- Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, IRCCS, Rome, Italy
While individually rare, disorders affecting development collectively represent a substantial clinical, psychological, and socioeconomic burden to patients, families, and society. Insights into the molecular mechanisms underlying these disorders are required to speed up diagnosis, improve counseling, and optimize management toward targeted therapies. Genome sequencing is now unveiling previously unexplored genetic variations in undiagnosed patients, which require functional validation and mechanistic understanding, particularly when dealing with novel nosologic entities. Functional perturbations of key regulators acting on signals’ intersections of evolutionarily conserved pathways in these pathological conditions hinder the fine balance between various developmental inputs governing morphogenesis and homeostasis. However, the distinct mechanisms by which these hubs orchestrate pathways to ensure the developmental coordinates are poorly understood. Integrative functional genomics implementing quantitative in vivo models of embryogenesis with subcellular precision in whole organisms contribute to answering these questions. Here, we review the current knowledge on genes and mechanisms critically involved in developmental syndromes and pediatric cancers, revealed by genomic sequencing and in vivo models such as insects, worms and fish. We focus on the monomeric GTPases of the RAS superfamily and their influence on crucial developmental signals and processes. We next discuss the effectiveness of exponentially growing functional assays employing tractable models to identify regulatory crossroads. Unprecedented sophistications are now possible in zebrafish, i.e., genome editing with single-nucleotide precision, nanoimaging, highly resolved recording of multiple small molecules activity, and simultaneous monitoring of brain circuits and complex behavioral response. These assets permit accurate real-time reporting of dynamic small GTPases-controlled processes in entire organisms, owning the potential to tackle rare disease mechanisms.
Introduction
Rare diseases are individually uncommon but collectively frequent, affecting approximately 25 million people in Europe and impacting between 263 million and 446 million people worldwide (Wakap et al., 2020), with a significant proportion of cases awaiting diagnosis (sources: Orphanet, Eurordis, and WHO, Kaplan et al., 2013). They are often chronic, degenerative, and disabling conditions, which in approximately 70% of cases have a pediatric onset and show high morbidity and mortality. As estimated by the BURQOL-RD project (“Social Economic Burden and Health-Related Quality of Life in Patients with Rare Diseases in Europe”), a high level of socioeconomic burden is associated with these conditions (Angelis et al., 2015), which challenges health care systems globally, as well as the quality of life of the patients and their families. Particularly dramatic is the situation for pediatric cancers, which, despite their rarity, represent a significant disease burden nowadays. Yearly, more than 500,000 new cases of rare cancers are diagnosed (Gatta et al., 2017), causing approximately 6000 deaths in children, according to the European Society for Pediatric Oncology. The World Health Organization estimates that half of these tumors are malignant hematological cancers (e.g., leukemia) or solid nervous system tumors (e.g., neuroblastoma) (Gupta et al., 2015; Steliarova-Foucher et al., 2017).
A significant proportion of these disorders underlie one or more genetic alterations causing functional dysregulation of master regulators involved at various levels and stages of complex and dynamic developmental programs (e.g., cell proliferation, migration, differentiation, and developmental competence) of virtually any growing tissue or organ. Molecularly, a relatively small number of signaling pathways and networks (Wingless/Integrated (Wnt), hedgehog (Hh), Notch, Bone morphogenetic protein (BMP), fibroblast growth factor (FGF), mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK), etc.) are responsible for directing developmental programs. The crosstalk among these pathways, together with positive and negative control loop stations mediated by highly conserved molecular nodes, accounts for the pleiotropy of signaling, which ultimately shapes organismal development. These pathways’ interplays ensure differential responses to converging – and sometimes conflicting – messages and thereby multiorgan morphogenesis and homeostasis (Basson, 2012). Therefore, it is not surprising that from various alterations of core signaling hubs mastering multiple developmental networks, both developmental syndromes and malignancies arise. Yet, our biological knowledge on the key genes, the regulated signaling pathways, and the intracellular nodes differentially involved in development and disease remains poor. The current lack of a case-specific mechanistic understanding further hinders the disease identification, leaving many of them “orphan” of an accurate “diagnosis” and therefore targeted cure. This knowledge gap is particularly challenging, given the short life expectancy associated with a large fraction of rare conditions (Courbier and Berjonneau, 2017).
Following the EU call for action, revolutionary sophistication and rapid implementation of next-generation sequencing (NGS) techniques, especially whole-exome sequencing (WES), have allowed a considerable boost in the identification of genomic modifications and signaling pathways’ alterations in the field of rare diseases. Recently, the National Institutes of Health (NIH)–supported Centers for Mendelian Genomics noted an unprecedented increase in the number of novel diseases discovered per year, estimated to be more than 200 (Posey et al., 2019). Clearly, besides identifying the genes, precise fingerprints of disease mechanisms would help create a new “taxonomy of the disease” with immediate benefit on patient care specialization. Yet, for many of the newly discovered genetic conditions, even the physiological activity of the proteins involved remains poorly known. To resolve this gap, it is beneficial to invest into the smart combination between (i) in silico wide genome search for disease–gene/pathogenic variants in undiagnosed patients enrolled in international networks and (ii) functional genomics approaches using ad hoc in vitro systems (i.e., iPS, patient-specific induced pluripotent stem cells), supported and enhanced by (iii) both vertebrate and invertebrate animal disease models. Along this line, in the past decade, others and we have contributed to decisive advancements in the understanding of the pathophysiological role of a number of small GTPases belonging to the large RAS superfamily. In an international research framework dedicated to undiagnosed patients started at the “Ospedale Pediatrico Bambino Gesù” children’s hospital, functional genomic studies employing WES analysis and complementary in vitro experimental approaches, as well as invertebrate and vertebrate models, have allowed us to identify new nosologic entities caused by mutations affecting several genes, including a subset of which encode small GTPases. For instance, we associated a uniquely behaving genetic alteration in CDC42 [OMIM: 116952] with a severe autoinflammatory condition (Lam et al., 2019). The specific molecular profiling of these patients allowed prompt lifesaving treatment, whereas validation of the pathogenicity in nematodes and human immune cells unraveled the impact on development, hematological cell maturation, and motility (Lam et al., 2019). Of note, we previously identified a different class of mutations affecting the same genes as the cause of a clinically variable neurodevelopmental disorder (Martinelli et al., 2018), emphasizing the requirement of functional characterization analyses to casually associate genomic variants with disease and decipher the underlying mechanisms. More recently, we identified activating mutations in the gene encoding the key effector of the MAPK signaling cascade, MAPK1 [OMIM: 176948], as cause of a neurodevelopmental disease within the RASopathies spectrum (Motta et al., 2020). Again, in vivo assays in the context of cell differentiation and morphogenesis contributed to the validation of the pathogenicity of the mutations during embryonic development. The work provided evidence for a differential impact of germline inherited (found in developmental disorders) and somatically acquired (cancer-associated) mutations in this gene. The expansion of such paradigm in modern biomedical research clearly represents a valid tool for deepening our understanding of healthy and diseased mechanisms as well as core developmental hubs.
Here, we review recent functional genomics findings proving mutations in some among the large group of small GTPases molecules to be critically involved in rare developmental syndromes and cancers. We next discuss the current knowledge on the interplay with signaling pathways networks, whose tightly regulated activity is essential in many developmental processes, from lateral inhibition (which differentiates cell fate from initial equivalence fields) to cell polarity mechanisms (instructing gastrulation cell movements) and is being implicated in pediatric diseases. The cost reductions in sequencing and the extraordinary progress in functional imaging, signal biosensors optimization, and genome engineering in living organisms are opening long-awaited possibilities to combine in a single workflow (a) analyses directed to identify new disease genes with (b) sophisticated functional approaches in vivo to validate the putative pathogenic variants. Global conservation of the genes exists across taxa such that smartly chosen tractable model systems and ad hoc in vivo tools are and will be crucial for the majority of the newly discovered diseases for which we still fail to understand the impact on the signaling networks during development. In this context, we briefly discuss the advantages in using zebrafish for functional genomics of rare diseases and examine the latest tools, which enable highly resolved in vivo whole-embryo real-time reporting of dynamic small GTPase-regulated processes during development.
The RAS Superfamily of Small GTPases in Development and Disease
The RAS superfamily of small GTPases comprises five main protein families grouped by structure and function. They include proteins belonging to the (i) RAS family, involved in cell proliferation, specification, and differentiation (Figure 1); (ii) RHO family, known to influence actin and microtubule (MT) cytoskeleton and thereby cell migration and morphology (Figure 2); (iii) RAN family, which control nuclear transport (Figure 3); and (iv) ADP-ribosylation factor (ARF) and RAB families, involved in various steps of vesicle trafficking and organelles’ dynamics (Wennerberg et al., 2005). They all regulate their activity by cycling between an active (GTP-bound) and an inactive (GDP-bound) form, a switch determined by a number of extracellular signals and effectors (Bourne et al., 1991). By this dynamic activity and the myriad of effectors, these small GTPases function as molecular hubs at the crossroad between morphogenetic inputs, crucial for signal integration to determine cell precursors’ state, behavior, and specification in developing organisms. Given the plethora of cell processes that they assist, these proteins and their related signaling components have long emerged as major players in developmental disorders and malignancies (Schubbert et al., 2007; Simanshu et al., 2017; Qu et al., 2019; Figures 1–3). Indeed, the list of disease-causing mutations affecting signalings modulated by proteins of the RAS superfamily is increasing. However, we only begin to understand the complexity of their role in developmental pathways and their relevance for the onset of disease. Here, we focus on the emerging rare disease–causing genes encoding proteins of the RAS, RHO, ARF, and RAB families and the known mechanistic consequences altering developmental processes and relevant for pathogenesis, also emerging from in vivo models.
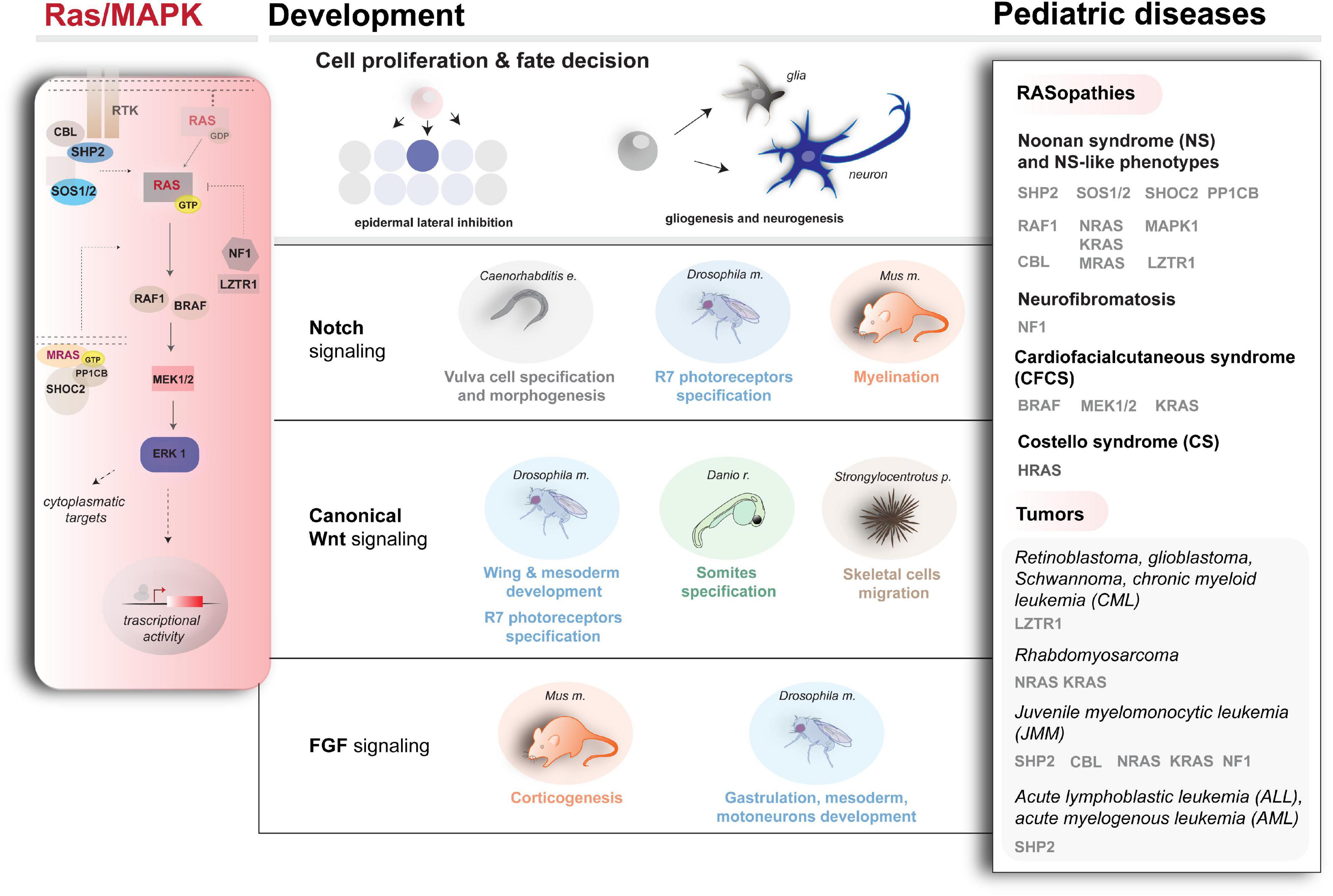
Figure 1. Schematic representation of Ras/MAPK cascade (left), Ras/MAPK-influenced pathways, and developmental processes (center) and examples of genetic conditions underlying a dysregulated cascade (right). For the diseases and disease–genes depicted here, besides the literature cited in the text, refer to Roberts et al. (2007) and Tartaglia et al. (2007) (SOS1 mutations in NS), Carta et al. (2006); Pandit et al. (2007); Cordeddu et al. (2009); Cirstea et al. (2010) (KRAS, NRAS, RAF1, and SHOC2 mutations in NS and related conditions), Aoki et al. (2005, 2013) (HRAS mutations in CS), Flex et al. (2014) (RRAS mutations in a RASopathy condition prone to cancer), Yamamoto et al. (2015) (SOS2, LZTR1 mutations in NS), Martinelli et al. (2010, 2015); Pérez et al., 2010 (CBL mutations in a developmental syndrome prone to cancer), Urosevic et al. (2011); Aoidi et al. (2018) (BRAF, MEK1, or MEK2 mutations in CFCS), and Capri et al. (2019) (RRAS2 mutations in NS).
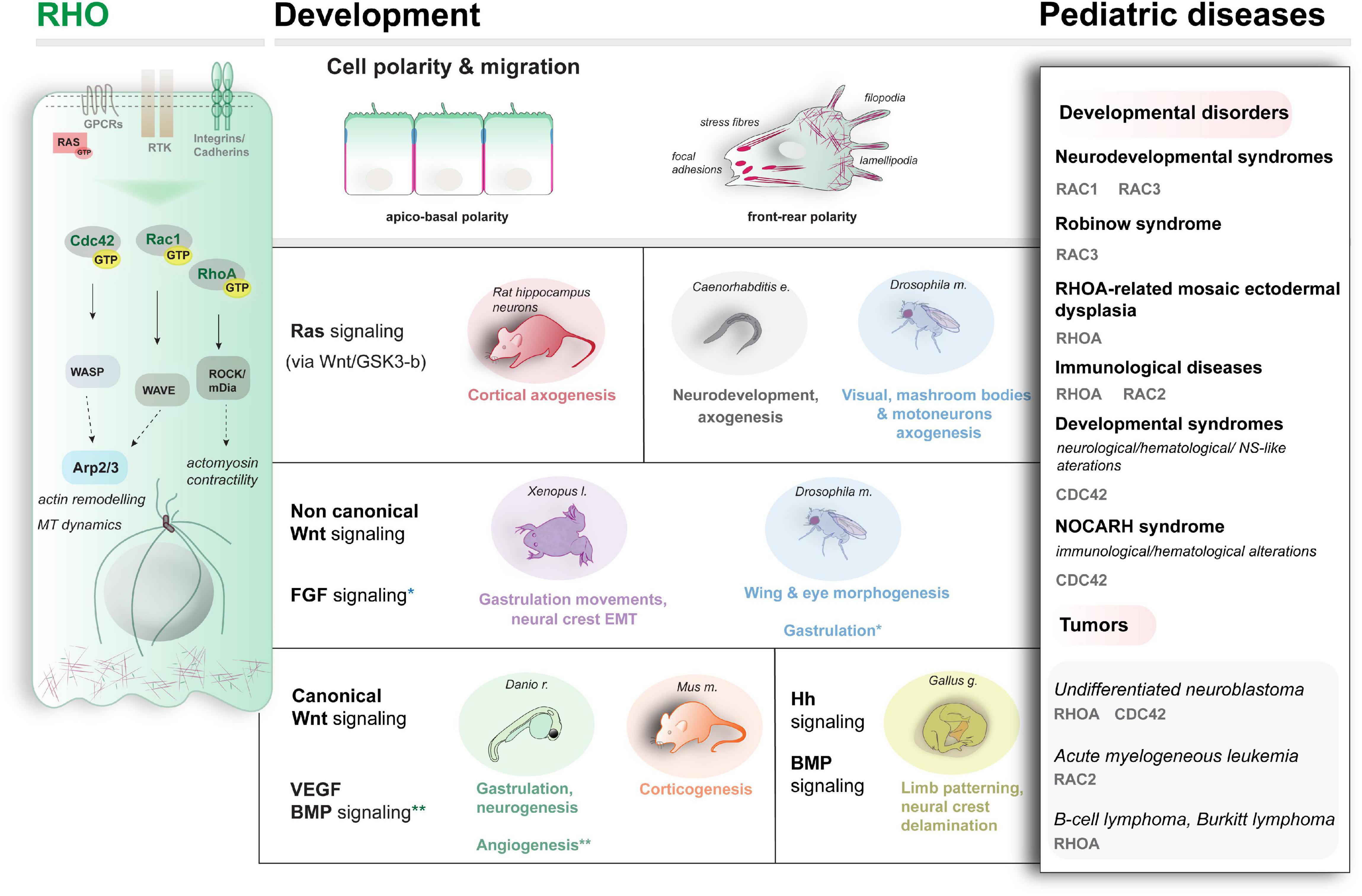
Figure 2. Schematic representation of the main RHO proteins’ activity (left), examples of the developmental pathways and processes modulated by RHO (center) and examples of genetic conditions associated with altered Rho activity (right).
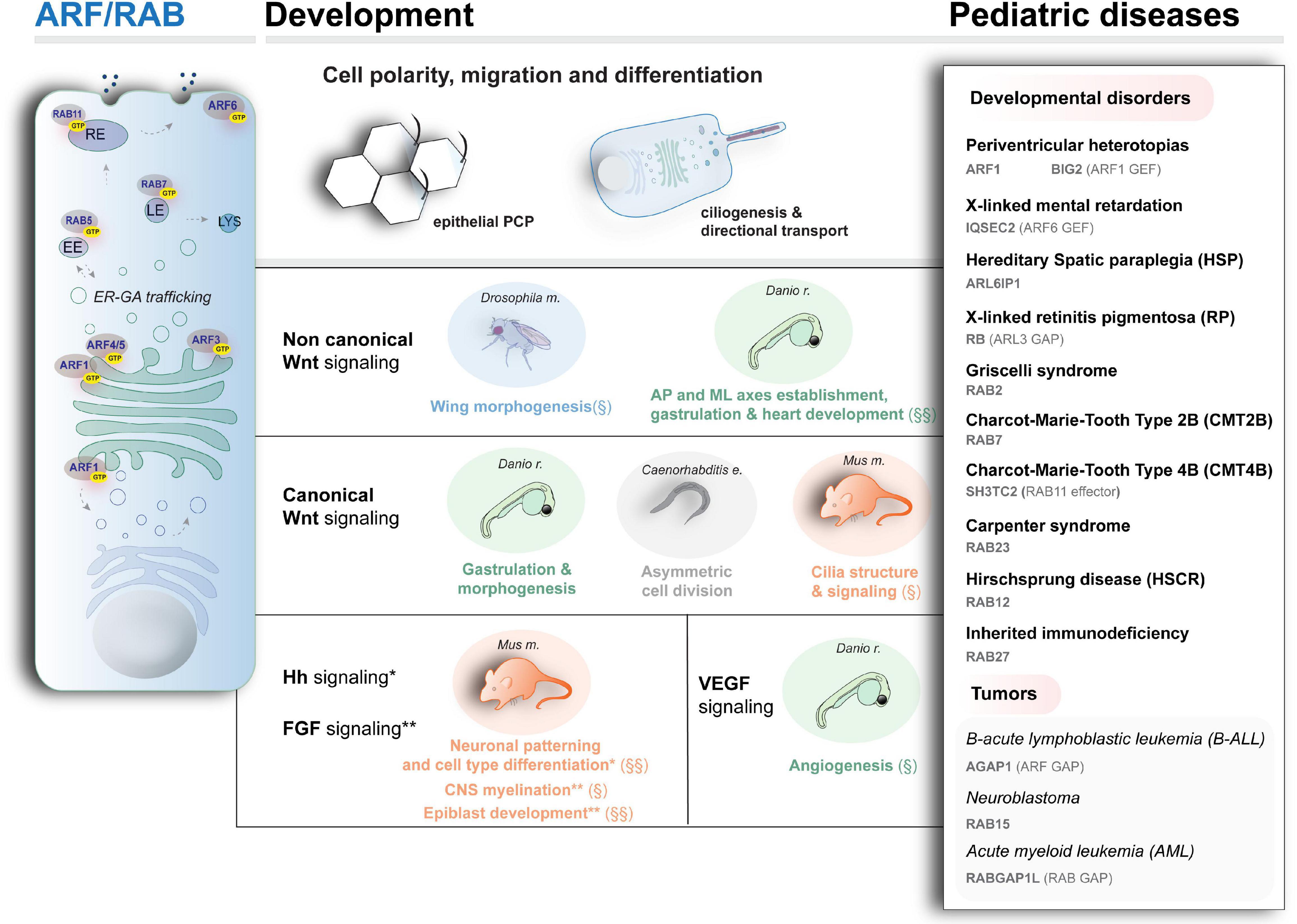
Figure 3. Schematic representation of intracellular trafficking and organelles associated with ARF and RAB activity (left), the main developmental pathways and processes modulated by vesicular trafficking (center), and examples of rare diseases caused by mutant ARF and RAB proteins. § and §§ indicate processes associated with ARF and RAB proteins, respectively. No symbol indicates evidence for processes involving both ARF and RAB activity. EE, LE, RE: early, late, and recycling endosomes, respectively. LYS: lysosomes. For the disease genes and diseases depicted here, besides the literature cited in the text, refer to Novarino et al. (2014); Wakil et al. (2019) (ARF-like 6 interacting protein 1 mutations in neuropathy with spastic paraplegia, microcephaly, leukoencephalopathy, and seizures); Griscelli and Prunieras (1978) (mutations in RAB2 in Griscelli syndrome affecting the immune system); Cogli et al. (2009); Lupo et al. (2009); Stendel et al. (2010), for the motor and sensory neuropathies Charcot–Marie–Tooth type 2B and type 4 (CMT2B and CMT4), caused by mutations in RAB7 and RAB11 effector SH3TC2, respectively; Harvey et al. (2010) (mutations in the ARF-specific GAP encoding gene AGAP1 in pediatric high-risk B-cell ALL); Roberti et al. (2009) (gene rearrangement in the RAB-specific GAP encoding gene RABGAP1L in patient with Klinefelter syndrome who developed AML).
RAS/MAPK Signaling Cascade and Its Dysregulation in Developmental Disorders and Cancer
Since the identification of the RAS proteins in the 1980s, the biochemistry of their signaling and their role as regulators of multiple cellular processes (e.g., proliferation, survival, differentiation, and apoptosis) in development and homeostasis has been intensively examined (Downward, 1998; Drosten et al., 2010; Kang and Lee, 2019). Various growth factors, cytokines, and hormones activate the Ras signaling network leading to the MAPK cascade and other pathways (Schlessinger, 2000) in a highly conserved manner (Rojas et al., 2012). A schematic overview of the Ras/MAPK signaling pathway is shown in Figure 1, left. Briefly, autophosphorylation of cell surface receptor tyrosine kinases promotes membrane recruitment of guanine nucleotide exchange factors (GEFs, e.g., SOS). This triggers the binding of RAS to GTP, activating the signaling (Cox and Der, 2010). The phosphatase SHP2 further contributes to RAS activation by inactivating regulatory tyrosines in receptors and scaffolding proteins (Matozaki et al., 2009). On the other hand, deactivation of RAS is promoted by GTPase-activating proteins (GAPs), such as neurofibromin 1 (NF1), via positive regulation of GTP hydrolysis (Vigil et al., 2010) or via ubiquitination directly by a recently identified cullin3-RING ubiquitin ligase complex (Steklov et al., 2018; Motta et al., 2019; Abe et al., 2020) and via the E3 ubiquitin ligase CBL by functional downmodulation of the activated cell surface receptors (Mohapatra et al., 2013). Downstream kinases (e.g., RAF1) are responsible for translating Ras signaling into the activation of MAPK ERK kinases (MEKs), resulting in the activation of ERKs (extracellular signal–regulated kinases), which phosphorylate various cytoplasmic and nuclear targets to mediate cellular responses. A phosphatase complex (MRAS, SHOC2, and PP1CB) dephosphorylates a single inhibitory site on RAF kinase, activating signal flow through the MAPK cascade. Depending on the cellular context, the strength and length of signaling, proliferation, apoptotic, or differentiation signals can be triggered (Downward, 1998; Murphy et al., 2004; Drosten et al., 2010; Kang and Lee, 2019).
