- 1Key Laboratory of Animal Genetics, Breeding and Reproduction of Shaanxi Province, College of Animal Science and Technology, Northwest A&F University, Yangling, China
- 2Shaanxi Kingbull Livestock Co., Ltd., Yangling, China
The level of muscle development in livestock directly affects the production efficiency of livestock, and the contents of intramuscular fat (IMF) is an important factor that affects meat quality. However, the molecular mechanisms through which circular RNA (circRNA) affects muscle and IMF development remains largely unknown. In this study, we isolated myoblasts and intramuscular preadipocytes from fetal bovine skeletal muscle. Oil Red O and BODIPY staining were used to identify lipid droplets in preadipocytes, and anti-myosin heavy chain (MyHC) immunofluorescence was used to identify myotubes differentiated from myoblasts. Bioinformatics, a dual-fluorescence reporter system, RNA pull-down, and RNA-binding protein immunoprecipitation were used to determine the interactions between circINSR and the micro RNA (miR)-15/16 family. Molecular and biochemical assays were used to confirm the roles played by circINSR in myoblasts and intramuscular preadipocytes. We found that isolated myoblasts and preadipocytes were able to differentiate normally. CircINSR was found to serve as a sponge for the miR-15/16 family, which targets CCND1 and Bcl-2. CircINSR overexpression significantly promoted myoblast and preadipocyte proliferation and inhibited cell apoptosis. In addition, circINSR inhibited preadipocyte adipogenesis by alleviating the inhibition of miR-15/16 against the target genes FOXO1 and EPT1. Taken together, our study demonstrated that circINSR serves as a regulator of embryonic muscle and IMF development.
Introduction
In livestock production, the development of muscle and intramuscular fat (IMF) are important factors that determine meat quality. Bovine muscle development begins during the early embryonic stage, including the proliferation of myoblast progenitor cells and the proliferation and fusion of mononuclear myoblasts to form multinucleated myotubes. After birth, the number of muscle fibers does not change, but the fibers become thicker (Rehfeldt et al., 2000; Picard et al., 2010). Therefore, the level of muscle development that occurs during pregnancy directly affects meat production. Fat begins to develop during the second trimester (Sanchez-Gurmaches and Guertin, 2014). The IMF content determines the tenderness and juiciness of beef, and the optimization of this factor is a research hotspot. Increasing the number of preadipocytes in fetal muscle contributes to improved fat deposition and marbling after birth (Rajesh et al., 2010). However, the mechanisms that control muscle and IMF development remain unclear. In addition, pre-mature adipogenesis and the maturation of intramuscular preadipocytes in the fetus can result in muscle tissue dysfunction (Taga et al., 2011). Therefore, exploring the molecular mechanisms that regulate muscle and IMF development is of great significance to the livestock industry.
The development of biotechnology has greatly promoted the ability to screen and study key genes involved in muscle and IMF development. Several critical genes have been demonstrated to mediate muscle development and adipogenesis, including peroxisome proliferative activated receptor gamma (PPARγ) and CCAAT/enhancer-binding protein alpha (C/EBPα), which have been characterized as adipogenesis regulators (Lin and Lane, 1994; Tontonoz et al., 1994). The myogenic regulatory factors (MRFs), myocyte enhancer factor (MEF2), PAX3/PAX7, and myostatin (MSTN) have been reported to be effective for the induction of myoblasts proliferation and differentiation (Sassoon et al., 1989; McPherron and Lee, 1997; Soumillion et al., 1997; Relaix et al., 2005; Potthoff and Olson, 2007).
In addition to coding genes, a large number of non-coding RNAs have also been shown to regulate muscle and fat development. For example, a large number of micro RNAs (miRNAs) and circular RNAs (circRNAs) have been reported to participate in the physiological regulation of muscle and fat. CircRNAs have a covalent closed-loop structure, with neither 5′-3′ polarity nor a polyadenylated tail (Hansen et al., 2013a). They can participate in physiological regulation by sponging miRNAs or binding regulatory proteins (Meng et al., 2017). CircFUT10 and circFGFR4 have been reported to regulate the muscle development-related genes through the sponging of miR-133a and miR-107 (Li et al., 2018a,b). CircINSR has been shown to adsorb miR-34a to promote the proliferation of bovine myoblasts (Shen et al., 2020). CircTshz2-1 and circArhgap5-2 have been shown to be indispensable regulators of fat formation (Arcinas et al., 2019). Despite these findings, the roles played by circRNAs in muscle and IMF development continues to require additional research. Based on our previous studies, we have focused on a circRNA derived from the insulin receptor gene (INSR) in this study. CircINSR is named after the INSR gene and is formed by the cyclization of the second exon of INSR (Shen et al., 2020). A previous study indicated that circINSR could affect the proliferation and apoptosis of bovine embryonic myoblasts by adsorbing miR-34a; however, whether circINSR interacts with other miRNAs and whether circINSR plays a role in preadipocyte development remains unclear.
In this study, we isolated fetal bovine myoblasts and intramuscular preadipocytes. The targeting relationship between circINSR and the miR-15/16 family and the effects of circINSR on the proliferation and apoptosis of myoblasts and preadipocytes were analyzed in vitro. Importantly, we demonstrated that circINSR could target miR-15/16 to inhibit the pre-mature differentiation of intramuscular preadipocytes, ensuring normal muscle function.
Materials and Methods
Progenitor Cell Isolation and Cell Lines
Bovine fetuses, from 120 to 180 days, were collected from the slaughterhouse and transported immediately to the laboratory. All animal experiments and study protocols were approved by the Animal Care Commission of the College of Veterinary Medicine, Northwest A&F University. Using enzyme digestion, combined with the differential adhesion method, primary myoblasts and intramuscular preadipocytes were isolated from the longissimus dorsi, as previously described (Rando and Blau, 1994; Conboy and Rando, 2002; Miyake et al., 2012). The longissimus dorsi was isolated from the fetus, washed with phosphate-buffered saline (PBS), and minced into small fragments. It was then digested in Dulbecco's modified Eagle's medium (DMEM, HyClone, USA) containing type II collagenase (w/v, 2%; C5138, Sigma, USA) at 37°C, with continuous shaking for 2 h. The cell plasma was filtered through a 200-μm filter, collected by centrifugation, and resuspended in DMEM. The cells were grown in high-glucose DMEM supplemented with 20% fetal bovine serum (FBS, Gibco, USA) and 1% penicillin-streptomycin solution (Gibco, USA), and incubated at 37°C with 5% CO2. Within 2 h of inoculation, the adherent cells were collected for adipogenesis induction. The supernatant containing non-adherent cells was cultured for an additional 24 h, after which the supernatant was collected and cultured in a new dish. After repeating this process for 2 days, the adherent cells were collected for myogenesis induction. HEK-293T cells were purchased from the American Type Culture Collection (ATCC) and were verified to be negative for mycoplasma contamination.
Differentiation of Myoblasts and Preadipocytes
Myoblasts were cultured in high-glucose DMEM, supplemented with 20% FBS. Two days after cells reached confluence, DMEM containing 2% horse serum (HyClone, USA) and 1% penicillin-streptomycin solution was used for myogenesis induction. Myotube identification was performed on the 4th day of induction. Intramuscular preadipocytes were cultured in DMEM/F12 (HyClone, USA) supplemented with 10% FBS and 1% penicillin-streptomycin solution. Adipocyte differentiation was induced by M1 medium [DMEM/F12 containing 10% FBS, 1% penicillin-streptomycin solution, 0.5 mM 3-isobutyl-1-methylxanthine (Sigma, USA), 1 μM dexamethasone (Sigma, USA), and 1.5 μg/mL insulin (Sigma, USA)]. Two days later, the M1 medium was replaced with M2 medium (DMEM/F12 containing 10% FBS, 1% penicillin-streptomycin solution, and 1.5 μg/mL insulin). Then, differentiation was induced for 8 days, during which time the medium was changed once every 2 days. Oil Red O staining was performed on the 8th day of adipogenic differentiation.
RNA Extraction and Real-Time qPCR
Total RNA was isolated with Trizol reagent (Invitrogen, Carlsbad, CA, USA) and cDNA was synthesized with PrimeScript™ RT reagent kit with gDNA Eraser (Takara, Tokyo, Japan). Real-time qualitative polymerase chain reaction (qPCR) for RNA analyses were performed using the SYBR Green PCR Master Mix (Takara, Tokyo, Japan). MiRNA-specific stem-loop primers were used for reverse transcription. The level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used to normalize the expression levels of circRNAs and mRNAs, and the level of small nuclear U6 was used to normalize the expression levels of miRNAs. The primers used in the analysis are shown in Supplementary Table 1.
Vector Construction and Cell Transfection
The second exon sequence of the INSR gene was inserted into the pCD2.1 vector (Geneseed Biotech, Guangzhou, China) and psi-CHECK2 vector (Promega, Fitchburg, WI, USA). Small interfering RNA (siRNA) oligonucleotides were designed to combine with the back-splice region of circINSR (RiboBio, Guangzhou, China). These siRNAs inhibited the expression of circINSR after transfection into the cell and was named si-circINSR. The mimics of bta-miR-15a, bta-miR-15b, bta-miR-16a, and bta-miR-16b were purchased from RiboBio (Guangzhou, China). The 3′-untranslated regions (UTRs) of the cyclin D1 (CCND1) and B-cell lymphoma 2 (Bcl-2) genes containing the miR-15/16 binding sites were amplified using the PCR enzyme mix (Platinum II Taq Hot-Start DNA Polymerase, Invitrogen). The wild-type and mutant 3'-UTR gene sequences were cloned into the psi-CHECK2 vector. The Renilla: Firefly ratio was measured and compared against that for the non-treated control. The mimics (50 nM) or vectors (2 μg/mL) were transfected into cells using a transfection reagent (R0531, Thermo Fisher Scientific, USA). For the overexpression of the miR-15/16 family, the miR-15a, miR-15b, miR-16a, and miR-16b mimics were mixed in equal amounts for transfection.
Oil Red O and BODIPY Staining
After 8 days of differentiation, the intramuscular preadipocytes were stained with Oil Red O (#O0625, Sigma, USA) and 4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene (BODIPY 493/503; D3922, Thermo Fisher Scientific). Oil Red O staining was performed according to the manufacturer's instructions. To quantify the staining of fat droplets, 100% isopropanol was used to dissolve the lipid droplets, and the absorbance was measured at 510 nm. For BODIPY staining, the cells were washed twice with PBS to remove residual 4% paraformaldehyde. Hank's Balanced Salt Solution containing 10 μM BODIPY 493/503 was added to the cells and then incubated at 37°C for 30 min in the dark. The samples were washed three times with PBS and photographed immediately.
