
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 18 January 2021
Sec. Signaling
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.632632
This article is part of the Research Topic With the Eyes on Non-Coding RNAs View all 13 articles
The use of model systems that are capable of robust, spontaneous retina regeneration has allowed for the identification of genetic pathways and components that are required for retina regeneration. Complemented by mouse models in which retina regeneration can be induced after forced expression of key factors, altered chromatin accessibility, or inhibition of kinase/signaling cascades, a clearer picture of the key regulatory events that control retina regeneration is emerging. In all cases, Müller glia (MG) serve as an adult retinal stem cell that must be reprogrammed to allow for regeneration, with the end goal being to understand why regenerative pathways are blocked in mammals, but spontaneous in other vertebrates such as zebrafish. miRNAs have emerged as key gene regulatory molecules that control both development and regeneration in vertebrates. Here, we focus on a small subset of miRNAs that control MG reprogramming during retina regeneration and have the potential to serve as therapeutic targets for treatment of visual disorders and damage.
In mammals and humans, the extent of spontaneous repair after retina injury or disease is either non-existent or extremely limited (Karl and Reh, 2010). Rather than regenerate, damaged mammalian retinas commonly undergo reactive gliosis and scar formation (Bringmann et al., 2006). This lack of a complete regenerative response to damage directly limits the treatment options for retinal based diseases such as age-related macular degeneration or Stargardt's disease (Link and Collery, 2015; Zarbin, 2016). Numerous strategies are currently being tested to address this limitation, including gene therapy approaches and transplantation of stem cell-derived progenitor cells (MacLaren et al., 2006; Pearson et al., 2012; Cehajic-Kapetanovic et al., 2015; Roska and Sahel, 2018; Stern et al., 2018). An attractive alternative strategy for treatment is to induce endogenous MG-derived regeneration of the retina as is observed in fish and amphibians (Hamon et al., 2016; Lahne et al., 2020). Zebrafish have the ability to regenerate a large array of tissues and organs (Gemberling et al., 2013). One goal for these studies is to determine the factors and pathways that allow for persistent and spontaneous regeneration. Focusing on the retina, knowledge gained from zebrafish studies (Wan and Goldman, 2016; Yao et al., 2018; Hoang et al., 2020; VandenBosch et al., 2020; Zhou et al., 2020) can be applied to identify common mechanisms that induce mammalian retina regeneration. Here, we will focus on the explicit role of miRNAs during MG reprogramming.
The retina forms from the central nervous system (CNS) and develops into a three-layered structure consisting of seven main types of cells and numerous other cell types identified by single cell RNAseq (Macosko et al., 2015). The structure, function, cell types, and genes expressed in the retina are largely conserved among vertebrates, supporting the notion that information gained from models capable of spontaneous regeneration might apply to mammals whose regenerative capacity isn't clear (Hitchcock and Raymond, 2004; Stenkamp, 2007; Hamon et al., 2016).
The three main layers that constitute the retina include the outer nuclear layer (ONL), inner nuclear layer (INL), and ganglion cell layer (GCL). The INL and ONL are separated by a thin synaptic layer called the outer plexiform layer (OPL), and the INL and GCL are separated by a thick synaptic layer called the inner plexiform layer (IPL). The ONL contains rod and cone photoreceptors. The INL contains three types of interneurons: bipolar cells (BCs), horizontal cells (HCs), and amacrine cells (ACs). Ganglion cells (GCs) populate the GCL, collect information from BCs, and send signals to the brain for higher order visual processing. In addition to these neuronal cell types, Müller glia (MG) constitute the main glial cell type spanning all three layers of the retina.
The unique behavior and placement of MG following damage led to hypotheses that they play an integral role in retina regeneration. Multiple lines of evidence, largely from zebrafish, strongly support MG as the source of retina progenitors after damage. First, new retinal progenitor cells (RPCs) formed in the INL migrate along MG processes to the ONL (Raymond and Rivlin, 1987; Vihtelic and Hyde, 2000; Wu et al., 2001; Raymond et al., 2006). Second, MG become mitotic after damage (Braisted et al., 1994; Vihtelic and Hyde, 2000; Wu et al., 2001; Faillace et al., 2002; Yurco and Cameron, 2005; Raymond et al., 2006). Third, gene expression profiles of MG and RPCs are very similar (Hoang et al., 2020). Most directly, the Raymond lab showed that zebrafish MG produce rod precursors during development and also produce RPCs that can differentiate into any retina cell type following damage (Bernardos et al., 2007). The mechanism by which MG produce RPCs is by dedifferentiation of the MG, owing to the fact that shortly after damage, zebrafish MG begin to produce markers of neural progenitors such as Pax6, α-tubulin, and BLBP (Fausett and Goldman, 2006; Raymond et al., 2006; Thummel et al., 2010).
Following damage in zebrafish, signaling cascades induce MG to dedifferentiate to a stem cell-like state and reenter the cell cycle, followed by asymmetric division for self-renewal and for the generation of proliferating RPCs (Nagashima et al., 2013). These cells cluster along MG processes and then migrate to sites of damage where they exit the cell cycle and differentiate into new cells that can replace any damaged cell type (Figure 1; Fausett and Goldman, 2006; Bernardos et al., 2007; Thummel et al., 2008b, 2010; Montgomery et al., 2010; Ramachandran et al., 2010; Qin et al., 2011; Powell et al., 2012; Taylor et al., 2012). Though the current understanding of retina regeneration is ongoing, a number of factors have been identified that transition the retina through the various stages of MG-derived retina regeneration (Wan and Goldman, 2016; Lahne et al., 2020; Figure 2). Here, we will focus on the identification and role of miRNAs during MG reprogramming and retina regeneration.
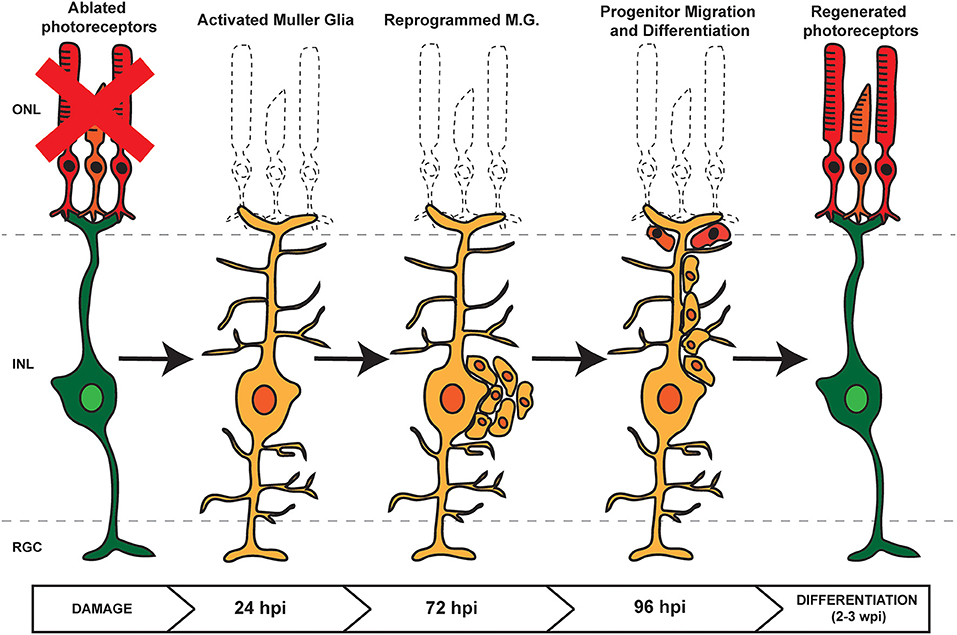
Figure 1. Retina Regeneration in Zebrafish. In response to retinal damage or cell loss, zebrafish Müller Glia (MG) are activated and undergo dedifferentiation. Asymmetric division allows for self renewal and the generation of proliferating retinal progenitor cells (RPCs) which cluster along MG processes. As regeneration proceeds, the RPCs migrate to the site of damage before differentiating into any lost or damaged cell types.