Considering the plethora of functions of Ras/MAPK signaling during development, it is clear that mutations affecting signaling backbone’s core components have various deleterious effects in terms of development. Indeed, germline mutations affecting different components of signaling cascade are responsible for RASopathies, a group of developmental disorders comprising Noonan syndrome (NS), LEOPARD syndrome (NS with multiple lentigines), Costello syndrome (CS), cardiofaciocutaneous syndrome (CFCS), neurofibromatosis type 1 (NF-1), and other clinically related disorders, displaying high genetic and clinical heterogeneity (for a comprehensive review of the work in the field, refer to Cox and Der, 2010; Tartaglia et al., 2011; Rauen, 2013; Simanshu et al., 2017; Tajan et al., 2018; and more recently Kang and Lee, 2019). It is also equally established that activating mutations in genes encoding members of the Ras/MAPK signaling are commonly associated with cancers (Malumbres and Barbacid, 2003). They are among the primary causes of several pediatric malignancies affecting the nervous system, such as gliomas and astrocytomas, which show the highest degree of mortality in children. Myeloproliferative syndromes, such as juvenile myelomonocytic leukemia (JMML) and pediatric acute lymphoblastic leukemia (ALL) are also characterized by hyperactivated Ras/MAPK signaling (Zhang et al., 2011; Rauen, 2013). Of note, pediatric patients affected by NS, CS and NF show increased cancer predisposition (Brems et al., 2009; Kratz et al., 2015) with a high incidence of leukemia (Emanuel, 2004; Strullu et al., 2014) and other cancers [i.e., neuroblastoma or rhabdomyosarcoma (RMS); Moschovi et al., 2007].
After the discovery of germline gain-of-function mutations in PTPN11 (encoding SHP2), the first gene involved in NS and LS (Tartaglia et al., 2001; Digilio et al., 2002; Legius et al., 2002), and of somatic activating mutations in contributing to JMML and acute leukemia (Tartaglia et al., 2003; Jongmans et al., 2011), enormous work was carried out via NGS, which disclosed a plethora of new disease-causing genes and mutations. Combined with increasingly sophisticated functional investigations, this approach is contribuiting to depict a complex mechanism of action of Ras/MAPK signaling in the pathophysiology of diseases (Kang and Lee, 2019). Besides the cases discussed here, an overview of the main members of Ras/MAPK signaling for which mutations have been described over the years that associate with developmental diseases and malignancies is shown in Figure 1 (right). For clinical and genetic review, refer to Tartaglia and Gelb (2010); Tidyman and Rauen (2016); Tajan et al. (2018). For mechanistic investigations, several animal models are available, which recapitulate various aspects of RASopathies (Jindal et al., 2015; Tajan et al., 2018). Among these, NS mice models showcase the impact of PTPN11 mutations on neuronal and glial cell development (Araki et al., 2004; Gauthier et al., 2007; Ehrman et al., 2014), whereas in zebrafish an NRAS-depending NS phenotype rescued by MEK inhibitors was successfully modeled (Runtuwene et al., 2011). Moreover, besides rodents (Schuhmacher et al., 2008), fish models for CS caused by hyperactive HRAS-G12V variant are also available, which recapitulate the human condition and associate with tumorigenesis (Santoriello et al., 2009).
Recent Genetic Findings in Rasopathies and Pediatric Tumors
A large repertoire of genetic conditions described since the discovery of PTPN11 mutations, and more genes and mutations linked to the Ras/MAPK cascade and impacting developmental programs as well as cancer onset continue to be characterized by NGS and functional genomics efforts (Figure 1, right). Among the recent findings, loss of function and dominant negative mutations in LZTR1, which disables the ubiquitination of RAS and thereby the suppression of the signaling, were found in NS and pediatric cancers (Bigenzahn et al., 2018; Steklov et al., 2018; Motta et al., 2019; Abe et al., 2020). Noteworthy, Gröbner et al. (2018) have recently established LZTR1 mutations as a hereditary factor predisposing to pediatric retinoblastoma, hypodiploid B-cell ALL, and high-grade glioma K27wt. Inactivating mutations of LZTR1 have also been associated with drug resistance in RAS-induced chronic myeloid leukemia (Bigenzahn et al., 2018), whereas another class of both loss of function and dominantly acting LZTR1 mutations seems to predispose to the development of glioblastoma and adult-onset schwannomatosis (a rare cancer-prone disorder) (Piotrowski et al., 2014; Paganini et al., 2015; Motta et al., 2019).
Of note, Drosophila and murine models exist for this condition, which showed the involvement in morphogenesis (Bigenzahn et al., 2018) and in Schwann cells’ behavior to shift from quiescent supporting cells into a highly dedifferentiated and proliferating state (Steklov et al., 2018). The recent establishment of vital CRISPR/CAS-based zebrafish lztr1 null models expands further the possible comparative work (Nakagama et al., 2020).
Lastly, WES sequencing in a group of patients showing neurodevelopmental alterations within the RASopathy spectrum coupled to functional validation in nematodes has more recently established the pathogenicity of de novo mutations affecting MAPK1 (ERK2) directly and possibly their ability to interact with regulators and effectors (Motta et al., 2020). The underlying mechanism and plausible perturbance of fine signaling balances within developmental programs remain to be characterized.
Functional Relevance of Developmental Signalings’ Interplay Involving RAS/MAPK for Cell-Type Specification, Rasopathies, and Pediatric Tumors
The differential impact of the Ras/MAPK signaling during embryogenesis likely depends largely on the cross-modulatory signaling interplays in which RAS activity is involved, which contribute to determine combinatorial codes that stir developmental competencies into specific cell fate (Halfon et al., 2000). A schematic overview of some of the main developmental signalings and processes involving the Ras/MAPK pathway is shown in Figure 1 (center). Functional studies in convenient model systems (such as the developing vulva in nematodes, the compound eye in insects, the somites’ development in zebrafish and the rodent brain) are contributing to tease apart some of these interplays with relevant developmental pathways such as Notch (Sundaram, 2005) and Wnt (Jeong et al., 2018) and others, as shown by the examples below.
Notch Signaling
Compelling evidence demonstrates that Ras/MAPK signaling is able to modulate Notch pathway both positively and negatively in various embryonic precursor fields, contributing to the critical balance between inductive and inhibitory inputs, which instruct cell-type specification within a single equivalence domain. For instance, this mechanism is instrumental in generating lateral inhibition in progenitor cells during vulva patterning in developing Caenorhabditis elegans (Levitan and Greenwald, 1998; Chen and Greenwald, 2004; Sternberg, 2005).
The sequential interplay and various modes of crosstalk between Ras/MAPK and Notch inputs influence also progenitor specification during the development of prospective R photoreceptor cells (Tsuda et al., 2002; Roignant and Treisman, 2009), wing (Marygold et al., 2011), muscle and cardiac tissue in Drosophila (Carmena et al., 2002). Specifically, the activity of epidermal growth factor (EGF)-triggered Ras/MAPK cascade induces photoreceptor identity in the developing eye while promoting the expression of the Notch ligand Delta in the same cells, via the control of the transcriptional corepressor complex Ebi (transducin β-like 1, TBL1, in mammals). This mechanism based on inhibitory and inductive signals contributes to the acquisition of non-neuronal identity by neighboring cells and therefore to the global functional patterning of the differentiating cell clusters within the developing compound eye (Tsuda et al., 2002; Roignant and Treisman, 2009).
Moreover, in Drosophila wing and zebrafish somites’ development, EGF-dependent Ras/MAPK signaling induces reduction of the repressor activity of Groucho (Gro) to downmodulate Notch-controlled transcriptional output and likely that of other developmental pathways (Hasson et al., 2005).
A mammalian example is offered by rodent brain development. Here the establishment of cell identity is normally influenced by Ras/MAPK signaling, which contributes to balance neuronal and glial differentiation (elegantly summarized by Kang and Lee, 2019), regulating directly the expression of distinct proneural genes (e.g., Neurog2 or Ascl1) during corticogenesis (Li et al., 2014). In this developmental context, a correct interplay between Ras/MAPK and Notch is likely crucial for myelinogenesis and relevant for pathology. Indeed, patients affected by NF-1, caused by dominant inactivating mutations in NF1 resulting in an increase the active GTP-bound RAS (Cawthon et al., 1990; Wallace et al., 1990), show pronounced myelin damage, and decompaction is observed, as well a predisposition to life-threatening tumors (Brems et al., 2009; Ratner and Miller, 2015). Accordingly, recent in vivo experiments in mice demonstrated that NF1 loss of function, resulting in sustained Ras/MAPK signaling, causes myelin defects underlying a deregulation of Notch activity (López-Juárez et al., 2017).
On the other hand, an extensive collection of studies indicates the importance of a complex pathway interplay involving Ras/MAPK and Notch also in tumorigenesis (Fitzgerald et al., 2000). For instance, in neuroblastoma, which represents at least 8% of all pediatric cancers (Colon and Chung, 2011), transforming growth factor β–induced Ras signaling positively regulates the Notch pathway (Stockhausen et al., 2005). Mechanistically, work in rodents traced back RAS and Notch’s activity during early development of nestin + glial cells, which, if dysregulated, might trigger cancerogenic lesions at the level of the subventricular zone in gliomas (Shih and Holland, 2006). Lastly, rodent models also offered evidence that a mild hyperactivation of Notch1, behind the dose required for normal T-cell differentiation during development, contributes to leukemia onset, synergically with RAS activation (Chiang et al., 2008).
Wnt Signaling
Evidence for a modulatory function of Ras/MAPK on Wnt in development is available from various animal models. Different modes of crosstalk between these two signalings have been described that contribute to adequate cellular response in different developmental contexts and timings. In the insect imaginal disk of the developing wing, for instance, tissue patterning is controlled via a conserved MAPK cascade downstream insulin-like growth factor, which regulates canonical Wnt pathway by stabilizing the β-catenin effector, thanks to a direct interaction with the ortholog of MEK1/2 (Hall and Verheyen, 2015).
Offering another type of example of the impact of cross-modulatory activity between Ras/MAPK and Wnt signaling, experiments in the invertebrate sea urchin suggested that maternally deposited β-catenin drives transcription of MEK and of the RAS target Ets1 during ectodermal-to-mesenchymal transition (EMT) in migrating skeletal precursor cells (Rottinger et al., 2004).
On the other hand, as suggested by zebrafish disease models, Ras/MAPK pathway seems to mediate the activity also of non-canonical Wnt signaling during vertebrate gastrulation (Kilian et al., 2003; Jopling et al., 2007). As an example, overexpression of shp2 mutant variants in zebrafish embryos, recapitulating human NS and LS traits, indicates a Wnt-dependent effect of Shp2 on embryonic convergence and extension (CE) movements, resembling phenotypes found by downregulating the non-canonical Wnt (Kilian et al., 2003; Jopling et al., 2007). Zebrafish work further linked Wnt-dependent Shp2 activity even to RhoA signaling (Jopling et al., 2007), similarly to an interplay described already in frog development (O’Reilly et al., 2000). Of note, the activity of SHP2 seems to be crucial to influence also other signaling pathways relevant to developmental diseases and tumorigenesis, such as Hippo and Shh (Huang et al., 2014).
Moreover, Drosophila mesoderm specification shows a good example of a rather complex cross-modulation of the Ras/MAPK cascade on Wnt pathway for signal integration. In eve + domain, the muscle prepatterning signaling of the Wnt and TGF-β orthologs induces activation of Tin and Twi, which function together with Ras-activated Ets protein in tissue-specific enhancer domains to establish muscle and cardiac identity (Halfon et al., 2000).
Further manifesting the complexity of the interplays and multilevel integration of these signalings during development, the insect PDZ domain-containing protein called Canoe (Cno) mediates the crosstalk between Ras-, Notch-, and Wnt-induced pathways via interacting with disheveled (Dsh/Dvl). This Wnt-dependent mechanism seems to facilitate Ras induction and Notch signaling to finely modulate their relative signal intensities throughout mesoderm specification (Carmena et al., 2006). Similar interactions to induce R7 photoreceptors were observed in the developing compound eye (Cooper and Bray, 2000). Demonstrating the relevance of this crosstalk for pathology, in severe forms of acute myeloid leukemia (AML), a translocation event involving AF6, the human ortholog of Cno, triggers RAS activation (Manara et al., 2014; Smith et al., 2017). Nevertheless, a clear perturbation of Wnt and Notch remains to be proven in this context.
FGF Signaling
The interaction between Ras/MAPK on the FGF signaling is also normally necessary in various developmental contexts during cell proliferation, migration, and differentiation (Tsang and Dawid, 2004). For instance, in Drosophila, multiple interplays were shown that instruct mesoderm migration and muscle specification (Lin et al., 1999). In addition, an intermediary role of FGF signaling in the Ras/MAPK-dependent activity on suppression of Notch-induced HES transcription factors was demonstrated in the context of both insect wings and zebrafish somites’ development (Kawamura et al., 2005). A crucial integration of Ras, FGF, and Notch signaling was also shown for muscle and cardiomyocyte specification (Carmena et al., 2002). Pointing to the importance of a possible dysfunctional interplay between FGF and Ras/MAPK in early embryological events for developmental disease etiology, studies in zebrafish models of CFCS in vivo demonstrate that perturbation of gastrulation movements due to hyperactive Ras is rescued by inhibiting FGF signaling (Anastasaki et al., 2009).
On the other hand, a clear crosstalk of FGF on Ras/MAPK also contributes to the balance between cell-type specifications instrumental to brain development. Li et al. (2012) demonstrated that neural stem cells lacking MEK1/2 activity fail to produce glial cells, a mechanism likely acting via modulating an stromal derived factor (SDF-1) input and FGF signaling, as shown for astrocyte development (Bajetto et al., 2001; Song and Ghosh, 2004; Dinh Duong et al., 2019). In addition, it was proven that the regulatory activity of FGF on Ras/MAPK via SOS/Grb2 and Stump seems particularly important for the correct neuromuscular junction (NMJ) formation during Drosophila nervous system development via synergistic action of Smn1, which positively regulates FGF pathway components (Sen et al., 2011). In insect models of human spinal muscular atrophy (SMA), a severe autosomal recessive neurodegenerative disease caused by mutation in SMN1 and a primary cause of death in children (Lefebvre et al., 1995), alteration of this interplay seems to contribute to the NMJ defects observed. The impact of a possible dysregulation of Ras/MAPK and FGF for the human SMA condition remains to be assessed.
RHO Proteins and Their Involvement in Heterogeneous Neurodevelopmental and Hematological Disorders
The RHO family of small GTPases is a large group of proteins (>20) within the RAS superfamily. RAC1, RAC2, RHOA, and CDC42 are classical members regulated by cycling between an active and inactive state via hydrolysis of GTP (Haga and Ridley, 2016; Etienne-Manneville and Hall, 2002; Heasman and Ridley, 2008). Atypical proteins with no intrinsic GTPase activity also exist (for a general survey, refer to Heasman and Ridley, 2008; Ji and Rivero, 2016). By interacting with a myriad of effectors and other small GTPases (Etienne-Manneville and Hall, 2002; Schmidt and Hall, 2002; Ridley et al., 2003; Nouri et al., 2020), RHO proteins control cell polarity establishment and trafficking, cell shape, and motility in health and disease (Ellis and Mellor, 2000; Etienne-Manneville and Hall, 2002; Hall, 2005; Ridley, 2006; Govek et al., 2011, Haga and Ridley, 2016; Lawson and Ridley, 2018; Ueyama, 2019; Boueid et al., 2020), via direct actin-cytoskeletal and MT rearrangements, to generate protrusive and contractile forces by means of filopodia (CDC42), lamellipodia, and stress fibers (RHOA and RAC1) (Etienne-Manneville and Hall, 2002; Heasman and Ridley, 2008; Figure 2). Molecularly, active RHO proteins exert their role by regulating their spatial distribution in the cell (e.g., by shuttling between the cell membrane and the Golgi). Among RHO small GTPases, CDC42 has been extensively studied. By promoting actin-rich filopodia formation via direct activation of N-WASP, Arp2/3, and formin (Etienne-Manneville and Hall, 2002; Mellor, 2010), CDC42 generates polarized cell migration (Govek et al., 2011; Zhang et al., 2014), contributes to various polarized processes underlying morphogenesis, as shown in yeast (Adams et al., 1990; Evangelista et al., 1997; Wedlich-Soldner et al., 2004), nematodes (Kay and Hunter, 2001) and vertebrates (Stanganello et al., 2015). Of note, CDC42 seems to regulate also polar vescicular trafficking as shown in various organisms (Harris and Tepass, 2010).
The importance of RHO proteins in early developmental schemes is illustrated by the embryo lethality often observed in mutant mice models (Sugihara et al., 1998; Liu et al., 2017). As discussed below, particularly important is the impact of RHO proteins on both brain and hematological development. Thanks to the advances of NGS, aberrant Rho signaling caused by mutations affecting multiple genes is emerging also as a prominent cause of clinically heterogeneous neurodevelopmental and hematological rare disorders, which include pediatric cancers. In addition, the numbers of genes (>20) encoding key players of Rho signaling were recently classified as risk factors for autism spectrum disorders by the Simons Foundation Autism Research Initiative, which was backed up by mice models (Guo et al., 2020).
At least 30% of neuroblastoma cases are indeed due to mutations that alter RHO and RAC activity (Dyberg et al., 2017), involved in the migration of neural crest (NC) cells from which neuroblastoma originates. An overview of the genetic conditions linked to RHO proteins is summarized in Figure 2, right (refer to Boueid et al., 2020 for an up-to-date review).
Brain Formation and Neurodevelopmental Diseases
Extensive work in rodent models highlighted the importance of RHO-dependent neuronal precursors’ mobility and radial glia expansion processes for neuronal circuit establishment and maturation (i.e., corticogenesis) (Govek et al., 2011; Ito et al., 2014; Azzarelli et al., 2015; Xu et al., 2019; Heide et al., 2020). Developmental pathways used by growing axons for initiation, extension, and target innervation depend on Rho signaling in vivo (Hall and Lalli, 2010). Accordingly, classical studies using inactive or constitutively active mutants demonstrated the requirements of RAC1-dependent actin remodeling in axon guidance for insect motoneurons innervation (Kaufmann et al., 1998), and similar functions were confirmed in the visual and mushroom body circuits (Hakeda-Suzuki et al., 2002; Ng et al., 2002), as well as in nematode development (Shakir et al., 2006). Rac1 forebrain knockout (KO) mice showing microcephaly further confirmed a function in vertebrate neuronal migration, proliferation, and dendritic arborization (Chen et al., 2009; Leone et al., 2010).
Of note, the fruitful combination of WES carried on a large cohort of heterogeneous undiagnosed conditions coupled to in vitro and in vivo functional approaches has recently contributed to map and validate a number of de novo mutations in RAC1 in patients affected by a range of developmental defects, including brain malformations (Reijnders et al., 2017). Similarly, independent studies employing exome and genome sequencing have recently identified also dominant RAC3 mutations as causative of neurodevelopmental diseases with divergent clinical features, such as the rare Robinow syndrome–like disorder, which shows also impaired skeletal development (White et al., 2018; Costain et al., 2019).
Moreover, experimental evidence shows a fundamental role also of RHOA in cell-type specification of developing brains (Dupraz et al., 2019) and neurite outgrowth (Kouchi et al., 2011; Tan et al., 2020), and this protein was recently involved in a newly discovered syndrome, the “RHOA-related mosaic ectodermal dysplasia,” with clear signs of leukoencephalopathy and anomalies in NC derivatives (Vabres et al., 2019).
Among the other RHO proteins, also CDC42 participates in brain development. The protein regulates polarization and motility in neuronal precursors (Govek et al., 2011; Govek et al., 2018). By acting directly on PAR complex, numb, and E-cadherin, CDC42 orchestrates apicobasal trafficking, spindle orientation, and influences adherens junction integrity, as shown during Drosophila neuroepithelium development (Georgiou et al., 2008; Harris and Tepass, 2008) but also in C. elegans and other eukaryotes (Gotta et al., 2001; Kay and Hunter, 2001; Harris and Tepass, 2010), and during the normal development of various tissues and organs in mammals (Melendez et al., 2013; Elias et al., 2015). In addition, conditional mice models have been already useful to prove the importance of CDC42 and RAC in the establishment of cell polarity for acquiring the specialized cell morphology in the context of cochlear hair cells (Ueyama et al., 2014; Kirjavainen et al., 2015) and in hippocampal axonal formation (Garvalov et al., 2007).
Lastly, unexplored signaling via MT during migration is seemingly able to activate RHO molecules within a feedback loop, which has a great impact on neuronal polarity establishment (Wojnacki et al., 2014). However, the mechanism awaits confirmatory in vivo analysis.
Hematological Development and Disease
A large body of evidence in vitro and in animal model systems demonstrates a crucial function of RHO proteins also in immune cell development and physiology (Nayak et al., 2013).
Rodent models illustrate indeed a unique role of RAC2 for chemoattractant-dependent neutrophil migration and oxygen radical production during immune response to infections (Troeger and Williams, 2013). CDC42 has an equally important role in this developmental context, by controlling the events at the front and back of migratory immune cells via the integration of integrins, WASP protein, CD11b, and MT signaling (Kumar et al., 2012). Confirming the relevance of RHO proteins in hematopoietic cell development and disease, by using WES, several authors have recently established the pathogenicity of various dominantly acting mutations in RAC2, which cause pediatric immunodeficiencies affecting T, B, and myeloid cells (including Hsu et al., 2019; Sharapova et al., 2019; Lagresle-Peyrou et al., 2020). These results were also substantiated by mice models (Hsu et al., 2019). Fish models also exist, which showed an involvement of the ortholog version of RAC2 in controlling neutrophils and leukocytes’ behavior (Rosowski et al., 2016). Given the importance of cytoskeletal dynamics in modulating immune development and response, and cell migration in general, it is not surprising that mutations in RHO proteins alter important features of hematopoietic stem cells (HSCs) and contribute to tumorigenic and metastatic processes involving these cells (Maldonado and Dharmawardhane, 2018). RAC2 genetic lesions were indeed observed in acute myelogenous leukemia (Ross et al., 2004; Thomas et al., 2008). Independent mice models show that active CDC42 causes aging in HSC with impairment in cell polarity and function, a condition that might be linked to aging and myeloid tumorigenesis (Kerber et al., 2009; Geiger and Zheng, 2013). Based on these considerations, inhibiting CDC42 function has been recently proposed as a valid approach to ensure long-term HSC mobilization as a therapeutic tool for various blood diseases (Liu et al., 2019).
Newly Discovered Syndromes Linked to Aberrant CDC42
Multiple lines of evidence from human disease studies and in vivo models are also pointing to a broad impact of dysregulated CDC42 function in various processes, which impact both brain and hematological development. Indeed, Martinelli et al. (2018) linked dominantly acting missense mutations causing variably malfunctioning of CDC42 to an unusually heterogeneous group of developmental conditions (including RASopathy traits), mainly characterized by variable growth dysregulation, neurodevelopmental defects with impaired hearing and vision, and immunological and hematological anomalies. The mutations altering variably the interaction with regulatory and signaling effectors impacted cell migration and nematode vulva morphogenesis, likewise in RASopathies models caused by altered Ras/MAPK signaling.
Noteworthy, a peculiar mutation in CDC42, which locks the protein in the Golgi, was more recently proven to be causative of a complex and previously undiagnosed life-threatening autoinflammatory condition, NOCARH syndrome (neonatal onset cytopenia with dyshematopoiesis, autoinflammation, rash, and hemophagocytic lymphohistiocytosis), impairing hematological development (Lam et al., 2019). HSCs of NOCARH patients had reduced responsiveness to proliferation stimuli and immune response due to altered cell polarity (Lam et al., 2019). It would be interesting to test the effect of CDC42 inhibition approach (Liu et al., 2019) on HSC cell behavior of future NOCARH in vivo experimental settings.
RHO-Mediated Developmental Signals Integration in Cell Polarity, Morphogen Distribution, and Relevance for Pathology
Although more investigation is needed, we now know that RHO proteins exert their function also by acting as molecular switches on several signaling pathways during embryogenesis. A schematic overview of some of the main signaling and events influenced by RHO small GTPases is illustrated in Figure 2 (center). Indeed, the clinically broad spectrum of RHO-linked diseases might reflect the pleiotropic impact of RHO proteins’ functions on modulating and interpreting the different signalings. However, the implications for disease etiology are poorly investigated, which calls for systematic functional profiling of the mutations in the context of vertebrate development, currently lacking. We examine here some examples of the available evidence for signaling interplay also in the context of pediatric diseases.
PCP and Wnt Signaling
RHO requirement for signal integration on non-canonical Wnt signaling–meditated planar cell polarity (PCP) was shown in Drosophila mutants defective in wing and eye morphogenesis, as well as in fish, frogs, and other models (Schlessinger et al., 2009). In addition, RHO-dependent actin rearrangement and polarity establishment for PCP-dependent CE cell movements during gastrulation were demonstrated in mammalian cells and in the context of frog and zebrafish gastrulation (Habas et al., 2001, 2003; Marlow et al., 2002). On the other hand, a growing body of data in animal models proves the interplay between RHO and Wnt during embryonic NC migration (reviewed by Mayor and Theveneau, 2013). In this context, Kratzer et al. (2020) have recently provided evidence for a novel and a complex modulatory mechanism acting during frog development, where Rho GEF Trio activates Rac1 at the level of cell protrusions of migratory cranial NC via interaction with Dvl, a major player of PCP signaling (Gao and Chen, 2010), similar to other Dvl-dependent mechanisms seen for Rho activation in Xenopus, Drosophila, and zebrafish development (Schlessinger et al., 2009). It is also becoming clear that RAC1 is involved in a positive regulation of canonical Wnt signaling by enabling the nuclear accumulation of β-catenin (Wu et al., 2008; Schlessinger et al., 2009).