Immunofluorescence Analysis
After 4 days of myogenic differentiation, myoblasts were fixed with 4% paraformaldehyde. After washing with PBS, myosin heavy chain (MyHC) antibody (1:250, Heavy chain cardiac Myosin antibody, GTX20015, GeneTex, USA) was incubated with the cells overnight at 4°C. The goat anti-mouse IgG (H&L) -Alexa Fluor 594 (1:500; RS3608; Immunoway Biotechnology, USA) was used as a secondary antibody. After the secondary antibody was diluted with PBS containing 1% bovine serum albumin, it was added to the cells and incubated for 2 h at room temperature. The nucleus was stained with 4′,6-diamidino-2-phenylindole (DAPI, 1:500, #62248, Thermo, USA). Finally, we washed the cells three times with PBS and observed them under a fluorescence microscope (DM5000B; Leica, Germany). The myotube coverage area was analyzed by Image-Pro Plus software.
Dual-Luciferase Reporter Assay
HEK-293T cells were co-transfected with miR-15/16 mimics and plasmids. After 24 h of transfection, luciferase activity was detected using the Dual-Luciferase Reporter Assay Kit (#E2920, Promega, Fitchburg, WI, USA). The optical density of the resulting solution was assessed using an automatic microplate reader (Molecular Devices, Sunnyvale, USA).
RNA Fish
RNA fluorescence in situ hybridization (RNA-FISH) probes that specifically recognize the back-splicing junction region of circINSR were designed (RiboBio, Guangzhou, China). Primary myoblasts and preadipocytes were fixed with in situ hybridization fixative. After pre-hybridization, the cells were incubated with the circINSR probes at 37°C overnight. Nuclei were stained with DAPI. Laser confocal microscopy was used to observe the localization of circINSR in cells (Nikon, Tokyo, Japan).
RNA-Binding Protein Immunoprecipitation (RIP)
RNA-binding protein immunoprecipitation (RIP) assay was performed using the EZ-Magna RIP kit (#17-701, Millipore, Billerica, USA), according to the manufacturer's instructions. The cells were pelleted and resuspended in an equal pellet volume of RIP Lysis Buffer (~100 mL). The cell lysates (100 mL) were incubated with 5 mg beads coated with control mouse IgG or an antibody against Argonaute2 (Ago2, Abcam, UK) with rotation at 4°C overnight. After treatment with proteinase K, the RNAs were extracted with phenol: chloroform: isoamyl alcohol (125:24:1; pH, 4.5; #PD0419, Huyu Biotechnology, China) according to the steps in the Magna RIP kit. RNA was reverse transcribed using the Prime-Script RT Master Mix (TaKaRa, Japan), and the abundance of circINSR and miR-15/16 was detected by real-time qPCR.
CircRNA Pull-Down
Biotin-labeled circINSR probe and negative control probe (NC probe) (RiboBio, Guangzhou, China) were used for circRNAs pull-down. We purchased the Pierce™ magnetic RNA-protein pull-down kit (#20164, Thermo, USA) and performed the experiment according to the manufacturer's instructions. In brief, the biotin-labeled probe was bound to streptavidin magnetic beads for 30 min. Then, the probe-labeled magnetic beads were incubated with the cell lysates from myogenic cells and preadipocytes for 1 h. Then, the RNA in the immunoprecipitates were eluted, extracted, and purified. Finally, real-time qPCR was used to detect the expression levels of circINSR and miR-15/16 in the immunoprecipitates.
5-Ethynyl-2′-deoxyuridine (EdU) and Cell Counting Kit-8 (CCK-8) Assay
When the density of myoblasts and preadipocytes reached 40–50%, transfection was performed with overexpression plasmid, siRNA, or miRNA mimics. After 24 h of transfection, cell proliferation was tested using an EdU assay kit (RiboBio, Guangzhou, China). The nucleus was stained with Hoechst 33342 (RiboBio, Guangzhou, China), and a fluorescence microscope was used to obtain images immediately after staining (AMG EVOS, Seattle, WA, USA). Similarly, we also used the CCK-8 (Multisciences, Hangzhou, China) assay to detect the level of cell proliferation after transfection. The optical density of CCK-8 at 450 nm was measured using an automatic microplate reader (Synergy4, BioTek, Winooski, USA).
Cell Cycle and Apoptosis Assay
We used flow cytometry and the Cell Cycle Testing Kit (Multisciences, Hangzhou, China) to analyze the cell cycle. Myoblasts and preadipocytes were transfected when the cell growth density reached 50%. After transfection for 24 h the cells were collected and fixed overnight in 75% ethanol at 4°C. Subsequently, the kit instructions were followed to perform staining. Flow cytometry analysis was performed on a BD Accuri C6 now cytometer (FACS Canto™ II, BD Biosciences, USA), and data were processed using FlowJo7.6 software. Cell apoptosis assays were performed with the Annexin V-PE/7-AAD Apoptosis Detection Kit (RiboBio, Guangzhou, China) according to the manufacturer's recommendations. Afterward, the apoptosis rate was analyzed using flow cytometry (FACS Canto™ II, BD BioSciences, USA). Each treatment group was performed using three independent replicates.
Western Blot Analysis
Proteins from cultured myoblasts and preadipocytes were lysed with RIPA buffer (Solarbio, Beijing, China). Proteins were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes (Thermo Fisher Scientific). The membranes were incubated overnight with primary antibodies specific for anti-GAPDH (1:1,000, #ab9485, Abcam, Cambridge, UK), anti-CyclinD1 (CCND1, 1:1,000, #ab226977, Abcam, Cambridge, UK), anti-Bcl-2 (1:500, #bs-0032R, Bioss, Beijing, China), anti-caspase-9 (1:500, #bs-0049R, Bioss, Beijing, China), anti-Bax (1:500, #bs-0127M, Bioss, Beijing, China), anti-fatty acid-binding protein (FABP4, 1:500, #bsm-51247M, Bioss, Beijing, China), anti-proliferating cell nuclear antigen (PCNA, 1:500, #WL01804, Wanlei Bio, Shenyang, China), anti-cyclin-dependent kinase (CDK2, 1:500, #WL01543, Wanlei Bio, Shenyang, China), anti-PPARγ (1:500, #WL01800, Wanlei Bio, Shenyang, China) and anti-C/EBPα (1:500, #WL01899, Wanlei Bio, Shenyang, China) at 4°C. The goat anti-mouse IgG (H&L)-horseradish peroxidase (HRP, 1:5,000, #bs-40296G, Bioss, China), and goat anti-rabbit IgG (H&L)-HRP (1:5,000, #bs-40295G, Bioss, China) were used as secondary antibodies. Protein bands were visualized using an enhanced chemiluminescence visualization system (ECL Plus, Amersham Life Sciences). The membranes were quantified with the chemiluminescence system (Bio-Rad, Hercules, CA, USA).
Statistical Analyses
Data are expressed as the mean ± standard error (SEM) of at least three independent experiments. Statistical analyses were performed using SPSS 19.0 statistical software (SPSS, Chicago, IL, USA). Significant differences were determined using Student's t-test. A probability of 0.05 or less was considered statistically significant.
Results
Myoblasts and Intramuscular Preadipocytes Were Isolation From the Bovine Fetus
Skeletal muscle and intramuscular adipose tissue differentiate from mesodermal mesenchymal stem cells (MSCs) (Westerweel and Verhaar, 2008; Du et al., 2010a). The muscle begins to develop during the early embryonic period, and adipose tissue begins to develop during the second trimester (Du et al., 2010b) (Figure 1A). Enzymatic digestion, combined with the differential adhesion method, was able to roughly separate myoblasts from intramuscular preadipocytes. In this study, these isolated preadipocytes were spindle-shaped and possessed characteristics common to fibroblasts. After 8 days of adipogenic induction, small lipid droplets accumulated in some cells. The Oil Red O staining results also intuitively indicated that the isolated cells had undergone adipogenic differentiation (Figure 1B). The results of BODIPY staining showed that 8 days of adipogenic induction resulted in lipid accumulation in intramuscular preadipocytes (Figure 1C). In addition, the real-time qPCR results indicated the significantly increased expression levels of adipogenesis marker genes PPARγ and C/EBPα in these cells (Figure 1E). Similarly, MyHC immunofluorescence showed that the isolated myoblasts were able to be induced into myotubes. The proportion of myotubes was analyzed by ImageJ software, showing ~35% differentiation (Figure 1D). Myogenic differentiation marker genes were also significantly overexpressed in these differentiated myotubes (Figure 1F).
