
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 25 February 2021
Sec. Molecular and Cellular Pathology
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.605686
This article is part of the Research Topic Emerging Roles of Circular RNAs in the Tumor: Functions and Potential Applications View all 25 articles
 John Greene1,2*
John Greene1,2* Anne-Marie Baird3
Anne-Marie Baird3 Marvin Lim1,2
Marvin Lim1,2 Joshua Flynn3
Joshua Flynn3 Ciara McNevin1,2
Ciara McNevin1,2 Lauren Brady1
Lauren Brady1 Orla Sheils1,3
Orla Sheils1,3 Steven G. Gray3,4
Steven G. Gray3,4 Raymond McDermott2,5
Raymond McDermott2,5 Stephen P. Finn1,4,6
Stephen P. Finn1,4,6Circular RNAs (circRNAs), a recently discovered non-coding RNA, have a number of functions including the regulation of miRNA expression. They have been detected in a number of malignancies including prostate cancer (PCa). The differential expression pattern of circRNAs associated with PCa and androgen receptor (AR) status was investigated in this study. circRNA profiling was performed using a high throughout microarray assay on a panel of prostate cell lines, which consisted of normal, benign, and malignant cells (n = 9). circRNAs were more commonly significantly up-regulated (p < 0.05) than downregulated in malignant cell lines (n = 3,409) vs. benign cell lines (n = 2,949). In a grouped analysis based on AR status, there were 2,127 down-regulated circRNAs in androgen independent cell lines compared to 2,236 in androgen dependent cell lines, thus identifying a potential circRNA signature reflective of androgen dependency. Through a bioinformatics approach, the parental genes associated with the top 10 differentially expressed circRNAs were identified such as hsa_circ_0064644, whose predicted parental gene target is RBMS3, and hsa_circ_0060539, whose predicted gene target is SDC4. Furthermore, we identified three circRNAs associated with the parental gene Caprin1 (hsa_circ_0021652, hsa_circ_0000288, and hsa_circ_0021647). Other studies have shown the importance of Caprin1 in PCa cell survival and drug resistance. Given the modified circRNA expression signatures identified here, these hypothesis generating results suggest that circRNAs may serve as potential putative diagnostic and predictive markers in PCa. However, further validation studies are required to assess the true potential of these markers in the clinical setting.
Non-coding RNAs (ncRNAs), which include microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) (Shih et al., 2015; Lin et al., 2017; Misawa et al., 2017), play an important role in gene regulation (Warner, 1999; Esteller, 2011; Jeck et al., 2013). circular RNAs (circRNAs), a recently discovered type of ncRNA, are generated from the backsplicing of exons, introns, or both to form exonic or intronic circRNAs (Jeck et al., 2013). circRNAs are covalently joined by their 3′- and 5′- ends, which are formed by back-splice events, thus presenting as closed continuous structures, making them highly stable and resistant to degradation (Salzman et al., 2012; Barrett et al., 2015). circRNAs have many postulated functions such as the regulation of miRNA function by controlling the expression of miRNAs through a “sponging effect,” however this appears to apply to only a small number of circRNAs, such as ciRS-7 acting as a miR-7 sponge (Bahn et al., 2015). It has also been proposed that circRNAs may have a role in protein synthesis, with a number of circRNAs implicated in the translation of peptides, such as ZNF609 (Legnini et al., 2017). As research in this space increases, it is now clear that the circRNA-miRNA-mRNA network, plays an important role in both gene regulation and carcinogenesis (Su et al., 2019). circRNAs have also been detected in prostate cancer, which make them an attractive research target (Greene et al., 2019). PCa growth and development is primarily dependent on the androgen receptor (AR), which is the target of therapeutic agents such as enzalutamide and abiraterone (Antonarakis et al., 2014). AR copy number gain and generation of variants such as AR-V7 are associated with the development of castration-resistant disease and drug resistance, however, despite significant advances; no suitable assay has become routinely available in the clinic to identify resistance mediators of AR targeting therapeutics (Antonarakis et al., 2014; Romanel et al., 2015). Currently, PSA is used to screen men for PCa, yet when used in isolation, it is sub-optimal given issues with specificity and sensitivity (O’Sullivan, 2017). Similarly, PSA cannot reliably detect early relapse or resistance to drug treatments, nonetheless earlier detection of castration-resistant disease would allow for more appropriate management of these patients and therefore significantly improve outcomes (Di Nunno et al., 2018). Thus, there is a need to identify new diagnostic and predictive biomarkers, as well as new therapeutic agents for use in the clinical setting. circRNAs may fulfill these roles, given their expression in PCa, in addition to studies showing circRNA such as circFOXO3 (hsa_circ_0006404) acting as an miRNA sponge in this disease (Kong et al., 2020).
The aim of this study was to profile circRNAs in a panel of prostate cell lines and identify potential signatures associated with malignancy and androgen dependency. We propose that circRNAs have the potential to serve as useful biomarkers to improve diagnostic screening for PCa and/or identify men who are at risk of developing castration-resistant disease.