Similar to what was observed in mammalian cells, classical Drosophila wing and eye systems, as well as Xenopus embryo models, were useful to demonstrate the necessity of several RHO GTPases in mediating actin cytoskeleton modifications via non-canonical Wnt signaling during development (Strutt et al., 1997; Habas et al., 2003; Mezzacappa et al., 2012).
When it comes to relevance for pathology, the involvement of Wnt pathway alteration in RHO-associated diseases begins now to emerge. For instance, although the molecular mechanism of the RAC3-linked Robinow syndrome–like disorder (Costain et al., 2019) remains unsolved, the disease is normally associated with Wnt signaling alterations, which would be interesting to validate in vivo functionally in relation to disease etiology (White et al., 2018). In addition, lack of CDC42 results in loss of apical molecules’ distribution throughout rodent telencephalon development, including the canonical Wnt effector β-catenin, and determines Shh-independent holoprosencephaly (Chen et al., 2006).
Lastly, a role for RHOA-dependent kinase alteration underlying non-canonical Wnt (PCP) pathway is emerging also for neuroblastoma (Becker and Wilting, 2019) and B-cell precursor ALL (Karvonen et al., 2019). Recent work on KRAS-G12D–induced zebrafish models of embryonal RMS and in vitro human RMS has demonstrated a crucial role of hyperactive RHOA in the promotion of tumor propagating cell self-renewal downstream Vangl2, a classical non-canonical Wnt regulator, under a similar molecular axis known in embryogenesis (Hayes et al., 2018).
FGF and VEGF Signaling
RHO proteins are able to integrate a number of other signalings relevant in different contexts during normal embryonic development. Their activity is essential for orchestrating the molecular dynamics needed for cell shape changes, for example, by assisting actin–myosin ring formation for apical cell constriction during gastrulation (Nikolaidou and Barrett, 2004; Dawes-Hoang et al., 2005). Experiments in Drosophila models showed that the modulatory activity on FGF signaling contributes substantially to these processes and ultimately to mesodermal cell motility (Smallhorn et al., 2004). Specifically, it was demonstrated that insect Rho GEF pebble, normally involved in Rac1 and Rac2 activation (van Impel et al., 2009), can modulate FGF-mediated Ras/MAPK signaling to establish EMT conversion and mesodermal cells migration (Smallhorn et al., 2004).
On the other hand, RHOA is fundamental for the reorganization of the F-actin during cytoskeleton remodeling in endothelial cells and angiogenesis, which is highly relevant for pathology onset and progression (Merajver and Usmani, 2005). Importantly, both in vitro and in vivo models showed that cytoskeletal dynamics tuned by RHO molecules act under direct VEGF signaling to impact migratory movements and trafficking of endothelial cells during angiogenesis (Soga et al., 2001; van Nieuw Amerongen et al., 2003; Garret et al., 2007; Lamalice et al., 2007; El Baba et al., 2020). In this regard, inhibitors acting on the RHO/ROCK-mediated pathway are becoming a promising therapeutic approach for vascular endothelial growth factor (VEGF)-induced angiogenesis in the context of tumor progression and invasion (van der Meel et al., 2011; Chen et al., 2014).
PI3K Signaling
Experiments with photoactivable RHO versions in zebrafish demonstrated a direct involvement of Rho proteins in cell polarity, actin dynamic, and migration in neutrophils by the activity of PI3K signaling (Yoo et al., 2010). While demonstrating a crucial involvement of SDF-1/CXCR4-mediated Rac2 activity in limiting neutrophil mobilization, zebrafish models of primary immune deficiency caused by human RAC2 mutations or morpholino approaches indicate that the pathogenic role for RAC2 in immune cell physiology (Hsu et al., 2019; Sharapova et al., 2019; Lagresle-Peyrou et al., 2020) is linked to an altered PI3K-mediated cell polarity signaling in neutrophil migration during an inflammatory response (Deng et al., 2011). On the other hand, confirming the relevance of RHO modulatory activity on developmental signaling also for cancer, mutations in RHOA have been recently linked to B-cell lymphoma and Burkitt lymphoma via impaired PI3K pathway (Svensmark and Brakebusch, 2019; Voena and Chiarle, 2019).
Even a complex interplay between Ras and Rho signaling involving PI3K/AKT pathway and likely also canonical Wnt via GSK3-β modulation was shown in rat hippocampus neurons. A fine balance of signaling output in this crosstalk ensures the activity of CDC42 and RAC for regulating MT dynamics during axon initiation (Schwamborn and Püschel, 2004; Hall and Lalli, 2010).
The modulation of PI3K seems relevant also for brain tumor onset, as demonstrated by the increased expression of CDC42 through the PI3K/AKT/N-myc signaling pathway, which correlates with undifferentiated childhood neuroblastoma (Lee et al., 2014).
CDC42-Controlled Morphogen Distribution
Of particular interest for development and disease are in vivo findings demonstrating a unique mechanism by which CDC42 acts as a special signaling node for pathways directly regulating morphogen distribution in different developmental contexts. In vivo fish studies have demonstrated that Cdc42/N-Wasp filopodia act as “signaling extensions,” allowing fine control of morphogen propagation during development. Elegant genetic and imaging experiments in zebrafish embryos show that Cdc42-dependent filopodia determine short- and long-range propagation of canonical Wnt signaling and paracrine signal activation during vertebrate gastrulation, with major impact directly on the anterior–posterior (AP) axis and neurogenesis (Stanganello et al., 2015). This unique function of CDC42-induced filopodia was shown to contribute also to Hh signaling during avian tissue patterning (Sanders et al., 2013). In addition, work with zebrafish transgenic tools labeling intracellular structures and reporting and modulating Cdc42 activity carried at the single-cell precursor level in vivo suggested a BMP control of Cdc42-enriched filopodia necessary for in vivo endothelial cell motility during angiogenic sprouting (Wakayama et al., 2015). It is worth mentioning that control of morphogen asymmetric distribution, polarity, and signaling modulation seem to depend on RHO-like proteins also during plants’ root hair formation, suggesting a deeply conserved function in organismal development (Wu et al., 2011).
ARF and RAB-Mediated Biosynthetic Trafficking and Involvement in Pediatric Diseases
Biosynthetic trafficking is a highly conserved process crucial for setting signaling coordinates during organismal development and physiology (Biechele et al., 2011; Fernandez-Valdivia et al., 2011; Shimokawa et al., 2011). A number of proteins regulate and participate in this process, among which the small ARFs, including several classes, ARF-like (ARL) and RAB GTPases (>70 proteins in humans), which we discuss here, as well as related SAR proteins (Kahn et al., 2006). Mechanistically, like the other small GTPases, the function of ARF and RAB small GTPases is controlled via cycling between the GTP and GDP-bound forms by the action of specific GEFs and GAPs (Barr and Lambright, 2010; Sztul et al., 2019). Many of these proteins show a GTP hydrolysis-dependent spatial shuffling between cytoplasm and Golgi apparatus (GA), which is crucial to regulate important steps in the endoplasmic reticulum (ER)–GA network together with RAB proteins, such as coat proteins recruitment, vesicles biogenesis, cargo sorting, and signaling (Palmer et al., 1993; Reinhard et al., 2003; Figure 3). For a comprehensive review of ARF and RAB biochemistry, function, and on the role of membrane dynamics in development, refer to Wandinger-Ness and Zerial, 2014; Wada et al., 2016; Sztul et al., 2019; Marwaha et al., 2019.
Several ARF proteins, divided in three major classes based on their sequence homology, are involved in various steps of the intracellular trafficking, recruiting effectors for vesicle formation, budding, tethering, and cargo sorting as modulating actin and MT-based cytoskeleton (Sztul et al., 2019). On the other hand, RAB proteins distribute to specific subcellular compartments in combination with other proteins (e.g., tethering complex proteins or SNARE) to control vesicles formation and fusion via interaction with cytoskeleton components essential in development (Hutagalung and Novick, 2011; Wandinger-Ness and Zerial, 2014). These small GTPases have broad functions during development, ranging from gastrulation events to differentiation. Manifesting the importance of ARF and RAB during these processes, mutations affecting the function of these proteins, their regulators, and effectors have a deleterious effect on development and contribute to human pathology, with an important impact on the nervous system formation, as discussed below. In Figure 3, right, an overview is shown of the main genes (encoding key proteins belonging to or interacting with members of the ARF and RAB family) whose mutations have been associated with developmental diseases in which organelle’s dynamic and morphogen distributions are altered.
Further highlighting the extended domain of action in development and physiology, mutant RAB proteins also cause inherited pediatric immunodeficiencies (Griscelli and Prunieras, 1978) and in cancer. As an example, alterations of RAB15 alternative splicing, for instance, were linked to neuroblastoma tumor-initiating cells (Nishimura et al., 2011; Pham et al., 2012). In cancer, the aberrant activity of these proteins can modulate negatively various tumorigenic steps (including metastasis), where they can work both as oncogenes and tumor suppressors (a role recently reviewed by Casalou et al., 2020 and Gopal Krishnan et al., 2020). Besides the cases discussed below, the role of ARF and ARF-related proteins in animal development was recently reviewed in pathological contexts by Rodrigues and Harris (2019).
Maintenance of GA Integrity and Cytoskeleton Physiology by ARF
Of particular interest is the role of ARF proteins in the maintenance of organelle integrity, cytoskeleton remodeling and dynamics (Myers and Casanova, 2008; Kondo et al., 2012), highly relevant processes for organism development, brain formation, and neuronal circuits’ function, and which are involved in the onset of neurodevelopmental pathologies. Indeed, hyperactive ARF1 mutations induce loss of GA structure and fragmentation, which is likely mediated by COPI + vesicle budding in both healthy and diseased tissues (Zhang et al., 1994; Xiang et al., 2007). In fact, a series of genetic diseases affecting the developing nervous system caused by ARF and GA structural and functional alterations are emerging as “golgipathies” (Dupuis et al., 2015; Rasika et al., 2018) whose mechanisms need to be explored. Cancer-related GA fragmentation, often coupled to an increase of Ras/MAPK signaling and likely to alteration of ARF function, might even be a promising therapeutic target (Petrosyan, 2015).
Interestingly, GA alterations are also a hallmark of common forms of adult neurodegenerative processes (Rabouille and Haase, 2016), and cancers (Petrosyan, 2015) and ARF-mediated ER–GA trafficking perturbation were demonstrated in amyotrophic lateral sclerosis (ALS) (Zhai et al., 2015; Atkin et al., 2017). Of note, in nematode, superoxide dismutase 1–ALS disease models Arf proteins might even have a protective function on neurons (Zhai et al., 2015).
Moreover, employing rodent disease models, Bellouze et al. (2014) proved clear crosstalk between ARF-dependent trafficking and MT, which causes GA disintegration and is likely responsible for early onset neurodegeneration with progressive motor neuropathy (Schaefer et al., 2007; Sferra et al., 2016). Despite that further investigation is needed, it is tempting to speculate that a lack of function in the tubulin cofactor proteins (i.e., TBCE) in the affected children (with a possible loss of interaction with ARF1) (Bellouze et al., 2014) contributes to motoneuron degeneration (Schaefer et al., 2007; Sferra et al., 2016).
Via a specific GAP protein (RP2), also ARL2 and ARL3 influence MT dynamics and GA stability, as well as protein trafficking to the cilium, with a major impact on photoreceptor development in mice (Evans et al., 2010; Schrick et al., 2006). Accordingly, mutations affecting the ARL3 GAP protein RP were long shown to cause a severe form of X-linked retinitis pigmentosa, linked to altered GA stability and impaired trafficking to the photoreceptors’ cilia (Schwahn et al., 1998). Consistently, retinal degeneration was observed in Arl3 KO mice (Schrick et al., 2006), and mutant arl3 alters ciliogenesis in C. elegans (Li et al., 2010), with an important consequence on cilia signaling as discussed below.
Lastly, evidence for a crosstalk between ARF1 and actin–cytoskeleton regulators of RHO family was also shown. ARF1-mediated assembly of COPI complex is crucial for recruitment of CDC42 to the GA and for the local activation of N-WASP, Arp2/3, and actin polymerization, necessary to promote vesicle formation and scission (Wu et al., 2000; Myers and Casanova, 2008). An ARF1–RAC interaction was also shown for recruiting to the membrane WAVE (WASP family) and actin polymerization (Koronakis et al., 2011).
ARF and RAB Proteins’ Contributions to Set Signaling Coordinates Across Developmental Fields and Relevance for Pathology
Acting on shaping developmental signals strength and distribution and regulating cell behavior, ARF and RAB-controlled biosynthetic trafficking globally modulates morphogens’ distribution and function in developmental programs. The regulated process of polarized vesicular transport of morphogenes guarantees coordinated cell–cell signals and movements during development, ultimately leading to cell specification and organogenesis (Eaton and Martin-Belmonte, 2014; Wada et al., 2016; Rodrigues and Harris, 2019). Indeed, an aberrant function of ER and GA enzymes impairs proteins’ modification, with a deleterious impact on Wnt, Notch, and FGF signaling, as shown in insects and rodents (Biechele et al., 2011; Fernandez-Valdivia et al., 2011; Shimokawa et al., 2011). Furthermore, endosomes are emerging as important signaling platforms, mediating canonical Wnt signaling (Blitzer and Nusse, 2006) and sorting TGF-β signaling outcomes (Di Guglielmo et al., 2003; Felici et al., 2003). Examples of the direct influence of ARF and RAB function on developmental signaling in organismal context is also available (some of the main examples are schematized in Figure 3, center).
Here, we examine established and mounting evidence on the direct action of ARF and RAB-mediated intracellular trafficking on crucial developmental pathways, relevant for morphogenesis and disease. In general, these proteins have an active role in intracellular trafficking and cytoskeleton reorganization in cilia formation, directly regulating cilia development (Fisher et al., 2020), their general function, and cilium-dependent signaling pathways, including Shh and Wnt during organogenesis, as proven in different model systems (Huangfu and Anderson, 2005; Eggenschwiler et al., 2006).
PCP and Wnt Signaling
Despite poor mechanistic understanding, a clear contribution of ARF function in the modulation of both canonical and non-canonical Wnt signaling is emerging to be directly relevant for pathology.
Many developmental processes including proliferation and differentiation require controlled ARF-dependent biosynthetic pathways for establishing cell polarity. This was demonstrated in several developmental models and timings, i.e., gastrulation events (Lee et al., 2015), dendritic spine formation and growth in vertebrate hippocampal neurons (Jain et al., 2012), insect neuronal maturation (Chang et al., 2015), photoreceptor differentiation (Mazelova et al., 2009; Wang et al., 2012), and bone formation (Unlu et al., 2014), and is confirmed by genomic studies of developmental conditions.
Activating mutations affecting ARF1 result in a complex neurodevelopmental condition called “periventricular nodular heterotopia.” This neuronal migration disorder is characterized by microcephaly with brain malformations and progressive cerebral atrophy and spasticity (Ge et al., 2016), and invertebrate and vertebrate embryo models expressing dominant ARF1 exist, which show typical non-canonical Wnt-dependent PCP defects (Carvajal-Gonzalez et al., 2015). Specifically, solid experiments in the Drosophila wing model showed that Arf1, together with the Ap-1 adaptor complex, is instrumental for setting PCP during cell specification. Via direct control of Frizzled trafficking, Arf1 is majorly responsible for the restricted polarized accumulation of the signaling complexes formed by frizzled/disheveled/Diego and Van Gogh/prickle (Vang/Pk) within a single precursor cell, which guarantees correct morphogenesis. Furthermore, although the exact mechanism remains unproven in vertebrates, suggestive of the decisive impact on complex vertebrate embryogenesis events, constitutively active Arf1 (obtained by overexpressing the human variants) results in typical PCP-dependent phenotypes in zebrafish, i.e., body shortening and morphological alterations of the AP axis, likely caused by perturbed gastrulation cell movements (Carvajal-Gonzalez et al., 2015). Furthermore, work in C. elegans suggests a novel mechanism for both ARF and RAB small GTPases involving the modulation of a special non-canonical Wnt signaling that uses β-catenin for asymmetric divisions during development (Hardin and King, 2008).
On the other hand, canonical Wnt signaling might also be mediated by ARF-trafficking activity during development. Active ARF1 and ARF6 stimulate the production of PtdIns (4,5) P2 (Godi et al., 1999; Honda et al., 1999), which activates the Wnt coreceptor LRP6 (Zeng et al., 2005), resulting in hyperactivation of canonical Wnt signaling (Zhang et al., 2007). Consistently, it was shown that the function of specific ARF GEFs (such as BIG2) is essential for β-catenin distribution and activation in human cortical development (Sheen et al., 2004). Vice versa, even a positive control of canonical Wnt on ARF was demonstrated (Kim et al., 2013). An involvement of Wnt signaling modulation might underlie the X-linked mental retardation (Shoubridge et al., 2010) caused by missense mutations in IQSEC2 (encoding an ARFGEF specific for ARF6) and the complex and pleiotropic ciliopathy Bardet–Biedl syndrome showing polarity defects, associated with ARL6 (Wiens et al., 2010). Mutations affecting directly the ARFGEF BIG2 protein, fundamental for β-catenin action during brain development, were observed in children with autosomal recessive periventricular heterotopia manifesting severe cerebral cortex malformations and microcephaly, also likely underlying impaired Wnt signaling and impaired neuronal cell migration (Sheen et al., 2004).
Evidence exists also for a modulation of various aspects of Wnt signaling by members of the RAB family. It was shown that, by regulating the internalization of LRP6 receptor, RAB8B can control Wnt signaling. Confirming the in vitro data, a lack of Rab8b was found to block Wnt signaling during fish development (Demir et al., 2013). Moreover, RAB23 was implicated in positively regulating Wnt11/AP-1 signaling in a mechanism mediating C-Jun N-terminal kinase, contributing to cardiomyocyte differentiation in fish models (Jenkins et al., 2012).
By controlling the generation of endocytic compartments, precursor cells can regulate their fate in embryonic developmental fields to shape tissue formation. Demonstrating further the importance of RAB-cargo transport in embryogenesis and signaling, Winter et al. (2012) showed a role of Rab11-enriched recycling endosomes for regulating epithelial Par5-dependent polarity in nematodes, whereas Ulrich et al. (2005) clarified a new mechanism for non-canonical Wnt11 activity during zebrafish gastrulation, which functioned via E-cadherin–mediated cell cohesion and establishment of PCP through Rab5-dependent recycling. In the context of pathology, among the genes recently associated with Hirschsprung disease (HSCR), showing impaired enteric nervous system development (Gui et al., 2017), WES analysis identified mutations affecting the GEF DENND3, typically involved in intracellular trafficking by activation of RAB12. Functional investigation using zebrafish morpholino and CRISPR/Cas approach already exists, which supported the function of the fish ortholog in enteric nervous system development (Gui et al., 2017). It would be interesting in this context to also test the functional link between perturbation of RAB activity and Wnt signaling during NC migration for the onset of the pathology, as suggested by zebrafish ovo1 mutants (Piloto and Schilling, 2010).
FGF, EGF, and VEGF Signaling
During nervous system development, Schwann cells have a crucial role in responding to a number of signaling and reshape their morphology to form myelin. Mice models show that a specific ARF1 and ARF6 GEF (cytohesin) is involved in this morphogenetic process (Yamauchi et al., 2012), as well as RAB proteins (Stendel et al., 2010). Mechanistically, conditional KO mice provided evidence for ARF6-controlled FGF signaling, which impacted central nervous system (CNS) morphogenesis and myelin formation itself. Indeed, specific lack of ARF6 in rodent neurons resulted in a reduced size of the corpus callosum and of the hippocampal fimbria, underlying impaired secretion of the guidance factor FGF2. This results in defective oligodendrocytes migration and thereby axonal myelination (Akiyama and Kanaho, 2015). Moreover, experiments in mice models of KIF16B loss of function, which recapitulate FGFR2 KO animals, demonstrate that a KINESIN/RAB14 complex mediates Golgi-to-endosome trafficking of the FGFR and that this is crucial for epiblast development (Ueno et al., 2011). Evidence for an involvement in regulating also EGF- and VEGF-mediated signaling events during development emerges from animal models. Indeed, beyond its participation in insect insulin signaling pathway (Fuss et al., 2006), the Drosophila ortholog of the ARF-GEF cytohesin was shown to modulate EGF-mediated Ras/MAPK signaling in the context of wing growth and vein morphogenesis, as well as in and eye formation (Hahn et al., 2013). On the other hand, a recent study employing in vitro systems and zebrafish has proven the importance of Big2 in angiogenesis, likely depending on the Arf1-controlled VEGF signaling (Lu et al., 2019). The exact mechanism underlying these signalings’ interplays and the relevance for pathology remain to be addressed.
Shh Signaling
Alteration of cilium-related structure, function by perturbed ARF and RAB-related activity can impact on a number of other signaling pathways influencing brain formation and likely involved in neurodevelopmental diseases. As an example, we know that mutations in ARL13b are lined to altered Shh signaling and underlie Joubert syndrome, a condition causing midbrain–hindbrain developmental abnormalities and various other defects typical of impaired Shh signaling (Doherty, 2009). Importantly, mouse and zebrafish models for this condition are available, which can be employed to further investigate the mechanism (Larkins et al., 2011; Zhu et al., 2020). Null mice models for arl3 have a defective cilium-dependent signaling influencing different pathways, including Shh (Horner and Caspary, 2011) and show retinal degeneration (Schrick et al., 2006). A large amount of data strongly support the involvement of many RAB proteins and effectors in pathogenic alteration of cilium-mediated signaling, which is worth to investigate further (for a comprehensive review on the topic, refer to Oro, 2007; Yoshimura et al., 2007; Banworth and Li, 2018). In this context, the Carpenter syndrome, which harbors prominent neurological features and craniofacial and cardiac malformation, is an example. This condition is caused by mutations in RAB23, which is also known to regulate Shh signaling via controlled trafficking to the primary signaling center of the cilium (Boehlke et al., 2010). Accordingly, altered RAB23 in mouse models shows Shh-dependent ventralization defects and altered patterning of neural cell types during spinal cord development (Eggenschwiler et al., 2001; Eggenschwiler et al., 2006). It remains to be proven whether an impaired Shh signaling is involved also in the etiology of the Bardet–Biedl syndrome via the activity of Rabin 8 (a specific RAB8 GEF) (Oro, 2007). Of note, given that alteration of Shh signaling is common in various serious cancer conditions also in the adult, it would be interesting to test the potential of blocking the signaling acting directly on RAB proteins activity.
Notch Signaling
Lastly, it is well known that RAB-dependent endocytosis contributes to the regulation of the number of Notch/Delta molecules present on precursor cells’ surface and thereby of Notch directional signaling, which is normally fundamental for cell commitment and cell identity, as discussed above. Various mechanisms have been implicated in the regulation of Notch signaling during development. To mention few examples, Notch receptor activation is mediated by RAB5-positive early endosomes in dividing sensory organ precursors of Drosophila during asymmetric cell division, which instructs cell specification, tissue growth, and morphogenesis (Coumailleau et al., 2009). Also Rab11-dependent recycling of the specific Notch effector Delta is involved in this process in insects and mammalian cells (Emery et al., 2005) and in general, both Rab1 and Rab11 seem to regulate Notch signaling in Drosophila (Charng et al., 2014). Insect mutant screening based on wing morphogenesis identified also RAB7 and RAB8 orthologs as major positive modulators of Notch signaling activity (Court et al., 2017).
Established and Emerging Tools for in vivo Interrogation of Small GTPases Shaping Developmental Dynamics in the Vertebrate Zebrafish Model
To progress our knowledge on rare diseases and boost precision therapy, appropriate in vivo tools are required to assess the impact of the identified genetic lesions and map the spatiotemporal alterations of developmental pathways at an organismal level. Animal models, now equipped with unprecedented genomic and imaging-based possibilities, are irreplaceable. When zebrafish or C. elegans are used, the workflow is even time- and cost-efficient. As shown, animal models allow us to validate the impact of the identified genetic lesion on a global pathophysiological level, inferring readily altered pathways via established developmental paradigms (e.g., the fly wing system or the CE movements of vertebrate gastrulation), while discovering novel mechanisms on multiple developmental contexts and tissues simultaneously. The utility of animal models also relies on the possibility of setting up preclinical systems for assessing potential targeted treatments, identifying development and physiology principles that can uncover evolutionary rules. Moreover, sophisticated xenografting in vivo models enable innovative studies of cancer cells’ heterogeneity (Kim et al., 2017) and investigation of pediatric tumors (Rokita et al., 2019).
Besides the aforementioned assets, different biosensors, fluorescent-based reporters, and actuators superior to classical biochemical approaches are becoming available to use in vivo for a highly resolved real-time investigation of small GTPases dynamics. These tools allow the visualization and manipulation of small GTPases’ activity in a controllable manner, directly in the developing tissues of entire organisms, thus expanding the possibilities to answer mechanistic questions. Concurrent advances in the development of fluorescent proteins are rapidly accumulating toward the development of near-infrared (NIR) emitting molecules that improve the light penetration in deep tissues with little scattering (Shcherbakova et al., 2018b). Furthermore, advanced modalities for deep tissue imaging are also exponentially becoming available (i.e., two- and three-photon microscopy and a range of optoacoustic modalities) (Helmchen and Denk, 2005; Deán-Ben et al., 2016; Shcherbakova et al., 2018b). Nevertheless, the application and visualization of reporters’ dynamics and the use of genetically encoded actuators in large species remain challenging.