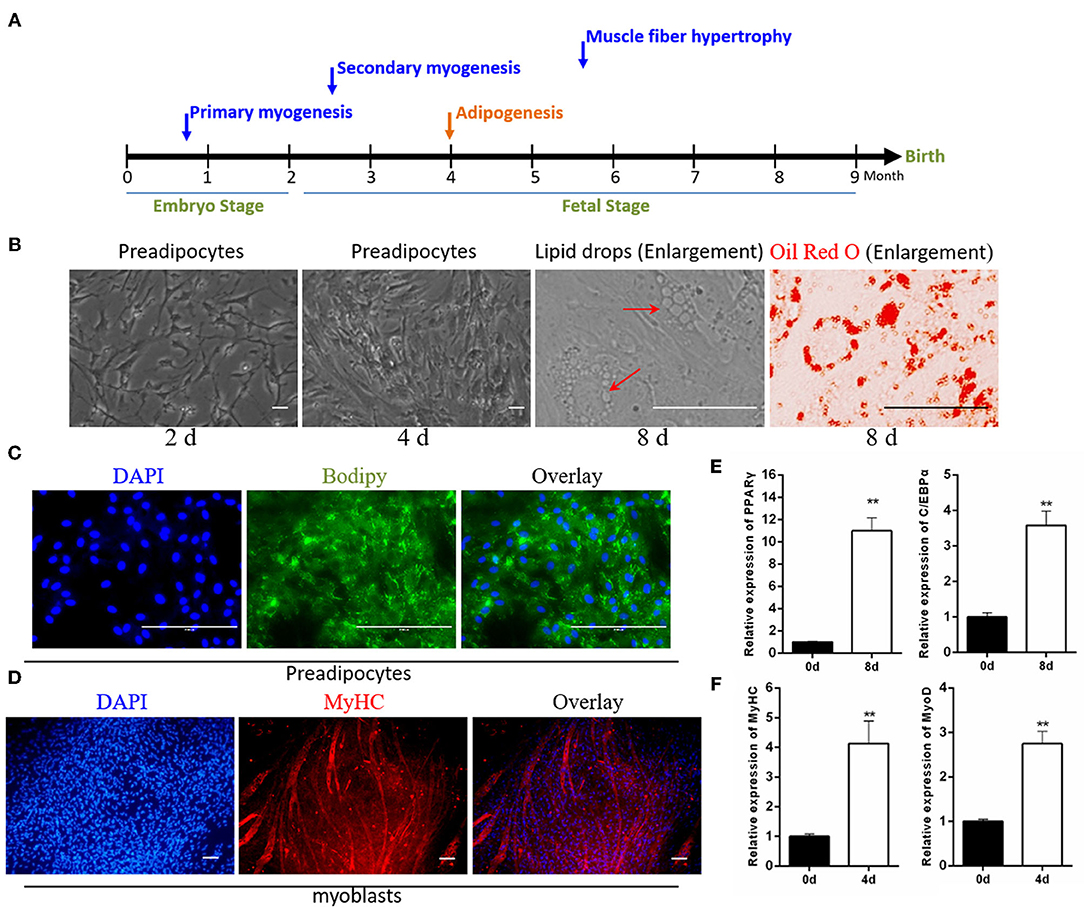
Figure 1. Identification of isolated myoblasts and intramuscular preadipocytes. (A) The growth pattern of muscle and fat during the embryonic development of cattle. (B) The growth morphology of isolated preadipocytes on days 2 and 4, and the Oil Red O staining of intracellular lipid droplets after 8 days of adipogenic differentiation. (C) BODIPY staining of preadipocytes after 8 days of adipogenic differentiation. (D) Anti-MyHC immunofluorescence staining of myoblasts after 4 days of myogenic differentiation. (E) The expression of adipogenic marker genes PPARγ and C/EBPα after the differentiation of preadipocytes. (F) The expression of muscle differentiation marker genes MyHC and MyoD after the differentiation of myoblasts. Data are presented as the mean ± SEM. n = 3. **P < 0.01.
CircINSR Serves as a Sponge for miR-15/16
Previous studies have shown that circINSR is able to adsorb miR-34a (Shen et al., 2020). According to the principle of adsorption between circRNAs and miRNAs, combined with prediction results based on high-throughput sequencing data, we found that circINSR also has the seed region binding site for the miR-15/16 family. In this study, after the overexpression of circINSR in myoblasts and preadipocytes, real-time qPCR results showed that the expression levels of miR-15a, miR-15b, miR-16a, and miR-16b were significantly reduced (Figures 2A,B). MiR-195 also belongs to the miR-15/16 family, but the expression level of miR-195 did not change following the overexpression of circINSR (Figure 2A). The binding sites for the miR-15/16 family on circINSR were predicted using Target Scan 7.0 and miRanda (Figure 2C). The results of the dual fluorescence reporter system showed that the overexpression of the miR-15/16 family significantly inhibited Renilla luciferase activity in the psi-CHECK2-circINSRWT vector (Figure 2D). Because mature miRNA functions in the cytoplasm, circRNAs located in the cytoplasm are more likely to absorb miRNA (Hansen et al., 2013b; Barrett and Salzman, 2016). We used RNA-fluorescence in situ hybridization (FISH) probes targeting circINSR to analyze the subcellular localization of circINSR in myoblasts and preadipocytes. The results showed that circINSR was highly expressed in the cytoplasm of both cell types (Figure 2E). To determine the adsorption relationship between circINSR and miR-15/16, we designed a biotin-labeled circINSR probe. By taking advantage of the affinity between streptavidin magnetic beads and biotin probes, we conducted an RNA-pull down test. The results of the RNA-pull down assay revealed the high expression levels of circINSR and the presence of miR-15/16 family members in the circINSR immunoprecipitates from both myogenic cells and preadipocytes. A negative probe (NC probe) was used as a control (Figures 2F,G). To verify this adsorption, we performed an Ago2-RIP assay in both myoblasts and preadipocytes to detect whether both endogenous circINSR and miR-15/16 were bound to Ago2 protein. The results showed that the miR-15/16 family could be enriched by Ago2 protein pull-down from both myoblasts and preadipocytes (Figure 2I). The expression of circINSR was significantly higher in the Ago2 protein immunoprecipitation than in the IgG group (Figure 2H). To determine the accuracy of subsequent cell function tests, we overexpressed circINSR in myogenic cells and preadipocytes and examined the overexpression level of circINSR in cells co-transfected with circINSR and miR-15/16. The results show that the pCD2.1 vector can successfully overexpress circINSR in cells (Figure 2J).
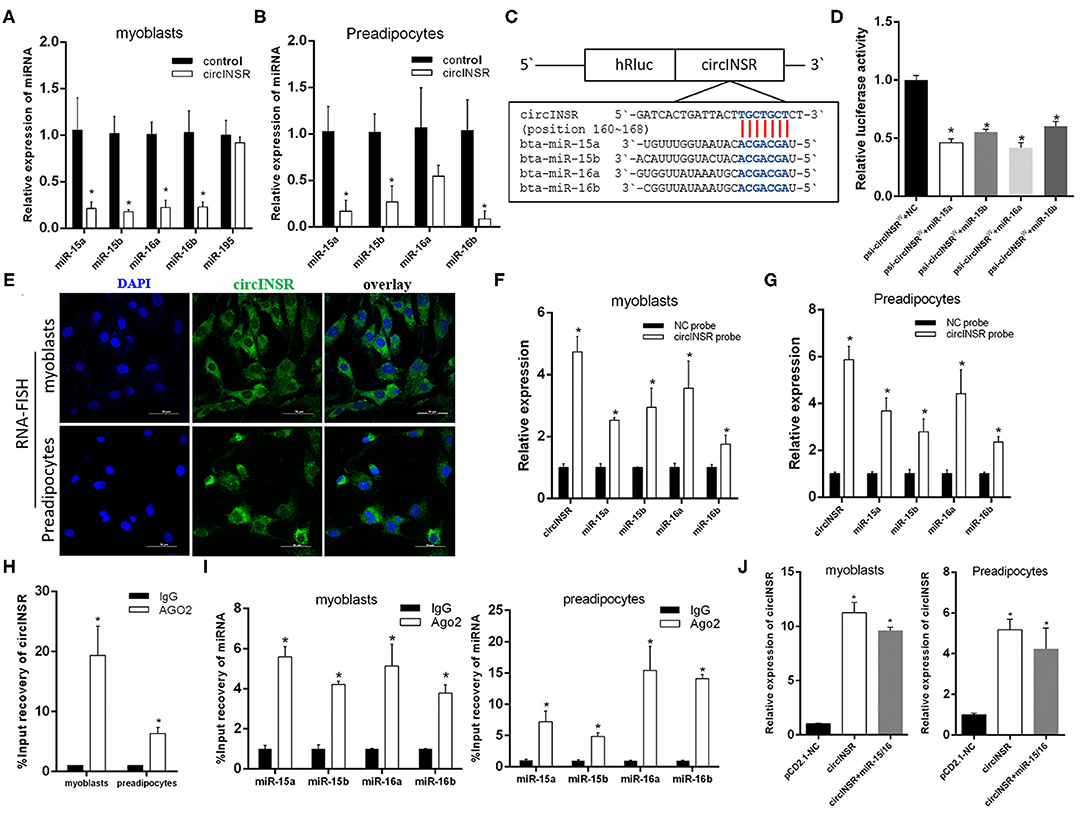
Figure 2. CircINSR sponges the miR-15/16 family in myoblasts and preadipocytes. (A) Changes in miR-15/16 family expression levels in myoblasts after the overexpression of circINSR. (B) Changes of miR-15/16 family expression levels in preadipocytes after the overexpression of circINSR. (C,D) Luciferase reporter activity of circINSR-WT in HEK-293T cells co-transfected with miR-15/16 mimics or mimic NC. (E) FISH detection of circINSR in myoblasts and preadipocytes. Scale bars, 50 μm. (F,G) CircRNA pull-down assays were performed using a specific biotin-labeled circINSR probe in myoblasts and preadipocytes. Real-time qPCR was used to detect the expression levels of circINSR and miR-15/16 family members in immunoprecipitates. n = 3. *P < 0.05 compared to negative control (NC) probe. (H) Ago2-RIP assay to assess the quantity of circINSR in myoblasts and preadipocytes. (I) Ago2-RIP assay to assess the quantity of miR-15/16 family members in myoblasts and preadipocytes. n = 3. *P < 0.05. (J) After overexpressing circINSR or co-transfecting circINSR with miR-15/16 in myoblasts and preadipocytes, real-time qPCR was used to detect the expression level of circINSR in cells. Data are presented as the mean ± SEM. n = 3. *P < 0.05.
CircINSR Promotes Myoblast Proliferation and Inhibits Apoptosis
Our results showed that the expression level of circINSR in myoblasts gradually increases during in vitro culture, which implied that circINSR plays an important role in myoblast development (Figure 3A). Previous studies have reported that CCND1 and Bcl-2 are the target genes of the miR-15/16 family (Cimmino et al., 2005; Cai et al., 2012; Pekarsky et al., 2018; Cao et al., 2019). We inserted wild-type and mutant forms of the 3'-UTR sequence containing the miR-15/16 family binding site into the psi-CHECK2 vector to verify this binding using the dual fluorescent reporter gene system (Figure 3B). The results showed that the overexpression of the miR-15/16 family could significantly inhibit the activity of Renilla luciferase in the wild-type vector (Figure 3C). CCND1 gene is a key regulator of cell proliferation. The real-time qPCR results showed that the overexpression of miR-15/16 mixed mimics could significantly reduce the expression levels of CCND1 and other cell proliferation marker genes. However, the co-transfection of circINSR and miR-15/16 was able to alleviate this inhibition (Figure 3D). The EdU assay results showed that the transfection of miR-15/16 mixed mimics was able to significantly reduce the number of proliferating cells, and co-transfection with circINSR was able to rescue this anti-proliferation effect (Figures 3E,F). CCK-8 cell proliferation analysis also obtained similar results (Figure 3G). The cell cycle results indicated that miR-15/16 blocked the cell cycle and reduced the number of cells that entered the S phase, whereas co-transfection with circINSR alleviated this inhibition (Figures 3H,I).
