
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 11 January 2021
Sec. Epigenomics and Epigenetics
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.615071
This article is part of the Research Topic Epigenetics in Cancer: Mechanisms and Drug Development View all 15 articles
Accumulating evidence indicates that N6-methyladenosine (m6A), which directly regulates mRNA, is closely related to multiple biological processes and the progression of different malignancies, including breast cancer (BC). Studies of the aberrant expression of m6A mediators in BC revealed that they were associated with different BC subtypes and functions, such as proliferation, apoptosis, stemness, the cell cycle, migration, and metastasis, through several factors and signaling pathways, such as Bcl-2 and the PI3K/Akt pathway, among others. Several regulators that target m6A have been shown to have anticancer effects. Fat mass and obesity-associated protein (FTO) was identified as the first m6A demethylase, and a series of inhibitors that target FTO were reported to have potential for the treatment of BC by inhibiting cell proliferation and promoting apoptosis. However, the exact mechanism by which m6A modifications are regulated by FTO inhibitors remains unknown. m6A modifications in BC have only been preliminarily studied, and their mechanisms require further investigation.
Understanding the origins of cancer has changed significantly in recent decades, from being considered solely a genetic disease to being considered a genetic and/or epigenetic disease. Traditional epigenetic modifications, including the dysregulation of DNA methylation and histone modification, are causes of cancer, but chemical modifications of RNA were recently discovered to also cause cancer (Esteve-Puig et al., 2020). Breast cancer (BC) is the most common malignancy among women in the world (Siegel et al., 2020). Aberrant RNA modifications open a new era for studying the development and progression of BC (Chen et al., 2019).
Since the first modified nucleotides in RNAs were discovered in 1960 (Cohn, 1960), more than 170 distinct cellular RNA chemical modifications have been identified (Esteve-Puig et al., 2020), including pseudouridine (Ψ), N1-methyladenosine (m1A), 5-methylcytidine (m5C), N6-methyladenosine (m6A), 5-hydroxymethylcytosine (hm5C), etc. Among these, m6A is the most common and abundant posttranscriptional modification of messenger RNA (mRNA) and non-coding RNA (ncRNA). High-throughput sequencing revealed that one-third to one-half of mRNA transcripts had m6A modifications in human and mouse transcriptomes (Dominissini et al., 2012). Although m6A was first reported in 1974 (Desrosiers et al., 1974), little progress was made for decades (Reichel et al., 2019). Recently, the function of m6A has been gradually unveiled through advances in m6A detection techniques, such as liquid chromatography-tandem mass spectrometry (Fu et al., 2015), methylated RNA immunoprecipitation sequencing (Meyer et al., 2012), methylation individual nucleotide-resolution crosslinking immunoprecipitation (Linder et al., 2015), and single-molecule real-time sequencing (Zhu et al., 2016).
The m6A modification mainly identifies the conserved sequence RRACH (R = G/A, H = A/C/U). GGACU is one of the most common motifs (Harper et al., 1990). m6A modifications were previously believed to be mainly concentrated in the 3′-untranslated region (UTR), especially around the stop codon (Meyer et al., 2012). In recent years, researchers confirmed that the 5′-UTR (Meyer et al., 2015) and coding sequence (CDS; Mao et al., 2019) were also important for m6A modifications. Many studies have shown that they can regulate almost every stage of RNA metabolism, including alternative RNA splicing (Bartosovic et al., 2017; Louloupi et al., 2018), localization (Roundtree et al., 2017), translation efficiency (Wang et al., 2015b), mRNA stability (Wang et al., 2014), and protein expression (Yue et al., 2015).
N6-methyladenosine is involved in the regulation of many biological processes, such as the transition fate of mammalian embryonic stem cells (Batista et al., 2014) and circadian rhythms (Fustin et al., 2013; Zhong et al., 2018), and various diseases, such as obesity (Dina et al., 2007), infertility (Ding et al., 2018), type 2 diabetic mellitus (Shen et al., 2015), and many kinds of cancers (Chen et al., 2019). Compared with various studies that explored the interplay between m6A modifications and several types of cancers, studies of the role of m6A in BC are still in their infancy. Some conclusions of existing studies are controversial, thus underscoring the necessity to review and discuss the functions of m6A modifications and therapeutic strategies for BC.
This review focuses on the mechanism of m6A modifications, the roles of different m6A modulators in BC, especially their effect on proliferation, apoptosis, the cell cycle, and stemness, and therapeutic strategies for BC. This review will advance our understanding of the role of m6A modifications in the development and progression of BC.
There are mainly three kinds of mediator proteins that regulate m6A modifications: methyltransferase (writer), demethylase (eraser), and binding protein (reader). The regulation of m6A modifications is dynamically reversible, depending on the activity of the “writer” and “eraser.”
N6-methyladenosine modification is installed by the methyltransferase complex (MTC), which mainly consists of the methyltransferase like 3 (METTL3)/METTL14 heterodimer and Wilms’ tumor 1-associated protein (WTAP; Scholler et al., 2018; Figure 1). In this m6A MTC, METTL3 acts as the core catalytic subunit and transfers the methyl donor S-adenosylmethionine (SAM) to the adenine acceptor within the RRACH consensus motif. METTL14 colocalizes with METTL3 in nuclear speckles at a stoichiometric 1:1 ratio (Liu et al., 2014). METTL14 is responsible for stabilizing the structure of MTC, recognizing the substrate RNA sequence, and providing a binding platform (Wang P. et al., 2016; Wang X. et al., 2016). Huang et al. recently demonstrated that METTL14 can also recognize and bind H3K36me3. Thus, METTL14 facilitates m6A MTC binding to RNA polymerase II to mediate the m6A methylation of nascent RNA during transcription elongation (Huang H. et al., 2019). The combination of these histone and RNA modifications opens up new directions for epigenetics research. WTAP alone does not have any catalytic activity because it lacks the catalytic methylation domain, but its knockdown prominently decreases m6A levels (Liu et al., 2014). It regulates the level of m6A modification by interacting with the METTL3–METTL14 complex and promoting accumulation of the complex into nuclear speckles to efficiently methylate target RNAs (Ping et al., 2014). Other identified regulatory factors of MTC include KIAA1429/VIRMA (Yue et al., 2018), RBM15/15B (Patil et al., 2016; Knuckles et al., 2018), HAKAI (Ruzicka et al., 2017), and ZC3H13 (Knuckles et al., 2018).
