
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 04 February 2021
Sec. Membrane Traffic and Organelle Dynamics
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.627700
This article is part of the Research TopicCoupling and Uncoupling: Dynamic Control of Membrane ContactsView all 15 articles
The endoplasmic reticulum (ER) forms direct membrane contact sites with the plasma membrane (PM) in eukaryotic cells. These ER-PM contact sites play essential roles in lipid homeostasis, ion dynamics, and cell signaling, which are carried out by protein-protein or protein-lipid interactions. Distinct tethering factors dynamically control the architecture of ER-PM junctions in response to intracellular signals or external stimuli. The physiological roles of ER-PM contact sites are dependent on a variety of regulators that individually or cooperatively perform functions in diverse cellular processes. This review focuses on proteins functioning at ER-PM contact sites and highlights the recent progress in their mechanisms and physiological roles.
Intracellular trafficking between membrane-bound organelles is divided into two types, vesicular trafficking and non-vesicular trafficking. Vesicular trafficking is the predominant pathway to transport macromolecular substances and exchange information between organelles. The cargo is wrapped by or integrated into the membrane to form a vesicle, and exchange proteins or lipids between organelles through membrane fusion (Bonifacino and Glick, 2004; Südhof and Rothman, 2009). However, recent studies demonstrated that non-vesicular trafficking is another critical trafficking approach among intracellular membranous organelles, directly communicating through a close gap (typically within 10–30 nm) formed by two opposed membranes (Wong et al., 2019). This kind of intracellular communication is ensured by particular regions within the cell, defined as membrane contact sites (MCSs), structures mediated by protein-protein or protein-lipid interactions.
The largest membrane-bound organelle in eukaryotic cells is the endoplasmic reticulum (ER). It is the primary place for the synthesis of proteins and lipids, which are needed to maintain and propagate other membranous organelles and plasma membrane (PM) (Bonifacino and Glick, 2004). The ER extends throughout the whole cell and engages in broad communications with PM and other organelles by MCSs. ER-PM contact sites were first observed in muscle cells in the 1950s (Porter and Palade, 1957), and later were demonstrated as a general feature in eukaryotes. The MCSs formed between the ER and the PM provide an ideal platform for non-vesicular transport of lipids, ions, and many other signaling molecules (Gallo et al., 2016; Saheki and De Camilli, 2017a; Stefan, 2020). The architecture of ER-PM junctions is dynamically controlled by distinct tethering factors, and the cellular functions of ER-PM contact sites are highly dependent on those regulators located in these regions (Gallo et al., 2016). However, a variety of proteins localized at the crowded ER-PM junctions, frequently resulting in the co-existence of multiple regulators with similar or partially similar functions (Manford et al., 2012; Hoffmann et al., 2019; Johnson et al., 2019; Kang et al., 2019). They act synergistically to maintain the local microenvironment, which largely increases the difficulties of identifying their individual functions and mechanisms. Therefore, it is important to clarify how these proteins act in concert to play roles in the MCSs, especially under physiological or pathological conditions. This review focuses on the representative regulators localized at ER-PM contact sites, highlighting their physiological functions, molecular mechanisms as well as conservations in eukaryotes.
The extensive cortical ER network is highly dynamic in eukaryotic cells. The tethering factors build and maintain the ER-PM contact sites demanded by diverse biological processes. Most proteins localized at the ER-PM contact sites can span and tether the two opposed membranes. It often happens that multiple proteins coordinate to tether the same MCSs and perform more than one physiological functions (Manford et al., 2012; Fernandez-Busnadiego et al., 2015; Kang et al., 2019). While some protein tethers are constitutively localized at ER-PM contact sites, the locations of others are dynamically regulated by stimuli such as calcium ions and phosphoinositides (Figure 1A) (Gallo et al., 2016; Okeke et al., 2016; Saheki and De Camilli, 2017a; Stefan, 2020). How these protein tethers accurately modulate the structure and plasticity of ER-PM contact sites has not been completely understood. Comprehensive understanding of the tethering mechanisms will shed important light on the dynamical control of the cortical ER network. In this section, we summarized the major ER-PM tethering proteins and discussed their membrane-targeting mechanisms.
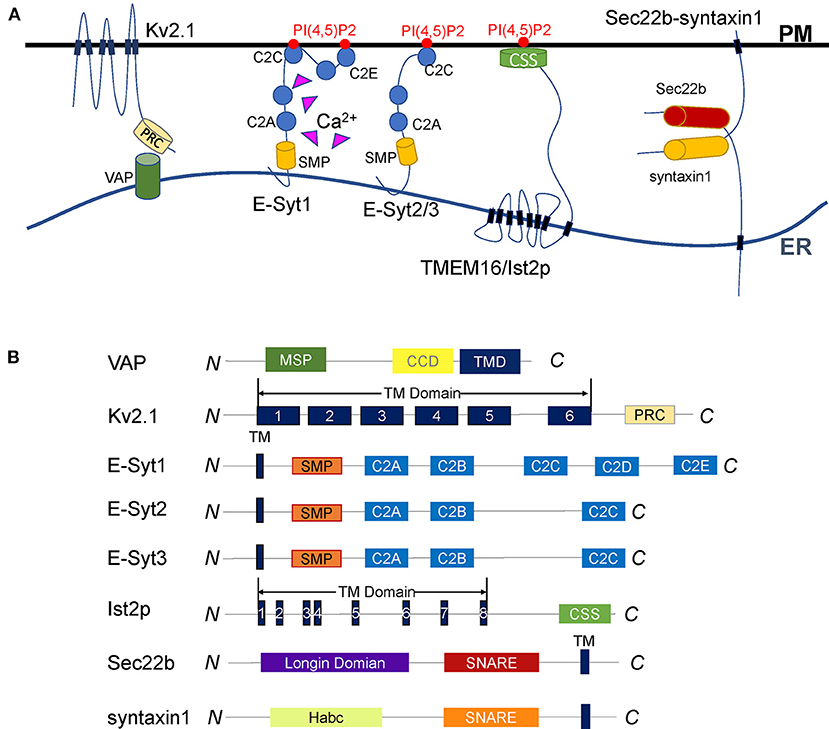
Figure 1. Diverse tethering mechanisms of proteins at ER-PM contact sites. (A) Representative illustration of the proteins that tether the ER and the PM. VAP anchors to the ER surface through the transmembrane domain and interacts with its binding partners to tether the ER and PM. The Kv2.1 channel is a VAP-binding partner. It anchors to the PM by six transmembrane domains and interacts with VAP through the C-terminal PRC domain. The Kv2.1-VAP interaction bridges the ER-PM junctions regulated by PRC domain phosphorylation. E-Syts are ER membrane proteins anchored to the ER via an N-terminal hydrophobic hairpin. E-Syt1 dynamically tethers the ER to the PM through a Ca2+-dependent interaction between the C2C domain and PI(4,5)P2. E-Syt2/3 constitutively maintains ER-PM junctions regardless of Ca2+. TMEM16/Ist2p is an eight-span integrin protein in the ER and connects the ER to the PM by its C-terminal CSS domain binding to PI(4,5)P2. The ER SNARE protein Sec22b forms an incomplete trans-SNARE complex with syntaxin1 on the PM that does not mediate membrane fusion but promotes the ER and PM tethering. (B) Diagrams of the membrane tethering proteins described in (A). The major functional domains are shown in each of the proteins.
Vesicle-associated membrane protein (VAMP)-associated protein (VAP) is an evolutionarily conserved ER membrane protein in all eukaryotes. It plays a vital role in many ER processes, especially in maintaining ER-PM contact sites. Loss of VAP by mutations leads to neurodegeneration, such as sporadic ALS or Parkinson's disease (Anagnostou et al., 2010; Kun-Rodrigues et al., 2015). There are mainly 2 VAPs (VAP-A and VAP-B) in mammals, 2 homologs (Scs2p and Scs22p) in yeast, and 10 homologs (VAP27-1 to VAP27-10) in Arabidopsis. VAP contains a major sperm protein (MSP) domain, a coiled-coil linker domain, and a C-terminal transmembrane domain required for ER surface location (Figure 1B) (Lev et al., 2008).
Although named by interaction with VAMP, VAP's primary function is not as a soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) regulator. It binds more than 100 peripheral proteins, including those anchored into other organelles (Murphy and Levine, 2016). By interacting with those partners, VAP dynamically controls the junctions formed between the ER and other organelles such as Golgi, lipid droplets, mitochondria, endosomes, and PM (Murphy and Levine, 2016). By localization at particular contact sites, VAPs and their binding partners coordinate to mediate diverse cellular processes (Lev et al., 2008). However, the distribution of VAP in mammals is not limited to the MCSs but throughout the ER, suggesting it has other functions than membrane tethering. For example, VAP interacts with secernin-1 at the ER membrane to regulate dynamic ER remodeling (Lindhout et al., 2019).
Unlike in mammals, the homologs of VAPs in fission yeast and Arabidopsis are more concentrated at MCSs (Zhang et al., 2012; Wang et al., 2014). Scs2p and Scs22p were discovered as two major tethering factors at the ER-PM contact sites (Manford et al., 2012; Zhang et al., 2012). Scs2p was reported as an inositol binding protein that responds to phospholipid composition (Kagiwada and Hashimoto, 2007). However, there is no direct evidence showing Scs2p connects the ER to the PM through the interaction with phosphoinositides. In Arabidopsis, the tethering function of VAP27s at ER-PM contact sites was also identified. VAP27 co-localizes with NET3C and forms a tetra complex with microtubules and actin filaments to tether the ER to the PM (Wang et al., 2014). Recent studies suggested VAP27s-mediated ER-PM contact sites regulate plant endocytosis (Stefano et al., 2018; Wang et al., 2019). However, the detailed tethering mechanisms of VAP27s remain largely unknown.
The location of VAP largely relies on its binding partners. It is generally accepted that most of the VAP-mediated ER-PM tethering requires at least one PM-targeting partner. One primary class of the VAP-binding partners are cytoplasmic proteins containing an FFAT motif, which binds specifically to the MSP domain (Kamemura and Chihara, 2019). At MCSs, these FFAT-containing proteins interact with the PM through a membrane-targeting domain, for example, the pleckstrin homology domain (PHD). The lipid-transfer proteins (LTPs) are the most well-studied FFAT-containing proteins at MCSs. For instance, as an Scs2p/Scs22p-binding partner, yeast LTP Osh3p dynamically targets the PM by its PHD under the regulation of PM PI(4)P levels (Jansen et al., 2011; Kamemura and Chihara, 2019). Nir2, another type of FFAT-containing LTP, connects the PM by its C-terminal LNS2 domain binding to phosphatidic acid (PA) (Kim et al., 2013; Balla, 2018). The mechanisms of how these LTPs coordinate with VAP to regulate lipid metabolism will be discussed in the following corresponding section.
Voltage-gated potassium (Kv) channel is a tetramer composed of 4α subunits (70 kDa), and each subunit monomer contains six transmembrane helix segments (Figure 1B) (Christie, 1995; Yellen, 1998; Shah and Aizenman, 2014; Fu et al., 2017; Jedrychowska and Korzh, 2019). Kv2 channels Kv2.1 and Kv2.2, also named KCNB1 and KCNB2, are abundantly expressed in the brain and present in other tissues like muscle and pancreatic islets. The central part of the Kv2 channel is cytosolic, which forms large clusters in the ER-PM interface. The Kv2 is a delayed rectifier potassium channel, participates in the repolarization of neural action potentials (Murakoshi and Trimmer, 1999; Bishop et al., 2015). However, the clustered Kv2 channels do not readily conduct potassium (Lim et al., 2000; O'Connell et al., 2010), but involved in reshaping the ER-PM connections (Fox et al., 2015; Kirmiz et al., 2018a).
Kv2 channels are VAP-binding partners. They interact with VAPs through the C-terminal proximal restriction and clustering (PRC) domain (Lim et al., 2000; Johnson et al., 2019). This Kv2-VAP interaction mediates the ER-PM junctions responsible for Kv2 clustering (Johnson et al., 2018, 2019). The phosphorylation of serine residues in the PRC domain produces negative charges to enable VAP binding and control the clustering of Kv2 channels, which is the prerequisites for Kv2 channels-mediated ER-PM connections (Redman et al., 2007; Cobb et al., 2015; Johnson et al., 2018, 2019; Kirmiz et al., 2018b).
The clustering enables Kv2 channels to play a structural role in forming ER-PM junctions, and the non-conductive state is essential for avoiding electrically silencing neuronal activity (Fox et al., 2015). Hence, Kv2 clusters-induced ER-PM junctions could serve as a scaffold for other cell activities such as Ca2+ signaling and membrane trafficking. It has been reported Kv2.1 cluster promotes the coupling of PM L-type Ca2+ channels (LTCCs) and ER ryanodine receptor (RyR) Ca2+ release channels to generate partial Ca2+ release without the requirement of action potentials (Vierra et al., 2019). Recent studies revealed the Kv2.1 channels facilitate insulin exocytosis in pancreatic beta cells by their structural role of the clustering rather than the ability to conduct K+. Kv2.1 clusters could be applied as a target for insulin secretion (Fu et al., 2017; Greitzer-Antes et al., 2018). Currently, the localization mechanism of Kv2 channels at ER-PM junctions has been primarily uncovered, but the physiological function is still unclear.
E-Syts are integral membrane proteins anchored on the ER membrane. They are named by the similarity with synaptotagmins, key regulators in calcium-dependent vesicle fusion (Min et al., 2007). E-Syts are identified as a conserved family of tethering proteins at ER-PM contact sites. All E-Syts contain an N-terminal membrane anchor, followed by a synaptotagmin-like mitochondrial lipid-binding protein (SMP) domain and multiple C2 domains (Figure 1B) (Lee and Hong, 2006; Manford et al., 2012; Yu et al., 2016). While they are anchored to the ER membrane by the hydrophobic hairpin region, E-Syts can associate with the inner leaflet of the PM through their C-terminal C2 domains. The SMP domain is capable of harboring lipids, which we will discuss separately in the lipid exchange section.
The C2 domains are membrane-binding molecules representing a family of proteins with diverse functions (Rizo and Südhof, 1998). As for E-Syts, the C2 domains are connected in series to interact with acidic phospholipids on PM to mediate the ER-PM tethering. In mammals, E-Syt1 has five C2 domains, while E-Syt2 and E-Syt3 have three. The difference in numbers and characteristics of C2 domains among E-Syts leads to their distinct subcellular localization and tethering functions. E-Syt2 and E-Syt3 are located mainly at cortical ER. E-Syt1, by contrast, is broadly localized to the ER but migrate to ER-PM MCSs in response to elevated cytosolic Ca2+ (Min et al., 2007; Chang et al., 2013; Giordano et al., 2013; Idevall-Hagren et al., 2015). Cryo-ET studies indicated ER-PM contact sites mediated by E-Syts are structurally different from those bridged by STIM1 (Fernandez-Busnadiego et al., 2015). The average ER-PM distance at E-Syt3-mediated junctions is shorter than that observed at E-Syt1-mediated contact sites, although the latter could be shortened about 30% when cytosolic Ca2+ increases (Fernandez-Busnadiego et al., 2015). The C2C domain of E-Syt2/3 binds to PI(4,5)P2 through a conserved basic patch to constitutively maintain ER-PM contact sites. In contrast, the C2C domain of E-Syt1 interacts with PI(4,5)P2 upon Ca2+ binding to dynamically control the MCSs (Giordano et al., 2013; Idevall-Hagren et al., 2015; Saheki et al., 2016; Yu et al., 2016). In addition to ER-PM connections, E-Syts are also involved in the tethering of peroxisome-ER membrane contacts. They regulate cholesterol transport employing a similar C2C domain-PI(4,5)P2-binding mechanism (Xiao et al., 2019).
