- Unit of Cell and Developmental Biology, Department of Biology, University of Pisa, Pisa, Italy
Glioblastoma represents an aggressive form of brain cancer characterized by poor prognosis and a 5-year survival rate of only 3–7%. Despite remarkable advances in brain tumor research in the past decades, very little has changed for patients, due in part to the recurrent nature of the disease and to the lack of suitable models to perform genotype-phenotype association studies and personalized drug screening. In vitro culture of cancer cells derived from patient biopsies has been fundamental in understanding tumor biology and for testing the effect of various drugs. These cultures emphasize the role of in vitro cancer stem cells (CSCs), which fuel tumor growth and are thought to be the cause of relapse after treatment. However, it has become clear over the years that a 2D monolayer culture of these CSCs has certain disadvantages, including the lack of heterogeneous cell-cell and cell-environment interactions, which can now be partially overcome by the introduction of 3D organoid cultures. This is a novel and expanding field of research and in this review, I describe the emerging 3D models of glioblastoma. I also discuss their potential to advance our knowledge of tumor biology and CSC heterogeneity, while debating their current limitations.
Introduction
The idea that tumor initiation, progression and regrowth after treatment are sustained by a subpopulation of cancer cells, the glioblastoma stem cells (GSCs), has been crucial to our current understating of glioblastoma (GBM) biology (Swartling et al., 2015; Alcantara Llaguno et al., 2016; Azzarelli et al., 2018; Hakes and Brand, 2019; Lu et al., 2019). Glioblastoma is a highly aggressive brain tumor characterized by elevated intratumor heterogeneity, which could be potentially attributed to variations in GSC behavior and stochastic consequences of their hierarchical growth pattern. Recent studies provided evidence for a proliferative hierarchy in GBM, by using a combination of experimental approaches, such as quantitative lineage tracing, clonal size dependences, mutational signature analysis, and single cell RNA sequencing (Patel et al., 2014; Tirosh et al., 2016; Lan et al., 2017; Neftel et al., 2019). Not only do these works indicated that tumor expansion follows a hierarchical lineage progression, but they also demonstrated that tumor cell fate decisions are rooted in a developmental program of neurogenesis. As such, early tumorigenesis is not primarily driven by genetic evolution, although genetic variations could still modulate the patterns self-renewal and differentiation of tumor cells, especially during later stages of disease progression. Evidence in another brain tumor originating in the cerebellum also showed that targeting the stem cell at the apex of a conserved developmental hierarchy could block tumor regrowth (Vanner et al., 2014). Thus, the cancer stem cell (CSC) model applied to GBM provided a framework to understand tumor heterogeneity, predict tumor evolution, and might contribute to the identification of novel therapeutic targets aimed at eliminating the GSC in order to eradicate the tumor.
The simultaneous presence of different stem, progenitor, and differentiated cells along the developmental hierarchy and the high degree of intra-tumor heterogeneity render in vitro modeling of GBM particularly challenging. GSCs have been isolated from primary tumor biopsies and have been show to recapitulate in vivo tumor heterogeneity when forced to differentiate in culture or upon xenotransplantation (Galli et al., 2004; Singh et al., 2004; Pollard et al., 2009; Figure 1). However, when GSCs are grown in adherent 2D monolayer cultures, they lack intrinsic heterogeneity and 3D relative spatial organization, and lose interactions with the diverse components of the tumor extracellular matrix and the microenvironment. Moreover, these cells scarcely predict treatment efficacy, as drugs that initially proved effective in the context of cultured cell lines did not result in clinical applications (Zanders et al., 2019).
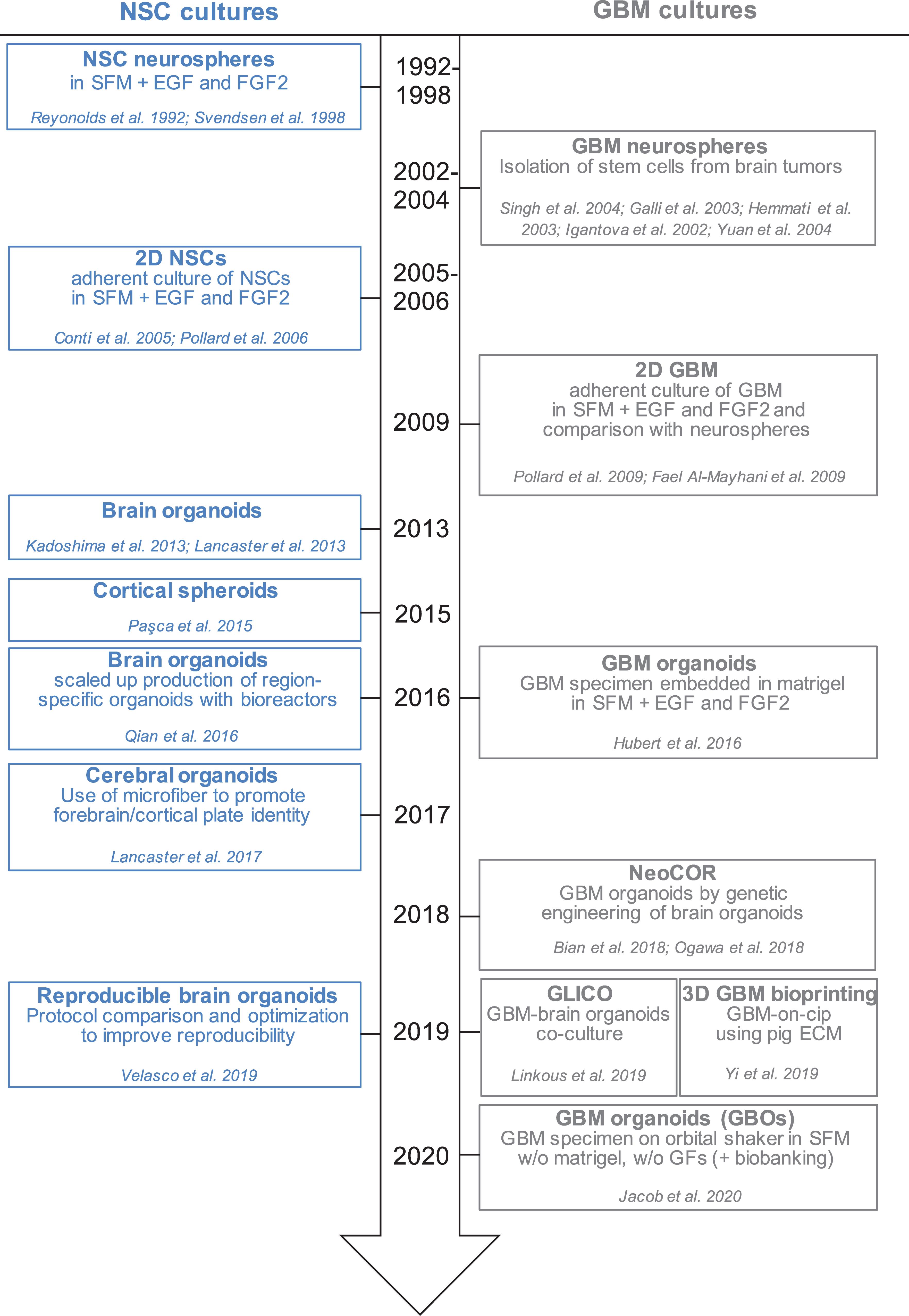
Figure 1. Timeline of in vitro method to culture neural stem cells and glioblastoma cells in 2D and 3D. Schematic representation of the development of different protocols to culture NSCs (neural stem cells) and GBM (glioblastoma) cells in monolayer, spheres and organoids. SFM, serum free medium; EGF, epidermal growth factor; FGF2, fibroblast growth factor 2; GFs, growth factors; NeoCOR, neoplastic cerebral organoids; GLICO, GLIoma cerebral organoids.