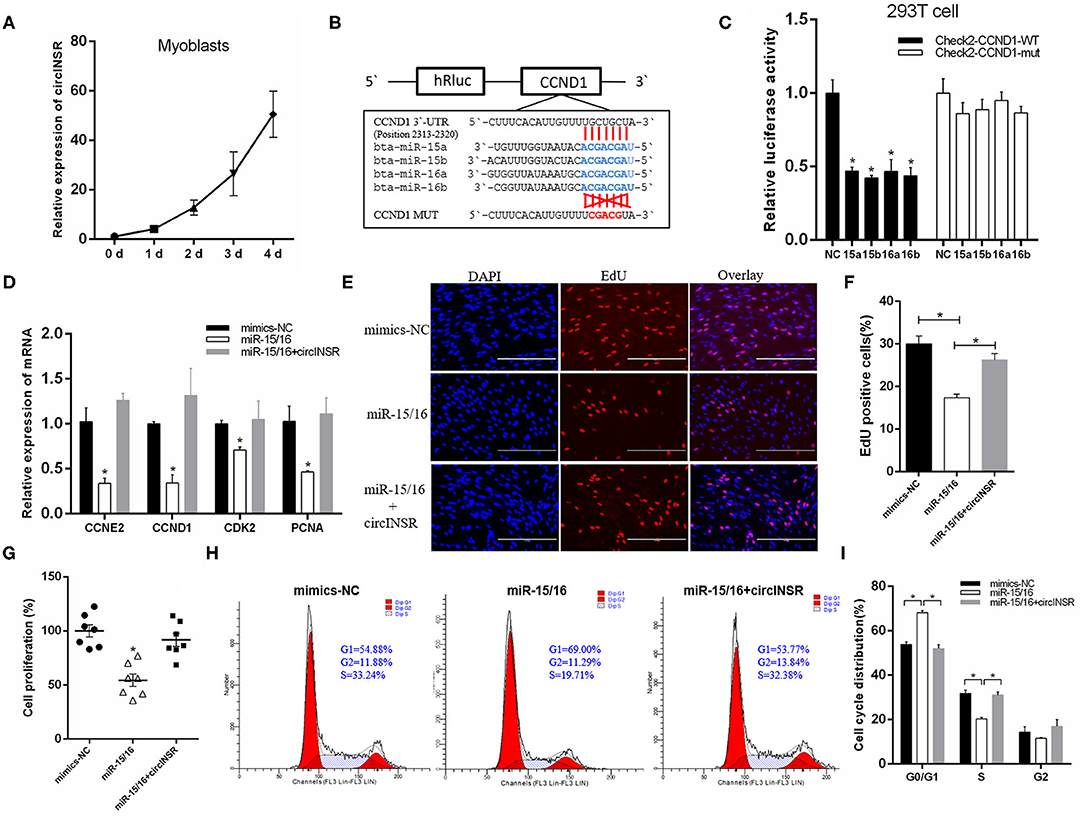
Figure 3. CircINSR promotes myoblasts proliferation by sponging the miR-15/16 family. (A) The expression of circINSR was detected by real-time qPCR during in vitro culture of myoblasts. (B) TargetScan predicted that CCND1 3'-UTRs contained binding sites for miR-15/16. (C) The fluorescence activity changes after co-transfection with dual fluorescent reporter vectors and miR-15/16. n = 3. (D) The expression of marker genes associated with cell proliferation after co-transfection with circINSR and miR-15/16 in myoblasts. n = 3. (E,F) EdU assay for myoblasts transfected with miR-15/16 mimics, either alone or with circINSR. Scale bars, 200 μm. n = 3. (G) CCK-8 assay for myoblasts transfected with miR-15/16 mimics, either alone or with circINSR. n = 6. (H,I) Cell cycle assay for myoblasts transfected with miR-15/16 mimics, either alone or with circINSR. n = 3. Data are presented as the mean ± SEM. *P < 0.05.
To verify the targeting relationship between miR-15/16 and Bcl-2, we constructed wild-type and mutant psi-CHECK2 vectors (Figure 4A). The results of the fluorescence report analysis indicate that Bcl-2 was a potential target gene of miR-15/16 (Figure 4B). Real-time qPCR results showed that miR-15/16 significantly inhibited the expression of Bcl-2 and promoted the expression of apoptosis marker genes Bax and Caspase9 (Figure 4C). The subsequent flow cytometry analysis results showed that overexpression of miR-15/16 promoted myoblast apoptosis, whereas the co-transfection with circINSR reduced the number of apoptotic cells (Figures 4D,E).
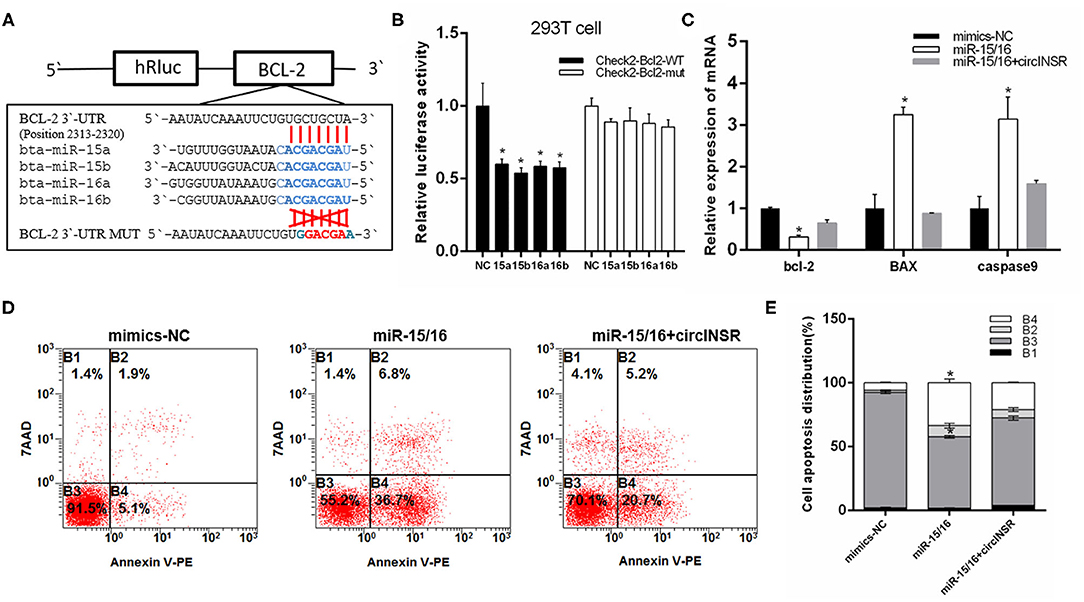
Figure 4. CircINSR inhibits the apoptosis of bovine primary myocytes by sponging the miR-15/16 family. (A) TargetScan predicted that Bcl-2 3'-UTRs contained binding sites for miR-15/16. (B) The fluorescence activity changes after co-transfection with dual fluorescent reporter vectors and miR-15/16. (C) The expression of marker genes associated with cell apoptosis after the co-transfection of circINSR and miR-15/16 in myoblasts. (D,E) Cell apoptosis was determined by Annexin V/7-AAD dual staining, followed by flow cytometry. B1: Annexin V−/7-AAD+, B2: Annexin V+/7-AAD+, B3: Annexin V−/7-AAD−, B4: Annexin V+/7-AAD−. n = 3. Data are presented as the mean ± SEM. n = 3. *P < 0.05.
CircINSR Promotes Preadipocytes Proliferation by Sponging miR-15/16
To investigate the role played by circINSR during adipogenesis, preadipocytes were transfected with either the circINSR overexpression vector or si-circINSR. The results of real-time qPCR showed that the overexpression of circINSR significantly promoted the expression of cell proliferation marker genes (Figure 5A), whereas circINSR interference with circINSR inhibited the expression of these genes (Figure 5B). Western blot analysis revealed similar results (Figure 5C). In addition, the transfection of mixed miR-15/16 mimics in preadipocytes was able to significantly inhibit the expression of cell proliferation-related genes, whereas the co-transfection with circINSR was able to restore gene expression (Figure 5D). The results of the EdU (Figures 5E,F) and CCK-8 (Figure 5G) assays also showed that miR-15/16 inhibited cell proliferation, whereas the co-transfection of miR-15/16 and circINSR alleviated this inhibitory effect. Cell cycle assay showed that miR-15/16 inhibited preadipocytes from entering the S phase, whereas the co-transfection with circINSR promoted cell proliferation (Figures 5H,I).
