- 1Shandong Provincial Key Laboratory of Animal Cells and Developmental Biology, School of Life Sciences, Shandong University, Qingdao, China
- 2Department of Biomedical Engineering, Southern University of Science and Technology, Shenzhen, China
- 3Key Laboratory of Neuroregeneration of Jiangsu and Ministry of Education, Co-Innovation Center of Neuroregeneration, School of Life Sciences, Nantong University, Nantong, China
- 4Shandong Provincial Collaborative Innovation Center of Cell Biology, Shandong Normal University, Jinan, China
Hair cells in the inner ear and lateral lines are mechanosensitive receptor cells whose development and function are tightly regulated. Several transcription factors as well as splicing factors have been identified to play important roles in hair cell development, whereas the role of RNA stability in this process is poorly understood. In the present work, we report that RNA-binding motif protein 24a (Rbm24a) is indispensable for hair cell development in zebrafish. Rbm24a expression is detected in the inner ear as well as lateral line neuromasts. Albeit rbm24a deficient zebrafish do not survive beyond 9 days post fertilization (dpf) due to effects outside of the inner ear, rbm24a deficiency does not affect the early development of inner ear except for delayed otolith formation and semicircular canal fusion. However, hair cell development is severely affected and hair bundle is disorganized in rbm24a mutants. As a result, the auditory and vestibular function of rbm24a mutants are compromised. RNAseq analyses identified several Rbm24a-target mRNAs that are directly bound by Rbm24a and are dysregulated in rbm24a mutants. Among the identified Rbm24a-target genes, lrrc23, dfna5b, and smpx are particularly interesting as their dysregulation might contribute to the inner ear phenotypes in rbm24a mutants. In conclusion, our data suggest that Rbm24a affects hair cell development in zebrafish through regulating mRNA stability.
Introduction
Hair cells are mechanosensitive sensory receptor cells in the inner ear and fish lateral line, characterized by the hairy-looking protrusions called hair bundles on their apical surface. The hair bundle of each hair cell consists of dozens to hundreds of actin-based stereocilia and one tubulin-based kinocilium. The stereocilia are organized into several rows of increasing height, with the mechano-electrical transduction (MET) channels localized at the tips of shorter row stereocilia (Beurg et al., 2009). The kinocilium is juxtaposed next to the tallest row stereocilia and plays an essential role in the development of hair bundle (Jones et al., 2008). As highly differentiated sensory receptor cells, hair cells are very sensitive to genetic mutations and environmental assaults, and deficits in hair cell development or function are the main reasons for hearing loss (Müller and Barr-Gillespie, 2015). Several key transcription factors such as Atoh1, Pou4f3, Gfi1, and Rfxs have been identified to play important roles in hair cell development (Erkman et al., 1996; Bermingham et al., 1999; Wallis et al., 2003; Elkon et al., 2015). At the post-transcriptional level, alternative splicing regulated by Srrm4, Sfswap, and Esrp1 has been shown to be involved in hair cell development (Nakano et al., 2012; Moayedi et al., 2014; Rohacek et al., 2017). However, the role of RNA stability regulation in hair cell development/function is less understood.
RNA-binding proteins (RBPs) play pivotal roles in post-transcriptional RNA processing, from pre-mRNA alternative splicing to mRNA stability, localization and translation (Hentze et al., 2018). Dysfunction of RBPs is associated with various types of developmental diseases (Brinegar and Cooper, 2016). RBPs usually bind to target RNAs through their so-called RNA-recognition motifs (RRMs). RNA-binding motif protein 24 (Rbm24) is an RBP that contains a single RRM and an alanine-rich low-complexity region (Fetka et al., 2000). It regulates alternative splicing through binding to the intronic splicing enhancer (ISE) sites in its target pre-mRNAs (Yang et al., 2014; Ito et al., 2016; Zhang et al., 2016; Lin et al., 2018; van den Hoogenhof et al., 2018; Liu et al., 2019). Moreover, Rbm24 regulates the stability or translation of its target mRNAs through binding to AU/U-rich elements (AREs; Jiang Y. et al., 2014; Xu et al., 2014; Zhang et al., 2018).
In situ hybridization results reveal that Rbm24 is expressed in the otic vesicle during early embryonic development in zebrafish, frog, chick, and mouse (Fetka et al., 2000; Poon et al., 2012; Grifone et al., 2014; Maragh et al., 2014). Rbm24 is also expressed during later developmental stages of mouse inner ear (Cai et al., 2015; Grifone et al., 2018). Immunostaining and in situ hybridization reveal that Rbm24 is specifically expressed in the hair cells of embryonic and neonatal mice (Cai et al., 2015; Grifone et al., 2018). Rbm24 expression in the developing mouse hair cells is further supported by the transcriptome data (Scheffer et al., 2015; Shen et al., 2015). The specific expression of Rbm24 in the otic vesicle during early development as well as in the hair cells at a later developmental stage suggests that Rbm24 might play important roles in the inner ear.
In the present work, we investigate the role of Rbm24 in hair cells using the zebrafish as a model. Zebrafish sensory hair cells are located in the five sensory epithelia (two maculae and three cristae) of the otic vesicle and the two lateral line systems (anterior lateral lines (aLL) and posterior lateral lines (pLL)) at the body surface (Nicolson, 2005). Each of the anterior and posterior macula is associated with a calcium carbonate-based otolith, and their hair cells sense sound and linear acceleration. Hair cells in the anterior, lateral and posterior cristae sense angular acceleration, whereas the lateral line hair cells sense water movement. Our present data suggest that inactivation of the rbm24a gene affects the development of hair cells in both the otic vesicle and the lateral line systems. Further investigations show that Rbm24a affects hair cell development through regulating the stability of its target mRNAs.
Materials and Methods
Zebrafish
All zebrafish animal procedures were carried out following the institutional guidelines approved by the Animal Ethics Committee of Shandong University School of Life Sciences. The rbm24a mutant zebrafish was generated using TALENs (transcription activator-like effector nucleases) as described previously (Shao et al., 2020). The brn3c:GFP transgenic zebrafish were generated as described previously (He et al., 2017).
In situ Hybridization
Whole-mount in situ hybridization was carried out according to a standard protocol (Thisse and Thisse, 2008). For each target gene, a corresponding cDNA fragment was cloned into a pEASY Blunt Zero Cloning vector (Tiangen) and used as DNA template for synthesis of antisense RNA probe. The probes were labeled with digoxygenin-labeled rNTP mix (Roche Diagnostics), and NBT/BCIP was used as substrate. The probes for dlx3, pax2a, and lrrc23 are the same as reported previously (Han et al., 2018; Xing et al., 2018). Primers for other probes are listed in Supplementary Table 1.
Startle Response Measurement
Startle response was measured as described previously (Wang et al., 2017). Briefly, 10–20 zebrafish larvae at 5 dpf were maintained in an 8-cm Petri dish containing a thin layer (2 mm) of water. Tone bursts of 400 Hz at different sound intensity were delivered to the Petri dish through a mini vibrator (QY50R-Z). The movement of each larva was recorded using a digital camera (Basler acA1300–200 μm) at 120-frame per second (fps) and analyzed using a customized software developed in MATLAB (MathWorks, MA, United States). The distance of larvae’s C-shape movement upon sound stimulation was used as a measure of its auditory startle response.
Vestibular Head Tilt Response Measurement
Vestibular head tilt response was measured as described previously (Sun et al., 2018). Briefly, individual zebrafish larva at 5 dpf was placed in a customized holder, where its tail was glued to immobilize the fish. The head of the fish was merged in the water for comfortable accommodation. The holder was then placed on a rotary platform with the fish head-up. The rotary platform was driven by a stepper motor (model TSM17Q-3AG, MOONS’, Shanghai, China) running in a sinusoidal profile of ±75 degrees around the vertical location. Larval eye movement stimulated by rotation was recorded using a monochrome IR camera (Point Gray, Richmond, Canada) at 30 fps and analyzed using a customized imaging analysis program written in MATLAB (Mathworks). The ratio of the maximum projection area change during eye movement to the maximum projection area of the eye in the larval frontal plane was used as a measure of the vestibular head tilt response.
FM1-43FX Uptake Assay
Zebrafish larvae at 3 dpf were treated with 1.2 μM FM1-43FX (Molecular Probes, Invitrogen) in embryo medium for 30 s. After rinsing three times in fresh embryo medium, the larvae were anesthetized with 0.17 mg/mL–1 Tricaine (MS-222, Sigma) and the labeled lateral line hair cells were visualized using a florescent microscope (Olympus IX53).
Paraffin Section and HE Staining
Embryos were anesthetized with 0.17 mg/mL–1 Tricaine (MS-222, Sigma), then fixed in 4% paraformaldehyde (PFA) at 4°C overnight. After that, embryos were embedded in paraffin and sliced into 5 μm-thick sections. Hematoxylin (Solarbio, H8070) and Eosin Y (Solarbio, G1100) staining was performed subsequently according to the manufacturer’s user guide. Images were taken using a light microscope (Leica DM2000).
Confocal Microscopy
Embryos in brn3c:GFP transgenic background were anesthetized with 0.17 mg/mL–1 Tricaine (MS-222, Sigma), followed by fixation in 4% PFA at 4°C overnight. After washing with phosphate-buffered saline with Tween-20 (PBST), the samples were incubated with phalloidin (4 μg/ml, YEASEN, 40734ES80) at 37°C for 15 min. The samples were washed with PBST again, then mounted in 1% low-melting agarose. Images were taken using a confocal microscope (Zeiss, LSM700), and Z-stack projections were obtained by using the z-projection function. Image volumes were acquired at 0.5-μm intervals along the z-axis. Crista hair cells were imaged with a 0.95NA/20× Kort M27 objective lens. Neuromast hair cells were imaged with a 0.95NA/40× Kort M27 objective lens. Higher resolution images of crista and macula hair cells were taken with a 0.95NA/63× Kort M27 objective lens. The x-y pixel size is 1024 × 1024 for all images.
Scanning Electronic Microscopy
Scanning electronic microscopy (SEM) was performed as previously described with modifications (Du et al., 2020). Briefly, embryos after 100% epiboly were treated with 0.06–0.08 g/L PTU (P110661, Aladdin Industrial Corporation, China) to block pigment synthesis. Then larvae at 72 hpf were anesthetized with 0.17 mg/mL–1 Tricaine (MS-222, Sigma) for 30 s and fixed with 2.5% glutaraldehyde at 4°C over night, followed by post-fixation with 1% osmium tetroxide at 4°C for 2 h. After dehydration in ethanol, samples were critically point dried using a Leica EM CPD300 (Leica, Germany), then mounted and sputter coated with 10-nm platinum using a Cressington 108 sputter coater (Cressington, United Kingdom). Images were taken using a Quanta250 field-emission scanning electron microscope (FEI, Netherlands) with a beam strength of 3 kV.
