- 1Jiangsu Key Laboratory of Neuropsychiatric Diseases, Jiangsu Key Laboratory of Preventive and Translational Medicine for Geriatric Diseases, College of Pharmaceutical Sciences, Soochow University, Suzhou, China
- 2Department of Medicine, Weill Cornell Medicine, New York, NY, United States
- 3Department of Pharmacology, Weill Cornell Medicine, New York, NY, United States
- 4Department of Pathology and Laboratory Medicine, Weill Cornell Medicine, New York, NY, United States
Cereblon (CRBN), a substrate receptor of cullin 4-RING E3 ligase (CRL4), mediates the ubiquitination and degradation of constitutive substrates and immunomodulatory drug-induced neo-substrates including MEIS2, c-Jun, CLC1, IKZF1/3, CK1α, and SALL4. It has been reported that CRBN itself could be degraded through the ubiquitin-proteasome system by its associated or other cullin-RING E3 ligases, thus influencing its biological functions. However, it is unknown whether the CRBN stability and its biological function could be modulated by caspases. In this study, using model cell lines, we found that activation of the death receptor using tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) leads to the decreased CRBN protein level. Through pharmacological inhibition and activation of caspase-8 (CASP-8), we disclosed that CASP-8 regulates CRBN cleavage in cell lines. Site mapping experiments revealed that CRBN is cleaved after Asp9 upon CASP-8 activation, resulting in the reduced stability. Using myeloma as a model system, we further revealed that either inhibition or genetic depletion of CASP-8 enhances the anti-myeloma activity of lenalidomide (Len) by impairing CRBN cleavage, leading to the attenuated IKZF1 and IKZF3 protein levels and the reduced viability of myeloma cell lines and primary myeloma cells from patients. The present study discovered that the stability of the substrate receptor of an E3 ligase can be modulated by CASP-8 and suggested that administration of CASP-8 inhibitors enhances the overall effectiveness of Len-based combination therapy in myeloma.
Introduction
Cereblon (CRBN) interacts with damage-specific DNA-binding protein 1 (DDB1) and thus forms a cullin 4-RING E3 ligase (CRL4CRBN) with cullin 4A/B and RING-box protein ROC1 (Angers et al., 2006; Jackson and Xiong, 2009; Ito et al., 2010; Xu et al., 2013). Therefore, CRBN functions as a substrate receptor and recruits proteins for ubiquitination and their subsequent proteasomal degradation. It has been discovered that the primary target of immunomodulatory drug thalidomide (Thal) is CRBN (Ito et al., 2010). Thal and its structural analogs lenalidomide (Len) and pomalidomide (Pom) bind to CRBN and thus recruit new substrates that would otherwise not bind to CRBN. These substrates, termed as “neo-substrates,” are ubiquitinated by the CRL4CRBN E3 ligase leading to their degradation. Two of the most studied neo-substrates of this E3 ligase are transcription factors IKZF1 (Ikaros) and IKZF3 (Aiolos). Their degradation suppresses the proliferation of myeloma cells (Krönke et al., 2014; Lu et al., 2014). This is regarded as the major mechanism by which Len is used to treat myeloma patients.
Low CRBN expression is associated with the Len resistance of myeloma cells, suggesting that high CRBN protein level is required for the anti-myeloma activity of IMiDs (Zhu et al., 2011). After 2–6 months of Len treatment, drug resistance frequently develops as a result of down-regulation of CRBN mRNA and protein levels (Lopez-Girona et al., 2012; Gandhi et al., 2014), which also indicates that CRBN protein levels regulate the sensitivity of myeloma cells to IMiDs. CRBN is targeted for ubiquitination-mediated degradation by SCFFbxo7 ubiquitin ligase (Song et al., 2018). CSN9 signalosome inhibits SCFFbxo7-mediated CRBN degradation, thereby promoting the sensitivity of myeloma cells to IMiDs (Sievers et al., 2018; Liu et al., 2019). Several caspases are activated when myeloma cells are treated with proteasomal inhibitor bortezomib (Btz) (Hideshima et al., 2003). However, it is largely unknown whether CRBN stability and its functions are affected by caspase activity.
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is an inducer of apoptosis through binding to death receptor 4/5 (Gazitt, 1999), which results in the cleavage and activation of caspase-8 (CASP-8), a caspase in the extrinsic apoptotic pathway (Galluzzi et al., 2018). Because activated CASP-8 could cleave BID (Li et al., 1998; Luo et al., 1998), BID cleavage or reduction could serve as an indicator to demonstrate the activation of death receptor and CASP-8 (Chou et al., 1999). In our previous study, we found that CRBN inhibits the etoposide-induced intrinsic apoptosis (Zhou and Xu, 2019). However, it is unknown whether CRBN is involved in the death receptor-induced extrinsic apoptotic pathway and whether modulation of caspase activity could regulate the biological function of CRBN.
In this work, we examined CRBN stability in cervical cancer cell line HeLa and small cell lung cancer cell line NCI-H1688 upon the activation of the death receptor by TRAIL. Surprisingly, we discovered that TRAIL down-regulates CRBN protein level. The combination treatment of HeLa and NCI-H1688 cells with TRAIL and Btz results in the observation of the cleaved CRBN. This CRBN cleavage could be blocked by CASP-8 inhibition. Interestingly, CASP-8 activation by TRAIL and Btz also leads to CRBN cleavage in myeloma cells. Using myeloma as a model system, we further demonstrated that blockage of the CRBN cleavage by pharmacological inhibition or genetic depletion of CASP-8 potentiates the anti-myeloma activity of Len in both myeloma cell lines and bone marrow primary myeloma cells. Database analysis showed that CASP-8 mRNA expression is inversely correlated with the overall survival rate of myeloma patients. Therefore, this work reveals a novel molecular mechanism by which the CRBN cleavage and stability is modulated. Using this discovery, we further disclosed that the anti-myeloma activity of IMiDs can be augmented by inhibiting the CASP-8 activation and suggests a potential new combination therapy that might benefit myeloma patients.
Materials and Methods
Materials
Bortezomib (S1013), CASP-3 inhibitor z-DEVD-fmk (S7312), CASP-8 inhibitor z-IETD-fmk (S7314), Len (CC-5013), MG132 (S8410), MLN4924 (S7109), and Pom (S1567) were purchased from Selleck; TRAIL (abs04233) was obtained from Absin; pan-caspase inhibitor z-VAD-fmk (C1202) was ordered from Beyotime Biotechnology; and cycloheximide (CHX, C104450) was obtained from Sigma.
The antibodies used in this work were purchased from the following companies: anti-CASP-8 antibody (BA2143) was purchased from Boster Biological Technology; anti-ubiquitin (Ub, sc-8017) and anti-HA (sc-7392) antibodies were from Santa Cruz Biotechnology; anti-Flag (0912-1) and anti-GST (ET1611-47) antibodies were from HuaAn Biotechnology; anti-PARP1 (9532S), anti-CRBN (71810S), and anti-cleaved CASP-8 (9496T) antibodies were from Cell Signaling Technology; anti-GAPDH (60004-1-Ig) and anti-IKZF3 (13561-1-AP) antibodies were from ProteinTech Group; anti-BID (CPA4351) antibody was from Cohesion Biosciences; and anti-IKZF1 (YM1278) antibody was from Immunoway. Mouse anti-CRBN antibody (Xu et al., 2016) was a kind gift from Dr. Xiu-Bao Chang (Mayo Clinic College of Medicine, United States). Secondary antibodies (sheep anti-mouse IgG-HRP and anti-rabbit IgG-HRP) were from Thermo Fisher.
shRNA and CRBN Plasmids
To make CASP-8 shRNA (shCASP-8), CASP-8 forward oligonucleotide (5′-CCGGCACCAGGCAGGGCTCAAATTTCTGC AGAAATTTGAGCCCTGCCTGGTGTTTTTG-3′) and CASP-8 reverse complementary oligonucleotide (5′-AATTCAAAAACA CCAGGCAGGGCTCAAATTTCTGCAGAAATTTGAGCCCT GCCTGGTG-3′) were annealed and ligated to the pLKO.1 TRC cloning vector (a gift from David Root, Addgene plasmid #10878) using a published procedure (Moffat et al., 2006). A digestion with PstI and BamHI was performed to identify the positive clone, which was further validated by Sanger sequencing.
shNC or shCRBN lentiviruses were purchased from GeneChem (Shanghai, China). The target sequence of shNC was TTCTCCGAACGTGTCACGT, and the target sequence of shCRBN was CCCAGACACTGAAGATGAAAT.
Plasmids for CRBN-Flag, D-to-A, and Del9 mutants were subcloned or constructed using standard point mutagenesis.
Generation of Stable Knockdown Cells
The shLacZ (The RNAi Consortium), shCRBN, and shCASP-8 lentiviruses were produced as described in a previous publication (Moffat et al., 2006). Myeloma cells MM1.S and CAG were infected with lentiviruses and selected with puromycin (1 μg/ml) for 2 weeks to generate stable knockdown cells.
