
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 21 January 2021
Sec. Cell Death and Survival
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.637162
This article is part of the Research TopicNovel Insights into FerroptosisView all 6 articles
The induction and consequences of regulated cell death (RCD) are accompanied by changes in gene and protein expression, biochemical pathways, as well as cell morphology and size. Such RCDs have a significant impact on development, tissue homeostasis, and the occurrence and progression of disease. Among different forms of RCD, ferroptosis appears to be the main cause of tissue damage driven by iron overload and lipid peroxidation. In fact, the dysfunctional ferroptotic response is implicated in a variety of pathological conditions and diseases, such as neurodegenerative diseases, tissue ischemia-reperfusion injury, tumorigenesis, infections, and immune diseases. Ferroptotic response can be fine-tuned through various oxidative stress and antioxidant defense pathways, coupling with metabolism, gene transcription, and protein degradation machinery. Accordingly, a series of ferroptosis inducers or inhibitors targeting redox- or iron metabolism-related proteins or signal transduction have been developed. Although this kind of RCD has recently attracted great interest in basic and clinical research, detecting and monitoring a ferroptotic response still faces challenges. In this mini-review, we not only summarize the latest knowledge about the characteristics of ferroptosis in vitro and in vivo, but also discuss the specificity and limitations of current biomarkers of ferroptosis.
Cell death is a basic biological process that regulates cell fate, tissue regeneration, and the body’s immune response. Accidental cell death (ACD) and regulated cell death (RCD) are two major subcategories of cell death (Galluzzi et al., 2018). Unlike ACD, which is an uncontrolled rapid process, RCD usually utilizes clear molecular machinery involving multiple genes or proteins, and can be intervened on at multiple levels, especially using drugs or small molecular compounds (Tang et al., 2019). According to the publication record in PubMed,1 apoptosis, necroptosis, pyroptosis, and ferroptosis may be the most studied forms of RCD, involved in multiple pathological conditions and diseases. In some cases, several rare RCDs [e.g., alkaliptosis (Song et al., 2018b) and oxeiptosis (Holze et al., 2018)] have also been observed in cell cultures or mouse models, although their physiological or pathological significance remains largely unknown.
Ferroptosis is a non-apoptotic and oxidative damage-related RCD (Dixon et al., 2012), mainly driven by iron accumulation, lipid peroxidation, and subsequent plasma membrane rupture (Tang and Kroemer, 2020). The process of ferroptosis is further controlled through a variety of molecular signaling achieved using epigenetic, transcription, and post-translational mechanisms (Chen et al., 2020b). After treatment with reagents or suffering environmental stresses, an increase in ferroptotic cell death can be observed in experimental models by activating the intrinsic or extrinsic pathways, which is discussed in depth in recent reviews (Chen et al., 2020b; Tang et al., 2020). Notably, two small molecular compounds, namely erastin (Dixon et al., 2012) and RSL3 (Yang et al., 2014), are the most commonly used reagents for triggering ferroptosis to study its molecular mechanisms. Excessive or deficient ferroptotic response also occurs in tissue samples of human diseases (Ashraf et al., 2020), which highlight the potential of pathological relevance of ferroptosis (Stockwell et al., 2017). In this min-review, we used some examples and their corresponding regulatory mechanisms to focus on the characteristics and biomarkers of ferroptosis (Figure 1).
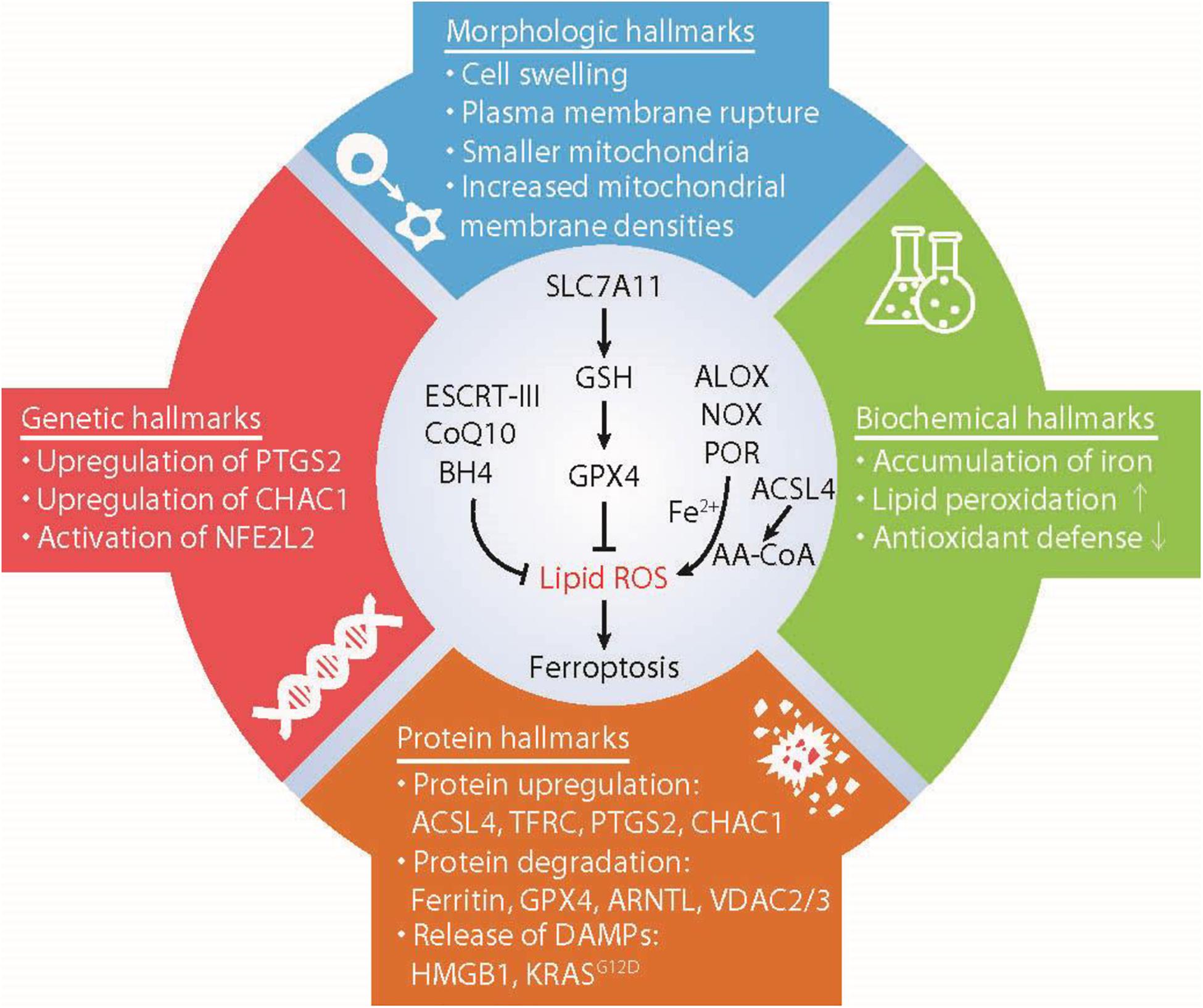
Figure 1. Hallmarks of ferroptosis. Ferroptosis is a type of iron-dependent regulated cell death mainly caused by unrestricted lipid peroxidation and subsequent membrane damage. Ferroptotic cell death may show some morphological and biochemical characteristics as well as common changes in gene and protein levels. AA-COA, arachidonoyl-coenzyme A; ACSL4, acyl-CoA synthetase long-chain family member 4; ALOX, lipoxygenases; ARNTL, aryl hydrocarbon receptor nuclear translocator like; BH4, tetrahydrobiopterin; CHAC1, ChaC glutathione specific gamma-glutamylcyclotransferase 1; COQ10, Coenzyme Q10; ESCRT-III, endosomal sorting complex required for transport-III; GPX4, glutathione peroxidase 4; GSH, glutathione; HMGB1, high mobility group box 1; NFE2L2, nuclear factor erythroid 2-like 2; NOX, NADPH oxidases; POR, cytochrome P450 oxidoreductase; PTGS2, prostaglandin-endoperoxide synthase 2; PUFA, polyunsaturated fatty acids; SLC7A11, solute carrier family 7 member 11; TFRC, transferrin receptor; VDAC2/3, voltage dependent anion channel 2/3.