In yeast, the homologs of E-Syts are called tricalbins (Tcb1, Tcb2, and Tcb3) (Creutz et al., 2004; Schulz and Creutz, 2004; Lee and Hong, 2006). All the three tricalbins are major tethering contributors for ER-PM contact sites (Manford et al., 2012; Toulmay and Prinz, 2012). Recent studies showed tricalbins form curved cortical ER membrane with a requirement of C2 domains (Collado et al., 2019; Hoffmann et al., 2019). However, the detailed molecular mechanism of tricalbins in maintaining MCSs is still missing.
Plant SYT1 (synaptotagmin 1), the homolog of E-Syts in Arabidopsis, is enriched at ER-PM contact sites, especially the MCSs between immobile ER tubules and the PM (Yamazaki et al., 2010; Perez-Sancho et al., 2015; Ishikawa et al., 2018). The cortical ER network maintained by SYT1 correlates with the C2 domains, identical to E-Syts in mammals and tricalbins in yeast (Yamazaki et al., 2010). A recent study discovered that ionic stress could increase SYT1-mediated ER-PM connectivity by promoting the accumulation of PI(4,5)P2 on PM (Lee et al., 2019). These data suggest that the interaction between negatively charged lipids on PM and the C2 domains represents an evolutionarily conserved mechanism for E-Syt family proteins.
Ist2p is the yeast homolog of the TMEM16, an eight-span integrin in the ER. The structure of Ist2p contains a specific ion channel followed by a long cytoplasmic C-terminal region riches in lysine and histidine residues (Figure 1B) (Juschke et al., 2005; Maass et al., 2009; Brach et al., 2011). The cortical localization of Ist2p relies on its C-terminal region, which was defined as the cortical sorting signal (CSS) (Brach et al., 2011). The CSS fragment regulates Ist2p expression and transports the protein to the PM, where it interacts with PI(4,5)P2 to bring the cortical ER and the PM closer to 15–50 nm (Juschke et al., 2005; Fischer et al., 2009; Maass et al., 2009; Wolf et al., 2012). The loss of Ist2p leads to an increase in the distance between the ER and the PM, suggesting Ist2p is a determinant for the span of ER-PM connections (Ercan et al., 2009).
The sorting mechanism of Ist2p from the ER to PM-associated domains is somewhat similar to the recruitment of STIM. They both bind to phospholipids on PM through the C-terminal domain, suggesting the recruitment of integral membrane proteins to PM through specific protein-lipid interactions represents a common mechanism. Ist2p was reported to be associated with the H+ pump Pma1 in the PM, allowing cells to adapt to different growth stages (Wolf et al., 2012). A recent study showed Ist2p and the LTP Osh6p are co-localized at ER-PM connections. Ist2p interacts with Osh6p to target the latter to the ER-PM contact sites, and they jointly participate in the lipid transport between the ER and the PM (D'Ambrosio et al., 2020).
While the function of Ist2p in yeast has been extensively studied, we currently still know little about its mammalian homolog TMEM16 (Hartzell et al., 2009). The two isoforms TMEM16A and TMEM16B, have recently been identified as calcium-activated chloride channels (Ercan et al., 2009; Xiao et al., 2011). However, whether TMEM16 and Ist2p have conserved functions at ER-PM contact sites remains to be clarified (Kunzelmann et al., 2016).
SNARE proteins represent a superfamily in which the members share a conserved SNARE motif with about 60–70 residues. They are the core engine of intracellular vesicle fusion. SNAREs can be classified as Q-SNAREs and R-SNAREs. Membrane fusion is initiated when one R-SNARE on the vesicle pairs with three t-SNAREs on the target membrane to form a four-helix trans-SNARE complex (Sutton et al., 1998; Weber et al., 1998). Sec22 has three isoforms in mammals as Sec22a, Sec22b, and Sec22c. Only Sec22b has a SNARE motif and is conserved in yeast (Sun et al., 2020). Sec22b belongs to the R-SNARE family. It anchored to ER through a C-terminal transmembrane domain right after the SNARE motif. In addition to the coiled-coil SNARE motif and transmembrane domain, Sec22b contains an N-terminal longin domain conserved with a profilin-like folded structure (Figure 1B) (Fasshauer, 2003; Hong, 2005; Jahn and Scheller, 2006). The SNARE motif and longin domain of Sec22b may play essential roles in vesicular transport between the ER and the Golgi apparatus. While the SNARE motif forms four helixes with its cognate t-SNAREs, the longin domain regulates the membrane fusion by interaction with the SNARE motif (Daste et al., 2015).
Distinct to its traditional function on membrane fusion, Sec22b has another non-fusogenic role in PM expansion (Petkovic et al., 2014). It can interact with syntaxin1 to form a partial but tight SNARE complex, which could not drive the membrane fusion due to the absence of SNAP25. However, this kind of non-fusogenic SNARE bridge tethers the ER to the PM, and more interesting, this tethering function is conserved in yeast. The yeast Sec22p and Sso1p (the homolog of syntaxin1) interact with Osh2p and Osh3p to regulate non-vesicular lipid transport between the ER and the PM. The existence of these SNARE-mediated junctions can shorten the distances and improve the efficiency of lipid transport (Prinz, 2010; Petkovic et al., 2014). In the mammalian nervous system, one latest research found the Sec22b-syntaxin1 complex can interact with E-Syts and form a ternary complex that plays a vital role in PM expansion and axon growth (Gallo et al., 2020). Together, the Sec22b-syntaxin1 complex plays a role in the tethering of the ER to the PM, from which it indirectly participate in the regulation of lipid metabolism and contribute to PM extension and other physiological processes (Petkovic et al., 2014; Gallo et al., 2016).
Besides these representative tethering factors, there are many other versatile regulators at ER-PM contact sites. These regulators tether the ER and the PM when they perform their critical cellular functions, for example, the Ca2+ dynamics regulator stromal interaction molecule 1 (STIM1). Interestingly, STIM1-mediated membrane tethering is Ca2+-dependent. In response to the low concentration of Ca2+, The ER-anchored STIM1 oligomerizes and recognizes the PM polyphosphoinositides and Orai1 (Liou et al., 2007; Zhou et al., 2013). This action coordinatively regulates the membrane tethering and Ca2+ homeostasis. Another type of versatile-tethering protein is LTPs, which couple the membrane tethering and lipid metabolisms. Most LTPs anchor the ER through the transmembrane domain or VAP interactions while target the PM using protein-lipid interactions (Kim et al., 2015; Ghai et al., 2017; Naito et al., 2019). The mechanisms of these regulators will be discussed in the following sections.
Together, a variety of regulators have the ability of membrane tethering at ER-PM contact sites. They bridge the two membranes via diverse connections. Some connections are constant to maintain the primary cortical ER network, while the others are dynamically regulated to perform demanded functions. As the foundation of ER-PM contacts, all these tethering molecules coordinate to modulate the cellular processes through the fine tune of MCSs.
As an important second messenger, Ca2+ is essential for many cellular and physiological processes, including gene transcription, protein modification, lipid metabolism, cell growth, and apoptosis (Stathopulos et al., 2006; Soboloff et al., 2012). So that precise and dynamic controls are needed to ensure calcium ions play proper functions at a specific time or place (Stathopulos et al., 2006). The cytoplasmic Ca2+ signals are generated by releasing Ca2+ from the calcium pool or the extracellular Ca2+ influx. The store-operated calcium entry (SOCE), a concept proposed in the 1990s, is a ubiquitous Ca2+ influx pathway at the ER-PM contact sites (Putney, 1986, 1990; Patterson et al., 1999; Yao et al., 1999). The Ca2+ entry is triggered when Ca2+ stores in the ER lumen depleted and the cytosolic Ca2+ concentration is at a low level. STIM proteins and Orai channels (Figure 2A) are the foundation proteins in the regulation of SOCE and Ca2+ signals (Liou et al., 2005; Roos et al., 2005; Feske et al., 2006; Vig et al., 2006; Zhang et al., 2006).
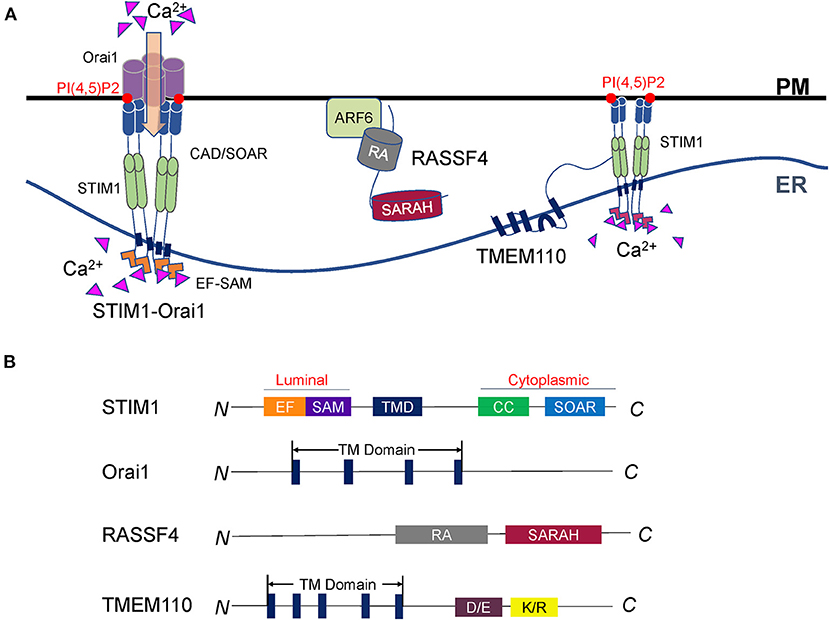
Figure 2. Regulation of Ca2+ dynamics at ER-PM contact sites. (A) Representative illustration of the STIM-Orai complex and its regulators. STIM1 in the ER and Orai1 on the PM compose the core machinery of SOCE. STIM1 senses the Ca2+ change in the ER lumen and forms oligomers. The STIM1 oligomers bind to polyphosphoinositides and activate Orai1 to import extracellular Ca2+. RASSF4 regulates SOCE and ER-PM junction through the control of the PM PI(4,5)P2 level, which is essential for the localization of STIM1. TMEM110 is an ER membrane protein that physically interacts with STIM1 to reshape ER-PM connections and facilitate STIM1 conformational conversion. Future studies are required to discover more STIM interacting partners, which is important to understand STIM proteins' sensing and coupling mechanisms. (B) Diagrams of the SOCE regulators described in A. The major functional domains are shown in each of the proteins.
STIM crosses the ER membrane and senses Ca2+ in the cavity of the ER (Williams et al., 2001; Zhang et al., 2005; Feske et al., 2006). Orai is a Ca2+ release-activated Ca2+ channel on PM (Chakrabarti and Chakrabarti, 2006; Prakriya et al., 2006; Soboloff et al., 2006a). Upon Ca2+ depletion, STIM protein interacts with Orai and initiates SOCE (Carrasco and Meyer, 2011; Zhou et al., 2013; Balla, 2018). There are two STIM proteins in mammals: STIM1 and STIM2. Both of them are mainly located at the ER with a similar structure (Hogan and Rao, 2015). STIM is anchored to the ER via a transmembrane domain, with an N-terminal domain in the ER lumen and a C-terminal domain in the cytoplasm (Figure 2B). The expression pattern of STIM1 and STIM2 are different. In most tissues, the expression level of STIM1 is higher than STIM2 in support that STIM1 is the predominant STIM protein that regulates the influx of Ca2+ in non-excitable cells (Soboloff et al., 2006b; Collins and Meyer, 2011; Hogan and Rao, 2015; Prakriya and Lewis, 2015).
STIM has the EF-hand and stereo alpha motif (SAM) domains in the lumen of the ER. Its cytoplasmic side contains several coiled-coil domains and a CRAC activation domain (CAD, also known as the STIM-Orai activation region, Or SOAR) (Manji et al., 2000; Yang et al., 2012; Prakriya and Lewis, 2015). In the presence of Ca2+, the EF-hand domains are tightly bound to the SAM domain. Therefore, the STIM protein exists as a monomer and in an inactive state, in which the nearby coiled-coil domain blocks CAD/SOAR. When the ER calcium pool is exhausted, the STIM protein senses Ca2+ change (Yuan et al., 2009). The structure of the EF-SAM domain becomes loose and stretched. STIM proteins dimerize from the ER lumen side to the cytoplasm and further form oligomers. These activated STIM1 oligomers interact with PI(4,5)P2 and the Orai channel through the released CAD/SOAR domain to accurately mediate the Ca2+ influx (Park et al., 2009; Ma et al., 2015; Prakriya and Lewis, 2015).
Although occupying a similar structure, the EF-SAM domains from STIM1 and STIM2 show different affinity to Ca2+. STIM1 is in a state of autoinhibition at rest but highly active upon Ca2+ depletion. STIM2 is more sensitive to tiny changes in the ER calcium store due to its low Ca2+ affinity, thus could be activated in response to less Ca2+ change (Zheng et al., 2011; Soboloff et al., 2012; Hogan and Rao, 2015). The different biophysical characteristics of STIM1 and STIM2 enable cells to sense changes in intracellular Ca2+ concentration accurately and take further actions (Brandman et al., 2007; Prakriya and Lewis, 2015). It seems reasonable that the basal Ca2+ homeostasis maintenance at rest is the primary responsibility of STIM2 (Wang et al., 2009; Kar et al., 2012). However, no significant change of Ca2+ level was observed in the calcium store of STIM2 KO cells at rest. In some tissues, for example, the neuronal cells and dendritic cells, the expression of STIM2 is significantly higher than STIM1, suggesting STIM2 may have other functions than the regulation of basal Ca2+ homeostasis (Williams et al., 2001; Oh-Hora et al., 2008; Prakriya and Lewis, 2015).
Many proteins around the STIM-Orai complex participate in SOCE. It is worth noted that SOCE recruits E-Syt1 to ER-PM junctions and rearranges adjacent ER structures into circular MCSs, which in turn stabilizes STIM-Orai clusters and accelerates Ca2+ replenishment (Kang et al., 2019). Another recent study showed ER protein Anoctamin 8 (ANO8) is translocated to STIM1-Orai1-mediated contact sites in a PI(4,5)P2-dependent manner. ANO8 further recruits the ER-localized SERCA (Sarco/endoplasmic reticulum Ca2+-ATPase) Ca2+ pump to replenish the Ca2+ reservoir, which may inactivate SOCE and regulate the receptor-stimulated Ca2+ signaling (Jha et al., 2019; Stefan, 2020).
The RAS association domain family (RASSF) consists of 10 members (RASSF1-10) localized at the cytoplasmic side of the PM. RASSF4 contains a C-terminal RAS association (RA) domain linked to a Sav-RASSF-Hpo (SARAH) domain (Figure 2B) (Chan et al., 2013; Iwasa et al., 2013). While the RA domain mediates the interactions with RAS GTPases, the SARAH domain was reported to facilitate dimerization between SARAH domain-containing proteins (Chan et al., 2013).