A panel of malignant and benign prostate cell lines were used in this project, all of which were purchased from the ATCC (LGC standards, Middlesex, United Kingdom). The benign prostate cell lines (PWR-1E, RWPE-1) were cultured in Keratinocyte Serum Free Media (Thermo Fisher Scientific, CA, US), containing 0.05 mg/mL bovine pituitary extract (BPE) and 5 ng/mL Epidermal growth factor (EGF) (Thermo Fisher Scientific) and 1% Penicillin Streptomycin (P/S) (Merck KGaA, Darmstadt, Germany). The benign prostatic hyperplasia (BPH-1) and PCa cell lines (22Rv1, VCaP, LNCaP, DU145) were cultured in RPMI-1640 (Merck KGaA) supplemented with 10% Fetal Bovine Serum (FBS) (Merck KGaA) and 1% P/S. PC-3 was cultured in ATCC-formulated F-12K Medium (Thermo Fisher Scientific) supplemented with 10% FBS and 1% P/S.
Total RNA was prepared from cell lines (biological triplicates) according to manufacturer’s instructions. Briefly, cells were lysed directly with 2 mL TRIzol® Reagent (Thermo Fisher Scientific). For phase separation, 200 μL chloroform (Merck KGaA) was added to the cell lysate. The sample were vortexed vigorously for 15 s, incubated at RT for 5 min and centrifuged at 12,000 × g for 15 min at 4°C. The upper aqueous phase was transferred into a fresh tube and the RNA precipitated from the aqueous phase, by the addition of 400 μL isopropyl alcohol (Merck KGaA). The sample was mixed, incubated for 5 min at RT and centrifuged at 12,000 × g for 15 min at 4°C. One milliliter 75% EtOH (Merck KGaA) was added to pellet, and centrifuged at 12,000 × g for 10 min at 4°C. The EtOH was removed and a repeat pulse centrifuge was performed. Surplus EtOH was removed and the pellet air-dried. The sample was re-suspended in 30 μL molecular grade H2O before DNase treatment with Ambion® TURBOTM DNase (Thermo Fisher Scientific), and an additional RNA clean-up using standard EtOH precipitation.
The panel of cell lines (three biological replicates for each) were profiled using the Arraystar Human circRNA Array version 2.0 (Arraystar, MD, United States). The sample preparation and microarray hybridization were performed according to manufacturer’s instructions. Briefly, total RNA was digested with RNAse R (Epicenter, Illumina, CA, United States) to remove linear RNAs and enrich for circRNAs. The enriched circRNAs were amplified and transcribed into fluorescent cRNA using a random priming method with Arraystar Super RNA Labeling Kit (Arraystar). The labeled cRNAs were hybridized onto the Arraystar Human circRNA Array V2 (8 × 15 K). The array slides were washed and scanned on the Agilent Scanner G2505C. Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images.
Quantile normalization and subsequent data processing were executed using the R software package (R Core Team, 2020). circRNAs with at least 4 out of 8 samples that were flagged as present or marginal (an attribute that denotes the quality of the entities) were considered as target circRNAs according to GeneSpring software’s definitions and instructions. circRNA and miRNA interactions were predicted with the Arraystar’s miRNA target prediction software based on TargetScan (Agarwal et al., 2015) and miRanda (John et al., 2004). These target circRNAs were used for further differential analysis. A student’s paired t-test was used to identify significantly altered circRNAs. The false discovery rate (FDR) was applied to determine the threshold of p-value. An FDR of < 0.05 was used. Changes in expression were identified using unsupervised clustering analysis (euclidean distance measure and the “average” agglomeration method) and associated heat-maps generated.
The human circular RNA microarray (Arraystar) version 2.0 covers 13,617 previously discovered human circRNAs. Our cell line panel consisted of malignant (22Rv1, LNCaP, VCAP, DU145, and PC-3), benign (BPH-1), and normal (PWR1, RWPE-1) cell lines. AR expression in 22Rv1, LNCaP, and VCaP was confirmed using qPCR (Supplementary Methods and Supplementary Figure S1). In total 9,805 circRNAs were classified as present across the complete panel of cell lines. Initially, circRNA expression levels between cell lines based on malignancy was investigated. Differentially expressed circRNAs were examined by computing the fold change (FC) (i.e., the ratio of the group averages) for each circRNA between malignant, normal and benign cell lines. Associated parental genes for the top 10 altered circRNAs were predicted from circBase, which uses the co-ordinates from each detected circRNA to identify the associated gene from the UCSC genome database (Glazar et al., 2014).
CircRNAs were significantly differentially expressed between these groups (BPH vs. normal; malignant vs. BPH/normal) according to FC and p-value (Figure 1). We first examined the difference in expression of circRNAs between normal cell lines (PWR1, RWPE-1) and the benign cell line, BPH-1. CircRNAs were more often down-regulated in normal cell lines compared to the benign cell line (5,429 vs. 4,344). Next, we examined the expression of circRNAs between the normal/benign cells compared to malignant cells. In total, a higher number of circRNAs were down-regulated in normal/benign cells compared to malignant cells (5,300 vs. 4,457). The top 10 down and up-regulated circRNAs according to FC, along with their associated parental gene, are given in Tables 1, 2, respectively.