What we call morphological features usually refers to cell changes observed through light or electron microscopy, and does not involve staining of special protein markers. Ferroptosis is generally a type of regulated necrosis, which is devoid of the morphological characteristics of apoptosis (e.g., cell shrinkage and plasma membrane blistering; Conrad et al., 2016). In contrast, necrotic morphology, such as cell enlargement and plasma membrane rupture, are commonly observed in most ferroptotic cells (Conrad et al., 2016). Consequently, the activation of cell membrane repair pathways, such as endosomal sorting complex required for transport-III (ESCRT-III) machinery, prevents ferroptotic cell death (Dai et al., 2020c). Electron microscopy shows that ferroptotic cells represent a vast change in mitochondrial ultrastructure, such as reduction in mitochondrial volume, increase in mitochondrial membrane density, and disappearance of mitochondrial cristae (Yagoda et al., 2007; Dixon et al., 2012). Since elevated autophagy promotes ferroptosis (Liu J. et al., 2020), autophagy-related ultrastructures (e.g., double-membrane autophagosomes and various lysosome-related vesicles) are often observed in ferroptotic cells or tissues (Friedmann Angeli et al., 2014). Although activators of ferroptosis lead to oxidative damage in DNA (Song et al., 2016), the nucleus of ferroptotic cells seem to be normal and lack chromatin concentration (Yagoda et al., 2007), which is a morphological marker of apoptotic cells. As a type of inflammatory RCD, immune cell infiltration is observed in tissues affected by the ferroptotic damage. For example, acute pancreatitis is a sterile inflammatory disease caused by the death of acinar cells. Ferroptotic acinar death contributes to experimental pancreatitis in mice, especially in cases of impaired circadian rhythms (Liu et al., 2020b). This ferroptosis-related pancreatitis is associated with pancreatic histological damage and leukocyte infiltration that can be detected using hematoxylin and eosin stain (Liu et al., 2020b). In general, the necrotic morphology caused by ferroptosis has been observed in vitro or in tissues, but it is difficult to distinguish ferroptosis from other types of regulated necrosis based on these changes alone.
Since the initial study defined ferroptosis as an iron-dependent RCD, iron accumulation-mediated biochemical events (e.g., elevated Fenton reaction or activated iron-containing enzymes) seem to be a biochemical sign of ferroptosis (Chen et al., 2020c). Iron is an essential trace element, distributed in various subcellular organelles (e.g., mitochondria and lysosomes). The level of intracellular or mitochondrial ferrous iron (Fe2+) is increased in ferroptotic cells or tissues, which can be monitored by a biochemical assay kit (Yuan et al., 2016a), Prussian blue staining (Lu et al., 2020), or probes [e.g., FerroFarRed (Homma et al., 2020) or Phen Green SK (Song et al., 2018a)]. In contrast, iron chelators (e.g., deferoxamine) limit ferroptosis in vitro (Dixon et al., 2012) or in vivo (Lu et al., 2020). The level of iron in cells is controlled by a complex network that involves the absorption, storage, utilization, and outflow of iron (Chen et al., 2020c). As expected, certain molecular regulators related to iron homeostasis control ferroptosis sensitivity. For example, iron uptake mediated by transferrin (TF; Gao et al., 2015), lactotransferrin (LTF; Wang et al., 2020), and transferrin receptor (TFRC; Gao et al., 2015), as well as nuclear receptor coactivator 4 (NCOA4)-dependent ferritin degradation (Hou et al., 2016) facilities ferroptotic cell death. It is worth noting that TFRC is considered as a biomarker of ferroptosis in cell cultures or tissues, and an anti-TFRC antibody (called 3F3-FMA) especially plays a role in indicating ferroptotic death or damage (Feng et al., 2020).
In contrast, heat shock protein family B small member 1 (HSPB1/HSP25/HSP27; Sun et al., 2015) or prominin-2 (PROM2; Brown et al., 2019) diminishes ferroptosis by preventing cytoskeleton-mediated iron uptake or promoting ferritin export to extracellular space, respectively. Moreover, iron chaperones, such as poly(RC) binding protein 1 (PCBP1), reduce ferroptosis sensitivity in hepatocytes (Protchenko et al., 2020). Whether different iron metabolism regulators play an equivalent role in ferroptosis is still inconclusive. More importantly, given the numerous types of iron-dependent enzymes present in different subcellular organelles, the process of enzyme coordination and their regulation of ferroptosis remain unclear.
Lipid peroxidation plays a central role in mediating ferroptosis (Yang and Stockwell, 2016). In particular, the oxidation of polyunsaturated fatty acids (PUFAs) by reactive oxygen species (ROS) to produce lipid hydroperoxides is the most important hallmark of ferroptosis (Kuang et al., 2020). In addition to iron-triggered ROS production by the Fenton reaction, mitochondria- or NADPH oxidase (NOX)-mediated ROS production (Dixon et al., 2012; Gao et al., 2015; Xie et al., 2017; Yang W. H. et al., 2019) also play a cell type-dependent role in initiating lipid peroxidation. The key enzymes of lipid peroxidation that causes ferroptosis are the isoforms of arachidonate lipoxygenase (ALOX), including ALOX5, ALOX12, ALOX15, ALOX15B, and ALOXE3 in humans (Yang et al., 2016; Wenzel et al., 2017; Chu et al., 2019; Li et al., 2020). Alternatively, cytochrome P450 oxidoreductase (POR) may mediate lipid peroxidation in an ALOX-independent manner (Zou et al., 2020).
