- 1Faculty of Medicine and Faculty of Life Sciences, Institute of Biomedical Sciences and FONDAP Center for Genome Regulation, Universidad Andres Bello, Santiago, Chile
- 2Faculty of Medicine, Universidad de los Andes, Santiago, Chile
- 3Molecular Biology and Bioinformatic Lab, Program in Molecular Biology and Bioinformatics, Center for Biomedical Research and Innovation (CIIB), Universidad de los Andes, Santiago, Chile
Within the eukaryotic nucleus the genomic DNA is organized into chromatin by stably interacting with the histone proteins as well as with several other nuclear components including non-histone proteins and non-coding RNAs. Together these interactions distribute the genetic material into chromatin subdomains which can exhibit higher and lower compaction levels. This organization contributes to differentially control the access to genomic sequences encoding key regulatory genetic information. In this context, epigenetic mechanisms play a critical role in the regulation of gene expression as they modify the degree of chromatin compaction to facilitate both activation and repression of transcription. Among the most studied epigenetic mechanisms we find the methylation of DNA, ATP-dependent chromatin remodeling, and enzyme-mediated deposition and elimination of post-translational modifications at histone and non-histone proteins. In this mini review, we discuss evidence that supports the role of these epigenetic mechanisms during transcriptional control of osteoblast-related genes. Special attention is dedicated to mechanisms of epigenetic control operating at the Runx2 and Sp7 genes coding for the two principal master regulators of the osteogenic lineage during mesenchymal stem cell commitment.
Introduction
Osteoblast lineage commitment is regulated by a coordinated set of extra cellular stimuli and developmentally-regulated signaling pathways, including those mediated by bone morphogenic proteins (BMPs), Wnt-ligands, steroid hormones, and growth factors, among others (Li et al., 1998; Nishimura et al., 1998; Drissi et al., 2000; Yamaguchi et al., 2000; Zhang et al., 2008). Following activation in pre-osteogenic cells, these signaling pathways modulate the expression and function of osteoblast master transcription factors, which in turn control the expression of downstream bone-phenotypic genes, thus establishing the osteoblastic cell component of the mammalian skeleton (Stein et al., 2004; Long, 2012). Although the precise molecular mechanisms associated with transcriptional control of these osteoblast-related genes are still far from being completely understood, significant progress has been made during the last two decades.
We discuss research demonstrating the role of epigenetic mechanisms in controlling gene transcription during mesenchymal cell commitment to the osteogenic lineage. We first overview key components of epigenetic mechanisms that control gene expression in mammals and then describe how these epigenetic processes contribute to regulate transcription of the Runx2 and Sp7 genes, two critical osteogenic master regulators.
General Overview of Epigenetic Control in Mammals
Nuclear DNA is organized as chromatin. Nucleosomes are the fundamental units of this chromatin and are composed of an approximately 147 bp DNA segment wrapped around an octamer of histone proteins (two each of histones H2A, H2B, H3, and H4) (Ramakrishnan, 1997; Richmond and Davey, 2003). A portion of these histone proteins (N-terminus tail) extends beyond the limits of the particle core, providing additional surfaces for interaction with nuclear proteins that regulate transcription (Strahl and Allis, 2000; Bannister and Kouzarides, 2011; Voigt et al., 2013). As specific residues (e.g., lysines and arginines) within these histone tails are subject to extensive enzymatic post-translational modifications, the potential for modifying the chemical environment surrounding the chromatin fibers represents a key regulatory component during gene expression control (Bannister and Kouzarides, 2011). The genomic DNA can be also enzymatically modified affecting the degree of compaction of chromatin. Increased condensation reduces the possibility that specific regulatory DNA sequence motifs are recognized by transcription factors that control transcription (Franchini et al., 2012; Bogdanović and Lister, 2017).
Molecular components that regulate chromatin organization and transcription are critical players during cell differentiation. Among them the large group of “histone post-translational modifications” (HPTMs), which are considered a principal “epigenetic” mechanism (Bannister and Kouzarides, 2011; Shilatifard, 2012; Voigt et al., 2013). HPTMs may function as docking sites on the chromatin fiber surface that can be recognized by regulatory proteins (“epigenetic readers”) that contain specific complementary domains (e.g., chromo-domains interact with methylated histone lysine residues) (Voigt et al., 2013; Venkatesh and Workman, 2015). A number of protein complexes that are capable of mediating deposition (“epigenetic writers”) or elimination (“epigenetic erasers”) of HPTMs have been identified in eukaryotic cells. Importantly, most of the core subunits of these complexes are evolutionary conserved (Dimitrova et al., 2015; Piunti and Shilatifard, 2016; Jambhekar et al., 2019), indicating that their functions during gene expression control are also conserved across species.
Genetic studies allowed the identification of the Polycomb Group (PcG) and the Trithorax Group (TrxG) complexes, which mediate inhibition and activation of transcription, respectively, during embryogenesis (Ringrose and Paro, 2007). One signature property of the evolutionary conserved PcG complexes PRC1 and PRC2 (Polycomb Repressive Complex 1 and 2) is to mediate the formation of repressed chromatin. In mammals, PRC2 includes as main subunits Enhancer of Zeste Homolog 2 or 1 (Ezh2/Ezh1), Suppressor of Zeste 12 (Suz12), and Embryonic Ectoderm Development (Eed) (Piunti and Shilatifard, 2016; Yu et al., 2019). Ezh2 (or alternatively Ezh1) is the main catalytic component of PRC2 mediating tri-methylation of the lysine 27 residue of histone H3 (H3K27me3), modification that is associated with transcriptionally silent chromatin (Voigt et al., 2013; Piunti and Shilatifard, 2016).
Several mammalian TrxG complexes, including COMPASS (Complex of Proteins Associated with Set1a/b) and the Mixed Lineage Leukemia (MLL1–5)-containing COMPASS-like complexes have been reported (Shilatifard, 2012). TrxG-mediated activity involves mono-, di-, and tri-methylation of the lysine 4 residue of histone H3 (H3K4me1, H3K4me2, and H3K4me3). H3K4me3 is often found enriched at transcriptionally active chromatin (euchromatin), mainly around the transcription start sites (TSSs) of promoters. Moreover, H3K4me3 can be recognized by the RNA polymerase II complex, hence facilitating transcriptional activity at H3K4me3-marked gene promoters (Vermeulen et al., 2007). Set1-COMPASS and MLL-COMPASS-like complexes include the Wdr5 (WD Repeat Domain 5) subunit which has been shown necessary for assembly, stability and optimal enzymatic activity of these complexes (Shilatifard, 2012). Interestingly, some of these histone-methylating complexes also include enzymatic activities that can remove other histone marks. For instance, MLL3/4-COMPASS-like that also contains the H3K27me3 demethylase Utx/Kdm6a (Piunti and Shilatifard, 2016). Hence, binding of MLL3/4-COMPASS-like to a particular genomic region can also result in reduced H3K27me3, an epigenetic signature associated with decreased transcription (Voigt et al., 2013; Piunti and Shilatifard, 2016).