RNAseq and RT-PCR
RNAseq analysis was performed by Genewiz (Suzhou, China) following the manufacturer’s protocols. Briefly, total RNAs were extracted from the otic vesicles of zebrafish larvae at 34 hpf (3 samples from wild-type and 3 samples from rbm24a mutants) using TRIzol reagent (Invitrogen), then mRNAs were enriched using the Poly(A) mRNA Magnetic Isolation Module (NEB). After mRNA fragmentation and cDNA synthesis, cDNA libraries were constructed using Ultra RNA Library Prep Kit for Illumina (NEB), followed by purification using Agencourt AMPure XP beads (Beckman) and quantification using Agilent 2100 (Agilent Technologies). The libraries were then multiplexed and clustered, and 150 bp paired-end sequencing was performed on Illumina HiSeq 2000. The sequencing results were filtered using Cutadapt and aligned with reference genome using Hisat2. Differential expression was analyzed using Htseq and DESeq2, and differential alternative splicing was analyzed using ASprofile and String Tie. RT-PCR and qPCR were then performed to confirm the sequencing results using primers specific for candidate genes (Supplementary Table 2).
RNA Immunoprecipitation
RNA immunoprecipitation (RIP) analysis was performed as described previously (Li et al., 2020). Briefly, HEK293T cells were transfected with expression vectors using LipoMax transfection reagents (Sudgen, Cat. No. 32012) to express Rbm24a-Myc together with its target mRNAs. The transfected cells were lysed in lysis buffer containing 100 mM KCl, 5 mM MgCl2, 10 mM HEPES-NaOH, 0.5% NP-40, 1 mM DTT, 200 units/mL RNase inhibitor (Takara, Cat. No. 2313A), and EDTA-free protease inhibitor cocktail (Sigma-Aldrich, Cat. No. S8830). After centrifugation, the supernatant was collected and incubated with immobilized anti-Myc antibody (Sigma-Aldrich, Cat. No. E6654). The immunoprecipitated RNA was used as template for RT-PCR analysis using specific primers for Rbm24a-target mRNAs (Supplementary Table 3).
Western Blot
The coding sequence (CDS) of rbm24a with or without the 8-bp deletion was inserted into modified pEGFP-N2 vector with EGFP-CDS replaced by Myc-CDS. HEK293T cells were transfected with expression vectors using LipoMax transfection reagents (Sudgen, Cat. No. 32012). 24 h after transfection, cells were lysed in ice-cold lysis buffer containing 150 mM NaCl, 50 mM Tris at pH 7.5, 1% (vol/vol) Triton X-100, 1 mM PMSF, and 1 × protease inhibitor cocktail (Roche). The supernatant was then collected after centrifugation and separated by polyacrylamide gel electrophoresis (PAGE), then transferred to PVDF membrane. After blocking in TBS (20 mM Tris–HCl, 150 mM NaCl) containing 5% non-fat dry milk and 0.05% Tween-20, the membrane was incubated with anti-Myc antibody (Abclonal, Cat. No. AE010) or anti β-Actin antibody (Abmart, Cat. No. P30002) at 4°C overnight, followed by incubation with HRP-conjugated secondary antibody (Bio-Rad, Cat. No. 170-6515 and 170-6516) at room temperature for an hour. The signals were detected with the ECL system (Thermo Fisher Scientific).
Statistical Analysis
Each experiment was repeated at least three times. Student’s t-test was used to determine the statistical significance, and p < 0.05 was considered statistically significant. Data were shown as means ± SEM.
Results
Rbm24a Is Expressed in the Inner Ear and Lateral Line Neuromasts
There are two rbm24 homologs in the zebrafish, rbm24a and rbm24b. Rbm24a shows higher sequence homology with mammalian Rbm24 compared to rbm24b. At the protein level, the similarity of mouse Rbm24 to zebrafish Rbm24a and Rbm24b is 87% and 72%, respectively. Meanwhile, rbm24a has been detected in the otic vesicles, whereas rbm24b has not (Poon et al., 2012; Maragh et al., 2014). Hence we focus on rbm24a in the present study. We first examined the spatial-temporal expression pattern of rbm24a by performing in situ hybridization. The results show that rbm24a transcripts could be readily detected in the otic vesicle and lateral line neuromasts at 36 hours post fertilization (hpf; Figures 1A,A’,A”). Rbm24a expression persists at 48 hpf and 72 hpf (Figures 1B–C’). By 96 hpf, rbm24a expression is still present in the otic vesicle and lateral lines (Figures 1D,D’). This specific expression pattern suggests that rbm24a might play an important role in the development of inner ear and lateral line neuromasts.
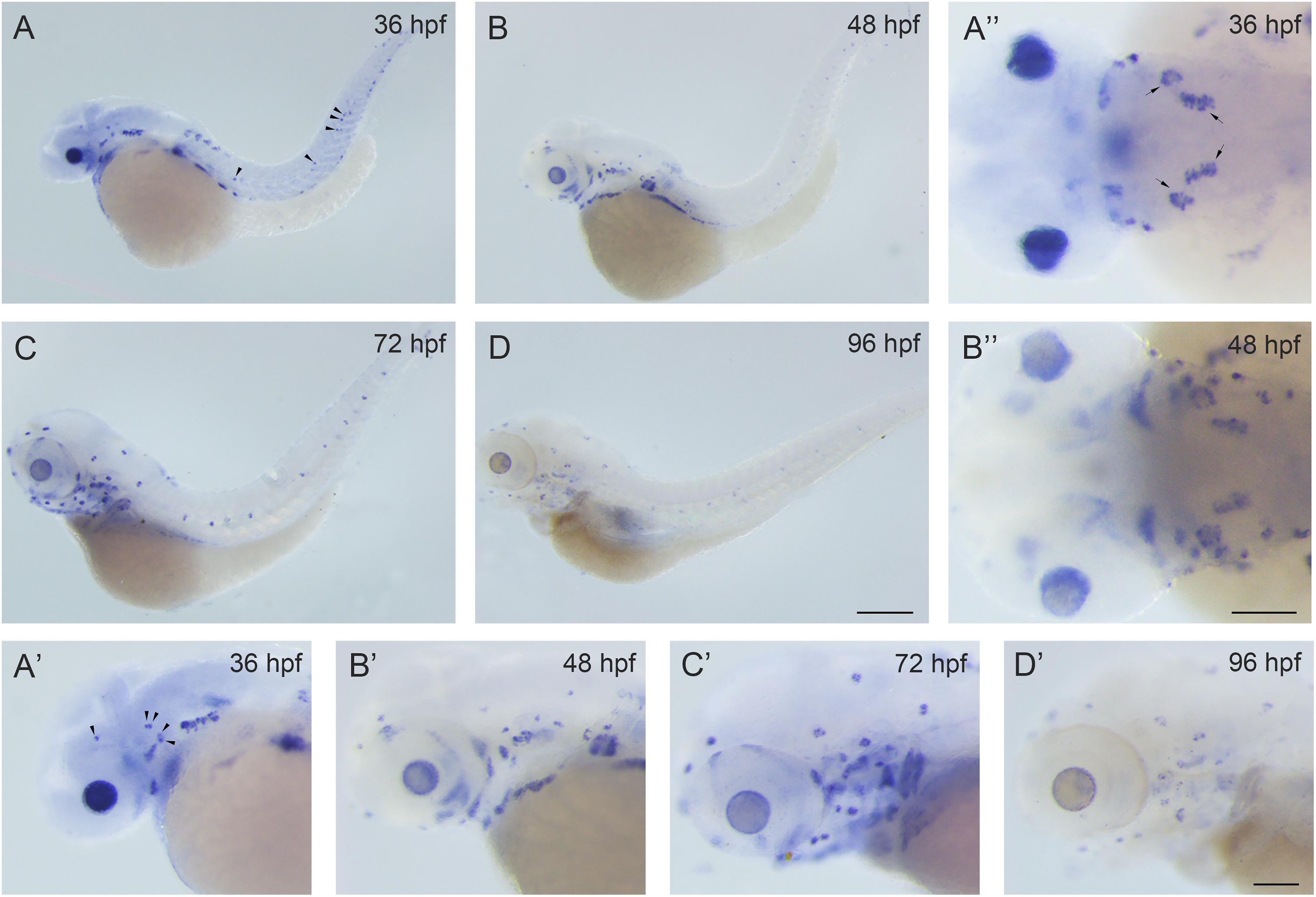
Figure 1. Rbm24a is expressed in the otic vesicles and lateral line neuromasts of the zebrafish. Expression pattern of rbm24a during development was examined by performing in situ hybridization of zebrafish embryos at 36 hpf (A–A”), 48 hpf (B–B”), 72 hpf (C,C’), and 96 hpf (D,D’). Otic vesicles are indicated with arrows, and lateral line neuromasts are indicated with arrowheads. Scale bars, 0.25 mm in (A–D), 0.1 mm in (A’–D’) and (A”,B”).
Rbm24a Deficiency Affects Hair Cell Function
We investigated the function of rbm24a in hair cells using a rbm24a mutant zebrafish line, which contains a deletion of 8 base pairs (bp) in exon 1 of the rbm24a gene (Shao et al., 2020; Supplementary Figure 1A). The 8 bp-deletion will result in premature translational termination, giving rise to a truncated Rbm24a protein of only 28 amino acids (aa) instead of the full-length 230 aa (Supplementary Figure 1B). RT-PCR and in situ hybridization results show that rbm24a mRNA is still present in the homozygous rbm24a mutants (rbm24a–/–; Supplementary Figures 1C,D). There are several downstream AUG codons in rbm24a CDS, among which M59 is the first one (Supplementary Figure 1B). Translation started from these AUG codons in the mutant rbm24a mRNA might produce truncated Rbm24a proteins. Because Rbm24a-specific antibodies are not available, we expressed wild type or mutant Rbm24a with a Myc tag at the C-terminus in cultured HEK293T cells and examined their expression by performing western blot with anti-Myc antibody. From the cells expressing wild-type Rbm24a, we detected a band with a molecular weight of around 30 kDa. However, from the cells expressing mutant Rbm24a, we detected a band with a molecular weight of lower than 25 kDa, possibly corresponding to a truncated Rbm24a protein started from M59 (Supplementary Figure 1E). This mutant Rbm24a lacks most part of the RRM domain, suggesting that it is not functional. Consistent with this hypothesis, heterozygous rbm24a mutants are morphologically and behaviorally indistinguishable from wild type zebrafish, whereas homozygous rbm24a mutants suffer from severe heart defects and do not survive beyond 9 days post fertilization (dpf; Supplementary Figure 1F). Moreover, the body length of rbm24a mutants is slightly shorter than that of wild type (wild type at 48 hpf, 3.19 ± 0.02 mm; rbm24a mutant at 48 hpf, 2.85 ± 0.01 mm; wild type at 96 hpf, 3.55 ± 0.03 mm; rbm24a mutant at 96 hpf, 3.17 ± 0.02 mm. For each genotype, n = 18. P value < 0.000001).
We first evaluated the auditory function of mutant zebrafish larvae by measuring startle response (Kimmel et al., 1974; Zeddies and Fay, 2005). Various levels of sound stimulus were given to the fish larvae at 5 dpf, and the moving distance in a characteristic C-shape motion of the larvae was used as a measure of its auditory startle response. Compared to control wild-type larvae, rbm24a mutants show significant reduced startle response at each sound level, suggesting that rbm24a deficiency affects the auditory function of zebrafish (Figure 2A). We also evaluated the vestibular function of the mutants by measuring vestibular head tilt response (Mo et al., 2010). In this experiment, eye movement of 5 dpf larvae evoked by the rotational motion was used as a measure of the vestibular function. The results show that compared to wild-type control, rbm24a mutants show significantly reduced vestibular head tilt response, suggesting that rbm24a deficiency affects the vestibular function of zebrafish (Figure 2B).
