
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 04 February 2021
Sec. Molecular and Cellular Oncology
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.625730
Accumulating evidence has shown that long non-coding RNAs (lncRNAs) can be used as biological markers and treatment targets in cancer and play various roles in cancer-related biological processes. However, the lncRNA expression profiles and their roles and action mechanisms in ovarian cancer (OC) are largely unknown. Here, we assessed the lncRNA expression profiles in OC tissues from The Cancer Genome Atlas (TCGA) database, and one upregulated lncRNA, LINC01969, was selected for further study. LINC01969 expression levels in 41 patients were verified using quantitative real-time polymerase chain reaction (qRT-PCR). The in vitro effects of LINC01969 on OC cell migration, invasion, and proliferation were determined by the CCK-8, ethynyl-2-deoxyuridine (EdU), wound healing, and Transwell assays. Epithelial–mesenchymal transition (EMT) was evaluated using qRT-PCR and Western blotting. The molecular mechanisms of LINC01969 in OC were assessed through bioinformatics analysis, RNA-binding protein immunoprecipitation (RIP), dual luciferase reporter gene assays, and a rescue experiment. Finally, in vivo experiments were conducted to evaluate the functions of LINC01969. The results of the current study showed that LINC01969 was dramatically upregulated in OC, and patients with lower LINC01969 expression levels tended to have better overall survival. Further experiments demonstrated that LINC01969 promoted the migration, invasion, and proliferation of OC cells in vitro and sped up tumor growth in vivo. Additionally, LINC01969, which primarily exists in the cytoplasm, boosted LARP1 expression by sponging miR-144-5p and promoted the malignant phenotypes of OC cells. In conclusion, the LINC01969/miR-144-5p/LARP1 axis is a newly identified regulatory signaling pathway involved in OC progression.
Ovarian cancer (OC) ranks second among lethal gynecological cancers worldwide (Lheureux et al., 2019). According to estimates, 238,719 women worldwide were newly diagnosed with OC in 2012, and 151,917 died from OC (Iversen et al., 2018). Despite progress in surgical treatment and chemoradiotherapy, the survival rate of OC patients has only changed modestly, and the 5-year survival rate is only 47%, even in the USA, Canada, and other wealthy countries (Burki, 2017; Torre et al., 2018). As the anatomical characteristics of the ovary complicate diagnosis, the disease is often diagnosed in late stage when the survival rate is low (Dafni et al., 2021). Hence, much attention has been paid to the molecular biological mechanisms underlying the occurrence and progression of OC.
Long non-coding RNAs (lncRNAs) are ncRNAs more than 200 nt in length that were recently discovered (Li et al., 2017; Xie et al., 2018; Liu et al., 2019b), and their functions are just now being explored. It has been reported that lncRNAs play important roles in multiple biological and pathogenic processes, such as genomic imprinting (Liu et al., 2017; Sanli et al., 2018), cancer metastasis (Kim et al., 2018; Zhuo et al., 2019; Qiu et al., 2020), stem cell differentiation (Daneshvar et al., 2016; Hou et al., 2017; Zhang et al., 2019), and X chromosome inactivation (Tian et al., 2010; Furlan and Rougeulle, 2016; Carter et al., 2020) among many others. LncRNAs function via diverse mechanisms, including chromatin modification and cell signaling (Satpathy and Chang, 2015). One of the major functions of lncRNAs is as competing endogenous RNAs (ceRNAs), which sequester microRNAs (miRNAs) from their messenger RNA (mRNA) targets, thereby increasing target gene expression (Ransohoff et al., 2018). For example, in lung cancer, lncRNA LCAT1 functions as a ceRNA by sponging miR-4715-5p to control RAC1 function (Yang et al., 2019). LncRNA MNX1-AS1 induces lung cancer progression through the miR-527/BRF2 pathway (Liu et al., 2019a). LncRNA AK002107 inversely controls miR-140-5p and triggers EMT in OC by targeting TGFBR1 (Tang et al., 2019). LncRNA AC010789.1 promotes colorectal cancer progression by targeting the microRNA-432-3p/ZEB1 axis and the Wnt/β-catenin signaling pathway (Duan et al., 2020). A study of the BRCA1/2 ceRNA network in OC patients with wild-type BRCA1/2 revealed a novel three-lncRNA signature that could predict both prognosis and chemo-response (Zhang et al., 2020).
An assessment of the differentially expressed lncRNAs in OC using The Cancer Genome Atlas (TCGA) database showed that LINC01969 is expressed at high levels in OC and is associated with patient survival. However, whether LINC01969 correlates with the onset and development of OC is unknown. In the current study, LINC01969 expression levels in OC and non-cancer tissues were compared, which showed that LINC01969 expression was markedly higher in OC. Functional studies showed that LINC01969 acts as an oncogene to promote OC through the miR-144-5p/LARP1 axis.
In 2016–2018, 41 paired fresh tumor and non-tumor tissues were harvested from patients at Liaoning Cancer Hospital and Institute and snap frozen at −80°C (Table 1). Written informed consent was obtained from all patients, and the study was approved by the Ethnics Committees of Liaoning Cancer Hospital and Institute.
Human OC cell lines (A2780, OVCAR-3, SKOV3, and TOV112D) and a normal human ovarian cell line (IOSE-80) (ATCC, Manassas, VA, USA) were cultured in Dulbecco's modified Eagle's medium (DMEM) (HyClone, Beijing, China) containing 10% fetal bovine serum (FBS) (HyClone).
A LINC01969 short hairpin RNA (shRNA) (sh-LINC01969) and negative control shRNA (sh-NC) were designed, synthesized, and inserted into plasmid vector PLKO.1-puro (BioVector NTCC Inc., Beijing, China). To overexpress LINC01969, the LINC01969 genomic sequence was amplified and inserted into pLV-CMV-EF1a-GFP-T2A-Puro vector. LARP1 small-interfering (si-LARP1), negative control (si-NC), and miRNA mimics and inhibitors were provided by RiboBio (Guangzhou, China). After 24 h of culture, the cells underwent a 48-h transient transfection with the corresponding vector using Lipofectamine 3000 transfection reagent (Invitrogen, Shanghai, China) according to the manufacturer's instructions. The transfected cells were analyzed by quantitative real-time PCR (qRT-PCR). Assays were carried out at least thrice.
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Shanghai, China) and was reverse transcribed using the PrimerScript RT-PCR kit (Takara, Dalian, China) according to the manufacturer's instructions. Then, RNA levels were assessed by qRT-PCR using the TaqMan MiRNA Assay Kit (Applied Biosystems). The relative expression of targets was determined in triplicate on an ABI 7500 RT-PCR system (Applied Biosystems). β-Actin or U6 small nuclear RNA (snRNA) was used as a reference gene for normalization of miRNA or mRNA expression. The delta Ct method was used to calculate the relative expression. The primers used in this study are shown in Supplementary Table 1.
Cytoplasmic and nuclear RNA were extracted with the PARIS Kit (Life Technologies, USA), and cytoplasm and nuclear RNA levels were assessed by qRT-RCR using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 as internal references, respectively.
The Cell Counting Kit-8 from Dojindo (Beijing, China) was utilized to assess cell proliferation according to the manufacturer's instructions. The cells were seeded in 96-well plates (2 × 103 cells/well) in 100 μl of medium in quintuplicate. After 1, 2, 3, and 4 days, Cell Counting Kit-8 (CCK8) reagent was added to the wells and incubated for 2.5 h. Then, the absorbance was measured at 450 nm.
Cell proliferation was evaluated using the ethynyl-2-deoxyuridine (EdU) Apollo DNA in vitro kit (RiboBio, Guangzhou, China) according to the manufacturer's instructions. OC cells (1 × 104) were seeded in each well of 96-well plates for transfection. After incubation at 37°C and 5% CO2 for 48 h, cells were added with 20 μM EdU and incubated for another 2 h. OC cells were fixed with 4% paraformaldehyde and dyed with Apollo 567 and Hoechst 33342 (Hu et al., 2017). The number of cells was counted using Image J software (NIH, Bethesda, MD, USA). The cell proliferation rate was calculated according to the manufacturer's instructions.
Wounds were generated with a 200-μl pipette tip in a cell monolayer at 80% confluence. After washing with phosphate-buffered saline (PBS), medium containing 20 μg/ml mitomycin C without FBS was added to assess the impact of various genes on cell proliferation. At 0 and 24 h after wounding, pictures were taken to assess healing.