At ER-PM contact sites, RASSF4 acts in concert with ARF6, the upstream regulator of type I phosphatidylinositol phosphokinase (PIP5K), to regulate PI(4,5)P2 levels on the PM (Chen et al., 2017). Since PI(4,5)P2 is essential to position STIM1 and E-Syts, RASSF4 participates in the regulation of SOCE and ER-PM junctions indirectly through the regulation of PI(4,5)P2 homeostasis (Dickson, 2017).
ER-resident transmembrane protein 110 (TMEM110) is a STIM-activating enhancer (STIMATE). It contains 4–5 transmembrane domains, co-localized with STIM (Figure 2B). TMEM110 can remodel the short-term physiological junctions and relocate STIM1. Overexpression of TMEM110 leads to the formation of large STIM aggregates, while knockdown of this gene reduces STIM1 puncta at the ER-PM junctions (Jing et al., 2015; Quintana et al., 2015).
Furthermore, TMEM110 could physically interact with STIM1 and interfere with the autoinhibition of CAD/SOAR. When the Ca2+ storage is exhausted, STIM1 converts its conformation, facilitating the TMEM110 C-terminus interaction with the coiled-coil domain of STIM1. It releases the autoinhibition of CAD/SOAR and activates Ca2+ channel Orai (Hooper and Soboloff, 2015; Jing et al., 2015). Overall, TMEM110 is an ER protein that cooperates with STIM to reshape ER-PM connections and regulate calcium signaling dynamically. These studies indicate the STIM-Orai signaling heavily relies on proteins that regulate the ER-PM connections or the STIM conformation.
MCSs are ideal platforms to exchange molecules and local signals between organelles, dependent on their carriers or enzymes. Many protein enzymes, especially the phosphatases, play critical regulatory roles in ER-PM contact sites (Figure 3A) (Saheki and De Camilli, 2017a). They can catalyze substrates either in cis or in trans to participate in the regulatory network of many cellular processes such as cyclic adenosine 3′,5′-adenosine monophosphate (cAMP) signaling, calcium dynamics, and lipid metabolism.
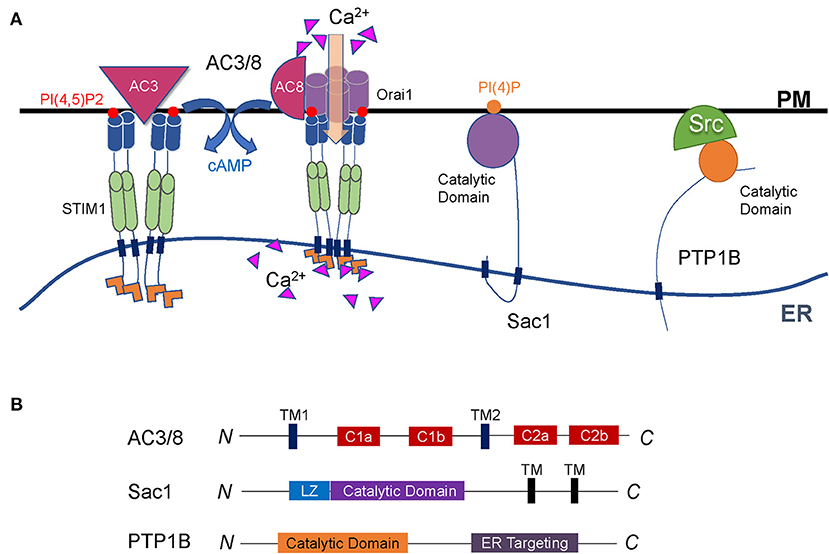
Figure 3. Regulation of cell signaling by enzymes at ER-PM contact sites. (A) Representative illustration of enzymes functioning at the interface between the ER and the PM. Both AC3 and AC8 are located on the PM through multiple transmembrane domains. AC3 interacts with STIM1, and AC8 binds directly to Orai1. They are both regulated by Ca2+ signals to produce cAMP. Sac1 is a conserved PI(4)P phosphatase anchored to the ER and can dephosphorylate PI(4)P both on the PM and in the ER membrane. PTP1B is anchored to the ER membrane via a C-terminal fragment and dephosphorylates its substrates on the PM through the cytosolic catalytic domain. (B) Diagrams of the enzymes described in A. The major functional domains are shown in each of the proteins.
Adenylate cyclase (AC) is an important signaling molecule downstream of G protein-coupled receptors. It is located on the PM via two multi-transmembrane domains and contains two catalytic domains (Figure 3B) (Cooper et al., 1995; Cooper and Crossthwaite, 2006; Dessauer et al., 2017). AC regulates cAMP, thereby participating in various physiological processes. Nine AC subtypes have been identified in mammals. Among them, the AC3 and AC8 are located at ER-PM junctions, and both of their activities are regulated by Ca2+. AC3 regulates blood glucose homeostasis, which makes it a new target for the development of anti-obesity drugs. However, AC8 plays a crucial role in neuroplasticity rather than in glucose regulation (Zachariou et al., 2008; Bogard et al., 2014; Wu et al., 2016).
Inside the cell, many signals transmit between the ER and the PM. In addition to the SOCE-regulated Ca2+ signal, cAMP is another vital signal which usually functions as a second messenger. At the ER-PM junctions, AC3 is an enzyme that relies on STIM1. STIM1 interacts with AC3 and generates cAMP, and this process is called storage operational cAMP signaling (SOcAMPS) (Lefkimmiatis et al., 2009; Maiellaro et al., 2012; Willoughby et al., 2012). AC8 directly interacts with Orai1 to alter the Ca2+ microenvironment under the PM, which in turn activates AC8 and produces cAMP (Willoughby et al., 2010, 2012). The dynamic interaction between Ca2+ and cAMP signals at the ER-PM junctions represents an important scenario of cell homeostasis and plays vital roles in physiology and pathology (Lefkimmiatis et al., 2009; Maiellaro et al., 2012; Willoughby et al., 2012; Okeke et al., 2016).
Localized on the ER and Golgi apparatus, suppressor of actin 1 (Sac1) is a phosphoinositide phosphatase whose protein sequence and function are both highly conserved in yeast and mammals. In mammals, Sac1 protein commonly expresses in adult and embryonic tissues (Del Bel and Brill, 2018). The C-terminal region anchored Sac1 to the ER. A conserved catalytic CX5R (T/S) motif in the N-terminal domain enables Sac1 to have a catalytic function (Figure 3B) (Manford et al., 2010; Saheki and De Camilli, 2017a). The role of Sac1 is to remove phosphoric acid from the inositol ring to balance the level of PI(4)P (Del Bel and Brill, 2018).
Primarily as a PI(4)P phosphatase between the ER and the PM, Sac1 was proposed to either act in trans on the opposed PM PI(4)P or act in cis on the ER PI(4)P (Manford et al., 2010; Stefan et al., 2011; Mesmin et al., 2013). In the latter, ORP5/8 (Osh6p/7p in yeast) transfers PM PI(4)P to the ER, where Sac1 dephosphorylates the lipid on the same ER membrane. Sac1-catalyzed PI(4)P hydrolysis is essential to maintain PI(4)P concentration gradient, which facilitates the continuous exchange of PI(4)P/PS between the ER and the PM (Chung et al., 2015; Moser von Filseck et al., 2015b; Del Bel and Brill, 2018). Therefore, Sac1 indirectly controls the lipid metabolism where the ER and the PM are in close contact.
However, PI(4)P alone is insufficient to localize Sac1 to the ER-PM contact sites. Other proteins that connect the ER and the PM may jointly participate in the Sac1 localization. For example, the activated SOCE increases the amount of Sac1 in contact with the PM, while disruption of E-Syt2-mediated ER-PM junctions reduces the access of Sac1 on the PM (Dickson et al., 2016). Sac1 was found co-localized with E-Syt2 at ER-PM contact sites. E-Syt2 narrows ER and PM distance, which may restrict Sac1 to the right position. Sac1 consumes PI(4)P in this microdomain and thus produces the PI(4)P gradient (Dickson et al., 2016). However, it is still uncertain whether E-Syt2 directly interacts with Sac1.
Protein tyrosine phosphatase 1B (PTP1B) is a non-receptor phosphatase and belongs to the PTP family. First isolated from the human placenta, PTP1B is anchored to the surface of the ER membrane via a C-terminal fragment composed of 35 proline-rich residues (Frangioni et al., 1992). The N-terminus of PTP1B protein contains the catalytic domain with two proline-rich motifs (Figure 3B). PTP1B plays a catalytic function at ER-PM junctions by dephosphorylation of its substrates located on the PM through the cytosolic catalytic domain (Anderie et al., 2007).
PTP1B has several identified substrates. These substrates have diverse functions that make PTP1B play various roles in cellular physiology. For example, ER-bound PTP1B dynamically interacts with the protein tyrosine kinase Src on the PM, controls Src activation, and recruits adhesion complexes (Monteleone et al., 2012). PTP1B also plays roles in tumor growth, metastasis, and metabolism. It has double-sided effects with either promoting or suppressing cancer in tumor tissues, depending on the active substrate and cell environment (Lessard et al., 2010). Exploring the roles of PTP1B in individual tumors will provide new ideas for the diagnosis and treatment of tumors. The insulin signaling pathway and glucose metabolism is another process that PTP1B negatively regulates. The tyrosine-phosphorylated insulin receptor, insulin receptor substrate-1, and AKT are all possible targets of PTP1B (Abdelsalam et al., 2019). Targeting PTP1B is considered a strategy to treat insulin resistance and type 2 diabetes by improving insulin sensitivity (Hussain et al., 2019).
The ER is the central organelle that synthesizes various lipids such as phospholipids and cholesterol, which need to be transported to or exchanged with other organelles and PM. Unlike the bulk lipid transports mediated by vesicle fusion, LTPs are able to sense and transport particular lipids between organelles that are mostly happened within a short distance like at the MCS regions (Wong et al., 2019). The ER forms extensive membrane junctions with the PM where LTPs play vital roles in the regulation of lipid metabolism as well as other physiological processes (Figures 4A,B) (Kentala et al., 2016; Saheki and De Camilli, 2017a; Cockcroft and Raghu, 2018; Jeyasimman and Saheki, 2019; Stefan, 2020).
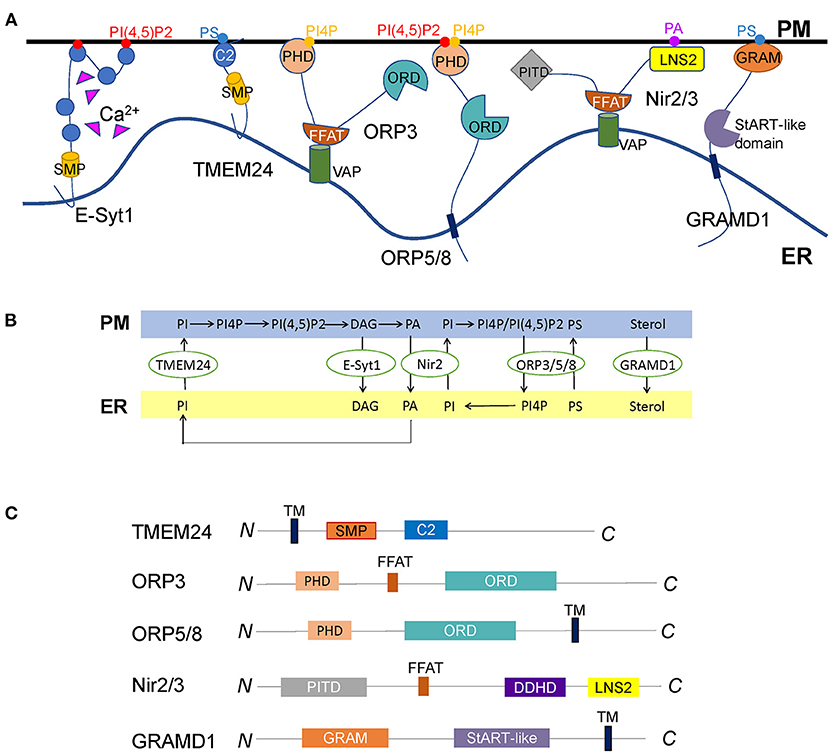
Figure 4. Diverse LTPs functioning at the ER-PM contact sites. (A) Representative illustration of LTPs located at the ER-PM junctions. E-Syt1 transfers phospholipids and DAG directly through the SMP domain between the ER and the PM, regulated by interactions of the C2 domains with Ca2+. TMEM24 is another SMP domain protein localized to the ER through a transmembrane domain. It transfers PI from the ER to the PM controlled by the dephosphorylation of the C-terminal polybasic region. Whether other SMP domain proteins, for example, E-Syt2/3 and tricalbins, mediate lipid transport at ER-PM contact sites remains unknown. ORP3, ORP5, and ORP8 are so far discovered ORPs at ER-PM contact sites. While ORP5 and ORP8 are ER membrane proteins, ORP3 is anchored to the ER through the interaction of its FFAT motif with VAP. All the three ORPs contain a PHD to interact with the PM lipids and an ORD to exchange PI(4)P/PI(4,5)P2 and PS between the ER and the PM. It remains elusive why the cell has three ORPs at ER-PM contact sites to mediate the same lipids. The discovery of the initial triggers of these LTPs might solve this problem. Nir2 and Nir3 are anchored to the ER via interactions of FFAT motifs with VAPs and connect to the PM by LNS2 domains, facilitating the PI and PA exchange between the two membranes. How other tethering molecules, such as E-Syts and Kv2s, couple with Nir2/3 to mediate PA/PI exchange need to be further explored. GRAMD1s are anchored to the ER via a C-terminal transmembrane domain and interact with cholesterol and PS on the PM through the N-terminal GRAM domain to connect the two opposed membranes. Thus, the accessible cholesterol is transported from the PM to the ER by StART-like domains. Whether there are other sterol transfer proteins transporting PM cholesterol to the ER or other organelles remains to be discovered. (B) The summary of LTPs-mediated lipid exchange at ER-PM contact sites. (C) Diagrams of the lipid transfer proteins described in A, except E-Syt1, are shown in Figure 1B. The major functional domains are shown in each of the proteins.
SMP domain proteins are evolutionarily conserved in eukaryotes. The SMP domain was first discovered in 2006 by the sequence analysis of a mitochondrial integral membrane protein. Later it was identified as a member of the superfamily of tubular Lipid-binding (TULIP) domain-containing proteins that have the ability to harbor lipids in the hydrophobic cavity (Kopec et al., 2010; Alva and Lupas, 2016). SMP domain proteins are commonly localized at MCSs formed by organelles and play versatile functions such as lipid transport, Ca2+ homeostasis, and signaling (Saheki and De Camilli, 2017a).