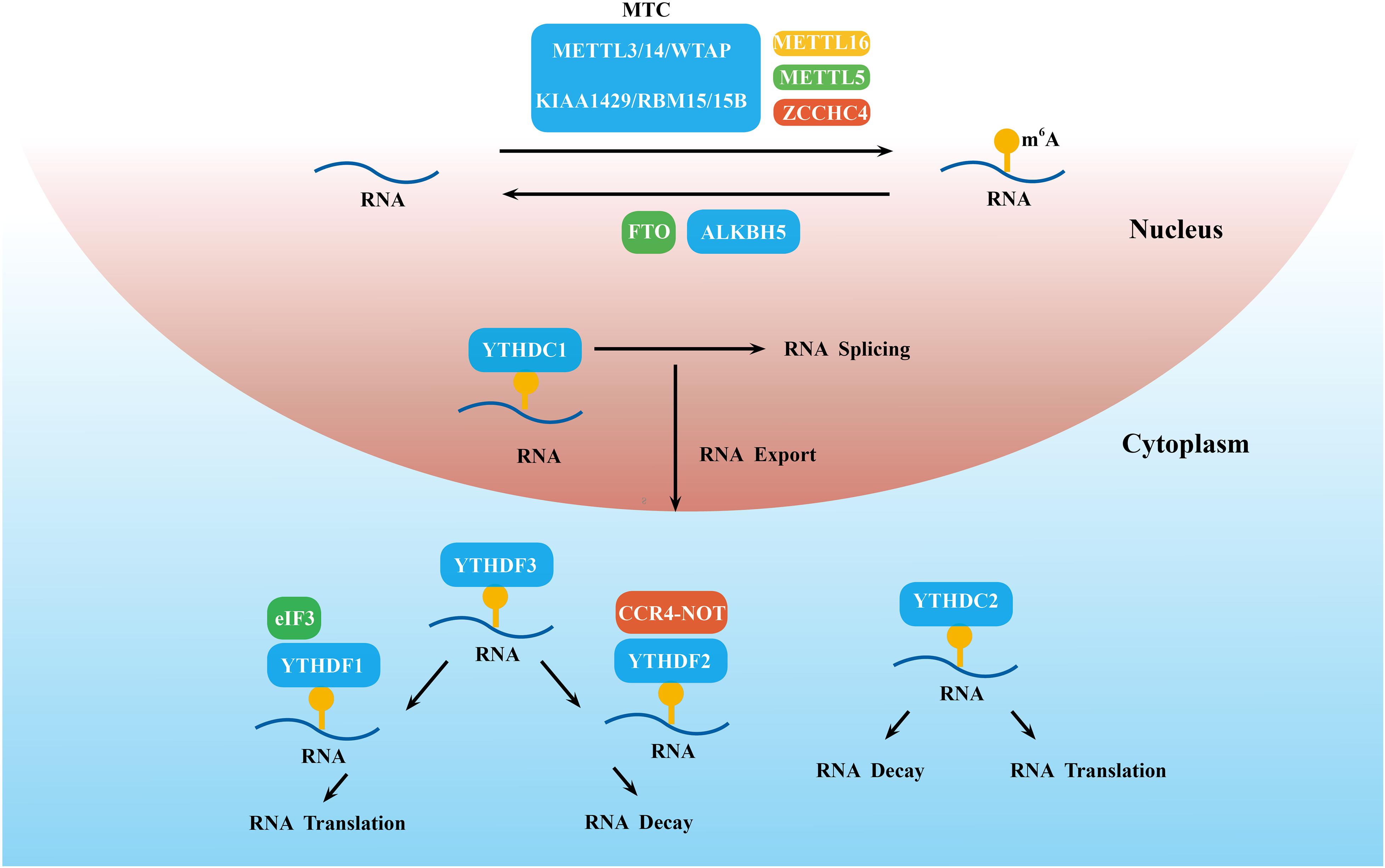
Figure 1. Detailed molecular mechanism of m6A modifications. These modifications are regulated by “writers” (the MTC, METTL5/16 and ZCCHC4), “erasers” (FTO and ALKBH5), and “readers” (YTHDC1-2 and YTHDF1-3), which install, remove, and recognize m6A and thereby regulate RNA splicing, export, decay, translation, and so on.
Some newly discovered methyltransferase, such as METTL16, METTL5, and zinc finger CCHC-type containing 4 (ZCCHC4), can work alone and catalyze m6A on some structured RNAs, such as U6 snRNA (Aoyama et al., 2020), 18S rRNA (Leismann et al., 2020), and 28S rRNA (Ren et al., 2019), respectively (Figure 1).
N6-methyladenosine demethylases, including Fat mass and obesity-associated protein (FTO; Jia et al., 2012) and ALKB family protein 5 (ALKBH5; Zheng et al., 2013), can selectively remove the m6A modification and reverse the methylation process (Figure 1). The coordination between m6A methyltransferase and demethylase indicates that the m6A modification is dynamic and reversible. Although both FTO and ALKBH5 are members of the ALKB family, they have different substrates. Emerging evidence demonstrates that FTO is able to mediate the demethylation of m6A, m6Am, and m1A in the cell nucleus and cytoplasm (Wei et al., 2018). To date, m6A in mRNA was the main zymolytic substrate of FTO. Different from FTO, ALKBH5 is mainly localized in the nucleus and selectively removes the m6A methyl group (Mauer et al., 2017). This phenomenon reflects the complexity and specificity of the mechanism of m6A modifications. Additionally, ALKBH5 accelerated the process of both mRNA transfer from intranuclear to extranuclear and promoted translation initiation (Zheng et al., 2013), whereas METTL3 downregulation delayed this process (Fustin et al., 2013).
Other types of m6A regulatory proteins, including YT521-B homology domain-containing family protein 1/2/3 (YTHDF1/2/3), YTH domain-containing proteins 1/2 (YTHDC1/2) (Haussmann et al., 2016), eukaryotic initiation factor 3 (elF3; Meyer and Jaffrey, 2017), the insulin-like growth factor-2 mRNA-binding protein (IGF2BP) family (Muller et al., 2019), and heterogeneous nuclear ribonucleoprotein (hnRNP) family (Zhao et al., 2017), recognize the m6A modification site and directly determine the fate of m6A-modified RNA (Casella et al., 2019; Figure 1).
YT521-B homology domain-containing family protein 1/2/3 and YTHDC1/2 share the same YTH domain and have a 50-times higher affinity for m6A mRNA than unmethylated mRNA (Theler et al., 2014). YTHDC1 was shown to be the major reader of nuclear m6A modifications and accelerated mature mRNA transportation from the nucleus to the cytoplasm by affecting mRNA splicing (Roundtree et al., 2017). To date, however, little is known about the function of YTHDC2 in m6A modifications. Hsu reported that YTHDC2 preferentially bound to m6A-marked RNA with the RRACH consensus motif and then increased translation efficiency by 52% but also reduced mRNA abundance (Hsu et al., 2017).
When mRNA arrives at the cytoplasm, m6A-methylated mRNA is mainly regulated by YTHDF1-3. YTHDF1 mainly binds to m6A sites near the stop codon in the 3′-UTR and then interacts with the translation initiation factor eIF3 to improve the translation efficiency of target mRNA (Wang et al., 2015b). In 2019, Lin et al. (2019) reported that YTHDF1, by binding with eEF-2 at m6A sites of the Snail CDS but not the 3′-UTR, promoted the translation of Snail. YTHDF2 was the first identified m6A-binding protein, which triggered mRNA degradation by recruiting the CCR4-NOT deadenylase complex (Du et al., 2016). The role of YTHDF3 is more complex. YTHDF3 acts in concert with YTHDF2 to accelerate mRNA decay (Shi et al., 2017), but it promoted the translation of m6A-modified RNA by cooperating with YTHDF1 (Li A. et al., 2017). IGF2BP family proteins (IGF2BP1, IGF2BP2, and IGF2BP3) are located in the cytoplasm and employ their K homology domains to identify the GG (m6A) C sequence. They promote the stability and storage of target mRNAs under both normal and stress conditions (Huang et al., 2018). Some other m6A readers, such as the HNRNP family and elF3, are under current exploration.
The majority of BC is sporadic and associated with alterations of genetics and epigenetics (Byler et al., 2014). Genetic alterations include mutations and copy number variations of certain genes. Conventional epigenetic remodeling consists of microRNA regulation, histone modification, and DNA methylation in BC (Rahman et al., 2019). m6A modifications open new directions for studying epigenetics. An increasing number of studies have revealed the importance of m6A in the development and progression of BC.
In various cancer cells, the abnormal expression of METTL3 affects proliferation, invasion, metastasis, and the cell cycle (Zheng et al., 2019). In BC, METTL3 mainly acts as an oncogene (Figure 2). For example, METTL3 overexpression in transformed cells enhanced proliferation and migration ability, suggesting that the downregulation of m6A modification acts as a brake during malignant transformation (Fry et al., 2018). B-cell lymphoma 2 (Bcl-2) is an anti-apoptotic protein (Konig et al., 2019) that has been shown to be a potential target of METTL3 in BC tissues and cells. The knockdown of METTL3 reduced the expression of Bcl-2, repressed the proliferation of MDA-MB-231 and MCF-7 cells, accelerated their apoptosis, and inhibited the growth of transplanted tumors in vivo (Wang et al., 2020).