Set1-COMPASS mediates global genomic deposition of H3K4me3 in most mammalian cells and therefore its function is tightly associated with transcriptional activation of a large number of genes. MLL2-COMPASS-like has been shown responsible for H3K4me3 deposition at promoters in embryonic stem cells (ESCs), contributing to transcriptional upregulation of homeobox genes (Denissov et al., 2014). Interestingly, MLL3/4-COMPASS-like can catalyze the deposition of H3K4me1 at enhancer elements in mammalian cells (Hu et al., 2013; Yan et al., 2018). Recent results from several teams, however, also show that MLL3/4 complexes can mediate the maintenance of the H3K4me1 mark at proximal promoter regions of repressed, but poised for expression, genes (Cheng et al., 2014; Rojas et al., 2015, 2019; Aguilar et al., 2016; Sepulveda et al., 2017a; Local et al., 2018). Moreover, MLL3/4 depletion results in transcriptional activation of some MLL3/4-target genes as both Set1a/b-COMPASS and MLL1-COMPASS-like gain access to these genes to mediate the transition from H3K4me1 to H3K4me3 that accompanies transcription (Cheng et al., 2014). Together, these studies imply that different COMPASS and COMPASS-like complexes bind to target sequences in a highly coordinately manner to first maintain a gene silent, but poised for transcription, and subsequently to activate its expression.
Chromatin domains with decreased enrichment of H3K4me3 or H3K27me3 can also be maintained through the activity of lysine demethylases (Dimitrova et al., 2015; Jambhekar et al., 2019). In particular, demethylation of H3K4me3 in mammals is mediated by members of the Jarid1/Kdm5 family (Jarid1a, b, c, and d) (Kooistra and Helin, 2012), which convert H3K4me3 and H3K4me2 to H3K4me1 (Christensen et al., 2007; Iwase et al., 2007; Yamane et al., 2007). Absence of these enzymes can result in enrichment of H3K4me3 at target genes and transcriptional activation (Albert et al., 2013; Rojas et al., 2015). On the other hand, histone demethylases Utx/Kdm6a and Jmjd3/Kdm6b can catalyze the removal of methyl groups from H3K27me3 and therefore counteract the silencing activity of PRC2 (Agger et al., 2007; Hong et al., 2007; Lan et al., 2007; Santa et al., 2007; Xiang et al., 2007).
Methylation at H3K9 (H3K9me1, H3K9me2, and H3K9me3) is strongly associated with the formation of highly compacted and transcriptionally repressed chromatin (heterochromatin) (Saksouk et al., 2015; Nicetto and Zaret, 2019). The methyl-transferases that deposit this modification (“H3K9me writers”) include Suv39H1/Kmt1a, Suv39H2/Kmt1b and Setdb1/ESET/Kmt1e, which can mediate mono-, di-, and tri-methylation. Also, G9a/Ehmt2/Kmt1c and GLP/Ehmt1/Kmt1d, which mediate H3K9me1 and H3K9me2 (Saksouk et al., 2015; Nicetto et al., 2019). All of these enzymes are critical components of several transcription repressive complexes that operate during cell differentiation (Nicetto et al., 2019). The H3K9 methyl-transferase activity is counteracted by H3K9 demethylases, that remove these marks (“H3K9me1/2/3 erasers”) (Janssen et al., 2018). Among them, Lsd1/Kdm1, Jmjd1a/Kdm3a, Jmjd1c/Kdm3c, which can eliminate H3K9me1 or H3K9me2, and Jmjd2a/Kdm4a, Jmjd2b/Kdm4b, Jmjd2c/Kdm4c, and Jmjd2d/Kdm4d, which erase H3K9me1, H3K9me2, and H3K9me3.
Chromatin organization and transcriptional activity is also mediated by ATP-dependent remodelers (Horn and Peterson, 2001; Bakshi et al., 2010; Längst and Manelyte, 2015), which are multi-subunit complexes that include a catalytic core (e.g., Brg1 in the mammalian SWI/SNF and INO80 in the INO80 complex) mediating binding and hydrolysis of ATP (ATPase activity) (Tsukiyama, 2002; Martens and Winston, 2003). These remodelers alter chromatin structure by mobilizing nucleosomes in cis or by transferring histone octamers in an ATP-dependent manner. This changes the exposure of regulatory motifs thereby facilitating or preventing recognition by cognate factors (Liu et al., 2011). The contribution of ATP-dependent remodelers during development and differentiation is well-established (Srivastava et al., 2010; Ruijtenberg and van den Heuvel, 2016). Whereas these complexes can be specifically recruited to gene promoters by tissue-specific transcription factors (Armstrong et al., 1998; Serna et al., 2001), several reports also indicate that their targeting is modulated by HPTMs, including histone lysine acetylation and histone arginine methylation (Kanno et al., 2004; Pal et al., 2004). This regulation is due to the presence of bromo- and chromo-domains at subunits of these complexes (e.g., SWI/SNF and CHD3/4) that recognize these modified histone residues (Becker and Workman, 2013).
Genomic DNA can be methylated at cytosines that are followed by guanosines (CpG dinucleotides). The DNA methyl-transferases that mediate this modification belong to a well-conserved family of proteins that include both maintenance (Dnmt1) and de novo (Dnmt3a and Dnmt3b) activities (Franchini et al., 2012; Bogdanović and Lister, 2017). Most reports demonstrate that methylated CpG is associated with higher chromatin compaction and reduced transcriptional activity (Franchini et al., 2012; Bogdanović and Lister, 2017). DNA demethylation in mammalian cells involves the conversion from 5-methyl-CpG (5mCpG) to 5-hydroxymethyl-CpG (5hmCpG) by the activity of the Ten Eleven Transformation (Tet) family of dioxygenases (Tahiliani et al., 2009; Ito et al., 2010; Bogdanović and Lister, 2017). Importantly, Tet proteins form regulatory complexes with chromatin remodelers like SWI/SNF as well as with histone methyl-transferases and histone demethylases (Williams et al., 2011; Yildirim et al., 2011; Neri et al., 2013).
Enrichment of H3K9me3/2 at transcriptionally-inactive chromatin can be associated with the presence of 5mCpG. This is due to the ability of the proteins that “write” and “read” these two epigenetic marks to form complexes (Janssen et al., 2018; Nicetto and Zaret, 2019) thereby providing a means for both repressive epigenetic modifications (5mCpG and H3K9me3) to collaborate in generating a condensed chromatin structure that reduces transcription. These findings further indicate that different epigenetic mechanisms leading to chromatin remodeling and transcriptional control can function in a coordinated and complementary manner within eukaryotic cells, allowing an effective regulation of gene expression in response to physiological cues.
Epigenetic Control of the Expression of Master Regulators of Osteogenesis
Differentiation of mesenchymal stem cells (MSCs) to the osteogenic lineage requires the expression and function of two master transcription factors: Runx2 (Runt Related Transcription Factor 2) and Sp7 (also known as Osterix). These factors bind to and control the expression of numerous downstream target genes that code for proteins that are necessary for establishing the osteoblast phenotype (Zhang, 2010; Sinha and Zhou, 2013; Meyer et al., 2014; Wu et al., 2014). Runx2 and Sp7 are expressed at early embryonic stages of the mesenchyme-osteoblast lineage commitment process and optimal concentrations of both factors in pre-osteogenic cells is essential for proper osteoblast differentiation (Long, 2012). Runx2 binds to and activates the Sp7 promoter (Yoshida et al., 2012), indicating that Runx2 is an upstream transcription factor during osteogenesis, but that this differentiation process requires the expression of both master regulators (Long, 2012). In recent years, several groups have demonstrated that changes in chromatin structure and expression of the Runx2 and Sp7 genes are epigenetically-controlled (Yang et al., 2013, 2015; Tai et al., 2014; Dudakovic et al., 2015; Rojas et al., 2015, 2019; Zhang et al., 2015; Aguilar et al., 2016, 2020; Park-Min, 2016; Zhou et al., 2016; Sepulveda et al., 2017a; Cakouros and Gronthos, 2020; Figure 1).