Because it embodies all these advances in vivo, zebrafish is once more forcefully becoming a convenient system for rare disease research. Numerous illustrative examples for zebrafish models of diseases underlying a functional dysregulation of small GTPases exist. Among those, notorious RASopathy models are available (Jindal et al., 2015) and RHO-associated developmental syndromes (Boueid et al., 2020). Typical advantages of zebrafish include the high fecundity, rapid development, and a rich community distributing transgenic lines and forward and reverse genetics mutants of various players involved in developmental signalings. Sophisticated genetics and a range of synthetic biology applications as well as imaging innovations discussed below, which allow monitoring of fast subcellular events at nanoresolution, are being quickly implemented in this model. Altogether, these tools are uniquely valuable to dissect real-time altered signaling dynamics throughout embryogenesis with single-cell precision, which is directly translatable to humans.
Genetic and in in vivo Imaging Advances
Zebrafish is especially amenable to gene perturbation for both loss- or gain-of-function genetic alterations via transient approaches (morpholino-based gene knockdown and gene overexpression) or strategies for stable modifications (TALENs and CRISPR-Cas9-based genetic engineering, Hwang et al., 2013). This allows the generation of models for the genetic diseases with a fast phenotyping at different levels, which can be obtained even in 2 days from the microinjection in F0 animals (Wu et al., 2018). Noteworthy, thanks to the continuous optimization of the Base Editor–CRISPR/Cas technology, it is now becoming possible in zebrafish to refine diseases’ modeling even toward patient-specific endeavors, obtaining inheritable precise single-nucleotide conversions (Qin et al., 2018; Rosello et al., 2021).
As far as in vivo functional imaging is concerned, zebrafish embryos show far fewer constraints as compared to rodents. They develop externally and are mostly transparent such that cellular dynamics can be readily resolved under fast microscopes in the whole-organism (Wolf et al., 2015; Abu-Siniyeh and Al-Zyoud, 2020), allowing, for instance, accurate brain-wide mapping of calcium fluxes (Renninger and Orger, 2013), even at the level of the whole adult brain (Deán-Ben et al., 2016; Chow et al., 2020). Moreover, a repertoire of behavioral readouts is available that can be implemented to evaluate intellectual delays and complex cognitive deficit, modeling, for instance, RASopathies traits (Wolman et al., 2014). Cancer models based on live imaging of xeno-transplanted malignant cells are also being successfully employed (Cayuela et al., 2019), whereas optimization of the Nobel-worth super-resolution structured illumination microscopy is now being experimented to image live brains in zebrafish (Turcotte et al., 2019), and more advanced imaging possibilities are currently unrolling.
Combining genetics and imaging advances, it is possible to follow molecular and cellular dynamics of virtually any developing tissue. mRNAs encoding fluorescent markers, which emit in a wide range of wavelengths and label-specific cell compartment, can be readily coinjected at early embryonic stages and offer the possibility for multiplexing live imaging, from the very early blastula and gastrula stages, to create mosaic expression both for overexpression and cell-labeling studies. Testifying the efficacy of these simple tools for investigation of in vivo developmental signaling, coinjection of mRNA encoding the membrane marker mCherry-GPI together with Wnt8-eGFP permitted to infer canonical Wnt transport and its paracrine activity mediated by Cdc42/N-Wasp + filipodia during early zebrafish development (Stanganello et al., 2015). Moreover, a variety of transgenic and enhancer trap lines are available, and effective fluorescent reporters for major signaling pathways are routinely utilized in zebrafish, including Wnt (Facchinello et al., 2016) and Hh (Mich et al., 2014). Semitransparent pigment mutants used as background in imaging applications (Antinucci and Hindges, 2016) are useful for dissecting the impact of disease-causing mutations on specific anatomical districts (Tabor et al., 2019) even in juvenile and adult fish.
Cell Lineage Tracing Tools and Signal Perturbation With Photosensitive Proteins
Special transgenic fish exist for specific and dynamic cell lineage tracing, which employ a large variety of photosensitive proteins controlled by light (Chow and Vermot, 2017). Among these, photoconvertible fluorescent proteins such as KikGR (Lombardo et al., 2012) and Kaede are employed to trace various neurons, axons, and circuits (Sato et al., 2006) or migratory cells in the context of retina development and morphogenesis (Kwan et al., 2012). Together with sophisticated Cre-based multicolor (multibow) barcoding strategies (Xiong et al., 2015), these tools expand the available palette and allow following movements and ontogeny of a specific subset of cells and their derivatives throughout development in fish models of genetic diseases. Thereby, the origin of cellular organization in rather complex organs and tissues and their alterations can be tackled in vivo.
Proof of the possibility to capture even the signaling pathway history in a subset of zebrafish cells during development using photoconvertible proteins exists. The PHotoconvertible REporter of Signaling History (PHRESH) method using Kaede under the control of ptch1 regulatory sequence allowed indeed mapping temporal dynamics of Hh signaling during cell fate decision in fish spinal cord (Huang et al., 2012). Moreover, sophisticated photochemistry approaches using synthetic and genetically encoded photoactivable probes are being vastly implemented to study several developmental biology mechanisms in animal models, including zebrafish (Kowalik and Chen, 2017).
In vivo Reporters, Actuators, and Transgenic Lines to Study Small GTPASES Activity
Reporters and actuators to monitor and manipulate small GTPases are being developed for fish, which can be combined with the aforementioned tools. In this context, different strategies have been established, with remarkable applications in vivo (schematized in Table 1).
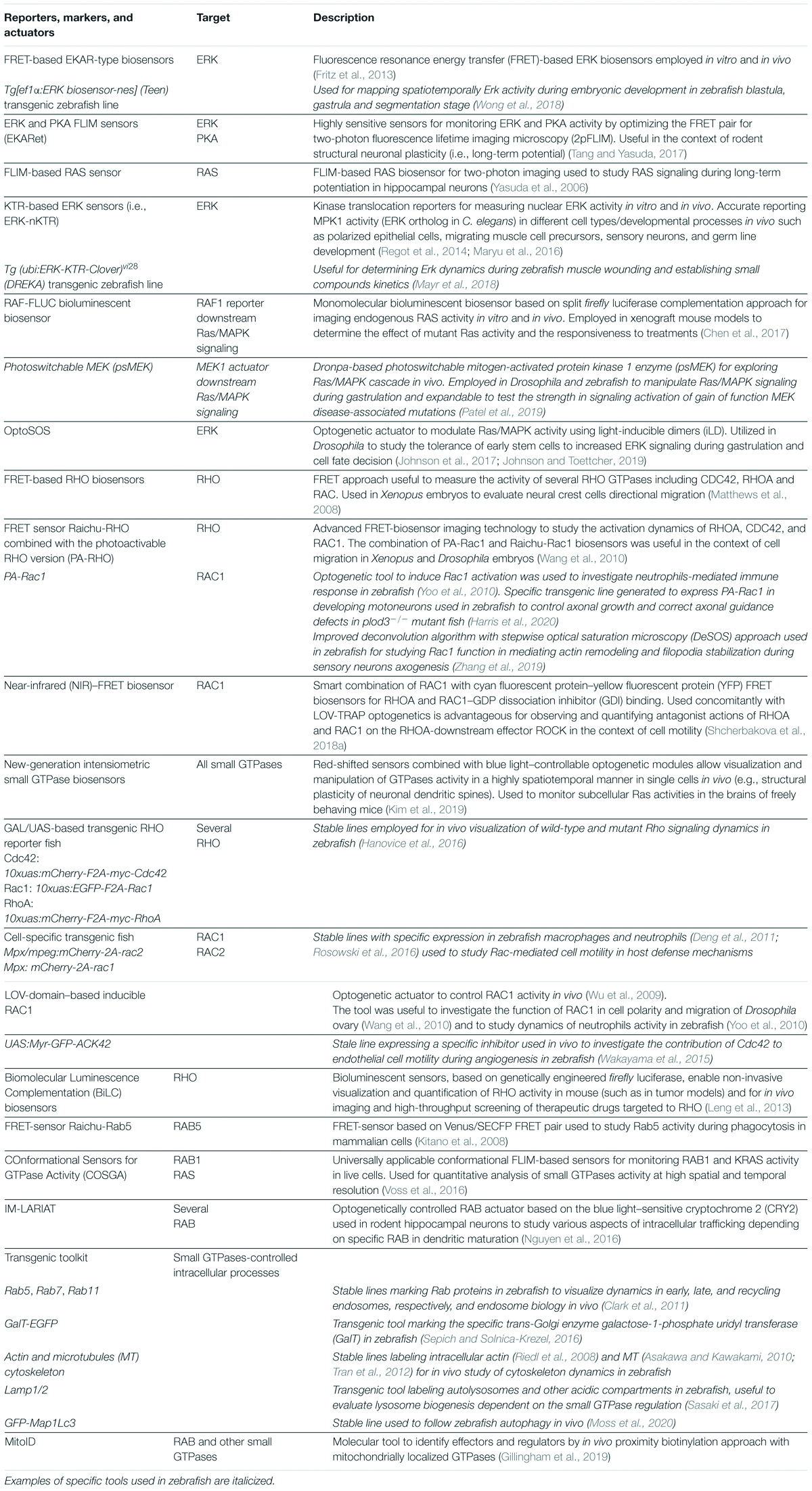
Table 1. Advanced genetic reporters, markers, and actuators to study RAS-, RHO-, and ARF/RAB-dependent processes in the context of live cells and animal development.
RAS/MAPK Biosensors and Actuators
A variety of biosensors for the Ras/MAPK signaling are used in vivo (i.e., rodents, insects, and zebrafish; Hirata and Kiyokawa, 2019) to capture spatiotemporally cell dynamics, depending on ERK activity in the context of cell growth, differentiation, and migration. Classical fluorescence resonance energy transfer (FRET)–based extracellular-regulated kinase activity reporter (EKAR)–types biosensors exploit the ability of activated ERK to phosphorylate a substrate that triggers a conformational change of the sensor, such as to bring the FRET pair of fluorophores in close physical proximity (FRET pairs classically employing a GFP-like fluorescent protein as a donor and a red-emitting protein as an acceptor). This results in an increase of the FRET efficiency quantified by ratiometric measurements, in which the ratio between the intensities of the donor and acceptor fluorescence is calculated and correlates with the ERK activity (Harvey et al., 2008; Fritz et al., 2013). Alternatively, sensitive sensors have also been developed for fluorescent lifetime measurement (FLIM) quantification on the donor protein, which shortens as the FRET efficiency increases. Originally, Yasuda et al., 2006, established a 2-p (two photon microscopy)–FLIM–based RAS biosensor in which the FRET efficiency increased when active RAS-EGFP was recruited to the membrane to bind the RAS-binding domain of RAF-RFP. This tool was employed already to study the RAS signaling spreading dynamics for local long-term potentiation (LTP) in organotypic preparation of rodent hippocampal neurons. Moreover, FRET-FLIM ERK sensors (named EKA-Ret-cyto), which used a highly absorbent acceptor, were established and used in a deep 2-p confocal setting to capture ERK activity in subcellular compartments such as the dendritic spines. The sensor further contributed to clarify the role of Ras/MAPK signaling during LTP-dependent plasticity of hippocampal neuronal circuit (Tang and Yasuda, 2017).
Novel genetically encoded ERK sensors have been developed by innovative processes of dimerization-dependent fluorescent protein exchange, permitting a green-to-red shift in fluorescent intensity (Ding et al., 2015). On the other hand, sensitive kinase translocation reporters (KTRs) are able to convert the protein phosphorylation state into a nuclear–cytoplasmic shift in fluorescence that enables visualizing single-cell signaling dynamics of multiple events (e.g., to differentiate ERK and AKT activities downstream the RAS signaling, Maryu et al., 2016). In fact, these sensors are based on the ability of ERK to phosphorylate a substrate and mask, by conformational change, the nuclear localization signal, while unmasking the nuclear export signal. This results in an increase of cytoplasmic fluorescent upon ERK activation. Given that the fluorescent readout of KTR sensors depends on the shuttling between the nucleus and cytoplasm, it does not suffer from delays due to protein stability and expression level. The tool has been recently optimized for nematodes, to assess in vivo the dynamic function of Ras-dependent ERK activity in establishing cell competence and fate decision (de La Cova et al., 2017). The authors established a precise protocol for image analysis of ERK-nKTR in different developmental stages and contexts, including vulva morphogenesis, migratory muscle, sensory neurons, and gonad precursor cells. The work allowed mapping EGF-dependent modulation of frequency of ERK activity fluxes in real time with a specific spatial pattern in the different precursor cells, not possible with standard methods.
Similarly, a KTR dynamic reporter of ErK activity (DREKA) sensor has been established and successfully employed in zebrafish (Mayr et al., 2018). Dynamic Erk activity was registered in the dividing cells in the developing fish embryo during wound healing and for studying the uptake kinetics of chemical compounds influencing Ras/MAPK signaling relevant for preclinical applications. However, given that several factors might influence the readout based on the sensor nucleus–cytoplasm shuttling (i.e., nucleus morphology, expression levels of exportin proteins), resulting in inhomogeneous labeling of different cells, optimization of the DREKA sensor would be required for broader applications in the context of developmental dynamics and neuronal precursor cells.
Wong et al. (2018) have developed in parallel a stable transgenic fish expressing the FRET-based ERK biosensor Teen, proving already its usefulness to unravel ERK dynamics in a number of Ras/MAPK-controlled processes throughout developmental stages. A spatiotemporal map of Erk activity was generated in the entire developing organism from the early blastula until segmentation stages. Teen allowed discovering an overlooked domain of action of Ras/MAPK signaling during fish development in the caudal region of the neural tube, where the regulated activity of FGF-Erk, Wnt, and Bmp signaling contributes to shape the embryo axes and stem cell differentiation (neuronal and mesodermal fate). This zebrafish toolkit is of great potential to link precisely healthy and pathogenic molecular dynamics, involving Ras/MAPK signaling and cellular events in vivo, with considerable applications for understanding of RASopathies.
In parallel, other smart methods to visualize more dynamics are being developed (Ross et al., 2018), which will likely be translated in vivo in the future.
A deep understanding of signaling networks benefits also from genetically controlled actuators that can be used to perturb signaling and infer signaling rules. Tunable optogenetic control of Ras/MAPK activity was achieved employing optimized light-inducible dimers. In the OptoSOS system, the membrane-bound SsrA peptide of the α-helix of the plant-derived light-oxygen-voltage 2 (LOV2) domain is masked and cannot bind its receptor SsrB fused to a fluorescently tag SOS activator in the dark. The binding can be, however, triggered by blue light allowing strict spatiotemporal control of SOS membrane recruitment and thereby Ras/MAPK signaling activation. The use of this tool in vivo demonstrated the sensitivity of early Drosophila blastula and gastrula stages to ectopic Erk expression for axial morphogenesis (Johnson et al., 2017). Further investigations using OptoSOSO clarified the importance of a correct dosage and timing of the Ras/MAPK signaling for stem cell fate decision during these crucial developmental windows (Johnson and Toettcher, 2019). Furthermore, coupling precise optogenetic actuators of the Ras/MAPK signaling and quantitative reporting of ERK activity in a single cell line by CRISPR/Cas have allowed to describe the Ras/MAPK-dependent ERK pulses inducing immediate early gene transcription (Wilson et al., 2017).
Of note, S. Y. Shvartsman’s laboratory recently optimized a photoswitchable MEK device (psMEK) to manipulate the Ras/MAPK signaling using microscopy and Dronpa-based photo-dimerizable protein domains, already applicable to Drosophila and zebrafish (Patel et al., 2019). In the “off” state, the active site of constitutively active psMEK is blocked by Dronpa dimers. Upon illumination at 500 nm, the site is exposed via dissociation of the Dronpa dimers, and activation of Erk can occur (“on” state). Importantly, the signal can be reversibly switched off by using 400-nm light. The authors already used pMEK in Drosophila and zebrafish development and demonstrated that this actuator can be smartly utilized to classify mutations for an in vivo genotype–phenotype correlation analysis of RASopathies, based on the strength of signaling perturbation and known morphogenesis and organogenesis phenotypic readouts. Experiments in zebrafish already demonstrated the usefulness of this tool to probe the relevant time window in which the impact of the various MEK mutations causing gastrulation and axes defects influences other signaling networks that must be tightly controlled during the same processes (Patel et al., 2019).
RHO Biosensors and Actuators
Optimized versions of genetically encoded RHO sensors are also beingintensely utilized to observe RHO GTPases dynamics in various contexts since many years (Hodgson et al., 2010; Fritz et al., 2013; Donnelly et al., 2014; Boueid et al., 2020), including developments for cell-based high-throughput applications (Koraïchi et al., 2018). Classically, frogs were widely used to study RHO dynamics via simple fluorescent effector translocation probes and FRET biosensors. A first use involved the study of the modulation of Cdc42, Rac, and RhoA on proteoglycan and non-canonical Wnt signaling during NC migration in vivo (Matthews et al., 2008) and the observation of characteristic flares of RHO (dynamic accumulation of the active GTPases at the cell–cell junction) indispensable throughout embryo cell motility (Stephenson and Miller, 2017).
Ontogenetically induced actuators based on the LOV domains were engineered also to generate a photoactivable RAC1 by fusing the PHOT1 LOV2 to a constitutively active RAC1. Upon blue light illumination, the small GTPase is unmasked and able to constitutively bind its effectors and thereby trigger the downstream signal (Wu et al., 2009). By using the FRET sensor Raichu-Rac1, based on a CFP/YPF-mediated FRET upon the activation of the GTPase, combined with this type of actuator, it was possible to highlight the contribution of Rac1 in cell polarization and collective migration in Drosophila ovary development (Wang et al., 2010) and in neutrophil immune response of zebrafish (Yoo et al., 2010). Recently, a specific transgenic tool was generated to control optogenetically growth cone guidance mechanisms of growing motoneuron axons in zebrafish via the tissue-specific expression of PA-Rac1. The tool was employed to rescue innervation defects observed in mutant animals (Harris et al., 2020), demonstrating validity for study healthy and diseased neuronal circuit wiring.
Interestingly, genetically encoded in the GAL4/UAS versatile system, specific inhibitors of Cdc42 (Myr-GFP-ACK42) have been already used in combination to specific Cdc42 FRET sensor to assess the function of this small GTPases during angiogenesis (Wakayama et al., 2015).
Elegant functional studies in zebrafish exist, combining overexpression and knockdown approaches together with FRET sensors to model the dynamic activities of both Rac1 and RhoA in establishing actin-rich blebbing and retrograde actin flow for E-cadherin–dependent traction forces, respectively, in the context of early germ cells migration (Kardash et al., 2010). More recently, sophisticated deconvolution algorithms to obtain super-resolved images from PA-Rac1 experiments have allowed Zhang et al. (2019) to confirm a crucial role of Rac1 in mediating actin remodeling and filopodia stabilization for zebrafish pioneer axon formation of sensory neurons. Utility of FRET-based sensors is also clear from transgenic rodent models (Johnsson et al., 2014), for instance, to monitor RHO-dependent invasiveness of engrafted glioblastoma cells (Hirata et al., 2012). Noteworthy, an NIR FRET RAC1 biosensor for deep multiplexing imaging and signaling manipulation has been recently developed. The implementation of the most NIR FRET pair miRFP670–miRFP720 for this sensor enables the combinatorial use with classical CFP-YFP FRET pair for RHOA sensor upon optogenetic signal activation to study their concurrent dynamics during cell motility (Shcherbakova et al., 2018a).
In addition, exploiting the versatile GAL/UAS genetic system, Hanovice et al. (2016) established useful zebrafish GAL4-inducible transgenic lines for tissue and temporally tuned modulation and visualization of mutant (i.e., dominant negative) and wild-type Rac, RhoA, and Cdc42 (10xuas:mCherry-F2A-myc-Rac, RhoA, or Cdc42). The system is expandable to a range of mutants and allows systematic functional investigation of RHO protein–specific cells and developmental windows of interest and possible crosstalks in fish cancer models (Chew et al., 2014).
Specific transgenic lines to study the role of RHO proteins during immune response have also been developed (e.g., mpeg:mcherry-2A-rac2; Rosowski et al., 2016). Smart multiplexing transgenic tools in zebrafish permitted the discovery of Cdc42-induced “filopodia extensions” for mediating paracrine and large-range Wnt signaling in the context of zebrafish development, as discussed earlier (Stanganello et al., 2015).
Noteworthy, Kim et al. (2019) have established highly sensitive and expandable intensiometric biosensors for the simultaneous detection of smaller GTPases combined with optogenetic signaling actuation in vivo, previously used to describe the regulated activity of CDC42 and RAS in the context of rodent structural dendritic changes upon neurotrophic signaling activation. Furthermore, optogenetic activation of CDC42 was used to study the immune cell migration (O’Neill et al., 2016), which can be further exploited for counteracting the invasiveness of cancer cells (Palucka and Coussens, 2016).
Vital Dyes and Transgenic Tools for Monitoring ARF and RAB-Regulated Intracellular Trafficking and Organelle’s Dynamics
At present, a series of tools are readily available in animal models to map intracellular trafficking events and investigate the role of small GTPase in these processes. Among vertebrate models, fish harbors good molecular devices not only for visualizing but also for manipulating organelle dynamics and cell biology of developing tissues in virtually any developmental stage.
The use of vital dyes and advanced imaging techniques is largely used to label intracellular compartments, molecules, and dynamics. For instance, lifeact allows actin network dynamic visualization via fluorescence recovery after photobleaching (Riedl et al., 2008). The rhodamine-labeled phalloidin, which selectively binds to F-actin, can be readily used to label actin filaments in zebrafish developmental studies (Li et al., 2008). Other dyes to label subcellular structures are widely employed to study organelles’ dynamics in disease models (e.g., lysosomes, Tseng et al., 2018). Besides these tools, all relevant intracellular organelles can be stably labeled by transgenic lines and/or transiently by libraries of constructs expressing fluorescently tagged markers for multiplexed imaging. In addition to standard subcellular markers labeling nuclei and cell membranes, specific organelles and vesicle markers are available in zebrafish (reviewed by Vacaru et al., 2014). Among those, GalT-GFP fish expressing galactose-1-phosphate uridyl transferase (Gerhart et al., 2012) are useful to map trans-Golgi, whereas Lamp2-mCherry can be used to visualize lysosomes (Sasaki et al., 2017), and GFP-Map1Lc3 has been already employed to image disease-associated autophagy in vivo (Moss et al., 2020).
A number of transgenic lines and constructs for labeling Rab and endosomal vesicles of different types (such as fluorescently labeled Rab5, Rab7, and Rab11 for early, late, and recycling endosomes, respectively) are successfully used for dynamic analysis of recycling endosomes in vivo (Clark et al., 2011), as well as transgenic fish lines marking actin, i.e., Tg (uas:lifeact-GFP) (Riedl et al., 2008) and a number of fish labeling MT such as the Tg (UAS:EGFP-tuba2) (Asakawa and Kawakami, 2010), the MT-associated doublecortin-like kinase Tg (XlEef1a1:dclk2DeltaK-GFP), the Eb3 + growing tip of the MT (Tran et al., 2012), and Tg (bactin2:HsENSCONSIN17–282-3xEGFP) fish expressing GFP-tagged MT-binding region of ensconsin (Wühr et al., 2011).
Notably, protein engineering translated into transgenic fish allows now to map with a substantial spatiotemporal resolution also highly complex phenomena, such as the site of neuronal protein synthesis during early zebrafish CNS development (Garcez Palha et al., 2018). More recently, Verweij et al. (2019) managed to develop a transgenic fish line expressing CD63-pHluorin for direct monitoring with high spatiotemporal accuracy of extracellular vesicles (EVs) dynamics secreted by multivesicular endosomes at an interorgan level. The work utilizes the in vivo whole-embryo reporter and demonstrated the dynamics of formation, transport, and function and the trophic role of the EV secreted by cells of yolk syncytial layer during development. In addition, the FRET type of RAB biosensors also exists and has been developed and used to monitor, for instance, Rab5 activity in phagosome maturation of immune cells (Kitano et al., 2008). Of note, Gillingham et al. (2019) have reported the development of MitoID, an innovative methodology for identifying a wide range of small GTPases’ effectors and regulators employing in vivo proximity biotinylation of mitochondrial-restricted GTPases and found several RAB effectors. Lastly, via advanced protein design and chemical strategies, Conformational sensors for GTPase activity (COSGAs) awaiting in vivo applications, allowing direct observation of GTPase activation state, were recently used to detect RAB1 and K-RAS activity in vitro and quantification of RAB1 GTP-/GDP ratio at high spatial and temporal resolution (Voss et al., 2016).
The development of optogenetic devices to manipulate intracellular trafficking has been notoriously more challenging. Nevertheless, plant photoreceptors responsive to UV light have been genetically engineered for controlling protein secretion and used to investigate dendritic cargo secretion (Chen et al., 2013). More recently light-induced actuators to perturb RAB-dependent intracellular trafficking have also been established, which permit fast, tunable, and reversible interference of membrane dynamics, protein sorting, and endosomes signaling. In the IM-LARIAT system engineered by Nguyen et al. (2016), a subdomain of the blue light–sensitive cryptochrome 2 (CRY2) from Arabidopsis thaliana self-oligomerizes immediately together with CIB1 fused to RAB upon blue light illumination. This results in aggregation and perturbation of the GTPases’ activity during intracellular trafficking at various levels, depending on the targeted RAB protein (early, late, or recycling endosomes, ER–GA transport, and secretion). The tool was already implemented in rodent hippocampal neurons to study the contribution of RAB5 and RAB11 in influencing dendritic growth’s rate.