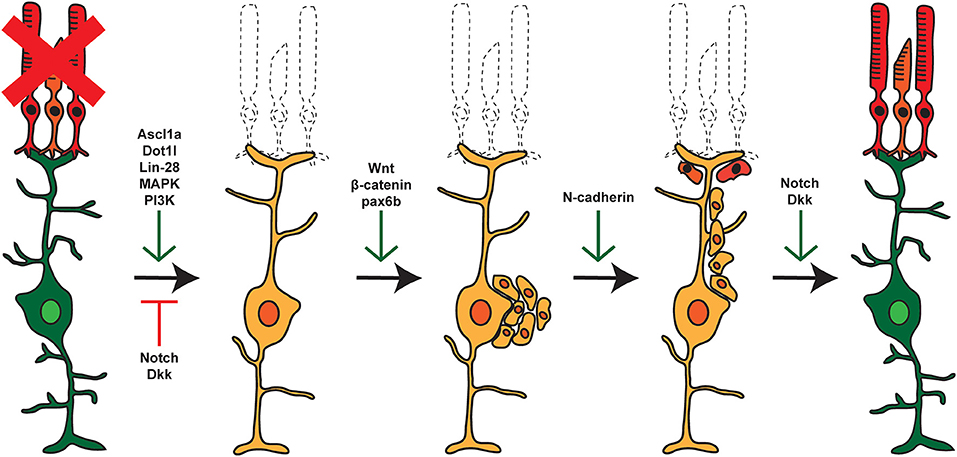
Figure 2. Pathways and factors involved in retina regeneration. Following damage in the zebrafish retina, multiple pathways are activated controlling dedifferentiation of MG, re-entry into the cell cycle, asymmetric cell division, generation of proliferating RPCs, and eventual differentiation into replacement cell types. Green arrows represent factors that are active at the given step, whereas red arrows represent factors that are repressed or inactive at that time.
miRNAs are highly conserved ~22 nucleotide (nt) RNAs that post-transcriptionally regulate gene expression (Krol et al., 2010; Bartel, 2018; Gebert and MacRae, 2019). Primary miRNA transcripts are initially processed in the nucleus into ~70 nt precursor structures by a multi-protein complex referred to as the Microprocessor, the main component of which is Drosha (Kim, 2005). After export from the nucleus, cytoplasmic processing is accomplished by another multi-protein complex that includes the enzyme Dicer, which yields ~22 nt double stranded RNAs. One of the strands is subsequently assembled into an RNA Induced Silencing Complex (RISC) (Schwarz et al., 2003; Filipowicz, 2005) containing one or more members of the Argonaute protein family (Peters and Meister, 2007). miRNA-mediated gene silencing occurs by pairing between miRNAs and their target mRNAs, usually in the 3′ UTR. Once paired, miRNAs inhibit translation and induce deadenylation leading to mRNA degradation (Giraldez et al., 2006; Guo et al., 2010).
miRNAs were first discovered in C. elegans where they control development (Lee et al., 1993; Wightman et al., 1993), but they have now been shown to play important roles in a number of biological processes including metabolism, cancer, metastasis, and regeneration (Alvarez-Garcia and Miska, 2005). miRNAs have been implicated in regeneration in a number of biological models ranging from planaria to mice (Yin and Poss, 2008; Williams et al., 2009; Thatcher and Patton, 2010) and in zebrafish have been shown to regulate regeneration of the heart, fin, muscle, liver, lens, and inner ear hair cells (Tsonis et al., 2007; Liu et al., 2008, 2012; Thatcher et al., 2008; Yin et al., 2008; Song et al., 2010). Knockdown of Dicer in the adult zebrafish retina prior to constant intense light damage reduced the ability of MG to produce proliferating RPCs in response to damage (Rajaram et al., 2014a). This indicated a general requirement for miRNAs during retina regeneration. After damage, most miRNA expression levels remain unchanged or undergo only small changes during regeneration. However, specific subsets of zebrafish miRNAs show both up- and down-regulation throughout the regenerative process (Figure 3; Rajaram et al., 2014a). Similarly, a small subset of miRNAs have been implicated in controlling the reprogramming of mammalian Müller glia (Wohl and Reh, 2016b; Wohl et al., 2019).
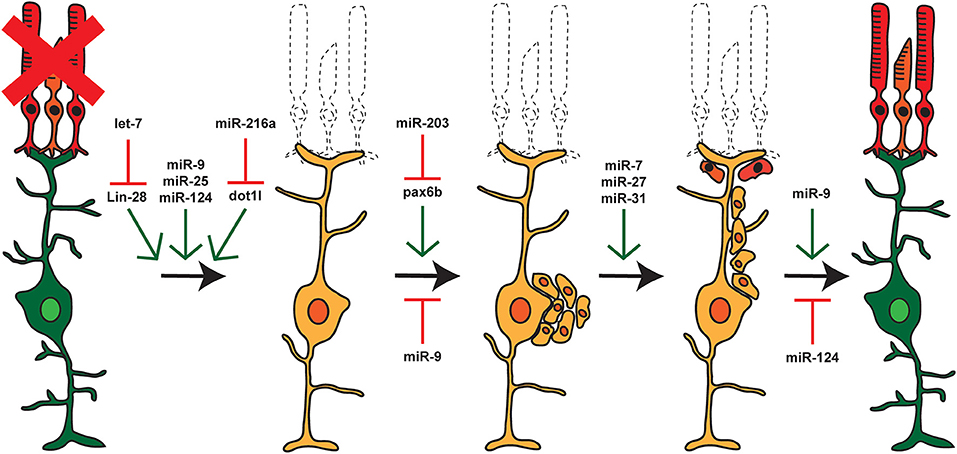
Figure 3. miRNAs and Retina Regeneration. The adult zebrafish retina expresses ~200 miRNAs but only a small subset of these miRNAs are differentially expressed during regeneration. RNAseq and other analyses after damage or during distinct stages of regeneration have identified miRNAs and select target mRNAs, as indicated. Green arrows represent miRNAs that are expressed or active at that given step, and red arrows represent miRNAs that are not expressed or inactive.
