- 1Central Laboratory, Huazhong University of Science and Technology Union Shenzhen Hospital and the Affiliated Shenzhen Sixth Hospital of Guangdong Medical University, Shenzhen, China
- 2Shenzhen Key Lab of Endogenous Infection, Huazhong University of Science and Technology Union Shenzhen Hospital and the Affiliated Shenzhen Sixth Hospital of Guangdong Medical University, Shenzhen, China
- 3Department of Stomotology, Huazhong University of Science and Technology Union Shenzhen Hospital and the Affiliated Shenzhen Sixth Hospital of Guangdong Medical University, Shenzhen, China
- 4Department of Pathology, Huazhong University of Science and Technology Union Shenzhen Hospital and the Affiliated Shenzhen Sixth Hospital of Guangdong Medical University, Shenzhen, China
- 5Shenzhen Key Laboratory for Neuronal Structural Biology, Biomedical Research Institute, Shenzhen Peking University–The Hong Kong University of Science and Technology Medical Center, Shenzhen, China
- 6Division of Life Science, The Hong Kong University of Science and Technology, Kowloon, Hong Kong
- 7Department of Endocrinology, Huazhong University of Science and Technology Union Shenzhen Hospital and the Affiliated Shenzhen Sixth Hospital of Guangdong Medical University, Shenzhen, China
Organogenesis, including renal development, requires an appropriate retinoic acid concentration, which is established by differential expression of aldehyde dehydrogenase 1 family member A2 (ALDH1A2) and cytochrome P450 family 26 subfamily A/B/C member 1 (CYP26A1/B1/C1). In the fetal kidney, ALDH1A2 expresses in the developing stroma and renal vesicle and its derivatives but does not present in the ureteric bud. It remains unclear what may contribute to this expression pattern. Here we show that the glycogen synthase kinase 3 alpha/beta (GSK3A/B) inhibitor CHIR99021 significantly represses ALDH1A2 expression in WiT49, which is a Wilms’ tumor cell line that exhibits “triphasic” differential potential and is used as a fetal kidney cell model. CHIR99021 fails to suppress ALDH1A2 as β-catenin is inhibited, suggesting that the downregulation of ALDH1A2 by CHIR99021 is through Wnt/β-catenin signaling. Ectopic expression of mouse Wnt1, Wnt3a, Wnt4, and Wnt9b represses ALDH1A2 expression in WiT49 cells. Using immunohistochemistry, we show an inverse correlation of Aldh1a2 expression with β-catenin in rat E18.5 kidney. ChIP demonstrated that β-catenin is recruited to the ALDH1A2 promoter, the conserved intron1G, and another site within intron 1 of ALDH1A2. Using a luciferase assay, we further show that the ALDH1A2 promoter and the intron1G element are involved in the repression of ALDH1A2 expression by CHIR99021. Our work demonstrates that ALDH1A2 expression can be directly repressed by the Wnt/β-catenin signaling in fetal kidney cells, suggesting that Wnt/β-catenin may play a role in maintaining the expression pattern of ALDH1A2 in the fetal kidney, thus controlling the availability and localization of retinoic acid and regulating aspects of kidney development.
Introduction
Renal development is initiated by mutual signaling interactions between the ureteric bud derived from the Wolffian duct and its surrounding mesenchyme. The Wolffian duct extends caudally and interacts with its adjacent mesenchyme to generate a pronephric, mesonephric, and, finally, the metanephric kidney. The metanephros grows and differentiates to make up the functional kidney in adult. Metanephric mesenchyme differentiates and becomes the condensed mesenchyme (cap mesenchyme), which forms lineages of epithelial, stromal, and endothelial cells. The epithelial cell lineage differentiates into structures that include the renal vesicle, pretubular aggregate, comma-shaped and S-shaped bodies, and renal nephron, including podocyte on the renal vesicle layer of the glomerulus, proximal convoluted tubule, loop of Henle, and distal convoluted tubule. Ureteric buds develop to form collecting duct, renal calyces, pelvis, and ureter (Davidson, 2008; Dressler, 2009). Kidney development is a continuous process in the outer cortex of the developing kidney, where signaling interaction among cell types, including mesenchyme, ureteric bud, and stroma, continues until birth in human or after birth in mouse (Cullen-Mcewen et al., 2016).
Renal development depends on all-trans retinoic acid (atRA) signaling. atRA mainly acts as a ligand for nuclear retinoic acid receptors (RARs), which consists of RARα, RARβ, and RARγ, resulting in a change in subsequent biological procedures and gene transcription (Dolle, 2009). atRA can also act through non-canonical pathways including binding to the PPARδ/β (peroxisome proliferator-activated receptor delta/beta) receptor in the nucleus (Napoli, 2017) or binding to the RARα/Gαq (G protein alpha Q) complex on the cell membrane lipid raft to activate the p38MAPK (Piskunov and Rochette-Egly, 2011) and the PI3K pathway (Masiá et al., 2007). The knockout of both Rara and Rarb results in the hypoplasia/agenesis of the kidney (Mendelsohn et al., 1999). The impact of retinoic acid in renal development is known to mediate through the upregulation of ret proto-oncogene (Ret) expression on the cell membrane of the ureter bud. Ret is a receptor that binds its ligand, glial cell line-derived neurotrophic factor (Gdnf), secreted from the cap mesenchyme (Batourina et al., 2001; Rosselot et al., 2010) and stromal cells (Magella et al., 2018). Endogenous atRA is produced by two stages of oxidation of vitamin A (all-trans-retinol) via an intermediate state, all-trans retinal (Duester, 2008; Napoli, 2012). The second stop of retinoic acid synthesis is catalyzed by enzymes including aldehyde dehydrogenase 1 family member A1 (ALDH1A1) (Fan et al., 2003), aldehyde dehydrogenase 1 family member A2 (ALDH1A2) (Wang et al., 1996; Zhao et al., 1996), and aldehyde dehydrogenase 1 family member A3 (ALDH1A3) (Grun et al., 2000; Sima et al., 2009). Aldh1a2–/– mutant mouse kidney exhibits a significant reduction of ureteric buds and nephrons (Rosselot et al., 2010). Aldh1a3–/– has no or little effect on renal development (Rosselot et al., 2010). Aldh1a1–/– mutant is non-lethal, adults are fertile, and no abnormality has been reported (Fan et al., 2003; Matt et al., 2005). These data suggest that Aldh1a2 activity is the primary source of retinoic acid during kidney development (Rosselot et al., 2010). The retinoic acid can be inactivated by cytochrome P450 family 26 (CYP26) subfamily A/B/C member 1 (CYP26A1/B1/C1). Excessive retinoic acid is teratogenic (Piersma et al., 2017) and causes hypoplastic and polycystic kidney (Fantel et al., 1977; Lee et al., 2012). Therefore, the deficiency or excess of retinoic acid signaling leads to abnormalities in kidney development, suggesting that the proper regulation of retinoic acid concentration and retinoic acid synthetic enzymes is critical.
Regulation of Aldh1a2 expression is important for pronephros development. In zebrafish pronephros, cdx mutation leads to Aldh1a2 upregulation and disrupts pronephros positioning and distal segment formation (Wingert et al., 2007), suggesting that inhibition of Aldh1a2 expression by cdx is critical for pronephros development. Recently, it was shown that anterior kidney fates are induced by retinoic acid on the dorsal side of the embryo with Aldh1a2 expression, while posterior kidney progenitors are protected by the Cyp26a1 enzyme (Naylor et al., 2016). These studies suggest that Aldh1a2 expression determines the precise localization of retinoic acid signaling and that the repression of the Aldh1a2 is necessary for the pronephros formation of zebrafish. However, studies of the regulation of Aldh1a2 expression within the metanephric kidney remains rare.
In the metanephric kidney, as well as being strongly expressed in the stromal cells of the outer cortex, Aldh1a2 is weakly presented in the proximal section of the comma-shaped body, vigorously but transiently expressed in glomerular anlagen in the S-shaped body and the visceral layer of the glomerulus in stage III nephron, and declines aggressively in the podocyte of stage IV nephron. Its expression in collecting tubules during E18-P4 has also been observed (Batourina et al., 2001; Niederreither et al., 2002; Marlier and Gilbert, 2004; Rosselot et al., 2010). This intricate expression pattern of Aldh1a2 should be tightly controlled. It was reported that Foxd1 knockout led to the repression of Aldh1a2 expression in stromal cells of the fetal kidney (Levinson et al., 2005; Fetting et al., 2013). Recently we provide in vitro evidence that ectopic expression of WT1 transcription factor (WT1) represses ALDH1A2 expression in the human fetal kidney cell line HEK293 (Li et al., 2017). It is important to note that, as the retinoic acid concentration is dependent on the expression of enzymes like ALDH1A2, repression of ALDH1A2 is as vital as its upregulation for the formation of the correct local concentration of retinoic acid in kidney development. The regulator responsible for the lack of ALDH1A2 expression in ureteric bud and cap mesenchyme remains unclear.
Wnt signaling is essential for fetal kidney development (Halt and Vainio, 2014). The canonical Wnt/β-catenin pathway is the most well-characterized Wnt pathway (Nusse and Clevers, 2017). The signal is executed by translocation of β-catenin from plasma into the nucleus and acts together with TCF or other unknown factors to regulate gene expression. Observations show that Wnt-2b, -4, -5a, -6, -7b, -9b, and -11 are expressed during kidney development (Kispert et al., 1996; Kispert et al., 1998; Lin et al., 2001; Itaranta et al., 2002; Carroll et al., 2005). Wnt1, -3a, -4, -7a, and -7b were reported to trigger nephrogenesis in the metanephric mesenchyme (Kispert et al., 1998). Wnt4 was secreted by metanephric mesenchyme. The loss of Wnt4 leads to the failure of the formation of pretubular cell aggregates (Stark et al., 1994). Wnt9b is expressed in ureteric buds and activates the canonical Wnt signaling pathway in the surrounding metanephric mesenchyme (Kispert et al., 1998; Carroll et al., 2005; Park et al., 2007). Loss of Wnt9a leads to failure of nephron formation. It also plays an important role in the convergent extension of renal tubular epithelial cells. Wnt6, Wnt7b, and Wnt11 are generated by ureteric bud cells. Wnt2b is secreted by developing stromal cells (Halt and Vainio, 2014).
Recently, increasing evidence suggests that the canonical Wnt signaling may regulate Aldh1a2 expression. It was shown that, in 293T cells, β-catenin and TCF4 activate mouse Aldh1a2 promoter (Hong et al., 2015). In the Six2 positive pre-tubular aggregate of the embryonic kidney, β-catenin/LEF/TCF complex and Six2 were recruited to a region within the first intron of the Aldh1a2 gene (chr9:71065207-71065805) (Park et al., 2012). This region partially overlapped with a TCF7L2 (TCF4) bound region identified in HEK293 cells using ChIP-sequencing (Frietze et al., 2012). It was previously found that in the first intron of the Aldh1a2 gene, there is an enhancer termed intron1G that can activate Aldh1a2 expression in the mouse developing neuron cells. Intron1G contains at least three predicted LEF/TCF binding motif (Castillo et al., 2010).
As Wnt signaling and retinoic acid signaling are critically involved in kidney development and disease and the above data suggest that the canonical Wnt signaling may regulate Aldh1a2, we hypothesized that Wnt/β-catenin signaling might regulate Aldh1a2 expression in the developing kidney. In this study, we focus on examining whether the canonical Wnt signaling can control Aldh1a2 expression in the fetal kidney context. We report for the first time that Aldh1a2 expression can be suppressed by CHIR99021 via the canonical Wnt signaling. We confirm that Aldh1a2 repression by Wnt signaling involves the Aldh1a2 promoter and intron1G element. Our data indicate that Wnt signaling could be upstream of retinoic acid signaling during kidney development. Our findings have implications for linking the Wnt and retinoic acid pathways during kidney development.
Materials and Methods
Chemicals
The GSK3 inhibitor CHIR99021 (Cat. SML1046, Sigma-Aldrich Corp., St. Louis, MO, United States) was obtained from Sigma. CHIR99021 was dissolved in dimethyl sulfoxide (DMSO) (Cat.196055, MP Biomedicals). CHIR99021 is an aminopyrimidine derivative that is a highly potent inhibitor of GSK3. It inhibits GSK3β (IC50 = 6.7 nM) and GSK3α (IC50 = 10 nM), acting as a Wnt activator.
Primers
The primers for real-time PCR used in this study are listed in Table 1.
Plasmids
M50 Super 8x TOPFlash (Addgene #12456) and M51 Super 8x FOPFlash (TOPFlash mutant) (Addgene #12457) were gifts from Randall Moon (Veeman et al., 2003). The HSV-thymidine kinase promoter (pRL-TK) (Promega), kindly provided by Dr. Kun Du, was used as a Renilla luciferase control reporter.