Benefited from the SMP domain, E-Syts are considered as LTPs at ER-PM contact sites (Giordano et al., 2013; Saheki and De Camilli, 2017b). The crystal structure of E-Syt2 showed SMP domain forms a dimer of approximately nine nm-long cylinders that harbors glycerolipids without selectivity (Schauder et al., 2014). E-Syt1, E-Syt-2, and E-Syt-3 have been demonstrated to form homo- and heterodimers inside the cell (Giordano et al., 2013). By the in vitro reconstituted system, E-Syt1 was identified as a Ca2+ dependent LTP, which directly transfers glycerophospholipids and DAG between the ER and the PM (Saheki et al., 2016; Yu et al., 2016). The SMP dimer is indispensable for E-Syt1 to transfer lipids. Considering the length of SMP dimer is far less than the average distance between the ER and the PM, the “shuttle model” in which SMP dimer shuttles between the ER and the PM to transport lipids is more acceptable. This shuttle model was further supported by an artificially designed assay using the DNA-origami nanostructures to define precise distances between membranes (Bian et al., 2019).
In addition to the SMP domain, the C2 domains in E-Syt1 are also essential to regulate lipid transfer (Saheki et al., 2016; Yu et al., 2016; Bian et al., 2018). At least, both the C2A and C2C domains are indispensable in E-Syt1-mediated lipid transport (Saheki et al., 2016; Yu et al., 2016; Bian et al., 2018). The C2C domain is the predominant region for membrane tethering, facilitating lipid transport by shortening the distance. The C2A domain was proposed as an autoinhibitory domain that inactivates the SMP domain. Ca2+ releases this autoinhibition to enable lipid transport (Bian et al., 2018). Recent studies in yeast demonstrated SMP domain and C2 domains of tricalbins act in concert to form highly curved ER peaks on the cortical ER membrane facing the PM, raising a possibility that the C2A domain involves in the interaction with the ER membrane to facilitate the lipid transport (Collado et al., 2019; Hoffmann et al., 2019).
Although the membrane tethering function of the E-Syt family is conserved in eukaryotes, it was unclear whether all family members are capable of transporting lipids between the ER and PM. Only E-Syt1 in mammalian cells was identified to mediate lipid exchange directly. Although the locations of E-Syt2 and E-Syt3 at ER-PM contact sites are not affected by Ca2+, Ca2+ does bind to their C2A domains and induce a local protein conformational transition (Xu et al., 2014). Whether E-Syt2 and E-Syt3 directly mediate lipid exchange are still open questions to the field.
As a highly conserved protein family, E-Syts likely play essential roles in cell physiology. Unexpectedly, they're non-essential proteins. No obvious defects were observed in E-Syts triple knockout mice (Sclip et al., 2016; Tremblay and Moss, 2016). Neither mammalian cells lacking E-Syts nor yeast cells lacking tricalbins showed significant abnormalities (Manford et al., 2012; Toulmay and Prinz, 2012; Saheki et al., 2016). Given that many factors coordinate at ER-PM junctions, one possible explanation could be the functional redundancy. Other factors may replace the function of E-Syts once the latter is omitted (Saheki, 2017). However, E-Syts and tricalbins do have physiological effects. For example, E-Syts can maintain PM lipid homeostasis, promote nerve transmission and synaptic growth, modulate virus-induced membrane fusion, mediate the endocytosis of FGFR, and play roles in insulin secretion and diet-induced obesity development (Saheki et al., 2016; Tremblay et al., 2016; Kikuma et al., 2017; El Kasmi et al., 2018; Xie et al., 2019; Nath et al., 2020; Zhang et al., 2020). Tricalbins act in maintaining PM integrity (Toulmay and Prinz, 2012; Collado et al., 2019). More interesting, plant SYT1 is required for withstanding mechanical stress, maintaining cell membrane integrity and virus movement, suggesting this protein family is of importance to cell physiology (Min et al., 2007; Chang et al., 2013; Giordano et al., 2013; Idevall-Hagren et al., 2015).
Transmembrane protein 24 (TMEM24) is another SMP domain protein localized at ER-PM contact sites. It is anchored to the ER membrane through an N-terminal transmembrane domain, followed by an SMP domain, a C2 domain, and a polybasic C-terminal region (Figure 4C). TMEM24 is involved in regulating insulin secretion and neuronal excitability (Pottekat et al., 2013; Lees et al., 2017; Sun et al., 2019). Like E-Syts, the SMP domain forms a dimer in TMEM24, and each SMP domain binds one lipid molecule, one less than that in E-Syt2 (Schauder et al., 2014; Lees et al., 2017). TMEM24 selectively transports PI from the ER to the PM, supplying PM with PI(4,5)P2 during signal transductions. This process is regulated by protein kinase C (PKC)-dependent phosphorylation of the C-terminal PM binding regions in response to cytosolic Ca2+ (Lees et al., 2017).
Glucose-stimulated insulin secretion is regulated by the inositol phosphate signaling pathway and Ca2+. While TMEM24 transfers the PI(4,5)P2 precursor PI from the ER to the PM, E-Syt1 clears the PI(4,5)P2 metabolite DAG on PM. Since the dissociation of TMEM24 from PM is controlled by PKC-mediated phosphorylation, and E-Syt1 regulates PKC activity, E-Syt1 is considered an indirect regulator of TMEM24 (Xie et al., 2019). Both of them play a role in phosphoinositide metabolism and Ca2+ homeostasis, thus indirectly regulate insulin secretion in pancreatic β cells (Xie et al., 2019).
The oxysterol-binding protein (OSBP) and its related proteins (ORPs) compose a conserved family that mediates non-vesicular lipid transports at the MCSs (Im et al., 2005; de Saint-Jean et al., 2011; Olkkonen and Li, 2013; Du et al., 2015). There are two conserved domains in this protein family, the PHD and OSBP-related domain (ORD). The PHD plays a role in membrane docking by interactions with anionic lipids such as phosphatidylserine (PS), PI(4)P, and PI(4,5)P2. The ORD is a ligand binding and lipid exchange domain (Kentala et al., 2016; Cockcroft and Raghu, 2018). Besides, ORPs have either an FFAT domain or a transmembrane domain through which they are located on the surface of membranous organelles, including the ER (Mesmin et al., 2013; Pulli et al., 2018).
ORP3, ORP5, and ORP8 are ORPs mainly localized at ER-PM contact sites in mammals. ORP5 and ORP8 possess a single C-terminal transmembrane domain that anchors them on the ER surface and a PHD to interact with the PM (Figure 4C). They were reported to mediate the PI(4)P/PS exchange cycle that transfers PS from the ER to the PM and move PI(4)P from the PM to the ER. Sac1 depletes PI(4)P and generates a PI(4)P gradient to drive this exchange (Chung et al., 2015; Moser von Filseck et al., 2015b; Dickson et al., 2016). However, another study showed PI(4,5)P2, rather than PI(4)P is the critical lipid for the targeting of ORP5 and ORP8 to the PM (Ghai et al., 2017). Besides ER-PM contact sites, ORP5 and ORP8 are also localized at other MCSs such as ER-mitochondria and ER-lipid droplet contact sites (Galmes et al., 2016; Du et al., 2020). Unlike ORP5 and ORP8, ORP3 is anchored to the ER via its FFAT motif that interacts with VAP protein (Figure 4C). Recent studies uncovered that ORP3 is capable of regulating PI(4)P homeostasis and Ca2+ dynamics by activating PKC (Dong et al., 2020; D'Souza et al., 2020; Gulyás et al., 2020).
In yeast, the conserved ORP family is called oxysterol-binding homology (Osh) protein. At ER-PM contact sites, Osh6p and Osh7p mediate the exchange of PI(4)P and PS fueled by PI(4)P metabolism, similar to the ORP5 and ORP8 in mammals (Maeda et al., 2013; Moser von Filseck et al., 2015a). Osh3p is more like ORP3 in mammals, which binds PI(4)P and recruits the Sac1p to ER-PM contact sites to regulate the PM PI(4)P levels (Stefan et al., 2011; Tong et al., 2013; Omnus et al., 2020). These studies demonstrated that phosphoinositides, mainly the PI(4)P and PI(4,5)P2, are common lipid ligands for the ORP family localized at ER-PM contact sites. Although 12 conserved members have been identified in Arabidopsis, the functions of ORPs in plant lipid metabolism are still poorly understood (Skirpan et al., 2006). Whether the functions and mechanisms of ORPs discovered in mammals and yeast are conserved in plants remain to be determined.
ORPs localized at ER-PM contact sites play multiple roles in cell physiology. For example, Osh2p and Osh3p interact with Myo5p and Scs2p to bridge the ER contact with endocytic areas and facilitate actin polymerization (Encinar Del Dedo et al., 2017). ORP3 participates in Ca2+ homeostasis and cell adhesion (Lehto et al., 2008). ORP5 and ORP8 can regulate cancer growth, making them potential drug targets for cancer therapy (Ishikawa et al., 2010; Guo et al., 2017).
Nir2 and Nir3 belong to the PI transfer protein (PITP) family, a class of central players involved in phospholipid homeostasis at ER-PM contact sites. They are the mammalian ortholog proteins of Drosophila retinal degeneration B (rdgB), which was proven to transfer PI and phosphatidylcholine (PC) between membrane bilayers (Amarilio et al., 2005). The structure of Nir2/3 contains a PI-transfer domain at the very N-terminus, then followed by an FFAT motif, six hydrophobic stretches, and a C-terminal LNS2 domain (Figure 4C). Nir2/3 is anchored to the ER through the binding of the FFAT sequence to the VAP protein and connects with PM by the interaction of the LNS2 domain with phosphatidic acid (PA) (Kim et al., 2013; Balla, 2018).
PA and PI are interconverted lipid second messengers that play roles in many signaling pathways, coupled by the Nir2/rdgB family. Phospholipase C (PLC) hydrolyses PI(4,5)P2 to generate DAG and its phosphorylated lipid PA. After binding PA, Nir2/rdgB is translocated to ER-PM contact sites to exchange PI and PA between the two opposed membranes (Chang et al., 2013; Kim et al., 2013, 2015; Balla, 2018). This process coordinates local lipid metabolism with downstream signaling at ER-PM contact sites (Kim et al., 2013). A recent study found that Nir2 and Kv2.1 are co-localized at ER-PM contact sites in neuronal cells, indicating Kv2-VAP tethers may regulate Nir2 localization and PI homeostasis (Kirmiz et al., 2019). Different from Nir2, the ability of Nir3 is to sense subtle PA production and sustain basal PM PI(4,5)P2 levels (Chang and Liou, 2015). They cooperatively regulate PI(4,5)P2 homeostasis at ER-PM contact sites.
The protein containing the Glucosyltransferases, Rab-like GTPase activators, and myotubularins (GRAM) domain was named GRAMD, a recently discovered class of conserved ER proteins. Bioinformatics studies identified six GRAMD proteins (Ysp1p, Sip3p, Ysp2p, Lam4p, Lam5p, and Lam6p) in yeast and five members (GRAMD1a, GRAMD1b, GRAMD1c, GRAMD2, and GRAMD3) in mammals (Gatta et al., 2015). However, only three GRAMD1s in mammals contain StART-like lipid transfer domains. GRAMD2 and GRAMD3 are supposed not to mediate lipid transport (Naito et al., 2019). GRAMD proteins are anchored to ER via its C-terminal transmembrane domain and target the PM using the N-terminal GRAM domain (Figure 4C) (Stefan et al., 2011; Chu et al., 2015; Besprozvannaya et al., 2018).
The existence of the StART-like domain makes GRAMD1 proteins contribute to PM sterol homeostasis by recognizing accessible PM cholesterol and transporting it to the ER (Holthuis and Levine, 2005; van Meer et al., 2008; Sandhu et al., 2018). A recent study showed GRAMD1s form homo- and heteromeric complexes that interact with the free cholesterol and PS on the PM by the GRAM domain. Thus, the accessible cholesterol is transported to the ER through StART-like domains. Loss of the three GRAMD1s leads to a significant expansion of the available PM cholesterol pool, suggesting GRAMD1s are major cholesterol transporters between the ER and the PM (Sandhu et al., 2018; Naito et al., 2019). Different from GRAMD1, GRAMD2 co-localizes with E-Syts and help to maintain the ER-PM contacts. Based on this tethering function, GRAMD2 may play a role in SOCE and Ca2+ homeostasis by the recruitment of STIM1 (Besprozvannaya et al., 2018).
The trafficking of low-density-lipoprotein (LDL)-cholesterol is essential in cholesterol metabolism. Imbalance in this process could cause diseases, for example, the Niemann-Pick type C (NPC). The ER is the central organelle senses and synthesizes endogenous cholesterol. After endocytosis, the vast majority of LDL-cholesterol is transported to the PM (Pfisterer et al., 2016). The PM cholesterol could then supply the ER by GRAMD1s (Sandhu et al., 2018). However, about 30% of LDL-cholesterol is directly transported from endosomes/lysosomes to the ER, mediated by ORP1L and STARD3. When ER cholesterol is excessive, these sterol-transfer proteins can act in the opposite direction, transporting the cholesterol to the endosomes (Eden et al., 2016; Wilhelm et al., 2017). Recent studies indicate NPC1 tethers the ER to the endocytic organelles and facilitates cholesterol egress through ORP5 and GRAMD1s. Both of the LTPs localized at the ER-endocytic organelles MCSs in response to the cholesterol levels (Du et al., 2011; Höglinger et al., 2019).
CERT (ceramide transport protein) is another LTP containing a StART-like domain. In addition to glycerophospholipids and sterols, ceramide is also synthesized in the ER. CERT anchors to the ER via the FFAT motif-VAP interaction. It contains a PHD that interacts with PI(4)P to build the ER-Golgi contacts. Through the StART-like domain, CERT transports ceramide to the Golgi apparatus, where glucosylceramide and sphingomyelin are synthesized (Hanada et al., 2003). Glycolipid-transfer protein, another VAP-interacting LTP, transports glucosylceramide from cis-Golgi to trans-Golgi or the ER (Smith et al., 2006; Halter et al., 2007; Backman et al., 2018). These processes are indispensable for sphingomyelin and glycosphingolipid homeostasis (Breslow, 2013; Hanada, 2018).
Many regulators coordinate to form an extensive protein network at ER-PM contact sites, regulating intracellular signal transductions coupled with Ca2+ and phospholipid signaling. The elevated cytoplasmic Ca2+ triggers the enrichment of E-Syt1 at ER-PM contact sites (Giordano et al., 2013). In another way, the ER-Ca2+ depletion induces STIM1 translocation to ER-PM contact sites (Liou et al., 2007). Both the Ca2+-regulated processes enhance the ER-PM connections, which subsequently promotes the recruitment of Nir2 to the ER-PM interface (Chang et al., 2013). Nir2 binds and transfers PA from the PM to the ER (Kim et al., 2015). In turn, it moves PI from the ER to the PM and generates PI(4)P and PI(4,5)P2, in which step RASSF4 and ARF6 participate (Stathopulos et al., 2006; Chen et al., 2017; Dickson, 2017). PI(4,5)P2 reinforces relocations of E-Syts and STIM1 at the contact sites and further regulates Ca2+ dynamics. The consumption of excess PI(4,5)P2 generates DAG and PA, which facilitate Nir2 enter the next circle. Overall, this network combines SOCE regulators with LTPs to develop a synergistic effect between Ca2+ signaling and PI(4,5)P2 metabolism at the ER-PM junctions and extend the duration of signal transductions (Dickson, 2017; Ong and Ambudkar, 2020).