One of the first demonstrations of a role for miRNA involvement in retina regeneration was by the Goldman lab focusing on let-7 (Ramachandran et al., 2010). Using a puncture damage model in zebrafish, they used a candidate gene approach to identify pluripotency factors whose expression changes during retina regeneration. One of those factors was Lin-28, an RNA-binding protein that was first discovered to regulate development in C. elegans as part of a double negative feedback loop with let-7 (Moss et al., 1997; Reinhart et al., 2000). After retina damage, Lin-28 is induced downstream of the transcription factor Ascl1 and knockdown of Lin-28 inhibits retina regeneration (Ramachandran et al., 2010; Zhao et al., 2017). Lin-28 activation leads to the repression of let-7 expression which in turn derepresses the expression of multiple regeneration and pluripotency factors including Ascl1a (Ramachandran et al., 2010). Let-7 has 12 family members; Lin-28 inhibits the production of most let-7 family members by recruiting a uridylyl transferase to pre-let-7 transcripts leading to uridylation, inhibition of processing, and subsequent decay (Hagan et al., 2009; Heo et al., 2009). For MG, it appears that expression of let-7 maintains the differentiated state, but that induction of Lin-28 after injury allows dedifferentiation by reducing let-7 levels. This is consistent with a role for regulation of Lin-28 by let-7 during development as well (La Torre et al., 2013; Fairchild et al., 2019).
In a screen to identify differentially expressed miRNAs during zebrafish MG-derived retina regeneration, miR-203 was found to be downregulated and that artificially maintaining its expression blocked retina regeneration (Rajaram et al., 2014b). Previously, miR-203 downregulation had been shown to be required for caudal fin regeneration and it is similarly downregulated during mouse skin regeneration (Lena et al., 2008; Thatcher et al., 2008; Yi et al., 2008). These data supported a role for miR-203 in promoting differentiation and repressing stemness. Elevated levels of miR-203 inhibit proliferation of RPCs and are essential for the formation of clusters of RPCs that are commonly observed along MG processes (Figure 1; Rajaram et al., 2014b). However, miR-203 does not play a role in dedifferentiation of MG during earlier stages of regeneration, indicating a temporal expression pattern throughout the regenerative processes.
In a search for potential mRNA targets for miR-203, bioinformatic and reporter analyses identified the Paired-box gene 6b (Pax6b) (Rajaram et al., 2014b). Pax6 expression is essential for eye development across species (Shaham et al., 2012; Baker et al., 2018). There are two Pax6 paralogs in zebrafish (Pax6a and Pax6b) with distinct roles during retina regeneration (Thummel et al., 2010). Misregulation or loss of Pax6 expression can lead to multiple mammalian visual system defects, most commonly aniridia or nystagmus (Lima Cunha et al., 2019). Consistent with a role for miR-203 controlling progenitor proliferation and cluster formation, the Hyde lab had previously shown that pax6b in zebrafish is expressed in NPCs and is required for the formation of clusters (Thummel et al., 2008a). miR-203 must be repressed during regeneration to allow for pax6b expression and the formation of RPC clusters on MG.
The Fu laboratory discovered that repression of the hnRNP protein Polypyrimidine tract-binding protein 1 (PTB1) can convert fibroblasts into a neuronal cell type fate (Xue et al., 2013). PTB1 is a ubiquitously expressed regulator of alternative splicing that itself undergoes alternative splicing to autoregulate its levels (Wollerton et al., 2004). A neuronal paralog of PTB (PTB2 or nPTB) controls multiple neuronal alternative splicing events and its levels are regulated by an alternative splicing event controlled by PTB1 (Boutz et al., 2007; Makeyev et al., 2007). PTB1 is expressed in neuronal precursor cells and glia; nPTB is expressed during neuronal induction and maturation, with the expression of both paralogs decreasing during neuronal differentiation (Boutz et al., 2007; Hu et al., 2018). Two regulatory loops control PTB and nPTB expression through the action of miR-9 and miR-124, respectively, along with the transcription factors REST and BRN2 (Makeyev et al., 2007; Hu et al., 2018). Intriguingly, it was recently shown that targeted destruction of mRNAs encoding PTB in the mouse retina can cause MG to dedifferentiate (Zhou et al., 2020). Using an NMDA damage model in mice, targeting of mRNAs encoding PTB by CRISPR/CasRx led to MG dedifferentiation and replacement of damaged ganglion cells. Also, targeted depletion of mRNAs encoding PTB by antisense oligonucleotides led to the conversion of astrocytes to dopaminergic neurons (Qian et al., 2020). Together, the data support the surprising finding that targeting a single, widely expressed regulator of alternative splicing can drive the conversion of glia to neurons.
Even though targeting of PTB provides an attractive single gene approach for retina regeneration, an alternative would be to deliver miR-9 and miR-124 to not only regulate PTB and nPTB expression, but also to upregulate the transcription factors REST and BRN2 (Hu et al., 2018). It will be crucial to determine whether the expression of PTB and nPTB are controlled in the retina by the miR-9 and miR-124 regulatory loops. If so, regulation of REST by these regulatory loops could control expression of NeuroD which plays a role in neuronal cell fate, and Ascl1 which is required for retina regeneration (Ramachandran et al., 2010; Cherry et al., 2011). Similarly, regulation of BRN2 could in turn control expression of neuronal maturation genes including NEUN and NLGN2. Gene profiling experiments did not observe significant changes in PTB between control and neurogenic MG (Hoang et al., 2020) so it may be that increased expression or delivery of miR-9 and miR-124 mimics might drive broader overall gene expression changes to induce retina regeneration rather than just targeting PTB (Wohl and Reh, 2016b).
The Reh lab used dissociated mouse MG cultures to identify factors and miRNAs that can stimulate reprogramming of retinal cell fate (Pollak et al., 2013; Wohl and Reh, 2016b). Induced overexpression of the transcription factor Asc1 can reprogram mouse MG into neurogenic RPCs and the effects of Ascl1 overexpression can be augmented by parallel overexpression of both miR-124 and miR-9 (Wohl and Reh, 2016b). This agrees nicely with the regulatory loops controlling neuronal induction and maturation controlled by miR-124 and miR-9 but to date, whether these loops control gene expression in the retina is not clear (Hu et al., 2018).
In a follow up study to the effects of miR-9 and miR-124 on reprogramming of mammalian MG, the Reh lab profiled miRNA expression patterns in sorted mouse MG and RPCs, and also utilized a conditional mouse model with a MG-specific deletion of Dicer (Wohl and Reh, 2016a; Wohl et al., 2017, 2019). After Dicer knockdown, the most significantly altered gene was Brevican (BCAN) which is targeted by miR-9. More broadly, loss of Dicer led to a dramatic loss of retinal architecture indicating an important role for miRNAs in the maintenance of homeostasis, and also supporting the overall importance of miRNAs in regeneration, a process which is blocked by the loss of Dicer (Rajaram et al., 2014a). When comparing sorted MG and RPCs, Ascl1 expression was found to be enhanced by either overexpression of miR-25 and miR-124 or by downregulation of let-7 (Wohl et al., 2019). Targeting of Lin-28 by let-7 can explain indirect regulation of Ascl1.
miR-25 is expressed as part of the highly conserved miR-106b/25 cluster with roles in DNA damage response, cell cycle regulation, cell proliferation, migration, and differentiation (Sarkozy et al., 2018). Additional potential target mRNAs for miR-25 include REST, Tpm1, Itgb1, Ctdsp1, Rcor1, and Ccnd2 (Sarkozy et al., 2018; Wohl et al., 2019). Besides targeting the protein components of the REST complex, another key predicted target of miR-25 (and let-7) is the Wnt inhibitor Dickkopf 3 (Dkk3) (Huo et al., 2016; Wohl et al., 2019). This is consistent with the requirement for Wnt activation during retina regeneration in zebrafish and possibly mice (Osakada et al., 2007; Ramachandran et al., 2011; Kara et al., 2019).
