3 deletion on the mRNA level of Hox PGs during body axis patterning. Interestingly, Jmjd3 knockout by tamoxifen at E8.0 reduced the mRNA level of Hox3–9 PGs in E8.5 embryos (Figure 4A and Supplementary Figure 5B). However, GSK-J4 treatment at E8.0 only decreased the mRNA level of Hox8–9 PGs in E8.5 embryos (Figure 2A and Supplementary Figure 5A). Knockout of Jmjd3 at E9.0 by tamoxifen significantly decreased the expression level of Hox8–13 PGs (Supplementary Figures 5B, 7A) in E9.5 embryos, while GSK-J4 treatment at E9.0 only reduced the transcriptional level of Hox11–13 PGs in E9.5 embryos (Supplementary Figures 5A, 6A). Jmjd3 deletion in chondrogenic cells obviously affected more Hox PG expression than those by GSK-J4 treatment at the same time points. These results were consistent with the fact that Jmjd3 deletion by tamoxifen produced more broadness and frequency of anterior transformation of axial skeleton than those by GSK-J4 treatment at the same time points (Figures 1A,B, 3A,B). Consistently, tamoxifen instead of GSK-J4 treatment at E8.0 significantly reduced the level of Jmjd3 protein on the Hox3–9 PGs in E8.5 embryos (Figures 2C, 4C and Supplementary Figures 5A,B, 13). However, Jmjd3 deletion only increased the H3K27me3 level on Hox8–9 PGs instead of Hox3–7 PGs (Figure 4B and Supplementary Figures 5B, 13). Because GSK-J4 treatment also only increased the H3K27me3 level on Hox8–9 PGs but not on Hox3–7 PGs (Figure 2B and Supplementary Figures 5A, 13), these results support that Jmjd3 activates Hox8–9 PGs dependent on its H3K27m3 demethylase activity, but maintains the expression of Hox3–7 PGs independent on H3K27m3 demethylase activity. In the same way, tamoxifen rather than GSK-J4 treatment at E9.0 significantly reduced the Jmjd3 protein level on the Hox8–13 PGs (Supplementary Figures 5B,C, 7C, 13) but only increased the H3K27me3 level on Hox10–13 PGs instead of on Hox8–10 PGs in E9.5 embryos (Supplementary Figures 5B, 6B, 7B, 13). Coherently, GSK-J4 treatment at E9.0 only increased the H3K27me3 level on Hox10–13 PGs but not on Hox8–10 PGs (Supplementary Figures 5A, 6B, 13). These results support that Jmjd3 activates Hox10–13 PGs dependent on H3K27m3 demethylase activity but maintains the expression of Hox8–10 PGs independent on H3K27m3 demethylase activity at E9.5 days. Therefore, we conclude that Jmjd3 temporally activates and maintains Hox gene expression in mice.
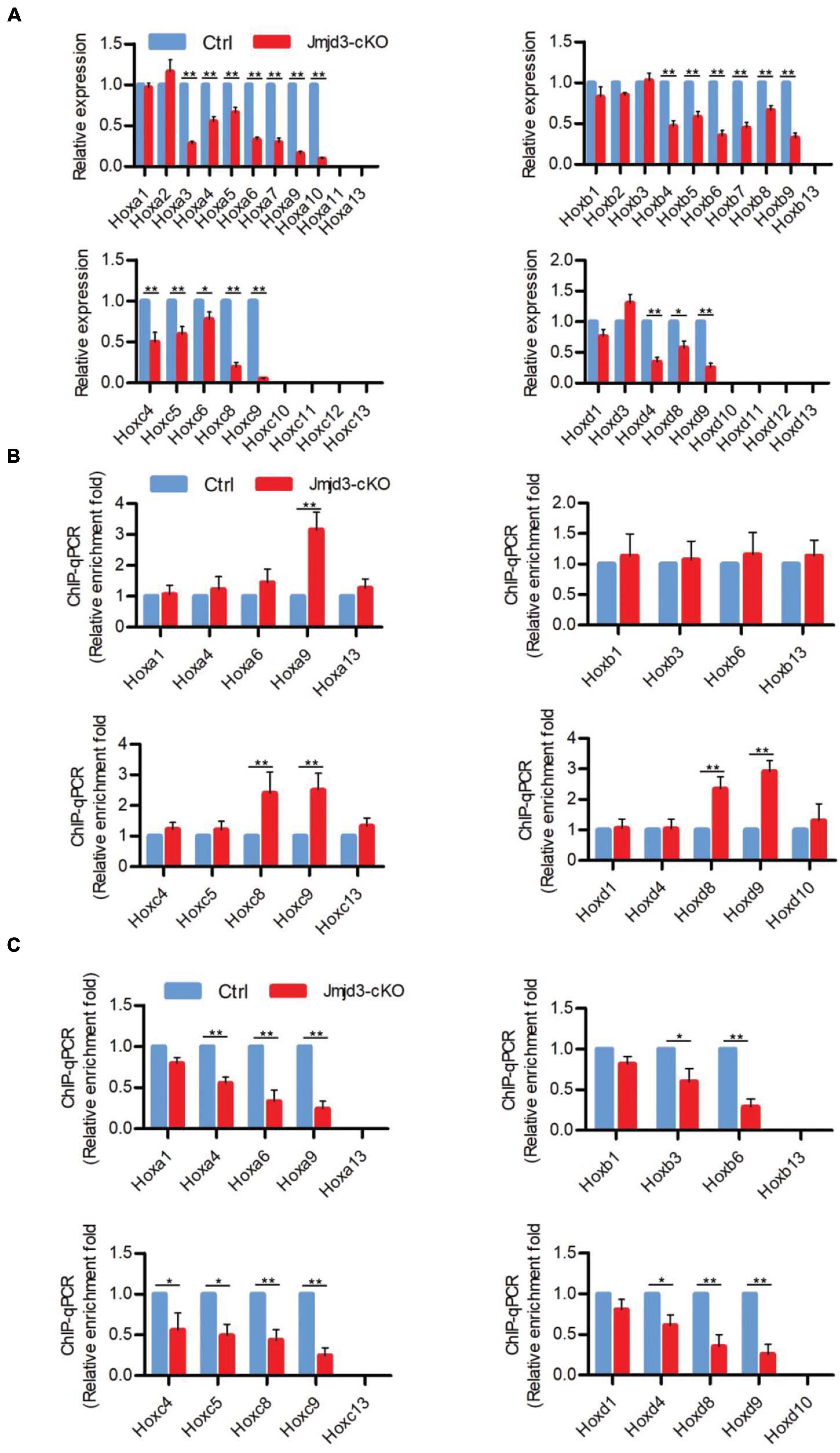
Figure 4. Jmjd3 temporally activated and maintained Hox gene expression in mice. (A) The mRNA levels of Hox genes were determined by RT-qPCR in E8.5 control or Jmjd3-cKO embryos, which were treated by tamoxifen (50 mg/kg) 12 h before. (B,C) ChIP-qPCR assays with antibodies specific for H3K27me3 (B) or Jmjd3 (C) on the Hox genes in E8.5 control or Jmjd3-cKO embryos treated by tamoxifen (50 mg/kg) 12 h before. Bars indicate triplicate PCR reactions ± SD. Representative results of three independent experiments are shown. *p < 0.05, **p < 0.01.