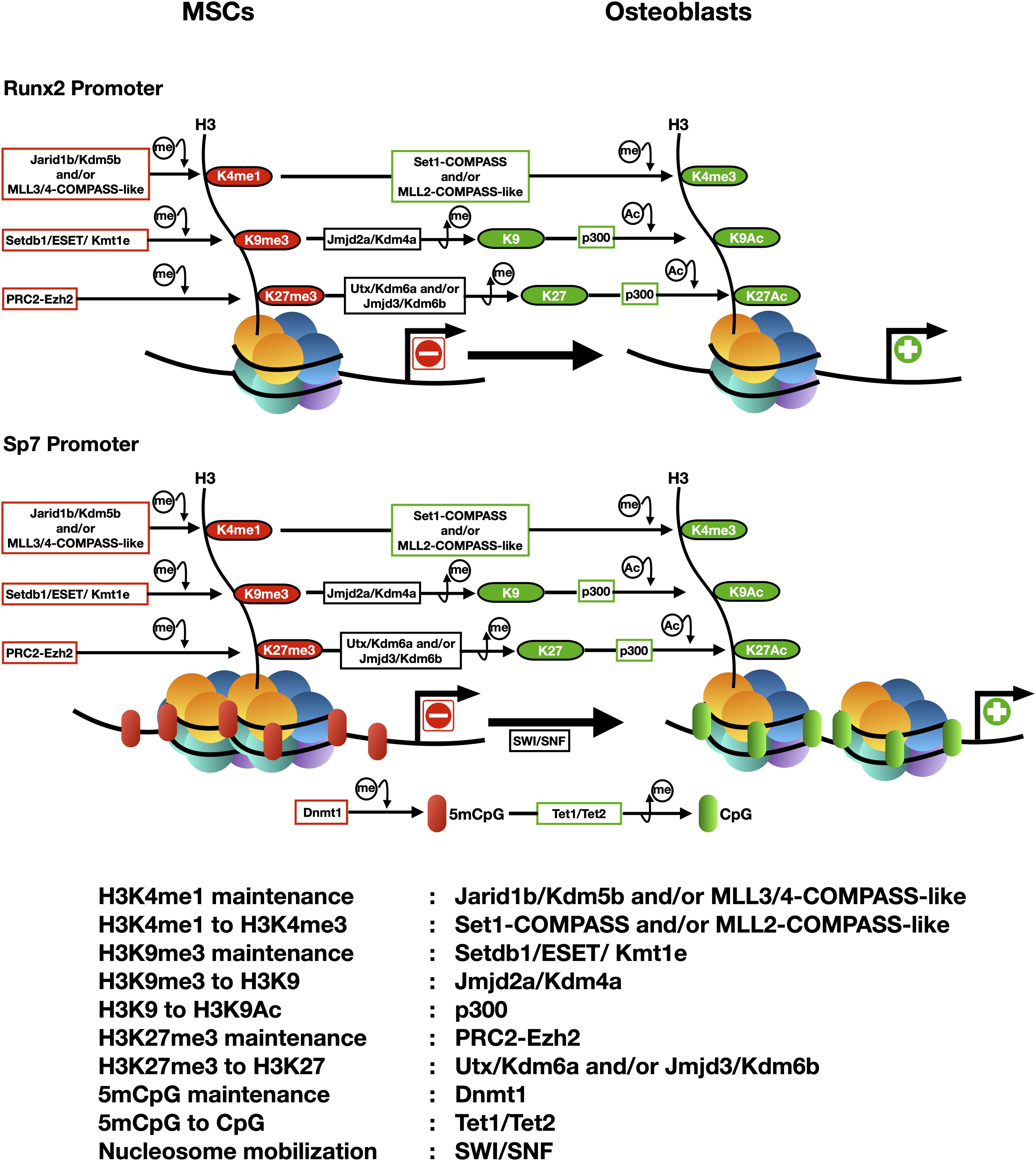
Figure 1. Epigenetic landscapes at the transcriptionally active and repressed Runx2 and Sp7 gene promoters during osteogenic differentiation of mesenchymal stem cells (e.g., CD10 positive hMSCs). The arrowhead indicates the transcriptional start site (TSS). Principal epigenetic features and each enzyme associated are included and summarized below.
Methylation at Histone H3K4 and H3K27 Residues
Active transcription of the Runx2 and Sp7 genes is accompanied by an epigenetic landscape that includes increased H3K4me3 enrichment at both promoter regions (Figure 1). Hence, transcription of these master genes requires binding and activity of COMPASS and COMPASS-like complexes (Shilatifard, 2012). Studies initiated two decades ago first demonstrated that over-expression of Wdr5, a critical subunit of these complexes, accelerates osteoblast and chondrocyte differentiation programs (Gori et al., 2001, 2006; Zhu et al., 2008). Wdr5 binds to the Runx2-P1 promoter in osteoblastic cells (Rojas et al., 2015) where it can mediate transcriptional upregulation. It was recently found that transcription of both Runx2 and Sp7 during osteogenesis requires that Set1-COMPASS and MLL2-COMPASS-like complexes mediate deposition of the H3K4me3 mark at both promoters (Rojas et al., 2015, 2019; Sepulveda et al., 2017b).
It has been determined that an enrichment of H3K4me1 accompanies transcriptional silence at the Runx2 and Sp7 genes in non-osteoblastic and mesenchymal uncommitted cells (Rojas et al., 2015; Aguilar et al., 2016, 2020; Sepulveda et al., 2017a,b). As MLL3/4-COMPASS can mediate maintenance of H3K4me1 at repressed, but poised for expression, genes (Cheng et al., 2014; Rojas et al., 2015, 2019; Aguilar et al., 2016; Sepulveda et al., 2017a; Local et al., 2018), one possibility is that H3K4me1 is deposited at the Runx2 and Sp7 promoters by the MLL3/4 complexes that have been found associated with both genes in MSCs prior to engage osteogenesis (Sepulveda et al., 2017b; Rojas et al., 2019).
Maintenance of chromatin with low H3K4me3 enrichment (and hence elevated H3K4me1) at the Runx2 promoter can be also achieved by binding and activity of the H3K4 demethylase Jarid1b/Kdm5b (Figure 1; Rojas et al., 2015, 2019; Bustos et al., 2017; Sepulveda et al., 2017a), which converts H3K4me3 to H3K4me1 (Christensen et al., 2007; Iwase et al., 2007; Yamane et al., 2007). Targeted depletion of Jarid1b/Kdm5b in mice results in elevated lethality at early post-natal stages, exhibiting these animals cranial dysmorphic parameters and skeletal alterations (Albert et al., 2013). Moreover, silencing of Runx2 expression during myogenic cell differentiation is mediated by Jarid1b/Kdm5b bound to the Runx2 promoter (Rojas et al., 2015, 2019). Accordingly, knockdown of Jarid1b/Kdm5b expression is accompanied by enrichment of H3K4me3 at the Runx2 promoter and active Runx2 transcription in mouse mesenchymal pluripotent cells differentiating to non-osteoblastic lineages (Rojas et al., 2015; Bustos et al., 2017; Sepulveda et al., 2017a).