Expression plasmids of multiple Wnts, including pRK5-mWnt1 (Addgene #42273), pRK5-mWnt2b (Addgene #42275), pRK5-mWnt3a (Addgene #42277), pRK5-mWnt4 (Addgene #42278), pRK5-mWnt5a (Addgene #42279), pRK5-mWnt6 (Addgene #42281), pRK5-mWnt7a (Addgene #42282), pRK5-mWnt7b (Addgene #42283), pRK5-mWnt9b (Addgene #42287) and pRK5-mWnt11 (Addgene #42290), were gifts from Chris Garcia and Jeremy Nathans (Yu et al., 2012). pRK5 empty vector was made by re-ligation of the backbone of pRK5-mWnt4 (Addgene #42278) digested by PstI.
ALDH1A2 promoter or intron1G was cloned using the primers shown in Table 2. Promoter construct pGL3B-1A2ps (889 bp) was made by digestion of pGL3B-1A2p with KpnI/AvrII, followed by ligation of the promoter sequence to the pGL3-basic vector, which was digested with KpnI/NheI. The MF2 and PF2 reporter vectors were made by joining the minus strand or the plus strand of ALDH1A2 intron1G with the ALDH1A2 promoter F2 using the overlap extension polymerase chain reaction method (OE-PCR) (Figure 6A) followed by ligation with pGL3-basic plasmid using KpnI/HindIII enzyme sites.
siRNAs
siRNAs targeting human CTNNB1 exons were designed and synthesized by GenePharma (Shanghai, China). The siRNA oligos include CTNNB1 siRNA-1 5′-GGA CAC AGC AGC AAU UUG UTT-3′, CTNNB1 siRNA-2 5′-GCU GCU UUA UUC UCC CAU UTT-3′, and the negative control siRNA oligo 5′-UUC UCC GAA CGU GUC ACG UTT-3′. The final concentration of siRNA for transfection is 50 nmol/L. Lipofectamine® RNAiMAX reagent (Invitrogen) was used for siRNA transfection according to the manufacturer’s instruction.
Cell Culture and Transfection
The WiT49 and 293 cells were cultured as described previously (Li et al., 2017). The WiT49 cell line was donated by Dr. Herman Yeger. It is a Wilms’ tumor (WT) cell line that is derived from the first-generation xenograft of a human WT lung metastasis. Some differentiation potential is retained by WiT49 cells, displaying the so-called “triphasic” histology (epithelioid, stromal-like, and mesenchymal) when grown in tissue culture plates (Alami et al., 2003). It is also triphasic (Mengelbier et al., 2014) or biphasic with stromal and epithelial components dominant (Li et al., 2010) in orthotopic xenograft models, suggesting that WiT49 may mimic certain aspects of the developing kidney (Li et al., 2002; Rivera and Haber, 2005).
Animals
The pregnant Sprague-Dawley rats (SD rats) were obtained from Shenzhen Peking University–The Hong Kong University of Science and Technology Medical Center. Animal welfare and experimental procedures were carried out following the Guide for the Care and Use of Laboratory Animals (Ministry of Science and Technology of China, 2006) and were approved by the animal ethics committee of Nanshan People’s Hospital.
Real-Time RT-PCR
RNA was purified using TRIZOL reagent (Invitrogen), and its concentration and purity were assessed at 260/280 nm by applying Nanodrop (Thermo). A sample of 1 μg total RNA was reverse-transcribed with oligo (dT)20 at 50°C for 1 h using the ThermoScript RT-PCR system (Invitrogen). Comparative quantitative real-time PCR was carried out employing SYBR Green qPCR supermix universal (Invitrogen) using the ABI7500 real-time PCR system. The reaction mixture was comprised of 10 μl SYBR (Invitrogen), 50 nmol/l ROX reference dye, 0.2 μmol/l forward primer, 0.2 μmol/l reverse primer, and 5 μl of 1:10 diluted cDNA template. PCR cycling was as follows: 50°C for 2 min, a denaturing step of 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 58°C for 30 s, and 72°C for 30 s. Gene expression was quantified by applying the comparative Ct method, normalizing to the housekeeping gene TATA box binding protein (TBP). Assays were accomplished in triplicate.
Western Blot
Western blot was performed using polyclonal antibodies against β-catenin (Cat. 51067-2-AP, Proteintech), ALDH1A2 (Cat. ab75674, Abcam), and GAPDH (Goodhere, China). Secondary antibodies, including anti-rabbit IgG (whole molecule) – anti-mouse IgG and peroxidase antibody (whole molecule) – peroxidase antibody, were obtained from Sigma. In brief, 1 × 106 cells were lysed in 150 μl 1 × Cell Lysis Buffer (Cell Signaling Technology, Inc.) plus EDTA-free protease inhibitor cocktail (Roche). Protein concentrations were measured by employing the DC Protein Assay Kit (Bio-Rad). For each lane, 50 μg proteins were denatured in 30 μl sample buffer (60 mmol/l Tris pH 6.8, 10% glycerol, 2% SDS, 5% mercaptoethanol), followed by incubation at 100°C for 5 min, and cooling on ice to denature proteins. Protein samples were loaded for electrophoresis and transferred to Immobilon-P Transfer Membrane (Millipore) with a wet transfer apparatus (Bio-Rad). The Immobilon-P membrane was blocked with 1% bovine serum albumin (BSA) (Sigma) in phosphate-buffered saline (PBS) for 2 h at room temperature. The membrane was then probed with primary antibodies overnight at 4°C at a dilution of 1:1000 or 1:200, followed by incubation with secondary antibody at room temperature for 1.5 h. Protein bands were visualized with ECLTM Prime Western Blotting Detection Reagent (Amersham, GE Healthcare). Gel images were obtained employing the MiniChemi professional machine (SageCreation Science Co., Ltd., Beijing, China). The semi-quantification of protein bands was performed using ImageJ software.
Chromatin Immunoprecipitation (ChIP)
A Magna CHIPTM A chromatin immunoprecipitation kit (Millipore) was used according to the manufacturer’s protocol. Briefly, WiT49 cells were treated with 5 μM CHIR99021 for 48 h, and then the cells (5.4 × 106/T75) were fixed with 1% formaldehyde for 20 min at room temperature. Sonication was performed for 5 min (with cycles of 30 s on/30 s off) in a Bioruptor® Pico (Diagenode) on 300 μl cell lysates in 1.5-ml Bioruptor Pico microtubes with Caps (Diagenode, Cat. No. C30010016) at a concentration of 18 × 106 cells/ml of nuclear lysis buffer. Acetyl-Histone 3 and Rabbit IgG in the Magna CHIPTM A kit were used as positive and negative controls, respectively. Mouse Pol II antibody from the Magna CHIPTM G kit (Millipore) was also used as a control. Rabbit polyclonal anti-β-catenin (Cat. 51067-2-AP, Proteintech) was used. The DNA yielded was then amplified by PCR with primers specific to different regions, using HotStarTaq Plus DNA polymerase (Qiagen) with the primers shown in Table 1. The PCR temperature program for these ChIP DNA samples is 95°C for 5 min, 40 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 35 s, followed by an extension step of 72°C for 5 min. ChIP PCR products were loaded to the 1.5% agarose gel, and electrophoresis was performed.
Luciferase Assay
FuGENE® HD transfection reagent (Roche) and luciferase assay system with reporter lysis buffer (Promega) were used for the promoter transcriptional activity assay according to the manufacturer’s instructions. Briefly, 105 cells were seeded to each well of a 24-well plate 18–20 h before transfection and maintained in 1 ml media. Serum-free DMEM media was mixed with FuGENE® HD, followed by incubation at room temperature for 5 min. Plasmids were then added to this mixture and mixed, followed by incubation for 20 min. The ratio between the FuGENE® HD volume and total plasmid quantity was 3 μl:1 μg. A volume of 40 μl of the serum-free DMEM/FuGENE® HD/plasmids mixture was added to each well. Cells were harvested 48 h after transfection using 100 μl of 1 × reporter lysis buffer in each well of the 24-well plate on a Belly Dancer (Stovall Life Science, Inc) for 20 min at room temperature. Luciferase assay was performed by adding 10 μl of cell lysate to 50 μl of lyophilized luciferase assay reagent II (LAR II) (Promega) and tapping the reaction mixture for 10 s. The reaction was then measured by a ModulusTM single tube multimode reader (Turner Biosystems). The renilla activity of the same tube was also measured. The value of luciferase activity relative to the renilla activity of the same sample was used for data analysis. Student’s t-tests (two tails, unpaired) were performed using Excel software (Microsoft), with p < 0.05 considered statistically significant.
Immunofluorescence
Sterile coverslips were placed in 6-well plates before WiT49 cell seeding at a density of 100,000 cells/cm2. Cells were allowed to grow for 2 days before fixing with 8% paraformaldehyde in 1 × PBS (pH 7.0, 500 μl/well) for 15 min. The fixation was stopped with 100 mM glycine, followed by 0.1% Triton X-100 treatment. Cells were blocked with 1% BSA (Sigma, 2 ml/well) in PBS (pH 7.5) for 30 min at room temperature. Afterward, the coverslip was lifted, and the side with cells attached was placed face down directly in contact with 100 μl of primary antibody (1/200 diluted) on plastic at 4°C overnight. The coverslips were then incubated with 100 μl of secondary antibodies (1/200 diluted) in 1% BSA dissolved in PBS (pH 7.5) at room temperature for 60 min in the dark. Anti-rabbit secondary antibody AlexaFluor 546 (red) F(ab)2 GαR (Invitrogen) or anti-mouse secondary antibody AlexaFluor 488 (green) F(ab)2 GαM (Invitrogen) was used. Afterward, the cells on each coverslip were stained with ProLong Gold (Invitrogen), which contains 4′-6-diamidino-2-phenylindole (DAPI). The glass slides were examined under an Olympus BX51 fluorescent microscope and analyzed.
Immunohistochemistry (IHC) and Hematoxylin and Eosin (H&E) Staining
Histology was performed to quantify Aldh1a2 and Ctnnb1 (β-catenin) expression in paraffin-embedded rat E18.5 embryonic kidney samples. IHC was performed on sections using rabbit polyclonal anti-β-catenin (Cat. 51067-2-AP, Proteintech) and rabbit polyclonal anti-ALDH1A2 (Cat. ab75674, Abcam). H&E staining was performed using Mayer’s hematoxylin solution.
Software
Primers were designed using the PrimerQuest Design Tool1.
Statistical Analysis
Student’s t-test was used to compare the significance of the difference between every two groups. Statistical analysis was performed using Excel software (Microsoft). Results were presented as mean ± standard error of the mean. All P-values were two-tailed, and P < 0.05 was considered to be a statistically significant difference.
Results
GSK3 Inhibitor CHIR99021 Inhibits Aldh1a2 Expression in WiT49 Cells
Previous studies have used CHIR99021 in a range of 1–10 μM in experiments with fetal kidney cells (Francipane and Lagasse, 2015) or kidney organoids generated from iPSC (Takasato et al., 2016; Taguchi and Nishinakamura, 2017; Przepiorski et al., 2018). We used a concentration of 5 μM, which is the median value of the concentration range that has been used in these experiments. WiT49 cells were treated with 0.1% DMSO or 5 μM CHIR99021 for 48 h. Real-time PCR results show that ALDH1A2 mRNA levels were reduced to 25% of that measured in DMSO control cultures (Figure 1A). Western blot shows that ALDH1A2 protein levels were also significantly repressed in the CHIR99021 group compared to the DMSO control (Figure 1B).