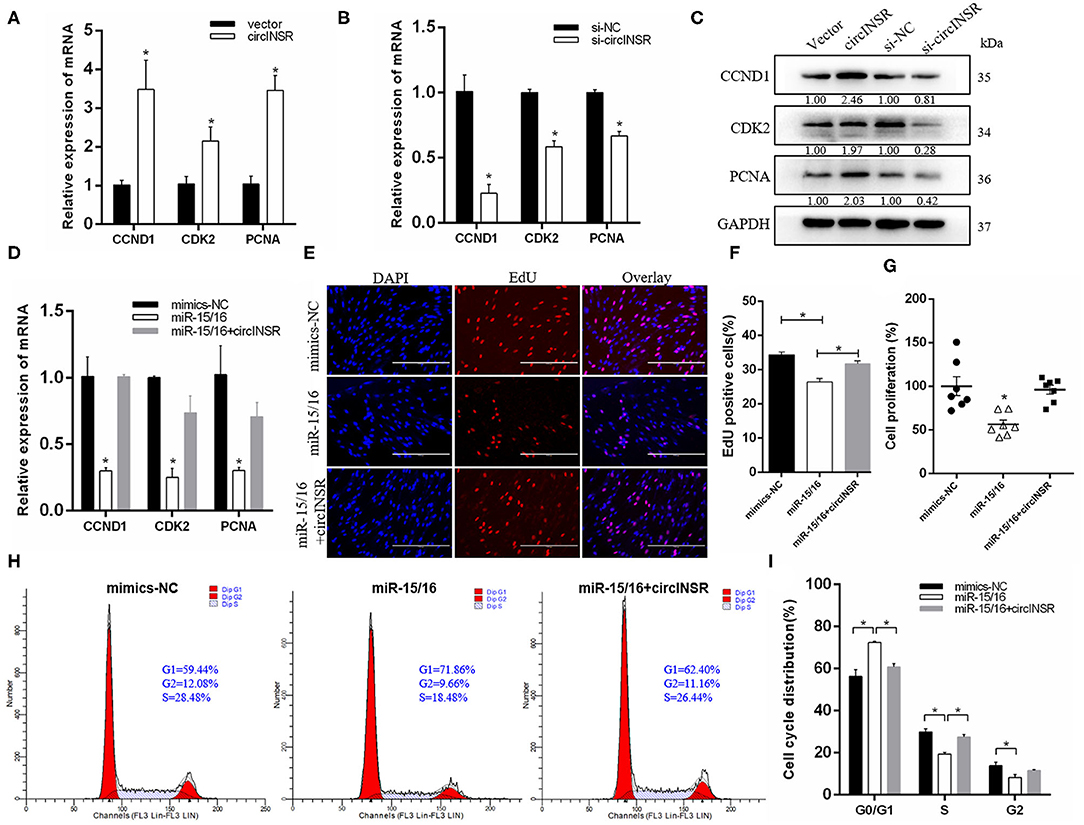
Figure 5. CircINSR promotes preadipocyte proliferation by sponging the miR-15/16 family. (A,B) The effects of circINSR overexpression and interference on proliferation marker genes in preadipocyte. n = 3. (C) The expression of CCND1, CDK2, and PCNA was detected by western blot analysis. (D) The expression of marker genes related to cell proliferation after co-transfection with circINSR and miR-15/16 in preadipocytes. n = 3. (E,F) EdU assay for preadipocytes transfected with miR-15/16 mimics, either alone or with circINSR. Scale bars, 200 μm. n = 3. (G) CCK-8 assay for preadipocytes transfected with miR-15/16 mimics, either alone or with circINSR. n = 6. (H,I) Cell cycle assay for preadipocytes transfected with miR-15/16 mimics, either alone or with circINSR. n = 3. Data are presented as the mean ± SEM. *P < 0.05.
CircINSR Inhibits Preadipocyte Apoptosis by Sponging miR-15/16
To further explore the function of circINSR in preadipocytes apoptosis, real-time qPCR was used to detect the expression of apoptosis-related genes after either the overexpression or interference of circINSR. The results showed that circINSR promoted the expression of the anti-apoptotic gene Bcl-2 and inhibited the expression of the pro-apoptotic genes BAX and Caspase9 (Figures 6A,B). The western blots analysis revealed a similar trend (Figure 6C). In contrast, real-time qPCR results showed that the overexpression of miR-15/16 inhibited Bcl-2 gene expression and promoted BAX and caspase9 expression. The co-transfection of miR-15/16 with circINSR inhibited the apoptosis of preadipocytes (Figure 6D). To further verify that circINSR could inhibit cell apoptosis by adsorbing the miR-15/16 family, we used Annexin V-PE/7-AAD staining combined with flow cytometry to analyze the effects of the co-transfection of miR-15/16 and circINSR on cell apoptosis. The results showed that miR-15/16 promoted cell apoptosis, whereas the co-transfection with circINSR rescued this anti-apoptotic effect (Figures 6E,F).
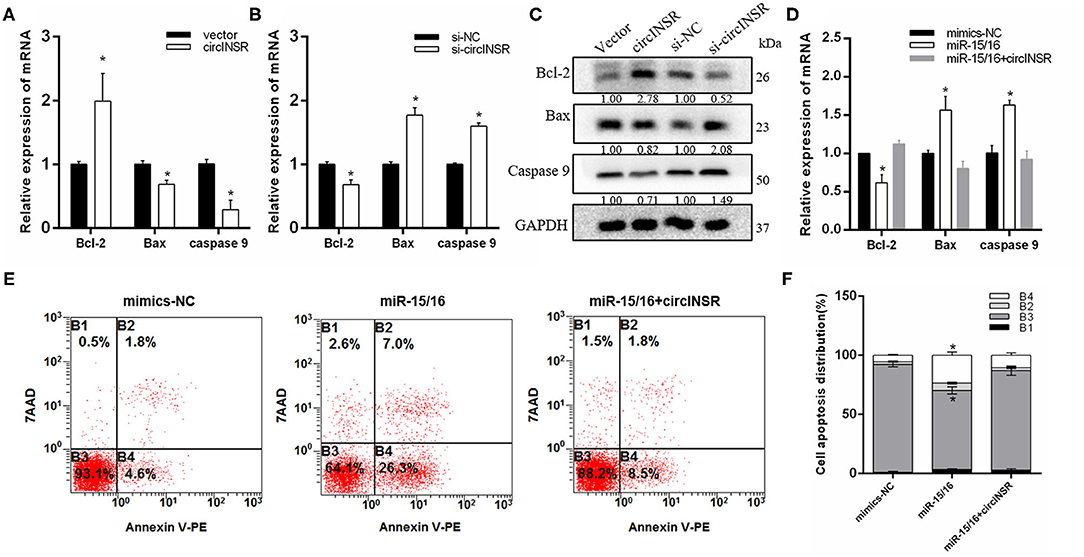
Figure 6. CircINSR inhibits preadipocytes apoptosis by sponging the miR-15/16 family. (A,B) The mRNA levels of cell apoptosis markers, including Bcl-2, Bax, and Caspase9, were detected by real-time qPCR in preadipocytes transfected with circINSR or siRNA. (C) The protein expression levels of Bcl-2, Bax, and Caspase9 were detected by western blot analysis. (D) The expression of marker genes associated with cell apoptosis after the co-transfection of circINSR and miR-15/16 in preadipocytes. (E,F) Cell apoptosis was determined by Annexin V/7-AAD dual staining, followed by flow cytometry. B1: Annexin V−/7-AAD+, B2: Annexin V+/7-AAD+, B3: Annexin V−/7-AAD, B4: Annexin V+/7-AAD. Data are presented as the mean ± SEM. n = 3. *P < 0.05.
CircINSR Inhibits Preadipocytes Differentiation
The above results suggested that circINSR could promote the proliferation of myoblasts and preadipocytes and inhibit cell apoptosis. Our results showed that the expression of circINSR gradually decreased during preadipocyte differentiation (Figure 7A). However, the expression of miR-15/16 family increased during the differentiation process (Supplementary Figure 1). We further studied the function of circINSR during preadipocyte differentiation. The results showed that the overexpression of circINSR was able to inhibit the expression of adipogenesis-related genes, including PPARγ (Castillo et al., 1999), C/EBPα (Westerweel and Verhaar, 2008), and FABP4 (Hoashi et al., 2008) (Figures 7B,C). In contrast, interference with circINSR promoted the expression of these genes (Figures 7D,E). BODIPY staining facilitates the direct observation of lipid droplet formation in adipocytes by monitoring green fluorescence. Because the circINSR overexpression vector carries a GFP cassette, we could only analyze BODIPY staining after circINSR interference and not under conditions of circINSR overexpression. Eight days after induction, the cells were subjected to BODIPY staining. The results showed that si-circINSR significantly increased the intensity of green fluorescence in preadipocytes (Figure 7F). In addition, Oil Red O staining results showed that overexpression of circINSR inhibited the lipogenesis of precursor fat, whereas the accumulation of lipid droplets increased after interference with circINSR (Figures 7G,H).
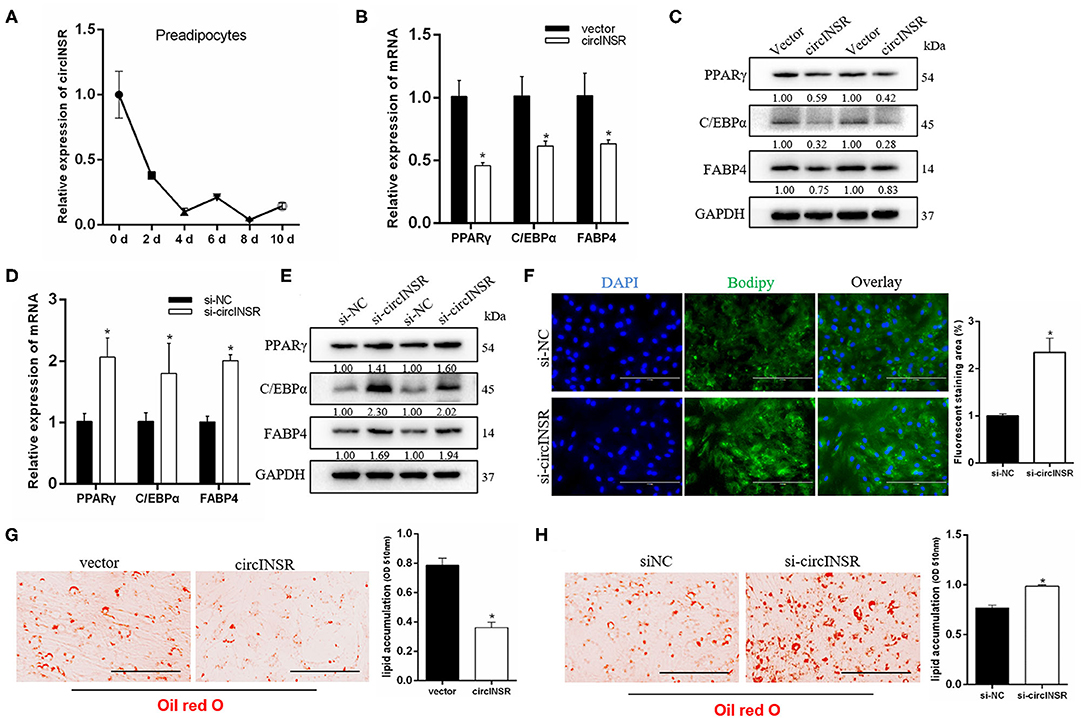
Figure 7. CircINSR inhibits adipogenic differentiation of preadipocytes. (A) The expression of circINSR was detected by real-time qPCR during preadipocytes differentiation. (B,C) The expression of adipogenic marker genes after the overexpression of circINSR in preadipocytes was detected by real-time qPCR and western blot analyses. (D,E) The expression of adipogenic marker genes after interference with circINSR in preadipocytes was detected by real-time qPCR and western blot analyses. (F) Interference with circINSR in preadipocytes, followed by BODIPY staining to analyze lipid droplet deposition. The fluorescence signal was analyzed by ImageJ software. (G,H) Lipid droplets in preadipocytes were stained with Oil Red O. Lipid contents were measured by spectrophotometric analysis after dissolution in isopropanol. Data are presented as the mean ± SEM. n = 3. *P < 0.05.