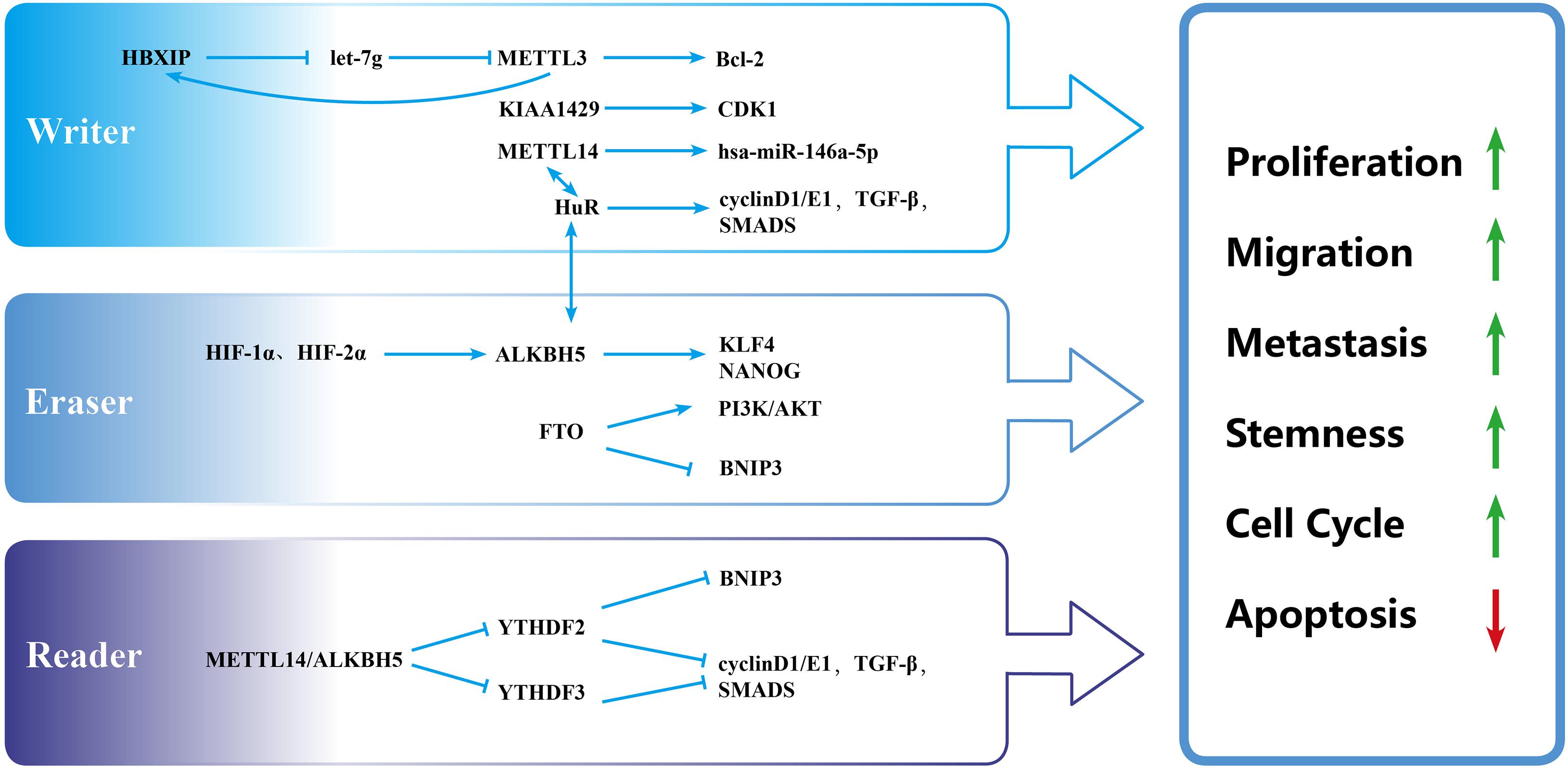
Figure 2. Biological function of m6A modulators in breast cancer. “Writers,” “erasers,” and “readers” promote cell proliferation, migration, metastasis, and stemness and inhibit apoptosis by regulating different targets.
A few studies have shown that hepatitis B x-interacting protein (HBXIP) is highly expressed in BC as an oncogene (Yue et al., 2013; Liu et al., 2014). METTL3 was positively linked with HBXIP in BC tissues and cells. In BC cells, HBXIP inhibited the expression of let-7g, which repressed the expression of METTL3 by targeting the 3′-UTR of METTL3 (Cai et al., 2018). Additionally, METTL3 upregulated the expression of HBXIP by stimulating the m6A modification of HBXIP and thus formed a positive feedback regulatory loop of HBXIP/let-7g/METTL3/HBXIP, leading to the proliferation of BC cells (Cai et al., 2018). Cui et al. (2017) reported that the knockout of METTL3 and METTL14 promoted the growth, self-renewal, and tumorigenesis of glioblastoma stem cells (GCS) partly by upregulating BRCA2. BRCA2 was tightly related to hereditary BC (Paul and Paul, 2014). However, whether METTL3 and METTL14 regulate the development and progression of BC through BRCA2 is unknown. Altogether, Bcl-2, HBXIP, and BRCA2 are all potential targets of METTL3. We speculate that other targets of METTL3 will be uncovered as regulators of BC.
METTL14 is also closely related to the progression of BC (Figure 2). The knockdown of METTL14 reduced the long-term viability, migration, and invasion of BC cell lines (MDA-MB-231, MDA-MB-468, and BT549) and inhibited tumor growth in tumor xenograft models (Panneerdoss et al., 2018). BC cells were arrested in the G1–S phase when METTL14 was silenced. Mechanistically, METTL14 and the RNA-binding protein HuR form a potential positive feedback loop that regulates transforming growth factor-β (TGF-β) signaling pathway genes and cell cycle-associated genes (cyclin D1 and cyclin E1) and exhibit hyper m6A with lower expression compared with scrambled control cells (Panneerdoss et al., 2018). m6 A also affects BC cells through regulating the noncoding RNAs expression. Yi et al. found that METTL14 overexpression promoted the migration and invasion of MDA-MB-231 and MCF-7 cells through the upregulation of hsa-mir-146a-5p (Yi et al., 2020). However, Wu et al. reported that the overexpression of METTL14 in MDA-MB-231 cells inhibited cell viability, clone formation, and cell migration (Wu et al., 2019). METTL14 appears to play opposing roles in the same cell line. Whether other factors, such as the tumor microenvironment, impact METTL14 function deserves further exploration.
The role of methyltransferase KIAA1429 in BC had been preliminarily elucidated. A previous study (Qian et al., 2019) showed that KIAA1429 promoted BC cell proliferation and the epithelial–mesenchymal transition (EMT) in MCF-7 and SUM1315 cell lines. This may be caused by an increase in stability of the cell cycle regulator cyclin-dependent kinase 1 (CDK1) mRNA in an m6A-independent manner (Qian et al., 2019). The roles of other methyltransferases in BC have rarely been reported and deserve further study.
Breast cancer tissues exhibit the dysregulation of m6A methyltransferase compared with normal breast tissues, although m6A methyltransferase levels vary (Table 1). The mRNA expression of classic methyltransferases (i.e., METTL3, METTL14, and WTAP) was reported to be either upregulated (Cai et al., 2018; Wang et al., 2020; Yi et al., 2020) or downregulated (Liu et al., 2019; Wu et al., 2019). Moreover, their mRNA levels were sometimes inconsistent with protein levels. Liu performed immunohistochemical staining in 20 matched BC and adjacent normal tissues with a BC tissue microarray (TMA) and found the upregulation of WTAP (p = 0.002), KIAA1429 (p < 0.001), and RBM15 (p = 0.012) but no significant changes in METTL3, METTL14, METTL16, or RBM15B in BC specimens (Liu et al., 2019), which was different from mRNA expression.
