
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Cell Dev. Biol. , 08 October 2020
Sec. Stem Cell Research
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.575382
This article is part of the Research Topic Hair Follicle Stem Cell Regeneration in Aging View all 21 articles
Reciprocal interactions between hair-inductive dermal cells and epidermal cells are essential for de novo genesis of hair follicles. Recent studies have shown that outer root sheath (ORS) follicular keratinocytes can be expanded in vitro, but the cultured cells often lose receptivity to hair-inducing dermal signals. In this study, we first investigated whether the hair-inductive activity (trichogenicity) of cultured human ORS follicular keratinocytes was correlated with the cultivation period. ORS follicular keratinocytes from the scalp were cultured for 3, 4, 5, or 6 weeks and were then implanted into nude mice along with freshly isolated neonatal mouse dermal cells. We observed that the trichogenicity of the implanted ORS cells was inversely correlated with their cultivation period. These initial findings prompted us to investigate the differentially expressed genes between the short-term (20 days) and long-term (42 days) cultured ORS cells, trichogenic and non-trichogenic, respectively, by microarray analysis. We found that forkhead box protein A2 (FOXA2) was the most up-regulated transcription factor in the trichogenic ORS cells. Thus, we investigated whether the trichogenicity of the cells was affected by FOXA2 expression. We found a significant decrease in the number of induced hair follicles when the ORS cells were transfected with a FOXA2 small interfering RNA versus control small interfering RNA. Taken together, our data strongly suggest that FOXA2 significantly influences the trichogenicity of human ORS cells.
Reciprocal interactive events between the dermal mesenchyme and the overlying epithelium are essential for hair follicle (HF) morphogenesis (Paus and Cotsarelis, 1999; Millar, 2002). Likewise, reciprocal interactions between hair-inductive dermal cells and epithelial cells are needed for de novo genesis or neogenesis of HFs (Yang and Cotsarelis, 2010). However, these cells often lose hair-inductive potency (trichogenicity) after conventional in vitro culture (Ohyama et al., 2010; Yang and Cotsarelis, 2010). Therefore, to generate HFs by implantation of cells in recipient skin, restoration or maintenance of the trichogenicity of those cells together with expansion of cell numbers is required.
Several strategies have been attempted to restore the trichogenicity of cultured follicular dermal cells, especially dermal papilla (DP) cells, and at least partial restoration was achieved, particularly by spheroid formation and complementation of activators of DP signature pathways (Kang et al., 2012; Ohyama et al., 2012; Higgins et al., 2013; Ohyama and Veraitch, 2013). Regarding epithelial cells for neogenesis of HFs, follicular outer root sheath (ORS) keratinocytes containing hair follicle stem cells (HFSCs) are considered the optimal cells. However, thus far, reports on the restoration of trichogenicity of cultured ORS cells are rare. Chan et al. from Lin’s laboratory have shown that coculture of high-passage rat ORS cells with rat vibrissae DP cells restores trichogenicity (Chan et al., 2015). More recently, we have also shown that the trichogenicity of human ORS cells from occipital scalp can be restored by coculturing with human DP cells (Bak et al., 2018).
In this study, we observed that forkhead box protein A2 (FOXA2), a member of the forkhead/winged-helix family of transcription factors, is the most differentially expressed transcription factor between the short-term and long-term cultured ORS cells by microarray analysis. This, along with inverse correlation of hair-inducing capacity of ORS cells with their cultivation period, led us to investigate whether the trichogenicity of the cells was affected by FOXA2 expression. Using a hair reconstitution assay in combination with a FOXA2 small interfering RNA (siRNA)-mediated knockdown of ORS cells, we show that FOXA2 augments the trichogenicity of cultured human follicular keratinocytes.
Biopsy specimens were obtained from non-balding (occipital) scalp of male patients undergoing hair transplantation surgery for androgenetic alopecia. The medical ethical committee of the Kyungpook National University Hospital (Daegu, South Korea) approved all described studies. Written informed consent was obtained from study participants. HFs were isolated using a previously described method with minor modifications (Philpott et al., 1994; Magerl et al., 2002). Briefly, the subcutaneous fat portion of the scalp skin, including the lower HFs, was dissected from the epidermis and dermis. HFs were then isolated under a binocular microscope by forceps.
The hair shaft and hair bulb regions of the HFs were cut off to prevent contamination with other cells. The ORS cells were cultured as described before (Kwack et al., 2008). Trimmed hair follicles were immersed in Dulbecco’s modified eagle medium supplemented with 20% fetal bovine serum in Biocoat collagen type I-coated tissue culture dishes (Corning, Kennebunk, ME, United States). On the third day of culture, the medium was changed to EpiLife (Gibco-BRL, Gaithersburg, MD, United States) containing 1% antibiotic-antimycotic solution and 1% EpiLife defined growth supplement. Once subconfluent, the cells were harvested with 0.25% trypsin/10 mM EDTA in phosphate-buffered saline (PBS), split at a 1:5 ratio, and maintained in EpiLife medium. ORS cells in passages 1–4 were used in this study.