The ER communicates with the PM through direct physical contacts, which are regulated by various proteins. These key players cooperatively mediate the reactions between the opposed membranes and drive diverse fundamental cellular processes. ER-PM contact sites are involved in the regulation of ion and lipid transports, signal transductions, ER morphology and remodeling, membrane trafficking, and yeast polarized growth (Encinar Del Dedo et al., 2017; Ng et al., 2018, 2020; Kang et al., 2019; Kirmiz et al., 2019; Weber-Boyvat et al., 2020). Substantial progress has been made toward understanding their functions and mechanisms (Table 1) (Saheki and De Camilli, 2017a; Wang et al., 2017; Ong and Ambudkar, 2020; Stefan, 2020). However, all proteins located at the MCSs seem to tether the membranes. Multiple regulators perform the same functions at the ER-PM contact sites in some cellular processes (Manford et al., 2012; Collado et al., 2019). The unique significance of these proteins needs to be further explored. For example, why are three E-Syts in yeast and mammals but only one in plants and flies?
Given that most of the regulators at ER-PM contact sites are highly conserved, it seems unlikely the cell keeps redundant proteins at this narrow and crowded place during evolution. These proteins are more likely of great significance to the cell. However, the general functional redundancy gives us trouble understanding their exact physiological functions. One speculation is each of these functional redundant proteins still has its features. Take tethering as an example. Although all protein tethers are capable of bridging the ER and the PM, the ER-PM junctions formed by distinct tethers have variable architectures and different mechanisms (Petkovic et al., 2014; Fernandez-Busnadiego et al., 2015; Johnson et al., 2018). These differences make them feasible to regulate the diverse cellular processes or play roles in specific conditions such as ER stress or cell damage. The same situation also happens in LTPs. It remains unclear why multiple LTPs transfer one specific lipid at MCSs. Perhaps the future discovery of their regulation and triggering mechanisms will give us the answer.
Tissue specificity is another possibility. Proteins with similar functions at ER-PM contact sites may individually play a predominant role in the specific type of cells, depending on their expression enrichments or stimuli. Studies of those proteins in specific tissues will be a way to identify their physiological roles. Recent studies have already confirmed this possibility (Guo et al., 2017; Kirmiz et al., 2018a; Zhang et al., 2020). On the other hand, some regulators' functions are not related to ER-PM contact sites (Tremblay et al., 2016; El Kasmi et al., 2018). Further efforts are needed to study their correlations.
To better understand the functions and mechanisms of ER-PM contact sites, high-resolution structures of protein-membrane complexes mimicking MCSs or partially mimicking MCSs will be expected. The development of advanced Cryo-EM and live-cell imaging technology will enable us to comprehensively understand the protein network-mediated ER-PM contact sites. Many regulators at these sites are implicated in disease pathologies. Studies of these proteins for their physiological and pathological functions will be of great significance for understanding the disease occurrence, new drug developments, and clinical applications.
CL, TQ, RH, CW, YL, and HY wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (NSFC) grants (Nos. 91854117 and 31871425), Natural Science Foundation of Jiangsu Province (BK20200036), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), Six talent peaks project in Jiangsu Province, and Jiangsu Distinguished Professor Funding.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Jingshi Shen for the Comments and critically reading of the manuscript and all Yu lab members for helpful discussions.
Abdelsalam, S. S., Korashy, H. M., Zeidan, A., and Agouni, A. (2019). The role of protein tyrosine phosphatase (PTP)-1B in cardiovascular disease and its interplay with insulin resistance. Biomolecules 9:9070286. doi: 10.3390/biom9070286
Alva, V., and Lupas, A. N. (2016). The TULIP superfamily of eukaryotic lipid-binding proteins as a mediator of lipid sensing and transport. Biochim. Biophys. Acta 1861, 913–923. doi: 10.1016/j.bbalip.2016.01.016
Amarilio, R., Ramachandran, S., Sabanay, H., and Lev, S. (2005). Differential regulation of endoplasmic reticulum structure through VAP-Nir protein interaction. J. Biol. Chem. 280, 5934–5944. doi: 10.1074/jbc.M409566200
Anagnostou, G., Akbar, M. T., Paul, P., Angelinetta, C., Steiner, T. J., and de Belleroche, J. (2010). Vesicle associated membrane protein B (VAPB) is decreased in ALS spinal cord. Neurobiol. Aging 31, 969–985. doi: 10.1016/j.neurobiolaging.2008.07.005
Anderie, I., Schulz, I., and Schmid, A. (2007). Direct interaction between ER membrane-bound PTP1B and its plasma membrane-anchored targets. Cell Signal. 19, 582–592. doi: 10.1016/j.cellsig.2006.08.007
Backman, A. P. E., Halin, J., Nurmi, H., Möuts, A., Kjellberg, M. A., and Mattjus, P. (2018). Glucosylceramide acyl chain length is sensed by the glycolipid transfer protein. PLoS ONE 13:0209230. doi: 10.1371/journal.pone.0209230
Balla, T. (2018). Ca(2+) and lipid signals hold hands at endoplasmic reticulum-plasma membrane contact sites. J. Physiol. 596, 2709–2716. doi: 10.1113/JP274957
Besprozvannaya, M., Dickson, E., Li, H., Ginburg, K. S., Bers, D. M., Auwerx, J., et al. (2018). GRAM domain proteins specialize functionally distinct ER-PM contact sites in human cells. Elife 7:31019. doi: 10.7554/eLife.31019
Bian, X., Saheki, Y., and De Camilli, P. (2018). Ca(2+) releases E-Syt1 autoinhibition to couple ER-plasma membrane tethering with lipid transport. EMBO J. 37, 219–234. doi: 10.15252/embj.201797359
Bian, X., Zhang, Z., Xiong, Q., De Camilli, P., and Lin, C. (2019). A programmable DNA-origami platform for studying lipid transfer between bilayers. Nat. Chem. Biol. 15, 830–837. doi: 10.1038/s41589-019-0325-3
Bishop, H. I., Guan, D., Bocksteins, E., Parajuli, L. K., Murray, K. D., Cobb, M. M., et al. (2015). Distinct cell- and layer-specific expression patterns and independent regulation of Kv2 channel subtypes in cortical pyramidal neurons. J. Neurosci. 35, 14922–14942. doi: 10.1523/JNEUROSCI.1897-15.2015
Bogard, A. S., Birg, A. V., and Ostrom, R. S. (2014). Non-raft adenylyl cyclase 2 defines a cAMP signaling compartment that selectively regulates IL-6 expression in airway smooth muscle cells: differential regulation of gene expression by AC isoforms. Naunyn. Schmiedebergs. Arch. Pharmacol. 387, 329–339. doi: 10.1007/s00210-013-0950-4
Bonifacino, J. S., and Glick, B. S. (2004). The mechanisms of vesicle budding and fusion. Cell 116, 153–166. doi: 10.1016/S0092-8674(03)01079-1
Brach, T., Specht, T., and Kaksonen, M. (2011). Reassessment of the role of plasma membrane domains in the regulation of vesicular traffic in yeast. J. Cell Sci. 124, 328–337. doi: 10.1242/jcs.078519
Brandman, O., Liou, J., Park, W. S., and Meyer, T. (2007). STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels. Cell 131, 1327–1339. doi: 10.1016/j.cell.2007.11.039
Breslow, D. K. (2013). Sphingolipid homeostasis in the endoplasmic reticulum and beyond. Cold Spring Harb. Perspect. Biol. 5:a013326. doi: 10.1101/cshperspect.a013326
Carrasco, S., and Meyer, T. (2011). STIM proteins and the endoplasmic reticulum-plasma membrane junctions. Annu. Rev. Biochem. 80, 973–1000. doi: 10.1146/annurev-biochem-061609-165311
Chakrabarti, R., and Chakrabarti, R. (2006). Calcium signaling in non-excitable cells: Ca2+ release and influx are independent events linked to two plasma membrane Ca2+ entry channels. J. Cell Biochem. 99, 1503–1516. doi: 10.1002/jcb.21102
Chan, J. J., Flatters, D., Rodrigues-Lima, F., Yan, J., Thalassinos, K., and Katan, M. (2013). Comparative analysis of interactions of RASSF1-10. Adv. Biol. Regul. 53, 190–201. doi: 10.1016/j.jbior.2012.12.001
Chang, C. L., Hsieh, T. S., Yang, T. T., Rothberg, K. G., Azizoglu, D. B., Volk, E., et al. (2013). Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep. 5, 813–825. doi: 10.1016/j.celrep.2013.09.038
Chang, C. L., and Liou, J. (2015). Phosphatidylinositol 4,5-bisphosphate homeostasis regulated by Nir2 and Nir3 proteins at endoplasmic reticulum-plasma membrane junctions. J. Biol. Chem. 290, 14289–14301. doi: 10.1074/jbc.M114.621375
Chen, Y. J., Chang, C. L., Lee, W. R., and Liou, J. (2017). RASSF4 controls SOCE and ER-PM junctions through regulation of PI(4,5)P2. J. Cell Biol. 216, 2011–2025. doi: 10.1083/jcb.201606047
Christie, M. J. (1995). Molecular and functional diversity of K+ channels. Clin. Exp. Pharmacol. Physiol. 22, 944–951. doi: 10.1111/j.1440-1681.1995.tb02331.x
Chu, B. B., Liao, Y. C., Qi, W., Xie, C., Du, X., Wang, J., et al. (2015). Cholesterol transport through lysosome-peroxisome membrane contacts. Cell 161, 291–306. doi: 10.1016/j.cell.2015.02.019
Chung, J., Torta, F., Masai, K., Lucast, L., Czapla, H., Tanner, L. B., et al. (2015). PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER–plasma membrane contacts. Science 349, 428–432. doi: 10.1126/science.aab1370
Cobb, M. M., Austin, D. C., Sack, J. T., and Trimmer, J. S. (2015). Cell cycle-dependent changes in localization and phosphorylation of the plasma membrane Kv2.1 K+ channel impact endoplasmic reticulum membrane contact sites in COS-1 cells. J. Biol. Chem. 290, 29189–29201. doi: 10.1074/jbc.M115.690198
Cockcroft, S., and Raghu, P. (2018). Phospholipid transport protein function at organelle contact sites. Curr. Opin. Cell Biol. 53, 52–60. doi: 10.1016/j.ceb.2018.04.011
Collado, J., Kalemanov, M., Campelo, F., Bourgoint, C., Thomas, F., Loewith, R., et al. (2019). Tricalbin-mediated contact sites control ER curvature to maintain plasma membrane integrity. Dev. Cell. 51, 476–487 e477. doi: 10.1016/j.devcel.2019.10.018
Collins, S. R., and Meyer, T. (2011). Evolutionary origins of STIM1 and STIM2 within ancient Ca2+ signaling systems. Trends Cell Biol. 21, 202–211. doi: 10.1016/j.tcb.2011.01.002
Cooper, D. M., and Crossthwaite, A. J. (2006). Higher-order organization and regulation of adenylyl cyclases. Trends Pharmacol. Sci. 27, 426–431. doi: 10.1016/j.tips.2006.06.002
Cooper, D. M., Mons, N., and Karpen, J. W. (1995). Adenylyl cyclases and the interaction between calcium and cAMP signalling. Nature 374, 421–424. doi: 10.1038/374421a0
Creutz, C. E., Snyder, S. L., and Schulz, T. A. (2004). Characterization of the yeast tricalbins: membrane-bound multi-C2-domain proteins that form complexes involved in membrane trafficking. Cell Mol. Life Sci. 61, 1208–1220. doi: 10.1007/s00018-004-4029-8
D'Ambrosio, J. M., Albanèse, V., Lipp, N. F., Fleuriot, L., Debayle, D., Drin, G., et al. (2020). Osh6 requires Ist2 for localization to ER-PM contacts and efficient phosphatidylserine transport in budding yeast. J. Cell Sci. 133:243733. doi: 10.1242/jcs.243733
Daste, F., Galli, T., and Tareste, D. (2015). Structure and function of longin SNAREs. J. Cell Sci. 128, 4263–4272. doi: 10.1242/jcs.178574
de Saint-Jean, M., Delfosse, V., Douguet, D., Chicanne, G., Payrastre, B., Bourguet, W., et al. (2011). Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J. Cell Biol. 195, 965–978. doi: 10.1083/jcb.201104062
Del Bel, L. M., and Brill, J. A. (2018). Sac1, a lipid phosphatase at the interface of vesicular and nonvesicular transport. Traffic 19, 301–318. doi: 10.1111/tra.12554
Dessauer, C. W., Watts, V. J., Ostrom, R. S., Conti, M., Dove, S., and Seifert, R. (2017). International union of basic and clinical pharmacology. CI. structures and small molecule modulators of mammalian adenylyl cyclases. Pharmacol. Rev. 69, 93–139. doi: 10.1124/pr.116.013078
Dickson, E. J. (2017). RASSF4: Regulator of plasma membrane PI(4,5)P2. J. Cell Biol. 216, 1879–1881. doi: 10.1083/jcb.201706042
Dickson, E. J., Jensen, J. B., Vivas, O., Kruse, M., Traynor-Kaplan, A. E., and Hille, B. (2016). Dynamic formation of ER-PM junctions presents a lipid phosphatase to regulate phosphoinositides. J. Cell Biol. 213, 33–48. doi: 10.1083/jcb.201508106
Dong, X., Wang, Z., Ye, S., and Zhang, R. (2020). The crystal structure of ORP3 reveals the conservative PI4P binding pattern. Biochem. Biophys. Res. Commun. 529, 1005–1010. doi: 10.1016/j.bbrc.2020.06.090
D'Souza, R. S., Lim, J. Y., Turgut, A., Servage, K., Zhang, J., Orth, K., et al. (2020). Calcium-stimulated disassembly of focal adhesions mediated by an ORP3/IQSec1 complex. Elife 9:e54113. doi: 10.7554/eLife.54113.sa2
Du, X., Brown, A. J., and Yang, H. (2015). Novel mechanisms of intracellular cholesterol transport: oxysterol-binding proteins and membrane contact sites. Curr. Opin. Cell Biol. 35, 37–42. doi: 10.1016/j.ceb.2015.04.002