Findings from several teams revealed that regulating the repressive mark H3K27me3 represents an important step during the differentiation of MSCs toward osteoblasts. It was demonstrated that the activity of the H3K27 demethylases Utx/Kdm6a and Jmjd3/Kdm6b is critical for erasing this mark from the promoter of the Runx2 and Sp7 genes (Figure 1) and hence to induce their expression during osteogenesis (Ye et al., 2012; Hemming et al., 2014; Rojas et al., 2015; Park-Min, 2016; Sepulveda et al., 2017b; Cakouros and Gronthos, 2020; Sen et al., 2020). Additionally, an increased activity of the PRC2 complex that “writes” the H3K27me3 mark, can significantly limit the ability of MSCs to engage osteogenic differentiation (Wei et al., 2010). Moreover, forced expression of Ezh2 in pre-osteoblastic cells results in significant downregulation of osteogenic genes, including Runx2 and its downstream targets (Hemming et al., 2014). Accordingly, knockdown of Ezh2 expression in MSCs and pre-osteoblastic cells results in increased expression of the Runx2 (Rojas et al., 2015, 2019; Hemming et al., 2016) and Sp7 (Sepulveda et al., 2017b) genes.
Methylation at the Histone H3K9 Residue
The repressive mark H3K9me3 plays an important role during differentiation of MSCs to the osteoblastic lineage (Ye et al., 2012, 2018; Lawson et al., 2013; Tai et al., 2014; Rojas et al., 2015; Zhang et al., 2015; Sepulveda et al., 2017a,b). H3K9me3 is found enriched at the Runx2 and Sp7 promoters in cells that do not express these genes (Figure 1) and binding of proteins that can write and read this mark has been detected in non-osseous cells (Zhang et al., 2015; Aguilar et al., 2016; Sepulveda et al., 2017b). The H3K9 methyl transferase Setdb1/ESET/Kmt1c binds to the Sp7 promoter and maintains a H3K9me3-enriched transcriptionally-silent environment in mouse MSCs and other non-osseous cells (Sepulveda et al., 2017b). Accordingly, during BMP2-induced osteogenic differentiation of MSCs, Setdb1/ESSET/Kmt1c is released from this promoter and replaced by the H3K9me3 demethylase Jmjd2a/Kdm4a, an exchange that is accompanied by decreased enrichment of H3K9me3 and transcription of the Sp7 gene (Sepulveda et al., 2017b). Interestingly, it was also established that a different member of this family of demethylases, Jmjd2b/Kdm2b, plays a critical role during formation of the mouse skeleton (Ye et al., 2012, 2018). However, this demethylase neither directly binds nor decreases the H3K9me3 enrichment at the Sp7 and Runx2 promoters (Ye et al., 2012, 2018).
Although most studies support the role of the H3K27me3 and H3K9me3 marks during repression of both master osteogenic genes, recent analyses indicate that for certain cells, this role may have to be re-evaluated. Thus, analysis of uncommitted human umbilical cord-derived Wharton Jelly MSCs (WJ-MSCs) showed that the Runx2-P1 promoter region does not exhibit significant enrichment of H3K9me3 and H3K27me3 (Bustos et al., 2017; Sepulveda et al., 2017a). Interestingly, both marks are detected at the Sp7 gene promoter in these cells and represent a relevant component of the epigenetic barrier preventing the expression of Sp7 when the cells are induced to differentiate to the osteoblastic lineage (Bustos et al., 2017; Sepulveda et al., 2017a). Hence additional mechanisms are preventing the expression of Runx2-p57 in these uncommitted WJ-MSCs in the absence of H3K27me3 and H3K9me3. One proposed component is the H3K4 demethylase Jarid1b/Kdm5b which was found enriched at this Runx2 P1 promoter concomitant with elevated H3K4me1 (Bustos et al., 2017; Sepulveda et al., 2017a). Knockdown of Jarid1b/Kdm5b expression or selective inhibition of Jarid1b/Kdm5b activity using small molecules, is accompanied by enrichment of H3K4me3 at the Runx2 promoter and Runx2 upregulation in these WJ-MSCs (Bustos et al., 2017; Sepulveda et al., 2017a). Future studies need to carefully address what alternative mechanisms control the expression of Runx2 and Sp7 in the different sources of human MSCs available.
Histone H3 and H4 Acetylation
Histone lysine acetylation also plays a relevant role during bone formation as it supports increased expression of critical genes for osteoblast differentiation (Schroeder and Westendorf, 2005; McGee-Lawrence and Westendorf, 2011). Chromatin remodeling and transcription of the Runx2 gene is accompanied by acetylation of histones H3 and H4 (Figure 1) in nucleosomes associated with the P1 promoter region (Lee H. W. et al., 2006; Hassan et al., 2007; Cruzat et al., 2009; Gordon et al., 2011). This acetylation is correlated with decreased expression of HDACs (HDAC1, 2, and 3) during BMP2-induced osteoblast differentiation (McGee-Lawrence and Westendorf, 2011). Accordingly, knockdown of HDAC1 and HDAC3 expression facilitates osteogenic differentiation (Maroni et al., 2012) and a number of HDAC inhibitors [Sodium Butyrate, Trichostatin A, Valproic acid, and Suberoyl Anilide Hydroxamic Acid (SAHA)] can promote osteogenesis in models including primary calvarial cells and MSCs (Cho et al., 2005; Schroeder and Westendorf, 2005; Lee H. W. et al., 2006; Haberland et al., 2010; Hu et al., 2012; Dudakovic et al., 2013; Zych et al., 2013). Moreover, the histone acetyl transferase (HAT) p300 plays a major role in maintaining histone acetylation at the Runx2-P1 (Rojas et al., 2015) and Sp7 (Sepulveda et al., 2017b) gene promoters (Figure 1).
DNA Methylation
DNA methylation (5mCpG) can regulate osteogenic differentiation of MSCs. Addition of the DNA methyl-transferase inhibitor 5-Azacytidine (5-Aza) to human and murine mesenchymal cells enhances their ability to engage osteogenesis by reducing 5mCpG enrichment at regulatory regions of osseous genes (Lee J. Y. et al., 2006; Zhou et al., 2009; Zych et al., 2013; Sepulveda et al., 2017b; Cakouros et al., 2019). In particular, an active Sp7 gene promoter exhibits reduced 5mCpG at its proximal promoter (Figure 1), whereas this region shows increased 5mCpG in non-osseous cells that do not express Sp7 (Lee J. Y. et al., 2006; Sepulveda et al., 2017b). Moreover, during BMP2-induced osteoblast differentiation DNA demethylation of the Sp7 promoter is mediated by a Tet1/Tet2-containing complex which transforms 5mCpG to 5hmCpG (Sepulveda et al., 2017b). This process is tightly coordinated with nucleosome remodeling mediated by SWI/SNF, “erasing” of the H3K9me3 and H3K27me3 repressive marks and “writing” of the H3ac and H3K4me3 active marks (Sepulveda et al., 2017b; Figure 1). A recent report indicates that in mouse bone marrow-derived MSCs (BM-MSCs), Tet1 is pre-bound to osteogenic gene promoters in the absence of Tet2, playing an initial repressor role. This may result from the recruitment of repressor activities, including Sin3A, Hdacs, and PRC2 (Cakouros et al., 2019). Interestingly, Tet1 can also play a key role during transcriptional activation of osteogenic genes. Tet1 recruits Tet2 to mediate 5mCpG demethylation and gene transcription during differentiation of BM-MSCs (Cakouros et al., 2019). Together these results support a model where binding of Tet1 may initially contribute to maintain target osteogenic genes (including Runx2) silent but poised for expression as the MSCs engage osteoblast lineage commitment.