The siRNAs used in this study were purchased from Bioneer (Daejeon, South Korea). Cultured human ORS cells were transfected with a control siRNA or a FOXA2 siRNA (Bioneer) at a final concentration of 10 nM in the presence of RNAiMAX reagent (Invitrogen, Carlsbad, CA, United States) for 24 h. Transfected cells (104) were harvested 48 h after transfection and were used for a hair reconstitution assay after the confirmation of a significant knockdown of the genes by real-time polymerase chain reaction (PCR) analysis.
Patch hair reconstitution assays were used in this study (Zheng et al., 2005). Dorsal skin from C57BL/6 neonates was collected and incubated overnight with 1 mg/ml collagenase/dispase (Roche, Mannheim, Germany). The dermis and epidermis were separated by incubating the skin with 0.25% trypsin/10 mM EDTA in PBS at 37°C for 15 min. Dermal cells were filtered through 100 μm cell strainers (Becton Dickinson, Franklin, NJ, United States), and then centrifuged at 1,500 rpm for 5 min. Cultured human ORS cells (1 × 106 cells) were combined with freshly isolated neonatal mouse dermal cells (1 × 106 cells) and then subcutaneously implanted into the skin on the backs of 7-week-old female nude mice. After 2 weeks, photos were taken of excised skin samples under a stereoscope, and the reconstituted HFs were quantified. All animal procedures were approved by the Animal Care and Use Committee at Kyungpook National University (Daegu, South Korea).
Five micrograms of total RNA from short-term (20 days) and long-term (42 days) ORS cell cultures was used for labeling. Probe synthesis, hybridization, detection, and scanning were performed as described previously (Bak et al., 2018) according to standard protocols from Affymetrix, Inc (Santa Clara, CA, United States). Microarray data were analyzed as we conducted in previous studies (Choi et al., 2017; Wang et al., 2017). Briefly, a bubble chart was used to present biological processes revealed by gene ontology (GO) analysis. The size of circle is dependent on total number of the associated differentially expressed genes (DEGs), while the rank of the circle is set according to P-value of analysis. Genes that were significantly associated with these processes were used to compare DEGs with gene set enrichment analysis (GSEA) information related to the skin-associated gene sets as well. Kyoto Encyclopedia of Genes and Genomes (KEGG) 2019 was used for pathway analysis.
Total RNA was isolated using RNeasy Mini Kit (Qiagen, Austin, TX, United States) according to the manufacturer’s protocol. The purity and integrity of the RNA was measured using a NanoDrop (NanoDrop Technologies, Wilmington, DE, United States). Complementary DNA (cDNA) was synthesized from 3 μg total RNA using a cDNA synthesis kit containing Improm-II reverse transcriptase and oligo-dT primers according to the manufacturer’s instructions (Promega, Madison, WI, United States). Real-time PCR was performed using a StepOnePlus Real-time PCR System (Applied Biosystems, Foster City, CA, United States). All reactions were performed using Power SYBR Green premix (Applied Biosystems), 10 ng cDNA, and 10 μM primers. Amplification was performed under the following cycling conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Primer sequences used were as follows: FOXA2, forward primer 5′-CCCCACAAAATGGACCTCAAG-3′ and reverse primer 5′-GAGTACACCCCCTGGTAGTAG-3′; and GAPDH, forward primer 5′-TGGAAATCCCATCACCATCTTC-3′ and reverse primer 5′-CGCCCCACTTGATTTTGG-3′. Differences between samples and controls were calculated using StepOnePlus Real-Time PCR analysis software (Applied Biosystems).
Cultured human ORS cells were plated in each eight-chamber slide (Nunc, Rosklide, Denmark) at a density of 100,000 cells per well. After cultivation in EpiLife medium for 24 h, the chamber slides were fixed with methanol for 20 min at −20°C and equilibrated in PBS for 10 min at 25°C. After blocking with 4% bovine serum albumin (Jackson) for 1 h at 25°C, the slides were incubated with a rabbit polyclonal antibody to SOX9 (1:100 dilution; Abcam) at 4°C overnight, washed four times with PBS, and incubated with donkey anti-rabbit horseradish peroxidase-conjugated antibody (1:100 dilution; Jackson ImmunoResearch, West Grove, PA, United States) for 1 h at 25°C. AEC substrate chromogen (DAKO, Glostrup, Denmark) was used as a color formation reagent for horseradish peroxidase. Finally, slides were counterstained with hematoxylin for 2 min. We used normal rabbit IgG (R&D Systems, Minneapolis, CA, United States) for immunostaining negative controls.
Data are expressed as mean ± standard deviation (SD). Differences between pairs of groups were analyzed by Student’s t-test. A P-value of less than 0.05 was considered statistically significant.
Trimmed HFs (25–30 HFs/10 cm2 dish) were immersed in Dulbecco’s modified eagle medium supplemented with fetal bovine serum in collagen type I-coated tissue culture dishes. Culture medium was then changed to EpiLife medium on the third day of culture (Figure 1A). Once subconfluent, ORS cells were harvested and split at a 1:5 ratio, and maintained in EpiLife medium. Cells were cultured up to 6 weeks and immunostained with cytokeratin 8 antibodies (Figure 1B).