miR-124 is one of the most abundant miRNAs in the adult brain and is thought to be a master regulator of neuronal differentiation, including its role in regulating PTB expression (Yeom et al., 2018) and targeting of REST (Wohl et al., 2019). miR-124 is known to reduce the expression of a small phosphatase specific for phosphoserines in the C-terminus of RNA Polymerase II called SCP1, which is a repressor of neuron-specific transcription in nonneuronal cells and is also a component of REST (Cao et al., 2007; Makeyev et al., 2007; Visvanathan et al., 2007).
miR-216a is another well-known miRNA that plays a role in gliogenesis during retinal development by indirectly regulating Notch signaling (Olena et al., 2015). For MG-derived regeneration, miR-216a can be thought of as a gatekeeper miRNA in reprogramming events, as its expression holds MG in a quiescent state until the retina is damaged (Kara et al., 2019). One mechanism for how miR-216a can serve as a gatekeeper controlling the early steps of regeneration is by targeting mRNAs encoding the Disruptor of telomeric silencing 1-like (DOT1l) gene (Kara et al., 2019). Dot1l plays a role in many chromatin-associated functions such as gene-transcription, heterochromatin formation, and DNA repair, as well as the response to DNA damage and chemotherapy responsiveness (McLean et al., 2014). After retinal damage in zebrafish, miR-216a is downregulated allowing increased expression of Dot1l which leads to activation of Wnt target genes, presumably by altering chromatin accessibility surrounding these genes (Kara et al., 2019). The idea that dedifferentiation of MG involves changes in chromatin accessibility is expected and has been experimentally supported (Jorstad et al., 2017; Mitra et al., 2018; VandenBosch et al., 2020). When combined with Ascl1 overexpression in an NMDA damage model in adult mice, the addition of the general histone deactylase inhibitor trichostatin A (TSA) stimulated neuronal regeneration which was otherwise only observed in developing mice <12 days old (Jorstad et al., 2017). Also, after puncture damage in zebrafish, inhibition of histone deacetylases by valproic acid suppressed the formation of MG-derived NPCs (Mitra et al., 2018). Further work is needed to identify specific genes whose chromatin accessibility changes during MG dedifferentiation and eventual re-differentiation, but miRNA control is an attractive regulatory mechanism that might allow for fine-tuned control of signaling cascades that are induced after cellular damage.
In a screen to identify differentially expressed miRNAs during zebrafish retina regeneration, miR-7, miR-27, and miR-31 were all found to be upregulated at 72 h post light damage and targeted knockdown of these miRNAs led to decreased numbers of proliferating cells (Rajaram et al., 2014a). The timing of overexpression and the effects of loss of function of these miRNAs during regeneration suggest that they function during continued RPC proliferation and migration, similar to the proposed role for Pax6a (Thummel et al., 2010; Rajaram et al., 2014b). Exact targets for these miRNAs remain to be identified but related experiments summarized below might provide hints to possible mRNA targets for these miRNAs.
miR-7 regulates multiple signaling pathways including epidermal growth factor receptor (EGFR), insulin-like growth factor (IGF), Hedgehog, Notch, and the mammalian target of rapamycin (mTOR) pathways, as well as being a key regulator of pax6a in mice (Needhamsen et al., 2014; Baba et al., 2015; Zhao et al., 2015). In the forebrain, miR-7 regulates pax6 to spatially control the origin of dopaminergic neurons (de Chevigny et al., 2012).
miR-27 promotes blood vessel development, particularly in the eye (Liu et al., 2020). miR-27 also plays an important role in mitochondrial dynamics as it inhibits degradation of damaged mitochondria by regulating PINK1 and also by inhibiting mitochondrial fission factor (MFF) expression, which increases mitochondrial membrane potential (Tak et al., 2014; Kim et al., 2016). Loss of miR-27c in the retina decreases proliferation of MG-derived RPCs during regeneration, a similar phenotype to what occurs with loss of miR-27a and miR-27b in muscle progenitor cell proliferation (Crist et al., 2009; Lozano-Velasco et al., 2011; Rajaram et al., 2014a).
miR-31 is a well-studied miRNA with a major target being transcripts encoding the myogenic determining factor Myf5 (Crist et al., 2012). miR-31 levels affect both satellite cell differentiation ex vivo and muscle regeneration in vivo, making miR-31 a miRNA of great interest in regard to stem cell research. miR-31 is a regulator of many signaling pathways relevant to developmental biology and cancer including the Prlr/Stat5, TGFβ, and Wnt/β-catenin pathways (Lv et al., 2017). Additionally, miR-31 has been shown to coordinate signals from BMP, TGFβ, and Wnt pathways in intestinal stem cells to regulate their proliferation, regeneration, and homeostasis, further reinforcing its impact in progenitor proliferation during regeneration (Tian et al., 2017).
miRNAs regulate gene expression by binding to 3′ UTR elements leading to deadenylation and subsequent degradation of mRNA targets (Giraldez et al., 2006; Guo et al., 2010). Target recognition typically involves imperfect base pairing, often within the seed region (nucleotides 2-8) that is commonly used to predict miRNA targets (Li et al., 2008; Broughton et al., 2016; Bartel, 2018). Because the base pairing interaction is imperfect, miRNA target prediction algorithms can identify candidate mRNAs, but experimental validation is necessary to confirm direct silencing. Thus, for all of the miRNAs discussed above, there are likely additional mRNA targets that could affect the same processes, ranging from regeneration to signaling cascades. Further work is required to identify the complete set of miRNAs that regulate retina regeneration and the target genes they control.
Because miRNAs are largely conserved among vertebrates, the expection is that discoveries across species will illustrate general principles and uncover common mechanisms. Fortunately, it does not appear that there are hundreds of miRNAs that regulate MG reprogramming and, for the subset that has been identified, they comprise an attractive class of regulatory molecules, especially because retinal architecture and MG gene expression patterns are evolutionarily conserved suggesting that elucidating overall gene regulation will help to understand the inability of mammalian MG to initiate regeneration. It remains possible that species-specific networks or species-specific factors might control the passage of MG from quiescence to reactivity and further to the generation of proliferating RPCs, but the weight of evidence thus far seems to suggest that activating MG-derived regeneration cascades in mammals will be possible by derepression of existing pathways as opposed to delivery of species-specific genes (Ahmad et al., 2011; Lust and Wittbrodt, 2018; Hoang et al., 2020).
The accessibility of the eye and the small size of miRNAs raises the possibility of delivering miRNA mimics or antisense RNAs (antagomirs) that block miRNA function for therapeutic purposes. The challenges for such experiments are at least three-fold: (1) how to target injected miRNAs to MG; (2) whether single injections will be sufficient to induce a regenerative response; and (3), avoidance of off-target effects if high concentrations are required. While direct injections of miRNAs or antagomirs are possible, the discovery that extracellular vesicles (EVs) can be used to deliver therapeutic cargo opens an exciting possibility for cell-specific delivery (Mead and Tomarev, 2020). Recently, it has become increasingly clear that miRNAs can engage in cell-cell signaling via EVs (Maas et al., 2017; O'Brien et al., 2020). Transfer of miRNAs or other cargo by EVs might play a role in patterning the retina during development and may also be a key part of degeneration and regeneration (Bian et al., 2020). Indeed, retina regeneration can be induced by delivery of EVs (Didiano et al., 2020). Although EVs were shown to induce the early stages of retina regeneration, the effects were quite modest. However, as a therapeutic tool, it may be possible to load EVs with specific miRNAs or other small molecules for delivery to MG after intravitreal or subretinal injection. The miRNAs described in this review may be candidate miRNAs for the development of designer EVs that could be targeted to MG to induce retina regeneration.