To further analyze whether the effect of circINSR on adipogenesis was related to miR-15/16, we identified two reported target genes for miR-15/16, Forkhead box protein O1 (Foxo1), (Dong et al., 2014) and Ethanolamine phosphotransferase 1 (EPT1) (Xu et al., 2019) (Figure 8A). Real-time qPCR results showed that the expressions of FOXO1 and EPT1 were significantly reduced after miR-15/16 overexpression, whereas the co-transfection with circINSR rescued this inhibition (Figure 8B). In addition, miR-15/16 expression promoted the expression of adipogenic genes, whereas circINSR expression inhibited the expression of adipogenic genes (Figure 8C). The results of Oil Red O staining showed that miR-15/16 expression promoted lipid accumulation in preadipocytes, whereas the co-transfection with circINSR inhibited adipogenesis (Figures 8D,E).
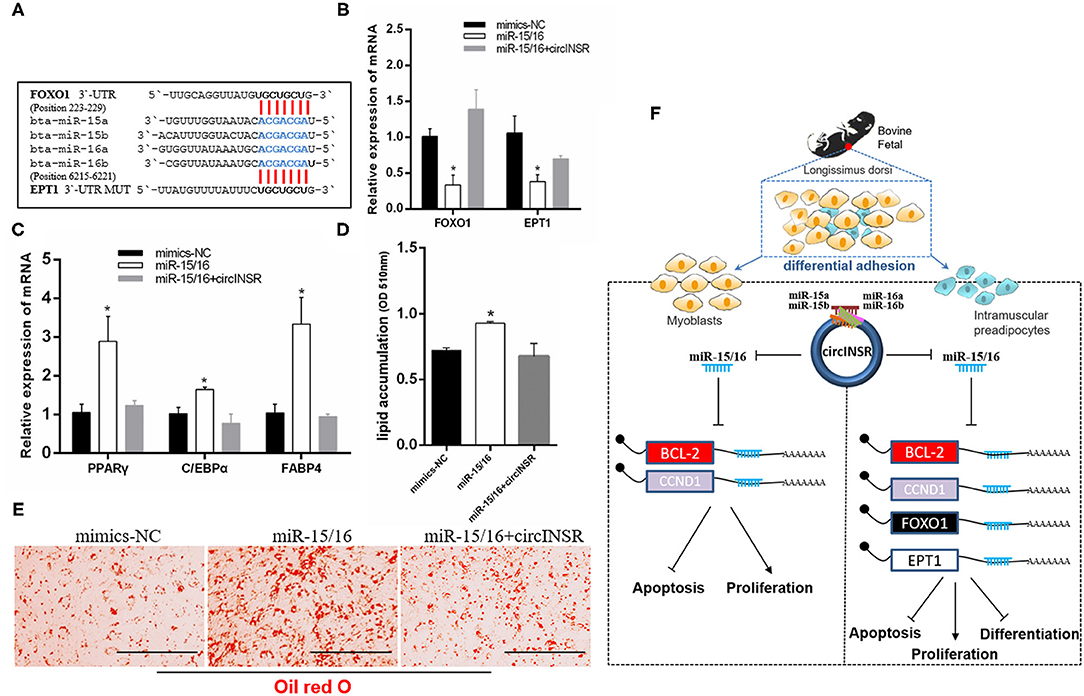
Figure 8. Adipogenic differentiation was regulated by circINSR through miR-15/16. (A) The reported target genes of the miR-15/16 family. (B) The effects of miR-15/16 transfection, either alone or with circINSR, on target genes in preadipocytes. (C) The expression of adipogenic marker genes in preadipocytes was detected by real-time qPCR. (D,E) Lipid droplets in preadipocytes were stained with Oil Red O. Lipid contents were measured by spectrophotometric analysis after dissolution in isopropanol. Data are presented as the mean ± SEM. n = 3. *P < 0.05. (F) Schematic diagram of the mechanisms through which circINSR regulates the proliferation, apoptosis, and differentiation in myoblasts and preadipocytes.
Discussion
Myoblasts and intramuscular adipocytes are derived from mesenchymal stem cells (MSCs) (Du et al., 2010a). As a result of complex signaling regulatory pathways, some MSCs will differentiate into myogenic and non-myogenic cell lines. Myogenic cells enter the process of muscle development, whereas non-myogenic cells enter the process of adipogenesis or fibroblast development (Du et al., 2010b; Stachecka et al., 2018). At ~180 days of gestation in bovine fetuses, IMF begins to appear, suggesting that adipogenic progenitor cells have already begun to differentiate into preadipocytes at this stage (Du et al., 2013). Therefore, we can separate myoblasts and intramuscular preadipocytes by using enzyme digestion combined with differential adhesion screening.
In this study, by strictly following the isolation and induced differentiation protocols that have been reported by previous authors (Beier et al., 2011; Miyake et al., 2012; Wang et al., 2015), we obtained myoblasts with high purity. The method of myoblast separation in this study has been widely used (Wei et al., 2017; Song et al., 2019; Wang et al., 2019). The paired box transcription factor Pax7 is a key regulator of skeletal muscle stem cells and is required along with Pax3 to generate skeletal muscle precursors (Seale et al., 2000; Relaix et al., 2005). Wang et al. reported that the primary myoblasts isolated using the same method could be stained with anti-PAX7 fluorescent antibody (Yi-Min et al., 2014). In this study, the thick myotubes could be labeled with anti-MyHC fluorescent antibody after 4 days of differentiation. However, the isolated preadipocytes contained MSCs, fibroblasts, and adipogenic progenitor cells, which affected cell purity. In studies of bovine intramuscular fat, adult beef is typically used as the starting material for preadipocyte isolation (Aso et al., 1995; Li et al., 2011). However, in our research, to study the parallel roles played by circINSR in the development of embryonic muscle and intramuscular fat, we chose to perform the cell isolation procedure during the embryonic stage. Existing research has indicated that the overall mass of adipose tissue increases with age in livestock (Hausman et al., 2009). Therefore, the number of embryonic preadipocytes was likely very small. Combining cell surface marker proteins (such as CD140a) and flow cytometry sorting might improve the purity of preadipocytes in future experiments (Guan et al., 2017). However, in view of the pluripotency of embryonic cell differentiation, under the adipogenic induction conditions in this study (100% cell density before differentiation), about 30% of the cells undergo adipogenic differentiation and form lipid droplets (Figure 8E). The preadipocytes were also successfully induced to differentiate, and lipid droplets could be identified using Oil Red O and BODIPY staining. Therefore, the cells obtained through our isolation and differentiation method could be used to explain the function of circINSR to a certain extent.
The development of muscle and fat is regulated by a complex signal network that involves both coding genes and non-coding RNAs. In this study, the expression levels of PPARγ and C/EBPa in differentiated preadipocytes increased significantly. After 4 days of myogenic differentiation, MyoD and MyHC expression increased significantly. These coding genes play important roles in the differentiation of muscle and fat. In addition, an increasing number of reports have indicated that non-coding RNAs are also involved in the regulation of muscle and fat development (Sun et al., 2018; Li et al., 2019; Jiang et al., 2020).
CircINSR is highly homologous to the human has_circ_0048966, which is formed by the head-to-tail splicing of INSR exon 2 (552 bp), and is primarily expressed in the cytoplasm. In previous studies, circINSR has been shown to regulate cell proliferation and apoptosis through miR-34a-modulated Bcl-2 and CyclinE2 expression (Shen et al., 2020). In this study, we found that circINSR was able to sponge the miR-15/16 family. The real-time qPCR results showed that the overexpression of circINSR in myoblasts and preadipocytes could significantly inhibit the expression of the miR-15/16 family. After the co-transfection of the psi-CHECK2-circINSRWT vector and miR-15/16, the circINSR sequence, which contained in the 3'-UTR region of Renilla luciferase, was recognized by miR-15/16, affecting the translation of the Renilla gene and decreasing the Renilla: Firefly ratio in the final system, which indirectly supports a targeting relationship between circINSR and miR-15/16.
In addition, we used RNA pull-down and RNA-RIP technologies to verify the adsorption relationship between circINSR and the miR-15/16 family. First, we designed a biotin-labeled oligonucleotide probe for the specific linker sequence of circINSR. Using streptavidin-coated magnetic beads, we could precipitate and separate the protein and RNA complexes bound to circINSR. In this study, we detected high levels of miR-15/16 in the immunoprecipitation complex, further supporting that circINSR could adsorb the miR-15/16 family in cells. Ago2 (Argonaute 2) protein is the core component of the RNA-induced silencing complex (RISC), which acts on mature miRNA (Chendrimada et al., 2005). The Ago2 protein immunoprecipitation method is an important method used to isolate and identify miRNA target genes and is now widely used to verify the targeting relationship between circRNA and miRNA (Wang et al., 2018; Tang et al., 2019). In our study, the results of Ago2-RIP also illustrated the targeting relationship between circINSR and miR-15/16. Therefore, according to the molecular mechanism of sponging miRNAs, circRNAs should have the same targeting ability in view of the same seed sequence of miRNAs family.
In animals, single-stranded miRNAs bind to specific mRNAs through sequences that are imperfectly complementary to the target mRNAs, particularly those found in the 3'-UTR (Carthew and Sontheimer, 2009). Existing studies have reported the regulatory mechanism of Bcl-2 and CCND1 in cancer cells, including the post-transcriptional downregulation by miR-15 and miR-16 (Cimmino et al., 2005; Pekarsky and Croce, 2015; Pekarsky et al., 2018). In this study, we verified the interaction between miR-15/16 with Bcl-2 and CCND1. The overexpression of miR-15/16 in myoblasts and preadipocytes inhibited cell proliferation and promoted apoptosis. Additionally, the effects of miR-15/16 were counteracted when circINSR was co-transfected. These results indicated that during the embryonic stage, circINSR could promote muscle development and increase the number of intramuscular preadipocytes.