Cell Culture
Cervical cancer cell line HeLa, human embryonic kidney cell line HEK293T, multiple myeloma cell lines MM1.S and RPMI8226, and small cell lung cancer cell line NCI-H1688 were obtained from American Type Culture Collection (ATCC). Multiple myeloma CAG cells (Borset et al., 2000) were a kind gift from Dr. Joshua Epstein (University of Arkansas for Medical Sciences, Little Rock, AK, United States). MM1.S, RPMI8226, CAG, and NCI-H1688 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium. HeLa and HEK293T cells were cultured in Dulbecco′s Modified Eagle’s Medium (DMEM; HyClone). Growth medium was supplemented with 10% fetal bovine serum (FBS; Gibco and Lonsera), 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco).
Bone marrow specimens were obtained from deidentified multiple myeloma patients at the Weill Cornell Medicine under informed consent as part of an Institutional Review Board approved study. CD138+ primary myeloma cells were isolated from bone marrow and co-cultured with a layer of HS-5 cells and cytokines as previously described (Huang et al., 2012).
Western Blotting Analysis
Cell lysates or immunoprecipitates were analyzed by Western blotting according to a previously described method (Hou et al., 2015) using NcmECL Ultra substrate (NCM Biotech) for visualization (Tao et al., 2018).
Affinity Purification
The HEK293T cell lysates were incubated with the anti-Flag affinity gel (Sigma) at 4°C for 3–4 h. The gel was then washed three times with TBST (TBS with 0.1% Tween 20). The Flag-tagged proteins were eluted with the 2× sample loading buffer for Western blotting analysis.
CASP-8 Activity Assay
RPMI8226 cells were first treated with DMSO or 40 μM CASP-8 inhibitor z-IETD-fmk for 30 min and then treated with DMSO or 10 μM Len for another 24 h. CASP-8 activity was measured using a CASP-8 fluorometric assay (Beyotime Biotechnology) according to the manufacturer’s instruction.
Flow Cytometry Analysis
Myeloma cells expressing shLacZ or shCASP-8 were cultured and treated with DMSO or Len (10 μM) for 4 days. Cells were stained with ToPro-3 (Life Technologies, United States) and analyzed in a BD flow cytometry according to a previously used method (Liu et al., 2015). The data were processed with FlowJo.
Cell Viability Measurement
Cells were treated with DMSO or the indicated compounds and stained with trypan blue (Beyotime Biotechnology) or analyzed with cell counting kit-8 assay (CCK-8, Beyotime Biotechnology). Live cells were counted under the microscope, and optical density at 460 nm was measured. The percentage of live cells was calculated.
Analysis of CASP-8 mRNA and Patient Overall Survival Rate
Kaplan–Meier plot of overall survival rate in patients with low (<20 FPKM, Fragments Per Kilobase of exon model per Million mapped fragments) and high (>20 FPKM) CASP-8 mRNA expression levels in the CoMMpass trial (IA14) of single Len or combination of Btz, Len, and dexamethasone (Dex) was generated, and statistical analysis for the pairwise comparison was performed using log-rank test integrated with the tool available at Multiple Myeloma Research Foundation (MMRF) Researcher Gateway1.
Results
CRBN Is Decreased in HeLa and NCI-H1688 Cells Upon TRAIL Treatment
Recently, it has been discovered that CRBN inhibits DNA damage-induced apoptosis (Zhou and Xu, 2019). However, it is unknown whether and how death receptors regulate CRBN and its function. To explore this possibility, we treated HeLa cells with TRAIL to activate the death receptor, which was confirmed by immunoblotting for a death receptor-related biomarker BID (Figures 1A,B). We further discovered that TRAIL clearly led to the down-regulation of CRBN (Figures 1A,B). Similar results were also observed in NCI-H1688 cells (Figures 1C,D), indicating that CRBN reduction upon the activation of death receptor may be a general phenomenon.
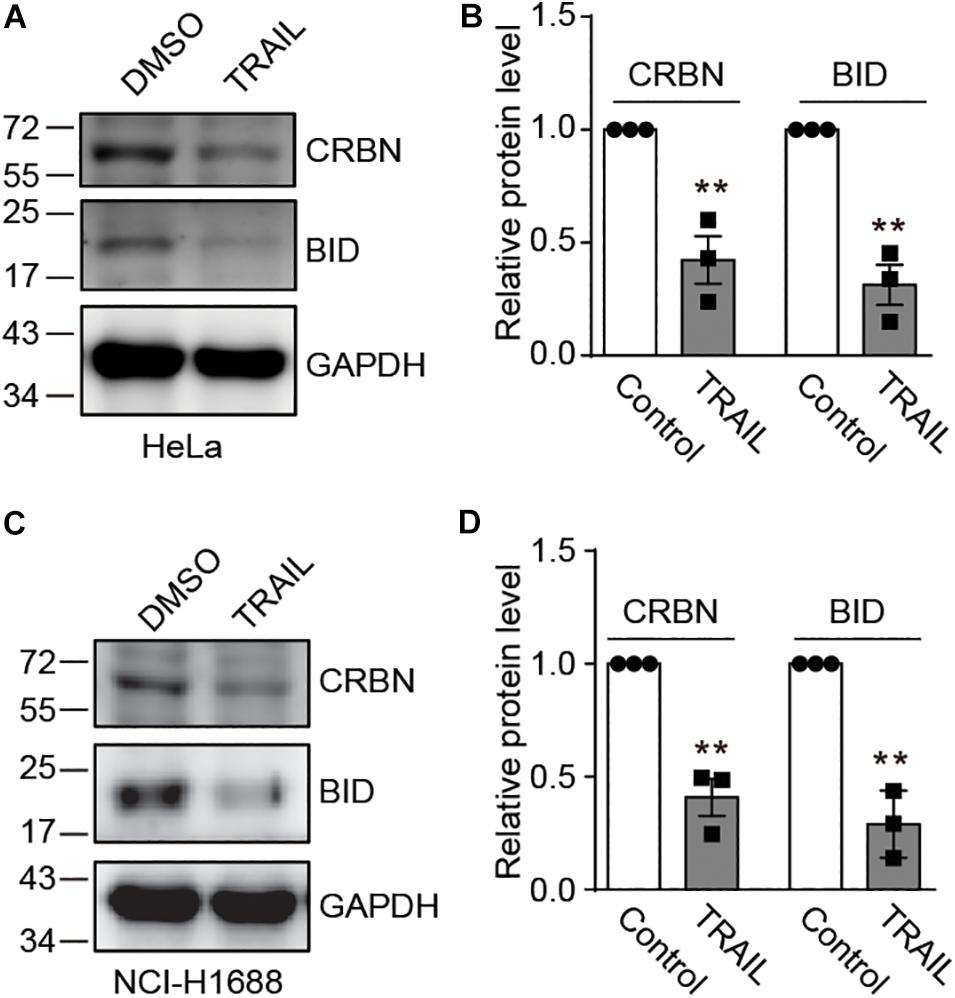
Figure 1. TRAIL decreases cereblon (CRBN) protein level in HeLa and NCI-H1688 cell lines. (A–D) HeLa (A,B) and NCI-H1688 (C,D) cells were treated with vehicle or TRAIL (100 ng/ml) for 24 h, and the resulting cell lysates were immunoblotted with the indicated antibodies. Quantification (mean ± SEMs) was performed for cells from three biological replicates. SEM, standard error of measurements; Student’s t-test, **P < 0.01.
CRBN Is Cleaved Upon TRAIL and Btz Co-treatment
We next sought to investigate the possible molecular mechanisms underlying TRAIL-induced down-regulation of CRBN. Since CRBN undergoes proteasomal degradation, we used the proteasome inhibitors Btz and MG132 to treat HeLa and NCI-H1688 cells in the presence of TRAIL (Figures 2A–D). Surprisingly, immunoblotting of CRBN showed that the band for the full length CRBN disappeared, whereas a new band appeared at about 1–5 kDa below the full length CRBN. This result indicates that CRBN is cleaved and the cleaved fragment is stable in HeLa and NCI-H1688 cells after TRAIL-Btz and TRAIL-MG132 treatment (Figures 2A–D). Similar results were found in MM1.S myeloma cells (Supplementary Figure 1). These data suggested that the cleaved CRBN is most probably degraded through the ubiquitin-proteasome system. The previous study demonstrated that combination use of TRAIL and Btz could dramatically activate CASP-8 and cause apoptosis in the lung cancer cell line through enhancing the surface expression of TRAIL receptor (Voortman et al., 2007). We then examine whether CASP-8 is responsible for the CRBN cleavage. Pharmacological inhibition experiments demonstrated that the CASP-8 inhibitor z-IETD-fmk but not the CASP-3 specific inhibitor z-DEVD-fmk and the NEDD8-activating enzyme inhibitor MLN4924 block synergistic TRAIL-Btz-induced CRBN cleavage in HeLa cells (Figure 2E and Supplementary Figure 2), indicating that CASP-8 is required for the cleavage of CRBN upon TRAIL-Btz treatment.