Thus, more refined model systems that allow the recapitulation of complex cancer phenotypes and yet retain the amenability to perform detailed analysis are needed, especially in view of the need to provide more accurate predictions of the therapeutic potential of new treatments. Encouraged by promising results in other cancer fields (Boj et al., 2015; Van De Wetering et al., 2015; Sachs et al., 2018; Tuveson and Clevers, 2019), several laboratories have directed their efforts to generate organoid models of glioblastoma, which consist, by definition, of 3D structures in which different cell types self-organize to establish appropriate cell–cell contacts and to create a microenvironment (Huch and Koo, 2015). As such, GBM organoids could better mimic tumor complexity and heterogeneity in growth potential and treatment responsiveness. This review describes the existing organoid GBM models that have just started to be developed and compares them to other 3D models, such as neurospheres and 3D bioprinted GBMs. It also discusses their potential to advance our understanding of GBM biology and to predict clinical outcome, while also considering their current limitations.
Three-Dimensional Models of Glioblastoma
Tumorspheres and Glioblastoma Organoids From Primary Tissue
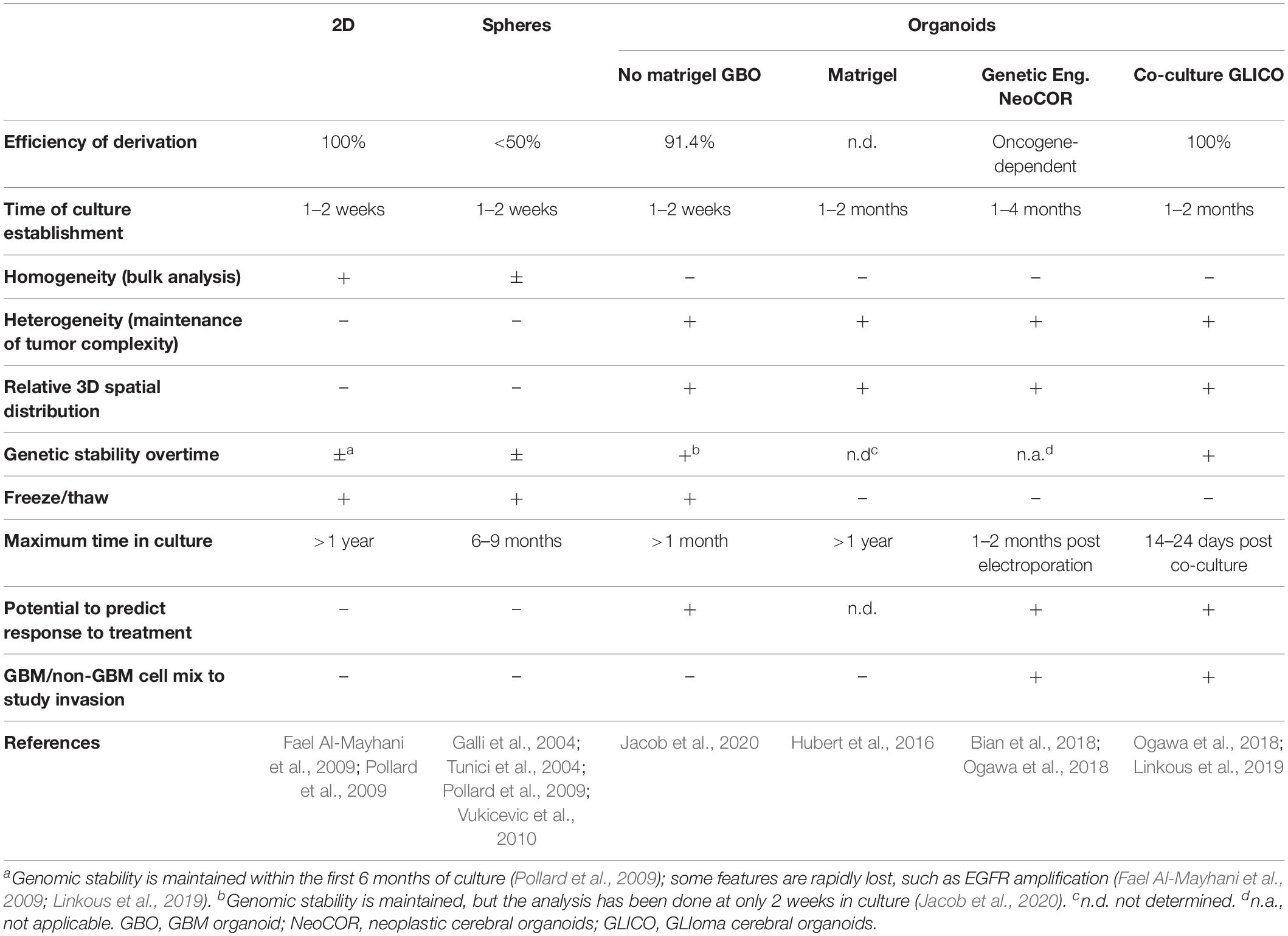
Glioblastoma stem cells can be isolated from primary tumors and can be grown in culture for an extended period of time (Ignatova et al., 2002; Hemmati et al., 2003; Galli et al., 2004; Singh et al., 2004; Tunici et al., 2004; Yuan et al., 2004; Fael Al-Mayhani et al., 2009; Pollard et al., 2009; Vukicevic et al., 2010; Figure 1 and Table 1). In vitro expansion of GSC is sustained by growth factors like EGF (Epidermal Growth Factor) and FGF2 (Fibroblast Growth Factor), conditions that also expand neural stem cells, highlighting the close relationship between GSC and their normal counterpart (Conti et al., 2005; Pollard et al., 2006, 2009). GSC can be grown in 2D adherent culture or as 3D neurospheres: the latter can be considered the very first “3D model” of GBM, as cells maintain a certain degree of polarization and 3D spatial organization (Galli et al., 2004; Azari et al., 2011). However, neurospheres are characterized by a necrotic core and can thus be able to achieve a maximum size of around 300 μm, before needing disruption and replating to survive (Reynolds and Weiss, 1992; Svendsen et al., 1998; Reynolds and Rietze, 2005). In addition, cells in neurospheres have lost their interaction with components of the extracellular matrix, and thus hardly mimic in vivo GSC behavior (Table1).