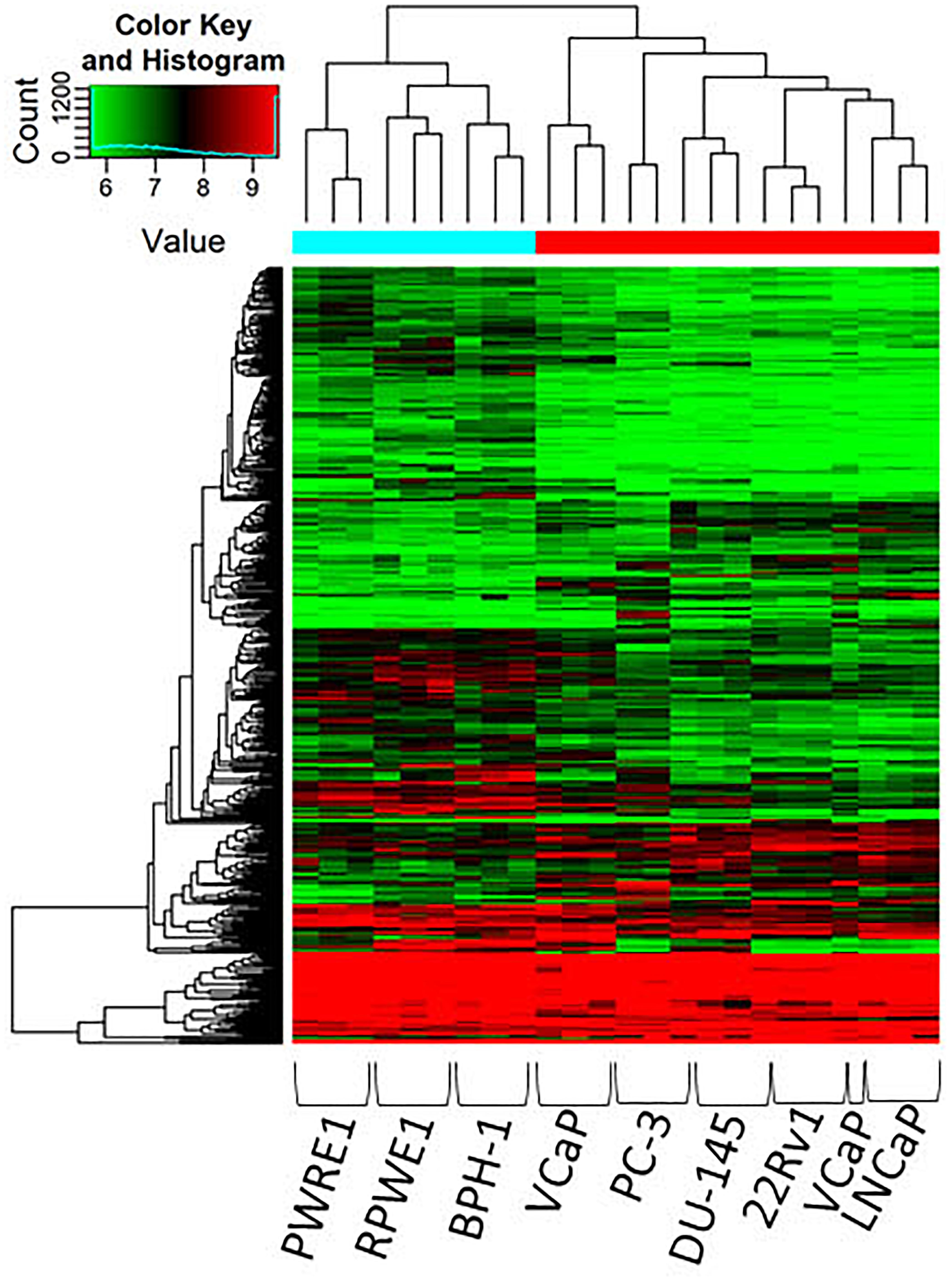
Figure 1. Clustering heatmap of microarray data showing differential expression of circRNAs between malignant, benign and normal cell lines. Unsupervised clustering (euclidean distance measure and the “average” agglomeration method) was used for analysis (n = 3). circRNAs were more likely to be down-regulated in normal and benign cell lines compared to malignant cells.
A number of circRNAs, that could play a role in PCa, were identified through their interaction with their associated parental genes. hsa_circ_0064644 was the highest down-regulated circRNA (FC 5, p = 0.001), located on chromosome 3 (Table 1). Its’ predicted parental gene target using circBase is RBMS3, which has previously been identified to suppress cell proliferation, migration, invasion, and angiogenesis in a number of different cancers (Zhu et al., 2019). RBMS3 has also been shown to act as a miRNA sponge in PCa via lncRNAs. Other genes of interest include HIPK2 (hsa_circ_0082672) and NFIX (hsa_circ_0049657), both known to be involved in the regulation of the AR (Imberg-Kazdan et al., 2013). hsa_circ_0005402, is linked to ANXA2, which has been suggested to play a role in epithelial-mesenchymal transition (EMT) in PCa (Yang et al., 2018) via transcriptional repression by ERG (Griner et al., 2015). hsa_circ_0060539, located on chromosome 20, was the highest up-regulated circRNA. The predicted gene target for this circRNA is SDC4, a known active gene in PCa (Griner et al., 2015). Similarly, KRT5 (hsa_circ_0026457) has been shown to be involved in the inhibition of PCa stem cell invasion, migration and proliferation, as has KRT7 (hsa_circ_0026358), which has been identified along with FAM129A, MME and SOD2 as part of a four-gene androgen regulated panel (Thomas et al., 2016).