Previous studies indicated that sequential activation of the Hox gene relies on dynamic degradation of transcription repression hallmark H3K27me3 (Soshnikova and Duboule, 2009), which is established by histone methyltransferase Ezh2 (Cao et al., 2002; Muller et al., 2002) and erased by Kdm6 (Utx and Jmjd3) (De Santa et al., 2007; Lan et al., 2007; Lee et al., 2007). However, it is not clear whether an antagonistic interplay between Ezh2 and Jmjd3 occurs in a dynamic fashion during axial skeletal patterning. To explore this question, we crossed Jmjd3fl/fl;Col2a1-CreERT2 mice and Ezh2fl/fl mice to generate Jmjd3fl/fl;Ezh2fl/fl;Col2a1-CreERT2 mice, in which both Jmjd3 and Ezh2 could be concurrently deleted in chondrogenic cells after tamoxifen treatment. One dose of tamoxifen treatment (50 mg/kg) at E8.5 induced homeotic transformations including anterior transformation of C2 to C1 (5/12), T1 to C7 (8/12), T3 to T2 (5/12), T8 to T7 (10/12), L1 to T13 (12/12), and S1 to T6 (9/12) at the cervical, thoracic, lumbar, and sacral regions (Supplementary Figures 8A,B, 9A and Table 1) in E18.5 Jmjd3fl/fl;Ezh2fl/fl;Col2a1-CreERT2 mouse embryos, respectively. However, these abnormalities of axial skeletal patterning were very similar to the phenotypes of Jmjd3-deleted mice which were treated by tamoxifen at E8.5 (Figures 3A,B, Supplementary Figures 8A,B, 9A, and Table 1). Consistently, double-knockout Jmjd3 and Ezh2 by tamoxifen at E9.5 or E10.5 in chondrogenic cells produced similar patterning abnormalities of axial skeletons to those of Jmjd3-deleted mice by tamoxifen at E9.5 or E10.5, respectively (Figures 3A,B, Supplementary Figures 8A,B, 9A, and Table 1). Therefore, these indicated that Jmjd3 and Ezh2 do not antagonistically and dynamically control the axial skeletal patterning of mice. Consistently, GSK-126—an inhibitor of Ezh2—treatment (100 mg/kg) at E8.5, E9.5, or E10.5 produced no broad homeotic transformations in the vertebrae along the entire body axis, except for mild increasing frequency of posterior transformation of L6 to S1 at the sacral region (Supplementary Figure 10 and Table 2). Similarly, posterior transformation of L6 to S1 was the only detected homeotic transformation in Ezh2fl/fl;Prx1-Cre (7/15) and Ezh2fl/+;Prx1-Cre mice (10/17) (Supplementary Figure 11 and Table 1). Furthermore, L6 to S1 was the only detected posterior homeotic transformation in E18.5 Ezh2fl/fl;Col2a1-CreERT2 embryos after tamoxifen treatment at E8.5, E9.5, or E10.5, respectively (Supplementary Figures 12A,B and Table 1). Consistently, Ezh2 deletion cannot rescue the mRNA level of Hox6–10 PGs by Jmjd3 knockout (Supplementary Figures 14A,B). Therefore, these results strongly suggested that Jmjd3 and Ezh2 do not antagonistically control the axial skeletal patterning in mice.