The production of PUFA for subsequent lipid peroxidation requires the activation of several upstream lipid synthesis and metabolism pathways, especially acyl-CoA synthetase long-chain family member 4 (ACSL4)-mediated conversion of arachidonic acid (AA) to AA-CoA (Yuan et al., 2016b; Doll et al., 2017; Kagan et al., 2017). Critically, the up-regulation of ACSL4 expression, but not the expression of other ACSL members, is a biomarker indicative of ferroptosis sensitivity (Yuan et al., 2016b). While acyl-CoA synthetase long-chain family member 3 (ACSL3) is not a biomarker of ferroptosis, ACSL3-mediated monounsaturated fatty acids (MUFAs) production limits oxidative PUFA-mediated ferroptosis in certain cancer cells (Magtanong et al., 2019), indicating that ACSL4- or ACSL3-dependent fatty acid metabolism plays opposite roles in ferroptosis.
Several assays can be used to detect and quantitate the extent of lipid peroxidation of ferroptotic cells in vitro and in vivo. First, C11 BODIPY 581/591 (Dixon et al., 2012) and LiperFluo (Kagan et al., 2017) are lipid-soluble fluorescent probes, which are widely used to monitor lipid peroxidation in cell cultures by microscope, plate reader, or flow cytometry. Functionally, LiperFluo is better than C11-BODIPY in indicating lipid peroxidation during ferroptosis, because it directly interacts with (phosphate) lipid hydroperoxide (Kagan et al., 2017). Second, liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis may be used to analyze changes in lipid profile, quantitative lipid peroxide species (PUFA-OOH and PL-OOH) or identification of oxidative proteins in the process of ferroptosis (Kagan et al., 2017). Third, through biochemical, enzyme-linked immunosorbent assay (ELISA) or staining methods, the detection of the final product of lipid peroxidation [e.g., malonyl dialdehyde (MDA; Ye et al., 2020) and 4-hydroxynon-enal (4-HNE; Shintoku et al., 2017)] or oxidative DNA damage biomarkers [e.g., phosphorylated H2A.X variant histone (γH2AX; Song et al., 2016) and 8-hydroxydeoxyguanosine (8-OH-dG; Zhang et al., 2019; Dai et al., 2020b)] has been applied in various samples in vitro or in vivo. However, the specificity and sensitivity of each measurement are different, which may affect the interpretation of the results.
In support of the role of lipid peroxidation in ferroptosis, the classic ferroptosis inducers, such as erastin (Dixon et al., 2012) and RSL3 (Yang et al., 2014), are indeed inhibitors of the antioxidant system. The three antioxidant defense systems [referred to as glutathione (GSH; Dixon et al., 2012), coenzyme Q10 (CoQ10; Bersuker et al., 2019; Doll et al., 2019), or tetrahydrobiopterin (BH4) system (Kraft et al., 2020; Soula et al., 2020)] can work together or separately to limit ferroptotic death mediated by oxidative damage. The GSH system is the main pathway limiting ferroptosis. The inhibition of the upstream regulator system xc– (an amino acid antiporter) or the downstream effector glutathione peroxidase 4 (GPX4) of GSH by drugs or small molecule compounds is recognized as the classic external or internal pathway of ferroptosis (Tang and Kroemer, 2020). In addition to blocking the system xc– activity on the cell membrane, erastin is also a potential activator of mitochondrial voltage dependent anion channel 2/3 (VDAC2/3) (Yagoda et al., 2007), highlighting the participation of mitochondria dysfunction in erastin-induced ferroptosis. GPX4-independent anti-ferroptosis pathway relies on the production of CoQ10 (Bersuker et al., 2019; Doll et al., 2019) or BH4 (Kraft et al., 2020; Soula et al., 2020), which is further regulated by apoptosis inducing factor mitochondria associated 2 (AIFM2/FSP1) or GTP cyclohydrolase 1 (GCH1), respectively. Interestingly, AIFM2 was previously thought to be a pro-apoptotic protein in mitochondria (Wu et al., 2002). In contrast, the translocation of AIFM2 from the mitochondria to the membrane converts this pro-apoptotic activity into an anti-ferroptotic effect (Bersuker et al., 2019; Doll et al., 2019). The location and function switch of AIFM2 from apoptosis to ferroptosis depends on its post-translational modification, such as myristoylation (Bersuker et al., 2019; Doll et al., 2019). Other studies have shown that the anti-ferroptotic effect of AIFM2 may depend on its function of promoting membrane repair, rather than the production of reduced CoQ10 (Dai et al., 2020d). Since GSH, CoQ10, and BH4 are broad-spectrum antioxidants, these studies have also raised questions about the specific molecular mechanisms of ferroptosis. Indeed, conditional depletion of GPX4 in mice not only mediates ferroptosis (Friedmann Angeli et al., 2014), but also triggers apoptosis (Ran et al., 2003), necroptosis (Canli et al., 2016), or pyroptosis (Kang et al., 2018). Therefore, testing the levels of these antioxidants alone may not be enough to indicate changes in ferroptosis levels.
The well-known function of prostaglandin-endoperoxide synthase 2 (PTGS2/COX2) is to metabolize AA into prostaglandins. PTGS2 is the most upregulated gene among 83 oxidative stress-associated genes in BJeLR cells following treatment with erastin or RSL3 (Yang et al., 2014). The up-regulation of PTGS2 mRNA is used as a pharmacodynamic marker of ferroptotic tissues in mice exposed to erastin or RSL3 (Yang et al., 2014; Sun et al., 2015). Although it is a widely used biomarker of ferroptosis in vitro or in vivo, PTGS2 inhibitor (e.g., indomethacin) fails to affect ferroptotic cell death (Yang et al., 2014), indicating it is not a contributor of ferroptosis. In contradiction, MIR212-mediated the downregulation of PTGS2 mRNA prevents ferroptotic neuronal death in a traumatic brain injury mouse model (Xiao et al., 2019), suggesting a cell type-dependent role of PTGS2 in ferroptosis. Further mechanism studies suggest that the up-regulation of PTGS2 gene expression in ferroptosis requires lipid peroxidation, because antioxidant vitamin E or toxic 4-HNE can inhibit or induce PTGS2 expression in cancer cells or macrophages, respectively (Kumagai et al., 2004; Yang et al., 2014). The great challenge of PTGS2 as a biomarker of ferroptosis is that the up-regulation of PTGS2 is observed under various inflammatory conditions (FitzGerald, 2003), at least some of which are non-ferroptotic conditions.
ChaC glutathione specific gamma-glutamylcyclotransferase 1 (CHAC1/BOTCH) has γ-glutamyl cyclotransferase activity and reduces intracellular GSH levels by digesting glutathione into 5-oxoproline and cysteinylglycine dipeptide (Kumar et al., 2012). RNA sequencing studies show that CHAC1 is the most up-regulated gene after treatment with systemic xc– inhibitors (e.g., erastin and sorafenib) in vitro (Dixon et al., 2014). Later, the upregulation of CHAC1 mRNA is confirmed in tissues from mice treated with certain ferroptosis inducers (e.g., erastin and artesunate; Xie et al., 2017; Wang N. et al., 2019). However, CHAC1 is not a biomarker of ferroptosis caused by the GPX4 inhibitor RSL3 or the GSH synthesis inhibitor buthionine sulphoximine (Dixon et al., 2014). Further functional studies have shown that CHAC1-mediated GSH degradation acts as a promoter of ferroptosis induced by erastin or artesunate, which is downstream of the activation of the endoplasmic reticulum (ER) stress pathway [especially the eukaryotic translation initiation factor 2 alpha (EIF2A)-activating transcription factor 4 (ATF4) pathways] (Mungrue et al., 2009; Dixon et al., 2014; Wang N. et al., 2019). Therefore, the upregulation of CHAC1 gene expression provides a selective pharmacodynamic marker for ferroptosis induced by system xc– inhibitors.