The invasion ability of OC cells was probed using a Transwell apparatus. Briefly, OC cells in DMEM containing 1% FBS were seeded at 105 cells/well in the upper chamber of a Transwell apparatus (8-μm pore size; Corning, Corning, NY, USA) precoated with Matrigel (1:6 dilution; Corning). DMEM containing 10% FBS was added to the lower chamber. After a 24-h incubation at 37°C, the migrated or invaded cells in the lower chamber were fixed with 4% methanol and stained with crystal violet. The cells in five random fields of a microscope (100×) were counted. All assays were performed thrice.
Total cell lysates were prepared in a 1 × sodium dodecyl sulfate buffer, and total proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes. The membranes were blocked with skim milk (5%) and incubated with primary antibody at 4°C overnight. The primary antibodies used were against β-actin (1:1,000, ab8226), LARP1 (1:1,000, ab86359), E-cadherin (1:50, ab1416), Snail (1:1000, ab216347), and Vimentin (1:1,000, ab8069) (Abcam, Shanghai, China). The membranes were then incubated with antirabbit or antimouse secondary antibodies (1:1,000, Abcam, Shanghai, China) and visualized by ECL. All assays were performed at least thrice.
A fluorescence in situ hybridization (FISH) assay was performed using the Ribo™ FISH Kit (RiboBio, China) according to the manufacturer's instructions. The Cy3-conjugated LINC01969 probe was designed and synthesized by RiboBio. Fluorescence was detected with a confocal laser-scanning microscope (Leica, Germany).
An RNA-binding protein immunoprecipitation (RIP) assay was performed using the EZ-magna RIP kit (Millipore, China) according to the manufacturer's instructions. First, A2780 and SKOV3 cells were harvested and lysed in complete RIP lysis buffer. Then, the cell extracts were incubated with RIP buffer containing anti-Argonaute-2 antibody-conjugated magnetic beads (ab32381, Abcam, Shanghai, China); an anti-immunoglobulin G (anti-IgG) antibody (ab6702, Abcam) was used as a control. Then, the samples were incubated with protease K and oscillated to digest proteins and isolate immunoprecipitated RNAs. The RNA concentration was determined by using a NanoDrop spectrophotometer, and the purified RNAs were evaluated by RT-PCR.
LINC01969 and LARP1 fragments containing the miR-144-5p binding sites were amplified by PCR and cloned into the pmirGLO vector downstream of the luciferase reporter gene to generate LINC01969 Wt and LARP1 Wt. Then, the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene) was utilized to generate LINC01969 Mut and LARP1 Mut (with mutations in the miR-144-5p binding sites) according to the manufacturer's guidelines. HEK293T cells were cotransfected with either miR-NC or miR-144-5p mimic and either LINC01969 Wt/Mut or LARP1 Wt/Mut. After 48 h of transfection, cells were collected, and a luciferase assay was conducted using the Dual-Luciferase Reporter Assay System (Promega, Beijing, China).
Four-week-old female athymic BALB/c nude mice were randomly divided into two groups (n = 3/group) and cultured under aseptic conditions with sterile food and water. A xenograft model of OC was generated by subcutaneously injecting A2780 cells into the mice. Tumor volume was measured for 4 weeks and was calculated according to the following formula: volume = (length) × (width)2/2. After 4 weeks, the mice were killed, and tumor weight was measured. All animal experiments were approved by the Liaoning Cancer Hospital and Institute and were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (NIH publication 80-23).
Statistical analyses were conducted using SPSS 16.0 (SPSS, Beijing, China) and GraphPad Prism 5.0 (GraphPad, La Jolla, CA, USA). Data were collected from at least three separate assays and are expressed as means ± SEM. Intergroup differences were evaluated using Student's t-test. Normally distributed data were assessed using one-way ANOVA, and non-normally distributed data were assessed using the Mann–Whitney U test. A P-value < 0.05 was considered statistically significant.
We first assessed the lncRNA expression levels in OC tissues in TCGA database. We divided the OC tissues into two groups: a stage I + II group and a stage III + IV group. The heat and volcano maps revealed all differentially expressed lncRNAs with a log2FC >2 or < −2, P < 0.05 (Figures 1A,B). We chose the top 15 most highly expressed lncRNAs in stage III + IV compared to stage I + II and analyzed their impact on survival in TCGA. We found that only LINC01969 was associated with survival in OC, and higher LINC01969 expression was associated with lower overall survival of OC patients (Figure 1C). These findings indicate that LINC01969 is probably involved in the progression of OC.
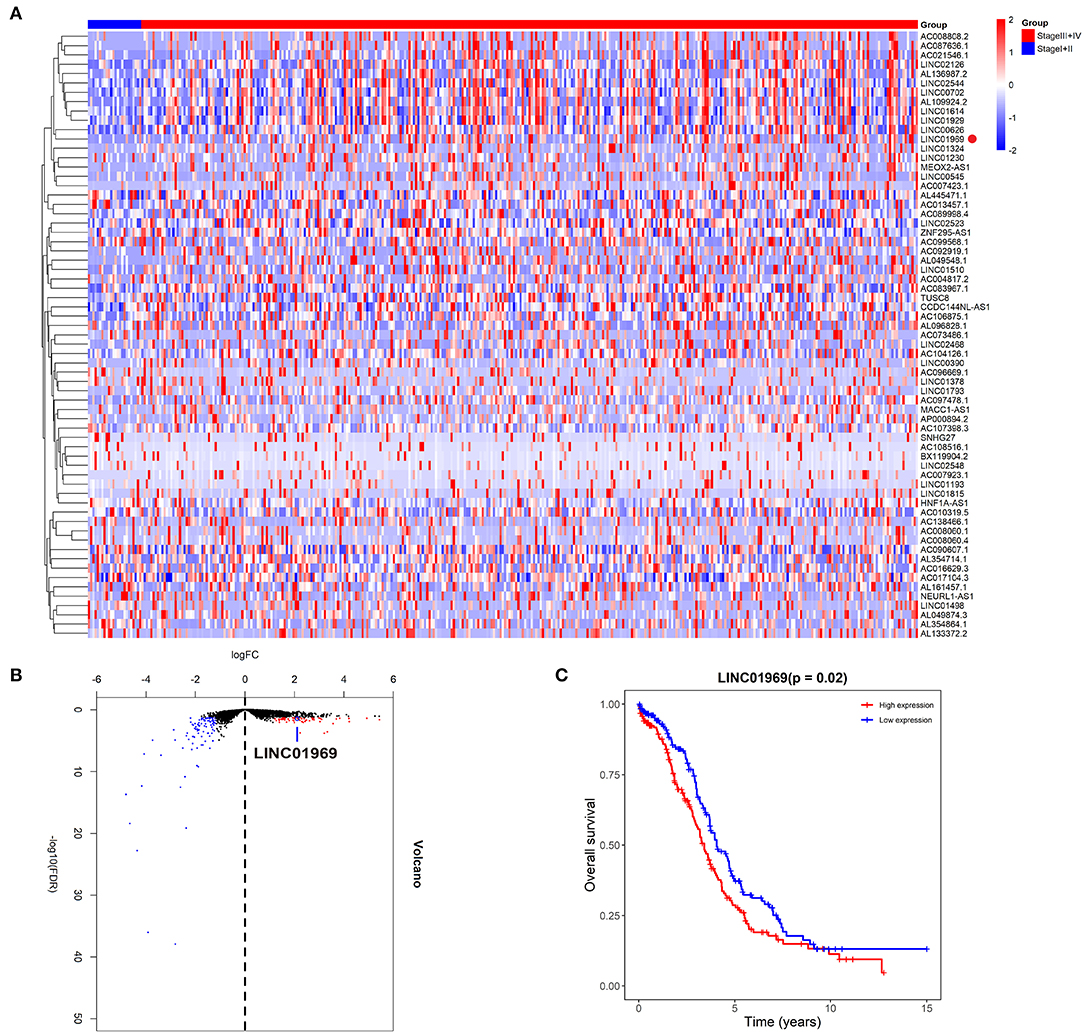
Figure 1. Upregulated LINC01969 correlates with unsatisfactory prognosis of ovarian cancer (OC) cases revealed by The Cancer Genome Atlas (TCGA) database. As per the Neoplasm American Joint Committee on Cancer Clinical Group Stage, we allocated OC tissues into stage I + II group and stage III + IV group. (A,B) The heat and volcano maps revealed all differentially expressed long non-coding RNAs (lncRNAs) of OC tissues in the TCGA database with log2FC >2 or < −2, P < 0.05. (C) Kaplan–Meier survival analysis of LINC01969 expression in OC tissues from TCGA database.