Conclusion
Next-generation sequencing of previously unrecognized pediatric conditions disclosed an unforeseen impact of small GTPases of the RAS superfamily on the pathogenicity of a growing number of developmental disorders. The precision of this approach is bringing up amazing possibilities for investigating unexplored mechanistic principles of developmental biology, re-employing classical animal models. Continuous advances in the field of high-resolution microscopy, genetic engineering, and synthetic biology for optimized biosensors and actuators for in vivo studies are now unfolding, well exemplified by the zebrafish model. We anticipate that the expansion and optimization of these tools for multiplexing in vivo signal visualization and manipulations will have an unprecedented impact for the spatiotemporal investigation of developmental signaling networks modulated by small GTPases in health and disease. In conclusion, an integrated pipeline from patients back to precise organismal biology in the global context of embryo development represents the blueprint for a modern global health care response to the burden of the ever-increasing pediatric genetic diseases, critical for developing tailored measures in the rapidly emerging field of precision medicine.
Author Contributions
AL, GF, MT, and BD wrote the manuscript. AL conceived and generated all the illustrations. AL and GF conceived the table. GF and MV generated the table. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from European Commission (individual fellowship Marie Skłodowska-Curie, 844636 – Innervate, AL), Fondazione Umberto Veronesi (individual research fellowship, AL), Ministero della Salute (Ricerca 2019 5 × 1000, CCR-2017-23669081 and RCR-2020-23670068_001, MT), Fondazione Bambino Gesù (Vite Coraggiose, MT), and Ministero dell’Università e della Ricerca (Sviluppo di protocolli innovativi e applicazione di nuovi strumenti-omici nei pazienti orfani di diagnosi, MT).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors apologize to colleagues whose work was not cited due to limited space are gratefully acknowledge all the collaborators who have contributed to the original research works cited here.
References
Abe, T., Umeki, I., Kanno, S., Inoue, S., Niihori, T., and Aoki, Y. (2020). LZTR1 facilitates polyubiquitination and degradation of RAS-GTPases. Cell Death Differ. 27, 1023–1035. doi: 10.1038/s41418-019-0395-5
Abu-Siniyeh, A., and Al-Zyoud, W. (2020). Highlights on selected microscopy techniques to study zebrafish developmental biology. Lab. Anim. Res. 36:12. doi: 10.1186/s42826-020-00044-2
Adams, A. E., Johnson, D. I., Longnecker, R. M., Sloat, B. F., and Pringle, J. R. (1990). CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111, 131–142. doi: 10.1083/jcb.111.1.131
Akiyama, M., and Kanaho, Y. (2015). Physiological functions of the small GTPase Arf6 in the nervous system. Small GTPases 6, 160–164. doi: 10.1080/21541248.2015.1043041
Anastasaki, C., Estep, A. L., Marais, R., Rauen, K. A., and Patton, E. E. (2009). Kinase-activating and kinase-impaired cardio-facio-cutaneous syndrome alleles have activity during zebrafish development and are sensitive to small molecule inhibitors. Hum. Mol. Genet. 18, 2543–2554. doi: 10.1093/hmg/ddp186
Angelis, A., Tordrup, D., and Kanavos, P. (2015). Socio-economic burden of rare diseases: a systematic review of cost of illness evidence. Health Policy. 119, 964–979. doi: 10.1016/j.healthpol.2014.12.016
Antinucci, P., and Hindges, R. (2016). A crystal-clear zebrafish for in vivo imaging. Sci. Rep. 6:29490. doi: 10.1038/srep29490
Aoidi, R., Houde, N., Landry-Truchon, K., Holter, M., Jacquet, K., Charron, L., et al. (2018). Mek1Y130C mice recapitulate aspects of human cardio-facio-cutaneous syndrome. Dis. Model. Mech. 11:dmm031278. doi: 10.1242/dmm.031278
Aoki, K., Kumagai, Y., Sakurai, A., Komatsu, N., Fujita, Y., Shionyuet, C., et al. (2013). Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol. Cell 52, 529–540. doi: 10.1016/j.molcel.2013.09.015
Aoki, Y., Niihori, T., Kawame, H., Kurosawa, K., Ohashi, H., Tanaka, Y., et al. (2005). Germlinemutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet. 37, 1038–1040. doi: 10.1038/ng1641
Araki, T., Mohi, M. G., Ismat, F. A., Bronson, R. T., Williams, I. R., Kutok, J. L., et al. (2004). Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat. Med. 10, 849–857. doi: 10.1038/nm1084
Asakawa, K., and Kawakami, K. A. (2010). transgenic zebrafish for monitoring in vivo microtubule structures. Dev. Dyn. 239, 2695–2699. doi: 10.1002/dvdy.22400
Atkin, J. D., Farg, M. A., Turner, B. J., Tomas, D., Lysaght, J. A., Nunan, J., et al. (2017). Induction of the unfolded protein response in familial amyotrophic lateral sclerosis and association of protein-disulfide isomerase with superoxide dismutase 1. J. Biol. Chem. 292:12007. doi: 10.1074/jbc.a117.603393
Azzarelli, R., Kerloch, T., and Pacary, E. (2015). Regulation of cerebral cortex development by Rho GTPases: insights from in vivo studies. Front. Cell. Neurosci. 8:445. doi: 10.3389/fncel.2014.00445
Bajetto, A., Barbero, S., Bonavia, R., Piccioli, P., Pirani, P., Florio, T., et al. (2001). Stromal cell-derived factor-1alpha induces astrocyte proliferation through the activation of extracellular signal-regulated kinases 1/2 pathway. J. Neurochem. 77, 1226–1236. doi: 10.1046/j.1471-4159.2001
Banworth, M. J., and Li, G. (2018). Consequences of Rab GTPase dysfunction in genetic or acquired human diseases. Small GTPases 9, 158–181. doi: 10.1080/21541248.2017.1397833
Barr, F., and Lambright, D. G. (2010). Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22, 461–470. doi: 10.1016/j.ceb.2010.04.007
Basson, M. A. (2012). Signaling in cell differentiation and morphogenesis. Cold Spring Harb. Perspect. Biol. 4:a008151. doi: 10.1101/cshperspect.a008151
Becker, J., and Wilting, J. (2019). WNT signaling in neuroblastoma. Cancers 11:1013. doi: 10.3390/cancers11071013
Bellouze, S., Schäfer, M. K., Buttigieg, D., Baillat, G., Rabouille, C., and Haase, G. (2014). Golgi fragmentation in pmn mice is due to a defective ARF1/TBCE cross-talk that coordinates COPI vesicle formation and tubulin polymerization. Hum. Mol. Genet. 23, 5961–5975. doi: 10.1093/hmg/ddu320
Biechele, S., Cox, B. J., and Rossant, J. (2011). Porcupine homolog is required for canonical Wnt signaling and gastrulation in mouse embryos. Dev. Biol. 355, 275–285. doi: 10.1016/j.ydbio.2011.04.029
Bigenzahn, J. W., Collu, G. M., Kartnig, F., Pieraks, M., Vladimer, G. I., Heinz, L. X., et al. (2018). LZTR1 is a regulator of RAS ubiquitination and signaling. Science 362, 1171–1177. doi: 10.1126/science.aap8210
Blitzer, J. T., and Nusse, R. (2006). A critical role for endocytosis in Wnt signaling. BMC Cell Biol. 7:28. doi: 10.1186/1471-2121-7-28
Boehlke, C., Bashkurov, M., Buescher, A., Krick, T., John, A. K., Nitschke, R., et al. (2010). Differential role of Rab proteins in ciliary trafficking: Rab23 regulates smoothened levels. J. Cell Sci. 123, 1460–1467. doi: 10.1242/jcs.058883
Boueid, M. J., Mikdache, A., Lesport, E., Degerny, C., and Tawk, M. (2020). Rho GTPases signaling in Zebrafish development and disease. Cells 9:2634. doi: 10.3390/cells9122634
Bourne, H. R., Sanders, D. A., and McCormick, F. (1991). The GTPase superfamily: conserved structure and molecular mechanism. Nature 349, 117–127. doi: 10.1038/349117a0
Brems, H., Beert, E., de Ravel, T., and Legius, E. (2009). Mechanisms in the pathogenesis of malignant tumours in neurofibromatosis type 1. Lancet Oncol. 10, 508–515. doi: 10.1016/S1470-2045(09)70033-6
Capri, Y., Flex, E., Krumbach, O. H. F., Carpentieri, G., Cecchetti, S., Lißewski, C., et al. (2019). Activating mutations of RRAS2 are a rare cause of noonan syndrome. Am. J. Hum. Genet. 104, 1223–1232. doi: 10.1016/j.ajhg.2019.04.013
Carmena, A., Buff, E., Halfon, M. S., Gisselbrecht, S., Jimenez, F., and Baylies, M. K. (2002). Reciprocal regulatory interactions between the Notch and Ras signaling pathways in the Drosophila embryonic mesoderm. Dev. Biol. 244, 226–242. doi: 10.1006/dbio.2002.0606
Carmena, A., Speicher, S., and Baylies, M. (2006). The PDZ protein Canoe/AF-6 links Ras-MAPK, Notch and Wingless/Wnt signaling pathways by directly interacting with Ras, Notch and Dishevelled. PLoS One 1:e66. doi: 10.1371/journal.pone.0000066
Carta, C., Pantaleoni, F., Bocchinfuso, G., Stella, L., Vasta, I., Sarkozy, A., et al. (2006). Germline missense mutations affecting KRAS isoform b are associated with a severe Noonan syndrome phenotype. Am. J. Hum. Genet. 79, 129–135. doi: 10.1086/504394
Carvajal-Gonzalez, J. M., Balmer, S., Mendoza, M., Dussert, A., Collu, G., Roman, A. C., et al. (2015). The clathrin adaptor AP-1 complex and Arf1 regulate planar cell polarity in vivo. Nat. Commun. 6:6751. doi: 10.1038/ncomms7751
Casalou, C., Ferreira, A., and Barral, D. C. (2020). The role of ARF family proteins and their regulators and effectors in cancer progression: a therapeutic perspective. Front. Cell. Dev. Biol. 8:217. doi: 10.3389/fcell.2020.00217
Cawthon, R. M., Weiss, R., Xu, G. F., Viskochil, D., Culver, M., Stevens, J., et al. (1990). A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell 62, 193–201. doi: 10.1016/0092-8674(90)90252-a
Cayuela, M. L., Claes, K. B. M., Ferreira, M. G., Henriques, C. M., van Eeden, F., Varga, M., et al. (2019). The Zebrafish as an emerging model to study DNA damage in aging, cancer and other diseases. Front. Cell Dev. Biol. 6:178. doi: 10.3389/fcell.2018.00178
Chang, L., Kreko-Pierce, T., and Eaton, B. A. (2015). The guanine exchange factor Gartenzwerg and the small GTPase Arl1 function in the same pathway with Arfaptin during synapse growth. Biol. Open 4, 947–953. doi: 10.1242/bio.011262
Charng, W. L., Yamamoto, S., Jaiswal, M., Bayat, V., Xiong, B., Zhang, K., et al. (2014). Drosophila Tempura, a novel protein prenyltransferaseα subunit, regulates notch signaling via Rab1 and Rab11. PLoS Biol. 12:e1001777. doi: 10.1371/journal.pbio.1001777
Chen, D., Gibson, E. S., and Kennedy, M. J. (2013). A light-triggered protein secretion system. J. Cell Biol. 201, 631–640. doi: 10.1083/jcb.201210119
Chen, L., Leng, W. B., Li, D. Z., Xia, H. W., Ren, M., Tang, Q. L., et al. (2017). Noninvasive imaging of Ras activity by monomolecular biosensor based on Split-Luciferase complementary assay. Sci. Rep. 7:9945. doi: 10.1038/s41598-017-08358-3
Chen, L., Liao, G., Yang, L., Campbell, K., Nakafuku, M., Kuan, C. Y., et al. (2006). Cdc42 deficiency causes Sonic hedgehog-independent holoprosencephaly. Proc. Natl. Acad. Sci. U.S.A. 103, 16520–16525. doi: 10.1073/pnas.0603533103
Chen, L., Melendez, J., Campbell, K., Kuan, C. Y., and Zheng, Y. (2009). Rac1 deficiency in the forebrain results in neural progenitor reduction and microcephaly. Dev. Biol. 325, 162–170. doi: 10.1016/j.ydbio.2008.10.023
Chen, N., and Greenwald, I. (2004). The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev. Cell 6, 183–192. doi: 10.1016/s1534-5807(04)00021-8
Chen, W., Mao, K., Liu, Z., and Dinh-Xuan, A. T. (2014). The role of the RhoA/Rho kinase pathway in angiogenesis and its potential value in prostate cancer. Oncol. Lett. 8, 1907–1911. doi: 10.3892/ol.2014.2471
Chew, T. W., Liu, X. J., Liu, L., Spitsbergen, J. M., Gong, Z., and Low, B. C. (2014). Crosstalk of Ras and Rho: activation of RhoA abates Kras-induced liver tumorigenesis in transgenic zebrafish models. Oncogene 33, 2717–2727. doi: 10.1038/onc.2013.240
Chiang, M. Y., Xu, L., Shestova, O., Histen, G., L’heureux, S., Romany, C., et al. (2008). Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J. Clin. Invest. 118, 3181–3194. doi: 10.1172/JCI35090
Chow, D. M., Sinefeld, D., Kolkman, K. E., Ouzounov, D. G., Akbari, N., Tatarsky, R., et al. (2020). Deep three-photon imaging of the brain in intact adult zebrafish. Nat. Methods 17, 605–608. doi: 10.1038/s41592-020-0819-7
Chow, R. W., and Vermot, J. (2017). The rise of photoresponsive protein technologies applications in vivo: a spotlight on zebrafish developmental and cell biology. F1000Res. 6:F1000 Faculty Rev-459. doi: 10.12688/f1000research.10617.1
Cirstea, I., Kutsche, K., Dvorsky, R., Gremer, L., Carta, C., and Horn, D. (2010). A restricted spectrum of NRAS mutations causes Noonan syndrome. Nat. Genet. 42, 27–29. doi: 10.1038/ng.497
Clark, B. S., Winter, M., Cohen, A. R., and Link, B. A. (2011). Generation of Rab-based transgenic lines for in vivo studies of endosome biology in zebrafish. Dev. Dyn. 240, 2452–2465. doi: 10.1002/dvdy.22758
Cogli, L., Piro, F., and Bucci, C. (2009). Rab7 and the CMT2B disease. Biochem. Soc. Trans. 37, 1027–1031. doi: 10.1042/BST0371027
Colon, N. C., and Chung, D. H. (2011). Neuroblastoma. Adv. Pediatr. 58, 297–311. doi: 10.1016/j.yapd.2011.03.011
Cooper, M. T., and Bray, S. J. (2000). R7 photoreceptor specification requires Notch activity. Curr. Biol. 10, 1507–1510. doi: 10.1016/s0960-9822(00)00826-5
Cordeddu, V., Di Schiavi, E., Pennacchio, L. A., Ma’ayan, A., Sarkozy, A., Fodale, V., et al. (2009). Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nat. Genet. 41, 1022–1026. doi: 10.1038/ng.425
Costain, G., Callewaert, B., Gabriel, H., Tan, T. Y., Walker, S., Christodoulou, J., et al. (2019). Do novo missense variants in RAC3 cause a novel neurodevelopmental syndrome. Genet. Med. 21, 1021–1026. doi: 10.1038/s41436-018-0323-y
Coumailleau, F., Fürthauer, M., Knoblich, J., and González-Gaitán, M. (2009). Directional Delta and Notch trafficking in Sara endosomes during asymmetric cell division. Nature 458, 1051–1055. doi: 10.1038/nature07854
Courbier, S., and Berjonneau, E. (2017). Juggling Care and Daily life – the Balancing Act of the Rare Disease Community. A Rare Barometer Survey. Paris: EURODIS.
Court, H., Ahearn, I. M., Amoyel, M., Bach, E. A., and Philips, M. R. (2017). Regulation of NOTCH signaling by RAB7 and RAB8 requires carboxyl methylation by ICMT. J. Cell Biol. 216, 4165–4182. doi: 10.1083/jcb.201701053
Cox, A. D., and Der, C. J. (2010). Ras history: the saga continues. Small GTPases 1, 22–27. doi: 10.4161/sgtp.1.1.12178
Dawes-Hoang, R. E., Parmar, K. M., Christiansen, A. E., Phelps, C. B., Brand, A. H., and Wieschaus, E. F. (2005). folded gastrulation, cell shape change and the control of myosin localization. Development 132, 4165–4178. doi: 10.1242/dev.01938
de La Cova, C., Townley, R., Regot, S., and Greenwald, I. (2017). A real-timeBiosensor for ERK activity reveals signaling dynamics during C. elegans cell fatespecification. Dev. Cell 42, 542–553.e4. doi: 10.1016/j.devcel.2017.07.014
Deán-Ben, X. L., Sela, G., Lauri, A., Kneipp, M., Ntziachristos, V., Westmeyer, G. G., et al. (2016). Functional optoacoustic neuro-tomography for scalable whole-brain monitoring of calcium indicators. Light Sci. Appl. 5:e16201. doi: 10.1038/lsa.2016.201
Demir, K., Kirsch, N., Beretta, C. A., Erdmann, G., Ingelfinger, D., Moro, E., et al. (2013). RAB8B is required for activity and caveolar endocytosis of LRP6. Cell Rep. 4, 1224–1234. doi: 10.1016/j.celrep.2013.08.008
Deng, Q., Yoo, S. K., Cavnar, P. J., Green, J. M., and Huttenlocher, A. (2011). Dual roles for Rac2 in neutrophil motility and active retention in zebrafish hematopoietic tissue. Dev. Cell 21, 735–745. doi: 10.1016/j.devcel.2011.07.013
Di Guglielmo, G. M., Le Roy, C., Goodfellow, A. F., and Wrana, J. L. (2003). Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat. Cell Biol. 5, 410–421. doi: 10.1038/ncb975
Digilio, M. C., Conti, E., Sarkozy, A., Mingarelli, R., Dottorini, T., Marino, B., et al. (2002). Grouping of multiple-lentigines/LEOPARD and Noonan syndromes on the PTPN11 gene. Am. J. Hum. Genet. 71, 389–394. doi: 10.1086/341528
Ding, Y., Li, J., Enterina, J. R., Shen, Y., Zhang, I., Tewson, P. H., et al. (2015). Ratiometric biosensors based on dimerization-dependent fluorescent protein exchange. Nat. Methods 12, 195–198. doi: 10.1038/nmeth.3261
Dinh Duong, T. A., Hoshiba, Y., Saito, K., Kawasaki, K., Ichikawa, Y., Matsumoto, N., et al. (2019). FGF signaling directs the cell fate switch from neurons to astrocytes in the developing mouse cerebral cortex. J. Neurosci. 39, 6081–6094. doi: 10.1523/JNEUROSCI.2195-18.2019
Doherty, D. (2009). Joubert syndrome: insights into brain development, cilium biology, and complex disease. Semin. Pediatr. Neurol. 16, 143–154. doi: 10.1016/j.spen.2009.06.002
Donnelly, S. K., Bravo-Cordero, J. J., and Hodgson, L. (2014). Rho GTPase isoforms in cell motility: don’t fret, we have FRET. Cell Adh. Migr. 8, 526–534. doi: 10.4161/cam.29712
Downward, J. (1998). Ras signalling and apoptosis. Curr. Opin. Genet. Dev. 8, 49–54. doi: 10.1016/s0959-437x(98)80061-0
Drosten, M., Dhawahir, A., Sum, E. Y., Urosevic, J., Lechuga, C. G., Esteban, L. M., et al. (2010). Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J. 29, 1091–1104. doi: 10.1038/emboj.2010.7
Dupraz, S., Hilton, B. J., Husch, A., Santos, T. E., Coles, C. H., Stern, S., et al. (2019). RhoA controls axon extension independent of specification in the developing brain. Curr. Biol. 29, 3874–3886.e9. doi: 10.1016/j.cub.2019.09.040
Dupuis, N., Fafouri, A., Bayot, A., Kumar, M., Lecharpentier, T., Ball, G., et al. (2015). Dymeclin deficiency causes postnatal microcephaly, hypomyelination and reticulum-to-Golgi trafficking defects in mice and humans. Hum. Mol. Genet. 24, 2771–2783. doi: 10.1093/hmg/ddv038
Dyberg, C., Fransson, S., Andonova, T., Sveinbjörnsson, B., Lännerholm-Palm, J., Olsen, T. K., et al. (2017). Rho-associated kinase is a therapeutic target in neuroblastoma. Proc. Natl. Acad. Sci. U.S.A. 114, E6603–E6612. doi: 10.1073/pnas.1706011114
Eaton, S., and Martin-Belmonte, F. (2014). Cargo sorting in the endocytic pathway: a key regulator of cell polarity and tissue dynamics. Cold Spring Harb. Perspect. Biol. 6:a016899. doi: 10.1101/cshperspect.a016899
Eggenschwiler, J. T., Bulgakov, O. V., Qin, J., Li, T., and Anderson, K. V. (2006). Mouse Rab23 regulates hedgehog signaling from smoothened to Gli proteins. Dev. Biol. 290, 1–12. doi: 10.1016/j.ydbio.2005.09.022
Eggenschwiler, J. T., Espinoza, E., and Anderson, K. V. (2001). Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature 412, 194–198. doi: 10.1038/35084089
Ehrman, L. A., Nardini, D., Ehrman, S., Rizvi, T. A., Gulick, J., Krenz, M., et al. (2014). The protein tyrosine phosphatase Shp2 is required for the generation of oligodendrocyte progenitor cells and myelination in the mouse telencephalon. J. Neurosci. Res. 34, 3767–3778. doi: 10.1523/JNEUROSCI.3515-13.2014
El Baba, N., Farran, M., Khalil, E. A., Jaafar, L., Fakhoury, I., and El-Sibai, M. (2020). The Role of Rho GTPases in VEGF signaling in cancer cells. Anal. Cell Pathol. 2020:2097214. doi: 10.1155/2020/2097214
Elias, B. C., Das, A., Parekh, D. V., Mernaugh, G., Adams, R., Yang, Z., et al. (2015). Cdc42 regulates epithelial cell polarity and cytoskeletal function during kidney tubule development. J. Cell. Sci. 128, 4293–4305. doi: 10.1242/jcs.164509
Ellis, S., and Mellor, H. (2000). Regulation of endocytic traffic by rho family GTPases. Trends Cell Biol. 10, 85–88. doi: 10.1016/s0962-8924(99)01710-9
Emery, G., Hutterer, A., Berdnik, D., Mayer, B., Wirtz-Peitz, F., Gaitan, M. G., et al. (2005). Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell 122, 763–773. doi: 10.1016/j.cell.2005.08.017
Etienne-Manneville, S., and Hall, A. (2002). Rho GTPases in cell biology. Nature 420, 629–635. doi: 10.1038/nature01148
Evangelista, M., Blundell, K., Longtine, M. S., Chow, C. J., Adames, N., Pringle, J. R., et al. (1997). Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276, 118–122. doi: 10.1126/science.276.5309.118
Evans, R. J., Schwarz, N., Nagel-Wolfrum, K., Wolfrum, U., Hardcastle, A. J., and Cheetham, M. E. (2010). The retinitis pigmentosa protein RP2 links pericentriolar vesicle transport between the Golgi and the primary cilium. Hum. Mol. Genet. 19, 1358–1367. doi: 10.1093/hmg/ddq012
Facchinello, N., Schiavone, M., Vettori, A., Argenton, F., and Tiso, N. (2016). Monitoring Wnt signaling in Zebrafish using fluorescent biosensors. Methods Mol. Biol. 1481, 81–94. doi: 10.1007/978-1-4939-6393-5_9
Felici, A., Wurthner, J. U., Parks, W. T., Giam, L. R., Reiss, M., Karpova, T. S., et al. (2003). TLP, a novel modulator of TGF-beta signaling, has opposite effects on Smad2- and Smad3-dependent signaling. EMBO J. 22, 4465–4477. doi: 10.1093/emboj/cdg428
Fernandez-Valdivia, R., Takeuchi, H., Samarghandi, A., Lopez, M., Leonardi, J., Haltiwanger, R. S., et al. (2011). Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development. 138, 1925–1934. doi: 10.1242/dev.060020
Fisher, S., Kuna, D., Caspary, T., Kahn, R. A., and Sztul, E. (2020). ARF family GTPases with links to cilia. Am. J. Physiol. Cell Physiol. 319, C404–C418. doi: 10.1152/ajpcell.00188.2020
Fitzgerald, K., Harrington, A., and Leder, P. (2000). Ras pathway signals are required for notch-mediated oncogenesis. Oncogene 37, 4191–4198. doi: 10.1038/sj.onc.1203766
Flex, E., Jaiswal, M., Pantaleoni, F., Martinelli, S., Strullu, M., Fansa, E. K., et al. (2014). Activating mutations in RRAS underlie a phenotype within the RASopathy spectrum and contribute to leukaemogenesis. Hum. Mol. Genet. 23, 4315–4327. doi: 10.1093/hmg/ddu148
Fritz, R. D., Letzelter, M., Reimann, A., Martin, K., Fusco, L., and Ritsma, L. (2013). A versatile toolkit to produce sensitive FRET biosensors to visualize signaling in time and space. Sci. Signal. 6:rs12. doi: 10.1126/scisignal.2004135
Fuss, B., Becker, T., Zinke, I., and Hoch, M. (2006). The cytohesinSteppke is essential for insulin signalling in Drosophila. Nature 444, 945–948. doi: 10.1038/nature05412
Gao, C., and Chen, Y. G. (2010). Dishevelled: the hub of Wnt signaling. Cell. Signal. 22, 717–727. doi: 10.1016/j.cellsig.2009.11.021
Garcez Palha, I., Anselme, I., Schneider-Maunoury, S., and Giudicelli, F. (2018). An in vivo translation-reporter system for the study of protein synthesis in zebrafish embryos. Biol. Open 7:bio039362. doi: 10.1242/bio.039362
Garret, T. A., Van Buul, J. D., and Burridge, K. (2007). VEGF-induced Rac1 activation in endothelial cells is regulated by the guanine nucleotide exchange factor Vav2. Exp. Cell Res. 313, 3285–3297. doi: 10.1016/j.yexcr.2007.05.027
Garvalov, B. K., Flynn, K. C., Neukirchen, D., Meyn, L., Teusch, N., Wu, X., et al. (2007). Cdc42 regulates cofilin during the establishment of neuronal polarity. J. Neurosci. 27, 13117–13129. doi: 10.1523/JNEUROSCI.3322-07.2007
Gatta, G., Capocaccia, R., Botta, L., Mallone, S., De Angelis, R., Ardanaz, E., et al. (2017). Burden and centralised treatment in Europe of rare tumours: results of RARECAREnet-a population-based study. Lancet Oncol. 18, 1022–1039. doi: 10.1016/S1470-2045(17)30445-X
Gauthier, A. S., Furstoss, O., Araki, T., Chan, R., Neel, B. G., Kaplan, D. R., et al. (2007). Control of CNS cell-fate decisions by SHP-2 and its dysregulation in Noonan syndrome. Neuron 54, 245–262. doi: 10.1016/j.neuron.2007.03.027
Ge, X., Gong, H., Dumas, K., Litwin, J., Phillips, J. J., Waisfisz, Q., et al. (2016). Missense-depleted regions in population exomes implicate ras superfamily nucleotide-binding protein alteration in patients with brain malformation. NPJ Genomic Med. 1:16036. doi: 10.1038/npjgenmed.2016.36
Geiger, H., and Zheng, Y. (2013). Cdc42 and aging of hematopoietic stem cells. Curr. Opin. Hematol. 20, 295–300. doi: 10.1097/MOH.0b013e3283615aba
Georgiou, M., Marinari, E., Burden, J., and Baum, B. (2008). Cdc42, Par6, and aPKC regulate Arp2/3-mediated endocytosis to control local adherens junction stability. Curr. Biol. 18, 1631–1638. doi: 10.1016/j.cub.2008.09.029
Gerhart, S. V., Eble, D. M., Burger, R. M., Oline, S. N., Vacaru, A., Sadler, K. C., et al. (2012). The Cx43-like connexin protein Cx40.8 is differentially localized during fin ontogeny and fin regeneration. PLoS One 7:e31364. doi: 10.1371/journal.pone.0031364
Gillingham, A. K., Bertram, J., Begum, F., and Munro, S. (2019). In vivo identification of GTPase interactors by mitochondrial relocalization and proximity biotinylation. eLife 8:e45916. doi: 10.7554/eLife.45916
Godi, A., Pertile, P., Meyers, R., Marra, P., Di Tullio, G., Iurisci, C., et al. (1999). ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1, 280–287. doi: 10.1038/12993
Gopal Krishnan, P. D., Golden, E., Woodward, E. A., Pavlos, N. J., and Blancafort, P. (2020). Rab GTPases: emerging oncogenes and tumor suppressive regulators for the editing of survival pathways in cancer. Cancers 12:259.
