
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Cell Dev. Biol. , 25 February 2021
Sec. Cell Growth and Division
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.612539
This article is part of the Research Topic Cytoskeletal Regulation of Immune Response View all 18 articles
 Marco Fritzsche1,2*
Marco Fritzsche1,2*Mechanobiology is a critical frontier in the biomedical sciences. Across many of its fields a new perspective is emerging to perceive the immune response as a single multi-scale super-organism continuously interacting and interpreting the biochemical and biomechanical micro-environment. Large numbers of immune cells communicate through a combination of chemical and mechanical signals to organise and orchestrate their behaviour and function against an immunological threat. However, disease often circumvents and even exploits mechanobiological features of this defence machinery, highlighting the need to better understand the intimate coupling between biology and mechanics.
Mechanical force underpins the immune response at the multi-scale (Fritzsche, 2020). While almost all physical forces relevant to immune cell biology are practically restricted to the sub-cellular level such as electrostatics (e.g., receptor ligand binding) and thermodynamics (e.g., molecule diffusion), biomechanics takes on a special importance as mechanical force influences many functional features and behavior of immune cells over multiple scales in space and time (Dumont and Prakash, 2014; Egan et al., 2015). Mechanics contributes to the dynamics of single molecules, cells, tissues, and entire organisms (Blanchard and Adams, 2011; Chen and Zhu, 2013). The effects on the biology result from contributions of mechanics that are combination of those generated locally and those that influence from a distance. Importantly, both mechanical force and mechanical properties differ at distinct spatio-temporal frequencies such as constant and oscillatory forces, tension, or elasticity and viscosity (see Figure 1), respectively. For example, living cells can be elastic, viscous, or visco-elastic depending on the spatial and temporal measurement frequency (see Figure 1A). Adequate quantification promises thus a deeper understanding of the multi-scale spatio-temporal coupling of biology and mechanics and its control over the immune response (Fritzsche, 2020).
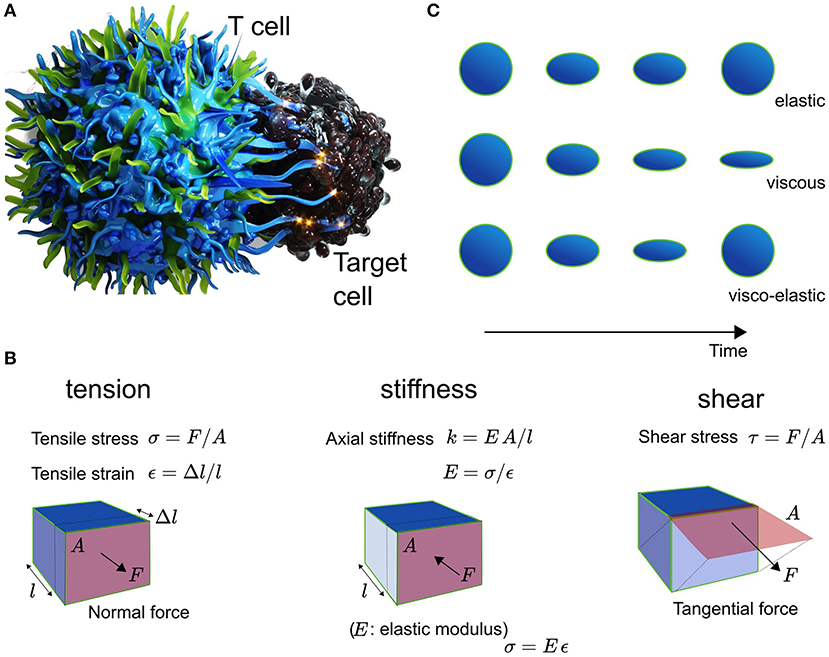
Figure 1. (A) Schematic of the T cell (left) and Target cell (right) interplay. (B) Explanation of the mechanical metric tension with tensile stress σ in units of N/m2 and tensile strain ε being dimenionless, and shear with shear stress τ in N/m2, as well as the elasticity with Youngst modulus E, which equals the ratio of stress over strain. The stiffness of a material is given by EA/l. (C) Cells are elastic, viscous, or visco-elastic.