The number of intramuscular preadipocytes determines the degree of marbling that can occur during fattening. However, the pre-mature maturation of the IMF can result in fetal muscle insufficiency and the wasting of nutrition during pregnancy. For example, the early muscle tissue that develops during Duchenne muscular dystrophy is manifested as muscle fiber regeneration and mild lipid droplets; however, the late muscle fibers are gradually replaced by fat and connective tissue, with deleterious effects on muscle function (Foxley et al., 1991; Wren et al., 2008; Gaeta et al., 2012). Studies have indicated that miR-15/16 promotes adipogenesis by targeting Foxo1 (Dong et al., 2014) and EPT1 (Xu et al., 2019). Existing studies have revealed that Foxo1 can inhibit the expression of PPARγ to regulate the differentiation of adipocytes (Armoni et al., 2006; Wang and Tong, 2008). Dong et al. (2014) reported that miR-15a/b could promote adipogenesis by inhibiting the target gene Foxo1. EPT1, also known as selenoprotein 1 (SELENO1), encodes an enzyme that transfers phosphor-ethanolamine to produce ethanolamine glycerophospholipids (Horibata, 2018). Xu et al. (2019) reported that miR-16 regulates 3T3-L1 adipocyte differentiation through its target gene EPT1. In our results, the overexpression of circINSR in preadipocytes inhibited the expression of key genes associated with adipogenic differentiation and reduced lipid droplet formation. This circINSR-mediated inhibition of adipogenesis was achieved by sponging miR-15/16. Therefore, one function of circINSR may be to inhibit adipogenesis during the fetal period to maintain the normal muscle function while ensuring the development of a sufficient number of intramuscular preadipocytes.
In conclusion, our results revealed that circINSR could negatively regulate miR-15/16 family expression (Figure 8F). CircINSR promotes the proliferation of myoblasts and preadipocytes and inhibits apoptosis. CircINSR also serves to inhibit the differentiation of intramuscular preadipocytes from ensuring the normal development of embryonic muscle. These results provide potential molecular targets for improving beef production and molecular breeding.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by the Animal Care Commission of the College of Veterinary Medicine, Northwest A&F University.
Author Contributions
HC and XS designed research. XS, XZ, WR, and YH performed experiments and analyzed data. XS wrote the paper. CL and XL contributed new analytic tools. HC and JT helped modify the language of this manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China [Grant Nos. 31772574 and 31872331], Major project of collaborative innovation of industry, university, research and application in Yangling Demonstration Zone [2018CXY-05]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This manuscript has been released as a pre-print at Research Square (Xuemei et al., 2020).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.615638/full#supplementary-material
References
Arcinas, C., Tan, W., Fang, W., Desai, T. P., Teh, D. C. S., Degirmenci, U., et al. (2019). Adipose circular RNAs exhibit dynamic regulation in obesity and functional role in adipogenesis. Nature Metab. 1, 688–703. doi: 10.1038/s42255-019-0078-z
Armoni, M., Harel, C., Karni, S., Chen, H., Bar-Yoseph, F., Ver, M. R., et al. (2006). FOXO1 represses peroxisome proliferator-activated receptor-gamma1 and -gamma2 gene promoters in primary adipocytes. A novel paradigm to increase insulin sensitivity. J. Biol. Chem. 281, 19881–19891. doi: 10.1074/jbc.M600320200
Aso, H., Abe, H., Nakajima, I., Ozutsumi, K., Yamaguchi, T., Takamori, Y., et al. (1995). A Preadipocyte clonal line from bovine intramuscular adipose-tissue-nonexpression of glut-4 protein during adipocyte differentiation. Biochem. Biophys. Res. Commun. 213, 369–375. doi: 10.1006/bbrc.1995.2141
Barrett, S. P., and Salzman, J. (2016). Circular RNAs: analysis, expression and potential functions. Development 143, 1838–1847. doi: 10.1242/dev.128074
Beier, J. P., Bitto, F. F., Lange, C., Klumpp, D., Arkudas, A., Bleiziffer, O., et al. (2011). Myogenic differentiation of mesenchymal stem cells co-cultured with primary myoblasts. Cell Biol. Int. 35, 397–406. doi: 10.1042/CBI20100417
Cai, C. K., Zhao, G. Y., Tian, L. Y., Liu, L., Yan, K., Ma, Y. L., et al. (2012). miR-15a and miR-16-1 downregulate CCND1 and induce apoptosis and cell cycle arrest in osteosarcoma. Oncol. Rep. 28, 1764–1770. doi: 10.3892/or.2012.1995
Cao, P., Feng, Y., Deng, M., Li, J., Cai, H., Meng, Q., et al. (2019). MiR-15b is a key regulator of proliferation and apoptosis of chondrocytes from patients with condylar hyperplasia by targeting IGF1, IGF1R and BCL2. Osteoarthr. Cartil. 27, 336–346. doi: 10.1016/j.joca.2018.09.010
Carthew, R. W., and Sontheimer, E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655. doi: 10.1016/j.cell.2009.01.035
Castillo, G., Brun, R. P., Rosenfield, J. K., Hauser, S., Park, C. W., Troy, A. E., et al. (1999). An adipogenic cofactor bound by the differentiation domain of PPARγ. EMBO J. 18, 3676–3687. doi: 10.1093/emboj/18.13.3676
Chendrimada, T. P., Gregory, R. I., Kumaraswamy, E., Norman, J., Cooch, N., Nishikura, K., et al. (2005). TRBP recruits the dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740–744. doi: 10.1038/nature03868
Cimmino, A., Calin, G. A., Fabbri, M., Iorio, M. V., Ferracin, M., Shimizu, M., et al. (2005). miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U. S.A. 102, 13944–13949. doi: 10.1073/pnas.0506654102
Conboy, I. M., and Rando, T. A. (2002). The regulation of notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell. 3, 397–409. doi: 10.1016/S1534-5807(02)00254-X
Dong, P., Mai, Y., Zhang, Z., Mi, L., Wu, G., Chu, G., et al. (2014). MiR-15a/b promote adipogenesis in porcine pre-adipocyte via repressing FoxO1. Acta Biochim. Biophys. Sin. 46, 565–571. doi: 10.1093/abbs/gmu043
Du, M., Huang, Y., Das, A. K., Yang, Q., Duarte, M. S., Dodson, M. V., et al. (2013). Meat science and muscle biology symposium: manipulating mesenchymal progenitor cell differentiation to optimize performance and carcass value of beef cattle. J. Anim. Sci. 91, 1419–1427. doi: 10.2527/jas.2012-5670
Du, M., Tong, J., Zhao, J., Underwood, K. R., Zhu, M., Ford, S. P., et al. (2010b). Fetal programming of skeletal muscle development in ruminant animals. J. Anim. Sci. 88, 51–60. doi: 10.2527/jas.2009-2311
Du, M., Yan, X., Tong, J. F., Zhao, J., and Zhu, M. J. (2010a). Maternal obesity, inflammation, and fetal skeletal muscle development. Biol. Reprod. 82:4. doi: 10.1095/biolreprod.109.077099
Foxley, A., Edwards, R. H., and Jackson, M. J. (1991). Enhanced lipid peroxidation in Duchenne dystrophy muscle may be secondary to muscle damage. Biochem. Soc. Trans. 19, 180S. doi: 10.1042/bst019180s
Gaeta, M., Messina, S., Mileto, A., Vita, G. L., Ascenti, G., Vinci, S., et al. (2012). Muscle fat-fraction and mapping in duchenne muscular dystrophy: evaluation of disease distribution and correlation with clinical assessments. Skeletal Radiol. 41, 955–961. doi: 10.1007/s00256-011-1301-5
Guan, L., Hu, X., Liu, L., Xing, Y., Zhou, Z., Liang, X., et al. (2017). bta-miR-23a involves in adipogenesis of progenitor cells derived from fetal bovine skeletal muscle. Sci. Rep. 7:43716. doi: 10.1038/srep43716
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013a). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi: 10.1038/nature11993
Hansen, T. B., Kjems, J., and Damgaard, C. K. (2013b). Circular RNA and miR-7 in cancer. Cancer Res. 73, 5609–5612. doi: 10.1158/0008-5472.CAN-13-1568
Hausman, G. J., Dodson, M. V., Ajuwon, K., Azain, M., Barnes, K. M., Guan, L. L., et al. (2009). Board-invited review: the biology and regulation of preadipocytes and adipocytes in meat animals. J. Anim. Sci. 87, 1218–1246. doi: 10.2527/jas.2008-1427
Hoashi, S., Hinenoya, T., Tanaka, A., Ohsaki, H., and Mannen, H. (2008). Association between fatty acid compositions and genotypes of FABP4 and LXR-alpha in Japanese black cattle. BMC Genet. 9:84. doi: 10.1186/1471-2156-9-84
Horibata, Y.asuhiro, Elpeleg, O.rly, and Eran, A.yelet. (2018). EPT1 (selenoprotein I) is critical for the neural development and maintenance of plasmalogen in humans. J. Lipid Res. 59, 1015–1026. doi: 10.1194/jlr.P081620
Jiang, R., Li, H., Yang, J. M., Shen, X. M., Song, C. C., Yang, Z. X., et al. (2020). circRNA Profiling reveals an abundant circFUT10 that promotes adipocyte proliferation and inhibits adipocyte differentiation via sponging let-7. Mol. Ther. Nucleic Acids 20, 491–501. doi: 10.1016/j.omtn.2020.03.011
Li, H., Wei, X., Yang, J., Dong, D., Hao, D., Huang, Y., et al. (2018b). circFGFR4 promotes differentiation of myoblasts via binding miR-107 to relieve its inhibition of Wnt3a. Mol. Ther. Nucleic Acids 11, 272–283. doi: 10.1016/j.omtn.2018.02.012