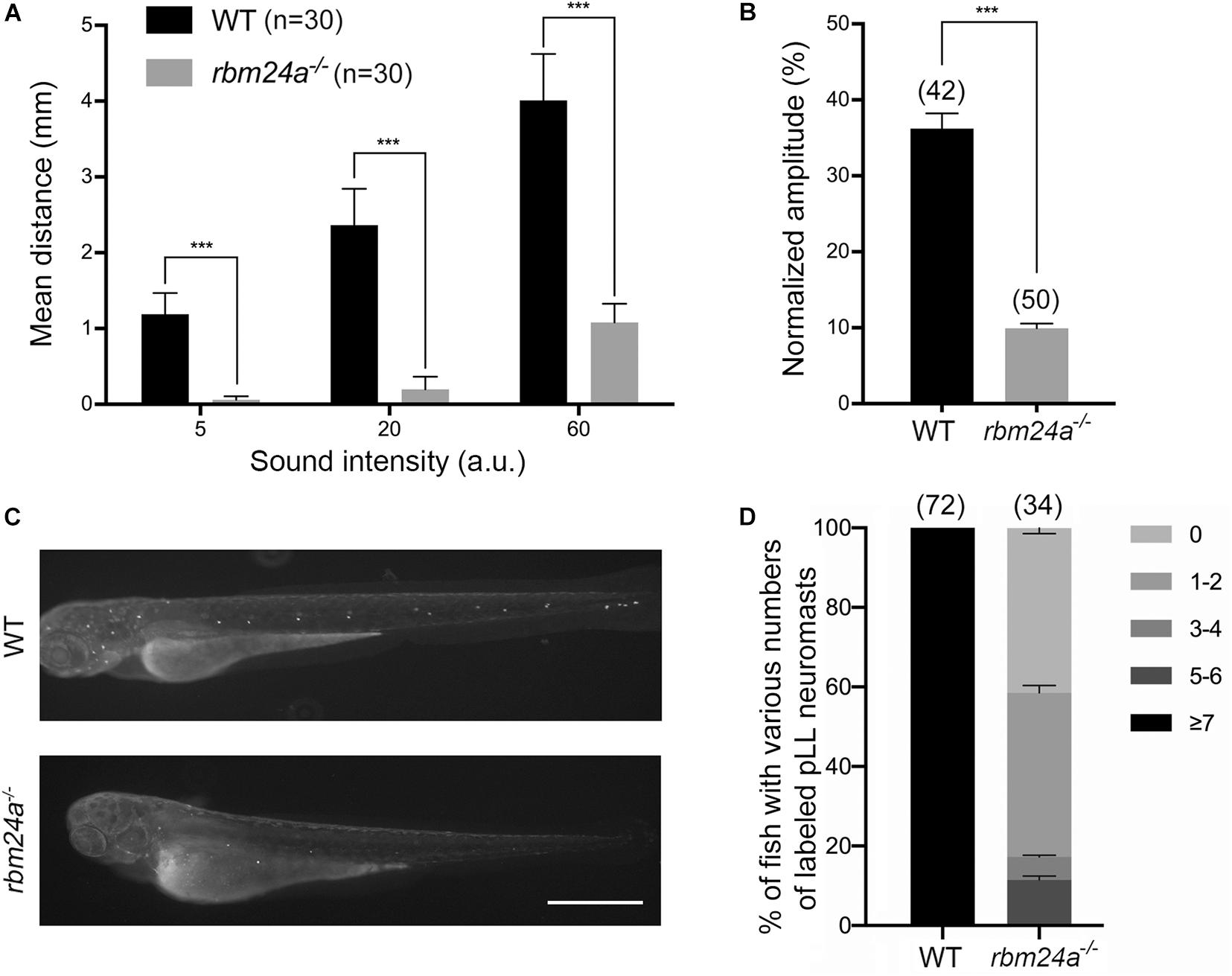
Figure 2. Hair cell function is affected in rbm24a mutants. (A) The auditory function of zebrafish larvae at 5 dpf is evaluated by examining the startle response to various sound intensity. ***p < 0.001. (B) The vestibular function of zebrafish larvae at 5 dpf is evaluated by vestibular head tilt response measurement. ***p < 0.001. (C) The function of hair cells is evaluated by performing FM1-43FX uptake experiment at 3 dpf. Scale bar, 0.5 mm. (D) The numbers of labeled pLL neuromasts per larvae are calculated according to the results from (C). P value < 0.001. The numbers of larvae for each group are indicated in brackets.
Considering that rbm24a deficiency might affect muscle function, which could in turn affect the performance of the larvae in startle response and vestibular head tilt response, we then directly examined the functional integrity of hair cells by performing FM1-43FX uptake experiment. Florescent dye FM1-43 or its fixable analog FM1-43FX can enter hair cells through the MET channels, which provides an indicator of the functional integrity of hair cells (Gale et al., 2001; Meyers et al., 2003). The results show that FM1-43FX uptake by both aLL and pLL neuromasts is significantly decreased in 3 dpf rbm24a mutant larvae (Figures 2C,D). Taken together, our data suggest that hair cell function is severely affected in rbm24a mutant zebrafish.
Early Inner Ear Development Is Largely Unaffected by rbm24a Deficiency
Given the expression of rbm24a in the otic vesicles during early embryonic development, we wonder whether rbm24a deficiency affects early inner ear development. The morphology of otic vesicles was examined by performing hematoxylin-eosin (HE) staining, which does not reveal any significant difference between wild-type control and rbm24a mutants (Supplementary Figures 2A–C). However, examination with a light microscope shows that otolith formation in rbm24a mutants is delayed during early development. In each otic vesicle of early zebrafish embryo, otolith precursor particles are first distributed throughout the otic vesicle lumen, then tethered to form two otoliths at the otic vesicle poles, which are readily detectable by 24 hpf (Riley et al., 1997). Consistently, our data show that at 24 hpf, most wild-type embryos contain 4 otoliths (2 for each otic vesicle). In contrast, less than 40% of rbm24a mutants contain 4 otoliths, and all the others contain 5 or 6 otoliths (Figures 3A,B). The ectopic otolith gradually fuses to the regular posterior otolith, and there is no visible difference between the otoliths of the mutant and control larvae after 48 hpf (Figure 3A).
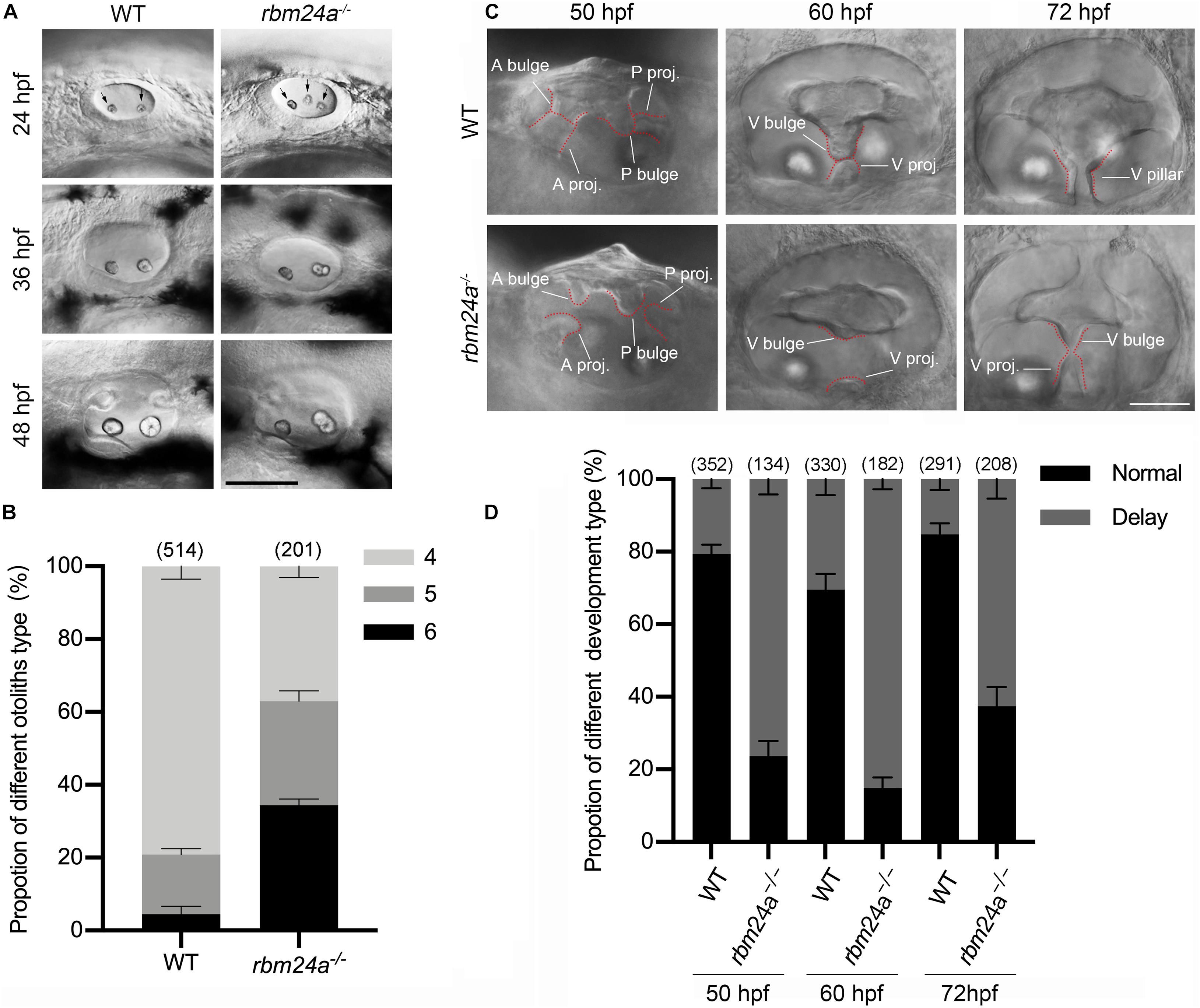
Figure 3. Otolith formation and semicircular canal fusion are delayed in rbm24a mutants. (A) Otolith formation in zebrafish larvae is examined using a light microscope at 24, 36, and 48 hpf as indicated. Otoliths are indicated with arrows. Scale bar, 0.1 mm. (B) The numbers of otic vesicles per larvae at 24 hpf are calculated according to the results from (A). P values are 0.000833 (larvae with 4 otoliths), 0.020678 (larvae with 5 otoliths), and 0.00044 (larvae with 6 otoliths). (C) Fusion of the semicircular canals is examined using a light microscope at 50, 60, and 72 hpf as indicated. Scale bar, 50 μm. (D) The numbers of larvae with normal or delayed semicircular canal fusion are calculated according to the results from (C). P values are 0.000001 (50 hpf), 0.00005 (60 hpf), and 0.000034 (72 hpf). The numbers of larvae for each genotype are indicated in brackets. A bulge, anterior bulge; A proj., anterior projection; P bulge, posterior bulge; P proj., posterior projection; V bulge, ventral bulge; V proj., ventral projection; and V pillar, ventral pillar.