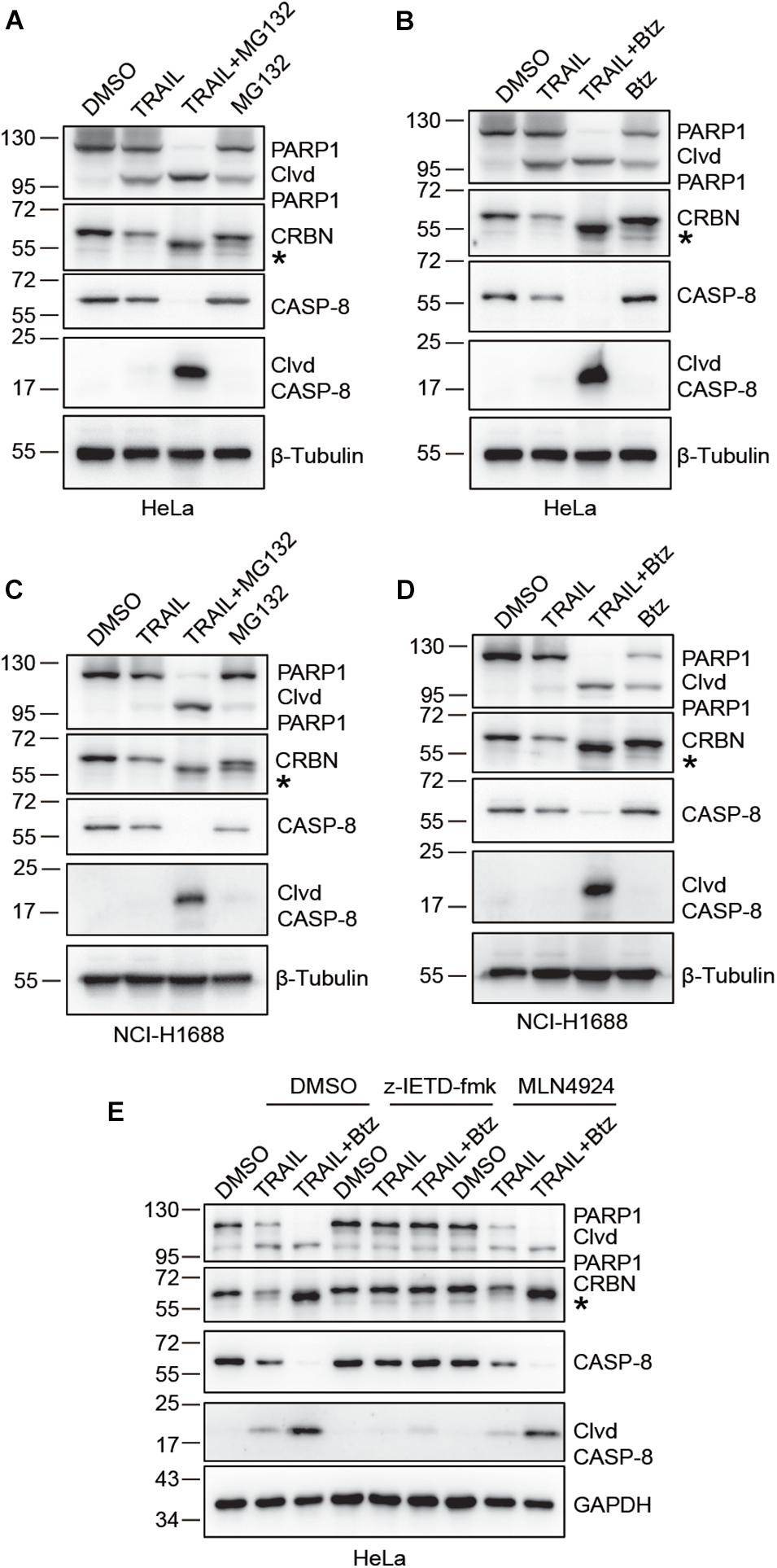
Figure 2. CRBN is cleaved upon co-treatment with TRAIL and proteasome inhibitors bortezomib (Btz) and MG132, and CASP-8 inhibition prevents the cleavage. (A–D) CRBN cleavage is detected when treated with TRAIL and MG132 or Btz. HeLa cells (A,B) or NCI-H1688 cells (C,D) were treated with DMSO, TRAIL (100 ng/ml), TRAIL (100 ng/ml) and MG132 (10 μM)/Btz (0.5 μM), or MG132/Btz for 24 h. (E) CASP-8 inhibitor (z-IETD-fmk) but not NEDD8-activating enzyme inhibitor (MLN4924) prevents CRBN cleavage. HeLa cells were pretreated with DMSO, z-IETD-fmk (40 μM), or MLN4924 (1 μM) for 30 min and then treated with DMSO, TRAIL (100 ng/ml), or TRAIL (100 ng/ml) and Btz (0.5 μM) for 24 h. After the treatment, cells were washed, harvested, and lysed, and the resulting cell lysates were subjected to immunoblotting analysis. Clvd, cleaved; *, cleaved CRBN.
CASP-8 Activation Cleaves CRBN at Asp9 and This Cleavage Reduces CRBN Stability
Next, we sought to determine the cleavage site in CRBN upon CASP-8 activation. To do so, we first constructed a CRBN-Flag plasmid and transfected this plasmid into HeLa cells, which were further treated with TRAIL and Btz. Immunoblotting of cell lysates with both anti-Flag and anti-CRBN antibodies resulted in two bands, the full length CRBN and the cleaved CRBN in HeLa cells upon CASP-8 activation (Figure 3A). This result indicates that the cleavage site on CRBN is located at its N-terminus because the Flag tag is fused to the CRBN C-terminus. Sequence alignment analysis of CRBN indicates that the Asp (D) residues at N-termini are highly conserved among human, mouse, rat, and western clawed frog (Figure 3B). To further determine the exact cleavage site, we mutated five Asp residues at the N-terminus to Ala, one at a time, and carried out the same experiment. Immunoblotting of cell lysates demonstrated that CASP-8 activation resulted in the cleavage of the WT, D29A, D35A, and D37A CRBN mutants but not the D6A and D9A mutants (Figure 3C). The sequence of the 6–10 amino acids in CRBN (DQQDA) is the CASP-8 preferred cleavage sequence (L/D/V)XXD(G/S/A) (Stennicke et al., 2000), and the D6A mutation disrupts this sequence. Therefore, these data demonstrate that CRBN is cleaved by CASP-8 after Asp9. This cleavage site was also detected previously by a quantitative N-terminomics (Shimbo et al., 2012).
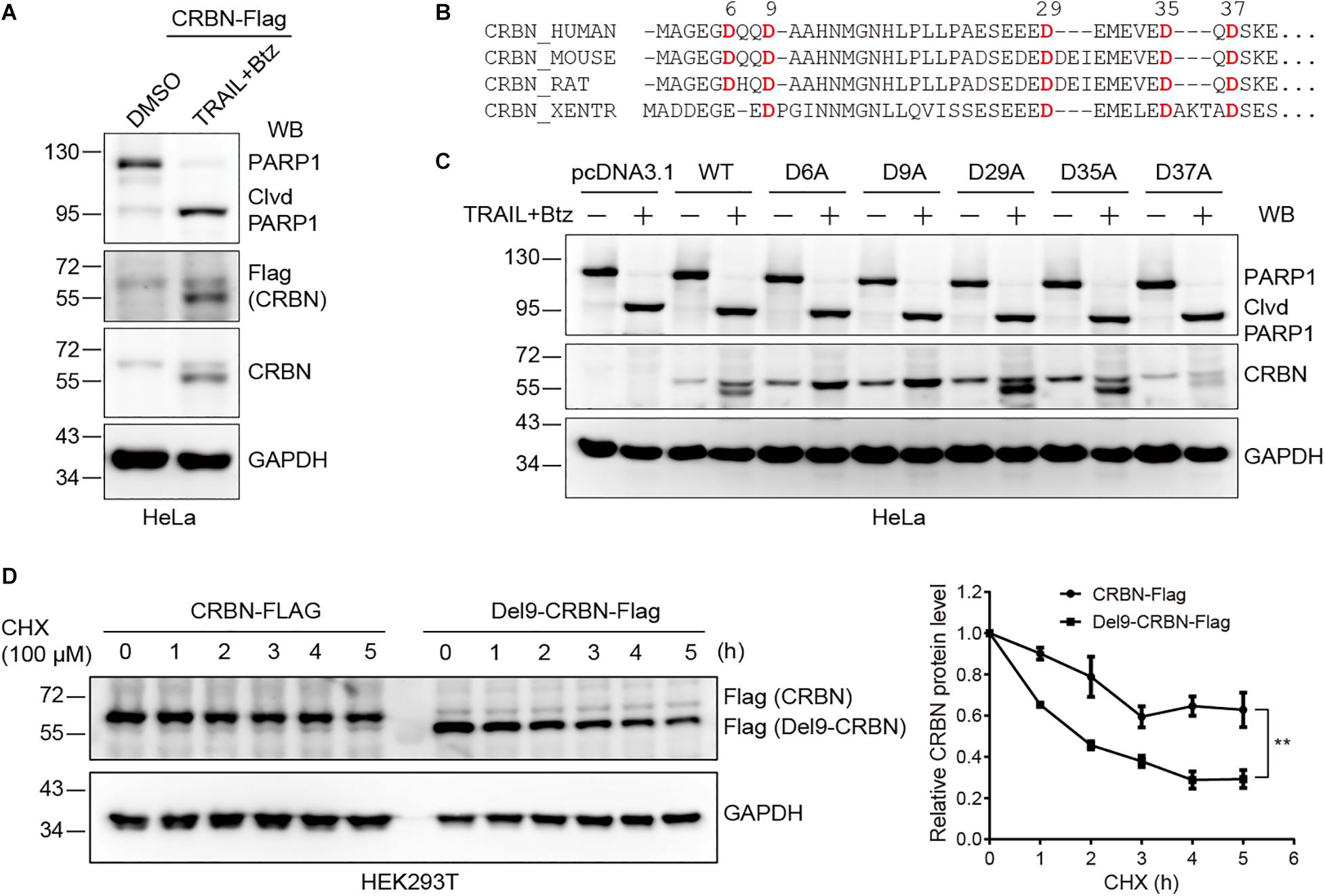
Figure 3. Cleavage of CRBN at Asp9 (D9) reduces its stability. (A) CRBN is cleaved at the N-terminus upon CASP-8 activation. HeLa cells were transfected with CRBN-Flag plasmid for 48 h and treated with DMSO or TRAIL (100 ng/ml) and Btz (0.5 μM) for 24 h. The resulting cell lysates were subjected to immunoblotting analysis. (B) Amino acid sequence alignment for CRBN from human, mouse, rat, and western clawed frog (Xentr). Conserved Asp (D) residues were indicated in red. (C) CRBN is cleaved at Asp9 (D9) upon CASP-8 activation. HeLa cells were transfected with the WT and CRBN Asp (D) to Ala (A) mutants for 48 h and then treated with DMSO or TRAIL (100 ng/ml) and Btz (0.5 μM) for 24 h. Cell lysates were used for immunoblotting. (D) Deletion of the N-terminal nine amino acids in CRBN reduces it stability. WT CRBN and Del9-CRBN mutants were expressed in HEK293T cells and split to 24-well plates. At 48 h post-transfection, cells were further treated with cycloheximide (CHX, 100 μM) for the indicated time. The cell lysates were immunoblotted with the indicated antibodies. Mean ± SEMs were from three independent biological replicates. Two-way ANOVA, **P < 0.01; Clvd, cleaved.