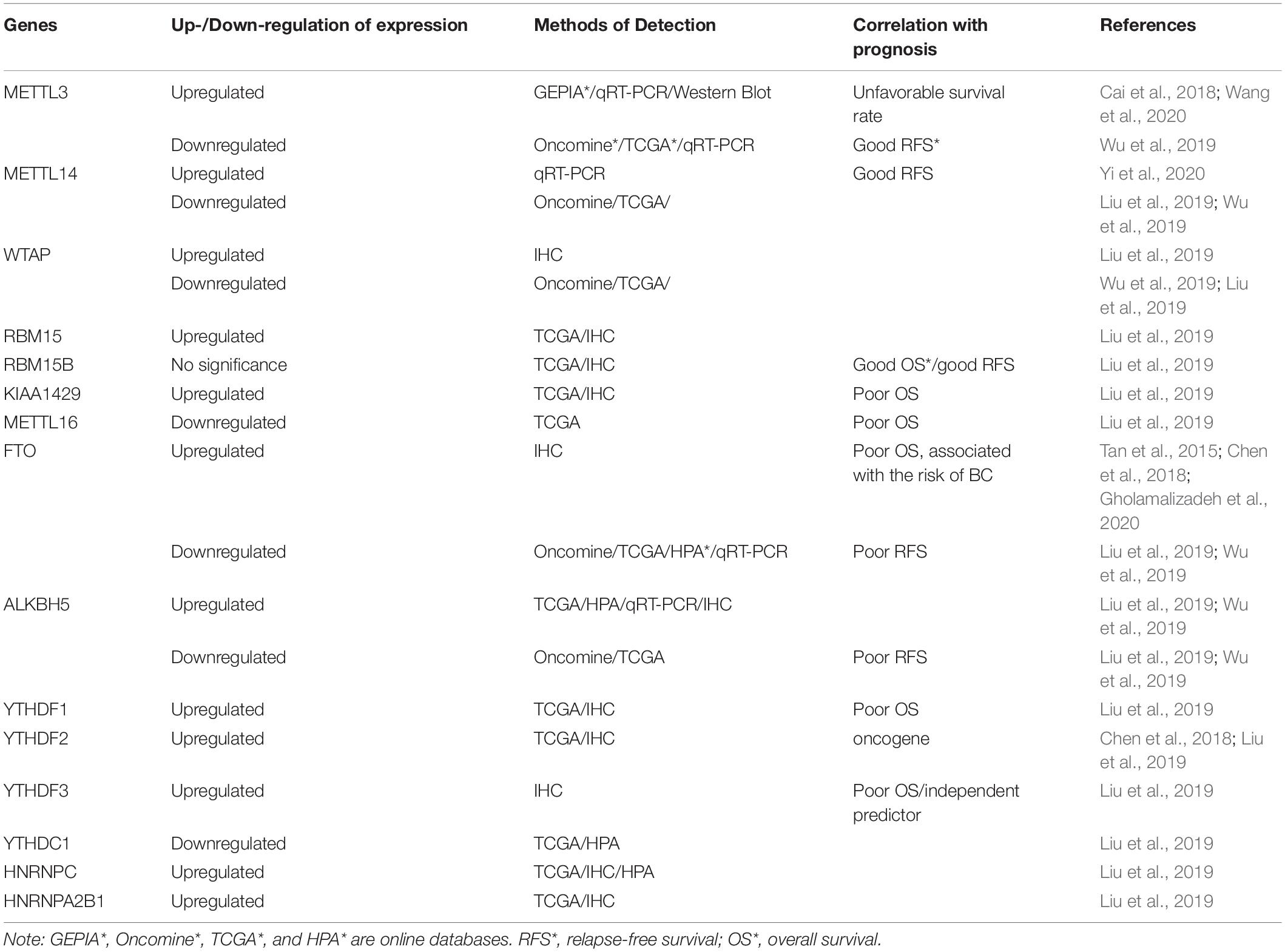
Table 1. Different expression levels and potential prognostic value of m6A modulators in breast cancer (BC).
The expression of m6A methyltransferase also correlated with molecular subtypes of BC (Table 2). METTL3 and METTL14 were highly expressed in normal breast-like and luminal A/B BC, but WTAP was mainly expressed in basal-like BC, based on results from the Oncomine and The Cancer Genome Atlas (TCGA) database (Wu et al., 2019). Inflammatory BC (IBC) is a rare and aggressive form of BC. The triple-negative subtype of IBC (TN-IBC) is substantially more metastatic and fatal than TN-non-IBC. In an analysis of differentially regulated genes in TN-IBC and TN-non-IBC, ZC3H13 (i.e., a member of the MTC) was among the top 10 genes and downregulated in TN-IBC (Funakoshi et al., 2019). These data are summarized in Table 2.
Studies have reported conflicting reports on the role of m6A methyltransferase in the prognosis of BC (Table 1). Based on data from the TCGA-BC cohort, the higher expression of RBM15B (p = 0.014, 95% confidence interval: 0.48–0.94) significantly correlated with favorable overall survival (OS), whereas the high expression of KIAA1429 (p = 0.032, 95% confidence interval: 1.03–1.96) and METTL16 (p = 0.02, 95% confidence interval: 1.06–2.02) correlated with poor OS. With regard to relapse-free survival (RFS), the overexpression of RBM15B (p = 0.021, 95% confidence interval: 0.54–0.96) correlated with good RFS. However, other “writers” did not critically affect the survival rate (Liu et al., 2019). Another study, based on the bc-GenExMiner 4.0 database, found that the high expression of METTL3, METTL14, and WTAP correlated with good metastasis relapse (MR)-free survival in all BC patients (Wu et al., 2019). However, a separate study that conducted a Kaplan–Meier test revealed that BC patients with high METTL3 expression had unfavorable survival rates (Wang et al., 2020).
At the cellular level, FTO promoted the proliferation and mammosphere formation and suppressed cell apoptosis by inhibiting BCL2/adenovirus E1B 19-kDa protein-interacting protein 3 (BNIP3; i.e., a pro-apoptosis protein of the Bcl-2 family) in BC (Niu et al., 2019) (Figure 2). FTO demethylated the 3′-UTR of BNIP3 mRNA and promoted the degradation of BNIP3 mRNA in a YTHDF2-dependent manner. In MDA-MB-231 and MCF-7 cell lines, silencing BNIP3 alleviated the inhibition of cell proliferation that was mediated by FTO knocking down. In 4T1 cells, BNIP3 knockdown significantly weakened FTO-accelerated tumor growth and metastasis in a subcutaneous implantation model and tumor metastasis model in Balb/c mice. Additionally, FTO upregulated glycolysis and energy metabolism through the PI3K/AKT pathway in MDA-MB-231 and MCF-7 cells (Liu et al., 2017). The PI3K/AKT signaling pathway has a close relationship with proliferation, metabolism, immune response regulation, motility, and survival (Ortega et al., 2020). Therefore, FTO may exert important actions through the PI3K/AKT pathway, which deserves further study.
ALKB family protein 5 can promote the growth and metastasis of BC cells (Figure 2). The silencing of ALKBH5 inhibited the viability, migration, and invasion of BC cell lines and tumor growth in a tumor xenograft model in mice (Panneerdoss et al., 2018), similar to METTL14 knockdown that was mentioned above. In MDA-MB-231 cells, the knockdown of ALKBH5 promoted the m6A modification of mRNA and inhibited the ability of survival, clone formation, and cell migration (Wu et al., 2019). In a mouse model of the orthotopic transplantation of BC tumors, ALKBH5-deficient MDA-MB-231 cells developed fewer tumors compared with the control group (43% vs. 100%), and only a few small lung metastases were found in the ALKBH5 knockdown group, thus confirming that ALKBH5 promoted the initiation of BC and lung metastasis (Zhang et al., 2016b). Under hypoxic conditions, hypoxia-inducible factor 1α (HIF-1α) and HIF-2α induced overexpression of the m6A demethylase ALKBH5, which increased demethylation of the 3′-UTR of pluripotent factor NANOG mRNA and enhanced its protein stability (Zhang et al., 2016a). Therefore, ALKBH5 mediates BC stem cell (BCSC) transformation by NANOG in a HIF-dependent manner. Moreover, Zhang et al. (2016b) showed that hypoxia induced expression of the important oncogene ZNF217, which restrained m6A RNA methylation by blocking METTL3. The knockdown of ZNF217 and ALKBH5 increased m6A modification and inhibited hypoxia-induced expression of the pluripotent stem cell factors NANOG and KLF4, thereby suppressing the pluripotency of BC cells (Zhang et al., 2016b). Based on these studies, we speculate that silencing ALKBH5 may be an effective therapeutic strategy that can inhibit proliferation, metastasis, and stemness in BC.