Central to the immune defense against an invading threat is a well-orchestrated sequence of events carried out by specialized cells of the multi-scale super-organism of the immune system (Chaplin, 2010). The Success of the process relies on the quality of the spatiotemporal organization of the cellular responses in the tissue micro-environment of the host organism for instance against cancer or an invading pathogenic threat (Swartz and Lund, 2012). Cytotoxic immune cells circulate through tissue, track soluble cytokines and chemokines, respond to antigens, and kill diverse immuno-targets (Andersen et al., 2006; Grivennikov et al., 2010). Among many of these events, they involve a combination of biological and mechanical spheres of influence, as immune cells continuously interact and interpret the physical surroundings (Colin-York et al., 2016; Schwarz, 2017). This environment is also mechanically diverse over scales of space and time. It is comprised of different types of molecules, cells, and tissues (Needleman and Dogic, 2017). Mechanical aspects of the tissue environment are known to influence both the function and behavior of cells (Schwarz, 2017; Colin-York and Fritzsche, 2018). The complex multi-scale nature of mechanical force may have evolutionary been the reason for immune cells to develop the ability to adjust their own biomechanics to their physiological needs in response to the ever-changing physical world. Strikingly, when cells loose mechanosensation, the immune response is hampered to robustly and/or reliably achieve its protective function, which has been demonstrated for example during cell-cell interactions such as activation and cytotoxicity (Huse, 2017; Kumari et al., 2019), as well as during cellular interactions with the tumor micro-environment (Mohammadi and Sahai, 2018; Majedi et al., 2020). Consequently, this gives a special significance to mechanobiology of being important for the understanding of the functioning of the immune response as a whole in health and disease.
One of the most illustrative and visually impressive examples for the impact of mechanobiology is the activation of T cells and antigen presenting cells (APCs) (Chen and Zhu, 2013; Harrison et al., 2019). Micron-scale ruffles protruding from the T cell initiate contact and binding between T cell receptors (TCRs) and the APC's peptide-loaded major hist-compatibility complexes (pMHCs) (Fritz-Laylin et al., 2017; Fritzsche et al., 2017). In the event of recognition and binding of pMHCs by a TCR, the T cell rearranges, assisted by its cytoskeleton, the totality of its metabolism, inner organelles, membrane, and its surface receptors and ligands (English and Voeltz, 2013; Maciver et al., 2013; Carlton et al., 2020). Strikingly, physical symmetry plays a major factor in these re-arrangements, possibly because of the need to balance and direct mechanical force between the T cell and the APC (Sims et al., 2007; Dustin, 2009; Arsenio et al., 2015). Calcium release in response to TCR-pMHC binding leads to the depolymerisation of microtubules, which in turn facilitates the rearrangement of organelles such as the nucleus and endoplasmic reticulum to the center of the T-cell body volume (Joseph et al., 2014; Ilan-Ber and Ilan, 2019). Cytoskeletal ruffles depolymerise and actin-rich lamellum and lamellipodium polymerise at the contact between both cells. The lamellipodium constantly propels freshly forming TCR clusters to the center of the contact (Fritzsche et al., 2017), whose function is thought to amplify the pMHC binding and recognition (Harrison et al., 2019). At the interface between the T cell and the APC, the immunological synapse (IS) takes shape involving a complex spatio-temporal orchestration of receptors, positive and negative co-stimulatory co-receptors, and integrins (Dustin, 2009). They contribute in concert to the force balance between the T cell and the APC. As antigens are processed at the IS, the T cell ensures a mechanically stable and flat contact interface in the form of a ramified actin network (Fritzsche et al., 2017). Visual inspection suggested, that this network is shaped under mechanical tension and held stable over time through the actively polymerising actin-rich lamellipodium and its shear force producing actin retrograde flow (Colin-York et al., 2019b). These interfacial processes are further supported by mechanically active protrusions (Tamzalit et al., 2019). On one side of IS, the T cell constantly assembles and disassembles short-lived actin foci facilitating and perhaps ensuring localized contact between both cells (Kumari et al., 2019, 2020). On the other side of the interaction, the APC forms mechano-transducing podosomes in the outer periphery of the IS (Malinova et al., 2016), which may serve to monitor the mechanical stiffness at the contact interface. The mechanical properties of the cytoskeletal actin architectures and protrusions are also time-dependent with distinct viscosities depending on the observation frequency, because they and their crosslinkers are constantly turning over (Fritzsche et al., 2016; Gat et al., 2020). Loss of symmetry at the IS, and thus loss of mechanical force balance at the contact, has been demonstrated to influence instability and success of immune cell activation (Sims et al., 2007; Lee et al., 2017). These outlined processes are further underpinned by dynamic molecular changes in the nanoscale organization and turnover of actin filaments in the actin cortex and lamellipodium as a function of the antigen affinity (Billadeau et al., 2007; Colin-York et al., 2019b; Wahl et al., 2019), suggesting mechanical feedback on multiple length- and time-scales. Over the years, a multitude of measurements in the study of immune cell activation reported the importance of different mechanical metrics such as stiffness, tension, shear, and structural integrity, highlighting the diversity of current mechanical quantifications (Figure 1B) (Egan et al., 2015).