Du, X., Kumar, J., Ferguson, C., Schulz, T. A., Ong, Y. S., Hong, W., et al. (2011). A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J. Cell Biol. 192, 121–135. doi: 10.1083/jcb.201004142
Du, X., Zhou, L., Aw, Y. C., Mak, H. Y., Xu, Y., Rae, J., et al. (2020). ORP5 localizes to ER-lipid droplet contacts and regulates the level of PI(4)P on lipid droplets. J. Cell Biol, 219:201905162. doi: 10.1083/jcb.201905162
Eden, E. R., Sanchez-Heras, E., Tsapara, A., Sobota, A., Levine, T. P., and Futter, C. E. (2016). Annexin A1 tethers membrane contact sites that mediate ER to endosome cholesterol transport. Dev. Cell. 37, 473–483. doi: 10.1016/j.devcel.2016.05.005
El Kasmi, I., Khadivjam, B., Lackman, M., Duron, J., Bonneil, E., Thibault, P., et al. (2018). Extended synaptotagmin 1 interacts with herpes simplex virus 1 glycoprotein M and negatively modulates virus-induced membrane fusion. J. Virol. 92:17. doi: 10.1128/JVI.01281-17
Encinar Del Dedo, J., Idrissi, F. Z., Fernandez-Golbano, I. M., Garcia, P., Rebollo, E., Krzyzanowski, M. K., et al. (2017). ORP-Mediated ER contact with endocytic sites facilitates actin polymerization. Dev. Cell 43, 588–602.e586. doi: 10.1016/j.devcel.2017.10.031
Ercan, E., Momburg, F., Engel, U., Temmerman, K., Nickel, W., and Seedorf, M. (2009). A conserved, lipid-mediated sorting mechanism of yeast Ist2 and mammalian STIM proteins to the peripheral ER. Traffic 10, 1802–1818. doi: 10.1111/j.1600-0854.2009.00995.x
Fasshauer, D. (2003). Structural insights into the SNARE mechanism. Biochim. Biophys. Acta 1641, 87–97. doi: 10.1016/S0167-4889(03)00090-9
Fernandez-Busnadiego, R., Saheki, Y., and De Camilli, P. (2015). Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum-plasma membrane contact sites. Proc. Natl. Acad. Sci. U.S.A. 112, E2004–2013. doi: 10.1073/pnas.1503191112
Feske, S., Gwack, Y., Prakriya, M., Srikanth, S., Puppel, S. H., Tanasa, B., et al. (2006). A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 441, 179–185. doi: 10.1038/nature04702
Fischer, M. A., Temmerman, K., Ercan, E., Nickel, W., and Seedorf, M. (2009). Binding of plasma membrane lipids recruits the yeast integral membrane protein Ist2 to the cortical ER. Traffic 10, 1084–1097. doi: 10.1111/j.1600-0854.2009.00926.x
Fox, P. D., Haberkorn, C. J., Akin, E. J., Seel, P. J., Krapf, D., and Tamkun, M. M. (2015). Induction of stable ER-plasma-membrane junctions by Kv2.1 potassium channels. J. Cell Sci. 128, 2096–2105. doi: 10.1242/jcs.166009
Frangioni, J. V., Beahm, P. H., Shifrin, V., Jost, C. A., and Neel, B. G. (1992). The nontransmembrane tyrosine phosphatase PTP-1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence. Cell 68, 545–560. doi: 10.1016/0092-8674(92)90190-N
Fu, J., Dai, X., Plummer, G., Suzuki, K., Bautista, A., Githaka, J. M., et al. (2017). Kv2.1 Clustering contributes to insulin exocytosis and rescues human beta-cell dysfunction. Diabetes 66, 1890–1900. doi: 10.2337/db16-1170
Gallo, A., Danglot, L., Giordano, F., Hewlett, B., Binz, T., Vannier, C., et al. (2020). Role of the Sec22b–E-Syt complex in neurite growth and ramification. J. Cell Sci. 133:jcs247148. doi: 10.1242/jcs.247148
Gallo, A., Vannier, C., and Galli, T. (2016). Endoplasmic reticulum-plasma membrane associations:structures and functions. Annu. Rev. Cell Dev. Biol. 32, 279–301. doi: 10.1146/annurev-cellbio-111315-125024
Galmes, R., Houcine, A., van Vliet, A. R., Agostinis, P., Jackson, C. L., and Giordano, F. (2016). ORP5/ORP8 localize to endoplasmic reticulum-mitochondria contacts and are involved in mitochondrial function. EMBO Rep. 17, 800–810. doi: 10.15252/embr.201541108
Gatta, A. T., Wong, L. H., Sere, Y. Y., Calderon-Norena, D. M., Cockcroft, S., Menon, A. K., et al. (2015). A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. Elife 4:253. doi: 10.7554/eLife.07253.024
Ghai, R., Du, X., Wang, H., Dong, J., Ferguson, C., Brown, A. J., et al. (2017). ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane. Nat. Commun. 8:757. doi: 10.1038/s41467-017-00861-5
Giordano, F., Saheki, Y., Idevall-Hagren, O., Colombo, S. F., Pirruccello, M., Milosevic, I., et al. (2013). PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 153, 1494–1509. doi: 10.1016/j.cell.2013.05.026
Greitzer-Antes, D., Xie, L., Qin, T., Xie, H., Zhu, D., Dolai, S., et al. (2018). Kv2.1 clusters on beta-cell plasma membrane act as reservoirs that replenish pools of newcomer insulin granule through their interaction with syntaxin-3. J. Biol. Chem. 293, 6893–6904. doi: 10.1074/jbc.RA118.002703
Gulyás, G., Sohn, M., Kim, Y. J., Várnai, P., and Balla, T. (2020). ORP3 phosphorylation regulates phosphatidylinositol 4-phosphate and Ca(2+) dynamics at plasma membrane-ER contact sites. J. Cell Sci. 133:jcs.237388. doi: 10.1242/jcs.237388
Guo, X., Zhang, L., Fan, Y., Zhang, D., Qin, L., Dong, S., et al. (2017). Oxysterol-binding protein-related protein 8 inhibits gastric cancer growth through induction of ER stress, inhibition of Wnt signaling, and activation of apoptosis. Oncol. Res. 25, 799–808. doi: 10.3727/096504016X14783691306605
Halter, D., Neumann, S., van Dijk, S. M., Wolthoorn, J., de Maziere, A. M., Vieira, O. V., et al. (2007). Pre- and post-Golgi translocation of glucosylceramide in glycosphingolipid synthesis. J. Cell. Biol. 179, 101–115. doi: 10.1083/jcb.200704091
Hanada, K. (2018). Lipid transfer proteins rectify inter-organelle flux and accurately deliver lipids at membrane contact sites. J. Lipid Res. 59, 1341–1366. doi: 10.1194/jlr.R085324
Hanada, K., Kumagai, K., Yasuda, S., Miura, Y., Kawano, M., Fukasawa, M., et al. (2003). Molecular machinery for non-vesicular trafficking of ceramide. Nature 426, 803–809. doi: 10.1038/nature02188
Hartzell, H. C., Yu, K., Xiao, Q., Chien, L.-T., and Qu, Z. (2009). Anoctamin/TMEM16 family members are Ca2+-activated Cl–channels. J. Physiol. 587, 2127–2139. doi: 10.1113/jphysiol.2008.163709
Hoffmann, P. C., Bharat, T. A. M., Wozny, M. R., Boulanger, J., Miller, E. A., and Kukulski, W. (2019). Tricalbins contribute to cellular lipid flux and form curved ER-PM contacts that are bridged by rod-shaped structures. Dev. Cell 51, 488–502.e488. doi: 10.1016/j.devcel.2019.09.019
Hogan, P. G., and Rao, A. (2015). Store-operated calcium entry: mechanisms and modulation. Biochem. Biophys. Res. Commun. 460, 40–49. doi: 10.1016/j.bbrc.2015.02.110
Höglinger, D., Burgoyne, T., Sanchez-Heras, E., Hartwig, P., Colaco, A., Newton, J., et al. (2019). NPC1 regulates ER contacts with endocytic organelles to mediate cholesterol egress. Nat. Commun. 10:4276. doi: 10.1038/s41467-019-12152-2
Holthuis, J. C. M., and Levine, T. P. (2005). Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6, 209–220. doi: 10.1038/nrm1591
Hong, W. (2005). SNAREs and traffic. Biochim. Biophys. Acta 1744, 120–144. doi: 10.1016/j.bbamcr.2005.03.014
Hooper, R., and Soboloff, J. (2015). STIMATE reveals a STIM1 transitional state. Nat. Cell Biol. 17, 1232–1234. doi: 10.1038/ncb3245
Hussain, H., Green, I. R., Abbas, G., Adekenov, S. M., Hussain, W., and Ali, I. (2019). Protein tyrosine phosphatase 1B (PTP1B) inhibitors as potential anti-diabetes agents: patent review (2015-2018). Exp. Opin. Ther. Pat. 29, 689–702. doi: 10.1080/13543776.2019.1655542
Idevall-Hagren, O., Lu, A., Xie, B., and De Camilli, P. (2015). Triggered Ca2+ influx is required for extended synaptotagmin 1-induced ER-plasma membrane tethering. EMBO J. 34, 2291–2305. doi: 10.15252/embj.201591565
Im, Y. J., Raychaudhuri, S., Prinz, W. A., and Hurley, J. H. (2005). Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature 437, 154–158. doi: 10.1038/nature03923
Ishikawa, K., Tamura, K., Ueda, H., Ito, Y., Nakano, A., Hara-Nishimura, I., et al. (2018). Synaptotagmin-associated endoplasmic reticulum-plasma membrane contact sites are localized to immobile ER tubules. Plant Physiol. 178, 641–653. doi: 10.1104/pp.18.00498
Ishikawa, S., Nagai, Y., Masuda, T., Koga, Y., Nakamura, T., Imamura, Y., et al. (2010). The role of oxysterol binding protein-related protein 5 in pancreatic cancer. Cancer Sci. 101, 898–905. doi: 10.1111/j.1349-7006.2009.01475.x
Iwasa, H., Kuroyanagi, H., Maimaiti, S., Ikeda, M., Nakagawa, K., and Hata, Y. (2013). Characterization of RSF-1, the Caenorhabditis elegans homolog of the Ras-association domain family protein 1. Exp. Cell Res. 319, 1–11. doi: 10.1016/j.yexcr.2012.10.008
Jahn, R., and Scheller, R. H. (2006). SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7, 631–643. doi: 10.1038/nrm2002
Jansen, M., Ohsaki, Y., Rega, L. R., Bittman, R., Olkkonen, V. M., and Ikonen, E. (2011). Role of ORPs in sterol transport from plasma membrane to ER and lipid droplets in mammalian cells. Traffic 12, 218–231. doi: 10.1111/j.1600-0854.2010.01142.x
Jedrychowska, J., and Korzh, V. (2019). Kv2.1 voltage-gated potassium channels in developmental perspective. Dev. Dyn. 248, 1180–1194. doi: 10.1002/dvdy.114
Jeyasimman, D., and Saheki, Y. (2019). SMP domain proteins in membrane lipid dynamics. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 1865:158447. doi: 10.1016/j.bbalip.2019.04.007
Jha, A., Chung, W. Y., Vachel, L., Maleth, J., Lake, S., Zhang, G., et al. (2019). Anoctamin 8 tethers endoplasmic reticulum and plasma membrane for assembly of Ca(2+) signaling complexes at the ER/PM compartment. EMBO J. 38:e101452. doi: 10.15252/embj.2018101452
Jing, J., He, L., Sun, A., Quintana, A., Ding, Y., Ma, G., et al. (2015). Proteomic mapping of ER-PM junctions identifies STIMATE as a regulator of Ca(2)(+) influx. Nat. Cell Biol. 17, 1339–1347. doi: 10.1038/ncb3234
Johnson, B., Leek, A. N., Sole, L., Maverick, E. E., Levine, T. P., and Tamkun, M. M. (2018). Kv2 potassium channels form endoplasmic reticulum/plasma membrane junctions via interaction with VAPA and VAPB. Proc. Natl. Acad. Sci. U.S.A. 115, E7331–E7340. doi: 10.1073/pnas.1805757115
Johnson, B., Leek, A. N., and Tamkun, M. M. (2019). Kv2 channels create endoplasmic reticulum/plasma membrane junctions: a brief history of Kv2 channel subcellular localization. Channels (Austin) 13, 88–101. doi: 10.1080/19336950.2019.1568824
Juschke, C., Wachter, A., Schwappach, B., and Seedorf, M. (2005). SEC18/NSF-independent, protein-sorting pathway from the yeast cortical ER to the plasma membrane. J. Cell Biol. 169, 613–622. doi: 10.1083/jcb.200503033
Kagiwada, S., and Hashimoto, M. (2007). The yeast VAP homolog Scs2p has a phosphoinositide-binding ability that is correlated with its activity. Biochem. Biophys. Res. Commun. 364, 870–876. doi: 10.1016/j.bbrc.2007.10.079
Kamemura, K., and Chihara, T. (2019). Multiple functions of the ER-resident VAP and its extracellular role in neural development and disease. J. Biochem. 165, 391–400. doi: 10.1093/jb/mvz011
Kang, F., Zhou, M., Huang, X., Fan, J., Wei, L., Boulanger, J., et al. (2019). E-syt1 re-arranges STIM1 clusters to stabilize ring-shaped ER-PM contact sites and accelerate Ca(2+) store replenishment. Sci. Rep. 9:3975. doi: 10.1038/s41598-019-40331-0
Kar, P., Bakowski, D., Di Capite, J., Nelson, C., and Parekh, A. B. (2012). Different agonists recruit different stromal interaction molecule proteins to support cytoplasmic Ca2+ oscillations and gene expression. Proc. Natl. Acad. Sci. U.S.A. 109, 6969–6974. doi: 10.1073/pnas.1201204109
Kentala, H., Weber-Boyvat, M., and Olkkonen, V. M. (2016). OSBP-related protein family: mediators of lipid transport and signaling at membrane contact sites. Int. Rev. Cell Mol. Biol. 321, 299–340. doi: 10.1016/bs.ircmb.2015.09.006
Kikuma, K., Li, X., Kim, D., Sutter, D., and Dickman, D. K. (2017). Extended synaptotagmin localizes to presynaptic ER and promotes neurotransmission and synaptic growth in Drosophila. Genetics 207, 993–1006. doi: 10.1534/genetics.117.300261
Kim, S., Kedan, A., Marom, M., Gavert, N., Keinan, O., Selitrennik, M., et al. (2013). The phosphatidylinositol-transfer protein Nir2 binds phosphatidic acid and positively regulates phosphoinositide signalling. EMBO Rep. 14, 891–899. doi: 10.1038/embor.2013.113
Kim, Y. J., Guzman-Hernandez, M. L., Wisniewski, E., and Balla, T. (2015). Phosphatidylinositol-phosphatidic acid exchange by Nir2 at ER-PM contact sites maintains phosphoinositide signaling competence. Dev. Cell 33, 549–561. doi: 10.1016/j.devcel.2015.04.028
Kirmiz, M., Gillies, T. E., Dickson, E. J., and Trimmer, J. S. (2019). Neuronal ER-plasma membrane junctions organized by Kv2-VAP pairing recruit Nir proteins and affect phosphoinositide homeostasis. J. Biol. Chem. 294, 17735–17757. doi: 10.1074/jbc.RA119.007635
Kirmiz, M., Palacio, S., Thapa, P., King, A. N., Sack, J. T., and Trimmer, J. S. (2018a). Remodeling neuronal ER-PM junctions is a conserved nonconducting function of Kv2 plasma membrane ion channels. Mol. Biol. Cell 29, 2410–2432. doi: 10.1091/mbc.E18-05-0337