Lastly, the differing expression of circRNAs in relation to androgen sensitivity was examined, which we hypothesized would reflect the development of castration resistant PCa. Cell lines were stratified into two groups based on AR status: androgen dependent/castration-sensitive (22Rv1, LNCaP, VCaP) and androgen independent/castration resistant) cell lines (DU145, PC-3). Response to androgen ablation was demonstrated by confirming the presence or absence of AR-FL across cell lines (see Supplementary Figure S1). CircRNAs were more often down-regulated in cell lines that were androgen independent compared to androgen-dependent cell lines (5,067 down-regulated vs. 4,693 up-regulated, respectively) (Figure 2). The top 10 down and up-regulated circRNAs and their associated parental genes are given in Tables 3, 4, respectively. The up-regulated hsa_circ_0000361 (FC 19.2, p < 0.001) is an exonic circRNA located on chromosome 11 and is associated with the gene SIAE (Table 3). mRNA expression of SIAE has previously been shown to be reduced in patients with cancer (Oh et al., 2020). We identified three circRNAs associated with the parental gene Caprin1 (hsa_circ_0021652, hsa_circ_0000288, and hsa_circ_0021647). Caprin1 plays an important role in PCa cell survival and its upregulation has been shown to be associated with drug resistance, and is increased in SPOP mutated cell lines (Shi et al., 2019).
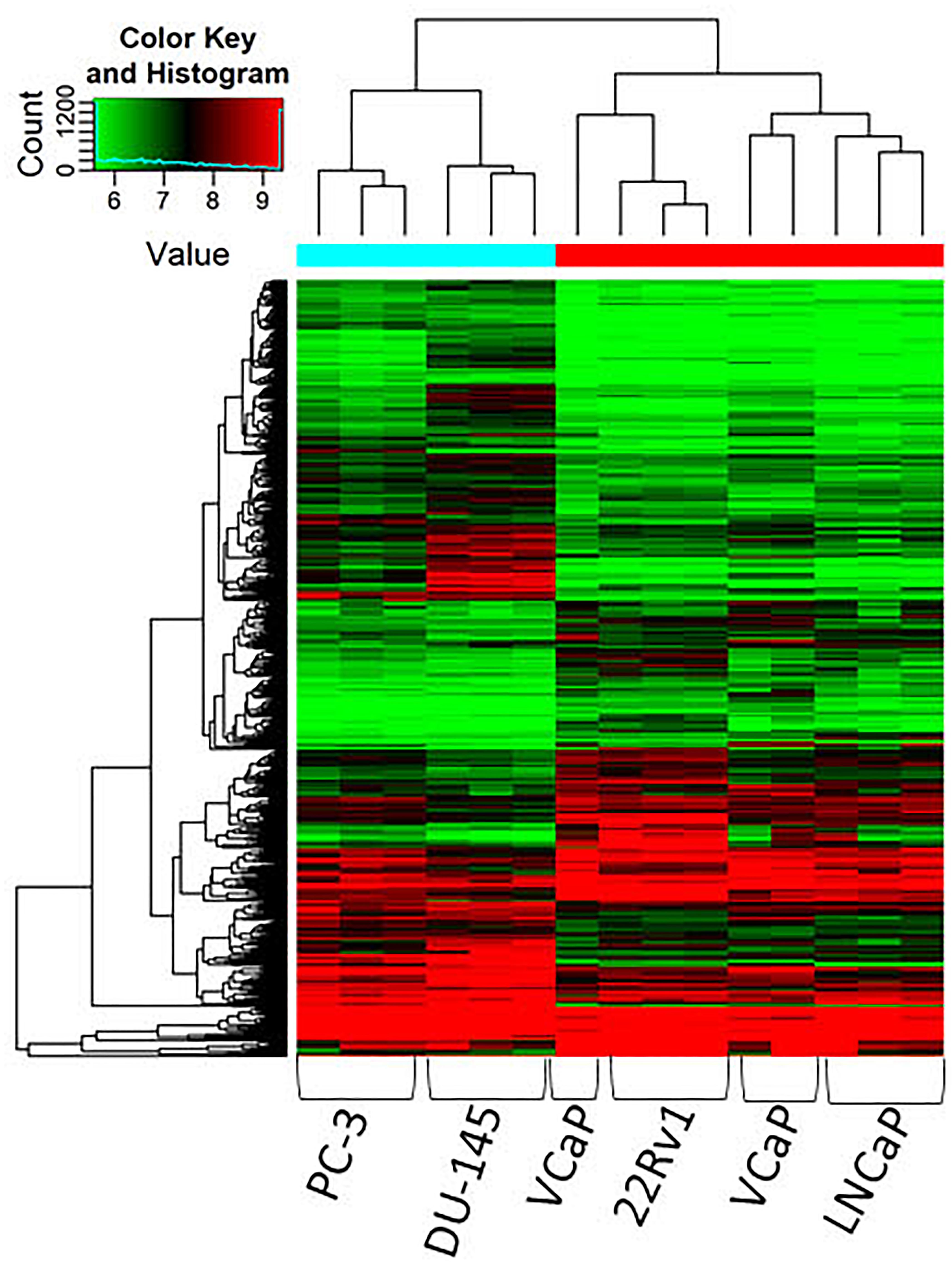
Figure 2. Clustering heatmap showing differential expression of circRNAs between AR dependent cells LNCaP, 22Rv1 and VCaP (hormone sensitive) and AR independent cells DU145 and PC-3 (castration resistant). Unsupervised clustering (euclidean distance measure and the “average” agglomeration method) was used for analysis (n = 3).