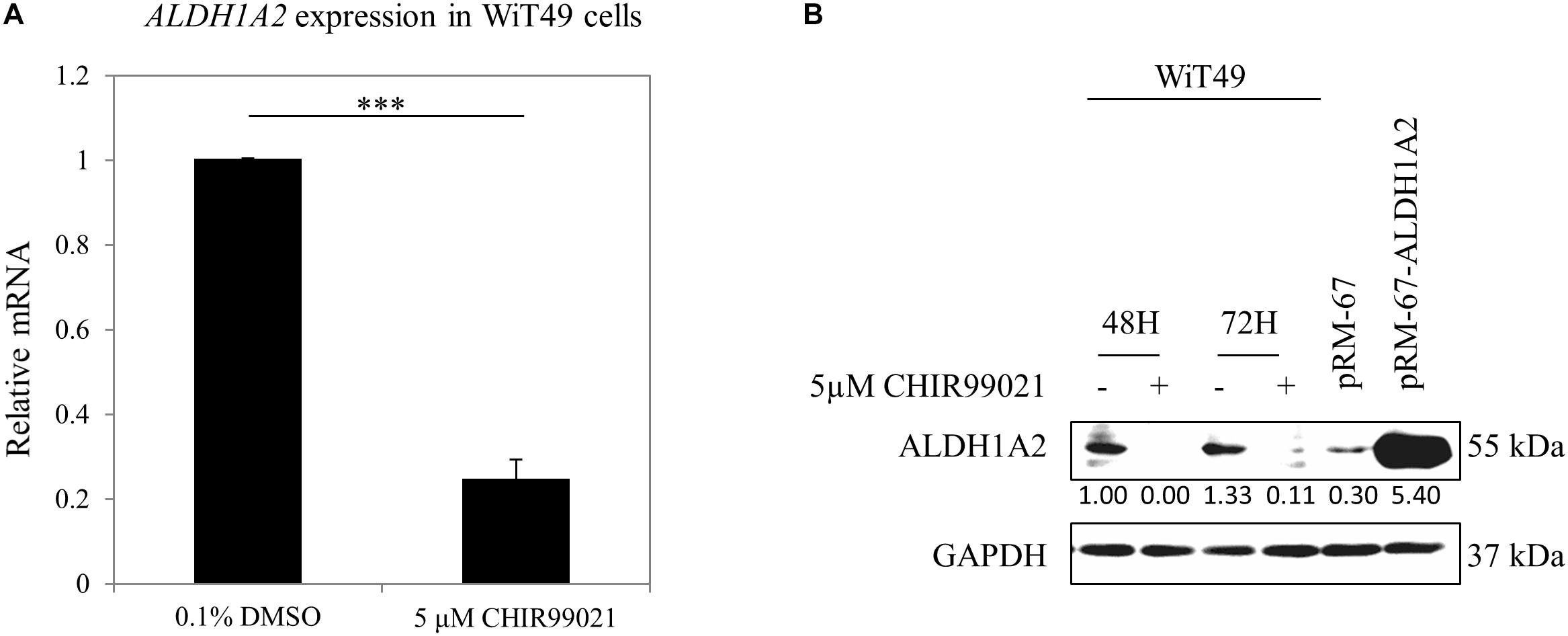
Figure 1. GSK3 inhibitor CHIR99021 inhibits Aldh1a2 expression in WiT49 cells. (A) Effect of CHIR99021 on ALDH1A2 mRNA expression in WiT49 cells. WiT49 cells were treated with 0.1% DMSO (control vehicle) or 5 μM CHIR99021 for 48 h. ALDH1A2 expression relative to TBP was measured by real-time RT-PCR. Values are the mean ± SD of three determinations; representative results from at least three separate experiments. ∗∗∗P < 0.001 compared with the DMSO control group (without CHIR99021 addition). (B) Effect of CHIR99021 on ALDH1A2 protein expression in WiT49 cells. WiT49 cells were treated with 0.1% DMSO or 5 μM CHIR99021 for 48 or 72 h. ALDH1A2 and GAPDH protein expression were measured by Western blot. pRM-67-ALDH1A2, positive control of ALDH1A2 protein made by transfection of mammalian expression vector pReceiver-M67-ALDH1A2 to WiT49 cells. pRM-67, negative control of ALDH1A2 protein generated by transfection of pReceiver-M67 empty vector. Values under protein bands were calculated by the density of bands of ALDH1A2 divided by the density of bands of GAPDH.
Inhibition of ALDH1A2 Expression by GSK3 Inhibitor CHIR99021 Is via β-Catenin Signaling
To confirm that the GSK3 inhibitor CHIR99021 affects Wnt/β-catenin signaling, we measured whether CHIR99021 affects the β-catenin nuclear translocation and the activity of TCF reporter. WiT49 cells were treated with 0.1% DMSO or 5 μM CHIR99021 for 48 h. Immunofluorescence detection of β-catenin shows that β-catenin protein mainly presented on the membrane/cytoplasm of WiT49 cells of the 0.1% DMSO control. With 5-μM CHIR99021 treatment, the majority of β-catenin translocated to the nucleus of WiT49 cells (Figure 2A). Luciferase assay results show that CHIR99021 activated TOPFlash TCF reporter (Figure 2B), suggesting that CHIR99021 treatment can activate Wnt/β-catenin signaling in WiT49 cells.
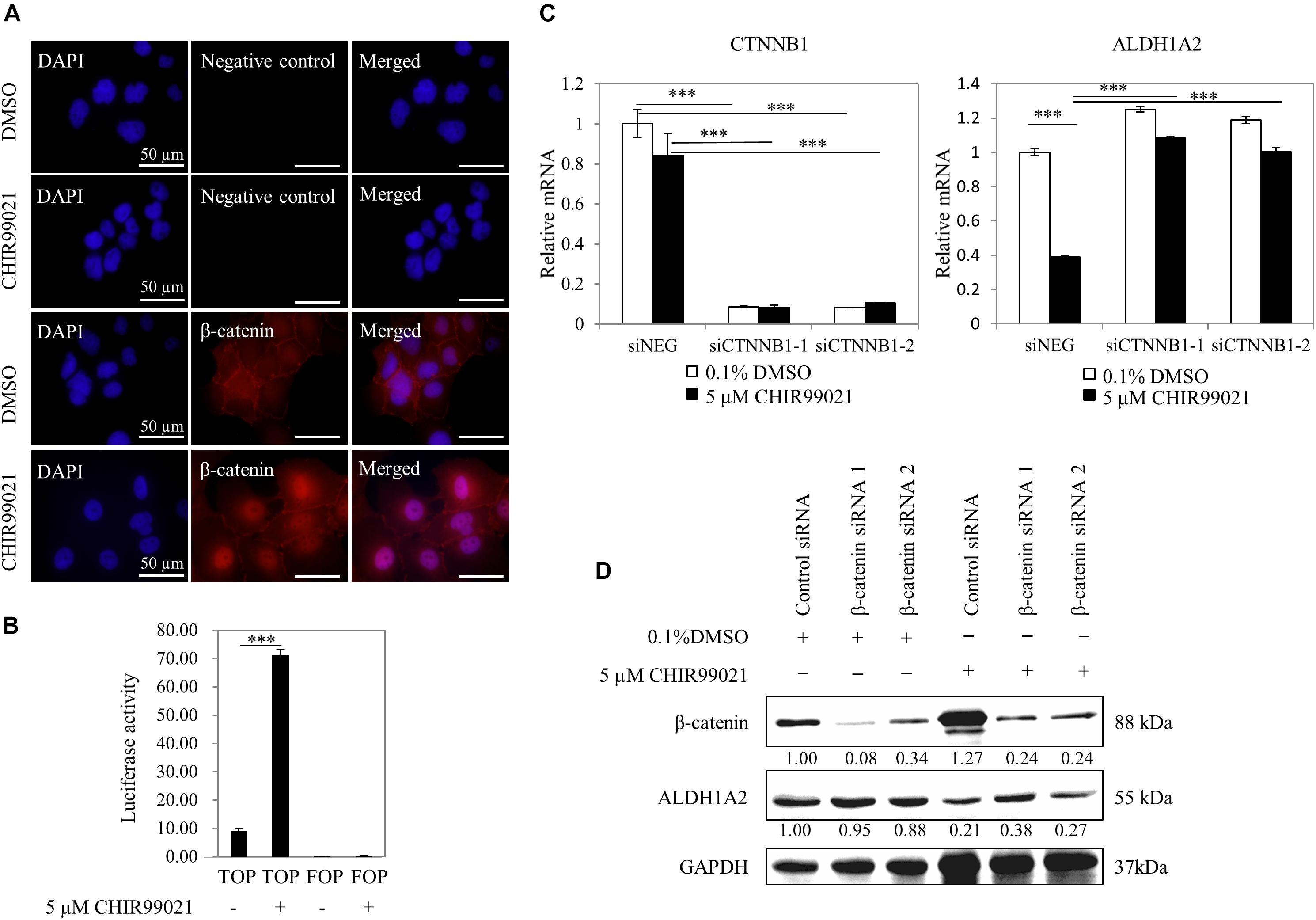
Figure 2. Inhibition of ALDH1A2 expression by GSK3 inhibitor CHIR99021 is via β-catenin signaling. (A) Effect of CHIR99021 on β-catenin nuclear translocation in WiT49 cells. WiT49 cells were treated with 5 μM CHIR99021 or 0.1% DMSO for 48 h, and then immunofluorescence was performed with β-catenin antibody and DAPI staining. Negative control represents no primary antibody control. Bar length equals 50 μm. (B) Effect of CHIR99021 on the TCF reporter in WiT49 cells. A 500-ng sample of TOPFlash or FOPFlash constructs, together with 50 ng of the pRL-TK renilla control vector, was transiently transfected into WiT49 cells. Cells were treated with 0.1% DMSO or 5 μM CHIR99021 after 24 h of transfection. Luciferase reporter activities relative to renilla (pRL-TK) are shown in percentage terms. Values are the mean ± SD of three determinations from separate experiments. ∗∗∗P < 0.001 compared with the DMSO control group (without CHIR99021 addition). (C,D) Effect of CTNNB1 knockdown on the downregulation of ALDH1A2 by CHIR99021 treatment. WiT49 was transfected with 50 nM CTNNB1 siRNA or negative control siRNA for 24 h, followed by treatment of 5 μM CHIR99021 for another 48 h. CTNNB1 and ALDH1A2 mRNA expression relative to TBP control in WiT49 cells were measured by real-time RT-PCR (C). Values are the mean ± SD of three determinations from separate experiments. ∗∗∗P < 0.001 compared with the siNEG control group. β-catenin (CTNNB1), ALDH1A2 and GAPDH protein expression in WiT49 cells were measured by Western blot (D). Values under bands were calculated by the density of bands of β-catenin or ALDH1A2 divided by the density of bands of GAPDH.
Next, we tested whether repression of ALDH1A2 by CHIR99021 requires β-catenin. The expression of CTNNB1 (β-catenin) in WiT49 cells was knocked down with siRNA for 24 h, followed by treatment of cells with 0.1% DMSO or 5 μM CHIR99021 for another 48 h. Real-time RT-PCR shows that when CTNNB1 was knocked down, CHIR99021 failed to represses ALDH1A2 (Figure 2C). Suppression of CTNNB1 alone slightly upregulated ALDH1A2 expression (Figure 2C), probably because β-catenin is mainly located on the membrane/cytoplasm in WiT49 cells. Western blot shows that in the 0.1% DMSO control group, CTNNB1 knockdown had little effect on ALDH1A2 expression. In the 5-μM CHIR99021 treatment group, CTNNB1 knockdown led to ALDH1A2 upregulation (Figure 2D). These data suggest that the repression of ALDH1A2 expression by CHIR99021 requires CTNNB1. Taken together, these results suggest that canonical Wnt signaling represses ALDH1A2 expression.
Distinct Wnt Family Members Can Repress ALDH1A2 Expression in WiT49 Cells
Before measuring whether Wnt molecules may affect ALDH1A2 expression, we tested whether Wnts that have been shown to express or have an effect in kidney development (Halt and Vainio, 2014) may activate the TCF reporter. Luciferase assay using TOPFlash/FOPFlash reporter shows that transient transfection of mouse Wnt1, Wnt3a, Wnt4, and Wnt9b expression vector to WiT49 cells for 48 h led to significant activation of the TOPFlash reporter, suggesting that these four Wnts can activate the canonical Wnt signaling in WiT49 cells. Six other mouse Wnts, namely Wnt2b, Wnt5a, Wnt6, Wnt7a, Wnt7b, and Wnt11, did not affect TOPFlash reporter activity (Figure 3A).