In this study, we demonstrated that (1) Jmjd3 regulates axial skeletal patterning and Hox gene temporal collinear activation with its H3K27me3 demethylase activity. (2) There is no continuously antagonistic interplay between Jmjd3 and Ezh2 in the control of body axis patterning. The results revealed that Jmjd3 rather than Ezh2 is an essential epigenetic regulator of Hox gene temporal collinear activation.
Hox gene temporal collinear activation is essential for the development of the body axis (Deschamps and Duboule, 2017). During the past decades, important progress has been made in the molecular regulatory mechanism of Hox temporal collinearity activation. For example, genetic knockout experiments confirmed that there are different cis regulators at the 3′ and 5′ ends to regulate the temporal collinear activation of Hox genes (Tschopp and Duboule, 2011). Moreover, it was found that the 3′ activated Hox and the 5′ silenced Hox were in different chromatin domains (Noordermeer et al., 2011, 2014). However, it has long been speculated that signal molecules are involved in the temporal collinear activation of Hox genes, such as Wnt3a (Ikeya and Takada, 2001), Fgfr1 (Partanen et al., 1998), and Gdf11 (McPherron et al., 1999). However, because these reports were not based on conditional real-time gene knockout experiments, whether these signal molecules are really required for Hox gene temporal collinear activation remains in need of further research. For the same reason, Naruse et al. (2017) showed that knockout of Jmjd3 in germ cells can cause homeotic transformations in mice. However, their methods cannot prove that Jmjd3 is the regulator of Hox gene temporal collinear activation. In addition, it has been reported that retinoids regulate Hox gene temporal collinear activation in vitro (Chambeyron and Bickmore, 2004). However, in vivo experiments show that retinoids promote Hox gene activation at E7.5 but inhibit Hox gene expression at E8.5 (Kessel and Gruss, 1991). Thus, it is impossible for retinoids to regulate the whole process of Hox gene temporal collinear activation. In the present experiment, we demonstrated that the H3K27me3 demethylase Jmjd3 is the first identified epigenetic molecule regulating the temporal collinear activation of Hox genes. The evidence is as follows: among 28 histone demethylases and 17 histone methyltransferases, only Jmjd3 is successively activated and overexpressed in the development of the body axis during the period of Hox gene temporal activation (Supplementary Figures 1A,B). GSK-J4 treatment at different time points induced an alteration of the corresponding parts of the axial bone (Figures 1A,B, Supplementary Figure 9A, and Table 1). Finally, Jmjd3 could initiate Hox gene temporal collinear activation by erasing H3K27me3 (Figures 2B, 4B and Supplementary Figure 9B).