The nuclear factor erythroid 2-like 2 (NFE2L2/NRF2) is a key transcription factor for cell survival during oxidative stress by activating the expression of detoxification and antioxidant genes. While the activation of the NFE2L2 pathway relies on the inhibition of the kelch like ECH associated protein 1 (KEAP1)-mediated degradation by ubiquitin-proteasome system (UPS) pathway, the excessive up-regulation of the NFE2L2 target gene may reflect the increase in oxidative damage during the activation of ferroptosis. This notion was first described in sorafenib-induced ferroptosis in hepatocellular carcinoma cells identifying metallothionein 1 G (MT1G) as a new NFE2L2-target gene responsible for ferroptosis resistance (Sun et al., 2016a,b). Increasing evidence points out that NFE2L2 plays a critical role in protecting damage under various ferroptotic conditions in vitro or in vivo. Many NFE2L2 targeted genes are upregulated in ferroptosis and these genes are involved in iron metabolism [e.g., ferritin heavy chain 1 (FTH1; Sun et al., 2016b; Shin et al., 2018), solute carrier family 40 member 1 (SLC40A1; Chang et al., 2018; Shin et al., 2018), heme oxygenase 1 (HMOX1; Sun et al., 2016b; Chang et al., 2018; Shin et al., 2018; Fang et al., 2019), and MT1G (Sun et al., 2016a)], GSH metabolism [e.g., solute carrier family 7 member 11 (SLC7A11; Chen et al., 2017), cystathionine beta-synthase (CBS; Liu N. et al., 2020), CHAC1 (Gagliardi et al., 2019), and ATP binding cassette subfamily C member 1 (ABCC1/MRP1; Cao et al., 2019)], and detoxification or antioxidant responses [e.g., NAD(P)H quinone dehydrogenase 1 (NQO1; Sun et al., 2016b; Telorack et al., 2016; Shin et al., 2018), thioredoxin reductase 1 (TXNRD1; Shin et al., 2018; Takahashi et al., 2020), aldo-keto reductase family 1 member C1/2/3 (AKR1C1/2/3; Gagliardi et al., 2019)]. However, it is difficult to distinguish the role of NFE2L2 in ferroptotic- and non-ferroptotic RCD if only relying on the detection of the expression of NFE2L2 target gene.
Genes are used to guide protein synthesis. Thus, the proteins corresponding to the genes mentioned above can theoretically be used to evaluate the sensitivity to ferroptosis. Western blot analysis of ACSL4, TFRC, PTGS2, or CHAC1 protein expression and immunohistochemical or immunofluorescence analysis of their signal or location distribution has been used to monitor ferroptotic response in vitro and/or in vivo. Since excessive autophagy promotes ferroptosis, the detection of the protein conversion of microtubule associated protein 1 light chain 3 (MAP1LC3)-I to MAP1LC3-II may reflect the degree of damage caused by ferroptosis activators (Zhou et al., 2020). Combining the use of various lysosomal inhibitors (e.g., chloroquine) to analyze changes in autophagic flux through various protein probes (e.g., RFP-GFP-LC3B or GFP-LC3-RFP-LC3ΔG) is also a commonly used detection method in vitro (Sun et al., 2018; Li et al., 2020).
In addition to control by gene transcription, the intracellular level of protein is affected by protein degradation. Both UPS and autophagy pathway participate in the regulation of ferroptosis sensitivity in a context-dependent manner. In particular, autophagic degradation of anti-ferroptotic protein appears to be a good indicator of ferroptosis sensitivity. This type of autophagy-dependent ferroptosis was first described in erastin-induced ferritin degradation in mouse embryonic fibroblasts and pancreatic ductal adenocarcinoma (PDAC) cells (Hou et al., 2016). Ferritinophagy, namely autophagic degradation of ferritin, is mediated by the cargo receptor NCOA4 (Gao et al., 2016; Hou et al., 2016), which may be a useful marker for this process. The degradation of GPX4 protein can also be observed in ferroptosis-sensitive cells in response to various reagents, such as erastin (Zhu et al., 2017; Wu et al., 2019), RSL3 (Liu et al., 2020a), FIN56 (Shimada et al., 2016), and PdPT (Yang L. et al., 2020). The ER molecular chaperone heat shock protein family A (hsp70) member 5 (HSPA5) prevents GPX4 degradation in PDAC cells by the protein-protein interaction (Zhu et al., 2017). In contrast, heat shock protein 90 (HSP90)-dependent chaperone-mediated autophagy (CMA) promotes GPX4 degradation in neuronal cells or breast cancer cells (Wu et al., 2019). In addition to autophagy, UPS also mediates GPX4 degradation, but the mechanism remains unclear (Yang L. et al., 2020). Clockophagy, a type of selective autophagy for the degradation of clock circadian regulator aryl hydrocarbon receptor nuclear translocator like protein 1 (ARNTL/BMAL1) through sequestosome 1 (SQSTM1/p62), promotes ferroptosis by inhibiting hypoxia inducible factor 1 subunit alpha (HIF1A)-dependent lipid uptake and storage (Yang M. et al., 2019). On the contrary, UPS-mediated degradation of VDAC2/3 may limit the anticancer activity of erastin in melanoma cells (Yang Y. et al., 2020). These findings indicate that protein degradation pathways play a dual role in determining the sensitivity of ferroptosis, relying on its degrading substrates.
The immune characteristics of cell death are not only factors affecting the occurrence and development of inflammatory diseases, but also involved in the regulation of tumor immunity (Galluzzi et al., 2020). This process usually depends on the release of damage associated molecular patterns (DAMPs) and subsequent activation of DAMP receptors. DAMPs are endogenous molecules, including protein and non-protein subgroups (Tang et al., 2012). High mobility group box 1 (HMGB1) is a typical nuclear DAMP (Kang et al., 2014), which triggers an immune response during various types of RCD, including ferroptosis (Wen et al., 2019). Advanced glycosylation end-product specific receptor (AGER/RAGE) has been recognized as a receptor for HMGB1, responsible for the inflammatory response caused by ferroptotic cell death in macrophages (Wen et al., 2019). Another protein-related DAMP marker involved in ferroptosis includes mutated KRAS protein (KRASG12D) (Dai et al., 2020a). The release of KRASG12D by ferroptotic cancer cells is then taken up by macrophages through its receptor AGER (Dai et al., 2020a). This cell–cell communication results in M2 macrophage polarization and subsequent tumor formation in xenograft models (Dai et al., 2020a). Conversely, ferroptotic cell death may promote anti-tumor immunity by activating cytotoxic T cell responses (Wang W. et al., 2019), although the key DAMP mediator of this process is unidentified. There are still many uncertainties in the interaction of protein and non-protein (e.g., oxidative lipid or host DNA) DAMPs in shaping ferroptosis-associated inflammation and immune response, including their receptors and target immune cells.