Moreover, delayed fusion of the semicircular canals is observed in rbm24a mutant larvae. The development of semicircular canals begins with the formation of three protrusions (the anterior, posterior, and lateral projections) from the otic vesicle between 42 and 48 hpf (Waterman and Bell, 1984). Afterward, the lateral projection bifurcates into the anterior and posterior bulges, which then fuse with the anterior and posterior projections to form the anterior and posterior pillars, respectively. Finally, the lateral projection gives rise to a ventral bulge, which then fuses with the newly developed ventral projection to form the ventral pillar by 72 hpf (Waterman and Bell, 1984). We found that in most wild-type larvae at 50 hpf, the anterior and posterior projections fuse with the anterior and posterior bulges, respectively, which is not observed in most rbm24a mutant larvae until 60 hpf (Figures 3C,D). Similarly, the ventral projection is fused with the ventral bulge in most wild-type larvae at 60 hpf, but not in most rbm24a mutant larvae until 72 hpf (Figures 3C,D).
We then performed in situ hybridization to examine the expression of early otic vesicle markers in rbm24a mutants. Dlx3 is expressed in the otic vesicle at an early developmental stage (Ekker et al., 1992). Our results show that dlx3 expression in the otic vesicles of rbm24a mutant larvae is comparable to that of control at 24 and 36 hpf (Figures 4A,B). Similar results were obtained for another early otic vesicle marker, pax2a (Pfeffer et al., 1998; Figure 4C). Taken together, our present data suggest that early inner ear development is largely unaffected by rbm24a deficiency except for delayed otolith formation and semicircular canal fusion.
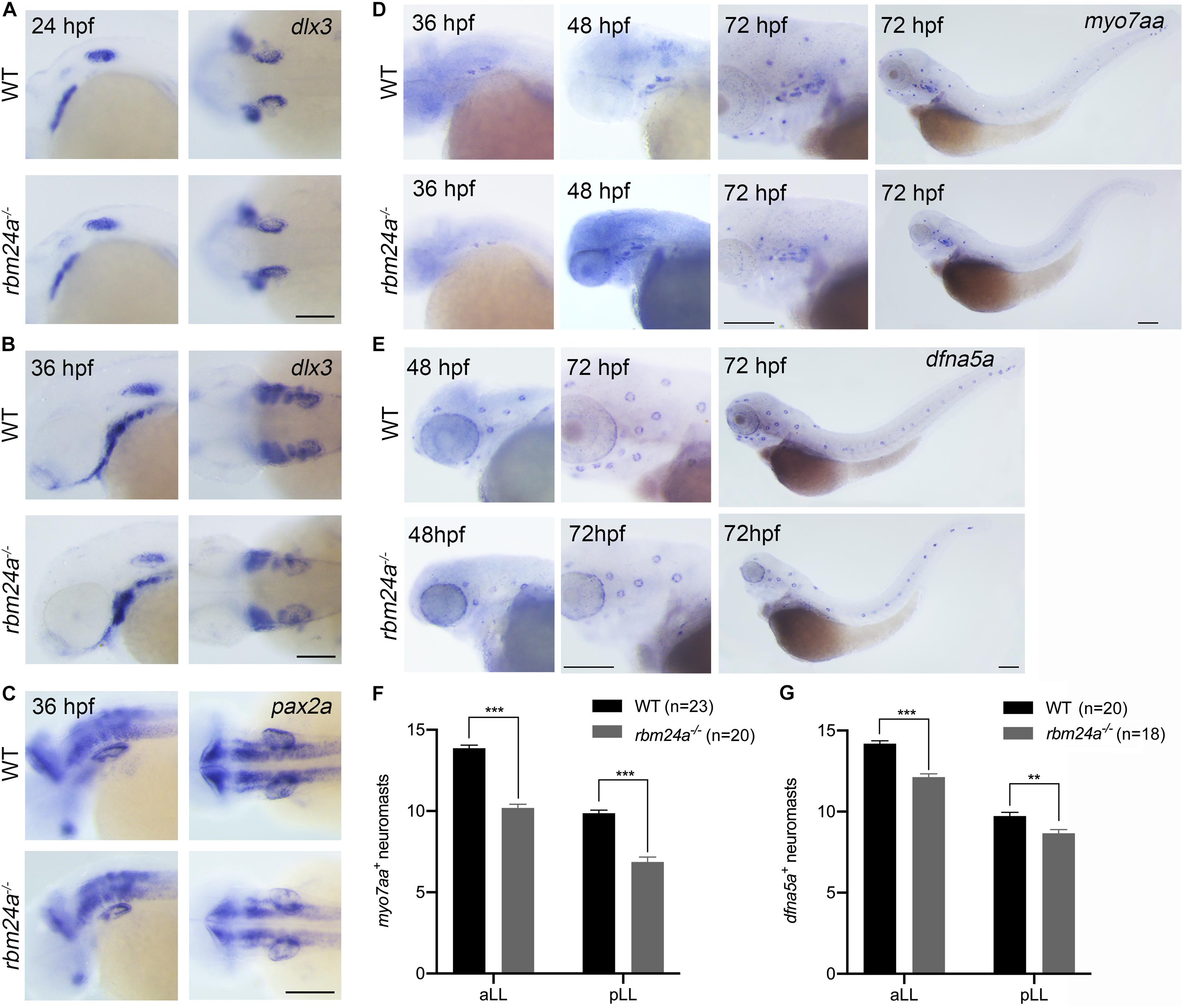
Figure 4. Early inner ear development is largely unaffected in rbm24a mutants. Inner ear development is examined by performing in situ hybridization of dlx3 (A,B), pax2a (C), myo7aa (D), and dfna5a (E) at different developmental stages as indicated. (F) The numbers of myo7aa+ neuromasts are calculated according to the results from (D). (G) The numbers of dfna5a+ neuromasts are calculated according to the results from (E). The numbers of larvae for each group are indicated in brackets. Scale bars, 0.2 mm. **p < 0.01; ***p < 0.001.
Rbm24a Deficiency Leads to Reduced Hair Cell Number and Disorganized Hair Bundle
We moved on to examine the hair cells in rbm24a mutants by performing in situ hybridization of a hair cell marker myo7aa (Ernest et al., 2000). At 36 and 48 hpf, myo7aa is expressed in the otic vesicles of rbm24a mutant larvae similarly to that in controls (Figure 4D). By 72 hpf, robust expression of myo7aa in aLL and pLL neuromasts is detected in wild-type larvae; however, the number of myo7aa+ neuromasts is significantly decreased in rbm24a mutants (Figures 4D,F). The reduced expression of myo7aa in lateral lines of rbm24a mutants suggest that hair cell development is affected by rbm24a deficiency. We also performed in situ hybridization of dfna5a, a gene that is specifically expressed in the lateral line neuromasts (Jiang L. et al., 2014). The results show that the number of dfna5a+ neuromasts is slightly reduced in rbm24a mutants (Figures 4E,G), which could be explained by the shortened body length.
To visualize the hair cells directly, we crossed rbm24a mutant zebrafish with brn3c:GFP transgenic zebrafish, which express GFP in the hair cells of both inner ear and lateral lines. The numbers of GFP-positive hair cells in each pLL neuromast are similar in rbm24a mutants and wild-type controls at 48 hpf (Figures 5A,B). However, by 72 hpf, the hair cell numbers per pLL neuromast in rbm24a mutants are less than that in controls, which becomes more obvious at 96 hpf (Figures 5A,C). At 96 hpf, there are 6–8 hair cells per pLL neuromast in wild-type controls, whereas no more than 4 hair cells per neuromast could be found in rbm24a mutants (Figures 5A,C). Meanwhile, similar to the in situ hybridization results of dfna5a, the total numbers of pLL neuromasts in rbm24a mutants are slightly less than that in controls (Figures 5B,C). Decreased hair cell numbers are also observed in the maculae and the cristae of the inner ear in rbm24a mutants (Supplementary Figures 3, 4).
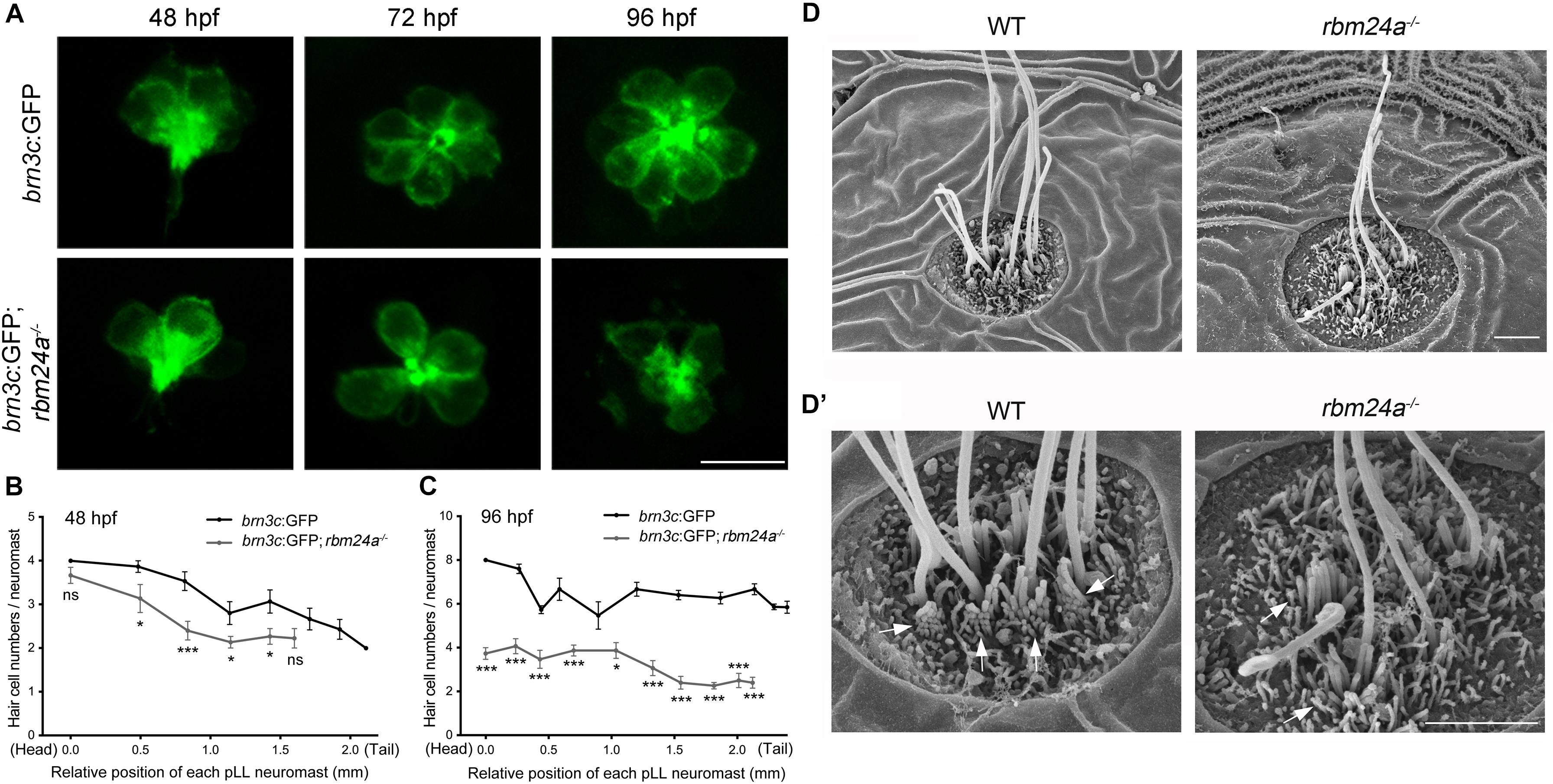
Figure 5. Development of pLL hair cells is affected in rbm24a mutants. (A) Confocal microscopic imaging analysis of pLL neuromasts in wild-type or rbm24a mutant brn3c:GFP line at different developmental stages as indicated. (B,C) The numbers of GFP-positive hair cells of each pLL neuromasts at 48 and 96 hpf are calculated according to the results from (A). The numbers of larvae for each group is 15. (D,D’) Hair bundles of pLL neuromasts at 72 hpf are examined using SEM. Stereocilia are indicated with arrows. Scale bars, 10 μm in (A), 2 μm in (D,D’). ns, not significant; *p < 0.05; and ***p < 0.001.