Although Jmjd3 and Utx have proved to be H3K27m3 demethylases (Agger et al., 2007; De Santa et al., 2007; Lan et al., 2007; Lee et al., 2007), in some cases, their function did not rely on H3K27m3 demethylase activity. For example, Jmjd3 and Utx play demethylase-independent roles in chromatin remodeling in differentiated cells where the epigenetic profile is already established (Miller et al., 2010). Shpargel et al. (2014) reported that early embryonic H3K27me3 repression can be alleviated in the absence of active demethylation of Jmjd3 and Utx. Here, we found that the activation rather than maintenance of Hox gene expression depended on the H3K27m3 demethylase activity of Jmjd3. For instance, we found that the phenotypes induced by GSK-J4 treatment or conditional deletion of Jmjd3 by tamoxifen were very different (Figures 1A,B, 3A,B, Supplementary Figure 9A, and Tables 1, 2). At E8.5, GSK-J4 induced only anterior transformation of T8 to T7 (6/28) in the middle part of the trunk of mice (Figures 1A,B and Table 2). However, Jmjd3 deletion at E8.5 by tamoxifen induced more extensive homeotic transformations across the whole-body axis, including C2 to C1, T1 to C7, T3 to T2, T8 to T7, L1 to T13, and S1 to T6 (Figures 3A,B and Table 1). RT-qPCR revealed that GSK-J4 treatment at E8.0 only reduced the mRNA level on Hox8–9 PGs (Figure 2A and Supplementary Figure 5A), while Jmjd3 deletion by tamoxifen at E8.0 decreased the mRNA level of Hox3–10 PGs in E8.5 mouse embryos (Figure 4A and Supplementary Figure 5B). Increasing the level of H3K27me3 on Hox8–9 PGs in mouse embryos by both GSK-J4 treatment and Jmjd3 deletion indicated that the removal of H3K27me3 on Hox8–9 PGs depends on the Jmjd3 demethylase activity at the time point of E8.5 (Figures 2B, 4B and Supplementary Figures 5A,B). However, the Jmjd3 deletion instead of GSK-J4 treatment can reduce the mRNA level of Hox 3–5 PGs, on which no alteration of the H3K27me3 level was detected at this time point (Figures 2A,B, 4A,B and Supplementary Figures 5A,B), indicating that the maintaining expression of these Hox genes did not require the H3K27me3 demethylase of Jmjd3. In the same way, at E9.5, GSK-J4 only decreased the mRNA level of Hox11–13 PGs and the corresponding caudal homeotic transformations (Figures 1A,B, 2A and Supplementary Figure 5A). However, Jmjd3 knockout can reduce the mRNA level of not only Hox11–13 PGs but also Hox8–10 PGs (Figure 4A and Supplementary Figure 5B), thus inducing homeotic transformations of the thoracic, lumbar, and sacral regions (Figures 3A,B and Table 2). Accordingly, these results showed that the removal of H3K27me3 on Hox11–13 PGs instead of on Hox8–10 PGs depends on the H3K27me3 demethylase activity of Jmjd3 (Figures 2B, 4B and Supplementary Figure 5B). Therefore, the maintaining expression of Hox8–10 PGs requires Jmjd3 protein rather than its enzyme activity.
PcG or TrxG group proteins played critical roles in maintaining the expression pattern of Hox genes (Mallo and Alonso, 2013; Geisler and Paro, 2015). Mutations in PcG genes lead to ectopic Hox gene expression and consequent posterior homeotic transformations, while deletions of TrxG genes result in delayed Hox gene expression and consequent anterior homeotic transformations in both Drosophila and vertebrates (Mallo and Alonso, 2013; Geisler and Paro, 2015). Therefore, Hox gene expression was proposed to be dynamically and antagonistically regulated by PcG and TrxG (Hanson et al., 1999; Klymenko and Muller, 2004; Sheikh et al., 2015). Here, we detected that the homolog of TrxG group Jmjd3 loss results in anterior transformation along the axial skeleton (Figures 1A,B, 3A,B, Supplementary Figure 9A, and Tables 1, 2). However, the subunit of PcG group protein Ezh2 inactivation only produced posterior transformation of L6 to S1 (Supplementary Figures 10–12 and Tables 1, 2). Furthermore, the phenotypes of double Jmjd3 and Ezh2 gene-deleted mice were similar to those of Jmjd3 knockout mice (Figures 2A,B, Supplementary Figures 8A,B, 9A, and Table 2). Consistently, whole-mount in situ hybridization of mouse embryos at E9.5 showed that Jmjd3 was expressed more obviously in the head and trunk, while Ezh2 was expressed more strongly in the tail (Supplementary Figure 15). Therefore, these results strongly indicated that there is no continuous dynamic interplay between Jmjd3 and Ezh2 in the control of body axis patterning.