Analysis of TCGA data revealed that LINC01969 was expressed at a dramatically lower level in OC stage I + II tumors than in OC stage III + IV tumors (Figure 2A). qRT-PCR analysis of 41 OC samples revealed significantly higher LINC01969 levels in OC tissues than in paired non-normal tissues (Figure 2B). LINC01969 expression was also assessed in OC cell lines, which showed that LINC01969 was expressed at markedly higher levels in OC cell lines (A2780, TOV112D, OVCAR-3, and SKOV3) than in normal human cells (IOSE-80) (Figure 2C). The role of LINC01969 as a biomarker for prognosis was determined by dividing the 41 OC tissues into high and low LINC01969 expression groups based on the average LINC01969 expression. Kaplan–Meier analysis revealed that LINC01969 overexpression was related to lower overall survival (Figure 2D). These findings imply that LINC01969 might be a useful biomarker for OC prognosis.
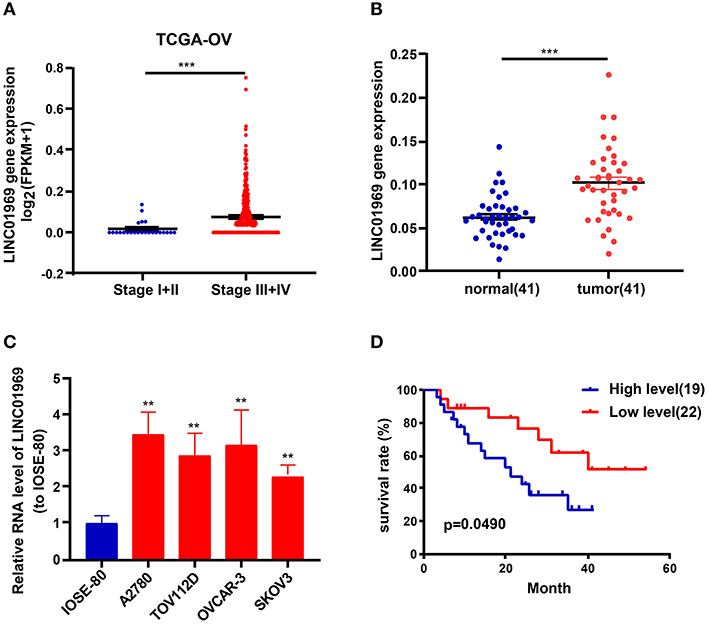
Figure 2. LINC01969 expression is upregulated in ovarian cancer (OC) tissues and cell lines. (A) LINC01969 expression in OC stage I + II and stage III + IV from TCGA database. (B) Quantitative real-time PCR (qRT-PCR) results indicated that LINC01969 was upregulated in OC tissues (n = 41) compared with paired non-normal tissues (n = 41). (C) LINC01969 expression in OC cell lines and IOSE-80 cell line was analyzed by qRT-PCR. (D) Kaplan–Meier curves of overall survival (OS) in 41 OC patients based on LINC01969 level (average expression value was the cutoff). Data represent the mean ± SD; **P < 0.01, ***P < 0.001.
LINC01969 was expressed at higher levels in OC tissues and was related to unsatisfactory prognosis of OC patients. These findings indicate that LINC01969 may promote the occurrence and development of OC. To verify this, LINC01969 expression in A2780 OC cells was knocked down with a shRNA (Figure 3A), and LINC01969 was overexpressed in SKOV3 OC cells with a LINC01969-overexpressing lentivirus (Figure 3A). Then, the CCK8 and EdU assays were implemented to assess the proliferation of A2780 and SKOV3 cells. The CCK-8 assay showed that LINC01969 knockdown significantly suppressed proliferation in A2780 cells, while overexpression of LINC01969 markedly promoted proliferation in SKOV3 cells (Figure 3B). The results of the EdU assay confirmed the impact of LINC01969 on OC proliferation (Figure 3C).
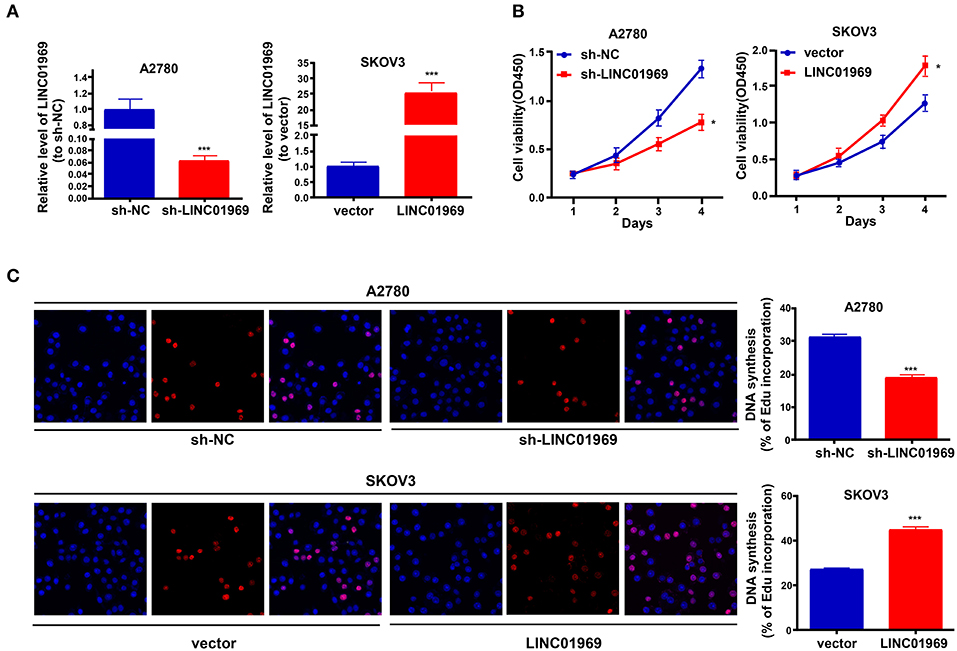
Figure 3. LINC01969 promotes cell growth in ovarian cancer (OC) cells. (A) Transfection efficiency of sh-LINC01969 and LINC01969-overexpressing vector. (B) Cell viability was analyzed by CCK-8 assay. (C) EdU assay indicated that LINC01969 silencing delayed OC cell proliferation. Data were presented as represent the mean ± SD of three independent experiments; *P < 0.05, ***P < 0.001.
We used wound healing and Transwell assays to test whether LINC01969 modulated the migration and invasion ability of OC cells. LINC01969-overexpressing SKOV3 cells migrated at a higher rate than the control cells, whereas LINC01969 knockdown A2780 cells barely migrated (Figure 4A). As revealed by the Transwell assay results, cells that express high levels of LINC01969 had stronger invasion capability (Figure 4B). Next, we evaluated the effect of LINC01969 on OC cell EMT by measuring EMT marker expression using qRT-PCR and Western blotting. The results showed that knockdown of LINC01969 increased E-cadherin expression but decreased Snail and Vimentin expression levels in OC cells (Figures 4C,D). These results indicate that LINC01969 probably affects OC cell invasion and migration.