These results demonstrated that treatment of HeLa and NCI-H1688 cells with TRAIL and Btz led to CRBN cleavage. However, we only observed reduced CRBN levels but not the cleaved fragments in these cells upon the activation of CASP-8 by TRAIL, suggesting that the stability of CRBN might be reduced after cleavage. To test this hypothesis, we measured the stability of the WT and Del9 CRBN in HEK293T cells treated with a protein synthesis inhibitor CHX. Immunoblotting of cell lysates showed that Del9 CRBN was diminished at a much faster rate than the WT counterpart upon CHX treatment (Figure 3D), confirming its reduced stability. Inhibition of neddylation by MLN4924 significantly increased the WT and Del9 CRBN protein levels (Supplementary Figure 3), indicating that they are ubiquitinated by the cullin RING E3 ligases and subsequently degraded by the proteasome. This is in concert with the fact that the cleaved CRBN was observed in HeLa cells only in the presence of proteasome inhibitor (Figure 2). Furthermore, we found that TRAIL-induced apoptosis was not affected in the CRBN deficient HeLa cells (Supplementary Figure 4), suggesting that the cleavage of CRBN did not regulate CASP-8-dependent apoptosis.
CASP-8 Inhibition Enhances the Anti-myeloma Activity of Len in Cell Lines
After the discovery of CRBN cleavage by CASP-8, we thought to investigate how this cleavage affects its biological function. It has been reported that IMiDs can activate CASP-8 (Mitsiades et al., 2002; Chauhan and Anderson, 2003; Martiniani et al., 2012). Therefore, we thought to use myeloma cells as a model system to explore whether CASP-8 regulates CRBN levels after addition of Len. We found that CASP-8 activity is increased upon Len treatment in RPMI8226 cells (Figure 4A). Immunoblotting of CRBN showed that Len increases CRBN protein level at 24 h, which is consistent with previous studies (Liu et al., 2015). CASP-8 inhibitor z-IETD-fmk further increases CRBN protein levels (Figures 4B,C). These results suggest that the effect of Len and CASP-8 inhibitors on CRBN protein levels might be additive. Consequently, z-IETD-fmk further down-regulates the IKZF1 and IKZF3 protein levels mediated by Len (Figures 4B,C). To further investigate the effect of CRBN and CASP-8 on the IKZF1 and IKZF3 upon Len treatment, we obtained the stable shNC, shCRBN, and shCASP-8 knockdown MM1.S cell lines. We found the same results that CASP-8 inhibitor z-IETD-fmk could further down-regulate IKZF1 and IKZF3 upon Len treatment, which was mediated by CRBN (Figures 4D,E). Furthermore, the CRBN protein level was increased in the CASP-8 deficient cells (Figures 4D,E).
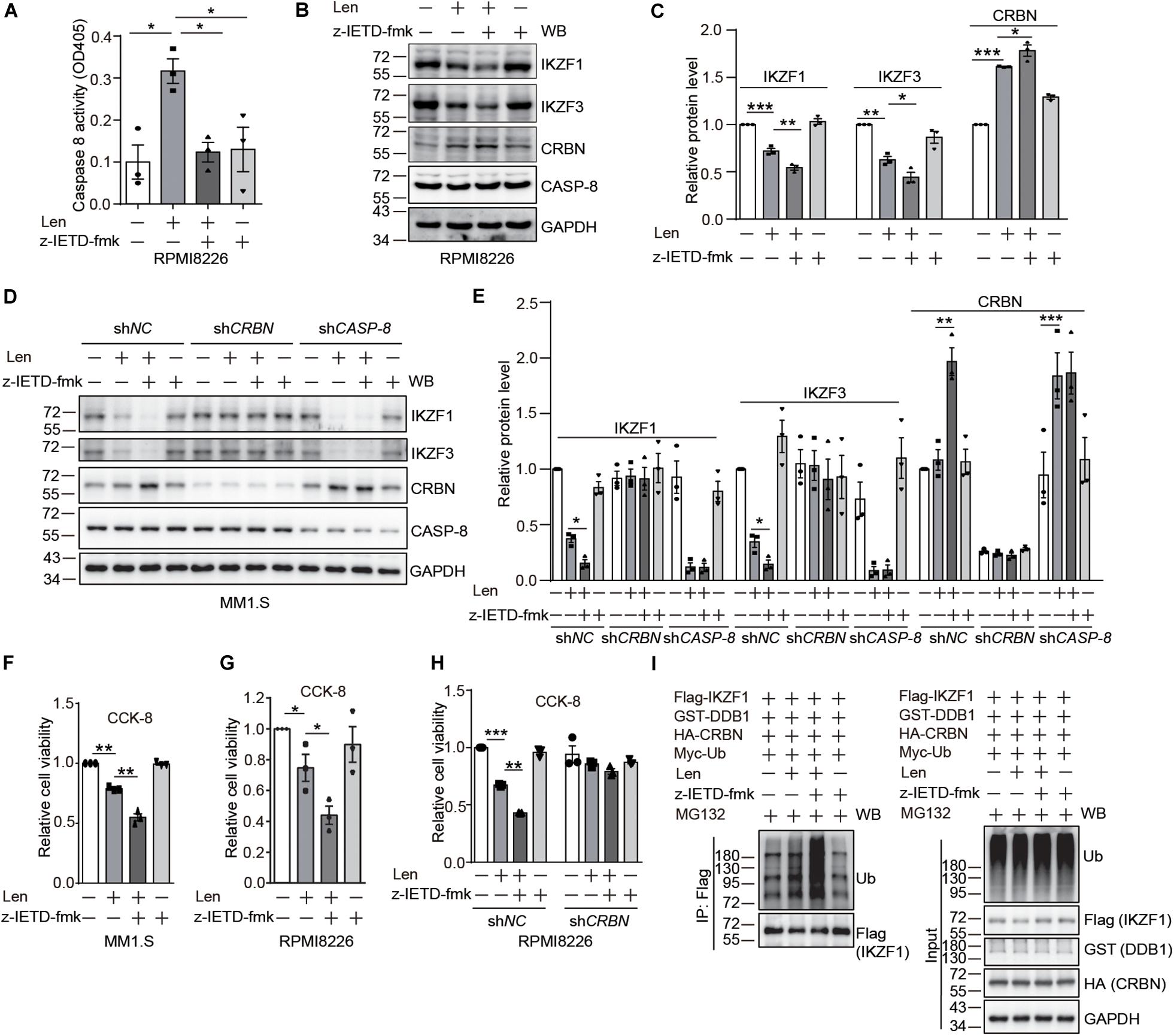
Figure 4. CASP-8 inhibition potentiates the anti-myeloma activity of Len. (A) Len enhances CASP-8 activity. RPMI8226 cells were pretreated with DMSO or z-IETD-fmk (40 μM) for 30 min and then treated with DMSO or Len (10 μM) for 24 h. CASP-8 activity was measured using a CASP-8 fluorometric assay kit. Mean ± SEMs from three independent biological replicates were plotted. Student’s t-test, *P < 0.05. (B) CASP-8 inhibitor z-IETD-fmk enhances the Len-mediated reduction of transcription factors IKZF1 and IKZF3 in RPMI8226 cells. RPMI8226 cells were pretreated with DMSO or CASP-8 inhibitor z-IETD-fmk (40 μM) for 30 min and then treated with DMSO or Len (10 μM) for 24 h. The resulting cell lysates were immunoblotted with the indicated antibodies. (C) Quantitative data (mean ± SEMs) for (B) were from three independent biological replicates. Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001. (D) CASP-8 inhibitor z-IETD-fmk enhances the Len-mediated reduction of transcription factors IKZF1 and IKZF3 in stable MM1.S knockdown cell lines. The stable shNC, shCRBN, and shCASP-8 MM1.S cell lines were pretreated with DMSO or CASP-8 inhibitor z-IETD-fmk (40 μM) for 30 min and then treated with DMSO or Len (10 μM) for 3 h. The resulting cell lysates were immunoblotted with the indicated antibodies. (E) Quantitative data (mean ± SEMs) for (D) were from three independent biological replicates. Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001. (F) CASP-8 inhibitor enhances Len-mediated reduction of cell viability in MM1.S cell line. MM1.S cells were pretreated with DMSO or z-IETD-fmk (40 μM) for 30 min and then treated with DMSO or Len (10 μM) for 48 h. The relative cell viability was measured with CCK-8 assay. Mean ± SEMs from three independent biological replicates. Student’s t-test, **P < 0.01. (G) CASP-8 inhibitor enhances Len-mediated reduction of cell viability in RPMI8226 cell line. RPMI8226 cells were pretreated with DMSO or z-IETD-fmk (40 μM) for 30 min and then treated with DMSO or Len (10 μM) for 48 h. The relative cell viability was measured with CCK-8 assay. Mean ± SEMs from three independent biological replicates. Student’s t-test, *P < 0.05. (H) CASP-8 inhibitor enhances Len-mediated reduction of cell viability in a CRBN-dependent manner. RPMI8226 cells were infected with shNC or shCRBN lentiviruses for 16 h and then treated as described in (G). Student’s t-test, **P < 0.01, ***P < 0.001. (I) CASP-8 inhibitor z-IETD-fmk enhances the Len-mediated ubiquitination of IKZF1. HEK293T cells were transfected with Flag-IKZF1, HA-CRBN, GST-DDB1, and Myc-Ub and split to four 6-cm plates. At 48 h post-transfection, cells were pretreated with DMSO or z-IETD-fmk (40 μM) for 30 min, then with DMSO or Len (10 μM) for 1 h, and again with MG132 (10 μM) for 12 h. The Flag tagged IKZF1 was purified with anti-Flag affinity gel, and the purified samples and whole cell lysates were immunoblotted with the indicated antibodies.
It has been demonstrated that high CRBN protein levels enhance the anti-myeloma activity of Len (Zhu et al., 2011; Liu et al., 2015). Therefore, we examined whether z-IETD-fmk increases the anti-myeloma activity of Len. Results from trypan blue staining and cell counting kit-8 (CCK-8) assay indicated that treatment with Len and z-IETD-fmk indeed further suppresses the viability of myeloma cells compared with Len treatment alone (Figures 4F,G and Supplementary Figures 5, 6). We further knocked down CRBN in RPMI8226 cells with lentivirus and treated the cells with z-IETD-fmk to investigate the anti-myeloma activity of Len. The data showed that z-IETD-fmk potentiated the anti-myeloma activity of Len in the mock knockdown and this effect disappeared when CRBN was knocked down (Figure 4H and Supplementary Figure 7). We also purified the Flag tagged IKZF1 and determined its ubiquitination upon z-IETD-fmk and Len treatment. We found that z-IETD-fmk could enhance the ubiquitination of IKZF1 upon Len treatment (Figure 4I), which further supported our conclusion that CASP-8 inhibition promotes their degradation through upregulating CRBN. This indicates that CRBN is required for the enhanced anti-myeloma activity of Len upon CASP-8 inhibition.
CASP-8 Knockdown Potentiates the Anti-myeloma Activity of Len in Cell Lines
To further determine the role of CASP-8 on the anti-myeloma activity of Len, we established MM1.S and CAG cell lines stably expressing control shLacZ or shCASP-8. The flow cytometry analysis demonstrated that ToPro-3+ dead cells are significantly increased upon Len treatment in the CASP-8 knockdown cells (Figures 5A,B). Consistent with this, the percentage of live cells determined by trypan blue exclusion in the CASP-8 knockdown cells is markedly reduced upon Len treatment (Figure 5C). Similar results were obtained for CAG myeloma cells (Figures 5D–F). Taken together, our results demonstrated that both inhibition and knockdown of CASP-8 enhance the anti-myeloma activity of Len in cell lines.
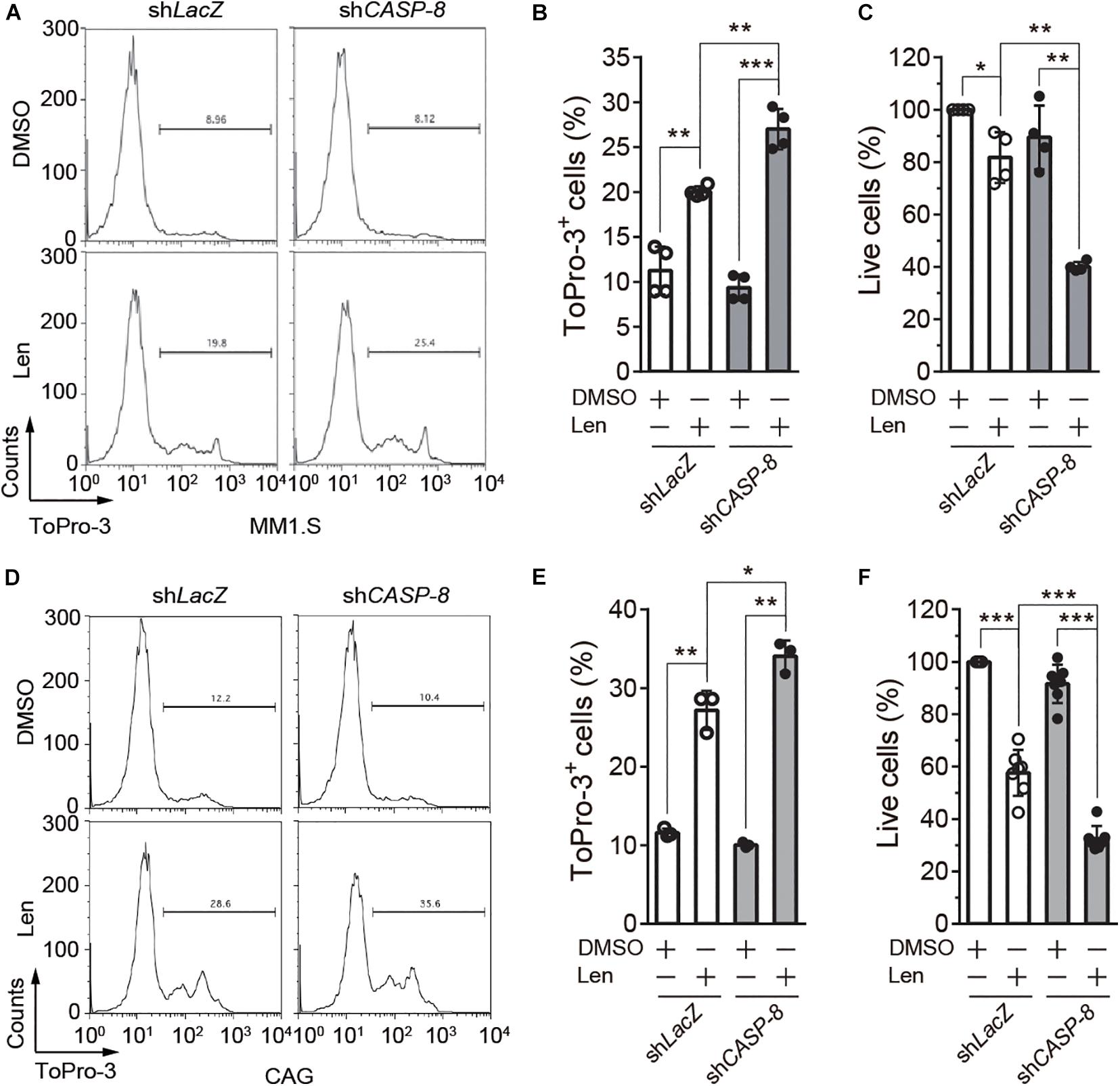
Figure 5. CASP-8 knockdown enhances the anti-myeloma activity of Len in myeloma cell lines. (A,B) CASP-8 knockdown increases Len-induced ToPro3+ cells. MM1.S cells with LacZ (mock) or CASP-8 knockdown were treated with DMSO or Len (10 μM) for 4 days and stained with ToPro-3. Cells were analyzed with flow cytometry, and ToPro-3+ cells (mean ± SEMs) were quantified from four biological replicates. Student’s t-test, **P < 0.01, ***P < 0.001. (C) CASP-8 knockdown potentiates the anti-myeloma activity of Len. MM1.S cells stably expressing shLacZ or shCASP-8 were treated with DMSO or Len (10 μM) for 4 days. Live cells were stained with trypan blue and counted under the microscope. Quantification was performed for cells (mean ± SEMs) from four biological replicates and normalized to the DMSO treated sample. Student’s t-test, *P < 0.05, **P < 0.01. (D–F) Same experiments as (A–C) were performed for CAG myeloma cells. In (E,F), the numbers of biological replicates were three and seven, respectively. Mean ± SEMs, Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001.