Scanning electrical microscopy was then employed to examine the morphology of hair bundles of the mutants. The results show that the stereocilia of pLL hair cells in wild type zebrafish are organized into a nice staircase pattern, which is largely lost in rbm24a mutants (Figures 5D,D’). To examine the hair bundles of inner ear hair cells, we used phalloidin to stain the F-actin core of stereocilia. Meanwhile, stereocilia and kinocilia could also be visualized by the strong GFP fluorescence in brn3c:GFP zebrafish. The results show that at 72 hpf, the length of hair cell stereocilia in rbm24a mutant crista is decreased to about half of that in wild-type control (Figures 6A,B). The length of kinocilia is also decreased in rbm24a mutants although to a lesser extent (Figure 6C). This phenotype remains in rbm24a mutants at 96 hpf (Figures 6D–F). Similar results are also observed in the macula (Supplementary Figures 5A–D). Taken together, our data suggest that rbm24a deficiency leads to reduced hair cell number and disorganized hair bundle.
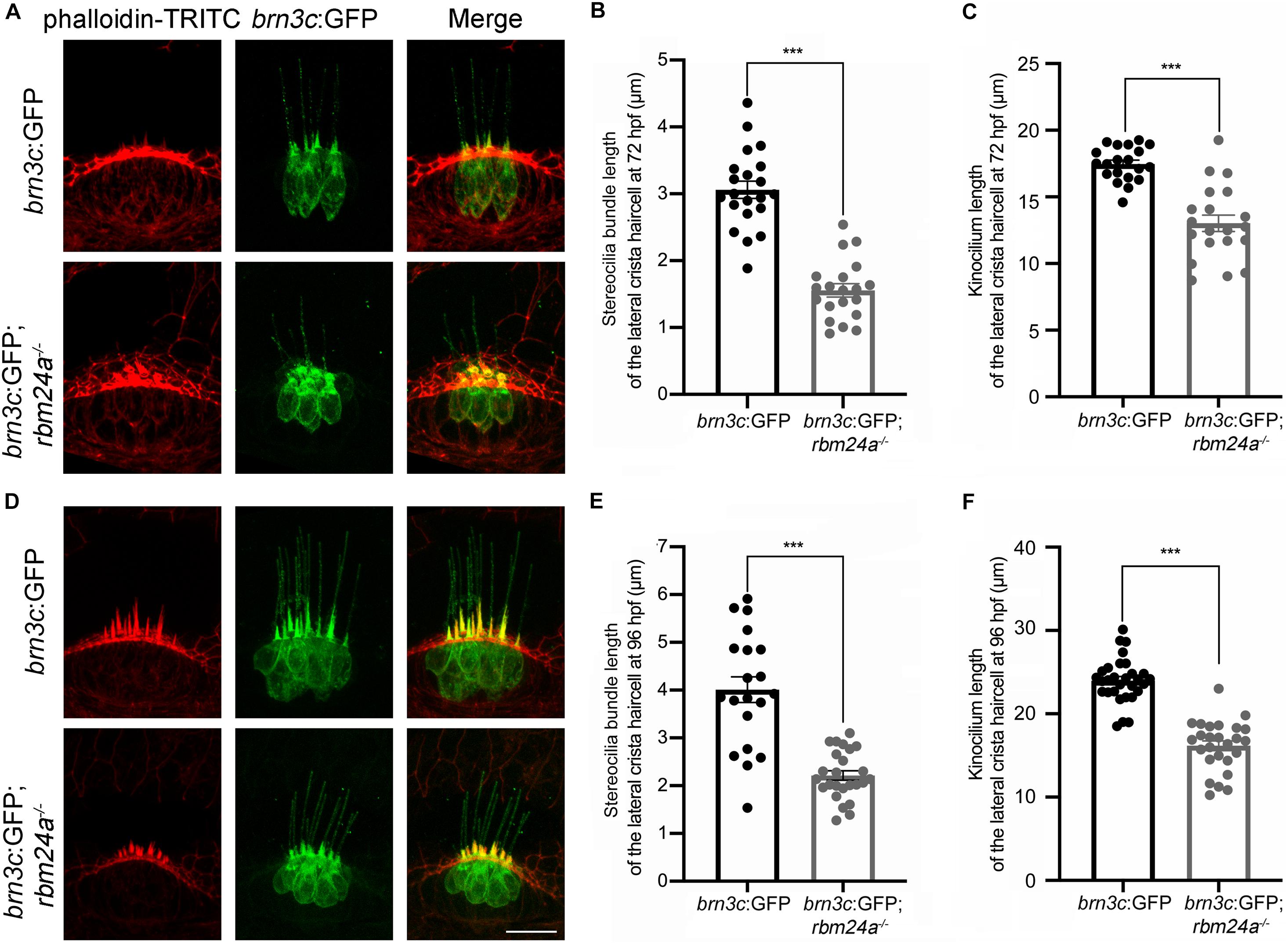
Figure 6. Development of lateral crista hair cells is affected in rbm24a mutants. Confocal microscopic imaging analysis of rbm24a mutant brn3c:GFP line at 72 hpf (A) and 96 hpf (D). (B,E) Stereocilia length is calculated according to the results from (A,D), respectively. (C,F) Kinocilia length is calculated according to the results from (A,D), respectively. For stereocilia length measurement at 72 hpf, brn3c:GFP, n = 21 (from 3 larvae); brn3c:GFP;rbm24a– /–, n = 20 (from 4 larvae). For stereocilia length measurement at 96 hpf, brn3c:GFP, n = 20 (from 3 larvae); brn3c:GFP;rbm24a– /–, n = 25 (from 4 larvae). For kinocilia length measurement at 72 hpf, brn3c:GFP, n = 20 (from 3 larvae); brn3c:GFP;rbm24a– /–, n = 20 (from 4 larvae). For kinocilia length measurement at 96 hpf, brn3c:GFP, n = 30 (from 3 larvae); brn3c:GFP;rbm24a– /–, n = 26 (from 3 larvae). Scale bar, 10 μm. ***p < 0.001.
Rbm24a Deficiency Leads to Dysregulated Expression of Several Inner Ear-Expressed Genes
Previously, we performed RNA sequencing (RNAseq) of rbm24a mutant embryos to analyze the Rbm24a-regulated mRNAs (Shao et al., 2020). In the present work, we also prepared polyadenylated mRNA samples from the otic vesicle of rbm24a mutants at 34 hpf and performed RNAseq analysis. Neither of the analyses reveals meaningful differences in alternative splicing events between rbm24a mutants and controls. However, the RNAseq results reveal that the mRNA level of several genes such as lrrc23, smpx, casp6a, and dfna5b is significantly decreased in the rbm24a mutants (Figure 7A). On the other hand, the mRNA level of a few genes such as cyp2aa7 and dnah7 is upregulated in the rbm24a mutants (Figure 7A). The dysregulation was further validated by performing RT-PCR and qPCR (Figures 7B,C). Furthermore, RNA immunoprecipitation (RIP) experiments confirm that Rbm24a directly binds to these target mRNAs (Figure 7D).
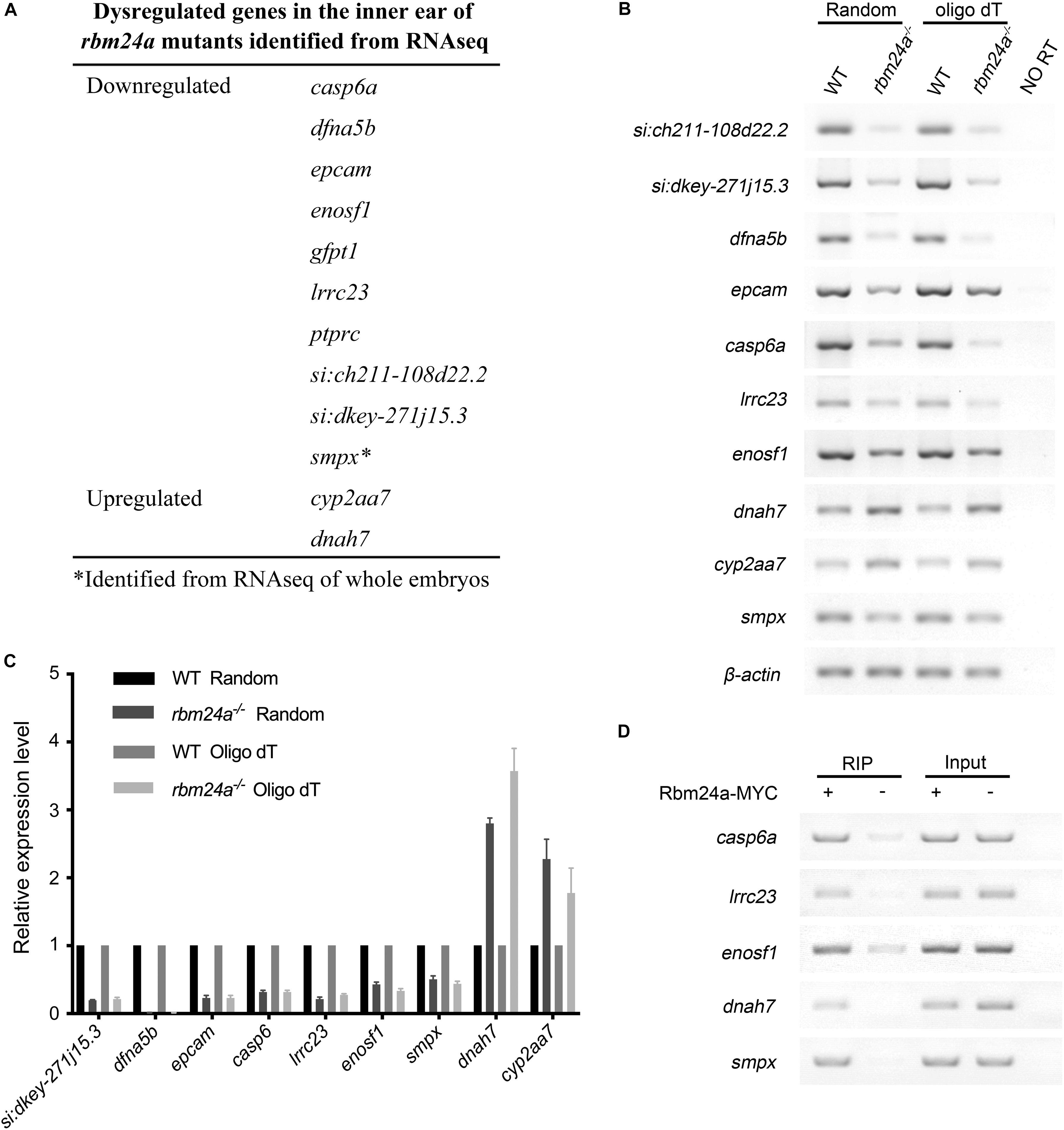
Figure 7. Dysregulated genes in the inner ear of rbm24a mutants. (A) Dysregulated genes in the inner ear of rbm24a mutants identified from RNAseq analysis. RT-PCR (B) and qPCR (C) were performed to confirm the RNAseq results. Random primers or oligo dT were used as primers for reverse transcription as indicated. (D) RIP experiments were performed to examine the binding of Rbm24a with its target mRNAs. Statistical analysis of the qPCR results shows that the expression difference is significant (P value < 0.000001, except that P value of cyp2aa7 is 0.001315).