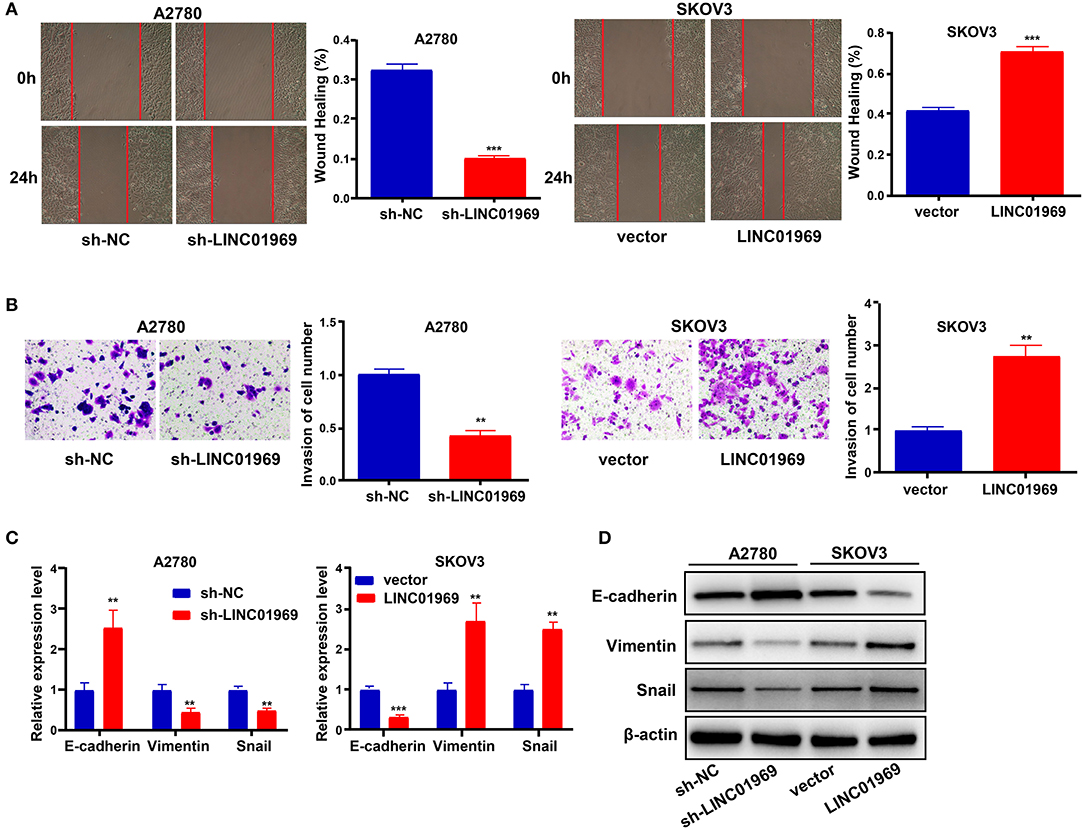
Figure 4. LINC01969 promotes the cell migration, invasion, and epithelial–mesenchymal transition (EMT) of ovarian cancer (OC) cells. (A) Wound healing assay showed that LINC01969 silencing suppressed the ability of OC cell migration. (B) Transwell assay showed that LINC01969 silencing suppressed the ability of OC cell invasion. (C,D) The expression of EMT markers were determined using quantitative real-time PCR (qRT-PCR) and Western blot assay. Data were presented as mean ± SD of three independent experiments; **P < 0.01, ***P < 0.001.
The subcellular location of an lncRNA is intimately correlated with its biological and molecular functions (Wen et al., 2018). Hence, nucleo-cytoplasmic sequestering and RNA-FISH assays were used to determine the subcellular distribution of LINC01969. The results showed that most of the LINC01969 signal is located within the cytoplasm, but some signal was detected in the nucleus (Figures 5A,B). Then, we looked for possible LINC01969 targets to determine its functional mechanism using starBase (http://starbase.sysu.edu.cn), which identified miR-144-5p as a potential target. The miR-144-5p binding sites in LINC01969-Wt and LINC01969-Mut are shown in Figure 5C. According to the starBase results, the binding site is located at chr17:48292600-48292621 within the lncRNA. The binding region is exon-3.
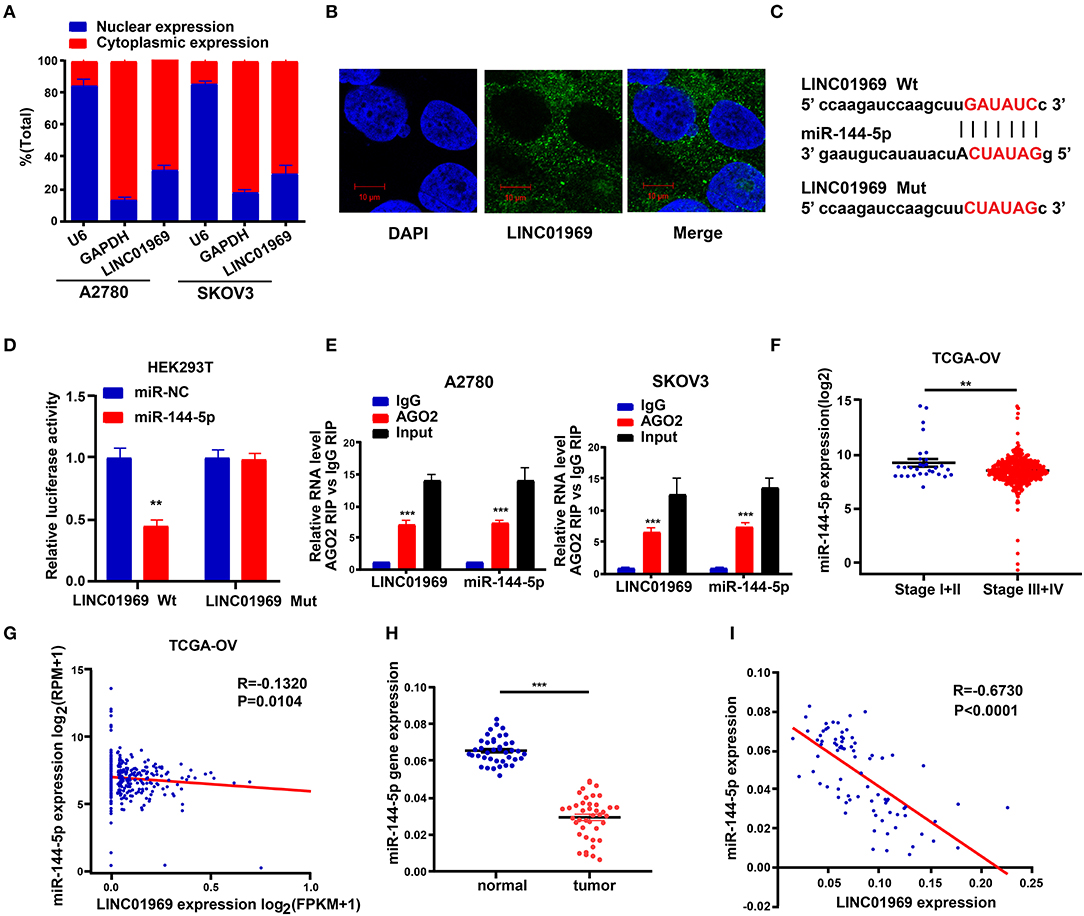
Figure 5. LINC01969 is targeted by miR-144-5p. (A) Localization of LINC01969 by nucleocytoplasmic separation experiment. (B) RNA-FISH in A2780 cells. (C) Putative miR-144-5p binding sequence and mutation sequence of LINC01969 messenger RNA (mRNA) were as shown. (D) Dual luciferase reporter assay was used to confirm the direct target between LINC01969 and miR-144-5p. (E) RNA-binding protein immunoprecipitation (RIP) assay was used to detect whether miR-144-5p could bind with LINC01969. (F) MiR-144-5p expression in ovarian cancer (OC) stage I + II and stage III + IV, from The Cancer Genome Atlas (TCGA) database. (G) The correlation analysis between miR-144-5p expression and LINC01969 expression in OC from TCGA database. (H) MiR-144-5p expression in OC tissues and paired non-normal tissues. (I) The correlation analysis between miR-144-5p expression and LINC01969 expression in 41 OC tissues and 41 adjacent no normal tissues. Data were presented as mean ± SD; **P < 0.01, ***P < 0.001.
A dual luciferase reporter assay showed that cotransfection of the LINC01969-Wt luciferase reporter construct with miR-144-5p mimics significantly reduced luciferase activity, while cotransfection of the LINC01969-Mut luciferase reporter construct with miR-144-5p mimics did not affect luciferase activity in HEK293T cells (Figure 5D). The interaction of miR-144-5p and LINC01969 was confirmed via RIP assay, which showed that, relative to those in IgG, LINC01969 and miR-144-5p were enriched in the AGO2-containing miRNA ribonucleoprotein complexes (Figure 5E). TCGA database analysis showed that miR-144-5p expression was higher in stage I + II OC than in stage III + IV OC (Figure 5F), and LINC01969 expression levels were inversely correlated with miR-144-5p expression in OC samples (Figure 5G). Consistently, miR-144-5p gene expression levels in the 41 OC samples in our dataset were higher than in the non-cancer samples (Figure 5H), and LINC01969 expression was inversely related to miR-144-5p expression in the OC samples (Figure 5I). Based on these data, we concluded that LINC01969 is targeted by miR-144-5p.