The highest down-regulated circRNA by FC was hsa_circ_0000825 (FC 40, p < 0.0001) located on chromosome 18 and is associated with the gene MTCL1 (Table 4). This gene has previously been identified to play a role in the development of castration-resistant PCa (Zhang et al., 2018). Other parental genes of interest include FLNA (hsa_circ_0091934 and hsa_circ_0091894) located on the X chromosome, which plays a significant role in PCa development and progression, and has been highlighted as a possible biomarker for disease screening and detection (Ravipaty et al., 2017).
CircRNAs are a novel type of ncRNA which have been identified across a range of cancers including PCa (Lim et al., 2018; Greene et al., 2019). The most abundant class of ncRNA are lncRNAs which have been extensively investigated in PCa (Ramnarine et al., 2019). A number of lncRNAs have been identified as potential predictive and prognostic biomarkers including SChLAP1, UCA1 and PCAT14 (Mehra et al., 2016; Fotouhi Ghiam et al., 2017; White et al., 2017). Further work is exploring lncRNAs as therapeutic targets in PCa using small interfering RNAs (siRNAs), antisense oligonucleotides (ASOs) and small molecule inhibitors (Fu et al., 2020). Therefore, there is significant interest in the role ncRNAs may play in the future management of PCa. The exact role of circRNAs in cancer has yet to be fully elucidated, however initial studies suggest circRNAs may play a role in cancer development, progression and the development of resistance to therapeutic agents (Greene et al., 2017). CircRNAs have been identified as potential biomarkers in cancer due to their abundance, stability and disease specific activity (Dong et al., 2017). The identification of circRNAs has been complicated by bioinformatics challenges, however the development of improved detection methodologies and statistical methods has aided circRNA research, particularly with improvements in reducing false-positive detection rates (Szabo and Salzman, 2016). In PCa, a number of circRNAs have been identified which have been identified as potential biomarkers (Tucker et al., 2020). Our laboratory previously identified the circRNA, hsa_circ_0004870, to be associated with resistance to the androgen receptor inhibitor enzalutamide (Greene et al., 2019). Other circRNAs of note that have been identified in PCa include circAMACR, circAURKA and circAR3 which is encoded by the AR gene (Chen et al., 2019).
We identified a number of circRNAs that could represent a unique gene signature of aggressive PCa. Initial bioinformatics analysis identified a significant number of differentially expressed circRNAs between malignant vs. benign/normal cell lines. There was a trend for circRNAs to be down-regulated in normal compared to benign cells. This trend continued when we compared normal/benign cells to malignant cells. When we examined circRNA expression in cell lines according to androgen dependency, we found circRNAs again to be more often down-regulated in androgen independent cell lines, reflective of aggressive PCa.
Parental genes of interest were RBMS3 (hsa_circ_0064644) and SDC4 (hsa_circ_0060539). Interestingly, we identified three circRNAs associated with the parental gene Caprin1 (hsa_circ_0021652, hsa_circ_0000288, and hsa_circ_0021647) and two circRNAs associated with FLNA (hsa_circ_0091934 and hsa_circ_0091894). Many of these genes are currently under active investigation as biomarkers in PCa, and identifying their associated circRNAs may contribute to ongoing research in this field (Ravipaty et al., 2017; Shi et al., 2019; Zhu et al., 2019; Jiang et al., 2020).
There is evidence to suggest that circRNAs regulate miRNA function by competing for the pool of miRNA binding sites to influence the activities of miRNAs in regulating gene expression (Hansen et al., 2013). Interestingly, a number of miRNAs that have been detected in PCa are linked to the AR pathway (Budd et al., 2015; Fabris et al., 2016). miRNA binding sites can be predicted using a bioinformatics pipeline for each detected circRNA that identified an associated gene. For instance, for hsa_circ_0064644, the predicted miRNA is miR-15a, which has been show to play a role in PCa progression and metastases (Jin et al., 2018). Therefore, predicted miRNAs could be used to identify circRNAs that play a role in gene regulation in PCa, and further our understanding of the importance of the circRNA-miRNA-mRNA network in this disease.
This study had several limitations. Firstly, we used a circRNA microarray platform to detect circRNAs compared to RNA-seq methods which may have higher sensitivity and accuracy to detect circRNAs. Secondly, this was a preliminary in vitro study which identified multiple potential genes and ongoing validation is required in cell lines and in vivo to confirm our findings here.
In conclusion, circRNAs were modified between benign and malignant cell lines, as well as androgen dependent and independent cell lines, which suggests a role for circRNAs in PCa initiation and progression. This study highlights the importance of circRNAs in PCa and their potential role in identifying the disease and monitoring response to treatment. The dysregulated signature identified here may prove useful for the development of a blood-based assay with both diagnostic and predictive value in this disease. However, this would require validation in larger patient cohorts.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.605686/full#supplementary-material
Agarwal, V., Bell, G. W., Nam, J.-W., and Bartel, D. P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife 4:e05005.
Antonarakis, E. S., Lu, C., Wang, H., Luber, B., Nakazawa, M., Roeser, J. C., et al. (2014). AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. New Engl. J. Med. 371, 1028–1038.