We then performed in situ hybridization to analyze the expression pattern of these Rbm24a-target mRNAs. The results show that the examined mRNAs are all expressed in the otic vesicles of wild-type controls (Figures 8A,B). Meanwhile, the expression of dfna5b, epcam, casp6a, lrrc23, enosf1, and smpx is decreased in the rbm24a mutants, whereas the expression of dnah7 is increased in the mutants, which is consistent with the RNAseq, RT-PCR, and qPCR results (Figures 8A,B).
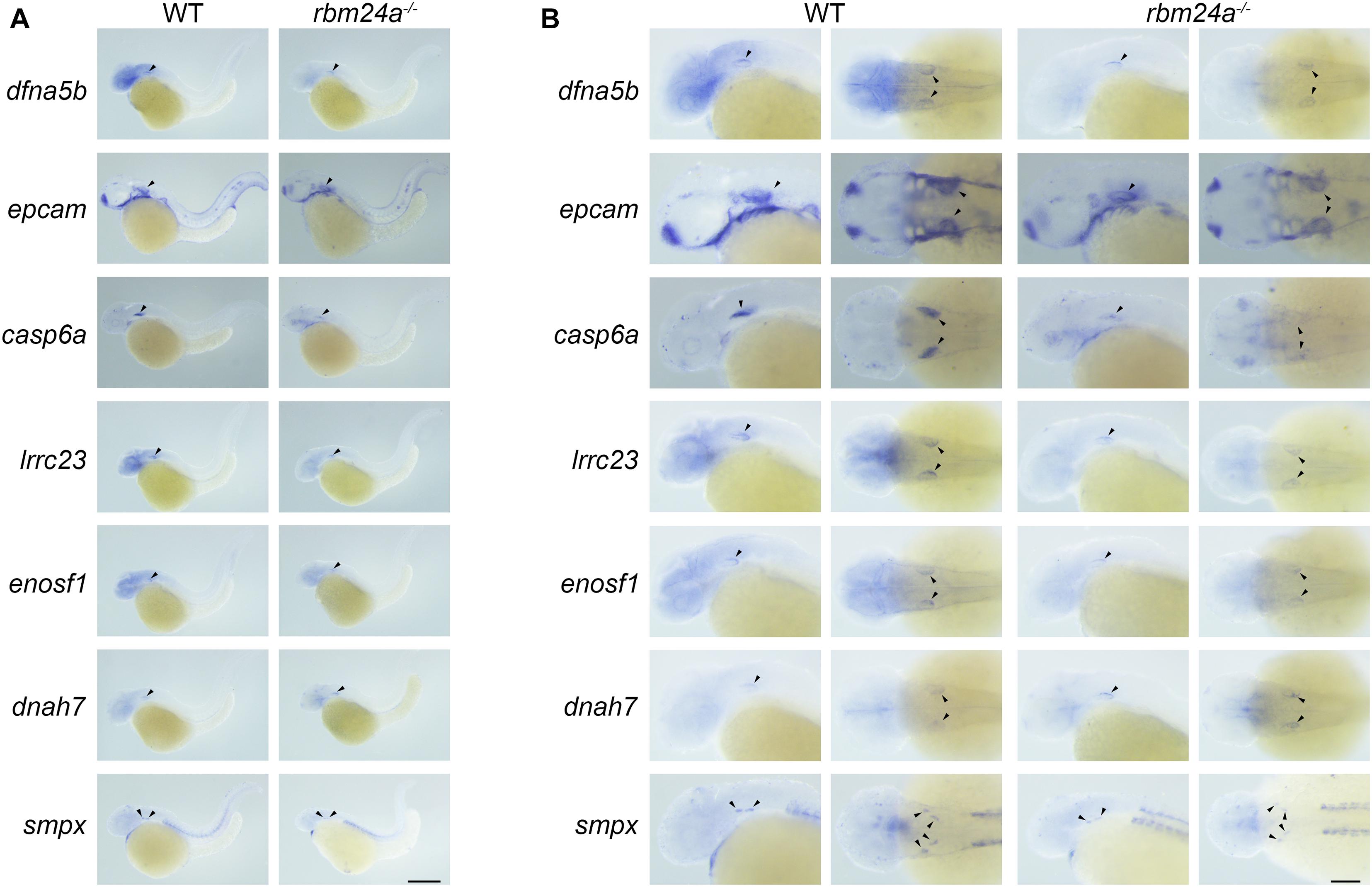
Figure 8. Expression pattern of important Rbm24a target mRNAs in the zebrafish. In situ hybridization of zebrafish embryos at 34 hpf was performed to examine the expression of dfna5b, epcam, casp6a, lrrc23, enosf1, dnah7, and smpx. (A) Lower magnification images. (B) Higher magnification images. Scale bars, 0.25 mm in (A) and 0.1 mm in (B).
Discussion
The physiological role of Rbm24 has been intensively investigated in the heart and skeletal muscles. Rbm24 knockdown in C2C12 cells, Xenopus, or chick somatic myogenic progenitor cells affects myogenic differentiation (Miyamoto et al., 2009; Li et al., 2010; Grifone et al., 2014). Rbm24 knockdown in zebrafish leads to deficits in cardiovascular development and somitogenesis (Maragh et al., 2011, 2014; Poon et al., 2012). Furthermore, inactivation of Rbm24 in mice disrupts cardiac development through activating aberrant p53-dependent apoptosis (Yang et al., 2014; Zhang et al., 2018). Recently, we and others reported that Rbm24a plays pivotal roles in eye development of zebrafish through regulating mRNA stability and translation efficiency (Dash et al., 2020; Shao et al., 2020). In the present work, we report that Rbm24a is indispensable for hair cell development through regulating mRNA stability in zebrafish.
Expression of rbm24a could be detected in the otic visicles during early embryonic development. However, the morphology of otic visicle is largely unaffected in rbm24a mutants, suggesting that Rbm24a is dispensable for early inner ear development in the zebrafish. Nevertheless, otolith formation and semicircular canal fusion are delayed in rbm24a mutants. Otoliths are biomineralized aggregates of calcium carbonate and proteins sitting over hair cells in the maculae, embedded in the otolithic membrane (Schulz-Mirbach et al., 2019). Otolith formation in the zebrafish requires kinocilia and motile cilia, and defects in otolith formation is a characteristic phenotype of ciliary mutants (Stooke-Vaughan et al., 2012; Whitfield, 2020). For example, otolith formation is delayed in zebrafish deficient for lrrc23 gene that encodes a radial spoke protein of the cilia (Han et al., 2018). Our data show that lrrc23 mRNA is bound with Rbm24a and its level is decreased in rbm24a mutants, suggesting that Rbm24a might affect otolith formation through regulating lrrc23 mRNA stability. Another potential Rbm24a-target that might contribute to the otolith phenotype is dnah7, which encodes dynein heavy chain 7, an inner arm component of cilia (Zhang et al., 2002). Interestingly, dnah7 is one of the few genes whose expression is upregulated in rbm24a mutants.
Another important Rbm24a-target is dfna5b. Mutations in human DFNA5 gene are associated with nonsyndromic hearing loss (Van Laer et al., 1998). There are two DFNA5 orthologs in the zebrafish genome, dfna5a and dfna5b, among which dfna5b shows higher homology with human DFNA5. Dfna5b is expressed in the inner ear, and dfna5b knockdown leads to malformation of the semicircular canals (Busch-Nentwich et al., 2004). Given that both rbm24a mutation and dfna5b knockdown affect the development of semicircular canals, and that dfna5b expression is downregulated in rbm24a mutant larvae, we hypothesize that Rbm24a regulates semicircular canal development through affecting the stability of dfna5b mRNA. Meantime, dfna5a shows different expression pattern from dfna5b. Our in situ hybridization results show that dfna5a is expressed in a circular pattern at the lateral line neuromasts, and its expression level is largely unaffected in rbm24a mutants.
Rbm24a is also expressed in the hair cells of the inner ear and the lateral line neuromasts. Our data show that rbm24a deficiency results in decreased hair cell numbers and disorganized hair bundle, suggesting that Rbm24a is indispensable for hair cell development. Consistently, the MET function of hair cells is compromised in rbm24a mutants. As a result, both the auditory and vestibular function are affected in rbm24a mutants. RNAseq reveals that many inner ear-expressed genes are dysregulated in rbm24a mutants, which might contribute to the hair cell phenotypes. An interesting candidate is smpx, which is highly expressed in the hair cells of both mice and zebrafish (Yoon et al., 2011; Ghilardi et al., 2020). SMPX gene mutations are associated with nonsyndromic hearing loss DFNX4 (Huebner et al., 2011; Schraders et al., 2011). Smpx protein has been localized at the actin-based cuticular plate, a specialized structure at the apical surface of hair cells (Ghilardi et al., 2020). The cuticular plate is important for stabilizing the stereocilia, and its deficits could lead to stereocilia abnormalities (Pollock and McDermott, 2015; Du et al., 2019). Our present data suggest that Rbm24a might affect stereocilia development through regulating the stability of smpx mRNA. Further investigations are warranted to learn more about the underlying mechanism.
Our in situ hybridization results show that the number of dfna5a+ neuromasts is slightly decreased in rbm24a mutants. Similar pattern is also observed when GFP-positive neuromast hair cells are examined in brn3c:GFP background zebrafish. The slightly decreased pLL neuromast number could be explained by the shortened body length of rbm24a mutants. Rbm24a is indispensable for heart development, and rbm24a mutants suffer from severe heart defects and do not survive beyond 9 dpf (Shao et al., 2020). The severe overall sickness of rbm24a mutants might lead to the shortened body length before their eventual death. In situ hybridization results also reveal more significant decrease of myo7aa+ neuromast number in rbm24a mutants, which might be a result of reduced myo7aa+ hair cell numbers in each neuromast.