Ferroptosis is implicated in many pathological conditions of iron overload, including cancer (Chen et al., 2020a; Conrad et al., 2020; Tang et al., 2020). On the one hand, ferroptosis caused by small molecule compounds can inhibit tumor growth in mouse models. For example, in a genetically engineered mouse model of pancreatic cancer, pancreatic tumors treated with cyst(e)inase show a ferroptotic morphological phenotype, with mitochondrial defects and extensive lipid droplet formation (Badgley et al., 2020), which may act as a source of PUFA for lipid peroxidation (Bai et al., 2019). Immunohistochemical staining of 4-HNE further indicates that the level of lipid peroxidation in pancreatic tumors is increased (Badgley et al., 2020). On the other hand, DAMP released by ferroptotic cells can promote tumor growth by maintaining an immunosuppressive microenvironment. For example, conditional depletion of pancreatic Gpx4 or a high-iron diet triggers the release of mutant KRASG12D protein or nuclear DNA, thereby inducing the tumor-promoting effects of macrophages (Dai et al., 2020a,b). The expression level of KRASG12D in macrophages is negatively correlated with the survival of pancreatic cancer patients (Dai et al., 2020a). Therefore, these findings suggest that the simultaneous detection of intracellular and extracellular markers may help diagnose and treat diseases related to ferroptotic damage.
In the past several years, we have witnessed the rapid development of ferroptosis research. This trend provides more opportunities to think deeply about the differences in the molecular mechanisms of RCD. A key unresolved question is how ferroptotic response causes cell death. Although the onset and intermediate signals and processes of ferroptosis have been described, the executioner of ferroptosis is still unknown. Unfortunately, the biomarkers of ferroptosis discussed in this review also present in other types of RCD or pathological conditions. A more precise understanding of specific biomarkers and contributors of ferroptosis (not non-ferroptotic deaths) may provide new opportunities for designing treatments for iron overload-related diseases. A combination of multiple biomarkers may help detect ferroptotic cell death in time. The challenge remains how to transform basic research findings into clinical applications. Solving these challenges requires further understanding of the molecular mechanisms and signal transduction of ferroptosis, as well as the use of new technologies to discover specific biomarkers.
XC, RK, and DT conceived the topic for this review. All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ashraf, A., Jeandriens, J., Parkes, H. G., and So, P. W. (2020). Iron dyshomeostasis, lipid peroxidation and perturbed expression of cystine/glutamate antiporter in Alzheimer’s disease: evidence of ferroptosis. Redox Biol. 32:101494. doi: 10.1016/j.redox.2020.101494
Badgley, M. A., Kremer, D. M., Maurer, H. C., DelGiorno, K. E., Lee, H. J., Purohit, V., et al. (2020). Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science 368, 85–89. doi: 10.1126/science.aaw9872
Bai, Y., Meng, L., Han, L., Jia, Y., Zhao, Y., Gao, H., et al. (2019). Lipid storage and lipophagy regulates ferroptosis. Biochem. Biophys. Res. Commun. 508, 997–1003. doi: 10.1016/j.bbrc.2018.12.039
Bersuker, K., Hendricks, J. M., Li, Z., Magtanong, L., Ford, B., Tang, P. H., et al. (2019). The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575, 688–692. doi: 10.1038/s41586-019-1705-2
Brown, C. W., Amante, J. J., Chhoy, P., Elaimy, A. L., Liu, H., Zhu, L. J., et al. (2019). Prominin2 drives ferroptosis resistance by stimulating iron export. Dev. Cell 51, 575–586e4. doi: 10.1016/j.devcel.2019.10.007
Canli, O., Alankus, Y. B., Grootjans, S., Vegi, N., Hultner, L., Hoppe, P. S., et al. (2016). Glutathione peroxidase 4 prevents necroptosis in mouse erythroid precursors. Blood 127, 139–148. doi: 10.1182/blood-2015-06-654194
Cao, J. Y., Poddar, A., Magtanong, L., Lumb, J. H., Mileur, T. R., Reid, M. A., et al. (2019). A genome-wide haploid genetic screen identifies regulators of glutathione abundance and ferroptosis sensitivity. Cell Rep. 26, 1544–1556e8. doi: 10.1016/j.celrep.2019.01.043
Chang, L. C., Chiang, S. K., Chen, S. E., Yu, Y. L., Chou, R. H., and Chang, W. C. (2018). Heme oxygenase-1 mediates BAY 11-7085 induced ferroptosis. Cancer Lett. 416, 124–137. doi: 10.1016/j.canlet.2017.12.025
Chen, D., Tavana, O., Chu, B., Erber, L., Chen, Y., Baer, R., et al. (2017). NRF2 is a major target of ARF in p53-independent tumor suppression. Mol. Cell 68, 224–232e4. doi: 10.1016/j.molcel.2017.09.009
Chen, X., Kang, R., Kroemer, G., and Tang, D. (2020a). Broadening horizons: the role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. doi: 10.1038/s41571-020-00462-0 [Epub ahead of print].
Chen, X., Li, J., Kang, R., Klionsky, D. J., and Tang, D. (2020b). Ferroptosis: machinery and regulation. Autophagy 1–28. doi: 10.1080/15548627.2020.1810918 [Epub ahead of print].
Chen, X., Yu, C., Kang, R., and Tang, D. (2020c). Iron metabolism in ferroptosis. Front. Cell Dev. Biol. 8:590226. doi: 10.3389/fcell.2020.590226
Chu, B., Kon, N., Chen, D., Li, T., Liu, T., Jiang, L., et al. (2019). ALOX12 is required for p53-mediated tumour suppression through a distinct ferroptosis pathway. Nat. Cell Biol. 21, 579–591. doi: 10.1038/s41556-019-0305-6
Conrad, M., Angeli, J. P., Vandenabeele, P., and Stockwell, B. R. (2016). Regulated necrosis: disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 15, 348–366. doi: 10.1038/nrd.2015.6
Conrad, M., Lorenz, S. M., and Proneth, B. (2020). Targeting ferroptosis: new hope for as-yet-incurable diseases. Trends Mol. Med. doi: 10.1016/j.molmed.2020.08.010 [Epub ahead of print].