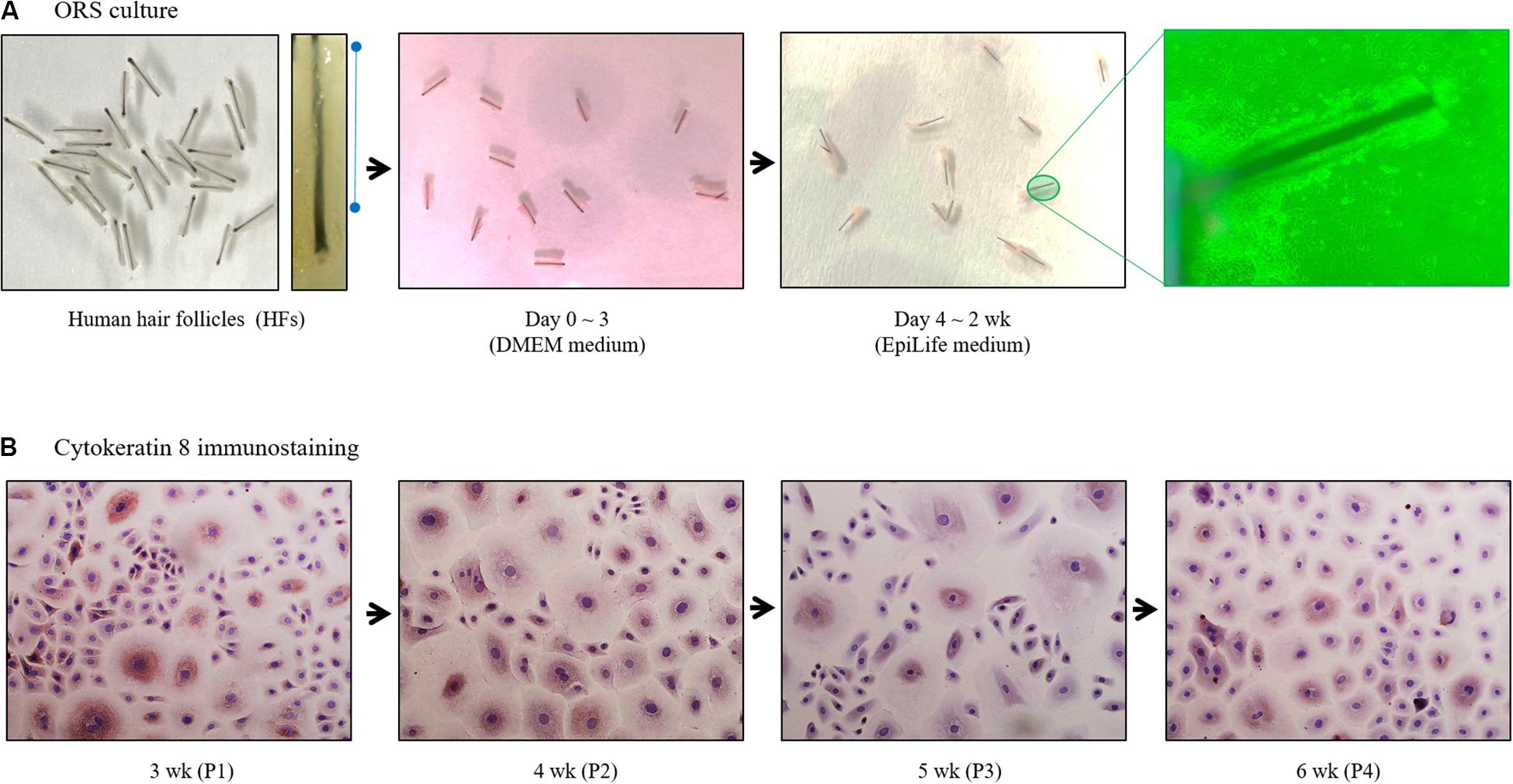
Figure 1. Diagram of human outer root sheath (ORS) follicular keratinocytes cultivation. (A) Human ORS cells were cultured as described in the Materials and Methods section. Representative images from a patient (male, age 37 years) are shown. (B) Cells in various culture periods (3, 4, 5, and 6 weeks) from a patient (male, age 28 years) were immunostained with cytokeratin 8 antibody.
To investigate whether the hair-inductive activity (trichogenicity) of ORS cells was correlated with the cultivation period, cells in various cultivation periods (3, 4, 5, and 6 weeks) were implanted into nude mice along with freshly isolated neonatal mouse dermal cells (Figure 2A). When quantified, induced hairs were inversely correlated with the cultivation period, and no HF induction was observed in co-implants containing 6-week-old ORS cells and mouse dermal cells (Figures 2B,C and Supplementary Figure S1). The human origin of epithelial cells of reconstituted hair follicles and the mouse origin of mesenchymal part of follicles remains to be proved.