Gotta, M., Abraham, M. C., and Ahringer, J. (2001). CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr. Biol. 11, 482–488. doi: 10.1016/s0960-9822(01)00142-7
Govek, E. E., Hatten, M. E., and Van Aelst, L. (2011). The role of Rho GTPase proteins in CNS neuronal migration. Dev. Neurobiol. 71, 528–553. doi: 10.1002/dneu.20850
Govek, E. E., Wu, Z., Acehan, D., Molina, H., Rivera, K., Zhu, X., et al. (2018). Cdc42 regulates neuronal polarity during cerebellar axon formation and glial-guided migration. iScience 1, 35–48. doi: 10.1016/j.isci.2018.01.004
Griscelli, C., and Prunieras, M. (1978). Pigment dilution and immunodeficiency: a new syndrome. Int. J. Dermatol. 17, 788–791. doi: 10.1111/j.1365-4362.1978.tb05980.x
Gröbner, S. N., Worst, B. C., Weischenfeldt, J., Buchhalter, I., Kleinheinz, K., Rudnava, V. A., et al. (2018). The landscape of genomic alterations across childhood cancers. Nature 555, 321–327. doi: 10.1038/nature25480
Gui, H., Schriemer, D., Cheng, W. W., Chauhan, R. K., Antiňolo, G., Berrios, C., et al. (2017). Whole exome sequencing coupled with unbiased functional analysis reveals new Hirschsprung disease genes. Genome Biol. 8:48. doi: 10.1186/s13059-017-1174-6
Guo, D., Yang, X., and Shi, L. (2020). Rho GTPase regulators and effectors in autism spectrum disorders: animal models and insights for therapeutics. Cells 9:835. doi: 10.3390/cells9040835
Gupta, S., Howard, S., Hunger, S., Antillon, F., Metzger, M., Israels, T., et al. (2015). “Treating childhood cancers in low- and middle-income countries,” in Disease Control Priorities, 3rd Edn, eds H. Gelband, P. Jha, R. Sankaranarayanan, and S. Horton (Washington, DC: The International Bank for Reconstruction and Development).
Habas, R., Dawid, I. B., and He, X. (2003). Coactivation of Rac and Rho by Wnt/Frizzled signaling is required for vertebrate gastrulation. Genes Dev. 17, 295–309. doi: 10.1101/gad.1022203
Habas, R., Kato, Y., and He, X. (2001). Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell 107, 843–854. doi: 10.1016/s0092-8674(01)00614-6
Haga, R. B., and Ridley, A. J. (2016). Rho GTPases: regulation and roles in cancer cell biology. Small GTPases 7, 207–221. doi: 10.1080/21541248.2016.1232583
Hahn, I., Fuss, B., Peters, A., Werner, T., Sieberg, A., Gosejacob, D., et al. (2013). The Drosophila Arf GEF Steppke controls MAPK activation in EGFR signaling. J. Cell Sci. 126, 2470–2479. doi: 10.1242/jcs.120964
Hakeda-Suzuki, S., Ng, J., Tzu, J., Dietzl, G., Sun, Y., Harms, M., et al. (2002). Rac function and regulation during Drosophila development. Nature 416, 438–442. doi: 10.1038/416438a
Halfon, M. S., Carmena, A., Gisselbrecht, S., Sackerson, C. M., Jiménez, F., and Baylies, M. K. (2000). Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell 103, 63–74. doi: 10.1016/s0092-8674(00)00105-7
Hall, A. (2005). Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 33, 891–895. doi: 10.1042/BST20050891
Hall, A., and Lalli, G. (2010). Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb. Perspect. Biol. 2:a001818. doi: 10.1101/cshperspect.a001818
Hall, E. T., and Verheyen, E. M. (2015). Ras-activated Dsor1 promotes Wnt signaling in Drosophila development. J. Cell Sci. 128, 4499–4511. doi: 10.1242/jcs.175240
Hanovice, N. J., McMains, E., and Gross, J. M. (2016). A GAL4-inducible transgenic tool kit for the in vivo modulation of Rho GTPase activity in zebrafish. Dev. Dyn. 245, 844–853. doi: 10.1002/dvdy.24412
Hardin, J., and King, R. S. (2008). The long and the short of Wnt signaling in C. elegans. Curr. Opin. Genet. Dev. 18, 362–367. doi: 10.1016/j.gde.2008.06.006
Harris, J. M., Wang, A. Y., Boulanger-Weill, J., Santoriello, C., Foianini, S., Lichtman, J. W., et al. (2020). Long-range optogenetic control of axon guidance overcomes developmental boundaries and defects. Dev. Cell 53, 577–588.e7. doi: 10.1016/j.devcel.2020.05.009
Harris, K. P., and Tepass, U. (2008). Cdc42 and Par proteins stabilize dynamic adherens junctions in the Drosophila neuroectoderm through regulation of apical endocytosis. J. Cell. Biol. 183, 1129–1143. doi: 10.1083/jcb.200807020
Harris, K. P., and Tepass, U. (2010). Cdc42 and vesicle trafficking in polarized cells. Traffic 11, 1272–1279. doi: 10.1111/j.1600-0854.2010.01102.x
Harvey, C. D., Ehrhardt, A. G., Cellurale, C., Zhong, H., Yasuda, R., Davis, R. J., et al. (2008). A genetically encoded fluorescent sensor of ERK activity. Proc. Natl. Acad. Sci. USA 105, 19264–19269. doi: 10.1073/pnas.0804598105
Harvey, R. C., Mullighan, C. G., Wang, X., Dobbin, K. K., Davidson, G. S., Bedrick, E. J., et al. (2010). Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood 116, 4874–4884. doi: 10.1182/blood-2009-08-239681
Hasson, P., Egoz, N., Winkler, C., Volohonsky, G., Jia, S., Dinur, T., et al. (2005). EGFR signaling attenuates Groucho-dependent repression to antagonize Notch transcriptional output. Nat. Genet. 37, 101–105. doi: 10.1038/ng1486
Hayes, M. N., McCarthy, K., Jin, A., Oliveira, M. L., Iyer, S., Garcia, S. P., et al. (2018). Vangl2/RhoA signaling pathway regulates stem cell self-renewal programs and growth in rhabdomyosarcoma. Cell Stem Cell 22, P414–427.E6. doi: 10.1016/j.stem.2018.02.002
Heasman, S. J., and Ridley, A. J. (2008). Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9, 690–701. doi: 10.1038/nrm2476
Heide, M., Haffner, C., Murayama, A., Kurotaki, Y., Shinohara, H., Okano, H., et al. (2020). Human-specific ARHGAP11B increases size and folding of primate neocortex in the fetal marmoset. Science 369, 546–550. doi: 10.1126/science.abb2401
Helmchen, F., and Denk, W. (2005). Deep tissue two-photon microscopy. Nat. Methods 2, 932–940. doi: 10.1038/nmeth818
Hirata, E., and Kiyokawa, E. (2019). ERK activity imaging during migration of living cells in vitro and in vivo. Int. J. Mol. Sci. 20:679. doi: 10.3390/ijms20030679
Hirata, E., Yukinaga, H., Kamioka, Y., Arakawa, Y., Miyamoto, S., Okada, T., et al. (2012). In vivo fluorescence resonance energy transfer imagingreveals differential activation of Rho-family GTPases in glioblastoma cell invasion. J. Cell Sci. 125, 858–868. doi: 10.1242/jcs.089995
Hodgson, L., Shen, F., and Hahn, K. (2010). Biosensors for characterizing the dynamics of rho family GTPases in living cells. Curr. Protoc. Cell Biol. 14, 1–26. doi: 10.1002/0471143030.cb1411s46
Honda, A., Nogami, M., Yokozeki, T., Yamazaki, M., Nakamura, H., Watanabe, H., et al. (1999). Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell 99, 521–532. doi: 10.1016/s0092-8674(00)81540-8
Horner, V. L., and Caspary, T. (2011). Disrupted dorsal neural tube BMP signaling in the cilia mutant Arl13b hnn stems from abnormal Shh signaling. Dev. Biol. 355, 43–54.
Hsu, A. P., Donkó, A., Arrington, M. E., Swamydas, M., Fink, D., Das, A., et al. (2019). Dominant activating RAC2 mutation with lymphopenia, immunodeficiency, and cytoskeletal defects. Blood 133, 1977–1988. doi: 10.1182/blood-2018-11-886028
Huang, P., Xiong, F., Megason, S. G., and Schier, A. F. (2012). Attenuation of Notch and Hedgehog signaling is required for fate specification in the spinal cord. PLoS Genet. 8:e1002762. doi: 10.1371/journal.pgen.1002762
Huang, W. Q., Lin, Q., Zhuang, X., Cai, L. L., Ruan, R. S., Lu, Z. X., et al. (2014). Structure, function, and pathogenesis of SHP2 in developmental disorders and tumorigenesis. Curr. Cancer Drug Targets 14, 567–588. doi: 10.2174/1568009614666140717105001
Huangfu, D., and Anderson, K. V. (2005). Cilia and Hedgehog responsiveness in the mouse. Proc. Natl. Acad. Sci. U.S.A. 102, 11325–11330. doi: 10.1073/pnas.0505328102
Hutagalung, A. H., and Novick, P. J. (2011). Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91, 119–149. doi: 10.1152/physrev.00059.2009
Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., et al. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229. doi: 10.1038/nbt.2501
Ito, H., Morishita, R., Tabata, H., and Nagata, K. (2014). Roles of Rho small GTPases in the tangentially migrating neurons. Histol. Histopathol. 29, 871–879. doi: 10.14670/HH-29.871
Jain, S., Yoon, S. Y., Zhu, L., Brodbeck, J., Dai, J., Walker, D., et al. (2012). Arf4 determines dentate gyrus-mediated pattern separation by regulating dendritic spine development. PLoS One 7:e46340. doi: 10.1371/journal.pone.0046340
Jenkins, D., Beales, P. L., and Wilkie, A. (2012). Rab23 is required for cardiac progenitor cell differentiation and positively-regulates Wnt11/AP-1 signalling in zebrafish. Cilia 1:O6. doi: 10.1186/2046-2530-1-S1-O6
Jeong, W., Ro, E. J., and Choi, K. (2018). Interaction between Wnt/β-catenin and RAS-ERK pathways and an anti-cancer strategy via degradations of β-catenin and RAS by targeting the Wnt/β-catenin pathway. Adv. Biol. Regul. 68, 46–54. doi: 10.1016/j.jbior.2018.01.001
Ji, W., and Rivero, F. (2016). Atypical Rho GTPases of the RhoBTB subfamily: roles in vesicle trafficking and tumorigenesis. Cells 5:28. doi: 10.3390/cells5020028
Jindal, G. A., Goyal, Y., Burdine, R. D., Rauen, K. A., and Shvartsman, S. Y. (2015). RASopathies: unraveling mechanisms with animal models. Dis. Model. Mech. 8, 769–782. doi: 10.1242/dmm.020339
Johnson, H. E., Goyal, Y., Pannucci, N. L., Schüpbach, T., Shvartsman, S. Y., and Toettcher, J. E. (2017). The spatiotemporal limits of developmental Erk signaling. Dev. Cell 40, 185–192. doi: 10.1016/j.devcel.2016.12.002
Johnson, H. E., and Toettcher, J. E. (2019). Signaling dynamics control cell fate in the early Drosophila embryo. Dev. Cell 48, 361–370.e3. doi: 10.1016/j.devcel.2019.01.009
Johnsson, A. E., Dai, Y., Nobis, M., Baker, M. J., McGhee, E. J., Walker, S., et al. (2014). The Rac-FRET mouse reveals tight spatiotemporal control of Rac activity in primary cells and tissues. Cell Rep. 6, 1153–1164. doi: 10.1016/j.celrep.2014.02.024
Jongmans, M. C., van der Burgt, I., Hoogerbrugge, P. M., Noordam, K., Yntema, H. G., Nillesen, W. M., et al. (2011). Cancer risk in patients with Noonan syndrome carrying a PTPN11 mutation. Eur. J. Hum. Genet. 19, 870–874. doi: 10.1038/ejhg.2011.37
Jopling, C., van Geemen, D., and den Hertog, J. (2007). Shp2 knockdown and Noonan/LEOPARD mutant Shp2-induced gastrulation defects. PLoS Genet. 3:e225. doi: 10.1371/journal.pgen.0030225
Kahn, R. A., Cherfils, J., Elias, M., Lovering, R. C., Munro, S., and Schurmann, A. (2006). Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J. Cell Biol. 172, 645–650. doi: 10.1083/jcb.200512057
Kang, M., and Lee, Y. S. (2019). The impact of RASopathy-associated mutations on CNS development in mice and humans. Mol. Brain 12:96. doi: 10.1186/s13041-019-0517-5
Kaplan, W., Wirtz, V. J., Mantel-Teeuwisse, A., Stolk, P., Duthey, B., and Laing, R. (2013). Priority Medicines for Europe and the World 2013 Update. Geneva: World Health Organization.
Kardash, E., Reichman-Fried, M., Maître, J. L., Boldajipour, B., Papusheva, E., Messerschmidt, E. M., et al. (2010). A role for Rho GTPases and cell-cell adhesion in single-cell motility in vivo. Nat. Cell Biol. 12, 47–53. doi: 10.1038/ncb2003
Karvonen, H., Perttilä, R., Niininen, W., Hautanen, V., Barker, H., Murumägi, A., et al. (2019). Wnt5a and ROR1 activate non-canonical Wnt signaling via RhoA in TCF3-PBX1 acute lymphoblastic leukemia and highlight new treatment strategies via Bcl-2 co-targeting. Oncogene 38, 3288–3300. doi: 10.1038/s41388-018-0670-9
Kaufmann, N., Wills, Z. P., and Van Vactor, D. (1998). Drosophila Rac1 controls motor axon guidance. Development 125, 453–461.
Kawamura, A., Koshida, S., Hijikata, H., Sakaguchi, T., Kondoh, H., and Takada, S. (2005). Zebrafish hairy/enhancer of split protein links FGF signaling to cyclic gene expression in the periodic segmentation of somites. Genes Dev. 19, 1156–1161. doi: 10.1101/gad.1291205
Kay, A. J., and Hunter, C. P. (2001). CDC-42 regulates PAR protein localization and function to control cellular and embryonic polarity in C. elegans. Curr. Biol. 11, 474–481. doi: 10.1016/s0960-9822(01)00141-5
Kerber, R. A., O’Brien, E., and Cawthon, R. M. (2009). Gene expression profiles associated with aging and mortality in humans. Aging Cell 8, 239–250. doi: 10.1111/j.1474-9726.2009.00467.x
Kilian, B., Mansukoski, H., Barbosa, F. C., Ulrich, F., Tada, M., and Heisenberg, C. P. (2003). The role of Ppt/Wnt5 in regulating cell shape and movement during zebrafish gastrulation. Mech. Dev. 120, 467–476. doi: 10.1016/s0925-4773(03)00004-2
Kim, I. S., Heilmann, S., Kansler, E. R., Zhang, Y., Zimmer, M., Ratnakumar, K., et al. (2017). Microenvironment-derived factors driving metastatic plasticity in melanoma. Nat. Commun. 8:14343. doi: 10.1038/ncomms14343
Kim, J., Lee, S., Jung, K., Oh, W. C., Kim, N., Son, S., et al. (2019). Intensiometric biosensors visualize the activity of multiple small GTPases in vivo. Nat. Commun. 10:211. doi: 10.1038/s41467-018-08217-3
Kim, W., Kim, S. Y., Kim, T., Kim, M., Bae, D. J., Choi, H. I., et al. (2013). ADP-ribosylation factors 1 and 6 regulate Wnt/β-catenin signaling via control of LRP6 phosphorylation. Oncogene 32, 3390–3396. doi: 10.1038/onc.2012.373
Kirjavainen, A., Laos, M., Anttonen, T., and Pirvola, U. (2015). The Rho GTPase Cdc42 regulates hair cell planar polarity and cellular patterning in the developing cochlea. Biol. Open 4, 516–526. doi: 10.1242/bio.20149753
Kitano, M., Nakaya, M., Nakamura, T., Nagata, S., and Matsuda, M. (2008). Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature 453, 241–245. doi: 10.1038/nature06857
Kondo, Y., Hanai, A., Nakai, W., Katoh, Y., Nakayama, K., and Shin, H. W. (2012). ARF1 and ARF3 are required for the integrity of recycling endosomes and the recycling pathway. Cell Struct. Funct. 37, 141–154. doi: 10.1247/csf.12015
Koraïchi, F., Gence, R., Bouchenot, C., Grosjean, S., Lajoie-Mazenc, I., Favre, G., et al. (2018). High-content tripartite split-GFP cell-based assays to screen for modulators of small GTPase activation. J. Cell Sci. 131:jcs210419. doi: 10.1242/jcs.210419
Koronakis, V., Hume, P. J., Humphreys, D., Liu, T., Hørning, O., Jensen, O. N., et al. (2011). WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc. Natl. Acad. Sci. U.S.A. 108, 14449–14454. doi: 10.1073/pnas.1107666108
Kouchi, Z., Igarashi, T., Shibayama, N., Inanobe, S., Sakurai, K., Yamaguchi, H., et al. (2011). Phospholipase Cdelta3 regulates RhoA/Rho kinase signaling and neurite outgrowth. J. Biol. Chem. 286, 8459–8471. doi: 10.1074/jbc.M110.171223
Kowalik, L., and Chen, J. K. (2017). Illuminating developmental biology through photochemistry. Nat. Chem. Biol. 13, 587–598. doi: 10.1038/nchembio.2369
Kratz, C. P., Franke, L., Peters, H., Kohlschmidt, N., Kazmierczak, B., and Finckh, U. (2015). Cancer spectrum and frequency among children with Noonan, Costello, and cardio-facio-cutaneous syndromes. Br. J. Cancer 112, 1392–1397. doi: 10.1038/bjc.2015.75
Kratzer, M. C., Becker, S. F. S., Grund, A., Merks, A., Harnoš, J., Bryja, V., et al. (2020). The Rho guanine nucleotide exchange factor Trio is required for neural crest cell migration and interacts with Dishevelled. Development 147, dev186338. doi: 10.1242/dev.186338
Kumar, S., Xu, J., Perkins, C., Guo, F., Snapper, S., Finkelman, F. D., et al. (2012). Cdc42 regulates neutrophil migration via crosstalk between WASp, CD11b, and microtubules. Blood 120, 3563–3574.
Kwan, K. M., Otsuna, H., Kidokoro, H., Carney, K. R., Saijoh, Y., and Chien, C. B. (2012). A complex choreography of cell movements shapes the vertebrate eye. Development 139, 359–372. doi: 10.1242/dev.071407
Lagresle-Peyrou, C., Olichon, A., Sadek, H., Roche, P., Tardy, C., Da Silva, C., et al. (2020). A gain-of-function RAC2 mutation is associated with bone-marrow hypoplasia and an autosomal dominant form of severe combined immunodeficiency. Haematologica 9:haematol.2019.230250. doi: 10.3324/haematol.2019.230250
Lam, M. T., Coppola, S., Krumbach, O. H. F., Prencipe, G., Insalaco, A., Cifaldi, C., et al. (2019). A novel disorder involving dyshematopoiesis, inflammation, and HLH due to aberrant CDC42 function. J. Exp. Med. 216, 2778–2799. doi: 10.1084/jem.20190147
Lamalice, L., Le Boeuf, F., and Huot, J. (2007). Endothelial cell migration during angiogenesis. Circ. Res. 100, 782–794. doi: 10.1161/01.RES.0000259593.07661.1e
Larkins, C. E., Aviles, G. D., East, M. P., Kahn, R. A., and Caspary, T. (2011). Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol. Biol. Cell 22, 4694–4703. doi: 10.1091/mbc.E10-12-0994
Lawson, C. D., and Ridley, A. J. (2018). Rho GTPase signaling complexes in cell migration and invasion. J. Cell Biol. 217, 447–457. doi: 10.1083/jcb.201612069
Lee, D. M., Rodrigues, F. F., Yu, C. G., Swan, M., and Harris, T. J. (2015). PH Domain-Arf G protein interactions localize the Arf-GEF steppke for cleavage furrow regulation in Drosophila. PLoS One 10:e0142562. doi: 10.1371/journal.pone.0142562
Lee, S., Craig, B. T., Romain, C. V., Qiao, J., and Chung, D. H. (2014). Silencing of CDC42 inhibits neuroblastoma cell proliferation and transformation. Cancer Lett. 355, 210–216. doi: 10.1016/j.canlet.2014.08.033
Lefebvre, S., Burglen, L., Reboullet, S., Clermont, O., Burlet, P., Viollet, L., et al. (1995). Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80, 155–165. doi: 10.1016/0092-8674(95)90460-3
Legius, E., Schrander-Stumpel, C., Schollen, E., Pulles-Heintzberger, C., Gewillig, M., and Fryns, J. P. (2002). PTPN11 mutations in LEOPARD syndrome. J. Med. Genet. 39, 571–574. doi: 10.1136/jmg.39.8.571
Leng, W., Pang, X., Xia, H., Li, M., Chen, L., Tang, Q., et al. (2013). Novel split-luciferase-based genetically encoded biosensors for noninvasive visualization of Rho GTPases. PLoS One 8:e62230. doi: 10.1371/journal.pone.0062230
Leone, D. P., Srinivasan, K., Brakebusch, C., and McConnell, S. K. (2010). The rho GTPase Rac1 is required for proliferation and survival of progenitors in the developing forebrain. Dev. Neurobiol. 70, 659–678. doi: 10.1002/dneu.20804
Levitan, D., and Greenwald, I. (1998). LIN-12 protein expression and localization during vulval development in C. elegans. Development 125, 3101–3109.
Li, S., Mattar, P., Dixit, R., Lawn, S. O., Wilkinson, G., Kinch, C., et al. (2014). RAS/ERK signaling controls proneural genetic programs in cortical development and gliomagenesis. J. Neurosci. 34, 2169–2190. doi: 10.1523/JNEUROSCI.4077-13.2014
Li, W. M., Webb, S. E., Chan, C. M., and Miller, A. L. (2008). Multiple roles of the furrow deepening Ca2+ transient during cytokinesis in zebrafish embryos. Dev. Biol. 316, 228–248. doi: 10.1016/j.ydbio.2008.01.027
Li, X., Newbern, J. M., Wu, Y., Morgan-Smith, M., Zhong, J., Charron, J., et al. (2012). MEK is a key regulator of gliogenesis in the developing brain. Neuron 75, 1035–1050. doi: 10.1016/j.neuron.2012.08.031
Li, Y., Wei, Q., Zhang, Y., Ling, K., and Hu, J. (2010). The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J. Cell Biol. 189, 1039–1051. doi: 10.1083/jcb.200912001
Lin, X., Buff, E. M., Perrimon, N., and Michelson, A. M. (1999). Heparan sulfate proteoglycans are essential for FGF receptor signaling during Drosophila embryonic development. Development 126, 3715–3723.