CASP-8 Inhibition Enhances the Therapeutic Effect of Len in Primary Myeloma Cells
To further validate whether CASP-8 modulates the anti-myeloma activity of Len in primary cells, we cultured CD138+ primary myeloma cells from bone marrow of two patients and treated them with CASP-8 inhibitor z-IETD-fmk and/or Len. Trypan blue staining and cell counting analyses demonstrated that z-IETD-fmk reduces the percentage of live cells upon Len treatment, whereas z-IETD-fmk alone does not affect the cell viability in both primary myeloma cells (Figure 6A). It should be noted that Len alone exhibited different effects in two primary patient samples. The viability of primary myeloma cells from Patient 1 but not from Patient 2 was significantly reduced by Len treatment. This result suggests that the genetic backgrounds of these two patients might be different, resulting in different sensitivities to Len. Nevertheless, CASP-8 inhibition enhances the anti-myeloma activity of Len in both cases.
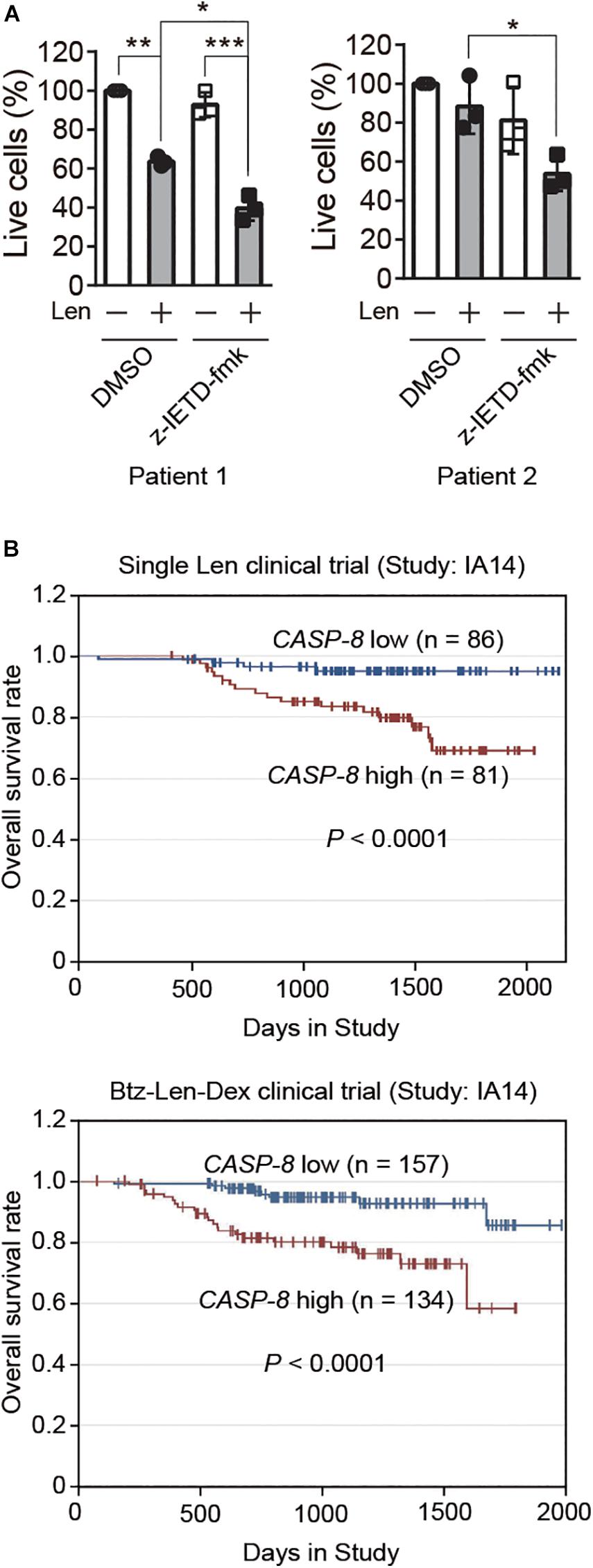
Figure 6. CASP-8 inhibition enhances the anti-myeloma activity of Len in primary myeloma cells, and low CASP-8 mRNA level increases the survival rate of myeloma patients. (A) CASP-8 inhibitor augments the anti-myeloma effect of Len in primary myeloma cells. CD138+ primary cells were isolated from myeloma Patient 1 and Patient 2, co-cultured with HS-5 cells, pretreated with DMSO or z-IETD-fmk (40 μM) for 1 h, and then treated with DMSO or Len (10 μM) for 4 and 5 days, respectively. The percentage of live cells was determined by staining with trypan blue and examined under the microscope. Experiments were carried out in triplicates, and pairwise Student’s t-test was used to obtain the P-value (mean ± SEMs). *P < 0.05, **P < 0.01, ***P < 0.001. (B) Low gene expression of CASP-8 increases the overall survival rate of myeloma patients. The data were obtained from patients in the MMRF CoMMpass trial (IA14) of combination therapy with Btz (Velcade), Len (Revlimid), and Dex (VRd). The numbers of patients for low (<20 FPKM) and high (>20 FPKM) CASP-8 mRNA levels were indicated in the images.
High CASP-8 Gene Expression Correlates With Poor Overall Survival in Myeloma Patients
Next, we would like to test whether the gene expression of CASP-8 is associated with the clinical outcome of myeloma patients. To do this, we analyzed the CASP-8 mRNA level and the overall survival rate of patients participating in a clinical trial of single Len or Btz, Len, and Dex combination therapy obtained from the datasets in MMRF. The result showed that those with lower CASP-8 mRNA levels exhibited a higher overall survival rate (Figure 6B), suggesting that CASP-8 expression may be an important factor in determining the clinical response to Len-based therapies likely through the regulation of CRBN protein level in myeloma. This result is also in concert with the data obtained from myeloma cell lines where CASP-8 inhibition or depletion further reduces the Len-mediated viability of myeloma cells (Figures 4–5).
Discussion
As a substrate receptor, CRBN mediates the ubiquitination and degradation of constitutive substrates and neo-substrates upon IMiD treatment (Krönke et al., 2014, 2015; An et al., 2017; Donovan et al., 2018; Matyskiela et al., 2018). CRBN can be regulated by SCFFbox7 and its associated E3 ligase CRL4CRBN, thus modulating its biological function. CRBN is a key modulator in the treatment of myeloma cells with Len and its structural analogs. However, whether CRBN and its function can be regulated by caspases was not explored. In this study, using three different types of cell lines (HeLa, NCI-H1688, and MM1.S cells), we discovered that CRBN can be cleaved upon TRAIL and Btz treatment (Figures 1, 2, and Supplementary Figure 1) and further demonstrated that this cleavage is blocked by CASP-8 inhibition (Figure 2).
Transcription factors IKZF1 and IKZF3 are required for myeloma cells to undergo proliferation. IMiDs bind to CRBN and recruit IKZF1 and IKZF3 for their ubiquitination and degradation, leading to the reduced proliferation of myeloma cells. This is the recently discovered major mechanism of action of IMiDs for the treatment of myeloma cells (Krönke et al., 2014; Lu et al., 2014). Using this model system, we showed that both pharmacological inhibition and genetic depletion of CASP-8 increase the level of full length CRBN, enhance the degradation of IKZF1 and IKZF3, and then suppress the proliferation of myeloma cells when treated with Len, which is consistent with the previous studies that CRBN protein levels control the sensitivity to IMiDs (Zhu et al., 2011; Liu et al., 2019). Two molecular mechanisms were discovered for the regulation of CRBN by Len. On the one hand, CRBN is required for the anti-myeloma activity of Len. However, the cleavage of CRBN can be induced by CASP-8 activation, which could be mediated by Len in myeloma cells, and the stability of the cleaved CRBN is reduced. On the other hand, our previous experiments detected the increase of CRBN protein level after 3 days treatment of myeloma cell lines with IMiDs (Liu et al., 2015). In that work, we also revealed that IMiDs can prevent CRBN from ubiquitination and subsequent degradation, leading to the increased CRBN protein level and enhanced CRL4CRBN E3 ligase activity, contributing to the anti-myeloma effect of IMiDs. In this work, we also observed the increase of CRBN protein level upon Len treatment. These two mechanisms of action of Len possibly result in two opposite effects, increase and decrease, on the CRBN protein level. Nevertheless, both mechanisms support the idea that inhibiting CASP-8 activity increases CRBN protein level and benefits to the therapeutic effect of Len for the treatment of myeloma. Therefore, combination of CASP-8 inhibitor with Len would most likely benefit to the treatment of myeloma, which was indeed confirmed in myeloma cell lines and primary myeloma cells.