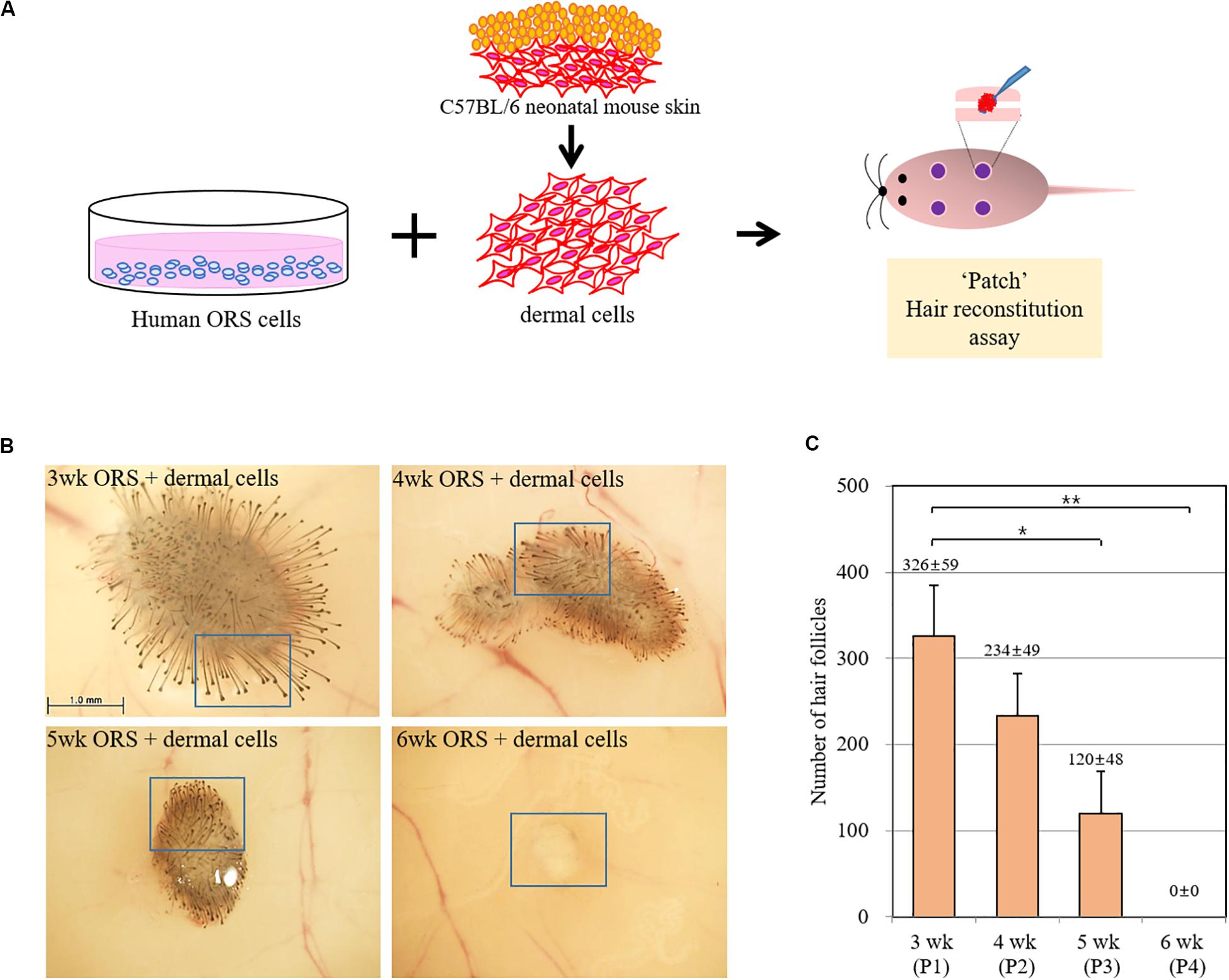
Figure 2. Inverse correlation of hair-inductive activity of ORS cells with their cultivation period. (A) Diagram of a “patch” hair reconstitution assay used in this study. Cultured human ORS cells (1 × 106 cells) were combined with freshly isolated neonatal mouse dermal cells (1 × 106 cells) and then subcutaneously implanted into the skin on the backs of 7-week-old female nude mice. (B) Hair follicle (HF) formation was examined from the back skin of the mice after 2 weeks and representative pictures are shown. High power images of boxed regions are shown in Supplementary Figure S2. (C) Total induced HFs were quantified and the data are the means ± standard deviation of four determinations using same batch ORS cells from a patient (male, age 26 years). *P < 0.05, **P < 0.01.