Together, having uncovered the presence of a variety of length- and time-scale dependent mechanical force regimes and properties during immune cell activation (Pageon et al., 2018), and moreover, knowing that T cells and APCs integrate a variety of mechanical readouts (Jain et al., 2019), leads to the question what mechanical measurements are necessary and sufficient to understand the mechanobiology of the immune response? More specifically, how do these mechanical signatures couple and feed into biology over space and time, and are thus integrated into cellular function and behavior (Harris et al., 2018)? To add further complexity to this picture, the T cell-APC interplay is usually not insulated but maintains processes of continuous communication and interpretation of the surrounding biochemical and biomechanical micro-environment. Consequently, without understanding comprehensively the dynamic relationship of these processes, it is challenging to determine the biological significance of mechanobiology in health and disease.
Hence, a grand challenge for the understanding of mechanbiology is the determination of the right mechanical readout. Ideally, from the theoretical physics point of view, one aims for a full quantitative parametrisation of the desired biological phenomena, which ultimately comes down to identification and determination of a well-defined control parameter (Bechhoefer, 2005). The determinant at which biomechanics regulates behavior and/or function of the immunological process of interest. The prospect of knowing the one (or the many) mechanical control parameter(s) against all other system parameters is of particular importance in the context of stability of the biological function (Bechhoefer, 2005; McEvoy, 2018). For example the stability of IS formation could be regulated through changes in mechanical feedback between the T cell and the APC (Harrison et al., 2019). Stability of such a biological feature is mathematically determined by the so-called eigenmodes of its stability matrix, which grow or shrink when the control parameter for example mechanical feedback changes (Bechhoefer, 2005). Crucially, while experimentally, changes in different mechanical parameters could be observed throughout a biological process (Dumont and Prakash, 2014; Egan et al., 2015), only the determination of the biological control parameter aids to the understanding of the biological mechanisms and concepts being at play (McEvoy, 2018). In other words, one may observe quantitative changes in mechanical metrics such as stiffness and/or tension but in practice misinterpret the observed phenomena, let it be for example mechanical feedback during IS formation, if the control parameter feedback has not been correctly identified and parameterized.
How to then find the right mechanical readout? The answer to this question is not trivial and practically challenging for many biological systems due to the numerous molecular players involved, the number and complexity of their interactions, but probably mostly due to a broad lack of quantitative technology with the right sensitivity (Polacheck and Chen, 2016; Roca-Cusachs et al., 2017).
Over the last 10 years, recent advances in quantitative technology have enabled the spatio-temporal sensitivity demanded by the immune response drawing a promising perspective for the future. These new methodologies will enable a complete quantitative characterization of biological processes to the best of the experimentalist's abilities allowing the full parameterization of theoretical physics descriptions (McEvoy, 2018). For this, a variety of different technologies are needed to quantify correlatively or co-incidentally the mechanical setting with the sensitivity demanded by the biology of interest. We and others have spent significant efforts in evolving the sensitivity of traction force microscopy, probably the most widely applied force quantification methodology, which offers the simultaneous quantification of mechanical force production and the dynamics of cells (Colin-York and Fritzsche, 2018; Colin-York et al., 2019a; Stubb et al., 2020; Vorselen et al., 2020). More recent efforts of combining different types of quantitative simultaneous measurements hold the promise to achieve a more complete understanding of how biomechanics feeds into immune cell physiology of the immune response (Skamrahl et al., 2019; Hobson et al., 2020; Moreno-Flores, 2020; Nelsen et al., 2020). Quantifying the mechanical settings of the immune response in full utilizing methodologies with the right sensitivity may thus be the route to enable broad recognition of mechanobiology in health and disease.
MF designed and wrote the perspective article.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past co-authorship with one of the authors MF.