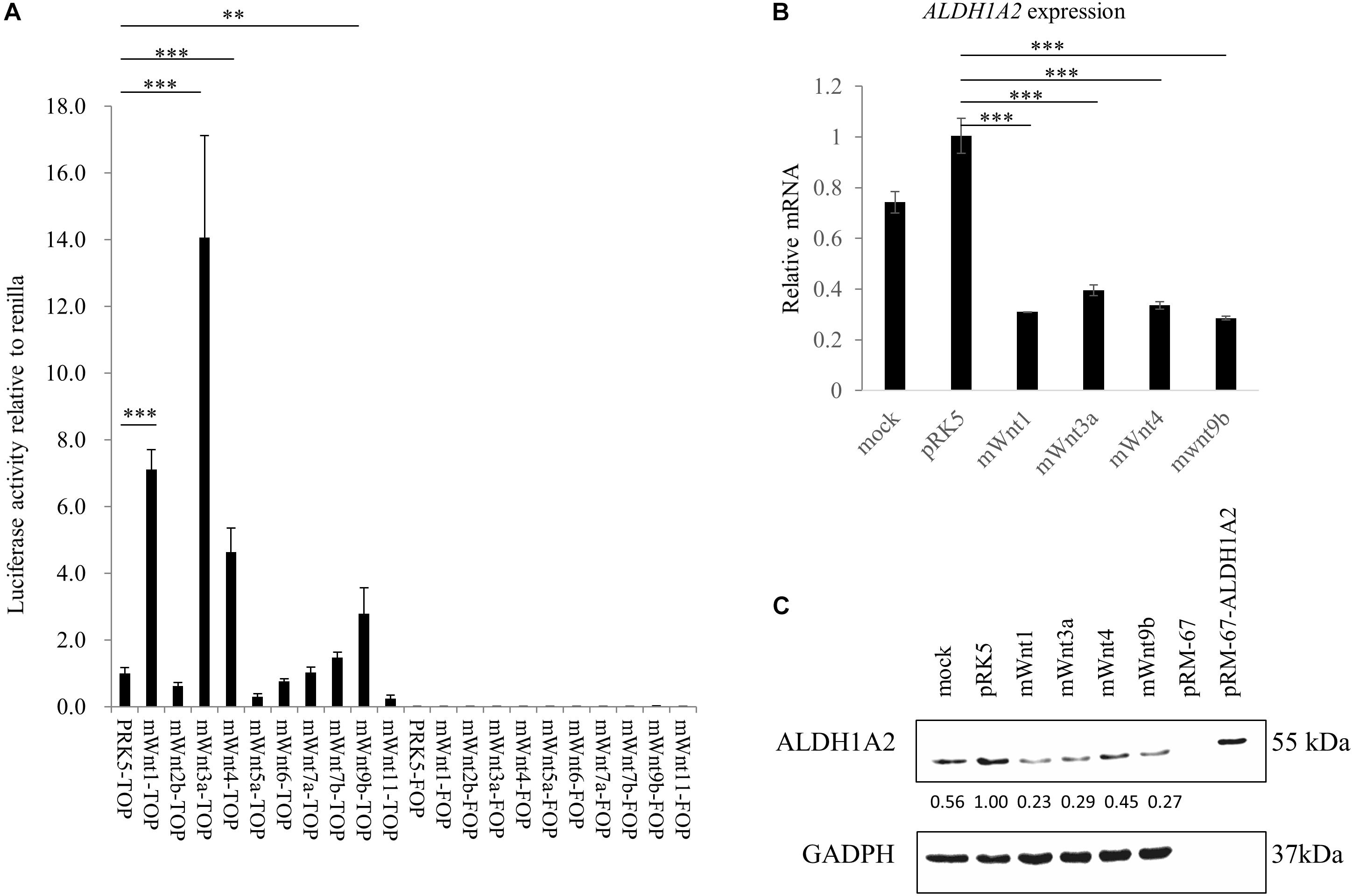
Figure 3. Wnts affect ALDH1A2 expression in WiT49 cells. (A) Effects of Wnts on TCF reporter activity in WiT49 cells. 400 ng TOPFlash or FOPFlash constructs with 100 ng pRK5-mWnts expression vectors or pRK-5 empty vector, as well as 25 ng of the pRL-TK renilla control vector, was transiently transfected into WiT49 cells for 48 h. The pRK5-mWnts include pRK5-mWnt1, pRK5-mWnt2b, pRK5-mWnt3a, pRK5-mWnt4, pRK5-mWnt5a, pRK5-mWnt6, pRK5-mWnt7a, pRK5-mWnt7b, pRK5-mWnt9b, and pRK5-mWnt11. Luciferase reporter activities relative to renilla (pRL-TK) are shown. Values are the mean ± SD of four determinations; representative results from at least three separate experiments are shown. ∗∗P < 0.01 and ∗∗∗P < 0.001 compared with their respective controls, calculated using the Student’s t-test. (B,C) Effect of Wnts on ALDH1A2 expression in WiT49 cells. WiT49 cells were transiently transfected with pRK5-mWnt1, pRK5-mWnt3a, pRK5-mWnt4, pRK5-mWnt9b, or the empty vector pRK5 for 48 h. The ALDH1A2 expression relative to TBP was measured by real-time RT-PCR (B). Values are the mean ± SD of three determinations; representative results from at least three separate experiments. ∗∗P < 0.01 and ∗∗∗P < 0.001 compared with the empty vector pRK5 control group. The ALDH1A2 protein expression was measured by Western blot (C). pRM-67-ALDH1A2, positive control of ALDH1A2 protein made by transfection of mammalian expression vector pReceiver-M67-ALDH1A2 to WiT49 cells. pRM-67, negative control of ALDH1A2 protein generated by transfection of pReceiver-M67 empty vector. Per lane, 50 μg of the sample was loaded, and 2 μg ALDH1A2 protein-positive control was loaded. Mock represents samples of cells treated with transfection reagent and without plasmid DNA. Values under bands were calculated by the density of bands of ALDH1A2 divided by the density of bands of GAPDH.
Next, we transiently transfected expression vectors of Wnts that have been shown to activate TCF reporter (Figure 3A) to WiT49 cells and measured whether ALDH1A2 expression was altered. Mouse Wnt1, Wnt3a, Wnt4, and Wnt9b expression vectors were transiently transfected to WiT49 cells for 48 h. Samples were tested for ALDH1A2 expression. Real-time RT-PCR and Western blot results both show that mouse Wnt1, Wnt3a, Wnt4, and Wnt9b transfection led to repression of ALDH1A2 expression (Figures 3B,C).
Taken together, the above data suggest that Wnt/β-catenin signaling can repress ALDH1A2 expression in the fetal kidney model WiT49 cells.
β-Catenin and Aldh1a2 Proteins Are Localized in Distinct, Adjacent Domains in vivo
Having shown negative regulation of ALDH1A2 expression by β-catenin in WiT49 cell line in vitro, we next address whether the localization pattern of Aldh1a2 and β-catenin in the fetal kidney is consistent with this repressive effect. We determined the Aldh1a2 and β-catenin protein localization in the rat E18.5 fetal kidney by immunohistochemistry. The results show that Aldh1a2 primarily localized in the stroma of the developing renal cortex but did not localize in the ureteric bud and cap mesenchyme (Figures 4C,F). β-catenin strongly localized in the ureteric bud and cap mesenchyme, but little was detectable in the developing stroma (Figures 4D,G). In the developing podocytes, Aldh1a2 was strongly localized (Figures 4E,I), while β-catenin was not detectable (Figures 4D,H). These data suggest an inverse relationship between β-catenin and Aldh1a2 protein localization in the fetal kidney.
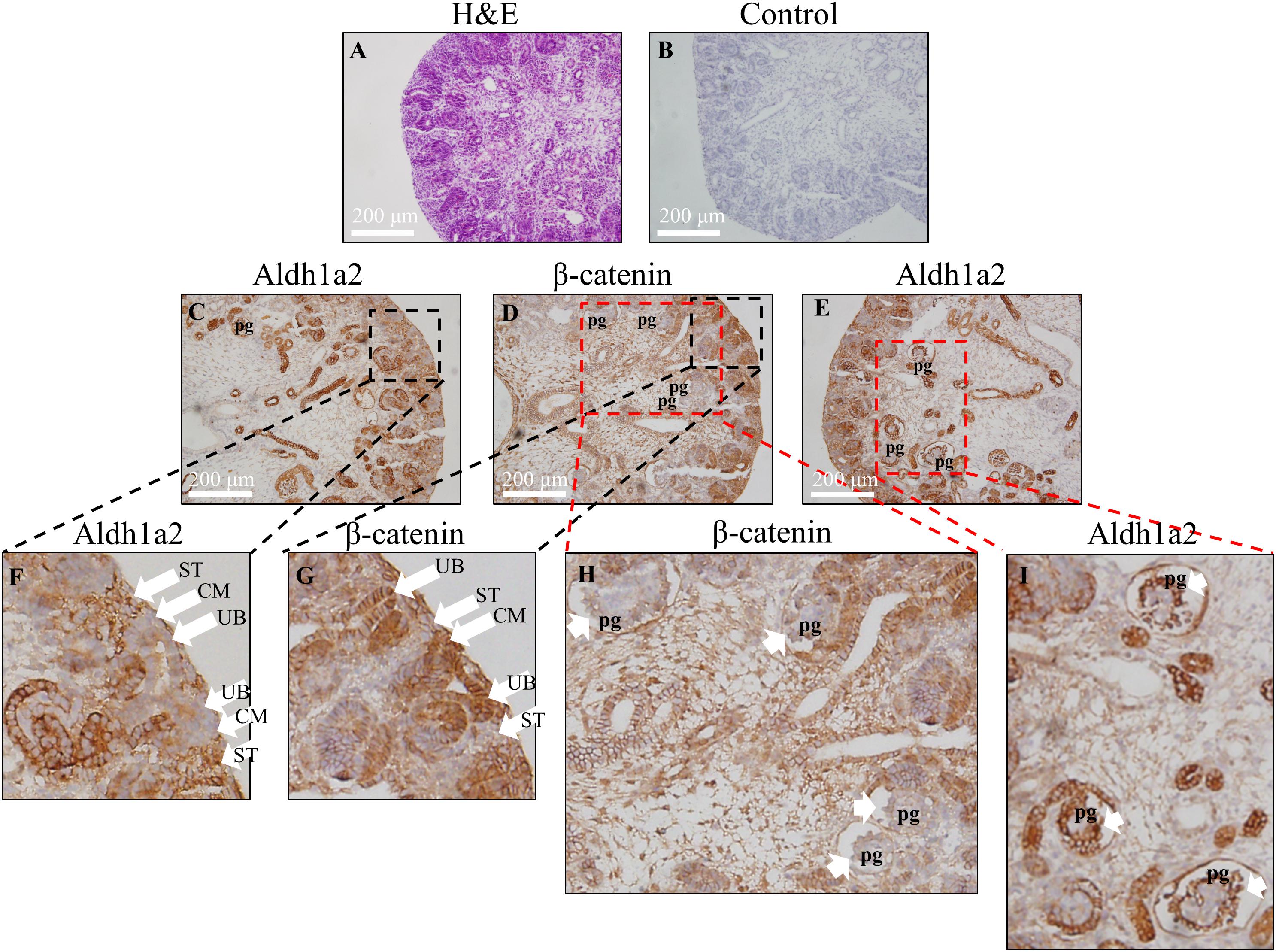
Figure 4. Inverse correlation of Aldh1a2 localization with β-catenin in E18.5 embryonic rat kidney. (A) Hematoxylin and eosin (H&E) staining of rat embryonic kidney (E18.5). Rat embryonic kidney (E18.5) were immunohistochemically stained with no antibody control (B), ALDH1A2 antibody (C,E,F,I), and β-catenin antibody (D,G,H). CM, cap mesenchyme; UB, ureteric bud; ST, stroma; pg, the primitive podocytes of the glomerulus. Length of the bar equals 200 μm.
β-Catenin Is Associated With ALDH1A2 Promoter and Intron1G
Given the ability of β-catenin to repress ALDH1A2 expression, we next determined whether β-catenin is associated with the ALDH1A2 promoter and two DNA elements within ALDH1A2 intron 1 using ChIP analysis. One DNA element, named “intron1G,” was previously shown to activate Aldh1a2 expression in the mouse spine (Castillo et al., 2010). Another DNA element, which we named the “β-catenin and Six2 co-bound region (BSCR),” was previously shown to recruit both β-catenin and Six2 in fetal kidney pretubular aggregate (Park et al., 2012) (Figure 5A). In our ChIP assay, control acetyl-histone 3 and pol II immunoprecipitations confirmed the presence of acetyl-histone 3 and RNA polymerase II at the promoter of GAPDH (Figure 5B) and also showed their recruitment to the MYC and ALDH1A2 promoters (Figure 5B). β-catenin is known to regulate the MYC promoter, and MYC promoter DNA was precipitated by the β-catenin antibody (Figure 5B), providing a positive control. Significantly, the β-catenin antibody also precipitated the ALDH1A2 promoter DNA (Figure 5B).