GK made the figures. All authors contributed to the writing.
This work was supported by a grant from the National Eye Institute within the National Institutes of Health, UO1 EY027265.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ahmad, I., Del Debbio, C. B., Das, A. V., and Parameswaran, S. (2011). Müller glia: a promising target for therapeutic regeneration. Invest. Ophthalmol. Vis. Sci. 52, 5758–5764. doi: 10.1167/iovs.11-7308
Alvarez-Garcia, I., and Miska, E. A. (2005). MicroRNA functions in animal development and human disease. Development 132, 4653–4662. doi: 10.1242/dev.02073
Baba, Y., Aihara, Y., and Watanabe, S. (2015). MicroRNA-7a regulates Muller glia differentiation by attenuating Notch3 expression. Exp. Eye Res. 138, 59–65. doi: 10.1016/j.exer.2015.06.022
Baker, L. R., Weasner, B. M., Nagel, A., Neuman, S. D., Bashirullah, A., and Kumar, J. P. (2018). Eyeless/Pax6 initiates eye formation non-autonomously from the peripodial epithelium. Development 145:dev163329. doi: 10.1242/dev.163329
Bernardos, R. L., Barthel, L. K., Meyers, J. R., and Raymond, P. A. (2007). Late-stage neuronal progenitors in the retina are radial Müller glia that function as retinal stem cells. J. Neurosci. 27, 7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007
Bian, B., Zhao, C., He, X., Gong, Y., Ren, C., Ge, L., et al. (2020). Exosomes derived from neural progenitor cells preserve photoreceptors during retinal degeneration by inactivating microglia. J. Extracell. Vesicles 9:1748931. doi: 10.1080/20013078.2020.1748931
Boutz, P. L., Stoilov, P., Li, Q., Lin, C. H., Chawla, G., Ostrow, K., et al. (2007). A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 21, 1636–1652. doi: 10.1101/gad.1558107
Braisted, J. E., Essman, T. F., and Raymond, P. A. (1994). Selective regeneration of photoreceptors in goldfish retina. Development 120, 2409–2419.
Bringmann, A., Pannicke, T., Grosche, J., Francke, M., Wiedemann, P., Skatchkov, S. N., et al. (2006). Müller cells in the healthy and diseased retina. Prog. Retin. Eye Res. 25, 397–424. doi: 10.1016/j.preteyeres.2006.05.003
Broughton, J. P., Lovci, M. T., Huang, J. L., Yeo, G. W., and Pasquinelli, A. E. (2016). Pairing beyond the seed supports MicroRNA targeting specificity. Mol. Cell 64, 320–333. doi: 10.1016/j.molcel.2016.09.004
Cao, X., Pfaff, S. L., and Gage, F. H. (2007). A functional study of miR-124 in the developing neural tube. Genes Dev. 21, 531–536. doi: 10.1101/gad.1519207
Cehajic-Kapetanovic, J., Eleftheriou, C., Allen, A. E., Milosavljevic, N., Pienaar, A., Bedford, R., et al. (2015). Restoration of vision with ectopic expression of human rod opsin. Curr. Biol. 25, 2111–2122. doi: 10.1016/j.cub.2015.07.029
Cherry, T. J., Wang, S., Bormuth, I., Schwab, M., Olson, J., and Cepko, C. L. (2011). NeuroD factors regulate cell fate and neurite stratification in the developing retina. J. Neurosci. 31, 7365–7379. doi: 10.1523/JNEUROSCI.2555-10.2011
Crist, C. G., Montarras, D., and Buckingham, M. (2012). Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell 11, 118–126. doi: 10.1016/j.stem.2012.03.011
Crist, C. G., Montarras, D., Pallafacchina, G., Rocancourt, D., Cumano, A., Conway, S. J., et al. (2009). Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. PNAS 106, 13383–13387. doi: 10.1073/pnas.0900210106
de Chevigny, A., Core, N., Follert, P., Gaudin, M., Barbry, P., Beclin, C., et al. (2012). miR-7a regulation of Pax6 controls spatial origin of forebrain dopaminergic neurons. Nat. Neurosci. 15, 1120–1126. doi: 10.1038/nn.3142
Didiano, D., Abner, J. J., Hinger, S. A., Flickinger, Z., Kent, M., Clement, M. A., et al. (2020). Induction of a proliferative response in the zebrafish retina by injection of extracellular vesicles. Exp. Eye Res. 200:108254. doi: 10.1016/j.exer.2020.108254
Faillace, M. P., Julian, D., and Korenbrot, J. I. (2002). Mitotic activation of proliferative cells in the inner nuclear layer of the mature fish retina: regulatory signals and molecular markers. J. Comp. Neurol. 451, 127–141. doi: 10.1002/cne.10333
Fairchild, C. L. A., Cheema, S. K., Wong, J., Hino, K., Simo, S., and La Torre, A. (2019). Let-7 regulates cell cycle dynamics in the developing cerebral cortex and retina. Sci. Rep. 9:15336. doi: 10.1038/s41598-019-51703-x
Fausett, B. V., and Goldman, D. (2006). A role for alpha1 tubulin-expressing Muller glia in regeneration of the injured zebrafish retina. J. Neurosci. 26, 6303–6313. doi: 10.1523/JNEUROSCI.0332-06.2006
Filipowicz, W. (2005). RNAi: the nuts and bolts of the RISC machine. Cell 122, 17–20. doi: 10.1016/j.cell.2005.06.023
Gebert, L. F. R., and MacRae, I. J. (2019). Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 20, 21–37. doi: 10.1038/s41580-018-0045-7
Gemberling, M., Bailey, T. J., Hyde, D. R., and Poss, K. D. (2013). The zebrafish as a model for complex tissue regeneration. Trends Genet. 29, 611–620. doi: 10.1016/j.tig.2013.07.003
Giraldez, A. J., Mishima, Y., Rihel, J., Grocock, R. J., Van Dongen, S., Inoue, K., et al. (2006). Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79. doi: 10.1126/science.1122689
Guo, H., Ingolia, N. T., Weissman, J. S., and Bartel, D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840. doi: 10.1038/nature09267
Hagan, J. P., Piskounova, E., and Gregory, R. I. (2009). Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 16, 1021–1025. doi: 10.1038/nsmb.1676
Hamon, A., Roger, J. E., Yang, X. J., and Perron, M. (2016). Müller glial cell-dependent regeneration of the neural retina: an overview across vertebrate model systems. Dev. Dyn. 245, 727–738. doi: 10.1002/dvdy.24375
Heo, I., Joo, C., Kim, Y. K., Ha, M., Yoon, M. J., Cho, J., et al. (2009). TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138, 696–708. doi: 10.1016/j.cell.2009.08.002
Hitchcock, P. F., and Raymond, P. A. (2004). The teleost retina as a model for developmental and regeneration biology. Zebrafish 1, 257–271. doi: 10.1089/zeb.2004.1.257