To explore the possible roles of miR-144-5p in OC progression, the target genes of miR-144-5p were screened using starBase, and we found that miR-144-5p may interact with LARP1. We then mutated the two potential miR-144-5p binding sites in LARP1 (Figure 6A), which, according to the starBase search results, are located within the 3′-untranslated region (UTR) of the LARP1 mRNA. Dual luciferase reporter assays revealed that cotransfection of the LARP1-Wt luciferase reporter construct with miR-144-5p mimics reduced luciferase activity, whereas cotransfection of the LARP1-Mut luciferase reporter construct with miR-144-5p mimics had no impact on luciferase activity in HEK293T cells (Figure 6B). TCGA database analysis showed that LARP1 expression was lower in stage I + II OC than in stage III + IV OC (Figure 6C). Our analysis also showed that LARP1 expression was higher in 41 OC samples than in the normal tissue samples (Figure 6F). Moreover, LARP1 expression was positively related to LINC01969 expression (Figures 6D,G) and negatively related to miR-144-5p expression in the OC samples (Figures 6E,H) in both TCGA database and our dataset. We also analyzed its expression in OC cell lines, and the results indicated that LARP1 expression is significantly higher in the tested OC cell lines (A2780, TOV112D, OVCAR-3, and SKOV3) than in the tested normal human cell line (IOSE-80) (Figure 6I). These data indicate that LARP1 is a target gene of miR-144-5p.
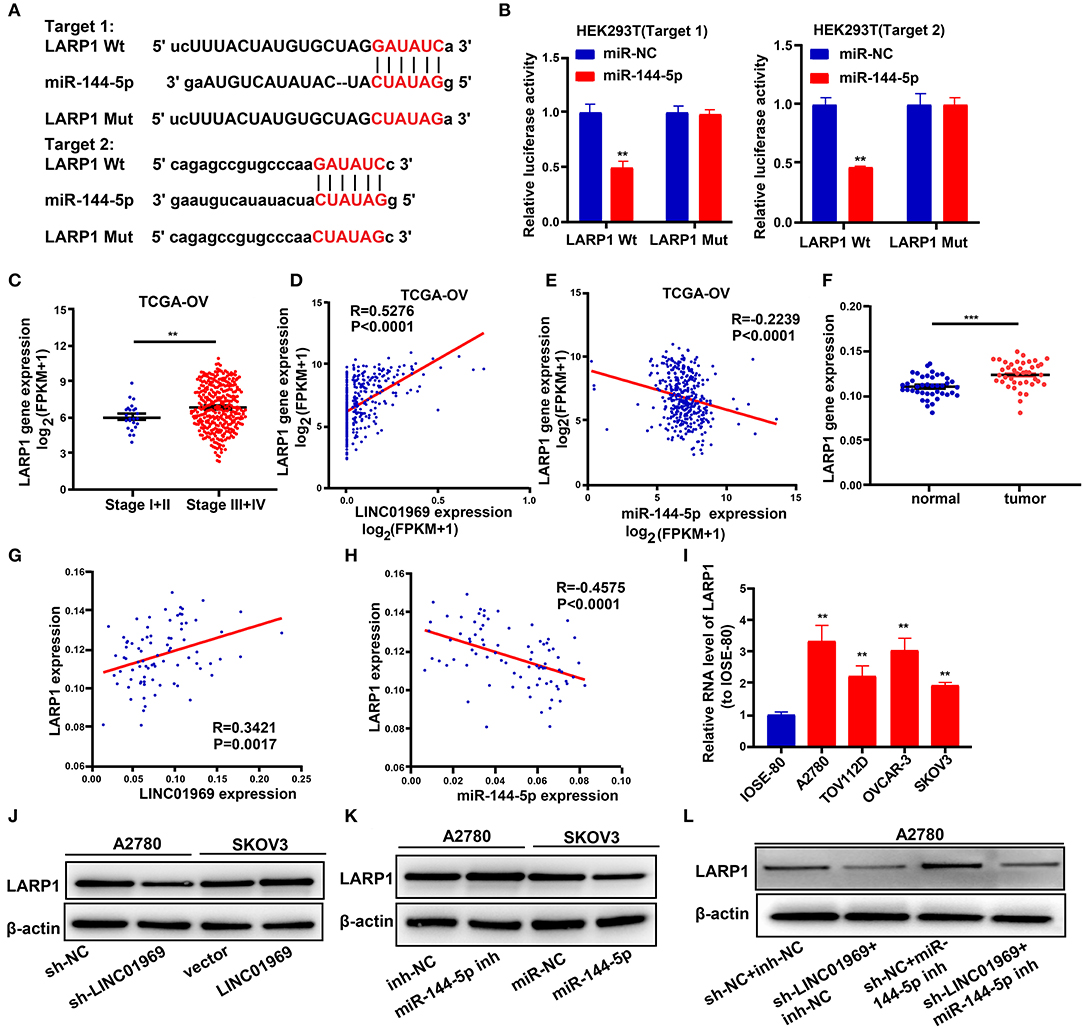
Figure 6. LINC01969 regulates LARP1 by acting as a competing endogenous RNA (ceRNA) for miR-144-5p. (A) Two potential miR-144-5p binding sequences and mutation sequences of LARP1 messenger RNA (mRNA) were shown. (B) Dual luciferase reporter assays were used to confirm the direct target between LARP1 and miR-144-5p. (C) LARP1 expression in ovarian cancer (OC) stage I + II and stage III + IV from The Cancer Genome Atlas (TCGA) database. (D) The correlation analysis between LINC01969 expression and LARP1 expression in OC from TCGA database. (E) The correlation analysis between miR-144-5p expression and LARP1 expression in OC from TCGA database. (F) LARP1 gene expression in OC tissues and paired non-normal tissues from our dataset. (G) The correlation analysis between LINC01969 expression and LARP1 expression in OC tissues and paired non-normal tissues from our dataset. (H) The correlation analysis between miR-144-5p expression and LARP1 expression in OC tissues and paired non-normal tissues from our dataset. (I) LARP1 expression in OC cell lines and IOSE-80 cell line was analyzed by quantitative real-time PCR (qRT-PCR). (J–L) The level of LARP1 was detected by Western blot assay. Data were presented as mean ± SD; **P < 0.01, ***P < 0.001.
Next, we used a Western blot assay to explore whether LINC01969 can regulate LARP1 expression in OC cells via miR-144-5p. The results showed that LARP1 expression was reduced by LINC01969 knockdown (sh-LINC01969) and miR-144-5p mimics (Figures 6J,K). In contrast, LARP1 expression was elevated by LINC01969 overexpression and a miR-144-5p inhibitor (Figures 6I,J). Additionally, LINC01969 silencing reduced LARP1 expression in A2780 cells, and this was reversed by knockdown of miR-144-5p (Figure 6L). These findings imply that LINC01969 controls LARP1 expression in OC cells in a miR-144-5p-dependent manner.
We then evaluated the effect of the LINC01969/miR-144-5p axis in OC. This was achieved by cotransfecting A2780 cells with the following combinations: sh-NC + inh-NC, sh-LINC01969 + inh-NC, sh-NC + miR-144-5p inh, and sh-LINC01969 + miR-144-5p inh. SKOV3 cells were cotransfected with the following combinations: vector + miR-NC, LINC01969 + miR-NC, vector + miR-144-5p, and LINC01969 + miR-144-5p. An EdU assay exhibited that transfection of sh-LINC01969 inhibited cell proliferation, whereas miR-144-5p inhibitor promoted cell proliferation and reversed the effect of LINC01969 silencing on A2780 cell proliferation (Figure 7A). Transfection of the miR-144-5p mimics reversed the effect of LINC01969 overexpression on SKOV3 cell proliferation (Figure 7A). The results presented in Figures 7B,C show that cell migration and invasion were increased by LINC01969 overexpression and miR-144-5p inhibitor but decreased by sh-LINC01969 and miR-144-5p mimics. Conversely, transfection of miR-144-5p mimics reversed the cell invasion- and migration-promoting effects of LINC01969 overexpression. Transfection of the miR-144-5p inhibitor reversed the effect of LINC01969 silencing on cell migration and invasion. Next, we determined whether LINC01969 regulates EMT of OC cells through LARP1. EMT marker expression was evaluated using a Western blot assay. Knockdown of LARP1 increased E-cadherin expression and decreased Vimentin and Snail expression in OC cells (Figure 7D). In addition, knockdown of LARP1 reversed the EMT-promoting effect of LINC01969 overexpression (Figure 7D).