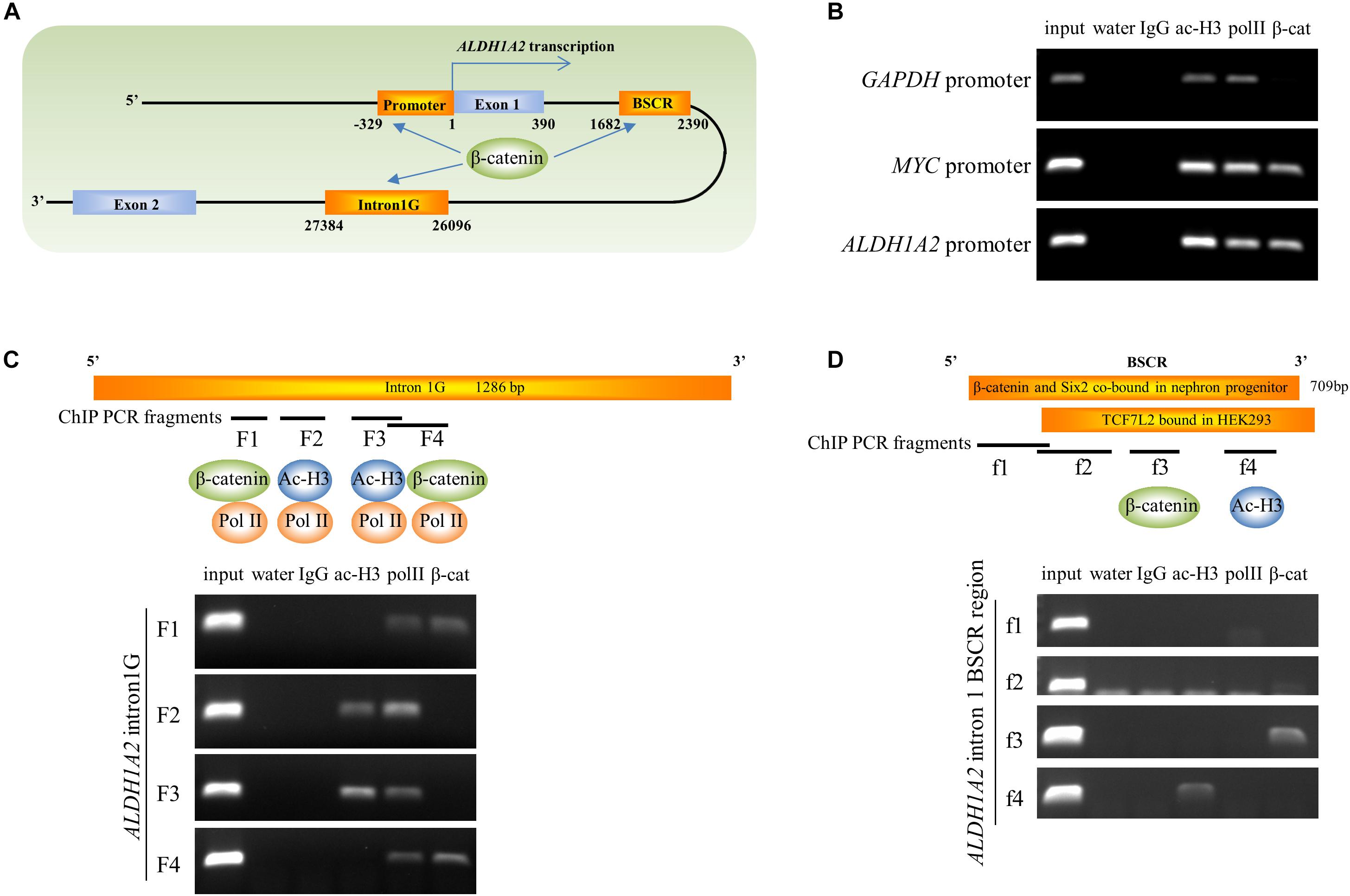
Figure 5. β-catenin is associated with the ALDH1A2 gene promoter and intron1G enhancer. (A) Diagram of 5′ terminal of the ALDH1A2 gene. DNA elements include ALDH1A2 promoter, intron1G, β-catenin, and the Six2 co-bound region (BSCR). (B) Chromatin immunoprecipitation demonstrating the binding of β-catenin to the ALDH1A2 promoter in WiT49 cells. β-catenin is associated with the promoter of the known β-catenin target C-MYC but not with the promoter of the housekeeping gene GAPDH. Acetyl histone 3 (ac-H3) and RNA polymerase II (pol II) were associated with all three gene promoters. (C) Chromatin immunoprecipitation demonstrating binding of β-catenin to the evolutionarily conserved element intron1G within intron 1 of the ALDH1A2 gene (Castillo et al., 2010) at two loci (F1 and F4) in WiT49 cells. (D) Chromatin immunoprecipitation showing binding of β-catenin to the f3 fragment of the “β-catenin and Six2 co-bound region (BSCR)” within intron 1 of the ALDH1A2 gene in WiT49 cells. BSCR was previously shown to co-bound by β-catenin and Six2 in nephron progenitor (chr9:71065207–71065805) (Park et al., 2012). This region partially overlapped with a TCF7L2 (TCF4) bound region in HEK293 previously suggested by a ChIP-seq result (Frietze et al., 2012).
In the ChIP assay of the ALDH1A2 intron1G DNA element, the presence of RNA polymerase II was confirmed by pol II immunoprecipitations at the F1, F2, F3, and F4 fragment of intron1G (Figure 5C). The intense presence of Pol II on intron1G suggests that this conserved DNA element is very likely to be involved in ALDH1A2 transcription. Acetyl-histone 3 only binds to F2 and F3 sites of intron1G (Figure 5C). β-catenin antibody precipitated the F1 and F4 fragments of intron1G (Figure 5C).
In the ChIP assay of the ALDH1A2 BSCR element, the Pol II antibody does not precipitate any fragment of this BSCR element (Figure 5D). Acetyl-histone 3 only binds to f4 pieces of the BSCR element (Figure 5D). β-catenin antibody precipitated the f3 fragment of the BSCR element (Figure 5D).
Taken together, ChIP analysis of ALDH1A2 promoter, intron1G, and BSCR elements suggests that β-catenin represses ALDH1A2 expression through direct recruitment of β-catenin to these DNA elements.
Canonical Wnt Signaling Activated by CHIR99021 Inhibits the Transcriptional Activity of ALDH1A2 Promoter and Intron1G Enhancer
Given that both the control acetyl-histone 3 and pol II antibodies immunoprecipitated ALDH1A2 promoter and multiple sites of intron1G, chromatin within these two sites is more likely to be in an open state and relevant to the ALDH1A2 transcription. To examine whether canonical Wnt signaling activated by CHIR99021 may affect the transcriptional activity of the ALDH1A2 promoter and intron1G element, we conducted luciferase assay. First, we cloned ALDH1A2 promoter of different lengths, intron1G with opposite orientations, and the combination of intron1G and ALDH1A2 promoter (Figure 6A). The luciferase assay results show that the ALDH1A2 promoter F2 (−1031 bp to 290 bp relative to the transcription start site of NM_003888) is the most active and essential region for ALDH1A2 promoter activation (Figure 6B). Comparison between ALDH1A2 promoters QF1 and 1A2ps suggests that a region from 113 to 290 bp (relative to the transcription start site) is the basal and most essential element for ALDH1A2 promoter activity (Figure 6B). Comparison between ALDH1A2 promoter QF1 and 1A2p suggests that a region between 290 and 380 bp has a strong inhibitory effect on the ALDH1A2 promoter activity. The luciferase activity of intron1G at minus strand (M) is higher than that of intron1G at the plus strand (P) (Figure 6B).
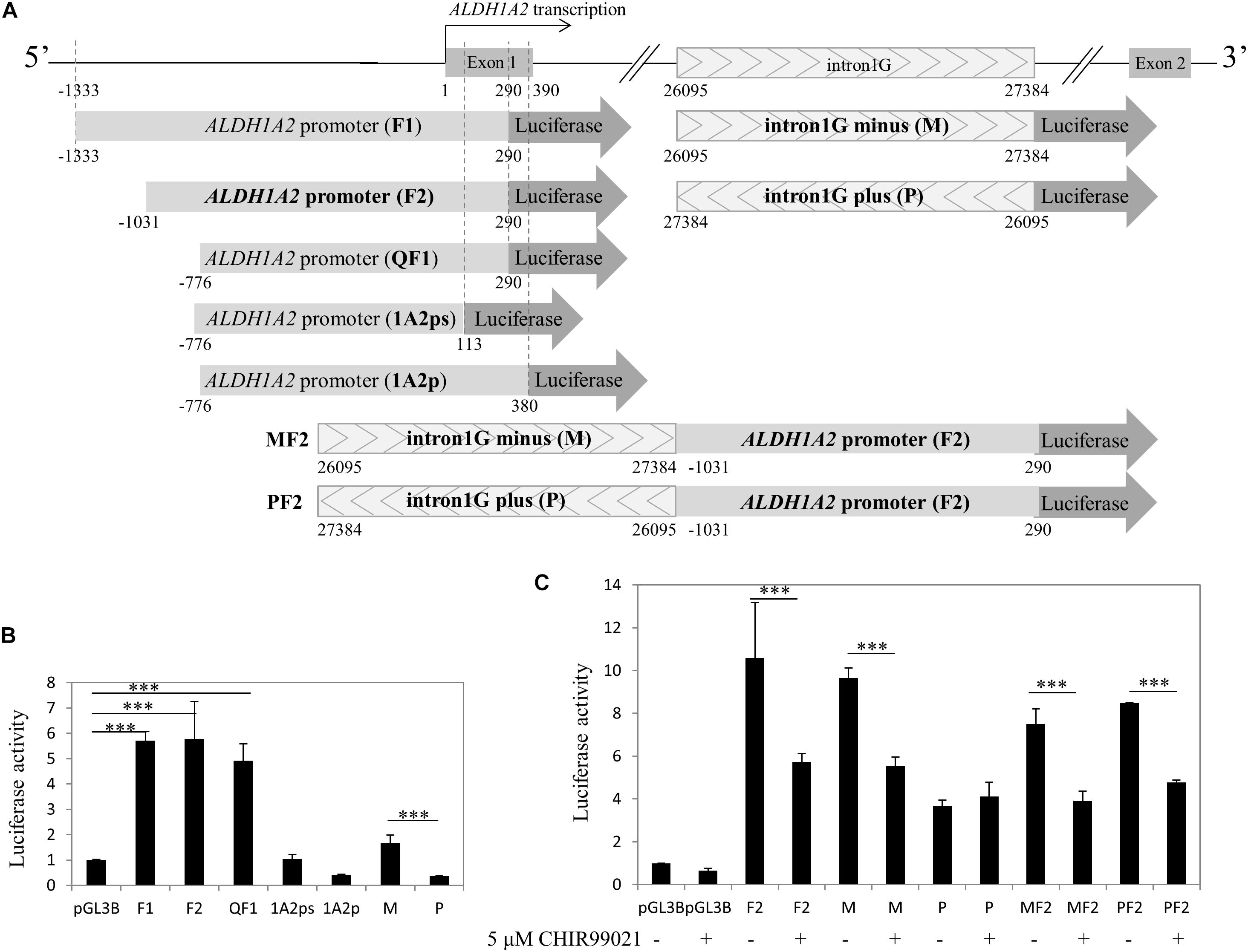
Figure 6. CHIR99021 represses ALDH1A2 promoter and the intron1G element. (A) Maps of ALDH1A2 promoter and intron1G DNA element, numbered relative to the ALDH1A2 transcriptional start. pGL3B luciferase assay reporter driven by ALDH1A2 promoter with different lengths (F1, F2, QF1, 1A2ps, 1A2p), intron1G with opposite orientations (M, P), and the combination of intron1G and ALDH1A2 promoter (MF2, PF2). (B) Identification of the essential promoter region of the ALDH1A2 gene. A 500-ng sample of pGL3-basic (pGL3B) or of pGL3B inserted with ALDH1A2 promoter of different lengths, together with 25 ng of the pRL-TK renilla control vector, was transiently transfected to HEK293 cells for 48 h. Luciferase reporter activities relative to renilla control are shown. Values are the mean ± SD of three determinations; representative results from at least three separate experiments. ∗∗∗P < 0.001 compared with the empty vector pGL3B control group. (C) The repressive effect of CHIR99021 to ALDH1A2 promoter and intron1G. A 500-ng sample of pGL3B, pGL3B construct driven by ALDH1A2 promoter, intron1G, or the combination of intron1G and ALDH1A2 promoter was transiently transfected to WiT49 cells. At 24 h of transfection, cells were treated with 0.1% DMSO or 5 μM CHIR99021 for another 48 h. Luciferase reporter activities relative to renilla (pRL-TK) are shown. Values are the mean ± SD of four determinations; representative results from at least three separate experiments. ∗∗∗P < 0.001 compared with their respective controls, calculated using the Student’s t-test.
Since the above results show that β-catenin binds to the ALDH1A2 promoter and enhancers within intron 1, we next used data mining on in situ Hi-C data (Rao et al., 2014) to see if these elements are within a chromatin loop, which is a chromatin structure enabling interaction of enhancer and promoter. We found that the ALDH1A2 promoter, intron1G, and BSCR are within a chromatin loop defined by CTCF binding sites (Supplementary Figure S1). To explore whether ALDH1A2 promoter, Intron1G, and BSCR sequences are in an open chromatin state, we aligned sequences of these three elements to the UCSC genome browser on human Feb. 2009 (GRCh37/hg19) assembly. The alignment of ALDH1A2 promoter, Intron1G, and BSCR sequences to the UCSC genome browser shows that the ALDH1A2 promoter and intron1G are relatively in an open chromatin state in multiple cell lines (Supplementary Figure S2) (Ernst et al., 2011), suggesting that they may potentially interact with each other. Although these results from data mining are not obtained from fetal kidney cells, they still provide the useful information that ALDH1A2 promoter, intron1G, and BSCR may be within the same chromatin loop and in an open chromatin state, considering that chromatin 3D structure is usually conserved (Dixon et al., 2012; Rao et al., 2014).