Concluding Remarks
Controlling lineage commitment in MSCs represents a critical challenge to overcome skeletal deficiencies in patients. Extensive research during the last 20 years has shed light into potential targeting of specific epigenetic mechanisms that control bone-related gene expression. Future studies will need to consider the influence of the mechanical environments at which MSCs are maintained and differentiated ex vivo. Recent results demonstrate that the substrate stiffness at which hMSCs are grown in culture can induce rapid and stable changes in chromatin organization and the associated epigenetic landscapes that permanently up- or down-regulate genes (Killaars et al., 2019), hence modifying the ability of these cells to engage osteogenesis. Future research also needs to develop approaches that include safe delivery strategies that limit undesired effects of treatments based on epidrugs on non-target tissues and organs, thereby decreasing potential negative secondary effects in treated patients.
Author Contributions
MM analyzed the data and wrote most parts of the manuscript. MC wrote parts of the text and prepared the figures. GN analyzed the data, wrote parts of the text and helped preparing the figures. All authors contributed to the article and approved the submitted version.
Funding
This work has been supported by FONDAP 15090007 (to MM); FONDECYT 1170878 (to MM); and FONDECYT 11190998 (to GN).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Agger, K., Cloos, P. A. C., Christensen, J., Pasini, D., Rose, S., Rappsilber, J., et al. (2007). UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731–734. doi: 10.1038/nature06145
Aguilar, R., Bustos, F. J., Nardocci, G., Zundert, B., and Montecino, M. (2020). Epigenetic silencing of the osteoblast-lineage gene program during hippocampal maturation. J. Cell. Biochem. [Epub ahead of print]. doi: 10.1002/jcb.29865
Aguilar, R., Bustos, F. J., Saez, M., Rojas, A., Allende, M. L., van Wijnen, A. J., et al. (2016). Polycomb PRC2 complex mediates epigenetic silencing of a critical osteogenic master regulator in the hippocampus. Biochim. Biophys. Acta 1859, 1043–1055. doi: 10.1016/j.bbagrm.2016.05.009
Albert, M., Schmitz, S. U., Kooistra, S. M., Malatesta, M., Torres, C. M., Rekling, J. C., et al. (2013). The histone demethylase jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet. 9:e1003461. doi: 10.1371/journal.pgen.1003461
Armstrong, J. A., Bieker, J. J., and Emerson, B. M. (1998). A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95, 93–104. doi: 10.1016/s0092-8674(00)81785-7
Bakshi, R., Hassan, M. Q., Pratap, J., Lian, J. B., Montecino, M. A., van Wijnen, A. J., et al. (2010). The human SWI/SNF complex associates with RUNX1 to control transcription of hematopoietic target genes. J. Cell. Physiol. 225, 569–576. doi: 10.1002/jcp.22240
Bannister, A. J., and Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381–395. doi: 10.1038/cr.2011.22
Becker, P. B., and Workman, J. L. (2013). Nucleosome remodeling and epigenetics. Cold Spring Harb. Perspect. Biol. 5:a017905. doi: 10.1101/cshperspect.a017905
Bogdanović, O., and Lister, R. (2017). DNA methylation and the preservation of cell identity. Curr. Opin. Genet. Dev. 46, 9–14. doi: 10.1016/j.gde.2017.06.007
Bustos, F., Sepúlveda, H., Prieto, C. P., Carrasco, M., Díaz, L., Palma, J., et al. (2017). Runt-related transcription factor 2 induction during differentiation of wharton’s jelly mesenchymal stem cells to osteoblasts is regulated by jumonji at-rich interactive domain 1b histone demethylase. Stem Cells 35, 2430–2441. doi: 10.1002/stem.2704
Cakouros, D., and Gronthos, S. (2020). Epigenetic regulators of mesenchymal stem/stromal cell lineage determination. Curr. Osteoporos. Rep. 18, 597–605. doi: 10.1007/s11914-020-00616-0
Cakouros, D., Hemming, S., Gronthos, K., Liu, R., Zannettino, A., Shi, S., et al. (2019). Specific functions of TET1 and TET2 in regulating mesenchymal cell lineage determination. Epigenet. Chromatin. 12:3. doi: 10.1186/s13072-018-0247-4
Cheng, J., Blum, R., Bowman, C., Hu, D., Shilatifard, A., Shen, S., et al. (2014). A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol. Cell. 53, 979–992. doi: 10.1016/j.molcel.2014.02.032
Cho, H. H., Park, H. T., Kim, Y. J., Bae, Y. C., Suh, K. T., and Jung, J. S. (2005). Induction of osteogenic differentiation of human mesenchymal stem cells by histone deacetylase inhibitors. J. Cell. Biochem. 96, 533–542. doi: 10.1002/jcb.20544
Christensen, J., Agger, K., Cloos, P. A. C., Pasini, D., Rose, S., Sennels, L., et al. (2007). RBP2 belongs to a family of demethylases, specific for Tri-and dimethylated lysine 4 on histone 3. Cell 128, 1063–1076. doi: 10.1016/j.cell.2007.02.003
Cruzat, F., Henriquez, B., Villagra, A., Hepp, M., Lian, J. B., van Wijnen, A. J., et al. (2009). SWI/SNF-independent nuclease hypersensitivity and an increased level of histone acetylation at the P1 promoter accompany active transcription of the bone master gene Runx2. Biochemistry 48, 7287–7295. doi: 10.1021/bi9004792
Denissov, S., Hofemeister, H., Marks, H., Kranz, A., Ciotta, G., Singh, S., et al. (2014). Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development 141, 526–537. doi: 10.1242/dev.102681
Dimitrova, E., Turberfield, A. H., and Klose, R. J. (2015). Histone demethylases in chromatin biology and beyond. EMBO Rep. 16, 1620–1639. doi: 10.15252/embr.201541113
Drissi, H., Luc, Q., Shakoori, R., Lopes, S. C. D. S., Choi, J. Y., Terry, A., et al. (2000). Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J. Cell. Physiol. 184, 341–350. doi: 10.1002/1097-4652(200009)184:3<341::aid-jcp8>3.0.co;2-z