Bahn, J. H., Zhang, Q., Li, F., Chan, T. M., Lin, X., Kim, Y., et al. (2015). The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 61, 221–230. doi: 10.1373/clinchem.2014.230433
Barboro, P., Benelli, R., Tosetti, F., Costa, D., Capaia, M., Astigiano, S., et al. (2020). Aspartate β-hydroxylase targeting in castration resistant prostate cancer modulates the NOTCH/HIF1α/GSK3β crosstalk. Carcinogenesis 41, 1246–1252. doi: 10.1093/carcin/bgaa053
Barrett, S. P., Wang, P. L., and Salzman, J. (2015). Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife 4:e07540.
Bii, V. M., Collins, C. P., Hocum, J. D., and Trobridge, G. D. (2018). Replication-incompetent gammaretroviral and lentiviral vector-based insertional mutagenesis screens identify prostate cancer progression genes. Oncotarget 9, 15451–15463. doi: 10.18632/oncotarget.24503
Budd, W. T., Seashols-Williams, S. J., Clark, G. C., Weaver, D., Calvert, V., Petricoin, E., et al. (2015). Dual Action of miR-125b as a tumor suppressor and OncomiR-22 promotes prostate cancer tumorigenesis. PLoS One 10:e0142373. doi: 10.1371/journal.pone.0142373
Chen, S., Huang, V., Xu, X., Livingstone, J., Soares, F., Jeon, J., et al. (2019). Widespread and functional RNA circularization in localized prostate cancer. Cell 176, 831–843.e22.
Di Nunno, V., Gatto, L., Santoni, M., Cimadamore, A., Lopez-Beltran, A., Cheng, L., et al. (2018). Recent advances in liquid biopsy in patients with castration resistant prostate cancer. Front. Oncol. 8:397. doi: 10.3389/fonc.2018.00397
Dong, Y., He, D., Peng, Z., Peng, W., Shi, W., Wang, J., et al. (2017). Circular RNAs in cancer: an emerging key player. J. Hematol. Oncol. 10:2.
Du, H., Wang, X., Dong, R., Hu, D., and Xiong, Y. (2019). miR-601 inhibits proliferation, migration and invasion of prostate cancer stem cells by targeting KRT5 to inactivate the Wnt signaling pathway. Int. J. Clin. Exp. Pathol. 12, 4361–4379.
Du, Y., Bi, W., Zhang, F., Wu, W., Xia, S., and Liu, H. G. - (2015). protein-coupled receptor 137 accelerates proliferation of urinary bladder cancer cells in vitro. Biotechnol. Appl. Biochem. 62, 855–860. doi: 10.1002/bab.1329
Fabris, L., Ceder, Y., Chinnaiyan, A. M., Jenster, G. W., Sorensen, K. D., Tomlins, S., et al. (2016). The potential of MicroRNAs as prostate cancer biomarkers. Eur. Urol. 70, 312–322.
Fotouhi Ghiam, A., Taeb, S., Huang, X., Huang, V., Ray, J., Scarcello, S., et al. (2017). Long non-coding RNA urothelial carcinoma associated 1 (UCA1) mediates radiation response in prostate cancer. Oncotarget 8, 4668–4689. doi: 10.18632/oncotarget.13576
Fu, D., Shi, Y., Liu, J.-B., Wu, T.-M., Jia, C.-Y., Yang, H.-Q., et al. (2020). Targeting long Non-coding RNA to therapeutically regulate gene expression in cancer. Mol. Ther. Nucleic Acids 21, 712–724. doi: 10.1016/j.omtn.2020.07.005
Glazar, P., Papavasileiou, P., and Rajewsky, N. (2014). CircBase: a database for circular RNAs. RNA 20, 1666–1670. doi: 10.1261/rna.043687.113
Grabowska, M. M., Elliott, A. D., DeGraff, D. J., Anderson, P. D., Anumanthan, G., Yamashita, H., et al. (2014). NFI transcription factors interact with FOXA1 to regulate prostate-specific gene expression. Mol. Endocrinol. 28, 949–964. doi: 10.1210/me.2013-1213
Greene, J., Baird, A. M., Brady, L., Lim, M., Gray, S. G., McDermott, R., et al. (2017). Circular RNAs: biogenesis, function and role in human diseases. Front. Mol. Biosci. 4:38. doi: 10.3389/fmolb.2017.00038
Greene, J., Baird, A. M., Casey, O., Brady, L., Blackshields, G., Lim, M., et al. (2019). Circular RNAs are differentially expressed in prostate cancer and are potentially associated with resistance to enzalutamide. Sci. Rep. 9:10739.
Griner, N. B., Young, D., Chaudhary, P., Mohamed, A. A., Huang, W., Chen, Y., et al. (2015). ERG oncoprotein inhibits ANXA2 expression and function in prostate cancer. Mol. Cancer Res. 13, 368–379. doi: 10.1158/1541-7786.mcr-14-0275-t
Hansen, T. B., Kjems, J., and Damgaard, C. K. (2013). Circular RNA and miR-7 in cancer. Cancer Res. 73, 5609–5612.
He, J. Y., Wei, X. H., Li, S. J., Liu, Y., Hu, H. L., Li, Z. Z., et al. (2018). Adipocyte-derived IL-6 and leptin promote breast cancer metastasis via upregulation of Lysyl Hydroxylase-2 expression. Cell Commun. Signal. 16:100.