In 2016, the lab of Jeremy Rich developed in vitro conditions to grow 3D organoids from human GBM cells and from GBM biopsies. When embedded in matrigel, finely minced GBM specimens grew up to 3–4 mm in 2 months and could be kept in culture for over a year (Table 1), even if their growth slows down after several months, probably due to the limited diffusion of nutrients as the organoids grow in size (Hubert et al., 2016; Figure 2A). An interesting feature of these GBM organoids is that they recapitulate the gradient of stem cell density in relation to hypoxic levels found in vivo. The authors reported a high number of Sox2+ stem cells at the periphery of the organoid, while the core was characterized by lower abundance of Sox2+ cells and increased levels of hypoxia. Sox2+ stem cells also exhibited different molecular properties when located at the core or at the periphery of the organoids (Hubert et al., 2016). Thus, multiple Sox2+ populations may co-exist in organoids, suggesting that the organoid microenvironment might be able to sustain the simultaneous growth of different CSCs and would allow the study of cellular hierarchies in tumors. While highly promising, this system would require further characterization and validation across several GBMs. Indeed, establishment rates have yet to be determined and may be very patient-specific. Moreover, this system suffers from a relatively low to medium throughput capability and the long time necessary to establish the cultures (1–2 months) (Table 1).
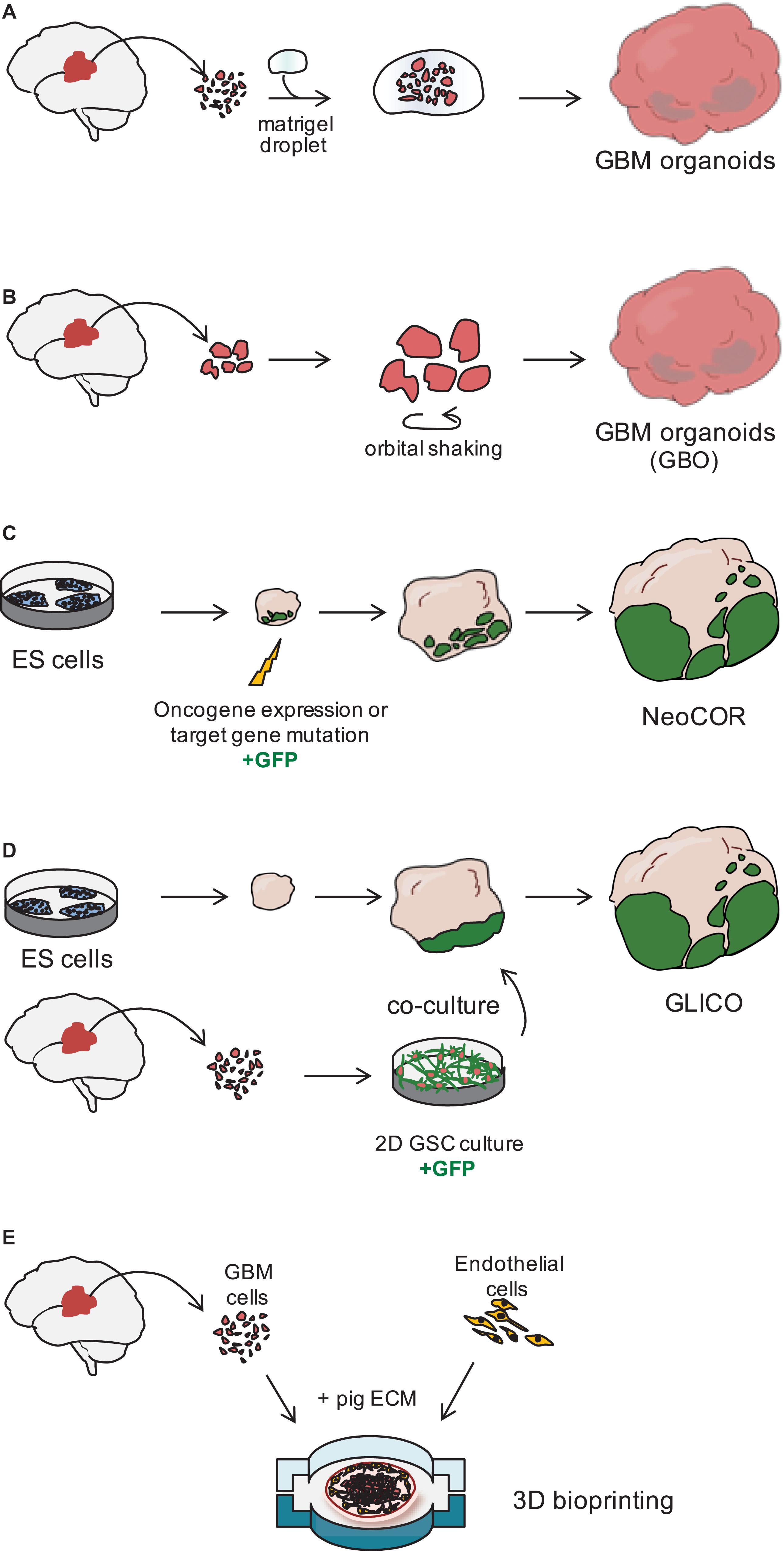
Figure 2. Three dimensional models of glioblastoma. (A) Glioblastoma (GBM) organoids have been derived by embedding finely minced GBM specimen in matrigel (Hubert et al., 2016) or (B) by culturing pieces of tumor biopsies in defined matrigel-free and serum-free conditions, on an orbital shaker. GBO. GBM Organoid (Jacob et al., 2020). (C) Embryonic stem (ES) cell-derived brain organoids can be nucleofected at early stages of the differentiation to introduce tumor-promoting genetic alterations. During nucleofection, cells are also marked with green fluorescent protein (GFP) to visualize tumor cell growth. NeoCOR, neoplastic cerebral organoids (Bian et al., 2018; Ogawa et al., 2018). (D) Patient-derived glioblastoma stem cells (GSCs) are initially cultured in 2D, before being co-cultured with brain organoids. GSCs are marked by GFP to visualize integration and growth in the organoid. GLICO, GLIoma cerebral organoids (Ogawa et al., 2018; Linkous et al., 2019). (E) Patient-derived GBM cells and endothelial cells are seeded on a chip using a pig extracellular matrix (ECM) bio-ink and a 3D bioprinter (Yi et al., 2019).