On the one hand, we discovered that the viability of myeloma cells is reduced by the addition of CASP-8 inhibitor during the Len treatment (Figures 4–6). On the other hand, PARP1 cleavage is not significantly altered by the addition of CASP-8 inhibitor. These phenomena suggest that CASP-8 inhibition might affect the proliferation of myeloma cells. This is in accordance with the fact that Len reduces the proliferation of myeloma cells (Krönke et al., 2014; Lu et al., 2014). Using a CASP-8 fluorometric assay, a previous study demonstrated that Len and Pom activate CASP-8 (Mitsiades et al., 2002; Das et al., 2015), although no apparent CASP-8 cleavage was observed in the immunoblotting analysis (Chauhan et al., 2010; Das et al., 2015). Using HeLa cells as a model system, we discovered that CRBN is cleaved at Asp9 upon CASP-8 activation (Figure 3C). However, the cleaved CRBN has much lower stability (Figure 3D), which could reduce the CRL4CRBN E3 ligase activity. In combination with CASP-8 inhibitor, Len further elevates the CRBN protein level, resulting in the enhanced degradation of IKZF1 and IKZF3 and enhanced anti-myeloma activity (Figures 4–7). Therefore, a strategy could be an alternative treatment of myeloma with the combination of IMiDs and CASP-8 inhibitors, which suppresses the proliferation of myeloma cells.
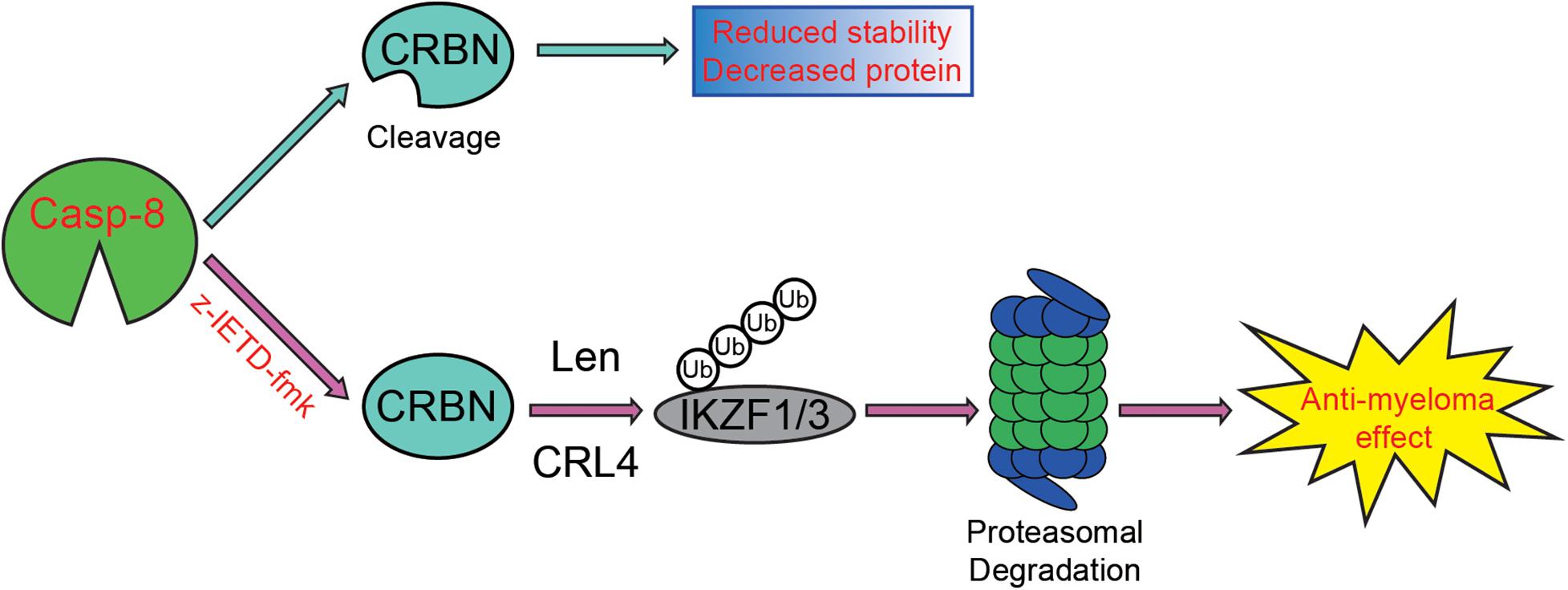
Figure 7. Proposed model for the enhancement of CASP-8 inhibition on the anti-myeloma effect of Len. In the absence of the CASP-8 inhibitor, CRBN is cleaved by CASP-8, and the cleaved CRBN has reduced stability, leading to the decreased CRBN protein level. However, in the presence of the CASP-8 inhibitor, CRBN cleavage is blocked, resulting in the increased CRBN protein level and enhanced Len-mediated ubiquitination and degradation of IKZF1/IKZF3. Therefore, the anti-myeloma activity of Len is augmented.
It should be noted that different myeloma cell lines and primary myeloma cells may have distinct genetic backgrounds, such as mutations and expression level of genes including CRBN, CASP-8, and DDB1, which affect cell proliferation and regulate cell death pathways. Indeed, truncation and point mutations in CRBN and DDB1 were discovered in myeloma cells and patient samples despite the fact that these mutations were rare (Thakurta et al., 2014). Although our conclusion was obtained from multiple cell lines and two primary patient samples, we cannot completely rule out the possibility that CASP-8 inhibition might not have a significant influence on the treatment of certain myeloma cell lines or some patient samples when CASP-8, CRBN, or other component of the CRL4CRBN E3 ligase is not expressed or is mutated.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by an Institutional Review Board of Weill Cornell Medicine. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
LZ, XH, SJ, and GX designed the research. LZ, WY, and XH performed the research. LZ, WY, XH, and GX analyzed the data. DJ and RN provided the patient samples. LZ, XH, and GX wrote the manuscript. WY, DJ, RN, and SJ reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (2019YFA0802400), National Natural Science Foundation of China (31670833 and 81703535), National Institutes of Health (NIH R01 CA186702), Open Project Program of the State Key Laboratory of Proteomics (SKLP-O201905), Jiangsu Key Laboratory of Neuropsychiatric Diseases (BM2013003), National Center for International Research (2017B01012), Suzhou Science and Technology Project (SYS2020093), and a project funded by the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Selina Chen-Kiang from Weill Medical College for kind support in experiments and fruitful discussion, Dr. Xiu-Bao Chang from the Mayo Clinic College of Medicine (United States) for generously providing the mouse anti-CRBN antibody, and Dr. Joshua Epstein from the University of Arkansas for Medical Sciences (United States) for kindly providing CAG cells.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.605989/full#supplementary-material
Footnotes
References
An, J., Ponthier, C. M., Sack, R., Seebacher, J., Stadler, M. B., Donovan, K. A., et al. (2017). pSILAC mass spectrometry reveals ZFP91 as IMiD-dependent substrate of the CRL4CRBN ubiquitin ligase. Nat. Commun. 8:15398. doi: 10.1038/ncomms15398
Angers, S., Li, T., Yi, X., MacCoss, M. J., Moon, R. T., and Zheng, N. (2006). Molecular architecture and assembly of the DDB1–CUL4A ubiquitin ligase machinery. Nature 443:590. doi: 10.1038/nature05175
Borset, M., Hjertner, O., Yaccoby, S., Epstein, J., and Sanderson, R. D. (2000). Syndecan-1 is targeted to the uropods of polarized myeloma cells where it promotes adhesion and sequesters heparin-binding proteins. Blood 96, 2528–2536.
Chauhan, D., and Anderson, K. C. (2003). Mechanisms of cell death and survival in multiple myeloma (MM): therapeutic implications. Apoptosis 8, 337–343.