MF would like to acknowledge generous funding from the Rosalind Franklin Institute and the Kennedy Trust for Rheumatology Research. MF also thanks the Wellcome Trust (212343/Z/18/Z) and EPSRC (EP/S004459/1).
Andersen, M. H., Schrama, D., Thor Straten, P., and Becker, J. C. (2006). Cytotoxic T cells. J. Invest. Dermatol. 126, 32–41. doi: 10.1038/sj.jid.5700001
Arsenio, J., Metz, P. J., and Chang, J. T. (2015). Asymmetric cell division in T lymphocyte fate diversification. Trends Immunol. 36, 670–683. doi: 10.1016/j.it.2015.09.004
Bechhoefer, J. (2005). Feedback for physicists: a tutorial essay on contro. Rev. Mod. Phys. 77, 783–836. doi: 10.1103/RevModPhys.77.783
Billadeau, D. D., Nolz, J. C., and Gomez, T. S. (2007). Regulation of T-cell activation by the cytoskeleton. Nat. Rev. Immunol. 7, 131–143. doi: 10.1038/nri2021
Blanchard, G. B., and Adams, R. J. (2011). Measuring the multi-scale integration of mechanical forces during morphogenesis. Curr. Opin. Genet. Dev. 21, 653–663. doi: 10.1016/j.gde.2011.08.008
Carlton, J. G., Jones, H., and Eggert, U. S. (2020). Membrane and organelle dynamics during cell division. Nat. Rev. Mol. Cell Biol. 21, 151–166. doi: 10.1038/s41580-019-0208-1
Chaplin, D. D. (2010). Overview of the immune response. J. Allergy Clin. Immunol. 125:S3. doi: 10.1016/j.jaci.2009.12.980
Chen, W., and Zhu, C. (2013). Mechanical regulation of T-cell functions. Immunol. Rev. 256, 160–176. doi: 10.1111/imr.12122
Colin-York, H., and Fritzsche, M. (2018). The future of traction force microscopy. Curr. Opin. Biomed. Eng. 5, 1–5. doi: 10.1016/J.COBME.2017.10.002
Colin-York, H., Javanmardi, Y., Barbieri, L., Li, D., Korobchevskaya, K., Guo, Y., et al. (2019a). Spatiotemporally super-resolved volumetric traction force microscopy. Nano Lett. 19, 4427–4434. doi: 10.1021/acs.nanolett.9b01196
Colin-York, H., Javanmardi, Y., Skamrahl, M., Cerundolo, V., Eggeling, C., and Correspondence, M. F. (2019b). Cytoskeletal control of antigen-dependent T cell activation. CellRep. 26, 3369–3379.e5. doi: 10.1016/j.celrep.2019.02.074
Colin-York, H., Shrestha, D., Felce, J. H., Waithe, D., Moeendarbary, E., Davis, S. J., et al. (2016). Super-resolved traction force microscopy (STFM). Nano Lett. 16, 2633–2638. doi: 10.1021/acs.nanolett.6b00273
Dumont, S., and Prakash, M. (2014). Emergent mechanics of biological structures. Mol. Biol. Cell 25, 3461–3465. doi: 10.1091/mbc.E14-03-0784
Dustin, M. L. (2009). The cellular context of T cell signaling. Immunity 30, 482–492. doi: 10.1016/j.immuni.2009.03.010
Egan, P., Sinko, R., Leduc, P. R., and Keten, S. (2015). The role of mechanics in biological and bio-inspired systems. Nat. Commun. 6, 1–12. doi: 10.1038/ncomms8418
English, A. R., and Voeltz, G. K. (2013). Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb. Perspect. Biol. 5, 1–16. doi: 10.1101/cshperspect.a013227
Fritz-Laylin, L. K., Riel-Mehan, M., Chen, B. C., Lord, S. J., Goddard, T. D., Ferrin, T. E., et al. (2017). Actin-based protrusions of migrating neutrophils are intrinsically lamellar and facilitate direction changes. Elife 6:e26990. doi: 10.7554/eLife.26990
Fritzsche, M. (2020). Thinking multi-scale to advance mechanobiology. Commun. Biol. 3, 1–2. doi: 10.1038/s42003-020-01197-5
Fritzsche, M., Erlenkämper, C., Moeendarbary, E., Charras, G., and Kruse, K. (2016). Actin kinetics shapes cortical network structure and mechanics. Sci. Adv. 2:e1501337. doi: 10.1126/sciadv.1501337