Recently, a novel and faster protocol (1–2 weeks) of 3D GBM organoid derivation that overcome such limitations has been reported by collaboration between Donald O’Rourke, Guo-li Ming, and Hongjun Song at the University of Pennsylvania (Table 1 and Figure 1). Instead of dissociating tumor biopsies to fine pieces, the authors cut the biopsies into around 1 mm fragments and culture them on an orbital shaker without matrigel and in serum free conditions not supplemented with EGF and FGF2 (Figure 2B; Jacob et al., 2020). These conditions should avoid selection of specific cell populations, thus better preserving inter- and intra-tumoral heterogeneity. These glioblastoma organoids, called GBOs, also developed hypoxic gradient and were further propagated in culture by cutting them into smaller pieces to avoid inner core necrosis. Importantly, these GBOs were cryopreserved and able to recover and continue their growth upon thawing. This is an essential step to generate GBM biobanks for subsequent recovery and analysis and the authors have currently biobanked around 70 GBOs from 53 patients (sometimes including different tumor subregions). GBOs largely maintain genetic and molecular signatures of the parental tumors. However, most analyses have been done within the first two weeks in culture, as GBOs maintenance over very long periods has been variable.
This novel protocol is fast and reproducible and provides enough material for RNA and exome sequencing, as well as drug sensitivity tests. It is thus suitable for genotype-drug association studies and opens new avenues to personalized medicine approaches, along the line of current advances in other cancer fields in which organoid biobanks have already been established or are currently being generated (Boj et al., 2015; Van De Wetering et al., 2015; Sachs et al., 2018; Yan et al., 2018; Kim et al., 2019).
The establishment of the cultures within 1–2 weeks from surgical resection is particularly important, because current treatments are initiated within a month post-surgery and having preclinical information about potentially effective treatments might be extremely useful and might also help refine patient enrolment in clinical trials. In this direction, GBO treatment with CAR-T cells against EGFRvIII variant, which is currently in clinical trials (O’Rourke et al., 2017; Goff et al., 2019), resulted in specific effect only on GBOs containing high percentage of EGFRvIII+ cells, thus showing the translational impact and future pre-clinical potential of this approach (Jacob et al., 2020).
Tumor Development by Genetic Engineering of Brain Organoids
The development of human brain organoids or “minibrians” have revolutionized the way we operate in developmental neurobiology, by providing unprecedented access to aspects of human brain development functioning and disorders (Kadoshima et al., 2013; Lancaster et al., 2013, 2017; Paşca et al., 2015; Qian et al., 2016, 2018, 2019; Amin and Paşca, 2018; Figure 1). The potential of brain organoids to recapitulate aspects of brain cancer is particularly valuable as neither patient derived xenotransplantation in mice nor human brain tumor stem cells in 2D culture behave in the same way as tumors in vivo.
The labs of Jürgen Knoblich and Inder Verma have recently genetically engineered organoids to develop tumors (Bian et al., 2018; Ogawa et al., 2018; Figures 1, 2C and Table 1). Bian et al. (2018) screened for genetic alterations that could lead to tumorigenesis and called the resulting tumor NeoCor (neoplastic cerebral organoids): the authors overexpressed known oncogenes by a transposase-based system and/or deleted tumor suppressor gene functions via CRIPSR Cas-9. Organoid cells have been targeted by nucleofection at very early stages of the differentiation and the cells carrying the genetic alterations have been marked by GFP, so that cell growth and tumor transformation could be easily followed. This screening identified MYC overexpression and a few more genetic combinations often found in human GBM to provide cells with a strong growth advantage. Transcriptomic profiling showed that MYC overexpressing tumors have a CNS-PNET-like identity (CNS-PNET: Primitive Neuro-Ectodermal Tumor of the Central Nervous System), while the other tumors resemble GBM more, suggesting that distinct genetic aberrations can induce tumors with specific cellular identities. Other works have also shown that mesenchymal GBM can be induced in organoids by defined genetic mutations, namely HRasG12V activation and p53 disruption (Ogawa et al., 2018).
Although these works show that a certain degree of GBM subtyping can be reproduced in organoids, whether all different GBM subtypes can be recapitulated in this system and how closely GBM-derived organoids resemble patient-derived GBM cells is still under investigated. Tumors other than GBM did not develop despite the genetic manipulation of genes classically altered in these tumors, such as the deletion of the inhibitory Sonic Hedgehog (SHH) receptor PTCH1 in SHH-group medulloblastoma (Bian et al., 2018). However, the oncogenic effect of PTCH1 deletion is known to be cell-type specific (Schüller et al., 2008) and might therefore necessitate organoid pre-differentiation down the cerebellar route for transformation to occur (Ballabio et al., 2020).