Next, we compared the gene expression profiles between the short-term (20 days) and long-term (42 days) cultured ORS cells from two individuals (#1 and #2) by microarray analysis. A number of genes were found to be differentially expressed (Figure 3). Among the 144 genes with a more than twofold increase in short-term cultured ORS cells (Supplementary Table S1), we found 6 transcription factors, including FOXA2, BARX2, and BHLHA15. Also, among the 278 genes with a more than twofold decrease in short-term cultured ORS cells (Supplementary Table S2), we found four transcription factors, including ZNF257and TBX3. Next, we visualized the expression value of DEGs to compare the up-regulated genes and down-regulated genes using Heatmap (Figure 3B) to show the global patterns of the DEGs dynamics between short-term and long-term culture. Then, we used GSEA information to look for the genes that are related to the potential skin associated pathway. Among our findings, FOXA2 is associated with the Hedgehog (HH) signaling pathway, while the other specific genes are related to each pathway (FGF, VEFG, Notch, BMP, TGF-b) (Figure 3C). Intriguingly, FGF5 is highly down-regulated in the short-term culture. At the same time, several genes related to the TGF-b signaling pathway are significantly down-regulated in the short-term culture as well. We also speculated the WNT-related genes between two groups. Total eight genes including WNT7b, WNT9a, WNT3, WNT5a, WNT6, FZD6, FZD1, and FZD10 were identified to be up-regulated, although they are not statistically significant, in WNT signaling pathway (Figure 3C’). To further analysis to find the potential mechanism, the DEGs were used to investigate GO biological process and KEGG pathway (Supplementary Figure S3 and Supplementary Tables S3–S6). The up-regulated genes were enriched in regulation of stem cell proliferation, positive regulation of endothelial cell proliferation, regulation of embryonic development, and negative regulation of cell death. The down-regulated genes were enriched in epidermal cell differentiation, keratinocyte differentiation, and epidermis development (Supplementary Figure S3 and Supplementary Tables S3–S6). The statistically enriched list is summarized in Supplementary Tables S3–S6.
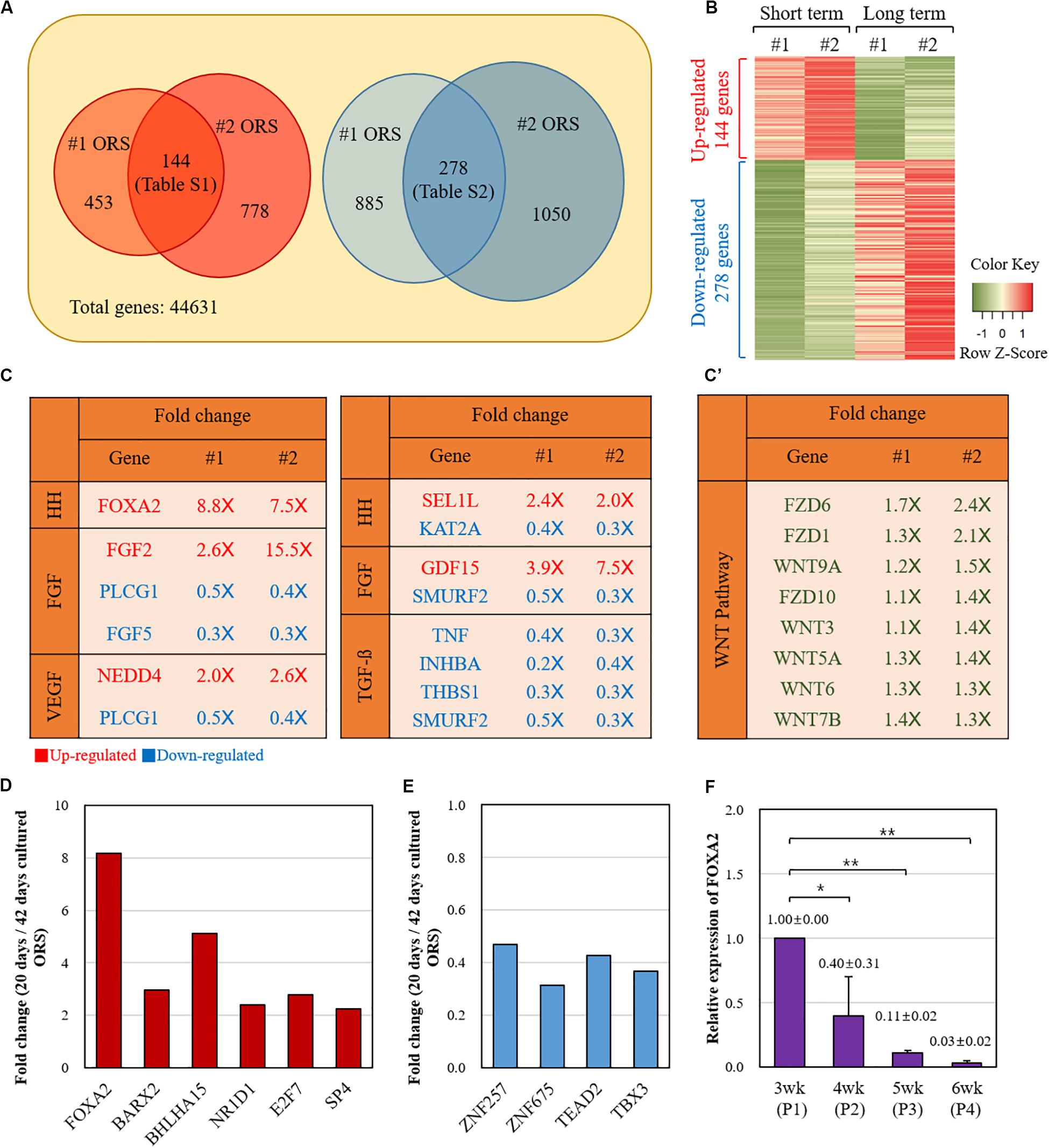
Figure 3. Comparison of the gene expression profiles between the short-term and long-term cultured ORS cells. (A) Diagram of twofold up- and down-regulated genes in short-term cultured ORS cells. Two independent cell lines (#1 and #2) from two male patients (ages 50 and 64 years) were cultured for 20 days and 42 days, and microarray analysis was performed with transcripts derived from those cells. The number of genes with a more than twofold increase and decrease in the short-term cultured ORS cells are shown on the left and right, respectively. A total of 144 genes had a more than twofold increase in both short-term cultured ORS cells (listed in Supplementary Table S1), and 278 