Fritzsche, M., Fernandes, R. A., Chang, V. T., Colin-York, H., Clausen, M. P., Felce, J. H., et al. (2017). Cytoskeletal actin dynamics shape a ramifying actin network underpinning immunological synapse formation. Sci. Adv. 3:e1603032. doi: 10.1126/sciadv.1603032
Gat, S., Simon, C., Campillo, C., Bernheim-Groswasser, A., and Sykes, C. (2020). Finger-like membrane protrusions are favored by heterogeneities in the actin network. Soft Matter 16, 7222–7230. doi: 10.1039/c9sm02444a
Grivennikov, S. I., Greten, F. R., and Karin, M. (2010). Immunity, inflammation, and cancer. Cell 140, 883–899. doi: 10.1016/j.cell.2010.01.025
Harris, M. J., Wirtz, D., and Wu, P.-H. (2018). Dissecting cellular mechanics: implications for aging, cancer, and immunity. Semin. Cell Dev. Biol. 93, 16–25. doi: 10.1016/j.semcdb.2018.10.008
Harrison, D. L., Fang, Y., and Huang, J. (2019). T-cell mechanobiology: force sensation, potentiation, and translation. Front. Phys. 7:45. doi: 10.3389/fphy.2019.00045
Hobson, C. M., Kern, M., O'Brien, E. T., Stephens, A. D., Falvo, M. R., and Superfine, R. (2020). Correlating nuclear morphology and external force with combined atomic force microscopy and light sheet imaging separates roles of chromatin and lamin A/C in nuclear mechanics. Mol. Biol. Cell 31, 1788–1801. doi: 10.1091/mbc.E20-01-0073
Huse, M. (2017). Mechanical forces in the immune system. Nat. Rev. Immunol. 17, 679–690. doi: 10.1038/nri.2017.74
Ilan-Ber, T., and Ilan, Y. (2019). The role of microtubules in the immune system and as potential targets for gut-based immunotherapy. Mol. Immunol. 111, 73–82. doi: 10.1016/j.molimm.2019.04.014
Jain, N., Moeller, J., and Vogel, V. (2019). Mechanobiology of macrophages: how physical factors coregulate macrophage plasticity and phagocytosis. Annu. Rev. Biomed. Eng. 21, 267–297. doi: 10.1146/annurev-bioeng-062117-121224
Joseph, N., Reicher, B., and Barda-Saad, M. (2014). The calcium feedback loop and T cell activation: how cytoskeleton networks control intracellular calcium flux. Biochim. Biophys. Acta - Biomembr. 1838, 557–568. doi: 10.1016/j.bbamem.2013.07.009
Kumari, S., Colin-York, H., Irvine, D. J., and Fritzsche, M. (2019). Not all T cell synapses are built the same way. Trends Immunol. 40, 977–980. doi: 10.1016/j.it.2019.09.009
Kumari, S., Mak, M., Poh, Y., Tohme, M., Watson, N., Melo, M., et al. (2020). Cytoskeletal tension actively sustains the migratory T-cell synaptic contact. EMBO J. 39:e102783. doi: 10.15252/embj.2019102783
Lee, A. M., Colin-York, H., and Fritzsche, M. (2017). CalQuo 2 : Automated Fourier-space, population-level quantification of global intracellular calcium responses. Sci. Rep. 7:5416. doi: 10.1038/s41598-017-05322-z
Maciver, N. J., Michalek, R. D., and Rathmell, J. C. (2013). Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 31, 259–283. doi: 10.1146/annurev-immunol-032712-095956
Majedi, F. S., Hasani-Sadrabadi, M. M., Thauland, T. J., Li, S., Bouchard, L. S., and Butte, M. J. (2020). T-cell activation is modulated by the 3D mechanical microenvironment. Biomaterials 252:120058. doi: 10.1016/j.biomaterials.2020.120058
Malinova, D., Fritzsche, M., Nowosad, C. R., Armer, H., Munro, P. M. G., Blundell, M. P., et al. (2016). WASp-dependent actin cytoskeleton stability at the dendritic cell immunological synapse is required for extensive, functional T cell contacts. J. Leukoc. Biol. 99, 699–710. doi: 10.1189/jlb.2a0215-050rr
McEvoy, P. B. (2018). Theoretical contributions to biological control success. BioControl 63, 87–103. doi: 10.1007/s10526-017-9852-6