Kirmiz, M., Vierra, N. C., Palacio, S., and Trimmer, J. S. (2018b). Identification of VAPA and VAPB as Kv2 channel-interacting proteins defining endoplasmic reticulum-plasma membrane junctions in mammalian brain neurons. J. Neurosci. 38, 7562–7584. doi: 10.1523/JNEUROSCI.0893-18.2018
Kopec, K. O., Alva, V., and Lupas, A. N. (2010). Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics 26, 1927–1931. doi: 10.1093/bioinformatics/btq326
Kun-Rodrigues, C., Ganos, C., Guerreiro, R., Schneider, S. A., Schulte, C., Lesage, S., et al. (2015). A systematic screening to identify de novo mutations causing sporadic early-onset Parkinson's disease. Hum. Mol. Genet. 24, 6711–6720. doi: 10.1093/hmg/ddv376
Kunzelmann, K., Cabrita, I., Wanitchakool, P., Ousingsawat, J., Sirianant, L., Benedetto, R., et al. (2016). Modulating Ca(2)(+) signals: a common theme for TMEM16, Ist2, and TMC. Pflugers Arch. 468, 475–490. doi: 10.1007/s00424-015-1767-4
Lee, E., Vanneste, S., Perez-Sancho, J., Benitez-Fuente, F., Strelau, M., Macho, A. P., et al. (2019). Ionic stress enhances ER-PM connectivity via phosphoinositide-associated SYT1 contact site expansion in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 116, 1420–1429. doi: 10.1073/pnas.1818099116
Lee, I., and Hong, W. (2006). Diverse membrane-associated proteins contain a novel SMP domain. FASEB J. 20, 202–206. doi: 10.1096/fj.05-4581hyp
Lees, J. A., Messa, M., Sun, E. W., Wheeler, H., Torta, F., Wenk, M. R., et al. (2017). Lipid transport by TMEM24 at ER-plasma membrane contacts regulates pulsatile insulin secretion. Science 355:aah6171. doi: 10.1126/science.aah6171
Lefkimmiatis, K., Srikanthan, M., Maiellaro, I., Moyer, M. P., Curci, S., and Hofer, A. M. (2009). Store-operated cyclic AMP signalling mediated by STIM1. Nat. Cell Biol. 11, 433–442. doi: 10.1038/ncb1850
Lehto, M., Mäyränpää, M. I., Pellinen, T., Ihalmo, P., Lehtonen, S., Kovanen, P. T., et al. (2008). The R-Ras interaction partner ORP3 regulates cell adhesion. J. Cell Sci. 121, 695–705. doi: 10.1242/jcs.016964
Lessard, L., Stuible, M., and Tremblay, M. L. (2010). The two faces of PTP1B in cancer. Biochim. Biophys. Acta 1804, 613–619. doi: 10.1016/j.bbapap.2009.09.018
Lev, S., Ben Halevy, D., Peretti, D., and Dahan, N. (2008). The VAP protein family: from cellular functions to motor neuron disease. Trends Cell Biol. 18, 282–290. doi: 10.1016/j.tcb.2008.03.006
Lim, S. T., Antonucci, D. E., Scannevin, R. H., and Trimmer, J. S. (2000). A novel targeting signal for proximal clustering of the Kv2.1 K+ channel in hippocampal neurons. Neuron 25, 385–397. doi: 10.1016/S0896-6273(00)80902-2
Lindhout, F. W., Cao, Y., Kevenaar, J. T., Bodzeta, A., Stucchi, R., Boumpoutsari, M. M., et al. (2019). VAP-SCRN1 interaction regulates dynamic endoplasmic reticulum remodeling and presynaptic function. EMBO J. 38:e101345. doi: 10.15252/embj.2018101345
Liou, J., Fivaz, M., Inoue, T., and Meyer, T. (2007). Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc. Natl. Acad. Sci. U.S.A. 104, 9301–9306. doi: 10.1073/pnas.0702866104
Liou, J., Kim, M. L., Heo, W. D., Jones, J. T., Myers, J. W., Ferrell, J. E. Jr., et al. (2005). STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 15, 1235–1241. doi: 10.1016/j.cub.2005.05.055
Ma, G., Wei, M., He, L., Liu, C., Wu, B., Zhang, S. L., et al. (2015). Inside-out Ca(2+) signalling prompted by STIM1 conformational switch. Nat. Commun. 6:7826. doi: 10.1038/ncomms8826
Maass, K., Fischer, M. A., Seiler, M., Temmerman, K., Nickel, W., and Seedorf, M. (2009). A signal comprising a basic cluster and an amphipathic alpha-helix interacts with lipids and is required for the transport of Ist2 to the yeast cortical ER. J. Cell Sci. 122, 625–635. doi: 10.1242/jcs.036012
Maeda, K., Anand, K., Chiapparino, A., Kumar, A., Poletto, M., Kaksonen, M., et al. (2013). Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature 501, 257–261. doi: 10.1038/nature12430
Maiellaro, I., Lefkimmiatis, K., Moyer, M. P., Curci, S., and Hofer, A. M. (2012). Termination and activation of store-operated cyclic AMP production. J. Cell Mol. Med. 16, 2715–2725. doi: 10.1111/j.1582-4934.2012.01592.x
Manford, A., Xia, T., Saxena, A. K., Stefan, C., Hu, F., Emr, S. D., et al. (2010). Crystal structure of the yeast Sac1: implications for its phosphoinositide phosphatase function. EMBO J. 29, 1489–1498. doi: 10.1038/emboj.2010.57
Manford, A. G., Stefan, C. J., Yuan, H. L., Macgurn, J. A., and Emr, S. D. (2012). ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell 23, 1129–1140. doi: 10.1016/j.devcel.2012.11.004
Manji, S. S., Parker, N. J., Williams, R. T., van Stekelenburg, L., Pearson, R. B., Dziadek, M., et al. (2000). STIM1: a novel phosphoprotein located at the cell surface. Biochim. Biophys. Acta 1481, 147–155. doi: 10.1016/S0167-4838(00)00105-9
Mesmin, B., Bigay, J., Moser von Filseck, J., Lacas-Gervais, S., Drin, G., and Antonny, B. (2013). A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell 155, 830–843. doi: 10.1016/j.cell.2013.09.056
Min, S. W., Chang, W. P., and Südhof, T. C. (2007). E-Syts, a family of membranous Ca2+-sensor proteins with multiple C2 domains. Proc. Natl. Acad. Sci. U.S.A. 104, 3823–3828. doi: 10.1073/pnas.0611725104
Monteleone, M. C., Gonzalez Wusener, A. E., Burdisso, J. E., Conde, C., Caceres, A., and Arregui, C. O. (2012). ER-bound protein tyrosine phosphatase PTP1B interacts with Src at the plasma membrane/substrate interface. PLoS ONE 7:e38948. doi: 10.1371/journal.pone.0038948
Moser von Filseck, J., Copič, A., Delfosse, V., Vanni, S., Jackson, C. L., Bourguet, W., et al. (2015a). Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science 349:aab1346. doi: 10.1126/science.aab1346
Moser von Filseck, J., Vanni, S., Mesmin, B., Antonny, B., and Drin, G. (2015b). A phosphatidylinositol-4-phosphate powered exchange mechanism to create a lipid gradient between membranes. Nat. Commun. 6:6671. doi: 10.1038/ncomms7671
Murakoshi, H., and Trimmer, J. S. (1999). Identification of the Kv2.1 K+ channel as a major component of the delayed rectifier K+ current in rat hippocampal neurons. J. Neurosci. 19, 1728–1735. doi: 10.1523/JNEUROSCI.19-05-01728.1999
Murphy, S. E., and Levine, T. P. (2016). VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta 1861, 952–961. doi: 10.1016/j.bbalip.2016.02.009
Naito, T., Ercan, B., Krshnan, L., Triebl, A., Koh, D. H. Z., Wei, F. Y., et al. (2019). Movement of accessible plasma membrane cholesterol by the GRAMD1 lipid transfer protein complex. Elife 8:e51401. doi: 10.7554/eLife.51401.sa2
Nath, V. R., Mishra, S., Basak, B., Trivedi, D., and Raghu, P. (2020). Extended synaptotagmin regulates membrane contact site structure and lipid transfer function in vivo. EMBO Rep. 21:e50264. doi: 10.15252/embr.202050264
Ng, A. Q. E., Ng, A. Y. E., and Zhang, D. (2020). Plasma membrane furrows control plasticity of ER-PM contacts. Cell Rep. 30, 1434–1446.e1437. doi: 10.1016/j.celrep.2019.12.098
Ng, A. Y. E., Ng, A. Q. E., and Zhang, D. (2018). ER-PM contacts restrict exocytic sites for polarized morphogenesis. Curr. Biol. 28, 146–153.e145. doi: 10.1016/j.cub.2017.11.055
O'Connell, K. M., Loftus, R., and Tamkun, M. M. (2010). Localization-dependent activity of the Kv2.1 delayed-rectifier K+ channel. Proc. Natl. Acad. Sci. U.S.A. 107, 12351–12356. doi: 10.1073/pnas.1003028107
Oh-Hora, M., Yamashita, M., Hogan, P. G., Sharma, S., Lamperti, E., Chung, W., et al. (2008). Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat. Immunol. 9, 432–443. doi: 10.1038/ni1574
Okeke, E., Dingsdale, H., Parker, T., Voronina, S., and Tepikin, A. V. (2016). Endoplasmic reticulum-plasma membrane junctions: structure, function and dynamics. J. Physiol. 594, 2837–2847. doi: 10.1113/JP271142
Olkkonen, V. M., and Li, S. (2013). Oxysterol-binding proteins: sterol and phosphoinositide sensors coordinating transport, signaling and metabolism. Prog. Lipid Res. 52, 529–538. doi: 10.1016/j.plipres.2013.06.004
Omnus, D. J., Cadou, A., Thomas, F. B., Bader, J. M., Soh, N., Chung, G. H. C., et al. (2020). A heat-sensitive Osh protein controls PI4P polarity. BMC Biol. 18:28. doi: 10.1186/s12915-020-0758-x
Ong, H. L., and Ambudkar, I. S. (2020). The endoplasmic reticulum-plasma membrane junction: a hub for agonist regulation of Ca(2+) entry. Cold Spring Harb. Perspect. Biol. 12:a035253. doi: 10.1101/cshperspect.a035253
Park, C. Y., Hoover, P. J., Mullins, F. M., Bachhawat, P., Covington, E. D., Raunser, S., et al. (2009). STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell 136, 876–890. doi: 10.1016/j.cell.2009.02.014
Patterson, R. L., van Rossum, D. B., and Gill, D. L. (1999). Store-operated Ca2+ entry: evidence for a secretion-like coupling model. Cell 98, 487–499. doi: 10.1016/S0092-8674(00)81977-7
Perez-Sancho, J., Vanneste, S., Lee, E., McFarlane, H. E., Esteban Del Valle, A., Valpuesta, V., et al. (2015). The Arabidopsis synaptotagmin1 is enriched in endoplasmic reticulum-plasma membrane contact sites and confers cellular resistance to mechanical stresses. Plant Physiol. 168, 132–143. doi: 10.1104/pp.15.00260
Petkovic, M., Jemaiel, A., Daste, F., Specht, C. G., Izeddin, I., Vorkel, D., et al. (2014). The SNARE Sec22b has a non-fusogenic function in plasma membrane expansion. Nat. Cell Biol. 16, 434–444. doi: 10.1038/ncb2937
Pfisterer, S. G., Peränen, J., and Ikonen, E. (2016). LDL-cholesterol transport to the endoplasmic reticulum: current concepts. Curr. Opin. Lipidol. 27, 282–287. doi: 10.1097/MOL.0000000000000292
Porter, K. R., and Palade, G. E. (1957). Studies on the endoplasmic reticulum. III. Its form and distribution in striated muscle cells. J. Biophys. Biochem. Cytol. 3, 269–300. doi: 10.1083/jcb.3.2.269
Pottekat, A., Becker, S., Spencer, K. R., Yates, J. R. III., Manning, G., Itkin-Ansari, P., et al. (2013). Insulin biosynthetic interaction network component, TMEM24, facilitates insulin reserve pool release. Cell Rep. 4, 921–930. doi: 10.1016/j.celrep.2013.07.050
Prakriya, M., Feske, S., Gwack, Y., Srikanth, S., Rao, A., and Hogan, P. G. (2006). Orai1 is an essential pore subunit of the CRAC channel. Nature 443, 230–233. doi: 10.1038/nature05122
Prakriya, M., and Lewis, R. S. (2015). Store-operated calcium channels. Physiol. Rev. 95, 1383–1436. doi: 10.1152/physrev.00020.2014
Prinz, W. A. (2010). Lipid trafficking sans vesicles: where, why, how? Cell. 143, 870–874. doi: 10.1016/j.cell.2010.11.031
Pulli, I., Lassila, T., Pan, G., Yan, D., Olkkonen, V. M., and Tornquist, K. (2018). Oxysterol-binding protein related-proteins (ORPs) 5 and 8 regulate calcium signaling at specific cell compartments. Cell Calcium. 72, 62–69. doi: 10.1016/j.ceca.2018.03.001
Putney, J. W. Jr. (1986). A model for receptor-regulated calcium entry. Cell Calcium. 7, 1–12. doi: 10.1016/0143-4160(86)90026-6
Putney, J. W. Jr. (1990). Capacitative calcium entry revisited. Cell Calcium. 11, 611–624. doi: 10.1016/0143-4160(90)90016-N
Quintana, A., Rajanikanth, V., Farber-Katz, S., Gudlur, A., Zhang, C., Jing, J., et al. (2015). TMEM110 regulates the maintenance and remodeling of mammalian ER-plasma membrane junctions competent for STIM-ORAI signaling. Proc. Natl. Acad. Sci. U.S.A. 112, E7083–7092. doi: 10.1073/pnas.1521924112
Redman, P. T., He, K., Hartnett, K. A., Jefferson, B. S., Hu, L., Rosenberg, P. A., et al. (2007). Apoptotic surge of potassium currents is mediated by p38 phosphorylation of Kv2.1. Proc. Natl. Acad. Sci. U.S.A. 104, 3568–3573. doi: 10.1073/pnas.0610159104
Rizo, J., and Südhof, T. C. (1998). C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 273, 15879–15882. doi: 10.1074/jbc.273.26.15879
Roos, J., DiGregorio, P. J., Yeromin, A. V., Ohlsen, K., Lioudyno, M., Zhang, S., et al. (2005). STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 169, 435–445. doi: 10.1083/jcb.200502019
Saheki, Y. (2017). Endoplasmic reticulum—plasma membrane crosstalk mediated by the extended synaptotagmins. Adv. Exp. Med. Biol. 997, 83–93. doi: 10.1007/978-981-10-4567-7_6
Saheki, Y., Bian, X., Schauder, C. M., Sawaki, Y., Surma, M. A., Klose, C., et al. (2016). Control of plasma membrane lipid homeostasis by the extended synaptotagmins. Nat. Cell Biol. 18, 504–515. doi: 10.1038/ncb3339
Saheki, Y., and De Camilli, P. (2017a). Endoplasmic reticulum-plasma membrane contact sites. Annu. Rev. Biochem. 86, 659–684. doi: 10.1146/annurev-biochem-061516-044932
Saheki, Y., and De Camilli, P. (2017b). The extended-synaptotagmins. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1490–1493. doi: 10.1016/j.bbamcr.2017.03.013
Sandhu, J., Li, S., Fairall, L., Pfisterer, S. G., Gurnett, J. E., Xiao, X., et al. (2018). Aster proteins facilitate nonvesicular plasma membrane to ER cholesterol transport in mammalian cells. Cell 175, 514–529.e520. doi: 10.1016/j.cell.2018.08.033