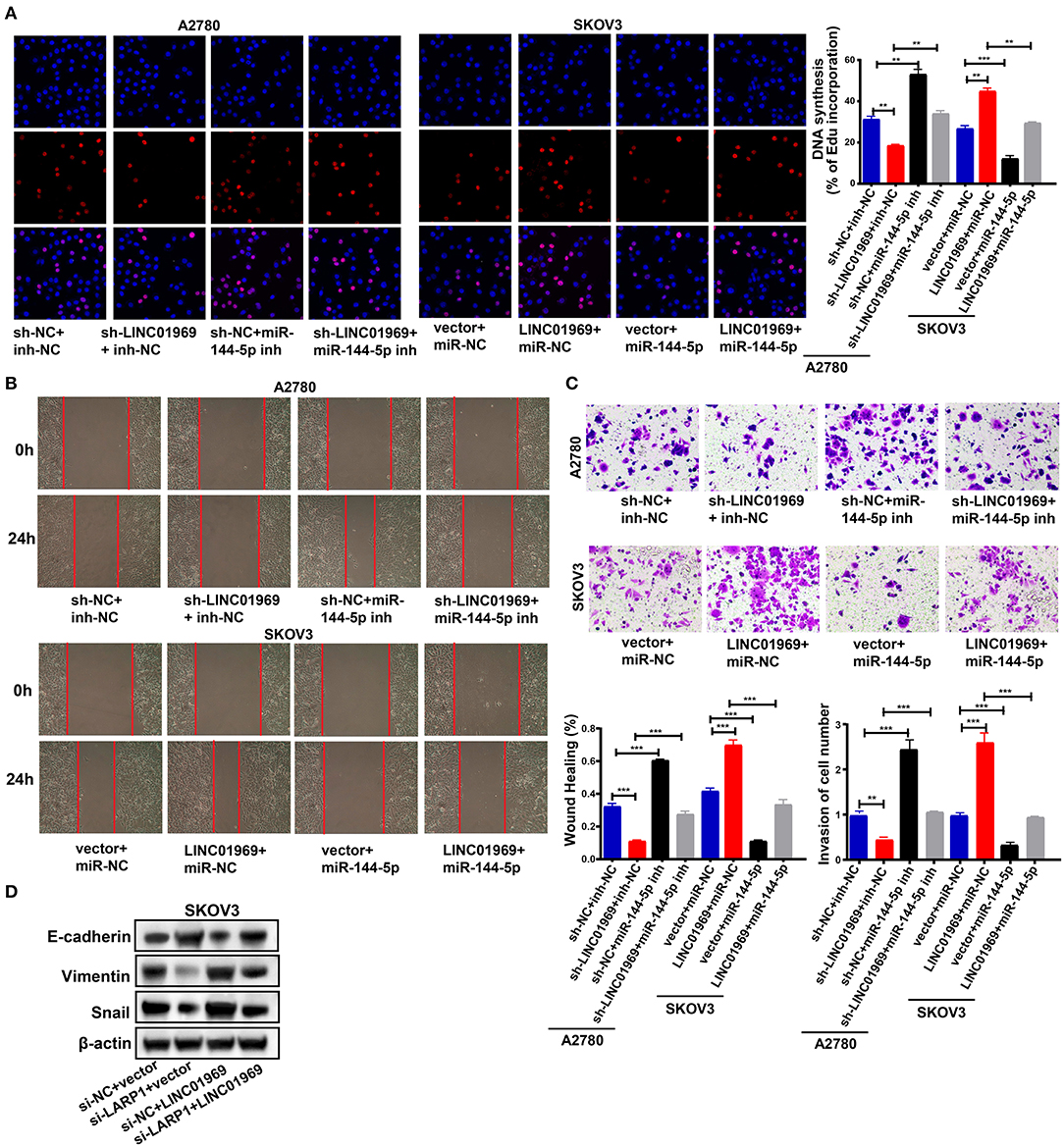
Figure 7. LINC01969/miR-144-5p axis regulates ovarian cancer (OC) progression. (A) Ethynyl-2-deoxyuridine (EdU) showed that miR-144-5p mimics treatment reversed the effect of LINC01969 overexpression on OC cell proliferation. (B) Wound healing assay showed that miR-144-5p inhibitor treatment reversed the effect of LINC01969 silencing on OC cell migration. (C) MiR-144-5p mimics treatment reversed the effect of LINC01969 overexpression on OC cell invasion. (D) The expression of epithelial–mesenchymal transition (EMT) markers were determined using Western blot assay. Data were presented as represent the mean ± SD of three independent experiments; **P < 0.01, ***P < 0.001.
All these findings imply that LINC01969 functions as a ceRNA for miR-144-5p to control LARP1 expression and promote the proliferation, migration, and invasion of OC cells.
We established xenograft models to confirm our in vitro findings in vivo. We injected LINC01969-silenced and non-silenced A2780 cells into nude mice. Then, tumor volume and weight were measured, and the tumors obtained from the mice are shown in Figure 8A. We found that LINC01969 knockdown greatly slowed tumor growth (Figure 8B), and the LINC01969 knockdown tumors were lighter at the end of the experiment (Figure 8C). qRT-PCR and Western blot assays showed that LARP1, Vimentin, and Snail expression levels were lower and E-cadherin expression levels were higher in mice with LINC01969-silenced tumors (Figures 8D,E), indicating that LINC01969 knockdown decreased LARP1 expression and OC cell EMT in vivo. These results show that LINC01969 can promote OC growth and metastasis in vivo.
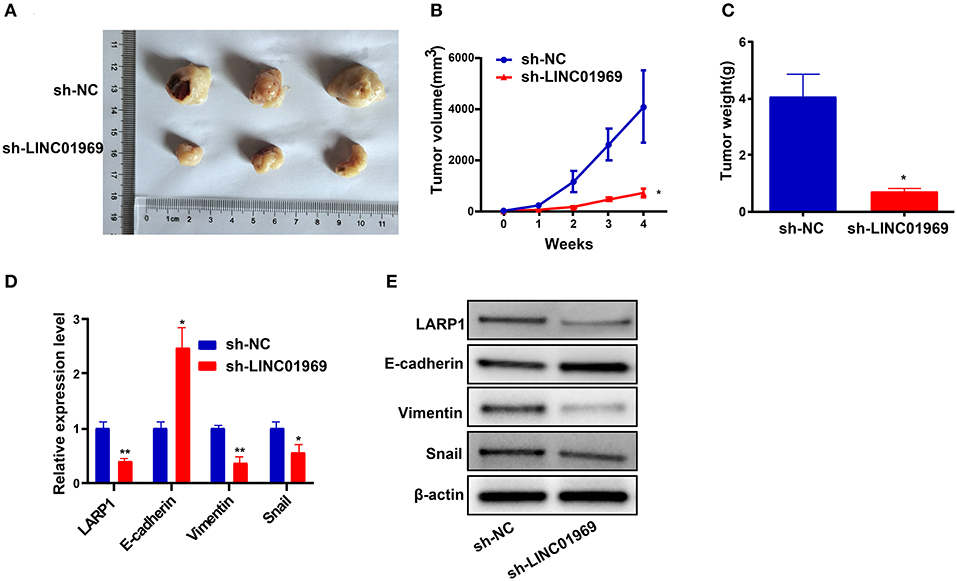
Figure 8. LINC01969 short hairpin RNA (shRNA) curbs ovarian cancer (OC) cells to proliferate and metastasize in vivo. (A) Representative images of tumors from indicated orthotopic xenografts. (B) Tumor volume and (C) weight data of indicated orthotopic xenografts. (D,E) Messenger RNA (mRNA) and protein expression levels of LARP1 and epithelial–mesenchymal transition (EMT)-related markers after LINC01969 knockdown. Data were presented as mean ± SD; *P < 0.05, **P < 0.01.