Co-culture of GBM Cells With Brain Organoids
Tumor models created by genetic engineering of organoids described above are particularly advantageous to effectively model GBM initiation, but they hardly recapitulate the genomic complexity of in vivo tumors, as the methodology requires genetic manipulation of the few known driver genes, which are not necessarily representative of the genomic GBM heterogeneity. The laboratory of Howard Fine and other laboratories have recently developed a novel approach that overcome this disadvantage, by co-culturing patient-derived GSCs with 3D brain organoids and called their model GLICO (GLIoma cerebral organoids; Linkous et al., 2019; Figures 1, 2D and Table 1). The authors co-cultured different GFP-marked GSC cell lines with fully grown cerebral brain organoids and demonstrated that GSCs proliferate over time and integrate into the organoids. Each line behaves in a unique way, with some lines showing diffuse invasion, others forming “honeycomb”-like structures and others forming small regional nodes of proliferation (Linkous et al., 2019). Interestingly, co-cultured GSCs that exhibit higher degree of invasiveness were also more lethal when transplanted in mice (Ogawa et al., 2018). Thus, the observed heterogeneity in growth and invasion in the GLICO model likely reflects certain intrinsic properties of that particular patient-derived GSC line.
Cancer cells in this system not only displayed a cellular behavior that closely mimics the original tumor, but they also maintained key genetic aberrations of the native tumor. EGFR amplification, which was identified in two of the analyzed cell lines, was maintained in the GLICO models, while often lost in 2D cultures (Linkous et al., 2019; Table 1), indicating that this model may provide a more suitable microenvironment to preserve the genetic background of the in vivo tumor.
Three-Dimensional GBM Model via Bioprinting
The use of advanced 3D bioprinting technologies could enhance the way we design 3D GBM models in vitro (van Pel et al., 2018; Yi et al., 2019). Yi et al. (2019) created a GBM-on-chip model, in which they used decellularized pig brain extracellular matrix as a bio-ink to seed patient-derived cancer cells together with vascular endothelial cells in separated compartments (Figure 2E). Compartmentalization by seeding endothelial cells on the outside and cancer cells in the core of the chip established a radial oxygen gradient, which recapitulated important pathological features of the tumor. This model indeed exhibited hypoxia induced necrotic core, a perivascular niche and maintained a degree of spatial heterogeneity of the different cell types, with the higher number of Sox2+ stem cells at the periphery of the seeded tumor.
The “biomimetic” conditions of this system provided a microenvironment comparable to that of the original in vivo tumor tissue, offering the advantage of promoting cell-cell and cell-matrix interactions, and of better predicting treatment responses in a shorter time frame than other models (1–2 weeks compared to 1–2 months). However, the system still lacks accurate 3D spatial organization that can be only generated using self-assembled 3D organoid cultures, and it requires advanced technologies and expertise not always available to common biological laboratories.
Advantages and Limitations: Which Model to Use?
The best GBM model would be one that is complex enough to recapitulate features of the original tumor and simple enough to support investigation of different aspects of carcinogenesis in isolation. While the focus of this review is to look at emerging 3D in vitro models of GBM, several other approaches, including engineered mouse models or xenotransplantation, have been particularly useful to address tumor biology in other contexts [for a review of GBM models in vivo and in vitro, see Robertson et al. (2019)]. Thus, researchers might have to balance pros and cons of the different models to find the best fit for their research question, and might have to combine more than one model to take advantage of their complementary strengths (Table 1).
The development of 3D in vitro models of GBM holds great potential to study GBM biology and predict response to treatment, as they more closely recapitulate the complexity and heterogeneity of the original tumor. Indeed, most of the 3D organoid models described here have also shown selective vulnerabilities for targeted therapies or radiation that closely resemble tumor sensitivity in vivo (Hubert et al., 2016; Bian et al., 2018; Linkous et al., 2019). In addition, the recent establishment of 3D GBM organoids from biopsies with a novel and faster protocol (1–2 weeks) promoted the generation of a live GBM biobank that can be used for genotype-drug association studies on a medium to high throughput capability (Jacob et al., 2020). Thus, 3D GBM models provide a powerful predicting tool that could be used one day to guide clinical decisions.
Some of the models described in this review (Bian et al., 2018; Ogawa et al., 2018; Linkous et al., 2019) and other similar models (da Silva et al., 2018; Plummer et al., 2019) allow the possibility to mix GBM and non-GBM brain cells (Table 1). This is particularly useful to study tumor invasion of the normal tissue and the interaction of tumor cells with normal brain cells. By targeting only one or the other compartment at a time, it will thus be possible to dissect the specific role of genes involved in cell-cell interaction, adhesion, guidance and migration, and this might identify novel therapeutical targets to block tumor infiltration.
The brain organoid tissue GBM cells interact with, however, resembles more an embryonic type of tissue, rather than the adult brain tissue of GBM derivation. At present, it is not clear how this might influence tumor properties. In the future, it would be interesting to develop a similar approach to model and study prenatal and childhood tumors, such as pediatric gliomas and medulloblastomas, which should maintain a closer link to their developmental origin (Liu and Zong, 2012; Azzarelli et al., 2018; Lu et al., 2019). As medulloblastoma did not develop in organoids, even when genetic alterations typical of this tumors were introduced, it might be necessary to generate regionalized organoids (Muguruma et al., 2015; Dias and Guillemot, 2017; Renner et al., 2017; Ballabio et al., 2020) tailored to the area of origin of that specific tumor, prior to transformation.
An aspect that has likely benefited from having this embryonic type of tissue, is the maintenance of the CSC compartment of the tumor. By providing cancer cells with the more appropriate environment that could support the simultaneous presence of different stem and progenitor cells, these 3D GBM organoid models will foster the investigation of CSC heterogeneity (Bhaduri et al., 2020). They will also open the possibility to study CSC developmental hierarchy in tumors and the influence of other cell types or of the environment on CSC fate decisions. While it is possible to incorporate non-neuronal cell types into organoids, such as microglia or other immune cells (Abud et al., 2017; Brownjohn et al., 2018; Ormel et al., 2018; Jacob et al., 2020), the main challenge still remains to recreate an environment that includes the vasculature and other cell types that could exhibit inflammatory and immunitary responses similar to an intact brain (Daviaud et al., 2018; Lancaster, 2018; Mansour et al., 2018).
The presence of different cell types and the high degree of heterogeneity is probably the main advantage and, at the same time, a disadvantage of the system because, while it reflects the complexity of the original tumor, it is probably the source of variability typical of these 3D cultures (Lancaster and Knoblich, 2014; Quadrato et al., 2017; Amin and Paşca, 2018; Qian et al., 2019; Velasco et al., 2019; Table 1). Thus, investigators might have to choose between growing cells in classical 2D monolayer or sphere cultures or in 3D organoids depending on whether they are more interested in performing bulk analysis on an homogenous cell population or whether they aim to investigate tumor aspects that requires maintenance of tumor complexity and heterogeneity.