Schauder, C. M., Wu, X., Saheki, Y., Narayanaswamy, P., Torta, F., Wenk, M. R., et al. (2014). Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature 510, 552–555. doi: 10.1038/nature13269
Schulz, T. A., and Creutz, C. E. (2004). The tricalbin C2 domains: lipid-binding properties of a novel, synaptotagmin-like yeast protein family. Biochemistry 43, 3987–3995. doi: 10.1021/bi036082w
Sclip, A., Bacaj, T., Giam, L. R., and Sudhof, T. C. (2016). Extended synaptotagmin (ESyt) triple knock-out mice are viable and fertile without obvious endoplasmic reticulum dysfunction. PLoS ONE 11:e0158295. doi: 10.1371/journal.pone.0158295
Shah, N. H., and Aizenman, E. (2014). Voltage-gated potassium channels at the crossroads of neuronal function, ischemic tolerance, and neurodegeneration. Transl. Stroke Res. 5, 38–58. doi: 10.1007/s12975-013-0297-7
Skirpan, A. L., Dowd, P. E., Sijacic, P., Jaworski, C. J., Gilroy, S., and Kao, T. H. (2006). Identification and characterization of PiORP1, a Petunia oxysterol-binding-protein related protein involved in receptor-kinase mediated signaling in pollen, and analysis of the ORP gene family in Arabidopsis. Plant Mol. Biol. 61, 553–565. doi: 10.1007/s11103-006-0030-y
Smith, D. C., Sillence, D. J., Falguières, T., Jarvis, R. M., Johannes, L., Lord, J. M., et al. (2006). The association of shiga-like toxin with detergent-resistant membranes is modulated by glucosylceramide and is an essential requirement in the endoplasmic reticulum for a cytotoxic effect. Mol. Biol. Cell. 17, 1375–1387. doi: 10.1091/mbc.e05-11-1035
Soboloff, J., Rothberg, B. S., Madesh, M., and Gill, D. L. (2012). STIM proteins: dynamic calcium signal transducers. Nat. Rev. Mol. Cell Biol. 13, 549–565. doi: 10.1038/nrm3414
Soboloff, J., Spassova, M. A., Dziadek, M. A., and Gill, D. L. (2006a). Calcium signals mediated by STIM and Orai proteins—a new paradigm in inter-organelle communication. Biochim. Biophys. Acta 1763, 1161–1168. doi: 10.1016/j.bbamcr.2006.09.023
Soboloff, J., Spassova, M. A., Hewavitharana, T., He, L. P., Xu, W., Johnstone, L. S., et al. (2006b). STIM2 is an inhibitor of STIM1-mediated store-operated Ca2+ entry. Curr. Biol. 16, 1465–1470. doi: 10.1016/j.cub.2006.05.051
Stathopulos, P. B., Li, G. Y., Plevin, M. J., Ames, J. B., and Ikura, M. (2006). Stored Ca2+ depletion-induced oligomerization of stromal interaction molecule 1 (STIM1) via the EF-SAM region: an initiation mechanism for capacitive Ca2+ entry. J. Biol. Chem. 281, 35855–35862. doi: 10.1074/jbc.M608247200
Stefan, C. J. (2020). Endoplasmic reticulum-plasma membrane contacts: principals of phosphoinositide and calcium signaling. Curr. Opin. Cell Biol. 63, 125–134. doi: 10.1016/j.ceb.2020.01.010
Stefan, C. J., Manford, A. G., Baird, D., Yamada-Hanff, J., Mao, Y., and Emr, S. D. (2011). Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 144, 389–401. doi: 10.1016/j.cell.2010.12.034
Stefano, G., Renna, L., Wormsbaecher, C., Gamble, J., Zienkiewicz, K., and Brandizzi, F. (2018). Plant endocytosis requires the ER membrane-anchored proteins VAP27-1 and VAP27-3. Cell Rep. 23, 2299–2307. doi: 10.1016/j.celrep.2018.04.091
Südhof, T. C., and Rothman, J. E. (2009). Membrane fusion: grappling with SNARE and SM proteins. Science 323, 474–477. doi: 10.1126/science.1161748
Sun, E. W., Guillen-Samander, A., Bian, X., Wu, Y., Cai, Y., Messa, M., et al. (2019). Lipid transporter TMEM24/C2CD2L is a Ca(2+)-regulated component of ER-plasma membrane contacts in mammalian neurons. Proc. Natl. Acad. Sci. U.S.A. 116, 5775–5784. doi: 10.1073/pnas.1820156116
Sun, W., Tian, B. X., Wang, S. H., Liu, P. J., and Wang, Y. C. (2020). The function of SEC22B and its role in human diseases. Cytoskeleton (Hoboken) 77, 303–312. doi: 10.1002/cm.21628
Sutton, R. B., Fasshauer, D., Jahn, R., and Brunger, A. T. (1998). Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353. doi: 10.1038/26412
Tong, J., Yang, H., Yang, H., Eom, S. H., and Im, Y. J. (2013). Structure of Osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins. Structure 21, 1203–1213. doi: 10.1016/j.str.2013.05.007
Toulmay, A., and Prinz, W. A. (2012). A conserved membrane-binding domain targets proteins to organelle contact sites. J. Cell Sci. 125, 49–58. doi: 10.1242/jcs.085118
Tremblay, M. G., Herdman, C., Guillou, F., Mishra, P. K., Baril, J., Bellenfant, S., et al. (2016). Extended synaptotagmin interaction with the fibroblast growth factor receptor depends on receptor conformation, not catalytic activity. J. Biol. Chem. 291:3173. doi: 10.1074/jbc.A115.656918
Tremblay, M. G., and Moss, T. (2016). Loss of all 3 extended synaptotagmins does not affect normal mouse development, viability or fertility. Cell Cycle 15, 2360–2366. doi: 10.1080/15384101.2016.1203494
van Meer, G., Voelker, D. R., and Feigenson, G. W. (2008). Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124. doi: 10.1038/nrm2330
Vierra, N. C., Kirmiz, M., van der List, D., Santana, L. F., and Trimmer, J. S. (2019). Kv2.1 mediates spatial and functional coupling of L-type calcium channels and ryanodine receptors in mammalian neurons. Elife 8:49953. doi: 10.7554/eLife.49953
Vig, M., Peinelt, C., Beck, A., Koomoa, D. L., Rabah, D., Koblan-Huberson, M., et al. (2006). CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 312, 1220–1223. doi: 10.1126/science.1127883
Wang, P., Hawes, C., and Hussey, P. J. (2017). Plant endoplasmic reticulum-plasma membrane contact sites. Trends Plant Sci. 22, 289–297. doi: 10.1016/j.tplants.2016.11.008
Wang, P., Hawkins, T. J., Richardson, C., Cummins, I., Deeks, M. J., Sparkes, I., et al. (2014). The plant cytoskeleton, NET3C, and VAP27 mediate the link between the plasma membrane and endoplasmic reticulum. Curr. Biol. 24, 1397–1405. doi: 10.1016/j.cub.2014.05.003
Wang, P., Pleskot, R., Zang, J., Winkler, J., Wang, J., Yperman, K., et al. (2019). Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat. Commun. 10:5132. doi: 10.1038/s41467-019-12782-6
Wang, Y., Deng, X., Zhou, Y., Hendron, E., Mancarella, S., Ritchie, M. F., et al. (2009). STIM protein coupling in the activation of Orai channels. Proc. Natl. Acad. Sci. U.S.A. 106, 7391–7396. doi: 10.1073/pnas.0900293106
Weber, T., Zemelman, B. V., McNew, J. A., Westermann, B., Gmachl, M., Parlati, F., et al. (1998). SNAREpins: minimal machinery for membrane fusion. Cell 92, 759–772. doi: 10.1016/S0092-8674(00)81404-X
Weber-Boyvat, M., Trimbuch, T., Shah, S., Jäntti, J., Olkkonen, V. M., and Rosenmund, C. (2020). ORP/Osh mediate cross-talk between ER-plasma membrane contact site components and plasma membrane SNAREs. Cell Mol. Life Sci. doi: 10.1007/s00018-020-03604-w. [Epub ahead of print].
Wilhelm, L. P., Wendling, C., Védie, B., Kobayashi, T., Chenard, M. P., Tomasetto, C., et al. (2017). STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites. EMBO J. 36, 1412–1433. doi: 10.15252/embj.201695917
Williams, R. T., Manji, S. S., Parker, N. J., Hancock, M. S., Van Stekelenburg, L., Eid, J. P., et al. (2001). Identification and characterization of the STIM (stromal interaction molecule) gene family: coding for a novel class of transmembrane proteins. Biochem. J. 357, 673–685. doi: 10.1042/bj3570673
Willoughby, D., Everett, K. L., Halls, M. L., Pacheco, J., Skroblin, P., Vaca, L., et al. (2012). Direct binding between Orai1 and AC8 mediates dynamic interplay between Ca2+ and cAMP signaling. Sci. Signal. 5:ra29. doi: 10.1126/scisignal.2002299
Willoughby, D., Wachten, S., Masada, N., and Cooper, D. M. (2010). Direct demonstration of discrete Ca2+ microdomains associated with different isoforms of adenylyl cyclase. J. Cell Sci. 123, 107–117. doi: 10.1242/jcs.062067
Wolf, W., Kilic, A., Schrul, B., Lorenz, H., Schwappach, B., and Seedorf, M. (2012). Yeast Ist2 recruits the endoplasmic reticulum to the plasma membrane and creates a ribosome-free membrane microcompartment. PLoS ONE 7:e39703. doi: 10.1371/journal.pone.0039703
Wong, L. H., Gatta, A. T., and Levine, T. P. (2019). Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes. Nat. Rev. Mol. Cell Biol. 20, 85–101. doi: 10.1038/s41580-018-0071-5
Wu, L., Shen, C., Seed Ahmed, M., Östenson, C. G., and Gu, H. F. (2016). Adenylate cyclase 3: a new target for anti-obesity drug development. Obes. Rev. 17, 907–914. doi: 10.1111/obr.12430
Xiao, J., Luo, J., Hu, A., Xiao, T., Li, M., Kong, Z., et al. (2019). Cholesterol transport through the peroxisome-ER membrane contacts tethered by PI(4,5)P(2) and extended synaptotagmins. Sci. China Life Sci. 62, 1117–1135. doi: 10.1007/s11427-019-9569-9
Xiao, Q., Yu, K., Perez-Cornejo, P., Cui, Y., Arreola, J., and Hartzell, H. C. (2011). Voltage- and calcium-dependent gating of TMEM16A/Ano1 chloride channels are physically coupled by the first intracellular loop. Proc. Natl. Acad. Sci. U.S.A. 108, 8891–8896. doi: 10.1073/pnas.1102147108
Xie, B., Nguyen, P. M., and Idevall-Hagren, O. (2019). Feedback regulation of insulin secretion by extended synaptotagmin-1. FASEB J. 33, 4716–4728. doi: 10.1096/fj.201801878R
Xu, J., Bacaj, T., Zhou, A., Tomchick, D. R., Sudhof, T. C., and Rizo, J. (2014). Structure and Ca(2)(+)-binding properties of the tandem C(2) domains of E-Syt2. Structure 22, 269–280. doi: 10.1016/j.str.2013.11.011
Yamazaki, T., Takata, N., Uemura, M., and Kawamura, Y. (2010). Arabidopsis synaptotagmin SYT1, a type I signal-anchor protein, requires tandem C2 domains for delivery to the plasma membrane. J. Biol. Chem. 285, 23165–23176. doi: 10.1074/jbc.M109.084046
Yang, X., Jin, H., Cai, X., Li, S., and Shen, Y. (2012). Structural and mechanistic insights into the activation of Stromal interaction molecule 1 (STIM1). Proc. Natl. Acad. Sci. U.S.A. 109, 5657–5662. doi: 10.1073/pnas.1118947109
Yao, Y., Ferrer-Montiel, A. V., Montal, M., and Tsien, R. Y. (1999). Activation of store-operated Ca2+ current in Xenopus oocytes requires SNAP-25 but not a diffusible messenger. Cell 98, 475–485. doi: 10.1016/S0092-8674(00)81976-5
Yellen, G. (1998). The moving parts of voltage-gated ion channels. Q. Rev. Biophys. 31, 239–295. doi: 10.1017/S0033583598003448
Yu, H., Liu, Y., Gulbranson, D. R., Paine, A., Rathore, S. S., and Shen, J. (2016). Extended synaptotagmins are Ca2+-dependent lipid transfer proteins at membrane contact sites. Proc. Natl. Acad. Sci. 113, 4362–4367. doi: 10.1073/pnas.1517259113
Yuan, J. P., Zeng, W., Dorwart, M. R., Choi, Y. J., Worley, P. F., and Muallem, S. (2009). SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat. Cell Biol. 11, 337–343. doi: 10.1038/ncb1842
Zachariou, V., Liu, R., LaPlant, Q., Xiao, G., Renthal, W., Chan, G. C., et al. (2008). Distinct roles of adenylyl cyclases 1 and 8 in opiate dependence: behavioral, electrophysiological, and molecular studies. Biol. Psychiatry. 63, 1013–1021. doi: 10.1016/j.biopsych.2007.11.021
Zhang, D., Vjestica, A., and Oliferenko, S. (2012). Plasma membrane tethering of the cortical ER necessitates its finely reticulated architecture. Curr. Biol. 22, 2048–2052. doi: 10.1016/j.cub.2012.08.047
Zhang, S. L., Yeromin, A. V., Zhang, X. H., Yu, Y., Safrina, O., Penna, A., et al. (2006). Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity. Proc. Natl. Acad. Sci. U.S.A. 103, 9357–9362. doi: 10.1073/pnas.0603161103
Zhang, S. L., Yu, Y., Roos, J., Kozak, J. A., Deerinck, T. J., Ellisman, M. H., et al. (2005). STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature 437, 902–905. doi: 10.1038/nature04147
Zhang, Y., Guan, Y., Pan, S., Yan, L., Wang, P., Chen, Z., et al. (2020). Hypothalamic extended synaptotagmin-3 contributes to the development of dietary obesity and related metabolic disorders. Proc. Natl. Acad. Sci. U.S.A. 117, 20149–20158. doi: 10.1073/pnas.2004392117
Zheng, L., Stathopulos, P. B., Schindl, R., Li, G. Y., Romanin, C., and Ikura, M. (2011). Auto-inhibitory role of the EF-SAM domain of STIM proteins in store-operated calcium entry. Proc. Natl. Acad. Sci. U.S.A. 108, 1337–1342. doi: 10.1073/pnas.1015125108
Keywords: membrane contact sites (MCSs), endoplasmic reticulum (ER), plasma membrane, tether, lipid transfer, enzyme
Citation: Li C, Qian T, He R, Wan C, Liu Y and Yu H (2021) Endoplasmic Reticulum–Plasma Membrane Contact Sites: Regulators, Mechanisms, and Physiological Functions. Front. Cell Dev. Biol. 9:627700. doi: 10.3389/fcell.2021.627700
Received: 10 November 2020; Accepted: 13 January 2021;
Published: 04 February 2021.
Edited by:
Dan Zhang, Temasek Life Sciences Laboratory, SingaporeReviewed by:
Tito Cali', University of Padua, ItalyCopyright © 2021 Li, Qian, He, Wan, Liu and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinghui Liu, eWluZ2h1aWxpdUBuam51LmVkdS5jbg==; Haijia Yu, eXVoYWlqaWFAbmpudS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.