Chauhan, D., Singh, A. V., Ciccarelli, B., Richardson, P. G., Palladino, M. A., and Anderson, K. C. (2010). Combination of novel proteasome inhibitor NPI-0052 and lenalidomide trigger in vitro and in vivo synergistic cytotoxicity in multiple myeloma. Blood 115, 834–845. doi: 10.1182/blood-2009-03-213009
Chou, J. J., Li, H., Salvesen, G. S., Yuan, J., and Wagner, G. (1999). Solution structure of BID, an intracellular amplifier of apoptotic signaling. Cell 96, 615–624. doi: 10.1016/s0092-8674(00)80572-3
Das, D. S., Ray, A., Song, Y., Richardson, P., Trikha, M., Chauhan, D., et al. (2015). Synergistic anti-myeloma activity of the proteasome inhibitor marizomib and the IMiD® immunomodulatory drug pomalidomide. Br. J. Haematol. 171, 798–812. doi: 10.1111/bjh.13780
Donovan, K. A., An, J., Nowak, R. P., Yuan, J. C., Fink, E. C., Berry, B. C., et al. (2018). Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane Radial Ray syndrome. eLife 7:e38430. doi: 10.7554/eLife.38430
Galluzzi, L., Vitale, I., Aaronson, S. A., Abrams, J. M., Adam, D., Agostinis, P., et al. (2018). Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 25, 486–541. doi: 10.1038/s41418-017-0012-14
Gandhi, A. K., Mendy, D., Waldman, M., Chen, G., Rychak, E., Miller, K., et al. (2014). Measuring cereblon as a biomarker of response or resistance to lenalidomide and pomalidomide requires use of standardized reagents and understanding of gene complexity. Br. J. Haematol. 164, 233–244. doi: 10.1111/bjh.12622
Gazitt, Y. (1999). TRAIL is a potent inducer of apoptosis in myeloma cells derived from multiple myeloma patients and is not cytotoxic to hematopoietic stem cells. Leukemia 13, 1817–1824. doi: 10.1038/sj.leu.2401501
Hideshima, T., Mitsiades, C., Akiyama, M., Hayashi, T., Chauhan, D., Richardson, P., et al. (2003). Molecular mechanisms mediating antimyeloma activity of proteasome inhibitor PS-341. Blood 101, 1530–1534. doi: 10.1182/blood-2002-08-2543
Hou, X., Si, J., Ren, H., Chen, D., Wang, H., Ying, Z., et al. (2015). Parkin represses 6-hydroxydopamine-induced apoptosis via stabilizing scaffold protein p62 in PC12 cells. Acta Pharmacol. Sin. 36, 1300–1307. doi: 10.1038/aps.2015.54
Huang, X., Di Liberto, M., Jayabalan, D., Liang, J., Ely, S., Bretz, J., et al. (2012). Prolonged early G1 arrest by selective CDK4/CDK6 inhibition sensitizes myeloma cells to cytotoxic killing through cell cycle-coupled loss of IRF4. Blood 120, 1095–1106. doi: 10.1182/blood-2012-03-415984
Ito, T., Ando, H., Suzuki, T., Ogura, T., Hotta, K., Imamura, Y., et al. (2010). Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350. doi: 10.1126/science.1177319
Jackson, S., and Xiong, Y. (2009). CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 34, 562–570. doi: 10.1016/j.tibs.2009.07.002
Krönke, J., Fink, E. C., Hollenbach, P. W., MacBeth, K. J., Hurst, S. N., Udeshi, N. D., et al. (2015). Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS. Nature 523, 183–188. doi: 10.1038/nature14610
Krönke, J., Udeshi, N. D., Narla, A., Grauman, P., Hurst, S. N., McConkey, M., et al. (2014). Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305. doi: 10.1126/science.1244851
Li, H., Zhu, H., Xu, C.-J., and Yuan, J. (1998). Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94, 491–501. doi: 10.1016/s0092-8674(00)81590-1
Liu, J., Song, T., Zhou, W., Xing, L., Wang, S., Ho, M., et al. (2019). A genome-scale CRISPR-Cas9 screening in myeloma cells identifies regulators of immunomodulatory drug sensitivity. Leukemia 33, 171–180. doi: 10.1038/s41375-018-0205-y
Liu, Y., Huang, X., He, X., Zhou, Y., Jiang, X., Chen-Kiang, S., et al. (2015). A novel effect of thalidomide and its analogs: suppression of cereblon ubiquitination enhances ubiquitin ligase function. FASEB J. 29, 4829–4839. doi: 10.1096/fj.15-274050
Lopez-Girona, A., Mendy, D., Ito, T., Miller, K., Gandhi, A. K., Kang, J., et al. (2012). Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia 26, 2326–2335. doi: 10.1038/leu.2012.119
Lu, G., Middleton, R. E., Sun, H., Naniong, M., Ott, C. J., Mitsiades, C. S., et al. (2014). The myeloma drug lenalidomide promotes the cereblon-dependent destruction of ikaros proteins. Science 343, 305–309. doi: 10.1126/science.1244917
Luo, X., Budihardjo, I., Zou, H., Slaughter, C., and Wang, X. (1998). Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94, 481–490. doi: 10.1016/s0092-8674(00)81589-5
Martiniani, R., Di Loreto, V., Di Sano, C., Lombardo, A., and Liberati, A. M. (2012). Biological activity of lenalidomide and its underlying therapeutic effects in multiple myeloma. Adv. Hematol. 2012:842945. doi: 10.1155/2012/842945
Matyskiela, M. E., Couto, S., Zheng, X., Lu, G., Hui, J., Stamp, K., et al. (2018). SALL4 mediates teratogenicity as a thalidomide-dependent cereblon substrate. Nat. Chem. Biol. 14, 981–987. doi: 10.1038/s41589-018-0129-x
Mitsiades, N., Mitsiades, C. S., Poulaki, V., Chauhan, D., Richardson, P. G., Hideshima, T., et al. (2002). Apoptotic signaling induced by immunomodulatory thalidomide analogs in human multiple myeloma cells: therapeutic implications. Blood 99, 4525–4530. doi: 10.1182/blood.V99.12.4525
Moffat, J., Grueneberg, D. A., Yang, X., Kim, S. Y., Kloepfer, A. M., Hinkle, G., et al. (2006). A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298. doi: 10.1016/j.cell.2006.01.040
Shimbo, K., Hsu, G. W., Nguyen, H., Mahrus, S., Trinidad, J. C., Burlingame, A. L., et al. (2012). Quantitative profiling of caspase-cleaved substrates reveals different drug-induced and cell-type patterns in apoptosis. Proc. Natl. Acad. Sci. U.S.A. 109, 12432–12437. doi: 10.1073/pnas.1208616109
Sievers, Q. L., Gasser, J. A., Cowley, G. S., Fischer, E. S., and Ebert, B. L. (2018). Genome-wide screen identifies cullin-RING ligase machinery required for lenalidomide-dependent CRL4CRBN activity. Blood 132, 1293–1303. doi: 10.1182/blood-2018-01-821769
Song, T., Liang, S., Liu, J., Zhang, T., Yin, Y., Geng, C., et al. (2018). CRL4 antagonizes SCFFbxo7-mediated turnover of cereblon and BK channel to regulate learning and memory. PLoS Genet. 14:e1007165. doi: 10.1371/journal.pgen.1007165
Stennicke, H. R., Renatus, M., Meldal, M., and Salvesen, G. S. (2000). Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem. J. 350, 563–568.
Tao, J., Yang, J., and Xu, G. (2018). The interacting domains in cereblon differentially modulate the immunomodulatory drug-mediated ubiquitination and degradation of its binding partners. Biochem. Biophys. Res. Commun. 507, 443–449.
Thakurta, A., Gandhi, A. K., Waldman, M. F., Bjorklund, C., Ning, Y., Mendy, D., et al. (2014). Absence of mutations in cereblon (CRBN) and DNA damage-binding protein 1 (DDB1) genes and significance for IMiD therapy. Leukemia 28, 1129–1131. doi: 10.1038/leu.2013.315
Voortman, J., Resende, T. P., Abou El Hassan, M. A. I., Giaccone, G., and Kruyt, F. A. E. (2007). TRAIL therapy in non–small cell lung cancer cells: sensitization to death receptor–mediated apoptosis by proteasome inhibitor bortezomib. Mol. Cancer Ther. 6, 2103–2112. doi: 10.1158/1535-7163.mct-07-0167
Xu, G., Jiang, X., and Jaffrey, S. R. (2013). A mental retardation-linked nonsense mutation in cereblon is rescued by proteasome inhibition. J. Biol. Chem. 288, 29573–29585. doi: 10.1074/jbc.M113.472092
Xu, Q., Hou, Y., Langlais, P., Erickson, P., Zhu, J., Shi, C., et al. (2016). Expression of the cereblon binding protein argonaute 2 plays an important role for multiple myeloma cell growth and survival. BMC Cancer 16:297. doi: 10.1186/s12885-016-2331-0
Zhou, L., and Xu, G. (2019). Cereblon attenuates DNA damage-induced apoptosis by regulating the transcription-independent function of p53. Cell Death Dis. 10:69. doi: 10.1038/s41419-019-1317-7
Keywords: cereblon, caspase-8, cleavage, TRAIL, multiple myeloma, lenalidomide, anti-myeloma activity, cell viability
Citation: Zhou L, Yu W, Jayabalan DS, Niesvizky R, Jaffrey SR, Huang X and Xu G (2020) Caspase-8 Inhibition Prevents the Cleavage and Degradation of E3 Ligase Substrate Receptor Cereblon and Potentiates Its Biological Function. Front. Cell Dev. Biol. 8:605989. doi: 10.3389/fcell.2020.605989
Received: 14 September 2020; Accepted: 20 November 2020;
Published: 17 December 2020.
Edited by:
Cui Hua Liu, Institute of Microbiology, Chinese Academy of Sciences, ChinaCopyright © 2020 Zhou, Yu, Jayabalan, Niesvizky, Jaffrey, Huang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoqiang Xu, Z3V4MjAwMkBzdWRhLmVkdS5jbg==; orcid.org/0000-0002-4753-4769; Xiangao Huang, eGloMjAwNEBtZWQuY29ybmVsbC5lZHU=
 Liang Zhou
Liang Zhou Wenjun Yu1
Wenjun Yu1 Samie R. Jaffrey
Samie R. Jaffrey Xiangao Huang
Xiangao Huang Guoqiang Xu
Guoqiang Xu