genes had a more than twofold decrease in both short-term cultured ORS cells (listed in Supplementary Table S2). (B) Heat map of differentially expressed genes (DEGs). The up-regulated genes were shown in red color and down-regulated genes in green color. (C) List of genes related to the skin signaling pathways from twofold up-regulated (2.0×) and down-regulated (0.5×) genes. Genes were selected based on gene-set enrichment analysis (GSEA) information in each pathway (HH, FGF, VEGF, Notch, BMP, and TGF-b). (C’) List of up-regulated Wnts and their receptor (FZD) among WNT signaling pathway. (D) Relative fold changes of six up-regulated transcription factors. (E) Relative fold changes of four down-regulated transcription factors. (F) Relative levels of FOXA2 expression of ORS cells in various cultivation periods (3, 4, 5, and 6 weeks). Data are expressed as means ± standard deviation of three different ORS cell lines from three male patients (ages 33, 38, and 67 years).
Relative fold changes of the transcription factors, averaged between two individual samples, are shown in Figures 3D,E. We next focused on FOXA2, the most up-regulated transcription factor in the short-term cultured ORS cells compared with long-term cultured ORS cells. In line with microarray data, quantitative reverse transcription-PCR showed that the FOXA2 expression levels gradually decreased as the cultivation period increased (Figure 3F).
We then investigated whether the trichogenicity of ORS cells was affected by the expression level of FOXA2. We adopted a siRNA-mediated knockdown of FOXA2 in ORS cells in combination with a patch hair reconstitution assay (Figure 4A). Short-term cultured ORS cells were plated for transfection with a FOXA2 siRNA. After transfection for 2 days, the decreased expression of a FOXA2 was confirmed by real-time PCR (Figure 4B). As expected, freshly isolated newborn mouse dermal cells (106 cells) induced HFs when co-transplanted with negative control siRNA-transfected ORS cells (106 cells) (Figure 4C). However, we found a significantly decreased number of induced HFs using FOXA2 siRNA-transfected ORS cells (Figure 4D).
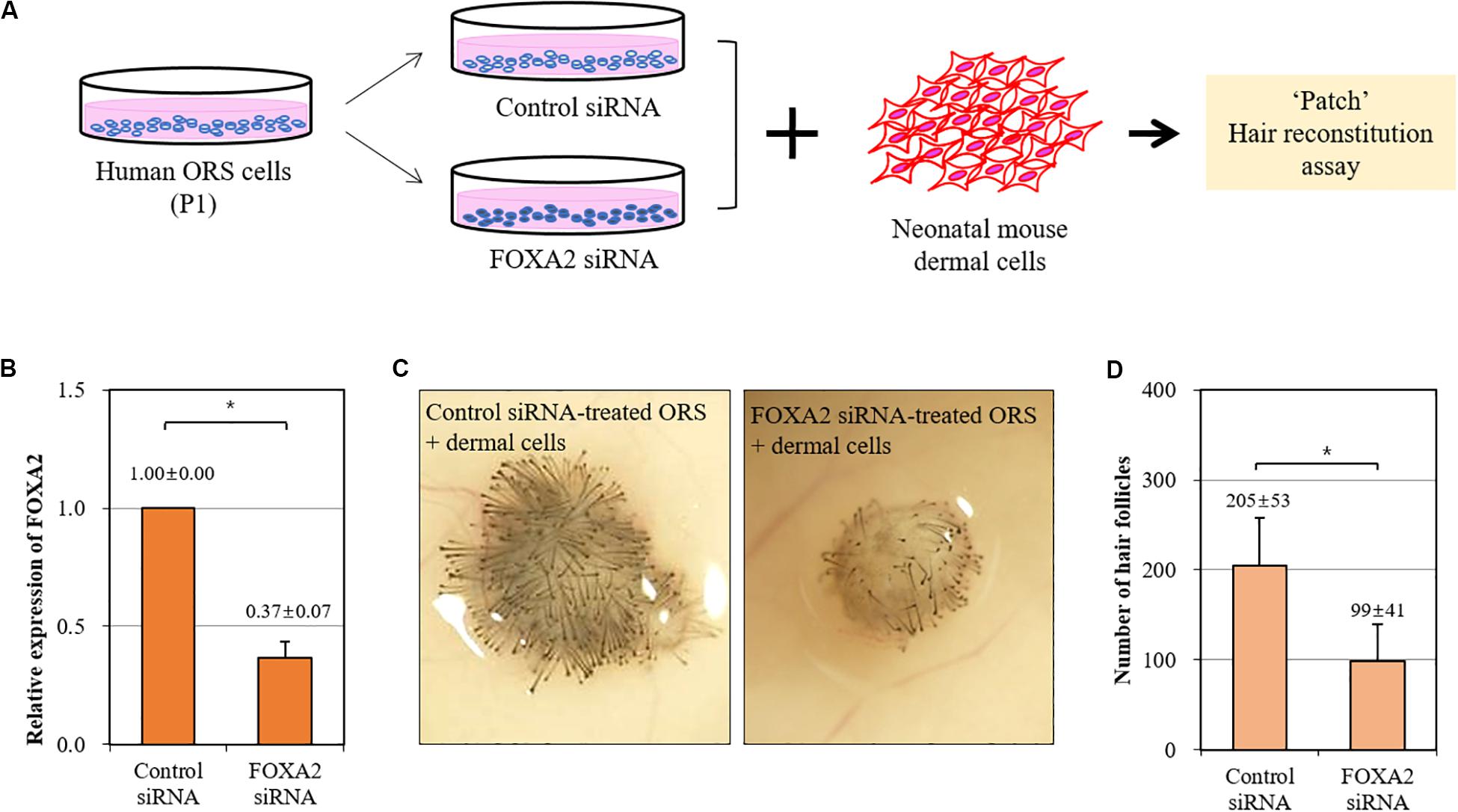
Figure 4. Impairment of hair-inductive activity of ORS cells by knockdown of FOXA2. (A) A schematic illustration of the experimental procedure. (B) The relative expression of FOXA2 in ORS cells transfected with control siRNA or FOXA2 siRNA was measured by qPCR. (C) Representative pictures of induced hair follicles from nude mice skin are shown. (D) The total number of induced hair follicles was counted at each injection site. Data are expressed as means ± standard deviation of eight determinations using three different ORS cell lines from three male patients (ages 38, 50, and 58 years). *P < 0.05.