With the advancement of high-throughput sequencing technology, a growing number of lncRNAs have been identified. The roles of lncRNAs in diverse biological processes have been widely explored, and how lncRNAs regulate tumorigenesis has become a hot research topic. Mounting evidence has implicated various lncRNAs in the modulation of cancer cell proliferation, migration, and apoptosis (Jiang et al., 2018; Wu et al., 2018; Zhao et al., 2019; Chen et al., 2020). Likewise, we found a lncRNA, LINC01969, that was highly expressed in OC samples and was related to clinical stage and prognosis. The role of LINC01969 in cancer has not yet been well-researched. Using tumor tissues from a population of OC patients (n = 41) and OC cell lines, we verified that LINC01969 was indeed expressed at high levels in OC tissues and cell lines. Using functional assays, we revealed that LINC01969 promoted the proliferation, migration, invasion, and EMT of OC cells. We then probed the action mechanism of LINC01969 in OC.
LncRNA–miRNA–mRNA regulatory networks, in which lncRNAs act as ceRNAs for miRNAs to boost expression of specific mRNAs, are well-recognized (Feng et al., 2018; Yang et al., 2019). For example, lncRNA FAM225A facilitates nasopharyngeal carcinoma (NPC) tumor formation and metastasis by sponging miR-590-3p/miR-1275 as a ceRNA and increasing ITGB3 expression (Zheng et al., 2019). In addition, the LINC01287/miR-298/STAT3 feedback loop modulates the growth and EMT phenotype of hepatocellular carcinoma (HCC) cells (Mo et al., 2018). Knockdown of lncRNA PVT1 reduces cell migration and invasion and increases cell apoptosis via miR-145-mediated inhibition of FSCN1 in esophageal carcinoma cell (Shen et al., 2019). Here, we identified MiR-144-5p as a target of LINC01969 using starBase. MiR-144-5p was previously reported to function as an antitumor miRNA in renal cell carcinoma (Yamada et al., 2018) and OC progression (Song et al., 2018). Assays showed that miR-144-5p mimics inhibited the proliferation, invasion, and migration of SKOV3 cells. Moreover, upregulation of miR-144-5p partially reversed the effects of sh-LINC01969 on the proliferation, invasion, and migration of OC cells.
LARP1 was predicted as a target gene of miR-144-5p using starBase. LARP1 encodes an RNA-binding protein that is necessary for cancer cell survival and ribosome biogenesis (Al-Ashtal et al., 2019). LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs (Hong et al., 2017) and posttranscriptionally regulates mTOR and contributes to cancer progression (Mura et al., 2015) by regulating cell division, apoptosis, and cell migration (Burrows et al., 2010). Hopkins TG et al. reported that LARP1 promotes OC progression and enhances resistance to chemotherapy by differentially modulating the stability of diverse mRNAs (Hopkins et al., 2016). The results of the present research confirmed that LARP1 is a downstream target gene of miR-144-5p and that LINC01969 competitively bound to miR-144-5p to release LARP1, thereby modulating LARP1 expression.
In conclusion, LINC01969 acts as an oncogene and promotes the migration, proliferation, invasion, and EMT of OC cells through the miR-144-5p/LARP1 axis. LINC01969 might be a useful prognostic biomarker in OC.
The original contributions generated for the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by the Ethics Committee of Liaoning Cancer Hospital & Institute. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Ethics Committee of Liaoning Cancer Hospital & Institute.
JZ made prosperous contributions to conception and design. JC and XL performed the experiments. JC wrote the draft manuscript. LY analyzed the data. XL made collection of data. All authors made contributions to the examination of the manuscript and approved the final manuscript for submission.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Editage (www.editage.com) for English language editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.625730/full#supplementary-material
Al-Ashtal, H. A., Rubottom, C. M., Leeper, T. C., and Berman, A. J. (2019). The LARP1 La-Module recognizes both ends of TOP mRNAs. RNA Biol. 1–11. doi: 10.1080/15476286.2019.1669404
Burki, T. K. (2017). Cancer incidence in Canadian prisoners. Lancet Oncol. 18:e200. doi: 10.1016/S1470-2045(17)30176-6
Burrows, C., Abd Latip, N., Lam, S. J., Carpenter, L., Sawicka, K., Tzolovsky, G., et al. (2010). The RNA binding protein Larp1 regulates cell division, apoptosis, and cell migration. Nucleic Acids Res. 38, 5542–5553. doi: 10.1093/nar/gkq294
Carter, A. C., Xu, J., Nakamoto, M. Y., Wei, Y., Zarnegar, B. J., Shi, Q., et al. (2020). Spen links RNA-mediated endogenous retrovirus silencing and X chromosome inactivation. Elife 9:e54508. doi: 10.7554/eLife.54508.sa2
Chen, Z., Pan, T., Jiang, D., Jin, L., Geng, Y., Feng, X., et al. (2020). The lncRNA-GAS5/miR-221-3p/DKK2 axis modulates ABCB1-mediated adriamycin resistance of breast cancer via the Wnt/beta-catenin signaling pathway. Mol. Ther. Nucleic Acids 19, 1434–1448. doi: 10.1016/j.omtn.2020.01.030
Dafni, U., Martin-Lluesma, S., Balint, K., Tsourti, Z., Vervita, K., Chenal, J., et al. (2021). Efficacy of cancer vaccines in selected gynaecological breast and ovarian cancers: a 20-year systematic review and meta-analysis. Eur. J. Cancer 142, 63–82. doi: 10.1016/j.ejca.2020.10.014
Daneshvar, K., Pondick, J. V., Kim, B. M., Zhou, C., York, S. R., Macklin, J. A., et al. (2016). DIGIT is a conserved long noncoding RNA that regulates GSC expression to control definitive endoderm differentiation of embryonic stem cells. Cell Rep. 17, 353–365. doi: 10.1016/j.celrep.2016.09.017
Duan, W., Kong, X., Li, J., Li, P., Zhao, Y., Liu, T., et al. (2020). LncRNA AC010789.1 promotes colorectal cancer progression by targeting microRNA-432-3p/ZEB1 axis and the Wnt/beta-catenin signaling pathway. Front. Cell Dev. Biol. 8:565355. doi: 10.3389/fcell.2020.565355
Feng, K., Liu, Y., Xu, L. J., Zhao, L. F., Jia, C. W., and Xu, M. Y. (2018). Long noncoding RNA PVT1 enhances the viability and invasion of papillary thyroid carcinoma cells by functioning as ceRNA of microRNA-30a through mediating expression of insulin like growth factor 1 receptor. Biomed. Pharmacother. 104, 686–698. doi: 10.1016/j.biopha.2018.05.078
Furlan, G., and Rougeulle, C. (2016). Function and evolution of the long noncoding RNA circuitry orchestrating X-chromosome inactivation in mammals. Wiley Interdiscip. Rev. RNA 7, 702–722. doi: 10.1002/wrna.1359
Hong, S., Freeberg, M. A., Han, T., Kamath, A., Yao, Y., Fukuda, T., et al. (2017). LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. Elife 6:e25237. doi: 10.7554/eLife.25237.026
Hopkins, T. G., Mura, M., Al-Ashtal, H. A., Lahr, R. M., Abd-Latip, N., Sweeney, K., et al. (2016). The RNA-binding protein LARP1 is a post-transcriptional regulator of survival and tumorigenesis in ovarian cancer. Nucleic Acids Res. 44, 1227–1246. doi: 10.1093/nar/gkv1515
Hou, J., Long, H., Zhou, C., Zheng, S., Wu, H., Guo, T., et al. (2017). Long noncoding RNA Braveheart promotes cardiogenic differentiation of mesenchymal stem cells in vitro. Stem Cell Res. Ther. 8:4. doi: 10.1186/s13287-016-0454-5
Hu, J., Meng, Y., Zhang, Z., Yan, Q., Jiang, X., Lv, Z., et al. (2017). MARCH5 RNA promotes autophagy, migration, and invasion of ovarian cancer cells. Autophagy 13, 333–344. doi: 10.1080/15548627.2016.1256520