Dudakovic, A., Camilleri, E. T., Xu, F., Riester, S. M., McGee-Lawrence, M. E., Bradley, E. W., et al. (2015). Epigenetic control of skeletal development by the histone methyltransferase Ezh2. J. Biol. Chem. 290, 27604–27617. doi: 10.1074/jbc.m115.672345
Dudakovic, A., Evans, J. M., Li, Y., Middha, S., McGee-Lawrence, M. E., van Wijnen, A. J., et al. (2013). Histone deacetylase inhibition promotes osteoblast maturation by altering the histone 4 (H4) epigenome and reduces Akt phosphorylation. J. Biol. Chem. 288, 28783–28791. doi: 10.1074/jbc.m113.489732
Franchini, D.-M., Schmitz, K.-M., and Petersen-Mahrt, S. K. (2012). 5-Methylcytosine DNA demethylation: more than losing a methyl group. Annu. Rev. Genet. 46, 419–441. doi: 10.1146/annurev-genet-110711-155451
Gordon, J. A. R., Hassan, M. Q., Koss, M., Montecino, M., Selleri, L., van Wijnen, A. J., et al. (2011). Epigenetic regulation of early osteogenesis and mineralized tissue formation by a HOXA10-PBX1-associated complex. Cells Tissues Organs 194, 146–150. doi: 10.1159/000324790
Gori, F., Divieti, P., and Demay, M. B. (2001). Cloning and characterization of a novel WD-40 repeat protein that dramatically accelerates osteoblastic differentiation. J. Biol. Chem. 276, 46515–46522. doi: 10.1074/jbc.m105757200
Gori, F., Friedman, L. G., and Demay, M. B. (2006). Wdr5, a WD-40 protein, regulates osteoblast differentiation during embryonic bone development. Dev. Biol. 295, 498–506. doi: 10.1016/j.ydbio.2006.02.031
Haberland, M., Carrer, M., Mokalled, M. H., Montgomery, R. L., and Olson, E. N. (2010). Redundant control of adipogenesis by histone deacetylases 1 and 2. J. Biol. Chem. 285, 14663–14670. doi: 10.1074/jbc.m109.081679
Hassan, M. Q., Tare, R., Lee, S. H., Mandeville, M., Weiner, B., Montecino, M., et al. (2007). HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol. Cell. Biol. 27, 3337–3352. doi: 10.1128/mcb.01544-06
Hemming, S., Cakouros, D., Isenmann, S., Cooper, L., Menicanin, D., Zannettino, A., et al. (2014). EZH2and KDM6AAct as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells 32, 802–815. doi: 10.1002/stem.1573
Hemming, S. E., Cakouros, D., and Vandyke, K. (2016). Identification of novel EZH2 targets regulating osteogenic differentiation in mesenchymal stem cells. Stem Cells Dev. 25, 909–921. doi: 10.1089/scd.2015.0384
Hong, S., Cho, Y.-W., Yu, L.-R., Yu, H., Veenstra, T. D., and Ge, K. (2007). Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. U.S.A. 104, 18439–18444. doi: 10.1073/pnas.0707292104
Horn, P. J., and Peterson, C. L. (2001). The bromodomain: a regulator of ATP-dependent chromatin remodeling? Front. Biosci. 6, D1019–D1023. doi: 10.2741/horn
Hu, D., Gao, X., Morgan, M. A., Herz, H.-M., Smith, E. R., and Shilatifard, A. (2013). The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell. Biol. 33, 4745–4754. doi: 10.1128/mcb.01181-13
Hu, X., Zhang, X., Dai, L., Zhu, J., Jia, Z., Wang, W., et al. (2012). Histone deacetylase inhibitor trichostatin A promotes the osteogenic differentiation of rat adipose-derived stem cells by altering the epigenetic modifications on Runx2 promoter in a BMP signaling-dependent manner. Stem Cells Dev. 22, 248–255. doi: 10.1089/scd.2012.0105
Ito, S., D’Alessio, A. C., Taranova, O. V., Hong, K., Sowers, L. C., and Zhang, Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133. doi: 10.1038/nature09303
Iwase, S., Lan, F., Bayliss, P., de la Torre-Ubieta, L., Huarte, M., Qi, H. H., et al. (2007). The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128, 1077–1088. doi: 10.1016/j.cell.2007.02.017
Jambhekar, A., Dhall, A., and Shi, Y. (2019). Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell. Biol. 20, 625–641. doi: 10.1038/s41580-019-0151-1
Janssen, A., Colmenares, S. U., and Karpen, G. H. (2018). Heterochromatin: guardian of the genome. Annu. Rev. Cell. Dev. Biol. 34, 265–288. doi: 10.1146/annurev-cellbio-100617-062653
Kanno, T., Kanno, Y., Siegel, R. M., Jang, M. K., Lenardo, M. J., and Ozato, K. (2004). Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell. 13, 33–43. doi: 10.1016/s1097-2765(03)00482-9
Killaars, A. R., Grim, J. C., Walker, C. J., Hushka, E. A., Brown, T. E., and Anseth, K. S. (2019). Extended exposure to stiff microenvironments leads to persistent chromatin remodeling in human mesenchymal stem cells. Adv. Sci. 6:1801483. doi: 10.1002/advs.201801483
Kooistra, S. M., and Helin, K. (2012). Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 13, 297–311. doi: 10.1038/nrm3327
Lan, F., Bayliss, P. E., Rinn, J. L., Whetstine, J. R., Wang, J. K., Chen, S., et al. (2007). A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449, 689–694. doi: 10.1038/nature06192
Längst, G., and Manelyte, L. (2015). Chromatin remodelers: from function to dysfunction. Genes 6, 299–324. doi: 10.3390/genes6020299
Lawson, K. A., Teteak, C. J., Gao, J., Li, N., Hacquebord, J., Ghatan, A., et al. (2013). ESET histone methyltransferase regulates osteoblastic differentiation of mesenchymal stem cells during postnatal bone development. FEBS Lett. 587, 3961–3967. doi: 10.1016/j.febslet.2013.10.028
Lee, H. W., Suh, J. H., Kim, A. Y., Lee, Y. S., Park, S. Y., and Kim, J. B. (2006). Histone deacetylase 1-mediated histone modification regulates osteoblast differentiation. Mol. Endocrinol. 20, 2432–2443. doi: 10.1210/me.2006-0061
Lee, J. Y., Lee, Y. M., Kim, M. J., Choi, J.-Y., Park, E. K., Kim, S.-Y., et al. (2006). Methylation of the mouse DIx5 and Osx gene promoters regulates cell type-specific gene expression. Mol. Cells 22, 182–188.