Hoang, T., Wang, J., Boyd, P., Wang, F., Santiago, C., Jiang, L., et al. (2020). Gene regulatory networks controlling vertebrate retinal regeneration. Science. 370:eabb8595. doi: 10.1126/science.abb8598
Hu, J., Qian, H., Xue, Y., and Fu, X. D. (2018). PTB/nPTB: master regulators of neuronal fate in mammals. Biophys. Rep. 4, 204–214. doi: 10.1007/s41048-018-0066-y
Huo, J., Zhang, Y., Li, R., Wang, Y., Wu, J., and Zhang, D. (2016). Upregulated MicroRNA-25 mediates the migration of melanoma cells by targeting DKK3 through the WNT/beta-catenin pathway. Int. J. Mol. Sci. 17:1124. doi: 10.3390/ijms17111124
Jorstad, N. L., Wilken, M. S., Grimes, W. N., Wohl, S. G., VandenBosch, L. S., Yoshimatsu, T., et al. (2017). Stimulation of functional neuronal regeneration from muller glia in adult mice. Nature 548, 103–107. doi: 10.1038/nature23283
Kara, N., Rajaram, K., Didiano, D., Kent, M., Summerbell, E. R., Zhao, A., et al. (2019). The miR-216a-Dot1l regulatory axis is necessary and sufficient for muller glia reprogramming during retina regeneration. Cell Rep. 28, 2037–2047. doi: 10.1016/j.celrep.2019.07.061
Karl, M. O., and Reh, T. A. (2010). Regenerative medicine for retinal diseases: activating endogenous repair mechanisms. Trends Mol. Med. 16, 193–202. doi: 10.1016/j.molmed.2010.02.003
Kim, J., Fiesel, F. C., Belmonte, K. C., Hudec, R., Wang, W. X., Kim, C., et al. (2016). miR-27a and miR-27b regulate autophagic clearance of damaged mitochondria by targeting PTEN-induced putative kinase 1 (PINK1). Mol. Neurodegener. 11:55. doi: 10.1186/s13024-016-0121-4
Kim, V. (2005). MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 6, 376–385. doi: 10.1038/nrm1644
Krol, J., Loedige, I., and Filipowicz, W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610. doi: 10.1038/nrg2843
La Torre, A., Georgi, S., and Reh, T. A.. (2013). Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc. Natl. Accad. Sic. U.S.A. 110:E2362–70. doi: 10.1073/pnas.1301837110
Lahne, M., Nagashima, M., Hyde, D. R., and Hitchcock, P. F. (2020). Reprogramming muller glia to regenerate retinal neurons. Annu. Rev. Vis. Sic. 6, 171–193. doi: 10.1146/annurev-vision-121219-081808
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. doi: 10.1016/0092-8674(93)90529-Y
Lena, A. M., Shalom-Feuerstein, R., Rivetti di Val Cervo, P., Aberdam, D., Knight, R. A., Melino, G., et al. (2008). miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 15, 1187–1195. doi: 10.1038/cdd.2008.69
Li, N., Flynt, A., Kim, H. R., Solnica-Krezel, L., and Patton, J. (2008). Dispatched homolog 2 is targeted by miR-214 through a combination of three weak microRNA recognition sites. Nucleic Acids Res. 36, 4277–4285. doi: 10.1093/nar/gkn388
Lima Cunha, D., Arno, G., Corton, M., and Moosajee, M. (2019). The spectrum of PAX6 mutations and genotype-phenotype correlations in the eye. Genes 10:1050. doi: 10.3390/genes10121050
Link, B. A., and Collery, R. F. (2015). Zebrafish models of retinal disease. Annu. Rev. Vis. Sci. 1, 125–153. doi: 10.1146/annurev-vision-082114-035717
Liu, C. H., Huang, S., Britton, W. R., and Chen, J. (2020). MicroRNAs in vascular eye diseases. Int. J. Mol. Sci. 21:649. doi: 10.3390/ijms21020649
Liu, N., Bezprozvannaya, S., Williams, A. H., Qi, X., Richardson, J. A., Bassel-Duby, R., et al. (2008). microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 22, 3242–3254. doi: 10.1101/gad.1738708
Liu, N., Williams, A. H., Maxeiner, J. M., Bezprozvannaya, S., Shelton, J. M., Richardson, J. A., et al. (2012). microRNA-206 promotes skeletal muscle regeneration and delays progression of duchenne muscular dystrophy in mice. J. Clin. Invest. 122, 2054–2065. doi: 10.1172/JCI62656
Lozano-Velasco, E., Contreras, A., Crist, C. G., Hernandez-Torres, F., Franco, D., and Aranega, A. E. (2011). Pitx2c modulates Pax3+/Pax7+ cell populations and regulates Pax3 expression by repressing miR27 expression during myogenesis. Dev. Biol. 357, 165–178. doi: 10.1016/j.ydbio.2011.06.039
Lust, K., and Wittbrodt, J. (2018). Activating the regenerative potential of muller glia cells in a regeneration-deficient retina. Elife 7:e32319. doi: 10.7554/eLife.32319
Lv, C., Li, F., Li, X., Tian, Y., Zhang, Y., Sheng, X., et al. (2017). MiR-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing Wnt signaling antagonists. Nat. Commun. 8:1036. doi: 10.1038/s41467-017-01059-5
Maas, S. L. N., Breakefield, X. O., and Weaver, A. M. (2017). Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 27, 172–188. doi: 10.1016/j.tcb.2016.11.003
MacLaren, R. E., Pearson, R. a, MacNeil, a., Douglas, R. H., Salt, T. E., et al. (2006). Retinal repair by transplantation of photoreceptor precursors. Nature 444, 203–207. doi: 10.1038/nature05161
Macosko, E. Z., Basu, A., Satija, R., Nemesh, J., Shekhar, K., Goldman, M., et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. doi: 10.1016/j.cell.2015.05.002
Makeyev, E. V., Zhang, J., Carrasco, M. A., and Maniatis, T. (2007). The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448. doi: 10.1016/j.molcel.2007.07.015
McLean, C. M., Karemaker, I. D., and van Leeuwen, F. (2014). The emerging roles of DOT1L in leukemia and normal development. Leukemia 28, 2131–2138. doi: 10.1038/leu.2014.169
Mead, B., and Tomarev, S. (2020). Extracellular vesicle therapy for retinal diseases. Prog Retin Eye Res. 79:100849. doi: 10.1016/j.preteyeres.2020.100849
Mitra, S., Sharma, P., Kaur, S., Khursheed, M. A., Gupta, S., Ahuja, R., et al. (2018). Histone deacetylase-mediated muller glia reprogramming through Her4.1-Lin28a axis is essential for retina regeneration in zebrafish. iScience 7, 68–84. doi: 10.1016/j.isci.2018.08.008
Montgomery, J. E., Parsons, M. J., and Hyde, D. R. (2010). A novel model of retinal ablation demonstrates that the extent of rod cell death regulates the origin of the regenerated zebrafish rod photoreceptors. J. Comp. Neurol. 518, 800–814. doi: 10.1002/cne.22243
Moss, E. G., Lee, R. C., and Ambros, V. (1997). The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88, 637–646. doi: 10.1016/S0092-8674(00)81906-6