Rbm24 has been shown to be a major regulator of alternative splicing (Yang et al., 2014). Recently we showed that Rbm24 regulates the alternative splicing of Cdh23 exon 68 in mice (Li et al., 2020). CDH23 forms the upper part of tip links that play a pivotal role in hair cell MET (Assad et al., 1991; Siemens et al., 2004; Kazmierczak et al., 2007). Mutations in CDH23 gene have been associated with hearing loss in human, mice and zebrafish (Bolz et al., 2001; Bork et al., 2001; Di Palma et al., 2001; Sollner et al., 2004). However, our RNAseq analyses do not reveal any significant changes of alternative splicing in rbm24a mutants. We therefore performed RT-PCR experiments to examine the alternative splicing of cdh23 gene in rbm24a mutants. The results show that the inclusion of cdh23 exon 68 in rbm24a mutants is indeed decreased than that in wild type zebrafish (Supplementary Figures 6A,B). However, the remained cdh23 exon 68 inclusion suggests that altered splicing of cdh23 gene is not a main contributor to the auditory and vestibular phenotypes in rbm24a mutants.
In conclusion, our present work reveal that Rbm24a is an important post-transcriptional regulator in the hair cells, and that Rbm24a deficiency affects hair cell development in zebrafish through regulating RNA stability.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by Animal Ethics Committee of Shandong University School of Life Sciences.
Author Contributions
ZX: study concept and design. YZ, YW, XY, and CW: acquisition of data. YZ, FC, DL, MS, and ZX: analysis and interpretation of data. YZ and ZX: drafting the manuscript. MS and ZX: study supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Key Basic Research Program of China (2018YFC1003600), the National Natural Science Foundation of China (81771001), and the Fundamental Research Funds of Shandong University (2018JC025).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Sen Wang, Xiaomin Zhao, and Haiyan Yu from the core facilities for life and environmental sciences, Shandong University for the technical support in SEM and confocal microscopy.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.604026/full#supplementary-material
References
Assad, J. A., Shepherd, G. M. G., and Corey, D. P. (1991). Tip-link integrity and mechanical transduction in vertebrate hair-cells. Neuron 7, 985–994. doi: 10.1016/0896-6273(91)90343-x
Bermingham, N. A., Hassan, B. A., Price, S. D., Vollrath, M. A., Ben-Arie, N., Eatock, R. A., et al. (1999). Math1: an essential gene for the generation of inner ear hair cells. Science 284, 1837–1841. doi: 10.1126/science.284.5421.1837
Beurg, M., Fettiplace, R., Nam, J. H., and Ricci, A. J. (2009). Localization of inner hair cell mechanotransducer channels using high-speed calcium imaging. Nat. Neurosci. 12, 553–558. doi: 10.1038/nn.2295
Bolz, H., von Brederlow, B., Ramirez, A., Bryda, E. C., Kutsche, K., Nothwang, H. G., et al. (2001). Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat. Genet. 27, 108–112. doi: 10.1038/83667
Bork, J. M., Peters, L. M., Riazuddin, S., Bernstein, S. L., Ahmed, Z. M., Ness, S. L., et al. (2001). Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am. J. Hum. Genet. 68, 26–37. doi: 10.1086/316954
Brinegar, A. E., and Cooper, T. A. (2016). Roles for RNA-binding proteins in development and disease. Brain Res. 1647, 1–8. doi: 10.1016/j.brainres.2016.02.050
Busch-Nentwich, E., Sollner, C., Roehl, H., and Nicolson, T. (2004). The deafness gene dfna5 is crucial for ugdh expression and HA production in the developing ear in zebrafish. Development 131, 943–951. doi: 10.1242/dev.00961
Cai, T., Jen, H. I., Kang, H., Klisch, T. J., Zoghbi, H. Y., and Groves, A. K. (2015). Characterization of the transcriptome of nascent hair cells and identification of direct targets of the Atoh1 transcription factor. J. Neurosci. 35, 5870–5883. doi: 10.1523/jneurosci.5083-14.2015
Dash, S., Brastrom, L. K., Patel, S. D., Scott, C. A., Slusarski, D. C., and Lachke, S. A. (2020). The master transcription factor SOX2, mutated in anophthalmia/microphthalmia, is post-transcriptionally regulated by the conserved RNA-binding protein RBM24 in vertebrate eye development. Hum. Mol. Genet. 29, 591–604. doi: 10.1093/hmg/ddz278
Di Palma, F., Holme, R. H., Bryda, E. C., Belyantseva, I. A., Pellegrino, R., Kachar, B., et al. (2001). Mutations in Cdh23, encoding a new type of cadherin, cause stereocilia disorganization in waltzer, the mouse model for Usher syndrome type 1D. Nat. Genet. 27, 103–107. doi: 10.1038/83660
Du, H., Zou, L., Ren, R., Li, N., Li, J., Wang, Y., et al. (2020). Lack of PDZD7 long isoform disrupts ankle-link complex and causes hearing loss in mice. FASEB J. 34, 1136–1149. doi: 10.1096/fj.201901657rr
Du, T. T., Dewey, J. B., Wagner, E. L., Cui, R., Heo, J., Park, J. J., et al. (2019). LMO7 deficiency reveals the significance of the cuticular plate for hearing function. Nat. Commun. 10:1117.
Ekker, M., Akimenko, M. A., Bremiller, R., and Westerfield, M. (1992). Regional expression of three homeobox transcripts in the inner ear of zebrafish embryos. Neuron 9, 27–35. doi: 10.1016/0896-6273(92)90217-2
Elkon, R., Milon, B., Morrison, L., Shah, M., Vijayakumar, S., Racherla, M., et al. (2015). RFX transcription factors are essential for hearing in mice. Nat. Commun. 6:8549.
Erkman, L., McEvilly, R. J., Luo, L., Ryan, A. K., Hooshmand, F., O’Connell, S. M., et al. (1996). Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature 381, 603–606. doi: 10.1038/381603a0
Ernest, S., Rauch, G. J., Haffter, P., Geisler, R., Petit, C., and Nicolson, T. (2000). Mariner is defective in myosin VIIA: a zebrafish model for human hereditary deafness. Hum. Mol. Genet. 9, 2189–2196. doi: 10.1093/hmg/9.14.2189
Fetka, I., Radeghieri, A., and Bouwmeester, T. (2000). Expression of the RNA recognition motif-containing protein SEB-4 during Xenopus embryonic development. Mech. Dev. 94, 283–286. doi: 10.1016/s0925-4773(00)00284-7
Gale, J. E., Marcotti, W., Kennedy, H. J., Kros, C. J., and Richardson, G. P. (2001). FM1-43 dye behaves as a permeant blocker of the hair-cell mechanotransducer channel. J. Neurosci. 21, 7013–7025. doi: 10.1523/jneurosci.21-18-07013.2001
Ghilardi, A., Diana, A., Prosperi, L., and Del Giacco, L. (2020). Expression pattern of the small muscle protein, X-linked (smpx) gene during zebrafish embryonic and larval developmental stages. Gene Expr. Patterns 36:119110. doi: 10.1016/j.gep.2020.119110
Grifone, R., Saquet, A., Xu, Z., and Shi, D. L. (2018). Expression patterns of Rbm24 in lens, nasal epithelium, and inner ear during mouse embryonic development. Dev. Dyn. 247, 1160–1169. doi: 10.1002/dvdy.24666
Grifone, R., Xie, X., Bourgeois, A., Saquet, A., Duprez, D., and Shi, D. L. (2014). The RNA-binding protein Rbm24 is transiently expressed in myoblasts and is required for myogenic differentiation during vertebrate development. Mech. Dev. 134, 1–15. doi: 10.1016/j.mod.2014.08.003
Han, X., Xie, H., Wang, Y., and Zhao, C. (2018). Radial spoke proteins regulate otolith formation during early zebrafish development. FASEB J. 32, 3984–3992. doi: 10.1096/fj.201701359r
He, Y. Z., Lu, X. L., Qian, F. P., Liu, D., Chai, R. J., and Li, H. W. (2017). Insm1a is required for zebrafish posterior lateral line development. Front. Mol. Neurosci. 10:241. doi: 10.3389/fnmol.2017.00241
Hentze, M. W., Castello, A., Schwarzl, T., and Preiss, T. (2018). A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327–341. doi: 10.1038/nrm.2017.130
Huebner, A. K., Gandia, M., Frommolt, P., Maak, A., Wicklein, E. M., Thiele, H., et al. (2011). Nonsense mutations in SMPX, encoding a protein responsive to physical force, result in X-chromosomal hearing loss. Am. J. Hum. Genet. 88, 621–627. doi: 10.1016/j.ajhg.2011.04.007
Ito, J., Iijima, M., Yoshimoto, N., Niimi, T., Kuroda, S., and Maturana, A. D. (2016). RBM20 and RBM24 cooperatively promote the expression of short enh splice variants. FEBS Lett. 590, 2262–2274. doi: 10.1002/1873-3468.12251
Jiang, L., Romero-Carvajal, A., Haug, J. S., Seidel, C. W., and Piotrowski, T. (2014). Gene-expression analysis of hair cell regeneration in the zebrafish lateral line. Proc. Natl. Acad. Sci. U.S.A. 111, E1383–E1392.
Jiang, Y., Zhang, M., Qian, Y., Xu, E., Zhang, J., and Chen, X. (2014). Rbm24, an RNA-binding protein and a target of p53, regulates p21 expression via mRNA stability. J. Biol. Chem. 289, 3164–3175. doi: 10.1074/jbc.m113.524413
Jones, C., Roper, V. C., Foucher, I., Qian, D., Banizs, B., Petit, C., et al. (2008). Ciliary proteins link basal body polarization to planar cell polarity regulation. Nat. Genet. 40, 69–77. doi: 10.1038/ng.2007.54
Kazmierczak, P., Sakaguchi, H., Tokita, J., Wilson-Kubalek, E. M., Milligan, R. A., Muller, U., et al. (2007). Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature 449, 87–91. doi: 10.1038/nature06091
Kimmel, C. B., Patterson, J., and Kimmel, R. O. (1974). The development and behavioral characteristics of the startle response in the zebra fish. Dev. Psychobiol. 7, 47–60. doi: 10.1002/dev.420070109
Li, H. Y., Bourdelas, A., Carron, C., and Shi, D. L. (2010). The RNA-binding protein Seb4/RBM24 is a direct target of MyoD and is required for myogenesis during Xenopus early development. Mech. Dev. 127, 281–291. doi: 10.1016/j.mod.2010.03.002
Li, N., Du, H., Ren, R., Wang, Y., and Xu, Z. (2020). Alternative splicing of Cdh23 exon 68 is regulated by RBM24, RBM38, and PTBP1. Neural Plast. 2020:8898811.