Li, J., Tsuji, K., Komori, T., Miyazono, K., Wrana, J. L., Ito, Y., et al. (1998). Smad2 overexpression enhances Smad4 gene expression and suppresses CBFA1 gene expression in osteoblastic osteosarcoma ROS17/2.8 cells and primary rat calvaria cells. J. Biol. Chem. 273, 31009–31015. doi: 10.1074/jbc.273.47.31009
Liu, N., Balliano, A., and Hayes, J. J. (2011). Mechanism(s) of SWI/SNF-induced nucleosome mobilization. Chem. Biol. Chem. 12, 196–204. doi: 10.1002/cbic.201000455
Local, A., Huang, H., Albuquerque, C. P., Singh, N., Lee, A. Y., Wang, W., et al. (2018). Identification of H3K4me1-associated proteins at mammalian enhancers. Nat. Genet. 50, 73–82. doi: 10.1038/s41588-017-0015-6
Long, F. (2012). Building strong bones: molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell. Biol. 13, 27–38. doi: 10.1038/nrm3254
Maroni, P., Brini, A. T., Arrigoni, E., de Girolamo, L., Niada, S., Matteucci, E., et al. (2012). Chemical and genetic blockade of HDACs enhances osteogenic differentiation of human adipose tissue-derived stem cells by oppositely affecting osteogenic and adipogenic transcription factors. Biochem. Biophys. Res. Comm. 428, 271–277. doi: 10.1016/j.bbrc.2012.10.044
Martens, J. A., and Winston, F. (2003). Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opn. Genets Dev. 13, 136–142. doi: 10.1016/s0959-437x(03)00022-4
McGee-Lawrence, M. E., and Westendorf, J. J. (2011). Histone deacetylases in skeletal development and bone mass maintenance. Gene 474, 1–11. doi: 10.1016/j.gene.2010.12.003
Meyer, M. B., Benkusky, N. A., and Pike, J. W. (2014). The RUNX2 cistrome in osteoblasts: characterization, down-regulation following differentiation, and relationship to gene expression. J. Biol. Chem. 289, 16016–16031. doi: 10.1074/jbc.m114.552216
Neri, F., Incarnato, D., Krepelova, A., Rapelli, S., Pagnani, A., Zecchina, R., et al. (2013). Genome-wide analysis identifies a functional association of Tet1 and Polycomb repressive complex 2 in mouse embryonic stem cells. Genome Biol. 14:R91. doi: 10.1186/gb-2013-14-8-r91
Nicetto, D., Donahue, G., Jain, T., Peng, T., Sidoli, S., Sheng, L., et al. (2019). H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science 363:eaau0583. doi: 10.1126/science.aau0583
Nicetto, D., and Zaret, K. S. (2019). Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. Curr. Opin. Genet. Dev. 55, 1–10. doi: 10.1016/j.gde.2019.04.013
Nishimura, R., Kato, Y., Chen, D., Harris, S. E., Mundy, G. R., and Yoneda, T. (1998). Smad5 and DPC4 are key molecules in mediating BMP-2-induced osteoblastic differentiation of the pluripotent mesenchymal precursor cell line C2C12. J. Biol. Chem. 273, 1872–1879. doi: 10.1074/jbc.273.4.1872
Pal, S., Vishwanath, S. N., Erdjument-Bromage, H., Tempst, P., and Sif, S. (2004). Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 24, 9630–9645. doi: 10.1128/mcb.24.21.9630-9645.2004
Park-Min, K. H. (2016). Epigenetic regulation of bone cells. Connect. Tissue Res. 58, 1–15. doi: 10.1080/03008207.2016.1177037
Piunti, A., and Shilatifard, A. (2016). Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 352:aad9780. doi: 10.1126/science.aad9780
Ramakrishnan, V. (1997). Histone structure and the organization of the nucleosome. Annu. Rev. Biophys. Biomol. Struct. 26, 83–112. doi: 10.1146/annurev.biophys.26.1.83
Richmond, T. J., and Davey, C. A. (2003). The structure of DNA in the nucleosome core. Nature 423, 145–150. doi: 10.1038/nature01595
Ringrose, L., and Paro, R. (2007). Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134, 223–232. doi: 10.1242/dev.02723
Rojas, A., Aguilar, R., Henriquez, B., Lian, J. B., Stein, J. L., Stein, G. S., et al. (2015). Epigenetic control of the bone-master Runx2 gene during osteoblast-lineage commitment by the histone demethylase JARID1B/KDM5B. J. Biol. Chem. 290, 28329–28342. doi: 10.1074/jbc.m115.657825
Rojas, A., Sepulveda, H., Henriquez, B., Aguilar, R., Opazo, T., Nardocci, G., et al. (2019). Mll-COMPASS complexes mediate H3K4me3 enrichment and transcription of the osteoblast master gene Runx2/p57 in osteoblasts. J. Cell. Physiol. 234, 6244–6253. doi: 10.1002/jcp.27355
Ruijtenberg, S., and van den Heuvel, S. (2016). Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 15, 196–212. doi: 10.1080/15384101.2015.1120925
Saksouk, N., Simboeck, E., and Déjardin, J. (2015). Constitutive heterochromatin formation and transcription in mammals. Epigenet. Chromatin. 8:3. doi: 10.1186/1756-8935-8-3
Santa, F. D., Totaro, M. G., Prosperini, E., Notarbartolo, S., Testa, G., and Natoli, G. (2007). The histone H3 lysine-27 demethylase jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130, 1083–1094. doi: 10.1016/j.cell.2007.08.019
Schroeder, T. M., and Westendorf, J. J. (2005). Histone deacetylase inhibitors promote osteoblast maturation. J. Bone. Miner. Res. 20, 2254–2263. doi: 10.1359/jbmr.050813
Sen, B., Paradise, C. R., Xie, Z., Sankaran, J., Uzer, G., Styner, M., et al. (2020). β-catenin preserves the stem state of murine bone marrow stromal cells through activation of EZH2. J. Bone. Miner. Res. 35, 1149–1162. doi: 10.1002/jbmr.3975
Sepulveda, H., Aguilar, R., Prieto, C. P., Bustos, F., Aedo, S., Lattus, J., et al. (2017a). Epigenetic signatures at the RUNX2-P1 and Sp7 gene promoters control osteogenic lineage commitment of umbilical cord-derived mesenchymal stem cells. J. Cell. Physiol. 232, 2519–2527. doi: 10.1002/jcp.25627
Sepulveda, H., Villagra, A., and Montecino, M. (2017b). Tet-mediated DNA demethylation is required for SWI/SNF-dependent chromatin remodeling and histone-modifying activities that trigger expression of the Sp7 osteoblast master gene during mesenchymal lineage commitment. Mol. Cell. Biol. 37:e00177-17. doi: 10.1128/mcb.00177-17
Serna, I. L., de la Carlson, K. A., and Imbalzano, A. N. (2001). Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 27, 187–190. doi: 10.1038/84826
Shilatifard, A. (2012). The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81, 65–95. doi: 10.1146/annurev-biochem-051710-134100
Sinha, K. M., and Zhou, X. (2013). Genetic and molecular control of osterix in skeletal formation. J. Cell. Biochem. 114, 975–984. doi: 10.1002/jcb.24439
Srivastava, S., Mishra, R. K., and Dhawan, J. (2010). Regulation of cellular chromatin state: insights from quiescence and differentiation. Organogenesis 6, 37–47. doi: 10.4161/org.6.1.11337
Stein, G. S., Lian, J. B., van Wijnen, A. J., Stein, J. L., Montecino, M., Javed, A., et al. (2004). Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene 23, 4315–4329. doi: 10.1038/sj.onc.1207676