Nagashima, M., Barthel, L. K., and Raymond, P. A. (2013). A self-renewing division of zebrafish Muller glial cells generates neuronal progenitors that require N-cadherin to regenerate retinal neurons. Development 140, 4510–4521. doi: 10.1242/dev.090738
Needhamsen, M., White, R. B., Giles, K. M., Dunlop, S. A., and Thomas, M. G. (2014). Regulation of Human PAX6 Expression by miR-7. Evol. Bioinform. Online 10, 107–113. doi: 10.4137/EBO.S13739
O'Brien, K., Breyne, K., Ughetto, S., Laurent, L. C., and Breakefield, X. O. (2020). RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 21, 585–606. doi: 10.1038/s41580-020-0251-y
Olena, A. F., Rao, M. B., Thatcher, E. J., Wu, S. Y., and Patton, J. G. (2015). miR-216a regulates snx5, a novel notch signaling pathway component, during zebrafish retinal development. Dev. Biol. 400, 72–81. doi: 10.1016/j.ydbio.2015.01.016
Osakada, F., Ooto, S., Akagi, T., Mandai, M., Akaike, A., and Takahashi, M. (2007). Wnt signaling promotes regeneration in the retina of adult mammals. J. Neurosci. 27, 4210–4219. doi: 10.1523/JNEUROSCI.4193-06.2007
Pearson, R. a, Barber, a,.C., Rizzi, M., Hippert, C., Xue, T., et al. (2012). Restoration of vision after transplantation of photoreceptors. Nature 485, 99–103. doi: 10.1038/nature10997
Peters, L., and Meister, G. (2007). Argonaute proteins: mediators of RNA silencing. Mol. Cell 26, 611–623. doi: 10.1016/j.molcel.2007.05.001
Pollak, J., Wilken, M. S., Ueki, Y., Cox, K. E., Sullivan, J. M., Taylor, R. J., et al. (2013). ASCL1 reprograms mouse Muller glia into neurogenic retinal progenitors. Development 140, 2619–2631. doi: 10.1242/dev.091355
Powell, C., Elsaeidi, F., and Goldman, D. (2012). Injury-dependent muller glia and ganglion cell reprogramming during tissue regeneration requires Apobec2a and Apobec2b. J. Neurosci. 32, 1096–1109. doi: 10.1523/JNEUROSCI.5603-11.2012
Qian, H., Kang, X., Hu, J., Zhang, D., Liang, Z., Meng, F., et al. (2020). Reversing a model of Parkinson's disease with in situ converted nigral neurons. Nature 582, 550–556. doi: 10.1038/s41586-020-2388-4
Qin, Z., Kidd, A. R., Thomas, J. L., Poss, K. D., Hyde, D. R., Raymond, P., et al. (2011). FGF signaling regulates rod photoreceptor cell maintenance and regeneration in zebrafish. Exp. Eye Res. 93, 726–734. doi: 10.1016/j.exer.2011.09.003
Rajaram, K., Harding, R. L., Bailey, T., Patton, J. G., and Hyde, D. R. (2014a). Dynamic miRNA expression patterns during retina regeneration in zebrafish: reduced dicer or miRNA expression suppresses proliferation of muller glia-derived neuronal progenitor cells. Dev. Dyn. 243, 1591–1605. doi: 10.1002/dvdy.24188
Rajaram, K., Harding, R. L., Hyde, D. R., and Patton, J. G. (2014b). miR-203 regulates progenitor cell proliferation during adult zebrafish retina regeneration. Dev. Biol. 392, 393–403. doi: 10.1016/j.ydbio.2014.05.005
Ramachandran, R., Fausett, B. V., and Goldman, D. (2010). Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat. Cell Biol. 12, 1101–1107. doi: 10.1038/ncb2115
Ramachandran, R., Zhao, X. F., and Goldman, D. (2011). Ascl1a/Dkk/beta-catenin signaling pathway is necessary and glycogen synthase kinase-3beta inhibition is sufficient for zebrafish retina regeneration. Proc. Natl. Acad. Sci. U.S.A. 108, 15858–15863. doi: 10.1073/pnas.1107220108
Raymond, P. A., Barthel, L. K., Bernardos, R. L., and Perkowski, J. J. (2006). Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev. Biol. 6, 36. doi: 10.1186/1471-213X-6-36
Raymond, P. A., and Rivlin, P. K. (1987). Germinal cells in the goldfish retina that produce rod photoreceptors. Dev. Biol. 122, 120–138. doi: 10.1016/0012-1606(87)90338-1
Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., et al. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. doi: 10.1038/35002607
Roska, B., and Sahel, J. A. (2018). Restoring vision. Nature 557, 359–367. doi: 10.1038/s41586-018-0076-4
Sarkozy, M., Kahan, Z., and Csont, T. (2018). A myriad of roles of miR-25 in health and disease. Oncotarget 9, 21580–21612. doi: 10.18632/oncotarget.24662
Schwarz, D. S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., and Zamore, P. D. (2003). Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208. doi: 10.1016/S0092-8674(03)00759-1
Shaham, O., Menuchin, Y., Farhy, C., and Ashery-Padan, R. (2012). Pax6: a multi-level regulator of ocular development. Prog. Retin. Eye Res. 31, 351–376. doi: 10.1016/j.preteyeres.2012.04.002
Song, G., Sharma, A. D., Roll, G. R., Ng, R., Lee, A. Y., Blelloch, R. H., et al. (2010). MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology 51, 1735–1743. doi: 10.1002/hep.23547
Stenkamp, D. L. (2007). Neurogenesis in the fish retina. Int. Rev. Cytol. 259, 173–224. doi: 10.1016/S0074-7696(06)59005-9
Stern, J. H., Tian, Y., Funderburgh, J., Pellegrini, G., Zhang, K., Goldberg, J. L., et al. (2018). Regenerating eye tissues to preserve and restore vision. Cell Stem Cell 22, 834–849. doi: 10.1016/j.stem.2018.05.013
Tak, H., Kim, J., Jayabalan, A. K., Lee, H., Kang, H., Cho, D. H., et al. (2014). miR-27 regulates mitochondrial networks by directly targeting the mitochondrial fission factor. Exp. Mol. Med. 46:e123. doi: 10.1038/emm.2014.73
Taylor, S., Chen, J., Luo, J., and Hitchcock, P. (2012). Light-induced photoreceptor degeneration in the retina of the zebrafish. Ret. Dev. Methods Protoc. 884, 247–254. doi: 10.1007/978-1-61779-848-1_17
Thatcher, E., Paydar, I., Anderson, K. K., and Patton, J. (2008). Regulation of zebrafish fin regeneration by microRNAs. Proc. Natl. Accad. Sic. U.S.A. 105, 18384–18389. doi: 10.1073/pnas.0803713105
Thatcher, E. J., and Patton, J. G. (2010). Small RNAs have a big impact on regeneration. RNA Biol. 7, 333–338. doi: 10.4161/rna.7.3.12085
Thummel, R., Enright, J. M., Kassen, S. C., Montgomery, J. E., Bailey, T. J., and Hyde, D. R. (2010). Pax6a and Pax6b are required at different points in neuronal progenitor cell proliferation during zebrafish photoreceptor regeneration. Exp. Eye Res. 90, 572–582. doi: 10.1016/j.exer.2010.02.001