Conclusion and Perspectives
The emerging development of 3D organoids of GBM adds on to an abundance of choices to model this aggressive brain tumor (Robertson et al., 2019; Figure 1 and Table 1). It provides researchers with an additional tool to understand GBM biology, and predict tumor progression and response to treatment. One of the main advantages of growing GBM in brain organoids is the possibility to simultaneously grow different stem cells, progenitors and their differentiated progeny within the same conditions (Hubert et al., 2016; Jacob et al., 2020). This not only mimics better the heterogeneity of the original tumor, but it also shows the persistence of developmental programs of neurogenesis in the tumor. By understanding how CSCs make fate decisions and by defining the aberrant developmental pathways that lead to tumorigenesis, it will be possible to exploit novel emerging vulnerabilities to kill or differentiate CSCs to eradicate the tumor.
Future challenges include the reduction of organoid variability, while maintaining tumor complexity and heterogeneity, and the incorporation of an appropriate microenvironment that could, for example, mimic inflammatory and immunological responses. This would be particularly relevant in view of current successes in cancer immunotherapy (Fesnak et al., 2016) and, once incorporated in organoids, it would help to understand how immunological response might influence CSC hierarchy, and tumor progression and regrowth (Weller et al., 2017; Lim et al., 2018).
Our knowledge on GBM has massively expanded in the past decades and future collaborations between oncologists, clinicians, and researchers in the cancer, stem cell, and developmental biology fields, together with the possibility to share different complementary models and tools are likely to bring the long-sought breakthroughs that will improve patient treatment and prognosis.
Author Contributions
RA wrote the manuscript and prepared the figures.
Funding
RA was supported by “Rita Levi Montalcini” funding from MIUR (Italian Ministry of Education, University and Research).
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I would like to thank Prof. Benjamin Simons and Prof. Anna Philpott (University of Cambridge) for discussions and critical reading of the manuscript and Dr. Christopher Hubert (Cleveland Clinic, Lerner Research Institute) for discussions.
References
Abud, E. M., Ramirez, R. N., Martinez, E. S., Healy, L. M., Nguyen, C. H. H., Newman, S. A., et al. (2017). iPSC-derived human microglia-like cells to study neurological diseases. Neuron 94, 278–293.e9. doi: 10.1016/j.neuron.2017.03.042.
Alcantara Llaguno, S. R., Xie, X., and Parada, L. F. (2016). Cell of origin and cancer stem cells in tumor suppressor mouse models of glioblastoma. Cold Spring Harb. Symp. Quant. Biol. 81, 31–36. doi: 10.1101/sqb.2016.81.030973
Amin, N. D., and Paşca, S. P. (2018). Building models of brain disorders with three-dimensional organoids. Neuron 100, 389–405. doi: 10.1016/j.neuron.2018.10.007
Azari, H., Millette, S., Ansari, S., Rahman, M., Deleyrolle, L. P., and Reynolds, B. A. (2011). Isolation and expansion of human glioblastoma multiforme tumor cells using the neurosphere assay. J. Vis. Exp. 56:e3633. doi: 10.3791/3633
Azzarelli, R., Simons, B. D., and Philpott, A. (2018). The developmental origin of brain tumours: a cellular and molecular framework. Development 145:dev162693. doi: 10.1242/dev.162693
Ballabio, C., Anderle, M., Gianesello, M., Lago, C., Miele, E., Cardano, M., et al. (2020). Modeling medulloblastoma in vivo and with human cerebellar organoids. Nat. Commun. 11, 1–18.
Bhaduri, A., Di Lullo, E., Jung, D., Müller, S., Crouch, E. E., Espinosa, C. S., et al. (2020). Outer radial glia-like cancer stem cells contribute to heterogeneity of glioblastoma. Cell Stem Cell 26, 48–63.e6. doi: 10.1016/j.stem.2019.11.015
Bian, S., Repic, M., Guo, Z., Kavirayani, A., Burkard, T., Bagley, J. A., et al. (2018). Genetically engineered cerebral organoids model brain tumor formation. Nat. Methods 15, 631–639. doi: 10.1038/s41592-018-0070-7
Boj, S. F., Hwang, C. I., Baker, L. A., Chio, I. I. C., Engle, D. D., Corbo, V., et al. (2015). Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338. doi: 10.1016/j.cell.2014.12.021
Brownjohn, P. W., Smith, J., Solanki, R., Lohmann, E., Houlden, H., Hardy, J., et al. (2018). Functional studies of missense TREM2 mutations in human stem cell-derived microglia. Stem Cell Rep. 10, 1294–1307. doi: 10.1016/j.stemcr.2018.03.003
Conti, L., Pollard, S. M., Gorba, T., Reitano, E., Toselli, M., Biella, G., et al. (2005). Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 3:1594–1606. doi: 10.1371/journal.pbio.0030283
da Silva, B., Mathew, R. K., Polson, E. S., Williams, J., and Wurdak, H. (2018). Spontaneous glioblastoma spheroid infiltration of early-stage cerebral organoids models brain tumor invasion. SLAS Discov. 23, 862–868. doi: 10.1177/2472555218764623
Daviaud, N., Friedel, R. H., and Zou, H. (2018). Vascularization and engraftment of transplanted human cerebral organoids in mouse cortex. eNeuro 5, 1–18. doi: 10.1523/ENEURO.0219-18.2018
Dias, C., and Guillemot, F. (2017). Revealing the inner workings of organoids. EMBO J. 36, 1299–1301. doi: 10.15252/embj.201796860
Fael Al-Mayhani, T. M., Ball, S. L. R., Zhao, J. W., Fawcett, J., Ichimura, K., Collins, P. V., et al. (2009). An efficient method for derivation and propagation of glioblastoma cell lines that conserves the molecular profile of their original tumours. J. Neurosci. Methods 176, 192–199. doi: 10.1016/j.jneumeth.2008.07.022
Fesnak, A. D., June, C. H., and Levine, B. L. (2016). Engineered T cells: the promise and challenges of cancer immunotherapy. Nat. Rev. Cancer 16, 566–581. doi: 10.1038/nrc.2016.97
Galli, R., Binda, E., Orfanelli, U., Cipelletti, B., Gritti, A., De Vitis, S., et al. (2004). Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma (Cancer Research (October 2004) 64 (7011-7021). Cancer Res. 64:8130.