Lin, Y., Tan, K. T., Liu, J., Kong, X., Huang, Z., and Xu, X. Q. (2018). Global profiling of Rbm24 bound RNAs uncovers a multi-tasking RNA binding protein. Int. J. Biochem. Cell Biol. 94, 10–21. doi: 10.1016/j.biocel.2017.11.002
Liu, J., Kong, X., Zhang, M., Yang, X., and Xu, X. (2019). RNA binding protein 24 deletion disrupts global alternative splicing and causes dilated cardiomyopathy. Protein Cell 10, 405–416. doi: 10.1007/s13238-018-0578-8
Maragh, S., Miller, R. A., Bessling, S. L., McGaughey, D. M., Wessels, M. W., de Graaf, B., et al. (2011). Identification of RNA binding motif proteins essential for cardiovascular development. BMC Dev. Biol. 11:62. doi: 10.1186/1471-213X-11-62
Maragh, S., Miller, R. A., Bessling, S. L., Wang, G., Hook, P. W., and McCallion, A. S. (2014). Rbm24a and Rbm24b are required for normal somitogenesis. PLoS One 9:e105460. doi: 10.1371/journal.pone.0105460
Meyers, J. R., MacDonald, R. B., Duggan, A., Lenzi, D., Standaert, D. G., Corwin, J. T., et al. (2003). Lighting up the senses: FM1-43 loading of sensory cells through nonselective ion channels. J. Neurosci. 23, 4054–4065. doi: 10.1523/jneurosci.23-10-04054.2003
Miyamoto, S., Hidaka, K., Jin, D., and Morisaki, T. (2009). RNA-binding proteins Rbm38 and Rbm24 regulate myogenic differentiation via p21-dependent and -independent regulatory pathways. Genes Cells 14, 1241–1252. doi: 10.1111/j.1365-2443.2009.01347.x
Mo, W., Chen, F., Nechiporuk, A., and Nicolson, T. (2010). Quantification of vestibular-induced eye movements in zebrafish larvae. BMC Neurosci. 11:110. doi: 10.1186/1471-2202-11-110
Moayedi, Y., Basch, M. L., Pacheco, N. L., Gao, S. S., Wang, R., Harrison, W., et al. (2014). The candidate splicing factor Sfswap regulates growth and patterning of inner ear sensory organs. PLoS Genet. 10:e1004055. doi: 10.1371/journal.pgen.1004055
Müller, U., and Barr-Gillespie, P. G. (2015). New treatment options for hearing loss. Nat. Rev. Drug Discov. 14, 346–365. doi: 10.1038/nrd4533
Nakano, Y., Jahan, I., Bonde, G., Sun, X., Hildebrand, M. S., Engelhardt, J. F., et al. (2012). A mutation in the Srrm4 gene causes alternative splicing defects and deafness in the Bronx waltzer mouse. PLoS Genet. 8:e1002966. doi: 10.1371/journal.pgen.1002966
Nicolson, T. (2005). The genetics of hearing and balance in zebrafish. Ann. Rev. Genet. 39, 9–22. doi: 10.1146/annurev.genet.39.073003.105049
Pfeffer, P. L., Gerster, T., Lun, K., Brand, M., and Busslinger, M. (1998). Characterization of three novel members of the zebrafish Pax2/5/8 family: dependency of Pax5 and Pax8 expression on the Pax2.1 (noi) function. Development 125, 3063–3074.
Pollock, L. M., and McDermott, B. M. Jr. (2015). The cuticular plate: a riddle, wrapped in a mystery, inside a hair cell. Birth Defects Res. C. Embryo Today 105, 126–139. doi: 10.1002/bdrc.21098
Poon, K. L., Tan, K. T., Wei, Y. Y., Ng, C. P., Colman, A., Korzh, V., et al. (2012). RNA-binding protein RBM24 is required for sarcomere assembly and heart contractility. Cardiovasc. Res. 94, 418–427. doi: 10.1093/cvr/cvs095
Riley, B. B., Zhu, C., Janetopoulos, C., and Aufderheide, K. J. (1997). A critical period of ear development controlled by distinct populations of ciliated cells in the zebrafish. Dev. Biol. 191, 191–201. doi: 10.1006/dbio.1997.8736
Rohacek, A. M., Bebee, T. W., Tilton, R. K., Radens, C. M., McDermott-Roe, C., Peart, N., et al. (2017). ESRP1 mutations cause hearing loss due to defects in alternative splicing that disrupt cochlear development. Dev. Cell 43, 318–331. doi: 10.1016/j.devcel.2017.09.026
Scheffer, D. I., Shen, J., Corey, D. P., and Chen, Z. Y. (2015). Gene expression by mouse inner ear hair cells during development. J. Neurosci. 35, 6366–6380. doi: 10.1523/jneurosci.5126-14.2015
Schraders, M., Haas, S. A., Weegerink, N. J., Oostrik, J., Hu, H., Hoefsloot, L. H., et al. (2011). Next-generation sequencing identifies mutations of SMPX, which encodes the small muscle protein, X-linked, as a cause of progressive hearing impairment. Am. J. Hum. Genet. 88, 628–634. doi: 10.1016/j.ajhg.2011.04.012
Schulz-Mirbach, T., Ladich, F., Plath, M., and Hess, M. (2019). Enigmatic ear stones: what we know about the functional role and evolution of fish otoliths. Biol. Rev. Camb. Philos. Soc. 94, 457–482. doi: 10.1111/brv.12463
Shao, M., Lu, T., Zhang, C., Zhang, Y. Z., Kong, S. H., and Shi, D. L. (2020). Rbm24 controls poly(A) tail length and translation efficiency of crystallin mRNAs in the lens via cytoplasmic polyadenylation. Proc. Natl. Acad. Sci. U.S.A. 117, 7245–7254. doi: 10.1073/pnas.1917922117
Shen, J., Scheffer, D. I., Kwan, K. Y., and Corey, D. P. (2015). SHIELD: an integrative gene expression database for inner ear research. Database Oxford 2015:bav071. doi: 10.1093/database/bav071
Siemens, J., Lillo, C., Dumont, R. A., Reynolds, A., Williams, D. S., Gillespie, P. G., et al. (2004). Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature 428, 950–955. doi: 10.1038/nature02483
Sollner, C., Rauch, G. J., Siemens, J., Geisler, R., Schuster, S. C., Muller, U., et al. (2004). Mutations in cadherin 23 affect tip links in zebrafish sensory hair cells. Nature 428, 955–959. doi: 10.1038/nature02484
Stooke-Vaughan, G. A., Huang, P., Hammond, K. L., Schier, A. F., and Whitfield, T. T. (2012). The role of hair cells, cilia and ciliary motility in otolith formation in the zebrafish otic vesicle. Development 139, 1777–1787. doi: 10.1242/dev.079947
Sun, P., Zhang, Y., Zhao, F., Wu, J. P., Pun, S. H., Peng, C., et al. (2018). An assay for systematically quantifying the vestibulo-ocular reflex to assess vestibular function in zebrafish larvae. Front. Cell. Neurosci. 12:257. doi: 10.3389/fncel.2018.00257
Thisse, C., and Thisse, B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69. doi: 10.1038/nprot.2007.514
van den Hoogenhof, M. M. G., van der Made, I., de Groot, N. E., Damanafshan, A., van Amersfoorth, S. C. M., Zentilin, L., et al. (2018). AAV9-mediated Rbm24 overexpression induces fibrosis in the mouse heart. Sci. Rep. 8:11696.
Van Laer, L., Huizing, E. H., Verstreken, M., van Zuijlen, D., Wauters, J. G., Bossuyt, P. J., et al. (1998). Nonsyndromic hearing impairment is associated with a mutation in DFNA5. Nat. Genet. 20, 194–197. doi: 10.1038/2503
Wallis, D., Hamblen, M., Zhou, Y., Venken, K. J., Schumacher, A., Grimes, H. L., et al. (2003). The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development 130, 221–232. doi: 10.1242/dev.00190
Wang, C., Zhong, Z., Sun, P., Zhong, H., Li, H., and Chen, F. (2017). Evaluation of the hair cell regeneration in zebrafish larvae by measuring and quantifying the startle responses. Neural Plast. 2017:8283075.
Waterman, R. E., and Bell, D. H. (1984). Epithelial fusion during early semicircular canal formation in the embryonic zebrafish, Brachydanio rerio. Anat. Rec. 210, 101–114. doi: 10.1002/ar.1092100113
Whitfield, T. T. (2020). Cilia in the developing zebrafish ear. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 375:20190163. doi: 10.1098/rstb.2019.0163
Xing, Y. Y., Cheng, X. N., Li, Y. L., Zhang, C., Saquet, A., Liu, Y. Y., et al. (2018). Mutational analysis of dishevelled genes in zebrafish reveals distinct functions in embryonic patterning and gastrulation cell movements. PLoS Genet. 14:e1007551. doi: 10.1371/journal.pgen.1007551
Xu, E., Zhang, J., Zhang, M., Jiang, Y., Cho, S. J., and Chen, X. (2014). RNA-binding protein RBM24 regulates p63 expression via mRNA stability. Mol. Cancer Res. 12, 359–369. doi: 10.1158/1541-7786.mcr-13-0526
Yang, J., Hung, L. H., Licht, T., Kostin, S., Looso, M., Khrameeva, E., et al. (2014). RBM24 is a major regulator of muscle-specific alternative splicing. Dev. Cell 31, 87–99. doi: 10.1016/j.devcel.2014.08.025
Yoon, H., Lee, D. J., Kim, M. H., and Bok, J. (2011). Identification of genes concordantly expressed with Atoh1 during inner ear development. Anat. Cell Biol. 44, 69–78. doi: 10.5115/acb.2011.44.1.69
Zeddies, D. G., and Fay, R. R. (2005). Development of the acoustically evoked behavioral response in zebrafish to pure tones. J. Exp. Biol. 208, 1363–1372. doi: 10.1242/jeb.01534
Zhang, M., Zhang, Y., Xu, E., Mohibi, S., de Anda, D. M., Jiang, Y., et al. (2018). Rbm24, a target of p53, is necessary for proper expression of p53 and heart development. Cell Death Differ. 25, 1118–1130. doi: 10.1038/s41418-017-0029-8
Zhang, T., Lin, Y., Liu, J., Zhang, Z. G., Fu, W., Guo, L. Y., et al. (2016). Rbm24 regulates alternative splicing switch in embryonic stem cell cardiac lineage differentiation. Stem Cells 34, 1776–1789. doi: 10.1002/stem.2366
Zhang, Y. J., O’Neal, W. K., Randell, S. H., Blackburn, K., Moyer, M. B., Boucher, R. C., et al. (2002). Identification of dynein heavy chain 7 as an inner arm component of human cilia that is synthesized but not assembled in a case of primary ciliary dyskinesia. J. Biol. Chem. 277, 17906–17915. doi: 10.1074/jbc.m200348200
Keywords: Rbm24a, hair cells, inner ear, lateral line, mRNA stability
Citation: Zhang Y, Wang Y, Yao X, Wang C, Chen F, Liu D, Shao M and Xu Z (2020) Rbm24a Is Necessary for Hair Cell Development Through Regulating mRNA Stability in Zebrafish. Front. Cell Dev. Biol. 8:604026. doi: 10.3389/fcell.2020.604026
Received: 08 September 2020; Accepted: 01 December 2020;
Published: 17 December 2020.
Edited by:
Qingyin Zheng, Case Western Reserve University, United StatesReviewed by:
Artur A. Indzhykulian, Massachusetts Eye and Ear Infirmary, Harvard Medical School, United StatesSuk-Won Jin, Gwangju Institute of Science and Technology, South Korea
Shan Sun, Fudan University, China
Copyright © 2020 Zhang, Wang, Yao, Wang, Chen, Liu, Shao and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Shao, c2hhb21pbmdAc2R1LmVkdS5jbg==; Zhigang Xu, eHV6Z0BzZHUuZWR1LmNu
 Yan Zhang
Yan Zhang Yanfei Wang
Yanfei Wang Xuebo Yao
Xuebo Yao Changquan Wang2
Changquan Wang2 Fangyi Chen
Fangyi Chen Ming Shao
Ming Shao Zhigang Xu
Zhigang Xu