Therefore, we combined the ALDH1A2 promoter and intron1G to see their transcriptional activity and whether canonical Wnt signaling activated by CHIR99021 affects their transcriptional activity. Luciferase assay was performed to measure the effect of canonical Wnt signaling activated by CHIR99021 on the ALDH1A2 promoter F2, intron1G with opposite orientations, or the combination of intron1G with F2. The luciferase assay results show that the canonical Wnt signaling activated by CHIR99021 represses the transcriptional activity of both the ALDH1A2 promoter F2 and the intron1G in WiT49 cells (Figure 6C). The canonical Wnt signaling activated by CHIR99021 also suppresses the combination of intron1G and ALDH1A2 promoter F2 (Figure 6C).
Taken together, the canonical Wnt signaling activated by CHIR99021 can repress the ALDH1A2 promoter and intron1G DNA element.
Discussion
The Wnt/β-catenin signaling and retinoic acid pathways are critical factors regulating fetal kidney development (Mendelsohn et al., 1999; Halt and Vainio, 2014). The retinoic acid synthetic enzyme ALDH1A2, which is essential for renal development (Rosselot et al., 2010), strongly expresses in the stroma of the renal outer cortex (Batourina et al., 2001; Niederreither et al., 2002; Marlier and Gilbert, 2004; Rosselot et al., 2010), while β-catenin predominantly expresses in the ureteric bud (Bridgewater et al., 2008; Marose et al., 2008; Wang et al., 2018), suggesting an inverse relationship between their expression in the fetal kidney. Recent data from several lines of evidence indicate that ALDH1A2 could be a novel target of β-catenin signaling (Castillo et al., 2010; Frietze et al., 2012; Park et al., 2012; Hong et al., 2015). Indeed, we show here for the first time that activation of the canonical Wnt signaling pathway using CHIR99021 can negatively regulate ALDH1A2 expression in WiT49 cells. We demonstrate that knockdown of CTNNB1 abrogates the down-regulation of ALDH1A2 by CHIR99021. Transient transfection of Wnt1, Wnt3a, Wnt4, and Wnt9b to WiT49 cells leads to ALDH1A2 downregulation. We confirm that Aldh1a2 and β-catenin expression in the fetal kidney are inversely correlated. Importantly, the ALDH1A2 promoter and the intron1G element recruit β-catenin and are transcriptionally repressed by canonical Wnt signaling activated by CHIR99021, suggesting that β-catenin directly represses ALDH1A2 expression via its promoter and intron1G. Taken together, we provide in vitro evidence showing that the Wnt/β-catenin pathway directly represses ALDH1A2 expression in a fetal kidney cell model.
Although Wnt/β-catenin signaling is known to activate gene expression, the expression of several target genes can also be inhibited by the Wnt/β-catenin signaling (including E-cadherin, Hath1, Sox9, 15-PGDH, RANKL and Osteocalcin) (Jamora et al., 2003; Kahler and Westendorf, 2003; Leow et al., 2004; Hill et al., 2005; Spencer et al., 2006; Smartt et al., 2012). It has also been shown that the GSK3 inhibitors (canonical Wnt signaling activator) can repress PEPCK and G6Pase gene expression (Lochhead et al., 2001). The exact mechanisms that are responsible for transcriptional repression are less well known than for transcriptional activation by Wnt/β-catenin signaling. As our understanding of the mechanisms of gene repression by β-catenin gets better, it will be of tremendous interest to elucidate the mechanism that was responsible for ALDH1A2 suppression.
Our work shows that β-catenin is strongly localized in the ureteric bud of the E18.5 rat kidney and does not present in the developing stroma, consistent with previous reports showing that β-catenin and its signaling primarily present in the ureteric bud (Bridgewater et al., 2008; Marose et al., 2008; Wang et al., 2018). Other literature also shows that β-catenin intensely presents in the branching ureteric bud (Iglesias et al., 2007; Billfeldt et al., 2016; Kovacs et al., 2017; Terada et al., 2017). Although our immunohistochemistry result shows that β-catenin is mainly localized to the membrane/cytosol, another study has documented the nuclear staining of β-catenin in ureteric bud cells using immunofluorescence method (Wang et al., 2018). Active canonical WNT signaling (TCF reporter activity) was obvious in the branching ureteric bud throughout its transition into renal tubules (Iglesias et al., 2007). The presence of Wnt/β-catenin signaling in the ureteric bud is also supported by the fact that Axin2, the Wnt/β-catenin signaling typical target, expresses in the ureteric bud of the fetal kidney (Mohri et al., 2011). Taken together, it is clear that Wnt/β-catenin signaling presents in the ureteric bud, supported by all three types of evidence from the literature and our result (Figure 4), including expression of β-catenin/activated β-catenin, TCF reporter activity, and conditional knockout of β-catenin driven by the Hoxb7 promoter (Iglesias et al., 2007; Bridgewater et al., 2008; Marose et al., 2008; Billfeldt et al., 2016; Kovacs et al., 2017; Terada et al., 2017; Wang et al., 2018).
Our result showing that β-catenin presents in the cap mesenchyme is consistent with previous reports showing that β-catenin expresses in Six2 positive cap mesenchyme cells (Park et al., 2007; Karner et al., 2011; Park et al., 2012), in Myc positive nephron progenitor cells (Pan et al., 2017), and in the nephrogenic mesenchyme (Iglesias et al., 2007; Billfeldt et al., 2016; Kovacs et al., 2017; Terada et al., 2017). Furthermore, active canonical WNT signaling (TCF reporter activity) was obvious in the epithelia of the nephrogenic mesenchyme (Iglesias et al., 2007). However, other works show an absence of β-catenin activity in nephron progenitor (Burn et al., 2011; Tanigawa et al., 2011). These works may lack evidence from the higher resolution pictures of β-catenin staining and TCF reporter activity on the cap mesenchyme that would be necessary to show a lack of β-catenin activity in cap mesenchyme. Taken together, it is very likely that cap mesenchyme also has β-catenin activity, considering that all three types of evidence present, namely the intense β-catenin staining, TCF reporter activity, and conditional knockout of β-catenin within Six2 expression positive cells, result in a phenotype (Iglesias et al., 2007; Park et al., 2007; Karner et al., 2011; Park et al., 2012; Billfeldt et al., 2016; Kovacs et al., 2017; Pan et al., 2017; Terada et al., 2017).
Our result showing that β-catenin is not present in the developing podocyte is consistent with a previous report showing that TCF reporter activity is quickly downregulated in maturing nephrons and becomes undetectable in the postnatal kidney (Iglesias et al., 2007).
Our work showing that β-catenin does not present in the developing stroma is also consistent with previous reports. However, it has been shown in other studies that β-catenin presents in the developing stroma (Yu et al., 2008; Boivin et al., 2015) and the medullar stroma of the developing human kidney (Billfeldt et al., 2016). This discrepancy may arise from differences in the specificity and sensitivity of different β-catenin antibodies and detection methods (IHC vs. immunofluorescence). Although conditional deletion of β-catenin in stroma driven by the Foxd1 promoter shows a phenotype of kidney development (Yu et al., 2008; Boivin et al., 2015), most of the literature supports β-catenin not being expressed in the developing stroma (Iglesias et al., 2007; Park et al., 2007; Bridgewater et al., 2008; Marose et al., 2008; Karner et al., 2011; Park et al., 2012; Kovacs et al., 2017; Pan et al., 2017; Terada et al., 2017; Wang et al., 2018). The presence of Wnt/β-catenin signaling in stroma also needs support from result of TCF luciferase reporter activity (Yu et al., 2008; Boivin et al., 2015). At least we can be sure that Wnt/β-catenin expression/signaling in the ureteric bud and cap mesenchyme is stronger than that in the developing stroma.
We show that activation of Wnt/β-catenin signaling by CHIR99021 represses ALDH1A2 expression. This result reveals for the first time that β-catenin can downregulate, rather than upregulate, Aldh1a2 expression, indicating that the canonical Wnt signaling is upstream of retinoic acid synthesis. This finding is different from reports suggesting that Wnt/β-catenin signaling may upregulate Aldh1a2 expression in Zebrafish development (Wehner et al., 2014) and dendritic cells (Suryawanshi and Manicassamy, 2015). This difference suggests that the regulation of Aldh1a2 expression by β-catenin may be tissue and developmental context-dependent, which is a common phenomenon for transcriptional regulation (Ma, 2005; Essafi et al., 2011). It would be of interest in the future to study what may contribute to this opposite regulation of Aldh1a2 expression by Wnt/β-catenin signaling in different contexts.
The result that Wnt1, Wnt3a, Wnt4, and Wnt9b activate TOPFlash activity in WiT49 cells (Figure 3A) is consistent with reports showing that Wnt3a and Wnt9b activate Wnt/β-catenin signaling (Dallosso et al., 2009; Karner et al., 2011; Pan et al., 2017). The mild activation of TCF reporter by Wnt4 shown by us (Figure 3A) is different from reports showing that Wnt4 does not activate TCF reporter in fetal kidney cells (Tanigawa et al., 2011; Billfeldt et al., 2016). This discrepancy may arise from the difference in the expression of Wnt receptors. Nonetheless, our results demonstrate that Wnt molecules, at least Wnt3a, Wnt9b, and Wnt1, can activate TCF reporter activity and repress ALDH1A2 expression in WiT49 cells. As Wnt9b is expressed in ureteric buds, we speculate that Wnt9b might directly repress Aldh1a2 expression via canonical Wnt signaling in the ureteric branching by autocrine, leading to a proper local concentration of retinoic acid.
We have defined the F2 region (−1031 to 290 bp) as the most active promoter region of the ALDH1A2 gene (Figure 6B). We found a region (290–380 bp) that has an inhibitory effect on the ALDH1A2 promoter activity, and another region (96–290 bp) has an essential effect on ALDH1A2 promoter activity (Figure 6B). This study also shows that Aldh1a2 intron1G, which was shown to have an enhancing effect on the expression of Aldh1a2 in the developing neuron cells (Castillo et al., 2010), can be repressed by activation of the canonical Wnt signaling pathway using CHIR99021 in WiT49 cells.
ChIP-PCR data show that pol II was recruited to all four regions within intron1G (Figure 5C), suggesting that this conserved element is very likely to be involved in pol II-dependent ALDH1A2 transcription, although the intron1G is 26095 bp away from the ALDH1A2 transcription start site. The basal luciferase activity of intron1G on the minus strand (M) is higher than that of intron1G on the plus strand (P) (Figure 6B). The luciferase assay result shows that the repressive effect of intron1G by CHIR99021 works on the minus strand (M) but not on the plus strand (P) (Figure 6C), suggesting that intron1G on the same stand of the ALDH1A2 gene is more likely to play a role in the repressive regulation by β-catenin. The BSCR element is a DNA region previously suggested to be co-bound by β-catenin and Six2 in nephron progenitor (Park et al., 2012) and bound by TCF7L2 in HEK293 (Frietze et al., 2012) based on ChIP-seq data. We show by ChIP-PCR that β-catenin is recruited to the BSCR element at the f3 region, where acetyl-histone 3 and pol II do not bind (Figure 5D), suggesting that the chromatin state of this region may not be as open as that of intron1G.
Taken together, our data suggest a novel role for Wnt/β-catenin signaling in direct repression of Aldh1a2 expression via the Aldh1a2 promoter and the intron1G conserved element, which may lead to alteration of retinoic acid synthesis in ureteric bud branching and cap mesenchyme. Because Aldh1a2–/– mutant mouse kidney exhibits a significant reduction of ureteric buds and nephrons (Rosselot et al., 2010) and excessive retinoic acid is teratogenic (Fantel et al., 1977; Lee et al., 2012; Piersma et al., 2017), our findings suggest that the regulation of Aldh1a2 by Wnt/β-catenin signaling may play a significant role in the limitation of retinoic acid production in ureteric bud and cap mesenchyme, where a proper control of retinoic acid concentration is critical for renal development.
Conclusion
Our findings of the direct repression of Aldh1a2 by Wnt/β-catenin signaling have a significant consequence for the regulation of retinoic acid synthesis, which may play roles in fetal kidney development. Future study is necessary to address how the Aldh1a2 expression and control of retinoic acid concentration are altered in the ureteric bud when Ctnnb1 is conditionally knocked out in ureteric bud of fetal kidney in animal models.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The animal study was reviewed and approved by the Ethics Committee of Huazhong University of Science and Technology Union Shenzhen Hospital.
Author Contributions
YL, JfD, and JhD designed and co-directed the research and wrote the manuscript. YL, HG, FZ, JZ, SL, and JfD performed the experiments, with the help of LW, JhD, and BZ in all assays involving cell cultures, and contributed to the interpretation of data. JW provided the pregnant rat kidney. HG, YL, FZ, JZ, and SL contributed to the immunohistochemistry experiment. JL provided reagents and knowledge of vitamin metabolisms. YL, JfD, HG, FZ, JhD, JW, JZ, SL, JL, LW, and BZ reviewed and contributed to manuscript editing. All authors read and approved the manuscript.