Liu, W., Du, W., Shang, X., Wang, L., Evelyn, C., Florian, M. C., et al. (2019). Rational identification of a Cdc42 inhibitor presents a new regimen for long-term hematopoietic stem cell mobilization. Leukemia 33, 749–761. doi: 10.1038/s41375-018-0251-5
Liu, Y., Wang, J., Li, J., Wang, R., Tharakan, B., Zhang, S. L., et al. (2017). Deletion of Cdc42 in embryonic cardiomyocytes results in right ventricle hypoplasia. Clin. Transl. Med. 6:40. doi: 10.1186/s40169-017-0171-4
Lombardo, V. A., Sporbert, A., and Abdelilah-Seyfried, S. (2012). Cell tracking using photoconvertible proteins during zebrafish development. J. Vis. Exp. 67:4350. doi: 10.3791/4350
López-Juárez, A., Titus, H. E., Silbak, S. H., Pressler, J. W., Rizvi, T. A., Bogard, M., et al. (2017). Oligodendrocyte Nf1 controls aberrant notch activation and regulates myelin structure and behavior. Cell Rep. 19, 545–557. doi: 10.1016/j.celrep.2017.03.073
Lu, F. I., Wang, Y. T., Wang, Y. S., Wu, C. Y., and Li, C. C. (2019). Involvement of BIG1 and BIG2 in regulating VEGF expression and angiogenesis. FASEB J. 33, 9959–9973. doi: 10.1096/fj.201900342
Lupo, V., Galindo, M. I., Martínez-Rubio, D., Sevilla, T., Vílchez, J. J., and Palau, F. (2009). Missense mutations in the SH3TC2 protein causing Charcot-Marie-Tooth disease type 4C affect its localization in the plasma membrane and endocytic pathway. Hum. Mol. Genet. 18, 4603–4614. doi: 10.1093/hmg/ddp427
Maldonado, M. D. M., and Dharmawardhane, S. (2018). Targeting Rac and Cdc42 GTPases in cancer. Cancer Res. 78, 3101–3111. doi: 10.1158/0008-5472.CAN-18-0619
Malumbres, M., and Barbacid, M. (2003). RAS oncogenes: the first 30 years. Nat. Rev. Cancer 3, 459–465. doi: 10.1038/nrc1097
Manara, E., Baron, E., Tregnago, C., Aveic, S., Bisio, V., Bresolin, S., et al. (2014). MLL-AF6 fusion oncogene sequesters AF6 into the nucleus to trigger RAS activation in myeloid leukemia. Blood 124, 263–272. doi: 10.1182/blood-2013-09-525741
Marlow, F., Topczewski, J., Sepich, D., and Solnica-Krezel, L. (2002). Zebrafish Rho kinase 2 acts downstream of Wnt11 to mediate cell polarity and effective convergence and extension movements. Curr. Biol. 12, 876–884. doi: 10.1016/s0960-9822(02)00864-3
Martinelli, S., De Luca, A., Stellacci, E., Rossi, C., Checquolo, S., Lepri, F., et al. (2010). Heterozygous germline mutations in the CBL tumor-suppressor gene cause a Noonan syndrome-like phenotype. Am. J. Hum. Genet. 87, 250–257. doi: 10.1016/j.ajhg.2010.06.015
Martinelli, S., Krumbach, O. H. F., Pantaleoni, F., Coppola, S., Amin, E., Pannone, L., et al. (2018). Functional dysregulation of CDC42 causes diverse developmental phenotypes. Am. J. Hum. Genet. 102, 309–320. doi: 10.1016/j.ajhg.2017.12.015
Martinelli, S., Stellacci, E., Pannone, L., D’Agostino, D., Consoli, F., Lissewski, C., et al. (2015). Molecular diversity and associated phenotypic spectrum of germline CBL mutations. Hum. Mutat. 36, 787–796. doi: 10.1002/humu.22809
Marwaha, R., Dwivedi, D., and Sharma, M. (2019). Emerging roles of Arf-Like GTP-binding proteins: from membrane trafficking to cytoskeleton dynamics and beyond. Proc. Indian Natl. Sci. Acad. 85, 189–212. doi: 10.16943/ptinsa/2019/49574
Marygold, S. J., Walker, C. D., Orme, M. H., and Leevers, S. J. (2011). Genetic characterization of ebi reveals its critical role in Drosophila wing growth. Fly 5, 291–303. doi: 10.4161/fly.5.4.18276
Maryu, G., Matsuda, M., and Aoki, K. (2016). Multiplexed fluorescence imaging of ERK and Akt activities and cell-cycle progression. Cell Struct. Funct. 41, 81–92. doi: 10.1247/csf.16007
Matozaki, T., Murata, Y., Saito, Y., Okazawa, H., and Ohnishi, H. (2009). Protein tyrosine phosphatase SHP-2: a proto-oncogene product that promotes Ras activation. Cancer Sci. 100, 1786–1793. doi: 10.1111/j.1349-7006.2009.01257.x
Matthews, H. K., Marchant, L., Carmona-Fontaine, C., Kuriyama, S., Larraín, J., Holt, M. R., et al. (2008). Directional migration of neural crest cells in vivo is regulated by Syndecan-4/Rac1 and non-canonical Wnt signaling/RhoA. Development 135, 1771–1780. doi: 10.1242/dev.017350
Mayor, R., and Theveneau, E. (2013). The neural crest. Development 140, 2247–2251. doi: 10.1242/dev.091751
Mayr, V., Sturtzel, C., Stadler, M., Grissenberger, S., and Distel, M. (2018). Fast dynamic in vivo monitoring of Erk activity at single cell resolution in DREKA Zebrafish. Front. Cell Dev. Biol. 6:111. doi: 10.3389/fcell.2018.00111
Mazelova, J., Astuto-Gribble, L., Inoue, H., Tam, B. M., Schonteich, E., Prekeris, R., et al. (2009). Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J. 28, 183–192. doi: 10.1038/emboj.2008.267
Melendez, J., Liu, M., Sampson, L., Akunuru, S., Han, X., Vallance, J., et al. (2013). Cdc42 coordinates proliferation, polarity, migration, and differentiation of small intestinal epithelial cells in mice. Gastroenterology 145, 808–819. doi: 10.1053/j.gastro.2013.06.021
Mellor, H. (2010). The role of formins in filopodia formation. Biochim. Biophys. Acta 1803, 191–200. doi: 10.1016/j.bbamcr.2008.12.018
Merajver, S. D., and Usmani, S. Z. (2005). Multifaceted role of Rho proteins in angiogenesis. J. Mammary Gland Biol. Neoplasia 10, 291–298. doi: 10.1007/s10911-006-9002-8
Mezzacappa, C., Komiya, Y., and Habas, R. (2012). Activation and function of small GTPases Rho, Rac, and Cdc42 during gastrulation. Methods Mol. Biol. 839, 119–131. doi: 10.1007/978-1-61779-510-7_10
Mich, J. K., Payumo, A. Y., Rack, P. G., and Chen, J. K. (2014). In vivo imaging of Hedgehog pathway activation with a nuclear fluorescent reporter. PLoS One 9:e103661. doi: 10.1371/journal.pone.0103661
Mohapatra, B., Ahmada, G., Nadeau, S., Zutshi, N., An, W., Scheffe, S., et al. (2013). Protein tyrosine kinase regulation by ubiquitination: critical roles of Cbl-family ubiquitin ligases. Biochim. Biophys. Acta 1833, 122–139. doi: 10.1016/j.bbamcr.2012.10.010
Moschovi, M., Touliatou, V., Papadopoulou, A., Mayakou, M. A., Nikolaidou-Karpathiou, P., and Kitsiou-Tzeli, S. (2007). Rhabdomyosarcoma in a patient with Noonan syndrome phenotype and review of the literature. J. Pediatr. Hematol. Oncol. 29, 341–344. doi: 10.1097/MPH.0b013e31805d8f57
Moss, J. J., Hammond, C. L., and Lane, J. D. (2020). Zebrafish as a model to study autophagy and its role in skeletal development and disease. Histochem. Cell Biol. 154, 549–564. doi: 10.1007/s00418-020-01917-2
Motta, M., Fidan, M., Bellacchio, E., Pantaleoni, F., Schneider-Heieck, K., Coppola, S., et al. (2019). Dominant Noonan syndrome-causing LZTR1 mutations specifically affect the Kelch domain substrate-recognition surface and enhance RAS-MAPK signaling. Hum. Mol. Genet. 28, 1007–1022. doi: 10.1093/hmg/ddy412
Motta, M., Pannone, L., Pantaleoni, F., Bocchinfuso, G., Radio, F. C., Cecchetti, S., et al. (2020). Enhanced MAPK1 function causes a neurodevelopmental disorder within the RASopathy clinical spectrum. Am. J. Hum. Genet. 107, 499–513. doi: 10.1016/j.ajhg.2020.06.018
Murphy, L. O., MacKeigan, J. P., and Blenis, J. (2004). A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Mol. Cell Biol. 24, 144–153.
Myers, K. R., and Casanova, J. E. (2008). Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends Cell Biol. 18, 184–192. doi: 10.1016/j.tcb.2008.02.002
Nakagama, Y., Takeda, N., Ogawa, S., Takeda, H., Furutani, Y., Nakanishi, T., et al. (2020). Noonan syndrome-associated biallelic LZTR1 mutations cause cardiac hypertrophy and vascular malformations in zebrafish. Mol. Genet. Genomic Med. 8:e1107. doi: 10.1002/mgg3.1107
Nayak, R. C., Chang, K. H., Vaitinadin, N. S., and Cancelas, J. A. (2013). Rho GTPases control specific cytoskeleton-dependent functions of hematopoietic stem cells. Immunol. Rev. 256, 255–268. doi: 10.1111/imr.12119
Ng, J., Nardine, T., Harms, M., Tzu, J., Goldstein, A., Sun, Y., et al. (2002). Rac GTPases control axon growth, guidance and branching. Nature 416, 442–447. doi: 10.1038/416442a
Nguyen, M. K., Kim, C. Y., Kim, J. M., Park, B. O., Lee, S., Park, H., et al. (2016). Optogenetic oligomerization of Rab GTPases regulates intracellular membrane trafficking. Nat. Chem. Biol. 12, 431–436. doi: 10.1038/nchembio.2064
Nikolaidou, K. K., and Barrett, K. (2004). A Rho GTPase signaling pathway is used reiteratively in epithelial folding and potentially selects the outcome of Rho activation. Curr. Biol. 14, 1822–1826. doi: 10.1016/j.cub.2004.09.080
Nishimura, N., Van Huyen Pham, T., Hartomo, T. B., Lee, M. J., Hasegawa, D., Takeda, H., et al. (2011). Rab15 expression correlates with retinoic acid-induced differentiation of neuroblastoma cells. Oncol. Rep. 26, 145–151. doi: 10.3892/or.2011.1255
Nouri, K., Timson, D. J., and Ahmadian, M. R. (2020). New model for the interaction of IQGAP1 with CDC42 and RAC1. Small GTPases 11, 16–22. doi: 10.1080/21541248.2017.1321169
Novarino, G., Fenstermaker, A. G., Zaki, M. S., Hofree, M., Silhavy, J. L., Heiberg, A. D., et al. (2014). Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science 343, 506–511. doi: 10.1126/science.1247363
O’Neill, P. R., Kalyanaraman, V., and Gautam, N. (2016). Subcellular optogenetic activation of Cdc42 controls local and distal signaling to drive immune cell migration. Mol. Biol. Cell 27, 1442–1450. doi: 10.1091/mbc.E15-12-0832
O’Reilly, A. M., Pluskey, S., Shoelson, S. E., and Neel, B. G. (2000). Activated mutants of SHP-2 preferentially induce elongation of Xenopus animal caps. Mol. Cell Biol. 20, 299–311. doi: 10.1128/mcb.20.1.299-311.2000
Oro, A. E. (2007). The primary cilia, a ‘Rab-id’ transit system for hedgehog signaling. Curr. Opin. Cell Biol. 19, 691–696. doi: 10.1016/j.ceb.2007.10.008
Paganini, I., Chang, V. Y., Capone, G. L., Vitte, J., Benelli, M., Barbetti, L., et al. (2015). Expanding the mutational spectrum of LZTR1 in schwannomatosis. Eur. J. Hum. Genet. 23, 963–968. doi: 10.1038/ejhg.2014.220
Palmer, D. J., Helms, J. B., Beckers, C. J., Orci, L., and Rothman, J. E. (1993). Binding of coatomer to Golgi membranes requires ADP-ribosylation factor. J. Biol. Chem. 268, 12083–12089.
Palucka, A. K., and Coussens, L. M. (2016). The basis of oncoimmunology. Cell 164, 1233–1247. doi: 10.1016/j.cell.2016.01.049
Pandit, B., Sarkozy, A., Pennacchio, L. A., Carta, C., Oishi, K., Martinelli, S., et al. (2007). Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat. Genet. 39, 1007–1012. doi: 10.1038/ng2073
Patel, A. L., Yeung, E., McGuire, S. E., Wu, A. Y., Toettcher, J. E., Burdine, R. D., et al. (2019). Optimizing photoswitchable MEK. Proc. Natl Acad. Sci. U.S.A. 116, 25756–25763. doi: 10.1073/pnas.1912320116
Pérez, B., Mechinaud, F., Galambrun, C., Ben Romdhane, N., Isidor, B., Philip, N., et al. (2010). Germline mutations of the CBL gene define a new genetic syndrome with predisposition to juvenile myelomonocytic leukaemia. J. Med. Genet. 47, 686–691. doi: 10.1136/jmg.2010.076836
Petrosyan, A. (2015). Onco-Golgi: Is fragmentation a gate to cancer progression? Biochem. Mol. Biol. J. 1:16. doi: 10.21767/2471-8084.100006
Pham, T. V., Hartomo, T. B., Lee, M. J., Hasegawa, D., Ishida, T., Kawasaki, K., et al. (2012). Rab15 alternative splicing is altered in spheres of neuroblastoma cells. Oncol. Rep. 27, 2045–2049. doi: 10.3892/or.2012.1731
Piloto, S., and Schilling, T. F. (2010). Ovo1 links Wnt signaling with N-cadherin localization during neural crest migration. Development 137, 1981–1990. doi: 10.1242/dev.048439
Piotrowski, A., Xie, J., Liu, Y. F., Poplawski, A. B., Gomes, A. R., Madanecki, P., et al. (2014). Germline loss-of-function mutations in LZTR1 predispose to an inherited disorder of multiple schwannomas. Nat. Genet. 46, 182–187. doi: 10.1038/ng.2855
Posey, J. E., O’Donnell-Luria, A. H., Chong, J. X., Harel, T., Jhangiani, S. N., CobanAkdemir, Z. H., et al. (2019). Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet. Med. 21, 798–812. doi: 10.1038/s41436-018-0408-7
Qin, W., Lu, X., and Lin, S. (2018). Programmable base editing in zebrafish using a modified CRISPR-Cas9 system. Methods 150, 19–23. doi: 10.1016/j.ymeth.2018.07.010
Qu, L., Pan, C., He, S. M., Lang, B., Gao, G. D., Wang, X. L., et al. (2019). The Ras superfamily of small GTPases in Non-neoplastic cerebral diseases. Front. Mol. Neurosci. 12:121. doi: 10.3389/fnmol.2019.00121
Rabouille, C., and Haase, G. (2016). Editorial: golgi pathology in neurodegenerative diseases. Front. Neurosci. 9:489. doi: 10.3389/fnins.2015.00489
Rasika, S., Passemard, S., Verloes, A., Gressens, P., and El Ghouzzi, V. (2018). Golgipathies in neurodevelopment: a new view of old defects. Dev. Neurosci. 40, 396–416. doi: 10.1159/000497035
Ratner, N., and Miller, S. J. (2015). A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat. Rev. Cancer 15, 290–301. doi: 10.1038/nrc3911
Rauen, K. A. (2013). The RASopathies. Annu. Rev. Genomics Hum. Genet. 14, 355–369. doi: 10.1146/annurev-genom-091212-153523
Regot, S., Hughey, J. J., Bajar, B. T., Carrasco, S., and Covert, M. W. (2014). High-sensitivity measurements of multiple kinase activities in live single cells. Cell 157, 1724–1734. doi: 10.1016/j.cell.2014.04.039
Reijnders, M., Ansor, N. M., Kousi, M., Yue, W. W., Tan, P. L., Clarkson, K., et al. (2017). RAC1 missense mutations in developmental disorders with diverse phenotypes. Am. J. Hum. Genet. 101, 466–477. doi: 10.1016/j.ajhg.2017.08.007
Reinhard, C., Schweikert, M., Wieland, F. T., and Nickel, W. (2003). Functional reconstitution of COPI coat assembly and disassembly using chemically defined components. Proc. Natl. Acad. Sci U.S.A. 100, 8253–8257. doi: 10.1073/pnas.1432391100
Renninger, S. L., and Orger, M. B. (2013). Two-photon imaging of neural population activity in zebrafish. Methods 62, 255–267. doi: 10.1016/j.ymeth.2013.05.016
Ridley, A. J. (2006). Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 16, 522–529. doi: 10.1016/j.tcb.2006.08.006
Ridley, A. J., Schwartz, M. A., Burridge, K., Firtel, R. A., Ginsberg, M. H., Borisy, G., et al. (2003). Cell migration: integrating signals from front to back. Science 302, 1704–1709. doi: 10.1126/science.1092053
Riedl, J., Crevenna, A. H., Kessenbrock, K., Yu, J. H., Neukirchen, D., Bista, M., et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5, 605–607. doi: 10.1038/nmeth.1220
Roberti, M. C., La Starza, R., Surace, C., Sirleto, P., Pinto, R. M., Pierini, V., et al. (2009). RABGAP1L gene rearrangement resulting from a der(Y)t(Y;1)(q12;q25) in acute myeloid leukemia arising in a child with Klinefelter syndrome. Virchows Arch. 454, 311–316. doi: 10.1007/s00428-009-0732-z
Roberts, A. E., Araki, T., Swanson, K. D., Montgomery, K. T., Schiripo, T. A., Joshi, V. A., et al. (2007). Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat. Genet. 39, 70–74. doi: 10.1038/ng1926
Rodrigues, F. F., and Harris, T. J. C. (2019). Key roles of Arf small G proteins and biosynthetic trafficking for animal development. Small GTPases 10, 403–410. doi: 10.1080/21541248.2017.1304854
Roignant, J. Y., and Treisman, J. E. (2009). Pattern formation in the Drosophila eye disc. Int. J. Dev. Biol. 53, 795–804. doi: 10.1387/ijdb.072483jr
Rojas, A. M., Fuentes, G., Rausell, A., and Valencia, A. (2012). The Ras protein superfamily: evolutionary tree and role of conserved amino acids. J. Cell Biol. 196, 189–201. doi: 10.1083/jcb.201103008
Rokita, J. L., Rathi, K. S., Cardenas, M. F., Upton, K. A., Jayaseelan, J., Cross, K. L., et al. (2019). Genomic profiling of childhood tumor patient-derived xenograft models to enable rational clinical trial design. Cell Rep. 29, 1675–1689.e9. doi: 10.1016/j.celrep.2019.09.071
Rosello, M., Vougny, J., Czarny, F., Mione, M. C., Concordet, J. P., Albadri, S., et al. (2021). Precise base editing for the in vivo study of developmental signaling and human pathologies in zebrafish. eLife 10:e65552. doi: 10.7554/eLife.65552
Rosowski, E. E., Deng, Q., Keller, N. P., and Huttenlocher, A. (2016). Rac2 functions in both neutrophils and macrophages to mediate motility and host defense in larval Zebrafish. J. Immunol. 197, 4780–4790. doi: 10.4049/jimmunol.1600928
Ross, B. L., Tenner, B., Markwardt, M. L., Zviman, A., Shi, G., Kerr, J. P., et al. (2018). Single-color, ratiometric biosensors for detecting signaling activities in live cells. eLife 7:e35458. doi: 10.7554/eLife.35458
Ross, M. E., Mahfouz, R., Onciu, M., Liu, H. C., Zhou, X., Song, G., et al. (2004). Gene expression profiling of pediatric acute myelogenous leukemia. Blood 104, 3679–3687. doi: 10.1182/blood-2004-03-1154
Rottinger, E., Besnardeau, L., and Lepage, T. (2004). A Raf/MEK/ERK signaling pathway is required for development of the sea urchin embryo micromere lineage through phosphorylation of the transcription factor Ets. Development 131, 1075–1087. doi: 10.1242/dev.01000
Runtuwene, V., van Eekelen, M., Overvoorde, J., Rehmann, H., Yntema, H. G., Nillesen, W. M., et al. (2011). Noonan syndrome gain-of-function mutations in NRAS cause zebrafish gastrulation defects. Dis. Model. Mech. 4, 393–399. doi: 10.1242/dmm.007112
Sanders, T. A., Llagostera, E., and Barna, M. (2013). Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nature 497, 628–632. doi: 10.1038/nature12157
Santoriello, C., Deflorian, G., Pezzimenti, F., Kawakami, K., Lanfrancone, L., d’Adda di Fagagna, F., et al. (2009). Expression of H-RASV12 in a zebrafish model of Costello syndrome causes cellular senescence in adult proliferating cells. Dis. Model. Mech. 2, 56–67. doi: 10.1242/dmm.001016
Sasaki, T., Lian, S., Khan, A., Llop, J. R., Samuelson, A. V., Chen, W., et al. (2017). Autolysosome biogenesis and developmental senescence are regulated by both Spns1 and v-ATPase. Autophagy 13, 386–403. doi: 10.1080/15548627.2016.1256934
Sato, T., Takahoko, M., and Okamoto, H. (2006). HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis 44, 136–142. doi: 10.1002/gene.20196
Schaefer, M. K., Schmalbruch, H., Buhler, E., Lopez, C., Martin, N., Guénet, J. L., et al. (2007). Progressive motor neuronopathy: a critical role of the tubulin chaperone TBCE in axonal tubulin routing from the Golgi apparatus. J. Neurosci. 27, 8779–8789. doi: 10.1523/JNEUROSCI.1599-07.2007
Schlessinger, J. (2000). Cell signaling by receptor tyrosine kinases. Cell 103, 211–225. doi: 10.1016/s0092-8674(00)00114-8
Schlessinger, K., Hall, A., and Tolwinski, N. (2009). Wnt signaling pathways meet Rho GTPases. Genes Dev. 23, 265–277. doi: 10.1101/gad.1760809
Schmidt, A., and Hall, A. (2002). Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 16, 1587–1609. doi: 10.1101/gad.1003302
Schrick, J. J., Vogel, P., Abuin, A., Hampton, B., and Rice, D. S. (2006). ADP-ribosylation factor-like 3 is involved in kidney and photoreceptor development. Am. J. Pathol. 168, 1288–1298. doi: 10.2353/ajpath.2006.050941
Schubbert, S., Shannon, K., and Bollag, G. (2007). Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer 7, 295–308. doi: 10.1038/nrc2109
Schuhmacher, A. J., Guerra, C., Sauzeau, V., Cañamero, M., Bustelo, X. R., and Barbacid, M. (2008). A mouse model for Costello syndrome reveals an Ang II-mediated hypertensive condition. J. Clin. Invest. 118, 2169–2179. doi: 10.1172/JCI34385
Schwahn, U., Lenzner, S., Dong, J., Feil, S., Hinzmann, B., van Duijnhoven, G., et al. (1998). Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat. Genet. 19, 327–332. doi: 10.1038/1214
Schwamborn, J. C., and Püschel, A. W. (2004). The sequential activity of the GTPases Rap1B and Cdc42 determines neuronal polarity. Nat. Neurosci. 7, 923–929. doi: 10.1038/nn1295
Sen, A., Yokokura, T., Kankel, M. W., Dimlich, D. N., Manent, J., Sanyal, S., et al. (2011). Modeling spinal muscular atrophy in Drosophila links Smn to FGF signaling. J. Cell Biol. 192, 481–495. doi: 10.1083/jcb.201004016
Sepich, D. S., and Solnica-Krezel, L. (2016). Intracellular Golgi Complex organization reveals tissue specific polarity during zebrafish embryogenesis. Dev. Dyn. 245, 678–691. doi: 10.1002/dvdy.24409
Sferra, A., Baillat, G., Rizza, T., Barresi, S., Flex, E., Tasca, G., et al. (2016). TBCE mutations cause early-onset progressive encephalopathy with distal spinal muscular atrophy. Am. J. Hum. Genet. 99, 974–983. doi: 10.1016/j.ajhg.2016.08.006
Shakir, M. A., Gill, J. S., and Lundquist, E. A. (2006). Interactions of UNC-34 Enabled with Rac GTPases and the NIK kinase MIG-15 in Caenorhabditis elegans axon pathfinding and neuronal migration. Genetics 172, 893–913. doi: 10.1534/genetics.105.046359
Sharapova, S. O., Haapaniemi, E., Sakovich, I. S., Kostyuchenko, L. V., Donkó, A., Dulau-Florea, A., et al. (2019). Heterozygous activating mutation in RAC2 causes infantile-onset combined immunodeficiency with susceptibility to viral infections. Clin. Immunol. 205, 1–5. doi: 10.1016/j.clim.2019.05.003
Shcherbakova, D. M., Cammer, N. C., Huisman, T. M., Verkhusha, V. V., and Hodgson, L. (2018a). Direct multiplex imaging and optogenetics of Rho GTPases enabled by near-infrared FRET. Nat. Chem. Biol. 14, 591–600. doi: 10.1038/s41589-018-0044-1
Shcherbakova, D. M., Stepanenko, O. V., Turoverov, K. K., and Verkhusha, V. V. (2018b). Near-infrared fluorescent proteins: multiplexing and optogenetics across scales. Trends Biotechnol. 36, 1230–1243. doi: 10.1016/j.tibtech.2018.06.011
Sheen, V. L., Ganesh, V. S., Topcu, M., Sebire, G., Bodell, A., Hill, R. S., et al. (2004). Mutations in ARFGEF2 implicate vesicle trafficking in neural progenitor proliferation and migration in the human cerebral cortex. Nat. Genet. 36, 69–76. doi: 10.1038/ng1276
Shih, A. H., and Holland, E. C. (2006). Notch signaling enhances nestin expression in gliomas. Neoplasia 8, 1072–1082. doi: 10.1593/neo.06526
Shimokawa, K., Kimura-Yoshida, C., Nagai, N., Mukai, K., Matsubara, K., Watanabe, H., et al. (2011). Cell surface heparan sulfate chains regulate local reception of FGF signaling in the mouse embryo. Dev. Cell 21, 257–272. doi: 10.1016/j.devcel.2011.06.027
Shoubridge, C., Tarpey, P. S., Abidi, F., Ramsden, S. L., Rujirabanjerd, S., Murphy, J. A., et al. (2010). Mutations in the guanine nucleotide exchange factor gene IQSEC2 cause nonsyndromic intellectual disability. Nat. Genet. 42, 486–488. doi: 10.1038/ng.588
Simanshu, D. K., Nissley, D. V., and McCormick, F. (2017). RAS proteins and their regulators in human disease. Cell 170, 17–33. doi: 10.1016/j.cell.2017.06.009
Smallhorn, M., Murray, M. J., and Saint, R. (2004). The epithelial-mesenchymal transition of the Drosophila mesoderm requires the Rho GTP exchange factor Pebble. Development 131, 2641–2651. doi: 10.1242/dev.01150
Smith, C. C., Paguirigan, A., Jeschke, G. R., Lin, K. C., Massi, E., Tarver, T., et al. (2017). Heterogeneous resistance to quizartinib in acute myeloid leukemia revealed by single-cell analysis. Blood 130, 48–58. doi: 10.1182/blood-2016-04-711820
Soga, N., Namba, N., McAllister, S., Cornelius, L., Teitelbaum, S. L., Dowdy, S. F., et al. (2001). Rho family GTPases regulate VEGF-stimulated endothelial cell motility. Exp. Cell Res. 269, 73–87. doi: 10.1006/excr.2001.5295
Song, M. R., and Ghosh, A. (2004). FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat. Neurosci. 7, 229–235. doi: 10.1038/nn1192
Stanganello, E., Hagemann, A. I., Mattes, B., Sinner, C., Meyen, D., Weber, S., et al. (2015). Filopodia-based Wnt transport during vertebrate tissue patterning. Nat. Commun. 6:5846. doi: 10.1038/ncomms6846
Steklov, M., Pandolfi, S., Baietti, M. F., Batiuk, A., Carai, P., Najm, P., et al. (2018). Mutations in LZTR1 drive human disease by dysregulating RAS ubiquitination. Science 362, 1177–1182. doi: 10.1126/science.aap7607
Steliarova-Foucher, E., Colombet, M., Ries, L. A. G., Moreno, F., Dolya, A., Brayet, F., et al. (2017). International incidence of childhood cancer, 2001-10: a population-based registry study. Lancet Oncol. 18, 719–731. doi: 10.1016/S1470-2045(17)30186-9
Stendel, C., Roos, A., Kleine, H., Arnaud, E., Ozçelik, M., Sidiropoulos, P. N., et al. (2010). SH3TC2, a protein mutant in Charcot-Marie-Tooth neuropathy, links peripheral nerve myelination to endosomal recycling. Brain 133, 2462–2474. doi: 10.1093/brain/awq168
Stephenson, R. E., and Miller, A. L. (2017). Tools for live imaging of active Rho GTPases in Xenopus. Genesis 55:10.1002/dvg.22998. doi: 10.1002/dvg.22998
Sternberg, P. W. (2005). Vulval Development, ed. WormBook, The C. elegans Research Community, WormBook. Available online at: doi: 10.1895/wormbook.1.6.1, http://www.wormbook.org (accessed June 25, 2005).