Dai, E., Han, L., Liu, J., Xie, Y., Kroemer, G., Klionsky, D. J., et al. (2020a). Autophagy-dependent ferroptosis drives tumor-associated macrophage polarization via release and uptake of oncogenic KRAS protein. Autophagy 16, 2069–2083. doi: 10.1080/15548627.2020.1714209
Dai, E., Han, L., Liu, J., Xie, Y., Zeh, H. J., Kang, R., et al. (2020b). Ferroptotic damage promotes pancreatic tumorigenesis through a TMEM173/STING-dependent DNA sensor pathway. Nat. Commun. 11:6339. doi: 10.1038/s41467-020-20154-8
Dai, E., Meng, L., Kang, R., Wang, X., and Tang, D. (2020c). ESCRT-III-dependent membrane repair blocks ferroptosis. Biochem. Biophys. Res. Commun. 522, 415–421. doi: 10.1016/j.bbrc.2019.11.110
Dai, E., Zhang, W., Cong, D., Kang, R., Wang, J., and Tang, D. (2020d). AIFM2 blocks ferroptosis independent of ubiquinol metabolism. Biochem. Biophys. Res. Commun. 523, 966–971. doi: 10.1016/j.bbrc.2020.01.066
Dixon, S. J., Lemberg, K. M., Lamprecht, M. R., Skouta, R., Zaitsev, E. M., Gleason, C. E., et al. (2012). Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072. doi: 10.1016/j.cell.2012.03.042
Dixon, S. J., Patel, D. N., Welsch, M., Skouta, R., Lee, E. D., Hayano, M., et al. (2014). Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife 3:e02523. doi: 10.7554/eLife.02523.018
Doll, S., Freitas, F. P., Shah, R., Aldrovandi, M., da Silva, M. C., Ingold, I., et al. (2019). FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698. doi: 10.1038/s41586-019-1707-0
Doll, S., Proneth, B., Tyurina, Y. Y., Panzilius, E., Kobayashi, S., Ingold, I., et al. (2017). ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 13, 91–98. doi: 10.1038/nchembio.2239
Fang, X., Wang, H., Han, D., Xie, E., Yang, X., Wei, J., et al. (2019). Ferroptosis as a target for protection against cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 116, 2672–2680. doi: 10.1073/pnas.1821022116
Feng, H., Schorpp, K., Jin, J., Yozwiak, C. E., Hoffstrom, B. G., Decker, A. M., et al. (2020). Transferrin receptor is a specific ferroptosis marker. Cell Rep. 30, 3411–3423e7. doi: 10.1016/j.celrep.2020.02.049
FitzGerald, G. A. (2003). COX-2 and beyond: approaches to prostaglandin inhibition in human disease. Nat. Rev. Drug Discov. 2, 879–890. doi: 10.1038/nrd1225
Friedmann Angeli, J. P., Schneider, M., Proneth, B., Tyurina, Y. Y., Tyurin, V. A., Hammond, V. J., et al. (2014). Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 16, 1180–1191. doi: 10.1038/ncb3064
Gagliardi, M., Cotella, D., Santoro, C., Cora, D., Barlev, N. A., Piacentini, M., et al. (2019). Aldo-keto reductases protect metastatic melanoma from ER stress-independent ferroptosis. Cell Death Dis. 10:902. doi: 10.1038/s41419-019-2143-7
Galluzzi, L., Vitale, I., Aaronson, S. A., Abrams, J. M., Adam, D., Agostinis, P., et al. (2018). Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 25, 486–541. doi: 10.1038/s41418-017-0012-4
Galluzzi, L., Vitale, I., Warren, S., Adjemian, S., Agostinis, P., Martinez, A. B., et al. (2020). Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer 8:e000337. doi: 10.1136/jitc-2019-000337
Gao, M., Monian, P., Pan, Q., Zhang, W., Xiang, J., and Jiang, X. (2016). Ferroptosis is an autophagic cell death process. Cell Res. 26, 1021–1032. doi: 10.1038/cr.2016.95
Gao, M., Monian, P., Quadri, N., Ramasamy, R., and Jiang, X. (2015). Glutaminolysis and transferrin regulate ferroptosis. Mol. Cell 59, 298–308. doi: 10.1016/j.molcel.2015.06.011
Holze, C., Michaudel, C., Mackowiak, C., Haas, D. A., Benda, C., Hubel, P., et al. (2018). Oxeiptosis, a ROS-induced caspase-independent apoptosis-like cell-death pathway. Nat. Immunol. 19, 130–140. doi: 10.1038/s41590-017-0013-y
Homma, T., Kobayashi, S., and Fujii, J. (2020). Cysteine preservation confers resistance to glutathione-depleted cells against ferroptosis via CDGSH iron sulphur domain-containing proteins (CISDs). Free Radic. Res. 54, 397–407. doi: 10.1080/10715762.2020.1780229
Hou, W., Xie, Y., Song, X., Sun, X., Lotze, M. T., and Zeh, H. J. III, et al. (2016). Autophagy promotes ferroptosis by degradation of ferritin. Autophagy 12, 1425–1428. doi: 10.1080/15548627.2016.1187366
Kagan, V. E., Mao, G., Qu, F., Angeli, J. P., Doll, S., Croix, C. S., et al. (2017). Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 13, 81–90. doi: 10.1038/nchembio.2238
Kang, R., Chen, R., Zhang, Q., Hou, W., Wu, S., Cao, L., et al. (2014). HMGB1 in health and disease. Mol. Aspects Med. 40, 1–116. doi: 10.1016/j.mam.2014.05.001
Kang, R., Zeng, L., Zhu, S., Xie, Y., Liu, J., Wen, Q., et al. (2018). Lipid peroxidation drives gasdermin D-mediated pyroptosis in lethal polymicrobial sepsis. Cell Host Microbe 24, 97–108e4. doi: 10.1016/j.chom.2018.05.009
Kraft, V. A. N., Bezjian, C. T., Pfeiffer, S., Ringelstetter, L., Muller, C., Zandkarimi, F., et al. (2020). GTP cyclohydrolase 1/tetrahydrobiopterin counteract ferroptosis through lipid remodeling. ACS Cent. Sci. 6, 41–53. doi: 10.1021/acscentsci.9b01063
Kuang, F., Liu, J., Tang, D., and Kang, R. (2020). Oxidative damage and antioxidant defense in ferroptosis. Front. Cell Dev. Biol. 8:586578. doi: 10.3389/fcell.2020.586578
Kumagai, T., Matsukawa, N., Kaneko, Y., Kusumi, Y., Mitsumata, M., and Uchida, K. (2004). A lipid peroxidation-derived inflammatory mediator: identification of 4-hydroxy-2-nonenal as a potential inducer of cyclooxygenase-2 in macrophages. J. Biol. Chem. 279, 48389–48396. doi: 10.1074/jbc.m409935200
Kumar, A., Tikoo, S., Maity, S., Sengupta, S., Sengupta, S., Kaur, A., et al. (2012). Mammalian proapoptotic factor ChaC1 and its homologues function as gamma-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep. 13, 1095–1101. doi: 10.1038/embor.2012.156
Li, C., Zhang, Y., Liu, J., Kang, R., Klionsky, D. J., and Tang, D. (2020). Mitochondrial DNA stress triggers autophagy-dependent ferroptotic death. Autophagy 1–13. doi: 10.1080/15548627.2020.1739447 [Epub ahead of print].