Funding
This work was funded by the Natural Science Foundation of Guangdong Province, China (No. 2017A030313170), the National Natural Science Foundation of China (No. 31100943), the Science, Technology, and Innovation Commission of Shenzhen Municipality (No. JCYJ20180302144937970), and Shenzhen Nanshan Science and Technology Research Funds (Nos. 2018002 and 2018001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Herman Yeger (Hospital for Sick Children, Toronto, ON, Canada) for permitting us to use the WiT49 cell line. We also thank Dr. Mei-Hong Li and Dr. Fernando Ferrer (Center for Vascular Biology, University of Connecticut Health Center, Farmington, CT, United States) for shipping us this cell line.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.00094/full#supplementary-material
FIGURE S1 | CCCTC-binding factor (CTCF) binding to ALDH1A2 gene locus and formation of DNA loops in GM12878 cells. The arrows show the two distal DNA sites where the chromatin loops bind to each other (the straight line extending from the point directs to two DNA sites; in most cases, the CTCF binding site). The red (a, b, c) and blue (d, e) arcs indicate the extent of each chromatin loop, and the red chromatin loop (a, b, c) totally or partially covers the ALDH1A2 gene. This figure was based on a previous report using the Hi-C method (Rao et al., 2014). The report detailed a Hi-C experiment conducted to examine the genome-wide chromatin loop in the GM12878 cell line. The data were integrated with the ChIP-seq data of CTCF, H3K36me3 (active mark), H3K27ac (active mark), H3K27me3 (repressive mark), H3K4me3 (active mark), H3K9me3 (repressive mark), H3K9ac (active mark), and Dnase Seq data. Since the chromatin loop is relatively stable among species and different tissues, the data of chromatin loop at ALDH1A2 locus obtained from the lymphoblastoid cell line GM12878 give us clues on how the chromatin at ALDH1A2 gene locus is organized. The data show that ALDH1A2 promoter, intron1G, and the BSCR element are likely to be within the same chromatin loop. Thus, they may have close spatial proximity and less physical segmentation, suggesting that they have the potential to interact with each other and may be relevant to β-catenin recruitment and regulation of ALDH1A2 expression.
FIGURE S2 | The chromatin state of the ALDH1A2 gene. Intron1G, BSCR, and ALDH1A2 promoter sequences were searched on genome on the UCSC browser and highlighted in a light blue color. The lane “100 vertebrates Basewise Conservation by PhyloP” shows that the ALDH1A2 promoter, intron1G, and BSCR element are all evolutionally conserved. The lane “Chromatin State Segmentation by HMM from ENCODE/Broad” shows that ALDH1A2 promoter and intron1G are in an open chromatin state in nine cell lines, namely GM12878, H1-hESC, K562, HepG2, HUVEC, HMEC, HSMM, NHEK, and NHLF. The lane “DNaseI Hypersensitivity Clusters in 125 cell types from ENCODE (V3)” also shows that the ALDH1A2 promoter and intron1G are in an open chromatin state. The lane “Transcription Factor ChIP-seq Clusters (161 factors) from ENCODE with Factorbook Motifs” shows moderate TCF7L2 binding on the intron1G and BSCR site.
Footnotes
References
Alami, J., Williams, B. R., and Yeger, H. (2003). Derivation and characterization of a Wilms’ tumour cell line, WiT 49. Int. J. Cancer 107, 365–374. doi: 10.1002/ijc.11429
Batourina, E., Gim, S., Bello, N., Shy, M., Clagett-Dame, M., Srinivas, S., et al. (2001). Vitamin A controls epithelial/mesenchymal interactions through Ret expression. Nat. Genet. 27, 74–78. doi: 10.1038/83792
Billfeldt, N. K., Banyai, D., and Kovacs, G. (2016). Absence of canonical WNT signaling in adult renal cell tumors of embryonal origin. Anticancer Res. 36, 2169–2173.
Boivin, F. J., Sarin, S., Lim, J., Javidan, A., Svajger, B., Khalili, H., et al. (2015). Stromally expressed beta-catenin modulates Wnt9b signaling in the ureteric epithelium. PLoS One 10:e0120347. doi: 10.1371/journal.pone.0120347
Bridgewater, D., Cox, B., Cain, J., Lau, A., Athaide, V., Gill, P. S., et al. (2008). Canonical WNT/β-catenin signaling is required for ureteric branching. Dev. Biol. 317, 83–94. doi: 10.1016/j.ydbio.2008.02.010
Burn, S. F., Webb, A., Berry, R. L., Davies, J. A., Ferrer-Vaquer, A., Hadjantonakis, A. K., et al. (2011). Calcium/NFAT signalling promotes early nephrogenesis. Dev. Biol. 352, 288–298. doi: 10.1016/j.ydbio.2011.01.033
Carroll, T. J., Park, J. S., Hayashi, S., Majumdar, A., and Mcmahon, A. P. (2005). Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev. Cell 9, 283–292. doi: 10.1016/j.devcel.2005.05.016
Castillo, H. A., Cravo, R. M., Azambuja, A. P., Simões-Costa, M. S., Sura-Trueba, S., Gonzalez, J., et al. (2010). Insights into the organization of dorsal spinal cord pathways from an evolutionarily conserved raldh2 intronic enhancer. Development 137, 507–518. doi: 10.1242/dev.043257
Cullen-Mcewen, L., Sutherland, M. R., and Black, M. J. (2016). “Chapter 3 – The human kidney: parallels in structure, spatial development, and timing of nephrogenesis,” in Kidney Development, Disease, Repair and Regeneration, ed. M. H. Little (San Diego: Academic Press), 27–40.
Dallosso, A. R., Hancock, A. L., Szemes, M., Moorwood, K., Chilukamarri, L., Tsai, H. H., et al. (2009). Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms’ tumor. PLoS Genet. 5:e1000745. doi: 10.1371/journal.pgen.1000745
Davidson, A. (2008). Mouse Kidney Development. Available at: www.stembook.org [accessed April 7, 2015].
Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., et al. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. doi: 10.1038/nature11082
Dolle, P. (2009). Developmental expression of retinoic acid receptors (RARs). Nucl. Recept. Signal. 7:e006. doi: 10.1621/nrs.07006
Dressler, G. R. (2009). Advances in early kidney specification, development and patterning. Development 136, 3863–3874. doi: 10.1242/dev.034876
Duester, G. (2008). Retinoic acid synthesis and signaling during early organogenesis. Cell 134, 921–931. doi: 10.1016/j.cell.2008.09.002
Ernst, J., Kheradpour, P., Mikkelsen, T. S., Shoresh, N., Ward, L. D., Epstein, C. B., et al. (2011). Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49. doi: 10.1038/nature09906
Essafi, A., Webb, A., Berry, R. L., Slight, J., Burn, S. F., Spraggon, L., et al. (2011). A Wt1-controlled chromatin switching mechanism underpins tissue-specific Wnt4 activation and repression. Dev. Cell 21, 559–574. doi: 10.1016/j.devcel.2011.07.014
Fan, X., Molotkov, A., Manabe, S.-I., Donmoyer, C. M., Deltour, L., Foglio, M. H., et al. (2003). Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol. Cell. Biol. 23, 4637–4648. doi: 10.1128/mcb.23.13.4637-4648.2003
Fantel, A. G., Shepard, T. H., Newell-Morris, L. L., and Moffett, B. C. (1977). Teratogenic effects of retinoic acid in pigtail monkeys (Macaca nemestrina) I, General features. Teratology 15, 65–71. doi: 10.1002/tera.1420150109
Fetting, J. L., Guay, J. A., Karolak, M. J., Iozzo, R. V., Adams, D. C., Maridas, D. E., et al. (2013). FOXD1 promotes nephron progenitor differentiation by repressing decorin in the embryonic kidney. Development 141, 17–27. doi: 10.1242/dev.089078
Francipane, M. G., and Lagasse, E. (2015). The lymph node as a new site for kidney organogenesis. Stem Cells Transl. Med. 4, 295–307. doi: 10.5966/sctm.2014-0208
Frietze, S., Wang, R., Yao, L., Tak, Y. G., Ye, Z., Gaddis, M., et al. (2012). Cell type-specific binding patterns reveal that TCF7L2 can be tethered to the genome by association with GATA3. Genome Biol. 13:R52. doi: 10.1186/gb-2012-13-9-r52
Grun, F., Hirose, Y., Kawauchi, S., Ogura, T., and Umesono, K. (2000). Aldehyde dehydrogenase 6, a cytosolic retinaldehyde dehydrogenase prominently expressed in sensory neuroepithelia during development. J. Biol. Chem. 275, 41210–41218. doi: 10.1074/jbc.M007376200
Halt, K., and Vainio, S. (2014). Coordination of kidney organogenesis by Wnt signaling. Pediatr. Nephrol. 29, 737–744. doi: 10.1007/s00467-013-2733-z
Hill, T. P., Später, D., Taketo, M. M., Birchmeier, W., and Hartmann, C. (2005). Canonical Wnt/β-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev. Cell 8, 727–738. doi: 10.1016/j.devcel.2005.02.013
Hong, Y., Manoharan, I., Suryawanshi, A., Majumdar, T., Angus-Hill, M. L., Koni, P. A., et al. (2015). Beta-catenin promotes regulatory T-cell responses in tumors by inducing vitamin A metabolism in dendritic cells. Cancer Res. 75, 656–665. doi: 10.1158/0008-5472.can-14-2377
Iglesias, D. M., Hueber, P. A., Chu, L., Campbell, R., Patenaude, A. M., Dziarmaga, A. J., et al. (2007). Canonical WNT signaling during kidney development. Am. J. Physiol. Renal Physiol. 293, F494–F500. doi: 10.1152/ajprenal.00416.2006
Itaranta, P., Lin, Y., Perasaari, J., Roel, G., Destree, O., and Vainio, S. (2002). Wnt-6 is expressed in the ureter bud and induces kidney tubule development in vitro. Genesis 32, 259–268. doi: 10.1002/gene.10079
Jamora, C., Dasgupta, R., Kocieniewski, P., and Fuchs, E. (2003). Links between signal transduction, transcription and adhesion in epithelial bud development. Nature 422, 317–322. doi: 10.1038/nature01458
Kahler, R. A., and Westendorf, J. J. (2003). Lymphoid enhancer factor-1 and beta-catenin inhibit Runx2-dependent transcriptional activation of the osteocalcin promoter. J. Biol. Chem. 278, 11937–11944. doi: 10.1074/jbc.M211443200
Karner, C. M., Das, A., Ma, Z., Self, M., Chen, C., Lum, L., et al. (2011). Canonical Wnt9b signaling balances progenitor cell expansion and differentiation during kidney development. Development 138, 1247–1257. doi: 10.1242/dev.057646
Kispert, A., Vainio, S., and Mcmahon, A. P. (1998). Wnt-4 is a mesenchymal signal for epithelial transformation of metanephric mesenchyme in the developing kidney. Development 125, 4225–4234.
Kispert, A., Vainio, S., Shen, L., Rowitch, D. H., and Mcmahon, A. P. (1996). Proteoglycans are required for maintenance of Wnt-11 expression in the ureter tips. Development 122, 3627–3637.