Goff, S. L., Morgan, R. A., Yang, J. C., Sherry, R. M., Robbins, P. F., Restifo, N. P., et al. (2019). Pilot trial of adoptive transfer of chimeric antigen receptor-Transduced t cells targeting egfrviii in patients with glioblastoma. J. Immunother. 42, 126–135. doi: 10.1097/CJI.0000000000000260
Hakes, A. E., and Brand, A. H. (2019). Neural stem cell dynamics: the development of brain tumours. Curr. Opin. Cell Biol. 60, 131–138. doi: 10.1016/j.ceb.2019.06.001
Hemmati, H. D., Nakano, I., Lazareff, J. A., Masterman-Smith, M., Geschwind, D. H., Bronner-Fraser, M., et al. (2003). Cancerous stem cells can arise from pediatric brain tumors. Proc. Natl. Acad. Sci. U.S.A. 100, 15178–15183. doi: 10.1073/pnas.2036535100
Hubert, C. G., Rivera, M., Spangler, L. C., Wu, Q., Mack, S. C., Prager, B. C., et al. (2016). A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 76, 2465–2477. doi: 10.1158/0008-5472.can-15-2402
Huch, M., and Koo, B. K. (2015). Modeling mouse and human development using organoid cultures. Development 142, 3113–3125. doi: 10.1242/dev.118570
Ignatova, T. N., Kukekov, V. G., Laywell, E. D., Suslov, O. N., Vrionis, F. D., and Steindler, D. A. (2002). Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia 39, 193–206. doi: 10.1002/glia.10094
Jacob, F., Salinas, R. D., Zhang, D. Y., and Rourke, D. M. O. (2020). Resource a patient-derived glioblastoma organoid model and resource a patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell 180, 188–204.e22. doi: 10.1016/j.cell.2019.11.036
Kadoshima, T., Sakaguchi, H., Nakano, T., Soen, M., Ando, S., Eiraku, M., et al. (2013). Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. U.S.A. 110, 20284–20289. doi: 10.1073/pnas.1315710110
Kim, M., Mun, H., Sung, C. O., Cho, E. J., Jeon, H. J., Chun, S. M., et al. (2019). Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 10:3991.
Lan, X., Jörg, D. J., Cavalli, F. M. G., Richards, L. M., Nguyen, L. V., Vanner, R. J., et al. (2017). Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature 549, 227–232. doi: 10.1038/nature23666
Lancaster, M. A. (2018). Brain organoids get vascularized. Nat. Biotechnol. 36, 407–408. doi: 10.1038/nbt.4133
Lancaster, M. A., Corsini, N. S., Wolfinger, S., Gustafson, E. H., Phillips, A. W., Burkard, T. R., et al. (2017). Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 35, 659–666. doi: 10.1038/nbt.3906
Lancaster, M. A., and Knoblich, J. A. (2014). Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9, 2329–2340. doi: 10.1038/nprot.2014.158
Lancaster, M. A., Renner, M., Martin, C. A., Wenzel, D., Bicknell, L. S., Hurles, M. E., et al. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. doi: 10.1038/nature12517
Lim, M., Xia, Y., Bettegowda, C., and Weller, M. (2018). Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 15, 422–442. doi: 10.1038/s41571-018-0003-5
Linkous, A., Balamatsias, D., Snuderl, M., Edwards, L., Miyaguchi, K., Milner, T., et al. (2019). Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep. 26, 3203–3211.e5. doi: 10.1016/j.celrep.2019.02.063
Liu, C., and Zong, H. (2012). Developmental origins of brain tumors. Curr. Opin. Neurobiol. 22, 844–849. doi: 10.1016/j.conb.2012.04.012
Lu, Q. R., Qian, L., and Zhou, X. (2019). Developmental origins and oncogenic pathways in malignant brain tumors. Wiley Interdiscip. Rev. Dev. Biol. 8, 1–23. doi: 10.1002/wdev.342
Mansour, A. A., Gonçalves, J. T., Bloyd, C. W., Li, H., Fernandes, S., Quang, D., et al. (2018). An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 36, 432–441. doi: 10.1038/nbt.4127
Muguruma, K., Nishiyama, A., Kawakami, H., Hashimoto, K., and Sasai, Y. (2015). Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 10, 537–550. doi: 10.1016/j.celrep.2014.12.051
Neftel, C., Laffy, J., Filbin, M. G., Hara, T., Shore, M. E., Rahme, G. J., et al. (2019). An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178, 835–849.e21. doi: 10.1016/j.cell.2019.06.024
Ogawa, J., Pao, G. M., Shokhirev, M. N., and Verma, I. M. (2018). Glioblastoma model using human cerebral organoids. Cell Rep. 23, 1220–1229. doi: 10.1016/j.celrep.2018.03.105
Ormel, P. R., Vieira, de Sá, R., van Bodegraven, E. J., Karst, H., Harschnitz, O., et al. (2018). Microglia innately develop within cerebral organoids. Nat. Commun. 9:4167.
O’Rourke, D. M., Nasrallah, M. P., Desai, A., Melenhorst, J. J., Mansfield, K., Morrissette, J. J. D., et al. (2017). A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci. Transl. Med. 9:eaaa0984. doi: 10.1126/scitranslmed.aaa0984
Paşca, A. M., Sloan, S. A., Clarke, L. E., Tian, Y., Makinson, C. D., Huber, N., et al. (2015). Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12:671. doi: 10.1038/nmeth.3415
Patel, A. P., Tirosh, I., Trombetta, J. J., Shalek, A. K., Gillespie, S. M., Wakimoto, H., et al. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science (80-) 344, 1396–1401. doi: 10.1126/science.1254257
Plummer, S., Wallace, S., Ball, G., Lloyd, R., Schiapparelli, P., Quiñones-Hinojosa, A., et al. (2019). A Human iPSC-derived 3D platform using primary brain cancer cells to study drug development and personalized medicine. Sci. Rep. 9, 1–11.