Hudson, B. D., Hum, N. R., Thomas, C. B., Kohlgruber, A., Sebastian, A., Collette, N. M., et al. (2015). SOST inhibits prostate cancer invasion. PLoS One 10:e0142058. doi: 10.1371/journal.pone.0142058
Imberg-Kazdan, K., Ha, S., Greenfield, A., Poultney, C. S., Bonneau, R., Logan, S. K., et al. (2013). A genome-wide RNA interference screen identifies new regulators of androgen receptor function in prostate cancer cells. Genome Res. 23, 581–591. doi: 10.1101/gr.144774.112
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. doi: 10.1261/rna.035667.112
Jiang, Z., Zhang, Y., Chen, X., Wu, P., and Chen, D. (2020). Long noncoding RNA RBMS3-AS3 acts as a microRNA-4534 sponge to inhibit the progression of prostate cancer by upregulating VASH1. Gene Ther. 27, 143–156. doi: 10.1038/s41434-019-0108-1
Jin, W., Chen, F., Wang, K., Song, Y., Fei, X., and Wu, B. (2018). miR-15a/miR-16 cluster inhibits invasion of prostate cancer cells by suppressing TGF-β signaling pathway. Biomed. Pharmacother. 104, 637–644. doi: 10.1016/j.biopha.2018.05.041
John, B., Enright, A. J., Aravin, A., Tuschl, T., Sander, C., and Marks, D. S. (2004). Human MicroRNA targets. PLoS Biol. 2:e363. doi: 10.1371/journal.pbio.0020363
Knight, J. F., Shepherd, C. J., Rizzo, S., Brewer, D., Jhavar, S., Dodson, A. R., et al. (2008). TEAD1 and c-Cbl are novel prostate basal cell markers that correlate with poor clinical outcome in prostate cancer. Br. J. Cancer 99, 1849–1858. doi: 10.1038/sj.bjc.6604774
Kong, Z., Wan, X., Lu, Y., Zhang, Y., Huang, Y., Xu, Y., et al. (2020). Circular RNA circFOXO3 promotes prostate cancer progression through sponging miR-29a-3p. J. Cell. Mol. Med. 24, 799–813. doi: 10.1111/jcmm.14791
Kurozumi, A., Goto, Y., Matsushita, R., Fukumoto, I., Kato, M., Nishikawa, R., et al. (2016). Tumor-suppressive microRNA-223 inhibits cancer cell migration and invasion by targeting ITGA3/ITGB1 signaling in prostate cancer. Cancer Sci. 107, 84–94. doi: 10.1111/cas.12842
Leblanc, R., Sahay, D., Houssin, A., Machuca-Gayet, I., and Peyruchaud, O. (2018). Autotaxin-β interaction with the cell surface via syndecan-4 impacts on cancer cell proliferation and metastasis. Oncotarget 9, 33170–33185. doi: 10.18632/oncotarget.26039
Legnini, I., Di Timoteo, G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., et al. (2017). Circ-ZNF609 Is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66, 22–37.e9.
Lim, M. C. J., Baird, A.-M., Aird, J., Greene, J., Kapoor, D., Gray, S. G., et al. (2018). RNAs as candidate diagnostic and prognostic markers of prostate cancer—from cell line models to liquid biopsies. Diagnostics 8:60. doi: 10.3390/diagnostics8030060
Lin, H.-M., Mahon, K. L., Spielman, C., Gurney, H., Mallesara, G., Stockler, M. R., et al. (2017). Phase 2 study of circulating micicroRNA biomarkers in castration-resistant prostate cancer. Br. J. Cancer 116:1002. doi: 10.1038/bjc.2017.50
Martiny, P. B., Alcoba, D. D., Neto, B. S., Carvalho, P. C., and Brum, I. S. A. (2018). Proteomic glimpse into the oncogenesis of prostate cancer. J. Appl. Biomed. 16, 328–336. doi: 10.1016/j.jab.2018.05.003
Mehra, R., Udager, A. M., Ahearn, T. U., Cao, X., Feng, F. Y., Loda, M., et al. (2016). Overexpression of the long Non-coding RNA SChLAP1 independently predicts lethal prostate cancer. Eur. Urol. 70, 549–552. doi: 10.1016/j.eururo.2015.12.003
Misawa, A., Takayama, K. I., and Inoue, S. (2017). Long non-coding RNAs and prostate cancer. Cancer Sci. 108, 2107–2114. doi: 10.1111/cas.13352
Muharram, G., Sahgal, P., Korpela, T., De Franceschi, N., Kaukonen, R., Clark, K., et al. (2014). Tensin-4-dependent MET stabilization is essential for survival and proliferation in carcinoma cells. Dev. Cell. 29, 421–436. doi: 10.1016/j.devcel.2014.03.024
Oh, S. Y., Kang, S. M., Kang, S. H., Lee, H. J., Kwon, T. G., Kim, J. W., et al. (2020). Potential salivary mRNA biomarkers for early detection of oral cancer. J. Clin. Med. 9:243. doi: 10.3390/jcm9010243
O’Sullivan, J. (2017). Controversies in PSA screening. Evid. Based Med. 22:198. doi: 10.1136/ebmed-2017-110858
Ramnarine, V. R., Kobelev, M., Gibb, E. A., Nouri, M., Lin, D., Wang, Y., et al. (2019). The evolution of long noncoding RNA acceptance in prostate cancer initiation, progression, and its clinical utility in disease management. Eur. Urol. 76, 546–559. doi: 10.1016/j.eururo.2019.07.040
Ravipaty, S., Wu, W., Dalvi, A., Tanna, N., Andreazi, J., Friss, T., et al. (2017). Clinical validation of a serum protein panel (FLNA, FLNB and KRT19) for diagnosis of prostate cancer. J. Mol. Biomark. Diagn. 8:323.