To explore the clinical significance of FTO and ALKBH5 demethylases in BC, various studies have detected their expression levels based on three independent databases [Oncomine, TCGA, and the Human Protein Atlas (HPA)] in clinical BC specimens (Table 1). All of these databases indicated that FTO mRNA is significantly reduced in BC (Liu et al., 2019; Wu et al., 2019). ALKBH5 mRNA was either increased (Liu et al., 2019; Wu et al., 2019) or decreased (Wu et al., 2019) compared with normal tissue. At the protein level, immunohistochemical staining showed that ALKBH5 and FTO expression was high in BC (Tan et al., 2015; Liu et al., 2019; Niu et al., 2019). Notwithstanding these studies, no definitive conclusions can be drawn about the expression level of m6A demethylase in BC. We briefly summarize these data in Table 1.
A correlation was found between the expression level of demethylase and molecular subtypes of BC (Table 2). ALKBH5 and FTO mRNA expression significantly increased in estrogen receptor (ER)-positive or progesterone receptor (PR)-positive patients, whereas FTO mRNA expression decreased in human epidermal growth factor receptor 2 (HER2)-positive patients, based on clinicopathological parameters from the bc-GenExMiner 4.0 database (Wu et al., 2019). Tan reported that FTO was highly expressed in hormone receptor (HR)-negative and HER2-positive BC patients, based on the immunohistochemical staining of specimens from 79 infiltrating ductal breast cancer (IDBC) patients (Tan et al., 2015). In another study that analyzed data from Gene Expression Omnibus (GEO) database (GSE9014), FTO expression was upregulated in HER2-positive BC (Niu et al., 2019).
Findings of the prognostic value of FTO and ALKBH5 in BC have been variable (Table 1). Kaplan–Meier analysis, meta-analysis, and univariate Cox analysis indicated that decreases in FTO mRNA levels suggest poor RFS, based on data from the bc-GenExMiner 4.0 database, whereas ALKBH5 was not significantly related to prognosis (Wu et al., 2019). Another study found that high FTO expression was directly related to poor OS in ER-negative BC patients and advanced BC patients (Niu et al., 2019), based on the Genomics Analysis and Visualization Platform.1 FTO gene polymorphisms, such as rs9939609 and rs1477196, were associated with the risk of BC (Gholamalizadeh et al., 2020). This association may be affected by ER/PR status, tumor stage, and body mass index (BMI) (Gholamalizadeh et al., 2020).
Compared with other m6A modulators, less research on the effect of m6A-binding protein in BC has been conducted (Tables 1, 2). Liu et al. (2019) reported that YTHDF1 (p < 0.001), YTHDF2 (p = 0.022), HNRNPC (p < 0.001), and HNRNPA2B1 (p < 0.001) were upregulated, and YTHDC1 (p = 0.013) was downregulated in BC samples, based on RNA-seq data from TCGA. At the protein level, immunohistochemical staining from 20 pairs of BC and adjacent matched normal breast tissue samples showed that YTHDF1-3, HNRNPC, and HNRNPA2B1 were significantly overexpressed in neoplastic tissues. No significant difference in YTHDC1 was found (Liu et al., 2019). However, HPA (without YTHDF1 or YTHDF3 data) analysis showed that HNRNPC and YTHDC1 expression was high in BC, whereas YTHDF2 and HNRNPA2B1 expression was not significantly different between normal and BC tissues (Liu et al., 2019). Further investigations of the prognostic value of m6A-binding protein in BC revealed that the high expression of YTHDF1 (p = 0.049, 95% confidence interval: 1–1.91) and YTHDF3 (p < 0.001, 95% confidence interval: 1.28–2.49) was related to poor OS, based on clinical data from TCGA. Furthermore, YTHDF3 overexpression was deemed to be an independent predictor of poor OS in BC patients, based on univariate and multivariate analyses (Liu et al., 2019). Studies of the mechanism of regulation found that YTHDF2 participated in the degradation of BNIP3 by binding to potential m6A sites in the 3′-UTR in BC (Figure 2). BNIP3 acts as a tumor suppressor, and YTHDF2 may play an oncogenic role by regulating BNIP3 (Niu et al., 2019).
Furthermore, significant crosstalk among YTHDF3, METTL14, and ALKBH5 has been reported (Panneerdoss et al., 2018). YTHDF3 levels significantly increased in BC cells with METTL14 and ALKBH5 knockdown, detected by RNA-seq and Western blot. It was further confirmed that such crosstalk regulated cancer cell growth and progression in rescue experiments in which YTHDF3 was knocked down in MRTTL14- and ALKBH5-depleted BC cells. We speculate that additional crosstalk exists among other m6A molecules, which requires further study. Overall, m6A modifications are involved in the development and progression of BC, the relevant mechanisms of which are summarized in Figure 2.
The dysregulation of m6A modifications is linked to various diseases, especially cancer. The development of small-molecule m6A-targeting drugs is an attractive therapeutic strategy. Some m6A inhibitors, such as cycloleucine (Wang et al., 2015a) and 3-deazaadenosine (Chen et al., 2018; Yi et al., 2020), were shown to nonspecifically reduce m6A levels by dose-dependently inhibiting SAM activity. During tumor progression, m6A modification acts as a “dual-edged sword” with the controversial role of some m6A modulators. The discovery of specific m6A regulators may contribute to more effective treatment strategies for cancer.
Among the m6A regulators, FTO was the first m6A demethylase that was discovered in 2011 (Jia et al., 2011), which has attracted much interest because of its involvement in obesity and obesity-induced metabolic diseases (Zhao et al., 2014) and the occurrence, development, and prognosis of many kinds of cancer, such as melanoma (Yang et al., 2019), acute myeloid leukemia (AML; Li Z. et al., 2017), glioblastoma (Cui et al., 2017), lung carcinoma (Li et al., 2019a), hepatocellular carcinoma (Li et al., 2019b), and BC (Niu et al., 2019). FTO has been well studied, and several inhibitors of FTO have been developed with regard to their actions against m6A modifications (Deng et al., 2018). To date, such FTO inhibitors include entacapone (which lowers fasting blood glucose levels and reduces body weight in diet-induced obese mice) (Peng et al., 2019), R-2HG (which inhibits proliferation/viability and promotes cell cycle arrest and apoptosis in FTO-high cancer cells) (Su et al., 2018), rhein (which suppresses axon elongation in axons) (Yu et al., 2018), meclofenamic acid (MA; which inhibits the proliferation of BC cells) (Kovala-Demertzi et al., 2009), MA2 (the ethyl ester form of MA; which arrests the G1/S transition and inhibits cell proliferation in germ cells) (Huang T. et al., 2019), FB23 and FB23-2 (which suppress proliferation and promote the differentiation and apoptosis of AML cells in vitro and in vivo) (Huang Y. et al., 2019), and CS1 and CS2 (which attenuate the self-renewal ability and reprograming immune response of leukemia cells) (Huang et al., 2015). These inhibitors have been shown to have different mechanisms of action, and the effects of rhein and MA have been explored in BC.