Pollard, S. M., Conti, L., Sun, Y., Goffredo, D., and Smith, A. (2006). Adherent neural stem (NS) cells from fetal and adult forebrain. Cereb. Cortex 16, i112–i120. doi: 10.1093/cercor/bhj167
Pollard, S. M., Yoshikawa, K., Clarke, I. D., Danovi, D., Stricker, S., Russell, R., et al. (2009). Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 4, 568–580. doi: 10.1016/j.stem.2009.03.014
Qian, X., Jacob, F., Song, M. M., Nguyen, H. N., Song, H., and Ming, G. L. (2018). Generation of human brain region–specific organoids using a miniaturized spinning bioreactor. Nat. Protoc. 13, 565–580. doi: 10.1038/nprot.2017.152
Qian, X., Nguyen, H. N., Song, M. M., Hadiono, C., Ogden, S. C., Hammack, C., et al. (2016). Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238–1254. doi: 10.1016/j.cell.2016.04.032
Qian, X., Song, H., and Ming, G. L. (2019). Brain organoids: advances, applications and challenges. Development 146:dev166074. doi: 10.1242/dev.166074
Quadrato, G., Nguyen, T., Macosko, E. Z., Sherwood, J. L., Yang, S. M., Berger, D. R., et al. (2017). Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53. doi: 10.1038/nature22047
Renner, M., Lancaster, M. A., Bian, S., Choi, H., Ku, T., Peer, A., et al. (2017). Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 36, 1316–1329. doi: 10.15252/embj.201694700
Reynolds, B. A., and Rietze, R. L. (2005). Neural stem cells and neurospheres – Re-evaluating the relationship. Nat. Methods 2, 333–336. doi: 10.1038/nmeth758
Reynolds, B. A., and Weiss, S. (1992). Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255, 1707–1710. doi: 10.1126/science.1553558
Robertson, F. L., Marqués-Torrejón, M. A., Morrison, G. M., and Pollard, S. M. (2019). Experimental models and tools to tackle glioblastoma. DMM Dis. Model. Mech. 12:dmm040386. doi: 10.1242/dmm.040386
Sachs, N., de Ligt, J., Kopper, O., Gogola, E., Bounova, G., Weeber, F., et al. (2018). A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172, 373–386.e10. doi: 10.1016/j.cell.2017.11.010
Schüller, U., Heine, V. M., Mao, J., Kho, A. T., Dillon, A. K., Han, Y. G., et al. (2008). Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form shh-induced medulloblastoma. Cancer Cell 14, 123–134. doi: 10.1016/j.ccr.2008.07.005
Singh, S. K., Hawkins, C., Clarke, I. D., Squire, J. A., Bayani, J., Hide, T., et al. (2004). Identification of human brain tumour initiating cells. Nature 432, 396–401. doi: 10.1038/nature03128
Svendsen, C. N., ter Borg, M. G., Armstrong, R. J., Rosser, A. E., Chandran, S., Ostenfeld, T., et al. (1998). A new method for the rapid and long term growth of human neural precursor cells. J. Neurosci. Methods 85, 141–152. doi: 10.1016/s0165-0270(98)00126-5
Swartling, F. J., Èanèer, M., Frantz, A., Weishaupt, H., and Persson, A. I. (2015). Deregulated proliferation and differentiation in brain tumors. Cell Tissue Res. 359, 225–254. doi: 10.1007/s00441-014-2046-y
Tirosh, I., Venteicher, A. S., Hebert, C., Escalante, L. E., Patel, A. P., Yizhak, K., et al. (2016). Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313. doi: 10.1038/nature20123
Tunici, P., Bissola, L., Lualdi, E., Pollo, B., Cajola, L., Broggi, G., et al. (2004). Genetic alterations and in vivo tumorigenicity of neurospheres derived from an adult glioblastoma. Mol. Cancer 3:25. doi: 10.1186/1476-4598-3-25
Tuveson, D., and Clevers, H. (2019). Cancer modeling meets human organoid technology. Science (80-) 364, 952–955. doi: 10.1126/science.aaw6985
Van De Wetering, M., Francies, H. E., Francis, J. M., Bounova, G., Iorio, F., Pronk, A., et al. (2015). Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945. doi: 10.1016/j.cell.2015.03.053
van Pel, D. M., Harada, K., Song, D., Naus, C. C., and Sin, W. C. (2018). Modelling glioma invasion using 3D bioprinting and scaffold-free 3D culture. J. Cell Commun. Signal. 12, 723–730. doi: 10.1007/s12079-018-0469-z
Vanner, R. J., Remke, M., Gallo, M., Selvadurai, H. J., Coutinho, F., Lee, L., et al. (2014). Quiescent Sox2+ cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell 26, 33–47. doi: 10.1016/j.ccr.2014.05.005
Velasco, S., Kedaigle, A. J., Simmons, S. K., Nash, A., Rocha, M., Quadrato, G., et al. (2019). Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570, 523–527. doi: 10.1038/s41586-019-1289-x
Vukicevic, V., Jauch, A., Dinger, T. C., Gebauer, L., Hornich, V., Bornstein, S. R., et al. (2010). Genetic instability and diminished differentiation capacity in long-term cultured mouse neurosphere cells. Mech. Ageing Dev. 131, 124–132. doi: 10.1016/j.mad.2010.01.001
Weller, M., Roth, P., Preusser, M., Wick, W., Reardon, D. A., Platten, M., et al. (2017). Vaccine-based immunotherapeutic approaches to gliomas and beyond. Nat. Rev. Neurol. 13, 363–374. doi: 10.1038/nrneurol.2017.64
Yan, H. H. N., Siu, H. C., Law, S., Ho, S. L., Yue, S. S. K., Tsui, W. Y., et al. (2018). A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell 23, 882–897.e11. doi: 10.1016/j.stem.2018.09.016
Yi, H. G., Jeong, Y. H., Kim, Y., Choi, Y. J., Moon, H. E., Park, S. H., et al. (2019). A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat. Biomed. Eng. 3, 509–519. doi: 10.1038/s41551-019-0363-x
Yuan, X., Curtin, J., Xiong, Y., Liu, G., Waschsmann-Hogiu, S., Farkas, D. L., et al. (2004). Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene 23, 9392–9400. doi: 10.1038/sj.onc.1208311
Keywords: glioblastoma, brain tumors, organoids, 3D models, cancer stem cells, neural stem cells, neurogenesis
Citation: Azzarelli R (2020) Organoid Models of Glioblastoma to Study Brain Tumor Stem Cells. Front. Cell Dev. Biol. 8:220. doi: 10.3389/fcell.2020.00220
Received: 06 January 2020; Accepted: 16 March 2020;
Published: 16 April 2020.
Edited by:
Deema Hussein, King Abdulaziz University, Saudi ArabiaReviewed by:
Hiroaki Wakimoto, Massachusetts General Hospital and Harvard Medical School, United StatesRoberto Würth, German Cancer Research Center (DKFZ), Germany
Copyright © 2020 Azzarelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberta Azzarelli, cm9iZXJ0YS5henphcmVsbGlAdW5pcGkuaXQ=
 Roberta Azzarelli
Roberta Azzarelli