Axial skeletal formation is primarily through the process of endochondral bone formation, which starts from the differentiation of condensed mesenchymal cells to cartilaginous templates (Karsenty, 2008; Long and Ornitz, 2013). Hox genes are well-known patterning genes that confer identity to skeletogenic condensations (Krumlauf, 1994). There are few studies with the conditional knockout mice to explore the mechanism of spatiotemporal control of Hox gene transcription. Here, chondrogenic cell conditional gene knockout mice were used to address the roles of Jmjd3 during the process of mouse body axial patterning. Previously, the transcription factor Sox9 of Col2a1 was reported be observed in the mouse embryos at E8.0 (Zhao et al., 1997; Akiyama et al., 2005). Laz reporter gene of Col2a1-Cre mice (Ovchinnikov et al., 2000; Nakamura et al., 2006) and in situ hybridization with Col2a1 probe experiments showed that Col2a1 could be observed in mouse embryos at E8.5 (Ng et al., 1997). Consistently, the mRNA levels of Col2a1 and Sox9 could be detected by RT-qPCR in E8.5 embryos of mice (Supplementary Figure 1B). Jmjd3 deletion in Col2a1-expressing chondrogenic cells by tamoxifen treatment at E8.5 could produce homeotic transformations of the whole-body axis (Figures 3A,B and Table 1). Therefore, we suggested that axial bone patterning may synchronize with chondrogenic differentiation. This idea is further supported by a previous report that the expression of retinoic acid receptor driven by the Col2a1 promoter could induce homeotic transformations in mice (Yamaguchi et al., 1998).
In summary, we demonstrated that the temporal collinearity activation of Hox genes primarily relied on Jmjd3 to erasing H3K27me3 on Hox genes. Jmjd3 regulates not only the initiation but also the maintenance of the temporal collinear expression of Hox genes. No continuous dynamic interplay was detected between Jmjd3 and Ezh2 in the control of body axis patterning. Lastly, we demonstrated that chondrogenic cells were essential for axial skeletal patterning and proposed that axial bone patterning may synchronize with chondrogenic differentiation in mice.
Materials, methods, and associated references are described in Supplementary Materials and Methods.
The authors declare that data supporting the findings of this study are available within the article and its Supplementary Material files or from the corresponding author on request. The RNA sequencing data cited in Supplementary Figures 1, 2 have been deposited in the NCBI SRA database under the accession code PRJNA673586.
The animal study was reviewed and approved by Institutional Animal Care and Use Committee at the Fourth Military Medical University.
FZ conceived the project, performed most of the experiments, analyzed the data, and wrote the manuscript. XZ, RJ, YW, and XW help to raise the mice, collected the samples, and performed the molecular biology experiments. All the authors helped to analyze the data and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (81572631 and 31000559 to FZ, 81772865 to SG, 31370981 to DZo), Shaanxi Society Development Sci-Tech Research Project (2016SF-064 to FZ, 2016SF-028 to DZn), State Key Laboratory of Cancer Biology, Fourth Military Medical University (CBSKL2019ZZ28 to FZ), and Key Research Projects of Shaanxi Province (2019ZDLSF02-01 to DZo).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.642931/full#supplementary-material
Agger, K., Cloos, P. A., Christensen, J., Pasini, D., Rose, S., Rappsilber, J., et al. (2007). UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731–734. doi: 10.1038/nature06145
Akiyama, H., Kim, J. E., Nakashima, K., Balmes, G., Iwai, N., Deng, J. M., et al. (2005). Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc. Natl. Acad. Sci. U.S.A. 102, 14665–14670. doi: 10.1073/pnas.0504750102
Alexander, T., Nolte, C., and Krumlauf, R. (2009). Hox genes and segmentation of the hindbrain and axial skeleton. Annu. Rev. Cell Dev. Biol. 25, 431–456. doi: 10.1146/annurev.cellbio.042308.113423
Cao, R., Tsukada, Y., and Zhang, Y. (2005). Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell 20, 845–854. doi: 10.1016/j.molcel.2005.12.002
Cao, R., Wang, L., Wang, H., Xia, L., Erdjument-Bromage, H., Tempst, P., et al. (2002). Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043. doi: 10.1126/science.1076997
Casaca, A., Santos, A. C., and Mallo, M. (2014). Controlling Hox gene expression and activity to build the vertebrate axial skeleton. Dev. Dyn. 243, 24–36. doi: 10.1002/dvdy.24007
Chambeyron, S., and Bickmore, W. A. (2004). Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes. Dev. 18, 1119–1130. doi: 10.1101/gad.292104
Copur, O., and Muller, J. (2013). The histone H3-K27 demethylase Utx regulates HOX gene expression in Drosophila in a temporally restricted manner. Development 140, 3478–3485. doi: 10.1242/dev.097204
Copur, O., and Muller, J. (2018). Histone demethylase activity of Utx Is essential for viability and regulation of HOX Gene expression in Drosophila. Genetics 208, 633–637. doi: 10.1534/genetics.117.300421
Czermin, B., Melfi, R., McCabe, D., Seitz, V., Imhof, A., and Pirrotta, V. (2002). Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111, 185–196. doi: 10.1016/s0092-8674(02)00975-3
De Santa, F., Totaro, M. G., Prosperini, E., Notarbartolo, S., Testa, G., and Natoli, G. (2007). The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130, 1083–1094. doi: 10.1016/j.cell.2007.08.019
Deschamps, J., and Duboule, D. (2017). Embryonic timing, axial stem cells, chromatin dynamics, and the Hox clock. Genes. Dev. 31, 1406–1416. doi: 10.1101/gad.303123.117
Deschamps, J., van den Akker, E., Forlani, S., De Graaff, W., Oosterveen, T., Roelen, B., et al. (1999). Initiation, establishment and maintenance of Hox gene expression patterns in the mouse. Int. J. Dev. Biol. 43, 635–650.