Strahl, B. D., and Allis, C. D. (2000). The language of covalent histone modifications. Nature 403, 41–45. doi: 10.1038/47412
Tahiliani, M., Koh, K. P., Shen, Y., Pastor, W. A., Bandukwala, H., Brudno, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. doi: 10.1126/science.1170116
Tai, P. W. L., Wu, H., Gordon, J. A. R., Whitfield, T. W., Barutcu, A. R., van Wijnen, A. J., et al. (2014). Epigenetic landscape during osteoblastogenesis defines a differentiation-dependent Runx2 promoter region. Gene 550, 1–9. doi: 10.1016/j.gene.2014.05.044
Tsukiyama, T. (2002). The in vivo functions of ATP-dependent chromatin-remodelling factors. Nat. Rev. Mol. Cell Biol. 3, 422–429. doi: 10.1038/nrm828
Venkatesh, S., and Workman, J. L. (2015). Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell. Biol. 16, 178–189. doi: 10.1038/nrm3941
Vermeulen, M., Mulder, K. W., Denissov, S., Pijnappel, W. W. M. P., van Schaik, F. M. A., Varier, R. A., et al. (2007). Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69. doi: 10.1016/j.cell.2007.08.016
Voigt, P., Tee, W.-W., and Reinberg, D. (2013). A double take on bivalent promoters. Genes Dev. 27, 1318–1338. doi: 10.1101/gad.219626.113
Wei, Y., Chen, Y.-H., Li, L.-Y., Lang, J., Yeh, S.-P., Shi, B., et al. (2010). CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat. Cell Biol. 13, 87–94. doi: 10.1038/ncb2139
Williams, K., Christensen, J., Pedersen, M. T., Johansen, J. V., Cloos, P. A. C., Rappsilber, J., et al. (2011). TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348. doi: 10.1038/nature10066
Wu, H., Whitfield, T. W., Gordon, J. A. R., Dobson, J. R., Tai, P. W. L., van Wijnen, A. J., et al. (2014). Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome Biol. 15:R52. doi: 10.1186/gb-2014-15-3-r52
Xiang, Y., Zhu, Z., Han, G., Lin, H., Xu, L., and Chen, C. D. (2007). JMJD3 is a histone H3K27 demethylase. Cell Res. 17, 850–857. doi: 10.1038/cr.2007.83
Yamaguchi, A., Komori, T., and Suda, T. (2000). Regulation of osteoblast differentiation mediated by bone morphogenetic proteins, hedgehogs, and Cbfa1. Endocr. Rev. 21, 393–411. doi: 10.1210/edrv.21.4.0403
Yamane, K., Tateishi, K., Klose, R. J., Fang, J., Fabrizio, L. A., Erdjument-Bromage, H., et al. (2007). PLU-1 Is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell. 25, 801–812. doi: 10.1016/j.molcel.2007.03.001
Yan, J., Chen, S.-A. A., Local, A., Liu, T., Qiu, Y., Dorighi, K. M., et al. (2018). Histone H3 lysine 4 monomethylation modulates long-range chromatin interactions at enhancers. Cell Res. 28, 204–220. doi: 10.1038/cr.2018.1
Yang, D., Okamura, H., Nakashima, Y., and Haneji, T. (2013). Histone demethylase Jmjd3 regulates osteoblast differentiation via transcription factors Runx2 and osterix. J. Biol. Chem. 288, 33530–33541. doi: 10.1074/jbc.m113.497040
Yang, D., Okamura, H., Teramachi, J., and Haneji, T. (2015). Histone demethylase Utx regulates differentiation and mineralization in osteoblasts. J. Cell. Biochem. 116, 2628–2636. doi: 10.1002/jcb.25210
Ye, L., Fan, Z., Yu, B., Chang, J., Hezaimi, K. A., Zhou, X., et al. (2012). Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 11, 50–61. doi: 10.1016/j.stem.2012.04.009
Ye, L., Fan, Z., Yu, B., Chang, J., Hezaimi, K. A., Zhou, X., et al. (2018). Histone demethylases KDM4B and KDM6B promote osteogenic differentiation of human MSCs. Cell Stem Cell 23, 898–899. doi: 10.1016/j.stem.2018.11.002
Yildirim, O., Li, R., Hung, J.-H., Chen, P. B., Dong, X., Ee, L.-S., et al. (2011). Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147, 1498–1510. doi: 10.1016/j.cell.2011.11.054
Yoshida, C. A., Komori, H., Maruyama, Z., Miyazaki, T., Kawasaki, K., Furuichi, T., et al. (2012). SP7 inhibits osteoblast differentiation at a late stage in mice. PLoS One 7:e32364. doi: 10.1371/journal.pone.0032364
Yu, J.-R., Lee, C.-H., Oksuz, O., Stafford, J. M., and Reinberg, D. (2019). PRC2 is high maintenance. Genes Dev. 33, 903–935. doi: 10.1101/gad.325050.119
Zhang, C. (2010). Transcriptional regulation of bone formation by the osteoblast-specific transcription factor Osx. J. Orthop. Surg. Res. 5:37. doi: 10.1186/1749-799x-5-37
Zhang, Y., Hassan, M. Q., Xie, R. L., Hawse, J. R., Spelsberg, T. C., Montecino, M., et al. (2008). Co-stimulation of the bone-related Runx2 P1 promoter in mesenchymal cells by SP1 and ETS transcription factors at polymorphic purine-rich DNA sequences (Y-repeats). J. Biol. Chem. 284, 3125–3135. doi: 10.1074/jbc.m807466200
Zhang, Y.-X., Sun, H.-L., Liang, H., Li, K., Fan, Q.-M., and Zhao, Q.-H. (2015). Dynamic and distinct histone modifications of osteogenic genes during osteogenic differentiation. J. Biochem. 158, 445–457. doi: 10.1093/jb/mvv059
Zhou, C., Zou, J., Zou, S., and Li, X. (2016). INO80 is required for osteogenic differentiation of human mesenchymal stem cells. Sci. Rep. 6:35924. doi: 10.1038/srep35924
Zhou, G.-S., Zhang, X.-L., Wu, J.-P., Zhang, R.-P., Xiang, L.-X., Dai, L.-C., et al. (2009). 5-Azacytidine facilitates osteogenic gene expression and differentiation of mesenchymal stem cells by alteration in DNA methylation. Cytotechnology 60, 11–22. doi: 10.1007/s10616-009-9203-2
Zhu, E. D., Demay, M. B., and Gori, F. (2008). Wdr5 is essential for osteoblast differentiation. J. Biol. Chem. 283, 7361–7367. doi: 10.1074/jbc.m703304200
Zych, J., Stimamiglio, M. A., Senegaglia, A. C., Brofman, P. R. S., Dallagiovanna, B., Goldenberg, S., et al. (2013). The epigenetic modifiers 5-aza-2’-deoxycytidine and trichostatin A influence adipocyte differentiation in human mesenchymal stem cells. Braz. J. Med. Biol. Res. 46, 405–416. doi: 10.1590/1414-431x20132893
Keywords: epigenetic control, bone-related expression, osteoblast differentiation, histone marks, chromatin
Citation: Montecino M, Carrasco ME and Nardocci G (2021) Epigenetic Control of Osteogenic Lineage Commitment. Front. Cell Dev. Biol. 8:611197. doi: 10.3389/fcell.2020.611197
Received: 28 September 2020; Accepted: 11 December 2020;
Published: 08 January 2021.
Edited by:
Monica De Mattei, University of Ferrara, ItalyReviewed by:
Quamarul Hassan, University of Alabama at Birmingham, United StatesLiliana Burlibasa, University of Bucharest, Romania
Copyright © 2021 Montecino, Carrasco and Nardocci. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Montecino, bW1vbnRlY2lub0B1bmFiLmNs
 Martin Montecino
Martin Montecino Margarita E. Carrasco
Margarita E. Carrasco Gino Nardocci
Gino Nardocci