Kovacs, G., Billfeldt, N. K., Farkas, N., Dergez, T., Javorhazy, A., Banyai, D., et al. (2017). Cytoplasmic expression of beta-catenin is an independent predictor of progression of conventional renal cell carcinoma: a simple immunostaining score. Histopathology 70, 273–280. doi: 10.1111/his.13059
Lee, L. M. Y., Leung, C.-Y., Tang, W. W. C., Choi, H.-L., Leung, Y.-C., Mccaffery, P. J., et al. (2012). A paradoxical teratogenic mechanism for retinoic acid. Proc. Natl. Acad. Sci. U.S.A. 109, 13668–13673. doi: 10.1073/pnas.1200872109
Leow, C. C., Romero, M. S., Ross, S., Polakis, P., and Gao, W.-Q. (2004). Hath1, down-regulated in colon adenocarcinomas, inhibits proliferation and tumorigenesis of colon cancer cells. Cancer Res. 64, 6050–6057. doi: 10.1158/0008-5472.can-04-0290
Levinson, R. S., Batourina, E., Choi, C., Vorontchikhina, M., Kitajewski, J., and Mendelsohn, C. L. (2005). Foxd1-dependent signals control cellularity in the renal capsule, a structure required for normal renal development. Development 132, 529–539. doi: 10.1242/dev.01604
Li, C.-M., Guo, M., Borczuk, A., Powell, C. A., Wei, M., Thaker, H. M., et al. (2002). Gene expression in Wilms’ tumor mimics the earliest committed stage in the metanephric mesenchymal-epithelial transition. Am. J. Pathol. 160, 2181–2190. doi: 10.1016/s0002-9440(10)61166-2
Li, M. H., Yamase, H., and Ferrer, F. (2010). Characterization of a WiT49 cell line derived orthotopic model of Wilms tumor. Pediatr. Blood Cancer 54, 316–318. doi: 10.1002/pbc.22205
Li, Y., Wang, L., Ai, W., He, N., Zhang, L., Du, J., et al. (2017). Regulation of retinoic acid synthetic enzymes by WT1 and HDAC inhibitors in 293 cells. Int. J. Mol. Med. 40, 661–672. doi: 10.3892/ijmm.2017.3051
Lin, Y., Liu, A., Zhang, S., Ruusunen, T., Kreidberg, J. A., Peltoketo, H., et al. (2001). Induction of ureter branching as a response to Wnt-2b signaling during early kidney organogenesis. Dev. Dyn. 222, 26–39. doi: 10.1002/dvdy.1164
Lochhead, P. A., Coghlan, M., Rice, S. Q. J., and Sutherland, C. (2001). Inhibition of GSK-3 selectively reduces glucose-6-phosphatase and phosphoenolpyruvate carboxykinase gene expression. Diabetes 50, 937–946. doi: 10.2337/diabetes.50.5.937
Ma, J. (2005). Crossing the line between activation and repression. Trends Genet. 21, 54–59. doi: 10.1016/j.tig.2004.11.004
Magella, B., Adam, M., Potter, A. S., Venkatasubramanian, M., Chetal, K., Hay, S. B., et al. (2018). Cross-platform single cell analysis of kidney development shows stromal cells express Gdnf. Dev. Biol. 434, 36–47. doi: 10.1016/j.ydbio.2017.11.006
Marlier, A., and Gilbert, T. (2004). Expression of retinoic acid-synthesizing and -metabolizing enzymes during nephrogenesis in the rat. Gene Expr. Patterns 5, 179–185. doi: 10.1016/j.modgep.2004.08.005
Marose, T. D., Merkel, C. E., Mcmahon, A. P., and Carroll, T. J. (2008). β-Catenin is necessary to keep cells of ureteric bud/Wolffian duct epithelium in a precursor state. Dev. Biol. 314, 112–126. doi: 10.1016/j.ydbio.2007.11.016
Masiá, S., Alvarez, S., De Lera, A. R., and Barettino, D. (2007). Rapid, nongenomic actions of retinoic acid on phosphatidylinositol-3-kinase signaling pathway mediated by the retinoic acid receptor. Mol. Endocrinol. 21, 2391–2402. doi: 10.1210/me.2007-0062
Matt, N., Dupé, V., Garnier, J.-M., Dennefeld, C., Chambon, P., Mark, M., et al. (2005). Retinoic acid-dependent eye morphogenesis is orchestrated by neural crest cells. Development 132, 4789–4800. doi: 10.1242/dev.02031
Mendelsohn, C., Batourina, E., Fung, S., Gilbert, T., and Dodd, J. (1999). Stromal cells mediate retinoid-dependent functions essential for renal development. Development 126, 1139–1148.
Mengelbier, L. H., Bexell, D., Sehic, D., Ciornei, C. D., and Gisselsson, D. (2014). Orthotopic Wilms tumor xenografts derived from cell lines reflect limited aspects of tumor morphology and clinical characteristics. Pediatr. Blood Cancer 61, 1949–1954. doi: 10.1002/pbc.25131
Mohri, Y., Oyama, K., Akamatsu, A., Kato, S., and Nishimori, K. (2011). Lgr4-deficient mice showed premature differentiation of ureteric bud with reduced expression of Wnt effector Lef1 and Gata3. Dev. Dyn. 240, 1626–1634. doi: 10.1002/dvdy.22651
Napoli, J. L. (2012). Physiological insights into all-trans-retinoic acid biosynthesis. Biochim. Biophys. Acta 1821, 152–167. doi: 10.1016/j.bbalip.2011.05.004
Napoli, J. L. (2017). Cellular retinoid binding-proteins, CRBP, CRABP, FABP5: effects on retinoid metabolism, function and related diseases. Pharmacol. Ther. 173, 19–33. doi: 10.1016/j.pharmthera.2017.01.004
Naylor, R. W., Skvarca, L. B., Thisse, C., Thisse, B., Hukriede, N. A., and Davidson, A. J. (2016). BMP and retinoic acid regulate anterior–posterior patterning of the non-axial mesoderm across the dorsal–ventral axis. Nat. Commun. 7:12197. doi: 10.1038/ncomms12197
Niederreither, K., Fraulob, V., Garnier, J.-M., Chambon, P., and Dollé, P. (2002). Differential expression of retinoic acid-synthesizing (RALDH) enzymes during fetal development and organ differentiation in the mouse. Mech. Dev. 110, 165–171. doi: 10.1016/s0925-4773(01)00561-5
Nusse, R., and Clevers, H. (2017). Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999. doi: 10.1016/j.cell.2017.05.016
Pan, X., Karner, C. M., and Carroll, T. J. (2017). Myc cooperates with β-catenin to drive gene expression in nephron progenitor cells. Development 144, 4173–4182. doi: 10.1242/dev.153700
Park, J.-S., Ma, W., O’Brien, L. L., Chung, E., Guo, J.-J., Cheng, J.-G., et al. (2012). Six2 and Wnt regulate self-renewal and commitment of Nephron progenitors through shared gene regulatory networks. Dev. Cell 23, 637–651. doi: 10.1016/j.devcel.2012.07.008
Park, J.-S., Valerius, M. T., and Mcmahon, A. P. (2007). Wnt/β-catenin signaling regulates nephron induction during mouse kidney development. Development 134, 2533–2539. doi: 10.1242/dev.006155
Piersma, A. H., Hessel, E. V., and Staal, Y. C. (2017). Retinoic acid in developmental toxicology: teratogen, morphogen and biomarker. Reprod. Toxicol. 72, 53–61. doi: 10.1016/j.reprotox.2017.05.014
Piskunov, A., and Rochette-Egly, C. (2011). A retinoic acid receptor RARα pool present in membrane lipid rafts forms complexes with G protein αQ to activate p38MAPK. Oncogene 31, 3333–3345. doi: 10.1038/onc.2011.499
Przepiorski, A., Sander, V., Tran, T., Hollywood, J. A., Sorrenson, B., Shih, J. H., et al. (2018). A simple bioreactor-based method to generate kidney organoids from pluripotent stem cells. Stem Cell Rep. 11, 470–484. doi: 10.1016/j.stemcr.2018.06.018
Rao, S. S. P., Huntley, M. H., Durand, N. C., Stamenova, E. K., Bochkov, I. D., Robinson, J. T., et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. doi: 10.1016/j.cell.2014.11.021
Rivera, M. N., and Haber, D. A. (2005). Wilms’ tumour: connecting tumorigenesis and organ development in the kidney. Nat. Rev. Cancer 5, 699–712. doi: 10.1038/nrc1696
Rosselot, C., Spraggon, L., Chia, I., Batourina, E., Riccio, P., Lu, B., et al. (2010). Non-cell-autonomous retinoid signaling is crucial for renal development. Development 137, 283–292. doi: 10.1242/dev.040287
Sima, A., Parisotto, M., Mader, S., and Bhat, P. V. (2009). Kinetic characterization of recombinant mouse retinal dehydrogenase types 3 and 4 for retinal substrates. Biochim. Biophys. Acta 1790, 1660–1664. doi: 10.1016/j.bbagen.2009.09.004
Smartt, H. J. M., Greenhough, A., Ordóñez-Morán, P., Talero, E., Cherry, C. A., Wallam, C. A., et al. (2012). β-catenin represses expression of the tumour suppressor 15-prostaglandin dehydrogenase in the normal intestinal epithelium and colorectal tumour cells. Gut 61, 1306–1314. doi: 10.1136/gutjnl-2011-300817
Spencer, G. J., Utting, J. C., Etheridge, S. L., Arnett, T. R., and Genever, P. G. (2006). Wnt signalling in osteoblasts regulates expression of the receptor activator of NFκB ligand and inhibits osteoclastogenesis in vitro. J. Cell Sci. 119, 1283–1296. doi: 10.1242/jcs.02883
Stark, K., Vainio, S., Vassileva, G., and Mcmahon, A. P. (1994). Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature 372, 679–683. doi: 10.1038/372679a0
Suryawanshi, A., and Manicassamy, S. (2015). Tumors induce immune tolerance through activation of beta-catenin/TCF4 signaling in dendritic cells: a novel therapeutic target for cancer immunotherapy. Oncoimmunology 4:e1052932. doi: 10.1080/2162402X.2015.1052932
Taguchi, A., and Nishinakamura, R. (2017). Higher-order kidney organogenesis from pluripotent stem cells. Cell Stem Cell 21, 730–746.e6.
Takasato, M., Er, P. X., Chiu, H. S., Maier, B., Baillie, G. J., Ferguson, C., et al. (2016). Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 536:238. doi: 10.1038/nature17982
Tanigawa, S., Wang, H., Yang, Y., Sharma, N., Tarasova, N., Ajima, R., et al. (2011). Wnt4 induces nephronic tubules in metanephric mesenchyme by a non-canonical mechanism. Dev. Biol. 352, 58–69. doi: 10.1016/j.ydbio.2011.01.012
Terada, N., Karim, M. R., Izawa, T., Kuwamura, M., and Yamate, J. (2017). Immunolocalization of beta-catenin, E-cadherin and N-cadherin in neonate and adult rat kidney. J. Vet. Med. Sci. 79, 1785–1790. doi: 10.1292/jvms.17-0439
Veeman, M. T., Slusarski, D. C., Kaykas, A., Louie, S. H., and Moon, R. T. (2003). Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 13, 680–685. doi: 10.1016/s0960-9822(03)00240-9
Wang, P., Chen, Y., Yong, J., Cui, Y., Wang, R., Wen, L., et al. (2018). Dissecting the global dynamic molecular profiles of human fetal kidney development by single-cell RNA sequencing. Cell Rep. 24, 3554–3567.e3. doi: 10.1016/j.celrep.2018.08.056
Wang, X., Penzes, P., and Napoli, J. L. (1996). Cloning of a cDNA encoding an aldehyde dehydrogenase and its expression in Escherichia coli, Recognition of retinal as substrate. J. Biol. Chem. 271, 16288–16293. doi: 10.1074/jbc.271.27.16288
Wehner, D., Cizelsky, W., Vasudevaro, M. D., Ozhan, G., Haase, C., Kagermeier-Schenk, B., et al. (2014). Wnt/beta-catenin signaling defines organizing centers that orchestrate growth and differentiation of the regenerating zebrafish caudal fin. Cell Rep. 6, 467–481. doi: 10.1016/j.celrep.2013.12.036
Wingert, R. A., Selleck, R., Yu, J., Song, H. D., Chen, Z., Song, A., et al. (2007). The cdx genes and retinoic acid control the positioning and segmentation of the Zebrafish pronephros. PLoS Genet. 3:e189. doi: 10.1371/journal.pgen.0030189
Yu, H., Ye, X., Guo, N., and Nathans, J. (2012). Frizzled 2 and frizzled 7 function redundantly in convergent extension and closure of the ventricular septum and palate: evidence for a network of interacting genes. Development 139, 4383–4394. doi: 10.1242/dev.083352
Yu, J., Carroll, T. J., Rajagopal, J., Kobayashi, A., Ren, Q., and Mcmahon, A. P. (2008). A Wnt7b-dependent pathway regulates the orientation of epithelial cell division and establishes the cortico-medullary axis of the mammalian kidney. Development 136, 161–171. doi: 10.1242/dev.022087
Keywords: ALDH1A2, RALDH2, fetal kidney, Wnt signaling, β-catenin, GSK3 inhibitor, retinoic acid
Citation: Li Y, Gong H, Ding J, Zhao F, Du J, Wan J, Zhang J, Liu S, Li J, Wang L and Zhou B (2020) Inhibition of GSK3 Represses the Expression of Retinoic Acid Synthetic Enzyme ALDH1A2 via Wnt/β-Catenin Signaling in WiT49 Cells. Front. Cell Dev. Biol. 8:94. doi: 10.3389/fcell.2020.00094
Received: 03 October 2019; Accepted: 04 February 2020;
Published: 17 March 2020.
Edited by:
Gregory Kelly, University of Western Ontario, CanadaReviewed by:
Wen-Hui Lien, Catholic University of Leuven, BelgiumThomas Arthur Drysdale, University of Western Ontario, Canada
Copyright © 2020 Li, Gong, Ding, Zhao, Du, Wan, Zhang, Liu, Li, Wang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yifan Li, bGl5aWZhbjIwMDVAeWFob28uY29t; Jiangfeng Ding, dGludGluZGpmQDEyNi5jb20=; Jihui Du, amlodWlkdUAxNjMuY29t
†These authors have contributed equally to this work
 Yifan Li
Yifan Li Hui Gong1†
Hui Gong1† Jun Wan
Jun Wan