Li, H., Yang, J., Wei, X., Song, C., Dong, D., Huang, Y., et al. (2018a). CircFUT10 reduces proliferation and facilitates differentiation of myoblasts by sponging miR-133a. J. Cell. Physiol. 233, 4643–4651. doi: 10.1002/jcp.26230
Li, H. X., Zhang, Z., Zhou, X., Wang, Z. Y., Wang, G. L., and Han, Z. Y. (2011). Effects of MicroRNA-143 in the differentiation and proliferation of bovine intramuscular preadipocytes. Mol. Biol. Rep. 38, 4273–4280. doi: 10.1007/s11033-010-0550-z
Li, Z. H., Cai, B. L., Abdalla, B. A., Zhu, X. N., Zheng, M., Han, P. G., et al. (2019). LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway. J Caxhexia Sarcopenia Muscle 10, 391–410. doi: 10.1002/jcsm.12374
Lin, F. T., and Lane, M. D. (1994). CCAAT/enhancer binding protein alpha is sufficient to initiate the 3T3-L1 adipocyte differentiation program. Proc. Natl. Acad. Sci. U.S.A. 91, 8757–8761. doi: 10.1073/pnas.91.19.8757
McPherron, A. C., and Lee, S. J. (1997). Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. U.S.A. 94, 12457–12461. doi: 10.1073/pnas.94.23.12457
Meng, S., Zhou, H., Feng, Z., Xu, Z., Tang, Y., Li, P., et al. (2017). CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer 16:94. doi: 10.1186/s12943-017-0663-2
Miyake, M., Takahashi, H., Kitagawa, E., Watanabe, H., Sakurada, T., Aso, H., et al. (2012). AMPK activation by AICAR inhibits myogenic differentiation and myostatin expression in cattle. Cell Tissue Res. 349, 615–623. doi: 10.1007/s00441-012-1422-8
Pekarsky, Y., Balatti, V., and Croce, C. M. (2018). BCL2 and miR-15/16: from gene discovery to treatment. Cell Death Differ. 25, 21–26. doi: 10.1038/cdd.2017.159
Pekarsky, Y., and Croce, C. M. (2015). Role of miR-15/16 in CLL. Cell Death Differ. 22, 6–11. doi: 10.1038/cdd.2014.87
Picard, B., Berri, C., Lefaucheur, L., Molette, C., Sayd, T., and Terlouw, C. (2010). Skeletal muscle proteomics in livestock production. Brief. Funct. Genomics. 9, 259–278. doi: 10.1093/bfgp/elq005
Potthoff, M. J., and Olson, E. N. (2007). MEF2: a central regulator of diverse developmental programs. Development 134, 4131–4140. doi: 10.1242/dev.008367
Rajesh, R. V., Heo, G. N., Park, M. R., Nam, J. S., Kim, N. K., Yoon, D., et al. (2010). Proteomic analysis of bovine omental, subcutaneous and intramuscular preadipocytes during in vitro adipogenic differentiation. Comp. Biochem. Physiol. D Genomics Proteomics. 5, 234–244. doi: 10.1016/j.cbd.2010.06.004
Rando, T. A., and Blau, H. M. (1994). Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene-therapy. J. Cell Biol. 125, 1275–1287. doi: 10.1083/jcb.125.6.1275
Rehfeldt, C., Fiedler, I., Dietl, G., and Ender, K. (2000). Myogenesis and postnatal skeletal muscle cell growth as influenced by selection. Livestock Prod. Sci. 66, 177–188. doi: 10.1016/S0301-6226(00)00225-6
Relaix, F., Rocancourt, D., Mansouri, A., and Buckingham, M. (2005). A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 435, 948–953. doi: 10.1038/nature03594
Sanchez-Gurmaches, J., and Guertin, D. A. (2014). Adipocyte lineages: Tracing back the origins of fat. Biochim. Biophys. Acta Mol. Basis Dis. 1842, 340–351. doi: 10.1016/j.bbadis.2013.05.027
Sassoon, D., Lyons, G., Wright, W. E., Lin, V., Lassar, A., Weintraub, H., et al. (1989). Expression of two myogenic regulatory factors myogenin and MyoDl during mouse embryogenesis. Nature 341, 303–307. doi: 10.1038/341303a0
Seale, P., Sabourin, L. A., Girgis-Gabardo, A., Mansouri, A., Gruss, P., and Rudnicki, M. A. (2000). Pax7 is required for the specification of myogenic satellite cells. Cell 102, 777–786. doi: 10.1016/S0092-8674(00)00066-0
Shen, X. M., Zhang, X. Y., Ru, W. X., Huang, Y. Z., Lan, X. Z., Lei, C. Z., et al. (2020). circINSR promotes proliferation and reduces apoptosis of embryonic myoblasts by sponging miR-34a. Mol. Ther. Nucleic Acids 19, 986–999. doi: 10.1016/j.omtn.2019.12.032
Song, C. C., Yang, Z. X., Dong, D., Xu, J. W., Wang, J., Li, H., et al. (2019). miR-483 inhibits bovine myoblast cell proliferation and differentiation via IGF1/PI3K/AKT signal pathway. J. Cell. Physiol. 234, 9839–9848. doi: 10.1002/jcp.27672
Soumillion, A., Erkens, J. H., Lenstra, J. A., Rettenberger, G., and te Pas, M. F. (1997). Genetic variation in the porcine myogenin gene locus. Mamm. Genome 8, 564–568. doi: 10.1007/s003359900504
Stachecka, J., Walczak, A., Kociucka, B., Ruszczycki, B., Wilczyński, G., and Szczerbal, I. (2018). Nuclear organization during in vitro differentiation of porcine mesenchymal stem cells (MSCs) into adipocytes. Histochem. Cell Biol. 149, 113–126. doi: 10.1007/s00418-017-1618-9
Sun, L. J., Si, M. J., Liu, X. Y., Choi, J. M., Wang, Y. L., Thomas, S. S., et al. (2018). Long-noncoding RNA Atrolnc-1 promotes muscle wasting in mice with chronic kidney disease. J. Caxhexia Sarcopenia Muscle 9, 962–974. doi: 10.1002/jcsm.12321
Taga, H., Bonnet, M., Picard, B., Zingaretti, M. C., Cassar-Malek, I., Cinti, S., et al. (2011). Adipocyte metabolism and cellularity are related to differences in adipose tissue maturity between Holstein and Charolais or Blond d'Aquitaine fetuses. J. Anim. Sci. 89, 711–721. doi: 10.2527/jas.2010-3234
Tang, H., Huang, X., Wang, J., Yang, L., Kong, Y., Gao, G., et al. (2019). circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol. Cancer 18:23. doi: 10.1186/s12943-019-0946-x
Tontonoz, P., Hu, E., and Spiegelman, B. M. (1994). Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 79, 1147–1156. doi: 10.1016/0092-8674(94)90006-X
Wang, F., and Tong, Q. (2008). SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1's repressive interaction with PPARγ. Mol. Biol. Cell. 20, 801–808. doi: 10.1091/mbc.e08-06-0647
Wang, H., Chen, W., Jin, M., Hou, L., Chen, X., Zhang, R., et al. (2018). CircSLC3A2 functions as an oncogenic factor in hepatocellular carcinoma by sponging miR-490-3p and regulating PPM1F expression. Mol. Cancer. 17:165. doi: 10.1186/s12943-018-0909-7
Wang, X., Cao, X., Dong, D., Shen, X., Cheng, J., Jiang, R., et al. (2019). Circular RNA TTN acts as a miR-432 sponge to facilitate proliferation and differentiation of myoblasts via the IGF2/PI3K/AKT signaling pathway. Mol. Ther. Nucleic Acids 18, 966–980. doi: 10.1016/j.omtn.2019.10.019
Wang, Y. M., Ding, X. B., Dai, Y., Liu, X. F., Guo, H., and Zhang, Y. (2015). Identification and bioinformatics analysis of miRNAs involved in bovine skeletal muscle satellite cell myogenic differentiation. Mol. Cell. Biochem. 404, 113–122. doi: 10.1007/s11010-015-2371-9
Wei, X., Li, H., Yang, J., Hao, D., Dong, D., Huang, Y., et al. (2017). Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 8:e3153. doi: 10.1038/cddis.2017.541
Westerweel, P. E., and Verhaar, M. C. (2008). Directing myogenic mesenchymal stem cell differentiation. Circ. Res. 103, 560–561. doi: 10.1161/CIRCRESAHA.108.184374
Wren, T. A. L., Bluml, S., Tseng-Ong, L., and Gilsanz, V. (2008). Three-point technique of fat quantification of muscle tissue as a marker of disease progression in duchenne muscular dystrophy: preliminary study. Am. J. Roentgenol. 190, W8–W12. doi: 10.2214/AJR.07.2732
Xu, J., Zhang, L., Shu, G., and Wang, B. (2019). microRNA-16-5p promotes 3T3-L1 adipocyte differentiation through regulating EPT1. Biochem. Biophys. Res. Commun. 514, 1251–1256. doi: 10.1016/j.bbrc.2019.04.179
Xuemei, S., Jia, T., Wenxiu, R., Xiaoyan, Z., Yongzhen, H., Xianyong, L., et al. (2020). CircINSR regulates fetal bovine muscle and fat development. Res. Square [Preprint]. doi: 10.21203/rs.3.rs-79997/v1
Yi-Min, W., Yang, D., Xin-Feng, L., Zhong-Wei, L., Ji-Xia, L. I., Hong, G., et al. (2014). Isolation, identification and induced differentiation of bovine skeletal muscle satellite cells. China Anim. Husbandry Vet. Med 41, 142–147. Available online at: http://www.chvm.net/CN/Y2014/V41/I7/142
Keywords: bovine, muscle development, circular RNAs, cell proliferation, preadipocyte
Citation: Shen X, Tang J, Ru W, Zhang X, Huang Y, Lei C, Cao H, Lan X and Chen H (2021) CircINSR Regulates Fetal Bovine Muscle and Fat Development. Front. Cell Dev. Biol. 8:615638. doi: 10.3389/fcell.2020.615638
Received: 09 October 2020; Accepted: 07 December 2020;
Published: 06 January 2021.
Edited by:
Tomokazu Tomo Fukuda, Iwate University, JapanReviewed by:
Masayuki Kobayashi, Akita Prefectural University, JapanTetsuya Tani, Kindai University, Japan
Copyright © 2021 Shen, Tang, Ru, Zhang, Huang, Lei, Cao, Lan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Chen, Y2hlbmhvbmcxMjEyQDI2My5uZXQ=; Xianyong Lan, bGFueGlhbnlvbmc3OUBud3N1YWYuZWR1LmNu
 Xuemei Shen
Xuemei Shen Jia Tang
Jia Tang Wenxiu Ru
Wenxiu Ru Xiaoyan Zhang
Xiaoyan Zhang Yongzhen Huang
Yongzhen Huang Chuzhao Lei
Chuzhao Lei Hui Cao
Hui Cao Xianyong Lan
Xianyong Lan Hong Chen
Hong Chen