Rhein was the first discovered natural small-molecule inhibitor of FTO. It competitively binds the catalytic region of FTO and upregulates cellular m6A levels in vitro (Chen et al., 2012). In axons, FTO inhibition by rhein or FTO knockdown by siRNA downregulated the local translation of GAP-43 mRNA and resulted in the suppression of axon elongation (Yu et al., 2018). Before rhein was found to be an inhibitor of FTO, it was shown to have antitumor effects. Rhein inhibited MCF-7 and MDA-MB-435 BC cell viability and growth by suppressing vascular endothelial growth factor (VEGF)- and endothelial growth factor (EGF)-induced activation of the PTEN/PI3K/AKT/mTOR and MAPK/ERK pathways in BC (Fernand et al., 2011). Rhein also had anti-proliferative and pro-apoptosis ability in MCF-7 cells and HER2-overexpressing MCF-7 cells (MCF-7/HER2). The rhein-induced inhibition of cell growth was associated with caspase-9-mediated apoptosis, and reactive oxygen species-mediated activation of the NF-κB and p53 signaling pathways also participated in this process (Chang et al., 2012). In 2019, one study evaluated rhein’s function in vivo. Rhein enhanced the apoptotic effect of atezolizumab on 4T1 BC cells (Shen et al., 2019). Treatment with a combination of rhein (10 mg/kg) and atezolizumab (10 mg/kg) inhibited 4T1 xenograft tumor growth and increased serum levels of tumor necrosis factor α and interleukin-6, the CD8+ T-cell ratio, apoptotic factors (e.g., caspase-3, caspase-8, and caspase-9), and Bax/Bcl-2 mRNA levels compared with rhein or atezolizumab treatment alone. However, these experiments did not clarify the role of m6A modifications in the inhibitory effect of rhein in BC, which deserves further exploration.
Meclofenamic acid is a nonsteroidal anti-inflammatory drug that was originally approved by the United States Food and Drug Administration. It competes for FTO binding sites on single-stranded DNA (ssDNA) and directly interacts with FTO protein in HeLa cells to inhibit the catalytic activity of FTO and improve levels of m6A modifications in RNA (Huang et al., 2015). Meclofenamic acid inhibited the proliferation of MCF-7 BC cells, with low toxicity (Kovala-Demertzi et al., 2009). However, whether this result can also be achieved by changing the level of m6A modification remains unknown and requires further investigation.
Although numerous small-molecule m6A regulators have been identified, further studies are needed to define their specificity and adverse effects for the treatment of BC in vitro and in vivo. Furthermore, multiple combinations of existing medical approaches with m6A-targeting drugs may be viable options for the treatment of BC.
m6A modifications are one of the most common modifications of mRNA and, most importantly, dynamically reversible. Several studies have shown that m6A modifications are closely related to many human diseases, including cancer. However, the relationship between m6A modifications and BC has not been fully elucidated. m6A modifications play a complex role in expression levels, genotyping, prognosis, cell proliferation, and metastasis in BC. The controversial findings on the role of some m6A modulators in BC may be attributable to the different test platforms that were employed or inconsistent proteomics and transcriptomics analyses or selective bias of clinical specimens. Most studies of m6A modifications in BC have been performed in vitro. Therefore, more in vivo evidence is required to reveal the regulatory mechanism of m6A modifications in BC.
Based on the significant role of m6A modifications in oncogenesis and progression that has been reported, the development of novel m6A regulators may be the new therapeutic options for malignant tumors. FTO, which was originally found to be involved in obesity and fat metabolism, was the first discovered m6A demethylase. The antitumor effects of some agents that target FTO have been reported in BC. However, little is known about their exact mechanism of action and side effects. Whether these inhibitors influence other RNA modifications, such as m1A, m6Am, m5C, and hm5C, is unknown. More studies are also needed to demonstrate their effectiveness in vivo. Combination therapies, such as combining chemotherapy, radiotherapy, and immune checkpoint blockade with proper m6A inhibitors, may hold promise for the treatment of BC, particularly those that have failed in routine treatment. Based on the oncogenic role of FTO in several cancers, inhibitors that target FTO have attracted scientific interests, but more information about its role in BC is needed. YTHDF3, an independent prognostic factor in BC, may be another target that merits further investigation.
N6-methyladenosine modifications in the progression of BC are still being discovered. We expect that substantial progress will be made in further revealing the role of m6A modifications in BC. Further advances in technology will likely contribute to the development of new, promising, and unexpected therapeutic strategies for the treatment of BC in coming years.
MW and J-WB designed the project and wrote the manuscript. Y-QZ, LN, and H-YC performed the literature searches in PubMed, Medline, and Google Scholar. G-JZ arranged and supervised the overall project. All authors contributed to the article and approved the submitted version.
This work was supported in part by grants from the National Natural Science Foundation of China (nos. 81602345 and 91859120), the Research Team Project of the Natural Science Foundation of Guangdong Province (no. 2016A030312008), Xiamen Science and Technology Bureau funded project (nos. 3502Z20194040 and 3502Z20209101). Xiamen’s Key Laboratory of Precision Medicine for Endocrine-Related Cancers, start-up funds from Xiamen University, and the Youth Fund of Xiang’an Hospital of Xiamen University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aoyama, T., Yamashita, S., and Tomita, K. (2020). Mechanistic insights into m6A modification of U6 snRNA by human METTL16. Nucleic Acids Res. 48, 5157–5168. doi: 10.1093/nar/gkaa227
Bartosovic, M., Molares, H. C., Gregorova, P., Hrossova, D., Kudla, G., and Vanacova, S. (2017). N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 45, 11356–11370. doi: 10.1093/nar/gkx778
Batista, P. J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., et al. (2014). m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719. doi: 10.1016/j.stem.2014.09.019
Byler, S., Goldgar, S., Heerboth, S., Leary, M., Housman, G., Moulton, K., et al. (2014). Genetic and epigenetic aspects of breast cancer progression and therapy. Anticancer Res. 34, 1071–1077.
Cai, X., Wang, X., Cao, C., Gao, Y., Zhang, S., Yang, Z., et al. (2018). HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 415, 11–19. doi: 10.1016/j.canlet.2017.11.018
Casella, G., Tsitsipatis, D., Abdelmohsen, K., and Gorospe, M. (2019). mRNA methylation in cell senescence. Wiley Interdiscip. Rev. RNA 10:e1547.
Chang, C. Y., Chan, H. L., Lin, H. Y., Way, T. D., Kao, M. C., Song, M. Z., et al. (2012). Rhein induces apoptosis in human breast cancer cells. Evid Based Comp. Altern. Med. 2012:952504.
Chen, B., Ye, F., Yu, L., Jia, G., Huang, X., Zhang, X., et al. (2012). Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J. Am. Chem. Soc. 134, 17963–17971.
Chen, M., Wei, L., Law, C. T., Tsang, F. H., Shen, J., Cheng, C. L., et al. (2018). RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67, 2254–2270. doi: 10.1002/hep.29683
Chen, X. Y., Zhang, J., and Zhu, J. S. (2019). The role of m(6)A RNA methylation in human cancer. Mol. Cancer 18:103.
Cohn, W. E. (1960). Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: isolation, structure, and chemical characteristics. J. Biol. Chem. 235, 1488–1498.
Cui, Q., Shi, H., Ye, P., Li, L., Qu, Q., Sun, G., et al. (2017). m(6)A RNA Methylation Regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18, 2622–2634. doi: 10.1016/j.celrep.2017.02.059
Deng, X., Su, R., Stanford, S., and Chen, J. (2018). Critical enzymatic functions of FTO in obesity and cancer. Front. Endocrinol. 9:396. doi: 10.3389/fendo.2018.00396
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U.S.A. 71, 3971–3975. doi: 10.1073/pnas.71.10.3971
Dina, C., Meyre, D., Gallina, S., Durand, E., Korner, A., Jacobson, P., et al. (2007). Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 39, 724–726.
Ding, C., Zou, Q., Ding, J., Ling, M., Wang, W., Li, H., et al. (2018). Increased N6-methyladenosine causes infertility is associated with FTO expression. J. Cell Physiol. 233, 7055–7066. doi: 10.1002/jcp.26507
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Du, H., Zhao, Y., He, J., Zhang, Y., Xi, H., Liu, M., et al. (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7:12626.