Rodgers, J. J., McClure, R., Epis, M. R., Cohen, R. J., Leedman, P. J., Harvey, J. M., et al. (2019). ETS1 induces transforming growth factor β signaling and promotes epithelial-to-mesenchymal transition in prostate cancer cells. J. Cell. Biochem. 120, 848–860. doi: 10.1002/jcb.27446
Romanel, A., Gasi Tandefelt, D., Conteduca, V., Jayaram, A., Casiraghi, N., Wetterskog, D., et al. (2015). Plasma AR and abiraterone-resistant prostate cancer. Sci. Transl. Med. 7:312re10.
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N., and Brown, P. O. (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7:e30733. doi: 10.1371/journal.pone.0030733
Sawada, Y., Kikugawa, T., Iio, H., Sakakibara, I., Yoshida, S., Ikedo, A., et al. (2020). GPRC5A facilitates cell proliferation through cell cycle regulation and correlates with bone metastasis in prostate cancer. Intern. J. Cancer J. Intern. Cancer 146, 1369–1382. doi: 10.1002/ijc.32554
Shi, Q., Zhu, Y., Ma, J., Chang, K., Ding, D., Bai, Y., et al. (2019). Prostate cancer-associated SPOP mutations enhance cancer cell survival and docetaxel resistance by upregulating Caprin1-dependent stress granule assembly. Mol. Cancer 18:170.
Shih, J. W., Wang, L. Y., Hung, C. L., Kung, H. J., and Hsieh, C. L. (2015). Non-Coding RNAs in castration-resistant prostate cancer: regulation of androgen receptor signaling and cancer metabolism. Intern. J. Mol. Sci. 16, 28943–28978. doi: 10.3390/ijms161226138
Su, M., Xiao, Y., Ma, J., Tang, Y., Tian, B., Zhang, Y., et al. (2019). Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 18:90.
Szabo, L., and Salzman, J. (2016). Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 17, 679–692. doi: 10.1038/nrg.2016.114
Thomas, B. C., Kay, J. D., Menon, S., Vowler, S. L., Dawson, S. N., Bucklow, L. J., et al. (2016). Whole blood mRNA in prostate cancer reveals a four-gene androgen regulated panel. Endocr. Relat. Cancer 23, 797–812. doi: 10.1530/erc-16-0287
Tucker, D., Zheng, W., Zhang, D.-H., and Dong, X. (2020). Circular RNA and its potential as prostate cancer biomarkers. World J. Clin. Oncol. 11, 563–572. doi: 10.5306/wjco.v11.i8.563
Warner, J. R. (1999). The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440. doi: 10.1016/s0968-0004(99)01460-7
White, N. M., Zhao, S. G., Zhang, J., Rozycki, E. B., Dang, H. X., McFadden, S. D., et al. (2017). Multi-institutional analysis shows that low PCAT-14 expression associates with poor outcomes in prostate cancer. Eur. Urol. 71, 257–266. doi: 10.1016/j.eururo.2016.07.012
Yang, N., Wang, L., Liu, J., Liu, L., Huang, J., Chen, X., et al. (2018). MicroRNA-206 regulates the epithelial-mesenchymal transition and inhibits the invasion and metastasis of prostate cancer cells by targeting Annexin A2. Oncol. Lett. 15, 8295–8302.
Zhang, C., Xiong, J., Yang, Q., Wang, Y., Shi, H., Tian, Q., et al. (2018). Profiling and bioinformatics analyses of differential circular RNA expression in prostate cancer cells. Future Sci. OA 4:FSOA340-FSOA.
Zheng, X., Jia, B., Lin, X., Han, J., Qiu, X., Chu, H., et al. (2016). FRMD4A: a potential therapeutic target for the treatment of tongue squamous cell carcinoma. Int. J. Mol. Med. 38, 1443–1449. doi: 10.3892/ijmm.2016.2745
Keywords: prostate cancer, circRNA, non-coding RNA, biomarkers, androgen signaling
Citation: Greene J, Baird AM, Lim M, Flynn J, McNevin C, Brady L, Sheils O, Gray SG, McDermott R and Finn SP (2021) Differential CircRNA Expression Signatures May Serve as Potential Novel Biomarkers in Prostate Cancer. Front. Cell Dev. Biol. 9:605686. doi: 10.3389/fcell.2021.605686
Received: 12 September 2020; Accepted: 05 February 2021;
Published: 25 February 2021.
Edited by:
Mingli Liu, Morehouse School of Medicine, United StatesReviewed by:
Zoran Culig, Innsbruck Medical University, AustriaCopyright © 2021 Greene, Baird, Lim, Flynn, McNevin, Brady, Sheils, Gray, McDermott and Finn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John Greene, Z3JlZW5lam9AdGNkLmll
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.