Thummel, R., Kassen, S. C., Enright, J. M., Nelson, C. M., Montgomery, J. E., and Hyde, D. R. (2008a). Characterization of Müller glia and neuronal progenitors during adult zebrafish retinal regeneration. Exp. Eye Res 87, 433–444. doi: 10.1016/j.exer.2008.07.009
Thummel, R., Kassen, S. C., Montgomery, J. E., Enright, J. M., and Hyde, D. R. (2008b). Inhibition of muller glial cell division blocks regeneration of the light-damaged zebrafish retina. Dev. Neurobiol. 68, 392–408. doi: 10.1002/dneu.20596
Tian, Y., Ma, X., Lv, C., Sheng, X., Li, X., Zhao, R., et al. (2017). Stress responsive miR-31 is a major modulator of mouse intestinal stem cells during regeneration and tumorigenesis. Elife 6:e29538. doi: 10.7554/eLife.29538
Tsonis, P. A., Call, M. K., Grogg, M. W., Sartor, M. A., Taylor, R. R., Forge, A., et al. (2007). MicroRNAs and regeneration: let-7 members as potential regulators of dedifferentiation in lens and inner ear hair cell regeneration of the adult newt. Biochem. Biophys. Res. Commun. 362, 940–945. doi: 10.1016/j.bbrc.2007.08.077
VandenBosch, L. S., Wohl, S. G., Wilken, M. S., Hooper, M., Finkbeiner, C., Cox, K., et al. (2020). Developmental changes in the accessible chromatin, transcriptome and Ascl1-binding correlate with the loss in muller glial regenerative potential. Sci. Rep. 10:13615. doi: 10.1038/s41598-020-70334-1
Vihtelic, T. S., and Hyde, D. R. (2000). Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J. Neurobiol. 44, 289–307. doi: 10.1002/1097-4695(20000905)44:3<289::AID-NEU1>3.0.CO;2-H
Visvanathan, J., Lee, S., Lee, B., Lee, J. W., and Lee, S. K. (2007). The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 21, 744–749. doi: 10.1101/gad.1519107
Wan, J., and Goldman, D. (2016). Retina regeneration in zebrafish. Curr. Opin. Genet. Dev. 40, 41–47. doi: 10.1016/j.gde.2016.05.009
Wightman, B., Ha, I., and Ruvkun, G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. doi: 10.1016/0092-8674(93)90530-4
Williams, A. H., Liu, N., van Rooij, E., and Olson, E. N. (2009). MicroRNA control of muscle development and disease. Curr. Opin. Cell Biol. 21, 461–469. doi: 10.1016/j.ceb.2009.01.029
Wohl, S. G., Hooper, M. J., and Reh, T. A. (2019). MicroRNAs miR-25, let-7 and miR-124 regulate the neurogenic potential of Muller glia in mice. Development 146:dev179556. doi: 10.1242/dev.179556
Wohl, S. G., Jorstad, N. L., Levine, E. M., and Reh, T. A. (2017). Müller glial microRNAs are required for the maintenance of glial homeostasis and retinal architecture. Nat. Commun. 8:1603. doi: 10.1038/s41467-017-01624-y
Wohl, S. G., and Reh, T. A. (2016a). The microRNA expression profile of mouse Müller glia in vivo and in vitro. Sci. Rep 6:35423. doi: 10.1038/srep35423
Wohl, S. G., and Reh, T. A. (2016b). miR-124-9-9* potentiates Ascl1-induced reprogramming of cultured Muller glia. Glia 64, 743–762. doi: 10.1002/glia.22958
Wollerton, M. C., Gooding, C., Wagner, E. J., Garcia-Blanco, M. A., and Smith, C. W. (2004). Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13, 91–100. doi: 10.1016/S1097-2765(03)00502-1
Wu, D. M., Schneiderman, T., Burgett, J., Gokhale, P., Barthel, L., and Raymond, P. A. (2001). Cones regenerate from retinal stem cells sequestered in the inner nuclear layer of adult goldfish retina. Invest. Ophthalmol. Vis. Sic. 42, 2115–2124.
Xue, Y., Ouyang, K., Huang, J., Zhou, Y., Ouyang, H., Li, H., et al. (2013). Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 152, 82–96. doi: 10.1016/j.cell.2012.11.045
Yao, K., Qiu, S., Wang, Y. V., Park, S. J. H., Mohns, E. J., Mehta, B., et al. (2018). Restoration of vision after de novo genesis of rod photoreceptors in mammalian retinas. Nature 560, 484–488. doi: 10.1038/s41586-018-0425-3
Yeom, K. H., Mitchell, S., Linares, A. J., Zheng, S., Lin, C. H., Wang, X. J., et al. (2018). Polypyrimidine tract-binding protein blocks miRNA-124 biogenesis to enforce its neuronal-specific expression in the mouse. Proc. Natl. Acad. Sci. U.S.A. 115, E11061–E11070. doi: 10.1073/pnas.1809609115
Yi, R., Poy, M. N., Stoffel, M., and Fuchs, E. (2008). A skin microRNA promotes differentiation by repressing ‘stemness’. Nature 452, 225–229. doi: 10.1038/nature06642
Yin, V. P., and Poss, K. D. (2008). New regulators of vertebrate appendage regeneration. Curr. Opin. Genet. Dev. 18, 381–386. doi: 10.1016/j.gde.2008.06.008
Yin, V. P., Thomson, J. M., Thummel, R., Hyde, D. R., Hammond, S. M., and Poss, K. D. (2008). Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 22, 728–733. doi: 10.1101/gad.1641808
Yurco, P., and Cameron, D. A. (2005). Responses of muller glia to retinal injury in adult zebrafish. Vision Res. 45, 991–1002. doi: 10.1016/j.visres.2004.10.022
Zarbin, M. (2016). Cell-based therapy for degenerative retinal disease. Trends Mol. Med. 22, 115–134. doi: 10.1016/j.molmed.2015.12.007
Zhao, C., Tao, Z., Xue, L., Zeng, Y., Wang, Y., Xu, H., et al. (2017). Lin28b stimulates the reprogramming of rat muller glia to retinal progenitors. Exp. Cell Res. 352, 164–174. doi: 10.1016/j.yexcr.2017.02.010
Zhao, J., Tao, Y., Zhou, Y., Qin, N., Chen, C., Tian, D., et al. (2015). MicroRNA-7: a promising new target in cancer therapy. Cancer Cell Int. 15:103. doi: 10.1186/s12935-015-0259-0
Keywords: miRNA, Müller glia, retina, regeneration, zebrafish
Citation: Konar GJ, Ferguson C, Flickinger Z, Kent MR and Patton JG (2021) miRNAs and Müller Glia Reprogramming During Retina Regeneration. Front. Cell Dev. Biol. 8:632632. doi: 10.3389/fcell.2020.632632
Received: 23 November 2020; Accepted: 21 December 2020;
Published: 18 January 2021.
Edited by:
Ivan Conte, University of Naples Federico II, ItalyReviewed by:
Seth Blackshaw, Johns Hopkins University, United StatesCopyright © 2021 Konar, Ferguson, Flickinger, Kent and Patton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James G. Patton, SmFtZXMuRy5QYXR0b25AVmFuZGVyYmlsdC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.