Deschamps, J., and van Nes, J. (2005). Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development 132, 2931–2942. doi: 10.1242/dev.01897
Deschamps, J., and Wijgerde, M. (1993). Two phases in the establishment of HOX expression domains. Dev. Biol. 156, 473–480. doi: 10.1006/dbio.1993.1093
Dhar, S. S., Lee, S. H., Chen, K., Zhu, G., Oh, W., Allton, K., et al. (2016). An essential role for UTX in resolution and activation of bivalent promoters. Nucleic Acids Res. 44, 3659–3674. doi: 10.1093/nar/gkv1516
Favier, B., and Dolle, P. (1997). Developmental functions of mammalian Hox genes. Mol. Hum. Reprod. 3, 115–131. doi: 10.1093/molehr/3.2.115
Forlani, S., Lawson, K. A., and Deschamps, J. (2003). Acquisition of Hox codes during gastrulation and axial elongation in the mouse embryo. Development 130, 3807–3819. doi: 10.1242/dev.00573
Geisler, S. J., and Paro, R. (2015). Trithorax and Polycomb group-dependent regulation: a tale of opposing activities. Development 142, 2876–2887. doi: 10.1242/dev.120030
Hanson, R. D., Hess, J. L., Yu, B. D., Ernst, P., van Lohuizen, M., Berns, A., et al. (1999). Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc. Natl. Acad. Sci. U.S.A. 96, 14372–14377. doi: 10.1073/pnas.96.25.14372
Hashizume, R., Andor, N., Ihara, Y., Lerner, R., Gan, H., Chen, X., et al. (2014). Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat. Med. 20, 1394–1396. doi: 10.1038/nm.3716
Hong, S., Cho, Y. W., Yu, L. R., Yu, H., Veenstra, T. D., and Ge, K. (2007). Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. U.S. A. 104, 18439–18444. doi: 10.1073/pnas.0707292104
Ikeya, M., and Takada, S. (2001). Wnt-3a is required for somite specification along the anteroposterior axis of the mouse embryo and for regulation of cdx-1 expression. Mech. Dev. 103, 27–33. doi: 10.1016/s0925-4773(01)00338-0
Juan, A. H., and Ruddle, F. H. (2003). Enhancer timing of Hox gene expression: deletion of the endogenous Hoxc8 early enhancer. Development 130, 4823–4834. doi: 10.1242/dev.00672
Karsenty, G. (2008). Transcriptional control of skeletogenesis. Annu. Rev. Genomics Hum. Genet. 9, 183–196. doi: 10.1146/annurev.genom.9.081307.164437
Kennison, J. A., and Tamkun, J. W. (1988). Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 85, 8136–8140. doi: 10.1073/pnas.85.21.8136
Kessel, M., and Gruss, P. (1991). Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell 67, 89–104. doi: 10.1016/0092-8674(91)90574-i
Klymenko, T., and Muller, J. (2004). The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 5, 373–377. doi: 10.1038/sj.embor.7400111
Kmita, M., and Duboule, D. (2003). Organizing axes in time and space; 25 years of colinear tinkering. Science 301, 331–333. doi: 10.1126/science.1085753
Kondrashov, N., Pusic, A., Stumpf, C. R., Shimizu, K., Hsieh, A. C., Ishijima, J., et al. (2011). Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145, 383–397. doi: 10.1016/j.cell.2011.03.028
Krumlauf, R. (1994). Hox genes in vertebrate development. Cell 78, 191–201. doi: 10.1016/0092-8674(94)90290-9
Krumlauf, R. (2018). Hox genes, clusters and collinearity. Int. J. Dev. Biol. 62, 659–663. doi: 10.1387/ijdb.180330rr
Kuzmichev, A., Nishioka, K., Erdjument-Bromage, H., Tempst, P., and Reinberg, D. (2002). Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes. Dev. 16, 2893–2905. doi: 10.1101/gad.1035902