Esteve-Puig, R., Bueno-Costa, A., and Esteller, M. (2020). Writers, readers and erasers of RNA modifications in cancer. Cancer Lett. 474, 127–137. doi: 10.1016/j.canlet.2020.01.021
Fernand, V. E., Losso, J. N., Truax, R. E., Villar, E. E., Bwambok, D. K., Fakayode, S. O., et al. (2011). Rhein inhibits angiogenesis and the viability of hormone-dependent and -independent cancer cells under normoxic or hypoxic conditions in vitro. Chem. Biol. Interact. 192, 220–232. doi: 10.1016/j.cbi.2011.03.013
Fry, N. J., Law, B. A., Ilkayeva, O. R., Carraway, K. R., Holley, C. L., and Mansfield, K. D. N. (2018). (6)-methyladenosine contributes to cellular phenotype in a genetically-defined model of breast cancer progression. Oncotarget 9, 31231–31243. doi: 10.18632/oncotarget.25782
Fu, L., Amato, N. J., Wang, P., McGowan, S. J., Niedernhofer, L. J., and Wang, Y. (2015). Simultaneous quantification of methylated cytidine and adenosine in cellular and Tissue RNA by nano-flow liquid chromatography-tandem mass spectrometry coupled with the stable isotope-dilution method. Anal. Chem. 87, 7653–7659. doi: 10.1021/acs.analchem.5b00951
Funakoshi, Y., Wang, Y., Semba, T., Masuda, H., Hout, D., Ueno, N. T., et al. (2019). Comparison of molecular profile in triple-negative inflammatory and non-inflammatory breast cancer not of mesenchymal stem-like subtype. PLoS One 14:e0222336. doi: 10.1371/journal.pone.0222336
Fustin, J. M., Doi, M., Yamaguchi, Y., Hida, H., Nishimura, S., Yoshida, M., et al. (2013). RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806. doi: 10.1016/j.cell.2013.10.026
Gholamalizadeh, M., Jarrahi, A. M., Akbari, M. E., Bourbour, F., Mokhtari, Z., Salahshoornezhad, S., et al. (2020). Association between FTO gene polymorphisms and breast cancer: the role of estrogen. Expert Rev. Endocrinol. Metab. 15, 115–121. doi: 10.1080/17446651.2020.1730176
Harper, J. E., Miceli, S. M., Roberts, R. J., and Manley, J. L. (1990). Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Res. 18, 5735–5741. doi: 10.1093/nar/18.19.5735
Haussmann, I. U., Bodi, Z., Sanchez-Moran, E., Mongan, N. P., Archer, N., Fray, R. G., et al. (2016). m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540, 301–304. doi: 10.1038/nature20577
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127. doi: 10.1038/cr.2017.99
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295. doi: 10.1038/s41556-018-0045-z
Huang, H., Weng, H., Zhou, K., Wu, T., Zhao, B. S., Sun, M., et al. (2019). Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 567, 414–419. doi: 10.1038/s41586-019-1016-7
Huang, T., Guo, J., Lv, Y., Zheng, Y., Feng, T., Gao, Q., et al. (2019). Meclofenamic acid represses spermatogonial proliferation through modulating m(6)A RNA modification. J. Anim. Sci. Biotechnol. 10:63.
Huang, Y., Su, R., Sheng, Y., Dong, L., Dong, Z., Xu, H., et al. (2019). Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell 35, 677.e10–691.e10.
Huang, Y., Yan, J., Li, Q., Li, J., Gong, S., Zhou, H., et al. (2015). Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 43, 373–384. doi: 10.1093/nar/gku1276
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi: 10.1038/nchembio.687
Jia, G. F., Fu, Y., Zhao, X., Dai, Q., Zheng, G. Q., Yang, Y., et al. (2012). N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO (vol 7, pg 885, 2011). Nat. Chem. Biol. 8:1008. doi: 10.1038/nchembio1212-1008a
Knuckles, P., Lence, T., Haussmann, I. U., Jacob, D., Kreim, N., Carl, S. H., et al. (2018). Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 32, 415–429. doi: 10.1101/gad.309146.117
Konig, S. M., Rissler, V., Terkelsen, T., Lambrughi, M., and Papaleo, E. (2019). Alterations of the interactome of Bcl-2 proteins in breast cancer at the transcriptional, mutational and structural level. PLoS Comput. Biol. 15:e1007485. doi: 10.1371/journal.pcbi.1007485
Kovala-Demertzi, D., Dokorou, V., Primikiri, A., Vargas, R., Silvestru, C., Russo, U., et al. (2009). Organotin meclofenamic complexes: synthesis, crystal structures and antiproliferative activity of the first complexes of meclofenamic acid - novel anti-tuberculosis agents. J. Inorg. Biochem. 103, 738–744. doi: 10.1016/j.jinorgbio.2009.01.014
Leismann, J., Spagnuolo, M., Pradhan, M., Wacheul, L., Vu, M. A., Musheev, M., et al. (2020). The 18S ribosomal RNA m(6) A methyltransferase Mettl5 is required for normal walking behavior in Drosophila. EMBO Rep. 21:e49443.
Li, A., Chen, Y. S., Ping, X. L., Yang, X., Xiao, W., Yang, Y., et al. (2017). Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 27, 444–447. doi: 10.1038/cr.2017.10
Li, J., Han, Y., Zhang, H., Qian, Z., Jia, W., Gao, Y., et al. (2019a). The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem. Biophys. Res. Commun. 512, 479–485. doi: 10.1016/j.bbrc.2019.03.093
Li, J., Zhu, L., Shi, Y., Liu, J., Lin, L., and Chen, X. (2019b). m6A demethylase FTO promotes hepatocellular carcinoma tumorigenesis via mediating PKM2 demethylation. Am. J. Transl. Res. 11, 6084–6092.
Li, Z., Weng, H., Su, R., Weng, X., Zuo, Z., Li, C., et al. (2017). FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 31, 127–141. doi: 10.1016/j.ccell.2016.11.017
Lin, X., Chai, G., Wu, Y., Li, J., Chen, F., Liu, J., et al. (2019). RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 10:2065.
Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., and Jaffrey, S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772. doi: 10.1038/nmeth.3453
Liu, F., You, X., Wang, Y., Liu, Q., Liu, Y., Zhang, S., et al. (2014). The oncoprotein HBXIP enhances angiogenesis and growth of breast cancer through modulating FGF8 and VEGF. Carcinogenesis 35, 1144–1153. doi: 10.1093/carcin/bgu021
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Liu, L., Liu, X., Dong, Z., Li, J., Yu, Y., Chen, X., et al. (2019). N6-methyladenosine-related genomic targets are altered in breast cancer tissue and associated with poor survival. J. Cancer 10, 5447–5459. doi: 10.7150/jca.35053
Liu, Y., Wang, R., Zhang, L., Li, J., Lou, K., and Shi, B. (2017). The lipid metabolism gene FTO influences breast cancer cell energy metabolism via the PI3K/AKT signaling pathway. Oncol. Lett. 13, 4685–4690. doi: 10.3892/ol.2017.6038
Louloupi, A., Ntini, E., Conrad, T., and Orom, U. A. V. (2018). Transient N-6-Methyladenosine transcriptome sequencing reveals a regulatory role of m6A in splicing efficiency. Cell Rep. 23, 3429–3437. doi: 10.1016/j.celrep.2018.05.077
Mao, Y., Dong, L., Liu, X. M., Guo, J., Ma, H., Shen, B., et al. (2019). m(6)A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 10:5332.
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m(6)Am in the 5’ cap controls mRNA stability. Nature 541, 371–375. doi: 10.1038/nature21022
Meyer, K. D., and Jaffrey, S. R. (2017). Rethinking m(6)a readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342. doi: 10.1146/annurev-cellbio-100616-060758
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5’. UTR m(6)A promotes Cap-Independent translation. Cell 163, 999–1010. doi: 10.1016/j.cell.2015.10.012
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646. doi: 10.1016/j.cell.2012.05.003
Muller, S., Glass, M., Singh, A. K., Haase, J., Bley, N., Fuchs, T., et al. (2019). IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 47, 375–390. doi: 10.1093/nar/gky1012
Niu, Y., Lin, Z. Y., Wan, A., Chen, H. L., Liang, H., Sun, L., et al. (2019). RNA N6-methyladenosine demethylase FTO promotes breast tumor progression updates through inhibiting BNIP3. Mol. Cancer 18:46.