Liu, J., Kuang, F., Kroemer, G., Klionsky, D. J., Kang, R., and Tang, D. (2020). Autophagy-dependent ferroptosis: machinery and regulation. Cell Chem. Biol. 27, 420–435. doi: 10.1016/j.chembiol.2020.02.005
Liu, N., Lin, X., and Huang, C. (2020). Activation of the reverse transsulfuration pathway through NRF2/CBS confers erastin-induced ferroptosis resistance. Br. J. Cancer 122, 279–292. doi: 10.1038/s41416-019-0660-x
Liu, Y., Wang, Y., Liu, J., Kang, R., and Tang, D. (2020a). Interplay between MTOR and GPX4 signaling modulates autophagy-dependent ferroptotic cancer cell death. Cancer Gene Ther. doi: 10.1038/s41417-020-0182-y [Epub ahead of print].
Liu, Y., Wang, Y., Liu, J., Kang, R., and Tang, D. (2020b). The circadian clock protects against ferroptosis-induced sterile inflammation. Biochem. Biophys. Res. Commun. 525, 620–625. doi: 10.1016/j.bbrc.2020.02.142
Lu, J., Xu, F., and Lu, H. (2020). LncRNA PVT1 regulates ferroptosis through miR-214-mediated TFR1 and p53. Life Sci. 260:118305. doi: 10.1016/j.lfs.2020.118305
Magtanong, L., Ko, P. J., To, M., Cao, J. Y., Forcina, G. C., Tarangelo, A., et al. (2019). Exogenous monounsaturated fatty acids promote a ferroptosis-resistant cell state. Cell Chem. Biol. 26, 420–432e9. doi: 10.1016/j.chembiol.2018.11.016
Mungrue, I. N., Pagnon, J., Kohannim, O., Gargalovic, P. S., and Lusis, A. J. (2009). CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J. Immunol. 182, 466–476. doi: 10.4049/jimmunol.182.1.466
Protchenko, O., Baratz, E., Jadhav, S., Li, F., Shakoury-Elizeh, M., Gavrilova, O., et al. (2020). Iron chaperone poly rC binding protein 1 protects mouse liver from lipid peroxidation and steatosis. Hepatology doi: 10.1002/hep.31328 [Epub ahead of print].
Ran, Q., Van Remmen, H., Gu, M., Qi, W., Roberts, L. J. II, Prolla, T., et al. (2003). Embryonic fibroblasts from Gpx4+/- mice: a novel model for studying the role of membrane peroxidation in biological processes. Free Radic. Biol. Med. 35, 1101–1109. doi: 10.1016/s0891-5849(03)00466-0
Shimada, K., Skouta, R., Kaplan, A., Yang, W. S., Hayano, M., Dixon, S. J., et al. (2016). Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat. Chem. Biol. 12, 497–503. doi: 10.1038/nchembio.2079
Shin, D., Kim, E. H., Lee, J., and Roh, J. L. (2018). Nrf2 inhibition reverses resistance to GPX4 inhibitor-induced ferroptosis in head and neck cancer. Free Radic. Biol. Med. 129, 454–462. doi: 10.1016/j.freeradbiomed.2018.10.426
Shintoku, R., Takigawa, Y., Yamada, K., Kubota, C., Yoshimoto, Y., Takeuchi, T., et al. (2017). Lipoxygenase-mediated generation of lipid peroxides enhances ferroptosis induced by erastin and RSL3. Cancer Sci. 108, 2187–2194. doi: 10.1111/cas.13380
Song, X., Xie, Y., Kang, R., Hou, W., Sun, X., Epperly, M. W., et al. (2016). FANCD2 protects against bone marrow injury from ferroptosis. Biochem. Biophys. Res. Commun. 480, 443–449. doi: 10.1016/j.bbrc.2016.10.068
Song, X., Zhu, S., Chen, P., Hou, W., Wen, Q., Liu, J., et al. (2018a). AMPK-Mediated BECN1 Phosphorylation promotes ferroptosis by directly blocking system Xc(-) activity. Curr. Biol. 28, 2388–2399e5. doi: 10.1016/j.cub.2018.05.094
Song, X., Zhu, S., Xie, Y., Liu, J., Sun, L., Zeng, D., et al. (2018b). JTC801 induces pH-dependent death specifically in cancer cells and slows growth of tumors in mice. Gastroenterology 154, 1480–1493. doi: 10.1053/j.gastro.2017.12.004
Soula, M., Weber, R. A., Zilka, O., Alwaseem, H., La, K., Yen, F., et al. (2020). Metabolic determinants of cancer cell sensitivity to canonical ferroptosis inducers. Nat. Chem. Biol. 16, 1351–1360. doi: 10.1038/s41589-020-0613-y
Stockwell, B. R., Friedmann Angeli, J. P., Bayir, H., Bush, A. I., Conrad, M., Dixon, S. J., et al. (2017). Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171, 273–285. doi: 10.1016/j.cell.2017.09.021
Sun, X., Niu, X., Chen, R., He, W., Chen, D., Kang, R., et al. (2016a). Metallothionein-1G facilitates sorafenib resistance through inhibition of ferroptosis. Hepatology 64, 488–500. doi: 10.1002/hep.28574
Sun, X., Ou, Z., Chen, R., Niu, X., Chen, D., Kang, R., et al. (2016b). Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 63, 173–184. doi: 10.1002/hep.28251
Sun, X., Ou, Z., Xie, M., Kang, R., Fan, Y., Niu, X., et al. (2015). HSPB1 as a novel regulator of ferroptotic cancer cell death. Oncogene 34, 5617–5625. doi: 10.1038/onc.2015.32
Sun, Y., Zheng, Y., Wang, C., and Liu, Y. (2018). Glutathione depletion induces ferroptosis, autophagy, and premature cell senescence in retinal pigment epithelial cells. Cell Death Dis. 9:753. doi: 10.1038/s41419-018-0794-4
Takahashi, N., Cho, P., Selfors, L. M., Kuiken, H. J., Kaul, R., Fujiwara, T., et al. (2020). 3D culture models with CRISPR screens reveal hyperactive NRF2 as a prerequisite for spheroid formation via regulation of proliferation and ferroptosis. Mol. Cell 80, 828–844.e6. doi: 10.1016/j.molcel.2020.10.010
Tang, D., and Kroemer, G. (2020). Ferroptosis. Curr. Biol. 30, R1292–R1297. doi: 10.1016/j.cub.2020.09.068
Tang, D., Chen, X., Kang, R., and Kroemer, G. (2020). Ferroptosis: molecular mechanisms and health implications. Cell Res. doi: 10.1038/s41422-020-00441-1 [Epub ahead of print].