Lan, F., Bayliss, P. E., Rinn, J. L., Whetstine, J. R., Wang, J. K., Chen, S., et al. (2007). A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449, 689–694. doi: 10.1038/nature06192
Lee, M. G., Villa, R., Trojer, P., Norman, J., Yan, K. P., Reinberg, D., et al. (2007). Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318, 447–450. doi: 10.1126/science.1149042
Lewis, E. B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276, 565–570. doi: 10.1038/276565a0
Li, L., Liu, B., Wapinski, O. L., Tsai, M. C., Qu, K., Zhang, J., et al. (2013). Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 5, 3–12. doi: 10.1016/j.celrep.2013.09.003
Liu, P., Wakamiya, M., Shea, M. J., Albrecht, U., Behringer, R. R., and Bradley, A. (1999). Requirement for Wnt3 in vertebrate axis formation. Nat. Genet. 22, 361–365. doi: 10.1038/11932
Logan, M., Martin, J. F., Nagy, A., Lobe, C., Olson, E. N., and Tabin, C. J. (2002). Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33, 77–80. doi: 10.1002/gene.10092
Long, F., and Ornitz, D. M. (2013). Development of the endochondral skeleton. Cold Spring Harb. Perspect. Biol. 5:a008334. doi: 10.1101/cshperspect.a008334
Mallo, M., and Alonso, C. R. (2013). The regulation of Hox gene expression during animal development. Development 140, 3951–3963. doi: 10.1242/dev.068346
McCabe, M. T., Ott, H. M., Ganji, G., Korenchuk, S., Thompson, C., Van Aller, G. S., et al. (2012). EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112. doi: 10.1038/nature11606
McPherron, A. C., Lawler, A. M., and Lee, S. J. (1999). Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat. Genet. 22, 260–264. doi: 10.1038/10320
Miller, S. A., Mohn, S. E., and Weinmann, A. S. (2010). Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell 40, 594–605. doi: 10.1016/j.molcel.2010.10.028
Mongera, A., Michaut, A., Guillot, C., Xiong, F., and Pourquie, O. (2019). Mechanics of anteroposterior axis formation in vertebrates. Annu. Rev. Cell Dev. Biol. 35, 259–283. doi: 10.1146/annurev-cellbio-100818-125436
Montavon, T., and Soshnikova, N. (2014). Hox gene regulation and timing in embryogenesis. Semin. Cell Dev. Biol. 34, 76–84. doi: 10.1016/j.semcdb.2014.06.005
Muller, J., Hart, C. M., Francis, N. J., Vargas, M. L., Sengupta, A., Wild, B., et al. (2002). Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111, 197–208. doi: 10.1016/s0092-8674(02)00976-5
Nakamura, E., Nguyen, M. T., and Mackem, S. (2006). Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev. Dyn. 235, 2603–2612. doi: 10.1002/dvdy.20892
Naruse, C., Shibata, S., Tamura, M., Kawaguchi, T., Abe, K., Sugihara, K., et al. (2017). New insights into the role of Jmjd3 and Utx in axial skeletal formation in mice. FASEB J. 31, 2252–2266. doi: 10.1096/fj.201600642r
Neijts, R., Amin, S., van Rooijen, C., Tan, S., Creyghton, M. P., de Laat, W., et al. (2016). Polarized regulatory landscape and Wnt responsiveness underlie Hox activation in embryos. Genes. Dev. 30, 1937–1942. doi: 10.1101/gad.285767.116
Ng, L. J., Wheatley, S., Muscat, G. E., Conway-Campbell, J., Bowles, J., Wright, E., et al. (1997). SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev. Biol. 183, 108–121. doi: 10.1006/dbio.1996.8487
Noordermeer, D., Leleu, M., Schorderet, P., Joye, E., Chabaud, F., and Duboule, D. (2014). Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. Elife 3:e02557.