Mohammadi, H., and Sahai, E. (2018). Mechanisms and impact of altered tumour mechanics. Nat. Cell Biol. 20, 766–774. doi: 10.1038/s41556-018-0131-2
Moreno-Flores, S. (2020). Hallmarks of life in single cell contact mechanics: outstanding challenges and perspectives. Front. Mech. Eng. 6:58. doi: 10.3389/fmech.2020.00058
Needleman, D., and Dogic, Z. (2017). Active matter at the interface between materials science and cell biology. Nat. Rev. Mater. 2, 1–14. doi: 10.1038/natrevmats.2017.48
Nelsen, E., Hobson, C. M., Kern, M. E., Hsiao, J. P., O'Brien, E. T., Watanabe, T., et al. (2020). Combined atomic force microscope and volumetric light sheet system for correlative force and fluorescence mechanobiology studies. Sci. Rep. 10, 1–12. doi: 10.1038/s41598-020-65205-8
Pageon, S. V., Govendir, M. A., Kempe, D., and Biro, M. (2018). Mechanoimmunology: molecular-scale forces govern immune cell functions. Mol. Biol. Cell 29, 1919–1926. doi: 10.1091/mbc.E18-02-0120
Polacheck, W. J., and Chen, C. S. (2016). Measuring cell-generated forces: a guide to the available tools. Nat. Methods 13, 415–423. doi: 10.1038/nmeth.3834
Roca-Cusachs, P., Conte, V., and Trepat, X. (2017). Quantifying forces in cell biology. Nat. Cell Biol. 19, 742–751 doi: 10.1038/ncb3564
Schwarz, U. S. (2017). Mechanobiology by the numbers: a close relationship between biology and physics. Nat. Rev. Mol. Cell Biol. 18, 711–712. doi: 10.1038/nrm.2017.109
Sims, T. N., Soos, T. J., Xenias, H. S., Dubin-Thaler, B., Hofman, J. M., Waite, J. C., et al. (2007). Opposing effects of PKCθ and WASp on symmetry breaking and relocation of the immunological synapse. Cell 129, 773–785. doi: 10.1016/j.cell.2007.03.037
Skamrahl, M., Colin-York, H., Barbieri, L., and Fritzsche, M. (2019). Simultaneous quantification of the interplay between molecular turnover and cell mechanics by AFM–FRAP. Small 15:1902202. doi: 10.1002/smll.201902202
Stubb, A., Laine, R. F., Miihkinen, M., Hamidi, H., Guzmán, C., Henriques, R., et al. (2020). Fluctuation-based super-resolution traction force microscopy. Nano Lett. 20, 2230–2245. doi: 10.1021/acs.nanolett.9b04083
Swartz, M. A., and Lund, A. W. (2012). Lymphatic and interstitial flow in the tumour microenvironment: linking mechanobiology with immunity. Nat. Rev. Cancer 12, 210–219. doi: 10.1038/nrc3186
Tamzalit, F., Wang, M. S., Jin, W., Tello-Lafoz, M., Boyko, V., Heddleston, J. M., et al. (2019). Interfacial actin protrusions mechanically enhance killing by cytotoxic T cells. Sci. Immunol. 4:eaav5445. doi: 10.1126/sciimmunol.aav5445
Vorselen, D., Wang, Y., de Jesus, M. M., Shah, P. K., Footer, M. J., Huse, M., et al. (2020). Microparticle traction force microscopy reveals subcellular force exertion patterns in immune cell–target interactions. Nat. Commun. 11, 1–14. doi: 10.1038/s41467-019-13804-z
Keywords: mechanics, biophysics, mechanical force, mechanical properties, feedback, dynamics, stiffness, tension
Citation: Fritzsche M (2021) What Is the Right Mechanical Readout for Understanding the Mechanobiology of the Immune Response? Front. Cell Dev. Biol. 9:612539. doi: 10.3389/fcell.2021.612539
Received: 30 September 2020; Accepted: 02 February 2021;
Published: 25 February 2021.
Edited by:
Sudha Kumari, Massachusetts Institute of Technology, United StatesReviewed by:
Brian Gabrielli, The University of Queensland, AustraliaCopyright © 2021 Fritzsche. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marco Fritzsche, bWFyY28uZnJpdHpzY2hlQGtlbm5lZHkub3guYWMudWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.