Our data in this study suggest that FOXA2, a transcription factor, may regulate downstream target genes which, in turn, enhance the trichogenicity of ORS cells. Considering that FOXA2 binds to the promoter of WNT7b (Weidenfeld et al., 2002), we propose that FOXA2 might function through WNT7b. Consistent with this hypothesis, we found that the relative fold changes of WNT7b were 1.393 and 1.257 in two individual samples exhibiting 8.840- and 7.506-fold increases of FOXA2, respectively. We speculate that secreted WNT7b from follicular keratinocytes activates inductive dermal cells or makes follicular keratinocytes more susceptible to dermal signals. In this regard, it is interesting to note that loss of WNT7b results in perturbed HF development and differentiation with aberrant hair shaft production (Kandyba and Kobielak, 2014). Overexpression of FOXA2 in cultured ORS cells may help to determine whether this factor can enhance or restore trichogenicity of the cells; these experiments might yield intriguing results and are worthy of further consideration.
Our data strongly suggest that FOXA2 significantly influences the trichogenicity of human ORS cells. Our findings will provide a rationale for a new strategy for preparing competent human epidermal cells.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by The Medical Ethical Committee of the Kyungpook National University Hospital (Daegu, South Korea). The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Animal Care and Use Committee at Kyungpook National University (Daegu, South Korea).
S-SB and YS designed the research and wrote the manuscript. S-SB performed the experiments. S-SB, JP, and JO analyzed the data. JK and MK contributed the materials that are essential to perform the project. All authors contributed to the article and approved the submitted version.
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF-2018R1A2A2A05018574).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge other members of our laboratory (Hyun Woo Joo, Min Kyu Kim, Mee Sook Jun, Mi Hee Kwack, Eun Kyung Kim) for their technical help and valuable discussions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.575382/full#supplementary-material
Supplementary Figure 1 | Hair follicles quantified in Figure 2 from the nude mice skin.
Supplementary Figure 2 | High power images of boxed regions in Figure 2B.
Supplementary Figure 3 | GO analysis and KEGG pathway analysis of microarray data.
Supplementary Table 1 | List of genes with a more than twofold increase in short-term cultured ORS cells.
Supplementary Table 2 | List of genes with a more than twofold decrease in short-term cultured ORS cells.
Supplementary Table 3 | Detail list of up-regulated genes related to GO biological process.
Supplementary Table 4 | Detail list of down-regulated genes related to GO biological process.
Supplementary Table 5 | Detail list of up-regulated genes related to KEGG pathway.
Supplementary Table 6 | Detail list of down-regulated genes related to KEGG pathway.
DP, dermal papilla; FOXA2, forkhead box protein A2; HF, hair follicle; ORS, outer root sheath; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; siRNA, small interfering RNA.