Tang, D., Kang, R., Berghe, T. V., Vandenabeele, P., and Kroemer, G. (2019). The molecular machinery of regulated cell death. Cell Res. 29, 347–364. doi: 10.1038/s41422-019-0164-5
Tang, D., Kang, R., Coyne, C. B., Zeh, H. J., and Lotze, M. T. (2012). PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol. Rev. 249, 158–175. doi: 10.1111/j.1600-065x.2012.01146.x
Telorack, M., Meyer, M., Ingold, I., Conrad, M., Bloch, W., and Werner, S. (2016). A glutathione-Nrf2-thioredoxin cross-talk ensures keratinocyte survival and efficient wound repair. PLoS Genet. 12:e1005800. doi: 10.1371/journal.pgen.1005800
Wang, N., Zeng, G. Z., Yin, J. L., and Bian, Z. X. (2019). Artesunate activates the ATF4-CHOP-CHAC1 pathway and affects ferroptosis in Burkitt’s lymphoma. Biochem. Biophys. Res. Commun. 519, 533–539. doi: 10.1016/j.bbrc.2019.09.023
Wang, W., Green, M., Choi, J. E., Gijon, M., Kennedy, P. D., Johnson, J. K., et al. (2019). CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 569, 270–274. doi: 10.1038/s41586-019-1170-y
Wang, Y., Liu, Y., Liu, J., Kang, R., and Tang, D. (2020). NEDD4L-mediated LTF protein degradation limits ferroptosis. Biochem. Biophys. Res. Commun. 531, 581–587. doi: 10.1016/j.bbrc.2020.07.032
Wen, Q., Liu, J., Kang, R., Zhou, B., and Tang, D. (2019). The release and activity of HMGB1 in ferroptosis. Biochem. Biophys. Res. Commun. 510, 278–283. doi: 10.1016/j.bbrc.2019.01.090
Wenzel, S. E., Tyurina, Y. Y., Zhao, J., St Croix, C. M., Dar, H. H., Mao, G., et al. (2017). PEBP1 wardens ferroptosis by enabling lipoxygenase generation of lipid death signals. Cell 171, 628–641.e26. doi: 10.1016/j.cell.2017.09.044
Wu, M., Xu, L. G., Li, X., Zhai, Z., and Shu, H. B. (2002). AMID, an apoptosis-inducing factor-homologous mitochondrion-associated protein, induces caspase-independent apoptosis. J. Biol. Chem. 277, 25617–25623. doi: 10.1074/jbc.m202285200
Wu, Z., Geng, Y., Lu, X., Shi, Y., Wu, G., Zhang, M., et al. (2019). Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc. Natl. Acad. Sci. U.S.A. 116, 2996–3005. doi: 10.1073/pnas.1819728116
Xiao, X., Jiang, Y., Liang, W., Wang, Y., Cao, S., Yan, H., et al. (2019). miR-212-5p attenuates ferroptotic neuronal death after traumatic brain injury by targeting Ptgs2. Mol. Brain 12:78. doi: 10.1186/s13041-019-0501-0
Xie, Y., Zhu, S., Song, X., Sun, X., Fan, Y., Liu, J., et al. (2017). The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep. 20, 1692–1704. doi: 10.1016/j.celrep.2017.07.055
Yagoda, N., von Rechenberg, M., Zaganjor, E., Bauer, A. J., Yang, W. S., Fridman, D. J., et al. (2007). Smukste. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature 447, 864–868. doi: 10.1038/nature05859
Yang, L., Chen, X., Yang, Q., Chen, J., Huang, Q., Yao, L., et al. (2020). Broad spectrum deubiquitinase inhibition induces both apoptosis and ferroptosis in cancer cells. Front. Oncol. 10:949. doi: 10.3389/fonc.2020.00949
Yang, M., Chen, P., Liu, J., Zhu, S., Kroemer, G., Klionsky, D. J., et al. (2019). Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci. Adv. 5:eaaw2238. doi: 10.1126/sciadv.aaw2238
Yang, W. H., Ding, C. C., Sun, T., Rupprecht, G., Lin, C. C., Hsu, D., et al. (2019). The hippo pathway effector TAZ regulates ferroptosis in renal cell carcinoma. Cell Rep. 28, 2501–2508e4. doi: 10.1016/j.celrep.2019.07.107
Yang, W. S., and Stockwell, B. R. (2016). Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 26, 165–176. doi: 10.1016/j.tcb.2015.10.014
Yang, W. S., Kim, K. J., Gaschler, M. M., Patel, M., Shchepinov, M. S., and Stockwell, B. R. (2016). Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. U.S.A. 113, E4966–E4975. doi: 10.1073/pnas.1603244113
Yang, W. S., SriRamaratnam, R., Welsch, M. E., Shimada, K., Skouta, R., Viswanathan, V. S., et al. (2014). Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331. doi: 10.1016/j.cell.2013.12.010
Yang, Y., Luo, M., Zhang, K., Zhang, J., Gao, T., Connell, D. O., et al. (2020). Nedd4 ubiquitylates VDAC2/3 to suppress erastin-induced ferroptosis in melanoma. Nat. Commun. 11:433. doi: 10.1038/s41467-020-14324-x
Ye, L. F., Chaudhary, K. R., Zandkarimi, F., Harken, A. D., Kinslow, C. J., Upadhyayula, P. S., et al. (2020). Radiation-induced lipid peroxidation triggers ferroptosis and synergizes with ferroptosis inducers. ACS Chem. Biol. 15, 469–484. doi: 10.1021/acschembio.9b00939
Yuan, H., Li, X., Zhang, X., Kang, R., and Tang, D. (2016a). CISD1 inhibits ferroptosis by protection against mitochondrial lipid peroxidation. Biochem. Biophys. Res. Commun. 478, 838–844. doi: 10.1016/j.bbrc.2016.08.034
Yuan, H., Li, X., Zhang, X., Kang, R., and Tang, D. (2016b). Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem. Biophys. Res. Commun. 478, 1338–1343. doi: 10.1016/j.bbrc.2016.08.124
Zhang, Y., Tan, H., Daniels, J. D., Zandkarimi, F., Liu, H., Brown, L. M., et al. (2019). Imidazole ketone erastin induces ferroptosis and slows tumor growth in a mouse lymphoma model. Cell Chem. Biol. 26, 623–633e9. doi: 10.1016/j.chembiol.2019.01.008
Zhou, B., Liu, J., Kang, R., Klionsky, D. J., Kroemer, G., and Tang, D. (2020). Ferroptosis is a type of autophagy-dependent cell death. Semin. Cancer Biol. 66, 89–100. doi: 10.1016/j.semcancer.2019.03.002
Zhu, S., Zhang, Q., Sun, X., Zeh, H. J. III, Lotze, M. T., Kang, R., et al. (2017). HSPA5 regulates ferroptotic cell death in cancer cells. Cancer Res. 77, 2064–2077. doi: 10.1158/0008-5472.can-16-1979
Keywords: ferroptosis, cell death, biomarker, lipid perioxidation, iron metabolism
Citation: Chen X, Comish PB, Tang D and Kang R (2021) Characteristics and Biomarkers of Ferroptosis. Front. Cell Dev. Biol. 9:637162. doi: 10.3389/fcell.2021.637162
Received: 02 December 2020; Accepted: 05 January 2021;
Published: 21 January 2021.
Edited by:
Binfeng Lu, University of Pittsburgh, United StatesReviewed by:
Jingfang Ju, Stony Brook Medicine, United StatesCopyright © 2021 Chen, Comish, Tang and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daolin Tang, ZGFvbGluLnRhbmdAdXRzb3V0aHdlc3Rlcm4uZWR1; Rui Kang, cnVpLmthbmdAdXRzb3V0aHdlc3Rlcm4uZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.