Iversen, L., Fielding, S., Lidegaard, O., Morch, L. S., Skovlund, C. W., and Hannaford, P. C. (2018). Association between contemporary hormonal contraception and ovarian cancer in women of reproductive age in Denmark: prospective, nationwide cohort study. BMJ 362:k3609. doi: 10.1136/bmj.k3609
Jiang, C., Yang, Y., Yang, Y., Guo, L., Huang, J., Liu, X., et al. (2018). Long noncoding RNA (lncRNA) HOTAIR affects tumorigenesis and metastasis of non-small cell lung cancer by upregulating miR-613. Oncol. Res. 26, 725–734. doi: 10.3727/096504017X15119467381615
Kim, J., Piao, H. L., Kim, B. J., Yao, F., Han, Z., Wang, Y., et al. (2018). Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 50, 1705–1715. doi: 10.1038/s41588-018-0252-3
Lheureux, S., Braunstein, M., and Oza, A. M. (2019). Epithelial ovarian cancer: evolution of management in the era of precision medicine. CA Cancer J. Clin. 69, 280–304. doi: 10.3322/caac.21559
Li, J., Li, Z., Zheng, W., Li, X., Wang, Z., Cui, Y., et al. (2017). LncRNA-ATB: an indispensable cancer-related long noncoding RNA. Cell Prolif. 50:e12381. doi: 10.1111/cpr.12381
Liu, H., Han, L., Liu, Z., and Gao, N. (2019a). Long noncoding RNA MNX1-AS1 contributes to lung cancer progression through the miR-527/BRF2 pathway. J. Cell. Physiol. 234, 13843–13850. doi: 10.1002/jcp.28064
Liu, H., Shang, X., and Zhu, H. (2017). LncRNA/DNA binding analysis reveals losses and gains and lineage specificity of genomic imprinting in mammals. Bioinformatics 33, 1431–1436. doi: 10.1093/bioinformatics/btw818
Liu, X., Duan, X., Holmes, J. A., Li, W., Lee, S. H., Tu, Z., et al. (2019b). A long noncoding RNA regulates Hepatitis C virus infection through interferon alpha-inducible protein 6. Hepatology 69, 1004–1019. doi: 10.1002/hep.30266
Mo, Y., He, L., Lai, Z., Wan, Z., Chen, Q., Pan, S., et al. (2018). LINC01287/miR-298/STAT3 feedback loop regulates growth and the epithelial-to-mesenchymal transition phenotype in hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. 37:149. doi: 10.1186/s13046-018-0831-2
Mura, M., Hopkins, T. G., Michael, T., Abd-Latip, N., Weir, J., Aboagye, E., et al. (2015). LARP1 post-transcriptionally regulates mTOR and contributes to cancer progression. Oncogene 34, 5025–5036. doi: 10.1038/onc.2014.428
Qiu, J. J., Lin, X. J., Tang, X. Y., Zheng, T. T., Zhang, X. Y., and Hua, K. Q. (2020). Long noncoding RNA TC0101441 induces epithelial-mesenchymal transition in epithelial ovarian cancer metastasis by downregulating KiSS1. Int. J. Cancer 146, 2588–2598. doi: 10.1002/ijc.32692
Ransohoff, J. D., Wei, Y., and Khavari, P. A. (2018). The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 19, 143–157. doi: 10.1038/nrm.2017.104
Sanli, I., Lalevee, S., Cammisa, M., Perrin, A., Rage, F., Lleres, D., et al. (2018). Meg3 non-coding RNA expression controls imprinting by preventing transcriptional upregulation in cis. Cell Rep. 23, 337–348. doi: 10.1016/j.celrep.2018.03.044
Satpathy, A. T., and Chang, H. Y. (2015). Long noncoding RNA in hematopoiesis and immunity. Immunity 42, 792–804. doi: 10.1016/j.immuni.2015.05.004
Shen, S. N., Li, K., Liu, Y., Yang, C. L., He, C. Y., and Wang, H. R. (2019). Down-regulation of long noncoding RNA PVT1 inhibits esophageal carcinoma cell migration and invasion and promotes cell apoptosis via microRNA-145-mediated inhibition of FSCN1. Mol. Oncol. 13, 2554–2573. doi: 10.1002/1878-0261.12555
Song, L., Peng, L., Hua, S., Li, X., Ma, L., Jie, J., et al. (2018). miR-144-5p enhances the radiosensitivity of non-small-cell lung cancer cells via targeting ATF2. Biomed. Res. Int. 2018:5109497. doi: 10.1155/2018/5109497
Tang, Y. H., He, G. L., Huang, S. Z., Zhong, K. B., Liao, H., Cai, L., et al. (2019). The long noncoding RNA AK002107 negatively modulates miR-140-5p and targets TGFBR1 to induce epithelial-mesenchymal transition in hepatocellular carcinoma. Mol. Oncol. 13, 1296–1310. doi: 10.1002/1878-0261.12487
Tian, D., Sun, S., and Lee, J. T. (2010). The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143, 390–403. doi: 10.1016/j.cell.2010.09.049
Torre, L. A., Trabert, B., Desantis, C. E., Miller, K. D., Samimi, G., Runowicz, C. D., et al. (2018). Ovarian cancer statistics, 2018. CA Cancer J. Clin. 68, 284–296. doi: 10.3322/caac.21456
Wen, X., Gao, L., Guo, X., Li, X., Huang, X., Wang, Y., et al. (2018). lncSLdb: a resource for long non-coding RNA subcellular localization. Database (Oxford) 2018, 1–6. doi: 10.1093/database/bay085
Wu, Y., Yuan, T., Wang, W. W., Ge, P. L., Gao, Z. Q., Zhang, G., et al. (2018). Long noncoding RNA HOST2 promotes epithelial-mesenchymal transition, proliferation, invasion, and migration of hepatocellular carcinoma cells by activating the JAK2-STAT3 signaling pathway. Cell. Physiol. Biochem. 51, 301–314. doi: 10.1159/000495231
Xie, Y., Zhang, Y., Du, L., Jiang, X., Yan, S., Duan, W., et al. (2018). Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol. Oncol. 12, 648–658. doi: 10.1002/1878-0261.12188
Yamada, Y., Arai, T., Kojima, S., Sugawara, S., Kato, M., Okato, A., et al. (2018). Regulation of antitumor miR-144-5p targets oncogenes: direct regulation of syndecan-3 and its clinical significance. Cancer Sci. 109, 2919–2936. doi: 10.1111/cas.13722
Yang, J., Qiu, Q., Qian, X., Yi, J., Jiao, Y., Yu, M., et al. (2019). Long noncoding RNA LCAT1 functions as a ceRNA to regulate RAC1 function by sponging miR-4715-5p in lung cancer. Mol. Cancer 18:171. doi: 10.1186/s12943-019-1107-y
Zhang, M., Wang, G., Zhu, Y., and Wu, D. (2020). Characterization of BRCA1/2-directed ceRNA network identifies a novel three-lncRNA signature to predict prognosis and chemo-response in ovarian cancer patients with wild-type BRCA1/2. Front. Cell Dev. Biol. 8:680. doi: 10.3389/fcell.2020.00680
Zhang, W., Chen, L., Wu, J., Li, J., Zhang, X., Xiang, Y., et al. (2019). Long noncoding RNA TUG1 inhibits osteogenesis of bone marrow mesenchymal stem cells via Smad5 after irradiation. Theranostics 9, 2198–2208. doi: 10.7150/thno.30798
Zhao, Y., Zhao, L., Li, J., and Zhong, L. (2019). Silencing of long noncoding RNA RP11-476D10.1 enhances apoptosis and autophagy while inhibiting proliferation of papillary thyroid carcinoma cells via microRNA-138-5p-dependent inhibition of LRRK2. J. Cell. Physiol. 234, 20980–20991. doi: 10.1002/jcp.28702
Zheng, Z. Q., Li, Z. X., Zhou, G. Q., Lin, L., Zhang, L. L., Lv, J. W., et al. (2019). Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 79, 4612–4626. doi: 10.1158/0008-5472.CAN-19-0799
Zhuo, W., Liu, Y., Li, S., Guo, D., Sun, Q., Jin, J., et al. (2019). Long noncoding RNA GMAN, up-regulated in gastric cancer tissues, is associated with metastasis in patients and promotes translation of ephrin A1 by competitively binding GMAN-AS. Gastroenterology 156, 676–691. doi: 10.1053/j.gastro.2018.10.054
Keywords: lncRNA, ovarian cancer, epithelial-mesenchymal transition, cell proliferation, invasion
Citation: Chen J, Li X, Yang L and Zhang J (2021) Long Non-coding RNA LINC01969 Promotes Ovarian Cancer by Regulating the miR-144-5p/LARP1 Axis as a Competing Endogenous RNA. Front. Cell Dev. Biol. 8:625730. doi: 10.3389/fcell.2020.625730
Received: 03 November 2020; Accepted: 28 December 2020;
Published: 04 February 2021.
Edited by:
Peti Thuwajit, Mahidol University, ThailandReviewed by:
Daniele Vergara, University of Salento, ItalyCopyright © 2021 Chen, Li, Yang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingru Zhang, anJ6aGFuZ19sbnN6bEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.