Bak, S. S., Kwack, M. H., Shin, H. S., Kim, J. C., Kim, M. K., and Sung, Y. K. (2018). Restoration of hair-inductive activity of cultured human follicular keratinocytes by co-culturing with dermal papilla cells. Biochem. Biophys. Res. Commun. 505, 360–364. doi: 10.1016/j.bbrc.2018.09.125
Chan, C. C., Fan, S. M., Wang, W. H., Mu, Y. F., and Lin, S. J. (2015). A two-stepped culture method for efficient production of trichogenic keratinocytes. Tissue Eng. Part C Methods. 21, 1070–1079. doi: 10.1089/ten.TEC.2015.0033
Choi, J. W., Kim, Y. H., and Oh, J. W. (2017). Comparative analyses of signature genes in acute rejection and operational tolerance. Immune Network. 17, 237–249. doi: 10.4110/in.2017.17.4.237
Higgins, C. A., Chen, J. C., Cerise, J. E., Jahoda, C. A., and Christiano, A. M. (2013). Microenvironmental reprogramming by three-dimensional culture enables dermal papilla cells to induce de novo human hair-follicle growth. Proc. Natl. Acad. Sci. U.S.A. 110, 19679–19688. doi: 10.1073/pnas.1309970110
Kandyba, E., and Kobielak, K. (2014). Wnt7b is an important intrinsic regulator of hair follicle stem cell homeostasis and hair follicle cycling. Stem Cells 32, 886–901. doi: 10.1002/stem.1599
Kang, B. M., Kwack, M. H., Kim, M. K., Kim, J. C., and Sung, Y. K. (2012). Sphere formation increases the ability of cultured human dermal papilla cells to induce hair follicles from mouse epidermal cells in a reconstitution assay. J. Invest. Dermatol. 132, 237–239. doi: 10.1038/jid.2011.250
Kwack, M. H., Sung, Y. K., Chung, E. J., Im, S. U., Ahn, J. S., Kim, M. K., et al. (2008). Dihydrotestosterone-inducible dickkopf 1 from balding dermal papilla cells causes apoptosis in follicular keratinocytes. J. Invest. Dermatol. 128, 262–269. doi: 10.1038/sj.jid.5700999
Magerl, M., Kauser, S., Paus, R., and Tobin, D. J. (2002). Simple and rapid method to isolate and culture follicular papillae from human scalp hair follicles. Exp. Dermatol. 11, 381–385. doi: 10.1034/j.1600-0625.2002.110414.x
Millar, S. E. (2002). Molecular mechanisms regulating hair follicle development. J. Invest. Dermatol. 118, 216–225. doi: 10.1046/j.0022-202x.2001.01670.x
Ohyama, M., Kobayashi, T., Sasaki, T., Shimizu, A., and Amagai, M. (2012). Restoration of the intrinsic properties of human dermal papilla in vitro. J. Cell Sci. 125, 4114–4125. doi: 10.1242/jcs.105700
Ohyama, M., and Veraitch, O. (2013). Strategies to enhance epithelial-mesenchymal interactions for human hair follicle bioengineering. J. Dermatol. Sci. 70, 78–87. doi: 10.1016/j.jdermsci.2013.02.004
Ohyama, M., Zheng, Y., Paus, R., and Stenn, K. S. (2010). The mesenchymal component of hair follicle neogenesis: background, methods and molecular characterization. Exp. Dermatol. 19, 89–99. doi: 10.1111/j.1600-0625.2009.00935.x
Paus, R., and Cotsarelis, G. (1999). The biology of hair follicles. N. Engl. J. Med. 341, 491–497. doi: 10.1056/NEJM199908123410706
Philpott, M. P., Sanders, D., Westgate, G. E., and Kealey, T. (1994). Human hair growth in vitro: a model for the study of hair follicle biology. J. Dermatol. Sci. 7(Suppl.), S55–S72. doi: 10.1016/0923-1811(94)90036-1
Wang, Q., Oh, J. W., Lee, H. L., Dhar, A., Peng, T., Ramos, R., et al. (2017). A multi-scale model for hair follicles reveals heterogeneous domains driving rapid spatiotemporal hair growth patterning. eLife 6:e22772. doi: 10.7554/eLife.22772
Weidenfeld, J., Shu, W., Zhang, L., Millar, S. E., and Morrisey, E. E. (2002). The WNT7b promoter is regulated by TTF-1. GATA6, and Foxa2 in lung epithelium. J. Biol. Chem. 277, 21061–21070. doi: 10.1074/jbc.M111702200
Yang, C. C., and Cotsarelis, G. (2010). Review of hair follicle dermal cells. J. Dermatol. Sci. 57, 2–11. doi: 10.1016/j.jdermsci.2009.11.005
Keywords: forkhead box protein A2, hair induction, outer root sheath, transcription factor, trichogenicity
Citation: Bak S-S, Park JM, Oh JW, Kim JC, Kim MK and Sung YK (2020) Knockdown of FOXA2 Impairs Hair-Inductive Activity of Cultured Human Follicular Keratinocytes. Front. Cell Dev. Biol. 8:575382. doi: 10.3389/fcell.2020.575382
Received: 23 June 2020; Accepted: 17 September 2020;
Published: 08 October 2020.
Edited by:
Mingxing Lei, Chongqing University, ChinaReviewed by:
Yuhong Li, Army Medical University, ChinaCopyright © 2020 Bak, Park, Oh, Kim, Kim and Sung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Young Kwan Sung, eXN1bmdAa251LmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.