Ortega, M. A., Fraile-Martinez, O., Asunsolo, A., Bujan, J., Garcia-Honduvilla, N., and Coca, S. (2020). Signal transduction pathways in breast cancer: the important role of PI3K/Akt/mTOR. J. Oncol. 2020:9258396.
Panneerdoss, S., Eedunuri, V. K., Yadav, P., Timilsina, S., Rajamanickam, S., Viswanadhapalli, S., et al. (2018). Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci. Adv. 4:eaar8263. doi: 10.1126/sciadv.aar8263
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. doi: 10.1038/nature19342
Paul, A., and Paul, S. (2014). The breast cancer susceptibility genes (BRCA) in breast and ovarian cancers. Front. Biosci. 19:605–618. doi: 10.2741/4230
Peng, S., Xiao, W., Ju, D., Sun, B., Hou, N., Liu, Q., et al. (2019). Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci. Transl. Med. 11:eaau7116. doi: 10.1126/scitranslmed.aau7116
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189. doi: 10.1038/cr.2014.3
Qian, J. Y., Gao, J., Sun, X., Cao, M. D., Shi, L., Xia, T. S., et al. (2019). KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene 38, 6123–6141. doi: 10.1038/s41388-019-0861-z
Rahman, M. M., Brane, A. C., and Tollefsbol, T. O. (2019). MicroRNAs and epigenetics strategies to reverse Breast Cancer. Cells 8:1214. doi: 10.3390/cells8101214
Reichel, M., Koster, T., and Staiger, D. (2019). Marking RNA: m6A writers, readers, and functions in Arabidopsis. J. Mol. Cell Biol. 11, 899–910. doi: 10.1093/jmcb/mjz085
Ren, W., Lu, J., Huang, M., Gao, L., Li, D., Wang, G. G., et al. (2019). Structure and regulation of ZCCHC4 in m(6)A-methylation of 28S rRNA. Nat. Commun. 10:5042.
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife 6:e31311.
Ruzicka, K., Zhang, M., Campilho, A., Bodi, Z., Kashif, M., Saleh, M., et al. (2017). Identification of factors required for m(6) A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215, 157–172. doi: 10.1111/nph.14586
Scholler, E., Weichmann, F., Treiber, T., Ringle, S., Treiber, N., Flatley, A., et al. (2018). Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA 24, 499–512. doi: 10.1261/rna.064063.117
Shen, F., Huang, W., Huang, J. T., Xiong, J., Yang, Y., Wu, K., et al. (2015). Decreased N(6)-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J. Clin. Endocrinol. Metab. 100, E148–E154.
Shen, Z., Zhu, B., Li, J., and Qin, L. (2019). Rhein augments antiproliferative effects of atezolizumab based on Breast Cancer (4T1) regression. Planta Med. 85, 1143–1149. doi: 10.1055/a-1012-7034
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27, 315–328. doi: 10.1038/cr.2017.15
Siegel, R. L., Miller, K. D., and Jemal, A. (2020). Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30.
Su, R., Dong, L., Li, C., Nachtergaele, S., Wunderlich, M., Qing, Y., et al. (2018). R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell 172, 90.e23–105.e23.
Tan, A., Dang, Y., Chen, G., and Mo, Z. (2015). Overexpression of the fat mass and obesity associated gene (FTO) in breast cancer and its clinical implications. Int. J. Clin. Exp. Pathol. 8, 13405–13410.
Theler, D., Dominguez, C., Blatter, M., Boudet, J., and Allain, F. H. (2014). Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 42, 13911–13919. doi: 10.1093/nar/gku1116
Wang, H., Xu, B., and Shi, J. (2020). N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene 722:144076. doi: 10.1016/j.gene.2019.144076
Wang, P., Doxtader, K. A., and Nam, Y. (2016). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mo. l Cell 63, 306–317. doi: 10.1016/j.molcel.2016.05.041
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578. doi: 10.1038/nature18298
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730
Wang, X., Zhu, L., Chen, J., and Wang, Y. (2015a). mRNA m(6)A methylation downregulates adipogenesis in porcine adipocytes. Biochem. Biophys. Res. Commun. 459, 201–207. doi: 10.1016/j.bbrc.2015.02.048
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015b). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399. doi: 10.1016/j.cell.2015.05.014
Wei, J., Liu, F., Lu, Z., Fei, Q., Ai, Y., He, P. C., et al. (2018). Differential m(6)A, m(6)Am, and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71, 973.e5–985.e5.
Wu, L., Wu, D., Ning, J., Liu, W., and Zhang, D. (2019). Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer 19:326. doi: 10.1186/s12885-019-5538-z
Yang, S., Wei, J., Cui, Y. H., Park, G., Shah, P., Deng, Y., et al. (2019). m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 10:2782.
Yi, D., Wang, R., Shi, X., Xu, L., Yilihamu, Y., and Sang, J. (2020). METTL14 promotes the migration and invasion of breast cancer cells by modulating N6-methyladenosine and hsa-miR-146a-5p expression. Oncol. Rep. 43, 1375–1386.
Yu, J., Chen, M., Huang, H., Zhu, J., Song, H., Zhu, J., et al. (2018). Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 46, 1412–1423. doi: 10.1093/nar/gkx1182
Yue, L., Li, L., Liu, F., Hu, N., Zhang, W., Bai, X., et al. (2013). The oncoprotein HBXIP activates transcriptional coregulatory protein LMO4 via Sp1 to promote proliferation of breast cancer cells. Carcinogenesis 34, 927–935. doi: 10.1093/carcin/bgs399
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4:10.
Yue, Y., Liu, J., and He, C. R. N. A. (2015). N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355. doi: 10.1101/gad.262766.115
Zhang, C., Samanta, D., Lu, H., Bullen, J. W., Zhang, H., Chen, I., et al. (2016a). Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U.S.A. 113, E2047–E2056.
Zhang, C., Zhi, W. I., Lu, H., Samanta, D., Chen, I., Gabrielson, E., et al. (2016b). Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget 7, 64527–64542. doi: 10.18632/oncotarget.11743
Zhao, B. S., Roundtree, I. A., and He, C. (2017). Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42. doi: 10.1038/nrm.2016.132
Zhao, X., Yang, Y., Sun, B. F., Zhao, Y. L., and Yang, Y. G. (2014). FTO and obesity: mechanisms of association. Curr. Diab. Rep. 14:486.
Zheng, G. Q., Dahl, J. A., Niu, Y. M., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 Is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. doi: 10.1016/j.molcel.2012.10.015
Zheng, W., Dong, X., Zhao, Y., Wang, S., Jiang, H., Zhang, M., et al. (2019). Multiple functions and mechanisms underlying the role of METTL3 in human cancers. Front. Oncol. 9:1403. doi: 10.3389/fonc.2019.01403
Zhong, X., Yu, J., Frazier, K., Weng, X., Li, Y., Cham, C. M., et al. (2018). Circadian clock regulation of hepatic lipid metabolism by modulation of m(6)A mRNA methylation. Cell Rep. 25, 1816.e4–1828.e4.
Keywords: N6-methyladenosine, breast cancer, m6A modification regulator, mechanism pathways, FTO inhibitor
Citation: Wei M, Bai J-W, Niu L, Zhang Y-Q, Chen H-Y and Zhang G-J (2021) The Complex Roles and Therapeutic Implications of m6A Modifications in Breast Cancer. Front. Cell Dev. Biol. 8:615071. doi: 10.3389/fcell.2020.615071
Received: 08 October 2020; Accepted: 07 December 2020;
Published: 11 January 2021.
Edited by:
Xiao Zhu, Guangdong Medical University, ChinaReviewed by:
Ruiqiong Guo, Michigan State University, United StatesCopyright © 2021 Wei, Bai, Niu, Zhang, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guo-Jun Zhang, Z2p6aGFuZ0B4YWgueG11LmVkdS5jbg==; Z3Vval96aGFuZ0B5YWhvby5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.