Noordermeer, D., Leleu, M., Splinter, E., Rougemont, J., De Laat, W., and Duboule, D. (2011). The dynamic architecture of Hox gene clusters. Science 334, 222–225. doi: 10.1126/science.1207194
Ntziachristos, P., Tsirigos, A., Welstead, G. G., Trimarchi, T., Bakogianni, S., Xu, L., et al. (2014). Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature 514, 513–517. doi: 10.1038/nature13605
Ovchinnikov, D. A., Deng, J. M., Ogunrinu, G., and Behringer, R. R. (2000). Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis 26, 145–146. doi: 10.1002/(sici)1526-968x(200002)26:2<145::aid-gene14>3.0.co;2-c
Partanen, J., Schwartz, L., and Rossant, J. (1998). Opposite phenotypes of hypomorphic and Y766 phosphorylation site mutations reveal a function for Fgfr1 in anteroposterior patterning of mouse embryos. Genes. Dev. 12, 2332–2344. doi: 10.1101/gad.12.15.2332
Piunti, A., and Shilatifard, A. (2016). Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 352:aad9780. doi: 10.1126/science.aad9780
Sheikh, B. N., Downer, N. L., Phipson, B., Vanyai, H. K., Kueh, A. J., McCarthy, D. J., et al. (2015). MOZ and BMI1 play opposing roles during Hox gene activation in ES cells and in body segment identity specification in vivo. Proc. Natl. Acad. Sci. U.S.A. 112, 5437–5442. doi: 10.1073/pnas.1422872112
Shen, X., Liu, Y., Hsu, Y. J., Fujiwara, Y., Kim, J., Mao, X., et al. (2008). EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491–502. doi: 10.1016/j.molcel.2008.10.016
Shpargel, K. B., Starmer, J., Yee, D., Pohlers, M., and Magnuson, T. (2014). KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS Genet. 10:e1004507. doi: 10.1371/journal.pgen.1004507
Simon, J., Chiang, A., and Bender, W. (1992). Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 114, 493–505. doi: 10.1242/dev.114.2.493
Soshnikova, N., and Duboule, D. (2009). Epigenetic temporal control of mouse Hox genes in vivo. Science 324, 1320–1323. doi: 10.1126/science.1171468
Srivastava, S., Dhawan, J., and Mishra, R. K. (2015). Epigenetic mechanisms and boundaries in the regulation of mammalian Hox clusters. Mech. Dev. 138(Pt 2), 160–169. doi: 10.1016/j.mod.2015.07.015
Tschopp, P., and Duboule, D. (2011). A genetic approach to the transcriptional regulation of Hox gene clusters. Annu. Rev. Genet. 45, 145–166. doi: 10.1146/annurev-genet-102209-163429
Wellik, D. M. (2009). Hox genes and vertebrate axial pattern. Curr. Top. Dev. Biol. 88, 257–278. doi: 10.1016/s0070-2153(09)88009-5
Yamaguchi, M., Nakamoto, M., Honda, H., Nakagawa, T., Fujita, H., Nakamura, T., et al. (1998). Retardation of skeletal development and cervical abnormalities in transgenic mice expressing a dominant-negative retinoic acid receptor in chondrogenic cells. Proc. Natl. Acad. Sci. U.S.A. 95, 7491–7496. doi: 10.1073/pnas.95.13.7491
Ye, L., Fan, Z., Yu, B., Chang, J., Al Hezaimi, K., Zhou, X., et al. (2012). Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 11, 50–61. doi: 10.1016/j.stem.2012.04.009
Young, T., Rowland, J. E., van de Ven, C., Bialecka, M., Novoa, A., Carapuco, M., et al. (2009). Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev. Cell 17, 516–526. doi: 10.1016/j.devcel.2009.08.010
Zhang, F., Xu, L., Xu, L., Xu, Q., Li, D., Yang, Y., et al. (2015). JMJD3 promotes chondrocyte proliferation and hypertrophy during endochondral bone formation in mice. J. Mol. Cell Biol. 7, 23–34. doi: 10.1093/jmcb/mjv003
Zhao, Q., Eberspaecher, H., Lefebvre, V., and De Crombrugghe, B. (1997). Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev. Dyn. 209, 377–386. doi: 10.1002/(sici)1097-0177(199708)209:4<377::aid-aja5>3.0.co;2-f
Keywords: Jmjd3, Ezh2, H3K27me3, homeotic transformation, temporal collinearity of Hox gene activation, chondrogenic cells
Citation: Zhang F, Zhao X, Jiang R, Wang Y, Wang X, Gu Y, Xu L, Ye J, Chen CD, Guo S, Zhang D and Zhao D (2021) Identification of Jmjd3 as an Essential Epigenetic Regulator of Hox Gene Temporal Collinear Activation for Body Axial Patterning in Mice. Front. Cell Dev. Biol. 9:642931. doi: 10.3389/fcell.2021.642931
Received: 17 December 2020; Accepted: 23 June 2021;
Published: 21 July 2021.
Edited by:
Claire Rougeulle, UMR7216 Epigénétique et Destin Cellulaire, FranceReviewed by:
Toshiro Iwagawa, The University of Tokyo, JapanCopyright © 2021 Zhang, Zhao, Jiang, Wang, Wang, Gu, Xu, Ye, Chen, Guo, Zhang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Zhang, emhmMTk3NUBmbW11LmVkdS5jbg==; Dawei Zhang, emR3eXhqQDE2My5jb20=; Daqing Zhao, emhhb2RxNDMwQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.