Stockhausen, M. T., Sjolund, J., and Axelson, H. (2005). Regulation of the Notch target gene Hes-1 by TGFalpha induced Ras/MAPK signaling in human neuroblastoma cells. Exp. Cell Res. 1, 218–228. doi: 10.1016/j.yexcr.2005.07.011
Strullu, M., Caye, A., Lachenaud, J., Cassinat, B., Gazal, S., and Fenneteau, O. (2014). Juvenile myelomonocytic leukaemia and Noonan syndrome. J. Med. Genet. 51, 689–697. doi: 10.1136/jmedgenet-2014-102611
Strutt, D. I., Weber, U., and Mlodzik, M. (1997). The role of RhoA in tissue polarity and Frizzled signalling. Nature 387, 292–295. doi: 10.1038/387292a0
Sugihara, K., Nakatsuji, N., Nakamura, K., Nakao, K., Hashimoto, R., Otani, H., et al. (1998). Rac1 is required for the formation of three germ layers during gastrulation. Oncogene 17, 3427–3433. doi: 10.1038/sj.onc.1202595
Sundaram, M. V. (2005). The Love-Hate Relationship Between Ras and Notch. Genes Dev. 19, 1825–1839. doi: 10.1101/gad.1330605
Svensmark, J. H., and Brakebusch, C. (2019). Rho GTPases in cancer: Friend or foe? Oncogene 38, 7447–7456. doi: 10.1038/s41388-019-0963-7
Sztul, E., Chen, P. W., Casanova, J. E., Cherfils, J., Dacks, J. B., Lambright, D. G., et al. (2019). ARF GTPases and their GEFs and GAPs: concepts and challenges. Mol. Biol. Cell 30, 1249–1271. doi: 10.1091/mbc.E18-12-0820
Tabor, K. M., Marquart, G. D., Hurt, C., Smith, T. S., Geoca, A. K., Bhandiwad, A. A., et al. (2019). Brain-wide cellular resolution imaging of Cre transgenic zebrafish lines for functional circuit-mapping. eLife 8:e42687. doi: 10.7554/eLife.42687
Tajan, M., Paccoud, R., Branka, S., Edouard, T., and Yart, A. (2018). The RASopathy family: consequences of germline activation of the RAS/MAPK pathway. Endocr. Rev. 39, 676–700. doi: 10.1210/er.2017-00232
Tan, D., Zhang, H., Deng, J., Liu, J., Wen, J., Li, L., et al. (2020). RhoA-GTPase modulates neurite outgrowth by regulating the expression of spastin and p60-Katanin. Cells 9:230. doi: 10.3390/cells9010230
Tang, S., and Yasuda, R. (2017). Imaging ERK and PKA activation in single dendritic spines during structural plasticity. Neuron 93, 1315–1324.e3. doi: 10.1016/j.neuron.2017.02.032
Tartaglia, M., and Gelb, B. D. (2010). Disorders of dysregulated signal traffic through the RAS-MAPK pathway: phenotypic spectrum and molecular mechanisms. Ann. N. Y. Acad. Sci. 1214, 99–121. doi: 10.1111/j.1749-6632.2010.05790.x
Tartaglia, M., Gelb, B. D., and Zenker, M. (2011). Noonan syndrome and clinically related disorders. Best Pract. Res. Clin. Endocrinol. Metab. 25, 161–179. doi: 10.1016/j.beem.2010.09.002
Tartaglia, M., Mehler, E. L., Goldberg, R., Zampino, G., Brunner, H. G., Kremer, H., et al. (2001). Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 29, 465–468. doi: 10.1038/ng772
Tartaglia, M., Niemeyer, C. M., Fragale, A., Song, X., Buechner, J., Jung, A., et al. (2003). Somaticmutations in PTPN11 in juvenilemyelomonocyticleukemia, myelodysplasticsyndromes and acute myeloidleukemia. Nat. Genet. 34, 148–150. doi: 10.1038/ng1156
Tartaglia, M., Pennacchio, L. A., Zhao, C., Yadav, K. K., Fodale, V., Sarkozyet, A., et al. (2007). Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat. Genet. 39, 75–79. doi: 10.1038/ng1939
Thomas, E. K., Cancelas, J. A., Zheng, Y., and Williams, D. A. (2008). Rac GTPases as key regulators of p210-BCR-ABL-dependent leukemogenesis. Leukemia 22, 898–904. doi: 10.1038/leu.2008.71
Tidyman, W. E., and Rauen, K. A. (2016). Expansion of the RASopathies. Curr. Genet. Med. Rep. 4, 57–64. doi: 10.1007/s40142-016-0100-7
Tran, L. D., Hino, H., Quach, H., Lim, S., Shindo, A., Mimori-Kiyosue, Y., et al. (2012). Dynamic microtubules at the vegetal cortex predict the embryonic axis in zebrafish. Development 139, 3644–3652. doi: 10.1242/dev.082362
Troeger, A., and Williams, D. A. (2013). Hematopoietic-specific Rho GTPases Rac2 and RhoH and human blood disorders. Exp. Cell Res. 319, 2375–2383. doi: 10.1016/j.yexcr.2013.07.002
Tsang, M., and Dawid, I. B. (2004). Promotion and attenuation of FGF signaling through the Ras-MAPK pathway. Sci. STKE 2004:e17. doi: 10.1126/stke.2282004pe17
Tseng, W. C., Loeb, H. E., Pei, W., Tsai-Morris, C. H., Xu, L., Cluzeau, C. V., et al. (2018). Modeling Niemann-Pick disease type C1 in zebrafish: a robust platform for in vivo screening of candidate therapeutic compounds. Dis. Model. Mech. 11:dmm034165. doi: 10.1242/dmm.034165
Tsuda, L., Nagaraj, R., Zipursky, S. L., and Banerjee, U. (2002). An EGFR/Ebi/Sno pathway promotes delta expression by inactivating Su(H)/SMRTER repression during inductive notch signaling. Cell 110, 625–637. doi: 10.1016/S0092-8674(02)00875-9
Turcotte, R., Liang, Y., Tanimoto, M., Zhang, Q., Li, Z., Koyama, M., et al. (2019). Dynamic super-resolution structured illumination imaging in the living brain. Proc. Natl. Acad. Sci. U.S.A. 116, 9586–9591. doi: 10.1073/pnas.1819965116
Ueno, H., Huang, X., Tanaka, Y., and Hirokawa, N. (2011). KIF16B/Rab14 molecular motor complex is critical for early embryonic development by transporting FGF receptor. Dev. Cell 20, 60–71. doi: 10.1016/j.devcel.2010.11.008
Ueyama, T. (2019). Rho-family small GTPases: from highly polarized sensory neurons to cancer cells. Cells 8:92. doi: 10.3390/cells8020092
Ueyama, T., Sakaguchi, H., Nakamura, T., Goto, A., Morioka, S., Shimizu, A., et al. (2014). Maintenance of stereocilia and apical junctional complexes by Cdc42 in cochlear hair cells. J. Cell Sci. 127, 2040–2052. doi: 10.1242/jcs.143602
Ulrich, F., Krieg, M., Schotz, E. M., Link, V., Castanon, I., Schnabel, V., et al. (2005). Wnt11 functions in gastrulation by controlling cell cohesion through Rab5c and E-cadherin. Dev. Cell 9, 555–564. doi: 10.1016/j.devcel.2005.08.011
Unlu, G., Levic, D. S., Melville, D. B., and Knapik, E. W. (2014). Trafficking mechanisms of extracellular matrix macromolecules: insights from vertebrate development and human diseases. Int. J. Biochem. Cell Biol. 47, 57–67. doi: 10.1016/j.biocel.2013.11.005
Urosevic, J., Sauzeau, V., Soto-Montenegro, M. L., Reig, S., Desco, M., Wright, E. M., et al. (2011). Constitutive activation of B-Raf in the mouse germ line provides a model for human cardio-facio-cutaneous syndrome. Proc. Natl. Acad. Sci. U.S.A. 108, 5015–5020. doi: 10.1073/pnas.1016933108
Vabres, P., Sorlin, A., Kholmanskikh, S. S., Demeer, B., St-Onge, J., Duffourd, Y., et al. (2019). Postzygotic inactivating mutations of RHOA cause a mosaic neuroectodermal syndrome. Nat. Genet. 51, 1438–1441. doi: 10.1038/s41588-019-0498-4
Vacaru, A. M., Unlu, G., Spitzner, M., Mione, M., Knapik, E. W., and Sadler, K. C. (2014). In vivo cell biology in zebrafish - providing insights into vertebrate development and disease. J. Cell Sci. 127, 485–495. doi: 10.1242/jcs.140194
van der Meel, R., Symons, M. H., Kudernatsch, R., Kok, R. J., Schiffelers, R. M., Storm, G., et al. (2011). The VEGF/Rho GTPase signalling pathway: a promising target for anti-angiogenic/anti-invasion therapy. Drug Discov. Today 16, 219–228. doi: 10.1016/j.drudis.2011.01.005
van Impel, A., Schumacher, S., Draga, M., Herz, H. M., Grosshans, J., and Müller, H. A. (2009). Regulation of the Rac GTPase pathway by the multifunctional Rho GEF Pebble is essential for mesoderm migration in the Drosophila gastrula. Development 136, 813–822. doi: 10.1242/dev.026203
van Nieuw Amerongen, G. P., Koolwijk, P., Versteilen, A., and van Hinsbergh, V. W. (2003). Involvement of RhoA/Rho kinase signaling in VEGF-induced endothelial cell migration and angiogenesis in vitro. Arterioscler. Thromb. Vasc. Biol. 23, 211–217. doi: 10.1161/01.atv.0000054198.68894.88
Verweij, F. J., Revenu, C., Arras, G., Dingli, F., Loew, D., Pegtel, D. M., et al. (2019). Live tracking of inter-organ communication by endogenous exosomes in vivo. Dev. Cell 48, 573–589.e4. doi: 10.1016/j.devcel.2019.01.004
Vigil, D., Cherfils, J., Rossman, K. L., and Der, C. J. (2010). Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat. Rev. Cancer 10, 842–857. doi: 10.1038/nrc2960
Voena, C., and Chiarle, R. (2019). RHO family GTPases in the biology of lymphoma. Cells 8:646. doi: 10.3390/cells8070646
Voss, S., Krüger, D. M., Koch, O., and Wu, Y. W. (2016). Spatiotemporal imaging of small GTPases activity in live cells. Proc. Natl. Acad. Sci. U.S.A. 113, 14348–14353. doi: 10.1073/pnas.1613999113
Wada, Y., Sun-Wada, G. H., Kawamura, N., and Yasukawa, J. (2016). Membrane dynamics in mammalian embryogenesis: implication in signal regulation. Birth Defects Res. C Embryo Today 108, 33–44. doi: 10.1002/bdrc.21124
Wakap, N. S., Lambert, D. M., Olry, A., Rodwell, C., Gueydan, C., Lanneau, V., et al. (2020). Estimating cumulative point prevalence of rare diseases: analysis of the Orphanet database. Eur. J. Hum. Genet. 28, 165–173. doi: 10.1038/s41431-019-0508-0
Wakayama, Y., Fukuhara, S., Ando, K., Matsuda, M., and Mochizuki, N. (2015). Cdc42 mediates Bmp-induced sprouting angiogenesis through Fmnl3-driven assembly of endothelial filopodia in zebrafish. Dev. Cell 32, 109–122. doi: 10.1016/j.devcel.2014.11.024
Wakil, S. M., Alhissi, S., Al Dossari, H., Alqahtani, A., Shibin, S., Melaiki, B. T., et al. (2019). Truncating ARL6IP1 variant as the genetic cause of fatal complicated hereditary spastic paraplegia. BMC Med. Genet. 20:119. doi: 10.1186/s12881-019-0851-6
Wallace, M. R., Marchuk, D. A., Andersen, L. B., Letcher, R., Odeh, H. M., Saulino, A. M., et al. (1990). Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science 249, 181–186. doi: 10.1126/science.2134734
Wandinger-Ness, A., and Zerial, M. (2014). Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb. Perspect. Biol. 6:a022616. doi: 10.1101/cshperspect.a022616
Wang, J., Morita, Y., Mazelova, J., and Deretic, D. (2012). The Arf GAP ASAP1 provides a platform to regulate Arf4-and Rab11-Rab8-mediated ciliary receptor targeting. EMBO J. 31, 4057–4071. doi: 10.1038/emboj.2012.253
Wang, X., He, L., Wu, Y. I., Hahn, K. M., and Montell, D. J. (2010). Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol. 12, 591–597. doi: 10.1038/ncb2061
Wedlich-Soldner, R., Wai, S. C., Schmidt, T., and Li, R. (2004). Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J. Cell Biol. 166, 889–900. doi: 10.1083/jcb.200405061
Wennerberg, K., Rossman, K. L., and Der, C. J. (2005). The Ras superfamily at a glance. J. Cell Sci. 118, 843–846. doi: 10.1242/jcs.01660
White, J. J., Mazzeu, J. F., Coban-Akdemir, Z., Bayram, Y., Bahrambeigi, V., Hoischen, A., et al. (2018). WNT signaling perturbations underlie the genetic heterogeneity of Robinow syndrome. Am. J. Hum. Genet. 102, 27–43. doi: 10.1016/j.ajhg.2017.10.002
Wiens, C. J., Tong, Y., Esmail, M. A., Oh, E., Gerdes, J. M., Wang, J., et al. (2010). Bardet-Biedl syndrome-associated small GTPase ARL6 (BBS3) functions at or near the ciliary gate and modulates Wnt signaling. J. Biol. Chem. 285, 16218–16230. doi: 10.1074/jbc.M109.070953
Wilson, M. Z., Ravindran, P. T., Lim, W. A., and Toettcher, J. E. (2017). Tracing information flow from Erk to target gene induction reveals mechanisms of dynamic and combinatorial control. Mol. Cell 67, 757–769.e5. doi: 10.1016/j.molcel.2017.07.016
Winter, J. F., Höpfner, S., Korn, K., Farnung, B. O., Bradshaw, C. R., Marsico, G., et al. (2012). Caenorhabditis elegans screen reveals role of PAR-5 in RAB-11-recycling endosome positioning and apicobasal cell polarity. Nat. Cell Biol. 14, 666–676. doi: 10.1038/ncb2508
Wojnacki, J., Quassollo, G., Marzolo, M. P., and Cáceres, A. (2014). Rho GTPases at the crossroad of signaling networks in mammals: impact of Rho-GTPases on microtubule organization and dynamics. Small GTPases 5:e28430. doi: 10.4161/sgtp.28430
Wolf, S., Supatto, W., Debrégeas, G., Mahou, P., Kruglik, S. G., Sintes, J. M., et al. (2015). Whole-brain functional imaging with two-photon light-sheet microscopy. Nat. Methods 12, 379–380. doi: 10.1038/nmeth.3371
Wolman, M. A., de Groh, E. D., McBride, S. M., Jongens, T. A., Granato, M., and Epstein, J. A. (2014). Modulation of cAMP and ras signaling pathways improves distinct behavioral deficits in a zebrafish model of neurofibromatosis type 1. Cell Rep. 8, 1265–1270. doi: 10.1016/j.celrep.2014.07.054
Wong, K. L., Akiyama, R., Bessho, Y., and Matsui, T. (2018). ERK activity dynamics during Zebrafish embryonic development. Int. J. Mol. Sci. 20:109. doi: 10.3390/ijms20010109
Wu, H. M., Hazak, O., Cheung, A. Y., and Yalovsky, S. (2011). RAC/ROP GTPases and auxin signaling. Plant Cell 23, 1208–1218. doi: 10.1105/tpc.111.083907
Wu, R. S., Lam, I. I., Clay, H., Duong, D. N., Deo, R. C., and Coughlin, S. R. (2018). A Rapid method for directed gene knockout for screening in G0 zebrafish. Dev. Cell 46, 112–125.e4. doi: 10.1016/j.devcel.2018.06.003
Wu, W. J., Erickson, J. W., Lin, R., and Cerione, R. A. (2000). The gamma-subunit of the coatomer complex binds Cdc42 to mediate transformation. Nature 405, 800–804. doi: 10.1038/35015585
Wu, X., Tu, X., Joeng, K. S., Hilton, M. J., Williams, D. A., and Long, F. (2008). Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell 133, 340–353. doi: 10.1016/j.cell.2008.01.052
Wu, Y. I., Frey, D., Lungu, O. I., Jaehrig, A., Schlichting, I., Kuhlman, B., et al. (2009). A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108. doi: 10.1038/nature08241
Wühr, M., Obholzer, N. D., Megason, S. G., Detrich, H. W. III, and Mitchison, T. J. (2011). Live imaging of the cytoskeleton in early cleavage-stage zebrafish embryos. Methods Cell Biol. 101, 1–18. doi: 10.1016/B978-0-12-387036-0.00001-3
Xiang, Y., Seemann, J., Bisel, B., Punthambaker, S., and Wang, Y. (2007). Active ADP-ribosylation factor-1 (ARF1) is required for mitotic Golgi fragmentation. J. Biol. Chem. 282, 21829–21837. doi: 10.1074/jbc.M611716200
Xiong, F., Obholzer, N. D., Noche, R. R., and Megason, S. G. (2015). Multibow: digital spectral barcodes for cell tracing. PLoS One 10:e0127822. doi: 10.1371/journal.pone.0127822
Xu, Z., Chen, Y., and Chen, Y. (2019). Spatiotemporal regulation of Rho GTPases in neuronal migration. Cells 8:568. doi: 10.3390/cells8060568
Yamamoto, G. L., Aguena, M., Gos, M., Hung, C., Pilch, J., Fahiminiya, S., et al. (2015). Rare variants in SOS2 and LZTR1 are associated with Noonan syndrome. J. Med. Genet. 52, 413–421. doi: 10.1136/jmedgenet-2015-103018
Yamauchi, J., Miyamoto, Y., Torii, T., Takashima, S., Kondo, K., Kawahara, K., et al. (2012). Phosphorylation of cytohesin-1 by Fyn is required for initiation of myelination and the extent of myelination during development. Sci. Signal. 5:ra69. doi: 10.1126/scisignal.2002802
Yasuda, R., Harvey, C. D., Zhong, H., Sobczyk, A., van Aelst, L., and Svoboda, K. (2006). Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat. Neurosci. 9, 283–291. doi: 10.1038/nn1635
Yoo, S. K., Deng, Q., Cavnar, P. J., Wu, Y. I., Hahn, K. M., and Huttenlocher, A. (2010). Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev. Cell 18, 226–236. doi: 10.1016/j.devcel.2009.11.015
Yoshimura, S., Egerer, J., Fuchs, E., Haas, A. K., and Barr, F. A. (2007). Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 178, 363–369. doi: 10.1083/jcb.200703047
Zeng, X., Tamai, K., Doble, B., Li, S., Huang, H., Habas, R., et al. (2005). A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature 438, 873–877. doi: 10.1038/nature04185
Zhai, J., Zhang, L., Mojsilovic-Petrovic, J., Jian, X., Thomas, J., Homma, K., et al. (2015). Inhibition of cytohesins protects against genetic models of motor neuron disease. J. Neurosci. 35, 9088–9105. doi: 10.1523/JNEUROSCI.5032-13.2015
Zhang, C. J., Rosenwald, A. G., Willingham, M. C., Skuntz, S., Clark, J., and Kahn, R. A. (1994). Expression of a dominant allele of human ARF1 inhibits membrane traffic in vivo. J. Cell Biol. 124, 289–300. doi: 10.1083/jcb.124.3.289
Zhang, J., Mullighan, C. G., Harvey, R. C., Wu, G., Chen, X., and Edmonson, M. (2011). Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood 118, 3080–3087. doi: 10.1182/blood-2011-03-341412
Zhang, Q., Major, M. B., Takanashi, S., Camp, N. D., Nishiya, N., Peters, E. C., et al. (2007). Small-molecule synergist of the Wnt/beta-catenin signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 7444–7448. doi: 10.1073/pnas.0702136104
Zhang, Y., Nichols, E. L., Zellmer, A. M., Guldner, I. H., Kankel, C., Zhang, S., et al. (2019). Generating intravital super-resolution movies with conventional microscopy reveals actin dynamics that construct pioneer axons. Development 146:dev171512. doi: 10.1242/dev.171
Zhang, Z., Yang, M., Chen, R., Su, W., Li, P., Chen, S., et al. (2014). IBP regulates epithelial-to-mesenchymal transition and the motility of breast cancer cells via Rac1, RhoA and Cdc42 signaling pathways. Oncogene 33, 3374–3382. doi: 10.1038/onc.2
Keywords: small GTPases, rare diseases, undiagnosed patients, next generation sequencing, in vivo models, zebrafish, functional genomics
Citation: Lauri A, Fasano G, Venditti M, Dallapiccola B and Tartaglia M (2021) In vivo Functional Genomics for Undiagnosed Patients: The Impact of Small GTPases Signaling Dysregulation at Pan-Embryo Developmental Scale. Front. Cell Dev. Biol. 9:642235. doi: 10.3389/fcell.2021.642235
Received: 15 December 2020; Accepted: 12 March 2021;
Published: 25 May 2021.
Edited by:
Maria Pia Felli, Sapienza University of Rome, ItalyReviewed by:
Karl J. Clark, Mayo Clinic, United StatesMarcel Tawk, INSERM U1195 Petites Molécules de Neuroprotection, Neurogénération et Remyélinisation, France
Copyright © 2021 Lauri, Fasano, Venditti, Dallapiccola and Tartaglia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonella Lauri, YW50b25lbGxhLmxhdXJpQG9wYmcubmV0; Marco Tartaglia, bWFyY28udGFydGFnbGlhQG9wYmcubmV0
 Antonella Lauri
Antonella Lauri Giulia Fasano
Giulia Fasano Martina Venditti
Martina Venditti Bruno Dallapiccola
Bruno Dallapiccola Marco Tartaglia
Marco Tartaglia